
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 07 June 2017
Sec. Cell Growth and Division
Volume 5 - 2017 | https://doi.org/10.3389/fcell.2017.00061
This article is part of the Research TopicDeterminants of Cell SizeView all 8 articles
Cell size is amenable by genetic and environmental factors. The highly conserved nutrient-responsive Target of Rapamycin (TOR) signaling pathway regulates cellular metabolic status and growth in response to numerous inputs. Timing and duration of TOR pathway activity is pivotal for both cell mass built up as well as cell cycle progression and is controlled and fine-tuned by the abundance and quality of nutrients, hormonal signals, growth factors, stress, and oxygen. TOR kinases function within two functionally and structurally discrete multiprotein complexes, TORC1 and TORC2, that are implicated in temporal and spatial control of cell size and growth respectively; however, recent data indicate that such functional distinctions are much more complex. Here, we briefly review roles of the two complexes in cellular growth and cytoarchitecture in various experimental model systems.
The Target of Rapamycin (TOR) is an evolutionarily conserved Ser/Thr-protein kinase functioning as the heart of signaling networks toward nutrient and hormonal sensing. The role of these networks is to regulate anabolism and catabolism by coordinating numerous cellular metabolic processes, such as protein translation, formation of ribosome components, mRNA stability, autophagy, cell-cycle, transcription, and cellular architecture (Laplante and Sabatini, 2012; Rallis and Bahler, 2013; Rodland et al., 2014). Tor genes together with the FKBP12 homolog fpr1 were first isolated in Saccharomyces cerevisiae (Heitman et al., 1991; Kunz et al., 1993) as the mediators of the toxic effects of sirolimus or rapamycin, a macrolide from Streptomyces hygroscopicus bacteria living within the soil on the Rapa Nui or Easter Island (Sehgal et al., 1975; Sehgal, 2003). Rapamycin exhibits broad anti-proliferative properties and is a potent anti-tumor and immunosuppressant drug (Law, 2005). Rapamycin directly binds FKBP12 and the complex then binds and inhibits the TOR kinase (Yang et al., 2013). In all eukaryotes, TOR kinases are found in two distinct protein complexes, termed TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Huang and Fingar, 2014). Both complexes are implicated in cell growth. TORC1 is generally responsible for promoting ribosome biogenesis, protein anabolism and cell proliferation (Averous and Proud, 2006; Morita et al., 2015) and repressing cell differentiation (Alvarez and Moreno, 2006). The TSC1–TSC2 (hamartin and tuberin, respectively) protein complex can repress TORC1 by affecting Rheb, a G-protein that acts as positive regulator of this complex (Huang and Manning, 2008). TORC2 can have opposing or antagonistic functions to those of TORC1 (Weisman et al., 2007; Ikai et al., 2011). It gets input from carbon sources and insulin and regulates actin cytoskeleton (De Virgilio and Loewith, 2006). Fission and budding yeasts have two TOR kinases, Tor1 and Tor2 (Otsubo and Yamamato, 2008; Weisman, 2016). Fission yeast Tor1 protein is not essential and is found to be associated with both TORC1 and TORC2 (Hartmuth and Petersen, 2009). Tor1 is required for survival in stress response, appropriate G1 arrest, gene silencing, telomere integrity and sexual development. Conversely, the essential protein Tor2 is associated with TORC1 and is pivotal for growth by positively regulating protein synthesis, metabolism and transcription (Weisman et al., 2007; Otsubo and Yamamato, 2008; Ikai et al., 2011; Weisman, 2016).
TORC complexes are hubs of huge signaling networks that govern cellular growth in space in time and are heavily implicated in pathologies, such as cancer, obesity, type 2 diabetes, and neurodegeneration (Menon and Manning, 2008; Laplante and Sabatini, 2012) as well as in lifespan control and aging (Bjedov and Partridge, 2011; Rallis et al., 2013, 2014). In this mini review, we discuss roles of TORC1 and TORC2 in temporal and spatial control of cell growth and size. TORC1 while traditionally considered to be controlling temporal cell growth is also implicated in spatial control aspects with interdependent relationships with the cytoskeleton. Likewise, new interactions of TORC2 provide further links to TORC1 and temporal cell size regulation and cell cycle progression.
