
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 30 July 2020
Sec. Molecular and Cellular Oncology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00728
This article is part of the Research Topic Therapeutic Opportunities and Innovative Biomarkers in Tumor Microenvironment View all 23 articles
Immunotherapies that harness the immune system to kill cancer cells have showed significant therapeutic efficacy in many human malignancies. A growing number of studies have highlighted the relevance of neoantigens in recognizing cancer cells by intrinsic T cells. Cancer neoantigens are a direct consequence of somatic mutations presenting on the surface of individual cancer cells. Neoantigens are fully cancer-specific and exempt from central tolerance. In addition, neoantigens are important targets for checkpoint blockade therapy. Recently, technological innovations have made neoantigen discovery possible in a variety of malignancies, thus providing an impetus to develop novel immunotherapies that selectively enhance T cell reactivity for the destruction of cancer cells while leaving normal tissues unharmed. In this review, we aim to introduce the methods of the identification of neoantigens, the mutational patterns of human cancers, related clinical trials, neoantigen burden and sensitivity to immune checkpoint blockade. Moreover, we focus on relevant challenges of targeting neoantigens for cancer treatment.
The occurrence rate of cancer is increasing rapidly (Torre et al., 2015). New methods such as immunotherapy have become an optimal choice in cancer treatment, along with chemotherapy, radiation and surgery (Restifo et al., 2012; Hamid et al., 2013; Robert et al., 2015a). The foundation of cancer immunology was based on tumor transplantation studies in syngeneic mice (Prehn and Main, 1957; Old and Boyse, 1964). Concurrently, clinicians observed that in gastric carcinoma patients, tumor-infiltration lymphocytes (TILs) were correlated with longer post-operative survival (Black et al., 1954). Recently, it has demonstrated that autologous T cells can show profound tumoricidal activity and immunotherapies based on T cells are effective in multiple human malignancies (Rosenberg, 2011; Rosenberg et al., 2011). In particular, treatment of patients with checkpoint blockade has shown a significant effect on patient survival (Hodi et al., 2010; Brahmer et al., 2012; Topalian et al., 2012; Sharma and Allison, 2015). These data provide clear evidence that endogenous T cells can recognize antigenic determinants—epitopes that present on the tumor cell surface from the major histocompatibility complexes (MHCs). Such epitopes may be originated from three classes of antigens: (1) viral antigens that are encoded by viral open reading frames (ORF) in virus-infected tumor cells (Walboomers et al., 1999; Gillison et al., 2000; Feng et al., 2008), such as hepatitis B virus (HBV) (Merlo et al., 2010), Epstein-Barr virus (EBV) (Lin et al., 2002), and human papilloma virus (HPV) (Morrow et al., 2013); (2) tumor-associated antigens (TAAs) that expression levels are very low in some normal tissues but are overexpressed in malignant cells, including oncofetal antigens, cancer testis antigens (CTA), overexpressed oncogenic proteins and selected differentiation antigens (Caballero and Chen, 2009; Melero et al., 2014; Ward et al., 2016), for example NY-ESO-1 (D’Angelo et al., 2018) and CD19 on B cell malignancies (Sabatino et al., 2016); and (3) neoantigens, which are immunogenic products of somatic mutations that are fully specific to tumors. In the late 1980s, Boon and colleagues were among the first to report that aberrant peptides derived from tumor mutations were able to elicit a tumor specific T cell response in a mouse model (De Plaen et al., 1988; Lurquin et al., 1989). A few years later, it was also observed in human tumors that somatic mutations were a source of neoantigens recognized by T cells (Coulie et al., 1995; Monach et al., 1995; Wolfel et al., 1995). Recently, studies have demonstrated that neoantigens are able to recognize cancer cells by intrinsic T cells (Lennerz et al., 2005; Castle et al., 2012; Matsushita et al., 2012; Kvistborg et al., 2014; Rajasagi et al., 2014; Wick et al., 2014; Chan et al., 2015; Cohen et al., 2015; Rizvi et al., 2015). Neoantigens are fully tumor-specific and bypass central tolerance (Heemskerk et al., 2013), and thus they are not expected to induce autoimmune toxicity and they are a potential target for cancer immunotherapy. With the recent development of cancer genomics (Garraway and Lander, 2013; Vogelstein et al., 2013) and high-throughput immunologic screening (Lin et al., 2008; Zhang et al., 2011), the goal of the analysis of neoantigens based on individual patients has become achievable, which makes neoantigen-directed immunotherapy highly attractive. In this review, we aim to introduce the framework for the identification and prioritization of neoantigens, the mutational patterns of cancer, neoantigen-related trials, mutational burden and sensitivity to immune checkpoint blockade. More importantly, we highlight the relevant challenges of targeting neoantigens for cancer treatment.
Advances in next-generation sequencing (NGS) (Margulies et al., 2005; Shendure et al., 2005; Bentley et al., 2008; Wheeler et al., 2008; Drmanac et al., 2010) have allowed for the rapid and relatively inexpensive exhaustive sequencing of genomic changes across tens of thousands of human cancers (Li et al., 2011; Stratton, 2011; Gubin et al., 2015; Lu and Robbins, 2016). In conjunction, the innovation of high-throughput immunologic screening techniques has promoted the detection and isolation of neoantigens that can evoke specific T cell responses (Hadrup et al., 2009; Bentzen and Marquard, 2016; Bentzen and Hadrup, 2017) (Figure 1). In the human genome, only 1% codes for known expressed genes (the exomes) among the approximately 3 billion nucleotides (Choi et al., 2009; Ng et al., 2009). Therefore, it can significantly reduce time and costs to sequence only functional exomes. Large projects such as The Cancer Genome Atlas (TCGA) [Cancer Genome Atlas (TCGA) Research Network, 2008; Garraway and Lander, 2013] and The International Cancer Genome Consortium (ICGC) (Hudson et al., 2010) have identified cancer genomes across multiple tumor types. However, whole-exome sequencing (WES) provides limited information on non-coding regions of cancer genomes, including untranslated regions (UTRs), promoters, enhancers, introns, regulatory elements and diverse non-coding RNAs (ncRNAs) as well as unannotated regions (Garraway and Lander, 2013). In contrast, whole-genome sequencing (WGS) may be able to detect these events (Meyerson et al., 2010). Second-generation sequencing of the transcriptome (RNA-seq) is another powerful approach — as cDNA may derive from mRNA, total RNA or other RNAs, such as microRNAs and lncRNA (Morrissy et al., 2009). Recent studies have suggested that tumors harbor more abundant alternative splicing events than paired normal tissues by comprehensive analysis of WES with RNA-seq data and proteomic data, which is a potential source to generate tumor-specific neoantigens (Kahles et al., 2018; Park et al., 2018). In fact, a few studies have developed peptide arrays representing all possible frameshift peptides and detected the antibody responses to the frameshift peptides. This might be a useful method for cancer neoantigen screening (Zhang et al., 2018; Shen et al., 2019). In conclusion, somatic DNA mutations are usually computed from WGS, WES or RNA-seq data from comparisons between tumor and normal sequences (Ding et al., 2014; Tran et al., 2015).
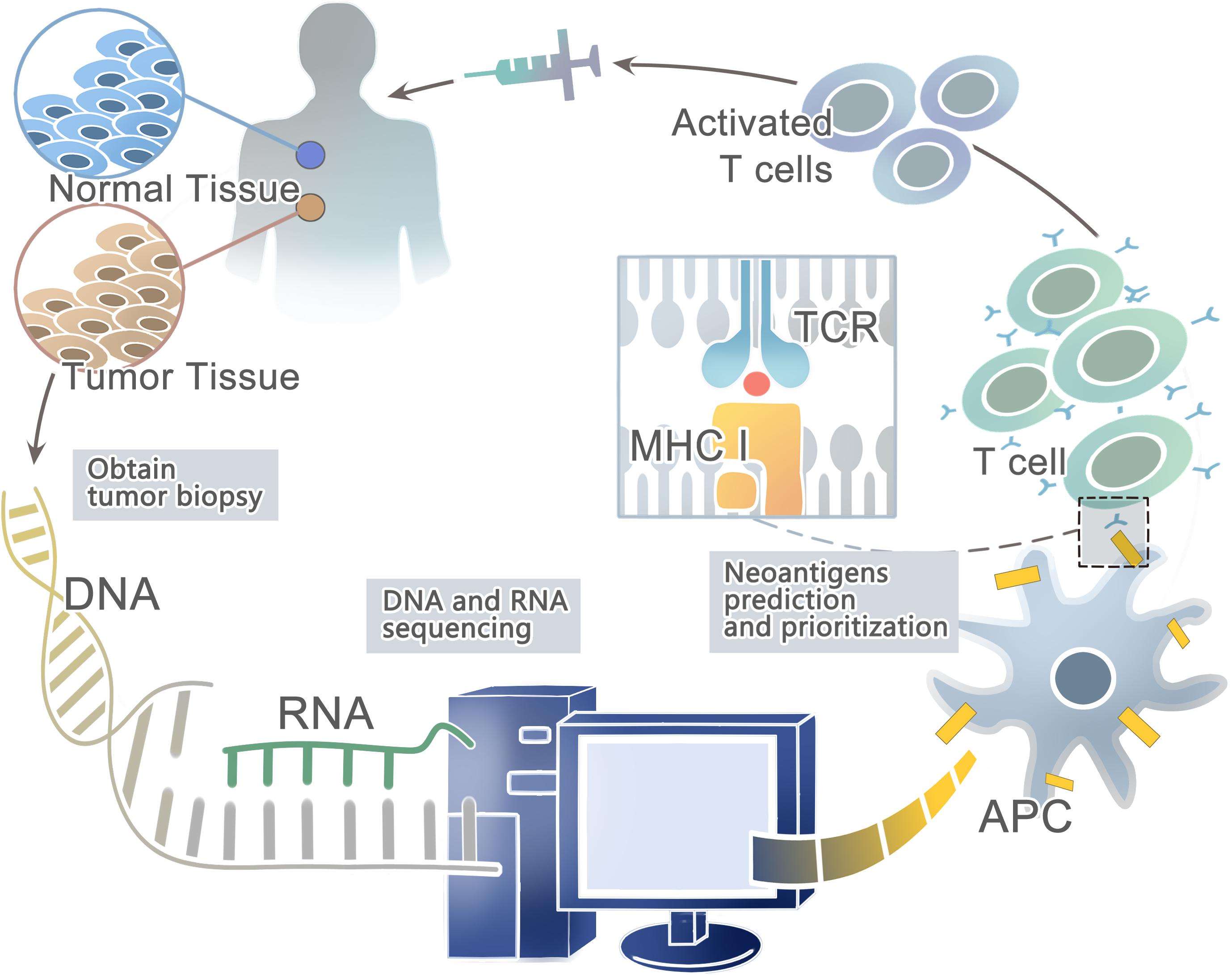
Figure 1. A framework for the identification and prioritization of neoantigens from computational analysis for an individual cancer sample.
Then the patient-specific NGS data from WGS, WES, or RNA-seq can be used to predict HLA types with computational tools such as Optiptype (Szolek et al., 2014) and Polysolver (polymorphic loci resolver) (Shukla et al., 2015). First, reads are selected from the NGS data that potentially derived from the HLA region; second, they are aligned to a full-length genomic library of all known HLA alleles (Robinson et al., 2013); and third, all the best-scoring alignments of each read are kept for further study. After predicting HLA types, computational algorithms such as NetMHC (Andreatta and Nielsen, 2016), NetMHCpan (Rammensee et al., 1999; Nielsen et al., 2007; Nielsen and Andreatta, 2016; Jurtz and Paul, 2017), and MHCflurry (O’Donnell et al., 2018) trained on large in vitro experimental datasets can be used to prioritize candidate neoantigens that bind to the predicted HLA types with high affinity. For example, Neopepsee and pVAC-Seq are representative analysis pipelines for tumor somatic mutations (Hundal et al., 2016; Kim et al., 2018). However, these algorithms are not a good predictor of actual HLA presentation (Bassani-Sternberg et al., 2016; Abelin et al., 2017), with only <5% of predicted peptides found on tumor cell surface (Yadav et al., 2014; Bassani-Sternberg et al., 2015). On the other hand, these prediction strategies do not consider proximal variants that can alter peptide sequences and affect neoantigen binding predictions (Hundal and Kiwala, 2019). Recently, a new prediction model-EDGE based on tumor HLA peptide mass spectrometry (MS) datasets has increased the positive predictive value up to nine-fold. However, it still does not incorporate TCR binding or predict HLA class II binding epitopes (Bulik-Sullivan et al., 2018).
Besides, a range of methodologies can be used to identify autoantibodies against tumor neoantigens based on B-cell response. The costimulatory molecules from CD4+ helper T cells and the neoepitopes presented on the surface of antigen-presenting cells (APCs) could activate the Naïve B cells in lymphoid organs. Most activated B cells will then differentiate into plasma cells to produce antibodies against tumor neoantigens (Zaenker et al., 2016). Protein microarrays are time-effective high-throughput tools (Figure 2). Sandwich immunoassays in the miniaturized system could successfully identify tumor antigens in sera samples extracted from patients (Pollard et al., 2007; Yang et al., 2013). First described by O’Farrell (1975), another tool named Serologic Proteome analysis (SERPA) or 2-D western blots, consists of the isoelectric focusing (IEF) gel run in the first dimension and SDS-PAGE gel run in the second dimension. SERPA separates the proteins in the gel by their isoelectric point (IP) and molecular mass and then transfers the proteins from the gel to a carrier membrane to screen antibodies. Finally, the antigenic protein spots can be identified by MS (Tjalsma et al., 2008). This approach has been used to identify antigens in different tumor types (Dai et al., 2017; Belousov et al., 2019). Serological analysis of recombinant cDNA expression libraries (SEREX), which combines serological analysis with antigen cloning techniques, is a widely used technique to explore tumors’ antigen repertoire. SEREX first construct a cDNA library from cancer cell lines or fresh tumor samples, then screen the cDNA library with autologous sera of cancer patients, and finally sequence the immune-reactive clones. Despite the laborious process, SEREX have identified a variety of tumor antigens including CTAs, differentiation antigens, mutational antigens, splice-variant antigens and over-expressed antigens (Chen et al., 1997; Scanlan et al., 1998; Türeci et al., 1998a, b; Brass et al., 1999; Jäger et al., 1999). Furthermore, other methods such as Multiple Affinity Protein Profiling (MAPPing) and nanoplasmonic biosensor have also been developed to identify tumor antigens (Lee et al., 2015; Jo et al., 2016).
