- 1CAD & Atherosclerosis Department, Republican Specialized Center of Cardiology, Tashkent, Uzbekistan
- 2Center for Advanced Technologies, Ministry of Innovative Development of Uzbekistan, Tashkent, Uzbekistan
Objective: To assess the distribution of PCSK9 E670G genetic polymorphism and PCSK9 levels in patients with Coronary Artery Disease (CAD) and Heterozygous Familial Hypercholesterolemia (HeFH), based on the presence of type 2 Diabetes Mellitus (T2DM).
Methods: The study included 201 patients with chronic CAD, including those with HeFH (n=57, group I) and without it (n=144, group II). DLCN was used to diagnose HeFH. The PCSK9 E670G (rs505151) polymorphism was genetically typed using the PCR-RFLP procedure. In both the patient and control groups, the genotype frequency matched the Hardy-Weinberg equilibrium distribution (P>0.05).
Results: There were twice more G alleles in group I (13, 11.4%) than in group II (17, 6.0%), and thrice more (1, 3.0%) than in the healthy control group; nevertheless, these differences weren’t statistically significant. Simultaneously, PCSK9 levels were higher in HeFH patients (P<0.05) compared to non-HeFH patients not taking statins (n=63). T2DM was equally represented in groups I and II (31.6% vs. 33.3%). But carriers of AG+GG genotypes in group I had a higher chance of having a history of T2DM (RR 4.18; 95%CI 2.19-8.0; P<0.001), myocardial infarction (RR 1.79; 95%CI 1.18-2.73; P<0.05), and revascularization (RR 12.6; 95%CI 4.06-38.8; P<0.01), than AA carriers. T2DM was also more common among G allele carriers (RR 1.85; 95% CI 1.11-3.06; P<0.05) in patients with non-HeFH.
Conclusion: T2DM in patients with CAD, both with HeFH and non-HeFH, in the Uzbek population was significantly more often associated with the presence of the “gain-of-function” G allele of the PCSK9 E670G genetic polymorphism.
1 Introduction
In our investigation, the distribution of the gain-of-function (GOF) E670G PCSK9 mutation in the patients with HeFH who had prematurely acquired CAD was analysed. As is known, LDL-Cholesterol (LDL-C) has been officially declared the primary cause of Atherosclerotic Cardiovascular diseases (1). PCSK9 GOF mutations significantly increase LDL-C and the risk of CAD through accelerating the degradation of LDL receptors (2). HeFH is a clear example demonstrating the burden of genetically elevated LDL in the world (3).
Another leading cause of CAD is type 2 diabetes mellitus (T2DM): the risk increases by two to four times (4). Up to two-thirds of people with T2DM have Atherosclerotic Cardiovascular Diseases (ASCVD) during their lifetimes (5) and pass away as a result (6). Therefore, the position of leading European guidelines seems well-founded, stating that patients with T2DM should always be considered high-risk or higher (7). Dyslipidemia is the main damaging factor in CAD and T2DM, and an incorrect risk scale leads to frequent underestimation of the disease, resulting in inadequate treatment (8).
As a result, a triangle of issues including CAD, T2DM, and PCSK9 has to be resolved. It is noteworthy that GOF PCSK9 mutations, which initially served as a key discovery in understanding the role of PCSK9 in DLP, including FH (9) subsequently took a backseat to LOF mutations (10), despite their rare occurrence - about 1.5-2.5% in the population (11, 12), and thus their infrequency in HeFH with CAD. However, LOF genetic variants became the prototype for the creation of a generation of targeted PCSK9 therapy drugs, which showed unprecedented efficacy in treating dyslipidemia both in CAD and in CAD with T2DM (13, 14). For the first time, Abifadel et al. (2009) established the modulating effect of the PCSK9 loss-of-function (LOF) genetic variant in FH patients with the LDLR mutation in the Lebanese population, leading to a reduction in LDL cholesterol levels (15). It has also been shown that in FH patients with the LDLR mutation in the Japanese population, the addition of the PCSK9 gain-of-function (GOF) genetic variant is accompanied by an increase in PCSK9 and LDL-C levels (16). Subsequently, a study by Tada et al. (2016) found that the modulatory effects of PCSK9 GOF genetic variants increased the incidence of T2DM (17) compared with patients in whom FH was caused by the LDLR mutation alone (P<0.05).
Warden et al. found a “poor” response to PCSK9 inhibitor therapy in 13.1% of patients, linked to various reasons, including mutations in the PCSK9, LDLR, and other genes (18). Currently, only 1% of patients in Europe receive PCSK9 inhibitors (19). However, the need to expand targeted PCSK9 therapy in the future requires an in-depth study of personalized approaches to their use in both CAD and CAD with diabetes.
The aim of our study was to assess the distribution of PCSK9 E670G genetic polymorphism and PCSK9 levels in patients with CAD and HeFH, based on the presence of T2DM.
2 Materials and methods
The study comprised 57 patients with chronic stable CAD and HeFH (I group), who were diagnosed for the first time, and were not taking statins. There were 144 CAD patients in the control group (group II) without HeFH. This group was further divided into subgroup A (n=63), composed of patients who had not taken statins prior to the study, and subgroup B (n=81), made up of patients who regularly took statins at doses prescribed by general practitioners in Uzbekistan: simvastatin 20 mg/day, atorvastatin 10–20 mg/day, rosuvastatin 10 mg/day.
To diagnose HeFH, we used the Dutch Lipid Clinic Network Criteria (DLCN), which is recommended by the European Atherosclerosis Society (EAS) (20). The study included 35 patients with clinically verified probable HeFH scores (6–8). In 21 cases, HeFH was confirmed by an LDL-R gene mutation and in 1 case by an ApoB mutation.
Patients with unstable angina, myocardial infarction in the previous 3 months, chronic renal and hepatic insufficiency, atrial fibrillation, life-threatening ventricular arrhythmias, chronic heart failure above functional class I (NYHA) were not included in the study.
The following clinical and functional parameters were studied in all patients to verify the diagnosis of chronic stable CAD and determine inclusion and exclusion criteria for the study: complaints, family history, medical history, physical examination, presence of T2DM, 12-lead ECG, echocardiography (EchoCG), 24-hour Holter ECG monitoring, exercise stress test.
