
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell Dev. Biol., 19 July 2016
Sec. Cell Growth and Division
Volume 4 - 2016 | https://doi.org/10.3389/fcell.2016.00075
Maintenance of the chromosomal copy number over generations and recombination between homologous chromosomes are hallmarks of meiotic cell division. This genetic exchange that take place during gamete formation leads to genetic diversity, the main driving force behind natural selection. Formation of chiasmata, the physical link between homologous chromosomes during meiosis, is a requisite for recombination. In addition, chiasmata also aid in proper segregation of homologous chromosomes and has a major impact on reproductive fitness. Given these facts it is intriguing that many insect species have forgone the need for genetic exchange between homologous chromosomes during meiosis. Geneticists for several decades knew that meiotic crossover and recombination is absent in Drosophila males and some female lepidopterans, a condition termed achiasmy. However, a good understanding of the mechanisms that cause achiasmy and the evolutionary benefits of achiasmy is currently lacking. In this article we will discuss possible genetic and molecular basis of achiasmy in male Drosophila.
Meiotic cell division, an essential step in sexual reproduction, helps in the segregation of homologous chromosomes and sister chromatids. In addition, a crucial task for meiotic cell division is the maintenance of recombination mediated genetic variability (Hunter, 2015). A “standard meiotic script” and maintenance of high-fidelity during chromosomal segregation is well conserved among eukaryotes (Nicklas, 1997; McKee, 2004). Mis-segregation of homologs during meiosis leads to aneuploidy which causes lethality or genetic disorders in the offsprings. Aneuploidy is a major cause for approximately one-third of spontaneous miscarriages in humans, developmental disabilities, and mental retardation (Hassold et al., 2007).
The greatest advantage of sexual reproduction, which otherwise is a bottle neck due to the complexities involved, is meiotic recombination. Recombination yields newer combinations of alleles, which helps in the genetic adaptability of the organism (Carvalho, 2003). The adverse effects of the absence of meiotic recombination is clear from Steinmann's analysis of genes in Drosophila miranda “neo-Y chromosome,” which resulted from the fusion of an autosome to the Y-chromosome estimated to have happened a million years ago (Bachtrog, 2005). The genes on the attached autosome (neo-Y) underwent degeneration due to the lack of recombination during meiosis in Drosophila males, while its homolog neo-X remained intact in females due to the existence of recombination. Extending this observation, Bachtrog et al. showed that deleterious mutations accumulate on a non-recombining chromosome (Bachtrog and Charlesworth, 2000, 2002). The human Y chromosome has been shown to reduce errors in the coding regions by having a self-recombination mechanism (Rozen et al., 2003; Skaletsky et al., 2003). An added advantage of meiotic recombination is that the chiasmata formation during crossover helps in proper alignment and segregation of chromosomes (Carpenter, 1994). Given the benefits, it is confounding that meiotic recombination is absent in some species.
Loss of meiotic recombination results in aneuploidy in plants, but with less deleterious consequences than in animals. This is a boon for plant breeders and farmers due to the obvious advantages (Caryl et al., 2003). Heterochiasmy, the dimorphism in meiotic recombination rates between sexes is seen in various divergent species. Several hypotheses have been proposed to explain the evolution of heterochiasmy (Lenormand, 2003; Lenormand and Dutheil, 2005). Achiasmy a form of heterochiasmy, where males or females of a species completely lack meiotic recombination, occurs frequently in Dipterans and in several orders of Lepidopterans. According to the Haldane-Huxley rule, it is the heterogametic sex (XY or WZ) that shows achiasmic meiosis. Morgan (1910) was the first to describe achiasmy in Drosophila males (Morgan, 1910). However, Drosophila females, like the majority of sexually reproducing organisms, generate crossovers between homologous chromosomes to direct segregation at the first meiotic division (Lindsley and Sandler, 1977; Puro and Nokkala, 1977; Lin et al., 1981; Orr-Weaver, 1995; Lichten, 2001; McKim et al., 2002; Figure 1). During meiosis the male germ-line cells of fruit flies undergo homolog pairing of chromosomes creating bivalents that can be sequestered to unique territories inside the Prophase nucleus (Hawley, 2002). However, no genetic exchange occurs during this process. Interestingly, there are rare reports of spontaneous meiotic recombination in male Drosophila melanogaster (Hiraizumi, 1971).
