
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 16 July 2020
Sec. Mitochondrial Research
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00595
 Ming Yang1
Ming Yang1 Chenrui Li1
Chenrui Li1 Shikun Yang2
Shikun Yang2 Ying Xiao1
Ying Xiao1 Xiaofen Xiong1
Xiaofen Xiong1 Wei Chen1
Wei Chen1 Hao Zhao1
Hao Zhao1 Qin Zhang1
Qin Zhang1 Yachun Han1
Yachun Han1 Lin Sun1*
Lin Sun1*Autophagy is a process of intracellular self-recycling and degradation that plays an important role in maintaining cell homeostasis. However, the molecular mechanism of autophagy remains to be further studied. Mitochondria-associated endoplasmic reticulum membranes (MAMs) are the region of the ER that mediate communication between the ER and mitochondria. MAMs have been demonstrated to be involved in autophagy, Ca2+ transport and lipid metabolism. Here, we discuss the composition and function of MAMs, more specifically, to emphasize the role of MAMs in regulating autophagy. Finally, some key information that may be useful for future research is summarized.
Autophagy is an evolutionarily conserved cellular process by which damaged organelles and excess proteins are degraded, and then, the decomposition products are recycled back to the cytoplasm (Mizushima and Komatsu, 2011; Kim and Lee, 2014; Onorati et al., 2018; Yang and Klionsky, 2020). Autophagy is classified into many types, such as mitophagy, lipophagy, and ribophagy, according to the cellular components or organelles that are degraded (Condello et al., 2019). Basal autophagy occurs in cells under normal conditions, and the level of autophagy changes during stress such as starvation (Fernandez et al., 2020). The development of a variety of diseases, such as inflammation (Racanelli et al., 2018) and cancer (Zeng and Ju, 2018), is accompanied by abnormal autophagy. Autophagy may be a potential target for disease treatment in the future. However, the molecular mechanism of autophagy, especially of the initiation and expansion of autophagosomes, has not been fully elucidated. Increasing evidence has shown that mitochondria-associated endoplasmic reticulum membranes (MAMs) are indispensable in the autophagy process, and many proteins that are directly involved in autophagy are located in MAMs (Betz et al., 2013). However, the relationship between MAMs and autophagy is not well understood. Here, we discuss the current evidence supporting the important role of MAMs in autophagy.
Previous studies have indicated that cellular organelles, such as mitochondria and the endoplasmic reticulum (ER), play independent biological roles. However, increasing evidence has shown that organelles are not independent structures, and it has been found that there is a physical connection between the ER and mitochondria, which has been named the MAMs (van Vliet and Agostinis, 2018). The relationship between mitochondria and the ER was observed in rat liver cells by Bernhard et al. (1952) and Bernhard and Rouiller (1956) and further observed by Copeland and Dalton in their studies of the pseudobranch gland of a teleost (Copeland and Dalton, 1959). It was not until 1990 when, due to the development of biological technology, Vance isolated “fraction X” from rat livers and named it the “MAMs” (Vance, 1990). Electron microscopy has shown that the ER and mitochondria can interact at a distance of approximately 10–20 nm (Csordas et al., 2006).
As a bridge between the ER and mitochondria, the MAMs are the dynamic connection that is composed of the subdomain of the ER, the outer mitochondrial membrane (OMM) and a series of proteins. Recently, more than 1,000 proteins have been found in MAMs fragments by mass spectrometric analysis (Sala-Vila et al., 2016). In addition, Del et al. (2017) used immunoprecipitation combined with a proteomic approach and revealed that the proteins that interact with AβPP on MAMs have the following main functions: mitochondrial function and lipid metabolism. Using peroxidase-mediated proximity biotinylation, Hung et al. identified 634 and 137 proteins on the ER and mitochondria, respectively. Upon Intersecting these proteins, 68 proteins were found to be localized to the MAMs (Hung et al., 2017). In addition to the above, mass spectrometric analysis of MAMs proteins has been performed by several laboratories (Zahedi et al., 2006; Poston et al., 2013; Ma et al., 2017; Wang et al., 2018). Since the MAMs are the signal communication platform, it mainly relies on proteins to perform its various functions, and the proteins located in MAMs are grouped according to their primary functions, for instance, Ca2+ transport: inositol 1,4,5-triphosphate receptor (IP3R) and voltage-dependent anion channel (VDAC1) (Tubbs et al., 2014; D’Eletto et al., 2018); lipid metabolism: acyl coenzyme A-cholesterol acyltransferase (ACAT) (Rusinol et al., 1994), acyl CoA:diacylgycerol acyltransferase 2 (DGAT2) (Stone et al., 2009); autophagy: autophagy related 14 (ATG14), autophagy related 5 (ATG5) (Hamasaki et al., 2013); and insulin signaling: protein kinase B (PKB), mammalian target of rapamycin complex (mTORC) (Betz et al., 2013; Rieusset, 2017). These multifunctional protein groups also suggest that the MAM play an important role in maintaining intracellular homeostasis and biological functions.
MAMs are the region of the ER that mediate communication between the ER and mitochondria (Annunziata et al., 2018; Lee and Min, 2018). The integrity of the MAMs is the basis of its biological function. Some proteins found in MAMs are involved in different biochemical reactions in the cell, while others are involved in maintaining the structural stability of the MAMs, and their absence destroys the integrity of the MAMs. The ER-mitochondria encounter structure (ERMES) is the protein complex that connects the ER and mitochondria in yeast cells (Kundu and Pasrija, 2020). The ERMES contains four core proteins: maintenance of mitochondrial morphology 1 (Mmm1), which is an anchoring ER protein; mitochondrial distribution and morphology protein 12 (Mdm12), which is a cytoplasmic junction protein; and Mdm34 and Mdm10, which are two OMM proteins (Lang et al., 2015). The physical Mmm1-Mdm12-Mdm34/Mdm10 interaction mediates efficient lipid transport, especially the transport of phospholipids between the ER and mitochondria (Kawano et al., 2018); an abnormal ERMES leads to dysregulated lipid exchange between the ER and mitochondria, resulting in abnormal cell growth (Kornmann et al., 2009).
The ER-mitochondria connections in mammalians are more complicated than those in yeast. The most important group of proteins involved in ER-mitochondria coupling is IP3R/Grp75/VDAC. IP3R is one of the most important calcium channels that is located in the ER and controls the release of Ca2+, thus affecting cellular metabolism and autophagy (Kania et al., 2017; Valladares et al., 2018). VDAC is a Ca2+-related protein located in the OMM that mediates the uptake of Ca2+ by mitochondria (Lipper et al., 2019). Grp75, a member of the heat shock protein 70 family, binds to IP3R and VDAC, improving the stability of the interaction and thus increasing the efficiency of Ca2+ transfer (Xu et al., 2018). Moreover, the sigma-1 receptor (Sig-1R) is a chaperone that is also located on MAMs and affects the transport of calcium ions between the ER and mitochondria by IP3R to increase the production of ATP (Hayashi and Su, 2007; Tagashira et al., 2014). In fact, the IP3R-VDAC1 complex is the core structure for calcium ion transport in MAMs, and this protein complex is also a marker of MAMs. We can use an in situ proximity ligation assay (PLA) to detect the integrity of the IP3R-VDAC1 complex to quantify MAMs (Tubbs and Rieusset, 2016; Zhu et al., 2017; Yang et al., 2019). In addition, the split-GFP-based contact site sensor (SPLICS) probe from the Cali laboratory can be used to measure the coupling between the ER and mitochondria (Cieri et al., 2018). The PTPIP51-VAPB interaction is also among the recently discovered set of interactions involved in ER-mitochondria coupling. Vesicle-associated membrane protein-associated protein B (VAPB) is a protein that is anchored to the ER membrane by a C-terminal transmembrane domain, and it plays an important role in the unfolded protein response and vesicle trafficking (Lee and Min, 2018). Protein tyrosine phosphatase-interacting protein 51 (PTPIP51) is a protein that is located in the OMM. PTPIP51 is a microtubule-associated protein, and it performs different biological functions by forming multiple protein structure complexes (Brobeil et al., 2017). Surprisingly, recent studies have shown that PTPIP51 forms protein complexes with VAPB in the MAMs that mediate calcium ion transport and autophagy (De Vos et al., 2012; Gomez-Suaga et al., 2017). The PTPIP51-VAPB complex can be regulated by other proteins. α-Synuclein is the central protein in the progression of Parkinson’s disease, and mutant α-synuclein disrupts the VAPB-PTPIP51 complex, resulting in the uncoupling of ER-mitochondria contacts and leading to dysregulated Ca2+ transfer and decreased mitochondrial ATP production in the development of Parkinson’s disease (Paillusson et al., 2017). TAR DNA-binding domain protein 43 (TDP-43) is a highly conserved and widely expressed nuclear protein (Chang et al., 2016). The accumulation of TDP-43 is associated with the development of various neurodegenerative diseases. Stoica et al. (2014) verified that overexpression of TDP-43 activated glycogen synthase kinase-3β (GSK-3β) by inhibiting its phosphorylation at serine-9, and that activated GSK-3β reduced binding of VAPB to PTPIP51, which resulted in disordered Ca2+ homeostasis. Another combination of proteins involved in ER-mitochondria conjugation is BAP31 and TOM40. B cell receptor-associated protein 31 (BAP31) is a transmembrane protein that is located in the ER and plays an important role in apoptosis and the endoplasmic reticulum-associated degradation (ERAD) pathway (Niu et al., 2017). The outer mitochondrial membrane 40 (TOM40) is the translocation enzyme complex on the OMM that promotes the translocation of external proteins into mitochondria (Gonzalez et al., 2018). Recently, Namba (2019) demonstrated that the connections formed by BAP31 and TOM40 facilitate pre-NDUFS4 transfer from the cytoplasm to the mitochondria, thereby increasing the activity of mitochondrial complex 1 and oxygen consumption. The protein combinations between the ER and mitochondria mentioned above play central roles in maintaining the structural integrity of MAMs.
