
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 07 August 2018
Sec. Cell Adhesion and Migration
Volume 6 - 2018 | https://doi.org/10.3389/fcell.2018.00086
This article is part of the Research Topic A Deep Look into the Mechanisms of Adhesion Molecule-Mediated Pathogenesis View all 6 articles
 Takeshi Ito1†
Takeshi Ito1† Yutaka Kasai1†
Yutaka Kasai1† Yuki Kumagai1
Yuki Kumagai1 Daisuke Suzuki1
Daisuke Suzuki1 Misaki Ochiai-Noguchi1
Misaki Ochiai-Noguchi1 Daisuke Irikura2
Daisuke Irikura2 Shiro Miyake2‡
Shiro Miyake2‡ Yoshinori Murakami1*
Yoshinori Murakami1*The cell adhesion molecule (CADM) family of the immunoglobulin superfamily (IgSF) comprises four members, CADM1–CADM4, and participates in the formation of epithelial and synaptic adhesion through cell–cell homophilic and heterophilic interactions. To identify the partners that interact with each member of the CADM family proteins, we set up a platform for multiple detection of the extracellular protein–protein interactions using surface plasmon resonance imaging (SPRi) and analyzed the interactions between the CADM family proteins and 10 IgSF of their structurally related cell adhesion molecules. SPRi analysis identified a new interaction between CADM1 and CADM4, where this heterophilic interaction was shown to be involved in morphological spreading of adult T-cell leukemia (ATL) cells expressing CADM1 when incubated on CADM4-coated glass. Moreover, class-I MHC-restricted T-cell-associated molecule (CRTAM) was identified to show the highest affinity to CADM1 among its binding partners by comparing the dissociation constants calculated from the SPR sensorgrams. These results suggest that the SPRi platform would provide a novel screening tool to characterize extracellular protein–protein interactions among cell-surface and secreted proteins, including IgSF molecules.
The immunoglobulin superfamily (IgSF) encompasses cell-surface and secreted proteins with immunoglobulin loops that are involved in a variety of physiological functions, including immune regulation, neural transmission, and epithelial tissue formation through interaction with IgSF molecules, integrins, and other proteins within the extracellular space (Aricescu and Jones, 2007). Most of IgSF molecules form both homophilic and heterophilic interactions to perform their functions; however, because of the limited approaches to identifying the partners in these interactions, their interaction network is not fully understood (Gonzalez, 2012). For example, immunoprecipitation coupled with mass spectrometry is generally not applicable for detecting extracellular interactions because of the low affinity of the membrane proteins. The yeast two-hybrid screening system is also unsuitable because extracellular proteins are often misfolded in the nucleus. Furthermore, comprehensive screening of molecular interaction using synthesized proteins from Escherichia coli or the wheat germ cell-free system is not appropriate for detecting interactions between extracellular proteins, like IgSF molecules (Wright et al., 2010) because post-translational modification such as glycosylation, which is critical to extracellular interactions, cannot be reconstituted.
A number of methods have been developed to screen the interactions between a pair of IgSF molecules. One such method is the avidity-based extracellular interaction screen (AVEXIS), a type of ELISA, used to detect the direct interactions between each bait–prey pair of recombinant protein fragments. AVEXIS was used to screen pairwise interactions between 249 of cell-surface and secreted IgSFs and leucine-rich repeat proteins from a protein library of a zebrafish (Danio rerio) and a number of molecular interactions were newly identified (Bushell et al., 2008; Martin et al., 2010). Similar findings were obtained for the interactions between IgSF, leucine-rich repeat, and fibronectin type III proteins in Drosophila melanogaster using the extracellular interactome assay, a modified AVEXIS method (Özkan et al., 2013). The protein microarray assay also succeeded in detecting several interaction pairs by screening a set of 89 human IgSF proteins against 686 highly diverse extracellular proteins (Ramani et al., 2012).
Surface plasmon resonance (SPR) is a label-free and real-time bio-sensing technology that is widely used for the validation of direct protein–protein interactions. Although SPR requires micrograms of purified proteins and so is not generally suitable for large-scale screening of protein–protein interactions, it was used to screen >2,000 proteins to identify the co-receptor for the B- and T-lymphocyte attenuator (BTLA), a kind of immune receptor of IgSF (Gonzalez et al., 2005). The interactions between 36 IgSF proteins and leucocyte surface proteins was examined using a 6 × 6 SPR interaction array (Jiang and Neil Barclay, 2010). Recently, SPR imaging (SPRi) instruments have been developed carrying a larger sensoring surface than conventional SPR to enable multiple detection of protein–protein interactions in combination with microarray spotting technology (Faye et al., 2009).
Here we developed a platform for screening the pairs of interacting IgSF molecules using a SPRi system. Our platform can detect up to 96 protein interactions in a single injection and the amount of protein solution required for spotting onto the chip surface is <10 nL, making it conducive to large-scale screening. Using the SPRi system, we examined the interactions among the IgSF molecules, including the cell adhesion molecule (CADM) and the Nectin families. The CADM and the Nectin families comprise four members each and share three Ig loops within the extracellular region and a short cytoplasmic domain. These molecules are involved in various types of cell–cell adhesion, such as that between epithelial cells or synaptic cells through homophilic and heterophilic interactions within the family molecules (Takai et al., 2008). Poliovirus receptor (PVR) is structurally related to the CADM and the Nectin families and forms heterophilic interactions with CADM1 and Nectin-3 (Ikeda et al., 2003; Wakayama et al., 2007). CADM1 also forms a heterophilic interaction with class-I MHC-restricted T-cell-associated molecule (CRTAM), another IgSF molecule (Arase et al., 2005; Boles et al., 2005; Galibert et al., 2005). By producing recombinant proteins of 10 IgSF molecules (CADM1–CADM4, Nectin-1–Nectin-4, PVR, and CRTAM), we examined their interactions with the CADM family proteins using SPRi.
