
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 12 June 2020
Sec. Molecular and Cellular Pathology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00474
This article is part of the Research Topic Molecular Mechanisms of Pathogen-Driven Infectious and Neoplastic Diseases View all 16 articles
The spliceosomal complex components, together with the heterogeneous nuclear ribonucleoproteins (hnRNPs) and serine/arginine-rich (SR) proteins, regulate the process of constitutive and alternative splicing, the latter leading to the production of mRNA isoforms coding multiple proteins from a single pre-mRNA molecule. The expression of splicing factors is frequently deregulated in different cancer types causing the generation of oncogenic proteins involved in cancer hallmarks. Cervical cancer is caused by persistent infection with oncogenic human papillomaviruses (HPVs) and constitutive expression of viral oncogenes. The aberrant activity of hnRNPs and SR proteins in cervical neoplasia has been shown to trigger the production of oncoproteins through the processing of pre-mRNA transcripts either derived from human genes or HPV genomes. Indeed, hnRNP and SR splicing factors have been shown to regulate the production of viral oncoprotein isoforms necessary for the completion of viral life cycle and for cell transformation. Target-therapy strategies against hnRNPs and SR proteins, causing simultaneous reduction of oncogenic factors and inhibition of HPV replication, are under development. In this review, we describe the current knowledge of the functional link between RNA splicing factors and deregulated cellular as well as viral RNA maturation in cervical cancer and the opportunity of new therapeutic strategies.
The large majority of human genes are transcribed as pre-mRNAs, containing non-coding (introns) and coding sequences (exons), that are processed by spliceosomal complexes to remove introns and produce mature mRNAs (Shi, 2017). The alternative removal of introns from pre-mRNAs and joining of exons into different mature transcripts enables the translation of multiple proteins from the transcription of a single gene (Bush et al., 2017). For this reason, the approximately 20,000 human genes are able to encode at least 100,000 different proteins (Wang et al., 2015).
Cell proteins generated by alternative splicing are selectively expressed in a tissue-specific and time-dependent manner and contribute to the regulation of numerous metabolic pathways involved in cell cycle control, differentiation and apoptosis (Baralle and Giudice, 2017). Aberrant splicing may cause the production of abnormal mRNA isoforms encoding mutated proteins with gain or loss of functions that are involved in neoplastic cell transformation, cancer development and metastasis (Oltean and Bates, 2014).
The role of spliceosome complexes and splicing regulatory factors in cancer has been widely investigated (Oltean and Bates, 2014). In particular, the snRNPs, the hnRNPs and the SR proteins have been shown to act either as oncoproteins or tumor suppressor proteins in different cancer types, including cervical neoplasia (Sorlie et al., 2001; Dvinge et al., 2016; Kohler et al., 2016; Cheng et al., 2017).
The aim of this review was to summarize the current studies on the role played by splicing factors in different cancer types with a particular focus on their peculiar activity in HPV-related cervical neoplasia. Indeed, in HPV infected cells the splicing factors are able to modulate the maturation either of cellular transcripts or of viral RNAs leading to the viral life cycle completion or production of viral and host cell transcripts encoding oncoproteins that cause transformation of cervical epithelium.
The removal of introns from pre-mRNAs is a process catalyzed by two large ribonucleoprotein complexes, namely major and minor spliceosomes, in cooperation with numerous splicing factors (Grabowski et al., 1985; Papasaikas and Valcarcel, 2016). The major spliceosome, responsible for more than 99% of splicing reactions in human cells, is composed of five uridine-rich small nuclear RNAs (snRNA U1, U2, U4, U5, and U6) and over 100 snRNA associated proteins (snRNPs) that undergo complex conformational changes during the different phases of splicing reactions (Frendewey and Keller, 1985; Wahl et al., 2009; Will and Luhrmann, 2011; Kastner et al., 2019). On the other hand, the minor spliceosome, including the U5 snRNA as well as functional analogs of the major spliceosome snRNAs (U11, U12, U4atac and U6atac), catalyzes the splicing of the less abundant U12-type introns (Tarn and Steitz, 1997; Turunen et al., 2013).
The splicing process consists of several sequential reactions involving recruitment of spliceosome components and interaction with cis-acting regulatory intronic sequences, such as the 5′ splice donor (SD) and 3′ splice acceptor (SA) sites, the intervening branch-point and the polypyrimidine tract (Aebi et al., 1986; Wang and Burge, 2008). Briefly, the formation of an early spliceosome complex involves the interaction of the U1 snRNP with the 5′ splice site through the base pairing of the U1 snRNA component, the binding of the U2 accessory factor (U2AF) to the polypyrimidine region and the connection of U2AF to U1 through the bridging splicing factor SF1. Binding of U2 snRNA to the branch sequence and 3′ splice site facilitates the U1 and U2 snRNPs interaction and the formation of the spliceosome complex A. Then, the U4/U5/U6 snRNP trimer interacts with the U1 and U2 snRNPs forming the spliceosome complex B, which releases the U1 and U4 snRNPs and becomes activated (complex C) (Wahl et al., 2009). The active spliceosome causes the final exclusion of the intron through: (1) the cleavage of the intron 5′-end; (2) the formation of a lariat (the 5′-end of the intron binds to the branch point); (3) the cleavage of the intron 3′-end; (4) the release of the lariat/U2/U5/U6 complex; and (5) the joining of exons (Lee and Rio, 2015; Papasaikas and Valcarcel, 2016) (Figure 1).
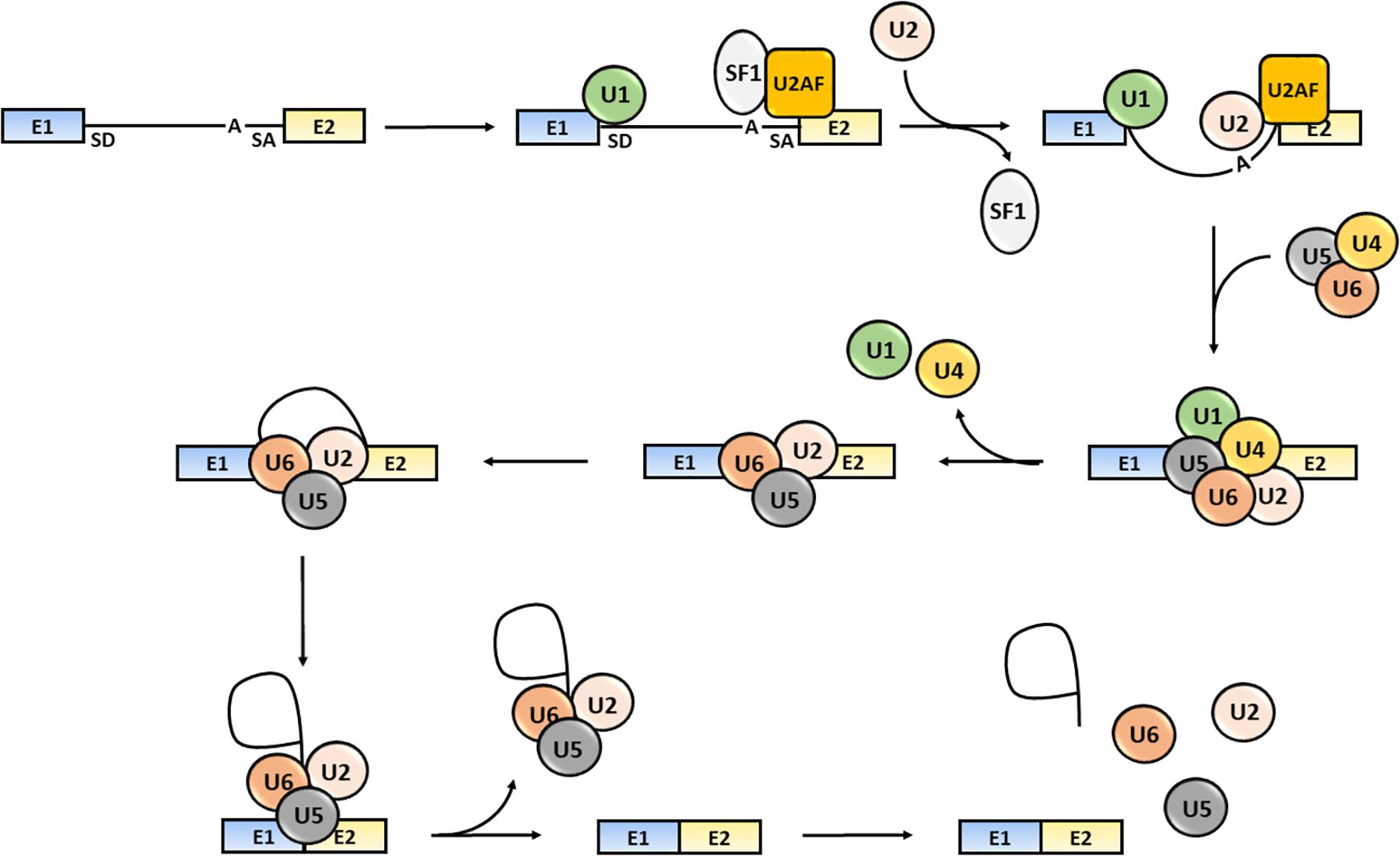
Figure 1. Steps of the splicing process. The snRNP U1, SF1 and U2AF bind to the intron 5′-end SD site, to an intronic branch point site (A) and to the intron 3′-end SA site, respectively. The U2 displaces SF1 and the U4/U5/U6 snRNPs trimer interacts with snRNPs U1 and U2 causing U1 and U4 release. The activated spliceosome catalyzes the cleavage of the intron 5′-end, the formation of a lariat, the intron the cleavage of the intron 3′-end, the release of the lariat/U2/U5/U6 complex release and joining of exons (Shi, 2017). Exons are defined as “E”; the intron is represented as a black solid line.
The alternative recognition of differential splice donor and acceptor sites allows the production of several mRNA isoforms according to five different figures: (1) the exon is totally or partially skipped; (2) mutually exclusive exons are included; (3) introns are retained; (4) alternative splice donor; or (5) splice acceptor sites are selectively chosen leading to the production of exons with different lengths (Lee and Rio, 2015; Wang et al., 2015) (Figure 2).
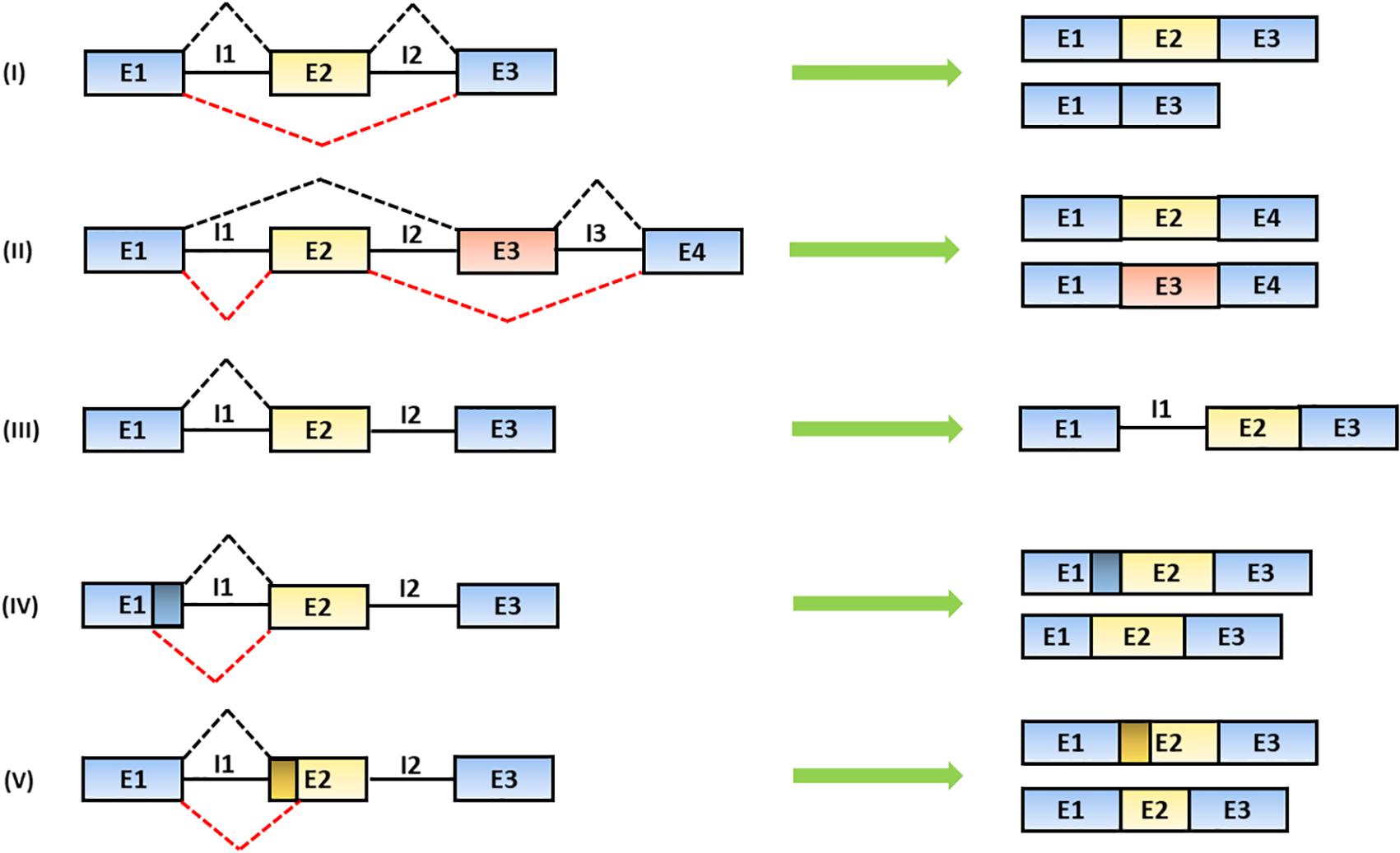
Figure 2. Mechanisms of alternative splicing. The spliceosome generates distinct mRNA isoforms by alternative usage of splice donor (SD) and acceptor (SA) sites, located at the 5′ and 3′ end of introns, respectively. The splicing products include: (I) exon skipping (the partial or total removal of exons), (II) inclusion of mutually exclusive exons, (III) intron retention and the use of alternative (IV) SD or (V) SA sites (Baralle and Giudice, 2017). In the figure, black solid lines represent introns; black and red dashed lines represent alternative splicing mechanisms. Exons are indicated as “E” and introns are indicated as “I.”
The fine tuning of pre-mRNA maturation is regulated by trans-acting splicing factors including hnRNPs, that generally inhibit the splicing by binding to exonic (ESSs) or intronic splicing silencers (ISSs) sequences, and SR proteins that typically activate the splicing by interacting with exonic (ESEs) and intronic (ISEs) splicing enhancers (Wang and Burge, 2008; Chen and Manley, 2009) (Figure 3).
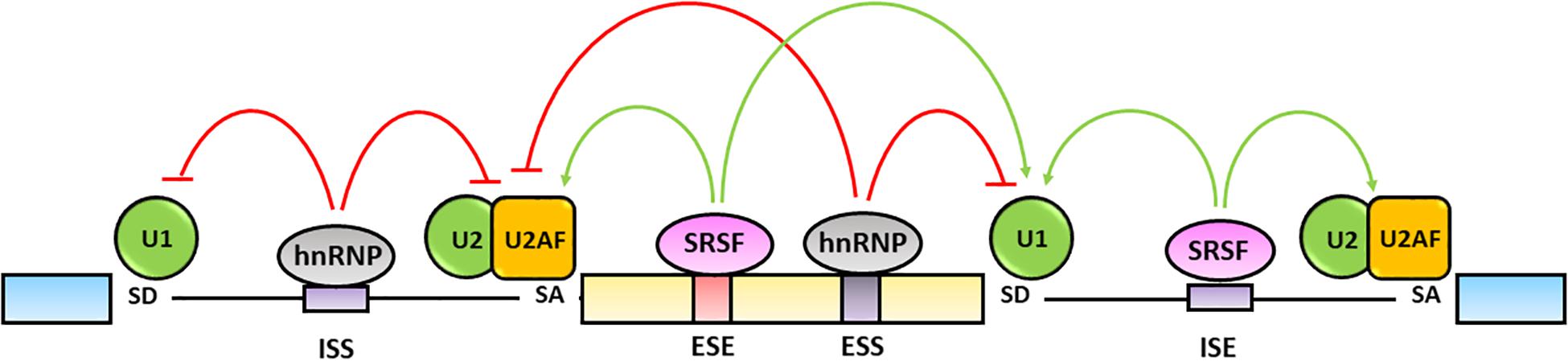
Figure 3. Regulation of alternative splicing. Alternative splicing is finely regulated by trans-acting hnRNPs and SRSFs protein families. The hnRNPs generally bind exonic (ESSs) and intronic splicing silencers (ISSs), antagonize SRs activity and inhibit spliceosome assembly on the splicing sites (red lines). The SRSFs bind to exonic (ESEs) and intronic (ISEs) splicing enhancers and increase the splicing efficiency by favoring the spliceosome recruitment and assembly on the splicing sites (green lines) (Chen and Manley, 2009; Wang et al., 2015).
The hnRNPs are a protein family comprising 20 major RNA-binding proteins that contain a RRM, a quasi-RRM domain and a K Homology (KH) domain binding to pre-mRNA sequences as well as a glycine-rich domain interacting with other hnRNPs (Busch and Hertel, 2012). The canonical function of hnRNPs is the inhibition of splicing reactions through their binding to cognate sites and repression of either the assembly of the spliceosome complex on the 5′-SD and 3′-SA sites or the recruitment of SR proteins on ESEs and ISEs following multimerization along exons or by looping out entire exons (Nasim et al., 2002; Geuens et al., 2016; Dvinge, 2018; Ule and Blencowe, 2019) (Figure 3). On the other hand, some hnRNPs have been shown to promote splicing through their interaction with ISE sequences containing G triplets. Indeed, hnRNP A1 and hnRNP F have been recognized to bind a AGGGA sequence in the 5′ GA-rich enhancer within the intron 10 of the insulin receptor gene (INSR) transcript as well as similar motifs located at the 3′ end of the same intron and to regulate the skipping or the inclusion of exon 11 causing the differential expression of the insulin receptor A or B isoforms, respectively (Talukdar et al., 2011).
Most of the hnRNPs, including hnRNP A1/A2, hnRNP B1/B2, hnRNP E, hnRNP J, and hnRNP K, are localized to the nucleus and after homomeric and heteromeric complexes formation they shuttle to the cytoplasm via the transportins binding (Siomi and Dreyfuss, 1995; Mili et al., 2001; Geuens et al., 2016). On the other hand, hnRNP C and hnRNP U possess a nuclear retention sequence that inhibits transfer to the cytoplasm (Nakielny and Dreyfuss, 1996; Mili et al., 2001). Post-translational modifications of hnRNPs, such as serine and threonine phosphorylation, arginines methylation, SUMOylation and ubiquitination, affect their sub-cellular localization and modulate positively or negatively their activity (Lee et al., 2012; Friend et al., 2013; Moujalled et al., 2015; Wang et al., 2017). For instance, the ERK kinase was shown to phosphorylate hnRNP K serines 284 and 353 in response to stress stimuli and to cause cytoplasmic accumulation and inhibition of hnRNP K-mediated translation in HeLa cells (Habelhah et al., 2001). In addition, the hnRNPs are able to modulate also the mRNA stability, capping, transport and poly-adenylation as well as to regulate the telomeres maintenance and chromatin organization (Han et al., 2010; Geuens et al., 2016).
