
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Built Environ. , 07 February 2023
Sec. Indoor Environment
Volume 9 - 2023 | https://doi.org/10.3389/fbuil.2023.1063804
This article is part of the Research Topic Adaptive Design Solutions to Buildings and the Built Environment after lessons learnt from the COVID-19 Pandemic View all 7 articles
Indoor residents are constantly exposed to dynamic microbiota that have significant health effects. In addition to hand hygiene, cleaning, and disinfection, antimicrobial coatings (AMCs) can prevent the spread of infectious diseases in public areas. The sustainable use of antimicrobial-coated products requires an assessment of their pros and cons for human health and the environment. The toxicity and resistance risks of AMCs have been considered, but large-scale genetic studies on the microbial community compositions and resistomes of AMCs are scarce. The use of an AMC can reduce the total number of microbes on a surface but poses the risk of dysbiosis, microbial imbalance, such as the polarized growth of metallophilic, metal- and antimicrobial-resistant, and other survivor bacteria, and the overall reduction of microbial diversity. Loss of diversity may lead to the enrichment of harmful bacteria and an increased risk of communicable or immunological non-communicable inflammatory diseases (NCDs). In public buildings, such as kindergartens and nursing homes for the elderly, the use of AMCs is likely to increase due to epidemics and pandemics in recent years. Therefore, comprehensive metagenomic research is needed to monitor the effects of AMCs on indoor microbial community compositions and functions. Although the determination of good indoor microbiota and homeostasis is difficult, microbial communities that have health-protective or harmful effects can and should be identified using a metagenomic sequencing approach before the large-scale implementation of AMCs.
The interactions between human hosts and microbes have changed due to population growth, diet change, urbanization, increased travel, habitat pollution, disinfectant use, synthetic construction, and utility material use, medications such as broad-spectrum antibiotics, and historical events in recent centuries (Davenport et al., 2017; Prescott et al., 2017; Parajuli et al., 2018; SanMiguel et al., 2018; Ahn and Hayes, 2021; Danko et al., 2021). The adaptation ability of humans to coexist with microbes in the natural environment has come a long way, raising the need for a comprehensive and long-term approach to understanding and combating challenges such as epidemics and pandemics. Our lives are completely dependent on microbes in and around us, and our coexistence with microbes usually involves a harmonious symbiosis of entities that maintain health and function (Kilian et al., 2016).
The human body is home to trillions of microbes residing in the small and large intestines, saliva, stomach, dental plaque, and skin. Each person has their own microbiota—a term that refers to all microbes within a certain area—and the organs in a human body are inhabited by different microbiota colonizing their niches (Sender et al., 2016; Scotti et al., 2017; Dekaboruah et al., 2020). For instance, the level of skin microbiota stays modulated throughout its lifetime, and the diversity of the gut microbiota increases and develops gradually during the first 3 years. However, factors such as birth mode, antibiotic exposure, and geographical location can change the species-specific abundance of gut microbiota. After early childhood, microbiota diversity continues to develop throughout an individual’s life. A Western lifestyle, which includes living indoors in a built environment with low exposure to natural microbes, can reduce microbial diversity in human habitats and bodies. (Yatsunenko et al., 2012; Moskovicz et al., 2020; Niu et al., 2020).
Indoor microbiota constitute an integral part of human life and are linked to health and wellbeing (Kumar et al., 2021). The community composition and dynamics of indoor microbes are affected by the features of the building (Table 1), such as heating, ventilation, water-damaged materials, and dust resuspension, the building’s occupants (e.g., humans, pets, and plants) and their behaviors, and the outdoor environment (Adams et al., 2015; Prussin and Marr, 2015). Microbial eradication is required to prevent the spread of infectious diseases in built environments, even though the need for beneficial microbes is known. Cleaning, disinfection, and hand hygiene are traditional methods of preventing the spread of infectious diseases. Antimicrobial coating (AMC) gained attention as a supplementary method during the COVID-19 pandemic, as contaminated surfaces provided considerable pathways for the spread of pathogens (Lei et al., 2017).
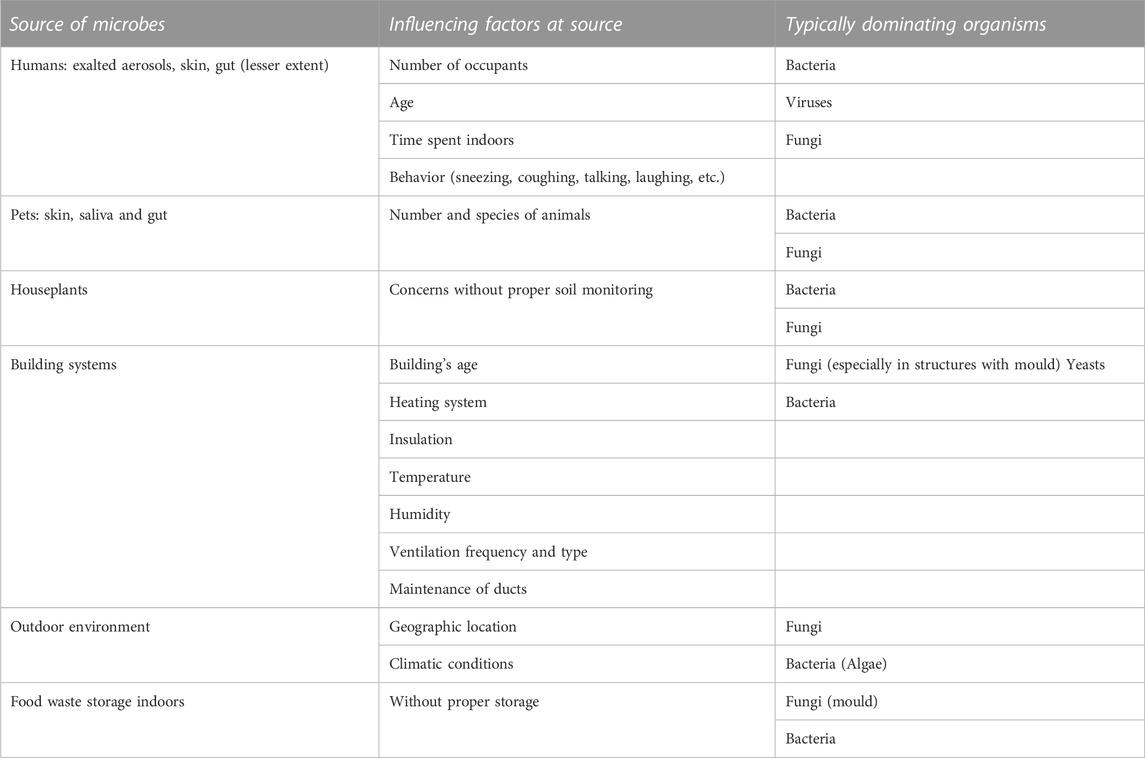
TABLE 1. Common microbial sources of surfaces and air in the indoor environment. The features of the building, the building’s occupants, and the outdoor environment affect the species and prevalence of the microbial organism (Prussin and Marr, 2015; Kumar et al., 2021). Fungal contamination with spores and mycotoxins, for example in damp houses, may cause increased health concerns. Some microbes are beneficial to health, especially from outdoors, and others are pathogenic.
Many important research results have already been published on COVID-19 infection prevention, but more solutions are needed. Riddell et al. (2020) showed that the viable SARS-CoV-2 virus could be isolated for up to 28 days at 20°C from common surfaces but the virus survived for less than 24 h at 40°C. Joonaki et al. (2020) called for research on the interaction between virus and surface. The molecular mechanisms that control the adsorption, desorption, and spreading of the virus from surfaces should be clarified. The effects of the chemistry of solid surfaces, such as pH values, relative humidity, and temperature, on virus adsorption and stability under varying conditions have not been adequately studied. The pandemic has shown that communities need to be prepared for emerging virus outbreaks. Social distancing, lockdown, case detection, isolation, contact tracing, and quarantine of exposed have been found to be the most efficient disease-controlling actions (Khanna et al., 2020). But the effectiveness of lockdowns is reduced due to insufficient or delayed action. Sweden’s strategy was initially to use the emergence of natural immunity to prevent the pandemic, which soon proved to be an ineffective measure (Stenseth et al., 2021). Global vaccination campaigns proved effective against COVID-19 disease spreading but the emergence of different variants of the virus (Figure 1) has weakened the effectiveness of vaccines (Corey et al., 2021; Planas et al., 2022) Intranasal administration of a prophylactic and therapeutic small-molecule compound or lipoprotein has shown promising effects in preventing and treating SARS-COVID-2 infection, but the studies are still incomplete. (de Vries et al., 2021; Shapira et al., 2022). Whether AMCs can be an effective, adaptive design solution for combating the spread of infections and keeping the indoor environment healthy remains to be determined.
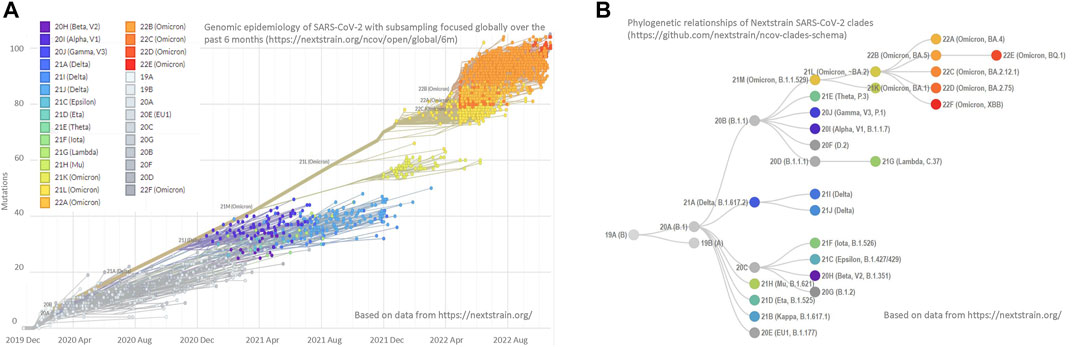
FIGURE 1. Distribution and phylogeny of SARS-Cov-2 variants (A) Data from GenBank has been used to visualize the genomic epidemiology of SARS-CoV-2 globally over the past 6 months. The graph shows mutations of 2,853 genomes sampled between December 2019 and October 2022 and is constructed by Nextstrain. (B) The chart visualizes the phylogenetic tree of SARS-CoV-2 variants. The phylogeny tree is enabled by Nextstrain (Hadfield et al., 2018).
