
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 30 March 2016
Sec. Membrane Traffic and Organelle Dynamics
Volume 4 - 2016 | https://doi.org/10.3389/fcell.2016.00020
This article is part of the Research Topic Tethering of transport vesicles in secretory and endocytic pathways: mechanisms and players View all 18 articles
TRAPP is a highly conserved modular multi-subunit protein complex. Originally identified as a “transport protein particle” with a role in endoplasmic reticulum-to-Golgi transport, its multiple subunits and their conservation from yeast to humans were characterized in the late 1990s. TRAPP attracted attention when it was shown to act as a Ypt/Rab GTPase nucleotide exchanger, GEF, in the 2000s. Currently, three TRAPP complexes are known in yeast, I, II, and III, and they regulate two different intracellular trafficking pathways: secretion and autophagy. Core TRAPP contains four small subunits that self assemble to a stable complex, which has a GEF activity on Ypt1. Another small subunit, Trs20/Sedlin, is an adaptor required for the association of core TRAPP with larger subunits to form TRAPP II and TRAPP III. Whereas the molecular structure of the core TRAPP complex is resolved, the architecture of the larger TRAPP complexes, including their existence as dimers and multimers, is less clear. In addition to its Ypt/Rab GEF activity, and thereby an indirect role in vesicle tethering through Ypt/Rabs, a direct role for TRAPP as a vesicle tether has been suggested. This idea is based on TRAPP interactions with vesicle coat components. While much of the basic information about TRAPP complexes comes from yeast, mutations in TRAPP subunits were connected to human disease. In this review we will summarize new information about TRAPP complexes, highlight new insights about their function and discuss current controversies and future perspectives.
Trafficking between intracellular compartments is mediated by vesicles and regulated by the highly conserved Ypt/Rab GTPases, their nucleotide exchangers, GEFs, and their downstream effectors (Lipatova et al., 2015a). Multi-protein complexes facilitate vesicle formation at donor compartments, subsequent targeting, tethering and fusion with acceptor compartments. One such complex, the transport protein particle TRAPP, acts as a Ypt/Rab GEF and possibly as a tether.
Yeast TRAPP was identified as a ten-subunit protein complex (~800 kDa) that co-precipitated with Bet3, which mediates endoplasmic reticulum (ER)-to-Golgi transport (Sacher et al., 1998, 2000). To date, eleven TRAPP subunits conserved between yeast and humans have been identified. Seven subunits are small, 140–283 amino acids, and the rest are larger, 417–1435 residues. These subunits are termed TrsN (N = size in kDa) or Bet3/5 in yeast, and TrappC (or TCP) 1–10 and 13, in mammalian cells. In yeast, these subunits assemble into three known complexes, TRAPP I, II, and III. Six of the small subunits form TRAPP I or attach to it for assembly of the two other complexes, while not much is known about the seventh small subunit Tca17. The large subunits are exclusive to either TRAPP II or TRAPP III complexes. Two additional subunits, TrappC/TCP 11-12, are metazoan specific and their TRAPP-related function will be discussed (Table 1).
Based on similarities between the purified Ypt1 GEF (Jones et al., 1998) and TRAPP, the activity of TRAPP as a Ypt GEF was determined (Jones et al., 2000; Wang et al., 2000). The idea that TRAPP acts as a tether to mediate vesicle docking was proposed at the same time based on the requirement of TRAPP for association of ER vesicles with the Golgi in a cell-free system (Barrowman et al., 2000). Since 2000, while evidence has accumulated regarding the importance of TRAPP subunits in human health (Brunet and Sacher, 2014), many uncertainties and controversies about the mechanisms of action of the TRAPP complexes have piled up.
Here, we summarize current views on TRAPP complexes structure, their function as Ypt/Rab GEFs and tethers, and their connection to human disease. Most of the evidence on the structure and function of TRAPP complexes comes from yeast and pertains to mammalian cells, while studies of human TRAPPs highlight their importance to human health.
In early reports about TRAPP, ten of the eleven yeast TRAPP subunits, except for Tca17, were found in a single high-molecular weight complex (~800 kDa) that co-precipitated with tagged Bet3 (Sacher et al., 2000). Later, two complexes I and II, were separated by gel filtration chromatography, where the larger complex (~1000 kDa) contains all the subunits of the smaller complex (~300 kDa), plus three large subunits (Sacher et al., 2001). Similarly, all the mammalian TRAPP subunits can be purified as a large complex (~670 kDa) by co-precipitation with a single core TRAPP subunit (Yamasaki et al., 2009).
The problem with assigning individual subunits to a specific TRAPP complex based solely on their purification from cell lysates is that TRAPP complexes oligomerize in cell lysates (Choi et al., 2011). The fact that this oligomerization depends on salt concentration or detergents suggests that it happens during cell lysis or purification (Brunet et al., 2012, 2013). Homotypic or heterotypic oligomerization can explain why the apparent molecular weights of TRAPP I and TRAPP II are higher than their expected sizes based on their composition; e.g., the expected size of TRAPP I is ~170 kDa, the size of recombinant TRAPP I purified from bacteria is ~170 kDa (Kim et al., 2006) as compared to ~300 kDa for the complex purified from yeast (Sacher et al., 2001). Oligomerization can also explain the observation that Trs85 fractionates with all the other TRAPP subunits (Sacher et al., 2000) even though it does not seem to function with TRAPP II. Therefore, assignment of individual subunits to specific TRAPP complexes should be based not only on their presence in purified complexes, but also on in vivo assays such as phenotypic and co-localization analyses.
Currently, in yeast all the small subunits are assigned to TRAPP I (Sacher et al., 2008). However, even though a recombinant yeast TRAPP I complex can be purified with six small subunits in stoichiometry of 2:1 Bet3 to each of the other five subunits (Kim et al., 2006), it is not clear whether two of these subunits, Trs20 and Trs33, function as part of this complex. Trs20 plays a role as an adaptor and is required for the assembly of the large TRAPP complexes: the association of TRAPP I with Trs120 for TRAPP II assembly (Taussig et al., 2013), and with Trs85 for TRAPP III assembly (Brunet et al., 2013; Taussig et al., 2014). However, it is currently unknown whether Trs20 is required for the function of TRAPP I. Tca17, a paralog of Trs20, also does not have a known function with TRAPP I (see below). Unlike the other small TRAPP subunits, Trs33 is not required for yeast cell viability or secretion. A role for Trs33 in the context of TRAPP I is currently unknown, and it is required for TRAPP II assembly only when Trs65 is absent (Tokarev et al., 2009). Therefore, in Table 1, Trs33 is listed as a TRAPP I-associated subunit. TRAPP I cannot be separated from the mammalian TRAPP II and its existence in vivo in mammalian cells is currently in question (Barrowman et al., 2010).
The assignment of Trs120, Trs130, and Trs65 as TRAPP II-specific subunits in yeast is based on their purification with this complex and on mutational analysis (Sacher et al., 2001; Liang et al., 2007). Trs85, in spite of its co-purification with TRAPP I, is considered as a TRAPP III-specific subunit in yeast (Sacher et al., 2008).
