- 1Unità di Biologia Cellulare e dello Sviluppo, Dipartimento di Biologia, Università di Pisa, Pisa, Italy
- 2Molecular Medicine, IRCCS Fondazione Stella Maris, Pisa, Italy
- 3Scuola Normale Superiore di Pisa, Pisa, Italy
Mex3A is an RNA binding protein that can also act as an E3 ubiquitin ligase to control gene expression at the post-transcriptional level. In intestinal adult stem cells, MEX3A is required for cell self-renewal and when overexpressed, MEX3A can contribute to support the proliferation of different cancer cell types. In a completely different context, we found mex3A among the genes expressed in neurogenic niches of the embryonic and adult fish brain and, notably, its expression was downregulated during brain aging. The role of mex3A during embryonic and adult neurogenesis in tetrapods is still unknown. Here, we showed that mex3A is expressed in the proliferative region of the developing brain in both Xenopus and mouse embryos. Using gain and loss of gene function approaches, we showed that, in Xenopus embryos, mex3A is required for neuroblast proliferation and its depletion reduced the neuroblast pool, leading to microcephaly. The tissue-specific overexpression of mex3A in the developing neural plate enhanced the expression of sox2 and msi-1 keeping neuroblasts into a proliferative state. It is now clear that the stemness property of mex3A, already demonstrated in adult intestinal stem cells and cancer cells, is a key feature of mex3a also in developing brain, opening new lines of investigation to better understand its role during brain aging and brain cancer development.
Introduction
In developmental processes, spatial and temporal control of gene expression occurs at transcriptional, post-transcriptional and post-translational levels. More than 1000 genes in the eukaryotic genome encode multifunctional RNA-binding proteins (RBPs) and 50% of these RBPs are expressed in the brain where they regulate all levels of RNA biogenesis at different levels (Bryant and Yazdani, 2016). The neural specific RBPs play a key role in post-transcriptional control, regulating RNA splicing, transport, surveillance, decay and translation (Glisovic et al., 2008).
By RNA-seq analysis we identified a set of evolutionarily conserved, age-regulated genes, expressed in adult neural stem cell niches (aNSCs), in the short-lived fish Nothobranchius furzeri, a well-established animal model for aging studies (Baumgart et al., 2014). Among them, the RNA-binding protein mex3A emerged as a putative new neurogenic regulator, down-regulated with age and expressed in neurogenic regions of the zebrafish embryo (Baumgart et al., 2014). This RNA-binding protein belongs to MEX3 family and vertebrates have four distinct mex-3 orthologs (mex-3A–D). All four proteins predominantly accumulate in the cytoplasm, and shuttle between the cytoplasm and the nucleus via CRM1-dependent export pathway (Fornerod et al., 1997). MEX3 genes encode proteins containing two heterogeneous nuclear ribonucleoprotein K homology (KH) domains and one carboxy-terminal RING finger module with E3 ubiquitin ligase activity (Draper et al., 1996; Buchet-Poyau et al., 2007) sharing the highest identity with Caenorhabditis elegans mex-3, a translational repressor involved in the maintenance of germline pluripotency (Ciosk et al., 2006; Hwang and Rose, 2010). The role of mex3 genes in mammals is poorly understood, though several studies suggest its putative involvement in self-renewal/differentiation decisions with implications for stem cell and cancer biology. In particular, human MEX3A was shown to play a key function in gastrointestinal context by impairing intestinal differentiation and simultaneously promoting an increased expression of intestinal stem cells markers such as LGR5, BML1, and MS1 (Pereira et al., 2013, 2020). In mice, mex3A is expressed in the crypt base and labels a slowly cycling subpopulation of Lgr5+ intestinal stem cell population (Barriga et al., 2017; Chatterji and Rustgi, 2018). MEX3A is overexpressed in pancreatic ductal adenocarcinoma (Wang et al., 2020) and strongly up-regulated in glioblastoma samples (Bufalieri et al., 2020). Despite this evidence, to our knowledge, there are no data available regarding the putative role of mex3a during embryonic and adult neurogenesis.
Here we used the clawed frog Xenopus laevis embryos to characterize the biological function of mex3A in the developing central nervous system (CNS). Xenopus embryos gave us the unique opportunity to perform functional experiments in a tissue specific manner without interfering with the normal development of all other tissues (Vitobello et al., 2011; Naef et al., 2018). We showed that mex3A is expressed in proliferative regions of Xenopus and mouse developing brain including the eye, the brain and neural crest cells. The results from gain and loss of gene function experiments suggested that mex3A plays key role in primary mechanisms of proliferation of neural precursors linking cell division and neuronal differentiation during embryonic neurogenesis.
Materials and Methods
Molecular Cloning of mex3A
The available Expressed Sequence Tag (EST) clone of X. laevis mex3A (ID_6638558, gene bank BC_130195) lacks the coding region at 5′-end. To isolate the 5′-end coding sequence, we used the SMARTTM RACE cDNA Amplification kit (Clontech). The final PCR product was purified and sequenced. We obtained the full-length coding sequence of X. laevis mex3A submitted to The National Center for Biotechnology Information (NCBI) (ID_2213511) (Gene bank: MK_800014). A fragment of 975 bp of mouse mex3a cDNA (Gene Bank NM_001029890) was amplified and cloned into pGEM-T vector (Promega). The full-length cDNA sequence of zebrafish mex3a (Gene Bank XM_009292667) was amplified and cloned into pCS2+ vector.
Multiple Sequence Alignments of MEX3A Amino Acid Sequences
Multiple sequence alignments of MEX3A amino acid sequences were performed using the NCBI GeneBank for the following organisms: X. laevis mex3A (MK_800014); zebrafish mex3a (XM_009292667); Homo sapiens MEX3A (NM_001093725.2); Mus musculus Mex3A (NM_001029890.2).
Embryo Collection
Animal handling and care were performed in strict compliance with protocols approved by Italian Ministry of Public Health and of local Ethical Committee of University of Pisa (authorization n. 99/2012-A, 19.04.2012). X. laevis embryos were obtained by hormone-induced laying and in vitro fertilization then reared in 0.1× Marc’s Modified Ringer’s Solution (MMR 1× : 0.1 M NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES pH 7.5) until the desired stage according to Nieuwkoop and Faber (Nieuwkoop, 1956).
