
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 07 March 2016
Sec. Cellular Biochemistry
Volume 4 - 2016 | https://doi.org/10.3389/fcell.2016.00014
This article is part of the Research Topic Mobile Genetic Elements in Cellular Differentiation, Genome Stability, and Cancer View all 12 articles
Long INterspersed Element-1 (LINE-1 or L1) retrotransposons form the only autonomously active family of transposable elements in humans. They are expressed and mobile in the germline, in embryonic stem cells and in the early embryo, but are silenced in most somatic tissues. Consistently, they play an important role in individual genome variations through insertional mutagenesis and sequence transduction, which occasionally lead to novel genetic diseases. In addition, they are reactivated in nearly half of the human epithelial cancers, contributing to tumor genome dynamics. The L1 element codes for two proteins, ORF1p and ORF2p, which are essential for its mobility. ORF1p is an RNA-binding protein with nucleic acid chaperone activity and ORF2p possesses endonuclease and reverse transcriptase activities. These proteins and the L1 RNA assemble into a ribonucleoprotein particle (L1 RNP), considered as the core of the retrotransposition machinery. The L1 RNP mediates the synthesis of new L1 copies upon cleavage of the target DNA and reverse transcription of the L1 RNA at the target site. The L1 element takes benefit of cellular host factors to complete its life cycle, however several cellular pathways also limit the cellular accumulation of L1 RNPs and their deleterious activities. Here, we review the known cellular host factors and pathways that regulate positively or negatively L1 retrotransposition at post-transcriptional level, in particular by interacting with the L1 machinery or L1 replication intermediates; and how they contribute to control L1 activity in somatic cells.
The Long INterspersed Element-1 (LINE-1 or L1) retrotransposon forms 17% of our genome (Lander et al., 2001). Most L1 copies present in the reference human genome are defective but ~100 copies could be retrotransposition-competent (Brouha et al., 2003). In addition, many polymorphic L1 elements, not included in the reference genome, also have the potential to mobilize (Beck et al., 2010; Ewing, 2015; Mir et al., 2015).
L1 elements can retrotranspose in the germline, in embryonic stem cells and in the early embryo (Kazazian et al., 1988; Garcia-Perez et al., 2007; van den Hurk et al., 2007). However, L1 retrotransposons are repressed in most tested normal somatic cells except in the brain (Coufal et al., 2009; Baillie et al., 2011; Evrony et al., 2012; Richardson et al., 2014b; Upton et al., 2015). L1 mobilization impacts human genome evolution through insertional mutagenesis and sequence transduction, which occasionally results in inherited genetic diseases (Hancks and Kazazian, 2012). Somatic retrotransposition in the brain could also contribute to the etiology of some mental disorders or disabilities, such as Rett Syndrome or Ataxia Telangiectasia, characterized by increased levels of L1 mobilization (Muotri et al., 2010; Coufal et al., 2011). Moreover, somatic L1 mobilization participates to the dynamics of many tumor genomes and can lead to driver mutations (Miki et al., 1992; Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Helman et al., 2014; Tubio et al., 2014; Doucet-O'Hare et al., 2015; Ewing et al., 2015; Rodić et al., 2015). Besides its impact as an insertional mutagen, L1 also triggers other forms of genomic alterations such as DNA double-strand breaks or chromosomal translocations, and these activities could participate to normal aging or tumorigenesis (Wallace et al., 2008; Lin et al., 2009; Belancio et al., 2010). Finally, the L1 machinery also drives the retrotransposition of Short INterspersed Elements (SINEs) and the formation of processed pseudogenes (Esnault et al., 2000; Dewannieux et al., 2003).
L1 elements and their host have co-evolved: L1s use the cellular machinery for their own replication, while the host cell has evolved multiple defense mechanisms limiting L1 deleterious effects. Silencing L1 expression, through CpG DNA methylation and histone modifications is a major repressive mechanism, which prevents the accumulation of mutagenic events (Bourc'His and Bestor, 2004; Castro-Diaz et al., 2014; Jacobs et al., 2014). Here we review post-transcriptional cellular pathways, which regulate positively or negatively L1 retrotransposition in somatic cells, in particular by interacting with the L1 machinery or L1 replicative intermediates.
An active L1 retrotransposon comprises a 5′ untranslated region (UTR), two open reading frames (ORF1 and ORF2) separated by a short inter-ORF spacer and a 3′ UTR (Figure 1). An antisense ORF0 of unknown function has also been recently described in the 5′ UTR (Denli et al., 2015). As a consequence of the reverse transcription and integration mechanism, L1 sequence ends with a poly(dA) stretch and is flanked by target site duplications (TSD) of variable size. The 5′ UTR contains RNA polymerase II sense and antisense promoters (Swergold, 1990; Speek, 2001; Nigumann et al., 2002). The translation of the bicistronic L1 mRNA by an unconventional mechanism produces two proteins, named ORF1p and ORF2p (Alisch et al., 2006; Dmitriev et al., 2007). ORF1p is a 40 kDa RNA-binding protein, forming trimers and with nucleic acid chaperone activity (Martin, 1991; Holmes et al., 1992; Martin and Bushman, 2001; Martin et al., 2003; Khazina et al., 2011). ORF2p is a ~150 kDa protein with endonuclease (EN) and reverse transcriptase (RT) activities, which are critical for L1 retrotransposition (Mathias et al., 1991; Feng et al., 1996; Moran et al., 1996). ORF2p also contains a C-terminal cysteine-rich region, potentially contributing to its RNA binding capability (Piskareva et al., 2013). ORF1p and ORF2p bind the L1 mRNA to form a ribonucleoprotein particle (RNP), considered as the core of the L1 replicative complex (Hohjoh and Singer, 1996; Kolosha and Martin, 1997; Kulpa and Moran, 2005, 2006; Doucet et al., 2010; Goodier et al., 2010). This assembly occurs preferentially in cis (Esnault et al., 2000; Wei et al., 2001; Kulpa and Moran, 2006), through binding of ORF2p to the L1 RNA poly(A) sequence (Doucet et al., 2015). L1 RNPs accumulate in cytoplasmic foci, which colocalize with stress granules (Goodier et al., 2007, 2010; Doucet et al., 2010). The functional importance of these cytoplasmic complexes remains to be elucidated. Although cell division seems to promote retrotransposition, it is not absolutely required (Kubo et al., 2006; Shi et al., 2007; Xie et al., 2013). Thus, access of L1 RNPs to chromatin can occur independently of mitotic nuclear envelope breakdown through an unknown nuclear import mechanism.
