- Department of Pediatrics, University of Chicago, Chicago, IL, United States
The zebrafish animal model is gaining increasing popularity as a tool for studying human disease. Over the past 15 years, many models of leukemia and other hematological malignancies have been developed in the zebrafish. These confer some significant advantages over similar models in other animals and systems, representing a powerful resource for investigation of the molecular basis of human leukemia. This review discusses the various zebrafish models of lymphoid and myeloid leukemia available, the major discoveries that have been made possible by them, and opportunities for future exploration.
Introduction
Leukemia
Leukemia is a broad designation encompassing hematological malignancies that produce the expansion of blood cells, typically starting in the bone marrow. In 2015, there were over 2.3 million patients suffering from leukemia, resulting in over 350,000 deaths worldwide (GBD 2015 Mortality and causes of death collaborators, 2016). In the United States, an estimated 62,130 new leukemia cases were diagnosed and 24,500 deaths caused by leukemia in 2017, with a 5 years survival rate of ∼63% (NCI SEER Cancer Stat Facts: Leukemia). Although the majority of leukemias affect adults, leukemia is also the most common cancer diagnosis in children.
Leukemias are categorized by two major criteria into four groups. The first criterion relates to the cell of origin: leukemias of lymphoid origin are classified “lymphocytic or lymphoblastic” and those of myeloid origin called “myelogenous or myeloid.” The second criterion deems rapidly growing leukemias as “acute” and those with more indolent growth as “chronic.” The causes of these different malignancies are varied. Some are directly linked to a chromosomal abnormality, such as the Philadelphia chromosome in chronic myelogenous leukemia (Bartram et al., 1983) or the increased incidence of leukemia in patients with trisomy 21 (down syndrome) (Evans and Steward, 1972). However, the etiology of most leukemias is less straightforward. Some leukemias involve mutations and/or translocations of multiple genes associated with growth, differentiation and survival of blood cells. Others have a normal karyotype and no known genetic mutations, highlighting the need for further studies in animal models to uncover these unknown drivers of hematopoietic malignancy.
Zebrafish as a Model Organism
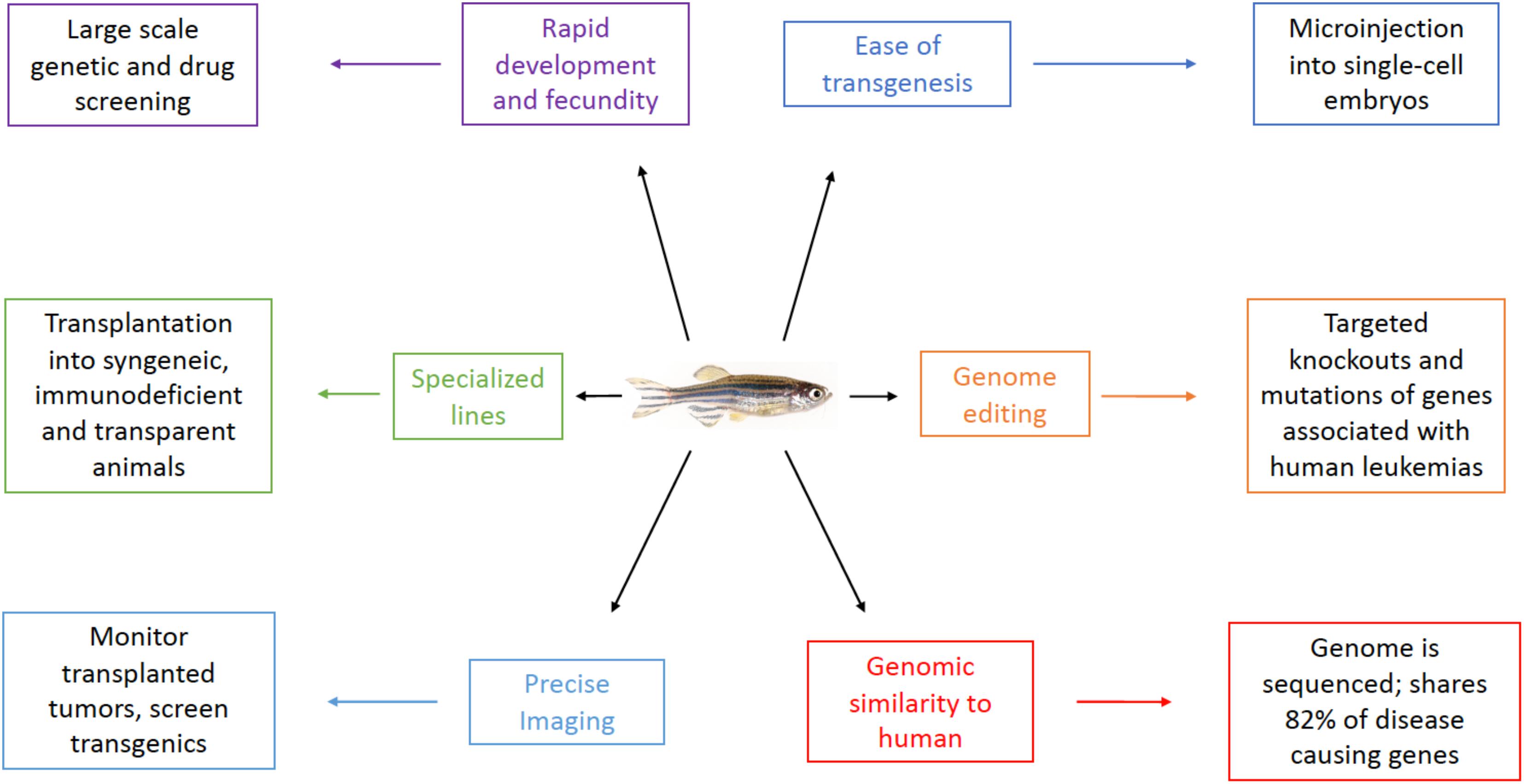
Danio rerio, commonly known as the zebrafish, is a small tropical fish popular in pet stores and aquariums. Since the 1970’s when George Streisinger first began using the zebrafish as a model organism (Walker and Streisinger, 1983), more and more labs have begun utilizing this powerful tool for studying development and disease due to numerous advantages over other model systems (Figure 1).
Zebrafish fertilization and development occurs externally in optically clear embryos that are easily observed and manipulated. Development is much faster than mammals, with most major organs forming by 2–3 days post-fertilization (dpf). Animals reach sexual maturity by 2–3 months of age (Kimmel et al., 1995) and a single breeding pair produces several hundred embryos weekly. This fecundity coupled with their rapid development makes the zebrafish an excellent model for large-scale screening. Forward and reverse genetic screens as well as toxicity and drug screens in zebrafish have been performed around the world over the past three decades (Ransom et al., 1996; Weinberg et al., 1996; Sood et al., 2006; North et al., 2007; Ridges et al., 2012), including significant work more recently to evaluate therapeutics in zebrafish leukemia models (Mizgirev and Revskoy, 2010; Deveau et al., 2017). Although teleosts (like the zebrafish) and mammals diverged from a common ancestor approximately 340 million years ago, they still share a remarkable amount of their genomes, with a zebrafish ortholog identified for 82% of known disease-causing genes in humans (Howe et al., 2013). Many of these zebrafish genes have already been shown to recapitulate human disease when affected in zebrafish, including several connected to hematopoiesis (Brownlie et al., 1998; Wang et al., 1998) and cancer, as we will discuss in this review.
Several systems have been developed within the zebrafish model to create transgenic and knockout animals. Because the zebrafish embryos are externally developed, it is possible to microinject directly into the single-cell for the first 15–30 min following fertilization. Although, the first transgenic zebrafish were created through injection of naked, linearized DNA (Stuart et al., 1988), more efficient systems of genomic incorporation are now available. The tol2 transposon system creates randomly inserted transgenes that heavily favor single copy insertions (Urasaki et al., 2006) and the I-SceI meganuclease system inserts one or more copies into double-stranded breaks in the genome (Grabher et al., 2004; Ogino et al., 2006). In addition to the ability to integrate transgenes, the advent of CRISPR/Cas9 technology has made it possible to directly edit the zebrafish genome; from creating knockouts to mimicking human mutations to introducing specific SNPs. The use of CRISPR/Cas9 in zebrafish was first described by Keith Joung’s lab in 2013 (Hwang et al., 2013), and has since spread throughout the field to become a common tool in many labs’ arsenal, just as it has throughout the biomedical community at large (Hruscha et al., 2013; Ablain et al., 2015).
Over the years, the zebrafish community has amassed a large number of inbred, transgenic, knockout, or other specialized lines that have been characterized and maintained for various applications. Important to leukemia models are several lines that allow for transplantation of tumors without the need for pre-transplant immune ablation. Generated by parthenogenesis, the clonal golden lines (CG1 and CG2) allow for syngeneic transplantation within a genetically identical line, similar to transplantation experiments using inbred mice (Mizgireuv and Revskoy, 2006; Smith et al., 2010). The rag2 (E450fs) mutant line has reduced numbers of functional T- and B-cells, and thus is unable to mount a significant immune response against transplanted cells (Tang et al., 2014). The c-mybI181N hypomorphic mutant is another immunocompromised line that has shown promise in xenograft experiments (Hess and Boehm, 2016). These lines allow for immunologically unmatched transplantation from other zebrafish lines as well as xenografts.