The pros and cons of AMCs should be carefully weighed before their widespread use. Continuous development and safety evaluations of AMCs are needed to create increasingly better health-promoting elements (Imani et al., 2020). We have previously contributed to many surveys, reports, and publications concerning the benefits of AMCs and different innovations to prevent the spread of infectious diseases, and we have also critically considered antimicrobial resistance (AMR) and ecotoxicological issues related to AMCs (Ahonen et al., 2017; Rosenberg et al., 2019b; Dunne et al., 2020; Pietsch et al., 2020; Blomberg et al., 2022). In this paper, we take a closer look at microbial ecology concerns with regard to hypothetical microbial community compositions and functional changes in AMCs, which are issues that have received little attention in AMC studies. To our knowledge, no AMC innovations for indoor surfaces have been developed to selectively degrade pathogens and leave vital, health-beneficial microbes undisturbed. By focusing on the skewed structures of microbial communities that affect human health, we address the risk of AMR in metal-based AMCs and the non-selective manipulation of microbial growth by AMCs. Further, we justify the benefits of metagenomic research on AMC monitoring and, based on evolutionarily important nature-derived microbes, provide future prospects, and suggestions for shaping indoor microbiota in hygiene design to support human health.
Prior to the use of indoor antimicrobial solutions, care should be taken not to create conditions that expose pathogenic microbes to positive selection pressure within the microbiota. Although the idea of a living and dynamic microbiota as a health promoter is not yet widely accepted, studies have utilized the deliberate addition of benign microbes to control the negative health effects of pathogenic microbes (Horve et al., 2020). When the abundance of benign microbes decreases indoors, the competition between benign and pathogenic microbes for resources and a safe habitat reduces, allowing pathogens to become enriched in the microbiota if they can outcompete benign strains (Anttila et al., 2013; Bauer et al., 2018; Paquette et al., 2018). As Stevens et al. (2021) reported in their review, the interactions within a microbiota are complex. Although resident microbial species can form a defense against pathogens, resident microbiota can aid pathogenic species in proliferating via metabolic cross-feeding. In terms of human host health, the substantial benefits of microbiota outweigh the potential harms (Stevens et al., 2021). Microbes can also act as host defenders against parasites; defensive microbes can, for example, provide a complementary or substitute repertoire for host immunity, reduce parasite infectivity, produce toxic compounds, and kill parasites or reduce parasitic growth. The complex tripartite coevolution between hosts, defensive microbes, and parasites can contribute to the health of the human host (Ford and King, 2016; King and Bonsall, 2017).
It is important to look at the complex evolutionary history of microbes and humans, as the interplay between the two groups is changing significantly due to overpopulation, industrialization, urbanization, travel, and the use of antimicrobial products. Throughout our existence, human host control and microbial competition have formed an interplay that can explain the human health benefits of microbiota. Population genetics and community genetics can be utilized to model the evolutionary processes of hosts and microbiomes (a term that refers to the collection of all microbial genomes in a certain habitat). Evolutionary views may also come together to provide a universal perspective for studying indoor microbiota and their effects on human health (Davenport et al., 2017; Foster et al., 2017). For example, genetic mutations can alter the complex population genetic processes and functional characteristics of a microbiome, such as its ability to obtain nutrients from food, metabolize drugs, avoid antibiotic effects, and form an immune response in the host—traits that affect the health of a human host (Garud and Pollard, 2020). The microbiome of a host codes more genes than the host itself and can increase the range of the host genome, providing the host with an advantage of adapting to a new environment. Microbiota’s contributions to host phenotypic variations and evolution have been neglected in past studies, and many challenges still exist in this field of research. However, advancements in genetic characterization, such as metagenomic long-read third-generation sequencing, offer new possibilities for host–microbiome research into processes that influence evolutionary dynamics (Amarasinghe et al., 2020; Henry et al., 2021).
The microbiota in human habitats have changed dramatically because of intense urbanization over the last 2 decades. A massive study involving 60 cities, nearly 5,000 metagenomic samples, and eight trillion bases resulted in a catalog of the urban microbial ecosystem (Danko et al., 2021). This study provided geospatial knowledge of the strains, functional properties, and AMR markers of microbes in addition to comprehensive information on other aspects. This knowledge can serve as a basis for or aid in the designing and molding of balanced, health-supporting microbiota in interior environments. It has been suggested that urbanization and reduced exposure to microbiota in the natural environment affect the human immune system and increase the risk of allergies and other immunological disorders (Hui et al., 2019). In addition to environmental and lifestyle changes, seasonal variations have also been found to influence the microbial compositions of built environments and, thus, the microbiota of inhabitants. Scientists have announced activities to facilitate the migration of non-pathogenic environmental microbes into urban interiors, an issue that is discussed in the “Looking to the future” section of this paper (Parajuli et al., 2018; Hui et al., 2019; Selway et al., 2020; Roslund et al., 2021).
When interactions between microbiota species and their functions are perturbed, a non-adaptive state of disturbance called dysbiosis may occur. Dysbiosis refers to a disruption in homeostasis because of an imbalance in microbiota (Huitzil et al., 2018). Skin is the organ that comes into direct contact with indoor and outdoor surfaces, so it is important to evaluate the health risks of microbial imbalances on antimicrobial-coated surfaces. The biodiversity and ecosystem of a human residence affect the microbial biodiversity of human skin. An imbalance in skin microbiota can lead to both skin diseases and inflammatory non-communicable diseases (NCDs). The skin microbiota plays an important part in the formation and regulation of the body’s immune response (Prescott et al., 2017; Li et al., 2021). Early childhood is an especially important period for the development of healthy microbiota and immunity (Tanaka and Nakayama, 2017; Ronan et al., 2021). Imbalanced microbiota in AMCs may lead to potential health risks on large surface areas such as floors if, for example, infant children crawl on such surfaces for several hours daily in a kindergarten or elderly people are exposed to biased microbiota due to an excessive use of AMCs in a nursing home. If microbial diversity in an indoor environment is reduced, changes are needed to restore microbial homeostasis. When evaluating the risk levels of AMC solutions and products, it is vital to consider skin contact time and frequency. Long daily contact on large skin areas poses a bigger risk than short exposure on small skin area.
In a real-life experiment on skin bacterial dynamics, the microenvironments of the forearms and backs of 13 healthy people were mapped using a targeted 16S ribosomal RNA (rRNA) metagenomic sequencing approach to observe the effects of topical antiseptics or water treatments on the skin microbiome. Skin samples were taken every 6 h for a period of 72 h (SanMiguel et al., 2018). The study results showed a real impact on microbial community diversity and resilience. The effects of the treatments largely depended on the personalized and site-specific characteristics of the microbial communities. Minor microbial members were found to be prone to displacement and replacement with abundant taxa prior to treatment. The study underlined that many microbial taxa can thrive on the skin for a short period, while some have uniquely adapted to long-term colonization. The study also raised the possibility of long-term disturbances in the microbial communities leading to considerable changes in the bacterial communities of the skin (SanMiguel et al., 2018).
Indoor solutions that utilize AMCs target the growth and community composition structures of all microbes, not just pathogenic species, so more attention should be given to the health impacts resulting from changes in microbial communities. Studies have emphasized that the loss of microbial diversity correlates with increased AMR problems (Mahnert et al., 2019). Metagenomic comparisons of surfaces in clinical settings and other built environments have shown that confinement and cleaning can convert beneficial microbiomes into harmful microbiomes as well as result in a loss of microbial diversity, which correlates with an increase in AMR. Regular, strict cleaning has been shown to direct microbiome functions toward increased degradation of anthropogenic cleaning agents, such as xenobiotics, geraniol, limonene, pinene, and naphthalene (Mahnert et al., 2019; Hu and Hartmann, 2021). Based on these findings, it can be hypothesized that various AMCs sustain surface-specific bacterial strains to which humans have not yet adapted. Real-life, large-scale microbial community studies are needed to determine whether the compositions and functions of microbiomes on antimicrobial-coated surfaces change dramatically compared to those of non-coated surfaces. Accurate and reliable research results can dispel prejudices and contribute to the development of safe AMCs.
AMCs enhance indoor hygiene by killing, inactivating, or repelling microorganisms on surfaces. The use of AMCs on a critical, frequently touched surface can decrease microbial load and stop the surface from functioning as a microbial reservoir, in turn reducing the risk of onward transmission of microbes (Dancer, 2014; Ejerhed et al., 2020). AMCs constitute a non-specific intervention that targets a wide spectrum of pathogens, such as bacteria, viruses, mold, and yeast (Cassidy et al., 2020). Thus, while inactivating pathogens, AMCs continuously reduce the total microbiota on surfaces. Antimicrobial surfaces are primarily classified by their functional properties as contact-active, biocide-release, light-activated, or anti-adhesive surfaces (Figure 2) or combinations of these; other categories to classify them can also be found in the literature (Ahonen et al., 2017; Rosenberg et al., 2019b; Mahanta et al., 2021). Biocide-release surface materials are preloaded with biocides that need to be released to destroy microorganisms. Most of the AMCs in the market are biocide-releasing agents and contain antimicrobial metals, especially silver and copper, from which ionic compounds are released (Vincent et al., 2016; Ahonen et al., 2017; Rosenberg et al., 2019b; 2020; Zou et al., 2021). Different AMCs also vary in their effects on microorganisms because of their differing properties and mechanisms of action. Depending on the antimicrobial material, environmental conditions, and microorganisms involved, the inactivation time of microbes ranges from minutes to hours after contamination (Querido et al., 2019; Bäumler et al., 2021). Antimicrobial efficiency is also dependent on the concentration and particle size of antimicrobial products (Linzner and Antelmann, 2021).
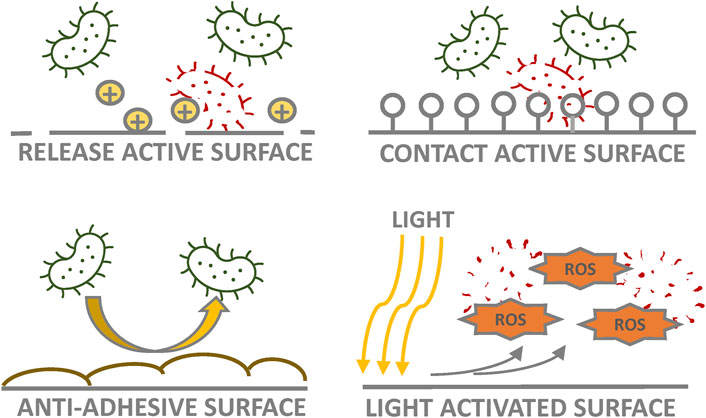
FIGURE 2. Four basic antimicrobial surface types. The biocide-release surface is preloaded with biocides to be released from the surface (e.g., metal ions) for antimicrobial action. On the contact-active surface, biocide is permanently immobilized on the surface (e.g., covalently bound quaternary ammonium compounds) and presents antimicrobial action when microbes come in close contact with the surface. The anti-adhesive surface (e.g., superhydrophilic polyethylene glycol) prevents microbial adhesion and accumulation on the surface, thus reducing the number of attached microorganisms. The light-activated antimicrobial material (e.g., titanium dioxide) destroys microorganisms via the production of reactive oxygen species under specific wavelengths of light.
