
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 23 January 2020
Sec. Mitochondrial Research
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00004
This article is part of the Research TopicMitochondrial Genetics and EpigeneticsView all 13 articles
In human beings, there is a ∼16,569 bp circular mitochondrial DNA (mtDNA) encoding 22 tRNAs, 12S and 16S rRNAs, 13 polypeptides that constitute the central core of ETC/OxPhos complexes, and some non-coding RNAs. Recently, mtDNA has been shown to have some covalent modifications such as methylation or hydroxylmethylation, which play pivotal epigenetic roles in mtDNA replication and transcription. Post-translational modifications of proteins in mitochondrial nucleoids such as mitochondrial transcription factor A (TFAM) also emerge as essential epigenetic modulations in mtDNA replication and transcription. Post-transcriptional modifications of mitochondrial RNAs (mtRNAs) including mt-rRNAs, mt-tRNAs and mt-mRNAs are important epigenetic modulations. Besides, mtDNA or nuclear DNA (n-DNA)-derived non-coding RNAs also play important roles in the regulation of translation and function of mitochondrial genes. These evidences introduce a novel concept of mitoepigenetics that refers to the study of modulations in the mitochondria that alter heritable phenotype in mitochondria itself without changing the mtDNA sequence. Since mitochondrial dysfunction contributes to carcinogenesis and tumor development, mitoepigenetics is also essential for cancer. Understanding the mode of actions of mitoepigenetics in cancers may shade light on the clinical diagnosis and prevention of these diseases. In this review, we summarize the present study about modifications in mtDNA, mtRNA and nucleoids and modulations of mtDNA/nDNA-derived non-coding RNAs that affect mtDNA translation/function, and overview recent studies of mitoepigenetic alterations in cancer.
Epigenetics is the study of mitotically and/or meiotically heritable phenotype alterations that do not entail a change in DNA sequence (Wu and Morris, 2001). Epigenetics in nuclear genome (nDNA) has been well described and characterized. Generally, epigenetic regulation contains three levels of biological actions, including covalent modifications in DNA bases, histone variants, post-translational modifications of histones, RNA modifications, and non-coding RNA (ncRNA) modulations (Dupont et al., 2009; Peschansky and Wahlestedt, 2014). Epigenetic regulation has been shown to be an important biological process that participates in tumorigenesis and cancer development, and epigenetic biomarkers or targets can be used in diagnosis, prognosis, and treatment of these diseases (Kondo et al., 2017; Dong and Cui, 2018; Nebbioso et al., 2018; Zhu et al., 2019).
As cellular organelles present in almost all eukaryotic cells, mitochondria are places where ATP is biosynthesized and are essential for various cellular biological processes, including reactive oxygen species (ROS) generation, intracellular Ca2+ signaling, heme metabolism, intrinsic apoptosis, mitophagy, metabolism and cell cycle progression (Yien et al., 2014; Picard et al., 2016; van der Bliek et al., 2017; Gómez-Durán et al., 2018). Genetic mutations in mitochondrial DNA (mtDNA) and perturbations in mitochondrial proteins can cause dysfunction of mitochondria that has been shown to be tightly associated with many mitochondrial diseases and cancer (Taylor and Turnbull, 2005; Garone et al., 2012a, b, 2013; Gómez-Durán et al., 2012; Ronchi et al., 2012; Kullar et al., 2017; Chinnery and Gómez-Durán, 2018; Garone and Viscomi, 2018; Andreazza et al., 2019). Therefore, mitochondria are promising targets for the treatment of these diseases (Gómez-Durán et al., 2010; Zong et al., 2016; Dong et al., 2019). Recently, in addition to mitogenetics, epigenetics in mitochondria (mitoepigenetics) also emerges as an important regulatory mode that is related to human physiology and disorders, such as stemness, drug addiction, neurodegenerative diseases, cardiovascular diseases and metabolic diseases (Wallace, 1992; Gómez-Durán et al., 2010; Manev and Dzitoyeva, 2013; Kalani et al., 2014; Sadakierska-Chudy et al., 2014; Ghosh et al., 2015; Gao D. et al., 2017; Stimpfel et al., 2018; Coppede and Stoccoro, 2019). Importantly, this kind of mode of action has also emerged to be associated with cancers (Ferreira et al., 2015; Lozano-Rosas et al., 2018). However, mitoepigenetics has not been well depicted.
Herein, we define the concept of mitoepigenetics as the study of modulations occurring in the mitochondria that induce heritable phenotype alterations in mitochondria without involving a change of mtDNA sequence. Based on current studies, mitoepigenetics comprises of four levels: mtDNA methylation/hydroxylmethylation, mitochondrial nucleoid modifications, mtRNA modifications, and mtDNA-derived or nDNA-derived non-coding RNA modulations during mtDNA-encoded gene translation/function. In this article, we review the current study on mitoepigenetics and its roles in cancers, so as to provide a new sight for the diagnosis and treatment of these disorders.
mtDNA exists in the matrix or the inner membrane of mitochondria. Genome signature comparisons reveal that mtDNA is analogous to prokaryotic genome (Campbell et al., 1999). Especially, alpha-proteobacteria seems to be the most likely bacterial ancestor of the mitochondria (Gray, 2012; Muñoz-Gómez et al., 2017; Martijn et al., 2018). mtDNA has a loop structure of guanine-rich heavy chains (H-strand) and cytosine-rich light chains (L-strand) (Chinnery and Hudson, 2013). The length of mtDNA ranges from 15,000 to 17,000 bp in different species (Chinnery and Hudson, 2013). Human mtDNA is a ∼16,569 bp circular DNA that encodes 37 genes (28 on the H-strand and 9 on the L-strand), including 2 rRNAs (12S and 16S rRNAs), 22 tRNAs, and 13 proteins in the electron transport chain (ETC)/oxidative phosphorylation (OxPhos) system (Table 1 and Figure 1) (Andrews et al., 1999; Brandon et al., 2005; Chinnery and Hudson, 2013). Besides, mtDNA also contains some pseudogenes (Woischnik and Moraes, 2002; Gunbin et al., 2017) and encodes non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs) (Rackham et al., 2011) and small non-coding RNAs (sncRNAs), such as microRNAs (Lung et al., 2006; Barrey et al., 2011; Bandiera et al., 2013; Ro et al., 2013; Duarte et al., 2014). Mitochondrial genome has some unique genetic characteristics, including high mutation, heteroplasmy, threshold effect, maternal inheritance and mitotic segregation (Wallace, 2005). The copy number of mtDNA varies between 100 and 10 000 per cell dependent upon cellular energy demand (Srinivasan et al., 2017).
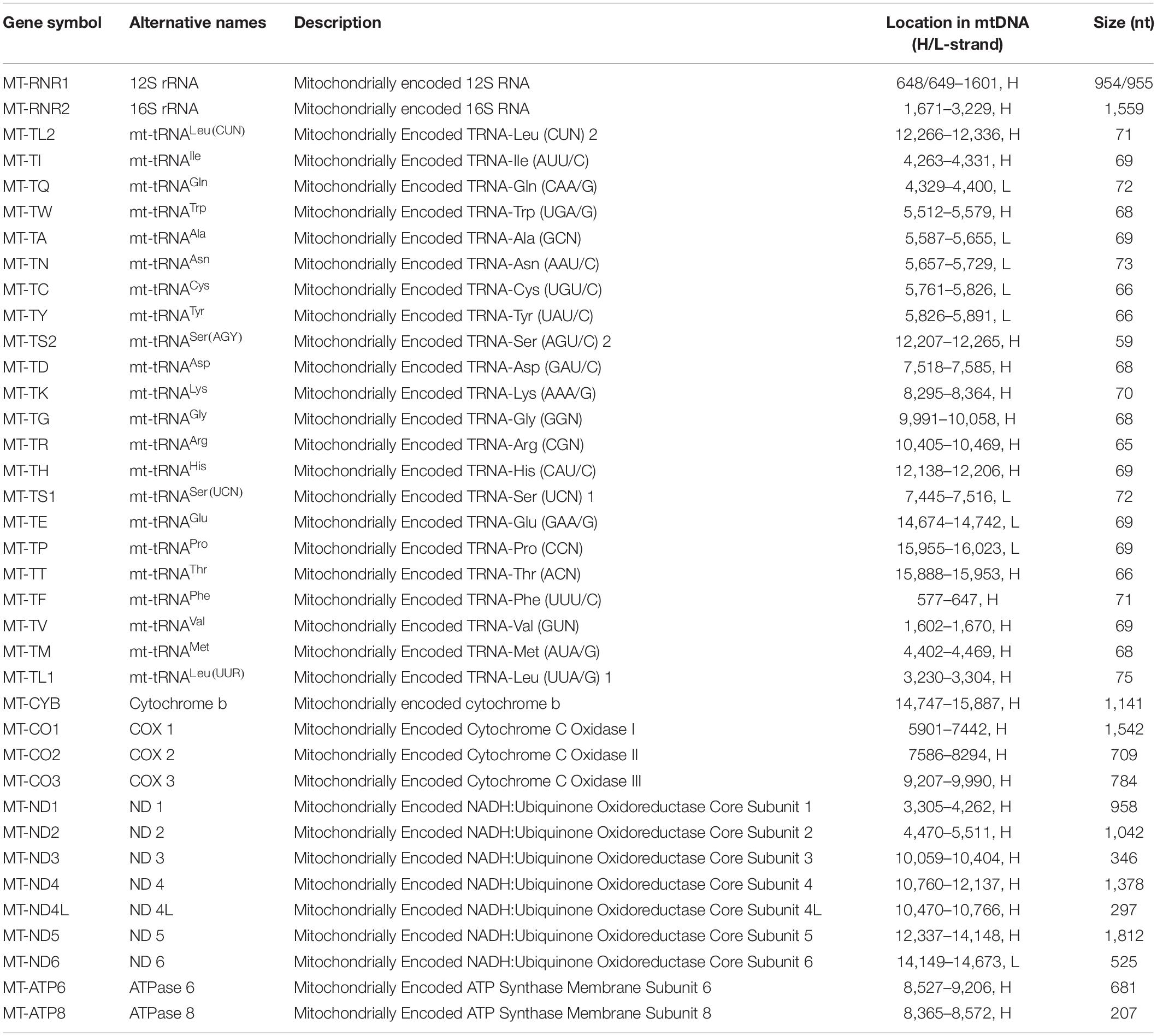
Table 1. Protein-coding and RNA genes that encoded by human mtDNA (Pseudogenes and non-coding RNA genes are not included in this table).
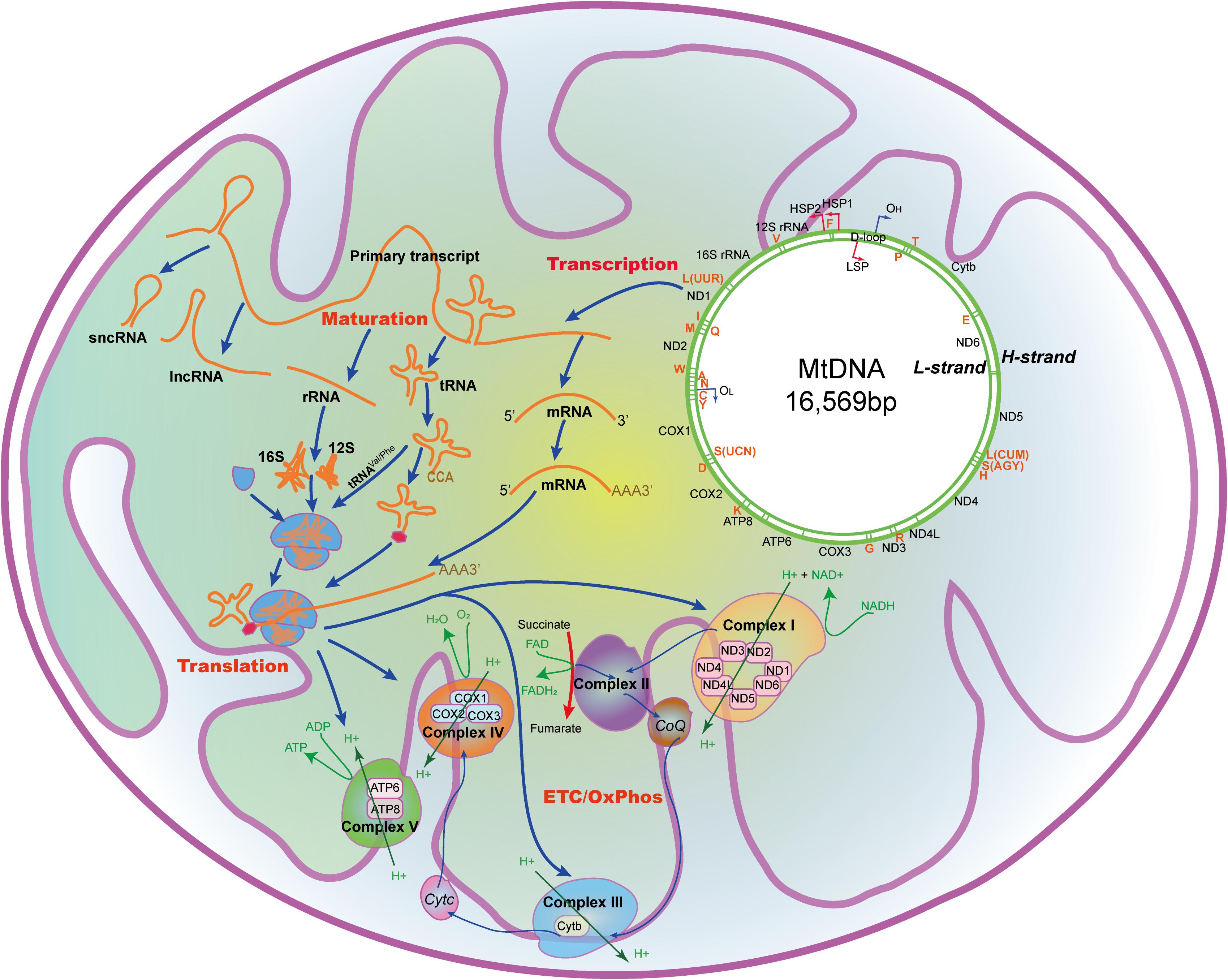
Figure 1. The mtDNA and the processing and function of its encoding genes in the mitochondria. mtDNA with 16,569 nucleotides encodes 22 tRNAs, 2 rRNAs, 13 peptides that constitutes the ETC/OxPhos, and some non-coding RNAs. A, mt-tRNAAla; C, mt-tRNACys; D, mt-tRNAAsp; E, mt-tRNAGlu; F, mt-tRNAPhe; G, mt-tRNAGly; H, mt-tRNAHis; I, mt-tRNAIle; K, mt-tRNALys; L(CUN), mt-tRNALeu(CUN); L(UUR), mt-tRNALeu(UUR); M, mt-tRNAMet; N, mt-tRNAAsn; P, mt-tRNAPro; Q, mt-tRNAGln; R, mt-tRNAArg; S(AGY), mt-tRNASer(AGY); S(UCN), mt-tRNASer(UCN); T, mt-tRNAThr; V, mt-tRNAVal; W, mt-tRNATrp; Y, mt-tRNATyr; ATP6/8, Mitochondrially encoded ATP synthase membrane subunit 6/8; CoQ, Coenzyme Q; COX1/2/3, Mitochondrially encoded cytochrome C oxidase I/II/III; Cytb, Cytochrome b; Cytc, Cytochrome c; ETC, Electron transport chain; FAD, Flavine adenine dinucleotide; FADH2, Flavine adenine dinucleotide, reduced; HSP1/2, H-strand promotor 1/2; H-strand, Heavy strand; LSP, L-strand promotor; L-strand, Light strand; NAD+, Nicotinamide adenine dinucleotide; NADH, Nicotinamide adenine dinucleotide, reduced; ND1/2/3/4/4L/5/6, Mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1/2/3/4/4L/5/6; OL, L-strand origin of replication; OH, H-strand origin of replication; OxPhos, Oxidative phosphorylation.
Unlike nDNA, mtDNA contains only one non-coding triple-helical region (16,024-576, ∼1000 bp, ∼7% of all sequence), the displacement loop (D-loop) formed by abortive initiation of replication (Yamamoto, 2001). Both replication and transcription are initiated from D-loop, which contains the H/L-strand promotor (HSP1/LSP1), and the H-strand origin of replication (OH) (Jemt et al., 2015). Sixty bp upstream of HSP1, there is a HSP2. There are also some specific sites (ITL, ITH1, ITH2) within the promotors, where mtDNA transcription initiates. Mitochondrial RNAs are firstly transcribed as primary transcripts, which are subsequently cleaved by enzymes and affected by specific nucleotide modifications to yield polycistronic precursors and finally mature RNAs (Van Haute et al., 2015). Besides, like prokaryotic cells, mtDNA lacks intronic regions, and intergenic sequences are either absent or only a few nucleotide bases long. Some genes, such as MT-ATP6/8 and MT-ND4/4L, have overlapping regions.
Although mitochondrial genome has a diminutive size, mutations in mtDNA occur frequently, because mtDNA lacks the error checking system that nDNA has. Some of mutations are tightly associated with inherited diseases (Taylor and Turnbull, 2005). Apart from mtDNA mutations, mtDNA is also shown to have some modifications, like that in nDNA.
As shown in Figure 2, DNA can be methylated by methyltransferases, which can transfer a methyl group from a methyl donor S-adenosylmethionine (SAM) onto the C5 position of the cytosine to form 5-methylcytosine (5mC) by the aid of DNA methyltransferases (DNMTs) such as DNMT1, DNMT3A, and DNMT3B (Moore et al., 2013). Methylation leads to changes of the molecular structure in DNA and affects transcription factor, RNA polymerase, topoisomerase, or inhibitory protein binding to DNA, resulting in dysfunction of gene transcription and expression (Moore et al., 2013). DNA 5mC can also be demethylated via either passive demethylation carried out by dilution by replication without de novo methylation or active demethylation carried out by oxidation or deamination.
