
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 07 August 2020
Sec. Molecular and Cellular Oncology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00755
This article is part of the Research TopicTumor Vessels as Directors of the Tumor Microenvironment: New Findings, Current Challenges & PerspectivesView all 19 articles
We previously demonstrated that vascular endothelial cells released VEGF-enriched exosomes to promote the tumor vasculogenesis and progression after anti-angiogenic therapies (AATs). To clarify how microRNA (miR)-9 promoted the angiogenesis of tumor-associated endothelial cells, in the present study, we investigated the association between miR-9 and sphingosine-1-phosphate (S1P) receptors in angiogenesis. The levels of miR-9 and S1P receptors in normal and tumor endothelial cells were compared with EndoDB database and their correlations were analyzed. The levels of S1P1, S1P2, and S1P3 were detected in miR-9 overexpressing endothelial cells by qRT-PCR and western blot. The binding sites of miR-9 on S1P1 and S1P3 were predicted and tested by dual-luciferase reporter assays. Then, angiogenesis in endothelial cells overexpressing both S1P1 and miR-9 was detected. The results showed that miR-9 is overexpressed in ECs from medulloblastoma and glioblastoma xenograft, which is negatively associated with S1P1 and S1P3. Overexpression of miR-9 significantly inhibited S1P1 and S1P3 in both mRNA and protein levels. We predicted that binding sites exist between miR-9 and S1P1, S1P3, but only S1P1 was directly targeted by miR-9. Overexpression of S1P1 significantly suppressed the miR-9-induced angiogenesis. Therefore, miR-9 induces angiogenesis via targeting on S1P1.
Angiogenesis is an important progress during physiological and pathophysiological development. Angiogenesis is a complex process of vessel growth but in the strictest sense denotes vessels sprouting from pre-existing ones (Potente et al., 2011). Inadequate angiogenesis causes ischemia in myocardial infarction, stroke, and neurodegenerative or obesity-associated disorders, whereas excessive angiogenesis promotes many ailments including cancer, inflammatory disorders such as atherosclerosis, and eye diseases. Anti-angiogenic therapies (AATs) have been developed to combat tumor metastasis (Hosein et al., 2020). Nowadays, AATs strategies include blood vessel pruning, disruption or normalization of the tumor vasculature, and tumor immunosensitization but did not yield satisfactory efficacy (Cully, 2017; Cloughesy et al., 2020). Bevacizumab antagonizes vascular endothelial growth factor (VEGF) to induce vascular normalization and therefore reduce edema. Vascular disruptive agents such as VB-111 disrupts the angiogenic vasculature via promoting tumor starvation and enhancing the vascular permeability in the tumor environment to increase edema and recruit the immune cells (Cloughesy et al., 2020). AATs and immune checkpoint inhibitors were combined to acquire promising outcomes of cancer patients (Yi et al., 2019). However, how and by which mechanism does the intratumoral vessel form remains unclear.
The angiogenetic process is rather complex involving localized breakdown of the basement membrane and extracellular matrix of a pre-existing vessel, proliferation, and migration of capillary endothelial cells (ECs) into the surrounding tissue, and new vessel formation. Stimulating by the proangiogenic signals such as hypoxia (Cantelmo et al., 2017) and cytokines (e.g., VEGF; Gerhardt et al., 2003), ECs become motile and invasive. MicroRNAs (miRNAs) have recently been shown to regulate gene expression associated with tumorigenesis and angiogenesis (Zhuang et al., 2012; Zeng et al., 2019). Oncogenic miR-9 is significantly elevated in breast cancer cells (Ma et al., 2010), hepatocellular carcinoma (Zhuang et al., 2012; Drakaki et al., 2015), squamous cell carcinomas (White et al., 2013), lung and colorectal carcinoma (Ma et al., 2010), and ovarian cancer (Laios et al., 2008). MiR-9 is significantly increased in ECs upon in vitro tumor-ECs co-cultures. We recently demonstrated that miR-9 promotes angiogenesis via activating the autophagy pathway (Zeng et al., 2019). MiR-9 also promotes the angiogenesis of endothelial progenitor cells via activating the autophagy pathway (Zhou et al., 2020). However, the molecular mechanism involved in miR-9-induced angiogenesis in ECs has not been fully explored.
G protein-coupled sphingosine-1-phosphate (S1P) receptors including S1P1 (formerly endothelial differentiation gene-1, EDG-1), S1P2 (EDG-5), and S1P3 (EDG-3) are abundant in ECs. During vascular development in vivo, S1P1 is required in ECs (Gaengel et al., 2012). Decrease of S1P1 increases angiogenic sprouting and destabilization of the endothelium, while the activation of S1P1 signaling inhibits angiogenic sprouting and enhances cell-to-cell adhesion (Gaengel et al., 2012; Cartier and Hla, 2019). It was reported that S1P2 inhibited tumor angiogenesis in ECs in mouse models (Du et al., 2010). S1P3 promotes EC migration and plays a critical role in developmental and pathological angiogenesis (Jin et al., 2018). Therefore, we hypothesis that miR-9 induces angiogenesis in ECs via targeting S1P receptors.
In the present study, we aimed to investigate which S1P receptor is regulated by miR-9 and aberrant expressed in ECs during angiogenesis. The miR-9 was transfected into the ECs to simulate the tumor-associated ECs as previously described (Zeng et al., 2019). The mRNA and protein levels of S1P1, S1P2, and S1P3 were detected. Then, miR-9 binding sites of the S1P receptors were predicted and verified by Dual-Luciferase Reporter Assay. The role of putative S1P receptor in cell migration, invasion, and angiogenesis was explored. A rescue assay was performed to validate that the putative S1P receptor is a bona fide antigenic target regulated by miR-9 in ECs.
Databases E-GEOD-73753 [normal hindbrain ECs, n = 8; Sonic hedgehog (Shh)-driven medulloblastoma ECs, n = 8; Wnt-driven medulloblastoma ECs, n = 8], E-MTAB-3949 (normal ECs, n = 2; angiogenic glioblastoma xenograft ECs, n = 2), GSE-77199 [colorectal cancer ECs (colon), n = 4; colon ECs, n = 3; ECs from colorectal metastasis to the liver, n = 4; liver ECs, n = 4; renal cell carcinoma ECs, n = 4; renal ECs, n = 4], E-GEOD-51401 (CD105+ ECs from hepatocellular carcinoma, n = 7; CD105+ normal ECs, n = 8; CD31+ ECs from hepatocellular carcinoma, n = 8; CD31+ normal ECs, n = 8) and E-GEOD-41614 (vessels isolated from bladder cancer tissues, n = 5; vessels isolated from bladder normal tissues, n = 5) in EndoDB1 from Khan et al. (2019) were obtained. In the human genome, three independent miR-9 loci including miR-9-1, miR-9-2, miR-9-3 reside at chromosomes 1, 5, and 15, respectively, which share an identical mature miR-9 sequence. The expression of miR-9-1, miR-9-2, miR-9-3, S1P1, S1P2, and S1P3 in those normal ECs and tumor ECs were compared. The relationships between the significant differentially expressed miR-9 loci and specific S1P receptors were analyzed.
HUVECs (Allcells, Shanghai, China) were cultured in HUVEC medium (HUVEC-004; Allcells) as previously described (Zeng et al., 2019). Cells (passages three to six) were maintained at 37°C with 5% CO2 in a humidified incubator. Lentivirus infections of miR-9 mimic (5′-TCTTTGGTTATCTAGCTGTATGA-3′) and negative control (NC; 5′-UUGUACUACACAAAAGUACUG-3′; B04001; GenePharma, Shanghai, China) were performed as previously described (Zeng et al., 2019). cDNAs for S1P1 were ligated into plasmid pcDNA3.1. For overexpression of S1P1, cells were transfected with pcDNA3.1 vector expressing S1P1 using Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, United States). Transfected cells after 48 h were used for the subsequent experiments. Recombinant human VEGF 165 Protein (50 ng/mL; hVEGF; 293-VE, R&D system, Minneapolis, MN, United States) was used to test the migration, invasion and in vitro angiogenesis of cells overexpressing S1P1.