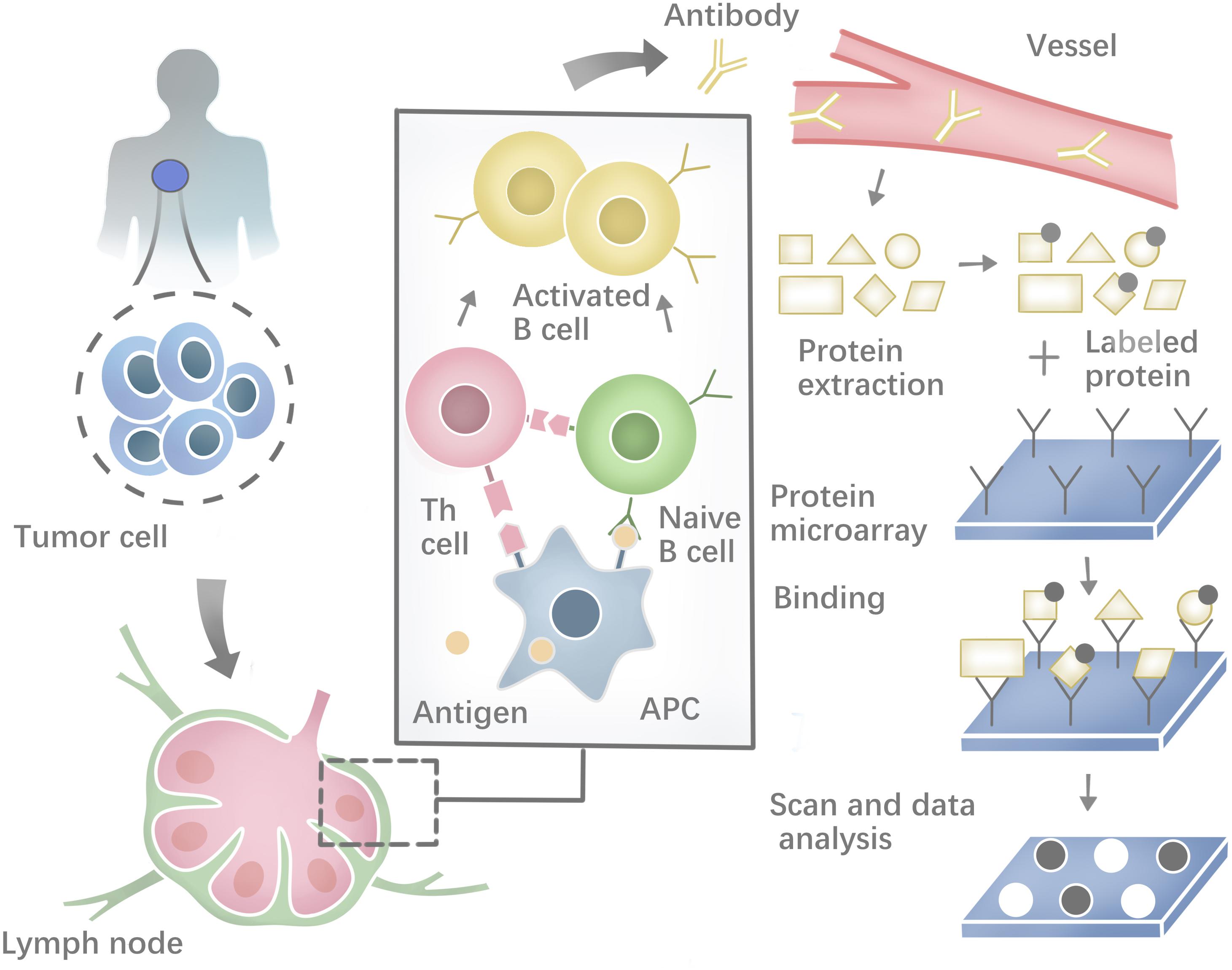
Figure 2. Profiling the humoral immune response and the process of protein microarray to capture autoantibodies against antigens.
Somatic mutations presenting in cancer cells but not in a patient’s germ line are the primary cause of cancers (Weir et al., 2004). The cancer genome alterations include single base substitutions, insertion-deletions (so-called indels), rearrangements (inversions, translocations, duplications, and deletions) (Stratton et al., 2009; Alexandrov et al., 2013; Barrick and Lenski, 2013; Vogelstein et al., 2013; Helleday et al., 2014), but also new DNA sequences acquired from exogenous sources, notably those of viruses such as HBV, HPV, and EBV (Talbot and Crawford, 2004). Somatic mutations can be classified as driver and passenger mutations, and driver mutations, such as BRAF, KRAS, EGFR, IDH1, and PIK3CA, can provide a selective growth advantage and promote cancer development while passenger mutations do not (Haber and Settleman, 2007). Even though cancer has a mere handful of driver mutations, they are still attractive targets for immunotherapy when they are shared between different cancers and individuals. The rates of mutations vary among tumors and cancer types. Some cancer types, such as medulloblastomas, carcinoids, acute leukemias, and testicular germ cell tumors, generally carry relatively few mutations (Greenman et al., 2007; Parsons et al., 2011). However, lung cancers and melanomas occasionally have more than 100,000 mutations (Ding et al., 2008; Kan et al., 2010; Pleasance et al., 2010a), most likely because of overwhelming exposure to mutagenic carcinogen such as ultraviolet (UV) light (Trucco et al., 2018) and tobacco carcinogens (Pleasance et al., 2010b). Except for exogenous mutagenic exposure, endogenous mutational processes, such as mismatch repair deficiency in some colorectal cancers (Pena-Diaz et al., 2017) or upregulation of APOBEC cytosine deaminases, can also contribute to mutation burden (Alexandrov et al., 2013; Helleday et al., 2014). More information about the somatic mutations in human cancer can be found in COSMIC, the Catalog Of Somatic Mutations In Cancer1 (Forbes et al., 2015). One can even explore how cancer mutations impact the structure and function of more than 8,000 human proteins in COSMIC-3D (No authors, 2017a). Non-synonymous somatic mutations, which can alter amino acid coding sequences, are the main cause of neo-epitopes (Wood et al., 2007). Here, we summarize the number of non-synonymous somatic mutations in different cancer types (Table 1). For example, Sjoblom et al. (2006) identified 1,307 somatic mutations among 13,023 genes in 11 breast and 11 colorectal cancers (83% were missense mutations, 6% were nonsense, and the remainder were insertions, deletions, duplications, and changes in non-coding regions). Neoantigens generated from tumor somatic DNA mutations can be identified by the approach described in the first section.
Except for somatic non-synonymous protein-altering mutations, tumor neoantigens can be generated from alternative splicing variations. Pre-mRNA splicing is a biological process that contains the removal of introns and the ligation of exons to form mature RNA products (Sharp, 1994). However, the splice site and exons are alternative, which means a single gene can produce numerous mRNA isoforms (Nilsen and Graveley, 2010). And genomic variants in splicing regulatory sequences can disrupt splicing and produce aberrant mRNA and protein products (Pagani and Baralle, 2004). Recently, studies have suggested that tumors harbor more abundant alternative splicing events than paired normal tissues by comprehensive analysis of WES with RNA-seq data and proteomic data. For example, Kahles et al. (2018) performed a comprehensive analysis of alternative splicing across 32 tumor types from 8,705 patients and they found that tumors have on average 20% more alternative splicing events than normal tissues. They mainly focused on five types of alternative splicing events including exon skipping, mutually exclusive exons, intron retention, as well as alternative 3′ and alternative 5′ splice site changes. What’s more, they found that tumors harbored more exon–exon junctions termed neojunctions than normal tissues. Besides, they confirmed neojunctions-derived peptides from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and predicted the underlying target, MHC-I. The evidence showed that neojunctions-derived peptides were potential neoantigens for cancer immunotherapy. Multiple computational methods and databases have been developed to identify alternative splicing events from RNA-seq data, such as SplAdder and CancerSplicingQTL2 (Kahles et al., 2016; Tian et al., 2019). Smart and Margolis (2018) developed a computational strategy to identify neoepitopes generated from intron retention events in tumor transcriptomes and confirmed that these neoepitopes were processed and presented on MHC-I. Their results suggested that RNA-derived neoantigens were promising candidates for a personalized cancer vaccine (Smart and Margolis, 2018). Another mechanism to promote proteome diversity is RNA editing, which will influence RNA metabolism and function. The most prevalent form of RNA editing is the deamination of adenosine (A) to inosine (I) which may be a source for neoantigen production (Peng et al., 2018). What’s more, databases of A-to-I RNA editing events in humans have been developed, such as REDIportal3 and RADAR4 (Ramaswami and Li, 2014; Picardi et al., 2017). Recently, Zhang and Fritsche (2018) demonstrated that epitopes derived from RNA editing were presented by HLA molecules and capable of inducing immune responses.
Promising results of neoantigen vaccines from preclinical studies have raised significant interest in clinical development (Castle et al., 2012; Gubin et al., 2014; Kreiter et al., 2015; Zhang et al., 2017). Recently, the clinical response in patients with advanced melanoma who received neoantigen vaccines treatment is quite encouraging in several phase I clinical trials (Yadav et al., 2014; Ott et al., 2017; Sahin et al., 2017). The main platforms for neoantigen vaccine are synthetic long peptide (SLP) vaccine, DNA vaccine, RNA vaccine, and dendritic cell (DC) vaccine. In addition, adoptive T cell therapy (ACT) targeting neo-epitopes has shown significant efficacy. Currently, there is an increasing number of clinical trials on neoantigen vaccines in a variety of cancers (Tables 2, 3).
The peptide vaccine is fast and flexible, and it is simple to make an individual cocktail for patients. Antigenic peptide has been widely developed in vaccination due to its strengths including low cost, low toxicity and direct function as pivotal T cell epitope (Li et al., 2014; Kumai et al., 2017). Ott et al. (2017) developed an immunogenic personal neoantigen vaccine (NCT01970358). In this phase I clinical study, 6 patients with resected high-risk melanoma (stage IIIB/C and IVM1a/b) received personalized long peptide vaccines targeting up to 20 neoantigens per patient after sequencing and prioritizing HLA class I prediction (toll-like receptor 3 [TLR3] and melanoma differentiation-associated protein 5 [MDA-5] poly-ICLC were also co-administered as adjuvants). The vaccination activated both CD8+ and CD4+ T cells responses against tumor. Treatment-related adverse events were fatigue, rash, injection site reactions and mild flu-like symptoms, with no major autoimmune toxicity. Encouragingly, four of these six vaccinated patients experienced no recurrence at 25 months after vaccination, and another two relapse patients with metastatic disease later experienced complete responses to anti- PD-1 therapy (Ott et al., 2017). However, the therapeutic efficacy of SLP vaccine is limited by inefficient delivery to desired lymphoid organs as it could rapidly diffuse into the peripheral blood vessels due to its small molecular size (Irvine et al., 2015; Zhu and Zhang, 2017).
DNA vaccine is stable, safe in handling, cost-efficient and easy to manufacture (Prazeres and Monteiro, 2014). Importantly, DNA vaccine can activate both CD4+ and CD8+ T cell response, as well as innate immune response due to the recognition of double stranded DNA structure by cytosolic sensors (Tang and Pietersz, 2009; Ori et al., 2017). Newer DNA vaccines have shown efficacy in the clinic (Trimble et al., 2015; Tebas et al., 2017; Aggarwal et al., 2019). Recently, one group utilized a DNA vaccine platform to target tumor neoantigens in a mouse model. They chose TC1, LLC, and ID8 as tumor models that hardly respond to immune-checkpoint blockade alone. After sequencing these tumors and identifying neoantigens, they designed 12 epitopes per plasmid and these synthetic neoantigen DNA vaccines (SNDVs) were tested in vivo. Not surprisingly, it showed robust T cell immunity. Intriguingly, a larger proportion of CD8+ T cell responses was generated during the SNDVs treatment (25% CD4+ and 75% CD8+ T cell responses) compared with other platforms such as SLP neoantigen vaccines (60% CD4+ and 16% CD8+ T cell responses). Although the epitopes were selected in silico for high MHC class I binding affinity, SLP and RNA neoantigen vaccines generated a higher proportion of MHC class II–restricted CD4+ T cells (Kreiter et al., 2015; Martin et al., 2016; Ott et al., 2017; Sahin et al., 2017). However, DNA vaccine shows poor immunogenicity in human trials (Jorritsma et al., 2016).
