- Division of Cell and Developmental Biology, School of Life Sciences, University of Dundee, Dundee, UK
In the developing vertebrate embryo, segmentation initiates through the formation of repeated segments, or somites, on either side of the posterior neural tube along the anterior to posterior axis. The periodicity of somitogenesis is regulated by a molecular oscillator, the segmentation clock, driving cyclic gene expression in the unsegmented paraxial mesoderm, from which somites derive. Three signaling pathways underlie the molecular mechanism of the oscillator: Wnt, FGF, and Notch. In particular, Notch has been demonstrated to be an essential piece in the intricate somitogenesis regulation puzzle. Notch is required to synchronize oscillations between neighboring cells, and is moreover necessary for somite formation and clock gene oscillations. Following ligand activation, the Notch receptor is cleaved to liberate the active intracellular domain (NICD) and during somitogenesis NICD itself is produced and degraded in a cyclical manner, requiring tightly regulated, and coordinated turnover. It was recently shown that the pace of the segmentation clock is exquisitely sensitive to levels/stability of NICD. In this review, we focus on what is known about the mechanisms regulating NICD turnover, crucial to the activity of the pathway in all developmental contexts. To date, the regulation of NICD stability has been attributed to phosphorylation of the PEST domain which serves to recruit the SCF/Sel10/FBXW7 E3 ubiquitin ligase complex involved in NICD turnover. We will describe the pathophysiological relevance of NICD-FBXW7 interaction, whose defects have been linked to leukemia and a variety of solid cancers.
Introduction
The formation of a segmented body plan is a conserved feature of embryogenesis for all vertebrate species. This process leads to the formation of transient embryonic segments, called somites. Somites are precursors of vertebrae and ribs, associated skeletal muscles, and some dermis (Christ et al., 2007). Their formation is regulated by a molecular oscillator called the segmentation clock (Gibb et al., 2010; Oates et al., 2012; Benazeraf and Pourquie, 2013). Aberrations in this mechanism lead to human developmental disorders, such as spondylocostal dysostosis (Pourquie, 2011; Eckalbar et al., 2012). Some of these malformations originate from defects in Notch signaling, suggesting that this pathway is essential in controlling and regulating vertebrate segmentation.
This review aims to give a general overview of the importance of the Notch signaling pathway in the segmentation clock in addition to a description of our current understanding of the Notch pathway, particularly focusing on the turnover and regulation of the Notch intracellular domain.
Somitogenesis
Somitogenesis has been the topic of several outstanding reviews (Pourquie, 2001; Maroto et al., 2012; Oates et al., 2012; Benazeraf and Pourquie, 2013; Hubaud and Pourquie, 2014; Bailey and Dale, 2015), thus we will provide a general overview.
Early in development, segmentation initiates through the formation of repeated segments, or somites (Christ et al., 2007; Gibb et al., 2010). Somitogenesis is a cyclical and gradual process such that somites are sequentially pinched off in pairs from the anterior end of two rods of paraxial mesoderm, the presomitic mesoderm (PSM), lying on either side of the caudal neural tube (Gossler and De Angelis, 1998; Cambray and Wilson, 2007; Dequeant and Pourquie, 2008; Gibb et al., 2010; Maroto et al., 2012). The PSM is continuously replenished with progenitor cells located initially in both the epiblast adjacent to the primitive streak and the rostral primitive streak and later in the tail bud (Iimura et al., 2007; Gomez and Pourquie, 2009; Henrique et al., 2015), and thus the presomitic mesoderm preserves its length (Dequeant and Pourquie, 2008; Figure 1A).
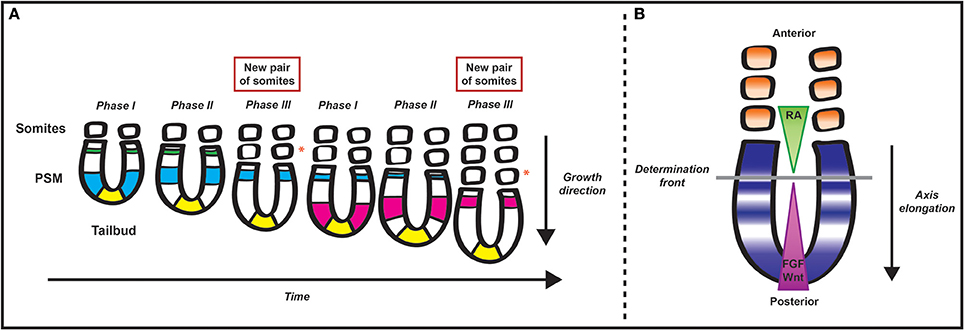
Figure 1. Schematic representation of somitogenesis and the segmentation clock. (A) Pairs of somites bud off from the rostral end of the presomitic mesoderm (PSM) progressively during early development. The tail bud, a site of gastrulation that lies at the posterior end of the embryo, continuously “replenishes” the posterior end of the PSM with progenitor cells. The periodicity of segmentation is regulated by a molecular oscillator that drives cyclic gene expression from the posterior to the anterior tip of the PSM. The different colors represent domains of clock gene expression in different cycles. As time progresses in each cycle, the domain of clock gene expression shifts anteriorly while narrowing until it reaches the anterior limit of the PSM. The periodicity of this cyclic gene expression matches that of somite formation. An orange asterisk lies adjacent to each of the new pairs of somites formed in the time series—the first pair is formed after the blue wave of clock gene expression traverses the PSM and the second pair is formed after the pink wave of clock genes expression traverses the PSM from the tail bud to the anterior limit of the tissue. (B) Two mutually opposing gradients of retinoic acid (RA) and FGF/Wnt regulate the maturation wavefront within the paraxial mesoderm. Due to somite formation anteriorly and gastrulation at the caudal end of the PSM, cells within the PSM become progressively more anteriorly displaced, and, as a result, they are exposed to progressively lower levels of FGF/Wnt. There is a position within the PSM, termed the determination front, where cells are released from the effect of FGF and can respond to the segmentation clock and RA, embarking on their segmentation programme.
The periodicity of this segmentation process is different from species to species: 30 min in zebrafish (Schroter et al., 2008), 90 min in chicken (Palmeirim et al., 1997), 2 h in mice (Tam, 1981), 6–8 h in human (William et al., 2007). Similarly, the total number of somites is a characteristic feature of each species: 31 pairs in zebrafish, 50 somite pairs in chicken, 65 in mice, and about 500 in some snakes.
The regulation of the periodicity of somitogenesis is governed by the segmentation clock, a molecular oscillator (Palmeirim et al., 1997) whose existence was first proposed in theoretical models such as the “Clock and Wavefront model” (Cooke and Zeeman, 1976). According to the model, a wavefront of maturation sweeps along the body axis concomitant with extension of the trunk and tail, governing maturation of the PSM to become somites. This positional information gradient within the PSM interacts with a smooth cellular oscillator (the clock), driving cells to oscillate between a permissive and a non-permissive state. Segmentation of the PSM only occurs when the maturation wavefront reaches a group of cells in a specific “permissive” clock phase (Cooke and Zeeman, 1976).