The SR proteins family includes twelve serine-arginine rich splicing factors (SRSF1 to 12) characterized by canonical RRM, that interact with the ESE and ISE elements, and C-terminal serine-arginine repeats, which facilitate the recruitment of the spliceosome components to the splice sites (i.e., the binding of U1 to the 5′-SD site) and promote splicing processes (Manley and Krainer, 2010; Shen and Mattox, 2012; Bradley et al., 2015; Jeong, 2017) (Figure 3). In a few cases the SR proteins have been shown to bind silencer elements and to repress splicing (Kanopka et al., 1996; Shin et al., 2004; Shen and Mattox, 2012). For example, SRSF1 (SF2/ASF), SRSF4 (SRp75), SRSF5 (SRp40), and SRSF6 (SRp55) are able to bind an ISS located in the intron 9 of the CFTR transcript and to skip the exon 9 causing the production of a nonfunctional protein associated with cystic fibrosis development (Pagani et al., 2000).
The localization of SR proteins is regulated by dynamic serine-arginine repeats phosphorylation/dephosphorylation cycles that also modify their protein–protein interaction and functional activities (Zhou and Fu, 2013; Long et al., 2019). For example, the SRSF1, SRSF3 (SRp20), and SRSF7 (9G8) phosphorylated proteins are bound and shuttled by transportin SR2 from the cytoplasm to the nucleus (Lai et al., 2000, 2001). The serine-arginine rich proteins are specifically phosphorylated by the serine/arginine-protein kinases (SRPKs) and the Cdc2-like kinase/Ser-Thr-Tyr (Clk/Sty) kinases (Colwill et al., 1996; Giannakouros et al., 2011). The SRPKs are constitutively active in normal cells and localize either in the cytoplasm or into the nucleus (Giannakouros et al., 2011). The nuclear kinase CLK1 has been shown to phosphorylate the RS2 domain of SRSF1 and to cause the re-localization from speckles to the nucleoplasm (Aubol et al., 2018). In particular conditions, the SR proteins are phosphorylated by other kinases such as topoisomerase 1 (TOPO1), which becomes active in response to extra-cellular stimuli, and by Akt kinase and MAP kinases (MAPKs) that are constitutively active in cancer cells (Rossi et al., 1996; Naro and Sette, 2013). The SRSFs, similarly to hnRNPs, have also a variety of non-canonical activities related to the stability, export and translation of mRNAs as well as to the chromatin remodeling, nucleolar stress response, genome stability and cell cycle regulation (Howard and Sanford, 2015).
Several studies demonstrated that deregulation of constitutive and alternative splicing plays a crucial role in carcinogenesis and could be considered a novel cancer hallmark (Oltean and Bates, 2014; El and Younis, 2018; Wang and Lee, 2018; Zhang Y. et al., 2019; Che and Fu, 2020). Indeed, recent transcriptome sequencing analyses demonstrated that splicing factors expression is commonly deregulated in a multitude of cancer types and is associated with alterations of the tissue-specific transcripts as well as modification of protein–protein interactions (Buljan et al., 2012; Singh and Eyras, 2017).
Characterization of known transcripts from RNA-seq data of 4,542 tumor samples from 11 cancer types recorded in TCGA revealed the presence of 8,122 switches in the RNA isoforms encoded by 6,442 genes, the most common being RAC1 gaining an extra Ras domain and TP53 losing coding capacity (Climente-Gonzalez et al., 2017). Such changes were found mainly associated with alterations in apoptosis, ubiquitin-mediated proteolysis, ERBB-signaling, RNA transcription and splicing pathways (Climente-Gonzalez et al., 2017). In addition, the comprehensive analysis of whole exome sequencing and RNA-seq data of 8,705 tumor samples and 640 normal control tissues from 32 cancer types, including cervical cancer, showed approximately 251,000 new exon–exon junctions in tumors (around 930 per sample), of which 18,000 were recurrent events detected in at least 100 samples (Kahles et al., 2018). Such novel exon–exon junctions were predicted to produce “neoantigens” and to affect the immune response (Kahles et al., 2018). David et al. (2020) extended the analysis of exon–exon junctions to 10,549 tumor samples across 33 TCGA cancer types and observed that more than 50% of new junctions were not shared among cancer samples. Importantly, in cervical cancer they identified a total of 14,086,434 exon–exon junctions of which 2,263,326 were specific to cervical cancer (David et al., 2020).
Several mechanisms are responsible for aberrant splicing processes in human cancers. First, the uncontrolled overexpression of splicing factors may cause anomalous splicing events in tumors (Sveen et al., 2016). The up-regulation of hnRNPs and SRSFs has shown to be commonly caused by gene rearrangements and copy number variations in a variety of cancer types (Shilo et al., 2015). For example, the HNRNPA2B1 gene was found amplified in glioblastoma and its copy number was demonstrated to be inversely correlated with patients survival (Golan-Gerstl et al., 2011). Moreover, the amplification of the chromosome 17q23 and chromosome 6p21 regions has been shown to cause the overexpression of SRSF1 in breast cancer and SRSF3 in lung and cervical carcinoma, respectively (Parssinen et al., 2007; Jia et al., 2010).
The expression of hnRNP A1, hnRNP A2 and hnRNP I (also known as polypyrimidine tract binding protein, PTB) was demonstrated to be trans-activated by Myc oncoprotein. Specifically, the Myc mediated up-regulation of hnRNP A1, hnRNP A2 and hnRNP I induces the production of the pyruvate kinase embryonic isoform PKM2 and the activation of the aerobic glycolysis in different cancer cells (Christofk et al., 2008; David et al., 2010; Chen H. et al., 2018). Myc was also demonstrated to bind E-boxes in the SRSF1 promoter and to transactivate the SRSF1 overexpression that cause production of MKNK2 +13B and TEAD1 +5 isoforms and enhanced proliferation of lung cancer cells (Das et al., 2012).
Altered processing of splicing regulator transcripts represents a further mechanism of abnormal pre-mRNA maturation (Czubaty and Piekielko-Witkowska, 2017). For instance, knockdown experiments of S6K2 kinase, that phosphorylates the serine6 residue in hnRNP A1 protein, caused enhanced production of the PKM2 isoform and increased glycolysis in colorectal cancer cells (Sun Y. et al., 2017). The overexpression of SRPK1 causes the hyper-phosphorylation of SRSF1, leading to increased production of Rac1b, an oncogenic variant of the GTPase Rac1 signaling protein that promotes survival and proliferation of colorectal cancer cells (Goncalves et al., 2014; Patel et al., 2019). The overexpression of Clk2 kinase was also shown to sustain cell growth in breast cancer, while its silencing was demonstrated to reduce the SRSF1 phosphorylation and to induce the production of the ENAH isoform, which is typical of the mesenchymal phenotype and associated with tumor invasion and metastasis (Yoshida et al., 2015). In vitro studies demonstrated that the up-regulation of the NEK2 kinase promotes the production of the anti-apoptotic BCL-XL variant through the anomalous phosphorylation of SRSF1 (Naro et al., 2014). In addition, the PI3K/Akt signaling pathway, constitutively activated by mutated EGFR, leads to SRSF1 hyper-phosphorylation and consequent reduction of the anti-apoptotic isoform Casp-9b in lung cancer (Shultz et al., 2010).
Other post-translational modifications, such as acetylation and ubiquitination, are important modulators of splicing factors activity. For example, the hyper-acetylation of hnRNP A1 induced by high glucose levels in hepatocellular carcinoma cells promotes the production of the PKM2 variant and consequent enhancement of glucose metabolism as well as transcription of genes responsible for cell proliferation and growth, such as GLUT1, LDHA, PDK1, CCND1, and MYC (Yang et al., 2019). In addition, the hyper-acetylation of SRSF5 by Tip60 acetyl-transferase causes the production of the CCAR1 isoform S, which favors tumor growth by promoting glucose consumption and acetyl-CoA production (Chen et al., 2018a). On the other hand, the hypo-acetylation and consequent degradation of SRSF5 by the Smurf1 ubiquitin ligase causes CCAR1S reduction under low intracellular glucose levels (Chen et al., 2018a).
The hyper-O-GlcNAcylation of hnRNP-K in cholangiocarcinoma cells induces its translocation to the nucleus, where it drives the expression of genes involved in the extracellular matrix composition, cell movement, angiogenesis and epithelial mesenchymal transition, such as CCK, MMP3, PTGS2, and CTGF as well as CCND1 and XIAP (Gao R. et al., 2013; Phoomak et al., 2019). The overexpression of the SUMO1 was shown to cause the sumoylation of hnRNP K and increased c-Myc transcription associated with cell proliferation in Burkitt lymphoma (Suk et al., 2015).
Recurrent somatic mutations in splicing factors encoding genes have also been shown to affect the splicing processes in cancer (Kandoth et al., 2013; Watson et al., 2013; Sveen et al., 2016). Specifically, oncogenic driver mutations have been identified in 119 genes encoding for splicing factors in a variety of tumors (Seiler et al., 2018). For instance, point mutations and deletions in the HNRNPK gene, causing hnRNP K down-regulation, have been suggested to have a role in the development of acute myeloid leukemia (Sweetser et al., 2005; Gallardo et al., 2015). Moreover, hotspot mutations in the SRSF2 gene, such as P95H, change the binding properties of the SRSF2 protein and cause genome-wide splicing network alteration and aberrant maturation of different hnRNPs, including HNRNPA2B1, HNRNPM, HNRNPH1, and HNRNPH3, especially in myelodysplastic syndromes (Komeno et al., 2015; Arbab et al., 2018; Aujla et al., 2018; Liang et al., 2018; Masaki et al., 2019).
Moreover, nucleotide changes within the exonic or intronic cis-acting splicing factors binding motifs may result in the disruption or creation of binding sites and aberrant splicing reactions (Wang and Cooper, 2007; Sterne-Weiler and Sanford, 2014). A recent whole-exome and transcriptome comprehensive study including 31 cancer types identified 14,438 splicing-associated variants, most of which were shown to disrupt donor and acceptor sites and to cause exon skipping, intron retention and alternative 5′ or 3′ splicing site usage particularly in TP53 and GATA3 genes (Shiraishi et al., 2018).
Additionally, non-coding RNAs, such as miRNAs, are able to regulate splicing and transcription both in normal and tumor tissues for their ability to target the 3′-UTR of hnRNPs and SRSFs mRNAs (Anczukow and Krainer, 2016). For instance, the miR-15a-5p and miR-25-3p have been described as negative regulators of hnRNP A1 expression, required for the maturation of miR-18a-3p, that in turn inhibits the K-RAS oncogene in ovarian cancer (Rodriguez-Aguayo et al., 2017). Besides, miR-183-5p and miR-200c-3p in renal cancer as well as miR-193a-3p in gastric cancer have been demonstrated to target the SRSF2 (SC35) 3′-UTR mRNA, affecting the maturation of several pre-mRNAs involved in the apoptosis (Sokol et al., 2018; Lee et al., 2019).
On the other hand, the hnRNP and SRSF proteins are able to control directly or indirectly the expression of several miRNAs and other non-coding RNAs involved in cancer development (Ratnadiwakara et al., 2018). Indeed, the hnRNP A1 has been shown to interact with pri-miR-18a conserved terminal loop and to induce a relaxation in the stem loop that facilitates the cleavage by Drosha and production of mature miR-18a in prostate, esophageal, pancreatic, hepatocellular, and colorectal cancer (Guil and Caceres, 2007; Komatsu et al., 2014; Kooshapur et al., 2018). Moreover, the hnRNP D reduces the Dicer1 levels by targeting the 3′-UTR of DICER1 mRNA, causing down-regulation of tumor suppressor miR-122 and increased viability of PLC/PRF/5 hepatoma as well as Huh7 liver derived cell lines (Wu et al., 2018). Among SRSF proteins, the SRSF3 was demonstrated to enhance the processing of miR-16-1, miR-30a and miR-223 through the binding of a CNNC motif located 17-18 nucleotides downstream the Drosha cleavage signal (Auyeung et al., 2013).
Several hnRNPs, especially hnRNP K, are also able to modulate the expression of genes involved in the carcinogenic processes through their interaction with lncRNAs (Sun X. et al., 2017). In particular, Li et al. (2018) observed that linc00460 was overexpressed in non-small cell lung tumors and was able to interact with hnRNP K promoting migration and invasion of H460 and A549 human lung cancer cell lines. Furthermore, the analysis of RNA binding proteins-lncRNAs interaction network in the POSTAR2 database and co-immunoprecipitation experiments showed that SRSF1 was able to interact with and to stabilize the lncRNA NEAT1 causing increased cell proliferation in U87MG glioma cell line (Zhou et al., 2019).
Recently, hnRNPs have also been demonstrated to modulate the production of circular RNAs (Kramer et al., 2015). Specifically, the profiling of circular RNAs transcriptome in LNCaP prostate cancer cells expressing HNRNPL compared to HNRNPL-knockdown cells revealed the differential production of circ-PRKAR1B, circ-ZMIZ1, circ-FOXJ3, and circ-CCNY that have been suggested to be involved in prostate cancer development (Fei et al., 2017).
Several hnRNP and SRSF proteins have been demonstrated to be overexpressed in tumors and to possess multiple oncogenic functions (El and Younis, 2018) (Table 1). In particular, hnRNP A1 is upregulated in lung cancer and its silencing in lung cancer cell line A549 causes cell cycle arrest in G0/G1 phase (Liu et al., 2016). The hnRNP A1 expression enhances the production of a cyclin dependent kinase 2 (CDK2) isoform, characterized by the exon 5 retention, that promotes cell growth, while its inhibition causes the cell cycle arrest at the G2/M phase in oral squamous cancer cell lines (Yu et al., 2015). The overexpression of hnRNP A1 in gastric cancer and hnRNP A2/B1 in lung cancer has been shown to enhance the metastatic potential of tumor cells by inducing a shift from the expression of epithelial markers (i.e., E-cadherin) to mesenchymal markers (i.e., vimentin and snail) phenotype (Boukakis et al., 2010; Tauler et al., 2010; Chen et al., 2018b). The hnRNP A1 also mediates the production of the CD44v6 isoform, associated with larger tumor size, microvascular invasion and tumor recurrence in hepatocellular carcinoma patients (Zhou et al., 2013).
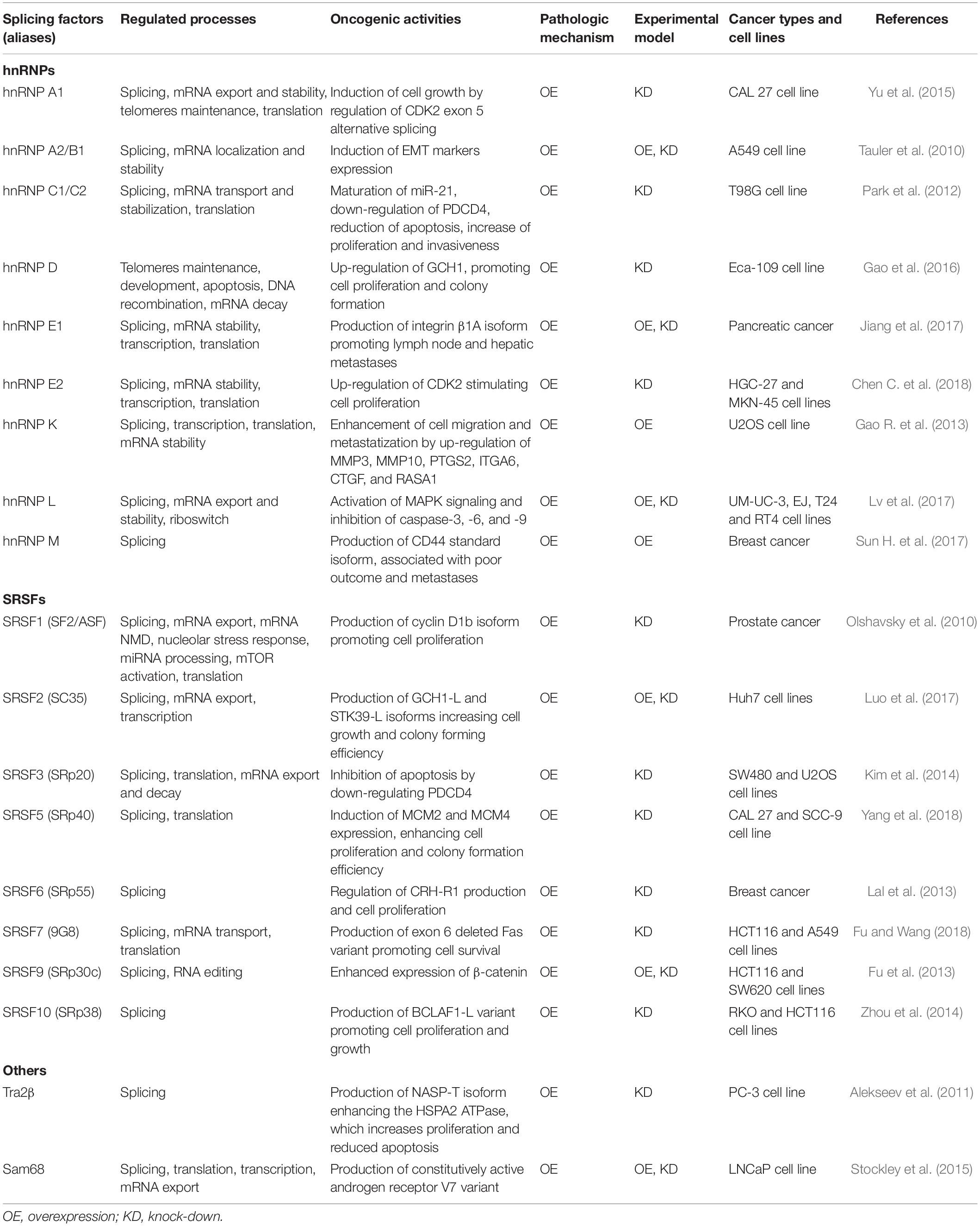
Table 1. Splicing factors and regulated oncogenic processes: physiological functions and oncogenic activities in different cancer types and cell lines.
The hnRNP D, that is overexpression in Eca-109 esophageal cancer cell line, has been recognized to bind the 3′-UTR AU-rich motifs of GTP cyclohydrolase (GCH1) transcripts causing GCH1 overexpression and enhanced cell proliferation and colony formation (Gao et al., 2016). Moreover, the hnRNP D protein can specifically bind to single-stranded d(TTAGGG)n human telomeric repeats through its C-terminal binding domain (BD2), thus impeding the formation of a DNA-quadruplex while favoring telomeres elongation and maintenance (Enokizono et al., 2005). The ectopic expression of p42 and/or p45 hnRNP D isoforms in hnRNP D-deleted mouse embryonic fibroblasts demonstrated that such isoforms are able to bind TERT promoter and to strongly transactivate TERT expression, thus reducing cell senescence (Pont et al., 2012).
The hnRNP E1 plays a major role in the production of integrin β1A isoform, whose expression has been found to correlate with lymph node and hepatic metastasis of pancreatic cancer, while the upregulation of hnRNP E2 was shown to promote the expression of CDK2 causing increased proliferation of gastric cancer cells (Jiang et al., 2017; Chen C. et al., 2018).
The hnRNP K expression has been demonstrated to enhance the transcription of genes involved in the extracellular matrix composition, especially MMP3 and MMP10, as well as of genes responsible for cell motility, such as PTGS2 and ITGA6, and angiogenesis, like CTGF and RASA1 (Gao R. et al., 2013).