A single TRAPP complex purified from mammalian cell lysate contains all the known subunits, including the Trs85 ortholog, TrappC8 (Scrivens et al., 2011). However, based on genetic interaction maps, two TRAPP complexes were suggested to exist in mammalian cell: TRAPP II, which contains core TRAPP plus TrappC9-10, and TRAPP III, which contains core TRAPP plus TrappC8, 11-13 (Bassik et al., 2013). The inclusion of TrappC13, the homolog of Trs65, in the mammalian TRAPP III differs from its inclusion in the yeast TRAPP II complex. Importantly, there is currently no evidence for the existence of TRAPP I in mammalian cells.
A number of proteins were suggested to be TRAPP subunits. Tca17 (TRAPP complex associated) and its mammalian ortholog TrappC2L (TrappC2-like) were identified as sub-stoichiometric components of purified TRAPP that have sequence similarity to Trs20/Sedlin, and may play a role in in TRAPP assembly or stability (Montpetit and Conibear, 2009; Scrivens et al., 2009). Two metazoan-specific subunits, TrappC11, which is distantly related to Trs130/TrappC10, and TrappC12, were identified based on their co-fractionation with TRAPP and effect of their depletion on ER-to-Golgi transport (Wendler et al., 2010; Scrivens et al., 2011). A role for TrappC12, renamed TRAMM, without the rest of the TRAPP complex, was recently shown in mitosis (Milev et al., 2015). While it is possible that TrappC12 has a moonlighting function in mitosis, it suggests that proteins that associate with TRAPP are not necessarily TRAPP subunits. Therefore, additional criteria should be considered before assigning a Trs or TrappC number for new yeast and mammalian subunits, respectively.
The crystal structure of all the small yeast and mammalian TRAPP subunits has been resolved. Structurally, the subunits fall into two groups. Members of the Bet5 group, which also includes Trs20, Trs23, and Tca17, have a longin-domain (LD) fold (α–β–α sandwich), which is involved in protein-protein interactions and was suggested to form a LD-LD platform for GTPase binding (Levine et al., 2013). Specifically, in TRAPP I, Bet5 and Trs20 form a LD-LD platform for Ypt1 binding (Cai et al., 2008). Members of the Bet3 group, which also includes Trs31 and Trs33, have similar structures (α/β-plait topology) despite low sequence similarity (Kümmel et al., 2005), and hetero-dimerize with Bet3 in the context of TRAPP I (Kim et al., 2006). Currently, the structures of the large TRAPP subunits are not resolved.
While the subunit organization of TRAPP I was solved by crystallography, the architecture of the larger complexes was suggested based on single-particle electron microscopy (EM) and mutational analyses (Figure 1).
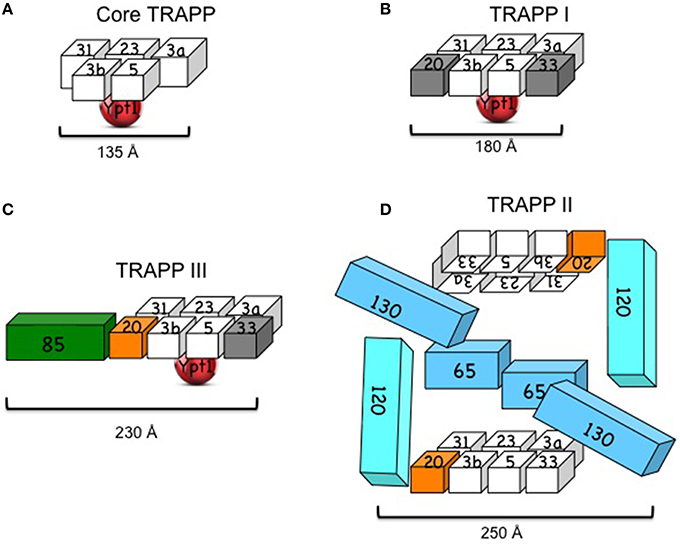
Figure 1. Yeast TRAPP complexes. (A) Core TRAPP, which contains four small subunits with two copies of Bet3 (3a and 3b), self assembles, binds Ypt1 through Bet5 and Trs23 and acts as a Ypt1-GEF. The diagram is based on the published structure of core TRAPP in a complex with Ypt1 (Cai et al., 2008). (B) TRAPP I contains core TRAPP, Trs20 and Trs33. However, it is not clear that the presence of the latter two subunits (in gray) is required for its function. The diagram is based on the published EM structure of core TRAPP subunits with Trs33 or Trs20 (Kim et al., 2006). (C) TRAPP III contains core TRAPP, Trs20 (orange), which is required for its assembly with Trs85 (green) and its function in autophagy. A role for Trs33 (in gray) in this complex has not been shown. The diagram is based on the published architecture and mutational analysis of TRAPP III (Tan et al., 2013; Taussig et al., 2014). (D) TRAPP II contains TRAPP I, including Trs20 (orange) and Trs33, and three large subunits: Trs120, Trs130, and the non-essential Trs65. Trs20 is required for the interaction of TRAPP I with Trs120, and either Trs65 or Trs33 are required for assembly of TRAPP II in vivo. TRAPP II is depicted here as a dimer (see discussion in the text). The cubes representing the subunits are roughly proportionate to their size; numbers stand for TrsN, except for Bet3 and Bet5. Approximate length (Å) is based on crystal structure for core TRAPP and negative staining EM for the other complexes (see text).
Recombinant core yeast TRAPP complex can self assemble when at least four subunits are co-expressed in bacterial cells: Bet3, Bet5, Trs23, and Trs31 (Table 1). The three-dimensional structure of core TRAPP has been solved by crystallography. The ~135 Å-long and 35–70 Å-wide cylinder contains two copies of Bet3 and one copy of each of the other three subunits (Cai et al., 2008). Two additional small subunits, Trs33 and Trs20, can attach at the two ends of the yeast core TRAPP, if co-expressed with the core subunits, to form the six-subunit TRAPP I. This complex is ~180 Å long and 65 Å wide. An atomic model for subunit organization of TRAPP I was suggested by single particle EM analysis and docking of crystal structures of two sub-complexes of mammalian TRAPP I. The two Bet3 copies have similar orientations to the same flat side of the complex, which contains both acidic and basic patches and is thought to face the membrane (Kim et al., 2006). Ypt1 binds the opposite, mostly acidic, side of TRAPP I (Kim et al., 2006; Cai et al., 2008).
The architecture of the yeast TRAPP III was suggested based on negative EM staining and docking of the atomic model of TRAPP I (see above) in the EM density map of TRAPP III. According to this model, Trs85 attaches to TRAPP I through Trs20, to form an elongated crescent shaped complex with approximate dimensions of 230 × 68 × 46 Å. Ypt1 binds to the same position as in TRAPP I (Tan et al., 2013). The EM structure is supported by mutational analysis of Trs20, which identified the Trs20 interface where Trs85 attaches to TRAPP I (Taussig et al., 2014). However, the structure of Trs85 and its interface with TRAPP I are not known.
