
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 22 January 2020
Sec. Stem Cell Research
Volume 7 - 2019 | https://doi.org/10.3389/fcell.2019.00360
This article is part of the Research TopicThe RNA Revolution in Embryonic Development and Cell Differentiation in Health and DiseaseView all 17 articles
The highly conserved Argonaute protein family members play a central role in the regulation of gene expression networks, orchestrating the establishment and the maintenance of cell identity throughout the entire life cycle, as well as in several human disorders, including cancers. Four functional Argonaute proteins (AGO1–4), with high structure similarity, have been described in humans and mice. Interestingly, only AGO2 is robustly expressed during human and mouse early development, in contrast to the other AGOs. Consequently, AGO2 is indispensable for early development in vivo and in vitro. Here, we review the roles of Argonaute proteins during early development by focusing on the interplay between specific domains of the protein and their function. Moreover, we report recent works highlighting the importance of AGO posttranslational modifications in cancer.
Historically, the Argonaute (AGO) protein family has been discovered in a plant mutagenesis screen, performed to identify new genes involved in Arabidopsis thaliana development (Bohmert et al., 1998). This first report already highlighted the conservation of the Argonaute gene family in multicellular organisms suggesting its important functions. It was later demonstrated that AGOs are conserved throughout all domains of life (Swarts et al., 2014). Eukaryotic AGOs are involved in many cellular processes and act as mediators of gene silencing (Bartel, 2018). In mammals, AGOs have been mainly described for their cytoplasmic role in small RNA (smRNA) biogenesis, as key components of the RNA-induced silencing complex (RISC) (Bodak et al., 2017a).
Two types of ∼22 nt smRNAs can be loaded into AGOs to induce translational inhibition or exonucleolytic messenger RNA (mRNA) decay of specific transcripts: small interfering RNAs (siRNAs) and microRNAs (miRNAs). Both species are processed in the cytoplasm by DICER, leading to the release of double-stranded RNA (dsRNA) duplexes, which will be loaded into the RISC complex to achieve its RNA interference (RNAi) functions [for reviews (Bodak et al., 2017a; Treiber et al., 2019)].
Furthermore, the regulatory role of smRNAs expands beyond the posttranscriptional regulation mediated by miRNAs. In fact, smRNAs with AGOs as their effector proteins have been described to be involved in transcriptional gene silencing or activation (Malecová and Morris, 2010), alternative splicing (Alló et al., 2009; Harel-Bellan et al., 2013), antiviral defense (Maillard et al., 2013), genome integrity control (Svoboda et al., 2004; Kanellopoulou et al., 2005; Bodak et al., 2017b), DNA repair (Hawley et al., 2017), and epigenetic modification of the chromatin (Li, 2014). Although the expression of new smRNA species such as small nucleolar RNA (sno-RNA)- and transfer RNA (tRNA)-derived fragments has been recently described to be altered in the context of cancer, their functions remain largely unexplored (Martens-Uzunova et al., 2013; Schorn et al., 2017; Kuscu et al., 2018). However, recent evidence shows that the functions and biogenesis of these new smRNA species are tightly connected to the RNAi pathway, functioning both in the cytoplasm and the nucleus (Huang and Li, 2014; Sarshad et al., 2018).
In this review, we highlight novel findings on the structures of AGO proteins since the description of the human AGO2 (Schirle and Macrae, 2012) and link these with their roles in mammalian early development and carcinogenesis.
Structures of prokaryotic and mammalian Argonaute proteins have been extensively studied in the past decades and have given revealing insights into the mechanism of translational inhibition by miRNAs. In this part, we only focus on the structural data of the human AGO proteins, which are highly conserved and share ∼85% of sequence identity (https://myhits.isb-sib.ch/cgi-bin/profile_search?data=5485215623128241).
Four Argonaute proteins (AGO1–4) are expressed in humans. AGO2 is described best and has long been thought to be the only Argonaute protein member having mRNA slicing activity, due to its unique structural characteristics (Liu et al., 2004; Meister et al., 2004). Nevertheless, AGO3 has recently been shown to slice target RNAs, however, only when loaded with certain miRNAs (Park et al., 2017). In these cases compared to AGO2, the slicing activity depended strongly on the pairing of the postseed region of the guide RNA as well as on the 5′ and 3′ flanking regions of the target RNA (Park et al., 2017).
The four human Argonaute proteins are structurally very similar but nevertheless contain few non-conserved amino acids in their functional domains. The AGO2 full-length protein structure was resolved first and was largely studied (Elkayam et al., 2012; Schirle and Macrae, 2012; Figure 1A). Since then, structural data on all the others, AGO1, 3, 4 full proteins have become available (Faehnle et al., 2013; Nakanishi et al., 2013; Park et al., 2017, 2019). These studies have revealed four conserved domains: the N-terminal domain (N), the PIWI/Argonaute/Zwille (PAZ) domain, the MID domain, and the P-element-induced whimpy tested (PIWI) domain. The PAZ domain, which is required for anchoring the 3′ end of guide RNAs, and the MID domain, which binds the 5′ phosphate of guide RNAs (Lingel et al., 2003; Song et al., 2003, 2004; Ma et al., 2004; Yan et al., 2004; Boland et al., 2010, 2011; Frank et al., 2010), are very similar between the four AGOs (Figure 1A).
