
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 22 October 2019
Sec. Developmental Epigenetics
Volume 7 - 2019 | https://doi.org/10.3389/fcell.2019.00241
This article is part of the Research TopicGene Regulation From the X-Chromosome During Development and DiseaseView all 8 articles
 Bronwyn J. Posynick
Bronwyn J. Posynick Carolyn J. Brown*
Carolyn J. Brown*Sex chromosomes originate as a pair of homologus autosomes that then follow a general pattern of divergence. This is evident in mammalian sex chromosomes, which have undergone stepwise recombination suppression events that left footprints of evolutionary strata on the X chromosome. The loss of genes on the Y chromosome led to Ohno’s hypothesis of dosage equivalence between XY males and XX females, which is achieved through X-chromosome inactivation (XCI). This process transcriptionally silences all but one X chromosome in each female cell, although 15–30% of human X-linked genes still escape inactivation. There are multiple evolutionary pathways that may lead to a gene escaping XCI, including remaining Y chromosome homology, or female advantage to escape. The conservation of some escape genes across multiple species and the ability of the mouse inactive X to recapitulate human escape status both suggest that escape from XCI is controlled by conserved processes. Evolutionary pressures to minimize dosage imbalances have led to the accumulation of genetic elements that favor either silencing or escape; lack of dosage sensitivity might also allow for the escape of flanking genes near another escapee, if a boundary element is not present between them. Delineation of the elements involved in escape is progressing, but mechanistic understanding of how they interact to allow escape from XCI is still lacking. Although increasingly well-studied in humans and mice, non-trivial challenges to studying escape have impeded progress in other species. Mouse models that can dissect the role of the sex chromosomes distinct from sex of the organism reveal an important contribution for escape genes to multiple diseases. In humans, with their elevated number of escape genes, the phenotypic consequences of sex chromosome aneuplodies and sexual dimorphism in disease both highlight the importance of escape genes.
Sex can be determined through various strategies, either environmentally or genetically. The evolution of chromosomal sex determination begins as the result of a sex-determining mutation on an autosome, favoring the loss of recombination at that location (reviewed in Bergero and Charlesworth, 2009; Bachtrog et al., 2014; Ponnikas et al., 2018). This nascent sex chromosome is then present in one sex (the heterogametic sex) but not the other (the homogametic sex), which retains the ancestral chromosome pair. Male heterogametic sex-determining systems (e.g., mammals), are termed XY, while female heterogametic systems (e.g., birds) are termed ZW. Y and W chromosomes are generally smaller and more heterochromatic, while X and Z chromosomes retain much of their ancestral gene complement (reviewed in Bachtrog et al., 2011). Progressive recombination suppression promotes further divergence between the sex chromosome pair: the Y or W chromosome rapidly degrades while the X or Z chromosome remains protected by recombination in the homogametic sex. This evolutionary divergence of the sex chromosomes has been well described in mammals (Figure 1).
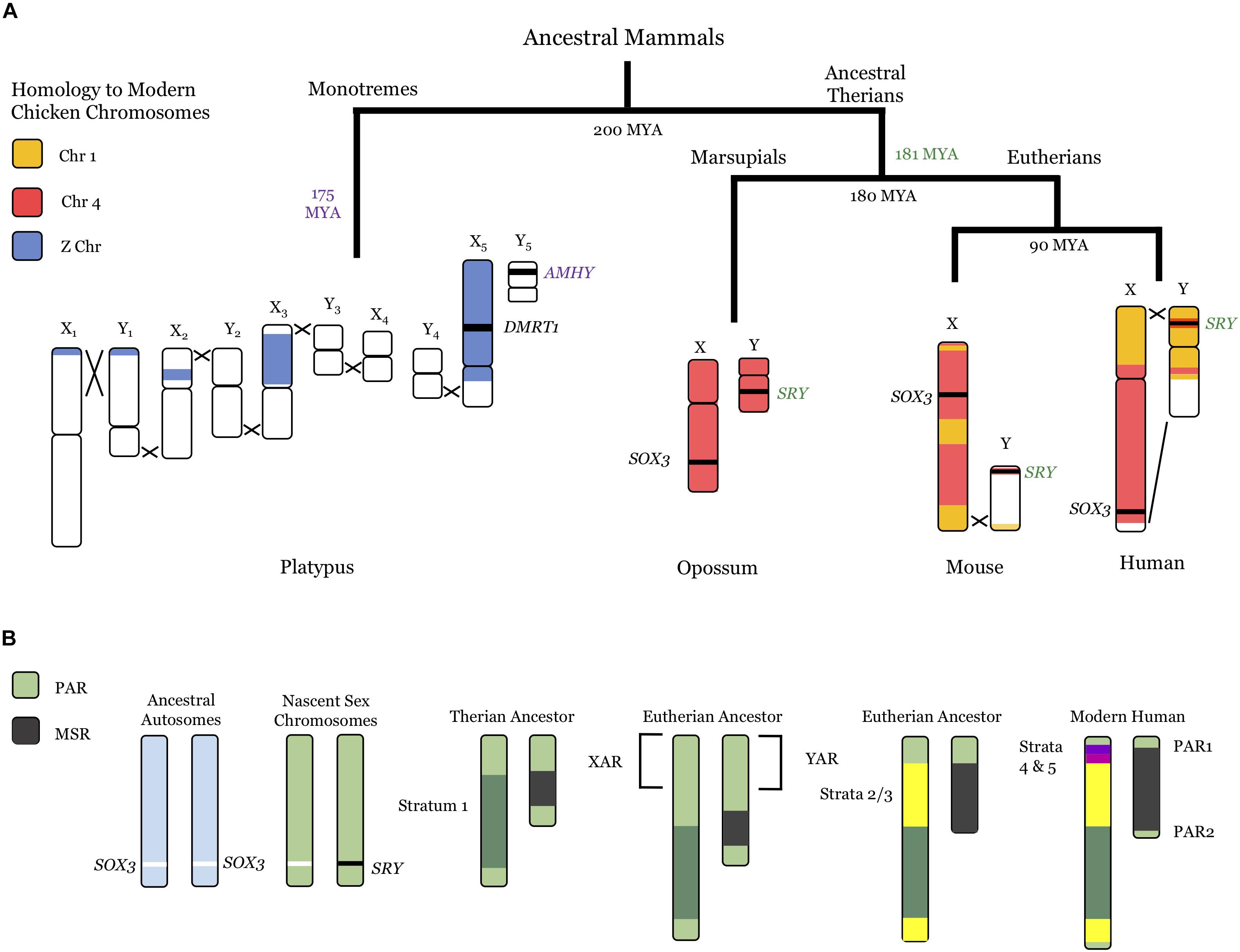
Figure 1. Evolution of mammalian sex chromosomes. (A) Current sex chromosomes of the platypus, opossum, mouse, and human, including recombination locations (black Xs) and homology to current chicken chromosomes (Z chromosome, blue; Chromosome 4, red; Chromosome 1, orange). Divergence times between lineages (black) and approximate dates of sex-determining mutations (monotremes, AMHY, purple text; marsupials and eutherians, SRY, green text) are also noted. There is variation in dates in the literature: those shown are from Cortez et al. (2014). See text for further references. (B) Evolutionary progression from autosomes (light blue, left) to modern human sex chromosomes (right). PARs, light green; Stratum 1, dark green; Strata 2/3, yellow; Stratum 4, fuchsia; Stratum 5, purple. The XAR/YAR regions from the ancestral chromosome 4 (A) are also noted. MYA, million years ago; PAR, pseudoautosomal region; MSR, male-specific region; XAR/YAR, X/Y added region.
