- 1Unidad de Genómica Avanzada (Langebio), Centro de Investigación y de Estudios Avanzados, Guanajuato, Mexico
- 2Department of Plant Pathology and Microbiology, Robert H. Smith Faculty of Agriculture, Food, and Environment, The Hebrew University of Jerusalem, Rehovot, Israel
Protein science has moved from a focus on individual molecules to an integrated perspective in which proteins emerge as dynamic players with multiple functions, rather than monofunctional specialists. Annotation of the full functional repertoire of proteins has impacted the fields of biochemistry and genetics, and will continue to influence basic and applied science questions – from the genotype-to-phenotype problem, to our understanding of human pathologies and drug design. In this review, we address the phenomena of pleiotropy, multidomain proteins, promiscuity, and protein moonlighting, providing examples of multitasking biomolecules that underlie specific mechanisms of human disease. In doing so, we place in context different types of multifunctional proteins, highlighting useful attributes for their systematic definition and classification in future research directions.
At the time when George Beadle and Edward Tatum proposed the one gene–one enzyme paradigm (Beadle and Tatum, 1941), the key and lock mechanism of substrate recognition in enzymes was used to explain how proteins accelerate chemical reactions. In the early days of molecular biology, enzymes and other proteins were thus seen as highly specific monofunctional molecules. This theoretical framework has influenced our functional annotations in biological systems: once a role is assigned to a given protein, other functions at any level are usually overlooked (McKay and Ashley, 2008). However, since the 1940s many different mechanisms of multifunctionality have been described.
Proteins with multiple functions can influence the evolution of organisms, as they contribute to complexity and robustness. For instance, in early forms of life and in organisms undergoing genome reduction, multifunctionality may increase the functional repertoire of a limited set of genes (Kelkar and Ochman, 2013; Ferla et al., 2017). In addition, multifunctional proteins may coordinate the crosstalk of dissimilar biological processes, which is specially relevant in organisms with large genomes and intricate metabolic and regulatory pathways (Flores and Gancedo, 2011; Irving et al., 2018). Thus, the presence of undetected multifunctionality is a confounding factor in functional association analyses based on guilt-by-association, genotype-phenotype maps, and homology prediction and annotation (Jeffery, 1999; Salathe et al., 2006; Gillis and Pavlidis, 2011; Espinosa-Cantu et al., 2018). Identification and understanding of protein multifunctionality is needed to grant a better understanding of living systems in health and disease. Indeed, multifunctional proteins are frequently determinants of virulence and disease (reviewed in Henderson and Martin, 2011; Jeffery, 2015, 2018b; Franco-Serrano et al., 2018c), as we will emphasize.
Other genetic mechanisms that are also known to contribute to the overall functional diversity of living systems are not herein addressed, such as alternative promoters or splicing in transcription, overlapping ORFs, alternative UTRs in translation, or post-translational modifications (Ayoubi and Van De Ven, 1996; Delaye et al., 2008; Kelemen et al., 2013; Fellner et al., 2015; Mayr, 2016). Here, we review alternative definitions of protein multifunctionality and provide examples of multifunctional proteins associated to human disease. We focus specifically on multifunctionality at the protein level.
Naming and Classifying Multiple Forms of Protein Multifunctionality
To date, several non-mutually exclusive terms have been used to describe recurrent phenomena in which proteins perform two or more functions, notably pleiotropy, multidomain proteins, promiscuity, and moonlighting. Although the terms discussed here are widely used in the literature, there is an underlying lack of consensus in their usage, which may consequently hinder our understanding of the extent and biological significance of each of the different forms of protein multifunctionality. For instance, a single enzyme can be pleiotropic, multidomain, and show promiscuous and moonlighting activities (Figure 1; Aigle and Lacroute, 1975; Bai et al., 2004; Gancedo and Flores, 2008; Zhang and Perrett, 2009; The UniProt Consortium, 2018).
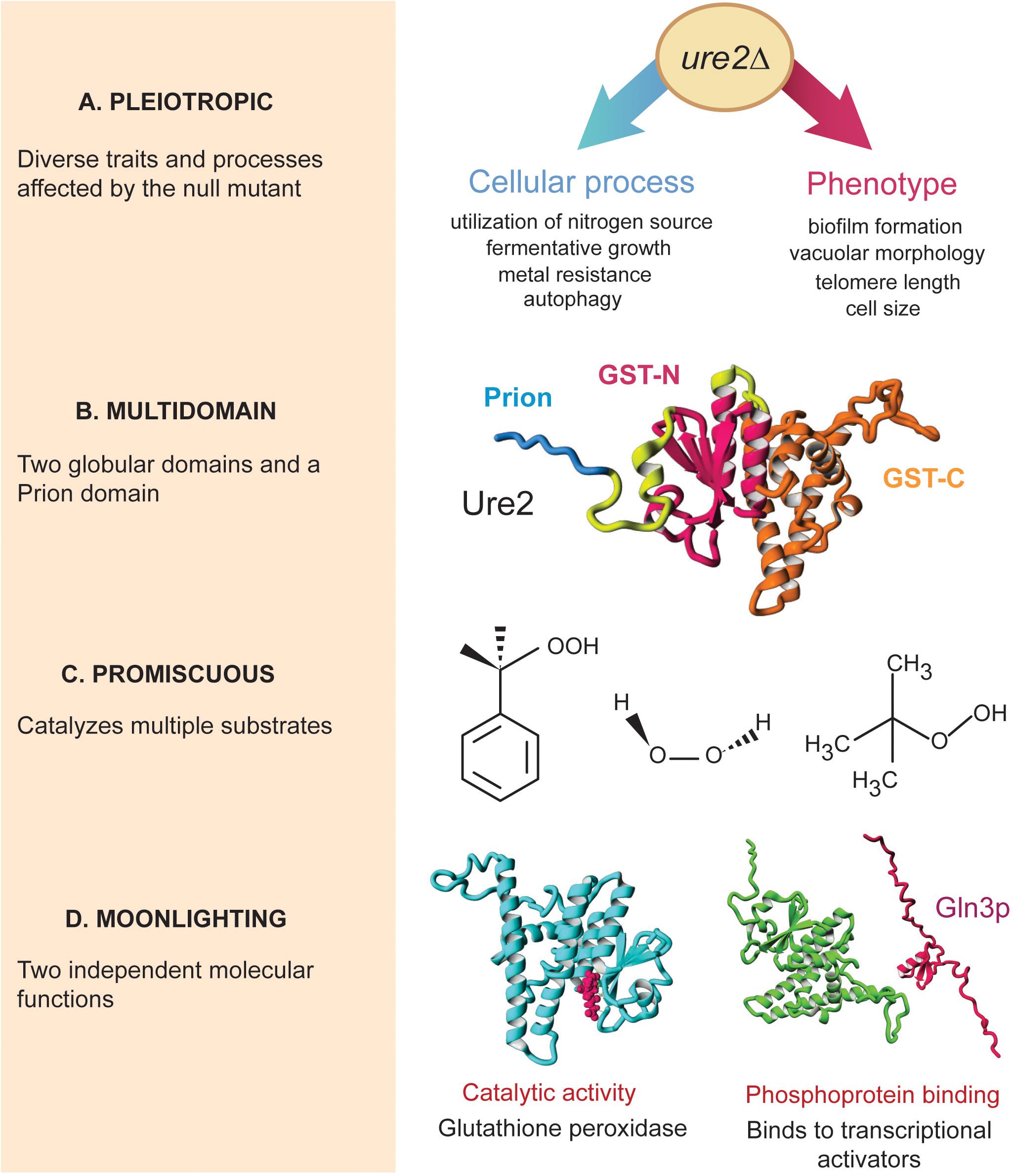
Figure 1. Different forms of protein multifunctionality in Ure2p. The Ure2 protein from budding yeast is a peroxidase enzyme that can form prions and is directly involved in nitrogen-catabolite repression by binding to and sequestering phosphorylated transcription factors in the cytoplasm, such as Gln3. (A) Deletion of URE2 results in numerous phenotypes and observable traits related to different cellular pathways, which makes Ure2 a pleiotropic protein; only representative phenotypes associated to ure2Δ are shown. (B) Ure2 is a multidomain protein composed of an unstructured prion domain in addition to two globular domains typical of glutathione S-transferases (GST-N and GST-C). (C) The enzymatic activity of Ure2 is highly promiscuous: in vitro, Ure2 catalyze reactions with diverse substrates, involving different reaction mechanisms, within a single active site. Three different substrates for the glutathione-peroxidase activity of Ure2 are shown (from left to right: cumene hydroperoxide, hydrogen peroxide, and tert-butyl hydroperoxide). (D) The catalytic and binding activities of Ure2 are independent from one another: mutants lacking the Gln3-regulatory activity maintain their catalytic activity and such protein moonlighting is not explained by gene fusion, alternative splicing, or differential proteolysis. Protein images were generated using structure data from Protein Data Bank entry 1GgY, or Raptor X-predicted structures from Uniprot accessions P18494 and P23202.