Morpholino Oligonucleotides, mRNA in vitro Transcription and Microinjections
All morpholinos (MOs) were obtained from Gene Tools, LLC (Philomath, OR, United States). The injections were performed into one side of the embryo in the dorsal blastomere at the 4 cells stage embryo to target neural tissue. The sequences of MOs used were mex3A MO1 sequence: 5′-CAGCAGG CTCGGCATGGCTAATAAC-3′; mex3A MO2 sequence: 5′ CATT CCTCTCCATCATCCCTGAGAG-3′; Control Standard Morpholino sequence: 5′-CCTCTTACCTCAGTTACAATTTA TA-3′. Microinjections were performed as described previously (Corsinovi et al., 2019). We injected 12ng per embryos of experimental and control morpholinos. To select properly injected embryos, we co-injected MOs with 250 pg of gfp mRNA and we proceeded with the analysis of the embryos that, at neurula stages (stage 15), showed a specific fluorescence in the neural plate of the injected site. The un-injected side represented an internal control in each embryo. We prepared capped mex3A and gfp mRNAs using the SP6 mMessage Machine in vitro transcription kit (Ambion), according to manufacturer’s instructions. For rescue experiments, we co-injected 12ng mex3A MO2 and 600ng of full-length Xenopus or zebrafish mex3A mRNA.
Whole Mount in situ Hybridization
Whole Mount in situ Hybridizations (WISHs) were performed as described (Naef et al., 2018). After color development, embryos were post-fixed and bleached over light to remove the pigment. The following plasmids were used for preparation of antisense RNA probes, enzyme used for linearization and the polymerases used for probe synthesis were and polymerases are indicated; X. laevis mex3A-pGEM-T (ClaI, Sp6); pcna-pBSK, sox2-pCS2+, N-tubulin-pBKS, elrC-pBKS, huD-pBSK,; twist-pcr2.1 topo (HindIII,T7); sox10-pBKS (EcoRI;T3); slug-sp72 (EcorV,SP6); foxd3-pBSK (EcoRI;T7); msi-1-pCMV-sport6 (EcoRI;T7) (a kind gift from Dr. Romualdo Ciau-Uitz); mouse mex3A-pGEM-T (NotI, T7).
In situ Hybridization on Frozen Tissue Sections (ISH)
For ISH on cryosections, Xenopus embryos were fixed in 4% paraformaldehyde in PBS, cryoprotected with 30% sucrose in PBS and embedded in Tissue-Tek OCT compound (Sakura, 4583). We prepared 12 μm cryosections and ISH was performed according to (Casini et al., 2012). Mouse embryo sections are a kind gift of Prof. Massimo Pasqualetti and were prepared as described in Pelosi et al. (2014). In situ Hybridization (ISH) on mouse embryo cryosections at 18 dpc was performed according to (Borello et al., 2014).
TUNEL and PH3 Staining in Xenopus and Statistical Analysis
TdT-mediated DUTP-dig nick end labeling (TUNEL) and PH3 (phospho histone 3) staining were performed according to established protocols (Ori et al., 2006). TUNEL and PH3 positive cells were counted within defined areas in control and injected sides of each manipulated embryo using (Ori et al., 2006) the ImageJ64 software. P-values were calculated by paired Student’s t-test using GraphPad Prism 6 software (San Diego, CA, United States). Statistical significance was indicated as: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ****p ≤ 0.0001.
Measurement of Brain Areas in Xenopus and Statistical Analysis
To determine the brain area, embryos at stage 41 (swimming larvae) were anesthetized with buffered tricaine methane sulfonate (MS222) and then fixed in 4% paraformaldehyde in PBS. Brains were isolated using fine forceps and areas of the un-injected and injected sides were calculated using the ImageJ64 software. P-values were calculated by paired Student’s t-test using GraphPad Prism 6 software (San Diego, CA, United States). Statistical significance was indicated as: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ****p ≤ 0.0001.
Quantitative Reverse Transcription Polymerase Chain Reaction and Statistical Analysis
Total RNA was extracted from 30 Xenopus morphants at neurula stage (stage 18) using Nucleospin® RNA (Macherey-Nagel) according to manufacturer’s instructions. cDNA was prepared by using iScriptTM cDNA Synthesis Kit (Bio-Rad) and quantitative real-time PCR was performed using GoTaq®qPCR master mix (Promega) according to the manufacturer’s instruction. Relative expression levels of each gene were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The results obtained in three independent experiments were normalized to the expression of housekeeping gene, gapdh. The mean of the Control-Morpholino was set at 1. Statistical analysis for qRT-PCR experiments was performed by Student’s t-test using GraphPad Prism 6 software (San Diego, CA, United States). Statistical significance was indicated as: ∗p ≤ 0.05. Following primers were used to perform qRT-PCR: pcna (Huyck et al., 2015); N-tubulin and sox2 (De Robertis’s lab, web site: http://www.hhmi.ucla.edu/derobertis/); elrC (Seo et al., 2007); Glyceraldehyde 3-phosphate dehydrogenase (gapdh) (Naef et al., 2018).
Statistical Analysis of Embryo Phenotype
Statistical analysis for phenotypes observed after the injection of the Control-Morpholino or the injection of mex3A-MO2 was performed by Student’s t-test using GraphPad Prism 6 software (San Diego, CA, United States). We compared the percentage of embryos with altered marker genes expression between Control-Morpholino injected embryos and mex3A-MO2 injected embryos. Statistical significance was indicated as: ∗p ≤ 0.5, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
Results
Mex3A Is Expressed in the Developing Xenopus laevis Brain
We compared X. laevis mex3A predicted protein sequence with the zebrafish, mouse and human homologs revealing a high degree of similarity, especially in RNA binding domains (96%) and C-terminal Ring finger domain with E3 ligase activity (95%) suggesting a conserved function of mex3A in vertebrates (Supplementary Figure 1). As a prerequisite for functional studies, firstly we analyzed the spatial expression pattern of mex3A during early embryogenesis. Whole mount in situ hybridization (WISH) revealed that mex3A is already present in early cleaving stage (four cells stage) before the midblastula transition suggesting that it is maternally supplied (Figure 1A). At mid neurula stage, mex3A could be detected in the neural plate, in presumptive eyes territory, in pre-placodal territory and in cranial neural crest cells (NCC) (Figure 1B). At later stages of development, mex3A mRNA is present in the eye, in the CNS and in NCC migrated in branchial arches (Figures 1C,D). In situ hybridization on cryosections at stage 41 showed the mex3A expression in brain areas with high proliferative activity such as the ciliary marginal zone (CMZ) in the retina, the ventricular zone of the midbrain and the subventricular zone of the hindbrain (Figures 1E,F).
