- Instituto de Neurociencias, Consejo Superior de Investigaciones Científicas and Universidad Miguel Hernández (CSIC-UMH), San Joan d Alacant, Spain
Cell death is an essential physiological process for the survival of multicellular organisms. Our understanding of programmed cell death in development, immune function maintenance, and adult tissue repair has significantly advanced over the past decade. However, there are still gaps in our knowledge about the induction, regulation, and checkpoints of this process due to the diverse forms of cellular suicide and the rapid nature of the process. Molecular advancements such as specific cell death sensors, RNA-seq, single-cell RNA-seq, and proteomics have allowed for identifying new factors and a better understanding of the molecular networks and pathways that regulate these processes. Programmed cell death also plays a role in cancer, both limiting and facilitating aspects of the malignant process, making its analysis and inhibition challenging. This review discusses the field’s advancements using the model organism Drosophila melanogaster, the types of cell death in development and adult tissues, the techniques for studying it, and its role in cancer.
Introduction
The ability of an organism to induce its own cells to die is an evolutionarily advantageous process that safeguards the organism and ensures only the fittest cells survive. Early studies characterizing the distinct morphologies associated with cell death processes revealed the inherently regulated and intentional nature of programmed cell death (PCD) (Kerr et al., 1972). PCD occurs under many normal physiological conditions and through many different means, playing an essential role in the removal of unfitted or surplus cells (Fuchs and Steller, 2011). The pioneering genetic and molecular studies on the regulation of PCD were conducted using C. elegans (Ellis and Horvitz, 1986). In C. elegans, PCD is essential for germ-cell death during oogenesis (Lettre and Hengartner, 2006) and protection against pathogen attack (Aballay and Ausubel, 2001). Examples of PCD during development are the hormonal signalling pathways selectively eliminating Müllerian ducts in males and Wolffian ducts in females (Arya and White, 2015), the developing of digits in higher vertebrates (Fuchs and Steller, 2011) or the regular lattice of the retina of insects (Cagan and Ready, 1989). PCD also acts in adult organisms to eliminate surplus progenitor cells in the mammalian immune system (John Cohen, 1991) and the excess enteroblast cells during the repair and homeostasis of the adult intestine of fruit fly (Reiff et al., 2019). PCD is also a critical process in cancer as evading cell death is one of the biological processes that allow cancer cells to thrive, survive, and facilitate invasion and migration of metastatic cells (Koren and Fuchs, 2021).
The fruit fly D. melanogaster is an excellent model for investigating the complex interplay between cell death, cell competition, and tumorigenesis (Adrados et al., 2024; Mirzoyan et al., 2019; Parvy et al., 2018; Pinal et al., 2019; Sollazzo et al., 2023). For instance, cell competition, initially discovered in D. melanogaster, plays a crucial role in both fly and human cancer development as an intrinsic tumour suppression mechanism (Morata and Ripoll, 1975; Simpson and Morata, 1981). Studies in the fruit fly of this evolutionarily conserved process provide valuable insights into clonal evolution and tumour heterogeneity in human cancers, potentially leading to novel therapeutic approaches (Cong and Cagan, 2024). The genetic tools available for this animal model allow for precise manipulation of genes involved in cell death and the cancer-related processes, making it an ideal system for studying the molecular pathways that regulate these phenomena (Hay et al., 2004; Munnik et al., 2022). Additionally, the conservation of signalling pathways between D. melanogaster and humans has contributed significantly to understanding cancer hallmarks such as genomic instability, resistance to cell death, altered metabolism, inflammation, and immune evasion (Baonza et al., 2022; García-López et al., 2021; Jiang et al., 2022; Shan and Mollereau, 2024; Stefanatos and Vidal, 2011).
This review aims to critically analyse the most recent advancements for investigating cell death and cancer-related cell death in the D. melanogaster model. By integrating and synthesizing the latest developments in techniques and methodologies, we aim to provide researchers with a comprehensive interpretation of the cutting-edge tools available to uncover the fundamental biological processes that underlie PCD. Here we summarize and exemplify the roles of PCD to equip researchers with the knowledge and insights necessary to advance the field and make significant contributions to understanding cell death and cancer in D. melanogaster.
The multifaceted roles of PCD in D. melanogaster
Cell death during development
Studies on cell death in D. melanogaster have offered valuable insights into the complex mechanisms that regulate this fundamental process in development and biological homeostasis [reviewed in Yalonetskaya et al. (2018)]. From embryonic stages to metamorphosis, D. melanogaster has proven to be an exceptional model for unravelling the mysteries of PCD in diverse physiological contexts orchestrating essential processes for maintaining cellular equilibrium. The D. melanogaster life cycle comprises four distinct stages: embryo, larva (first, second and third instar), pupa (prepupal and pupal stage) and adult. Notably, cell death manifests early in embryogenesis, with apoptotic cells appearing approximately 7 hours after egg deposition and subsequently becoming more widespread throughout the embryo (Abrams et al., 1993). During the larval stages, PCD occurs in several tissues, with a pronounced presence in the peripheral and central nervous system (CNS). Crucially, specific signalling pathways tightly regulate cell death, playing a vital role in the selective elimination of cells during tissue morphogenesis (Rusconi et al., 2000). Examples of that are the waves of PCD among neurons, initiated during mid-to-late embryogenesis, moulding the CNS development (Abrams et al., 1993). The first wave of neurogenesis in the embryonic stages establishes the larval nervous system, while a second wave, which develops during the larval and pupal stages, shapes the remaining components of the CNS that will function in the adulthood (Rogulja-Ortmann et al., 2007). This process, however, is not static, as numerous larval neurons meet their fate in cell death during metamorphosis, underlining the dynamics and plasticity of these events (Truman and Bate, 1988). In addition to the death of neuroblasts, neurons and glia also die throughout development, both to establish appropriate cell numbers and to remove cells that are no longer required in later stages (Pinto-Teixeira et al., 2016).
The differentiation of the adult eye from imaginal tissue during pupal development is another clear example of PCD, which involves the precise patterning of the interommatidial cells surrounding the photoreceptor clusters. Here, apoptosis serves as the mechanism for eliminating superfluous cells, highlighting its role in the formation of the mature organ (Cagan and Ready, 1989; Wolff and Ready, 1991). The larval-to-pupal transition represents a crucial stage characterised by dramatic cell death, devised primarily by the steroid hormone ecdysone (Jiang et al., 1997). This process engages the elimination of many larval tissues, a phenomenon tightly controlled by nuclear hormone receptors and transcription factors that provide spatial and temporal regulation (Garelli et al., 2012). This massive cell death during the larval-to-pupal transition appears to involve mechanisms beyond canonical apoptosis, underscoring the complexity of the regulatory landscape. During the third instar larva stage of development, autophagy has been shown to function as a key cell death mechanism. Removing the obsolete larval midgut and fat body is a prime example of autophagy-dependent cell death through regulation of the PI3K pathway (Berry and Baehrecke, 2007; Rusten et al., 2004). In some cases, this process requires autophagy but occurs independently of apoptosis (Denton et al., 2009), but in others, like salivary gland degradation, it also involves both autophagy and apoptosis working in concert (Martin et al., 2007). This process, triggered by the steroid hormone ecdysone, involves the activation of autophagy-related genes as well as apoptotic machinery.
In the context of oogenesis, the D. melanogaster ovary offers a fascinating terrain for studying PCD. This process is also notable for its complexity, with hundreds of ovarian chambers progressing through defined stages of development. Here, nurse cells, essential for oogenesis, undergo PCD after transferring their cytoplasmic contents to oocytes, and are subsequently eliminated by a subset of follicular epithelial cells. This meticulous process reveals the interconnection between cell death and reproductive development (Timmons et al., 2016).
The D. melanogaster cell death machinery is primarily represented by apoptosis, a caspase-dependent cell death pathway highly conserved among metazoans. Caspases, cysteine proteases, play a central role in apoptosis, where the initiator caspases Dronc respond to apoptotic stimuli and the effector caspases Drice and Dcp-1 cleave substrates to induce cell death (Kumar and Cakouros, 2004). In D. melanogaster, several types of PCD have been identified, contributing to various developmental processes (Figure 1). Briefly, prominent forms of cell death and related processes include:
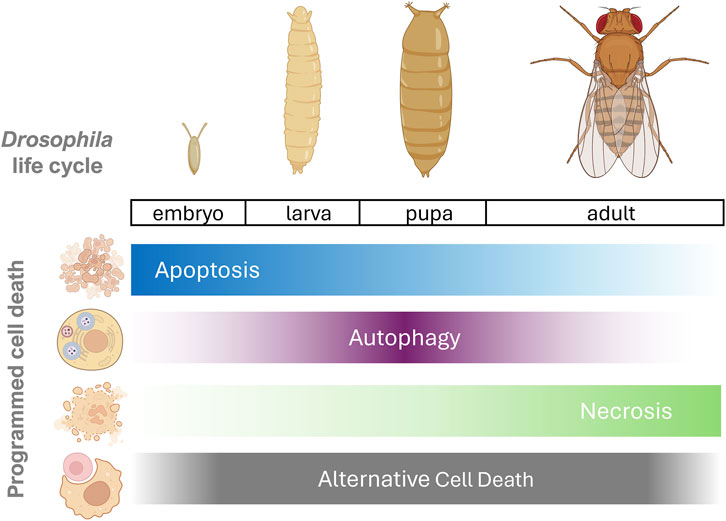
Figure 1. D. melanogaster developmental cell death. An illustration of main programmed cell death programs activated during the D. melanogaster life cycle. Apoptosis is initiated in early embryogenesis and continues during neurodevelopmental stages. Despite most Drosophila tissues dying via apoptosis, in the larval and pupal stages, autophagic-dependent cell death is activated in response to ecdysone. During adulthood, male spermatogonia cyst cells and female nurse cells die by necrotic process. Alternative cell death processes occur during all stages of D. melanogaster development, i.e., parthanatos at embryogenesis, ferroptosis during wing disc development and phagoptosis (also called phagocyte-driven cell death) during spermatogenesis. Created with BioRender.com.
