
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 17 March 2020
Sec. Molecular and Cellular Pathology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00162
This article is part of the Research TopicPerspectives in Mammary Gland Development and Breast Cancer ResearchView all 13 articles
The immune checkpoint blockade therapy has drastically advanced treatment of different types of cancer over the past few years. Female breast cancer is the second leading cause of death in the overall burden of cancers worldwide that is encouraging healthcare professionals to improve cancer care management. The checkpoint blockade therapies combined with novel agents become the recent focus of various clinical trials in breast cancer. However, identification of the patients who are responsive to these therapeutic strategies remained as a major issue for enhancing the efficacy of these treatments. This highlights the unmet need in discovery and development of novel biomarkers to add predictive values for prosperous personalized medicine. In this review we summarize the advances done in the era of biomarker studies and highlight their link in supporting breast cancer immunotherapy.
Recent advances in cancer therapy present immunotherapy as a prospect change in treating various cancers. The immune-checkpoint blockade (ICB), designated as a cutting edge therapy, is used in increasing number of advance cancer diseases with durable responses compared to most chemotherapy and targeted therapies (Ansell et al., 2015; Gettinger et al., 2015; Larkin et al., 2015; Spencer et al., 2016; Tray et al., 2018). Breast cancer is the most common malignancies among women worldwide and many breast cancers have been recently determined immunogenic and enriched in tumor-infiltrating lymphocytes (TILs) (Cimino-Mathews et al., 2016).
The ICB monotherapy, anti-programmed death-1/programmed death-ligand-1 (anti-PD1/PD-L1), has demonstrated promising outcomes in metastatic triple negative breast cancer (Nanda et al., 2016; Schmid et al., 2018; Adams et al., 2019a, b; Emens et al., 2019). There is a considerable attention for developing immunotherapy-based strategies to escalate anti-cancer responses and to reduce the side effects, such as trials on combination therapy of anti-PD-1/PD-L1 with chemotherapy agents or combination with targeted therapies in metastatic patients (Nolan et al., 2017; Domchek et al., 2018; Nicolas et al., 2018). Moreover, developing strategies on combining different ICBs are appealing in breast cancer treatment, such as combining anti-PD-1/PD-L1 with anti-cytotoxic T-lymphocyte associated protein-4 (anti-CTLA-4) (Wolchok et al., 2017) or other co-inhibitory molecules (Chester et al., 2018; Harding et al., 2019). One of the major challenges in this regard is to establish predictive biomarkers for the stratification of breast cancer patients benefiting from these ICBs therapeutic strategies.
In March 2019, the U.S. Food and Drug Administration (FDA) approved the first ICB drug, a PD-L1 antibody called Atezolizumab (Tecentriq) in combination with a chemotherapy agent, for the treatment of triple-negative metastatic breast cancer patients (TNBC) (NCT02425891) (Schmid et al., 2018; Emens et al., 2019). The TNBC is a subtype of the disease with frequency of 15% and lacks hormone receptors, estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor (HER2) (Anders et al., 2016). The Atezolizumab authorized to be applied only on metastatic TNBC patients whose tumors express the PD-L1 protein that is characterized by an FDA-approved test, VENTANA PD-L1 (SP142), as a predictive biomarker. This was a significant fundamental step in predicting clinical benefit of only one ICBs combination strategies in breast cancer treatments. However, different ICBs agents and strategies are going to bring new treatment modalities for this disease. Therefore, there is an unmet need in developing novel predictive biomarkers for proper selection of patients who are benefiting from ICBs treatments and for avoiding unnecessary toxicity in unresponsive patients. Furthermore, identifying predictive biomarkers are necessary for better management of the expensive health care costs, especially for those patients that are unlikely to be responsive to the ICBs therapies. Here we summarize the attempts that have been done on the discovery of major predictive biomarkers in liquid biopsies, tumor tissues and tumor microenvironment that might contribute into advancing prediction of therapeutic decisions as well as the future challenges in this era.
Over the past few years, considerable effort has been done in discovery and development of liquid biopsy-based biomarkers, as it is minimally invasive, cost effective and can be replicated during patients’ follow-up (Wan et al., 2017; Pantel and Alix-Panabieres, 2019; Rothwell et al., 2019). These biomarkers can be detected in blood, cerebrospinal fluid and urine of cancer patients but not in healthy individuals. Today, liquid biopsy-based biomarkers are defined as soluble proteins, exosomes or other vesicles transmitting proteins or nucleic acids driven from a tumor, circulating tumor cells (CTCs) and circulating tumor DNAs (ctDNAs) (Kohler et al., 2011; Siravegna et al., 2017). All these properties make liquid biopsy-based biomarkers attractive in immunotherapy for the assessment of predictive biomarkers at the baseline or in monitoring therapy response.
The plasma proteins such as soluble PD-L1, cytokines and exosomes’ bound proteins are considered as important source of information in biomarker discovery and development. A high level of soluble PD-L1 has been demonstrated a poor prognosis in ICB response (Okuma et al., 2018). Nonetheless, the plasma level of soluble PD-L1 can be increased due to the different physiological conditions and diseases e.g., pregnancy or autoimmune diseases (Yanaba et al., 2016; Jovanovic et al., 2018). Therefore, the utility of soluble PD-L1 as a cancer biomarker remained as a controversial issue that need more in depth studies to define proper cut-off to differentiate between cancer therapeutic response and other diseases or physiological conditions. The cytokine and chemokine signature in cancer indicated potential predictive value in ICB therapy (Arrieta et al., 2017). Development of a cytokine panel to evaluate ICB response for patient classification in breast cancer seems to be encouraging, for instance, it has been shown that IL-27 up-regulated PD-L1 and promoted breast cancer growth (Yan et al., 2019), yet more studies need to be done to develop a proper panel of cytokines with predictive values in breast cancer. Another plasma protein candidate biomarkers are exosomes, a detective level of RNA molecules and proteins including PDL-1 are packed in exosome and secreted from cancer cells into the blood and lymphatic systems. Exosomes’ transmitting PD-L1 can bind to PD-1 on T-lymphocytes and consequently inactivate immune system from attacking cancer cells (Yang et al., 2018; Poggio et al., 2019). Increased level of circulating exosomal PD-L1 is a predictive marker for patient clinical response, e.g., indicating poor prognosis in melanoma patients (Chen et al., 2018). Moreover, as it was mentioned, exosomes and other vesicles contain detective levels of different classes of RNA molecules including, protein-coding RNAs (mRNA) and non-coding RNAs (e.g., miRNA) (Umu et al., 2018). Recent studies demonstrated that miRNAs directly or indirectly regulate the expression of different immune checkpoints on T-cells and on Tumor cells or APCs (Zhang et al., 2019). Hence, miRNAs that specifically control one-target checkpoints are favored in biomarker development. Various miRNAs such as miR-34a, miR-17-5p, miR-15b, miR-193a-3p, miR-197miR-200c showed correlation with expression of PD-L1 in tumor tissues as well as in sera or plasmas, and purposed to have predictive values in ICBs therapy of different cancers (Chen et al., 2014; Cortez et al., 2016; Ahn et al., 2017; Audrito et al., 2017; Kao et al., 2017; Fan et al., 2019). In breast cancer, a panel of thirteen miRNAs has been identified that directly target and down-regulate B7-H3. Among these thirteen miRNAs, expression of miR-29 is associated with higher survival rate of breast cancer patients (Nygren et al., 2014). Therefore, circulating exosomal biomarkers are considerably perceived as a robust source of information – both proteins and nucleic acids e.g., miRNA – to be investigated in breast cancer (Meng et al., 2019).