Achiasmy in male fruit flies arose at least tens of million years ago as it is a common trait in the Drosophila clade, this raises several interesting questions:
• How can an evolutionarily conserved process like meiotic recombination be excluded in a sex-specific manner?
• How can a trait that helped in laying the foundation for natural selection get erased completely from one sex?
• In spite of the risks of accumulation of deleterious mutations how does heterochiasmic species benefit from forgoing meiotic recombination?
In addition to several invertebrates many vertebrates exhibit lower recombination frequency in the heterogametic sex, which is usually male. However, it has so far been hard to establish if the higher recombining sex would compensate for the low recombination rates in the other sex. The fact that achiasmy is observed in one sex (mostly in heterogametic sex) argues that there could be compensation of recombination rates in the other sex. In mice, the female sex chromosomes have more chiasmata for their length than the autosomes, this is probably to compensate for the lower levels of recombination in males (Burt et al., 1991). However, there is no direct evidence for compensation in the recombining sex.
We tried to figure out whether compensation of recombination rates exists in heterochiasmic species by comparing closely related chiasmic and achiasmic species. Among Drosophilids male recombination have been recorded from Drosophila ananassae and Drosophila willistoni, species closely related to achiasmic D. melanogaster. Comparison of recombination rates in autosomes of females of D. ananassae and D. melanogaster do not show significant differences. The only exception is the X-chromosome in D. melanogaster, which has lower recombination rates when compared to autosomes and to D. ananassae (Caceres et al., 1999). After taking the above fact in to consideration it could be concluded that at least in Drosophila melanogaster there is no obvious compensation of recombination rates in chiasmate females. Also, using simple mathematical calculations we found that the absence of recombination does not lead to an overall effect on genetic variability (see Supplementary Material for details).
One prevailing hypothesis regarding achiasmy in male Drosophila is the “Lazy Male hypothesis.” According to this hypothesis, the non-recombining males that has diverted the recombination task entirely to females could be more fit in comparison. This might allow the non-recombining sperms gain increased fitness resulting from the conservation of energy due to lack of recombination events and result in enhanced fecundity. Another suggestion based on Haldane's views is that the loss of recombination in males could have evolved as a mechanism for preventing recombination of the male sex-chromosome.
Our attempt in this article is not to explain why recombination is absent in male fruit flies, we will try to elaborate on the possible genetic and molecular reasons behind achiasmy in male Drosophila here. We propose that the absence of meiotic recombination in males is due to the absence of key recombination factors or is due to the presence of negative factors that prevent recombination from occurring during spermatogenesis.
Though the basic archetype of meiosis is met in Drosophila male meiosis, the chromosomal pairing events do not facilitate genetic exchange, but create bivalents that are assigned with discrete territories in the Prophase nucleus (Fabian and Brill, 2012). Near the apical end of the testis, are the cells that undergo Meiosis I, the spermatocytes. The Prophase I stage in Drosophila males show non-conventional phenotypes as the chromosomes of male fruit flies are indistinguishable from G2 phase and lack structural features of a traditional Prophase I. A trilobular nucleus is visible at this stage, corresponding to three major bivalents in the nucleus. Prominent axes normally decorated with cohesins and other lateral element proteins are missing from the Prophase I chromosomes. Bivalents are also not attached by classical chiasmata. Another deviation from the “standard script” is that the homologs enter the meiotic cycle already paired, abolishing the need for homolog search. Since there is no recombination during Drosophila male meiosis the synaptonemal complex is undetectable. However, the pairing sites on the homologs are bound by specific protein complexes that ensure the legitimate segregation of the homologs (Ault et al., 1982; Ault and Rieder, 1994; Vazquez et al., 2002; McKee et al., 2012). To find the possible mechanisms behind achisamy in male Drosophila, we compared the expression of genes between adult ovary and testis using GEO2R (Barrett et al., 2013). This analysis showed downregulation of several recombination specific genes in testis in comparison to ovary. The genes which show differential expression between sexes are listed down in Table 1, and the functions of some interesting candidate genes are discussed in detail.