Apoptosis
This is a well-characterized and extensively studied form of PCD in D. melanogaster. It plays a crucial role in development, tissue homeostasis, and the removal of unwanted or damaged cells (White et al., 1994). The proapoptotic genes grim, reaper, hid, and sickle, collectively known as RHG, initiate apoptosis in response to specific developmental cues (Bergmann et al., 1998; Goyal et al., 2000; Hay et al., 1995; White et al., 1996). Other routes besides the RHG motif could activate apoptosis. For instance, mitochondrial cytochrome c (Cyt c) that regulates apoptosis in the developing eye (Mendes et al., 2006) and is involved in caspase activation for spermatid individualization (Arama et al., 2006) and salivary gland degradation (Long et al., 2024).
Autophagy
This process involves the degradation and recycling of cellular components within lysosomes. Autophagy is essential for maintaining cellular homeostasis, and its dysregulation has been implicated in various diseases including cancer (Debnath et al., 2023). In D. melanogaster, autophagy plays a role in various developmental stages and responses to nutrient availability (Berry and Baehrecke, 2007; Denton et al., 2009). While autophagy is a well-studied mechanism of cell death, it can also contribute to the pro-survival mechanism, modulating necrotic cell death in Drosophila neurons and potentially acting as a protective mechanism against stress-induced necrosis (Lei et al., 2017). Furthermore, autophagy has been found to regulate necrosis in specific contexts, while necrosis signalling can influence autophagic activity in others (Park et al., 2020).
Necrosis
While apoptosis is the primary mode of PCD in D. melanogaster, instances of necrosis have also been observed, particularly in response to specific stress conditions (Park et al., 2020). Necrotic cell death involves rapid cellular swelling and membrane rupture, leading to inflammation (Yacobi-Sharon et al., 2013).
Efferocytosis
A process closely related to cell death, important for development and homeostasis, and responsible for removing apoptotic cells by phagocytes. This process is critical for maintaining tissue integrity and preventing inflammation. In D. melanogaster, efferocytosis occurs in nearly all tissues, including the CNS, where phagocytic glia and haemocytes play essential roles in clearing apoptotic cells (Davidson and Wood, 2020; Zheng et al., 2017; 2021).
Cancer cell death mechanisms
Cell death is an essential area of study for understanding the fundamental biological processes underlying cancer development and progression. D. melanogaster is a valuable model organism for studying these mechanisms due to its genetic similarity to humans and its well-characterized genetics and developmental biology (Jennings, 2011). Until now, cancer-related research has focused on understanding cell death evasion mechanisms and developing promising anticancer strategies that inhibit them. In this context, several forms of PCD have been identified and shown to play crucial roles in modulating the tumour microenvironment (TME), making their study attractive in cancer research (Parvy et al., 2018).
Studies carried out in D. melanogaster have made it possible to identify PCD associated with tumour initiation and progression processes. An example of this is the phenomenon called “cellular competition,” which plays a role in eliminating oncogenic cells or selecting fitter cells. This effect has been demonstrated in the fruit fly intestinal tumours, where oncogenic cells hack the system, competing with surrounding cells and inducing their elimination, creating a permissive environment for tumour growth (Suijkerbuijk et al., 2016). Furthermore, in different D. melanogaster cancer models, it has been shown that epithelial tumours exhibit a high level of cell death when grown under competitive stress. This pressure allows excessive growth of the cancerous mass, mainly dependent on the activation of caspases since their inhibition is sufficient to reduce the size of the tumours (Sollazzo et al., 2023).
In addition, autophagy can also play an important role in cancer cell death. In D. melanogaster, the induction of autophagy in the TME is mediated by ROS accumulation and activation of the JNK signalling pathway in tumour cells. Notably, the active transport of nutrients from cells surrounding the tumour sustains tumour growth, indicating that tumour cells proliferate and grow at the expense of their neighbouring normal cells through non-cell autonomous autophagy (Zhao et al., 2021). JNK signalling also contains tumour growth through necrosis, an alternative PCD activated when Egr/JNK-mediated apoptosis fails to inhibit the oncogenic growth of scrib mutant cells. This activation of necrosis is mediated by the initiator caspase Dronc (Li et al., 2019).
Despite the significant advances in the study of cell death in the context of cancer, there is still much to understand about this process and its dual role in suppressing and promoting tumours. Past and present studies highlight the power of D. melanogaster model to genetically dissect the complex interplay between the cell death pathways and oncogenic signalling. Inhibiting apoptosis can lead context and stage-dependent effects, sometimes promoting senescence and tumour growth while restraining malignant traits in other cases.
Techniques used to study cell death in D. melanogaster
The precise characterization of different types of cell death does not rely on a single technical method. Instead, it uses a multifaceted approach that combines various markers and techniques (Napoletano et al., 2019). Distinguishing between different cell death processes is achieved through the integration of both classical and modern methods. This comprehensive strategy includes the use of electron microscopy to observe ultrastructural changes, analysis of specific biochemical markers, real-time imaging techniques to track the progression of cell death, and advanced molecular methods such as single-cell RNA sequencing. Additionally, functional assays and genetic manipulations are employed to validate the signalling pathways involved. This combination of approaches allows for a more robust and reliable characterization of cell death mechanisms, overcoming the limitations of any individual method and providing a more complete understanding of these complex biological processes.
Classical approaches
In situ approaches have been utilized to study PCD during development of D. melanogaster, allowing for the visualization and quantification of cell death processes at the single-cell level (Denton et al., 2008; Denton and Kumar, 2015). These methods enable the detection of morphological and biochemical changes associated with PCD, such as DNA fragmentation, chromosome condensation, and nuclear deformation, providing insights into the dynamics of cell death during development (Richardson and Kumar, 2002).
The study of DNA fragmentation as a marker of apoptosis in D. melanogaster has been facilitated by several in situ techniques. The TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) assay is widely used to detect apoptosis by identifying DNA breaks (Figure 2A), labelling their 3′-hydroxyl termini and allowing visualization of apoptotic cells in D. melanogaster tissues like imaginal discs (Gavrieli et al., 1992). The vital dye acridine orange (AO) is another method that has been used for decades to detect apoptosis in D. melanogaster tissues and cells (Figure 2B; Abrams et al., 1993; Wolff and Ready, 1991), as AO is a fluorescent dye that intercalates with DNA and can identify cells undergoing apoptosis-associated DNA fragmentation (Spreij, 1970). These well-established assays have made it possible to catalogue the pattern of cell death during D. melanogaster embryogenesis and metamorphosis over the years (Abrams et al., 1993; Baechrecke, 2000; Pazdera et al., 1998; Peterson et al., 2002). These methods continue to be employed to confirm and differentiate apoptosis from other types of cell death, such as necrosis. Their enduring relevance is further enhanced by refinements in detection techniques, improving sensitivity and specificity (Chimata et al., 2022). In addition, immunohistochemistry assays have been used to study apoptosis, detecting proteins such as p53, annexin V, and caspases. These in situ techniques enable the detection and quantification of apoptotic cells undergoing DNA fragmentation in D. melanogaster tissues during development and in response to various stimuli (Dichtel-Danjoy et al., 2012; Robin et al., 2019; Sarkissian et al., 2014; Shklyar et al., 2013).
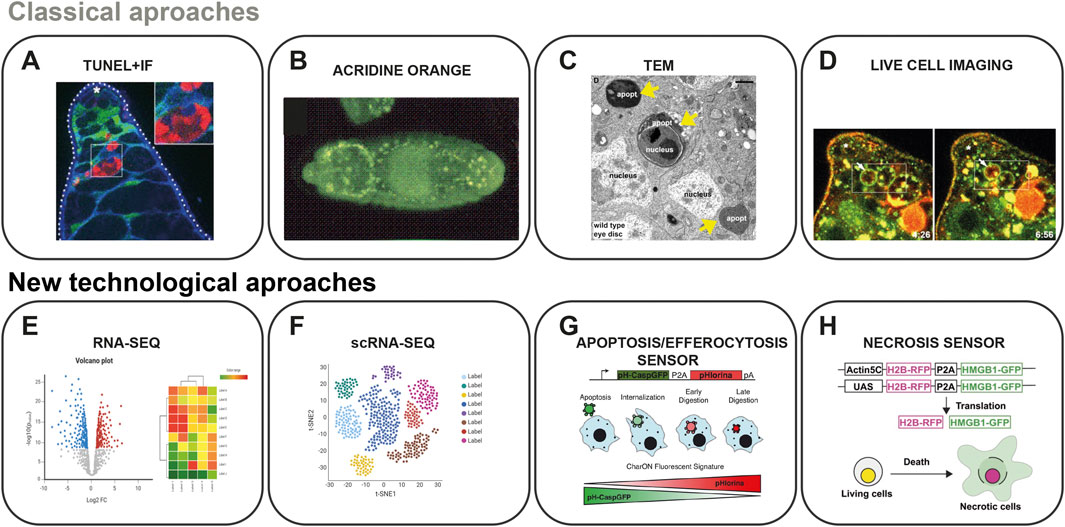
Figure 2. Approaches to study programmed cell death in D. melanogaster. This figure showcases the array of techniques available for studying PCD in D. melanogaster. Example results for classical approaches (A–D) and schematic representations for new technology approaches (E–H). (A) The apical tip of a testis showing the expression of cytGFP in cyst cells (green), the immunofluorescence (IF) of Drpr (blue) and the TUNEL staining (red, dying germ cells). Asterisk marks the hub.(B) Dorsal view of a stage 14 embryo stained with Acridine Orange. (C) TEM photography revealing apoptotic bodies (yellow arrows) induced by clonal overexpression of Hid in eye imaginal disc cells. Apoptotic cells appear darker than healthy cells. (D) Live-imaged testis from Rab7-YFP (green) marked with LysoTracker (red). Boxed region, highlight late endosomes (white arrow) surrounding live germ cells that are gradually filled with LysoTracker. (E) Common graphical tools for interpretation of the RNA-seq data. (F) Schematic representation for visualization of scRNA-seq spatial transcriptomic data. (G) Example diagram of the genetic sensor CharON. The construct design is shown on the top, and at the bottom is a representation of the mechanism of an apoptotic CharON-expressing cell (green or red) engulfed by a macrophage (blue). (H) Diagram of the genetic sensor Necrosensor 2. The necrosensor (HMGB1-GFP) is connected with H2B-RFP via P2A. HMGB1-GFP is released into the extracellular space upon necrotic stimuli (green), whereas H2B-RFP remains in the nucleus (red). Panels A and D adapted from Zohar-Fux et al. (2022), C from Nagy et al. (2015), G from Raymond et al. (2022) and H from Nishida et al. (2024) under Creative Commons CC-BY licenses. Panel B adapted with permission from Developmental Journal (Abrams et al., 1993). Panels E and F were created with BioRender.com.