2.1 Biochemical tests
Determination of lipid profile. Blood sampling was carried out on the next day after admission to hospital, in the morning, after 12 hours of fasting, from the cubital vein, in the horizontal position of the patient. Determination of blood lipid levels – total cholesterol (TC), high density lipoprotein (HDL), triglycerides (TG) - performed by enzymatic method on biochemical analyzer “Daytona” (RANDOX, UK). The concentration of LDL-C was determined by the Fridvald’s formula: LDL-C=TC-HDL-TG/5 (mg/dl).
Concentration of high-sensitive C-reactive protein (hsCRP) was determined by a highly sensitive particle-enhanced turbidimetric immunoassay method with latex amplification using “Daytona” (RANDOX, UK).
The level of PCSK9 in the blood was measured using an enzyme-linked immunoassay, utilizing a commercially available “Human Proprotein Convertase 9/PCSK9 ELISA Kit” (MULTI SCIENCE, China).
2.2 Determination of genotypic frequencies
The genotypic frequency at the PCSK9 E670G (rs505151) polymorphism was determined using the PCR-RFLP method (21). To amplify the polymorphic gene sequences of E670G of the PCSK9 gene, the following forward primer 5′- CAC GGT TGT GTC CCA AAT GG -3′ and reverse primer 5′- GAG AGG GAC AAG TCG GAA CC -3′ were used.
The PCR reaction was conducted in a 10 μl mixture containing 1X reaction buffer, 1.5mM MgCl2, 0.2mM dNTP, 0.3 μM primers, 25 ng template DNA, and 0.5 U Taq DNA polymerase (Applied Biosystems, Foster City, USA). The amplification was carried out using a GeneAmp 9700 (Applied Biosystems, Foster City, USA), with an initial denaturation at 94°C for 3 minutes, followed by 35 cycles each of 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C, plus a final extension of 5 minutes at 72°C. The specific amplified fragment was 440 bp long. To identify the E670G polymorphism of the PCSK9 gene, the amplified PCR product was subjected to restriction digestion. The reaction was carried out in a volume of 10 μl, containing 5 μl of the PCR product, 1 μl of the 10x restriction buffer, 0.2 U of the restriction endonuclease Bst6I (OOO SibEnzyme, Novosibirsk), and sterile, non-ionized water. Digestion was performed at 65°C for 16 hours.
After digestion, 5 μl of the reaction mixture was run on a 2% agarose gel for 30 minutes at a constant voltage of 100 V in 0.5x TBE buffer. After staining with ethidium bromide, the fragments were visualized using a gel photo-documentation system. Genotyping was performed as follows: a 440 bp fragment corresponded to the GG genotype, fragments of 150 bp and 290 bp represented the AA genotype, and heterozygotes (AG) had fragment sizes of 440, 290, and 150 bp.
2.3 Statistical analysis
Statistica 10.0 advanced statistical analysis package was used. The collected data was shown as mean and standard deviation (m ± SD). The statistical importance of the noted measurements for the compared mean values were determined using the Student’s t-test (t) with a calculated error probability (p) to confirm the normality of the distribution. If the distribution of studied variables varied from the normal distribution, then non-parametric analysis tests were made. Specifically, the Wilcoxon signed-rank test was used for comparing two related samples, matched samples, or repeated measurements, and the Mann-Whitney U test was used for comparing the two samples. For searching differences between qualitative statistical measures, the χ2 method was used in simultaneity with Fisher’s exact test for samples that were smaller. The empirical genotype frequency distribution’s conformity to the theoretically expected Hardy-Weinberg equilibrium was estimated using the χ2 test.
3 Results
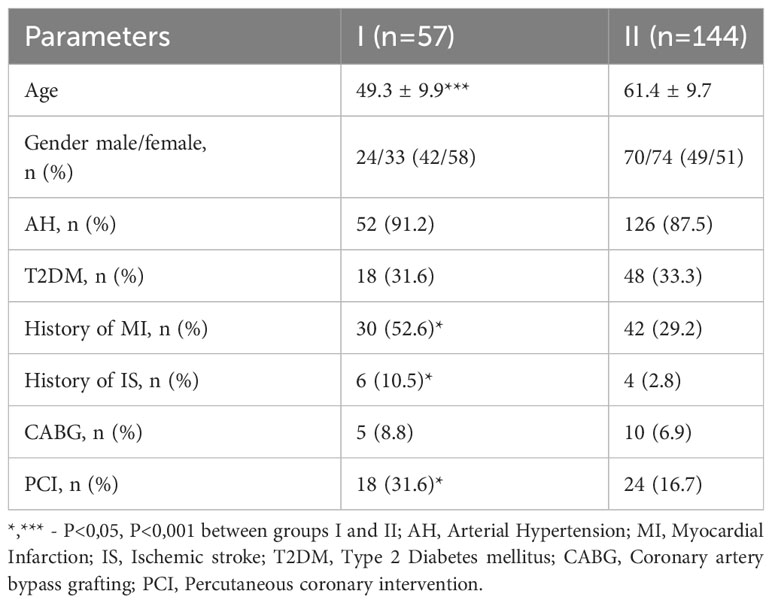
Patients with CAD and HeFH were younger than those in the comparison group (P<0.001), but they often had a history of MI (P<0.05), stroke (P<0.05), and PCI (P<0.05, Table 1). T2DM was present in 66 patients and was equally represented between groups I and II (31.6% vs. 33.3%).
