
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 18 October 2019
Sec. Stem Cell Research
Volume 7 - 2019 | https://doi.org/10.3389/fcell.2019.00228
This article is part of the Research TopicThe RNA Revolution in Embryonic Development and Cell Differentiation in Health and DiseaseView all 17 articles
The early mammalian embryo is characterized by the presence of three germ layers-the outer ectoderm, middle mesoderm and inner endoderm. The mesoderm is organized into paraxial, intermediate and lateral plate mesoderm. The musculature, vasculature and heart of the adult body are the major derivatives of mesoderm. Tracing back the developmental process to generate these specialized tissues has sparked much interest in the field of regenerative medicine focusing on generating specialized tissues to treat patients with degenerative diseases. Several Long Non-Coding RNAs (lncRNAs) have been identified as regulators of development, proliferation and differentiation of various tissues of mesodermal origin. A better understanding of lncRNAs that can regulate the development of these tissues will open potential avenues for their therapeutic utility and enhance our knowledge about disease progression and development. In this review, we aim to summarize the functions and mechanisms of lncRNAs regulating the early mesoderm differentiation, development and homeostasis of skeletal muscle and cardiovascular system with an emphasis on their therapeutic potential.
Gastrulation results in the formation of the three germ layers - ectoderm, mesoderm and endoderm. The mesoderm is a middle layer between the innermost, endoderm and the outer ectoderm. The transition from epithelial cells to mesenchymal cells marks the formation of mesoderm which is further organized into the paraxial, intermediate and lateral mesoderm (Nakaya and Sheng, 2008; Evseenko et al., 2010). The three parts of the mesoderm are acted upon by several lineage commitment programs and differentiate into the progenitor cells that give rise to musculoskeletal, urogenital and cardiovascular structures of the body (Doss et al., 2012). Cells of these organs have the same genome, but the differences in transcriptionally active and inactive regions of genome guide the precursor cells toward different cell fates (Iwafuchi-Doi and Zaret, 2016). The differences in genomic organization, followed by activation or silencing of genes, are the result of complex gene regulatory networks (GRNs) (Materna and Davidson, 2007). For many years these GRNs were thought to be controlled exclusively by protein coding genes until the discovery of functional non-coding RNA transcripts (ncRNAs) which form an integrated network to shape the cellular environment during different developmental and metabolic processes (Kim and Sung, 2012). These ncRNAs are divided into two categories based on the transcript length—small ncRNAs (<200 nucleotides) and long ncRNAs (>200 nucleotides) (Mercer et al., 2009). Currently, miRNAs are the best-characterized ncRNAs that are well conserved and repress the expression of target mRNA by binding to its 3′ UTR (Majoros and Ohler, 2007). On the other hand, long ncRNAs (lncRNAs) constitute a less characterized but highly diverse class of ncRNAs. lncRNAs are structurally similar to protein-coding genes as most of them are transcribed by RNA polymerase II, 5′ capped and polyadenylated at 3′ end (Bunch et al., 2016). Regardless of their close similarity to the protein-coding mRNAs, lncRNAs lack the potential to code functional proteins. Although there are many lncRNAs that contain putative open reading frames and indeed some have been re-classified to protein-coding genes (Anderson et al., 2015; Nelson et al., 2016; Matsumoto et al., 2017). The number of bona-fide lncRNAs identified in human genome is, in general, comparable to that of protein-coding genes, but only a few have been functionally characterized. Functionally, lncRNAs can either act in cis by regulating expression of neighboring genes, or in trans, regulating the expression of distant genes (Ulitsky and Bartel, 2013). lncRNAs regulate the gene expression by mending the 3-dimentional genome organization, mediating the binding of chromatin modifying proteins or by sequestering the bound regulatory factors or miRNAs by acting as molecular decoys or sponge (Morriss and Cooper, 2017). A summary of different mechanisms of lncRNA mediated genome regulation is represented in Figure 1. A further class of lncRNAs emerging from regulatory regions of the genome such as enhancers can initiate chromatin looping by recruiting chromatin modifying factors to activate or repress transcription at distant genomic location (Wang et al., 2011). In the past decade several research groups have speculated on the functions of lncRNAs in different biological and pathological systems. More specifically, many lncRNAs have been reported to play crucial roles in the development of skeletal muscle and cardiac lineages, and connected diseases. Here, we discuss the current understanding of the roles of lncRNAs in skeletal muscle and cardiac derivatives emphasizing on their therapeutic potential in the associated pathological conditions.
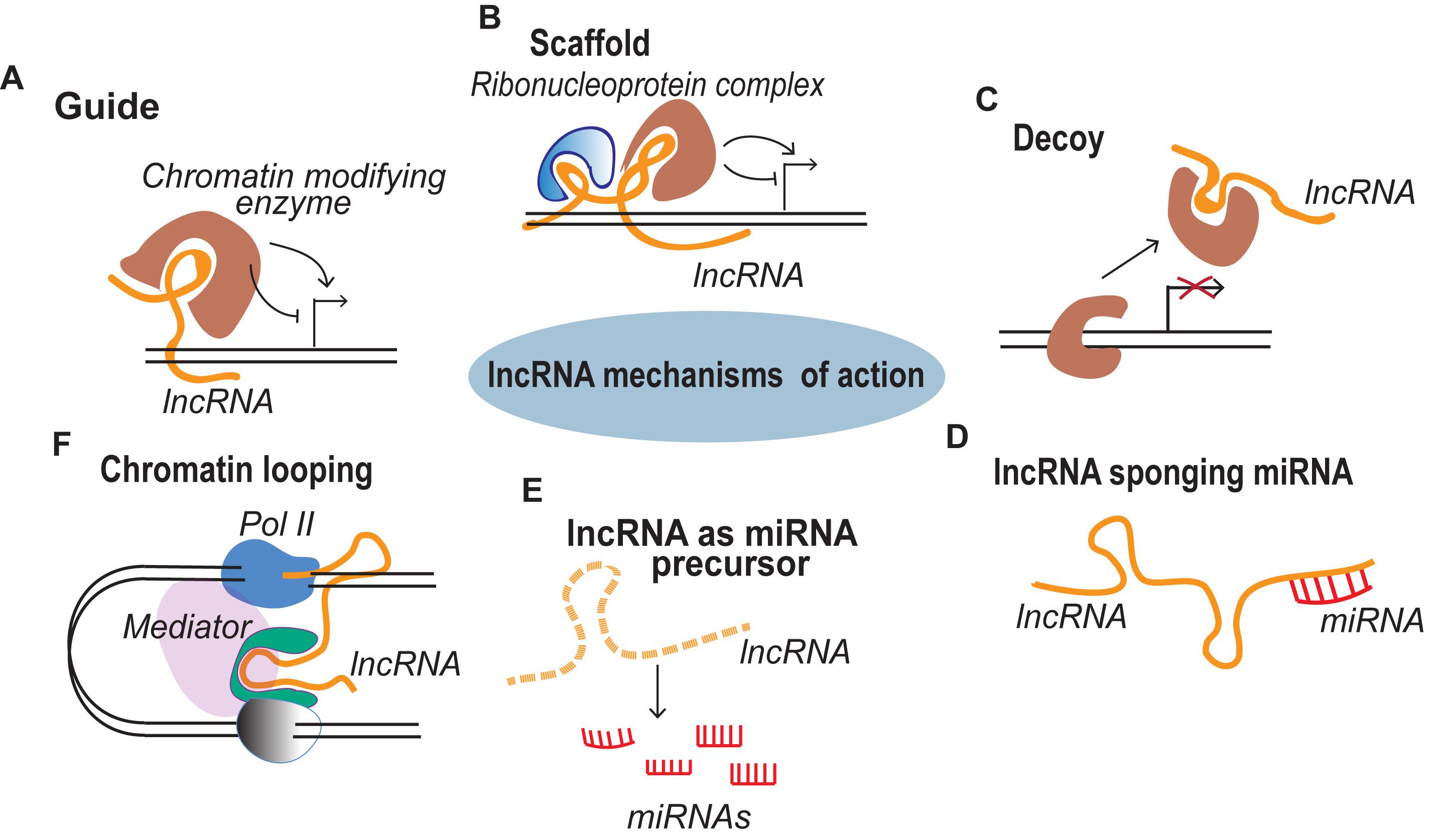
Figure 1. lncRNA mechanisms of action. (A) Guide lncRNAs activate or repress gene expression through relocalization of regulatory factors. (B) Scaffold lncRNAs aid in the formation of Ribonucleoprotein (RNP) complexes. (C) Decoy lncRNAs remove the regulatory factor bound to the genome thereby terminating its regulation. (D) lncRNAs sponge the miRNAs, thus inhibiting the miRNA mediated gene repression. (E) miRNA precursor lncRNAs function as primary miRNA precursors that are processed into mature miRNAs. (F) lncRNA transcription from regulatory regions of the genome initiate long range gene regulation.
Recent studies suggest that lncRNAs are important for mesodermal specification and further differentiation, development and function of mesodermal derivatives (Figure 2). For instance, antisense RNA Evx1as regulates mesendodermal differentiation toward the mesoderm/posterior streak fate through cis regulation of Even-skipped homeobox 1 (Evx1) (Luo et al., 2016). Evx1 is a homeodomain TF that promotes mesoderm differentiation by inhibiting the endoderm/anterior streak gene GSC (Kalisz et al., 2012). The expression of the divergent lncRNA Evx1as was highly correlated with the expression of Evx1. Interestingly, Evx1as knockdown exhibited a higher impact on the expression of mesendoderm markers than the knockdown of Evx1, suggesting the possible trans regulation by Evx1as independent of Evx1 (Luo et al., 2016). lncRNA HoxBlinc is involved in early differentiation and is transcribed from the Homeobox B (Hoxb) locus marking the formation of Flk+ mesoderm with the potential to form hematopoietic and cardiac cells (Deng et al., 2016). As in the case of Evx1as and HoxBlinc, genomic loci of many other key developmental regulators were found to transcribe divergent lncRNAs, collectively termed as Ying Yang lncRNAs (yylncRNAs). Ying Yang lncRNAs follow tissue-specific expression patterns similar to that of their protein-coding counterparts (Frank et al., 2019). The active locus of the mesoderm specifier BRACHUARY (T) encodes yylncT and the expression patterns of the two were nearly identical during mesodermal commitment. The depletion of yylncT specifically abolished the differentiation of human embryonic stem cells (hESCs) to mesoderm without affecting the differentiation toward ectoderm and endoderm, emphasizing the mesoderm specific role of yylncT (Frank et al., 2019). A summary of lncRNAs regulating early mesodermal differentiation is illustrated in Figure 2.
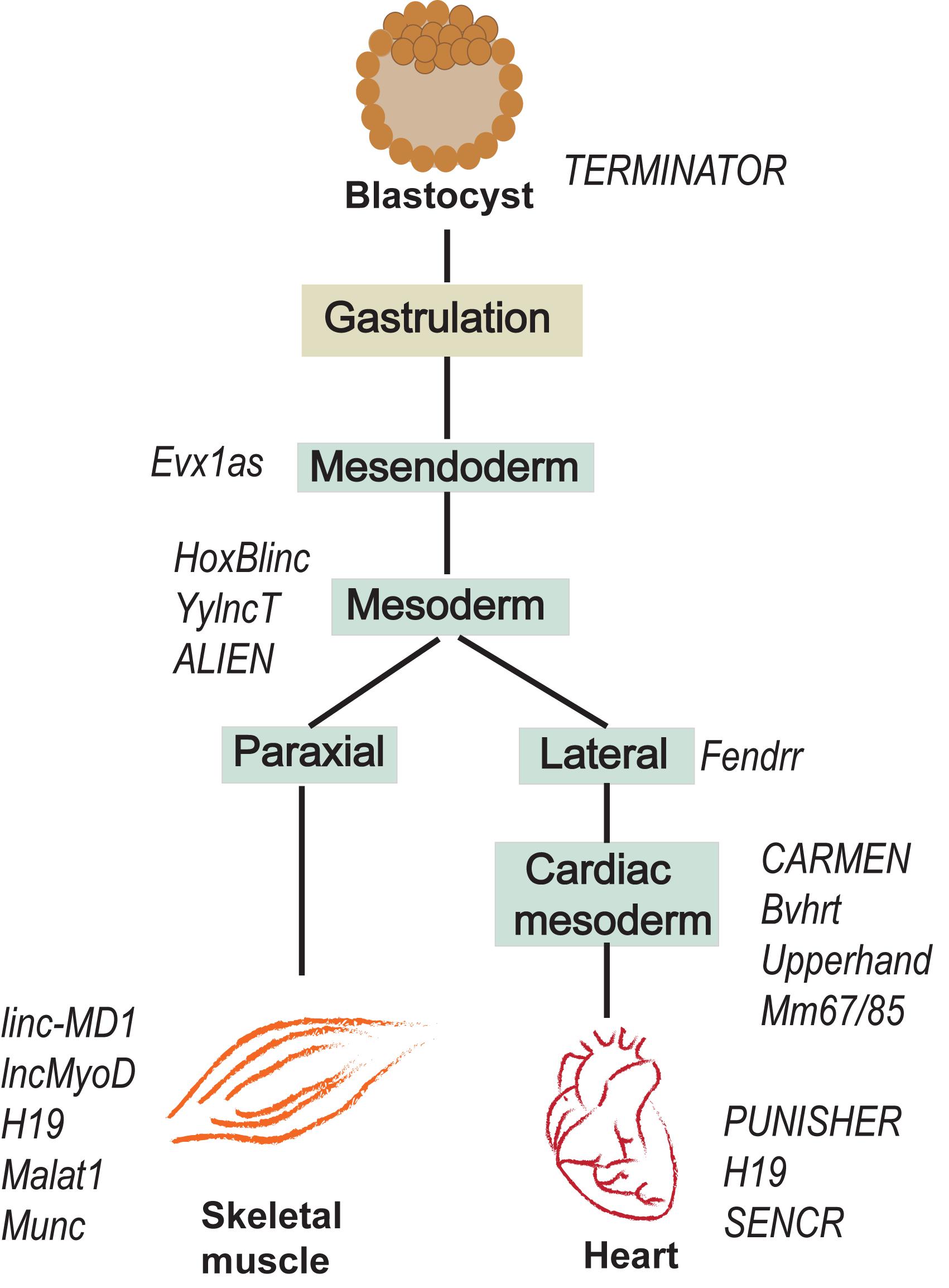
Figure 2. lncRNAs expressed during early development of mesoderm and further differentiation towards skeletal muscle and cardiac lineages.