FreeStyle 293-F cells were purchased from Thermo Fisher Scientific (Waltham, MA, USA) and cultured in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) supplemented with 100 units/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific). 293FT cells were purchased from Thermo Fisher Scientific and cultured in Dulbecco's modified Eagle's medium with 4.5 g/L glucose (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS) (BioWest, Nuaille, France), 100 units/mL penicillin, and 100 μg/mL streptomycin. ATN1 cells were obtained from RIKEN BioResource Center (Tsukuba, Japan), cultured in RPMI1640 medium (Nacalai Tesque) supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin, and the cells were maintained at 37°C in a humidified 5% CO2 incubator (Panasonic, Osaka, Japan).
The FreeStyle™ 293 Expression System (Thermo Fisher Scientific) was used to produce IgSF-Fc protein, and the expression vectors for protein production were prepared using Gateway™ Cloning Technology (Thermo Fisher Scientific), as described below.
Extracellular domains of the 10 IgSF molecules, CADM1 (NP_055148; 1–374 a.a.), CADM2 (NP_001161146; 1–367 a.a.), CADM3 (NP_001120645; 1–330 a.a.), CADM4 (NP_660339; 1–324 a.a.), Nectin-1 (NP_002846; 1–355 a.a.), Nectin-2 (NP_002847; 1–360 a.a.), Nectin-3 (NP_056295; 1–404 a.a.), Nectin-4 (NP_112178; 1–349 a.a.), PVR (NP_006496; 1–343 a.a.), and CRTAM (NP_062550; 1–287 a.a.), were cloned into the pENTR/D-TOPO vector (Thermo Fisher Scientific). To obtain a Gateway-based destination vector, a Gateway cassette containing attR recombination sites flanking a ccdB gene and a chloramphenicol-resistance gene was cloned into the SmaI site of pHEK293 Ultra Expression Vector II (TaKaRa Bio Inc., Shiga, Japan), and a human IgG2-Fc fragment from the pFUSE-hIgG2-Fc1 vector (InvivoGen, San Diego, CA, USA) was subsequently inserted into the vector's XhoI-SphI site. This destination vector was named “pHEK-Fc.” The expression vectors of the extracellular domains of the IgSF molecules with a C-terminal Fc tag were obtained using a Gateway recombination with LR clonase (Thermo Fisher Scientific) between the pENTR/D-TOPO and pHEK-Fc vectors.
For the production of IgSF-Fc proteins, 30 μg of each expression vector and 6 μg pHEK293 Enhancer Vector (TaKaRa Bio Inc.) were transiently transfected into 3 × 107 FreeStyle 293-F cells with 36 μL TransIT-PRO Reagent (Mirus Bio, Madison, WI, USA) in 30 mL Freestyle 293 Expression Medium (Thermo Fisher Scientific). The cells were cultured in a disposable Erlenmeyer flask (Thermo Fisher Scientific) that was shaken at 120 rpm using a rotary shaker (RemoteShake; Wakenbtech, Kyoto, Japan) for 5 d at 37°C in a humidified 8% CO2 incubator.
The supernatants from 293-F cells transfected with IgSF-Fc expression vectors were filtered through a 0.45-μm membrane filter (Merck Millipore, Darmstadt, Germany). Prewashed 50 μL of Protein G Sepharose (GE Healthcare, Little Chalfont, UK) was added to each supernatant and rotated overnight at 4°C. Protein G Sepharose was collected after brief centrifugation and placed into MicroSpin Empty Columns (GE Healthcare). After five washes with 400 μL wash buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5), the proteins were eluted by adding 200 μL elution buffer (0.1 M glycine, pH 3.0) and were subsequently neutralized with 30 μL 1 M Tris-HCl (pH 8.8). This elution step was performed twice. The obtained protein solution was dialyzed against phosphate buffered saline (PBS) using a Slide-A-Lyzer MINI Dialysis Device (Thermo Fisher Scientific). Each protein was stained with silver, and protein purity was analyzed using the Silver Stain MS Kit (Wako Pure Chemical Industries, Osaka, Japan). The Bradford method, the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA, USA), and a microplate reader (Model 680; Bio-Rad) were used to measure protein concentration. If needed, the protein solution was concentrated using Amicon Ultra Centrifugal Filter Units (Merck Millipore). We obtained ~10–700 μg purified protein from 30 mL of the supernatant from 293-F cells that were transiently transfected with the expression vectors.
Binding assays were performed using the SPRi system (OpenPlex; HORIBA France, Palaiseau, France) according to instructions in the previous report (Yamasaki et al., 2016) with some modifications.
For the preparation of sensor chips, purified IgSF-Fc proteins and normal human IgG (Sigma-Aldrich, St. Louis, MO, USA) in PBS were spotted onto an SPRi-Biochip (HORIBA France) at 10 nL/spot using a dedicated spotter in a 96-spot format at HORIBA Ltd. (Kyoto, Japan). The spotted proteins were immobilized on the chip by amine coupling. The prepared sensor chip was inserted into the instrument according to the manufacturer's instructions.
All the reactions were performed at 25°C. Running buffer [PBS with 0.2% bovine serum albumin (BSA) and 0.02% Tween20] continuously flowed into the reaction chamber at 50 μL/min using the MINIPULS 3 peristaltic pump (Gilson, Inc., Middleton, WI, USA). The prepared protein samples were first preserved in a sample loop (volume = 200 μL) and then allowed to flow into the chamber by switching the valve to the injection mode. Intensity of the light reflected from an 810-nm LED with attenuation by SPR phenomena was recorded every 3 s using an 8-bit charge-coupled device (CCD) camera. The reflectivity of each spot was altered when the protein samples or cells were bound to the immobilized proteins. Percent change in reflectivity (%ΔR) was calculated on the basis of the CCD signal and normalized by subtracting the reflectivity for the same concentration of normal human IgG. Data were processed and analyzed using ScrubberGen (HORIBA France). A schematic diagram of the SPRi experiment is shown in Supplementary Figure 1.