RNA was extracted from cells using TRIzol (Invitrogen) and qRT-PCR assays were performed using a SYBR Premix Ex Taq kit (TaKaRa, Shiga, Japan). The primer sequences were as follows: S1P1, 5′-TTTCTGCGGGAAGGGAGTATGT-3′ (forward) and 5′- GCAGGAAGAGGCGGAAGTTATT-3′ (reverse); S1P2, 5′-CTGTATGGCAGCGACAAGAGC-3′ (forward) and 5′-GAGGCAGGACAGTGGAGCAG-3′ (reverse); S1P3, 5′-TACGCACGCATCTACTTCCTGG-3′ (forward) and 5′-GCTCCGAGTTGTTGTGGTTGG-3′ (reverse); and GAPDH, 5′-TGTTCGTCATGGGTGTGAAC-3′ (forward) and 5′-ATGGCATGGACTGTGGTCAT-3′ (reverse). All primers and probes were obtained from TaKaRa. Gene expression was normalized to that of GAPDH using the 2–△△CT method, and data are presented as expression relative to the indicated controls. The stem-loop primers and probes for mature miR-9 and U6 small nuclear (sn)RNA were as follows: hsa-miR-9-5p, 5′-ACACTCCAGCTGGGTCTTTGGTTATCTAG-3′ (forward) and 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGT CATACAG-3′ (reverse); and U6 snRNA 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The relative expression level of miR-9 was normalized to that of U6 snRNA and shown as a ratio relative to the expression level in the control. Data are representative of three independent experiments.
Proteins were extracted from cells using radioimmunoprecipitation assay lysis buffer containing a protease inhibitor cocktail (Beyotime, Beijing, China). After determination of protein concentration using a protein determination kit (Cayman Chemical Company, Ann Arbor, MI, United States), equal amounts (20–30 μg) of protein samples were size-fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrotransferred onto a polyvinylidene fluoride membrane (Millipore, United States), blocked with 5% non-fat milk in PBS, and hybridized with antibodies against S1P1 (TA311878; Origene, Rockville, MD, United States), S1P2 (TA311287; Origene), and S1P3 (TA321693; Origene) at 4°C overnight. A 1:1000 dilution of each antibody was used for detection. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The blots were incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000; Beyotime), and enhanced chemiluminescence was performed using an Immobilon Western Chemiluminescent HRP Substrate (WBKLS0050; Millipore, Billerica, MA, United States) to visualize the bands. Densitometric quantification was performed using ImageJ software (version 1.52u; National Institutes of Health, Bethesda, MD, United States).
The wild-type (WT) and the corresponding mutation (MUT) sites at 3′-untranslated regions (3′UTRs) of S1P1 and S1P3 were cloned into the psiCHECK-2 vector (Promega, Madison, WI, United States), respectively. The recombinant reporter plasmids were validated by DNA sequencing and then transfected into the miR-9 overexpressing cells and NC cells using Lipofectamine 2000 reagent. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (E1910, Promega). Renilla luciferase activity was normalized to Firefly luciferase activity.
The in vitro angiogenesis assay was performed as previously described (Zeng et al., 2019). The 24-well plates were coated with Matrigel (300 μL/well; BD Biosciences, San Jose, CA, United States) without introducing air bubbles. After gelling of the Matrigel, 5 × 104 cells were plated into each Matrigel-coated well along with 200 μL of HUVEC basal medium containing 10% FBS. After 6 h incubation with or without 50 ng/mL hVEGF at 37°C with 5% CO2 in a humidified incubator, the medium was gently aspirated from each well and incubated with Diff-Quick fixative (Dade Behring, Deerfield, IL, United States) for 30 s and subsequently stained with solution II for 2 min. Tube structures were observed and imaged by microscopy. The pseudo-vascular organization of cells was analyzed by ImageJ software (version 1.52u) using the Angiogenesis Analyzer plugin2.
Cells were digested with 0.25% EDTA trypsin and resuspended in HUVEC basal medium (HUVEC-004B; Allcells) with or without 50 ng/mL hVEGF. Cells were seeded at a density of 1 × 105 cells per Transwell (BD Biosciences), and HUVEC basal medium containing 10% fetal bovine serum (FBS; Hyclone; GE Healthcare, Logan, UT, United States) was added to the lower chamber. The Transwell membrane was precoated with Matrigel for invasion and not precoated for migration. After 48 h, cells that had migrated or invaded through the membrane were quantified as previously described (Zeng et al., 2019).
Statistical significance was determined by Student’s t-test or one-way analysis of variance with either the least significant difference test or Tamhane’s T2 test (depending on Levene’s statistic for homogeneity of variance) using SPSS software (v25.0; IBM, Armonk, NY, United States). The relationships between miR-9 and S1P receptors were measured using Pearson correlation methods. Data were presented as Mean ± SEM. P < 0.05 was considered statistically significant.
The levels of miR-9 and S1P receptors in normal ECs and tumor ECs in multiple tumors were compared (Figure 1). As shown in Figure 1A, databases E-GEOD-73753 and E-MTAB-3949 show miR-9-1, miR-9-2., miR-9-3, S1P1, S1P2, and S1P3 levels in Shh-medulloblastoma ECs, Wnt-medulloblastoma ECs, and ECs from angiogenic glioblastoma xenograft. Databases GSE-77199, E-GEDO-51401, and E-GEOD-41614 show levels of S1P receptors in colorectal cancer ECs, renal cell carcinoma ECs, hepatocellular carcinoma ECs, and bladder cancer ECs. Specifically, miR-9-1 was significantly upregulated in Shh-medulloblastoma ECs (Figure 2B). miR-9-2 was significantly upregulated in Wnt-medulloblastoma ECs, Shh-medulloblastoma ECs, and ECs from glioblastoma xenograft. miR-9-3 was not significantly changed between normal ECs and those tumor ECs. S1P1 was significantly downregulated in Wnt-medulloblastoma ECs, ECs from colorectal metastasis to liver, and hepatocellular carcinoma ECs (Figure 2C). S1P2 was significantly upregulated in medulloblastoma and hepatocellular carcinoma ECs. S1P3 was significantly downregulated in Wnt-medulloblastoma and hepatocellular carcinoma ECs. Taken together, miR-9 was significantly upregulated in multiple tumor ECs. It was accompanied by significant downregulation of S1P1 and S1P3.
