
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 18 September 2020
Sec. Cellular Biochemistry
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.574034
 Rona Karahoda1†
Rona Karahoda1† Cilia Abad1†
Cilia Abad1† Hana Horackova1
Hana Horackova1 Petr Kastner2
Petr Kastner2 Jonas Zaugg3,4
Jonas Zaugg3,4 Lukas Cerveny1
Lukas Cerveny1 Radim Kucera2
Radim Kucera2 Christiane Albrecht3,4
Christiane Albrecht3,4 Frantisek Staud1*
Frantisek Staud1*L-Tryptophan is an essential amino acid and a precursor of several physiologically active metabolites. In the placenta, the serotonin and kynurenine metabolic pathways of tryptophan metabolism have been identified, giving rise to various molecules of neuroactive or immunoprotective properties, such as serotonin, melatonin, kynurenine, kynurenic acid, or quinolinic acid. Current literature suggests that optimal levels of these molecules in the fetoplacental unit are crucial for proper placenta functions, fetal development and programming. Placenta is a unique endocrine organ that, being equipped with a battery of biotransformation enzymes and transporters, precisely orchestrates homeostasis of tryptophan metabolic pathways. However, because pregnancy is a dynamic process and placental/fetal needs are continuously changing throughout gestation, placenta must adapt to these changes and ensure proper communication in the feto-placental unit. Therefore, in this study we investigated alterations of placental tryptophan metabolic pathways throughout gestation. Quantitative polymerase chain reaction (PCR) analysis of 21 selected genes was carried out in first trimester (n = 13) and term (n = 32) placentas. Heatmap analysis with hierarchical clustering revealed differential gene expression of serotonin and kynurenine pathways across gestation. Subsequently, digital droplet PCR, Western blot, and functional analyses of the rate-limiting enzymes suggest preferential serotonin synthesis early in pregnancy with a switch to kynurenine production toward term. Correspondingly, increased function and/or protein expression of serotonin degrading enzyme and transporters at term indicates efficient placental uptake and metabolic degradation of serotonin. Lastly, gene expression analysis in choriocarcinoma-derived cell lines (BeWo, BeWo b30, JEG-3) revealed dissimilar expression patterns and divergent effect of syncytialization compared to primary trophoblast cells isolated from human term placentas; these findings show that the commonly used in vitro placental models are not suitable to study placental handling of tryptophan. Altogether, our data provide the first comprehensive evidence of changes in placental homeostasis of tryptophan and its metabolites as a function of gestational age, which is critical for proper placental function and fetal development.
Placenta is a multifunctional organ providing the fetus with optimal conditions for its growth, development, and programming (Staud and Karahoda, 2018). As a continuously maturing organ, it undergoes structural (Kingdom et al., 2000), epigenetic, and transcriptomic (Uuskula et al., 2012; Cox et al., 2015) changes to adapt to its own as well as maternal and fetal demands. Correspondingly, a wide number of biological processes and molecular and metabolic pathways are differentially affected during gestation (Mikheev et al., 2008; Sitras et al., 2012).
Tryptophan (TRP) is an important amino acid necessary for protein synthesis as well as a precursor of several biologically active metabolites. During pregnancy, TRP and its metabolites are of crucial importance for placentation, fetal development, and immune regulation (Sedlmayr et al., 2014; Laurent et al., 2017). In the placenta, two main TRP metabolic pathways have been identified: the serotonin (5-HT) (Bonnin et al., 2011) and kynurenine (KYN) pathways (Sedlmayr et al., 2002; Goeden et al., 2017). The rate-limiting enzyme of the 5-HT pathway, tryptophan hydroxylase (TPH), gives rise to 5-HT, an important trophic factor early in gestation (Bonnin et al., 2011). Within the placenta a fraction of 5-HT is additionally metabolized to melatonin (Lanoix et al., 2008), which is involved in circadian rhythmicity, fetal growth, and placental function regulation (Iwasaki et al., 2005; Nagai et al., 2008; Seron-Ferre et al., 2012). Several studies have shown that maternal 5-HT also contributes to fetal 5-HT levels (Cote et al., 2007; Gleason et al., 2010; Muller et al., 2017). While early in pregnancy the fetus is dependent on placental/maternal 5-HT, from midgestation it synthesizes its own 5-HT from maternally derived TRP (Arevalo et al., 1991; Sano et al., 2016) suggesting that placental/maternal 5-HT is no longer needed. Indeed, in our latest study (Karahoda et al., 2020) we observed that at term, rat and human placenta does not provide 5-HT to the fetus; in contrast, it takes up fetal 5-HT across the basal membrane of the syncytiotrophoblast (STB) for subsequent degradation by monoamine oxidase-A (MAO-A). Together these findings indicate that placental handling of 5-HT changes throughout gestation.
The KYN pathway generally accounts for most of the TRP degrading activity via the rate-limiting enzymes, indoleamine 2,3-dioxygenase-1/2 (IDO1/2), and tryptophan 2,3-dioxygenase (TDO) (Sedlmayr et al., 2014). In the placenta, this pathway plays a crucial role in preventing fetal rejection by the maternal immune system (Munn et al., 1998). Extensive studies have been carried out to evaluate IDO1 expression/localization in the placenta, indicating that IDO1 expression/function increases during gestation (Sedlmayr et al., 2002; Ligam et al., 2005; Blaschitz et al., 2011; Murthi et al., 2017; Wakx et al., 2018), yet the exact localization in the placenta remains contradictory (Sedlmayr et al., 2014). Contrary to other studies (Sedlmayr et al., 2002; Honig et al., 2004; Kudo et al., 2004), it has been recently observed that IDO1 is not expressed in villous or extravillous trophoblast and the increasing IDO activity at term is exclusively due to expression in endothelial cells (playing a role in immunosuppression and placental tone relaxation) (Blaschitz et al., 2011). KYN is further metabolized to kynurenic acid (KYNA) and quinolinic acid (QUIN), which have neuroprotective and neurotoxic properties, respectively (Foster et al., 1984; Schwarcz et al., 2012). However, the importance of placental KYNA and QUIN remains to be fully elucidated. Recent studies in mouse term placenta report minimal placental contribution to fetal KYNA levels (Goeden et al., 2017; Notarangelo et al., 2019). Importantly, little is known about the effects of gestational age on expression and function of the enzymes down the KYN metabolic pathway, particularly those responsible for production of KYNA and QUIN.
Recently, the importance of gut microbiome metabolism of TRP for gut–brain axis has been described (Kaur et al., 2019; Gao et al., 2020). Similarly, placental metabolism of TRP might form a crucial component of the placenta–brain axis (Rosenfeld, 2020a,b). Considering the large spectrum of TRP metabolites and their roles in pregnancy, it is important to elucidate and understand the shifts in enzyme/transporter expression/activity occurring during gestation. Knowledge on the interplay between enzymes and transporters could provide a better understanding on the significance of a specific pathway at a certain point in pregnancy. Thus, in our study we investigated how advancing gestation affects expression and function of selected enzymes/transporters involved in placental homeostasis of TRP and its metabolites. In addition, we analyzed the effect of cell/trophoblast differentiation on gene expression patterns in isolated primary trophoblast cells and placenta-derived cell lines (BeWo, BeWo b30 clone, JEG-3) to assess their suitability for designated studies.
Serotonin hydrochloride, L-Tryptophan, and phenelzine (MAO inhibitor) were purchased from Sigma–Aldrich (St. Louis, MO, United States). Forskolin (proliferation-activating agent) was obtained from Scintila, s.r.o. (Jihlava, CZ). Bicinchoninic acid assay (BCA assay) reagents were purchased from Thermo Fisher Scientific (Waltham, MA, United States). All other chemicals were of analytical grade.
First-trimester placentas (n = 13) were obtained after elective interruption of healthy pregnancy between 8 and 11 weeks of gestation. Term placentas (n = 32 for mRNA/protein/functional analysis and n = 5 for primary trophoblast isolation) were obtained from uncomplicated pregnancies at 38 to 40 weeks of gestation immediately after delivery. Samples were collected at the University Hospital in Hradec Kralove, Czech Republic or at the Division of Gynecology and Obstetrics, Lindenhofgruppe, Bern, Switzerland. All experiments were performed in accordance with the Declaration of Helsinki and human placenta samples were obtained upon women’s written informed consent and with the approval of the University Hospital Research Ethics Committee (201006 S15P) and Ethics Committee of the Canton of Bern (Basec No. 2016-00250).
The human choriocarcinoma-derived BeWo and JEG-3 cell lines were obtained from the European Cell Culture Collection (ECACC; Salisbury, Wiltshire, United Kingdom). BeWo cells were cultured in Ham F-12 medium supplemented with 10% fetal bovine serum (FBS), whereas JEG-3 cells were cultured in MEM medium supplemented with 10% FBS.
The human choriocarcinoma-derived BeWo b30 cell line (known to form integral monolayers) was obtained from Dr. A. Schwartz (Washington University, St. Louis, United States). Cells were cultured in Dulbecco modified eagle medium (high glucose) supplemented with 10% FBS.
All cell lines were cultivated without antibiotics at 37°C/5% CO2. For differentiation induction, BeWo b30 cell line was treated with 100 μM forskolin for 72 h with daily change of medium.
Villous cytotrophoblast cells (CTBs) were isolated from term placental tissue by enzymatic digestion and Percoll gradient separation, as previously described, with minor modifications (Kallol et al., 2018). In brief, approximately 50 g of villous tissue was washed in 0.9% NaCl (Sigma–Aldrich) four times for 5 min. Thereafter, the tissue was minced and digested three times with 0.25% trypsin (Sigma–Aldrich) and 300 IU/mL deoxyribonuclease I (Sigma–Aldrich) at 37°C (20 min each). The cell suspension was filtered and overlaid on FBS (Seraglob, Switzerland). After centrifugation at 1,000 × g for 15 min at 10°C, the cell pellet was collected in Dulbecco modified eagle medium (high glucose) basic medium (without FBS) and filtered through 100-μm strainer (BD Biosciences, San Jose, CA, United States). Next, cells were overlaid on a discontinuous Percoll® (Sigma–Aldrich) density gradient. After centrifugation, CTBs were located at the layer corresponding to 1.046–1.065 g/mL (35–50%) density (Petroff et al., 2006). The isolated CTBs were cultured at a density of 1 × 106 cells/cm2 in 6-well CellBIND® plates (Corning, New York, NY, United States) in Dulbecco modified eagle medium (high glucose) supplemented with 10% FBS and 1% antibiotic–antimitotic (Thermo Fisher Scientific). Cells were cultured for 12 h (CTB stage) or 72 h (STB stage).