The architecture of TRAPP II purified from yeast cell lysates was proposed based on single-particle EM and docking the atomic model of TRAPP I (see above). The suggested ~225 × 250 Å diamond-like structure is of a dimer in which the TRAPP II-specific subunits are sandwiched between two anti-parallel TRAPP I complexes (Yip et al., 2010). Currently, there is no evidence that dimerization of TRAPP II is required for its role in secretion to maintain cell viability, since Trs65, which is required for this dimerization, is not essential for cell viability. Moreover, in this model, the assignments of Trs120 and Trs130 to the Trs33 and Trs20 sides of TRAPP I, respectively, do not agree with existing mutational analyses. We have found that whereas Trs20 is required for the attachment of Trs120 to TRAPP I, Trs33 plays a role in the interaction of Trs130 with TRAPP I (Tokarev et al., 2009; Taussig et al., 2013), thus placing Trs120 and Trs130 to the opposite sides of TRAPP I when compared to the EM-based structure (Figure 1D). Considering these disagreements, the uncertainty regarding the significance of the dimer, and the fact that structural information is currently not available for the large subunits of TRAPP II, we conclude that the architecture of TRAPP II is still unclear.
In yeast, based on effect of mutations in subunits of TRAPP I, II and III, it is now accepted that TRAPP I acts at early Golgi, TRAPP II in late Golgi, and TRAPP III in autophagy (Figure 2). The cellular function of the different TRAPP complexes can be inferred from whether they are essential for cell viability. In addition to the core TRAPP subunits, the Trs20 adaptor and the two TRAPP II-specific subunits are essential for cell viability, whereas the others, Trs33, Trs65, and Trs85, are not (Table 2).
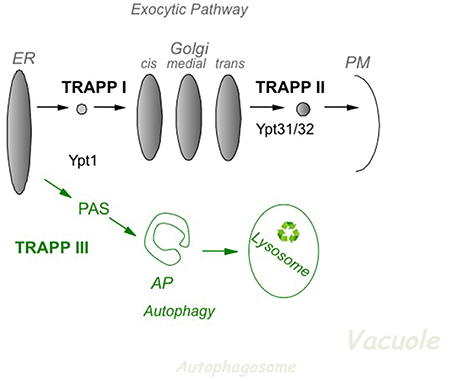
Figure 2. The role of TRAPP complexes and their Ypt substrates in yeast intra-cellular trafficking. In the exocytic pathway (top), TRAPP I activates Ypt1 to regulate ER-to-Golgi transport, whereas TRAPP II activates Ypt31/32 to regulate Golgi-to-PM transport. In autophagy (bottom), a cellular recycling pathway (in green), TRAPP III activates Ypt1 to regulate the assembly of PAS, the first step of autophagy. PAS is required for the formation of the double-membrane autophagosome (AP), which delivers cargo for degradation in the lysosome. See discussion in the text.
In the secretory pathway, cargo is delivered from the ER, through the Golgi apparatus, to the plasma membrane (PM). A set of Sec proteins that mediate the different steps of this pathway are essential for cell viability (Feyder et al., 2015). The essential TRAPP subunits make up the two TRAPP complexes, I and II, which function in the secretory pathway. The assignment of TRAPP I and TRAPP II to early and late Golgi, respectively, is based on mutation analysis. Specifically, whereas conditional mutations in Bet3 and Bet5, which are present in all TRAPP complexes, elicit an ER-to-Golgi as the first blocked transport step, mutations in the TRAPP II-specific subunits Trs130 and Trs120 result in a late Golgi transport block (Sacher et al., 2001).
Autophagy is a cellular recycling process in which excess or damaged cellular components are delivered for degradation in the lysosome via a double-membrane organelle termed autophagosome (AP). This process starts with the formation of the pre-autophagosomal structure, PAS, by a set of ~40 autophagy-specific proteins, Atgs. Usually there is one PAS per cell. Atgs are not required for cell viability during normal cell growth, but only during stress, e.g., nitrogen starvation (Weidberg et al., 2011). The TRAPP III-specific subunit, Trs85, which is not essential for cell viability, was instead shown to play a role in selective and non-selective autophagy (Meiling-Wesse et al., 2005; Nazarko et al., 2005).
While currently it is not clear how TRAPP complexes localize to the compartment on which they function, Golgi or AP, there are conflicting reports about the relationship between Trs85 and Atg9 and localization to PAS. Atg9 is the only integral membrane Atg and Atg9 vesicles are considered to bring the membrane component of the PAS (Weidberg et al., 2011). Trs85 and Atg9 co-localize on PAS, and also in multiple puncta in the cytosol (Lipatova et al., 2012). Based on co-immuno-precipitation and yeast-two hybrid interaction of Trs85 and Atg9 (neither shows a direct interaction), and the finding that Trs85 is not recruited to PAS in atg9Δ mutant cells, it was suggested that Atg9 brings TRAPP III to PAS (Kakuta et al., 2012). While this is a possibility, the problem is that in atg9Δ mutant cells there is no membrane in PAS and therefore no place for Trs85 or TRAPP to attach to. In contrast, it was proposed that TRAPP III plays a role in bringing Atg9 to PAS (Shirahama-Noda et al., 2013). Hopefully, future experiments will help resolve this issue.
In yeast, TRAPP subunits required for secretion are essential for cell viability. In three recent screens of ~20,000 human genes, about 2000 essential genes were identified (Blomen et al., 2015; Hart et al., 2015; Wang T. et al., 2015), with about a third having yeast essential orthologs (Blomen et al., 2015). We looked for the essentiality of TRAPP subunits in these three studies, which together analyzed 10 cell lines in 11 independent gene-trap or CRISPR/Cas9 screens (Table 2). The four core TRAPP complex subunits, TrappC1 (Bet5), TrappC3 (Bet3), TrappC4 (Trs23), and TrappC5 (Trs31), which are essential in yeast, are also essential in human cells. Two subunits not essential in yeast are also not essential in human cells: Trs33/TrappC6A/B (even though A and B could be supplementing for each other), and Trs65/TrappC13. The outliers are three genes essential in yeast but not in human cells: the adaptor Trs20/TrappC2, and the two TRAPP II-specific subunits, Trs120/TrappC9 and Trs130/TrappC10. In yeast, these three proteins assemble with core TRAPP to make TRAPP II, which is required for secretion. One subunit, Trs85, is not essential for yeast cell viability, whereas the human ortholog TrappC8 is; however, the similarity between these two subunits is very limited. Of the two metazoan-specific TRAPP subunits, one is essential, TrappC11, and one is not, TrappC12. Thus, more studies would be essential to interpret the reason for essentiality of the different TRAPP subunits in human cells.
Based on the migration of all the mammalian TRAPP subunits in a single high-molecular weight peak (~670 kDa) (Yamasaki et al., 2009), the currently dominating idea is that TRAPP II is the only mammalian Golgi TRAPP (Barrowman et al., 2010). However, because the expected size of individual TRAPP complexes is much smaller than the apparent ~670–800 kDa size of complexes purified from cell lysates, and because purified TRAPP complexes tend to oligomerize (Choi et al., 2011; Brunet et al., 2012), we suggest that the jury is still out regarding the existence of more than one mammalian Golgi TRAPP complex.