Cell size is regulated depending on nutrient availability, extracellular signals and stress (Perez-Hidalgo and Moreno, 2016). TORC1 senses diverse inputs, such as nitrogen- and carbon-containing nutrients, hormonal stimulation, various stresses, availability of energy within the cell and oxygen (De Virgilio and Loewith, 2006). Interestingly, various types of nutrients have diverse effects on the duration and strength of TORC1 activity that is reflected to the cellular phenotypes observed (De Virgilio and Loewith, 2006; Stracka et al., 2014). TORC1 boosts growth by promoting protein translation, ribosome biogenesis (Averous and Proud, 2006; Ma and Blenis, 2009), and glycolysis (Laplante and Sabatini, 2012) as well as suppressing stress responses (Lopez-Maury et al., 2008) as well as autophagy mechanisms (Ganley et al., 2009; Hosokawa et al., 2009).
TOR complex 1 (TORC1) has major contributions in temporal aspects of cell size control. For example, inhibition of TORC1 using rapamycin advances mitotic onset in fission yeast. The effect is similar to cell size reduction observed when cells sense and experience poor nitrogen sources. This is achieved through the stress MAPK pathway via the Pyp2 phosphatase (Petersen and Nurse, 2007). Interestingly, regulation of mitotic entry through the MAPK Sty1 upon rapamycin treatment involves the Tor1-containing TORC1 and not the complex that contains Tor2 (Hartmuth and Petersen, 2009). In budding yeast S. cerevisiae, TORC1 coordinates cell size by regulating timing of G1-S cell cycle progression. This is achieved by G1 cyclins and CDK activation as well as through destabilization of Sic1, a CDK inhibitor. When TORC1 is pharmacologically inhibited or following starvation, Sic1 is stabilized by c-terminal phosphorylation. The mechanism behind this involves endosulfines activated by the greatwall kinase. These in turn will stimulate Mpk1 and will inhibit the Cdc55 protein phosphatase 2A (Moreno-Torres et al., 2015). When ample nutrients are present, fission yeast TORC1 is highly active and inhibits Greatwall (Ppk18) protein kinase and resulting in PP2A-B55 activation. PP2A-B55 activity prevents Cdk1-Cyclin B action. Cells, therefore, increase in size during G2 before they commit to mitosis. In nutrient limitations, TORC1 activity lowers and Greatwall (Ppk18) activation results in endosulfine (Igo1) phosphorylation and PP2A-B55 inhibition. These events fully activate Cdk1·CyclinB and cells commit to mitosis at a small cell size (Chica et al., 2016). In pancreatic beta cells, TORC1 controls cell size by regulating cell cycle progression through modulation of the synthesis and stability of cyclin D2, an important regulator of beta cell proliferation and mass buildup (Balcazar et al., 2009).
TOR complex 1 (TORC1) promotes cell growth by stimulating synthesis of the primary building blocks of macromolecules, by inhibiting autophagy and influencing cell cycle progression (De Virgilio and Loewith, 2006). TOR functions on cell physiology are achieved through crosstalk with multiple other signaling pathways. The Hippo tumor suppressor pathway regulates tissue homeostasis, cell and organ size (Pfleger, 2017). Functional connections between TOR and Hippo pathways have started to emerge: YAP and TAZ are transcriptional co-activators and represent the major effectors of the Hippo pathway (Hansen et al., 2015a; Moroishi et al., 2015). YAP and TAZ are able to activate TORC1 transcriptional induction of the high affinity leucine transporter LAT1 in HEK293 cells (Hansen et al., 2015b). In addition, YAP downregulates PTEN, a negative regulator of TOR pathway, via a posttranscriptional mechanism that involves miR-29 in the inhibition of PTEN translation (Tumaneng et al., 2012). In multicellular organisms, TOR function is regulated by growth signals through the PI3K pathway. Insulin or Insulin-like factors binding to their receptors result in insulin receptor substrate phosphorylation (IRS) and activation of PI3K. The latter converts phosphatidylinositol-4,5-phosphate (PIP2) to phosphatidylinositol-3,4,5-phosphate (PIP3) which in turn activates Akt via PDK1. Akt inhibits TSC2, thus, activating TOR (Wullschleger et al., 2006). AMP-activated protein kinase signaling (AMPK) monitors energy levels within the cell. Upon nutritional or energy stress AMPK inhibits TOR pathway (Gwinn et al., 2008; Davie et al., 2015). This relationship is conserved from yeast to man. AMPK inhibitory action on TOR is coordinated with that of the Glycogen Synthase Kinase (Gsk3) an inhibitor of the Wnt signaling pathway in metazoan and a known regulator of protein translation (Rallis et al., 2017).