Most of the interactions among the purified proteins were dissociated spontaneously within 30 min by the continuous flow of running buffer without any regeneration reagents. If needed, 0.1 M acetic acid was injected into the chamber for 2 min at 25 μL/min and flushed out with running buffer at 1 mL/min. Removal efficiency was monitored by SPR signals and the sensor chip was reused for the next binding assay without any other treatments after the signals had sufficiently returned to baseline levels.
Mouse monoclonal anti-FLAG M2 and anti-GFP antibodies were purchased from Sigma-Aldrich and Roche (Basel, Switzerland), respectively. Rabbit polyclonal anti-CADM1 (C-18) antibody was previously described (Ito et al., 2011). Chicken monoclonal anti-CADM1 antibody (9D2) was purchased from Medical and Biological Laboratories Co., Ltd (Nagoya, Japan). Goat polyclonal anti-GAPDH (V-18) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Goat polyclonal anti-human IgG (Fc specific) antibody was purchased from Sigma-Aldrich.
293FT cells were transfected with CADM4-FLAG, CADM1-GFP, CADM3-GFP, and E-cadherin-GFP in four separate 6-cm dishes. After 24 h, the cells transfected with CADM4-FLAG were co-cultured with cells transfected with CADM1-GFP, CADM3-GFP, or E-cadherin-GFP in 10-cm dishes at a 1:1 ratio. After 2-day culture, the co-cultured cells were treated with 1 mM DTSSP/PBS for 30 min at room temperature, and any cross-linking was quenched with 20 mM Tris-HCl (pH 7.4). The cells were lysed with TNE buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton-X100, pH 7.8) containing a protease inhibitor cocktail (200 μM AEBSF, 10 μM leupeptin, and 1 μM pepstatin A) for 30 min at 4°C. The lysates were centrifuged at 12,000 rpm for 20 min at 4°C and the supernatants were immunoprecipitated using anti-FLAG M2 Affinity Gel (Sigma-Aldrich) overnight at 4°C. After washing the gel four times with TNE buffer, the FLAG-tagged CADM4 was eluted using 150 μg/mL of 3 × FLAG peptide (ApexBio, Houston, TX, USA) and subjected to Western blotting. The CADM1-GFP, E-cadherin-GFP, and CADM4-FLAG expression vectors were previously described (Kuramochi et al., 2001; Williams et al., 2006; Sakurai-Yageta et al., 2015). The CADM3-GFP expression vector was obtained by cloning CADM3 cDNA into the pEGFP-N3 BglII-SalI site (TaKaRa Bio Inc.).
CADM1 was knocked down by the lentiviral delivery of short-hairpin (sh)RNA. The oligonucleotides for shRNA against CADM1 or non-targeting shRNA were annealed and subcloned into the pENTR4-H1 BglII-XbaI site (RIKEN BioResource Center), and the fragment that included the H1 promoter and shRNA precursor was transferred to the CS-RfA-CG destination vector (RIKEN BioResource Center) using LR clonase (Thermo Fisher Scientific). The obtained shRNA-expressing lentiviral vector as well as pCAG-HIVgp and pCMV-VSV-G-RSV-Rev (RIKEN BioResource Center) were co-transfected into the 293FT cells using Polyethylenimine Max (Polysciences, Warrington, PA, USA). After 72 h, the culture supernatants containing the lentiviruses were collected, and the viruses were concentrated by ultracentrifugation at 50,000 × g for 2 h at 4°C. The virus titers were determined using qPCR Lentivirus Titration Kit (Applied Biological Materials, Richmond, Canada), SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and the ABI 7300 real-time PCR system (Applied Biosystems). ATN1 cells were infected with the lentiviruses at a multiplicity of infection of 5. The cells expressing shRNA were GFP-positive cells obtained using FACS Aria II (BD Biosciences, Franklin Lakes, NJ, USA). The target sequences of shRNA were as follows: shCADM1, 5,5′-CGAAAGACGTGACAGTGAT-3′; shCADM1, 8,5′-GCGCTTGAGTTAACATGT-3'; and shControl, 5'-ACTACCGTTGTTATAGGT-3′.
Western blotting was performed as previously described (Ito et al., 2011). Briefly, cell lysates were prepared using a lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing a protease inhibitor cocktail. Equal amounts of total protein were fractionated in 7.5% SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Merck Millipore), and incubated with primary antibodies. Primary antibody binding was detected using the Pierce Western Blotting Substrate (Thermo Fisher Scientific) with horseradish peroxidase-conjugated secondary antibodies, and the signals were visualized using ImageQuant LAS 4000 mini (GE Healthcare).
A cell-spreading assay was performed as previously described (Murakami et al., 2014). Coverslips were precoated with 50 μg/mL poly-L-lysine (Sigma-Aldrich), fixed with 0.5% glutaraldehyde (Sigma-Aldrich), and then coated with 7 μg/mL CADM4-Fc or normal human IgG (Sigma-Aldrich) in PBS. After blocking with 1% BSA (Wako Pure Chemical Industries) in HBSS (Thermo Fisher Scientific), 1 × 104 cells in culture medium were plated onto coverslips and incubated for 60 min at 37°C. The cells were fixed with 4% paraformaldehyde and labeled with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). The coverslips were mounted with Prolong Gold antifade reagent with DAPI (Thermo Fisher Scientific), and the cells were imaged using the Axio Observer D1 epifluorescence microscope (Carl Zeiss, Oberkochen, Germany). The cell area was measured using AutoMeasure, AxioVision Version 4 (Carl Zeiss). An area comprising 100 cells was examined in each experiment. Statistical significance was determined using Student's t-test.
