
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 05 February 2020
Sec. Molecular and Cellular Oncology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00032
 Eunbyul Yeom1,2
Eunbyul Yeom1,2 Dae-Woo Kwon1,3
Dae-Woo Kwon1,3 Jaemin Lee4
Jaemin Lee4 Seok-Ho Kim5
Seok-Ho Kim5 Ji-Hyeon Lee6
Ji-Hyeon Lee6 Kyung-Jin Min6
Kyung-Jin Min6 Kyu-Sun Lee1,3*
Kyu-Sun Lee1,3* Kweon Yu1,3,7*
Kweon Yu1,3,7*Aminoacyl-tRNA synthetases (ARSs), which are essential for protein translation, were recently shown to have non-translational functions in various pathological conditions including cancer. However, the molecular mechanism underlying the role of ARSs in cancer remains unknown. Here, we demonstrate that asparaginyl-tRNA synthetase (NRS) regulates Yorkie-mediated tumorigenesis by binding to the Hippo pathway component Salvador. NRS-RNAi and the NRS inhibitor tirandamycin B (TirB) suppressed Yorkie-mediated tumor phenotypes in Drosophila. Genetic analysis showed that NRS interacted with Salvador, and NRS activated Hippo target genes by regulating Yorkie phosphorylation. Biochemical analyses showed that NRS blocked Salvador-Hippo binding by interacting directly with Salvador, and TirB treatment inhibited NRS-Salvador binding. YAP target genes were upregulated in a mammalian cancer cell line with high expression of NRS, whereas TirB treatment suppressed cancer cell proliferation. These results indicate that NRS regulates tumor growth by interacting with Salvador in the Hippo signaling pathway.
Aminoacyl-tRNA synthetases (ARSs) are enzymes that catalyze the binding of amino acids to their cognate tRNAs with high fidelity (McClain, 1993; Delarue, 1995; Ogunbiyi et al., 1998). Once the tRNA is charged, ribosomes transfer the amino acid from the tRNA onto a growing peptide according to the specific genetic code (McClain, 1993; Delarue, 1995; Ogunbiyi et al., 1998). ARSs play an important role in the translation of RNA into protein. In addition to their canonical role in protein translation, ARSs function in metabolism, development, inflammatory responses, and tumorigenesis (Park et al., 2008). ARSs are composed of a highly conserved catalytic domain and an RNA-binding domain that recognizes tRNA anticodons (Arnez and Moras, 1997). In addition to these two domains, certain ARSs contain an amino acid-binding pocket that is essential for non-translational functions (Guo and Schimmel, 2013). The non-translational functions of ARSs have been expanded by the identification of new domains involved in protein-protein interactions (PPIs).
Aminoacyl-tRNA synthetases are involved in cancer cell survival and tumor progression. Low expression of tryptophanyl-tRNA synthetase (WRS) in colorectal cancer correlates with increased risk of metastasis and poor prognosis (Ghanipour et al., 2009). Nine ARSs in species from Drosophila to mammals [glutamyl-prolyl-tRNA synthetase, isoleucyl-tRNA synthetase, leucyl-tRNA synthetase, glutaminyl-tRNA synthetase (QRS), lysyl-tRNA synthetase (KRS), arginyl-tRNA synthetase, aspartyl-tRNA synthetase, and methionyl-tRNA synthetase] can form macromolecular multi-synthetase complexes (MSCs) with three scaffold proteins (Cerini et al., 1991; Quevillon and Mirande, 1996; Rho et al., 1999). Among them, KRS is involved in the development of melanoma (Levy and Fisher, 2011), and QRS inhibits apoptosis (Ko et al., 2001). A recent study suggests that methionyl-tRNA synthetase is overexpressed in non-small cell lung cancer (Kim et al., 2017). Although these ARSs have potential roles in tumorigenesis, the molecular mechanism underlying the role of ARSs in cancer remains unclear.
In this study, we used a Drosophila model system to identify candidate ARSs involved in tumorigenesis, and analyzed the tumorigenic effects of ARSs to elucidate the underlying molecular mechanisms. Drosophila is an excellent genetic model system because it has several conserved signaling pathways that are also present in mammals. Drosophila is therefore a powerful model to study human disorders such as cancer and neurodegenerative diseases. The Hippo (Hpo) signaling pathway is a well-studied pathway involved in growth regulation. The Hpo signaling pathway is evolutionary conserved from Drosophila to mammals, and evidence indicates that dysregulation of the Hpo pathway is involved in many types of human cancer (Halder and Johnson, 2011; Yu et al., 2015; Zanconato et al., 2016). The core components of the Hpo pathway are the Hpo/Mst1/2 (Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003) kinase, the scaffold protein Salvador (Sav/SAV1) (Kango-Singh et al., 2002; Tapon et al., 2002), and the Warts (Wts/Lats1/2) (Justice et al., 1995; Xu et al., 1995) kinase. The activated Hpo-Sav complex phosphorylates and activates the downstream Wts (Harvey et al., 2003; Wu et al., 2003), which phosphorylates and inactivates the transcription factor Yorkie (Yki/YAP) (Huang et al., 2005; Zhao et al., 2007). Inhibition of the core kinase cascade results in the translocation of unphosphorylated Yki to the nucleus, where it activates the expression of target genes of the Hpo pathway that regulate cell proliferation and survival (Huang et al., 2005).
Here, we established a fly cancer model by overexpressing Yki to investigate the tumorigenic effect of ARSs. The results showed that asparaginyl-tRNA synthetase (NRS) is highly expressed in cancer, and physically and genetically interacts with Salvador, a Hpo pathway component. NRS inhibited Sav-Hpo binding by sequestering Sav, and activated the expression of Yki target genes. Inhibition of NRS rescued the tumor phenotype in Drosophila and mammalian cancer cells. The present results demonstrate that NRS is a conserved growth regulator that modulates Hpo signaling.
Drosophila melanogaster was maintained at 25°C on standard media. GMR-Gal4, UAS-ykiWT-GFP, and UAS-ykiS168A-GFP were obtained from the Bloomington Stock Center (Bloomington, IN, United States), and all Drosophila ARS-RNAi lines were obtained from Vienna Drosophila RNAi Center (VDRC, Vienna, Austria). UAS-Sav was a gift from Nicholas Tapon (The Francis Crick Institute, United Kingdom), and en-Gal4-GFP was a gift from Kwang-Wook Choi (KAIST, South Korea). The pUAST-NRS transgenic fly was generated by p-element-mediated germline transformation (Rubin and Spradling, 1982).
Tirandamycin B (TirB) (ChemFaces, Hubei, China) solution was added to the standard medium during food preparation to a final TirB concentration of 0, 20, or 50 μM. TirB-supplemented food (3 mL) at the indicated concentrations was delivered to individual Drosophila polypropylene vials, and the flies were cultured.
The LLC and C26 cell lines were obtained directly from the ATCC and cultured according to the manufacturer’s guidelines. LLC and C26 cells were purchased in 2018. These cell lines were periodically authenticated by monitoring cell morphology, growth curve analysis, and inspection of Mycoplasma contamination using a Mycoplasma detection kit (Lonza). Cells were cultured at 37°C in a humidified 5% CO2 incubator in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS). Cells transfected with YAP-siRNA were considered as the siRNA group (siYAP) (siYAP-1: CUGCUAUGAUAACUACGUU, siYAP-2: CCGGCUCUAAAGAACCCGA), cells transfected with scrambled-siRNA were considered to be the negative control group (siScr), and cells without any treatment were considered to be the blank control group (cont).
The WST-1 measurement was performed according to the manufacturer’s standard protocol. LLC and C26 cells (1 × 104/well) were cultured in 96-well plates and incubated overnight. TirB was added to a final concentration of 10 ng/mL and incubated for 24, 48, and 72 h. After addition of 10 μL of WST-1, cells were incubated for an additional 2 h at 37°C. The absorbance was monitored at 450 nm.
