
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 14 July 2020
Sec. Cell Growth and Division
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00576
This article is part of the Research Topic Craniofacial Growth and Development: Novel Insights View all 17 articles
 Shu Lou1,2†
Shu Lou1,2† Lan Ma1,3†
Lan Ma1,3† Shiyi Kan1,2†
Shiyi Kan1,2† Xin Yu1,2
Xin Yu1,2 Yuting Wang1,2
Yuting Wang1,2 Fan Yang1,2
Fan Yang1,2 Guirong Zhu1,2
Guirong Zhu1,2 Liwen Fan1,2
Liwen Fan1,2 Dandan Li1,2
Dandan Li1,2 Hua Wang1,2
Hua Wang1,2 Wei Wang1,2
Wei Wang1,2 Weibing Zhang1,2
Weibing Zhang1,2 Lin Wang1,2,4*
Lin Wang1,2,4* Yongchu Pan1,2,4*
Yongchu Pan1,2,4*Although genetic variants in autophagy pathway genes were associated with the risk of oral cancers and early development in embryos, their associations with non-syndromic cleft lip with or without cleft palate (NSCL/P) risk remained unclear. A two-stage case-control study (2,027 NSCL/P cases and 1,843 controls) was performed to investigate the associations between single nucleotide polymorphisms (SNPs) in 23 autophagy pathway genes and NSCL/P susceptibility. The logistic regression model was used to calculate effects of SNPs on NSCL/P susceptibility. Gene-based analysis was performed via the sequence kernel association test (SKAT) and multi-marker analysis of genomic annotation (MAGMA) methods. Expression quantitative trait loci (eQTL) analysis was conducted using NSCL/P lip tissue samples. Gene expression during embryonic development was evaluated using RNA-Seq. Functional roles were explored by luciferase activity assay, cell apoptosis, proliferation, and cycle in vitro. Rs2301104 in HIF1A was significantly associated with NSCL/P susceptibility in the combined analysis (OR: 1.29, 95% CI: 1.09–1.29, P = 3.39 × 10–03), and showed strong evidence of association heterogeneity (P = 9.06 × 10–03) with obvious association in the female (OR: 1.80; 95% CI: 1.32–2.45; P = 1.79 × 10–04). The G allele of rs2301104 was associated with enhanced transcription activity and high expression of HIF1A compared with that of C allele. Moreover, rs2301104 exhibited an eQTL effect for HIF1A with its GC/CC genotypes associated with decreased HIF1A expression compared with those with GG genotypes (P = 3.1 × 10–2). Knockdown of HIF1A induced cell apoptosis and inhibited cell proliferation in human embryonic palate mesenchyme (HEPM) and human oral epithelium cells (HOEC). This study demonstrated that rs2301104 in autophagy pathway gene HIF1A was associated with susceptibility of NSCL/P.
Non-syndromic cleft lip with or without cleft palate (NSCL/P) is one of the most common human birth defects contributing huge health and financial burdens to the affected individuals, families, and societies (Mangold et al., 2010; Heike and Evans, 2016). It occurs in ~1 in 700 live births worldwide, with prevalence varying by population (Birnbaum et al., 2009).
The occurrence of NSCL/P results from failure of the facial processes to grow or fuse appropriately during early embryologic development (between the 4th and 12th week of gestation) (Beaty et al., 2016; Li et al., 2017). Signaling pathways, including Bmp, Fgf, Shh, and Wnt signaling pathways, are critical for proper lip fusion (Jiang et al., 2006). Similarly, the growth of the palatal shelves is also mediated by epithelial-mesenchymal interactions regulated by multiple signaling pathways and transcriptional factors (Lan et al., 2015). Cell migration, proliferation, and apoptosis in both epithelial cells and cranial neural crest (CNC)-derived mesenchymal cells are involved in mechanisms leading to cleft lip and palate (Tian et al., 2017).
The underlying etiology is complex and multifactorial with a wide range of influences including genetic and environmental factors. Extensive human genetic studies and animal studies have attempted to identify the genetic variants associated with NSCL/P risk (Leslie et al., 2016; Hammond et al., 2018; Reynolds et al., 2019). For instance, Leslie et al. (2016) conducted a multiethnic genome-wide association study in 6480 participants and revealed novel associations on 2p24 near FAM49A, and 19q13 near RHPN2. Hammond et al. demonstrated that ectopic Hh-Smo signaling downregulates Wnt/BMP pathways, resulting in cleft palate and defective osteogenesis (Hammond et al., 2018).
As a conserved lysosomal degradation process in eukaryotes, autophagy prevents cells from different kinds of stress, such as starvation, hypoxia, or exposure to toxic molecules (Huang and Klionsky, 2007). Defective autophagy is related to physiological and pathological conditions, such as cancer, metabolic, neurodegenerative disease, and aging (Netea-Maier et al., 2016). In the early development stage, autophagy has been shown to be essential in the transition of oocytes to embryos, postpartum survival, development, differentiation and aging in mouse models (Hale et al., 2013). Additionally, autophagy degrades cytoplasmic components, which is vital for embryonic development and human health (Fenouille et al., 2017). Susceptible genes may play essential roles in cell death and inflammation by influencing autophagy in neighboring cells during developmental periods (Lin et al., 2017). Previous studies showed that single nucleotide polymorphisms (SNPs) in autophagy pathway genes were related to the risk of melanoma, neck squamous cell carcinoma, and early embryo development (Capela et al., 2016; White et al., 2016; Fernandez-Mateos et al., 2017), however, their associations with the risk of NSCL/P have never been explored.
Highlighting these bases, with the aim of exploring associations between genetic variants in autophagy pathway and the risk of NSCL/P, we conducted a two-stage case-control study with 2,027 NSCL/P cases and 1,843 controls in the present study. We finally found that rs2301104 in autophagy pathway gene HIF1A was associated with susceptibility of NSCL/P. Moreover, we explored the functional roles of the SNP and the gene with in vivo and in vitro experiments. Our study provides a deeper insight into the roles of autophagy pathway-related genetic variants in the development of NSCL/P.
The key autophagy associated genes were selected from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and published studies. We obtained 109 genes from the autophagy pathway included in KEGG (map04140), and the following key words were used to search PubMed for publications on or before September 30th, 2018: gene name and craniofacial, craniofacial development, or facial development. Finally, 23 genes related to autophagy and craniofacial development were kept for further analysis (Supplementary Figure 1 and Supplementary Table 1).
The flow chart of selecting putative functional SNPs based on 23 genes we selected is shown in Figure 1. First, a total of 27,240 SNPs in gene regions was selected using the CHB data from the 1000 Genomes Project (Phase I, March 2012) and 3,156 SNPs remained after the following quality control criteria (Wang et al., 2019; Zhu et al., 2019): (a) minor allele frequency (MAF) ≥ 0.05, (b) Hardy-Weinberg equilibrium (HWE) ≥ 0.05, and (c) call rate ≥ 95%. Second, we used RegulomeDB and HaploReg to predict SNP functions. SNPs with Regulome DB Score below five and predicted to hit eQTL effects or bind transcription factors from HaploReg database were retained. Besides, SNPs with pairwise linkage disequilibrium (LD) r2≥ 0.5 were excluded using PLINK 1.09. Finally, a total of 146 putative functional SNPs in 23 autophagy pathway genes were identified and further explored in a two-stage case-control study.