Recent data indicate that TORC1 has roles in spatial cell size and growth control and organization. In budding yeast, Las24/Kog1 a TORC1 component regulates, as expected, processes that are directly related to the rapamycin-sensitive TORC1 complex. These include global protein translation as well as phosphorylation of the Ser/Thr Npr1p kinase and the Gln3p GATA transcription factor both involved in nitrogen catabolite repression (Loewith and Hall, 2011). Nevertheless, Las2/Kog1 is reported to also be implicated in the spatial arrangement of the actin cytoskeleton (Araki et al., 2005). The latter has been related so far mainly with TORC2 functions rather than TORC1. Cell growth and size in yeast have been shown to be highly dependent on actin cytoskeleton. Polarized cytoskeleton that results in morphological changes also leads to reduced cellular growth (Goranov et al., 2013). Polarization of the actin cytoskeleton inhibits TORC1 and the Iml1 complex is proved to be required for this inhibition. Interestingly, the same complex regulates the activity of TORC1 depending on the availability of nitrogen sources (Goranov et al., 2013). Both TORC1 and TORC2 protein complexes fractionate with membrane formations that are resistant to detergents and distinct from cell membrane rafts (Kunz et al., 2000; Chen and Kaiser, 2003; Wedaman et al., 2003; Aronova et al., 2007). Proteomics analysis of the TOR-containing membrane fractions was conducted revealing numerous endocytosis as well as actin cytoskeleton regulators. To validate the obtained results further genetic and biochemistry experiments were undertaken. These experiments demonstrated important numerous interactions between TORC1 and these regulators and proving the connection of TORC1 and actin cytoskeleton related functions (Aronova et al., 2007). Importantly, rapamycin is able to disrupt polarization of actin. Moreover, it can delay actin repolarization following glucose starvation and delay the localization and accumulation of Lucifer yellow -a tracer to observe kinetics of endocytosis and transportation through the vacuole membrane-within the vacuole (Aronova et al., 2007).
TOR complex 1 (TORC1) has also been implicated in special aspects of cell growth in mammals and fish. Mammalian injured mature CNS axons do not regenerate. PTEN inhibition results in TORC1-dependent, rather than TORC2, CNS axon regeneration and growth. The effect is mediated through the S6-kinase (Miao et al., 2016). Without TSC1 or TSC2 (Huang and Manning, 2008), an increase in the activity TORC1 causes regional neuronal cell growth with axons having multiple axons (Choi et al., 2008). In zebrafish, TORC1-dependent translation of ciliary precursors regulates cilia length, motility and cilia-directed flow. In particular, activation of TORC1 signaling increases ciliary length through S6 kinase 1 (S6K1)-mediated protein synthesis of cilia precursors. Inhibition of TORC1 with rapamycin, decreases ciliary length through suppression of S6K1-mediated protein translation (Yuan and Sun, 2012; Yuan S. et al., 2012). TORC1 can therefore regulate special aspects of cell growth and organization in mitotic and post mitotic cells.
TOR complex 2 (TORC2) is reported to have opposing roles to TORC1 in fission yeast (Ikai et al., 2011). TORC2 affects cell growth and size by regulating aspects of glucose metabolism (Garcia-Martinez and Alessi, 2008; Yuan M. et al., 2012) as well as the actin cytoskeleton (Oh and Jacinto, 2011) and apoptosis (Datta et al., 1997). TORC2 activates several kinases belonging to the AGC family, such as AKT/PKB, PKC, and SGK (serum and glucocorticoid-regulated kinase) (Zoncu et al., 2011; Aimbetov et al., 2012). Interestingly, studies have shown that there is a requirement for ribosomes for TORC2 signaling. Nevertheless, protein translation is not. TORC2 protein complexes can bind with the ribosome PI3K activated by insulin is found to promotes this physical association (Zinzalla et al., 2011). Ribosomal content is a determinant of cell growth and this physical interaction could therefore serve to ensure appropriate levels of TORC2 activity in growing cells (Xie and Guan, 2011). TORC1 and TORC2 are reported to work together to regulate S6 phosphorylation in both budding and fission yeasts (Du et al., 2012; Yerlikaya et al., 2016). These findings challenge the role of ribosomal S6 protein phosphorylation in global translation. In Drosophila, Lst8, a component of both TORC1 and TORC2, is found to regulate cell size but not cell cycle phasing in a cell-autonomous manner through TORC2 (Wang et al., 2012). Likewise in the same animal, TORC2 regulates cell size through Myc (Kuo et al., 2015) a known regulator of organ size that induces cell competition (de la Cova et al., 2004).