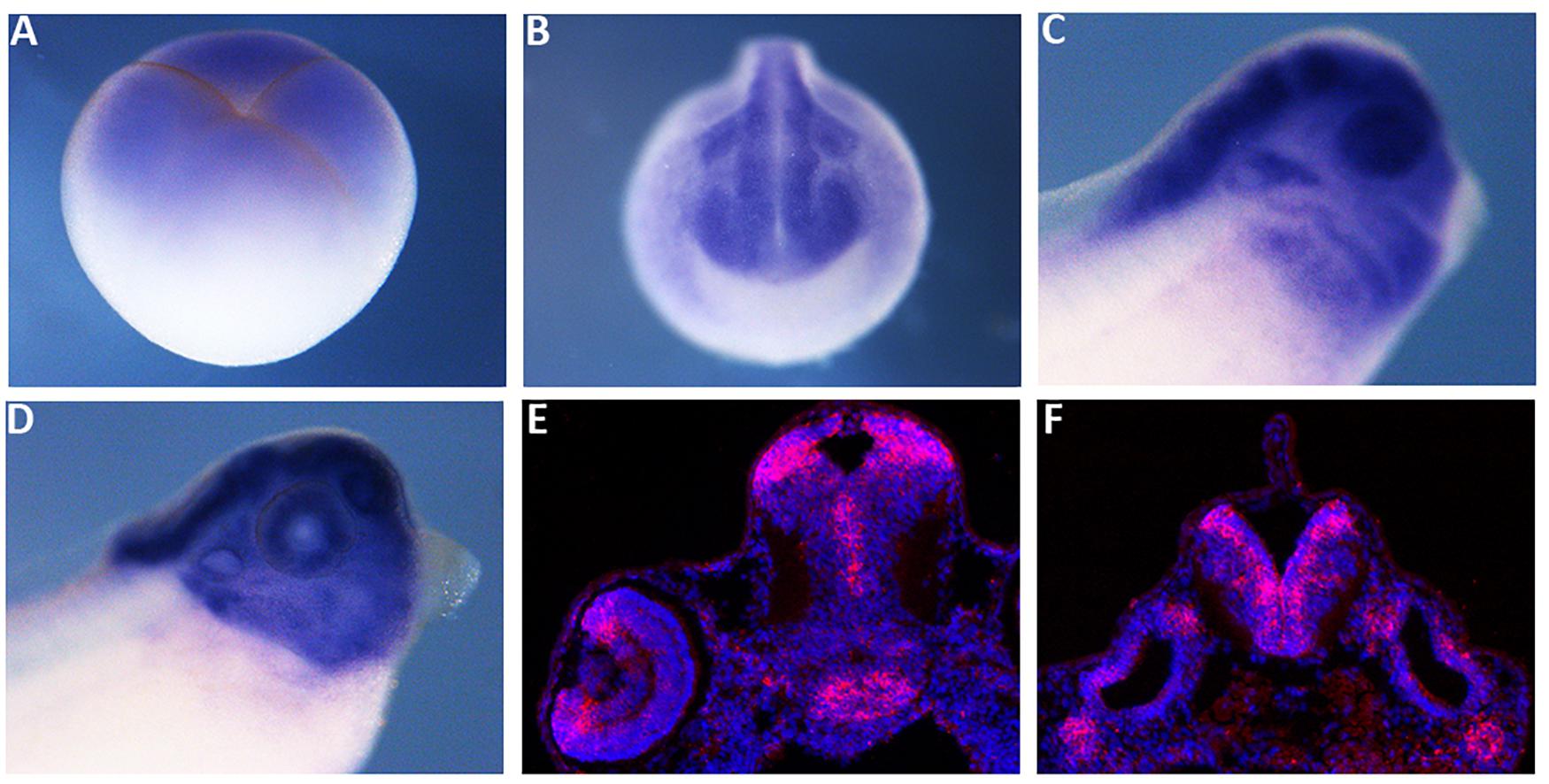
Figure 1. The spatial expression pattern of Xenopus mex3A. Whole mount in situ hybridization approaches show that mex3A is expressed in the central nervous system. (A) Mex3A expression at blastula stage (stage 3). (B,C) At neurula (stage 20) and at tadpole stages (stage 27), mex3A is expressed in: neural tube, developing eye, neural crest cells (white arrowhead) and otic vesicle (white arrow). At stage 37, an accumulation of mex3A transcript persisted in the most anterior region of the central nervous system (D). (E,F) In situ hybridization on frozen tissue of transverse sections on WT embryos at stage 41. ISH signal was revealed using fluorescent Fast Red and is visualized in red. Nuclei were revealed in blue with Hoechst. (E,F) Mex3A is expressed in CMZ of the retina (red arrow), in ventricular zone of the midbrain and in subventricular zone of the hindbrain.
Mex3A Supports Neuroblasts Proliferative State
Since the expression of mex3A suggested a role during primary neurogenesis, we overexpressed mex3A in X. laevis embryos to evaluate its possible impact on primary neuron formation. For all experiments described below, mex3A-mRNA injections were done unilaterally into the animal region of one dorsal blastomere at the four cells stage embryo to target neural tissue. The un-injected side served as internal injection control and the co-injection of gfp mRNA was used to select and analyze only embryos in which the transcripts correctly localized in the neural plate (Figure 2A). At neurula stage (stage 18), WISH experiments revealed that the overexpression of mex3A altered expression domains of sox2 and musashi-1 (msi-1). The expression domains of sox2, a neuroblast marker (Mizuseki et al., 1998), and msi-l, commonly considered a specific marker for stem/progenitor cells (Okano et al., 2005), were markedly expanded in the injected side of the embryo as compared to the un-injected side (Figures 2B,C). Furthermore, we examined the expression of elrC, a marker of cells undergoing a transition from proliferation to differentiation (Carruthers et al., 2003), at neurula and tailbud stages. The expression domain of elrC appeared dramatically down-regulated in injected side of the embryos compared to un-injected side (Figures 2D–E′). Given these preliminary results, well correlated with the function of human MEX3A as positive regulator of cell cycle progression of intestinal precursors (Pereira et al., 2013; Barriga et al., 2017), we hypothesized that mex3A might be involved in cell proliferation also in the neural context. To elucidate this possibility, we analyzed the number of mitotically active cells in mex3A overexpressing embryos by immunostaining for mitotic Ser-10-phosphorylated Histone 3 (pH3). We observed a significant increase in mitotic cell number in the injected side of the embryo compared to the control side (Figures 2F,G). These data suggested that mex3A could maintain the proliferative state of neuroblasts delaying or preventing the neuronal differentiation during embryonic neurogenesis.