The CADM and the Nectin family of IgSF cell-adhesion molecules contribute to cell–cell adhesion through homophilic and heterophilic interactions. We thus analyzed the interactions among IgSF molecules, focusing on the CADM and the Nectin family members and their related molecules using SPRi. Extracellular domains of 10 IgSF molecules, including the CADM family (CADM1–CADM4), Nectin family (Nectin-1–Nectin-4), PVR, and CRTAM, were produced as Fc-fused proteins in the supernatant of transfected 293-F cells. In SDS-PAGE, the purified proteins exhibited a shift toward higher molecular weight after their post-translational modifications such as glycosylation as observed in human tissues (Figure 1).
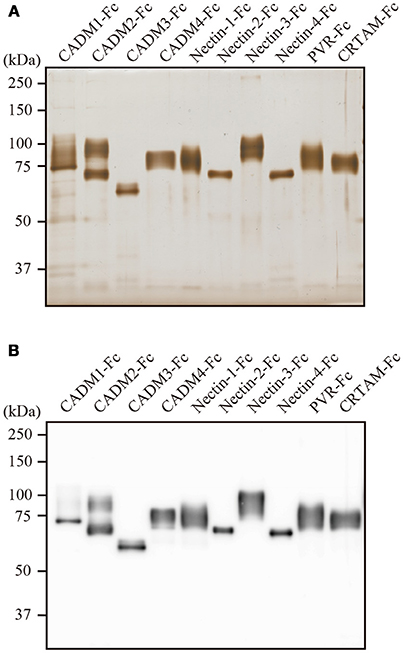
Figure 1. Characterization of the purified IgSF-Fc proteins. (A) Silver staining of the purified IgSF-Fc proteins. Each 100 ng of IgSF-Fc protein was fractionated in a 7.5% SDS-PAGE gel. (B) Western blotting of the purified IgSF-Fc proteins using an anti-human IgG-Fc antibody.
To examine the interactions between CADM1 and the 10 IgSF molecules using SPRi, IgSF-Fc proteins were prepared at a concentration of 7–24 μM as described in Supplementary Table 1 and spotted on an SPR sensor chip in hexaplicate. When CADM1-Fc was subjected to SPRi, the sensorgrams as well as the SPR image showed that CADM1 interacts with CADM1, CADM2, CADM3, and CRTAM (Figures 2A,B). Furthermore, the sensorgrams and SPR image demonstrated that CADM1 interacts with CADM4, which had not been reported previously. The association rate constants (kon), dissociation rate constants (koff), and dissociation constants (Kd) of the interactions were then calculated from the sensorgrams (Table 1). Notably, the response curve of CRTAM was quite different from that of the other molecules. The dissociation of CADM1 from CRTAM was much slower than that from the CADM family proteins with approximately a 100-fold difference in koff (58–173), suggesting that the interaction between CADM1 and CRTAM is stable and does not easily dissociate once it is formed.
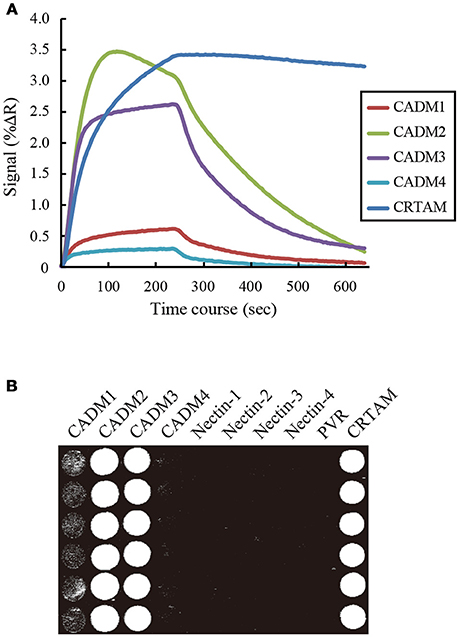
Figure 2. Surface plasmon resonance imaging analysis of the interactions between CADM1 and its binding partners. (A) SPRi analysis was performed by injecting 2.4 μM CADM1-Fc for 4 min onto the sensor chip where the 10 IgSF-Fc proteins were spotted. The analysis was performed in hexaplicate and the averaged signal of the six spots are shown. Only five molecules giving positive signals are shown. (B) The percent change in reflectivity (%ΔR) at 60 s was visualized as a bright spot on the image.
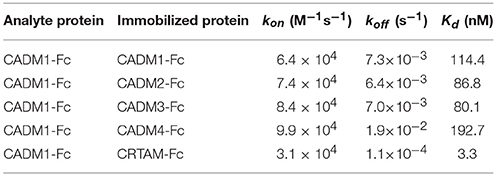
Table 1. Surface plasmon resonance analysis of the interaction between CADM1 and its binding partners.
We then subjected CADM2-Fc, CADM3-Fc, and CADM4-Fc to SPRi and examined their possible interaction with the 10 IgSF-Fc proteins. These analyses detected both the homophilic and heterophilic interactions of CADM2, CADM3, and CADM4 with all the CADM family proteins (Figure 3). However, some of the interactions previously reported were not detected in the present SPRi analysis. These include the heterophilic interactions of CADM1 with Nectin-3 or PVR and the heterophilic interactions of CADM3 with the Nectins 1 or 3 (Shingai et al., 2003; Kakunaga et al., 2005; Wakayama et al., 2007; Figure 3). Failure of the detection of these known interactions by SPRi might be caused by their lower affinity than the detection limit of SPRi in this experimental condition. Alternatively, when interactions previously reported in cell-based assay required some accessory proteins, SPRi could not detect such interactions because it could only detect direct interactions between two molecules.
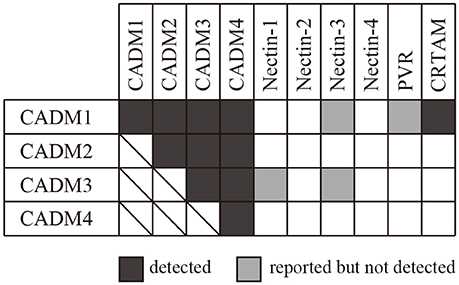
Figure 3. Combination of the interactions between the CADM family proteins and the 10 IgSF molecules including the CADM and the Nectin family proteins. The interactions detected by SPRi are shown in black and those reported but not detected by SPRi are shown in gray.