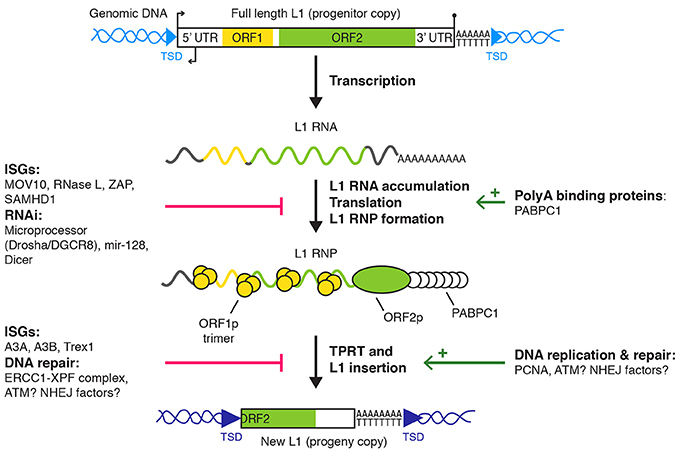
Figure 1. L1 retrotransposition and cellular regulators. L1 replication starts with L1 transcription into a full length bicistronic L1 mRNA, its translation into ORF1p and ORF2p, and the assembly of an L1 RNP. For the sake of simplicity, the recently described antisense ORF0 in the 5′ UTR is not depicted (Denli et al., 2015). The L1 RNP accumulates in stress granules and at least a fraction of it is imported to the nucleus (not shown) where target-primed reverse transcription (TPRT) occurs at the target DNA site. Finally, L1 insertion is resolved by an unknown mechanism (see main text for details). Only L1 regulators with a defined target/mechanism are depicted. Small broken arrows, L1 sense and antisense promoter activities; lollipop, L1 polyadenylation signal; light and dark blue arrowheads, target sites of L1 progenitor and progeny copies, respectively; red bars, negative regulation; green arrows, positive regulation. TSD, target site duplication; UTR, untranslated region; RNP, ribonucleoprotein particle; ISG, interferon-responsive genes; RNAi, RNA interference.
New L1 copies are directly synthesized and inserted in the genome by a process called TPRT (Luan et al., 1993; Feng et al., 1996; Cost et al., 2002; Christensen et al., 2006). During TPRT, ORF2p binds and nicks a consensus sequence of the form 5′-TTTT/A-3′ in the genomic DNA (Feng et al., 1996). This cleavage, potentially followed by additional processing steps, exposes a single-stranded T-rich DNA stretch able to partially or completely anneal to the L1 RNA poly(A) tail and to prime ORF2p-mediated reverse transcription (Kulpa and Moran, 2006; Monot et al., 2013; Viollet et al., 2014). A possible second nick, generally few nucleotides downstream of the first one, allows priming and synthesis of the second DNA strand. Finally, the L1 DNA ends are filled in and sealed, creating TSD (Luan et al., 1993; Feng et al., 1996; Cost et al., 2002). The molecular actors involved in these late stages are unknown. This process is frequently abortive, resulting in 5′ truncated L1 copies.
L1 activity is regulated at multiple stages of the L1 retrotransposition cycle (Figure 1). We focus here on post-transcriptional mechanisms and their molecular effectors acting in human or mammalian somatic cells and interacting with components of the L1 RNP or with L1 replication intermediates. L1 regulation in the germline, notably by Piwi-interacting RNA (piRNA), has been reviewed elsewhere (Zamudio and Bourc'his, 2010; Crichton et al., 2014) and is not detailed in the present article.
Several recent studies have identified cellular partners of L1 RNPs through tagging of ORF1p, ORF2p or L1 RNA, followed by affinity chromatography and mass-spectrometry (Goodier et al., 2013; Peddigari et al., 2013; Taylor et al., 2013; Moldovan and Moran, 2015). These experimental efforts differ by the cell line, the L1 clone, the tagged component in the complex and the chromatography method used, but eventually lead to a number of common host factors (Figure 2). It should be underlined that only a fraction of the hits has been validated by co-immunoprecipitation, and only a single study used quantitative mass-spectrometry to measure the specific enrichment of the detected proteins upon elution (Taylor et al., 2013). A first step toward functional characterization generally involves retrotransposition assays in cultured cells upon depletion or overexpression of the tested factor. The outcome of these genetic assays allows a first classification into positive or negative regulators. However, many binding partners only modestly impact the levels of L1 retrotransposition in these assays, or have pleiotropic effects preventing unambiguous interpretation. With few exceptions, the majority of the tested factors are RNA-binding proteins, which copurify with ORF1p through an indirect RNA bridge, colocalize with L1 RNPs in stress granules, and inhibit L1 retrotransposition.