During embryonic development, the paraxial mesoderm develops into segmented somites, organized into ventral sclerotome and dorsal dermomyotome which form the axial skeleton, the skeletal muscle and the dermis of the adult body, respectively (Chal and Pourquié, 2017). Myogenic progenitor cells (MPCs) are formed in the myotome and subsequently become myoblasts that proliferate and differentiate to form myotubes maturing into skeletal muscle fibers. The MPCs are also responsible for the formation of quiescent satellite cells that contribute to regeneration in adult muscles (Chal and Pourquié, 2017). Upon injury, satellite cells in the adult body get activated and become proliferative myoblasts further differentiating to form new muscle fibers. Alterations in regulatory circuitry of myogenesis leads to muscle disorders and diseases such as, hypertrophy and atrophy. This makes it advantageous to identify novel molecular regulators of myogenesis and injury induced regeneration which will aid to discern new therapeutic targets.
Before differentiating into mature myofibers, the myoblasts proliferate with the activation of cell cycle genes. lncRNA Sirt1AS is an antisense RNA that promotes myoblast proliferation by protecting Sirt1 mRNA—a suppressor of cell cycle inhibitors—from miR-34a mediated degradation (Wang G.-Q. et al., 2016). lnc-31 also promotes proliferation by maintaining the expression of critical cell cycle genes, cyclin D1 (Ccnd1), cyclin E (Ccne1) and Cdc25a (Ballarino et al., 2015). lnc-31 harbors miR-31 precursor sequence, but works independently of miR-31. Despite the poor sequence conservation, both lnc-31 and its human homologue, has-lnc-31, are upregulated during proliferation antagonizing the differentiation process. Furthermore, lnc-31 and has-lnc-31 are abundantly expressed in Duchenne muscular dystrophy (DMD) in mice and humans (Ballarino et al., 2015).
Recently, lncRNA Syisl was reported to supress myoblast differentiation, promoting cell proliferation and fusion. Syisl escorts the EZH2 of PRC2 to the promoter of cell cycle inhibitor p21 and core myogenic genes like MyoG, Mck, Myh4 (Jin et al., 2018). The same study reported increase in muscle density upon in vivo knockout of Syisl. lncRNA Syisl presents an example of lncRNAs activated in a stage specific manner in regulation of early myogenesis. A schematic representation of lncRNAs regulating the early and late phases of myogenesis with their downstream and upstream effector targets is provided in Figure 3.
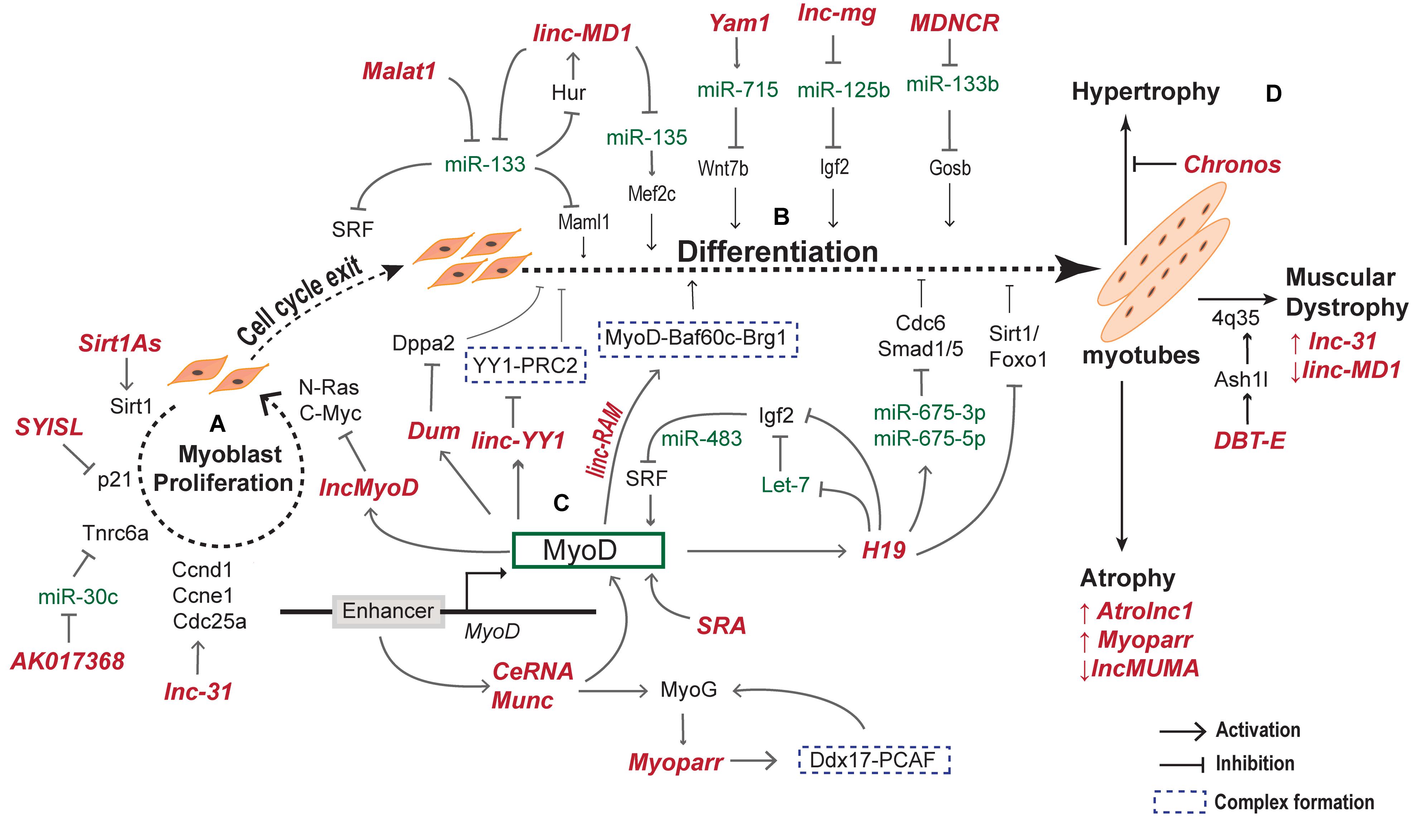
Figure 3. lncRNAs regulate myogenesis at different stages. During myogenesis, myoblast cells proliferate and differentiate into myocytes that fuse together to form multinucleate myotubes. The proliferative and differentiation stages of myogenesis are regulated by several lncRNAs. (A) lncRNAs regulating the myoblast proliferation by activating downstream cell cycle genes. (B) lncRNAs expression and function during cell cycle exit and differentiation to form myotubes. (C) MyoD activated lncRNAs regulate other myogenic factors including MyoD and MyoG to form a complex MyoD–lncRNA–miRNA–mRNA regulatory network during differentiation of myoblasts. (D) Examples of lncRNA expressed and regulating various skeletal muscle disorders.
Myoblast differentiation leads to the formation of multinucleate myotubes that form the mature myofiber (Chal and Pourquié, 2017). A number of stage specific factors, such as MyoD, Myf5, MyoG, and MRF4 act in coordination with epigenetic and transcriptional regulators to regulate the myoblast differentiation (Braun and Gautel, 2011). Enhancer regions of MyoD and MyoG were shown to give rise to eRNAs (enhancer RNAs) which in turn regulate their expression. Two such examples are core enhancer eRNA (CEeRNA) and MUNC – both of which are transcribed from the upstream regulatory region of MyoD (Mousavi et al., 2013). CEeRNA is transcribed from the core enhancer of MyoD regulating its expression in cis by facilitating the chromatin accessibility to RNA polymerase ll. Whereas, MUNC (also called as DRRRNA) is transcribed from Distal Regulatory Region (DRR) of MyoD, enhances the expression of MyoD in cis and of distantly located myogenic genes, MyoG and Myh3, in trans (Mueller et al., 2015). Albeit MUNC overlaps the DRR of MyoD, MUNC acts on multiple myogenic gene promoters. Similar to eRNAs, the promoter region of mouse and human MyoG transcribes lncRNA Myoparr essential for cell cycle withdrawal by activating multiple myogenic factors including the neighboring TF MyoG (Hitachi et al., 2019). Myoparr regulates the interaction between MyoD coactivator Ddx17 and histone acetyl transferase and promotes denervation caused atrophy (Hitachi et al., 2019). The discovery of eRNAs and promoter associated RNAs highlight an additional role of regulatory regions of genome, such as enhancers, in genome regulation.
lncRNA and mRNA microarray analysis identified 997 differentially expressed lncRNAs upon MyoD knockdown in C2C12 cells (Guo et al., 2017). Gene ontology predicted that most of these lncRNAs are associated with pathways involved in muscle formation and cell cycle regulation. The study also identified lncRNA-AK143003 to be significantly regulated by MyoD. In silencing and overexpression experiments AK143003 acts as differentiation antagonist, but further investigation is required to assess its role and mechanism during myogenesis. This study provides a repertoire of lncRNAs in MyoD network for further validation. Another lncRNA involving MyoD in the regulatory circuitry is linc-RAM (Linc-RNA Activator of Myogenesis). It binds to MyoD and facilitates the assemblage of MyoD-Baf60c–Brg1 complex onto the regulatory regions of myogenic genes (Yu et al., 2017). In addition, lncRNA SRA acts as a coactivator for master regulator of muscle differentiation, MyoD and SRA Knockdown prevented proper expression of muscle genes and differentiation (Caretti et al., 2006). Taken together, these lncRNAs present a second layer of regulation in the MyoD regulatory network. Figure 3C, depicts an overview of MyoD regulated lncRNAs and in turn the lncRNAs modulating MyoD and other myogenic genes.
lncRNAs are known to act as guides and scaffolds by recruiting chromatin or DNA modifying complexes. DUM silences Dppa2, an anti-myogenic regulator, by recruiting Dnmts to CpG sites of Dppa2 promoter (Wang L. et al., 2015). Dum acts as a promyogenic factor transcriptionally induced by MyoD with highest expression during proliferation and early myogenesis. Ectopic overexpression of DUM improved regeneration of muscle mass (Wang L. et al., 2015). lncMyoD is another lncRNA activated by MyoD in terminal muscle differentiation suppressing the IGF2-mRNA-binding protein 2 (IMP2) mediated translation of proliferation genes (Gong et al., 2015).
The maternally imprinted lncRNA H19 is expressed exclusively in embryonic tissues and strongly repressed after birth in all tissue types but skeletal muscles (Brannan et al., 1990; Poirier et al., 1991). H19 knockdown decreased the differentiation of myoblast cells and mouse satellite cells by derepressing the mRNAs targets of miR-675-3p and miR-675-5p, encoded by H19, emphasizing a crucial role of H19 during skeletal muscle differentiation (Dey et al., 2014). During myoblast differentiation, miR-675-3p downregulated the BMP pathway by targeting anti-differentiation smad1 and smad5, whereas miR-675-5p repressed the DNA replication initiation factor Cdc6 (Dey et al., 2014). Another study suggests that H19 decrease the expression of myoblast inhibitory genes Sirt1/FoxO1, thus reinforcing the role of H19 in muscle differentiation (Xu et al., 2017). In addition, H19 has been shown to act as a molecular sponge for microRNAs belonging to let-7 family, preventing premature myoblast differentiation (Kallen et al., 2013). More recently, a study of double mutant mice lacking MyoD and Igf2 genes elucidates a complex loop of H19 mediated Igf2 repression by MyoD in development of diaphragm muscle (Borensztein et al., 2013). The authors demonstrate that MyoD stabilized the interaction of CS9 mesodermal enhancer with H19 promoter that accounts for increased expression of H19 in the presence of MyoD. Furthermore, H19 represses Igf2 expression in trans (Wilkin et al., 2000). MyoD is in turn negatively regulated due to downregulation of SRF by Igf2 encoded miR-483. Conclusively, H19-Igf2-MyoD are tightly regulated in a negative feedback loop during embryonic myogenesis (Qiao et al., 2011; Borensztein et al., 2013). Overall, it appears that H19 has a crucial role in myoblast differentiation by mechanistically regulating the key genes such as IGF-2, Sirt1/FoxO1, and microRNAs miR-675 and let-7 during adult as well as embryonic myogenesis.
Similar to H19, lncRNA MALAT1 targets multiple factors during myogenesis. Malat1 is incessantly upregulated during the differentiation of myoblasts to myotubes and its downregulation results in cell cycle arrest in G0/G1 phase, suppressing myoblast proliferation (Watts et al., 2013). A recent study demonstrated a new mechanism of microRNA mediated degradation of MALAT1 transcripts in myoblast nucleus by miR-181 through Ago2-dependent nuclear RNA-induced silencing complex (RISC) (Chen et al., 2017). MALAT1 was also found to influence miR-133 mediated SRF targeting during myogenesis (Han et al., 2015). Conclusively, these mechanistic studies provide evidence that a single lncRNA can act at different levels in GRN through multiple of modes of action.
As seen in H19 and MALAT1, sponging of miRNA appears to be a common mechanism of lncRNA action in muscle differentiation. For instance, lnc-mg (myogenesis associated lncRNA) promoted Igf2 mediated myogenesis by titrating miR-125b (Zhu et al., 2017). lnc-mg was enriched in skeletal muscle and its silencing caused muscle atrophy and loss of muscular endurance during exercise and its overexpression led to hypertrophy. Furthermore, its expression was dynamically induced during differentiation of muscle stem cells (Zhu et al., 2017). Similarly, linc-MD1 modulates the time of muscle differentiation by favoring the expression of MAML1 and MEF2C by sponging miR-133 and miR-135, respectively (Cesana et al., 2011). In addition to sponging of miR-133, linc-MD1 is also the host of miR-133 enabling its alternate synthesize from linc-MD1, controlled by a feedforward positive loop of HuR protein and linc-MD1 (Legnini et al., 2014). linc-MD1 sponges miR-133b derepressing the expression of HuR protein which in turn physically interacts with linc-MD1 to prevent Drosha cleavage of pri-miR-133 sequence (Legnini et al., 2014).