The purity of the isolated trophoblast (PHT) cells was evaluated by staining with specific cell markers followed by flow cytometry analysis as previously described with minor modifications (Kallol et al., 2018). Cells were grown on CellBIND® plates, detached by Accutase® (Sigma–Aldrich), and fixed in 4% formaldehyde (Thermo Fisher Scientific) for 10 min on ice. After washing with Dulbecco phosphate-buffered saline (DPBS; Sigma–Aldrich) cells were centrifuged at 200 g for 10 min at 4°C and then permeabilized with 0.5% Tween-20 (wt/vol) (Sigma–Aldrich) in DPBS for 15 min at room temperature. For evaluation of cell purity, dual staining of CTB and STB with directly labeled antibodies (Novus Biologicals, CO, United States) prepared in staining buffer (5% FBS, 0.1% Tween-20 (wt/vol) in DPBS), was performed. Antibody cocktails comprised (1) anti–cytokeratin 7 (CK-7; AF 488®) plus anti-vimentin (Vim; AF 647®); (2) anti–E-cadherin (E-cad; AF 488®) plus anti-von Willebrand Factor (vWF; AF 647®). Cells were incubated with the respective antibody cocktail for 45 min on ice, followed by two times washing in DPBS (1 min each). After centrifugation at 200 g for 10 min at 4°C, pelleted cells were suspended in DPBS and acquired by flow cytometry (BD FACS LSRII; BD Biosciences). Data acquisition and analysis for each staining were based on at least 10,000 events and performed by using BD FACSDivaTM (BD Biosciences) and FlowJo® software version 10 (FlowJo LLC, Ashland, OR, United States). Because CTB and STB are epithelial cells, staining for CK-7 and E-cad served as positive cell markers (Maldonado-Estrada et al., 2004; Li and Schust, 2015). Anti-vim, known to predominantly stain mesenchymal cells, fibroblast, and stromal cells and anti-vWF staining endothelial cells (Zanetta et al., 2000; Maldonado-Estrada et al., 2004; Li and Schust, 2015) served to quantify potential cellular contaminations by other cell types.
RNA isolation was performed using Tri Reagent (Molecular Research Centre, Cincinnati, OH, United States) or Trizol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. RNA concentration was calculated by measuring absorbance (A) at 260 nm and purity by the A260/280 and A260/230 ratios measured on NanoDropTM 1000 Spectrophotometer (Thermo Fisher Scientific). RNA integrity was confirmed by electrophoresis on a 1.5% agarose gel. 1 μg of total RNA was reversely transcribed to cDNA in a total volume of 20 μL using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States) on a Bio-Rad T100TM Thermal Cycler; for primary trophoblast cells using oligo(dT)15 primers and GoScriptTM Reverse Transcriptase System (Promega, Madison, WI, United States) according to the manufacturer’s instructions.
cDNA (12.5 ng/μL) was amplified in QuantStudioTM 6 (Thermo Fisher Scientific) using the TaqMan® Universal Master Mix II without UNG (Thermo Fisher Scientific) and predesigned TaqMan® Real Time Expression polymerase chain reaction (PCR) assays (listed in Supplementary Table 1, Additional File 1). PCR analysis was run in 5 μL volume, in 384-well plate format. Each sample was amplified in triplicate, following the thermal conditions according to the manufacturer’s instructions.
Prior to quantitative analysis, we evaluated several reference genes for their stable expression during gestation/upon differentiation. Target gene expression in choriocarcinoma-derived cell cultures and primary trophoblast cells was normalized against the predesigned reference gene tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ; Thermo Fisher Scientific) using the 2ΔΔCt method, whereby ΔCt = Ctref – Cttarget and ΔΔCt = ΔCtdifferentiated – ΔCtundifferentiated. On the other hand, gene expression of target genes in human placenta samples was normalized against the predesigned reference gene TATA-box binding protein (TBP; Thermo Fisher Scientific) using the ΔCt method, whereby ΔCt = Ctref – Cttarget. These values were used to generate a gene expression heat map, through the freely available web server Heatmapper1 (Babicki et al., 2016). Hierarchical clustering (Average linkage, Euclidean distance) was applied to group samples with similar expression levels. The scatter plot was constructed in GraphPad Prism 8.3.1 software (GraphPad Software, Inc., San Diego, CA, United States) using the average 2ΔCt values for first-trimester and term placentas.
Absolute quantification of SLC6A4, SLC22A3, MAO-A, TPH1, TPH2, IDO1, and IDO2 in human first-trimester and term placentas was performed using duplex droplet digital PCR (ddPCR) analysis, as described previously (Karahoda et al., 2020). Briefly, the duplex reaction mixture consisted of 10 μL of ddPCRTM Supermix for Probes (Bio-Rad), 1 μL of each of the predesigned probe assays (target – FAM and reference – HEX) (listed in Supplementary Table 1, Additional File 1), and 0.5 μL of cDNA (50 ng/μL), in a total volume of 20 μL. Droplets were generated using QX200 Droplet Generator and subsequently amplified to end-point using T100TM Thermal Cycler following the thermal conditions recommended by the manufacturer. Droplet counting was performed in QX200TM Droplet Reader and the concentration of the target gene was calculated using the QuantaSoftTM Software. For final data evaluation, only wells in which the number of droplets obtained was higher than 13,000 were used. Expression levels are reported in number of transcripts/ng of transcribed RNA. The QX200TM Droplet DigitalTM PCR System, T100TM Thermal Cycler, and all consumables and reagents were obtained from Bio-Rad (unless otherwise stated).
Human first-trimester and term placentas were washed with 0.9% NaCl at 4°C. After weighing and cleaning, the decidua and the chorionic plate were removed, and the placentas were cut in small pieces and homogenized at 4°C in a buffer containing 50 mM Tris-HEPES (pH 7.2), 5 mM EGTA, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 250 mM sucrose. The homogenates were filtered through gauze and centrifuged at 15,000 g for 10 min. The supernatant was collected and stored in the freezer at −80°C until use. Protein concentration was determined using the BCA protein assay kit.
Aliquots of placenta homogenates (30 μg total protein) were mixed with loading buffer under reducing conditions (Laemmli, 1970), heated at 96°C for 5 min, and separated by SDS-PAGE on polyacrylamide gels (10% for SLC6A4, SLC22A3, and MAO-A; 15% for IDO and TPH). Electrophoresis was performed at 150 V and proteins were transferred to polyvinylidene fluoride (PVDF) membranes (SERVA, Heidelberg, DE). The membranes were blocked in 20 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1% Tween 20 (TBS-T) containing 5% non-fat milk for 1 h at room temperature and washed with TBS-T buffer. Incubation with primary antibodies against SLC6A4, SLC22A3, MAO-A, IDO, and TPH (listed in Supplementary Table 2, Additional File 1) was performed overnight at 4°C. After washing with TBS-T buffer, the membranes were incubated with a specific secondary antibody (listed in Supplementary Table 2, Additional File 1) for 1 h at room temperature. Membranes were developed using Chemiluminescence HRP Substrate Kit (SERVA Light Vega). Band intensity was visualized and quantified by densitometric analysis using ChemiDoc MP, Imaging systemTM (Bio-Rad). To ensure equal loading of proteins, membranes were probed for β-actin and specific secondary antibody (listed in Supplementary Table 2, Additional File 1).
MAO-A activity was determined by the method of Carrasco et al. (2000). Briefly, 180 μL placenta homogenate (1.5–2 mg/mL) was pre-incubated with or without MAO-A inhibitor, phenelzine (100 μM) for 5 min at 37°C, and then the reaction was initiated by incubation with 20 μL of 5-HT (0.5 mM) for an indicated time period. The reaction was stopped by adding 40 μL of HClO4 (3.4 M) and placed on ice for 5 min. Samples were centrifuged at 5,000 g for 10 min, and the supernatant was used for 5-HT determination by high-performance liquid chromatography (HPLC).
IDO activity was determined by the method of Takikawa et al. (1988). The incubation media (50 mM potassium phosphate buffer pH 6.5, 20 mM ascorbate, 0.01 mM methylene blue, 100 units/mL catalase) was pre-incubated for 5 min at 37°C, with or without 0.4 mM TRP. The reaction was initiated by adding the placenta homogenate and terminated after 30 min with 200 μL of trichloroacetic acid 30%. Samples were further incubated for an additional 30 min at 50°C to assure complete hydrolysis of N-formyl KYN to KYN. The reaction mixture was then centrifuged for 20 min at 3,000 g, 20°C and supernatant was collected for HPLC measurement of KYN. The IDO enzymatic activity was calculated as the difference between the amount of KYN produced in the media with and without TRP. The results are expressed as nmol KYN/μg protein per min.
TPH enzymatic activity was determined by the method by Goeden et al. (2016). Human placenta homogenates were supplemented with 1 mM dithiothreitol as a reducing agent ensuring complete enzymatic activity (Fitzpatrick, 1999). The enzymatic reaction was carried out at 37°C, pH 7.5, with ∼1.5 to 2 mg protein/mL. The incubation media contained (final concentrations): 50 mM Tris buffer, 1 mM EGTA, 100 units/mL catalase, 0.1 mM ammonium iron (II) sulfate, 0.1 mM tetrahydrobiopterin (BH4, a cofactor required for TPH activity), either in the absence or in the presence of 0.25 mM TRP. Briefly, placenta homogenate was incubated with the incubation media for 30 min at 37°C. Reaction was terminated by adding 200 μL of HClO4 with 100 μM EDTA. Samples were incubated on ice for 15 min for complete protein denaturation and then centrifuged for 15 min at 21,000 g. Supernatants were collected for determination by HPLC of 5-hydroxytryptophan (5-OH-TRP), a metabolic intermediate in 5-HT synthesis. The results were calculated as the difference between the amount of 5-OH-TRP liberated in samples with and without TRP and are expressed as nmol 5-OH-TRP/μg protein per min.
The HPLC analyses were performed using Shimadzu LC20 Performance HPLC chromatograph (Shimadzu, Kyoto, Japan) equipped with UV and fluorescence detector. For simultaneous chromatographic separation of all tested compounds, Phenomenex Kinetex 5 μm EVO C18 100 A 150 × 3 mm with a guard column was used. An isocratic elution, at a flow rate of 0.5 mL/min, was performed with mobile phase consisting of 0.1 M acetic acid, pH 4.5 (adjusted with NaOH), and methanol 97 + 3. All analytes were eluted within 8.5 min.
Excitation and emission wavelengths of fluorescence detector were set for individual compounds: 275/333 nm for 5-OH-TRP from 0 to 3.1 min and 280/334 nm for 5-HT and TRP from 3.1 min. KYN was detected by UV detection with wavelength set to 369 nm. Additionally, in cases of TRP concentrations higher than the range of fluorescence detection, UV detection was used with wavelength set to 300 nm.