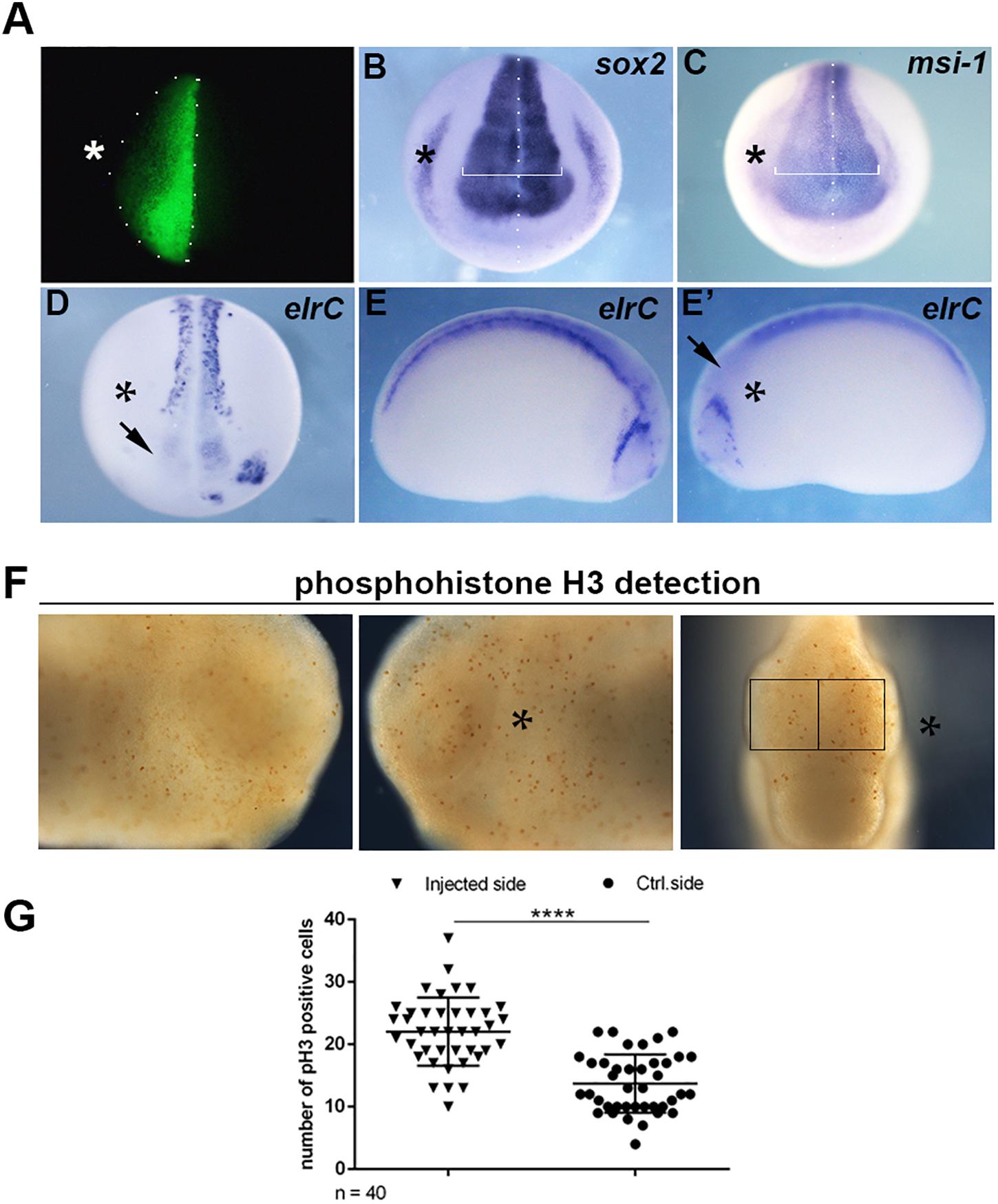
Figure 2. (A) Embryos injected with gfp (250 pg) and mex3A (500 pg) mRNA in one dorsal blastomere at the four-cells stage showing fluorescence only in the neural plate at neurula stage were cultured till different stages of development for WISH analysis. In each panel the asterisk (*) indicates the injected side of the embryo. (B,C) mRNA distribution of sox2 and msi-1 (sox2 phenotype 54%, n = 116; msi-1 phenotype 50%, n = 80) in mex3A overexpressing embryos. The arrow in (D) showed the lack of elrC expression in the anterior neural plate. (D–E′) mRNA distribution of elrC at 18 (phenotype 54%, n = 114, D) and at 23 (phenotype 57%, n = 70, E,E′) stages in mex3A overexpressing embryos. The arrow in (E′) shows the lack of neurons in the anterior neural tube. (F,G) pH3 positive cells were counted in the areas defined by the black rectangles. Statistical evaluation of the data is shown (n = 40). Abbreviations: n total number of processed embryos; error bars indicate standard error of the means (SEM); ***p ≤ 0.001.
Mex3A Depletion Impairs Primary Neurogenesis
To study the role of mex3A in primary neurogenesis context, we also performed experiments of gene loss of function by using a specific morpholino oligo designed to block mRNA translation. However, by analyzing the sequence of the unique mex3A exon, we found that there are two possible translation start codons in frame (Supplementary Figure 1). Because both codons can be used as translation initiation sites, if we block the first translation start site using a specific morpholino oligo there is the possibility that the second start site could be used to translate a protein identical to the native one except for the first eight amino acids. The presence of a second ATG in frame and in the same position is conserved in vertebrate orthologs of mex3A (Supplementary Figure 1). We designed two specific morpholinos to inject them individually or in combination in the same embryo: morpholino 1 (MO1) designed to block the first ATG and morpholino 2 (MO2) designed to block the second ATG of the Xenopus mex3A mRNA. Since the injection of the MO1 did not generate any type of phenotype and the combination of the MO1 and MO2 increased the mortality rate without any synergic or additive effect, we used MO2 alone for subsequent analyses (Figure 3A). A standard control morpholino (CoMO) was used to evaluate non-specific embryo responses. By WISH experiments we showed that the expression domain of sox2 was reduced in mex3A-MO injected side of the embryo whereas both un-injected and CoMO injected sides were unaffected (Figures 3B,C). These data were confirmed by qRT-PCR analysis that showed a significant down-regulation of sox2 mRNA in mex3A morphants (Figure 3D). To further verify whether the loss of mex3A function could alter the regulation of neuroblast proliferation, we also examined the mRNA expression of pcna (proliferating cell nuclear antigen) (Strzalka and Ziemienowicz, 2011). Mex3A morphants showed a reduced pcna expression as detected by WISH (Figures 3B,C) and qRT-PCR experiments (Figure 3D). As a consequence of the impairment in the maintenance of neuronal progenitors pool, we observed that the lateral stripe of N-tubulin and elrC expression domains, the future sensory neurons, appear expanded on the injected side of the embryos compared to control side and CoMO injected embryos (Figures 3E,F). This phenotype might be due to an altered density and/or number of primary neurons. Hence, we performed qRT-PCR analysis that revealed a significant raise of N-tubulin and elrC mRNA level in mex3A morphants (Figure 3G). In order to verify the specificity of the mex3A-MO, we designed functional rescue experiments by co-injecting mex3A-MO together with the full-length mex3A mRNA. As the mex3A-MO could target not only the endogenous mex3A but also the in vitro transcribed Xenopus mex3A mRNA, for rescue experiments we cloned the zebrafish mex3A mRNA that is not recognized by mex3A-MO (Supplementary Figure 3). We already showed that the zebrafish mex3A is localized in proliferating region of the developing brain (Baumgart et al., 2014). We further showed that the overexpression of zebrafish mex3A, in Xenopus embryos, reproduced the same phenotype obtained by the Xenopus mex3A mRNA injection, thus confirming its functional conservation (Supplementary Figure 3). We then analyzed 123 co-injected embryos (mex3A-MO plus zebrafish mex3A mRNA) and we observed a restoration of the phenotype at neurula stage (stage 18) (Supplementary Figure 3).