The definition of multifunctionality fundamentally relies on the definition of protein function itself. Protein function is the potential activity with a given physiological role that a polypeptide with a single primary sequence can partake by interacting with other biomolecules or chemical compounds. Based on Gene Ontology (GO) annotations, protein multifunctionality may be described at different overlapping levels (Figure 2). The GO Consortium describes the function of a gene or gene product using three attributes: molecular function, biological process, and cellular component (Ashburner et al., 2000).
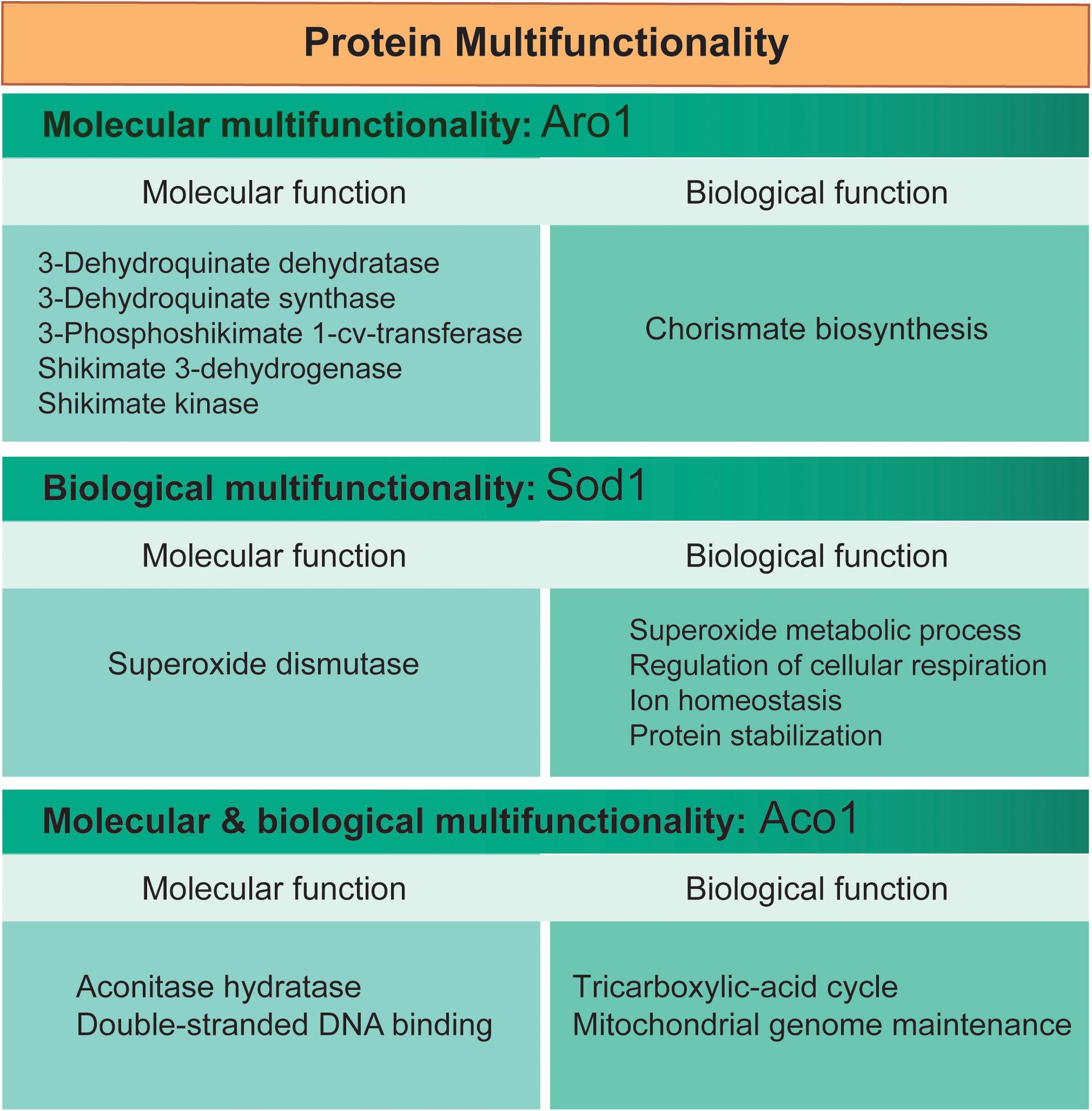
Figure 2. Different kinds of multifunctional proteins. Different kinds of multifunctional proteins may be described based on their “molecular” or “biological” functions. Examples are provided for three multifunctional proteins from S. cerevisiae (Aro1, Sod1, and Aco1); functional annotations (GO terms) are from the Saccharomyces Genome Database (www.yeastgenome.org).
Pritykin et al. (2015) analyzed the genome-wide multifunctionality of proteins in yeast, fly, and humans. Between 20 and 26% of the characterized genes in these organisms are multifunctional proteins annotated in two or more significantly different GO biological process terms. A smaller percentage of proteins are annotated within two or more GO molecular function terms, but this observation can be partly explained by the fact that the number of annotations belonging to the molecular function classification is lower relative to the biological process class. Compared to other proteins in the restricted set of analyzed organisms, proteins annotated with more than one GO molecular function – which may include pleiotropic, multidomain, promiscuous, and moonlighting proteins – are typically longer in sequence, are frequently more broadly expressed, have more protein domains and higher fractions of disordered regions, and tend to be conserved and essential (Pritykin et al., 2015). Importantly, such multifunctional genes are more often involved in human disorders. These results highlight the convenience of a systematic classification of multifunctionality to uncover its prevalence, characteristics, and relevance in biological systems.