Mammalian TRAPP II was suggested to function in ER-to-Golgi transport based on the localization and knockdown effect of the TRAPP II-specific subunit mTrs130 (Yamasaki et al., 2009). On the other hand, roles for mTRAPP II and Rab11 (a Ypt31 homolog), together with the Golgi-to-PM regulators Rab8 and Rabin8 (a Sec2 homolog), were shown in human cells during primary cilia assembly (Westlake et al., 2011). These conflicting findings highlight the uncertainty regarding the roles of mammalian Golgi TRAPP complexes.
Mutations in SEDL, the gene that encodes Sedlin, were implicated in the cartilage disease SEDT (Table 1). Knockout of Sedlin/TRAPPC2 showed that the gene is not essential for cell viability (Table 2). Sedlin knockdown specifically affects export of procollagen, a large cargo that cannot fit into COPII vesicles, from the ER into megacarriers. While Sedlin probably does it in the context of TRAPP because mBet3 is also required for this process (Venditti et al., 2012), it is not clear which TRAPP complex is involved in this process.
While there is currently no direct evidence for the existence of TRAPP III or for a role of TrappC8 in autophagy, TrappC8 was identified in a proteomic study as a component of the human autophagy interaction network (Behrends et al., 2010). Unlike yeast, based on genetic interaction maps and co-IP, a role for the mammalian TRAPP III was proposed in ER-to-Golgi transport together with COPII vesicles (Bassik et al., 2013). In addition, TrappC8 was shown to be required for cell entry of the human papilloma virus, implying its role in endocytosis (Ishii et al., 2013) and for ciliogenesis (Schou et al., 2014). Thus, the conservation of a role for TRAPP complexes in autophagy is still unclear.
Plants have orthologs for all the TRAPP I and TRAPP II subunits essential in yeast and the TRAPP III-specific subunit Trs85 (Thellmann et al., 2010). As in yeast, TRAPP II was implicated in a late Golgi transport step together with the exocyst (Rybak et al., 2014). The functions of TRAPP I and TRAPP III in plants are currently unknown.
Identification of GEFs for individual Ypt/Rabs is not straightforward because, although individual GEFs are conserved through evolution, they do not share obvious similarities with GEFs for other Ypt/Rabs. The implication of TRAPP as a GEF for Ypt1 was inspired by biochemical similarities observed independently in different labs between a purified Ypt1 GEF and the TRAPP complex (Jones et al., 1998; Sacher et al., 1998). Soon after, TRAPP was identified as a Ypt1 GEF (Jones et al., 2000; Wang et al., 2000). While the idea that TRAPP acts as a Ypt/Rab GEF is well-established, the Ypt-GEF specificity of individual TRAPP complexes has been under debate.
Using biochemical and genetic analyses, including GEF activity, co-precipitation assays, the effect of TRAPP mutations on Ypt cellular localization and high-copy suppression of mutations, we have shown that TRAPP I acts as a GEF on Ypt1 whereas TRAPP II is a Ypt31/32 GEF (Morozova et al., 2006). In contrast, based on lack of GEF activity of purified TRAPP II on Ypt31/32 (Wang and Ferro-Novick, 2002), inability of GST-tagged Ypt31/32 to pull-down a TRAPP II-specific subunit (Yip et al., 2010), and a proposed localization and function of Ypt1 at the late Golgi (Sclafani et al., 2010), the view that all TRAPP complexes act on Ypt1 throughout the Golgi was promoted (Barrowman et al., 2010). However, recently this view was disputed at multiple levels. First, the observed phenotypes of YPT1 mutations assigning it a role in the late Golgi were shown to result from a defect in ER-phagy and not in late Golgi transport (Lipatova et al., 2013). Second, localization of Ypt1 and Ypt31 to opposite sides of the Golgi, early and late, respectively, was recently established (Kim et al., 2016). Third, the activity of TRAPP II as a GEF for the Ypt31 ortholog RabERAB11 at the trans Golgi was shown in Aspargilus nidulans, a filamentous fungus, both in vivo and in vitro (Pinar et al., 2015). Therefore, the current model for TRAPP complexes and their Ypt substrates in yeast depicts that TRAPP I and TRAPP II activate Ypt1 and Ypt31/32, respectively, on the two ends of the Golgi (Figure 2).
Support for the current model comes from two metazoan model systems. First, in Drosophila, the Trs120 homolog bru was shown to function in the late Golgi, to interact with Rab11, a Ypt31/32 homolog, and to affect the latter's cellular localization (Robinett et al., 2009). Second, in Arabidopsis, TRAPP II was functionally linked to the Ypt31 plant ortholog Rab-A, but not to the Ypt1 ortholog Rab-D (Qi et al., 2011). These results support the idea that TRAPP II acts as a Ypt31/32 GEF not only in yeast. The mammalian Golgi TRAPP II (see above) was shown to act in vitro as a Rab1, but not Rab11, GEF (Yamasaki et al., 2009). While the GEF specificity of yeast and mammalian TRAPPs might differ, we suggest that more research is needed to resolve this issue.
In yeast, Ypt1 is required for stress-induced and the selective autophagy pathway CVT (cytoplasm-to-vacuole) (Segev and Botstein, 1987; Lynch-Day et al., 2010; Lipatova and Segev, 2012; Lipatova et al., 2013). The idea that the Trs85-containing TRAPP III complex activates Ypt1 in autophagy is based on the following evidence: First, Trs85 is required for bringing Ypt1 to PAS (Lynch-Day et al., 2010). Second, using bimolecular fluorescence complementation (BiFC), Ypt1 and Trs85 were shown to interact on PAS (Lipatova et al., 2012, 2015b). Third, overexpression of Ypt1 can suppress the autophagy defects of bet3 and trs85Δ mutant cells (Lipatova et al., 2012; Zou et al., 2015). In addition to the BiFC interaction of Ypt1 and Trs85 on PAS, they also interact on Atg9 membranes. Once Ypt1 is activated on Atg9 membranes, it recruits Atg proteins to form PAS (Lipatova et al., 2012). Recruitment of two Atgs requires Ypt1, Atg1 and Atg11; only the last was shown to be a bona fide Ypt1 effector (Lipatova et al., 2012; Wang et al., 2013).
In mammalian cells, the orthologs of Ypt1 and Trs85, Rab1 and TrappC8, respectively, were implicated in autophagy (Behrends et al., 2010; Zoppino et al., 2010). However, because the similarity between Trs85 and TrappC8 is very low, and because TrappC8 was also suggested to play a role in ER-to-Golgi transport (Bassik et al., 2013), the connection of the mammalian TRAPP III complex to autophagy is currently not clear.