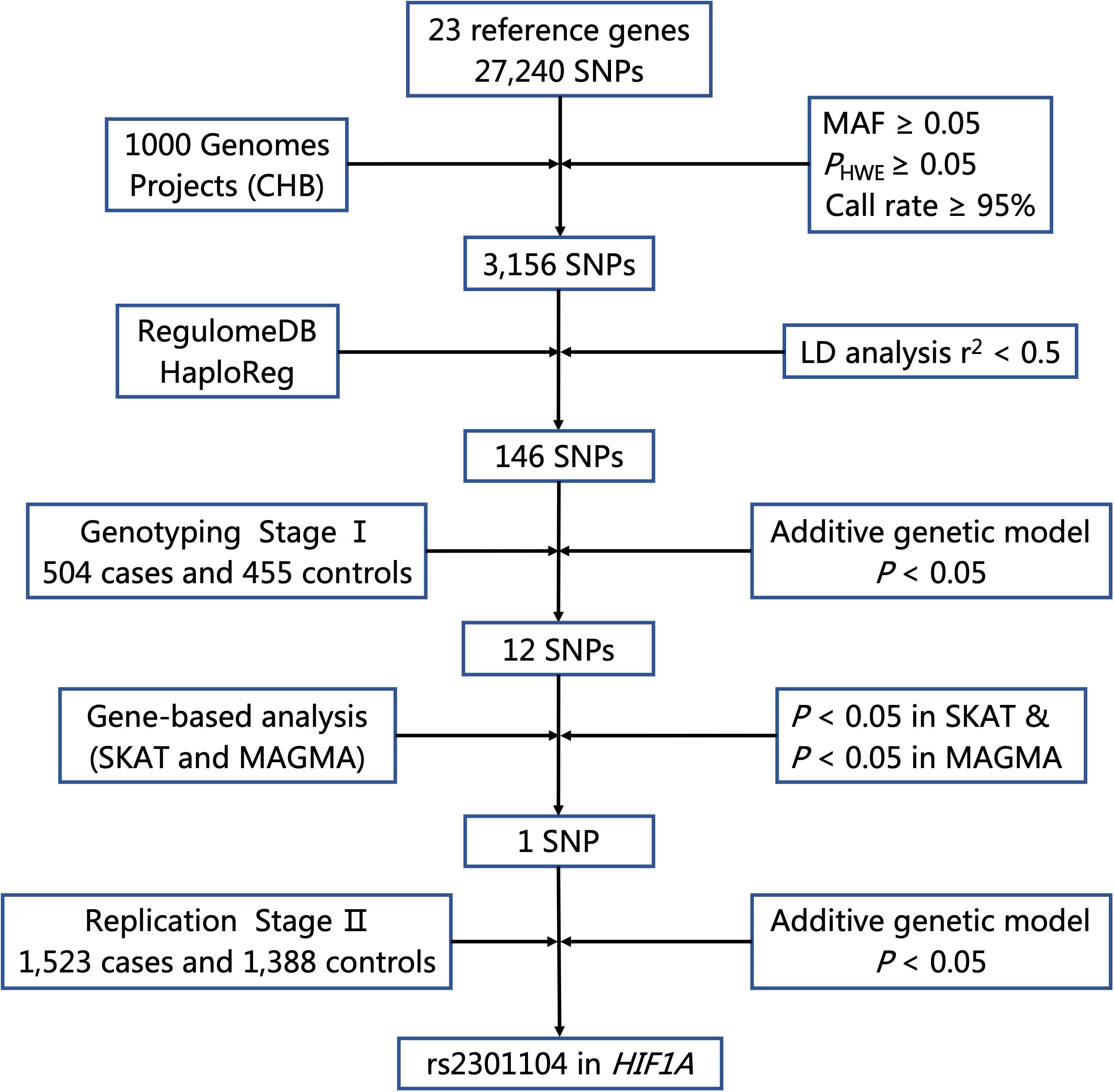
Figure 1. Flow chart for selecting putative functional SNP in autophagy pathway genes. MAF, minor allele frequency; HWE, Hardy–Weinberg Equilibrium; LD, linkage disequilibrium.
We performed a two-stage case-control analysis of NSCL/P in the Chinese Han population. Two stages were totally independent from each other: the stage I was comprised of 504 NSCL/P cases, and 455 newborn controls who were recruited from West China Hospital of Stomatology Sichuan University and the stage II included 1,523 NSCL/P cases and 1,388 controls who were recruited from Affiliated Stomatological Hospital of Nanjing Medical University, Nanjing Children’s Hospital, Xuzhou First People’s Hospital, and Huai’an First People’s Hospital (Supplementary Table 2).
All patients were interviewed and clinically evaluated by an experienced oral surgeon based on detailed diagnostic information from medical records and physical examinations to ensure that individuals with other congenital anomalies, identified CL/P syndromes or developmental delays were excluded from this study. Venous blood samples were collected from all subjects for genetic analysis. The present study was approved by the Ethics Committee of Nanjing Medical University [NJMUERC (2008) No. 20]. At recruitment, informed written consent was obtained from all the participants or their guardians.
All samples in stage I were genotyped using Affymetrix Axiom CHB1 & CHB2 arrays. Systematic quality control (Wang et al., 2019; Zhu et al., 2019) was performed to filter SNPs, including PHWE ≥ 0.05, MAF ≥ 0.05, and call rate ≥ 95%. Genotyping in stage II was performed by TaqMan-MGB assays on an ABI-Prism 7900 instrument (Applied Biosystems, Foster City, CA). The primers of the Taqman probes are listed in Supplementary Table 3.
SNPs were annotated for potential regulatory functions using RegulomeDB1 and HaploReg v4.12. We also evaluated the SNPs in the integrative analysis of 127 reference human epigenomes based on NIH Roadmap Epigenomics database3.
We purchased six adult male and twelve adult female C57BL/6 mice from Animal Center of Yangzhou University. Two female mice and one male mouse were placed together in the same cage at 8 p.m. and separated at 8 a.m. the next morning. Embryos were counted as E0.5d in the morning that vaginal plug was checked. Lip and palate tissues of E10.5d, E11.5d, E12.5d, E13.5d, E14.5d, and E15.5d embryos were collected. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) from embryonic tissues of mice, which were approved in quantity and quality by Nanodrop and 1% agarose electrophoresis. mRNA sequencing was performed on Hiseq3000 platform in 10M and 6G depth, respectively. RNA reads were aligned to the mouse genome (MM10) by gSNAP, and the average fragments per kilobase of exon model per million fragments mapped (FPKM) value of all samples was used to normalize mRNA expression.
