
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 07 August 2020
Sec. Stem Cell Research
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00635
This article is part of the Research Topic Context-Dependent Regulation of Neurogenesis: Common Themes and Unique Features of the Neurogenic Process in Different Model Systems View all 21 articles
The neural crest (NC) is a transient multipotent cell population that originates in the dorsal neural tube. Cells of the NC are highly migratory, as they travel considerable distances through the body to reach their final sites. Derivatives of the NC are neurons and glia of the peripheral nervous system (PNS) and the enteric nervous system as well as non-neural cells. Different signaling pathways triggered by Bone Morphogenetic Proteins (BMPs), Fibroblast Growth Factors (FGFs), Wnt proteins, Notch ligands, retinoic acid (RA), and Receptor Tyrosine Kinases (RTKs) participate in the processes of induction, specification, cell migration and neural differentiation of the NC. A specific set of signaling pathways and transcription factors are initially expressed in the neural plate border and then in the NC cell precursors to the formation of cranial nerves. The molecular mechanisms of control during embryonic development have been gradually elucidated, pointing to an important role of transcriptional regulators when neural differentiation occurs. However, some of these proteins have an important participation in malformations of the cranial portion and their mutation results in aberrant neurogenesis. This review aims to give an overview of the role of cell signaling and of the function of transcription factors involved in the specification of ganglia precursors and neurogenesis to form the NC-derived cranial nerves during organogenesis.
During the embryonic development of vertebrates, one of the main events after the gastrulation process is neurulation, which allows the formation of the neural tube (NT). The neural ectoderm generates not only the central nervous system (CNS) but also another set of cells between the NT and the non-neural ectoderm located in the most dorsal part of the NT, called the neural crest (NC) (Hall, 2008; Simões-Costa et al., 2015). This versatile and plastic cell population was first described by Wilhelm His 150 years ago (Hall, 1999). The NC is one of the most important features that separate vertebrates from other chordate organisms. It arises at the posterior and lateral borders of the neural and non-neural ectoderm, the neural plate border (Figure 1) (Cerrizuela et al., 2020).
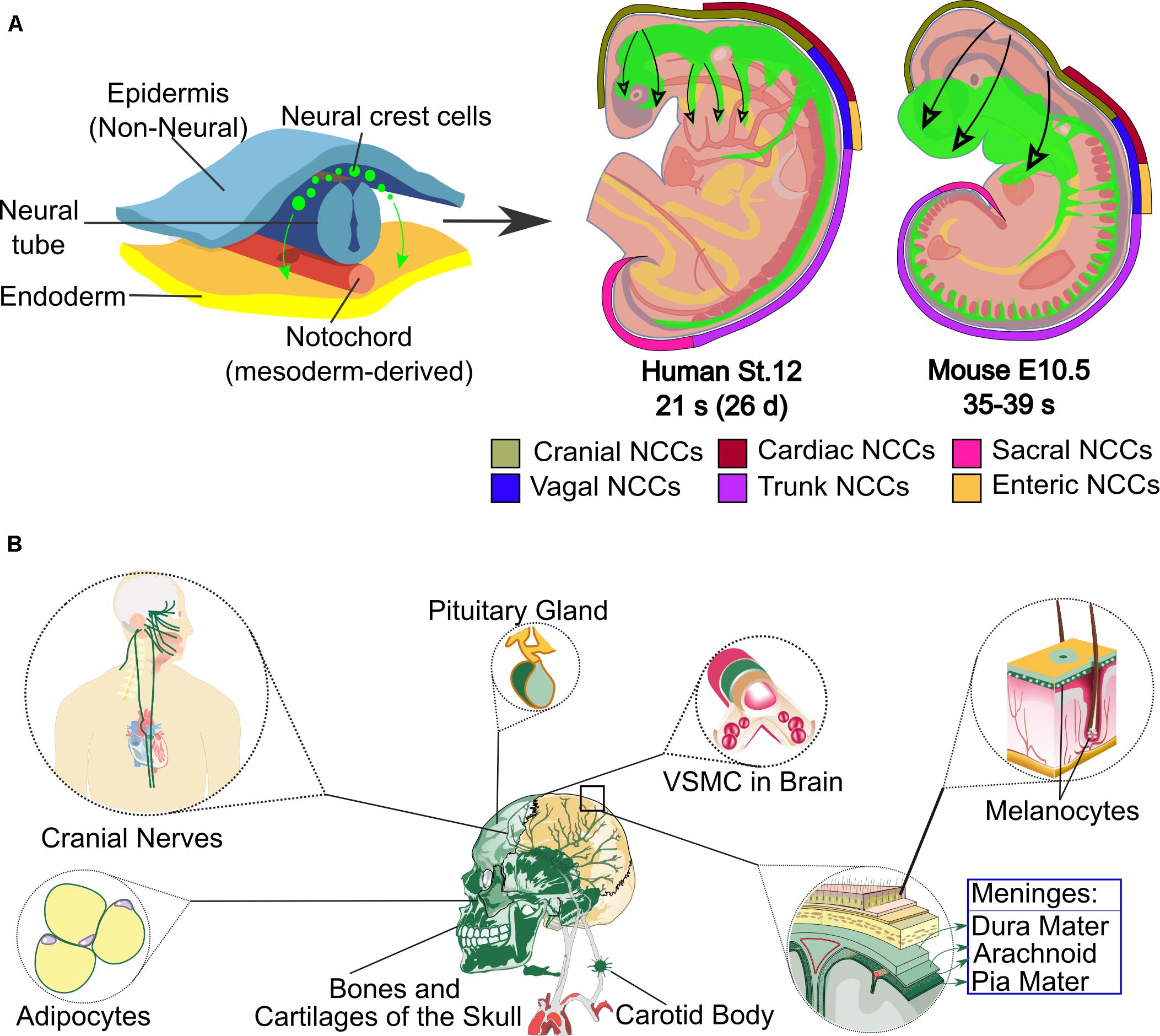
Figure 1. Neural crest origin, regions in human and mouse embryos and some of its cranial derivatives. (A) The top-left part of the scheme shows the origin of the neural crest cells (green) that migrate through the embryo. On the top-right side, the level of axial origin (see axial color key) of different regions of the neural crest is represented in developing mouse or equivalent human embryos; the migration of neural crest is represented in green inside the embryos and the direction of migration is marked with black arrows. Depending on their axial level of origin and migratory pathways, neural crest cells adopt different fates and contribute to various tissues and organs. (B) The main cranial derivatives, labeled in green, are shown. Abbreviations: d, days, E, mouse embryonic stage; NCCs, Neural Crest Cells; s, somite; St, human stage; VSMC, vascular smooth muscle cells.
NC cells (NCCs) are multipotent and give rise to several cell types, depending on the site of origin along the anteroposterior axis of the embryo. NCCs are divided into cranial, trunk (including cardiac), vagal and sacral (Figure 1A) (Minoux and Rijli, 2010; Simões-Costa and Bronner, 2013; Vega-Lopez et al., 2018). Cranial nerves (CN) transmit sensory and motor information between the brain and tissues of the head and cervical region. The CN are formed from the contribution of two specialized embryonic cell populations, cranial NC and ectodermal placodes.
NCCs, which are multipotent, delaminate from their origin and migrate throughout the body to differentiate into several cell types including cells of the peripheral nervous system (PNS), melanocytes, cranial cartilage and bone, neuroendocrine cells, and several other phenotypes (Figure 1B). In humans, at least 47 cell types have been defined as NC derivatives (Vickaryous and Hall, 2006). Proper NC migration relies on environmental cues such as Eph-Ephrins (Smith et al., 1997), Semaphorin-3F (Gammill et al., 2007), Versican (Szabó et al., 2016), the chemokine Stromal cell-derived factor 1 (Theveneau et al., 2013) or Robo2 (Shiau et al., 2008). The migration patterns of NCCs have been clearly described for model organisms like birds, frogs and mice. In all vertebrates, cranial NCCs emerge from the forebrain, midbrain and hindbrain regions (Couly and Le Douarin, 1987; Serbedzija et al., 1992). Depending on their axial origin, cranial NCCs will either migrate through the facial mesenchyme and into the frontonasal process, or will populate the branchial arches (Noden, 1975; Lumsden et al., 1991; Serbedzija et al., 1992). The sensory module of the PNS in the cranial region is composed of an array of paired ganglia adjacent to the hindbrain that transduce the perception of touch, pain, temperature, position and special sensory information from the periphery to the CNS. Cranial NCCs migrate to form sensory ganglia such as the trigeminal (V), the facial (VII), the glossopharyngeal (IX), the vagus (X) CN, and also to form the motor ganglia for the oculomotor (III) and accesory (XI) CN (Table 1 and Figures 2, 3).
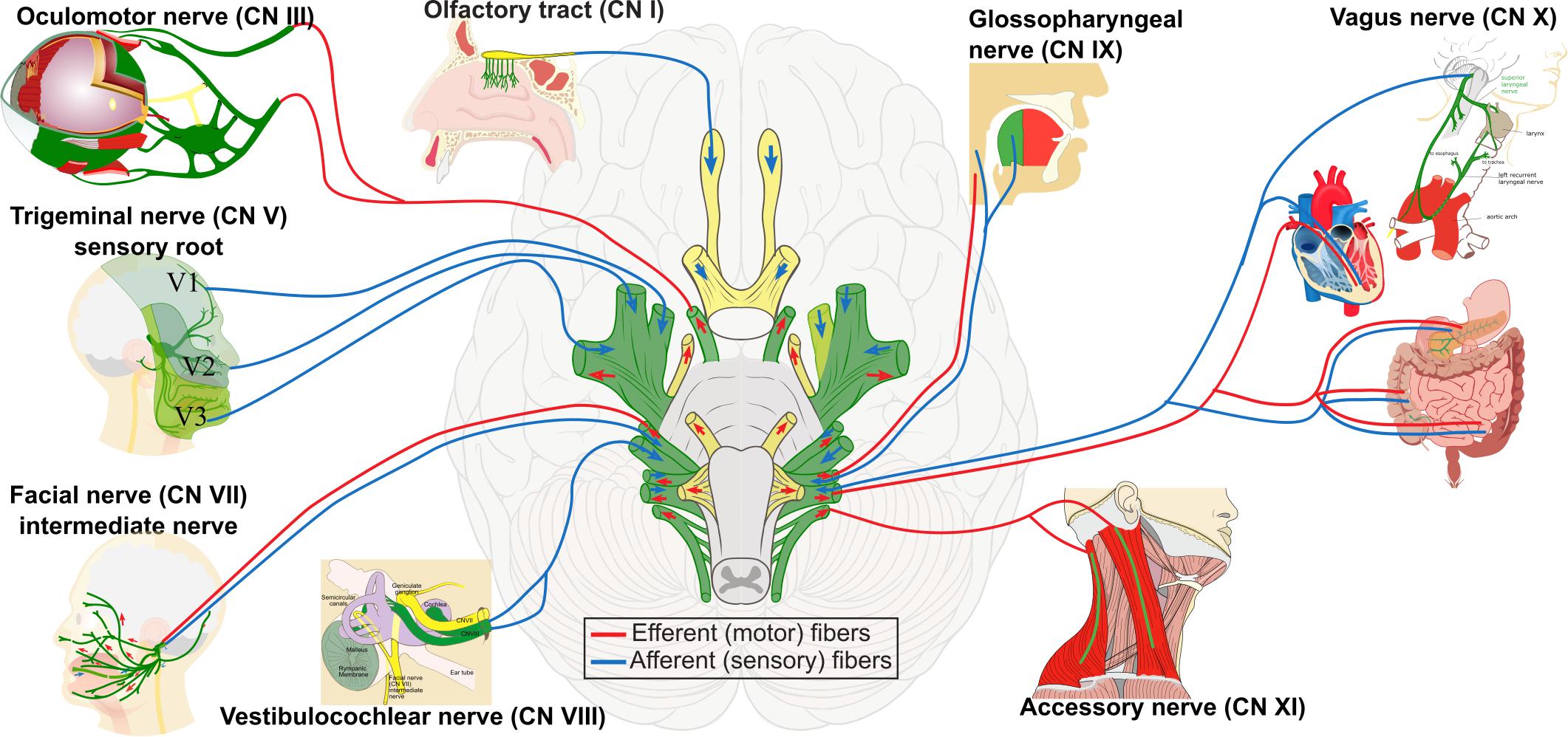
Figure 2. Contribution of neural crest cells to the formation of cranial nerves I, III, V, VII, VIII, IX, X, and XI. These selected cranial nerves are formed by the contribution of cranial placodes and neural crest cells, indicated in green. Neural crest-derived Schwann cells produce peripheral myelination of cranial nerves III–XII. The sensory nerves are the olfactory (I), the optic (II), and the vestibulocochlear (VIII). The motor nerves are the oculomotor (III), the trochlear (IV), the abducens (VI), and the accessory (XI). The remaining are mixed nerves.
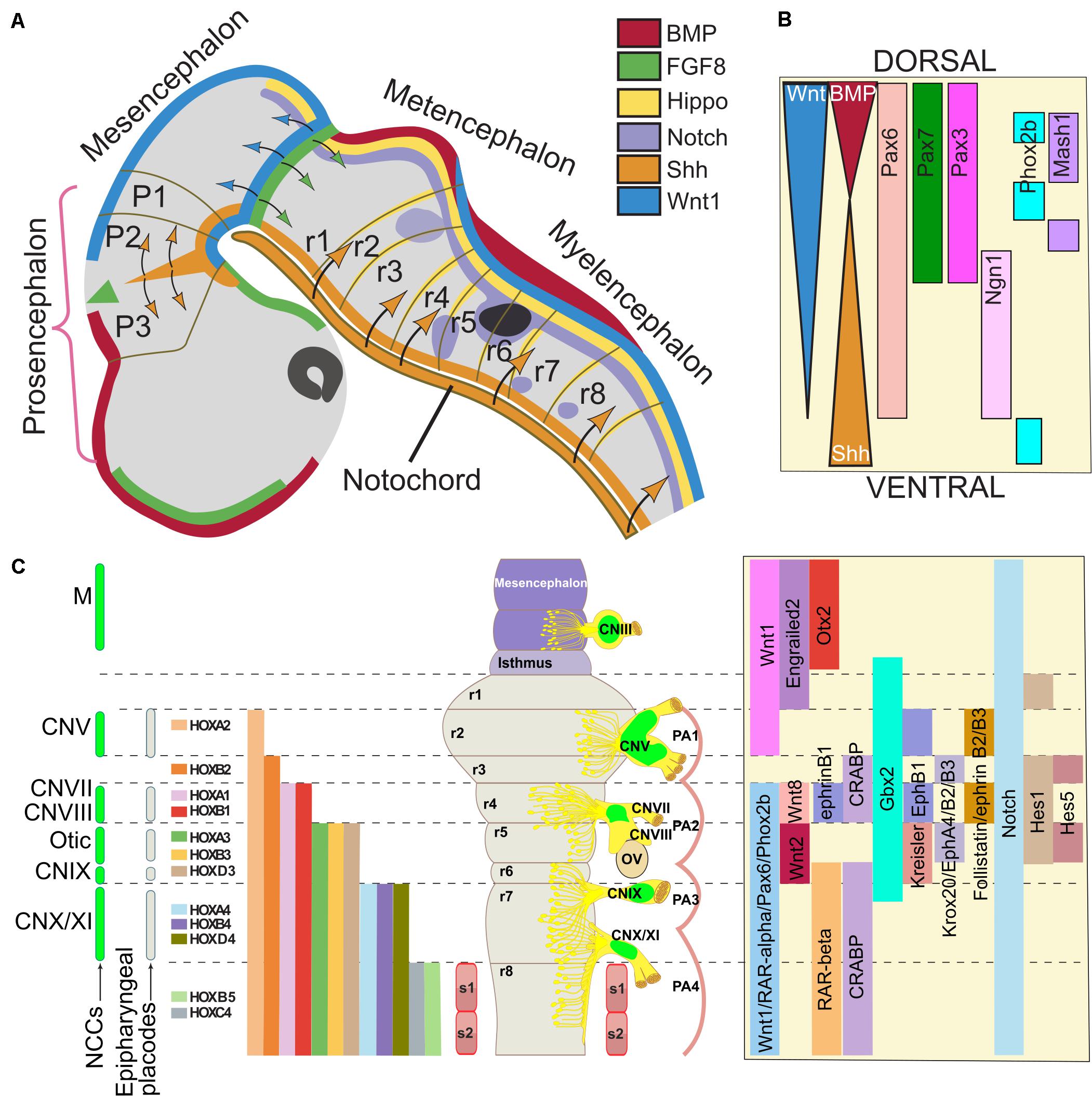
Figure 3. Gene regulatory network involved in neural crest contribution to the formation of cranial nerves. The cranial ganglia and cranial nerves are formed in precise positions along the dorso-ventral and antero-posterior axes of the midbrain/hindbrain region. (A) The drawing represents a human embryo at stage 13 (30 days, 32 somites), equivalent to mouse day 9.5-10 (E9.5-10, 20 somites) and chick stage 14 (50 h, 22 somites). The cell signaling pathways that provide developmental cues to neural crest precursors are color-coded; when these factors diffuse, the target regions are indicated with arrows with the same color. In panel (B), an idealized scheme of the hindbrain shows the cell signaling gradients and the genes that establish the dorsoventral pattern. (C) The illustration of the human (33 days, stage 14) and chick (stage 21) hindbrain rendered flat to eliminate cerebral flexures. The levels of origin of the neural crest cells (NCCs) and placodes, which contribute to the formation on cranial nerves, are indicated on the left. NCCs from the corresponding rhombomeres also populate other embryo structures in a segmental fashion and generate different craniofacial derivatives. The positions of the cranial ganglia and the otic vesicles are indicated on the right side; the contribution of NCCs is indicated in green. The segmental nested expression of HOX genes is color-coded. On the right, signaling pathways and the expression of transcription factors involved in cranial nerve (CN) formation are indicated. Adapted from Lumsden and Keynes (1989), Noden (1991), Yamamoto and Schwarting (1991), Bally-Cuif and Wassef (1995), Takahashi and Osumi (2002), and Müller and O’Rahilly (2011). Abbreviations: CN, cranial nerve; FP, floor plate; M, mesencephalon; NCCs, neural crest cells; OV, otic vesicle; r, rhombomere; PA, pharyngeal arches.
NC formation is a complex and multistep process initially directed by cell signaling molecules including Bone Morphogenetic Proteins (BMPs), Wnts (Wingless and Int-1), Fibroblast Growth Factor (FGF), and retinoic acid (RA). These signals reveal the tissue interactions into the ectodermal cell populations, the neural plate, the non-neural ectoderm, and the underlying mesoderm in a highly coordinated manner (Vega-Lopez et al., 2017). It has been proposed that NC specification occurs during gastrulation as a consequence of the action of two successive gradients of secreted signals. A combination of intermediate levels of activity of BMP and Wnt signaling acting on the ectoderm to induce and specify NC precursors at the neural plate border, and a subsequent requirement of both signals is needed for maintenance of specification during neurulation (Aybar and Mayor, 2002; Steventon et al., 2009). In chick embryos, it was shown that NCCs are specified as early as the blastula stage (Prasad et al., 2020). It was demonstrated that, during gastrulation, Pax7 expression is restricted to cells located in a region in the medial epiblast, which are NC-fated and contribute to the neural folds and later to migrating NCCs (Basch et al., 2006). The inhibition of Pax7 function in chicks inhibited the expression of key NC markers such as Snai2 (OMIM 602150), Sox9 (Sry-box 9, OMIM 608160), Sox10 (OMIM 602229), and HNK1 (beta-1,3-glucuronyltransferase 1 like, OMIM 151290) (Basch and Bronner-Fraser, 2006). This evidence suggests that the neural plate-prospective ectoderm interaction at the neural plate border might not be a requisite for NC specification or induction, and that neural plate border formation and NC induction might be separable events.