Quantitative PCR (qPCR) results were assessed using Mann–Whitney tests. ddPCR analyses, protein expression, and functional studies were evaluated using unpaired t-test. All statistical analyses were implemented in GraphPad Prism 8.3.1 software (GraphPad Software, Inc.). Asterisks in the figures indicate significance levels: ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001.
Characteristics of the first-trimester and term pregnancies are listed in Table 1. No statistical differences were found in the mean maternal age and maternal BMI before pregnancy between the two groups. In the term group (n = 32) only healthy, non-medicated and non-smoking mothers were included. On the other hand, 5 of 13 women from the first trimester samples were smokers (frequency: three women < 10 cigarettes/day; two women > 10 cigarettes/day). Placenta samples from these women were included in the PCR analysis only if no association between smoking and gene expression was found. These samples have been marked with an asterisks (∗) in the heatmap representing gene expression data (Figure 1).
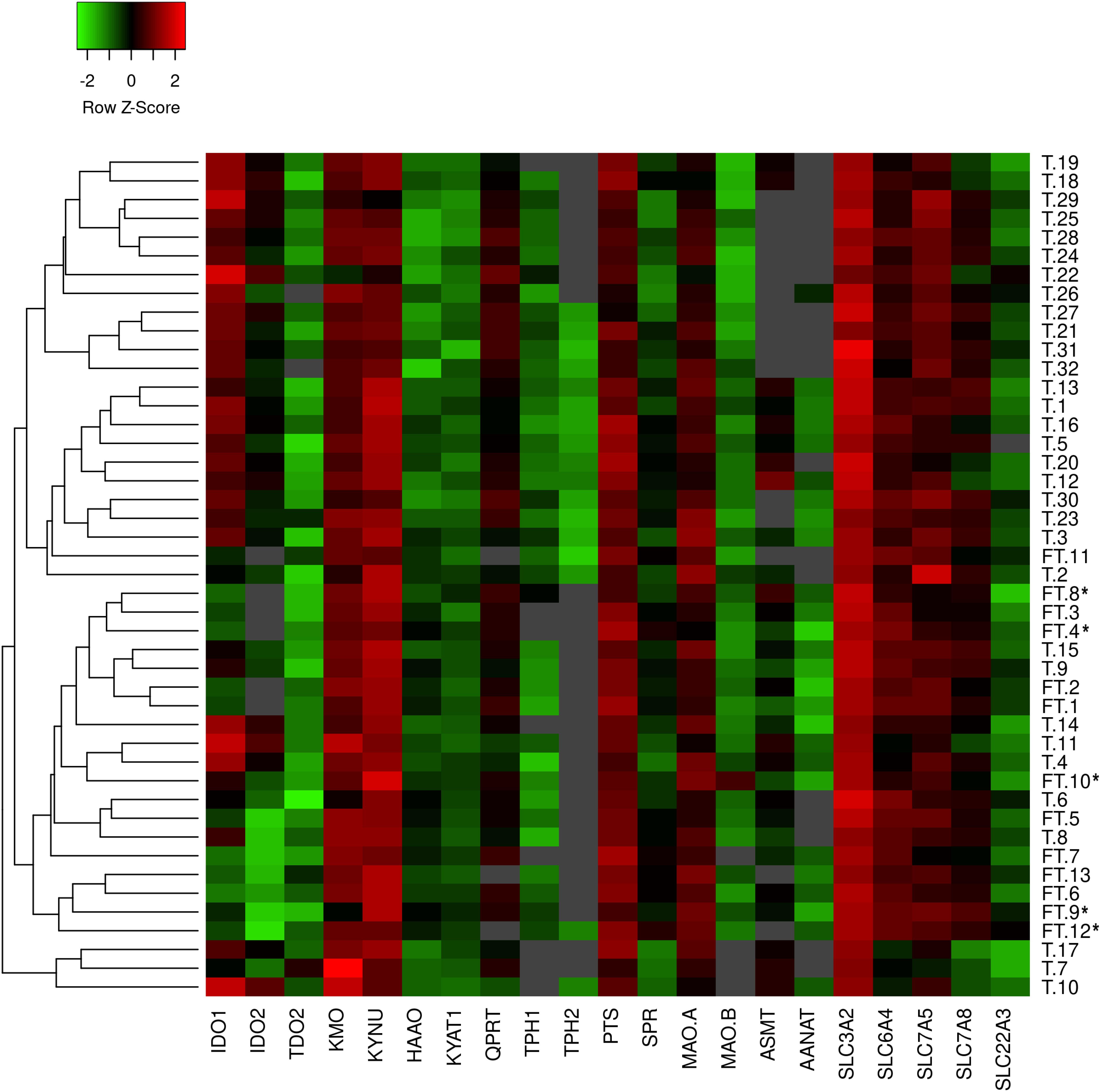
Figure 1. Heatmap generated from qPCR gene expression analysis of TRP pathway in human placenta. Expression of 21 key transporters/enzymes was analyzed in first trimester (FT; n = 13) and term (T; n = 32) human placenta samples. Hierarchical clustering (average linkage, Euclidean distance) was applied to group samples with similar expression levels, emphasizing the overall changed expression during gestation. The color intensity indicates expression levels; red: up-regulation, green: down-regulation, gray: not detected. *Placentas from smokers.
Human placenta fully expresses the enzymatic machinery for TRP metabolism to 5-HT and KYN pathways. A heatmap with hierarchical clustering revealed sample distribution into three main clusters, whereby the first-trimester placentas were clustered predominantly into one cluster, whereas term placenta samples were distributed into the remaining two clusters (Figure 1); the pattern of distribution indicated differential expression of the pathways across gestation. Subsequently, individual analysis of genes in first-trimester and term placentas showed several enzymes/transporters to be significantly up- or down-regulated at term.
Of the 5-HT pathway, gene expression of the main enzymes (PTS, SPR) responsible for BH4 production, a co-factor necessary for TPH function, were found to be significantly decreased at term, compared with first-trimester placenta. On the other hand, the rate-limiting enzymes of the KYN pathway, IDO1/2, were negligibly expressed in the first-trimester placenta, whereas we observed significant expression at term. Interestingly, the subsequent enzymes of the KYN pathway (specifically KMO, KYNU, HAAO, KYAT1, QPRT) were found to be expressed in lower amounts at term, compared with first-trimester placenta. Lastly, of the transport proteins tested, SLC3A2, SLC6A4, and SLC7A8 revealed higher expression in the first-trimester placenta. Scatter plots of the log10-expression in first-trimester and term placenta were used to display the data and visualize the gene expression differences (Figure 2).
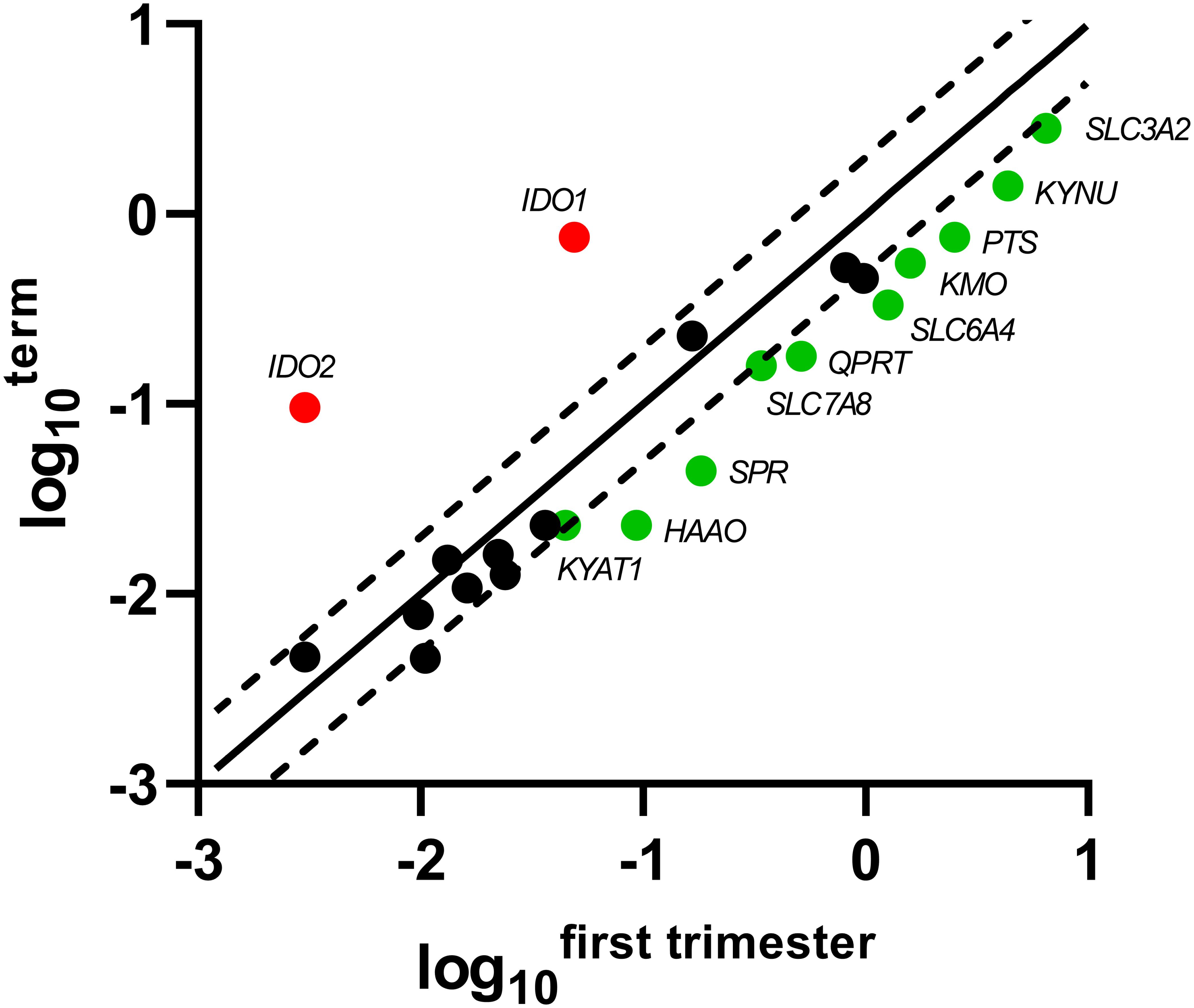
Figure 2. Scatter plot of the TRP pathway enzymes/transporters showing significant changes in gene expression during gestation. Log10 of each gene-specific 2ΔCt in the first-trimester placenta is plotted against those of term placenta. The central diagonal line shows unchanged gene expression, whereas the dotted lines depict the upper and lower limit of the threshold fold regulation, which was set to 2. Data were further evaluated using non-parametric Mann–Whitney test on ΔCt values, and those that exceed the threshold fold regulation and were statistically significant (p ≤ 0.05) are named and shown in color: red – up-regulation, green – down-regulation.
ddPCR analysis was conducted in 13 first-trimester and 25 term placentas for absolute quantification of transcripts of the rate-limiting enzymes and main transporters of 5-HT and KYN pathways in the human placenta. We observed statistically significant down-regulation of TPH1 (Figure 3A), MAO-A (Figure 3B), and SLC6A4 (Figure 4A) at term, whereas IDO1 (Figure 3C) and IDO2 (Supplementary Figure 1B, Additional File 1) gene expression was up-regulated. SLC22A3 (Figure 4B) and TPH2 (Supplementary Figure 1A, Additional File 1) expression remained unchanged during gestation.