This experiment used the CS10 (w1118 outcrossed 10 times to Canton-S) D. melanogaster strain, which was obtained from the Bloomington Drosophila Stock Center at Indiana University. Flies were reared at 25°C and 65% humidity on a 12:12 h light:dark cycle. Fly larvae were reared in standard cornmeal-sugar-yeast with agar (CSY) medium (5.2% cornmeal, 11% sugar, 2.6% baker’s yeast, 0.5% propionic acid, 0.04% methyl-4-hydroxybenzoate, and 0.8% agar). Newly eclosed male and female adult fruit flies (100 of each) were transferred to a 500 cm2 demography cage. Three replicate cages were set up for each experimental group. Vials containing fresh SY food with/without TirB were affixed to the cages and changed every 2 days, at which time dead flies were removed and recorded.
For immunostaining, larval discs were dissected in PBS, fixed in 4% paraformaldehyde, blocked in 5% BSA, and incubated in primary antibodies overnight at 4°C, followed by secondary antibody incubation for 2 h at room temperature. The tissues were mounted with Vectashield mounting medium (Vectashield), and fluorescence images were acquired with a FluoView confocal microscope (Olympus). Primary antibodies used were rabbit anti-phospho-histone3 (Santa Cruz Biotechnology, Santa Cruz, CA, United States; 1:200), mouse anti-CD2 (AbD Serotec, 1:200), rabbit anti-CycE (Santa Cruz Biotechnology, 1:200), and goat anti-Diap1 (Santa Cruz Biotechnology, 1:200); and secondary antibodies were anti-mouse IgG Alexa 488, anti-rabbit IgG Alexa 594, and anti-goat IgG Alexa 594 (Life Technologies, 1:200).
Drosophila S2 cells were cultured in Schneider’s Insect media (Sigma) with 10% FBS (Gibco). Transfection was carried out with Effectene reagent (Qiagen) according to the manufacturer’s instructions. Each transfection was performed using 1–2 μg of DNA. For the immunoprecipitation assay, cells were lysed on ice for 30 min with lysis buffer containing 1 M Tris pH 7.4, 5 M NaCl, 0.5 M EDTA, 0.1% NP-40, and 5% glycerol. The cell lysates were incubated with Anti-FLAG M2 magnetic beads (Sigma) overnight at 4°C. Immunoprecipitates were washed three times with lysis buffer and subjected to SDS-PAGE. Western blot analysis was performed using a standard protocol. Yki phosphorylation was examined using a 12.5% acrylamide phos-tag gel (50 μM) (Wako). Primary antibodies used were mouse anti-HA (Invitrogen, 1:5000), rabbit anti-Sav (Dr. Jiang, 1:2000) (Yue et al., 2012), rabbit anti-Yki (Dr. Irvine, 1:4000) (Oh and Irvine, 2008), mouse anti-V5 (Invitrogen, 1:5000), mouse anti-Myc (Santa Cruz Biotechnology, 1:1000), guinea pig anti-Hpo (Dr. Halder, 1:2000), rabbit anti-pS6K (Invitrogen, 1:1000), and mouse β-actin (1:3000, DSHB). Secondary antibodies were anti-rabbit IgG (Santa Cruz Biotechnology, 1:5000), anti-mouse IgG (Pierce, 1:5000), and anti-guinea pig IgG (Jackson, 1:5000). Western blot bands were quantified using ImageJ software.
Total RNA was extracted using the easy-BLUETM reagent (iNtRON Biotechnology). All RNA samples were treated with RNase-free DNase (Promega), and cDNA was synthesized using a SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative RT-PCR analysis was performed using an ABI Prism 7900 Sequence Detection System (Applied Biosystems) and SYBR Green PCR Core reagents (Applied Biosystems). Data were analyzed using the comparative cycle threshold (Ct) method (User Bulletin 2, Applied Biosystems). All experiments were repeated at least three times, and the data are presented as the mean and error bar (±SEM).
All statistical significance were tested by Microsoft Excel-based application for the Student’s t-test statistical analysis. Values in this paper are presented as the mean ± SEM p-values were as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To identify candidate ARSs involved in growth regulation, we first established a Drosophila cancer model by overexpressing yki, a downstream transcription factor of the Hpo pathway, in the adult eye using GMR-Gal4. Expression of wild-type yki (GMR > ykiWT) caused mild rough eye phenotype (Figure 1A, middle panel), whereas the constitutively active form of yki (GMR > ykiS168A) resulted in abnormal overgrowth of the eye by inducing massive cell proliferation (Figure 1A, right panel). GMR > ykiS168A flies were crossed with each ARS-RNAi line, and the anticancer effect by knockdown of ARSs was analyzed in the Drosophila cancer model. Nine ARS-RNAi lines (ArgRS, CysRS, GlnRS, GluproRS, HisRS, LysRS, ThrRS, TyrRS, and SerRS) showed pupal lethality, indicating a deleterious effect on survival in the yki-induced tumor model. Five ARS-RNAi lines (LeuRS, MetRS, PheRS, TrpRS, and VarRS) showed no significant changes (Supplementary Figure S1A), whereas the AsnRS-, AlaRS-, AspRS-, and GlyRS-RNAi lines showed suppression of the overgrowth tumor phenotype (Figure 1B). Among them, Asn-tRNA synthetase (NRS) knockdown most effectively suppressed the yki-overexpression phenotype in the adult eye (Figure 1B, red box). We then tested whether NRS expression was altered in cancer, and the results showed significant upregulation of Drosophila NRS in the yki-overexpression background (Supplementary Figure S1B).
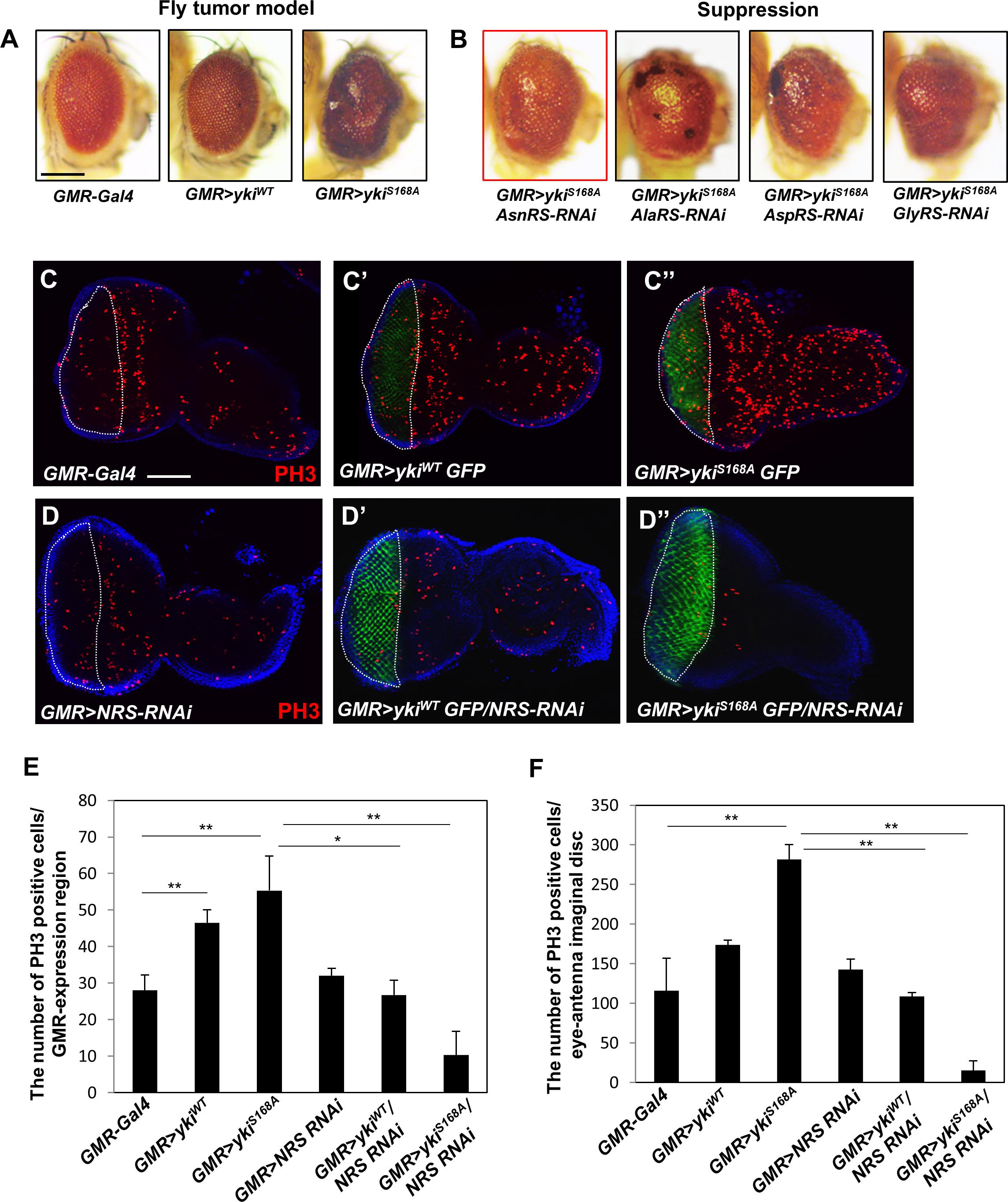
Figure 1. Knockdown of NRS suppresses the yki-induced tumor phenotype in the adult eye and larval eye disc. (A) Overexpression of the wild-type form of yki caused mild roughness and overexpression of the active form of yki caused abnormal overgrowth of the eye. (B) Library screening showed that AsnRS-RNAi strongly suppressed the eye phenotype induced by yki overexpression. Scale bar, 200 μm. (C–C”) Overexpression of ykiWT and ykiS168A in the larval eye disc increased cell proliferation, as determined by detection of phospho-H3 expression in the GMR-expressing region compared with the control. (D–D”) Consistent with the adult eye phenotype, knockdown of NRS significantly rescued cell proliferation caused by yki overexpression. Blue, DAPI; green, GFP; red, phospho-H3. Scale bar, 50 μm. (E) Quantification of phospho-H3-positive cells in the GMR-expressing region [white-dashed line in panels (C,D)]. (F) Quantification of phospho-H3-positive cells in the eye-antenna disc. (GMR-Gal4 n = 6, GMR > ykiWT n = 7, GMR > ykiSA n = 9, GMR > NRS RNAi n = 6, GMR > ykiWT/NRS RNAi n = 8, GMR > ykiS168A/NRS RNAi n = 5). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01.