The advantages of RNA vaccine include low risk of insertional mutagenesis, direct translation into the cytoplasm and simple and inexpensive manufacturing procedure (Sahin et al., 2014). Kreiter et al. (2015) developed synthetic poly-neo-epitope messenger RNA vaccines after exome sequencing and bioinformatic prioritization in three independent murine tumor models. This vaccination induced complete rejection of established tumors and reshaped the tumor microenvironment (Kreiter et al., 2015). As RNA is the genetic material in many viruses, the human immune system tends to be on high alert for it, which gives an RNA vaccine a unique advantage. “It is its own adjuvant,” Sahin says (Sahin et al., 2017), so he implemented the RNA-based poly-neo-epitope vaccines in 13 patients with stage III-IV melanoma (NCT02035956). Two patients had a vaccine-related objective response among the five patients with metastatic disease, and the other eight patients who had no detectable disease mostly experienced prolonged disease-free survival. Two-thirds of vaccination developed de novo in addition to pre-existing immunity. Weide et al. (2009) also showed that direct injection of protamine-protected mRNA vaccine is feasible and safe; it can also increase the T cell response and decrease immunosuppressive cells in metastatic melanoma patients (NCT00204607). However, the translational efficiency of RNA vaccine remains challenging as only a small portion of administered mRNA can be captured and presented by APCs. Therefore, Sahin and his group administered the RNA vaccine directly into lymph nodes through ultrasound-guided percutaneous injection, noted as intranodal injection (i.n.).
Dendritic cells have a key role in presenting antigens to the immune system. DCs are often recognized as the most potent APCs, which are capable of acquiring and processing antigens for presentation to T cells and expressing high levels of costimulatory molecules (Sabado et al., 2017). Therefore, vaccination based on DCs is a promising platform for neoantigen vaccine. Carreno et al. (2015) were the first to report autologous DC vaccines directed at tumor amino acid substitutions (AAS) in three melanoma patients (NCT00683670). They filtered the candidate HLA-A∗ 02: 01 epitopes containing mutations residues after whole exome sequencing and HLA binding prediction and evaluated the MHC-epitope binding using mass spectrometry. Then they filtered precursors of DCs from patients’ bloodstream, matured them and exposed them to synthetic epitopes. The peptide-loaded DCs were then returned to the patients by intravenous infusion. It increased the breadth and diversity of anti-tumor immunity after receiving the DC neoantigen vaccine (Carreno et al., 2015). However, DC vaccine is laborious, costly and need highly skilled technicians for manufacturing (Chen et al., 2016).
T cell therapy targeting driver mutations is quite attractive, since they are not only specific and biologically important, but also shared between different patients (McGranahan et al., 2015). Currently, KRAS mutations are hot-spot driver mutations and the most frequent KRAS mutant is KRAS G12D that is expressed in ∼45% of pancreatic adenocarcinomas (Bryant et al., 2014) and ∼13% of colorectal cancers (Vaughn et al., 2011). Rosenberg et al. (2016) administered cytotoxic T cells targeting mutant KRAS G12D into a patient with metastatic colorectal cancer (NCT01174121). After whole-exome and transcriptome sequencing of three resected lung lesions, they found that CD8+ T cells in TILs specifically recognized mutant KRAS G12D. Then they selected and expanded TILs that were reactive to the mutant KRAS G12D. The patient received a single infusion of 1.48 × 1011 TILs, which contained 1.11 × 1011 HLA-C∗08:02–restricted CD8+ T cells that specifically targeted KRAS G12D. The objective regression of all seven lung metastases was observed at the first follow-up visit. However, one of these metastatic lesions had progressed when evaluating after 9 months of therapy. The loss of the chromosome 6 haplotype encoding the HLA-C∗08:02 class I MHC molecule resulted in progression after resecting this lesion and sequencing, which provides direct evidence of tumor immune evasion (Tran et al., 2016). Furthermore, the group used this approach to demonstrate that CD4+ T helper 1 (TH1) cells in TILs recognized a mutation in erbb2 interacting protein (ERBB2IP) in a patient with metastatic cholangiocarcinoma. This patient was treated with mutation-reactive TH1 cells twice and experienced tumor regression (NCT01174121) (Tran et al., 2014).
Antibodies targeting two immune checkpoints, PD-1 and CTLA-4, represent the greatest success of cancer immunotherapy, which can elicit durable antitumor responses in a wide range of malignancies (Hodi et al., 2010; Hamid et al., 2013; Chen and Han, 2015; Postow et al., 2015; Robert et al., 2015a; Zou et al., 2016). The treatment of immune checkpoint inhibitors has improved OS and PFS in many different cancers (Robert et al., 2011; Topalian et al., 2014; Borghaei et al., 2015; Brahmer et al., 2015; Garon et al., 2015; Larkin et al., 2015; Robert et al., 2015b; Sundar et al., 2015; George et al., 2016; Tomita et al., 2017). Accumulating evidence suggests that cancers with higher mutation burden are associated with more survival benefits from both anti- PD-1 and anti- CTLA-4 therapy (Table 4) (Hamid et al., 2013; Asaoka et al., 2015; Rizvi et al., 2015; Andor and Graham, 2016; Erratum for the Report “Genomic correlates of response to CTLA-4 blockade in metastatic melanoma” by Van Allen et al., 2016; Hugo et al., 2016; Matsushita et al., 2016; McGranahan et al., 2016; Morris et al., 2016; Rosenberg et al., 2016). Mutations may increase the possibility of generating immunogenic neoantigens, which facilitate the recognition of cancer cells as foreign (Schumacher and Schreiber, 2015; Riaz and Havel, 2016). Studies in melanoma patients have demonstrated that neoantigen-specific CD8+ and CD4+ T cells in TILs responded to checkpoint blockade therapy (Kvistborg et al., 2014; Lu et al., 2014; Tran et al., 2014; Linnemann et al., 2015), which provides testimony for the hypothesis. Furthermore, neoantigen loss may contribute to acquired resistance through tumor cell elimination or chromosomal deletions during immune checkpoint blockade therapy (Anagnostou et al., 2017); other mechanisms include upregulation of alternate immune checkpoints (Koyama et al., 2016), loss of HLA haplotypes (Maeurer et al., 1996), or somatic mutations in HLA or JAK1/JAK2 genes (Shukla et al., 2015; Garcia-Diaz et al., 2017). However, a proof-of-concept by Nicholas in which cytotoxic chemotherapy–induced subclonal neoantigens were enriched in certain poor responders to immune checkpoint inhibitors was presented (McGranahan et al., 2016). Additionally, gliomas that recurred after treatment with the DNA alkylating agent temozolomide were identified to carry numbers of mutations (Hunter et al., 2006; Cahill et al., 2007), which had a higher mutation burden generated from chemotherapy, but had less clinical benefit. As a result, the association between response to immune checkpoint blockade and neoantigen burden is not linear and clear (Le et al., 2015). Matsushita et al. (2017) demonstrated that the number of neoantigens per missense mutation (neoAg frequency) was an independent predictive factor for PFS in ovarian clear cell carcinoma (OCCC), and the low neoAg frequencies were correlated with increased PFS. High mutation and neoantigen load negatively influenced PFS in multiple myeloma (MM) patients, which had a lower mutational burden (Miller et al., 2017). Luksza et al. (2017) developed a neoantigen fitness model that could describe the evolutionary dynamics of cancer cells and predict tumor response to checkpoint blockade immunotherapy. Further studies are needed regarding the relationship between neoantigen load and checkpoint blockade immunotherapy.
High mutation burden is associated with survival benefits from ICB therapy, but autoantibodies may also predict the efficacy of immune checkpoint inhibitors (ICIs) (De Moel et al., 2019). However, recent studies have demonstrated that autoantibodies correlate with immune checkpoint therapy-induced toxicities (Da Gama et al., 2018; De Moel et al., 2019; Tahir et al., 2019). Since the mechanism of ICB therapy is to mediate non-specific suppression of T cells by negative costimulatory signals, it may break the balance of autoimmunity and lead to the activation of autoreactive B cells that produce autoantibodies (Pardoll, 2012; Oh et al., 2017). As we know, ICB therapy can cause immune-related adverse events (irAEs), referring to the release of distinctive toxicities including pneumonitis, dermatitis, hepatitis, colitis, and hypophysitis (Hodi et al., 2010; Weber et al., 2012; Gao et al., 2015). A recent study found that CD21lo B cells and plasmablasts increased in patients following ICB treatment, and these changes in B cells preceded and correlated with the frequency and timing of irAEs (Das et al., 2018). Besides, one phase I clinical trial including advanced metastatic melanoma patients who received BCG and ipilimumab treatment was suspended due to the occurrence of irAEs. Researchers found profound increases in the repertoire of autoantibodies directed against both selves- and cancer antigens at time points preceded the development of symptomatic toxicity (NCT01838200) (Da Gama et al., 2018). Furthermore, Tahir et al. (2019) also found that anti-GNAL and anti-ITM2B autoantibodies correlated with the development of ICI–related hypophysitis and that anti-CD74 autoantibodies were associated with ICB–induced pneumonitis development. They also tested additional patient samples by enzyme-linked immunosorbent assay to verify these findings (Tahir et al., 2019). These data suggest that autoantibodies under ICB treatment may serve as a predictive biomarker for irAEs.
Current neoantigen identification techniques are still time consuming and labourious (Tran et al., 2015; Gros et al., 2016). The predictors of immunogenicity are immature (Calis et al., 2013). In addition, since cytotoxic CD8+ T cells are the main killer of cancer cells, the available computational tools can only predict neo-epitopes that bind to MHC class I molecules presented on CD8+ T cells (Nielsen and Andreatta, 2016). Humans have approximately 5,000 alleles encoding MHC class I molecules, with expression of up to six MHC class I molecules (No authors, 2017b) and computational tools cannot predict them all. Intriguingly, even though the epitopes are selected for high MHC class I binding affinity, the neoantigen vaccine trials showed a higher proportion of MHC class II–restricted, CD4+ T cells (Kreiter et al., 2015; Ott et al., 2017). Furthermore, studies demonstrated that CD4+ T cells also recognize a higher number of neo-epitopes than was previously known and can confer potent antitumor activity (Tran et al., 2014; Kreiter et al., 2015). However, it will be more difficult to develop predictive algorithms for MHC class II molecules (Nielsen et al., 2010). First, MHC class II molecules are heterodimers of alpha and beta peptides encoded by four different loci, with three of them being highly polymorphic in the human genome (Robinson et al., 2003). Second, the MHC class II binding groove is open on both ends, presenting longer sequences of amino acids (11–20 amino acids or even longer) than MHC class I molecules (8–11 amino acids) (Babbitt et al., 1985; Bjorkman et al., 1987). Recently, Andreatta et al. (2015) described the method for the quantitative prediction of peptide binding affinity of MHC class II molecules of known sequence.
Primarily, the protein that contains the mutated residue is processed by the proteasome (Figure 3) (Nielsen et al., 2005) —a catalytic complex in the cytosol that can cleave the amino acid (AA) sequence to peptides ranging from 3 to 22 AA in length (Kisselev et al., 1999; Rock et al., 2002; Murata et al., 2009). A fraction of the peptides is further trimmed by aminopeptidases and endopeptidases in the cytosol and the endoplasmic reticulum(ER) (Beninga et al., 1998; York et al., 2002; Yan et al., 2006; Rock et al., 2010). Then, this peptide will be transported into the ER lumen by the TAP1/TAP2 transporter to assemble with MHC class I molecules (Peters et al., 2003; Larsen et al., 2007). Finally, the peptide-MHC class I complex can be presented on the cell surface. However, these computational tools hardly consider the endogenous processing and transport of peptides before HLA binding, which results in a high false-positive rate. As studied by Robbins and colleagues, 229 tumor-specific neo-epitopes were predicted in three melanoma patients, but only 11 (4.8%) of these neo-epitopes elicited a T cell response (Robbins et al., 2013). Neo-epitopes can also be produced by an altered MHC class I processing machinery in cancer cells (Van Hall et al., 2006; Seidel et al., 2012; Van Der Burg et al., 2016), which results in a high false-negative rate. Recently, Abelin et al. (2017) developed a new method of liquid chromatography-tandem mass spectrometry (LC–MS/MS) analysis of HLA-associated peptides which takes into account the endogenous processing of peptides, they also developed a new predictor with better algorithm since it is trained on peptide affinity data.
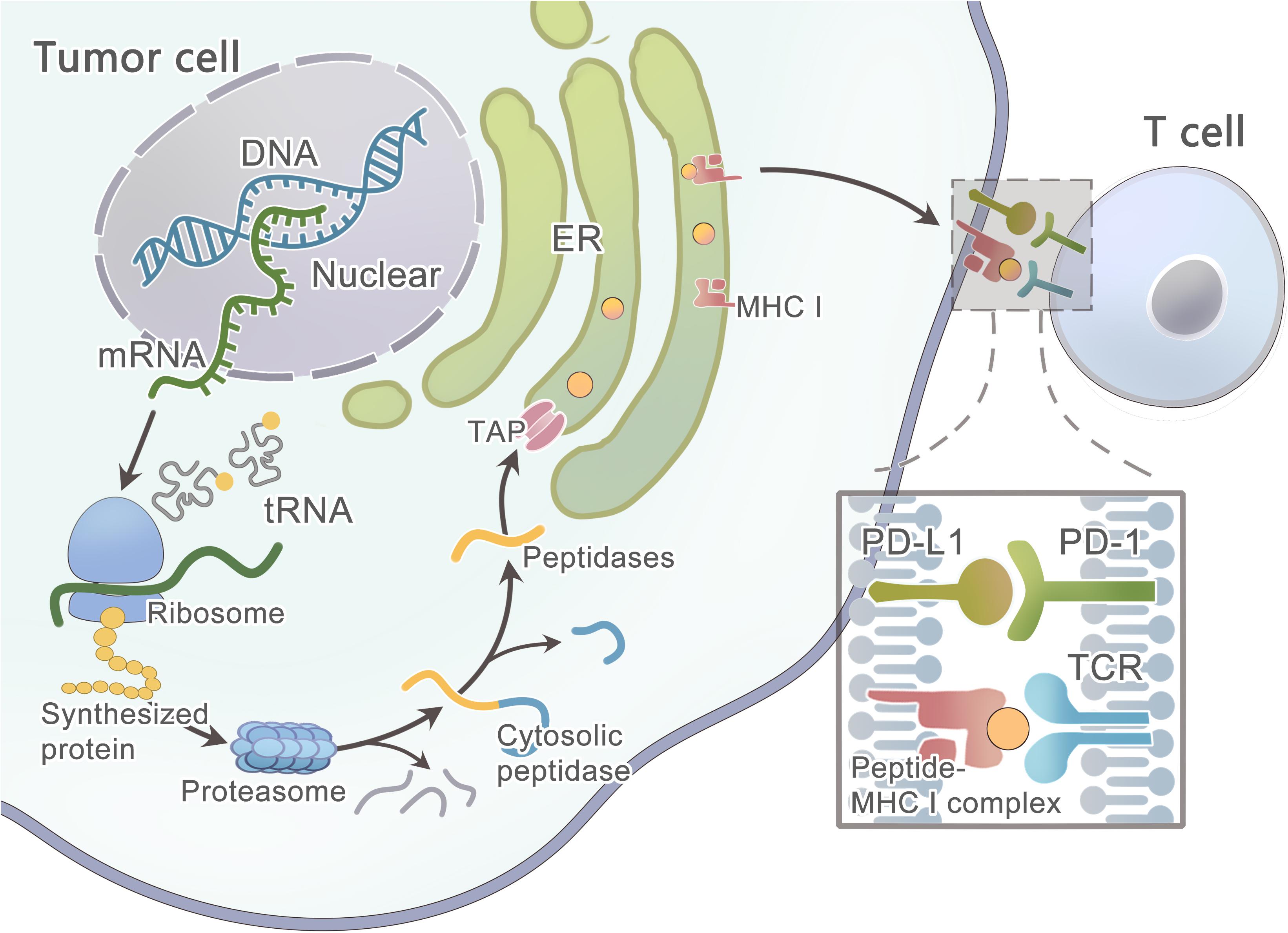
Figure 3. Schematic diagram illustrates the steps involved in tumor neoantigen processing and presentation on MHC class I molecules.