TF Yin Yang 1 (YY1) is an important regulator of myogenesis that silences multiple genes in myoblasts by recruiting Ezh2 (Enhancer of ZesteHomologue2) (Caretti et al., 2004; Wang et al., 2007). Promoter region of YY1 gives rise to linc-YY1 exhibiting low expression in proliferating myoblasts, increased at the beginning of the myogenic program with gradual decrease in the late stages of myogenesis (Zhou et al., 2015). Interestingly, YY1 follows a similar expression profile during the process. When myoblasts undergo differentiation, linc-YY1 is activated by MyoD, which then destabilizes the YY1/PRC2 complex activating pro-differentiation genes (Zhou et al., 2015). Knockdown of linc-YY1 delayed the expression of many myogenic markers which are direct targets of YY1/PRC2, such as MyoG, MyHC, Tnni2 and a-Actin, and miR-1 and miR-29. This indicates that linc-YY1 is a pro-myogenic factor whose knockdown in C2C12 cells delayed myogenic differentiation. Genome wide ChIP-Seq in myoblasts revealed that YY1 regulates several lincRNAs in skeletal muscle collectively called as Yams (YY1-associated muscle lincRNAs) (Lu et al., 2013). Yam1 is a lncRNA regulated by YY1 and acts as an anti-myogenic factor and exerts its function in cis through regulation of miR-715 and via targeting Wnt7b expression (Lu et al., 2013). Downregulation of Wnt7b inhibits myogenic differentiation (von Maltzahn et al., 2012). Among other Yams, Yam-3 also inhibits differentiation while, Yam-2 and Yam-4 facilitates early differentiation (Lu et al., 2013). These observations indicate that lncRNAs are not only regulated by key TFs but in turn regulate the function of other TFs. A list of lncRNAs regulating myogenesis is represented in Table 1.
Given the known importance of lncRNAs in skeletal muscle myogenesis, it is not surprising to know that they also regulate the process of muscle regeneration. lncRNAs such as H19, DUM, MUNC, Yam1 and lncMyoD have been shown to regulate regeneration in cardiotoxin (CTX) model of muscle injury (Shiekhattar, 2013; Dey et al., 2014; Gong et al., 2015; Mueller et al., 2015; Wang L. et al., 2015). Upon CTX mediated injury, the expression of H19 was decreased at days 1–3, followed by increase at days 5–7 and again decreased after the formation of new myofibers at day 14. Interestingly, miR-675-3p and miR-675-5p are co-expressed with H19 throughout the course of regeneration, making it evident that the pro-differentiation action of H19 is mediated by microRNAs generated from it (Dey et al., 2014). While H19 is implicated in regulating regeneration at several stages, expression of Yam1 appears to be stage specific with highest expression at day 2 followed by a sharp reduction for the rest of the regeneration process (Lu et al., 2013). Likewise, lncMyoD is upregulated at days 3–5 and decreased for the remaining regeneration process (Gong et al., 2015). In contrast, MUNC regulates late stage muscle regeneration as its knockdown decreased the average diameter of myofiber at day 14 (Gong et al., 2015; Mueller et al., 2015). Among others, the knockout of linc-RAM and lncRNA IRM in mice displayed impaired muscle regeneration, while the knockdown of MALAT1 enhanced the regeneration (Chen et al., 2017; Yu et al., 2017; Sui et al., 2019). Paradoxically, lncRNA LINC00961 encodes a conserved polypeptide, SPAR, downregulated during the regeneration process (Matsumoto et al., 2017). The downregulation of SPAR activates mTORC1 important for skeletal muscle regeneration and hypertrophy. The functional importance of SPAR emphasizes that not all lncRNA-encoded peptides are translational noise and also raises concern regarding the classification of lncRNAs.
Impaired regeneration process leads to conditions like atrophy and hypertrophy characterized by muscle wasting and increase in size of muscle cells, respectively. Cachexia is a condition that involves atrophy and muscle wasting, commonly associated with chronic kidney disease (CKD) (Morley et al., 2006). lncRNA Atrolnc-1 is elevated in atrophic muscles of mice with cachexia and its inhibition in mice with CKD attenuated muscle wasting (Sun et al., 2018). Targeting Atrolnc-1 could possibly help ameliorate the severity of CKD. lncMUMA was also regulated during atrophy with minimum expression during atrophy development in hindlimb suspension (HLS) mouse model (Zhang et al., 2018). A study on the role of lncRNA in age-associated atrophy identified lncRNA Chronos as a repressor of hypertrophic growth through negative regulation of BMP7 (Neppl et al., 2017). The dysregulation of lncRNAs during atrophy and hypertrophy further strengthens their importance in development of muscle fibers.
Similar to atrophy and hypertrophy, lncRNA expression is dysregulated in muscular dystrophy. DMD is the most prevalent type of dystrophy caused by lack of functional dystrophin protein that connects muscle fibers to extracellular matrix (Hoffman et al., 1987). A study using tilling array designed for dystrophin locus identified novel lncRNAs with expression profiles similar to those of dystrophin gene (Bovolenta et al., 2012). These lncRNAs have a repressive role on the full-length dystrophin isoform and their expression is inversely correlated with dystrophin long isoform in the muscle of female dystrophinopathy carriers. Among other lncRNAs, lnc-31 is upregulated in mdx mice muscle and human DMD patients (Ballarino et al., 2015). On the contrary, linc-MD1 was reduced in DMD patients (Cesana et al., 2011). Facioscapulohumeral muscular dystrophy (FSHD) is another type of dystrophy characterized by wasting of facial, upper arm and shoulder girdle muscle. In 95% of FSHD cases the defect is a deletion in D4Z4 microsatellite repeat, leading to loss of repressive mark (Wijmenga et al., 1990, 1992). This de-repression is coordinated by a chromatin associated lncRNA DBT-E transcribed from D4Z4 repeat through the recruitment of Trithorax group protein at FSHD locus (Cabianca et al., 2012). Although, a few lncRNAs have been reported and studied in muscular dystrophies, they haven’t been studied in non-dystrophic muscle diseases.
Given that lncRNAs are dysregulated in various muscular diseases, they can possibly be novel biomarkers or targets for therapeutic interventions. However, studies investigating lncRNAs in patients with muscular diseases are much languished. Genome wide association studies on different cohorts of muscular disorders may help identify lncRNA loci closely associated with genetic disorders. lncRNAs regulating myogenesis has been well explored in in vitro and in vivo mouse models, nonetheless their role in humans needs to be investigated in depth.
The heart is the first organ to be formed during mammalian embryogenesis. It consists of a multitude of cell types that are formed through complex lineage commitment programs acting upon lateral plate and paraxial mesoderm (Bruneau, 2013; Stone and Stainier, 2019). The intricate network of signaling pathways and the core transcriptional networks in cardiovascular biology have been extensively investigated for many years. On par with the skeletal muscle development and function, lncRNA discovery has unraveled a new layer of regulation in cardiac biology. A number of studies have identified several lncRNAs crucial for cardiac commitment, differentiation and dysfunction leading to diseased conditions. Many groups have discovered new platforms to identify and catalog lncRNAs that regulate cardiac commitment and pathologies (Grote et al., 2013; Kumarswamy et al., 2014; Ounzain et al., 2014; Viereck et al., 2016). Furthermore, the involvement of lncRNAs as therapeutic targets for cardiovascular diseases (CVDs) is beginning to be understood. In this section, we discuss the role of lncRNAs that specifies and regulates the function of cardiomyocytes, smooth muscle cells and endothelial cells and their roles in CVDs and therapeutics.
The minimal regenerative capacity of cardiomyocytes makes it difficult to overcome the damage caused by cardiac diseases. Hence, novel strategies are needed that can improve the regenerative potential of the damaged myocardium. lncRNAs have emerged as novel modulators in cardiac development and regeneration in recent years. Braveheart (Bvht) is the first lncRNA identified in mouse cardiac commitment and its depletion in differentiating mouse embryonic stem cells (mESCs) reduced the potential to form cardiac tissue (Klattenhoff et al., 2013). More specifically, Bvht functions upstream of Mesp1, a master regulator of cardiac differentiation and depletion of Bvht decreased the expression of early cardiac cell surface markers (such as PdgfRa and Flk-1) with consistent increase in hematopoietic markers, suggesting its involvement in regulating cell fate decisions (Klattenhoff et al., 2013). However, the role of Bvht in hematopoietic differentiation is unclear and needs further investigation. Bvht lacks a human ortholog and this loss in the due course of evolution represents the species-specific differences in heart development.
lncRNA Fendrr with its expression restricted to the nascent lateral plate mesoderm, regulates cardiac differentiation by interacting with the PRC complex (Grote et al., 2013). Fendrr is imperative for the development of the heart and the body wall. Mechanistically, Fendrr binds to two major chromatin modifiers, PRC2 and TrxG/MLL, and recruits these complexes to the promoters of genes involved in inception and differentiation of the lateral plate mesoderm, thus regulating cardiac lineage commitment (Grote et al., 2013).
CARMEN, (CAR)diac (M)esoderm (E)nhancer-associated (N)oncoding RNA) is a super enhancer (SE)-associated lncRNA identified in the transcriptome of cardiac precursor cells (CPCs) obtained from the human fetal heart (Ounzain et al., 2014, 2015). The CARMEN locus is upstream of two microRNAs known to direct differentiation toward SMC—MiR-143 and -145. CARMEN is a conserved lncRNA and shelters a highly active, notch-repressive, SRF/NKX2.5 bound cardiac enhancer, responsible for restricted expression of miR-143 and miR-145 during cardiac development. Notably, the mouse ortholog of Carmen and previously mentioned lncRNA Bvht are co-located in the mouse genome. Both of the lncRNAs showed maximum expression between cardiac mesoderm and cardiac precursor stages during induced cardiac differentiation of P19CL6 cells, indicative of their involvement in early cardiac specification. While Bvht works in trans, CARMEN functions in cis as well as in trans and both were found to be essential for maintaining the cardiac identity in neonatal cardiomyocytes (Klattenhoff et al., 2013; Ounzain et al., 2015). Thus, CARMEN represents a SE-associated lncRNA that can potentially be manipulated for initiating neocardiogenesis for treating a heart damage.
Global transcriptomic profiling of enhancer transcribed lncRNAs during ESC differentiation into cardiomyocytes in mouse and human reveal co-expression of many of lncRNAs and their predicted downstream targets (Ounzain et al., 2014). The human enhancer transcripts of mm-67, -85, and -130 were significantly upregulated at different time points of cardiac differentiation. More specifically, knockdown of mm-85 derived lncRNA in P19CL6 mouse embryonic carcinoma cells significantly decreased myocardin expression and upregulated mm-67 present within the myocardin gene (Ounzain et al., 2014). This shows the crucial role of mm-85 in regulating myocardin expression in mouse P19CL6 embryonic carcinoma cells.
The growing body of experimental evidence suggests that enhancer derived lncRNAs are important for expression of proximal target genes in cardiac development. lncRNA Uph (also named Upperhand or Hand2os1) is transcribed from upstream enhancer of HAND2, a regulator of heart development and reprogramming of fibroblast to cardiomyocytes (Anderson et al., 2016). Knockout of Uph in mouse embryos resulted in the inability of the embryo to develop a right ventricular chamber and the cardiac phenotype emulated by HAND2 knockout embryos (Anderson et al., 2016). More recently, deletion of Uph in the mouse upregulated the expression of Hand2 accompanied by the dysregulated cardiac gene program, congenital heart defects and prenatal lethality (Han et al., 2019). While Uph is transcribed upstream, lncRNA Handsdown (Hdn) is located downstream of Hand2 (Ritter et al., 2019). The genetic analysis in mice demonstrated that Hdn is haploinsufficient and Hdn-heterozygous mice presented right ventricular hyperplasia with increased levels of Hand2 (Ritter et al., 2019). Thus, Uph and Hdn regulate Hand2 expression in cis thereby playing a crucial role in cardiac development.
A study in human ESCs and zebrafish developmental models identified three lncRNAs implicated at different stages of mesoderm and cardiovascular development, namely, TERMINATOR, PUNISHER and ALIEN (Kurian et al., 2015). These three lncRNAs are conserved from zebrafish to humans and manifest similar stage-specific expression. TERMINATOR is essential for early embryonic survival, pluripotency and early mesendodermal differentiation. TERMINATOR knockdown in zebrafish resulted in >70% lethal embryos, and developmental arrest and severe cardiovascular defects in the surviving embryos (Kurian et al., 2015). Mesodermal specification is marked by the expression of ALIEN. Loss of ALIEN resulted in mesoderm-related defects including defective vascular patterning and cardiac chamber formation, alluding toward specific role of ALIEN in the early developmental of the progenitor stage common to vascular and cardiomyocyte fates. Silencing of PUNISHER was also accompanied by severe impairments in vasculature and cardiac development and function (Kurian et al., 2015). These results were extrapolated to mouse embryos and human ESCs by knockdown of the three lncRNAs followed by microarray hybridization to check for the differential expression of different genes at different stages upon knockdown of these individual lncRNAs (Kurian et al., 2015). Albeit the mechanisms employed by these lncRNAs in controlling the developmental processes remains elusive.
lncRNA H19 besides its role in early embryonic development, is also critical for late stage cardiac differentiation. During the late stage cardiac differentiation of P19CL6 mouse cells, H19 knockdown promoted cell proliferation and inhibited apoptosis (Han et al., 2016). H19 curbs the expression of miR-19b, thereby increasing the expression of miR-19b target Sox-6. Thus, H19 presents a classic example of lncRNAs having tissue specific roles and targets and provides an explanation for how lncRNAs are capable of regulating a wide variety of cellular processes at different stages in various tissues. Table 2 consolidates the function of lncRNAs involved in cardiovascular development, homeostasis and their relevance to CVDs.
