- Center for Biochemistry and Microbial Science, School of Basic and Applied Science, Central University of Punjab, Bathinda, India
A commentary on
HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition
by Sakuma, K., Sasaki, E., Kimura, K., Komori, K., Shimizu, Y., Yatabe, Y., et al. (2017). Gut. doi: 10.1136/gutjnl-2016-312927
Metastasis, the spread of a tumor from its primary site to a distant organ, is the leading cause of cancer morbidity and mortality (Lambert et al., 2017). Metastasis Suppressors (MS) and Metastasis Inducers (MI) genes are critical mediators of this process. By definition, MS gene inhibits metastasis without affecting in-vitro cell proliferation and in-vivo primary tumor growth (Smith and Theodorescu, 2009). To date, more than 40 metastasis suppressor genes have been reported in colorectal cancer (Zhao et al., 2015); restoration of MS gene and its activity is an attractive strategy to prevent metastasis and disease-free survival of cancer patients.
Aberrant splicing generates diversity in human proteome and emerges as a key mediator in the differentiation and epithelial-mesenchymal transition (EMT) (Biamonti et al., 2012). Differential splicing of CD44 imparts an instrumental role in metastasis and drug resistance in several cancers. CD44v6, a marker for cancer stem cells, has been shown to promote cancer metastasis and drive rewiring (reprogramming) of Cancer Stem Cell (CSC) in colon cancer (Todaro et al., 2014).
Heterogeneous nuclear ribonucleoprotein L-like (HNRNPLL), located on the chromosome 2p22.1, is a RNA-binding protein that functions as a regulator of alternative splicing for multiple mRNAs target. Genome-wide screening using shRNA library in the orthotropic and lung metastasis mouse model has identified several MS genes in breast and colon cancer (Gumireddy et al., 2009; Duquet et al., 2014). Using a genome-wide silencing approach in non-metastatic CMT93 mouse cell line expressing Venus fluorescent protein in a syngeneic mouse model, Sakuma and colleagues have delineated the inverse relationship between HNRNPLL and CD44v6 expression in the metastasis of colon cancer (Sakuma et al., 2017).
shRNA mediated silencing of HNRNPLL inhibits in-vitro cell proliferation and enhances matrigel invasion to promote metastasis in colon cancer cells. Further, knocking down HNRNPLL inhibits in-vivo primary tumor growth, and lung metastasis of colon cancer which is reverted by shRNA resistant HNRNPLL plasmid. RNA immunoprecipitation (RIP) analysis in HNRNPLL silencing SW480 cells increases expression of CD44 exon v3-10 while exogenous expression of FLAG-tagged HNRNPLL-infected cells shows a significant increase in the CD44v6 expression. Further, Western blot analysis revealed the significantly higher expression of CD44v6 in the HNRNPLL-depleting CMT93 cells. Interestingly, pre-incubation with 2F10 and 9A4 neutralizing antibodies for CD44v6 reduced the matrigel invasion in control and HNRNPLL-knocked down cells confirming its function in promoting invasion of colon cancer cells. Moreover, knocking down HNRNPLL promotes CD44v6 mediated HGF induced c-Met phosphorylation to promotes invasion in colon cancer cells.
Epithelial-mesenchymal transition emerges as a driving force for the tumor metastasis, drug resistance, and cancer stem cell generation (Shibue and Weinberg, 2017). Mechanistically, HNRNPLL expression was significantly reduced both at the mRNA and protein level in the HGF/FGF-induced in-vitro epithelial-mesenchymal transition in SW480 colon cells which is recovered by mesenchymal-epithelial transition (MET). Moreover, immunostaining analysis of tumor samples shows the substantially decreased expression of HNRNPLL at the invasive front with E-cadherin suggesting its function in regulating early event in metastasis. Further, Induction of EMT specifically downregulated expression of HNRNPLL without much change in closely related splicing factors such as Heterogeneous Nuclear Ribonucleoprotein M (HNRNPM), Heterogeneous Nuclear Ribonucleoprotein L (HNRNPL) and epithelial splicing regulatory protein 1 (ESRP1). In summary, their data provide the critical role of HNRNPLL in regulating tumor cell invasion and epithelial-mesenchymal transition to regulate metastasis of colon cancer cells (Figure 1).