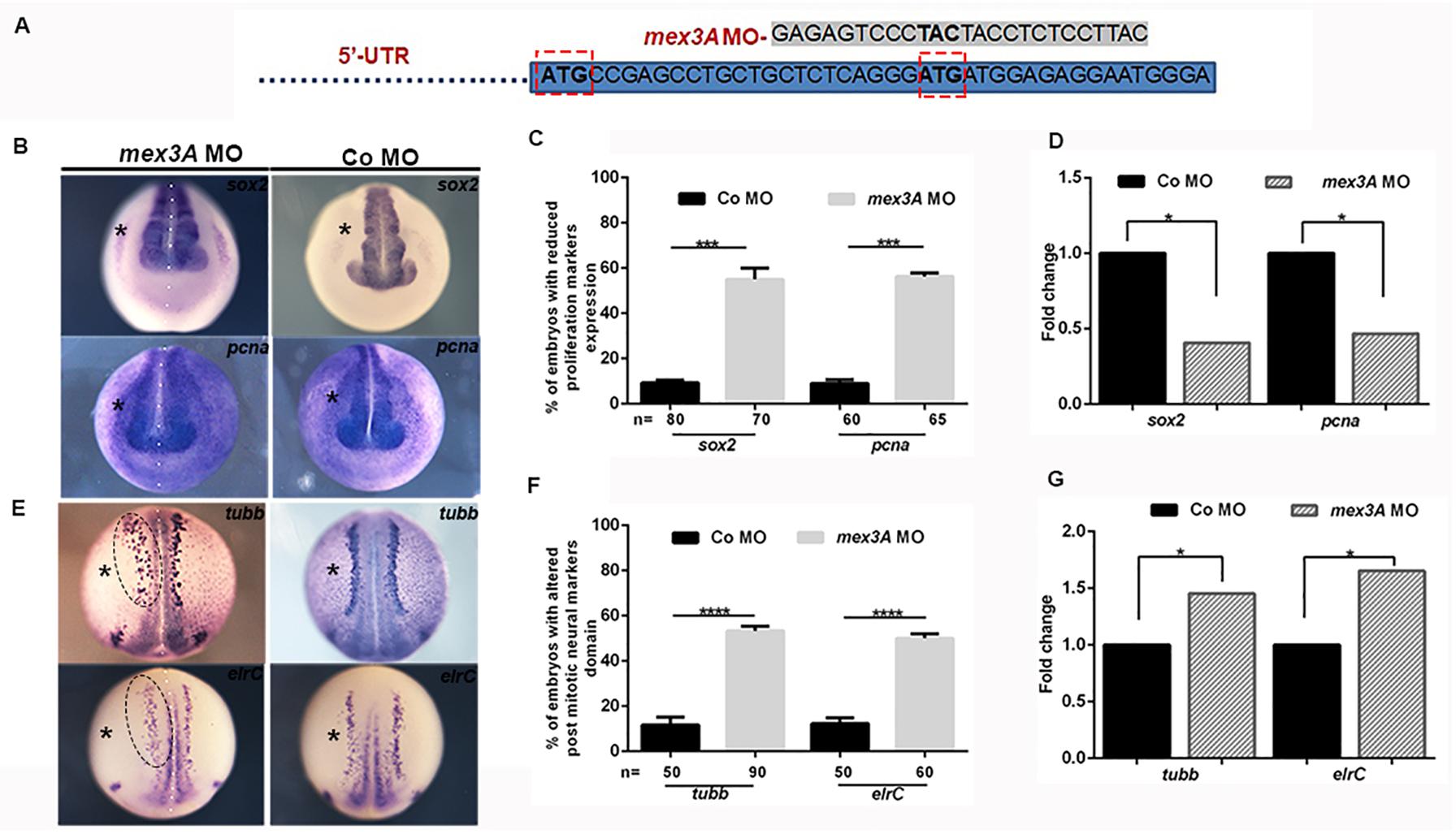
Figure 3. Loss of mex3A alters neuronal specification and differentiation (A) Structure of mex3A-morpholino oligonucleotide. The MO targets the second translation start site. (B) mRNA distribution of sox2 and pcna in mex3A morphants and controls. (C) Quantification of the data in B. (D) qRT-PCR analysis. Relative expression levels of each gene are normalized to gapdh expression. (E) mRNA distribution of N-tubulin and elrC in mex3A morphants and controls. (F) Quantification of the data in E. (G) qRT PCR analysis. (Abbreviations: n number of evaluated embryos in total; error bars indicate standard error of the means (SEM); *p ≤ 0,05, ***p ≤ 0.001, ****p ≤ 0.0001.
Mex3A Is Required for Anterior Neuronal Development in Xenopus laevis
The analysis of gene expression profile of mex3A showed a specific mex3A expression in the anterior neural tissue in Xenopus larvae including eye and brain (Figure 1). Therefore, to investigate in more details the putative biological function of mex3A during anterior neural development, we analyzed embryos at later stages of development. We observed in mex3A morphants, at larval stage 41, smaller and deformed eye with variable penetrance (Figures 4A,B). In contrast, in control side, as well as in CoMO injected embryos, the eye was always normal (Figure 4A). To test the specificity of mex3A-MO to induce eye phenotype, we performed rescue experiments co-injecting the mex3A-MO with the zebrafish mex3A mRNA, observing a restoration of the eye phenotype (Figures 4A,B). To better show possible alteration in larval brain development, we dissected morphants and control brains from larvae at stage 41 and we measured the areas of both brain hemispheres of injected versus un-injected side. We calculated brain area as described in Kiem et al., 2017. In comparison to the CoMO hemisphere (Figures 4C,D), the mex3A-depleted hemisphere exhibited a significant size reduction (Figures 4C,D). This phenotype could be due to a decrease in the cell proliferation rate. To examine this hypothesis, we performed pH3 immunohistochemistry (to visualize mitotic cells) experiments using mex3A-depleted embryos at tailbud stage (stage 24). pH3 staining showed a significant reduction in cell proliferation in mex3A morphants compared to un-injected control side and to CoMO injection (Figures 4F–I). These results suggested a requirement for mex3A in the control of cell proliferation at both neurula and tailbud stages.