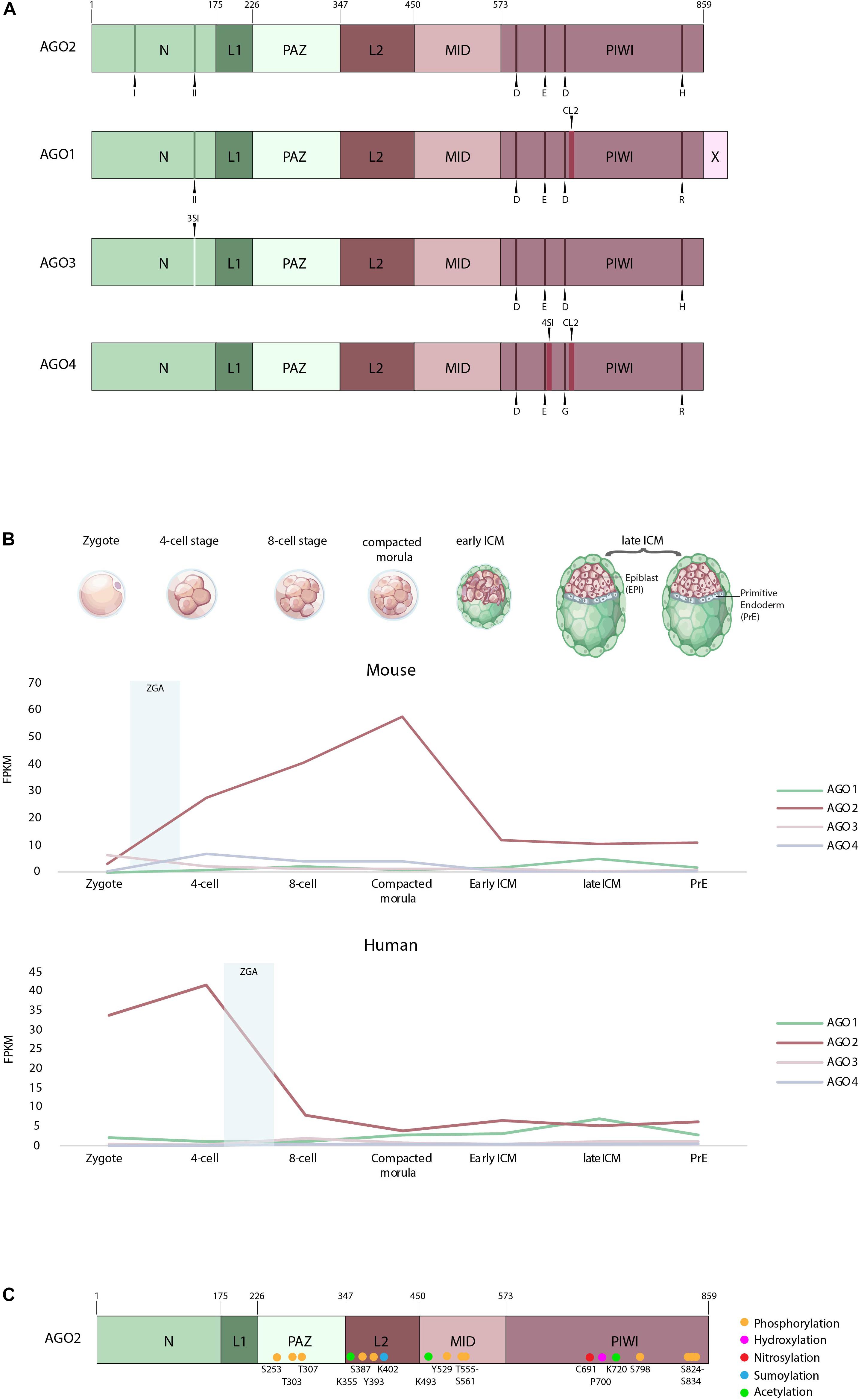
Figure 1. (A) Domain organization of AGO1–4 (adapted from Elkayam et al., 2012). Indicated are the two N-terminal motifs labeled I and II and the residues of the catalytic tetrad in the PIWI domain D, E, D, H, R, and G. Also highlighted in AGO1 and AGO4 is cluster 2 (CL2), the AGO3-specific insertion (3SI) and the AGO4-specific insertion (4SI). N, N-terminus; L1, linker domain 1; PAZ, PIWI/Argonaute/Zwille domain; L2, linker domain 2; MID, MID domain; PIWI, P-element-induced whimpy testes domain; X, AGO1x additional 33 aa; D, aspartate; E, glutamate; H, histidine; R, arginine; G, glycine. (B) Expression of human and mouse AGO1–4 in the zygote, four-cell, eight-cell, compacted morula, early inner cell mass (ICM) and late ICM, according to single-cell expression data from Boroviak et al. (2018). ZGA, first major wave of zygotic gene activation. (C) Posttranslational modifications of the human Argonaute 2 protein.
The N-terminal domain, however, differs between AGO1–4. In AGO2, the N-terminal domain comprises two motifs (residues 44–48 and 134–166), which are required for its full catalytic activity. Upon mutation of these motifs, AGO2 fails to initiate RISC activation and mRNA cleavage. During protein folding, these residues are located in the vicinity of the PIWI domain and hence are required for correct guide-target positioning (Hauptmann et al., 2013; Schürmann et al., 2013; Figure 1A). On the other hand, AGO1 harbors only one of the N-terminal motifs, required for full catalytic activity (Faehnle et al., 2013; Hauptmann et al., 2013, 2014; Figure 1A), whereas AGO3 and AGO4 possess none, which was thought to render them catalytically inactive (Faehnle et al., 2013; Hauptmann et al., 2013, 2014; Nakanishi et al., 2013; Schürmann et al., 2013; Park et al., 2019). In addition, AGO3 has a specific insertion (3SI) in the N-terminal domain, which leads to a wider and imperfect nucleic-acid binding channel compared to AGO2 (Park et al., 2017).
The PIWI domain is similar to an RNAse H domain, harboring the catalytic triad DDH, which is critical for the slicing activity of AGO2 (Parker et al., 2004, 2005; Song et al., 2004; Ma et al., 2005; Rivas et al., 2005; Yuan et al., 2005). This work has later been challenged by Nakanishi et al. (2012), who demonstrated that not only is a catalytic triad but also a catalytic tetrad (DEDH) is essential for the AGO2 slicing activity (Figure 1A). Indeed, mutation of the glutamate in this catalytic tetrad abolishes the ability of the protein to induce RNAi (Nakanishi et al., 2012). AGO3, like AGO2, has a fully functional PIWI domain with a DEDH. The slicing activity of this domain has been proven by domain swap experiments, showing that AGO3 PIWI domain introduced in an AGO2–AGO3 PIWI chimeric protein can be catalytically active (Hauptmann et al., 2013; Schürmann et al., 2013). AGO1 comprises also several domain changes, the first one being a residue change in the catalytic tetrad of the PIWI domain (Figure 1A). Second, two proline residues at position 670 and 675 in the unique structural element, called cluster 2 (CL2) [also known as conserved segment (CS7)] can bend the protein, which sterically hinders the positioning of the guide/target complex (Nakanishi et al., 2013). In the same conserved segment, another mutated residue, L674, has been shown to decrease the slicing efficiency of AGO1 (Faehnle et al., 2013). Similar to AGO1, AGO4 lacks key catalytic residues and has an additional AGO4-specific insertion (4SI) in the PIWI domain, together with the cluster 2, already observed in AGO1. Only swapping of these domains with their AGO2 counterparts has enabled AGO4 to be catalytically active. Therefore, the native AGO4 is thought to be slicing incompetent (Nakanishi et al., 2013; Hauptmann et al., 2014; Park et al., 2019; Figure 1A). In addition, in the recently published AGO4 structure, the so-called LAKEs were observed, which are an accumulation of water molecules below the nucleic acid binding channel. This formation is conserved in all human AGOs. LAKE formation aids to establish the RISC assembly and is important for smRNA duplex loading (Park et al., 2019).
Finally, AGO1 has also been detected as a candidate for programed translational readthrough, a process generating longer isoforms by continuing translation beyond the stop codon (Eswarappa et al., 2014). Two recent studies demonstrated the presence of this translational readthrough product of AGO1 in cells, termed AGO1x (Ghosh et al., 2019; Singh et al., 2019). AGO1x is a protein isoform, which is 33 amino acids longer than AGO1 (Figure 1A). Initially, it was shown in HeLa cells that AGO1x can interact with miRNAs and their mRNA targets; however, no interaction with GW182 has been observed. GW182 proteins normally interact with the Argonautes, mediating translational repression (Eulalio et al., 2009). Since AGO1x is incapable of interacting with GW182, it cannot induce translational repression. It is therefore thought that AGO1x competes with the canonical miRNA pathway and thereby leads to reduced posttranscriptional repression of target mRNAs (Singh et al., 2019). Second, in breast cancer cells, AGO1x has been shown to prevent the accumulation of dsRNAs and thereby suppresses the interferon response in these cells, a function independent of the canonical miRNA pathway (Ghosh et al., 2019).