In addition to the protein-protein interactions described above, some independent proteins in MAMs are also essential for maintaining ER-mitochondria conjugation. PDZD8 is an ER protein in metazoans that is functionally orthologous to Mmm1, and this protein is essential for maintaining the stability of the MAMs structure and contributes to calcium ion homeostasis in neurons (Hirabayashi et al., 2017). MFN2 is a GTPase that mediates mitochondrial fusion. In addition, MFN2 is also involved in respiration, autophagy and mitochondrial movement regulation, and in particular, MFN2 mediates the coupling of the mitochondria and the ER (Filadi et al., 2018). MFN2 in mitochondria is assembled into homo- or heterodimeric complexes with MFN2 in the ER when the mitochondrial fusion process is initiated (de Brito and Scorrano, 2008). When the expression of MFN2 is suppressed, the structure and function of the mitochondria are destroyed (Filadi et al., 2018). However, when MFN2 is overexpressed, the interaction of the ER and mitochondria is enhanced (Filadi et al., 2018), implying that MFN2 plays a significant role in the connection of the ER and mitochondria. Phosphofurin acidic cluster sorting 2 protein (PACS-2) is another protein that is involved in MAMs integrity, and it is also involved in apoptosis and autophagy (Herrera-Cruz and Simmen, 2017; Moulis et al., 2019). It has demonstrated that when PACS-2 is absent from cells, p20 is generated from BAP31 through a caspase-dependent pathway; p20 then induces mitochondrial fission by regulating Drp1, thus causing the destruction of MAMs integrity (Simmen et al., 2005). In contrast, overexpression of PACS-2 increases ER-mitochondria coupling (Herrera-Cruz and Simmen, 2017; Moulis et al., 2019). In fact, reducing the expression of PACS-2 will decrease the integrity of MAMs and inhibit the lipidation of LC3-II, thereby inhibiting autophagy (Hamasaki et al., 2013). Fetal and adult testis expressed 1 (FATE-1) is a testicular cancer antigen that is involved in uncoupling ER-mitochondria interactions and disrupting Ca2+ transfer from the ER to mitochondria, suggesting that it plays a role in regulating apoptosis (Doghman-Bouguerra et al., 2016). Because of the specific expression of FATE-1 in the testis and its low expression in other cells, the use of plasmids overexpressing FATE-1 is now a beneficial method for studying the effects of structural and functional changes of the MAMs on cell growth and metabolism. Parkin is also a protein that is involved in maintaining the integrity of MAMs; it is an E3 ubiquitin ligase and is associated with mitophagy (Wang et al., 2019). Ziviani (2018) demonstrated that Parkin alters MAMs integrity by affecting the ubiquitination of MFN2 (Basso et al., 2018). Recently, our group also verified that disulfide-bond A oxidoreductase-like protein (DsbA-L), which is a 25-kDa antioxidant enzyme that is also located in MAMs, inhibits the apoptosis of tubular cells in diabetic nephropathy by maintaining MAMs integrity (Yang et al., 2019; Figure 1). Under pathological conditions, the increase or decrease in ER-mitochondria connections caused by various factors leads to dysregulated intracellular communication signaling. However, the precise mechanism of MAMs regulation remains to be further studied.
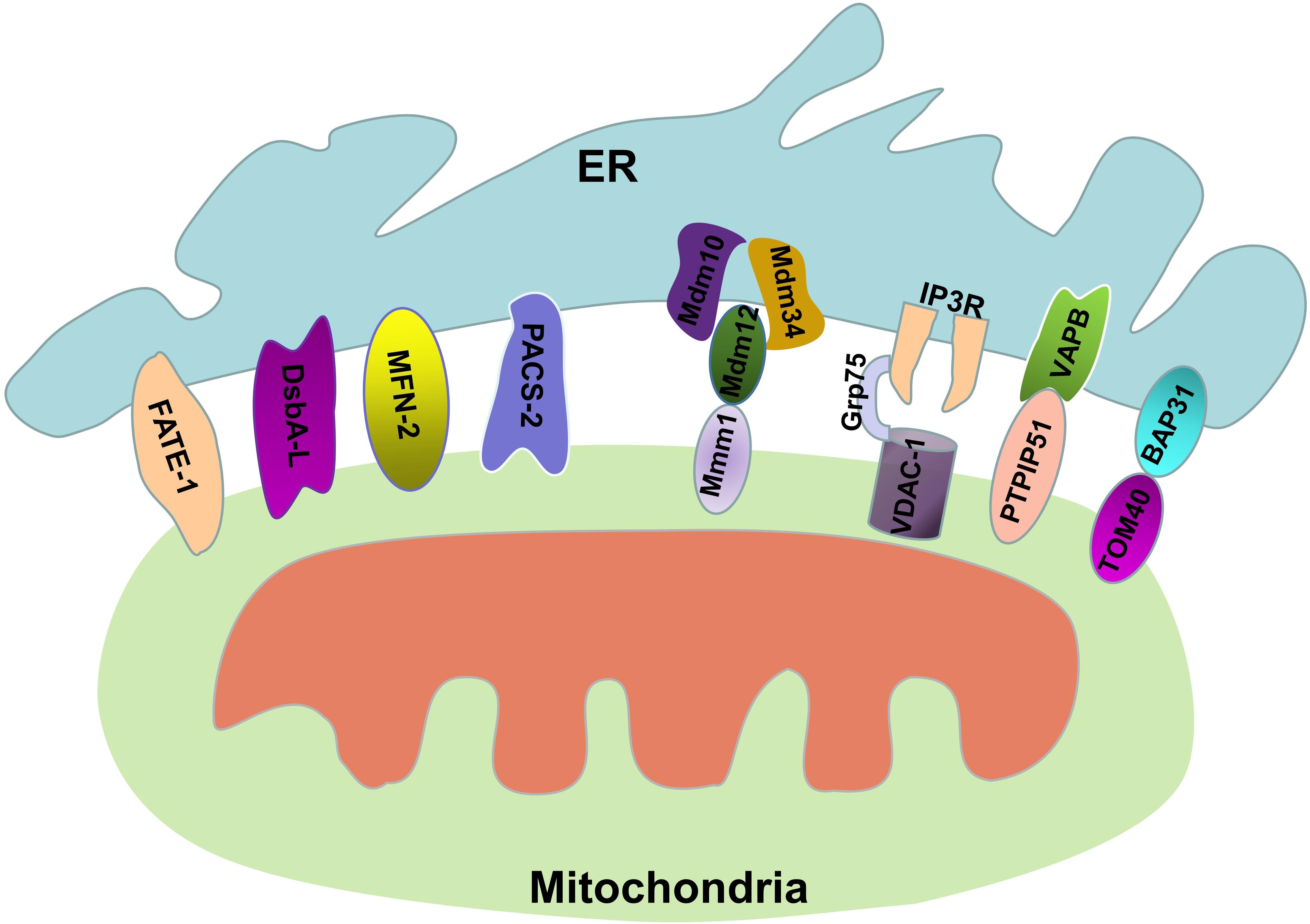
Figure 1. Protein connections involved in maintaining MAM stability. The MAM consists of parts of the OMM, ER subdomain, and some proteins. The structural stability of the MAM is maintained by these proteins.