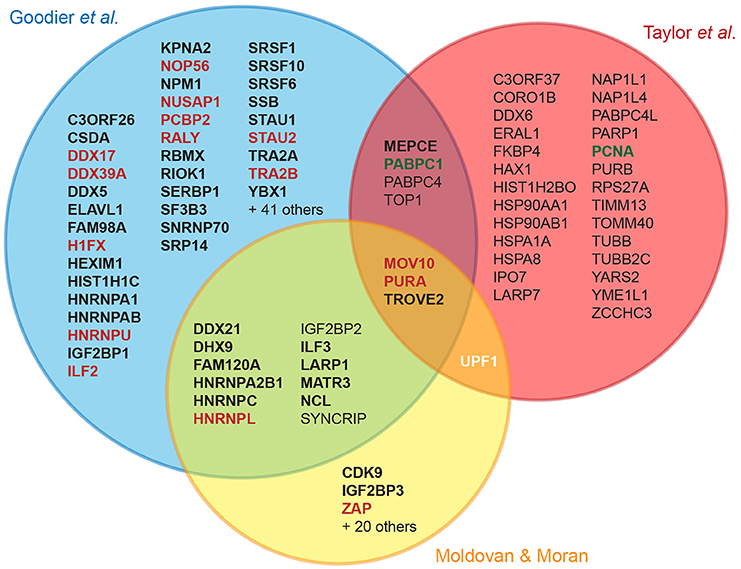
Figure 2. Cellular L1 interactors discovered in recent proteomic studies. The Venn diagram displays the overlap between three major proteomic studies designed to identify L1 cellular partners (Goodier et al., 2013; Taylor et al., 2013; Moldovan and Moran, 2015). For the sake of simplicity, the overlap with a more limited fourth study is not shown, but includes NCL and HNRNPL (Peddigari et al., 2013). For hits found in a single study, only those confirmed by coimmunoprecipitation (coIP) or by quantitative mass-spectrometry are depicted. Bold, confirmed by coIP; red and green, L1 negative and positive regulators, respectively; white, potential dual role: UPF1 knockdown decreases overall L1 retrotransposition but increases L1 RNA levels, suggesting that it could act at several stages with opposing effects (Taylor et al., 2013). Upf1 overexpression is not impacting retrotransposition (Moldovan and Moran, 2015).
Due to the scarcity of L1 endogenous complexes in cells, all proteomic studies rely on the overexpression of engineered L1 constructs. It is conceivable that: (i) some of the discovered partners become associated with L1 components as a result of L1 overexpression beyond physiological levels. (ii) L1 RNP stoichiometry is altered; (iii) the retrotransposition reporter cassette, which contains an intron, modifies L1 RNA cellular processing, and thus its binding partners.
Poly(A) binding proteins (PABPs) bind mRNA poly(A) tails and are involved in mRNA stability and translation initiation (Goss and Kleiman, 2013). Short hairpin RNA (shRNA)-mediated knockdown of PABPC1, reduces L1 retrotransposition with minimal effects on L1 RNA and proteins accumulation, or poly(A) tail length (Dai et al., 2012). This effect is associated with reduced L1 RNP levels and reduced nuclear accumulation of this complex, suggesting a possible—direct or indirect—role of PABPC1 in the assembly or the subcellular trafficking of the L1 RNP. Consistently, PABPC1 associates with the L1 RNP in an RNA-dependent manner, they colocalize in stress granules (Goodier et al., 2013; Taylor et al., 2013), and moderate PABPC1 overexpression stimulates retrotransposition (Dai et al., 2012). Other PABPs have been found to associate with the L1 RNA (PABPN1, PABPC4) but addressing their specific role in L1 retrotransposition has been hampered by pleiotropic effects, or not yet tested (Dai et al., 2012; Goodier et al., 2013; Taylor et al., 2013).
PCNA is a DNA sliding clamp acting as a processivity factor for many DNA polymerases during DNA replication or DNA damage repair (Moldovan et al., 2007). ORF2p binds PCNA through a PCNA-interacting protein (PIP) box, located between the EN and RT domains of ORF2p (Taylor et al., 2013). Mutations in ORF2p PIP box disrupt PCNA-ORF2p interaction and inhibit L1 retrotransposition. Interestingly, ORF2p mutations abrogating its EN or RT activity also disrupt PCNA-ORF2p interaction, suggesting that PCNA binding to ORF2p occurs downstream or concomitantly with TPRT.
ORF1p contains several (S/T)-P putative phosphorylation sites for proline-directed protein kinases (PDPKs), such as mitogen-activated protein kinases and cyclin-dependent kinases. Mutations of S18, S27, T203, and T213, which are potential PDPK targets, decrease L1 retrotransposition; and these residues were found phosphorylated by mass-spectrometry in human cells (Cook et al., 2015). Interestingly, several protein kinases associate with the L1 RNP (Goodier et al., 2013; Taylor et al., 2013; Moldovan and Moran, 2015), however it remains to be demonstrated if one or several of them might directly target ORF1p. Interestingly, S18/S27 sites in ORF1p are required for binding by Pin1 prolyl isomerase (Cook et al., 2015), suggesting a scenario in which binding of Pin1 promotes ORF1p conformational change, which could affect its stability, activity or localization, or its subsequent ability to be dephosphorylated (Yeh et al., 2004; Liou et al., 2011).
L1 RNA serves both as an mRNA to produce the L1 machinery and as a template for reverse transcription. Multiple RNA interference (RNAi) pathways act in somatic or embryonic cells to prevent the accumulation of L1 RNA, and eventually retrotransposition.
First, the Microprocessor complex (Drosha/DGCR8), a major nuclear complex implicated in microRNA (miRNA) biosynthesis through pri-miRNA processing, is also able to bind L1 RNA in vivo, to reduce its abundance and to limit L1 retrotransposition. In addition, it can cleave various L1 RNA fragments derived from the L1 5′ UTR region in vitro, indicating that L1 RNA can be a direct Microprocessor substrate (Heras et al., 2013, 2014). Moreover, the miRNA pathway could also act downstream of Microprocessor to inhibit retrotransposition. Indeed, miR-128 in complex with the Argonaute (Ago) protein binds the L1 RNA in the ORF2 region leading to L1 transcript degradation (Hamdorf et al., 2015).