Prototherians (monotremes) and therians (marsupials and placental mammals) appear to have developed their modern sex chromosome systems in tandem, following the divergence between the two lineages about 200 million years ago (MYA) (Cortez et al., 2014). Monotremes developed their current sex chromosomes roughly 175 MYA (Cortez et al., 2014): the male platypus has five X chromosomes (X1–5) and five Y chromosomes (Y1–5), while the female has two copies of each X1–5 (Grützner et al., 2004; Rens et al., 2007). Four of the platypus X chromosomes share homology with the bird Z chromosome (Veyrunes et al., 2008), with the bird sex-determining gene (DMRT1) present on X5 (Smith et al., 2009). The best candidate for the monotreme sex-determining gene is the anti-mullerian hormone (AMHY) gene on Y5 (Cortez et al., 2014), which is important in other vertebrate sex-determining pathways (Hattori et al., 2012). Notably, the therian sex-determining system is entirely divergent from monotremes, and likely arose just prior to the split between marsupials and placental mammals (Potrzebowski et al., 2008; Cortez et al., 2014).
Modern therian sex chromosomes originated from a pair of autosomes homologous to the chicken chromosome 4 (Lahn and Page, 1999; Ross et al., 2005). A SOX3 mutation likely resulted in the sex-determining male gene (SRY), creating de facto sex chromosomes (Sinclair et al., 1990; Stevanovlć et al., 1993). Strong selection for linkage between SRY and other male-specific genes resulted in stepwise recombination suppression between the ancestral X and Y chromosomes and created strata of different divergence eras (Rice, 1984; Lahn and Page, 1999; Skaletsky et al., 2003; Ross et al., 2005; Bergero and Charlesworth, 2009). Throughout evolution, the male-specific region (MSR) present only on the Y chromosome expanded, while the X–Y homologous pseudoautosomal region (PAR) that is essential for proper pairing with the X chromosome diminished (reviewed in Bergero and Charlesworth, 2009). The first recombination suppression event appears to have been an inversion in the last common therian ancestor, shown as Stratum 1. In placental mammals, an autosomal region homologous to chicken chromosome 1 (X added region, XAR) was added prior to the placental radiation, apparently around the same time as Strata 2/3 were created through further recombination suppression (Spencer et al., 1991; Ross et al., 2005; Cortez et al., 2014). Interestingly, more sophisticated bioinformatic approaches have found evidence for seven distinct strata within Strata 1–3 (Pandey et al., 2013), indicative of numerous recombination-suppressing events within those more distant time frames. The XAR also underwent successive inversion events, resulting in Stratum 4 and 5 (Lemaitre et al., 2009), and leaving the modern human PAR (PAR1) (Lahn and Page, 1999; Ross et al., 2005). A second pseudoautosomal region, PAR2, is present in humans (Charchar et al., 2003).
As these recombination suppressing events occurred, the Y chromosome became progressively degraded, with the human only retaining about 3% of its ancestral genes (Skaletsky et al., 2003; Bellott et al., 2010). Although there has been speculation about an impending Y chromosome extinction (e.g., Aitken and Graves, 2002), the human Y has generally conserved its remaining genes over the last 25 million years (Hughes et al., 2012). Not surprisingly, many Y-linked genes are involved in male-specific processes such as spermatogenesis (Skaletsky et al., 2003); surviving X–Y pairs (gametologs) are largely involved in dosage-sensitive processes (Bellott et al., 2014), further discussed below.
Erosion of the Y chromosome leads to gene imbalance in males compared to the ancestral autosomal gene dosage. Susumu Ohno hypothesized that male mammals must upregulate their single X chromosome to match autosomal expression levels (Ohno, 1967), and that females balance dosage with males through X-chromosome inactivation (XCI, discussed below). While Ohno originally referred to XCI as a mechanism of dosage compensation, some research groups argue that dosage compensation occurs only in the heterogametic sex (Mank et al., 2011). Therefore, in this review we will refer to expression balance between the heterogametic sex chromosomes and the autosomes as dosage compensation, and that between male and female sex chromosomes as dosage equivalance.
There have been contradictory findings about dosage compensation in mice and humans, largely influenced by the filtering of genes examined (Castagné et al., 2011). Early microarray studies in mice supported Ohno’s hypothesis (Gupta et al., 2006; Nguyen and Disteche, 2006), but RNA-sequencing (RNA-seq) studies in mice and humans have been inconsistent (reviewed in Mank, 2013; Pessia et al., 2014; Disteche, 2016). RNA-seq studies that only analyzed highly expressed genes have supported Ohno’s hypothesis (Deng et al., 2011), although it has been noted that this filtering may enrich for dosage-sensitive genes, such as those in large protein complexes (see Pessia et al., 2012, for further discussion). Similarly, comparisons including testis-specific genes showed no evidence of global dosage compensation (Xiong et al., 2010), arguably since average transcription was decreased (Kharchenko et al., 2011). Notably, none of these studies considered the evolutionary age of the genes in question (He et al., 2011). Ohno’s hypothesis specifically refered to compensation between genes on the ancestral sex chromosome and their autosomal partners, and so ampliconic or elsewise added genes (Bellott et al., 2010) might not be expected to be dosage compensated. Additionally, particularly dosage-sensitive ancestral genes may have either been translocated to autosomes or duplicated into paralogous gene families (Hurst et al., 2015), obviating the need for dosage compensation. When RNA-seq data was compared between X-linked genes and their autosomal chicken homologs (Julien et al., 2012), there was no evidence for global X-upregulation. Intriguingly, evidence for downregulation of autosomal genes interacting with X-linked genes was observed, consistent with the need to maintain balance of expression between the sex chromosomes and the autosomes, as well as between the sexes.
X-chromosome inactivation is an epigenetic process that occurs in the early development of therian females, transcriptionally silencing all but one of the X chromosomes present in each cell. First proposed in the 1960s (Lyon, 1961), silencing is initiated by the expression of a long non-coding RNA (lncRNA) on what will become the inactive X (Xi). The lncRNA spreads in cis to coat the future Xi, beginning a cascade of epigenetic changes that results in the formation of a dense heterochromatic Barr body (recently reviewed in Galupa and Heard, 2018). Once formed, the Xi is stably inherited through subsequent mitotic cell divisions. XCI is an imprinted process in marsupials (Richardson et al., 1971), mediated by expression of the lncRNA Rsx on the paternal X chromosome (Grant et al., 2012). In placental mammals, XCI is initiated by the lncRNA XIST (Brown et al., 1991), which likely evolved from the protein-coding gene Lnx2b (Duret et al., 2006). The region syntenic to the XIST-containing X inactivation center is rearranged in marsupials (Davidow et al., 2007) and Lnx2b remains protein-coding, suggesting that XIST arose after divergence between marsupials and placentals. The origin of Rsx is unknown; the syntenic region in eutheria is flanked by HPRT and FAM122B, and contains the placenta-specific gene PLAC1.
In placental mammals, XCI is random in embryonic tissues, although imprinted XCI occurs in the extraembryonic tissues of mice (Takagi and Sasaki, 1975). Therefore, females are usually mosaics of cells with either the paternal or the maternal active X chromosome (Xa). Random XCI requires the existence of a counting mechanism to ensure that one X chromosome is always active; this mechanism must distinguish between two functionally equivalent X chromosomes in the same nuclear environment. Although numerous models have been proposed, the factor(s) involved in this mechanism have not yet been identified (Loos et al., 2016; Migeon et al., 2017; Mutzel et al., 2019).