A common challenge for many model systems is the ability to visualize and trace the fate of a cancer cell within an animal over time. These issues are often circumvented by euthanizing, sectioning, and staining multiple animals at different timepoints, however, this increases the number of animals required, increases time commitment, and may blur inter-individual variability. In the zebrafish, fluorescently tagged proteins or cells can be clearly imaged from embryo to adulthood in live animals by confocal or lightsheet microscopy (Kaufmann et al., 2012) and at even greater resolutions in the pigment-less Casper line (White et al., 2008). An excellent example of this utility was described by Kaufman et al. (2016), when they used a crestin-EGFP line to show melanoma initiation and progression from a single cell. Also, with the macroscope developed by the Langeneau lab, high-throughput imaging of adult fish is possible for transgenic lines or screening for tumor engraftment in transplantation models (Blackburn et al., 2011).
Hematopoiesis: Zebrafish and Human
Many of the transcription factors and major signaling pathways controlling hematopoietic differentiation are mutated or dysregulated in the transformation and progression of leukemia. Therefore, in addition to the general advantages described above, the zebrafish is an appealing model for studying leukemia because of the close parallels to mammalian hematopoiesis (de Jong and Zon, 2005). Though the locations of hematopoiesis are not perfectly shared between species, the ontogeny of the different hematopoietic cells from progenitors to maturity, as well as the genes and pathways driving differentiation are well conserved (Paik and Zon, 2010). There are two distinct waves of hematopoiesis in all vertebrates; a transient primitive wave supplying necessary macrophages and erythrocytes for early embryonic development, followed by the definitive wave that gives rise to the full complement of blood cells throughout an animal’s lifetime. In mammalian development, the primitive hematopoietic stem cells (HSCs) appear within the blood islands in the embryonic yolk sac (Palis and Yoder, 2001). In zebrafish, these limited HSCs instead arise from the intermediate cell mass (ICM) within the ventral mesoderm, and, similarly, produce erythrocytes and other myeloid cells (Detrich et al., 1995). Expression of the transcription factors scl, gata2, lmo2, tif1γ, and fli1 promotes the primitive HSC lineage (Liao et al., 1998; Thompson et al., 1998; Ransom et al., 2004), while gata1 and spi1 (also known as pu.1) drive their differentiation into the erythroid and myeloid lineages, respectively (Detrich et al., 1995; Lieschke et al., 2002).
The mammalian definitive wave of hematopoiesis begins with true multipotent HSCs emerging from the ventral wall of the dorsal aorta in the aorta-gonad-mesenephros (AGM) region that then migrate to the fetal liver to proliferate and differentiate, and ultimately migrate to seed the bone marrow (Cumano and Godin, 2007). This process is mirrored in the zebrafish with the definitive HSCs also arising from the ventral wall of the dorsal aorta, and migrating to the caudal hematopoietic tissue (CHT) before seeding the kidney marrow, which is the zebrafish adult hematopoietic tissue (Burns et al., 2002; Jin et al., 2007). Definitive HSCs are true multipotent hematopoietic progenitors and are marked by their expression of the transcription factors runx1, c-myb, lmo-2, and scl (Thompson et al., 1998; Burns et al., 2002). Similar to primitive hematopoiesis, gata1 and tif1γ drive erythropoiesis (Detrich et al., 1995; Ransom et al., 2004) and spi1 and c/ebp1 drive myelopoiesis (Lyons et al., 2001; Lieschke et al., 2002). Unlike the primitive lineages, definitive HSCs also produce lymphoid cells through expression of rag1, rag2, ikaros, lck, and gata3 (Willett et al., 1997, 2001; Langenau et al., 2004). There are several functional and structural differences in the hematopoietic system of zebrafish compared to mammals, namely the location of the marrow, the lack of lymph nodes, and the rapid development and early reliance on the innate immune system (Novoa and Figueras, 2012; Renshaw and Trede, 2012). However, ultimately the blood cells of the zebrafish and human are molecularly very similar and thus have common genetic drivers of leukemia.
The conservation of the genes and pathways regulating hematopoiesis between humans and zebrafish, combined with the significant technical advantages provided by this model animal, make the zebrafish an ideal system for investigating hematological malignancies. In this review, we will examine the many leukemia models that have been developed within the zebrafish, and discuss the major findings made possible by each model that have advanced our understanding of human leukemia.
Zebrafish Leukemia Models: Lymphoid Origin
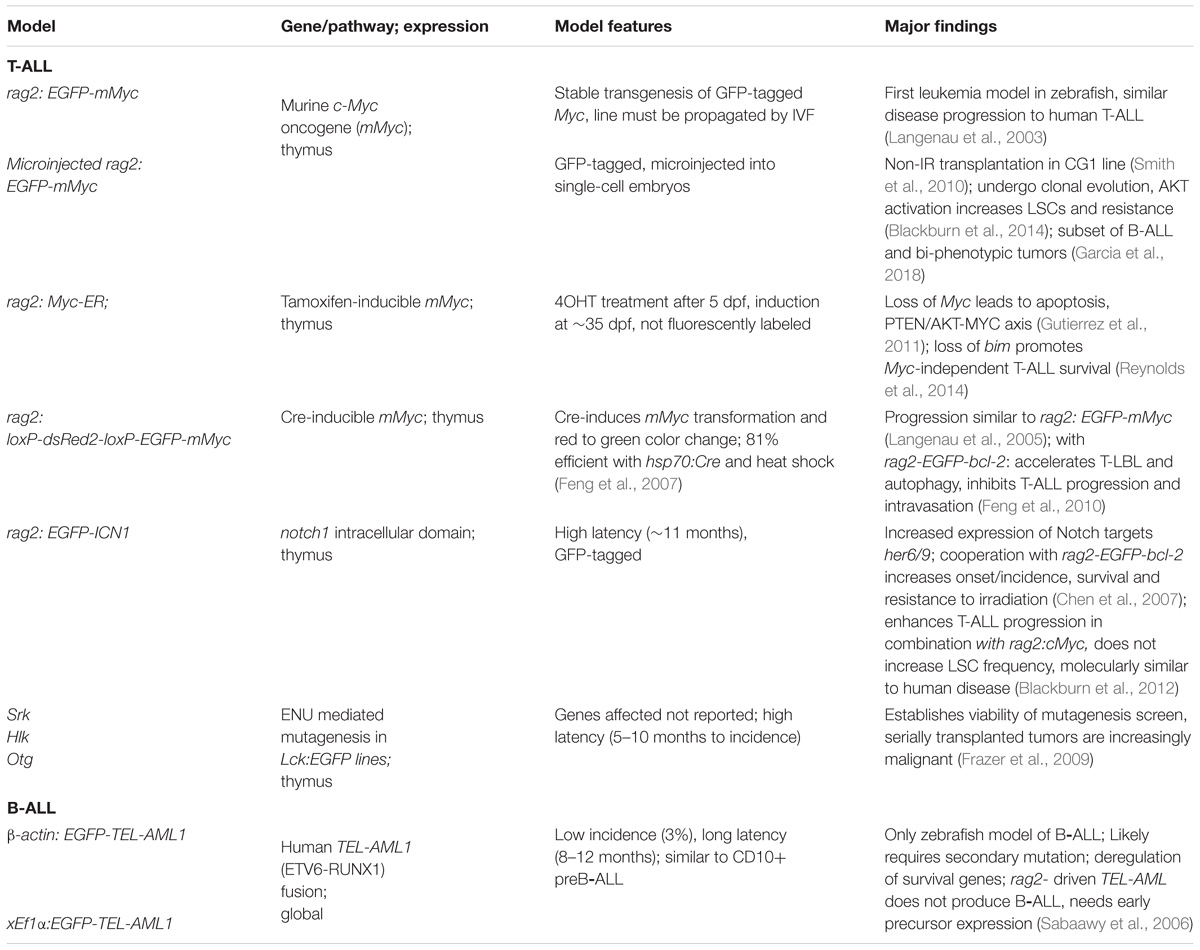
The first leukemia model in zebrafish was developed over 15 years ago using the lymphocyte-specific rag2 promoter driving the murine c-Myc oncogene to produce T-cell acute lymphoblastic leukemia (T-ALL) (Langenau et al., 2003). The success of that first step has spawned a variety of other models tied to different types of leukemia. Over time, these models have been altered and improved to fit the particular investigations of each project, and there are now multiple similar models available, each with their own strengths and weaknesses (Table 1). This section discusses models of lymphoid origin, and the major discoveries made possible by them.
T-cell Acute Lymphoblastic Leukemia (T-ALL)
The majority of lymphoid leukemia models in zebrafish replicate T-ALL, partially due to the success of the rag2 promoter in driving that particular malignancy. Although rag2 is expressed in both T- and B-cell precursors in zebrafish (Langenau et al., 2004), only T-cell leukemias were initially identified from models utilizing this promoter. Interestingly, the Langenau lab has recently published a brief communication describing a subset of B-cell derived and bi-phenotypic leukemias produced from a rag2 promoter (Garcia et al., 2018), suggesting some of the research done on these T-ALL models may have unknown contributions from B-ALL as well.