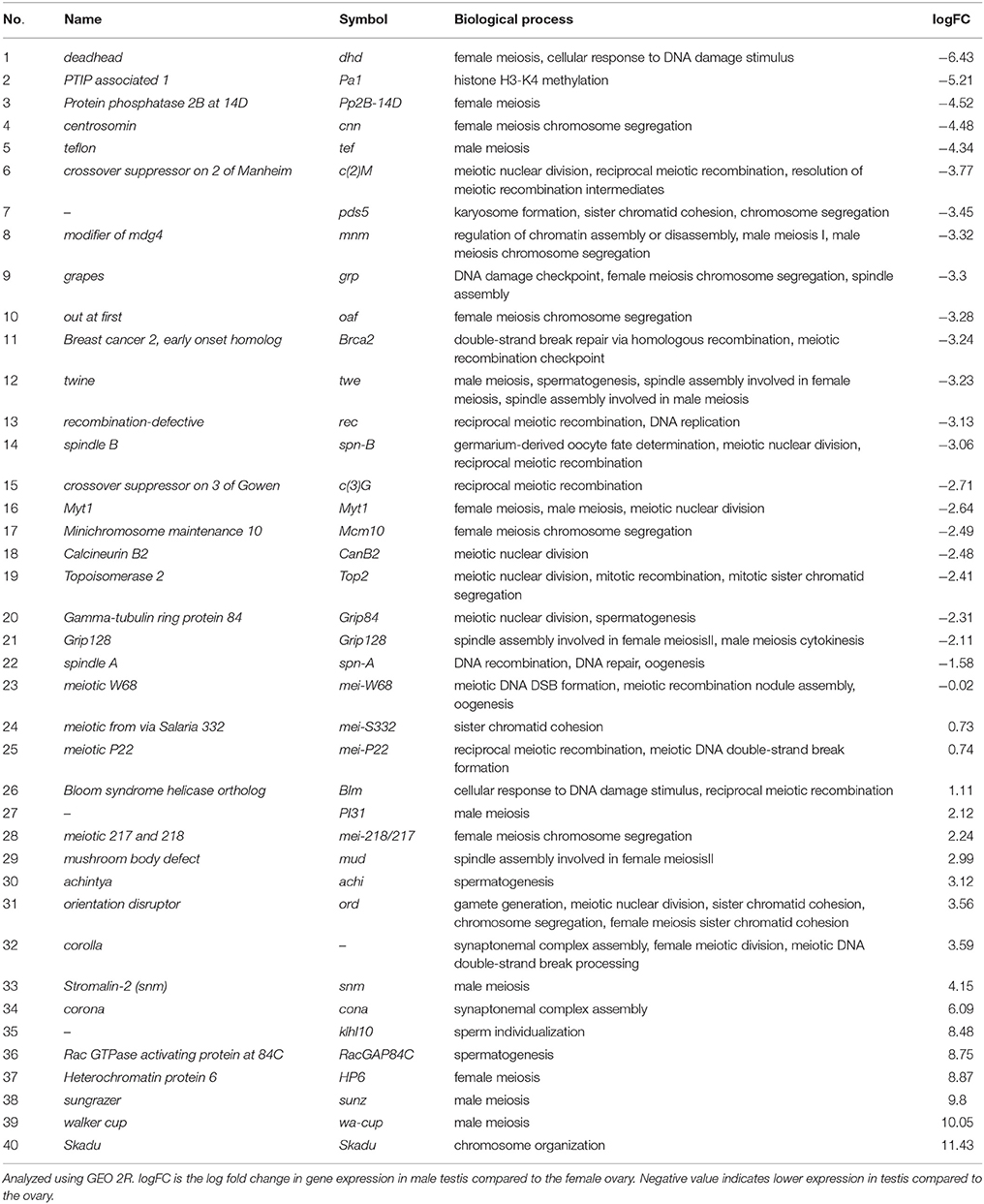
Table 1. Genes playing a role during meiosis that show differential expression between testis and ovary (Data acquired from GEO Dataset GSE7763, contributed by Chintapalli VR, Wang J, and Dow JA available at flyatlas.org; Chintapalli et al., 2007).