Diverse stimuli may trigger action of different functions of a multifunctional protein (reviewed in Jeffery, 1999, 2018b; Nobeli et al., 2009). Cellular conditions such as temperature, pH, concentration of metabolites, and presence or absence of ligands have been observed to regulate the functions of multitasking proteins. Differential expression, post-transcriptional modifications, intra- or extra-cellular localization, protein structural changes – from flexibility of small regions to complete refolding of an entire domain – and homo-or hetero oligomeric conformations are frequent molecular mechanisms of protein multifunctionality. Such mechanisms are not mutually exclusive; actually, they tend to take place concertedly in the occurrence and alternation of multiple functions.
Pleiotropy May Reflect Multifunctionality at Different Levels
Pleiotropy refers to phenomena in which a single genetic factor contributes to different cellular, physiological, or organismic traits (Stearns, 2010; Paaby and Rockman, 2013; Zhang and Wagner, 2013). In genetic analysis, pleiotropy is assessed by describing the effects of mutations in measurable phenotypic traits. For example, the complete deletion of the URE2 gene in the budding yeast Saccharomyces cerevisiae results in abnormal autophagy, decreased resistance to several cellular stresses, decreased telomere length, and increased cell size, among other phenotypes (Figure 1A). Likewise, mutations in Swc6, a chromatin-remodeling protein from Arabidopsis thaliana, result in different leaf, flower, and fruit morphologies (Lazaro et al., 2008).
The mutant phenotypes of a pleiotropic multifunctional protein may arise from the perturbation of a single molecular function that partakes in different biological processes, or multiple molecular functions, each with impact on a different biological process (Dudley et al., 2005; He and Zhang, 2006). A major challenge in genetics, cell biology, and biomedical research is to identify the molecular mechanisms underlying pleiotropy (van de Peppel and Holstege, 2005; He and Zhang, 2006). For example, the multiple phenotypes observed in phenylketonuria – behavioral and mental disorders along with specific physical traits – result from the inactivation of a single catalytic activity in the phenylalanine hydroxylase enzyme (reviewed in Hodgkin, 1998; Sarkissian and Gamez, 2005). Conversely, numerous non-enzymatic activities have been described for the pleiotropic enzyme glyceraldehyde-3-phosphate dehydrogenase, some involved in human pathologies including different types of cancer and Huntington’s, Parkinson’s, and Alzheimer’s diseases (reviewed in Tarze et al., 2007; Sirover, 2018).
Multiple Protein Domains May Results in Manifold Functions
A protein domain is a polypeptide fragment with independent evolutionary history, structure, or function (Ekman et al., 2005). Multidomain proteins typically arise from gene fusion, whereby two protein fragments become linked together in a single open reading frame (Kirschner and Bisswanger, 1976). For example, in Escherichia coli, hydroxyethylthiazole kinase and thiamine-phosphate synthase are expressed in different proteins. However, in S. cerevisiae both catalytic activities are present in a single multidomain protein whereby each domain performs one enzymatic function (Jardine et al., 2002). The analysis of protein domain composition depends on the definition of domain and identification methods used, either based on protein-sequence comparison or structural properties (Ekman et al., 2005). For instance, the two domains of Ure2 are sometimes considered as subdomains of a single protein domain (Figure 1B), while the limits of each structural region vary frequently among protein databases. Importantly, not all multidomain proteins are multifunctional. The shikimate dehydrogenase from E. coli is a two-domain NAD(P)-oxidoreductase: while one domain binds the cofactor, the other couples with the substrate, in such a way that the two domains catalyze a single reaction in the aromatic amino-acid biosynthesis pathway (Michel et al., 2003).
An example of a multidomain protein involved in cardiovascular disease is the G-protein coupled receptor kinase 2 (GRK2, also known as beta-adrenergic receptor kinase). The role of GRK2 on the regulation of cardiac receptors is the result of successful coordination among the three protein domains that compose the enzyme: a regulator of G-protein signaling homology domain, a protein-kinase domain, and a pleckstrin-homology domain (Lodowski et al., 2003, 2005). While the disruption of the kinase gene can be lethal in mice, GRK2 overexpression has been linked to several cardiovascular diseases in humans (Ungerer et al., 1994; Akhter et al., 1999). Interestingly, the expression of the pleckstrin-homology domain in mice with GRK2 overexpression can revert the disease phenotype, and could therefore be used as a treatment strategy to improve compromised heart function (Akhter et al., 1999).
Promiscuity Is (Or Is Not) True Protein Multifunctionality
Protein “promiscuity” describes a wide variety of related phenomena, all of which describe multiple catalytic activities or binding to multiple molecules (Hult and Berglund, 2007; Nobeli et al., 2009; Copley, 2015, 2017). The meaning and implications of promiscuity will depend on who one asks (Copley, 2015). From the evolutionary and biochemistry perspective, promiscuity usually involves physiologically irrelevant activities, by means of either inefficient or fortuitous activities in non-natural cellular contexts (Khersonsky et al., 2006; Copley, 2017). In other fields, however, promiscuity typically describes broad specificity or binding to multiple ligands, substrates, or other macromolecules with no consideration to the relevance of the activities (Copley, 2015). In the former scenario pertaining enzyme activity, promiscuity is not a form of molecular multifunctionality, while in the latter cases it actually is.
The use of the term promiscuity also depends on the type of activity being described: in non-enzymatic activities, promiscuity is used to describe binding to several different molecules in a single or multiple structural locations (Humphris and Kortemme, 2007; Patil et al., 2010; Schreiber and Keating, 2011). For instance, thioredoxin from E. coli or human Ras proteins have been described as promiscuous hub proteins given their many protein-protein interactions. While thioredoxin interactions take place in the same protein interface, Ras interactions occur in different binding sites (Humphris and Kortemme, 2007). For catalytic activities, promiscuity refers to a single active site – although not necessarily within the exact same residues – that either binds to a diverse set of substrates (substrate promiscuity or ambiguity) or catalyzes different chemical transformations (catalytic promiscuity) (Khersonsky et al., 2006; Nobeli et al., 2009; Khersonsky and Tawfik, 2010; Copley, 2015, 2017). For example, yeast Ure2 has glutathione peroxidase activity on different hydroperoxide molecules in vitro (Bai et al., 2004) and it can also oxidize a wide variety of substrates through alternative reaction mechanisms (Zhang and Perrett, 2009; Figure 1C).
The altered efficiency of enzymes catalyzing different substrates has been linked to health complications, for instance those having to do with perturbed metabolism of exogenous compounds. Such is the case of serum paraoxonase 1 (PON1), a glycoprotein with broad substrate specificity capable of hydrolyzing organophosphates, lactones, and aromatic esters (Costa et al., 2003). PON1 is involved in a variety of roles, from pesticide sensitivity to a protective role against atherosclerosis by hydrolyzing oxidized lipids and preventing their accumulation in artery walls. Frequent human polymorphic variants of PON1 show different catalytic efficiencies to alternative organophosphorus compounds and are related to different pesticide susceptibilities: while variant PON1R192 hydrolyzes paraoxon faster than PON1Q192, PON1Q192 hydrolyzes diazoxon, sarin, and soman faster than the other isozyme (Davies et al., 1996). The altered hydrolysis efficiency toward other lipidic compounds may also explain why the PON1R192 variant has been associated with cardiovascular disease (Aviram et al., 2000; Costa et al., 2003).