The oncogene c-Myc is associated with many cancers and is one of the most frequently affected gene pathways in lymphoid leukemia (La Starza et al., 2014). The first T-ALL model was developed by Langenau et al. by expressing the murine c-Myc oncogene under the zebrafish rag2 promoter, with an EGFP tag for easy monitoring by fluorescent microscopy (Langenau et al., 2003). Tumors were generated in microinjected mosaic F0 fish at similar rates to EGFP expression in control animals injected with rag2: EGFP, suggesting complete penetrance of tumor induction upon successful integration of the c-Myc transgene. These tumors grew rapidly, with a mean latency of 52 days post-fertilization (dpf), extensively infiltrating the entirety of the fish. Analysis of the expression profiles and T-cell receptor (TCR) rearrangements confirmed that the tumor cells derived from clonal expansion of transformed T lymphocyte precursors and originated in the thymus. Tumor cells could be transplanted into irradiated recipients and quickly grew new tumors that homed to the thymus before spreading throughout the animal. Overall, the tumors progressed, similarly, to human T-ALL, at an accelerated pace. However, most F1 progeny developed advanced disease well before reaching sexual maturity (mean latency 32 dpf), necessitating sperm collection and in vitro fertilization (IVF) to continue the stable transgenic line. Subsequent characterization of this model showed that the tumors express tal1/scl and lmo2, genes associated with a molecular subgroup of Myc-induced T-ALL in humans (Langenau et al., 2005).
To circumvent the necessity of IVF, Langenau et al. sought to create an inducible version of their model. They achieved this by inserting a loxP-DsRed2-loxP sequence cassette between the rag2 promoter and EGFP-mMyc oncogene, creating the rag2:loxP-dsRed2-loxP-EGFP-mMyc line (rag2: LDL-Emyc) (Langenau et al., 2005). This allowed for default red fluorescent expression with a switch to EGFP-mMyc expression in the presence of Cre recombinase. The disease in these animals was morphologically similar to that in the rag2:EGFP-mMyc model, but with significantly decreased incidence (6.5%) and delayed latency (mean 151 dpf). This was presumed to be due to incomplete recombination of the transgene, as evidenced by the persistence of red fluorescent expression within the tumor. To combat this, they developed a heat shock-inducible Cre line, hsp70: Cre (Feng et al., 2007). When combined with their Cre-inducible rag2: LDL-Emyc line and subjected embryos to heat shock at 3 dpf, the penetrance (81%) and latency (120 dpf) were closer to those of the original rag2: EGFP-mMyc model. This improved model allowed them to explore the molecular events governing the progression of the disease from the localized T-lymphoblastic lymphoma (T-LBL) to disseminated T-ALL. All of the Myc-induced models of T-ALL in zebrafish begin as T-LBL with thymic hyperplasia and localized outgrowth before advancing to T-ALL and expanding into the circulation and other tissues. The investigation into this transition led them to combine the rag2:LDL-Emyc; hsp70:Cre model with a line overexpressing the survival gene bcl2 (Feng et al., 2010). This combination accelerated T-LBL induction by suppressing Myc-induced apoptosis. However, it also promoted homotypic cell adhesion through s1p1 and icam1 that prevented intravasation into the vascular space and restricted the tumor to the thymus. The tumor cells then proliferated until they exhausted their nutrient supply and underwent autophagy. Because AKT-signaling is known to promote T-cell migration and to suppress autophagy (Sotsios and Ward, 2000; Lum et al., 2005), they hypothesized that addition of constitutively active AKT could force progression to T-ALL. Indeed, when their Myc;Cre;bcl-2 embryos were injected with a myristolated-akt2 transgene, the resulting tumors rapidly advanced to T-ALL (Feng et al., 2010).
The importance of AKT signaling in zebrafish T-ALL progression is not surprising, given the PTEN-PI3K-AKT pathway is frequently disrupted in human T-ALL (Palomero et al., 2008; Gutierrez et al., 2009). Gutierrez et al. further investigated this connection with the aid of another Myc-induced model, the tamoxifen inducible rag2: Myc-ER line (Gutierrez et al., 2011). This model allows for conditional expression of the c-Myc oncogene only in the presence of 4-hydroxytamoxifen (4-OHT). When continually treated with 4-OHT these fish develop T-ALL, but upon cessation of treatment and loss of c-Myc expression, the tumor cells undergo apoptosis and the tumor rapidly regresses. However, when AKT signaling was increased through either loss-of-function mutations in pten or constitutively active akt2, the tumors lost their dependence on Myc expression and were able to continue progression after removal of 4-OHT treatment. Further investigation into the relationship between Myc and the AKT pathway revealed that Myc drove the expression of the proapoptotic protein bim, while the constitutively active myr-akt2 blocked that induction (Reynolds et al., 2014). Additionally, loss-of-function bim mutations allowed for increased persistence of T-ALL after cessation of 4-OHT treatment and Myc expression. Overall, these results suggest AKT-signaling enhances Myc-induced T-ALL progression via promotion of T-cell migration, suppression of autophagy, and inhibition of apoptosis.
Due to difficulty maintaining stable transgenic lines expressing c-Myc, an alternative approach was developed involving co-injection of the rag2-EGFP and rag2-mMyc transgenes into single-cell embryos (Langenau et al., 2008; Smith et al., 2010). In this model, the two transgenes randomly integrated into the genome to be co-expressed such that GFP expression was observed only in tumors and tumor induction only with GFP+ thymocytes. The resulting tumors followed the same pathology as the stable Myc-induced models. Smith et al. (2010) used this method to create tumors in clonal CG1 fish, demonstrating that they could be transplanted into syngeneic recipient CG1 fish without irradiation. This also allowed them to determine the frequency of leukemia stem cells (LSCs) present in these tumors through limit dilution analysis of the transplanted tumors. Each successful engraftment requires at least one LSC, and by transplanting different doses of cells, they were able to determine that 0.1–1.4% of the primary T-ALL tumor cells were LSCs. Transplantation of T-ALLs generated using this co-injection model was further investigated by Blackburn et al. (2014) who demonstrated that serial transplantation of T-ALL tumors led to spontaneous clonal evolution of monoclonal tumor subclones. As tumors were passaged from primary to secondary to tertiary recipients, some subclones evolved increased LSC frequency, growth, and/or resistance to therapy. Subclones with increased LSC frequency also displayed increased AKT phosphorylation, and treatment with an AKT inhibitor dramatically reduced their engraftment after transplant. Co-expression of myr-akt2 with Myc significantly increased proliferation of tumor cells, decreased latency after transplantation, and increased LSC frequency sixfold, and these effects are at least partially due to AKT’s induction of mtorc1 expression. Additionally, the subclones that had evolved glucocorticoid resistance were resensitized to dexamethasone treatment by AKT inhibitors. Altogether, these results provided further evidence of the connection between Myc and AKT in T-ALL.
Another major player in the transformation of T-cell precursors to T-ALL is Notch1, which has activating mutations in over 65% of T-ALL patients (Weng et al., 2004). To further study the role of Notch1 in T-ALL, Chen et al. (2007) created a transgenic line expressing rag2:ICN1-EGFP, a GFP-tagged Notch1 intracellular domain which acts as a constitutively active transcription factor to drive Notch target gene expression. This line develops T-ALL, but at a lower incidence (40%) and higher latency (>11 months) than the Myc-driven tumors. However, in the presence of bcl2 overexpression, the incidence (60–80%) and latency (40 dpf to induction; 3 months to dissemination) were significantly enhanced and apoptosis was decreased. Blackburn et al. further demonstrated this by combining the rag2:ICN1-EGFP and rag2:cMyc models which accelerated leukemia onset and incidence (Blackburn et al., 2012). They concluded that Notch signaling expanded pre-leukemic clones that required Myc (or acquired secondary mutations) to transform, and that Notch signaling did not increase the overall frequency of LSCs. They also used this model to make cross-species microarray comparisons with mouse and human T-ALL to identify a common T-ALL gene signature and novel Notch gene expression profile present in humans that is regulated independently of Myc. These two studies suggest that Notch1 activation alone is not sufficient for induction of T-ALL and requires additional oncogene activation and/or tumor suppressor mutations.
Taking advantage of the ability to perform large-scale forward-genetic screens in zebrafish to identify genetic modifiers of disease, Frazer et al. (2009) developed one such screen for causative mutations in T-ALL using ENU-mediated mutagenesis of an lck-EGFP line. This screen identified three mutant lines that developed outgrowth of the GFP-tagged thymus and subsequently T-ALL. Two of these lines, shrek (srk) and hulk (hlk), contained dominant mutations and one dubbed Oscar-the-grouch (otg) contained a recessive mutation. Homozygous fish from all 3 lines had incidences around 50% and time to tumor induction between 6 and 8 months. The mutated genes in these lines have not yet been reported, but the screen demonstrates the potential for identification of genes driving different leukemias. The lab also developed a chemical screen to identify small molecules capable of eradicating immature T-cells, using the same lck-EGFP line (Ridges et al., 2012). They identified Lenaldekar (LDK; 1H-indole-3-carbaldehyde 8-quinolinylhydrazone) as a compound capable of killing both normal and T-ALL blasts in zebrafish, and showed it was effective in mouse xenograft and human primary leukemia cells as well.