Second, the combined expression of sense and antisense L1 transcripts driven by L1 5′ UTR promoters reduces L1 RNA stability and L1 retrotransposition (Yang and Kazazian, 2006). This process is associated with the synthesis of rasiRNA (repeat-associated small interfering RNA) consistent with a possible processing of L1 RNA duplexes, and is modestly inhibited by Dicer knockdown, suggesting an additional layer of L1 repression mediated by siRNA mechanisms. In agreement with a role of RNAi pathways in somatic L1 regulation, L1 RNPs tend to accumulate in stress granules where they colocalize with several RNAi factors and often interact with them (Goodier et al., 2007, 2013).
The cellular innate immune response is one of the first lines of defense against a broad range of viral infections. It involves cellular factors with antiviral activities, among which the interferon (IFN) response pathway plays a central role (MacMicking, 2012; Ivashkiv and Donlin, 2014). This pathway leads to the activation of IFN-stimulated genes (ISG) acting as effectors and reinforcing IFN-signaling itself. A significant proportion of ISG are viral restriction factors (MacMicking, 2012), which also appear to counteract L1 retrotransposition (Goodier et al., 2015), and are described below.
Upon overexpression, several members of the APOBEC3 (A3) cytidine deaminase family inhibit L1 retrotransposition (A3A, A3B, A3C and A3F) (Bogerd et al., 2006; Chen et al., 2006; Muckenfuss et al., 2006; Stenglein and Harris, 2006; Kinomoto et al., 2007; Niewiadomska et al., 2007). A3A is a nuclear protein predominantly expressed in peripheral blood mononuclear cells (PBMCs) and is induced by IFN-β (Chen et al., 2006; Muckenfuss et al., 2006; Stenglein et al., 2010). A3A-mediated L1 inhibition depends on A3A deaminase activity and on the subsequent processing of the deaminated DNA by uracil DNA glycosylase (UNG) and apurinic/apyrimidinic endonuclease (APE) (Richardson et al., 2014a). A3B is also a nuclear protein. It is endogenously expressed in embryonic stem cells, in induced-pluripotent stem cells and in a number of cancer cell lines. Its depletion stimulates L1 retrotransposition (Wissing et al., 2011; Marchetto et al., 2013); however, catalytically dead A3B mutants still inhibit L1 retrotransposition (Wissing et al., 2011). Thus, the mechanism by which A3B represses L1 mobilization remains unknown. Similarly, reducing the expression of A3C moderately increases retrotransposition in cancer cell lines that express detectable levels of endogenous A3C (Muckenfuss et al., 2006). As for A3B, A3C- and A3F-mediated L1 repression is deaminase-independent (Muckenfuss et al., 2006; Stenglein and Harris, 2006; Kinomoto et al., 2007; Horn et al., 2014). A3C might interfere with L1 reverse transcription or the activity of ORF2p in the L1 RNP (Horn et al., 2014).
Several other ISG products, such as MOV10, ZAP or RNase L, limit L1 replication by limiting L1 RNA accumulation. The RNA helicase MOV10 robustly copurifies with the L1 complex, colocalizes with L1 RNPs in stress granules, reduces L1 RNA half-life, and ultimately strongly inhibits retrotransposition (Arjan-Odedra et al., 2012; Goodier et al., 2012, 2013; Li et al., 2013; Taylor et al., 2013; Moldovan and Moran, 2015). Similarly, the zinc-finger antiviral protein (ZAP) associates with the L1 RNP and accumulates with it in stress granules (Goodier et al., 2015; Moldovan and Moran, 2015). Its overexpression reduces full-length L1 RNA levels, and L1 retrotransposition levels. ZAP zinc finger domain is necessary and sufficient for its anti-L1 activity. Inversely, knocking down endogenous ZAP increases L1 retrotransposition. The ribonuclease L (RNase L) degrades L1 RNA and inhibits retrotransposition although no association or colocalization was detected with the L1 RNP (Zhang et al., 2014). Other ISGs with known viral restriction activities (e.g., BST2, ISG20, MAVS, and MX2) are also strong inhibitors of L1 retrotransposition (Goodier et al., 2015), but their mechanism of action has not yet been explored.
Finally, SAMHD1 and TREX1 are ISGs involved in a negative feedback loop, acting as repressors of the interferon response itself. Loss-of-function mutations in these genes lead to the Aicardi-Goutières syndrome, an autoimmune disease. Both factors inhibit L1 retrotransposition (Stetson et al., 2008; Zhao et al., 2013). Trex1 (Three-prime-repair exonuclease 1) is an abundant 3′-5′ DNA exonuclease and its overexpression inhibits engineered L1 retrotransposition in cultured cells (Stetson et al., 2008). Trex1-deficient cells accumulate ssDNA fragments derived from various retroelements including L1, suggesting that Trex1 can metabolize reverse transcribed L1 cDNA (Stetson et al., 2008). SAMHD1 (SAM Domain And HD Domain 1) impairs lentivirus replication in non-dividing cells by depleting the intracellular pool of dNTPs and thereby inhibiting reverse transcription (Lahouassa et al., 2012). In contrast, SAMHD1 inhibits L1 retrotransposition in dividing cells, through a dNTPase-independent mechanism, which might directly affect ORF2p levels, and thus inhibit L1 reverse transcription (Zhao et al., 2013).
EN-mediated cleavage of the target DNA or other TPRT intermediates could lead to DNA double-strand break (DSB) or DNA lesion signaling, and activation of subsequent DNA repair pathways. Conversely, these cellular processes could also participate in the resolution of L1 integration, through L1 second strand DNA synthesis or DNA ligation.
