- 1Institute of Cancer and Genomic Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham, United Kingdom
- 2Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru, India
- 3Cellular Immunology Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
- 4Department of Physiological Science, Santa Casa de São Paulo School of Medical Sciences, São Paulo, Brazil
Autophagy is an intracellular degradation pathway for malfunctioning aggregation-prone proteins, damaged organelles, unwanted macromolecules and invading pathogens. This process is essential for maintaining cellular and tissue homeostasis that contribute to organismal survival. Autophagy dysfunction has been implicated in the pathogenesis of diverse human diseases, and therefore, therapeutic exploitation of autophagy is of potential biomedical relevance. A number of chemical screening approaches have been established for the drug discovery of autophagy modulators based on the perturbations of autophagy reporters or the clearance of autophagy substrates. These readouts can be detected by fluorescence and high-content microscopy, flow cytometry, microplate reader and immunoblotting, and the assays have evolved to enable high-throughput screening and measurement of autophagic flux. Several pharmacological modulators of autophagy have been identified that act either via the classical mechanistic target of rapamycin (mTOR) pathway or independently of mTOR. Many of these autophagy modulators have been demonstrated to exert beneficial effects in transgenic models of neurodegenerative disorders, cancer, infectious diseases, liver diseases, myopathies as well as in lifespan extension. This review describes the commonly used chemical screening approaches in mammalian cells and the key autophagy modulators identified through these methods, and highlights the therapeutic benefits of these compounds in specific disease contexts.
Introduction
Macroautophagy, herein referred to as autophagy, is an intracellular degradation process essential for ensuring cellular homeostasis. This well-conserved catabolic process mediates the targeted degradation of unwanted or excess cytoplasmic materials, such as aggregation-prone proteins, pathogens and damaged organelles like mitochondria, amongst others (Ravikumar et al., 2010). This process is also involved in the bulk degradation of cytoplasmic macromolecules and recycling of the breakdown products especially during nutrient deprivation to provide energy homeostasis, thereby forming a crucial connection between anabolism and catabolism (Boya et al., 2013; Kaur and Debnath, 2015). Due to its vital function as a homeostatic regulator, impairment of the autophagy is implicated in several human pathologies including certain cancer, metabolic syndromes, infectious diseases, liver diseases, myopathies, aging and neurodegenerative disorders (Mizushima et al., 2008). Therefore, therapeutic modulation of autophagy holds great potential in the development of treatment strategies for these diseases (Rubinsztein et al., 2012).
Autophagy is evolutionarily-conserved from yeast to humans. The de novo formation of phagophores, the double-membrane structures that expand to form double-membrane vesicles called autophagosomes, require multiple autophagy-related (Atg) genes in the autophagic machinery, such as the Atg5-Atg12-Atg16 complex and the phosphatidylethanolamine-conjugated microtubule-associated protein 1 light chain 3 (LC3-II) (Kabeya et al., 2000; Mizushima et al., 2011; Ktistakis and Tooze, 2016). Maturation of autophagosomes into the degradative autolysosomes occurs either via the multi-step route involving the fusion of autophagosomes with late endosomes to form amphisomes which subsequently fuse with the lysosomes, or via the direct route involving the fusion between autophagosomes and the lysosomes (Nakamura and Yoshimori, 2017). The autophagic cargo engulfed by the autophagosomes are ultimately degraded in the acidic autolysosomes by the lysosomal hydrolases, which are only active at the low pH maintained by the vacuolar-type H+-ATPase (V-ATPase) on the lysosomal membrane (Saftig and Klumperman, 2009). Finally, the breakdown products are recycled and utilized as inputs to cellular metabolism for energy generation (Rabinowitz and White, 2010). The rate at which this dynamic turnover of cellular contents occurs through the process of autophagy is referred to as autophagic flux. Autophagic flux encompasses all stages of autophagy which includes autophagosome formation, fusion with the lysosomes and cargo degradation in the autolysosomes (Figure 1).
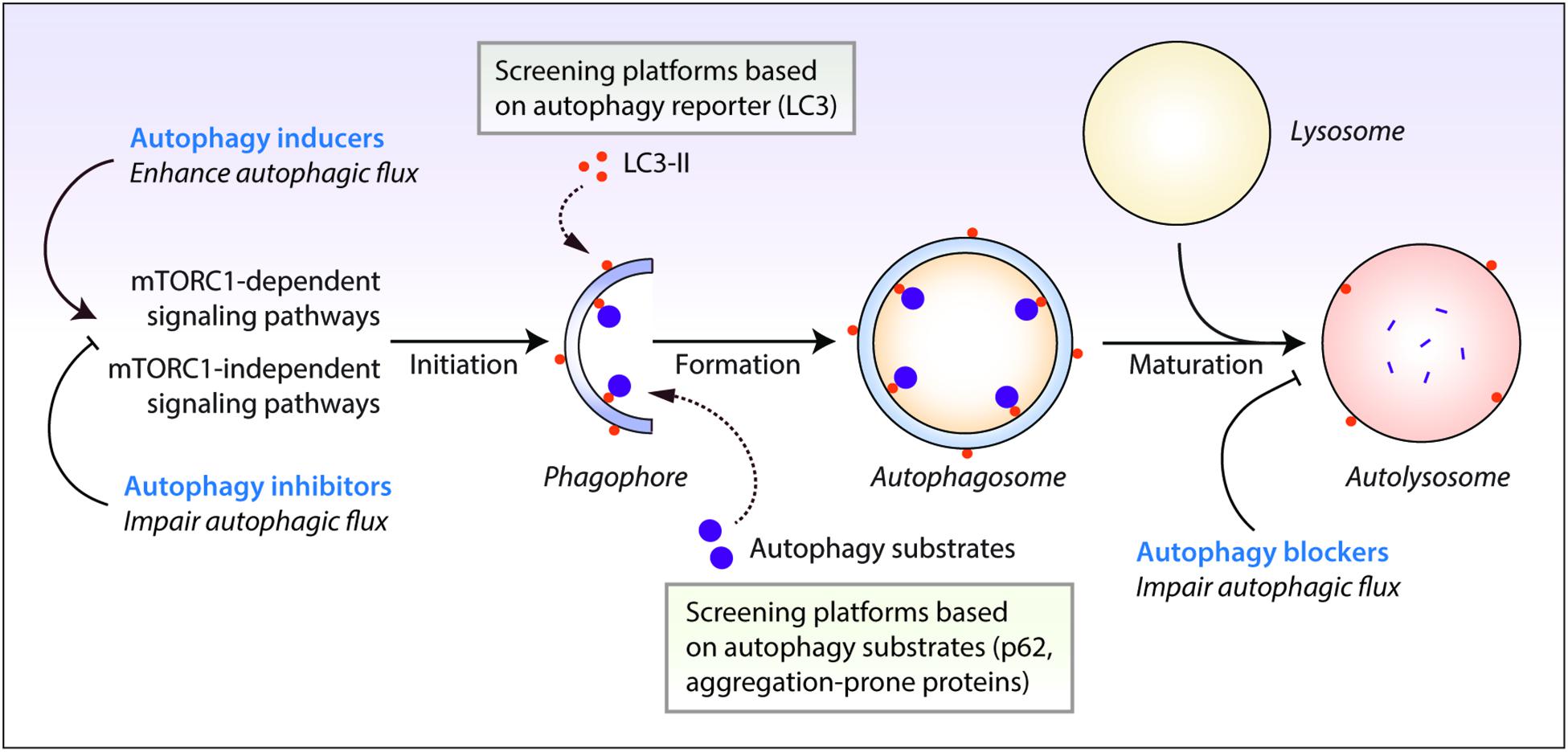
Figure 1. Autophagy reporter and substrate based screening strategies and the impact of autophagy modulators at different stages of the autophagy process. Autophagy is regulated by the mechanistic target of rapamycin complex 1 (mTORC1) or mTORC1-independent pathways. This process initiates by the formation of phagophores that expand and engulf autophagy substrates to form autophagosomes, which then fuse with the lysosomes to form autolysosomes where the autophagic cargo is degraded. Autophagy inducers and inhibitors increase or decrease autophagosome formation, respectively, at the early stages of autophagy, whereas autophagy blockers prevent lysosomal degradation and/or autophagosome maturation at late stages of autophagy. Autophagic flux is thus enhanced by autophagy inducers but is retarded by autophagy inhibitors and blockers. Chemical screening methods for identifying autophagy modulators are commonly based on the readouts of perturbations in autophagy reporters such as LC3-II, or autophagy substrate clearance such as aggregation-prone proteins or p62/SQSTM1.
Key upstream modulators of autophagy include the mechanistic target of rapamycin complex 1 (mTORC1) pathway, which promotes cellular biosynthesis and inhibits autophagy (Saxton and Sabatini, 2017). Regulation of autophagosome formation by mTORC1 is mediated via the ULK1–Atg13–FIP200 complex; mTORC1 suppresses autophagy under nutrient-rich conditions by phosphorylation-dependent inactivation of ULK1 and Atg13 (Mizushima, 2010; Zachari and Ganley, 2017). Various signals such as growth factors and nutrients impinge on mTORC1 to negatively influence autophagy (Kim and Guan, 2015). Conversely, during nutrient starvation, autophagy is promoted by inhibition of the mTORC1 activity (Carroll et al., 2014; Russell et al., 2014). Furthermore, ULK1 can be directly phosphorylated and activated by the energy sensor AMPK to stimulate autophagy (Egan et al., 2011; Kim et al., 2011). In addition, several mTORC1-independent pathways have been described where autophagy is negatively regulated by the elevation in intracellular inositol, Ca2+ and nitric oxide levels, amongst others (Sarkar, 2013b). Molecular mediators of the late stage of autophagy involving autophagosome maturation include Rab7, SNAREs (N-ethylmaleimide-sensitive factor-attachment protein receptors), GABARAPs, BRUCE and Beclin1-interacting partners such as Atg14L, UVRAG and Ambra1 (He and Levine, 2010; Nguyen et al., 2016; Wang et al., 2016; Reggiori and Ungermann, 2017; Ebner et al., 2018). At a transcriptional level, autophagy is governed by the transcription factor EB (TFEB) (Settembre et al., 2011), which in itself is activated by lysosomal Ca2+ (Medina et al., 2015).
Chemical modulation of autophagy by targeting the mTOR-dependent and mTOR-independent pathways has proven to be of potential biomedical relevance due to therapeutic advantages, especially in neurodegenerative disorders as well as in diverse human pathological conditions such as in certain liver diseases, myopathies, infectious diseases, metabolic diseases, cancer and aging (Rubinsztein et al., 2012; Sarkar, 2013b; Levine et al., 2015). Hence, the discovery of potent small molecules regulating autophagy is of great interest. Here we review the chemical screening strategies for autophagy drug discovery, and highlight the potential benefits of autophagy modulators in human diseases.
Chemical Screening Strategies for Identifying Autophagy Modulators
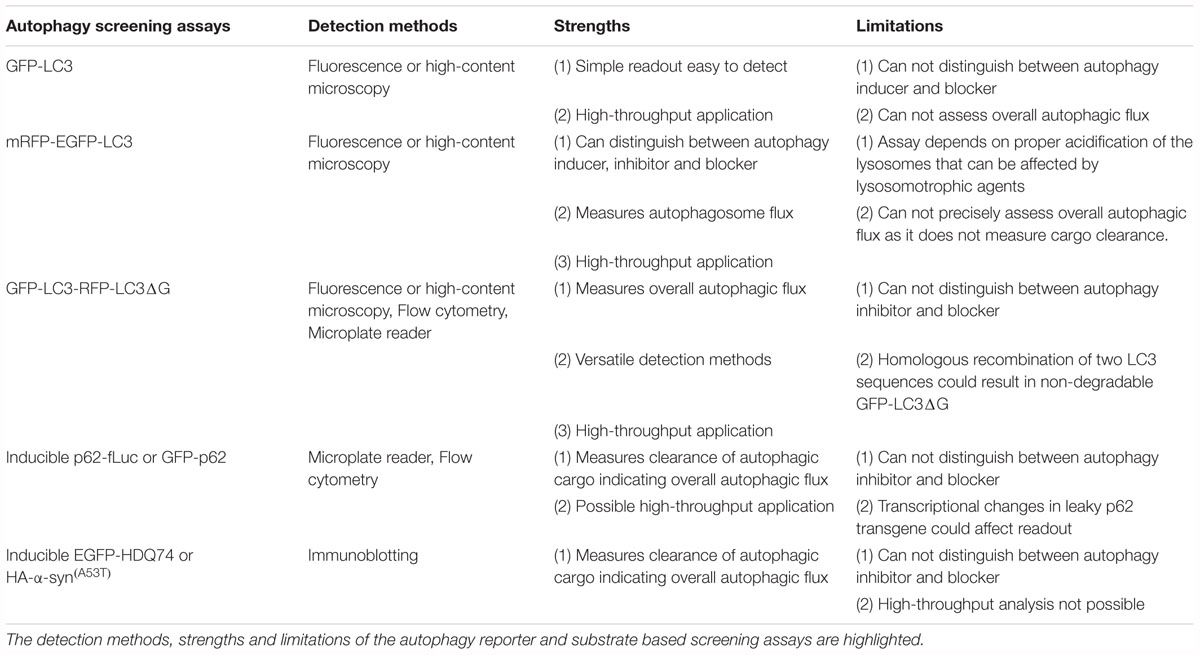
A number of in vitro screening methods have been designed for identifying compounds (Sarkar, 2013a; Joachim et al., 2015; Seranova et al., 2019). The assays are primarily based on the perturbations of autophagy reporters or autophagy cargoes as readouts (Figure 1), which can be measured via fluorescence or high-content imaging, immunoblotting, flow cytometry and microplate reader (Mizushima et al., 2010; Klionsky et al., 2016; Figure 2 and Table 1). Some of these screening methods can be subjected to high-throughput applications. Below are descriptions of the common screening approaches in mammalian cells, and the identification and therapeutic benefits of key autophagy modulators.