Although structurally very similar, the expression of AGOs can differ greatly during early development. We focus in this part on the difference in expression of mammalian AGO1–4 during early embryonic development. The mouse and human AGOs are highly conserved with almost identical protein sequences [99% for AGO2, 3, and 4 and 100% for AGO1 between mouse and human (https://myhits.isb-sib.ch/cgi-bin/profile_search?data=5485215623128241)].
The expression of the four AGOs during mouse early development was originally monitored using reverse transcription followed by PCR approaches and revealed the expression of the four transcripts in oocytes and at early stages of development (Lykke-Andersen et al., 2008). Nowadays, newer technologies allow to determine the expression of certain transcripts on a single-cell level (Stuart and Satija, 2019). A recent study, using this single-cell sequencing technology at different stages of preimplantation development in humans and mice allowed us to look into the detailed expression of the Argonautes throughout preimplantation development (Boroviak et al., 2018). Two cell fate decision events occur during preimplantation development (Niakan and Eggan, 2013). At the blastocyst stage, two populations of cells are segregating to create two distinct lineages: the trophectoderm, an extraembryonic tissue at the origin of the placenta, and the inner cell mass (ICM), the future epiblast at the origin of the three germ layers of the embryo. This first cell fate choice takes place 3 days postfertilization (dpf) in mice and 5 dpf in humans. The second cell fate specification event allows the segregation of the ICM and another extraembryonic layer: the primitive endoderm at the origin of the yolk sac, which appears 4 and 7 dpf in mice and humans, respectively (Niakan and Eggan, 2013).
For both species, the monitored expression of Ago1, 3, and 4 mRNAs during preimplantation stages is low compared to Ago2 transcripts (Figure 1B). However, it is to note that, in both species, Ago2 represents still <1% of all detected transcripts (Boroviak et al., 2018). In mouse, Ago1, 3, and 4 mRNAs are lowly expressed from the zygote to the early ICM and the primitive endoderm, compared to Ago2 (Figure 1B). Interestingly, at the late blastocyst stage, a decrease in Ago2 is observed, in parallel with an increase in Ago1, suggesting a possible novel role for AGO1 just before implantation (Figure 1B). The expression profiles of the AGOs seem very different in human early embryos. AGO1 mRNAs increase continuously from the eight-cell stage to reach comparable levels of AGO2 expression, or even slightly higher at the late ICM stage (Figure 1B). In both species, however, AGO2 is the most expressed Argonaute mRNA during early preimplantation development, in embryonic and primitive endoderm lineages. However, maximal expression occurs at different stages in human and mouse. Human AGO2 (hAGO2) peaks at the four-cell stage, whereas mouse Ago2 (mAgo2) peaks at the morula stage (Figure 1B). As AGOs are actually required in early stages to degrade maternally deposited transcripts (Lykke-Andersen et al., 2008), the question arises whether the expression of AGO2 coincides with zygotic gene activation (ZGA). In humans, the first major wave of ZGA occurs at the 4- to 8-cell stages followed by a second one at the 8- to 16-cell stages [for review, see Jukam et al. (2017)]. This, however, does not anymore correlate with the expression of hAGO2 transcripts, which decreases after the four-cell stage. Moreover, it raises the question whether most of the hAGO2 in early development is actually maternally deposited. On the contrary, the first major wave of ZGA in mice is already detectable at the two-cell stage, followed by a second wave at the four- to eight-cell stages [for review, see Jukam et al. (2017)]. This correlates with the increasing expression of mAgo2, which reaches its peak at the morula stage. Interestingly, recent studies in mice preimplantation development have demonstrated that mRNA levels do not always correlate with the protein levels (Gao et al., 2017). Moreover, protein expression often lags behind the mRNA expression in the process of ZGA. Therefore, we do not know whether the mRNA expression levels of the Argonautes discussed above reflect the protein levels within each cell (Gao et al., 2017).
Posttranslational modifications (PTMs) of proteins mediate a huge range of signaling events within a cell and are therefore critical for distinct processes such as developmental timing (Seet et al., 2006). In fact, in Caenorhabditis elegans, phosphorylation of the ALG-1 protein, an ortholog of the human Argonaute proteins, is required for miRNA-mediated gene silencing and the proper animal development (Quévillon Huberdeau et al., 2017). Animals expressing a phosphomutant ALG-1 display developmental defects and die at the adult stage, exemplifying the importance of posttranslational modification of Argonaute proteins during development (Quévillon Huberdeau et al., 2017).
AGO2 has been shown to be highly posttranslationally modified, which affects its protein stability and miRNA activity (Figure 1C) (Meister, 2013). Most of the posttranslational modifications have been observed in human cancer cells, yet their amino acids are conserved between mice and humans. The regulation of PTM of AGO proteins was recently related to the activity of well-characterized oncogenes, underlining the relevance of AGO-dependent pathways deregulation in cancer development. However, their importance in early development has not been assessed in mammals. In Table 1, we highlight the reported PTM sites of AGO2 and their molecular consequences.