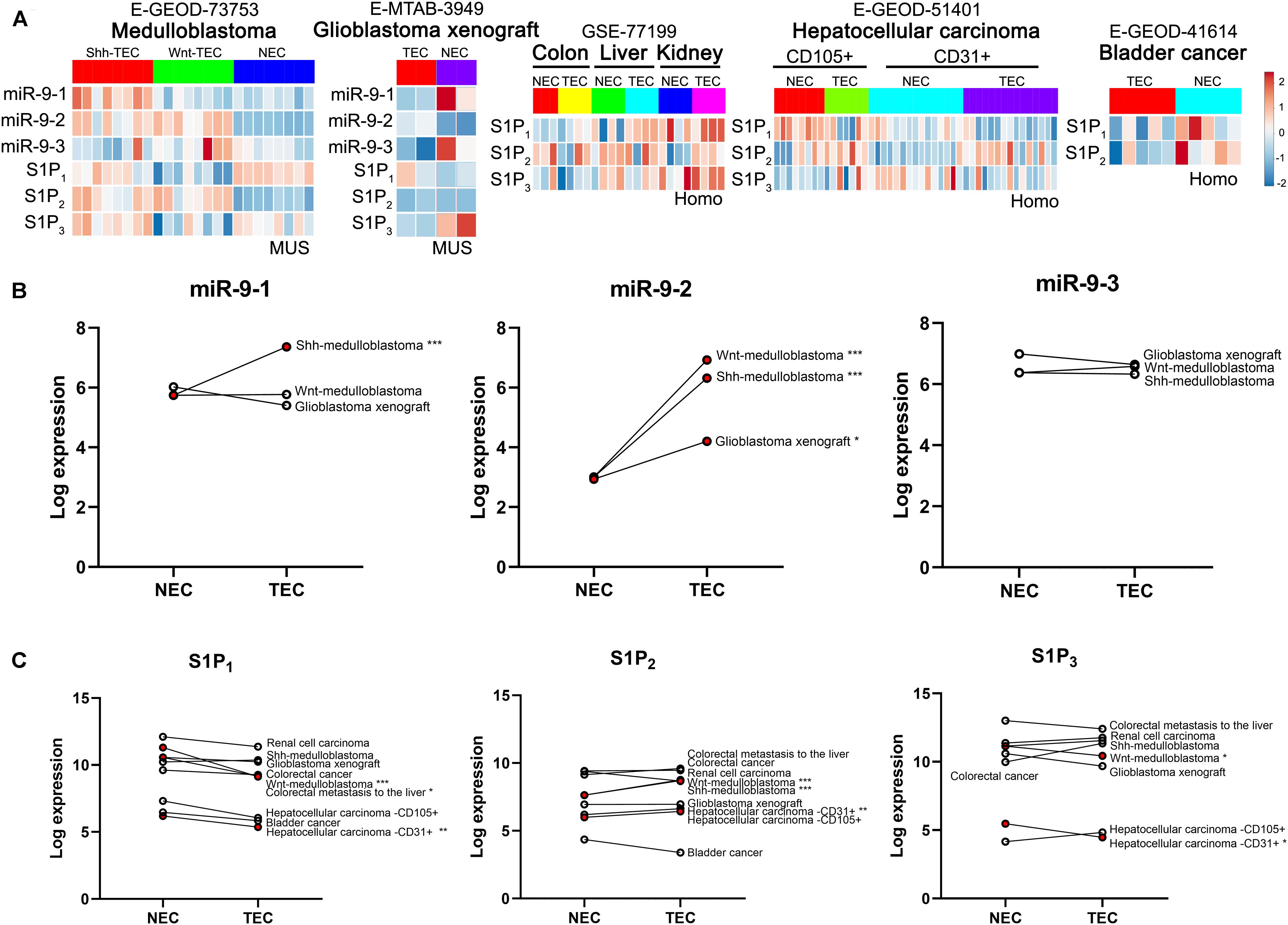
Figure 1. Expression analysis of miR-9 and S1P receptors in tumor ECs and normal ECs basing on EndoDB databases. (A) Gene expression heatmap of normal ECs (NEC) vs. tumor ECs (TEC). Red indicates high-expression levels, and blue indicates low-expression levels. (B) Expression of the miR-9-1, miR-9-2, and miR-9-3 in NECs and TECs. MUS: musculus; Homo: Homo sapiens. *P < 0.05, **P < 0.01, ***P < 0.001.
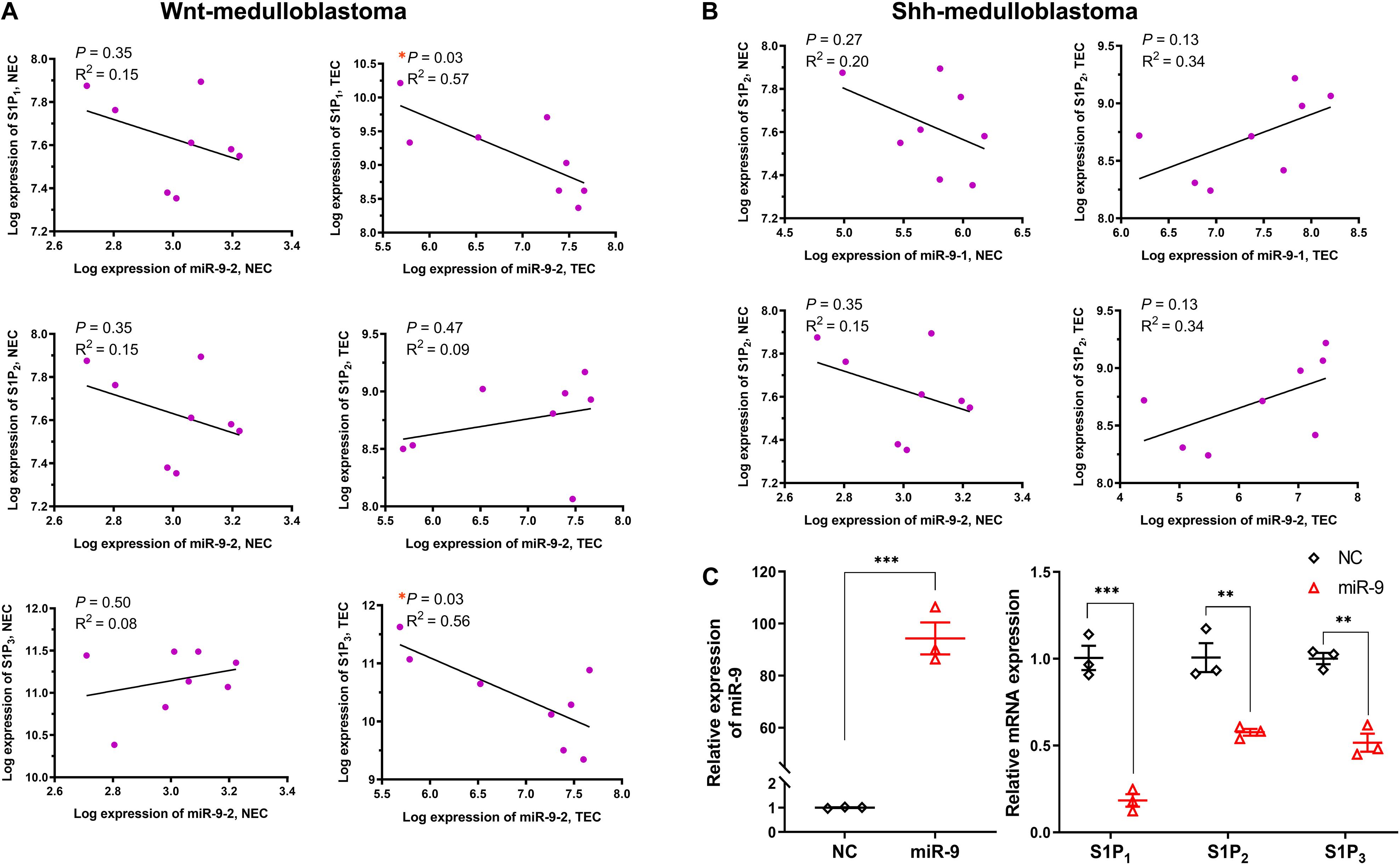
Figure 2. Relationship between miR-9 and S1P receptors. The correlation between the significant differentially expressed miR-9 loci and specific S1P receptors in normal ECs (NEC) and tumor ECs (TEC) were performed. (A) Correlation between miR-9-2 and S1P receptors in Wnt-medulloblastoma; (B) correlation between miR-9-1, miR-9-2, and S1P2 in Shh-medulloblastoma. Then, the levels of S1P receptors in HUVECs upon miR-9 mimics transfection were detected by qRT-PCR. (C) miR-9 levels in HUVECs overexpressing miR-9 (left) and mRNA expression of S1P1, S1P2, and S1P3 (right). NC: miR-9 mimics negative control. Mean ± SEM. n = 4, *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, the correlations between miR-9-2 and S1P1, S1P2, and S1P3 in normal ECs and medulloblastoma ECs were analyzed (Figures 2A,B). Results have shown miR-9-2 negatively correlated to S1P1 and S1P3 in Shh-medulloblastoma ECs, but there was no significant correlation in normal ECs. There was no significant correlation between miR-9-2 and S1P2 in both Shh- and Wnt-medulloblastoma ECs (Figures 2A,B). Thus, the upregulation of miR-9 in tumor ECs is significantly associated with the downregulation of S1P1 and S1P3.