Over the last 20 years the theoretical “Clock and Wavefront model” has received significant experimental support. The wavefront of maturation is thought to rely on the intersecting gradients and cross-regulatory activities of three signal pathways, namely a caudo-rostral gradient of FGF and Wnt and rostro-caudal gradient of retinoic acid (RA). The determination front marks the point of intersection of these gradients, where the next prospective somite boundary will form (Figure 1B). These cross-regulatory activities thereby regulate somite size. The activity of Wnt and FGF also controls cell maturation in the PSM. These roles have been reviewed elsewhere, thus will not be covered here (Aulehla et al., 2003; Dubrulle and Pourquie, 2004; Wahl et al., 2007; Aulehla and Pourquie, 2010; Hubaud and Pourquie, 2014).
It is well established that the rhythmicity of somitogenesis is regulated by the segmentation clock driving cyclic and dynamic expression of “clock genes” in the PSM, with a periodicity that matches somite formation. This feature is conserved among a variety of vertebrate species (Jiang et al., 2000; Cinquin, 2007; Dequeant and Pourquie, 2008; Gomez et al., 2008; Ozbudak and Lewis, 2008; Krol et al., 2011). The clock genes are components of the Notch, Wnt, and FGF pathways (Aulehla et al., 2003; Dequeant and Pourquie, 2008; Yabe and Takada, 2016), playing a reciprocal regulatory role in oscillatory gene expression (reviewed in Gibb et al., 2010; Maroto et al., 2012). While the specific genes which oscillate may vary among species, the most highly represented pathway among the clock genes is the Notch (Krol et al., 2011).
Stemming from the observation that the proteins encoded by clock genes are predominantly unstable negative regulators of the pathway that activates them, it is believed that oscillatory gene expression relies on negative feedback loops of these unstable regulators, such as the two Notch target clock genes, Hes7, and Lunatic Fringe (Lfng) (Bessho et al., 2001a,b, 2003; Cole et al., 2002; Hirata et al., 2002; Dale et al., 2003; Serth et al., 2003; Kageyama et al., 2012; Okubo et al., 2012). It is particularly interesting that blocking Lfng oscillations disturbs somitogenesis in the thoracic and lumbar areas but not in more posterior areas of the embryo (Shifley et al., 2008), implying the role of Notch signaling in segmentation is not uniform along the axis.
In addition to negative feedback, oscillatory gene expression in the PSM also invokes positive feedback; Notch signaling regulates dynamic expression of Notch1 itself, whereas Wnt regulates dynamic expression of Dll1 (Bone et al., 2014).
As the most highly conserved pathway involved in the segmentation clock, a wealth of studies have focused on elucidating the fundamental role of Notch in somitogenesis and in the segmentation clock mechanism (Barrantes et al., 1999; Jiang et al., 2000; Bessho et al., 2001b, 2003; Dale et al., 2003; Julich et al., 2005; McGrew et al., 2008; Hubaud and Pourquie, 2014; Wahi et al., 2014; Liao and Oates, 2016). Notch is clearly required to synchronize oscillations between neighboring cells (Jiang et al., 2000; Shimojo et al., 2016). A question that arises is whether oscillations are actually necessary for the segmentation process to occur or whether just non-oscillatory activity of the Notch pathway is sufficient. Mutant mice or fish lacking Notch components all display severe segmentation defects (Conlon et al., 1995; Barrantes et al., 1999; Jiang et al., 2000; Liao and Oates, 2016). For example, the lack of the obligate transcription factor RBP-Jκ, in mouse, leads to lethality before day E10.5 and only the first few cervical somites are formed (Oka et al., 1995). A pivotal study conducted by Ferjentsik et al. pointed out that Notch activity, per se, is indeed essential for somite formation. Mutating crucial Notch pathway components, or using a complementary pharmacological approach, they demonstrated that in mouse Notch activity is crucial for the oscillatory activity of all clock genes, and thus essential for the formation of a segmented body axis (Ferjentsik et al., 2009) (see also Huppert et al., 2005).
Notch Signaling Pathway
The Notch pathway is highly conserved among metazoans and mediates short range juxtacrine communication. The Notch locus was first cloned in Drosophila and it was found to encode a large single pass type I transmembrane protein (Wharton et al., 1985), whose epidermal growth factor (EGF) repeats mediate interaction with their canonical activators—two ligands, Delta, and Serrate, in the Delta-Serrate-Lag2 (DSL) family. Drosophila studies have contributed hugely to our current understanding of Notch (Artavanis-Tsakonas et al., 1999). The role of Notch in developmental processes of multicellular species has been extensively elucidated (Dumortier et al., 2005; Radtke et al., 2005; Aster, 2014). Notch signaling outcome mostly relies on the cellular context, and thus Notch affects stem cell maintenance, cell fate choice, cell differentiation, lineage progression, and apoptosis in a context-dependent fashion (Bray, 2006; Hori et al., 2013).
Despite its multiple roles and versatility, the Notch pathway is relatively simple and conserved across species (Artavanis-Tsakonas et al., 1999; Bray, 2006; Kopan and Ilagan, 2009). In mammals, there are four Notch receptors (NOTCH1-4) and five DSL ligands (JAG1-2 and Delta-like 1-3-4). Both receptors and ligands are single transmembrane proteins and thus to trigger the signaling cascade, cell-cell contact is required (D'souza et al., 2010; Andersson et al., 2011; Greenwald and Kovall, 2013).
The Notch receptor is typically comprised of: (i) 29–36 EFG-like repeats in its extracellular domain, involved in ligand interaction; (ii) three juxtamembrane repeats (Lin-12-Notch, LIN), required for extra-intracellular domain interaction (located within the Negative Regulatory Region (NRR); (iii) the intracellular region, including seven ankyrin (ANK) repeats flanked by a PEST [rich in proline (P), glutamic acid (E), serine (S) and threonine (T) residues] and a transactivation (TAD) domain (Figure 2C).