RNA-Seq data on craniofacial structures during mouse embryo development were downloaded from the FaceBase consortium at FaceBase.org4, an online database that describes the overall gene expression during mouse craniofacial development. Count data were normalized using the regularized logarithmic transformation in the DESeq2 R package.
Gene expression in NSCL/P cases and controls of dental pulp stem cells (DPSCs), and mesenchymal stem cells (MSCs) in lip muscle were obtained from Gene Expression Omnibus (GEO) repository (GSE42589 and GSE85748, respectively).
The human embryonic palatal mesenchymal (HEPM) cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, United States), and cultured in Eagle’s Minimum Essential Medium (ATCC), supplemented with 10% fetal bovine serum (FBS, Gibco), 100 units/ml antibiotics at 37°C under 5% CO2. The human oral epithelial cells (HOEC) were obtained from BeNa Culture Collection (Beijing, China), and cultured in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% FBS (Gibco) and 100 units/ml antibiotics and maintained at 37°C under 5% CO2.
In order to evaluate whether there is a difference in the enhancer activity in HIF1A of different alleles, we performed the luciferase activity assay. A sequence of 1,000-bp containing rs2301104 G or C allele and HIF1A promoter region were synthesized and cloned into the NheI and XhoI restriction sites of the pGL3-basic vector (Promega). HEPM and HOEC cells were seeded at in 24-well culture plates, and each well was transiently transfected with reporter plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. All plasmids were co-transfected with 10 ng pRL-SV40, which contained the Renilla luciferase gene. The luciferase activities were measured 48 h after transfection with a dual-luciferase reporter assay system (Promega). The ratio of Firefly luciferase to Renilla luciferase activity was assessed. The transfection experiments were performed in triplicate.
To compare gene expression of cells and tissues with different genotypes, we constructed the pcDNA3.1-rs2301104 G and pcDNA3.1-rs2301104 C vectors, which were transfected into cells and harvested for 48 h. Total RNA was extracted from cells and 68 NSCL/P lip tissue samples to reverse transcription and PCR reactions using PrimeScriptTM RT-PCR kit (TaKaRa, Shiga, Japan). The relative mRNA expression level of HIF1A and the internal control GAPDH were quantified using ABI 7900 Real-Time PCR system (Applied Biosystems). The primers were listed as follows: HIF1A (forward: TCAGGACACAGATTTAGACTTGGAG, reverse: AGTGGTAG TGGTGGCATTAGCA) and GAPDH (forward: GGACCTGAC CTGCCGTCTAG, reverse: GTAGCCCAGGATGCCCTTGA). All reactions were conducted in triplicate, and the data were analyzed by the 2–ΔΔCt method. Data are shown as the mean ± standard deviations (SD).
The small interference RNA (siRNA) oligonucleotides including HIF1A siRNA-378 (5′-CCAGAUCUCGGCGAAGUAATT-3′, 5′-UUACUUCGCCGAGAUCUGGTT-3′), HIF1A siRNA-2107 (5′-CCAGCAGACUCAAAUACAATT-3′, 5′-UUGUAUUUGAG UCUGCUGGTT-3′), HIF1A siRNA-2637 (5′-GCUACUACAUC ACUUUCUUTT-3′, 5′-AAGAAAGUGAUGUAGUAGCTT-3′) and control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′) were designed and purchased from GenePharma (Shanghai, China). Transfection of siRNAs with final concentration 100 nM was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. Cells were harvested 48 h later for further experiments.
Cells were treated with trypsin-EDTA (Gibco) and resuspended as single-cell suspension 48 h after transfection. We stained cells with Annexin V:PE Apoptosis Detection Kit (BD Biosciences, San Jose, CA, United States) for cell apoptosis analysis and analyzed using a Fluorescence Activated Cell Sorting (FACS) System by BD Biosciences (San Jose, CA, United States). As for cell cycle analysis, cells were then fixed in 70% ethanol overnight at 4°C after digested. Propidium iodide (PI)/RNase staining was conducted on fixed cells for flow cytometric analysis. Data were analyzed with FlowJo software (TreeStar, Ashland, OR, United States). Cell proliferation was assessed by absorbance using a Cell Counting Kit-8 assay (CCK8, Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. Cells were seeded in 96-well plates at a density of 3 × 103 cells per well, approximately. CCK-8 reaction solution was added into each well for 2 h incubation in new medium containing. The absorbance was measured on a spectrophotometer microplate reader (Multiskan MK3, Thermo) at a wavelength of 450 nm. All data are shown as the mean ± SD.
For evaluating the association between NSCL/P risk and genetic variants, logistic regression analysis under additive model with adjustment for gender was implemented to calculate the crude and adjusted odds ratios (ORs) and their 95% confidence intervals (CIs) (Wang et al., 2016; Zhu et al., 2019). Haplotype analysis were performed using Haploview software with parameters of MAF ≥ 5% and pairwise r2 threshold of 0.8 (Barrett et al., 2005). The epistasis test between SNPs in the candidate gene was performed using R software, which was computed by an unconditional logistic regression model specifies the log-odds as: logit(P) = β0 + βG(Gi) + βG×G(Gi*Gi). Goodness-of-fit Chi-square test was performed to test HWE in control groups. We used the sequence kernel association test (SKAT) (Timbers et al., 2016) and Multi-marker Analysis of GenoMic Annotation (MAGMA) tool (de Leeuw et al., 2015) for gene-based analysis. Combined analysis of two stages was performed using a fixed-effect model in R software (Dai et al., 2019). The measure of heterogeneity was tested using Cochran’s Q statistics and I2. Gene-by-sex (G × S) interaction was computed by an unconditional logistic regression model specifies the log-odds as: logit(P) = β0 + βG(Gi) + βG×E(Gi*Ei). For all graphs, statistical analyses were carried out using two-tailed, unpaired Student’ s t-test. Before t-test, normal distribution of all the data were checked using normality test and equality of variances were checked using F-test. All statistical analyses were performed by R software 3.4.25 and PLINK 1.09. Data were considered statistically significant when P-value < 0.05.
As shown in Figure 1, 27,240 SNPs in 23 genes related to autophagy pathway were identified. Among them, 146 putative functional SNPs were picked out, and their associations with risk of NSCL/P were examined in the stage I that was comprised of 504 NSCL/P cases and 455 newborn controls. Twelve SNPs (rs77141447, rs16822638, rs7625881, rs14016, rs12813551, rs147920828, rs2301104, rs4776786, rs80225705, rs3784605, rs7182342, and rs2283791) were significantly associated with the risk of NSCL/P in additive logistic model with the adjustment of gender (Table 1).
