
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 01 October 2019
Sec. Developmental Epigenetics
Volume 7 - 2019 | https://doi.org/10.3389/fcell.2019.00219
This article is part of the Research TopicGene Regulation From the X-Chromosome During Development and DiseaseView all 8 articles
X inactivation represents a complex multi-layer epigenetic mechanism that profoundly modifies chromatin composition and structure of one X chromosome in females. The heterochromatic inactive X chromosome adopts a unique 3D bipartite structure and a location close to the nuclear periphery or the nucleolus. X-linked lncRNA loci and their transcripts play important roles in the recruitment of proteins that catalyze chromatin and DNA modifications for silencing, as well as in the control of chromatin condensation and location of the inactive X chromosome. A subset of genes escapes X inactivation, raising questions about mechanisms that preserve their expression despite being embedded within heterochromatin. Escape gene expression differs between males and females, which can lead to physiological sex differences. We review recent studies that emphasize challenges in understanding the role of lncRNAs in the control of epigenetic modifications, structural features and nuclear positioning of the inactive X chromosome. Second, we highlight new findings about the distribution of genes that escape X inactivation based on single cell studies, and discuss the roles of escape genes in eliciting sex differences in health and disease.
Evolution of the mammalian sex chromosomes from a pair of autosomes resulted in the emergence of distinct heteromorphic chromosomes that govern sex determination (Graves, 2016). The Y chromosome contains few genes (∼70) and is present only in males, while the X chromosome contains many genes (∼900–1500) and is present in two copies in females and one copy in males. This contributes to gene dosage imbalance between X-linked and autosomal genes and between the sexes (Disteche, 2016). To relieve these imbalances two mechanisms of dosage compensation evolved: X upregulation of expressed genes in males and females, and X inactivation or silencing of one X chromosome in females (Deng et al., 2014).
Here we focus on X chromosome inactivation (XCI), a mechanism that results in silencing of a randomly chosen X chromosome in early female embryogenesis (Lyon, 1961). XCI is characterized by a cascade of molecular events beginning shortly after embryo implantation, and is faithfully maintained throughout somatic cells in an organism, providing a robust model to study epigenetic and structural changes associated with gene silencing (Galupa and Heard, 2018). This complex process starts with the cis-coating of the future inactive X chromosome (Xi) by the long-non-coding RNA (lncRNA) Xist (Borsani et al., 1991; Brockdorff et al., 1991; Brown et al., 1991). Layers of chromatin and DNA modifications catalyzed by proteins initially recruited by Xist RNA are then put in place over several days during early development for stable transcriptional silencing of each gene on the Xi (Froberg et al., 2013; Mira-Bontenbal and Gribnau, 2016). These modifications are associated with profound changes in the 3D structure and location of the Xi, both processes depending on X-linked lncRNA loci. The Xi adopts a bipartite structure consisting of two superdomains of chromatin condensation separated by the lncRNA locus Dxz4, and the Xi visits the nucleolus, a process facilitated by the lncRNA Firre (Zhang et al., 2007; Rao et al., 2014; Deng et al., 2015; Minajigi et al., 2015; Yang et al., 2015; Giorgetti et al., 2016; Fang et al., 2019).
Despite the multiple layers of gene repression that stabilize XCI, a subset of developmentally critical genes remains expressed, albeit at a lower level, from the Xi (Carrel and Willard, 2005; Berletch et al., 2011). These escape genes adopt chromatin signatures and structural features more akin to those found in regions of active transcription (Balaton and Brown, 2016). Such genes can have higher expression in females, leading to sex differences in normal physiology and in susceptibility to disease. Abnormal escape gene dosage contributes to a milieu of deleterious phenotypes including infertility, intellectual disability, immune diseases, and cancer (Disteche, 2016; Balaton et al., 2018).
This review focuses first on mechanisms that govern X chromosome structure and nuclear location in relation to XCI, with a specific emphasis on the role of X-linked lncRNAs in these processes. We next discuss mechanisms that allow a select subset of genes to escape silencing in the context of the repressed Xi environment and how single-cell RNA sequencing has been used to identify novel escape genes. Lastly, we review new data on the role of escape gene dosage in sex differences in health and disease.
The importance of lncRNAs in controlling nuclear structure and gene expression has become increasingly clear (Engreitz et al., 2016). Here we consider three X-linked lncRNAs, Xist, Dxz4, and Firre, which have been implicated in various aspects of the onset and maintenance of XCI.
Recent reviews have considered the role of Xist in great detail (Mira-Bontenbal and Gribnau, 2016; Galupa and Heard, 2018). Thus, we will focus our discussion on issues related to structural changes on the Xi. One of those is a localized chromatin conformation change at the X inactivation center (XIC), which is essential for the correct initiation of XCI. The XIC harbors both the Xist locus and its antisense transcription unit Tsix, together with multiple other loci that regulate Xist (Froberg et al., 2013; Galupa and Heard, 2018). The Xist and Tsix promoters lie in separate but adjacent regions of local chromatin interactions called topologically associated domains (TADs) (Nora et al., 2012). Interestingly, swapping the Xist/Tsix transcriptional unit and placing their promoters in each other’s TADs leads to a switch in their expression dynamics, indicating the topological partitioning of the XIC is critical for proper initiation of XCI (van Bemmel et al., 2019).
Once XCI is initiated, chromosome-wide structural changes give rise to the condensed Barr body coated by Xist RNA (Figure 1). Imaging studies show the rapid formation of a nuclear compartment devoid of transcriptional machinery and euchromatic marks, in which X-linked genes, initially located at the periphery of the Xist RNA cloud, adopt a more internal position when silenced (Chaumeil et al., 2006; Clemson et al., 2006). Subsequent chromatin conformation analyses by Hi-C demonstrate that Xist is essential for the formation of the unique Xi bipartite structure further discussed below (see section “Dxz4”) (Minajigi et al., 2015; Giorgetti et al., 2016). Most of the Xi shows attenuation of local TADs and of large A/B compartments of active and inactive chromatin, normally evident on autosomes and the active X chromosome (Lieberman-Aiden et al., 2009; Dixon et al., 2012; Minajigi et al., 2015; Giorgetti et al., 2016). Xist RNA pull-down studies have identified two RNA-binding proteins implicated in liquid-to-solid phase transition, FUS and hnRNPA2, suggesting the possibility that phase separation facilitates heterochromatin formation and Xi silencing (Calabrese et al., 2012; Patel et al., 2015; Ryan et al., 2018). Indeed, most proteins within the Xist interactome are predicted to be prone to phase separation. By high-resolution RNA-FISH about one hundred Xist foci can be identified on the Xi, these foci having a comparable shape, size, and morphology to other phase-separated condensates such as paraspeckles and stress granules (Cerase et al., 2019).