In the limited species examined to date, not all X-linked genes are silenced by XCI. Genes that are still expressed from the Xi are considered to “escape,” while genes that are silenced are “subject.” Some escape genes consistently escape inactivation, while others exhibit variable escape and are only expressed in some cell types or individuals. Conventionally, genes are considered to escape if they are expressed at a level that is at least 10% of the Xa allele (Carrel and Willard, 2005). The exclusively paternal marsupial XCI has 14–30% of X-linked genes escaping inactivation (Al Nadaf et al., 2011; Wang et al., 2014; Whitworth and Pask, 2016). Humans also have considerable expression from the Xi, with at least 15% consistently escaping and another 15% variably escaping XCI (Balaton et al., 2015). Mice appear to be more stringent in their silencing with only 3–7% of X-linked genes consistently escaping XCI, although more variable escape genes are being described (reviewed in Carrel and Brown, 2017). When discussing escape from XCI, it is generally unknown if escape occurs from reactivation or avoidance of an initial silencing signal.
There is no single model that satisfactorily explains the variability in both extent and specific genes that escape inactivation across species. There are likely multiple evolutionary paths that can result in ongoing expression from the Xi. Those X-linked genes that retain Y homology are argued to have critical dosage sensitivity that did not allow loss of expression from either the Y or the Xi (Bellott et al., 2014). However, not all genes that escape XCI need to be dosage-sensitive. Other X–Y gametologues with a more recent addition to the sex chromosomes, or a more recent divergence between them, may have neither lost Y expression nor gained X silencing capacity. Additionally, over half of the genes that escape from XCI lack functional Y gametologs (Balaton et al., 2015). Escape for these genes could simply reflect that the acquisition of features favoring silencing may not have occurred. If escape genes have DNA sequences that promote expression from the Xi then a flanking dosage-insensitive gene may also be expressed as a bystander effect. Additionally, selection might favor expression from the Xi in females. Below, and in Figure 2, we outline aspects of these scenarios, and conjecture which escape genes might be so explained.
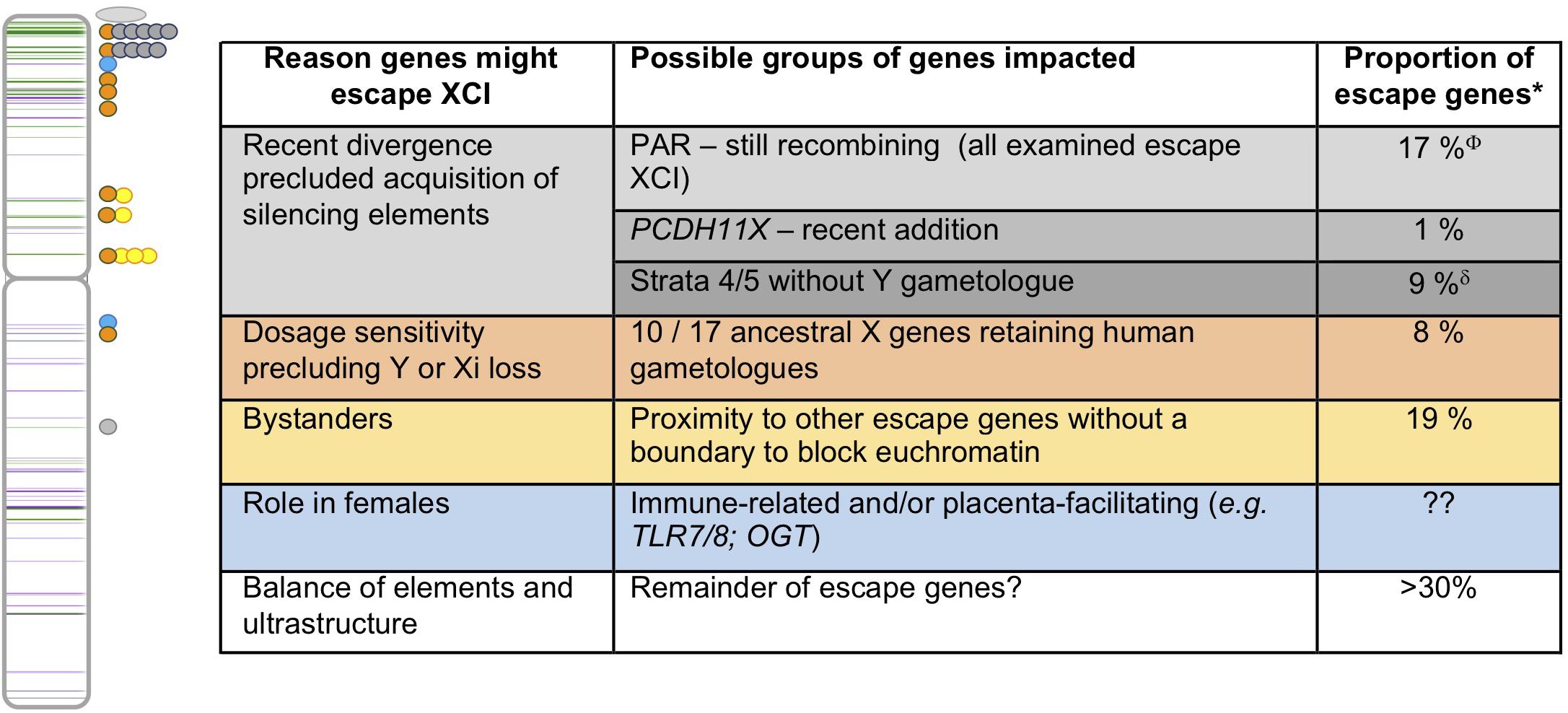
Figure 2. Human escape genes and possible underlying evolutionary reasons for their ongoing expression from the Xi. On the left is an ideogram of the X chromosome showing the genes that escape XCI (green) or variably escape XCI (purple) with lines appearing darker when multiple genes are nearby (data from Balaton et al., 2015). Circles to the right of the chromosome reflect locations of escape genes with features highlighted in the table to the right. The color of the dots matches the table, with PAR1 genes shown as an oval, given their abundance. ∗As tabulated in Balaton et al. (2015); ΦIncludes three ancestral X genes; δIncludes four ancestral X genes.
The presence of Y homology is clearly a strong driver for escape, albeit for only a subset of genes. The most complete block of Y homology is the PAR, and in humans there is both a short arm PAR (PAR1) and a smaller long arm PAR (PAR2) (Figure 1). As initially proposed by Lyon, escape for these genes would allow dosage equivalence between males and females (Lyon, 1962). All of the genes examined to date within PAR1 do escape from XCI, although they generally show some male-bias in expression (Balaton et al., 2015; Tukiainen et al., 2017). Surprisingly, some PAR2 genes are silenced on both the Xi and the Y (Ciccodicola et al., 2000), highlighting another route to equalize expression between the sexes.
Outside of the PARs, ongoing Y homology still strongly correlates with escape from XCI, most strikingly for those genes that have retained functional Y gametologs (Carrel and Willard, 2005). In an analysis of eight mammals, convergent survival of 36 ancestral X–Y genes was observed (Bellott et al., 2014). The remarkable longevity of these pairs was argued to result from either acquisition of male-specific function, or the need to preserve dosage of critical regulatory genes. Indeed, many of the long-lived X-Y gametologous genes are involved in transcription/translation or chromatin modification, and 10 of the 17 ancestral gametologs in humans are known to escape XCI (Bellott et al., 2014; Balaton et al., 2015). These findings have been supported by a recent study that used the conservation of miRNA targeting to demonstrate that remaining X–Y pairs are more dosage sensitive than escape genes without gametologs (Naqvi et al., 2018).
