- 1CNRS-LIA Hematology and Cancer, Sino-French Research Center for Life Sciences and Genomics, State Key Laboratory of Medical Genomics, RuiJin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Prenatal Diagnosis Center, Shanghai Jiao Tong University Affiliated First People’s Hospital, Shanghai, China
A variety of cardiac transcription factors/cofactors, signaling pathways, and downstream structural genes integrate to form the regulatory hierarchies to ensure proper cardiogenesis in vertebrate. Major interaction proteins of the transcription cofactor vestigial like family member 4 (VGLL4) include myocyte enhancer factor 2 (MEF2) and TEA domain transcription factors (TEAD), both of which play important roles in embryonic cardiac development and in adulthood. In this study, we identified that the deficiency of zebrafish vgll4b paralog, a unique family member expressed in developing heart, led to an impaired valve development. Mechanistically, in vgll4b mutant embryos the disruption of Vgll4b-Mef2c complex, rather than that of Vgll4b-Tead complex, resulted in an aberrant expression of krüppel-like factor 2a (klf2a) in endocardium. Such misexpression of klf2a eventually evoked the valvulogenesis defects. Our findings suggest that zebrafish Vgll4b plays an important role in modulating the transcription activity of Mef2c on klf2a during valve development in a blood-flow-independent manner.
Introduction
Four TONDU (TDU) domain-containing proteins, named VGLL1 to VGLL4 exist in mammals. As transcription cofactor, VGLL proteins do not contain a DNA binding domain (DBD), their transcriptional activities could only be mediated through the TDU domain which interacts with different transcription factors such as TEADs (also named TEFs) and MEF2s (Chen et al., 2004a). Structurally, unlike VGLL1 to 3 which only possess one TDU domain, there are two tandem TDU domains located in VGLL4. VGLL genes play different roles in various physiological processes. For example, VGLL2 acts as a cofactor of TEAD1 during skeletal muscle development (Maeda et al., 2002; Chen et al., 2004b; Honda et al., 2017) while VGLL4 functions as a positive regulator in survival of human embryonic stem cells (hESCs) (Tajonar et al., 2013). Numerous studies indicate that VGLL genes participate in tumorigenesis. VGLL1 is highly expressed in basal-like breast cancer and promotes cell proliferation (Castilla et al., 2014). Similarly, VGLL3 is overexpressed in a subset of soft tissue sarcomas (Hélias-Rodzewicz et al., 2009). Interestingly, the expression of VGLL3 was absent in high-grade serous ovarian carcinomas (HGSC), suggesting VGLL3 might also be involved in tumor suppressor pathways (Gambaro et al., 2013). Among all of the family members, VGLL4 is the most frequently tumorigenesis-related one. Decreased expression level of VGLL4 has been observed in multiple types of tumors including gastric cancer (Jiao et al., 2014), lung cancer (Zhang et al., 2014), breast cancer (Zhang et al., 2017), colorectal cancer (Jiao et al., 2017), and esophageal squamous cell carcinoma (ESCC) (Jiang et al., 2015). The Hippo signaling pathway is highly conserved in mammals and implicated in regulation of cell growth and organ size. The transcription coactivator YAP is one major downstream target of Hippo pathway which binds to transcription factors to activate gene expression and regulate cell growth or death (Kim et al., 2019). In the malignancies mentioned above the balance between TEAD-VGLL4 and TEAD-YAP complexes is disrupted (Jiao et al., 2014; Zhang et al., 2014, 2017), thus VGLL4 is regarded as a tumor suppressor by negatively regulating the Hippo pathway to inhibit cell proliferation.
Since the presence of an extra TDU domain, VGLL4 is believed to be functionally different with other VGLLs. Actually their expression patterns are quite different. VGLL1 and VGLL3 are enriched in placenta, whereas VGLL2 is detected only in skeletal muscle (Maeda et al., 2002). VGLL4 is the unique VGLL gene expressed in heart (Chen et al., 2004a), moreover, it has been reported that from the post-endothelial to mesenchyme transition (EMT) stage to adult stage murine Vgll4 is notably expressed in heart valve endothelial and interstitial cells, and endothelial loss of Vgll4 results in valve malformation (Yu et al., 2019). Mechanistic studies reveal that VGLL4 competes with YAP for TEADs binding to regulate valvulogenesis (Yu et al., 2019).
The heart is the first functional organ in vertebrate embryo (Moorman et al., 2003). Heart development begins with the formation of the primitive heart tube, that loops and septates into four chambers and paired arterial trunks to form the mature heart in human (Moorman et al., 2003). During heart development the formation of valve is a complicated process. Defective valvulogenesis generally leads to impaired cardiac function and congenital heart diseases (CHD) (Combs and Yutzey, 2009). In addition to mouse, xenopus and chick, zebrafish has emerged as a powerful model organism in genetic studies of cardiac development in recent decades. Unlike human counterpart, three vgll4 paralog genes named vgll4a, vgll4b and vgll4l exist in zebrafish genome. We have demonstrated in our previous study that only vgll4b, but not vgll4a and vgll4l, is expressed in the developing heart (Xue et al., 2018). Except for TEADs, MEF2 family members are also major interactants of VGLL4 (Chen et al., 2004a), which are enriched in heart and play a variety of functions during embryonic cardiac development and in adulthood (Chen et al., 1994; Naya et al., 2002; Durham et al., 2006; Huang et al., 2006; Ewen et al., 2011; Lockhart et al., 2013; Dong et al., 2017). In the current work we took advantage of a knockout line to provide in vivo evidence that the zebrafish vgll4b paralog is indispensable for normal valvulogenesis, of which the mechanism differs from that in mice. Upon loss of Vgll4b, the Vgll4b-Mef2c complex, rather than Vgll4b-Tead complex, is disrupted in endocardium. The released Mef2c in turns aberrantly activates klf2a, which eventually evokes the defective valvulogenesis.
Materials and Methods
Zebrafish Maintenance and Mutant Generation
Zebrafish were raised, bred, and staged according to standard protocols (Kimmel et al., 1995). All animal works were strictly conducted following the guidelines of the Animal Care and Use Committee of Shanghai Jiao Tong University. For CRISPR/Cas9 system mediated vgll4b knockout zebrafish generation, guide RNA targeting exon1 of vgll4b was designed using an online tool ZiFiT Targeter software1, which was synthesized by cloning the annealed oligonucleotides into the sgRNA transcription vector. Cas9 mRNA and sgRNA were co-injected into one-cell stage zebrafish embryos. The injected F0 founder embryos were raised to adulthood and then outcrossed with wild type zebrafish. F1 embryos carrying potential indel mutations were raised to adulthood. Then PCR amplification and sequencing were performed on genomic DNA isolated from tail clips of F1 zebrafish to identify mutants.
Plasmid Construction
Zebrafish vgll4b, mef2cb, tead, sumo1, klf2a genes and their serial mutants were cloned into PCS2+ vector. Vgll4b ΔTDU1 and ΔTDU2 mutants were constructed by PCR-mediated deletion of vgll4b plasmid. Primers used were listed in Table 1. Tol2-plasmid was constructed by insertion of zebrafish mef2cb DN or klf2a DN under cmlc2 promoter (0.9 k) or flk1 promoter (6.4 k) (Huang et al., 2003; Jin, 2005). Transgenes were transiently expressed by co-injecting 80 pg of Tol2-plasmid and 120 pg of Tol2 transpose mRNA at one-cell stage.
Whole-Mount mRNA in situ Hybridization (WISH)
Digoxigenin-labeled RNA probes were transcribed with T7, T3 or SP6 polymerase (Ambion, Life Technologies, United States). WISH was performed as described previously (Thisse and Thisse, 2008). The probes labeled by digoxigenin were detected using alkaline phosphatase coupled anti-digoxigenin Fab fragment antibody (Roche) with 5-bromo-4-chloro-3-indolyl-phosphate nitro blue tetrazolium staining (Vector Laboratories, Burlingame, CA, United States).
Morpholinos and mRNA Synthesis for Microinjection
Zebrafish vgll4b (5′ ACAGGTCCATTTTGGTAAAAAGCAT 3′) morpholino oligonucleotides (MO) targeting the transcriptional initiation ATG of vgll4b was designed and purchased from Gene Tools. Full-length capped mRNA samples were all synthesized from linearized plasmids using the mMessage mMachine SP6 kit (Invitrogen, Thermo Fisher, United States). Microinjection concentration of mRNA was between 50 and 200 ng/μl and 2 nl of mRNA was injected at one-cell stage embryos. All injections were performed with a Harvard Apparatus micro-injector.
Heart Rates Measurement and Ventricular Contractility Analysis
Zebrafish embryos were incubated at 28.5°C (VWR Scientific incubator). Cardiac function of sibling WT control and vgll4b mutants were quantitatively assayed by the Optical Heartbeat analysis under an Olympus IX71 microscope with HC Image software. The lengths of ventricles in diastolic and systolic conditions were measured to calculate the ventricular shortening fraction (VSF). Values are presented as mean ± SD. VSF = (short axis of ventricle in end-diastole − short axis of ventricle in end-systole)/short axis of ventricle in end-diastole.