Beyond its roles in spatial aspects of cell growth and size, TORC2 is also implicated in temporal aspects. Rictor null mice exhibit mild hyperglycemia and glucose intolerance that originates from the reduction in both β-cell cellular mass and multiplication (Gu et al., 2011). A known role for TORC2 is the maintenance of genome stability during S phase and its requirement for return to cell cycle progression following stress (Schonbrun et al., 2013). TORC2 has been linked to cell size upon division and the timing of mitosis: Fission yeast cells mutant for Sin1, a TORC2 component (Yang et al., 2006), divide at a longer cell size. This implies that timing of mitotic initiation is delayed (Wilkinson et al., 1999). Elongated cell morphology has also been reported in cells mutated for Tor1 (Kawai et al., 2001) and for Gad8 kinase (Matsuo et al., 2003), a known direct substrate of TORC2 related to human AKT. These data show that TORC2–Gad8 pathway promotes the initiation of mitosis. Double mutants of Tor1 and Gad8 with cdc25-22, a temperature-sensitive mutation of the Cdc25 phosphatase that activates Cdc2 kinase at G2/M (Russell and Nurse, 1986), exhibit a cell cycle arrest phenotype with highly elongated morphology. Similar phenotypes are observed when the cdc25-22 mutation is introduced to cells lacking components of the TORC2 complex, such as Sin1 and Ste20 strains (Ikeda et al., 2008). These results support the model in which TORC2 activates Gad8 which in turn advances mitosis.
Fission yeast TORC2 and Gad8 can be localized within the nucleus and bound to chromatin. Gad8 can physically interact with the MBF transcription complex which is implicated in the regulation of DNA stress response as well as the G1/S cell cycle progression. Gad8 deletion or genetic inactivation of TORC2 function (by deleting its core components) results in reduced binding of MBF to its target promoters. Moreover, MBF target genes fail to be induced in the aforementioned mutants upon DNA replication stress. Finally, the protein levels of Cdt2 and Cig2 (both of them MBF targets) are reduced when TORC2 or Gad8 function is abolished. These data highlight pivotal roles of TORC2 within the cell nucleus and reveal a TORC2-dependent mechanism in DNA replication stress response through the control of MBF transcriptional targets (Cohen et al., 2016).
Beyond its connections with cell cycle-related proteins, TORC2 is implicated in the timing of cell growth and division through interactions with the cytoskeleton. Fission yeast TORC2 regulates the timing and fidelity of cytokinesis: Disruption of TORC2 intracellular localisation or function leads in defects in cytokinetic actomyosin ring (CAR) morphology and constriction (Baker et al., 2016). Interestingly, myosin II protein Myp2 and the myosin V protein Myo51 recruit TORC2 to the CAR. TORC2 controls the fidelity of cell division and CAR stability through phosphorylation of the actin-capping protein 1 (Acp1, a regulator of cytokinesis) (Baker et al., 2016).
Roles of TORC2 in temporal aspects of cell size and growth are emerging in systems as diverse as protozoa and human cancer cells. In the protozoan parasite Trypanosoma brucei four TOR kinase orthologues TbTOR1, TbTOR2, TbTOR3, and TbTOR4 have been identified. TbTOR1 and TbTOR2 resemble the structure of all TOR kinases (Saldivia et al., 2013). However, TbTOR3 contains an additional PDZ domain while TbTOR4 has no FRB domain (Barquilla et al., 2008). TbTOR proteins differ in their functions, subcellular localization, and rapamycin sensitivity. Additionally, each TbTOR protein has distinct partners forming four TORC complexes (Barquilla et al., 2008, 2012; Barquilla and Navarro, 2009; de Jesus et al., 2010). TORC1 and TORC2 complexes contain KOG1/raptor and AVO3/rictor orthologues, respectively. TbTOR1 controls cell growth by regulating cell cycle, nucleolus structure, and protein synthesis, whereas TbTOR2 coordinates cell polarization and cytokinesis (Saldivia et al., 2013). Rapamycin treatment of bloodstream trypanosomes results in a profound reduction of cell proliferation. Nevertheless, this effect of rapamycin is due to exclusive action on TORC2 inhibition, with no effect on TORC1 (Barquilla et al., 2008). In tumor cells, TORC2 is required for cell cycle progression (Hietakangas and Cohen, 2008): TORC2 inhibition reduced proliferation of two cancer cell lines MCF7 and PC3. Cells lacking Rictor accumulate in G1 phase and consistently exhibit reduced levels of Cyclin D1 (Hietakangas and Cohen, 2008).