The various research works carried out to study the origin of NCCs have identified genes organized into a gene regulatory network that participate in and control the induction, specification and differentiation of NC (Simões-Costa et al., 2015). An example of this are the transcription factors involved in induction such as FoxD3 (Forkhead Box D3, OMIM 611539), Snai2 and Sox9 (Sauka-Spengler and Bronner-Fraser, 2006). García-Castro and co-workers identified a novel pre-neural border state characterized by early Wnt/β-catenin signaling targets that displayed different responses to BMP and FGF signaling from the neural border genes in human cells (Leung et al., 2016). These pre-border genes Gbx2 (Gastrulation brain homeobox 2, OMIM 601135), SP5 (OMIM 609391), Zic3 (OMIM 300265) and Zeb2 (OMIM 605802) had their induction and peak of expression before the classical neural plate border specifier genes such Msx1/2 (Muscle segment-related homeobox 1/2, OMIM 142983/123101), Pax3/7 (OMIM 606597/167410) and Zic1 (OMIM 600470). Such specifier genes, together with signaling molecules, direct the expression of NC-specific genes like AP-2 (OMIM 107580), FoxD3, Snai2, Sox9, and Sox10. Specifiers regulate NC effector genes involved in migration (Sox9, Sox10, Cad7) and differentiation [Col1a, (Collagen, type I alpha, OMIM 120150); Ngn1 (Neurogenin 1, OMIM 601726); Mitf (Microphthalmia-associated transcription factor, OMIM 156845)] in human NC development (Betters et al., 2010). The NC population migrates to different regions of the mouse embryo from the NT after the epithelial-mesenchymal transition, maintaining its multipotential character until completing differentiation in its final destination (Baggiolini et al., 2015).
To study the ontogeny of the NC, different model organisms, both in vivo and in vitro, have been used. Several proteins including transcription factors as well as epigenetic modifiers that take part in the specification and differentiation of the NC have been described. The study of transcription factors and of the signaling pathways in which they participate is important to understand the differentiation programs and how these multipotent cells are committed to a specific destination. On the other hand, transcriptome analysis during the development of the NC from specification to migration (Meulemans and Bronner-Fraser, 2004), and a more recent study covering the migration to the differentiation of the NC, show the importance of the interaction between the different transcription factors and the signaling pathways at every stage of NC development (Simões-Costa et al., 2015). However, these authors acknowledge that it is difficult to have a complete global map since only a few transcriptional regulators have been characterized, and little is known about the function of the products of the effector genes acting on NC migration (Betancur et al., 2010; Simões-Costa and Bronner, 2013; Vega-Lopez et al., 2017).
NC and cranial placodes are thought to appear together during the evolution of vertebrates to give rise to specific sensory structures of the head (Northcutt and Gans, 1983; Northcutt, 2005). The components of the sensory nervous system of the head are derived from the NC and from an embryonic cell population developing in close proximity, the cranial sensory placodes (the olfactory, lens, otic, trigeminal, epibranchial and paratympanic placodes). A series of events induce, develop and organize these cell precursors which, through reciprocal interactions with NCCs, build the functional sensory system in vertebrates (Steventon et al., 2014; Singh and Groves, 2016). Migrating NCCs arrive first at the site of ganglia development (i.e., the trigeminal ganglion), but the differentiation of these cells is delayed until the migration and differentiation of the corresponding placodal cells in chicks (Covell and Noden, 1989). Placodal specification and development, as well as its contribution to the assembly of placodal derivatives, is a complex and wide-ranging topic that is beyond the scope of this review. We will focus on discussing the main signaling pathways and relevant transcription factors involved in the specification of cranial NCCs precursors, their differentiation to form CNs and ganglia that are exclusively NC-derived, and the alterations caused by the mutations of certain genes that are important for the neurogenesis of NC derivatives.
There are several signaling pathways and transcription factors that are known to regulate NC and CN formation during development. We discuss some important pathways involved in cranial NCCs induction and specification, in close relationship with the cranial ganglia and nerves derived from the NC (Figure 3).
Bone morphogenetic proteins are proteins that control several important steps in the formation and differentiation of the CNS of vertebrates. These proteins act in different regions of the CNS to regulate fate, proliferation and differentiation. After gastrulation, the presence of BMPs and the activation of this signaling pathway are essential for the differentiation of the non-neural ectoderm whereas the inhibition of this pathway is required for the proper formation of the neural plate. It has been proposed that the later activation of BMPs receptors participates in the induction of the NC through a very fine regulation where the presence of BMPs at a specific time will give rise to the NC in mouse and human Embryonic Stem Cells (ESCs) (Figure 3B) (Mizuseki et al., 2003; Leung et al., 2016).
Seminal studies in Xenopus have shown that there is an activity gradient of BMPs controlled by their antagonists and that an intermediate level is needed to induce the formation of the NC (LaBonne and Bronner-Fraser, 1998; Marchant et al., 1998; Barth et al., 1999; Tribulo et al., 2003). Thus, the BMP antagonists Chordin (OMIM 603475) and Noggin (OMIM 602991) are expressed in a spatio-temporal manner that influences the formation of the NC. In mouse, at embryonic day (E) 8.0, Noggin is expressed in the neural folds and in the dorsal region after the closure of the NT. The expression of Chordin is low at the level of the neural plate and in the paraxial mesoderm. These antagonists participate in the induction of NC as well as in delamination, but also protect from apoptosis induced by BMP during migration and differentiation of NCCs. Importantly, it was observed that the decrease in the expression of these BMP antagonists alters the PNS derived from the NC and craniofacial skeletal elements. Noggin knockout mice presented all cranial nerves, but the vagus (X) and glossopharyngeal (IX) are disorganized and fused. Double-knockout mice of Noggin and Chordin lack CN and only a structure similar to the trigeminal ganglion (V) is present (Anderson et al., 2006). In the chick embryo, the activity of BMP signaling during the formation of NC precursors is modulated by CKIP/Smurf factors through the regulation of Smad degradation, resulting in intermediate levels of BMP activity required for proper NC formation (Piacentino and Bronner, 2018). In contrast, placode progenitors have differential BMP signaling requirements as they can be specified under low or no BMP signaling (Thiery et al., 2020).
A study of human ESCs (hESC) showed that if BMPs are blocked with Noggin for 24 h on days 0, 1, or 2 of the differentiation protocol, there is a dramatic decrease in the induction of human NCCs. However, if the inhibition is made on day 3, the inhibition is partial, so the participation of BMPs at the beginning of the induction of the NC is very important, while the inhibition of this pathway promoted the expression of neural genes such as SOX1 (OMIM 602148), HES5 (OMIM 607348), and SOX2 (OMIM 184429) (Leung et al., 2016). This protocol produced sensory peripheral neurons, and it will be of interest to investigate if such neurons can contribute to the sensory CN after grafting them in experimental animals, as well as the effect of modulating BMPs on peripheral neuron differentiation. Interestingly, BMP antagonism upregulates these neural stem cell markers, but several reports indicated that Sox1, Hes5, and Sox2 are involved in the suppression of neuronal differentiation by maintaining neural stem and progenitor cells in an undifferentiated state in mammalian cells (Kan et al., 2004; Bani-Yaghoub et al., 2006). The generation of neurons from stem cells depends on the decrease of Sox1-3 expression caused by proneural proteins. However, if Sox1-3 target genes were repressed, independently of proneural activity, neural progenitor cells differentiated prematurely, and some neuronal features emerged. These results demonstrate a dual role of proneural proteins in the acquisition of a definitive neuronal fate and indicate that the proneural protein-directed repression of Sox1-3 expression is a required and irreversible step in the commitment to neuronal differentiation in several species, including mammals (Guillemot, 1999; Farah et al., 2000; Bylund et al., 2003).
BMP4 (OMIM 112262) and Smad proteins have been involved in an interesting mechanism called retrograde signaling in trigeminal ganglia from rats (Ji and Jaffrey, 2012). This mechanism elicits a specific transcriptional response that contributes to the specification of different subpopulations of sensory neurons in the trigeminal ganglia (CN V). As axons from the neurons of trigeminal ganglia grow and extend into their three main peripheral axonal branches (ophthalmic, maxillary and mandibular) that innervate the corresponding regions of the face, they encounter BMP4, which results in a retrograde signal that leads to transport back transcription factors SMAD-1, -5, and -8 from axons to the somata, where nuclear accumulation of the phosphorylated and transcriptionally active Smad forms contributes to neuronal specification and ganglia patterning (Nohe et al., 2004; Ji and Jaffrey, 2012). BDNF (Brain-Derived Neurotrophic Factor, OMIM 113505) signaling was also found to regulate axonal levels of SMAD-1, -5, and -8 in concert with BMP4, for patterning of the trigeminal ganglia (Ji and Jaffrey, 2012).
Genetic studies have demonstrated that Hippo signaling is crucial in organ size regulation, controlling cell number by modulating cell proliferation and apoptosis processes (Huang et al., 2005). Hippo is a critical factor for proliferation and epithelial-mesenchymal transition during embryonic development and cancer. In the neural tube of the mouse, chicken, and frog, YAP (Yes-Associated Protein, OMIM 606608) is expressed in the ventricular zone progenitor cells and co-localizes with the neural progenitor cell marker Sox2 (Milewski et al., 2004; Cao et al., 2008). It has been observed that the ectopic expression of one of the transcriptional regulators of this pathway, TAZ (Transcriptional Coactivator With PDZ-Binding Motif, OMIM 607392) in mammalian cells, stimulates cell proliferation, reduces the inhibition by contact and promotes the epithelial-mesenchymal transition (Lei et al., 2008).
A relationship between this signaling pathway and the classical NC genes, such as interaction with Pax3 is through TAZ and the phosphoprotein YAP65. These proteins participate as co-activators of Pax3. It has been suggested, using transgenic mice, that Tead2 (TEA Domain Family Member 2, OMIM 601729) is an endogenous activator of Pax3 in NCCs (Milewski et al., 2004). Through expression assays, Pax3 and Yap65 were co-localized in the nucleus of NC progenitors in the dorsal region of the NT. Hippo/TAZ/YAP are critical for Schwann cell proliferation and differentiation in a stage-dependent manner. Nuclear TAZ/YAP complexes activate cell cycle regulators to promote Schwann cell proliferation while directing differentiation regulators in cooperation with Sox10 for myelination in rodents (Deng et al., 2017).
Neurofibromatosis 2 (Nf2, OMIM 101000) is a tumor suppressor that inhibits YAP during dorsal root ganglia (DRG) development. Merlin, encoded by the NF2 tumor-suppressive gene, was identified through genetic studies in mouse embryos and proved to be an important upstream regulator of the Hippo-Yap pathway. Neurofibromatosis is an inherited disease characterized by the development of bilateral Schwann cell tumors originated from CN VIII. Mouse with specific Schwann cell-inactivated Nf2 alleles developed schwannomas and SC hyperplasia (McClatchey et al., 1998; Giovannini et al., 1999, 2000). Merlin has also been shown to act as a suppressor of mouse neural progenitor proliferation, by inhibiting TAZ/YAP pathway activity (Lavado et al., 2013). The mechanism by which Merlin regulates YAP activity might involve p21 Protein-activated kinase 1 (PAK1, OMIM 602590) activation, which induces phosphorylation of Merlin, thus abrogating its scaffold function for YAP and LATS1/2 (OMIM 603473/604861), and thereby attenuates YAP phosphorylation by LATS1/2 in mouse cells (Sabra et al., 2017); it has been suggested that nuclear export signals of Merlin mediate YAP nuclear export in epithelial mammalian cells (Furukawa et al., 2017).
Hindley and co-workers investigated the role of Hippo/YAP signaling in NC development and neural differentiation. They showed that the activity of YAP promotes an early NC phenotype accompanied by premature migratory behavior, and that Hippo/YAP interacts with RA signaling in hESCs (Hindley et al., 2016). A recent study demonstrates that YAP is necessary for the migration of a premigratory pool of NCCs, since they incorporated YAP signaling into a BMP/Wnt-dependent molecular network responsible for the migration of trunk-level NC in avians (Kumar et al., 2019).
Notch is a family of conserved receptors whose activation is induced by specific ligands, Delta-1 (OMIM 606582), Delta-3 (OMIM 602768), Delta-4 (OMIM 605185), Jagged-1 (OMIM 601920), and Jagged-2 (OMIM 602570), through interaction with four possible receptors (Notch1-4) (Perdigoto and Bardin, 2013). Once the Notch receptors are activated through the cell-cell interaction, proteolytic cuts are carried out resulting in the release of the Notch Intracellular Domain (NICD) (Mumm et al., 2000). NICD translocate to the nucleus and forms a transcriptional complex together with the DNA binding protein CBF1 (C promoter binding factor 1, OMIM 147183). This complex recognizes the specific sequence (C/T)GTGGGAA in its target genes, for example Hes1 (OMIM 139605) (Kageyama et al., 2000).
Notch1 receptor is present during development of the rhomboencephalon at E9.5 in mice, showing strong expression within the hindbrain, including the trigeminal, geniculate, petrosum and nodose placodes, which give rise to CN V, VII, IX and X, respectively, and is also expressed in the otic and olfactory vesicle (Reaume et al., 1992). A study where human induced pluripotent stem cells were induced toward NC differentiation showed that when Notch signaling is blocked using a γ-secretase inhibitor (DAPT) or shRNA for JAGGED-1, the genes specifying NC [DLX5 (Distal-less homeobox 5, OMIM 600028), PAX3, SNAI2, SOX10, and TWIST1 (OMIM 601622)] are down-regulated. However, the ectopic expression of NICD1 increased its expression, demonstrating that Notch also participates significantly in NC induction (Noisa et al., 2014). Mead and Yutzey evaluated the function of Notch signaling in murine NC-derived cell lineages in vivo. They demonstrated that cell-autonomous Notch has an essential role in proper NCCs migration, proliferation and differentiation, with critical implications in craniofacial, cardiac and neurogenic development (Mead and Yutzey, 2012).
Sonic Hedgehog (Shh) signaling is involved in the correct development of NC and therefore in the generation of its cellular derivatives (Figure 3B). Shh is a member of the family of the secreted Hedgehog proteins: Sonic (Shh, OMIM 600725), Indian (Ihh, OMIM 600726), and Desert Hedgehog (Dhh, OMIM 605423). Shh regulation during NC differentiation is crucial during head and face morphogenesis. Mutant mice and humans lacking Shh present holoprosencephaly and cyclopia due to the lack of separation of the forebrain lobes (Chiang et al., 1996). It is suggested that Shh inhibition maintains Pax3 expression, so the lack of Shh-mediated regulation for Pax3 induction promotes the constitutive induction of NC, generating the aforementioned phenotypes. A subset of Fox genes regulated by Shh signaling is important during lip morphogenesis in mice. Either Shh addition or Foxf2 (OMIM 603250) overexpression was shown to be sufficient to induce cranial NCCs proliferation (Everson et al., 2017).
On the other hand, enhanced Shh signaling in mouse, mediated by loss-of-function (Ptch1Wig/Wig) of the Shh receptor Patched1 (Ptch1, OMIM 601309), suppressed canonical Wnt signaling in the CN region. This critically affected the survival and migration of cranial NCCs and the development of placodes, as well as the integration between NC and placodes (Kurosaka et al., 2015). Ptch1Wig/Wig mutants exhibited severely disorganized trigeminal (CNV) and facial nerves (CNVII) that did not develop properly and failed to project to their appropriate target tissues (Kurosaka et al., 2015). High levels of Shh signaling have been correlated with Moebius Syndrome, which is characterized by cranial nerve defects including trigeminal, abducens (CNVI) and facial alterations concurrent with other craniofacial defects (Verzijl et al., 2003; Vega-Lopez et al., 2018). NCCs migration is particularly sensitive to Shh levels since in mice lacking Shh, these cells continue their migration beyond the normal position and fuse medially, condensing into a single midline ganglion (Fedtsova et al., 2003). Mutation in the mouse Hedgehog acyltransferase (Hhat, OMIM 605743) gene produced hypoplasia and aberrant fusion of cranial ganglia (CN V, VII, IX, and X) and affected NC and placode gene markers expression, suggesting that a regionalized action of the Hedgehog signaling is required for proper cranial ganglia and nerve development and patterning (Dennis et al., 2012). In vitro analyses showed that Shh increased the number of cranial NC progenitors, from quail embryos, yielding neural and mesenchymal lineages. Shh can decrease the neural-restricted precursors without affecting survival or proliferation. These data also suggest that the mesenchymal-neural precursor was able to yield both the PNS and superficial skeleton (Calloni et al., 2007).
Humans have 58 known RTKs, which fall into 20 subfamilies. A few years ago, a systematic work summarized the contribution of the mouse model to the understanding of the role of a subset of RTKs in regulating the activity of NCCs in development (Fantauzzo and Soriano, 2015). With respect to its downstream signaling, RTKs induce the activation of various pathways, including PLC-γ, PI3K, MAPK, JNK, Shc, Erk, and the JAK/STAT pathways. In this section, we discuss insights pointing to mechanisms of action of some RTK families in relation to the development of the cranial NC that have emerged from recent evidence.
Ephrin ligands and Eph (erythropoietin-producing human hepatocellular carcinoma) receptors comprise an increasingly well studied family of signaling molecules. Ephrins bind to two families of transmembrane tyrosine kinase receptors, EphA and EphB. While A-type Ephrins preferentially bind to EphA receptors, B-type Ephrins do so to EphB receptors. In Xenopus, the streams of NCCs going to the second branchial arch express Ephrin-B2, whereas cells reaching the third arch express EphB1; disruption of Eph-Ephrin signaling results in aberrant migration of NCCs, causing mixing of the streams in the branchial pouches (Smith et al., 1997). Eph receptor functions are best characterized in the mouse nervous system, where they are involved in neuronal development and axon guidance (Wilkinson, 2001; Xu and Henkemeyer, 2012), migration and proliferation (Conover et al., 2000; Holmberg et al., 2006; Jurek et al., 2016) as well as inflammation (Coulthard et al., 2012).
The Ephrin ligand/Eph receptor proteins are widely expressed in embryonic tissues. Eph receptors participate in the development of several NC-derivatives in mouse: teeth and the establishment of tooth nerves (CN V) (Luukko et al., 2005; Stokowski et al., 2007; Arthur et al., 2009; Diercke et al., 2011; Matsumura et al., 2017) and participate in cochlear innervation patterns (Zhou et al., 2011). Eph receptors play a role in mouse segmentation and boundary formation of the developing hindbrain, which results in the formation of rhombomeres (r), which are crucial for the orderly formation of CN and specification of NCCs (Flenniken et al., 1996; Flanagan and Vanderhaeghen, 1998; Merrill et al., 2006; Mellott and Burke, 2008; Klein, 2012). Mouse EphA5–/– (OMIM 600004) had only <15% of the normal complement of Gonadotropin-releasing hormone neurons in the brain (Gamble, 2005). This also produced infertility in adult female homozygous GNR23 mice, providing a causal link between Ephrin-related mutations and human hypogonadotropic hypogonadism such as Kallman syndrome. It has been shown through genetic labeling that a fraction of GnRH neurons are derived from NCCs (Forni et al., 2011).