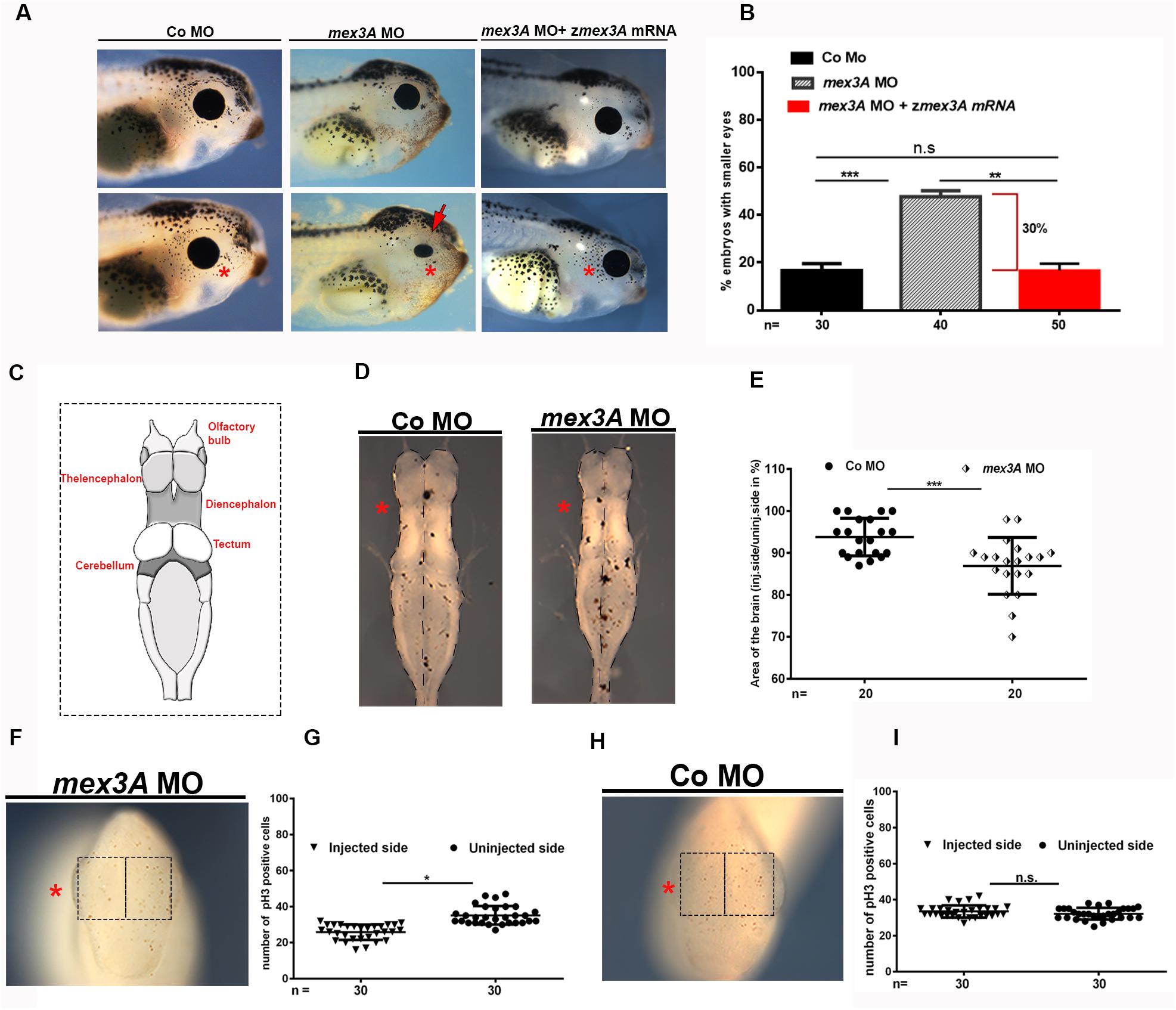
Figure 4. The inhibition of mex3A function causes defects in anterior neuronal development. In each panel, the asterisk indicates the injected side of the embryo (A) Representative images and (B) quantification of the effect of injection of mex3A-MO and co-injection of mex3A-MO with zebrafish mex3A mRNA on the eye phenotype. (C) Image showing the anatomy of Xenopus brain. Diagrams showing a dorsal view of isolated brains. (D) Bright field images of Xenopus brains at stage 41, anterior to the top after unilateral injection of mex3A-MO or CoMO. (E) Statistical evaluation of the brain size in injected embryos. (F–I) pH3 staining in mex3A-deficient embryos at stage 24. Mex3A depletion leads to a significant reduction of proliferating cells compared to the un-injected side, whereas the CoMO injection does not influence on proliferation. pH3 positive cells were counted in the areas defined by the black rectangles. Statistical evaluations of the data are shown. Statistical quantifications of the data are given. Abbreviations: n, total number of evaluated embryos in total; SEM, error bars indicate standard error of the means; ns, not significant. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Mex3A Is Expressed in Developing Mouse Brain
The hypothesis that the intestinal stemness-related gene mex3A could be considered as a regulator of neuroblast proliferation in the CNS is intriguing but no data are available for the expression of mex3a in mammalian CNS. For this reason, we performed a preliminary analysis of mouse mex3A mRNA distribution in the developing mouse brain. We revealed that at 18 dpc mex3A mRNA is present in proliferating regions of the mouse embryonic CNS such as telencephalic ventricular and sub-ventricular zone, developing hippocampus, olfactory bulbs and olfactory epithelium (Supplementary Figure 2) strongly suggesting a conserved role of mex3A in tetrapods CNS.
Discussion
Mex-3 family members are mediators of post-transcriptional regulation in different organisms (Pereira et al., 2013). Several studies highlighted their involvement in different physiological processes, including the maintenance of the balance between stem cell self-renewal and differentiation. In particular, human MEX3A is necessary to post-transcriptionally regulate the levels of CDX2, mRNA coding for an intestinal transcription factor required in gastrointestinal homeostasis (Pereira et al., 2013). Mex3A appears crucial for the maintenance of the slowly cycling subpopulation of lgr5+ gut stem cells (Chatterji and Rustgi, 2018), and lgr5 absence in Mex3A–/– mice leads to growth retardation, postnatal mortality, and severe impairment of intestinal crypt development (Pereira et al., 2020).
Recent data showed that MEX3A is up-regulated in glioblastoma specimens (Bufalieri et al., 2020). In glioblastoma cells, MEX3A interacts with the tumor suppressor RIG-I inducing its ubiquitinylation and the proteasome-dependent degradation, supporting tumor growth (Bufalieri et al., 2020). Although MEX3A has a key role in gastrointestinal homeostasis and tumor progression, its putative role in neural context is not yet defined.