The goal of an AMC is to continuously reduce the microbial load on a surface and achieve a health-promoting effect without human involvement. The aim of these self-disinfecting surfaces is the same as traditional infection prevention and control methods, such as hand washing, cleaning, and disinfecting, which can prevent the spread of microbes in an episodic manner (Imani et al., 2020; Birkett et al., 2022). AMCs do not replace but, rather, support traditional methods. (Dunne et al., 2018; Cassidy et al., 2020; Blomberg et al., 2022). Clean and hygienic environments need to be maintained in public hospital destinations, such as intensive care wards, premature infant care wards, infectious disease wards, and immunocompromised patient rooms, and the use of antimicrobial products can function as obligatory lifelines for residents. In these destinations, the use of AMCs is highly justified to limit the spread of healthcare-associated infections and infections caused by antibiotic-resistant bacteria. In other public destinations, such as schools, kindergartens, and leisure places, the use of AMCs is optional. Microbial ecology studies could determine the need for use of antimicrobial surfaces.
The transmission of pathogens can be mediated by contaminated surfaces (Otter et al., 2013). As many microbes can stay viable for a long time on different surfaces, indirect contact infection is a significant risk. For example, noroviruses and coronaviruses can remain viable on dry surfaces from several days to several weeks, depending on environmental conditions (Warnes et al., 2015; Cook et al., 2016; Kwan et al., 2018; Kampf et al., 2020). Pathogenic bacteria, including antibiotic-resistant bacteria, can persist for up to several months on dry surfaces, and bacterial spores are even more resistant to environmental conditions than vegetative bacteria (Dancer, 2014; Cassidy et al., 2020). Different antimicrobial coatings and materials have been developed to reduce the lifetime of pathogens on surfaces. The benefits and mechanisms of action of such coatings have already been thoroughly elucidated in several articles (Salwiczek et al., 2014; Bäumler et al., 2021; Lin et al., 2021; Birkett et al., 2022). New antimicrobial coatings are developed with various innovative mechanisms. Durable High-entropy alloys (HEAs) antimicrobial coatings containing many different metals have recently received attention, for example in biomedical applications and in the prevention of marine biofilms. Due to their properties and diverse composition design, these coatings are competitive alternatives for many applications (Zhou et al., 2020; Chen et al., 2021; Rashidy Ahmady et al., 2023).
As AMCs can control and prevent the spread of bacterial and viral pathogens, they have become even more important since the outbreak of the COVID-19 pandemic (Minoshima et al., 2016; Imani et al., 2020; Rakowska et al., 2021; Birkett et al., 2022; Vijayan et al., 2022). Considering this, attention needs to be paid to the challenges of using AMCs on indoor surfaces, such as floors, tables, cupboards, doorframes, mattresses, soft furnishing, and walls. The microbial changes that occur on these surfaces cover a large area and presumably contribute more to the overall changes in microbial community composition and the effects on human health than those on small surfaces. Small, high-touch areas, such as door handles, handrails, counters, taps, touchscreens, and computer mice, can also be substantial infection-spreading surfaces, and AMCs can act as significant inhibitors of pathogens on these surfaces (Adlhart et al., 2018; Reynolds et al., 2019; Rakowska et al., 2021).
Biocidal chemicals used in AMCs are toxic by nature, as they are intended to destroy, prevent the action of, or otherwise exert a controlling effect on harmful or undesired microbes. In the European Union, the relevant regulations for antimicrobial coatings include the Biocidal Product Regulation (BPR, Regulation (EU) 528/2012) and Regulation (EC) No 1907/2006 of the European Parliament and of the Council on the Registration, Evaluation, Authorization, and Restriction of Chemicals (REACH). A safe-by-design (SbD) approach has been developed for AMCs for the timely identification of risks related to industrial innovation processes and the value chain of antimicrobial materials and antimicrobial-coated products. The idea is to ensure safety for three different but interrelated communities—that is, workplace, consumer, and environmental communities. For example, antiadhesive surfaces can be considered SbD because they do not involve the use of biocidal molecules that can induce toxic effects or lead to resistance (reviewed in Ahonen et al., 2017; Adlhart et al., 2018).
Recent antimicrobial metal-based mechanisms that affect microbial attachment, lifetime, death, and spread have been comprehensively reviewed by Birkett et al. (2022). The article provides information on binary and ternary coatings for which different elements are combined. TiO2 photocatalytic surfaces combined with active Cu and Ag were found to have significant long-term antimicrobial efficacies. Superhydrophobic nanoporous and nanopillar coatings were proven to reduce the attachment of bacteria to surfaces. Further, TiO2 surface nanopillars were reported to induce cell impedance and oxidative stress, thus inhibiting bacterial proliferation. However, unlike the use of superhydrophobic coatings, the use of effective metal-based surfaces may lead to a decrease in performance and AMR over time (Birkett et al., 2022).
Antimicrobial resistance is considered to be one of the greatest risks to human health worldwide (Murray et al., 2022). Predictive statistical models have estimated a total of 4.95 million bacterial AMR-associated deaths across 204 countries and territories in 2019, of which 1.27 million deaths were attributable to bacterial AMR. Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa were the most prevalent pathogens for deaths associated with resistance in 2019 (Murray et al., 2022). In the United States, almost three million people get ill because of antibiotic resistance every year and about 35,000 of them die (Vassallo et al., 2021). These numbers clearly lighten the global burden of bacterial resistance and the seriousness of the global health threat.
An important aim of using AMCs is to reduce the global health risk of AMR, however, the heavy metals contained in the products themselves can induce the emergence and proliferation of resistance via the co-selection mechanism (Knapp et al., 2011; Wang et al., 2021). We have contributed to a publication mapping the risks associated with the use of antimicrobial surfaces made of copper, silver, and antimicrobial peptides (Pietsch et al., 2020). Evolution through de novo mutations, horizontal gene transfers (HGTs), and species sorting of inherently resistant bacteria are potential processes to increase AMR on antimicrobial surfaces. The mechanisms, evolution, and spread of AMR as well as cross-resistance and co-resistance events have been discussed in Pietsch et al.‘s report, which are important considerations with respect to AMC resistance. Briefly, cross-resistance refers to the phenomenon in which AMR mechanism (for example metal resistance) against AMC decreases the sensitivity of bacteria to antibiotics, and co-resistance involves the genetic element, such as plasmid, possessing resistance genes against both metals and antibiotics and acting as a mechanism for co-selection (Baker-Austin et al., 2006; Pal et al., 2017; Mustafa et al., 2021). Pietsch et al. (2020) pointed out the rarity of studies on the development of resistance to antimicrobial silver and copper coatings. Research data, especially in healthcare settings, are needed.
Mustafa et al. (2021) studied the prevalence of metal, antibiotic, and disinfectant resistances in 300 isolates of Salmonella Typhimurium, as metals are widely used in animal feed due to their antimicrobial properties but may cause the proliferation of antibiotic resistance through co-selection. Metal and heavy metal resistance genes were reported to be linked to resistance to antibiotics and disinfectants. In particular, copper resistance genes seemed to enable the development of resistance to several antibiotics and disinfectants. The transfer of the resistance genes czcD, pcoC, and cnrA to Escherichia coli and the increased copper resistance of the transconjugants indicated that these resistance genes were present in the conjugative plasmids. In the context of water and waste treatment, nanoalumina has been shown to cause an over 200-fold increase in the plasmid-mediated HGT of antibiotic multiresistance genes (Qiu et al., 2012). Using both shotgun metagenome and 16S rRNA gene amplicon sequencing, Hartmann et al. (2016) showed that antimicrobial chemicals are linked to increased antibiotic resistance genes (ARGs) in the indoor dust microbiome. Li et al. (2017) used a large complete genome collection from databases and bioinformatics analysis to study the genetic linkages between ARGs and metal resistance genes (MRGs). They emphasized the potential risk that ARG and MRG co-selection poses to both the environmental and medical relevance. These and other resistance research findings should be considered when planning and implementing the use of metal-containing AMCs, as metals and disinfectants have the potential to promote antibiotic resistance.
Studies have found that, in addition to metals, other non-antibiotic compounds such as biocides, can promote antibiotic resistance via co-selection (Pal et al., 2017). Non-antibiotic metalloids, such as selenate, have been shown to enhance the spread of ARGs in bacteria (Shi et al., 2021). Shi et al. (2021) applied metagenomic sequencing to identify ARGs whose abundances increased upon selenate addition. Multidrug-resistant species and the mechanisms of ARG spreading via vertical transfer and HGT in bacteria were identified. Adding selenate was found to increase the ratios of key genes engaged in intracellular reactive oxygen species production, bacterial membrane permeability, type IV pilus formation, and DNA repair and recombination, all of which enhance HGT and ARG spreading (Shi et al., 2021). When designing new AMCs and selecting materials, the risk of resistance due to metal and metalloid components should be considered (Tran et al., 2022).
It is likely that microbes have faced selective pressure due to the antimicrobial effects of metals and metalloids since the early days of their existence. Microbes that evolved and developed resistance to metals gained a competitive advantage over other non-resistant microbes (Li et al., 2021). This phenomenon occurs according to the principles of natural selection and adaptation; evolution favors microbes that tolerate and survive disturbing treatments, and these are enriched in the microbial population (Foster et al., 2017). Antimicrobial resistance is an inevitable continuum in microbial evolution, so developing effective methods to limit the harmfulness of the phenomenon should be prioritized (Imchen et al., 2020). Wu et al. (2022) described the development of microbial resistance to modern antimicrobial nanotechnologies in their extensive review. Since microbial resistance to current antibiotic therapies is a major cause of implant failure in orthopedic surgery, the authors have focused on generating new insights that can prevent resistance development. Wu et al. (2022) explored the design of nanomaterials that are bactericidal, antifouling, immunomodulating, and capable of producing an antibacterial compound, and they provided comprehensive information on the development process and key mechanisms of AMR. The same principles can be largely applied to AMCs with the knowledge that resistance problems do not arise immediately after the use of antimicrobial products; only time will tell if the microbial strains on surfaces are tough and resistant to destruction attempts (Wu et al., 2022). Therefore, the continuous monitoring and mapping of microbial communities on AMCs should be included in the process of product development and use.
In the early 1970s, it was observed that environmental ARGs resembled clinical ARGs, and following this, ARGs were studied for over 30 years mainly from a clinical perspective until a decade ago when resistome (i.e., the collection of AMR genes in a certain area) studies shed light on the origins, evolution, and spread mechanisms of ARGs (Benveniste and Davies, 1973; Rovira et al., 2019; Kim and Cha, 2021). Metagenomic research methods enable the precise and extensive mapping of the resistome and mobilome (i.e., the collection of mobile genetic elements [MGEs] of the genome, namely integrons, plasmids, transposons, and insertion sequences) in a certain area.
Yin et al. (2022) investigated the temporal and spatial variations of resistomes and mobilomes through a metagenomic analysis of wastewater treatment plants. Samples collected monthly for 13 months from three local sewage treatment plants in Hong Kong were compared with corresponding samples from North American urban wastewater. The analysis revealed valuable results on the long-term proliferation of ARGs, showing that the resistome samples from urban sewage in North America differed significantly from each other, while the ARGs within Hong Kong differed only slightly. Although research methods such as mathematical equations based on bacterial mass could not be directly applied in resistome studies of AMCs with a low mass of bacteria, the goal of both studies is the same: evaluating the health risks of the resistome. The development of more precise genetic-based measurement scales for the risk assessment of AMC resistances would be useful in this regard, and clear, consistent lines of research and modeling would facilitate routine risk assessments.