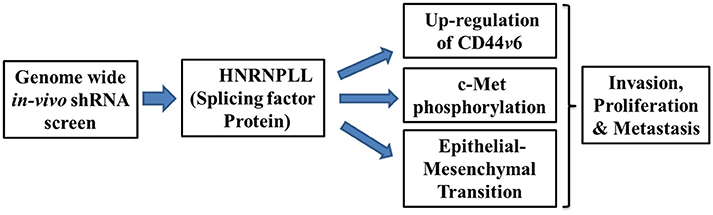
Figure 1. Schematic illustration showing mechanisms of metastatic suppressor function of HNRNPLL. Silencing of HNRNPLL promotes alternate splicing of CD44v6 by exon inclusion, c-Met phosphorylation and epithelial-mesenchymal transition to regulates invasion, proliferation and metastasis in colon cancer.
HNRNPLL expressed ubiquitously with strong nuclear and cytoplasmic expression with at least 9 predicted transcript variant of molecular weight ranging from 16.1 to 60.1 kDa (Uhlen et al., 2015). HNRNPLL predicted to undergo post-translational modification such as ubiquitination and phosphorylation1. Future studies will elucidate the transcript-specific roles of HNRNPLL in the regulation of metastasis, and exact function of post-translational modification in the stability and function of HNRNPLL.
Despite these, the exact mechanism of how and when HNRNPLL expression is lost or suppressed, during colon cancer progression remains to be characterized. Further, given its critical importance in EMT transition, it will be plausible to see whether HNRNPLL regulates CSC growth, self-renewal, and drug resistance in colon cancer. Moreover, how expression level of HNRNPLL in a large cohort will serve as a prognostic marker and whether HNRNPLL expression can be used as tools for predicting drug treatment for colon cancer metastasis needs to be determined. In summary, future studies will unleash the detailed molecular mechanisms of HNRNPLL on the regulation of cancer progression and provide a way for targeting the early events in the colorectal cancer metastasis.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
SB is thankful for financial support from DBT-Ramalingaswami Fellowship grant (BT/HRD/02/09/2013), DST-SERB (ECR/2016/000903) grant, and Research Seed Money (GP-25) grant from Central University of Punjab, Bathinda.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Biamonti, G., Bonomi, S., Gallo, S., and Ghigna, C. (2012). Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell. Mol. Life Sci. 69, 2515–2526. doi: 10.1007/s00018-012-0931-7
Duquet, A., Melotti, A., Mishra, S., Malerba, M., Seth, C., Conod, A., et al. (2014). A novel genome-wide in vivo screen for metastatic suppressors in human colon cancer identifies the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol. Med. 6, 882–901. doi: 10.15252/emmm.201303799
Gumireddy, K., Li, A., Gimotty, P. A., Klein-Szanto, A. J., Showe, L. C., Katsaros, D., et al. (2009). KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat. Cell Biol. 11, 1297–1304. doi: 10.1038/ncb1974
Lambert, A. W., Pattabiraman, D. R., and Weinberg, R. A. (2017). Emerging biological principles of metastasis. Cell 168, 670–691. doi: 10.1016/j.cell.2016.11.037
Sakuma, K., Sasaki, E., Kimura, K., Komori, K., Shimizu, Y., Yatabe, Y., et al. (2017). HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Gut. doi: 10.1136/gutjnl-2016-312927. [Epub ahead of print].
Shibue, T., and Weinberg, R. A. (2017). EMT, CSC, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 4, 611–629. doi: 10.1038/nrclinonc.2017.44
Smith, S. C., and Theodorescu, D. (2009). Learning therapeutic lessons from metastasis suppressor proteins. Nat. Rev. Cancer 9, 253–264. doi: 10.1038/nrc2594
Todaro, M., Gaggianesi, M., Catalano, V., Benfante, A., Iovino, F., Biffoni, M., et al. (2014). CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 14, 342–356. doi: 10.1016/j.stem.2014.01.009
Uhlen, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419
Keywords: metastasis, shRNA, cell adhesion, mouse model, alternative splicing
Citation: Baranwal S (2017) Commentary: HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Front. Cell Dev. Biol. 5:91. doi: 10.3389/fcell.2017.00091
Received: 18 August 2017; Accepted: 02 October 2017;
Published: 16 October 2017.
Edited by:
Uwe Knippschild, University of Ulm, GermanyReviewed by:
Georgios Giamas, University of Sussex, United KingdomCopyright © 2017 Baranwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Somesh Baranwal, c29tZXNoYmFyYW53YWxAZ21haWwuY29t
 Somesh Baranwal
Somesh Baranwal