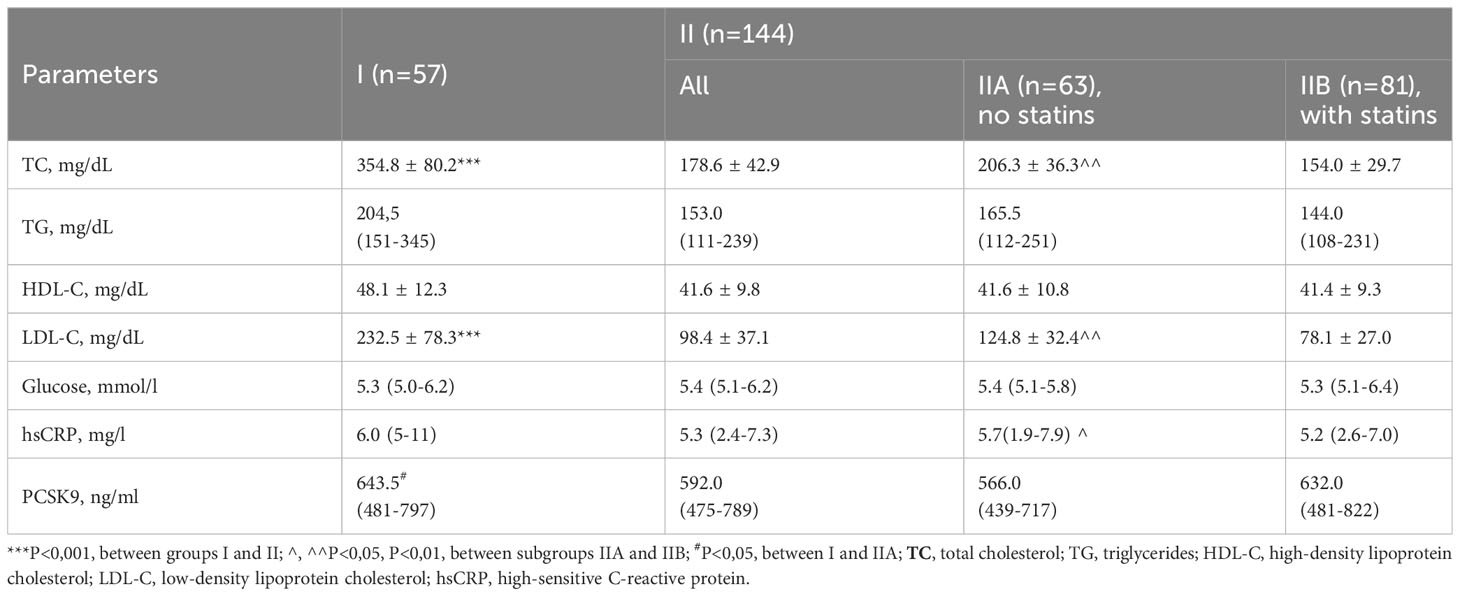
In group I, among patients with HeFH, Total (P<0.001) and LDL cholesterol (P<0.001) were significantly higher than in group II. Additionally, the PCSK9 level was higher (P<0.05) in I group as opposed to subgroup IIA, which consisted of patients who had not used statins before the study began. Subgroup IIA demonstrated increased levels of Total cholesterol (P<0.01), LDL-C (P<0.01), and hsCRP (P<0.05) when set against subgroup IIB, comprising patients who were on statins before entering the study (Table 2).
Both the patient group and the healthy individual’s genotype frequencies for the PCSK9 E670G (rs505151) polymorphism were consistent with the Hardy-Weinberg equilibrium (P>0.05).
The occurrence of G alleles was doubled in group I (13, representing 11.4%) when compared to group II (17, at 6.0%), and tripled in comparison to the healthy Uzbek population (1, or 3.0%). These differences, however, weren’t statistically significant (Table 3).
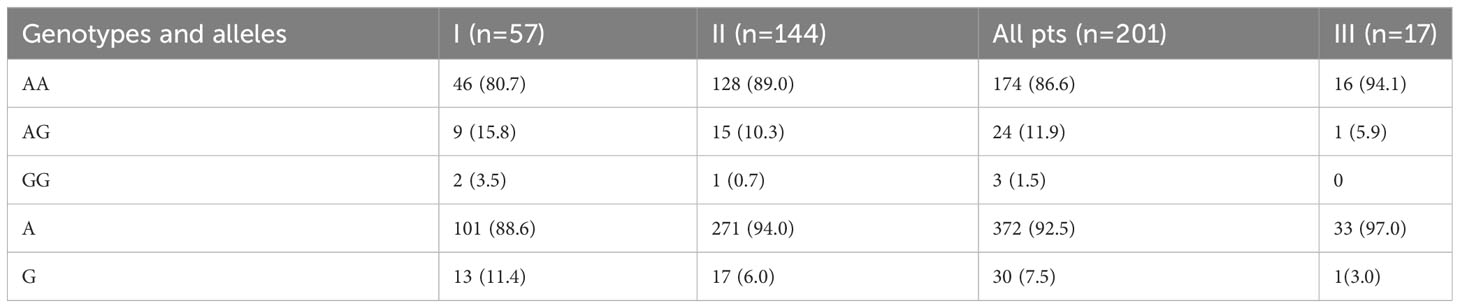
Table 3 Distribution of genotypes and alleles of PCSK9 E670G (rs505151) polymorphism in I (HeFH), II (non-HeFH) and III (healthy Uzbek persons) groups, n (%).
We looked at the clinical traits of patients based on whether they carried the “damaging” G allele since it is a “gain-of-function” mutation that is uncommon (Table 4). Among HeFH patients who were carriers of the AG+GG genotypes, a presence of T2DM (RR 4.18; 95% CI 2.19-8.0; P<0.001), history of myocardial infarction (RR 1.79; 95% CI 1.18-2.73; P<0.05), and any revascularization (RR 12.6; 95% CI 4.06-38.8; P<0.01) were significantly more common. T2DM was also more common among G allele carriers (RR 1.85; 95% CI 1.11-3.06; P<0.05) in patients with non-HeFH.