B-Cell Acute Lymphoblastic Leukemia (B-ALL)
The TEL-AML1 (also known as ETV6-RUNX1) fusion protein results from t(12;21), the most common translocation in childhood cancer, present in ∼25% of B-cell acute lymphoblastic leukemia (B-ALL) (Romana et al., 1995). However, attempts to produce a model of B-ALL from this fusion gene were unsuccessful in mice (Andreasson et al., 2001). Sabaawy et al. (2006) created multiple lines expressing human TEL-AML1 from different promoters in zebrafish and were able to produce the only zebrafish model of B-ALL. Three different promoters were tested: the Xenopus ef1a (Xef1a) and zebrafish beta-actin (zba) for global expression, and zebrafish rag2 for lymphocyte specific expression. Both of the global promoters produced B-ALL tumors in ∼3% of fish with 8–12 months latency and similar molecular and morphological features to pediatric CD10+ B-ALL. The low incidence likely indicates the need for a secondary mutation for oncogenic transformation. They surmised that the rag2: TEL-AML1 fish did not develop tumors because the transformation occurs prior to the expression of Rag2 in the common lymphoid progenitor, and instead occurs in an earlier multipotent progenitor or hematopoietic stem cell in the global promoter lines. With the apparent T-cell bias of the rag2 promoter in zebrafish, it also seems possible that a different promoter of common lymphoid or B-cell progenitors may have more success. However, the recent discovery of B-ALL in the rag2: cMyc fish provides an opportunity for studying B-ALL in a more accessible model, with much shorter latency and higher incidence (Garcia et al., 2018).
Zebrafish Leukemia Models: Myeloid Origin
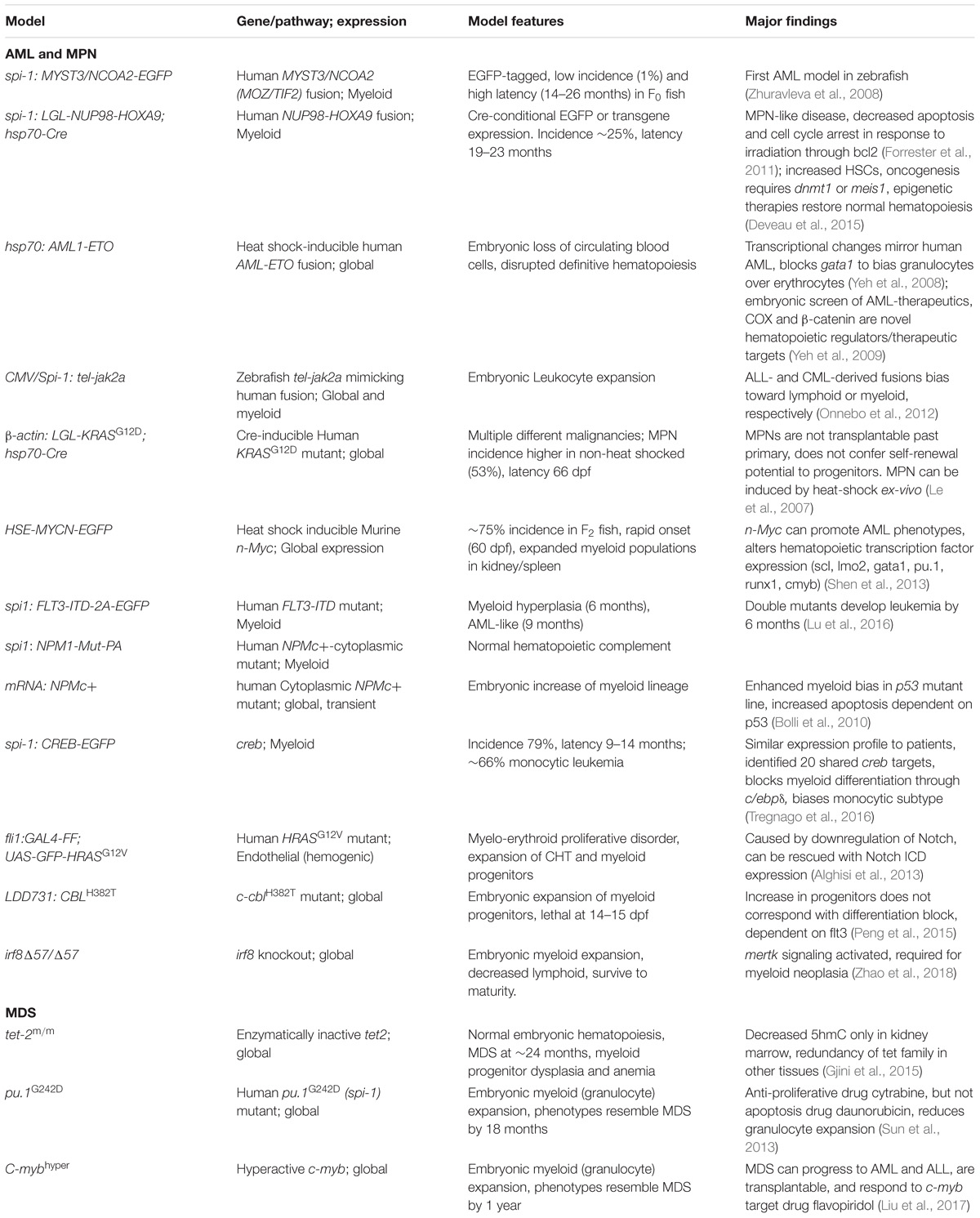
Following the initial success of the zebrafish ALL models, serious efforts began to recapitulate myeloid leukemias including myeloproliferative neoplasms (MPN) and acute myeloid leukemia (AML) in zebrafish. This was done largely through creating transgenic lines that expressed oncogenic fusion genes and mutations commonly found in patients with MPN and AML. This section discusses the features and major findings of the myeloid leukemia models developed to date in zebrafish (Table 2).
Acute Myeloid Leukemia (AML) and Myeloproliferative Neoplasms (MPN)
Many hematological malignancies are driven by oncogenic fusion genes created after chromosomal translocations and these fusions can often be expressed in animal models or cell lines to drive transformation and oncogenesis. Zhuravleva et al. were the first to do so with a myeloid malignancy in zebrafish by creating transgenic fish expressing the MYST3/NCOA2 (MOZ/TIF2) fusion product under the spi-1 (pu.1) early myeloid promoter (Hsu et al., 2004) along with EGFP (Zhuravleva et al., 2008). This fusion protein is the result of the inv(8)(p11q13) chromosome abnormality found in human AML, and fuses two histone acetyltransferases (HATs). A small number of F0 fish (1.1%) expressing the transgene developed AML after 14–26 months, characterized by expansion of myeloid blast cells and invasion of the kidney. This low incidence and long latency suggest that secondary mutations may be necessary to induce transformation.
Another model using the NUP98-HOXA9 (NHA9) fusion gene [t(7;11)(p15;p15)] was developed by Forrester et al. (2011) with an spi-1 promoter driving conditional expression of either EGFP or the transgene after heat shock by the hsp70-Cre line. This oncogenic fusion product is associated with poor prognosis in AML and chronic myeloid leukemia (CML) (Gough et al., 2011). Following heat shock at 24 hpf, NUP98-HOXA9;Cre embryos had perturbed hematopoiesis promoting myeloid fates, and also showed reduced apoptosis and cell cycle arrest in response to irradiation, correlating with increased levels of bcl2. 23% of NUP98-HOXA9;Cre fish developed myeloid tumors with a latency of 19–23 months. These tumors closely resembled the pathology of the polyclonal MPN found in NUP98-HOXA9-transgenic mice (Kroon et al., 2001). Further investigation into the model uncovered an increase in HSCs, as well as a dependency on meis1, the prostaglandin/cyclooxygenase pathway, and genome hypermethylation via dnmt1 for the fusion gene’s oncogenic potential (Deveau et al., 2015). This dependency could be exploited through treatment with DNMT or COX inhibitors, or sub-therapeutic doses of either in combination with HDAC inhibitors. This study both revealed mechanistic details of the NHA9 oncogene and demonstrated the potential of zebrafish leukemia models in identification of new treatment combinations.
Because most leukemia oncogenes produce early detectable effects on hematopoiesis, along with the inherent advantages of the zebrafish model, it is possible to develop drug screens in preleukemic embryonic models. One such model was developed by Yeh et al. (2008) using the AML1(RUNX1)-ETO fusion oncogene under the heat shock responsive hsp-70 promoter. After heat shock, embryos accumulated non-circulating immature blast cells, with disruption of definitive hematopoiesis via loss of runx1 and cmyb expression, loss of gata1-expressing erythroid cells, and were ultimately driven to a myeloid-granulocytic fate. These effects were all downstream of AML1-ETO’s suppression of scl, and could be reversed with scl overexpression. The transcriptional signature of AML1-ETO-expressing embryos closely paralleled that of human AML. Using the perturbation of embryonic hematopoiesis and the AML transcriptional signature as a readout of AML1-ETO oncogenic activity, they were able to develop a chemical screen for inhibitors that can rescue AML1-ETO’s effects (Yeh et al., 2009). This screen identified the COX and β-catenin pathways as vital to the function of AML1-ETO.