The most commonly used in situ approaches to study PCD in D. melanogaster include in situ hybridization and in situ optical imaging. In situ hybridization is used to determine gene expression patterns by detecting RNA transcripts within cells or tissues (Van De Corput et al., 1998). However, this method has been optimized to target specifically cell death genes in D. melanogaster during development (Lécuyer et al., 2008; Tan et al., 2011). On the other hand, in situ optical imaging techniques, such as fluorescence, confocal, or multiphoton microscopy, offer a powerful approach to studying the metabolic dynamics of lipids and proteins during ageing, allowing for the visualization of cellular processes associated with PCD at a high resolution. These methods are particularly useful for studying PCD during D. melanogaster oogenesis, which is regulated by mechanisms different from those that control cell death in other tissues (Foley and Cooley, 1998; Nezis et al., 2000). New approaches have emerged combining traditional ones with advanced imaging systems such as direct image of lipid metabolic changes by Raman spectroscopy (DO-SRS) or mitochondrial dynamics by fluorescence wide-field microscopy (LaJeunesse et al., 2004; Li et al., 2022). This emerging technology has allowed the identification of unsaturated lipids and Cyt c protein accumulated simultaneously in egg chambers from old flies that could induce cell death, as demonstrated during salivary gland degradation and neuronal apoptotic cell death (Hung et al., 2021; Long et al., 2024).
PCD is also observed in patches of cells during the larval stages, but during metamorphosis most larval tissues undergo this process mainly regulated by the steroid hormone 20-hydroxyecdysone (ecdysone). This process plays a crucial role in eliminating obsolete larval tissues and organs, allowing for the formation of the adult body structure (Jiang et al., 1997). In addition to apoptosis, autophagy contributes to this tissue remodelling during a developmental programmed 5-day starvation period. It involves the degradation of cellular components through the formation of autophagosomes, which can have survival and death functions depending on the context (Berry and Baehrecke, 2007; Denton et al., 2009; Rusten et al., 2004). Analysis of autophagy in D. melanogaster encompasses various techniques to monitor and analyse the PCD at different stages. Fluorescent markers, such as GFP-tagged Atg8a, allow visualization of autophagosome formation through fluorescence microscopy (Juhasz and Neufeld, 2008). Transmission electron microscopy (TEM; Figure 2C) provides high-resolution ultrastructural analysis of autophagic vesicles (Eskelinen et al., 2011). Autophagic flux assays, using lysosomal inhibitors like chloroquine or bafilomycin A1, measure the dynamic process of autophagy by blocking autophagosome-lysosome fusion (Nagy et al., 2015).
Necrosis, traditionally viewed as a response to severe damage and stress, can also occur as a physiological event in the absence of external insult in organisms. Drosophila spermatogenesis provided the first evidence of physiologically programmed necrosis controlled by p53, a crucial mechanism in tumour suppression. This discovery illustrates a p53-dependent mechanism that is evolutionarily preserved in mammals (Napoletano et al., 2017; Yacobi-Sharon et al., 2013). Necrosis also plays a crucial role in regulating female nurse cell death during the developmental processes of oogenesis (Bass et al., 2009). The main method for studying necrosis in these models has been the propidium iodide (PI) staining or PI/TUNEL double labelling combined with TEM and immunohistochemistry (Bass et al., 2009; Napoletano et al., 2017).
Alternative PCD forms have also been studied in D. melanogaster, which are activated during early embryogenesis. One of these processes is parthanatos, triggered by the overexpression or activation of the enzyme poly (ADP-ribose) polymerase-1 (PARP-1) after DNA damage caused by genotoxic stress or excitotoxicity (Wang and Ge, 2020). In D. melanogaster, parthanatos-like cell death is activated in 30% of primordial germ cells, usually eliminated during embryogenesis and is detected using a Top I-mediated ligation assay, allowing the visualization of DNase II-induced DNA cleavage (Tarayrah-Ibraheim et al., 2021). Phagoptosis, another form of PCD, is also activated in D. melanogaster germ cells (Kanaan et al., 2023; Zohar-Fux et al., 2022). In this process, phagocytes engulf and degrade viable cells in response to an “eat-me” signal or the loss of “don’t-eat-me” signals (Brown and Neher, 2012). Ferroptosis also emerged as a new PCD that has been studied in the fruit fly, a nonapoptotic form of cell death that results from iron accumulation and lipid peroxidation in cells (Saini and Owusu-Ansah, 2023). This alternative form has been demonstrated during the development of wing disc cells through morphological changes in the mitochondria and ROS accumulation (Mumbauer et al., 2019). Ferroptosis can be detected through direct measurement of lipid peroxidation by assessing the absorbance of samples at 532 nm, as well as measuring Fe levels in haemolymph using a colourimetric assay (Gomes et al., 2023). On the other hand, erebosis, a unique type of cell death, is found in the gut cells of adult fruit flies. Unlike apoptosis or necrosis, it involves a gradual loss of cell components and structure without triggering typical stress or immune responses, helping maintain gut health by replacing old cells with new ones (Bergmann, 2022; Ciesielski et al., 2022). A new proposed form of autophagy-associated cell death is karyoptosis, identified in a D. melanogaster model of neurodegenerative disease (Baron et al., 2017). Karyoptosis is triggered by chronic inhibition of autophagy and can be identified by detecting Lamin B1 in the cytoplasm colocalizing with autophagic markers, such as LC3 or p62 (Napoletano et al., 2019). The ancestral origin of mammalian pyroptosis, termed proto-pyroptosis, has also been described in D melanogaster. This inflammatory form of cell death is characterized by the recruitment of crystal cells to sites of injury and was first observed through live imaging of wounded Drosophila larvae (Dziedziech and Theopold, 2022). All these alternative forms of cellular death have been studied using TEM, in vivo live imaging and immunofluorescence techniques with fluorescent dyes such as lysotracker and Hoechst (Figure 2D).
Other alternative cell death mechanisms include entosis, a form of non-apoptotic cell death that occurs when one cell actively invades and becomes engulfed by a neighbouring cell (Overholtzer et al., 2007). Entosis has been linked to earlier studies on cell competition in D. melanogaster, but this mechanism requires apoptotic programs. This phenomenon is typically investigated using clonal analysis and tissue mosaics (Morata, 2021), and it has generated considerable attention in cancer research, where it is thought to play a critical role in tumour progression and the regulation of cell populations (Cong and Cagan, 2024). Alternative cell context-dependent PCD, such as excitotoxicity, have also been studied in the fruit fly. The overstimulation of neurons triggers excitotoxicity, leading to cell death through excessive excitatory neurotransmitters, especially glutamate (Sattler and Tymianski, 2001). In D. melanogaster, this process has been studied using calcium and glutamate live imaging combined with locomotion experiments to detect behavioural patterns after neuronal death (Peng et al., 2019; Xu and Xu, 2018).
There are also key determinants that support PCD during D. melanogaster development, such as efferocytosis. During embryogenesis, efferocytosis is essential for the removal of apoptotic cells from the nervous system, allowing for the proper formation and function of neurons (Kurant et al., 2008). Similarly, in the germline cells, efferocytosis helps to remove apoptotic cells and maintain tissue homeostasis during D. melanogaster development (Etchegaray et al., 2012; Timmons et al., 2016). The process of efferocytosis in the fruit fly has traditionally been assessed using a combination of methods, including TEM, AO staining, TUNEL assay, and immunohistochemistry.
Molecular techniques
New molecular technologies have allowed the advancement of our understanding of cell death in the fly model (Figures 2E, F). For instance, gene expression comparisons using the Gene Chip D. melanogaster Genome 2.0 arrays in young (two-day-old) and old (45-day-old) flies have revealed upregulation of genes promoting cell death in older flies, including the caspase genes Damm, Strica and Decay, as well as changes in apoptosis regulation in ageing tissues, suggesting that tissue-specific changes occur in the regulation of apoptosis as the organism ages, rather than a generalised increase in programmed cell death across all tissues (Bordet et al., 2021).
Additionally, RNA-sequencing (RNA-seq) analysis performed on the eyes of fruit fly pupae at two developmental stages, 21 and 40 h after pupa formation has provided insights into the regulation of cell death during development (DeAngelis et al., 2021). Comparing the temporal and spatial gene expression in apoptotic and non-apoptotic tissues during metamorphosis enabled the identification of a reduction in the ecdysone-induced gene E93, a critical regulator of cell death, in non-apoptotic tissues despite caspase activation (Ojha and Tapadia, 2020). These studies compared larval and pupal salivary glands, which undergo cell death during metamorphosis, with Malpighian tubules, which avoid apoptosis, utilising the Affymetrix D. melanogaster Genome 2.0 microarray chip.
Omics technologies have also addressed PCD in cancer and other pathological states. A major gene linking PCD and cancer is TP53, whose mutations are highly prevalent in human tumours. (Olivier et al., 2010). The p53 transcription factor coordinates various cellular responses to stress, including the initiation of apoptosis (Vousden and Prives, 2009). Using RNA-seq combined with chromatin immunoprecipitation sequencing (ChIP-Seq), the function of p53 was interrogated in postmitotic and embryonic D. melanogaster tissues (Kurtz et al., 2019). In the developing embryo, p53 robustly activates key apoptotic genes in response to radiation-induced DNA damage. The p53 enhancer near the cell death gene reaper forms chromatin contacts, facilitating the activation of p53 targets over long genomic distances. Interestingly, this typical p53 apoptotic response is absent in adult heads, a postmitotic tissue, and this lack of response is not associated with changes in chromatin contacts.