To evaluate the joint effects of common SNPs in a gene unit, we further performed gene-based analysis using SKAT and MAGMA methods to identify potentially associated genes and found that HIF1A (PSKAT = 9.42 × 10–03, PMAGMA = 2.74 × 10–02), BCL2L1 (PSKAT = 1.05 × 10–02) and KRAS (PMAGMA = 1.76 × 10–02) were significantly associated with NSCL/P susceptibility. Only HIF1A achieved consistent associations in both methods (Supplementary Table 4); thus, rs2301104 in HIF1A remained for further analysis.
Rs2301104 was genotyped for validation with an additional 1,523 NSCL/P case and 1f88 control subjects and showed significant association with the same direction of effect as that observed in stage I (OR: 1.39; 95% CI: 1.01–1.92; P = 4.55 × 10–02 in stage I, OR: 1.25; 95% CI: 1.03–1.53; P = 2.72 × 10–02 in stage II, Table 2). Similar results were also observed in the combined analysis from the two stages (OR: 1.29; 95% CI: 1.09–1.53; P = 3.39 × 10–03, Table 2). No significant heterogeneity for rs2301104 was observed among different stages (Phet = 0.70, Table 2). Further, to explore whether rs2301104 participated in development of NSCL/P alone or not, we also performed haplotype and epistasis analysis. No significant haplotype and SNPs interacted with rs2301104 was found, which indicated rs2301104 may play an independent role in development of NSCL/P (Supplementary Figure 2 and Supplementary Table 5).
As shown in Figure 2, the association of rs2301104 with the risk of NSCL/P showed strong evidence of heterogeneity (Phet = 9.06 × 10–03) among males and females. Significant association between rs2301104 and NSCL/P risk were observed in the female (OR: 1.80; 95% CI: 1.32–2.45; P = 1.79 × 10–04), rather than in the male (OR: 1.10; 95% CI: 0.90–1.35; P = 0.362, Figure 2). Further, rs2301104 had a significant interaction with gender (P = 1.13 × 10–03), constituting highly statistically significant evidence of the involvement of rs2301104 with gender preference in NSCL/P.
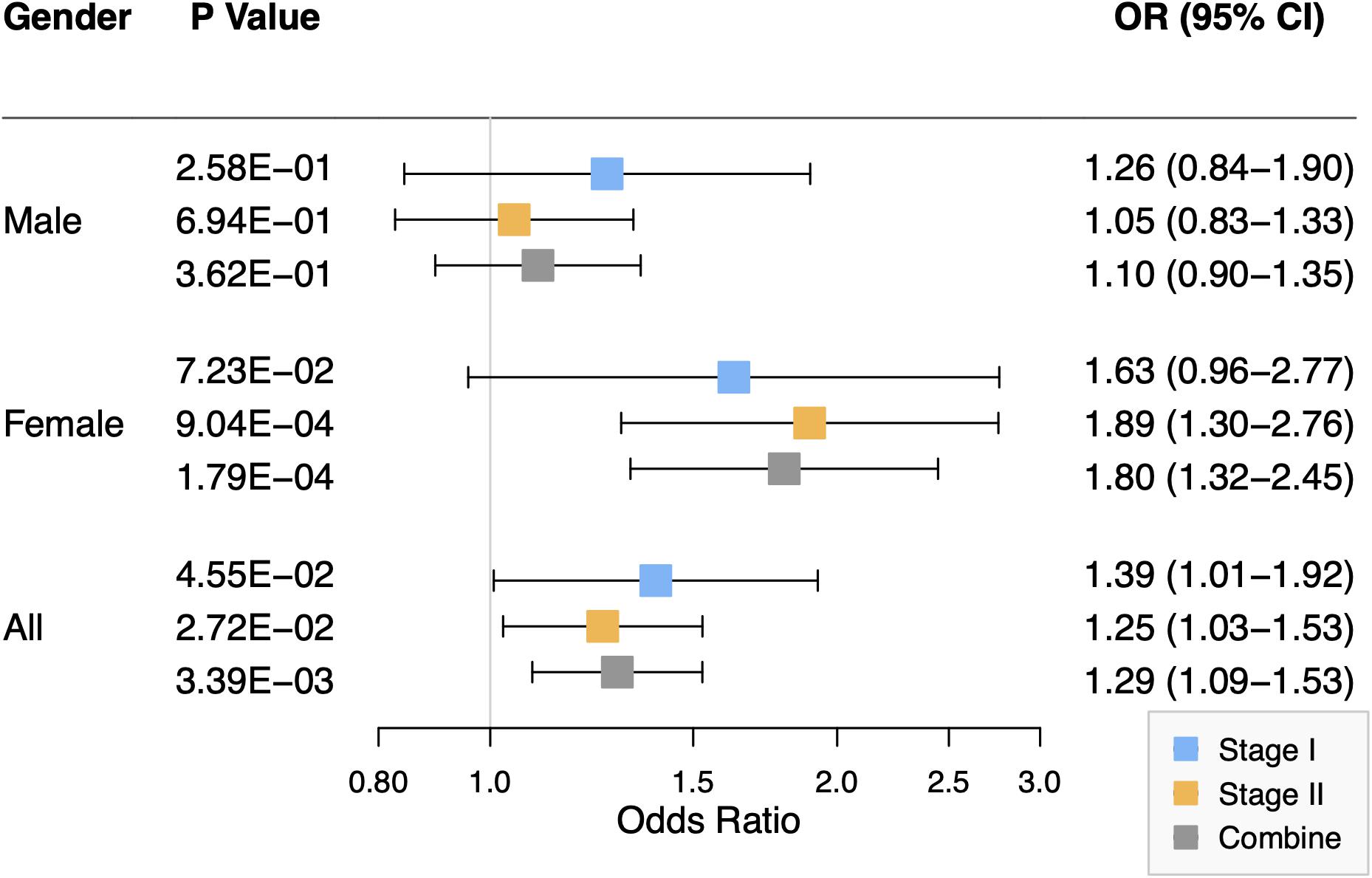
Figure 2. Forest plot of the association between rs2301104 with risk of NSCL/P in stage I, stage II, and combined analysis, stratified according to the gender of study participants.
To investigate the underlying mechanisms of rs2301104, we conducted functional annotation analysis on it. It was found that rs2301104 resided within chromatin regions expressing marks indicative of enhancer activity, promoter activity, transcription regulatory by predictions from Haploreg, RegulomeDB, UCSC Genome Browser, and Roadmap Epigenomics Project (Supplementary Table 6 and Supplementary Figure 3).
We thereby constructed enhancer luciferase reporter vectors containing the rs2301104-centered region and the HIF1A promoter and then tested the luciferase activity 48 h after transfecting different plasmids in HEPM and HOEC cells. The rs2301104 G allele revealed a significantly increased enhancer activity compared with that of C allele (Figures 3A,B). This result was also supported in HEPM and HOEC cells that the mRNA expression of HIF1A was lower with C allele constructs, compared with G allele constructs (Figures 3C,D).