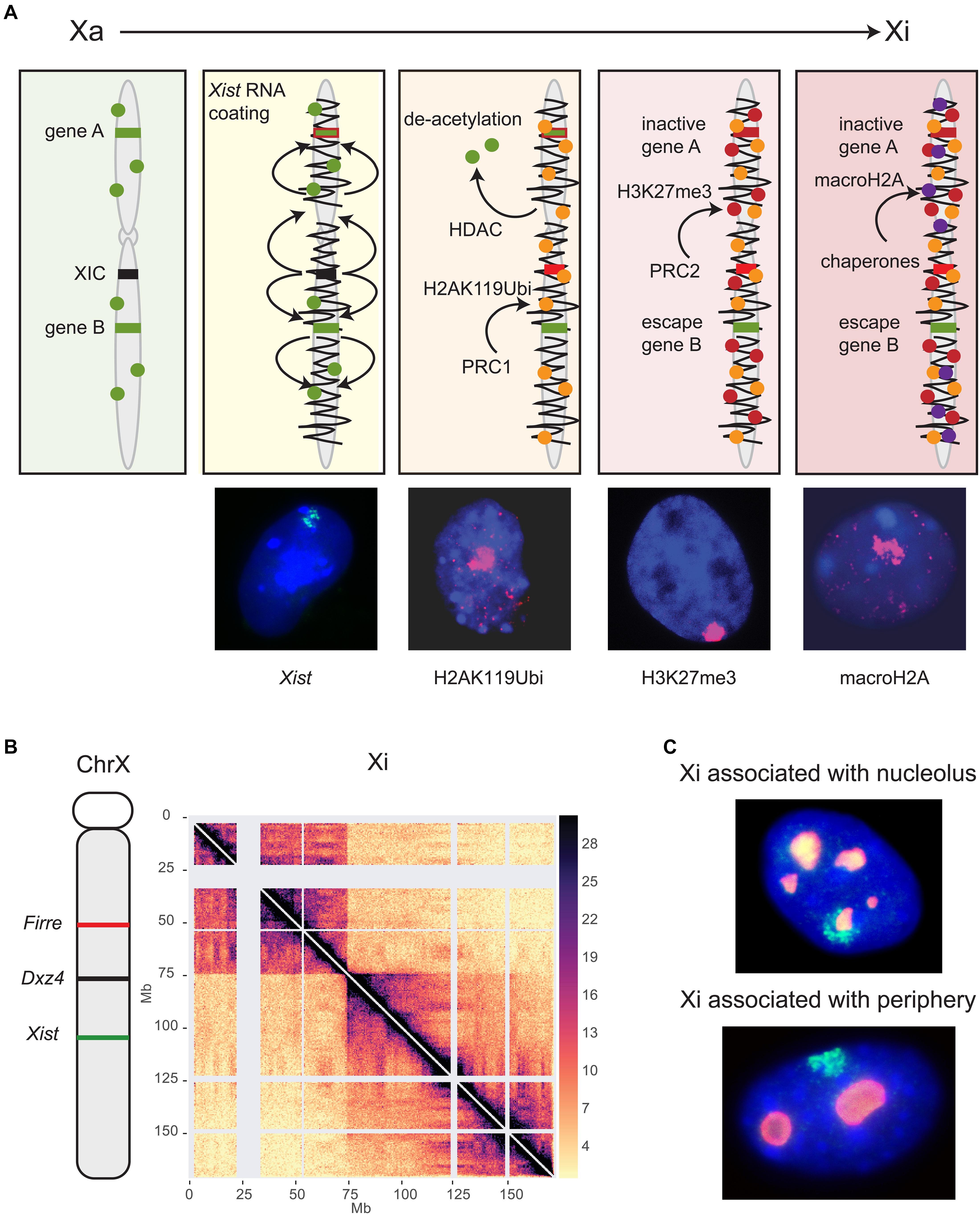
Figure 1. Long non-coding RNAs control epigenetic and structural characteristics of the Xi. (A) XCI begins with the expression of Xist located in the XIC (black). Xist RNA spreads (squiggly black line) along the X chromosome and recruits several protein complexes (see text). Major histone modifications take place, including histone deacetylation by HDAC3, followed by ubiquitination of H2AK119 mediated by the PRC1 complex, methylation of H3K27 mediated by the PRC2 complex, and finally incorporation of histone macroH2A. Gene A represents a gene that becomes inactivated (red) while gene B is an escape gene (green) that remains unchanged. This schematic focuses on histone modifications and does not show recruitment of other proteins or other repressive epigenetic modifications such as DNA methylation. Shown below the schematic are examples of female mouse nuclei showing an Xist cloud (green) after RNA-FISH, and enrichment of histone modifications (pink) by immunostaining of H2AK119Ubi, H3K27me3, and macroH2A. Nuclei are counterstained by Hoechst 33342. (B) The genomic location of Firre, Dxz4 and Xist is indicated on a schematic of the mouse X chromosome along a Hi-C contact map of the Xi in mouse Patski cells. Two superdomains of frequent contacts are separated by the Dxz4 region. The color scale shows normalized contact counts [adapted from a published figure (Bonora et al., 2018) in Nature Communications, under Springer Nature Publishing License: http://creativecommons.org/licenses/by/4.0/. (C) The Xi preferred locations are near the nucleolus or the nuclear periphery, as shown in examples of mouse fibroblast nuclei after RNA-FISH for Xist (green) to locate the Xi and immunostaining for nucleophosmin (red) to locate the nucleolus. Nuclei are counterstained by Hoechst 33342.
Xist RNA-mediated gene silencing is accomplished through recruitment of proteins that establish epigenetic and structural modifications on the Xi (Figure 1A; Chu et al., 2015; McHugh et al., 2015; Minajigi et al., 2015; Moindrot et al., 2015; Monfort et al., 2015). Originally, spreading of Xist RNA and its protein complexes was thought to be linear, but new studies have revealed complex interactions between epigenetic and genomic features, such as genomic distance from the Xist locus, gene density, and proximity to long interspersed nuclear elements (LINE) that act as waystations to enhance Xist RNA coating throughout the Xi 3D space (Sousa et al., 2019). Sequence analysis of Xist RNA reveals that a significant proportion of the primary RNA sequence is comprised of blocks of local tandem repeats (named A-F) with different functions in XCI. The utmost importance of the A-repeat in X silencing but not in Xist RNA coating was recognized early on, and has been further strengthened by demonstrating that it recruits the transcriptional repressor SPEN (Wutz et al., 2002; Zhao et al., 2008; Nesterova et al., 2019). The B-repeat together with a short part of the C-repeat are crucial for spreading of Xist RNA and for attracting the polycomb silencing complexes PRC1 and PRC2 (Pintacuda et al., 2017; Nesterova et al., 2019). Conversely, ablating PRC1 or PRC2 impairs Xist spreading, supporting interactive roles for this mega-protein-RNA complex (Colognori et al., 2019).
A new study has now clarified the precise order of appearance of histone modifications relative to X silencing (Żylicz et al., 2019). Importantly, loss of histone acetylation, in particular H3K27ac, is clearly one of the very first events following Xist RNA accumulation during XCI initiation (Figure 1A). Indeed, the histone deacetylase HDAC3 is pre-loaded at putative enhancers and is vital for efficient silencing of most genes on the Xi. Ubiquitination of histone H2K119 is then initiated by the nuclear matrix-PRC1 protein complex (hnRNPK-PCGF3/5-PRC1), signaling subsequent recruitment of other PRC1 complexes and of PRC2 (Figures 1A,B; Pintacuda et al., 2017). Tri-methylation of histone H3K27 is mediated by PRC2 and appears slightly later, even after gene silencing (Żylicz et al., 2019). The chromatin scaffolding protein SMCHD1 (structural maintenance of chromosomes flexible hinge domain-containing protein 1) plays an important role in gene silencing and structure of the Xi (Blewitt et al., 2008). A recent study proposes the Xi bipartite structure forms via an intermediate condensation step mediated by SMCHD1, in which A/B compartments initially fuse into S1/S2 compartments that subsequently merge into the compartment-less architecture of the Xi (Wang et al., 2018). Consistent with this finding, SMCHD1 loss of function results in the appearance of sub-megabase domains, A/B compartments, and a partial restoration of TAD boundaries on the Xi (Gdula et al., 2019). These changes are associated with de-repression of some X-inactivated genes and a local decrease in H3K27me3, but this reactivation is not observed in immortalized mouse embryonic fibroblasts, suggesting SMCHD1 facilitates H3K27me3 enrichment during XCI (Sakakibara et al., 2018; Gdula et al., 2019). At other heterochromatic regions of the genome SMCHD1 co-localizes with the repressive histone mark, H3K9me3, a process mediated by LRIF1 (Ligand Dependent Nuclear Receptor Interacting Factor 1). However, SMCHD1 remains enriched over the Xi even when SMCHD1-LRIF1 interactions are perturbed, suggesting an alternative mechanism by which SMCHD1 is targeted to the Xi, possibly via histone H2AK119 ubiquitination (Brideau et al., 2015; Jansz et al., 2018). Two later events that lock in silencing of the Xi are replacement of histone H2A by macrohistone H2A and DNA methylation of CpG islands by DNMT3B (Figure 1A; Gartler and Riggs, 1983; Costanzi and Pehrson, 1998; Gendrel et al., 2012).
During the establishment of XCI the Xi condenses in two superdomains of long-range contacts separated by a region that contains the conserved Dxz4 lncRNA microsatellite repeat (Figure 1B; Deng et al., 2015; Minajigi et al., 2015; Darrow et al., 2016; Giorgetti et al., 2016). This bipartite configuration is present in both mouse and human, albeit with different superdomain sizes, and deletion of Dxz4 specifically from the Xi results in disruption of the bipartite structure in both species, indicating a conserved function (Darrow et al., 2016; Giorgetti et al., 2016; Bonora et al., 2018). The deleted Xi acquires a configuration that resembles the Xa with enhanced TADs and compartments, but only in part, suggesting that, in addition to Dxz4, other factors control the Xi configuration.