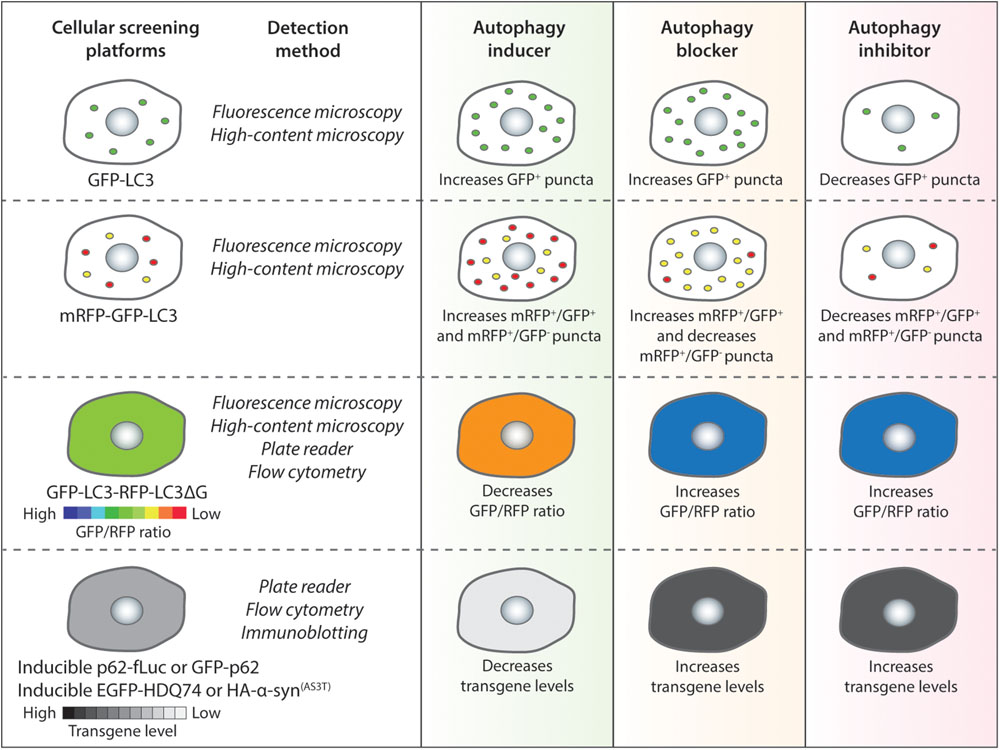
Figure 2. Autophagy chemical screening strategies in mammalian cells. Chemical screening methods that are commonly used for identifying autophagy modulators are based on autophagy reporters (LC3) or autophagy substrates (p62 or aggregation-prone proteins). The detection methods for the respective assays and the expected readouts for autophagy inducers, blockers or inhibitors are indicated as a general guidance.
Chemical Screening Methods Based on Autophagy Reporters
Screening methods based on autophagy reporters are the most commonly used approaches to detect changes in the numbers of autophagosomes and autolysosomes (Table 1). The protein reporter that is widely used to study autophagy is microtubule-associated protein 1 (MAP1) light chain 3 (LC3). The nascent LC3 is cleaved at its C-terminal arginine residue by Atg4 to form the cytoplasmic LC3-I, which is then post-translationally conjugated with phosphatidylethanolamine at its C-terminal glycine residue by Atg7 to form the autophagosome-associated LC3-II (Kabeya et al., 2000). The lipidated LC3-II remains associated to the autophagosomes throughout their lifespan, and is present on both the outer and inner membranes. Following the maturation of autophagosomes with lysosomes to form autolysosomes, the LC3-II on the inner surface is degraded whereas the LC3-II on the outer surface is delipidated and removed by Atg4B for recycling (Tanida et al., 2004). A number of fluorescent-tagged reporters of LC3, such as GFP-LC3 (Kabeya et al., 2000), mRFP-GFP-LC3 (Kimura et al., 2007) and GFP-LC3-RFP-LC3ΔG (Kaizuka et al., 2016), have been used to study autophagy and undertake chemical screening.
Identification of Autophagy Modulators by GFP-LC3 Screening Method
The most common LC3-based reporter that has been used in several studies is GFP-LC3, which labels autophagosomes, autolysosomes as well as phagophores (Kabeya et al., 2000). For the GFP-LC3 screening method, image-based analysis is done by quantifying the GFP+ puncta per cell to measure perturbations in autophagosome number. In general, an autophagy inducer as well as an autophagy blocker will increase GFP-LC3 puncta whereas an autophagy inhibitor will decrease GFP-LC3 puncta (Figure 2). A number of high-throughput and small-scale screens have been undertaken with this strategy that has been also utilized to assess the key hits arising from other screening methods; and some of the primary chemical screens utilizing GFP-LC3 readout are highlighted below.
Using GFP-LC3 as the primary screening method in a stable human glioblastoma H4 cell line, an image-based chemical screen with 480 bioactive compounds was performed wherein the number, size and intensity of GFP-LC3 spots were taken into consideration while selecting potent autophagy modulators (Zhang et al., 2007). Compounds were treated at 3–12 μM concentrations for 24 h. This screen identified 8 autophagy inducers, which included a number of FDA-approved drugs such as fluspirilene, trifluoperazine, pimozide (antipsychotic drugs), niguldipine, nicardipine, amiodarone (drugs used for cardiovascular conditions) and loperamide (used in diarrhea). While fluspirilene, trifluoperazine are dopamine antagonists, the other drugs are Ca2+ channel antagonists that lower intracellular Ca2+; all of which induced autophagy independently of mTOR (Zhang et al., 2007). Another image-based chemical screen was performed with a library of 3584 pharmacologically active compounds in human breast cancer MCF-7 cells stably expressing GFP-LC3 (Balgi et al., 2009). Treatment of compounds was done at ∼15 μM concentration for 4 h. This screen identified 3 FDA-approved drugs such as perhexilene, niclosamide and amiodarone, as well as rottlerin, as autophagy inducers; all of which were shown to inhibit mTORC1 (Balgi et al., 2009). However, other screens have reported amiodarone (Ca2+ channel antagonist) to act independently of mTORC1 for inducing autophagy at a much lower dose than what is required to inhibit mTORC1 (Williams et al., 2008); and likewise, perhexilene is a Ca2+ channel blocker that could be also mTOR-independent. Furthermore, one of the largest chemical screens for identifying autophagy modulators was undertaken in HeLa cells stably expressing GFP-LC3 with 59541 stereochemically and skeletally diverse compounds derived from diversity-oriented synthesis (Kuo et al., 2015). Compounds were treated for 4 h in 8-point dose with a maximal concentration of 10 μM. Several hits were subjected to a secondary screen at 10 μM concentration from which BRD5631 was identified as the potent autophagy inducer along with other hits like BRD2716 and BRD34009; all of which did not affect mTOR activity. Interestingly, the hit rate in the primary screen for compounds having an alkyl amine was higher than that for all of the compounds. This effect was augmented by the additional presence of a single lipophilic group, such as diphenyl alkyne, biphenyl, cyclohexane or naphthalene (Kuo et al., 2015). While the above screens were undertaken in immortalized human cell lines, another chemical screen was done with 1280 pharmacologically active compounds in mouse embryonic fibroblasts (MEFs) stably expressing GFP-LC3 (Li et al., 2016). Compounds were treated at 0.02–46 μM concentrations for 16 h in the presence or absence of chloroquine (autophagy blocker) to determine their effects on autophagic flux. Out of the 27 autophagy inducers identified, few were characterized further. These include anti-psychotic drugs such as indatraline hydrochloride (dopamine inhibitor), chlorpromazine hydrochloride and fluphenazine dihydrochloride (dopamine receptor antagonists). Fluphenazine was found to inhibit mTORC1 whereas indatraline and chlorpromazine were mTOR-independent (Li et al., 2016).
Although GFP-LC3 is a straightforward, widely-used screening assay, its inability to distinguish between autophagosomes and autolysosomes is a major inadequacy of this reporter. Accumulation of autophagosomes can occur either due to induction of autophagosome formation (by autophagy inducers) or due to block in autophagosome maturation (by autophagy blockers) in the early and late stages of autophagy, respectively (Rubinsztein et al., 2009). Since autophagy is a dynamic, multi-step process, it is imperative to measure autophagosome flux in order to assess the status of autophagy. Therefore, the hits from the primary GFP-LC3 screen are subjected to rigorous secondary assays (such as autophagosome formation and maturation, and autophagic substrate clearance, amongst others) (Mizushima et al., 2010; Klionsky et al., 2012) for characterizing autophagy modulators.
Identification of Autophagy Modulators by mRFP-GFP-LC3 Screening Method
In order to overcome the problem of the GFP-LC3 reporter, a tandem fluorescent-tagged mRFP-GFP-LC3 reporter can be employed to determine autophagosome maturation for distinguishing between the autophagosomes and the autolysosomes. This mRFP-GFP-LC3 reporter is pH-sensitive. When overexpressed in cells, the autophagosomes exhibit both mRFP and GFP signals, whereas the autolysosomes emit only mRFP signal because the acid-labile GFP signal is quenched in the acidic environment (Kimura et al., 2007). For the mRFP-GFP-LC3 screening method, image-based analysis is done by quantifying the mRFP+ and GFP+ puncta per cell to measure perturbations in the number of autophagosomes (mRFP+/GFP+) and autolysosomes (mRFP+/GFP-). In general, an autophagy inducer (acting at early stage) will increase autophagosomes and autolysosomes, an autophagy inhibitor (acting at early stage) will decrease both these compartments, whereas an autophagy blocker (acting at late stage) will increase autophagosomes and decrease autolysosomes (Figure 2). Alternative versions of the mRFP-GFP-LC3 reporter have been described that may provide better readouts. These include replacing mRFP with mCherry that has superior photostability over mRFP (Pankiv et al., 2007), and substituting GFP with mWasabi that is more acid-sensitive than GFP (Zhou et al., 2012).
This pH-sensitive reporter has been primarily utilized as a secondary screening strategy following primary screens utilizing the more simpler GFP-LC3 method. In a high-throughput screen with 59541 compounds in GFP-LC3 platform, 400 screen hits were subjected to additional screening in stable HeLa cells expressing mCherry-GFP-LC3 (Kuo et al., 2015). These compounds were treated at 10 μM concentration for 24 h, after which 250 compounds increased (putative inducers) and 80 compounds decreased (putative inhibitors/blockers) the number of mCherry+/GFP- autolysosomes. Following further characterization, potent mTOR-independent autophagy inducers identified were BRD5631, BRD2716, and BRD34009 (Kuo et al., 2015). In another study, HeLa cells stably expressing mRFP-GFP-LC3 was subjected to three drug libraries such as the Prestwick Chemical Library, Microsource Spectrum 2000 library and Johns Hopkins Library that encompass 3791 compounds including FDA-approved drugs and bioactive molecules (Chauhan et al., 2015). Compounds were treated at 10 μM concentration for 4 h. However, high-content image analysis was done based only on GFP-LC3 puncta and total integrated area per cell, but not together with mRFP-LC3 that was utilized later during secondary characterization. 80 compounds were identified, out of which 55 were novel and 25 were previously reported as autophagy modulators. Further characterization of the hits including the mRFP-GFP-LC3 analysis identified flubendazole as a novel autophagy inducer that is also an antihelminthic drug. Flubendazole was shown to impact on dynamic and acetylated microtubules to inhibit mTOR and disrupt Bcl2-Beclin 1 complex for inducing autophagy (Chauhan et al., 2015). More recently, a primary screen with mRFP-GFP-LC3 has been performed in U343 glioma cell spheroids (3D tumor spheroids) by dynamic live-cell imaging (Pampaloni et al., 2017). A subset of the Enzo Life Sciences Screen-Well Natural Compounds library comprising of 94 compounds were used at 1, 12.5, and 50 μM concentrations, followed by long-term time-lapse fluorescence imaging over 24 h at an interval of 1 h. Instead of measuring puncta formation, this study quantified the readout based on the ratio of mRFP and GFP emission intensities over time. Apart from validating this approach with the Enzo Life Sciences Screen-Well Autophagy library consisting of known autophagy modulators, the screen with selected natural compounds identified six potent autophagy inducers and four inhibitors. The autophagy-inducing natural compounds include PI-103, nonactin, valinomycin, quercetin, ivermectin, and harmine (Pampaloni et al., 2017).
The mRFP-GFP-LC3 reporter or its alternative versions can be subjected to high-throughput image-based screens to analyse autophagosome flux. This assay requires proper acidification of the lysosomes that could be affected by lysosomotrophic agents. However, autophagic substrate clearance along with other secondary assays should be assessed following the primary screen in order to assess the overall autophagic flux.
Identification of Autophagy Modulators by GFP-LC3-RFP-LC3ΔG Screening Method
A novel autophagy probe, GFP-LC3-RFP-LC3ΔG, has been recently developed for evaluating autophagic flux that can be used for high-throughput screening approaches (Kaizuka et al., 2016). When overexpressed in cells, the Atg4 family proteases can cleave this reporter into equimolar amounts of GFP-LC3 and RFP-LC3ΔG. While GFP-LC3 on the autophagosomes is degraded or recycled after fusion with the lysosomes, RFP-LC3ΔG cannot be lipidated due to a deletion in its C-terminal glycine and thus remains in the cytosol serving as an internal control. This GFP-LC3-RFP-LC3ΔG reporter can be subjected to both qualitative (by ratiometric imaging via fluorescence microscopy) and quantitative (via microplate reader or flow cytometry) analyses by measuring the fluorescence of GFP-LC3 and RFP-LC3ΔG, and then calculating the GFP/RFP ratio (Kaizuka et al., 2016). Autophagy inducers are expected to decrease GFP/RFP ratio by enhancing autophagic flux, whereas autophagy inhibitors or blockers will increase GFP/RFP ratio by reducing autophagic flux (Figure 2).