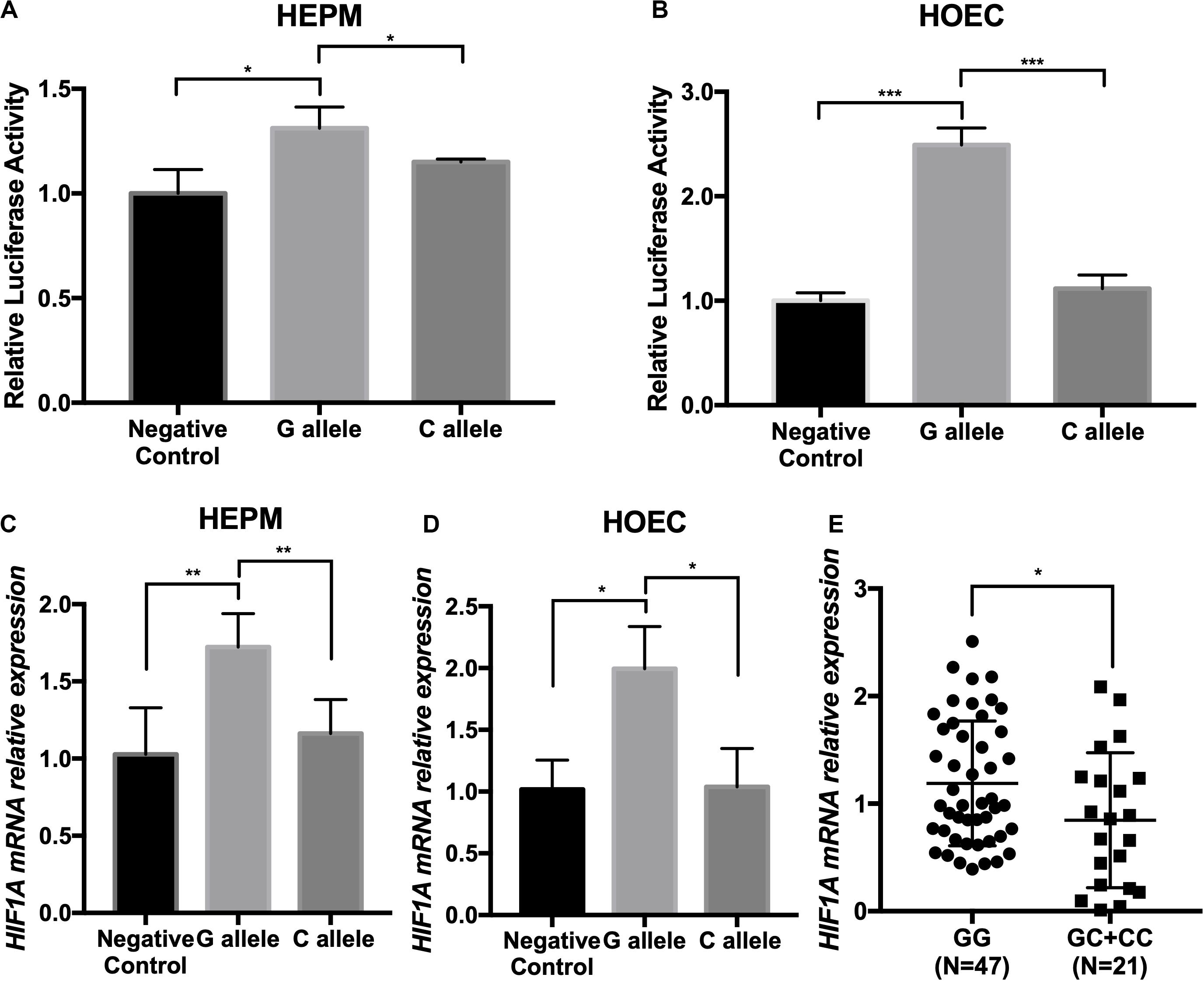
Figure 3. The rs2301104 alleles affect the enhancer activity and expression of HIF1A. (A) HEPM and (B) HOEC cells were transiently transfected with rs2301104 [G], rs2301104 [C], and HIF1A promoter with PGL3-basic luciferase reporter vector. pRL-SV40 was co-transfected into cells. The activity of luciferase was assayed 48h later. qRT-PCR analysis of HIF1A expression at 48 h after plasmids transfection in (C) HEPM and (D) HOEC cells. It was indicated that the expression of HIF1A was lower with C allele constructs, compared with G allele constructs. Results are shown as mean values with the standard deviation (SD) normalized to GAPDH. (E) HIF1A mRNA expression was detected by qRT-PCR in 68 NSCL/P lip tissue samples. The HIF1A expression was significantly lower in samples with GC/CC genotypes than those with GG genotypes. The results were normalized to GAPDH. The P-value was calculated with two-side t-test (n = 6; *P < 0.05, **P < 0.01, ***P < 0.001).
To evaluate the effect on rs2301104 in HIF1A, we further performed an eQTL analysis in lip tissue samples from 68 NSCL/P patients and found that rs2301104 was significantly associated with the expression of HIF1A (P = 3.1 × 10–2). Expression of HIF1A was higher in samples of GG genotypes compared with those of GC/CC genotypes (Figure 3E).
We detected continuous expression of HIF1A in the mouse lip and palate development during E10.5d to E15.5d as well as craniofacial structures from E10.5d to E14.5d (Supplementary Figure 4), implicating its essential role in the craniofacial development. In addition, we found that the expression of HIF1A was decreased in the DPSCs, and MSCs in lip muscle of NSCL/P cases, compared with those in controls (P = 8.37 × 10–2 and P = 3.74 × 10–1, respectively, Supplementary Figure 5).
To further address the function of HIF1A, we knocked down the expression of HIF1A in HEPM and HOEC cells through the transfection of HIF1A siRNA-2107 given its highest efficiency (Figures 4A,B). Flow cytometric analysis revealed a significantly increased apoptosis rate in cells transfected with HIF1A siRNA-2107 (Figures 4E–H). The results of CCK-8 assays showed that knockdown of HIF1A significantly inhibited cell proliferation (Figures 4C,D). Moreover, the cell cycle assay showed that knockdown of HIF1A increased the number of G0/G1 phase cells, with a significant reduction in the number of S phase cells (Figures 4I,J).