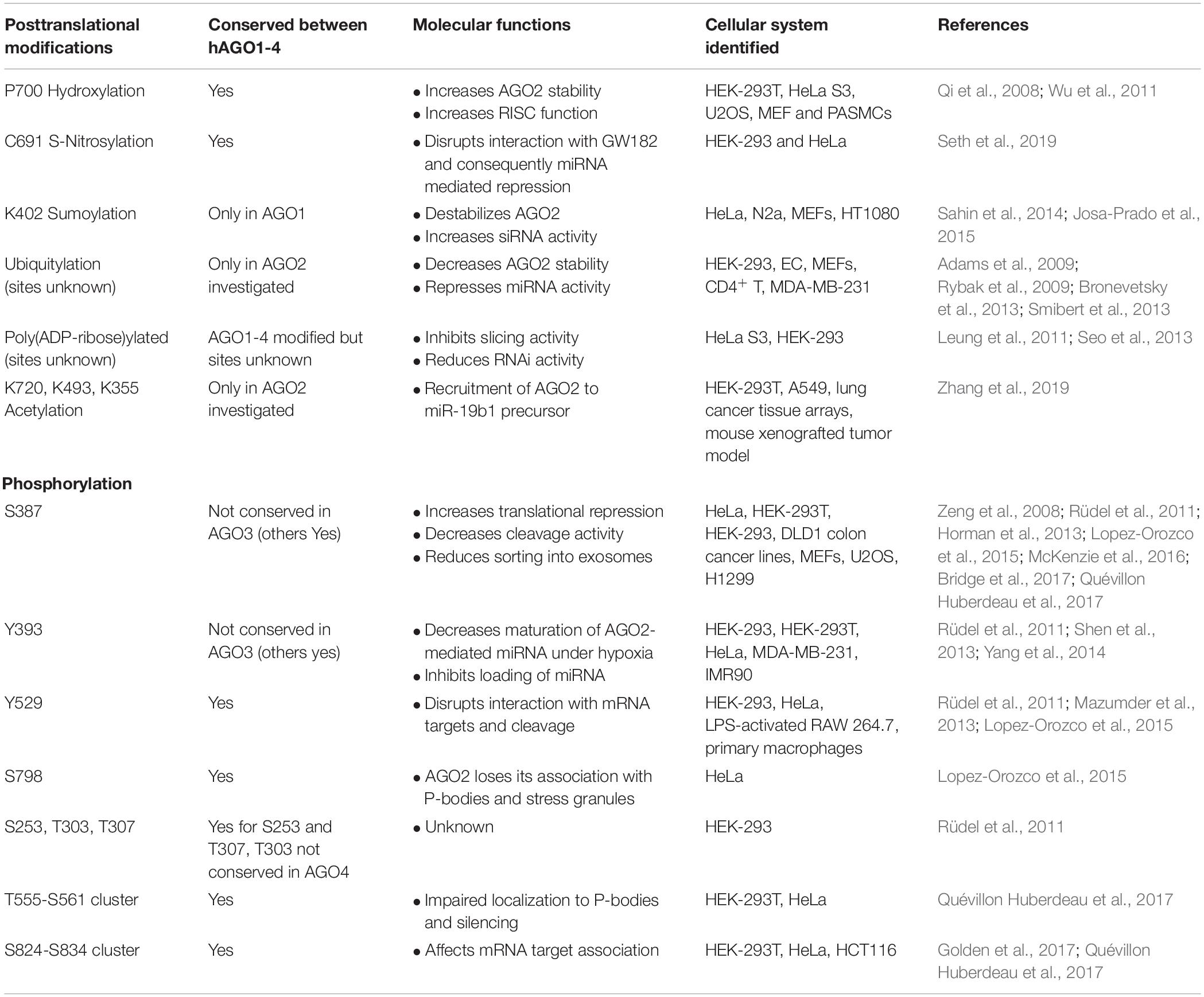
Table 1. Posttranslational modifications of the human Argonaute 2 protein (Jee and Lai, 2014; Gebert and MacRae, 2019).
As previously described, Argonaute proteins, through the formation of a RISC complex, enable miRNAs to downregulate partially complementary target mRNAs, making them relevant in normal physiology and disease. PTMs of AGOs can impact several features of RISC-mediated silencing. For example, a rapid cycle of AGO2 phosphorylation and dephosphorylation of a serine/threonine cluster located on a loop on the surface of the PIWI domain is relevant for miRNA binding to target mRNAs and for miRNA-mediated gene silencing. The dissection of the upstream signaling pathways that impact on AGO2 PTM and, consequently, on its cyclic functional activity would represent a relevant advance in the understanding of AGO2 activity and might possibly provide new ways to modulate the global activity of miRNAs (Golden et al., 2017).
The phosphorylation status of AGOs is also critical for the regulation of the miRNA function in humans (Quévillon Huberdeau et al., 2017). In particular, AGOs are hyperphosphorylated at a C-terminal serine/threonine cluster upon miRNA binding and repression of the mRNA target. The negative charge of phosphates within this region impairs the mRNA/AGO interaction and favors the release of target mRNA. The balance between the phosphorylated and dephosphorylated status of AGO may be relevant also for redirecting AGO to a new target mRNA and for modulation of its degradation (Quévillon Huberdeau et al., 2017).
Furthermore, PTMs of AGOs are involved in miRNA processing. AGO2 phosphorylation has been related to certain cancer phenotypes. In these cases, specific AGO2 phosphorylation leads to reduced interaction between DICER and AGO2 with consequent impairment of miRNA processing (Shen et al., 2013).
In addition, the acetylation of specific lysine residues of AGO2 has been reported. This represents a relevant modification for the recruitment of AGO2 to the miR-19b1 precursor, resulting in the enhancement of oncogenic miR-19b processing. Notably, in lung cancer patients, high levels of both miR-19b and AGO2 acetylation correlate with a poor prognosis (Zhang et al., 2019).
Finally, AGO2 phosphorylation also impacts its localization within the cell. Specific AGO2 phosphorylation has been reported to be essential for its localization into processing bodies (P-bodies), impinging on AGO2-dependent regulation of RNA-silencing activity (Zeng et al., 2008). Horman et al. (2013) subsequently show that AGO2 phosphorylation is critical for the interaction with GW182 protein and AGO2 localization in P-bodies. Furthermore, this modification was also shown to regulate localization of AGO2 into multivesicular endosomes resulting in the suppression of AGO2 secretion and influencing the sorting of specific miRNAs into exosomes (McKenzie et al., 2016).
In summary, PTMs affect several AGO exerted functions. In this paragraph, we have given only a few examples. A broader overview can be found in Table 1.
Several studies have examined the roles of AGOs during mouse early development. Earliest works demonstrated that the knockout (KO) of Ago2 is lethal during early mouse development at postimplantation stages (Liu et al., 2004; Alisch et al., 2007; Morita et al., 2007; Cheloufi et al., 2010). In contrast Ago1, 3, and 4 KO mice are viable (Modzelewski et al., 2012; Van Stry et al., 2012). These studies have shown that Ago2-deficient embryos are growth retarded and developmentally delayed. In addition, they display severe phenotypic defects, such as cardiac failure and impaired neuronal tube closure (Liu et al., 2004; Alisch et al., 2007; Cheloufi et al., 2010).
Interestingly, the phenotype of the Ago2-deficient mice compared to other RNAi-deficient mice is not identical. In addition to other phenotypic differences, Dicer- or Drosha-deficient mice, for example, display earlier embryonic lethality compared to Ago2-deficient mice, suggesting individual roles for the RNAi effector proteins in regulating embryonic development (Bernstein et al., 2003; Chong et al., 2010).
The function of AGO2 in the embryonic development has already been given ample attention with the help of several mouse models (Liu et al., 2004; Alisch et al., 2007; Morita et al., 2007; Cheloufi et al., 2010). However, detailed analyses of AGO2 in the development of the extraembryonic lineages are still missing. Recently, it has been reported that early mice lethality is often associated with placental defects (Perez-Garcia et al., 2018). Interestingly, previous histological analyses have already indicated that Ago2-deficient mice display extraembryonic defects. Supplementing these mice with wild-type extraembryonic tissue is able to rescue the mid-gestation death of Ago2 KO mice (Liu et al., 2004; Cheloufi et al., 2010). These defects might explain why Ago2-deficient mice die at the postimplantation stage; however, this has not been assessed yet.
Ngondo et al. (2018) have recently demonstrated a novel function of AGO2 in the development of the extraembryonic endoderm, in vitro. Using mouse embryonic stem cells (mESCs), which are derived from the blastocyst stage, they generated Ago2 KO mESCs, by CRISPR/Cas9 genome engineering (Wettstein et al., 2016). Upon in vitro differentiation, Ago2 KO mESCs were able to form all three embryonic germ layers; however, they showed impaired differentiation toward the extraembryonic endoderm (Ngondo et al., 2018). This differentiation defect was rescued by the reintroduction of a wild-type AGO2 or a catalytic dead AGO2 in mESCs, but not by an RNA-binding-deficient AGO2 (Ngondo et al., 2018). In line with these results, Ago2 catalytic dead mice were previously shown to be viable until a few hours after birth and subsequently died of anemia (Cheloufi et al., 2010; Jee et al., 2018). In these cases, the slicing activity of AGO2 is needed to process the pre-miRNA-451 and miRNA-486-3p, two miRNAs required in the development of the erythroblasts (Cheloufi et al., 2010; Papapetrou et al., 2010; Jee et al., 2018).