A key step in epigenetic control of expression is gene silencing by hypermethylation of CpG islands present at promoter regions (Nakao, 2001). Both specific enzymes and methyl-CpG-binding proteins (MBPs) play a major role in the epigenetic control of gene expression through the recognition and binding to methylated DNA, as well as by the recruitment of remodeling complexes (Defossez and Stancheva, 2011). During development, EphA5 receptor controls the axonal mapping of retinal ganglion cells in the visual system (Zhou, 1997). Recent findings showed site-specific differences in methylation of CpG islands in the EphA5 promoter, which could account for the activation or repression of this promoter and might influence the graded EphA5 expression in the mesencephalic tectum (Petkova et al., 2011). During mouse embryonic development, high levels of EphA5 protein were also found in cranial nerve ganglia V, VIII, X, and XII (Cooper et al., 2009). Therefore, it seems reasonable to speculate that this epigenetic methylation may regulate the neurogenesis of these cranial nerves as it does in the myencephalic region.
The Epidermal Growth Factor Receptor (EGFR, OMIM 131550) and the related ErbB (v-erb-b2 avian erythroblastic leukemia viral oncogene B) proteins transduce after EGF (OMIM 131530) binding. ErbB2–/– (OMIM 164870) mice die around midgestation due to cardiac defects. Cranial ganglia are also morphologically aberrant and these embryos show an altered pattern of ErbB3 (OMIM 190151) staining (Meyer and Birchmeier, 1995; Erickson et al., 1997; Britsch et al., 1998; Garratt et al., 2000). ErbB3 mutant mice embryos die at a later stage as they have reduced numbers of Schwann cell precursors derived from NCCs and therefore lack cranial ganglia nerves, caused by the death of around 80% of both motor and sensory neurons (Riethmacher et al., 1997). Chick NCCs from the hindbrain and ectodermal cells from placodes, participate in the development of cranial ganglia (D’amico-Martel and Noden, 1983; Le Douarin et al., 1986). A chemical mutagenesis screen in Sox10-reporter mice identified an amino acid substitution in the extracellular portion of ErbB3 that resulted in alterations in homozygotic mutants similar to those reported in ErbB3 knock-outs (Buac et al., 2008).
ErbB4 (OMIM 190151) null mouse die at mid-gestation, at E11, due to cardiac defects (Gassmann et al., 1995). In order to overcome this lethality, ErbB4 mutant mice were engineered to express ErbB4 only in the heart. The embryos survived, but presented aberrant cranial nerve architecture, such as ectopic nerve projections of trigeminal (V) and facial (VII) ganglia (Tidcombe et al., 2003). These results suggested the participation of ErbB4 in the control of NCCs migration and axon extension. ErbB4 (alongside Ephrin) is expressed in r3 while one of its ligands, Neuregulin 1 (OMIM 142445), is expressed in r2 and r4 (Golding et al., 2000, 2004).
Fibroblast growth factor signaling is composed of 22 members, although only eighteen FGFs signal via FGF Receptor (FGFR) interactions (FGF1–10 and 16–23). There are seven signaling receptors, encoded by four FGFR genes, FGFR1–4 (Zhang et al., 2006). FGFs exert their cellular effects by interacting with FGFRs, but FGF-FGFR complexes can only be formed in the presence of heparan sulfate (Pellegrini et al., 2000; Schlessinger et al., 2000). FGFRs, a class of RTK, dimerize and undergo transphosphorylation of the kinase domain upon ligand binding. Four signaling pathways can be activated to transduce intracellularly: MAP Kinase (MAPK), PI3K/AKT, PLC-γ, and STAT (Ornitz and Itoh, 2015).
In mice, FGF signaling is necessary for cell survival during the development of tissues, including the embryonic telencephalon and the mid-hindbrain junction (Sato et al., 2004; Zervas et al., 2005; Paek et al., 2009). In zebrafish and chick, FGF3 (OMIM 164950) and FGF8 (OMIM 600483) emanating from r4 are both necessary and sufficient to promote the development of the adjacent r5 and r6 by regulating the expression of transcription factors including Krox20 (Maves et al., 2002; Walshe et al., 2002; Waskiewicz et al., 2002; Wiellette and Sive, 2003; Hernandez et al., 2004; Aragón et al., 2005; Labalette et al., 2011).
Fibroblast growth factor activating FGFR-2(IIIb) (OMIM 176943) at placodal sites (Pirvola et al., 2000), and RA, primarily associated with NC-derived mesenchyme (LaMantia et al., 2000), modulate multiple aspects of sensory neuronal differentiation, including cranial sensory neuron survival, neurogenesis and cranial nerve differentiation. FGFR-2(IIIb) knock-in mouse shows severe dysgenesis of the cochleovestibular membranous labyrinth and sensory patches of the vestibulocochlear ganglion (CN VIII) remain small and poorly developed (Pirvola et al., 2000).
MBD1 (Methyl-CpG-binding domain protein 1, OMIM 156535)-null neural stem cells display impaired neurogenesis and increased genomic stability. A possible mechanism is the direct binding of MBD1 to the hypermethylated promoter region of the important neural growth factor FGF2. In agreement, MBD1 loss-of-function induces the FGF2 promoter hypomethylation, thus increasing its expression in mouse adult neural stem cells, which prevents differentiation (Li et al., 2008). Ma et al. showed that Gadd45b (Growth arrest and DNA damage-inducible gene 45 beta, OMIM 604948) could induce demethylation in promoters of several genes that participate in mouse neurogenesis, including Bdnf (region IX) and FGF1 (promoter B, OMIM 131220) (Alam et al., 1996). Interestingly, attenuated dendritic growth was found in Gadd45b knock-out mice after electro-convulsive treatment, compared to wild-type animals, indicating that Gadd45b is required for DNA demethylation in adult neurogenesis (Ma et al., 2009). Whether or not these mechanisms are shared in NC differentiation to CN is an interesting research topic.
Protein tyrosine kinase 7 (PTK7, OMIM 601890), also named Colon Carcinoma Kinase 4 (CCK4) and Kinase-Like Gene (KLG) in chicken, is the only member of this RTK family (Jung et al., 2002). PTK7 null mice die perinatally (Lu et al., 2004). Interestingly, Chuzhoi mice, which are homozygous for an ENU-induced splice site mutation in the PTK7 gene, also die perinatally and similarly to null individuals, exhibit severe neural tube closure defects, have abnormal NCCs distribution and display altered morphology of cranial ganglia and DRG, cardiac outflow tract and ventricular septal defects (Paudyal et al., 2010). PTK7 regulates NC migration via β-Catenin-independent Wnt signaling, and it has been shown that ROR2 (RTK-like orphan, OMIM 602337) is capable of replacing PTK7 function in this process (Podleschny et al., 2015). The human PTK7 gene has a promoter with 420-bp-long CpG islands (Jung et al., 2002), but epigenetic regulation is unclear at this point.
Trks (tropomyosin-related kinases) receptors are a subfamily of TRKs activated by neurotrophins (McDonald and Hendrickson, 1993; Murray-Rust et al., 1993). Three types of Trks receptors have been identified during vertebrate development: TrkA (OMIM 191315), TrkB (OMIM 600456) and TrkC (OMIM 191316), activated by NGF (OMIM 162030), BDNF/NT-4 (OMIM 162662) and NT-3 (OMIM 162660), respectively (Hempstead et al., 1991).
The mouse deficiency of NT-3 (Huang et al., 1999), TrkA, TrkB or TrkC (Lewin and Barde, 1996) causes variable loss (39–82%) or decrease of nociceptors and low-threshold mechanoreceptors in the trigeminal ganglion (CN V). TrkB has been found to directly interact with ErbB2 (also known as Her2) for signal transduction in human cells (Choy et al., 2017). Mouse TrkB and p75NTR (OMIM 162010) serve as co-receptors of Ephrin-A (Lim et al., 2008; Marler et al., 2008; Barton et al., 2014). Trks have been detected in all classes of PNS neurons with the notable exception of parasympathetic neurons of the ciliary ganglion. With regards to sensory neurons, TrkA is expressed only in DRG and other neural crest-derived ganglia, whereas TrkB and TrkC are expressed to some extent in all sensory ganglia. During embryogenesis, up to 70% of DRG neurons express TrkA but this number declines to around 40% in the adult rat. Co-expression in a single neuron of two members of the Trk family is common, e.g., in adult rat DRG few cells express TrkB alone, while the combinations TrkA + TrkB or TrkB + TrkC are more common (McMahon et al., 1994; Lindsay, 1996). TrkB expression is Ca2+ dependent in mouse cortical neurons (Kingsbury et al., 2003), but thyroid hormone T3 down-regulates the expression of TrkB through a negative response element located downstream of its transcription initiation site, during the development of rat brain (Pombo et al., 2000). TrkB was shown to be transcriptionally repressed by Runx3, a Runt domain transcription factor, in mouse and human cells (Inoue et al., 2007).
BDNF and NGF signals emanating from chicken sensory ganglia stimulate cranial motor axon growth (Li et al., 2020). MeCP2 (Methyl-CpG-Binding Protein 2, OMIM 300005) acts with REST/NRSF (Re1-Silencing Transcription factor/Neuron-Restrictive Silencer Factor, OMIM 600571) to recruit Histone Deacetylases, causing a decrease in the expression of BDNF. On the other hand, MeCP2 is released from the BDNF promoter in mouse neurons as a consequence of membrane depolarization, thereby allowing its transcription (Ballas et al., 2005; Zhou et al., 2006). Neuronal activity promotes MeCP2 phosphorylation at specific sites, which differentially changes its binding to gene promoters such as BDNF, a step that is decisive for proper neuronal development and synaptic plasticity in mice (Na and Monteggia, 2011).
Vascular endothelial growth factor receptors (VEGFRs) are important in the formation of the vascular system during embryonic development. The mammalian VEGFR are three related type III RTKs known as VEGFR1, VEGFR2, and VEGFR3. These receptors, which bind to VEGF ligands, consist of five glycoproteins referred to as VEGFA, VEGFB, VEGFC, VEGFD and placenta growth factor (PlGF) (Ferrara et al., 2003). The transmembrane protein neuropilin 1 (NRP1, OMIM 602069) is essential for the patterning of the facial nerve (VII) in mouse, as it binds the secreted Semaphorin SEMA3A (OMIM 603961) to guide facial branchiomotor axons in invading the second branchial arch. However, NRP1 can also be activated by the VEGF isoform VEGF164 to control the position of facial branchiomotor neuron cell bodies within the chick hindbrain (Anderson et al., 2003; Schwarz et al., 2004). Cranial NCCs express VEGFR2 and its co-receptor NRP1 as they migrate from the hindbrain at the level of r4 to invade pharyngeal arch 2 in response to chemoattraction by VEGF also in chicken (McLennan et al., 2010).
Wnts proteins are secreted glycoproteins that participate in a wide variety of cellular processes in development and disease. Binding of Wnts to receptors composed of Frizzled and Lrp5/6 triggers a canonical pathway that results in the stabilization of β-catenin (OMIM 116806), which otherwise is phosphorylated by GSK3β (OMIM 605004) and undergoes constant degradation by the proteasome. Stabilized β-catenin interacts with TCF to activate the expression of target genes (Nusse and Clevers, 2017). Non-canonical signaling pathways are associated with Wnts, namely the Planar cell polarity in Drosophila and the Wnt/Ca2+ pathway in vertebrates. The latter involves at least two branches: Ror1/2 activation of Phospholipase C, associated with Wnt binding to Frizzled receptors, produces IP3 and DAG, which increases cytoplasmic Ca2+ concentrations; the second mechanism is the direct activation of Ror1/2 by Wnts, resulting in increases in cytoplasmic Ca2+, which activates Calpain (De, 2011). The interaction of Wnt with other signaling pathways, e.g., with the Smad pathway, has been demonstrated in hESCs (Menendez et al., 2011). An efficient method was described for the generation of NCCs from human pluripotent stem cells through the sustained activation of Wnt signaling combined with low Smad signaling, accomplished by the inhibition of the Activin/Nodal pathway. After 12 days, this constant inhibition of Smad considerably inhibited the formation of CNS Pax6 (OMIM 607108)-positive cells and increased the percentage of cells positive for the low affinity neurotrophin receptor, p75NTR, which is expressed in the migratory NC (Heuer et al., 1990; Wislet et al., 2018). Within the population of p75 positive cells, authors found cells with intermediate levels of p75, but positive for Pax6; in contrast, the cell population that expresses high levels of p75 was positive for Ap-2α (OMIM 107580), characteristic of NCCs (Menendez et al., 2011). Whether or not a chronic inhibition of Smads has a similar effect in vivo remains to be tested.
The activities of genes that influence the morphogenesis of the head are related to Wnt signaling through the expression of Wnt antagonist proteins, the main one being Dkk1 (OMIM 605189). Loss of expression of Dkk1 promotes an ectopic activation of Wnt/β-catenin signaling during gastrulation. Using in vivo assays, it was demonstrated that Dkk1 and Wnt3a (OMIM 606359) are regulated in a negative feedback loop. In agreement with this, 51% of double heterozygous mice for Dkk1 and Wnt3a showed reduced forebrain while 30% were normal. A small percentage of mice had malformations of eyes and pharyngeal arches as well as defects in the trunk. Therefore, regulation of Wnt signaling participates in the formation of the head but also in several mouse NC derivatives, although there are other pathways and transcription factors involved in the morphogenesis of the head (Lewis et al., 2008).
The canonical Wnt pathway prominently participates in the induction, lineage specification, delamination and differentiation of NC derivatives (Figure 3B). Differentiation into several cell types of the mouse NC is dependent on the sequential activation of Wnt signaling, which indicates that the decision of the cellular differentiation is regulated by the activation state of Wnt/β-Catenin (Hari et al., 2012). In vitro, Wnt/β-Catenin signaling centrally participates during differentiation to NC, inducing transcriptional factors that are expressed before factors expressed in neural borders, such PAX3, PAX7, MSX1, and TFAP2A. These pre-border transcriptional regulators are GBX2, SP5, ZIC3 and ZEB2 (Leung et al., 2016). In the case of Gbx2 and its role in CN formation, the initial characterization of Gbx2 mutants in mice demonstrated defects, specifically the absence of the trigeminal nerve (CN V) (Byrd and Meyers, 2005). In addition to the several transcription factors that are important in the induction and specification of NC, there are some proteins, such as Heat Shock Proteins, that participate in these processes. An example of this is the heat shock binding protein 1 (HSBP1). A study in mouse and zebrafish showed that HSBP1 participates in both the pre-implantation status of the blastocyst and the development of the NC. This was demonstrated by the deletion of HSBP1, where its absence promoted a cell arrest or degeneration before reaching the blastocyst stage. With respect to NC, mice deficient in Hsbp1 showed an increase in the expression of inducers of NC, Snai2, Tfap2α and FoxD3, suggesting that HSBP1 has a potential role in the Wnt pathway (Eroglu et al., 2014). The participation of Heat Shock Proteins in neuronal differentiation to form CN has not been explored yet and, given the importance of Wnt signaling for NC, represents an area of opportunity. Some of the functions associated to molecules in NC development are summarized in Table 2.
The basic helix-loop-helix (bHLH) DNA binding protein Hand2 (dHand, Thing-2, Hed, OMIM 602407) is expressed in a subset of NC-derived cells where it participates in various aspects of cell specification, lineage segregation, and cell type-specific gene expression (Hendershot et al., 2007, 2008). Loss of Hand2 results in embryonic lethality by E9.5. In order to study the role of Hand2 in NC, a specific deletion of Hand2 was engineered by crossing floxed Hand2 mice with Wnt1-Cre transgenic mice. Hand2 knock-out in NC-derived cells caused severe effects on development in all NC-derived structures and tissues where Hand2 is expressed. In the autonomic nervous system, conditional interruption of Hand2 function results in a marked and progressive loss of neurons concomitant with a loss of Tyrosine Hydroxylase (TH) expression in mice (Hendershot et al., 2008). There are few studies tackling the importance of Hand2 in NC development and differentiation and none about its importance in CN formation.
The Hes genes are homologs of the Drosophila hairy and Enhancer of Split gene. The Hes family is composed of seven members, Hes4 being absent in the mouse genome. Hes genes encode nuclear proteins that repress transcription, either actively or passively (Kageyama and Ohtsuka, 1999). These genes have conserved domains that confer the transcriptional function to all Hes factors. The bHLH domain contains the DNA binding site and the dimerization region. Hes factors can form homo- and heterodimers with Hes-related bHLH repressors, such as Hey factors, Mash1 (OMIM 100790), E47 and Ids. The Orange domain regulates the selection of the bHLH heterodimer, and the WRPW Groucho-binding domain at the C-terminus consists of a tetrapeptide Trp-ArgPro-Trp that represses transcription. This sequence also acts as a polyubiquitination signal for the degradation of Hes by the proteasome (Akazawa et al., 1992; Sasai et al., 1992; Ohsako et al., 1994; Kobayashi and Kageyama, 2014). The Hes transcription factors are essential effectors of Notch signaling that regulate the maintenance of progenitor cells and the time of their differentiation into various tissues and organs (Kageyama and Ohtsuka, 1999). Hes1 (OMIM 139505) is a negative regulator of neural differentiation, since it represses the expression of pro-neural genes such as Mash1, Neurogenin-2 (OMIM 606624) and Math. Mice deficient for Hes1 show a severe neural hypoplasia due to accelerated neural differentiation and the consequent depletion of neural precursor cells (Ishibashi et al., 1995). In agreement with the above, Hatakeyama and co-workers demonstrated that the absence of Hes1 and Hes5 caused severe alterations in the size, shape and cytoarchitecture of the mouse CNS. They also found that in Hes1;Hes5 double-mutant mice, the cranial and spinal nerve systems were also severely disorganized, pointing to dysregulation of these NC derivatives (Hatakeyama et al., 2006). These results indicate that Hes1 and Hes5 play an important role in the formation of both CN and spinal nerves.
Id proteins are inhibitors of DNA binding and cell differentiation; four members of this family have been described, Id1-Id4. They are negative regulators of bHLH transcriptional factors that are involved in various processes such as neurogenesis, hematopoiesis, myeloid differentiation, and bone morphogenesis, among others. It has been reported that gene expression of Id is present in undifferentiated cells, highly proliferating cells, embryonic cells and cancer cells (Roschger and Cabrele, 2017). One of the Id proteins, Id2 (OMIM 600386), directs the ectodermal precursors to NC commitment and neuronal differentiation. It is expressed in the trunk and in cranial folds, and therefore also in cranial NCCs. The ectopic expression of Id2 in chick promoted a switch of ectodermal cells to NC fate. Overexpression of Id2 increases growth and causes premature neurogenesis in the dorsal region of the NT (Martinsen and Bronner-Fraser, 1998). Conversely, loss of Id2 in mice caused a decrease in newborn neurons while increasing the number of astrocytes (Havrda et al., 2008). It was recently shown that Id2a expression decreased in the forebrain, midbrain and hindbrain as a consequence of blocking Mecp2 expression with a morpholino oligonucleotide. This was consistent with the activation of Notch signaling in such morphants. Mechanistically, in Mecp2 morphants, her2 (the zebrafish ortholog of mammalian Hes5), was upregulated in an Id1-dependent manner (Gao et al., 2015).