PCD in other diseases has also been investigated employing omics techniques. One example is the autosomal dominant retinitis pigmentosa (ADRP), an age-related degenerative retinal disease (Sung et al., 1991), in which the chronic perturbation of the endoplasmic reticulum induces apoptosis. To understand the pathways that mediate apoptosis related to ER stress, Park and collaborators used an ADRP D. melanogaster model, and found that Wg/Wnt1 signalling mediates this process. Subsequent analysis by RNA-seq of eye imaginal discs showed that the ER stress-associated serine protease (Erasp) is a downstream target of Wg/Wnt1 during ER perturbation (Park et al., 2023).
RNA-seq analysis can be performed on whole D. melanogaster, dissected tissues, body parts, or at the single-cell level to examine healthy and disease cells and the PCD process. Using single-cell RNA sequencing (scRNA-seq), the impact of an Rbf mutation during D. melanogaster eye development was investigated (Ariss et al., 2018). The Rbf gene encodes the retinoblastoma protein (pRB) D. melanogaster orthologue, a tumour suppressor that blocks cell-cycle progression and is inactivated in human cancers (Dick et al., 2018). Analysis of the transcriptome profiles of wild-type and Rbf mutant eye imaginal disc cells revealed a mutant-specific cell population exhibiting intracellular acidification due to increased glycolytic activity. These metabolic changes, confined to this Rbf mutant population, sensitise cells to apoptosis and define the pattern of cell death in the Rbf mutant (Ariss et al., 2018).
The proteomic tools have also been instrumental in elucidating the molecular mechanisms underlying steroid-triggered autophagic cell death of the dying D. melanogaster salivary glands. These studies have confirmed the caspase-dependent autophagic transcriptional cascade, and additionally uncovered novel regulators, such as the cell cycle protein Warts, which participate in caspase-independent degradation pathways (Martin et al., 2007). By comparing the proteomes of salivary glands undergoing developmental versus stress-induced autophagic cell death, researchers have identified additional factors required for proper cell degradation (McPhee et al., 2012), highlighting the power of integrating genomic and proteomic approaches to obtain a comprehensive understanding of the complex cell death programs.
The proteome of D. melanogaster ovary, a robust system for investigating physiological cell death related to cell migration and other critical cell behaviours using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis uncovered critical regulator factors (Velentzas et al., 2015). This study identified signalling pathways previously analysed in mammals and showed a more comprehensive network of known factors such as p53, IGF, and PI3K. The study also contributed to linking distinct cell death sub-routines involved in the D. melanogaster ovary, indicating the co-expression and probably synergistic effects of cell death programs in the egg chamber compartments, during development and under stress conditions.
Autophagy plays a crucial role in cell survival and death, and its progression and resolution depend on lysosome function. A study by Xu et al., used a label-free LC-MS/MS approach to identify a group of proteins involved in the autophagy-dependent cell death program during degradation of D. melanogaster larval midguts (Xu et al., 2021). The study clarified how the lysosome contributes to this process through the essential function of cathepsins in the regulation of autophagic flux by maintaining a degradative environment inside the lysosome. Similarly, an optimised method for isolating autophagic structures from adult flies, with subsequent lipidomic analysis using MS/MS-based method, has contributed to pointing out the critical lipid transport function of the Atg2 protein in the de novo synthesis of early autophagic organelles (Laczkó-Dobos et al., 2021).
In a 2020 study, researchers used hydrophilic interaction LC-MS method to investigate controlled overexpression of Atg1 in specific tissues of D. melanogaster larvae. The study found that this overexpression increases mild autophagy and extends the lifespan of the flies. Although these flies were more sensitive to starvation, they also have an increased mitochondrial metabolism, which could be related to their longevity (Bjedov et al., 2020).
Furthermore, a study by Gao and collaborators combined RNA-seq and co-immunoprecipitation coupled LC-MS/MS of S2 cells to demonstrate that Wunen2 (Wun2) protein is required for efferocytosis both in vitro and in vivo (Gao et al., 2022). The study also revealed that Wun2 has a role in preventing the lysosomal degradation and transport of βν integrin from recycling endosomes to the plasma membrane to promote apoptotic cell clearance in D. melanogaster.
Genetically tractable systems
Genetically encoded sensors are powerful tools for studying PCD in the D. melanogaster model system (Figures 2G, H). Given the evolutionary conservation of core PCD mechanisms, insights gained from D. melanogaster PCD sensors are highly relevant for understanding cell death processes in human diseases like cancer.
There are multiple genetically encoded molecular sensors based on fluorescence for monitoring cell death in the fruit fly (Baena-Lopez et al., 2018; Nishida et al., 2024; Raymond et al., 2022; Schott et al., 2017) even though dying cells could be labelled in situ with vital dyes (Gavrieli et al., 1992). The sensors include the possibility of detecting cell death and associated processes in real-time (Raymond et al., 2022; Schott et al., 2017; To et al., 2015), overcoming the limitation of antibody staining. The fluorescence-based probes enable high-fidelity recording of processes with single-cell and subcellular spatial resolution (Greenwald et al., 2018). The genomic encoding of the sensors allows consistency and reliability in the detection of programmed cell death, making it easier to achieve accurate conclusions about this process (Table 1).
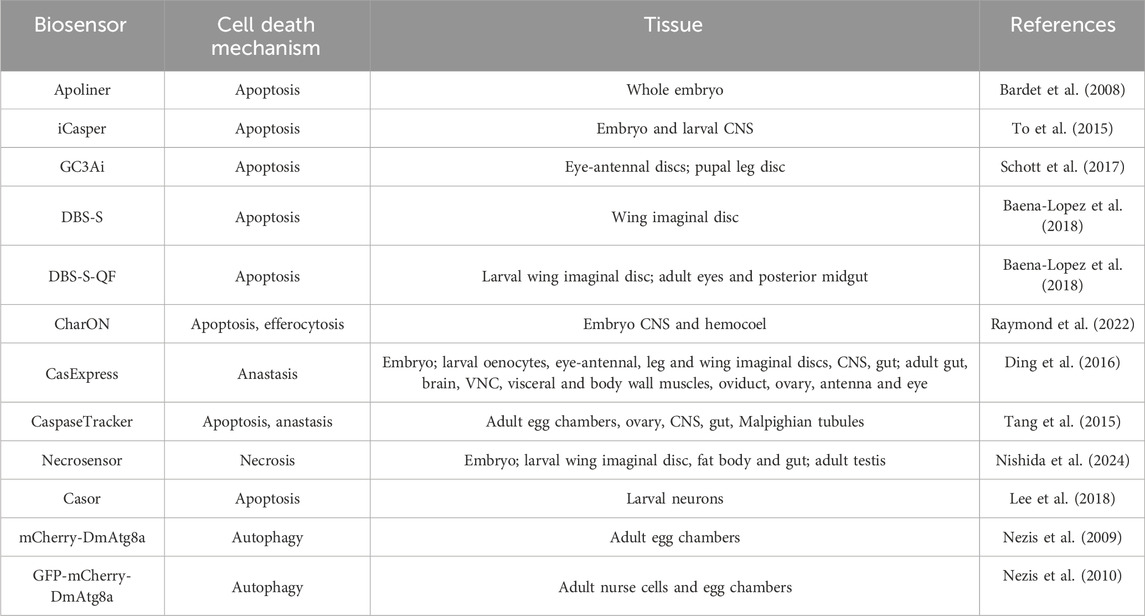
Table 1. Available genetic sensors to study programmed cell death mechanisms across different D. melanogaster tissues.
Many genetically encoded sensors engineered for studying cell death in D. melanogaster rely on caspase activity. These sensors become fluorescent upon the activation of caspases (Baena-Lopez et al., 2018; Lee et al., 2018; Raymond et al., 2022; Schott et al., 2017; To et al., 2015). For instance, the Apoliner sensor consists of two fused fluorescent proteins, mRFP and eGFP, separated upon caspase activity. When caspases are activated, the sensor is cleaved, causing eGFP to move to the nucleus while mRFP remains in the membranes (Bardet et al., 2008). Similarly, the infrared fluorescent executioner-caspase reporter iCasper becomes infrared fluorescent when apoptotic mechanisms start in cells. The iCasper apoptotic sensor has been used to measure apoptosis throughout developmental stages, demonstrating a spatiotemporal correlation between apoptosis and embryonic morphogenesis. Furthermore, it has been employed to investigate the dynamics of apoptosis during D. melanogaster brain tumour formation (To et al., 2015).
Another example of the development of sensors based on caspase activity and fluorescent proteins is the GFP-based variant of caspase 3-like protease activity indicator (GC3Ai). The expression of GC3Ai produces a non-fluorescent GFP that contains a caspase-1 recognizing sequence. After cleavage by active caspases, GFP becomes fluorescent and allows the visualization of apoptotic cells (Schott et al., 2017). Transgenic UAS-GC3Ai flies are available, as well as other alternative transgenic lines with different fluorescent proteins, including Cerulean and Venus, that are also appropriate for apoptosis detection in both live and fixed tissues. The use of the GC3Ai sensor permitted the description of the first apoptosis-inducing BH3-only protein (sayonara) in D. melanogaster (Ikegawa et al., 2023).
For its part, the Drice-based sensor (DBS) is another useful PCD sensor. Drice is a critical effector of apoptotic caspases, and after a two-step enzymatic process involving Dronc-mediated cleavage, forms two subunits—large and small—that associate to create the active caspase (Lannan et al., 2007). Baena-Lopez and collaborators developed a genetically encoded reporter system to detect early cell death stages, termed CD8-DriceC211A-short-Histone-GFP (DBS-S). Without caspase activation, DBS-S remains outside the nucleus. Upon cell death induction, the Drice subunit excision allows Histone-GFP to translocate to the nucleus, correlating with cleaved caspase-3 immunoreactivity (Baena-Lopez et al., 2018). The sensor allows studying temporal dynamics of cell proliferation and apoptosis after DNA damage (Ruiz-Losada et al., 2022), as well as investigating the involvement of different caspases in the cell death process (Aggarwal et al., 2022). To further increase the uses of the DBS sensor, Histone-GFP was replaced with the transcriptional activator QF to generate the DBS-S-QF, enabling genetic manipulation of caspase-activating cells using the QUAS-Gal4/UAS system, and lineage tracing of cells activating by Dronc (Baena-Lopez et al., 2018; Reiff et al., 2019).