Immunofluorescence
Hearts were harvested manually from the embryos at 52 hpf and fixed with 4% paraformaldehyde (PFA) in PBS overnight at 4°C, followed by the permeabilization with PBS containing 0.1% Tween-20 and 0.5% Triton-X 100 for 10 min. Then the hearts were blocked in blocking buffer with 1% BSA and 10% normal goat serum for 3 h at room temperature. Mouse anti-Alcam antibody (Developmental Studies Hybridoma Bank, United States) was added in the blocking buffer and incubated for 16 h at 4°C. Goat anti-mouse Alexa-488 secondary antibody (Thermo Fisher Scientific, United States) was added in blocking buffer after thorough washing and incubated overnight at 4°C. After washing with PBS, all the samples were stained with 100 nM Tetramethylrhodamine (TRITC) labeled phalloidin solution (Solarbio, Beijing, China) containing 1% BSA. AVC cells were recognized based on their characteristic cuboidal morphologies. Images were taken using an Olympus FV1000 scanning confocal microscope. The confocal images were captured with an UPLSAPO 40 × objective.
Quantitative RT-PCR
Quantitative PCR was carried out using a SYBR Green Real-Time PCR Master Mix (Toyobo, Osaka, Japan) with an ABI 7900HT real-time PCR machine. Three duplicated experiments were made independently for each group, β-actin served as the internal control. The final results were expressed as the means ± SD. All data were analyzed with GraphPad Prism software (GraphPad Software, La Jolla, CA, United States). The primers used were listed in Table 2.
Cell Culture and Luciferase Reporter Assay
HEK293T cells were maintained in DMEM (Life technologies, Grand Island, NY, United States) with 10% Fetal Bovine Serum (Life technologies, Grand Island, NY, United States). Plasmid transfection was carried out with Effectene Transfection Reagent (QIAGEN) according to manufacturer’s instruction. For the luciferase reporter assay, cells were harvested 48 h after transfection and analyzed using the Dual Luciferase Reporter Assay Kit (Promega, Maddison, WI, United States), according to the manufacturer’s protocols.
Statistical Analysis
Data were presented as mean ± standard deviation (SD). For comparison of two means, statistical significance was evaluated by unpaired Student’s t-test. For multiple comparisons, one-way analysis of variance (ANOVA) test was used (SPSS 17.0 software, IBM, Chicago, IL, United States). Differences were considered to be significant at P < 0.05.
Study Approval
The animal protocol listed above has been reviewed and approved by the Animal Ethical and Welfare Committee, Rui-Jin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Results
Generation of a vgll4b-Deficient Zebrafish Line
Zebrafish vgll4b gene shares high homology with its human counterpart (Xue et al., 2018). Among all of the three vgll4 paralogs, vgll4b is the only one expressed in the developing heart (Xue et al., 2018). To address the role of vgll4b in cardiogenesis, a vgll4b mutant line was generated using CRISPR/Cas9 technology. Eight nucleotides were deleted, which led to a truncated form of Vgll4b without TDU domains (Figures 1A,B). The mutant vgll4b gene was amplified by RT-PCR from the homozygous mutant, then cloned into an HA tagged expressing vector and transfected into HEK293T cells. As anticipated, a short protein with the predicted molecular weight was detected by western blot analysis (Figure 1C). The function of the Vgll4b mutant was assessed by a dual luciferase reporter assay performed in HEK293T cells expressing TEAD, YAP, as well as wild type or the truncated Vgll4b mutant constructs. Since VGLL4 can negatively regulate the TEAD-YAP pathway, the results showed that the expression of either human VGLL4 or zebrafish vgll4b could pronouncedly reduce the TEAD-YAP activity on a TEAD responsive reporter, whereas the zebrafish vgll4b mutant lacking two TDU domains could not, thus confirming the efficacy of the established mutant line (Figure 1D).
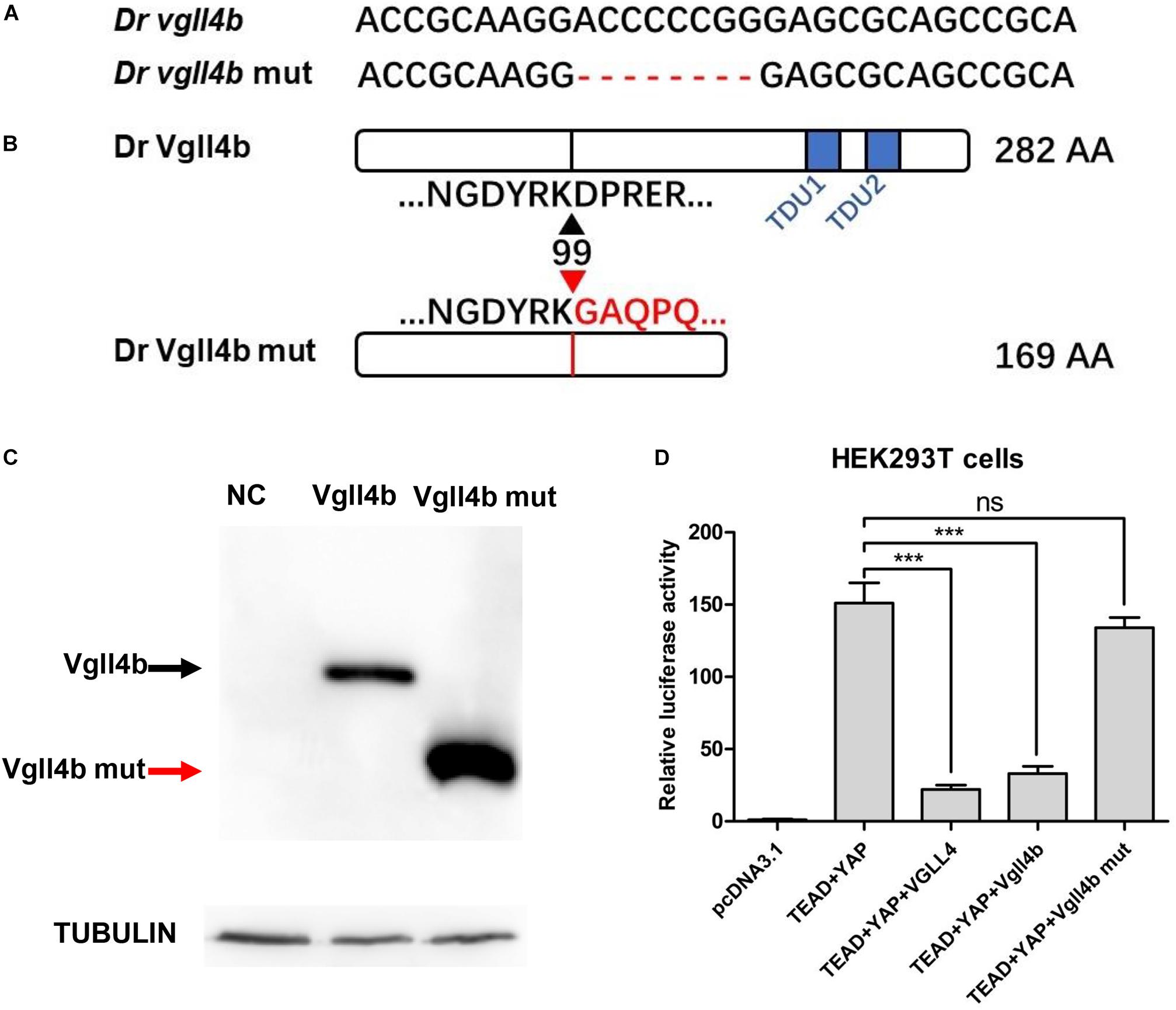
Figure 1. The establishment of a zebrafish vgll4b knockout line. (A) Schematic representation of Cas9 target site in the first exon of zebrafish vgll4b. Dr: Danio rerio. The deleted nucleotides in the mutant gene are marked by hyphens. (B) Schematic representation of wild type (282 amino acids) and mutant Vgll4b proteins (169 amino acids). The site where the frameshift was introduced is marked by triangles. (C) Western blot analysis of HA-tagged wild type and mutant Vgll4b proteins. NC: non-specific control. (D) Luciferase analysis of HEK293T cells transfected with TEAD1, YAP, VGLL4, and wild type or mutant Vgll4b expressing plasmids. Luciferase activity was normalized to empty vector pcDNA3.1 which was set to 1.0. Error bars represent ± SD of at least three replicates. p values are denoted by asterisks. ∗∗∗P < 0.001 (ANOVA test).