In avian and mammalian embryos, the proneural transcription factors Ngn1 and Ngn2 are expressed in NCCs during migration previous to their neuronal differentiation into sensory neurons. In mouse embryos, the functional inactivation of both Ngn genes led to a total absence of neurons of the DRG (Ma et al., 1999; Perez et al., 1999). In zebrafish, blocking with a morpholino for Ngn1 leads to a complete loss of neurons in the cranial ganglia and DRG neurons (Andermann et al., 2002; Cornell and Eisen, 2002). Recently, McGraw et al. (2008) demonstrated in zebrafish that, in the absence of Ngn1, the sensory neuron-restricted lineage of NC gives rise only to glial cells.
Hox genes play a central role in NC patterning, particularly in the cranial region (Figure 3C). These genes are essential for specifying segmental identity in the developing brain in several vertebrate species. The mechanism responsible for Hox genes expression at higher relative levels in specific rhombomeres is independent of the process that establishes the axial expression patterns found in the neural tube. Hox genes are organized into four distinct clusters (Hoxa-Hoxd) located on different chromosomes in higher vertebrates (McGinnis and Krumlauf, 1992).
It has been long proposed that the Hox “collinear expression” is the result of a unidirectional chromatin opening from 3′ to 5′ during development (Lewis, 1978; Duboule and Dollé, 1989; Graham et al., 1989). As a result of collinearity, Hox genes expressed in the hindbrain are from paralog groups 1–4. Members from groups 5 to 13 have anterior boundaries of expression which map to the spinal cord (Nolte and Krumlauf, 2007). Hox paralog group 1 genes have been suggested to influence early cranial NC development through NCC precursors by interacting with factors in the neural plate border or NC specification modules, although direct gene interactions remain to be determined. The expression of Hox paralog groups 2–4 genes in mouse cranial NCCs is modified by Hox auto- or cross-regulation in addition to other inputs from NC transcription factors such as AP-2 in the case of Hoxa2 (Parker et al., 2018).
Hoxa1 (OMIM 142955) mouse null mutants die at birth from anoxia and exhibit marked reductions in the sizes of r4 and r5, hypoplasia of the inner ear and specifically in CNIII. The embryonic phenotype is characterized by the absence of facial nerve and abducens motor nerve (Lufkin et al., 1991; Chisaka et al., 1992). In agreement, a homozygous truncating mutation of HOXA1 in humans causes severe congenital cardiovascular malformation, craniofacial and inner-ear defects, as well as brainstem abnormalities (Tischfield et al., 2005; Bosley et al., 2008).
Hoxb1 (OMIM 142968) loss-of-function mouse mutants exhibit alterations in the molecular markers associated with r4 identity, although no overt changes in the anatomy of the developing hindbrain are present (Goddard et al., 1996; Studer et al., 1996). These and previous results demonstrate that Hoxb1 has a normal role in regulating rhombomere identity, and also participates in controlling migratory properties of motor neurons in the hindbrain. In Hoxb1 mutant animals, the facial branchiomotor neurons (CNVII) and contralateral vestibular acoustic efferent (CNVIII), which are specific to r4, are incorrectly specified (Goddard et al., 1996; Studer et al., 1996). Hoxb1 deficiency in mouse also results in facial paralysis due to developmental defects in CNVII, originating from r4 (Figure 3C). In mouse lacking both Hoxa1 and Hoxb1 expression, the migration and development of NCCs derived from r4 fail, causing the loss of all second arch derivatives (Rossel and Capecchi, 1999; Arenkiel et al., 2004). These Hoxa1/Hoxb1 double mutants exhibit a wide range of phenotypes, which are not present in each of individual mutants, demonstrating that specification of r4 cell precursors and patterning of the CN VII-XI strongly requires cooperation between these 2 genes (Gavalas et al., 1998; Studer et al., 1998).
Hoxa2 (OMIM 604685) is the only member of the Hox family expressed in r2; this fact explains why Hoxa2 null mutations in mouse result in homeotic changes transforming second arch elements of NC origin into first arch derivatives, which was correlated with perinatal lethality. Patterning of the hindbrain rostral region also depends on Hoxa2 activity for the establishment of r2 identity and influencing the migration of trigeminal motor axons (CN V) originated from r2/3. In mutant embryos, this CN V, normally derived from r2/3, migrates caudally to exit the hindbrain from r4, the normal site for facial nerve (CN VII), rather than from r2. Hoxa2 is required for the maintenance of EphA4 (OMIM 602188) as its expression results selectively abolished in Hoxa2 mutants (Rijli et al., 1993). The loss of Hoxb2 (OMIM 142967) in mouse embryos results in impaired development of the facial nerve, CN VII, affecting its somatic motor component (Bailey et al., 1997).
Hoxa3 (OMIM 142954) null mutant mice show mesenchymal NCCs defects in the formation of CN IX and also fusions between CN IX and X. In addition, Hoxa3–/– mouse are athymic, aparathyroid, and have malformations in cartilage of the throat (Chisaka and Capecchi, 1991; Manley and Capecchi, 1995, 1997). Hoxb3–/– (OMIM 142966) embryos revealed similar cranial ganglia defects, but at a lower penetrance than in the Hoxa3 mutants (Manley and Capecchi, 1997). Hoxb3/Hoxd3 (OMIM 142980) double mutants have a clear increase in the presence of aberrant ganglionic phenotypes in CN IX compared to those reported in the Hoxb3 single mutant, even though the Hoxd3–/– does not show defects in these structures (Manley and Capecchi, 1998).
In conclusion, Hox patterning genes are crucial for NC development by interacting with signaling pathways that induce NC, but also to regulate expression of several genes involved in these essential cell and developmental processes. Some studies have shown that Polycomb group proteins are decisive in epigenetic silencing Hox genes by promoting changes in the chromatin structure. Dynamic patterns of histone modifications and 3D chromatin organization are also relevant regulators of Hox gene expression and function (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Noordermeer et al., 2011). The transcription factors Krox20 (OMIM 129010) and Kreisler (OMIM 608968), as well as the vitamin A derivative RA are the three main upstream regulators of Hox gene expression during hindbrain development.
The transcription factor Krox20 binds to specific DNA sequences located at 5′ flanking region of Hoxa2, Hoxb2, Hoxb3, and EphA4 genes, to directly control their expression (Lemaire et al., 1988; Nardelli et al., 1991). Targeted mutation of Krox20 in mouse embryos causes perinatal death and fusions of the trigeminal ganglia with facial and vestibular ganglia as a consequence of alterations on hindbrain patterning and morphogenesis. Krox20 is expressed in r3 and r5 at E8.0 in mouse embryos (Wilkinson et al., 1989; Schneider-Maunoury et al., 1993, 1997; Swiatek and Gridley, 1993; Nieto et al., 1995).
Kreisler expression, first detected at E8.5 in the prospective r5 region and later located in r5 and r6, is sharply downregulated afterward in these rhombomeres (Cordes and Barsh, 1994). Gene expression analyses in Kreisler mutant embryos and regulatory regions strongly pointed that this transcription factor could directly control the expression of genes required for inner ear and hindbrain development, in particular Hoxa3 and Hoxb3, which increase its expression in r5 and r6 (McKay et al., 1997; Manzanares et al., 1999). The primary defect in Kreisler mutant mouse embryos is an alteration in segmentation at the otic region of the hindbrain, resulting in defective rhombomeres since the borders that normally separate r4, r5, and r6 disappear. Consequently, in r6 important alterations are detected: the normal expression domains of FGF3 and CRABP1 (OMIM 180230) are lost, and Hoxa3 is not upregulated (Frohman et al., 1993). Although Krox20 expression in the prospective r3 is conserved, it is absent in r5. Similarly, the expression of Hoxb2, Hoxb3, and Hoxb4 in r5 are completely abolished. The expression pattern analysis of EphA7 and EphrinB2 indicates that only a single region that would correspond to r5 is absent. Thus, loss-of-function of Kreisler causes a segmentation defect which results in the precise loss of r5 patterning; furthermore, although the r6 territory forms, it fails to mature (Manzanares et al., 1999).
RA is a morphogen derived from Vitamin A (retinol) that reaches the cell nucleus after diffusing through cell membranes to act on histone acetylation and mediates transcriptional activation of target genes. RA is another important regulator of NC development. As mentioned earlier, Hox gene expression patterns specify AP identity in the hindbrain and this is transferred to NC migration (Briscoe and Wilkinson, 2004; Simkin et al., 2013). The “collinear pattern” of Hox gene expression in the hindbrain is partially dependent on RA control. Cellular retinoid-binding proteins (CRBPs) participate in controlling RA concentration locally, and hence facilitate its function. CRBPs might sequester RA and thus limit its availability to bind nuclear RA receptors (RARs and RXRs) that recognize a particular element on target genes, the RARE sequence (Mangelsdorf et al., 1992; Mangelsdorf and Evans, 1995). Members of the Hox family that harbor RAREs include Hoxa1, Hoxb1, Hoxa4 (OMIM 142953), Hoxb4 (OMIM 142965), Hoxd4 (OMIM 142981), and Hoxb5 (OMIM 142960) (Kesseland and Gruss, 1991; Langston and Gudas, 1992; Marshall et al., 1992, 1994; Studer et al., 1994; Dupé et al., 1997; Gould et al., 1998; Packer et al., 1998; Pera et al., 1999; Power et al., 1999).
Vitamin A-deficient pregnant rats were produced by feeding dams with low levels of all-trans RA. Such embryos presented loss of CN IX, X, XI, and XII and the associated sensory ganglia IX and X, as well as perturbations in hindbrain segmentation and otic vesicle development (White et al., 2000). These embryos have Hoxb1 protein in the NT, but caudal to the r3/r4 border at a time when its expression should be present only in r4, suggesting that RA is essential for neurogenesis, patterning, and segmentation in the posterior hindbrain. Neuron navigator 2 (Nav2) was first identified as an RA-responsive gene required for RA-mediated neurite outgrowth or survival of CN IX and X (McNeill et al., 2010). Nav2–/– mouse embryos showed an overall reduction in neurofilament density in the region of CN V to XII.
It was recently found that YAP (a Hippo signaling transcriptional co-activator, see above) regulates the expression of Hoxa1 and Hoxc13 in mouse oral and dental epithelial tissues as well as in embryonic and adult epidermal tissue (human keratinocytes) (Liu et al., 2015). Since Yap transcript was detected in the rhombencephalon and dorsal NT and also in NCCs that migrate from the dorsal region of the NT to the pharyngeal arches, Yap could regulate the activity of the Hoxa1 gene expression in the hindbrain.
The muscle segment-related homeobox (Msx) genes belong to the homeodomain family. These genes code for transcriptional factors with repressor activity. Proteins with homeodomains have various functions during embryonic development, from the formation of expression patterns to more specific functions such as differentiation toward a specific cell type (Catron et al., 1995). Msx genes are expressed in a range of vertebrate-specific tissues including NC, cranial sensory placodes, bones, and teeth (Davidson, 1995).
In vertebrates, there are three members of this family, Msx1-3; Msx1, and Msx2 being the best characterized ones with respect to their expression pattern and biochemical properties (Bendall and Abate-Shen, 2000). Msx1 and Msx2 are expressed in various regions of the mouse embryo such as the NT, in the primordial limbs and in derivatives of the cranial NC (Catron et al., 1996). The expression of Msx1 and Msx2 marks the area from which the cranial NC will migrate. Msx genes participate in the early specification of NCCs and in the control of apoptotic process under the control of BMP signaling (Tribulo et al., 2003, Tribulo et al., 2004; Ishii et al., 2005).
Pax genes, which encode transcription factors that contain a highly conserved DNA binding domain called PD, can be considered as the broad regulators of gene expression since they can repress pluripotency genes such as Oct4, Nanog and Myc, or induce the expression of genes involved in the differentiation of NC such as Snail1 and FoxD3. There are nine Pax genes (Pax1-Pax9), which have been characterized in mammals. There is a great diversity of studies on Pax genes in the early specification of cell fate and in the morphogenesis of various tissues and organs. The important participation of Pax genes in NC induction is discussed next. Pax3 participates in the early ontogenesis of the NT and NC; it is expressed in pre-migratory NCCs. The loss of Pax3 generates severe defects in embryonic development, leading to embryonic death (Goulding et al., 1991). A study in mouse evaluated the participation of the Wnt signaling pathway in the regulation of Pax3. It was demonstrated that the Wnt pathway induces expression of Pax3 indirectly, using Cdx1 as an intermediary that binds the PD domain of Pax3 (Sanchez-Ferras et al., 2012).
On the other hand, transcriptional enhancers are primary determinants of the specific gene expression of a cell type. Recently, an NC enhancer-2 (NCE2) in the 5′ region of Pax3 was identified as a cis regulatory element that is dependent on Cdx as a cofactor. Pax3 and Zic2 are expressed in the dorsal region of the NT when it closes. Therefore, the inductive Cdx-Zic2 interaction is integrated by NCE2, allowing the specific binding of the neural transcription factor Sox2 (Buecker and Wysocka, 2012; Sanchez-Ferras et al., 2014). This shows that not only NCE2, but also the transcription factor Zic 2 participate in the regulation of Pax3. Such data suggests that Zic2 is involved in NC induction as an activator of Pax3-NCE2 and as a Cdx co-factor. Mouse Pax3 mutants (Sp and Spd alleles) additionally exhibit malformations of ganglia of the PNS. The importance of Pax3 in the development of NC-derived structures has been shown, especially with respect to cranial ganglia and nerves. In the homozygous state, Sp and Spd alleles impair the development of the trigeminal (CN V), superior (CN IX), and jugular (CN X) ganglia, suggesting that the function of Pax3 is crucial for NC migration and proliferation, as well as for its differentiation into neurons capable of sending out axons (Tremblay et al., 1995). In Xenopus and zebrafish embryos, Pax3 has been proposed as a key player in the gene regulatory network as a neural plate border specifier controlling early specification of NCC (Hong and Saint-Jeannet, 2007; Minchin and Hughes, 2008; Milet et al., 2013).
Another crucial Pax gene in NC formation is Pax7, which has been described as necessary for NC development in birds. Pax7 is required for the expression of NC markers such as Sox9, Snai2, HNK1 and Sox10 (Basch and Bronner-Fraser, 2006). In human embryos, Pax7 is expressed in the dorsal NT and in cells of the migratory NC at early stages. In mouse, Pax7 is expressed in the rostral region, which includes a subpopulation of presumptive NC precursors. Pax7 contributes more to the formation of cranial lineages than to the cardiac or trunk regions. The expression of Pax7 is extensive, since it is detected in mesencephalon, rhombencephalon, dorsal NT, fronto-nasal region and NCCs that migrate from the dorsal region of the NT to the pharyngeal arches (Betters et al., 2010; Murdoch et al., 2012). A mutation of Pax7 (isoform 3) was recently found in patients, causing a phenotype of neurodevelopmental delay during development and promoting microcephaly, irritability and self-mutilation among others symptoms (Proskorovski-Ohayon et al., 2017). Therefore, Pax7 is a crucial gene in the induction of NC and in its migration.
Paired-like Homeobox 2b (Phox2b, OMIM 603851) is a transcription factor known to play a key role in the development of the autonomic nervous system. Phox2b is expressed in differentiating neurons of the mouse central and PNS as well as in motor nuclei of the hindbrain. Phox2a (OMIM 602753) and Phox2b are co-expressed at multiple sites, suggesting a broader role for Phox2 genes in the specification of autonomic neurons and cranial motor nuclei. The co-expression of these Phox proteins at various sites suggested positive crosstalk (Pattyn et al., 1997). Mash1 has been demonstrated to control the expression of Phox2a (but not of Phox2b) in autonomic ganglionic precursors and NC-stem cells (Lo et al., 1998), while Phox2b is required for the maintenance but not for the induction of Mash1 expression (Pattyn et al., 1999). Epistatic analyses have shown that, in cranial ganglia development, Phox2b is a downstream effector of Phox2a (Pattyn et al., 1999). The mutant Phox2bLacZ/LacZ mouse showed atrophic cranial ganglia formation that correlated with increased apoptotic cell death and decreased Ret and DBH expression in ganglionic anlages (Pattyn et al., 1999). The effect of the Phox2b null mutation on cranial ganglia cells was a phenotypical change on their molecular transcriptional signature to Tlx3+/Islet+/Phox2b–/Phox2a–/Brn3a+ profile, which means that the sensory neurons present in the cranial nerves VII, IX, and X change to a somatic sensory neuron-like, thus highlighting the role of Phox2b as a molecular switch that commands the somatic-to-visceral phenotype in the cranial sensory genetic cascade (D’Autréaux et al., 2011). Many cranial nerve-associated NCCs co-expressed the pan-autonomic determinant Phox2b and markers of Schwann cell precursors. Such cranial NCC precursors are the source of parasympathetic neurons during normal development (Espinosa-Medina et al., 2014). In humans, PHOX2B over-expression has been linked to the formation of tumors arising from the sympathetic nervous system such as neuroblastomas. Heterozygous PHOX2B mutations cause Congenital Central Hypoventilation Syndrome, a life-threatening neurocristopathy characterized by the defective autonomic control of breathing and involving altered CO2/H+ chemosensivity (Cardani et al., 2018; Vega-Lopez et al., 2018).
Otx1 and Otx2 genes are the mouse cognates of the Drosophila head gap genes. Orthologs have also been identified in human, chick, Xenopus and zebrafish. Otx2 may act as a key head organizer during the primitive streak stage. At subsequent neurula to pharyngula stages, those genes participate in the patterning of the forebrain and midbrain. The haplo-insufficiency mutation of Otx2 in the mouse affects the mandible and pre-mandibular skull elements, as well as the ophthalmic branch of the trigeminal nerve, and the differentiation of mesencephalic trigeminal neurons, all of which correspond to derivatives originated from mesencephalic NC (Puelles and Rubenstein, 1993; Matsuo et al., 1995). In chick and Xenopus embryos, Otx2 establishes cross-regulatory interactions with Gbx2 during the early specification of placodal precursors; by mutual repression, both genes pattern the territory, segregating trigeminal progenitors (Steventon et al., 2012). Additionally, Gbx2 is expressed early in the preplacodal region of Xenopus embryos and is required for NCCs formation as an effector of Wnt signaling (Li et al., 2009).
The proteins encoded by the Sox genes belong to the superfamily of the High Mobility Group transcriptional factors that bind to the DNA sequence (A/T)(A/T)CAA(A/T)G. They have a DNA binding domain of 80 amino acids. Based on phylogenetic analyses of their domains, Sox genes are divided into subgroups A-H in mouse (Bowles et al., 2000). Some are transcriptional activators, others are repressors, and a third group lacks the transactivation domain.
The subgroup of SoxE genes (Sox8, Sox9 and Sox10) has a prominent participation in NC differentiation. In mouse, Sox9 and Sox10 are among the first expressed genes in the NC progenitors overlapping with FoxD3 (Hong and Saint-Jeannet, 2005). A study showed that the defects in the expression of this subgroup affects many lineages of the NC, so these genes are important regulators in the formation of this multipotent population (Kelsh, 2006). However, it is not known if SoxE genes are also involved in NC induction in the mouse. Knock-out mice for Sox9 show expression of Snai1 in the NC; nevertheless, these cells undergo apoptosis either before or immediately after migrating, which suggests that Sox9 participates in the epithelial-mesenchymal transition, before delamination (Cheung et al., 2005).