Accurate homolog segregation depends on the pairing of homologs that form bivalents that interact with the meiotic spindle as a unit (Roeder, 1990). During meiosis, Drosophila females utilize chiasmata to pair up their three major homologous chromosomes (McKim et al., 1998). By contrast, even in the absence of chiasmata or synaptonemal complex formation and recombination, all the four chromosome pairs form stable bivalents in males (Ren et al., 1997).
The 200–250 copies of rRNA genes share homology in the Drosophila X and Y chromosomes. Stromalin in Meiosis (SNM) and Modifier of Mdg4 in Meiosis (MNM) are present in the X–Y pairing sites and are required for stable homolog pairing and segregation in male and not for female meiosis (Thomas et al., 2005). On the contrary, recruitment of SNM and MNM to autosomes depends on another protein TEFLON (TEF). Flies that lack tef, snm, and mnm show phenotypes during male meiosis, but not in female meiotic cells (Thomas et al., 2005). Each autosomal homolog is seen in a common territory till the late-Prophase I, SNM and MNM localize to these homolog territories and at pro-Metaphase I they start condensing into well aligned bivalents. This is lost in the mnm and snm mutants, which suggests that they help in bringing the homologs together into a common territory. Based on the FlyAtlas data snm transcript levels are more than 20-fold higher in testis compared to ovary. On the contrary, mnm and tef transcripts are very low in testis, though they are essential for the formation of chromosomal territories during male meiosis. One possible explanation for the lack of meiotic recombination in Drosophila males is that high levels of SNM might prevent recombination factors from acting. This can be addressed by expressing SNM in female germ cells and check effects on meiotic recombination. A change in the expression or binding of the recombination initiators in these females will answer a part of our query.
In most organisms, meiosis proceeds with synaptonemal complex (SC) formation in a DSB-dependent fashion. Quite contrary to this, SC formation precedes DSB formation and recombination events, and is a necessary step in Drosophila females, evident from mutant analyses (Jang et al., 2003; Mehrotra and McKim, 2006). A marker for SC showed that SC is present before DSB protein MEI-P22 foci appear on the chromosomes (Liu et al., 2002). Several components of SC in Drosophila females have been identified. C(3)G constitutes the transverse filaments (TFs) of SC (Page and Hawley, 2001). The central elements (CEs) are bound by the N-termini of C(3)G homodimers, while the C-termini help the TFs to form connections via axial/lateral elements (AEs/LEs). c(3)G mutants lack both SC formation and MEI-P22 foci, suggesting that DSB formation is dependent on SC formation in Drosophila females. In addition, these mutants completely lack genetic exchange during meiosis (Jeffress et al., 2007; Page et al., 2008). C(2)M helps the TFs to bind to the chromosomes and co-localizes with C(3)G. Corona (Cona) is a component of AEs, that is found to co-localize with C(3)G and is essential for the polymerization of C(3)G monomers. Recombination frequency in corona mutant females is found to be 50-to-200 fold lower. Another component of AEs is Corolla, which interacts with Cona, to stabilize the SC structure. corolla mutants show increased non-disjunction compared to the wildtype females. c(2)M mutants also show reduced meiotic crossover frequency (Manheim and McKim, 2003). Though levels of corolla and corona are high in testis, levels of c(3)G and c(2)M transcripts are very low in male testis, and could be key contributors to the lack of meiotic recombination (Anderson et al., 2005).
DSBs have been shown to be adept in initiating recombination in meiotic cells of Baker's yeast. Experiments in Drosophila females show that DSBs can act as recombination initiators during meiotic division. mei-W68 encodes the Baker's yeast recombinase spo11 homolog, which is required for the DSB initiation during meiotic recombination in Drosophila females (McKim and Hayashi-Hagihara, 1998). mei-P22 produces another factor for DSB formation, and have mutant phenotypes similar to mei-W68 (Liu et al., 2002). MEI-P22 foci is present for a short time during early meiotic prophase. However, our analysis do not reveal a huge difference in the levels of mei-P22 and mei-W68 levels in testis compared to ovary. Lack of expression of MEI-P22 and MEI-W68 proteins due to post-transcriptional regulation or lack of activation of these DSB proteins by post-transcriptional modifications could be responsible for achiasmy in male Drosophila. As synaptonemal complex formation is a pre-requisite for MEI-P22 association with the chromosomes in Drosophila as seen from the c(3)G mutants, it is not hard to assume that the MEI-P22 and MEI-W68 proteins are unable to generate DSBs in male Drosophila.