As discussed above, the step-wise loss of recombination left footprints of evolutionary strata on the X chromosome. There is a clear correlation between these strata and the likelihood of escape from inactivation, with the more recently diverged genes being more likely to escape (Carrel and Willard, 2005). It has been suggested that elements that favor silencing accumulated on the X chromosome once it diverged from the Y chromosome. The potential nature of these elements will be discussed in section “Elements That Influence Escape From XCI.”
Selection could also favor escape from XCI if there is a female-specific benefit to ongoing expression from the Xi. Notably, the acquisition of an invasive placenta coincides evolutionarily with the addition of the XAR, which harbors an excess of escape genes. This change in reproductive biology required female eutherian immune systems to both be able to tolerate fetal antigens and defend against parasites and pathogens during pregnancy. The “pregnancy compensation hypothesis” recently proposed that sex-specific differences in both autoimmune diseases (female-biased) and most cancers (male-biased) in industrialized environments are due to adaptions that occured to faciliate successive pregnancies in ancestral conditions (Natri et al., 2019).
The progressive sequestering of recombination between the sex chromosomes likely coincided with different rates of acquisition for genetic elements that favor the spread of silencing. Escape genes may be more prevalent in the XAR due to a paucity of silencing elements in this region. Notably, escape genes are still expressed from older areas of the X chromosome and so must have also acquired genetic elements that still allow expression from the Xi. Domains of silencing and escape require boundaries between the different regions of closed versus open chromatin. Here we discuss the evidence for silencing, escape, and boundary elements, and the potential effects of the physical structure of the Xi. To date, these features have only been studied in humans and mice.
The autosomal portion of an unbalanced X;autosome translocation is able to establish stable silencing, although less efficiently than the X chromosome itself, which led to the proposal of “waystations” that propagate the silencing signal along the X chromosome (Gartler and Riggs, 1983). These waystations would need to be present in greater frequency on the X chromosome compared to the autosomes; Mary Lyon was the first to suggest that waystations could be repeat elements such as LINEs (Lyon, 1998). Bioinformatic studies have supported this “repeat hypothesis,” demonstrating that LINEs are generally depleted around escape genes and enriched near subject genes, relative to the rest of the genome (Bailey et al., 2000; Wang et al., 2006). This pattern has held for autosomal genes on X;autosome translocations, suggesting a functional relationship rather than an evolutionary coincidence of both acquisition of repetitive elements and silencing (Bala Tannan et al., 2014; Cotton et al., 2014). LINEs are densest in the oldest strata of the human X (Ross et al., 2005), consistent with the notion of the accumulation of silencing elements over evolutionary time. The mouse, which has fewer escape genes, also has a greater density of LINEs (Ngamphiw et al., 2014). Additionally, pre-existing features of heterochromatin such as polycomb repressive complex marks are strongly associated with genes that become silenced and therefore may also act as waystations (Cotton et al., 2014; Loda et al., 2017).
A number of genes, particularly those with Y gametologs, consistently escape from XCI across multiple species, arguing that their escape is controlled by conserved elements that were present in the ancestral sex chromosomes. Definitive support for intrinsic escape elements was obtained from integrating a BAC transgene containing an escape gene (Kdm5c) in four random locations on the mouse Xi (Li and Carrel, 2008). Escape status was recapitulated in that Kdm5c always escaped, while the flanking genes present in the BAC remained subject to XCI. The ongoing expression of a primate-specific escape gene (RPS4X) that was observed upon integrating a human BAC transgene onto the mouse Xi demonstrated that such elements could be recognized across species (Peeters et al., 2018). This finding validates the use of mouse models to delineate escape elements present in humans or other mammals.
Boundaries between subject and escape regions on the X have been argued to be established by CTCF sites (Giorgetti et al., 2016), which are well established to create topologically associating domain (TAD) boundaries, marking regions of the genome that are enriched for within-domain interactions (Gómez-Marín et al., 2015). However, since there are over 2000 CTCF sites on the X, there must also be other factors involved in determining what designates escape regions (Ding et al., 2014). The BAC transgene studies mentioned above provided evidence for sequences inhibiting the spread of euchromatin: when the integrated escape region was truncated, spread of escape to flanking genes that are normally subject to XCI was observed (Horvath et al., 2013). In addition to serving as boundaries, CTCF has been reported to associate with promoters of escape genes (Loda et al., 2017), as has the YY1 transcription factor (Chen et al., 2016; Loda et al., 2017).
Boundary elements may be present between the clusters of escape and subject genes in humans; this clustering is an important contributor to the greater number of genes that escape inactivation in humans relative to mice. These escape clusters are notably larger on the human short arm and roughly coincide with TADs (Marks et al., 2015). In mice, only one or two genes within these regions escape XCI (reviewed in Balaton and Brown, 2016). The additional escape genes in these TADs in humans may be due to a “bystander” effect in which adjacent genes also escape because they lack proximal silencing elements or boundaries, features presumably acquired within the mouse TADs. To date, it is unknown whether the clustering patterns in humans or mice is the norm in other mammals.
The physical structure of the Xi likely also influences which genes escape from inactivation. The Xi is comprised of a gene-rich outer rim and repeat-rich internal core (Clemson et al., 2006). Upon silencing, subject genes transition inward, whereas the escape genes remain more peripheral (Chaumeil, 2006). Chromosome conformation capture techniques have allowed determination of the detailed structure of the Xi (Rao et al., 2014; Deng et al., 2015). Both mouse and human have an Xi that forms two major “superdomains,” with the “hinge” region containing the microsatellite Dxz4/DXZ4 (Deng et al., 2015). DXZ4 has been shown to bind CTCF exclusively on the Xi (Chadwick, 2008; Horakova et al., 2012). Intriguingly, deletion of the Dxz4/DXZ4 region leads to loss of Xi superstructure but has a limited effect on transcription from the Xi in either species; paradoxically, the mouse deletion resulted in a tendency for less escape from XCI for variable escape genes (Darrow et al., 2016; Giorgetti et al., 2016). It is possible that the different locations of the hinge between humans and mice might allow greater escape from XCI in humans, where the superdomains are less symmetrical. Additionally, the mouse X chromosome is acrocentric whereas the human X is submetacentric (Figure 1); the need to “traverse” the centric heterochromatin might reduce the strength of the X inactivation signal on the human short arm. Other species have differing centromere locations, including an acrocentric configuration in buffalo (Raudsepp et al., 2012), reinforcing that determining XCI status across eutheria has the potential to provide considerable insight into how genes escape from XCI.
Currently the inactivation status of genes has only been extensively studied in humans and mice. Challenges to studying escape in bulk cell samples across species include random XCI in eutherian females, and the lower expression from the Xi compared to the Xa. We will discuss approaches used in humans and mice, and their potential for studies across eutheria.