Sox10 is a protein that participates in maintaining multipotency in NCCs; it also contributes to proliferation and inhibits differentiation, so this transcriptional factor is expressed in the pre-migratory progenitor cells of the NC and its expression decreases at the beginning of the differentiation process (Kim et al., 2003). Several studies have shown that Sox10 controls the fate of the NC by activating critical genes for the differentiation of different cell types such as melanocytes, Schwann cells, autonomic and sensory neurons in different species (Mitfa, ErbB3, Phox2b, Mash1, and Ngn1, respectively) (Britsch et al., 2001; Elworthy, 2003; Kim et al., 2003; Elworthy et al., 2005; Kelsh, 2006). Sox10 is regulated by post-translational modifications. For example, changes in the state of SUMOylation affect its function, because it regulates interactions with different proteins and promotes the activation of different genes. Sox10 expression can be regulated by multiple enhancer elements such as U3, known as MCS4. The stimulation of the U3 enhancer activity promoted Sox10 transcription, which had a synergistic activity with other transcriptional factors involved in NC development, including Pax3, FoxD3, AP-2α, Krox20, and Sox2 (Taylor and LaBonne, 2005). Sox10 can be self-regulated as well as regulated by synergistic interactions during NC development (Wahlbuhl et al., 2012).
Zic genes are transcription factors with zinc fingers that contribute to different processes during embryonic development (Aruga et al., 1998). It has been proposed that Zic1-3 participate in lateral segmentation, NC induction and inhibition of neurogenesis (Nagai et al., 1997; Nakata et al., 1997). This gene family consists of five members, Zic1-Zic5 in the mouse. Zic genes are co-expressed in some cells during embryonic development, which gives the opportunity for heterogeneous protein-protein interactions and/or functional redundancy among family members. Zic2 (OMIM 603073) is expressed in the cells of the inner mass of the blastocyst and is required for the synchronization of neurulation. Zic2 mutants showed delayed production and decreased numbers of NCCs. Zic2 is also necessary for the formation of r3 and r5 and participates in the normal pattern of the mouse rhombencephalon (Elms et al., 2003).
Mouse homozygous mutants of Zic1 exhibit ataxia during development and die within the first month after birth. These mutants also show a hypoblastic cerebellum and absence of the anterior lobe (Elms et al., 2003). In Zic mutants, the expression of Msx1 in the region of the dorsal NT was not altered; however, its expression was lost in this region when the NT was closing, suggesting that signals from the floor plate are required for the maintenance of dorsal expression (Sanchez-Ferras et al., 2014). Zic5 (OMIM 617896) is expressed in the dorsal part of the NT and its mutation in humans produces holoprosencephalia, a severe brain malformation. A decrease in Zic5 promotes insufficient NT closure in the rostral-most part, which was also observed with Zic2 (Inoue et al., 2004). Zic2 mutant embryos showed affected CN V, VII and VII (Elms et al., 2003).
Deficient Zic5 mice show malformations of the facial bones derived from the NC, mainly the mandible, due to decreased generation of NCCs. During embryonic stages, there were also delays in the development of the first branchial arch and extension of the trigeminal and facial nerves. On the other hand, deletion of Zic2 promotes congenital malformation of the brain and digits in humans (Inoue et al., 2004). Cranial NCCs are also known to contribute to the development of the PNS. In both mutants, a reduction in the axonal projections from the trigeminal and facial ganglions was reported. These findings suggest that cephalic NC derivatives are selectively affected in these mutants (Inoue et al., 2004).
Some of the most relevant pathways and genes involved in CN formation are represented in Figure 3 and Table 2. Gene regulation during embryonic development as well as during induction, specification, delamination, migration, survival and differentiation of the NC is a very complex process that leads to a strict expression of genetic information. A remarkable conservation of many genes, signals and mechanisms between different vertebrate organisms, but also its repeated use at different places and times in NC development and cranial nerve/ganglia formation, contributes to the complexity of these processes. Adequate transduction of the signals is equally important for the development and differentiation of each of the cell types derived from the NC. The integration of knowledge from the various studies on such signaling pathways and the different types of proteins that participate in the sequential processes as well as their post-translational modifications will lead to a better understanding of neurogenesis and cranial nerve formation. The microenvironment in which these cells develop is of great importance in order to understand the mechanisms involved in proper NC induction and CN development.
All authors listed have made substantial, direct and intellectual contribution to the work and approved it for publication. KM-M did molecular neuroscience data, conception and design, compiling and summarizing articles, manuscript writing, and final approval of the manuscript. GV-L did cell and developmental biology data, conception and design, compiling and summarizing articles, figures elaboration, manuscript writing, and final approval of the manuscript. MA involved in financial support, cell and developmental biology data, conception and design, compiling and summarizing articles, manuscript writing, and final approval of manuscript. IV involved in financial support, molecular neuroscience data, conception design, correction of style, manuscript writing, and final approval of the manuscript.
This work was supported by PICT2015-1207, PICT2018-1370, PIUNT D605 (MA), PICT-2016-0835 (GV-L), and PAPIIT-UNAM IN213719 (IV).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to colleagues from our research groups for helpful discussions, especially Dr. Itzel Escobedo-Avila for logistical support. We thank Mrs. Virginia Méndez for manuscript proofreading. We wish to thank the postdoctoral training program of the Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (DGAPA-UNAM) for the fellowship awarded to KM-M. We regret that numerous publications dealing with the principal topic of this review could not be cited due to space limitations.
BMP, bone morphogenetic proteins; CN, cranial nerve; CNS, central nervous system; DRG, dorsal root ganglia; FP, floor plate; hESCs, human embryonic stem cells; Msx, Muscle segment-related homeobox; NC, neural crest; NCCs, neural crest cells; NT, neural tube; PA, pharyngeal arches; Pax, Paired box; PNS, peripheral nervous system; r, rhombomere; RA, retinoic acid; RTK, receptor tyrosine kinase; Shh, Sonic hedgehog; Sox, Sry-box; VSMC, vascular smooth muscle cells; Wnt, Wingless and Int-1; WRPW, Trp-Arg-Pro-Trp; Zic, Zinc finger protein of cerebellum.
Abe, M., Ruest, L. B., and Clouthier, D. E. (2007). Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor-deficient mice. Int. J. Dev. Biol. 51, 97–105. doi: 10.1387/ijdb.062237ma
Adams, R. H., Diella, F., Hennig, S., Helmbacher, F., Deutsch, U., and Klein, R. (2001). The cytoplasmic domain of the ligand ephrinB2 is required for vascular morphogenesis but not cranial neural crest migration. Cell 104, 57–69.
Aguero, T. H., Fernandez, J. P., Lopez, G. A., Tribulo, C., Aybar, M. J., Agüero, T. H., et al. (2012). Indian hedgehog signaling is required for proper formation, maintenance and migration of Xenopus neural crest. Dev. Biol. 364, 99–113. doi: 10.1016/j.ydbio.2012.01.020
Ahlgren, S. C., and Bronner-Fraser, M. (1999). Inhibition of Sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr. Biol. 9, 1304–1314. doi: 10.1016/S0960-9822(00)80052-4
Akazawa, C., Sasai, Y., Nakanishi, S., and Kageyama, R. (1992). Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J. Biol. Chem. 267, 21879–21885.
Alam, K. Y., Frostholm, A., Hackshaw, K. V., Evans, J. E., Rotter, A., and Chiu, I. M. (1996). Characterization of the 1B promoter of fibroblast growth factor 1 and its expression in the adult and developing mouse brain. J. Biol. Chem. 271, 30263–30271. doi: 10.1074/jbc.271.47.30263
Andermann, P., Ungos, J., and Raible, D. W. (2002). Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev. Biol. 251, 45–58. doi: 10.1006/dbio.2002.0820
Anderson, C. N. G., Ohta, K., Quick, M. M., Fleming, A., Keynes, R., and Tannahill, D. (2003). Molecular analysis of axon repulsion by the notochord. Development 130, 1123–1133. doi: 10.1242/dev.00327
Anderson, R. M., Stottmann, R. W., Choi, M., and Klingensmith, J. (2006). Endogenous bone morphogenetic protein antagonists regulate mammalian neural crest generation and survival. Dev. Dyn. 235, 2507–2520. doi: 10.1002/dvdy.20891
Aragón, F., Vázquez-Echeverría, C., Ulloa, E., Reber, M., Cereghini, S., Alsina, B., et al. (2005). vHnf1 regulates specification of caudal rhombomere identity in the chick hindbrain. Dev. Dyn. 234, 567–576. doi: 10.1002/dvdy.20528
Arenkiel, B. R., Tvrdik, P., Gaufo, G. O., and Capecchi, M. R. (2004). Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 18, 1539–1552. doi: 10.1101/gad.1207204
Arthur, A., Koblar, S., Shi, S., and Gronthos, S. (2009). Eph/ephrinB mediate dental pulp stem cell mobilization and function. J. Dent. Res. 88, 829–834. doi: 10.1177/0022034509342363
Aruga, J., Minowa, O., Yaginuma, H., Kuno, J., Nagai, T., Noda, T., et al. (1998). Mouse Zic1 is involved in cerebellar development. J. Neurosci. 18, 284–293. doi: 10.1523/jneurosci.5203-05.2006
Aybar, M. J., and Mayor, R. (2002). Early induction of neural crest cells: lessons learned from frog, fish and chick. Curr. Opin. Genet. Dev. 12, 452–458.
Baggiolini, A., Varum, S., Mateos, J. M., Bettosini, D., John, N., Bonalli, M., et al. (2015). Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell 16, 314–322. doi: 10.1016/j.stem.2015.02.017
Bailey, W. J., Kim, J., Wagner, G. P., and Ruddle, F. H. (1997). Phylogenetic reconstruction of vertebrate Hox cluster duplications. Mol. Biol. Evol. 14, 843–853. doi: 10.1093/oxfordjournals.molbev.a025825
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657. doi: 10.1016/j.cell.2005.03.013
Bally-Cuif, L., and Wassef, M. (1995). Determination events in the nervous system of the vertebrate embryo. Curr. Opin. Genet. Dev. 5, 450–458. doi: 10.1016/0959-437X(95)90048-L
Bani-Yaghoub, M., Tremblay, R. G., Lei, J. X., Zhang, D., Zurakowski, B., Sandhu, J. K., et al. (2006). Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 295, 52–66. doi: 10.1016/j.ydbio.2006.03.007
Barlow, L. A. (2002). Cranial nerve development: placodal neurons ride the crest. Curr. Biol. 12, R171–R173. doi: 10.1016/S0960-9822(02)00734-0
Barlow, L. A., and Northcutt, R. G. (1997). Taste buds develop autonomously from endoderm without induction by cephalic neural crest or paraxial mesoderm. Development 124, 949–957.
Barraud, P., Seferiadis, A. A., Tyson, L. D., Zwart, M. F., Szabo-Rogers, H. L., Ruhrberg, C., et al. (2010). Neural crest origin of olfactory ensheathing glia. Proc. Natl. Acad. Sci. U.S.A. 107, 21040–21045. doi: 10.1073/pnas.1012248107
Barth, K. A., Kishimoto, Y., Rohr, K. B., Seydler, C., Schulte-Merker, S., and Wilson, S. W. (1999). Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development 126, 4977–4987. doi: 10.1074/jbc.M112.386052
Barton, W. A., Dalton, A. C., Seegar, T. C. M., Himanen, J. P., and Nikolov, D. B. (2014). Tie2 and Eph receptor tyrosine kinase activation and signaling. Cold Spring Harb. Perspect. Biol. 6, 1–17. doi: 10.1101/cshperspect.a009142
Basch, M. L., and Bronner-Fraser, M. (2006). Neural crest inducing signals. Adv. Exp. Med. Biol. 589, 24–31. doi: 10.1007/978-0-387-46954-6_2
Basch, M. L., Bronner-Fraser, M., and García-Castro, M. I. (2006). Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218–222. doi: 10.1038/nature04684
Begbie, J., and Graham, A. (2001). Integration between the epibranchial placodes and the hindbrain. Science 294, 595–598. doi: 10.1126/science.1062028
Bendall, A. J., and Abate-Shen, C. (2000). Roles for Msx and Dlx homeoproteins in vertebrate development. Gene 247, 17–31. doi: 10.1016/S0378-1119(00)00081-0
Betancur, P., Bronner-Fraser, M., and Sauka-Spengler, T. (2010). Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 26, 581–603. doi: 10.1146/annurev.cellbio.042308.113245
Betters, E., Liu, Y., Kjaeldgaard, A., Sundström, E., and García-Castro, M. I. (2010). Analysis of early human neural crest development. Dev. Biol. 344, 578–592. doi: 10.1016/j.ydbio.2010.05.012
Bonano, M., Tríbulo, C., De Calisto, J., Marchant, L., Sánchez, S. S., Mayor, R., et al. (2008). A new role for the Endothelin-1/Endothelin-A receptor signaling during early neural crest specification. Dev. Biol. 323, 114–129. doi: 10.1016/j.ydbio.2008.08.007
Bosley, T. M., Alorainy, I. A., Salih, M. A., Aldhalaan, H. M., Abu-Amero, K. K., Oystreck, D. T., et al. (2008). The clinical spectrum of homozygous HOXA1 mutations. Am. J. Med. Genet. Part A 146, 1235–1240. doi: 10.1002/ajmg.a.32262
Bowles, J., Schepers, G., and Koopman, P. (2000). Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227, 239–255. doi: 10.1006/dbio.2000.9883
Boyd, J. G., Skihar, V., Kawaja, M., and Doucette, R. (2003). Olfactory ensheathing cells: historical perspective and therapeutic potential. Anat. Rec. Part B New Anat. 27, 49–60. doi: 10.1002/ar.b.10011
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. doi: 10.1038/nature04733
Bracken, A. P., Dietrich, N., Pasini, D., Hansen, K. H., and Helin, K. (2006). Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136. doi: 10.1101/gad.381706
Brault, V., Moore, R., Kutsch, S., Ishibashi, M., Rowitch, D. H., McMahon, A. P., et al. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264.
Briscoe, J., and Wilkinson, D. G. (2004). Establishing neuronal circuitry: hox genes make the connection. Genes Dev. 18, 1643–1648. doi: 10.1101/gad.1227004
Britsch, S., Goerich, D. E., Riethmacher, D., Peirano, R. I., Rossner, M., Nave, K. A., et al. (2001). The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66–78. doi: 10.1101/gad.186601
Britsch, S., Li, L., Kirchhoff, S., Theuring, F., Brinkmann, V., Birchmeier, C., et al. (1998). The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 12, 1825–1836. doi: 10.1101/gad.12.12.1825
Buac, K., Watkins-Chow, D. E., Loftus, S. K., Larson, D. M., Incao, A., Gibney, G., et al. (2008). A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet. 4:e1000177. doi: 10.1371/journal.pgen.1000177
Buecker, C., and Wysocka, J. (2012). Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 28, 276–284. doi: 10.1016/j.tig.2012.02.008
Bylund, M., Andersson, E., Novitch, B. G., and Muhr, J. (2003). Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6, 1162–1168. doi: 10.1038/nn1131
Byrd, N. A., and Meyers, E. N. (2005). Loss of Gbx2 results in neural crest cell patterning and pharyngeal arch artery defects in the mouse embryo. Dev. Biol. 284, 233–245. doi: 10.1016/j.ydbio.2005.05.023
Calloni, G. W., Glavieux-Pardanaud, C., Le Douarin, N. M., and Dupin, E. (2007). Sonic Hedgehog promotes the development of multipotent neural crest progenitors endowed with both mesenchymal and neural potentials. Proc. Natl. Acad. Sci. U.S.A. 104, 19879–19884. doi: 10.1073/pnas.0708806104
Cao, X., Pfaff, S. L., and Gage, F. H. (2008). YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 22, 3320–3334. doi: 10.1101/gad.1726608
Cardani, S., Di Lascio, S., Belperio, D., Di Biase, E., Ceccherini, I., Benfante, R., et al. (2018). Desogestrel down-regulates PHOX2B and its target genes in progesterone responsive neuroblastoma cells. Exp. Cell Res. 370, 671–679. doi: 10.1016/j.yexcr.2018.07.032
Catron, K. M., Wang, H., Hu, G., Shen, M. M., and Abate-Shen, C. (1996). Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech. Dev. 55, 185–199. doi: 10.1016/0925-4773(96)00503-5
Catron, K. M., Zhang, H., Marshall, S. C., Inostroza, J. A., Wilson, J. M., Abate, C., et al. (1995). Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol. Cell Biol. 15, 861–871.
Cerrizuela, S., Vega-Lopez, G. A., and Aybar, M. J. (2020). The role of teratogens in neural crest development. Birth Defects Res. 12, 584–632. doi: 10.1002/bdr2.1644
Cerrizuela, S., Vega-López, G. A., Palacio, M. B., Tríbulo, C., and Aybar, M. J. (2018). Gli2 is required for the induction and migration of Xenopus laevis neural crest. Mech. Dev. 154, 219–239. doi: 10.1016/j.mod.2018.07.010
Cheung, M., Chaboissier, M. C., Mynett, A., Hirst, E., Schedl, A., and Briscoe, J. (2005). The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell 8, 179–192. doi: 10.1016/j.devcel.2004.12.010
Chew, L. J., and Gallo, V. (2009). The Yin and Yang of Sox proteins: activation and repression in development and disease. J. Neurosci. Res. 87, 3277–3287. doi: 10.1002/jnr.22128
Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H., et al. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. doi: 10.1038/383407a0
Chisaka, O., and Capecchi, M. R. (1991). Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 350, 473–479. doi: 10.1038/350473a0
Chisaka, O., Musci, T. S., and Capecchi, M. R. (1992). Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature 355, 516–520. doi: 10.1038/355516a0
Choy, C., Ansari, K. I., Neman, J., Hsu, S., Duenas, M. J., Li, H., et al. (2017). Cooperation of neurotrophin receptor TrkB and Her2 in breast cancer cells facilitates brain metastases. Breast Cancer Res. 19:51. doi: 10.1186/s13058-017-0844-3
Clouthier, D. E., Williams, S. C., Hammer, R. E., Richardson, J. A., and Yanagisawa, M. (2003). Cell-autonomous and nonautonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Dev. Biol. 261, 506–519. doi: 10.1016/S0012-1606(03)00128-3
Clouthier, D. E., Williams, S. C., Yanagisawa, H., Wieduwilt, M., Richardson, J. A., and Yanagisawa, M. (2000). Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 217, 10–24.
Conover, J. C., Doetsch, F., Garcia-Verdugo, J.-M., Gale, N. W., Yancopoulos, G. D., and Alvarez-Buylla, A. (2000). Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat. Neurosci. 3, 1091–1097. doi: 10.1038/80606
Cooper, M. A., Crockett, D. P., Nowakowski, R. S., Gale, N. W., and Zhou, R. (2009). Distribution of EphA5 receptor protein in the developing and adult mouse nervous system. J. Comp. Neurol. 514, 310–328. doi: 10.1002/cne.22030
Cordes, S. P., and Barsh, G. S. (1994). The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79, 1025–1034. doi: 10.1016/0092-8674(94)90033-7
Cornell, R. A., and Eisen, J. S. (2002). Delta/Notch signaling promotes formation of zebrafish neural crest by repressing neurogenin 1 function. Development 129, 2639–2648.