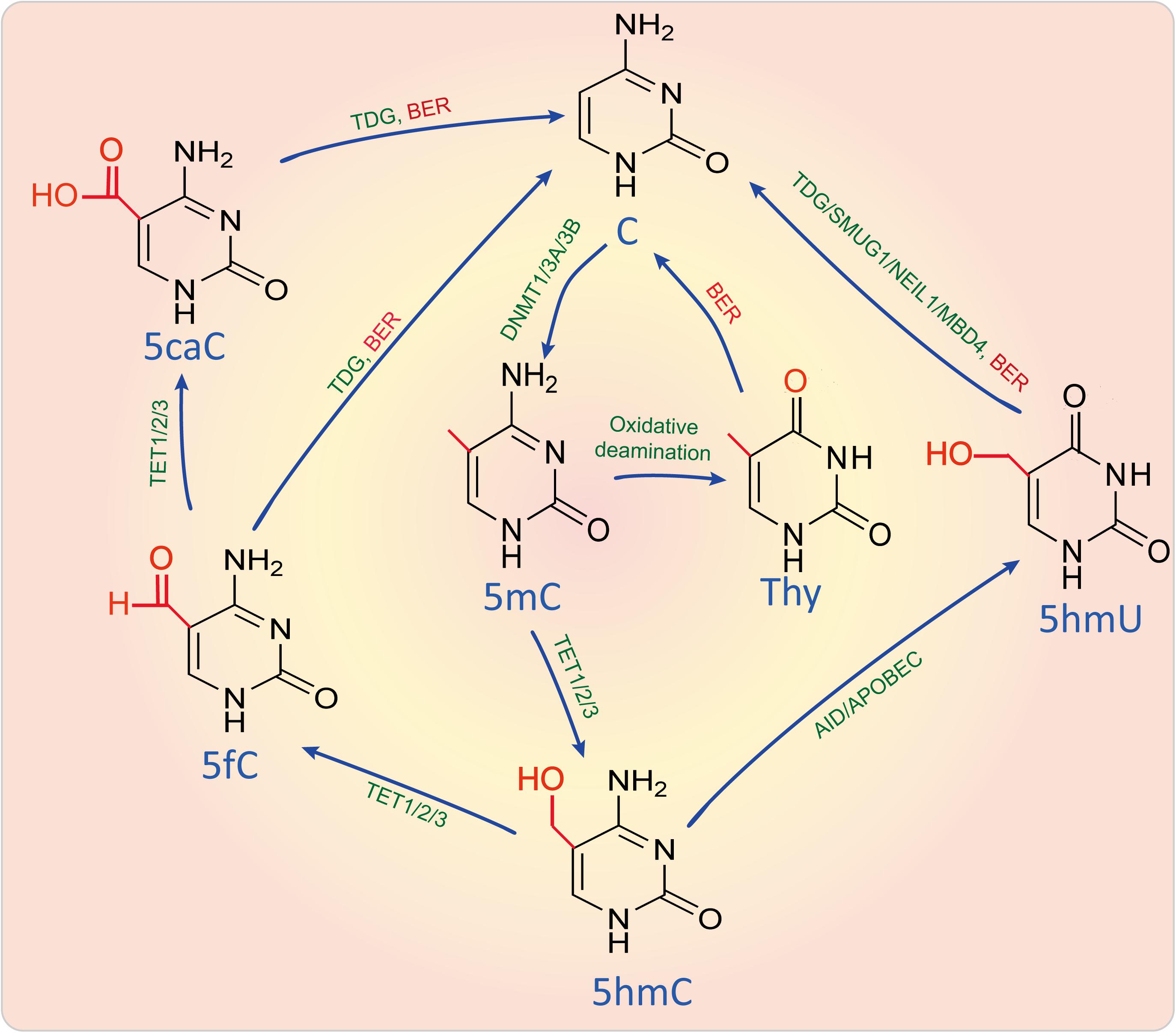
Figure 2. DNA methylation and active demethylation. DNA can be methylated by DNMTs and demethylated by active demethylation through oxidizing, deaminating and base-excision repair. Enzymes were marked in green, metabolites were marked in blue, while biological process like BER was marked in red. 5caC, 5-Carboxylcytosine; 5fC, 5-Formylcytosine; 5hmC, 5-Hydroxymethylcytosine; 5hmU, 5-Hydroxymethyluracil; 5-AID, activation induced cytidine deaminase; APOBEC, Apolipoprotein B mRNA editing enzyme catalytic subunit; BER, Base-excision repair; DNMT1/3A/3B, DNA methyltransferase 1/3A/3B; MBD4, Methyl-CpG binding domain 4, DNA glycosylase; NEIL1, Nei like DNA glycosylase 1; SMUG1, Single-strand-selective monofunctional uracil-DNA glycosylase 1; TET1/2/3, Tet methylcytosine dioxygenase 1/2/3; C, Cytosine; TDG, Thymine DNA glycosylase; Thy, Thymine.
During active demethylation pathway, some of 5mC sites can also be catalyzed and oxidized by 2-oxoglutarate and Fe(II)-dependent oxygenases of the ten-eleven-translocation (TET) proteins, including TET1, TET2, and TET3, to form 5-hydroxymethylcytosine (5hmC), which is considered as a possible intermediate in a replication-independent DNA demethylation pathway (Richa and Sinha, 2014). 5hmC is enriched in active genes that have a strong depletion of 5mC (Mellen et al., 2012). With the aid of TET1/2/3, 5hmC is further catalyzed into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which can be subsequently excised and replaced via base excision repair (BER). Besides, 5mC and 5hmC can also be deaminated to yield thymine and 5-hydroxymethyluracil (5hmU) by the aid of activation induced cytidine deaminase (AID)/apolipoprotein B mRNA editing enzyme and catalytic polypeptide (APOBEC). This results in a thymine-guanine mismatch that can lead to a DNA repair in which thymine and 5hmU can be replaced by unmethylated cytosine (Jang et al., 2017). However, 5hmC seems to be not only the intermediate of DNA demethylation, but is also a major element in the modulation of chromatin structure and gene expression through binding with methyl-CpG-binding protein 2 (MeCP2) (Mellen et al., 2012).
With the development of technology for detecting methylation, this kind of modification was also found in mtDNA. Distribution of 5mC seems to be conserved in mitochondrial genomes across all cell and tissue types (Ghosh et al., 2014). mtDNA methylation is usually found within the non-coding D-loop and gene start sites (GSS) (Mposhi et al., 2017), implying that methylation in mtDNA can affect mtDNA replication and transcription. Stimulating mtDNA replication results in increasing methylation (Rebelo et al., 2009), confirming that methylation can also be a feedback regulatory mode that maintains mtDNA copy number.
CpG dinucleotides are the most prominent regions where methylation occurs, however, non-CpG sites, such as CpA, CpT, and CpC also have methylations (Jang et al., 2017). The abundance of CpG sites varies in animal, fungal, protist, and plant mitochondrial genomes. Like nDNA, human mtDNA contains a relatively low frequency of CpG sites (435 in 16 659 nucleotides, 2.61%) (Cardon et al., 1994). Methylation of CpG in the H-strand promoter (HSP1) induces TFAM multimerization to augment cooperativity and enhances its binding affinity to mtDNA, compared to that of the non-methylated DNA. Although TFAM-dependent DNA compaction is not affected by methylation of CpG sites, transcription initiation from the three mitochondrial promoters is significantly impaired by CpG methylation (Dostal and Churchill, 2019). However, a study shows that mtDNA methylation mainly occurs within non-CpG sites of the promoter region of the H-strand, which is essential for mtDNA replication and transcription (Bellizzi et al., 2013).
5mC in mtDNA is catalyzed by mtDNMT1, an isoform of DNMT1. mtDNMT contains a mitochondrial targeting sequence, which can make it translocated into mitochondria (Shock et al., 2011). However, DNA methyltransferases seem to contribute to CpG methylation in the D-loop, while not non-CpG methylation (Bellizzi et al., 2013), because MtDNMT1 binding to the mtDNA is observed to be associated with the density of CpG sites (60).
5hmC is also found in mtDNA and it seems to promote demethylation through impairing mtDNMT1-mediated remethylation during replication (Manev and Dzitoyeva, 2013). However, the detailed process and the enzymes involved are not identified yet.
Similarly to nDNA, mtDNA is also packed by proteins to form a protein-DNA structure referred to as a nucleoid. It is located in mitochondrial pseudocompartments, and some of its proteins may play histone-like architectural roles (Kanki et al., 2004). There are more than 50 nucleoid-associated proteins that can either temporarily or permanently associate with mtDNA or other nucleoid-associated proteins to maintain mtDNA and regulate gene expression (Figure 3). In nucleoid, mitochondrial transcription factor A (TFAM), mitochondrial polymerase γ (POLG), ATPase family AAA-domain-containing protein 3 (ATAD3), mitochondrial AAA protease (LONP1), and mitochondrial single-stranded DNA-binding protein (mtSSB) possibly directly interact with the D-loop region of mtDNA (Lee and Han, 2017). Mitochondrial RNA polymerase (POLRMT), TFAM, mitochondrial transcription factor B2 (TFB2M) and mitochondrial transcriptional elongation factor (TEFM) are key components of the mitochondrial transcription (Shokolenko and Alexeyev, 2017). POLG, Twinkle, mtSSB are key components of the mitochondrial replication (Young and Copeland, 2016). Besides them, nucleoid-associated proteins also include RNA helicases (e.g., SUV3), RNA-binding proteins (e.g., FASTKD2), quality-control proteases (e.g., lon-like peptidase LONP1 and caseionlytic peptidase CLPXP), as well as mitochondrial RNA processing proteins (Koc and Spremulli, 2003; Szczesny et al., 2013; Popow et al., 2015; Levytskyy et al., 2016; Lee and Han, 2017). These evidences suggest that nucleoid may be a place where mtRNAs are processed and mitoribosomes are assembled. Recently, post-transcriptional mtRNA processing and ribosome biogenesis including, mtRNA maturation, ribosome assembly, and translation initiation may occur within mitochondrial RNA granules (MRGs), dynamic structures that juxtapose to nucleoids (Jourdain et al., 2013; Antonicka and Shoubridge, 2015; Hensen et al., 2019). MRGs are transiently associated with active nucleoids where they are assembled around the newly synthesized primary transcripts, then MRGs become detached and locate within the inner mitochondrial matrix and subsequent events in mitochondrial gene expression take place (Jourdain et al., 2016).
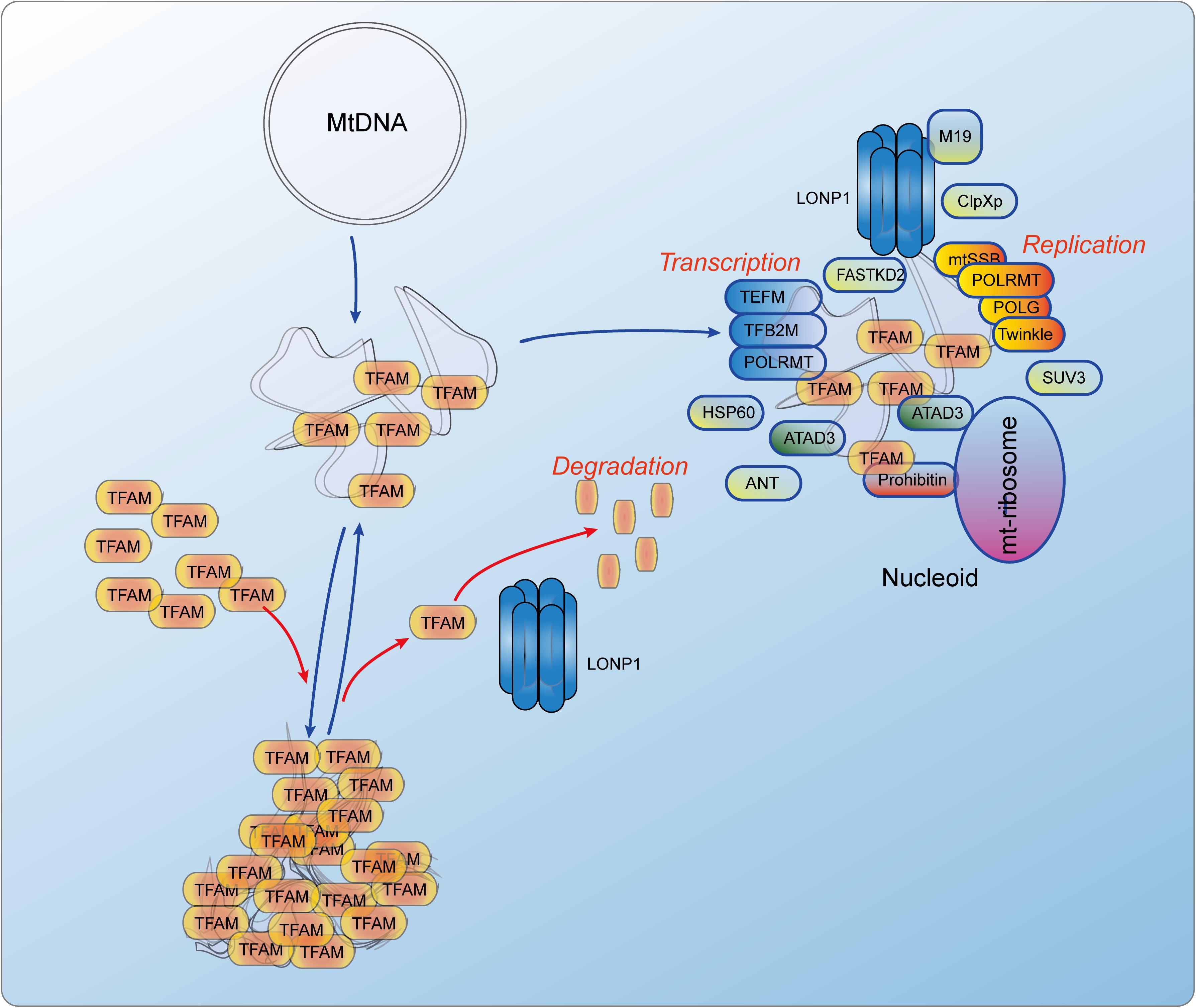
Figure 3. The dynamics of TFAM controlled mitochondrial nucleoid and the constitutions of nucleiod. TFAm can directly bind to mtDNA and functions as a histone-like protein. Its degradation is mediated by LONP1, an AAA + Lon protease. There are more than 50 nucleoid-associated proteins including POLRMT, TFAM, TFB2M and TEFM that initiate the mtDNA transcription, and POLG, Twinkle and mtSSB that initiate the mtDNA replication. There are also some proteins that are related to RNA processing and nucleoid regulation. ANT, Adenine nucleotide translocator; ATAD3, ATPase family AAA domain containing 3; ClpXp, ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial; FASTKD2, FAST kinase domains 2; HSP60, Short heat shock protein 60; LONP1, Lon peptidase 1, mitochondrial; M19, Mitochondrial protein M19; mtSSB, Single-stranded DNA binding protein 1, mitochondrial; POLG, DNA polymerase gamma; POLRMT, RNA polymerase mitochondrial; SUV3, ATP-dependent RNA helicase SUV3, mitochondrial; TEFM, Transcription elongation factor, mitochondrial; TFAM, Transcription factor A, mitochondrial; TFB2M, Transcription factor B2, mitochondrial.
TFAM is a member of the high-mobility group domain proteins family and can form a U-turn with an overall bend of 180° on unspecific mtDNA sequence, functioning as a transcription and packaging factor (Ngo et al., 2011). In mammalian cells, this protein is very abundant. Per each mtDNA molecule, there are about 1,000 molecules of TFAM protein, which means that there is a TFAM molecule in every 16 bp of mtDNA (Farge and Falkenberg, 2019). Importantly, only with TFAM nucleoid compaction of mtDNA can sufficiently complete (Kaufman et al., 2007). In addition, single TFAM protein can also bridge neighboring mtDNA duplexes to form a cross-strand binding and looping out (Kukat et al., 2015). In the condition of high TFAM/mtDNA ratio, the combination of duplex bending and cross-strand binding result into mtDNA full compaction, which leads to the blockade of mtDNA transcription and replication (Ngo et al., 2014). Besides, TFAM can also act as a homodimer, which promotes looping of the DNA (Ngo et al., 2014). In summary, TFAM is the only nucleoid-associated protein that can stringently fulfill the criteria of a true mtDNA packaging factor (Bonekamp and Larsson, 2018). Unbalanced levels (low or high) of TFAM result in decreasing mtDNA methylation (Rebelo et al., 2009).
Nucleoids have a large size and are dynamically distributed throughout the mitochondrial network. Therefore, nucleoids are unlikely to move freely within mitochondrial matrix (Bonekamp and Larsson, 2018). It may be anchored at the inner membrane of mitochondria and its distribution may depend on mitochondrial fusion and fission (Elgass et al., 2013). There are evidences showing that deficiency of the large GTPase dynamin-related protein 1 (Drp1), a major regulator during mitochondrial fission, leads to remodeling of nucleoid clustering (Ban-Ishihara et al., 2013). Mitochondrial topoisomerase 3a (Top3a) also plays an essential role in genome separation and nucleoid distribution because it can deconcatenate newly replicated mtDNA (Goffart et al., 2019).
The protein scaffold of mtDNA nucleoids can also be epigenetically and post-translationally modified, just like the histones in the nDNA nucleosomes. There are more than 50 nucleoid-related proteins, in which TFAM is the only protein whose behavior is highly similar with that of histones. Therefore, mitochondrial nucleoid modifications refer to TFAM modifications. Based on current studies, TFAM can be acetylated, phosphorylated and ubiquitinated, thereby affecting its function in mtDNA packaging.
A recent study showed that TFAM is lysine acetylated within its high-mobility-group box 1 (HMGB1), which reduces TFAM to interact with non-specific DNA through distinct kinetic pathways (King et al., 2018). Another study showed that TFAM was acetylated at a single lysine residue and the level of acetylation in rat liver did not change with age (Dinardo et al., 2003). SIRT3 is the deacetylase that mediates the deacetylation of K154 of TFAM (Bagul et al., 2018; Liu et al., 2018).