Intracellular calcium homeostasis is the basis of cell metabolism. Concentrations of calcium ions in mitochondria that are too low can cause cellular energy metabolism disorders, while a high concentration of Ca2+ can cause cell death (Vakifahmetoglu-Norberg et al., 2017; Rossi et al., 2019). Normally, the ER releases Ca2+, which is then transported to the mitochondrial matrix, where it activates the tricarboxylic acid (TCA) cycle to stimulate ATP synthesis (Hurst et al., 2017). However, the transfer of excessive Ca2+ to the mitochondria leads to mitochondrial calcium overload, and to the opening of the mitochondrial permeability transition pore, leading to apoptosis (Pinton et al., 2008). Therefore, the MAMs act as the bridge between the ER and mitochondria, providing a buffer area for the transfer of calcium ions between the ER and mitochondria. It was Rizzuto et al. (1998) first uncovered the Ca2+ transfer function of the MAMs when they observed the spatial relation between the ER and mitochondria. When Ca2+ is released from the ER, the Ca2+ concentrations in some parts of the mitochondrial surface are much higher than those in most of the cytoplasm (Rizzuto et al., 1998); therefore, there is a structure between the ER and mitochondria that specifically transports calcium ions from the ER to mitochondria. To date, the importance of MAMs as calcium ion exchange platforms has been well established. After a cell is stimulated, the ER releases calcium ions through IP3R or ryanodine receptors (RyRs), which are the main Ca2+ channels of the ER (Wang et al., 2017; Laver, 2018). Because of the presence of MAMs, the ER and mitochondria have a spatial relationship with each other, thus allowing calcium ions to enter the mitochondria through VDAC1 on the OMM. Unlike the passive movement of Ca2+ through the OMM via the high-conductance protein VDAC1, Ca2+ movement through the inner mitochondrial membrane (IMM) is driven by an electrical gradient, and Ca2+ enters through the mitochondrial calcium uniporter (MCU). High Ca2+ concentrations on the mitochondrial membrane activate MCU to mediate the entry of Ca2+ into the mitochondrial matrix, which subsequently participates in a series of metabolic reactions (Nemani et al., 2018). As a regulatory protein, Grp75 maintains the stability of the interaction between IP3R and VDAC1, thus promoting the absorption of calcium ions by mitochondria (Xu et al., 2018).
In addition to the proteins mentioned above, there are many other proteins in the MAMs that are involved in Ca2+ transport. Sigma-1 receptor (Sig-1R) is a non-G-protein coupled chaperone of the ER that maintains the stability of IP3R to ensure appropriate Ca2+ signaling between the ER and mitochondria (Su et al., 2010). RNA-dependent protein kinase(PKR)-like ER kinase (PERK) is a key protein that is associated with ER stress, and is also abundant in MAMs. When this protein is knocked out, cells exhibit abnormal ER morphology, MAMs destruction, and calcium dysregulation (Verfaillie et al., 2012). Moreover, calnexin (CNX) is another MAMs protein that not only is involved in protein folding but also interacts with ER calcium pumps (Lynes et al., 2012). In summary, MAMs play an irreplaceable role in maintaining calcium homeostasis, and abnormal MAMs lead to dysregulation of intracellular signals and cause metabolic disorders.
In addition to Ca2+ transport, participation in lipid metabolism is also an important biological function of MAMs. Many of the enzymes involved in lipid metabolism, such as phosphatidylserine synthase (PSS) (Vance, 1990), ACAT (Rusinol et al., 1994), and DGAT2 (Stone et al., 2009), are located on MAMs. MAMs participate in lipid synthesis and transport by connecting the ER and mitochondria. However, this role of MAMs is not our focus here; please refer to Vance (2014).
Autophagy is a process of intracellular degradation in which redundant proteins or damaged components within the cell are isolated by a double-membrane vesicle called an autophagosome. Then, autophagosomes fuse with lysosomes to form autolysosomes, which are eventually degraded by lysosomal enzymes (Scrivo et al., 2018). The formation and development of autophagosomes involve a series of conserved genes called autophagy-related genes (ATGs), which encode proteins that regulate autophagy (Yu et al., 2018). The initiation of the isolation membrane is activated by the unc-51-like autophagy-activating kinase (ULK) complex, and then, the ULK complex activates the Vps34 complex by phosphorylating serine 14 of BECN1 (Ravikumar et al., 2010). This process occurs in the phosphatidylinositol 3-phosphate (PI3P)-rich subdomains of the ER. Subsequently, PI3P promotes the aggregation of various proteins (ATG18, ATG20, ATG21, and ATG24) in the assembly area, resulting in the growth of the phagophore. As the phagophore grows, some substances in the cell that need to be degraded are gradually enveloped by the phagophore (Sciarretta et al., 2018). Furthermore, the phagophore eventually matures and closes to form autophagosomes, and this process requires the participation of the ATG conjugation system. First, the ubiquitin-like protein ATG12 interacts with ATG5, and this interaction is mediated by ATG7, ATG10, and ATG3 (Sciarretta et al., 2018). The subsequent extension of the autophagosome is the result of the interaction of the ATG12-ATG5 complex with ATG16. This interaction promotes the lipidation of LC3 through its conjugation to phosphatidylethanolamine (Sciarretta et al., 2018), and LC3 promotes the maturation of the autophagosome. The mature autophagosome fuses with a lysosomes, and this fusion is mediated by soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) proteins, such as STX17, SNAP29, and VAMP8; this fusion forms the autolysosome (Nair et al., 2011; Diao et al., 2015).
The origin of autophagosomal membranes is still controversial, and the major source of membranes for autophagosome formation is unknown. Ge et al. (2017) demonstrated that the ER-Golgi intermediate compartment (ERGIC) is one of the membrane sources of autophagosomes. In addition, there is ample evidence that autophagy begins at the ER-mitochondria coupling site. As we described earlier, ERMES is the ER-mitochondria coupling complex in yeast that assembles via the Mmm1-Mdm12-Mdm34/Mdm10 interaction (Kawano et al., 2018). Surprisingly, ubiquitination of Mdm34 and Mdm12 is necessary for autophagy (Belgareh-Touze et al., 2017). This observation suggests that ER-mitochondria coupling is involved in the initiation of phagophore expansion. ATG14 is a component of the PI3K complex that is involved in autophagosome formation and is also a preautophagosome marker (Diao et al., 2015). Under starvation conditions, the content of ATG14 transferred to the MAMs increases, while ATG5, another marker of autophagosome formation, translocates to the MAMs until the autophagosome is formed, as observed by time lapse images in HeLa cells (Hamasaki et al., 2013). Further observation showed that when ER-mitochondria coupling is disrupted, ATG14 cannot be correctly localized in the MAMs, and the formation of autophagosomes is also inhibited (Hamasaki et al., 2013). This evidence fully demonstrated that MAMs play an irreplaceable role in the formation of autophagosomes, and the molecular mechanism underlying this phenomenon can also be explained by the functional roles of MAMs. Although existing studies have mainly focused on the effects of proteins on autophagosome formation, it is undeniable that lipids, especially phospholipids and sterols, also play an important role in the formation of autophagosomes. ATG8 (LC3), a ubiquitin-like protein, binds to the membrane and is a marker of isolation membrane expansion. Phosphatidylethanolamine (PE, the second most abundant phospholipid in mammalian cells) plays a very important role in this process. During isolation membrane expansion, ATG8 (LC3) is connected to PE by its C-terminal glycine residue. In vivo, PE is the main target of ATG8 (LC3), and a high level of PE can promote the connection between PE and ATG8, thereby facilitating the ATG8-mediated fusion and closure of the phagophore (Nair et al., 2012). Therefore, PE may be indispensable for the formation of autophagosomes. Moreover, another phospholipid, phosphatidylserine (PS, an important constituent of membrane structure in cells), can also act as a receptor of ATG8, and a small portion of ATG8 binds to PS (Sou et al., 2006). In addition, lipid droplets (LDs), which are the lipid storage organelles in cells, have been shown to be a critical source of lipids for the synthesis of autophagosomes (Li et al., 2015; Shpilka et al., 2015). In turn, the fatty acids (FAs) released in the process of autophagy are transferred into new LDs via DGAT1 to prevent FA-induced damage to the cell (Nguyen et al., 2017).