The role of DSB signaling and non-homologous end-joining (NHEJ) pathways remains controversial. Ataxia-telangiectasia mutated (ATM) protein, a kinase activated upon DSB, was initially proposed to be required for L1 retrotransposition and L1-induced DSBs (Gasior et al., 2006; Wallace et al., 2013). However, independent studies using ATM-deficient mice or human cell models rather suggest that ATM is a repressor of retrotransposition (Coufal et al., 2011). Similarly, knocking out NHEJ genes (e.g., Ku70/80, DNA Ligase IV or Artemis) decreases L1 retrotransposition in chicken cells (Suzuki et al., 2009). However, loss-of-function of DNA-PKcs or DNA Ligase IV in mammalian cells does not impair L1 retrotransposition (Coufal et al., 2011), indicating that NHEJ is not absolutely required for L1 retrotransposition. An interesting possibility could be that DSB signaling and repair pathways compete with the L1 machinery or other cellular factors for the resolution of L1 insertion during—or after—cDNA synthesis, leading to 5′ truncated insertions (Zingler et al., 2005; Suzuki et al., 2009; Coufal et al., 2011).
Other DNA repair pathways can also antagonize L1 replication. The ERCC1-XPF complex, which plays a role in nucleotide excision, base excision and interstrand crosslink repair pathways is a potent inhibitor of L1 retrotransposition (Gasior et al., 2008). ERCC1-XPF is an endonuclease able to specifically cleave DNA at junctions between single-stranded and double-stranded regions, a predicted structure produced by the TPRT process. Thus, it has been hypothesized that ERCC1-XPF might cut off L1 cDNA at the target site during reverse transcription.
• How is unspliced L1 RNA exported to the cytosol and the L1 RNP imported back to the nucleus?
• How many distinct L1 RNP forms exist in the cell and throughout the L1 replication cycle?
• Do L1 components have a life outside of the L1 RNP and retrotransposition?
• How is L1 RNP assembly regulated?
• Does L1 component accumulation in stress granules reflect a host defense mechanism or an intermediate step during retrotransposition?
• Which restriction factors are the dominant ones and do they cooperate?
JGP drafted the manuscript. GC revised the manuscript.
GC is funded by the Fondation ARC pour la recherche sur le cancer, the European Research Council (ERC-2010-StG 243312, RETROGENOMICS), the French Government (National Research Agency, ANR) through the “Investments for the Future” (LABEX SIGNALIFE, ANR-11-LABX-0028-01), and the Fondation pour la Recherche Médicale (FRM DEP20131128533).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Aurelien J. Doucet for critical reading.
Alisch, R. S., Garcia-Perez, J. L., Muotri, A. R., Gage, F. H., and Moran, J. V. (2006). Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20, 210–224. doi: 10.1101/gad.1380406
Arjan-Odedra, S., Swanson, C. M., Sherer, N. M., Wolinsky, S. M., and Malim, M. H. (2012). Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 9:53. doi: 10.1186/1742-4690-9-53
Baillie, J. K., Barnett, M. W., Upton, K. R., Gerhardt, D. J., Richmond, T. A., De Sapio, F., et al. (2011). Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537. doi: 10.1038/nature10531
Beck, C. R., Collier, P., Macfarlane, C., Malig, M., Kidd, J. M., Eichler, E. E., et al. (2010). LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170. doi: 10.1016/j.cell.2010.05.021
Belancio, V. P., Roy-Engel, A. M., Pochampally, R. R., and Deininger, P. (2010). Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 38, 3909–3922. doi: 10.1093/nar/gkq132
Bogerd, H. P., Wiegand, H. L., Hulme, A. E., Garcia-Perez, J. L., O'shea, K. S., Moran, J. V., et al. (2006). Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785. doi: 10.1073/pnas.0603313103
Bourc'His, D., and Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. doi: 10.1038/nature02886
Brouha, B., Schustak, J., Badge, R. M., Lutz-Prigge, S., Farley, A. H., Moran, J. V., et al. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285. doi: 10.1073/pnas.0831042100
Castro-Diaz, N., Ecco, G., Coluccio, A., Kapopoulou, A., Yazdanpanah, B., Friedli, M., et al. (2014). Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 28, 1397–1409. doi: 10.1101/gad.241661.114
Chen, H., Lilley, C. E., Yu, Q., Lee, D. V., Chou, J., Narvaiza, I., et al. (2006). APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16, 480–485. doi: 10.1016/j.cub.2006.01.031
Christensen, S. M., Ye, J., and Eickbush, T. H. (2006). RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. U.S.A. 103, 17602–17607. doi: 10.1073/pnas.0605476103
Cook, P. R., Jones, C. E., and Furano, A. V. (2015). Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 112, 4298–4303. doi: 10.1073/pnas.1416869112
Cost, G. J., Feng, Q., Jacquier, A., and Boeke, J. D. (2002). Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910. doi: 10.1093/emboj/cdf592
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Marchetto, M. C., Muotri, A. R., Mu, Y., et al. (2011). Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 20382–20387. doi: 10.1073/pnas.1100273108