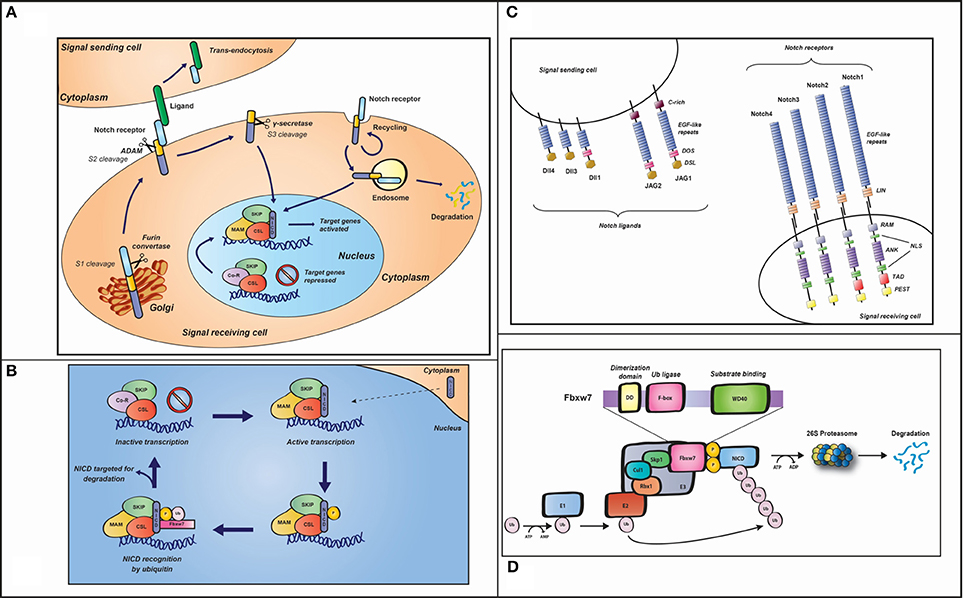
Figure 2. (A) The Notch signaling pathway. In the Golgi, after being glycosylated by members of the Fringe family, pre-Notch is cleaved by a Furin-like convertase into the extracellular and intracellular domains (termed the S1 cleavage), resulting in a heterodimeric receptor with non-covalently associated domains that is transported to the plasma membrane. The Fringe modifications introduced in the Golgi affect sensitivity of the receptor on the signal-receiving cell to the DSL (Delta-Serrate-Lag2) ligands, in the signal-sending cell. Following ligand-receptor interaction, trans-endocytosis of the Notch extracellular domain, by the signal-sending cell, exposes the second so called S2 cleavage site, facilitating intramembranous cleavage by an ADAM (a disintegrin and metalloproteinase domain) protease, in the extracellular domain. S2 cleavage, in turns, exposes the S3 proteolytic cleavage site within the transmembrane domain, which is cleaved by the γ-secretase complex and liberates the intracellular domain of Notch (NICD), allowing it to translocate to the nucleus and thus activate transcription of target genes. In order to prevent inappropriate signaling from the pool of Notch that has not been activated by ligand, Notch receptor is continuously internalized into early endosomes and thus degraded. (B) Zoom-in into the nucleus of the signal-receiving cell (A). Once released into the nucleus, NICD binds the DNA-binding protein CSL as well as the SKIP protein. The trimeric complex thus recruits Mastermind-like protein (MAM), which recruits additional co-activators (not shown), required for the transcriptional regulation of Notch target gene expression. Kinases, such as CDK8 and GSK3β, phosphorylate (p) NICD on its PEST domain, rendering it susceptible to recognition by Fbxw7 E3 ligase, leading to ubiquitination (Ub) and subsequent degradation by the proteasome. In the absence of NICD, CSL associates with transcriptional co-repressors blocking Notch target gene activation. Target genes are repressed until more NICD is produced to re-initiate a new cycle of target gene expression. (C) Notch ligands and receptors. In the signal-receiving cell, the four mammalian Notch receptors (Notch1-4) are represented. They are expressed on the cell surface as heterodimers and characterized by epidermal growth factor (EGF)-like and LIN repeats in their extracellular region. The intracellular domain includes an RBP-Jκ-associated molecule (RAM) domain, seven ankyrin (ANK) repeats, two nuclear localization signals (NLS), a transactivation (TAD) domain (lacking in Notch3 and Notch4), and a Proline-, Glutamate-, Serine-, and Threonine-rich (PEST) domain. The five Notch ligands (Delta-like 1, 3, and 4 and Jagged 1 and 2) are represented on the surface of the signal-sending cell. Each ligand contains an EGF-like repeat region and a conserved domain DSL (Delta/Serrate/Lag). A conserved cysteine-rich (CR) domain is also present on Jagged1 and Jagged2. The DOS (Delta and OSM-11) domain, containing two atypical EGF repeats, is part of Dll1, JAG1 and JAG2 ligands. (D) SCFFbxw7 E3 ubiquitin ligase complex representation. The upper part of the figure shows Fbxw7 domains: a conserved dimerization motif, which mediates dimerization of the SCF complex and thus facilitates ubiquitin conjugation; the F-box, which binds the SCF complex through Skp1; the WD40, containing three specific amino acid residues, which binds the phosphorylated substrate. In the bottom part of the figure the SCFFbxw7 complex is schematically represented. In general, in the ubiquitin system, three enzymes are involved in the signaling cascade: the ubiquitin-activating E1, the ubiquitin-conjugating E2 and an E3 ubiquitin ligase. The first step is ATP-dependent and involves the binding of ubiquitin to E1. Ubiquitin is then activated and transferred to E2. The ubiquitin-E2 complex then interacts with a specific E3 (SCFFbxw7, refer to main text for a description), which recognizes the substrate (phosphorylated NICD, in this case) and facilitates transfer of the ubiquitin molecules to the substrate, leading to substrate degradation by the 26S proteasome.
During its maturation, Notch undergoes ligand-independent cleavage by a furin-like convertase in the trans-Golgi (Artavanis-Tsakonas et al., 1999; Fiuza and Arias, 2007; Hori et al., 2013). This first cleavage (the S1) results in the production of a heterodimeric receptor comprised of a transmembrane/intracellular fragment non-covalently bound to the Notch extracellular domain (NECD). Notch is thus presented to the cell surface as a heterodimer. The non-activated Notch receptor is constitutively internalized, ubiquitinated by Itch/AIP4 (a member of the Nedd4 family of HECT domain E3 ubiquitin ligases), and thus targeted for lysosomal degradation (Chastagner et al., 2008; Moretti and Brou, 2013).
To ensure correct folding and activity, during synthesis and secretion in the Golgi, NECD undergoes O-linked glycosylation and fucosylation (Rana and Haltiwanger, 2011). These two modifications on the EGF repeats modulate Notch activity by modulating interaction with the Delta or Serrate ligands. The reaction is catalyzed by three Fringe homologs (Lunatic, Manic, and Radical Fringe), recognizing specific amino acids in individual EGF repeats (Rampal et al., 2005). In vitro, in the signal-receiving cell, all Fringes enhance Dll1-Notch1 interactions with comparable effects in both trans- and cis-(Lebon et al., 2014). Rfng also enhances trans- and cis-interactions between JAG1 and Notch1, but these interactions are weakened by Lfng and Mfng. By contrast, JAG1 activation of Notch2 is potentiated by Lfng, thereby expanding the ligand-receptor combinations that are differentially modified by the Fringe enzymes (Hicks et al., 2000). In the context of somitogenesis, Lfng is the only family member expressed in the PSM. In most systems, Lfng acts in the receiving-cell to potentiate receptor activation by Delta-like ligands while reducing activation by Jagged ligands (Hicks et al., 2000; Yang et al., 2005; Kato et al., 2010). However, it has been suggested that LFNG protein may synchronize clock oscillations between neighboring cells by acting in the signal-sending cell to inhibit Notch1 activation by Dll1 (Okubo et al., 2012). Ligand binding in an adjacent cell triggers a second cleavage, mediated by the metalloprotease ADAM10 (A disintegrin and metalloprotease) at S2 site in the juxtamembrane extracellular domain, proximal to the Notch transmembrane domain (Mumm et al., 2000; Dyczynska et al., 2007; Bozkulak and Weinmaster, 2009; Gordon et al., 2009; Van Tetering et al., 2009; Weber et al., 2011; Groot et al., 2014). The cleaved NECD product, bound to the ligand, undergoes trans-endocytosis into the ligand-expressing cell (Kramer, 2000; Parks et al., 2000; Meloty-Kapella et al., 2012). The second cleavage exposes the third cleavage site, S3, within the membrane-tethered Notch fragment, and is thus a rate-limiting step for the third and final cleavage (Brou et al., 2000; Mumm et al., 2000). Upon cleavage at the S3 site by a γ-secretase complex, the Notch intracellular domain (NICD) is then released (Schroeter et al., 1998) and translocates into the nucleus to activate transcription of target genes (Figure 2A). Notch can be activated in the endosomal pathway, independently of its ligands, through the activity of Deltex, a Ring-domain ubiquitin ligase that binds to NICD. However, it is unclear how the Deltex-activation mechanism relates to that of ligand-induced signaling.