To examine the possible interaction between CADM1 and CADM4 in cell-based analyses, 293FT cells independently transfected with either CADM4-FLAG or CADM1-GFP were co-cultured and subjected to immunoprecipitation. The signal from CADM1-GFP was detected in the precipitates using an anti-FLAG antibody (Figure 4A), suggesting that CADM1 and CADM4 form a trans-heterophilic interaction with neighboring cells. Next, we examined the interaction between CADM1 and CADM4 in cell spreading assay using a cell line, ATN1, which is derived from adult T-cell leukemia (ATL) expressing a high amount of CADM1 protein. This assay can detect the interaction of CADM1 expressed in an ATL cell with purified CADM4-Fc protein immobilized on the glass. ATN1 cells show CADM1-dependent morphological spreading when incubated on endothelial cells or fibroblasts (Masuda et al., 2010). Similarly, when incubated on CADM4-Fc-coated glass, ATN1 cells exhibited spread morphology with increased cell area. In contrast, CADM1 knockdown by two independent shRNAs inhibited the spreading of ATN1 cells on the CADM4-Fc-coated glass (Figures 4B–D). Furthermore, a neutralizing CADM1 antibody blocked the spreading of ATN1 cells on CADM4-Fc-coated glass (Figure 4E). These results indicate that the heterophilic interaction between CADM1 and CADM4 induces the spreading of ATN1 cells. It is known that CADM1 promotes adhesion of ATL cells to vascular endothelial cells (Sasaki et al., 2005; Masuda et al., 2010). However, the partners for CADM1 binding onto endothelial cells have not been identified yet. Since CADM4 expression is observed on endothelial cells (Yamana et al., 2015), the trans-heterophilic interaction between CADM1 and CADM4 might be involved in ATL cell adhesion to endothelial cells.
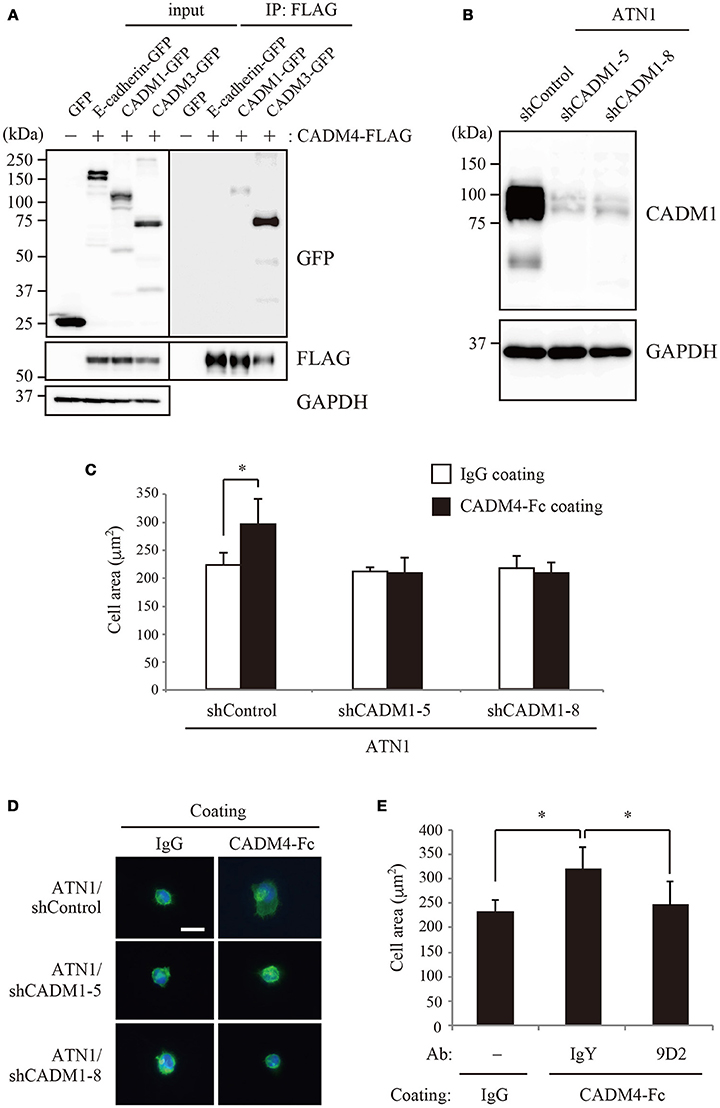
Figure 4. The interaction between CADM1 and CADM4 is involved in the cell spreading of ATN1 cells. (A) The trans-interaction between CADM1 and CADM4 in the extracellular region was examined by immunoprecipitation assay. E-cadherin and CADM3 were used as the negative and positive controls, respectively. (B) Knockdown of CADM1 in ATN1 cells by two independent shRNAs, shCADM1-5, and shCADM1-8. (C) ATN1 cells with control shRNA or shCADM1 were incubated on coverslips coated with control IgG or CADM4-Fc. The area of 100 cells was measured in an assay and the average of three independent experiments is shown. *p < 0.05 by t-test. (D) Representative images of the spread morphology of ATN1 cells with each shRNA incubated on IgG- or CADM4-Fc-coated glasses. The cells were visualized by staining the actin cytoskeleton with Alexa Fluor 488-labeled phalloidin. The nuclei were stained with DAPI. Magnification, × 200. Scale bar, 20 μm. (E) ATN1/shControl cells were incubated on IgG or CADM4-Fc for 60 min in the presence of control chicken IgY (10 μg/mL) or anti-CADM1 antibody, 9D2 (10 μg/mL). The average cell area of three independent experiments is shown. *p < 0.05 by t-test.