The key player of recombination in prokaryotes is RecA, which catalyses the pairing and strand invasion between homologous DNA strands, during both DNA repair and crossover recombination (Shinohara and Shinohara, 2004). RAD51 and DMC1, two RecA like proteins, in yeast are required for meiotic recombination. In Drosophila, SpnA (DmRAD51), shares strong sequence similarity with RAD51 protein of yeast, chicken, mouse, and human. spnA mutant females show a significant elevation in the frequency of DSBs during meiotic recombination (Staeva-Vieira et al., 2003) and also show single-strand annealing (SSA) repair than DSB repair through crossover and recombination during meiosis (Yoo and McKee, 2004). The levels of spnA transcript is significantly lower in testis during spermatogenesis and could be a limiting factor responsible for the absence of meiotic cross-over in male Drosophila.
Unlike meiotic crossovers that are beneficial, mitotic crossovers can lead to the loss of heterozygosity, possibly increasing the chances of tumorigenesis (Andersen and Sekelsky, 2010; Kohl and Sekelsky, 2013). As a safeguard mechanism mitotic crossovers are prevented by anti-crossover proteins like the helicase BLM, which unwinds the recombination intermediates during mitosis to generate non-crossover products, across metazoans. Generation of meiotic crossovers in most eukaryotes requires the removal of the anti-crossover proteins by Msh4–Msh5 complex. Drosophila lost msh4 and msh5 genes, which is functionally replaced by Mini-Chromosome Maintenance (MCM) complex proteins. Mutants of rec, mei-217 and mei-218, genes which encode for Drosophila MCM complex, show reduced female meiotic crossovers, which can be rescued by the removal of blm gene (Kohl et al., 2012). Transcript levels of blm is 2-fold higher in male testis; this and the MCM loss of function phenotypes in females suggest a role for BLM helicase in inhibiting meiotic crossovers in male fruit flies. However, while mei-217 and mei-218 transcript levels are significantly high in the testis, rec transcript levels are very low. An increased rate of non-crossover recombination was observed in rec mutant females, about 2-fold, not surprising as it is an MCM complex protein (Carpenter, 1975, 1989; Bojko, 1989; Matsubayashi and Yamamoto, 2003). As mei-217 and mei-218 expression is enhanced in testis according to the microarray data [GEO: Dataset GSE7763], it is possible that the low levels of rec transcripts in Drosophila males leads to the lack of inhibition of BLM anti-crossover proteins. Drosophila males mutant for blm gene could provide an answer to this. Overexpression of REC protein in testis would also reveal whether lack of functional MCM complex is responsible for lack of meiotic recombination in males.
We have not covered all the possible factors that could be responsible for achiasmy in male Drosophila here due to various limitations. Other meiotic genes like grapes (grp) and out at first (oaf), both needed for chromosome segregation (Dobie et al., 2001); deadhead (dhd), a thioredoxin homolog (Salz et al., 1994) and topoisomerase 2 (top2; Hughes and Hawley, 2014); show very low expression in the male testis. These genes are enriched in the germ line progenitors, the pole cells, during embryogenesis (Mukai et al., 2006). It is possible that some of these gene products could also play a prominent role in the absence of meiotic cross over in Drosophila males.