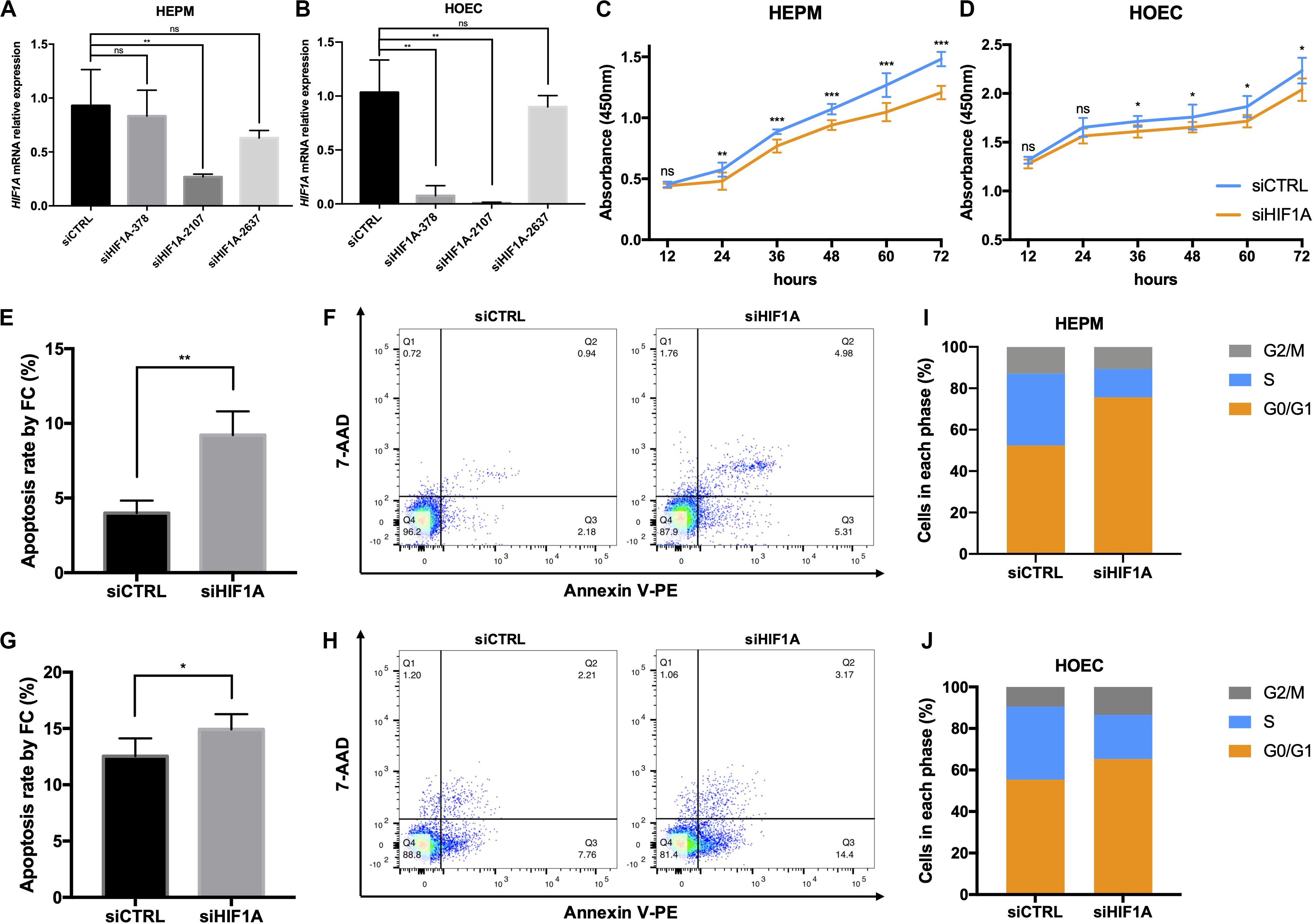
Figure 4. In vitro functional roles of HIF1A in HEPM and HOEC. (A,B) qRT-PCR analysis of the HIF1A knockdown efficiency in HEPM and HOEC cells, respectively (n = 6). (C,D) Cell counting kit-8 assay was used to assess the proliferation of HEPM and HOEC cells after transfection with HIF1A siRNAs (n = 6). (E–H) Quantitative analysis of cell apoptosis by flow cytometry between the two groups (n = 6). The rate of apoptosis is higher in (E,F) HEPM cells, and (G,H) HOEC cells with HIF1A knockdown than in the corresponding negative controls. (I,J) Effects of HIF1A knockdown on the cell cycle in HEPM and HOEC cells (n = 6). (ns, no significance, *P < 0.05, **P < 0.01, ***P < 0.001).
Autophagy participates in many pathological conditions, such as neurodegeneration, autoimmune disease, cancer, and Crohn’s disease (Nyfeler and Eng, 2016). In the present study, we evaluated the relationships between SNPs on core genes of autophagy pathway with risk of NSCL/P by a two-stage case-control study. Using association and gene-based analysis, we found that rs2301104 in HIF1A was significantly associated with NSCL/P risk. Notably, gender stratification analysis indicated that rs2301104 exhibited a significant association in the females but not males. Given several possible mechanisms, the underlying biochemical causes might involve sex hormones, sex-biased methylation (McCarthy et al., 2014). Moreover, autophagy is differentially regulated by sex, and the interaction between sex, sex-hormones, and tissue variability in regulating the heat shock response pathway was also observed (Tower et al., 2020).
Rs2301104 located in the first intron of HIF1A with enriched enhancer histone marks (including H3K4me1, H3K4me3, and H3K27ac) and previous studies had proven that SNPs in this region played essential roles in gene transcription and expression. For instance, Wang et al. identified a variant at 12p13.2 with enriched histone marks associated with colorectal cancer risk by affecting the binding affinity of transcriptional factor (Wang et al., 2016). Here, luciferase activity assay showed that rs2301104 C allele significantly decreased transcription activity, compared with that of G allele. Further, the rs2301104 C allele was associated with lower expression level of HIF1A mRNA in HEPM and HOEC cells. These findings suggested that rs2301104 was involved in transcriptional regulation of HIF1A.
We observed expression of HIF1A during mouse craniofacial development from E10.5d to E14.5d, implicating its involvement in the development of lip and palate. Notably, individuals carrying rs2301104 GC/CC genotypes (risk genotype) showed significantly lower expression of HIF1A than GG genotypes in lip tissue samples and a similar trend was also observed in DPSCs, and MSCs in lip muscle of NSCL/P cases and controls, highlighting the association between low expression of HIF1A and increased susceptibility of NSCL/P. We further knockdown of HIF1A to evaluate its role in the function of HOEC and HEPM. Promoted cell apoptosis, inhibited cell proliferation, and increase in G0/G1 phase cells, accompanied by a decrease in the number of S phase cells were detected and all of events commonly occurred during the development of NSCL/P (Tian et al., 2017; Lukacs et al., 2019). For instance, Lu et al. reported that cell proliferation increased and apoptosis decreased in the E13d mutant rabbit model related to NSCL/P, which would lead to the persistence of seams in the facial processes (Lu et al., 2019).
HIF1A played an important role in terms of embryo development. Severe congenital malformations including heart defects, retinopathy, brain lesions could be detected by culturing embryos at an inappropriate oxygen concentration level, which is mainly regulated by hypoxia-inducible factors (HIFs) (Bohuslavova et al., 2013; Feliciano et al., 2013; Nunes et al., 2015). Hif1a-deficient mice exhibited abnormal development of heart morphology and embryonic lethality between E11.0d and E12.0d (Krishnan et al., 2008; Forristal et al., 2010). According to the DECIPHER database, deletion of HIF1A resulted in congenital developmental defects, including microcephaly, aplasia of the tongue, and facial asymmetry (Firth et al., 2009).