Traditionally TORC1 is linked to temporal aspects of cell size and growth while TORC2 with spatial growth. Nevertheless, emerging data indicate more complex relationships (Figure 1). TORC protein complexes collect information from both intracellular and extracellular signals and are regulated in multiple levels including expression of their components and subcellular localization. Interesting directions and currently conducted work within the field include the interactions of TORC1 with cytoskeletal elements and vesicle mediated transport and their relationships with gene expression programs in cell size and growth control and during different nutritional regimes or stress. Another direction with interesting emerging results is the relationship of TORC2 with the cell cycle machinery and chromatin organization. These data will significantly enrich our knowledge on the control of cell size, growth and survival and will be pivotal for the understanding and treatment of diseases, such as cancer, diabetes and neurodegeneration.
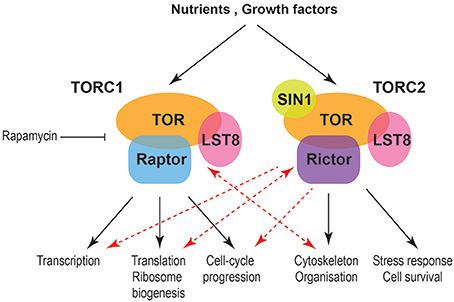
Figure 1. Schematic indicating processes affecting cell size and growth downstream of mammalian TORC complexes. Black arrows indicate well-established relationships between TORC1 and TORC2 with temporal and spatial aspects of cell growth, respectively. Red arrows indicate new functional connections while double red arrows indicate a cross-regulation between TORC1 and cytoskeleton as well as TORC2 and ribosomes.
CR conceptualized the review, decided on the content, drafted the work and prepared the figures. SG provided reference searching, proofreading and editing. Both authors approve the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work is supported by internal grants from the University of East London (UEL) to CR and a fully-funded UEL PhD studentship to SG. We apologize to all the authors that their work is not cited here due to space restrictions.
Aimbetov, R., Chen, C. H., Bulgakova, O., Abetov, D., Bissenbaev, A. K., Bersimbaev, R. I., et al. (2012). Integrity of mTORC2 is dependent on the rictor Gly-934 site. Oncogene 31, 2115–2120. doi: 10.1038/onc.2011.404
Alvarez, B., and Moreno, S. (2006). Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119(Pt 21), 4475–4485. doi: 10.1242/jcs.03241
Araki, T., Uesono, Y., Oguchi, T., and Toh-E, A. (2005). LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet. Syst. 80, 325–343. doi: 10.1266/ggs.80.325
Aronova, S., Wedaman, K., Anderson, S., Yates, J., and Powers, T. (2007). Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2779–2794. doi: 10.1091/mbc.E07-03-0274
Averous, J., and Proud, C. G. (2006). When translation meets transformation: the mTOR story. Oncogene 25, 6423–6435. doi: 10.1038/sj.onc.1209887
Baker, K., Kirkham, S., Halova, L., Atkin, J., Franz-Wachtel, M., Cobley, D., et al. (2016). TOR complex 2 localises to the cytokinetic actomyosin ring and controls the fidelity of cytokinesis. J. Cell Sci. 129, 2613–2624. doi: 10.1242/jcs.190124
Balcazar, N., Sathyamurthy, A., Elghazi, L., Gould, A., Weiss, A., Shiojima, I., et al. (2009). mTORC1 activation regulates beta-cell mass and proliferation by modulation of cyclin D2 synthesis and stability. J. Biol. Chem. 284, 7832–7842. doi: 10.1074/jbc.M807458200
Barquilla, A., Crespo, J. L., and Navarro, M. (2008). Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc. Natl. Acad. Sci. U.S.A. 105, 14579–14584. doi: 10.1073/pnas.0802668105
Barquilla, A., and Navarro, M. (2009). Trypanosome TOR complex 2 functions in cytokinesis. Cell Cycle 8, 697–699. doi: 10.4161/cc.8.5.7808
Barquilla, A., Saldivia, M., Diaz, R., Bart, J. M., Vidal, I., Calvo, E., et al. (2012). Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 109, 14399–14404. doi: 10.1073/pnas.1210465109
Bjedov, I., and Partridge, L. (2011). A longer and healthier life with TOR down-regulation: genetics and drugs. Biochem. Soc. Trans. 39, 460–465. doi: 10.1042/BST0390460
Chen, E. J., and Kaiser, C. A. (2003). LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161, 333–347. doi: 10.1083/jcb.200210141
Chica, N., Rozalen, A. E., Perez-Hidalgo, L., Rubio, A., Novak, B., and Moreno, S. (2016). Nutritional control of cell size by the Greatwall-Endosulfine-PP2A.B55 Pathway. Curr. Biol. 26, 319–330. doi: 10.1016/j.cub.2015.12.035