Circulating tumor cells are considered as important source of liquid biopsy-based biomarkers because they are driven from different sites of a tumor and could provide with more information about overall tumor characteristic. In breast cancer CTC clusters are recognized as a valuable prognostic biomarker and are associated with increasing metastatic potential (Aceto et al., 2014). Importantly, CTCs can interact with neutrophils and form CTC-neutrophil clusters that are proliferative and highly efficient metastatic precursors in breast cancer (Szczerba et al., 2019). The first study on expression of PD-L1 on CTCs was reported in patients with metastatic breast carcinoma (Mazel et al., 2015), and later it was investigated in other types of cancer. Further study on CTC/PD-L1 indicated that the frequency of PD-L1 positive CTCs are significantly higher in metastatic breast cancer patients compared to non-metastatic patients (Schott et al., 2017). These findings suggested CTC/PD-L1 assay as a potential non-invasive marker for stratification of patients benefiting from anti-PD-L1 therapies in clinical trials (Mazel et al., 2015; Schott et al., 2017). The expression of another important immune checkpoint member, B7-H3, on CTCs of breast cancer patients has been also reported. The B7-H3 positive CTCs showed pronounced correlation with Ki-67 expression, a tumor proliferation marker, and were proposed to be a potential biomarker or target for immune checkpoint blockade therapies in breast cancer (Pizon et al., 2018). CTCs are valuable source of tumor information to develop new biomarkers, however, there are some challenges on the reproducibility of the results which are linked to the applying various techniques for isolation and enriching of these cells. The techniques should be standardized among health service laboratories with the aim to provide qualified analysis of both the epithelial CTCs and the cells undergoing epithelial to mesenchymal transition (EMT), and at the same time to minimize contaminations with other cells such as circulating macrophages with the same surface marker expression. This will enable investigators to establish the CTC biomarkers with predictive or prognostic value in immune therapy.
The ctDNAs have been for decades center of attentions as a crucial source of liquid biopsy-based biomarkers. Today advances in the ctDNAs extraction and enrichments combined with next-generation sequencing (NGS) have contributed to development of valuable biomarkers such as genome instability number (GIN) (Jensen et al., 2019), tumor mutational burden (TMB) and aberrant DNA methylation (Sina et al., 2018) for cancer immunotherapy response stratification of patients.
Different attempts on identification of ctDNA GIN have indicated some merit in therapeutic decision-making (Ahlborn et al., 2019) and in monitoring of breast cancer recurrence (Yang et al., 2019). Nevertheless, the genome-wide analysis of GIN using ctDNA demonstrated GIN dynamic changes upon ICB therapy that enabled monitoring treatment outcome in cancer patients (Jensen et al., 2019). On the contrary, analysis of GIN using ctDNA marked no predictive value in response to ICB therapy for breast cancer (Jensen et al., 2019). Nevertheless, this study was done on a small breast cancer cohort; therefore, to have a meaningful closure about the GIN clinical values, the larger clinical trials should be conducted in breast cancer patients. The TMB assessment in tumor tissues is considered as a promising biomarker, solo or in combination with PD-L1 immunohistochemistry (IHC), with predictive value in immunotherapy efficacy in various types of cancer (Danilova et al., 2016; Ready et al., 2019; Samstein et al., 2019). The utility of ctDNA for assessment of TMBs delivers support for limited or inaccessible tissue samples to improve therapeutic decisions for some cancers (Gandara et al., 2018); however, this is highly dependent on the selection of the gene-targeted panel to evaluate TMBs, and most probably needs to be customized for each cancer type. In addition, the efficiency and precision of the assay using ctDNA to depict TMB is dependent on high coverage of the assay (Gandara et al., 2018; Georgiadis et al., 2019; Pasini and Ulivi, 2019). For breast cancer a NGS panel of mutations associated with 76 target genes, MammaSeqTM, have been recently developed with applicable use of ctDNA. MammaSeqTM showed encouraging result in detection of somatic mutations and monitoring disease burden. However, the assay has limitation in capturing all known mutations associated with cancer and is not specific for rare events in ctDNA (Smith et al., 2019).
DNA methylation changes are key to the development and progression of certain cancers (Chatterjee et al., 2018). Aberrant DNA methylation signature demonstrated great potential to be used as a ctDNA biomarker in cancer (Barekati et al., 2010; Radpour et al., 2011; Sina et al., 2018). It has been shown that DNA hypermethylation of promoter or distal enhancer regions play role in low expression of PD-L1 (Y. Zhang et al., 2018) and demethylation of PDCD1 promoter activates PD-1 expression (Mishra and Verma, 2018). Treating cancer cells with the DNA methyltransferase (DNMT) inhibitors resulted in a better response of anti-CTLA-4 immunotherapy (Chiappinelli et al., 2015). Moreover, other studies indicated that the expression of PD-L1 on cancer cells is associated with global hypomethylation that could play a role in the regulation of PD-L1 expression. This information is emphasizing on potential indication of DNA methylation signatures as biomarkers, which might suggest additional treatments or combination therapies to modulate responsiveness to PD-1 inhibitor treatment (Emran et al., 2019). Furthermore, the genome-wide technology and their corresponding data analysis illustrated a signature that might enable guiding the prediction of ICB immunotherapy response in cancer (Duruisseaux et al., 2018; Guo et al., 2019). However, relatively little attention has been given to develop a customized panel of genomic regions with aberrant methylation patterns for breast cancer.