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., et al. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131. doi: 10.1038/nature08248
Crichton, J. H., Dunican, D. S., Maclennan, M., Meehan, R. R., and Adams, I. R. (2014). Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell. Mol. Life Sci. 71, 1581–1605. doi: 10.1007/s00018-013-1468-0
Dai, L., Taylor, M. S., O'Donnell, K. A., and Boeke, J. D. (2012). Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol. Cell. Biol. 32, 4323–4336. doi: 10.1128/MCB.06785-11
Denli, A. M., Narvaiza, I., Kerman, B. E., Pena, M., Benner, C., Marchetto, M. C., et al. (2015). Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–593. doi: 10.1016/j.cell.2015.09.025
Dewannieux, M., Esnault, C., and Heidmann, T. (2003). LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48. doi: 10.1038/ng1223
Dmitriev, S. E., Andreev, D. E., Terenin, I. M., Olovnikov, I. A., Prassolov, V. S., Merrick, W. C., et al. (2007). Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 27, 4685–4697. doi: 10.1128/MCB.02138-06
Doucet, A. J., Hulme, A. E., Sahinovic, E., Kulpa, D. A., Moldovan, J. B., Kopera, H. C., et al. (2010). Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6:e1001150. doi: 10.1371/journal.pgen.1001150
Doucet, A. J., Wilusz, J. E., Miyoshi, T., Liu, Y., and Moran, J. V. (2015). A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell 60, 728–741. doi: 10.1016/j.molcel.2015.10.012
Doucet-O'Hare, T. T., Rodić, N., Sharma, R., Darbari, I., Abril, G., Choi, J. A., et al. (2015). LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 112, E4894–E4900. doi: 10.1073/pnas.1502474112
Esnault, C., Maestre, J., and Heidmann, T. (2000). Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. doi: 10.1038/74184
Evrony, G. D., Cai, X., Lee, E., Hills, L. B., Elhosary, P. C., Lehmann, H. S., et al. (2012). Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496. doi: 10.1016/j.cell.2012.09.035
Ewing, A. D. (2015). Transposable element detection from whole genome sequence data. Mob. DNA 6, 24. doi: 10.1186/s13100-015-0055-3
Ewing, A. D., Gacita, A., Wood, L. D., Ma, F., Xing, D., Kim, M. S., et al. (2015). Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 25, 1536–1545. doi: 10.1101/gr.196238.115
Feng, Q., Moran, J. V., Kazazian, H. H., and Boeke, J. D. (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916. doi: 10.1016/S0092-8674(00)81997-2
Garcia-Perez, J. L., Doucet, A. J., Bucheton, A., Moran, J. V., and Gilbert, N. (2007). Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17, 602–611. doi: 10.1101/gr.5870107
Gasior, S. L., Roy-Engel, A. M., and Deininger, P. L. (2008). ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst). 7, 983–989. doi: 10.1016/j.dnarep.2008.02.006
Gasior, S. L., Wakeman, T. P., Xu, B., and Deininger, P. L. (2006). The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393. doi: 10.1016/j.jmb.2006.01.089
Goodier, J. L., Cheung, L. E., and Kazazian, H. H. (2012). MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8:e1002941. doi: 10.1371/journal.pgen.1002941
Goodier, J. L., Cheung, L. E., and Kazazian, H. H. (2013). Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 41, 7401–7419. doi: 10.1093/nar/gkt512
Goodier, J. L., Mandal, P. K., Zhang, L., and Kazazian, H. H. (2010). Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum. Mol. Genet. 19, 1712–1725. doi: 10.1093/hmg/ddq048
Goodier, J. L., Pereira, G. C., Cheung, L. E., Rose, R. J., and Kazazian, H. H. (2015). The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 11:e1005252. doi: 10.1371/journal.pgen.1005252
Goodier, J. L., Zhang, L., Vetter, M. R., and Kazazian, H. H. (2007). LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483. doi: 10.1128/MCB.00332-07
Goss, D. J., and Kleiman, F. E. (2013). Poly(A) binding proteins: are they all created equal? Wiley Interdiscip. Rev. RNA 4, 167–179. doi: 10.1002/wrna.1151
Hamdorf, M., Idica, A., Zisoulis, D. G., Gamelin, L., Martin, C., Sanders, K. J., et al. (2015). miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat. Struct. Mol. Biol. 22, 824–831. doi: 10.1038/nsmb.3090
Hancks, D. C., and Kazazian, H. H. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203. doi: 10.1016/j.gde.2012.02.006
Helman, E., Lawrence, M. S., Stewart, C., Sougnez, C., Getz, G., and Meyerson, M. (2014). Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 24, 1053–1063. doi: 10.1101/gr.163659.113
Heras, S. R., Macias, S., Cáceres, J. F., and Garcia-Perez, J. L. (2014). Control of mammalian retrotransposons by cellular RNA processing activities. Mob. Genet. Elements 4:e28439. doi: 10.4161/mge.28439
Heras, S. R., Macias, S., Plass, M., Fernandez, N., Cano, D., Eyras, E., et al. (2013). The Microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 20, 1173–1181. doi: 10.1038/nsmb.2658
Hohjoh, H., and Singer, M. F. (1996). Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639.
Holmes, S. E., Singer, M. F., and Swergold, G. D. (1992). Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267, 19765–19768.