Dysfunction of catalytic-promiscuous enzymes can also influence Alzheimer’s disease and cancer incidence. The omega class gluthatione s-transferases GSTO1 and GSTO2 can carry out two different chemical reactions in the same active site: gluthatione-dependent thiol transfer and dehydroascorbate reduction (Board and Menon, 2016). The functions of GSTO1 and GSTO2 confer their role in oxidative stress and xenobiotic detoxification; glutathionylation prevents proteins from being oxidized and dehydroascorbate reduction takes part on the recycling pathway of the antioxidant ascorbic acid (Board and Menon, 2016; Kim et al., 2017). Altered function of these gluthatione transferases may underlie the physiological conditions of several diseases: a GSTO2 variant with low expression levels has been found to be associated with an increased risk of Alzheimer disease at older age, while increased GSTO1 levels have been linked to a number of cancers such as pancreatic, esophageal, colorectal, and ovarian cancer, among others (Allen et al., 2012; Board and Menon, 2016).
Moonlighting Proteins Multitask in Disparate Functions
Protein “moonlighting” was first defined as the phenomenon whereby a polypeptide performs two or more molecular functions that are not the result of gene fusion, splice variants, or multiple proteolytic fragments (Jeffery, 1999). “Gene sharing” was originally coined to refer to the phenomena where a gene specialized in a function performed alternative biological roles. While this term is sometimes used as a protein-moonlighting synonym, both phenomena may actually differ in their mechanisms and implications (Copley, 2012). The concept of moonlighting protein has continued to expand in a less restrictive definition, and is nowadays also used to name proteins showing unrelated roles in the organisms, irrespective of their evolutionary history (Chapple et al., 2015; Jungblut et al., 2016; Khan and Kihara, 2016).
The molecular activities of a moonlighting protein are usually described as unrelated to each other and independently of the primary sequence (Huberts and van der Klei, 2010; Flores and Gancedo, 2011). Indeed, experimental evidence for the characterization of a moonlighting protein is mostly provided by mutants that are defective in one molecular function without affecting the others (Jeffery, 2009; Gancedo et al., 2016; Espinosa-Cantu et al., 2018). In the case of yeast Ure2, mutants without nitrogen-catabolite repression activity maintain their enzymatic capacities (Figure 1D). One of the first examples of a moonlighting protein was quite astonishing: the delta-crystallin of birds and reptiles is the exact same polypeptide as the argininosuccinate lyase enzyme; with its molecular function depending on the expression level and cellular context (Piatigorsky et al., 1988; Piatigorsky and Horwitz, 1996). In the lens, the protein is expressed at a high level and has a structural role, while in other tissues it is expressed at lower quantities and has an enzymatic function.
Moonlighting proteins with different molecular and functional characteristics have been described in a wide variety of organisms and the number of examples has increased in the last few years to almost 700 carefully curated moonlighting proteins (Hernández et al., 2014; Mani et al., 2015; Chen et al., 2018; Franco-Serrano et al., 2018b). Even though experimental setups have been aimed at revealing protein moonlighting in an unbiased manner (Su and Pillus, 2016; Espinosa-Cantu et al., 2018), still most known cases have been described by serendipity, either by studying a single protein in an exhaustive manner (e.g., the delta-crystalline Piatigorsky and Horwitz, 1996) or as rather surprising results from functional screens (Zelenaya-Troitskaya et al., 1995; Hall et al., 2004; Chen et al., 2005; Scott and Pillus, 2010; Scherrer et al., 2010; Chang et al., 2012).
Computational strategies have been developed to characterize moonlighting proteins and to identify novel candidates for further experimental confirmation. Alignment algorithms evaluating similarity to other proteins, functional domains, or motifs have been tested to predict the nature of alternative functions within a protein (Gomez et al., 2003). Protein-protein interaction databases can also reveal potential cases of protein moonlighting and provide suggestions to further test multifunctionality (Gomez et al., 2011). The topological properties of a protein-protein interaction network and GO annotations were used to identify proteins exhibiting signs of so-called “extreme multifunctionality” (Chapple et al., 2015). Khan and Kihara developed a method based on machine learning of different “omic” data, such as expression profiles, phylogenetic profiles, protein-protein interactions, genetic interactions and gene ontology (Khan and Kihara, 2016). More recently, a method based on text-mining was developed to identify moonlighting protein candidates within publications hinting at semantically different functions (Jain et al., 2018).
As the number of known moonlighting proteins has increased, efforts have been made to understand how these molecules evolve and to guide in looking for more such multifunctional biomolecules. One of the first observations was that moonlighting proteins are enriched in conserved, highly expressed, proteins; however, this could also represent a bias where highly expressed and conserved proteins are also more likely of being studied (Huberts and van der Klei, 2010). In addition to their expression patterns, sequence and structural properties may be related to protein moonlighting. For example, it has been proposed that moonlighting proteins could be enriched in proteins with well-packed, independent structural domains with little evolutionary trade-offs between the functions. Disordered regions could also facilitate the accumulation of functional mutations and shifts between activities (Tompa et al., 2005). Yet, a correlation between domain composition, disordered regions, and moonlighting has not been documented unambiguously (Hernandez et al., 2011; Amblee and Jeffery, 2015; Chapple et al., 2015). With regards to the interactions, candidate moonlighting proteins interact with more functionally diverse proteins and more connected to other moonlighting proteins (Khan et al., 2014; Chapple et al., 2015). Importantly, moonlighting proteins tend to be involved in mechanisms of disease (Chapple et al., 2015; Franco-Serrano et al., 2018c).
Diseases and pathologies related to moonlighting phenomena are being constantly described (Sriram et al., 2005; Jeffery, 2011; Zanzoni et al., 2015; Henderson et al., 2016; Franco-Serrano et al., 2018c). It has been estimated that up to 78% of candidate moonlighting proteins are associated to a human disease, in contrast to a 17.9% disease association for proteins in general (Franco-Serrano et al., 2018c). Among the diseases with the highest representation among moonlighting candidate proteins are those related to cancer (27%), immune system (17%), and nervous system (14%). As moonlighting proteins can have unrelated functions, it is expected that dysfunction of one or more of the protein functions gives rise to complex phenotypes. So far, moonlighting candidates have been found to be enriched in proteins that are involved in more than one disease and associated to complex instead of monogenic diseases (Zanzoni et al., 2015). These proteins are associated to phenotypically dissimilar diseases that share co-morbidity patterns, hinting this could result from the perturbation of different processes of a single moonlighting protein.
The essential enzyme glucose-6-phosphate isomerase (GPI) is an example of a moonlighting protein that has been linked to anemia and neurological pathologies due to its multifuncional nature. The dimeric form of GPI catalyzes the reversible conversion of fructose-6-phosphate to glucose-6-phosphate in glycolysis (Kugler and Lakomek, 2000), while the monomeric proteins show a neurotrophic function, independently of catalytic activity, which promotes the survival of embryonic spinal neurons, skeletal motor neurons, and sensory neurons (Gurney et al., 1986). Mutations in GPI cause a disorder known as GPI-deficiency, characterized by breakage of red blood cells (hemolytic anemia) due to impairment of glycolysis (Kugler and Lakomek, 2000). Most forms of GPI-deficiency are associated to mutations affecting only the isoenzyme activity, but severe forms of the disease, including neurological pathologies – muscular weakness and intellectual disability – are associated to mutations speculated to prevent correct protein folding disrupting both its catalytic and neurotrophic functions (Kugler et al., 1998).