The exact reason that HIF1A contributed to these developmental anomalies remained obscure. However, autophagy, induced by hypoxia whose activation was dependent on HIF1A (Liu et al., 2017), may be a possible way. Lu et al. demonstrated that the inhibition of autophagy might cause delayed development by affecting the epithelial-mesenchymal transitions process during chick development (Lu et al., 2014). Previous study showed that the loss of autophagy function in both osteoblasts and osteocytes might lead to a decrease in craniofacial bone mass in mouse models (Thomas et al., 2019). It had been reported that impaired autophagy could cause the loss of the protein quality control mechanism that is at the basis of inherited cardiomyopathies (Dorsch et al., 2019). Based on these studies, we hypothesized that decreased expression of HIF1A might break the oxygen homeostatic response during embryonic development, thereby inhibit autophagy and further exacerbate susceptibility to various developmental anomalies, including NSCL/P. A schematic model is shown in Supplementary Figure 6.
Taken together, we identified rs2301104 in autophagy pathway genes HIF1A that contributed to the risk of NSCL/P, providing further knowledge about the etiology of the disease. Further studies were warranted to explore the underlying mechanisms.
All datasets generated for this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Ethics Committee of Nanjing Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. The animal study was reviewed and approved by the Ethics Committee of Nanjing Medical University.
SL analyzed the data, drafted the manuscript, and performed in vivo and in vitro experiments with SK. LM, WZ, YP, and LW designed and directed the study, obtained the financial support, and critically revised the manuscript. XY performed the genotyping in stage II. YW, LF, and DL were responsible for sample processing. FY and GZ performed the statistical analysis. HW and WW were responsible for subject recruitment and sample collection. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (81970969 and 81830031), the State Key Lab of Reproductive Medicine of Nanjing Medical University (JX116GSP 20171416), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD-2018-87), the Natural Science Foundation of Jiangsu Province (BL2014073 and 15KJA320002 to LW), the Qing Lan Project (to YP), Six Distinguished Talent (2016-WSW-008), Jiangsu Provincial Medical Talent (ZDRCC2016023 to YP), the Jiangsu Provincial Key Medical Discipline (zdxka2016026), and the Graduate Student Scientific Research Innovation Projects in Jiangsu Province (KYCX19_1148).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all the study participants, research staff, and students who contributed to this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00576/full#supplementary-material
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi: 10.1093/bioinformatics/bth457
Beaty, T. H., Marazita, M. L., and Leslie, E. J. (2016). Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Research 5:2800. doi: 10.12688/f1000research.9503.1
Birnbaum, S., Ludwig, K. U., Reutter, H., Herms, S., Steffens, M., Rubini, M., et al. (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 41, 473–477. doi: 10.1038/ng.333
Bohuslavova, R., Skvorova, L., Sedmera, D., Semenza, G. L., and Pavlinkova, G. (2013). Increased susceptibility of HIF-1alpha heterozygous-null mice to cardiovascular malformations associated with maternal diabetes. J. Mol. Cell. Cardiol. 60, 129–141. doi: 10.1016/j.yjmcc.2013.04.015
Capela, C., Dossou, A. D., Silva-Gomes, R., Sopoh, G. E., Makoutode, M., Menino, J. F., et al. (2016). Genetic variation in autophagy-related genes influences the risk and phenotype of buruli ulcer. PLoS Negl. Trop. Dis. 10:e0004671. doi: 10.1371/journal.pntd.0004671
Dai, J., Lv, J., Zhu, M., Wang, Y., Qin, N., Ma, H., et al. (2019). Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir. Med. 7, 881–891. doi: 10.1016/S2213-2600(19)30144-4
de Leeuw, C. A., Mooij, J. M., Heskes, T., and Posthuma, D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11:e1004219. doi: 10.1371/journal.pcbi.1004219
Dorsch, L. M., Schuldt, M., Knezevic, D., Wiersma, M., Kuster, D. W. D., van der Velden, J., et al. (2019). Untying the knot: protein quality control in inherited cardiomyopathies. Pflugers Arch. 471, 795–806. doi: 10.1007/s00424-018-2194-0
Feliciano, D. M., Zhang, S., Quon, J. L., and Bordey, A. (2013). Hypoxia-inducible factor 1a is a Tsc1-regulated survival factor in newborn neurons in tuberous sclerosis complex. Hum. Mol. Genet. 22, 1725–1734. doi: 10.1093/hmg/ddt018
Fenouille, N., Nascimbeni, A. C., Botti-Millet, J., Dupont, N., Morel, E., and Codogno, P. (2017). To be or not to be cell autonomous? Autophagy says both. Essays Biochem. 61, 649–661. doi: 10.1042/EBC20170025
Fernandez-Mateos, J., Seijas-Tamayo, R., Klain, J. C. A., Borgonon, M. P., Perez-Ruiz, E., Mesia, R., et al. (2017). Analysis of autophagy gene polymorphisms in Spanish patients with head and neck squamous cell carcinoma. Sci. Rep. 7:6887. doi: 10.1038/s41598-017-07270-0
Firth, H. V., Richards, S. M., Bevan, A. P., Clayton, S., Corpas, M., Rajan, D., et al. (2009). DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533. doi: 10.1016/j.ajhg.2009.03.010
Forristal, C. E., Wright, K. L., Hanley, N. A., Oreffo, R. O., and Houghton, F. D. (2010). Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction 139, 85–97. doi: 10.1530/REP-09-0300
Hale, A. N., Ledbetter, D. J., Gawriluk, T. R., and Rucker, E. B. III (2013). Autophagy: regulation and role in development. Autophagy 9, 951–972. doi: 10.4161/auto.24273
Hammond, N. L., Brookes, K. J., and Dixon, M. J. (2018). Ectopic hedgehog signaling causes cleft palate and defective osteogenesis. J. Dent. Res. 97, 1485–1493. doi: 10.1177/0022034518785336
Heike, C. L., and Evans, K. N. (2016). Evaluation of adults born with an oral cleft: aren’t adults just big kids? JAMA Pediatr. 170, 1045–1046. doi: 10.1001/jamapediatrics.2016.2639
Huang, J., and Klionsky, D. J. (2007). Autophagy and human disease. Cell Cycle 6, 1837–1849. doi: 10.4161/cc.6.15.4511