However, some patients may have no expression or aberrated expression of MHC molecules as immune evasion mechanisms (Marincola et al., 2000; Garrido et al., 2010; Leone et al., 2013; Textor et al., 2017). The loss of MHC class I can be detected in the early stage of some cancers (Vermeulen et al., 2005; Van Esch et al., 2014). As studied by Cabrera et al. (2003), β2m mutations and LMP7/TAP2 downregulation are the two main mechanisms in colorectal cancer that are responsible for the loss of MHC surface expression, and thus T cells fail to recognize cancer cells during an immune response. In some tumors, the MAPK pathway may regulate MHC I presentation (Mimura et al., 2013; Bradley et al., 2015; Ebert et al., 2016). After the mutated peptide and MHC class I complex presenting on the tumor cell surface, T cell recognition can occur only when TCRs that have the ability to recognize the mutant epitope exist within the T cell repertoire. Luckily, prior data showed that the T cell repertoire had a diversity of ∼2.5 × 107 (Arstila et al., 1999), and a single TCR was able to recognize up to 106 different MHC/peptide complexes, and thus the immune system could recognize ∼1012 possible foreign epitopes (Mason, 1998), which means the immune system has a strong recognition ability to distinguish even minor variations in MHC/peptide complexes. A study has shown that only certain types of mutations can be missed, such as conservative substitutions at other positions or alterations at the N-terminal peptide residue (Kessels et al., 2004).
Genomic instability and mutational processes can result in extensive tumor heterogeneity in each patient (Gerlinger et al., 2012). First, spontaneous mutations occur during the stages of tumor progression. Second, tumor microenvironments such as T cells can mediate neoantigen immunoediting (Verdegaal et al., 2016) or neoantigen loss. Third, metastatic lesions can involve the distal outgrowth of tumor cells originated from a subclone of the primary tumor; although there is little heterogeneity in driver mutations, there is still considerable epigenetic reprogramming between primary and metastatic tumors as studied in pancreatic ductal adenocarcinoma (PDAC) (Alderton, 2017). Therefore, a single site tumor biopsy may not adequately capture the total number of antigen clones present in the tumors (Sankin et al., 2014). It is well established that intratumor heterogeneity (ITH) correlates with the response of cancer patients to treatments with targeted therapies (Diaz et al., 2012; Shi et al., 2014; Landau et al., 2015). This is only tumor heterogeneity for each individual patient. For different patients and different cancers, the neo-epitopes were rarely shared except for driver mutations, which account for a fraction of mutations. Charoentong et al. (2017) analyzed more than 8,000 patients comprising 20 solid cancers from the TCGA (results available at https://tcia.at/). The pan-cancer analysis showed that the fraction of neo-epitopes generated from driver genes was 7.6%. Only 24 of 911,548 unique predicted neo-epitopes were common in more than 5% of patients (Charoentong et al., 2017). Therefore, neoantigen immunotherapy will probably need to be fully personalized for each patient, and this will be the next generation of precision medicine.
Cancer immunotherapy can offer limited clinical benefit without mitigating the immunosuppressive microenvironment of tumors. It has been increasingly recognized that tumors develop a specialized niche termed the tumor microenvironment (TME) in which tumor cells are protected from therapeutic interventions (Quail and Joyce, 2013). This niche includes fibroblasts (Kalluri and Zeisberg, 2006; Chen and Song, 2018), myeloid suppressor cells (MDSCs) (Goedegebuure et al., 2011; Kumar et al., 2016), regulatory T (Treg) cells (Van Der Burg et al., 2007; Bonertz et al., 2009), tumor-associated macrophages (TAM) (Qian and Pollard, 2010; Panni et al., 2013), lymphocytes, the extracellular matrix (ECM) and abnormal blood and lymphatic vessels (Jain, 2013). For example, high stromal density can limit T cell access to tumor cells and delivery of cytotoxic agents that provide a barrier (Provenzano et al., 2012; Feig et al., 2013; Ozdemir et al., 2014). Cancer cells can also express ligands for inhibitory receptors on T cells and secrete a multitude of chemokines and cytokines to affect antitumor immunity (Walker et al., 2003; Thomas and Massague, 2005). The vaccine strategies can successfully increase the frequency and activity of T cells, but they fail to guarantee that these T cells can exert their function within the tumors. The most important reason is the immune escape mechanisms in cancers, and thus proper co-treatment during vaccination is needed (Arens et al., 2013; Van Der Burg et al., 2016). Other than ICIs, there are multiple inhibitors targeting tumor immunosuppressive factors, including IDO1 inhibitors (Spranger et al., 2014), MEK inhibitors (Ebert et al., 2016), colony-stimulating factor-1 receptor (CSF1R) and chemokine (C-C motif) receptor 2 (CCR2) inhibitors (Mitchem et al., 2013), tumor extracellular matrix and stromal inhibitors (Provenzano et al., 2012), adenosine signaling through the adenosine A2a receptor (A2aR) (Leone et al., 2015), and other metabolic signaling pathways (Pardoll, 2015). In addition, combining such TME modulators with neoantigen-specific therapy may augment antitumor immunity (Melief et al., 2015). As studied by Zhu et al. (2014) CSF1R blockade could significantly improve the efficacy of PD-1 or CTLA-4 antagonists on tumor regressions. Furthermore, among the large number of predicted epitopes, only a minority can be recognized by autologous T cells (Robbins et al., 2013; Van Rooij et al., 2013; Rizvi et al., 2015); therefore, one group has proved that T cells redirected with T cell repertoires of healthy blood donors can efficiently recognize cancer epitopes that are neglected by a patient’s autologous T cells (Stronen et al., 2016).
Personal neoantigen vaccines can elicite strong T cell responses, which not only expand existing neoantigen-specific T cell populations, but also induce a new proportion of specific T cells in cancer patients (Ott et al., 2017). Hence, the identification of neoantigens is of utmost importance to improve cancer immunotherapy and broaden its efficacy to a larger number of patients. Khodadoust et al. (2017) discovered that immunoglobulin neoantigens in human mantle-cell lymphomas and CD4+ T cells specific for the neoantigens could mediate killing of autologous lymphoma cells. Keskin et al. (2019) also demonstrated that a strategy using multi-epitope, personalized neoantigen vaccination is feasible not only for high-risk melanoma (Rizvi et al., 2015; Ott et al., 2017; Sahin et al., 2017) but also for glioblastoma (Keskin et al., 2019), which has a relatively low mutation load. Furthermore, Balachandran et al. (2017) identified MUC16 as immunogenic hotspots in long-term survivors of pancreatic ductal adenocarcinoma, which is a presumed poorly immunogenic and checkpoint blockade-refractory tumor. These results are encouraging, but efforts are needed to tackle those challenges, and we will be very likely to witness these exciting developments in the near future.
X-JH and XM was a major writer of the manuscript and designed the figures, tables. XW and YW developed the structure of the article, reviewed and edited the manuscript. LY and YP researched the appropriate references and reviewed the manuscript. All the authors read and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China under (Grant Numbers 2016YFC1303502 and 2016YFA0201402).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abelin, J. G., Keskin, D. B., Sarkizova, S., Hartigan, C. R., Zhang, W., Sidney, J., et al. (2017). Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326. doi: 10.1016/j.immuni.2017.02.007
Aggarwal, C., Cohen, R. B., Morrow, M. P., Kraynyak, K. A., Sylvester, A. J., Knoblock, D. M., et al. (2019). Immunotherapy targeting HPV16/18 generates potent immune responses in HPV-associated head and neck cancer. Clin. Cancer Res. 25, 110–124. doi: 10.1158/1078-0432.ccr-18-1763
Alderton, G. K. (2017). Tumour evolution: epigenetic and genetic heterogeneity in metastasis. Nat. Rev. Cancer 17:141. doi: 10.1038/nrc.2017.11
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Aparicio, S. A., Behjati, S., Biankin, A. V., et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. doi: 10.1038/nature12477
Anagnostou, V., Smith, K. N., Forde, P. M., Niknafs, N., Bhattacharya, R., White, J., et al. (2017). Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276. doi: 10.1158/2159-8290.cd-16-0828
Andor, N., and Graham, T. A. (2016). Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113. doi: 10.1038/nm.3984
Andreatta, M., Karosiene, E., Rasmussen, M., Stryhn, A., Buus, S., and Nielsen, M. (2015). Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67, 641–650. doi: 10.1007/s00251-015-0873-y
Andreatta, M., and Nielsen, M. (2016). Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517. doi: 10.1093/bioinformatics/btv639
Arens, R., Van Hall, T., Van Der Burg, S. H., Ossendorp, F., and Melief, C. J. (2013). Prospects of combinatorial synthetic peptide vaccine-based immunotherapy against cancer. Semin Immunol. 25, 182–190. doi: 10.1016/j.smim.2013.04.008
Arstila, T. P., Casrouge, A., Baron, V., Even, J., Kanellopoulos, J., and Kourilsky, P. (1999). A direct estimate of the human alphabeta T cell receptor diversity. Science 286, 958–961. doi: 10.1126/science.286.5441.958
Asaoka, Y., Ijichi, H., and Koike, K. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 373:1979. doi: 10.1056/NEJMc1510353
Babbitt, B. P., Allen, P. M., Matsueda, G., Haber, E., and Unanue, E. R. (1985). Binding of immunogenic peptides to Ia histocompatibility molecules. Nature 317, 359–361. doi: 10.1038/317359a0
Balachandran, V. P., Luksza, M., Zhao, J. N., Makarov, V., Moral, J. A., Remark, R., et al. (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. doi: 10.1038/nature24462
Balar, A. V., Galsky, M. D., Rosenberg, J. E., Powles, T., Petrylak, D. P., Bellmunt, J., et al. (2017). Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 389, 67–76. doi: 10.1016/s0140-6736(16)32455-2
Barrick, J. E., and Lenski, R. E. (2013). Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839. doi: 10.1038/nrg3564
Bassani-Sternberg, M., Braunlein, E., Klar, R., Engleitner, T., Sinitcyn, P., Audehm, S., et al. (2016). Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7:13404. doi: 10.1038/ncomms13404
Bassani-Sternberg, M., Pletscher-Frankild, S., Jensen, L. J., and Mann, M. (2015). Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteomics 14, 658–673. doi: 10.1074/mcp.M114.042812
Belousov, P. V., Afanasyeva, M. A., Gubernatorova, E. O., Bogolyubova, A. V., Uvarova, A. N., Putlyaeva, L. V., et al. (2019). Multi-dimensional immunoproteomics coupled with in vitro recapitulation of oncogenic NRAS(Q61R) identifies diagnostically relevant autoantibody biomarkers in thyroid neoplasia. Cancer Lett. 467, 96–106. doi: 10.1016/j.canlet.2019.07.013
Beninga, J., Rock, K. L., and Goldberg, A. L. (1998). Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 273, 18734–18742. doi: 10.1074/jbc.273.30.18734
Bentley, D. R., Balasubramanian, S., Swerdlow, H. P., Smith, G. P., Milton, J., Brown, C. G., et al. (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59. doi: 10.1038/nature07517
Bentzen, A. K., and Hadrup, S. R. (2017). Evolution of MHC-based technologies used for detection of antigen-responsive T cells. Cancer Immunol. Immunother. 66, 657–666. doi: 10.1007/s00262-017-1971-5
Bentzen, A. K., and Marquard, A. M. (2016). Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34, 1037–1045. doi: 10.1038/nbt.3662
Bjorkman, P. J., Saper, M. A., Samraoui, B., Bennett, W. S., Strominger, J. L., and Wiley, D. C. (1987). Structure of the human class I histocompatibility antigen. HLA-A2. Nature 329, 506–512. doi: 10.1038/329506a0
Black, M. M., Opler, S. R., and Speer, F. D. (1954). Microscopic structure of gastric carcinomas and their regional lymph nodes in relation to survival. Surg. Gynecol. Obstet. 98, 725–734.