Generation of haploid gametes from diploid precursors by meiosis is a crucial step during sexual reproduction. During this process genetic exchange occurs, introducing variability in the population by mixing genotypes and also safeguards the segregation of the homologous chromosome pairs. Recent studies have thrown light on the possible mechanisms by which homologs are efficiently segregated in the absence of chiasmata and cross-over recombination during Drosophila spermatogenesis. What are the mechanisms that control sex-specific shutdown of meiotic recombination in one sex of several species like Drosophila melanogaster? And, why there is sex-specific shutdown of meiotic recombination in some species? A number of genes important for steps of crossover and meiotic recombination show differential expression when microarray data for gene expression between testis and ovary of fruit flies were compared. There are significant changes in the levels of transcripts of genes that encode for recombination factors in the testis compared to the ovary. The precise molecular pathways that regulate the expression of these recombination factors still remain to be discovered. We speculate that the absence or inactivation of key players of genetic recombination during spermatogenesis might be the cause for achiasmy in Drosophila males. This can be tested by attempting to restore meiotic recombination in male flies by the germ-line specific expression of these factors. As achiasmy could be a result of absence of multiple recombination-specific factors this may not be easy. Another factor to consider, which is a limitation of our analysis, is the lack of proteomics data or information related to post-translational modification of various recombination factors in testis and ovary. The discovery of a strain isolated from the wild that exhibit recombination in males is suggestive of the fact that few factors/genes could also be responsible for the lack of this genetic event in male Drosophila (Hiraizumi, 1971). The question remains whether there is a master regulator of meiotic recombination in male Drosophila. The genes that show very high expression in testis could be tested for this role. Research in this direction could possibly unravel reasons for fertility defects and disorders associated with aneuploidy.
AJ, KV, and JV conceptualized the idea behind this manuscript. AJ and KV performed the bioinformatics analysis and wrote the initial draft of this manuscript. JV generated the final draft of this manuscript. JV also generated the figures and tables for this manuscript.
AJ and KV were supported by the DST-Inspire Scholarship program while working toward this manuscript. JV was supported by DST-SERB Ramanujan Fellowship while working toward this manuscript. JV would like to thank IISER TVM and Ministry of Human Resources and Development, India for the generous financial support to his laboratory.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the members of D.R.E.A.M lab for their support during the preparation of this manuscript, especially Smitha Vishnu who did proof readings and suggested several changes.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcell.2016.00075
Andersen, S. L., and Sekelsky, J. (2010). Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32, 1058–1066. doi: 10.1002/bies.201000087
Anderson, L. K., Royer, S. M., Page, S. L., Mckim, K. S., Lai, A., Lilly, M. A., et al. (2005). Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U.S.A. 102, 4482–4487. doi: 10.1073/pnas.0500172102
Ault, J. G., Lin, H. P., and Church, K. (1982). Meiosis in Drosophila melanogaster. IV. The conjunctive mechanism of the XY bivalent. Chromosoma 86, 309–317. doi: 10.1007/BF00292259
Ault, J. G., and Rieder, C. L. (1994). Meiosis in Drosophila males. I. The question of separate conjunctive mechanisms for the XY and autosomal bivalents. Chromosoma 103, 352–356. doi: 10.1007/BF00417883
Bachtrog, D. (2005). Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila. Genome Res. 15, 1393–1401. doi: 10.1101/gr.3543605
Bachtrog, D., and Charlesworth, B. (2000). Reduced levels of microsatellite variability on the neo-Y chromosome of Drosophila miranda. Curr. Biol. 10, 1025–1031. doi: 10.1016/S0960-9822(00)00656-4
Bachtrog, D., and Charlesworth, B. (2002). Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416, 323–326. doi: 10.1038/416323a
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995. doi: 10.1093/nar/gks1193
Bojko, M. (1989). Two kinds of “recombination nodules” in Neurospora crassa. Genome 32, 309–317. doi: 10.1139/g89-446
Burt, A., Bell, G., and Harvey, P. H. (1991). Sex differences in recombination. J. Evol. Biol. 4, 259–277. doi: 10.1046/j.1420-9101.1991.4020259.x
Caceres, M., Barbadilla, A., and Ruiz, A. (1999). Recombination rate predicts inversion size in Diptera. Genetics 153, 251–259.
Carpenter, A. T. (1975). Electron microscopy of meiosis in Drosophila melanogaster females: II. The recombination nodule–a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. U.S.A. 72, 3186–3189. doi: 10.1007/bf00319833
Carpenter, A. T. (1989). Are there morphologically abnormal early recombination nodules in the Drosophila melanogaster meiotic mutant mei-218? Genome 31, 74–80.