Upregulation of miR-9 in tumor-associated ECs was simulated by the transfection of miR-9-expressing lentivirus into HUVECs. A 94.3-fold increase in miR-9 was shown in the ECs overexpressing miR-9, compared with NC (Figure 2C). The mRNA levels of S1P1, S1P2 and S1P3 in the ECs overexpressing miR-9 were significantly downregulated (0.18 ± 0.04 vs. 1.01 ± 0.07 for S1P1, P < 0.001; 0.58 ± 0.02 vs. 1.01 ± 0.02 for S1P2, P < 0.01; 0.52 ± 0.05 vs. 1.00 ± 0.03 for S1P3, P < 0.01, Figure 2C). Levels of S1P1, S1P2, and S1P3 in the ECs overexpressing miR-9 were also detected by Western Blot assay (Figures 3A,B). There were no significant changes in levels of S1P1, S1P2, and S1P3 between normal and NC cells, while miR-9 overexpression significantly downregulated the levels of S1P1 (0.30 ± 0.11 vs. 1.20 ± 0.07, miR-9 vs. NC, P < 0.001) and S1P3 (0.13 ± 0.05 vs. 0.96 ± 0.07, miR-9 vs. NC, P < 0.001) in the ECs. The expression of S1P2 was not significantly changed in HUVECs overexpressing miR-9 (1.45 ± 0.44 vs. 1.53 ± 0.27, miR-9 vs. NC, P > 0.05). These results suggest that miR-9 significantly inhibited S1P1 and S1P3 in both mRNA and protein levels.
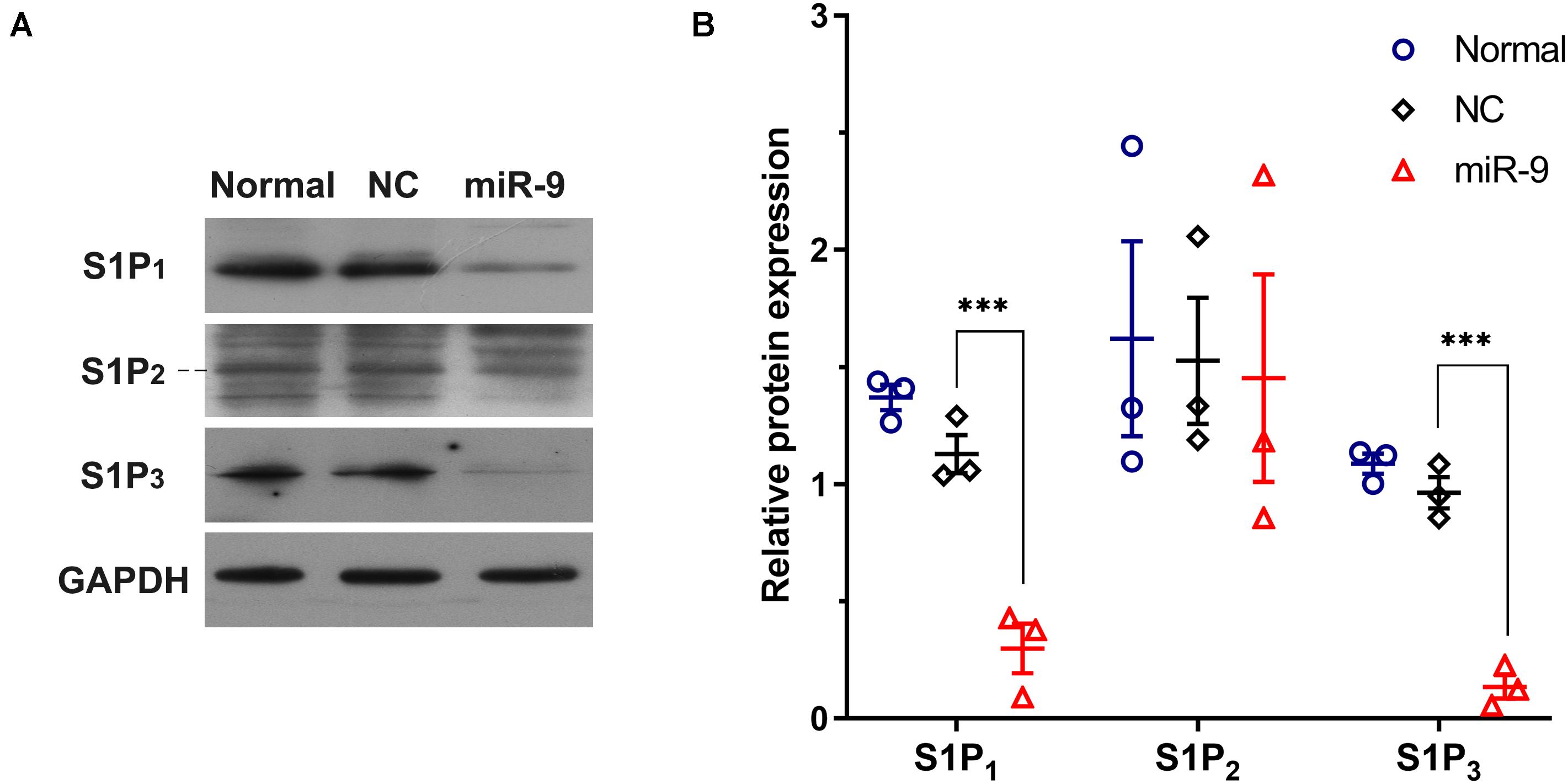
Figure 3. Western blot detection of S1P1, S1P2, and S1P3 in HUVECs overexpressing miR-9. Cell lysates were collected from HUVECs with or without NC or miR-9 transfection, and then 20 μg of protein samples were subjected to western blot analysis. (A) Western blot of cell lysates using anti-S1P1, anti-S1P2, and anti-S1P3. (B) The quantification was performed with ImageJ. The protein expression was normalized to the GAPDH level. NC: miR-9 mimics negative control. Mean ± SEM. n = 3, ***P < 0.001.