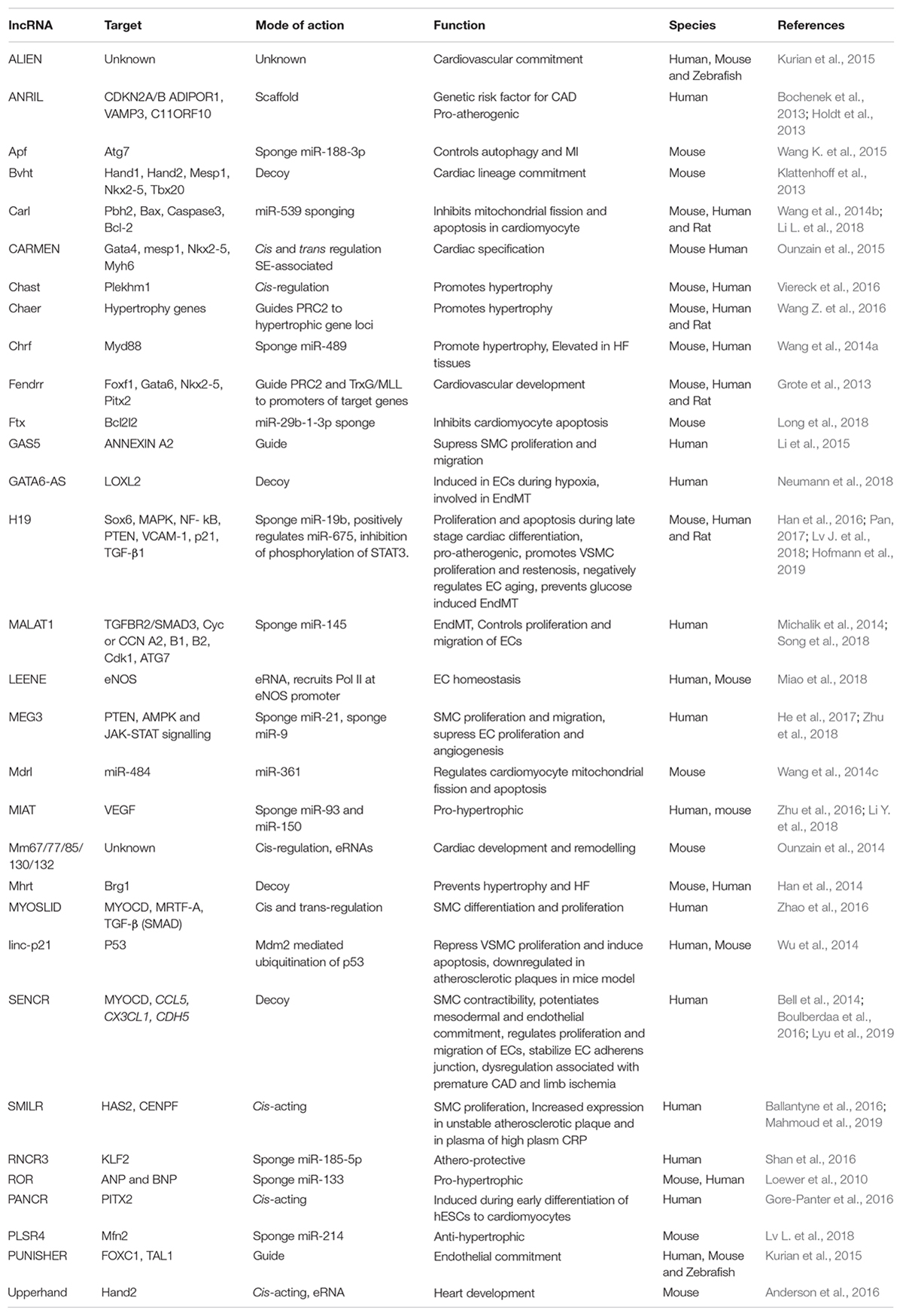
Table 2. List of lncRNAs with their regulatory mechanisms and physiological impact in cardiovascular biology.
With the expanding knowledge about the importance of lncRNAs in cell fate decisions and heart development, it is evident that they are operational in maintaining the homeostasis of cardiovascular system. High throughput RNA-sequencing has steered the identification of differentially regulated lncRNAs in cardiac pathologies. One of the common heart conditions is cardiac hypertrophy, characterized by the increase in cardiomyocyte size to compensate for inappropriate cardiac function leading to heart failure (HF). lncRNA CHAST was upregulated in murine and human cardiac hypertrophy and in vivo depletion of this lncRNA in mouse hypertrophic model prevented and reverted the condition (Viereck et al., 2016). CHAST promotes hypertrophy by blocking autophagy and its inhibition prevented HF, thus presenting a promising target for treatment. Chaer is another pro-hypertrophic lncRNA which transiently interacts with PRC2 complex and attenuates hypertrophy upon its silencing in pre-stressed heart (Wang Z. et al., 2016). Unlike CHAST and Chaer, Mhrt is a cardioprotective lncRNA regulated by the stress activated Brg1-Hdac-Parp chromatin repressive complex (Han et al., 2014). Pathological stress activates Brg1 leading to aberrant gene expression including inhibition of Mhrt. Ultimately, this leads to cardiomyopathy and hypertrophy (Hang et al., 2010; Han et al., 2014). Mhrt forms a feedback loop with chromatin remodeling factor Brg1 and its repression results in cardiomyopathy. Restoring the pre-stress levels of Mhrt prevented hypertrophy and heart failure. Human ortholog of lncRNA Mhrt is repressed in various myopathic hearts demonstrating a conserved lncRNA dependent mechanism in cardiomyopathy (Han et al., 2014). These studies not only highlight the role of lncRNAs in diseases but also emphasize their therapeutic potential.
New blood vessel formation is obviously an important aspect of heart regeneration. In the adult body, perturbed vessel formation may lead to inappropriate blood supply accompanied by shortage of oxygen to the heart resulting in diseased conditions such as myocardial infarction (MI). Endothelial cells (ECs) are important for the formation of the blood vessels and EC dysfunction is one of the early steps in the development of vascular pathologies. Many studies have identified lncRNAs regulating ECs function (Figure 4A) and lncRNAs differentially regulated during vascular diseases. One such example is MALAT1 which is elevated in TGF-β1-induced endothelial-to-mesenchymal transition (EndMT) where it regulates TGFBR2 and SMAD3 by negatively regulating miR-145 (Xiang et al., 2017). EndMT is a hallmark of various pathological conditions including CVDs, fibrosis and cancer and MALAT1 regulation of EndMT is a potent target for the development of gene therapy approaches. MALAT1 is also involved in the regulation of proliferation of ECs. MALAT1 knockdown increased basal sprouting and migration, inhibiting the proliferation. The reduced proliferation was due to the switch of ECs from a proliferative to promigratory phenotype. In addition, there was a simultaneous increase in the expression of cell cycle inhibitors, such as p21 (Michalik et al., 2014). Furthermore, Malat1 expression was elevated in hypoxic conditions and its inhibition in a hind limb ischemia mouse model showed reduced capillary density and blood flow recovery (Michalik et al., 2014). Similarly, the inhibition of lncRNA MANTIS also prevented the angiogenic sprouting and alignment of ECs subjected to sheer stress (Leisegang et al., 2017). MANTIS controls the angiogenesis by activating the ATPase activity of BRG1, facilitating the assemblage of RNA polymerase II onto key endothelial genes like SOX18, SMAD6, and COUP-TFII (Leisegang et al., 2017). Unlike MALAT1 and MANTIS, overexpression of lncRNA MEG3 suppressed EC proliferation and in vitro angiogenesis through negative regulation of miR-9 (He et al., 2017).
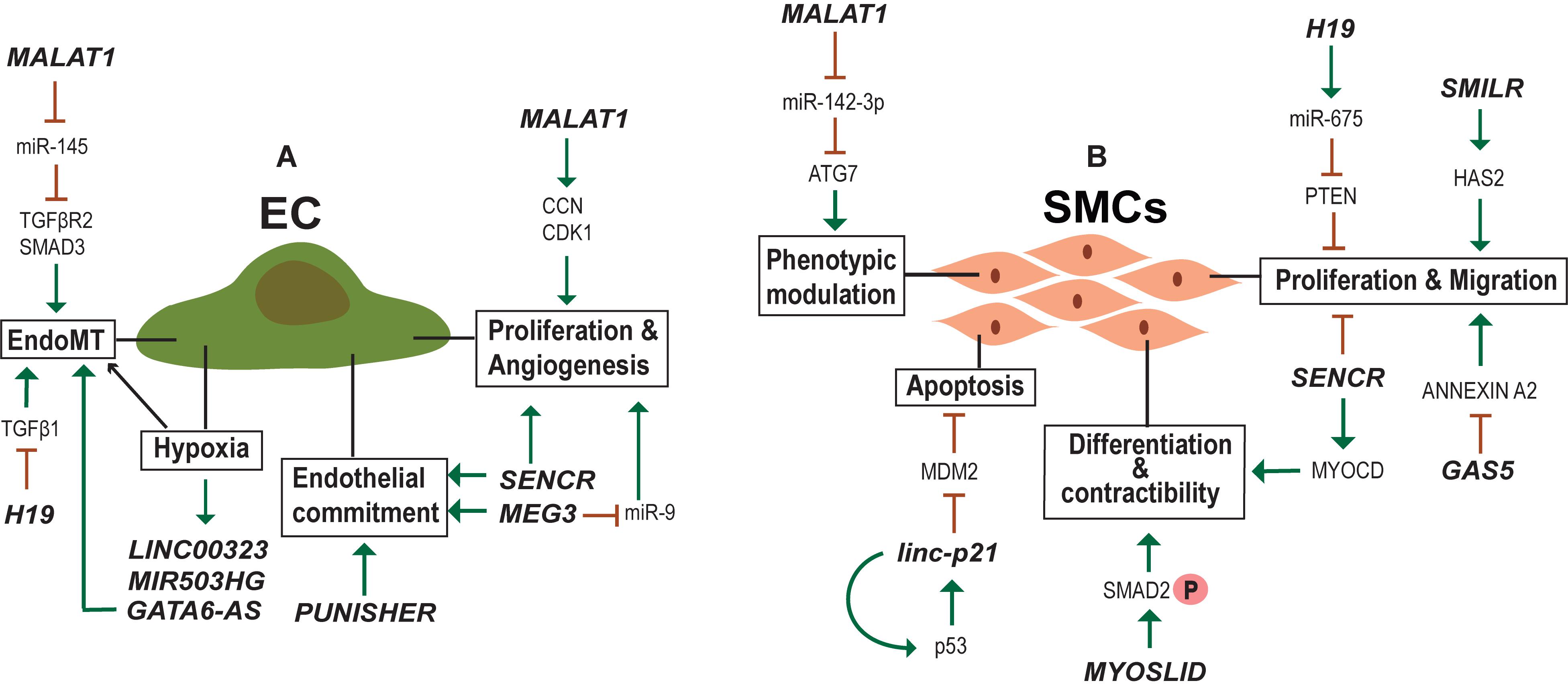
Figure 4. lncRNAs regulating endothelial (EC) and smooth muscle cell (SMC) biology. (A) Examples of lncRNA regulating Endothelial cell identity, physiology and activated in hypoxic conditions. (B) Examples of lncRNA regulating SMC proliferation, migration, contractibility and phenotypic modulation between contractile and synthetic phenotype.
Recently, Fiedler et al. (2015) generated an expression atlas of human hypoxia-sensitive lncRNAs with identification of two lncRNAs—LINC00323 and MIR503HG—important for sustaining EC function. The expression levels of growth factor signaling and endothelial TF GATA2 were alleviated upon silencing of these two lncRNAs, accompanied by impaired cell cycle control and blocked capillary formation (Fiedler et al., 2015). Another study aiming to determine hypoxia influence on lncRNA expression in HUVECs identified lncRNA GATA6-AS to be upregulated during hypoxia (Neumann et al., 2018). In vitro EndMT was reduced upon GATA6-AS silencing along with impaired vascular sprouting and endothelial cell migration (Neumann et al., 2018). Additionally, EndMT was regulated by H19 during diabetic retinopathy where overexpression of H19 prevented glucose mediated EndMT through regulation of TGF-β1 signaling in a Smad-independent manner (Thomas et al., 2019). Moreover, H19 was shown to regulate EC aging via negative regulation of age induced inflammatory activation (Hofmann et al., 2019).
lncRNA SENCR is important for endothelial cell commitment and function. SENCR overexpression substantially enhanced the mesodermal and endothelial commitment of hESC (Boulberdaa et al., 2016). In HUVECs, the upregulation of SENCR instigated migration, proliferation and formation of capillary-like structures. Concomitantly, silencing of SENCR had reducing effects on these processes. Known migratory and angiogenesis genes were downregulated upon SENCR silencing, with no effect on the expression of neighboring FLI1 gene. Vascular cells derived from patients with limb ischemia and premature coronary artery disease (CAD) showed a reduced level of SENCR as compared to control samples (Boulberdaa et al., 2016). Recently, SENCR was found to be important for maintaining the membrane integrity of ECs to control the vascular permeability (Lyu et al., 2019). Another important aspect of EC biology is Nitric Oxide synthesis (eNOS) that controls the vasodilation. RNA-seq and chromatin capture study identified eRNA LEENE enhancing the eNOS expression by recruiting RNA pol II to the eNOS promoter (Miao et al., 2018). These results suggest the importance of lncRNA in EC homeostasis and endothelial dysfunction which is one of the key triggers of vascular diseases.
In addition to ECs, VSMC development and function is important in vascular setting. In contrast to terminally differentiated skeletal muscles, VSMCs can undergo reversible phenotypic change between contractile and synthetic phenotypes (Rensen et al., 2007; Davis-Dusenbery et al., 2011). The phenotypic diversity of VSMCs provide the necessary flexibility to the blood vessels to function under different physiological (Figure 4B) and pathological conditions. In normal adult animals, VSMCs exist as highly specialized and differentiated cells with a contractile phenotype. During several vascular pathologies the differentiated VSMCs switch to a proliferative phenotype with surged synthetic activity (Iyemere et al., 2006). Recent studies have identified lncRNA MALAT1 and a novel lncRNA SMILR as important regulators of the switch from a differentiated to a proliferative phenotype of VSMCs (Ballantyne et al., 2016; Song et al., 2018; Mahmoud et al., 2019). Both were shown to promote proliferation and migration of VSMCs, but the mechanism employed and target genes are different for each. MALAT1 knockdown facilitated the conversion of SMCs from a proliferative phenotype to a differentiated one by inhibiting autophagy (Song et al., 2018). Mechanistically MALAT1 was found to compete with miR-142-3p to regulate ATG7 mediated activation of autophagy that results in the conversion of VSMCs from a contractile to a proliferative state. While MALAT1 acts in trans, SMILR influences cellular proliferation by regulating the expression of proximal gene HAS2, involved in atherosclerotic lesion formation (Ballantyne et al., 2016). More specifically, SMILR affects the late mitotic and cytokinesis phases of cell cycle by interacting with CENPF and this SMILR:CENPF interaction is in turn regulated by Staufen1 RNA binding proteins (Mahmoud et al., 2019). Alteration of mature VSMC from a contractile phenotype to an osteoblastic phenotype is another major aspect of VSMC biology, which leads to vascular calcification (Iyemere et al., 2006). The transcriptome analysis during calcification of rat VSMCs identified lncRNA Lrrc75a-as1 as a negative regulator of vascular calcification (Jeong et al., 2019).