Bonertz, A., Weitz, J., Pietsch, D. H., Rahbari, N. N., Schlude, C., Ge, Y., et al. (2009). Antigen-specific Tregs control T cell responses against a limited repertoire of tumor antigens in patients with colorectal carcinoma. J. Clin. Invest. 119, 3311–3321. doi: 10.1172/jci39608
Borghaei, H., Paz-Ares, L., Horn, L., Spigel, D. R., Steins, M., Ready, N. E., et al. (2015). Nivolumab versus Docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639. doi: 10.1056/NEJMoa1507643
Bradley, S. D., Chen, Z., Melendez, B., Talukder, A., Khalili, J. S., Rodriguez-Cruz, T., et al. (2015). BRAFV600E Co-opts a conserved MHC class I internalization pathway to diminish antigen presentation and CD8+ T-cell recognition of melanoma. Cancer Immunol. Res. 3, 602–609. doi: 10.1158/2326-6066.cir-15-0030
Brahmer, J., Reckamp, K. L., Baas, P., Crino, L., Eberhardt, W. E., Poddubskaya, E., et al. (2015). Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135. doi: 10.1056/NEJMoa1504627
Brahmer, J. R., Tykodi, S. S., Chow, L. Q., Hwu, W. J., Topalian, S. L., Hwu, P., et al. (2012). Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465. doi: 10.1056/NEJMoa1200694
Brass, N., Rácz, A., Bauer, C., Heckel, D., Sybrecht, G., and Meese, E. (1999). Role of amplified genes in the production of autoantibodies. Blood 93, 2158–2166. doi: 10.1182/blood.v93.7.2158.407a34_2158_2166
Bryant, K. L., Mancias, J. D., Kimmelman, A. C., and Der, C. J. (2014). KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 39, 91–100. doi: 10.1016/j.tibs.2013.12.004
Bulik-Sullivan, B., Busby, J., Palmer, C. D., Davis, M. J., Murphy, T., Clark, A., et al. (2018). Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 18:4313. doi: 10.1038/nbt.4313
Caballero, O. L., and Chen, Y. T. (2009). Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 100, 2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x
Cabrera, C. M., Jimenez, P., Cabrera, T., Esparza, C., Ruiz-Cabello, F., and Garrido, F. (2003). Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens 61, 211–219. doi: 10.1034/j.1399-0039.2003.00020.x
Cahill, D. P., Levine, K. K., Betensky, R. A., Codd, P. J., Romany, C. A., Reavie, L. B., et al. (2007). Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin. Cancer Res. 13, 2038–2045. doi: 10.1158/1078-0432.ccr-06-2149
Calis, J. J., Maybeno, M., Greenbaum, J. A., Weiskopf, D., De Silva, A. D., Sette, A., et al. (2013). Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 9:e1003266. doi: 10.1371/journal.pcbi.1003266
Cancer Genome Atlas (TCGA) Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. doi: 10.1038/nature07385
Carbone, D. P., Reck, M., Paz-Ares, L., Creelan, B., Horn, L., Steins, M., et al. (2017). First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med 376, 2415–2426. doi: 10.1056/NEJMoa1613493
Carreno, B. M., Magrini, V., Becker-Hapak, M., Kaabinejadian, S., Hundal, J., Petti, A. A., et al. (2015). Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808. doi: 10.1126/science.aaa3828
Castle, J. C., Kreiter, S., Diekmann, J., Lower, M., van de Roemer, N., De Graaf, J., et al. (2012). Exploiting the mutanome for tumor vaccination. Cancer Res. 72, 1081–1091. doi: 10.1158/0008-5472.can-11-3722
Chan, T. A., Wolchok, J. D., and Snyder, A. (2015). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 373:1984. doi: 10.1056/NEJMc1508163
Charoentong, P., Finotello, F., Angelova, M., Mayer, C., Efremova, M., Rieder, D., et al. (2017). Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18, 248–262. doi: 10.1016/j.celrep.2016.12.019
Chen, L., and Han, X. (2015). Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 125, 3384–3391. doi: 10.1172/jci80011
Chen, P., Liu, X., Sun, Y., Zhou, P., Wang, Y., and Zhang, Y. (2016). Dendritic cell targeted vaccines: recent progresses and challenges. Hum. Vaccin. Immunother. 12, 612–622. doi: 10.1080/21645515.2015.1105415
Chen, X., and Song, E. (2018). Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discovery 18, 99–115. doi: 10.1038/s41573-018-0004-1
Chen, Y. T., Scanlan, M. J., Sahin, U., Türeci, O., Gure, A. O., Tsang, S., et al. (1997). A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. U.S.A. 94, 1914–1918. doi: 10.1073/pnas.94.5.1914
Choi, M., Scholl, U. I., Ji, W., Liu, T., Tikhonova, I. R., Zumbo, P., et al. (2009). Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 106, 19096–19101. doi: 10.1073/pnas.0910672106
Cohen, C. J., Gartner, J. J., Horovitz-Fried, M., Shamalov, K., Trebska-McGowan, K., Bliskovsky, V. V., et al. (2015). Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest. 125, 3981–3991. doi: 10.1172/jci82416
Coulie, P. G., Lehmann, F., Lethe, B., Herman, J., Lurquin, C., Andrawiss, M., et al. (1995). A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. U.S.A. 92, 7976–7980. doi: 10.1073/pnas.92.17.7976
Da Gama, D. J., Parakh, S., Andrews, M. C., Woods, K., Pasam, A., et al. (2018). Autoantibodies may predict immune-related toxicity: results from a phase I study of intralesional bacillus calmette-guérin followed by ipilimumab in patients with advanced metastatic melanoma. Front. Immunol. 9:411. doi: 10.3389/fimmu.2018.00411
Dai, L., Li, J., Tsay, J. J., Yie, T. A., Munger, J. S., Pass, H., et al. (2017). Identification of autoantibodies to ECH1 and HNRNPA2B1 as potential biomarkers in the early detection of lung cancer. Oncoimmunology 6:e1310359. doi: 10.1080/2162402x.2017.1310359
D’Angelo, S. P., Melchiori, L., Merchant, M. S., Bernstein, D., Glod, J., Kaplan, R., et al. (2018). Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 (c259)T cells in synovial sarcoma. Cancer Discov. 8, 944–957. doi: 10.1158/2159-8290.cd-17-1417
Das, R., Bar, N., Ferreira, M., Newman, A. M., Zhang, L., Bailur, J. K., et al. (2018). Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Invest. 128, 715–720. doi: 10.1172/jci96798
De Moel, E. C., Rozeman, E. A., Kapiteijn, E. H., Verdegaal, E. M. E., Grummels, A., and Bakker, J. A. (2019). Autoantibody development under treatment with immune-checkpoint inhibitors. Cancer Immunol. Res. 7, 6–11. doi: 10.1158/2326-6066.cir-18-0245
De Plaen, E., Lurquin, C., Van Pel, A., Mariame, B., Szikora, J. P., Wolfel, T., et al. (1988). Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum- antigen P91A and identification of the tum- mutation. Proc. Natl. Acad. Sci. U.S.A. 85, 2274–2278. doi: 10.1073/pnas.85.7.2274
Diaz, L. A. Jr., Williams, R. T., Wu, J., Kinde, I., Hecht, J. R., Berlin, J., et al. (2012). The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540. doi: 10.1038/nature11219
Ding, L., Getz, G., Wheeler, D. A., Mardis, E. R., McLellan, M. D., Cibulskis, K., et al. (2008). Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075. doi: 10.1038/nature07423
Ding, L., Wendl, M. C., McMichael, J. F., and Raphael, B. J. (2014). Expanding the computational toolbox for mining cancer genomes. Nat. Rev. Genet. 15, 556–570. doi: 10.1038/nrg3767
Drmanac, R., Sparks, A. B., Callow, M. J., Halpern, A. L., Burns, N. L., Kermani, B. G., et al. (2010). Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81. doi: 10.1126/science.1181498
Ebert, P. J. R., Cheung, J., Yang, Y., McNamara, E., Hong, R., Moskalenko, M., et al. (2016). MAP kinase inhibition promotes T cell and anti-tumor activity in combination with pd-l1 checkpoint blockade. Immunity 44, 609–621. doi: 10.1016/j.immuni.2016.01.024
Feig, C., Jones, J. O., Kraman, M., Wells, R. J., Deonarine, A., Chan, D. S., et al. (2013). Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 20212–20217. doi: 10.1073/pnas.1320318110
Feng, H., Shuda, M., Chang, Y., and Moore, P. S. (2008). Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319, 1096–1100. doi: 10.1126/science.1152586
Forbes, S. A., Beare, D., Gunasekaran, P., Leung, K., Bindal, N., Boutselakis, H., et al. (2015). COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811. doi: 10.1093/nar/gku1075
Galsky, M., Saci, A., Szabo, P. M., Han, G. C., Grossfeld, G. D., Collette, S., et al. (2020). Nivolumab in patients with advanced platinum-resistant urothelial carcinoma: efficacy, safety, and biomarker analyses with extended follow-up from checkmate 275. Clin. Cancer Res. [Epub ahead of print].
Gao, J., He, Q., Subudhi, S., Aparicio, A., Zurita-Saavedra, A., Lee, D. H., et al. (2015). Review of immune-related adverse events in prostate cancer patients treated with ipilimumab: MD Anderson experience. Oncogene 34, 5411–5417. doi: 10.1038/onc.2015.5
Garcia-Diaz, A., Shin, D. S., Moreno, B. H., Saco, J., Escuin-Ordinas, H., Rodriguez, G. A., et al. (2017). Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201. doi: 10.1016/j.celrep.2017.04.031
Garon, E. B., Rizvi, N. A., Hui, R., Leighl, N., Balmanoukian, A. S., Eder, J. P., et al. (2015). Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028. doi: 10.1056/NEJMoa1501824
Garraway, L. A., and Lander, E. S. (2013). Lessons from the cancer genome. Cell 153, 17–37. doi: 10.1016/j.cell.2013.03.002
Garrido, F., Cabrera, T., and Aptsiauri, N. (2010). “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. Int. J. Cancer 127, 249–256. doi: 10.1002/ijc.25270
George, S., Motzer, R. J., Hammers, H. J., Redman, B. G., Kuzel, T. M., Tykodi, S. S., et al. (2016). Safety and efficacy of nivolumab in patients with metastatic renal cell carcinoma treated beyond progression: a subgroup analysis of a randomized clinical trial. JAMA Oncol. 2, 1179–1186. doi: 10.1001/jamaoncol.2016.0775
Gerlinger, M., Rowan, A. J., Horswell, S., Math, M., Larkin, J., Endesfelder, D., et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892. doi: 10.1056/NEJMoa1113205
Gillison, M. L., Koch, W. M., Capone, R. B., Spafford, M., Westra, W. H., Wu, L., et al. (2000). Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92, 709–720. doi: 10.1093/jnci/92.9.709
Goedegebuure, P., Mitchem, J. B., Porembka, M. R., Tan, M. C., Belt, B. A., Wang-Gillam, A., et al. (2011). Myeloid-derived suppressor cells: general characteristics and relevance to clinical management of pancreatic cancer. Curr. Cancer Drug Targets 11, 734–751. doi: 10.2174/156800911796191024
Goodman, A. M., Kato, S., Bazhenova, L., Patel, S. P., Frampton, G. M., Miller, V., et al. (2017). Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608. doi: 10.1158/1535-7163.mct-17-0386
Greenman, C., Stephens, P., Smith, R., Dalgliesh, G. L., Hunter, C., Bignell, G., et al. (2007). Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158. doi: 10.1038/nature05610
Gros, A., Parkhurst, M. R., Tran, E., Pasetto, A., Robbins, P. F., Ilyas, S., et al. (2016). Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438. doi: 10.1038/nm.4051
Gubin, M. M., Artyomov, M. N., Mardis, E. R., and Schreiber, R. D. (2015). Tumor neoantigens: building a framework for personalized cancer immunotherapy. J. Clin. Invest. 125, 3413–3421. doi: 10.1172/jci80008
Gubin, M. M., Zhang, X., Schuster, H., Caron, E., Ward, J. P., Noguchi, T., et al. (2014). Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581. doi: 10.1038/nature13988
Haber, D. A., and Settleman, J. (2007). Cancer: drivers and passengers. Nature 446, 145–146. doi: 10.1038/446145a
Hadrup, S. R., Bakker, A. H., Shu, C. J., Andersen, R. S., van Veluw, J., Hombrink, P., et al. (2009). Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6, 520–526. doi: 10.1038/nmeth.1345
Hamid, O., Robert, C., Daud, A., Hodi, F. S., Hwu, W. J., Kefford, R., et al. (2013). Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144. doi: 10.1056/NEJMoa1305133
Heemskerk, B., Kvistborg, P., and Schumacher, T. N. (2013). The cancer antigenome. Embo J. 32, 194–203. doi: 10.1038/emboj.2012.333
Helleday, T., Eshtad, S., and Nik-Zainal, S. (2014). Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598. doi: 10.1038/nrg3729
Hellmann, M. D., Callahan, M. K., Awad, M. M., Calvo, E., Ascierto, P. A., Atmaca, A., et al. (2018a). Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 33, 853–861.e854. doi: 10.1016/j.ccell.2018.04.001853-861.e854
Hellmann, M. D., Ciuleanu, T. E., Pluzanski, A., Lee, J. S., Otterson, G. A., Audigier-Valette, C., et al. (2018b). Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378, 2093–2104. doi: 10.1056/NEJMoa1801946
Hellmann, M. D., Nathanson, T., Rizvi, H., Creelan, B. C., Sanchez-Vega, F., Ahuja, A., et al. (2018c). Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33, 843–852.e844. doi: 10.1016/j.ccell.2018.03.018843-852.e844
Hodi, F. S., O’Day, S. J., McDermott, D. F., Weber, R. W., Sosman, J. A., Haanen, J. B., et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723. doi: 10.1056/NEJMoa1003466
Hudson, T. J., Anderson, W., Artez, A., Barker, A. D., Bell, C., Bernabe, R. R., et al. (2010). International network of cancer genome projects. Nature 464, 993–998. doi: 10.1038/nature08987
Hugo, W., Zaretsky, J. M., Sun, L., Song, C., Moreno, B. H., Hu-Lieskovan, S., et al. (2016). Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44. doi: 10.1016/j.cell.2016.02.065
Hundal, J., Carreno, B. M., Petti, A. A., Linette, G. P., Griffith, O. L., Mardis, E. R., et al. (2016). pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 8, 11. doi: 10.1186/s13073-016-0264-5
Hundal, J., and Kiwala, S. (2019). Accounting for proximal variants improves neoantigen prediction. Nat. Genet. 51, 175–179. doi: 10.1038/s41588-018-0283-9
Hunter, C., Smith, R., Cahill, D. P., Stephens, P., Stevens, C., Teague, J., et al. (2006). A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 66, 3987–3991. doi: 10.1158/0008-5472.can-06-0127
Irvine, D. J., Hanson, M. C., Rakhra, K., and Tokatlian, T. (2015). Synthetic Nanoparticles for Vaccines and Immunotherapy. Chem. Rev. 115, 11109–11146. doi: 10.1021/acs.chemrev.5b00109
Jäger, D., Stockert, E., Scanlan, M. J., Güre, A. O., Jäger, E., Knuth, A., et al. (1999). Cancer-testis antigens and ING1 tumor suppressor gene product are breast cancer antigens: characterization of tissue-specific ING1 transcripts and a homologue gene. Cancer Res. 59, 6197–6204.