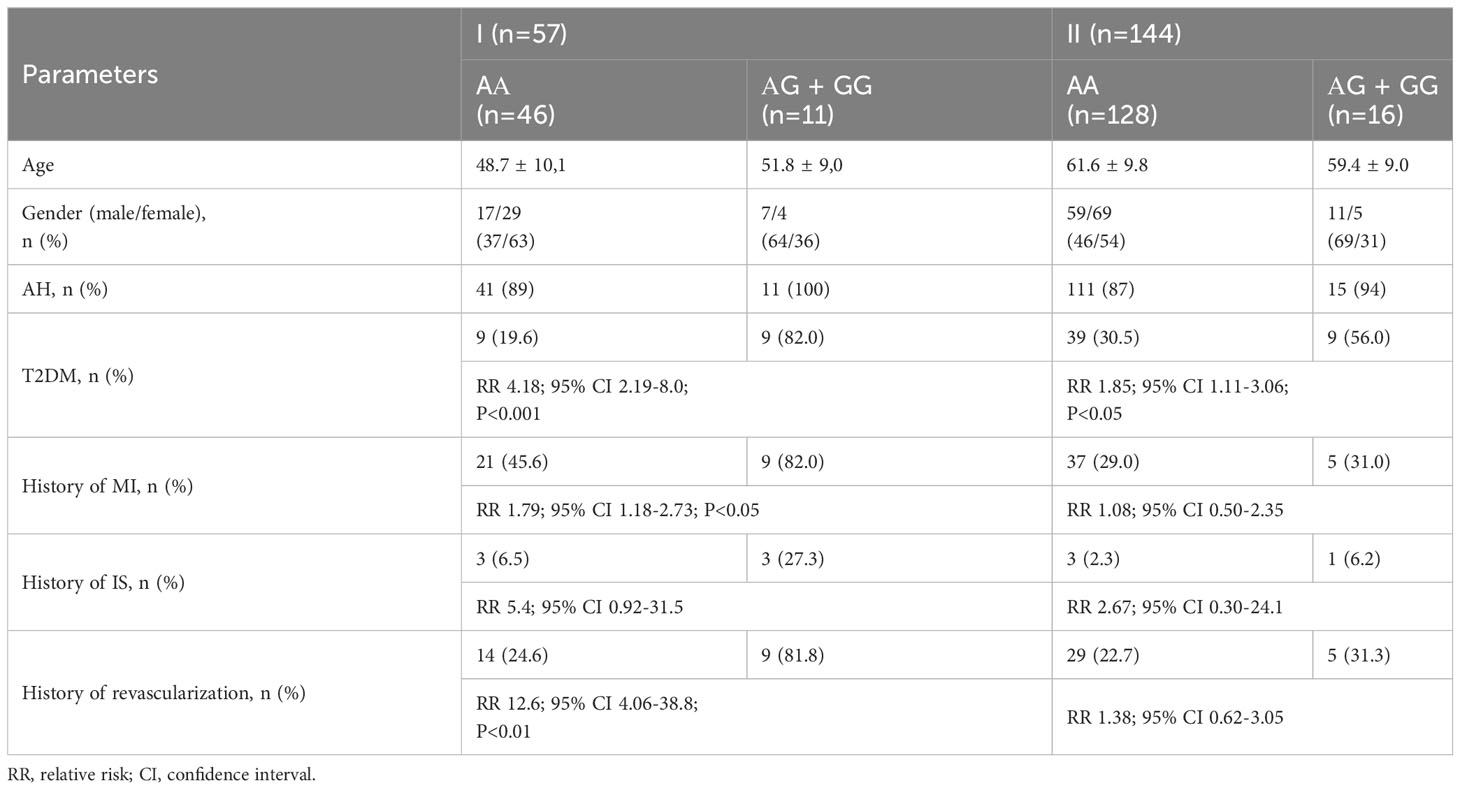
Table 4 Baseline clinical and demographic parameters of patients in I (HeFH) and II (non-HeFH) groups depending on the carriage of the G allele.
For an objective comparison of lipid and PCSK9 levels depending on the distribution of genotypes, we selected subgroup IIA of patients with non-HeFH (n=63) who did not take lipid-lowering therapy before inclusion in the study (Table 5). This decision was made because statins can contribute to an increase in PCSK9 levels by lowering lipid levels.
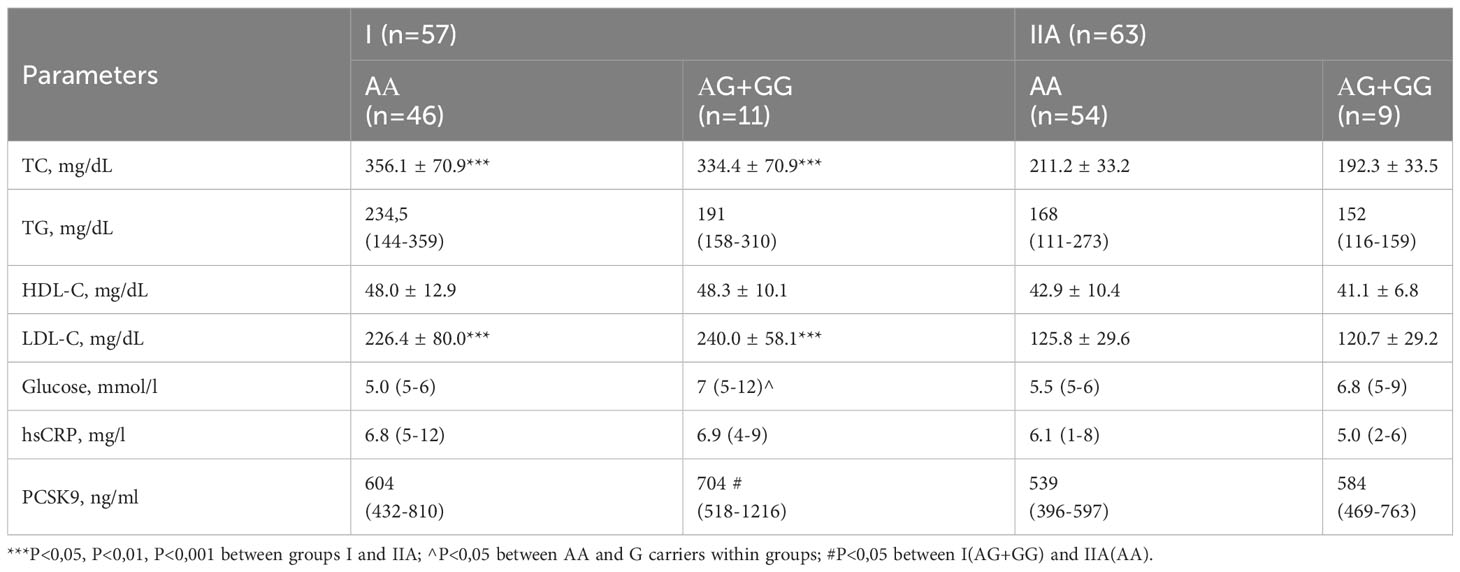
Table 5 Baseline levels of lipids and studied biomarkers in I (HeFH) and IIA (non-HeFH, statin free) groups depending on the carriage of the G allele.
The levels of Total (P<0.001) and LDL cholesterol were, as expected, higher in group I than in group IIA. In group I, the median glucose level was higher in AG+GG carriers, as they were significantly more likely to have T2DM.
The variability of the PCSK9 level and the relatively small number of carriers of the AG+GG genotype in groups I and II made it challenging to identify significant differences in PCSK9 concentration within the groups. However, the distribution median was nominally higher in the carriers of the G allele. Importantly, the level of PCSK9 was significantly higher in carriers of the AG+GG genotype in group I compared to AA carriers in subgroup IIA, although AA carriers in group I did not demonstrate such an effect.