Vgll4b-Deficient Zebrafish Embryos Display an Impaired Heart Valve Development
As in other vertebrates, the heart in zebrafish is the first organ to form and function. The linear heart tube (LHT) forms and rhythmically contracts by 24 h post fertilization (hpf). Looping and ballooning occur at ∼36 hpf, and LHT transforms into one atrium and one ventricle by 48 hpf (Glickman and Yelon, 2002). Each chamber is composed of two tissue layers: the inner endothelial endocardium and outer muscular myocardium (Keegan, 2004). To elucidate the role of vgll4b in zebrafish heart development, firstly, a series of cardiomyocyte markers were examined by whole-mount mRNA in situ hybridization (WISH). While cmlc2 (cardiac myosin light chain 2) marks pan myocardial cells, the two chambers are marked by two distinct myosin genes vmhc (ventricular myosin heavy chain) and amhc (atrial myosin heavy chain) (Chen et al., 1996; Yelon et al., 1999; Yelon, 2001; Berdougo, 2003). Tracking heart formation from 22 to 36 hpf, cmlc2 staining indicated that the primary LHT and chambers formed normally in vgll4b-deficient embryos (Figures 2A,B), whereas a slight defective S shaped heart looping compared with sibling embryos was observed, which lasted to 96 hpf (Figures 2C,D). At 48 hpf, cmlc2, vmhc and amhc showed generally regular temporo-spatial expressions in vgll4b mutants (Figures 2D–F). Physiologically, the expression of anf (natriuretic peptide type A) is confined to the outer curvature (OC) of the atrium and ventricle (Auman et al., 2007), such regionalized expression of anf was still kept in vgll4b-deficient embryos, further indicating that the specification of chambers occurred independently of vgll4b (Figure 2G). The myocardium contracts to drive circulation. To evaluate the function of the myocardium, we assessed the heart rate (HR) and cardiac contractile function by VSF measurements (Figures 2L,M). The results showed that both HR and VSF were comparable between siblings and vgll4b mutants. Moreover, visual inspection analysis showed a normal blood circulation (n > 100 embryos analyzed). These phenomena suggest that the myocardium still functions normally upon loss of vgll4b.
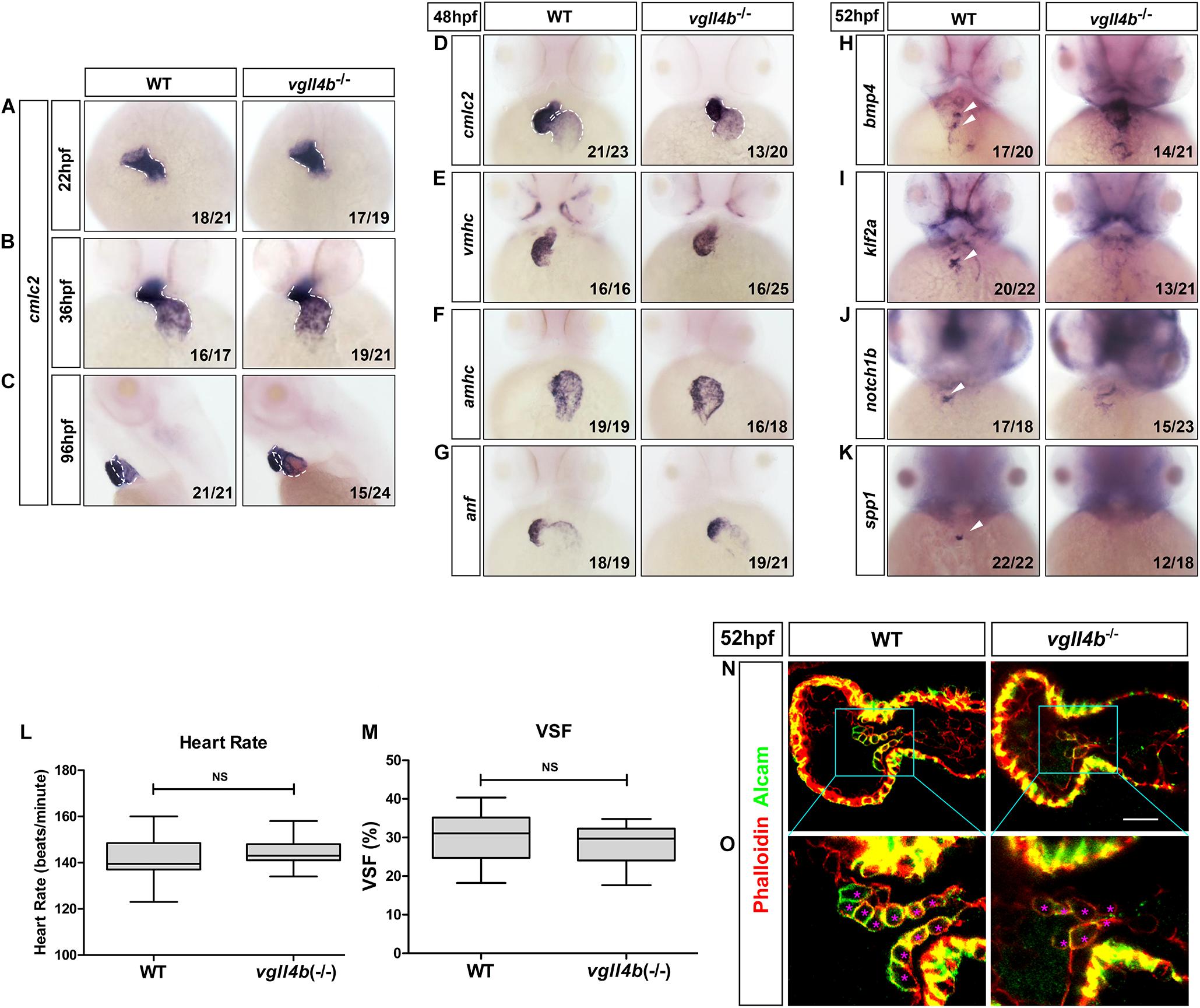
Figure 2. Deficiency of zebrafish vgll4b leads to impaired valvulogenesis. (A–D) WISH analysis of cmcl2 expression in zebrafish embryos at 22, 36, 48, and 96 hpf, a defective S shaped heart looping was clearly observed from 36 hpf. White dashed lines indicate the heart morphology outlined by cmlc2 expression. Lateral view, (C). (E–K) WISH analysis of vmhc, amhc, anf at 48 hpf, as well as bmp4, notch1b, klf2a, spp1 at 52 hpf. (L,M) Optical Heartbeat analysis of cardiac function. Comparison of the heart rate at 48 hpf (L) and ventricular shortening fraction at 52∼54 hfp (M). (N,O) Representative single confocal z-section images showing the endocardial cushion cells expressing Alcam (asterisks) of wild type sibling and vgll4b–/– embryos at 52 hpf. Scale bar: 10 μm.
The formation of the valves is a crucial event during cardiogenesis. The valvulogenesis process in zebrafish could be divided into three successive steps: (i) AV canal (AVC) patterning; (ii) formation of endocardial cushions; (iii) remodeling of endocardial cushions into mature valve leaflets (Peal et al., 2011). During the heart looping and ballooning at ∼36 hpf, the atrium and ventricle are spatially separated by the AVC. Proper AVC development not only divides the heart into two chambers, but also builds the morphological milieu allowing subsequent formation of endocardial cushions and valves. At 48hpf, on each side of the AVC the single-layered early endocardial cushion cells have developed to acquire cuboidal shapes and express Alcam (activated leukocyte cell adhesion molecule). Eventually the endocardial cushions transform through cellular rearrangement into valve leaflets by 96 hpf (Beis, 2005; Peal et al., 2011; Pestel et al., 2016). Certain signaling pathways such as endocardial Notch (Timmerman et al., 2004) and myocardial Bmp (Ma, 2005) have been shown to be indispensable to ensure proper valvulogenesis. To monitor this process, several well-characterized valve marker genes were examined by WISH. During normal valvulogenesis, the flow-responsive gene bmp4 gradually become stronger in myocardial cells in the AVC from 48hpf. At the same time, two other flow-dependent genes, klf2a and its downstream target notch1b, become progressively restricted to express in endocardial cells in the AVC. By 54 hpf, klf2a and notch1b are both significantly expressed within the endocardial cushions in the AVC, whereas in other domains of the endocardium their expression levels are lower or undetectable (Vermot et al., 2009; Donat et al., 2018). Nevertheless, in vgll4b-deficient embryos, the expression of bmp4 and notch1b was misexpressed in the entire ventricle, whereas that of klf2a was almost disappeared in the AVC at 52 hpf (Figures 2H–J). All of these observations together with the absence of the cell migration marker spp1 (Osteopontin or secreted phosphoprotein 1), a previously known factor that is upregulated in the AVC endocardial cushion in zebrafish (Peal et al., 2009), suggested the impairment of valvulogenesis (Figure 2K).
To further characterize the AVC defects in vgll4b mutant heart, we stained vgll4b mutant embryos with an Alcam-specific antibody (green) and phalloidin (red), which labels Actin within the entire heart. Compared with the wild type siblings, single confocal z-section images revealed that the endocardial cells within the AVC region were irregular shaped and disorganized in vgll4b mutants, further indicating that the valvulogenesis was affected due to the loss of vgll4b (Figures 2N,O).
All of these phenotypes could be recapitulated in the vgll4b morpholino (MO) antisense oligonucleotide injected morphants (data not shown). Specific in vivo rescue experiment was carried out and wild type vgll4b mRNA could effectively rescue all of the abnormalities, confirming the specificity of the phenotype observed.