Jain, R. K. (2013). Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J. Clin. Oncol. 31, 2205–2218. doi: 10.1200/jco.2012.46.3653
Jo, N. R., Lee, K. J., and Shin, Y. B. (2016). Enzyme-coupled nanoplasmonic biosensing of cancer markers in human serum. Biosens. Bioelectron. 81, 324–333. doi: 10.1016/j.bios.2016.03.009
Jones, S., Zhang, X., Parsons, D. W., Lin, J. C., Leary, R. J., Angenendt, P., et al. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806. doi: 10.1126/science.1164368
Jorritsma, S. H. T., Gowans, E. J., Grubor-Bauk, B., and Wijesundara, D. K. (2016). Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine 34, 5488–5494. doi: 10.1016/j.vaccine.2016.09.062
Jurtz, V., and Paul, S. (2017). NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368. doi: 10.4049/jimmunol.1700893
Kahles, A., Lehmann, K. V., Toussaint, N. C., Hüser, M., Stark, S. G., Sachsenberg, T., et al. (2018). Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e216. doi: 10.1016/j.ccell.2018.07.001
Kahles, A., Ong, C. S., Zhong, Y., and Rätsch, G. (2016). SplAdder: identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics 32, 1840–1847. doi: 10.1093/bioinformatics/btw076
Kalluri, R., and Zeisberg, M. (2006). Fibroblasts in cancer. Nat. Rev. Cancer 6, 392–401. doi: 10.1038/nrc1877
Kan, Z., Jaiswal, B. S., Stinson, J., Janakiraman, V., Bhatt, D., Stern, H. M., et al. (2010). Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873. doi: 10.1038/nature09208
Keskin, D. B., Anandappa, A. J., Sun, J., Tirosh, I., Mathewson, N. D., Li, S., et al. (2019). Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239. doi: 10.1038/s41586-018-0792-9
Kessels, H. W., de Visser, K. E., Tirion, F. H., Coccoris, M., Kruisbeek, A. M., and Schumacher, T. N. (2004). The impact of self-tolerance on the polyclonal CD8+ T cell repertoire. J. Immunol. 172, 2324–2331. doi: 10.4049/jimmunol.172.4.2324
Khodadoust, M. S., Olsson, N., Wagar, L. E., Haabeth, O. A., Chen, B., Swaminathan, K., et al. (2017). Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727. doi: 10.1038/nature21433
Kim, S., Kim, H. S., Kim, E., Lee, M. G., Shin, E. C., Paik, S., et al. (2018). Neopepsee: accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann. Oncol. 29, 1030–1036. doi: 10.1093/annonc/mdy022
Kisselev, A. F., Akopian, T. N., Woo, K. M., and Goldberg, A. L. (1999). The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 274, 3363–3371. doi: 10.1074/jbc.274.6.3363
Koyama, S., Akbay, E. A., Li, Y. Y., Herter-Sprie, G. S., Buczkowski, K. A., Richards, W. G., et al. (2016). Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 7:10501. doi: 10.1038/ncomms10501
Kreiter, S., Vormehr, M., van de Roemer, N., Diken, M., Lower, M., Diekmann, J., et al. (2015). Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696. doi: 10.1038/nature14426
Kumai, T., Kobayashi, H., Harabuchi, Y., and Celis, E. (2017). Peptide vaccines in cancer-old concept revisited. Curr. Opin. Immunol. 45, 1–7. doi: 10.1016/j.coi.2016.11.001
Kumar, V., Patel, S., Tcyganov, E., and Gabrilovich, D. I. (2016). The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 37, 208–220. doi: 10.1016/j.it.2016.01.004
Kvistborg, P., Philips, D., Kelderman, S., Hageman, L., Ottensmeier, C., Joseph-Pietras, D., et al. (2014). Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci. Transl. Med. 6:254ra128. doi: 10.1126/scitranslmed.3008918
Landau, D. A., Tausch, E., Taylor-Weiner, A. N., Stewart, C., Reiter, J. G., Bahlo, J., et al. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530. doi: 10.1038/nature15395
Larkin, J., Hodi, F. S., and Wolchok, J. D. (2015). Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 1270–1271. doi: 10.1056/NEJMc1509660
Larsen, M. V., Lundegaard, C., Lamberth, K., Buus, S., Lund, O., and Nielsen, M. (2007). Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8:424. doi: 10.1186/1471-2105-8-424
Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520. doi: 10.1056/NEJMoa1500596
Lee, J. U., Nguyen, A. H., and Sim, S. J. (2015). A nanoplasmonic biosensor for label-free multiplex detection of cancer biomarkers. Biosens. Bioelectron. 74, 341–346. doi: 10.1016/j.bios.2015.06.059
Lee, W., Jiang, Z., Liu, J., Haverty, P. M., Guan, Y., Stinson, J., et al. (2010). The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465, 473–477. doi: 10.1038/nature09004
Lennerz, V., Fatho, M., Gentilini, C., Frye, R. A., Lifke, A., Ferel, D., et al. (2005). The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc. Natl. Acad. Sci. U.S.A. 102, 16013–16018. doi: 10.1073/pnas.0500090102
Leone, P., Shin, E. C., Perosa, F., Vacca, A., Dammacco, F., and Racanelli, V. (2013). MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J. Natl. Cancer Inst. 105, 1172–1187. doi: 10.1093/jnci/djt184
Leone, R. D., Lo, Y. C., and Powell, J. D. (2015). A2aR antagonists: Next generation checkpoint blockade for cancer immunotherapy. Comput. Struct. Biotechnol. J. 13, 265–272. doi: 10.1016/j.csbj.2015.03.008
Ley, T. J., Mardis, E. R., Ding, L., Fulton, B., McLellan, M. D., Chen, K., et al. (2008). DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72. doi: 10.1038/nature07485
Li, L., Goedegebuure, P., Mardis, E. R., Ellis, M. J., Zhang, X., Herndon, J. M., et al. (2011). Cancer genome sequencing and its implications for personalized cancer vaccines. Cancers (Basel) 3, 4191–4211. doi: 10.3390/cancers3044191
Li, W., Joshi, M. D., Singhania, S., Ramsey, K. H., and Murthy, A. K. (2014). Peptide vaccine: progress and challenges. Vaccines (Basel) 2, 515–536. doi: 10.3390/vaccines2030515
Lin, C. L., Lo, W. F., Lee, T. H., Ren, Y., Hwang, S. L., Cheng, Y. F., et al. (2002). Immunization with Epstein-Barr Virus (EBV) peptide-pulsed dendritic cells induces functional CD8+ T-cell immunity and may lead to tumor regression in patients with EBV-positive nasopharyngeal carcinoma. Cancer Res. 62, 6952–6958.
Lin, H. H., Ray, S., Tongchusak, S., Reinherz, E. L., and Brusic, V. (2008). Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 9:8. doi: 10.1186/1471-2172-9-8
Linnemann, C., van Buuren, M. M., Bies, L., Verdegaal, E. M., Schotte, R., Calis, J. J., et al. (2015). High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med. 21, 81–85. doi: 10.1038/nm.3773
Lu, Y. C., and Robbins, P. F. (2016). Cancer immunotherapy targeting neoantigens. Semin Immunol. 28, 22–27. doi: 10.1016/j.smim.2015.11.002
Lu, Y. C., Yao, X., Crystal, J. S., Li, Y. F., El-Gamil, M., Gross, C., et al. (2014). Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res. 20, 3401–3410. doi: 10.1158/1078-0432.ccr-14-0433
Luksza, M., Riaz, N., Makarov, V., Balachandran, V. P., Hellmann, M. D., Solovyov, A., et al. (2017). A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520. doi: 10.1038/nature24473
Lurquin, C., Van Pel, A., Mariame, B., De Plaen, E., Szikora, J. P., Janssens, C., et al. (1989). Structure of the gene of tum- transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ld by cytolytic T cells. Cell 58, 293–303. doi: 10.1016/0092-8674(89)90844-1
Maeurer, M. J., Gollin, S. M., Storkus, W. J., Swaney, W., Karbach, J., Martin, D., et al. (1996). Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6. Clin. Cancer Res. 2, 641–652.
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. doi: 10.1038/nature03959
Marincola, F. M., Jaffee, E. M., Hicklin, D. J., and Ferrone, S. (2000). Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv. Immunol. 74, 181–273. doi: 10.1016/s0065-2776(08)60911-6
Martin, S. D., Brown, S. D., Wick, D. A., Nielsen, J. S., Kroeger, D. R., Twumasi-Boateng, K., et al. (2016). Low mutation burden in ovarian cancer may limit the utility of neoantigen-targeted vaccines. PLoS One 11:e0155189. doi: 10.1371/journal.pone.0155189
Mason, D. (1998). A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404. doi: 10.1016/s0167-5699(98)01299-7
Matsushita, H., Hasegawa, K., Oda, K., Yamamoto, S., Nishijima, A., Imai, Y., et al. (2017). The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 6:e1338996. doi: 10.1080/2162402x.2017.1338996
Matsushita, H., Sato, Y., Karasaki, T., Nakagawa, T., Kume, H., Ogawa, S., et al. (2016). Neoantigen load, antigen presentation machinery, and immune signatures determine prognosis in clear cell renal cell carcinoma. Cancer Immunol. Res. 4, 463–471. doi: 10.1158/2326-6066.cir-15-0225
Matsushita, H., Vesely, M. D., Koboldt, D. C., Rickert, C. G., Uppaluri, R., Magrini, V. J., et al. (2012). Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404. doi: 10.1038/nature10755
McGranahan, N., Favero, F., de Bruin, E. C., Birkbak, N. J., Szallasi, Z., and Swanton, C. (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7:283ra254. doi: 10.1126/scitranslmed.aaa1408
McGranahan, N., Furness, A. J., Rosenthal, R., Ramskov, S., Lyngaa, R., Saini, S. K., et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. doi: 10.1126/science.aaf1490
Melero, I., Gaudernack, G., Gerritsen, W., Huber, C., Parmiani, G., Scholl, S., et al. (2014). Therapeutic vaccines for cancer: an overview of clinical trials. Nat. Rev. Clin. Oncol. 11, 509–524. doi: 10.1038/nrclinonc.2014.111
Melief, C. J., van Hall, T., Arens, R., Ossendorp, F., and Van Der Burg, S. H. (2015). Therapeutic cancer vaccines. J. Clin. Invest. 125, 3401–3412. doi: 10.1172/jci80009
Merlo, A., Turrini, R., Dolcetti, R., Martorelli, D., Muraro, E., Comoli, P., et al. (2010). The interplay between Epstein-Barr virus and the immune system: a rationale for adoptive cell therapy of EBV-related disorders. Haematologica 95, 1769–1777. doi: 10.3324/haematol.2010.023689
Meyerson, M., Gabriel, S., and Getz, G. (2010). Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696. doi: 10.1038/nrg2841
Miller, A., Asmann, Y., Cattaneo, L., Braggio, E., Keats, J., Auclair, D., et al. (2017). High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 7:e612. doi: 10.1038/bcj.2017.94
Mimura, K., Shiraishi, K., Mueller, A., Izawa, S., Kua, L. F., So, J., et al. (2013). The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J. Immunol. 191, 6261–6272. doi: 10.4049/jimmunol.1301597
Mitchem, J. B., Brennan, D. J., Knolhoff, B. L., Belt, B. A., Zhu, Y., Sanford, D. E., et al. (2013). Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 73, 1128–1141. doi: 10.1158/0008-5472.can-12-2731
Monach, P. A., Meredith, S. C., Siegel, C. T., and Schreiber, H. (1995). A unique tumor antigen produced by a single amino acid substitution. Immunity 2, 45–59. doi: 10.1016/1074-7613(95)90078-0
Morris, L. G., Riaz, N., Desrichard, A., Senbabaoglu, Y., Hakimi, A. A., Makarov, V., et al. (2016). Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7, 10051–10063. doi: 10.18632/oncotarget.7067
Morrissy, A. S., Morin, R. D., Delaney, A., Zeng, T., McDonald, H., Jones, S., et al. (2009). Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 19, 1825–1835. doi: 10.1101/gr.094482.109
Morrow, M. P., Yan, J., and Sardesai, N. Y. (2013). Human papillomavirus therapeutic vaccines: targeting viral antigens as immunotherapy for precancerous disease and cancer. Expert Rev. Vaccines 12, 271–283. doi: 10.1586/erv.13.23
Murata, S., Yashiroda, H., and Tanaka, K. (2009). Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115. doi: 10.1038/nrm2630
Ng, S. B., Turner, E. H., Robertson, P. D., Flygare, S. D., Bigham, A. W., Lee, C., et al. (2009). Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276. doi: 10.1038/nature08250
Nielsen, M., and Andreatta, M. (2016). NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8:33. doi: 10.1186/s13073-016-0288-x
Nielsen, M., Lund, O., Buus, S., and Lundegaard, C. (2010). MHC class II epitope predictive algorithms. Immunology 130, 319–328. doi: 10.1111/j.1365-2567.2010.03268.x
Nielsen, M., Lundegaard, C., Blicher, T., Lamberth, K., Harndahl, M., Justesen, S., et al. (2007). NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796. doi: 10.1371/journal.pone.0000796
Nielsen, M., Lundegaard, C., Lund, O., and Kesmir, C. (2005). The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57, 33–41. doi: 10.1007/s00251-005-0781-7
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. doi: 10.1038/nature08909
No authors (2017a). Modeling cancer mutations in 3-D. Cancer Discov. 7, 787–788. doi: 10.1158/2159-8290.cd-nb2017-091
No authors (2017b). The problem with neoantigen prediction. Nat. Biotechnol. 35:97. doi: 10.1038/nbt.3800
O’Donnell, T. J., Rubinsteyn, A., Bonsack, M., Riemer, A. B., Laserson, U., and Hammerbacher, J. (2018). MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132.e124. doi: 10.1016/j.cels.2018.05.014129-132.e124
O’Farrell, P. H. (1975). High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021.