Serine phosphorylation in HMGB1 domain of TFAM also regulates mtDNA transcription by blocking TFAM to locate at the promoter sites of target genes (King et al., 2018). Extracellular signal-regulated protein kinases (ERK1/2) can mediate phosphorylation of serine 177 in TFAM, thereby downregulating mitochondrial transcription (Wang et al., 2014). The AAA + LONP1, an ssDNA-binding protein, is responsible for the degradation of DNA-free TFAM (Chen et al., 2008). When Lon binds to heavy-strand sequences upstream of light-strand promoter (LSPHS) or dsDNA-TFAM, its protease activity is directly blocked, resulting in TFAM stabilization (Liu et al., 2004). When TFAM is phosphorylated within its HMGB1 domain by cAMP-dependent protein kinase in mitochondria, the ability of TFAM to bind DNA is impaired and gene transcription is activated, then TFAM is degraded by LONP1 (Lu et al., 2013).
In addition to the modifications above, TFAM can also be ubiquitinated and its stability or sub-location may be affected. For instance, in the retina from diabetic rats, high glucose can lead to TFAM ubiquitination, which impedes its transport to the mitochondria, resulting in subnormal mtDNA transcription and mitochondria dysfunction (Santos et al., 2014).
RNA modifications are present in almost all the cellular RNAs in archaeobacteria, bacteria, plants, fungi, and animals. There are almost 150 kinds of modifications that have been found in RNA (Boccaletto et al., 2018). Mitochondrial RNAs (mtRNAs) are processed from single polycistronic precursor RNAs, however, there are also multiple different types of mt-RNAs, including 2 rRNAs, 22 tRNAs, 13 mRNAs and many ncRNAs, present in the mitochondrial matrix. This suggests that the post-transcriptional mechanisms are very important for the translation, maturation, stability and assembly of mtRNAs.
A study on m6A patterns in the transcriptomes of Arabidopsis mitochondria and chloroplast reveals that more than 86% of the transcripts are m6A methylated. Over 350 m6A sites were with ∼4.6 to ∼4.9 m6A sites per transcript are identified in mitochondrial genome. The extent of overall m6A methylation in mitochondria is much higher than that in the nucleus, but lower than that in the chloroplast. However, the m6A motif sequences in the transcriptome of mitochondria are similar to those of the nucleus and chloroplast, which means that m6A motif is conserved among them. Besides, the m6A patterns of rRNAs and tRNAs are also similar. However, the mitochondrial and chloroplastic m6A patterns in mRNAs are different from those of the nucleus. Methylated transcripts in mitochondria and chloroplast are shown to be associated with rRNA, ribosomal proteins, photosystem reaction proteins, tRNA, NADH dehydrogenase and redox systems. Different organs of the leaves, flowers and roots have differential m6A methylation, suggesting that m6A methylation plays an important role during development and differentiation (Wang et al., 2017). Besides which, more than 20 m1A sites are also identified in mitochondrial genes via using a transcriptome-wide analysis (Zhang and Jia, 2018).
These findings unravel a new concept of mitoepitranscriptome, referring to dynamic regulation of gene expression by the modified mtRNAs. Some mutations in mtDNA or nuclear-encoded mitochondrial modification enzymes can cause RNA modification defects, which are shown to be associated with various mitochondrial diseases. mtRNA modifications in mt-mRNAs, mt-rRNAs and mt-tRNAs are suspected to play essential roles, too. Besides, ncRNAs encoded by mtDNA may also be modified as that reported in the nDNA-derived ncRNAs (Romano et al., 2018). However, reports about mt-ncRNA modifications are absent at present, because mt-ncRNAs are not well identified yet.
The protein/DNA ratio of mitoribosomes is higher than that of all other ribosomes, indicating that mitochondria need a more stable structure to make sure that mt-rRNA is correctly scaffolded and accurately folded (Greber and Ban, 2016). Therefore, mt-rRNA modifications seem to be important for mitochondria. Currently, there are only eight different types of modifications in 10 nucleotide sites (including m5U429, m4C839, m5C841, m62A936, m62A937 in 12S rRNA and m1A947, Gm1145, Um1369, Gm1370 and ψ1397 in 16S rRNA) identified in mammalian mt-rRNAs (reviewed by Bohnsack and Sloan, 2018), which is lower than that of cytoplasmic and bacterial rRNAs. These sites are clustered at peptidyl transferase center (PTC) in 16S rRNA and decoding (DSC) sites in 12S rRNA, respectively, which are similar with the ribosomal modification features in bacteria and eukaryotic cytoplasm (Amunts et al., 2015).
The study of mitochondrial rRNA modification can be traced since 1970s. A study of methylation in a fungus Neurospora crassa shows that the mitochondrial rRNAs have 0.05–0.16 methyl groups per 100 nucleotides (Lambowitz and Luck, 1975). The 25S and 19S rRNAs of this species have methyl contents of approximately 70 and 55%, respectively (Kuriyama and Luck, 1974). Unlike highly modified cytoplasmic rRNA, mitochondrial rRNAs (15S and 21S) of the yeast Saccharomyces cerevisiae only contain three modified nucleotides: a pseudouridine (Ψ2918) and two 2′-O-methylated riboses (Gm2270 and Um2791) located at the peptidyl transferase center of 21S rRNA. Mrm2p, a yeast nuclear genome encoding mitochondrial protein, is required for methylating U2791 of 21S rRNA. Mrm2p belongs to a new class of three eukaryotic RNA-modifying enzymes and is the ortholog of Escherichia coli FtsJ/RrmJ that can methylate a nucleotide of the peptidyl transferase center of 23S rRNA (Pintard et al., 2002). Nuclear gene-encoded PET56 catalyzes the site-specific formation of 2’-O-methylguanosine on in vitro transcripts of both Saccharomyces cerevisiae mitochondrial large ribosomal RNA (21S rRNA) and Escherichia coli 23S rRNA. This modification is essential for the formation of functional large subunits of the mitoribosome (Sirum-Connolly and Mason, 1993).
In mitochondrial ribosomes of mammals, such as hamster, the large ribosomal subunit RNA (17S rRNA) contains UmpGmpUp, in which Um residue is methylated relatively later than Gm residue (Dubin and Taylor, 1978). The small ribosomal subunit RNA (13S rRNA) contains, on average, approximately one residue of m4Cp, m5Cp and m5Up, and two residues of m26Ap. m4Cp in 13 S rRNA is homologous to its ribose-methylated congener, m4Cmp of bacterial 16S ribosomal RNA. Neither m4Cp nor m4Cmp exists in cell cytoplasmic ribosomal RNA (Dubin et al., 1978).
There are three rRNA 2′-O-methyltransferase family members RNMTL1, MRM1, and MRM2 in mammals. MRM1 and MRM2 are bacterial and yeast homologs, whereas RNMTL1 is only found in eukaryotes. They also localize to the mitochondria, especially near mtDNA nucleoids. MRM1, MRM2, and RNMTL1 are responsible for modification of G1145, U1369, and G1370 residues of human 12S rRNA, respectively (Lee and Bogenhagen, 2014). Defective MRM2 can lead to mitochondrial encephalopathy, lactic acidosis and stroke-like episodes (MELAS)-like clinical syndrome possibly through reducing of the 2′-O-methyl modification at specific uracil position of 12S rRNA (Garone et al., 2017).
However, mitochondrial 16S rRNA can also be methylated (m1A) by tRNA methyltransferase TRMT61B in all vertebrates (Bar-Yaacov et al., 2016). Pseudouridine synthase RPUSD4 plays a role in the pseudouridylation (ψ) of a single residue in the 16S rRNA, a modification that is essential for its stability and assembly into the mitochondrial ribosome (Antonicka et al., 2017). In addition to mono-methylation, the 3′ end of the rRNA of the 12S rRNA of mouse also contains two dimethylated adenines (m62A) that are extremely highly conserved. TFB1M is a mammalian mitochondrial dimethyltransferase homologous to bacterial that is responsible for these two dimethylated adenines. Loss of TFB1M is embryonic lethal and deletion of TFB1M in heart results in complete demethylation of these two adenines of the 12S rRNA, thereby impairing mitochondrial ribosome assembly and abolishing mitochondrial translation (Metodiev et al., 2009). Besides, m5C911 in the mouse 12S rRNA is catalyzed by NSUN4 without forming a complex with MTERF4, which is essential in mitochondrial ribosomal biogenesis (Metodiev et al., 2014).
Modifications in mt-rRNAs are essential for their stability to ensure the normal functions of the mitoribosome. Abnormal modifications in mt-rRNAs can be associated with the dysfunction of the mitoribosome. For instance, mitochondrial mutation 1584A 12S rRNA N6, N6-dimethyladenosine (m62A) methylation is associated with hearing loss with 1555A > G mutation (O’Sullivan et al., 2014). Lack of the two dimethylated adenines (m62A) in 12S rRNA is associated with the pathogenesis of type 2 diabetes (Koeck et al., 2011; Sharoyko et al., 2014).
The genetic code used during mammal mitochondrial gene expression is non-universal. Mitochondria use only 22 mt-tRNAs to decode 60 different codons. Therefore, flexible decoding is needed for mt-tRNAs. Moreover, mt-tRNA modifications are important processes for the biogenesis of the mature tRNA. Based on the current studies, mt-tRNA contains several types of modifications, such as m1A, m1G, m2G, m5C, m3C, τm5U, τm5s2U, f5C, Q, t6A, i6A, ms2i6A, D, m22G and ψ (summarized by Bohnsack and Sloan, 2018), each of which is essential for biogenesis of mature mt-tRNA. These modifications are mediated by different kinds of enzymes such as MRPP1/2, PUS1, GTPBP3, MTO1, MTU1, NSUN2/3, ABH1, TRIT1/5, CDK5RAP1 and TRMT61B (Bohnsack and Sloan, 2018).
For instance, m1A9 can disfavor the non-functional conformation and shifts the observed equilibrium toward the functional cloverleaf (Voigts-Hoffmann et al., 2007). RNase P, a subcomplex in human mitochondria, is the endonuclease that removes tRNA 5′ extensions and is the methyltransferase responsible for m1G9 and m1A9 formation. The ability of the mitochondrial tRNA:m1R9 (R = G/A) methyltransferase (TRMT10C and SDR5C1) to modify both purines is uncommon among nucleic acid modification enzymes. In this process, PRORP, a short-chain dehydrogenase, is required as a partner protein (Vilardo et al., 2012).
NOP2/Sun RNA methyltransferase family member 2 (NSUN2) is an RNA methyltransferase previously shown to introduce m5C in tRNAs, mRNAs and microRNAs encoded by nucleic genome. However, NSUN2 can also be imported into mitochondrial matrix and introduces m5C at positions 48, 49, and 50 of several mitochondrial tRNAs, including mt-tRNATyr, mt-tRNAHis, mt-tRNALeu(UUR), mt-tRNAPhe, and mt-tRNAGlu. However, NSUN2 inactivation does not remarkably affect mitochondrial tRNA stability and OxPhos in differentiated cells (Van Haute et al., 2019).
NSUN3, a RNA methyltransferase that localizes to mitochondria and methylates cytosine 34 (C34) at the wobble position of mt-tRNAMet via specifically recognizing the anticodon stem loop (ASL) of the tRNA. Meanwhile, a dioxygenase ALKBH1/ABH1 can oxidize m5C34 mt-tRNAMet to yield an f5C34 mt-tRNAMet. During translation initiation, mt-tRNAMet recognizes AUG, AUA and AUU codons and mediates incorporation of methionine on these codons, whereas mt-tRNAMet recognizes AUA codons and introduces methionine incorporation during elongation. In fact, mitochondrial translation factors prefer to use m5C34 mt-tRNAMet during translation initiation. NSUN3 or ABH1 depletion remarkably affects mitochondrial translation (Haag et al., 2016).
mt-tRNA modifications are essential for their normal functions. Modifications in the anticodon loop can expand the decoding capacity of mt-tRNAs and make sure the fidelity of translation. Core modifications can enable the structural stability of mt-tRNAs, however, this kind of modifications can also affect its recognition by aminoacyl-tRNA synthetases in some cases (Degoul et al., 1998).
In addition to modifications in rRNAs and tRNAs, the post-transcriptional alterations in mRNA are also important for its maturation and function, as well as regulation. There are 8 H-strand-derived mitochondrial genes (COX3, ND1/2/3/4/4L, Cytb and ATP6/8) that lack translational termination codons, but can be stopped by the addition of a polyadenine (polyA) tail to a terminal U with the help of a mitochondrial PAP poly(A) polymerase (mtPAP) and a polynucleotide phosphorylase (PNPase), thereby generating sequence (UAA)TERAn (Chang and Tong, 2012; Shokolenko and Alexeyev, 2015). Primary transcript from the H-stand has an approximately 45 nt polyA extension. However, the length of the polyA tail varies in different cell types and different transcripts (Temperley et al., 2010). PolyA in the 3′-termini can mediate the stability or instability of the transcript possibly depending on additional polyA-binding factors or sequence-specific proteins, which may affect the translation of mt-DNA-encoded genes (Rorbach and Minczuk, 2012).
Recently, a transcriptome-wide analysis revealed that other post-transcriptional modifications also exist in mitochondrial mRNAs (Bohnsack and Sloan, 2018). Intriguingly, m1A in the coding region of mitochondrial transcripts can block the corresponding protein translation (Zhang and Jia, 2018). In addition to methylation, specific residues in mitochondrial mRNAs can also be pseudouridylated by TRUB2/RPUSD3, thereby affecting mitochondrial protein synthesis and cell viability (Antonicka et al., 2017). These results mean that m1A and ψ in mt-mRNA are essential for the regulation of protein translation.
Except for mRNAs, rRNAs and tRNAs, recent reports show that mtDNA can also encode non-coding RNAs (ncRNAs), including lncRNAs and small non-coding RNAs (sncRNAs) (Table 2 and Figure 4), which are termed mtDNA-encoded lncRNAs (lncRNAsmtDNA) and mtDNA-encoded sncRNAs (sncRNAsmtDNA), respectively. However, circular RNAs (circRNAs) seem to be absent in the mitochondria (Zhang et al., 2019).
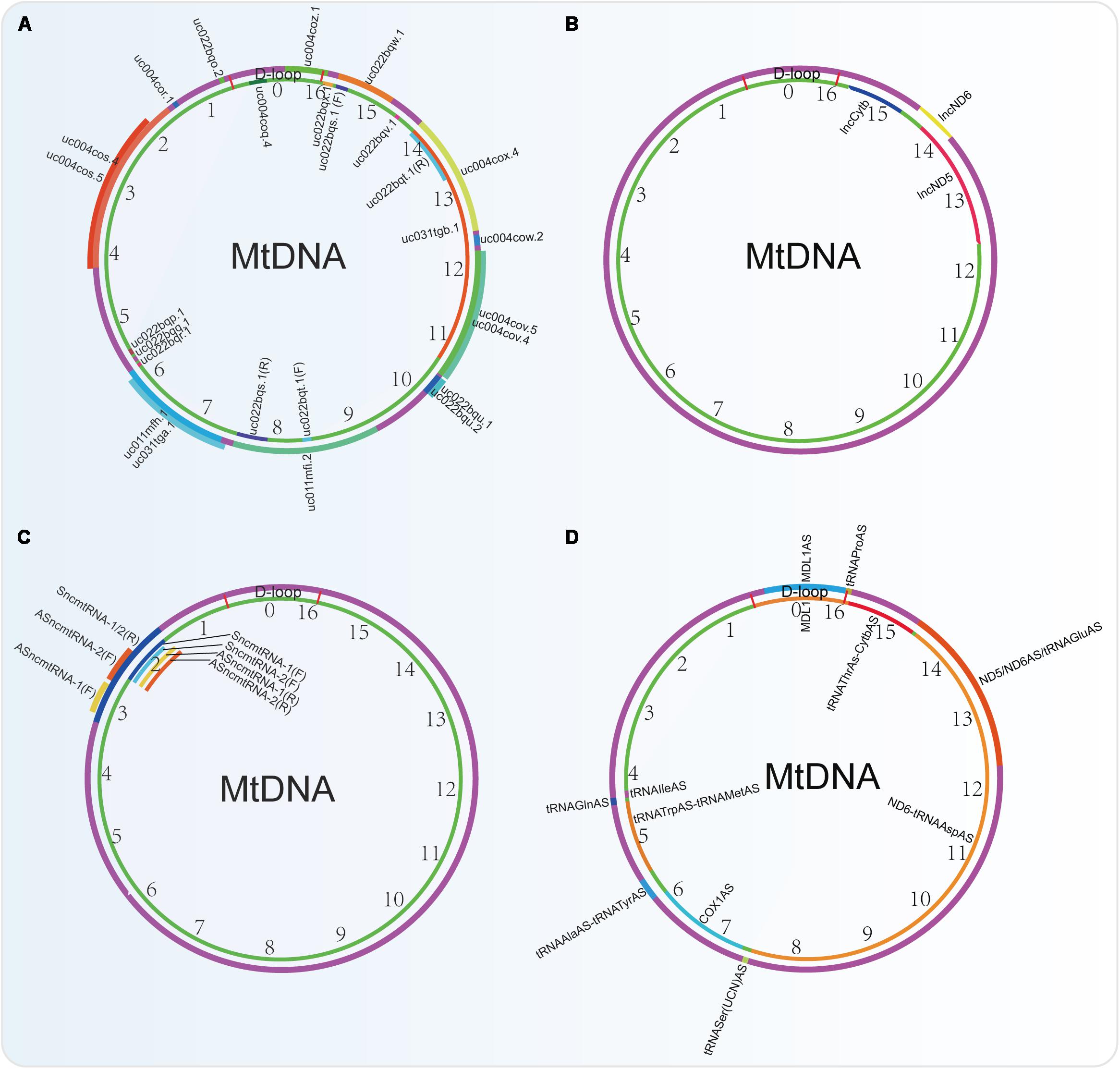
Figure 4. Non-coding RNAs encoded by the mtDNA identified by four different research groups. (A) ncRNAs obtained from the UCSC Genome Bioinformatics (2016 version). (B) lncRNAs identified by Rackham et al., 2011. (C) lncRNAs indented by researchers from Fundación Ciencia para la Vida, Chile. (D) ncRNAs identified by using PacBio full-length transcriptome data.