Thirty one lncRNAs were identified from RNA-Seq in human coronary artery SMCs (HCASMC), one of which was SENCR, that helps maintain the normal SMC differentiated state (Bell et al., 2014). SENCR is transcribed antisense to the Friend Leukemia Integration virus1 (FLI) gene, whereas, SENCR knockdown had no effect on the expression of FLI1 or other neighboring genes ruling out cis-acting effects of SENCR on local gene expression. Attenuation of SENCR significantly reduced the expression of many SMC contractile genes, including MYOCD, at both mRNA and protein levels with significant increase in genes inducing motility (Bell et al., 2014). Collectively, these results confirm the regulatory importance of SENCR in human coronary artery SMC (HCASMC) differentiation and migration. A study searching for lncRNAs regulated by myocardin (MYOCD/SRF), the master switch for VSMC differentiation, identified lncRNA MYOSLID (MYOcardin-induced Smooth muscle Long non-coding RNA, Inducer of Differentiation) as a direct target for MYOCD/SRF and TGFβ/SMAD pathways (Zhao et al., 2016). MYOSLID promoted VSMC differentiation and inhibited VSMC proliferation. lncRNAs regulating SMC phenotypic modulation, proliferation and migration might be used as molecular targets in therapies for diseases aggravated by vascular remodeling.
Excessive proliferation of VSMCs is a major attribute of restenosis. In vitro overexpression of H19 accelerated VSMC proliferation by positively regulating miR-675, which in turn downregulates PTEN expression (Lv J. et al., 2018). H19 and miR-675 were upregulated in injured arterial walls in a rat balloon injury model (Lv J. et al., 2018). lnc-GAS5 is another lncRNA implicated in proliferation and migration of SMCs. Overexpression of lnc-GAS5 inhibited the proliferation, migration and reduced apoptosis of human saphenous vein SMCs (HSVSMCs) and conversely, its silencing promoted these cellular behaviors (Li et al., 2015). lnc-GAS5 function is mediated through a Ca2+-dependent RNA binding protein, Annexin A2. Thus, low expression of lnc-GAS5 increases proliferation and migration of HSVSMCs through AnnexinA2 facilitating in the pathogenesis of the Great Saphenous Veins. linc-p21 is also shown to regulate SMC proliferation but its expression is downregulated during atherosclerosis (Wu et al., 2014). linc-p21 silencing in a carotid artery injury mouse model resulted in neonatal hyperplasia due to dysregulation of multiple P53 targets (Wu et al., 2014). The angiotensin II regulated lnc-Ang362 is a host for miR-221 and miR-222, two known miRNAs implicated in SMC proliferation (Leung et al., 2013). lnc-Ang362, miR-221 and miR-222 were increased in the lung tissues derived from pulmonary arterial hypertension (PAH) patients (Wang et al., 2019). Thus, lnc-Ang362 could be a target for treating PAH.
lncRNA with prognostic and diagnostic potential are of particular interest in the clinical arena. A Genome wide association study (GWAS) has identified chromosome 9p21 locus exhibiting the highest association with atherosclerosis (Holdt et al., 2013). This risk locus encodes for lncRNA ANRIL, associated with atherosclerotic severity. Further investigations revealed that ANRIL functions in trans, leading to pro-atherogenic effects such as cell proliferation, increased cell adhesion and decreased apoptosis (Holdt et al., 2013). The association of increased ANRIL level with LV dysfunction further highlights its diagnostic and prognostic importance. Similarly, another GWAS study identified that the single nucleotide polymorphism (SNP) associated with MI altered the expression of lncRNA MIAT (Ishii et al., 2006).
Circulating lncRNAs can serve as biomarkers of the diseases making the diagnosis much easier. A transcriptome study identified lncRNA LIPCAR in the plasma of patients with heart failure as a novel biomarker which can predict heart remodeling and future death in the patients (Kumarswamy et al., 2014). Another study performed the microarray analysis in the plasma of CAD patients and reported that lncRNA CoroMarker as a sensitive biomarker for CAD (Yang et al., 2015). Likewise, circulating ZFAS1 and CDR1AS were found to be decreased and increased in acute myocardial infarction (AMI), respectively (Zhang et al., 2016). Another study reported low levels of lncRNA GAS5 in the plasma of 30 CAD patients as compared to 30 healthy individuals (Yin et al., 2017).
Promising results for several kinds of lncRNAs were reported similarly (Schlosser et al., 2016). However, the authors observed difficulties in reproducibility of detection. This could be due to the low sensitivity of the conventional lncRNA detection methods. Regardless of the technical challenges the low abundancy of lncRNAs cannot undermine their reported functional importance in many instances highlighted in this review. A summary of lncRNAs as potential biomarkers and their roles in cardiac diseases is represented in Figure 5.
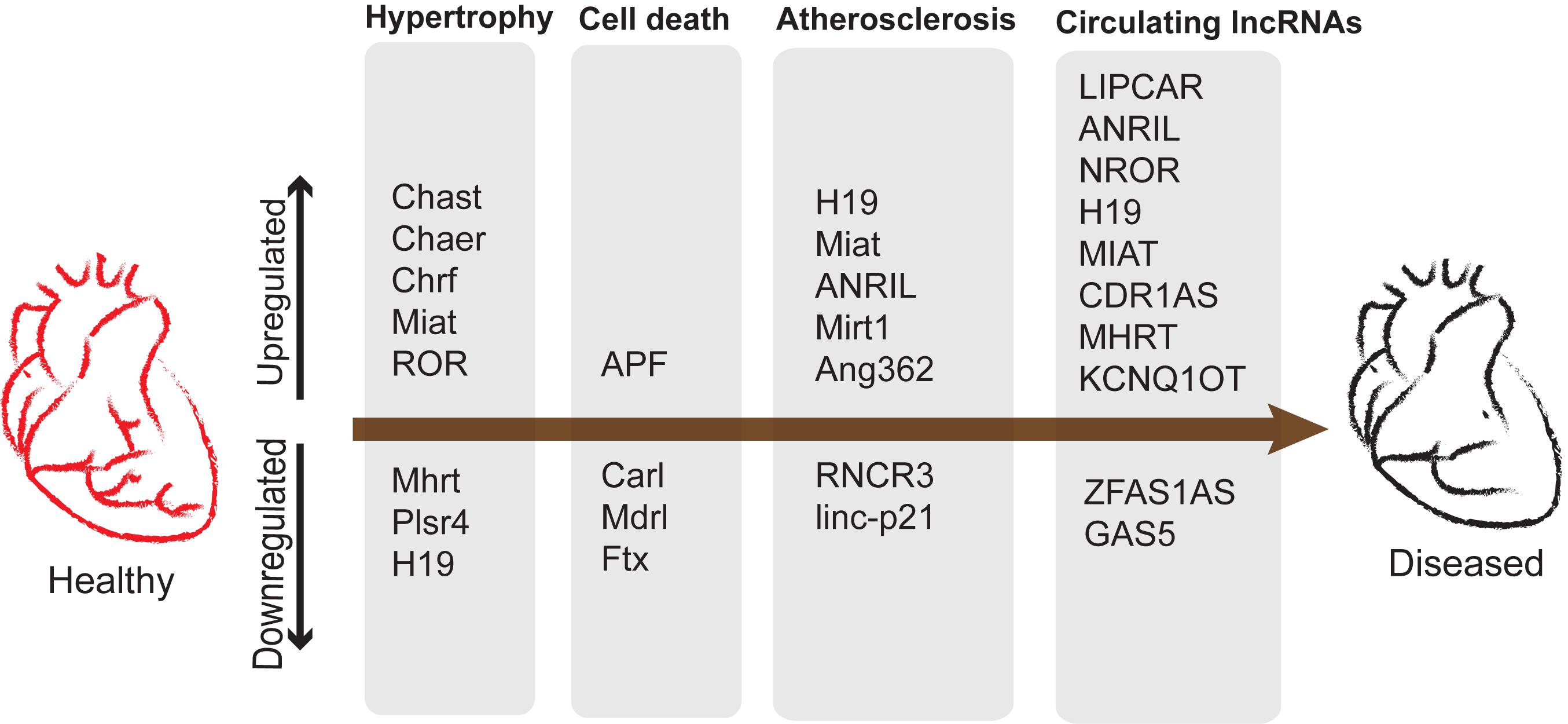
Figure 5. lncRNAs upregulated and downregulated during development of cardiac diseases and potential biomarkers. Cardiac remodeling is characterized by aberrant myocardium growth along with apoptosis and vascular remodeling leading to atherosclerosis.
The crosstalk between lncRNA–miRNA–mRNA appears to be common in different facets discussed so forth. lncRNAs inhibit the function of miRNA by binding to them in a sequence specific manner, thereby increasing the number of target mRNAs that would otherwise be suppressed by miRNAs. In this regard, comprehensive knowledge of complex lncRNA–miRNA–mRNA-networks would help develop novel RNA-based therapeutics for different diseases (Figure 6).
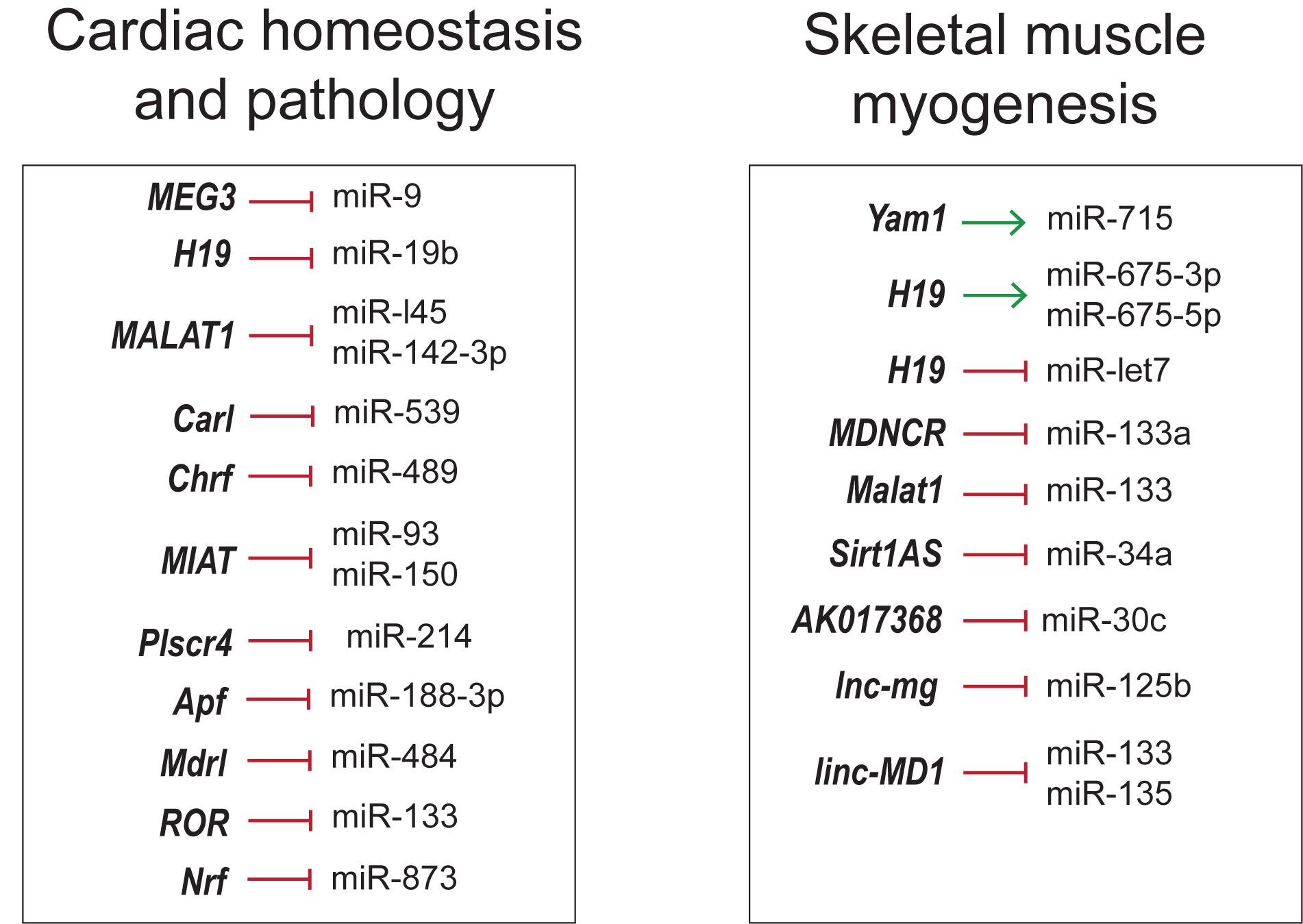
Figure 6. Overview of lncRNA–miRNA interactions in skeletal muscle and cardiovascular development and disease.
As seen in skeletal muscle proliferation and differentiation, lncRNA H19, linc-MD1, lnc-mg and Malat1 inhibit the action of miRNAs by the sponging mechanism (Cesana et al., 2011; Kallen et al., 2013; Han et al., 2015; Chen et al., 2017; Zhu et al., 2017). However, lncRNAs can regulate the function of miRNAs in many different ways. For instance, lncRNA Sirt1As shields the miR-34a binding site on Sirt1 mRNA by binding to the transcripts (Wang G.-Q. et al., 2016). lncRNAs can positively regulate miRNAs by acting as precursor miRNAs as in the case of H19 giving rise to miR-675-3p and miR-675-5p (Dey et al., 2014).