Notch signaling does not require the use of second messengers. The activity is exclusively driven by nuclear concentration of NICD (Struhl and Adachi, 1998; Ehebauer et al., 2006). In the nucleus, NICD binds a bi-functional transcription factor CSL [CBF1, Su(H), Lag-1], a DNA binding complex Mastermind (MAM), and a variety of other co-activators involved in the transcriptional activation of Notch target gene expression (Fryer et al., 2004; Kopan and Ilagan, 2009; Hori et al., 2013). The transcriptional co-regulator SKIP (Ski-interaction protein) and the histone acetylase p300 are recruited concomitantly to the promoter region of target genes promoting the assembly of the initiation and elongation complexes (Zhou et al., 2000; Wallberg et al., 2002; Fryer et al., 2004; Bray, 2006; Figure 2B). MAM also engages kinases that phosphorylate NICD (Wu et al., 2000; Kitagawa et al., 2001; Nam et al., 2003; Fryer et al., 2004), a crucial step in the regulation of NICD stability and activity (Ingles-Esteve et al., 2001; Espinosa et al., 2003; Fryer et al., 2004; Jin et al., 2009). The domain targeted is the C-terminal PEST domain that is phosphorylated by the cyclin C cyclin-dependent kinase-8 complex (Cyc:CDK8) and glycogen synthase kinase 3β (GSK-3β) (Espinosa et al., 2003; Fryer et al., 2004; Jin et al., 2009).
FBXW7 and Its Role in NICD Turnover
NICD phosphorylation leads to its ubiquitination, turnover, and degradation by the proteasome, defining the half-life of Notch signaling, allowing the cell once again to become ligand-competent and resetting the signaling for a new cycle of activation (Le Bras et al., 2011). In the prevailing model, the ubiquitin ligase involved is the SCFFbxw7 [S phase kinase-associated protein 1 (SKP1)-Cullin 1 (CUL1)-F-box] protein complex (Wu et al., 2001; Tsunematsu et al., 2004; Crusio et al., 2010). SCFFbxw7 is part of the RING-finger domain E3 family (Petroski and Deshaies, 2005). Briefly, Cullin 1 acts as a scaffold on which SKP1 and RBX1 subunits assemble. SKP1 is involved in the recruitment of F box proteins (FBXW7, in the case of NICD), and RBX1 recruits a cognate E2 (Hao et al., 2007; Skaar et al., 2013). Fbxw7 consists of three isoforms (α, β, and γ) generated by alternative splicing and the isoform α, shown to ubiquitinate NICD, is localized to the nucleus (Matsumoto et al., 2006; O'neil et al., 2007; Welcker and Clurman, 2008; Crusio et al., 2010). Two domains are functionally important in the FBXW7 protein: the F-box domain, binding SKP1 (Bai et al., 1996), and the seven WD40 repeats mediating recognition/binding to the target protein in a specific consensus phospho-motif, the Cdc4 phospho-degron (Thr-Pro-Pro-Xaa-Ser, in which Thr and Ser residues are phosphorylated; Koepp et al., 2001; Welcker et al., 2003; Hao et al., 2007; Skaar et al., 2013; Figure 2D). A number of these phospho-degrons have been identified in the NICD PEST domain. Intriguingly, an additional hNICD1-specific degron has recently been identified within the N-terminal region, distinct from the PEST domain that is not recognized by FBXW7 (Broadus et al., 2016). Moreover, the E3 ligase, Itch, promoting PEST domain-independent NICD1 degradation (Qiu et al., 2000), does not mediate NICD1 degradation through the N1-Box (Broadus et al., 2016).
NICD-FBXW7 Interaction
Given the importance of Notch signaling in cell fate determination and cell cycle progression, it is not surprising that aberrations in the pathway lead to cancers and other diseases (Roy et al., 2007; Simpson et al., 2011; Wang et al., 2011; Kamath et al., 2012; Bolos et al., 2013; Huang et al., 2013; Lobry et al., 2013). Moreover, the pleiotropic nature of the pathway means the various Notch receptors can act as tumor suppressors for example in epithelial tumors or as oncogenes in leukemia and a variety of solid cancers (Radtke and Raj, 2003; Miele et al., 2006; Lobry et al., 2014; Alketbi and Attoub, 2015; Habets et al., 2015; Bonyadi Rad et al., 2016). From this vast literature we will focus here on activating mutations in Notch1 which are predominantly located in the extracellular heterodimerization (HD) domain resulting in ligand-independent exposure of the S2 cleavage site (Malecki et al., 2006; Van Tetering et al., 2009), or in the PEST domain, leading to constitutive activation of the pathway through increased NICD stability or in FBXW7, in line with its fundamental role in restricting the signaling strength/duration of the Notch pathway (Oberg et al., 2001; Tetzlaff et al., 2004; O'neil et al., 2007; Thompson et al., 2007; Wang et al., 2012; Bolos et al., 2013). For instance, Notch1 mutations occur in over 50% of both pediatric and adult T-cell acute lymphoblastic leukemia (T-ALL) cases (Malyukova et al., 2007; Erbilgin et al., 2010), while Fbxw7 mutations are found in up to 20% of T-ALL cases (Baldus et al., 2009; Mullighan, 2009). Furthermore, Notch1 mutations were found in diffuse large B-cell lymphoma (DLBCL), splenic marginal zone lymphoma (SMZL), Hadju-Cheney syndrome (Isidor et al., 2011; Simpson et al., 2011; Kiel et al., 2012), breast cancer (Wang et al., 2015), and in 12% of non-small-cell lung carcinomas (NSCLCs), of which half were in the PEST domain (Westhoff et al., 2009). In these conditions, Notch target genes are highly upregulated.