Two chemical screens employing the GFP-LC3-RFP-LC3ΔG screening method have been undertaken using a selected library of 34 known autophagy-regulating compounds and 1054 approved drugs under basal or starvation conditions in HeLa cells stably expressing this reporter (Kaizuka et al., 2016). The GFP/RFP ratio was calculated from fluorescence measurement via a microplate reader. For the first screen with known autophagy-regulating compounds, cells were treated for 6, 12 or 24 h with concentrations previously shown to modulate autophagy. A number of known autophagy modulators, but not all, acted as expected primarily after 12 or 24 h treatment. Specifically, autophagy inducers such as rapamycin (Blommaart et al., 1995) and Torin 1 (Thoreen et al., 2009) decreased GFP/RFP ratio whereas autophagy blockers like bafilomycin A1 (Yamamoto et al., 1998) and chloroquine (Seglen et al., 1979) increased GFP/RFP ratio (Kaizuka et al., 2016). For the second screen with approved drug library, cells were treated for 24 h at 10 μM concentration with few exceptions at 5 μM. The screen hits included 47 autophagy-inducing drugs (comprising of certain anti-cancer drugs, antibiotics and cardiotonic drugs) and 43 autophagy inhibitory drugs. Although many of these hits were previously reported, 13 inducers and 18 inhibitors/blockers were identified as novel autophagy modulators, of which some of the novel autophagy inducers were adefovir pivoxil, methyltestosterone, norethisterone, oxaprozin, and zidovudine (Kaizuka et al., 2016). This GFP-LC3-RFP-LC3ΔG probe has been demonstrated to be capable of measuring basal and induced autophagic flux in Zebrafish and in tissues of transgenic mice (Kaizuka et al., 2016), and is thus valuable for monitoring autophagic flux in vivo.
Although this reporter can be used for high-throughput applications and in vivo studies to measure the overall autophagic flux, it is not ideal for investigating the distinct stages of autophagy such as autophagosome formation and maturation. Importantly, the two LC3 sequences of GFP-LC3-RFP-LC3ΔG in retrovirally transfected cells can undergo homologous recombination, which will generate GFP-LC3ΔG that is incapable of being degraded by autophagy. In addition, the expression levels of this reporter define the accuracy of the readout, and hence analysis in different cell lines or tissues will require comparable expression (Kaizuka et al., 2016; Geng and Klionsky, 2017).
Chemical Screening Methods Based on Autophagy Substrates
In addition to the screening approaches based on LC3 reporters, autophagy substrate clearance has also been utilized as a primary screening assay for identifying autophagy modulators (Table 1). This method measures the autophagic cargo flux, which together with LC3-based secondary assays for autophagosome flux can indicate the overall autophagic flux.
Identification of Autophagy Modulators by Clearance of Aggregation-Prone Proteins
A number of neurodegeneration-associated aggregation-prone proteins are predominantly degraded by autophagy (Menzies et al., 2017), and hence screening methods can be based on their clearance as readouts (Sarkar, 2013a). The well-established substrates undergoing autophagic degradation include mutant huntingtin (with expanded polyglutamine repeats) and mutant α-synuclein (A53T or A30P mutants) associated with Huntington’s and Parkinson’s disease, respectively (Webb et al., 2003; Ravikumar et al., 2004). Since the steady-state level of proteins is not ideal for accurately reflecting any impact on their degradation, stable inducible cell lines are required for analyzing autophagic substrate clearance where the transgene product is temporally synthesized by doxycycline followed by treatment with compounds after the expression is turned off (Wyttenbach et al., 2001; Webb et al., 2003; Sarkar et al., 2009). In general, autophagy inducers will enhance the clearance of aggregation-prone proteins, whereas autophagy inhibitors or blockers will retard their clearance (Figure 2).
Independent studies using a stable inducible PC12 cell line expressing EGFP-tagged mutant huntingtin (EGFP-HDQ74) identified mTOR-independent autophagy inducers such as trehalose (Sarkar et al., 2007a) as well as inositol-lowering agents (lithium, carbamazepine, valproic acid, L-690330) (Sarkar et al., 2005) and nitric oxide synthase inhibitors (L-NAME) (Sarkar et al., 2011). These studies also identified autophagy inhibitory compounds such as agents increasing inositol or inositol 1,4,5-trisphosphate (IP3) levels (myo-inositol, prolyl endopeptidase inhibitor 2) (Sarkar et al., 2005) and nitric oxide donors (DEA NONOate, DETA NONOate) (Sarkar et al., 2011). Utilizing stable inducible PC12 cell line expressing hemagglutinin (HA)-tagged A53T α-synuclein (HA-α-syn(A53T)) as the primary screening method, a chemical screen was undertaken with 72 hits arising from an yeast screen involving 50729 compounds (Sarkar et al., 2007b). Cells were treated with compounds at 2 mg mL-1 concentration for 24 h after the initial doxycycline-induced synthesis of the transgene product (A53T α-synuclein), followed by immunoblotting analysis to measure its clearance. A number of novel autophagy modulators were identified which enhanced the autophagy substrate clearance. These include 4 small molecule enhancers of rapamycin (SMERs) and 13 small molecule inhibitors of rapamycin (SMIRs), of which SMER10, SMER18, and SMER28 were characterized to be autophagy inducers acting independently of mTOR. Further screening of the chemical analogs of these SMERs identified 18 additional autophagy inducers, such as 1 SMER10, 7 SMER18 and 10 SMER28 analogs that are capable of enhancing substrate clearance; although not substantially better than the respective parent compounds (Sarkar et al., 2007b). Another screen also utilizing a stable inducible PC12 cell line expressing HA-tagged A30P α-synuclein (HA-α-syn(A30P)) was undertaken with a library of 253 compounds including FDA-approved drugs and pharmacological probes (Williams et al., 2008). Drug treatment was done at 1 μM for 24 h after the synthesis of the transgene product, followed by immunoblotting analysis. This study elucidated a cyclic mTOR-independent autophagy pathway with multiple drug targets, in which cAMP regulates IP3 levels that impact on calpain activity, which in turn activates Gsα that regulates cAMP levels. Some of the autophagy-inducing compounds identified include L-type Ca2+ channel blockers (verapamil, loperamide, amiodarone), calpain inhibitors (calpastatin), ATP-sensitive K+ channel agonist (minoxidil), cAMP reducing agents (rilmenidine, clonidine) and inositol lowering agents (valproic acid), whereas Ca2+ channel openers [(±)-Bay K8644] and agents elevating cAMP (dibutyryl cAMP, forskolin) and cytosolic Ca2+ (thapsigargin) levels were autophagy inhibitory (Williams et al., 2008). In addition to these immunoblotting based methods, the effects of autophagy modulators on autophagy-dependent clearance of EGFP-tagged mutant huntingtin aggregates can be validated by fluorescence microscopy in wild-type (Atg5+/+) and autophagy-deficient (Atg5-/-) mouse embryonic fibroblasts (MEFs) (Kuma et al., 2004; Sarkar et al., 2009).
Although autophagic clearance of aggregation-prone proteins is informative for autophagic flux, only low-throughput approaches are possible that creates a major hurdle for high-throughput applications. Nonetheless, this method could be used as a secondary assay for characterization of selected hits arising from screens with LC3-based reporters.
Identification of Autophagy Modulators by p62/SQSTM1 Clearance
An alternative approach to the clearance of aggregation-prone proteins is to monitor the autophagic degradation of a known autophagy substrate, p62/SQSTM1, which also functions as an adaptor protein during selective autophagy for recruiting specific autophagic cargo to the autophagosomes (Bjorkoy et al., 2005; Pankiv et al., 2007). Similarly, to the method involving aggregation-prone proteins, screening approaches based on p62 clearance would ideally require a stable inducible cell line where the transgene product is temporally expressed before the treatment with compounds. The p62 reporters, such as GFP-p62 (Larsen et al., 2010) or luciferase-tagged p62 (Brown et al., 2016; Min et al., 2018), could be utilized for medium- to high-throughput screens by flow cytometry or microplate reader (for analyzing p62 levels) or by fluorescence imaging (for analyzing p62 aggregates). Genetic screens have been undertaken with p62-based reporters (Pietrocola et al., 2015; Strohecker et al., 2015; DeJesus et al., 2016; Hale et al., 2016), and therefore, similar chemical screening approaches are also possible. In addition, analyzing the steady-state levels of endogenous p62 by immunoblotting is often used as a secondary assay for characterization of autophagy modulators (Klionsky et al., 2012). It is expected that an autophagy inducer will decrease p62 levels or aggregates, whereas an autophagy inhibitor or blocker will cause its accumulation (Figure 2). Recently, an assay based on LC3B-II and p62 time-resolved fluorescence resonance energy transfer (TR-FRET) has been described to monitor autophagy independent of any exogenous labels. This method is based on the proximity of the donor and the acceptor antibodies of LC3-II and p62, in which autophagy inducers increase LC3-II signal and decrease p62 signal, autophagy inhibitors do not display any turnover of either signals, whereas autophagy blockers will increase LC3-II signal without any turnover of p62 signal (Bresciani et al., 2018).
Although p62 is a specific autophagy substrate in most mammalian cell lines (Klionsky et al., 2012), its autophagic degradation should be confirmed in the cell-type and the time-points to be used in the screens. Moreover, transcriptional upregulation of p62 has been reported during some instances of autophagy activation, such as under prolonged starvation or with certain pharmacological inducers (Klionsky et al., 2012; Sahani et al., 2014; Kuo et al., 2015), and therefore, any perturbation in p62 protein levels needs to be accompanied by qPCR assessment of its mRNA levels.
Biomedical Applications of Autophagy Modulators in Human Diseases
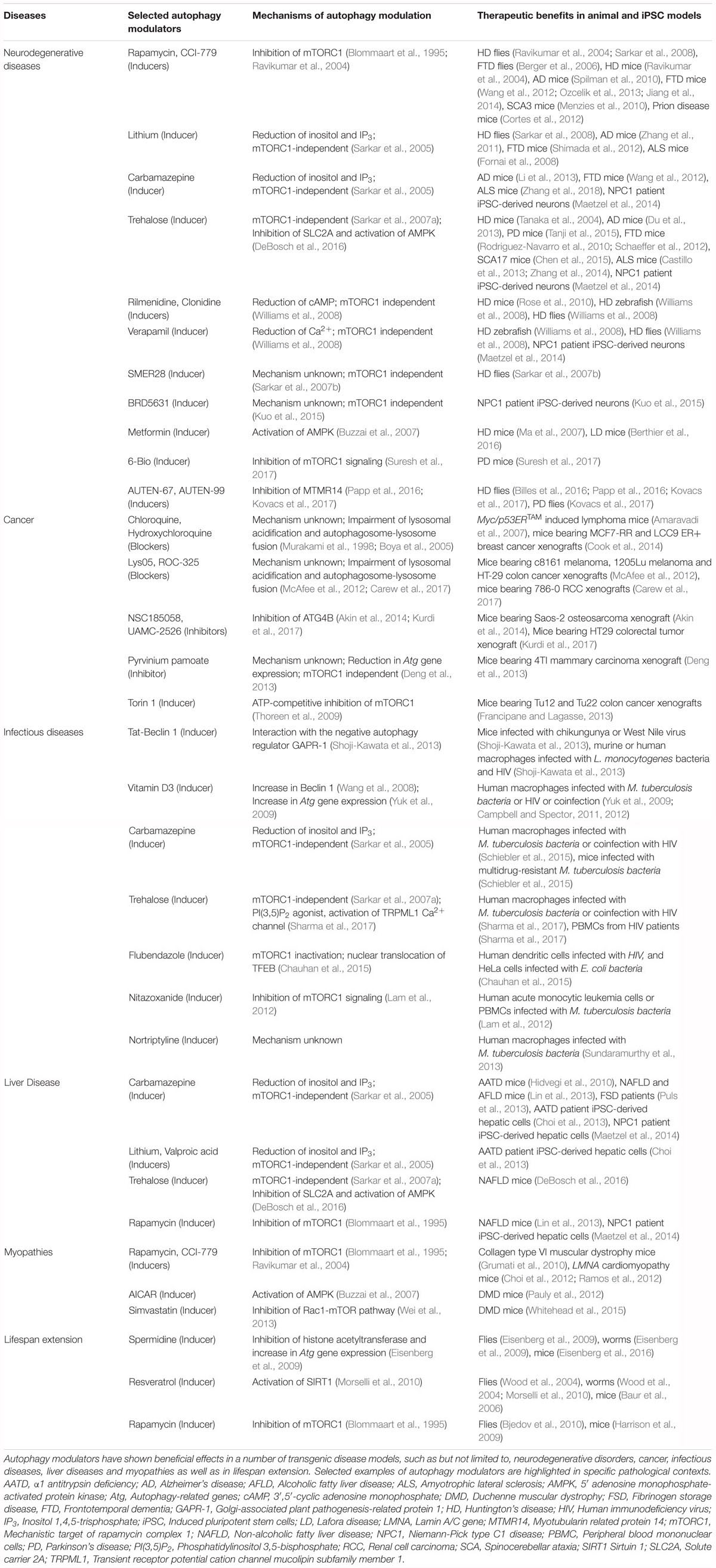
Autophagy plays an essential role for tissue homeostasis and cellular survival by removing unwanted materials like malfunctioning aggregated proteins and damaged organelles from the cells; however, deregulation of this process could contribute to cytotoxicity (Mizushima et al., 2008). Autophagy dysfunction has been implicated in the pathogenesis of diverse human diseases (Levine and Kroemer, 2008; Jiang and Mizushima, 2014), and therefore, therapeutic exploitation of autophagy is of potential biomedical relevance (Figure 3). A number of independent studies and chemical screens have identified several autophagy modulators, which have been shown to impart beneficial effects in various transgenic disease models (Table 2; Rubinsztein et al., 2012; Sarkar, 2013b; Levine et al., 2015). Some of the key studies in specific disease contexts are highlighted below.