These RNA networks are not restricted to skeletal muscle but are also noticeable in cardiovascular physiology and disease. CHRF, Plscr4, MIAT and ROR were involved in miRNA mediated cardiac hypertrophy regulation. lncRNA CHRF triggers cardiomyocyte hypertrophy by sponging antihypertrophic miR-489, which targets Myd88 (Wang et al., 2014a). Similarly, pro-hypertrophic lncRNA MIAT regulates anti-hypertrophic miR-93 and miR-150 (Zhu et al., 2016; Li Y. et al., 2018). ROR is another hypertrophic inducer that sponges miR-133 (Jiang et al., 2016). In contrast, Plscr4 is an anti-hypertrophic lncRNA downregulating miR-214 to promote the expression of Mfn2 (Lv L. et al., 2018). Apoptosis and autophagy are among other pathways targeted by competing-endogenous RNAs (ceRNAs). lncRNA CARL has been proposed to inhibit cardiomyocyte apoptosis by sequestering miR-539, a microRNA targeting PHB2 (Wang et al., 2014b), a subunit of mitochondrial membrane protein Prohibitin involved in mitochondrial homeostasis (Tatsuta et al., 2005). Cardiomyocyte apoptosis and mitochondrial fission was also inhibited by Mdrl via downregulation of miR-361, counteracting the inhibition of miR-484 processing (Wang et al., 2014c). lncRNA Ftx inhibited cardiomyocyte apoptosis by preventing miR-29b-1-5p mediated downregulation of Bcl2 (Long et al., 2018). Likewise, Apf triggers autophagy by targeting Atg7 through negative regulation of miR-188-3p, an inhibitor of autophagy and myocardial infarction (Wang K. et al., 2015). It is interesting to note that some of the above mentioned networks are well conserved across species, suggesting the crucial roles, they have in cardiovascular biology. Hence, modulating lncRNA-miRNA-mRNA pathways could possibly be a novel strategy to suppress cardiomyocyte loss in order to protect against myocardial infarction (MI) and tailor therapies for hypertrophy. Thorough understanding of these networks in development and disease could help design better therapeutic strategies in the field of regenerative medicine.
There has been an increased interest in the understanding of lncRNA regulation in disease systems due to their relatively restricted expression patterns and different possible actions on cellular function(s). Given that several of these ncRNAs are dysregulated in disease conditions, modulating the levels of such lncRNAs appears to be a promising approach for therapeutics and preclinical testing. However, only a few lncRNAs have been studied in depth in relevant ex vivo and in vivo systems. Upregulation or downregulation/inhibition of lncRNA function has been the most rigorously adopted methods to understand their therapeutic potential in muscular defects and cardiovascular damage. The most commonly used gene delivery methods for RNA based therapeutics are recombinant viral systems such as adenovirus, lentivirus and adeno-associated viruses (AAVs), which are employed either to inhibit or to overexpress the mRNA, miRNA, lncRNA whole transcripts (Eulalio et al., 2012; Gomes et al., 2017; Gabisonia et al., 2019). The use of RNAi and antisense oligonucleotides (ASOs) present the most commonly used approaches for selective downregulation of potential lncRNAs. On the contrary, the upregulation of lncRNAs can be achieved by viral vectors that are very efficient for muscle delivery. Using the above mentioned strategies, there have been promising results toward miRNA targeting and protein coding transcripts in preclinical systems, which reached clinical trials. However, using the same delivery methods the therapeutic relevance of lncRNAs remain to be resolved.
The siRNA mediated pharmacological inhibition of cytoplasmic lncRNA SMILR in the ex vivo vein graft model significantly reduced the SMC proliferation to ameliorate the effects of vascular remodeling (Mahmoud et al., 2019). In another approach using nanoparticle mediated transfection of siRNA against lncRNA Chaer directly into mouse heart decreased the cardiac hypertrophy and fibrosis and restored cardiac function (Wang Z. et al., 2016). The inhibition level achieved by shRNAs and siRNAs are, in general, limited to the cytoplasmic lncRNA molecules. Hence, on the other hand, ASOs or GapmeRs are more suitable for nuclear lncRNAs that direct Dicer independent degradation of the target lncRNA by RNase H activity (Fluiter et al., 2009; Bennett and Swayze, 2010; Lennox and Behlke, 2016). The ASO technology has fewer off target effects than the small RNA mediated approach. One such example is GapmeR mediated inhibition of lncRNA Chast for the prevention and regression of cardiac hypertrophy in vivo (Viereck et al., 2016). No noticeable side effects of GapmeR treatment were reported. In another study, GapmeR mediated knockdown of Malat1 in mice muscle resulted in poor blood flow recovery and diminished capillary density (Michalik et al., 2014). In addition, in vivo therapeutic intervention using GapmeRs targeting lncRNA Wisper suppressed cardiac fibrosis and improved function (Micheletti et al., 2017). So far, none of the antisense based lncRNA drugs have been tested in clinical studies but, this strategy has been proven to be effective and safe in a clinical trial targeting liver miR-122 with LNA-based antimiRs (mirvarsen) (Zumla et al., 2013). The newer generation of ASOs provide spatial control for target delivery and with further improvements, this could be translated for lncRNA inhibition in future clinical trials.
In comparison to inhibition, overexpression of cardioprotective lncRNAs appear to be much more challenging attributing to their length and locus complexity, and sometimes incomplete knowledge of the complete sequence identity and isoform structure in the current genome annotation. In this context, the major challenge is efficient transportation of these large transcripts across the membrane barriers and determining the potential toxicity. The transgenic gene activation of the cardioprotective lncRNA Mhrt demonstrates the first overexpression study highlighting the translational importance of delivery of lncRNA as a drug (Han et al., 2014). In another example, Chast overexpression using AAV9 (adeno-associated-virus serotype 9) viral particles promoted cardiac hypertrophy in mouse heart, suggesting it to be a crucial target for cardiac hypertrophy (Viereck et al., 2016).
Advancements in RNA-sequencing technologies have led to the identification of several lncRNAs in different diseases suggesting them to be crucial targets for therapeutics. In addition, due to their distinct expression patterns they can be powerful tools for diagnostics, personalized medicine and drug development. A brief description of lncRNA-based diagnostic and therapeutic strategies is depicted in Figure 7. There are several key challenges in any lncRNA-focused therapy. Firstly, the lncRNAs due to their pleiotropic actions regulate multiple targets and hence may not be target-specific and can act via more than one mechanism in a diseased state. In addition, low conservation of lncRNAs across species, is another challenge, for example a human specific lncRNA lacking conservation in mouse, makes its preclinical testing inappropriate (Lu and Thum, 2019). A further challenge in lncRNA therapeutics is the fact that lncRNAs are often integrated in complexes which make them inaccessible for access. Small molecules such as aptamers may help the lncRNA binding with the interacting protein complexes or induce conformational changes in the secondary or tertiary structures of lncRNA (Lunse et al., 2010). This is mainly important, where the lncRNA expression does not attribute to the disease phenotype but, the lncRNA interaction with other molecules accounts for the disease. Hence, the secondary and tertiary structure of lncRNAs and their structure-function relationship should to be resolved before lncRNAs enter into therapeutics.
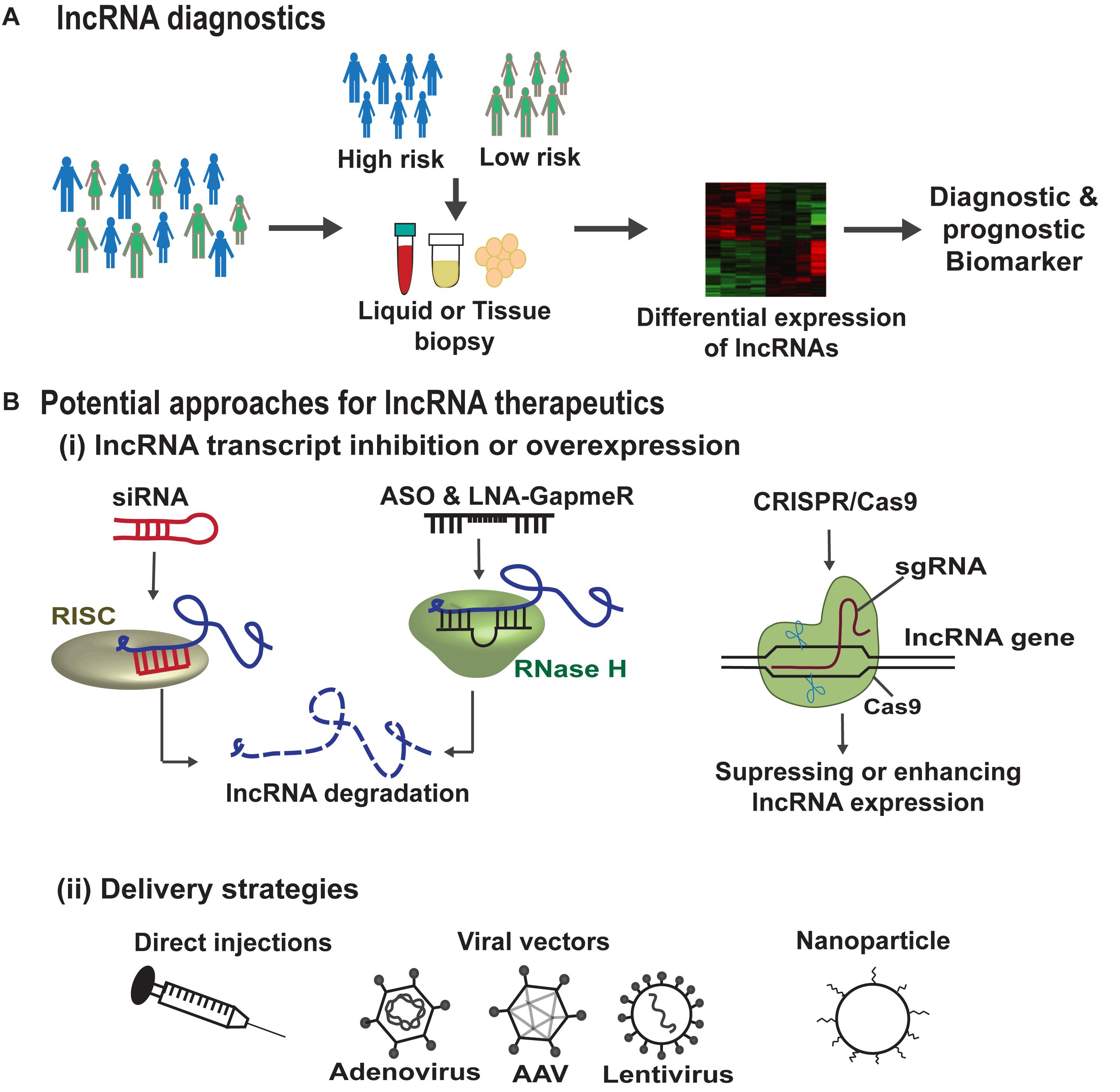
Figure 7. lncRNA based diagnostics and therapeutic strategies. (A) Genome wide association studies (GWAS) help in the identification of lncRNA single nucleotide polymorphisms (SNPs) associated with disease susceptibility. These SNPs can alter the lncRNA expression in the body fluids or tissues which can serve as diagnostic or prognostic markers. In addition, improvements in RNA-sequencing technologies can lead to the identification of lncRNAs as biomarkers/disease targets in different diseases. (B) (i) Summarizes the potential approaches for lncRNA therapeutics, such as siRNAs that are associated with RNA-induced silencing complex (RISC) bind and cleave the target lncRNA. ASOs and LNA-GapmeRs bind to the target lncRNA in sequence specific manner leading to RNase H mediated lncRNA degradation. CRISPR/Cas9 gene editing tool can make deletion or insertion in the DNA sequence of the target lncRNA to either enhance or abrogate its expression. (B) (ii) Demonstrates the different lncRNA delivery strategies such as direct injections, viral particles, or via encapsulation in nanoparticles.
The other translational limitations of lncRNA therapeutics is the lack of efficient and safe delivery systems for a controlled and targeted release. The delivery vehicle should ensure high transfection efficiency with minimal cytotoxicity but also controlled release of the lncRNA based drug during the complete process. lncRNA levels can be manipulated using viral vectors, however, the use of viral delivery methods is restricted due to the associated issues including off-target effects, activation of host immune response and the risk of insertional mutagenesis, although clinical evidence in gene therapy to date demonstrates a favorable situation. To overcome these risks, researchers believe non-viral vectors with chemical modifications and/or nanoparticles targeted to specific cell type would be advantageous in clinical trials, however, are relatively inefficient dampening down the possibility for clear efficacy signals. Finally, the use of gene editing technique such as CRISPR/Cas9 has provided both loss- or gain-of-function of lncRNA in in vitro and in vivo studies (Aparicio-Prat et al., 2015; Leisegang et al., 2017; Ballarino et al., 2018). Although, similar delivery issues need to be addressed with the use of gene editing tools in vivo too.
Several lncRNAs have been identified to be involved in development and pathophysiology by regulating gene expression at DNA, RNA and protein levels. Their specific expression during differentiation and disease helps qualify them as key regulators and potential therapeutic targets. However, at present the use of lncRNAs in therapeutics is in its inception. There are still certain technical issues that need to be addressed, such as the difficulties in targeting, accurate outcome predictions and discrepancies in knockdown phenotypes in in vitro and in vivo. Further improvement in detection, silencing approaches, mechanistic understanding and development of in vivo models would uncover the full potential of lncRNAs as diagnostic and possibly therapeutic targets.
SwS collected and analyzed the data, prepared the figures, and wrote the manuscript. TD and SmS corrected and gave suggestions to improve the manuscript. RB conceived, planned, analyzed, wrote, coordinated, and finalized the manuscript. AB wrote, reviewed, coordinated, and finalized the manuscript.
This work was supported by (1) Yenepoya (Deemed to be University) seed grant (YU/Seed grant/071-2018) to RB. (2) British Heart Foundation (BHF) Chair of Translational Cardiovascular Sciences grant (CH/11/2/28733), BIRAX and ERC Advanced grant VASCMIR to AB. (3) Department of Science & Technology, Science and Engineering Research Board grant (ECR/2017/001216) to SmS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Our sincere apologies to colleagues, whose work was not discussed or cited due to space constraints.