In certain circumstances, some cells survive PCD despite caspase activation, a phenomenon known as anastasis. The CasExpress sensor facilitates the identification of these cells by driving the expression of fluorescent proteins, transiently or permanently, in cells that persist after caspase activation (Ding et al., 2016). The use of this anastasis biosensor led to comprehend that cell survival after caspase activation is a physiological tissue repair mechanism that can be disrupted in an oncogene-driven overgrowth context (Sun et al., 2020). Another system to detect anastasis in D. melanogaster tissues is CaspaseTracker, a biosensor based on a caspase-activatable Gal4 and the G-TRACE fluorescent protein system (Tang et al., 2015).
PCD can also occur independently of caspase activity, as it occurs in autophagy or necrosis. For autophagy, the mCherry-DmAtg8a reporter was engineered to detect autophagosomes and autolysosomes (Nezis et al., 2009). A step forward is the double-tagged GFP-mCherry-DmAtg8a sensor, that is effective for detecting and discriminating the autophagosomes in yellow (red and green merged) and the autolysosomes in red fluorescence due to the acidic environment of the latter (Nezis et al., 2010).
In contrast, no genetic biosensor has been available to detect necrosis in vivo in any organism until 2024 when Necrosensor (Figure 2H) was developed (Nishida et al., 2024). This necrosis sensor employs the nuclear protein HMGB1 (high-mobility group box 1) as a marker, because HMGB1 is released during necrosis in tissue cultures (Scaffidi et al., 2002). By fusing HMGB1 with GFP, it enables the detection of necrosis in vivo without the need for live staining (Nishida et al., 2024).
As mentioned before, PCD is followed by the engulfment and degradation of dead cells by phagocytes through efferocytosis (Davidson and Wood, 2020; Zheng et al., 2017). One of the major limitations of fluorescent-based sensors in studying cell death is the high pH sensitivity of fluorescent proteins and their weak resistance to photo-quenching in the acidic conditions of lysosomes during phagocytosis (Shinoda et al., 2018). Mutating the CG3Ai sensor led to the creation of ph-CaspGFP, a GFP-based apoptosis sensor designed to resist photo-quenching. The red fluorescent pHlorina sensor tracks apoptotic corpses during phagosome acidification, increasing fluorescence as pH decreases. The combination of both generates the CharON sensor (Figure 2G), which shows GFP-positive cells undergoing apoptosis and increased fluorescence in phagocytes during efferocytosis in real-time (Raymond et al., 2022).
Conclusion and future perspective
The field of programmed cell death (PCD) has made significant strides, expanding beyond the classical apoptosis model to recognize a diverse array of pathways. This comprehensive understanding demands the refinement and adaptation of traditional techniques used to study PCD. The fruit fly has emerged as an invaluable model organism in this endeavour, providing insights into the intricate mechanisms of PCD in development, tissue homeostasis, and cancer biology. Its genetic malleability and decades of accumulated knowledge have facilitated the development of new methodologies for studying PCD.
Throughout the D. melanogaster life cycle, from embryonic stages to metamorphosis and adult tissues, PCD plays multifaceted roles. Researchers have identified and characterized various forms of cell death, including apoptosis, autophagy, necrosis, and emerging types such as parthanatos and phagoptosis. These findings highlight the complex regulation of PCD in diverse physiological contexts. Moreover, D. melanogaster has provided crucial insights into cancer-related cell death mechanisms, unveiling cellular competition, tumour microenvironment modulation, and the dual role of PCD in suppressing and promoting tumours (Diwanji and Bergmann, 2019). The progression of techniques used to study PCD, from traditional approaches to advanced genetic sensors, has equipped researchers with a powerful toolkit to investigate cell death processes at an unprecedented resolution. As new technologies emerge and our understanding deepens, the D. melanogaster model continues to offer immense potential for unravelling the complexities of PCD and its implications in development and disease, promising future breakthroughs in cell death research.
Author contributions
DT-L: Writing–original draft, Writing–review and editing. MD: Writing–review and editing. MU: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Work from our lab is supported by the Spanish National Grants PID2022-136859NB-I00, funded by MICIN/AEI/10.13039/501100011033/ and co-funded by FEDER, UE; the Proof-of-concept Grant R + D + i PDC2022-133387-I00, funded by MICIU/AEI/10.13039/501100011033 and European Union Next Generation EU/PRTR; the Severo Ochoa Grant CEX2021-001165-S, funded by MICIU/AEI/10.13039/501100011033; the IN.Pulse project INNVA1/2023/25, funded by IVACE+i from Comunitat Valenciana and FEDER; and the Excellence Grant PROMETEO/2021/027 from the Generalitat Valenciana. MU is supported by Fundación General CSIC’s ComFuturo programme, which has received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Sklodowska-Curie grant agreement No. 101034263. DT-L is a Spanish doctoral FPI fellow (PRE 2020-094990) funded by MICIU/AEI/10.13039/501100011033 and FSE invests in your future.
Acknowledgments
Figure 1 and panels E and F on Figure 2 were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aballay, A., and Ausubel, F. M. (2001). Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium -mediated killing. Proc. Natl. Acad. Sci. 98 (5), 2735–2739. doi:10.1073/pnas.041613098
Abrams, J. M., White, K., Fessier, L. I., and Steller, H. (1993). Programmed cell death during Drosophila embryogenesis. Development 117 (1), 29–43. doi:10.1242/dev.117.1.29
Adrados, I., García-López, L., Aguilar-Aragon, M., Maranillo, E., and Domínguez, M. (2024). Modeling childhood cancer in Drosophila melanogaster. In Methods cell biol., (185, 35–48). Academic Press. doi:10.1016/bs.mcb.2024.02.003
Aggarwal, P., Liu, Z., Cheng, G. Q., Singh, S. R., Shi, C., Chen, Y., et al. (2022). Disruption of the lipolysis pathway results in stem cell death through a sterile immunity-like pathway in adult Drosophila. Cell Rep. 39 (12), 110958. doi:10.1016/j.celrep.2022.110958
Arama, E., Bader, M., Srivastava, M., Bergmann, A., and Steller, H. (2006). The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 25 (1), 232–243. doi:10.1038/sj.emboj.7600920
Ariss, M. M., Islam, A., Critcher, M., Zappia, M. P., and Frolov, M. V. (2018). Single cell RNA-sequencing identifies a metabolic aspect of apoptosis in Rbf mutant. Nat. Commun. 9 (1), 5024. doi:10.1038/s41467-018-07540-z
Arya, R., and White, K. (2015). Cell death in development: signaling pathways and core mechanisms. Seminars Cell and Dev. Biol. 39, 12–19. doi:10.1016/j.semcdb.2015.02.001
Baechrecke, E. H. (2000). Steroid regulation of programmed cell death during Drosophila development. Cell Death Differ. 2000 7 (11), 1057–1062. doi:10.1038/sj.cdd.4400753
Baena-Lopez, L. A., Arthurton, L., Bischoff, M., Vincent, J.-P., Alexandre, C., and McGregor, R. (2018). Novel initiator caspase reporters uncover previously unknown features of caspase-activating cells. Development 145 (23), dev170811. doi:10.1242/dev.170811
Baonza, A., Tur-Gracia, S., Pérez-Aguilera, M., and Estella, C. (2022). Regulation and coordination of the different DNA damage responses in Drosophila. Front. Cell Dev. Biol. 10, 993257. doi:10.3389/fcell.2022.993257
Bardet, P.-L., Kolahgar, G., Mynett, A., Miguel-Aliaga, I., Briscoe, J., Meier, P., et al. (2008). A fluorescent reporter of caspase activity for live imaging. Proc. Natl. Acad. Sci. 105 (37), 13901–13905. doi:10.1073/pnas.0806983105
Baron, O., Boudi, A., Dias, C., Schilling, M., Nölle, A., Vizcay-Barrena, G., et al. (2017). Stall in canonical autophagy-lysosome pathways prompts nucleophagy-based nuclear breakdown in neurodegeneration. Curr. Biol. 27 (23), 3626–3642. doi:10.1016/j.cub.2017.10.054
Bass, B. P., Tanner, E. A., Mateos San Martín, D., Blute, T., Kinser, R. D., Dolph, P. J., et al. (2009). Cell-autonomous requirement for DNaseII in nonapoptotic cell death. Cell Death Differ. 2009 16 (10), 1362–1371. doi:10.1038/cdd.2009.79
Bergmann, A. (2022). Erebosis is a new type of cell death for tissue homeostasis in the Drosophila intestine. PLOS Biol. 20 (4), e3001614. doi:10.1371/journal.pbio.3001614
Bergmann, A., Agapite, J., McCall, K., and Steller, H. (1998). The Drosophila gene hid is a direct molecular target of ras-dependent survival signaling. Cell 95 (3), 331–341. doi:10.1016/S0092-8674(00)81765-1