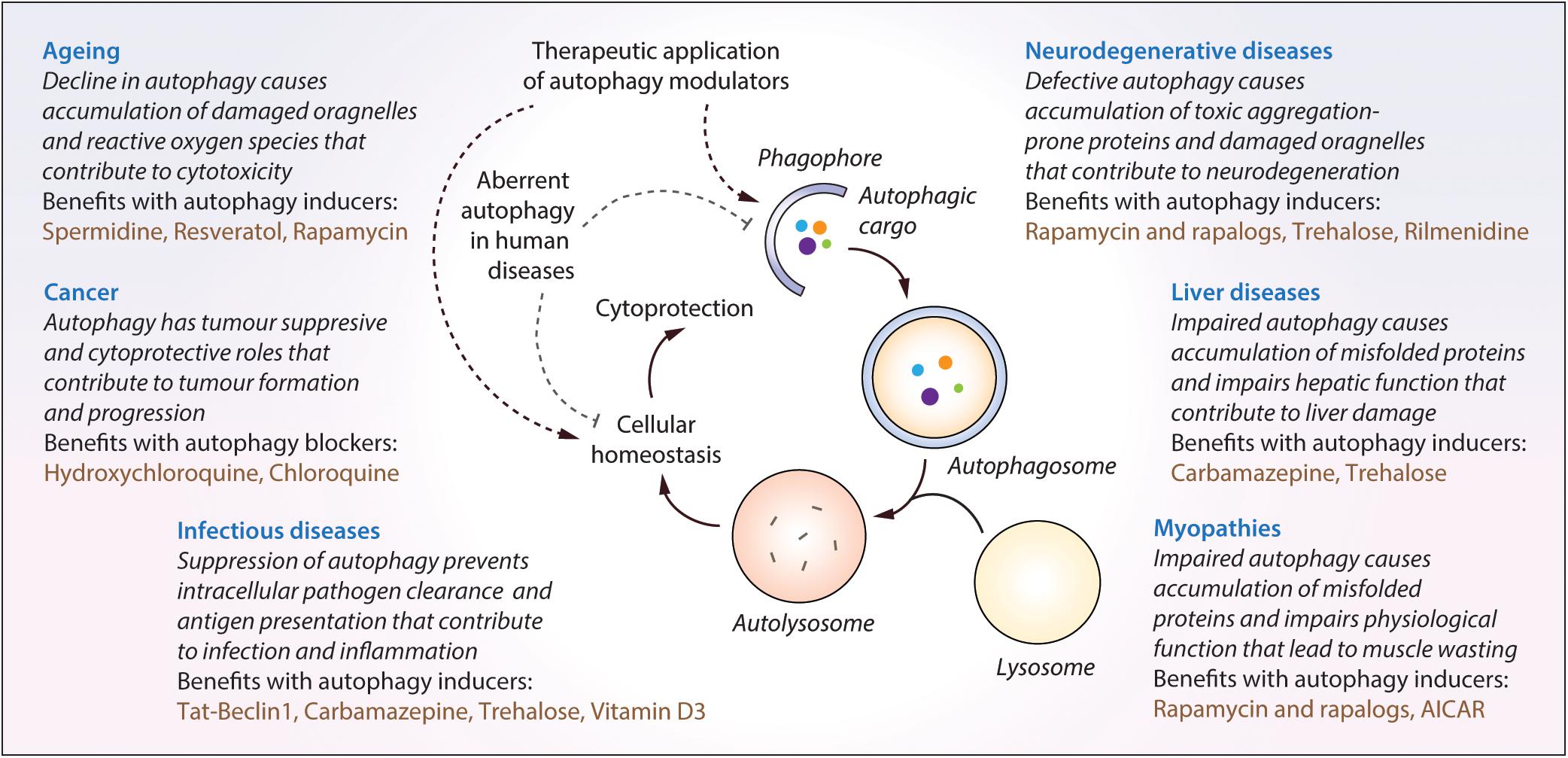
Figure 3. The impact of malfunctioning autophagy and the therapeutic benefits of autophagy modulators in diverse human diseases. Autophagy is implicated in diverse human diseases due to its vital role in maintaining cellular homeostasis. Defective or aberrant autophagy contributes to the cytotoxicity underlying many pathological conditions whereas pharmacological upregulation of autophagy is beneficial in various transgenic models. Key autophagy modulators exerting therapeutic benefits in neurodegenerative disorders, cancer, infectious diseases, liver diseases, myopathies and lifespan extension, as well as the impact of malfunctioning autophagy in these contexts, are highlighted.
Autophagy Modulators in Neurodegenerative Diseases
Basal autophagy in the brain is critical for maintaining cellular homeostasis in post-mitotic cells like neurons, which is evident from the genetic studies in mice where brain-specific deletion of essential autophagy genes resulted in neurodegenerative phenotypes (Hara et al., 2006; Komatsu et al., 2006). Particularly, autophagy is the primary degradation pathway for several aggregation-prone proteins associated with neurodegeneration (Rubinsztein, 2006; Nixon, 2013). However, defective autophagy has been reported in several neurodegenerative diseases, including neurodegenerative lysosomal storage disorders, and is considered a major causative factor for neurodegeneration (Nixon, 2013; Sarkar, 2013b; Menzies et al., 2017; Seranova et al., 2017). Therefore, induction of autophagy for enhancing the clearance of mutant aggregation-prone proteins is considered a potential treatment strategy. The therapeutic benefits of autophagy inducers have been robustly demonstrated in the context of neurodegeneration where upregulation of autophagy was protective in several in vitro and in vivo transgenic models of neurodegenerative diseases (Rubinsztein et al., 2012; Sarkar, 2013b; Levine et al., 2015; Seranova et al., 2017). Stimulating autophagy with mTOR inhibitors like rapamycin or its analogs had beneficial effects in fly and mouse models of Huntington’s disease, Alzheimer’s disease (AD), Parkinson’s disease (PD), frontotemporal dementia (FTD), spinocerebellar ataxia type 3 (SCA3) and prion disease (Ravikumar et al., 2004; Berger et al., 2006; Sarkar et al., 2008; Menzies et al., 2010; Spilman et al., 2010; Cortes et al., 2012; Wang et al., 2012; Ozcelik et al., 2013; Jiang et al., 2014). Likewise, several mTOR-independent autophagy inducers such as, but not limited to, lithium, carbamazepine (inositol lowering agents), rilmenidine (cAMP reducing agent), trehalose (AMPK activator), SMERs and BRD5631 have been shown to be protective in fly, Zebrafish, mouse or induced pluripotent stem cell (iPSC) models of AD, FTD, HD, amyotrophic lateral sclerosis (ALS) and Niemann-Pick type C1 (NPC1) disease (Sarkar et al., 2005, 2007a,b; Fornai et al., 2008; Williams et al., 2008; Rose et al., 2010; Zhang et al., 2011, 2018; Shimada et al., 2012; Wang et al., 2012; Li et al., 2013; Maetzel et al., 2014; Kuo et al., 2015). The most widely used mTOR-independent autophagy inducer in vivo is trehalose (Sarkar et al., 2007a), a disaccharide that stimulates autophagy by inhibiting SLC2A family of glucose transporters and activating AMPK (DeBosch et al., 2016), which in turn can directly influence the phosphorylation of the autophagy-initiating kinase ULK1 (Egan et al., 2011; Kim et al., 2011). Remarkably, trehalose had beneficial effects in mouse models of AD, PD, HD, FTD, SCA17, ALS, as well as cellular and iPSC-derived neuronal models of prion and NPC1 disease, respectively (Tanaka et al., 2004; Aguib et al., 2009; Rodriguez-Navarro et al., 2010; Schaeffer et al., 2012; Castillo et al., 2013; Du et al., 2013; Zhang et al., 2014; Chen et al., 2015; Tanji et al., 2015). Additional autophagy-inducing agents reported to be cytoprotective in neurodegenerative models such as HD, PD, ALS, FTD and Lafora disease include Tat-Beclin 1 peptide, calpastatin, verapamil, metformin, AUTEN-67, AUTEN-99, 6-Bio and fluphenazine (Ma et al., 2007; Williams et al., 2008; Shoji-Kawata et al., 2013; Barmada et al., 2014; Berthier et al., 2016; Billes et al., 2016; Papp et al., 2016; Kovacs et al., 2017; Suresh et al., 2017). A combinatorial approach in enhancing autophagy has been shown with rapamycin and mTOR-independent autophagy inducers such as lithium, trehalose or SMERs. Higher efficacy was achieved via the additive effects of dual treatment on autophagy induction and cytoprotection in cell and fly models of HD than the effects of single compounds (Sarkar et al., 2007a,b, 2008).
Autophagy Modulators in Cancer
The ability of autophagy in the maintenance of metabolic homeostasis has drawn considerable attention as a potential target for cancer therapy via its pro-survival and pro-death mechanisms (Rabinowitz and White, 2010; Levy et al., 2017). Autophagy plays tumor suppressive role by mitigating oxidative stress, removing superfluous mitochondria and preventing DNA damage and genome instability; and on the other hand, shows pro-tumor activity by preventing the induction of tumor suppressors, increasing resistance to apoptosis and maintaining tumor metabolism through recycling of nutrients (Mathew et al., 2007; Galluzzi et al., 2015; Kimmelman and White, 2017). Depending on the cancer context and the opposing effects of autophagy, either inhibitors or inducers of autophagy could be exploited for cancer therapy (Galluzzi et al., 2017; Levy et al., 2017). Since autophagy promotes tumorigenesis in most contexts, inhibition of autophagy has gathered considerable interest for cancer therapy. Accumulating evidence demonstrate that autophagy inhibitors/blockers exerted therapeutic benefits in cancer models. The clinically- approved autophagy inhibitors chloroquine or hydroxychloroquine (HCQ), which impair lysosomal acidification and block autophagic flux (Murakami et al., 1998; Boya et al., 2005), caused tumor shrinkage in preclinical studies; and thus HCQ being more potent with lesser side-effects is used in ongoing clinical trials either alone or in combination with other treatments (Briceno et al., 2003; Amaravadi et al., 2007; Cook et al., 2014; Chude and Amaravadi, 2017; Levy et al., 2017; Onorati et al., 2018). Autophagy inhibitory compounds, such as Lys05 and ROC-325, which exhibited anti-tumor activity in mice have been suggested to be more potent than HCQ (McAfee et al., 2012; Carew et al., 2017). In addition, autophagy inhibitors preventing autophagosome formation such as ATG4B antagonists (compounds NSC185058 and UAMC-2526), Vps34 (vacuolar protein sorting protein 34) inhibitor (compound SAR405), ULK1 (Unc-51-like kinase 1) inhibitor (compound SBI-0206965), USP10/USP13 (ubiquitin-specific peptidases) inhibitor (Spautin-1) and agents causing transcriptional inhibition of autophagy genes (pyrvinium pamoate), also exerted anti-proliferative and anti-tumor effects in cellular and in vivo models of cancer (Liu et al., 2011; Deng et al., 2013; Akin et al., 2014; Ronan et al., 2014; Shao et al., 2014; Egan et al., 2015; Kurdi et al., 2017). On the contrary, various chemical agents or natural products exerting antiproliferative or anti-tumor activity either alone or in combination with chemotherapeutic agents could induce autophagy or autophagic cell death, which include Torin 1, AC-73, MC-4, metformin, silibinin, Abrus agglutinin, curcumin, liensinine, spermidine, vitamin D3, and imatinib (Buzzai et al., 2007; Ertmer et al., 2007; Wang et al., 2008; Thoreen et al., 2009; Qian et al., 2011; Francipane and Lagasse, 2013; Law et al., 2014; Jiang et al., 2016; Pietrocola et al., 2016; Panda et al., 2017; Son et al., 2018; Spinello et al., 2018).