Post-translational modifications involved in the modulation of moonlighting activities have been associated to the development of human disease. Phosphorylation, acetylation, and ubiquitination – among other protein modifications – affect the cellular localization and the binding affinity of proteins to other molecules (Jeffery, 2016; Zanzoni et al., 2019). For example, the S-nitrosylation of the glyceraldehyde 3-phosphate dehydrogenase (G3PDH) protein promotes the shift from the catalytic function to a protein-binding function (Hara et al., 2005). In this non-catalytic activity, G3PDH binds to a E3-ubiquitin ligase, Siah1, which promotes enter to the nucleus, where this protein complex initiates apoptosis. This signaling cascade has been proposed to take part in the neurodegeneration observed in Parkinson’s and Alzheimer’s diseases. Importantly, inhibition of S-nitrosylation of G3PDH by the drug L-deprenyl prevents cell death in cerebellar granule neurons in mice and delays the progression of Parkinson’s disease in humans (Parkinson Study Group, 1993; Hara et al., 2006).
Disease can also be triggered by a protein acquiring a novel activity due to a structural perturbation (mutation or conformational change), in which case the new function is known as “neomorphic moonlighting function” (Jeffery, 2011). For example, the beta-2-microglobulin acquires a new aggregation function besides its normal function in the major histocompatibility complex of presenting peptide antigens (Miyata et al., 1993; Jeffery, 2011). In the context of diminished kidney function or patients in long-term dialysis, beta-2-microglobulin is accumulated in the blood. This condition favors the conformational change to the amyloid fiber form and further protein aggregation, which ultimately leads to the development of amyloidosis characterized by arthropathy, lytic bone lesions, and chronic kidney disease.
Moonlighting proteins are not relevant to disease only due to their malfunction in humans, but also due to their role in pathogenesis and interaction with a human host (Henderson and Martin, 2011; Franco-Serrano et al., 2018a; Jeffery, 2018a). Intracellular proteins of pathogenic bacteria with previously described roles as metabolic enzymes (G3PDH), chaperones (Hsp60), or elongation factors (EF-Tu) have been found to exert a secondary function on the surface of bacterial cells as adhesins binding directly to host cells or extracellular matrix to initiate the infection process (Amblee and Jeffery, 2015; Jeffery, 2018a; Kopeckova et al., 2020). Moonlighting proteins can also influence virulence by providing antibiotic resistance to bacteria (Sengupta et al., 2008) and influencing nutrient uptake of the pathogen (Modun and Williams, 1999). Overall, 25% of known moonlighting proteins are classified as virulence factors and are found in some pathogens of interest such as Staphylococcus aureus, Helicobacter pylori, Haemophilus influenza and Neisseria meningitides, among others (Franco-Serrano et al., 2018a; Jeffery, 2018a).
Toward a Systematic Description of Protein Multifunctionality
Huge amounts of data are being produced from experimental and computational analysis in functional genomics and systems biology, making it urgent to unify definitions of protein multifunctionality in a more systematic manner. Recent advances in large-scale genetic and genetic-interaction screens (Bellay et al., 2011; Martin et al., 2015; Espinosa-Cantu et al., 2018), proteomics and protein-protein interaction databases (Gomez et al., 2011; Becker et al., 2012; Chapple et al., 2015; Wang and Jeffery, 2016), and metabolite screens (Sevin et al., 2017; Piazza et al., 2018), along with computational analyses of functional data (Ekman et al., 2005; Nam et al., 2012; Pritykin et al., 2015) have huge potential to reveal patterns of protein multifunctionality in living systems.
In this context, we note that certain features are good descriptors of the types of protein multifunctionality. Structural similarity, biological relatedness, structural autonomy, and physiological relevance of the different functions of a single protein will certainly influence the evolutionary scenarios and outcomes involved in the gain and loss of multifunctionality. For example, the lesser the structural autonomy between the activities in a protein, one would expect evolution of more similar molecular functions or the appearance of stronger evolutionary trade-offs. Duplication and divergence of the gene copies of multifunctional proteins with evolutionary trade-offs could result in single monofunctional proteins (Piatigorsky, 2007; Espinosa-Cantú et al., 2015). Unrelated biological functions may be more prone to evolve separately if they are antagonistic between each other. Therefore, it is possible that less structural autonomous functions would have more biological coherence. Functions with strong – or relevant – phenotypic effects will typically be subjected to strong selective pressures. Therefore, in proteins from mild to highly structurally dependent and relevant activities, one would expect co-evolution of functions. In contrast, those cases in which only one of the functions is under strong selection may be prone to mild or little structural autonomy, allowing mutations of the residues involved in the activity.
Pleiotropy, multi-domain proteins, promiscuity, and moonlighting are different biological phenomena related to protein multifunctionality. The description of different recurrent phenomena has been – and will certainly continue to be – useful to the understanding of protein multifunctionality. However, as often seen in biology, huge diversity in the mechanisms and related phenomena will most likely exist. Moreover, the terms and limits we assign to each phenomenon are also prone to evolve, thus developing a natural lack of consensus. Therefore, it is possible that an alternative, straightforward, annotation of protein multifunctionality may enable future endeavors to comprehend the diversity, extent, and significance of protein multifunctionality in a more systematic manner.
Author Contributions
AE-C, EC-B, LN-G, and AD wrote the manuscript, read, and approved the final manuscript.
Funding
Research in the DeLuna lab is funded by the Consejo Nacional de Ciencia y Tecnología de México (Grants CB2015/254365 and PN2016/2370). AE-C received funding from the Alexander von Humboldt Foundation during the writing of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank L. Delaye and N. Selem for critical reading of the manuscript.
References
Aigle, M., and Lacroute, F. (1975). Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136, 327–335. doi: 10.1007/bf00341717
Akhter, S. A., Eckhart, A. D., Rockman, H. A., Shotwell, K., Lefkowitz, R. J., and Koch, W. J. (1999). In vivo inhibition of elevated myocardial beta-adrenergic receptor kinase activity in hybrid transgenic mice restores normal beta-adrenergic signaling and function. Circulation 100, 648–653. doi: 10.1161/01.cir.100.6.648
Allen, M., Zou, F., Chai, H. S., Younkin, C. S., Miles, R., Nair, A. A., et al. (2012). Glutathione S-transferase omega genes in Alzheimer and Parkinson disease risk, age-at-diagnosis and brain gene expression: an association study with mechanistic implications. Mol. Neurodegener. 7:13. doi: 10.1186/1750-1326-7-13
Amblee, V., and Jeffery, C. J. (2015). Physical features of intracellular proteins that moonlight on the cell surface. PLoS One 10:e0130575. doi: 10.1371/journal.pone.0130575
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29.