4 Discussion
The results of our study showed an assoсiation between the carriage of “gain-of-function” G allele of the PCSK9 E670G genetic polymorphism and the incidence of T2DM in CAD patients with HeFH and non-HeFH, in the Uzbek population. Obviously, this assoсiation may indicate the influence of genetically elevated PCSK9 on the development of coronary artery disease and T2DM simultaneously.
In the Dallas Heart Study (22), which was a large multi-ethnic study, PCSK9 levels varied over a substantial 100-fold range (33–2988 ng/ml; median, 487 ng/ml). This variation depended on factors such as gender, metabolic status, and genetics. Specifically, PCSK9 levels were found to be higher in individuals with metabolic syndrome, insulin resistance, and elevated fasting glucose levels. A wealth of subsequent studies confirmed that an increase in circulating PCSK9 is associated not only with hypercholesterolemia but also with insulin resistance, increased glucose levels, and diabetes mellitus (23–25).
However, Saavedra et al. (26) genotyped 724 patients with FH and found “loss-of-function” (LOF) PCSK9-InsLEU variant in 26%. At that, pre-diabetes and diabetes mellitus were more often observed in InsLEU-carrier group compared with non-carriers (10% vs. 6%, P=0.04). InsLEU carriage was not accompanied by a decrease in lipids, however, FH patients with LOF mutation had fewer coronary events (22.1% vs. 30.6%, P=0.04). The assessment of pre-diabetes and diabetes was extracted from an electronic patient database, where the diagnoses were formulated by the responsible physicians, following the ADA’s protocols (American Diabetes Association). Impaired fasting glucose (IFG) was evaluated according to the same guidelines, with a glucose cut-off value of ≥5.6 mmol/l.
Lorenzo Da Dalt et al. (27) have shown in the experimental study that genetic deletion of PCSK9 in mice contributes to impaired glucose tolerance due to impaired insulin secretion, but not insulin resistance. This could possibly be explained by an increase in LDL receptor activity in pancreatic beta-cells in response to decreased PCSK9, with subsequent cholesterol accumulation, cellular dysfunction, and impaired insulin secretion.
In contrast, a group of French researchers by Langhi et al. (28) showed that PCSK9 is localized in human pancreatic d-cells, is not expressed in beta-cells, and is also not secreted by isolated murine islets in culture. Fasting glucose levels were similar in their experiment between PCSK9+/+ and PCSK9_/_ mice. Also, in some other studies, LOF PCSK9 variants InsLEU (1745 healthy French-Canadian individuals) and R46L (5972 French individuals) did not demonstrate an effect on glucose homeostasis and insulin resistance (29, 30).
Randomized controlled trials investigating the safety and efficacy of evolocumab (31) and alirocumab (32) in patients with and without T2DM also do not indicate an increase in glucose level, glycated hemoglobin and an increased incidence of T2DM compared to standard therapy, however, it may be necessary to wait for long-term results of studies.
In 2005, Chen et al. (31) studied GOF genetic polymorphisms PCSK9 in patients who participated in LCAS study of fluvastatin efficacy (n=372) and in another independent population in TexGen study (n=319). They found that carriage of G-allele of the E670G was associated with an increase in LDL-C and coronary atherosclerosis. In the LCAS study, which predominantly involved Caucasians (90%), the G allele frequency stood at 7.4%. Meanwhile, in the TexGen study with 79% Caucasians, the G allele frequency was 4.4%.
Another study by Evans and Beil (32), which encompassed 506 patients attending the University of Hamburg’s Lipid Clinic, found G allele carriers at an average frequency of 5%. However, in individuals with LDL-C levels above the 95th percentile, which consisted of patients with FH and polygenic hypercholesterolemia, the G allele frequency rose to 6.4%. CAD in G-carriers was significantly more common among men, but not among women.
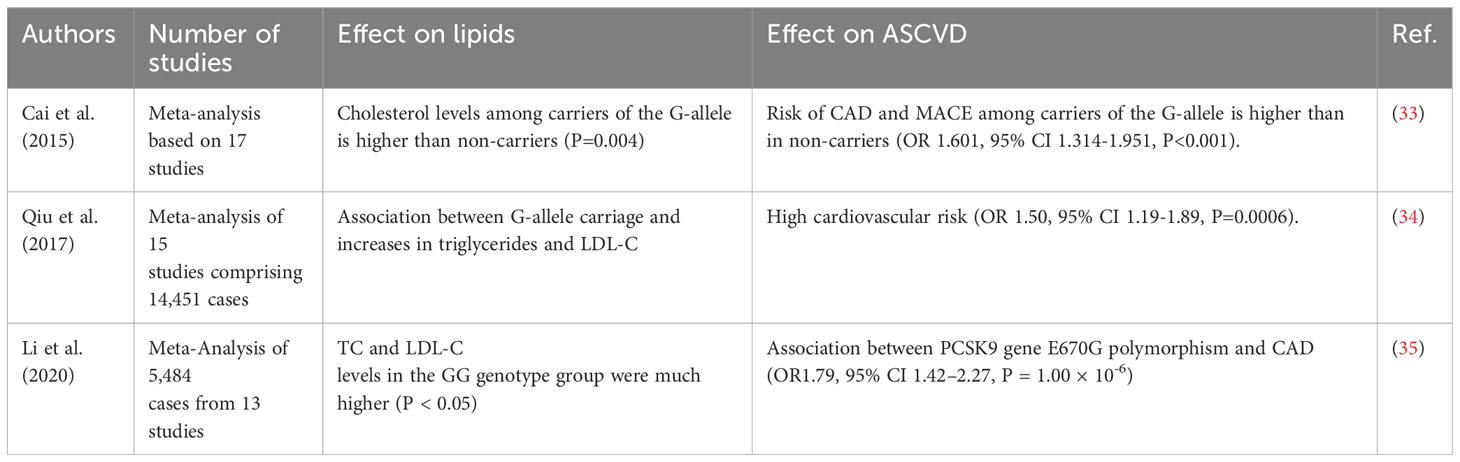
In our research involving an Uzbek population of 144 CAD patients, the G allele frequency was 6.0% (17 pts) but, this rate was doubled to 11.4% (13, including 2 GG) among the 57 FH patients. Data from large meta-analyses conducted in recent years confirm the association of carriage of the E670G G allele with increased lipid levels and the risk of CAD and MACE (Table 6).