Jiang, R., Bush, J. O., and Lidral, A. C. (2006). Development of the upper lip: morphogenetic and molecular mechanisms. Dev. Dyn. 235, 1152–1166. doi: 10.1002/dvdy.20646
Krishnan, J., Ahuja, P., Bodenmann, S., Knapik, D., Perriard, E., Krek, W., et al. (2008). Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ. Res. 103, 1139–1146. doi: 10.1161/01.RES.0000338613.89841.c1
Lan, Y., Xu, J., and Jiang, R. (2015). Cellular and molecular mechanisms of palatogenesis. Curr. Top. Dev. Biol. 115, 59–84. doi: 10.1016/bs.ctdb.2015.07.002
Leslie, E. J., Carlson, J. C., Shaffer, J. R., Feingold, E., Wehby, G., Laurie, C. A., et al. (2016). A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 25, 2862–2872. doi: 10.1093/hmg/ddw104
Li, C., Lan, Y., and Jiang, R. (2017). Molecular and cellular mechanisms of palate development. J. Dent. Res. 96, 1184–1191. doi: 10.1177/0022034517703580
Lin, L., Rodrigues, F., Kary, C., Contet, A., Logan, M., Baxter, R. H. G., et al. (2017). Complement-related regulates autophagy in neighboring cells. Cell 170, 158.e8–171.e8. doi: 10.1016/j.cell.2017.06.018
Liu, H., Zhang, Z., Xiong, W., Zhang, L., Xiong, Y., Li, N., et al. (2017). Hypoxia-inducible factor-1alpha promotes endometrial stromal cells migration and invasion by upregulating autophagy in endometriosis. Reproduction 153, 809–820. doi: 10.1530/REP-16-0643
Lu, W. H., Wang, G., Li, Y., Li, S., Song, X. Y., Wang, X. Y., et al. (2014). Autophagy functions on EMT in gastrulation of avian embryo. Cell Cycle 13, 2752–2764. doi: 10.4161/15384101.2015.945850
Lu, Y., Liang, M., Zhang, Q., Liu, Z., Song, Y., Lai, L., et al. (2019). Mutations of GADD45G in rabbits cause cleft lip by the disorder of proliferation, apoptosis and epithelial-mesenchymal transition (EMT). Biochim. Biophys. Acta Mol. Basis Dis. 1865, 2356–2367. doi: 10.1016/j.bbadis.2019.05.015
Lukacs, M., Roberts, T., Chatuverdi, P., and Stottmann, R. W. (2019). Glycosylphosphatidylinositol biosynthesis and remodeling are required for neural tube closure, heart development, and cranial neural crest cell survival. eLife 8:45248. doi: 10.7554/eLife.45248
Mangold, E., Ludwig, K. U., Birnbaum, S., Baluardo, C., Ferrian, M., Herms, S., et al. (2010). Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 42, 24–26. doi: 10.1038/ng.506
McCarthy, N. S., Melton, P. E., Cadby, G., Yazar, S., Franchina, M., Moses, E. K., et al. (2014). Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics 15:981. doi: 10.1186/1471-2164-15-981
Netea-Maier, R. T., Plantinga, T. S., van de Veerdonk, F. L., Smit, J. W., and Netea, M. G. (2016). Modulation of inflammation by autophagy: consequences for human disease. Autophagy 12, 245–260. doi: 10.1080/15548627.2015.1071759
Nunes, D. N., Dias-Neto, E., Cardo-Vila, M., Edwards, J. K., Dobroff, A. S., Giordano, R. J., et al. (2015). Synchronous down-modulation of miR-17 family members is an early causative event in the retinal angiogenic switch. Proc. Natl. Acad. Sci. U.S.A. 112, 3770–3775. doi: 10.1073/pnas.1500008112
Nyfeler, B., and Eng, C. H. (2016). Revisiting autophagy addiction of tumor cells. Autophagy 12, 1206–1207. doi: 10.1080/15548627.2016.1170265
Reynolds, K., Kumari, P., Sepulveda Rincon, L., Gu, R., Ji, Y., Kumar, S., et al. (2019). Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis. Model. Mech. 12:dmm037051. doi: 10.1242/dmm.037051
Thomas, N., Choi, H. K., Wei, X., Wang, L., Mishina, Y., Guan, J. L., et al. (2019). Autophagy regulates craniofacial bone acquisition. Calcif. Tissue Int. 105, 518–530. doi: 10.1007/s00223-019-00593-2
Tian, H., Feng, J., Li, J., Ho, T. V., Yuan, Y., Liu, Y., et al. (2017). Intraflagellar transport 88 (IFT88) is crucial for craniofacial development in mice and is a candidate gene for human cleft lip and palate. Hum. Mol. Genet. 26, 860–872. doi: 10.1093/hmg/ddx002
Timbers, T. A., Garland, S. J., Mohan, S., Flibotte, S., Edgley, M., Muncaster, Q., et al. (2016). Accelerating gene discovery by phenotyping whole-genome sequenced multi-mutation strains and using the sequence kernel association test (SKAT). PLoS Genet. 12:e1006235. doi: 10.1371/journal.pgen.1006235
Tower, J., Pomatto, L. C. D., and Davies, K. J. A. (2020). Sex differences in the response to oxidative and proteolytic stress. Redox Biol. 31:101488. doi: 10.1016/j.redox.2020.101488
Wang, M., Gu, D., Du, M., Xu, Z., Zhang, S., Zhu, L., et al. (2016). Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat. Commun. 7:11478. doi: 10.1038/ncomms11478
Wang, M. H., Cordell, H. J., and Van Steen, K. (2019). Statistical methods for genome-wide association studies. Semin. Cancer Biol. 55, 53–60. doi: 10.1016/j.semcancer.2018.04.008
White, K. A. M., Luo, L., Thompson, T. A., Torres, S., Hu, C.-A. A., Thomas, N. E., et al. (2016). Variants in autophagy-related genes and clinical characteristics in melanoma: a population-based study. Cancer Med. 5, 3336–3345. doi: 10.1002/cam4.929
Zhu, M., Xu, K., Chen, Y., Gu, Y., Zhang, M., Luo, F., et al. (2019). Identification of novel T1D risk loci and their association with age and islet function at diagnosis in autoantibody-positive T1D individuals: based on a two-stage genome-wide association Study. Diabetes Care 42, 1414–1421. doi: 10.2337/dc18-2023
Keywords: autophagy, non-syndromic cleft lip with or without cleft palate, single nucleotide polymorphism, molecular genetics, HIF1A
Citation: Lou S, Ma L, Kan S, Yu X, Wang Y, Yang F, Zhu G, Fan L, Li D, Wang H, Wang W, Zhang W, Wang L and Pan Y (2020) Association Study of Genetic Variants in Autophagy Pathway and Risk of Non-syndromic Cleft Lip With or Without Cleft Palate. Front. Cell Dev. Biol. 8:576. doi: 10.3389/fcell.2020.00576
Received: 08 May 2020; Accepted: 15 June 2020;
Published: 14 July 2020.
Edited by:
Erika Kuchler, Universidade Positivo, BrazilReviewed by:
Renato Assis Machado, Campinas State University, BrazilCopyright © 2020 Lou, Ma, Kan, Yu, Wang, Yang, Zhu, Fan, Li, Wang, Wang, Zhang, Wang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Wang, bHc2MDNAbmptdS5lZHUuY24=; Yongchu Pan, cGFueW9uZ2NodUBuam11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.