Choi, Y. J., Di Nardo, A., Kramvis, I., Meikle, L., Kwiatkowski, D. J., Sahin, M., et al. (2008). Tuberous sclerosis complex proteins control axon formation. Genes Dev. 22, 2485–2495. doi: 10.1101/gad.1685008
Cohen, A., Kupiec, M., and Weisman, R. (2016). Gad8 protein is found in the nucleus where it interacts with the MluI Cell Cycle Box-binding Factor (MBF) transcriptional complex to regulate the response to DNA replication stress. J. Biol. Chem. 291, 9371–9381. doi: 10.1074/jbc.M115.705251
Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y., et al. (1997). Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91, 231–241. doi: 10.1016/S0092-8674(00)80405-5
Davie, E., Forte, G. M., and Petersen, J. (2015). Nitrogen regulates AMPK to control TORC1 signaling. Curr. Biol. 25, 445–454. doi: 10.1016/j.cub.2014.12.034
de Jesus, T. C., Tonelli, R. R., Nardelli, S. C., da Silva Augusto, L., Motta, M. C., Girard-Dias, W., et al. (2010). Target of rapamycin (TOR)-like 1 kinase is involved in the control of polyphosphate levels and acidocalcisome maintenance in Trypanosoma brucei. J. Biol. Chem. 285, 24131–24140. doi: 10.1074/jbc.M110.120212
de la Cova, C., Abril, M., Bellosta, P., Gallant, P., and Johnston, L. A. (2004). Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116. doi: 10.1016/S0092-8674(04)00214-4
De Virgilio, C., and Loewith, R. (2006). The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 38, 1476–1481. doi: 10.1016/j.biocel.2006.02.013
Du, W., Halova, L., Kirkham, S., Atkin, J., and Petersen, J. (2012). TORC2 and the AGC kinase Gad8 regulate phosphorylation of the ribosomal protein S6 in fission yeast. Biol. Open 1, 884–888. doi: 10.1242/bio.20122022
Ganley, I. G., Lam du, H., Wang, J., Ding, X., Chen, S., and Jiang, X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305. doi: 10.1074/jbc.M900573200
Garcia-Martinez, J. M., and Alessi, D. R. (2008). mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385. doi: 10.1042/BJ20081668
Goranov, A. I., Gulati, A., Dephoure, N., Takahara, T., Maeda, T., Gygi, S. P., et al. (2013). Changes in cell morphology are coordinated with cell growth through the TORC1 pathway. Curr. Biol. 23, 1269–1279. doi: 10.1016/j.cub.2013.05.035
Gu, Y., Lindner, J., Kumar, A., Yuan, W., and Magnuson, M. A. (2011). Rictor/mTORC2 is essential for maintaining a balance between beta-cell proliferation and cell size. Diabetes 60, 827–837. doi: 10.2337/db10-1194
Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., et al. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226. doi: 10.1016/j.molcel.2008.03.003
Hansen, C. G., Moroishi, T., and Guan, K. L. (2015a). YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25, 499–513. doi: 10.1016/j.tcb.2015.05.002
Hansen, C. G., Ng, Y. L., Lam, W. L., Plouffe, S. W., and Guan, K. L. (2015b). The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 25, 1299–1313. doi: 10.1038/cr.2015.140
Hartmuth, S., and Petersen, J. (2009). Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J. Cell Sci. 122(Pt 11), 1737–1746. doi: 10.1242/jcs.049387
Heitman, J., Movva, N. R., and Hall, M. N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. doi: 10.1126/science.1715094
Hietakangas, V., and Cohen, S. M. (2008). TOR complex 2 is needed for cell cycle progression and anchorage-independent growth of MCF7 and PC3 tumor cells. BMC Cancer 8:282. doi: 10.1186/1471-2407-8-282
Hosokawa, N., Hara, T., Kaizuka, T., Kishi, C., Takamura, A., Miura, Y., et al. (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991. doi: 10.1091/mbc.E08-12-1248
Huang, J., and Manning, B. D. (2008). The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412, 179–190. doi: 10.1042/BJ20080281
Huang, K., and Fingar, D. C. (2014). Growing knowledge of the mTOR signaling network. Semin. Cell Dev. Biol. 36, 79–90. doi: 10.1016/j.semcdb.2014.09.011
Ikai, N., Nakazawa, N., Hayashi, T., and Yanagida, M. (2011). The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe. Open Biol. 1:110007. doi: 10.1098/rsob.110007
Ikeda, K., Morigasaki, S., Tatebe, H., Tamanoi, F., and Shiozaki, K. (2008). Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7, 358–364. doi: 10.4161/cc.7.3.5245