In rat mitochondrial genome, there are some unidentified RNAs on both the H/L-strand, such as precursors of the ND2 mRNA plus the tRNATrp and the tRNAs clustered in the Ori L region. Besides them, antisense RNA species in the region of L-strand replication and D-loop region are also observed (Sbisà et al., 1992). A deep RNA sequencing study in human cardiac tissues also shows that there is a high relative abundance (71%) of lncRNAsmtDNA (Yang et al., 2014). Another analysis in data sets from strand-specific deep sequencing shows that there is a significant proportion (15.02%, excluding rRNA and tRNA) of lncRNAs in the transcriptome of mitochondria in cervical cancer cell HeLa. Among them, 3 lncRNAs (lncND5, lncND6, lncCytb) transcripts are highly abundant (Figure 4B). These lncRNAs can form intermolecular duplexes and they are expressed in a cell- and tissue-specific manner. Their levels may be regulated by the nuclear-encoded proteins such as ELAC2, MRPP1, MRPP3, PTCD1, and PTCD2 (Rackham et al., 2011).
Recently, the PacBio full-length transcriptome data revealed that there are 6 lncRNAs in the H-strand and 6 lncRNAs in the L-strand of the mtDNA (Figure 4D). Among them, 2 novel lncRNAs of MDL1 and MDL1AS from the D-loop region exist ubiquitously in animal mtDNA (Gao et al., 2018).
However, from the UCSC Genome Bioinformatics (2016 version), 24 ncRNAs (including 6 microRNAs) derived from human mtDNA were provided, which are very different from those identified by the PacBio full-length transcriptome (Figure 4A). Actually, many researches that focus on the mtDNA-derived lncRNAs used this database to perform their studies. Among them, the mitochondrial lncRNA uc022bqs.1 (long intergenic non-coding RNA predicting cardiac remodeling, LIPCAR) is the best studied. This lncRNA is shown to be downregulated early after myocardial infarction but upregulated during later stages in the circulating blood of the patients (Kumarswamy et al., 2014). In addition, circulating LIPCAR also acts as a biomarker in patients with ST-segment elevation myocardial infarction (Li et al., 2018). Circulating LIPCAR is also inversely associated with diastolic function in patients with type 2 diabetes (de Gonzalo-Calvo et al., 2016). However, LIPCAR does not increase in human cardiac tissue after transcoronary ablation of septal hypertrophy, suggesting that this lncRNA is not originated from the cardiac tissues (Schulte et al., 2019). Besides, another two mtDNA-derived lnRNAs uc004cov.4 and uc022bqu.1 are also upregulated in serum of patients with hypertrophic obstructive cardiomyopathy (HOCM) but not hypertrophic non-obstructive cardiomyopathy (HNCM) (Kitow et al., 2016).
Other researchers also identified 4 lnRNAsmtDNA, termed SncmtRNA-1/2 and ASncmtRNA-1/2 (Figure 4C). Among them, lncRNA ASncmtRNA-2 is induced in during aging and replicative senescence in endothelial cells, but not in vascular smooth muscle cells (VSMC). Mechanically, ASncmtRNA-2 may be the non-canonical precursor of hsa-miR-4485 and hsa-miR-1973, which are originated at least in part from a mitochondrial transcript. These two microRNAs target p16 and induce a cell cycle arrest at G2M phase, thereby affecting replicative senescence establishment (Bianchessi et al., 2015). Besides, ASncmtRNA-2 is also upregulated in diabetic kidneys and high glucose-treated mesangial cells and can promote glomerular fibrosis via modulating the expression of pro-fibrotic factors in diabetic nephropathy (Gao Y. et al., 2017).
In conclusion, lncRNAsmtDNA are present in the mitochondria, and may function as important epigenetic regulators for the regulation of mitochondrial function. However, a systemic study is needed to clarify the lncRNAsmtDNA, and their mechanisms of biogenesis and processing, as well as their functions and mode of actions.
Small non-coding RNAs (sncRNAs) are highly structured, less than 200 nt ncRNA fragments that found in bacteria, mitochondria and eukaryotes. According to their functions and characteristics, they can be classified into at least 9 types, such as microRNAs (miRNAs, ∼22 nt), Piwi-interacting RNAs (piRNA), tiny non-coding RNAs (tncRNAs), short interfering RNAs (siRNAs), repeat-associated small interfering RNAs (rasiRNAs), small modulatory RNAs (smRNAs), palindrome small RNAs (psRNAs), guide RNAs (gRNAs) and transcription initial RNAs (tiRNAs). Most of them are encoded by the nucleus genome. However, at present, mtDNA is reported to encode some sncRNAs such as miRNAs, psRNAs, tiRNAs and gRNAs. Among them, only mitochondrial-derived sncRNAs (sncRNAmtDNA) are widely reported.
A comprehensive analysis of the human mitochondrial transcriptome across multiple cell lines and tissues shows that sncRNAs exist in the mitochondria (Mercer et al., 2011). Remarkably, mt-tRNALys and mt-tRNAMe can be exported to the cytoplasm and bind to argonaute-2 (AGO2), an essential component of the RNA-induced silencing complex (RISC), suggesting that mt-tRNAs may act as miRNA (Maniataki and Mourelatos, 2005; Beitzinger et al., 2007). Mature tRNAs can also be cleaved by stress-activated ribonuclease angiogenin to generate 5′- and 3′-tRNA halves: a novel class of 30–40 nt small non-coding RNAs (Saikia and Hatzoglou, 2015). Besides, miR-1974, miR-1977, and miR-1978 may be encoded by the tRNA and rRNA genes in mtDNA (Bandiera et al., 2011). Some sequences of pre-miR-let7b and pre-miR-302a located in the mitochondria of human muscle can be aligned with the mtDNA, implying that these miRNAs may be derived from mtDNA (Barrey et al., 2011). These evidences suggest that the mtDNA can be a source of microRNAs (Bandiera et al., 2013).
An analysis on the mouse mtDNA identified 6 sncRNAsmtDNA, among which Mt-5 RNA is transcribed in antisense orientation to ND4 and Mt-6 RNA is transcribed in antisense orientation to ND6 (Lung et al., 2006). Further study shows that thousands of sncRNAs are encoded in murine and human mtDNA and most of them derived from the sense transcripts (Ro et al., 2013). The processing of sncRNAsmtDNA is not only dependent on Dicer but also some mitochondrial ribonucleases that are currently unidentified (Ro et al., 2013). Overexpression of mitochondrial-derived sncRNAs can significantly enhance the expression levels of their host genes (Ro et al., 2013). Six miRNAs that are termed as miR-mit1 to miR-mit6 are identified in mouse mtDNA. Among them, miR-mit3 and miR-mit4 can target MT-RNR2 (16S rRNA) in skeletal muscles (Shinde and Bhadra, 2015). Furthermore, Mt-1 can exhibit variable length due to polyadenylation, in which contains a microRNA-like small RNA, mmu-mir-805, that is encoded in the termination association sequence (TAS) of the mtDNA, and it is upregulated in hippocampus during olfactory discrimination training in the mice (Smalheiser et al., 2011).
Besides that, mtDNA also produces psRNAs, tiRNAs (Gao et al., 2018), and gRNAs (Ochsenreiter and Hajduk, 2006). These RNAs also play essential roles in mitochondria. For instance, precise insertion and deletion of numerous uridines are required to make full-length mitochondrial mRNAs. Guide RNA responsible for COX3 mRNA can be alternatively edited to yield a stable mRNA in the mitochondria of Trypanosoma brucei brucei (Ochsenreiter and Hajduk, 2006).
Apart from the ncRNAsmtDNA, nuclear DNA-derived non-coding RNAs (ncRNAsnDNA) including mainly lncRNAsnDNA and miRNAsnDNA may also mediate mitoepigenetics referring to ncRNAsmtDNA that affect the translation and function of mtDNA-encoded genes. These RNAs encoded by nuclear DNA may be imported into mitochondria. In Trypanosoma brucei, RNA import into mitochondria has been studied well. Nuclear encoded tRNA of T. brucei can be imported into mitochondria (Schneider et al., 1994). In fact, T. brucei imports all mitochondrial tRNAs from the cytosol, and an in vitro study showed that there were possible some membrane-bound receptors that can mediate the import of small ribosomal RNAs (srRNAs) and tRNA in protozoon Leishmania mitochondria (Mahapatra et al., 1994; Mahapatra and Adhya, 1996). Subsequently, tRNA import into the kinetoplast mitochondrion of the Leishmania tropica were shown to be organized into a multiprotein RNA import complex (RIC) that contained 3 mitochondrion- and 8 nuclear−encoded subunits, such as Tim17 and mitochondrial heat-shock protein 70 (mtHSP70) at the inner membrane (Mukherjee et al., 2007; Tschopp et al., 2011).
However, the RNA import systems in different species exhibit some unique features (Schneider and Maréchal-Drouard, 2000). For instance, Saccharomyces cerevisiae imports cytoplasmic tRNAGln into mitochondria without any added protein factors (Rinehart et al., 2005). The RNA import of plant mitochondria is dependent on the voltage-dependent anion channel (Salinas et al., 2006). The study of RNA import into mammalian mitochondria is not so clear. A microarray analysis of highly purified rat liver-derived mitochondria identified 15 miRNAsnDNA, 5 of which were further confirmed by TaqMan 5′ nuclease assays. These miRNAs may be associated with the expression of some genes related to apoptosis, cell proliferation, and differentiation (Kren et al., 2009). Further studies also show that there are possible several ATP-dependent import pathways of nucleus-encoded RNAs to human mitochondria (Duarte et al., 2015). For example, polynucleotide phosphorylase (PNPase) is shown as a contributor to mitochondrial RNases P (MRP), 5S rRNA, tRNAs and miRNAs import into mitochondria (Wang et al., 2010; Shepherd et al., 2017). Mitochondrial enzyme rhodanese was also shown to be responsible for 5 S rRNA import into human mitochondria (Smirnov et al., 2010). Both pre-miRNAsnDNA and mature miRNAsnDNA were shown to be present in the human mitochondria (Barrey et al., 2011; Bandiera et al., 2013), implying that there is possible a miRNA synthesis system in the mitochondria. However, it is also reported that tRNA import into human mitochondria does not take place under normal physiological conditions, but it is possible for mutant human mitochondria to take in nucleus-encoded tRNAs (Kolesnikova et al., 2004; Mahata et al., 2006; Rubio et al., 2008). However, the RNA import to mitochondria needs to be further confirmed and its mechanism investigated.
Selected nucleus-derived ncRNAs are possibly imported into the mitochondria, where they can be involved in multiple mitochondrial biological processes to act as “messengers” between the nucleus and the mitochondria (Vendramin et al., 2017; Jeandard et al., 2019). All the microRNAs present in mitochondria are also called mitomiRNAs (mitomiRs), which is commonly used in some reports (Rippo et al., 2014; Duarte et al., 2015; Giuliani et al., 2018). Interestingly, all the mitomiRs seemed to preferentially target multiple mtDNA sites, other than nuclear-encoded mitochondrial genes, compared with a set of cytosolic miRNAs (Duarte et al., 2015).
We summarize the lncRNAnDNA that may affect mtDNA-encoded genes in Table 3. The behaviors of lncRNAs are multi-faced and can be implicated in various regulatory levels of gene expression, from transcription to post-translation. For instance, Cerox1 promotes the levels of mitochondrial OxPhos by upregulating mitochondrial complex I subunit transcripts through binding to microRNA-488-3p, thereby leading to a decrease of ROS production (Sirey et al., 2019). Besides, a skeletal muscle- and heart-enriched lncRNA LINC00116 can encode a highly conserved 56-AA mitoregulin (Mtln) that localizes in inner mitochondrial membrane. Mtln can interact with ND5, thereby bolstering protein complex assembly and/or stability. Overexpression of Mtln results in increasing mitochondrial membrane potential and Ca2+ retention capacity, decreasing fatty acid oxidation, mitochondrial ROS and matrix-free Ca2+, promoting respiratory complex I activity and oxygen consumption and maintaining lipid composition of the cell (Stein et al., 2018; Chugunova et al., 2019). These evidences show that lncRNAsnDNA act as messengers between nDNA and mitochondria.
AGO2, a Dicer1-interacting protein that plays an essential role in short interfering RNA-mediated gene silencing, is also found to localize to mitochondria and bind to the mitochondrial transcripts COX3 and tRNAMet (Bandiera et al., 2011), suggesting the activities of microRNA-mediated biological processes in the mitochondria. In addition to the miRNAsmtDNA, mitochondria also contain miRNAsnDNA. Table 4 summarizes different microRNAsnDNA that can target mtDNA-encoded genes in multiple species, thereby affecting various biological or pathological processes. For instance, miR-181c was shown to target mt-COX1 in cardiomyocytes of rat (Das et al., 2012). In addition, miR-181c-5p is predicted to have 12 potential targets (12S RNA, 16S RNA, ND1, ND4, ND5, ND6, COX1, COX2, COX3, ATP6, Cytb, tRNAGly) encoded by mtDNA, whereas miR-146a-5p is predicted to have 12 potential targets [16S RNA, ND1, ND2, ND4, ND5, ND6, ATP8, tRNAAla, tRNAGlu, tRNASer(UCN), tRNASer(AGY)] on mtRNAs (Dasgupta et al., 2015). Some microRNAs, including miR-1275, miR-1246, miR-328-5p, miR-1908, miR-1972, miR-1977, miR-638, miR-1974, miR-1978 and miR-1201, are also predicted to target mtDNA-derived RNAs (Bandiera et al., 2011). These predictions need to be further validated.
However, functional analysis of miRNAs identified in highly purified rat liver-derived mitochondria showed that they were not targeted to the mitochondrial genome nor nuclear RNAs encoding mitochondrial proteins (Kren et al., 2009). This result implies that there are may be other mechanisms independent of miRNA-mediated mRNA degradation. Interestingly, microRNAs are also shown to directly affect mtRNA translation. For example, miR-1 can promote MT-ND1 and MT-CO1 translation but not their mRNA stability in an AGO2-dependent manner, thereby affecting muscle differentiation in mice (Zhang et al., 2014).
Mitochondria are important organelles that are essential for functional eukaryotic cells. In fact, cancer cells also depend much on mitochondria. There are commonly many alterations in mitochondria induced by both extra-mitochondrial or intra-mitochondrial influencing factors that sustain excessive proliferation of cancer cells via providing energy and metabolites (Brandon et al., 2006; Wallace, 2012). For the extra-mitochondrial influencing factors, there are more than 1,000 of nucleus-encoded proteins and thousands of nucleus-encoded non-coding RNAs that can be imported into mitochondria and play a role in function, metabolism, regulation and the production, fission, fusion, trafficking and degradation of the mitochondria. Besides, many extra-mitochondrial signaling pathways such as apoptosis and mitophagy, as well as some factors can directly regulate the function of mitochondria. Intra-mitochondrial influencing factors include mutations in the mtDNA. However, most of these mutations in mtDNA do not inhibit energy metabolism in mitochondria but rather change the mitochondrial bioenergetic and biosynthetic state, which can communicate with the nucleus via modulating signaling pathways, transcriptional circuits and/or chromatin structural remodeling to meet the requirements of the cancer cells (Wallace, 2012). For instance, certain control region mitochondrial single-nucleotide variants (mtSNVs) highly co-occur with MYC oncogene amplification in prostate cancer, and predict a poorer patient survival (Hopkins et al., 2017).
Importantly, besides genetic mutations in mtDNA, mitoepigenetics is emerging as an important regulatory mode. All the mitoepigenetic networks including mtDNA methylation, mitochondrial nucleoid modifications, mtRNA methylation, mtDNA-encoded and nucleus-encoded ncRNAs, have been shown to play essential roles in tumor development and pathogenesis.
Cancer is often related with a low level of total nDNA methylation, hypermethylation of tumor suppressor gene promoters and hypomethylation of oncogene promoters (Kulis and Esteller, 2010). Therefore, it is suspected that methylation in mtDNA should be accurately modulated. The copy number of mtDNA is strictly regulated during cellular differentiation in cancer cells. In breast cancer, mtDNA methylation is maternally inherited in D-loop region, in which 8 aberrant mtDNA methylation sites are tightly dysregulated (Han et al., 2017). mtDNA copy number and ND-2 expression in colorectal cancer tissues are higher than that of the corresponding non-cancerous tissues. Methylation on the D-loop region in colorectal cancer tissues was lower than that of their corresponding non-cancerous tissues. Meanwhile, methylation on the D-loop region in stage III/IV colorectal cancer tissues is also significantly decreased, compared with that in stages I/II colorectal cancer tissues. Furthermore, D-loop region de-methylation is tightly correlated with a high mtDNA copy number and a high ND-2 expression. DNA methylation inhibitor 5-aza-deoxycytidine treatment also increases the mtDNA copy number and ND-2 expression in Caco-2 cells (Gao et al., 2015). Further study reveals that de-methylation of 4th and 6th/7th CpG islands of D-loop promoter can lead to the elevation of mtDNA copy number in colorectal cancer, thereby triggering cell proliferation, cell cycle progression and reducing apoptosis (Tong et al., 2017). These results indicate that mtDNA methylation is negatively correlated with mtDNA number and tumor progression.
Similarly, the level of 5mC at several sites of mtDNA is negatively correlated with mtDNA copy number in 143B osteosarcoma cells (Sun et al., 2018a). 5mC in mtDNA D-loop is low during tumor progression and may potentially contribute to the increase in mtDNA copy number observed in these tumor cells, including osteosarcoma and glioblastoma cells (Sun et al., 2018a). 5mC levels of D-loop also negatively correlate with ND5 and ND6 transcription during the tumorigenesis of 143B osteosarcoma cells (Sun et al., 2018a).
However, mtDNA of cancer stem-like cells is also hypermethylated and the mtDNA copy number is low, which makes them to use glycolysis for cell proliferation (Lee and St John, 2015). After sufficient mtDNA is restored in tumors to initiate tumorigenesis, 5mC in D-loop is increased to restrict further mtDNA replication (Sun et al., 2018a). That is the reason why cancer cells have a lower mtDNA copy number to maintain a “pseudo-differentiated” state and why global DNA demethylation can induce cellular differentiation and expansion of mtDNA copy number (Sun et al., 2018a).