Coulthard, M. G., Morgan, M., Woodruff, T. M., Arumugam, T. V., Taylor, S. M., and Carpenter, T. C. (2012). Eph/Ephrin Signaling in Injury and Inflammation. Am. J. Pathol. 181, 1493–1503. doi: 10.1016/j.ajpath.2012.06.043
Couly, G. F., Coltey, P. M., and Le Douarin, N. M. (1993). The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development 117, 409–429.
Couly, G. F., and Le Douarin, N. M. (1987). Mapping of the early neural primordium in quail-chick chimeras. II. The prosencephalic neural plate and neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev. Biol. 120, 198–214. doi: 10.1016/0012-1606(87)90118-7
Covell, D. A., and Noden, D. M. (1989). Embryonic development of the chick primary trigeminal sensory-motor complex. J. Comp. Neurol. 286, 488–503. doi: 10.1002/cne.902860407
d’Amico-Martel, A., and Noden, D. M. (1980). An autoradiographic analysis of the development of the chick trigeminal ganglion. J. Embryol. Exp. Morphol. 55, 167–182.
D’amico-Martel, A., and Noden, D. M. (1983). Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am. J. Anat. 166, 445–468. doi: 10.1002/aja.1001660406
D’Autréaux, F., Coppola, E., Hirsch, M. R., Birchmeier, C., and Brunet, J. F. (2011). Homeoprotein Phox2b commands a somatic-to-visceral switch in cranial sensory pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 20018–20023. doi: 10.1073/pnas.1110416108
Davidson, D. (1995). The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 11, 405–411. doi: 10.1016/S0168-9525(00)89124-6
Davy, A., Aubin, J., and Soriano, P. (2004). Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 18, 572–583.
De, A. (2011). Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim. Biophys. Sin. 43, 745–756. doi: 10.1093/abbs/gmr079
Defossez, P. A., and Stancheva, I. (2011). Biological functions of methyl-CpG-binding proteins. Prog. Mol. Biol. Transl. Sci. 101, 377–398. doi: 10.1016/B978-0-12-387685-0.00012-3
Deng, Y., Wu, L. M. N., Bai, S., Zhao, C., Wang, H., Wang, J., et al. (2017). A reciprocal regulatory loop between TAZ/YAP and G-protein Gas regulates Schwann cell proliferation and myelination. Nat. Commun. 8, 1–15. doi: 10.1038/ncomms15161
Dennis, J. F., Kurosaka, H., Iulianella, A., Pace, J., Thomas, N., Beckham, S., et al. (2012). Mutations in hedgehog acyltransferase (Hhat) perturb hedgehog signaling, resulting in severe acrania-holoprosencephaly-agnathia craniofacial defects. PLoS Genet. 8:e1002927. doi: 10.1371/journal.pgen.1002927
Diercke, K., Kohl, A., Lux, C. J., and Erber, R. (2011). Strain-dependent Up-regulation of Ephrin-B2 protein in periodontal ligament fibroblasts contributes to osteogenesis during tooth movement. J. Biol. Chem. 286, 37651–37664. doi: 10.1074/jbc.m110.166900
Duboule, D., and Dollé, P. (1989). The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8, 1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x
Dupé, V., Davenne, M., Brocard, J., Dollé, P., Mark, M., Dierich, A., et al. (1997). In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3’RARE). Development 124, 399–410.
Dupé, V., and Pellerin, I. (2009). Retinoic acid receptors exhibit cell-autonomous functions in cranial neural crest cells. Dev. Dyn. 238, 2701–2711. doi: 10.1002/dvdy.22087
Eickholt, B. J., Mackenzie, S. L., Graham, A., Walsh, F. S., and Doherty, P. (1999). Evidence for collapsin-1 functioning in the control of neural crest migration in both trunk and hindbrain regions. Development 126, 2181–2189.
Elms, P., Siggers, P., Napper, D., Greenfield, A., and Arkell, R. (2003). Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev. Biol. 264, 391–406. doi: 10.1016/j.ydbio.2003.09.005
Elworthy, S. (2003). Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 130, 2809–2818. doi: 10.1242/dev.00461
Elworthy, S., Pinto, J. P., Pettifer, A., Cancela, M. L., and Kelsh, R. N. (2005). Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech. Dev. 122, 659–669. doi: 10.1016/j.mod.2004.12.008
Erickson, S. L., O’Shea, K. S., Ghaboosi, N., Loverro, L., Frantz, G., Bauer, M., et al. (1997). ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124, 4999–5011.
Eroglu, B., Min, J. N., Zhang, Y., Szurek, E., Moskophidis, D., Eroglu, A., et al. (2014). An essential role for heat shock transcription factor binding protein 1 (HSBP1) during early embryonic development. Dev. Biol. 386, 448–460. doi: 10.1016/j.ydbio.2013.12.038
Espinosa-Medina, I., Outin, E., Picard, C. A., Chettouh, Z., Dymecki, S., Consalez, G. G., et al. (2014). Neurodevelopment. Parasympathetic ganglia derive from Schwann cell precursors. Science 345, 87–90. doi: 10.1126/science.1253286
Everson, J. L., Fink, D. M., Yoon, J. W., Leslie, E. J., Kietzman, H. W., Ansen-Wilson, L. J., et al. (2017). Sonic hedgehog regulation of Foxf2 promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development 144, 2082–2091. doi: 10.1242/dev.149930
Fantauzzo, K. A., and Soriano, P. (2015). Receptor tyrosine kinase signaling: regulating neural crest development one phosphate at a time. Curr. Top. Dev. Biol. 111, 135–182. doi: 10.1016/bs.ctdb.2014.11.005
Farah, M. H., Olson, J. M., Sucic, H. B., Hume, R. I., Tapscott, S. J., and Turner, D. L. (2000). Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127, 693–702.
Fedtsova, N., Perris, R., and Turner, E. E. (2003). Sonic hedgehog regulates the position of the trigeminal ganglia. Dev. Biol. 261, 456–469. doi: 10.1016/S0012-1606(03)00316-6
Ferrara, N., Gerber, H. P., and LeCouter, J. (2003). The biology of VEGF and its receptors. Nat. Med. 9, 669–676. doi: 10.1038/nm0603-669
Flanagan, J. G., and Vanderhaeghen, P. (1998). The ephrins and Eph receptors in neural development. Annu. Rev. Neurosci. 21, 309–345. doi: 10.1146/annurev.neuro.21.1.309
Flenniken, A. M., Gale, N. W., Yancopoulos, G. D., and Wilkinson, D. G. (1996). Distinct and overlapping expression patterns of ligands for Eph-Related receptor tyrosine kinases during mouse embryogenesis. Dev. Biol. 179, 382–401. doi: 10.1006/dbio.1996.0269
Forbes, D. J., and Welt, C. (1981). Neurogenesis in the trigeminal ganglion of the albino rat: a quantitative autoradiographic study. J. Comp. Neurol. 199, 133–147. doi: 10.1002/cne.901990111
Forni, P. E., Taylor-Burds, C., Melvin, V. S., Williams, T., and Wray, S. (2011). Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH-1 neurons, sensory neurons, and olfactory ensheathing cells. J. Neurosci. 31, 6915–6927. doi: 10.1523/jneurosci.6087-10.2011
Frohman, M. A., Martin, G. R., Cordes, S. P., Halamek, L. P., and Barsh, G. S. (1993). Altered rhombomere-specific gene expression and hyoid bone differentiation in the mouse segmentation mutant, kreisler (kr). Development 117, 925–936.
Furukawa, K. T., Yamashita, K., Sakurai, N., and Ohno, S. (2017). The epithelial circumferential actin belt regulates YAP/TAZ through Nucleocytoplasmic shuttling of merlin. Cell Rep. 20, 1435–1447. doi: 10.1016/j.celrep.2017.07.032
Gamble, J. A. (2005). Disruption of Ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J. Neurosci. 25, 3142–3150. doi: 10.1523/jneurosci.4759-04.2005
Gammill, L. S., Gonzalez, C., and Bronner-Fraser, M. (2006). Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. J. Neurobiol. 67, 47–56.
Gammill, L. S., Gonzalez, C., and Bronner-Fraser, M. (2007). Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. Dev. Neurobiol. 67, 47–56. doi: 10.1002/dneu.20326
Gao, H., Bu, Y., Wu, Q., Wang, X., Chang, N., Lei, L., et al. (2015). Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J. Cell Sci. 128, 2340–2350. doi: 10.1242/jcs.167874
Garratt, A. N., Voiculescu, O., Topilko, P., Charnay, P., and Birchmeier, C. (2000). A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J. Cell Biol. 148, 1035–1046. doi: 10.1083/jcb.148.5.1035
Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R., et al. (1995). Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378, 390–394. doi: 10.1038/378390a0
Gavalas, A., Studer, M., Lumsden, A., Rijli, F. M., Krumlauf, R., and Chambon, P. (1998). Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development 125, 1123–1136.
Gavalas, A., Trainor, P., Ariza-McNaughton, L., and Krumlauf, R. (2001). Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development 128, 3017–3027.
Giovannini, M., Robanus-Maandag, E., Niwa-Kawakita, M., Van Der Valk, M., Woodruff, J. M., Goutebroze, L., et al. (1999). Schwann cell hyperplasia and tumors in transgenic mice expressing a naturally occurring mutant NF2 protein. Genes Dev. 13, 978–986. doi: 10.1101/gad.13.8.978
Giovannini, M., Robanus-Maandag, E., Van Der Valk, M., Niwa-Kawakita, M., Abramowski, V., Goutebroze, L., et al. (2000). Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 14, 1617–1630. doi: 10.1101/gad.14.13.1617
Goddard, J. M., Rossel, M., Manley, N. R., and Capecchi, M. R. (1996). Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development 122, 3217–3228.
Golding, J. P., Sobieszczuk, D., Dixon, M., Coles, E., Christiansen, J., Wilkinson, D., et al. (2004). Roles of erbB4, rhombomere-specific, and rhombomere-independent cues in maintaining neural crest-free zones in the embryonic head. Dev. Biol. 266, 361–372. doi: 10.1016/j.ydbio.2003.11.003
Golding, J. P., Trainor, P., Krumlauf, R., and Gassmann, M. (2000). Defects in pathfinding by cranial neural crest cells in mice lacking the neuregulin receptor ErbB4. Nat. Cell Biol. 2, 103–109.
Gould, A., Itasaki, N., and Krumlauf, R. (1998). Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron 21, 39–51. doi: 10.1016/S0896-6273(00)80513-9
Goulding, M. D., Chalepakis, G., Deutsch, U., Erselius, J. R., and Gruss, P. (1991). Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 10, 1135–1147.
Graham, A., Papalopulu, N., and Krumlauf, R. (1989). The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57, 367–378. doi: 10.1016/0092-8674(89)90912-4
Guillemot, F. (1999). Vertebrate bHLH genes and the determination of neuronal fates. Exp. Cell Res. 253, 357–364. doi: 10.1006/excr.1999.4717
Hall, B. K. (1999). The Neural Crest in Development and Evolution. Berlin: Springer Science & Business Media.
Hall, B. K. (2008). The neural crest and neural crest cells: discovery and significance for theories of embryonic organization. J. Biosci. 33, 781–793. doi: 10.1007/s12038-008-0098-4
Han, J., Ito, Y., Yeo, J. Y., Sucov, H. M., Maas, R., and Chai, Y. (2003). Cranial neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19(INK4d) expression during odontogenesis. Dev. Biol. 261, 183–196. doi: 10.1016/s0012-1606(03)00300-2
Hari, L., Miescher, I., Shakhova, O., Suter, U., Chin, L., Taketo, M., et al. (2012). Temporal control of neural crest lineage generation by Wnt/ -catenin signaling. Development 139, 2107–2117. doi: 10.1242/dev.073064
Hatakeyama, J., Sakamoto, S., and Kageyama, R. (2006). Hes1 and Hes5 regulate the development of the cranial and spinal nerve systems. Dev. Neurosci. 28, 92–101. doi: 10.1159/000090756
Havrda, M. C., Harris, B. T., Mantani, A., Ward, N. M., Paolella, B. R., Cuzon, V. C., et al. (2008). Id2 is required for specification of dopaminergic neurons during adult olfactory neurogenesis. J. Neurosci. 28, 14074–14086. doi: 10.1523/JNEUROSCI.3188-08.2008
Hempstead, B. L., Martin-Zanca, D., Kaplan, D. R., Parada, L. F., and Chao, M. V. (1991). High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature 350, 678–683. doi: 10.1038/350678a0
Hendershot, T. J., Liu, H., Clouthier, D. E., Shepherd, I. T., Coppola, E., Studer, M., et al. (2008). Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev. Biol. 319, 179–191. doi: 10.1016/j.ydbio.2008.03.036
Hendershot, T. J., Liu, H., Sarkar, A. A., Giovannucci, D. R., Clouthier, D. E., Abe, M., et al. (2007). Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev. Dyn. 236, 93–105. doi: 10.1002/dvdy.20989
Hernandez, R. E., Rikhof, H. A., Bachmann, R., and Moens, C. B. (2004). vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development 131, 4511–4520. doi: 10.1242/dev.01297
Heuer, J. G., Fatemie-Nainie, S., Wheeler, E. F., and Bothwell, M. (1990). Structure and developmental expression of the chicken NGF receptor. Dev. Biol. 137, 287–304. doi: 10.1016/0012-1606(90)90255-H
Hindley, C. J., Condurat, A. L., Menon, V., Thomas, R., Azmitia, L. M., Davis, J. A., et al. (2016). The Hippo pathway member YAP enhances human neural crest cell fate and migration. Sci. Rep. 6, 1–9. doi: 10.1038/srep23208
Hoch, R. V., Lindtner, S., Price, J. D., and Rubenstein, J. L. R. (2015). OTX2 transcription factor controls regional patterning within the medial ganglionic eminence and regional identity of the septum. Cell Rep. 12, 482–494. doi: 10.1016/j.celrep.2015.06.043
Holmberg, J., Genander, M., Halford, M. M., Annerén, C., Sondell, M., Chumley, M. J., et al. (2006). EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125, 1151–1163. doi: 10.1016/j.cell.2006.04.030
Hong, C. S., and Saint-Jeannet, J. P. (2005). Sox proteins and neural crest development. Semin. Cell Dev. Biol. 16, 694–703. doi: 10.1016/j.semcdb.2005.06.005
Hong, C. S., and Saint-Jeannet, J. P. (2007). The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell 18, 2192–2202. doi: 10.1091/mbc.E06-11-1047
Huang, E. J., Wilkinson, G. A., Fariñas, I., Backus, C., Zang, K., Wong, S. L., et al. (1999). Expression of Trk receptors in the developing mouse trigeminal ganglion: in vivo evidence for NT-3 activation of TrkA and TrkB in addition to TrkC. Development 126, 2191–2203.
Huang, J., Wu, S., Barrera, J., Matthews, K., and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434. doi: 10.1016/j.cell.2005.06.007
Hunt, P., Gulisano, M., Cook, M., Sham, M. H., Faiella, A., Wilkinson, D., et al. (1991). A distinct Hox code for the branchial region of the vertebrate head. Nature 353, 861–864.
Inoue, K.-I., Ito, K., Osato, M., Lee, B., Bae, S.-C., and Ito, Y. (2007). The transcription factor Runx3 represses the neurotrophin receptor TrkB during lineage commitment of dorsal root ganglion neurons. J. Biol. Chem. 282, 24175–24184. doi: 10.1074/jbc.M703746200
Inoue, T., Hatayama, M., Tohmonda, T., Itohara, S., Aruga, J., and Mikoshiba, K. (2004). Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 270, 146–162. doi: 10.1016/j.ydbio.2004.02.017
Ishibashi, M., Ang, S. L., Shiota, K., Nakanishi, S., Kageyama, R., and Guillemot, F. (1995). Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9, 3136–3148. doi: 10.1101/gad.9.24.3136
Ishii, M., Han, J., Yen, H. Y., Sucov, H. M., Chai, Y., and Maxson, R. E. (2005). Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 132, 4937–4950. doi: 10.1242/dev.02072
Jay, P., Iman, S., Gozé, C., Taviaux, S., Poulat, F., Couly, G., et al. (1997). SOX22 is a new member of the SOX gene family, mainly expressed in human nervous tissue. Hum. Mol. Genet. 6, 1069–1077. doi: 10.1093/hmg/6.7.1069
Jeong, J., Mao, J., Tenzen, T., Kottmann, A. H., and McMahon, A. P. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937–951.
Ji, S. J., and Jaffrey, S. R. (2012). Intra-axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron 74, 95–107. doi: 10.1016/j.neuron.2012.02.022
Jung, J. W., Ji, A. R., Lee, J., Kim, U. J., and Lee, S. T. (2002). Organization of the human PTK7 gene encoding a receptor protein tyrosine kinase-like molecule and alternative splicing of its mRNA. Biochim. Biophys. Acta 1579, 153–163. doi: 10.1016/s0167-4781(02)00536-5
Jurek, A., Genander, M., Kundu, P., Catchpole, T., He, X., Strååt, K., et al. (2016). Eph receptor interclass cooperation is required for the regulation of cell proliferation. Exp. Cell Res. 348, 10–22. doi: 10.1016/j.yexcr.2016.08.017
Kageyama, R., and Ohtsuka, T. (1999). The Notch-Hes pathway in mammalian neural development. Cell Res. 9, 179–188. doi: 10.1038/sj.cr.7290016
Kageyama, R., Ohtsuka, T., and Tomita, K. (2000). The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cell 10, 1–7. doi: 10.1007/s10059-000-0001-0
Kan, L., Israsena, N., Zhang, Z., Hu, M., Zhao, L. R., Jalali, A., et al. (2004). Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 269, 580–594. doi: 10.1016/j.ydbio.2004.02.005
Kelsh, R. N. (2006). Sorting out Sox10 functions in neural crest development. BioEssays 28, 788–798. doi: 10.1002/bies.20445
Kesseland, M., and Gruss, P. (1991). Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104. doi: 10.1016/0092-8674(91)90574-I
Kim, J., Lo, L., Dormand, E., and Anderson, D. J. (2003). SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 38, 17–31. doi: 10.1016/S0896-6273(03)00163-6
Kingsbury, T. J., Murray, P. D., Bambrick, L. L., and Krueger, B. K. (2003). Ca(2+)-dependent regulation of TrkB expression in neurons. J. Biol. Chem. 278, 40744–40748. doi: 10.1074/jbc.M303082200
Klein, R. (2012). Eph/ephrin signalling during development. Development 139, 4105–4109. doi: 10.1242/dev.074997
Kobayashi, T., and Kageyama, R. (2014). Expression dynamics and functions of hes factors in development and diseases. Curr. Top. Dev. Biol. 110, 263–283. doi: 10.1016/B978-0-12-405943-6.00007-5
Krimm, R. F. (2007). Factors that regulate embryonic gustatory development. BMC Neurosci. 8, (Suppl. 3):S4. doi: 10.1186/1471-2202-8-S3-S4
Kumar, D., Nitzan, E., and Kalcheim, C. (2019). YAP promotes neural crest emigration through interactions with BMP and Wnt activities. Cell Commun. Signal. 17, 1–17. doi: 10.1186/s12964-019-0383-x
Kurosaka, H., Trainor, P. A., Leroux-Berger, M., and Iulianella, A. (2015). Cranial nerve development requires Co-ordinated shh and canonical Wnt signaling. PLoS One 10:e0120821. doi: 10.1371/journal.pone.0120821
Labalette, C., Bouchoucha, Y. X., Wassef, M. A., Gongal, P. A., Men, J., and Le Becker, T. (2011). Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development 138, 317–326. doi: 10.1242/dev.057299
LaBonne, C., and Bronner-Fraser, M. (1998). Induction and patterning of the neural crest, a stem cell-like precursor population. J. Neurobiol. 36, 175–189. doi: 10.1002/(SICI)1097-4695(199808)36:2<175::AID-NEU6<3.0.CO;2-Z
LaMantia, A.-S., Bhasin, N., Rhodes, K., and Heemskerk, J. (2000). Mesenchymal/Epithelial induction mediates olfactory pathway formation. Neuron 28, 411–425. doi: 10.1016/s0896-6273(00)00121-5
Langston, A. W., and Gudas, L. J. (1992). Identification of a retinoic acid responsive enhancer 3’ of the murine homeobox gene Hox-1.6. Mech. Dev. 38, 217–227. doi: 10.1016/0925-4773(92)90055-O
Lavado, A., He, Y., Paré, J., Neale, G., Olson, E. N., Giovannini, M., et al. (2013). Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development 140, 3323–3334. doi: 10.1242/dev.096537
Le Douarin, N. M., Le Douarin, N. M., Fontaine-Pérus, J., and Couly, G. (1986). Cephalic ectodermal placodes and neurogenesis. Trends Neurosci. 9, 175–180. doi: 10.1016/0166-2236(86)90055-x
Lee, K. F., Simon, H., Chen, H., Bates, B., Hung, M. C., and Hauser, C. (1995). Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378, 394–398. doi: 10.1038/378394a0
Lee, T. I., Jenner, R. G., Boyer, L. A., Guenther, M. G., Levine, S. S., Kumar, R. M., et al. (2006). Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125, 301–313. doi: 10.1016/j.cell.2006.02.043
Lee, V. M., Sechrist, J. W., Luetolf, S., and Bronner-Fraser, M. (2003). Both neural crest and placode contribute to the ciliary ganglion and oculomotor nerve. Dev. Biol. 263, 176–190. doi: 10.1016/j.ydbio.2003.07.004
Lee, Y. M., Osumi-Yamashita, N., Ninomiya, Y., Moon, C. K., Eriksson, U., and Eto, K. (1995). Retinoic acid stage-dependently alters the migration pattern and identity of hindbrain neural crest cells. Development 121, 825–837.