In a recently published review, Vassallo et al. (2022) argued that, with increasing urbanization, resistome elements are likely to become homogenized, and dominate the entire global resistome. This could result in a loss of natural diversity, ecological disturbances, and a worsening antibiotic resistance crisis. Different environmental, animal, and human resistomes as well as up-to-date research methods and critical ARGs and their hosts have been characterized in the comprehensive articles of Rovira et al. (2019) and Kim and Cha (2021). Although current metagenomic methods can theoretically map resistomes from antimicrobial surfaces to our knowledge, these mappings have not been published (Imchen et al., 2020). Notably, the risk of AMR development due to AMC usage can be reduced by using 1) reactive biocides with fast killing mechanisms 2) rapidly disappearing biocides 3) AMCs only on specified high-touch surfaces, and 4) AMCs with the anti-adhesive or photocatalytic mechanism.
Both, traditional culture-based methods and advanced genome-based methods are used when microbes are detected on surfaces. So far, research methods that are commonly employed for all types of AMCs cannot be identified because the diversity of AMCs and microbes that occupy surfaces would require coating- and microbe-specific testing methods and reagents (Rosenberg et al., 2020). Nevertheless, international standard test protocols are available to evaluate the antimicrobial efficacy of surfaces in laboratory conditions. The ISO testing standards include ISO 22196:2011 for antibacterial activity on hard non-porous surfaces (ISO 22196:2011, 2011) and ISO 21702:2019 for antiviral activity on non-porous surfaces (ISO 21702:2019, 2019). Antimicrobial efficacy of textiles used in furnishing can be evaluated using standards such as ISO 20743:2021 for antibacterial activity (ISO 20743:2021, 2021) and ISO 13629–2:2014 for antifungal activity (ISO 13629-2:2014, 2014). The observed drawbacks in the test protocols for hard non-porous surfaces, such as the humidity protocols not mimicking the values in real-life indoor environments, led to the development of a new test protocol in the ISO TC330 group. The new standards for antibacterial, antifungal, and antiviral activity are to be finalized within a couple of years. Notably, no standard protocols are available for testing the antimicrobial efficacy of AMCs in real-life settings.
When testing the effectiveness of an antimicrobial surface, it is important to estimate the number of dead and live microbes on it. The DNA intercalating agent propidium iodide (PI) can get inside a cell only if plasma membrane integrity is lost; in other words, PI staining detects dead cells. However, Rosenberg et al. (2019a) found that extracellular nucleic acids can cause false PI staining results in biofilm viability studies. To obtain accurate viability estimates, they proposed validating the results obtained via alternative methods of staining, such as by using the culture-based method, determining metabolic activity, and minimizing the extracellular matrix collection, as it can contain nucleic acids. Emerson et al. (2017) extensively described the methods used to separate living and dead cells, stating that this is a tricky task. The following methods are described comprehensively in Emerson et al. (2017) article: cultivation, propidium iodide (PI), propidium monoazide (PMA), ethidium monoazide (EMA), Alexa Fluor hydrazide (AFH), RNA analyses (e.g., metatranscriptomics), cellular energy measurements, bioorthogonal non-canonical amino acid tagging (BONCAT) with click chemistry, isothermal microcalorimetry (IMC), stable-isotope probing (SIP), and proteomics/metaproteomics.
When culture methods and genetic next-generation sequencing (NGS) methods were compared in the diagnosis of bacterial and fungal infections, NGS methods showed outstanding advantages (Chen et al., 2020). A disadvantage of NGS was that it could not be used to distinguish living pathogens from dead ones, while an excellent feature was its ability to take into account all types of pathogens, such as bacteria, fungi, viruses, mycoplasmas, and sporozoites. However, if RNA-based metatranscriptomics analysis of total rRNA is applied, only live, metabolically active microbes can be detected and classified. In DNA-based metagenomics, extracellular relic DNA and DNA from dead or inactive cells can cover up to 40%–90% of the total DNA in a sample distorting the results of microbial community structure (Carini et al., 2016; Mäki and Tiirola, 2018; Hempel et al., 2022). By comparing the NGS community composition results of RNA-based and DNA-based analyses, insights into the situation of living microbes versus that of a combination of living microbes and extracellular DNA/dead microbes could be gained. In eukaryotic microbes, community structure mapping using an RNA-based 18S rRNA approach is more accurate than using a DNA-based 18S rRNA marker gene approach because, in nucleated cells, the thousand-fold differences in 18S rRNA gene copy numbers between species can distort the community composition results. However, prokaryotic microbes such as bacteria have only one to several 16S rRNA gene copies per cell, so DNA-based results tend to be accurate enough for common environmental studies (Mäki et al., 2017).
Rawlinson et al. (2019) compiled a comprehensive overview of microbiological sampling of healthcare environment surfaces and guidelines for routine protocols that can be applied to surface sampling. Their review included the factors that influence variations in sampling efficiency, suitable sampling methods for different surfaces and target organisms, a flow diagram for molecular methods, and a collection of articles related to the use of different methods. According to the authors, the variations in the different methods were large, and some of the studies were carried out in laboratories and not in real hospital environments; due to this the results were not comparable, and it was difficult to draw overall conclusions. Guidelines for the examination of surfaces without antimicrobial properties may also be applicable to the study of AMCs, but it would likely be necessary to optimize and test the methods separately for each object.
Common detection methods for microbes on real-life touch surfaces include plating microbes on different agar plates and counting culturable colony-forming units (CFU), using pre-enrichment methods for culturable pathogens, adenosine triphosphate (ATP) bioluminescence testing, visualizing microbes with a light, laser, or electron microscope, and quantitative real-time polymerase chain reaction (qPCR) testing to determine bacterial load and microbial diversity (Landers et al., 2010; Johani et al., 2018; Inkinen et al., 2020). Swab methods and contact plate methods are basic standardized methods for surface sampling. The use of neutralizer agents to neutralize sanitizer residues and antimicrobial activity in AMCs can improve the recovery of bacteria (ISO 18593:2018, 2018; Li et al., 2020). Pathogenic bacteria such as Staphylococcus aureus can appear only in small amounts. In these cases, pre-enrichment can be used to improve detection (Landers et al., 2010).
A recently published study investigated the antimicrobial performance of a commercial laminate coating with silver-doped phosphate glass as an antimicrobial additive against Escherichia coli and Bacillus subtilis bacteria. The culture-based plating results showed that antimicrobial efficiency was highly dependent on the Gram-positive or Gram-negative property of the bacteria and the amount of silver added (Blomberg et al., 2022). In another study, the effect of photodynamic AMC on the microbial burden of frequently touched surfaces in public buses was investigated using the plating method. The results showed that photodynamic AMC kept the microbial load low and significantly reduced the risk of microbial transfer (Kalb et al., 2022). Numerous studies have applied traditional, culture-based microbial surveys, and the results have provided basic information about the microbial burden levels on different surfaces. Traditional methods are inexpensive and widely available; however, plating methods, for instance, detect only viable cells, and ATP activity testing primarily shows the cleanliness of the surface. Bacteria can be identified with the use of different selective agar plates and biochemical testing, but this is laborious and time-consuming.
Rosenberg et al. (2020) investigated the effectiveness of nano-ZnO and nano-ZnO/Ag composite AMCs against biofilm formation in an application-suitable oligotrophic environment by monitoring dissolution-induced toxicities to planktonic cells. Physical methods, such as vortexing, and ultrasonication, with chemical methods, such as high salt concentrations, were applied for biofilm collection. The culturable CFU counting method and statistical analyses were used to quantify the results. After staining, biofilms were evaluated using epifluorescence and confocal microscopy, and electron microscopy was also applied. The results showed, among other things, that bacterial biofilm formation was prevented by the coatings in a dose-dependent manner. The highest level of broad-spectrum inhibition was identified in the ZnO/Ag composite coating. A highlight of this study was that the antimicrobial effect was noticeable in the oligotrophic environment rather than in the often-used microbial growth medium. (Rosenberg et al., 2020). UVA exposure may also be required for testing AMCs with photocatalytic activity (Visnapuu et al., 2018).
Using the qPCR-based method with data and statistical analyses, Chen et al. (2019) revealed the levels of contamination of multiple metals, the main MRG types and distributions, and the co-occurrence mechanisms of MRGs, ARGs, and MGEs in a copper tailing dam area. They reported that the multimetal-polluted soil contained both MRGs and ARGs and then characterized the abundances of different resistance genes and their correlations to the different metals. The results of a versatile network analysis showed, among other things, that the copper resistance gene copB occurred with ARGs. Targeted MRGs, ARGs, and MGEs were amplified using a multitude of appropriate primers.
Bankier et al. (2018) compared three common methods—plate counts of vital cells, flow cytometry (LIVE/DEAD BacLight viability assay), and qPCR (viability qPCR)—to determine the antimicrobial activities of engineered nanoparticle combinations on Staphylococcus aureus and Pseudomonas aeruginosa bacteria. The best results were obtained using a flow cytometer due to its high throughput and rapid and quantifiable data. The plate count method revealed >8-log reduction for bacteria after exposure to high nanoparticle combination concentrations. qPCR did not yield results due to the interference of nanoparticles with qPCR amplification.
Since reports on amplification-based genetic microbial community mappings of AMCs are scarcely available in the published literature, it can be expected that PCR-based analyses of AMCs are not simplistic. Amplification of microbes from AMCs may require a large amount of optimization due to low microbial numbers and inhibited reactions due to antimicrobial products such as metals. In a recent study, the inhibition of PCR by metals was tested by comparing cycle threshold values in the reactions of 4-log metal ion concentrations (Kuffel et al., 2021). Zinc, tin, iron (II), and copper ions were potential disturbing factors but only in elevated concentrations. The tolerance level of different polymerases to metal ions in the PCR reaction also varies, so choosing the right polymerase can be a significant way to improve amplification efficiency (Kuffel et al., 2021). Metal ion chelating agents, such as ethylenediaminetetraacetic acid (EDTA) and ethylene glycol tetraacetic acid (EGTA), are valuable reagents for optimizing DNA recovery and PCR. For example, EDTA chelates a wide range of metals, improves DNA recovery efficiency when used in optimized concentrations, and does not interfere with downstream reactions (Nkuna et al., 2022). In forensic research, the recovery and amplification of touch DNA from metal surfaces is challenging due to metal ions binding to the DNA (Bonsu et al., 2020). Bonsu et al.’s (2020) review provided valuable insights into DNA binding to metal substrates and the merits and limitations of current methods, which can be considered when planning and performing PCR of microbial DNA from metal-based AMCs. Nucleic acid amplification can be a bottleneck in the NGS analysis of AMCs. If the extracted microbial DNA is cleaned carefully enough, PCR inhibition due to metals gets neutralized, and the PCR conditions and reagents are carefully tested, presumably, optimization of PCR conditions would facilitate successful amplification of microbiome on AMCs.