Oh, D. Y., Cham, J., Zhang, L., Fong, G., Kwek, S. S., Klinger, M., et al. (2017). Immune toxicities elicted by CTLA-4 blockade in cancer patients are associated with early diversification of the T-cell repertoire. Cancer Res. 77, 1322–1330. doi: 10.1158/0008-5472.can-16-2324
Old, L. J., and Boyse, E. A. (1964). IMMUNOLOGY OF EXPERIMENTAL TUMORS. Annu. Rev. Med. 15, 167–186. doi: 10.1146/annurev.me.15.020164.001123
Ori, D., Murase, M., and Kawai, T. (2017). Cytosolic nucleic acid sensors and innate immune regulation. Int. Rev. Immunol. 36, 74–88. doi: 10.1080/08830185.2017.1298749
Ott, P. A., Hu, Z., Keskin, D. B., Shukla, S. A., Sun, J., Bozym, D. J., et al. (2017). An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. doi: 10.1038/nature22991
Ozdemir, B. C., Pentcheva-Hoang, T., Carstens, J. L., Zheng, X., Wu, C. C., Simpson, T. R., et al. (2014). Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 25, 719–734. doi: 10.1016/j.ccr.2014.04.005
Pagani, F., and Baralle, F. E. (2004). Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 5, 389–396. doi: 10.1038/nrg1327
Panni, R. Z., Linehan, D. C., and DeNardo, D. G. (2013). Targeting tumor-infiltrating macrophages to combat cancer. Immunotherapy 5, 1075–1087. doi: 10.2217/imt.13.102
Pardoll, D. (2015). Cancer and the immune system: basic concepts and targets for intervention. Semin. Oncol. 42, 523–538. doi: 10.1053/j.seminoncol.2015.05.003
Pardoll, D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264. doi: 10.1038/nrc3239
Park, E., Pan, Z., Zhang, Z., Lin, L., and Xing, Y. (2018). The expanding landscape of alternative splicing variation in human populations. Am. J. Hum. Genet. 102, 11–26. doi: 10.1016/j.ajhg.2017.11.002
Parsons, D. W., Jones, S., Zhang, X., Lin, J. C., Leary, R. J., Angenendt, P., et al. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812. doi: 10.1126/science.1164382
Parsons, D. W., Li, M., Zhang, X., Jones, S., Leary, R. J., Lin, J. C., et al. (2011). The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439. doi: 10.1126/science.1198056
Pena-Diaz, J., Bregenhorn, S., Ghodgaonkar, M., Follonier, C., Artola-Boran, M., Castor, D., et al. (2017). Noncanonical mismatch repair as a source of genomic instability in human cells. Mol Cell 67:162. doi: 10.1016/j.molcel.2017.06.026
Peng, X., Xu, X., Wang, Y., Hawke, D. H., Yu, S., Han, L., et al. (2018). A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell 33:817–828.e817. doi: 10.1016/j.ccell.2018.03.026817-828.e817
Peters, B., Bulik, S., Tampe, R., Van Endert, P. M., and Holzhutter, H. G. (2003). Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 171, 1741–1749. doi: 10.4049/jimmunol.171.4.1741
Picardi, E., D’Erchia, A. M., Lo, G. C., and Pesole, G. (2017). REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45, D750–D757. doi: 10.1093/nar/gkw767
Pleasance, E. D., Cheetham, R. K., Stephens, P. J., McBride, D. J., Humphray, S. J., Greenman, C. D., et al. (2010a). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196. doi: 10.1038/nature08658
Pleasance, E. D., Stephens, P. J., O’Meara, S., McBride, D. J., Meynert, A., Jones, D., et al. (2010b). A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190. doi: 10.1038/nature08629
Pollard, H. B., Srivastava, M., Eidelman, O., Jozwik, C., Rothwell, S. W., Mueller, G. P., et al. (2007). Protein microarray platforms for clinical proteomics. Proteomics Clin. Appl. 1, 934–952. doi: 10.1002/prca.200700154
Postow, M. A., Chesney, J., Pavlick, A. C., Robert, C., Grossmann, K., McDermott, D., et al. (2015). Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 372, 2006–2017. doi: 10.1056/NEJMoa1414428
Powles, T., Durán, I., van der Heijden, M. S., Loriot, Y., Vogelzang, N. J., De Giorgi, U., et al. (2018). Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet 391, 748–757. doi: 10.1016/s0140-6736(17)33297-x
Prazeres, D. M. F., and Monteiro, G. A. (2014). Plasmid Biopharmaceuticals. Microbiol. Spectr. 2:1128. doi: 10.1128/microbiolspec.PLAS-0022-2014
Prehn, R. T., and Main, J. M. (1957). Immunity to methylcholanthrene-induced sarcomas. J. Natl. Cancer Inst. 18, 769–778.
Provenzano, P. P., Cuevas, C., Chang, A. E., Goel, V. K., Von Hoff, D. D., and Hingorani, S. R. (2012). Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21, 418–429. doi: 10.1016/j.ccr.2012.01.007
Qian, B. Z., and Pollard, J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51. doi: 10.1016/j.cell.2010.03.014
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. doi: 10.1038/nm.3394
Rajasagi, M., Shukla, S. A., Fritsch, E. F., Keskin, D. B., DeLuca, D., Carmona, E., et al. (2014). Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124, 453–462. doi: 10.1182/blood-2014-04-567933
Ramaswami, G., and Li, J. B. (2014). RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–D113. doi: 10.1093/nar/gkt996
Rammensee, H., Bachmann, J., Emmerich, N. P., Bachor, O. A., and Stevanovic, S. (1999). SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219. doi: 10.1007/s002510050595
Restifo, N. P., Dudley, M. E., and Rosenberg, S. A. (2012). Adoptive immunotherapy for cancer: harnessing the T cell response. Nat. Rev. Immunol. 12, 269–281. doi: 10.1038/nri3191
Riaz, N., and Havel, J. J. (2016). Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat. Genet. 48, 1327–1329. doi: 10.1038/ng.3677
Riaz, N., Havel, J. J., Makarov, V., Desrichard, A., Urba, W. J., Sims, J. S., et al. (2017). Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e916. doi: 10.1016/j.cell.2017.09.028934-949.e916
Rizvi, H., Sanchez-Vega, F., La, K., Chatila, W., Jonsson, P., Halpenny, D., et al. (2018). Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641. doi: 10.1200/jco.2017.75.3384
Rizvi, N. A., Hellmann, M. D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J. J., et al. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. doi: 10.1126/science.aaa1348
Robbins, P. F., Lu, Y. C., El-Gamil, M., Li, Y. F., Gross, C., Gartner, J., et al. (2013). Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752. doi: 10.1038/nm.3161
Robert, C., Long, G. V., Brady, B., Dutriaux, C., Maio, M., Mortier, L., et al. (2015a). Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330. doi: 10.1056/NEJMoa1412082
Robert, C., Schachter, J., Long, G. V., Arance, A., Grob, J. J., Mortier, L., et al. (2015b). Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372, 2521–2532. doi: 10.1056/NEJMoa1503093
Robert, C., Thomas, L., Bondarenko, I., O’Day, S., Weber, J., Garbe, C., et al. (2011). Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 364, 2517–2526. doi: 10.1056/NEJMoa1104621
Robinson, J., Halliwell, J. A., McWilliam, H., Lopez, R., Parham, P., and Marsh, S. G. (2013). The IMGT/HLA database. Nucleic Acids Res. 41, D1222–D1227. doi: 10.1093/nar/gks949
Robinson, J., Waller, M. J., Parham, P., de Groot, N., Bontrop, R., Kennedy, L. J., et al. (2003). IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31, 311–314. doi: 10.1093/nar/gkg070
Rock, K. L., Farfan-Arribas, D. J., and Shen, L. (2010). Proteases in MHC class I presentation and cross-presentation. J. Immunol. 184, 9–15. doi: 10.4049/jimmunol.0903399
Rock, K. L., York, I. A., Saric, T., and Goldberg, A. L. (2002). Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–70. doi: 10.1016/s0065-2776(02)80012-8
Rosenberg, J. E., Hoffman-Censits, J., Powles, T., van der Heijden, M. S., Balar, A. V., Necchi, A., et al. (2016). Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920. doi: 10.1016/s0140-6736(16)00561-4
Rosenberg, S. A. (2011). Cell transfer immunotherapy for metastatic solid cancer–what clinicians need to know. Nat. Rev. Clin. Oncol. 8, 577–585. doi: 10.1038/nrclinonc.2011.116
Rosenberg, S. A., Yang, J. C., Sherry, R. M., Kammula, U. S., Hughes, M. S., Phan, G. Q., et al. (2011). Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557. doi: 10.1158/1078-0432.ccr-11-0116
Sabado, R. L., Balan, S., and Bhardwaj, N. (2017). Dendritic cell-based immunotherapy. Cell Res. 27, 74–95. doi: 10.1038/cr.2016.157
Sabatino, M., Hu, J., Sommariva, M., Gautam, S., Fellowes, V., Hocker, J. D., et al. (2016). Generation of clinical-grade CD19-specific CAR-modified CD8+ memory stem cells for the treatment of human B-cell malignancies. Blood 128, 519–528. doi: 10.1182/blood-2015-11-683847
Sahin, U., Derhovanessian, E., Miller, M., Kloke, B. P., Simon, P., Löwer, M., et al. (2017). Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226. doi: 10.1038/nature23003
Sahin, U., Karikó, K., and Türeci, Ö (2014). mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780. doi: 10.1038/nrd4278
Sankin, A., Hakimi, A. A., Mikkilineni, N., Ostrovnaya, I., Silk, M. T., Liang, Y., et al. (2014). The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 3, 1485–1492. doi: 10.1002/cam4.293
Scanlan, M. J., Chen, Y. T., Williamson, B., Gure, A. O., Stockert, E., Gordan, J. D., et al. (1998). Characterization of human colon cancer antigens recognized by autologous antibodies. Int. J. Cancer 76, 652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7<3.0.co;2-p
Schumacher, T. N., and Schreiber, R. D. (2015). Neoantigens in cancer immunotherapy. Science 348, 69–74. doi: 10.1126/science.aaa4971
Segal, N. H., Parsons, D. W., Peggs, K. S., Velculescu, V., Kinzler, K. W., Vogelstein, B., et al. (2008). Epitope landscape in breast and colorectal cancer. Cancer Res. 68, 889–892. doi: 10.1158/0008-5472.can-07-3095
Seidel, U. J., Oliveira, C. C., Lampen, M. H., and Hall, T. (2012). A novel category of antigens enabling CTL immunity to tumor escape variants: Cinderella antigens. Cancer Immunol. Immunother. 61, 119–125. doi: 10.1007/s00262-011-1160-x
Sharma, P., and Allison, J. P. (2015). The future of immune checkpoint therapy. Science 348, 56–61. doi: 10.1126/science.aaa8172
Sharp, P. A. (1994). Split genes and RNA splicing. Cell 77, 805–815. doi: 10.1016/0092-8674(94)90130-9
Shen, L., Zhang, J., and Lee, H. (2019). RNA transcription and splicing errors as a source of cancer frameshift neoantigens for vaccines. Sci. Rep. 9:14184. doi: 10.1038/s41598-019-50738-4
Shendure, J., Porreca, G. J., Reppas, N. B., Lin, X., McCutcheon, J. P., Rosenbaum, A. M., et al. (2005). Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732. doi: 10.1126/science.1117389
Shi, H., Hugo, W., Kong, X., Hong, A., Koya, R. C., Moriceau, G., et al. (2014). Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 4, 80–93. doi: 10.1158/2159-8290.cd-13-0642
Shukla, S. A., Rooney, M. S., Rajasagi, M., Tiao, G., Dixon, P. M., Lawrence, M. S., et al. (2015). Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 33, 1152–1158. doi: 10.1038/nbt.3344
Sjoblom, T., Jones, S., Wood, L. D., Parsons, D. W., Lin, J., Barber, T. D., et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274. doi: 10.1126/science.1133427
Smart, A. C., and Margolis, C. A. (2018). Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058. doi: 10.1038/nbt.4239
Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J. M., Desrichard, A., et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199. doi: 10.1056/NEJMoa1406498
Spranger, S., Koblish, H. K., Horton, B., Scherle, P. A., Newton, R., and Gajewski, T. F. (2014). Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. J. Immunother. Cancer. 2:3. doi: 10.1186/2051-1426-2-3
Stratton, M. R. (2011). Exploring the genomes of cancer cells: progress and promise. Science 331, 1553–1558. doi: 10.1126/science.1204040
Stratton, M. R., Campbell, P. J., and Futreal, P. A. (2009). The cancer genome. Nature 458, 719–724. doi: 10.1038/nature07943
Stronen, E., Toebes, M., Kelderman, S., van Buuren, M. M., Yang, W., van Rooij, N., et al. (2016). Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341. doi: 10.1126/science.aaf2288