Disruption of Vgll4b-Mef2c Complex in Endocardium, Rather Than That of Vgll4b-Tead Complex, Is Responsible for the Defective Valvulogenesis Upon Loss of vgll4b
During the preparation of this manuscript, a research work showing that the murine VGLL4 also plays a role in valvulogenesis was published. Mechanistic studies indicate that endothelial VGLL4 controls valve development through negatively regulating the formation of TEAD-YAP complex (Yu et al., 2019). Nevertheless, it seems that similar mechanism is not utilized in zebrafish. In human genome, there are four TEAD genes known as TEAD1 to TEAD4, whereas in zebrafish only tead1 and tead3 orthologs exist. However, the introduction of the dominant negative form of tead1 or tead3 (tead DN, in which the C-terminal Yap-binding domain was deleted) (Zhao et al., 2008) mRNA into vgll4b-deficient embryos displayed no rescue effects (Figures 3A–D). Similarly, treatment with verteporfin (a small molecule can interrupt Tead-Yap association) failed to rescue, either (Figures 3E–G). Moreover, it has been reported that TDU1 of VGLL4 preferentially interacts with TEF1 (TEAD1), whereas TDU2 tends to interact with MEF2 (Chen et al., 2004a). Yet, Vgll4b ΔTDU1 mutant mRNA displayed an effective rescue effect, whereas ΔTDU2 exhibited a much weaker rescue effect than that of ΔTDU1, suggesting that Mef2, rather than Tead, would be responsible for the impairment of valve development in zebrafish (Figures 3H–L and Supplementary Figure 1).
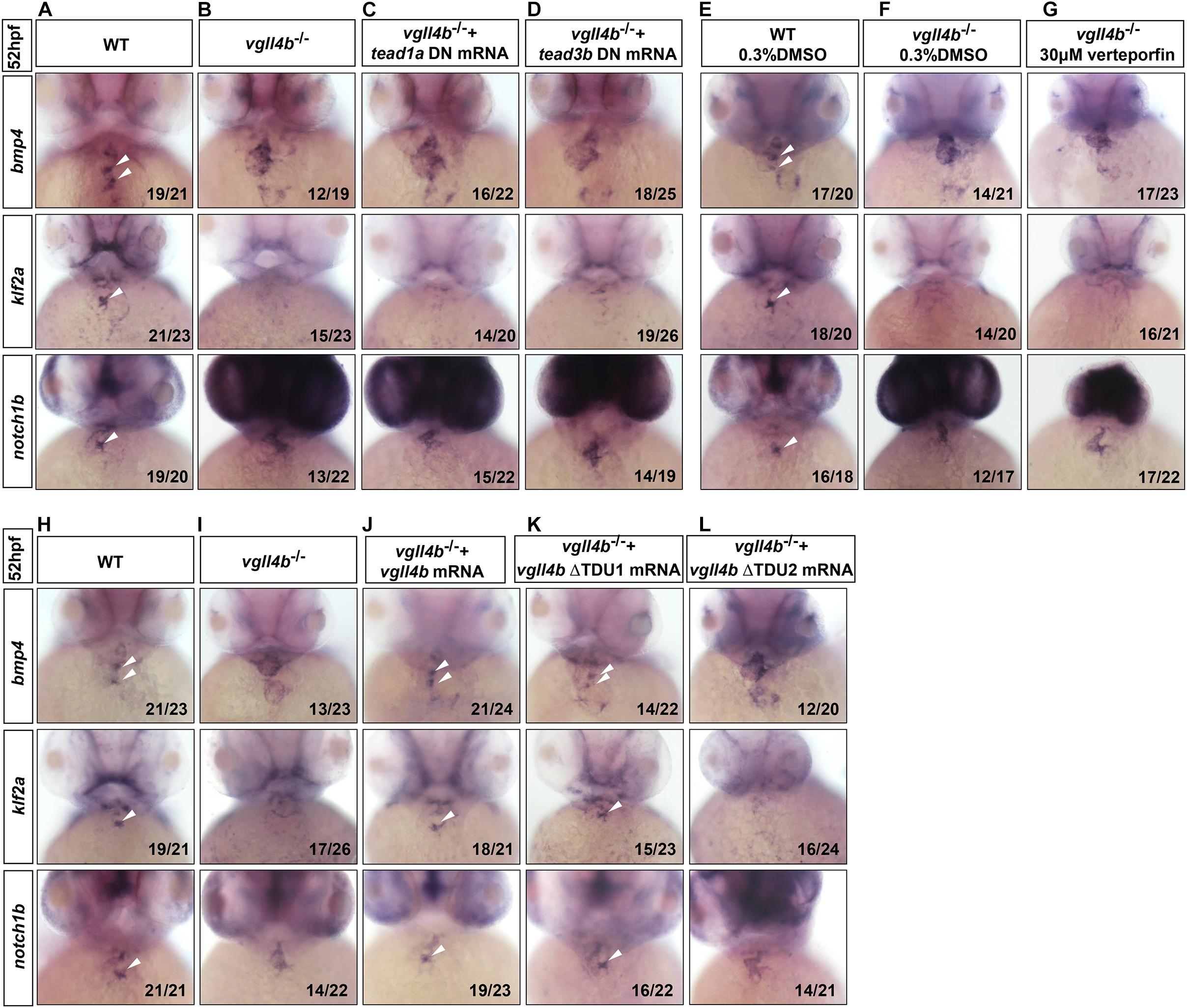
Figure 3. Tead is not involved in the impaired valvulogenesis in vgll4b-deficient embryos. (A–D) Tead1a and tead3b DN mRNA rescue assays in vgll4b–/– embryos. Bmp4, klf2a, and notch1b probes were used in WISH. (E–G) Vgll4b–/– embryos were treated with verteporfin (30 μM) from 32 hpf, but no rescue effect was observed. (H–L) Vgll4b mutants mRNA rescue assays in vgll4b–/– embryos.
In human or mouse, there are four conserved MEF2 genes named MEF2A to MEF2D, of which MEF2A and MEF2C are enriched in heart and play a variety of functions during embryonic cardiac development and in adulthood (Chen et al., 1994; Naya et al., 2002; Durham et al., 2006; Huang et al., 2006; Ewen et al., 2011; Lockhart et al., 2013; Dong et al., 2017), thus Vgll4b might participate in cardiogenesis through a broader mechanism. Although the four mef2 genes are all encoded in zebrafish genome, the main mef2 genes expressed in the developing heart are mef2a, mef2ca and mef2cb (Hinits et al., 2012). The deficiency of mef2a leads to sarcomere assembly defects and severely impedes the cardiac contractility (Wang et al., 2005). Mef2ca and mef2cb are two alternatively spliced variants which play indispensable but redundant roles in myocardial cell differentiation (Ganassi et al., 2014). Among all of the MEF2 family members, MEF2C is the unique one which is expressed not only in cardiomyocytes, but also in endocardial cells to regulate valvulogenesis (Lockhart et al., 2013). Since in vgll4b-deficient embryos the most prominent abnormality was the defective valvulogenesis, we inferred that the Vgll4b-Mef2c complex might be disrupted upon loss of Vgll4b, and released free Mef2c protein in endocardial cells would impair valve development. To validate this hypothesis, firstly, mef2cb mRNA (400 pg) was introduced into one-cell stage wild type embryos, and the overexpression did phenocopy the aberrant expression patterns of klf2a and notch1b as observed in vgll4b mutants (Figures 4A,B,D). It’s worth noting that once 600 pg of mef2cb mRNA was introduced, an obvious misexpression of klf2a could be detected (Figure 4C). These results suggested that the severity of phenotype would be dose-dependent. Moreover, in vgll4b mutant embryos, the overexpression of mef2cb mRNA could cause more severe phenotype (data not shown). To demonstrate that Mef2c was overactivated, a dominant negative Mef2cb mutant mRNA (mef2cb DN) containing only the MADS and MEF2 domains (Molkentin et al., 1996; Ornatsky et al., 1997) was injected into vgll4b mutants, and a profound rescue effect emerged as expected (Figures 4E,H). To further demonstrate that the transcriptional activity of Mef2c was increased in endothelial cells, we put the same mef2cb DN gene under the control of cmlc2 (cmlc2:mef2cb DN) or flk1 (flk1:mef2cb DN) gene’s promoter (in Tol2 backbone) to limit its expression in cardiomyocytes or endothelial cells (Supplementary Figure 2). As anticipated, only the latter could restore the normal expression of klf2a and notch1b, as well as the pattern of the endocardial cells within the AVC region in vgll4b-defecient embryos (Figures 4I–P). It’s worth noting that the flk1:mef2cb DN construct could even rescue the morphological abnormality of the developing heart (Figures 4I–L), further indicating that the primary cause of the cardiac defects exists in endocardium.