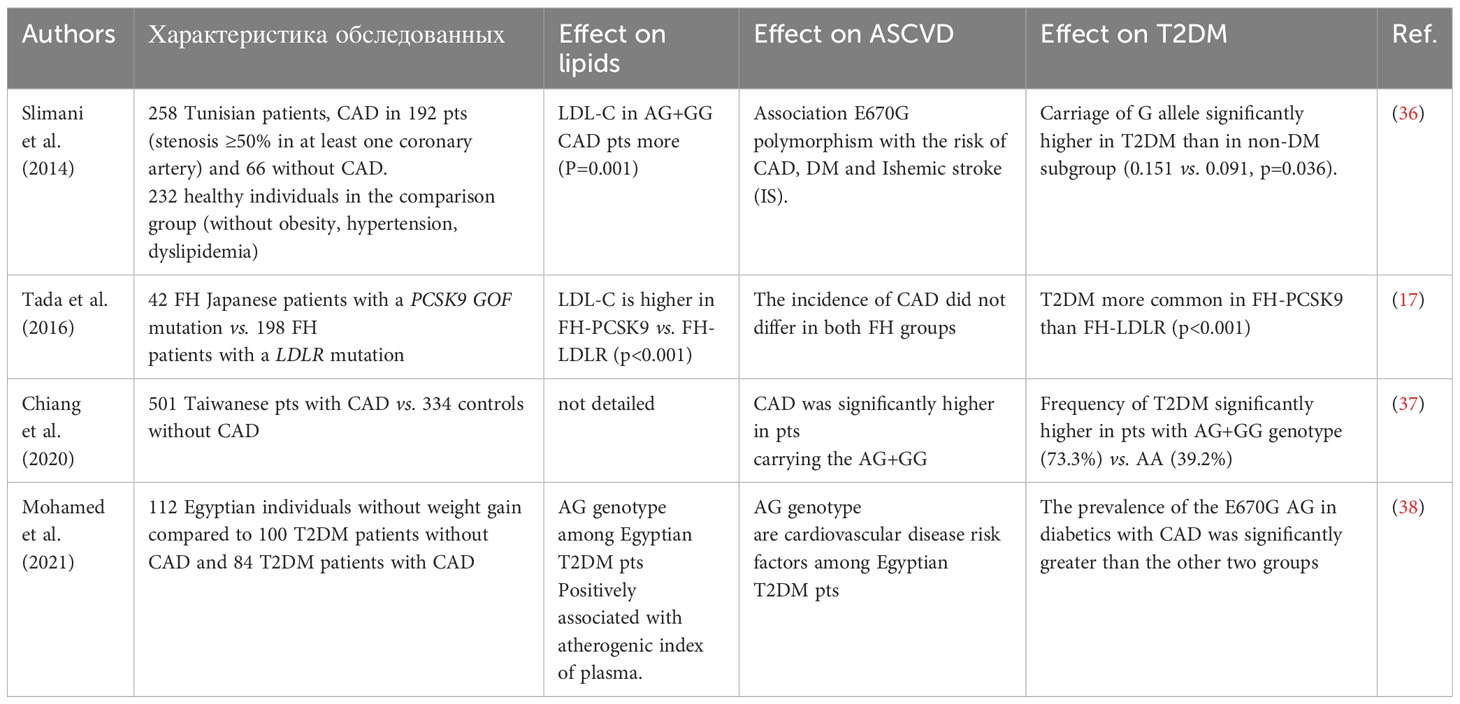
The association of E670G PCSK9 Polymorphism with T2DM has been established in several studies in patients of Asian ethnicity including with FH in recent years (Table 7).
In patients with CAD, in the Nothern Afrika and Asian populations (Tunisia, Egypt, Taiwan, Uzbekistan) in addition to elevated LDL-C and TG, the carriage of PCSK9 G-allele is associated with T2DM in some studies. This is consistently associated with increases in LDL cholesterol, triglycerides (TG), and CAD. Recent research confirms that an increase in circulating PCSK9 levels not only accompanies hypercholesterolemia but also leads to higher glucose levels, insulin resistance, and T2DM.
It can be observed that, unlike in the European population, the carriage of the G-allele E670G (rs505151) of PCSK9 genetic polymorphism in the Asian population occurs 2-3 times more frequently. Krittanawong et al. (2022) suggest that to optimize doses of PCSK9 targeted therapy, the PCSK9 R46L alleles should be tested in Europeans, while the PCSK9 E670G polymorphism should be considered in Asian populations (39). Considering recent research, T2DM should be considered another essential player in the selection of personalized therapy.
4.1 Conclusion
T2DM in patients with CAD, both with HeFH and non-HeFH, in the Uzbek population was significantly more often associated with the presence of the “gain-of-function” G allele of the PCSK9 E670G genetic polymorphism. This may have implications for the development of personalized PCSK9 targeted therapy in patients with CAD and T2DM.
4.2 Study limitation
The present study does have some limitations. It includes a relatively small number of patients with CAD (n=201), comprised of 57 patients with FH and 144 without FH. Among these, 30 G alleles were identified in 27 carriers (13.4%). Further risk stratification for this genetic marker is warranted, particularly among patients with coronary artery disease, diabetes mellitus, and FH.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethical Committee of the Republican Scientific and Practical Medical Center for Cardiology of the Ministry of Health of the Republic of Uzbekistan. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RA: Writing – review & editing, Conceptualization, Data curation, Funding acquisition, Project administration, Visualization, Writing – original draft. AS: Writing – original draft, Conceptualization, Validation. AA: Writing – review & editing, Methodology. KF: Writing – review & editing, Supervision. SK: Writing – original draft, Project administration, Validation. GA: Investigation, Writing – original draft. DZ: Writing – original draft, Methodology. RK: Writing – original draft, Funding acquisition, Project administration. LK: Data curation, Writing – original draft, Investigation. AK: Software, Writing – original draft, Data curation, Investigation.