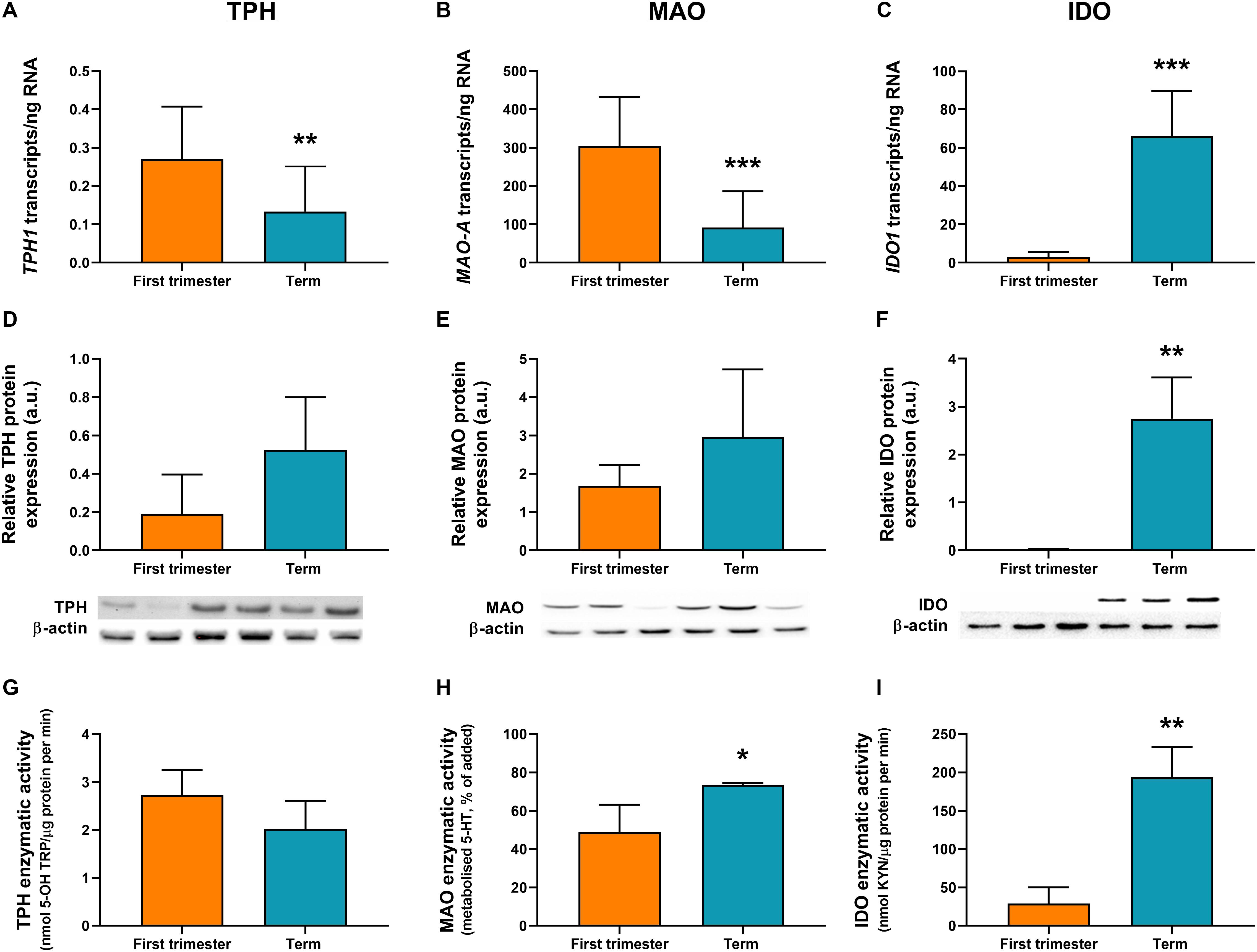
Figure 3. Gene/protein expression and functional analyses of the rate-limiting enzymes of 5-HT and KYN pathways. Expression and function of tryptophan hydroxylase-1 (TPH1/TPH1), monoamine oxidase-A (MAO-A/MAO-A), and indoleamine 2,3-dioxygenase-1 (IDO1/IDO1) between the first-trimester and term human placenta was analyzed by digital droplet PCR (A–C), Western blot (D–F), and enzymatic activity (G–I) analyses. Protein expression was normalized to β-actin as a loading control; representative immunoblots for target proteins and β-actin are shown. For ddPCR analysis n = 13 (first trimester) and n = 25 (term placentas); for protein/functional analysis n = 3 for each. Data are presented as mean ± SD and statistical significance was evaluated using unpaired t-test; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
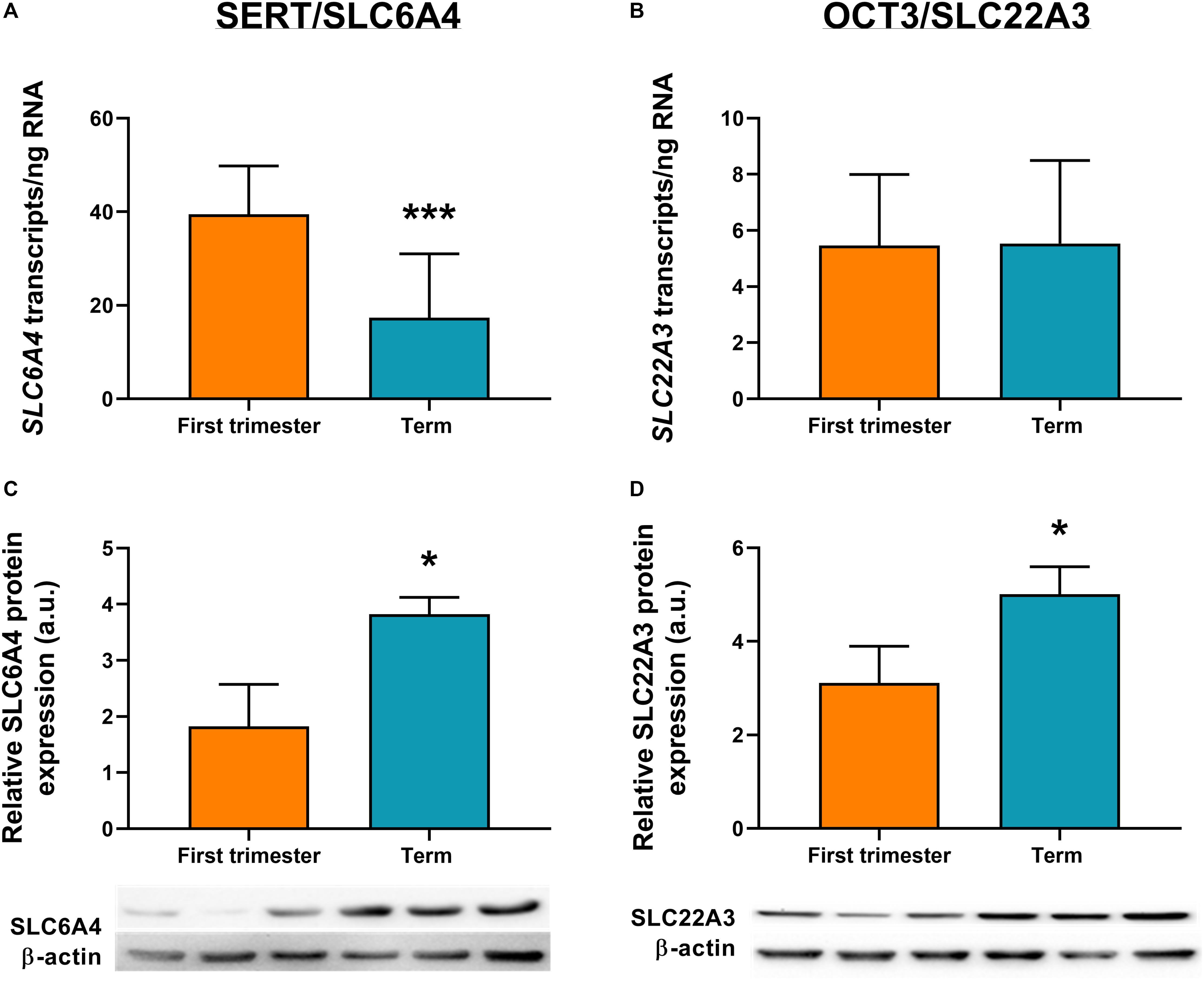
Figure 4. Gene and protein expression of the main transporters of 5-HT pathway. Comparison of gene and protein expression of 5-HT transporter (SLC6A4/SERT) and organic cation transporter 3 (SLC22A3/OCT3) between the first-trimester and term human placenta was analyzed by digital droplet PCR (A,B) and Western blot (C,D). Protein expression was normalized to β-actin as a loading control; representative immunoblots for target proteins and β-actin are shown. For ddPCR analysis n = 13 (first trimester) and n = 25 (term placentas); for protein analysis n = 3 for each. Data are presented as mean ± SD and statistical significance was evaluated using unpaired t-test; *p ≤ 0.05, ***p ≤ 0.001.
While human placenta expresses both isoforms (1 and 2) of TPH and IDO (Sedlmayr et al., 2014; Laurent et al., 2017; Ranzil et al., 2019), we found that TPH1 and IDO1 are predominant throughout gestation. Specifically, IDO1 (Figure 3C) levels in the first-trimester and term placenta exceeded those of IDO2 (Supplementary Figure 1B, Additional File 1) by 30- and 11-fold, respectively. On the other hand, TPH1 (Figure 3A) showed three to six times higher expression compared to TPH2 (Supplementary Figure 1A, Additional File 1), during gestation.
Moreover, we observed that MAO-A transcripts outnumbered those of TPH1 transcripts by more than 1,000-fold in the first trimester and almost 700-fold at term (Figures 3A,B). On the other hand, while TPH1 expression remained unchanged during pregnancy, we observed a 20-fold increase in IDO1 transcripts at term (Figure 3C), suggesting a shift/preferential TRP metabolism toward the KYN pathway at term.