A limited number of tumor biomarkers were already assessed in clinical trials of the ICBs approved by the FDA (Table 1). In tumor cells both genomic and non-genomic factors are studied as potential biomarkers to predict the response or resistance to ICB therapies. Genomic factors include tumor immunogenicity, mutation/neoantigen-load (Snyder et al., 2014), increased TMB (Keenan et al., 2019), increased PD-L1 level (Havel et al., 2019), interferon gamma (FNγ) response (Ayers et al., 2017), human leukocyte antigen (HLA) diversity, deficient DNA mismatch repair (dMMR) (Zhao et al., 2019), high microsatellite instability (MSI-hi), copy-number alterations, checkpoint regulators e.g., CMTM4/6 (Mezzadra et al., 2017), up-regulation of checkpoint receptors and oncogenic signaling (Havel et al., 2019; Keenan et al., 2019). Non-genomic factors such as gut microbiome (Gopalakrishnan et al., 2018; Routy et al., 2018), metabolic pathway and the activity of lactate dehydrogenase (LDH) (Wein et al., 2018) have been shown to strongly modulate immune responses and success in ICB therapy. Some biomarkers from Table 1 and other relevant tumor tissue biomarkers for ICB therapies in breast cancer are explained below.
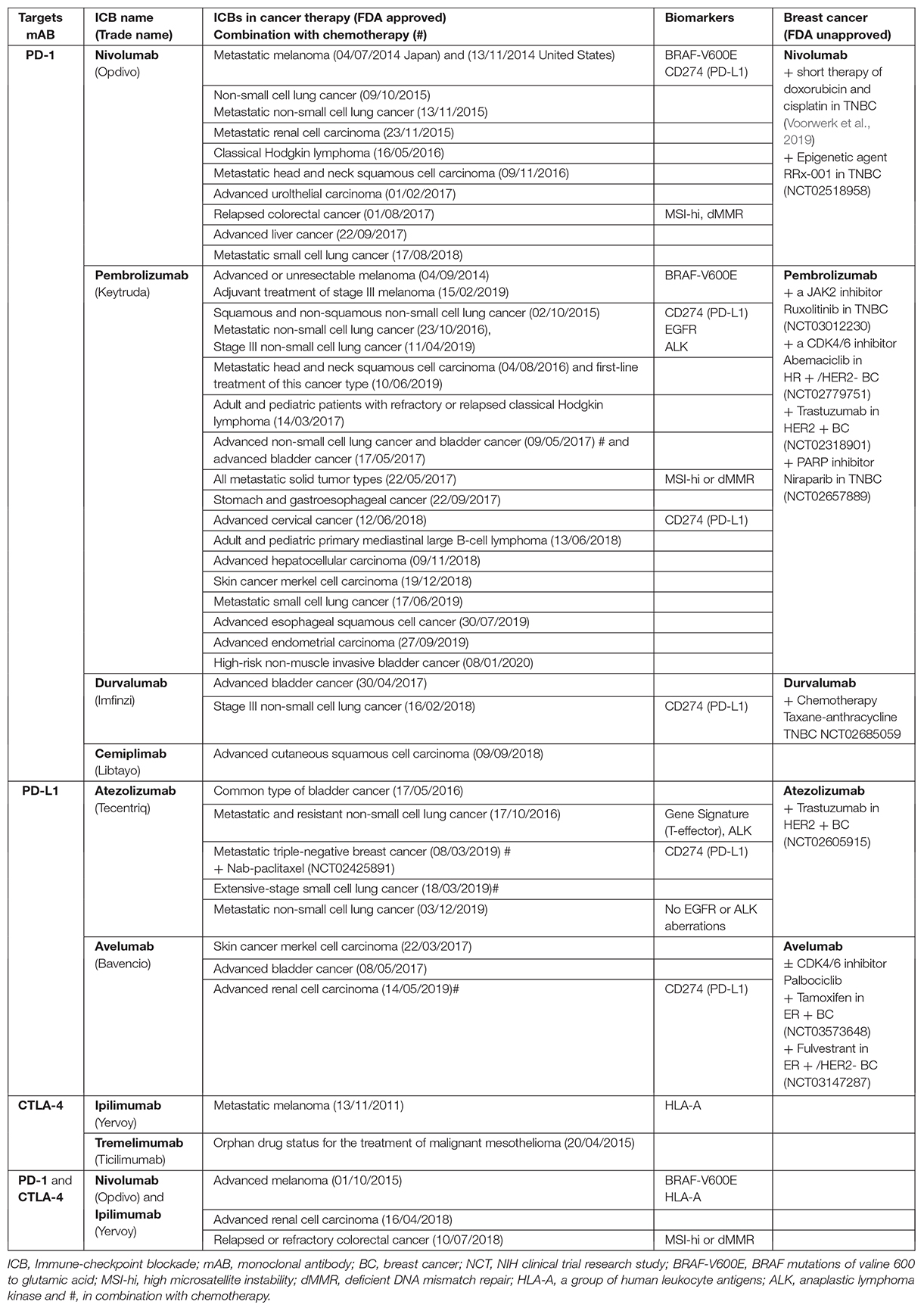
Table 1. A list of FDA-approved and validated ICBs targeting PD-1, PD-L1 or CTLA-4 axis with investigational biomarkers to predict an efficient patient response to the immunotherapy.
Mutational landscape of the tumor and the neoantigen load are associated with increased immunogenicity which are recognized by T cells. Several studies show that high TMB correlates with enhanced ICB response rates (Keenan et al., 2019). A whole-genome sequencing (WGS) analysis of 442 patients’ tumor tissue biopsies from metastatic breast cancer revealed two fold higher TMB compared to primary breast cancer (Angus et al., 2019). They could identify 11% of patient (threshold of ≥10 mutations per Mbp) with a high TMB as a potential biomarker to identify clinically relevant subgroups for immunotherapy. Interestingly, high TMB was only associated with metastatic tissue and it was equal between breast cancer subtypes and biopsy sites.
The DNA mismatch repair (MMR) system is crucial for genomic integrity and stability and it prevents microsatellite instability (MSI). Tumors with dMMR and MSI-hi are more sensitive to anti-PD-1/PD-L1 therapies (Zhao et al., 2019). This occurs due to mutation or loss of function of DNA repair proteins. Some data show that dMMR is more frequent in early stage cancers than in metastatic cancers, which is important for the selection of the best time point for ICB therapy. Although dMMR and MSI-hi are used as predictive biomarkers for Pembrolizumab therapy of all metastatic solid tumor types, both biomarkers are rarely present in most breast tumors (Mills et al., 2018), except BRCA-deficient TNBC. BRCA1- and BRCA2-deficient breast cancers are characterized by vast genomic instability and T cell-inflamed signature. BRCA1-deficient tumors indicated high expression of PD1 and PD-L1 (Wen and Leong, 2019) and similar to TNBC, seems to have best response to ICBs, especially in combination with cytotoxic agents.