Considering the variety of pathological conditions associated with alterations of NICD and FBXW7, there is a limited understanding of the regulation of this interaction. Our current understanding stems from a study on Sel-10, the nematode homolog of Fbxw7, showing the two proteins bind directly to each other and FBXW7 negatively regulates Notch signaling (Hubbard et al., 1997). Using cell models, human and murine homologs of Sel-10 were shown to play a key role in regulating Notch signaling by driving NICD to ubiquitin-proteasome mediated degradation (Gupta-Rossi et al., 2001; Oberg et al., 2001; Wu et al., 2001). NICD ubiquitination relies on the PEST domain. Studies on three NOTCH4 variants suggested that Sel-10 preferentially binds to phosphorylated forms of the C-terminal domain of NOTCH4 (Oberg et al., 2001; Wu et al., 2001). However, the NICD-Sel10 interaction has only been observed under overexpression conditions in vitro. It remains to be shown if this interaction occurs in vivo, if NICD interacts with any other E3 ligases, how this interaction is regulated and whether it is context-dependent. The FBXW7 null mutant mice exhibit elevated levels of Notch4 intracellular domain and/or Notch1 intracellular domain alongside defects that are in alignment with a variety of roles identified for Notch in different developmental process such as cardiogenesis and vascular development. However, intriguingly, with respect to the segmentation clock, the absence of Fbxw7 seems to play a less major role in this process, at least according to the mutant phenotypes—although a detailed analysis has yet to be conducted (Tetzlaff et al., 2004; Tsunematsu et al., 2004). The results of these reports suggest that the mechanisms of NICD1 degradation during the somitogenesis process might actually rely on alternative (or redundant) mechanisms, highlighting again the need to further study alternative means of regulation of stability NICD1/degradation.
Conclusions
In this review we provided a general overview of the critical role of Notch signaling in regulating the segmentation clock involved in somitogenesis. Notch activity is based on stability and turnover of its intracellular domain, NICD. This stability is regulated by phosphorylation of the PEST domain, targeting NICD to proteasome degradation upon recognition by the E3 ligase FBXW7. Mutations in the PEST domain, leading to aberrations in NICD stability, are the underlying cause of a number of solid and non-solid cancers and different genetic disorders. Therefore, uncovering the finer details of Notch pathway regulation merits attention, particularly because a wider comprehension of this process would provide further insights into the mechanisms involved in the onset of Notch-related diseases.
Author Contributions
FC and JD conceived the structure and content. FC wrote the initial draft document. FC designed and produced the figures. JD corrected and edited the document.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Laura D'Ignazio, Lucas Morales Moya and Ioanna Mastromina for helpful and constructive feedback. This work was supported by a Wellcome Trust PhD studentship to FC and a Wellcome Trust Strategic award [097945/Z/11/Z].
References
Alketbi, A., and Attoub, S. (2015). Notch signaling in cancer: rationale and strategies for targeting. Curr. Cancer Drug Targets 15, 364–374. doi: 10.2174/156800961505150710113353
Andersson, E. R., Sandberg, R., and Lendahl, U. (2011). Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612. doi: 10.1242/dev.063610
Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770–776. doi: 10.1126/science.284.5415.770
Aster, J. C. (2014). In brief: Notch signalling in health and disease. J. Pathol. 232, 1–3. doi: 10.1002/path.4291
Aulehla, A., and Pourquie, O. (2010). Signaling gradients during paraxial mesoderm development. Cold Spring Harb. Perspect. Biol. 2:a000869. doi: 10.1101/cshperspect.a000869
Aulehla, A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B., et al. (2003). Wnt3A plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395–406. doi: 10.1016/S1534-5807(03)00055-8
Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., et al. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. doi: 10.1016/S0092-8674(00)80098-7
Bailey, C., and Dale, K. (2015). “Somitogenesis in vertebrate development,” in eLS (John Wiley & Sons, Ltd), 1–15. doi: 10.1002/9780470015902.a0003820.pub2
Baldus, C. D., Thibaut, J., Goekbuget, N., Stroux, A., Schlee, C., Mossner, M., et al. (2009). Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica 94, 1383–1390. doi: 10.3324/haematol.2008.005272
Barrantes, I. B., Elia, A. J., Wunsch, K., Hrabe De Angelis, M. H., Mak, T. W., Rossant, J., et al. (1999). Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9, 470–480. doi: 10.1016/S0960-9822(99)80212-7
Benazeraf, B., and Pourquie, O. (2013). Formation and segmentation of the vertebrate body axis. Annu. Rev. Cell Dev. Biol. 29, 1–26. doi: 10.1146/annurev-cellbio-101011-155703
Bessho, Y., Hirata, H., Masamizu, Y., and Kageyama, R. (2003). Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456. doi: 10.1101/gad.1092303
Bessho, Y., Miyoshi, G., Sakata, R., and Kageyama, R. (2001a). Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6, 175–185. doi: 10.1046/j.1365-2443.2001.00409.x
Bessho, Y., Sakata, R., Komatsu, S., Shiota, K., Yamada, S., and Kageyama, R. (2001b). Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647. doi: 10.1101/gad.930601
Bolos, V., Mira, E., Martinez-Poveda, B., Luxan, G., Canamero, M., Martinez, A. C., et al. (2013). Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 15:R54. doi: 10.1186/bcr3447
Bone, R. A., Bailey, C. S., Wiedermann, G., Ferjentsik, Z., Appleton, P. L., Murray, P. J., et al. (2014). Spatiotemporal oscillations of Notch1, Dll1 and NICD are coordinated across the mouse PSM. Development 141, 4806–4816. doi: 10.1242/dev.115535
Bonyadi Rad, E., Hammerlindl, H., Wels, C., Popper, U., Ravindran Menon, D., Breiteneder, H., et al. (2016). Notch4 signaling induces a mesenchymal-epithelial-like transition in melanoma cells to suppress malignant behaviors. Cancer Res. 76, 1690–1697. doi: 10.1158/0008-5472.CAN-15-1722
Bozkulak, E. C., and Weinmaster, G. (2009). Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695. doi: 10.1128/MCB.00406-09
Bray, S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. doi: 10.1038/nrm2009
Broadus, M. R., Chen, T. W., Neitzel, L. R., Ng, V. H., Jodoin, J. N., Lee, L. A., et al. (2016). Identification of a paralog-specific notch1 intracellular domain degron. Cell Rep. 15, 1920–1929. doi: 10.1016/j.celrep.2016.04.070
Brou, C., Logeat, F., Gupta, N., Bessia, C., Lebail, O., Doedens, J. R., et al. (2000). A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216. doi: 10.1016/S1097-2765(00)80417-7
Cambray, N., and Wilson, V. (2007). Two distinct sources for a population of maturing axial progenitors. Development 134, 2829–2840. doi: 10.1242/dev.02877
Chastagner, P., Israel, A., and Brou, C. (2008). AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3:e2735. doi: 10.1371/journal.pone.0002735
Christ, B., Huang, R., and Scaal, M. (2007). Amniote somite derivatives. Dev. Dyn. 236, 2382–2396. doi: 10.1002/dvdy.21189
Cinquin, O. (2007). Understanding the somitogenesis clock: what's missing? Mech. Dev. 124, 501–517. doi: 10.1016/j.mod.2007.06.004
Cole, S. E., Levorse, J. M., Tilghman, S. M., and Vogt, T. F. (2002). Clock regulatory elements control cyclic expression of Lunatic fringe during somitogenesis. Dev. Cell 3, 75–84. doi: 10.1016/S1534-5807(02)00212-5
Conlon, R. A., Reaume, A. G., and Rossant, J. (1995). Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545.