To confirm that NRS knockdown suppressed the yki-induced tumor phenotype, we performed immunostaining of the larval eye disc using the mitotic marker phospho-H3. Overexpression of yki markedly increased cell proliferation in the GMR-expressing region marked by GFP (Figures 1C–C”,E,F). Consistent with the results of the adult eye phenotype, NRS knockdown by NRS-RNAi significantly suppressed cell proliferation induced by yki overexpression (Figures 1D–D”,E–F). As the control, KRS knockdown had no significant effect on cell proliferation (Supplementary Figures S1C,D), suggesting that the suppression of the tumor phenotype was NRS-specific.
To test the effect of chemical inhibition of NRS on the yki-induced tumor phenotype, GMR > ykiS168A flies were treated with the NRS inhibitor TirB, which was originally isolated from natural products and identified as an anti-filarial drug targeting Brugia malayi NRS (Yu et al., 2011). TirB treatment suppressed the yki-overexpression phenotype in a dose-dependent manner. TirB at 50 μM effectively rescued the tumor phenotype (Figures 2A–C). In the larval eye discs, TirB also dose-dependently suppressed cell proliferation, as determined by detection of the phospho-H3 marker (Figures 2D–D”). TirB treatment was not cytotoxic to either male or female flies at 10, 20, or 50 μM. TirB had no obvious effect on fly lifespan even at 50 μM, an effective concentration for tumor inhibition (Supplementary Figures S2A,B). Since, NRS functions mainly in protein translation, we tested whether NRS knockdown affected translation in our cancer model. NRS knockdown partially restored the increased phosphorylation of S6K induced by yki overexpression (Supplementary Figures S2C,D). To determine whether the rescue effect was caused by inhibition of protein translation, we tested the effect of inhibitors of protein synthesis or translation. Among inhibitors we tested, TirB strongly rescued the eye size of GMR > ykiS168A flies (Figures 2E,F). These data suggest that the NRS inhibitor TirB has a specific anticancer effect in the Drosophila tumor model.
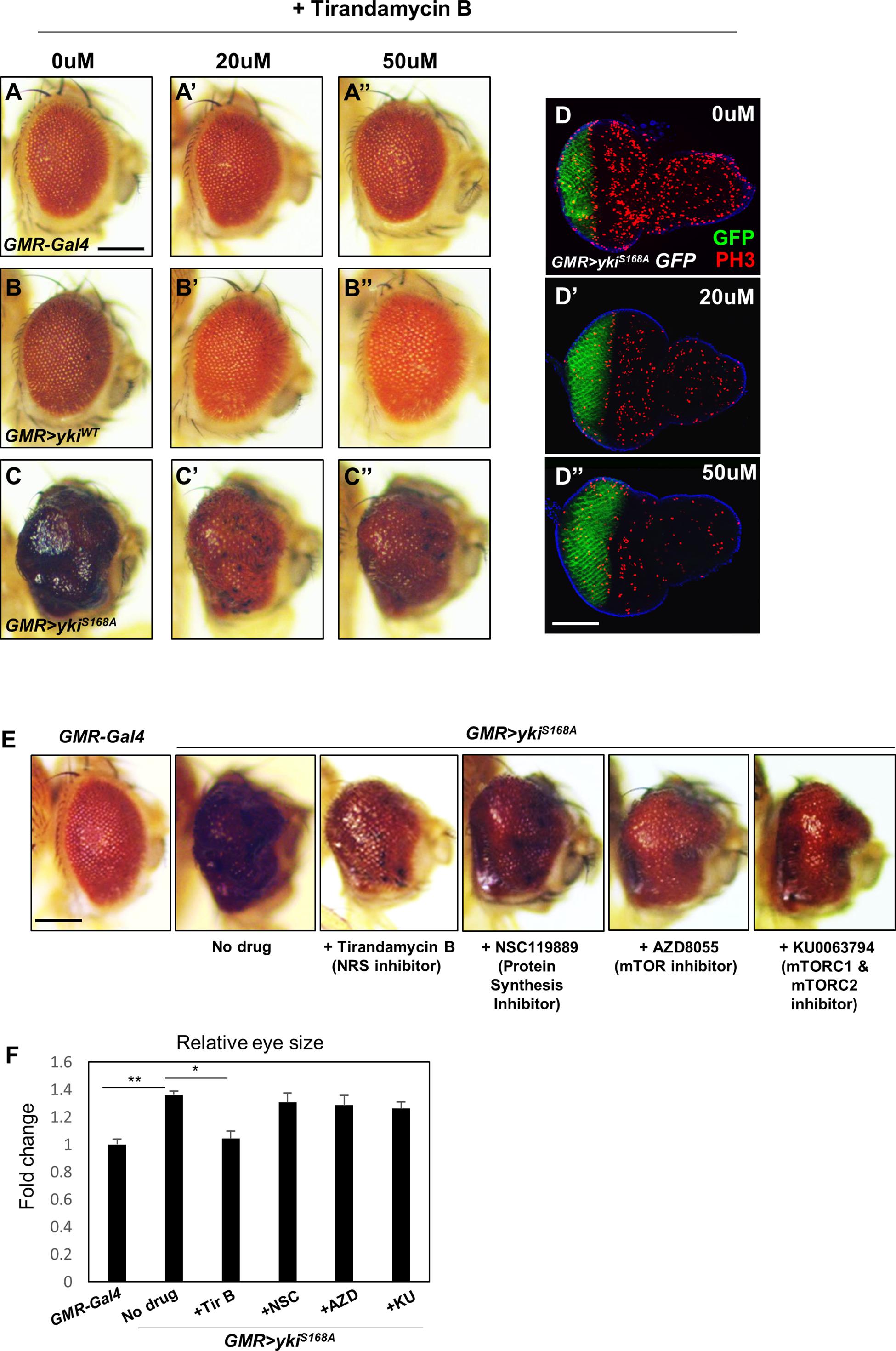
Figure 2. The NRS inhibitor tirandamycin B (TirB) suppresses the yki-induced tumor phenotype. (A–C) TirB treatments dose-dependently rescued the tumor growth phenotypes induced by yki overexpression. (A–A”) GMR-Gal4 control flies were treated with 0, 20, or 50 μM TirB. (B–B”) GMR > ykiWT flies were treated with 0, 20, or 50 μM TirB. (C–C”) GMR > ykiS168A flies were treated with 0, 20, or 50 μM TirB. Scale bar, 200 μm. (D) TirB decreased cell proliferation detected by phospho-H3 expression in the larval eye disc of GMR > ykiS168A flies. Blue, DAPI; green, GFP; red, phospho-H3. Scale bar, 50 μm. (E) Inhibitors blocking protein synthesis or translation do not suppress the yki-mediated tumor phenotype. Scale bar, 200 μm. (F) Quantification of relative eye size of panel (E). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01.