Notably, the molecular mechanism by which AGO2 regulates the formation of the extraembryonic endoderm still remains elusive. Interestingly, the differentiation defect of the Ago2 KO mESCs is comparable to what was observed previously for Gata6 KO mESCs, a key transcription factor required for the formation of the primitive endoderm lineage in vivo (Capo-Chichi et al., 2005). Together, these reports indicate a function of AGO2 not only in the development of the embryo proper but also in the extraembryonic lineages.
Mouse embryonic stem cells are a very informative in vitro culture system to mimic mouse early development at the blastocyst stage. Nevertheless, a stable in vitro system mimicking the earliest stages of development is still missing. Most of the studies focusing on the first cell fate decisions in early mouse development were performed by imaging wild-type or mutant embryos and relied on specific antibodies for the protein of interest, which were not available for a long time for the mouse AGO proteins. Furthermore, single-cell bulk analysis requires a lot of material, which is hard to obtain from early embryos. However, a powerful tool to study the earliest cell fate decision, the two-cell stage-like (2C) ESCs, has been discovered a few years ago (Macfarlan et al., 2012). Two-cell like cells are totipotent and therefore can still differentiate into the extraembryonic as well as embryonic lineages (Baker and Pera, 2018). MESCs have been shown to present a heterogeneous population, where a small subpopulation (<1%) cycles in and out of a two-cell stage (Macfarlan et al., 2012; Morgani et al., 2013; Ishiuchi et al., 2015). The totipotent two-cell stage subpopulation might provide a powerful way to study the impact of AGO2 on the early stages of mouse development, not only for the epiblast but also for the trophoblast lineage, where the impact of AGO2 loss has not been assessed yet. So far, we still do not know whether AGO2 is the only Argonaute protein well expressed in this lineage and whether it impacts trophoblast differentiation.
Since AGO2-deficient mice only die at the postimplantation stage, the question is raised, whether AGO2 is dispensable for preimplantation development or, whether maternally supplied AGO2 regulates these early stages. One important in vivo study assessed the requirement of AGO proteins before the blastocyst stage (Lykke-Andersen et al., 2008). Using injection of dsRNAs against maternally supplied Argonautes, they demonstrated that only AGO2 is essential for the development of mouse oocytes to the two-cell stage. Nevertheless, the molecular mechanism by which AGO2 regulates this early transition is still unknown. Strikingly, the loss of another RNAi effector protein in oocytes, DGCR8, displays a very different phenotype compared to the loss of AGO2. Dgcr8 KO oocytes are able to develop beyond the 2-cell stage to blastocysts. As DGCR8 is only involved in the processing of canonical miRNAs, this suggests that canonical miRNAs might be dispensable for early development (Suh et al., 2010). This is in line with the previous assumptions that miRNA function is lost in oocytes. One reason for the loss of miRNA function in oocytes was proposed to be due to an AGO2-specific oocyte isoform (Freimer et al., 2018). However, a recent report shows that miRNA activity might not be lost in oocytes but that the miRNA/mRNA stoichiometry is impaired in oocytes due to the low abundance of miRNAs (Kataruka et al., 2019). Furthermore, Ago2 KO oocytes seem very similar to Dicer KO oocytes. Both show abnormal spindle and chromosome positioning and fail to undergo the first cleavage to the two-cell stage (Murchison et al., 2007; Tang et al., 2007; Kaneda et al., 2009). Moreover, changes in gene transcripts in Dicer KO oocytes are claimed to be provoked by endo-siRNAs (Watanabe et al., 2008), which are the most prominently expressed smRNAs in oocytes and not preprocessed by DGCR8 (Tam et al., 2008; Watanabe et al., 2008; Suh et al., 2010). The loss of DICER and AGO2 in oocytes decreases siRNAs (Watanabe et al., 2008). It is therefore possible that the phenotype observed in Ago2 KO oocytes is a result of the loss of endo-siRNA-induced target silencing.
Hence, the exact function of AGO2 in early development still needs to be elucidated, as it is undoubtedly the only one leading to a lethal phenotype.
In this review, we highlight various differences and similarities between the Argonaute proteins to better understand their specialized roles within the cell, especially in regard to AGO2.
With the structural information available nowadays, it is possible to pinpoint the exact residues responsible for the catalytic function or disfunction of the AGOs. This has clarified why AGO2 specifically was thought for a long time to be the only slicer molecule of this family.
Interestingly, however, from available sequencing data, it seems that AGO2 is the only AGO protein well expressed in early mice or human development, at least at the transcriptional level (Boroviak et al., 2018). This, when reflected on protein levels, might also explain the severe defects observed upon the loss of AGO2 in early embryos when compared to AGO1, 3, and 4. Strikingly, the Ago2 KO phenotypes observed are not just linked to the embryonic development but also cause impairments in extraembryonic development, as studies show placental defects associated with the loss of AGO2 (Liu et al., 2004; Cheloufi et al., 2010). We argue that a deeper exploration in the early development of extraembryonic tissues is warranted in the context of AGO2 loss in vivo and in vitro.
Lastly, we present an overview of multiple to date known posttranslational modifications of AGO2. These modifications have so far been studied in several cancer models and furthermore have been linked to disease phenotypes. From such studies, we know that PTMs can impact the RISC activity as well as AGO2 stability, either positively or negatively. However, a detailed analysis of such modifications during early development is still missing. We still do not know which modifications are present in early embryos nor whether there is a switch of modification when going through different stages of embryonic or extraembryonic development. Hence, to better understand how AGO2 functions in these early stages of embryonic development, the PTMs of AGO2 must also be taken into consideration.
MM and CC wrote the sections related to Argonaute expression and structure in early development. MM drew the figure and established the Table 1. FF contributed to the writing of the section on the PTM of AGO in cancer. All authors read and approved the final version of the manuscript.
This work was supported by a core grant from ETH-Z (supported by Roche). MM is supported by a Swiss National Science Foundation Grant (31003A_173120).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Ciaudo lab and Dr. Tobias Beyer for the critical reading of the manuscript and for fruitful discussions. We apologize for not directly citing many crucial references; these references can, however, be found in the cited previous reviews.