The hnRNP L was shown to activate MAPK signaling while inhibiting caspase-3, -6 and -9 in bladder cancer cells as well as to suppress p53 expression and bcl-2/caspase-9/3 signaling via TP53 mRNA and bcl-2 binding, respectively, in prostate cancer cells (Lv et al., 2017; Zhou et al., 2017).
Overexpressed hnRNP M has been demonstrated to mediate the reduction of CD44v6 and increase of the CD44 standard isoform, that is associated with shorter overall survival and axillary lymph node metastases in breast cancer patients (Sun H. et al., 2017).
Among the SRSFs family, the SRSF1 gene was the first to be identified as a proto-oncogene with overexpression in colon, thyroid, small intestine, kidney and lung tumors (Karni et al., 2007). In particular, Karni et al. (2007) showed that SRSF1 regulates the alternative splicing of the tumor suppressor BIN1 and kinases MNK2 and S6K1 causing the production of a BIN1 isoform lacking tumor-suppressor activity, a MNK2 isoform promoting MAP kinase-independent eIF4E phosphorylation, and an oncogenic S6K1 isoform in transformed NIH 3T3 and Rat1 cell lines. The overexpression of SRSF1 was also shown to increase cell proliferation through the production of the exon 5 lacking-variant cyclin D1b in prostate cancer and to induce epithelial mesenchymal transition by stimulating the production of the ΔRon variant in breast cancer (Ghigna et al., 2005; Olshavsky et al., 2010).
Thereafter, several other SR proteins have been demonstrated to possess oncogenic activities in several tumor types (Table 1). For example, the knockdown of SRSF2 in Huh7 liver cancer cell line revealed 966 splicing alterations in cancer-related gene transcripts, favoring the production of oncogenic mRNA variants, such as the GCH1-L and STK39-L isoforms, that are involved in cell-cycle control and DNA repair (Luo et al., 2017).
The overexpression of SRSF3 into “normal” lung cell line WI-38 was able to modify the alternative splicing of the interleukin enhancer-binding factor 3 (ILF3) pre-mRNA and to increase the production of isoform-1 and isoform-2, both required for cell proliferation (Jia et al., 2019). Moreover, SRSF3 is able to deregulate the apoptotic pathway by binding to 5′-UTR of PDCD4 mRNA and by inhibiting the expression of the PDCD4, a critical suppressor of apoptosis (Kim et al., 2014).
The SRSF 5-7 proteins are particularly abundant in small cell lung cancer and associated with pleural metastasis (Kim et al., 2016). The SRSF5 enhances the expression of the MCM family members MCM2 and MCM4, that are involved in the initiation and elongation processes of DNA replication, as well as cell growth and colony formation when transfected in CAL 27 and SCC-9 oral squamous cell carcinoma cell lines (Yang et al., 2018). The SRSF6 is involved in the abnormal alternative splicing of the corticotropin-releasing hormone receptor type 1 (CRH-R1) causing indirect perturbation of oncogenic kinases, such as p38 MAPK, Akt and GSK3β, and accumulation of β-catenin in breast cancer (Lal et al., 2013). Knockdown experiments of SRSF7 in HCT116 colon and A549 lung cancer cell lines revealed the production of an exon 6-lacking isoform of the Fas receptor mRNA, that promotes cell survival (Fu and Wang, 2018).
The SRSF9 protein is frequently overexpressed in glioblastoma, colon adenocarcinoma, squamous cell lung carcinoma and malignant melanoma (Fu et al., 2013). The enhanced co-expression of SRSF9 and SRSF1 has been shown to promote tumourigenesis by Wnt signaling activation in a mTOR-dependent manner and to enhance translation of β-catenin mRNA (Fu et al., 2013).
The SRSF10 (SRp38) is an atypical SR protein acting as a potent general splicing repressor in its dephosphorylated form and as a sequence-specific splicing activator in its phosphorylated status (Feng et al., 2008). It was shown to mediate the expression of the BCLAF1 exon5a isoform (BCLAF1-L) that promotes cell proliferation and growth in colon cancer cell lines RKO and HCT116 (Zhou et al., 2014).
Splicing factors other than hnRNPs and SR family members, such as Tra2β, Brm and Sam68, have been also demonstrated to play a role in cancer development. The Tra2β is an SR-like protein characterized by a double N- and C-terminal RS domains that is able to promote alternative exons inclusion in a dose-dependent manner (Tacke et al., 1998; Stoilov et al., 2004; Venables et al., 2005; Grellscheid et al., 2011; Elliott et al., 2012). The TRA2B gene is amplified in several tumor types, including lung, head and neck, ovary, stomach and uterus cancers (Gabriel et al., 2009; Gao J. et al., 2013). Increased levels of Tra2β promote the production of the NASP-T isoform, enhances the ATPase activity of heat shock protein HSPA2 and causes increased proliferation as well as reduced apoptosis in PC-3 prostate cancer cell line (Alekseev et al., 2011; Best et al., 2013).
The Brm is a member of SWI/SNF (mating-type switch/sucrose non-fermentable) proteins family, involved in chromatin structure remodeling and DNA-damage response (Sudarsanam and Winston, 2000; Roberts and Orkin, 2004; Smith-Roe et al., 2015). The Brm factor interacts with U1, U3, and U5 snRNPs of the major spliceosome complex and favors the inclusion of alternative exons in gene transcripts involved in carcinogenesis (Smith-Roe et al., 2015; Pulice and Kadoch, 2016). In particular, the overexpression of BRM gene enhances the inclusion of exon 9 in the E-caderin mRNA in the MCF-7 human breast cancer cell line, while the Brm knockdown promotes the production of a cyclin D1b variant lacking exon 27 in Caco2 cell lines (Batsche et al., 2006).
The Sam68 is the prototypic member of the STAR family proteins that regulates alternative splicing and RNA processing in response to signaling pathways (Taylor et al., 1995). This factor contains a 200 amino acids long domain (GRP33/SAM68/GLD-1, GSG) that bind to RNA sequences as well as C-terminal six-proline-rich and tyrosine-rich sequences that interact with SRC homology 2 (SH2) and 3 (SH3) domains of BRK, FYN and Itk/Tec/BTK kinases (Andreotti et al., 1997; Derry et al., 2000; Paronetto et al., 2003; Najib et al., 2005). Sam68 was found overexpressed and associated with poor prognosis in several cancer types, including non-small cell lung carcinoma and breast, hepatocellular, renal, prostate and gastric cancers (Busa et al., 2007; Zhang et al., 2009, 2014, 2015; Song et al., 2010; Xiao et al., 2018). In addition, it was shown to modulate the splicing of transcripts encoded by genes involved in oncogenic pathways (Bielli et al., 2011). In particular, Paronetto et al. (2010) demonstrated that ERK-phosphorylated Sam68 was able to inhibit the U1 snRNP recruitment to the intron-exon 4 junction of CCND1 mRNA in the presence of the rs9344 (870A>G) polymorphism, thus favoring the production of the cyclin D1b. Moreover, Sam68 was also demonstrated to bind an ESE located near the cryptic exon 3b 3′ splice site of the androgen receptor mRNA, promoting the inclusion of exon 3b in the transcript and its translation into the androgen receptor V7 variant in the LNCaP prostate cancer cell line (Stockley et al., 2015).
The cell splicing factors contribute to cervical neoplasia by two distinct but converging mechanisms: (1) by mediating the differential maturation of HPV RNA isoforms, required for virus replication and for viral oncoprotein expression; and (2) by promoting the production of cell mRNA variants and proteins with oncogenic functions that may have roles in cervical neoplasia development and in the progression to later stages of cervical carcinogenesis (Mole et al., 2009a; Johansson and Schwartz, 2013; Oltean and Bates, 2014) (Table 2).
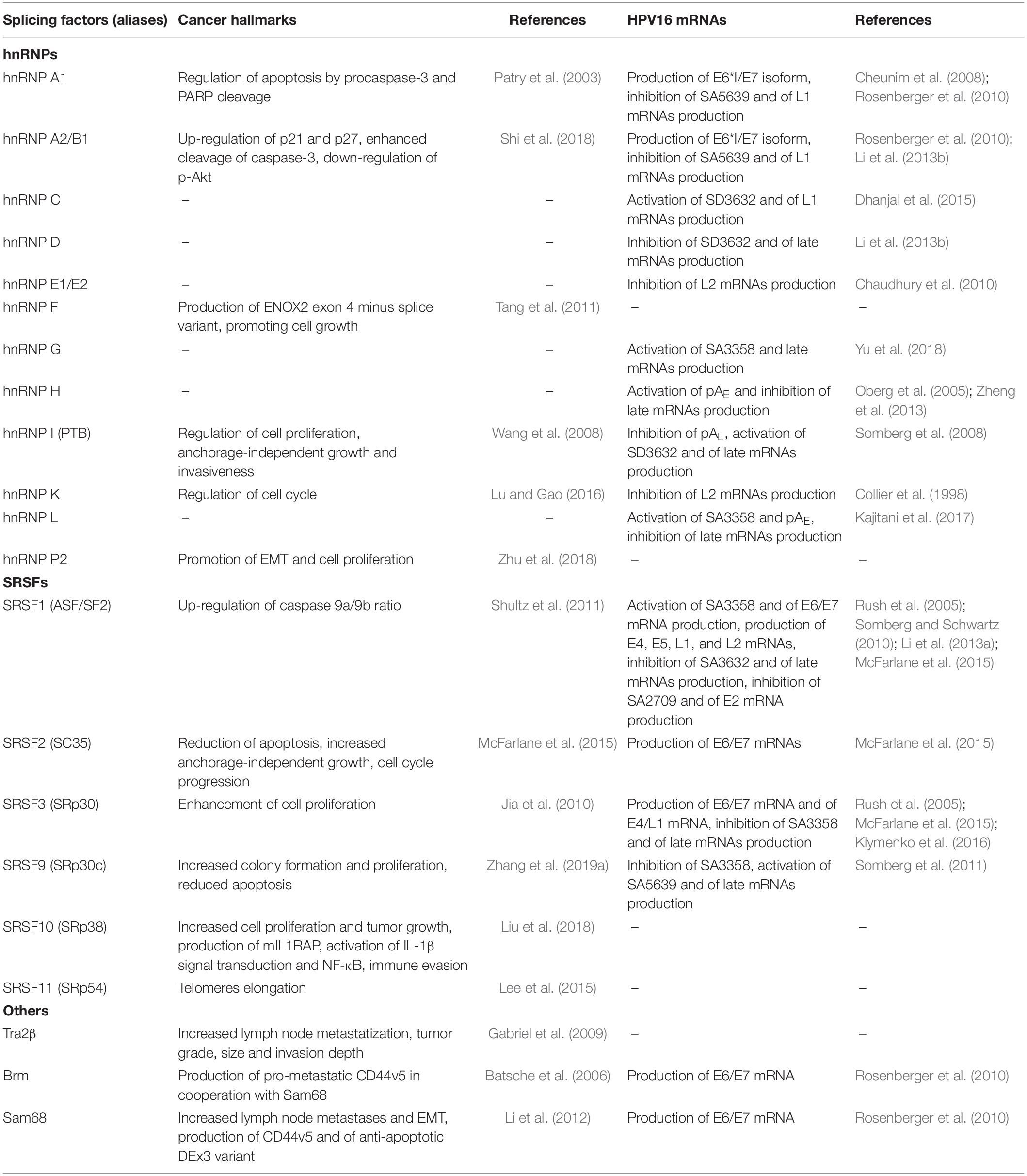
Table 2. Splicing factors that have been recognized to affect splicing of HPV16 and host cell transcripts in cervical cancer.
Persistent infection with high-risk HPVs, most commonly HPV16 and HPV18, is the main risk factor for development of almost all cases of cervical cancer (Doorbar, 2006; Schiffman et al., 2016), and of significant fraction of penile and anal cancers (Moscicki and Palefsky, 2011), vulvar cancer (Preti et al., 2020), as well as tumors of the upper respiratory tract, including head and neck cancers (Bzhalava et al., 2020). Most HPV infections of the cervix are successfully cleared by the host immune system in 2–3 years, but in several cases the infection may persist over the time leading to CIN graded 1 to 3 that eventually evolve during a long lasting period in invasive cervical carcinoma (McCredie et al., 2008; Doorbar et al., 2012; Oyervides-Munoz et al., 2018).
HPV genomes are double stranded circular DNAs of approximately 8000 bp which can be divided into three main regions: (1) the early region, containing the E1, E2, E3, E4, E5, E6 and E7 ORFs, encoding for regulatory proteins; (2) the late region, including L1 and L2 ORFs, encoding for viral capsid proteins and 3) the LCR, comprising the origin of replication and multiple transcription factor binding sites (Zheng and Baker, 2006; Doorbar et al., 2015). An early polyadenylation site (pAE) is located between the early and the late ORFs, while a late polyadenylation site (pAL) is positioned between the late genes and the LCR sequence (Zheng and Baker, 2006; Graham, 2010) (Figure 4).
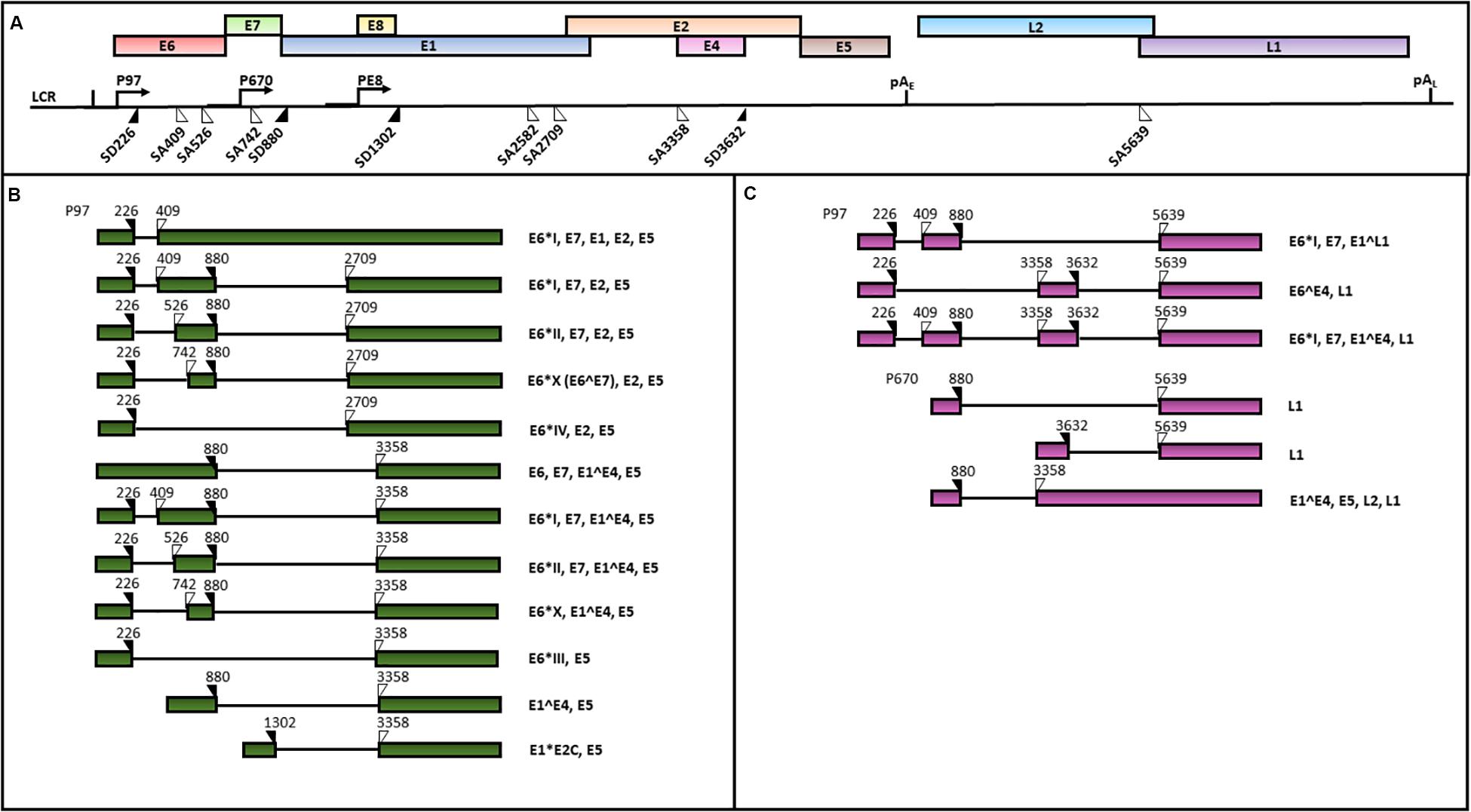
Figure 4. A schematic representation of the HPV16 genome and transcripts. The scheme is based on the HPV episteme (van et al., 2017). (A) The P97 derived early mRNAs are polyadenylated at the early polyadenylation site (pAE). The P670 and PE8 derived late mRNAs are polyadenylated at the late polyadenylation site (pAL). Both the early (B) and late (C) transcripts are polycistronic and subjected to alternative splicing (Zheng and Baker, 2006; Graham, 2010; Graham and Faizo, 2017). The donor (DS) and the acceptor (AS) splice sites in the HPV16 genome are indicated as black and white triangles, respectively. The coding potential of each transcript is also indicated.
The HPV 16 genome contains three promoters: 1) the early promoter P97 located in the LCR region driving the transcription of early polycistronic RNAs; 2) the late promoter P670 within the E7 ORF driving the expression of late polycistronic transcripts and 3) the recent discovered E8 promoter within the ORF E8, regulating the production of the E8^E2 transcript which has been demonstrated to control the viral copy number in undifferentiated keratinocytes (Schmitt and Pawlita, 2011; Chen et al., 2014; Straub et al., 2015) (Figure 4).
The HPV gene expression during the viral life cycle is dependent on cellular differentiation stages (Doorbar, 2005). Briefly, the E1 and E2 proteins as well as the E6 and E7 proteins are expressed at low levels in the basal cell stratum of the cervical epithelium and become overexpressed once the keratinocytes move from the basal to the mid layers (Doorbar, 2006; Xue et al., 2010; Doorbar et al., 2012). The L1 and L2 genes, encoding viral capsid proteins, are only produced in the terminally differentiated layers of the cervical epithelium (Doorbar et al., 1997, 2012). In high grade CIN and cervical cancer the viral genome usually integrates into host genome, causing disruption of the E2 ORF, E6 and E7 up-regulation as well as loss of late proteins expression (Collins et al., 2009; Oyervides-Munoz et al., 2018).
The balanced production of HPV16 early and late proteins is necessary for completion of the viral life cycle. The viral polycistronic transcripts are processed by cell splicing factors in order to remove introns through the differential usage of alternative splice sites and the production of viral proteins isoforms (Graham and Faizo, 2017). In particular, splicing sites SD226, SD880, SA409, SA526, SA742, SA2582, SA2709, and SA3358 in viral pre-mRNAs generated from the P97 promoter are recognized by the major spliceosome complex of the epithelial cells (Johansson and Schwartz, 2013). The SA3358 is the most used 3′ splice site, since these spliced mRNAs can either be polyadenylated at pAE to generate mRNAs encoding E6, E7, E4, or E5, or polyadenylated at pAL to produce mRNAs encoding L2 structural protein (Schmitt et al., 2010; Wu et al., 2017) (Figures 4B,C).
At late stages of productive viral infection, the binding of splicing factors to SD880, SD3632 and SA5639 sites in the transcripts generated from the P670 regulates late splicing events to produce E1^E4^L1 mRNAs, encoding both E4 and L1 proteins as well as the E1^E4 mRNAs (Johansson and Schwartz, 2013).