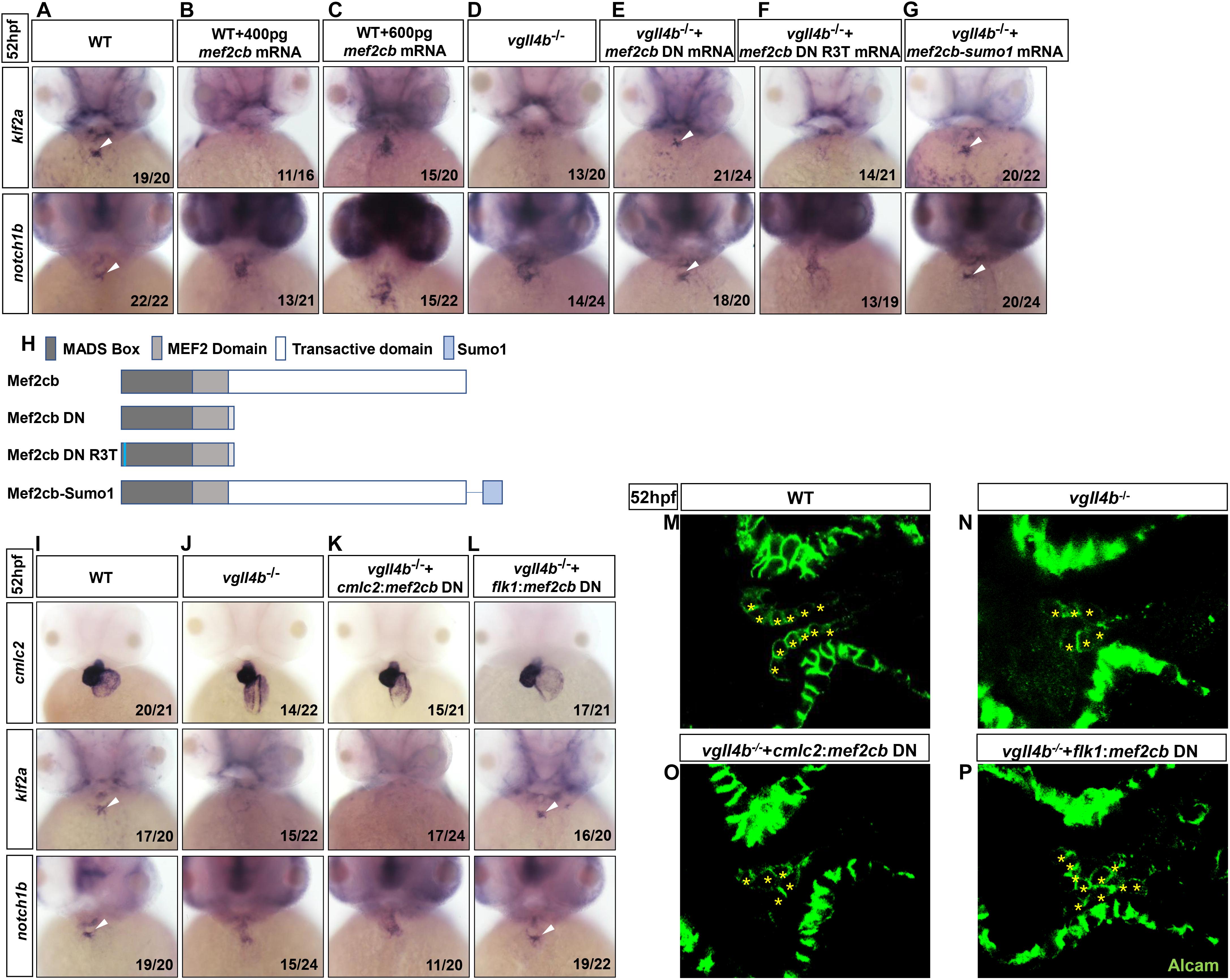
Figure 4. Aberrant activation of Mef2c due to the disruption of Vgll4b-Mef2c complex, accounts for the valvulogenesis defects in vgll4b mutants. (A–C) Mef2cb mRNA was injected into one-cell stage wild type embryos. (D–G) Mef2cb DN, mef2cb DN R3T, and mef2cb-sumo1 mRNA rescue assays in vgll4b–/– embryos. (H) Schematic representation of variant forms of Mef2cb, including WT, DN, R3T, and Sumo1 fusion mutants. (I–L) Cmlc2:mef2cb DN or flk1:mef2cb DN plasmid rescue assays in vgll4b–/– embryos. (M–P) Representative images show Alcam staining of sibling and vgll4b–/– embryos rescued with cmlc2:mef2cb DN or flk1:mef2cb DN plasmid at 52 hpf. The endocardial cushion cells expressing Alcam were indicated by asterisks. Scale bar: 10 μm.
MEF2C could not only function as a transcriptional activator but also a repressor through interaction with different cofactors which serve as positive or negative regulators of transcription (Dong et al., 2017). The post-translational modification SUMOylation mediates the MEF2-dependent repression (Gregoire and Yang, 2006; Kang et al., 2006; Riquelme et al., 2006). SUMO attachment to MEF2 occurs at a conserved lysine residue located in the C-terminal activation domain (Gregoire and Yang, 2006; Kang et al., 2006; Riquelme et al., 2006). Fusion of a SUMO1 molecule to the C-terminus of MEF2 turns it to be a repressor, which has been demonstrated for MEF2A, MEF2C, and MEF2D (Zhao et al., 2005; Gregoire and Yang, 2006; Kang et al., 2006; Shalizi et al., 2007). Hence, to clarify Mef2c functions as an activator or a repressor in valvulogenesis, mef2cb-sumo1 fusion mRNA was used in rescue assay. The results showed that this repression form of Mef2c effectively restored the normal expression pattern of klf2a and notch1b in vgll4b-deficient embryos, suggesting Mef2c acts as an activator (Figures 4G,H).
MEF2 proteins can work by themselves or synergistically along with other transcription factor such as GATA4 (Morin, 2000). The MADS domain of MEF2 protein interacts with the zinc finger domain of GATA4, and the activation domain of both two proteins are indispensable for the functional synergy. However, the DNA binding capacity of MEF2 is not required (Morin, 2000). To elucidate whether Mef2c functions alone or through another transcription factor, a critical arginine which is necessary for DNA binding in MADS domain of Mef2cb (Mef2cb R3T) was mutated (Molkentin et al., 1996). We found that the rescue effect of this mutant was abolished, suggesting Mef2c directly binds to DNA (Figures 4F,H).
Overall, we conclude that in vgll4b mutants the increased free Mef2c protein in endocardium would affect the valvulogenesis through over-activating certain Mef2c downstream targets.
Aberrant Activation of klf2a by Mef2c in Endocardium Contributes to the Impaired Valvulogenesis
As the heart develops, the reversing (or oscillatory) flows between the ventricle and atrium becomes increasingly pronounced, which are most significant in the AVC precede valves formation (Vermot et al., 2009). The reversing flows have been shown to be an important factor in stimulating the expression of the flow-responsive genes klf2a and its downstream target notch1b in the AVC and guiding endocardial cell fate toward valvulogenesis (Vermot et al., 2009). As a key mediator of the effect of the reversing flows on valve development, once the reversing flows are reduced, klf2a is dramatically downregulated and subsequently affects valvulogenesis (Vermot et al., 2009). Injection of gata1 (a gene controlling early hematopoiesis in zebrafish) MO can effectively increase the reversing flow fraction (RFF) through lowering blood viscosity (Vermot et al., 2009). Thus, to make sure whether the decreased klf2a expression in the AVC was caused by reduced RFF, we knocked down gata1 gene in vgll4b-deficient embryos. However, even with a high dose of gata1 MO depleting almost all circulating blood cells, no rescue effect of klf2a and notch1b could be found (Figures 5A–C). In addition, since it has also been reported that the endocardium senses the oscillatory flow via the mechanosensitive channel Trpv4 (transient receptor potential cation channel, subfamily V, member 4) to regulate the expression of klf2a (Heckel et al., 2015), trpv4 mRNA was also introduced into vgll4b-deficient embryos. Nevertheless, neither klf2a nor notch1b could be rescued (Figure 5D). Moreover, the combined injection with gata1 MO and trpv4 mRNA had no obvious rescue effect, either (Figure 5E). Collectively, these results suggest that vgll4b deficiency leads to the decreased expression of klf2a in the AVC through a blood-flow-independent manner.