Berry, D. L., and Baehrecke, E. H. (2007). Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131 (6), 1137–1148. doi:10.1016/j.cell.2007.10.048
Bjedov, I., Cochemé, H. M., Foley, A., Wieser, D., Woodling, N. S., Castillo-Quan, J. I., et al. (2020). Fine-tuning autophagy maximises lifespan and is associated with changes in mitochondrial gene expression in Drosophila. PLOS Genet. 16 (11), e1009083. doi:10.1371/journal.pgen.1009083
Bordet, G., Lodhi, N., Kossenkov, A., and Tulin, A. (2021). Age-related changes of gene expression profiles in Drosophila. Genes. 12 (12), 1982. doi:10.3390/genes12121982
Brown, G. C., and Neher, J. J. (2012). Eaten alive! Cell death by primary phagocytosis: ‘phagoptosis. Trends Biochem. Sci. 37 (8), 325–332. doi:10.1016/j.tibs.2012.05.002
Cagan, R. L., and Ready, D. F. (1989). The emergence of order in the Drosophila pupal retina. Dev. Biol. 136 (2), 346–362. doi:10.1016/0012-1606(89)90261-3
Chimata, A. V., Deshpande, P., Mehta, A. S., and Singh, A. (2022). Protocol to study cell death using TUNEL assay in Drosophila imaginal discs. Star. Protoc. 3 (1), 101140. doi:10.1016/j.xpro.2022.101140
Ciesielski, H. M., Nishida, H., Takano, T., Fukuhara, A., Otani, T., Ikegawa, Y., et al. (2022). Erebosis, a new cell death mechanism during homeostatic turnover of gut enterocytes. PLOS Biol. 20 (4), e3001586. doi:10.1371/JOURNAL.PBIO.3001586
Cong, B., and Cagan, R. L. (2024). Cell competition and cancer from Drosophila to mammals. Oncogenesis 13 (1), 1–7. doi:10.1038/s41389-023-00505-y
Davidson, A. J., and Wood, W. (2020). Phagocyte responses to cell death in flies. Cold Spring Harb. Perspect. Biol. 12 (4), a036350. doi:10.1101/cshperspect.a036350
DeAngelis, M. W., Coolon, J. D., and Johnson, R. I. (2021). Comparative transcriptome analyses of the Drosophila pupal eye. G3 Genes.|Genomes|Genetics 11 (1), jkaa003. doi:10.1093/g3journal/jkaa003
Debnath, J., Gammoh, N., and Ryan, K. M. (2023). Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 24 (8), 560–575. doi:10.1038/s41580-023-00585-z
Denton, D., and Kumar, S. (2015). Studying apoptosis in Drosophila. Cold Spring Harb. Protoc. 2015 (7), 609–613. pdb.top070433. doi:10.1101/pdb.top070433
Denton, D., Mills, K., and Kumar, S. (2008). Methods and protocols for studying cell death in Drosophila. In Methods enzymol., 446, 17–37. Academic Press. doi:10.1016/S0076-6879(08)01602-9
Denton, D., Shravage, B., Simin, R., Mills, K., Berry, D. L., Baehrecke, E. H., et al. (2009). Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 19 (20), 1741–1746. doi:10.1016/j.cub.2009.08.042
Dichtel-Danjoy, M. L., Ma, D., Dourlen, P., Chatelain, G., Napoletano, F., Robin, M., et al. (2012). Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ. 2013 20 (1), 108–116. doi:10.1038/cdd.2012.100
Dick, F. A., Goodrich, D. W., Sage, J., and Dyson, N. J. (2018). Non-canonical functions of the RB protein in cancer. Nat. Rev. Cancer 18 (7), 442–451. doi:10.1038/s41568-018-0008-5
Ding, A. X., Sun, G., Argaw, Y. G., Wong, J. O., Easwaran, S., and Montell, D. J. (2016). CasExpress reveals widespread and diverse patterns of cell survival of caspase-3 activation during development in vivo. ELife 5, e10936. doi:10.7554/eLife.10936
Diwanji, N., and Bergmann, A. (2019). Two sides of the same coin – compensatory proliferation in regeneration and cancer. Adv. Exp. Med. Biol. 1167, 65–85. doi:10.1007/978-3-030-23629-8_4
Dziedziech, A., and Theopold, U. (2022). Proto-pyroptosis: an ancestral origin for mammalian inflammatory cell death mechanism in Drosophila melanogaster. J. Mol. Biol. 434 (4), 167333. doi:10.1016/J.JMB.2021.167333
Ellis, H. M., and Horvitz, H. R. (1986). Genetic control of programmed cell death in the nematode C. elegans. Cell 44 (6), 817–829. doi:10.1016/0092-8674(86)90004-8
Eskelinen, E.-L., Reggiori, F., Baba, M., Kovács, A. L., and Seglen, P. O. (2011). Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 7 (9), 935–956. doi:10.4161/auto.7.9.15760
Etchegaray, J. I., Timmons, A. K., Klein, A. P., Pritchett, T. L., Welch, E., Meehan, T. L., et al. (2012). Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Dev. Camb. 139 (21), 4029–4039. doi:10.1242/dev.082776
Foley, K., and Cooley, L. (1998). Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development 125 (6), 1075–1082. doi:10.1242/dev.125.6.1075
Fuchs, Y., and Steller, H. (2011). Programmed cell death in animal development and disease. Cell 147 (4), 742–758. doi:10.1016/j.cell.2011.10.033
Gao, N., Zheng, Q., Wang, Y., Li, X., Li, Z., and Xiao, H. (2022). Wun2-mediated integrin recycling promotes apoptotic cell clearance in Drosophila melanogaster. Cell Death and Differ. 29 (12), 2545–2561. doi:10.1038/s41418-022-01039-3
García-López, L., Adrados, I., Ferres-Marco, D., and Dominguez, M. (2021). A Blueprint for cancer-related inflammation and host innate immunity. Cells 10 (11), 3211. doi:10.3390/cells10113211
Garelli, A., Gontijo, A. M., Miguela, V., Caparros, E., and Dominguez, M. (2012). Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Sci. (New York, N.Y.) 336 (6081), 579–582. doi:10.1126/science.1216735
Gavrieli, Y., Sherman, Y., and Ben-Sasson, S. A. (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119 (3), 493–501. doi:10.1083/jcb.119.3.493
Gomes, K. K., Dos Santos, A. B., Dos Anjos, J. S., Leandro, L. P., Mariano, M. T., Pinheiro, F. L., et al. (2023). Increased iron levels and oxidative stress mediate age-related impairments in male and female Drosophila melanogaster. Oxidative Med. Cell. Longev. 2023 (1), 7222462. doi:10.1155/2023/7222462
Goyal, L., McCall, K., Agapite, J., Hartwieg, E., and Steller, H. (2000). Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19 (4), 589–597. doi:10.1093/emboj/19.4.589
Greenwald, E. C., Mehta, S., and Zhang, J. (2018). Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118 (24), 11707–11794. doi:10.1021/acs.chemrev.8b00333
Hay, B. A., Huh, J. R., and Guo, M. (2004). The genetics of cell death: approaches, insights and opportunities in Drosophila. Nat. Rev. Genet. 5 (12), 911–922. doi:10.1038/nrg1491
Hay, B. A., Wassarman, D. A., and Rubin, G. M. (1995). Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83 (7), 1253–1262. doi:10.1016/0092-8674(95)90150-7
Hung, Y.-C., Huang, K.-L., Chen, P.-L., Li, J.-L., Lu, S. H.-A., Chang, J.-C., et al. (2021). UQCRC1 engages cytochrome c for neuronal apoptotic cell death. Cell Rep. 36 (12), 109729. doi:10.1016/j.celrep.2021.109729
Ikegawa, Y., Combet, C., Groussin, M., Navratil, V., Safar-Remali, S., Shiota, T., et al. (2023). Evidence for existence of an apoptosis-inducing BH3 -only protein, sayonara, in Drosophila. EMBO J. 42 (8), e110454. doi:10.15252/embj.2021110454
Jennings, B. H. (2011). Drosophila – a versatile model in biology and medicine. Mater. Today 14 (5), 190–195. doi:10.1016/S1369-7021(11)70113-4
Jiang, C., Baehrecke, E. H., and Thummel, C. S. (1997). Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124 (22), 4673–4683. doi:10.1242/dev.124.22.4673
Jiang, H., Kimura, T., Hai, H., Yamamura, R., and Sonoshita, M. (2022). Drosophila as a toolkit to tackle cancer and its metabolism. Front. Oncol. 12, 982751. doi:10.3389/fonc.2022.982751
John Cohen, J. (1991). Programmed cell death in the immune system. Adv. Immunol. 50, 55–85. doi:10.1016/S0065-2776(08)60822-6
Juhasz, G., and Neufeld, T. P. (2008). Experimental control and characterization of autophagy in Drosophila. In Methods mol. Biol., 445, 125–133). Totowa, NJ, United States: Humana Press. doi:10.1007/978-1-59745-157-4_8
Kanaan, D., Shklyar, B., Porat-Kuperstein, L., and Toledano, H. (2023). Live imaging of phagoptosis in ex vivo Drosophila testis. BIO-PROTOCOL 13 (6), e4637. doi:10.21769/BioProtoc.4637
Kerr, J. F. R., Wyllie, A. H., and Currie, A. R. (1972). Apoptosis: a basic biological phenomenon with wideranging implications in tissue kinetics. Br. J. Cancer 26 (4), 239–257. doi:10.1038/bjc.1972.33
Koren, E., and Fuchs, Y. (2021). Modes of regulated cell death in cancer. Cancer Discov. 11 (2), 245–265. doi:10.1158/2159-8290.CD-20-0789
Kumar, S., and Cakouros, D. (2004). Transcriptional control of the core cell-death machinery. Trends Biochem. Sci. 29 (4), 193–199. doi:10.1016/j.tibs.2004.02.001
Kurant, E., Axelrod, S., Leaman, D., and Gaul, U. (2008). Six-microns-under acts upstream of draper in the glial phagocytosis of apoptotic neurons. Cell 133 (3), 498–509. doi:10.1016/j.cell.2008.02.052
Kurtz, P., Jones, A. E., Tiwari, B., Link, N., Wylie, A., Tracy, C., et al. (2019). Drosophila p53 directs nonapoptotic programs in postmitotic tissue. Mol. Biol. Cell 30 (11), 1339–1351. doi:10.1091/mbc.E18-12-0791