The effect of NRS knockdown on rescuing the yki-mediated tumor phenotype led us to hypothesize that NRS may interact with a Hpo signaling component. Screening of the database of protein interactions in Flybase (yeast two-hybrid screening from Curagen) (Giot et al., 2003) identified Sav as a potential NRS-interacting protein. We first examined the genetic interaction between NRS and Sav. Overexpression of Sav by en-Gal4 in the adult wing resulted in the formation of slightly smaller wings (Figures 3B,E), whereas NRS overexpression led to increased wing size (Figures 3C,E) compared with that of the en > + control (Figure 3A). The enlarged wing phenotype induced by NRS overexpression was rescued by overexpression of Sav (Figures 3D,E). The genetic interaction was confirmed in the larval wing disc by immunostaining for the PH3 mitotic cell marker. Consistently, overexpression of Sav and NRS decreased (Figures 3G,J) and increased (Figures 3H,J) the number of PH3-positive mitotic cells, respectively, in the posterior compartment compared with that in the anterior control compartment (Figures 3F,J). Co-expression of Sav and NRS slightly rescued the NRS phenotype (Figures 3I,J). The effect of Sav and NRS overexpression was tested in the adult eye using eyeless-Gal4, and the results were similar to those observed in the wing (Supplementary Figure S3). These genetic interactions suggested that NRS regulates Sav.
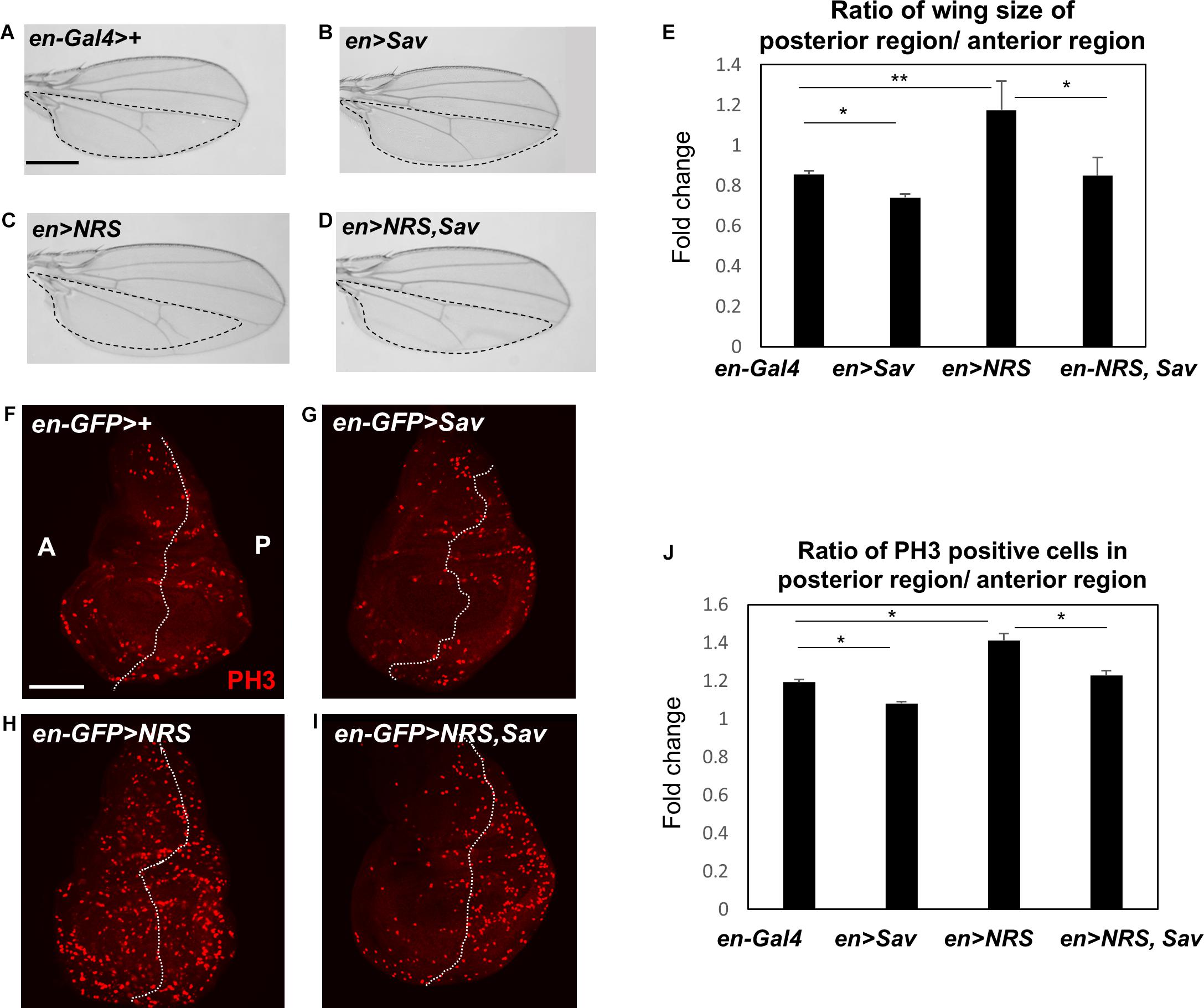
Figure 3. NRS genetically regulates the Hippo pathway component Salvador. (A) en-Gal4>+, the control wing (n = 7). (B) Overexpression of Salvador (Sav) (en > Sav) resulted in a slightly smaller wing (n = 9). (C) Overexpression of NRS (en > NRS) resulted in a larger wing (n = 10). (D) Co-expression of NRS and Sav (en > NRS, Sav) rescued the enlarged wing size induced by NRS overexpression (n = 9). Scale bar, 100 μm. (E) Quantification of relative wing size from panels (A–D). (F) en-GFP > +, the control wing disc. (G) Sav overexpression (en-GFP > Sav) in the wing disc decreased cell proliferation. (H) Overexpression of NRS (en-GFP > NRS) induced a dramatic increase of cell proliferation in the posterior wing disc. (I) NRS-induced cell proliferation was rescued by addition of Sav (en-GFP > NRS, Sav). Scale bar, 50 μm. (J) Quantification of PH3-positive cells in the posterior wing disc. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01.
The genetic interactions between NRS and Sav suggested that NRS positively regulates Yki signaling. Downregulation of Hpo or Sav increases Yki activity and activates the transcription of Yki target genes such as cyclin E and diap1, promoting cell cycle progression and inhibiting cell death, respectively (Huang et al., 2005). We therefore examined the effect of NRS overexpression on the transcription of Yki target genes. NRS overexpression by hs-Gal4 upregulated cyclin E and diap1 mRNA expression (Figures 4A,B). NRS overexpression clones in wing discs upregulated Cyclin E and Diap1 protein expression (Figures 4C–C”,D–D”). Because upregulation of Hpo or Sav increases the phosphorylation of Yki and inhibits Yki nuclear localization (Oh and Irvine, 2008), we examined the effect of NRS on the phosphorylation of Yki. In S2 cells, Sav induced a mobility shift of V5 tag-Yki on a phos-tag gel (Nagy et al., 2018), and NRS co-expression decreased Yki phosphorylation (Figure 4E). These results indicate that NRS activates Yki target genes by regulating Yki phosphorylation.