Alternative splicing plays a major role in the regulation of the HPV16 E6 and E7 oncogene expression. In fact, the E6 and E7 ORFs are transcribed as E6/E7 bicistronic mRNAs that can be spliced into diverse isoforms, such as E6∗I, E6∗II, E6∗III, E6∗IV, E6∗V, E6∗VI, E6^E7 (E6∗X), E6^E7∗I, and E6^E7∗II (Ajiro and Zheng, 2015; Brant et al., 2018; Olmedo-Nieva et al., 2018). Among these, the E6∗I and E6∗II, resulting from the SD226^SA409 and SD226^SA526 splicing, respectively, are the two major E6 isoforms expressed in cervical cancer (Zheng and Baker, 2006; Ajiro et al., 2012; Cerasuolo et al., 2017) (Figure 4B). McFarlane et al. (2015) analyzed the E6/E7 mRNA splicing pattern in cell subclones with transformed or differentiated phenotype obtained from the W12 cell line, originally derived from a HPV16-positive low grade cervical lesion (Doorbar et al., 1990). The analysis revealed the concomitant presence of E6, E6∗I and E6∗II in both transformed and differentiated W12 cell clones. However, the E6∗X isoform was only detected in W12 transformed cells, suggesting that the E6/E7 RNA splicing pattern may depend on specific cell transformation stages (McFarlane et al., 2015).
The alternative splicing of viral transcripts is also important to maximize the coding potential of the HPV16 genome. In fact, while the unspliced E6/E7 mRNA encodes for the full length E6 protein, the E6∗I and E6∗II mRNAs may favor the E7 expression by a termination-reinitiation process or leaky scanning mechanisms and the production of shorter E6 peptides (Zheng et al., 2004; Tang et al., 2006; Filippova et al., 2014). The E6 and E7 oncoproteins play a major role in cervical carcinogenesis for their ability to abrogate the functions of p53 and pRb oncosuppressors, respectively (Moody and Laimins, 2010; Tornesello et al., 2018; Yeo-Teh et al., 2018). Moreover, recent studies showed that the HPV16 E6∗I isoform have oncogenic activities, such as the disruption of mitochondrial functions and promotion of ROS production (Williams et al., 2014; Evans et al., 2016). Such activities are abrogated by the full-length E6 protein suggesting that the HPV-related cell transformation is regulated by the concerted expression of diverse E6 isoforms (Paget-Bailly et al., 2019). Further studies are needed to understand the precise role of viral mRNA splicing processes, especially E6/E7 mRNA, in high grade cervical neoplasia and cervical carcinoma.
The splicing of HPV RNAs is modulated by cellular hnRNPs and SRSFs factors, that recognize specific enhancer and silencers motifs in the viral transcripts (Graham and Faizo, 2017; Wu et al., 2017) (Table 3).
The hnRNP A1 was shown to favor the production of the E6∗I isoform in the absence of EGFR, although its binding site on the E6/E7 transcript has not yet been identified (Rosenberger et al., 2010). On the other hand, the hnRNP A1 was shown to bind an ESS in the L1 coding region and to inhibit the production of L1 protein (Zhao et al., 2004, 2007; Cheunim et al., 2008; Zhao and Schwartz, 2008) (Figure 5). Recently, Ajiro et al. (2016b) demonstrated that hnRNP A1 regulates the splicing of HPV18 transcripts by binding to the ESS located at nucleotides 612-639 thus inhibiting the removal of the intron comprised between SD233 and SA416 in the E6/E7 mRNA.
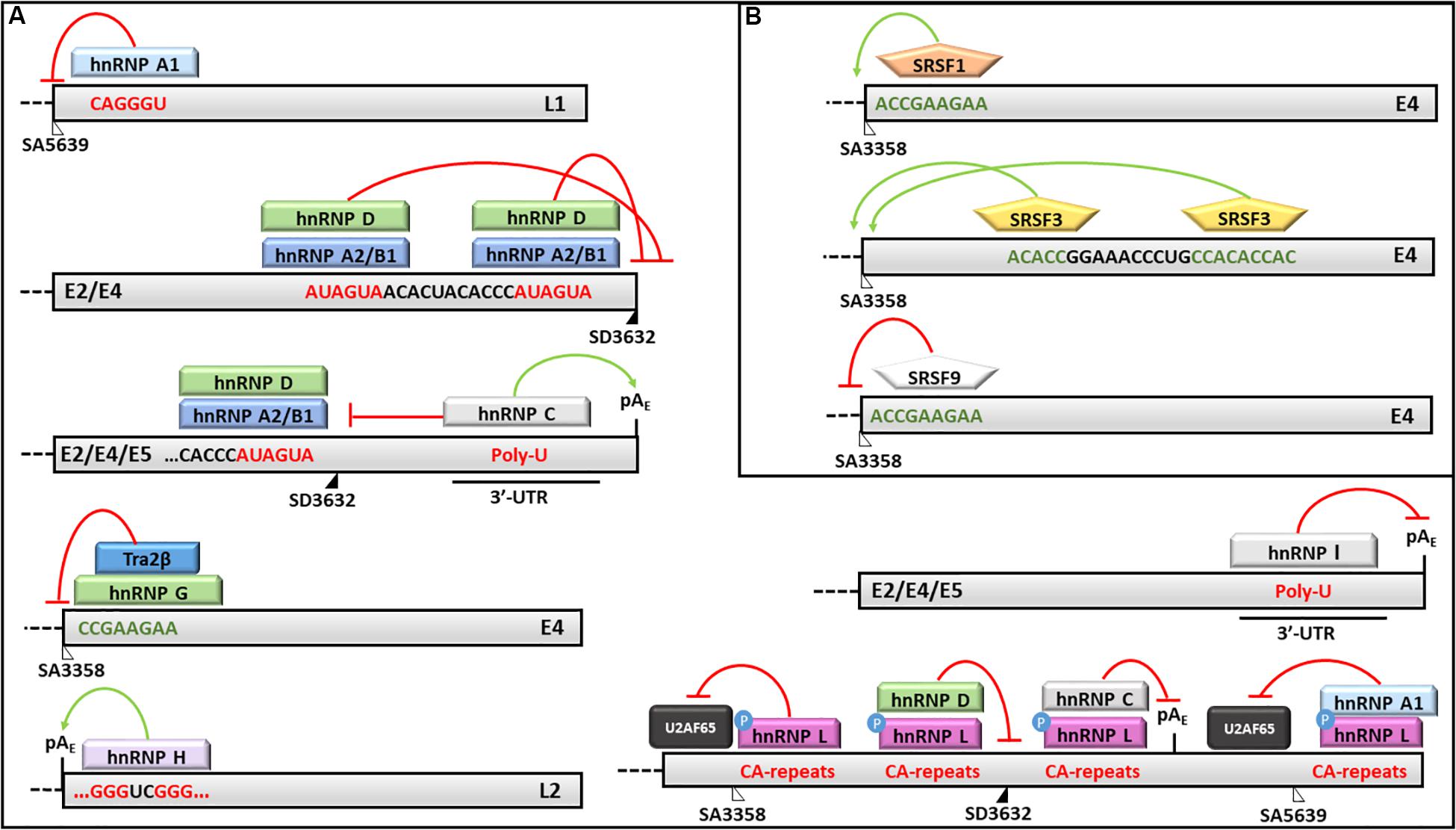
Figure 5. Splicing regulation of HPV16 mRNAs. Activities of (A) hnRNPs and (B) SRSFs and their binding motifs on HPV16 transcripts are shown. In particular: red lines indicate silencing activities; green arrows indicate enhancing activities; red sequences are silencing elements; while green sequences are enhancer elements. The donor (SD) and acceptor (AS) splice sites are indicated as black and white triangles, respectively.
Similarly, the hnRNP A2/B1 has been shown to mediate the production of the E6∗I isoform and to inhibit the splicing at SA5639 by preventing the binding of the U2AF65 splicing factor (Rosenberger et al., 2010; Kajitani et al., 2017). Moreover, the hnRNP A2/B1 and hnRNP D factors were demonstrated to bind two AUAGUA motifs located upstream the SD3632 and to inhibit L1 mRNA maturation and protein production in HeLa and C33A2 cervical cancer cell lines transfected with HPV16 complete genome (Li et al., 2013b) (Figure 5).
On the other hand, the hnRNP C1 has been demonstrated to increase the L1 and L2 mRNA levels through its binding to the HPV16 early untranslated region and to the AUAGUA silencer upstream the SD3632 in HPV16 -transfected HeLa and C33A cells (Dhanjal et al., 2015) (Figure 5). Recently, Nilsson et al. (2018) showed that the alkylating cancer drug melphalan induces the inhibition of HPV16 transcripts early poly-adenylation, by inducing hnRNP C interaction with the pAE site, and the expression of HPV16 late gene by inhibiting CPSF30-mediated suppression of the pAL.
Furthermore, the hnRNP E1/E2 were shown to interact with hnRNP K and to block the L2 mRNA translation by binding to a cis-acting inhibitory sequence located at the 3′-end of the L2 transcript (Collier et al., 1998; Chaudhury et al., 2010).
The hnRNP G has recently been shown to bind a 8-nucleotide sequence (CCGAAGAA) located downstream the SA3358 in HPV16, thus promoting the production of late mRNAs and skipping of the exon comprised between SA3358 and SD3632 in the L1 mRNA (Yu et al., 2018) (Figure 5).
The hnRNP H was shown to promote the polyadenylation at pAE site and to prevent late genes transcription in HeLa cells transfected with sub-genomic HPV16 plasmids through its interaction with the cut stimulating factor (CStF-64) and with G-rich motifs located in the L2 ORF (Oberg et al., 2005) (Figure 5). The late protein L1 was shown to interact with hnRNP H and to activate late genes transcription in differentiated keratinocytes (Zheng et al., 2013).
The hnRNP I binds a U-rich region in the 3′-UTR of HPV16 genome and induces L1/L2 mRNAs production by interfering with a polyadenylation signal at the pAE and with splicing inhibitory elements located upstream and downstream the SD3632 (Zhao et al., 2005; Somberg et al., 2008) (Figure 5).
Kajitani et al. (2017) showed that the hnRNP L phosphorylation by Akt causes its association with HPV16 late splice sites (SA3358, SD3632) and pAE, the inhibition of U2AF65 binding and decreased L1 mRNA production. Accordingly, the siRNA mediated knockdown of hnRNP L restored the expression of HPV16 L1 and L2 proteins confirming the this splicing factor performs a fine tuning of HPV16 late transcripts (Kajitani et al., 2017) (Figure 5).
The SRSF1 factor has been shown to bind the 8-nucleotide purine-rich enhancer downstream the SA3358 site in the E4 ORF and to activate the splicing of early transcripts (except E1 and E2) as well as late mRNAs in different phases of the viral life cycle (Rush et al., 2005; Milligan et al., 2007; Somberg and Schwartz, 2010; Li et al., 2013a) (Figure 5). In addition, the SRSF1 is able to bind the exon between SA3358 and SD3632 sites causing partial inhibition of the splicing at SA3632 and production of L1 mRNAs (Somberg and Schwartz, 2010). The SRSF1 has also been shown to reduce the splicing at SA2709 causing inhibition of E2 mRNA production while favoring production of E6 and E7 transcripts in the early phases of HPV16 life cycle (Somberg and Schwartz, 2010).
The SRSF2 and SRSF3 factors promote the early viral transcripts maturation, while their knockdown induces a strong reduction of E6 mRNA isoforms and E6 and E7 oncoproteins expression in CaSki cell lines (McFarlane et al., 2015; Klymenko et al., 2016). Interestingly, the transcriptional activity of the P97 promoter was not affected by SRSF2 levels suggesting it may favor the accumulation of E6/E7 RNAs by regulating the nonsense mediated decay (McFarlane et al., 2015). In addition, SRSF3 depletion causes reduction of E4/L1 mRNA and L1 protein production in W12E cells (McFarlane et al., 2015; Klymenko et al., 2016). Moreover, the SRSF3 is able to bind an A/C-rich enhancer downstream the SA3358 and to promote early genes expression and polyadenylation while inhibiting the late genes expression in CaSki and HeLa cells (Rush et al., 2005; Jia et al., 2009) (Figure 5).
The SRSF9 was also shown to inhibit the HPV16 SA3358 enhancer and to promote the production of L1 mRNAs through the skipping of the exon between SA3358 and SD3632 sites as well as through the activation of the splicing at SA5639 (Somberg et al., 2011) (Figure 5).
Finally, the cooperation of Sam68 and Brm has been shown to promote the production of full-length E6/E7 transcripts following the activation of Sam68 by Erk1/2 kinase in the presence of EGF in E6-transfected immortalized foreskin keratinocytes (Rosenberger et al., 2010).
The HPV E2 protein has been shown to possess SRSF-like activities and to regulate viral transcription and replication thus playing a key role in the HPV life cycle (Hegde, 2002; Graham, 2016). An interactome analysis showed that E2 protein binds to several RNA processing proteins and spliceosome components, such as EFTUD2, EIF4A3, SRPK1 and SRPK2, suggesting that it may be involved in the mRNA maturation processes (Jang et al., 2015). Bodaghi et al. (2009) showed that E2 binds to SA408 in the E6/E7 pre-mRNA and interacts with SRSF5, SRSF4, SRSF6 and SRSF9 through its C-terminal domain causing intron 1 exclusion from E6 and E7 mRNAs. Moreover, in mid layers HPV infected keratinocytes the levels of E2 correlate with the expression of SR proteins and both of them increase in cervical neoplasia during CIN progression from grade 1 to grade 3 (Coupe et al., 2012).
On the other hand, chromatin immunoprecipitation assays showed that E2 binds a region comprised between nucleotides 565-363 upstream the SRSF1 AUG starting site through its N-terminal domain and transactivates the SRSF1 expression in the U2OS human osteosarcoma cell line (Mole et al., 2009b). The E2 protein has also been shown to highly transactivate the SRSF3 promoter in the E2-gene transfected U2OS cells (Klymenko et al., 2016). Mole et al. (2020) recently observed that E2-transfected keratinocytes showed an increased expression of SRPK1 and hyper-phosphorylation of SRSF1, that translocate from the nucleus to the cytoplasm in a keratinocyte differentiation-specific manner.
Furthermore, the HPV16 E6 oncoprotein was demonstrated to interact with SRSF4, SRSF6, SRSF9 and with the E6/E7 pre-mRNA promoting intron 1 retention through a NLS3 sequence localized within its C-terminal domain (Bodaghi et al., 2009).
In HPV infected cells the release of E2F1 transcription factor, caused by the E7 binding to pRb, and its recruitment to binding sites in the SRSF10 promoter region determined increased expression of SRSF10 (Liu et al., 2018). Accordingly, the silencing of E6/E7 mRNA was shown to cause downregulation of SRSF10 in cervical cancer as well as in cervical cancer cell line SiHa (Liu et al., 2018).
The hnRNPs and SRSFs are generally upregulated in the basal and middle layers of the cervical epithelium and are downregulated in the terminal differentiated layers, while several of them have been found hyper-expressed in cervical carcinoma and suggested to play important roles in cervical carcinogenesis (Fay et al., 2009; Johansson and Schwartz, 2013).
In particular, the hnRNP A1 is significantly overexpressed either in high grade CIN or in cervical carcinoma compared to normal cervical epithelium representing a candidate diagnostic biomarker of cervical cancer (Kim et al., 2017; Qing et al., 2017). Interestingly, a transcriptome analysis of HPV16 E6 and E7 immortalized epithelial cells showed up-regulation of hnRNP A1 in cells cultured under hypoxic condition, suggesting that its expression could be induced by stress (Denko et al., 2000). The hnRNP A1 facilitates the production of mature miR-18a thus promoting proliferation and invasion of SiHa cells (Dong P. et al., 2019). The miR-18a suppresses the expression of PTEN, WNK2, SOX6, BTG3, and RBSP3 genes by binding to their 3′-UTR transcripts, and causes PD-L1 expression and Wnt/β-catenin pathway activation in cervical carcinoma (Dong P. et al., 2019). Moreover, the siRNA-mediated hnRNP A1/A2 knockdown caused shortening of telomeres, procaspase-3 and PARP cleavage as well as increased apoptosis in HeLa cells, suggesting that it plays a significant role in cervical cancer cell viability (Patry et al., 2003).
The role of hnRNP A2/B1 in cervical cancer has been recently investigated (Li et al., 2014). Interestingly, Li et al. (2014) observed that lobaplatin induced cell cycle arrest and apoptosis via downregulation of hnRNP A2/B1 in Caski cells. Moreover, the hnRNP A2/B1 knockdown was shown to suppress cell proliferation, migration and invasion, while increasing the apoptosis and the sensitivity to irinotecan or lobaplatin through the up-regulation of p21, p27 and cleaved caspase-3 as well as p-AKT down-regulation in HeLa and CaSki cell lines (Shi et al., 2018).
The hnRNP E1 and E2 expression decreases during the progress from low grade to high grade cervical neoplasia with an inverse correlation with HPV16 E6 protein levels (Pillai et al., 2003). Indeed, both factors may act as oncosuppressors, being under-expressed in CIN 2-3 and cervical carcinoma compared to CIN 1 (Gao et al., 2020).
Tang et al. (2011) demonstrated that hnRNP F binds to a GGGA ESS motif, located within ENOX2 exon 4, causing enhanced production of ENOX2 exon 4 minus splicing variant, that is overexpressed in cervical cancer and involved in increased cell growth.
The levels of hnRNP H and I are higher in cervical cancer and high grade CIN compared to low grade CIN and normal tissues (Oberg et al., 2005; Fay et al., 2009). The hnRNP I knockdown reduces the proliferation and anchorage-independent growth while increasing invasiveness of HeLa cells (Wang et al., 2008). The hnRNP I was also shown to regulate gene expression by forming a dimer with PTB-associated splicing factor (PSF) in SiHa and CaSki cells (Zhang et al., 2020). Moreover, Zhang et al. (2020) demonstrated that lncRNA ARAP1-AS1 interacts with PSF causing the release of hnRNP I, which in turns enhances the c-Myc IRES-dependent translation by binding to c-Myc mRNA 5′-UTR enhancing SiHa and Caski cell proliferation.
The hnRNP P2 (FUS/TLS) is highly expressed in cervical cancer as well as in SiHa and HeLa cells, dependently from the activation of XIST lncRNA/miR-200a axis, and induces the epithelial mesenchymal transition phenotype and proliferation while inhibiting apoptosis in cervical cancer cells (Zhu et al., 2018).
The SRSF1, SRSF2 and SRSF3 proteins are overexpressed in CIN 1 upper differentiated layers, in the CIN3 entire epithelium as well as in W12 cells (Mole et al., 2009a). The SRSF1 down-regulation causes the caspase 9a/9b decreased ratio and increased sensitivity of cancer cells to DNA damaging agents (Shultz et al., 2011). Similarly, the SRSF2 depletion causes increased p53 expression and apoptosis in W12 transformed subclones as well as decreased anchorage-independent growth and cell cycle arrest at G2/M checkpoint both in W12 and C33A cells (McFarlane et al., 2015). Wu et al. (2010) first demonstrated that SRSF1 was able to bind to CGGACAC motifs in the pri-miR-7-1, pri-miR-221 and pri-miR-222 stem loops thus enhancing Drosha cleavage and accumulation of miR-7, miR-221 and miR-222, promoting cervical cancer development (Pan et al., 2019). Moreover, Dong M. et al. (2019) demonstrated that the lncRNA MIR205HG, which is overexpressed in cervical cancer, targets SRSF1 and up-regulates KRT17 causing cell proliferation and apoptosis inhibition in CaSKi, Hela, MS751 and SiHa cells. The silencing of SRSF3 in HeLa cells was shown to promote the production of ILF3 isoforms 1 and 2 as well as cell cycle progression to the S and G2/M phases and proliferation enhancement (Jia et al., 2019). In addition, a miRNA array analysis showed that SRSF3 knockdown in HeLa cells caused the down-regulation of miR-16, miR-18a, miR-21, miR-92b, miR-128, miR-182, miR-629, miR-629, miR-1180, and miR-1308, and the up-regulation of miR-7, miR-26a, miR-30a, miR-99a, miR100, miR-125b, miR-181a, miR-206, miR-378, and miR-923, all involved in carcinogenic processes (Ajiro et al., 2016a; Tornesello et al., 2020).