Anderson, D. M., Anderson, K. M., Chang, C. L., Makarewich, C. A., Nelson, B. R., McAnally, J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606. doi: 10.1016/j.cell.2015.01.009
Anderson, K. M., Anderson, D. M., McAnally, J. R., Shelton, J. M., Bassel-Duby, R., and Olson, E. N. (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. doi: 10.1038/nature20128
Aparicio-Prat, E., Arnan, C., Sala, I., Bosch, N., Guigo, R., and Johnson, R. (2015). DECKO: single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics 16:846. doi: 10.1186/s12864-015-2086-z
Ballantyne, M. D., Pinel, K., Dakin, R., Vesey, A. T., Diver, L., Mackenzie, R., et al. (2016). Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation 133, 2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019
Ballarino, M., Cazzella, V., D’Andrea, D., Grassi, L., Bisceglie, L., Cipriano, A., et al. (2015). Novel long noncoding RNAs (lncRNAs) in myogenesis: amiR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 35, 728–736. doi: 10.1128/MCB.01394-14
Ballarino, M., Cipriano, A., Tita, R., Santini, T., Desideri, F., Morlando, M., et al. (2018). Deficiency in the nuclear long noncoding RNACharme causes myogenic defects and heart remodeling in mice. EMBO J. 37:e99697. doi: 10.15252/embj.201899697
Bell, R. D., Long, X., Lin, M., Bergmann, J. H., Nanda, V., Cowan, S. L., et al. (2014). Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 34, 1249–1259. doi: 10.1161/ATVBAHA.114.303240
Bennett, C. F., and Swayze, E. E. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293. doi: 10.1146/annurev.pharmtox.010909.105654
Bochenek, G., Hasler, R., El Mokhtari, N. E., Konig, I. R., Loos, B. G., Jepsen, S., et al. (2013). The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 22, 4516–4527. doi: 10.1093/hmg/ddt299
Borensztein, M., Monnier, P., Court, F., Louault, Y., Ripoche, M. A., Tiret, L., et al. (2013). Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development 140, 1231–1239. doi: 10.1242/dev.084665
Boulberdaa, M., Scott, E., Ballantyne, M., Garcia, R., Descamps, B., Angelini, G. D., et al. (2016). A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol. Ther. 24, 978–990. doi: 10.1038/mt.2016.41
Bovolenta, M., Erriquez, D., Valli, E., Brioschi, S., Scotton, C., Neri, M., et al. (2012). The DMD locus harbours multiple long non-coding RNAs which orchestrate and control transcription of muscle dystrophin mRNA isoforms. PLoS One 7:e45328. doi: 10.1371/journal.pone.0045328
Brannan, C. I., Dees, E. C., Ingram, R. S., and Tilghman, S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36. doi: 10.1128/mcb.10.1.28
Braun, T., and Gautel, M. (2011). Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12, 349–361. doi: 10.1038/nrm3118
Bruneau, B. G. (2013). Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 5:a008292. doi: 10.1101/cshperspect.a008292
Bunch, H., Lawney, B. P., Burkholder, A., Ma, D., Zheng, X., Motola, S., et al. (2016). RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics 108, 64–77. doi: 10.1016/j.ygeno.2016.07.003
Cabianca, D. S., Casa, V., Bodega, B., Xynos, A., Ginelli, E., Tanaka, Y., et al. (2012). A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149, 819–831. doi: 10.1016/j.cell.2012.03.035
Caretti, G., Di Padova, M., Micales, B., Lyons, G. E., and Sartorelli, V. (2004). The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18, 2627–2638. doi: 10.1101/gad.1241904
Caretti, G., Schiltz, R. L., Dilworth, F. J., Padova, M. D., Zhao, P., Ogryzko, V., et al. (2006). The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 11, 547–560. doi: 10.1016/j.devcel.2006.08.003
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chal, J., and Pourquié, O. (2017). Making muscle: skeletal myogenesisin vivo andin vitro. Development 144, 2104–2122. doi: 10.1242/dev.151035
Chen, X., He, L., Zhao, Y., Li, Y., Zhang, S., Sun, K., et al. (2017). Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 3:17002. doi: 10.1038/celldisc.2017.2
Davis-Dusenbery, B. N., Wu, C., and Hata, A. (2011). Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation. Arterioscler. Thromb. Vasc. Biol. 31, 2370–2377. doi: 10.1161/ATVBAHA.111.226670
Deng, C., Li, Y., Zhou, L., Cho, J., Patel, B., Terada, N., et al. (2016). HoxBlinc RNA recruits Set1/MLL complexes to activate hox gene expression patterns and mesoderm lineage development. Cell Rep. 14, 103–114. doi: 10.1016/j.celrep.2015.12.007
Dey, B. K., Pfeifer, K., and Dutta, A. (2014). The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 28, 491–501. doi: 10.1101/gad.234419.113
Doss, M. X., Gaspar, J. A., Winkler, J., Hescheler, J., Schulz, H., and Sachinidis, A. (2012). Specific gene signatures and pathways in mesodermal cells and their derivatives derived from embryonic stem cells. Stem Cell Rev. 8, 43–54. doi: 10.1007/s12015-011-9263-5
Eulalio, A., Mano, M., Dal Ferro, M., Zentilin, L., Sinagra, G., Zacchigna, S., et al. (2012). Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381. doi: 10.1038/nature11739
Evseenko, D., Zhu, Y., Schenke-Layland, K., Kuo, J., Latour, B., Ge, S., et al. (2010). Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 13742–13747. doi: 10.1073/pnas.1002077107
Fiedler, J., Breckwoldt, K., Remmele, C. W., Hartmann, D., Dittrich, M., Pfanne, A., et al. (2015). Development of long noncoding RNA-Based strategies to modulate tissue vascularization. J. Am. Coll. Cardiol. 66, 2005–2015. doi: 10.1016/j.jacc.2015.07.081
Fluiter, K., Mook, O. R., Vreijling, J., Langkjaer, N., Hojland, T., Wengel, J., et al. (2009). Filling the gap in LNA antisense oligo gapmers: the effects of unlocked nucleic acid (UNA) and 4′-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. Biosyst. 5, 838–843. doi: 10.1039/b903922h
Frank, S., Ahuja, G., Bartsch, D., Russ, N., Yao, W., Kuo, J. C., et al. (2019). yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24, 318–327.e8. doi: 10.1016/j.stem.2018.11.005
Gabisonia, K., Prosdocimo, G., Aquaro, G. D., Carlucci, L., Zentilin, L., Secco, I., et al. (2019). MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 569, 418–422. doi: 10.1038/s41586-019-1191-6
Gomes, C. P. C., Spencer, H., Ford, K. L., Michel, L. Y. M., Baker, A. H., Emanueli, C., et al. (2017). The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol. Ther. Nucleic Acids 8, 494–507. doi: 10.1016/j.omtn.2017.07.014
Gong, C., Li, Z., Ramanujan, K., Clay, I., Zhang, Y., Lemire-Brachat, S., et al. (2015). A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-Mediated mRNA translation. Dev. Cell 34, 181–191. doi: 10.1016/j.devcel.2015.05.009
Gore-Panter, S. R., Hsu, J., Barnard, J., Moravec, C. S., Van Wagoner, D. R., Chung, M. K., et al. (2016). PANCR, the PITX2 adjacent noncoding RNA, is expressed in human left atria and regulates PITX2c expression. Circ. Arrhythm. Electrophysiol. 9:e003197. doi: 10.1161/CIRCEP.115.003197
Grote, P., Wittler, L., Hendrix, D., Koch, F., Wahrisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi: 10.1016/j.devcel.2012.12.012
Guo, Y., Wang, J., Zhu, M., Zeng, R., Xu, Z., Li, G., et al. (2017). Identification of MyoDResponsive transcripts reveals a novel long non-coding RNA (lncRNA-AK143003) that negatively regulates myoblast differentiation. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-03071-7
Han, P., Li, W., Lin, C. H., Yang, J., Shang, C., Nuernberg, S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. doi: 10.1038/nature13596
Han, X., Yang, F., Cao, H., and Liang, Z. (2015). Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 29, 3054–3064. doi: 10.1096/fj.14-259952
Han, X., Zhang, J., Liu, Y., Fan, X., Ai, S., Luo, Y., et al. (2019). The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 146:dev176198. doi: 10.1242/dev.176198
Han, Y., Xu, H., Cheng, J., Zhang, Y., Gao, C., Fan, T., et al. (2016). Downregulation of long non-coding RNA H19 promotes P19CL6 cells proliferation and inhibits apoptosis during late-stage cardiac differentiation via miR-19b-modulated Sox6. Cell Biosci. 6:58. doi: 10.1186/s13578-016-0123-5
Hang, C. T., Yang, J., Han, P., Cheng, H. L., Shang, C., Ashley, E., et al. (2010). Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62–67. doi: 10.1038/nature09130
He, C., Yang, W., Yang, J., Ding, J., Li, S., Wu, H., et al. (2017). Long noncoding RNA MEG3 negatively regulates proliferation and angiogenesis in vascular endothelial cells. DNA Cell Biol. 36, 475–481. doi: 10.1089/dna.2017.3682
Hitachi, K., Nakatani, M., Takasaki, A., Ouchi, Y., Uezumi, A., Ageta, H., et al. (2019). Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 20:e47468. doi: 10.15252/embr.201847468
Hoffman, E. P., Brown, R. H. Jr., and Kunkel, L. M. (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928. doi: 10.1016/0092-8674(87)90579-4
Hofmann, P., Sommer, J., Theodorou, K., Kirchhof, L., Fischer, A., Li, Y., et al. (2019). Long non-coding RNA H19 regulates endothelial cell aging via inhibition of STAT3 signalling. Cardiovasc. Res. 115, 230–242. doi: 10.1093/cvr/cvy206
Holdt, L. M., Hoffmann, S., Sass, K., Langenberger, D., Scholz, M., Krohn, K., et al. (2013). Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 9:e1003588. doi: 10.1371/journal.pgen.1003588
Ishii, N., Ozaki, K., Sato, H., Mizuno, H., Saito, S., Takahashi, A., et al. (2006). Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 51, 1087–1099. doi: 10.1007/s10038-006-0070-9
Iwafuchi-Doi, M., and Zaret, K. S. (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. doi: 10.1242/dev.133900
Iyemere, V. P., Proudfoot, D., Weissberg, P. L., and Shanahan, C. M. (2006). Vascular smooth muscle cell phenotypic plasticity and the regulation of vascular calcification. J. Intern. Med. 260, 192–210. doi: 10.1111/j.1365-2796.2006.01692.x
Jeong, G., Kwon, D. H., Shin, S., Choe, N., Ryu, J., Lim, Y. H., et al. (2019). Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcification. Sci. Rep. 9:5848. doi: 10.1038/s41598-019-42283-x
Jiang, F., Zhou, X., and Huang, J. (2016). Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS One 11:e0152767. doi: 10.1371/journal.pone.0152767
Jin, J. J., Lv, W., Xia, P., Xu, Z. Y., Zheng, A. D., Wang, X. J., et al. (2018). Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U.S.A. 115, E9802–E9811. doi: 10.1073/pnas.1801471115
Kalisz, M., Winzi, M., Bisgaard, H. C., and Serup, P. (2012). EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression. Dev. Biol. 362, 94–103. doi: 10.1016/j.ydbio.2011.11.017
Kallen, A. N., Zhou, X. B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The imprinted H19 LncRNA antagonizes Let-7 MicroRNAs. Mol. Cell. 52, 101–112. doi: 10.1016/j.molcel.2013.08.027
Kim, E.-D., and Sung, S. (2012). Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 17, 16–21. doi: 10.1016/j.tplants.2011.10.008
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Kumarswamy, R., Bauters, C., Volkmann, I., Maury, F., Fetisch, J., Holzmann, A., et al. (2014). Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575. doi: 10.1161/CIRCRESAHA.114.303915
Kurian, L., Aguirre, A., Sancho-Martinez, I., Benner, C., Hishida, T., Nguyen, T. B., et al. (2015). Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131, 1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303
Legnini, I., Morlando, M., Mangiavacchi, A., Fatica, A., and Bozzoni, I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 53, 506–514. doi: 10.1016/j.molcel.2013.12.012
Leisegang, M. S., Fork, C., Josipovic, I., Richter, F. M., Preussner, J., Hu, J., et al. (2017). Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 136, 65–79. doi: 10.1161/CIRCULATIONAHA.116.026991
Lennox, K. A., and Behlke, M. A. (2016). Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877. doi: 10.1093/nar/gkv1206
Leung, A., Trac, C., Jin, W., Lanting, L., Akbany, A., Saetrom, P., et al. (2013). Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ. Res. 113, 266–278. doi: 10.1161/CIRCRESAHA.112.300849
Li, H., Yang, J., Jiang, R., Wei, X., Song, C., Huang, Y., et al. (2018). Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol. Ther. Nucleic Acids 12, 610–625. doi: 10.1016/j.omtn.2018.07.003
Li, L., Wang, J. J., and Zhang, H. S. (2018). LncRNA-CARl in a rat model of myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 22, 4332–4340. doi: 10.26355/eurrev_201807_15430
Li, Y., Wang, J., Sun, L., and Zhu, S. (2018). LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur. J. Pharmacol. 818, 508–517. doi: 10.1016/j.ejphar.2017.11.031
Li, L., Li, X., The, E., Wang, L. J., Yuan, T. Y., Wang, S. Y., et al. (2015). Low expression of lncRNA-GAS5 is implicated in human primary varicose great saphenous veins. PLoS One 10:e0120550. doi: 10.1371/journal.pone.0120550
Liang, T., Zhou, B., Shi, L., Wang, H., Chu, Q., Xu, F., et al. (2018). lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 32, 377–389. doi: 10.1096/fj.201700560RR
Loewer, S., Cabili, M. N., Guttman, M., Loh, Y. H., Thomas, K., Park, I. H., et al. (2010). Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117. doi: 10.1038/ng.710
Long, B., Li, N., Xu, X. X., Li, X. X., Xu, X. J., Guo, D., et al. (2018). Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 495, 312–318. doi: 10.1016/j.bbrc.2017.11.030
Lu, D., and Thum, T. (2019). RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. doi: 10.1038/s41569-019-0218-x
Lu, L., Sun, K., Chen, X., Zhao, Y., Wang, L., Zhou, L., et al. (2013). Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 32, 2575–2588. doi: 10.1038/emboj.2013.182
Lunse, C. E., Michlewski, G., Hopp, C. S., Rentmeister, A., Caceres, J. F., Famulok, M., et al. (2010). An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew. Chem. Int. Ed. Engl. 49, 4674–4677. doi: 10.1002/anie.200906919
Luo, S., Lu, J. Y., Liu, L., Yin, Y., Chen, C., Han, X., et al. (2016). Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652. doi: 10.1016/j.stem.2016.01.024
Lv, J., Wang, L., Zhang, J., Lin, R., Wang, L., Sun, W., et al. (2018). Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem. Biophys. Res. Commun. 497, 1154–1161. doi: 10.1016/j.bbrc.2017.01.011
Lv, L., Li, T., Li, X., Xu, C., Liu, Q., Jiang, H., et al. (2018). The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol. Ther. Nucleic Acids 10, 387–397. doi: 10.1016/j.omtn.2017.12.018
Lyu, Q., Xu, S., Lyu, Y., Choi, M., Christie, C. K., Slivano, O. J., et al. (2019). SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc. Natl. Acad. Sci. U.S.A. 116, 546–555. doi: 10.1073/pnas.1810729116
Mahmoud, A. D., Ballantyne, M. D., Miscianinov, V., Pinel, K., Hung, J., Scanlon, J. P., et al. (2019). The human- and smooth muscle cell-enriched lncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circ Res. 125, 535–551. doi: 10.1161/CIRCRESAHA.119.314876
Majoros, W. H., and Ohler, U. (2007). Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics 8:152. doi: 10.1186/1471-2164-8-152
Materna, S. C., and Davidson, E. H. (2007). Logic of gene regulatory networks. Curr. Opin. Biotechnol. 18, 351–354.