The binding sites between S1P1, S1P3, and miR-9 were predicted using TargetScan (Release 7.13, Whitehead Institute for Biomedical Research, Cambridge, MA, United States) and microRNA.org (Release August 20104, the Computational Biology Center at Memorial Sloan-Kettering Cancer Center, Rockville, MD, United States). There was a predicted miR-9 binding site (7mer-A1) at position 2380–2386 of S1P1 3′UTR and two predicted miR-9 binding sites at position 1587–1593 (7mer-m8) of S1P3 3′UTR and 816-822 (7mer-A1) (Figures 4A,B). However, the miR-9 binding site at 816-822 is within the coding region of S1P3 (NM_005226.4). Therefore, the luciferase reporter plasmids containing the WT and the corresponding MUT sites at 3′UTR of S1P1 (position 2380–2386) and S1P3 (position 1587–1593) were generated. The luciferase activity of cells with WT-3′UTR of S1P1 was significantly inhibited by miR-9, while the luciferase activity of cells with MUT-3′UTR of S1P1 was not significantly inhibited by miR-9 (Figure 4A). It was not observed a significant decrease in luciferase activity of cells with WT-3′UTR of S1P3 by miR-9, indicating that miR-9 was not directly targeted the 3′UTR of S1P3 (Figure 4B). These results suggest that S1P1 but not S1P3 is a bona fide miR-9 target.
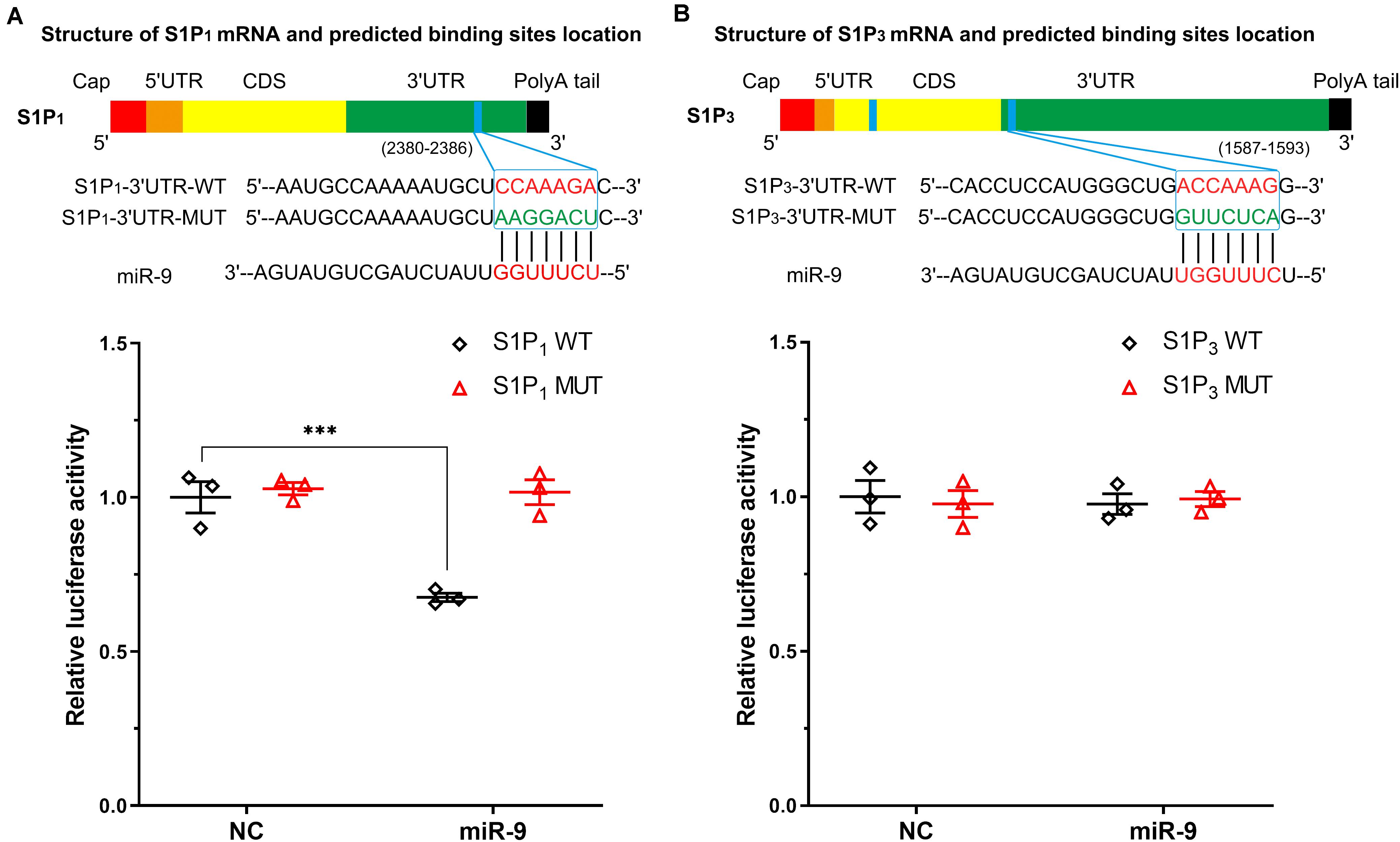
Figure 4. Associations between S1P1, S1P3, and miR-9 were detected by dual-luciferase reporter assay. The structures of S1P1 mRNA (transcript variant 1 M_001400.5, and transcript variant 2 NM_001320730.1; the variant 2 differs in the 5′UTR compared to variant 1, both variants 1 and 2 encode the same protein) and S1P3 mRNA (NM_005226.4) including coding DNA sequence (CDS) and untranslated regions were depicted. The blue band indicated the putative miR-9 binding sequence in the target mRNA molecules. The binding sites located at 3′UTR were screened for Dual-Luciferase Reporter Assay analysis. (A) Putative wild-type (WT) binding sequence and its mutation (MUT) sequence in the 3′UTR of S1P1. Luciferase activity was measured. (B) Putative WT binding sequence and its MUT sequence in the 3′UTR of S1P3. Luciferase activity was measured. NC: miR-9 mimics negative control. Mean ± SEM. n = 3, ***P < 0.001.
To investigate the role of S1P1 in angiogenesis, S1P1 was significantly overexpressed in ECs (2.17 ± 0.11 vs. 0.89 ± 0.03, S1P1 overexpression vs. vector, P < 0.05, Figure 5A). Overexpression of S1P1 significantly suppressed 50 ng/mL VEGF-induced migration and invasion (Figures 5B–D). As a consequence, the overexpression of S1P1 significantly inhibited the VEGF-induced tube formation. The tube length and number of junctions in the network were significantly reduced by S1P1 overexpression (Figures 5E–G). These results suggest that overexpression of S1P1 inhibits the VEGF-induced angiogenesis in HUVECs.