Aviram, M., Hardak, E., Vaya, J., Mahmood, S., Milo, S., Hoffman, A., et al. (2000). Human serum paraoxonases (PON1) Q and R selectively decrease lipid peroxides in human coronary and carotid atherosclerotic lesions: PON1 esterase and peroxidase-like activities. Circulation 101, 2510–2517. doi: 10.1161/01.cir.101.21.2510
Ayoubi, T. A., and Van De Ven, W. J. (1996). Regulation of gene expression by alternative promoters. FASEB J. 10, 453–460. doi: 10.1096/fasebj.10.4.8647344
Bai, M., Zhou, J.-M., and Perrett, S. (2004). The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J. Biol. Chem. 279, 50025–50030. doi: 10.1074/jbc.m406612200
Beadle, G. W., and Tatum, E. L. (1941). Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. U.S.A. 27, 499–506. doi: 10.1073/pnas.27.11.499
Becker, E., Robisson, B., Chapple, C. E., Guenoche, A., and Brun, C. (2012). Multifunctional proteins revealed by overlapping clustering in protein interaction network. Bioinformatics 28, 84–90. doi: 10.1093/bioinformatics/btr621
Bellay, J., Atluri, G., Sing, T. L., Toufighi, K., Costanzo, M., Ribeiro, P. S. M., et al. (2011). Putting genetic interactions in context through a global modular decomposition. Genome Res. 21, 1375–1387. doi: 10.1101/gr.117176.110
Board, P. G., and Menon, D. (2016). Structure, function and disease relevance of Omega-class glutathione transferases. Arch. Toxicol. 90, 1049–1067. doi: 10.1007/s00204-016-1691-1
Chang, C. S., Clarke, A., and Pillus, L. (2012). Suppression analysis of esa1 mutants in Saccharomyces cerevisiae links NAB3 to transcriptional silencing and nucleolar functions. G3 2, 1223–1232. doi: 10.1534/g3.112.003558
Chapple, C. E., Robisson, B., Spinelli, L., Guien, C., Becker, E., and Brun, C. (2015). Extreme multifunctional proteins identified from a human protein interaction network. Nat. Commun. 6:7412. doi: 10.1038/ncomms8412
Chen, C., Zabad, S., Liu, H., Wang, W., and Jeffery, C. (2018). MoonProt 2.0: an expansion and update of the moonlighting proteins database. Nucleic Acids Res. 46, D640–D644. doi: 10.1093/nar/gkx1043
Chen, X. J., Wang, X., Kaufman, B. A., and Butow, R. A. (2005). Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307, 714–717. doi: 10.1126/science.1106391
Copley, S. D. (2012). Moonlighting is mainstream: paradigm adjustment required. Bioessays 34, 578–588. doi: 10.1002/bies.201100191
Copley, S. D. (2015). An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 40, 72–78. doi: 10.1016/j.tibs.2014.12.004
Copley, S. D. (2017). Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 47, 167–175. doi: 10.1016/j.sbi.2017.11.001
Costa, L. G., Cole, T. B., Jarvik, G. P., and Furlong, C. E. (2003). Functional genomic of the paraoxonase (PON1) polymorphisms: effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annu. Rev. Med. 54, 371–392. doi: 10.1146/annurev.med.54.101601.152421
Davies, H. G., Richter, R. J., Keifer, M., Broomfield, C. A., Sowalla, J., and Furlong, C. E. (1996). The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat. Genet. 14, 334–336. doi: 10.1038/ng1196-334
Delaye, L., DeLuna, A., Lazcano, A., and Becerra, A. (2008). The origin of a novel gene through overprinting in Escherichia coli. BMC Evol. Biol. 8:31. doi: 10.1186/1471-2148-8-31
Dudley, A. M., Janse, D. M., Tanay, A., Shamir, R., and Church, G. M. (2005). A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 1:2005.0001.
Ekman, D., Bjorklund, A. K., Frey-Skott, J., and Elofsson, A. (2005). Multi-domain proteins in the three kingdoms of life: orphan domains and other unassigned regions. J. Mol. Biol. 348, 231–243. doi: 10.1016/j.jmb.2005.02.007
Espinosa-Cantú, A., Ascencio, D., Barona-Gómez, F., and De Luna, A. (2015). Gene duplication and the evolution of moonlighting proteins. Front. Genet. 6:227. doi: 10.3389/fgene.2015.00227
Espinosa-Cantu, A., Ascencio, D., Herrera-Basurto, S., Xu, J., Roguev, A., Krogan, N. J., et al. (2018). Protein moonlighting revealed by noncatalytic phenotypes of yeast enzymes. Genetics 208, 419–431. doi: 10.1534/genetics.117.300377
Fellner, L., Simon, S., Scherling, C., Witting, M., Schober, S., Polte, C., et al. (2015). Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evol. Biol. 15:283. doi: 10.1186/s12862-015-0558-z
Ferla, M. P., Brewster, J. L., Hall, K. R., Evans, G. B., and Patrick, W. M. (2017). Primordial-like enzymes from bacteria with reduced genomes. Mol. Microbiol. 105, 508–524. doi: 10.1111/mmi.13737
Flores, C. L., and Gancedo, C. (2011). Unraveling moonlighting functions with yeasts. IUBMB Life 63, 457–462. doi: 10.1002/iub.454
Franco-Serrano, L., Cedano, J., Perez-Pons, J. A., Mozo-Villarias, A., Piñol, J., Amela, I., et al. (2018a). A hypothesis explaining why so many pathogen virulence proteins are moonlighting proteins. Pathog. Dis. 76:fty046. doi: 10.1093/femspd/fty046
Franco-Serrano, L., Hernández, S., Calvo, A., Severi, M. A., Ferragut, G., Pérez-Pons, J., et al. (2018b). MultitaskProtDB-II: an update of a database of multitasking/moonlighting proteins. Nucleic Acids Res. 46, D645–D648. doi: 10.1093/nar/gkx1066
Franco-Serrano, L., Huerta, M., Hernández, S., Cedano, J., Perez-Pons, J., Piñol, J., et al. (2018c). Multifunctional proteins: involvement in human diseases and targets of current drugs. Protein J. 37, 444–453. doi: 10.1007/s10930-018-9790-x
Gancedo, C., and Flores, C. L. (2008). Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72, 197–210. doi: 10.1128/mmbr.00036-07
Gancedo, C., Flores, C.-L., and Gancedo, J. M. (2016). The expanding landscape of moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 80, 765–777. doi: 10.1128/MMBR.00012-16
Gillis, J., and Pavlidis, P. (2011). The impact of multifunctional genes on guilt “by association “analysis. PLoS One 6:e17258. doi: 10.1371/journal.pone.0017258
Gomez, A., Domedel, N., Cedano, J., Piñol, J., and Querol, E. (2003). Do current sequence analysis algorithms disclose multifunctional (moonlighting) proteins? Bioinformatics 19, 895–896. doi: 10.1093/bioinformatics/btg111
Gomez, A., Hernandez, S., Amela, I., Pinol, J., Cedano, J., and Querol, E. (2011). Do protein-protein interaction databases identify moonlighting proteins? Mol. Biosyst. 7, 2379–2382. doi: 10.1039/c1mb05180f
Gurney, M. E., Heinrich, S. P., Lee, M. R., and Yin, H. S. (1986). Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science 234, 566–574. doi: 10.1126/science.3764429
Hall, D. A., Zhu, H., Zhu, X., Royce, T., Gerstein, M., and Snyder, M. (2004). Regulation of gene expression by a metabolic enzyme. Science 306, 482–484. doi: 10.1126/science.1096773
Hara, M. R., Agrawal, N., Kim, S. F., Cascio, M. B., Fujimuro, M., Ozeki, Y., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674. doi: 10.1038/ncb1268
Hara, M. R., Thomas, B., Cascio, M. B., Bae, I., Hester, L. D., Dawson, V. L., et al. (2006). Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc. Natl. Acad. Sci. U.S.A. 103, 3887–3889. doi: 10.1073/pnas.0511321103
He, X., and Zhang, J. (2006). Toward a molecular understanding of pleiotropy. Genetics 173, 1885–1891. doi: 10.1534/genetics.106.060269
Henderson, B., Fares, M. A., and Martin, A. C. (2016). Protein Moonlighting in Biology and Medicine. Hoboken, NJ: John Wiley & Sons.