TEL(ETV6)-JAK2 fusion genes have been identified in both ALL and atypical chronic myelogenous leukemia (aCML), with slightly different translocations driving each, t(9;12)(p24;p13) and t(9;15;12)(p24;q15;p13), respectively (Peeters et al., 1997). Onnebo et al. (2012) created transgenic zebrafish lines expressing these different fusions under CMV or spi-1 promoters to better understand how they drive oncogenesis distinctly. These lines differ from other fusion gene transgenic lines in that the fusion proteins were generated from the zebrafish tel and jak2a genes combined to mimic two different human translocations found in T-ALL and aCML. Overall, the different genes behaved true to form, with the T-ALL fusion gene disrupting embryonic lymphopoiesis and the aCML fusion gene disrupting myelopoiesis similar to an MPN, driven by either CMV or spi-1. They were also able to demonstrate subtle differences in activity, with the T-ALL fusion gene showing greater enzymatic activity, but reduced downstream STAT activation and decreased sensitivity to JAK2 inhibition.
Although Zhuravleva et al. were the first to claim production of AML in a zebrafish model, the first myeloid malignancy was created in the Zon lab. Le et al. (2007) generated β-actin: LGL-KRASG12D; hsp70-Cre zebrafish, with conditional global expression of an oncogenic KRAS inducible by heat shock. This model produced a variety of tumors following heat shock, including rhabdomyosarcoma, myeloproliferative neoplasm, intestinal hyperplasia, and malignant peripheral nerve sheath tumor. Although the heat-shocked fish had juvenile lethality, they discovered that the non-heat shocked adult fish developed MPN, likely due to the well-known “leakiness” of heat-shock promoters. The MPN-affected fish displayed classic disease characteristics such as expansion of myeloid progenitors, invasion of the marrow (kidney), and depletion of erythroid cells. Interestingly, these MPN cells could engraft after primary transplantation into irradiated recipients, but were unable to engraft after secondary transplantation, suggesting that they lack self-renewal capabilities. Another model utilizing an oncogenic RAS mutation was developed by Alghisi et al. (2013) inducing expression in the hemogenic endothelium prior to hematopoietic emergence. This fli1:GAL4-FF; UAS-GFP-HRASG12V line developed an MPN characterized by prominent expansion of the CHT, increased number of immature hematopoietic cells, and a block of myeloid differentiation in the kidney marrow. The Notch pathway was significantly downregulated and overexpression of the active NICD rescued the MPN phenotypes. They used this model to identify candidate genes both downregulated by Notch and upregulated by RAS that could be involved myeloid oncogenesis.
Similar to the connection of c-Myc to T-ALL, n-Myc is frequently upregulated in AML and is a poor prognostic marker. Shen et al. (2013) created a heat shock responsive zebrafish line expressing murine n-Myc, MYCN:HSE:EGFP, that simultaneously drives expression of EGFP. Following heat shock, n-Myc overexpression promoted immature myeloid blast cell expansion and enhanced the repopulating activity of myeloid cells. N-Myc enhanced primitive hematopoiesis by upregulating scl and lmo2 expression and promoted myelopoiesis by inhibiting gata1 expression and inducing spi1 and mpo expression. Many major cancer pathways were upregulated, such as cell cycle, glycolysis/gluconeogenesis, MAPK/Ras, and p53-mediated apoptosis. In contrast, mismatch repair and transforming growth factor β (TGFβ) signaling were downregulated. Overall, the model faithfully recapitulates AML phenotypes with high incidence (∼75%) and rapid onset (∼60 dpf).
Internal tandem duplications of the receptor tyrosine kinase FLT3 (FLT3-ITD) is a common mutation in AML and associated with poor prognosis and increased risk of relapse (Takahashi, 2011; Hou et al., 2013). It frequently coincides with mutations to the nucleophosmin NPM1 that restrict it to the cytoplasm (NPMc+). Lu et al. (2016) sought to investigate the interaction of these two mutations in AML by making transgenic lines expressing each under the myeloid spi1 promoter, spi1:FLT3-ITD-2A-EGFP and spi1:NPM1-Mut-PA. The FLT3-ITD mutant fish alone developed moderate myeloid hyperplasia at 6 months and some of these progressed to leukemia at 9 months. NPMc+ mutants had grossly normal hematopoietic composition. However, double mutants for both FLT3-ITD and NPMc+ progressed to leukemia by 6 months, demonstrating their synergistic effect in driving AML. In a different model using NPMc+ mRNA embryonic microinjections, Bolli et al. (2010) saw an increase in spi1+ early myeloid progenitors, with a more pronounced effect in a p53 mutant line. NPMc+ expression resulted in increased erythromyeloid progenitors in the posterior blood island and c-myb/cd41+ cells in the ventral wall of the aorta. They suggest these results may be relevant to human NPMc+ AML, where a multilineage expression pattern implies transformation of a multipotent HSPC.
Using a large-scale ENU mutagenesis screen, Peng et al. (2015) identified a line with a significant increase in HSPCs in hematopoietic organs, designated LDD731:CBLH382T. They determined the causal mutation was in the c-cbl gene, which is found frequently mutated in human MPN and acute leukemias and acts as a tumor suppressor by depressing growth factor and cytokine signals. The mutation was homozygous lethal at ∼15 dpf and led to an expansion of the myeloid/erythroid lineages in definitive hematopoiesis. Flt3 was necessary for this expansion, consistent with that observed in both mice and humans, suggesting flt3 signaling promotes HSPC proliferation and is regulated by c-cbl.
cAMP response element binding protein (CREB) is another frequently overexpressed gene in AML, however, it is unclear whether overactivation alone is sufficient to induce leukemia. Tregnago et al. (2016) generated a zebrafish model overexpressing creb with the spi1 myeloid promoter, which resulted in a disruption of myelopoiesis in 79% of adult fish with 66% progressing to a monocytic leukemia (latency 9–14 months) mirroring the human counterpart. These fish showed a transcriptional signature with 20 differentially expressed genes in common with pediatric AML, including the CCAAT-enhancer-binding protein-δ (c/ebpδ). Increased c/ebpδ expression impaired myeloid differentiation which could be reversed through silencing of the creb-c/ebpδ axis. Identification of this creb-c/ebpδ axis in zebrafish AML led Tregnago et al to classify C/EBPδ expression as a new pediatric AML subgroup after validation in publicly available patient databases.
To study the role of interferon regulatory factor 8 (IRF8) in the pathogenesis of myeloid neoplasia, Zhao et al. (2018) created a missense mutation, irf8Δ57 that acted as a functional knockout. IRF8 is a critical transcription regulator for myeloid lineage commitment and closely tied to myeloid leukemia. irf8 mutants quickly developed MPN with expansion of myeloid precursors, which recurred after transplantation, and invasion of kidney marrow. Myeloid expansion was caused by both increased proliferation and decreased apoptosis. mertk expression was increased in irf8 mutants leading to hyperactivation of the ERK pathway. Transgenic mertk overexpression recapitulated the myeloid neoplasia and knockdown of mertk rescued irf8 mutant myeloid expansion. These results support mertk signaling as critical in the irf8-mediated regulation of myeloid proliferation and survival.
Myelodysplastic Syndrome
Myelodysplastic syndromes (MDS) are a group of diseases characterized by aberrant hematopoietic differentiation leading to cytopenias and increased blasts, and often splenomegaly and cytogenetic abnormalities (Gangat et al., 2016). Approximately 30% of MDS patients will eventually progress to AML or other leukemias, which are frequently more resistant to conventional therapies.
Somatic loss-of-function mutations of the 10–11 translocation 2 gene TET2 are frequently observed in patients with MDS. TET2 encodes a DNA methylcytosine oxidase that converts 5-methylcytosine (5 mC) to 5-hydroxymethylcytosine (5 hmC) to initiate the demethylation (and activation) of DNA. Gjini et al. (2015) created an enzymatically inert tet2 mutant zebrafish line through genome-editing technology. These fish had normal embryonic hematopoiesis, but developed progressive clonal myelodysplasia as they aged, eventually progressing to MDS by 24 months, with myeloid progenitor cell dysplasia and anemia. Decreased levels of 5 hmC were present in hematopoietic cells of the kidney marrow but not in other cell types, likely a result of compensation in non-hematopoietic tissues by other Tet family members.
The c-myb transcription factor is vital to hematopoietic proliferation and differentiation, and is closely associated with an array of hematological disorders. Liu et al. (2017) sought to better define its pathogenic role through characterization of a zebrafish model expressing a GFP-tagged c-myb mutant with increased activity, c-mybhyper. This hyperactive c-myb resulted in the dysregulation of cell cycle genes and subsequent proliferation of hematopoietic progenitor cells. Abnormal granulocyte expansion began embryonically and was maintained through adulthood, ultimately resulting in MDS. A small number of c-mybhyper fish developed AML or ALL and treatment with c-myb target drug flavopiridol relieved the MDS-like symptoms in both embryos and adult fish.
In addition to its previously discussed role in early myeloid progenitors, spi1 (pu.1) is an Ets-family transcription factor important in leukemogenesis. It is frequently impaired in AML either through decreased expression or loss-of-function mutations (Mueller et al., 2002; Dakic et al., 2007). Sun et al. (2013) used the Targeting Induced Local Lesions IN Genomes (TILLING) approach to create a hypomorphic spi1 mutant allele, dubbed pu.1G242D. These fish have expanded myelopoiesis by 3 dpf, with increased immature granulocytes within the CHT. By 18 months, immature myeloid cells were increased at the expense of the lymphoid population in both the kidney marrow and peripheral blood, consistent with an MDS or AML-like disorder. The antiproliferative drug cytarabine was able to relieve the myeloid expansion, while apoptosis-inducing daunorubicin could not. This may indicate that spi1-associated neoplasms are more susceptible to drugs limiting their proliferation.