Adams, B. D., Claffey, K. P., and White, B. A. (2009). Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology 150, 14–23. doi: 10.1210/en.2008-0984
Alisch, R. S., Jin, P., Epstein, M., Caspary, T., and Warren, S. T. (2007). Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet. 3:e227. doi: 10.1371/journal.pgen.0030227
Alló, M., Buggiano, V., Fededa, J. P., Petrillo, E., Schor, I., De La Mata, M., et al. (2009). Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724. doi: 10.1038/nsmb.1620
Baker, C. L., and Pera, M. F. (2018). Capturing totipotent stem cells. Cell Stem Cell 22, 25–34. doi: 10.1016/j.stem.2017.12.011
Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., et al. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215–217. doi: 10.1038/ng1253
Bodak, M., Cirera-Salinas, D., Luitz, J., and Ciaudo, C. (2017a). The role of RNA interference in stem cell biology: beyond the mutant phenotypes. J. Mol. Biol. 429, 1532–1543. doi: 10.1016/j.jmb.2017.01.014
Bodak, M., Cirera-Salinas, D., Yu, J., Ngondo, R. P., and Ciaudo, C. (2017b). Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Biol. 7, 204–220. doi: 10.1002/2211-5463.12174
Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. doi: 10.1093/emboj/17.1.170
Boland, A., Huntzinger, E., Schmidt, S., Izaurralde, E., and Weichenrieder, O. (2011). Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. U.S.A. 108, 10466–10471. doi: 10.1073/pnas.1103946108
Boland, A., Tritschler, F., Heimstädt, S., Izaurralde, E., and Weichenrieder, O. (2010). Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 11, 522–527. doi: 10.1038/embor.2010.81
Boroviak, T., Stirparo, G. G., Dietmann, S., Hernando-Herraez, I., Mohammed, H., Reik, W., et al. (2018). Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145:dev167833. doi: 10.1242/dev.167833
Bridge, K. S., Shah, K. M., Li, Y., Foxler, D. E., Wong, S. C. K., Miller, D. C., et al. (2017). Argonaute utilization for miRNA silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 20, 173–187. doi: 10.1016/j.celrep.2017.06.027
Bronevetsky, Y., Villarino, A. V., Eisley, C. J., Barbeau, R., Barczak, A. J., Heinz, G. A., et al. (2013). T cell activation induces proteasomal degradation of argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210, 417–432. doi: 10.1084/jem.20111717
Capo-Chichi, C. D., Rula, M. E., Smedberg, J. L., Vanderveer, L., Parmacek, M. S., Morrisey, E. E., et al. (2005). Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 286, 574–586. doi: 10.1016/j.ydbio.2005.07.037
Cheloufi, S., Dos Santos, C. O., Chong, M. M. W., and Hannon, G. J. (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589. doi: 10.1038/nature09092
Chong, M. M. W., Zhang, G., Cheloufi, S., Neubert, T. A., Hannon, G. J., and Littman, D. R. (2010). Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24:2228. doi: 10.1101/gad.1953310
Elkayam, E., Kuhn, C. D., Tocilj, A., Haase, A. D., Greene, E. M., Hannon, G. J., et al. (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110. doi: 10.1016/j.cell.2012.05.017
Eswarappa, S. M., Potdar, A. A., Koch, W. J., Fan, Y., Vasu, K., Lindner, D., et al. (2014). Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 157, 1605–1618. doi: 10.1016/j.cell.2014.04.033
Eulalio, A., Tritschler, F., and Izaurralde, E. (2009). The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 15, 1433–1442. doi: 10.1261/rna.1703809
Faehnle, C. R., Elkayam, E., Haase, A. D., Hannon, G. J., and Joshua-Tor, L. (2013). The making of a slicer: activation of human Argonaute-1. Cell Rep. 3, 1901–1909. doi: 10.1016/j.celrep.2013.05.033
Frank, F., Sonenberg, N., and Nagar, B. (2010). Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822. doi: 10.1038/nature09039
Freimer, J. W., Krishnakumar, R., Cook, M. S., and Blelloch, R. (2018). Expression of alternative Ago2 isoform associated with loss of microRNA-driven translational repression in mouse oocytes. Curr. Biol. 28, 296.e–302.e. doi: 10.1016/j.cub.2017.11.067
Gao, Y., Liu, X., Tang, B., Li, C., Kou, Z., Li, L., et al. (2017). Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969. doi: 10.1016/j.celrep.2017.11.111
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37. doi: 10.1038/s41580-018-0045-7
Ghosh, S., Guimaraes, J., Lanzafame, M., Schmidt, A., Syed, A. P., Dimitriades, B., et al. (2019). AGO1x prevents dsRNA-induced interferon signaling to promote breast cancer cell proliferation. bioRxiv [Preprint]
Golden, R. J., Chen, B., Li, T., Braun, J., Manjunath, H., Chen, X., et al. (2017). An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202. doi: 10.1038/nature21025
Harel-Bellan, A., Zazoua-Ameyar, M., Rachez, C., Muchardt, C., and Batsché, E. (2013). 10-million-years AGO: argonaute on chromatin in yeast and human, a conserved mode of action? Transcription 4, 89–91. doi: 10.4161/trns.24582
Hauptmann, J., Dueck, A., Harlander, S., Pfaff, J., Merkl, R., and Meister, G. (2013). Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 20, 814–817. doi: 10.1038/nsmb.2577
Hauptmann, J., Kater, L., Löffler, P., Merkl, R., and Meister, G. (2014). Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. RNA 20, 1532–1538. doi: 10.1261/rna.045203.114
Hawley, B. R., Lu, W. T., Wilczynska, A., and Bushell, M. (2017). The emerging role of RNAs in DNA damage repair. Cell Death Differ. 24, 580–587. doi: 10.1038/cdd.2017.16
Horman, S. R., Janas, M. M., Litterst, C., Wang, B., MacRae, I. J., Sever, M. J., et al. (2013). Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol. Cell 50, 356–367. doi: 10.1016/j.molcel.2013.03.015
Huang, V., and Li, L. C. (2014). Demystifying the nuclear function of Argonaute proteins. RNA Biol. 11, 18–24. doi: 10.4161/rna.27604
Ishiuchi, T., Enriquez-Gasca, R., Mizutani, E., Boškoviä, A., Ziegler-Birling, C., Rodriguez-Terrones, D., et al. (2015). Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662–671. doi: 10.1038/nsmb.3066
Jee, D., and Lai, E. C. (2014). Alteration of miRNA activity via context-specific modifications of Argonaute proteins. Trends Cell Biol. 24, 546–553. doi: 10.1016/j.tcb.2014.04.008
Jee, D., Yang, J. S., Park, S. M., Farmer, D. T., Wen, J., Chou, T., et al. (2018). Dual strategies for Argonaute2-mediated biogenesis of erythroid miRNAs underlie conserved requirements for slicing in mammals. Mol. Cell 69:265-278.e6. doi: 10.1016/j.molcel.2017.12.027