Tethers are proteins that can bridge two membranes and thereby capture vesicles (~500–1000 Å diameter) by an appropriate acceptor compartment. They are thought to bring two membranes into ~300 to >2000 Å proximity (Bröcker et al., 2010) and to facilitate vesicle uncoating and SNARE complex formation. SNAREs bring membranes to ~140 Å proximity and mediate membrane fusion (Hanson et al., 1997). Tethers can be long coiled-coil proteins or multisubunit-tethering complexes (MTCs), which typically interact with GTPases in their GTP-bound form, namely, they are downstream effectors of GTPases (Yu and Hughson, 2010).
While accumulation of vesicles when a putative tether is depleted agrees with the idea that this protein could be a tether, it does not prove this idea because the effect can be indirect or the protein might act in any sub step between vesicle formation and tethering, e.g., vesicle motility. Two types of assays have been used to establish tethering activity. First, an in vitro reconstitution assay was used to show that the HOPS complex is a tether (Hickey and Wickner, 2010). Second, an in vivo assay was used to show that directing the exocyst subunit Sec3 to the mitochondria, results in tethering of secretory vesicles to this compartment (Luo et al., 2014). Such assays have not been used for TRAPP yet.
The notion that TRAPP belongs to the MTC tether group was based on accumulation of ER vesicles in a cell-free system when TRAPP was depleted (Sacher et al., 2001). However, as mentioned above, this can result from an indirect effect, e.g., due to its GEF activity for Ypt1, which is required for recruitment of the Golgi tethers. If TRAPP is a tether, it is expected to bridge a distance >140 Å between two membranes. The length of TRAPP complexes ranges between ~180 Å for TRAPP I to ~250 Å for TRAPP II (Figure 1). Therefore, in principle, TRAPP complexes could act as tethers, especially in potential cooperation with other proteins. Like other known membrane tethers, TRAPP complexes interact with GTPases. However, unlike all other tethers, TRAPP complexes act as GEFs and not as downstream effectors of GTPases. The interaction of GEFs with their GTPase substrates is not stable and is interrupted once the GTPase binds GTP. Thus, it is unlikely that TRAPP GEFs and Ypts together create a bridge between two membranes. It is also not very likely that TRAPP GEFs cooperate with Ypt/Rab effectors to create membrane bridges, because the two bind to the two alternate forms of the GTPases, GDP, and GTP, respectively.
In yeast, the interaction of Bet3, a core TRAPP subunit, with Sec23, a component of the inner layer of COPII coat, was shown to be required for TRAPP binding to ER-derived vesicles (Cai et al., 2007). Interactions of TRAPP II and the non-essential TRAPP II subunit Trs65 with components of the COPI coat, Ret1/Cop1 and Gea2 (and Sec21), respectively, was used to implicate TRAPP II in tethering of COPI vesicles to the Golgi (Cai et al., 2005; Chen et al., 2011). In mammalian cells, it was suggested that TRAPP II interacts with COPI vesicles, the TRAPP II subunit TrappC9 interacts with a motor that moves COP II vesicle on microtubules, and COPII vesicles were suggested to interact with TRAPP III (Zong et al., 2012; Bassik et al., 2013). In addition to the inconsistency between yeast and mammalian cells, these findings provide information of TRAPP interactions with one membrane, while as a bridge it is expected to interact with two membranes. Interaction of the two Bet3 copies of a single TRAPP I complex with membranes of two separate vesicles was suggested to tether ER-derived vesicles during their homotypic fusion (Cai et al., 2008). However, the distance that one TRAPP I could bridge two membranes in this model is ~40 Å, which is shorter than a SNARE complex. Moreover, homotypic fusion of ER vesicles has not been shown in yeast, and the existence of TRAPP I in mammalian cells is questionable. Therefore, evidence that any TRAPP complex acts as a membrane tether is currently inconclusive.
To understand a possible mechanism by which TRAPP could act as a membrane tether, it is crucial to know how TRAPP complexes are positioned on membranes. Currently, the topology of TRAPP complexes on membranes is not known. TRAPP I was proposed to lie flat on one membrane through its two Bet3 copies, which would yield a distance shorter than expected for a tether. The idea that TRAPP I attaches to membranes through Bet3 was based on its hydrophobic channel (Kim et al., 2006), and/or palmitoylation. However, mutational analysis showed that neither is needed for TRAPP I function (Cai et al., 2008). The ways in which TRAPP II and TRAPP III are positioned on membranes are also unknown. Thus, additional evidence is needed to support the idea that TRAPP complexes act as membrane tethers and to elucidate mechanisms by which they might do that.
Because the secretory pathway is implicated in a broad range of human diseases (Aridor and Hannan, 2000), it was expected that it would be similar for TRAPP, a complex key for transport through the Golgi. Indeed, mutations in TRAPP subunits were linked to a wide range of human diseases (Brunet and Sacher, 2014) and their number keeps growing. Here, we highlight new disease connections (Table 1) and comment on the surprising fact that different TRAPP subunits seem to be linked to different diseases.
Examples of newly added disease connections for TRAPP subunits include miscarriage for TrappC2 (Wen et al., 2015), cancer for TrappC4 (Weng et al., 2014), and TrappC9 (Wetterskog et al., 2014; Zhang et al., 2015), intellectual disability and schizophrenia for TrappC9 (Khattak and Mir, 2014; McCarthy et al., 2014), and Alzheimer's disease for TrappC6 (Hamilton et al., 2011; Chang et al., 2015).
If there is really only one mammalian TRAPP complex (Barrowman et al., 2010), it is expected that mutations in individual subunits would have overlapping effects. Therefore, the most bewildering question about the involvement of TRAPP in human disease is why mutations in individual subunits result in such divergent diseases. One possible explanation is that there is more than one human TRAPP complex with different tissue specificities or cellular functions, and the different complexes are involved in different diseases. Another possible explanation is that “the screen is not saturated”; namely, the full range of representative diseases for individual TRAPP subunits has not been uncovered yet.
Recent findings connecting certain individual TRAPP subunits to multiple diseases support this idea. For example, only recently TrappC9 (Trs120) was connected to cancer and schizophrenia in humans (Khattak and Mir, 2014; McCarthy et al., 2014) and inflammatory disease in cows (Wang X. et al., 2015), thus broadening its disease connection from the previously known association with intellectual disability (Brunet and Sacher, 2014). Likewise, Alzheimer's disease was recently added (Hamilton et al., 2011; Chang et al., 2015) to the previously known association of TrappC6 (Trs33) with hypopigmentation (Brunet and Sacher, 2014). Another example is the addition of miscarriage (Wen et al., 2015) to the tissue specific disease X-linked SEDL associated with TrappC2 (Trs20) (Brunet and Sacher, 2014).
Recent findings also connect multiple TRAPP subunits to a single disease. For example, to date four TRAPP subunits are implicated in cancer: TrappC1 (Bet5), TrappC4 (Trs23) (Weng et al., 2014), TrappC9 (Zhang et al., 2015), and TrappC10 (Pongor et al., 2015). Likewise, two TRAPP subunits were recently implicated in brain-related diseases: TrappC6a (Trs33) (Hamilton et al., 2011; Chang et al., 2015), TrappC9 (Khattak and Mir, 2014; McCarthy et al., 2014).