CTCF-mediated interactions between the Dxz4/DXZ4 loci and other X-linked loci appear to be integral to forming chromatin loops for packaging the Xi. Indeed, Dxz4/DXZ4 bind the zinc-finger protein CTCF and components of the ring-shaped cohesin complex only on the Xi (Horakova et al., 2012a, b). Elsewhere in the genome convergent CTCF binding motifs at the base of a chromatin loop clearly favor strong interactions, and the inversion of CTCF sites disrupts loop formation (de Wit et al., 2015). The mouse Dxz4 locus contains a bank of CTCF motifs arranged in tandem orientation, while the human locus contains two banks of motifs with a different orientation (Horakova et al., 2012a, b). Our group reported that inversion of the mouse Dxz4 locus results in a massive reversal in long-range contacts, indicating that the Dxz4 locus itself acts as a structural platform for frequent long-range contacts with multiple X-linked loci in a direction dictated by the orientation of its CTCF motifs (Bonora et al., 2018). The anchoring of megabase size chromatin loops at Dxz4 causes the appearance of a line (or flame) emanating from Dxz4 in the contact map (Figure 1B). Whether contacts between Dxz4 and other X-linked loci rapidly fluctuate in individual cells remain to be determined. Surprisingly, deletion of Dxz4 causes only minor reactivation of X-linked genes and few changes in escape gene expression (Giorgetti et al., 2016; Bonora et al., 2018). A more recent study reported no changes in gene expression at all, suggesting inconsistencies between cell lines (Froberg et al., 2018). Furthermore, mice with a deletion of Dxz4 on the Xi show no apparent phenotype (Andergassen et al., 2019). Thus, the bipartite structure of the Xi has no clear function in gene regulation of the Xi at this point. However, conservation of the locus and of the bipartite structure between human and mouse suggest preservation of function.
Firre is another X-linked lncRNA locus that influences the epigenetic features and 3D structure of the Xi. We and others have shown that Firre is transcribed only from the active X chromosome (Calabrese et al., 2012; Froberg et al., 2018; Andergassen et al., 2019; Fang et al., 2019). Multiple isoforms of the Firre transcripts including lncRNAs and circular RNAs have been reported (Izuogu et al., 2018). Like Dxz4, the Firre region harbors many local repeats including the R0 repeat that recruits the chromatin organizers CTCF and YY1, as well as RAD21, a component of the cohesin ring complex (Yang et al., 2015; Hacisuleyman et al., 2016). The Firre locus recruits the nuclear matrix protein hnRNPU and interacts with many genomic regions, which might explain why depletion of Firre RNA causes widespread autosomal gene dysregulation (Hacisuleyman et al., 2014; Andergassen et al., 2019; Fang et al., 2019; Lewandowski et al., 2019). In regards to XCI, depletion of Firre RNA in differentiated fibroblasts does not disrupt Xist coating or gene silencing; however, a loss of H3K27me3 is observed on the Xi (Yang et al., 2015; Fang et al., 2019). In contrast, embryonic stem cells depleted of Firre prior to differentiation show no changes in H3K27me3 (Froberg et al., 2018). Together, these results support a role for Firre RNA in maintenance but not initiation of H3K27me3 enrichment on the Xi in differentiated cells.
Firre RNA is also important for maintenance of Xi location within the nucleus. Indeed, depletion of Firre in differentiated cells causes a decrease in perinucleolar and nuclear periphery association of the Xi (Yang et al., 2015; Fang et al., 2019). The perinucleolar and periphery compartments are often associated with heterochromatin and nucleolar location of the Xi has been proposed to be important for faithful replication of its epigenetic state (Figure 1C; Zhang et al., 2007; de Wit and van Steensel, 2009). Association of the Xi with the lamina may be facilitated by the lamin B receptor (LBR) recruited by Xist RNA (Chen C.K. et al., 2016). However, both the active and inactive X chromosomes are preferentially located near the nuclear periphery, suggesting that factors unrelated to XCI may control positioning (Bischoff et al., 1993). Surprisingly, tethering of one or both XIC alleles to the nuclear lamina via a TetR-EGFP-LaminB1 fusion protein does not disrupt XCI initiation, which would imply that XIC pairing and a visit to the nucleolus are not essential for XCI initiation (Pollex and Heard, 2019).
Deletion of Firre on the Xi does not disrupt the bipartite organization of the Xi, but causes localized changes in contact distribution, which may reflect disruption of the long-range contacts between Firre and Dxz4 thought to secure Xi compaction (Darrow et al., 2016; Barutcu et al., 2018; Bonora et al., 2018; Froberg et al., 2018; Fang et al., 2019). Long-range contacts between loci may help isolate the Xi in a specific compartment or phase of the nucleus either near the nuclear periphery or the nucleolus. The lncRNA loci Xist, Firre, and Dxz4 could play a concerted role in condensation and isolation of the Xi in a specific phase, which would ensure differential regulation of the two X chromosomes. Few studies have directly tested such a hypothesis due to the challenges in experimental design (Heard et al., 2004).
Despite being located in a constitutively repressed environment, a select subset of genes has evolved mechanisms to avoid silencing and thereby remain expressed from the Xi. A number of genes escape XCI in an individual-, tissue-, and cell type-specific manner, which can cause sex differences in gene expression (Berletch et al., 2015; Cotton et al., 2015; Tukiainen et al., 2017; Balaton et al., 2018). In mouse, 3–7% of X-linked genes escape transcriptional silencing, while that number increases to 20–30% in human (Berletch et al., 2015; Balaton and Brown, 2016). Comparisons between species show that a core set of genes escapes XCI in most cells and tissues in mammals, while other genes vary between cell types, tissues and species. New approaches by single-cell RNA sequencing (scRNA-seq) combined with SNP (single nucleotide polymorphism) analyses of allelic gene expression provides data on thousands of individual cells of varying types and thus promises a more complete picture of the variation in escape status between cell types (Figure 2A). Assigning escape status to a given gene formally requires finding biallelic reads in single cells or in a tissue with completely skewed XCI. A main advantage of single cell approaches is that tissues with random XCI can be analyzed, but allelic dropout can cause problems for genes with low expression (Reinius and Sandberg, 2015). A limited scRNA-seq analysis of about 1,000 cells representing two human cell types has uncovered genes (e.g., FHL1 and ATP6AP2) with incomplete XCI in a subset of cells, and also confirmed heterogeneity of XCI for TIMP1 (Tukiainen et al., 2017). Another allelic scRNA-seq in human fibroblasts shows little overlap with other studies in terms of identified escape genes, highlighting the difficulty of building a consensus (Wainer Katsir and Linial, 2019). Variability in escape from XCI between individuals has been confirmed in a single-cell allelic expression analysis from five individuals (Garieri et al., 2018).