Funding
The study was supported by a research grant from the Ministry of Innovative Development of Uzbekistan (State Registration No. PZ-202007041). The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ference BA, Ginsberg HN, Graham I, Ray KK, Packard CJ, Bruckert E, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur Heart J (2017) 38:2459–72. doi: 10.1093/eurheartj/ehx144
2. Guo Q, Feng X and Zhou Y. PCSK9 variants in familial hypercholesterolemia: A comprehensive synopsis. Front Genet (2020) 11:1020. doi: 10.3389/fgene.2020.01020
3. Vallejo-Vaz AJ, Stevens CAT, Lyons ARM, Dharmayat KI, Freiberger T, Hovingh GK, et al. EAS Familial Hypercholesterolaemia Studies Collaboration (FHSC). Global perspective of familial hypercholesterolaemia: a cross-sectional study from the EAS Familial Hypercholesterolaemia Studies Collaboration (FHSC). Lancet (2021) 398:1713–25. doi: 10.1016/S0140-6736(21)01122-3
4. Gregg EW, Sattar N, Ali MK. The changing face of diabetes complications. Lancet Diabetes Endocrinol (2016) 4(6):537–47. doi: 10.1016/S2213-8587(16)30010-9
5. Bancks MP, Ning H, Allen NB, Bertoni AG, Carnethon MR, Correa A, et al. Long-term absolute risk for cardiovascular disease stratified by fasting glucose level. Diabetes Care (2019) 42(3):457–65. doi: 10.2337/dc18-1773
6. Low Wang CC, Hess CN, Hiatt WR, Goldfine AB. ClinicalUpdate: Cardiovascular disease in diabetes mellitus: atherosclerotic cardiovascular disease and heart failure in type 2diabetes mellitus - mechanisms, management, and clinical considerations. Circulation (2016) 133(24):2459–502. doi: 10.1161/CIRCULATIONAHA.116.022194
7. Banach M, Burchardt P, Chlebus K, Dobrowolski P, Dudek D, Dyrbuś K, et al. PoLA/CFPiP/PCS/PSLD/PSD/PSH guidelines on diagnosis and therapy of lipid disorders in Poland 2021. Arch Med Sci (2021) 17(6):1447–547. doi: 10.5114/aoms/141941
8. Banach M, Surma S, Reiner Z, Katsiki N, Penson PE, Fras Z, et al. Personalized management of dyslipidemias in patients with diabetes-it is time for a new approach (2022). Cardiovasc Diabetol (2022) 21(1):263. doi: 10.1186/s12933-022-01684-5
9. Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet (2003) 34:154–6. doi: 10.1038/ng1161
10. Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet (2005) 37:161–5. doi: 10.1038/ng1509
11. Hopewell JC, Malik R, Valdes-Marquez E, Worrall BB, Collins R. ISGC MCot: Differential effects of PCSK9 variants on risk of coronary disease and ischaemic stroke. Eur Heart J (2018) 39:354–9. doi: 10.1093/eurheartj/ehx373
12. Cohen JC, Boerwinkle E, Mosley TH Jr., Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med (2006) 354(12):1264–72. doi: 10.1056/NEJMoa054013
13. Sabatine MS, Leiter LA, Wiviott SD, Giugliano RP, Deedwania P, Ferrari GM, et al. Cardiovascular safety and efficacy of the PCSK9 inhibitor evolocumab in patients with and without diabetes and the effect of evolocumab on glycaemia and risk of new-onset diabetes: a prespecified analysis of the FOURIER randomised controlled trial. Lancet Diabetes Endocrinol (2017) 5(12):941–50. doi: 10.1016/S2213-8587(17)30313-3
14. Ray KK, Colhoun HM, Szarek M, Cohen JC, Baccara-Dinet M, Bhatt DL, Bittner VA, et al. Effects of alirocumab on cardiovascular and metabolic outcomes after acute coronary syndrome in patients with or without diabetes: a prespecified analysis of the ODYSSEY OUTCOMES randomised controlled trial. Lancet Diabetes Endocrinol (2019) 7(8):618–28. doi: 10.1016/S2213-8587(19)30158-5
15. Abifadel M, Rabès JP, Jambart S, Halaby G, Gannagé-Yared MH, Sarkis A, et al. The molecular basis of familial hypercholesterolemia in Lebanon: spectrum of LDLR mutations and role of PCSK9 as a modifier gene. Hum Mutat (2009) 30(7):E682–91. doi: 10.1002/humu.21002
16. Noguchi T, Katsuda S, Kawashiri M, Tada H, Nohara A, Inazu A, et al. The E32K variant of PCSK9 exacerbates the phenotype of familial hypercholesterolaemia by increasing PCSK9 function and concentration in the circulation. Atherosclerosis (2010) 210(1):166–72. doi: 10.1016/j.atherosclerosis.2009.11.018
17. Tada H, Kawashiri MA, Yoshida T, Teramoto R, Nohara A, Konno T, et al. Lipoprotein(a) in familial hypercholesterolemia with proprotein convertase subtilisin/kexin type 9 (PCSK9) gain-of-function mutations. Circ J (2016) 80:512–8. doi: 10.1253/circj.CJ-15-0999
18. Warden BA, Miles JR, Oleaga C, Ganda OP, Duell PB, Purnell JQ, et al. Unusual responses to PCSK9 inhibitors in a clinical cohort utilizing a structured follow-up protocol. Am J Prev Cardiol (2020) 1:100012. doi: 10.1016/j.ajpc.2020.100012
19. Ray KK, Molemans B, Schoonen WM, Giovas P, Bray S, Kiru G, et al. DA VINCI study. EU-wide cross-sectional observational study of lipid-modifying therapy use in secondary and primary care: the DA VINCI study. Eur J Prev Cardiol (2020) 28(11): 1279–89. doi: 10.1093/eurjpc/zwaa047
20. Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease; consensus statement of the European Atherosclerosis Society. Eur Heart J (2013) 34:3478–90. doi: 10.1093/eurheartj/eht273
21. He X-M, Chen L, Wang T-S, Zhang Y-B, Luo J-B, Feng X-X. E670G polymorphism of PCSK9 gene of patients with coronary heart disease among Han population in Hainan and three provinces in the northeast of China. Asian Pac. J Trop Med (2016) 9(2):172–6. doi: 10.1016/j.apjtm.2016.01.008