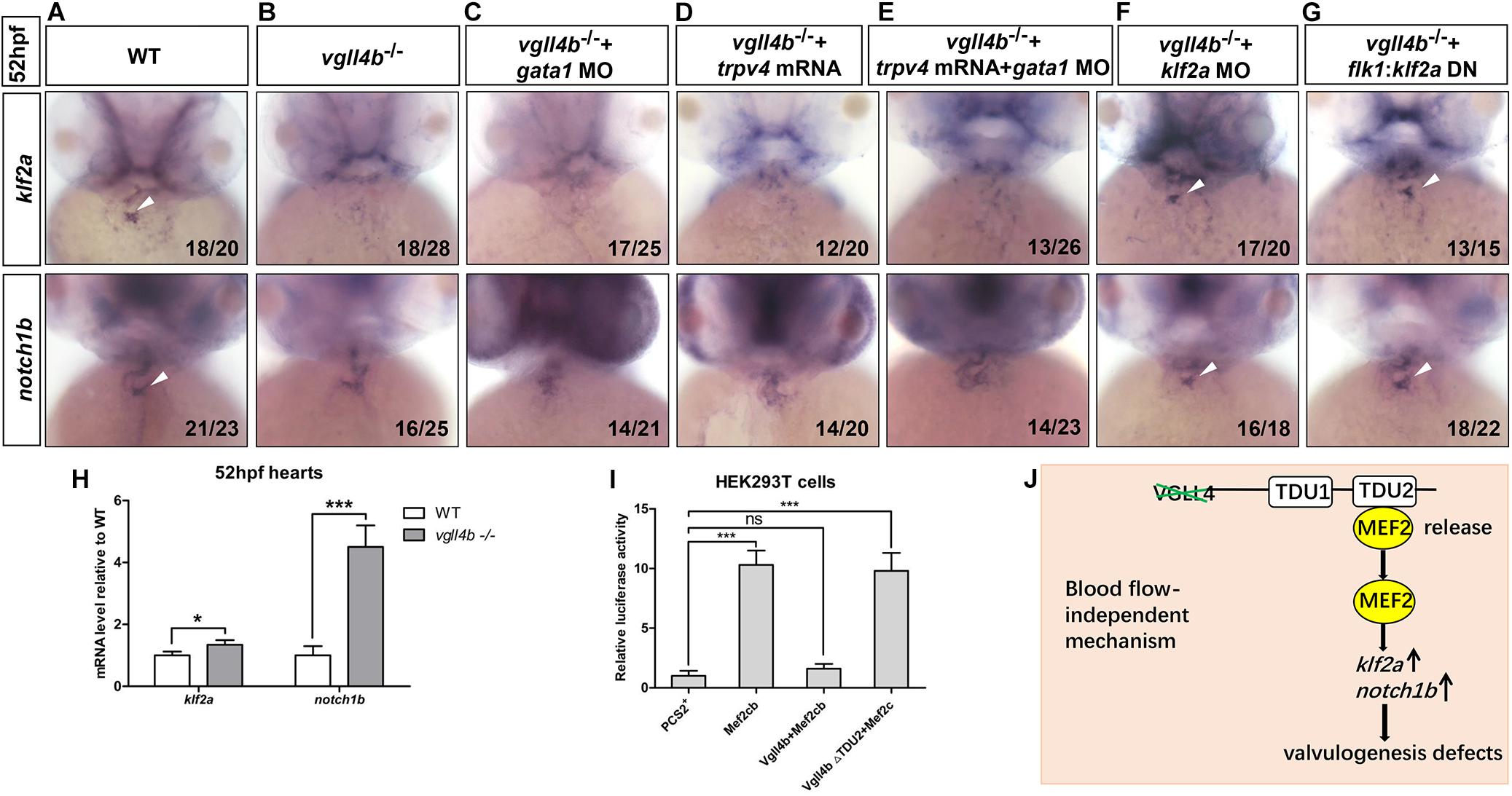
Figure 5. The failure of klf2a expression in the AVC is blood flow-independent, and klf2a is aberrantly activated by Mef2c in the endocardium, which ultimately impedes valvulogenesis. (A–C) Vgll4b–/– embryos were treated with a high dose of gata1 MO, but no rescue effect was observed. (D,E) Trpv4 mRNA was injected alone or in combined with gata1 MO into vgll4b–/– embryos. (F,G) Klf2a MO and flk1:klf2a DN rescue assays in vgll4b–/– embryos. (H) The hearts were harvested respectively from 35 wild type and 35 vgll4b–/– embryos at 52∼54 hfp. Q-PCR analysis revealed that the expression level of klf2a and notch1b in vgll4b–/– hearts was elevated (0.34-fold and 3.2-fold inductions compared to controls). Error bars represent ± SD of at least three replicates. p values are denoted by asterisks; ∗P < 0.1, ∗∗∗P < 0.001 (Student’s t test). (I) Dual luciferase vectors each with a fragment of the zebrafish klf2a promoter (–1.5 kb) were co-transfected into HEK293T cells with empty vector pCS2+, a mef2cb expressing vector, mef2cb combined with wild type vgll4b or vgll4b ΔTDU2 expressing vector. Luciferase activity was detected and normalized to empty vector pCS2+ which was set to 1.0. Error bars represent ± SD of at least three replicates. p values are denoted by asterisks; ∗∗∗P < 0.001 (ANOVA test). (J) Schematic depiction of the aberrant Vgll4b-Mef2c regulation in valvulogenesis in vgll4b–/– zebrafish.
The cerebral cavernous malformation (CCM) pathway is necessary for cardiovascular development. CCM1 (CCM gene 1) and HEG1 (heart of glass) are two major components of the CCM protein complex, which inhibit the endocardial and endothelial expressions of Klf2 during normal cardiovascular development in mice (Zhou et al., 2015, 2016). The loss of zebrafish Krit1 or Heg1 results in an increased expression of klf2a and notch1b throughout the endocardium in a blood-flow-independent manner, and as a consequence, the high levels of klf2a and Notch activity in endocardial cells ultimately impedes cardiac valve development (Donat et al., 2018). In our vgll4b-deficient embryos a drastic ectopic expression of notch1b did emerge at 52hpf, whereas no obvious misexpression of klf2a could be detected by WISH (Figures 3A,B). To evaluate more precisely the expression level of klf2a, the hearts were harvested from vgll4b-deficient embryos, and quantitative reverse transcription PCR (RT-qPCR) analyses were then performed. The results indicated that the transcription level of klf2a was even a little bit higher than that in the wild type siblings (Figure 5H), the discrepancy of the results obtained from the two methods could be explained by the low sensitivity of WISH to detect a weak increase of diffused signals in endocardium. Based on the observation that the activation of klf2a by Mef2cb is dose-dependent, we believe that the amount of released Mef2cb might not be enough to intensively activate klf2a transcription. To further demonstrate that the elevated expression of klf2a was the cause for the defective valvulogenesis, a mild dose of klf2a MO was injected, and a re-enrichment of both klf2a and notch1b in the AVC was observed (Figure 5F). Moreover, an flk1 promoter driven dominant-negative klf2a DBD construct (flk1:klf2a DN, in Tol2 backbone) displayed a similar rescue effect (Figure 5G), confirming the relationship between the increased klf2a expression and valvulogenesis defects.
Next we set out to investigate the reason underlying the upregulation of klf2a. We mentioned above that in vgll4b mutants the released Mef2c would affect the valvulogenesis through over-activating certain downstream targets. It has been reported that MEF2 transcription factors can bind and transactivate KLF2 promoter (Kumar et al., 2005). Furthermore, in endothelial cells the expression of KLF2 was significantly activated by MEF2C (Xu et al., 2015). Hence, we speculated that the released Mef2c from Vgll4b-Mef2c complex would stimulate klf2a expression in vgll4b mutants. To test this hypothesis, a fragment of zebrafish klf2a promoter was cloned into a luciferase reporter vector, and the luciferase activity was increased when co-transfected with a mef2cb expressing plasmid in HEK293T cells. The results also showed that the ectopic expression of vgll4b, but not vgll4bΔTDU2 effectively abolished the transcriptional activity of Mef2cb on klf2a promoter (Figure 5I).
Taken together, our findings indicate that the expression of klf2a is aberrantly activated by Mef2c upon loss of vgll4b in endocardial cells, which in turn impairs valvulogenesis (Figure 5J).
Discussion
Heart valve defects are the most common causes of CHDs in approximately 2% of newborns (Hoffman and Kaplan, 2002). The cardiac gene regulatory network (GRN) contains a core set of evolutionary conserved transcription factors such as NKX2, MEF2, GATA, TBX, and HAND (Olson, 2006), along with multiple other regulators serving as accessory factors for these core transcription factors to contribute to cardiogenesis. To better understand the cardiogenesis, it is important to identify and characterize the novel regulators in this process. Among all of the transcription cofactor VGLL family members, VGLL4 is the only one expressed in human heart (Chen et al., 2004a). Interestingly, VGLL4 is located within the CHD sensitive region of 3p25 deletion syndrome which is frequently related to atrioventricular septal defect (AVSD) (OMIM: 613792), highlighting its role in valvulogenesis.
The genes involved in Notch and Bmp pathways must get spatiotemporally from heart-wide to AVC-specific to set up endocardial cushion formation. Although reversing flows have been shown to play a critical role in this process (Vermot et al., 2009), the mechanism directing such restriction is still poorly understood. Therefore, it is important to define the molecular details controlling AVC patterning and subsequent valve development in order to gain further insights toward the etiology of valve diseases. In this study, we identified a pivotal role of Vgll4b in controlling valvulogenesis through Mef2c sequestration. As a consequence of vgll4b deficiency, the Mef2c target klf2a is activated, which ultimately impairs valve development. Thus the mechanism governing the restricted expression of these flow-responsive genes in the AVC is not merely stimulation by blood flow, but the repression throughout the endocardium by certain regulators. In addition, although altered expression patterns were observed for bmp4, klf2a, and notch1b in the AVC in vgll4b-deficient mutants, some other valve markers such as tbx2b and has2 (hyaluronan synthase 2) kept intact (data not shown). Transcription factor Tbx5 has been shown to restrict the expression of tbx2b and has2 in the AVC to elicit the formation of the endocardial cushion and valves (Camarata et al., 2010). These observations suggested that the valvulogenesis is a very sophisticated process in which retrograde blood flow and multiple regulators function coordinately to ensure proper formation of valves.