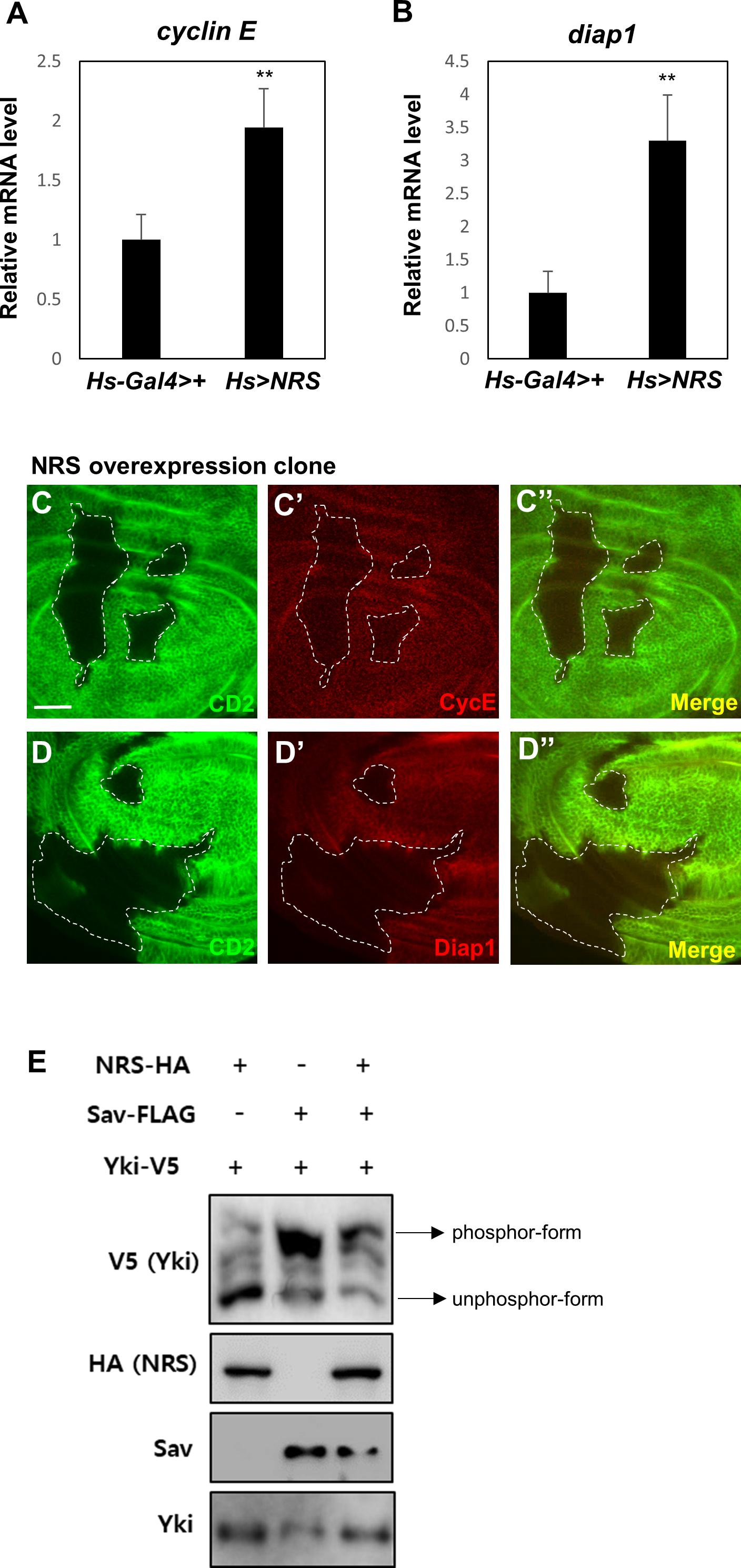
Figure 4. NRS overexpression activates Hippo pathway target genes. Adult whole body NRS overexpression (Hs > NRS) significantly upregulated cyclin E (A) and diap1 (B) target gene expression. (C,D) Consistent with mRNA levels, the CycE and Diap1 proteins were upregulated in the NRS-overexpressing clone marked by membrane marker CD2. The genotype of the NRS overexpression clone was yw hs-FLP; act > y + > Gal4 (Ay-Gal4) UAS-CD2/UAS-NRS. Scale bar, 20 μm. (E) NRS overexpression decreased the phosphorylation level of Yki in S2 cells, as detected by the phos-tag gel. Data are presented as the mean ± SEM. **p < 0.01.
To further understand the mechanism underlying the role of NRS in Hpo signaling-mediated growth regulation, the PPI of NRS with Hpo signaling components (Hpo, Sav, and Yki) was tested by co-immunoprecipitation assays in S2 cells. The results showed that NRS binds directly to Sav (Figures 5A,B), but not to Hpo (Supplementary Figure S3F) or Yki (Supplementary Figure S3G). Sav is a scaffold protein in which the stabilization effect depends on binding to Hpo (Pantalacci et al., 2003). We confirmed that Sav is stabilized when Hpo is co-transfected with Sav (Figure 5C, fourth lane, E). Addition of NRS significantly decreased the protein level of Sav (Figure 5C, fifth lane, E). This result suggests that NRS overexpression interferes with the interaction between Sav and Hpo, resulting in the destabilization of Sav. To test this hypothesis, we examined the binding of Sav and Hpo in cells co-transfected with NRS, which showed that the interaction between Sav and Hpo was significantly reduced (Figure 5D). This suggested that NRS acts as a competitive inhibitor of Hpo-Sav binding. To confirm this inhibition of Hpo-Sav binding by NRS regulates ykiS168A-induced tumor growth, we showed that over-expression of Hpo suppressed the overgrowth eye tumor phenotype of GMR > ykiS168A (Supplementary Figure S3I). The NRS inhibitor TirB effectively rescued tumor growth induced by yki overexpression in fly eyes in a dose-dependent manner (Figures 2C–C”). In S2 cells, TirB dose-dependently inhibited NRS-Sav binding and also partially restored the Sav level (Figure 5F and Supplementary Figure S3H). These results indicated that inhibition of NRS rescued tumor growth by blocking the interaction between NRS and Sav.
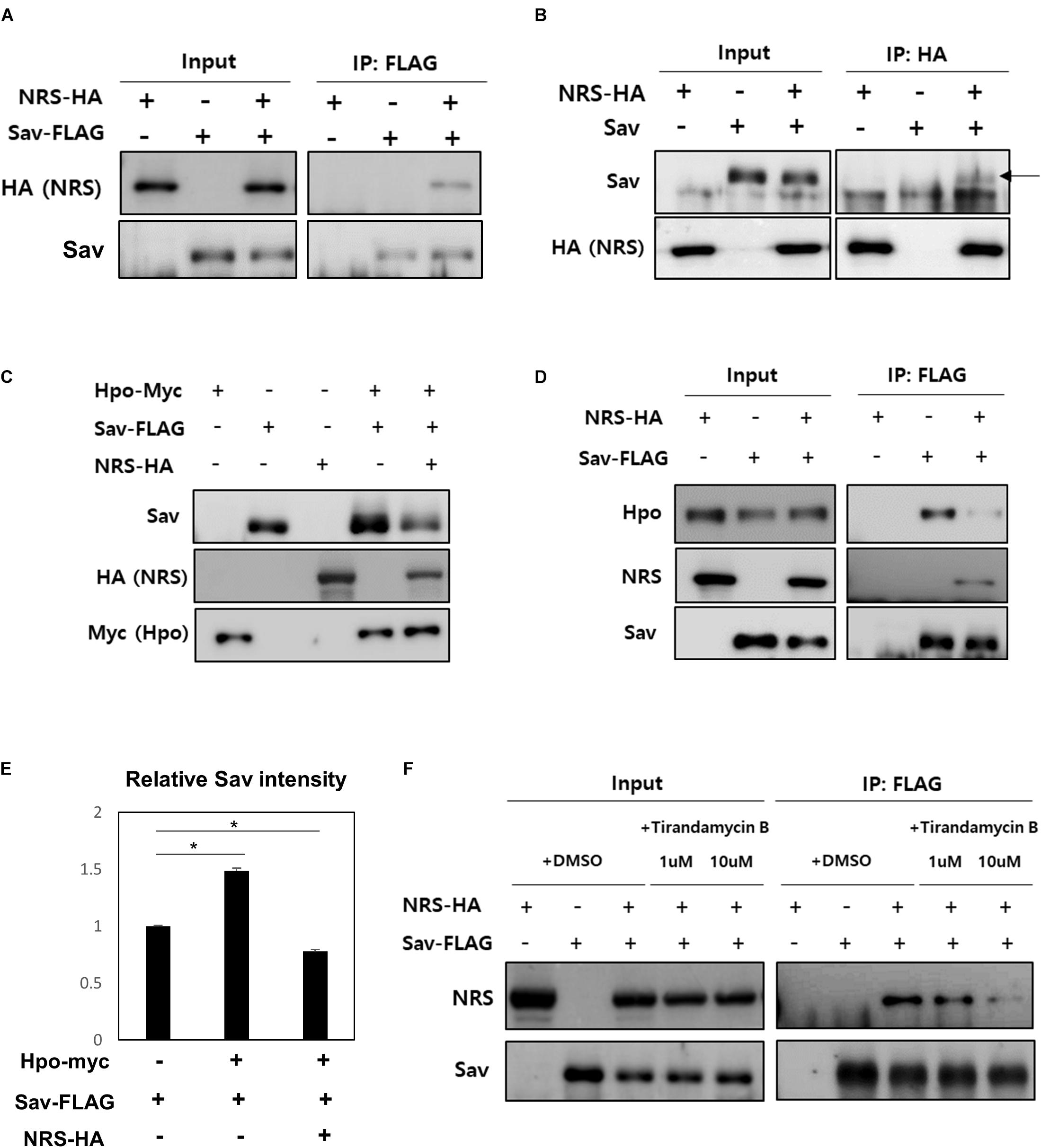
Figure 5. NRS promotes the dissociation of the Hippo-Salvador complex by interacting with Salvador. (A) Co-immunoprecipitation (Co-IP) assays showed that Sav physically interacts with NRS, as detected by Sav immunoprecipitation and NRS immunoblotting. (B) Co-IP between Sav and NRS was confirmed by immunoprecipitating NRS. (C) Hippo (Hpo) co-expression upregulated Sav expression, whereas Sav level was strongly reduced by addition of NRS. (D) Co-IP assays showed that binding of NRS to Sav dissociated the Sav-Hpo complex. (E) Quantification of Sav protein levels in panel (C). (F) Direct binding of NRS to Sav was reduced by TirB treatment in a dose-dependent manner. Data are presented as the mean ± SEM. *p < 0.05.