Kawai, M., Nakashima, A., Ueno, M., Ushimaru, T., Aiba, K., Doi, H., et al. (2001). Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39, 166–174. doi: 10.1007/s002940100198
Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N. R., and Hall, M. N. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596. doi: 10.1016/0092-8674(93)90144-F
Kunz, J., Schneider, U., Howald, I., Schmidt, A., and Hall, M. N. (2000). HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275, 37011–37020. doi: 10.1074/jbc.M007296200
Kuo, Y., Huang, H., Cai, T., and Wang, T. (2015). Target of Rapamycin complex 2 regulates cell growth via Myc in Drosophila. Sci. Rep. 5:10339. doi: 10.1038/srep10339
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
Law, B. K. (2005). Rapamycin: an anti-cancer immunosuppressant? Crit. Rev. Oncol. Hematol. 56, 47–60. doi: 10.1016/j.critrevonc.2004.09.009
Loewith, R., and Hall, M. N. (2011). Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201. doi: 10.1534/genetics.111.133363
Lopez-Maury, L., Marguerat, S., and Bahler, J. (2008). Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9, 583–593. doi: 10.1038/nrg2398
Ma, X. M., and Blenis, J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318. doi: 10.1038/nrm2672
Matsuo, T., Kubo, Y., Watanabe, Y., and Yamamoto, M. (2003). Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22, 3073–3083. doi: 10.1093/emboj/cdg298
Menon, S., and Manning, B. D. (2008). Common corruption of the mTOR signaling network in human tumors. Oncogene 27(Suppl. 2), S43–S51. doi: 10.1038/onc.2009.352
Miao, L., Yang, L., Huang, H., Liang, F., Ling, C., and Hu, Y. (2016). mTORC1 is necessary but mTORC2 and GSK3β are inhibitory for AKT3-induced axon regeneration in the central nervous system. Elife 5:e14908. doi: 10.7554/eLife.14908
Moreno-Torres, M., Jaquenoud, M., and De Virgilio, C. (2015). TORC1 controls G1-S cell cycle transition in yeast via Mpk1 and the greatwall kinase pathway. Nat. Commun. 6:8256. doi: 10.1038/ncomms9256
Morita, M., Gravel, S. P., Hulea, L., Larsson, O., Pollak, M., St-Pierre, J., et al. (2015). mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 14, 473–480. doi: 10.4161/15384101.2014.991572
Moroishi, T., Hansen, C. G., and Guan, K. L. (2015). The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79. doi: 10.1038/nrc3876
Oh, W. J., and Jacinto, E. (2011). mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316. doi: 10.4161/cc.10.14.16586
Otsubo, Y., and Yamamato, M. (2008). TOR signaling in fission yeast. Crit. Rev. Biochem. Mol. Biol. 43, 277–283. doi: 10.1080/10409230802254911
Perez-Hidalgo, L., and Moreno, S. (2016). Nutrients control cell size. Cell Cycle 15, 1655–1656. doi: 10.1080/15384101.2016.1172471
Petersen, J., and Nurse, P. (2007). TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat. Cell Biol. 9, 1263–1272. doi: 10.1038/ncb1646
Pfleger, C. M. (2017). The hippo pathway: a master regulatory network important in development and dysregulated in disease. Curr. Top. Dev. Biol. 123, 181–228. doi: 10.1016/bs.ctdb.2016.12.001
Rallis, C., and Bahler, J. (2013). Inhibition of TORC1 signaling and increased lifespan: gained in translation? Aging 5, 335–336. doi: 10.18632/aging.100560
Rallis, C., Codlin, S., and Bahler, J. (2013). TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 12, 563–573. doi: 10.1111/acel.12080
Rallis, C., Lopez-Maury, L., Georgescu, T., Pancaldi, V., and Bahler, J. (2014). Systematic screen for mutants resistant to TORC1 inhibition in fission yeast reveals genes involved in cellular ageing and growth. Biol. Open 3, 161–171. doi: 10.1242/bio.20147245
Rallis, C., Townsend, S., and Bahler, J. (2017). Genetic interactions and functional analyses of the fission yeast gsk3 and amk2 single and double mutants defective in TORC1-dependent processes. Sci. Rep. 7:44257. doi: 10.1038/srep44257
Rodland, G. E., Tvegard, T., Boye, E., and Grallert, B. (2014). Crosstalk between the Tor and Gcn2 pathways in response to different stresses. Cell Cycle 13, 453–461. doi: 10.4161/cc.27270
Russell, P., and Nurse, P. (1986). cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145–153. doi: 10.1016/0092-8674(86)90546-5