In addition to lipid synthesis, the dysregulation of Ca2+ in MAMs can also lead to abnormal autophagy (Ahumada-Castro et al., 2019). Elimination of etoposide-induced protein 2.4 (EI24), which is a protein located in the ER that is involved in regulating autophagy, destroys the integrity of the MAMs and inhibits autophagy in primary pancreatic β cells (Yuan et al., 2019). Further study has shown that when autophagy occurs, EI24 translocates to MAMs and interacts with the IP3R-Grp75-VDAC complex to maintain structural stability (Yuan et al., 2019). The disruption of Ca2+ signaling between the ER and mitochondria can interfere with the biological energy of cells and induce prosurvival autophagy (Cardenas et al., 2010). When ER-mitochondria Ca2+ transport is disrupted, AMPK translocates to MAMs and activates autophagy through BECN (Ahumada-Castro et al., 2019), which further confirms that the MAMs are the platform for the formation of autophagosomes. However, another group reached the opposite conclusion about the degree of MAMs integrity and autophagy. Disruption of the VAPB-PTPIP51 interaction through siRNA decreases the integrity of the MAMs, activates autophagy and overexpression of a synthetic protein that artificially increases ER-mitochondria coupling, which reduces the formation of autophagosomes (Gomez-Suaga et al., 2017). In terms of mechanism, VAPB-PTPIP51 coupling was found to affect autophagy by disrupting the transport of Ca2+ between the ER and mitochondria (Gomez-Suaga et al., 2017). Despite the seemingly opposite conclusion, we cannot deny the relationship between MAMs-mediated Ca2+ transport and autophagy.
ATG2 is also a key protein that regulates the expansion of phagophores; this protein has two subtypes, namely, ATG2A and ATG2B (Tang et al., 2017). During expansion, ATG2A translocates from the MAMs to the phagophore. ATG2A is anchored to the MAMs by a C-terminal, 45-amino-acid domain, which we called the MAMs localization domain (MLD), and TOM40 and TOM70 are responsible for the localization of ATG2A on the MAMs (Tang et al., 2019). Thus, a model was proposed in which the TOM40-TOM70 complex recruits ATG2 to the MAMs to transfer vesicular and/or non-vesicular lipids to the phagophore to enlarge the autophagosome and enhance autophagic flux (Tang et al., 2019). Similarly, promyelocytic leukemia protein (PML), which is a tumor suppressor, is located at the MAMs and controls the formation of autophagosomes by regulating the activity of the AMPK/mTOR/ULK1 pathway via affecting the transport of calcium ions from the ER to mitochondria (Missiroli et al., 2016).
Currently, one of the best understood and most well-studied pathways of mitophagy is the PTEN-induced putative kinase 1 (PINK1) and Parkin(PARK2) pathway, which is associated with the development of Parkinson’s disease (Barodia et al., 2017; Truban et al., 2017). Under physiological conditions and through a mitochondrial targeting sequence, PINK is continuously transported to the mitochondria and degraded by matrix processing peptidases (MPPs). The degradation product is then cleaved by a protease located in the mitochondrial inner membrane, namely, presenilin-associated rhomboid-like (PARL), and cleaved PINK is transported back to the cytoplasm, where it is finally degraded in lysosomes (Ashrafi and Schwarz, 2013; Wang et al., 2020). In the pathological state, the cleavage of PINK is reduced because of mitochondrial damage. The non-cleaved PINK, via a process mediated by the OMM protein translocase of the outer membrane (TOM), accumulates on the outer membrane of the mitochondria. In addition, the level of mitochondrial pyruvate can also influence the aggregation of PINK on the OMM by promoting the direct interaction between PINK1 and TOM (Park et al., 2015). The accumulated PINK on the OMM phosphorylates serine 65 (Ser65) of ubiquitin, thereby recruiting Parkin. Subsequently, PINK phosphorylates and activates Parkin on the OMM, and then, activated Parkin polyubiquitinates proteins such as VDAC1 and p62/SQSTM1 (Wang et al., 2020). The ubiquitinated substrates bind to LC3 through LIR to recruit the autophagosomal membrane around the mitochondria, and then, further extension of the autophagosomal membrane leads to fusion with lysosomes to form mature mitochondrial autophagosomes and initiate the process of mitochondrial degradation (Tanida et al., 2008; Schaaf et al., 2016). Normal PINK/Parkin pathway-mediated mitophagy is the basis for intracellular homeostasis, and defects in this process are associated with many diseases, such as Parkinson’s disease (Nardin et al., 2016) and acute kidney injury (Lin et al., 2019). As the bridge between the ER and mitochondria, the most active organelle in the cell, what is the role of the MAMs in this process?
Yang and Yang (2013) demonstrated that ubiquitinated sites gradually undergo Parkin-mediated mitophagy, and the region between the ER and damaged mitochondria is where LC3 is recruited. Consistent with this observation, BECN1, the core component of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, is also found in MAMs, where it strengthens the connection between the ER and mitochondria and promotes the formation of autophagosome precursors (Gelmetti et al., 2017); therefore, MAMs are the site of the initiation of PINK/Parkin-dependent mitophagy. However, even though PINK, Parkin and BECN1 are found in MAMs, loss of PINK prevents BECN1 from accumulating in MAMs, and this process is independent of PARK (Gelmetti et al., 2017), which suggests a new role for PINK in regulating mitophagy. In addition, the overexpression of Parkin enhances the structure and function of the ER-mitochondria connection, promotes the transfer of Ca2+ from the ER to mitochondria and increases the production of ATP in mitochondria (Cali et al., 2013). Similarly, in Parkin mutant human fibroblasts, the integrity of the MAMs was destroyed, and further research has shown that PINK-mediated destruction of MAMs integrity was achieved by affecting the ubiquitination of MFN2 (Gautier et al., 2016; Basso et al., 2018). Glycoprotein 78 (gp78), a ubiquitin ligase (E3) anchored in the ER membrane that is associated with mitophagy (Guardia-Laguarta et al., 2019), has been confirmed to be located in MAMs (Wang et al., 2000). The available evidence suggests that the core protein involved in PINK/Parkin-mediated mitophagy is located in MAMs and is involved in the regulation of MAMs integrity and function.
In mammalian cells, FUN14 domain containing 1 (FUNDC1) is involved in the receptor-mediated mitophagy pathway and is a highly conserved protein containing 155 amino acids (Zhang et al., 2017). FUNDC1 is located in the mitochondrial outer membrane protein-containing LC3-binding regions (LIR), and FUNDC1 recruits LC3 through LIR to initiate mitophagy during hypoxia (Liu et al., 2012). FUNDC1-dependent mitophagy is regulated by a variety of stress factors and cellular proteins. Under normoxic conditions, Tyr18 and Ser13 of FUNDC1 are phosphorylated by Src and casein kinase 2 (CK2), respectively, preventing it from binding to LC3 to induce autophagy (Liu et al., 2012; Chen et al., 2014). Under hypoxic conditions, the mitochondrial protein phosphatase PGAM5 mediates the dephosphorylation of Ser13, thus enabling FUNDC1 to bind to LC3 to form autophagosomes (Ma et al., 2020). In addition to Src and CK2, ULK1, which is a Ser/Thr kinase that participates in the formation of early autophagosomes, is also closely related to the mitophagy of FUNDC1. Under hypoxic conditions or treatment with FCCP, the expression of ULK1 increases, and ULK1 is recruited to the fragmented mitochondria; Moreover, transposable ULK1 interacts with FUNDC1 and promotes the phosphorylation of FUNDC1 at Ser17 to initiate autophagy (Wu et al., 2014). MARCH5 is a mitochondrial E3 ligase that can regulate mitophagy; Chen et al. demonstrated that MARCH5 directly interacts with FUNDC1, and degrades FUNDC1 by promoting its ubiquitination at lysine 119 and that the presence of MARCH5 induces the insensitivity of FUNDC1 to hypoxia-induced autophagy (Chen et al., 2017). FUNDC1 plays an essential role in receptor-mediated mitophagy, but what role does the MAMs play in this physiological process?