Carvalho, A. B. (2003). The advantages of recombination. Nat. Genet. 34, 128–129. doi: 10.1038/ng0603-128
Caryl, A. P., Jones, G. H., and Franklin, F. C. (2003). Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54, 25–38. doi: 10.1093/jxb/erg041
Chintapalli, V. R., Wang, J., and Dow, J. A. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720. doi: 10.1038/ng2049
Dobie, K. W., Kennedy, C. D., Velasco, V. M., Mcgrath, T. L., Weko, J., Patterson, R. W., et al. (2001). Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics 157, 1623–1637.
Fabian, L., and Brill, J. A. (2012). Drosophila spermiogenesis: big things come from little packages. Spermatogenesis 2, 197–212. doi: 10.4161/spmg.21798
Hassold, T., Hall, H., and Hunt, P. (2007). The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 2, R203–R208. doi: 10.1093/hmg/ddm243
Hawley, R. S. (2002). Meiosis: how male flies do meiosis. Curr. Biol. 12, R660–R662. doi: 10.1016/S0960-9822(02)01161-2
Hiraizumi, Y. (1971). Spontaneous recombination in Drosophila melanogaster males. Proc. Natl. Acad. Sci. U.S.A. 68, 268–270. doi: 10.1073/pnas.68.2.268
Hughes, S. E., and Hawley, R. S. (2014). Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet. 10:e1004650. doi: 10.1371/journal.pgen.1004650
Hunter, N. (2015). Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7:a016618. doi: 10.1101/cshperspect.a016618
Jang, J. K., Sherizen, D. E., Bhagat, R., Manheim, E. A., and Mckim, K. S. (2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116, 3069–3077. doi: 10.1242/jcs.00614
Jeffress, J. K., Page, S. L., Royer, S. K., Belden, E. D., Blumenstiel, J. P., Anderson, L. K., et al. (2007). The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N- and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics 177, 2445–2456. doi: 10.1534/genetics.107.078717
Kohl, K. P., Jones, C. D., and Sekelsky, J. (2012). Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338, 1363–1365. doi: 10.1126/science.1228190
Kohl, K. P., and Sekelsky, J. (2013). Meiotic and mitotic recombination in meiosis. Genetics 194, 327–334. doi: 10.1534/genetics.113.150581
Lenormand, T., and Dutheil, J. (2005). Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3:e63. doi: 10.1371/journal.pbio.0030063
Lichten, M. (2001). Meiotic recombination: breaking the genome to save it. Curr. Biol. 11, R253–R256. doi: 10.1016/S0960-9822(01)00131-2
Lin, H. P., Ault, J. G., and Church, K. (1981). Meiosis in Drosophila melanogaster. I. Chromosome identification and kinetochore microtubule numbers during the first and second meiotic divisions in males. Chromosoma 83, 507–521. doi: 10.1007/BF00328276
Lindsley, D. L., and Sandler, L. (1977). The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277, 295–312. doi: 10.1098/rstb.1977.0019
Liu, H., Jang, J. K., Kato, N., and Mckim, K. S. (2002). mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162, 245–258.