PD-L1 expression in tumor cells and TILs using immunostaining-scoring methods have been associated with response to blockade of the PD-1/PD-L1 axis (Dolled-Filhart et al., 2016; Torlakovic et al., 2020). However, there is no clear-cut between separation of responders and non-responders patients. A portion of PD-L1– tumors still responds to ICB therapy and on the other hand not all PD-L1+ tumors are responsive to ICB therapy. PD-L1 overexpression is correlated with copy-number alteration of 9p24.1 locus containing PD-L1, PD-L2 and JAK2 (Green et al., 2010). In addition, PD-L1 protein levels and stability in tumors can be increased using inhibitors of cyclin-dependent kinases 4 and 6 (CDK4/6). It has been shown that PD-L1 protein abundance is regulated by CDK4 and cullin 3-SPOP E3 ligase via proteasome-mediated degradation (Zhang et al., 2018). To increase the ICBs efficiency in breast cancer, some clinical studies used a combination of JAK2 or CDK4/6 inhibitors with anti-PD1 therapy and evaluated the safety and efficacy of these combinational therapies in patients (NCT03012230, NCT02779751, Table 1).
Oncogenes such as mutated BRAF, EGFR and KRAS and amplified HER2 and loss of tumor suppressor genes e.g., PTEN often regulate inflammatory and immune suppressive cytokines like IL-6 and affect ICB response rates (Keenan et al., 2019). IFNγ is released by activated T cells upon recognition of tumor neoantigens and activates IFNγ-JAK-STAT-IRF1 axis in tumor cells. Alteration of this pathway affects the response to ICBs via different mechanisms such as increasing the expression levels of HLA and induction of PD-1 and PD-L1 gene expression by direct binding of IRF1 and STAT3 to their promoters (Garcia-Diaz et al., 2017; Keenan et al., 2019).
Besides TMB and the expression levels of IFNγ and PD-L1 as dynamic biomarkers for ICB therapy in breast cancer, multi-gene based assays to develop combinational ICB biomarkers are required. Similar assays e.g., Oncotype DX (Paik et al., 2004), MammaPrint (van’t Veer et al., 2002) and Prosigna (Parker et al., 2009) have been previously used for the prediction of chemo and targeted therapy benefit. The current assays such as Enzyme-Linked Immunosorbent Spot (ELISpot) for the detection of cytokine and IFNγ secreting cells show limited sensitivity in assessing tumor-specific T-cell responses. However, a new assay “Mutation-Associated Neoantigen Functional Expansion of Specific T cells” (MANAFEST) allows a sensitive measurement of antigen-specific TCR clonotypic amplifications following treatment in blood, tumor, and normal tissue of patients receiving immunotherapy (Danilova et al., 2018).
Extracellular matrix (ECM) changes are predicted as prognosis factors that are correlated with immunological activity. ECM dysregulation is often linked to the presence of cancer-associated fibroblasts (CAFs) expressing activated TGFβ signaling (Chakravarthy et al., 2018). The cytokine TGFβ is the major mediator of immune suppression in the tumor microenvironment and has a central role in inhibition of the both adaptive and innate immune responses during tumor progression (Batlle and Massague, 2019). Reprogramming of CAFs and anti-TGFβ therapies can enhance checkpoint blockade. A recent preclinical study has shown that the lack of response to Atezolizumab therapy was associated with TGFβ signature in fibroblasts and exclusion of CD8+ T-effector cells from tumor parenchyma in metastatic urothelial tumors (Mariathasan et al., 2018). Several studies revealed that combinational therapy of both TGFβ and PD-L1 resulted in synergistic anti-tumor effect in both breast and colorectal cancers (Knudson et al., 2018; Tauriello et al., 2018). A novel bifunctional anti-PD-L1/TGFβ Trap fusion protein (M7824) was tested in EMT6 and 4T1 syngeneic mouse breast cancer models. M7824 decreased TMB and promoted CD8+ T cell and NK cell activation (Knudson et al., 2018). In a neoadjuvant setting, M7824 is used in treating patients with stage II-III HER2+ Breast Cancer (NCT03620201). Besides T-cell exclusion based on TGFβ-activated stroma, Wnt-β-catenin signaling plays a role in T-cell activation and CD8+ T-effector cells migration by decreasing CD103+ dendritic cell (DC) recruitment (Spranger et al., 2015). In a melanoma model, this migration depends on the presence of DC producing CXCL10 (Spranger et al., 2017). Importantly, anti-PD-1 efficacy depends on CXCR3 activity, which is a receptor for CXCL9, CXCL10, and CXCL11 chemokines. These CXCR3 ligands are identified as positive predictive biomarkers and their induction in non-responsive mouse tumors could restore the sensitivity to anti-PD-1 (Chow et al., 2019).
Vascular endothelial growth factor A (VEGFA) and angiopoietin-2 (ANG-2) produced by endothelial cells affect antitumor immunity. In a metastatic mouse mammary tumor model (MMTV-PyMT), Schmittnaegel et al. (2017) could show that a dual inhibition of both angiogenic factors resulted in an increase of tumor antigen presentation, activation of tumor-infiltrating CD8 + T cells, and induction of endothelial PD-L1 expression through IFNγ. Simultaneous blocking of PD-L1/PD-1 signaling in this tumor model improved antitumoral activity and increased survival rate by 30% in mice (Schmittnaegel et al., 2017).
Infiltrating and tumor-associated immune cells are the major component of tumor-associated stroma with both protumor and antitumor activities. An increase in peripheral ICOS+CD4+ T cells has been also shown as a good clinical ICB responses in patients with hormone-responsive advanced breast cancer, which were treated with the anti-CTLA4 tremelimumab in combination with exemestane (Vonderheide et al., 2010).
The efficiency of ICBs is highly based on TILs and we need a better understanding of molecular determinants of TILs phenotype in tumor and tumor microenvironment. Pre-existing of immune response in tumors and localization and density of TILs are strong prognostic indicators for selection of ICB-responsive patients in different cancer types including breast cancer (Wein et al., 2018). In addition to immunohistochemistry methods to study TILs density, single cell RNA sequencing provides high-resolution to study the immune cell diversity and tumor heterogeneity related to ICB responses. Recently, Jiang et al. (2018) have developed a computational tool for dysfunctional T-cell signature, called “Tumor Immune Dysfunction and Exclusion” (TIDE). They could show that TIDE signatures predict ICB immunotherapy response in melanoma patients treated with first-line anti-PD1 or anti-CTLA4. The TIDE tool predicts only intrinsic ICB resistance and models two distinct mechanisms of tumor immune evasion: (i) T cell dysfunction in tumors with high infiltration of cytotoxic T lymphocytes (CTL), and (ii) T cell exclusion and prevention of T cell infiltration in tumors with low CTL level.