The SRSF9 is overexpressed in cervical carcinoma, due to the down regulation of miRNA-802 that in normal epithelium binds the 3′-UTR region of SRSF9 transcript inhibiting translation, and causes increase in cell proliferation (Zhang et al., 2019b).
Similarly, the SRSF10 is increasingly overexpressed in CIN and cervical cancer cells. Its expression is higher in HPV16 and HPV18-positive compared to HPV-negative cervical cancers, but becomes strongly downregulated following the E6/E7 silencing in CaSki cells (Liu et al., 2018). Moreover, the injection of SRSF10-knockdown SiHa cells in nude mice reduced tumor growth suggesting that this factor promotes cell proliferation (Liu et al., 2018). The overexpression of SRSF10 has shown to favor the transcription of the CD47 immune evasion signal and the production of the mIL1RAP isoform, which induces the NF-κB activation and the IL-1β signal transduction associated with production of pro-inflammatory cytokines (Liu et al., 2018).
Finally, the SRSF11 (SRp54) has been observed to interact with TERC and TRF2 factors and to promote the telomerase association with telomeric repeats and telomeres elongation in HeLa cells (Lee et al., 2015).
Other proteins involved in splicing processes, including Tra2β, Brm and Sam68, have been found deregulated in cervical cancer and derived cell lines and proposed to have significant roles in cervical carcinogenesis. Specifically, the over expression of Tra2β protein, which localizes to the cervical carcinoma cell nuclei, was demonstrated to correlate with lymph node metastasis, higher tumor grade, size and depth of invasion (Gabriel et al., 2009; Best et al., 2013). The Brm factor is able to interact with ERK phosphorylated Sam68 causing the production of the CD44v5 variant by its direct binding to the exon 5 splice-regulatory elements and exon 5 retention in CD44 mRNA in HeLa cell line (Doorbar, 2005).
The Sam68 is highly expressed in cervical carcinoma and its cytoplasmic localization is shown to be associated with pelvic lymph node metastasis and poor prognosis in patients with early-stage cervical cancer (Li et al., 2012). Li et al. (2012) also demonstrated that down-regulation of Sam68 in cervical cancer cells reduced the cell motility and invasion as well as reversed the epithelial mesenchymal transition phenotype through the inhibition of the Akt/GSK-3β/Snail pathway. In HeLa cells the Sam68 was shown to bind a UAAAAAGCAU sequence within the exon 3 of survivin mRNA thus promoting the production of the anti-apoptotic DEx3 variant. Such variant has been found over-expressed in cervical adenocarcinoma HeLa cell line as well as in cervical carcinoma tissues (Futakuchi et al., 2007; Gaytan-Cervantes et al., 2017). Moreover, Sam68 was observed to interact with SRm160, a SR-rich splicing co-activator, to stimulate the inclusion of the v5 exon into the CD44 mRNA in a Ras-signaling dependent manner causing increased invasiveness of HeLa cells (Cheng and Sharp, 2006).
The relevant role of aberrant splicing in carcinogenesis highlights the need for novel splicing-targeted therapies. Currently, different strategies have been adopted to target deregulated splicing factors and abnormal splicing variants, including the usage of small bacteria-derived molecules against the core spliceosome, splicing regulators inhibitors and anti-sense oligonucleotides against oncogenic mRNA isoforms (Lee and Abdel-Wahab, 2016; Bates et al., 2017; Lin, 2017; Di et al., 2019; Bonnal et al., 2020) (Table 4).
Several bacterial derived small molecules or their synthetic analogs have been used to inhibit the spliceosome assembly or the post-translational modifications of the core spliceosome components (Effenberger et al., 2017). Particularly, spliceostatins and pladienolides derived from Pseudomonas and Streptomyces, respectively, have shown to bind the SF3b component of U2 snRNP causing the inhibition of the spliceosome assembly and cell cycle arrest at G1 and G2/M phases (Nakajima et al., 1996; Sakai et al., 2002, 2004). The binding of spliceostatin A to SF3b1 has been demonstrated to stimulate the production of unspliced p27 mRNA encoding the C-terminal truncated p27∗ variant which causes cell cycle arrest at G1 phase by inhibiting CDK2 in HeLa cells (Kaida et al., 2007; Satoh and Kaida, 2016). Treatment with pladienolide B caused reduction of SF3b1 expression in a dose-dependent manner, cell cycle arrest at the G2/M phase and induction of apoptosis by increasing the expression of Tap73, cytochrome C and pro-apoptotic Bax, while decreasing the levels of ΔNp73 and anti-apoptotic Bcl-2 in HeLa cells (Zhang et al., 2019a).
Targeting of kinases regulating the splicing factors activity, such as SRPK1 and CLK2, represents a further promising therapeutic model (Urbanski et al., 2018). Either SRPK1 siRNA knockdown or treatment by SPHINX and SRPIN340 inhibitors reduced the SRSF1, SRSF2 and SRSF5 phosphorylation causing increased production of the anti-apoptotic VEGF165b isoform in PC-3 prostate cancer cell line (Mavrou et al., 2015). Araki et al. (2015) showed that Cpd-1, Cpd-2, and Cpd-3 compounds inhibit the Clk1 and Clk2 kinases activity causing reduction of SRSF4 and SRSF6 phosphorylation and modifying genome-wide splicing patterns. In particular, the transcriptome analyses performed after treatment with Cpd-2 showed changes in the splicing patterns of gene transcripts involved in growth and survival, such as EGFR, CD44, EIF3H, AURKA, HDAC1, and PARP, leading to reduced proliferation and increased apoptosis in colorectal cancer COLO205 and COLO320DM, lung cancer NCI-H23 as well as breast cancer MDA-MB-468 cell lines (Araki et al., 2015).
Splice-switching antisense oligonucleotides (SSOs) are synthetic 15–30 nucleotide long sequences that bind matched splice sites and inhibit the splicing factors interaction by steric hindrance (Havens and Hastings, 2016). Chemical modifications, such as 2′-O-methylation and 2′-O-methoxyethylation are necessary to prevent degradation of pre-mRNA-SSO complexes by RNase H (Rigo et al., 2014). The 2′-O-methoxyethylated Bcl-x SSO oligonucleotide is able to target the 5′-splice site in exon 2 of Bcl-x mRNA causing switch from anti-apoptotic Bcl-xL to pro-apoptotic Bcl-xS in a dose dependent manner and apoptosis via PARP cleavage in B16F10 mouse melanoma cells (Bauman et al., 2010). Furthermore, 2′-O-methylated SSO, targeting the 3′-splice site in exon 14b of MKNK2 mRNA, enhances the tumor-suppressor Mnkn2a isoform levels, leading to increased phosphorylation and activation of p38α–MAPK and overexpression of cFOS, COX2 and IL-6 target genes (Mogilevsky et al., 2018). The treatment with 2′-O-methylated SSO causes reduction of anchorage independent growth and survival in glioblastoma U87MG, hepatocellular carcinoma HuH7 and breast cancer MDA–MB–231 cell lines (Mogilevsky et al., 2018).
The resveratrol has been shown to regulate the miR-424 and miR-503 expression that inhibit hnRNP A1 production and cell proliferation in human breast cancer cell line MCF7 (Otsuka et al., 2018). In addition, bortezomib has found to reduce the expression of hnRNP K, hnRNP H, Hsp90α, Grp78, and Hsp7C in Burkytt lymphoma cell lines such as CA46 and Daudi, as determined by a proteomic analysis (Suk et al., 2015).
A natural compound, caffeine, was demonstrated to reduce the SRSF3 levels by causing increased expression of the p53β variant, reduced expression of SRSF3 targets, such as HIF-1α, SREBP1c, COX-2, FASN, and EGFR, and enhanced cell senescence (Lu et al., 2014). Similarly, theophylline down-regulates SRSF3 expression and switches the production from p53α to p53β isoform in HeLa and MCF-7 cell lines causing apoptosis, senescence, and decreased colony formation (Chang et al., 2017). Another natural drug namely indacaterol was shown to rescue SRSF6 associated aberrant splicing events, such as ZO-1 exon 23 skipping, and to reduce the viability of RKO, HCT116, and HCT8 colorectal cancer cells as well as to inhibit cancer growth in a colorectal cancer mouse model (Wan et al., 2019).
Aberrant protein isoforms have also been proposed as potential therapeutic targets (Le et al., 2015). A comprehensive genomic analysis of 38’028 tumors allowed the discovery of recurrent somatic mutations in the splice donor and acceptor sites in MET transcripts causing skipping of exon 14 in lung adenocarcinoma (Frampton et al., 2015). The expression of exon 14-lacking MET in NIH3T3 cells conferred higher sensitivity to MET-specific inhibitors, such as capmatinib, compared to cells expressing wild type MET (Frampton et al., 2015).
In recent years, there is a growing experimental evidence of the key role played by deregulated splicing in cancer development and progression. Several genome-wide studies have revealed tumor specific splicing profiles at mRNA and protein level, identifying a multitude of aberrant splice variants with oncogenic functions. Different protein isoforms have been demonstrated to have a prognostic value and to modify the sensitivity of cancer cells to chemotherapeutic agents. On the other hand, therapies targeting deregulated splicing factors or pathogenic splicing isoforms are under development. Several data have been published on the role of splicing factors in cervical cancer development and further studies are needed to clarify the relevance of such factors in the production of viral or cell genome encoded oncogenic isoforms as well as in the promotion of pre-neoplastic lesions progression to invasive cancer. Much more needs to be done for the development and the clinical application of splicing-targeted therapeutic agents that would inhibit cervical cancer as well as other HPV-related tumors by affecting both cell oncogenic pathways as well as HPV life cycle.
AC performed bibliography analysis and wrote the manuscript. LB and FB supervised the whole project. MT designed the study and drafted the manuscript. All authors read and approved the final manuscript.
This work and the publication costs were supported by the research grants Ricerca Corrente (N. 2611892) and Ministero della Salute (RF-2018-12366163).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Clorinda Annunziata, Francesca Pezzuto, and Noemy Starita for helpful discussion.
BARD1, BRCA1 associated RING domain 1; BCLAF1, BCL2 associated transcription factor 1; BIN1, bridging integrator-1; BRCA1, breast cancer 1, early onset; BRM, brahma; BTG3, BTG anti-proliferation factor 3; Casp, caspase; CCAR1, cell cycle and apoptosis regulator 1; CCK, cholecystokinin; CCND1, cyclin D1; CDK, cyclin dependent kinase; CFTR, cystic fibrosis transmembrane conductance regulator; CIN, cervical intraepithelial neoplasia; Clk/Sty kinases, Cdc2-like kinase/Ser-Thr-Tyr kinase; COX-2, cyclooxygenase 2; CPSF30, cleavage polyadenylation specificity factor 30; CRH-R1, corticotropin-releasing hormone receptor type 1; CTGF, connective tissue growth factor; E2F1, E2F transcription factor 1; EFTUD2, elongation factor Tu GTP binding domain containing 2; EGFR, epidermal growth factor receptor; EIF3H, eukaryotic translation initiation factor 3 subunit H; EIF4A3, eukaryotic translation initiation factor 4A3; eIF4E, eukaryotic translation initiation factor 4E; EMT, epithelial-mesenchymal transition; ERK, extracellular signal-regulated kinase; ESE, exonic splicing enhancer; ESS, exonic splicing silencer; FASN, fatty acid synthase; FOXO4, forkhead box O4; GATA3, GATA binding protein 3; GCH1, GTP cyclohydrolase 1; GLUT1, glucose transporter; GSK3 β, glycogen synthase kinase 3 beta; HDAC1, histone deacetylase 1; HIF-1 α, hypoxia-inducible factor-1; hnRNP, heterogeneous nuclear ribonucleoprotein; HPV, human papillomavirus; Hsp90 α /7C, heat shock protein 90 α /7C; HSPA2, heat shock protein family A; IL1RAP, interleukin 1 receptor accessory protein; ILF3, interleukin enhancer-binding factor 3; ISE, intronic splicing enhancer; ISS, intronic splicing silencer; ITGA6, integrin subunit alpha 6; KH domain, K homology domain; LCR, long control region; LDHA, lactate dehydrogenase A; lncRNA, long non-coding RNA; MAPK, mitogen-activated protein kinase; MCM2/4, minichromosome maintenance complex component 2/4; miR, micro-RNA; MKNK2, MAPK interacting serine/threonine kinase 2; MMP3, matrix metallopeptidase 3; mTOR, mechanistic target of rapamycin kinase; NASP, nuclear autoantigenic sperm protein; NCL, nucleolin; NEK2, NIMA related kinase 2; NF- κ B, nuclear factor kappa-light-chain-enhancer of activated B cells; NLS, nuclear localization signal; NMD, non-sense mediated decay; ORF, open reading frame; pAE/pAL, early/late polyadenylation signal; PARP, poly ADP-ribose polymerase; PCNA, proliferating cell nuclear antigen; PDCD4, programmed cell death 4; PDK1, pyruvate dehydrogenase kinase 1; pRb, retinoblastoma protein; Pre-mRNA, pre-messenger RNA; PTEN, phosphatase and tensin homolog; PTGS2, prostaglandin-endoperoxide synthase 2; RAC1, ras-related C3 botulinum toxin substrate 1; RASA1, RAS protein activator 1; Ron, recepteur d’origine nantais; RRM, RNA recognition motif; Sam68, Src-associated in mitosis 68 kDa; S6K2, S6 kinase 2; SD/SA, splice donor/acceptor site; SH2/SH3, Sarc homology 2/3; snRNP, small nuclear ribonucleoprotein; SR protein, serine/arginine-rich protein; SREBP1c, sterol regulatory element-binding transcription factor 1; SRPK, serine/arginine kinase; SRSF, serine/arginine-rich splicing factor; STAR, signal transduction and activation of RNA; STAT, signal transducer and activator of transcription; STK, serine/threonine kinase; SUMO1, small ubiquitin-like modifiers 1; SWI/SNF, SWItch/sucrose non-fermentable; TCGA, The Cancer Genome Atlas; TERC, telomerase RNA component; TOPO1, topoisomerase 1; TRA2B, transformer-2 protein homolog beta; TRF2, telomeric repeat-binding factor 2; U2AF65, U2 small nuclear RNA auxiliary factor 2; UTR, untranslated region; VEGF, vascular endothelial growth factor; WNK2, WNK lysine deficient protein kinase 2; XIAP, X-linked inhibitor of apoptosis; XIST, X inactive specific transcript; ZO-1, tight junction protein-1.
Aebi, M., Hornig, H., Padgett, R. A., Reiser, J., and Weissmann, C. (1986). Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47, 555–565. doi: 10.1016/0092-8674(86)90620-3
Ajiro, M., Jia, R., Yang, Y., Zhu, J., and Zheng, Z. M. (2016a). A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 44, 1854–1870. doi: 10.1093/nar/gkv1500
Ajiro, M., Jia, R., Zhang, L., Liu, X., and Zheng, Z. M. (2012). Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6∗I and E7 expression. PLoS One 7:e46412. doi: 10.1371/journal.pone.0046412
Ajiro, M., Tang, S., Doorbar, J., and Zheng, Z. M. (2016b). Serine/Arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoprotein A1 regulate alternative RNA splicing and gene expression of human papillomavirus 18 through two functionally distinguishable cis elements. J. Virol. 90, 9138–9152. doi: 10.1128/jvi.00965-16
Ajiro, M., and Zheng, Z. M. (2015). E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. mBio 6:e02068-14.
Alekseev, O. M., Richardson, R. T., Tsuruta, J. K., and O’Rand, M. G. (2011). Depletion of the histone chaperone tNASP inhibits proliferation and induces apoptosis in prostate cancer PC-3 cells. Reprod. Biol. Endocrinol. 9:50. doi: 10.1186/1477-7827-9-50
Anczukow, O., and Krainer, A. R. (2016). Splicing-factor alterations in cancers. RNA 22, 1285–1301. doi: 10.1261/rna.057919.116
Andreotti, A. H., Bunnell, S. C., Feng, S., Berg, L. J., and Schreiber, S. L. (1997). Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature 385, 93–97. doi: 10.1038/385093a0
Araki, S., Dairiki, R., Nakayama, Y., Murai, A., Miyashita, R., Iwatani, M., et al. (2015). Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PLoS One 10:e0116929. doi: 10.1371/journal.pone.0116929
Arbab, J. P., Ayatollahi, H., Sadeghi, R., Sheikhi, M., and Asghari, A. (2018). Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: a meta-analysis. Hematology 23, 778–784. doi: 10.1080/10245332.2018.1471794
Aubol, B. E., Keshwani, M. M., Fattet, L., and Adams, J. A. (2018). Mobilization of a splicing factor through a nuclear kinase-kinase complex. Biochem. J. 475, 677–690. doi: 10.1042/bcj20170672
Aujla, A., Linder, K., Iragavarapu, C., Karass, M., and Liu, D. (2018). SRSF2 mutations in myelodysplasia/myeloproliferative neoplasms. Biomark. Res. 6:29.
Auyeung, V. C., Ulitsky, I., McGeary, S. E., and Bartel, D. P. (2013). Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152, 844–858. doi: 10.1016/j.cell.2013.01.031
Baralle, F. E., and Giudice, J. (2017). Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451. doi: 10.1038/nrm.2017.27
Bates, D. O., Morris, J. C., Oltean, S., and Donaldson, L. F. (2017). Pharmacology of modulators of alternative splicing. Pharmacol. Rev. 69, 63–79. doi: 10.1124/pr.115.011239
Batsche, E., Yaniv, M., and Muchardt, C. (2006). The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 13, 22–29. doi: 10.1038/nsmb1030
Bauman, J. A., Li, S. D., Yang, A., Huang, L., and Kole, R. (2010). Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 38, 8348–8356. doi: 10.1093/nar/gkq731
Best, A., Dagliesh, C., Ehrmann, I., Kheirollahi-Kouhestani, M., Tyson-Capper, A., and Elliott, D. J. (2013). Expression of Tra2 beta in cancer cells as a potential contributory factor to neoplasia and metastasis. Int. J. Cell Biol. 2013:843781.
Bielli, P., Busa, R., Paronetto, M. P., and Sette, C. (2011). The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer 18, R91–R102.
Bodaghi, S., Jia, R., and Zheng, Z. M. (2009). Human papillomavirus type 16 E2 and E6 are RNA-binding proteins and inhibit in vitro splicing of pre-mRNAs with suboptimal splice sites. Virology 386, 32–43. doi: 10.1016/j.virol.2008.12.037
Bonnal, S. C., Lopez-Oreja, I., and Valcarcel, J. (2020). Roles and mechanisms of alternative splicing in cancer – implications for care. Nat. Rev. Clin. Oncol. doi: 10.1038/s41571-020-0350-x [Epub ahead of print].