Therefore, unless the disease divergence is due to moonlighting activities of individual TRAPP subunits unrelated to TRAPP, it is expected that association of individual subunits with more diseases will be uncovered and that some overlap between the different subunits, if not complete, will emerge.
While plenty of information has been accumulated about TRAPP since its identification in yeast and mammalian cells and its coming to the limelight as a Ypt/Rab GEF, there are still multiple uncertainties and unanswered questions. Structure is known only for the small TRAPP subunits, but not for the large ones. Complex architecture is known for TRAPP I and to some extent for TRAPP III, but not for TRAPP II. It is also not clear whether dimerization of TRAPP II occurs in vivo and has a function. It is not known whether the cellular functions of the three known TRAPP complexes are conserved from yeast to human cells. The Ypt/Rab GEF specificity of TRAPP complexes is still under debate even in yeast. The topology of TRAPP complexes on membranes is not known and evidence for a direct role for TRAPP complexes in membrane tethering is lacking. Finally, the apparent divergence of human disease associated to different TRAPP subunits is unclear. Future experiments will hopefully help fill in these multiple holes in the current picture.
All authors listed have made substantial, direct, and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Our research was supported by grant GM-45444 from NIH to NS.
Aridor, M., and Hannan, L. A. (2000). Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic 1, 836–851. doi: 10.1034/j.1600-0854.2000.011104.x
Barrowman, J., Bhandari, D., Reinisch, K., and Ferro-Novick, S. (2010). TRAPP complexes in membrane traffic: convergence through a common rab. Nat. Rev. Mol. Cell Biol. 11, 759–763. doi: 10.1038/nrm2999
Barrowman, J., Sacher, M., and Ferro-Novick, S. (2000). TRAPP stably associates with the golgi and is required for vesicle docking. EMBO J. 19, 862–869. doi: 10.1093/emboj/19.5.862
Bassik, M. C., Kampmann, M., Lebbink, R. J., Wang, S., Hein, M. Y., Poser, I., et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922. doi: 10.1016/j.cell.2013.01.030
Behrends, C., Sowa, M. E., Gygi, S. P., and Harper, J. W. (2010). Network organization of the human autophagy system. Nature 466, 68–76. doi: 10.1038/nature09204
Blomen, V. A., Májek, P., Jae, L. T., Bigenzahn, J. W., Nieuwenhuis, J., Staring, J., et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. doi: 10.1126/science.aac7557
Bröcker, C., Engelbrecht-Vandré, S., and Ungermann, C. (2010). Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20, R943–R952. doi: 10.1016/j.cub.2010.09.015
Brunet, S., Noueihed, B., Shahrzad, N., Saint-Dic, D., Hasaj, B., Guan, T. L., et al. (2012). The sms domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in saccharomyces cerevisiae. Cell. Logist. 2, 28–42. doi: 10.4161/cl.19414
Brunet, S., and Sacher, M. (2014). In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic 15, 803–818. doi: 10.1111/tra.12183
Brunet, S., Shahrzad, N., Saint-Dic, D., Dutczak, H., and Sacher, M. (2013). A Trs20 mutation that mimics an sedt-causing mutation blocks selective and non-selective autophagy: a model for TRAPP III organization. Traffic 14, 1091–1104. doi: 10.1111/tra.12095
Cai, H., Yu, S., Menon, S., Cai, Y., Lazarova, D., Fu, C., et al. (2007). TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature 445, 941–944. doi: 10.1038/nature05527
Cai, H., Zhang, Y., Pypaert, M., Walker, L., and Ferro-Novick, S. (2005). Mutants in Trs120 disrupt traffic from the early endosome to the late golgi. J. Cell Biol. 171, 823–833. doi: 10.1083/jcb.200505145
Cai, Y., Chin, H. F., Lazarova, D., Menon, S., Fu, C., Cai, H., et al. (2008). The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133, 1202–1213. doi: 10.1016/j.cell.2008.04.049
Chang, J. Y., Lee, M. H., Lin, S. R., Yang, L. Y., Sun, H. S., Sze, C. I., et al. (2015). Trafficking protein particle complex 6a delta (TRAPPC6ADELTA) is an extracellular plaque-forming protein in the brain. Oncotarget 6, 3578–3589. doi: 10.18632/oncotarget.2876
Chen, S., Cai, H., Park, S. K., Menon, S., Jackson, C. L., and Ferro-Novick, S. (2011). Trs65p, a subunit of the Ypt1p gef TRAPPII, interacts with the Arf1p exchange factor Gea2p to facilitate copi-mediated vesicle traffic. Mol. Biol. Cell 22, 3634–3644. doi: 10.1091/mbc.E11-03-0197
Choi, C., Davey, M., Schluter, C., Pandher, P., Fang, Y., Foster, L. J., et al. (2011). Organization and assembly of the TRAPPII complex. Traffic 12, 715–725. doi: 10.1111/j.1600-0854.2011.01181.x
Feyder, S., De Craene, J. O., Bär, S., Bertazzi, D. L., and Friant, S. (2015). Membrane trafficking in the yeast saccharomyces cerevisiae model. Int. J. Mol. Sci. 16, 1509–1525. doi: 10.3390/ijms16011509
Hamilton, G., Harris, S. E., Davies, G., Liewald, D. C., Tenesa, A., Starr, J. M., et al. (2011). Alzheimer's Disease genes are associated with measures of cognitive ageing in the lothian birth cohorts of 1921 and 1936. Int. J. Alzheimer Dis. 2011:505984. doi: 10.4061/2011/505984
Hanson, P. I., Roth, R., Morisaki, H., Jahn, R., and Heuser, J. E. (1997). Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535. doi: 10.1016/S0092-8674(00)80512-7
Hart, T., Chandrashekhar, M., Aregger, M., Steinhart, Z., Brown, K. R., Macleod, G., et al. (2015). High-resolution crispr screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526. doi: 10.1016/j.cell.2015.11.015
Hickey, C. M., and Wickner, W. (2010). Hops initiates vacuole docking by tethering membranes before trans-snare complex assembly. Mol. Biol. Cell 21, 2297–2305. doi: 10.1091/mbc.E10-01-0044
Ishii, Y., Nakahara, T., Kataoka, M., Kusumoto-Matsuo, R., Mori, S., Takeuchi, T., et al. (2013). Identification of TRAPPC8 as a host factor required for human papillomavirus cell entry. PLoS ONE 8:E80297. doi: 10.1371/journal.pone.0080297
Jones, S., Newman, C., Liu, F., and Segev, N. (2000). The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell 11, 4403–4411. doi: 10.1091/mbc.11.12.4403