Although defective heart valve development was observed in both Vgll4-deficient mice and vgll4b-deficient zebrafish, different mechanism seems to be implicated. While imbalance of VGLL4-TEAD and TEAD-YAP complexes in Hippo pathway is responsible for valvulogenesis defects in mice (Yu et al., 2019), abnormal expression of klf2a in endocardium caused by disruption of Vgll4b-Mef2c complex impairs the valvulogenesis in zebrafish. Beyond that, since there is only one wildly expressed Vgll4 gene exists in mice, whereas three vgll4 paralog genes in zebrafish, more differences were observed. For example, the size of Vgll4–/– mice is much more less than that of siblings, but such phenomenon could not be found in vgll4b–/– zebrafish (data not shown). Moreover, a majority of Vgll4–/– mice died shortly after birth (Yu et al., 2019), whereas the vgll4b-deficient zebrafish could survive into adulthood. In addition, an incomplete penetrance (∼65%) was found for both vgll4b–/– and vgll4b morphants. These differences would be explained by the compensation of vgll4a and/or vgll4l paralogs in zebrafish.
The relationship between VGLL4 and its interacting proteins is intriguing. On one side, the acetylation of VGLL4 at lysine 225 in TDU1 can regulate TEAD1 degradation through a cysteine protease-dependent pathway (Lin et al., 2016). On the other side, VGLL4 can interact with IRF2BP2 independent of TDU domains and enhance its protein stability through preventing proteasome-mediated degradation (Wu et al., 2019). Nevertheless, the protein level of Mef2cb was not affected in HEK293T cells co-transfected with Vgll4b (data not shown). Thus the possibility that Vgll4b controlling the stability of Mef2cb could be excluded.
Most partners of VGLL4 are unable to induce transcription on their own. For example, TEAD proteins have to interact with transcriptional cofactors to activate transcription (Vassilev et al., 2001; Pobbati et al., 2012); IRF2BP2 is frequently described as a corepressor (Childs and Goodbourn, 2003; Carneiro et al., 2011). We thus believe that VGLL4 might be a reservoir of these transcription regulators, once Vgll4 is mutated, its partners would be released and lead to pathogenesis.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Animal Ethical and Welfare Committee, Rui-Jin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Author Contributions
CX, XL, BW, and SG performed the experiments and analyzed the data. RY established the vgll4b knockout line. JT and JZ participated in the preparation of the manuscript. JZ designed the research, analyzed the data, and wrote the manuscript.
Funding
This work was supported by research funding from the National Natural Science Foundation of China (81570280).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Yi Chen and Yi Jin (both from Shanghai Jiao Tong University School of Medicine, Shanghai, China), Jiaming Lou (Shanghai No. 4 High School, Shanghai, China) for technical support. We thank Dr. Lei Zhang (State Key Laboratory of Cell Biology, CAS Center for Excellence in Molecular Cell Science, Innovation Center for Cell Signaling Network, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai, China) for providing TEAD and YAP expressing plasmids, and Dr. Julien Vermot (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) for providing the zebrafish klf2a promoter. We thank Dr. X. Jiao and Dr. Sunny Sharma (all from Department of Cell Biology and Neuroscience, Rutgers University, Piscataway, NJ, United States) for their critical manuscript reading.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2019.00277/full#supplementary-material
Footnotes
References
Auman, H. J., Coleman, H., Riley, H. E., Olale, F., Tsai, H. J., and Yelon, D. (2007). Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 5:0604–0615. doi: 10.1371/journal.pbio.0050053
Beis, D. (2005). Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 132, 4193–4204. doi: 10.1242/dev.01970
Berdougo, E. (2003). Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130, 6121–6129. doi: 10.1242/dev.00838
Camarata, T., Krcmery, J., Snyder, D., Park, S., Topczewski, J., and Simon, H. G. (2010). Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrio-ventricular boundary and valve formation. Dev. Biol. 337, 233–245. doi: 10.1016/j.ydbio.2009.10.039
Carneiro, F. R. G., Ramalho-Oliveira, R., Mognol, G. P., and Viola, J. P. B. (2011). Interferon Regulatory factor 2 binding protein 2 Is a New NFAT1 partner and represses its transcriptional activity. Mol. Cell. Biol. 31, 2889–2901. doi: 10.1128/mcb.00974-10
Castilla, M. Á, López-García, M. Á, Atienza, M. R., Rosa-Rosa, J. M., Diáz-Martín, J., Pecero, M. L., et al. (2014). VGLL1 expression is associated with a triple-negative basal-like phenotype in breast cancer. Endocr. Relat. Cancer 21, 587–599. doi: 10.1530/ERC-13-0485
Chen, H. H., Mullett, S. J., and Stewart, A. F. R. (2004a). Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J. Biol. Chem. 279, 30800–30806. doi: 10.1074/jbc.M400154200
Chen, H. H., Maeda, T., Mullett, S. J., and Stewart, A. F. R. (2004b). Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis 39, 273–279. doi: 10.1002/gene.20055
Chen, J. N., Haffter, P., Odenthal, J., Vogelsang, E., Brand, M., van Eeden, F. J., et al. (1996). Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development 123, 293–302.
Chen, Z., Friedrich, G. A., and Soriano, P. (1994). Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8, 2293–2301. doi: 10.1101/gad.8.19.2293
Childs, K. S., and Goodbourn, S. (2003). Identification of novel co-repressor molecules for Interferon Regulatory Factor-2. Nucleic Acids Res. 31, 3016–3026. doi: 10.1093/nar/gkg431
Combs, M. D., and Yutzey, K. E. (2009). Heart valve development: regulatory networks in development and disease. Circ. Res. 105, 408–421. doi: 10.1161/CIRCRESAHA.109.201566
Donat, S., Lourenço, M., Paolini, A., Otten, C., Renz, M., and Abdelilah-Seyfried, S. (2018). Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. eLife 7:e28939. doi: 10.7554/eLife.28939
Dong, C., Yang, X. Z., Zhang, C. Y., Liu, Y. Y., Zhou, R. B., Cheng, Q. D., et al. (2017). Myocyte enhancer factor 2C and its directly-interacting proteins: a review. Prog. Biophys. Mol. Biol. 126, 22–30. doi: 10.1016/j.pbiomolbio.2017.02.002
Durham, J. T., Brand, O. M., Arnold, M., Reynolds, J. G., Muthukumar, L., Weiler, H., et al. (2006). Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, α-actinin-interacting, costamere-localized protein. J. Biol. Chem. 281, 6841–6849. doi: 10.1074/jbc.M510499200
Ewen, E. P., Snyder, C. M., Wilson, M., Desjardins, D., and Naya, F. J. (2011). The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J. Biol. Chem. 286, 29644–29653. doi: 10.1074/jbc.M111.268094
Gambaro, K., Quinn, M. C. J., Wojnarowicz, P. M., Arcand, S. L., de Ladurantaye, M., Barrès, V., et al. (2013). VGLL3 expression is associated with a tumor suppressor phenotype in epithelial ovarian cancer. Mol. Oncol. 7, 513–530. doi: 10.1016/j.molonc.2012.12.006
Ganassi, M., Badodi, S., Polacchini, A., Baruffaldi, F., Battini, R., Hughes, S. M., et al. (2014). Distinct functions of alternatively spliced isoforms encoded by zebrafish mef2ca and mef2cb. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 559–570. doi: 10.1016/j.bbagrm.2014.05.003
Glickman, N. S., and Yelon, D. (2002). Cardiac development in zebrafish: coordination of form and function. Semin. Cell Dev. Biol. 13, 507–513. doi: 10.1016/S1084952102001040
Gregoire, S., and Yang, X.-J. (2006). association with class iia histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25, 2273–2287. doi: 10.1128/mcb.26.8.3335.2006
Heckel, E., Boselli, F., Roth, S., Krudewig, A., Belting, H. G., Charvin, G., et al. (2015). Oscillatory flow modulates mechanosensitive klf2a expression through trpv4 and trpp2 during heart valve development. Curr. Biol. 25, 1354–1361. doi: 10.1016/j.cub.2015.03.038
Hélias-Rodzewicz, Z., Pérot, G., Chibon, F., Ferreira, C., Lagarde, P., Terrier, P., et al. (2009). YAP1 and VGLL3, encoding two cofactors of TEAD transcription factors, are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosom. Cancer. 49, 1161–1171. doi: 10.1002/gcc.20825
Hinits, Y., Pan, L., Walker, C., Dowd, J., Moens, C. B., and Hughes, S. M. (2012). Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 369, 199–210. doi: 10.1016/j.ydbio.2012.06.019
Hoffman, J. I. E., and Kaplan, S. (2002). The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900. doi: 10.1016/S0735-1097(02)01886-7
Honda, M., Hidaka, K., Fukada, S. I., Sugawa, R., Shirai, M., Ikawa, M., et al. (2017). Vestigial-like 2 contributes to normal muscle fiber type distribution in mice. Sci. Rep. 7:7168. doi: 10.1038/s41598-017-07149-0