Manheim, E. A., and McKim, K. S. (2003). The Synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13, 276–285. doi: 10.1016/S0960-9822(03)00050-2
Matsubayashi, H., and Yamamoto, M. T. (2003). REC, a new member of the MCM-related protein family, is required for meiotic recombination in Drosophila. Genes Genet. Syst. 78, 363–371. doi: 10.1266/ggs.78.363
McKee, B. D. (2004). Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677, 165–180. doi: 10.1016/j.bbaexp.2003. 11.017
McKee, B. D., Yan, R., and Tsai, J. H. (2012). Meiosis in male Drosophila. Spermatogenesis 2, 167–184. doi: 10.4161/spmg.21800
McKim, K. S., Green-Marroquin, B. L., Sekelsky, J. J., Chin, G., Steinberg, C., Khodosh, R., et al. (1998). Meiotic synapsis in the absence of recombination. Science 279, 876–878. doi: 10.1126/science.279.5352.876
McKim, K. S., and Hayashi-Hagihara, A. (1998). mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932–2942. doi: 10.1101/gad.12.18.2932
McKim, K. S., Jang, J. K., and Manheim, E. A. (2002). Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36, 205–232. doi: 10.1146/annurev.genet.36.041102.113929
Mehrotra, S., and McKim, K. S. (2006). Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2:e200. doi: 10.1371/journal.pgen.0020200
Morgan, T. H. (1910). Sex limited inheritance in Drosophila. Science 32, 120–122. doi: 10.1126/science.32.812.120
Mukai, M., Kitadate, Y., Arita, K., Shigenobu, S., and Kobayashi, S. (2006). Expression of meiotic genes in the germline progenitors of Drosophila embryos. Gene Expr. Patterns 6, 256–266. doi: 10.1016/j.modgep.2005.08.002
Nicklas, R. B. (1997). How cells get the right chromosomes. Science 275, 632–637. doi: 10.1126/science.275.5300.632
Orr-Weaver, T. L. (1995). Meiosis in Drosophila: seeing is believing. Proc. Natl. Acad. Sci. U.S.A. 92, 10443–10449. doi: 10.1073/pnas.92.23.10443
Page, S. L., and Hawley, R. S. (2001). c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143. doi: 10.1101/gad.935001
Page, S. L., Khetani, R. S., Lake, C. M., Nielsen, R. J., Jeffress, J. K., Warren, W. D., et al. (2008). Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4:e1000194. doi: 10.1371/journal.pgen.1000194
Puro, J., and Nokkala, S. (1977). Meiotic segregation of chromosomes in Drosophila melanogaster oocytes. Chromosoma 63, 273–286. doi: 10.1007/BF00 327454
Ren, X., Eisenhour, L., Hong, C., Lee, Y., and Mckee, B. D. (1997). Roles of rDNA spacer and transcription unit-sequences in X-Y meiotic chromosome pairing in Drosophila melanogaster males. Chromosoma 106, 29–36. doi: 10.1007/s004120050221
Roeder, G. S. (1990). Chromosome synapsis and genetic recombination: their roles in meiotic chromosome segregation. Trends Genet. 6, 385–389. doi: 10.1016/0168-9525(90)90297-J
Rozen, S., Skaletsky, H., Marszalek, J. D., Minx, P. J., Cordum, H. S., Waterston, R. H., et al. (2003). Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876. doi: 10.1038/nature01723
Salz, H. K., Flickinger, T. W., Mittendorf, E., Pellicena-Palle, A., Petschek, J. P., and Albrecht, E. B. (1994). The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics 136, 1075–1086.
Shinohara, A., and Shinohara, M. (2004). Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 107, 201–207. doi: 10.1159/000080598
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., et al. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837. doi: 10.1038/nature01722
Staeva-Vieira, E., Yoo, S., and Lehmann, R. (2003). An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22, 5863–5874. doi: 10.1093/emboj/cdg564
Thomas, S. E., Soltani-Bejnood, M., Roth, P., Dorn, R., Logsdon, J. M. Jr., and Mckee, B. D. (2005). Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123, 555–568. doi: 10.1016/j.cell.2005.08.043
Vazquez, J., Belmont, A. S., and Sedat, J. W. (2002). The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12, 1473–1483. doi: 10.1016/S0960-9822(02)01090-4
Keywords: Drosophila meiotic recombination, crossover, homologous recombination, achiasmy
Citation: John A, Vinayan K and Varghese J (2016) Achiasmy: Male Fruit Flies Are Not Ready to Mix. Front. Cell Dev. Biol. 4:75. doi: 10.3389/fcell.2016.00075
Received: 17 May 2016; Accepted: 29 June 2016;
Published: 19 July 2016.
Edited by:
Eiman Aleem, Phoenix Childrens Hospital and University of Arizona College of Medicine-Phoenix, GermanyReviewed by:
Douglas Mark Ruden, Wayne State University, USACopyright © 2016 John, Vinayan and Varghese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jishy Varghese, amlzaHlAaWlzZXJ0dm0uYWMuaW4=
†These authors are BS-MS Students and they have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.