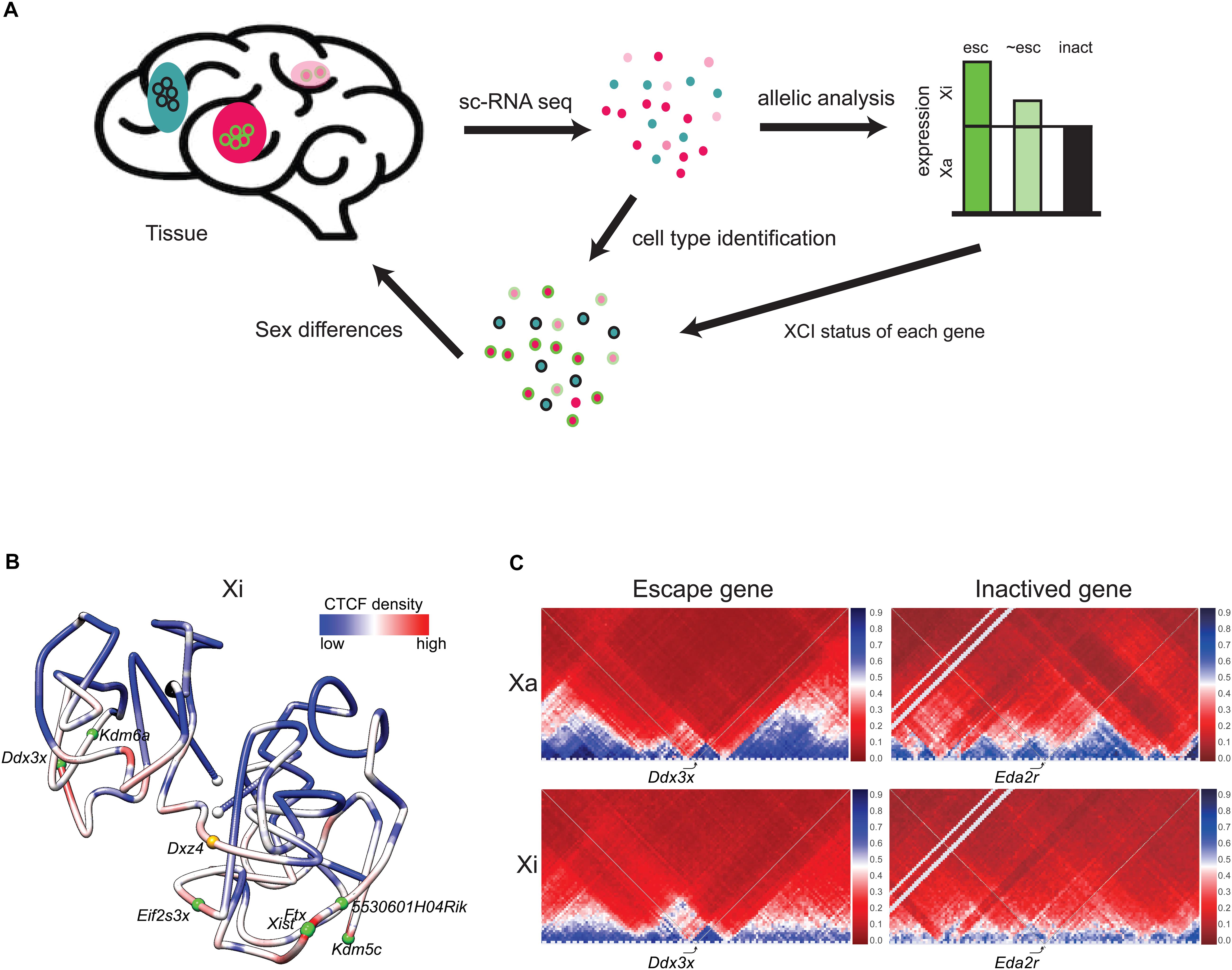
Figure 2. Escape genes distribution and structure. (A) Single-cell RNA-seq (scRNA-seq) enables determination of patterns of escape from XCI in cell types within a tissue. Beginning with a tissue, for example brain, individual cell types (colored pink, light pink, and blue) can be identified by scRNA-seq. Subsequent SNP analysis determines the escape status of each X-linked gene based on reads from the Xa and the Xi (escape gene, esc, colored green; inactivated gene, inact, colored black; gene that partially escapes, ∼esc, colored light green). Each cell type can be associated with the escape status of an ensemble of X-linked genes. For example, a specific cell type (pink) shows escape of the example gene (circled green), while another cell type (blue) shows inactivation of this same gene (circled black). Combination of data on an ensemble of genes can potentially inform sex differences in a particular cell type. (B) 3D model of the Xi at 1 Mb resolution in mouse brain colored to display the density of allelic CTCF binding (red indicates more binding and blue, less binding). Two domains of condensation are seen separated by Dxz4 at the hinge. The white dots indicate chromosome ends, the orange dot, Dxz4, and the green dots, escape genes. Escape genes tend to be located at the outside of the Xi 3D structure [adapted from a published figure (Deng et al., 2015) in Genome Biology, under Springer Nature Publishing License: http://creativecommons.org/licenses/by/4.0/]. (C) Partial Hi-C contact maps (4 Mb resolution) of the active (Xa) and inactive (Xi) X chromosomes in a 4 Mb region around the escape gene Ddx3x and the inactivated gene Eda2r highlight the attenuation of TADs (blue) on the mouse Xi, except at the escape gene Ddx3x where TADs are visible on both the Xi and Xa. The color scale shows normalized contact counts (blue, higher contact count, red, lower contact count) [adapted from a figure (Bonora et al., 2018) published in Nature Communications, under Springer Nature Publishing License: http://creativecommons.org/licenses/by/4.0/].
Developmental scRNA-seq studies have followed XCI progression during embryogenesis. A study of human preimplantation embryos reports that bi-allelic X-linked gene expression may persist until the blastocyst stage, together with dampening of both alleles (Petropoulos et al., 2016). However, re-analysis of these and additional data has provided a more detailed panorama of random XCI from human oocyte to blastocyst, demonstrating progressive establishment of mono-allelic X-linked gene expression and of X upregulation to maintain balance of expression throughout the genome (Moreira de Mello et al., 2017; Zhou et al., 2019). Unfortunately, very little information on escape genes is included in these studies, despite findings of extensive sex differences in overall gene expression. In mouse, XCI dynamics during embryonic stem cell differentiation show a gradual decrease in expression of escape genes, consistent with partial spreading of silencing (Chen G. et al., 2016).
Expectedly, genes expressed from the Xi lack epigenetic signatures characteristic of inactivated genes and appear to be located away from repressive genomic elements. A recent study using allele-specific PRO-seq and predictive machine learning shows that primary determinants of escape from XCI include distance from Xist and density of LINE elements (Sousa et al., 2019). Interestingly, escape genes often cluster in domains, a common finding in human, while mouse escape genes are often isolated (Tsuchiya et al., 2004; Prothero et al., 2009; Berletch et al., 2011). In general, escape regions lack repressive histone marks such as H3K27me3, and are enriched in active histone marks such as acetylation, and in transcription elongation marks including RNA PolII S2P and H3K36me3 (Figure 1A; Disteche and Berletch, 2015; Sousa et al., 2019). DNA hypomethylation of CpG islands is a reliable predictor of escape status, which has been successfully used to identify escape genes in an array of human tissues where allelic analyses are difficult due to few SNPs and/or absence of XCI skewing (Cotton et al., 2015; Duncan et al., 2018). Surprisingly, escape genes in brain and liver adopt specific DNA methylation signatures that include enrichment in non-CG hypermethylation (mCH) throughout their gene body, which may help maintain an open chromatin structure (Keown et al., 2017; Duncan et al., 2018).
Escape genes tend to reside toward the outside of the compacted inactivated interior of the Xi (Figure 2B; Chaumeil et al., 2006; Heard and Bickmore, 2007; Splinter et al., 2011; Deng et al., 2015; Bonora and Disteche, 2017). However, it remains to be determined exactly how the 3D structure of the Xi influences the propensity for escape from XCI. One factor may be repeat E of Xist, which is required for localization of ASH2L, a component of the histone methyltransferase that methylates H3K4 for increased expression (Yue et al., 2017). Other factors may be involved, for example, the lncRNAs often found near escape genes (Reinius et al., 2010). Escape genes often co-localize with clusters of CTCF binding and with TADs, suggesting local Xi compartmentalization (Figure 2C; Giorgetti et al., 2016; Bonora et al., 2018). However, deletion of Dxz4 and loss of the bipartite structure of the Xi causes little or no disruption in escape gene expression (Giorgetti et al., 2016; Bonora et al., 2018; Froberg et al., 2018).
It is increasingly clear that intrinsic genetic escape elements act in cis to facilitate expression from the Xi (Balaton et al., 2018). We and others have proposed that spreading of silencing into the escape region or vice versa spreading of gene activity into a silenced gene may be prevented by insulator elements such as CTCF or YY1, but these elements may not be sufficient and mechanisms may differ between escape genes (Filippova et al., 2005; Li and Carrel, 2008; Horvath et al., 2013; Chen C.Y. et al., 2016). Interestingly, a BAC harboring the human escape gene RPS4X inserted at the silenced Hprt locus in mouse retains escape status in vivo, both through the onset of XCI and the life of the mouse (Peeters et al., 2018). It will be interesting to identify the apparently conserved insulator elements involved in this process and to test their role in shaping regions of escape, for example by isolating them into separate chromatin loops or phases within the nucleus.
One main consequence of escape from XCI is differential gene expression between males and females (Mele et al., 2015). A recent study based on thousands of transcriptomes spanning 29 human tissues provides a detailed survey of sex-biased gene expression in humans and demonstrates that expression of escape genes is usually female-biased (Tukiainen et al., 2017). However, a subset of escape genes located in the pseudoautosomal region shared between the X and Y chromosomes is male-biased, probably due to lower expression in females due to spreading of silencing on the Xi (Tukiainen et al., 2017). The biological implications of these sex bias remain largely unexplored. While it is clear that certain escape genes, e.g., Kdm6a, and its Y-paralog Uty are expressed in different parts of the mouse brain, their role in phenotypic sex differences has not been clarified (Xu et al., 2008). In fact, few traits have been linked to sex bias in X-linked gene expression in normal healthy individuals. One example is longevity, with recent evidence suggesting that having two X chromosomes prolongs lifespan, independent of gonadal sex. This was demonstrated by using the four core genotype (FCG), a mouse model capable of differentiating the effects of hormones versus sex chromosome complement, which showed that XX mice with either ovaries or testes lived longer than XY mice of either gonadal phenotype (Davis et al., 2019). In addition, having two X chromosomes leads to improved blood pressure regulation and an increase in the capacity to blunt the effects of brain injuries (Pessoa et al., 2015; McCullough et al., 2016).