New genetic technologies have revolutionized identification of indoor microbes’ impacts on human wellbeing. The health effects of the compositions, functions, and dynamics of indoor microbiota are being studied to an increasing extent. Over the past decade, widespread and affordable large-scale, culture-independent, high-throughput metagenomic, and metatranscriptomic sequencing of DNA and RNA, respectively, has opened up new possibilities for identifying microbial communities that are beneficial or harmful to health, especially with the addition of immune response data gathered from indoor residents exposed to certain microbial communities (Kelley and Gilbert, 2013; Byrd et al., 2018; Boers et al., 2019; Soininen et al., 2022). Microbial ecology tools, such as microbial community diversity metrics of targeted and shotgun metagenomic sequencing data, have improved the possibility of understanding microbiota dynamics (Birtel et al., 2015; Horve et al., 2020). Microbial abundance and diversity measurements and visualizations, including species richness and evenness (α-diversity), non-metric multidimensional scaling (NMDS) plots of UniFrac and Bray-Curtis dissimilarities (β-diversity), and temporal and spatial variations in microbial compositions, offer valuable metrics for characterizing the safety, applicability, and efficiency of AMCs (Birtel et al., 2015; Kim et al., 2017; Avolio et al., 2019; Willis, 2019). Metagenomics can be used to detect known species as well as unknown microbes (Ko et al., 2022). In addition, multiomics approaches that can help identify connections between the microbiome of an antimicrobial surface and human gene expression can be beneficial for studies on AMC health risks (Lobanov et al., 2022; Park et al., 2022) The microbial metabolomics-based approach can provide an additional methodological repertoire for surface hygiene studies, complementing metagenomic, metatranscriptomic, and proteomic analyses (Tang, 2011; Vernocchi et al., 2016; Ahn and Hayes, 2021). Although metagenomic mappings of interior surfaces have been carried out for a decade, genetic real-life studies of AMCs are scarce. Figure 3 presents a workflow proposal for the molecular monitoring of AMCs.

FIGURE 3. Workflow for a targeted metagenomic and metatranscriptomic NGS of 16S rRNA. After isolation and amplification of nucleic acids, the concentrations, and quality of the products are checked using, for example, a Qubit fluorometer and TapeStation (or Bioanalyzer), respectively. Long-read sequencer NanoPore MinION has easier steps for library preparation.
In a recent study, skin microbiota was characterized by applying shotgun metagenomic sequencing; this method produces significantly more information than phylogenic marker gene-based microbiota assays because all microbiota kingdoms, including bacteria, fungi, and viruses, are revealed with high resolution from species to strain level and their functional diversities are revealed with same sequencing data (Li et al., 2021). This comprehensive study involved the construction of an integrated Human Skin Microbial Gene Catalog using 1,360 skin samples with over 10 million genes and the human skin resistome data of pathogenic and non-pathogenic bacteria. As homeostasis of skin microbiota results in the promotion of skin health, high-resolution characterizations of the skin microbiome and resistome were essential (Li et al., 2021). In a 2-year study, nasal and oral colonization and transmission dynamics of methicillin-susceptible Staphylococcus aureus (MSSA) bacteria were investigated in 326 healthcare workers and 388 patients as well as samples taken from a hospital environment (from nine departments, a near-patient environment, high-frequency touch sites, and air) using whole genome sequencing (WGS) approach (Kinnevey et al., 2022). The WGS method identified numerous potential MSSA transmission events in workers, patients, and the hospital environment under non-outbreak conditions. Many principles of this research can be applied when planning to monitor the health benefits and hazards resulting from AMC use, specifically regarding the dynamics of bacterial transfers from surfaces to humans. By taking samples from indoor residents in addition to surface samples, the health effects of AMCs can be mapped even more comprehensively. Bacterial shotgun metagenomic and whole genome sequencing approaches are massive data-generating analysis tools for clinical, agricultural, and environmental research. Metagenomic research results are significant, especially when investigating the type and range of AMR genes in AMCs (Raphenya et al., 2022). However, getting reliable results requires a vast amount of computing power and a lot of experience in using different bioinformatics software and pipelines.
Metagenomic NGS methods have evolved extensively over the past decade. New NGS approaches can produce real-time sequencing data of long reads; DNA molecules of over 10 kilobases, and they are designated third-generation sequencing (TGS) methods, produced mainly by Oxford Nanopore Technology (ONT) and Pacific Biosciences (Fukasawa et al., 2020; Xiao and Zhou, 2020; ben Khedher et al., 2022; Schiavone et al., 2022; Tan et al., 2022). The ONT MinION sequencer has some valuable features for TGS: it does not necessarily require template amplification, is portable and designed to work with a laptop, detects several kinds of modified bases, and can sequence RNA directly. The sequence accuracy of ONT has continuously improved due to better sequencing chemistry and pore design, and due to improved algorithms for basecalling; nevertheless, errors still occur, particularly due to homopolymers, which account for about 47% of errors. The accuracy of ONT can be improved significantly by using error correction tools and new chemistry to obtain over 99% (Q20) accuracy of basecalls. Accuracy is also dependent on guanine-cytosine (GC) content (van Dijk et al., 2018; Delahaye and Nicolas, 2021; Kerkhof, 2021; Sahlin and Medvedev, 2021). In research on plant viruses, the results of traditional Sanger sequencing were compared with the results of two different protocols of nanopore sequencing, targeting a known and an unknown virus genome. Nanopore sequencing was found to assemble genomic results that corresponded to 99.5%–99.7% identity to Sanger sequencing consensus results. The study showed that nanopore sequencing has a homopolymer issue that has been shown in Ion Torrent and Illumina sequencing—that is, homopolymer length variation causes sequencing inaccuracy (Marine et al., 2020; Chehida et al., 2021; Stoler and Nekrutenko, 2021).
Long-read sequencing greatly increases the resolution of microbial phylogenic classifications and is especially applicable when using approximately 1,550 bp length 16S rRNA marker gene-based classification of microbes. Traditionally, the targeted metagenomic approach, such as the amplicon sequencing of 16S rRNA gene fragments, has proven to be an efficient and reliable choice for microbial classification and requires significantly less computer power than WGS. In addition, the availability of increasingly complemented public 16S rRNA databases (e.g., SILVA, RDP, GenBank, and Greengenes) improves the reliability of the results. The 16S rRNA gene contains conserved sequences that are typical for all taxa and to which the primers are annealed as well as variable regions (V1–V9) from which microbial taxa can be classified with the help of bioinformatics (Matsuo et al., 2021). Before the era of TGS, only short, 150–600 bp hypervariation fragments were sequenced with the commonly used Illumina or Ion Torrent platforms. However, the taxonomic resolution of short fragments remains insufficient because the selection of region-specific primers affects resolution, and public databases do not yet contain full-length sequences of all known microbes (Myer et al., 2020) When the short-read method was compared to the full-length sequencing of 16S rRNA using the nanopore sequencer MinION, the relative abundances of dominant bacterial genera were highly similar, but long-read sequencing using MinION resulted in a better resolution and more accurate bacterial composition of the sample at the species level (Matsuo et al., 2021). For the microbiome monitoring of AMCs, MinION could be a good choice due to its capability for full-length sequencing of 16S rRNA. When the method was tested with simple and complex microbial communities, even from a low-biomass dog skin sample, full-length 16S rRNA sequencing successfully revealed the composition of the microbiome (Cuscó et al., 2019).
An approach to meeting the challenges of urbanization and decreased microbial diversity is derived from the natural selectivity of microbiota produced by soil and vegetation, which humankind has adapted to throughout history (Figure 4). Specifically, indoor microbiota can be shaped according to the diversity and composition of soil and plants by transferring outdoor microbes indoors and adding houseplants to built-in interiors (Roviello et al., 2021). To an increasing extent, studies have shown that the interaction between human and environmental microbiomes affects human health and the prevalence of NCDs (Ahn and Hayes, 2021; Panthee et al., 2022).

FIGURE 4. Living indoors, limited contact with natural microbes, strict cleaning with hygiene products, and the use of antimicrobial products can reduce the diversity of indoor microbiota and that of the inhabitants, impacting human health. The diversity of indoor microbiota increases when residents and domestic animals bring microbes from nature indoors and when green vegetation and domestic animals are found in the habitat. Farm life and time spent in nature are significant health-promoting factors.
Roslund et al. (2022) investigated the impact of biodiversity on immune tolerance using a double-blind and placebo-controlled intervention trial. The results showed that the immune responses of children in the intervention group differed from those of children in the placebo group. According to the article, children’s immune regulation can be supported by adding microbiological diversity to everyday life. The authors suggested that sustainable nature-based opportunities for the prevention of immune-mediated diseases should be further explored among urban residents. However, it should be kept in mind that some houseplants may create unfavorable effects, such as allergies, among vulnerable persons (Lamminpää et al., 1996; Paulsen et al., 2014; Pesonen and Aalto-Korte, 2020).
In a recent study, the skin microbiota of 57 healthy schoolchildren were studied by disturbing and swabbing skin before and after exposure to one of three environments (which were also sampled for comparison to the skin samples) for 45 min: a classroom, a sports field, or a biodiversity-rich forest (Mills et al., 2022). The results of the targeted metagenomic 16S rRNA gene-based sequencing, bioinformatics tools, and statistics calculations showed that moderately short exposures to biodiversity-rich areas may have a long-term diversifying effect on human skin. The study also showed that the microbial diversity and variability of skin microbes may decrease over time if the skin microbiota is repeatedly disturbed by cleansing. Microbiota of indoor air dust were also studied by characterizing the effects of houseplants on microbiota diversity using rRNA gene-targeted metagenomic sequencing. Houseplants were found to increase microbial diversity but not microbial numbers (Dockx et al., 2022).
Further, it has been proposed that the rich microbiota of traditional farm homes can protect against asthma, and a farm home-like composition of indoor microbiota may be used as a health-promoting model of indoor microbiota (Ege et al., 2011; Kirjavainen et al., 2019). Air-circulating green walls have been shown to alter bacterial abundance and diversity in human skin and the immune responses of blood cytokines. Working in an office with a green wall was associated with an increased relative abundance of beneficial skin bacteria, such as the genus Lactobacillus, and an increase in the Shannon diversity of phylum Proteobacteria and class Gammaproteobacteria. These findings support the idea that plant elements in an urban environment can alter residents’ microbiomes and immune responses and may promote health in built environments (Chapat et al., 2004; Soininen et al., 2022). However, more research is needed to provide sufficient evidence of the effects of nature-derived microbes on human health, taking allergic reactions into consideration (Tischer et al., 2022). Although nature-derived microbes can act as health promoters, the use of antimicrobial products is justified in the fight against pathogenic microbes. Shaping the indoor microbiota so that it supports the health of indoor residents is a great challenge in an urbanizing society.