The first extensive survey of escape from inactivation in humans utilized somatic cell hybrids (Carrel and Willard, 2005), in which expression from the human Xi could be distinguished from the Xa by separation of the human chromosomes in a mouse background. While this could be used for other eutheria, hybrids bring their own challenges, including loss of XIST localization (Clemson et al., 1998; Hansen et al., 1998) and reactivation of repetitive elements (Ward et al., 2013). Carrel and Willard (2005) also took advantage of expressed single nucleotide polymorphisms (SNPs) to examine allele-specific expression in clonal fibroblast cell lines. More recently, while surveying post-mortem tissue samples [the Genotype-Tissue Expression (GTEx) study], a female with completely non-random inactivation was identified, allowing a similar study of multiple tissues (albeit in a single female) (Tukiainen et al., 2017). In mice, to obtain samples with non-random inactivation, groups have studied extra-embryonic tissues (Mugford et al., 2014), clonal cell lines (Berletch et al., 2015), or engineered Xist mutations (Marks et al., 2015). The GTex study also investigated escape using single-cell RNA-seq (scRNA-seq). As each cell has only a single Xa, the presence of biallelic expression indicates escape from XCI, and clonal cell populations are not required. Therefore, this approach could be applied to other species. Currently scRNA-seq is often focused on cell identification rather than deep gene-wide reads that would be informative for XCI status, but new bioinformatic and sequencing approaches are being developed. In mice defined crosses between divergent strains or species with unique polymorphisms allows for greater informativity from allelic expression analyses. Limited heterogeneity in livestock, such as cows, could reduce the informativity of allelic expression assessment of XCI in other species (Couldrey et al., 2017).
In the absence of polymorphisms, comparisons of expression in females relative to males correlates with escape (Tukiainen et al., 2017), although this approach is limited in sensitivity, and hormonally induced gene expression reduces robustness. Extension to aneuploidies can improve sensitivity, but is confounded by impacts of the aneuploidies themselves (discussed further below). By using DNA methylation as a surrogate for inactivation, comparison of males to females has been quite informative, particularly for those genes with CpG-rich promoters that become highly methylated on the Xi (Cotton et al., 2015). Such analyses, while not directly examining expression, need neither clonal populations nor polymorphisms, and thus would be amenable to exploration of additional species. DNA methylation of candidate escape genes provided evidence for escape in moles and cow (Yen et al., 2007). As a caveat, marsupial DNA methylation is not differential at promoters, but rather shows hypomethylation in flanking regions (Waters et al., 2018). Other “marks” of an Xi can also be used to identify XCI status – such as RNA polymerase occupancy (Kucera et al., 2011) or ATAC-seq open chromatin (Qu et al., 2015) – but these have not been used frequently. Assessment of expression by in situ hybridization (RNA-FISH) allows the examination of expression from the Xa or Xi within individual nuclei. Biallelic signals (indicative of escape from XCI) were observed in elephant, human, and mouse for several candidate escape genes, although a substantial proportion of cells showed only a single focus of expression (Al Nadaf et al., 2011). This could be a result of the challenge of detecting low levels of expression from the Xi, or a reflection of heterogeneous Xi expression as observed in other studies (e.g., Garieri et al., 2018).
Sex chromosome aneuplodies occur in approximately 1/500 births and so are among the most common chromosomal abnormalities in humans (Nussbaum et al., 2001). This is presumably due to reduced phenotypic consequences, as all but one X chromosome is silenced, and the Y chromosome has both a reduced number and testis-specialization of genes.
Turner syndrome (TS) results from the absence of a second sex chromosome, although mosaicism and rearranged X chromosomes (such as an isochromosome) are observed in a significant proportion of cases. Strikingly, the vast majority of 45,X conceptuses are spontaneously aborted, which has been argued to reflect a need for some level of mosaicism (Kelly et al., 1992). In spite of this loss, TS is observed in ∼1/2,000 newborn females, leading to infertility, cardiovascular defects, and other neurological and physical abnormalities (Granger et al., 2016). Mice show far less phenotypic consequence of the absence of a second sex chromosome, which may be due to the reduced number of genes that escape inactivation (reviewed in Berletch et al., 2011).
Klinefelter syndrome, usually 47,XXY but sometimes 48,XXXY or 49,XXXXY, occurs in ∼1/650 male births and is accompanied by the inactivation of all but a single X. While infertility and tall stature are common, additional phenotypic consequences vary widely, contributing to an overall increase in morbidity and mortality (Kanakis and Nieschlag, 2018). In mice, the presence of imprinted X inactivation in the extraembryonic tissues causes an extra maternally derived X chromosome to be detrimental to early development (Shao and Takagi, 1990).
There are reports of sex chromosome aneuploidy in other mammals, and by reviewing this literature in comparison to genome assemblies, Raudsepp et al. suggest the viability of XO individuals is strongly correlated with a smaller PAR (Raudsepp et al., 2012). They fail to detect a similar correlation with X chromosome trisomy, implicating the PAR genes in early developmental failure rather than a PAR contribution to non-disjunction. In addition to the rare literature reports of cats with sex-chromosome monosomy, there is also a scientific and lay literature describing male tortoiseshell or calico cats. As black and orange coat color are X-linked alleles, these felines are therefore presumably XX. These have been shown to be both 39,XXY and chimeras (reviewed in Lyons, 2015). Interestingly, the karyotyping of intersex marsupials has suggested that while the Y chromosome is testis-determining, the development of a scrotum or pouch is more likely controlled by a gene(s) on the X chromosome, likely through dosage of a gene that escapes X inactivation (reviewed in Deakin et al., 2012).
Due to the complexity of comparing XX females and XY males, the X chromosome is often excluded from genome-wide association studies, limiting our understanding of its relationship to complex diseases. Conversely, X-linked mutations in hemizygous males are readily detected, and thus a high density of monogenic traits have been assigned despite the relatively low gene density. The X chromosome is subject to unique evolutionary pressures: it spends more time in females, yet the hemizygosity in males allows immediate expression (and therefore positive or negative selection) of new mutations. Thus, the X chromosome has a unique repertoire of genes, and has been described as a “smart and sexy” chromosome due to genes involved in brain and gametogenesis (Graves et al., 2002, reviewed in Disteche, 2016). There are over 141 known X-linked intellectual disability genes, with potential for another 80 currently only mapped as syndromes (Neri et al., 2018). Strikingly, duplications and consequent phenotypes have been observed for the majority of these genes, suggesting that there is considerable dosage sensitivity.
The X chromosome is home to a variety of immune-related genes. Auto-immunity is often more common in females (reviewed in Natri et al., 2019), and is also more prevalent in Klinefelter males than XY males. Furthermore, most cancers are more prevalent in males than females, and immunological differences have been argued to be a contributor to this sex differential (Natri et al., 2019). Additionally, for X-linked genes with tumor-suppression function, only a single mutation in males would result in loss of function. If the gene is subject to XCI, females would also lose function through a single Xa mutation. However, if this gene escapes XCI, then females would be protected through their Xi copy resulting in a lower prevalence in females. This scenario was recently developed into the EXITS hypothesis (escape from X inactivation for tumor suppressors) to explain the male predominance for many cancers (Dunford et al., 2017).
The evolution of divergent sex chromosomes led to dosage imbalances – both between the ancestral autosomes and the sex chromosomes, and between males and females. While Ohno proposed up-regulation of the residual X-linked genes (dosage compensation) and silencing of one X chromosome in females (dosage equivalence), studies in the last 50 years have revealed unexpected alternatives. In humans over 25% of genes escape from XCI, and over half of these do not have Y gametologs. Although multiple controlling elements have been suggested, it is currently unknown what DNA sequences, chromatin features, or ultrastructural contributions allow genes to escape from XCI. Identification of these elements will provide insights into heterochromatin, and may inform therapeutics aimed at reactivating X-linked genes, silencing autosomes, or avoiding host silencing of gene therapies. In mice, the use of sex-reversed mice to assess the “four core genotypes” allows researchers to distinguish the impact of the sex chromosomes from sex of the animal, and has revealed chromosomal contributions to many diseases (Burgoyne and Arnold, 2016). Since humans have more escape genes, these contributions could be even larger and thus understanding expression from the sex chromosomes is an important facet in delineating the abundant sex differences in human disease. As the ancestry of X-linked genes led to their ability to escape, exploring dosage compensation and dosage equivalence in more species could provide clues to the balance of features involved.