Cooke, J., and Zeeman, E. C. (1976). A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476. doi: 10.1016/S0022-5193(76)80131-2
Crusio, K. M., King, B., Reavie, L. B., and Aifantis, I. (2010). The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene 29, 4865–4873. doi: 10.1038/onc.2010.222
Dale, J. K., Maroto, M., Dequeant, M. L., Malapert, P., McGrew, M., and Pourquie, O. (2003). Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421, 275–278. doi: 10.1038/nature01244
Dequeant, M. L., and Pourquie, O. (2008). Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9, 370–382. doi: 10.1038/nrg2320
D'souza, B., Meloty-Kapella, L., and Weinmaster, G. (2010). Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 92, 73–129. doi: 10.1016/S0070-2153(10)92003-6
Dubrulle, J., and Pourquie, O. (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427, 419–422. doi: 10.1038/nature02216
Dumortier, A., Wilson, A., Macdonald, H. R., and Radtke, F. (2005). Paradigms of notch signaling in mammals. Int. J. Hematol. 82, 277–284. doi: 10.1532/IJH97.05099
Dyczynska, E., Sun, D., Yi, H., Sehara-Fujisawa, A., Blobel, C. P., and Zolkiewska, A. (2007). Proteolytic processing of delta-like 1 by ADAM proteases. J. Biol. Chem. 282, 436–444. doi: 10.1074/jbc.M605451200
Eckalbar, W. L., Fisher, R. E., Rawls, A., and Kusumi, K. (2012). Scoliosis and segmentation defects of the vertebrae. Wiley Interdiscip. Rev. Dev. Biol. 1, 401–423. doi: 10.1002/wdev.34
Ehebauer, M., Hayward, P., and Martinez-Arias, A. (2006). Notch signaling pathway. Sci. STKE 2006:cm7. doi: 10.1126/stke.3642006cm7
Erbilgin, Y., Sayitoglu, M., Hatirnaz, O., Dogru, O., Akcay, A., Tuysuz, G., et al. (2010). Prognostic significance of NOTCH1 and FBXW7 mutations in pediatric T-ALL. Dis. Markers 28, 353–360. doi: 10.1155/2010/740140
Espinosa, L., Ingles-Esteve, J., Aguilera, C., and Bigas, A. (2003). Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278, 32227–32235. doi: 10.1074/jbc.M304001200
Ferjentsik, Z., Hayashi, S., Dale, J. K., Bessho, Y., Herreman, A., De Strooper, B., et al. (2009). Notch is a critical component of the mouse somitogenesis oscillator and is essential for the formation of the somites. PLoS Genet. 5:e1000662. doi: 10.1371/journal.pgen.1000662
Fiuza, U. M., and Arias, A. M. (2007). Cell and molecular biology of Notch. J. Endocrinol. 194, 459–474. doi: 10.1677/JOE-07-0242
Fryer, C. J., White, J. B., and Jones, K. A. (2004). Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520. doi: 10.1016/j.molcel.2004.10.014
Gibb, S., Maroto, M., and Dale, J. K. (2010). The segmentation clock mechanism moves up a notch. Trends Cell Biol. 20, 593–600. doi: 10.1016/j.tcb.2010.07.001
Gomez, C., Ozbudak, E. M., Wunderlich, J., Baumann, D., Lewis, J., and Pourquie, O. (2008). Control of segment number in vertebrate embryos. Nature 454, 335–339. doi: 10.1038/nature07020
Gomez, C., and Pourquie, O. (2009). Developmental control of segment numbers in vertebrates. J. Exp. Zool. B Mol. Dev. Evol. 312, 533–544. doi: 10.1002/jez.b.21305
Gordon, W. R., Vardar-Ulu, D., L'heureux, S., Ashworth, T., Malecki, M. J., Sanchez-Irizarry, C., et al. (2009). Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE 4:e6613. doi: 10.1371/journal.pone.0006613
Greenwald, I., and Kovall, R. (2013). Notch signaling: genetics and structure. WormBook 1–28. doi: 10.1895/wormbook.1.10.2
Groot, A. J., Habets, R., Yahyanejad, S., Hodin, C. M., Reiss, K., Saftig, P., et al. (2014). Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol. Cell. Biol. 34, 2822–2832. doi: 10.1128/MCB.00206-14
Gupta-Rossi, N., Le Bail, O., Gonen, H., Brou, C., Logeat, F., Six, E., et al. (2001). Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276, 34371–34378. doi: 10.1074/jbc.M101343200
Habets, R. A., Groot, A. J., Yahyanejad, S., Tiyanont, K., Blacklow, S. C., and Vooijs, M. (2015). Human NOTCH2 is resistant to ligand-independent activation by metalloprotease Adam17. J. Biol. Chem. 290, 14705–14716. doi: 10.1074/jbc.M115.643676
Hao, B., Oehlmann, S., Sowa, M. E., Harper, J. W., and Pavletich, N. P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143. doi: 10.1016/j.molcel.2007.02.022
Henrique, D., Abranches, E., Verrier, L., and Storey, K. G. (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875. doi: 10.1242/dev.119768
Hicks, C., Johnston, S. H., Disibio, G., Collazo, A., Vogt, T. F., and Weinmaster, G. (2000). Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2, 515–520. doi: 10.1038/35019553
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843. doi: 10.1126/science.1074560
Hori, K., Sen, A., and Artavanis-Tsakonas, S. (2013). Notch signaling at a glance. J. Cell Sci. 126, 2135–2140. doi: 10.1242/jcs.127308
Huang, J., Song, H., Liu, B., Yu, B., Wang, R., and Chen, L. (2013). Expression of Notch-1 and its clinical significance in different histological subtypes of human lung adenocarcinoma. J. Exp. Clin. Cancer Res. 32:84. doi: 10.1186/1756-9966-32-84
Hubaud, A., and Pourquie, O. (2014). Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721. doi: 10.1038/nrm3891
Hubbard, E. J., Wu, G., Kitajewski, J., and Greenwald, I. (1997). sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11, 3182–3193. doi: 10.1101/gad.11.23.3182
Huppert, S. S., Ilagan, M. X., De Strooper, B., and Kopan, R. (2005). Analysis of Notch function in presomitic mesoderm suggests a gamma-secretase-independent role for presenilins in somite differentiation. Dev. Cell 8, 677–688. doi: 10.1016/j.devcel.2005.02.019
Iimura, T., Yang, X., Weijer, C. J., and Pourquie, O. (2007). Dual mode of paraxial mesoderm formation during chick gastrulation. Proc. Natl. Acad. Sci. U.S.A. 104, 2744–2749. doi: 10.1073/pnas.0610997104
Ingles-Esteve, J., Espinosa, L., Milner, L. A., Caelles, C., and Bigas, A. (2001). Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J. Biol. Chem. 276, 44873–44880. doi: 10.1074/jbc.M104703200
Isidor, B., Lindenbaum, P., Pichon, O., Bezieau, S., Dina, C., Jacquemont, S., et al. (2011). Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 43, 306–308. doi: 10.1038/ng.778