A high diversity of microbes is needed to promote health and as a prophylactic measure for communicable and non-communicable diseases. Antimicrobial products, such as antibiotics, disinfectants, and AMCs, may lead to selection pressure in microbiota, resulting in the spread of harmful microbes even when the total number of microbes in an environment decreases. Due to the COVID-19 pandemic, interest in AMC usage has increased remarkably. However, whether microbial diversity in AMCs decreases because of antimicrobial actions and whether pathogens or microbes with resistance have a selection advantage in AMCs has not yet been studied. The danger is that reduced microbial diversity increases the risk of NCDs and of AMR-related health issues. Since AMCs are potential modifiers of microbial community compositions, the taxonomic and functional diversity and resistance elements of microbiomes on AMCs should be effectively elucidated to avoid a state of ignorance, as was the case with the excessive use of antibiotics at the onset of resistance problems. Risk assessments focusing on the development of AMR and polarized growth of microbes that are detrimental to health should be conducted considering the ecological aspects of microbiota. In addition, new approaches, and adaptive design solutions for the built environment are needed to combat infectious diseases, such as COVID-19. Since aerosols and fomite-mediated transmission of viruses and other harmful microbes have been acknowledged, new innovations in AMC materials and genetic testing of microbes on AMCs are needed to control diseases from spreading in indoor environments.
Over the last decade, tremendous improvement in research methods has allowed for metagenomic analyses of microbial communities using powerful NGS and TGM methods and bioinformatic tools that accurately characterize microbiota. These can be used to find ways to reduce microbial load and control diseases. Although metagenomic analyses have been in use for a decade, the available standardized test protocols for antimicrobial surface efficacy rely mainly on culture-based techniques. When selecting antimicrobial-coated products for sustainable use, empirical, genetic-based research data on microbial communities is significant: which microbes thrive on antimicrobial-coated surfaces compared to non-coated surfaces, what functional genes or ARG microbes contain, which genes are expressed, and whether AMCs cause the gene pool of indoor microbes to become significantly homogeneous in the long term. These findings facilitate comparisons of microbiota diversity and composition in different coatings to predict the microbiome’s potential impact on human health. Shotgun metagenomic sequencing in particular offers possibilities for the versatile analysis of microbiota functionality compared to the phylogenetic marker-gene-based, targeted metagenomic sequencing approach. In addition to environmental microbial data, metagenomic data of microbiota from indoor occupants’ skin, saliva, and gut can provide comprehensive, precise knowledge about the effects of AMCs on health.
AM designed and wrote the original draft. NS contributed to the writing of Section 3 and offered valuable information on the properties of antimicrobial coatings. MK provided important information on indoor hygiene research methods and contributed to writing Section 5.1. MA offered remarkable information on the history and use of antimicrobial products and wrote down the safety regulations of antimicrobial coatings. MA and ML supervised the writing project. All authors contributed to the reviewing and editing of the manuscript as well as intellectual discussions on the research topic.
This study was funded by the Ministry of Education and Culture in Finland and Satakunta University of Applied Sciences and is a part of the HEAL project—Healthier life with comprehensive indoor hygiene concept.
We appreciate and thank all members of the WANDER research team for their information and assistance during the writing project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, R. I., Bhangar, S., Pasut, W., Arens, E. A., Taylor, J. W., Lindow, S. E., et al. (2015). Chamber bioaerosol study: Outdoor air and human occupants as sources of indoor airborne microbes. PLoS One 10, e0128022. doi:10.1371/journal.pone.0128022
Adlhart, C., Verran, J., Azevedo, N. F., Olmez, H., Keinänen-Toivola, M. M., Gouveia, I., et al. (2018). Surface modifications for antimicrobial effects in the healthcare setting: A critical overview. J. Hosp. Infect. 99, 239–249. doi:10.1016/j.jhin.2018.01.018
Ahn, J., and Hayes, R. B. (2021). Environmental influences on the human microbiome and implications for noncommunicable disease. Annu. Rev. Public Health 42, 277–292. doi:10.1146/annurev-publhealth-012420-105020
Ahonen, M., Kahru, A., Ivask, A., Kasemets, K., Kõljalg, S., Mantecca, P., et al. (2017). Proactive approach for safe use of antimicrobial coatings in healthcare settings: Opinion of the COST action network AMiCI. Int. J. Environ. Res. Public Health 14, 366. doi:10.3390/ijerph14040366
Amarasinghe, S. L., Su, S., Dong, X., Zappia, L., Ritchie, M. E., and Gouil, Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30. doi:10.1186/s13059-020-1935-5
Anttila, J., Ruokolainen, L., Kaitala, V., and Laakso, J. (2013). Loss of competition in the outside host environment generates outbreaks of environmental opportunist pathogens. PLoS One 8, e71621. doi:10.1371/journal.pone.0071621
Avolio, M. L., Carroll, I. T., Collins, S. L., Houseman, G. R., Hallett, L. M., Isbell, F., et al. (2019). A comprehensive approach to analyzing community dynamics using rank abundance curves. Ecosphere 10, e02881. doi:10.1002/ecs2.2881
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi:10.1016/j.tim.2006.02.006
Bankier, C., Cheong, Y., Mahalingam, S., Edirisinghe, M., Ren, G., Cloutman-Green, E., et al. (2018). A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLoS One 13, e0192093. doi:10.1371/journal.pone.0192093
Bauer, M. A., Kainz, K., Carmona-Gutierrez, D., and Madeo, F. (2018). Microbial wars: Competition in ecological niches and within the microbiome. Microb. Cell. 5, 215–219. doi:10.15698/mic2018.05.628
Bäumler, W., Eckl, D., Holzmann, T., and Schneider-Brachert, W. (2021). Antimicrobial coatings for environmental surfaces in hospitals: A potential new pillar for prevention strategies in hygiene. Crit rev microbiol 1, 35. doi:10.1080/1040841X.2021.1991271
ben Khedher, M., Ghedira, K., Rolain, J. M., Ruimy, R., and Croce, O. (2022). Application and challenge of 3rd generation sequencing for clinical bacterial studies. Int. J. Mol. Sci. 23, 1395. doi:10.3390/ijms23031395
Benveniste, R., and Davies, J. (1973). Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. 70, 2276–2280. doi:10.1073/pnas.70.8.2276
Birkett, M., Dover, L., Cherian Lukose, C., Wasy Zia, A., Tambuwala, M. M., and Serrano-Aroca, Á. (2022). Recent advances in metal-based antimicrobial coatings for high-touch surfaces. Int. J. Mol. Sci. 23, 1162. doi:10.3390/ijms23031162
Birtel, J., Walser, J. C., Pichon, S., Bürgmann, H., and Matthews, B. (2015). Estimating bacterial diversity for ecological studies: Methods, metrics, and assumptions. PLoS One 10, e0125356. doi:10.1371/journal.pone.0125356
Blomberg, E., Herting, G., Kuttuva Rajarao, G., Mehtiö, T., Uusinoka, M., Ahonen, M., et al. (2022). Weathering and antimicrobial properties of laminate and powder coatings containing silver phosphate glass used as high-touch surfaces. Sustainability 14, 7102. doi:10.3390/su14127102
Boers, S. A., Jansen, R., and Hays, J. P. (2019). Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1059–1070. doi:10.1007/s10096-019-03520-3
Bonsu, D. O. M., Higgins, D., and Austin, J. J. (2020). Forensic touch DNA recovery from metal surfaces - a review. Sci. Justice 60, 206–215. doi:10.1016/j.scijus.2020.01.002
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi:10.1038/nrmicro.2017.157
Carini, P., Marsden, P. J., Leff, J. W., Morgan, E. E., Strickland, M. S., and Fierer, N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 16242. doi:10.1038/nmicrobiol.2016.242
Cassidy, S. S., Sanders, D. J., Wade, J., Parkin, I. P., Carmalt, C. J., Smith, A. M., et al. (2020). Antimicrobial surfaces: A need for stewardship? PLoS Pathog. 16, e1008880. doi:10.1371/journal.ppat.1008880
Chapat, L., Chemin, K., Dubois, B., Bourdet-Sicard, R., and Kaiserlian, D. (2004). Lactobacillus casei reduces CD8+ T cell-mediated skin inflammation. Eur. J. Immunol. 34, 2520–2528. doi:10.1002/eji.200425139
Chehida, S. B., Filloux, D., Fernandez, E., Moubset, O., Hoareau, M., Julian, C., et al. (2021). Nanopore sequencing is a credible alternative to recover complete genomes of geminiviruses. Microorganisms 9, 903. doi:10.3390/microorganisms9050903
Chen, J., Li, J., Zhang, H., Shi, W., and Liu, Y. (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 10, 1916. doi:10.3389/fmicb.2019.01916
Chen, P., Sun, W., and He, Y. (2020). Comparison of the next-generation sequencing (NGS) technology with culture methods in the diagnosis of bacterial and fungal infections. J. Thorac. Dis. 12, 4924–4929. doi:10.21037/jtd-20-930
Chen, Y., Munroe, P., Xie, Z., and Zhang, S. (2021). “High-entropy alloy-based coatings,” in Protective thin coatings Technology (New York: CRC Press), 205–232. doi:10.1201/9781003088349-6
Cook, N., Knight, A., and Richards, G. P. (2016). Persistence and elimination of human norovirus in food and on food contact surfaces: A critical review. J. Food Prot. 79, 1273–1294. doi:10.4315/0362-028x.jfp-15-570
Corey, L., Beyrer, C., Cohen, M. S., Michael, N. L., Bedford, T., and Rolland, M. (2021). SARS-CoV-2 variants in patients with immunosuppression. N. Engl. J. Med. 385, 562–566. doi:10.1056/NEJMsb2104756
Cuscó, A., Catozzi, C., Viñes, J., Sanchez, A., and Francino, O. (2019). Microbiota profiling with long amplicons using nanopore sequencing: Full-length 16S rRNA gene and the 16S-ITS-23S of the rrn operon. F1000Res 7, 1755. doi:10.12688/f1000research.16817.2