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/IAI.00179-11
Hernandez, S., Amela, I., Cedano, J., Piñol, J., Perez-Pons, J., Mozo-Villarias, A., et al. (2011). Do moonlighting proteins belong to the intrinsically disordered protein class. Proteom. Bioinform. 5:262.
Hernández, S., Ferragut, G., Amela, I., Perez-Pons, J., Piñol, J., Mozo-Villarias, A., et al. (2014). MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res. 42, D517–D520. doi: 10.1093/nar/gkt1153
Huberts, D. H. E. W., and van der Klei, I. J. (2010). Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta Mol. Cell Res. 1803, 520–525. doi: 10.1016/j.bbamcr.2010.01.022
Hult, K., and Berglund, P. (2007). Enzyme promiscuity: mechanism and applications. Trends Biotechnol. 25, 231–238. doi: 10.1016/j.tibtech.2007.03.002
Humphris, E. L., and Kortemme, T. (2007). Design of multi-specificity in protein interfaces. PLoS Comput. Biol. 3:e164. doi: 10.1371/journal.pcbi.0030164
Irving, H. R., Cahill, D. M., and Gehring, C. (2018). Moonlighting proteins and their role in the control of signaling microenvironments, as exemplified by cGMP and phytosulfokine receptor 1 (PSKR1). Front. Plant Sci. 9:415. doi: 10.3389/fpls.2018.00415
Jain, A., Gali, H., and Kihara, D. (2018). Identification of moonlighting proteins in genomes using text mining techniques. Proteomics 18:1800083. doi: 10.1002/pmic.201800083
Jardine, O., Gough, J., Chothia, C., and Teichmann, S. A. (2002). Comparison of the small molecule metabolic enzymes of Escherichia coli and Saccharomyces cerevisiae. Genome Res. 12, 916–929. doi: 10.1101/gr.228002
Jeffery, C. J. (2009). Moonlighting proteins–an update. Mol. Biosyst. 5, 345–350. doi: 10.1039/b900658n
Jeffery, C. J. (2011). Proteins with neomorphic moonlighting functions in disease. IUBMB Life 63, 489–494. doi: 10.1002/iub.504
Jeffery, C. J. (2015). Why study moonlighting proteins? Front. Genet. 6:211. doi: 10.3389/fgene.2015.00211
Jeffery, C. J. (2016). Protein species and moonlighting proteins: very small changes in a protein’s covalent structure can change its biochemical function. J. Proteomics 134, 19–24. doi: 10.1016/j.jprot.2015.10.003
Jeffery, C. J. (2018a). Intracellular proteins moonlighting as bacterial adhesion factors. AIMS Microbiol. 4, 362–376. doi: 10.3934/microbiol.2018.2.362
Jeffery, C. J. (2018b). Protein moonlighting: what is it, and why is it important? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 373:20160523. doi: 10.1098/rstb.2016.0523
Jungblut, P. R., Thiede, B., and Schluter, H. (2016). Towards deciphering proteomes via the proteoform, protein speciation, moonlighting and protein code concepts. J. Proteomics 134, 1–4. doi: 10.1016/j.jprot.2016.01.012
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083
Kelkar, Y. D., and Ochman, H. (2013). Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193, 303–307. doi: 10.1534/genetics.112.145656
Khan, I., Chen, Y., Dong, T., Hong, X., Takeuchi, R., Mori, H., et al. (2014). Genome-scale identification and characterization of moonlighting proteins. Biol. Direct 9:30. doi: 10.1186/s13062-014-0030-9
Khan, I. K., and Kihara, D. (2016). Genome-scale prediction of moonlighting proteins using diverse protein association information. Bioinformatics 32, 2281–2288. doi: 10.1093/bioinformatics/btw166
Khersonsky, O., Roodveldt, C., and Tawfik, D. S. (2006). Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508. doi: 10.1016/j.cbpa.2006.08.011
Khersonsky, O., and Tawfik, D. S. (2010). Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505. doi: 10.1146/annurev-biochem-030409-143718
Kim, Y., Cha, S. J., Choi, H. J., and Kim, K. (2017). Omega class glutathione S-transferase: antioxidant enzyme in pathogenesis of neurodegenerative diseases. Oxid. Med. Cell. Longev. 2017:5049532.
Kirschner, K., and Bisswanger, H. (1976). Multifunctional proteins. Annu. Rev. Biochem. 45, 143–166.
Kopeckova, M., Pavkova, I., and Stulik, J. (2020). Diverse localization and protein binding abilities of glyceraldehyde-3-phosphate dehydrogenase in pathogenic bacteria: the key to its multifunctionality? Front. Cell. Infect. Microbiol. 10:89. doi: 10.3389/fcimb.2020.00089
Kugler, W., Breme, K., Laspe, P., Muirhead, H., Davies, C., Winkler, H., et al. (1998). Molecular basis of neurological dysfunction coupled with haemolytic anaemia in human glucose-6-phosphate isomerase (GPI) deficiency. Hum. Genet. 103, 450–454. doi: 10.1007/s004390050849
Kugler, W., and Lakomek, M. (2000). Glucose-6-phosphate isomerase deficiency. Baillieres Best Pract. Res. Clin. Haematol. 13, 89–101. doi: 10.1053/beha.1999.0059
Lazaro, A., Gomez-Zambrano, A., Lopez-Gonzalez, L., Pineiro, M., and Jarillo, J. A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666. doi: 10.1093/jxb/erm332
Lodowski, D. T., Barnhill, J. F., Pyskadlo, R. M., Ghirlando, R., Sterne-Marr, R., and Tesmer, J. J. G. (2005). The role of G beta gamma and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry 44, 6958–6970. doi: 10.1021/bi050119q
Lodowski, D. T., Pitcher, J. A., Capel, W. D., Lefkowitz, R. J., and Tesmer, J. J. G. (2003). Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science 300, 1256–1262. doi: 10.1126/science.1082348
Mani, M., Chen, C., Amblee, V., Liu, H., Mathur, T., Zwicke, G., et al. (2015). MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 43, D277–D282. doi: 10.1093/nar/gku954
Martin, H., Shales, M., Fernandez-Pinar, P., Wei, P., Molina, M., Fiedler, D., et al. (2015). Differential genetic interactions of yeast stress response MAPK pathways. Mol. Syst. Biol. 11:800. doi: 10.15252/msb.20145606
Mayr, C. (2016). Evolution and biological roles of alternative 3’UTRs. Trends Cell Biol. 26, 227–237. doi: 10.1016/j.tcb.2015.10.012
McKay, S., and Ashley, B. (2008). Gene sharing and evolution: the diversity of protein functions. By Joram Piatigorsky. Q. Rev. Biol. 83:99.