In the present study, we developed a platform for multiple and sensitive detection of extracellular protein–protein interactions using an SPRi system. In comparison with other high-throughput techniques for detecting protein–protein interactions such as protein microarrays or AVEXIS, the throughput of SPRi might be limited because SPRi requires a large amount of purified proteins. In fact, protein microarrays are mainly designed for screening interacting partners from a large-scale of protein library. For example, a commercially available protein microarray (ProtoArray, Thermo Fisher Scientific) contains more than 9,000 proteins immobilized on a glass slide. By contrast, SPRi have a great advantage in detecting protein–protein interaction sensitively and quantitatively on the basis of the reaction rate constants of the interactions. This advantage is attributed to the unique detection principle of SPRi, in which protein–protein interaction is quantitatively monitored in real time on the basis of the changes in the intensity of reflected light with attenuation by SPR phenomena. Thus, SPRi is more sensitive in detecting the molecular interactions than protein microarrays or AVEXIS, which monitors direct interaction of two molecules by fluorescence- or chemiluminescence-based techniques. As to the instruments, SPRi and protein microarrays need dedicated detectors as well as microarray spotters if customizing protein arrays are necessary, whereas AVEXIS is relatively a low-cost system except for a microplate reader.
In this study, we focused on CADM family proteins and examined possible protein–protein interactions among 10 IgSF molecules, including CADMs 1–4 and their structurally related 6 proteins, using an SPRi system. Then, we newly identified an interaction between CADM1 and CADM4, although previous analyses using affinity chromatography (Fogel et al., 2007) or cell-adhesion assay (Maurel et al., 2007; Thomas et al., 2008) could not detect this interaction. The discrepant results suggest that CADM1–CADM4 interaction might not provide enough activity of cell adhesion in the previous study. On the other hand, several interactions reported by previous studies were not detected in the present study. These include interactions between CADM1–Nectin-3, CADM1–PVR, CADM3–Nectin-1, and CADM3–Nectin-3, which were detected essentially by cell-based assay (Kakunaga et al., 2005; Wakayama et al., 2007; Masuda et al., 2010). This discrepancy may be caused if previously detected interactions are only formed in the presence of other accessary interactions between unidentified molecules in cell-based assay. Alternatively, we might detect these interactions by increasing the concentration of proteins in SPRi analysis, because extraordinary high concentration of analyte protein (>44 μM) was used to detect homophilic interactions between Nectin family proteins in the previous study (Harrison et al., 2012).
In SPRi analysis, we had the privilege of analyzing the kinetics of the interaction between CADM1 and each binding partner. Among them, the kinetics of the interaction between CADM1 and CRTAM was quite different from that between CADM1 and CADMs 1–4 (Figure 2). In particular, the koff of the interaction between CADM1 and CRTAM is much smaller than those of CADM1 and CADMs 1–4 (Table 1). Small koff indicated that the interaction between CADM1 and CRTAM is stable and dissociates very slowly. In fact, this interaction is found physiologically between epithelial cells expressing CADM1 and CD8+ T or natural killer (NK) cells expressing CRTAM (Arase et al., 2005; Boles et al., 2005; Galibert et al., 2005). Stable interaction with very slow dissociation would be suitable for CD8+ T or NK cells to exhibit efficient cytotoxic effects on epithelial target cells expressing CADM1 through specific cell–cell adhesion. On the other hand, homophilic interaction of CADM1 has a large koff, which makes it more susceptible to dissociation. Therefore, CADM1 appears to be engaged in cell–cell adhesion with continuous association–dissociation processes. This is consistent with a previous report by fluorescence recovery after photobleaching, demonstrating continuous renewal of CADM1 on the plasma membrane of epithelial cells even in a static state (Sakurai-Yageta et al., 2015). The difference in the kinetics of each interaction appears to be caused by the difference in the higher order structures of the molecules. The binding mode between CADM1 and CRTAM is a double “lock-and-key” structure (Zhang et al., 2013), whereas the homophilic binding of CADM1 appears to be mediated by a single hydrophobic interaction (Dong et al., 2006).
In this study, the Kd of the interaction between CADM1-Fc and CRTAM-Fc was estimated to be 3.3 nM. This constant was obtained by SPRi analysis using the whole extracellular domains fused with Fc that were purified from 293-F cells. On the other hand, the Kd of the interaction between the IgV domain of CADM1 and that of CRTAM purified from E. coli was 12.5 μM with kon = 6.27 × 102 M−1s−1 and koff = 7.84 × 10−3 s−1 in a previous study (Zhang et al., 2013). The marked difference in the Kd between two experiments suggest that the IgC-loops of CADM1 and CRTAM facilitate a part of interaction, although the N-terminal IgV-loops of each molecule are primarily involved in the interaction (Arase et al., 2005). In addition, dimerization by the Fc-tag used in our study may mimic and enhance the cis-dimer formation of both CADM1 and CRTAM (Masuda et al., 2002; Shingai et al., 2003; Arase et al., 2005). Moreover, the N-glycosylation of CADM1 and CRTAM which are only observed in our analysis using 293-F cells but not in the study using E. coli would enhance the interaction between CADM1 and CRTAM as was shown in the interaction between CADM1 and CADM2 (Fogel et al., 2010). These findings suggest that Kd obtained in our SPRi analysis would reflect the physiological condition more correctly.
Using SPRi analysis, we newly identified an interaction between CADM1 and CADM4. Furthermore, we showed that CADM1–CADM4 interaction was involved in the cell spreading activity of an ATL cell line, ATN-1 (Figure 4). CADM1 is ectopically expressed in ATL and promotes tumor growth and infiltration (Sasaki et al., 2005; Dewan et al., 2008), whereas CADM4 is expressed in endothelial cells. Therefore, interaction of CADM1 in ATL cells with CADM4 in endothelial cells may trigger infiltration of ATL cells into various organs. As this case illustrates, our platform using the SPRi system has the potential to identify a low-affinity but physiologically significant molecular interactions that have not been detected by other assays. Taking into account that SPR can determine the kinetics of the on and off rates for molecular interactions, our platform would provide an excellent and useful tool for both detecting and characterizing extracellular protein–protein interactions.