Huang, C. J., Tu, C. T., Hsiao, C. D., Hsieh, F. J., and Tsai, H. J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30–40. doi: 10.1002/dvdy.10356
Huang, H. T., Brand, O. M., Mathew, M., Ignatiou, C., Ewen, E. P., McCalmon, S. A., et al. (2006). Myomaxin is a novel transcriptional target of MEF2A that encodes a Xin-related α-actinin-interacting protein. J. Biol. Chem. 281, 39370–39379. doi: 10.1074/jbc.M603244200
Jiang, W., Yao, F., He, J., Lv, B., Fang, W., Zhu, W., et al. (2015). Downregulation of VGLL4 in the progression of esophageal squamous cell carcinoma. Tumor Biol. 36, 1289–1297. doi: 10.1007/s13277-014-2701-7
Jiao, S., Li, C., Hao, Q., Miao, H., Zhang, L., Li, L., et al. (2017). VGLL4 targets a TCF4–TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat. Commun. 8:14058. doi: 10.1038/ncomms14058
Jiao, S., Wang, H., Shi, Z., Dong, A., Zhang, W., Song, X., et al. (2014). A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180. doi: 10.1016/j.ccr.2014.01.010
Jin, S.-W. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209. doi: 10.1242/dev.02087
Kang, J., Gocke, C. B., and Yu, H. (2006). Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 7:5. doi: 10.1186/1471-2091-7-5
Keegan, B. R. (2004). Organization of cardiac chamber progenitors in the zebrafish blastula. Development 131, 3081–3091. doi: 10.1242/dev.01185
Kim, C.-L., Choi, S.-H., and Mo, J.-S. (2019). Role of the hippo pathway in fibrosis and cancer. Cells 8:468. doi: 10.3390/cells8050468
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kumar, A., Lin, Z., SenBanerjee, S., and Jain, M. K. (2005). Tumor necrosis factor alpha-mediated reduction of KLF2 Is due to inhibition of MEF2 by NF- B and histone deacetylases. Mol. Cell. Biol. 25, 5893–5903. doi: 10.1128/mcb.25.14.5893-5903.2005
Lin, Z., Guo, H., Cao, Y., Zohrabian, S., Zhou, P., Ma, Q., et al. (2016). Acetylation of VGLL4 regulates hippo-YAP signaling and postnatal cardiac growth. Dev. Cell 39, 466–479. doi: 10.1016/j.devcel.2016.09.005
Lockhart, M. M., Wirrig, E. E., Phelps, A. L., Ghatnekar, A. V., Barth, J. L., Norris, R. A., et al. (2013). Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. PLoS One 8:e57073. doi: 10.1371/journal.pone.0057073
Ma, L. (2005). Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132, 5601–5611. doi: 10.1242/dev.02156
Maeda, T., Chapman, D. L., and Stewart, A. F. R. (2002). Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 277, 48889–48898. doi: 10.1074/jbc.M206858200
Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1996). Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16, 2627–2636. doi: 10.1128/MCB.16.6.2627
Moorman, A., Webb, S., Brown, N. A., Lamers, W., and Anderson, R. H. (2003). Development of the heart (1) formation of the cardiac chambers and arterial trunks. Heart 89, 806–814. doi: 10.1136/heart.89.7.806
Morin, S. (2000). GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 19, 2046–2055. doi: 10.1093/emboj/19.9.2046
Naya, F. J., Black, B. L., Wu, H., Bassel-Duby, R., Richardson, J. A., Hill, J. A., et al. (2002). Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8, 1303–1309. doi: 10.1038/nm789
Olson, E. N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927. doi: 10.1126/science.1132292
Ornatsky, O. I., Andreucci, J. J., and McDermott, J. C. (1997). A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272, 33271–33278. doi: 10.1074/jbc.272.52.33271
Peal, D. S., Burns, C. G., Macrae, C. A., and Milan, D. (2009). Chondroitin sulfate expression is required for cardiac atrioventricular canal formation. Dev. Dyn. 238, 3103–3110. doi: 10.1002/dvdy.22154
Peal, D. S., Lynch, S. N., and Milan, D. J. (2011). Patterning and development of the atrioventricular canal in zebrafish. J. Cardiovasc. Transl. Res. 4, 720–726. doi: 10.1007/s12265-011-9313-z
Pestel, J., Ramadass, R., Gauvrit, S., Helker, C., Herzog, W., and Stainier, D. Y. R. (2016). Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development 143, 2217–2227. doi: 10.1242/dev.133272
Pobbati, A. V., Chan, S. W., Lee, I., Song, H., and Hong, W. (2012). Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure 20, 1135–1140. doi: 10.1016/j.str.2012.04.004
Riquelme, C., Barthel, K. K. B., and Liu, X. (2006). SUMO-1 modification of MEF2A regulates its transcriptional activity. J. Cell. Mol. Med. 10, 132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x
Shalizi, A., Bilimoria, P. M., Stegmuller, J., Gaudilliere, B., Yang, Y., Shuai, K., et al. (2007). PIASx Is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 27, 10037–10046. doi: 10.1523/jneurosci.0361-07.2007
Tajonar, A., Maehr, R., Hu, G., Sneddon, J. B., Rivera-Feliciano, J., Cohen, D. E., et al. (2013). Brief report: VGLL4 is a novel regulator of survival in human embryonic stem cells. Stem. Cells 31, 2833–2841. doi: 10.1002/stem.1445
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Timmerman, L. A., Grego-Bessa, J., Raya, A., Bertrán, E., Pérez-Pomares, J. M., Díez, J., et al. (2004). Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115. doi: 10.1101/gad.276304
Vassilev, A., Kaneko, K. J., Shu, H., Zhao, Y., and DePamphilis, M. L. (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 7:7168.
Vermot, J., Forouhar, A. S., Liebling, M., Wu, D., Plummer, D., Gharib, M., et al. (2009). Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 7:e1000246. doi: 10.1371/journal.pbio.1000246
Wang, Y. X., Qian, L. X., Yu, Z., Jiang, Q., Dong, Y. X., Liu, X. F., et al. (2005). Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 579, 4843–4850. doi: 10.1016/j.febslet.2005.07.068
Wu, A., Wu, Q., Deng, Y., Liu, Y., Lu, J., Liu, L., et al. (2019). Loss of VGLL4 suppresses tumor PD-L1 expression and immune evasion. EMBO J. 38:e99506. doi: 10.15252/embj.201899506
Xu, Z., Yoshida, T., Wu, L., Maiti, D., Cebotaru, L., and Duh, E. J. (2015). Transcription factor MEF2C suppresses endothelial cell inflammation via regulation of NF-κB and KLF2. J. Cell. Physiol. 230, 1310–1320. doi: 10.1002/jcp.24870
Xue, C., Wang, H. H., Zhu, J., and Zhou, J. (2018). The expression patterns of vestigial like family member 4 genes in zebrafish embryogenesis. Gene Expr. Patterns. 28, 34–41. doi: 10.1016/j.gep.2018.02.001
Yelon, D. (2001). Cardiac patterning and morphogenesis in zebrafish. Dev. Dyn. 222, 552–563. doi: 10.1002/dvdy.1243
Yelon, D., Horne, S. A., and Stainier, D. Y. R. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23–37. doi: 10.1006/dbio.1999.9406
Yu, W., Ma, X., Xu, J., Heumüller, A. W., Fei, Z., Feng, X., et al. (2019). VGLL4 plays a critical role in heart valve development and homeostasis. PLoS Genet. 15:e1007977. doi: 10.1371/journal.pgen.1007977
Zhang, W., Gao, Y., Li, P., Shi, Z., Guo, T., Li, F., et al. (2014). VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res 24, 331–343. doi: 10.1038/cr.2014.10
Zhang, Y., Shen, H., Withers, H. G., Yang, N., Denson, K. E., Mussell, A. L., et al. (2017). VGLL4 selectively represses YAP-dependent gene induction and tumorigenic phenotypes in breast cancer. Sci. Rep. 7:6190. doi: 10.1038/s41598-017-06227-7
Zhao, B., Ye, X., Yu, J., Li, L., Li, W., Li, S., et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971. doi: 10.1101/gad.1664408
Zhao, X., Sternsdorf, T., Bolger, T. A., Evans, R. M., and Yao, T.-P. (2005). Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 25, 8456–8464. doi: 10.1128/mcb.25.19.8456-8464.2005
Zhou, Z., Rawnsley, D. R., Goddard, L. M., Pan, W., Cao, X. J., Jakus, Z., et al. (2015). The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell 32, 168–180. doi: 10.1016/j.devcel.2014.12.009
Keywords: vgll4b, valvulogenesis, zebrafish, Mef2c, klf2a, endocardium, tead
Citation: Xue C, Liu X, Wen B, Yang R, Gao S, Tao J and Zhou J (2019) Zebrafish Vestigial Like Family Member 4b Is Required for Valvulogenesis Through Sequestration of Transcription Factor Myocyte Enhancer Factor 2c. Front. Cell Dev. Biol. 7:277. doi: 10.3389/fcell.2019.00277
Received: 27 August 2019; Accepted: 28 October 2019;
Published: 12 November 2019.
Edited by:
Tinte Itinteang, Ministry of Health & Medical Services, KiribatiReviewed by:
Chunliang Li, St. Jude Children’s Research Hospital, United StatesYuhua Sun, Chinese Academy of Sciences, China
Copyright © 2019 Xue, Liu, Wen, Yang, Gao, Tao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiong Tao, dGFvamlvbmdAaG90bWFpbC5jb20=; Jun Zhou, emoxMDgwMkByamguY29tLmNu
†These authors have contributed equally to this work
 Chang Xue
Chang Xue Xiaohui Liu
Xiaohui Liu Bin Wen
Bin Wen Ruimeng Yang
Ruimeng Yang Shuo Gao
Shuo Gao Jun Zhou
Jun Zhou