In terms of disease susceptibility there is ample evidence suggesting that sex bias in X-linked gene expression play a role. For example, having two X chromosomes increases the risk of developing autoimmunity [reviewed in Syrett and Anguera (2019)]. This phenomenon may be a result of unusual XCI patterns in immune cells, which could leave certain X-linked genes involved in immune response susceptible to reactivation (Wang et al., 2016; Syrett et al., 2019). This is supported by studies demonstrating that the Toll-like receptor seven gene, which escapes XCI in human lymphocytes, causes systemic lupus erythematosus when overexpressed in mouse models (Deane et al., 2007; Souyris et al., 2018). The X chromosome harbors a high number of genes important in brain function, and dosage of some escape genes has been implicated in neurological phenotypes such as seizures and Autism Spectrum Disorder (Shoubridge et al., 2019). For example, loss of function mutations in the escape gene, IQSEC2, contribute to the manifestation of phenotypes that include moderate to severe intellectual disability (Shoubridge et al., 2019). Another example is KDM5C, an escape gene that encodes a histone demethylase and regulates neuronal development and function (Iwase et al., 2016; Scandaglia et al., 2017; Kim et al., 2018). Mutations in KDM5C cause intellectual disability in males and females, illustrating the dosage sensitivity of this gene (Brookes et al., 2015).
Variability in expression of escape genes may contribute to sex differences in predisposition to certain cancers (Arnold and Disteche, 2018). Many types of cancers are sex-biased in nature and some of those skewed toward a male prevalence may be explained by mutations in X-linked escape genes called EXITS (Escape from X Inactivation Tumor Suppressor) (Table 1; Dunford et al., 2017). One example is bladder cancer, with incident rates ranging from three to five times higher in men than women (Edgren et al., 2012). Recent studies in FCG mice show that XX mice with bladder cancer survive at increased rates compared to XY mice regardless of gonadal sex, suggesting that X-linked gene dosage is an intrinsic determinant of survival (Kaneko and Li, 2018). Interestingly, the female-biased escape gene KDM6A is a strong tumor suppressor that acts through demethylation-dependent and -independent mechanisms to reduce bladder cancer cell proliferation (Kaneko and Li, 2018).
Disorders of sex chromosome number such as Turner syndrome (45,X) and Klinefelter syndrome (47,XXY) directly implicate escape genes in abnormal phenotypes, since such genes would be haplo-insufficient and overexpressed, respectively, in these conditions. One of the hallmark of Turner syndrome is premature ovarian failure and infertility. The X chromosome is three-fold more enriched for genes expressed in female reproductive organs when compared to autosomes, indicating a role in female fertility (Liu, 2019). Defining the contribution of specific X-linked genes in abnormal Turner phenotypes is a work in progress. For example, the escape genes KDM6A and TIMP1 are hypothesized to be involved in premature ovarian failure and aortic aneurysm formation, respectively (Trolle et al., 2016; Viuff et al., 2019). Recent screens of X-linked copy number variation (CNV) in cohorts of healthy women and those with primary ovarian insufficiency (POI) show a high prevalence of deletions encompassing escape genes as well as lncRNAs (Yatsenko et al., 2019). Of course, escape genes would not be the only X-linked genes involved in Turner infertility, since all X-linked genes become reactivated in female primordial germ cells, a process mediated by PR-domain containing protein 14 (PRDM14) for removal of H3K27me3 on the Xi (Mallol et al., 2019). The infertility observed in Klinefelter individuals could potentially also result from abnormal escape gene dosage, but another large factor would be abnormal meiotic pairing of the sex chromosomes. Interestingly, any abnormal X chromosome numbers (XXY, XXX or X) cause a general disruption of DNA methylation patterns at autosomal genes, demonstrating widespread epigenetic effects of X aneuploidy (Trolle et al., 2016; Skakkebaek et al., 2018). Furthermore, the number of X and/or Y chromosomes influences spatial chromosome conformation, particularly of the active X chromosome, but the role of this structural change is not elucidated (Jowhar et al., 2018). Together, these findings implicate improper X-linked gene dosage as a causative factor in disease phenotypes ranging from brain function, cancer susceptibility, to impaired fertility.
While a great deal has been learned about the various lncRNAs and proteins that control structural and epigenetic features of the Xi and its silencing, their exact modes of action remain to be further studied. In terms of the 3D structure of the Xi it will be of great interest to define factors involved in nuclear compartmentalization and phase separation. Additional experiments are needed in order to link the specific 3D structure and nuclear location of the Xi with its distinct epigenetic landscape. Moreover, little is known about contacts between each X chromosome with the rest of the genome in female cells and tissues, and about such contacts with the heterochromatic Y chromosome in male cells and tissues. Tissue-specific differences in the arrangement of chromosomes within the nucleus are poorly understood, and few functional studies have examined the consequences of manipulating chromosomal location. The epigenetic controls of escape from XCI also warrant further functional studies. Unfortunately, current analyses toward identifying escape genes in specific cell types and tissues are limited due to the relatively small number of informative polymorphisms in human. However, single-cell analyses are progressing at a fast rate, with some methods allowing analyses in thousands of cells in tissues to establish maps of gene expression or accessibility in a whole organism, as shown for example in a recently published atlas of mouse tissues/cell types (Cusanovich et al., 2018). While there is compelling evidence of sex differences in susceptibility to disease, understanding the role of individual sex-linked genes will require careful manipulation of their dosage.
JB, HF, and CD outlined and wrote the review. All authors approved the final manuscript.
The authorship of this review was supported by grants GM131745 from the National Institutes of General Medicine and DK107979 from the National Institutes of Health Common Fund 4D Nucleome Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Gala Filippova, Xinxian Deng, and Di Kim Nguyen for critical reading of the manuscript and for providing constructive editorial comments. We also thank Wenxiu Ma and Giancarlo Bonora for originally making the Hi-C contact maps and 3D model for the figures (see references in figure legends).