Josa-Prado, F., Henley, J. M., and Wilkinson, K. A. (2015). SUMOylation of Argonaute-2 regulates RNA interference activity. Biochem. Biophys. Res. Commun. 464, 1066–1071. doi: 10.1016/j.bbrc.2015.07.073
Jukam, D., Shariati, S. A. M., and Skotheim, J. M. (2017). Zygotic genome activation in vertebrates. Dev. Cell 42, 316–332. doi: 10.1016/j.devcel.2017.07.026
Kaneda, M., Tang, F., O’Carroll, D., Lao, K., and Surani, M. A. (2009). Essential role for Argonaute2 protein in mouse oogenesis. Epigenet. Chroma. 2, 1–8. doi: 10.1186/1756-8935-2-9
Kanellopoulou, C., Muljo, S. A., Kung, A. L., Ganesan, S., Drapkin, R., Jenuwein, T., et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501. doi: 10.1101/gad.1248505
Kataruka, S., Modrak, M., Kinterova, V., Zeitler, D. M., Malik, R., Kanka, J., et al. (2019). Low miRNA abundance disables microRNA pathway in mammalian oocytes. bioRxiv [Preprint]
Kuscu, C., Kumar, P., Kiran, M., Su, Z., Malik, A., and Dutta, A. (2018). tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105. doi: 10.1261/rna.066126.118
Leung, A. K. L., Vyas, S., Rood, J. E., Bhutkar, A., Sharp, P. A., and Chang, P. (2011). Poly(ADP-Ribose) regulates stress responses and MicroRNA activity in the cytoplasm. Mol. Cell 42, 489–499. doi: 10.1016/j.molcel.2011.04.015
Li, L. C. (2014). Chromatin remodeling by the small rna machinery in mammalian cells. Epigenetics 9, 45–52. doi: 10.4161/epi.26830
Lingel, A., Simon, B., Sattler, M., and Izaurralde, E. (2003). Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426, 465–469. doi: 10.1038/nature02123
Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J.-J., et al. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. doi: 10.1126/science.1102513
Lopez-Orozco, J., Pare, J. M., Holme, A. L., Chaulk, S. G., Fahlman, R. P., and Hobman, T. C. (2015). Functional analyses of phosphorylation events in human Argonaute 2. RNA 21, 2030–2038. doi: 10.1261/rna.053207.115
Lykke-Andersen, K., Gilchrist, M. J., Grabarek, J. B., Das, P., Miska, E., and Zernicka-Goetz, M. (2008). Maternal Argonaute 2 is essential for early mouse development at the Maternal-Zygotic transition. Mol. Biol. Cell 19, 4383–4392. doi: 10.1091/mbc.E08-02-0219
Ma, J. B., Ye, K., and Patel, D. J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322. doi: 10.1038/nature02519
Ma, J. B., Yuan, Y. R., Meister, G., Pei, Y., Tuschl, T., and Patel, D. J. (2005). Structural basis for 5′ -end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670. doi: 10.1038/nature03514
Macfarlan, T. S., Gifford, W. D., Driscoll, S., Lettieri, K., Rowe, H. M., Bonanomi, D., et al. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. doi: 10.1038/nature11244
Maillard, P., Ciaudo, C., Marchais, A., Li, Y., Jay, F., Ding, S., et al. (2013). Antiviral RNA interference in mammalian cells. Science 342, 235–238. doi: 10.1126/science.1241930
Malecová, B., and Morris, K. V. (2010). Transcriptional gene silencing mediated by non-coding RNAs. Curr. Opin. Mol. Ther. 12, 214–222.
Martens-Uzunova, E. S., Olvedy, M., and Jenster, G. (2013). Beyond microRNA - Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 340, 201–211. doi: 10.1016/j.canlet.2012.11.058
Mazumder, A., Bose, M., Chakraborty, A., Chakrabarti, S., and Bhattacharyya, S. N. (2013). A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep. 14, 1008–1016. doi: 10.1038/embor.2013.149
McKenzie, A. J., Hoshino, D., Hong, N. H., Cha, D. J., Franklin, J. L., Coffey, R. J., et al. (2016). KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15, 978–987. doi: 10.1016/j.celrep.2016.03.085
Meister, G. (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459. doi: 10.1038/nrg3462
Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197. doi: 10.1016/j.molcel.2004.07.007
Modzelewski, A. J., Holmes, R. J., Hilz, S., Grimson, A., and Cohen, P. E. (2012). AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell 23, 251–264. doi: 10.1016/j.devcel.2012.07.003
Morgani, S. M., Canham, M. A., Nichols, J., Sharov, A. A., Migueles, R. P., Ko, M. S. H., et al. (2013). Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 3, 1945–1957. doi: 10.1016/j.celrep.2013.04.034
Morita, S., Horii, T., Kimura, M., Goto, Y., Ochiya, T., and Hatada, I. (2007). One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89, 687–696. doi: 10.1016/j.ygeno.2007.01.004
Murchison, E. P., Stein, P., Xuan, Z., Pan, H., Zhang, M. Q., Schultz, R. M., et al. (2007). Critical roles for Dicer in the female germline. Genes Dev. 21, 682–693. doi: 10.1101/gad.1521307
Nakanishi, K., Ascano, M., Gogakos, T., Ishibe-Murakami, S., Serganov, A. A., Briskin, D., et al. (2013). Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 3, 1893–1900. doi: 10.1016/j.celrep.2013.06.010
Nakanishi, K., Weinberg, D. E., Bartel, D. P., and Patel, D. J. (2012). Structure of yeast Argonaute with guide RNA. Nature 486, 368–374. doi: 10.1038/nature11211
Ngondo, R. P., Cirera-Salinas, D., Yu, J., Wischnewski, H., Bodak, M., Vandormael-Pournin, S., et al. (2018). Argonaute 2 is required for extra-embryonic endoderm differentiation of mouse embryonic stem cells. Stem Cell Rep. 10, 461–476. doi: 10.1016/j.stemcr.2017.12.023
Niakan, K. K., and Eggan, K. (2013). Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 375, 54–64. doi: 10.1016/j.ydbio.2012.12.008
Papapetrou, E. P., Korkola, J. E., and Sadelain, M. (2010). A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 28, 287–296. doi: 10.1002/stem.257
Park, M. S., Araya-Secchi, R., Brackbill, J. A., Phan, H.-D., Kehling, A. C., Abd El-Wahab, E. W., et al. (2019). Multidomain convergence of Argonaute during RISC assembly correlates with the formation of internal water clusters. Mol. Cell 75, 725–740. doi: 10.1016/j.molcel.2019.06.011
Park, M. S., Phan, H. D., Busch, F., Hinckley, S. H., Brackbill, J. A., Wysocki, V. H., et al. (2017). Human Argonaute3 has slicer activity. Nucleic Acids Res. 45, 11867–11877. doi: 10.1093/nar/gkx916
Parker, J. S., Roe, S. M., and Barford, D. (2004). Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23, 4727–4737. doi: 10.1038/sj.emboj.7600488
Parker, J. S., Roe, S. M., and Barford, D. (2005). Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666.