The crosstalk between cancer cells and immune cells at the primary tumor site, in the circulation and in the metastatic niche has a strong influence on cancer progression that affects patients’ response to ICBs (Saini et al., 2019). In a recent study, Wagner et al. (2019) performed large-scale mass cytometry profiling of 144 human breast tumor and 50 non-tumor tissue samples and characterized features of cancer ecosystems, inter-patient variations in tumor-associated immune cells and their associations with clinical data. They could show that PD-L1+ tumor-associated macrophages (TAMs) and exhausted T cells are abundant in high-grade ER– and ER+ tumors. This single-cell mass spectrometric approach called mass cytometry (CyTOFTM) can be combined with immunohistochemical methods, which were used for multiplexed imaging of tumor tissues and with subcellular resolution (Giesen et al., 2014).
Reprogramming of tumor immune microenvironment presents a powerful strategy to enhance the response to anti-PD1/PD-L1 in different type of breast cancer. Recently, Voorwerk et al. (2019) showed that a short-term treatment with doxorubicin and cisplatin was able to reprogram tumor microenvironment. This caused up-regulation of inflammatory JAK-STAT and TNF-α signaling and increased the sensitivity to the Nivolumab in metastatic TNBC (Voorwerk et al., 2019).
Scientist and healthcare professionals have gained and explained a vast knowledge about potential predictive biomarkers for ICB patient’s classification. The most promising biomarkers have been presented as proteomic and transcriptomic signatures of exosomes and CTCs, genomic analysis of ctDNA (genome instability number, specific tumor mutations, and aberrant DNA methylation signature). The next crucial step is the clinical verification of these candidate biomarkers that requires a consensus on methodological standardization of the assays and in parallel to investigate these biomarkers in large patient populations.
The interaction between tumor cells and immune cells in TME leads to the dynamic change of immunotherapy targets. This is a challenging factor for the identification of appropriate biomarkers for the selection of drug responsive patients. Biological understanding of multigene-based biomarkers and combinational strategies for ICB biomarkers will give healthcare providers the opportunity to increase the effectiveness of immune therapy in breast cancer. Advanced technologies such as single cell RNA sequencing and CyTOF-based immune profiling provide high-resolution of tumor immune microenvironment. Enhancing the drug response by remodeling of dynamic tumor ecosystem is fundamental for a successful personalized cancer therapy.
Both authors wrote the manuscript without external help. VV prepared the table.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aceto, N., Bardia, A., Miyamoto, D. T., Donaldson, M. C., Wittner, B. S., Spencer, J. A., et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. doi: 10.1016/j.cell.2014.07.013
Adams, S., Loi, S., Toppmeyer, D., Cescon, D. W., De Laurentiis, M., Nanda, R., et al. (2019a). Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann. Oncol. 30, 405–411. doi: 10.1093/annonc/mdy518
Adams, S., Schmid, P., Rugo, H. S., Winer, E. P., Loirat, D., Awada, A., et al. (2019b). Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann. Oncol. 30, 397–404. doi: 10.1093/annonc/mdy517
Ahlborn, L. B., Rohrberg, K. S., Gabrielaite, M., Tuxen, I. V., Yde, C. W., Spanggaard, I., et al. (2019). Application of cell-free DNA for genomic tumor profiling: a feasibility study. Oncotarget 10, 1388–1398. doi: 10.18632/oncotarget.26642
Ahn, H., Yang, J. M., Kim, H., Chung, J. H., Ahn, S. H., Jeong, W. J., et al. (2017). Clinicopathologic implications of the miR-197/PD-L1 axis in oral squamous cell carcinoma. Oncotarget 8, 66178–66194. doi: 10.18632/oncotarget.19842
Anders, C. K., Abramson, V., Tan, T., and Dent, R. (2016). The evolution of triple-negative breast cancer: from biology to novel therapeutics. Am. Soc. Clin. Oncol. Educ. Book 35, 34–42. doi: 10.14694/EDBK_159135
Angus, L., Smid, M., Wilting, S. M., van Riet, J., Van Hoeck, A., Nguyen, L., et al. (2019). The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat. Genet. 51, 1450–1458. doi: 10.1038/s41588-019-0507-7
Ansell, S. M., Lesokhin, A. M., Borrello, I., Halwani, A., Scott, E. C., Gutierrez, M., et al. (2015). PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 372, 311–319. doi: 10.1056/NEJMoa1411087
Arrieta, O., Montes-Servin, E., Hernandez-Martinez, J. M., Cardona, A. F., Casas-Ruiz, E., Crispin, J. C., et al. (2017). Expression of PD-1/PD-L1 and PD-L2 in peripheral T-cells from non-small cell lung cancer patients. Oncotarget 8, 101994–102005. doi: 10.18632/oncotarget.22025
Audrito, V., Serra, S., Stingi, A., Orso, F., Gaudino, F., Bologna, C., et al. (2017). PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget 8, 15894–15911. doi: 10.18632/oncotarget.15213
Ayers, M., Lunceford, J., Nebozhyn, M., Murphy, E., Loboda, A., Kaufman, D. R., et al. (2017). IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940. doi: 10.1172/JCI91190
Barekati, Z., Radpour, R., Kohler, C., Zhang, B., Toniolo, P., Lenner, P., et al. (2010). Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum. Mol. Genet. 19, 2936–2946. doi: 10.1093/hmg/ddq199
Batlle, E., and Massague, J. (2019). Transforming growth factor-beta signaling in immunity and cancer. Immunity 50, 924–940. doi: 10.1016/j.immuni.2019.03.024
Chakravarthy, A., Khan, L., Bensler, N. P., Bose, P., and De Carvalho, D. D. (2018). TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 9:4692. doi: 10.1038/s41467-018-06654-8
Chatterjee, A., Rodger, E. J., and Eccles, M. R. (2018). Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol. 51, 149–159. doi: 10.1016/j.semcancer.2017.08.004
Chen, G., Huang, A. C., Zhang, W., Zhang, G., Wu, M., Xu, W., et al. (2018). Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386. doi: 10.1038/s41586-018-0392-8
Chen, L., Gibbons, D. L., Goswami, S., Cortez, M. A., Ahn, Y. H., Byers, L. A., et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5:5241. doi: 10.1038/ncomms6241
Chester, C., Sanmamed, M. F., Wang, J., and Melero, I. (2018). Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood 131, 49–57. doi: 10.1182/blood-2017-06-741041
Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., et al. (2015). Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986. doi: 10.1016/j.cell.2015.07.011
Chow, M. T., Ozga, A. J., Servis, R. L., Frederick, D. T., Lo, J. A., Fisher, D. E., et al. (2019). Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498.e5–1512.e5. doi: 10.1016/j.immuni.2019.04.010
Cimino-Mathews, A., Thompson, E., Taube, J. M., Ye, X., Lu, Y., Meeker, A., et al. (2016). PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum. Pathol. 47, 52–63. doi: 10.1016/j.humpath.2015.09.003
Cortez, M. A., Ivan, C., Valdecanas, D., Wang, X., Peltier, H. J., Ye, Y., et al. (2016). PDL1 regulation by p53 via miR-34. J. Natl. Cancer Inst. 108:djv303. doi: 10.1093/jnci/djv303
Danilova, L., Anagnostou, V., Caushi, J. X., Sidhom, J. W., Guo, H., Chan, H. Y., et al. (2018). The mutation-associated neoantigen functional expansion of specific T cells (MANAFEST) assay: a sensitive platform for monitoring antitumor immunity. Cancer Immunol. Res. 6, 888–899. doi: 10.1158/2326-6066.CIR-18-0129
Danilova, L., Wang, H., Sunshine, J., Kaunitz, G. J., Cottrell, T. R., Xu, H., et al. (2016). Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. Proc. Natl. Acad. Sci. U.S.A. 113, E7769–E7777. doi: 10.1073/pnas.1607836113
Dolled-Filhart, M., Locke, D., Murphy, T., Lynch, F., Yearley, J. H., Frisman, D., et al. (2016). Development of a prototype immunohistochemistry assay to measure programmed death ligand-1 expression in tumor tissue. Arch. Pathol. Lab. Med. 140, 1259–1266. doi: 10.5858/arpa.2015-0544-OA
Domchek, S., Postel-Vinay, S., Bang, Y.-J., Park, Y., Alexandre, J., Delord, J.-P., et al. (2018). An open-label, multitumor, phase II basket study of olaparib and durvalumab (MEDIOLA): results in germline BRCA-mutated (gBRCAm) HER2-negative metastatic breast cancer (MBC). Cancer Res. 78(Suppl. 4), D6–D11. doi: 10.1158/1538-7445.SABCS17-PD6-11
Duruisseaux, M., Martinez-Cardus, A., Calleja-Cervantes, M. E., Moran, S., Castro, de Moura, M., et al. (2018). Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis. Lancet Respir. Med. 6, 771–781. doi: 10.1016/S2213-2600(18)30284-4
Emens, L. A., Cruz, C., Eder, J. P., Braiteh, F., Chung, C., Tolaney, S. M., et al. (2019). Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: a phase 1 study. JAMA Oncol. 5, 74–82. doi: 10.1001/jamaoncol.2018.4224
Emran, A. A., Chatterjee, A., Rodger, E. J., Tiffen, J. C., Gallagher, S. J., Eccles, M. R., et al. (2019). Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 40, 328–344. doi: 10.1016/j.it.2019.02.004
Fan, J., Yin, Z., Xu, J., Wu, F., Huang, Q., Yang, L., et al. (2019). Circulating microRNAs predict the response to anti-PD-1 therapy in non-small cell lung cancer. Genomics 112, 2063–2071. doi: 10.1016/j.ygeno.2019.11.019
Gandara, D. R., Paul, S. M., Kowanetz, M., Schleifman, E., Zou, W., Li, Y., et al. (2018). Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 24, 1441–1448. doi: 10.1038/s41591-018-0134-3
Garcia-Diaz, A., Shin, D. S., Moreno, B. H., Saco, J., Escuin-Ordinas, H., Rodriguez, G. A., et al. (2017). Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201. doi: 10.1016/j.celrep.2017.04.031
Georgiadis, A., Durham, J. N., Keefer, L. A., Bartlett, B. R., Zielonka, M., Murphy, D., et al. (2019). Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin. Cancer Res. 25, 7024–7034. doi: 10.1158/1078-0432.CCR-19-1372
Gettinger, S. N., Horn, L., Gandhi, L., Spigel, D. R., Antonia, S. J., Rizvi, N. A., et al. (2015). Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J. Clin. Oncol. 33, 2004–2012. doi: 10.1200/JCO.2014.58.3708
Giesen, C., Wang, H. A., Schapiro, D., Zivanovic, N., Jacobs, A., Hattendorf, B., et al. (2014). Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422. doi: 10.1038/nmeth.2869
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. doi: 10.1126/science.aan4236
Green, M. R., Monti, S., Rodig, S. J., Juszczynski, P., Currie, T., O’Donnell, E., et al. (2010). Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116, 3268–3277. doi: 10.1182/blood-2010-05-282780
Guo, W., Zhu, L., Zhu, R., Chen, Q., Wang, Q., and Chen, J. Q. (2019). A four-DNA methylation biomarker is a superior predictor of survival of patients with cutaneous melanoma. eLife 8:e44310. doi: 10.7554/eLife.44310
Harding, J. J., Patnaik, A., Moreno, V., Stein, M., Jankowska, A. M., Velez de Men dizabal, N., et al. (2019). A phase Ia/Ib study of an anti-TIM-3 antibody (LY3321367) monotherapy or in combination with an anti-PD-L1 antibody (LY3300054): Interim safety, efficacy, and pharmacokinetic findings in advanced cancers. JCO 37:12. doi: 10.1200/jco.2019.37.8_suppl.12
Havel, J. J., Chowell, D., and Chan, T. A. (2019). The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150. doi: 10.1038/s41568-019-0116-x
Jensen, T. J., Goodman, A. M., Kato, S., Ellison, C. K., Daniels, G. A., Kim, L., et al. (2019). Genome-wide sequencing of cell-free DNA identifies copy-number alterations that can be used for monitoring response to immunotherapy in cancer patients. Mol. Cancer Ther. 18, 448–458. doi: 10.1158/1535-7163.MCT-18-0535
Jiang, P., Gu, S., Pan, D., Fu, J., Sahu, A., Hu, X., et al. (2018). Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558. doi: 10.1038/s41591-018-0136-1
Jovanovic, D., Roksandic Milenkovic, M., Kotur Stevuljevic, J., Markovic, J., Ceriman, V., Kontic, M., et al. (2018). Membrane PD-L1 expression and soluble PD-L1 plasma levels in idiopathic pulmonary fibrosis-a pilot study. J. Thorac. Dis. 10, 6660–6669. doi: 10.21037/jtd.2018.11.16
Kao, S. C., Cheng, Y. Y., Williams, M., Kirschner, M. B., Madore, J., Lum, T., et al. (2017). Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J. Thorac. Oncol. 12, 1421–1433. doi: 10.1016/j.jtho.2017.05.024
Keenan, T. E., Burke, K. P., and Van Allen, E. M. (2019). Genomic correlates of response to immune checkpoint blockade. Nat. Med. 25, 389–402. doi: 10.1038/s41591-019-0382-x
Knudson, K. M., Hicks, K. C., Luo, X., Chen, J. Q., Schlom, J., and Gameiro, S. R. (2018). M7824, a novel bifunctional anti-PD-L1/TGFbeta Trap fusion protein, promotes anti-tumor efficacy as monotherapy and in combination with vaccine. Oncoimmunology 7:e1426519. doi: 10.1080/2162402X.2018.1426519
Kohler, C., Barekati, Z., Radpour, R., and Zhong, X. Y. (2011). Cell-free DNA in the circulation as a potential cancer biomarker. Anticancer Res. 31, 2623–2628.