Autophagy Modulators in Infectious Diseases
Autophagy plays an important role in innate defense mechanism by removing intracellular pathogens; a process termed xenophagy (Levine et al., 2011; Deretic et al., 2013). The role of autophagy in regulating intracellular infections initially emerged through studies on Mycobacterium tuberculosis (Mtb) (Gutierrez et al., 2004; Singh et al., 2006). Subsequently, several other bacterial pathogens like Salmonella and Listeria, and viral pathogens like HIV and Dengue were shown to utilize host autophagy pathways for their own advantage (Jia et al., 2009; Kyei et al., 2009; Yoshikawa et al., 2009; Heaton and Randall, 2010). A genome-wide siRNA screen to identify host factors required for intracellular Mtb survival within macrophages revealed that a large number of host factors acted via regulation of autophagy to help the bacteria (Kumar et al., 2010). Induction of autophagy with rapamycin, carbamazepine, SMER28, and vitamin D3 were shown to prevent bacterial survival or HIV replication in macrophages (Gutierrez et al., 2004; Floto et al., 2007; Yuk et al., 2009; Kumar et al., 2010; Campbell and Spector, 2011, 2012; Schiebler et al., 2015). Notably, carbamazepine reduced bacterial burden, improved lung pathology and stimulated adaptive immunity in mice infected with multidrug-resistant Mtb (Schiebler et al., 2015). Rapamycin also controlled viral and bacterial pathogens both in vitro and in vivo (Donia et al., 2010). In an integrated chemical and RNAi screening for modulators of intracellular mycobacteria, one of the top three compounds was nortriptyline which significantly suppressed Mtb survival within macrophages and induced autophagy (Sundaramurthy et al., 2013). Other compounds limiting bacterial or HIV infections through activation of autophagic flux were nitazoxanide (anti-protozoan drug) and flubendazole (antihelminthic drug) (Lam et al., 2012; Chauhan et al., 2015). Similarly, the naturally occurring disaccharide trehalose, a potent mTOR-independent enhancer of autophagy in diverse cell-types (Sarkar et al., 2007a), can also induce autophagy and xenophagy in Mtb-infected macrophages that resulted in the killing of bacteria (Sharma et al., 2017). In this study, trehalose was found to act as a PI(3,5)P2 (phosphatidylinositol 3,5-bisphosphate) agonist for activating the lysosomal Ca2+ channel TRPML1 (Sharma et al., 2017), which in turn released lysosomal Ca2+ that caused nuclear translocation of TFEB to induce autophagy (Medina et al., 2015). Excitingly, trehalose also seemed to be effective during HIV-Mtb co-infection and limits Mtb survival by reversing the HIV-mediated block in autophagy flux (Sharma et al., 2017). Similarly, vitamin D3 could also kill Mtb during HIV co-infection by inducing autophagy (Campbell and Spector, 2012). Several host factors currently being tested for anti-Mtb therapeutics function by regulating host autophagy and xenophagy. For example, inhibition of host Src kinase by the compound AZD0530 induced autophagy and lysosomal maturation to clear Mtb (Chandra et al., 2016). A pioneering anti-infective, autophagy-inducing agent is Tat-Beclin 1, which is a peptide representing a region of the autophagy regulator Beclin 1 that interacts with the HIV-1 accessory protein NEF, and this domain is linked with the HIV-1 Tat transduction domain to make it cell permeable (Shoji-Kawata et al., 2013). Tat-Beclin 1 prevented the replication of a number of viral and bacterial pathogens in vitro in autophagy-dependent manner, as well as induced autophagy and anti-viral activity in mice infected with chikungunya or West Nile virus (Shoji-Kawata et al., 2013). Thus, it is evident that regulators of autophagy and xenophagy have tremendous potential for novel therapeutics against various infectious diseases. It is now clear that within an infected host cell, there is a possibility of uncoupling between homeostatic autophagy and anti-bacterial xenophagy (Chandra et al., 2015; Sharma et al., 2018). Therefore, it is desirable to perform chemical screening pertaining to infection-specific xenophagy flux for identifying novel regulators of bacterial/viral survival within the host cells through the autophagy pathway.
Autophagy Modulators in Liver Diseases
Liver autophagy is essential for various hepatic functions and is implicated in various liver conditions including α1-antitrypsin (AAT) deficiency, non-alcoholic fatty liver disease (NAFLD), hepatocellular carcinoma and viral hepatitis (Rautou et al., 2010; Ueno and Komatsu, 2017). Chemical modulation of autophagy has been shown to have beneficial effects in some of these diseases. Carbamazepine, an mTOR independent autophagy inducer acting by reducing inositol levels (Sarkar et al., 2005), reduced hepatic load of mutant α1-antitrypsin Z and hepatic fibrosis in a mouse model of AAT deficiency (Hidvegi et al., 2010), as well as decreased hepatocellular aggregate-related toxicity in patients suffering from fibrinogen storage disease (Puls et al., 2013). A high-throughput drug screen in hepatocyte-like cells derived from iPSC lines of patients with AAT deficiency also revealed inositol-lowering autophagy-inducing agents, such as carbamazepine, lithium, and valproic acid, in facilitating the clearance mutant AAT (Choi et al., 2013). Carbamazepine as well as the mTOR inhibitor rapamycin also rescued dysfunctional autophagic flux and improved cell viability in hepatic-like cells differentiated from patient-derived iPSC lines of Niemann-Pick type C1 (NPC1) disease (Maetzel et al., 2014). In addition, autophagy induction with trehalose, carbamazepine, rapamycin or hydrogen sulfide reduced steatosis, lipid accumulation and liver injury in high-fat diet-induced NAFLD in mice (Lin et al., 2013; Sun et al., 2015; DeBosch et al., 2016). Furthermore, the anti-diabetic drug metformin, which indirectly inhibits mTOR, induced SIRT1-mediated autophagy in primary hepatocytes and ameliorated hepatic steatosis in vivo (Song et al., 2015). Overall, these studies indicate that activation of autophagy via inhibition of mTOR, lowering inositol levels or with trehalose are effective modes of inducing autophagy in the liver.
Autophagy Modulators in Myopathies
Basal autophagy is required for maintaining muscle mass and myofiber integrity (Masiero et al., 2009), and thus deregulation of muscle autophagy is implicated in myopathies and muscular dystrophies (Sandri et al., 2013). Sustained activation of mTORC1 in skeletal muscle of TSC1-deficient mice could cause late-onset myopathy related to suppression of autophagy (Castets et al., 2013). Upregulation of autophagy, primarily by inhibiting the mTORC1 pathway, has been reported to have beneficial effects in certain transgenic disease models. Autophagy induction by rapamycin or low-protein diet increased myofiber survival and attenuated dystrophic phenotype in a mouse model of collagen type VI muscular dystrophy (Grumati et al., 2010). Likewise, activation of autophagy by dietary changes or with the AMP-activated protein kinase (AMPK) agonist, AICAR (5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside), improved dystrophic phenotypes in mouse models of Duchenne muscular dystrophy (DMD) (De Palma et al., 2012; Pauly et al., 2012). A potential role of simvastatin, which has been reported to induce autophagy by inhibiting the Rac1-mTOR pathway (Wei et al., 2013), has been suggested in improving the physiological function of skeletal muscle in DMD transgenic mice (Whitehead et al., 2015). In addition, rapamycin or its analog, temsirolimus, ameliorated cardiomyopathy and improved skeletal and cardiac muscle function in mouse models of LMNA (lamin A/C gene) cardiomyopathy that recapitulate Emery-Dreifuss muscular dystrophy (EDMD) (Choi et al., 2012; Ramos et al., 2012).
Autophagy Modulators in Lifespan Extension
The functionality of autophagy declines with aging (Rubinsztein et al., 2011), and thus restoring adequate autophagy is considered a possible anti-aging strategy for lifespan extension. There are a number of lifespan expanding strategies, and in many of such approaches, autophagy acts as a common denominator for promoting longevity (Madeo et al., 2010; Hansen et al., 2018). Pharmacological treatment with autophagy inducers has been linked to increasing longevity in transgenic in vivo models (Madeo et al., 2015). Lifespan extension via induction of autophagy with naturally- occurring polyamines such as spermidine, which is an acetyltransferase inhibitor, was shown in yeast, flies, worms and mice (Eisenberg et al., 2009, 2016); and likewise also reported with the natural phenol resveratrol, which is a deacetylase activator, in yeast, flies, worms as well as in mice on high-fat diet (Howitz et al., 2003; Wood et al., 2004; Baur et al., 2006; Morselli et al., 2010). Although both spermidine and resveratrol impacts on the acetylproteome, stimulation of autophagy by resveratrol requires the nicotinamide adenine dinucleotide-dependent deacetylase sirtuin 1 (SIRT1) whereas the effect of spermidine was SIRT1 independent (Morselli et al., 2010, 2011). Inhibition of mTOR by rapamycin also extended lifespan in yeast, flies and mice (Alvers et al., 2009; Harrison et al., 2009; Bjedov et al., 2010; Lamming et al., 2013). In addition, lifespan extension in multiple organisms including mice and apes could be achieved by caloric restriction, which is a physiological inducer of autophagy via AMPK activation, mTORC1 inhibition and SIRT1 activation (Mair and Dillin, 2008; Colman et al., 2009; Mercken et al., 2014; Mattison et al., 2017). In some of these studies reporting lifespan extension by autophagy activation, the role of autophagy has been specifically determined by abolishing the anti-aging effects via knockdown of essential autophagy genes (Madeo et al., 2015; Nakamura and Yoshimori, 2018).
Conclusion
The methodologies for measuring autophagy have evolved over the past decade and it is now feasible to undertake high-throughput chemical screens for identifying modulators of autophagic flux. A number of pharmacological modulators of autophagy have been identified via screening approaches or individual studies; some of which have been demonstrated to exert therapeutic benefits in diverse human diseases. Most of the key autophagy modulators have been identified either by the GFP-LC3 screening method in HeLa cells or via assessing the clearance of aggregation-prone proteins in inducible PC12 cell lines. While analysis of changes in autophagosome number with GFP-LC3 reporter requires shorter treatment period (such as 8–24 h), analysis of clearance of aggregation-prone proteins requires longer treatment duration (such as 24–72 h) depending on the nature of the transgene product. Following the primary screen, it is pertinent to characterize the high-confidence screen hits with secondary autophagy assays because there are no single assays to determine autophagic flux. These normally include analysis of autophagosome formation with bafilomycin A1 via immunoblotting with anti-LC3 antibody, analysis of autophagosome maturation with mRFP-GFP-LC3 reporter, and analysis of autophagy substrate (p62) clearance via immunoblotting with anti-p62 antibody (Mizushima et al., 2010; Klionsky et al., 2016).
Although the methods described in this review are those that have been generally used in the field, alternative autophagy assays could also be employed for chemical screening. One potential approach is the use of Keima, a fluorescent acid-stable protein that exhibits bimodal excitation spectra in neutral and acidic pH, such as in autophagosomes and autolysosomes, respectively (Katayama et al., 2011). The cumulative fluorescence readout can be used to measure bulk autophagic flux. This protein can also be utilized for selective autophagic flux, such as with mitochondria-targeted Keima to measure mitophagy (Katayama et al., 2011; Sun et al., 2017). However, Keima-based assays solely depend upon the lysosomal acidity and thus cannot be performed in fixed cells where the pH gradient across lysosomal membranes is lost. In addition, other screening approaches could be based on fluorescent-tagged early markers of autophagy initiation, such as with WIPI-1 (Proikas-Cezanne and Pfisterer, 2009) and DFCP1 (Axe et al., 2008); however, these methods will not capture the late events of autophagy pathway involving autophagosome maturation and cargo degradation.
For the therapeutic exploitation of autophagy modulators, mTOR-independent autophagy inducers are generally favorable and considered to have lesser side-effects than the mTOR inhibitors like rapamycin. This is because mTOR controls vital cellular functions like cell growth and translation and thus its inhibition can lead to undesirable side-effects unrelated to autophagy induction. For clinical translation to patients, it is important to determine the efficacy and penetrance of the autophagy modulators in the target organs. Future directions could include identifying specific inducers of autophagy acting at the level of autophagic machinery rather than the upstream signaling pathways.
Author Contributions
PP, AF, SV, DK, and SS wrote the manuscript. ES and SS made the figures. PP and SS made the tables. PP, AF, SV, ES, VS, MC, PD, JT, TR, DK, and SS reviewed the manuscript.
Funding
SS is funded by Wellcome Trust Seed Award (109626/Z/15/Z), UKIERI (UK-India Education and Research Initiative) DST Thematic Partnership Award (2016-17-0087) with DK, FAPESP-Birmingham-Nottingham Strategic Collaboration Fund with TR, and Birmingham Fellowship from the University of Birmingham (UoB). SV is also a Newton Bhabha Ph.D. Placement Fellow (funded by British Council) and TR is also a Brazil Visiting Fellow (funded by UoB) and Rutherford Fellow in SS lab at UoB. SS is also a Former Fellow for life at Hughes Hall, University of Cambridge, United Kingdom.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the funding agencies for supporting our research.