Next, we investigated the novel role of NRS in growth regulation through the Hpo pathway in mammalian cancer models. We used two mouse cancer cell lines, a Lewis lung carcinoma cell line (LLC) and a colon carcinoma cell line (C26). First, we checked the expression of NRS in both cell lines. NRS expression was considerably higher in C26 cells than in LLC cells (Figure 6A). Consistent with our data, a gene expression database of normal and tumor tissues from human samples (U133A datasets) showed that NRS is expressed at higher levels in colon cancer patients than in healthy controls, whereas no significant changes in NRS expression are observed in lung cancer patient samples (Supplementary Figure S4)1. To determine whether NRS also regulates the Hpo pathway in mammals, we examined the expression of YAP, a mammalian homolog of Yki, target genes in the two cell lines. YAP target genes were significantly upregulated in the C26 cell line, which has high expression of NRS (Figure 6B). Also, the upregulated YAP target gene level was suppressed by NRS inhibitor (TirB) treatment in C26 cells (Supplementary Figure S5A). In addition, the SAV1 protein level in C26 cells was significantly down-regulated compared to that in LLC cells (Supplementary Figure S5B), suggesting that upregulated NRS interacts with SAV1 to destabilize and inhibit Hpo signaling which is consistent to our fly data (Figures 5D,E). We then tested the effect of the NRS inhibitor TirB on tumor growth in mammalian cells. TirB had no effect in the LLC cell line (Figure 6C), whereas it significantly decreased cell proliferation in C26 cells (Figure 6D). Like NRS inhibition, YAP knockdown also showed reduced cell proliferation in C26 cells, but not in LLC cells (Supplementary Figures S5C–E), suggesting that NRS is required for YAP-induced proliferation in mammalian cells. Taken together, these results suggest that the modulation of NRS expression is critical for Hpo signaling-mediated tumorigenesis in different tumor types, providing a potential strategy to develop anticancer drugs targeting NRS.
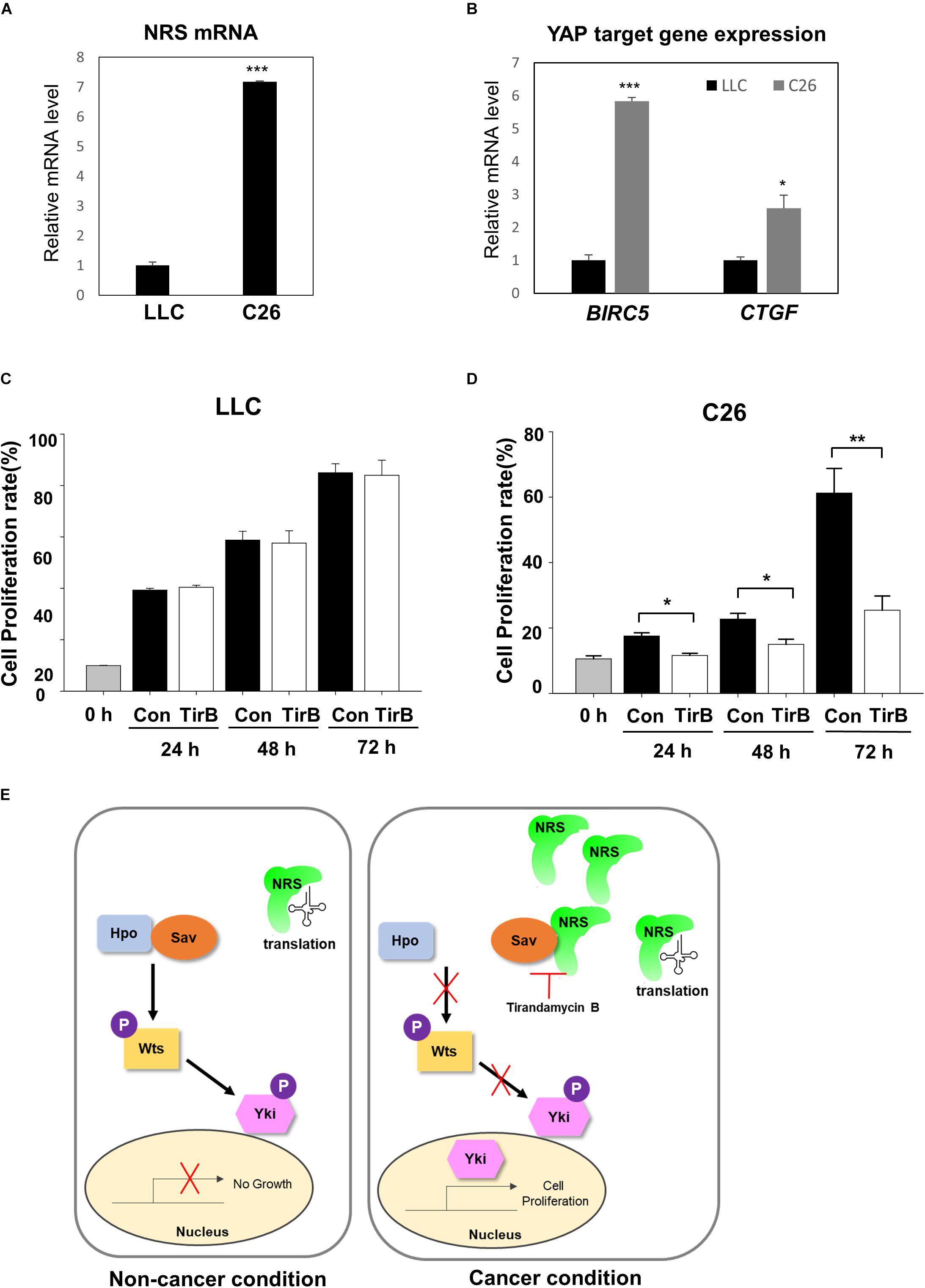
Figure 6. NRS-overexpressing mammalian cancer cells activate Hippo signaling. (A) NRS expression was higher in the C26 (colon carcinoma) cell line than in the LLC (Lewis lung carcinoma) cell line. (B) Expression of YAP target genes (BIRC5 and CTGF) was significantly higher in C26 cells than in LLC cells. (C,D) Cell proliferation assays in cells treated with TirB showed that cell proliferation was strongly suppressed in the C26 cell line (D), but not in the LLC cell line (C). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. (E) Proposed model of NRS function: NRS has a canonical translational function in the non-cancer condition. In cancer, NRS is upregulated and binds to Sav, which inhibits the interaction between Sav and Hpo. This inhibits the Hpo-Wts-Yki phosphorylation cascade, leading to Yki translocation to the nucleus to activate target genes.