Larkin, J., Lao, C. D., Urba, W. J., McDermott, D. F., Horak, C., Jiang, J., et al. (2015). Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAF wild-type advanced melanoma: a pooled analysis of 4 clinical trials. JAMA Oncol. 1, 433–440. doi: 10.1001/jamaoncol.2015.1184
Mariathasan, S., Turley, S. J., Nickles, D., Castiglioni, A., Yuen, K., Wang, Y., et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. doi: 10.1038/nature25501
Mazel, M., Jacot, W., Pantel, K., Bartkowiak, K., Topart, D., Cayrefourcq, L., et al. (2015). Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 9, 1773–1782. doi: 10.1016/j.molonc.2015.05.009
Meng, Y., Sun, J., Wang, X., Hu, T., Ma, Y., Kong, C., et al. (2019). Exosomes: a promising avenue for the diagnosis of breast cancer. Technol. Cancer Res. Treat. 18:1533033818821421. doi: 10.1177/1533033818821421
Mezzadra, R., Sun, C., Jae, L. T., Gomez-Eerland, R., de Vries, E., Wu, W., et al. (2017). Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110. doi: 10.1038/nature23669
Mills, A. M., Dill, E. A., Moskaluk, C. A., Dziegielewski, J., Bullock, T. N., and Dillon, P. M. (2018). The relationship between mismatch repair deficiency and PD-L1 expression in breast carcinoma. Am. J. Surg. Pathol. 42, 183–191. doi: 10.1097/PAS.0000000000000949
Mishra, A., and Verma, M. (2018). Epigenetic and genetic regulation of PDCD1 gene in cancer immunology. Methods Mol. Biol. 1856, 247–254. doi: 10.1007/978-1-4939-8751-1_14
Nanda, R., Chow, L. Q., Dees, E. C., Berger, R., Gupta, S., Geva, R., et al. (2016). Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J. Clin. Oncol. 34, 2460–2467. doi: 10.1200/JCO.2015.64.8931
Nicolas, E., Bertucci, F., Sabatier, R., and Goncalves, A. (2018). Targeting BRCA deficiency in breast cancer: what are the clinical evidences and the next perspectives? Cancers (Basel) 10:e506. doi: 10.3390/cancers10120506
Nolan, E., Savas, P., Policheni, A. N., Darcy, P. K., Vaillant, F., Mintoff, C. P., et al. (2017). Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci. Transl. Med. 9:aal4922. doi: 10.1126/scitranslmed.aal4922
Nygren, M. K., Tekle, C., Ingebrigtsen, V. A., Makela, R., Krohn, M., Aure, M. R., et al. (2014). Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br. J. Cancer 110, 2072–2080. doi: 10.1038/bjc.2014.113
Okuma, Y., Wakui, H., Utsumi, H., Sagawa, Y., Hosomi, Y., Kuwano, K., et al. (2018). Soluble programmed cell death ligand 1 as a novel biomarker for nivolumab therapy for non-small-cell lung cancer. Clin. Lung Cancer. 19, 410.e1–417.e1. doi: 10.1016/j.cllc.2018.04.014
Paik, S., Shak, S., Tang, G., Kim, C., Baker, J., Cronin, M., et al. (2004). A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826. doi: 10.1056/NEJMoa041588
Pantel, K., and Alix-Panabieres, C. (2019). Liquid biopsy and minimal residual disease – latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424. doi: 10.1038/s41571-019-0187-3
Parker, J. S., Mullins, M., Cheang, M. C., Leung, S., Voduc, D., Vickery, T., et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167. doi: 10.1200/JCO.2008.18.1370
Pasini, L., and Ulivi, P. (2019). Liquid biopsy for the detection of resistance mechanisms in NSCLC: comparison of different blood biomarkers. J. Clin. Med. 8:998. doi: 10.3390/jcm8070998
Pizon, M., Schott, D. S., Pachmann, U., and Pachmann, K. (2018). B7-H3 on circulating epithelial tumor cells correlates with the proliferation marker, Ki-67, and may be associated with the aggressiveness of tumors in breast cancer patients. Int. J. Oncol. 53, 2289–2299. doi: 10.3892/ijo.2018.4551
Poggio, M., Hu, T., Pai, C. C., Chu, B., Belair, C. D., Chang, A., et al. (2019). Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177, 414.e13–427.e13. doi: 10.1016/j.cell.2019.02.016
Radpour, R., Barekati, Z., Kohler, C., Lv, Q., Burki, N., Diesch, C., et al. (2011). Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One 6:e16080. doi: 10.1371/journal.pone.0016080
Ready, N., Hellmann, M. D., Awad, M. M., Otterson, G. A., Gutierrez, M., Gainor, J. F., et al. (2019). First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J. Clin. Oncol. 37, 992–1000. doi: 10.1200/JCO.18.01042
Rothwell, D. G., Ayub, M., Cook, N., Thistlethwaite, F., Carter, L., Dean, E., et al. (2019). Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–743. doi: 10.1038/s41591-019-0380-z
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillere, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706
Saini, M., Szczerba, B. M., and Aceto, N. (2019). Circulating tumor cell-neutrophil tango along the metastatic process. Cancer Res. 79, 6067–6073. doi: 10.1158/0008-5472.CAN-19-1972
Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206. doi: 10.1038/s41588-018-0312-8
Schmid, P., Adams, S., Rugo, H. S., Schneeweiss, A., Barrios, C. H., Iwata, H., et al. (2018). Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121. doi: 10.1056/NEJMoa1809615
Schmittnaegel, M., Rigamonti, N., Kadioglu, E., Cassara, A., Wyser Rmili, C., Kiialainen, A., et al. (2017). Dual angiopoietin-2 and VEGFA inhibition elicits antitumor immunity that is enhanced by PD-1 checkpoint blockade. Sci. Transl. Med. 9:eaak9670 doi: 10.1126/scitranslmed.aak9670
Schott, D. S., Pizon, M., Pachmann, U., and Pachmann, K. (2017). Sensitive detection of PD-L1 expression on circulating epithelial tumor cells (CETCs) could be a potential biomarker to select patients for treatment with PD-1/PD-L1 inhibitors in early and metastatic solid tumors. Oncotarget 8, 72755–72772. doi: 10.18632/oncotarget.20346
Sina, A. A., Carrascosa, L. G., Liang, Z., Grewal, Y. S., Wardiana, A., Shiddiky, M. J. A., et al. (2018). Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat. Commun. 9:4915. doi: 10.1038/s41467-018-07214-w
Siravegna, G., Marsoni, S., Siena, S., and Bardelli, A. (2017). Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548. doi: 10.1038/nrclinonc.2017.14