Jones, S., Richardson, C. J., Litt, R. J., and Segev, N. (1998). Identification of regulators for Ypt1 Gtpase nucleotide cycling. Mol. Biol. Cell 9, 2819–2837. doi: 10.1091/mbc.9.10.2819
Kakuta, S., Yamamoto, H., Negishi, L., Kondo-Kakuta, C., Hayashi, N., and Ohsumi, Y. (2012). Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 287, 44261–44269. doi: 10.1074/jbc.M112.411454
Khattak, N. A., and Mir, A. (2014). Computational analysis of TRAPPC9: candidate gene for autosomal recessive non-syndromic mental retardation. CNS Neurol. Disord. Drug Targets 13, 699–711. doi: 10.2174/18715273113129990105
Kim, J. J., Lipatova, Z., Majumdar, U., and Segev, N. (2016). Regulation of golgi cisternal progression by Ypt/Rab GTPases. Dev. Cell 36, 440–452. doi: 10.1016/j.devcel.2016.01.016
Kim, Y. G., Raunser, S., Munger, C., Wagner, J., Song, Y. L., Cygler, M., et al. (2006). the architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 127, 817–830. doi: 10.1016/j.cell.2006.09.029
Kümmel, D., Müller, J. J., Roske, Y., Misselwitz, R., Büssow, K., and Heinemann, U. (2005). The structure of the TRAPP subunit Tpc6 suggests a model for a TRAPP subcomplex. EMBO Rep. 6, 787–793. doi: 10.1038/sj.embor.7400463
Levine, T. P., Daniels, R. D., Wong, L. H., Gatta, A. T., Gerondopoulos, A., and Barr, F. A. (2013). Discovery of new longin and roadblock domains that form platforms for small GTPases in ragulator and TRAPP-II. Small GTPases 4, 62–69. doi: 10.4161/sgtp.24262
Liang, Y., Morozova, N., Tokarev, A. A., Mulholland, J. W., and Segev, N. (2007). The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol. Biol. Cell 18, 2533–2541. doi: 10.1091/mbc.E07-03-0221
Lipatova, Z., Belogortseva, N., Zhang, X. Q., Kim, J., Taussig, D., and Segev, N. (2012). Regulation of selective autophagy onset by a Ypt/Rab Gtpase module. Proc. Natl. Acad. Sci. U.S.A. 109, 6981–6986. doi: 10.1073/pnas.1121299109
Lipatova, Z., Hain, A. U., Nazarko, V. Y., and Segev, N. (2015a). Ypt/Rab GTPases: principles learned from yeast. Crit. Rev. Biochem. Mol. Biol. 50, 203–211. doi: 10.3109/10409238.2015.1014023
Lipatova, Z., Kim, J. J., and Segev, N. (2015b). Ypt1 and TRAPP interactions: optimization of multicolor bimolecular fluorescence complementation in yeast. Methods Mol. Biol. 1298, 107–116. doi: 10.1007/978-1-4939-2569-8_9
Lipatova, Z., and Segev, N. (2012). A Ypt/Rab GTPase module makes a PAS. Autophagy 8, 1271–1272. doi: 10.4161/auto.20872
Lipatova, Z., Shah, A. H., Kim, J. J., Mulholland, J. W., and Segev, N. (2013). Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol. Biol. Cell 24, 3133–3144. doi: 10.1091/mbc.E13-05-0269
Luo, G., Zhang, J., and Guo, W. (2014). The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. Mol. Biol. Cell 25, 3813–3822. doi: 10.1091/mbc.E14-04-0907
Lynch-Day, M. A., Bhandari, D., Menon, S., Huang, J., Cai, H., Bartholomew, C. R., et al. (2010). Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 7811–7816. doi: 10.1073/pnas.1000063107
McCarthy, S. E., Gillis, J., Kramer, M., Lihm, J., Yoon, S., Berstein, Y., et al. (2014). De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 19, 652–658. doi: 10.1038/mp.2014.29
Meiling-Wesse, K., Epple, U. D., Krick, R., Barth, H., Appelles, A., Voss, C., et al. (2005). Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280, 33669–33678. doi: 10.1074/jbc.M501701200
Milev, M. P., Hasaj, B., Saint-Dic, D., Snounou, S., Zhao, Q., and Sacher, M. (2015). TRAMM/TRAPPC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment. J. Cell Biol. 209, 221–234. doi: 10.1083/jcb.201501090
Montpetit, B., and Conibear, E. (2009). Identification of the novel TRAPP associated protein Tca17. Traffic 10, 713–723. doi: 10.1111/j.1600-0854.2009.00895.x
Morozova, N., Liang, Y., Tokarev, A. A., Chen, S. H., Cox, R., Andrejic, J., et al. (2006). TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8, 1263–1269. doi: 10.1038/ncb1489
Nazarko, T. Y., Huang, J., Nicaud, J. M., Klionsky, D. J., and Sibirny, A. A. (2005). Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in yarrowia lipolytica and saccharomyces cerevisiae. Autophagy 1, 37–45. doi: 10.4161/auto.1.1.1512
Pinar, M., Arst, H. N. Jr., Pantazopoulou, A., Tagua, V. G., De Los Ríos, V., Rodríguez-Salarichs, J., et al. (2015). TRAPPII regulates exocytic golgi exit by mediating nucleotide exchange on the Ypt31 ortholog raberab11. Proc. Natl. Acad. Sci. U.S.A. 112, 4346–4351. doi: 10.1073/pnas.1419168112
Pongor, L., Kormos, M., Hatzis, C., Pusztai, L., Szabó, A., and Gyorffy, B. (2015). A genome-wide approach to link genotype to clinical outcome by utilizing next generation sequencing and gene chip data of 6697 breast cancer patients. Genome Med. 7:104. doi: 10.1186/s13073-015-0228-1
Qi, X., Kaneda, M., Chen, J., Geitmann, A., and Zheng, H. (2011). A specific role for arabidopsis TRAPPII in post-golgi trafficking that is crucial for cytokinesis and cell polarity. Plant J. 68, 234–248. doi: 10.1111/j.1365-313X.2011.04681.x
Robinett, C. C., Giansanti, M. G., Gatti, M., and Fuller, M. T. (2009). TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of drosophila. J. Cell Sci. 122, 4526–4534. doi: 10.1242/jcs.054536
Rybak, K., Steiner, A., Synek, L., Klaeger, S., Kulich, I., Facher, E., et al. (2014). Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev. Cell 29, 607–620. doi: 10.1016/j.devcel.2014.04.029
Sacher, M., Barrowman, J., Schieltz, D., Yates, J. R. III, and Ferro-Novick, S. (2000). Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 79, 71–80. doi: 10.1078/S0171-9335(04)70009-6
Sacher, M., Barrowman, J., Wang, W., Horecka, J., Zhang, Y., Pypaert, M., et al. (2001). TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell 7, 433–442. doi: 10.1016/S1097-2765(01)00190-3
Sacher, M., Jiang, Y., Barrowman, J., Scarpa, A., Burston, J., Zhang, L., et al. (1998). Trapp, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17, 2494–2503. doi: 10.1093/emboj/17.9.2494
Sacher, M., Kim, Y. G., Lavie, A., Oh, B. H., and Segev, N. (2008). The TRAPP complex: insights into its architecture and function. Traffic 9, 2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x