Dancer, S. J. (2014). Controlling hospital-acquired infection: Focus on the role of the environment and new technologies for decontamination. Clin. Microbiol. Rev. 27, 665–690. doi:10.1128/cmr.00020-14
Danko, D., Bezdan, D., Afshin, E. E., Ahsanuddin, S., Bhattacharya, C., Butler, D. J., et al. (2021). A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell. 184, 3376–3393.e17. doi:10.1016/j.cell.2021.05.002
Davenport, E. R., Sanders, J. G., Song, S. J., Amato, K. R., Clark, A. G., and Knight, R. (2017). The human microbiome in evolution. BMC Biol. 15, 127. doi:10.1186/s12915-017-0454-7
de Vries, R. D., Schmitz, K. S., Bovier, F. T., Predella, C., Khao, J., Noack, D., et al. (2021). Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science 371, 1379–1382. doi:10.1126/science.abf4896
Dekaboruah, E., Suryavanshi, M. V., Chettri, D., and Verma, A. K. (2020). Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 202, 2C47–2167. doi:10.1007/s00203-020-01931-x
Delahaye, C., and Nicolas, J. (2021). Sequencing DNA with nanopores: Troubles and biases. PLoS One 16, e0257521. doi:10.1371/journal.pone.0257521
Dockx, Y., Täubel, M., Bijnens, E. M., Witters, K., Valkonen, M., Jayaprakash, B., et al. (2022). Indoor green can modify the indoor dust microbial communities. Indoor Air 32, e13011. doi:10.1111/ina.13011
Dunne, C. P., Askew, P. D., Papadopoulos, T., Gouveia, I. C., Ahonen, M., Modic, M., et al. (2020). Antimicrobial coating innovations to prevent infectious disease: A consensus view from the AMiCl COST action. J. Hosp. Infect. 105, 116–118. doi:10.1016/j.jhin.2020.04.006
Dunne, S. S., Ahonen, M., Modic, M., Crijns, F. R. L., Keinänen-Toivola, M. M., Meinke, R., et al. (2018). Specialized cleaning associated with antimicrobial coatings for reduction of hospital-acquired infection: Opinion of the COST action network AMiCI (CA15114). J. Hosp. Infect. 99, 250–255. doi:10.1016/j.jhin.2018.03.006
Ege, M. J., Mayer, M., Normand, A. C., Genuneit, J., Cookson, W. O. C. M., Braun-Fahrländer, C., et al. (2011). Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 364, 701–709. doi:10.1056/NEJMoa1007302
Ejerhed, L., Roshani, L., and Andersson, A. E. (2020). Antimicrobial coating is associated with significantly lower aerobic colony counts in high-touch areas in an orthopedic ward environment. Ann. Clin. Microbiol. Antimicrob. 19, 62. doi:10.1186/s12941-020-00406-7
Emerson, J. B., Adams, R. I., Román, C. M. B., Brooks, B., Coil, D. A., Dahlhausen, K., et al. (2017). Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86. doi:10.1186/s40168-017-0285-3
Ford, S. A., and King, K. C. (2016). Harnessing the power of defensive microbes: Evolutionary implications in nature and disease control. PLoS Pathog. 12, e1005465. doi:10.1371/journal.ppat.1005465
Foster, K. R., Schluter, J., Coyte, K. Z., and Rakoff-Nahoum, S. (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. doi:10.1038/nature23292
Fukasawa, Y., Ermini, L., Wang, H., Carty, K., and Cheung, M. S. (2020). LongQC: A quality control tool for third generation sequencing long read data. G3 Genes.|Genomes|Genetics 10, 1193–1196. doi:10.1534/g3.119.400864
Garud, N. R., and Pollard, K. S. (2020). Population genetics in the human microbiome. Trends Genet. 36, 53–67. doi:10.1016/j.tig.2019.10.010
Hadfield, J., Megill, C., Bell, S. M., Huddleston, J., Potter, B., Callender, C., et al. (2018). Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123. doi:10.1093/bioinformatics/bty407
Hartmann, E. M., Hickey, R., Hsu, T., Betancourt Román, C. M., Chen, J., Schwager, R., et al. (2016). Antimicrobial chemicals are associated with elevated antibiotic resistance genes in the indoor dust microbiome. Environ. Sci. Technol. 50, 9807–9815. doi:10.1021/acs.est.6b00262
Hempel, C. A., Wright, N., Harvie, J., Hleap, J. S., Adamowicz, S. J., and Steinke, D. (2022). Metagenomics versus total RNA sequencing: Most accurate data-processing tools, microbial identification accuracy and perspectives for ecological assessments. Nucleic Acids Res. 50, 9279–9293. doi:10.1093/nar/gkac689
Henry, L. P., Bruijning, M., Forsberg, S. K. G., and Ayroles, J. F. (2021). The microbiome extends host evolutionary potential. Nat. Commun. 12, 5141. doi:10.1038/s41467-021-25315-x
Horve, P. F., Lloyd, S., Mhuireach, G. A., Dietz, L., Fretz, M., MacCrone, G., et al. (2020). Building upon current knowledge and techniques of indoor microbiology to construct the next era of theory into microorganisms, health, and the built environment. J. Expo. Sci. Environ. Epidemiol. 30, 219–235. doi:10.1038/s41370-019-0157-y
Hu, J., and Hartmann, E. M. (2021). Anthropogenic chemicals and their impacts on microbes living in buildings. Microb. Biotechnol. 14, 798–802. doi:10.1111/1751-7915.13676
Hui, N., Parajuli, A., Puhakka, R., Grönroos, M., Roslund, M. I., Vari, H. K., et al. (2019). Temporal variation in indoor transfer of dirt-associated environmental bacteria in agricultural and urban areas. Environ. Int. 132, 105069. doi:10.1016/j.envint.2019.105069
Huitzil, S., Sandoval-Motta, S., Frank, A., and Aldana, M. (2018). Modeling the role of the microbiome in evolution. Front. Physiol. 9, 1836. doi:10.3389/fphys.2018.01836
Imani, S. M., Ladouceur, L., Marshall, T., Maclachlan, R., Soleymani, L., and Didar, T. F. (2020). Antimicrobial nanomaterials and coatings: Current mechanisms and future perspectives to control the spread of viruses including SARS-CoV-2. ACS Nano 14, 12341–12369. doi:10.1021/acsnano.0c05937
Imchen, M., Moopantakath, J., Kumavath, R., Barh, D., Tiwari, S., Ghosh, P., et al. (2020). Current trends in experimental and computational approaches to combat antimicrobial resistance. Front. Genet. 11, 563975. doi:10.3389/fgene.2020.563975
Inkinen, J., Ahonen, M., Iakovleva, E., Karppinen, P., Mielonen, E., Mäkinen, R., et al. (2020). Contamination detection by optical measurements in a real-life environment: A hospital case study. J. Biophot. 13, e201960069. doi:10.1002/jbio.201960069
ISO 13629-2:2014 (2014). Textiles – determination of antifungal activity of textile products Part 2: Plate count method. Available at: https://www.iso.org/standard/57949.html (Accessed October 4, 2022).
ISO 18593:2018 (2018). Microbiology of the food chain – horizontal methods for surface sampling. Available at: https://www.iso.org/standard/64950.html (Accessed July 7, 2022).
ISO 20743:2021 (2021). Textiles – determination of antibacterial activity of textile products. Available at: https://www.iso.org/standard/79819.html (Accessed October 4, 2022).
ISO 21702:2019 (2019). Measurement of antiviral activity on plastics and other non-porous surfaces. Available at: https://www.iso.org/standard/71365.html (Accessed October 4, 2022).
ISO 22196:2011 (2011). Measurement of antibacterial activity on plastics and other non-porous surfaces. Available at: https://www.iso.org/standard/54431.html (Accessed October 4, 2022).
Johani, K., Abualsaud, D., Costa, D. M., Hu, H., Whiteley, G., Deva, A., et al. (2018). Characterization of microbial community composition, antimicrobial resistance and biofilm on intensive care surfaces. J. Infect. Public Health 11, 418–424. doi:10.1016/j.jiph.2017.10.005
Joonaki, E., Hassanpouryouzband, A., Heldt, C. L., and Areo, O. (2020). Surface chemistry can unlock drivers of surface stability of SARS-CoV-2 in a variety of environmental conditions. Chem 6, 2135–2146. doi:10.1016/j.chempr.2020.08.001
Kalb, L., Bäßler, P., Schneider-Brachert, W., and Eckl, D. B. (2022). Antimicrobial photodynamic coatings reduce the microbial burden on environmental surfaces in public transportation-A field study in buses. Int. J. Environ. Res. Public Health 19, 2325. doi:10.3390/ijerph19042325
Kampf, G., Todt, D., Pfaender, S., and Steinmann, E. (2020). Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 104, 246–251. doi:10.1016/j.jhin.2020.01.022
Kelley, S. T., and Gilbert, J. A. (2013). Studying the microbiology of the indoor environment. Genome Biol. 14, 202. doi:10.1186/gb-2013-14-2-202
Kerkhof, L. J. (2021). Is Oxford Nanopore sequencing ready for analyzing complex microbiomes? FEMS Microbiol. Ecol. 97, fiab001. doi:10.1093/femsec/fiab001
Khanna, R., Cicinelli, M., Gilbert, S., Honavar, S., and Murthy, G. V. (2020). COVID-19 pandemic: Lessons learned and future directions. Indian J. Ophthalmol. 68, 703. doi:10.4103/ijo.IJO_843_20
Kilian, M., Chapple, I. L. C., Hannig, M., Marsh, P. D., Meuric, V., Pedersen, A. M. L., et al. (2016). The oral microbiome - an update for oral healthcare professionals. Br. Dent. J. 221, 657–666. doi:10.1038/sj.bdj.2016.865
Kim, B. R., Shin, J., Guevarra, R. B., Lee, J. H., Kim, D. W., Seol, K. H., et al. (2017). Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi:10.4014/jmb.1709.09027
Kim, D. W., and Cha, C. J. (2021). Antibiotic resistome from the one-health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 53, 301–309. doi:10.1038/s12276-021-00569-z
King, K. C., and Bonsall, M. B. (2017). The evolutionary and coevolutionary consequences of defensive microbes for host-parasite interactions. BMC Evol. Biol. 17, 190. doi:10.1186/s12862-017-1030-z
Kinnevey, P. M., Kearney, A., Shore, A. C., Earls, M. R., Brennan, G. I., Poovelikunnel, T. T., et al. (2022). Meticillin-susceptible Staphylococcus aureus transmission among healthcare workers, patients and the environment in a large acute hospital under non-outbreak conditions investigated using whole-genome sequencing. J. Hosp. Infect. 127, 15–25. doi:10.1016/j.jhin.2022.05.004
Kirjavainen, P. v., Karvonen, A. M., Adams, R. I., Täubel, M., Roponen, M., Tuoresmäki, P., et al. (2019). Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 25, 1089–1095. doi:10.1038/s41591-019-0469-4
Knapp, C. W., McCluskey, S. M., Singh, B. K., Campbell, C. D., Hudson, G., and Graham, D. W. (2011). Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS One 6, e27300. doi:10.1371/journal.pone.0027300