However, there are also some studies that do not support the results above. For instance, colorectal adenomas have a low-level methylation of specific sites in mtDNA, but it is not associated with changes of mitochondrial gene transcription (Morris et al., 2018). Maekawa et al. (2004) showed that methylation of mtDNA was a rare event in the CpG sites in cancer cell lines and tissues of gastric and colorectal cancer. Hong et al. (2013) also confirmed that CpG methylation was absent in HCT116 colerectal cancer cell lines. van der Wijst et al. (2017) showed that CpG mtDNA methylation didn’t affect mtDNA gene expression, whereas 5mC in the GpC context decreased mtDNA gene transcription.
In conclusion, the relationship between mtDNA methylation and cancer should be studied further. To solve the problems, a more precise method to detect the methylated sites of methylation in mtDNA is needed. A systematic study on mtDNA methylation in both normal tissues and tumor tissues should be performed to show differences between them. Finally, the clinical significance, function and mode of actions of mtDNA methylation should be further elucidated.
In fact, these explorations will not only benefit the study of cancer biology, but also would be useful for studying other mitochondrial diseases. mtDNA methylation is also shown to be connected with various human diseases, such as Down’s syndrome and Alzheimer’s disease. The levels of mitochondrial SAM is downregulated in Down’s syndrome compared to control cells, suggesting that there is a low level of 5mC level in the mtDNA of this disease (Infantino et al., 2011). 5mC in mtDNA D-loop is observed in the blood of patients with late-onset Alzheimer’s disease patients (Stoccoro et al., 2017). Besides, 5mC in mitochondrial the transfer RNA phenylalanine (MT-TF), MT-RNR1 gene while not D-loop region is shown to be associated with metal-rich particulate matter (PM1) exposure and mtDNA copy number (Byun et al., 2013). In umbilical cord blood, 5mC in the 12S rRNA (MT-RNR1) or the D-loop control region of mtDNA is positively correlated with the level of free thyroid hormones (FT3 and FT4) and mitochondrial DNA copy numbers regulated by these two hormones (Janssen et al., 2017).
TFAM is the only nucleoid-associated protein that functions as a histone-like factor. Its expression is shown to be positively correlated with the progression of multiple cancers, including melanoma (Araujo et al., 2018), hepatocellular carcinoma (Qiao et al., 2017), non-small cell lung cancer (Xie et al., 2016), colon cancer (Lin et al., 2018), bladder cancer (Mo et al., 2013), epithelial ovarian carcinoma (Gabrielson et al., 2014), glioma (Lee et al., 2017), and breast cancer (Fan et al., 2017). These phenomena may be the results of higher mtDNA copy number that is found in tumors compared to normal tissues (Sun et al., 2018b), which makes tumor cells produce more TFAM for sufficient compaction. Otherwise, mtDNA in cancer cells may be tightly wrapped by more TFAM, which leads to a lower expression of mtDNA encoding ETC/OxPhos-related genes, thereby promoting tumor cells to use aerobic glycolysis.
TFAM can also be regulated by post-translational modifications including acetylation, phosphorylation and ubiquitination. However, there is no direct evidence showing that TFAM modifications are correlated with cancer. Since post-translational modifications of TFAM significantly affect its stability or function, and its modifications may also be tightly associated with tumor progression.
mtDNA is transcribed to produce RNA with continuous polycistrons, implying that post-transcriptional modulations are essential for RNA processing. Recently, mtRNA transcripts were shown to be differently accumulated in tumor tissues (Stewart et al., 2015). Mutation of mtRNA processing enzymes, such as ELAC2, which has RNase Z activity and functions in the maturation of mt-tRNA by removing a 3′-trailer from tRNA precursors to generate 3′ termini of tRNAs, is associated with prostate cancer incidence (Tavtigian et al., 2001). mt-tRNAs are heavily post-transcriptionally modified mtRNAs, mutations within which are also related to cancer (Brandon et al., 2006). A tRNA-dihydrouridine synthases, DUS2, which catalyzes the conversion of uridine residues to dihydrouridine in the D-loop of tRNA, is also commonly upregulated in pulmonary cancer (Kato et al., 2005). These results imply that mtRNA processing is important for cancer.
Recent studies show that mtRNA modifications, which are essential for mtRNA processing, are also major regulatory factors in tumors. In tumor tissues across 12 cancer types, there are remarkable alterations in methylation levels of m1A and m1G RNA in mitochondrial tRNAs. In normal tissues, RNA processing pathways are specifically related to mt-tRNAs methylation levels, however, these connections are lost in tumors (Idaghdour and Hodgkinson, 2017). High mt-tRNAs methylation difference predicts a poorer prognosis in a cohort of patients with kidney renal clear cell carcinoma (Idaghdour and Hodgkinson, 2017). The level of m1A and m1G methylation in mtRNAs can significantly affect mitochondria-mediated metabolism (Hodgkinson et al., 2014). In conclusions, mt-tRNAs methylation affects their maturation and thus plays emerging roles in tumorigenesis.
There are emerging evidences showing that lncRNAsmtDNA may be involved in tumorigenesis by promoting cell proliferation and tumor growth. A lncRNA with an 815 nt inverted repeat (IR) and a stem-loop structure resistant to RNase A is covalently linked to the 5′ end of 16S rRNA in human cells. It is expressed in highly normal proliferating cells while not in resting cells. The expression of this lncRNA can be induced in phytohemagglutinin (PHA)-treated resting lymphocytes, while can be reversibly blocked in aphidicolin-treat DU145 pancreatic cancer cells, in which cell proliferation is also reversibly inhibited (Villegas et al., 2007). Besides which, two antisense mtRNA transcripts that contain stem-loop structures are expressed in normal proliferating cells but significantly downregulated in tumor cells (Burzio et al., 2009). Further study shows that a family of mitochondrial ncRNAs (ncmtRNAs) with stem-loop structures can be divided into sense (SncmtRNAs) and antisense (ASncmtRNAs) members. Both of the SncmtRNAs and ASncmtRNAs are expressed in normal proliferating cells, whereas ASncmtRNAs are downregulated in various types of tumor cells. ASncmtRNAs knockdown induces cell cycle arrest and apoptosis via inhibiting survivin expression in cancer cell lines without impairing cell viability of normal cells. MicroRNAs generated by dicing of the double-stranded stem of the ASncmtRNAs can downregulate survivin. Mechanically, ASncmtRNAs binds to Dicer to recruit to the 3′-UTR of survivin mRNA, resulting in degradation of this mRNA (Vidaurre et al., 2014). Preclinical studies also show that ASncmtRNAs knockdown blocks tumor growth in melanoma and renal cancer models (Olavarria et al., 2018). Immortalization of human keratinocytes with HPV-16/18 downregulates the expression of the ASncmtRNAs and induces the expression of SncmtRNA-2. Furthermore, E6 and E7 are shown to be responsible for SncmtRNA-2 upregulation, whereas E2 oncogene is responsible for ASncmtRNAs downregulation (Villota et al., 2012).
In addition, some specific lncRNAsmtDNA are highly upregulated in tumors or cancer patients’ urine and predict poor prognosis of these diseases. For instance, SncmtRNAs are upregulated and ASncmtRNAs are downregulated in the urine of patients with bladder cancer (Rivas et al., 2012). Higher level of lncRNAmtDNA uc004cox.4 in urine is associated with poorer recurrence-free survival (RFS) of non-muscle invasive BC (NMIBC) and act as an independent prognostic factor for RFS of this disease (Du et al., 2018).
In conclusion, ncRNAsmtDNA are important regulators during tumorigenesis and can be promising prognostic markers for cancers. Therefore, it is urgent to identify the mtDNA-encoded ncRNAs to provide a better understanding of this area.
Emerging evidences also show that ncRNAs derived from nDNA also act as messengers to regulate mitochondrial function in cancer cells. For instance, lncRNA MALAT1 can be transported into mitochondria by RNA-binding protein HuR and mitochondria transmembrane protein mitochondrial carrier 2 (MTCH2) in HepG2 hepatocellular carcinoma cells. Then the 3′-fragment of this lncRNA interacts with multiple mtDNA loci, including D-loop, COX2, ND3, and CYTB. MALAT1 knockdown results in low OxPhos, reduced ATP production, inhibited mitophagy, declined mtDNA copy number, and upregulated intrinsic apoptotic pathway (Zhao et al., 2019).
In addition to lncRNAsnDNA, miRNAsnDNA seem to play roles that are more important in epigenetic regulations of tumor cells. For instance, miR-24 targets ND2 in human Lewis lung carcinoma (LLC) cells, resulting in mitochondrial dysfunction and growth inhibition (Michael et al., 2017). miR-26a targets COX 2 in human prostate cancer cells, inhibiting cell proliferation and inducing apoptosis (Zhang et al., 2016). miR-4485 targets 16S rRNA in human breast cancer cells, leading to the decrease of mitochondrial complex I activity, the production of ATP, and inducing high ROS levels that activates caspase-3/7-dependent apoptosis (Sripada et al., 2017). miR-2392 localizes to mitochondria, silences mtDNA transcription through an AGO2-dependent mechanism, thereby inhibiting ND4, CYTB, and COX1 expression, and promotes cancer cells to chemosensitivity (Fan et al., 2019).
The puzzle of mitoepigenetics has been uncovered gradually in recent years. New findings also significantly alter the concept of mitoepigenetics. Manev and Dzitoyeva (2013) first proposed the concept of mitoepigenetics in 2013 referring to all epigenetic regulations that are related to mitochondria. Ferreira et al. (2015) also use this concept. According to their definition, mitoepigenetics is comprised of four levels: (i) epigenetic controls of expression of nDNA-encoded mitochondrial genes; (ii) a cell-specific mtDNA content and mitochondrial activity-determined epigenetic alterations in nuclear genes expression; (iii) mtDNA variants-influenced nuclear gene expression patterns and ncDNA methylation levels; (iv) epigenetic modifications in mtDNA like 5mC and 5hmC marks. However, Manev and Dzitoyeva (2013) suggested a restricted usage of mitoepigenetics as the last ones, which also was used by subsequent researchers such as Sadakierska-Chudy et al. (2014). However, van der Wijst and Rots (2015) and Coppede and Stoccoro (2019) used a more restricted definition of mitoepigenetics as 5mC or 5hmC in mtDNA. The concept of mitoepigenetics described by Ghosh et al. (2015) includes 5mC/5hmC in mtDNA, mitochondrial modulation of nuclear DNA methylation and non-coding RNAs regulatory epigenetics in mitochondria.
In this review, we definite the concept of mitoepigenetics as a study of molecular modifications occurring in mitochondria that affect mitochondrial inheritance without involving mtDNA changes. According to this definition, mitoepigenetics refers to mtDNA modifications, mitochondrial nucleoid modifications, mtRNA modifications as well as non-coding RNAs that affect the translation and function of mtDNA-encoded genes (Figure 5). This definition is narrower than the concept defined by Manev and Dzitoyeva (2013), but is an extension of the concept used by Ghosh et al. (2015) as it includes mtRNA modifications, mitochondrial nucleoid modifications and a new definition of non-coding RNAs-regulated mitoepigenetics. In fact, according to our definition, all the mitoepigenetic alterations seem to alter the expression and function of mtDNA-encoded proteins, which mainly play essential roles in ETC/OxPhos and participate in mitochondrial cellular metabolism including glucose, lipid and amino acid metabolism. Therefore, mitoepigenetics is tightly related to multiple mitochondria-mediated biological processes, such as intrinsic apoptosis (Dong et al., 2017), mitophagy (Dong and Cui, 2018; Li et al., 2019), ROS generation (Kausar et al., 2018), Ca2+ signaling (Bravo-Sagua et al., 2017) and hemoglobin synthesis (Fleming and Hamza, 2012).
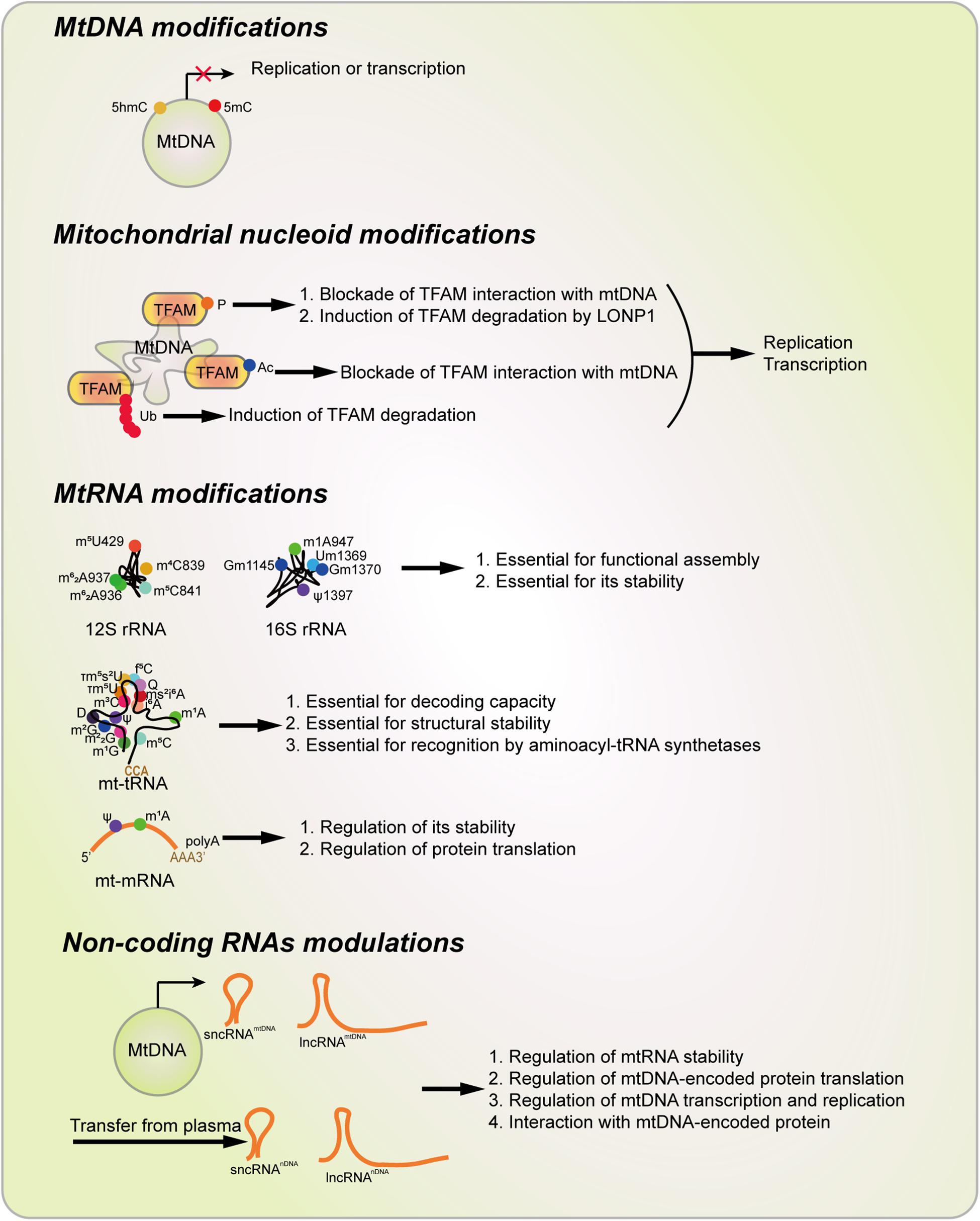
Figure 5. Types of Mitoepigenetics and their functions. Mitoepigenetics constitutes with four different types, including mtDNA modifications, nucleoid modifications, mtRNA modifications, and non-coding RNA modulations. Ac, acetylation; D, Dihydrouridine; f5C, 5-Formylcytosine; 5hmC 5-Hydroxymethylcytosine; 5mC, 5-methylcytosine; i6A, N6-Isopentenyladenosine; m1A, 1-Methyladenosine; m1G, 1-Methylguanosine; m2G, N2-Methylguanosine; m22G, N2,N2-Dimethylguanosine; m3C, 3-Methylcytosine; m4C, N4-Methylcytosine; m5C, 5-Methylcytosine; m5U, 5-Methyluridinep; m6A, N6-Methyladenosine; m62A, N6,N6-Dimethyladenosine; ms2i6A, 2-Methylthio-N6-isopentenyladenosine; P, phosphorylation; Ψ, Pseudouridine; Q, Queosine; t6A, N6-Threonylcarbamoyladenosine; TFAM, Transcription factor A, mitochondrial; τm5U, 5-Taurinomethyluridin; τm5s2U, 5-Taurinomethyl-2-thiouridine, Ub, Ubiquitination.
Since dysfunctional mitochondria are tightly related to cancer initiation and cancer progression (Wallace, 2012; Vyas et al., 2016), mitoepigenetics can also involve important modulations that occur in the pathological processes of cancer. 5mC in specific sites of mtDNA seems to be decreased during tumorigenesis. This phenomenon suggests that 5mC in these sites may be prognostic markers for cancers. Besides, since recent report shows that 5hmC and 5fC contents are decreased significantly in the very early stage of HCC (Liu et al., 2019), 5hmC found in mtDNA may also make some senses during cancer initiation and progression. Besides, TFAM is also shown to be positively related to malignant progression of multiple cancers, post-translational modifications that found in this protein may also be essential modulations in cancer progression. NcRNAs derived from both the nDNA and mtDNA are also promising prognostic factors that regulate tumorigenesis. Mitoepigenetic alterations may be one of the reasons for carcinogenesis, otherwise they are results of tumorigenesis. These alterations cannot be a main reason for tumorigenesis, because cancer cells without mitochondria (ρ0 cells) still can form tumors in vivo (Magda et al., 2008; Sun et al., 2018b). However, epigenetic alterations in mitochondrial indeed affect the development of tumors and mitochondrial Achilles’ heel in cancer can also be targeted by mitoepigenetic modulation (Hockenbery, 2002). Anyway, these findings about the connections between cancers and mitoepigenetics may provide some new clues for the prognosis, prevention and even therapeutic strategies for these diseases.