Boukakis, G., Patrinou-Georgoula, M., Lekarakou, M., Valavanis, C., and Guialis, A. (2010). Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer 10:434. doi: 10.1186/1471-2407-10-434
Bradley, T., Cook, M. E., and Blanchette, M. (2015). SR proteins control a complex network of RNA-processing events. RNA 21, 75–92. doi: 10.1261/rna.043893.113
Brant, A. C., Menezes, A. N., Felix, S. P., de Almeida, L. M., Sammeth, M., and Moreira, M. A. M. (2018). Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: a descriptive study in invasive cervical cancer. Genomics 111, 1853–1861. doi: 10.1016/j.ygeno.2018.12.008
Buljan, M., Chalancon, G., Eustermann, S., Wagner, G. P., Fuxreiter, M., Bateman, A., et al. (2012). Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883. doi: 10.1016/j.molcel.2012.05.039
Busa, R., Paronetto, M. P., Farini, D., Pierantozzi, E., Botti, F., Angelini, D. F., et al. (2007). The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene 26, 4372–4382. doi: 10.1038/sj.onc.1210224
Busch, A., and Hertel, K. J. (2012). Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3, 1–12. doi: 10.1002/wrna.100
Bush, S. J., Chen, L., Tovar-Corona, J. M., and Urrutia, A. O. (2017). Alternative splicing and the evolution of phenotypic novelty. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017:372.
Bzhalava, Z., Arroyo Muhr, L. S., and Dillner, J. (2020). Transcription of human papillomavirus oncogenes in head and neck squamous cell carcinomas. Vaccine 38, 4066–4070. doi: 10.1016/j.vaccine.2020.04.049
Cerasuolo, A., Annunziata, C., Tortora, M., Starita, N., Stellato, G., Greggi, S., et al. (2017). Comparative analysis of HPV16 gene expression profiles in cervical and in oropharyngeal squamous cell carcinoma. Oncotarget 8, 34070–34081. doi: 10.18632/oncotarget.15977
Chang, Y. L., Hsu, Y. J., Chen, Y., Wang, Y. W., and Huang, S. M. (2017). Theophylline exhibits anti-cancer activity via suppressing SRSF3 in cervical and breast cancer cell lines. Oncotarget 8, 101461–101474. doi: 10.18632/oncotarget.21464
Chaudhury, A., Chander, P., and Howe, P. H. (2010). Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1’s multifunctional regulatory roles. RNA 16, 1449–1462. doi: 10.1261/rna.2254110
Che, Y., and Fu, L. (2020). Aberrant expression and regulatory network of splicing factor-SRSF3 in tumors. J. Cancer 11, 3502–3511. doi: 10.7150/jca.42645
Chen, C., Lei, J., Zheng, Q., Tan, S., Ding, K., and Yu, C. (2018). Poly(rC) binding protein 2 (PCBP2) promotes the viability of human gastric cancer cells by regulating CDK2. FEBS Open Bio. 8, 764–773. doi: 10.1002/2211-5463.12408
Chen, H., Liu, H., and Qing, G. (2018). Targeting oncogenic Myc as a strategy for cancer treatment. Signal. Transduct. Target Ther. 3:5.
Chen, J., Xue, Y., Poidinger, M., Lim, T., Chew, S. H., Pang, C. L., et al. (2014). Mapping of HPV transcripts in four human cervical lesions using RNAseq suggests quantitative rearrangements during carcinogenic progression. Virology 46, 14–24. doi: 10.1016/j.virol.2014.05.026
Chen, M., and Manley, J. L. (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754. doi: 10.1038/nrm2777
Chen, Y., Huang, Q., Liu, W., Zhu, Q., Cui, C. P., Xu, L., et al. (2018a). Mutually exclusive acetylation and ubiquitylation of the splicing factor SRSF5 control tumor growth. Nat. Commun. 9:2464.
Chen, Y., Liu, J., Wang, W., Xiang, L., Wang, J., Liu, S., et al. (2018b). High expression of hnRNPA1 promotes cell invasion by inducing EMT in gastric cancer. Oncol. Rep. 39, 1693–1701.
Cheng, C., and Sharp, P. A. (2006). Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell Biol. 26, 362–370. doi: 10.1128/mcb.26.1.362-370.2006
Cheng, Z., Sun, Y., Niu, X., Shang, Y., Ruan, J., Chen, Z., et al. (2017). Gene expression profiling reveals U1 snRNA regulates cancer gene expression. Oncotarget 8, 112867–112874. doi: 10.18632/oncotarget.22842
Cheunim, T., Zhang, J., Milligan, S. G., McPhillips, M. G., and Graham, S. V. (2008). The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res. 131, 189–198. doi: 10.1016/j.virusres.2007.09.006
Christofk, H. R., Vander Heiden, M. G., Harris, M. H., Ramanathan, A., Gerszten, R. E., Wei, R., et al. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233. doi: 10.1038/nature06734
Climente-Gonzalez, H., Porta-Pardo, E., Godzik, A., and Eyras, E. (2017). The functional impact of alternative splicing in cancer. Cell Rep. 20, 2215–2226. doi: 10.1016/j.celrep.2017.08.012
Collier, B., Goobar-Larsson, L., Sokolowski, M., and Schwartz, S. (1998). Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273, 22648–22656. doi: 10.1074/jbc.273.35.22648
Collins, S. I., Constandinou-Williams, C., Wen, K., Young, L. S., Roberts, S., Murray, P. G., et al. (2009). Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res. 69, 3828–3832. doi: 10.1158/0008-5472.can-08-3099
Colwill, K., Pawson, T., Andrews, B., Prasad, J., Manley, J. L., Bell, J. C., et al. (1996). The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275. doi: 10.1002/j.1460-2075.1996.tb00357.x
Coupe, V. M., Gonzalez-Barreiro, L., Gutierrez-Berzal, J., Melian-Boveda, A. L., Lopez-Rodriguez, O., Alba-Dominguez, J., et al. (2012). Transcriptional analysis of human papillomavirus type 16 in histological sections of cervical dysplasia by in situ hybridisation. J. Clin. Pathol. 65, 164–170. doi: 10.1136/jclinpath-2011-200330
Czubaty, A., and Piekielko-Witkowska, A. (2017). Protein kinases that phosphorylate splicing factors: roles in cancer development, progression and possible therapeutic options. Int. J. Biochem. Cell Biol. 91(Pt B), 102–115. doi: 10.1016/j.biocel.2017.05.024
Das, S., Anczukow, O., Akerman, M., and Krainer, A. R. (2012). Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117. doi: 10.1016/j.celrep.2011.12.001
David, C. J., Chen, M., Assanah, M., Canoll, P., and Manley, J. L. (2010). HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368. doi: 10.1038/nature08697
David, J., Maden, S., Weeder, B., Thompson, R., and Nellore, A. (2020). Putatively cancer-specific exonΓÇôexon junctions are shared across patients and present in developmental and other non-cancer cells. NAR Cancer 1:2.
Denko, N., Schindler, C., Koong, A., Laderoute, K., Green, C., and Giaccia, A. (2000). Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin. Cancer Res. 6, 480–487.
Derry, J. J., Richard, S., Valderrama, C. H., Ye, X., Vasioukhin, V., Cochrane, A. W., et al. (2000). Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol. Cell Biol. 20, 6114–6126. doi: 10.1128/mcb.20.16.6114-6126.2000
Dhanjal, S., Kajitani, N., Glahder, J., Mossberg, A. K., Johansson, C., and Schwartz, S. (2015). Heterogeneous nuclear ribonucleoprotein C proteins interact with the human papillomavirus type 16 (HPV16) Early 3′-untranslated region and alleviate suppression of HPV16 late L1 mRNA splicing. J. Biol. Chem. 290, 13354–13371. doi: 10.1074/jbc.m115.638098
Di, C., Syafrizayanti Zhang, Q, Chen, Y., Wang, Y., Zhang, X., et al. (2019). Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 26, 1181–1194. doi: 10.1038/s41418-018-0231-3
Dong, M., Dong, Z., Zhu, X., Zhang, Y., and Song, L. (2019). Long non-coding RNA MIR205HG regulates KRT17 and tumor processes in cervical cancer via interaction with SRSF1. Exp. Mol. Pathol. 111:104322. doi: 10.1016/j.yexmp.2019.104322
Dong, P., Xiong, Y., Yu, J., Chen, L., Tao, T., Yi, S., et al. (2019). Correction: control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene 38:3972. doi: 10.1038/s41388-019-0677-x
Doorbar, J. (2006). Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 110, 525–541. doi: 10.1042/cs20050369
Doorbar, J., Egawa, N., Griffin, H., Kranjec, C., and Murakami, I. (2015). Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 25(Suppl. 1) 2–23. doi: 10.1002/rmv.1822
Doorbar, J., Foo, C., Coleman, N., Medcalf, L., Hartley, O., Prospero, T., et al. (1997). Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238, 40–52. doi: 10.1006/viro.1997.8768
Doorbar, J., Parton, A., Hartley, K., Banks, L., Crook, T., Stanley, M., et al. (1990). Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178, 254–262. doi: 10.1016/0042-6822(90)90401-c
Doorbar, J., Quint, W., Banks, L., Bravo, I. G., Stoler, M., Broker, T. R., et al. (2012). The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl. 5) F55–F70.
Dvinge, H. (2018). Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett. 592, 2987–3006. doi: 10.1002/1873-3468.13119
Dvinge, H., Kim, E., Abdel-Wahab, O., and Bradley, R. K. (2016). RNA splicing factors. Nat. Rev. Cancer 16, 413–430.
Effenberger, K. A., Urabe, V. K., and Jurica, M. S. (2017). Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip. Rev. RNA 8:10.1002/wrna.1381.
El, M. E., and Younis, I. (2018). The cancer spliceome: reprograming of alternative splicing in cancer. Front. Mol. Biosci. 5:80. doi: 10.3389/fmolb.2018.00080
Elliott, D. J., Best, A., Dalgliesh, C., Ehrmann, I., and Grellscheid, S. (2012). How does Tra2beta protein regulate tissue-specific RNA splicing? Biochem. Soc. Trans. 40, 784–788. doi: 10.1042/bst20120036
Enokizono, Y., Konishi, Y., Nagata, K., Ouhashi, K., Uesugi, S., Ishikawa, F., et al. (2005). Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 280, 18862–18870. doi: 10.1074/jbc.m411822200
Evans, W., Filippova, M., Filippov, V., Bashkirova, S., Zhang, G., Reeves, M. E., et al. (2016). Overexpression of HPV16 E6∗ alters beta-integrin and mitochondrial dysfunction pathways in cervical cancer cells. Cancer Genomics Proteomics 13, 259–273.
Fay, J., Kelehan, P., Lambkin, H., and Schwartz, S. (2009). Increased expression of cellular RNA-binding proteins in HPV-induced neoplasia and cervical cancer. J. Med. Virol. 81, 897–907. doi: 10.1002/jmv.21406
Fei, T., Chen, Y., Xiao, T., Li, W., Cato, L., Zhang, P., et al. (2017). Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. U.S.A. 114, E5207–E5215.
Feng, Y., Chen, M., and Manley, J. L. (2008). Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 15, 1040–1048. doi: 10.1038/nsmb.1485
Filippova, M., Evans, W., Aragon, R., Filippov, V., Williams, V. M., Hong, L., et al. (2014). The small splice variant of HPV16 E6, E6, reduces tumor formation in cervical carcinoma xenografts. Virology 45, 153–164. doi: 10.1016/j.virol.2013.12.011
Frampton, G. M., Ali, S. M., Rosenzweig, M., Chmielecki, J., Lu, X., Bauer, T. M., et al. (2015). Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5, 850–859. doi: 10.1158/2159-8290.cd-15-0285
Frendewey, D., and Keller, W. (1985). Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42, 355–367. doi: 10.1016/s0092-8674(85)80131-8
Friend, L. R., Landsberg, M. J., Nouwens, A. S., Wei, Y., Rothnagel, J. A., and Smith, R. (2013). Arginine methylation of hnRNP A2 does not directly govern its subcellular localization. PLoS One 8:e75669. doi: 10.1371/journal.pone.0075669
Fu, Y., Huang, B., Shi, Z., Han, J., Wang, Y., Huangfu, J., et al. (2013). SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO Mol. Med. 5, 737–750. doi: 10.1002/emmm.201202218
Fu, Y., and Wang, Y. (2018). SRSF7 knockdown promotes apoptosis of colon and lung cancer cells. Oncol. Lett. 15, 5545–5552.
Futakuchi, H., Ueda, M., Kanda, K., Fujino, K., Yamaguchi, H., and Noda, S. (2007). Transcriptional expression of survivin and its splice variants in cervical carcinomas. Int. J. Gynecol. Cancer 17, 1092–1098. doi: 10.1111/j.1525-1438.2007.00833.x
Gabriel, B., Zur, H. A., Bouda, J., Boudova, L., Koprivova, M., Hirschfeld, M., et al. (2009). Significance of nuclear hTra2-beta1 expression in cervical cancer. Acta Obstet. Gynecol. Scand. 88, 216–221. doi: 10.1080/00016340802503021
Gallardo, M., Lee, H. J., Zhang, X., Bueso-Ramos, C., Pageon, L. R., McArthur, M., et al. (2015). hnRNP K is a haploinsufficient tumor suppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell 28, 486–499. doi: 10.1016/j.ccell.2015.09.001
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. doi: 10.1126/scisignal.2004088
Gao, R., Yu, Y., Inoue, A., Widodo, N., Kaul, S. C., and Wadhwa, R. (2013). Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 288, 15046–15056. doi: 10.1074/jbc.m113.466136
Gao, W., Ding, L., Song, Z. C., Feng, M. J., Liu, C. L., Li, X. X., et al. (2020). [The role of human papillomavirus 16 early genes E2 and E6 and heterogeneous nuclear ribonucleoprotein E2 in cervical carcinogenesis and their interaction effect]. Zhonghua Yu Fang Yi Xue Za Zhi 54, 92–98.
Gao, Y., Wang, W., Cao, J., Wang, F., Geng, Y., Cao, J., et al. (2016). Upregulation of AUF1 is involved in the proliferation of esophageal squamous cell carcinoma through GCH1. Int. J. Oncol. 49, 2001–2010. doi: 10.3892/ijo.2016.3713
Gaytan-Cervantes, J., Gonzalez-Torres, C., Maldonado, V., Zampedri, C., Ceballos-Cancino, G., and Melendez-Zajgla, J. (2017). Protein Sam68 regulates the alternative splicing of survivin DEx3. J. Biol. Chem. 292, 13745–13757. doi: 10.1074/jbc.m117.800318
Geuens, T., Bouhy, D., and Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. doi: 10.1007/s00439-016-1683-5
Ghigna, C., Giordano, S., Shen, H., Benvenuto, F., Castiglioni, F., Comoglio, P. M., et al. (2005). Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890. doi: 10.1016/j.molcel.2005.10.026
Giannakouros, T., Nikolakaki, E., Mylonis, I., and Georgatsou, E. (2011). Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 278, 570–586. doi: 10.1111/j.1742-4658.2010.07987.x
Golan-Gerstl, R., Cohen, M., Shilo, A., Suh, S. S., Bakacs, A., Coppola, L., et al. (2011). Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 71, 4464–4472. doi: 10.1158/0008-5472.can-10-4410
Goncalves, V., Henriques, A. F., Pereira, J. F., Neves, C. A., Moyer, M. P., Moita, L. F., et al. (2014). Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 20, 474–482. doi: 10.1261/rna.041376.113
Graham, S. V. (2010). Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 5, 1493–1506. doi: 10.2217/fmb.10.107
Graham, S. V. (2016). Human papillomavirus E2 protein: linking replication, transcription, and RNA processing. J. Virol. 90, 8384–8388. doi: 10.1128/jvi.00502-16
Graham, S. V., and Faizo, A. A. A. (2017). Control of human papillomavirus gene expression by alternative splicing. Virus Res. 231, 83–95. doi: 10.1016/j.virusres.2016.11.016
Grellscheid, S., Dalgliesh, C., Storbeck, M., Best, A., Liu, Y., Jakubik, M., et al. (2011). Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2beta in development. PLoS Genet. 7:e1002390. doi: 10.1371/journal.pgen.1002390
Guil, S., and Caceres, J. F. (2007). The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 14, 591–596. doi: 10.1038/nsmb1250
Habelhah, H., Shah, K., Huang, L., Ostareck-Lederer, A., Burlingame, A. L., Shokat, K. M., et al. (2001). ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3, 325–330. doi: 10.1038/35060131
Han, S. P., Tang, Y. H., and Smith, R. (2010). Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392. doi: 10.1042/bj20100396
Havens, M. A., and Hastings, M. L. (2016). Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 44, 6549–6563. doi: 10.1093/nar/gkw533
Hegde, R. S. (2002). The papillomavirus E2 proteins: structure, function, and biology. Annu. Rev. Biophys. Biomol. Struct. 31, 343–360. doi: 10.1146/annurev.biophys.31.100901.142129
Howard, J. M., and Sanford, J. R. (2015). The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110. doi: 10.1002/wrna.1260
Jang, M. K., Anderson, D. E., van, D. K., and McBride, A. A. (2015). A proteomic approach to discover and compare interacting partners of papillomavirus E2 proteins from diverse phylogenetic groups. Proteomics 15, 2038–2050. doi: 10.1002/pmic.201400613
Jeong, S. (2017). SR proteins: binders, regulators, and connectors of RNA. Mol. Cells 40, 1–9. doi: 10.14348/molcells.2017.2319
Jia, R., Ajiro, M., Yu, L., McCoy, P. Jr., and Zheng, Z. M. (2019). Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA 25, 630–644. doi: 10.1261/rna.068619.118
Jia, R., Li, C., McCoy, J. P., Deng, C. X., and Zheng, Z. M. (2010). SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int. J. Biol. Sci. 6, 806–826. doi: 10.7150/ijbs.6.806
Jia, R., Liu, X., Tao, M., Kruhlak, M., Guo, M., Meyers, C., et al. (2009). Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J. Virol. 83, 167–180. doi: 10.1128/jvi.01719-08
Jiang, P., Li, Z., Tian, F., Li, X., and Yang, J. (2017). Fyn/heterogeneous nuclear ribonucleoprotein E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin beta1. Int. J. Oncol. 51, 169–183. doi: 10.3892/ijo.2017.4018
Johansson, C., and Schwartz, S. (2013). Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat. Rev. Microbiol. 11, 239–251. doi: 10.1038/nrmicro2984
Kahles, A., Lehmann, K. V., Toussaint, N. C., Huser, M., Stark, S. G., Sachsenberg, T., et al. (2018). Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.
Kaida, D., Motoyoshi, H., Tashiro, E., Nojima, T., Hagiwara, M., Ishigami, K., et al. (2007). Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583. doi: 10.1038/nchembio.2007.18
Kajitani, N., Glahder, J., Wu, C., Yu, H., Nilsson, K., and Schwartz, S. (2017). hnRNP L controls HPV16 RNA polyadenylation and splicing in an Akt kinase-dependent manner. Nucleic Acids Res. 45, 9654–9678. doi: 10.1093/nar/gkx606
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. doi: 10.1038/nature12634
Kanopka, A., Muhlemann, O., and Akusjarvi, G. (1996). Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381, 535–538. doi: 10.1038/381535a0
Karni, R., de, S. E., Lowe, S. W., Sinha, R., Mu, D., and Krainer, A. R. (2007). The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193. doi: 10.1038/nsmb1209
Kastner, B., Will, C. L., Stark, H., and Luhrmann, R. (2019). Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol. 11:a032417. doi: 10.1101/cshperspect.a032417
Kim, H. R., Lee, G. O., Choi, K. H., Kim, D. K., Ryu, J. S., Hwang, K. E., et al. (2016). SRSF5: a novel marker for small-cell lung cancer and pleural metastatic cancer. Lung Cancer 99, 57–65. doi: 10.1016/j.lungcan.2016.05.018
Kim, J., Park, R. Y., Chen, J. K., Kim, J., Jeong, S., and Ohn, T. (2014). Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell Death Differ. 21, 481–490. doi: 10.1038/cdd.2013.171
Kim, Y. J., Kim, B. R., Ryu, J. S., Lee, G. O., Kim, H. R., Choi, K. H., et al. (2017). HNRNPA1, a splicing regulator, is an effective target protein for cervical cancer detection: comparison with conventional tumor markers. Int. J. Gynecol. Cancer 27, 326–331. doi: 10.1097/igc.0000000000000868
Klymenko, T., Hernandez-Lopez, H., MacDonald, A. I., Bodily, J. M., and Graham, S. V. (2016). Human papillomavirus E2 regulates SRSF3 (SRp20) to promote capsid protein expression in infected differentiated keratinocytes. J. Virol. 90, 5047–5058. doi: 10.1128/jvi.03073-15
Kohler, J., Schuler, M., Gauler, T. C., Nopel-Dunnebacke, S., Ahrens, M., Hoffmann, A. C., et al. (2016). Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J. Cancer Res. Clin. Oncol. 142, 795–805. doi: 10.1007/s00432-015-2095-y
Komatsu, S., Ichikawa, D., Takeshita, H., Morimura, R., Hirajima, S., Tsujiura, M., et al. (2014). Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In Vivo 28, 293–297.