Smith, N. G., Gyanchandani, R., Shah, O. S., Gurda, G. T., Lucas, P. C., Hartmaier, R. J., et al. (2019). Targeted mutation detection in breast cancer using MammaSeq. Breast Cancer Res. 21:22. doi: 10.1186/s13058-019-1102-7
Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J. M., Desrichard, A., et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199. doi: 10.1056/NEJMoa1406498
Spencer, K. R., Wang, J., Silk, A. W., Ganesan, S., Kaufman, H. L., and Mehnert, J. M. (2016). Biomarkers for immunotherapy: current developments and challenges. Am. Soc. Clin. Oncol. Educ. Book 35, e493–e503. doi: 10.14694/EDBK_160766
Spranger, S., Bao, R., and Gajewski, T. F. (2015). Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235. doi: 10.1038/nature14404
Spranger, S., Dai, D., Horton, B., and Gajewski, T. F. (2017). Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711.e14–723.e14. doi: 10.1016/j.ccell.2017.04.003
Szczerba, B. M., Castro-Giner, F., Vetter, M., Krol, I., Gkountela, S., Landin, J., et al. (2019). Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566, 553–557. doi: 10.1038/s41586-019-0915-y
Tauriello, D. V. F., Palomo-Ponce, S., Stork, D., Berenguer-Llergo, A., Badia-Ramentol, J., Iglesias, M., et al. (2018). TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543. doi: 10.1038/nature25492
Torlakovic, E., Lim, H. J., Adam, J., Barnes, P., Bigras, G., Chan, A. W. H., et al. (2020). “Interchangeability” of PD-L1 immunohistochemistry assays: a meta-analysis of diagnostic accuracy. Mod. Pathol. 33, 4–17. doi: 10.1038/s41379-019-0327-4
Tray, N., Weber, J. S., and Adams, S. (2018). Predictive biomarkers for checkpoint immunotherapy: current status and challenges for clinical application. Cancer Immunol. Res. 6, 1122–1128. doi: 10.1158/2326-6066.CIR-18-0214
Umu, S. U., Langseth, H., Bucher-Johannessen, C., Fromm, B., Keller, A., Meese, E., et al. (2018). A comprehensive profile of circulating RNAs in human serum. RNA Biol. 15, 242–250. doi: 10.1080/15476286.2017.1403003
van’t Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A. M., Mao, M., et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536. doi: 10.1038/415530a
Vonderheide, R. H., LoRusso, P. M., Khalil, M., Gartner, E. M., Khaira, D., Soulieres, D., et al. (2010). Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment-associated modulation of inducible costimulator expression on patient T cells. Clin. Cancer Res. 16, 3485–3494. doi: 10.1158/1078-0432.CCR-10-0505
Voorwerk, L., Slagter, M., Horlings, H. M., Sikorska, K., van de Vijver, K. K., de Maaker, M., et al. (2019). Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat. Med. 25, 920–928. doi: 10.1038/s41591-019-0432-4
Wagner, J., Rapsomaniki, M. A., Chevrier, S., Anzeneder, T., Langwieder, C., Dykgers, A., et al. (2019). A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330.e18–1345.e18. doi: 10.1016/j.cell.2019.03.005
Wan, J. C. M., Massie, C., Garcia-Corbacho, J., Mouliere, F., Brenton, J. D., Caldas, C., et al. (2017). Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238. doi: 10.1038/nrc.2017.7
Wein, L., Luen, S. J., Savas, P., Salgado, R., and Loi, S. (2018). Checkpoint blockade in the treatment of breast cancer: current status and future directions. Br. J. Cancer 119, 4–11. doi: 10.1038/s41416-018-0126-6
Wen, W. X., and Leong, C. O. (2019). Association of BRCA1- and BRCA2-deficiency with mutation burden, expression of PD-L1/PD-1, immune infiltrates, and T cell-inflamed signature in breast cancer. PLoS One 14:e0215381. doi: 10.1371/journal.pone.0215381
Wolchok, J. D., Chiarion-Sileni, V., Gonzalez, R., Rutkowski, P., Grob, J.-J., Cowey, C. L., et al. (2017). Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 377, 1345–1356. doi: 10.1056/NEJMoa1709684
Yan, H., Viswanadhapalli, S., Chupp, D., Fernandez, M., Wu, S., Wang, J., et al. (2019). B cell-produced IL-27 up-regulates PD-L1 expression in the tumor microenvironment to promote breast cancer development. Cancer Res. 79(Suppl. 13):Abstractnr3250.
Yanaba, K., Hayashi, M., Yoshihara, Y., and Nakagawa, H. (2016). Serum levels of soluble programmed death-1 and programmed death ligand-1 in systemic sclerosis: association with extent of skin sclerosis. J. Dermatol. 43, 954–957. doi: 10.1111/1346-8138.13339
Yang, X., Zhang, K., Zhang, C., Peng, R., and Sun, C. (2019). Accuracy of analysis of cfDNA for detection of single nucleotide variants and copy number variants in breast cancer. BMC Cancer 19:465. doi: 10.1186/s12885-019-5698-x
Yang, Y., Li, C. W., Chan, L. C., Wei, Y., Hsu, J. M., Xia, W., et al. (2018). Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 28, 862–864. doi: 10.1038/s41422-018-0060-4
Zhang, J., Bu, X., Wang, H., Zhu, Y., Geng, Y., Nihira, N. T., et al. (2018). Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95. doi: 10.1038/nature25015
Zhang, Y., Tanno, T., and Kanellopoulou, C. (2019). Cancer therapeutic implications of microRNAs in the regulation of immune checkpoint blockade. ExRNA 1:19. doi: 10.1186/s41544-019-0022-0
Zhang, Y., Yang, L., Kucherlapati, M., Chen, F., Hadjipanayis, A., Pantazi, A., et al. (2018). A pan-cancer compendium of genes deregulated by somatic genomic rearrangement across more than 1,400 cases. Cell Rep. 24, 515–527. doi: 10.1016/j.celrep.2018.06.025
Keywords: immunotherapy, biomarker, breast cancer, immune checkpoint blockade, anti-PD-1 and anti-PD-L1
Citation: Vafaizadeh V and Barekati Z (2020) Immuno-Oncology Biomarkers for Personalized Immunotherapy in Breast Cancer. Front. Cell Dev. Biol. 8:162. doi: 10.3389/fcell.2020.00162
Received: 04 December 2019; Accepted: 28 February 2020;
Published: 17 March 2020.
Edited by:
Hasan Korkaya, Augusta University, United StatesReviewed by:
Raziye Piranlioglu, Augusta University, United StatesCopyright © 2020 Vafaizadeh and Barekati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vida Vafaizadeh, VmlkYVZhZmFpemFkZWhAZ21haWwuY29t; Zeinab Barekati, WmJhcmVrYXRpQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.