Michel, G., Roszak, A. W., Sauvé, V., Maclean, J., Matte, A., Coggins, J. R., et al. (2003). Structures of shikimate dehydrogenase AroE and its paralog YdiB. A common structural framework for different activities. J. Biol. Chem. 278, 19463–19472. doi: 10.1074/jbc.m300794200
Miyata, T., Oda, O., Inagi, R., Iida, Y., Araki, N., Yamada, N., et al. (1993). beta 2-Microglobulin modified with advanced glycation end products is a major component of hemodialysis-associated amyloidosis. J. Clin. Investig. 92, 1243–1252. doi: 10.1172/jci116696
Modun, B., and Williams, P. (1999). The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67, 1086–1092. doi: 10.1128/iai.67.3.1086-1092.1999
Nam, H., Lewis, N. E., Lerman, J. A., Lee, D.-H., Chang, R. L., Kim, D., et al. (2012). Network context and selection in the evolution to enzyme specificity. Science 337, 1101–1104. doi: 10.1126/science.1216861
Nobeli, I., Favia, A. D., and Thornton, J. M. (2009). Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 27, 157–167. doi: 10.1038/nbt1519
Paaby, A. B., and Rockman, M. V. (2013). The many faces of pleiotropy. Trends Genet. 29, 66–73. doi: 10.1016/j.tig.2012.10.010
Parkinson Study Group (1993). Effects of tocopherol and deprenyl on the progression of disability in early Parkinson’s disease. N. Engl. J. Med. 328, 176–183. doi: 10.1056/nejm199301213280305
Patil, A., Kinoshita, K., and Nakamura, H. (2010). Hub promiscuity in protein-protein interaction networks. Int. J. Mol. Sci. 11, 1930–1943. doi: 10.3390/ijms11041930
Piatigorsky, J. (2007). Gene Sharing and Evolution: The Diversity of Protein Functions. Cambridge, MA: Harvard University Press.
Piatigorsky, J., and Horwitz, J. (1996). Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens. Biochim. Biophys. Acta 1295, 158–164. doi: 10.1016/0167-4838(96)00030-1
Piatigorsky, J., O’Brien, W. E., Norman, B. L., Kalumuck, K., Wistow, G. J., Borras, T., et al. (1988). Gene sharing by delta-crystallin and argininosuccinate lyase. Proc. Natl. Acad. Sci. U.S.A. 85, 3479–3483. doi: 10.1073/pnas.85.10.3479
Piazza, I., Kochanowski, K., Cappelletti, V., Fuhrer, T., Noor, E., Sauer, U., et al. (2018). A map of protein-metabolite interactions reveals principles of chemical communication. Cell 172, 358–372.e23. doi: 10.1016/j.cell.2017.12.006
Pritykin, Y., Ghersi, D., and Singh, M. (2015). Genome-wide detection and analysis of multifunctional genes. PLoS Comput. Biol. 11:e1004467. doi: 10.1371/journal.pcbi.1004467
Salathe, M., Ackermann, M., and Bonhoeffer, S. (2006). The effect of multifunctionality on the rate of evolution in yeast. Mol. Biol. Evol. 23, 721–722. doi: 10.1093/molbev/msj086
Sarkissian, C. N., and Gamez, A. (2005). Phenylalanine ammonia lyase, enzyme substitution therapy for phenylketonuria, where are we now? Mol. Genet. Metab. 86(Suppl. 1), S22–S26.
Scherrer, T., Mittal, N., Janga, S. C., and Gerber, A. P. (2010). A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS One 5:e15499. doi: 10.1371/journal.pone.0015499
Schreiber, G., and Keating, A. E. (2011). Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 21, 50–61. doi: 10.1016/j.sbi.2010.10.002
Scott, E. M., and Pillus, L. (2010). Homocitrate synthase connects amino acid metabolism to chromatin functions through Esa1 and DNA damage. Genes Dev. 24, 1903–1913. doi: 10.1101/gad.1935910
Sengupta, S., Ghosh, S., and Nagaraja, V. (2008). Moonlighting function of glutamate racemase from Mycobacterium tuberculosis: racemization and DNA gyrase inhibition are two independent activities of the enzyme. Microbiology 154, 2796–2803. doi: 10.1099/mic.0.2008/020933-0
Sevin, D. C., Fuhrer, T., Zamboni, N., and Sauer, U. (2017). Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat. Methods 14, 187–194. doi: 10.1038/nmeth.4103
Sirover, M. A. (2018). Pleiotropic effects of moonlighting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in cancer progression, invasiveness, and metastases. Cancer Metastasis Rev. 37, 665–676. doi: 10.1007/s10555-018-9764-7
Sriram, G., Martinez, J. A., McCabe, E. R. B., Liao, J. C., and Dipple, K. M. (2005). Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 76, 911–924. doi: 10.1086/430799
Stearns, F. W. (2010). One hundred years of pleiotropy: a retrospective. Genetics 186, 767–773. doi: 10.1534/genetics.110.122549
Su, X. B., and Pillus, L. (2016). Functions for diverse metabolic activities in heterochromatin. Proc. Natl. Acad. Sci. U.S.A. 113, E1526–E1535. doi: 10.1073/pnas.1518707113
Tarze, A., Deniaud, A., Le Bras, M., Maillier, E., Molle, D., Larochette, N., et al. (2007). GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 26, 2606–2620. doi: 10.1038/sj.onc.1210074
Tompa, P., Szasz, C., and Buday, L. (2005). Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 30, 484–489. doi: 10.1016/j.tibs.2005.07.008
The UniProt Consortium (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46:2699. doi: 10.1093/nar/gky092
Ungerer, M., Parruti, G., Bohm, M., Puzicha, M., DeBlasi, A., Erdmann, E., et al. (1994). Expression of beta-arrestins and beta-adrenergic receptor kinases in the failing human heart. Circ. Res. 74, 206–213. doi: 10.1161/01.res.74.2.206
van de Peppel, J., and Holstege, F. C. P. (2005). Multifunctional genes. Mol. Syst. Biol. 1:2005.0003.
Wang, W., and Jeffery, C. J. (2016). An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol. Biosyst. 12, 1420–1431. doi: 10.1039/c5mb00550g
Zanzoni, A., Chapple, C. E., and Brun, C. (2015). Relationships between predicted moonlighting proteins, human diseases, and comorbidities from a network perspective. Front. Physiol. 6:171. doi: 10.3389/fphys.2015.00171
Zanzoni, A., Ribeiro, D. M., and Brun, C. (2019). Understanding protein multifunctionality: from short linear motifs to cellular functions. Cell. Mol. Life Sci. 76, 4407–4412. doi: 10.1007/s00018-019-03273-4
Zelenaya-Troitskaya, O., Perlman, P. S., and Butow, R. A. (1995). An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 14, 3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x
Zhang, J., and Wagner, G. P. (2013). On the definition and measurement of pleiotropy. Trends Genet. 29, 383–384. doi: 10.1016/j.tig.2013.05.002
Keywords: multidomain proteins, protein promiscuity, moonlighting proteins, gene ontology, pleiotropy, mechanisms of disease
Citation: Espinosa-Cantú A, Cruz-Bonilla E, Noda-Garcia L and DeLuna A (2020) Multiple Forms of Multifunctional Proteins in Health and Disease. Front. Cell Dev. Biol. 8:451. doi: 10.3389/fcell.2020.00451
Received: 03 December 2019; Accepted: 14 May 2020;
Published: 10 June 2020.
Edited by:
Malgorzata Wygrecka, University of Giessen, GermanyReviewed by:
Enrique Querol, Autonomous University of Barcelona, SpainTiina Annikki Jokela, City of Hope National Medical Center, United States
Copyright © 2020 Espinosa-Cantú, Cruz-Bonilla, Noda-Garcia and DeLuna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander DeLuna, YWxleGFuZGVyLmRlbHVuYUBjaW52ZXN0YXYubXg=
†Present address: Adriana Espinosa-Cantú, Institute for Biological Physics, University of Cologne, Cologne, Germany
 Adriana Espinosa-Cantú
Adriana Espinosa-Cantú Erika Cruz-Bonilla
Erika Cruz-Bonilla Lianet Noda-Garcia
Lianet Noda-Garcia Alexander DeLuna
Alexander DeLuna