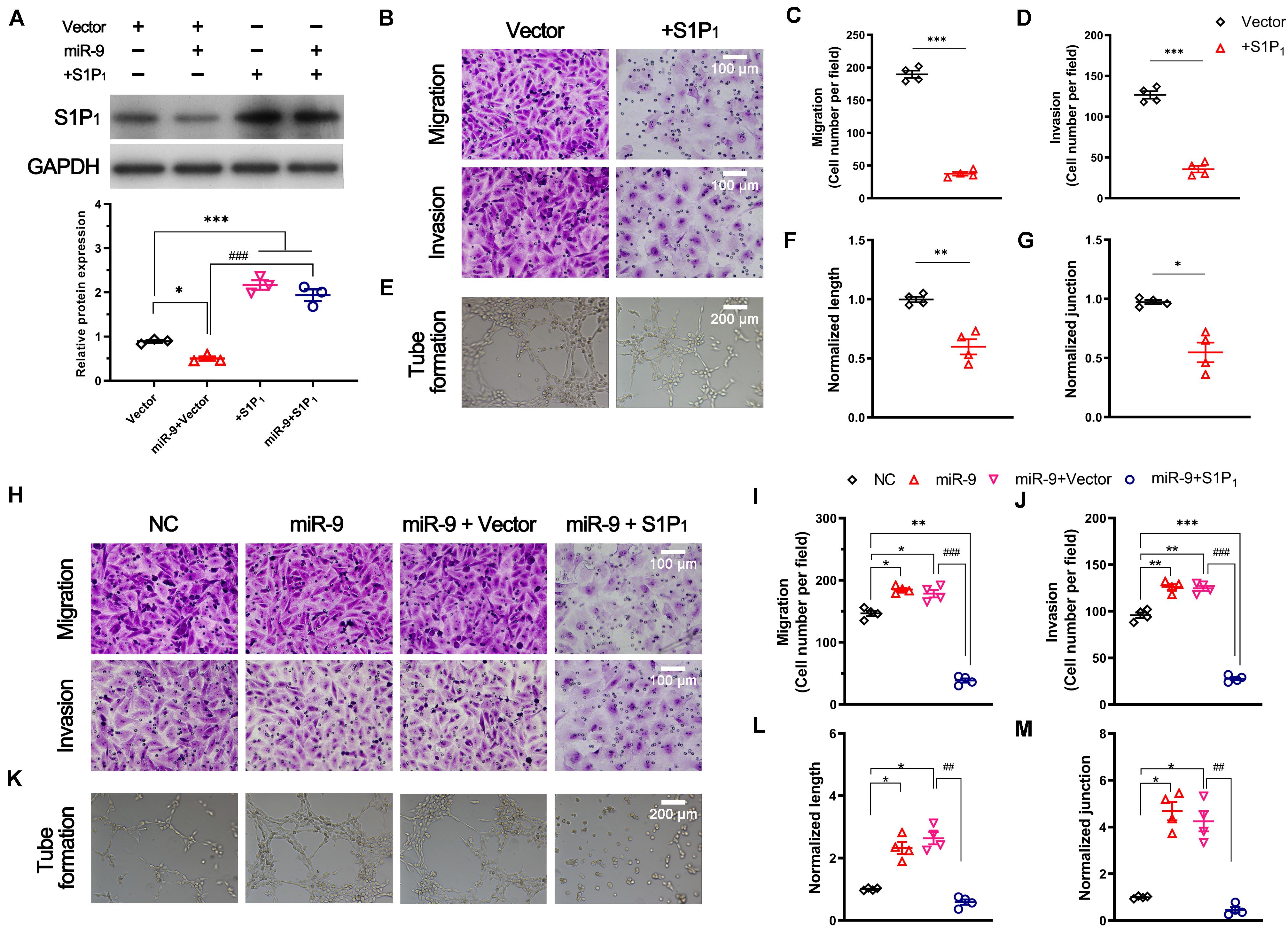
Figure 5. Effect of S1P1 on angiogenesis. Cells were transfected with pcDNA3.1 vector expressing S1P1 (+S1P1) or pcDNA3.1 vector (vector). For the rescue assay, cells overexpressing miR-9 were transfected with pcDNA3.1 vector expressing S1P1 (miR-9 + S1P1) or pcDNA3.1 vector (miR-9 + vector). After transfection for 48 h, the protein level, migration, invasion and in vitro angiogenesis were performed. (A) Western blot of S1P1 and quantification. Mean ± SEM. n = 3, *P < 0.05, ***P < 0.001. ###P < 0.001. (B–D) Transwell assays (B) of migration (top) and invasion (bottom) of HUVECs overexpressing S1P1 in response to 50 ng/mL hVEGF. Mean ± SEM, n = 4. ***P < 0.001. (E–G) Tube formation (E), normalized tube length (F), and number of junctions (J) formed by HUVECs overexpressing S1P1 in response to 50 ng/mL hVEGF. Mean ± SEM, n = 4. *P < 0.05, **P < 0.01. (H–J) Transwell assays (H) of migration (top) and invasion (bottom) of HUVECs overexpressing miR-9 and S1P1. NC: miR-9 mimics negative control. Mean ± SEM, n = 4. *P < 0.05, **P < 0.01, ***P < 0.001; ###P < 0.001. (K–M) Tube formation (K), normalized tube length (L), and the number of junctions (M) formed by HUVECs overexpressing miR-9 and S1P1. NC: miR-9 mimics negative control. Mean ± SEM, n = 4. *P < 0.05; ##P < 0.01.
We then investigated the role of S1P1 in miR-9-induced angiogenesis. A rescue assay demonstrated the significant upregulation of S1P1 in ECs overexpressing miR-9 (1.94 ± 0.23 vs. 0.50 ± 0.08, miR-9 + S1P1 vs. miR-9 + vector, P < 0.001, Figure 5A). The restoration of S1P1 expression significantly reversed the miR-9-induced migration, invasion, and angiogenesis in HUVECs (Figures 5H–M). The tube formation, tube length, and the number of junctions in the network induced by miR-9 were significantly reduced by the restoration of S1P1 (Figures 5K–M). These results suggest that miR-9 promotes angiogenesis via direct downregulation of S1P1.
Angiogenesis is considered one of the critical pathophysiological events in multiple disorders including atherosclerotic plaque rupture (Perrotta et al., 2020), cancer, and so on (Hosein et al., 2020). MiR-9 is elevated in atherosclerosis and many cancers such as breast cancer, hepatocellular carcinoma, squamous cell carcinomas, lung and colorectal carcinoma, and ovarian cancer (Laios et al., 2008; Ma et al., 2010; Zhuang et al., 2012; White et al., 2013; Drakaki et al., 2015). Our previous study has shown miR-9 induced angiogenesis via activating of the autophagy pathway (Zeng et al., 2019). Our new findings in the present study demonstrate that upregulation of miR-9-2 locus in tumor ECs is significantly associated with downregulation of S1P1 and S1P3, and further validate that S1P1 acts as a bona fide target for miR-9, and the downregulation of S1P1 contributes to EC migration, invasion, and angiogenesis.
The role of miR-9 in angiogenesis is a promising therapy for many diseases. MiR-9 could promote migration, invasion, and angiogenesis of endothelial progenitor cells via downregulating transient receptor potential melastatin 7 (TRPM7) and activating PI3K/Akt/autophagy pathway, which might facilitate thrombi recanalization in deep vein thrombosis (Zhou et al., 2020). Translocation of miR-9 from bone marrow-derived mesenchymal stem cells into vascular ECs could induce angiogenesis via activating PI3K/AKT pathway to repair the severe acute pancreatitis (Qian et al., 2018). Moreover, miR-9 promotes angiogenesis via the downregulation of CXC chemokine receptor-4 (CXCR4) and inhibition of PI3K/AKT/mammalian target of rapamycin (mTOR) pathway in HUVECs, thereby suppressing the high glucose-induced injury in HUVECs (Yi and Gao, 2019). It is unclear why both activation and inhibition of PI3K/AKT regulated by miR-9, but both of them involved in promoting angiogenesis in HUVECs. PI3K/AKT/mTOR pathway is an important regulator of autophagy. We previously described that miR-9 activated autophagy in HUVECs and reviewed that miR-9 may induce autophagy via targeted suppression of FOXO1, CUL4A, CK1α, GSK3β, Notch2, cyclin D1, and MCPIP1 (Zeng et al., 2019). In the present study, we verified that miR-9 directly bound to 3′UTR of S1P1. The overexpression of S1P1 inhibited the VEGF-induced angiogenesis. A rescue assay also demonstrated that restoration of S1P1 inhibited the miR-9 induced angiogenesis. For the rescue assay, the transfection efficacy of the pcDNA3.1 vector in EC is more than 80%. After transfected with pcDNA3.1 vector, S1P1 expression in ECs was reduced less by miR-9 compared with that without pcDNA3.1 vector transfection, without significant difference (by 43.4 ± 7.7% miR-9/NC vs. 74.3 ± 8.9% miR-9 + vector/vector, Mean ± SEM, P > 0.05, Figures 2, 4), suggesting the change in endogenous S1P1 could be disregarded. The slight change in reduction of S1P1 might be caused by too many vectors transfection into ECs overexpressing miR-9 or miR-9 trapped somewhere else by other cellular elements such as long non-coding RNAs.