22. Lakoski SG, Lagace TA, Cohen JC, Horton JD, Hobbs HH. Genetic and metabolic determinants of plasma PCSK9 levels. J Clin Endocrinol Metab (2009) 94(7):2537–43. doi: 10.1210/jc.2009-0141
23. Yang SH, Li S, Zhang Y, Xu RX, Guo YL, Zhu CG, et al. Positive correlation of plasma PCSK9 levels with HbA1c in patients with type 2 diabetes. Diabetes Metab Res Rev (2016) 32(2):193–9. doi: 10.1002/dmrr.2712
24. Ibarretxe D, Girona J, Plana N, Cabré A, Raimón Ferré R, Amigó N, et al. Circulating PCSK9 in patients with type 2 diabetes and related metabolic disorders. Clin Investig Arterioscler (2016) 28:71–8. doi: 10.1016/j.arteri.2015.11.001
25. Peng J, Liu MM, Jin JL, Cao YX, Guo YL, Wu NQ, et al. Association of circulating PCSK9 concentration with cardiovascular metabolic markers and outcomes in stable coronary artery disease patients with or without diabetes: a prospective, observational cohort study. Cardiovasc Diabetol (2020) 19(1):167. doi: 10.1186/s12933-020-01142-0
26. Saavedra YGL, Dufour R, Baass A. Familial hypercholesterolemia: PCSK9 InsLEU genetic variant and prediabetes/diabetes risk. J Clin Lipidology (2015) 9(6):786–793.e1. doi: 10.1016/j.jacl.2015.08.005
27. Da Dalt L, Ruscica M, Bonacina F, Balzarotti G, Dhyani A, Di Cairano E, et al. PCSK9 deficiency reduces insulin secretion and promotes glucose intolerance: the role of the low-density lipoprotein receptor. Eur Heart J (2019) 40:357–68. doi: 10.1093/eurheartj/ehy357
28. Langhi C, Le May C, Gmyr V, Vandewalle B, Kerr-Conte J, Krempf M, et al. PCSK9 is expressed in pancreatic delta-cells and does not alter insulin secretion. Biochem Biophys Res Commun (2009) 390:1288–93. doi: 10.1016/j.bbrc.2009.10.138
29. Awan Z, Delvin EE, Levy E, Genest J, Davignon J, Seidah NG, et al. Regional distribution and metabolic effect of PCSK9 insLEU and R46L gene mutations and apoE genotype. Can J Cardiol (2013) 29:927–33. doi: 10.1016/j.cjca.2013.03.004
30. Bonnefond A, Yengo L, Le May C, Fumeron F, Marre M, Balkau B, et al. The loss-of-function PCSK9 p.R46L genetic variant does not alter glucose homeostasis. Diabetologia (2015) 58(9):2051–5. doi: 10.1007/s00125-015-3659-8
31. Chen SN, Ballantyne CM, Gotto AM, Tan Y, Willerson JT, Marian AJ. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and the severity of coronary atherosclerosis. J Am Coll Cardiol (2005) 45:1611–9. doi: 10.1016/j.accreview.2005.09.096
32. Evans D, Beil FU. The E670G SNP in the PCSK9 gene is associated with polygenic hypercholesterolemia in men but not in women. BMC Med Genet (2006) 7:66. doi: 10.1186/1471-2350-7-66
33. Cai G, Zhang B, Shi G, Weng W, Ma C, Song Y. Zhang J.The associations between proprotein convertase subtilisin/kexin type 9 E670G polymorphism and the risk of coronary artery disease and serum lipid levels: a meta-analysis. Lipids Health Dis (2015) 14:149. doi: 10.1186/s12944-015-0154-7
34. Qiu C, Zeng P, Li X, Zhang Z, Pan B, Peng ZYF, et al. What is the impact of PCSK9 rs505151 and rs11591147 polymorphisms on serum lipids level and cardiovascular risk: a metaanalysis. Lipids Health Dis (2017) 16(1):111. doi: 10.1186/s12944-017-0506-6
35. Li Y-y, Wang H, Yang X-x, Geng H-y, Gong G, Lu X-z. PCSK9 gene E670G polymorphism and coronary artery disease: an updated meta-analysis of 5,484 subjects. Front Cardiovasc Med (2020) 7:582865. doi: 10.3389/fcvm.2020.582865
36. Slimani A, Harira Y, Trabelsi I, Jomaa W, Maatouk F, Hamda KB. Effect of E670G polymorphism in PCSK9 gene on the risk and severity of Coronary heart disease and ischemic stroke in a Tunisian cohort. J Mol Neurosci (2014) 53:150–7. doi: 10.1007/s12031-014-0238-2
37. Chiang SM, Yang YS, Yang SF, Tsai CF, Ueng KC. Variations of the proprotein convertase subtilisin/kexin type 9 gene in coronary artery disease. J Int Med Res (2020) 48:300060519839519. doi: 10.1177/0300060519839519
38. Mohamed SH, Hassaan MMM, Basma AI, Sabbah NA. PCSK9 E670G (rs505151) variant and coronary artery disease risk among diabetics. Genet Test Mol Biomarkers (2021) 25(9):615–23. doi: 10.1089/gtmb.2021.0010
Keywords: heterozygous familial hypercholesterolemia, CAD, type 2 diabetes mellitus, pcsk9, Uzbek population
Citation: Alieva R, Shek A, Abdullaev A, Fozilov K, Khoshimov S, Abdullaeva G, Zakirova D, Kurbanova R, Kan L and Kim A (2023) E670G PCSK9 polymorphism in HeFH & CAD with diabetes: is the bridge to personalized therapy within reach? Front. Clin. Diabetes Healthc. 4:1277288. doi: 10.3389/fcdhc.2023.1277288
Received: 14 August 2023; Accepted: 11 October 2023;
Published: 01 November 2023.
Edited by:
Belma Pojskic, Cantonal Hospital Zenica, Bosnia and HerzegovinaReviewed by:
Katarina Lalic, University of Belgrade, SerbiaErkin Mirrakhimov, Kyrgyz State Medical Academy, Kyrgyzstan
Maciej Banach, Polish Mother’s Memorial Hospital Research Institute, Poland
Copyright © 2023 Alieva, Shek, Abdullaev, Fozilov, Khoshimov, Abdullaeva, Zakirova, Kurbanova, Kan and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rano Alieva, cmFub2FsaWV2YUBtYWlsLnJ1
 Rano Alieva
Rano Alieva Aleksandr Shek
Aleksandr Shek Alisher Abdullaev
Alisher Abdullaev Khurshid Fozilov1
Khurshid Fozilov1