Komeno, Y., Huang, Y. J., Qiu, J., Lin, L., Xu, Y., Zhou, Y., et al. (2015). SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol. Cell Biol. 35, 3071–3082. doi: 10.1128/mcb.00202-15
Kooshapur, H., Choudhury, N. R., Simon, B., Muhlbauer, M., Jussupow, A., Fernandez, N., et al. (2018). Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. Nat. Commun. 9:2479.
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29, 2168–2182. doi: 10.1101/gad.270421.115
Lai, M. C., Lin, R. I., Huang, S. Y., Tsai, C. W., and Tarn, W. Y. (2000). A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275, 7950–7957. doi: 10.1074/jbc.275.11.7950
Lai, M. C., Lin, R. I., and Tarn, W. Y. (2001). Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159. doi: 10.1073/pnas.181354098
Lal, S., Allan, A., Markovic, D., Walker, R., Macartney, J., Europe-Finner, N., et al. (2013). Estrogen alters the splicing of type 1 corticotropin-releasing hormone receptor in breast cancer cells. Sci. Signal. 6:ra53. doi: 10.1126/scisignal.2003926
Le, K. Q., Prabhakar, B. S., Hong, W. J., and Li, L. C. (2015). Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol. Sin. 36, 1212–1218. doi: 10.1038/aps.2015.43
Lee, J. H., Jeong, S. A., Khadka, P., Hong, J., and Chung, I. K. (2015). Involvement of SRSF11 in cell cycle-specific recruitment of telomerase to telomeres at nuclear speckles. Nucleic Acids Res. 43, 8435–8451. doi: 10.1093/nar/gkv844
Lee, S. C., and Abdel-Wahab, O. (2016). Therapeutic targeting of splicing in cancer. Nat. Med. 22, 976–986. doi: 10.1038/nm.4165
Lee, S. D., Yu, D., Lee, D. Y., Shin, H. S., Jo, J. H., and Lee, Y. C. (2019). Upregulated microRNA-193a-3p is responsible for cisplatin resistance in CD44(+) gastric cancer cells. Cancer Sci. 110, 662–673. doi: 10.1111/cas.13894
Lee, S. W., Lee, M. H., Park, J. H., Kang, S. H., Yoo, H. M., Ka, S. H., et al. (2012). SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 31, 4441–4452. doi: 10.1038/emboj.2012.293
Lee, Y., and Rio, D. C. (2015). Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323.
Li, K., Sun, D., Gou, Q., Ke, X., Gong, Y., Zuo, Y., et al. (2018). Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 420, 80–90. doi: 10.1016/j.canlet.2018.01.060
Li, X., Johansson, C., Cardoso, P. C., Mossberg, A., Dhanjal, S., Bergvall, M., et al. (2013a). Eight nucleotide substitutions inhibit splicing to HPV-16 3′-splice site SA3358 and reduce the efficiency by which HPV-16 increases the life span of primary human keratinocytes. PLoS One 8:e72776. doi: 10.1371/journal.pone.0072776
Li, X., Johansson, C., Glahder, J., Mossberg, A. K., and Schwartz, S. (2013b). Suppression of HPV-16 late L1 5′-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs. Nucleic Acids Res. 41, 10488–10508. doi: 10.1093/nar/gkt803
Li, X., Ran, L., Fang, W., and Wang, D. (2014). Lobaplatin arrests cell cycle progression, induces apoptosis and alters the proteome in human cervical cancer cell Line CaSki. Biomed. Pharmacother. 68, 291–297. doi: 10.1016/j.biopha.2013.10.004
Li, Z., Yu, C. P., Zhong, Y., Liu, T. J., Huang, Q. D., Zhao, X. H., et al. (2012). Sam68 expression and cytoplasmic localization is correlated with lymph node metastasis as well as prognosis in patients with early-stage cervical cancer. Ann. Oncol. 23, 638–646. doi: 10.1093/annonc/mdr290
Liang, Y., Tebaldi, T., Rejeski, K., Joshi, P., Stefani, G., Taylor, A., et al. (2018). SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia 32, 2659–2671. doi: 10.1038/s41375-018-0152-7
Lin, J. C. (2017). Therapeutic applications of targeted alternative splicing to cancer treatment. Int. J. Mol. Sci. 19:75. doi: 10.3390/ijms19010075
Liu, F., Dai, M., Xu, Q., Zhu, X., Zhou, Y., Jiang, S., et al. (2018). SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-kappaB-CD47 axis. Oncogene 37, 2394–2409. doi: 10.1038/s41388-017-0119-6
Liu, X., Zhou, Y., Lou, Y., and Zhong, H. (2016). Knockdown of HNRNPA1 inhibits lung adenocarcinoma cell proliferation through cell cycle arrest at G0/G1 phase. Gene 576(2 Pt 2), 791–797. doi: 10.1016/j.gene.2015.11.009
Long, Y., Sou, W. H., Yung, K. W. Y., Liu, H., Wan, S. W. C., Li, Q., et al. (2019). Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 294, 1312–1327. doi: 10.1074/jbc.ra118.003392
Lu, G. Y., Huang, S. M., Liu, S. T., Liu, P. Y., Chou, W. Y., and Lin, W. S. (2014). Caffeine induces tumor cytotoxicity via the regulation of alternative splicing in subsets of cancer-associated genes. Int. J. Biochem. Cell Biol. 47, 83–92. doi: 10.1016/j.biocel.2013.12.004
Lu, J., and Gao, F. H. (2016). Role and molecular mechanism of heterogeneous nuclear ribonucleoprotein K in tumor development and progression. Biomed. Rep. 4, 657–663. doi: 10.3892/br.2016.642
Luo, C., Cheng, Y., Liu, Y., Chen, L., Liu, L., Wei, N., et al. (2017). SRSF2 Regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 77, 1168–1178. doi: 10.1158/0008-5472.can-16-1919
Lv, D., Wu, H., Xing, R., Shu, F., Lei, B., Lei, C., et al. (2017). HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget 8, 13586–13599. doi: 10.18632/oncotarget.14600
Manley, J. L., and Krainer, A. R. (2010). A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074. doi: 10.1101/gad.1934910
Masaki, S., Ikeda, S., Hata, A., Shiozawa, Y., Kon, A., Ogawa, S., et al. (2019). Myelodysplastic syndrome-associated SRSF2 mutations cause splicing changes by altering binding motif sequences. Front. Genet. 10:338. doi: 10.3389/fgene.2019.00338
Mavrou, A., Brakspear, K., Hamdollah-Zadeh, M., Damodaran, G., Babaei-Jadidi, R., Oxley, J., et al. (2015). Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene 34, 4311–4319. doi: 10.1038/onc.2014.360
McCredie, M. R., Sharples, K. J., Paul, C., Baranyai, J., Medley, G., Jones, R. W., et al. (2008). Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 9, 425–434. doi: 10.1016/s1470-2045(08)70103-7
McFarlane, M., MacDonald, A. I., Stevenson, A., and Graham, S. V. (2015). Human papillomavirus 16 oncoprotein expression is controlled by the cellular splicing factor SRSF2 (SC35). J. Virol. 89, 5276–5287. doi: 10.1128/jvi.03434-14
Mili, S., Shu, H. J., Zhao, Y., and Pinol-Roma, S. (2001). Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell Biol. 21, 7307–7319. doi: 10.1128/mcb.21.21.7307-7319.2001
Milligan, S. G., Veerapraditsin, T., Ahamet, B., Mole, S., and Graham, S. V. (2007). Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology 360, 172–181. doi: 10.1016/j.virol.2006.10.012
Mogilevsky, M., Shimshon, O., Kumar, S., Mogilevsky, A., Keshet, E., Yavin, E., et al. (2018). Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 46, 11396–11404. doi: 10.1093/nar/gky921
Mole, S., Faizo, A. A. A., Hernandez-Lopez, H., Griffiths, M., Stevenson, A., Roberts, S., et al. (2020). Human papillomavirus type 16 infection activates the host serine arginine protein kinase 1 (SRPK1) – splicing factor axis. J. Gen. Virol. doi: 10.1099/jgv.0.001402 [Epub ahead of print].
Mole, S., McFarlane, M., Chuen-Im, T., Milligan, S. G., Millan, D., and Graham, S. V. (2009a). RNA splicing factors regulated by HPV16 during cervical tumour progression. J. Pathol. 219, 383–391. doi: 10.1002/path.2608
Mole, S., Milligan, S. G., and Graham, S. V. (2009b). Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J. Virol. 83, 357–367. doi: 10.1128/jvi.01414-08
Moody, C. A., and Laimins, L. A. (2010). Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10, 550–560. doi: 10.1038/nrc2886
Moscicki, A. B., and Palefsky, J. M. (2011). Human papillomavirus in men: an update. J. Low. Genit. Tract. Dis. 15, 231–234. doi: 10.1097/lgt.0b013e318203ae61
Moujalled, D., James, J. L., Yang, S., Zhang, K., Duncan, C., Moujalled, D. M., et al. (2015). Phosphorylation of hnRNP K by cyclin-dependent kinase 2 controls cytosolic accumulation of TDP-43. Hum. Mol. Genet. 24, 1655–1669. doi: 10.1093/hmg/ddu578
Najib, S., Martin-Romero, C., Gonzalez-Yanes, C., and Sanchez-Margalet, V. (2005). Role of Sam68 as an adaptor protein in signal transduction. Cell. Mol. Life Sci. 62, 36–43. doi: 10.1007/s00018-004-4309-3
Nakajima, H., Sato, B., Fujita, T., Takase, S., Terano, H., and Okuhara, M. (1996). New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 49, 1196–1203. doi: 10.7164/antibiotics.49.1196
Nakielny, S., and Dreyfuss, G. (1996). The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134, 1365–1373. doi: 10.1083/jcb.134.6.1365
Naro, C., Barbagallo, F., Chieffi, P., Bourgeois, C. F., Paronetto, M. P., and Sette, C. (2014). The centrosomal kinase NEK2 is a novel splicing factor kinase involved in cell survival. Nucleic Acids Res. 42, 3218–3227. doi: 10.1093/nar/gkt1307
Naro, C., and Sette, C. (2013). Phosphorylation-mediated regulation of alternative splicing in cancer. Int. J. Cell Biol. 2013:151839.
Nasim, F. U., Hutchison, S., Cordeau, M., and Chabot, B. (2002). High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8, 1078–1089. doi: 10.1017/s1355838202024056
Nilsson, K., Wu, C., Kajitani, N., Yu, H., Tsimtsirakis, E., Gong, L., et al. (2018). The DNA damage response activates HPV16 late gene expression at the level of RNA processing. Nucleic Acids Res. 46, 5029–5049. doi: 10.1093/nar/gky227
Oberg, D., Fay, J., Lambkin, H., and Schwartz, S. (2005). A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol. 79, 9254–9269. doi: 10.1128/jvi.79.14.9254-9269.2005
Olmedo-Nieva, L., Munoz-Bello, J. O., Contreras-Paredes, A., and Lizano, M. (2018). The role of E6 spliced isoforms (E6∗) in human papillomavirus-induced carcinogenesis. Viruses 10:45. doi: 10.3390/v10010045
Olshavsky, N. A., Comstock, C. E., Schiewer, M. J., Augello, M. A., Hyslop, T., Sette, C., et al. (2010). Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res. 70, 3975–3984. doi: 10.1158/0008-5472.can-09-3468
Oltean, S., and Bates, D. O. (2014). Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318. doi: 10.1038/onc.2013.533
Otsuka, K., Yamamoto, Y., and Ochiya, T. (2018). Regulatory role of resveratrol, a microRNA-controlling compound, in HNRNPA1 expression, which is associated with poor prognosis in breast cancer. Oncotarget 9, 24718–24730. doi: 10.18632/oncotarget.25339
Oyervides-Munoz, M. A., Perez-Maya, A. A., Rodriguez-Gutierrez, H. F., Gomez-Macias, G. S., Fajardo-Ramirez, O. R., Trevino, V., et al. (2018). Understanding the HPV integration and its progression to cervical cancer. Infect. Genet. Evol. 61, 134–144. doi: 10.1016/j.meegid.2018.03.003
Pagani, F., Buratti, E., Stuani, C., Romano, M., Zuccato, E., Niksic, M., et al. (2000). Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275, 21041–21047. doi: 10.1074/jbc.m910165199
Paget-Bailly, P., Meznad, K., Bruyere, D., Perrard, J., Herfs, M., Jung, A. C., et al. (2019). Comparative RNA sequencing reveals that HPV16 E6 abrogates the effect of E6∗I on ROS metabolism. Sci. Rep. 9:5938.
Pan, Z. X., Zhang, X. Y., Chen, S. R., and Li, C. Z. (2019). Upregulated exosomal miR-221/222 promotes cervical cancer via repressing methyl-CpG-binding domain protein 2. Eur. Rev. Med. Pharmacol. Sci. 23, 3645–3653.
Papasaikas, P., and Valcarcel, J. (2016). The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem. Sci. 41, 33–45. doi: 10.1016/j.tibs.2015.11.003
Park, Y. M., Hwang, S. J., Masuda, K., Choi, K. M., Jeong, M. R., Nam, D. H., et al. (2012). Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol. Cell Biol. 32, 4237–4244. doi: 10.1128/mcb.00443-12
Paronetto, M. P., Cappellari, M., Busa, R., Pedrotti, S., Vitali, R., Comstock, C., et al. (2010). Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 70, 229–239. doi: 10.1158/0008-5472.can-09-2788
Paronetto, M. P., Venables, J. P., Elliott, D. J., Geremia, R., Rossi, P., and Sette, C. (2003). Tr-kit promotes the formation of a multimolecular complex composed by Fyn. PLCgamma1 and Sam68. Oncogene 22, 8707–8715. doi: 10.1038/sj.onc.1207016
Parssinen, J., Kuukasjarvi, T., Karhu, R., and Kallioniemi, A. (2007). High-level amplification at 17q23 leads to coordinated overexpression of multiple adjacent genes in breast cancer. Br. J. Cancer 96, 1258–1264. doi: 10.1038/sj.bjc.6603692
Patel, M., Sachidanandan, M., and Adnan, M. (2019). Serine arginine protein kinase 1 (SRPK1): a moonlighting protein with theranostic ability in cancer prevention. Mol. Biol. Rep. 46, 1487–1497. doi: 10.1007/s11033-018-4545-5
Patry, C., Bouchard, L., Labrecque, P., Gendron, D., Lemieux, B., Toutant, J., et al. (2003). Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 63, 7679–7688.
Phoomak, C., Park, D., Silsirivanit, A., Sawanyawisuth, K., Vaeteewoottacharn, K., Detarya, M., et al. (2019). O-GlcNAc-induced nuclear translocation of hnRNP-K is associated with progression and metastasis of cholangiocarcinoma. Mol. Oncol. 13, 338–357. doi: 10.1002/1878-0261.12406
Pillai, M. R., Chacko, P., Kesari, L. A., Jayaprakash, P. G., Jayaram, H. N., and Antony, A. C. (2003). Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. J. Clin. Pathol. 56, 569–574. doi: 10.1136/jcp.56.8.569
Pont, A. R., Sadri, N., Hsiao, S. J., Smith, S., and Schneider, R. J. (2012). mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell 47, 5–15. doi: 10.1016/j.molcel.2012.04.019
Preti, M., Rotondo, J. C., Holzinger, D., Micheletti, L., Gallio, N., McKay-Chopin, S., et al. (2020). Role of human papillomavirus infection in the etiology of vulvar cancer in Italian women. Infect. Agent Cancer 15:20.
Pulice, J. L., and Kadoch, C. (2016). Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb. Symp. Quant. Biol. 81, 53–60. doi: 10.1101/sqb.2016.81.031021
Qing, S., Tulake, W., Ru, M., Li, X., Yuemaier, R., Lidifu, D., et al. (2017). Proteomic identification of potential biomarkers for cervical squamous cell carcinoma and human papillomavirus infection. Tumour Biol. 39, 1010428317697547.
Ratnadiwakara, M., Mohenska, M., and Anko, M. L. (2018). Splicing factors as regulators of miRNA biogenesis - links to human disease. Semin. Cell Dev. Biol. 79, 113–122. doi: 10.1016/j.semcdb.2017.10.008
Rigo, F., Seth, P. P., and Bennett, C. F. (2014). Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol. 825, 303–352. doi: 10.1007/978-1-4939-1221-6_9
Roberts, C. W., and Orkin, S. H. (2004). The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer 4, 133–142.
Rodriguez-Aguayo, C., Monroig, P. D. C., Redis, R. S., Bayraktar, E., Almeida, M. I., Ivan, C., et al. (2017). Regulation of hnRNPA1 by microRNAs controls the miR-18a-K-RAS axis in chemotherapy-resistant ovarian cancer. Cell Discov. 3:17029.
Rosenberger, S., De-Castro, A. J., Langbein, L., Steenbergen, R. D., and Rosl, F. (2010). Alternative splicing of human papillomavirus type-16 E6/E6∗ early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc. Natl. Acad. Sci. U.S.A. 107, 7006–7011. doi: 10.1073/pnas.1002620107
Rossi, F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J. F., et al. (1996). Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature 381, 80–82. doi: 10.1038/381080a0
Rush, M., Zhao, X., and Schwartz, S. (2005). A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J. Virol. 79, 12002–12015. doi: 10.1128/jvi.79.18.12002-12015.2005
Sakai, T., Asai, N., Okuda, A., Kawamura, N., and Mizui, Y. (2004). Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. II. Physico-chemical properties and structure elucidation. J. Antibiot. 57, 180–187. doi: 10.7164/antibiotics.57.180
Sakai, Y., Yoshida, T., Ochiai, K., Uosaki, Y., Saitoh, Y., Tanaka, F., et al. (2002). GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. 55, 855–862. doi: 10.7164/antibiotics.55.855
Satoh, T., and Kaida, D. (2016). Upregulation of p27 cyclin-dependent kinase inhibitor and a C-terminus truncated form of p27 contributes to G1 phase arrest. Sci. Rep. 6:27829.
Schiffman, M., Doorbar, J., Wentzensen, N., de, S. S., Fakhry, C., Monk, B. J., et al. (2016). Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2:16086.