The direct relationship of the MAMs and FUNDC1-mediated mitophagy was demonstrated by Zhou et al., who showed that FUNDC1 is a MAM-localized protein that interacts with another MAMs protein, IP3R2, to mediate IP3R-dependent Ca2+ signaling from the ER to the mitochondria and cytosol (Wu et al., 2017). When the expression of FUNDC1 is decreased, the decreased intracellular Ca2+ levels inhibit the expression of Fis1 through Ca2+-sensitive cAMP-response element binding protein (CREB), thus causing mitochondrial dysfunction (Wu et al., 2017). Moreover, the decreased expression of FUNDC1 disrupts the interaction between the ER and mitochondria and reduces the protein abundance in MAMs (Wu et al., 2017). Another study of MAMs and FUNDC1 demonstrated that under normoxic conditions, there is a small amount of FUNDC1 in MAMs, and in response to hypoxia, FUNDC1 substantially accumulates in MAMs (Wu et al., 2016). What is the molecular mechanism by which FUNDC1 translocates to MAMs? CNX may play an indispensable role in this process. Immunoprecipitation experiments have shown that there is an interaction between the N terminus of CNX and the hydrophilic domain of FUNDC1. However, due to structural reasons, the N terminus of CNX is located in the lumen of the ER, and it is unlikely that the hydrophilic domain of FUNDC1 penetrates the lumen of the ER to interact with CNX; thus, there must be an unknown protein that mediates the interaction between CNX and FUNDC1. Depletion of CNX can inhibit FUNDC1 translocation to the MAMs under hypoxic conditions, which further confirms the role of CNX in FUNDC1 translocation (Wu et al., 2016; Figure 2). Although further studies on the role of MAMs in FUNDC1-mediated mitophagy are needed, the available evidence suggests that the MAMs provides a platform for FUNDC1 to perform its biological functions.
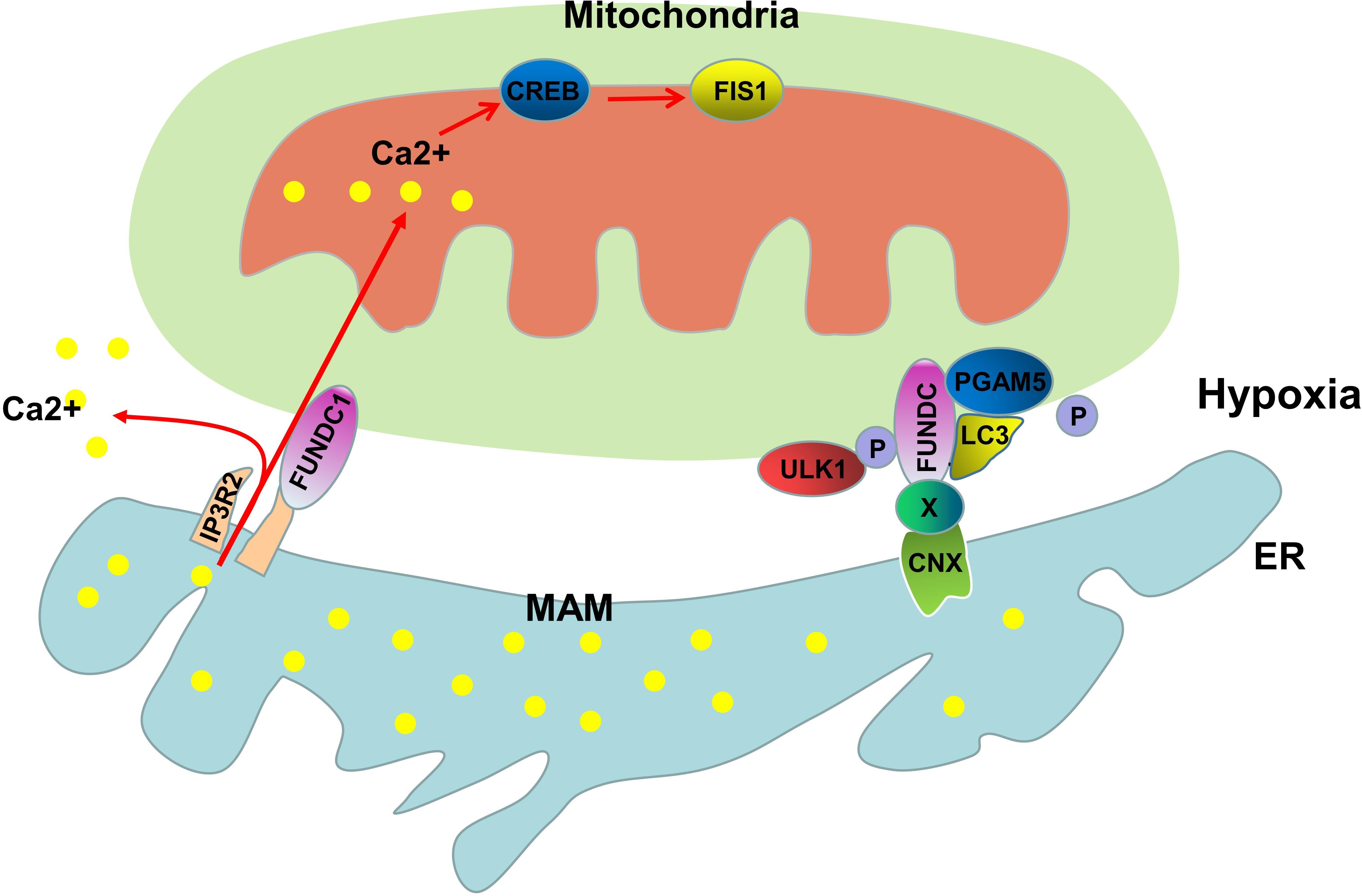
Figure 2. Molecular mechanisms of FUNDC1-mediated mitophagy. FUNDC1 is a MAM-associated protein that interacts with IP3R2 to regulate the expression of Fis1 through CREB. Under hypoxic conditions, FUNDC1 is transported to the MAM by CNX and unknown proteins (X). PGAM5-mediated dephosphorylation of Ser13 and ULK1-mediated phosphorylation of Ser17 promote the binding of FUNDC1 to LC3, leading to the initiation of mitophagy.
In addition to the proteins mentioned above, another protein involved in ER-mitochondria coupling, PACS2, also plays an important role in mitophagy. Coyne demonstrated that PACS2 mediates the integrity of the ER-mitochondria connection during stimulation with atherogenic lipids. The loss of PACS2 leads to MAMs destruction and dysregulated mitophagosome formation and mitophagy (Moulis et al., 2019).
Autophagy is an important physiological process for the maintainance of cellular homeostasis, and defects in this process are associated with many diseases, such as Parkinson’s disease, cancer and acute kidney injury (AKI). As a bridge between the ER and mitochondria, the MAM also play an important role in Ca2+ transport, lipid metabolism and autophagy. On the one hand, the MAMs serve as the platform for autophagy-related proteins to perform their biological functions. On the other hand, the Ca2+ transport and lipid metabolism functions of MAMs may be involved in autophagosome development. Disruption of the structure and function of MAMs leads to abnormal autophagy. Although existing studies have fully demonstrated the correlation between the MAMs and autophagy, there are still many questions for us to explore. What proteins mediate the involvement of MAMs in the expansion of autophagosomes? What is the relationship between MAMs and diseases caused by abnormal autophagy? The available evidence supporting a relationship between MAMs and autophagy was obtained in vitro, and experiments are needed to further verify this phenomenon in vivo. It is believed that with the development of cellular and molecular biology technology, the regulatory mechanisms of MAMs and autophagy will be continuously elucidated, and the MAMs are expected to be an important target for the treatment of diseases related to autophagy.
MY, CL, YX, YH, XX, and WC designed and performed the study. HZ conceived the project. QZ and SY supervised the work. LS wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81730018) and the National Key R&D Program of China (2018YFC1314002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahumada-Castro, U., Silva-Pavez, E., Lovy, A., Pardo, E., Molgomicron, J., and Cardenas, C. (2019). MTOR-independent autophagy induced by interrupted endoplasmic reticulum-mitochondrial Ca2+ communication: a dead end in cancer cells. Autophagy 15, 358–361. doi: 10.1080/15548627.2018.1537769
Annunziata, I., Sano, R., and D’Azzo, A. (2018). Mitochondria-associated ER membranes (MAMs) and lysosomal storage diseases. Cell Death Dis. 9:328. doi: 10.1038/s41419-017-0025-4
Ashrafi, G., and Schwarz, T. L. (2013). The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. doi: 10.1038/cdd.2012.81
Barodia, S. K., Creed, R. B., and Goldberg, M. S. (2017). Parkin and PINK1 functions in oxidative stress and neurodegeneration. Brain Res. Bull. 133, 51–59. doi: 10.1016/j.brainresbull.2016.12.004
Basso, V., Marchesan, E., Peggion, C., Chakraborty, J., von Stockum, S., Giacomello, M., et al. (2018). Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol. Res. 138, 43–56. doi: 10.1016/j.phrs.2018.09.006
Belgareh-Touze, N., Cavellini, L., and Cohen, M. M. (2017). Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy 13, 114–132. doi: 10.1080/15548627.2016.1252889
Bernhard, W., Haguenau, F., Gautier, a, and Oberling, C. (1952). Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope. Z. Zellforsch. Mikrosk. Anat. 37, 281–300.