Previously, we showed mex3A expression in aNSCs niches in N. furzeri and in proliferating areas of the developing brain in zebrafish embryos (Baumgart et al., 2014). In the last years, the single cell technologies allowed us to query publicly available datasets and to obtain precious clues on gene expression and possible gene function in different animal models. Transcriptomic analysis of the ventricular-subventricular zone (V-SVZ) of lateral ventricles of male mice at 2, 6, 18, and 22 months revealed mex3A among the genes that significantly change their expression, being down regulated, during aging (Apostolopoulou et al., 2017). Benayoun and collaborators included Mex3A among the top genes down regulated in olfactory bulbs, another neurogenic niche in the adult brain, during mouse aging (Benayoun et al., 2019). These data nicely correlated with our previous observation of an age-related decline of mex3a expression in aNSC niches during N. furzeri brain aging (Baumgart et al., 2014) strongly suggesting a functional conservation of the role of mex3a in brain aging among vertebrates. Despite these suggestive clues, nothing is known about mex3A function in the vertebrate nervous system. Here we revealed, for the first time, the expression and function of mex3A during early neural development using X. laevis as model system. We showed that, besides its widely described role in gastrointestinal context, mex3A is additionally involved in CNS development of tetrapods. Mex3A is expressed in the neural tissue of the early X. laevis embryo including the eye field and neural crest cells. Mex3A mRNA is localized in areas with high proliferative activity such as the ciliary marginal zone (CMZ) of the retina, the ventricular zone of the midbrain and the subventricular zone of the hindbrain strengthening the hypothesis that mex3A could promote proliferation of progenitor cells also in neural context. In order to verify possible evolutionary conservation of mex3A role in the developing CNS, we visualized mouse Mex3A expression in 18 dpc embryos. We confirmed that Mex3A is expressed in proliferative areas of the developing mouse brain, such as in the ventricular-subventricular zone of the lateral ventricles and in the olfactory bulbs. These data suggested a mex3A involvement in the context of primary neurogenesis conserved among vertebrates.
Gene gain and loss of function approaches in Xenopus revealed that this gene was able to keep the undifferentiated and proliferative state of neuroblasts increasing the expression of proliferation markers and decreasing the expression of marker such as elrC (huC) and elrD (huD) during neurogenesis. This evidence suggests that mex3A could function as a potential regulator of proliferation rate of neural progenitor cells and this hypothesis is also supported by the increased expression of musashi-1 in mex3A overexpressing embryos. Msi-1 was first reported to be required for the proper development of the neural sensory organ in Drosophila (Nakamura et al., 1994), whereas it is commonly considered a specific marker for stem/progenitor cells in mammals (Kaneko et al., 2000). Msi-1 maintains stem cell proliferation state by acting as a translational repressor (Ratti et al., 2006). Interestingly, Msi-1 is regulated by Mex3A in mammalian gut cell (Pereira et al., 2013). In Xenopus another member of Mex gene family, mex3b, is expressed during early development and neurogenesis (Takada et al., 2009). Even if the expression pattern of the mex3A and mex3B are not overlapping, they seem to be both expressed in the neural plate and then in the neural tube during neurulation. Comparing our data with that obtained by Takada and collaborators, mex3A and mex3B seem to act not redundantly. The overexpression of mex3B in the neuroectoderm did not affect the expression profile of sox2 (Takada et al., 2009) and the gain or loss of mex3B function suggested an involvement of the gene in antero-posterior patterning of the neural tube (Takada et al., 2009). Our results showed that the overexpression, or the knockdown of mex3A, did not affect the antero-posterior axis formation or the regionalization of the neural tube supporting the idea that the two genes could act independently and in different time windows during CNS development.
Several neural-specific RNA-binding proteins are key inducers of neuronal proliferation and/or differentiation through the stabilization and/or translational enhancement of target transcripts. Additionally, Mex3A seems to have an important role as post-translational regulators also acting as E3 ubiquitin ligase in glioblastoma cells (Bufalieri et al., 2020).
In conclusion, we showed a key role of mex3A as a new post-transcriptional regulator able to influence neuroblast proliferation during neurogenesis. Mex3A gene function is necessary and sufficient to support the expression of sox2 and msi1, required for neuroblast self-renewal.
In light of this, in the future, it will be interesting to focus on the possible mex3A targets in neuroblast and adult neural stem cells to better clarify its role in development and aging of the CNS with possible translational implications in brain cancer research.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MK800014.1.
Ethics Statement
The animal study was reviewed and approved by Ministry of Public Health and local Ethical Committee of University of Pisa (authorization n. 99/2012-A, 19.04.2012).
Author Contributions
VN, MD, and GT performed Xenopus experiments. DC and UB cloned mouse mex3a and performed ISH on mouse embryo cryosections. VN and RA contributed in the manuscript discussion and writing. VN performed the data analysis. MO contributed to conceptualization, provided necessary financial resources, experimental supervision, data analysis, discussion, and writing. All authors contributed to the article and approved the submitted version.
Funding
RA was supported by “Rita Levi Montalcini” funding from MIUR (Italian Ministry of Education, University and Research).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Guglielma De Matienzo for technical support and Salvatore Di Maria for Xenopus care. We would like to thank Prof. Massimo Pasqualetti for mouse embryo cryosections. This work was supported by funding from University of Pisa (Michela Ori).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.549533/full#supplementary-material
References
Apostolopoulou, M., Kiehl, T. R., Winter, M., Cardenas De La Hoz, E., Boles, N. C., Bjornsson, C. S., et al. (2017). Non-monotonic changes in progenitor cell behavior and gene expression during aging of the adult V-SVZ neural stem cell niche. Stem Cell Rep. 9, 1931–1947. doi: 10.1016/j.stemcr.2017.10.005
Barriga, F. M., Montagni, E., Mana, M., Mendez-Lago, M., Hernando-Momblona, X., Sevillano, M., et al. (2017). Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell 20, 801.e7–816.e7.