NRS has been studied mostly for its role in translation, and there is little evidence of the non-canonical function of NRS in tumorigenesis. In the present study, we identified a novel role of NRS in growth regulation mediated by the Hpo signaling pathway. In a proposed model (Figure 6E), we suggest that NRS is mainly involved in translation under normal conditions, in which Hpo signaling is active and growth is regulated. Because the core components of the Hpo pathway (Hpo, Sav, and Wts) are tumor suppressors, functional mutation of the core components induces tumor growth (Pan, 2010; Zhao et al., 2010). We propose that, under these tumorigenic conditions, NRS is upregulated and physically interacts with Sav, thereby inhibiting Sav-Hpo binding. This interaction suppresses the Hpo-Wts-Yki phosphorylation cascade, promotes Yki nuclear localization, and activates Yki target genes, which are involved in growth regulation. We showed that TirB (NRS inhibitor) rescued cancer cell proliferation in Drosophila and mammals (Figures 2, 6). These findings suggest that NRS negatively regulates the Hpo-Sav-Wts cascade to induce cell proliferation.
The Drosophila NRS protein is well conserved with the mammalian system, sharing more than 80% sequence similarity. The proteins also share conserved domains, such as the aminoacyl synthetase-like catalytic core domain and the anticodon recognition domain, which are both critical for the canonical function of NRS. However, recent evidence supports that amino acid-binding sites are also essential for non-translational functions associated with signaling pathways such as mTORC1 activation through KRS (Han et al., 2012; Segev and Hay, 2012) and anti-apoptosis signaling through QRS (Ko et al., 2001). In some cases, such as the WHEP domain of WRS (Wakasugi et al., 2002) or the UNE-S domain of SRS (Xu et al., 2012), additional new domains that are not essential for catalytic activity regulate non-translational functions. A recent study suggests that the unique N-terminal extension domain of human NRS (UNE-N) interacts with CCR3, mediating proinflammatory signaling in interstitial lung disease (Park et al., 2018). Although we demonstrated a new non-translational function of NRS associated with tumorigenesis, the specific domain of NRS mediating the regulation of Hpo signaling was not identified. Future studies should identify the NRS domain involved in binding to Sav. We also showed that mammalian cancer cells with high expression of NRS regulate YAP target gene expression (Figure 6). This introduces the question of whether mammalian NRS also directly binds to SAV1, leading to the activation of YAP in the C26 cell line. Whether mammalian NRS regulates the Hpo signaling pathway through a mechanism similar to that in Drosophila remains to be determined.
Based on our results that NRS level was increased in yki overexpression flies (Supplementary Figure S1B) and this could raise the possibility that NRS plays a role in downstream of Yki. Many Hpo pathway components act as not only upstream regulators but also Yki target genes (e.g., ex, wts, ft, ds, and kba) for negative feedback loop. Similar to these components, there is a possibility that NRS might act as both upstream Hpo pathway component and Yki target gene for positive feedback loop.
The Hpo pathway is a key regulator of tumor development in both Drosophila and mammals. We identified NRS as a novel component of the Hpo pathway. A previous study identified KRS as a binding candidate of Wts, as demonstrated by mass spectrometry and co-IP (Kwon et al., 2013). These findings support the non-translational roles of ARSs in Hpo signaling-mediated tumorigenesis.
Studies in animal models led to the emergence of ARSs as potential therapeutic agents for the treatment of various diseases. For example, administration of glycyl-tRNA synthetase to colon cancer-bearing mice significantly suppresses tumor formation by dephosphorylating and deactivating ERK (Park et al., 2012). Our data indicated that the NRS inhibitor TirB rescued tumor growth in Drosophila as well as cell proliferation in mammalian cancer cells. NRS expression was high in colon carcinoma, but low in LLC cells (Figure 6A), supporting that NRS is expressed at higher levels in colon cancer patients, whereas no significant changes in NRS expression are observed in lung cancer patient samples from human database (Supplementary Figure S4). In addition, analysis of the human transcriptome shows that high expression of NRS in liver cancer is correlated with poor survival rates (Uhlen et al., 2017), suggesting that NRS expression can be used as a prognostic marker in certain cancer types.
Recently, small molecule inhibitors against PPIs have emerged as new anticancer drugs (Nero et al., 2014). For example, chemical or peptide inhibitors that block the PPI between p53 and MDM2 restore p53 function and can be used as anticancer drugs (Liu et al., 2010). Taken together, the present results suggest that the physical interaction of NRS and Sav is a potential target that may lead to the development of new anticancer drugs based on PPI inhibition.
All datasets generated for this study are included in the article/Supplementary Material.
EY, K-SL, and KY designed the research and wrote the manuscript. EY, D-WK, JL, and J-HL performed the experiments. EY, S-HK, K-JM, K-SL, and KY analyzed the data.
This work was supported by grants from the KRIBB Research Initiative Program, the National Research Council of Science and Technology (CRC-15-04-KIST), and the National Research Foundation of Korea (2014M3A6A4075062, 2015R1A5A1009024, and 2019R1A2C2004149).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank N. Tapon, K. W. Choi, K. D. Irvine, G. Halder, and J. Jiang for providing reagents. Drosophila stocks were obtained from the Bloomington Stock Center (Bloomington, IN, United States) and Vienna Drosophila RNAi Center (VDRC, Vienna, Austria).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00032/full#supplementary-material
FIGURE S1 | Suppression of the tumor growth phenotype is NRS-specific. (A) LeuRS-RNAi, MetRS-RNAi, PheRS-RNAi, TrpRS-RNAi, and ValRS-RNAi did not suppress the tumor growth phenotype induced by yki overexpression. Scale bar, 200 μm. (B) yki overexpression upregulated NRS mRNA expression. (C) As a negative control, a cell proliferation marker was not significantly changed by KRS-RNAi. Blue, DAPI; green, GFP; red, phospho-H3. Scale bar, 50 μm. (D) Quantification of phospho-H3-positive cells in the GMR-expressing region. Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
FIGURE S2 | Cytotoxicity analysis of TirB. Analysis of lifespan in flies treated with TirB showed no cytotoxic effects in male (A) and female flies (B). (C) yki overexpression increased the phosphorylation activity of S6K. NRS knockdown reduced the phosphorylation activity of S6K in the yki-overexpression background. (D) Quantification of the intensity of western blot bands in panel (C). Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
FIGURE S3 | Genetic interaction between NRS and Sav. (A) ey > Gal4/+ was used as the control, n = 6. (B) Sav overexpression resulted in slightly smaller eye size, n = 7. (C) NRS overexpression resulted in bigger eye size n = 7. (D) Co-expression of NRS and Sav rescued the NRS-induced enlarged eye n = 8. Scale bar, 200 μm. (E) Quantification of relative eye size from panels (A–D). Co-IP assay showed that NRS did not directly bind to Hpo (F) and Yki (G). (H) Quantification of Sav input protein levels in Figure 5F. (I) Overexpression of Hpo suppressed the overgrowth eye phenotype of GMR > ykiS168A. Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
FIGURE S4 | A gene expression database of normal and tumor tissues from human samples. A gene expression database across normal and tumor tissue (U133A datasets) (http://medical-genome.kribb.re.kr/GENT) showed that NRS is highly expressed in colon cancer patients than in healthy controls, but not in lung cancer patient samples (C – cancer tissue, N – normal tissue).