Bernhard, W., and Rouiller, C. (1956). Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J. Biophys. Biochem. Cytol. 2, 73–78. doi: 10.1083/jcb.2.4.73
Betz, C., Stracka, D., Prescianotto-Baschong, C., Frieden, M., Demaurex, N., and Hall, M. N. (2013). Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. U.S.A. 110, 12526–12534. doi: 10.1073/pnas.1302455110
Brobeil, A., Dietel, E., Gattenlohner, S., and Wimmer, M. (2017). Orchestrating cellular signaling pathways-the cellular “conductor” protein tyrosine phosphatase interacting protein 51 (PTPIP51). Cell Tissue Res. 368, 411–423. doi: 10.1007/s00441-016-2508-5
Cali, T., Ottolini, D., Negro, A., and Brini, M. (2013). Enhanced parkin levels favor ER-mitochondria crosstalk and guarantee Ca2+ transfer to sustain cell bioenergetics. Biochim. Biophys. Acta 1832, 495–508. doi: 10.1016/j.bbadis.2013.01.004
Cardenas, C., Miller, R. A., Smith, I., Bui, T., Molgo, J., Muller, M., et al. (2010). Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142, 270–283. doi: 10.1016/j.cell.2010.06.007
Chang, X. L., Tan, M. S., Tan, L., and Yu, J. T. (2016). The Role of TDP-43 in Alzheimer’s Disease. Mol. Neurobiol. 53, 3349–3359. doi: 10.1007/s12035-015-9264-5
Chen, G., Han, Z., Feng, D., Chen, Y., Chen, L., Wu, H., et al. (2014). A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362–377. doi: 10.1016/j.molcel.2014.02.034
Chen, Z., Liu, L., Cheng, Q., Li, Y., Wu, H., Zhang, W., et al. (2017). Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep. 18, 495–509. doi: 10.15252/embr.201643309
Cieri, D., Vicario, M., Giacomello, M., Vallese, F., Filadi, R., Wagner, T., et al. (2018). SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 25, 1131–1145. doi: 10.1038/s41418-017-0033-z
Condello, M., Pellegrini, E., Caraglia, M., and Meschini, S. (2019). Targeting autophagy to overcome human diseases. Int. J. Mol. Sci. 20:725. doi: 10.3390/ijms20030725
Copeland, D. E., and Dalton, A. J. (1959). An association between mitochondria and the endoplasmic reticulum in cells of the pseudobranch gland of a teleost. J. Biophys. Biochem. Cytol. 5, 393–396. doi: 10.1083/jcb.5.3.393
Csordas, G., Renken, C., Varnai, P., Walter, L., Weaver, D., Buttle, K. F., et al. (2006). Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921. doi: 10.1083/jcb.200604016
de Brito, O. M., and Scorrano, L. (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. doi: 10.1038/nature07534
De Vos, K. J., Morotz, G. M., Stoica, R., Tudor, E. L., Lau, K. F., Ackerley, S., et al. (2012). VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21, 1299–1311. doi: 10.1093/hmg/ddr559
Del, P. D., Suski, J. M., Oules, B., Debayle, D., Gay, A. S., Lacas-Gervais, S., et al. (2017). Localization and processing of the amyloid-beta protein precursor in mitochondria-associated membranes. J. Alzheimers Dis. 55, 1549–1570. doi: 10.3233/JAD-160953
D’Eletto, M., Rossin, F., Occhigrossi, L., Farrace, M. G., Faccenda, D., Desai, R., et al. (2018). Transglutaminase type 2 regulates ER-mitochondria contact sites by interacting with GRP75. Cell Rep. 25, 3573–3581.e4. doi: 10.1016/j.celrep.2018.11.094
Diao, J., Liu, R., Rong, Y., Zhao, M., Zhang, J., Lai, Y., et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566. doi: 10.1038/nature14147
Doghman-Bouguerra, M., Granatiero, V., Sbiera, S., Sbiera, I., Lacas-Gervais, S., Brau, F., et al. (2016). FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep. 17, 1264–1280. doi: 10.15252/embr.201541504
Fernandez, A. F., Liu, Y., Ginet, V., Shi, M., Nah, J., Zou, Z., et al. (2020). Interaction between the autophagy protein Beclin 1 and Na+,K+-ATPase during starvation, exercise, and ischemia. JCI Insight 5:e133282. doi: 10.1172/jci.insight.133282
Filadi, R., Greotti, E., and Pizzo, P. (2018). Highlighting the endoplasmic reticulum-mitochondria connection: focus on Mitofusin 2. Pharmacol. Res. 128, 42–51. doi: 10.1016/j.phrs.2018.01.003
Gautier, C. A., Erpapazoglou, Z., Mouton-Liger, F., Muriel, M. P., Cormier, F., Bigou, S., et al. (2016). The endoplasmic reticulum-mitochondria interface is perturbed in PARK2 knockout mice and patients with PARK2 mutations. Hum. Mol. Genet. 25, 2972–2984. doi: 10.1093/hmg/ddw148
Ge, L., Zhang, M., Kenny, S. J., Liu, D., Maeda, M., Saito, K., et al. (2017). Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18, 1586–1603. doi: 10.15252/embr.201744559
Gelmetti, V., De Rosa, P., Torosantucci, L., Marini, E. S., Romagnoli, A., Di Rienzo, M., et al. (2017). PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 13, 654–669. doi: 10.1080/15548627.2016.1277309
Gomez-Suaga, P., Paillusson, S., Stoica, R., Noble, W., Hanger, D. P., and Miller, C. (2017). The ER-mitochondria tethering complex VAPB-PTPIP51 regulates autophagy. Curr. Biol. 27, 371–385. doi: 10.1016/j.cub.2016.12.038
Gonzalez, M. A., Auffarth, K., Honscher, C., Bohnert, M., Becker, T., Warscheid, B., et al. (2018). Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole-mitochondria contact sites. Dev. Cell 45, 621. doi: 10.1016/j.devcel.2018.05.011
Guardia-Laguarta, C., Liu, Y., Lauritzen, K. H., Erdjument-Bromage, H., Martin, B., Swayne, T. C., et al. (2019). PINK1 content in mitochondria is regulated by ER-associated degradation. J. Neurosci. 39, 7074–7085. doi: 10.1523/JNEUROSCI.1691-18.2019
Hamasaki, M., Furuta, N., Matsuda, A., Nezu, A., Yamamoto, A., Fujita, N., et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. doi: 10.1038/nature11910
Hayashi, T., and Su, T. P. (2007). Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610. doi: 10.1016/j.cell.2007.08.036
Herrera-Cruz, M. S., and Simmen, T. (2017). Over six decades of discovery and characterization of the architecture at mitochondria-associated membranes (MAMs). Adv. Exp. Med. Biol. 997, 13–31. doi: 10.1007/978-981-10-4567-7_2
Hirabayashi, Y., Kwon, S. K., Paek, H., Pernice, W. M., Paul, M. A., Lee, J., et al. (2017). ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630. doi: 10.1126/science.aan6009
Hung, V., Lam, S. S., Udeshi, N. D., Svinkina, T., Guzman, G., Mootha, V. K., et al. (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife 6:e50707. doi: 10.7554/eLife.24463
Hurst, S., Hoek, J., and Sheu, S. S. (2017). Mitochondrial Ca2+ and regulation of the permeability transition pore. J. Bioenerg. Biomembr. 49, 27–47. doi: 10.1007/s10863-016-9672-x
Kania, E., Roest, G., Vervliet, T., Parys, J. B., and Bultynck, G. (2017). IP3 receptor-mediated calcium signaling and its role in autophagy in cancer. Front. Oncol. 7:140. doi: 10.3389/fonc.2017.00140