BP and CB each contributed to writing of the review.
Research in the Brown laboratory was supported by the Canadian Institutes of Health Research, including funds for open access publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aitken, R. J., and Graves, J. A. M. (2002). Human spermatozoa: the future of sex. Nature 415, 963–963. doi: 10.1038/415963a
Al Nadaf, S., Deakin, J. E., Gilbert, C., Robinson, T. J., Graves, J. A. M., and Waters, P. D. (2011). A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121, 71–78. doi: 10.1007/s00412-011-0343-8
Bachtrog, D., Kirkpatrick, M., Mank, J. E., McDaniel, S. F., Pires, J. C., Rice, W., et al. (2011). Are all sex chromosomes created equal? Trends Genet. 27, 350–357. doi: 10.1016/j.tig.2011.05.005
Bachtrog, D., Mank, J. E., Peichel, C. L., Kirkpatrick, M., Otto, S. P., Ashman, T.-L., et al. (2014). Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. doi: 10.1371/journal.pbio.1001899
Bailey, J. A., Carrel, L., Chakravarti, A., and Eichler, E. E. (2000). Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. U.S.A. 97, 6634–6639. doi: 10.1073/pnas.97.12.6634
Bala Tannan, N., Brahmachary, M., Garg, P., Borel, C., Alnefaie, R., Watson, C. T., et al. (2014). DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum. Mol. Genet. 23, 1224–1236. doi: 10.1093/hmg/ddt553
Balaton, B. P., and Brown, C. J. (2016). Escape artists of the X chromosome. Trends Genet. 32, 348–359. doi: 10.1016/j.tig.2016.03.007
Balaton, B. P., Cotton, A. M., and Brown, C. J. (2015). Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 6:35. doi: 10.1186/s13293-015-0053-7
Bellott, D. W., Hughes, J. F., Skaletsky, H., Brown, L. G., Pyntikova, T., Cho, T.-J., et al. (2014). Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499. doi: 10.1038/nature13206
Bellott, D. W., Skaletsky, H., Pyntikova, T., Mardis, E. R., Graves, T., Kremitzki, C., et al. (2010). Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616. doi: 10.1038/nature09172
Bergero, R., and Charlesworth, D. (2009). The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24, 94–102. doi: 10.1016/j.tree.2008.09.010
Berletch, J. B., Ma, W., Yang, F., Shendure, J., Noble, W. S., Disteche, C. M., et al. (2015). Escape from X inactivation varies in mouse tissues. PLoS Genet. 11:e1005079. doi: 10.1371/journal.pgen.1005079
Berletch, J. B., Yang, F., Xu, J., Carrel, L., and Disteche, C. M. (2011). Genes that escape from X inactivation. Hum. Genet. 130, 237–245. doi: 10.1007/s00439-011-1011-z
Brown, C. J., Ballabio, A., Rupert, J. L., Lafrenière, R. G., Grompe, M., Tonlorenzi, R., et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. doi: 10.1038/349038a0
Burgoyne, P. S., and Arnold, A. P. (2016). A primer on the use of mouse models for identifying direct sex chromosome effects that cause sex differences in non-gonadal tissues. Biol. Sex Differ. 7:68. doi: 10.1186/s13293-016-0115-5
Carrel, L., and Brown, C. J. (2017). When the Lyon(ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372:20160355. doi: 10.1098/rstb.2016.0355
Carrel, L., and Willard, H. F. (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. doi: 10.1038/nature03479
Castagné, R., Rotival, M., Zeller, T., Wild, P. S., Truong, V., Trégouët, D.-A., et al. (2011). The choice of the filtering method in microarrays affects the inference regarding dosage compensation of the active X-chromosome. PLoS One 6:e23956. doi: 10.1371/journal.pone.0023956
Chadwick, B. P. (2008). DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 18, 1259–1269. doi: 10.1101/gr.075713.107
Charchar, F. J., Svartman, M., El-Mogharbel, N., Ventura, M., Kirby, P., Matarazzo, M. R., et al. (2003). Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 13, 281–286. doi: 10.1101/gr.390503
Chaumeil, J. (2006). A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. doi: 10.1101/gad.380906
Chen, C., Shi, W., Balaton, B. P., Matthews, A. M., Li, Y., Arenillas, D. J., et al. (2016). YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 6:37324. doi: 10.1038/srep37324
Ciccodicola, A., D’Esposito, M., Esposito, T., Gianfrancesco, F., Migliaccio, C., Miano, M. G., et al. (2000). Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum. Mol. Genet. 9, 395–401. doi: 10.1093/hmg/9.3.395
Clemson, C. M., Chow, J. C., Brown, C. J., and Lawrence, J. B. (1998). Stabilization and localization of Xist RNA are controlled by separate mechanisms and are not sufficient for X inactivation. J. Cell Biol. 142, 13–23. doi: 10.1083/jcb.142.1.13
Clemson, C. M., Hall, L. L., Byron, M., McNeil, J., and Lawrence, J. B. (2006). The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. 103, 7688–7693. doi: 10.1073/pnas.0601069103
Cortez, D., Marin, R., Toledo-Flores, D., Froidevaux, L., Liechti, A., Waters, P. D., et al. (2014). Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493. doi: 10.1038/nature13151
Cotton, A. M., Chen, C., Lam, L. L., Wasserman, W. W., Kobor, M. S., and Brown, C. J. (2014). Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum. Mol. Genet. 23, 1211–1223. doi: 10.1093/hmg/ddt513
Cotton, A. M., Price, E. M., Jones, M. J., Balaton, B. P., Kobor, M. S., and Brown, C. J. (2015). Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539. doi: 10.1093/hmg/ddu564
Couldrey, C., Johnson, T., Lopdell, T., Zhang, I. L., Littlejohn, M. D., Keehan, M., et al. (2017). Bovine mammary gland X chromosome inactivation. J. Dairy Sci. 100, 5491–5500. doi: 10.3168/jds.2016-12490
Darrow, E. M., Huntley, M. H., Dudchenko, O., Stamenova, E. K., Durand, N. C., Sun, Z., et al. (2016). Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. U. S. A. 113, E4504–E4512. doi: 10.1073/pnas.1609643113
Davidow, L. S., Breen, M., Duke, S. E., Samollow, P. B., McCarrey, J. R., and Lee, J. T. (2007). The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 15, 137–146. doi: 10.1007/s10577-007-1121-6
Deakin, J. E., Graves, J. A. M., and Rens, W. (2012). The evolution of marsupial and monotreme chromosomes. Cytogenet. Genome Res. 137, 113–129. doi: 10.1159/000339433
Deng, X., Hiatt, J. B., Nguyen, D. K., Ercan, S., Sturgill, D., Hillier, L. W., et al. (2011). Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43, 1179–1185. doi: 10.1038/ng.948
Deng, X., Ma, W., Ramani, V., Hill, A., Yang, F., Ay, F., et al. (2015). Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16:152. doi: 10.1186/s13059-015-0728-8
Ding, Z., Ni, Y., Timmer, S. W., Lee, B.-K., Battenhouse, A., Louzada, S., et al. (2014). Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 10:e1004798. doi: 10.1371/journal.pgen.1004798
Disteche, C. M. (2016). Dosage compensation of the sex chromosomes and autosomes. Semin. Cell Dev. Biol. 56, 9–18. doi: 10.1016/j.semcdb.2016.04.013
Dunford, A., Weinstock, D. M., Savova, V., Schumacher, S. E., Cleary, J. P., Yoda, A., et al. (2017). Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16. doi: 10.1038/ng.3726
Duret, L., Chureau, C., Samain, S., Weissenbach, J., and Avner, P. (2006). The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655. doi: 10.1126/science.1126316
Galupa, R., and Heard, E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. doi: 10.1146/annurev-genet-120116-024611
Garieri, M., Stamoulis, G., Blanc, X., Falconnet, E., Ribaux, P., Borel, C., et al. (2018). Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 115, 13015–13020. doi: 10.1073/pnas.1806811115
Gartler, S. M., and Riggs, A. D. (1983). Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190. doi: 10.1146/annurev.ge.17.120183.001103
Giorgetti, L., Lajoie, B. R., Carter, A. C., Attia, M., Zhan, Y., Xu, J., et al. (2016). Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. doi: 10.1038/nature18589
Gómez-Marín, C., Tena, J. J., Acemel, R. D., López-Mayorga, M., Naranjo, S., de la Calle-Mustienes, E., et al. (2015). Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U.SA. 112, 7542–7547. doi: 10.1073/pnas.1505463112
Granger, A., Zurada, A., Zurada-Zielińska, A., Gielecki, J., and Loukas, M. (2016). Anatomy of turner syndrome: turner syndrome. Clin. Anat. 29, 638–642. doi: 10.1002/ca.22727
Grant, J., Mahadevaiah, S. K., Khil, P., Sangrithi, M. N., Royo, H., Duckworth, J., et al. (2012). Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258. doi: 10.1038/nature11171
Graves, J. A., Gécz, J., and Hameister, H. (2002). Evolution of the human X - a smart and sexy chromosome that controls speciation and development. Cytogenet. Genome Res. 99, 141–145. doi: 10.1159/000071585
Grützner, F., Rens, W., Tsend-Ayush, E., El-Mogharbel, N., O’Brien, P. C. M., Jones, R. C., et al. (2004). In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913–917. doi: 10.1038/nature03021
Gupta, V., Parisi, M., Sturgill, D., Nuttall, R., Doctolero, M., Dudko, O. K., et al. (2006). Global analysis of X-chromosome dosage compensation. J. Biol. Epub 2006 Feb 16.