Future Directions and Emerging Methods
The discoveries described in this review open numerous avenues for further research. Many of these models require further characterization and could uncover important pathways in leukemia initiation and progression. Drug screens utilizing these models can teach us much about the resistance and response to different therapies depending on the specific genetic drivers of the leukemia. The advent of effective CRISPR-Cas9 protocols has allowed for a rapid advancement in the creation of knockout and transgenic zebrafish investigating various genetic pathways and oncogene fusion products tied to human leukemia. This development will only continue to expand in scope, as ongoing research within the zebrafish field continues to uncover more genes and pathways associated with leukemia, as well as new discoveries made in the clinic that are converted into zebrafish models for further characterization.
One major emerging avenue of research is the identification and characterization of LSCs within hematopoietic malignancies. Cancer stem cells (CSCs), defined by their ability to regrow a tumor from a single cell, are implicated as the cause of cancer evolution, resistance to therapy, and relapse after therapy (Adorno-Cruz et al., 2015). Increased tumor heterogeneity and CSCs have been associated with resistance and relapse for many tumor types, including AML and ALL (Mullighan et al., 2008; Anderson et al., 2011; Notta et al., 2011; Ding et al., 2012). Identification and isolation of these cells is difficult because of a lack of defined surface markers, but there are some promising methods being developed. The side population assay has long been used to isolate normal tissue stem cells by exploiting their ability to export Hoechst dye (Goodell et al., 1996), and has more recently been shown to enrich for CSCs in many cancers (Hu et al., 2010; Britton et al., 2011; Richard et al., 2013). Side populations have also been defined in zebrafish hematopoietic cells and leukemia (Kobayashi et al., 2008; Pruitt et al., 2017), making it possible for LSCs to be further studied in zebrafishleukemia models. Another similar protocol of enriching for stem cell activity is through the Aldefluor assay, which utilizes the increased aldehyde dehydrogenase (ALDH) activity common in stem cells to produce increased fluorescence from the aldefluor reagent (Storms et al., 1999; Ma et al., 2010). This assay can also be combined with the side population assay to isolate an even greater enrichment of stem cells (Pearce and Bonnet, 2007; Pierre-Louis et al., 2009). Genetic and functional characterization of the combined ALDHbright and side population in zebrafish leukemia models could uncover significant contributors to leukemia resistance and relapse.
Another potential method for defining LSCs within a tumor is through single cell sequencing. Single-cell RNA sequencing techniques are capable of discerning expression profiles of each cell within a population, allowing small subpopulations like LSCs to be characterized within a tumor (Zhang et al., 2016). Multiple microfluidic systems have been developed to produce single-cell expression data, with the Fluidigm system already used by Moore et al. (2016) in a zebrafish T-ALL model to identify a small population with reactivated expression of putative stem cell genes. The DropSeq system is an alternative with much higher throughput that could potentially identify very small subpopulations in tumors with lower LSC frequencies (Macosko et al., 2015). Both systems allow for characterization of the expression profiles of LSCs in the various leukemia models, which opens up exciting possibilities in discovering what drives the LSC subpopulation and their unique functions within the leukemia.
Author Contributions
JB wrote the entirety of the review, with significant input from JdJ in the initial outline and also comments and revisions of the final version.
Funding
Investigator Award University of Chicago Women’s Board Discovering the genetic drivers of leukemia initiating cells in T-cell acute lymphoblastic leukemia. Program Pilot Project Grant University of Chicago Comprehensive Cancer Center Genetic drivers of leukemia initiating cells in the side population of zebrafish T-cell leukemia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the University of Chicago Women’s Board and the University of Chicago Comprehensive Cancer Center for helping to fund our work in this field.
References
Ablain, J., Durand, E. M., Yang, S., Zhou, Y., and Zon, L. I. (2015). A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 32, 756–764. doi: 10.1016/j.devcel.2015.01.032
Adorno-Cruz, V., Kibria, G., Liu, X., Doherty, M., Junk, D. J., Guan, D., et al. (2015). Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 75, 924–929. doi: 10.1158/0008-5472.CAN-14-3225
Alghisi, E., Distel, M., Malagola, M., Anelli, V., Santoriello, C., Herwig, L., et al. (2013). Targeting oncogene expression to endothelial cells induces proliferation of the myelo-erythroid lineage by repressing the Notch pathway. Leukemia 27, 2229–2241. doi: 10.1038/leu.2013.132
Anderson, K., Lutz, C., van Delft, F. W., Bateman, C. M., Guo, Y., Colman, S. M., et al. (2011). Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361. doi: 10.1038/nature09650
Andreasson, P., Schwaller, J., Anastasiadou, E., Aster, J., and Gilliland, D. G. (2001). The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer Genet. Cytogenet. 130, 93–104. doi: 10.1016/S0165-4608(01)00518-0
Bartram, C. R., de Klein, A., Hagemeijer, A., van Agthoven, T., van Kessel, A. G., Bootsma, D., et al. (1983). Translocation of c-abl oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature 306, 277–280. doi: 10.1038/306277a0
Blackburn, J. S., Liu, S., Raimondi, A. R., Ignatius, M. S., Salthouse, C. D., and Langenau, D. M. (2011). High-throughput imaging of adult fluorescent zebrafish with an LED fluorescence macroscope. Nat. Protoc. 6, 229–241. doi: 10.1038/nprot.2010.170
Blackburn, J. S., Liu, S., Raiser, D. M., Martinez, S. A., Feng, H., Meeker, N. D., et al. (2012). Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia 26, 2069–2078. doi: 10.1038/leu.2012.116
Blackburn, J. S., Liu, S., Wilder, J. L., Dobrinski, K. P., Lobbardi, R., Moore, F. E., et al. (2014). Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 25, 366–378. doi: 10.1016/j.ccr.2014.01.032
Bolli, N., Payne, E. M., Grabher, C., Lee, J. S., Johnston, A. B., Falini, B., et al. (2010). Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood 115, 3329–3340. doi: 10.1182/blood-2009-02-207225
Britton, K. M., Kirby, J. A., Lennard, T. W. J., and Meeson, A. P. (2011). Cancer stem cells and side population cells in breast cancer and metastasis. Cancers 3, 2106–2130. doi: 10.3390/cancers3022106
Brownlie, A., Donovan, A., Pratt, S. J., Paw, B. H., Oates, A. C., Brugnara, C., et al. (1998). Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat. Genet. 20, 244–250. doi: 10.1038/3049
Burns, C. E., DeBlasio, T., Zhou, Y., Zhang, J., Zon, L., and Nimer, S. D. (2002). Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp. Hematol. 30, 1381–1389. doi: 10.1016/S0301-472X(02)00955-4
Chen, J., Jette, C., Kanki, J. P., Aster, J. C., Look, A. T., and Griffin, J. D. (2007). NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 21, 462–471. doi: 10.1038/sj.leu.2404546
Cumano, A., and Godin, I. (2007). Ontogeny of the hematopoietic system. Annu. Rev. Immunol. 25, 745–785. doi: 10.1146/annurev.immunol.25.022106.141538
Dakic, A., Wu, L., and Nutt, S. L. (2007). Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis?. Trends Immunol. 28, 108–114. doi: 10.1016/j.it.2007.01.006
de Jong, J. L., and Zon, L. I. (2005). Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 39, 481–501. doi: 10.1146/annurev.genet.39.073003.095931
Detrich, H. W., Kieran, M. W., Chan, F. Y., Barone, L. M., Yee, K., Rundstadler, J. A., et al. (1995). Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 92, 10713–10717. doi: 10.1073/pnas.92.23.10713
Deveau, A. P., Bentley, V. L., and Berman, J. N. (2017). Using zebrafish models of leukemia to streamline drug screening and discovery. Exp. Hematol. 45, 1–9. doi: 10.1016/j.exphem.2016.09.012
Deveau, A. P., Forrester, A. M., Coombs, A. J., Wagner, G. S., Grabher, C., Chute, I. C., et al. (2015). Epigenetic therapy restores normal hematopoiesis in a zebrafish model of NUP98–HOXA9-induced myeloid disease. Leukemia 29, 2086–2097. doi: 10.1038/leu.2015.126
Ding, L., Ley, T. J., Larson, D. E., Miller, C. A., Koboldt, D. C., Welch, J. S., et al. (2012). Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510. doi: 10.1038/nature10738
Evans, D. I. K., and Steward, J. K. (1972). Down’s syndrome and leukemia. Lancet 300:1322. doi: 10.1016/S0140-6736(72)92704-3
Feng, H., Langenau, D. M., Madge, J. A., Quinkertz, A., Gutierrez, A., Neuberg, D. S., et al. (2007). Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br. J. Haematol. 138, 169–175. doi: 10.1111/j.1365-2141.2007.06625.x
Feng, H., Stachura, D. L., White, R. M., Gutierrez, A., Zhang, L., Sanda, T., et al. (2010). T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 18, 353–366. doi: 10.1016/j.ccr.2010.09.009
Forrester, A. M., Grabher, C., McBride, E. R., Boyd, E. R., Vigerstad, M. H., Edgar, A., et al. (2011). NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. Br. J. Haematol. 155, 167–181. doi: 10.1111/j.1365-2141.2011.08810.x
Frazer, J. K., Meeker, N. D., Rudner, L., Bradley, D. F., Smith, A. C., Demarest, B., et al. (2009). Heritable T-cell malignancy models established in a zebrafish phenotypic screen. Leukemia 23, 1825–1835. doi: 10.1038/leu.2009.116
Gangat, N., Patnaik, M. M., and Tefferi, A. (2016). Myelodysplastic syndromes: contemporary review and how we treat. Am. J. Hematol. 91, 76–89. doi: 10.1002/ajh.24253
Garcia, E. G., Iyer, S., Garcia, S. P., Loontiens, S., Sadreyev, R. I., Speleman, F., et al. (2018). Cell of origin dictates aggression and stem cell number in acute lymphoblastic leukemia. Leukemia doi: 10.1038/s41375-018-0130-0 [Epub ahead of print].