Kawano, S., Tamura, Y., Kojima, R., Bala, S., Asai, E., Michel, A. H., et al. (2018). Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J. Cell Biol. 217, 959–974. doi: 10.1083/jcb.201704119
Kim, K. H., and Lee, M. S. (2014). Autophagy–a key player in cellular and body metabolism. Nat. Rev. Endocrinol. 10, 322–337. doi: 10.1038/nrendo.2014.35
Kornmann, B., Currie, E., Collins, S. R., Schuldiner, M., Nunnari, J., Weissman, J. S., et al. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. doi: 10.1126/science.1175088
Kundu, D., and Pasrija, R. (2020). The ERMES (Endoplasmic Reticulum and Mitochondria Encounter Structures) mediated functions in fungi. Mitochondrion 52, 89–99. doi: 10.1016/j.mito.2020.02.010
Lang, A., John, P. A., and Kornmann, B. (2015). ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr. Opin. Cell Biol. 35, 7–12. doi: 10.1016/j.ceb.2015.03.002
Laver, D. R. (2018). Regulation of the RyR channel gating by Ca2+ and Mg(2). Biophys. Rev. 10, 1087–1095. doi: 10.1007/s12551-018-0433-4
Lee, S., and Min, K. T. (2018). The interface between ER and mitochondria: molecular compositions and functions. Mol. Cells 41, 1000–1007. doi: 10.14348/molcells.2018.0438
Li, D., Song, J. Z., Li, H., Shan, M. H., Liang, Y., Zhu, J., et al. (2015). Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett. 589, 269–276. doi: 10.1016/j.febslet.2014.11.050
Lin, Q., Li, S., Jiang, N., Shao, X., Zhang, M., Jin, H., et al. (2019). PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 26:101254. doi: 10.1016/j.redox.2019.101254
Lipper, C. H., Stofleth, J. T., Bai, F., Sohn, Y. S., Roy, S., Mittler, R., et al. (2019). Redox-dependent gating of VDAC by mitoNEET. Proc. Natl. Acad. Sci. U.S.A. 116, 19924–19929. doi: 10.1073/pnas.1908271116
Liu, L., Feng, D., Chen, G., Chen, M., Zheng, Q., Song, P., et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185. doi: 10.1038/ncb2422
Lynes, E. M., Bui, M., Yap, M. C., Benson, M. D., Schneider, B., Ellgaard, L., et al. (2012). Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 31, 457–470. doi: 10.1038/emboj.2011.384
Ma, J. H., Shen, S., Wang, J. J., He, Z., Poon, A., Li, J., et al. (2017). Comparative proteomic analysis of the mitochondria-associated ER membrane (MAM) in a long-term type 2 diabetic rodent model. Sci. Rep. 7:2062. doi: 10.1038/s41598-017-02213-1
Ma, K., Zhang, Z., Chang, R., Cheng, H., Mu, C., Zhao, T., et al. (2020). Dynamic PGAM5 multimers dephosphorylate BCL-xL or FUNDC1 to regulate mitochondrial and cellular fate. Cell Death Differ. 27, 1036–1051. doi: 10.1038/s41418-019-0396-4
Missiroli, S., Bonora, M., Patergnani, S., Poletti, F., Perrone, M., Gafa, R., et al. (2016). PML at mitochondria-associated membranes is critical for the repression of autophagy and cancer development. Cell Rep. 16, 2415–2427. doi: 10.1016/j.celrep.2016.07.082
Mizushima, N., and Komatsu, M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. doi: 10.1016/j.cell.2011.10.026
Moulis, M., Grousset, E., Faccini, J., Richetin, K., Thomas, G., and Vindis, C. (2019). The multifunctional sorting protein PACS-2 controls mitophagosome formation in human vascular smooth muscle cells through mitochondria-ER contact sites. Cells 8:638. doi: 10.3390/cells8060638
Nair, U., Jotwani, A., Geng, J., Gammoh, N., Richerson, D., Yen, W. L., et al. (2011). SNARE proteins are required for macroautophagy. Cell 146, 290–302. doi: 10.1016/j.cell.2011.06.022
Nair, U., Yen, W. L., Mari, M., Cao, Y., Xie, Z., Baba, M., et al. (2012). A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 8, 780–793. doi: 10.4161/auto.19385
Namba, T. (2019). BAP31 regulates mitochondrial function via interaction with Tom40 within ER-mitochondria contact sites. Sci. Adv. 5:eaaw1386. doi: 10.1126/sciadv.aaw1386
Nardin, A., Schrepfer, E., and Ziviani, E. (2016). Counteracting PINK/Parkin deficiency in the activation of mitophagy: a potential therapeutic intervention for Parkinson’s disease. Curr. Neuropharmacol. 14, 250–259. doi: 10.2174/1570159x13666151030104414
Nemani, N., Shanmughapriya, S., and Madesh, M. (2018). Molecular regulation of MCU: implications in physiology and disease. Cell Calcium 74, 86–93. doi: 10.1016/j.ceca.2018.06.006
Nguyen, T. B., Louie, S. M., Daniele, J. R., Tran, Q., Dillin, A., Zoncu, R., et al. (2017). DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e5. doi: 10.1016/j.devcel.2017.06.003
Niu, K., Xu, J., Cao, Y., Hou, Y., Shan, M., Wang, Y., et al. (2017). BAP31 is involved in T cell activation through TCR signal pathways. Sci. Rep. 7:44809. doi: 10.1038/srep44809
Onorati, A. V., Dyczynski, M., Ojha, R., and Amaravadi, R. K. (2018). Targeting autophagy in cancer. Cancer Am. Cancer Soc. 124, 3307–3318. doi: 10.1002/cncr.31335
Paillusson, S., Gomez-Suaga, P., Stoica, R., Little, D., Gissen, P., Devine, M. J., et al. (2017). alpha-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol. 134, 129–149. doi: 10.1007/s00401-017-1704-z
Park, S., Choi, S. G., Yoo, S. M., Nah, J., Jeong, E., Kim, H., et al. (2015). Pyruvate stimulates mitophagy via PINK1 stabilization. Cell. Signal. 27, 1824–1830. doi: 10.1016/j.cellsig.2015.05.020
Pinton, P., Giorgi, C., Siviero, R., Zecchini, E., and Rizzuto, R. (2008). Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27, 6407–6418. doi: 10.1038/onc.2008.308
Poston, C. N., Krishnan, S. C., and Bazemore-Walker, C. R. (2013). In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteomics 79, 219–230. doi: 10.1016/j.jprot.2012.12.018
Racanelli, A. C., Kikkers, S. A., Choi, A., and Cloonan, S. M. (2018). Autophagy and inflammation in chronic respiratory disease. Autophagy 14, 221–232. doi: 10.1080/15548627.2017.1389823
Ravikumar, B., Sarkar, S., Davies, J. E., Futter, M., Garcia-Arencibia, M., Green-Thompson, Z. W., et al. (2010). Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435. doi: 10.1152/physrev.00030.2009
Rieusset, J. (2017). Role of endoplasmic reticulum-mitochondria communication in type 2 diabetes. Adv. Exp. Med. Biol. 997, 171–186. doi: 10.1007/978-981-10-4567-7_13
Rizzuto, R., Pinton, P., Carrington, W., Fay, F. S., Fogarty, K. E., Lifshitz, L. M., et al. (1998). Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766. doi: 10.1126/science.280.5370.1763
Rossi, A., Pizzo, P., and Filadi, R. (2019). Calcium, mitochondria and cell metabolism: a functional triangle in bioenergetics. Biochim. Biophys. Acta Mol. Cell Res. 1866, 1068–1078. doi: 10.1016/j.bbamcr.2018.10.016
Rusinol, A. E., Cui, Z., Chen, M. H., and Vance, J. E. (1994). A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269, 27494–27502.