To investigate the expression at protein level, western blot analysis using specific antibodies for TPH, MAO, IDO, SLC6A4, and SLC22A3 was performed in homogenates from first-trimester (n = 3) and term (n = 3) human placenta samples. We observed no difference in MAO-A and TPH1 protein expression during gestation (Figures 3D,E). On the other hand, IDO1 protein band (45 kDa) was clearly detected in term placenta homogenates, whereas it was not visible in first trimester samples (Figure 3F), indicating that IDO1 protein is not expressed at early stages of pregnancy. As for the transport proteins, both SLC6A4 (Figure 4C) and SLC22A3 (Figure 4D) showed a significantly increased protein expression in term placentas compared with the first trimester ones.
TPH activity was evaluated in first-trimester and term placenta homogenates, using TRP as a substrate and measuring the production of 5-OH TRP. As shown in Figure 3G, TPH activity ranged between 2 and 3 nmol 5-OH TRP/μg protein per min, and it was not affected by gestational age.
MAO enzymatic activity was determined in first-trimester and term placenta homogenates by measuring the amount of 5-HT metabolized in the placental homogenate after 60 min of incubation with 5-HT. As shown in Figure 3H, after 60 min of incubation, 45% of 5-HT was metabolized by first-trimester placenta and nearly 75% by term placenta. Metabolism of 5-HT was completely inhibited by addition of phenelzine (100 μM) (Karahoda et al., 2020), indicating that 5-HT was metabolized specifically by MAO. These data suggest that placental metabolism of 5-HT increases towards the end of pregnancy.
IDO activity in first-trimester and term placentas was evaluated using TRP as a substrate. IDO activity showed a significant increase during human pregnancy, with levels as low as 29 nmol KYN/μg protein per min (±17.3) in the first-trimester placenta to 7-fold higher activity at term (193 ± 32.3 nmol KYN/μg protein per min) (Figure 3I).
Choriocarcinoma-derived cell lines (BeWo, BeWob30, JEG-3) and PHT cells isolated from human term placentas, were analyzed for expression of the main enzymes/transporters of the TRP pathway. Out of 21 genes tested, only 10 were found to be co-expressed in all cell types, with the PHT and BeWo b30 cells showing the highest similarity in gene expression (Figure 5A). Nonetheless, none of the choriocarcinoma-derived cell lines expressed the rate-limiting enzyme of the KYN pathway, IDO1 or the 5-HT uptake transporter (OCT3/SLC22A3), making the PHT cells the only suitable in vitro model for studies of the respective pathways.
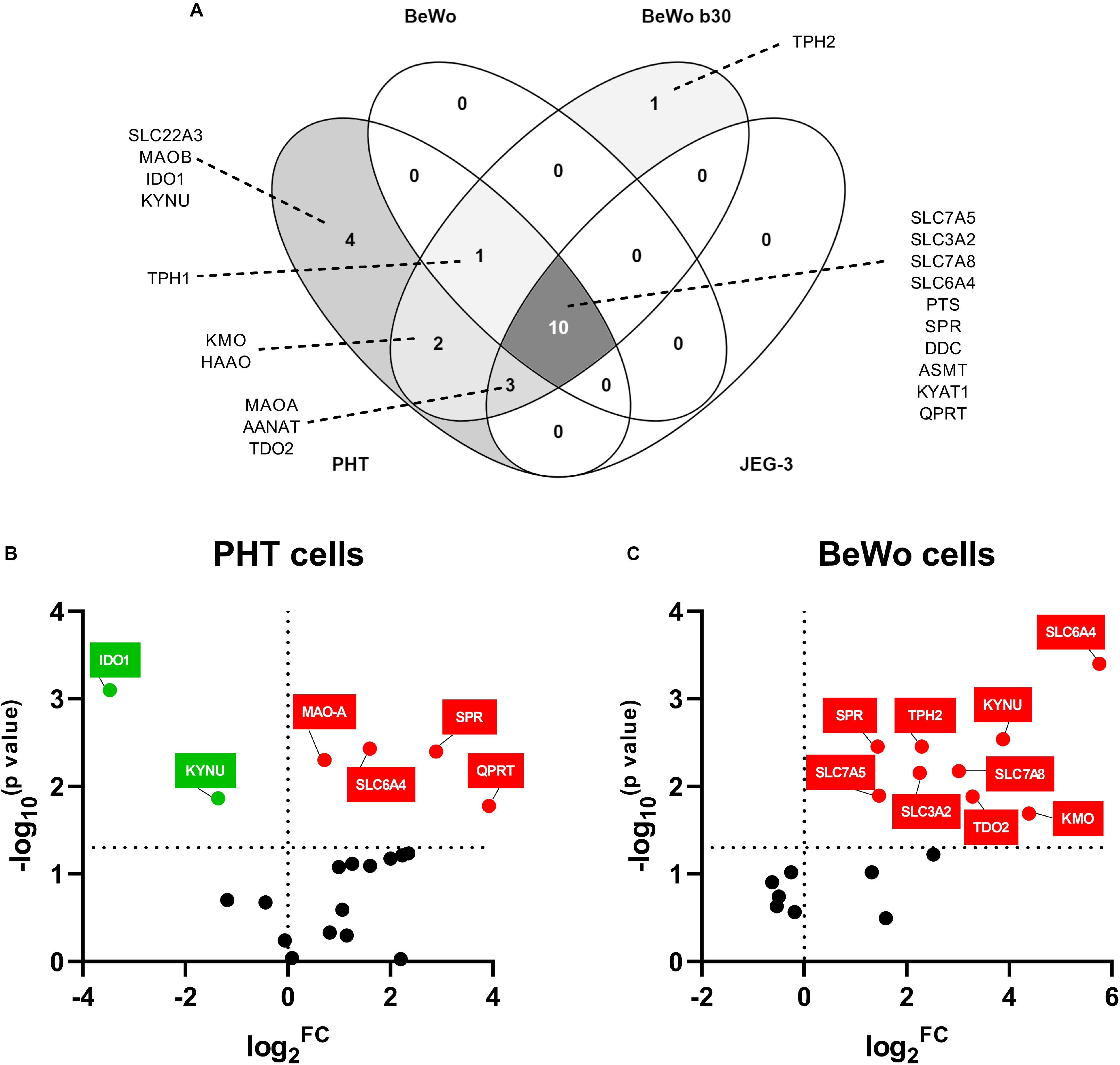
Figure 5. Gene expression of TRP pathway-related enzymes and transporters in placental-derived cells. Placental-derived cells were obtained from three different passages of BeWo, BeWo b30, and JEG cells and five isolations of primary human trophoblast cells (PHT). (A) Venn diagram of the overlap in gene expression among four cell lines. PHT and BeWo b30 cells share the highest gene homology, with the PHT cells additionally expressing four important genes, including the rate-limiting enzyme, IDO1, and the polyspecific organic transporter, SLC22A3. Subsequently, effect of differentiation in these two cell lines was tested, revealing dissimilarity between spontaneous syncytialization (B) and forskolin-induced differentiation (C). Data are visualized as volcano plots of fold changed expression in the differentiated cells, compared to the undifferentiated counterpart. Color indicates statistically significant changes: red – up-regulation, green – down-regulation. Statistical significance was evaluated using Mann–Whitney test.
Since the PHT and BeWo b30 cells shared the highest homology in genes expressed at mRNA level, we further studied the effect of differentiation in these cells. While CTB spontaneously syncytialized within 48–72 h, BeWo cell differentiation was induced with forskolin over a 72-h period. Only two of the tested genes (SLC6A4 and SPR), revealed similar gene expression changes, specifically up-regulation, in BeWo and PHT cells upon differentiation. However, altogether, we observed that the differentiation process in BeWo cells affects the gene expression of TRP pathway enzymes/transporters in a more profound manner than in PHT cells (Figures 5B,C). Specifically, in PTH cells IDO1 and KYNU were found to be down-regulated, whereas MAO-A and QPRT up-regulated (Figure 5B). Conversely, in BeWo cells we observed up-regulation for the following genes: TPH2, KYNU, KMO, TDO2, SLC3A2, SLC7A5, and SLC7A8 (Figure 5C).
In our recent study (Karahoda et al., 2020) we demonstrated that the term placenta no longer provides 5-HT to the fetus. In contrast, it extracts it from the fetal circulation via OCT3-mediated process for subsequent degradation by MAO-A. We thus hypothesized that fetoplacental homeostasis of TRP and 5-HT changes throughout gestation. In this follow up study, we investigated the gene expression of 16 enzymes and 5 transporters involved in the metabolism and transport of TRP and its metabolites in human placenta (first trimester and term) and placental-derived cells. Moreover, using a combination of gene/protein expression and functional analyses, we further report that the key enzymes and transporters involved in placental homeostasis of TRP, 5-HT, and KYN exhibit changes throughout gestation, likely as a result of adaptation to meet different placental/fetal needs with time.
Placental expression of several enzymes involved in TRP metabolism has been investigated before (Ligam et al., 2005; Lanoix et al., 2008; Bonnin et al., 2011; Laurent et al., 2017) and related to pathological conditions such as fetal growth restriction and preeclampsia (Carrasco et al., 2000; Murthi et al., 2017; Ranzil et al., 2019). Nevertheless, comprehensive characterization of the metabolic pathways in human placenta and physiological changes that may occur during gestation are poorly understood. As TRP is an essential amino acid, the placenta and fetus are dependent on maternal intake and placental transport from the maternal to fetal circulation. Therefore, apart from the metabolizing enzymes, it is also important to study the expression and function of placenta membrane transporters responsible for fetoplacental handling of TRP and its metabolites.
TRP is a substrate of L-type amino acid transporter-1 (LAT1/SLC7A5) on the maternal-facing membrane and L-type amino acid transporter-2 (LAT2/SLC7A8) on both maternal- and fetal-facing membranes; LAT1/2 functional activity is dependent on heterodimerization with the 4F2 heavy chain (SLC3A2) (Gaccioli et al., 2015). We observed that placental expression of SLC3A2 and SLC7A8 is down-regulated at term, which contrasts with a recent examination of publicly available gene expression array data, reporting no change at any stage of pregnancy for these transporters (Simner et al., 2017). However, it should be noted that the small sample size (n = 4 per gestational age) compared with our cohort (n = 13 for first-trimester and n = 32 for term placenta) could account for the different outcomes.
TRP metabolism to 5-HT is mediated by TPH, whose activity depends on tetrahydrobiopterin (BH4) as a cofactor (Mckinney et al., 2005). Two mechanisms of BH4 synthesis in the human placenta have been suggested, de novo synthesis and/or salvage pathway (Iwanaga et al., 2004). We observed that the expression of 6-pyruvoyltetrahydropterin synthase (PTS), involved in de novo synthesis, and of sepiapterin reductase (SPR), involved in both pathways, decreased significantly at term, which corresponds nicely with previous reports on decreased SPR activity with increasing gestational age (Iwanaga et al., 2004). With the importance of BH4 as a cofactor for endothelial nitric oxide synthase (necessary for nitric oxide production), we speculate that decreasing SPR expression and activity at term may decrease the availability of BH4 for TPH activity, thus 5-HT synthesis at term.