Epigenetic regulation is a kind of reversible mode for gene expression. Until now, there are several epigenetic drugs (epi-drugs), such as 5-azacytosine, decitabine, guadecitabine, belinostat, panobinostat, vorinostat, and romidepsin are approved by FDA in the clinic to treat some diseases including cancers (Jones et al., 2016; Nebbioso et al., 2018). Besides, other epigenetic drugs such as chidamide, givinostat, quisinostat, GSK2879552 and MAK683 are on clinical trials (Berdasco and Esteller, 2019). These studies have opened a new window for the treatment of cancers. Since mitoepigenetics also plays essential roles in mitochondrial function and processing, it may open a new window for cancer therapy. However, there are some questions need to be further solved. Firstly, mtDNA methylation should be systemically studied with high-resolution methylation sequencing. Secondly, core proteins of the nucleoid should be studied further and the post-translational modifications in TFAM and their connections with cancers should be validated. Thirdly, modifications in mtRNAs including mt-rRNAs, mt-tRNAs, mt-mRNAs and ncRNAsmtDNA should be further explored. Finally, both ncRNAsmtDNA and ncRNAsnDNA should be further characterized, their targets should be systemically identified and their clinical significances should be confirmed.
ZD wrote the manuscript, drew the figures, and made the tables. LP and HC reviewed and revised the manuscript.
We are grateful for funding support from the National Natural Science Foundation of China (Nos. 81902664, 81672502, and 81872071), the National Key Research and Development Program of China (Nos. 2017YFC1308600 and 2016YFC1302204), the Natural Science Foundation of Chongqing (No. cstc2019jcyj-zdxmX0033), the Fundamental Research Funds for the Central Universities (XDJK2019C013), and the project funded by Chongqing Special Postdoctoral Science Foundation (No. XmT2018080).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Amunts, A., Brown, A., Toots, J., Scheres, S. H. W., and Ramakrishnan, V. (2015). Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98. doi: 10.1126/science.aaa1193
Andreazza, S., Samstag, C., Sanchez-Martinez, A., Fernandez-Vizarra, E., Gomez-Duran, A., Lee, J., et al. (2019). Mitochondrially-targeted APOBEC1 is a potent mtDNA mutator affecting mitochondrial function and organismal fitness in Drosophila. Nat. Commun. 10:3280. doi: 10.1038/s41467-019-10857-y
Andrews, R. M., Kubacka, I., Chinnery, P. F., Lightowlers, R. N., Turnbull, D. M., and Howell, N. (1999). Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23:147. doi: 10.1038/13779
Antonicka, H., Choquet, K., Lin, Z. Y., Gingras, A. C., Kleinman, C. L., and Shoubridge, E. A. (2017). A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 18, 28–38. doi: 10.15252/embr.201643391
Antonicka, H., and Shoubridge, E. A. (2015). Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 10, 920–932. doi: 10.1016/j.celrep.2015.01.030
Araujo, L. F., Siena, A. D. D., Plaça, J. R., Brotto, D. B., Barros, I. I., Muys, B. R., et al. (2018). Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci. Rep. 8:14190. doi: 10.1038/s41598-018-31170-6
Bagul, P. K., Katare, P. B., Bugga, P., Dinda, A. K., and Banerjee, S. K. (2018). SIRT-3 modulation by resveratrol improves mitochondrial oxidative phosphorylation in diabetic heart through deacetylation of TFAM. Cells 7:E235. doi: 10.3390/cells7120235
Bai, M., Chen, H., Ding, D., Song, R., Lin, J., Zhang, Y., et al. (2019). MicroRNA-214 promotes chronic kidney disease by disrupting mitochondrial oxidative phosphorylation. Kidney Int. 95, 1389–1404. doi: 10.1016/j.kint.2018.12.028
Ban-Ishihara, R., Ishihara, T., Sasaki, N., Mihara, K., and Ishihara, N. (2013). Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 110, 11863–11868. doi: 10.1073/pnas.1301951110
Bandiera, S., Mategot, R., Girard, M., Demongeot, J., and Henrion-Caude, A. (2013). MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic. Biol. Med. 64, 12–19. doi: 10.1016/j.freeradbiomed.2013.06.013
Bandiera, S., Rüberg, S., Girard, M., Cagnard, N., Hanein, S., Chrétien, D., et al. (2011). Nuclear Outsourcing of RNA interference components to human mitochondria. PLoS One 6:e20746. doi: 10.1371/journal.pone.0020746
Bar-Yaacov, D., Frumkin, I., Yashiro, Y., Chujo, T., Ishigami, Y., Chemla, Y., et al. (2016). Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 14:e1002557. doi: 10.1371/journal.pbio.1002557
Barrey, E., Saint-Auret, G., Bonnamy, B., Damas, D., Boyer, O., and Gidrol, X. (2011). Pre-microRNA and Mature microRNA in Human Mitochondria. PLoS One 6:e20220. doi: 10.1371/journal.pone.0020220
Beitzinger, M., Peters, L., Zhu, J. Y., Kremmer, E., and Meister, G. (2007). Identification of Human microRNA targets from isolated argonaute protein complexes. RNA Biol. 4, 76–84. doi: 10.4161/rna.4.2.4640
Bellizzi, D., D’Aquila, P., Scafone, T., Giordano, M., Riso, V., Riccio, A., et al. (2013). The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 20, 537–547. doi: 10.1093/dnares/dst029
Berdasco, M., and Esteller, M. (2019). Clinical epigenetics: seizing opportunities for translation. Nat. Rev. Genet. 20, 109–127. doi: 10.1038/s41576-018-0074-2
Bianchessi, V., Badi, I., Bertolotti, M., Nigro, P., D’Alessandra, Y., Capogrossi, M. C., et al. (2015). The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J. Mol. Cell Cardiol. 81, 62–70. doi: 10.1016/j.yjmcc.2015.01.012
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Bohnsack, M. T., and Sloan, K. E. (2018). The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 75, 241–260. doi: 10.1007/s00018-017-2598-6
Bonekamp, N. A., and Larsson, N. G. (2018). SnapShot: mitochondrial nucleoid. Cell 172:388-388.e1. doi: 10.1016/j.cell.2017.12.039
Brandon, M., Baldi, P., and Wallace, D. C. (2006). Mitochondrial mutations in cancer. Oncogene 25, 4647–4662. doi: 10.1038/sj.onc.1209607
Brandon, M. C., Lott, M. T., Nguyen, K. C., Spolim, S., Navathe, S. B., Baldi, P., et al. (2005). MITOMAP: a human mitochondrial genome database–2004 update. Nucleic Acids Res. 33, D611–D613. doi: 10.1093/nar/gki079
Bravo-Sagua, R., Parra, V., Lopez-Crisosto, C., Diaz, P., Quest, A. F., and Lavandero, S. (2017). Calcium transport and signaling in mitochondria. Compr. Physiol. 7, 623–634. doi: 10.1002/cphy.c160013
Burzio, V. A., Villota, C., Villegas, J., Landerer, E., Boccardo, E., Villa, L. L., et al. (2009). Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 9430–9434. doi: 10.1073/pnas.0903086106
Byun, H. M., Panni, T., Motta, V., Hou, L., Nordio, F., Apostoli, P., et al. (2013). Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol. 10:18. doi: 10.1186/1743-8977-10-18
Campbell, A., Mrázek, J., and Karlin, S. (1999). Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 96:9184. doi: 10.1073/pnas.96.16.9184
Cardon, L. R., Burge, C., Clayton, D. A., and Karlin, S. (1994). Pervasive CpG suppression in animal mitochondrial genomes. Proc. Natl. Acad. Sci. U.S.A. 91, 3799–3803. doi: 10.1073/pnas.91.9.3799
Chang, J. H., and Tong, L. (2012). Mitochondrial poly(A) polymerase and polyadenylation. Biochim. Biophys. Acta 1819, 992–997. doi: 10.1016/j.bbagrm.2011.10.012
Chen, G., Guo, H., Song, Y., Chang, H., Wang, S., Zhang, M., et al. (2016). Long non-coding RNA AK055347 is upregulated in patients with atrial fibrillation and regulates mitochondrial energy production in myocardiocytes. Mol. Med. Rep. 14, 5311–5317. doi: 10.3892/mmr.2016.5893
Chen, S. H., Suzuki, C. K., and Wu, S. H. (2008). Thermodynamic characterization of specific interactions between the human Lon protease and G-quartet DNA. Nucleic Acids Res. 36, 1273–1287. doi: 10.1093/nar/gkm1140
Chinnery, P. F., and Hudson, G. (2013). Mitochondrial genetics. Br. Med. Bull. 106, 135–159. doi: 10.1093/bmb/ldt017
Chugunova, A., Loseva, E., Mazin, P., Mitina, A., Navalayeu, T., Bilan, D., et al. (2019). LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. J. Proc. Natl. Acad. Sci. U.S.A. 116, 4940–4945. doi: 10.1073/pnas.1809105116
Coppede, F., and Stoccoro, A. (2019). Mitoepigenetics and neurodegenerative diseases. Front. Endocrinol. 10:86. doi: 10.3389/fendo.2019.00086
Das, S., Ferlito, M., Kent, O. A., Fox-Talbot, K., Wang, R., Liu, D., et al. (2012). Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603. doi: 10.1161/circresaha.112.267732
Das, S., Kohr, M., Dunkerly-Eyring, B., Lee, D. I., Bedja, D., Kent, O. A., et al. (2017). Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J. Am. Heart Assoc. 6:e004694. doi: 10.1161/jaha.116.004694
Dasgupta, N., Peng, Y., Tan, Z., Ciraolo, G., Wang, D., and Li, R. (2015). miRNAs in mtDNA-less cell mitochondria. Cell Death Discov. 1:15004. doi: 10.1038/cddiscovery.2015.4
de Gonzalo-Calvo, D., Kenneweg, F., Bang, C., Toro, R., van der Meer, R. W., Rijzewijk, L. J., et al. (2016). Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 6:37354. doi: 10.1038/srep37354
Degoul, F., Brule, H., Cepanec, C., Helm, M., Marsac, C., Leroux, J., et al. (1998). Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum. Mol. Genet. 7, 347–354. doi: 10.1093/hmg/7.3.347
Dinardo, M. M., Musicco, C., Fracasso, F., Milella, F., Gadaleta, M. N., Gadaleta, G., et al. (2003). Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem. Biophys. Res. Commun. 301, 187–191. doi: 10.1016/S0006-291X(02)03008-5
Dong, Z., Abbas, M. N., Kausar, S., Yang, J., Li, L., Tan, L., et al. (2019). Biological functions and molecular mechanisms of antibiotic tigecycline in the treatment of cancers. Int. J. Mol. Sci. 20:E3577. doi: 10.3390/ijms20143577
Dong, Z., and Cui, H. (2018). The autophagy-lysosomal pathways and their emerging roles in modulating proteostasis in tumors. Cells 8:E4. doi: 10.3390/cells8010004
Dong, Z., Lei, Q., Yang, R., Zhu, S., Ke, X. X., Yang, L., et al. (2017). Inhibition of neurotensin receptor 1 induces intrinsic apoptosis via let-7a-3p/Bcl-w axis in glioblastoma. Br. J. Cancer 116, 1572–1584. doi: 10.1038/bjc.2017.126
Dostal, V., and Churchill, M. E. A. (2019). Cytosine methylation of mitochondrial DNA at CpG sequences impacts transcription factor A DNA binding and transcription. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 598–607. doi: 10.1016/j.bbagrm.2019.01.006
Du, L., Duan, W., Jiang, X., Zhao, L., Li, J., Wang, R., et al. (2018). Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J. Cell Mol. Med. 22, 2838–2845. doi: 10.1111/jcmm.13578
Duarte, F. V., Palmeira, C. M., and Rolo, A. P. (2014). The role of microRNAs in mitochondria: small players acting wide. Genes 5, 865–886. doi: 10.3390/genes5040865
Duarte, F. V., Palmeira, C. M., and Rolo, A. P. (2015). The emerging role of MitomiRs in the pathophysiology of human disease. Adv. Exp. Med. Biol. 888, 123–154. doi: 10.1007/978-3-319-22671-2-8
Dubin, D. T., and Taylor, R. H. (1978). Modification of mitochondrial ribosomal RNA from hamster cells: the presence of GmG and late-methylated UmGmU in the large subunit (17 S) RNA. J. Mol. Biol. 121, 523–540. doi: 10.1016/0022-2836(78)90398-4
Dubin, D. T., Taylor, R. H., and Davenport, L. W. (1978). Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N 4 -methylcytidine. Nucleic Acids Res. 5, 4385–4398. doi: 10.1093/nar/5.11.4385
Dupont, C., Armant, D. R., and Brenner, C. A. (2009). Epigenetics: definition, mechanisms and clinical perspective. Semin. Reprod. Med. 27, 351–357. doi: 10.1055/s-0029
Elgass, K., Pakay, J., Ryan, M. T., and Palmer, C. S. (2013). Recent advances into the understanding of mitochondrial fission. Biochim. Biophys. Acta 1833, 150–161. doi: 10.1016/j.bbamcr.2012.05.002
Chinnery, P. F., and Gómez-Durán, A. (2018). Oldies but Goldies mtDNA population variants and neurodegenerative diseases. Front. Neurosci. 12:682. doi: 10.3389/fnins.2018.00682
Fan, S., Tian, T., Chen, W., Lv, X., Lei, X., Zhang, H., et al. (2019). Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription. Cancer Res. 79, 1069–1084. doi: 10.1158/0008-5472.Can-18-2505
Fan, X., Zhou, S., Zheng, M., Deng, X., Yi, Y., and Huang, T. (2017). MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 88, 507–514. doi: 10.1016/j.biopha.2017.01.058
Farge, G., and Falkenberg, M. (2019). Organization of DNA in mammalian Mitochondria. Int. J. Mol. Sci. 20:2770. doi: 10.3390/ijms20112770
Ferreira, A., Serafim, T. L., Sardao, V. A., and Cunha-Oliveira, T. (2015). Role of mtDNA-related mitoepigenetic phenomena in cancer. Eur. J. Clin. Invest. 45(Suppl. 1), 44–49. doi: 10.1111/eci.12359
Fleming, M. D., and Hamza, I. (2012). Mitochondrial heme: an exit strategy at last. J. Clin. Invest. 122, 4328–4330. doi: 10.1172/JCI66607
Gabrielson, M., Björklund, M., Carlson, J., and Shoshan, M. J. P. O. (2014). Expression of mitochondrial regulators PGC1α and TFAM as putative markers of subtype and chemoresistance in epithelial ovarian carcinoma. PLoS One 9:e107109. doi: 10.1371/journal.pone.0107109
Gao, D., Zhu, B., Sun, H., and Wang, X. (2017). Mitochondrial DNA Methylation and related disease. Adv. Exp. Med. Biol. 1038, 117–132. doi: 10.1007/978-981-10-6674-0-9
Gao, J., Wen, S., Zhou, H., and Feng, S. (2015). De-methylation of displacement loop of mitochondrial DNA is associated with increased mitochondrial copy number and nicotinamide adenine dinucleotide subunit 2 expression in colorectal cancer. Mol. Med. Rep. 12, 7033–7038. doi: 10.3892/mmr.2015.4256
Gao, S., Tian, X., Chang, H., Sun, Y., Wu, Z., Cheng, Z., et al. (2018). Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion 38, 41–47. doi: 10.1016/j.mito.2017.08.002
Gao, Y., Chen, Z. Y., Wang, Y., Liu, Y., Ma, J. X., Li, Y. K. J. E., et al. (2017). Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp. Ther. Med. 13, 581–587. doi: 10.3892/etm.2017.4027
Garone, C., Alice Donati, M., Sacchini, M., Garcia-Diaz, B., Bruno, C., Calvo, S., et al. (2013). Mitochondrial encephalomyopathy due to a novel mutation in ACAD9. JAMA Neurol. 70, 1–3. doi: 10.1001/jamaneurol.2013.3197
Garone, C., Calvo, S., Emmanuele, V., Akman, H., Kaplan, P., Krishna, S., et al. (2012a). MitoExome sequencing reveals a mutation in the mitochondrial mrpl51 gene causing infantile encephalopathy (P05.139). Neurology 78:139. doi: 10.1212/WNL.78.1-MeetingAbstracts.P05.139
Garone, C., Carlos Rubio, J., Calvo, S., Naini, A., Tanji, K., Dimauro, S., et al. (2012b). MPV17 mutations causing adult-onset multisystemic disorder with multiple mitochondrial DNA deletions. Arch. Neurol. 69, 1–4. doi: 10.1001/archneurol.2012.405
Garone, C., D’Souza, A., Dallabona, C., Lodi, T., Guiomar, P., Rorbach, J., et al. (2017). Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum. Mol. Genet. 26, 1–10. doi: 10.1093/hmg/ddx314
Garone, C., and Viscomi, C. (2018). Towards a therapy for mitochondrial disease: an update. Biochem. Soc. Trans. 46:1247. doi: 10.1042/BST20180134
Garros, R. F., Paul, R., Connolly, M., Lewis, A., Garfield, B. E., Natanek, S. A., et al. (2017). MicroRNA-542 promotes mitochondrial dysfunction and SMAD activity and is elevated in intensive care unit-acquired weakness. Am. J. Respir. Crit. Care Med. 196, 1422–1433. doi: 10.1164/rccm.201701-0101OC
Ghosh, S., Sengupta, S., and Scaria, V. (2014). Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion 18, 58–62. doi: 10.1016/j.mito.2014.07.007
Ghosh, S., Singh, K. K., Sengupta, S., and Scaria, V. (2015). Mitoepigenetics: the different shades of grey. Mitochondrion 25, 60–66. doi: 10.1016/j.mito.2015.09.003
Giuliani, A., Micolucci, L., Olivieri, F., Procopio, A. D., and Rippo, M. R. (2018). “MitomiRs in human inflamm-aging,” in Handbook of Immunosenescence: Basic Understanding and Clinical Implications, eds T. Fulop, C. Franceschi, K. Hirokawa, and G. Pawelec, (Cham: Springer International Publishing), 1–29. doi: 10.1007/978-3-319-64597-1_121-1
Goffart, S., Hangas, A., and Pohjoismäki, J. L. O. (2019). Twist and turn-topoisomerase functions in mitochondrial DNA maintenance. Int. J. Mol. Sci. 20:2041. doi: 10.3390/ijms20082041