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Miao, Y., Ajami, N. E., Huang, T. S., Lin, F. M., Lou, C. H., Wang, Y. T., et al. (2018). Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat. Commun. 9:292. doi: 10.1038/s41467-017-02113-y
Michalik, K. M., You, X., Manavski, Y., Doddaballapur, A., Zornig, M., Braun, T., et al. (2014). Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 114, 1389–1397. doi: 10.1161/CIRCRESAHA.114.303265
Micheletti, R., Plaisance, I., Abraham, B. J., Sarre, A., Ting, C. C., Alexanian, M., et al. (2017). The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 9:eaai9118. doi: 10.1126/scitranslmed.aai9118
Morley, J. E., Thomas, D. R., and Wilson, M. M. (2006). Cachexia: pathophysiology and clinical relevance. Am. J. Clin. Nutr. 83, 735–743. doi: 10.1093/ajcn/83.4.735
Morriss, G. R., and Cooper, T. A. (2017). Protein sequestration as a normal function of long noncoding RNAs and a pathogenic mechanism of RNAs containing nucleotide repeat expansions. Hum. Genet. 136, 1247–1263. doi: 10.1007/s00439-017-1807-6
Mousavi, K., Zare, H., Dell’Orso, S., Grontved, L., Gutierrez-Cruz, G., Derfoul, A., et al. (2013). ERNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617. doi: 10.1016/j.molcel.2013.07.022
Mueller, A. C., Cichewicz, M. A., Dey, B. K., Layer, R., Reon, B. J., Gagan, J. R., et al. (2015). MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 35, 498–513. doi: 10.1128/MCB.01079-14
Nakaya, Y., and Sheng, G. (2008). Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev. Growth. Differ. 50, 755–766. doi: 10.1111/j.1440-169X.2008.01070.x
Nelson, B. R., Makarewich, C. A., Anderson, D. M., Winders, B. R., Troupes, C. D., Wu, F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. doi: 10.1126/science.aad4076
Neppl, R. L., Wu, C. L., and Walsh, K. (2017). lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 216, 3497–3507. doi: 10.1083/jcb.201612100
Neumann, P., Jae, N., Knau, A., Glaser, S. F., Fouani, Y., Rossbach, O., et al. (2018). The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 9:237. doi: 10.1038/s41467-017-02431-1
Ounzain, S., Micheletti, R., Arnan, C., Plaisance, I., Cecchi, D., Schroen, B., et al. (2015). CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 89(Pt A), 98–112. doi: 10.1016/j.yjmcc.2015.09.016
Ounzain, S., Pezzuto, I., Micheletti, R., Burdet, F., Sheta, R., Nemir, M., et al. (2014). Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 76, 55–70. doi: 10.1016/j.yjmcc.2014.08.009
Pan, J. X. (2017). LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 322–328.
Poirier, F., Chan, C. T. J., Timmons, P. M., Robertson, E. J., Evans, M. J., and Rigby, P. W. J. (1991). The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 113, 1105–1114.
Qiao, Y., Ma, N., Wang, X., Hui, Y., Li, F., Xiang, Y., et al. (2011). MiR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett. 585, 3095–3100. doi: 10.1016/j.febslet.2011.08.039
Rensen, S. S., Doevendans, P. A., and van Eys, G. J. (2007). Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 15, 100–108. doi: 10.1007/bf03085963
Ritter, N., Ali, T., Kopitchinski, N., Schuster, P., Beisaw, A., Hendrix, D. A., et al. (2019). The lncRNA locus handsdown regulates cardiac gene programs and is essential for early mouse development. Dev. Cell 50, 644–657.e8. doi: 10.1016/j.devcel.2019.07.013
Schlosser, K., Hanson, J., Villeneuve, P. J., Dimitroulakos, J., McIntyre, L., Pilote, L., et al. (2016). Assessment of circulating LncRNAs under physiologic and pathologic conditions in humans reveals potential limitations as biomarkers. Sci. Rep. 6:36596. doi: 10.1038/srep36596
Shan, K., Jiang, Q., Wang, X. Q., Wang, Y. N., Yang, H., Yao, M. D., et al. (2016). Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 7:e2248. doi: 10.1038/cddis.2016.145
Shiekhattar, R. (2013). The Yin and Yang of enhancer-like RNAs. EMBO J. 32, 2533–2534. doi: 10.1038/emboj.2013.185
Song, T. F., Huang, L. W., Yuan, Y., Wang, H. Q., He, H. P., Ma, W. J., et al. (2018). LncRNA MALAT1 regulates smooth muscle cell phenotype switch via activation of autophagy. Oncotarget 9, 4411–4426. doi: 10.18632/oncotarget.23230
Stone, O. A., and Stainier, D. Y. R. (2019). Paraxial Mesoderm is the major source of lymphatic endothelium. Dev. Cell 50, 247–255.e3. doi: 10.1016/j.devcel.2019.04.034
Sui, Y., Han, Y., Zhao, X., Li, D., and Li, G. (2019). Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D. Cell Death Dis. 10:181. doi: 10.1038/s41419-019-1399-2
Sun, L., Si, M., Liu, X., Choi, J. M., Wang, Y., Thomas, S. S., et al. (2018). Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle 9, 962–974. doi: 10.1002/jcsm.12321
Tatsuta, T., Model, K., and Langer, T. (2005). Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16, 248–259. doi: 10.1091/mbc.e04-09-0807
Thomas, A. A., Biswas, S., Feng, B., Chen, S., Gonder, J., and Chakrabarti, S. (2019). lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia 62, 517–530. doi: 10.1007/s00125-018-4797-6
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
Viereck, J., Kumarswamy, R., Foinquinos, A., Xiao, K., Avramopoulos, P., Kunz, M., et al. (2016). Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 8:326ra322. doi: 10.1126/scitranslmed.aaf1475
von Maltzahn, J., Chang, N. C., Bentzinger, C. F., and Rudnicki, M. A. (2012). Wnt signaling in myogenesis. Trends Cell Biol. 22, 602–609. doi: 10.1016/j.tcb.2012.07.008
Wang, G.-Q., Wang, Y., Xiong, Y., Chen, X.-C., Ma, M.-L., Cai, R., et al. (2016). Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 6:21865. doi: 10.1038/srep21865
Wang, Z., Zhang, X. J., Ji, Y. X., Zhang, P., Deng, K. Q., Gong, J., et al. (2016). The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139. doi: 10.1038/nm.4179
Wang, H., Hertlein, E., Bakkar, N., Sun, H., Acharyya, S., Wang, J., et al. (2007). NF- B Regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 27, 4374–4387. doi: 10.1128/MCB.02020-06
Wang, H., Qin, R., and Cheng, Y. (2019). LncRNA-Ang362 promotes pulmonary arterial hypertension by regulating miR-221 and miR-222. Shock [Epub ahead of print].
Wang, K., Liu, C. Y., Zhou, L. Y., Wang, J. X., Wang, M., Zhao, B., et al. (2015). APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 6:6779. doi: 10.1038/ncomms7779
Wang, L., Zhao, Y., Bao, X., Zhu, X., Kwok, Y. K., Sun, K., et al. (2015). LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25, 335–350. doi: 10.1038/cr.2015.21
Wang, K., Liu, F., Zhou, L. Y., Long, B., Yuan, S. M., Wang, Y., et al. (2014a). The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 114, 1377–1388. doi: 10.1161/CIRCRESAHA.114.302476
Wang, K., Long, B., Zhou, L. Y., Liu, F., Zhou, Q. Y., Liu, C. Y., et al. (2014b). CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 5:3596. doi: 10.1038/ncomms4596
Wang, K., Sun, T., Li, N., Wang, Y., Wang, J. X., Zhou, L. Y., et al. (2014c). MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genet. 10:e1004467. doi: 10.1371/journal.pgen.1004467
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–126. doi: 10.1038/nature09819
Watts, R., Johnsen, V. L., Shearer, J., and Hittel, D. S. (2013). Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Cell Physiol. 304:C995–C1001. doi: 10.1152/ajpcell.00392.2012
Wijmenga, C., Frants, R. R., Brouwer, O. F., Moerer, P., Weber, J. L., and Padberg, G. W. (1990). Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet 336, 651–653. doi: 10.1016/0140-6736(90)92148-b
Wijmenga, C., Hewitt, J. E., Sandkuijl, L. A., Clark, L. N., Wright, T. J., Dauwerse, H. G., et al. (1992). Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2, 26–30. doi: 10.1038/ng0992-26
Wilkin, F., Paquette, J., Ledru, E., Hamelin, C., Pollak, M., and Deal, C. L. (2000). H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. Eur. J. Biochem. 267, 4020–4027. doi: 10.1046/j.1432-1327.2000.01438.x
Wu, G., Cai, J., Han, Y., Chen, J., Huang, Z. P., Chen, C., et al. (2014). LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 130, 1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675
Xiang, Y., Zhang, Y., Tang, Y., and Li, Q. (2017). MALAT1 modulates TGF-beta1-induced endothelial-to-mesenchymal transition through downregulation of miR-145. Cell Physiol. Biochem. 42, 357–372. doi: 10.1159/000477479
Xu, X., Ji, S., Li, W., Yi, B., Li, H., Zhang, H., et al. (2017). LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol. Biol. Lett. 22, 1–10. doi: 10.1186/s11658-017-0040-6
Yang, Y., Cai, Y., Wu, G., Chen, X., Liu, Y., Wang, X., et al. (2015). Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 129, 675–685. doi: 10.1042/CS20150121
Yin, Q., Wu, A., and Liu, M. (2017). Plasma long non-coding RNA (lncRNA) GAS5 is a new biomarker for coronary artery disease. Med. Sci. Monit. 23, 6042–6048. doi: 10.12659/msm.907118
Yu, X., Zhang, Y., Li, T., Ma, Z., Jia, H., Chen, Q., et al. (2017). Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8:14016. doi: 10.1038/ncomms14016
Zhang, Y., Sun, L., Xuan, L., Pan, Z., Li, K., Liu, S., et al. (2016). Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci. Rep. 6:22384. doi: 10.1038/srep22384
Zhang, Z. K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018). Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 26, 2669–2680. doi: 10.1016/j.ymthe.2018.09.014
Zhao, J., Zhang, W., Lin, M., Wu, W., Jiang, P., Tou, E., et al. (2016). MYOSLID is a novel serum response factor-dependent long noncoding RNA that amplifies the vascular smooth muscle differentiation program. Arterioscler. Thromb. Vasc. Biol. 36, 2088–2099. doi: 10.1161/ATVBAHA.116.307879
Zhou, L., Sun, K., Zhao, Y., Zhang, S., Wang, X., Li, Y., et al. (2015). Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6:10026. doi: 10.1038/ncomms10026
Zhu, B., Gong, Y., Yan, G., Wang, D., Qiao, Y., Wang, Q., et al. (2018). Down-regulation of lncRNA MEG3 promotes hypoxia-induced human pulmonary artery smooth muscle cell proliferation and migration via repressing PTEN by sponging miR-21. Biochem. Biophys. Res. Commun. 495, 2125–2132. doi: 10.1016/j.bbrc.2017.11.185
Zhu, M., Liu, J., Xiao, J., Yang, L., Cai, M., Shen, H., et al. (2017). Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 8:14718. doi: 10.1038/ncomms14718
Zhu, X. H., Yuan, Y. X., Rao, S. L., and Wang, P. (2016). LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 20, 3653–3660.
Keywords: non-coding RNA, skeletal muscle, endothelial cell, vascular smooth muscle cell (VSMC), differentiation, mesoderm, myogenesis, cardiovascular diseases
Citation: Sweta S, Dudnakova T, Sudheer S, Baker AH and Bhushan R (2019) Importance of Long Non-coding RNAs in the Development and Disease of Skeletal Muscle and Cardiovascular Lineages. Front. Cell Dev. Biol. 7:228. doi: 10.3389/fcell.2019.00228
Received: 01 August 2019; Accepted: 26 September 2019;
Published: 18 October 2019.
Edited by:
Francesco Fazi, Sapienza University of Rome, ItalyReviewed by:
Tetsuya S. Tanaka, Elixirgen Scientific, Inc., United StatesCopyright © 2019 Sweta, Dudnakova, Sudheer, Baker and Bhushan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew H. Baker, QW5keS5CYWtlckBlZC5hYy51aw==; Raghu Bhushan, cmFnaHViaHVzaGFuaW5AeWFob28uY28uaW4=; cmFnaHViaHVzaGFuQHllbmVwb3lhLmVkdS5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.