Interestingly, we demonstrate that the first-trimester placentas show preferential expression of downstream enzymes of the KYN pathway, specifically KMO, KYNU, HAAO, and QPRT [involved in generation of 3-hydroxy-L-KYN (3-HK), 3-hydroxy anthranilic acid (3-HAA), and QUIN] and KYAT1 (involved in generation of KYNA). This was unexpected because the rate-limiting enzyme IDO1 is, in contrast, only modestly expressed in the first-trimester placenta. While in 1998, Munn et al. (1998) suggested IDO1-based suppression of immune reactions to mediate fetomaternal tolerance, in a follow-up study, they reported that pregnancy success rate is not affected in the IDO-deficient mouse model (Baban et al., 2004). The authors proposed involvement of alternative processes, such as TDO, which may compensate for IDO activity when low or absent. In our study, we observed that TDO expression, although at relatively low levels, remains stable throughout gestation. Our results thus support a concept proposed by Badawy (2015) in which TRP degradation in early-to-mid pregnancy is catalyzed by TDO, with IDO gaining a partial/transient role in midgestation. We speculate that in the first trimester, KYN synthesis via TDO serves mainly as a precursor of 3-HK, 3-HAA, QUIN, and KYNA. QUIN is important for NAD+ synthesis, necessary for numerous redox reactions and DNA repair. Similarly, 3-HK and 3-HAA are important metabolites with antioxidant and immunosuppressive properties. Lastly, KYNA, apart from its immunosuppressive function, plays a role in neuroprotection, probably through its action on the NDMA receptor (Foster et al., 1984). In accordance with our qPCR data, recent studies in mouse placenta showed limited placental KYNA synthesis at term (Goeden et al., 2017; Notarangelo et al., 2019). Thus, we believe that the impact of these metabolites may be of higher importance in the first trimester when the pro-inflammatory environment is less pronounced (Hannan et al., 2014; Holtan et al., 2015). On the other hand, the significant increase in IDO1 at term could account for high KYN production involved in the immune related activities. Indeed, this concept was previously discussed by Badawy who suggests preferential TRP utilization for protein, 5-HT and NAD+ synthesis in early pregnancy (Badawy, 2015).
Therefore, we investigated the expression profiles and metabolic activity of the rate-limiting enzymes of the 5-HT and KYN pathways, TPH and IDO, respectively, early in pregnancy and at term. Our results indicate that during the first trimester, placenta may preferentially metabolize TRP to 5-HT, an important trophic factor for fetal development. Indeed, Bonnin et al. (2011) showed that placental 5-HT synthesis occurs as early as E10.5 in mice and week 11 in humans. It is during this period when the fetus is not capable of synthesizing 5-HT, yet serotonergic neurons and receptors have been identified (Bonnin and Levitt, 2011). As the immature fetal blood–brain barrier is not fully functional (Daneman et al., 2010), it has been well-established that the placenta serves as the main source of fetal 5-HT in early gestation (Bonnin and Levitt, 2011; Bonnin et al., 2011). However, later in gestation, the fetus gains the capability of 5-HT synthesis (Arevalo et al., 1991; Sano et al., 2016). With increased IDO expression/activity at term, also reported before (Blaschitz et al., 2011; Murthi et al., 2017; Wakx et al., 2018), it seems plausible that at later stages of gestation, TRP is preferentially utilized for KYN production.
In our previous study (Karahoda et al., 2020) we described the importance of membrane transporters (SERT/OCT3) and metabolizing enzyme (MAO-A) for placental 5-HT homeostasis at term. Here we reveal that, at protein levels, both transporters and the metabolizing enzyme are up-regulated at term. Increased OCT3 protein expression in human term placenta was also described by Lee et al. (2013). These findings suggest that towards term, placental capacity to take up 5-HT from both maternal and fetal circulations increases. The parallel increase in MAO-A expression and activity toward term strengthens our hypothesis that an orchestration between SERT, OCT3, and MAO-A activity serves as a 5-HT detoxification mechanism, protecting the term placenta and the fetus from high 5-HT circulating levels.
In vitro cell-based approaches (e.g., BeWo, BeWo b30, and JEG-3) are often applied as alternative methods to investigate placental physiology. However, these cells are derived from first trimester choriocarcinoma and, correspondingly, we show that gene expression of TRP metabolic pathways differs largely from that of primary trophoblast cells isolated from human term placenta. Specifically, BeWo and JEG-3 cells lack expression of crucial proteins, IDO1 and SLC22A3; lack of IDO1 in BeWo cells was also reported before (Entrican et al., 2002). In contrast, isolated PHT cells show expression pattern similar to that of term placenta. Another advantage of isolated PHT cells is their spontaneous fusion in culture to form the syncytium (Huang et al., 2016) while in BeWo cells, syncytialization must be provoked by modulators of cAMP metabolism such as forskolin (Jiraskova et al., 2018) which, in the present study, resulted in a non-physiological up-regulation of several genes (see Figures 5B,C). Taken together, these results indicate that placenta-derived carcinoma cells, BeWo, BeWo b30, and JEG-3, are not optimal in vitro models for TRP-related placental research; instead, use of primary human trophoblast cells is recommended.
Inconsistency exists in the current literature on IDO1 localization in the placenta and its expression in trophoblast cells (Sedlmayr et al., 2014). In the present study, we observed higher expression of IDO1 in isolated CTB when compared to STB stage. While several studies report IDO1 in STB (Sedlmayr et al., 2002; Honig et al., 2004; Kudo et al., 2004) and CTB (Dong et al., 2008; Cvitic et al., 2013), recent papers (Ligam et al., 2005; Blaschitz et al., 2011) propose exclusive localization in vascular endothelium arguing that previous findings of IDO1 in trophoblast are a result of contaminating endothelial cells in isolated PHT cells (Sedlmayr et al., 2014). However, in our preparations, contamination with endothelial cells is routinely less than 1%, reflecting solely IDO1 expression in CTBs. Moreover, for a long time it was believed that as pregnancy proceeds, the CTB layer gradually disappears (Benirschke et al., 2016); however, latest research reveals increasing number of CTBs at term (Mori et al., 2007) and designates them as the most metabolically active cells in human term placenta (Kolahi et al., 2017). Thus, for certain enzymes such as IDO1, CTB layer may be more active in metabolism than STB, and functional studies in isolated CTB cells should not be neglected.
In conclusion, here we report that placental homeostasis of TRP is subject to strictly regulated developmental changes during pregnancy (Figure 6). Considering the manifold role of TRP metabolites in placenta function, fetal development, and programming, tight regulation is necessary to maintain its homeostasis in the fetoplacental unit and ensure optimal communication on the placenta–brain axis. Subsequently, any internal or external insults, such as polymorphisms, epigenetics, pharmaceuticals or diseases, may compromise this harmonized interplay of enzymes and transporters, and result in suboptimal in utero conditions, and subsequently poor pregnancy outcomes. Importantly, timing of these insults is critical for fetal development (Barker et al., 2010); thus, knowledge of TRP catabolic pathways in the placenta during pregnancy aids in understanding the biological roots of fetal programming.
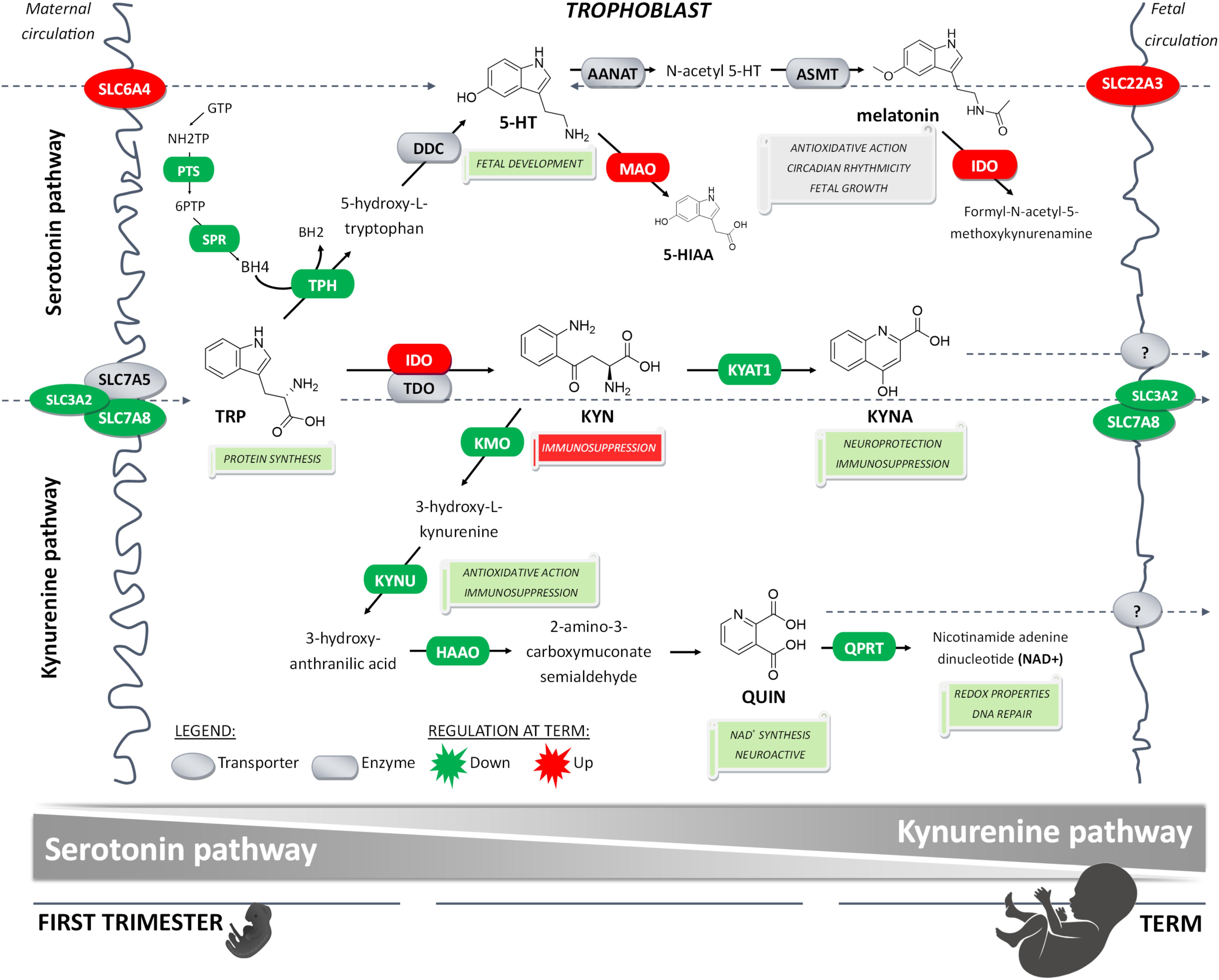
Figure 6. Proposed gestational age-dependent changes in placental TRP metabolic pathways. The schematic illustration is based on gene expression studies and, in the case of rate-limiting enzymes/key transporters, also on protein, and/or functional activity. The first-trimester placenta preferentially utilizes TRP for generation of 5-HT involved in fetal development and synthesis of KYN metabolites implicated with neuroprotection, antioxidative and redox effects, and DNA repair. On the other hand, the term placenta synthesizes significant amounts of KYN important for immune regulation activities. Collectively, our findings strongly indicate gestational age-related changes in placental handling of TRP and its main metabolites, 5-HT and KYN. This may reflect the changes in placental and fetal demands for metabolites of either pathway to ensure proper embryonic and fetal development throughout pregnancy.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the University Hospital Research Ethics Committee (201006 S15P) and the Ethics Committee of the Canton of Bern (Basec No. 2016-00250). The participants provided their written informed consent to participate in this study.