Gómez-Durán, A., Pacheu-Grau, D., López-Gallardo, E., Díez-Sánchez, C., Montoya, J., López-Pérez, M., et al. (2010). Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum. Mol. Genet. 19, 3343–3353. doi: 10.1093/hmg/ddq246
Gómez-Durán, A., Pacheu-Grau, D., Martinez-Romero, I., López-Gallardo, E., López-Pérez, M., Montoya, J., et al. (2012). Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber’s hereditary optic neuropathy. Biochim. Biophys. Acta 1822, 1216–1222. doi: 10.1016/j.bbadis.2012.04.014
Gómez-Durán, A., Yaobo, X., Golder, Z., Hudson, G., Santibanez-Koref, M., Ruiz-Pesini, E., et al. (2018). mtDNA polymorphic variants as metabolic hubs. Neuromuscul. Disord. 28, S35–S36. doi: 10.1016/S0960-8966(18)30397-3
Gray, M. W. (2012). Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4:a011403. doi: 10.1101/cshperspect.a011403
Greber, B. J., and Ban, N. (2016). Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 85, 103–132. doi: 10.1146/annurev-biochem-060815-0-14343
Gunbin, K., Peshkin, L., Popadin, K., Annis, S., Ackermann, R. R., and Khrapko, K. (2017). Integration of mtDNA pseudogenes into the nuclear genome coincides with speciation of the human genus A hypothesis. Mitochondrion 34, 20–23. doi: 10.1016/j.mito.2016.12.001
Haag, S., Sloan, K. E., Ranjan, N., Warda, A. S., Kretschmer, J., Blessing, C., et al. (2016). NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 35, 2104–2119. doi: 10.15252/embj.201694885
Han, X., Zhao, Z., Zhang, M., Li, G., Yang, C., Du, F., et al. (2017). Maternal trans-general analysis of the human mitochondrial DNA pattern. Biochem. Biophys. Res. Commun. 493, 643–649. doi: 10.1016/j.bbrc.2017.08.138
Hensen, F., Potter, A., van Esveld, S. L., Tarrés-Solé, A., Chakraborty, A., Solà, M., et al. (2019). Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 47, 3680–3698. doi: 10.1093/nar/gkz047
Hockenbery, D. M. (2002). A mitochondrial Achilles’ heel in cancer? Cancer Cell 2, 1–2. doi: 10.1016/S1535-6108(02)00087-9
Hodgkinson, A., Idaghdour, Y., Gbeha, E., Grenier, J. C., Hip-Ki, E., Bruat, V., et al. (2014). High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 344, 413–415. doi: 10.1126/science.1251110
Hong, E. E., Okitsu, C. Y., Smith, A. D., and Hsieh, C. L. (2013). Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell. Biol. 33, 2683–2690. doi: 10.1128/mcb.00220-13
Hopkins, J. F., Sabelnykova, V. Y., Weischenfeldt, J., Simon, R., Aguiar, J. A., Alkallas, R., et al. (2017). Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 8:656. doi: 10.1038/s41467-017-00377-y
Idaghdour, Y., and Hodgkinson, A. (2017). Integrated genomic analysis of mitochondrial RNA processing in human cancers. Genom. Med. 9:36. doi: 10.1186/s13073-017-0426-0
Infantino, V., Castegna, A., Iacobazzi, F., Spera, I., Scala, I., Andria, G., et al. (2011). Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down’s syndrome. Mol. Genet. Metab. 102, 378–382. doi: 10.1016/j.ymgme.2010.11.166
Jagannathan, R., Thapa, D., Nichols, C. E., Shepherd, D. L., Stricker, J. C., Croston, T. L., et al. (2015). Translational regulation of the mitochondrial genome following redistribution of mitochondrial MicroRNA in the diabetic heart. Circ. Cardiovasc. Genet. 8, 785–802. doi: 10.1161/CIRCGENETICS.115.001067
Jang, H. S., Shin, W. J., Lee, J. E., and Do, J. T. (2017). CpG and Non-CpG Methylation in epigenetic gene regulation and brain function. Genes 8:148. doi: 10.3390/genes8060148
Janssen, B. G., Byun, H. M., Roels, H. A., Gyselaers, W., Penders, J., Baccarelli, A. A., et al. (2017). Regulating role of fetal thyroid hormones on placental mitochondrial DNA methylation: epidemiological evidence from the ENVIRONAGE birth cohort study. Clin. Epigenet. 9:66. doi: 10.1186/s13148-017-0366-y
Jeandard, D., Smirnova, A., Tarassov, I., Barrey, E., Smirnov, A., and Entelis, N. (2019). Import of non-coding RNAs into human mitochondria: a critical review and emerging approaches. Cells 8:286. doi: 10.3390/cells8030286
Jemt, E., Persson, O., Shi, Y., Mehmedovic, M., Uhler, J. P., Dávila López, M., et al. (2015). Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 43, 9262–9275. doi: 10.1093/nar/gkv804
Jones, P. A., Issa, J. P., and Baylin, S. (2016). Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17, 630–641. doi: 10.1038/nrg.2016.93
Jourdain, A. A., Boehm, E., Maundrell, K., and Martinou, J. C. (2016). Mitochondrial RNA granules: compartmentalizing mitochondrial gene expression. J. Cell Biol. 212, 611–614. doi: 10.1083/jcb.201507125
Jourdain, A. A., Koppen, M., Wydro, M., Rodley, C. D., Lightowlers, R. N., Chrzanowska-Lightowlers, Z. M., et al. (2013). GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 17, 399–410. doi: 10.1016/j.cmet.2013.02.005
Kalani, A., Kamat, P. K., Voor, M. J., Tyagi, S. C., and Tyagi, N. (2014). Mitochondrial epigenetics in bone remodeling during hyperhomocysteinemia. Mol. Cell. Biochem. 395, 89–98. doi: 10.1007/s11010-014-2114-3
Kanki, T., Nakayama, H., Sasaki, N., Takio, K., Alam, T. I., Hamasaki, N., et al. (2004). Mitochondrial nucleoid and transcription factor A. Ann. N.Y. Acad. Sci. 1011, 61–68. doi: 10.1007/978-3-662-41088-2-7
Kato, T., Daigo, Y., Hayama, S., Ishikawa, N., Yamabuki, T., Ito, T., et al. (2005). A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 65, 5638–5646. doi: 10.1158/0008-5472.Can-05-0600
Kaufman, B. A., Durisic, N., Mativetsky, J. M., Costantino, S., Hancock, M. A., Grutter, P., et al. (2007). The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 18, 3225–3236. doi: 10.1091/mbc.e07-05-0404
Kausar, S., Wang, F., and Cui, H. (2018). The role of mitochondria in reactive oxygen species generation and its implications for Neurodegenerative diseases. Cells 7:E274. doi: 10.3390/cells7120274
King, G. A., Hashemi Shabestari, M., Taris, K. H., Pandey, A. K., Venkatesh, S., Thilagavathi, J., et al. (2018). Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res. 46, 3633–3642. doi: 10.1093/nar/gky204
Kitow, J., Derda, A. A., Beermann, J., Kumarswarmy, R., Pfanne, A., Fendrich, J., et al. (2016). Mitochondrial long noncoding RNAs as blood based biomarkers for cardiac remodeling in patients with hypertrophic cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 311, H707–H712. doi: 10.1152/ajpheart.00194.2016
Koc, E. C., and Spremulli, L. L. (2003). RNA-binding proteins of mammalian mitochondria. Mitochondrion 2, 277–291. doi: 10.1016/s1567-7249(03)00005-9
Koeck, T., Olsson, A. H., Nitert, M. D., Sharoyko, V. V., Ladenvall, C., Kotova, O., et al. (2011). A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 13, 80–91. doi: 10.1016/j.cmet.2010.12.007
Kolesnikova, O. A., Entelis, N. S., Jacquin-Becker, C., Goltzene, F., Chrzanowska-Lightowlers, Z. M., Lightowlers, R. N., et al. (2004). Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 13, 2519–2534. doi: 10.1093/hmg/ddh267
Kondo, Y., Shinjo, K., and Katsushima, K. (2017). Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 108, 1927–1933. doi: 10.1111/cas.13342
Kren, B. T., Wong, P. Y. P., Sarver, A., Zhang, X., Zeng, Y., and Steer, C. J. (2009). MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 6, 65–72. doi: 10.4161/rna.6.1.7534
Kukat, C., Davies, K. M., Wurm, C. A., Spåhr, H., Bonekamp, N. A., Kühl, I., et al. (2015). Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 112, 11288–11293. doi: 10.1073/pnas.1512131112
Kulis, M., and Esteller, M. (2010). DNA methylation and cancer. Adv. Genet. 70, 27–56. doi: 10.1016/b978-0-12-380866-0.60002-2
Kullar, P., Gomez-Duran, A., Gammage, P., Garone, C., Minczuk, M., Golder, Z., et al. (2017). Heterozygous SSBP1 start loss mutation co-segregates with hearing loss and the m. 1555A4G mtDNA variant in a large multigenerational family. Brain 141, 55–62. doi: 10.1093/brain/awx295
Kumarswamy, R., Bauters, C., Volkmann, I., Maury, F., Fetisch, J., Holzmann, A., et al. (2014). Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575. doi: 10.1161/circresaha.114.303915
Kuriyama, Y., and Luck, D. J. L. (1974). Methylation and processing of mitochondrial ribosomal RNAs in poky and wild-type Neurospora crassa. J. Mol. Biol. 83, 253–266. doi: 10.1016/0022-2836(74)90390-8
Lambowitz, A. M., and Luck, D. J. L. (1975). Methylation of mitochondrial RNA species in the wild-type and poky strains of Neurospora crassa. J. Mol. Biol. 96, 207–214. doi: 10.1016/0022-2836(75)90192-8
Lee, H., Park, J., Tran, Q., Kim, D., Hong, Y., Cho, H., et al. (2017). Mitochondrial transcription factor A (TFAM) is upregulated in glioma. Mol. Med. Rep. 15, 3781–3786. doi: 10.3892/mmr.2017.6467
Lee, K. W., and Bogenhagen, D. F. (2014). Assignment of 2′-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16 S ribosomal RNA (rRNA). J. Biol. Chem. 289, 24936–24942. doi: 10.1074/jbc.C114.581868
Lee, S. R., and Han, J. (2017). Mitochondrial nucleoid: shield and switch of the mitochondrial genome. Oxid. Med. Cell Longev. 2017:8060949. doi: 10.1155/2017/8060949
Lee, W. T., and St John, J. (2015). The control of mitochondrial DNA replication during development and tumorigenesis. Ann. N.Y. Acad. Sci. 1350, 95–106. doi: 10.1111/nyas.12873
Levytskyy, R. M., Germany, E. M., and Khalimonchuk, O. (2016). Mitochondrial quality control proteases in neuronal welfare. J. Neuroimmun. Pharmacol. 11, 629–644. doi: 10.1007/s11481-016-9683-8
Li, M., Wang, Y.-F., Yang, X.-C., Xu, L., Li, W.-M., Xia, K., et al. (2018). Circulating long noncoding RNA LIPCAR Acts as a novel biomarker in patients with ST-segment elevation myocardial infarction. Med. Sci. Monit. 24, 5064–5070. doi: 10.12659/MSM.909348
Li, Q., Dong, Z., Lian, W., Cui, J., Wang, J., Shen, H., et al. (2019). Ochratoxin A causes mitochondrial dysfunction, apoptotic and autophagic cell death and also induces mitochondrial biogenesis in human gastric epithelium cells. Arch. Toxicol. 93, 1141–1155. doi: 10.1007/s00204-019-02433-6
Lin, C. S., Liu, L. T., Ou, L. H., Pan, S. C., Lin, C. I., and Wei, Y. H. (2018). Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol. Rep. 39, 316–330. doi: 10.3892/or.2017.6087
Liu, H., Li, S., Liu, X., Chen, Y., and Deng, H. (2018). SIRT3 overexpression inhibits growth of kidney tumor cells and enhances mitochondrial biogenesis. J. Proteom. Res. 17, 3143–3152. doi: 10.1021/acs.jproteome.8b00260
Liu, J., Jiang, J., Mo, J., Liu, D., Cao, D., Wang, H., et al. (2019). Global DNA 5-hydroxymethylcytosine and 5-formylcytosine contents are decreased in the early stage of hepatocellular carcinoma. Hepatology 69, 196–208. doi: 10.1002/hep.30146
Liu, T., Lu, B., Lee, I., Ondrovicova, G., Kutejova, E., and Suzuki, C. K. (2004). DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J. Biol. Chem. 279, 13902–13910. doi: 10.1074/jbc.M309642200
Lozano-Rosas, M. G., Chávez, E., Aparicio-Cadena, A. R., Velasco-Loyden, G., and de Sánchez, V. C. J. (2018). Mitoepigenetics and hepatocellular carcinoma. Hepatoma Res. 4, 11–24.
Lu, B., Lee, J., Nie, X., Li, M., Morozov, Y. I., Venkatesh, S., et al. (2013). Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 49, 121–132. doi: 10.1016/j.molcel.2012.10.023
Lung, B., Zemann, A., Madej, M. J., Schuelke, M., Techritz, S., Ruf, S., et al. (2006). Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 34, 3842–3852. doi: 10.1093/nar/gkl448
Maekawa, M., Taniguchi, T., Higashi, H., Sugimura, H., Sugano, K., and Kanno, T. (2004). Methylation of mitochondrial DNA is not a useful marker for cancer detection. J. Clin. Chem. 50, 1480–1481. doi: 10.1373/clinchem.2004.035139
Magda, D., Lecane, P., Prescott, J., Thiemann, P., Ma, X., Dranchak, P. K., et al. (2008). mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC Genom. 9:521. doi: 10.1186/1471-2164-9-521
Mahapatra, S., and Adhya, S. (1996). Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J. Biol. Chem. 271, 20432–20437. doi: 10.1074/jbc.271.34.20432
Mahapatra, S., Ghosh, T., and Adhya, S. (1994). Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 22, 3381–3386. doi: 10.1093/nar/22.16.3381
Mahata, B., Mukherjee, S., Mishra, S., Bandyopadhyay, A., and Adhya, S. (2006). Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science 314, 471–474. doi: 10.1126/science.1129754
Manev, H., and Dzitoyeva, S. (2013). Progress in mitochondrial epigenetics. Biomol. Concepts 4, 381–389. doi: 10.1515/bmc-2013-0005
Maniataki, E., and Mourelatos, Z. (2005). Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11, 849–852. doi: 10.1261/rna.2210805
Martijn, J., Vosseberg, J., Guy, L., Offre, P., and Ettema, T. J. G. (2018). Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105. doi: 10.1038/s41586-018-0059-5
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S., and Heintz, N. (2012). MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. doi: 10.1016/j.cell.2012.11.022
Mercer, T. R., Neph, S., Dinger, M. E., Crawford, J., Smith, M. A., Shearwood, A. M., et al. (2011). The human mitochondrial transcriptome. Cell 146, 645–658. doi: 10.1016/j.cell.2011.06.051
Metodiev, M. D., Lesko, N., Park, C. B., Cámara, Y., Shi, Y., Wibom, R., et al. (2009). Methylation of 12S rRNA is necessary for In Vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9, 386–397. doi: 10.1016/j.cmet.2009.03.001
Metodiev, M. D., Spahr, H., Loguercio Polosa, P., Meharg, C., Becker, C., Altmueller, J., et al. (2014). NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 10:e1004110. doi: 10.1371/journal.pgen.1004110
Michael, J. V., Wurtzel, J. G. T., Mao, G. F., Rao, A. K., Kolpakov, M. A., Sabri, A., et al. (2017). Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood 130, 567–580. doi: 10.1182/blood-2016-11-751099
Mo, M., Peng, F., Wang, L., Peng, L., Lan, G., and Yu, S. (2013). Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol. Lett. 6, 617–623. doi: 10.3892/ol.2013.1419
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Morris, M. J., Hesson, L. B., Poulos, R. C., Ward, R. L., Wong, J. W. H., and Youngson, N. A. (2018). Reduced nuclear DNA methylation and mitochondrial transcript changes in adenomas do not associate with mtDNA methylation. Biomark Res. 6:37. doi: 10.1186/s40364-018-0151-x
Mposhi, A., Van der Wijst, M. G., Faber, K. N., and Rots, M. G. (2017). Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. 22, 1099–1113. doi: 10.2741/4535
Mukherjee, S., Basu, S., Home, P., Dhar, G., and Adhya, S. (2007). Necessary and sufficient factors for the import of transfer RNA into the kinetoplast mitochondrion. EMBO Rep. 8, 589–595. doi: 10.1038/sj.embor.7400979
Muñoz-Gómez, S. A., Wideman, J. G., Roger, A. J., and Slamovits, C. H. (2017). The origin of mitochondrial cristae from Alphaproteobacteria. Mol. Biol. Evol. 34, 943–956. doi: 10.1093/molbev/msw298
Nebbioso, A., Tambaro, F. P., Dell’Aversana, C., and Altucci, L. (2018). Cancer epigenetics: moving forward. PLoS Genet. 14:e1007362. doi: 10.1371/journal.pgen.1007362
Ngo, H. B., Kaiser, J. T., and Chan, D. C. (2011). The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 18, 1290–1296. doi: 10.1038/nsmb.2159
Ngo, H. B., Lovely, G. A., Phillips, R., and Chan, D. C. (2014). Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 5:3077. doi: 10.1038/ncomms4077
O’Sullivan, M., Rutland, P., Lucas, D., Ashton, E., Hendricks, S., Rahman, S., et al. (2014). Mitochondrial m. 1584A 12S m62A rRNA methylation in families with m. 1555A> G associated hearing loss. Hum. Mol. Genet. 24, 1036–1044. doi: 10.1093/hmg/ddu518
Ochsenreiter, T., and Hajduk, S. L. (2006). Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 7, 1128–1133. doi: 10.1038/sj.embor.7400817
Olavarria, J. V., Burzio, V. A., Borgna, V., Lobos-Gonzalez, L., Araya, M., Guevara, F., et al. (2018). “Long noncoding mitochondrial RNAs (LncmtRNAs) as targets for cancer therapy,” in Mitochondrial DNA - New Insights, ed. H. Seligmann, (London: Intechopen), 179.