GBD 2015 Mortality and causes of death collaborators (2016). Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544. doi: 10.1016/S0140-6736(16)31012-1
Gjini, E., Mansour, M. R., Sander, J. D., Moritz, N., Nguyen, A. T., Kesarsing, M., et al. (2015). A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol. Cell. Biol. 35, 789–804. doi: 10.1128/MCB.00971-14
Goodell, M. A., Brose, K., Paradis, G., Conner, A. S., and Mulligan, R. C. (1996). Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183, 1797–1806. doi: 10.1084/jem.183.4.1797
Gough, S. M., Slape, C. I., and Aplan, P. D. (2011). NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118, 6247–6257. doi: 10.1182/blood-2011-07-328880
Grabher, C., Joly, J. S., and Wittbrodt, J. (2004). Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 77, 381–401. doi: 10.1016/S0091-679X(04)77021-1
Gutierrez, A., Grebliunaite, R., Feng, H., Kozakewich, E., Zhu, S., Guo, F., et al. (2011). Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J. Exp. Med. 208, 1595–1603. doi: 10.1084/jem.20101691
Gutierrez, A., Sanda, T., Grebliunaite, R., Carracedo, A., Salmena, L., Ahn, Y., et al. (2009). High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood 114, 647–650. doi: 10.1182/blood-2009-02-206722
Hess, I., and Boehm, T. (2016). Stable multilineage xenogeneic replacement of definitive hematopoiesis in adult zebrafish. Sci. Rep. 6:19634. doi: 10.1038/srep19634
Hou, H. A., Lin, C. C., Chou, W. C., Liu, C. Y., Chen, C. Y., Tang, J. L., et al. (2013). Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia 28, 50–58. doi: 10.1038/leu.2013.236
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Hruscha, A., Krawitz, P., Rechenberg, A., Heinrich, V., Hecht, J., Haass, C., et al. (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982–4987. doi: 10.1242/dev.099085
Hsu, K., Traver, D., Kutok, J. L., Hagen, A., Liu, T.-X., Paw, B. H., et al. (2004). The pu.1 promoter drives myeloid gene expression in zebrafish. Blood 104, 1291–1297. doi: 10.1182/blood-2003-09-3105
Hu, L., McArthur, C., and Jaffe, R. B. (2010). Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br. J. Cancer 102, 1276. doi: 10.1038/sj.bjc.6605626
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Jin, H., Xu, J., and Wen, Z. (2007). Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 109, 5208–5214. doi: 10.1182/blood-2007-01-069005
Kaufman, C. K., Mosimann, C., Fan, Z. P., Yang, S., Thomas, A., Ablain, J., et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197–aad2197. doi: 10.1126/science.aad2197
Kaufmann, A., Mickoleit, M., Weber, M., and Huisken, J. (2012). Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247. doi: 10.1242/dev.082586
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kobayashi, I., Saito, K., Moritomo, T., Araki, K., Takizawa, F., and Nakanishi, T. (2008). Characterization and localization of side population (SP) cells in zebrafish kidney hematopoietic tissue. Blood 111, 1131–1137. doi: 10.1182/blood-2007-08-104299
Kroon, E., Thorsteinsdottir, U., Mayotte, N., Nakamura, T., and Sauvageau, G. (2001). NUP98–HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 20, 350–361. doi: 10.1093/emboj/20.3.350
La Starza, R., Borga, C., Barba, G., Pierini, V., Schwab, C., Matteucci, C., et al. (2014). Genetic profile of T-cell acute lymphoblastic leukemias with MYC translocations. Blood 124, 3577–3582. doi: 10.1182/blood-2014-06-578856
Langenau, D. M., Feng, H., Berghmans, S., Kanki, J. P., Kutok, J. L., and Look, A. T. (2005). Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 102, 6068–6073. doi: 10.1073/pnas.0408708102
Langenau, D. M., Ferrando, A. A., Traver, D., Kutok, J. L., Hezel, J. P., Kanki, J. P., et al. (2004). In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 101, 7369–7374. doi: 10.1073/pnas.0402248101
Langenau, D. M., Keefe, M. D., Storer, N. Y., Jette, C. A., Smith, A. C., Ceol, C. J., et al. (2008). Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 27, 4242–4248. doi: 10.1038/onc.2008.56
Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., et al. (2003). Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887–890. doi: 10.1126/science.1080280
Le, X., Langenau, D. M., Keefe, M. D., Kutok, J. L., Neuberg, D. S., and Zon, L. I. (2007). Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 9410–9415. doi: 10.1073/pnas.0611302104
Liao, E. C., Paw, B. H., Oates, A. C., Pratt, S. J., Postlethwait, J. H., and Zon, L. I. (1998). SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in?zebrafish. Genes Dev. 12, 621–626. doi: 10.1101/gad.12.5.621
Lieschke, G. J., Oates, A. C., Paw, B. H., Thompson, M. A., Hall, N. E., Ward, A. C., et al. (2002). Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev. Biol. 246, 274–295. doi: 10.1006/dbio.2002.0657
Liu, W., Wu, M., Huang, Z., Lian, J., Chen, J., Wang, T., et al. (2017). c-myb hyperactivity leads to myeloid and lymphoid malignancies in zebrafish. Leukemia 31, 222–233. doi: 10.1038/leu.2016.170
Lu, J. W., Hou, H. A., Hsieh, M. S., Tien, H. F., and Lin, L. I. (2016). Overexpression of FLT3-ITD driven by spi-1 results in expanded myelopoiesis with leukemic phenotype in zebrafish. Leukemia 30, 2098–2101. doi: 10.1038/leu.2016.132
Lum, J. J., DeBerardinis, R. J., and Thompson, C. B. (2005). Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6, 439–448. doi: 10.1038/nrm1660
Lyons, S. E., Shue, B. C., Oates, A. C., Zon, L. I., and Liu, P. P. (2001). A novel myeloid-restricted zebrafish CCAAT/enhancer-binding protein with a potent transcriptional activation domain. Blood 97, 2611–2617. doi: 10.1182/blood.V97.9.2611
Ma, A. C., Chung, M. I., Liang, R., and Leung, A. Y. (2010). A DEAB-sensitive aldehyde dehydrogenase regulates hematopoietic stem and progenitor cells development during primitive hematopoiesis in zebrafish embryos. Leukemia 24, 2090–2099. doi: 10.1038/leu.2010.206
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002
Mizgireuv, I. V., and Revskoy, S. Y. (2006). Transplantable tumor lines generated in clonal zebrafish. Cancer Res. 66, 3120–3125. doi: 10.1158/0008-5472.Can-05-3800
Mizgirev, I. V., and Revskoy, S. (2010). A new zebrafish model for experimental leukemia therapy. Cancer Biol. Ther. 9, 895–902. doi: 10.4161/cbt.9.11.11667
Moore, F. E., Garcia, E. G., Lobbardi, R., Jain, E., Tang, Q., Moore, J. C., et al. (2016). Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J. Exp. Med. 213, 979–992. doi: 10.1084/jem.20152013
Mueller, B. U., Pabst, T., Osato, M., Asou, N., Johansen, L. M., Minden, M. D., et al. (2002). Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood 100, 998–1007. doi: 10.1182/blood.V100.3.998
Mullighan, C. G., Phillips, L. A., Su, X., Ma, J., Miller, C. B., Shurtleff, S. A., et al. (2008). Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380. doi: 10.1126/science.1164266
North, T. E., Goessling, W., Walkley, C. R., Lengerke, C., Kopani, K. R., Lord, A. M., et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011. doi: 10.1038/nature05883
Notta, F., Mullighan, C. G., Wang, J. C., Poeppl, A., Doulatov, S., Phillips, L. A., et al. (2011). Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 469, 362–367. doi: 10.1038/nature09733
Novoa, B., and Figueras, A. (2012). “Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases,” in Current Topics in Innate Immunity II, eds J. D. Lambris and G. Hajishengallis (New York, NY: Springer), 253–275.