Perez-Garcia, V., Fineberg, E., Wilson, R., Murray, A., Mazzeo, C. I., Tudor, C., et al. (2018). Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468. doi: 10.1038/nature26002
Qi, H. H., Ongusaha, P. P., Myllyharju, J., Cheng, D., Pakkanen, O., Shi, Y., et al. (2008). Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455, 421–424. doi: 10.1038/nature07186
Quévillon Huberdeau, M., Zeitler, D. M., Hauptmann, J., Bruckmann, A., Fressigné, L., Danner, J., et al. (2017). Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 36, 2088–2106. doi: 10.15252/embj.201696386
Rivas, F. V., Tolia, N. H., Song, J. J., Aragon, J. P., Liu, J., Hannon, G. J., et al. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12, 340–349. doi: 10.1038/nsmb918
Rüdel, S., Wang, Y., Lenobel, R., Körner, R., Hsiao, H. H., Urlaub, H., et al. (2011). Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 39, 2330–2343. doi: 10.1093/nar/gkq1032
Rybak, A., Fuchs, H., Hadian, K., Smirnova, L., Wulczyn, E. A., Michel, G., et al. (2009). The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11, 1411–1420. doi: 10.1038/ncb1987
Sahin, U., Lapaquette, P., Andrieux, A., Faure, G., and Dejean, A. (2014). Sumoylation of human argonaute 2 at lysine-402 regulates its stability. PLoS One 9:e0102957. doi: 10.1371/journal.pone.0102957
Sarshad, A. A., Juan, A. H., Muler, A. I. C., Anastasakis, D. G., Wang, X., Genzor, P., et al. (2018). Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Mol. Cell 71:1040-1050.e8. doi: 10.1016/j.molcel.2018.07.020
Schirle, N. T., and Macrae, I. J. (2012). The crystal structure of human Argonaute2. Science 336, 1037–1040. doi: 10.1126/science.1221551
Schorn, A. J., Gutbrod, M. J., LeBlanc, C., and Martienssen, R. L. T. R. - (2017). Retrotransposon control by tRNA-derived small RNAs. Cell 170, 61–71.e11. doi: 10.1016/j.cell.2017.06.013
Schürmann, N., Trabuco, L. G., Bender, C., Russell, R. B., and Grimm, D. (2013). Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 20, 818–826. doi: 10.1038/nsmb.2607
Seet, B. T., Dikic, I., Zhou, M. M., and Pawson, T. (2006). Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7, 473–483. doi: 10.1038/nrm1960
Seo, G. J., Kincaid, R. P., Phanaksri, T., Burke, J. M., Pare, J. M., Cox, J. E., et al. (2013). Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14, 435–445. doi: 10.1016/j.chom.2013.09.002
Seth, P., Hsieh, P. N., Jamal, S., Wang, L., Gygi, S. P., Jain, M. K., et al. (2019). Regulation of MicroRNA Machinery and Development by Interspecies S-Nitrosylation. Cell 176, 1014.e–1025.e. doi: 10.1016/j.cell.2019.01.037
Shen, J., Xia, W., Khotskaya, Y. B., Huo, L., Nakanishi, K., Lim, S. O., et al. (2013). EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387. doi: 10.1038/nature12080
Singh, A., Manjunath, L. E., Kundu, P., Sahoo, S., Das, A., Suma, H. R., et al. (2019). Let-7a-regulated translational readthrough of mammalian AGO1 generates a micro RNA pathway inhibitor. EMBO J. 38:e100727. doi: 10.15252/embj.2018100727
Smibert, P., Yang, J. S., Azzam, G., Liu, J. L., and Lai, E. C. (2013). Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 20, 789–795. doi: 10.1038/nsmb.2606
Song, J., Smith, S. K., and Hannon, G. J. (2004). Crystal structure of argonaute its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Song, J. J., Liu, J., Tolia, N. H., Schneiderman, J., Smith, S. K., Martienssen, R. A., et al. (2003). The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10, 1026–1032. doi: 10.1038/nsb1016
Stuart, T., and Satija, R. (2019). Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272. doi: 10.1038/s41576-019-0093-7
Suh, N., Baehner, L., Moltzahn, F., Melton, C., Shenoy, A., Chen, J., et al. (2010). MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 20, 271–277. doi: 10.1016/j.cub.2009.12.044
Svoboda, P., Stein, P., Anger, M., Bernstein, E., Hannon, G. J., and Schultz, R. M. (2004). RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 269, 276–285. doi: 10.1016/j.ydbio.2004.01.028
Swarts, D. C., Makarova, K., Wang, Y., Nakanishi, K., Ketting, R. F., Koonin, E. V., et al. (2014). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753. doi: 10.1038/nsmb.2879
Tam, O. H., Aravin, A. A., Stein, P., Girard, A., Murchison, E. P., Cheloufi, S., et al. (2008). Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538. doi: 10.1038/nature06904
Tang, F., Kaneda, M., O’Carroll, D., Hajkova, P., Barton, S. C., Sun, Y. A., et al. (2007). Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 21, 644–648. doi: 10.1101/gad.418707
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20. doi: 10.1038/s41580-018-0059-1
Van Stry, M., Oguin, T. H., Cheloufi, S., Vogel, P., Watanabe, M., Pillai, M. R., et al. (2012). Enhanced Susceptibility of Ago1/3 Double-Null Mice to Influenza A Virus Infection. J. Virol. 86, 4151–4157. doi: 10.1128/JVI.05303-11
Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., et al. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543. doi: 10.1038/nature06908
Wettstein, R., Bodak, M., and Ciaudo, C. (2016). Generation of a knockout mouse embryonic stem cell line using a paired CRISPR/Cas9 genome engineering tool. Methods Mol. Biol. 1341, 321–343. doi: 10.1007/7651_2015_213
Wu, C., So, J., Davis-Dusenbery, B. N., Qi, H. H., Bloch, D. B., Shi, Y., et al. (2011). Hypoxia potentiates MicroRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 31, 4760–4774. doi: 10.1128/MCB.05776-11
Yan, K. S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M.-M. (2004). Structure and conserved RNA binding of the PAZ domain. Nature 427, 265–265. doi: 10.1038/nature02252
Yang, M., Haase, A. D., Huang, F. K., Coulis, G., Rivera, K. D., Dickinson, B. C., et al. (2014). Dephosphorylation of Tyrosine 393 in Argonaute 2 by Protein Tyrosine Phosphatase 1B Regulates Gene Silencing in Oncogenic RAS-Induced Senescence. Mol. Cell 55, 782–790. doi: 10.1016/j.molcel.2014.07.018
Yuan, Y. R., Pei, Y., Ma, J. B., Kuryavyi, V., Zhadina, M., Meister, G., et al. (2005). Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419. doi: 10.1016/j.molcel.2005.07.011
Zeng, Y., Sankala, H., Zhang, X., and Graves, P. R. (2008). Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 413, 429–436. doi: 10.1042/BJ20080599
Keywords: argonaute proteins, expression, structure, posttranslational modifications, development, cancer
Citation: Müller M, Fazi F and Ciaudo C (2020) Argonaute Proteins: From Structure to Function in Development and Pathological Cell Fate Determination. Front. Cell Dev. Biol. 7:360. doi: 10.3389/fcell.2019.00360
Received: 30 October 2019; Accepted: 12 December 2019;
Published: 22 January 2020.
Edited by:
Karthikeyan Narayanan, West Virginia University, United StatesReviewed by:
Anne Dueck, Technical University of Munich, GermanyCopyright © 2020 Müller, Fazi and Ciaudo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Fazi, ZnJhbmNlc2NvLmZhemlAdW5pcm9tYTEuaXQ=; Constance Ciaudo, Y2NpYXVkb0BldGh6LmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.