Horn, A. V., Klawitter, S., Held, U., Berger, A., Vasudevan, A. A., Bock, A., et al. (2014). Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 42, 396–416. doi: 10.1093/nar/gkt898
Iskow, R. C., McCabe, M. T., Mills, R. E., Torene, S., Pittard, W. S., Neuwald, A. F., et al. (2010). Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261. doi: 10.1016/j.cell.2010.05.020
Ivashkiv, L. B., and Donlin, L. T. (2014). Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49. doi: 10.1038/nri3581
Jacobs, F. M., Greenberg, D., Nguyen, N., Haeussler, M., Ewing, A. D., Katzman, S., et al. (2014). An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245. doi: 10.1038/nature13760
Kazazian, H. H., Wong, C., Youssoufian, H., Scott, A. F., Phillips, D. G., and Antonarakis, S. E. (1988). Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166. doi: 10.1038/332164a0
Khazina, E., Truffault, V., Büttner, R., Schmidt, S., Coles, M., and Weichenrieder, O. (2011). Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 18, 1006–1014. doi: 10.1038/nsmb.2097
Kinomoto, M., Kanno, T., Shimura, M., Ishizaka, Y., Kojima, A., Kurata, T., et al. (2007). All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35, 2955–2964. doi: 10.1093/nar/gkm181
Kolosha, V. O., and Martin, S. L. (1997). In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 94, 10155–10160. doi: 10.1073/pnas.94.19.10155
Kubo, S., Seleme, M. C., Soifer, H. S., Perez, J. L., Moran, J. V., Kazazian, H. H., et al. (2006). L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 103, 8036–8041. doi: 10.1073/pnas.0601954103
Kulpa, D. A., and Moran, J. V. (2005). Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14, 3237–3248. doi: 10.1093/hmg/ddi354
Kulpa, D. A., and Moran, J. V. (2006). Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13, 655–660. doi: 10.1038/nsmb1107
Lahouassa, H., Daddacha, W., Hofmann, H., Ayinde, D., Logue, E. C., Dragin, L., et al. (2012). SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228. doi: 10.1038/ni.2236
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Lee, E., Iskow, R., Yang, L., Gokcumen, O., Haseley, P., Luquette, L. J., et al. (2012). Landscape of somatic retrotransposition in human cancers. Science 337, 967–971. doi: 10.1126/science.1222077
Li, X., Zhang, J., Jia, R., Cheng, V., Xu, X., Qiao, W., et al. (2013). The MOV10 helicase inhibits LINE-1 mobility. J. Biol. Chem. 288, 21148–21160. doi: 10.1074/jbc.M113.465856
Lin, C., Yang, L., Tanasa, B., Hutt, K., Ju, B. G., Ohgi, K., et al. (2009). Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139, 1069–1083. doi: 10.1016/j.cell.2009.11.030
Liou, Y. C., Zhou, X. Z., and Lu, K. P. (2011). Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 36, 501–514. doi: 10.1016/j.tibs.2011.07.001
Luan, D. D., Korman, M. H., Jakubczak, J. L., and Eickbush, T. H. (1993). Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605. doi: 10.1016/0092-8674(93)90078-5
MacMicking, J. D. (2012). Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382. doi: 10.1038/nri3210
Marchetto, M. C., Narvaiza, I., Denli, A. M., Benner, C., Lazzarini, T. A., Nathanson, J. L., et al. (2013). Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529. doi: 10.1038/nature12686
Martin, S. L. (1991). Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11, 4804–4807. doi: 10.1128/MCB.11.9.4804
Martin, S. L., Branciforte, D., Keller, D., and Bain, D. L. (2003). Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 100, 13815–13820. doi: 10.1073/pnas.2336221100
Martin, S. L., and Bushman, F. D. (2001). Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21, 467–475. doi: 10.1128/MCB.21.2.467-475.2001
Mathias, S. L., Scott, A. F., Kazazian, H. H., Boeke, J. D., and Gabriel, A. (1991). Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810. doi: 10.1126/science.1722352
Miki, Y., Nishisho, I., Horii, A., Miyoshi, Y., Utsunomiya, J., Kinzler, K. W., et al. (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645
Mir, A. A., Philippe, C., and Cristofari, G. (2015). euL1db: the European database of L1HS retrotransposon insertions in humans. Nucleic Acids Res. 43, D43–D47. doi: 10.1093/nar/gku1043
Moldovan, G. L., Pfander, B., and Jentsch, S. (2007). PCNA, the maestro of the replication fork. Cell 129, 665–679. doi: 10.1016/j.cell.2007.05.003
Moldovan, J. B., and Moran, J. V. (2015). The zinc-finger antiviral protein ZAP inhibits LINE and alu retrotransposition. PLoS Genet. 11:e1005121. doi: 10.1371/journal.pgen.1005121
Monot, C., Kuciak, M., Viollet, S., Mir, A. A., Gabus, C., Darlix, J. L., et al. (2013). The specificity and flexibility of L1 reverse transcription priming at imperfect T-tracts. PLoS Genet. 9:e1003499. doi: 10.1371/journal.pgen.1003499
Moran, J. V., Holmes, S. E., Naas, T. P., DeBerardinis, R. J., Boeke, J. D., and Kazazian, H. H. (1996). High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927. doi: 10.1016/S0092-8674(00)81998-4
Muckenfuss, H., Hamdorf, M., Held, U., Perkovic, M., Lower, J., Cichutek, K., et al. (2006). APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281, 22161–22172. doi: 10.1074/jbc.M601716200
Muotri, A. R., Marchetto, M. C., Coufal, N. G., Oefner, R., Yeo, G., Nakashima, K., et al. (2010). L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446. doi: 10.1038/nature09544
Niewiadomska, A. M., Tian, C., Tan, L., Wang, T., Sarkis, P. T., and Yu, X. F. (2007). Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81, 9577–9583. doi: 10.1128/JVI.02800-06
Nigumann, P., Redik, K., Mätlik, K., and Speek, M. (2002). Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79, 628–634. doi: 10.1006/geno.2002.6758
Peddigari, S., Li, P. W., Rabe, J. L., and Martin, S. L. (2013). hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 41, 575–585. doi: 10.1093/nar/gks1075