FIGURE S5 | YAP target gene expression by NRS inhibition and cell proliferation assay by YAP knockdown. (A) Increased YAP target gene (BIRC5) level in C26 cell line was reduced by TirB treatment, but no reduction was observed in LLC cell line. (B) SAV1 protein expression level was decreased in C26 cells compared to LLC cells. (C) YAP-siRNA was efficiently knocked down in C26 cells. (D,E) Cell proliferation was reduced by YAP knockdown in C26 cells (D), but not in LLC cells (E). Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Arnez, J. G., and Moras, D. (1997). Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22, 211–216. doi: 10.1016/s0968-0004(97)01052-9
Cerini, C., Kerjan, P., Astier, M., Gratecos, D., Mirande, M., and Semeriva, M. (1991). A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 10, 4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x
Ghanipour, A., Jirstrom, K., Ponten, F., Glimelius, B., Pahlman, L., and Birgisson, H. (2009). The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 18, 2949–2956. doi: 10.1158/1055-9965.EPI-09-0456
Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736. doi: 10.1126/science.1090289
Guo, M., and Schimmel, P. (2013). Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153. doi: 10.1038/nchembio.1158
Halder, G., and Johnson, R. L. (2011). Hippo signaling: growth control and beyond. Development 138, 9–22. doi: 10.1242/dev.045500
Han, J. M., Jeong, S. J., Park, M. C., Kim, G., Kwon, N. H., Kim, H. K., et al. (2012). Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424. doi: 10.1016/j.cell.2012.02.044
Harvey, K. F., Pfleger, C. M., and Hariharan, I. K. (2003). The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467. doi: 10.1016/s0092-8674(03)00557-9
Huang, J., Wu, S., Barrera, J., Matthews, K., and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434. doi: 10.1016/j.cell.2005.06.007
Jia, J., Zhang, W., Wang, B., Trinko, R., and Jiang, J. (2003). The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519. doi: 10.1101/gad.1134003
Justice, R. W., Zilian, O., Woods, D. F., Noll, M., and Bryant, P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546. doi: 10.1101/gad.9.5.534
Kango-Singh, M., Nolo, R., Tao, C., Verstreken, P., Hiesinger, P. R., Bellen, H. J., et al. (2002). Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730. doi: 10.1242/dev.00168
Kim, E. Y., Jung, J. Y., Kim, A., Kim, K., and Chang, Y. S. (2017). Methionyl-tRNA synthetase overexpression is associated with poor clinical outcomes in non-small cell lung cancer. BMC Cancer 17:467. doi: 10.1186/s12885-017-3452-9
Ko, Y. G., Kim, E. Y., Kim, T., Park, H., Park, H. S., Choi, E. J., et al. (2001). Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem. 276, 6030–6036. doi: 10.1074/jbc.M006189200
Kwon, Y., Vinayagam, A., Sun, X., Dephoure, N., Gygi, S. P., Hong, P., et al. (2013). The Hippo signaling pathway interactome. Science 342, 737–740. doi: 10.1126/science.1243971
Levy, C., and Fisher, D. E. (2011). Dual roles of lineage restricted transcription factors: the case of MITF in melanocytes. Transcription 2, 19–22. doi: 10.4161/trns.2.1.13650
Liu, M., Li, C., Pazgier, M., Li, C., Mao, Y., Lv, Y., et al. (2010). D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc. Natl. Acad. Sci. U.S.A. 107, 14321–14326. doi: 10.1073/pnas.1008930107
McClain, W. H. (1993). Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234, 257–280. doi: 10.1006/jmbi.1993.1582
Nagy, Z., Comer, S., and Smolenski, A. (2018). Analysis of protein phosphorylation using phos-tag gels. Curr. Protoc. Protein Sci. 93:e64. doi: 10.1002/cpps.64
Nero, T. L., Morton, C. J., Holien, J. K., Wielens, J., and Parker, M. W. (2014). Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer 14, 248–262. doi: 10.1038/nrc3690
Ogunbiyi, O. A., Goodfellow, P. J., Herfarth, K., Gagliardi, G., Swanson, P. E., Birnbaum, E. H., et al. (1998). Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J. Clin. Oncol. 16, 427–433. doi: 10.1200/JCO.1998.16.2.427
Oh, H., and Irvine, K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088. doi: 10.1242/dev.015255
Pan, D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505. doi: 10.1016/j.devcel.2010.09.011
Pantalacci, S., Tapon, N., and Leopold, P. (2003). The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927. doi: 10.1038/ncb1051
Park, J. S., Park, M. C., Lee, K. Y., Goughnour, P. C., Jeong, S. J., Kim, H. S., et al. (2018). Unique N-terminal extension domain of human asparaginyl-tRNA synthetase elicits CCR3-mediated chemokine activity. Int. J. Biol. Macromol. 120(Pt A), 835–845. doi: 10.1016/j.ijbiomac.2018.08.171
Park, M. C., Kang, T., Jin, D., Han, J. M., Kim, S. B., Park, Y. J., et al. (2012). Secreted human glycyl-tRNA synthetase implicated in defense against ERK-activated tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 109, E640–E647. doi: 10.1073/pnas.1200194109
Park, S. G., Schimmel, P., and Kim, S. (2008). Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049. doi: 10.1073/pnas.0802862105
Quevillon, S., and Mirande, M. (1996). The p18 component of the multisynthetase complex shares a protein motif with the beta and gamma subunits of eukaryotic elongation factor 1. FEBS Lett. 395, 63–67. doi: 10.1016/0014-5793(96)01005-8
Rho, S. B., Kim, M. J., Lee, J. S., Seol, W., Motegi, H., Kim, S., et al. (1999). Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. U.S.A. 96, 4488–4493. doi: 10.1073/pnas.96.8.4488
Rubin, G. M., and Spradling, A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353. doi: 10.1126/science.6289436
Segev, N., and Hay, N. (2012). Hijacking leucyl-tRNA synthetase for amino acid-dependent regulation of TORC1. Mol. Cell 46, 4–6. doi: 10.1016/j.molcel.2012.03.028
Tapon, N., Harvey, K. F., Bell, D. W., Wahrer, D. C., Schiripo, T. A., Haber, D., et al. (2002). salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478. doi: 10.1016/s0092-8674(02)00824-3
Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C., and Halder, G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920. doi: 10.1038/ncb1050
Uhlen, M., Zhang, C., Lee, S., Sjostedt, E., Fagerberg, L., Bidkhori, G., et al. (2017). A pathology atlas of the human cancer transcriptome. Science 357:eaan2507. doi: 10.1126/science.aan2507
Wakasugi, K., Slike, B. M., Hood, J., Otani, A., Ewalt, K. L., Friedlander, M., et al. (2002). A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 173–177. doi: 10.1073/pnas.012602099
Wu, S., Huang, J., Dong, J., and Pan, D. (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456. doi: 10.1016/s0092-8674(03)00549-x
Xu, T., Wang, W., Zhang, S., Stewart, R. A., and Yu, W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063.
Xu, X., Shi, Y., Zhang, H. M., Swindell, E. C., Marshall, A. G., Guo, M., et al. (2012). Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 3:681. doi: 10.1038/ncomms1686
Yu, F. X., Zhao, B., and Guan, K. L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828. doi: 10.1016/j.cell.2015.10.044
Yu, Z., Vodanovic-Jankovic, S., Ledeboer, N., Huang, S. X., Rajski, S. R., Kron, M., et al. (2011). Tirandamycins from Streptomyces sp. 17944 inhibiting the parasite Brugia malayi asparagine tRNA synthetase. Org. Lett. 13, 2034–2037. doi: 10.1021/ol200420u
Yue, T., Tian, A., and Jiang, J. (2012). The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Dev. Cell 22, 255–267. doi: 10.1016/j.devcel.2011.12.011
Zanconato, F., Cordenonsi, M., and Piccolo, S. (2016). YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803. doi: 10.1016/j.ccell.2016.05.005
Zhao, B., Li, L., Lei, Q., and Guan, K. L. (2010). The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874. doi: 10.1101/gad.1909210
Keywords: asparaginyl-tRNA synthetase (NRS), Salvador, Hippo signaling, tirandamycin B, tumorigenesis
Citation: Yeom E, Kwon D-W, Lee J, Kim S-H, Lee J-H, Min K-J, Lee K-S and Yu K (2020) Asparaginyl-tRNA Synthetase, a Novel Component of Hippo Signaling, Binds to Salvador and Enhances Yorkie-Mediated Tumorigenesis. Front. Cell Dev. Biol. 8:32. doi: 10.3389/fcell.2020.00032
Received: 13 November 2019; Accepted: 15 January 2020;
Published: 05 February 2020.
Edited by:
Changshun Shao, Soochow University, ChinaReviewed by:
Gongping Sun, Shandong University, ChinaCopyright © 2020 Yeom, Kwon, Lee, Kim, Lee, Min, Lee and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyu-Sun Lee, ZWt1c2U3NEBrcmliYi5yZS5rcg==; Kweon Yu, a3dlb255dUBrcmliYi5yZS5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.