Andergassen, D., Smith, Z., Lewandowski, J., Gerhardinger, C., Meissner, A., and Rinn, J. (2019). In vivo firre and Dxz4 deletion elucidates roles for autosomal gene regulation. bioRxiv doi: 10.1101/612440
Arnold, A. P., and Disteche, C. M. (2018). Sexual inequality in the cancer cell. Cancer Res. 78, 5504–5505. doi: 10.1158/0008-5472.CAN-18-2219
Balaton, B. P., and Brown, C. J. (2016). Escape artists of the X chromosome. Trends Genet. 32, 348–359. doi: 10.1016/j.tig.2016.03.007
Balaton, B. P., Dixon-McDougall, T., Peeters, S. B., and Brown, C. J. (2018). The exceptional nature of the X chromosome. Hum. Mol. Genet. 27, R242–R249. doi: 10.1093/hmg/ddy148
Barutcu, A. R., Maass, P. G., Lewandowski, J. P., Weiner, C. L., and Rinn, J. L. (2018). A TAD boundary is preserved upon deletion of the CTCF-rich firre locus. Nat. Commun. 9:1444. doi: 10.1038/s41467-018-03614-3610
Berletch, J. B., Ma, W., Yang, F., Shendure, J., Noble, W. S., Disteche, C. M., et al. (2015). Escape from x inactivation varies in mouse tissues. PLoS Genet. 11:e1005079. doi: 10.1371/journal.pgen.1005079
Berletch, J. B., Yang, F., Xu, J., Carrel, L., and Disteche, C. M. (2011). Genes that escape from X inactivation. Hum. Genet. 130, 237–245. doi: 10.1007/s00439-011-1011-z
Bischoff, A., Albers, J., Kharboush, I., Stelzer, E., Cremer, T., and Cremer, C. (1993). Differences of size and shape of active and inactive X-chromosome domains in human amniotic fluid cell nuclei. Microsc. Res. Tech. 25, 68–77. doi: 10.1002/jemt.1070250110
Blewitt, M. E., Gendrel, A. V., Pang, Z., Sparrow, D. B., Whitelaw, N., Craig, J. M., et al. (2008). SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 40, 663–669. doi: 10.1038/ng.142
Bonora, G., Deng, X., Fang, H., Ramani, V., Qiu, R., Berletch, J. B., et al. (2018). Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat. Commun. 9:1445. doi: 10.1038/s41467-018-03694-y
Bonora, G., and Disteche, C. M. (2017). Structural aspects of the inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160357. doi: 10.1098/rstb.2016.0357
Borsani, G., Tonlorenzi, R., Simmler, M. C., Dandolo, L., Arnaud, D., Capra, V., et al. (1991). Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. doi: 10.1038/351325a0
Brideau, N. J., Coker, H., Gendrel, A. V., Siebert, C. A., Bezstarosti, K., Demmers, J., et al. (2015). Independent mechanisms target SMCHD1 to trimethylated histone H3 Lysine 9-modified chromatin and the inactive X chromosome. Mol. Cell Biol. 35, 4053–4068. doi: 10.1128/MCB.00432-415
Brockdorff, N., Ashworth, A., Kay, G. F., Cooper, P., Smith, S., McCabe, V. M., et al. (1991). Conservation of position and exclusive expression of mouse xist from the inactive X chromosome. Nature 351, 329–331. doi: 10.1038/351329a0
Brookes, E., Laurent, B., Ounap, K., Carroll, R., Moeschler, J. B., Field, M., et al. (2015). Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 24, 2861–2872. doi: 10.1093/hmg/ddv046
Brown, C. J., Lafreniere, R. G., Powers, V. E., Sebastio, G., Ballabio, A., Pettigrew, A. L., et al. (1991). Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349, 82–84. doi: 10.1038/349082a0
Calabrese, J. M., Sun, W., Song, L., Mugford, J. W., Williams, L., Yee, D., et al. (2012). Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151, 951–963. doi: 10.1016/j.cell.2012.10.037
Carrel, L., and Willard, H. F. (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. doi: 10.1038/nature03479
Cerase, A., Armaos, A., Neumayer, C., Avner, P., Guttman, M., and Tartaglia, G. G. (2019). Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 26, 331–334. doi: 10.1038/s41594-019-0223-220
Chaumeil, J., Le Baccon, P., Wutz, A., and Heard, E. (2006). A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. doi: 10.1101/gad.380906
Chen, C. K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. doi: 10.1126/science.aae0047
Chen, C. Y., Shi, W., Balaton, B. P., Matthews, A. M., Li, Y., Arenillas, D. J., et al. (2016). YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 6:37324. doi: 10.1038/srep37324
Chen, G., Schell, J. P., Benitez, J. A., Petropoulos, S., Yilmaz, M., Reinius, B., et al. (2016). Single-cell analyses of X Chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 26, 1342–1354. doi: 10.1101/gr.201954.115
Chu, C., Zhang, Q. C., da Rocha, S. T., Flynn, R. A., Bharadwaj, M., Calabrese, J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. doi: 10.1016/j.cell.2015.03.025
Clemson, C. M., Hall, L. L., Byron, M., McNeil, J., and Lawrence, J. B. (2006). The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. U.S.A. 103, 7688–7693. doi: 10.1073/pnas.0601069103
Colognori, D., Sunwoo, H., Kriz, A. J., Wang, C. Y., and Lee, J. T. (2019). Xist deletional analysis reveals an interdependency between xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell 74, 101.E–117.E. doi: 10.1016/j.molcel.2019.01.015
Costanzi, C., and Pehrson, J. R. (1998). Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601. doi: 10.1038/31275
Cotton, A. M., Ge, B., Light, N., Adoue, V., Pastinen, T., and Brown, C. J. (2013). Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 14:R122. doi: 10.1186/gb-2013-14-11-r122
Cotton, A. M., Price, E. M., Jones, M. J., Balaton, B. P., Kobor, M. S., and Brown, C. J. (2015). Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539. doi: 10.1093/hmg/ddu564
Cusanovich, D. A., Hill, A. J., Aghamirzaie, D., Daza, R. M., Pliner, H. A., Berletch, J. B., et al. (2018). A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18. doi: 10.1016/j.cell.2018.06.052
Darrow, E. M., Huntley, M. H., Dudchenko, O., Stamenova, E. K., Durand, N. C., Sun, Z., et al. (2016). Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. U.S.A. 113, E4504–E4512. doi: 10.1073/pnas.1609643113
Davis, E. J., Lobach, I., and Dubal, D. B. (2019). Female XX sex chromosomes increase survival and extend lifespan in aging mice. Aging Cell 18:e12871. doi: 10.1111/acel.12871
de Wit, E., and van Steensel, B. (2009). Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma 118, 25–36. doi: 10.1007/s00412-008-0186-180
de Wit, E., Vos, E. S., Holwerda, S. J., Valdes-Quezada, C., Verstegen, M. J., Teunissen, H., et al. (2015). CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684. doi: 10.1016/j.molcel.2015.09.023
Deane, J. A., Pisitkun, P., Barrett, R. S., Feigenbaum, L., Town, T., Ward, J. M., et al. (2007). Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27, 801–810. doi: 10.1016/j.immuni.2007.09.009
Deng, X., Berletch, J. B., Nguyen, D. K., and Disteche, C. M. (2014). X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378. doi: 10.1038/nrg3687
Deng, X., Ma, W., Ramani, V., Hill, A., Yang, F., Ay, F., et al. (2015). Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16:152. doi: 10.1186/s13059-015-0728-728
Disteche, C. M. (2016). Dosage compensation of the sex chromosomes and autosomes. Semin. Cell Dev. Biol. 56, 9–18. doi: 10.1016/j.semcdb.2016.04.013
Disteche, C. M., and Berletch, J. B. (2015). X-chromosome inactivation and escape. J. Genet. 94, 591–599.
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Duncan, C. G., Grimm, S. A., Morgan, D. L., Bushel, P. R., Bennett, B. D., Program, N. C. S., et al. (2018). Dosage compensation and DNA methylation landscape of the X chromosome in mouse liver. Sci. Rep. 8:10138. doi: 10.1038/s41598-018-28356-28353
Dunford, A., Weinstock, D. M., Savova, V., Schumacher, S. E., Cleary, J. P., Yoda, A., et al. (2017). Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16. doi: 10.1038/ng.3726
Edgren, G., Liang, L., Adami, H. O., and Chang, E. T. (2012). Enigmatic sex disparities in cancer incidence. Eur. J. Epidemiol. 27, 187–196. doi: 10.1007/s10654-011-9647-9645
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi: 10.1038/nrm.2016.126
Fang, H., Bonora, G., Lewandowski, J., Thakur, J., Filippova, G. N., Henikoff, S., et al. (2019). Trans- and cis-acting effects of the lncRNA firre on epigenetic and structural features of the inactive X chromosome. bioRxiv doi: 10.1101/687236
Filippova, G. N., Cheng, M. K., Moore, J. M., Truong, J. P., Hu, Y. J., Nguyen, D. K., et al. (2005). Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 8, 31–42.