Schmitt, M., Dalstein, V., Waterboer, T., Clavel, C., Gissmann, L., and Pawlita, M. (2010). Diagnosing cervical cancer and high-grade precursors by HPV16 transcription patterns. Cancer Res. 70, 249–256. doi: 10.1158/0008-5472.can-09-2514
Schmitt, M., and Pawlita, M. (2011). The HPV transcriptome in HPV16 positive cell lines. Mol. Cell Probes 25, 108–113. doi: 10.1016/j.mcp.2011.03.003
Seiler, M., Peng, S., Agrawal, A. A., Palacino, J., Teng, T., Zhu, P., et al. (2018). Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.
Shen, M., and Mattox, W. (2012). Activation and repression functions of an SR splicing regulator depend on exonic versus intronic-binding position. Nucleic Acids Res. 40, 428–437. doi: 10.1093/nar/gkr713
Shi, X., Ran, L., Liu, Y., Zhong, S. H., Zhou, P. P., Liao, M. X., et al. (2018). Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol. Rep. 39, 939–950.
Shi, Y. (2017). Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670. doi: 10.1038/nrm.2017.86
Shilo, A., Siegfried, Z., and Karni, R. (2015). The role of splicing factors in deregulation of alternative splicing during oncogenesis and tumor progression. Mol. Cell Oncol. 2:e970955. doi: 10.4161/23723548.2014.970955
Shin, C., Feng, Y., and Manley, J. L. (2004). Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558. doi: 10.1038/nature02288
Shiraishi, Y., Kataoka, K., Chiba, K., Okada, A., Kogure, Y., Tanaka, H., et al. (2018). A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28, 1111–1125. doi: 10.1101/gr.231951.117
Shultz, J. C., Goehe, R. W., Murudkar, C. S., Wijesinghe, D. S., Mayton, E. K., Massiello, A., et al. (2011). SRSF1 regulates the alternative splicing of caspase 9 via a novel intronic splicing enhancer affecting the chemotherapeutic sensitivity of non-small cell lung cancer cells. Mol. Cancer Res. 9, 889–900. doi: 10.1158/1541-7786.mcr-11-0061
Shultz, J. C., Goehe, R. W., Wijesinghe, D. S., Murudkar, C., Hawkins, A. J., Shay, J. W., et al. (2010). Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 70, 9185–9196. doi: 10.1158/0008-5472.can-10-1545
Singh, B., and Eyras, E. (2017). The role of alternative splicing in cancer. Transcription 8, 91–98. doi: 10.1080/21541264.2016.1268245
Siomi, H., and Dreyfuss, G. (1995). A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551–560. doi: 10.1083/jcb.129.3.551
Smith-Roe, S. L., Nakamura, J., Holley, D., Chastain, P. D., Rosson, G. B., Simpson, D. A., et al. (2015). SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget 6, 732–745. doi: 10.18632/oncotarget.2715
Sokol, E., Kedzierska, H., Czubaty, A., Rybicka, B., Rodzik, K., Tanski, Z., et al. (2018). microRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3′UTRs. Exp. Cell Res. 363, 208–217. doi: 10.1016/j.yexcr.2018.01.009
Somberg, M., Li, X., Johansson, C., Orru, B., Chang, R., Rush, M., et al. (2011). Serine/arginine-rich protein 30c activates human papillomavirus type 16 L1 mRNA expression via a bimodal mechanism. J. Gen. Virol. 92(Pt 10), 2411–2421. doi: 10.1099/vir.0.033183-0
Somberg, M., and Schwartz, S. (2010). Multiple ASF/SF2 sites in the human papillomavirus type 16 (HPV-16) E4-coding region promote splicing to the most commonly used 3′-splice site on the HPV-16 genome. J. Virol. 84, 8219–8230. doi: 10.1128/jvi.00462-10
Somberg, M., Zhao, X., Frohlich, M., Evander, M., and Schwartz, S. (2008). Polypyrimidine tract binding protein induces human papillomavirus type 16 late gene expression by interfering with splicing inhibitory elements at the major late 5′ splice site, SD3632. J. Virol. 82, 3665–3678. doi: 10.1128/jvi.02140-07
Song, L., Wang, L., Li, Y., Xiong, H., Wu, J., Li, J., et al. (2010). Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J. Pathol. 222, 227–237. doi: 10.1002/path.2751
Sorlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 98, 10869–10874. doi: 10.1073/pnas.191367098
Sterne-Weiler, T., and Sanford, J. R. (2014). Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol. 15:201. doi: 10.1186/gb4150
Stockley, J., Markert, E., Zhou, Y., Robson, C. N., Elliott, D. J., Lindberg, J., et al. (2015). The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci. Rep. 5:13426.
Stoilov, P., Daoud, R., Nayler, O., and Stamm, S. (2004). Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 13, 509–524. doi: 10.1093/hmg/ddh051
Straub, E., Fertey, J., Dreer, M., Iftner, T., and Stubenrauch, F. (2015). Characterization of the human papillomavirus 16 E8 promoter. J. Virol. 89, 7304–7313. doi: 10.1128/jvi.00616-15
Sudarsanam, P., and Winston, F. (2000). The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16, 345–351.
Suk, F. M., Lin, S. Y., Lin, R. J., Hsine, Y. H., Liao, Y. J., Fang, S. U., et al. (2015). Bortezomib inhibits Burkitt’s lymphoma cell proliferation by downregulating sumoylated hnRNP K and c-Myc expression. Oncotarget 6, 25988–26001. doi: 10.18632/oncotarget.4620
Sun, H., Liu, T., Zhu, D., Dong, X., Liu, F., Liang, X., et al. (2017). HnRNPM and CD44s expression affects tumor aggressiveness and predicts poor prognosis in breast cancer with axillary lymph node metastases. Genes Chromosomes Cancer 56, 598–607. doi: 10.1002/gcc.22463
Sun, X., Haider Ali, M. S. S., and Moran, M. (2017). The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 474, 2925–2935. doi: 10.1042/bcj20170280
Sun, Y., Luo, M., Chang, G., Ren, W., Wu, K., Li, X., et al. (2017). Phosphorylation of Ser6 in hnRNPA1 by S6K2 regulates glucose metabolism and cell growth in colorectal cancer. Oncol. Lett. 14, 7323–7331.
Sveen, A., Kilpinen, S., Ruusulehto, A., Lothe, R. A., and Skotheim, R. I. (2016). Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35, 2413–2427. doi: 10.1038/onc.2015.318
Sweetser, D. A., Peniket, A. J., Haaland, C., Blomberg, A. A., Zhang, Y., Zaidi, S. T., et al. (2005). Delineation of the minimal commonly deleted segment and identification of candidate tumor-suppressor genes in del(9q) acute myeloid leukemia. Genes Chromosomes Cancer 44, 279–291. doi: 10.1002/gcc.20236
Tacke, R., Tohyama, M., Ogawa, S., and Manley, J. L. (1998). Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93, 139–148. doi: 10.1016/s0092-8674(00)81153-8
Talukdar, I., Sen, S., Urbano, R., Thompson, J., Yates, J. R. III, and Webster, N. J. (2011). hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLoS One 6:e27869. doi: 10.1371/journal.pone.0027869
Tang, S., Tao, M., McCoy, J. P. Jr., and Zheng, Z. M. (2006). The E7 oncoprotein is translated from spliced E6∗I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80, 4249–4263. doi: 10.1128/jvi.80.9.4249-4263.2006
Tang, X., Kane, V. D., Morre, D. M., and Morre, D. J. (2011). hnRNP F directs formation of an exon 4 minus variant of tumor-associated NADH oxidase (ENOX2). Mol. Cell Biochem. 357, 55–63. doi: 10.1007/s11010-011-0875-5
Tarn, W. Y., and Steitz, J. A. (1997). Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 22, 132–137. doi: 10.1016/s0968-0004(97)01018-9
Tauler, J., Zudaire, E., Liu, H., Shih, J., and Mulshine, J. L. (2010). hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 70, 7137–7147. doi: 10.1158/0008-5472.can-10-0860
Taylor, S. J., Anafi, M., Pawson, T., and Shalloway, D. (1995). Functional interaction between c-Src and its mitotic target, Sam 68. J. Biol. Chem. 270, 10120–10124. doi: 10.1074/jbc.270.17.10120
Tornesello, M. L., Annunziata, C., Tornesello, A. L., Buonaguro, L., and Buonaguro, F. M. (2018). Human oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers 10:213. doi: 10.3390/cancers10070213
Tornesello, M. L., Faraonio, R., Buonaguro, L., Annunziata, C., Starita, N., Cerasuolo, A., et al. (2020). The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front. Oncol. 10:150. doi: 10.3389/fonc.2020.00150
Turunen, J. J., Niemela, E. H., Verma, B., and Frilander, M. J. (2013). The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4, 61–76. doi: 10.1002/wrna.1141
Ule, J., and Blencowe, B. J. (2019). Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell 76, 329–345. doi: 10.1016/j.molcel.2019.09.017
Urbanski, L. M., Leclair, N., and Anczukow, O. (2018). Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 9:e1476. doi: 10.1002/wrna.1476
van, D. K., Li, Z., Xirasagar, S., Maes, P., Kaminsky, D., Liou, D., et al. (2017). The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 45, D499–D506.
Venables, J. P., Bourgeois, C. F., Dalgliesh, C., Kister, L., Stevenin, J., and Elliott, D. J. (2005). Up-regulation of the ubiquitous alternative splicing factor Tra2beta causes inclusion of a germ cell-specific exon. Hum. Mol. Genet. 14, 2289–2303. doi: 10.1093/hmg/ddi233
Wahl, M. C., Will, C. L., and Luhrmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wan, L., Yu, W., Shen, E., Sun, W., Liu, Y., Kong, J., et al. (2019). SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut 68, 118–129. doi: 10.1136/gutjnl-2017-314983
Wang, B. D., and Lee, N. H. (2018). Aberrant RNA splicing in cancer and drug resistance. Cancers 10:458. doi: 10.3390/cancers10110458
Wang, C., Norton, J. T., Ghosh, S., Kim, J., Fushimi, K., Wu, J. Y., et al. (2008). Polypyrimidine tract-binding protein (PTB) differentially affects malignancy in a cell line-dependent manner. J. Biol. Chem. 283, 20277–20287. doi: 10.1074/jbc.m803682200
Wang, F., Fu, X., Chen, P., Wu, P., Fan, X., Li, N., et al. (2017). SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling. Cell Res. 27, 540–558. doi: 10.1038/cr.2017.7
Wang, G. S., and Cooper, T. A. (2007). Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8, 749–761. doi: 10.1038/nrg2164
Wang, Y., Liu, J., Huang, B. O., Xu, Y. M., Li, J., Huang, L. F., et al. (2015). Mechanism of alternative splicing and its regulation. Biomed. Rep. 3, 152–158.
Wang, Z., and Burge, C. B. (2008). Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813. doi: 10.1261/rna.876308
Watson, I. R., Takahashi, K., Futreal, P. A., and Chin, L. (2013). Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet. 14, 703–718. doi: 10.1038/nrg3539
Will, C. L., and Luhrmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707.
Williams, V. M., Filippova, M., Filippov, V., Payne, K. J., and Duerksen-Hughes, P. (2014). Human papillomavirus type 16 E6∗ induces oxidative stress and DNA damage. J. Virol. 88, 6751–6761. doi: 10.1128/jvi.03355-13
Wu, C., Kajitani, N., and Schwartz, S. (2017). Splicing and polyadenylation of human papillomavirus type 16 mRNAs. Int. J. Mol. Sci. 18:366. doi: 10.3390/ijms18020366
Wu, H., Sun, S., Tu, K., Gao, Y., Xie, B., Krainer, A. R., et al. (2010). A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell 38, 67–77. doi: 10.1016/j.molcel.2010.02.021
Wu, X., Yang, Y., Huang, Y., Chen, Y., Wang, T., Wu, S., et al. (2018). RNA-binding protein AUF1 suppresses miR-122 biogenesis by down-regulating Dicer1 in hepatocellular carcinoma. Oncotarget 9, 14815–14827. doi: 10.18632/oncotarget.24079
Xiao, J., Wang, Q., Yang, Q., Wang, H., Qiang, F., He, S., et al. (2018). Clinical significance and effect of Sam68 expression in gastric cancer. Oncol. Lett. 15, 4745–4752.
Xue, Y., Bellanger, S., Zhang, W., Lim, D., Low, J., Lunny, D., et al. (2010). HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 70, 5316–5325. doi: 10.1158/0008-5472.can-09-3789
Yang, H., Zhu, R., Zhao, X., Liu, L., Zhou, Z., Zhao, L., et al. (2019). Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 38, 4915–4931. doi: 10.1038/s41388-019-0764-z
Yang, S., Jia, R., and Bian, Z. (2018). SRSF5 functions as a novel oncogenic splicing factor and is upregulated by oncogene SRSF3 in oral squamous cell carcinoma. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1161–1172. doi: 10.1016/j.bbamcr.2018.05.017
Yeo-Teh, N. S. L., Ito, Y., and Jha, S. (2018). High-risk human papillomaviral oncogenes E6 and E7 target key cellular pathways to achieve oncogenesis. Int. J. Mol. Sci. 19:1706. doi: 10.3390/ijms19061706
Yoshida, T., Kim, J. H., Carver, K., Su, Y., Weremowicz, S., Mulvey, L., et al. (2015). CLK2 is an oncogenic kinase and splicing regulator in breast cancer. Cancer Res. 75, 1516–1526. doi: 10.1158/0008-5472.can-14-2443
Yu, C., Guo, J., Liu, Y., Jia, J., Jia, R., and Fan, M. (2015). Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J. Cell Physiol. 230, 2252–2261. doi: 10.1002/jcp.24956
Yu, H., Gong, L., Wu, C., Nilsson, K., Li-Wang, X., and Schwartz, S. (2018). hnRNP G prevents inclusion on the HPV16 L1 mRNAs of the central exon between splice sites SA3358 and SD3632. J. Gen. Virol. 99, 328–343. doi: 10.1099/jgv.0.001019
Zhang, Q., Di, C., Yan, J., Wang, F., Qu, T., Wang, Y., et al. (2019a). Inhibition of SF3b1 by pladienolide B evokes cycle arrest, apoptosis induction and p73 splicing in human cervical carcinoma cells. Artif Cells Nanomed. Biotechnol. 47, 1273–1280. doi: 10.1080/21691401.2019.1596922
Zhang, Q., Lv, R., Guo, W., and Li, X. (2019b). microRNA-802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine-rich splicing factor 9. J. Cell Biochem. 120, 10370–10379. doi: 10.1002/jcb.28321
Zhang, T., Wan, C., Shi, W., Xu, J., Fan, H., Zhang, S., et al. (2015). The RNA-binding protein Sam68 regulates tumor cell viability and hepatic carcinogenesis by inhibiting the transcriptional activity of FOXOs. J. Mol. Histol. 46, 485–497. doi: 10.1007/s10735-015-9639-y
Zhang, Y., Wu, D., and Wang, D. (2020). Long non-coding RNA ARAP1-AS1 promotes tumorigenesis and metastasis through facilitating proto-oncogene c-Myc translation via dissociating PSF/PTB dimer in cervical cancer. Cancer Med. 9, 1855–1866. doi: 10.1002/cam4.2860
Zhang, Y., Yan, L., Zeng, J., Zhou, H., Liu, H., Yu, G., et al. (2019). Pan-cancer analysis of clinical relevance of alternative splicing events in 31 human cancers. Oncogene 38, 6678–6695. doi: 10.1038/s41388-019-0910-7
Zhang, Z., Li, J., Zheng, H., Yu, C., Chen, J., Liu, Z., et al. (2009). Expression and cytoplasmic localization of SAM68 is a significant and independent prognostic marker for renal cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 18, 2685–2693. doi: 10.1158/1055-9965.epi-09-0097
Zhang, Z., Xu, Y., Sun, N., Zhang, M., Xie, J., and Jiang, Z. (2014). High Sam68 expression predicts poor prognosis in non-small cell lung cancer. Clin. Transl. Oncol. 16, 886–891. doi: 10.1007/s12094-014-1160-3
Zhao, X., Fay, J., Lambkin, H., and Schwartz, S. (2007). Identification of a 17-nucleotide splicing enhancer in HPV-16 L1 that counteracts the effect of multiple hnRNP A1-binding splicing silencers. Virology 369, 351–363. doi: 10.1016/j.virol.2007.08.002
Zhao, X., Oberg, D., Rush, M., Fay, J., Lambkin, H., and Schwartz, S. (2005). A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 79, 4270–4288. doi: 10.1128/jvi.79.7.4270-4288.2005
Zhao, X., Rush, M., and Schwartz, S. (2004). Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 78, 10888–10905. doi: 10.1128/jvi.78.20.10888-10905.2004
Zhao, X., and Schwartz, S. (2008). Inhibition of HPV-16 L1 expression from L1 cDNAs correlates with the presence of hnRNP A1 binding sites in the L1 coding region. Virus Genes 36, 45–53. doi: 10.1007/s11262-007-0174-0
Zheng, Z. M., and Baker, C. C. (2006). Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 11:2286–2302. doi: 10.2741/1971
Zheng, Z. M., Tao, M., Yamanegi, K., Bodaghi, S., and Xiao, W. (2004). Splicing of a cap-proximal human Papillomavirus 16 E6E7 intron promotes E7 expression, but can be restrained by distance of the intron from its RNA 5′ cap. J. Mol. Biol. 337, 1091–1108. doi: 10.1016/j.jmb.2004.02.023
Zheng, Z. Z., Sun, Y. Y., Zhao, M., Huang, H., Zhang, J., Xia, N. S., et al. (2013). Specific interaction between hnRNP H and HPV16 L1 proteins: implications for late gene auto-regulation enabling rapid viral capsid protein production. Biochem. Biophys. Res. Commun. 430, 1047–1053. doi: 10.1016/j.bbrc.2012.12.042
Zhou, X., Li, Q., He, J., Zhong, L., Shu, F., Xing, R., et al. (2017). HnRNP-L promotes prostate cancer progression by enhancing cell cycling and inhibiting apoptosis. Oncotarget 8, 19342–19353. doi: 10.18632/oncotarget.14258
Zhou, X., Li, X., Cheng, Y., Wu, W., Xie, Z., Xi, Q., et al. (2014). BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat. Commun. 5:4581.
Zhou, X., Li, X., Yu, L., Wang, R., Hua, D., Shi, C., et al. (2019). The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int. J. Biochem. Cell Biol. 113, 75–86. doi: 10.1016/j.biocel.2019.06.003
Zhou, Z., and Fu, X. D. (2013). Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207. doi: 10.1007/s00412-013-0407-z
Zhou, Z. J., Dai, Z., Zhou, S. L., Fu, X. T., Zhao, Y. M., Shi, Y. H., et al. (2013). Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int. J. Cancer 132, 1080–1089. doi: 10.1002/ijc.27742
Keywords: splicing factors, cervical cancer, human papillomavirus (HPV), RNA, heterogeneous nuclear ribonucleoproteins (hnRNPs), serine/arginine-rich proteins (SR)
Citation: Cerasuolo A, Buonaguro L, Buonaguro FM and Tornesello ML (2020) The Role of RNA Splicing Factors in Cancer: Regulation of Viral and Human Gene Expression in Human Papillomavirus-Related Cervical Cancer. Front. Cell Dev. Biol. 8:474. doi: 10.3389/fcell.2020.00474
Received: 24 March 2020; Accepted: 20 May 2020;
Published: 12 June 2020.
Edited by:
Giulia De Falco, Queen Mary University of London, United KingdomReviewed by:
Alok Chandra Bharti, University of Delhi, IndiaCopyright © 2020 Cerasuolo, Buonaguro, Buonaguro and Tornesello. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Lina Tornesello, bS50b3JuZXNlbGxvQGlzdGl0dXRvdHVtb3VyaS5uYS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.