Excessive tumor vessels were present in mice with endothelial cell-specific knockout of S1P1 (Cartier et al., 2020). Loss of S1P1 induced angiogenesis might be associated with increased VEGFR2 activity (Gaengel et al., 2012). Inhibition of S1P1 enhanced VEGFR2 activation in murine ECs upon murine VEGF injection (Fischl et al., 2019). The CD34 labeled vessels tended to increase with S1P1 inhibition, but the combined inhibition of S1P1 and VEGF pathways reduced blood flow in tumor, increased tumor cell apoptosis, and inhibited the tumor CD34 positive vessels in clear cell renal cell carcinoma tumor models (Fischl et al., 2019). We previously demonstrated that activation of S1P1 protects ECs against glycocalyx shedding and promotes the glycocalyx synthesis via activating PI3K signaling (Zeng et al., 2014, 2015). The association among S1P1, PI3K/AKT/mTOR and autophagy will be investigated in the future. In atherosclerosis, miR-9 upregulation is associated with inhibition of intracellular lipid accumulation and macrophage foam cell formation (Shao et al., 2020). Whether miR-9 contributed to the angiogenesis in atherosclerotic plaque rupture remains to be clarified by in vivo experiments.
Many biological functions of S1P1 on the vasculature and its expression are regulated by S1P. S1P acts primarily as an extracellular signaling molecule activated G protein-coupled S1P receptors including S1P1, S1P2, and S1P3 in ECs. S1P1 is the main S1P receptor expressed by ECs (Panetti, 2002). S1P induces migration of various ECs via S1P1 and S1P3 but does not induce the migration of nonendothelial cells (Panetti, 2002). It was demonstrated that S1P induces angiogenesis only in the presence of low levels of VEGF, providing a major caveat as an angiogenic factor (Panetti, 2002). S1P/S1P1 signaling activation inhibited VEGF-induced angiogenic responses (Jozefczuk et al., 2020). More than 50% of S1P is carried by high-density lipoprotein (HDL). HDL promotes angiogenesis via S1P/S1P3 mediated VEGFR2 activation (Jin et al., 2018). In the present study, we found that S1P3 was downregulated by miR-9. However, we failed to validate the predicted miR-9 binding sites within S1P3 3′UTR. There might be an unknown intermediate element such as a transcription factor in the miR-9/S1P3 pathway. In the future, it will be worth to find the intermediate element and test how is S1P3 regulated by miR-9 and associated with angiogenesis. S1P is significantly reduced in cardiovascular diseases (Jozefczuk et al., 2020), and significantly evaluated in hepatocellular carcinoma tissues (Zeng et al., 2016). The relationship between miR-9 and S1P in a special environment should be carefully considered. Although the S1P2 mRNA was downregulated by miR-9, the protein level of S1P2 was only slightly decreased without significant difference. This might be due to the poor sensitivity of the commercially available S1P2 antibody. Moreover, the miR-9 binding site was not predicted within the S1P2 3′UTR using the TargetScan and microRNA.org.
It was believed that most known miRNA target sites (miRNA seed) have 7 nt Watson-Crick seed matches (seed matches) (Lewis et al., 2003). The complete UTR sequence is most critical for mRNA recognition. After changing one or more base pairings between miRNAs and mRNAs, the target UTRs would not identify by miRNAs. We mutated 6 nt and had not tested other constructs with different mutations for S1P1. Moreover, it was demonstrated that the miRNA response elements might locate within the luciferase coding region (Campos-Melo et al., 2014). We predicted whether miR-9 targeting the mRNA sequence of Renilla luciferase gene (hRluc) or firefly luciferase gene (hluc+) by using custom prediction tools of miRbase (Release 22.15; managed by the Griffiths-Jones lab at the Faculty of Biology, Medicine and Health, University of Manchester, United Kingdom) and miRDB (Xiaowei Wang’s lab6 at the Department of Radiation Oncology, Washington University School of Medicine in St. Louis, United States) databases. The predicted miRNAs do not include miR-9, suggesting luciferase could not modify our results.
In HUVECs, an about 94-folds overexpression of miR-9 was achieved artificially. Nevertheless, the luminescence in the miR-9 cells with WT-3′UTR of S1P1 was decreased by only about 40%. Bioluminescence is a chemical process in which an enzyme such as luciferase breaks down a substrate such as luciferin and one of the by-products of this reaction is light. The transfection efficacy of the psiCHECK-2 vector and conversion efficiency of luciferin to oxyluciferin might limit the diminution of the luminescence. It was worth noting that the psiCHECK-2 vector contains both renilla and firefly luciferase genes. As Renilla transcript expressed with the 3′UTR sequence of interest gene, Renilla luciferase activity is used as a measure of the effect of the 3′UTR on transcript stability and translation efficiency. Firefly luciferase is used to normalize transfections and eliminates the need to transfect a second vector control. The transfection efficacy of the psiCHECK-2 vector was not detected in EC.
In summary, building on previous findings that miR-9 overexpression in HUVECs promotes angiogenesis, and AATs triggers VEGF-enriched exosomes to promote tumor vasculogenesis, we present data to show that S1P1 acts as a bona fide target for miR-9 to regulate the migration, invasion, and angiogenesis. The expressions of S1P1/S1P2/S1P3 between normal-associated ECs and tumor-associated ECs should be carried out by in vivo and in vitro experiments such as normal cells/tumor cells and ECs crosstalk assays in the future. The investigation on the molecular mechanism whereby miR-9 promotes angiogenesis in HUVECs might provide a critical cue to reveal the AATs triggered exosomes release and further to control the tumor vasculogenesis and progression following AATs.
All datasets generated for this study are included in the article.