Froberg, J. E., Pinter, S. F., Kriz, A. J., Jegu, T., and Lee, J. T. (2018). Megadomains and superloops form dynamically but are dispensable for X-chromosome inactivation and gene escape. Nat. Commun. 9:5004. doi: 10.1038/s41467-018-07446-w
Froberg, J. E., Yang, L., and Lee, J. T. (2013). Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 425, 3698–3706. doi: 10.1016/j.jmb.2013.06.031
Galupa, R., and Heard, E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. doi: 10.1146/annurev-genet-120116-124611
Garieri, M., Stamoulis, G., Blanc, X., Falconnet, E., Ribaux, P., Borel, C., et al. (2018). Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 115, 13015–13020. doi: 10.1073/pnas.1806811115
Gartler, S. M., and Riggs, A. D. (1983). Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190. doi: 10.1146/annurev.ge.17.120183.001103
Gdula, M. R., Nesterova, T. B., Pintacuda, G., Godwin, J., Zhan, Y., Ozadam, H., et al. (2019). The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 10:30. doi: 10.1038/s41467-018-07907-7902
Gendrel, A. V., Apedaile, A., Coker, H., Termanis, A., Zvetkova, I., Godwin, J., et al. (2012). Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell 23, 265–279. doi: 10.1016/j.devcel.2012.06.011
Giorgetti, L., Lajoie, B. R., Carter, A. C., Attia, M., Zhan, Y., Xu, J., et al. (2016). Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. doi: 10.1038/nature18589
Goldberg, J. M., Silverman, L. B., Levy, D. E., Dalton, V. K., Gelber, R. D., Lehmann, L., et al. (2003). Childhood T-cell acute lymphoblastic leukemia: the dana-farber cancer institute acute lymphoblastic leukemia consortium experience. J. Clin. Oncol. 21, 3616–3622. doi: 10.1200/JCO.2003.10.116
Graves, J. A. (2016). Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 17, 33–46. doi: 10.1038/nrg.2015.2
Hacisuleyman, E., Goff, L. A., Trapnell, C., Williams, A., Henao-Mejia, J., Sun, L., et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. doi: 10.1038/nsmb.2764
Hacisuleyman, E., Shukla, C. J., Weiner, C. L., and Rinn, J. L. (2016). Function and evolution of local repeats in the firre locus. Nat. Commun. 7:11021. doi: 10.1038/ncomms11021
Heard, E., and Bickmore, W. (2007). The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 19, 311–316. doi: 10.1016/j.ceb.2007.04.016
Heard, E., Chaumeil, J., Masui, O., and Okamoto, I. (2004). Mammalian X-chromosome inactivation: an epigenetics paradigm. Cold Spring Harb. Symp. Quant. Biol. 69, 89–102. doi: 10.1101/sqb.2004.69.89
Horakova, A. H., Calabrese, J. M., McLaughlin, C. R., Tremblay, D. C., Magnuson, T., and Chadwick, B. P. (2012a). The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 13:R70. doi: 10.1186/gb-2012-13-8-r70
Horakova, A. H., Moseley, S. C., McLaughlin, C. R., Tremblay, D. C., and Chadwick, B. P. (2012b). The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum. Mol. Genet. 21, 4367–4377. doi: 10.1093/hmg/dds270
Horesh, N., and Horowitz, N. A. (2014). Does gender matter in non-hodgkin lymphoma? Differences in epidemiology, clinical behavior, and therapy. Rambam Maimonides Med. J. 5:e0038. doi: 10.5041/RMMJ.10172
Horvath, L. M., Li, N., and Carrel, L. (2013). Deletion of an x-inactivation boundary disrupts adjacent gene silencing. PLoS Genet. 9:e1003952. doi: 10.1371/journal.pgen.1003952
Iwase, S., Brookes, E., Agarwal, S., Badeaux, A. I., Ito, H., Vallianatos, C. N., et al. (2016). A mouse model of x-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep. 14, 1000–1009. doi: 10.1016/j.celrep.2015.12.091
Izuogu, O. G., Alhasan, A. A., Mellough, C., Collin, J., Gallon, R., Hyslop, J., et al. (2018). Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular. BMC Genomics 19:276. doi: 10.1186/s12864-018-4660-4667
Jansz, N., Nesterova, T., Keniry, A., Iminitoff, M., Hickey, P. F., Pintacuda, G., et al. (2018). Smchd1 targeting to the inactive x is dependent on the xist-hnrnpK-PRC1 pathway. Cell Rep. 25, 1912–1923.e9. doi: 10.1016/j.celrep.2018.10.044
Jowhar, Z., Shachar, S., Gudla, P. R., Wangsa, D., Torres, E., Russ, J. L., et al. (2018). Effects of human sex chromosome dosage on spatial chromosome organization. Mol. Biol. Cell 29, 2458–2469. doi: 10.1091/mbc.E18-06-0359
Kaneko, S., and Li, X. (2018). X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci. Adv. 4:eaar5598. doi: 10.1126/sciadv.aar5598
Keown, C. L., Berletch, J. B., Castanon, R., Nery, J. R., Disteche, C. M., Ecker, J. R., et al. (2017). Allele-specific non-CG DNA methylation marks domains of active chromatin in female mouse brain. Proc. Natl. Acad. Sci. U.S.A. 114, E2882–E2890. doi: 10.1073/pnas.1611905114
Kim, Y., Jeong, Y., Kwon, K., Ismail, T., Lee, H. K., Kim, C., et al. (2018). Physiological effects of KDM5C on neural crest migration and eye formation during vertebrate development. Epigenetics Chromatin 11:72. doi: 10.1186/s13072-018-0241-x
Lewandowski, J., Lee, J. C., Hwang, T., Sunwoo, H., Goldstein, J. M., Groff, A. F., et al. (2019). The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. bioRxiv doi: 10.1101/648279
Li, N., and Carrel, L. (2008). Escape from X chromosome inactivation is an intrinsic property of the jarid1c locus. Proc. Natl. Acad. Sci. U.S.A. 105, 17055–17060. doi: 10.1073/pnas.0807765105
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi: 10.1126/science.1181369
Liu, W. S. (2019). Mammalian sex chromosome structure, gene content, and function in male fertility. Annu. Rev. Anim. Biosci. 7, 103–124. doi: 10.1146/annurev-animal-020518-115332
Lyon, M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. doi: 10.1038/190372a0
Mallol, A., Guirola, M., and Payer, B. (2019). PRDM14 controls X-chromosomal and global epigenetic reprogramming of H3K27me3 in migrating mouse primordial germ cells. Epigenetics Chromatin 12:38. doi: 10.1186/s13072-019-0284-287
McCullough, L. D., Mirza, M. A., Xu, Y., Bentivegna, K., Steffens, E. B., Ritzel, R., et al. (2016). Stroke sensitivity in the aged: sex chromosome complement vs. gonadal hormones. Aging 8, 1432–1441. doi: 10.18632/aging.100997
McHugh, C. A., Chen, C. K., Chow, A., Surka, C. F., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi: 10.1038/nature14443
Mele, M., Ferreira, P. G., Reverter, F., DeLuca, D. S., Monlong, J., Sammeth, M., et al. (2015). Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665. doi: 10.1126/science.aaa0355
Minajigi, A., Froberg, J., Wei, C., Sunwoo, H., Kesner, B., Colognori, D., et al. (2015). Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349:aab2276. doi: 10.1126/science.aab2276
Mira-Bontenbal, H., and Gribnau, J. (2016). New xist-interacting proteins in X-chromosome inactivation. Curr. Biol. 26, R338–R342. doi: 10.1016/j.cub.2016.03.022
Moindrot, B., Cerase, A., Coker, H., Masui, O., Grijzenhout, A., Pintacuda, G., et al. (2015). A pooled shRNA screen identifies Rbm15, spen, and wtap as factors required for xist RNA-mediated silencing. Cell Rep. 12, 562–572. doi: 10.1016/j.celrep.2015.06.053
Monfort, A., Di Minin, G., Postlmayr, A., Freimann, R., Arieti, F., Thore, S., et al. (2015). Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. doi: 10.1016/j.celrep.2015.06.067
Moreira de Mello, J. C., Fernandes, G. R., Vibranovski, M. D., and Pereira, L. V. (2017). Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci Rep. 7:10794. doi: 10.1038/s41598-017-11044-z
Morton, L. M., Wang, S. S., Devesa, S. S., Hartge, P., Weisenburger, D. D., and Linet, M. S. (2006). Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood 107, 265–276. doi: 10.1182/blood-2005-06-2508
Nesterova, T. B., Wei, G., Coker, H., Pintacuda, G., Bowness, J. S., Zhang, T., et al. (2019). Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10:3129. doi: 10.1038/s41467-019-11171-11173
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi: 10.1016/j.cell.2015.07.047
Patrat, C., Okamoto, I., Diabangouaya, P., Vialon, V., Le Baccon, P., Chow, J., et al. (2009). Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 5198–5203. doi: 10.1073/pnas.0810683106
Peeters, S. B., Korecki, A. J., Simpson, E. M., and Brown, C. J. (2018). Human cis-acting elements regulating escape from X-chromosome inactivation function in mouse. Hum. Mol. Genet. 27, 1252–1262. doi: 10.1093/hmg/ddy039