RKa, CAb, and FS participated in the study concept and design. RKa, CAb, HH, PK, and JZ participated in the data acquisition. RKa, CAb, HH, PK, LC, RKu, CAl, and FS performed the data analysis and participated in interpretation of the results. RKa, CAb, and FS wrote the article. All authors contributed to the article and approved the submitted version.
This study was financially supported by the Czech Science Foundation (Grant No. 20/13017S), by the Grant Agency of Charles University (SVV 2020/260414), and the EFSA-CDN project (No. CZ.02.1.01/0.0/0.0/16_019/0000841), which was co-funded by the ERDF. CAl and JZ were supported by the Swiss National Centre of Competence in Research, NCCR TransCure, University of Bern.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Prof. Marian Kacerovsky, Ph.D., Dr. Tomas Faist, Dr. Jaroslav Stranik, and Martina Hudeckova from the Department of Obstetrics and Gynecology, Faculty Hospital in Hradec Kralove for providing us the placental tissues as well as gathering all the available patient information data; physicians and midwives from the Lindenhofgruppe, Bern, Switzerland, are further acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.574034/full#supplementary-material
Arevalo, R., Afonso, D., Castro, R., and Rodriguez, M. (1991). Fetal brain serotonin synthesis and catabolism is under control by mother intake of tryptophan. Life Sci. 49, 53–66. doi: 10.1016/0024-3205(91)90579-z
Baban, B., Chandler, P., Mccool, D., Marshall, B., Munn, D. H., and Mellor, A. L. (2004). Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. J. Reprod. Immunol. 61, 67–77. doi: 10.1016/j.jri.2003.11.003
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153.
Badawy, A. A. (2015). Tryptophan metabolism, disposition and utilization in pregnancy. Biosci. Rep. 35:e00261.
Barker, D. J. P., Eriksson, J. G., Kajantie, E., Alwasel, S. H., Fall, C. H. D., Roseboom, T. J., et al. (2010). “The maternal and placental origins of chronic disease,” in The Placenta and Human Developmental Programming, eds G. J. Burton, D. J. P. Barker, A. Moffett, and K. Thornburg (Cambridge: Cambridge University Press), 5–17.
Benirschke, K., Kaufmann, P., and Baergen, R. (2016). Pathology of the Human Placenta. New York, NY: Springer.
Blaschitz, A., Gauster, M., Fuchs, D., Lang, I., Maschke, P., Ulrich, D., et al. (2011). Vascular endothelial expression of indoleamine 2,3-dioxygenase 1 forms a positive gradient towards the feto-maternal interface. PLoS One 6:e21774. doi: 10.1371/journal.pone.0021774
Bonnin, A., Goeden, N., Chen, K., Wilson, M. L., King, J., Shih, J. C., et al. (2011). A transient placental source of serotonin for the fetal forebrain. Nature 472, 347–350. doi: 10.1038/nature09972
Bonnin, A., and Levitt, P. (2011). Fetal, maternal, and placental sources of serotonin and new implications for developmental programming of the brain. Neuroscience 197, 1–7. doi: 10.1016/j.neuroscience.2011.10.005
Carrasco, G., Cruz, M. A., Dominguez, A., Gallardo, V., Miguel, P., and Gonzalez, C. (2000). The expression and activity of monoamine oxidase A, but not of the serotonin transporter, is decreased in human placenta from pre-eclamptic pregnancies. Life Sci. 67, 2961–2969. doi: 10.1016/s0024-3205(00)00883-3
Cote, F., Fligny, C., Bayard, E., Launay, J. M., Gershon, M. D., Mallet, J., et al. (2007). Maternal serotonin is crucial for murine embryonic development. Proc. Natl. Acad. Sci. U.S.A. 104, 329–334. doi: 10.1073/pnas.0606722104
Cox, B., Leavey, K., Nosi, U., Wong, F., and Kingdom, J. (2015). Placental transcriptome in development and pathology: expression, function, and methods of analysis. Am. J. Obstet Gynecol. 213, S138–S151.
Cvitic, S., Longtine, M. S., Hackl, H., Wagner, K., Nelson, M. D., Desoye, G., et al. (2013). The human placental sexome differs between trophoblast epithelium and villous vessel endothelium. PLoS One 8:e79233. doi: 10.1371/journal.pone.0079233
Daneman, R., Zhou, L., Kebede, A. A., and Barres, B. A. (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566. doi: 10.1038/nature09513
Dong, M., Ding, G., Zhou, J., Wang, H., Zhao, Y., and Huang, H. (2008). The effect of trophoblasts on T lymphocytes: possible regulatory effector molecules–a proteomic analysis. Cell Physiol. Biochem. 21, 463–472. doi: 10.1159/000129639
Entrican, G., Wattegedera, S., Chui, M., Oemar, L., Rocchi, M., and Mcinnes, C. (2002). Gamma interferon fails to induce expression of indoleamine 2,3-dioxygenase and does not control the growth of Chlamydophila abortus in BeWo trophoblast cells. Infect. Immun. 70, 2690–2693. doi: 10.1128/iai.70.5.2690-2693.2002
Fitzpatrick, P. F. (1999). Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 68, 355–381. doi: 10.1146/annurev.biochem.68.1.355
Foster, A. C., Vezzani, A., French, E. D., and Schwarcz, R. (1984). Kynurenic acid blocks neurotoxicity and seizures induced in rats by the related brain metabolite quinolinic acid. Neurosci. Lett. 48, 273–278. doi: 10.1016/0304-3940(84)90050-8
Gaccioli, F., Aye, I. L. M. H., Roos, S., Lager, S., Ramirez, V. I., Kanai, Y., et al. (2015). Expression and functional characterisation of System L amino acid transporters in the human term placenta. Reproduct. Biol. Endocrinol. 13, 57–57.
Gao, K., Mu, C. L., Farzi, A., and Zhu, W. Y. (2020). Tryptophan metabolism: a link between the gut microbiota and brain. Adv. Nutr. 11, 709–723. doi: 10.1093/advances/nmz127
Gleason, G., Liu, B., Bruening, S., Zupan, B., Auerbach, A., Mark, W., et al. (2010). The serotonin1A receptor gene as a genetic and prenatal maternal environmental factor in anxiety. Proc. Natl. Acad. Sci. U.S.A. 107, 7592–7597. doi: 10.1073/pnas.0914805107
Goeden, N., Notarangelo, F. M., Pocivavsek, A., Beggiato, S., Bonnin, A., and Schwarcz, R. (2017). Prenatal dynamics of kynurenine pathway metabolism in mice: focus on kynurenic acid. Dev. Neurosci. 39, 519–528. doi: 10.1159/000481168
Goeden, N., Velasquez, J., Arnold, K. A., Chan, Y., Lund, B. T., Anderson, G. M., et al. (2016). Maternal inflammation disrupts fetal neurodevelopment via increased placental output of serotonin to the fetal brain. J. Neurosci. 36, 6041–6049. doi: 10.1523/jneurosci.2534-15.2016
Hannan, N. J., Bambang, K., Kaitu’u-Lino, T. U. J., Konje, J. C., and Tong, S. (2014). A bioplex analysis of cytokines and chemokines in first trimester maternal plasma to screen for predictors of miscarriage. PLoS One 9:e93320. doi: 10.1371/journal.pone.0093320
Holtan, S. G., Chen, Y., Kaimal, R., Creedon, D. J., Enninga, E. A., Nevala, W. K., et al. (2015). Growth modeling of the maternal cytokine milieu throughout normal pregnancy: macrophage-derived chemokine decreases as inflammation/counterregulation increases. J. Immunol. Res. 2015:952571.
Honig, A., Rieger, L., Kapp, M., Sutterlin, M., Dietl, J., and Kammerer, U. (2004). Indoleamine 2,3-dioxygenase (IDO) expression in invasive extravillous trophoblast supports role of the enzyme for materno-fetal tolerance. J. Reprod. Immunol. 61, 79–86. doi: 10.1016/j.jri.2003.11.002
Huang, X., Luthi, M., Ontsouka, E. C., Kallol, S., Baumann, M. U., Surbek, D. V., et al. (2016). Establishment of a confluent monolayer model with human primary trophoblast cells: novel insights into placental glucose transport. Mol. Hum. Reprod. 22, 442–456. doi: 10.1093/molehr/gaw018
Iwanaga, N., Yamamasu, S., Tachibana, D., Nishio, J., Nakai, Y., Shintaku, H., et al. (2004). Activity of synthetic enzymes of tetrahydrobiopterin in the human placenta. Int. J. Mol. Med. 13, 117–120.
Iwasaki, S., Nakazawa, K., Sakai, J., Kometani, K., Iwashita, M., Yoshimura, Y., et al. (2005). Melatonin as a local regulator of human placental function. J. Pineal Res. 39, 261–265. doi: 10.1111/j.1600-079x.2005.00244.x
Jiraskova, L., Cerveny, L., Karbanova, S., Ptackova, Z., and Staud, F. (2018). Expression of concentrative nucleoside transporters (SLC28A) in the human placenta: effects of gestation age and prototype differentiation-affecting agents. Mol. Pharm. 15, 2732–2741. doi: 10.1021/acs.molpharmaceut.8b00238
Kallol, S., Moser-Haessig, R., Ontsouka, C. E., and Albrecht, C. (2018). Comparative expression patterns of selected membrane transporters in differentiated BeWo and human primary trophoblast cells. Placenta 7, 48–52. doi: 10.1016/j.placenta.2018.10.008
Karahoda, R., Horackova, H., Kastner, P., Matthios, A., Cerveny, L., Kucera, R., et al. (2020). Serotonin homeostasis in the materno-fetal interface at term: role of transporters (SERT/SLC6A4 and OCT3/SLC22A3) and monoamine oxidase A (MAO-A) in uptake and degradation of serotonin by human and rat term placenta. Acta Physiolo. 229:e13478.