Lei, Q.-Y., Zhang, H., Zhao, B., Zha, Z.-Y., Bai, F., Pei, X.-H., et al. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426–2436. doi: 10.1128/MCB.01874-07
Lemaire, P., Relevant, O., Bravo, R., and Charnay, P. (1988). Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 85, 4691–4695. doi: 10.1073/pnas.85.13.4691
Leung, A. W., Murdoch, B., Salem, A. F., Prasad, M. S., Gomez, G. A., and Garcia-Castro, M. I. (2016). WNT/ -catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 143, 398–410. doi: 10.1242/dev.130849
Lewin, G. R., and Barde, Y. A. (1996). Physiology of the neurotrophins. Annu. Rev. Neurosci. 19, 289–317. doi: 10.1146/annurev.ne.19.030196.001445
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi: 10.1038/276565a0
Lewis, S. L., Khoo, P.-L., De Young, R. A., Steiner, K., Wilcock, C., Mukhopadhyay, M., et al. (2008). Dkk1 and Wnt3 interact to control head morphogenesis in the mouse. Development 135, 1791–1801. doi: 10.1242/dev.018853
Li, B., Kuriyama, S., Moreno, M., and Mayor, R. (2009). The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development 136, 3267–3278. doi: 10.1242/dev.036954
Li, L., Pu, Q., Hintze, M., Wang, Y., Eckhardt, M., Gieselmann, V., et al. (2020). BDNF and NGF signals originating from sensory ganglia promote cranial motor axon growth. Exp. Brain Res. 238, 111–119. doi: 10.1007/s00221-019-05694-w
Li, X., Barkho, B. Z., Luo, Y., Smrt, R. D., Santistevan, N. J., Liu, C., et al. (2008). Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J. Biol. Chem. 283, 27644–27652. doi: 10.1074/jbc.M804899200
Lim, Y.-S., McLaughlin, T., Sung, T.-C., Santiago, A., Lee, K.-F., and O’Leary, D. D. M. (2008). p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron 59, 746–758. doi: 10.1016/j.neuron.2008.07.032
Lindsay, R. M. (1996). Role of neurotrophins and trk receptors in the development and maintenance of sensory neurons: an overview. Philos Trans R Soc L. B Biol Sci 351, 365–373.
Liu, M., Zhao, S., Lin, Q., and Wang, X.-P. (2015). YAP regulates the expression of Hoxa1 and Hoxc13 in mouse and human oral and skin epithelial tissues. Mol. Cell. Biol. 35, 1449–1461. doi: 10.1128/mcb.00765-14
Lo, L., Tiveron, M. C., and Anderson, D. J. (1998). MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of autonomic neuronal identity. Development 125, 609–620.
Lu, X., Borchers, A. G. M., Jolicoeur, C., Rayburn, H., Baker, J. C., and Tessier-Lavigne, M. (2004). PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature 430, 93–98. doi: 10.1038/nature02677
Lufkin, T., Dierich, A., LeMeur, M., Mark, M., and Chambon, P. (1991). Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66, 1105–1119.
Lumsden, A., and Keynes, R. (1989). Segmental patterns of neuronal development in the chick hindbrain. Nature 337, 424–428. doi: 10.1038/337424a0
Lumsden, A., Sprawson, N., and Graham, A. (1991). Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development 113, 1281–1291.
Luukko, K., Løes, S., Kvinnsland, I. H., and Kettunen, P. (2005). Expression of ephrin-A ligands and EphA receptors in the developing mouse tooth and its supporting tissues. Cell Tissue Res. 319, 143–152. doi: 10.1007/s00441-004-0951-1
Ma, D. K., Jang, M. H., Guo, J. U., Kitabatake, Y., Chang, M. L., Pow-anpongkul, N., et al. (2009). Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. doi: 10.1126/science.1166859
Ma, Q., Chen, Z., del Barco Barrantes, I., de la Pompa, J. L., and Anderson, D. J. (1998). neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20, 469–482.
Ma, Q., Fode, C., Guillemot, F., and Anderson, D. J. (1999). NEUROGENIN1 and NEUROGENIN2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 13, 1717–1728. doi: 10.1101/gad.13.13.1717
Mangelsdorf, D. J., Borgmeyer, U., Heyman, R. A., Zhou, J. Y., Ong, E. S., Oro, A. E., et al. (1992). Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 6, 329–344. doi: 10.1101/gad.6.3.329
Mangelsdorf, D. J., and Evans, R. M. (1995). The RXR heterodimers and orphan receptors. Cell 83, 841–850. doi: 10.1016/0092-8674(95)90200-7
Manley, N. R., and Capecchi, M. R. (1995). The role of Hoxa-3 in mouse thymus and thyroid development. Development 121, 1989–2003.
Manley, N. R., and Capecchi, M. R. (1997). Hox group 3 paralogous genes act synergistically in the formation of somitic and neural crest-derived structures. Dev. Biol. 192, 274–288.
Manley, N. R., and Capecchi, M. R. (1998). Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Dev. Biol. 195, 1–15.
Manzanares, M., Trainor, P. A., Nonchev, S., Ariza-McNaughton, L., Brodie, J., Gould, A., et al. (1999). The role of kreisler in segmentation during hindbrain development. Dev. Biol. 211, 220–237. doi: 10.1006/dbio.1999.9318
Marchant, L., Linker, C., Ruiz, P., Guerrero, N., and Mayor, R. (1998). The inductive properties of mesoderm suggest that the neural crest cells are specified by a BMP gradient. Dev. Biol. 198, 319–329.
Marler, K. J. M., Becker-Barroso, E., Martínez, A., Llovera, M., Wentzel, C., Poopalasundaram, S., et al. (2008). A TrkB/EphrinA interaction controls retinal axon branching and synaptogenesis. J. Neurosci. 28, 12700–12712. doi: 10.1523/JNEUROSCI.1915-08.2008
Marshall, H., Nonchev, S., Sham, M. H., Muchamore, I., Lumsden, A., and Krumlauf, R. (1992). Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature 360, 737–741.
Marshall, H., Studer, M., Pöpperl, H., Aparicio, S., Kuroiwa, A., Brenner, S., et al. (1994). A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 370, 567–571. doi: 10.1038/370567a0
Martinsen, B. J., and Bronner-Fraser, M. (1998). Neural crest specification regulated by the helix-loop-helix repressor Id2. Science 281, 988–991. doi: 10.1126/science.281.5379.988
Matsumura, S., Quispe-Salcedo, A., Schiller, C. M., Shin, J. S., Locke, B. M., Yakar, S., et al. (2017). IGF-1 mediates EphrinB1 activation in regulating tertiary dentin formation. J. Dent. Res. 96, 1153–1161. doi: 10.1177/0022034517708572
Matsuo, I., Kuratani, S., Kimura, C., Takeda, N., and Aizawa, S. (1995). Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 9, 2646–2658. doi: 10.1101/gad.9.21.2646
Maves, L., Jackman, W., and Kimmel, C. B. (2002). FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development 129, 3825–3837.
McClatchey, A. I., Saotome, I., Mercer, K., Crowley, D., Gusella, J. F., Bronson, R. T., et al. (1998). Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev. 12, 1121–1133. doi: 10.1101/gad.12.8.1121
McDonald, N. Q., and Hendrickson, W. A. (1993). A structural superfamily of growth factors containing a cystine knot motif. Cell 73, 421–424. doi: 10.1016/0092-8674(93)90127-c
McGinnis, W., and Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi: 10.1016/0092-8674(92)90471-N
McGraw, H. F., Nechiporuk, A., and Raible, D. W. (2008). Zebrafish dorsal root ganglia neural precursor cells adopt a glial fate in the absence of neurogenin1. J. Neurosci. 28, 12558–12569. doi: 10.1523/JNEUROSCI.2079-08.2008
McKay, I. J., Lewis, J., and Lumsden, A. (1997). Organization and development of facial motor neurons in the Kreisler mutant mouse. Eur. J. Neurosci. 9, 1499–1506. doi: 10.1111/j.1460-9568.1997.tb01504.x
McLennan, R., and Kulesa, P. M. (2007). In vivo analysis reveals a critical role for neuropilin-1 in cranial neural crest cell migration in chick. Dev Biol 301, 227–239.
McLennan, R., Teddy, J. M., Kasemeier-Kulesa, J. C., Romine, M. H., and Kulesa, P. M. (2010). Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Dev. Biol. 339, 114–125. doi: 10.1016/j.ydbio.2009.12.022
McMahon, S. B., Armanini, M. P., Ling, L. H., and Phillips, H. S. (1994). Expression and coexpression of Trk receptors in subpopulations of adult primary sensory neurons projecting to identified peripheral targets. Neuron 12, 1161–1171. doi: 10.1016/0896-6273(94)90323-9
McNeill, E. M., Roos, K. P., Moechars, D., and Clagett-Dame, M. (2010). Nav2 is necessary for cranial nerve development and blood pressure regulation. Neural Dev. 5, 1–14. doi: 10.1186/1749-8104-5-6
Mead, T. J., and Yutzey, K. E. (2012). Notch pathway regulation of neural crest cell development in vivo. Dev. Dyn. 241, 376–389. doi: 10.1002/dvdy.23717
Mellott, D. O., and Burke, R. D. (2008). Divergent roles for Eph and ephrin in avian cranial neural crest. BMC Dev. Biol. 8:56. doi: 10.1186/1471-213x-8-56
Menegola, E., Broccia, M. L., Di Renzo, F., Massa, V., and Giavini, E. (2004). Relationship between hindbrain segmentation, neural crest cell migration and branchial arch abnormalities in rat embryos exposed to fluconazole and retinoic acid in vitro. Reprod. Toxicol. 18, 121–130. doi: 10.1016/j.reprotox.2003.09.004
Menendez, L., Yatskievych, T. A., Antin, P. B., and Dalton, S. (2011). Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. U.S.A. 108, 19240–19245. doi: 10.1073/pnas.1113746108
Merrill, A. E., Bochukova, E. G., Brugger, S. M., Ishii, M., Pilz, D. T., Wall, S. A., et al. (2006). Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum. Mol. Genet. 15, 1319–1328. doi: 10.1093/hmg/ddl052
Meulemans, D., and Bronner-Fraser, M. (2004). Gene-regulatory interactions in neural crest evolution and development. Dev. Cell 7, 291–299. doi: 10.1016/j.devcel.2004.08.007
Meyer, D., and Birchmeier, C. (1995). Multiple essential functions of neuregulin in development. Nature 378, 386–390. doi: 10.1038/378386a0
Milet, C., MacZkowiak, F., Roche, D. D., and Monsoro-Burq, A. H. (2013). Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proc. Natl. Acad. Sci. U.S.A. 110, 5228–5233. doi: 10.1073/pnas.1219124110
Milewski, R. C., Chi, N. C., Li, J., Brown, C., Lu, M. M., and Epstein, J. A. (2004). Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development 131, 829–837. doi: 10.1242/dev.00975
Minchin, J. E. N., and Hughes, S. M. (2008). Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 317, 508–522. doi: 10.1016/j.ydbio.2008.02.058
Minoux, M., and Rijli, F. M. (2010). Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 137, 2605–2621. doi: 10.1242/dev.040048
Mizuseki, K., Sakamoto, T., Watanabe, K., Muguruma, K., Ikeya, M., Nishiyama, A., et al. (2003). Generation of neural crest-derived peripheral neurons and floor plate cells from mouse and primate embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 100, 5828–5833. doi: 10.1073/pnas.1037282100
Muller, F., and O’Rahilly, R. (1980). The early development of the nervous system in staged insectivore and primate embryos. J. Comp. Neurol. 193, 741–751.
Müller, F., and O’Rahilly, R. (2011). The initial appearance of the cranial nerves and related neuronal migration in staged human embryos. Cells Tissues Organs 193, 215–238. doi: 10.1159/000320026
Muller, F., O’Rahilly, R., Müller, F., and O’Rahilly, R. (2004). Olfactory structures in staged human embryos. Cells Tissues Organs 178, 93–116. doi: 10.1159/000081720
Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., et al. (2000). A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206. doi: 10.1016/S1097-2765(00)80416-5
Murdoch, B., DelConte, C., and García-Castro, M. I. (2012). Pax7 lineage contributions to the mammalian neural crest. PLoS One 7:e41089. doi: 10.1371/journal.pone.0041089
Murray-Rust, J., McDonald, N. Q., Blundell, T. L., Hosang, M., Oefner, C., Winkler, F., et al. (1993). Topological similarities in TGF-β2, PDGF-BB and NGF define a superfamily of polypeptide growth factors. Structure 1, 153–159. doi: 10.1016/0969-2126(93)90029-g
Na, E. S., and Monteggia, L. M. (2011). The role of MeCP2 in CNS development and function. Horm. Behav. 59, 364–368. doi: 10.1016/j.yhbeh.2010.05.014
Nagai, T., Aruga, J., Takada, S., Günther, T., Spörle, R., Schughart, K., et al. (1997). The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 182, 299–313. doi: 10.1006/dbio.1996.8449
Nakao, M. (2001). Epigenetics: interaction of DNA methylation and chromatin. Gene 278, 25–31. doi: 10.1016/S0378-1119(01)00721-1
Nakata, K., Nagai, T., Aruga, J., and Mikoshiba, K. (1997). Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. U.S.A. 94, 11980–11985. doi: 10.1073/pnas.94.22.11980
Narayanan, C. H., and Narayanan, Y. (1980). Neural crest and placodal contributions in the development of the glossopharyngeal-vagal complex in the chick. Anat. Rec. 196, 71–82. doi: 10.1002/ar.1091960108
Nardelli, J., Gibson, T. J., Vesque, C., and Charnay, P. (1991). Base sequence discrimination by zinc-finger DNA-binding domains. Nature 349, 175–178. doi: 10.1038/349175a0
Nie, X., Luukko, K., and Kettunen, P. (2006). BMP signalling in craniofacial development. Int. J. Dev. Biol. 50, 511–521.
Nieto, M. A. A., Sechrist, J., Wilkinson, D. G. G., and Bronner-Fraser, M. (1995). Relationship between spatially restricted Krox-20 gene expression in branchial neural crest and segmentation in the chick embryo hindbrain. Embo J. 14, 1697–1710. doi: 10.1002/j.1460-2075.1995.tb07159.x
Noden, D. M. (1975). An analysis of the migratory behavior of avian cephalic neural crest cells. Dev. Biol. 42, 106–130. doi: 10.1016/0012-1606(75)90318-8
Noden, D. M. (1978). The control of avian cephalic neural crest cytodifferentiation. I. Skeletal and connective tissues. Dev. Biol. 67, 296–312. doi: 10.1016/0012-1606(78)90201-4
Noden, D. M. (1991). Vertebrate craniofacial development: the relation between ontogenetic process and morphological outcome. Brain. Behav. Evol. 38, 190–225. doi: 10.1159/000114388
Nohe, A., Keating, E., Knaus, P., and Petersen, N. O. (2004). Signal transduction of bone morphogenetic protein receptors. Cell. Signal. 16, 291–299. doi: 10.1016/j.cellsig.2003.08.011
Noisa, P., Lund, C., Kanduri, K., Lund, R., and La, H. (2014). Notch signaling regulates the differentiation of neural crest from human pluripotent stem cells. J. Cell Sci. 127, 2083–2094. doi: 10.1242/jcs.145755
Nolte, C., and Krumlauf, R. (2007). “Expression of Hox Genes in the Nervous System of Vertebrates,” in HOX Gene Expression, ed. S. Papageorgiou (New York, NY: Springer), 14–41. doi: 10.1007/978-0-387-68990-6_2
Noordermeer, D., Leleu, M., Splinter, E., Rougemont, J., De Laat, W., and Duboule, D. (2011). The dynamic architecture of Hox gene clusters. Science 334, 222–225. doi: 10.1126/science.1207194
Northcutt, R. G. (2005). The new head hypothesis revisited. J. Exp. Zool. Part B Mol. Dev. Evol. 304, 274–297. doi: 10.1002/jez.b.21063
Northcutt, R. G., and Gans, C. (1983). The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q. Rev. Biol. 58, 1–28. doi: 10.1086/413055
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. doi: 10.1016/j.cell.2017.05.016
Ohsako, S., Hyer, J., Panganiban, G., Oliver, I., and Caudy, M. (1994). Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 8, 2743–2755. doi: 10.1101/gad.8.22.2743
O’Rahilly, R., and Müller, F. (1984). The early development of the hypoglossal nerve and occipital somites in staged human embryos. Am. J. Anat. 169, 237–257. doi: 10.1002/aja.1001690302
O’Rahilly, R., and Müller, F. (2007). The development of the neural crest in the human. J. Anat. 211, 335–351. doi: 10.1111/j.1469-7580.2007.00773.x
Ornitz, D. M., and Itoh, N. (2015). The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266. doi: 10.1002/wdev.176
Osborne, N. J., Begbie, J., Chilton, J. K., Schmidt, H., and Eickholt, B. J. (2005). Semaphorin/neuropilin signaling influences the positioning of migratory neural crest cells within the hindbrain region of the chick. Dev. Dyn. 232, 939–949.
Packer, A. I., Crotty, D. A., Elwell, V. A., and Wolgemuth, D. J. (1998). Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development 125, 1991–1998.