TI and SM conceived and designed the experiments. YKa and YKu performed vector construction, protein production and immunoprecipitation. DS performed the cell spreading assay. MO-N produced CADM1-knockdown cells. DI performed the SPR analyses. TI and YM wrote the manuscript.
This work was supported in part by the Project for Cancer Research And Therapeutics Evolution (P-CREATE) from the Japan Agency for Medical Research and Development to TI and YM, and by JSPS KAKENHI Grant Number 16K18412 and 17H05993 to TI.
DI and SM are employees of HORIBA Ltd., which is the company that sells the surface plasmon resonance imaging system used in this study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to M. Sakurai-Yageta and S. Kobayashi for their generous help for the lentiviral delivery of shRNAs. We also thank T. Maruyama and A. Nakamura for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2018.00086/full#supplementary-material
SPRi, surface plasmon resonance imaging; IgSF, immunoglobulin superfamily; CRTAM, class-I MHC-restricted T-cell-associated molecule; Fc, fragment crystallizable region; ATL, adult T-cell leukemia.
Arase, N., Takeuchi, A., Unno, M., Hirano, S., Yokosuka, T., Arase, H., et al. (2005). Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int. Immunol. 17, 1227–1237. doi: 10.1093/intimm/dxh299
Aricescu, A. R., and Jones, E. Y. (2007). Immunoglobulin superfamily cell adhesion molecules: zippers and signals. (1016) Curr. Opin. Cell biol. 19, 543–50. doi: 10./j.ceb.2007.09.010
Boles, K. S., Barchet, W., Diacovo, T., Cella, M., and Colonna, M. (2005). The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood 106. 779–786. doi: 10.1182/blood-2005-02-0817
Bushell, K. M., Söllner, C., Schuster-Boeckler, B., Bateman, A., and Wright, G. J. (2008). Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 18, 622–630. doi: 10.1101/gr.7187808
Dewan, M. Z., Takamatsu, N., Hidaka, T., Hatakeyama, K., Nakahata, S., Fujisawa, J., et al. (2008). Critical role for TSLC1 expression in the growth and organ infiltration of adult T-cell leukemia cells in vivo. J. Virol. 82, 11958–11963. doi: 10.1128/JVI.01149-08
Dong, X., Xu, F., Gong, Y., Gao, J., Lin, P., Chen, T., et al. (2006). Crystal structure of the V domain of human Nectin-like molecule-1/Syncam3/Tsll1/Igsf4b, a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule. J. Biol. Chem. 281, 10610–10617. doi: 10.1074/jbc.M513459200
Faye, C., Chautard, E., Olsen, B. R., and Ricard-Blum, S. (2009). The first draft of the endostatin interaction network. J. Biol. Chem. 284, 22041–22047. doi: 10.1074/jbc.M109.002964
Fogel, A. I., Akins, M. R., Krupp, A. J., Stagi, M., Stein, V., and Biederer, T. (2007). SynCAMs organize synapses through heterophilic adhesion. J. Neurosci. 27, 12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007
Fogel, A. I., Li, Y., Giza, J., Wang, Q., Lam, T. T., Modis, Y., et al. (2010). N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion. J. Biol. Chem. 285, 34864–34874. doi: 10.1074/jbc.M110.120865
Galibert, L., Diemer, G. S., Liu, Z., Johnson, R. S., Smith, J. L., Walzer, T., et al. (2005). Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 280, 21955–21964. doi: 10.1074/jbc.M502095200
Gonzalez, L. C. (2012). Protein microarrays, biosensors, and cell-based methods for secretome-wide extracellular protein-protein interaction mapping. Methods 57, 448–458. doi: 10.1016/j.ymeth.2012.06.004
Gonzalez, L. C., Loyet, K. M., Calemine-Fenaux, J., Chauhan, V., Wranik, B., Ouyang, W., et al. (2005). A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc. Nat. Acad. Sci. U.S.A. 102, 1116–1121. doi: 10.1073/pnas.0409071102
Harrison, O. J., Vendome, J., Brasch, J., Jin, X., Hong, S., Katsamba, P. S., et al. (2012). Nectin ectodomain structures reveal a canonical adhesive interface. Nat. Struct. Mol. Biol. 19, 906–915. doi: 10.1038/nsmb.2366
Ikeda, W., Kakunaga, S., Itoh, S., Shingai, T., Takekuni, K., Satoh, K., et al. (2003). Tage4/Nectin-like molecule-5 heterophilically trans-interacts with cell adhesion molecule Nectin-3 and enhances cell migration. J. Biol. Chem. 278, 28167–28172. doi: 10.1074/jbc.M303586200
Ito, T., Williams-Nate, Y., Iwai, M., Tsuboi, Y., Hagiyama, M., Ito, A., et al. (2011). Transcriptional regulation of the CADM1 gene by retinoic acid during the neural differentiation of murine embryonal carcinoma P19 cells. Genes Cells 16, 791–802. doi: 10.1111/j.1365-2443.2011.01525.x
Jiang, L., and Neil Barclay, A. N. (2010). Identification of leucocyte surface protein interactions by high-throughput screening with multivalent reagents. Immunology 129, 55–61. doi: 10.1111/j.1365-2567.2009.03153.x
Kakunaga, S., Ikeda, W., Itoh, S., Deguchi-Tawarada, M., Ohtsuka, T., Mizoguchi, A., et al. (2005). Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J. Cell Sci. 118(Pt 6), 1267–1277. doi: 10.1242/jcs.01656
Kuramochi, M., Fukuhara, H., Nobukuni, T., Kanbe, T., Maruyama, T., Ghosh, H. P., et al. (2001). TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat. Genet. 27, 427–430. doi: 10.1038/86934