References
Aguib, Y., Heiseke, A., Gilch, S., Riemer, C., Baier, M., Schatzl, H. M., et al. (2009). Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 5, 361–369. doi: 10.4161/auto.5.3.7662
Akin, D., Wang, S. K., Habibzadegah-Tari, P., Law, B., Ostrov, D., Li, M., et al. (2014). A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 10, 2021–2035. doi: 10.4161/auto.32229
Alvers, A. L., Wood, M. S., Hu, D., Kaywell, A. C., Dunn, W. A. Jr., and Aris, J. P. (2009). Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 5, 847–849. doi: 10.4161/auto.8824
Amaravadi, R. K., Yu, D., Lum, J. J., Bui, T., Christophorou, M. A., Evan, G. I., et al. (2007). Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336. doi: 10.1172/JCI28833
Axe, E. L., Walker, S. A., Manifava, M., Chandra, P., Roderick, H. L., Habermann, A., et al. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701. doi: 10.1083/jcb.200803137
Balgi, A. D., Fonseca, B. D., Donohue, E., Tsang, T. C., Lajoie, P., Proud, C. G., et al. (2009). Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4:e7124. doi: 10.1371/journal.pone.0007124
Barmada, S. J., Serio, A., Arjun, A., Bilican, B., Daub, A., Ando, D. M., et al. (2014). Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat, Chem. Biol. 10, 677–685. doi: 10.1038/nchembio.1563
Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., et al. (2006). Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342. doi: 10.1038/nature05354
Berger, Z., Ravikumar, B., Menzies, F. M., Oroz, L. G., Underwood, B. R., Pangalos, M. N., et al. (2006). Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 15, 433–442. doi: 10.1093/hmg/ddi458
Berthier, A., Paya, M., Garcia-Cabrero, A. M., Ballester, M. I., Heredia, M., Serratosa, J. M., et al. (2016). Pharmacological interventions to ameliorate neuropathological symptoms in a mouse model of lafora disease. Mol. Neurobiol. 53, 1296–1309. doi: 10.1007/s12035-015-9091-8
Billes, V., Kovacs, T., Hotzi, B., Manzeger, A., Tagscherer, K., Komlos, M., et al. (2016). AUTEN-67 (Autophagy Enhancer-67) hampers the progression of neurodegenerative symptoms in a Drosophila model of Huntington’s disease. J. Huntingtons Dis. 5, 133–147. doi: 10.3233/JHD-150180
Bjedov, I., Toivonen, J. M., Kerr, F., Slack, C., Jacobson, J., Foley, A., et al. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46. doi: 10.1016/j.cmet.2009.11.010
Bjorkoy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Overvatn, A., et al. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614. doi: 10.1083/jcb.200507002
Blommaart, E. F., Luiken, J. J., Blommaart, P. J., van Woerkom, G. M., and Meijer, A. J. (1995). Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326. doi: 10.1074/jbc.270.5.2320
Boya, P., Gonzalez-Polo, R. A., Casares, N., Perfettini, J. L., Dessen, P., Larochette, N., et al. (2005). Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 25, 1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005
Boya, P., Reggiori, F., and Codogno, P. (2013). Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720. doi: 10.1038/ncb2788
Bresciani, A., Spiezia, M. C., Boggio, R., Cariulo, C., Nordheim, A., Altobelli, R., et al. (2018). Quantifying autophagy using novel LC3B and p62 TR-FRET assays. PLoS One 13:e0194423. doi: 10.1371/journal.pone.0194423
Briceno, E., Reyes, S., and Sotelo, J. (2003). Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg Focus 14:e3. doi: 10.3171/foc.2003.14.2.4
Brown, A., Patel, S., Ward, C., Lorenz, A., Ortiz, M., DuRoss, A., et al. (2016). PEG-lipid micelles enable cholesterol efflux in Niemann-pick type C1 disease-based lysosomal storage disorder. Sci. Rep. 6:31750. doi: 10.1038/srep31750
Buzzai, M., Jones, R. G., Amaravadi, R. K., Lum, J. J., DeBerardinis, R. J., Zhao, F., et al. (2007). Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 67, 6745–6752. doi: 10.1158/0008-5472.CAN-06-4447
Campbell, G. R., and Spector, S. A. (2011). Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J. Biol. Chem. 286, 18890–18902. doi: 10.1074/jbc.M110.206110
Campbell, G. R., and Spector, S. A. (2012). Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog 8:e1002689. doi: 10.1371/journal.ppat.1002689
Carew, J. S., Espitia, C. M., Zhao, W., Han, Y., Visconte, V., Phillips, J., et al. (2017). Disruption of autophagic degradation with ROC-325 antagonizes renal cell carcinoma pathogenesis. Clin. Cancer Res. 23, 2869–2879. doi: 10.1158/1078-0432.CCR-16-1742
Carroll, B., Korolchuk, V. I., and Sarkar, S. (2014). Amino acids and autophagy: cross-talk and co-operation to control cellular homeostasis. Amino Acids 47, 2065–2088. doi: 10.1007/s00726-014-1775-2
Castets, P., Lin, S., Rion, N., Di Fulvio, S., Romanino, K., Guridi, M., et al. (2013). Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 17, 731–744. doi: 10.1016/j.cmet.2013.03.015
Castillo, K., Nassif, M., Valenzuela, V., Rojas, F., Matus, S., Mercado, G., et al. (2013). Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9, 1308–1320. doi: 10.4161/auto.25188
Chandra, P., Ghanwat, S., Matta, S. K., Yadav, S. S., Mehta, M., Siddiqui, Z., et al. (2015). Mycobacterium tuberculosis inhibits RAB7 recruitment to selectively modulate autophagy flux in macrophages. Sci. Rep. 5:16320. doi: 10.1038/srep16320
Chandra, P., Rajmani, R. S., Verma, G., Bhavesh, N. S., and Kumar, D. (2016). Targeting drug-sensitive and resistant strains of Mycobacterium tuberculosis by inhibition of src family kinases lowers disease burden and pathology. mSphere 1, e43–e15. doi: 10.1128/mSphere.00043-15
Chauhan, S., Ahmed, Z., Bradfute, S. B., Arko-Mensah, J., Mandell, M. A., Won Choi, S., et al. (2015). Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat. Commun. 6:8620. doi: 10.1038/ncomms9620
Chen, Z. Z., Wang, C. M., Lee, G. C., Hsu, H. C., Wu, T. L., Lin, C. W., et al. (2015). Trehalose attenuates the gait ataxia and gliosis of spinocerebellar ataxia type 17 mice. Neurochem. Res. 40, 800–810. doi: 10.1007/s11064-015-1530-4
Choi, J. C., Muchir, A., Wu, W., Iwata, S., Homma, S., Morrow, J. P., et al. (2012). Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 4:144ra102. doi: 10.1126/scitranslmed.3003875
Choi, S. M., Kim, Y., Shim, J. S., Park, J. T., Wang, R. H., Leach, S. D., et al. (2013). Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 57, 2458–2468. doi: 10.1002/hep.26237
Chude, C. I., and Amaravadi, R. K. (2017). Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18:1279. doi: 10.3390/ijms18061279
Colman, R. J., Anderson, R. M., Johnson, S. C., Kastman, E. K., Kosmatka, K. J., Beasley, T. M., et al. (2009). Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204. doi: 10.1126/science.1173635
Cook, K. L., Warri, A., Soto-Pantoja, D. R., Clarke, P. A., Cruz, M. I., Zwart, A., et al. (2014). Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin. Cancer Res. 20, 3222–3232. doi: 10.1158/1078-0432.CCR-13-3227
Cortes, C. J., Qin, K., Cook, J., Solanki, A., and Mastrianni, J. A. (2012). Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Straussler-Scheinker disease. J. Neurosci. 32, 12396–12405. doi: 10.1523/JNEUROSCI.6189-11.2012
De Palma, C., Morisi, F., Cheli, S., Pambianco, S., Cappello, V., Vezzoli, M., et al. (2012). Autophagy as a new therapeutic target in duchenne muscular dystrophy. Cell Death Dis. 3:e418. doi: 10.1038/cddis.2012.159
DeBosch, B. J., Heitmeier, M. R., Mayer, A. L., Higgins, C. B., Crowley, J. R., Kraft, T. E., et al. (2016). Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 9, ra21. doi: 10.1126/scisignal.aac5472
DeJesus, R., Moretti, F., McAllister, G., Wang, Z., Bergman, P., Liu, S., et al. (2016). Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. eLife 5:17290. doi: 10.7554/eLife.17290
Deng, L., Lei, Y., Liu, R., Li, J., Yuan, K., Li, Y., et al. (2013). Pyrvinium targets autophagy addiction to promote cancer cell death. Cell Death Dis. 4:e614. doi: 10.1038/cddis.2013.142
Deretic, V., Saitoh, T., and Akira, S. (2013). Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737. doi: 10.1038/nri3532
Donia, M., McCubrey, J. A., Bendtzen, K., and Nicoletti, F. (2010). Potential use of rapamycin in HIV infection. Br. J. Clin. Pharmacol. 70, 784–793. doi: 10.1111/j.1365-2125.2010.03735.x
Du, J., Liang, Y., Xu, F., Sun, B., and Wang, Z. (2013). Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J. Pharm. Pharmacol. 65, 1753–1756. doi: 10.1111/jphp.12108
Ebner, P., Poetsch, I., Deszcz, L., Hoffmann, T., Zuber, J., and Ikeda, F. (2018). The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat. Commun. 9:599. doi: 10.1038/s41467-018-02823-x
Egan, D. F., Chun, M. G., Vamos, M., Zou, H., Rong, J., Miller, C. J., et al. (2015). Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297. doi: 10.1016/j.molcel.2015.05.031
Egan, D. F., Shackelford, D. B., Mihaylova, M. M., Gelino, S., Kohnz, R. A., Mair, W., et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. doi: 10.1126/science.1196371
Eisenberg, T., Abdellatif, M., Schroeder, S., Primessnig, U., Stekovic, S., Pendl, T., et al. (2016). Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 22, 1428–1438. doi: 10.1038/nm.4222
Eisenberg, T., Knauer, H., Schauer, A., Buttner, S., Ruckenstuhl, C., Carmona-Gutierrez, D., et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314. doi: 10.1038/ncb1975
Ertmer, A., Huber, V., Gilch, S., Yoshimori, T., Erfle, V., Duyster, J., et al. (2007). The anticancer drug imatinib induces cellular autophagy. Leukemia 21, 936–942. doi: 10.1038/sj.leu.2404606
Floto, R. A., Sarkar, S., Perlstein, E. O., Kampmann, B., Schreiber, S. L., and Rubinsztein, D. C. (2007). Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy, reduce toxicity in Huntington’s disease models and enhance killing of mycobacteria by macrophages. Autophagy 3, 620–622. doi: 10.4161/auto.4898
Fornai, F., Longone, P., Cafaro, L., Kastsiuchenka, O., Ferrucci, M., Manca, M. L., et al. (2008). Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 105, 2052–2057. doi: 10.1073/pnas.0708022105
Francipane, M. G., and Lagasse, E. (2013). Selective targeting of human colon cancer stem-like cells by the mTOR inhibitor Torin-1. Oncotarget 4, 1948–1962. doi: 10.18632/oncotarget.1310
Galluzzi, L., Bravo-San Pedro, J. M., Levine, B., Green, D. R., and Kroemer, G. (2017). Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511. doi: 10.1038/nrd.2017.22
Galluzzi, L., Pietrocola, F., Bravo-San Pedro, J. M., Amaravadi, R. K., Baehrecke, E. H., Cecconi, F., et al. (2015). Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880. doi: 10.15252/embj.201490784
Geng, J., and Klionsky, D. J. (2017). Direct quantification of autophagic flux by a single molecule-based probe. Autophagy 13, 639–641. doi: 10.1080/15548627.2017.1280646
Grumati, P., Coletto, L., Sabatelli, P., Cescon, M., Angelin, A., Bertaggia, E., et al. (2010). Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 16, 1313–1320. doi: 10.1038/nm.2247
Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., and Deretic, V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766. doi: 10.1016/j.cell.2004.11.038
Hale, C. M., Cheng, Q., Ortuno, D., Huang, M., Nojima, D., Kassner, P. D., et al. (2016). Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy 12, 713–726. doi: 10.1080/15548627.2016.1147669
Hansen, M., Rubinsztein, D. C., and Walker, D. W. (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593. doi: 10.1038/s41580-018-0033-y
Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. doi: 10.1038/nature04724
Harrison, D. E., Strong, R., Sharp, Z. D., Nelson, J. F., Astle, C. M., Flurkey, K., et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395. doi: 10.1038/nature08221
He, C., and Levine, B. (2010). The Beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140–149. doi: 10.1016/j.ceb.2010.01.001
Heaton, N. S., and Randall, G. (2010). Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 8, 422–432. doi: 10.1016/j.chom.2010.10.006
Hidvegi, T., Ewing, M., Hale, P., Dippold, C., Beckett, C., Kemp, C., et al. (2010). An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science 329, 229–232. doi: 10.1126/science.1190354
Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., et al. (2003). Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196. doi: 10.1038/nature01960
Jia, K., Thomas, C., Akbar, M., Sun, Q., Adams-Huet, B., Gilpin, C., et al. (2009). Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U.S.A. 106, 14564–14569. doi: 10.1073/pnas.0813319106
Jiang, C., Jin, S., Jiang, Z., and Wang, J. (2016). Inhibitory effects of silibinin on proliferation and lung metastasis of human high metastasis cell line of salivary gland adenoid cystic carcinoma via autophagy induction. Oncol. Targets Ther. 9, 6609–6618. doi: 10.2147/OTT.S107101
Jiang, P., and Mizushima, N. (2014). Autophagy and human diseases. Cell Res. 24, 69–79. doi: 10.1038/cr.2013.161
Jiang, T., Yu, J. T., Zhu, X. C., Zhang, Q. Q., Cao, L., Wang, H. F., et al. (2014). Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance. Neuropharmacology 85, 121–130. doi: 10.1016/j.neuropharm.2014.05.032