Saldivia, M., Barquilla, A., Bart, J. M., Diaz-Gonzalez, R., Hall, M. N., and Navarro, M. (2013). Target of rapamycin (TOR) kinase in Trypanosoma brucei: an extended family. Biochem. Soc. Trans. 41, 934–938. doi: 10.1042/BST20130052
Schonbrun, M., Kolesnikov, M., Kupiec, M., and Weisman, R. (2013). TORC2 is required to maintain genome stability during S phase in fission yeast. J. Biol. Chem. 288, 19649–19660. doi: 10.1074/jbc.M113.464974
Sehgal, S. N. (2003). Sirolimus: its discovery, biological properties, and mechanism of action. Transplant. Proc. 35(3 Suppl), 7S–14S. doi: 10.1016/S0041-1345(03)00211-2
Sehgal, S. N., Baker, H., and Vezina, C. (1975). Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. 28, 727–732. doi: 10.7164/antibiotics.28.727
Stracka, D., Jozefczuk, S., Rudroff, F., Sauer, U., and Hall, M. N. (2014). Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289, 25010–25020. doi: 10.1074/jbc.M114.574335
Tumaneng, K., Schlegelmilch, K., Russell, R. C., Yimlamai, D., Basnet, H., Mahadevan, N., et al. (2012). YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 14, 1322–1329. doi: 10.1038/ncb2615
Wang, T., Blumhagen, R., Lao, U., Kuo, Y., and Edgar, B. A. (2012). LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2). Mol. Cell. Biol. 32, 2203–2213. doi: 10.1128/MCB.06474-11
Wedaman, K. P., Reinke, A., Anderson, S., Yates, J., McCaffery, J. M., and Powers, T. (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220. doi: 10.1091/mbc.E02-09-0609
Weisman, R. (2016). Target of Rapamycin (TOR) Regulates growth in response to nutritional signals. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.FUNK-0006-2016
Weisman, R., Roitburg, I., Schonbrun, M., Harari, R., and Kupiec, M. (2007). Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics 175, 1153–1162. doi: 10.1534/genetics.106.064170
Wilkinson, M. G., Pino, T. S., Tournier, S., Buck, V., Martin, H., Christiansen, J., et al. (1999). Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18, 4210–4221. doi: 10.1093/emboj/18.15.4210
Wullschleger, S., Loewith, R., and Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. doi: 10.1016/j.cell.2006.01.016
Xie, X., and Guan, K. L. (2011). The ribosome and TORC2: collaborators for cell growth. Cell 144, 640–642. doi: 10.1016/j.cell.2011.02.029
Yang, H., Rudge, D. G., Koos, J. D., Vaidialingam, B., Yang, H. J., and Pavletich, N. P. (2013). mTOR kinase structure, mechanism and regulation. Nature 497, 217–223. doi: 10.1038/nature12122
Yang, Q., Inoki, K., Ikenoue, T., and Guan, K. L. (2006). Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832. doi: 10.1101/gad.1461206
Yerlikaya, S., Meusburger, M., Kumari, R., Huber, A., Anrather, D., Costanzo, M., et al. (2016). TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol. Biol. Cell 27, 397–409. doi: 10.1091/mbc.E15-08-0594
Yuan, M., Pino, E., Wu, L., Kacergis, M., and Soukas, A. A. (2012). Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J. Biol. Chem. 287, 29579–29588. doi: 10.1074/jbc.M112.386854
Yuan, S., Li, J., Diener, D. R., Choma, M. A., Rosenbaum, J. L., and Sun, Z. (2012). Target-of-rapamycin complex 1 (Torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 2021–2026. doi: 10.1073/pnas.1112834109
Yuan, S., and Sun, Z. (2012). TORC1-mediated protein synthesis regulates cilia size and function: implications for organelle size control by diverse signaling cascades. Cell Cycle 11, 1750–1752. doi: 10.4161/cc.20312
Zinzalla, V., Stracka, D., Oppliger, W., and Hall, M. N. (2011). Activation of mTORC2 by association with the ribosome. Cell 144, 757–768. doi: 10.1016/j.cell.2011.02.014
Keywords: cell size, cell cycle, growth, rapamycin, signaling, nutrients, TORC1, TORC2
Citation: Gonzalez S and Rallis C (2017) The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front. Cell Dev. Biol. 5:61. doi: 10.3389/fcell.2017.00061
Received: 17 March 2017; Accepted: 22 May 2017;
Published: 07 June 2017.
Edited by:
Mikael Bjorklund, University of Dundee, United KingdomReviewed by:
Sandra Lopez, University of Oslo, NorwayCopyright © 2017 Gonzalez and Rallis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charalampos Rallis, Yy5yYWxsaXNAdWVsLmFjLnVr; Yy5yYWxsaXNAdWNsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.