Ogino, H., McConnell, W. B., and Grainger, R. M. (2006). High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 1, 1703–1710. doi: 10.1038/nprot.2006.208
Onnebo, S. M., Rasighaemi, P., Kumar, J., Liongue, C., and Ward, A. C. (2012). Alternative TEL-JAK2 fusions associated with T-cell acute lymphoblastic leukemia and atypical chronic myelogenous leukemia dissected in zebrafish. Haematologica 97, 1895–1903. doi: 10.3324/haematol.2012.064659
Paik, E. J., and Zon, L. I. (2010). Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 54, 1127–1137. doi: 10.1387/ijdb.093042ep
Palis, J., and Yoder, M. C. (2001). Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp. Hematol. 29, 927–936. doi: 10.1016/S0301-472X(01)00669-5
Palomero, T., Dominguez, M., and Ferrando, A. A. (2008). The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle 7, 965–970. doi: 10.4161/cc.7.8.5753
Pearce, D. J., and Bonnet, D. (2007). The combined use of Hoechst efflux ability and aldehyde dehydrogenase activity to identify murine and human hematopoietic stem cells. Exp. Hematol. 35, 1437–1446. doi: 10.1016/j.exphem.2007.06.002
Peeters, P., Raynaud, S. D., Cools, J., Wlodarska, I., Grosgeorge, J., Philip, P., et al. (1997). Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9; 12) in a lymphoid and t(9; 15; 12) in a myeloid leukemia. Blood 90, 2535–2540.
Peng, X., Dong, M., Ma, L., Jia, X. E., Mao, J., Jin, C., et al. (2015). A point mutation of zebrafish c-cbl gene in the ring finger domain produces a phenotype mimicking human myeloproliferative disease. Leukemia 29, 2355–2365. doi: 10.1038/leu.2015.154
Pierre-Louis, O., Clay, D., Brunet de la Grange, P., Blazsek, I., Desterke, C., Guerton, B., et al. (2009). Dual SP/ALDH functionalities refine the human hematopoietic Lin-CD34+CD38- stem/progenitor cell compartment. Stem Cells 27, 2552–2562. doi: 10.1002/stem.186
Pruitt, M. M., Marin, W., Waarts, M. R., and de Jong, J. L. O. (2017). Isolation of the side population in myc-induced T-cell acute lymphoblastic leukemia in zebrafish. J. Vis. Exp. 123:e55711. doi: 10.3791/55711
Ransom, D. G., Bahary, N., Niss, K., Traver, D., Burns, C., Trede, N. S., et al. (2004). The zebrafish moonshine gene encodes transcriptional intermediary factor 1γ, an essential regulator of hematopoiesis. PLoS Biol. 2:e237. doi: 10.1371/journal.pbio.0020237
Ransom, D. G., Haffter, P., Odenthal, J., Brownlie, A., Vogelsang, E., Kelsh, R. N., et al. (1996). Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development 123, 311–319.
Renshaw, S. A., and Trede, N. S. (2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38–47. doi: 10.1242/dmm.007138
Reynolds, C., Roderick, J. E., LaBelle, J. L., Bird, G., Mathieu, R., Bodaar, K., et al. (2014). Repression of BIM mediates survival signaling by MYC and AKT in high-risk T-cell acute lymphoblastic leukemia. Leukemia 28, 1819–1827. doi: 10.1038/leu.2014.78
Richard, V., Nair, M. G., Santhosh Kumar, T. R., and Pillai, M. R. (2013). Side population cells as prototype of chemoresistant, tumor-initiating cells. Biomed Res. Int. 2013:517237. doi: 10.1155/2013/517237
Ridges, S., Heaton, W. L., Joshi, D., Choi, H., Eiring, A., Batchelor, L., et al. (2012). Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631. doi: 10.1182/blood-2011-12-398818
Romana, S. P., Poirel, H., Leconiat, M., Flexor, M. A., Mauchauffe, M., Jonveaux, P., et al. (1995). High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood 86, 4263–4269.
Sabaawy, H. E., Azuma, M., Embree, L. J., Tsai, H. J., Starost, M. F., and Hickstein, D. D. (2006). TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 103, 15166–15171. doi: 10.1073/pnas.0603349103
Shen, L.-J., Chen, F.-Y., Zhang, Y., Cao, L.-F., Kuang, Y., Zhong, M., et al. (2013). MYCN transgenic zebrafish model with the characterization of acute myeloid leukemia and altered hematopoiesis. PLoS One 8:e59070. doi: 10.1371/journal.pone.0059070
Smith, A. C., Raimondi, A. R., Salthouse, C. D., Ignatius, M. S., Blackburn, J. S., Mizgirev, I. V., et al. (2010). High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood 115, 3296–3303. doi: 10.1182/blood-2009-10-246488
Sood, R., English, M. A., Jones, M., Mullikin, J., Wang, D.-M., Anderson, M., et al. (2006). Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods 39, 220–227. doi: 10.1016/j.ymeth.2006.04.012
Sotsios, Y., and Ward, S. G. (2000). Phosphoinositide 3-kinase: a key biochemical signal for cell migration in response to chemokines. Immunol. Rev. 177, 217–235. doi: 10.1034/j.1600-065X.2000.17712.x
Storms, R. W., Trujillo, A. P., Springer, J. B., Shah, L., Colvin, O. M., Ludeman, S. M., et al. (1999). Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. U.S.A. 96, 9118–9123. doi: 10.1073/pnas.96.16.9118
Stuart, G. W., McMurray, J. V., and Westerfield, M. (1988). Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 103, 403–412.
Sun, J., Liu, W., Li, L., Chen, J., Wu, M., Zhang, Y., et al. (2013). Suppression of Pu.1 function results in expanded myelopoiesis in zebrafish. Leukemia 27, 1913–1917. doi: 10.1038/leu.2013.67
Takahashi, S. (2011). Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J. Hematol. Oncol. 4, 13–13. doi: 10.1186/1756-8722-4-13
Tang, Q., Abdelfattah, N. S., Blackburn, J. S., Moore, J. C., Martinez, S. A., Moore, F. E., et al. (2014). Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods 11, 821–824. doi: 10.1038/nmeth.3031
Thompson, M. A., Ransom, D. G., Pratt, S. J., MacLennan, H., Kieran, M. W., Detrich, H. W., et al. (1998). Theclocheandspadetailgenes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 197, 248–269. doi: 10.1006/dbio.1998.8887
Tregnago, C., Manara, E., Zampini, M., Bisio, V., Borga, C., Bresolin, S., et al. (2016). CREB engages C/EBPdelta to initiate leukemogenesis. Leukemia 30, 1887–1896. doi: 10.1038/leu.2016.98
Urasaki, A., Morvan, G., and Kawakami, K. (2006). Functional dissection of the Tol2 transposable element identified the minimal cis sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649. doi: 10.1534/genetics.106.060244
Walker, C., and Streisinger, G. (1983). Induction of mutations by γ-rays in pregonial germ cells of zebrafish embryos. Genetics 103, 125–136.
Wang, H., Long, Q., Marty, S. D., Sassa, S., and Lin, S. (1998). A zebrafish model for hepatoerythropoietic porphyria. Nat. Genet. 20, 239–243. doi: 10.1038/3041
Weinberg, E. S., Allende, M. L., Kelly, C. S., Abdelhamid, A., Murakami, T., Andermann, P., et al. (1996). Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271–280.
Weng, A. P., Ferrando, A. A., Lee, W., Morris, J. P., Silverman, L. B., Sanchez-Irizarry, C., et al. (2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271. doi: 10.1126/science.1102160
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Willett, C. E., Cherry, J. J., and Steiner, L. A. (1997). Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics 45, 394–404. doi: 10.1007/s002510050221
Willett, C. E., Kawasaki, H., Amemiya, C. T., Lin, S., and Steiner, L. A. (2001). Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev. Dyn. 222, 694–698. doi: 10.1002/dvdy.1223
Yeh, J. R., Munson, K. M., Chao, Y. L., Peterson, Q. P., Macrae, C. A., and Peterson, R. T. (2008). AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development 135, 401–410. doi: 10.1242/dev.008904
Yeh, J. R., Munson, K. M., Elagib, K. E., Goldfarb, A. N., Sweetser, D. A., and Peterson, R. T. (2009). Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat. Chem. Biol. 5, 236–243. doi: 10.1038/nchembio.147
Zhang, X., Marjani, S. L., Hu, Z., Weissman, S. M., Pan, X., and Wu, S. (2016). Single-cell sequencing for precise cancer research: progress and prospects. Cancer Res. 76, 1305–1312. doi: 10.1158/0008-5472.CAN-15-1907
Zhao, F., Shi, Y., Huang, Y., Zhan, Y., Zhou, L., Li, Y., et al. (2018). Irf8 regulates the progression of myeloproliferative neoplasm-like syndrome via Mertk signaling in zebrafish. Leukemia 32, 149–158. doi: 10.1038/leu.2017.189
Keywords: zebrafish, leukemia, animal models, ALL, AML, MDS, MPN
Citation: Baeten JT and de Jong JLO (2018) Genetic Models of Leukemia in Zebrafish. Front. Cell Dev. Biol. 6:115. doi: 10.3389/fcell.2018.00115
Received: 30 April 2018; Accepted: 23 August 2018;
Published: 20 September 2018.
Edited by:
Rebecca Ann Wingert, University of Notre Dame, United StatesReviewed by:
Lasse Dahl Ejby Jensen, Linköping University, SwedenEirini Trompouki, Max-Planck-Institut für Immunbiologie und Epigenetik, Germany
Copyright © 2018 Baeten and de Jong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jill L. O. de Jong, amRlam9uZ0BwZWRzLmJzZC51Y2hpY2Fnby5lZHU=
 Jeremy T. Baeten
Jeremy T. Baeten Jill L. O. de Jong
Jill L. O. de Jong