Joachim, J., Jiang, M., McKnight, N. C., Howell, M., and Tooze, S. A. (2015). High-throughput screening approaches to identify regulators of mammalian autophagy. Methods 75, 96–104. doi: 10.1016/j.ymeth.2015.02.002
Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. doi: 10.1093/emboj/19.21.5720
Kaizuka, T., Morishita, H., Hama, Y., Tsukamoto, S., Matsui, T., Toyota, Y., et al. (2016). An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849. doi: 10.1016/j.molcel.2016.09.037
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T., and Miyawaki, A. (2011). A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052. doi: 10.1016/j.chembiol.2011.05.013
Kaur, J., and Debnath, J. (2015). Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472. doi: 10.1038/nrm4024
Kim, J., Kundu, M., Viollet, B., and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Kim, Y. C., and Guan, K. L. (2015). mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 125, 25–32. doi: 10.1172/JCI73939
Kimmelman, A. C., and White, E. (2017). Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043. doi: 10.1016/j.cmet.2017.04.004
Kimura, S., Noda, T., and Yoshimori, T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460. doi: 10.4161/auto.4451
Klionsky, D. J., Abdalla, F. C., Abeliovich, H., Abraham, R. T., Acevedo-Arozena, A., Adeli, K., et al. (2012). Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544. doi: 10.4161/auto.19496
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., et al. (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884. doi: 10.1038/nature04723
Kovacs, T., Billes, V., Komlos, M., Hotzi, B., Manzeger, A., Tarnoci, A., et al. (2017). The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci. Rep. 7:42014. doi: 10.1038/srep42014
Ktistakis, N. T., and Tooze, S. A. (2016). Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 26, 624–635. doi: 10.1016/j.tcb.2016.03.006
Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., et al. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. doi: 10.1038/nature03029
Kumar, D., Nath, L., Kamal, M. A., Varshney, A., Jain, A., Singh, S., et al. (2010). Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743. doi: 10.1016/j.cell.2010.02.012
Kuo, S. Y., Castoreno, A. B., Aldrich, L. N., Lassen, K. G., Goel, G., Dancik, V., et al. (2015). Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl. Acad. Sci. U.S.A. 112, E4281–E4287. doi: 10.1073/pnas.1512289112
Kurdi, A., Cleenewerck, M., Vangestel, C., Lyssens, S., Declercq, W., Timmermans, J. P., et al. (2017). ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem. Pharmacol. 138, 150–162. doi: 10.1016/j.bcp.2017.06.119
Kyei, G. B., Dinkins, C., Davis, A. S., Roberts, E., Singh, S. B., Dong, C., et al. (2009). Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186, 255–268. doi: 10.1083/jcb.200903070
Lam, K. K., Zheng, X., Forestieri, R., Balgi, A. D., Nodwell, M., Vollett, S., et al. (2012). Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog 8:e1002691. doi: 10.1371/journal.ppat.1002691
Lamming, D. W., Ye, L., Sabatini, D. M., and Baur, J. A. (2013). Rapalogs and mTOR inhibitors as anti-aging therapeutics. J. Clin. Invest. 123, 980–989. doi: 10.1172/JCI64099
Larsen, K. B., Lamark, T., Overvatn, A., Harneshaug, I., Johansen, T., and Bjorkoy, G. (2010). A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 6, 784–793. doi: 10.4161/auto.6.6.12510
Law, B. Y., Chan, W. K., Xu, S. W., Wang, J. R., Bai, L. P., Liu, L., et al. (2014). Natural small-molecule enhancers of autophagy induce autophagic cell death in apoptosis-defective cells. Sci. Rep. 4:5510. doi: 10.1038/srep05510
Levine, B., and Kroemer, G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. doi: 10.1016/j.cell.2007.12.018
Levine, B., Mizushima, N., and Virgin, H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323–335. doi: 10.1038/nature09782
Levine, B., Packer, M., and Codogno, P. (2015). Development of autophagy inducers in clinical medicine. J. Clin. Invest. 125, 14–24. doi: 10.1172/JCI73938
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542. doi: 10.1038/nrc.2017.53
Li, L., Zhang, S., Zhang, X., Li, T., Tang, Y., Liu, H., et al. (2013). Autophagy enhancer carbamazepine alleviates memory deficits and cerebral amyloid-beta pathology in a mouse model of Alzheimer’s disease. Curr. Alzheimer Res. 10, 433–441. doi: 10.2174/1567205011310040008
Li, Y., McGreal, S., Zhao, J., Huang, R., Zhou, Y., Zhong, H., et al. (2016). A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacol. Res. 110, 35–49. doi: 10.1016/j.phrs.2016.05.004
Lin, C. W., Zhang, H., Li, M., Xiong, X., Chen, X., Chen, X., et al. (2013). Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 58, 993–999. doi: 10.1016/j.jhep.2013.01.011
Liu, J., Xia, H., Kim, M., Xu, L., Li, Y., Zhang, L., et al. (2011). Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234. doi: 10.1016/j.cell.2011.08.037
Ma, T. C., Buescher, J. L., Oatis, B., Funk, J. A., Nash, A. J., Carrier, R. L., et al. (2007). Metformin therapy in a transgenic mouse model of Huntington’s disease. Neurosci. Lett. 411, 98–103. doi: 10.1016/j.neulet.2006.10.039
Madeo, F., Tavernarakis, N., and Kroemer, G. (2010). Can autophagy promote longevity? Nat. Cell Biol. 12, 842–846. doi: 10.1038/ncb0910-842
Madeo, F., Zimmermann, A., Maiuri, M. C., and Kroemer, G. (2015). Essential role for autophagy in life span extension. J. Clin. Invest. 125, 85–93. doi: 10.1172/JCI73946
Maetzel, D., Sarkar, S., Wang, H., Abi-Mosleh, L., Xu, P., Cheng, A. W., et al. (2014). Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Rep. 2, 866–880. doi: 10.1016/j.stemcr.2014.03.014
Mair, W., and Dillin, A. (2008). Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754. doi: 10.1146/annurev.biochem.77.061206.171059
Masiero, E., Agatea, L., Mammucari, C., Blaauw, B., Loro, E., Komatsu, M., et al. (2009). Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515. doi: 10.1016/j.cmet.2009.10.008
Mathew, R., Karantza-Wadsworth, V., and White, E. (2007). Role of autophagy in cancer. Nat. Rev. Cancer 7, 961–967. doi: 10.1038/nrc2254
Mattison, J. A., Colman, R. J., Beasley, T. M., Allison, D. B., Kemnitz, J. W., Roth, G. S., et al. (2017). Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8:14063. doi: 10.1038/ncomms14063
McAfee, Q., Zhang, Z., Samanta, A., Levi, S. M., Ma, X. H., Piao, S., et al. (2012). Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. U.S.A. 109, 8253–8258. doi: 10.1073/pnas.1118193109
Medina, D. L., Di Paola, S., Peluso, I., Armani, A., De Stefani, D., Venditti, R., et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299. doi: 10.1038/ncb3114
Menzies, F. M., Fleming, A., Caricasole, A., Bento, C. F., Andrews, S. P., Ashkenazi, A., et al. (2017). Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034. doi: 10.1016/j.neuron.2017.01.022
Menzies, F. M., Huebener, J., Renna, M., Bonin, M., Riess, O., and Rubinsztein, D. C. (2010). Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain 133(Pt 1), 93–104. doi: 10.1093/brain/awp292
Mercken, E. M., Hu, J., Krzysik-Walker, S., Wei, M., Li, Y., McBurney, M. W., et al. (2014). SIRT1 but not its increased expression is essential for lifespan extension in caloric-restricted mice. Aging Cell 13, 193–196. doi: 10.1111/acel.12151
Min, Z., Ting, Y., Mingtao, G., Xiaofei, T., Dong, Y., Chenguang, Z., et al. (2018). Monitoring autophagic flux using p62/SQSTM1 based luciferase reporters in glioma cells. Exp. Cell Res. 363, 84–94. doi: 10.1016/j.yexcr.2017.12.027
Mizushima, N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139. doi: 10.1016/j.ceb.2009.12.004
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075. doi: 10.1038/nature06639
Mizushima, N., Yoshimori, T., and Levine, B. (2010). Methods in mammalian autophagy research. Cell 140, 313–326. doi: 10.1016/j.cell.2010.01.028
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Morselli, E., Maiuri, M. C., Markaki, M., Megalou, E., Pasparaki, A., Palikaras, K., et al. (2010). Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 1:e10. doi: 10.1038/cddis.2009.8
Morselli, E., Marino, G., Bennetzen, M. V., Eisenberg, T., Megalou, E., Schroeder, S., et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615–629. doi: 10.1083/jcb.201008167
Murakami, N., Oyama, F., Gu, Y., McLennan, I. S., Nonaka, I., and Ihara, Y. (1998). Accumulation of tau in autophagic vacuoles in chloroquine myopathy. J. Neuropathol. Exp. Neurol. 57, 664–673. doi: 10.1097/00005072-199807000-00003
Nakamura, S., and Yoshimori, T. (2017). New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216. doi: 10.1242/jcs.196352
Nakamura, S., and Yoshimori, T. (2018). Autophagy and longevity. Mol. Cells 41, 65–72. doi: 10.14348/molcells.2018.2333
Nguyen, T. N., Padman, B. S., Usher, J., Oorschot, V., Ramm, G., and Lazarou, M. (2016). Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215, 857–874. doi: 10.1083/jcb.201607039
Nixon, R. A. (2013). The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997. doi: 10.1038/nm.3232
Onorati, A. V., Dyczynski, M., Ojha, R., and Amaravadi, R. K. (2018). Targeting autophagy in cancer. Cancer 124, 3307–3318. doi: 10.1002/cncr.31335
Ozcelik, S., Fraser, G., Castets, P., Schaeffer, V., Skachokova, Z., Breu, K., et al. (2013). Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS One 8:e62459. doi: 10.1371/journal.pone.0062459
Pampaloni, F., Mayer, B., Kabat Vel-Job, K., Ansari, N., Hotte, K., Kogel, D., et al. (2017). A novel cellular spheroid-based autophagy screen applying live fluorescence microscopy identifies nonactin as a strong inducer of autophagosomal turnover. SLAS Discov. 22, 558–570. doi: 10.1177/2472555217696798
Panda, P. K., Behera, B., Meher, B. R., Das, D. N., Mukhopadhyay, S., Sinha, N., et al. (2017). Abrus Agglutinin, a type II ribosome inactivating protein inhibits Akt/PH domain to induce endoplasmic reticulum stress mediated autophagy-dependent cell death. Mol. Carcinog. 56, 389–401. doi: 10.1002/mc.22502
Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J. A., Outzen, H., et al. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145. doi: 10.1074/jbc.M702824200
Papp, D., Kovacs, T., Billes, V., Varga, M., Tarnoci, A., Hackler, L., et al. (2016). AUTEN-67, an autophagy-enhancing drug candidate with potent antiaging and neuroprotective effects. Autophagy 12, 273–286. doi: 10.1080/15548627.2015.1082023
Pauly, M., Daussin, F., Burelle, Y., Li, T., Godin, R., Fauconnier, J., et al. (2012). AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am. J. Pathol. 181, 583–592. doi: 10.1016/j.ajpath.2012.04.004
Pietrocola, F., Lachkar, S., Enot, D. P., Niso-Santano, M., Bravo-San Pedro, J. M., Sica, V., et al. (2015). Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 22, 509–516. doi: 10.1038/cdd.2014.215
Pietrocola, F., Pol, J., Vacchelli, E., Rao, S., Enot, D. P., Baracco, E. E., et al. (2016). Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 30, 147–160. doi: 10.1016/j.ccell.2016.05.016
Proikas-Cezanne, T., and Pfisterer, S. G. (2009). Assessing mammalian autophagy by WIPI-1/Atg18 puncta formation. Methods Enzymol. 452, 247–260. doi: 10.1016/S0076-6879(08)03616-1
Puls, F., Goldschmidt, I., Bantel, H., Agne, C., Brocker, V., Dammrich, M., et al. (2013). Autophagy-enhancing drug carbamazepine diminishes hepatocellular death in fibrinogen storage disease. J. Hepatol. 59, 626–630. doi: 10.1016/j.jhep.2013.05.018
Qian, H., Yang, Y., and Wang, X. (2011). Curcumin enhanced adriamycin-induced human liver-derived Hepatoma G2 cell death through activation of mitochondria-mediated apoptosis and autophagy. Eur. J. Pharm. Sci. 43, 125–131. doi: 10.1016/j.ejps.2011.04.002
Rabinowitz, J. D., and White, E. (2010). Autophagy and metabolism. Science 330, 1344–1348. doi: 10.1126/science.1193497
Ramos, F. J., Chen, S. C., Garelick, M. G., Dai, D. F., Liao, C. Y., Schreiber, K. H., et al. (2012). Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl. Med. 4:144ra103. doi: 10.1126/scitranslmed.3003802
Rautou, P. E., Mansouri, A., Lebrec, D., Durand, F., Valla, D., and Moreau, R. (2010). Autophagy in liver diseases. J. Hepatol. 53, 1123–1134. doi: 10.1016/j.jhep.2010.07.006
Ravikumar, B., Sarkar, S., Davies, J. E., Futter, M., Garcia-Arencibia, M., Green-Thompson, Z. W., et al. (2010). Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435. doi: 10.1152/physrev.00030.2009
Ravikumar, B., Vacher, C., Berger, Z., Davies, J. E., Luo, S., Oroz, L. G., et al. (2004). Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595. doi: 10.1038/ng1362
Reggiori, F., and Ungermann, C. (2017). Autophagosome maturation and fusion. J. Mol. Biol. 429, 486–496. doi: 10.1016/j.jmb.2017.01.002
Rodriguez-Navarro, J. A., Rodriguez, L., Casarejos, M. J., Solano, R. M., Gomez, A., Perucho, J., et al. (2010). Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol. Dis. 39, 423–438. doi: 10.1016/j.nbd.2010.05.014
Ronan, B., Flamand, O., Vescovi, L., Dureuil, C., Durand, L., Fassy, F., et al. (2014). A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019. doi: 10.1038/nchembio.1681