Jiang, Y. J., Aerne, B. L., Smithers, L., Haddon, C., Ish-Horowicz, D., and Lewis, J. (2000). Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479. doi: 10.1038/35044091
Jin, Y. H., Kim, H., Oh, M., Ki, H., and Kim, K. (2009). Regulation of Notch1/NICD and Hes1 expressions by GSK-3alpha/beta. Mol. Cells 27, 15–19. doi: 10.1007/s10059-009-0001-7
Julich, D., Hwee Lim, C., Round, J., Nicolaije, C., Schroeder, J., Davies, A., et al. (2005). beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev. Biol. 286, 391–404. doi: 10.1016/j.ydbio.2005.06.040
Kageyama, R., Niwa, Y., Isomura, A., Gonzalez, A., and Harima, Y. (2012). Oscillatory gene expression and somitogenesis. Wiley Interdiscip. Rev. Dev. Biol. 1, 629–641. doi: 10.1002/wdev.46
Kamath, B. M., Bauer, R. C., Loomes, K. M., Chao, G., Gerfen, J., Hutchinson, A., et al. (2012). NOTCH2 mutations in Alagille syndrome. J. Med. Genet. 49, 138–144. doi: 10.1136/jmedgenet-2011-100544
Kato, T. M., Kawaguchi, A., Kosodo, Y., Niwa, H., and Matsuzaki, F. (2010). Lunatic fringe potentiates Notch signaling in the developing brain. Mol. Cell. Neurosci. 45, 12–25. doi: 10.1016/j.mcn.2010.05.004
Kiel, M. J., Velusamy, T., Betz, B. L., Zhao, L., Weigelin, H. G., Chiang, M. Y., et al. (2012). Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 209, 1553–1565. doi: 10.1084/jem.20120910
Kitagawa, M., Oyama, T., Kawashima, T., Yedvobnick, B., Kumar, A., Matsuno, K., et al. (2001). A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 21, 4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001
Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W., et al. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177. doi: 10.1126/science.1065203
Kopan, R., and Ilagan, M. X. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. doi: 10.1016/j.cell.2009.03.045
Kramer, H. (2000). RIPping notch apart: a new role for endocytosis in signal transduction? Sci. STKE 2000:pe1. doi: 10.1126/stke.2000.29.pe1
Krol, A. J., Roellig, D., Dequeant, M. L., Tassy, O., Glynn, E., Hattem, G., et al. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792. doi: 10.1242/dev.063834
Lebon, L., Lee, T. V., Sprinzak, D., Jafar-Nejad, H., and Elowitz, M. B. (2014). Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. Elife 3:e02950. doi: 10.7554/eLife.02950
Le Bras, S., Loyer, N., and Le Borgne, R. (2011). The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic 12, 149–161. doi: 10.1111/j.1600-0854.2010.01126.x
Liao, B. K., and Oates, A. C. (2016). Delta-notch signalling in segmentation. Arthropod. Struct. Dev. doi: 10.1016/j.asd.2016.11.007. [Epub ahead of print].
Lobry, C., Ntziachristos, P., Ndiaye-Lobry, D., Oh, P., Cimmino, L., Zhu, N., et al. (2013). Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J. Exp. Med. 210, 301–319. doi: 10.1084/jem.20121484
Lobry, C., Oh, P., Mansour, M. R., Look, A. T., and Aifantis, I. (2014). Notch signaling: switching an oncogene to a tumor suppressor. Blood 123, 2451–2459. doi: 10.1182/blood-2013-08-355818
Malecki, M. J., Sanchez-Irizarry, C., Mitchell, J. L., Histen, G., Xu, M. L., Aster, J. C., et al. (2006). Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26, 4642–4651. doi: 10.1128/MCB.01655-05
Malyukova, A., Dohda, T., Von Der Lehr, N., Akhoondi, S., Corcoran, M., Heyman, M., et al. (2007). The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 67, 5611–5616. doi: 10.1158/0008-5472.CAN-06-4381
Maroto, M., Bone, R. A., and Dale, J. K. (2012). Somitogenesis. Development 139, 2453–2456. doi: 10.1242/dev.069310
Matsumoto, A., Onoyama, I., and Nakayama, K. I. (2006). Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem. Biophys. Res. Commun. 350, 114–119. doi: 10.1016/j.bbrc.2006.09.003
McGrew, M. J., Sherman, A., Lillico, S. G., Ellard, F. M., Radcliffe, P. A., Gilhooley, H. J., et al. (2008). Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development 135, 2289–2299. doi: 10.1242/dev.022020
Meloty-Kapella, L., Shergill, B., Kuon, J., Botvinick, E., and Weinmaster, G. (2012). Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 22, 1299–1312. doi: 10.1016/j.devcel.2012.04.005
Miele, L., Golde, T., and Osborne, B. (2006). Notch signaling in cancer. Curr. Mol. Med. 6, 905–918. doi: 10.2174/156652406779010830
Moretti, J., and Brou, C. (2013). Ubiquitinations in the notch signaling pathway. Int. J. Mol. Sci. 14, 6359–6381. doi: 10.3390/ijms14036359
Mullighan, C. G. (2009). Mutations of NOTCH1, FBXW7, and prognosis in T-lineage acute lymphoblastic leukemia. Haematologica 94, 1338–1340. doi: 10.3324/haematol.2009.012047
Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., et al. (2000). A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206. doi: 10.1016/S1097-2765(00)80416-5
Nam, Y., Weng, A. P., Aster, J. C., and Blacklow, S. C. (2003). Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278, 21232–21239. doi: 10.1074/jbc.M301567200
Oates, A. C., Morelli, L. G., and Ares, S. (2012). Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development 139, 625–639. doi: 10.1242/dev.063735
Oberg, C., Li, J. H., Pauley, A., Wolf, E., Gurney, M., and Lendahl, U. (2001). The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian sel-10 homolog. J. Biol. Chem. 276, 35847–35853. doi: 10.1074/jbc.M103992200
Oka, C., Nakano, T., Wakeham, A., De La Pompa, J. L., Mori, C., Sakai, T., et al. (1995). Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 121, 3291–3301.
Okubo, Y., Sugawara, T., Abe-Koduka, N., Kanno, J., Kimura, A., and Saga, Y. (2012). Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of Notch signalling. Nat. Commun. 3:1141. doi: 10.1038/ncomms2133
O'neil, J., Grim, J., Strack, P., Rao, S., Tibbitts, D., Winter, C., et al. (2007). FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 204, 1813–1824. doi: 10.1084/jem.20070876
Ozbudak, E. M., and Lewis, J. (2008). Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 4:e15. doi: 10.1371/journal.pgen.0040015
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquie, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi: 10.1016/S0092-8674(00)80451-1
Parks, A. L., Klueg, K. M., Stout, J. R., and Muskavitch, M. A. (2000). Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373–1385.