Paek, H., Gutin, G., and Hébert, J. M. (2009). FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development 136, 2457–2465. doi: 10.1242/dev.032656
Parker, H. J., Pushel, I., and Krumlauf, R. (2018). Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 444, S67–S78. doi: 10.1016/j.ydbio.2018.03.016
Pattyn, A., Morin, X., Cremer, H., Goridis, C., and Brunet, J. F. (1997). Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development 124, 4065–4075.
Pattyn, A., Morin, X., Cremer, H., Goridis, C., and Brunet, J. F. (1999). The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 399, 366–370.
Paudyal, A., Damrau, C., Patterson, V. L., Ermakov, A., Formstone, C., Lalanne, Z., et al. (2010). The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC Dev. Biol. 10:87. doi: 10.1186/1471-213X-10-87
Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B., and Blundell, T. L. (2000). Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034. doi: 10.1038/35039551
Pera, E., Stein, S., and Kessel, M. (1999). Ectodermal patterning in the avian embryo: epidermis versus neural plate. Development 126, 63–73.
Perdigoto, C. N., and Bardin, A. J. (2013). Sending the right signal: notch and stem cells. Biochim. Biophys. Acta Gen. Subj. 1830, 2307–2322. doi: 10.1016/j.bbagen.2012.08.009
Perez, S. E., Rebelo, S., and Anderson, D. J. (1999). Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development 126, 1715–1728.
Petkova, T. D., Seigel, G. M., and Otteson, D. C. (2011). A role for DNA methylation in regulation of EphA5 receptor expression in the mouse retina. Vision Res. 51, 260–268. doi: 10.1016/j.visres.2010.09.022
Piacentino, M. L., and Bronner, M. E. (2018). Intracellular attenuation of BMP signaling via CKIP-1/Smurf1 is essential during neural crest induction. PLoS Biol. 16:e2004425. doi: 10.1371/journal.pbio.2004425
Pirvola, U., Spencer-Dene, B., Xing-Qun, L., Kettunen, P., Thesleff, I., Fritzsch, B., et al. (2000). FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J. Neurosci. 20, 6125–6134.
Podleschny, M., Grund, A., Berger, H., Rollwitz, E., and Borchers, A. (2015). A PTK7/Ror2 co-receptor complex affects xenopus neural crest migration. PLoS One 10:e0145169. doi: 10.1371/journal.pone.0145169
Pombo, P. M., Barettino, D., Espliguero, G., Metsis, M., Iglesias, T., and Rodriguez-Pena, A. (2000). Transcriptional repression of neurotrophin receptor trkB by thyroid hormone in the developing rat brain. J. Biol. Chem. 275, 37510–37517. doi: 10.1074/jbc.M006440200
Power, S. C., Lancman, J., and Smith, S. M. (1999). Retinoic acid is essential for shh/hoxd signaling during rat limb outgrowth but not for limb initiation. Dev. Dyn. 216, 469–480. doi: 10.1002/(sici)1097-0177(199912)216:4/5<469::aid-dvdy15<3.0.co;2-3
Prasad, M. S., Uribe-Querol, E., Marquez, J., Vadasz, S., Yardley, N., Shelar, P. B., et al. (2020). Blastula stage specification of avian neural crest. Dev. Biol. 458, 64–74. doi: 10.1016/j.ydbio.2019.10.007
Proskorovski-Ohayon, R., Kadir, R., Michalowski, A., Flusser, H., Perez, Y., Hershkovitz, E., et al. (2017). PAX7 mutation in a syndrome of failure to thrive, hypotonia, and global neurodevelopmental delay. Hum. Mutat. 38, 1671–1683. doi: 10.1002/humu.23310
Puelles, L., and Rubenstein, J. L. R. (1993). Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 16, 472–479. doi: 10.1016/0166-2236(93)90080-6
Reaume, A. G., Conlon, R. A., Zirngibl, R., Yamaguchi, T. P., and Rossant, J. (1992). Expression analysis of a Notch homologue in the mouse embryo. Dev. Biol. 154, 377–387. doi: 10.1016/0012-1606(92)90076-S
Riethmacher, D., Sonnenberg-Riethmacher, E., Brinkmann, V., Yamaai, T., Lewin, G. R., and Birchmeier, C. (1997). Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature 389, 725–730. doi: 10.1038/39593
Rijli, F. M., Mark, M., Lakkaraju, S., Dierich, A., Dolle, P., and Chambon, P. (1993). A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 75, 1333–1349.
Roschger, C., and Cabrele, C. (2017). The Id-protein family in developmental and cancer-associated pathways Fritz Aberger. Cell Commun. Signal. 15, 1–26. doi: 10.1186/s12964-016-0161-y
Rossel, M., and Capecchi, M. R. (1999). Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126, 5027–5040.
Sabra, H., Brunner, M., Mandati, V., Wehrle-Haller, B., Lallemand, D., Ribba, A. S., et al. (2017). β1 integrin–dependent Rac/group I PAK signaling mediates YAP activation of Yes-associated protein 1 (YAP1) via NF2/merlin. J. Biol. Chem. 292, 19179–19197. doi: 10.1074/jbc.M117.808063
Sanchez-Ferras, O., Bernas, G., Laberge-Perrault, E., and Pilon, N. (2014). Induction and dorsal restriction of Paired-box 3 (Pax3) gene expression in the caudal neuroectoderm is mediated by integration of multiple pathways on a short neural crest enhancer. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 546–558. doi: 10.1016/j.bbagrm.2014.04.023
Sanchez-Ferras, O., Coutaud, B., Samani, T. D., Tremblay, I., Souchkova, O., and Pilon, N. (2012). Caudal-related homeobox (Cdx) protein-dependent integration of canonical Wnt signaling on Paired-box 3 (Pax3) neural crest enhancer. J. Biol. Chem. 287, 16623–16635. doi: 10.1074/jbc.M112.356394
Sandell, L. L., Butler Tjaden, N. E., Barlow, A. J., and Trainor, P. A. (2014). Cochleovestibular nerve development is integrated with migratory neural crest cells. Dev. Biol. 385, 200–210. doi: 10.1016/j.ydbio.2013.11.009
Sarkar, S., Petiot, A., Copp, A., Ferretti, P., and Thorogood, P. (2001). FGF2 promotes skeletogenic differentiation of cranial neural crest cells. Development 128, 2143–2152.
Sasai, Y., Kageyama, R., Tagawa, Y., Shigemoto, R., and Nakanishi, S. (1992). Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 6, 2026–2034. doi: 10.1101/gad.6.12b.2620
Sato, T., Joyner, A. L., and Nakamura, H. (2004). How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev. Growth Differ. 46, 487–494. doi: 10.1111/j.1440-169x.2004.00769.x
Sauka-Spengler, T., and Bronner-Fraser, M. (2006). Development and evolution of the migratory neural crest: a gene regulatory perspective. Curr. Opin. Genet. Dev. 16, 360–366. doi: 10.1016/j.gde.2006.06.006
Schlessinger, J., Plotnikov, A. N., Ibrahimi, O. A., Eliseenkova, A. V., Yeh, B. K., and Yayon, A. (2000). Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750. doi: 10.1016/s1097-2765(00)00073-3
Schneider-Maunoury, S., Seitanidou, T., Charnay, P., and Lumsden, A. (1997). Segmental and neuronal architecture of the hindbrain of Krox-20 mouse mutants. Development 124, 1215–1226.
Schneider-Maunoury, S., Topilko, P., Seitanidou, T., Levi, G., Cohen-Tannoudji, M., Pournin, S., et al. (1993). Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75, 1199–1214. doi: 10.1016/0092-8674(93)90329-O
Schwarz, Q., Gu, C., Fujisawa, H., Sabelko, K., Gertsenstein, M., Nagy, A., et al. (2004). Vascular endothelial growth factor controls neuronal migration and cooperates with SemaSA to pattern distinct compartments of the facial nerve. Genes Dev. 18, 2822–2834. doi: 10.1101/gad.322904
Schwarz, Q., Vieira, J. M., Howard, B., Eickholt, B. J., and Ruhrberg, C. (2008). Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development 135, 1605–1613. doi: 10.1242/dev.015412
Serbedzija, G. N., Bronner-Fraser, M., and Fraser, S. E. (1992). Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 116, 297–307. doi: 10.1080/10408390802565889
Shiau, C. E., Lwigale, P. Y., Das, R. M., Wilson, S. A., and Bronner-Fraser, M. (2008). Robo2-Slit1 dependent cell-cell interactions mediate assembly of the trigeminal ganglion. Nat. Neurosci. 11, 269–276. doi: 10.1038/nn2051
Simkin, J. E., Zhang, D., Rollo, B. N., and Newgreen, D. F. (2013). Retinoic acid upregulates ret and induces chain migration and population expansion in vagal neural crest cells to colonise the embryonic gut. PLoS One 8:e64077. doi: 10.1371/journal.pone.0064077
Simões-Costa, M., and Bronner, M. E. (2013). Insights into neural crest development and evolution from genomic analysis. Genome Res. 23, 1069–1080. doi: 10.1101/gr.157586.113
Simões-Costa, M., Bronner, M. E., Simoes-Costa, M., and Bronner, M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. doi: 10.1242/dev.105445
Singh, S., and Groves, A. K. (2016). The molecular basis of craniofacial placode development. Wiley Interdiscip. Rev. Dev. Biol. 5, 363–376. doi: 10.1002/wdev.226
Smith, A., Robinson, V., Patel, K., and Wilkinson, D. G. (1997). The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr. Biol. 7, 561–570.
Sperber, S. M., Saxena, V., Hatch, G., and Ekker, M. (2008). Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev. Biol. 314, 59–70. doi: 10.1016/j.ydbio.2007.11.005
Steventon, B., Araya, C., Linker, C., Kuriyama, S., and Mayor, R. (2009). Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development 136, 771–779. doi: 10.1242/dev.029017
Steventon, B., and Mayor, R. (2012). Early neural crest induction requires an initial inhibition of Wnt signals. Dev. Biol. 365, 196–207. doi: 10.1016/j.ydbio.2012.02.029
Steventon, B., Mayor, R., and Streit, A. (2012). Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev. Biol. 367, 55–65. doi: 10.1016/j.ydbio.2012.04.025
Steventon, B., Mayor, R., and Streit, A. (2014). Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol. 389, 28–38. doi: 10.1016/j.ydbio.2014.01.021
Stokowski, A., Shi, S., Sun, T., Bartold, P. M., Koblar, S. A., and Gronthos, S. (2007). EphB/ephrin-B interaction mediates adult stem cell attachment, spreading, and migration: implications for dental tissue repair. Stem Cells 25, 156–164.
Studer, M., Gavalas, A., Marshall, H., Ariza-McNaughton, L., Rijli, F. M., Chambon, P., et al. (1998). Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125, 1025–1036.
Studer, M., Lumsden, A., Ariza-McNaughton, L., Bradley, A., and Krumlauf, R. (1996). Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384, 630–634. doi: 10.1038/384630a0
Studer, M., Pöpperl, H., Marshall, H., Kuroiwa, A., Krumlauf, R., Popperl, H., et al. (1994). Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science 265, 1728–1732. doi: 10.1126/science.7916164
Swiatek, P. J., and Gridley, T. (1993). Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 7, 2071–2084. doi: 10.1101/gad.7.11.2071
Szabó, A., Melchionda, M., Nastasi, G., Woods, M. L., Campo, S., Perris, R., et al. (2016). In vivo confinement promotes collective migration of neural crest cells. J. Cell Biol. 213, 543–555. doi: 10.1083/jcb.201602083
Takahashi, M., and Osumi, N. (2002). Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development 129, 1327–1338.
Taylor, K. M., and LaBonne, C. (2005). SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 9, 503–593. doi: 10.1016/j.devcel.2005.09.016
Theveneau, E., Steventon, B., Scarpa, E., Garcia, S., Trepat, X., Streit, A., et al. (2013). Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 15, 763–772. doi: 10.1038/ncb2772
Thiery, A., Buzzi, A. L., and Streit, A. (2020). Cell fate decisions during the development of the peripheral nervous system in the vertebrate head. Curr. Top. Dev. Biol. 139, 127–167. doi: 10.1016/bs.ctdb.2020.04.002
Tidcombe, H., Jackson-Fisher, A., Mathers, K., Stern, D. F., Gassmann, M., and Golding, J. P. (2003). Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 100, 8281–8286. doi: 10.1073/pnas.1436402100
Tischfield, M. A., Bosley, T. M., Salih, M. A. M., Alorainy, I. A., Sener, E. C., Nester, M. J., et al. (2005). Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat. Genet. 37, 1035–1037. doi: 10.1038/ng1636
Trainor, P. A., and Krumlauf, R. (2000). Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 1, 116–124. doi: 10.1038/35039056
Trainor, P. A., and Krumlauf, R. (2001). Hox genes, neural crest cells and branchial arch patterning. Curr. Opin. Cell Biol. 13, 698–705.
Tremblay, P., Kessel, M., and Gruss, P. (1995). A transgenic neuroanatomical marker identifies cranial neural crest deficiencies associated with the Pax3 mutant splotch. Dev. Biol. 17, 317–329. doi: 10.1006/dbio.1995.1284
Tribulo, C., Aybar, M. J., Nguyen, V. H., Mullins, M. C., and Mayor, R. (2003). Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development 130, 6441–6452. doi: 10.1242/dev.00878
Tribulo, C., Aybar, M. J., Sánchez, S. S., and Mayor, R. (2004). A balance between the anti-apoptotic activity of Slug and the apoptotic activity of msx1 is required for the proper development of the neural crest. Dev. Biol. 275, 325–342. doi: 10.1016/j.ydbio.2004.07.041
Trokovic, N., Trokovic, R., and Partanen, J. (2005). Fibroblast growth factor signalling and regional specification of the pharyngeal ectoderm. Int. J. Dev. Biol. 49, 797–805. doi: 10.1387/ijdb.051976nt
Vega-Lopez, G. A., Cerrizuela, S., and Aybar, M. J. (2017). Trunk neural crest cells: formation, migration and beyond. Int. J. Dev. Biol. 61, 5–15. doi: 10.1387/ijdb.160408gv
Vega-Lopez, G. A., Cerrizuela, S., Tribulo, C., and Aybar, M. J. (2018). Neurocristopathies: new insights 150 years after the neural crest discovery. Dev. Biol. 44, S110—-S143. doi: 10.1016/j.ydbio.2018.05.013
Vega-López, G. A., Bonano, M., Tríbulo, C., Fernández, J. P., Agüero, T. H., Aybar, M. J., et al. (2015). Functional analysis of Hairy genes in Xenopus neural crest initial specification and cell migration. Dev. Dyn. 244, 988–1013. doi: 10.1002/dvdy.24295
Verzijl, H. T. F. M., Van der Zwaag, B., Cruysberg, J. R. M., and Padberg, G. W. (2003). Möbius syndrome redefined: a syndrome of rhombencephalic maldevelopment. Neurology 61, 327–333. doi: 10.1212/01.WNL.0000076484.91275.CD
Vickaryous, M. K., and Hall, B. K. (2006). Human cell type diversity, evolution, development, and classification with special reference to cells derived from the neural crest. Biol. Rev. Camb. Philos. Soc. 81, 425–455. doi: 10.1017/S1464793106007068
Wahl, C. M., Noden, D. M., and Baker, R. (1994). Developmental relations between sixth nerve motor neurons and their targets in the chick embryo. Dev. Dyn. 201, 192–202. doi: 10.1002/aja.1002010209
Wahlbuhl, M., Reiprich, S., Vogl, M. R., Bösl, M. R., and Wegner, M. (2012). Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 40, 88–101. doi: 10.1093/nar/gkr734
Walshe, J., Maroon, H., McGonnell, I. M., Dickson, C., and Mason, I. (2002). Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr. Biol. 12, 1117–1123. doi: 10.1016/S0960-9822(02)00899-0
Waskiewicz, A. J., Rikhof, H. A., and Moens, C. B. (2002). Eliminating zebrafish Pbx proteins reveals a hindbrain ground state. Dev. Cell 3, 723–733. doi: 10.1016/S1534-5807(02)00319-2
White, J. C., Highland, M., Kaiser, M., and Clagett-Dame, M. (2000). Vitamin A deficiency results in the dose-dependent acquisition of anterior character and shortening of the caudal hindbrain of the rat embryo. Dev. Biol. 220, 263–284. doi: 10.1006/dbio.2000.9635
Wiellette, E. L., and Sive, H. (2003). vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development 130, 3821–3829. doi: 10.1242/dev.00572
Wilkinson, D. G. (2001). Multiple roles of eph receptors and ephrins in neural development. Nat. Rev. Neurosci. 2, 155–164. doi: 10.1038/35058515
Wilkinson, D. G., Bhatt, S., Chavrier, P., Bravo, R., and Charnay, P. (1989). Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337, 461–464. doi: 10.1038/337461a0
Wislet, S., Vandervelden, G., and Rogister, B. (2018). From neural crest development to cancer and vice versa: how p75NTR and (Pro)neurotrophins could act on cell migration and invasion? Front. Mol. Neurosci. 11:244. doi: 10.3389/fnmol.2018.00244
Xu, N.-J., and Henkemeyer, M. (2012). Ephrin reverse signaling in axon guidance and synaptogenesis. Semin. Cell Dev. Biol. 23, 58–64. doi: 10.1016/j.semcdb.2011.10.024
Yamamoto, M., and Schwarting, G. (1991). The formation of axonal pathways in developing cranial nerves. Neurosci. Res. 11, 229–260. doi: 10.1016/0168-0102(91)90008-M
Yu, H. H., and Moens, C. B. (2005). Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev. Biol. 280, 373–385. doi: 10.1016/j.ydbio.2005.01.029
Zervas, M., Blaess, S., and Joyner, A. L. (2005). Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr. Top. Dev. Biol. 69, 101–138. doi: 10.1016/S0070-2153(05)69005-9
Zhang, X., Ibrahimi, O. A., Olsen, S. K., Umemori, H., Mohammadi, M., and Ornitz, D. M. (2006). Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 281, 15694–15700. doi: 10.1074/jbc.m601252200
Zhou, C. Q., Lee, J., Henkemeyer, M. J., and Lee, K. H. (2011). Disruption of ephrin B/Eph B interaction results in abnormal cochlear innervation patterns. Laryngoscope 121, 1541–1547. doi: 10.1002/lary.21861
Zhou, R. (1997). Regulation of topographic projection by the Eph family receptor Bsk (EphA5) and its ligands. Cell Tissue Res. 290, 251–259. doi: 10.1007/s004410050929
Keywords: cranial nerve, peripheral nervous system, hindbrain, cell signaling, transcriptional regulatory network, trigeminal nerve, facial nerve, vagus nerve
Citation: Méndez-Maldonado K, Vega-López GA, Aybar MJ and Velasco I (2020) Neurogenesis From Neural Crest Cells: Molecular Mechanisms in the Formation of Cranial Nerves and Ganglia. Front. Cell Dev. Biol. 8:635. doi: 10.3389/fcell.2020.00635
Received: 06 April 2020; Accepted: 24 June 2020;
Published: 07 August 2020.
Edited by:
Michael Piper, The University of Queensland, AustraliaReviewed by:
Roberto Mayor, University College London, United KingdomCopyright © 2020 Méndez-Maldonado, Vega-López, Aybar and Velasco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel J. Aybar, bWFudWVsLmF5YmFyQGZicWYudW50LmVkdS5hcg==; Iván Velasco, aXZlbGFzY29AaWZjLnVuYW0ubXg=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.