Rose, C., Menzies, F. M., Renna, M., Acevedo-Arozena, A., Corrochano, S., Sadiq, O., et al. (2010). Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum. Mol. Genet. 19, 2144–2153. doi: 10.1093/hmg/ddq093
Rubinsztein, D. C. (2006). The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786. doi: 10.1038/nature05291
Rubinsztein, D. C., Codogno, P., and Levine, B. (2012). Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730. doi: 10.1038/nrd3802
Rubinsztein, D. C., Cuervo, A. M., Ravikumar, B., Sarkar, S., Korolchuk, V., Kaushik, S., et al. (2009). In search of an “autophagomometer”. Autophagy 5, 585–589. doi: 10.4161/auto.5.5.8823
Rubinsztein, D. C., Marino, G., and Kroemer, G. (2011). Autophagy and aging. Cell 146, 682–695. doi: 10.1016/j.cell.2011.07.030
Russell, R. C., Yuan, H. X., and Guan, K. L. (2014). Autophagy regulation by nutrient signaling. Cell Res. 24, 42–57. doi: 10.1038/cr.2013.166
Saftig, P., and Klumperman, J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635. doi: 10.1038/nrm2745
Sahani, M. H., Itakura, E., and Mizushima, N. (2014). Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 10, 431–441. doi: 10.4161/auto.27344
Sandri, M., Coletto, L., Grumati, P., and Bonaldo, P. (2013). Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 126(Pt 23), 5325–5333. doi: 10.1242/jcs.114041
Sarkar, S. (2013a). Chemical screening platforms for autophagy drug discovery to identify therapeutic candidates for Huntington’s disease and other neurodegenerative disorders. Drug Discov. Today Technol. 10, e137–e144. doi: 10.1016/j.ddtec.2012.09.010
Sarkar, S. (2013b). Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 41, 1103–1130. doi: 10.1042/BST20130134
Sarkar, S., Davies, J. E., Huang, Z., Tunnacliffe, A., and Rubinsztein, D. C. (2007a). Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 282, 5641–5652. doi: 10.1074/jbc.M609532200
Sarkar, S., Perlstein, E. O., Imarisio, S., Pineau, S., Cordenier, A., Maglathlin, R. L., et al. (2007b). Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 3, 331–338. doi: 10.1038/nchembio883
Sarkar, S., Floto, R. A., Berger, Z., Imarisio, S., Cordenier, A., Pasco, M., et al. (2005). Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170, 1101–1111. doi: 10.1083/jcb.200504035
Sarkar, S., Korolchuk, V. I., Renna, M., Imarisio, S., Fleming, A., Williams, A., et al. (2011). Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell 43, 19–32. doi: 10.1016/j.molcel.2011.04.029
Sarkar, S., Krishna, G., Imarisio, S., Saiki, S., O’Kane, C. J., and Rubinsztein, D. C. (2008). A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum. Mol. Genet. 17, 170–178. doi: 10.1093/hmg/ddm294
Sarkar, S., Ravikumar, B., and Rubinsztein, D. C. (2009). Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 453, 83–110. doi: 10.1016/S0076-6879(08)04005-6
Saxton, R. A., and Sabatini, D. M. (2017). mTOR signaling in growth. Metab. Dis. Cell 168, 960–976. doi: 10.1016/j.cell.2017.02.004
Schaeffer, V., Lavenir, I., Ozcelik, S., Tolnay, M., Winkler, D. T., and Goedert, M. (2012). Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain 135(Pt 7), 2169–2177. doi: 10.1093/brain/aws143
Schiebler, M., Brown, K., Hegyi, K., Newton, S. M., Renna, M., Hepburn, L., et al. (2015). Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol. Med. 7, 127–139. doi: 10.15252/emmm.201404137
Seglen, P. O., Grinde, B., and Solheim, A. E. (1979). Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur. J. Biochem. 95, 215–225. doi: 10.1111/j.1432-1033.1979.tb12956.x
Seranova, E., Connolly, K. J., Zatyka, M., Rosenstock, T. R., Barrett, T., Tuxworth, R. I., et al. (2017). Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem. 61, 733–749. doi: 10.1042/EBC20170055
Seranova, E., Ward, C., Chipara, M., Rosenstock, T. R., and Sarkar, S. (2019). In vitro screening platforms for identifying autophagy modulators in mammalian cells. Methods Mol. Biol. 1880, 389–428. doi: 10.1007/978-1-4939-8873-0_26
Settembre, C., Di Malta, C., Polito, V. A., Garcia Arencibia, M., Vetrini, F., Erdin, S., et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. doi: 10.1126/science.1204592
Shao, S., Li, S., Qin, Y., Wang, X., Yang, Y., Bai, H., et al. (2014). Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int. J. Oncol. 44, 1661–1668. doi: 10.3892/ijo.2014.2313
Sharma, V., Makhdoomi, M., Kumar, P., Khan, N., Singh, S., Verma, H. N., et al. (2017). Induction of autophagy by trehalose limits opportunistic mycobacterial infections in HIV-infected macrophages. bioRxiv [Preprint]. doi: 10.1101/202697
Sharma, V., Verma, S., Seranova, E., Sarkar, S., and Kumar, D. (2018). Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 6:147. doi: 10.3389/fcell.2018.00147
Shimada, K., Motoi, Y., Ishiguro, K., Kambe, T., Matsumoto, S. E., Itaya, M., et al. (2012). Long-term oral lithium treatment attenuates motor disturbance in tauopathy model mice: implications of autophagy promotion. Neurobiol. Dis. 46, 101–108. doi: 10.1016/j.nbd.2011.12.050
Shoji-Kawata, S., Sumpter, R., Leveno, M., Campbell, G. R., Zou, Z., Kinch, L., et al. (2013). Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206. doi: 10.1038/nature11866
Singh, S. B., Davis, A. S., Taylor, G. A., and Deretic, V. (2006). Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441. doi: 10.1126/science.1129577
Son, J. Y., Yoon, S., Tae, I. H., Park, Y. J., De, U., Jeon, Y., et al. (2018). Novel therapeutic roles of MC-4 in combination with everolimus against advanced renal cell carcinoma by dual targeting of Akt/pyruvate kinase muscle isozyme M2 and mechanistic target of rapamycin complex 1 pathways. Cancer Med. 7, 5083–5095. doi: 10.1002/cam4.1748
Song, Y. M., Lee, Y. H., Kim, J. W., Ham, D. S., Kang, E. S., Cha, B. S., et al. (2015). Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy 11, 46–59. doi: 10.4161/15548627.2014.984271
Spilman, P., Podlutskaya, N., Hart, M. J., Debnath, J., Gorostiza, O., Bredesen, D., et al. (2010). Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One 5:e9979. doi: 10.1371/journal.pone.0009979
Spinello, I., Saulle, E., Quaranta, M. T., Pasquini, L., Pelosi, E., Castelli, G., et al. (2018). The small-molecule compound AC-73 targeting CD147 inhibits leukemic cell proliferation, induces autophagy and increases the chemotherapeutic sensitivity of acute myeloid leukemia cells. Haematologica doi: 10.3324/haematol.2018.199661 [Epub ahead of print].
Strohecker, A. M., Joshi, S., Possemato, R., Abraham, R. T., Sabatini, D. M., and White, E. (2015). Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel autophagy regulator by high content shRNA screening. Oncogene 34, 5662–5676. doi: 10.1038/onc.2015.23
Sun, L., Zhang, S., Yu, C., Pan, Z., Liu, Y., Zhao, J., et al. (2015). Hydrogen sulfide reduces serum triglyceride by activating liver autophagy via the AMPK-mTOR pathway. Am. J. Physiol. Endocrinol. Metab. 309, E925–E935. doi: 10.1152/ajpendo.00294.2015
Sun, N., Malide, D., Liu, J., Rovira, I. I., Combs, C. A., and Finkel, T. (2017). A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat. Protoc. 12, 1576–1587. doi: 10.1038/nprot.2017.060
Sundaramurthy, V., Barsacchi, R., Samusik, N., Marsico, G., Gilleron, J., Kalaidzidis, I., et al. (2013). Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe. 13, 129–142. doi: 10.1016/j.chom.2013.01.008
Suresh, S. N., Chavalmane, A. K., Dj, V., Yarreiphang, H., Rai, S., Paul, A., et al. (2017). A novel autophagy modulator 6-Bio ameliorates SNCA/alpha-synuclein toxicity. Autophagy 13, 1221–1234. doi: 10.1080/15548627.2017.1302045
Tanaka, M., Machida, Y., Niu, S., Ikeda, T., Jana, N. R., Doi, H., et al. (2004). Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 10, 148–154. doi: 10.1038/nm985
Tanida, I., Sou, Y. S., Ezaki, J., Minematsu-Ikeguchi, N., Ueno, T., and Kominami, E. (2004). HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279, 36268–36276. doi: 10.1074/jbc.M401461200
Tanji, K., Miki, Y., Maruyama, A., Mimura, J., Matsumiya, T., Mori, F., et al. (2015). Trehalose intake induces chaperone molecules along with autophagy in a mouse model of Lewy body disease. Biochem. Biophys. Res. Commun. 465, 746–752. doi: 10.1016/j.bbrc.2015.08.076
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q., Zhang, J., Gao, Y., et al. (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. doi: 10.1074/jbc.M900301200
Ueno, T., and Komatsu, M. (2017). Autophagy in the liver: functions in health and disease. Nat. Rev. Gastroenterol. Hepatol. 14, 170–184. doi: 10.1038/nrgastro.2016.185
Wang, I. F., Guo, B. S., Liu, Y. C., Wu, C. C., Yang, C. H., Tsai, K. J., et al. (2012). Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc. Natl. Acad. Sci. U.S.A. 109, 15024–15029. doi: 10.1073/pnas.1206362109
Wang, J., Lian, H., Zhao, Y., Kauss, M. A., and Spindel, S. (2008). Vitamin D3 induces autophagy of human myeloid leukemia cells. J. Biol. Chem. 283, 25596–25605. doi: 10.1074/jbc.M801716200
Wang, Y., Li, L., Hou, C., Lai, Y., Long, J., Liu, J., et al. (2016). SNARE-mediated membrane fusion in autophagy. Semin. Cell Dev. Biol. 60, 97–104. doi: 10.1016/j.semcdb.2016.07.009
Webb, J. L., Ravikumar, B., Atkins, J., Skepper, J. N., and Rubinsztein, D. C. (2003). Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013. doi: 10.1074/jbc.M300227200
Wei, Y. M., Li, X., Xu, M., Abais, J. M., Chen, Y., Riebling, C. R., et al. (2013). Enhancement of autophagy by simvastatin through inhibition of Rac1-mTOR signaling pathway in coronary arterial myocytes. Cell Physiol. Biochem. 31, 925–937. doi: 10.1159/000350111
Whitehead, N. P., Kim, M. J., Bible, K. L., Adams, M. E., and Froehner, S. C. (2015). A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc. Natl. Acad. Sci. U.S.A. 112, 12864–12869. doi: 10.1073/pnas.1509536112
Williams, A., Sarkar, S., Cuddon, P., Ttofi, E. K., Saiki, S., Siddiqi, F. H., et al. (2008). Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 4, 295–305. doi: 10.1038/nchembio.79
Wood, J. G., Rogina, B., Lavu, S., Howitz, K., Helfand, S. L., Tatar, M., et al. (2004). Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689. doi: 10.1038/nature02789
Wyttenbach, A., Swartz, J., Kita, H., Thykjaer, T., Carmichael, J., Bradley, J., et al. (2001). Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Hum. Mol. Genet. 10, 1829–1845. doi: 10.1093/hmg/10.17.1829
Yamamoto, A., Tagawa, Y., Yoshimori, T., Moriyama, Y., Masaki, R., and Tashiro, Y. (1998). Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42. doi: 10.1247/csf.23.33
Yoshikawa, Y., Ogawa, M., Hain, T., Yoshida, M., Fukumatsu, M., Kim, M., et al. (2009). Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11, 1233–1240. doi: 10.1038/ncb1967
Yuk, J. M., Shin, D. M., Lee, H. M., Yang, C. S., Jin, H. S., Kim, K. K., et al. (2009). Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 6, 231–243. doi: 10.1016/j.chom.2009.08.004
Zachari, M., and Ganley, I. G. (2017). The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61, 585–596. doi: 10.1042/EBC20170021
Zhang, J. J., Zhou, Q. M., Chen, S., and Le, W. D. (2018). Repurposing carbamazepine for the treatment of amyotrophic lateral sclerosis in SOD1-G93A mouse model. CNS Neurosci. Ther. 24, 1163–1174. doi: 10.1111/cns.12855
Zhang, L., Yu, J., Pan, H., Hu, P., Hao, Y., Cai, W., et al. (2007). Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. U.S.A. 104, 19023–19028. doi: 10.1073/pnas.0709695104
Zhang, X., Chen, S., Song, L., Tang, Y., Shen, Y., Jia, L., et al. (2014). MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy 10, 588–602. doi: 10.4161/auto.27710
Zhang, X., Heng, X., Li, T., Li, L., Yang, D., Zhang, X., et al. (2011). Long-term treatment with lithium alleviates memory deficits and reduces amyloid-beta production in an aged Alzheimer’s disease transgenic mouse model. J. Alzheimers Dis. 24, 739–749. doi: 10.3233/JAD-2011-101875
Keywords: autophagy, autophagy reporter, autophagy substrate, autophagy modulator, screening method, neurodegenerative diseases, cancer, lifespan extension
Citation: Panda PK, Fahrner A, Vats S, Seranova E, Sharma V, Chipara M, Desai P, Torresi J, Rosenstock T, Kumar D and Sarkar S (2019) Chemical Screening Approaches Enabling Drug Discovery of Autophagy Modulators for Biomedical Applications in Human Diseases. Front. Cell Dev. Biol. 7:38. doi: 10.3389/fcell.2019.00038
Received: 12 December 2018; Accepted: 01 March 2019;
Published: 19 March 2019.
Edited by:
Brian Storrie, University of Arkansas for Medical Sciences, United StatesReviewed by:
Vytaute Starkuviene, Vilnius University, LithuaniaSatoshi Kametaka, Nagoya University, Japan
Copyright © 2019 Panda, Fahrner, Vats, Seranova, Sharma, Chipara, Desai, Torresi, Rosenstock, Kumar and Sarkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sovan Sarkar, cy5zYXJrYXJAYmhhbS5hYy51aw==
†These authors have contributed equally to this work
 Prashanta Kumar Panda
Prashanta Kumar Panda Alexandra Fahrner
Alexandra Fahrner Somya Vats
Somya Vats Elena Seranova
Elena Seranova Vartika Sharma
Vartika Sharma Miruna Chipara
Miruna Chipara Priyal Desai
Priyal Desai Jorge Torresi
Jorge Torresi Tatiana Rosenstock
Tatiana Rosenstock Dhiraj Kumar
Dhiraj Kumar Sovan Sarkar
Sovan Sarkar