Laczkó-Dobos, H., Maddali, A. K., Jipa, A., Bhattacharjee, A., Végh, A. G., and Juhász, G. (2021). Lipid profiles of autophagic structures isolated from wild type and Atg2 mutant Drosophila. Biochimica Biophysica Acta (BBA) - Mol. Cell Biol. Lipids 1866 (3), 158868. doi:10.1016/j.bbalip.2020.158868
LaJeunesse, D. R., Buckner, S. M., Lake, J., Na, C., Pirt, A., and Fromson, K. (2004). Three new Drosophila markers of intracellular membranes. BioTechniques 36 (5), 784–788. doi:10.2144/04365ST01
Lannan, E., Vandergaast, R., and Friesen, P. D. (2007). Baculovirus caspase inhibitors P49 and P35 block virus-induced apoptosis downstream of effector caspase DrICE activation in Drosophila melanogaster cells. J. Virology 81 (17), 9319–9330. doi:10.1128/JVI.00247-07
Lécuyer, E., Parthasarathy, N., and Krause, H. M. (2008). Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol. Biol. Clift. N.J. 420, 289–302. doi:10.1007/978-1-59745-583-1_18
Lee, G., Kim, J., Kim, Y., Yoo, S., and Park, J. H. (2018). Identifying and monitoring neurons that undergo metamorphosis-regulated cell death (metamorphoptosis) by a neuron-specific caspase sensor (Casor) in Drosophila melanogaster. Apoptosis 23 (1), 41–53. doi:10.1007/s10495-017-1435-6
Lei, Y., Liu, K., Hou, L., Ding, L., Li, Y., and Liu, L. (2017). Small chaperons and autophagy protected neurons from necrotic cell death. Sci. Rep. 7 (1), 5650. doi:10.1038/s41598-017-05995-6
Lettre, G., and Hengartner, M. O. (2006). Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 7 (2), 97–108. doi:10.1038/nrm1836
Li, M., Sun, S., Priest, J., Bi, X., and Fan, Y. (2019). Characterization of TNF-induced cell death in Drosophila reveals caspase- and JNK-dependent necrosis and its role in tumor suppression. Cell Death and Dis. 10 (8), 613. doi:10.1038/s41419-019-1862-0
Li, Y., Bagheri, P., Chang, P., Zeng, A., Hao, J., Fung, A., et al. (2022). Direct imaging of lipid metabolic changes in Drosophila ovary during aging using DO-SRS microscopy. Front. Aging 2, 819903. doi:10.3389/fragi.2021.819903
Long, S., Cao, W., Qiu, Y., Deng, R., Liu, J., Zhang, L., et al. (2024). The appearance of cytoplasmic cytochrome C precedes apoptosis during Drosophila salivary gland degradation. Insect Sci. 31 (1), 157–172. doi:10.1111/1744-7917.13240
Martin, D. N., Balgley, B., Dutta, S., Chen, J., Rudnick, P., Cranford, J., et al. (2007). Proteomic analysis of steroid-triggered autophagic programmed cell death during Drosophila development. Cell Death Differ. 2007 14 (5), 916–923. doi:10.1038/sj.cdd.4402098
McPhee, C. K., Balgley, B. M., Nelson, C., Hill, J. H., Batlevi, Y., Fang, X., et al. (2012). Identification of factors that function in Drosophila salivary gland cell death during development using proteomics. Cell Death Differ. 2013 20 (2), 218–225. doi:10.1038/cdd.2012.110
Mendes, C. S., Arama, E., Brown, S., Scherr, H., Srivastava, M., Bergmann, A., et al. (2006). Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep. 7 (9), 933–939. doi:10.1038/sj.embor.7400773
Mirzoyan, Z., Sollazzo, M., Allocca, M., Valenza, A. M., Grifoni, D., and Bellosta, P. (2019). Drosophila melanogaster: a model organism to study cancer. Front. Genet. 10, 51. doi:10.3389/fgene.2019.00051
Morata, G. (2021). Cell competition: a historical perspective. Dev. Biol. 476, 33–40. doi:10.1016/J.YDBIO.2021.02.012
Morata, G., and Ripoll, P. (1975). Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev. Biol. 42 (2), 211–221. doi:10.1016/0012-1606(75)90330-9
Mumbauer, S., Pascual, J., Kolotuev, I., and Hamaratoglu, F. (2019). Ferritin heavy chain protects the developing wing from reactive oxygen species and ferroptosis. PLOS Genet. 15 (9), e1008396. doi:10.1371/journal.pgen.1008396
Munnik, C., Xaba, M. P., Malindisa, S. T., Russell, B. L., and Sooklal, S. A. (2022). Drosophila melanogaster: a platform for anticancer drug discovery and personalized therapies. Front. Genet. 13, 949241. doi:10.3389/fgene.2022.949241
Nagy, P., Varga, Á., Kovács, A. L., Takáts, S., and Juhász, G. (2015). How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods 75, 151–161. doi:10.1016/j.ymeth.2014.11.016
Napoletano, F., Baron, O., Vandenabeele, P., Mollereau, B., and Fanto, M. (2019). Intersections between regulated cell death and autophagy. Trends Cell Biol. 29 (4), 323–338. doi:10.1016/j.tcb.2018.12.007
Napoletano, F., Gibert, B., Yacobi-Sharon, K., Vincent, S., Favrot, C., Mehlen, P., et al. (2017). p53-dependent programmed necrosis controls germ cell homeostasis during spermatogenesis. PLOS Genet. 13 (9), e1007024. doi:10.1371/journal.pgen.1007024
Nezis, I. P., Lamark, T., Velentzas, A. D., Rusten, T. E., Bjørkøy, G., Johansen, T., et al. (2009). Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 5 (3), 298–302. doi:10.4161/auto.5.3.7454
Nezis, I. P., Shravage, B. V., Sagona, A. P., Lamark, T., Bjørkøy, G., Johansen, T., et al. (2010). Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190 (4), 523–531. doi:10.1083/jcb.201002035
Nezis, I. P., Stravopodis, D. J., Papassideri, I., Robert-Nicoud, M., and Margaritis, L. H. (2000). Stage-specific apoptotic patterns during Drosophila oogenesis. Eur. J. Cell Biol. 79 (9), 610–620. doi:10.1078/0171-9335-00088
Nishida, H., Albero, A. B., Onoue, K., Ikegawa, Y., Sulekh, S., Sakizli, U., et al. (2024). Necrosensor: a genetically encoded fluorescent sensor for visualizing necrosis in Drosophila. Biol. Open 13 (1), bio060104. doi:10.1242/bio.060104
Ojha, S., and Tapadia, M. G. (2020). Transcriptome profiling identifies multistep regulation through E93, Forkhead and Ecdysone Oxidase in survival of Malpighian tubules during metamorphosis in Drosophila. Int. J. Dev. Biol. 64 (4-5–6), 331–341. doi:10.1387/ijdb.190190mt
Olivier, M., Hollstein, M., and Hainaut, P. (2010). TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2 (1), a001008. doi:10.1101/cshperspect.a001008
Overholtzer, M., Mailleux, A. A., Mouneimne, G., Normand, G., Schnitt, S. J., King, R. W., et al. (2007). A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131 (5), 966–979. doi:10.1016/J.CELL.2007.10.040
Park, J., Lee, J.-H., Lee, Y., Lee, D., Kim, M. J., and Choe, K.-M. (2020). Necrotic cell death induces melanotic mass formation in Drosophila. Biochem. Biophysical Res. Commun. 526 (4), 1106–1111. doi:10.1016/j.bbrc.2020.04.012
Park, J.-E., Lee, J., Ok, S., Byun, S., Chang, E.-J., Yoon, S.-E., et al. (2023). Wg/Wnt1 and Erasp link ER stress to proapoptotic signaling in an autosomal dominant retinitis pigmentosa model. Exp. and Mol. Med. 55 (7), 1544–1555. doi:10.1038/s12276-023-01044-7
Parvy, J. P., Hodgson, J. A., and Cordero, J. B. (2018). Drosophila as a model system to study nonautonomous mechanisms affecting tumour growth and cell death. BioMed Res. Int. 2018, 7152962. doi:10.1155/2018/7152962
Pazdera, T. M., Janardhan, P., and Minden, J. S. (1998). Patterned epidermal cell death in wild-type and segment polarity mutant Drosophila embryos. Development 125 (17), 3427–3436. doi:10.1242/dev.125.17.3427
Peng, J. J., Lin, S. H., Liu, Y. T., Lin, H. C., Li, T. N., and Yao, C. K. (2019). A circuit-dependent ROS feedback loop mediates glutamate excitotoxicity to sculpt the Drosophila motor system. ELife 8, e47372. doi:10.7554/ELIFE.47372
Peterson, C., Carney, G. E., Taylor, B. J., and White, K. (2002). Reaper is required for neuroblast apoptosis during Drosophila development. Development 129 (6), 1467–1476. doi:10.1242/dev.129.6.1467
Pinal, N., Calleja, M., and Morata, G. (2019). Pro-apoptotic and pro-proliferation functions of the JNK pathway of Drosophila: roles in cell competition, tumorigenesis and regeneration. Open Biol. 9 (3), 180256. doi:10.1098/rsob.180256
Pinto-Teixeira, F., Konstantinides, N., and Desplan, C. (2016). Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 590 (15), 2435–2453. doi:10.1002/1873-3468.12298
Raymond, M. H., Davidson, A. J., Shen, Y., Tudor, D. R., Lucas, C. D., Morioka, S., et al. (2022). Live cell tracking of macrophage efferocytosis during Drosophila embryo development in vivo. Science 375 (6585), 1182–1187. doi:10.1126/science.abl4430
Reiff, T., Antonello, Z. A., Ballesta-Illán, E., Mira, L., Sala, S., Navarro, M., et al. (2019). Notch and EGFR regulate apoptosis in progenitor cells to ensure gut homeostasis in Drosophila. EMBO J. 38 (21), e101346. doi:10.15252/embj.2018101346
Richardson, H., and Kumar, S. (2002). Death to flies: Drosophila as a model system to study programmed cell death. J. Immunol. Methods 265 (1–2), 21–38. doi:10.1016/S0022-1759(02)00068-6
Robin, M., Issa, A. R., Santos, C. C., Napoletano, F., Petitgas, C., Chatelain, G., et al. (2019). Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy 15 (5), 771–784. doi:10.1080/15548627.2018.1558001