Schou, K. B., Morthorst, S. K., Christensen, S. T., and Pedersen, L. B. (2014). Identification of conserved, centrosome-targeting ash domains in TRAPPII complex subunits and TRAPPC8. Cilia 3:6. doi: 10.1186/2046-2530-3-6
Sclafani, A., Chen, S., Rivera-Molina, F., Reinisch, K., Novick, P., and Ferro-Novick, S. (2010). Establishing a role for the GTPase Ypt1p at the late golgi. Traffic 11, 520–532. doi: 10.1111/j.1600-0854.2010.01031.x
Scrivens, P. J., Noueihed, B., Shahrzad, N., Hul, S., Brunet, S., and Sacher, M. (2011). C4orf41 and Ttc-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell 22, 2083–2093. doi: 10.1091/mbc.E10-11-0873
Scrivens, P. J., Shahrzad, N., Moores, A., Morin, A., Brunet, S., and Sacher, M. (2009). TRAPPC2L is a novel, highly conserved TRAPP-interacting protein. Traffic 10, 724–736. doi: 10.1111/j.1600-0854.2009.00906.x
Segev, N., and Botstein, D. (1987). The Ras-like yeast Ypt1 gene is itself essential for growth, sporulation, and starvation response. Mol. Cell. Biol. 7, 2367–2377. doi: 10.1128/MCB.7.7.2367
Shirahama-Noda, K., Kira, S., Yoshimori, T., and Noda, T. (2013). TRAPPIII is responsible for vesicular transport from early endosomes to golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 126, 4963–4973. doi: 10.1242/jcs.131318
Tan, D., Cai, Y., Wang, J., Zhang, J., Menon, S., Chou, H. T., et al. (2013). The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 110, 19432–19437. doi: 10.1073/pnas.1316356110
Taussig, D., Lipatova, Z., Kim, J. J., Zhang, X., and Segev, N. (2013). Trs20 is required for TRAPP II assembly. Traffic 14, 678–690. doi: 10.1111/tra.12065
Taussig, D., Lipatova, Z., and Segev, N. (2014). Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic 15, 327–337. doi: 10.1111/tra.12145
Thellmann, M., Rybak, K., Thiele, K., Wanner, G., and Assaad, F. F. (2010). Tethering factors required for cytokinesis in arabidopsis. Plant Physiol. 154, 720–732. doi: 10.1104/pp.110.154286
Tokarev, A. A., Taussig, D., Sundaram, G., Lipatova, Z., Liang, Y., Mulholland, J. W., et al. (2009). TRAPP II complex assembly requires Trs33 or Trs65. Traffic 10, 1831–1844. doi: 10.1111/j.1600-0854.2009.00988.x
Venditti, R., Scanu, T., Santoro, M., Di Tullio, G., Spaar, A., Gaibisso, R., et al. (2012). Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science 337, 1668–1672. doi: 10.1126/science.1224947
Wang, J., Menon, S., Yamasaki, A., Chou, H. T., Walz, T., Jiang, Y., et al. (2013). Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. U.S.A. 110, 9800–9805. doi: 10.1073/pnas.1302337110
Wang, T., Birsoy, K., Hughes, N. W., Krupczak, K. M., Post, Y., Wei, J. J., et al. (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. doi: 10.1126/science.aac7041
Wang, W., and Ferro-Novick, S. (2002). A Ypt32p exchange factor is a putative effector of Ypt1p. Mol. Biol. Cell 13, 3336–3343. doi: 10.1091/mbc.01-12-0577
Wang, W., Sacher, M., and Ferro-Novick, S. (2000). TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151, 289–296. doi: 10.1083/jcb.151.2.289
Wang, X., Ma, P., Liu, J., Zhang, Q., Zhang, Y., Ding, X., et al. (2015). Genome-wide association study in chinese holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 16:111. doi: 10.1186/s12863-015-0263-3
Weidberg, H., Shvets, E., and Elazar, Z. (2011). Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156. doi: 10.1146/annurev-biochem-052709-094552
Wen, J., Hanna, C. W., Martell, S., Leung, P. C., Lewis, S. M., Robinson, W. P., et al. (2015). Functional consequences of copy number variants in miscarriage. Mol. Cytogenet. 8:6. doi: 10.1186/s13039-015-0109-8
Wendler, F., Gillingham, A. K., Sinka, R., Rosa-Ferreira, C., Gordon, D. E., Franch-Marro, X., et al. (2010). A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 29, 304–314. doi: 10.1038/emboj.2009.350
Weng, Y. R., Kong, X., Yu, Y. N., Wang, Y. C., Hong, J., Zhao, S. L., et al. (2014). The role of Erk2 in colorectal carcinogenesis is partly regulated by TRAPPC4. Mol. Carcinog. 53(Suppl. 1), E72–E84. doi: 10.1002/mc.22031
Westlake, C. J., Baye, L. M., Nachury, M. V., Wright, K. J., Ervin, K. E., Phu, L., et al. (2011). Primary cilia membrane assembly is initiated by rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. U.S.A. 108, 2759–2764. doi: 10.1073/pnas.1018823108
Wetterskog, D., Shiu, K. K., Chong, I., Meijer, T., Mackay, A., Lambros, M., et al. (2014). Identification of novel determinants of resistance to lapatinib in Erbb2-amplified cancers. Oncogene 33, 966–976. doi: 10.1038/onc.2013.41
Yamasaki, A., Menon, S., Yu, S., Barrowman, J., Meerloo, T., Oorschot, V., et al. (2009). Mtrs130 is a component of a mammalian TRAPPII complex, A Rab1 gef that binds to copi-coated vesicles. Mol. Biol. Cell 20, 4205–4215. doi: 10.1091/mbc.E09-05-0387
Yip, C. K., Berscheminski, J., and Walz, T. (2010). Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat. Struct. Mol. Biol. 17, 1298–1304. doi: 10.1038/nsmb.1914
Yu, I. M., and Hughson, F. M. (2010). Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156. doi: 10.1146/annurev.cellbio.042308.113327
Zhang, Y., Liu, S., Wang, H., Yang, W., Li, F., Yang, F., et al. (2015). Elevated NIBP/TRAPPC9 mediates tumorigenesis of cancer cells through NFκB signaling. Oncotarget 6, 6160–6178. doi: 10.18632/oncotarget.3349
Zong, M., Satoh, A., Yu, M. K., Siu, K. Y., Ng, W. Y., Chan, H. C., et al. (2012). TRAPPC9 mediates the interaction between P150 and COPII vesicles at the target membrane. PLoS ONE 7:E29995. doi: 10.1371/journal.pone.0029995
Zoppino, F. C., Militello, R. D., Slavin, I., Alvarez, C., and Colombo, M. I. (2010). Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x
Keywords: Ypt, Rab, GTPase, GEF, secretion, autophagy, tether
Citation: Kim JJ, Lipatova Z and Segev N (2016) TRAPP Complexes in Secretion and Autophagy. Front. Cell Dev. Biol. 4:20. doi: 10.3389/fcell.2016.00020
Received: 08 December 2015; Accepted: 04 March 2016;
Published: 30 March 2016.
Edited by:
Anne Spang, University of Basel, SwitzerlandReviewed by:
Vladimir Lupashin, University of Arkansas for Medical Sciences, USACopyright © 2016 Kim, Lipatova and Segev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nava Segev, bmF2YUB1aWMuZWR1
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.