Hansen, R. S., Canfield, T. K., Stanek, A. M., Keitges, E. A., and Gartler, S. M. (1998). Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc. Natl. Acad. Sci. U.S.A. 95, 5133–5138. doi: 10.1073/pnas.95.9.5133
Hattori, R. S., Murai, Y., Oura, M., Masuda, S., Majhi, S. K., Sakamoto, T., et al. (2012). A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U.S.A. 109, 2955–2959. doi: 10.1073/pnas.1018392109
He, X., Chen, X., Xiong, Y., Chen, Z., Wang, X., Shi, S., et al. (2011). He et al. Author reply. Nat. Genet. 43, 1171–1172. doi: 10.1038/ng.1010
Horakova, A. H., Moseley, S. C., McLaughlin, C. R., Tremblay, D. C., and Chadwick, B. P. (2012). The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum. Mol. Genet. 21, 4367–4377. doi: 10.1093/hmg/dds270
Horvath, L. M., Li, N., and Carrel, L. (2013). Deletion of an X-inactivation boundary disrupts adjacent gene silencing. PLoS Genet. 9:e1003952. doi: 10.1371/journal.pgen.1003952
Hughes, J. F., Skaletsky, H., Brown, L. G., Pyntikova, T., Graves, T., Fulton, R. S., et al. (2012). Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86. doi: 10.1038/nature10843
Hurst, L. D., Ghanbarian, A. T., and Forrest, A. R. R., Fantom consortium, and Huminiecki, L. (2015). The constrained maximal expression level owing to haploidy shapes gene content on the mammalian X chromosome. PLoS Biol. 13:e1002315. doi: 10.1371/journal.pbio.1002315
Julien, P., Brawand, D., Soumillon, M., Necsulea, A., Liechti, A., Schütz, F., et al. (2012). Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10:e1001328. doi: 10.1371/journal.pbio.1001328
Kanakis, G. A., and Nieschlag, E. (2018). Klinefelter syndrome: more than hypogonadism. Metabolism 86, 135–144. doi: 10.1016/j.metabol.2017.09.017
Kelly, T. E., Ferguson, J. E., and Golden, W. (1992). Survival of fetuses with 45,X - an instructive case and an hypothesis. Am. J. Med. Genet. 42, 825–826. doi: 10.1002/ajmg.1320420616
Kharchenko, P. V., Xi, R., and Park, P. J. (2011). Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat. Genet. 43, 1167–1169. doi: 10.1038/ng.991
Kucera, K. S., Reddy, T. E., Pauli, F., Gertz, J., Logan, J. E., Myers, R. M., et al. (2011). Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum. Mol. Genet 20, 3964–3973. doi: 10.1093/hmg/ddr315
Lahn, B. T., and Page, D. C. (1999). Four evolutionary strata on the human X chromosome. Science 286, 964–967. doi: 10.1126/science.286.5441.964
Lemaitre, C., Braga, M. D. V., Gautier, C., Sagot, M.-F., Tannier, E., and Marais, G. A. B. (2009). Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol. Evol. 1, 56–66. doi: 10.1093/gbe/evp006
Li, N., and Carrel, L. (2008). Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc. Natl. Acad. Sci. U.S.A. 105, 17055–17060. doi: 10.1073/pnas.0807765105
Loda, A., Brandsma, J. H., Vassilev, I., Servant, N., Loos, F., Amirnasr, A., et al. (2017). Genetic and epigenetic features direct differential efficiency of Xist-mediated silencing at X-chromosomal and autosomal locations. Nat. Commun. 8:690. doi: 10.1038/s41467-017-00528-1
Loos, F., Maduro, C., Loda, A., Lehmann, J., Kremers, G.-J., ten Berge, D., et al. (2016). Xist and Tsix transcription dynamics is regulated by the X-to-autosome ratio and semistable transcriptional states. Mol. Cell. Biol. 36, 2656–2667. doi: 10.1128/MCB.00183-16
Lyon, M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135–148.