Rogulja-Ortmann, A., Lüer, K., Seibert, J., Rickert, C., and Technau, G. M. (2007). Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development 134 (1), 105–116. doi:10.1242/dev.02707
Ruiz-Losada, M., González, R., Peropadre, A., Gil-Gálvez, A., Tena, J. J., Baonza, A., et al. (2022). Coordination between cell proliferation and apoptosis after DNA damage in Drosophila. Cell Death and Differ. 29 (4), 832–845. doi:10.1038/s41418-021-00898-6
Rusconi, J. C., Hays, R., and Cagan, R. L. (2000). Programmed cell death and patterning in Drosophila. Cell Death and Differ. 7 (11), 1063–1070. doi:10.1038/sj.cdd.4400767
Rusten, T. E., Lindmo, K., Juhász, G., Sass, M., Seglen, P. O., Brech, A., et al. (2004). Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7 (2), 179–192. doi:10.1016/j.devcel.2004.07.005
Saini, S., and Owusu-Ansah, E. (2023). Dissecting the mechanism of regulation of a ferroptosis-like form of cell death in Drosophila melanogaster. Front. Cell Death 2, 1209641. doi:10.3389/fceld.2023.1209641
Sarkissian, T., Timmons, A., Arya, R., Abdelwahid, E., and White, K. (2014). Detecting apoptosis in Drosophila tissues and cells. Methods 68 (1), 89–96. doi:10.1016/j.ymeth.2014.02.033
Sattler, R., and Tymianski, M. (2001). Molecular mechanisms of glutamate receptor-mediated excitotoxic neuronal cell death. Mol. Neurobiol. 2001 24 (1), 107–129. doi:10.1385/MN:24:1-3:107
Scaffidi, P., Misteli, T., and Bianchi, M. E. (2002). Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418 (6894), 191–195. doi:10.1038/nature00858
Schott, S., Ambrosini, A., Barbaste, A., Benassayag, C., Gracia, M., Proag, A., et al. (2017). A fluorescent toolkit for spatiotemporal tracking of apoptotic cells in living Drosophila tissues. Development 144 (20), 3840–3846. doi:10.1242/dev.149807
Shan, Y., and Mollereau, B. (2024). Non-canonical functions of regulated cell death machinery regulate cellular growth, invasion and the interplay between cell death modalities. Front. Cell Death 3, 1423805. doi:10.3389/fceld.2024.1423805
Shinoda, H., Shannon, M., and Nagai, T. (2018). Fluorescent proteins for investigating biological events in acidic environments. Int. J. Mol. Sci. 19 (6), 1548. doi:10.3390/ijms19061548
Shklyar, B., Shklover, J., and Kurant, E. (2013). Live imaging of apoptotic cell clearance during Drosophila embryogenesis. J. Vis. Exp. JoVE 78, 50151. doi:10.3791/50151
Simpson, P., and Morata, G. (1981). Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev. Biol. 85 (2), 299–308. doi:10.1016/0012-1606(81)90261-X
Sollazzo, M., Paglia, S., Di Giacomo, S., and Grifoni, D. (2023). Apoptosis inhibition restrains primary malignant traits in different Drosophila cancer models. Front. Cell Dev. Biol. 10, 1043630. doi:10.3389/fcell.2022.1043630
Spreij, T. E. (1970). Cell death during the development of the imaginal disks of Calliphora erythrocephala. Neth. J. Zoology 21 (3), 221–265. doi:10.1163/002829670X00295
Stefanatos, R. K. A., and Vidal, M. (2011). Tumor invasion and metastasis in Drosophila: a bold past, a bright future. J. Genet. Genomics 38 (10), 431–438. doi:10.1016/j.jgg.2011.09.004
Suijkerbuijk, S. J. E., Kolahgar, G., Kucinski, I., and Piddini, E. (2016). Cell competition drives the growth of intestinal adenomas in Drosophila. Curr. Biol. 26 (4), 428–438. doi:10.1016/j.cub.2015.12.043
Sun, G., Ding, X. A., Argaw, Y., Guo, X., and Montell, D. J. (2020). Akt1 and dCIZ1 promote cell survival from apoptotic caspase activation during regeneration and oncogenic overgrowth. Nat. Commun. 11 (1), 5726. doi:10.1038/s41467-020-19068-2
Sung, C. H., Schneider, B. G., Agarwal, N., Papermaster, D. S., and Nathans, J. (1991). Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. 88 (19), 8840–8844. doi:10.1073/pnas.88.19.8840
Tan, Y., Yamada-Mabuchi, M., Arya, R., St Pierre, S., Tang, W., Tosa, M., et al. (2011). Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138 (11), 2197–2206. doi:10.1242/dev.058826
Tang, H. L., Tang, H. M., Fung, M. C., and Hardwick, J. M. (2015). In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci. Rep. 5 (1), 9015. doi:10.1038/srep09015
Tarayrah-Ibraheim, L., Maurice, E. C., Hadary, G., Ben-Hur, S., Kolpakova, A., Braun, T., et al. (2021). DNase II mediates a parthanatos-like developmental cell death pathway in Drosophila primordial germ cells. Nat. Commun. 12 (1), 2285. doi:10.1038/s41467-021-22622-1
Timmons, A. K., Mondragon, A. A., Schenkel, C. E., Yalonetskaya, A., Taylor, J. D., Moynihan, K. E., et al. (2016). Phagocytosis genes nonautonomously promote developmental cell death in the Drosophila ovary. Proc. Natl. Acad. Sci. U. S. A. 113 (9), E1246–E1255. doi:10.1073/pnas.1522830113
To, T.-L., Piggott, B. J., Makhijani, K., Yu, D., Jan, Y. N., and Shu, X. (2015). Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. 112 (11), 3338–3343. doi:10.1073/pnas.1502857112
Truman, J. W., and Bate, M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125 (1), 145–157. doi:10.1016/0012-1606(88)90067-X
Van De Corput, M. P. C., Dirks, R. W., Van Gijlswijk, R. P. M., Van De Rijke, F. M., and Raap, A. K. (1998). Fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification for sensitive DNA and mRNA detection. Histochem. Cell Biol. 110 (4), 431–437. doi:10.1007/S004180050304
Velentzas, A. D., Anagnostopoulos, A. K., Velentzas, P. D., Mpakou, V. E., Sagioglou, N. E., Tsioka, M. M., et al. (2015). Global proteomic profiling of Drosophila ovary: a high-resolution, unbiased, accurate and multifaceted analysis. Cancer Genomics and Proteomics 12 (6), 369–384. Available at: http://david.abcc.ncifcrf.gov/home.jsp.
Vousden, K. H., and Prives, C. (2009). Blinded by the light: the growing complexity of p53. Cell 137 (3), 413–431. doi:10.1016/j.cell.2009.04.037
Wang, X., and Ge, P. (2020). Parthanatos in the pathogenesis of nervous system diseases. Neuroscience 449, 241–250. doi:10.1016/j.neuroscience.2020.09.049
White, K., Grether, M. E., Abrams, J. M., Young, L., Farrell, K., and Steller, H. (1994). Genetic control of programmed cell death in Drosophila. Science 264 (5159), 677–683. doi:10.1126/science.8171319
White, K., Tahaoglu, E., and Steller, H. (1996). Cell killing by the Drosophila gene reaper. Science 271 (5250), 805–807. doi:10.1126/science.271.5250.805
Wolff, T., and Ready, D. F. (1991). Cell death in normal and rough eye mutants of Drosophila. Development 113 (3), 825–839. doi:10.1242/dev.113.3.825
Xu, T., Nicolson, S., Sandow, J. J., Dayan, S., Jiang, X., Manning, J. A., et al. (2021). Cp1/cathepsin L is required for autolysosomal clearance in Drosophila. Autophagy 17 (10), 2734–2749. doi:10.1080/15548627.2020.1838105
Xu, W., and Xu, J. (2018). C9orf72 dipeptide repeats cause selective neurodegeneration and cell-autonomous excitotoxicity in Drosophila glutamatergic neurons. J. Neurosci. 38 (35), 7741–7752. doi:10.1523/JNEUROSCI.0908-18.2018
Yacobi-Sharon, K., Namdar, Y., and Arama, E. (2013). Alternative germ cell death pathway in Drosophila involves HtrA2/omi, lysosomes, and a caspase-9 counterpart. Dev. Cell 25 (1), 29–42. doi:10.1016/j.devcel.2013.02.002
Yalonetskaya, A., Mondragon, A. A., Elguero, J., and McCall, K. (2018). I spy in the developing fly a multitude of ways to die. J. Dev. Biol. 6 (4), 26. doi:10.3390/jdb6040026
Zhao, H., Shi, L., Kong, R., Li, Z., Liu, F., Zhao, H., et al. (2021). Autophagy induction in tumor surrounding cells promotes tumor growth in adult Drosophila intestines. Dev. Biol. 476, 294–307. doi:10.1016/j.ydbio.2021.04.008
Zheng, Q., Gao, N., Sun, Q., Li, X., Wang, Y., and Xiao, H. (2021). bfc, a novel serpent co-factor for the expression of croquemort, regulates efferocytosis in Drosophila melanogaster. PLOS Genet. 17 (12), e1009947. doi:10.1371/journal.pgen.1009947
Zheng, Q., Ma, A., Yuan, L., Gao, N., Feng, Q., Franc, N. C., et al. (2017). Apoptotic cell clearance in Drosophila melanogaster. Front. Immunol. 8 (DEC), 1881. doi:10.3389/fimmu.2017.01881
Keywords: programmed cell death, sensors, methods, -omics, development, cancer, Drosophila
Citation: Tendero-Lopez D, Dominguez M and Uribe ML (2024) Exploring advanced Drosophila cell death techniques and cancer-related studies. Front. Cell. Death 3:1478258. doi: 10.3389/fceld.2024.1478258
Received: 09 August 2024; Accepted: 08 November 2024;
Published: 26 November 2024.
Edited by:
Eugenia Piddini, University of Bristol, United KingdomReviewed by:
Francesco Napoletano, University of Trieste, ItalyBertrand Mollereau, Université de Lyon, France
Copyright © 2024 Tendero-Lopez, Dominguez and Uribe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary Luz Uribe, bXVyaWJlQHVtaC5lcw==
 Daniel Tendero-Lopez
Daniel Tendero-Lopez Maria Dominguez
Maria Dominguez Mary Luz Uribe
Mary Luz Uribe