Baumgart, M., Groth, M., Priebe, S., Savino, A., Testa, G., Dix, A., et al. (2014). RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell 13, 965–974. doi: 10.1111/acel.12257
Benayoun, B. A., Pollina, E. A., Singh, P. P., Mahmoudi, S., Harel, I., Casey, K. M., et al. (2019). Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709. doi: 10.1101/gr.240093.118
Borello, U., Madhavan, M., Vilinsky, I., Faedo, A., Pierani, A., Rubenstein, J., et al. (2014). Sp8 and COUP-TF1 reciprocally regulate patterning and fgf signaling in cortical progenitors. Cereb. Cortex 24, 1409–1421. doi: 10.1093/cercor/bhs412
Bryant, C. D., and Yazdani, N. (2016). RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 15, 169–186. doi: 10.1111/gbb.12273
Buchet-Poyau, K., Courchet, J., Le Hir, H., Séraphin, B., Scoazec, J. Y., Duret, L., et al. (2007). Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 35, 1289–1300. doi: 10.1093/nar/gkm016
Bufalieri, F., Caimano, M., Lospinoso, Severini L, Basili, I., Paglia, F., Sampirisi, L., et al. (2020). The RNA-binding ubiquitin ligase MEX3A affects glioblastoma tumorigenesis by inducing ubiquitylation and degradation of RIG-I. Cancers 12:321. doi: 10.3390/cancers12020321
Carruthers, S., Mason, J., and Papalopulu, N. (2003). Depletion of the cell-cycle inhibitor p27Xic1 impairs neuronal differentiation and increases the number of ElrC+ progenitor cells in Xenopus tropicalis. Mech. Dev. 120, 607–616. doi: 10.1016/s0925-4773(03)00010-8
Casini, P., Nardi, I., and Ori, M. (2012). Hyaluronan is required for cranial neural crest cells migration and craniofacial development. Dev. Dyn. 241, 294–302. doi: 10.1002/dvdy.23715
Chatterji, P., and Rustgi, A. K. (2018). RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol. Med. 24, 490–506. doi: 10.1016/j.molmed.2018.03.008
Ciosk, R., DePalma, M., and Priess, J. R. (2006). Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851–853. doi: 10.1126/science.1122491
Corsinovi, D., Giannetti, K., Cericola, A., Naef, V., and Ori, M. (2019). PDGF-B: the missing piece in the mosaic of PDGF family role in craniofacial development. Dev. Dyn. 248, 603–612. doi: 10.1002/dvdy.47
Draper, B. W., Mello, C. C., Bowerman, B., Hardin, J., and Priess, J. R. (1996). MEX-3 Is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87, 205–216. doi: 10.1016/s0092-8674(00)81339-2
Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060. doi: 10.1016/s0092-8674(00)80371-2
Glisovic, T., Bachorik, J. L., Yong, J., and Dreyfuss, G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986. doi: 10.1016/j.febslet.2008.03.004
Huyck, R. W., Nagarkar, M., Olsen, N., Clamons, S. E., and Saha, M. S. (2015). Methylmercury exposure during early Xenopus laevis development affects cell proliferation and death but not neural progenitor specification. Neurotoxicol. Teratol. 47, 102–113. doi: 10.1016/j.ntt.2014.11.010
Hwang, S.-Y., and Rose, L. S. (2010). Control of asymmetric cell division in early C. elegans embryogenesis: teaming-up translational repression and protein degradation. BMB Rep. 43, 69–78. doi: 10.5483/bmbrep.2010.43.2.069
Kaneko, Y., Sakakibara, S., Imai, T., Suzuki, A., Nakamura, Y., Sawamoto, K., et al. (2000). Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 22, 139–153. doi: 10.1159/000017435
Kiem, L. M., Dietmann, P., Linnemann, A., Schmeisser, M. J., and Kühl, S. J. (2017). The Nedd4 binding protein 3 is required for anterior neural development in Xenopus laevis. Dev. Biol. 423, 66–76. doi: 10.1016/j.ydbio.2017.01.009
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mizuseki, K., Kishi, M., Matsui, M., Nakanishi, S., and Sasai, Y. (1998). Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 125, 579–587.
Naef, V., Monticelli, S., Corsinovi, D., Mazzetto, M. T., Cellerino, A., Ori, M., et al. (2018). The age-regulated zinc finger factor ZNF367 is a new modulator of neuroblast proliferation during embryonic neurogenesis. Sci. Rep. 8:11836.
Nakamura, M., Okano, H., Blendy, J. A., and Montell, C. (1994). Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13, 67–81. doi: 10.1016/0896-6273(94)90460-x
Okano, H., Kawahara, H., Toriya, M., Nakao, K., Shibata, S., Imai, T., et al. (2005). Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306, 349–356. doi: 10.1016/j.yexcr.2005.02.021
Ori, M., Nardini, M., Casini, P., Perris, R., and Nardi, I. (2006). XHas2 activity is required during somitogenesis and precursor cell migration in Xenopus development. Development 133, 631–640. doi: 10.1242/dev.02225
Pelosi, B., Migliarini, S., Pacini, G., Pratelli, M., and Pasqualetti, M. (2014). Generation of Pet1210-Cre transgenic mouse line reveals non-serotonergic expression domains of pet1 both in CNS and periphery. PLoS One 9:e104318. doi: 10.1371/journal.pone.0104318
Pereira, B., Amaral, A. L., Dias, A., Mendes, N., Muncan, V., Silva, A. R., et al. (2020). MEX3A regulates Lgr5 + stem cell maintenance in the developing intestinal epithelium. EMBO Rep. 21:e48938. doi: 10.15252/embr.201948938
Pereira, B., Sousa, S., Barros, R., Carreto, L., Oliveira, P., Oliveira, C., et al. (2013). CDX2 regulation by the RNA-binding protein MEX3A: impact on intestinal differentiation and stemness. Nucleic Acids Res. 41, 3986–3999. doi: 10.1093/nar/gkt087
Ratti, A., Fallini, C., Cova, L., Fantozzi, R., Calzarossa, C., Zennaro, E., et al. (2006). A role for the ELAV RNA-binding proteins in neural stem cells: stabilization of Msi1 mRNA. J. Cell Sci. 119, 1442–1452. doi: 10.1242/jcs.02852
Seo, S., Lim, J.-W., Yellajoshyula, D., Chang, L.-W., and Kroll, K. L. (2007). Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 26, 5093–5108. doi: 10.1038/sj.emboj.7601923
Strzalka, W., and Ziemienowicz, A. (2011). Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 107, 1127–1140. doi: 10.1093/aob/mcq243
Takada, H., Kawana, T., Ito, Y., Kikuno, R. F., Mamada, H., Araki, T., et al. (2009). The RNA-binding protein Mex3b has a fine-tuning system for mRNA regulation in early Xenopus development. Development 136, 2413–2422. doi: 10.1242/dev.029165
Vitobello, A., Ferretti, E., Lampe, X., Vilain, N., Ducret, S., Ori, M., et al. (2011). Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell 20, 469–482. doi: 10.1016/j.devcel.2011.03.011
Keywords: neurogenesis, neuroblast, RNA binding protein (RBP), E3 ubiquitin ligase, SOX2, Msi1, Musashi1, Mex3a
Citation: Naef V, De Sarlo M, Testa G, Corsinovi D, Azzarelli R, Borello U and Ori M (2020) The Stemness Gene Mex3A Is a Key Regulator of Neuroblast Proliferation During Neurogenesis. Front. Cell Dev. Biol. 8:549533. doi: 10.3389/fcell.2020.549533
Received: 06 April 2020; Accepted: 31 August 2020;
Published: 22 September 2020.
Edited by:
Giuseppe Lupo, Sapienza University of Rome, ItalyReviewed by:
Muriel Perron, Centre National de la Recherche Scientifique (CNRS), FranceSally Ann Moody, George Washington University, United States
Copyright © 2020 Naef, De Sarlo, Testa, Corsinovi, Azzarelli, Borello and Ori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michela Ori, bWljaGVsYS5vcmlAdW5pcGkuaXQ=
 Valentina Naef1,2
Valentina Naef1,2 Giovanna Testa
Giovanna Testa Roberta Azzarelli
Roberta Azzarelli Michela Ori
Michela Ori