Peschansky, V. J., and Wahlestedt, C. (2014). Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 9, 3–12. doi: 10.4161/epi.27473
Picard, M., Wallace, D. C., and Burelle, Y. (2016). The rise of mitochondria in medicine. Mitochondrion 30, 105–116. doi: 10.1016/j.mito.2016.07.003
Pintard, L., Bujnicki, J. M., Lapeyre, B., and Bonnerot, C. (2002). MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 21, 1139–1147. doi: 10.1093/emboj/21.5.1139
Popow, J., Alleaume, A.-M., Curk, T., Schwarzl, T., Sauer, S., and Hentze, M. W. (2015). FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 21, 1873–1884. doi: 10.1261/rna.052365.115
Qiao, L., Ru, G., Mao, Z., Wang, C., Nie, Z., Li, Q., et al. (2017). Mitochondrial DNA depletion, mitochondrial mutations and high TFAM expression in hepatocellular carcinoma. Oncotarget 8, 84373–84383. doi: 10.18632/oncotarget.21033
Rackham, O., Shearwood, A.-M. J., Mercer, T. R., Davies, S. M. K., Mattick, J. S., and Filipovska, A. (2011). Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093. doi: 10.1261/rna.029405.111
Rebelo, A. P., Williams, S. L., and Moraes, C. T. (2009). In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 37, 6701–6715. doi: 10.1093/nar/gkp727
Richa, R., and Sinha, R. P. (2014). Hydroxymethylation of DNA: an epigenetic marker. EXCLI J. 13, 592–610.
Rinehart, J., Krett, B., Rubio, M. A., Alfonzo, J. D., and Soll, D. (2005). Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 19, 583–592. doi: 10.1101/gad.1269305
Rippo, M. R., Olivieri, F., Monsurro, V., Prattichizzo, F., Albertini, M. C., and Procopio, A. D. (2014). MitomiRs in human inflamm-aging: a hypothesis involving miR-181a, miR-34a and miR-146a. Exp. Gerontol. 56, 154–163. doi: 10.1016/j.exger.2014.03.002
Rivas, A., Burzio, V., Landerer, E., Borgna, V., Gatica, S., Avila, R., et al. (2012). Determination of the differential expression of mitochondrial long non-coding RNAs as a noninvasive diagnosis of bladder cancer. BMC Urol. 12:37. doi: 10.1186/1471-2490-12-37
Ro, S., Ma, H. Y., Park, C., Ortogero, N., Song, R., Hennig, G. W., et al. (2013). The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23, 759–774. doi: 10.1038/cr.2013.37
Romano, G., Veneziano, D., Nigita, G., and Nana-Sinkam, S. P. (2018). RNA Methylation in ncRNA: classes, detection, and molecular associations. Front. Genet. 9:243. doi: 10.3389/fgene.2018.00243
Ronchi, D., Garone, C., Bordoni, A., Gutierrez Rios, P., Calvo, S., Ripolone, M., et al. (2012). Next-generation sequencing reveals DGUOK mutations in adult patients with mitochondrial DNA multiple deletions. Brain 135, 3404–3415. doi: 10.1093/brain/aws258
Rorbach, J., and Minczuk, M. (2012). The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J. 444, 357–373. doi: 10.1042/bj20112208
Rubio, M. A. T., Rinehart, J. J., Krett, B., Duvezin-Caubet, S., Reichert, A. S., Söll, D., et al. (2008). Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. U.S.A. 105, 9186–9191. doi: 10.1073/pnas.0804283105
Sadakierska-Chudy, A., Frankowska, M., and Filip, M. (2014). Mitoepigenetics and drug addiction. Pharmacol. Ther. 144, 226–233. doi: 10.1016/j.pharmthera.2014.06.002
Saikia, M., and Hatzoglou, M. (2015). The many virtues of tRNA-derived stress-induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J. Biol. Chem. 290, 29761–29768. doi: 10.1074/jbc.R115.694661
Salinas, T., Duchene, A. M., Delage, L., Nilsson, S., Glaser, E., Zaepfel, M., et al. (2006). The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 103, 18362–18367. doi: 10.1073/pnas.0606449103
Santos, J. M., Mishra, M., and Kowluru, R. A. (2014). Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp. Eye Res. 121, 168–177. doi: 10.1016/j.exer.2014.02.010
Sbisà, E., Tullo, A., Nardelli, M., Tanzariello, F., and Saccone, C. (1992). Transcription mapping of the Ori L region reveals novel precursors of mature RNA species and antisense RNAs in rat mitochondrial genome. FEBS Lett. 296, 311–316. doi: 10.1016/0014-5793(92)80311-4
Schneider, A., and Maréchal-Drouard, L. (2000). Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 10, 509–513. doi: 10.1016/S0962-8924(00)01854-7
Schneider, A., Martin, J., and Agabian, N. (1994). A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol. Cell. Biol. 14, 2317–2322. doi: 10.1128/mcb.14.4.2317
Schulte, C., Barwari, T., Joshi, A., Theofilatos, K., Zampetaki, A., Barallobre-Barreiro, J., et al. (2019). Comparative analysis of circulating Noncoding RNAs versus protein biomarkers in the detection of myocardial injury. Circ. Res. 125, 328–340. doi: 10.1161/circresaha.119.314937
Sharoyko, V. V., Abels, M., Sun, J., Nicholas, L. M., Mollet, I. G., Stamenkovic, J. A., et al. (2014). Loss of TFB1M results in mitochondrial dysfunction that leads to impaired insulin secretion and diabetes. Hum. Mol. Genet. 23, 5733–5749. doi: 10.1093/hmg/ddu288
Shepherd, D. L., Hathaway, Q. A., Pinti, M. V., Nichols, C. E., Durr, A. J., Sreekumar, S., et al. (2017). Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J. Mol. Cell Cardiol. 110, 15–25. doi: 10.1016/j.yjmcc.2017.06.012
Shinde, S., and Bhadra, U. (2015). A complex genome-microRNA interplay in human mitochondria. Biomed. Res. Int. 2015:206382. doi: 10.1155/2015/206382
Shock, L. S., Thakkar, P. V., Peterson, E. J., Moran, R. G., and Taylor, S. M. (2011). DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 3630–3635. doi: 10.1073/pnas.1012311108
Shokolenko, I. N., and Alexeyev, M. F. (2015). Mitochondrial DNA: a disposable genome? Biochim. Biophys. Acta 1852, 1805–1809. doi: 10.1016/j.bbadis.2015.05.016
Shokolenko, I. N., and Alexeyev, M. F. (2017). Mitochondrial transcription in mammalian cells. Front. Biosci. 22:835. doi: 10.2741/4520
Sirey, T. M., Roberts, K., Haerty, W., Bedoya-Reina, O., Rogatti-Granados, S., Tan, J. Y., et al. (2019). The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. eLife 8:e45051. doi: 10.7554/eLife.45051
Sirum-Connolly, K., and Mason, T. (1993). Functional requirement of a site-specific ribose methylation in ribosomal RNA. J. Sci. 262, 1886–1889. doi: 10.1126/science.8266080
Smalheiser, N. R., Lugli, G., Thimmapuram, J., Cook, E. H., and Larson, J. (2011). Mitochondrial small RNAs that are up-regulated in hippocampus during olfactory discrimination training in mice. Mitochondrion 11, 994–995. doi: 10.1016/j.mito.2011.08.014
Smirnov, A., Comte, C., Mager-Heckel, A. M., Addis, V., Krasheninnikov, I. A., Martin, R. P., et al. (2010). Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. J. Biol. Chem. 285, 30792–30803. doi: 10.1074/jbc.M110.151183
Srinivasan, S., Guha, M., Kashina, A., and Avadhani, N. G. (2017). Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim. Biophys. Acta Bioenerg. 1858, 602–614. doi: 10.1016/j.bbabio.2017.01.004
Sripada, L., Singh, K., Lipatova, A. V., Singh, A., Prajapati, P., Tomar, D., et al. (2017). hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J. Mol. Med. 95, 641–651. doi: 10.1007/s00109-017-1517-5
Stein, C. S., Jadiya, P., Zhang, X., McLendon, J. M., Abouassaly, G. M., Witmer, N. H., et al. (2018). Mitoregulin: a lncRNA-Encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 23:3710-3720.e8. doi: 10.1016/j.celrep.2018.06.002
Stewart, J. B., Alaei-Mahabadi, B., Sabarinathan, R., Samuelsson, T., Gorodkin, J., Gustafsson, C. M., et al. (2015). Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 11:e1005333. doi: 10.1371/journal.pgen.1005333
Stimpfel, M., Jancar, N., and Virant-Klun, I. (2018). New challenge: mitochondrial epigenetics? Stem Cell Rev. 14, 13–26. doi: 10.1007/s12015-017-9771-z
Stoccoro, A., Siciliano, G., Migliore, L., and Coppede, F. (2017). Decreased Methylation of the mitochondrial D-Loop region in late-onset Alzheimer’s disease. J. Alzheimers Dis. 59, 559–564. doi: 10.3233/jad-170139
Sun, X., Vaghjiani, V., Jayasekara, W. S. N., Cain, J. E., and St John, J. C. (2018a). The degree of mitochondrial DNA methylation in tumor models of glioblastoma and osteosarcoma. Clin. Epigenet. 10:157. doi: 10.1186/s13148-018-0590-0
Sun, X., Zhan, L., Chen, Y., Wang, G., He, L., Wang, Q., et al. (2018b). Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct. Target. Ther. 3:8. doi: 10.1038/s41392-018-0011-z
Szczesny, R. J., Wojcik, M. A., Borowski, L. S., Szewczyk, M. J., Skrok, M. M., Golik, P., et al. (2013). Yeast and human mitochondrial helicases. Biochim. Biophys. Acta 1829, 842–853. doi: 10.1016/j.bbagrm.2013.02.009
Tavtigian, S. V., Simard, J., Teng, D. H., Abtin, V., Baumgard, M., Beck, A., et al. (2001). A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 27, 172–180. doi: 10.1038/84808
Taylor, R. W., and Turnbull, D. M. (2005). Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6, 389–402. doi: 10.1038/nrg1606
Temperley, R. J., Wydro, M., Lightowlers, R. N., and Chrzanowska-Lightowlers, Z. M. (2010). Human mitochondrial mRNAs—like members of all families, similar but different. Biochim. Biophys. Acta. 1797, 1081–1085. doi: 10.1016/j.bbabio.2010.02.036
Thum, T., Kumarswamy, R., Pinet, F., Bauters, C., and De Groote, P. (2017). Mitochondrial Non-Coding RNAs for Predicting Disease Progression in Heart Failure and Myocardial Infarction Patients. France: Pasteur Institute of Lille.
Tong, H., Zhang, L., Gao, J., Wen, S., Zhou, H., and Feng, S. (2017). Methylation of mitochondrial DNA displacement loop region regulates mitochondrial copy number in colorectal cancer. Mol. Med. Rep. 16, 5347–5353. doi: 10.3892/mmr.2017.7264
Tschopp, F., Charrière, F., and Schneider, A. (2011). In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 12, 825–832. doi: 10.1038/embor.2011.111
van der Bliek, A. M., Sedensky, M. M., and Morgan, P. G. (2017). Cell biology of the Mitochondrion. Genetics 207, 843–871. doi: 10.1534/genetics.117.300262
van der Wijst, M. G., and Rots, M. G. (2015). Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet. 31, 353–356. doi: 10.1016/j.tig.2015.03.009
van der Wijst, M. G., van Tilburg, A. Y., Ruiters, M. H., and Rots, M. G. (2017). Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 7:177. doi: 10.1038/s41598-017-00263-z
Van Haute, L., Lee, S. Y., McCann, B. J., Powell, C. A., Bansal, D., Vasiliauskaite, L., et al. (2019). NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 47, 8720–8733. doi: 10.1093/nar/gkz559
Van Haute, L., Pearce, S. F., Powell, C. A., D’Souza, A. R., Nicholls, T. J., and Minczuk, M. (2015). Mitochondrial transcript maturation and its disorders. J. Inherit. Metab. Dis. 38, 655–680. doi: 10.1007/s10545-015-9859-z
Vendramin, R., Marine, J.-C., and Leucci, E. (2017). Non-coding RNAs: the dark side of nuclear-mitochondrial communication. EMBO J. 36, 1123–1133. doi: 10.15252/embj.201695546
Vidaurre, S., Fitzpatrick, C., Burzio, V. A., Briones, M., Villota, C., Villegas, J., et al. (2014). Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy. J. Biol. Chem. 289, 27182–27198. doi: 10.1074/jbc.M114.558841
Vilardo, E., Nachbagauer, C., Buzet, A., Taschner, A., Holzmann, J., and Rossmanith, W. (2012). A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583–11593. doi: 10.1093/nar/gks910
Villegas, J., Burzio, V., Villota, C., Landerer, E., Martinez, R., Santander, M., et al. (2007). Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res. 35, 7336–7347. doi: 10.1093/nar/gkm863
Villota, C., Campos, A., Vidaurre, S., Oliveira-Cruz, L., Boccardo, E., Burzio, V. A., et al. (2012). Expression of mitochondrial non-coding RNAs (ncRNAs) is modulated by high risk human papillomavirus (HPV) oncogenes. J. Biol. Chem. 287, 21303–21315. doi: 10.1074/jbc.M111.326694
Voigts-Hoffmann, F., Hengesbach, M., Kobitski, A. Y., van Aerschot, A., Herdewijn, P., Nienhaus, G. U., et al. (2007). A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys). J. Am. Chem. Soc. 129, 13382–13383. doi: 10.1021/ja075520
Vyas, S., Zaganjor, E., and Haigis, M. C. (2016). Mitochondria and cancer. Cell 166, 555–566. doi: 10.1016/j.cell.2016.07.002
Wallace, D. C. (1992). Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science 256:628. doi: 10.1126/science.1533953
Wallace, D. C. (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407. doi: 10.1146/annurev.genet.39.110304.095751
Wang, G., Chen, H.-W., Oktay, Y., Zhang, J., Allen, E. L., Smith, G. M., et al. (2010). PNPASE regulates RNA import into mitochondria. Cell 142, 456–467. doi: 10.1016/j.cell.2010.06.035
Wang, K. Z., Zhu, J., Dagda, R. K., Uechi, G., Cherra, S. J. III, Gusdon, A. M., et al. (2014). ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson’s disease. Mitochondrion 17, 132–140. doi: 10.1016/j.mito.2014.04.008
Wang, Z., Tang, K., Zhang, D., Wan, Y., Wen, Y., Lu, Q., et al. (2017). High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS One 12:e0185612. doi: 10.1371/journal.pone.0185612
Woischnik, M., and Moraes, C. T. (2002). Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genom. Res. 12, 885–893. doi: 10.1101/gr.227202
Wu, C., and Morris, J. R. (2001). Genes, genetics, and epigenetics: a correspondence. Science 293, 1103–1105. doi: 10.1126/science.293.5532.1103
Xie, D., Wu, X., Lan, L., Shangguan, F., Lin, X., Chen, F., et al. (2016). Downregulation of TFAM inhibits the tumorigenesis of non-small cell lung cancer by activating ROS-mediated JNK/p38MAPK signaling and reducing cellular bioenergetics. Oncotarget 7, 11609–11624. doi: 10.18632/oncotarget.7018
Yamamoto, Y. (2001). “D-Loop,” in Encyclopedia of Genetics, eds S. Brenner, and J. H. Miller, (New York, NY: Academic Press), 539–540.
Yan, K., An, T., Zhai, M., Huang, Y., Wang, Q., Wang, Y., et al. (2019). Mitochondrial miR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 10:500. doi: 10.1038/s41419-019-1734-7
Yang, K. C., Yamada, K. A., Patel, A. Y., Topkara, V. K., George, I., Cheema, F. H., et al. (2014). Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021. doi: 10.1161/circulationaha.113.003863
Yien, Y., Robledo, R., Schultz, J., Takahashi-Makise, N., Gwynn, B., Bauer, D., et al. (2014). TMEM14C is required for erythroid mitochondrial heme metabolism. J. Clin. Invest. 124, 4294–4304. doi: 10.1172/JCI76979
Young, M. J., and Copeland, W. C. (2016). Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 38, 52–62. doi: 10.1016/j.gde.2016.03.005
Zhang, C., and Jia, G. (2018). Reversible RNA Modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom. Proteom. Bioinform. 16, 155–161. doi: 10.1016/j.gpb.2018.03.003
Zhang, J., Liang, J., and Huang, J. (2016). Downregulated microRNA-26a modulates prostate cancer cell proliferation and apoptosis by targeting COX-2. Oncol. Lett. 12, 3397–3402. doi: 10.3892/ol.2016.5070
Zhang, J., Zhang, X., Li, C., Yue, L., Ding, N., Riordan, T., et al. (2019). Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 16, 220–232. doi: 10.1080/15476286.2019.1565284
Zhang, X., Zuo, X., Yang, B., Li, Z., Xue, Y., Zhou, Y., et al. (2014). MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158, 607–619. doi: 10.1016/j.cell.2014.05.047
Zhao, Y., Cui, J., Hu, J., and Hoffman, A. R. (2019). Effect of MALAT1 in the crosstalk between nucleus and mitochondria on mitochondrial reprogramming in hepatocellular carcinoma cells. J. Clin. Oncol. 37:e14711. doi: 10.1200/JCO.2019.37.15-suppl.e14711
Zhou, R., Wang, R., Qin, Y., Ji, J., Xu, M., Wu, W., et al. (2015). Mitochondria-related miR-151a-5p reduces cellular ATP production by targeting CYTB in asthenozoospermia. Sci. Rep. 5:17743. doi: 10.1038/srep17743
Zhu, S., Dong, Z., Ke, X., Hou, J., Zhao, E., Zhang, K., et al. (2019). The roles of sirtuins family in cell metabolism during tumor development. Semin. Cancer Biol. 57, 59–71. doi: 10.1016/j.semcancer.2018.11.003
Keywords: mtDNA methylation, mitoepigenetics, mtRNA modification, non-coding RNAs, cancer
Citation: Dong Z, Pu L and Cui H (2020) Mitoepigenetics and Its Emerging Roles in Cancer. Front. Cell Dev. Biol. 8:4. doi: 10.3389/fcell.2020.00004
Received: 07 August 2019; Accepted: 08 January 2020;
Published: 23 January 2020.
Edited by:
Joanna Rorbach, Karolinska Institutet (KI), SwedenReviewed by:
Yasuo Shinohara, Tokushima University, JapanCopyright © 2020 Dong, Pu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjuan Cui, aGN1aUBzd3UuZWR1LmNu; aG9uZ2p1YW4uY3VpQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.