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. doi: 10.1038/nrm1547
Pourquie, O. (2001). Vertebrate somitogenesis. Annu. Rev. Cell Dev. Biol. 17, 311–350. doi: 10.1146/annurev.cellbio.17.1.311
Pourquie, O. (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663. doi: 10.1016/j.cell.2011.05.011
Qiu, L., Joazeiro, C., Fang, N., Wang, H. Y., Elly, C., Altman, Y., et al. (2000). Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275, 35734–35737. doi: 10.1074/jbc.M007300200
Radtke, F., and Raj, K. (2003). The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3, 756–767. doi: 10.1038/nrc1186
Radtke, F., Schweisguth, F., and Pear, W. (2005). The Notch ‘gospel’. EMBO Rep. 6, 1120–1125. doi: 10.1038/sj.embor.7400585
Rampal, R., Li, A. S., Moloney, D. J., Georgiou, S. A., Luther, K. B., Nita-Lazar, A., et al. (2005). Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J. Biol. Chem. 280, 42454–42463. doi: 10.1074/jbc.M509552200
Rana, N. A., and Haltiwanger, R. S. (2011). Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 21, 583–589. doi: 10.1016/j.sbi.2011.08.008
Roy, M., Pear, W. S., and Aster, J. C. (2007). The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 17, 52–59. doi: 10.1016/j.gde.2006.12.001
Schroeter, E. H., Kisslinger, J. A., and Kopan, R. (1998). Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386. doi: 10.1038/30756
Schroter, C., Herrgen, L., Cardona, A., Brouhard, G. J., Feldman, B., and Oates, A. C. (2008). Dynamics of zebrafish somitogenesis. Dev. Dyn. 237, 545–553. doi: 10.1002/dvdy.21458
Serth, K., Schuster-Gossler, K., Cordes, R., and Gossler, A. (2003). Transcriptional oscillation of lunatic fringe is essential for somitogenesis. Genes Dev. 17, 912–925. doi: 10.1101/gad.250603
Shifley, E. T., Vanhorn, K. M., Perez-Balaguer, A., Franklin, J. D., Weinstein, M., and Cole, S. E. (2008). Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development 135, 899–908. doi: 10.1242/dev.006742
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., and Kageyama, R. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. doi: 10.1101/gad.270785.115
Simpson, M. A., Irving, M. D., Asilmaz, E., Gray, M. J., Dafou, D., Elmslie, F. V., et al. (2011). Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 43, 303–305. doi: 10.1038/ng.779
Skaar, J. R., Pagan, J. K., and Pagano, M. (2013). Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381. doi: 10.1038/nrm3582
Struhl, G., and Adachi, A. (1998). Nuclear access and action of notch in vivo. Cell 93, 649–660. doi: 10.1016/S0092-8674(00)81193-9
Tam, P. P. (1981). The control of somitogenesis in mouse embryos. J. Embryol. Exp. Morphol. 65(Suppl), 103–128.
Tetzlaff, M. T., Yu, W., Li, M., Zhang, P., Finegold, M., Mahon, K., et al. (2004). Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. U.S.A. 101, 3338–3345. doi: 10.1073/pnas.0307875101
Thompson, B. J., Buonamici, S., Sulis, M. L., Palomero, T., Vilimas, T., Basso, G., et al. (2007). The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204, 1825–1835. doi: 10.1084/jem.20070872
Tsunematsu, R., Nakayama, K., Oike, Y., Nishiyama, M., Ishida, N., Hatakeyama, S., et al. (2004). Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423. doi: 10.1074/jbc.M312337200
Van Tetering, G., Van Diest, P., Verlaan, I., Van Der Wall, E., Kopan, R., and Vooijs, M. (2009). Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027. doi: 10.1074/jbc.M109.006775
Wahi, K., Bochter, M. S., and Cole, S. E. (2014). The many roles of Notch signaling during vertebrate somitogenesis. Semin. Cell Dev. Biol. 49, 68–75. doi: 10.1016/j.semcdb.2014.11.010
Wahl, M. B., Deng, C., Lewandoski, M., and Pourquie, O. (2007). FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development 134, 4033–4041. doi: 10.1242/dev.009167
Wallberg, A. E., Pedersen, K., Lendahl, U., and Roeder, R. G. (2002). p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 22, 7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002
Wang, K., Zhang, Q., Li, D., Ching, K., Zhang, C., Zheng, X., et al. (2015). PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a gamma-secretase inhibitor. Clin. Cancer Res. 21, 1487–1496. doi: 10.1158/1078-0432.CCR-14-1348
Wang, N. J., Sanborn, Z., Arnett, K. L., Bayston, L. J., Liao, W., Proby, C. M., et al. (2011). Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 108, 17761–17766. doi: 10.1073/pnas.1114669108
Wang, Z., Inuzuka, H., Fukushima, H., Wan, L., Gao, D., Shaik, S., et al. (2012). Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep. 13, 36–43. doi: 10.1038/embor.2011.231
Weber, S., Niessen, M. T., Prox, J., Lullmann-Rauch, R., Schmitz, A., Schwanbeck, R., et al. (2011). The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development 138, 495–505. doi: 10.1242/dev.055210
Welcker, M., and Clurman, B. E. (2008). FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93. doi: 10.1038/nrc2290
Welcker, M., Singer, J., Loeb, K. R., Grim, J., Bloecher, A., Gurien-West, M., et al. (2003). Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12, 381–392. doi: 10.1016/S1097-2765(03)00287-9
Westhoff, B., Colaluca, I. N., D'ario, G., Donzelli, M., Tosoni, D., Volorio, S., et al. (2009). Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 106, 22293–22298. doi: 10.1073/pnas.0907781106
Wharton, K. A., Johansen, K. M., Xu, T., and Artavanis-Tsakonas, S. (1985). Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43, 567–581. doi: 10.1016/0092-8674(85)90229-6
William, D. A., Saitta, B., Gibson, J. D., Traas, J., Markov, V., Gonzalez, D. M., et al. (2007). Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev. Biol. 305, 172–186. doi: 10.1016/j.ydbio.2007.02.007
Wu, G. Y., Lyapina, S., Das, I., Li, J. H., Gurney, M., Pauley, A., et al. (2001). SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21, 7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001
Wu, L., Aster, J. C., Blacklow, S. C., Lake, R., Artavanis-Tsakonas, S., and Griffin, J. D. (2000). MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489. doi: 10.1038/82644
Yabe, T., and Takada, S. (2016). Molecular mechanism for cyclic generation of somites: Lessons from mice and zebrafish. Dev. Growth Differ. 58, 31–42. doi: 10.1111/dgd.12249
Yang, L. T., Nichols, J. T., Yao, C., Manilay, J. O., Robey, E. A., and Weinmaster, G. (2005). Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol. Biol. Cell 16, 927–942. doi: 10.1091/mbc.E04-07-0614
Keywords: somitogenesis, embryonic development, signalling pathway, notch, FBXW7
Citation: Carrieri FA and Dale JK (2017) Turn It Down a Notch. Front. Cell Dev. Biol. 4:151. doi: 10.3389/fcell.2016.00151
Received: 22 September 2016; Accepted: 16 December 2016;
Published: 18 January 2017.
Edited by:
Juan Jose Sanz-Ezquerro, Spanish National Research Council, SpainReviewed by:
Raj Ladher, TIFR National Centre for Biological Sciences, IndiaMarc Vooijs, Maastricht University, Netherlands
Moises Mallo, Instituto Gulbenkian de Ciência, Portugal
Copyright © 2017 Carrieri and Dale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacqueline Kim Dale, ai5rLmRhbGVAZHVuZGVlLmFjLnVr
 Francesca A. Carrieri
Francesca A. Carrieri Jacqueline Kim Dale
Jacqueline Kim Dale