Pessoa, B. S., Slump, D. E., Ibrahimi, K., Grefhorst, A., van Veghel, R., Garrelds, I. M., et al. (2015). Angiotensin II type 2 receptor- and acetylcholine-mediated relaxation: essential contribution of female sex hormones and chromosomes. Hypertension 66, 396–402. doi: 10.1161/HYPERTENSIONAHA.115.05303
Petropoulos, S., Edsgard, D., Reinius, B., Deng, Q., Panula, S. P., Codeluppi, S., et al. (2016). Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 167, 285. doi: 10.1016/j.cell.2016.08.009
Pintacuda, G., Wei, G., Roustan, C., Kirmizitas, B. A., Solcan, N., Cerase, A., et al. (2017). hnRNPK recruits PCGF3/5-PRC1 to the xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–969.e10. doi: 10.1016/j.molcel.2017.11.013
Pollex, T., and Heard, E. (2019). Nuclear positioning and pairing of X-chromosome inactivation centers are not primary determinants during initiation of random X-inactivation. Nat. Genet. 51, 285–295. doi: 10.1038/s41588-018-0305-307
Prothero, K. E., Stahl, J. M., and Carrel, L. (2009). Dosage compensation and gene expression on the mammalian X chromosome: one plus one does not always equal two. Chromosome Res. 17, 637–648. doi: 10.1007/s10577-009-9063-9069
Rao, S. S., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021
Reinius, B., and Sandberg, R. (2015). Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat. Rev. Genet. 16, 653–664. doi: 10.1038/nrg3888
Reinius, B., Shi, C., Hengshuo, L., Sandhu, K. S., Radomska, K. J., Rosen, G. D., et al. (2010). Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11:614. doi: 10.1186/1471-2164-11-614
Ricketts, C. J., and Linehan, W. M. (2015). Gender specific mutation incidence and survival associations in clear cell renal cell carcinoma (CCRCC). PLoS One 10:e0140257. doi: 10.1371/journal.pone.0140257
Ryan, V. H., Dignon, G. L., Zerze, G. H., Chabata, C. V., Silva, R., Conicella, A. E., et al. (2018). Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465.e–479.e. doi: 10.1016/j.molcel.2017.12.022
Sakakibara, Y., Nagao, K., Blewitt, M., Sasaki, H., Obuse, C., and Sado, T. (2018). Role of SmcHD1 in establishment of epigenetic states required for the maintenance of the X-inactivated state in mice. Development 145:dev166462. doi: 10.1242/dev.166462
Scandaglia, M., Lopez-Atalaya, J. P., Medrano-Fernandez, A., Lopez-Cascales, M. T., Del Blanco, B., Lipinski, M., et al. (2017). Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep. 21, 47–59. doi: 10.1016/j.celrep.2017.09.014
Shoubridge, C., Harvey, R. J., and Dudding-Byth, T. (2019). IQSEC2 mutation update and review of the female-specific phenotype spectrum including intellectual disability and epilepsy. Hum. Mutat. 40, 5–24. doi: 10.1002/humu.23670
Skakkebaek, A., Nielsen, M. M., Trolle, C., Vang, S., Hornshoj, H., Hedegaard, J., et al. (2018). DNA hypermethylation and differential gene expression associated with klinefelter syndrome. Sci. Rep. 8:13740. doi: 10.1038/s41598-018-31780-31780
Sousa, L. B. D. E., Jonkers, I., Syx, L., Dunkel, I., Chaumeil, J., Picard, C., et al. (2019). Kinetics of Xist-induced gene silencing can be predicted from combinations of epigenetic and genomic features. Genome Res. 29, 1087–1099. doi: 10.1101/gr.245027.118
Souyris, M., Cenac, C., Azar, P., Daviaud, D., Canivet, A., Grunenwald, S., et al. (2018). TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 3:eaa8855. doi: 10.1126/sciimmunol.aap8855
Splinter, E., de Wit, E., Nora, E. P., Klous, P., van de Werken, H. J., Zhu, Y., et al. (2011). The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25, 1371–1383. doi: 10.1101/gad.633311
Sun, T., Plutynski, A., Ward, S., and Rubin, J. B. (2015). An integrative view on sex differences in brain tumors. Cell Mol. Life Sci. 72, 3323–3342. doi: 10.1007/s00018-015-1930-1932
Syrett, C. M., and Anguera, M. C. (2019). When the balance is broken: X-linked gene dosage from two X chromosomes and female-biased autoimmunity. J. Leukoc. Biol. doi: 10.1002/JLB.6RI0319-094R [Epub ahead of print].
Syrett, C. M., Paneru, B., Sandoval-Heglund, D., Wang, J., Banerjee, S., Sindhava, V., et al. (2019). Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4:126751. doi: 10.1172/jci.insight.126751
Trolle, C., Nielsen, M. M., Skakkebaek, A., Lamy, P., Vang, S., Hedegaard, J., et al. (2016). Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci Rep. 6:34220. doi: 10.1038/srep34220
Tsuchiya, K. D., Greally, J. M., Yi, Y., Noel, K. P., Truong, J. P., and Disteche, C. M. (2004). Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res. 14, 1275–1284. doi: 10.1101/gr.2575904
Tukiainen, T., Villani, A. C., Yen, A., Rivas, M. A., Marshall, J. L., Satija, R., et al. (2017). Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248. doi: 10.1038/nature24265
Valentin-Vega, Y. A., Wang, Y. D., Parker, M., Patmore, D. M., Kanagaraj, A., Moore, J., et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 6:25996. doi: 10.1038/srep25996
van Bemmel, J. G., Galupa, R., Gard, C., Servant, N., Picard, C., Davies, J., et al. (2019). The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 51, 1024–1034. doi: 10.1038/s41588-019-0412-410
Van der Meulen, J., Sanghvi, V., Mavrakis, K., Durinck, K., Fang, F., Matthijssens, F., et al. (2015). The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21. doi: 10.1182/blood-2014-05-577270
Viuff, M., Skakkebaek, A., Nielsen, M. M., Chang, S., and Gravholt, C. H. (2019). Epigenetics and genomics in turner syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 181, 68–75. doi: 10.1002/ajmg.c.31683
Wainer Katsir, K., and Linial, M. (2019). Human genes escaping X-inactivation revealed by single cell expression data. BMC Genomics 20:201. doi: 10.1186/s12864-019-5507-5506
Wang, C. Y., Jegu, T., Chu, H. P., Oh, H. J., and Lee, J. T. (2018). SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174, 406–421e425. doi: 10.1016/j.cell.2018.05.007
Wang, J., Syrett, C. M., Kramer, M. C., Basu, A., Atchison, M. L., and Anguera, M. C. (2016). Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. U.S.A. 113, E2029–E2038. doi: 10.1073/pnas.1520113113
Wutz, A., Rasmussen, T. P., and Jaenisch, R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. doi: 10.1038/ng820
Xu, J., Deng, X., Watkins, R., and Disteche, C. M. (2008). Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 28, 4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008
Yang, F., Deng, X., Ma, W., Berletch, J. B., Rabaia, N., Wei, G., et al. (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16:52. doi: 10.1186/s13059-015-0618-610
Yatsenko, S. A., Wood-Trageser, M., Chu, T., Jiang, H., and Rajkovic, A. (2019). A high-resolution X chromosome copy-number variation map in fertile females and women with primary ovarian insufficiency. Genet. Med. doi: 10.1038/s41436-019-0505-502 [Epub ahead of print].
Yue, M., Ogawa, A., Yamada, N., Charles Richard, J. L., Barski, A., and Ogawa, Y. (2017). Xist RNA repeat E is essential for ASH2L recruitment to the inactive X and regulates histone modifications and escape gene expression. PLoS Genet. 13:e1006890. doi: 10.1371/journal.pgen.1006890
Zhang, L. F., Huynh, K. D., and Lee, J. T. (2007). Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129, 693–706. doi: 10.1016/j.cell.2007.03.036
Zhao, J., Sun, B. K., Erwin, J. A., Song, J. J., and Lee, J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. doi: 10.1126/science.1163045
Zhou, Q., Wang, T., Leng, L., Zheng, W., Huang, J., Fang, F., et al. (2019). Single-cell RNA-seq reveals distinct dynamic behavior of sex chromosomes during early human embryogenesis. Mol. Reprod. Dev. 86, 871–882. doi: 10.1002/mrd.23162
Keywords: X chromosome, X inactivation, 3D-structure, LncRNAs, escape gene, epigenetics, dosage
Citation: Fang H, Disteche CM and Berletch JB (2019) X Inactivation and Escape: Epigenetic and Structural Features. Front. Cell Dev. Biol. 7:219. doi: 10.3389/fcell.2019.00219
Received: 02 July 2019; Accepted: 18 September 2019;
Published: 01 October 2019.
Edited by:
Celine Morey, UMR7216 Epigénétique et Destin Cellulaire, FranceReviewed by:
Joseph Mauro Calabrese, The University of North Carolina at Chapel Hill, United StatesCopyright © 2019 Fang, Disteche and Berletch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine M. Disteche, Y2Rpc3RlY2hAdXcuZWR1; Joel B. Berletch, am9lbGIwN0B1dy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.