YZ contributed to the conception and design of the work. YZ, XY, and LX performed the acquisition, analysis, and interpretation of data for the work. YZ and XY drafted the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant nos. 11932014, 11402153).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Campos-Melo, D., Droppelmann, C. A., Volkening, K., and Strong, M. J. (2014). Comprehensive luciferase-based reporter gene assay reveals previously masked up-regulatory effects of miRNAs. Int. J. Mol. Sci. 15, 15592–15602. doi: 10.3390/ijms150915592
Cantelmo, A. R., Pircher, A., Kalucka, J., and Carmeliet, P. (2017). Vessel pruning or healing: endothelial metabolism as a novel target? Expert Opin. Ther. Targets 21, 239–247. doi: 10.1080/14728222.2017.1282465
Cartier, A., and Hla, T. (2019). Sphingosine 1-phosphate: lipid signaling in pathology and therapy. Science 366:6463. doi: 10.1126/science.aar5551
Cartier, A., Leigh, T., Liu, C. H., and Hla, T. (2020). Endothelial sphingosine 1-phosphate receptors promote vascular normalization and antitumor therapy. Proc. Natl. Acad. Sci. U.S.A. 117, 3157–3166. doi: 10.1073/pnas.1906246117
Cloughesy, T. F., Brenner, A., de Groot, J. F., Butowski, N. A., Zach, L., and Campian, J. L., et al. (2020). A randomized controlled phase III study of VB-111 combined with bevacizumab vs bevacizumab monotherapy in patients with recurrent glioblastoma (GLOBE). Neuro Oncol. 22, 705–717. doi: 10.1093/neuonc/noz232
Cully, M. (2017). Cancer: tumour vessel normalization takes centre stage. Nat. Rev. Drug Discov. 16:87. doi: 10.1038/nrd.2017.4
Drakaki, A., Hatziapostolou, M., Polytarchou, C., Vorvis, C., Poultsides, G. A., Souglakos, J., et al. (2015). Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer 15:542. doi: 10.1186/s12885-015-1562-1569
Du, W., Takuwa, N., Yoshioka, K., Okamoto, Y., Gonda, K., Sugihara, K., et al. (2010). S1P(2), the G protein-coupled receptor for sphingosine-1-phosphate, negatively regulates tumor angiogenesis and tumor growth in vivo in mice. Cancer Res. 70, 772–781. doi: 10.1158/0008-5472.Can-09-2722
Fischl, A. S., Wang, X., Falcon, B. L., Almonte-Baldonado, R., Bodenmiller, D., Evans, G., et al. (2019). Inhibition of sphingosine phosphate receptor 1 signaling enhances the efficacy of VEGF receptor inhibition. Mol. Cancer Ther. 18, 856–867. doi: 10.1158/1535-7163.Mct-18-0548
Gaengel, K., Niaudet, C., Hagikura, K., Lavina, B., Muhl, L., Hofmann, J. J., et al. (2012). The sphingosine-1-phosphate receptor S1PR1 restricts sprouting angiogenesis by regulating the interplay between VE-cadherin and VEGFR2. Dev. Cell. 23, 587–599. doi: 10.1016/j.devcel.2012.08.005
Gerhardt, H., Golding, M., Fruttiger, M., Ruhrberg, C., Lundkvist, A., Abramsson, A., et al. (2003). VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell. Biol. 161, 1163–1177. doi: 10.1083/jcb.200302047
Hosein, A. N., Brekken, R. A., and Maitra, A. (2020). Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. [Epub ahead of print]. doi: 10.1038/s41575-020-0300-1
Jin, F., Hagemann, N., Sun, L., Wu, J., Doeppner, T. R., Dai, Y., et al. (2018). High-density lipoprotein (HDL) promotes angiogenesis via S1P3-dependent VEGFR2 activation. Angiogenesis 21, 381–394. doi: 10.1007/s10456-018-9603-z
Jozefczuk, E., Guzik, T. J., and Siedlinski, M. (2020). Significance of sphingosine-1-phosphate in cardiovascular physiology and pathology. Pharmacol. Res. 156:104793. doi: 10.1016/j.phrs.2020.104793
Khan, S., Taverna, F., Rohlenova, K., Treps, L., Geldhof, V., de Rooij, L., et al. (2019). EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res. 47, D736–D744. doi: 10.1093/nar/gky997
Laios, A., O’Toole, S., Flavin, R., Martin, C., Kelly, L., Ring, M., et al. (2008). Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer 7:35. doi: 10.1186/1476-4598-7-35
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. doi: 10.1016/s0092-8674(03)01018-3
Ma, L., Young, J., Prabhala, H., Pan, E., Mestdagh, P., Muth, D., et al. (2010). miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256. doi: 10.1038/ncb2024
Panetti, T. S. (2002). Differential effects of sphingosine 1-phosphate and lysophosphatidic acid on endothelial cells. Biochim. Biophys. Acta 1582, 190–196. doi: 10.1016/s1388-1981(02)00155-5
Perrotta, P., Van der Veken, B., Van Der Veken, P., Pintelon, I., Roosens, L., Adriaenssens, E., et al. (2020). Partial inhibition of glycolysis reduces atherogenesis independent of intraplaque neovascularization in mice. Arterioscler. Thromb. Vasc. Biol. 40, 1168–1181. doi: 10.1161/atvbaha.119.313692
Potente, M., Gerhardt, H., and Carmeliet, P. (2011). Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887. doi: 10.1016/j.cell.2011.08.039
Qian, D., Song, G., Ma, Z., Wang, G., Jin, L., Hu, M., et al. (2018). MicroRNA-9 modified bone marrow-derived mesenchymal stem cells (BMSCs) repair severe acute pancreatitis (SAP) via inducing angiogenesis in rats. Stem Cell Res. Ther. 9:282. doi: 10.1186/s13287-018-1022-y
Shao, D., Di, Y., Lian, Z., Zhu, B., Xu, X., Guo, D., et al. (2020). Grape seed proanthocyanidins suppressed macrophage foam cell formation by miRNA-9 via targeting ACAT1 in THP-1 cells. Food Funct. 11, 1258–1269. doi: 10.1039/c9fo02352f
White, R. A., Neiman, J. M., Reddi, A., Han, G., Birlea, S., Mitra, D., et al. (2013). Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J. Clin. Invest. 123, 4390–4404. doi: 10.1172/jci65856
Yi, J., and Gao, Z. F. (2019). MicroRNA-9-5p promotes angiogenesis but inhibits apoptosis and inflammation of high glucose-induced injury in human umbilical vascular endothelial cells by targeting CXCR4. Int. J. Biol. Macromol. 130, 1–9. doi: 10.1016/j.ijbiomac.2019.02.003
Yi, M., Jiao, D., Qin, S., Chu, Q., Wu, K., and Li, A. (2019). Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment. Mol. Cancer 18:60. doi: 10.1186/s12943-019-0974-6
Zeng, Y., Adamson, R. H., Curry, F. R., and Tarbell, J. M. (2014). Sphingosine-1-phosphate protects endothelial glycocalyx by inhibiting syndecan-1 shedding. Am. J. Physiol. Heart Circ. Physiol. 306, H363–H372. doi: 10.1152/ajpheart.00687.2013
Zeng, Y., Liu, X. H., Tarbell, J., and Fu, B. (2015). Sphingosine 1-phosphate induced synthesis of glycocalyx on endothelial cells. Exp. Cell Res. 339, 90–95. doi: 10.1016/j.yexcr.2015.08.013
Zeng, Y., Yao, X., Chen, L., Yan, Z., Liu, J., Zhang, Y., et al. (2016). Sphingosine-1-phosphate induced epithelial-mesenchymal transition of hepatocellular carcinoma via an MMP-7/syndecan-1/TGF-beta autocrine loop. Oncotarget 7, 63324–63337. doi: 10.18632/oncotarget.11450
Zeng, Y., Yao, X., Liu, X., He, X., Li, L., Liu, X., et al. (2019). Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. J. Extracell. Vesicles 8:1629865. doi: 10.1080/20013078.2019.1629865
Zhou, D. M., Sun, L. L., Zhu, J., Chen, B., Li, X. Q., and Li, W. D. (2020). MiR-9 promotes angiogenesis of endothelial progenitor cell to facilitate thrombi recanalization via targeting TRPM7 through PI3K/Akt/autophagy pathway. J. Cell. Mol. Med. 24, 4624–4632. doi: 10.1111/jcmm.15124
Keywords: sphingosine-1-phosphate receptor, miR-9, angiogenesis, endothelial cell, microRNA
Citation: Yao X, Xie L and Zeng Y (2020) MiR-9 Promotes Angiogenesis via Targeting on Sphingosine-1- Phosphate Receptor 1. Front. Cell Dev. Biol. 8:755. doi: 10.3389/fcell.2020.00755
Received: 24 May 2020; Accepted: 20 July 2020;
Published: 07 August 2020.
Edited by:
Lucas Treps, VIB KU Leuven Center for Cancer Biology, BelgiumReviewed by:
Jiwei Guo, Binzhou Medical University, ChinaCopyright © 2020 Yao, Xie and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Zeng, eWVAc2N1LmVkdS5jbg==; eWVxZ3plbmdAR21haWwuY29t
†These authors have contributed equally to this work
‡ORCID: Ye Zeng, orcid.org/0000-0001-5440-6194
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.