Sala-Vila, A., Navarro-Lerida, I., Sanchez-Alvarez, M., Bosch, M., Calvo, C., Lopez, J. A., et al. (2016). Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 6:27351. doi: 10.1038/srep27351
Schaaf, M. B., Keulers, T. G., Vooijs, M. A., and Rouschop, K. M. (2016). LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 30, 3961–3978. doi: 10.1096/fj.201600698R
Sciarretta, S., Maejima, Y., Zablocki, D., and Sadoshima, J. (2018). The role of autophagy in the heart. Annu. Rev. Physiol. 80, 1–26. doi: 10.1146/annurev-physiol-021317-121427
Scrivo, A., Bourdenx, M., Pampliega, O., and Cuervo, A. M. (2018). Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 17, 802–815. doi: 10.1016/S1474-4422(18)30238-2
Shpilka, T., Welter, E., Borovsky, N., Amar, N., Mari, M., Reggiori, F., et al. (2015). Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 34, 2117–2131. doi: 10.15252/embj.201490315
Simmen, T., Aslan, J. E., Blagoveshchenskaya, A. D., Thomas, L., Wan, L., Xiang, Y., et al. (2005). PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729. doi: 10.1038/sj.emboj.7600559
Sou, Y. S., Tanida, I., Komatsu, M., Ueno, T., and Kominami, E. (2006). Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 281, 3017–3024. doi: 10.1074/jbc.M505888200
Stoica, R., De Vos, K. J., Paillusson, S., Mueller, S., Sancho, R. M., Lau, K. F., et al. (2014). ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5:3996. doi: 10.1038/ncomms4996
Stone, S. J., Levin, M. C., Zhou, P., Han, J., Walther, T. C., and Farese, R. J. (2009). The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284, 5352–5361. doi: 10.1074/jbc.M805768200
Su, T. P., Hayashi, T., Maurice, T., Buch, S., and Ruoho, A. E. (2010). The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 31, 557–566. doi: 10.1016/j.tips.2010.08.007
Tagashira, H., Bhuiyan, M. S., Shioda, N., and Fukunaga, K. (2014). Fluvoxamine rescues mitochondrial Ca2+ transport and ATP production through sigma(1)-receptor in hypertrophic cardiomyocytes. Life Sci. 95, 89–100. doi: 10.1016/j.lfs.2013.12.019
Tang, Z., Takahashi, Y., Chen, C., Liu, Y., He, H., Tsotakos, N., et al. (2017). Atg2A/B deficiency switches cytoprotective autophagy to non-canonical caspase-8 activation and apoptosis. Cell Death Differ. 24, 2127–2138. doi: 10.1038/cdd.2017.133
Tang, Z., Takahashi, Y., He, H., Hattori, T., Chen, C., Liang, X., et al. (2019). TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep. 28, 1744–1757.e5. doi: 10.1016/j.celrep.2019.07.036
Tanida, I., Ueno, T., and Kominami, E. (2008). LC3 and Autophagy. Methods Mol. Biol. 445, 77–88. doi: 10.1007/978-1-59745-157-4_4
Truban, D., Hou, X., Caulfield, T. R., Fiesel, F. C., and Springer, W. (2017). PINK1, Parkin, and mitochondrial quality control: What can we learn about Parkinson’s disease pathobiology? J. Parkinsons Dis. 7, 13–29. doi: 10.3233/JPD-160989
Tubbs, E., and Rieusset, J. (2016). Study of endoplasmic reticulum and mitochondria interactions by in situ proximity ligation assay in fixed cells J. Vis. Exp. 118:54899. doi: 10.3791/54899
Tubbs, E., Theurey, P., Vial, G., Bendridi, N., Bravard, A., Chauvin, M. A., et al. (2014). Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes 63, 3279–3294. doi: 10.2337/db13-1751
Vakifahmetoglu-Norberg, H., Ouchida, A. T., and Norberg, E. (2017). The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 482, 426–431. doi: 10.1016/j.bbrc.2016.11.088
Valladares, D., Utreras-Mendoza, Y., Campos, C., Morales, C., Diaz-Vegas, A., Contreras-Ferrat, A., et al. (2018). IP3 receptor blockade restores autophagy and mitochondrial function in skeletal muscle fibers of dystrophic mice. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3685–3695. doi: 10.1016/j.bbadis.2018.08.042
van Vliet, A. R., and Agostinis, P. (2018). Mitochondria-associated membranes and ER stress. Curr. Top. Microbiol. Immunol. 414, 73–102. doi: 10.1007/82_2017_2
Vance, J. E. (1990). Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256.
Vance, J. E. (2014). MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta 1841, 595–609. doi: 10.1016/j.bbalip.2013.11.014
Verfaillie, T., Rubio, N., Garg, A. D., Bultynck, G., Rizzuto, R., Decuypere, J. P., et al. (2012). PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 19, 1880–1891. doi: 10.1038/cdd.2012.74
Wang, H., Ni, H. M., Chao, X., Ma, X., Rodriguez, Y. A., Chavan, H., et al. (2019). Double deletion of PINK1 and Parkin impairs hepatic mitophagy and exacerbates acetaminophen-induced liver injury in mice. Redox Biol. 22:101148. doi: 10.1016/j.redox.2019.101148
Wang, H. J., Guay, G., Pogan, L., Sauve, R., and Nabi, I. R. (2000). Calcium regulates the association between mitochondria and a smooth subdomain of the endoplasmic reticulum. J. Cell Biol. 150, 1489–1498. doi: 10.1083/jcb.150.6.1489
Wang, X., Wen, Y., Dong, J., Cao, C., and Yuan, S. (2018). Systematic in-depth proteomic analysis of mitochondria-associated endoplasmic reticulum membranes in mouse and human testes. Proteomics 18:e1700478. doi: 10.1002/pmic.201700478
Wang, Y., Cai, J., Tang, C., and Dong, Z. (2020). Mitophagy in acute kidney injury and kidney repair. Cells 9:338. doi: 10.3390/cells9020338
Wang, Y. J., Huang, J., Liu, W., Kou, X., Tang, H., Wang, H., et al. (2017). IP3R-mediated Ca2+ signals govern hematopoietic and cardiac divergence of Flk1+ cells via the calcineurin-NFATc3-Etv2 pathway. J. Mol. Cell Biol. 9, 274–288. doi: 10.1093/jmcb/mjx014
Wu, S., Lu, Q., Wang, Q., Ding, Y., Ma, Z., Mao, X., et al. (2017). Binding of FUN14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation 136, 2248–2266. doi: 10.1161/CIRCULATIONAHA.117.030235
Wu, W., Li, W., Chen, H., Jiang, L., Zhu, R., and Feng, D. (2016). FUNDC1 is a novel mitochondrial-associated-membrane (MAM) protein required for hypoxia-induced mitochondrial fission and mitophagy. Autophagy 12, 1675–1676. doi: 10.1080/15548627.2016.1193656
Wu, W., Tian, W., Hu, Z., Chen, G., Huang, L., Li, W., et al. (2014). ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 15, 566–575. doi: 10.1002/embr.201438501
Xu, H., Guan, N., Ren, Y. L., Wei, Q. J., Tao, Y. H., Yang, G. S., et al. (2018). IP3R-Grp75-VDAC1-MCU calcium regulation axis antagonists protect podocytes from apoptosis and decrease proteinuria in an Adriamycin nephropathy rat model. BMC Nephrol. 19:140. doi: 10.1186/s12882-018-0940-3
Yang, J. Y., and Yang, W. Y. (2013). Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat. Commun. 4:2428. doi: 10.1038/ncomms3428
Yang, M., Zhao, L., Gao, P., Zhu, X., Han, Y., Chen, X., et al. (2019). DsbA-L ameliorates high glucose induced tubular damage through maintaining MAM integrity. EBioMedicine 43, 607–619. doi: 10.1016/j.ebiom.2019.04.044
Yang, Y., and Klionsky, D. J. (2020). Autophagy and disease: unanswered questions. Cell Death Differ. 27, 858–871. doi: 10.1038/s41418-019-0480-9
Yu, L., Chen, Y., and Tooze, S. A. (2018). Autophagy pathway: cellular and molecular mechanisms. Autophagy 14, 207–215. doi: 10.1080/15548627.2017.1378838
Yuan, L., Liu, Q., Wang, Z., Hou, J., and Xu, P. (2019). Correction to: EI24 tethers endoplasmic reticulum and mitochondria to regulate autophagy flux. Cell. Mol. Life Sci. 77, 2255–2256. doi: 10.1007/s00018-019-03355-3
Zahedi, R. P., Sickmann, A., Boehm, A. M., Winkler, C., Zufall, N., Schonfisch, B., et al. (2006). Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450. doi: 10.1091/mbc.e05-08-0740
Zeng, X., and Ju, D. (2018). Hedgehog signaling pathway and autophagy in cancer. Int. J. Mol. Sci. 19:E2279. doi: 10.3390/ijms19082279
Zhang, W., Siraj, S., Zhang, R., and Chen, Q. (2017). Mitophagy receptor FUNDC1 regulates mitochondrial homeostasis and protects the heart from I/R injury. Autophagy 13, 1080–1081. doi: 10.1080/15548627.2017.1300224
Keywords: autophagy, mitophagy, mitochondria-associated endoplasmic reticulum membranes (MAMs), mitochondria, endoplasmic reticulum
Citation: Yang M, Li C, Yang S, Xiao Y, Xiong X, Chen W, Zhao H, Zhang Q, Han Y and Sun L (2020) Mitochondria-Associated ER Membranes – The Origin Site of Autophagy. Front. Cell Dev. Biol. 8:595. doi: 10.3389/fcell.2020.00595
Received: 30 March 2020; Accepted: 18 June 2020;
Published: 16 July 2020.
Edited by:
Martin Van Der Laan, Saarland University, GermanyReviewed by:
Thomas Simmen, University of Alberta, CanadaCopyright © 2020 Yang, Li, Yang, Xiao, Xiong, Chen, Zhao, Zhang, Han and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Sun, c3VubGluQGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.