Lyon, M. F. (1998). X-chromosome inactivation: a repeat hypothesis. Cytogenet. Genome Res. 80, 133–137. doi: 10.1159/000014969
Lyons, L. A. (2015). DNA mutations of the cat: the good, the bad and the ugly. J. Feline Med. Surg. 17, 203–219. doi: 10.1177/1098612X15571878
Mank, J. E. (2013). Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29, 677–683. doi: 10.1016/j.tig.2013.07.005
Mank, J. E., Hosken, D. J., and Wedell, N. (2011). Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution 65, 2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x
Marks, H., Kerstens, H. H. D., Barakat, T. S., Splinter, E., Dirks, R. A. M., van Mierlo, G., et al. (2015). Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 16:149. doi: 10.1186/s13059-015-0698-x
Migeon, B. R., Beer, M. A., and Bjornsson, H. T. (2017). Embryonic loss of human females with partial trisomy 19 identifies region critical for the single active X. PLoS One 12:e0170403. doi: 10.1371/journal.pone.0170403
Mugford, J. W., Starmer, J., Williams, R. L., Calabrese, J. M., Mieczkowski, P., Yee, D., et al. (2014). Evidence for local regulatory control of escape from imprinted X chromosome inactivation. Genetics 197, 715–723. doi: 10.1534/genetics.114.162800
Mutzel, V., Okamoto, I., Dunkel, I., Saitou, M., Giorgetti, L., Heard, E., et al. (2019). A symmetric toggle switch explains the onset of random X inactivation in different mammals. Nat. Struct. 38 Mol. Biol. 26, 350–360. doi: 10.1038/s41594-019-0214-1
Naqvi, S., Bellott, D. W., Lin, K. S., and Page, D. C. (2018). Conserved microRNA targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Genome Res. 28, 474–483. doi: 10.1101/gr.230433.117
Natri, H., Garcia, A. R., Buetow, K. H., Trumble, B. C., and Wilson, M. A. (2019). The pregnancy pickle: evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends Genet. 35, 478–488. doi: 10.1016/j.tig.2019.04.008
Neri, G., Schwartz, C. E., Lubs, H. A., and Stevenson, R. E. (2018). X-linked intellectual disability update 2017. Am. J. Med. Genet. B Neuropsychiatr. Genet. 176, 1375–1388. doi: 10.1002/ajmg.a.38710
Ngamphiw, C., Tongsima, S., and Mutirangura, A. (2014). Roles of intragenic and intergenic L1s in mouse and human. PLoS One 9:e113434. doi: 10.1371/journal.pone.0113434
Nguyen, D. K., and Disteche, C. M. (2006). Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38, 47–53. doi: 10.1038/ng1705
Nussbaum, R. L., McInnes, R. R., and Willard, H. F. (2001). Thompson & Thompson Genetics in Medicine, 6th Edn. Philadelphia: W.B. Saunders Co.
Pandey, R. S., Wilson Sayres, M. A., and Azad, R. K. (2013). Detecting evolutionary strata on the human X chromosome in the absence of gametologous Y-linked sequences. Genome Biol. Evol. 5, 1863–1871. doi: 10.1093/gbe/evt139
Peeters, S. B., Korecki, A. J., Simpson, E. M., and Brown, C. J. (2018). Human cis-acting elements regulating escape from X-chromosome inactivation function in mouse. Hum. Mol. Genet. 27, 1252–1262. doi: 10.1093/hmg/ddy039
Pessia, E., Engelstädter, J., and Marais, G. A. B. (2014). The evolution of X chromosome inactivation in mammals: the demise of Ohno’s hypothesis? Cell. Mol. Life Sci. 71, 1383–1394. doi: 10.1007/s00018-013-1499-6
Pessia, E., Makino, T., Bailly-Bechet, M., McLysaght, A., and Marais, G. A. B. (2012). Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. U.S.A. 109, 5346–5351. doi: 10.1073/pnas.1116763109
Ponnikas, S., Sigeman, H., Abbott, J. K., and Hansson, B. (2018). Why do sex chromosomes stop recombining? Trends Genet. 34, 492–503. doi: 10.1016/j.tig.2018.04.001
Potrzebowski, L., Vinckenbosch, N., Marques, A. C., Chalmel, F., Jégou, B., and Kaessmann, H. (2008). Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6:e80. doi: 10.1371/journal.pbio.0060080
Qu, K., Zaba, L. C., Giresi, P. G., Li, R., Longmire, M., Kim, Y. H., et al. (2015). Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 1, 51–61. doi: 10.1016/j.cels.2015.06.003
Rao, S. S. P., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021
Raudsepp, T., Das, P. J., Avila, F., and Chowdhary, B. P. (2012). The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex. Dev. 6, 72–83. doi: 10.1159/000330627
Rens, W., O’Brien, P. C., Grutzner, F., Clarke, O., Graphodatskaya, D., Tsend-Ayush, E., et al. (2007). The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 8, R243. doi: 10.1186/gb-2007-8-11-r243
Rice, W. R. (1984). Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x
Richardson, B. J., Czuppon, A. B., and Sharman, G. B. (1971). Inheritance of glucose-6-phosphate dehydrogenase variation in kangaroos. Nature. New Biol. 230, 154–155. doi: 10.1038/newbio230154a0
Ross, M. T., Grafham, D. V., Coffey, A. J., Scherer, S., McLay, K., Muzny, D., et al. (2005). The DNA sequence of the human X chromosome. Nature 434, 325–337. doi: 10.1038/nature03440
Shao, C., and Takagi, N. (1990). An extra maternally derived X chromosome is deleterious to early mouse development. Development 110, 969–975.
Sinclair, A. H., Berta, P., Palmer, M. S., Hawkins, J. R., Griffiths, B. L., Smith, M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. doi: 10.1038/346240a0
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., et al. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837. doi: 10.1038/nature01722
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271. doi: 10.1038/nature08298
Spencer, J. A., Sinclair, A. H., Watson, J. M., and Marshall Graves, J. A. (1991). Genes on the short arm of the human X chromosome are not shared with the marsupial X. Genomics 11, 339–345. doi: 10.1016/0888-7543(91)90141-Z
Stevanovlć, M., Lovell-Badge, R., Collignon, J., and Goodfellow, P. N. (1993). SOX3 is an X-linked gene related to SRY. Hum. Mol. Genet. 2, 2013–2018. doi: 10.1093/hmg/2.12.2013
Takagi, N., and Sasaki, M. (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. doi: 10.1038/256640a0
Tukiainen, T., Villani, A.-C., Yen, A., Rivas, M. A., Marshall, J. L., Satija, R., et al. (2017). Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248. doi: 10.1038/nature24265
Veyrunes, F., Waters, P. D., Miethke, P., Rens, W., McMillan, D., Alsop, A. E., et al. (2008). Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18, 965–973. doi: 10.1101/gr.7101908
Wang, X., Douglas, K. C., VandeBerg, J. L., Clark, A. G., and Samollow, P. B. (2014). Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum. Monodelphis domestica. Genome Res. 24, 70–83. doi: 10.1101/gr.161919.113
Wang, Z., Willard, H. F., Mukherjee, S., and Furey, T. S. (2006). Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput. Biol. 2:e113. doi: 10.1371/journal.pcbi.0020113
Ward, M. C., Wilson, M. D., Barbosa-Morais, N. L., Schmidt, D., Stark, R., Pan, Q., et al. (2013). Latent regulatory potential of human-specific repetitive elements. Mol. Cell. 49, 262–272. doi: 10.1016/j.molcel.2012.11.013
Waters, S. A., Livernois, A. M., Patel, H., O’Meally, D., Craig, J. M., Marshall Graves, J. A., et al. (2018). Landscape of DNA methylation on the marsupial X. Mol. Biol. Evol. 35, 431–439. doi: 10.1093/molbev/msx297
Whitworth, D. J., and Pask, A. J. (2016). The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Semin. Cell Dev. Biol. 56, 117–121. doi: 10.1016/j.semcdb.2016.01.006
Xiong, Y., Chen, X., Chen, Z., Wang, X., Shi, S., Wang, X., et al. (2010). RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 42, 1043–1047. doi: 10.1038/ng.711
Keywords: dosage compensation, X-chromosome inactivation, mammalian evolution, escape from X-chromosome inactivation, sex chromosomes, gametologues
Citation: Posynick BJ and Brown CJ (2019) Escape From X-Chromosome Inactivation: An Evolutionary Perspective. Front. Cell Dev. Biol. 7:241. doi: 10.3389/fcell.2019.00241
Received: 28 June 2019; Accepted: 02 October 2019;
Published: 22 October 2019.
Edited by:
Celine Morey, UMR 7216 Epigénétique et Destin Cellulaire, FranceReviewed by:
Laura Carrel, Penn State Health Milton S. Hershey Medical Center, United StatesCopyright © 2019 Posynick and Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolyn J. Brown, Q2Fyb2x5bi5Ccm93bkBVQkMuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.