Sundar, R., Cho, B. C., Brahmer, J. R., and Soo, R. A. (2015). Nivolumab in NSCLC: latest evidence and clinical potential. Ther. Adv. Med. Oncol. 7, 85–96. doi: 10.1177/1758834014567470
Szolek, A., Schubert, B., Mohr, C., Sturm, M., Feldhahn, M., and Kohlbacher, O. (2014). OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. doi: 10.1093/bioinformatics/btu548
Tahir, S. A., Gao, J., Miura, Y., Blando, J., Tidwell, R. S. S., Zhao, H., et al. (2019). Autoimmune antibodies correlate with immune checkpoint therapy-induced toxicities. Proc. Natl. Acad. Sci U.S.A. 116, 22246–22251. doi: 10.1073/pnas.1908079116
Talbot, S. J., and Crawford, D. H. (2004). Viruses and tumours–an update. Eur. J. Cancer 40, 1998–2005. doi: 10.1016/j.ejca.2003.11.039
Tang, C. K., and Pietersz, G. A. (2009). Intracellular detection and immune signaling pathways of DNA vaccines. Expert. Rev. Vaccines 8, 1161–1170. doi: 10.1586/erv.09.79
Tebas, P., Roberts, C. C., Muthumani, K., Reuschel, E. L., Kudchodkar, S. B., Zaidi, F. I., et al. (2017). Safety and immunogenicity of an anti-zika virus DNA vaccine–preliminary report. N. Engl. J. Med. 4:1056. doi: 10.1056/NEJMoa1708120
Textor, A., Schmidt, K., Kloetzel, P. M., Weissbrich, B., Perez, C., Charo, J., et al. (2017). Correction: Preventing tumor escape by targeting a post-proteasomal trimming independent epitope. J. Exp. Med. 214:567. doi: 10.1084/jem.2016063601122017c
Thomas, D. A., and Massague, J. (2005). TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380. doi: 10.1016/j.ccr.2005.10.012
Tian, J., Wang, Z., Mei, S., Yang, N., Yang, Y., Ke, J., et al. (2019). CancerSplicingQTL: a database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res. 47, D909–D916. doi: 10.1093/nar/gky954
Tjalsma, H., Schaeps, R. M., and Swinkels, D. W. (2008). Immunoproteomics: from biomarker discovery to diagnostic applications. Proteomics Clin. Appl. 2, 167–180. doi: 10.1002/prca.200780012
Tomita, Y., Fukasawa, S., Shinohara, N., Kitamura, H., Oya, M., Eto, M., et al. (2017). Nivolumab versus everolimus in advanced renal cell carcinoma: Japanese subgroup analysis from the CheckMate 025 study. Jpn. J. Clin. Oncol. 47, 639–646. doi: 10.1093/jjco/hyx049
Topalian, S. L., Hodi, F. S., Brahmer, J. R., Gettinger, S. N., Smith, D. C., McDermott, D. F., et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454. doi: 10.1056/NEJMoa1200690
Topalian, S. L., Sznol, M., McDermott, D. F., Kluger, H. M., Carvajal, R. D., Sharfman, W. H., et al. (2014). Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol. 32, 1020–1030. doi: 10.1200/jco.2013.53.0105
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Totoki, Y., Tatsuno, K., Yamamoto, S., Arai, Y., Hosoda, F., Ishikawa, S., et al. (2011). High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469. doi: 10.1038/ng.804
Tran, E., Ahmadzadeh, M., Lu, Y. C., Gros, A., Turcotte, S., Robbins, P. F., et al. (2015). Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390. doi: 10.1126/science.aad1253
Tran, E., Robbins, P. F., Lu, Y. C., Prickett, T. D., Gartner, J. J., Jia, L., et al. (2016). T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262. doi: 10.1056/NEJMoa1609279
Tran, E., Turcotte, S., Gros, A., Robbins, P. F., Lu, Y. C., Dudley, M. E., et al. (2014). Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645. doi: 10.1126/science.1251102
Trimble, C. L., Morrow, M. P., Kraynyak, K. A., Shen, X., Dallas, M., Yan, J., et al. (2015). Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet 386, 2078–2088. doi: 10.1016/s0140-6736(15)00239-1
Trucco, L. D., Mundra, P. A., Hogan, K., Garcia-Martinez, P., Viros, A., Mandal, A. K., et al. (2018). Ultraviolet radiation-induced DNA damage is prognostic for outcome in melanoma. Nat. Med. 25, 221–224. doi: 10.1038/s41591-018-0265-6
Türeci, O., Sahin, U., Vollmar, E., Siemer, S., Göttert, E., Seitz, G., et al. (1998a). Human carbonic anhydrase XII: cDNA cloning, expression, and chromosomal localization of a carbonic anhydrase gene that is overexpressed in some renal cell cancers. Proc. Natl. Acad. Sci. U.S.A. 95, 7608–7613. doi: 10.1073/pnas.95.13.7608
Türeci, O., Sahin, U., Zwick, C., Koslowski, M., Seitz, G., and Pfreundschuh, M. (1998b). Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc. Natl. Acad. Sci. U.S.A. 95, 5211–5216. doi: 10.1073/pnas.95.9.5211
Van Allen, E. M., Miao, D., Schilling, B., Shukla, S. A., Blank, C., Zimmer, L., et al. (2016). Erratum for the Report Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 352:6283. doi: 10.1126/science.aaf8264
Van Der Burg, S. H., Arens, R., Ossendorp, F., van Hall, T., and Melief, C. J. (2016). Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat. Rev. Cancer 16, 219–233. doi: 10.1038/nrc.2016.16
Van Der Burg, S. H., Piersma, S. J., de Jong, A., van der Hulst, J. M., Kwappenberg, K. M., van den Hende, M., et al. (2007). Association of cervical cancer with the presence of CD4+ regulatory T cells specific for human papillomavirus antigens. Proc. Natl. Acad. Sci. U.S.A. 104, 12087–12092. doi: 10.1073/pnas.0704672104
Van Esch, E. M., Tummers, B., Baartmans, V., Osse, E. M., Ter Haar, N., Trietsch, M. D., et al. (2014). Alterations in classical and nonclassical HLA expression in recurrent and progressive HPV-induced usual vulvar intraepithelial neoplasia and implications for immunotherapy. Int. J. Cancer 135, 830–842. doi: 10.1002/ijc.28713
Van Hall, T., Wolpert, E. Z., van Veelen, P., Laban, S., van der Veer, M., Roseboom, M., et al. (2006). Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nat. Med. 12, 417–424. doi: 10.1038/nm1381
Van Rooij, N., van Buuren, M. M., Philips, D., Velds, A., Toebes, M., Heemskerk, B., et al. (2013). Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442. doi: 10.1200/jco.2012.47.7521
Vaughn, C. P., Zobell, S. D., Furtado, L. V., Baker, C. L., and Samowitz, W. S. (2011). Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer 50, 307–312. doi: 10.1002/gcc.20854
Verdegaal, E. M., de Miranda, N. F., Visser, M., Harryvan, T., van Buuren, M. M., Andersen, R. S., et al. (2016). Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 536, 91–95. doi: 10.1038/nature18945
Vermeulen, C. F., Jordanova, E. S., Zomerdijk-Nooijen, Y. A., Ter Haar, N. T., Peters, A. A., and Fleuren, G. J. (2005). Frequent HLA class I loss is an early event in cervical carcinogenesis. Hum. Immunol. 66, 1167–1173. doi: 10.1016/j.humimm.2005.10.011
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A. Jr., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339, 1546–1558. doi: 10.1126/science.1235122
Walboomers, J. M., Jacobs, M. V., Manos, M. M., Bosch, F. X., Kummer, J. A., Shah, K. V., et al. (1999). Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19. doi: 10.1002/(sici)1096-9896(199909)189:1<12::aid-path431<3.0.co;2-f
Walker, M. R., Kasprowicz, D. J., Gersuk, V. H., Benard, A., Van Landeghen, M., Buckner, J. H., et al. (2003). Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25- T cells. J. Clin. Invest. 112, 1437–1443. doi: 10.1172/jci19441
Ward, J. P., Gubin, M. M., and Schreiber, R. D. (2016). The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv. Immunol. 130, 25–74. doi: 10.1016/bs.ai.2016.01.001
Weber, J. S., Kähler, K. C., and Hauschild, A. (2012). Management of immune-related adverse events and kinetics of response with ipilimumab. J. Clin. Oncol. 30, 2691–2697. doi: 10.1200/jco.2012.41.6750
Wei, X., Walia, V., Lin, J. C., Teer, J. K., Prickett, T. D., Gartner, J., et al. (2011). Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 43, 442–446. doi: 10.1038/ng.810
Weide, B., Pascolo, S., Scheel, B., Derhovanessian, E., Pflugfelder, A., Eigentler, T. K., et al. (2009). Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 32, 498–507. doi: 10.1097/CJI.0b013e3181a00068
Weir, B., Zhao, X., and Meyerson, M. (2004). Somatic alterations in the human cancer genome. Cancer Cell 6, 433–438. doi: 10.1016/j.ccr.2004.11.004
Wheeler, D. A., Srinivasan, M., Egholm, M., Shen, Y., Chen, L., McGuire, A., et al. (2008). The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876. doi: 10.1038/nature06884
Wick, D. A., Webb, J. R., Nielsen, J. S., Martin, S. D., Kroeger, D. R., Milne, K., et al. (2014). Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin. Cancer Res. 20, 1125–1134. doi: 10.1158/1078-0432.ccr-13-2147
Wolfel, T., Hauer, M., Schneider, J., Serrano, M., Wolfel, C., Klehmann-Hieb, E., et al. (1995). A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269, 1281–1284. doi: 10.1126/science.7652577
Wood, L. D., Parsons, D. W., Jones, S., Lin, J., Sjoblom, T., Leary, R. J., et al. (2007). The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113. doi: 10.1126/science.1145720
Yadav, M., Jhunjhunwala, S., Phung, Q. T., Lupardus, P., Tanguay, J., Bumbaca, S., et al. (2014). Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576. doi: 10.1038/nature14001
Yan, J., Parekh, V. V., Mendez-Fernandez, Y., Olivares-Villagomez, D., Dragovic, S., Hill, T., et al. (2006). In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J. Exp. Med. 203, 647–659. doi: 10.1084/jem.20052271
Yang, Z., Chevolot, Y., Géhin, T., Solassol, J., Mange, A., Souteyrand, E., et al. (2013). Improvement of protein immobilization for the elaboration of tumor-associated antigen microarrays: application to the sensitive and specific detection of tumor markers from breast cancer sera. Biosens. Bioelectron. 40, 385–392. doi: 10.1016/j.bios.2012.08.019
York, I. A., Chang, S. C., Saric, T., Keys, J. A., Favreau, J. M., Goldberg, A. L., et al. (2002). The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 3, 1177–1184. doi: 10.1038/ni860
Zaenker, P., Gray, E. S., and Ziman, M. R. (2016). Autoantibody production in cancer–the humoral immune response toward autologous antigens in cancer patients. Autoimmun. Rev. 15, 477–483. doi: 10.1016/j.autrev.2016.01.017
Zhang, G. L., Ansari, H. R., Bradley, P., Cawley, G. C., Hertz, T., Hu, X., et al. (2011). Machine learning competition in immunology–Prediction of HLA class I binding peptides. J. Immunol. Methods 374, 1–4. doi: 10.1016/j.jim.2011.09.010
Zhang, J., Shen, L., and Johnston, S. A. (2018). Using frameshift peptide arrays for cancer neo-antigens screening. Sci. Rep. 8:17366. doi: 10.1038/s41598-018-35673-0
Zhang, M., and Fritsche, J. (2018). RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 9:3919. doi: 10.1038/s41467-018-06405-9
Zhang, X., Kim, S., Hundal, J., Herndon, J. M., Li, S., Petti, A. A., et al. (2017). Breast cancer neoantigens can induce CD8(+) T-cell responses and antitumor immunity. Cancer Immunol. Res. 5, 516–523. doi: 10.1158/2326-6066.cir-16-0264
Zhu, G., and Zhang, F. (2017). Efficient nanovaccine delivery in cancer immunotherapy. ACS Nano. 11, 2387–2392. doi: 10.1021/acsnano.7b00978
Zhu, Y., Knolhoff, B. L., Meyer, M. A., Nywening, T. M., West, B. L., Luo, J., et al. (2014). CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069. doi: 10.1158/0008-5472.can-13-3723
Keywords: neoantigens, cancer immunotherapy, clinical trial, checkpoint blockade, targeted cancer
Citation: Han X-J, Ma X, Yang L, Wei Y, Peng Y and Wei X (2020) Progress in Neoantigen Targeted Cancer Immunotherapies. Front. Cell Dev. Biol. 8:728. doi: 10.3389/fcell.2020.00728
Received: 22 April 2020; Accepted: 14 July 2020;
Published: 30 July 2020.
Edited by:
Kexin Xu, The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Jianjun Gao, The University of Texas MD Anderson Cancer Center, United StatesCopyright © 2020 Han, Ma, Yang, Wei, Peng and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia-wei Wei, eGlhd2Vpd2VpQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.