Piskareva, O., Ernst, C., Higgins, N., and Schmatchenko, V. (2013). The carboxy-terminal segment of the human LINE-1 ORF2 protein is involved in RNA binding. FEBS Open Bio 3, 433–437. doi: 10.1016/j.fob.2013.09.005
Richardson, S. R., Morell, S., and Faulkner, G. J. (2014b). L1 Retrotransposons and Somatic Mosaicism in the Brain. Annu. Rev. Genet. 48, 1–27. doi: 10.1146/annurev-genet-120213-092412
Richardson, S. R., Narvaiza, I., Planegger, R. A., Weitzman, M. D., and Moran, J. V. (2014a). APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 3:e02008. doi: 10.7554/elife.02008
Rodić, N., Steranka, J. P., Makohon-Moore, A., Moyer, A., Shen, P., Sharma, R., et al. (2015). Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064. doi: 10.1038/nm.3919
Shi, X., Seluanov, A., and Gorbunova, V. (2007). Cell divisions are required for L1 retrotransposition. Mol. Cell. Biol. 27, 1264–1270. doi: 10.1128/MCB.01888-06
Shukla, R., Upton, K. R., Muñoz-Lopez, M., Gerhardt, D. J., Fisher, M. E., Nguyen, T., et al. (2013). Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111. doi: 10.1016/j.cell.2013.02.032
Solyom, S., Ewing, A. D., Rahrmann, E. P., Doucet, T., Nelson, H. H., Burns, M. B., et al. (2012). Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22, 2328–2338. doi: 10.1101/gr.145235.112
Speek, M. (2001). Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001
Stenglein, M. D., Burns, M. B., Li, M., Lengyel, J., and Harris, R. S. (2010). APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17, 222–229. doi: 10.1038/nsmb.1744
Stenglein, M. D., and Harris, R. S. (2006). APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281, 16837–16841. doi: 10.1074/jbc.M602367200
Stetson, D. B., Ko, J. S., Heidmann, T., and Medzhitov, R. (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598. doi: 10.1016/j.cell.2008.06.032
Suzuki, J., Yamaguchi, K., Kajikawa, M., Ichiyanagi, K., Adachi, N., Koyama, H., et al. (2009). Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5:e1000461. doi: 10.1371/journal.pgen.1000461
Swergold, G. D. (1990). Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10, 6718–6729. doi: 10.1128/MCB.10.12.6718
Taylor, M. S., Lacava, J., Mita, P., Molloy, K. R., Huang, C. R., Li, D., et al. (2013). Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155, 1034–1048. doi: 10.1016/j.cell.2013.10.021
Tubio, J. M., Li, Y., Ju, Y. S., Martincorena, I., Cooke, S. L., Tojo, M., et al. (2014). Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345:1251343. doi: 10.1126/science.1251343
Upton, K. R., Gerhardt, D. J., Jesuadian, J. S., Richardson, S. R., Sánchez-Luque, F. J., Bodea, G. O., et al. (2015). Ubiquitous l1 mosaicism in hippocampal neurons. Cell 161, 228–239. doi: 10.1016/j.cell.2015.03.026
van den Hurk, J. A., Meij, I. C., Seleme, M. C., Kano, H., Nikopoulos, K., Hoefsloot, L. H., et al. (2007). L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592. doi: 10.1093/hmg/ddm108
Viollet, S., Monot, C., and Cristofari, G. (2014). L1 retrotransposition: the snap-velcro model and its consequences. Mob. Genet. Elements 4:e28907. doi: 10.4161/mge.28907
Wallace, N. A., Belancio, V. P., and Deininger, P. L. (2008). L1 mobile element expression causes multiple types of toxicity. Gene 419, 75–81. doi: 10.1016/j.gene.2008.04.013
Wallace, N. A., Gasior, S. L., Faber, Z. J., Howie, H. L., Deininger, P. L., and Galloway, D. A. (2013). HPV 5 and 8 E6 expression reduces ATM protein levels and attenuates LINE-1 retrotransposition. Virology 443, 69–79. doi: 10.1016/j.virol.2013.04.022
Wei, W., Gilbert, N., Ooi, S. L., Lawler, J. F., Ostertag, E. M., Kazazian, H. H., et al. (2001). Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001
Wissing, S., Montano, M., Garcia-Perez, J. L., Moran, J. V., and Greene, W. C. (2011). Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J. Biol. Chem. 286, 36427–36437. doi: 10.1074/jbc.M111.251058
Xie, Y., Mates, L., Ivics, Z., Izsvák, Z., Martin, S. L., and An, W. (2013). Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 4:10. doi: 10.1186/1759-8753-4-10
Yang, N., and Kazazian, H. H. (2006). L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771. doi: 10.1038/nsmb1141
Yeh, E., Cunningham, M., Arnold, H., Chasse, D., Monteith, T., Ivaldi, G., et al. (2004). A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308–318. doi: 10.1038/ncb1110
Zamudio, N., and Bourc'his, D. (2010). Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity (Edinb). 105, 92–104. doi: 10.1038/hdy.2010.53
Zhang, A., Dong, B., Doucet, A. J., Moldovan, J. B., Moran, J. V., and Silverman, R. H. (2014). RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 42, 3803–3820. doi: 10.1093/nar/gkt1308
Zhao, K., Du, J., Han, X., Goodier, J. L., Li, P., Zhou, X., et al. (2013). Modulation of LINE-1 and Alu/SVA retrotransposition by aicardi-goutières syndrome-Related SAMHD1. Cell Rep. 4, 1108–1115. doi: 10.1016/j.celrep.2013.08.019
Zingler, N., Willhoeft, U., Brose, H. P., Schoder, V., Jahns, T., Hanschmann, K. M., et al. (2005). Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 15, 780–789. doi: 10.1101/gr.3421505
Keywords: LINE-1, retrotransposon, genome evolution, repeated sequences, retrotransposition, structural variation (SV)
Citation: Pizarro JG and Cristofari G (2016) Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Front. Cell Dev. Biol. 4:14. doi: 10.3389/fcell.2016.00014
Received: 14 December 2015; Accepted: 18 February 2016;
Published: 07 March 2016.
Edited by:
Tammy A. Morrish, Independent Researcher, USAReviewed by:
Sandy Martin, University of Colorado School of Medicine, USACopyright © 2016 Pizarro and Cristofari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaël Cristofari, Z2FlbC5jcmlzdG9mYXJpQHVuaWNlLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.