Martin, S., Söllner, C., Charoensawan, V., Adryan, B., Thisse, B., Thisse, C., et al. (2010). Construction of a large extracellular protein interaction network and its resolution by spatiotemporal expression profiling. Mol. Cell. Proteomics 9, 2654–2665. doi: 10.1074/mcp.M110.004119
Masuda, M., Maruyama, T., Ohta, T., Ito, A., Hayashi, T., Tsukasaki, K., et al. (2010). CADM1 interacts with Tiam1 and promotes invasive phenotype of human T-cell leukemia virus type I-transformed cells and adult T-cell leukemia cells. J. Biol. Chem. 285, 15511–15522. doi: 10.1074/jbc.M109.076653
Masuda, M., Yageta, M., Fukuhara, H., Kuramochi, M., Maruyama, T., Nomoto, A., et al. (2002). The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J. Biol. Chem. 277, 31014–31019. doi: 10.1074/jbc.M203620200
Maurel, P., Einheber, S., Galinska, J., Thaker, P., Lam, I., Rubin, M. B., et al. (2007). Nectin-like proteins mediate axon–Schwann cell interactions along the internode and are essential for myelination. J. Cell Biol. 178, 861–874. doi: 10.1083/jcb.200705132
Murakami, S., Sakurai-Yageta, M., Maruyama, T., and Murakami, Y. (2014). Trans-homophilic interaction of CADM1 activates PI3K by forming a complex with MAGuK-family proteins MPP3 and Dlg. PLoS ONE 9:e82894. doi: 10.1371/journal.pone.0082894
Özkan, E., Carrillo, R. A., Eastman, C. L., Weiszmann, R., Waghray, D., Johnson, K. G., et al. (2013). An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239. doi: 10.1016/j.cell.2013.06.006
Ramani, S. R., Tom, I., Lewin-Koh, N., Wranik, B., DePalatis, L., Zhang, J., et al. (2012). A secreted protein microarray platform for extracellular protein interaction discovery. Anal. Biochem. 420, 127–138. doi: 10.1016/j.ab.2011.09.017
Sakurai-Yageta, M., Maruyama, T., Suzuki, T., Ichikawa, K., and Murakami, Y. (2015). Dynamic regulation of a cell adhesion protein complex including CADM1 by combinatorial analysis of FRAP with exponential curve-fitting. PLoS ONE 10:e0116637. doi: 10.1371/journal.pone.0116637
Sasaki, H., Nishikata, I., Shiraga, T., Akamatsu, E., Fukami, T., Hidaka, T., et al. (2005). Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 105, 1204–1213. doi: 10.1182/blood-2004-03-1222
Shingai, T., Ikeda, W., Kakunaga, S., Morimoto, K., Takekuni, K., Itoh, S., et al. (2003). Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J. Biol. Chem. 278, 35421–35427. doi: 10.1074/jbc.M305387200
Takai, Y., Miyoshi, J., Ikeda, W., and Ogita, H. (2008). Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell biol. 9, 603–615. doi: 10.1038/nrm2457
Thomas, L. A., Akins, M. R., and Biederer, T. (2008). Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J. Comp. Neurol. 510, 47–67. doi: 10.1002/cne.21773
Wakayama, T., Sai, Y., Ito, A., Kato, Y., Kurobo, M., Murakami, Y., et al. (2007). Heterophilic binding of the adhesion molecules poliovirus receptor and immunoglobulin superfamily 4A in the interaction between mouse spermatogenic and Sertoli cells. Biol. reprod. 76, 1081–1090. doi: 10.1095/biolreprod.106.058974
Williams, Y. N., Masuda, M., Sakurai-Yageta, M., Maruyama, T., Shibuya, M., and Murakami, Y. (2006). Cell adhesion and prostate tumor-suppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4. Oncogene 25, 1446–1453. doi: 10.1038/sj.onc.1209192
Wright, G. J., Martin, S., Bushell, K. M., and Söllner, C. (2010). High-throughput identification of transient extracellular protein interactions. Biochem. Soc. Trans. 38, 919–922. doi: 10.1042/BST0380919
Yamana, S., Tokiyama, A., Mizutani, K., Hirata, K. I., Takai, Y., and Rikitake, Y. (2015). The cell adhesion molecule Necl-4/CADM4 serves as a novel regulator for contact inhibition of cell movement and proliferation. PLoS ONE 10:e0124259. doi: 10.1371/journal.pone.0124259
Yamasaki, T., Miyake, S., Nakano, S., Morimura, H., Hirakawa, Y., Nagao, M., et al. (2016). Development of a surface plasmon resonance-based immunosensor for detection of 10 major O-antigens on shiga toxin-producing Escherichia coli, with a gel displacement technique to remove bound bacteria. Anal. Chem. 88, 6711–6717. doi: 10.1021/acs.analchem.6b00797
Keywords: surface plasmon resonance imaging, protein–protein interaction, cell adhesion molecule, immunoglobulin superfamily, adult T-cell leukemia
Citation: Ito T, Kasai Y, Kumagai Y, Suzuki D, Ochiai-Noguchi M, Irikura D, Miyake S and Murakami Y (2018) Quantitative Analysis of Interaction Between CADM1 and Its Binding Cell-Surface Proteins Using Surface Plasmon Resonance Imaging. Front. Cell Dev. Biol. 6:86. doi: 10.3389/fcell.2018.00086
Received: 15 May 2018; Accepted: 20 July 2018;
Published: 07 August 2018.
Edited by:
Mitsugu Fujita, Kindai University, JapanReviewed by:
Simone Diestel, Universität Bonn, GermanyCopyright © 2018 Ito, Kasai, Kumagai, Suzuki, Ochiai-Noguchi, Irikura, Miyake and Murakami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshinori Murakami, eW11cmFrYW1AaW1zLnUtdG9reW8uYWMuanA=
†These authors have contributed equally to this work.
‡Present Address: Shiro Miyake, Laboratory of Food and Hygiene, Department of Food and Life Science, School of Life and Environmental Science, Azabu University, Kanagawa, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.