Kaur, H., Bose, C., and Mande, S. S. (2019). Tryptophan metabolism by gut microbiome and gut-brain-axis: an in silico analysis. Front. Neurosci. 13:1365. doi: 10.3389/fnins.2019.01365
Kingdom, J., Huppertz, B., Seaward, G., and Kaufmann, P. (2000). Development of the placental villous tree and its consequences for fetal growth. Eur. J. Obstet. Gynecol. Reprod. Biol. 92, 35–43. doi: 10.1016/s0301-2115(00)00423-1
Kolahi, K. S., Valent, A. M., and Thornburg, K. L. (2017). Cytotrophoblast, not syncytiotrophoblast, dominates glycolysis and oxidative phosphorylation in human term placenta. Sci. Rep. 7:42941.
Kudo, Y., Boyd, C. A., Spyropoulou, I., Redman, C. W., Takikawa, O., Katsuki, T., et al. (2004). Indoleamine 2,3-dioxygenase: distribution and function in the developing human placenta. J. Reprod. Immunol. 61, 87–98. doi: 10.1016/j.jri.2003.11.004
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lanoix, D., Beghdadi, H., Lafond, J., and Vaillancourt, C. (2008). Human placental trophoblasts synthesize melatonin and express its receptors. J. Pineal Res. 45, 50–60. doi: 10.1111/j.1600-079x.2008.00555.x
Laurent, L., Deroy, K., St-Pierre, J., Cote, F., Sanderson, J. T., and Vaillancourt, C. (2017). Human placenta expresses both peripheral and neuronal isoform of tryptophan hydroxylase. Biochimie 140, 159–165. doi: 10.1016/j.biochi.2017.07.008
Lee, N., Hebert, M. F., Prasad, B., Easterling, T. R., Kelly, E. J., Unadkat, J. D., et al. (2013). Effect of gestational age on mRNA and protein expression of polyspecific organic cation transporters during pregnancy. Drug Metab. Disposit. 41, 2225–2232. doi: 10.1124/dmd.113.054072
Li, L., and Schust, D. J. (2015). Isolation, purification and in vitro differentiation of cytotrophoblast cells from human term placenta. Reprod. Biol. Endocrinol. 13:71.
Ligam, P., Manuelpillai, U., Wallace, E. M., and Walker, D. (2005). Localisation of indoleamine 2,3-dioxygenase and kynurenine hydroxylase in the human placenta and decidua: implications for role of the kynurenine pathway in pregnancy. Placenta 26, 498–504. doi: 10.1016/j.placenta.2004.08.009
Maldonado-Estrada, J., Menu, E., Roques, P., Barre-Sinoussi, F., and Chaouat, G. (2004). Evaluation of Cytokeratin 7 as an accurate intracellular marker with which to assess the purity of human placental villous trophoblast cells by flow cytometry. J. Immunol. Methods 286, 21–34. doi: 10.1016/j.jim.2003.03.001
Mckinney, J., Knappskog, P. M., and Haavik, J. (2005). Different properties of the central and peripheral forms of human tryptophan hydroxylase. J. Neurochem. 92, 311–320. doi: 10.1111/j.1471-4159.2004.02850.x
Mikheev, A. M., Nabekura, T., Kaddoumi, A., Bammler, T. K., Govindarajan, R., Hebert, M. F., et al. (2008). Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reproduct. Sci. (Thousand Oaks Calif). 15, 866–877. doi: 10.1177/1933719108322425
Mori, M., Ishikawa, G., Luo, S. S., Mishima, T., Goto, T., Robinson, J. M., et al. (2007). The cytotrophoblast layer of human chorionic villi becomes thinner but maintains its structural integrity during gestation. Biol. Reprod. 76, 164–172. doi: 10.1095/biolreprod.106.056127
Muller, C. L., Anacker, A. M., Rogers, T. D., Goeden, N., Keller, E. H., Forsberg, C. G., et al. (2017). Impact of maternal serotonin transporter genotype on placental serotonin, fetal forebrain serotonin, and neurodevelopment. Neuropsychopharmacology 42, 427–436. doi: 10.1038/npp.2016.166
Munn, D. H., Zhou, M., Attwood, J. T., Bondarev, I., Conway, S. J., Marshall, B., et al. (1998). Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281, 1191–1193. doi: 10.1126/science.281.5380.1191
Murthi, P., Wallace, E. M., and Walker, D. W. (2017). Altered placental tryptophan metabolic pathway in human fetal growth restriction. Placenta 52, 62–70. doi: 10.1016/j.placenta.2017.02.013
Nagai, R., Watanabe, K., Wakatsuki, A., Hamada, F., Shinohara, K., Hayashi, Y., et al. (2008). Melatonin preserves fetal growth in rats by protecting against ischemia/reperfusion-induced oxidative/nitrosative mitochondrial damage in the placenta. J. Pineal Res. 45, 271–276. doi: 10.1111/j.1600-079x.2008.00586.x
Notarangelo, F. M., Beggiato, S., and Schwarcz, R. (2019). Assessment of prenatal kynurenine metabolism using tissue slices: focus on the neosynthesis of kynurenic acid in mice. Dev. Neurosci. 41, 102–111. doi: 10.1159/000499736
Petroff, M. G., Phillips, T. A., Ka, H., Pace, J. L., and Hunt, J. S. (2006). “Isolation and culture of term human trophoblast cells,” in Placenta and Trophoblast: Methods and Protocols, eds M. J. Soares and J. S. Hunt (Totowa, NJ: Humana Press).
Ranzil, S., Ellery, S., Walker, D. W., Vaillancourt, C., Alfaidy, N., Bonnin, A., et al. (2019). Disrupted placental serotonin synthetic pathway and increased placental serotonin: potential implications in the pathogenesis of human fetal growth restriction. Placenta 84, 74–83. doi: 10.1016/j.placenta.2019.05.012
Rosenfeld, C. S. (2020a). The placenta-brain-axis. J. Neurosci. Res. doi: 10.1002/jnr.24603 [Epub ahead of print].
Rosenfeld, C. S. (2020b). Placental serotonin signaling, pregnancy outcomes, and regulation of fetal brain development†. Biol. Reprod. 102, 532–538. doi: 10.1093/biolre/ioz204
Sano, M., Ferchaud-Roucher, V., Kaeffer, B., Poupeau, G., Castellano, B., and Darmaun, D. (2016). Maternal and fetal tryptophan metabolism in gestating rats: effects of intrauterine growth restriction. Amino Acids 48, 281–290. doi: 10.1007/s00726-015-2072-4
Schwarcz, R., Bruno, J. P., Muchowski, P. J., and Wu, H. Q. (2012). Kynurenines in the mammalian brain: when physiology meets pathology. Nat. Rev. Neurosci. 13, 465–477. doi: 10.1038/nrn3257
Sedlmayr, P., Blaschitz, A., and Stocker, R. (2014). The role of placental tryptophan catabolism. Front. Immunol. 5:230–230.
Sedlmayr, P., Blaschitz, A., Wintersteiger, R., Semlitsch, M., Hammer, A., Mackenzie, C. R., et al. (2002). Localization of indoleamine 2,3-dioxygenase in human female reproductive organs and the placenta. Mol. Hum. Reprod. 8, 385–391. doi: 10.1093/molehr/8.4.385
Seron-Ferre, M., Mendez, N., Abarzua-Catalan, L., Vilches, N., Valenzuela, F. J., Reynolds, H. E., et al. (2012). Circadian rhythms in the fetus. Mol. Cell Endocrinol. 349, 68–75.
Simner, C., Novakovic, B., Lillycrop, K. A., Bell, C. G., Harvey, N. C., Cooper, C., et al. (2017). DNA methylation of amino acid transporter genes in the human placenta. Placenta 60, 64–73. doi: 10.1016/j.placenta.2017.10.010
Sitras, V., Fenton, C., Paulssen, R., Vårtun, Å, and Acharya, G. (2012). Differences in gene expression between first and third trimester human placenta: a microarray study. PLoS One 7:e33294. doi: 10.1371/journal.pone.0033294
Staud, F., and Karahoda, R. (2018). Trophoblast: The central unit of fetal growth, protection and programming. Int. J. Biochem. Cell Biol. 105, 35–40. doi: 10.1016/j.biocel.2018.09.016
Takikawa, O., Kuroiwa, T., Yamazaki, F., and Kido, R. (1988). Mechanism of interferon-gamma action. Characterization of indoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. J. Biol. Chem. 263, 2041–2048.
Uuskula, L., Mannik, J., Rull, K., Minajeva, A., Koks, S., Vaas, P., et al. (2012). Mid-gestational gene expression profile in placenta and link to pregnancy complications. PLoS One 7:e49248. doi: 10.1371/journal.pone.0049248
Wakx, A., Nedder, M., Tomkiewicz-Raulet, C., Dalmasso, J., Chissey, A., Boland, S., et al. (2018). Expression, localization, and activity of the aryl hydrocarbon receptor in the human placenta. Int. J. Mol. Sci. 19:3762. doi: 10.3390/ijms19123762
Zanetta, L., Marcus, S. G., Vasile, J., Dobryansky, M., Cohen, H., Eng, K., et al. (2000). Expression of Von Willebrand factor, an endothelial cell marker, is up-regulated by angiogenesis factors: a potential method for objective assessment of tumor angiogenesis. Int. J. Cancer 85, 281–288. doi: 10.1002/(SICI)1097-0215(20000115)85:23C281::AID-IJC213E3.0.CO;2-3
Keywords: fetal programming, trophoblast, tryptophan metabolism, placenta–brain axis, kynurenine pathway, serotonin pathway
Citation: Karahoda R, Abad C, Horackova H, Kastner P, Zaugg J, Cerveny L, Kucera R, Albrecht C and Staud F (2020) Dynamics of Tryptophan Metabolic Pathways in Human Placenta and Placental-Derived Cells: Effect of Gestation Age and Trophoblast Differentiation. Front. Cell Dev. Biol. 8:574034. doi: 10.3389/fcell.2020.574034
Received: 18 June 2020; Accepted: 21 August 2020;
Published: 18 September 2020.
Edited by:
Maria Barile, University of Bari Aldo Moro, ItalyReviewed by:
Luc Maroteaux, INSERM U839 Institut du Fer à Moulin (IFM), FranceCopyright © 2020 Karahoda, Abad, Horackova, Kastner, Zaugg, Cerveny, Kucera, Albrecht and Staud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frantisek Staud, ZnJhbnRpc2VrLnN0YXVkQGZhZi5jdW5pLmN6
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.