Ko, K. K. K., Chng, K. R., and Nagarajan, N. (2022). Metagenomics-enabled microbial surveillance. Nat. Microbiol. 7, 486–496. doi:10.1038/s41564-022-01089-w
Kuffel, A., Gray, A., and Daeid, N. N. (2021). Impact of metal ions on PCR inhibition and RT-PCR efficiency. Int. J. Leg. Med. 135, 63–72. doi:10.1007/s00414-020-02363-4
Kumar, P., Kausar, M. A., Singh, A. B., and Singh, R. (2021). Biological contaminants in the indoor air environment and their impacts on human health. Air Qual. Atmos. Health 14, 1723–1736. doi:10.1007/s11869-021-00978-z
Kwan, S. E., Shaughnessy, R. J., Hegarty, B., Haverinen-Shaughnessy, U., and Peccia, J. (2018). The reestablishment of microbial communities after surface cleaning in schools. J. Appl. Microbiol. 125, 897–906. doi:10.1111/jam.13898
Lamminpää, A., Estlander, T., Jolanki, R., and Kanerva, L. (1996). Occupational allergic contact dermatitis caused by decorative plants. Contact Dermat. 34, 330–335. doi:10.1111/j.1600-0536.1996.tb02217.x
Landers, T. F., Hoet, A., and Wittum, T. E. (2010). Swab type, moistening, and preenrichment for Staphylococcus aureus on environmental surfaces. J. Clin. Microbiol. 48, 2235–2236. doi:10.1128/jcm.01958-09
Lei, H., Li, Y., Xiao, S., Yang, X., Lin, C., Norris, S. L., et al. (2017). Logistic growth of a surface contamination network and its role in disease spread. Sci. Rep. 7, 14826. doi:10.1038/s41598-017-13840-z
Li, F., Xian, Z., Kwon, H. J., Yoo, J., Burall, L., Chirtel, S. J., et al. (2020). Comparison of three neutralizing broths for environmental sampling of low levels of Listeria monocytogenes desiccated on stainless steel surfaces and exposed to quaternary ammonium compounds. BMC Microbiol. 20, 333. doi:10.1186/s12866-020-02004-1
Li, L. G., Xia, Y., and Zhang, T. (2017). Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 11, 651–662. doi:10.1038/ismej.2016.155
Li, Y. P., ben Fekih, I., Chi Fru, E., Moraleda-Munoz, A., Li, X., Rosen, B. P., et al. (2021a). Antimicrobial activity of metals and metalloids. Annu. Rev. Microbiol. 75, 175–197. doi:10.1146/annurev-micro-032921-123231
Li, Z., Xia, J., Jiang, L., Tan, Y., An, Y., Zhu, X., et al. (2021b). Characterization of the human skin resistome and identification of two microbiota cutotypes. Microbiome 9, 47. doi:10.1186/s40168-020-00995-7
Lin, N., Verma, D., Saini, N., Arbi, R., Munir, M., Jovic, M., et al. (2021). Antiviral nanoparticles for sanitizing surfaces: A roadmap to self-sterilizing against COVID-19. Nano Today 40, 101267. doi:10.1016/j.nantod.2021.101267
Linzner, N., and Antelmann, H. (2021). The antimicrobial activity of the AGXX® surface coating requires a small particle size to efficiently kill Staphylococcus aureus. Front. Microbiol. 12, 731564. doi:10.3389/fmicb.2021.731564
Lobanov, V., Gobet, A., and Joyce, A. (2022). Ecosystem-specific microbiota and microbiome databases in the era of big data. Environ. Microbiome 17, 37. doi:10.1186/s40793-022-00433-1
Mahanta, U., Khandelwal, M., and Deshpande, A. S. (2021). Antimicrobial surfaces: A review of synthetic approaches, applicability and outlook. J. Mater Sci. 56, 17915–17941. doi:10.1007/s10853-021-06404-0
Mahnert, A., Moissl-Eichinger, C., Zojer, M., Bogumil, D., Mizrahi, I., Rattei, T., et al. (2019). Man-made microbial resistances in built environments. Nat. Commun. 10, 968. doi:10.1038/s41467-019-08864-0
Mäki, A., Salmi, P., Mikkonen, A., Kremp, A., and Tiirola, M. (2017). Sample Preservation, DNA or RNA extraction and data analysis for high-throughput phytoplankton community sequencing. Front. Microbiol. 8, 1848. doi:10.3389/fmicb.2017.01848
Mäki, A., and Tiirola, M. (2018). Directional high-throughput sequencing of RNAs without gene-specific primers. Biotechniques 65, 219–223. doi:10.2144/btn-2018-0082
Marine, R. L., Magaña, L. C., Castro, C. J., Zhao, K., Montmayeur, A. M., Schmidt, A., et al. (2020). Comparison of Illumina MiSeq and the Ion Torrent PGM and S5 platforms for whole-genome sequencing of picornaviruses and caliciviruses. J. Virol. Methods 280, 113865. doi:10.1016/j.jviromet.2020.113865
Matsuo, Y., Komiya, S., Yasumizu, Y., Yasuoka, Y., Mizushima, K., Takagi, T., et al. (2021). Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinIONTM nanopore sequencing confers species-level resolution. BMC Microbiol. 21, 35. doi:10.1186/s12866-021-02094-5
Mills, J. G., Selway, C. A., Thomas, T., Weyrich, L. S., and Lowe, A. J. (2022). Schoolyard biodiversity determines short-term recovery of disturbed skin microbiota in children. Microb. Ecol. 1, 1–12. doi:10.1007/s00248-022-02052-2
Minoshima, M., Lu, Y., Kimura, T., Nakano, R., Ishiguro, H., Kubota, Y., et al. (2016). Comparison of the antiviral effect of solid-state copper and silver compounds. J. Hazard Mater 312, 1–7. doi:10.1016/j.jhazmat.2016.03.023
Moskovicz, V., Gross, A., and Mizrahi, B. (2020). Extrinsic factors shaping the skin microbiome. Microorganisms 8, 1023. doi:10.3390/microorganisms8071023
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655. doi:10.1016/s0140-6736(21)02724-0
Mustafa, G. R., Zhao, K., He, X., Chen, S., Liu, S., Mustafa, A., et al. (2021). Heavy metal resistance in Salmonella Typhimurium and its association with disinfectant and antibiotic resistance. Front. Microbiol. 12, 702725. doi:10.3389/fmicb.2021.702725
Myer, P. R., McDaneld, T. G., Kuehn, L. A., Dedonder, K. D., Apley, M. D., Capik, S. F., et al. (2020). Classification of 16S rRNA reads is improved using a niche-specific database constructed by near-full length sequencing. PLoS One 15, e0235498. doi:10.1371/journal.pone.0235498
Niu, J., Xu, L., Qian, Y., Sun, Z., Yu, D., Huang, J., et al. (2020). Evolution of the gut microbiome in early childhood: A cross-sectional study of Chinese children. Front. Microbiol. 11, 439. doi:10.3389/fmicb.2020.00439
Nkuna, R., Ijoma, G. N., and Matambo, T. S. (2022). Applying EDTA in chelating excess metal ions to improve downstream DNA recovery from mine tailings for long-read amplicon sequencing of acidophilic fungi communities. J. Fungi 8, 419. doi:10.3390/jof8050419
Otter, J. A., Yezli, S., Salkeld, J. A. G., and French, G. L. (2013). Evidence that contaminated surfaces contribute to the transmission of hospital pathogens and an overview of strategies to address contaminated surfaces in hospital settings. Am. J. Infect. Control 41, S6–S11. doi:10.1016/j.ajic.2012.12.004
Pal, C., Asiani, K., Arya, S., Rensing, C., Stekel, D. J., Larsson, D. G. J., et al. (2017). Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 70, 261–313. doi:10.1016/bs.ampbs.2017.02.001
Panthee, B., Gyawali, S., Panthee, P., and Techato, K. (2022). Environmental and human microbiome for health. Life 12, 456. doi:10.3390/life12030456
Paquette, S. J., Zaheer, R., Stanford, K., Thomas, J., and Reuter, T. (2018). Competition among Escherichia coli strains for space and resources. Vet. Sci. 5, 93. doi:10.3390/vetsci5040093
Parajuli, A., Grönroos, M., Siter, N., Puhakka, R., Vari, H. K., Roslund, M. I., et al. (2018). Urbanization reduces transfer of diverse environmental microbiota indoors. Front. Microbiol. 9, 84. doi:10.3389/fmicb.2018.00084
Park, C. H., Hong, C., Lee, A., Sung, J., and Hwang, T. H. (2022). Multi-omics reveals microbiome, host gene expression, and immune landscape in gastric carcinogenesis. iScience 25, 103956. doi:10.1016/j.isci.2022.103956
Paulsen, E., Svendsen, M. T., and Frankild, S. (2014). Contact urticaria and contact sensitization to yucca (Yucca gigantea Lem.) in a plant keeper. Contact Dermat. 71, 119–121. doi:10.1111/cod.12202
Pesonen, M., and Aalto-Korte, K. (2020). Occupational allergic contact dermatitis and contact urticaria caused by indoor plants in plant keepers. Contact Dermat. 83, 515–518. doi:10.1111/cod.13647
Pietsch, F., O’Neill, A. J., Ivask, A., Jenssen, H., Inkinen, J., Kahru, A., et al. (2020). Selection of resistance by antimicrobial coatings in the healthcare setting. J. Hosp. Infect. 106, 115–125. doi:10.1016/j.jhin.2020.06.006
Planas, D., Saunders, N., Maes, P., Guivel-Benhassine, F., Planchais, C., Buchrieser, J., et al. (2022). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675. doi:10.1038/s41586-021-04389-z
Prescott, S. L., Larcombe, D. L., Logan, A. C., West, C., Burks, W., Caraballo, L., et al. (2017). The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 10, 29. doi:10.1186/s40413-017-0160-5
Prussin, A. J., and Marr, L. C. (2015). Sources of airborne microorganisms in the built environment. Microbiome 3, 78. doi:10.1186/s40168-015-0144-z
Qiu, Z., Yu, Y., Chen, Z., Jin, M., Yang, D., Zhao, Z., et al. (2012). Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. U. S. A. 109, 4944–4949. doi:10.1073/pnas.1107254109
Querido, M. M., Aguiar, L., Neves, P., Pereira, C. C., and Teixeira, J. P. (2019). Self-disinfecting surfaces and infection control. Colloids Surf. B Biointerfaces 178, 8–21. doi:10.1016/j.colsurfb.2019.02.009
Rakowska, P. D., Tiddia, M., Faruqui, N., Bankier, C., Pei, Y., Pollard, A. J., et al. (2021). Antiviral surfaces and coatings and their mechanisms of action. Commun. Mater 2, 53. doi:10.1038/s43246-021-00153-y
Raphenya, A. R., Robertson, J., Jamin, C., de Oliveira Martins, L., Maguire, F., McArthur, A. G., et al. (2022). Datasets for benchmarking antimicrobial resistance genes in bacterial metagenomic and whole genome sequencing. Sci. Data 9, 341. doi:10.1038/s41597-022-01463-7
Rashidy Ahmady, A., Ekhlasi, A., Nouri, A., Haghbin Nazarpak, M., Gong, P., and Solouk, A. (2023). High entropy alloy coatings for biomedical applications: A review. Smart Mater. Manuf. 1, 100009. doi:10.1016/j.smmf.2022.100009
Rawlinson, S., Ciric, L., and Cloutman-Green, E. (2019). How to carry out microbiological sampling of healthcare environment surfaces? A review of current evidence. J. Hosp. Infect. 103, 363–374. doi:10.1016/j.jhin.2019.07.015
Reynolds, K. A., Sexton, J. D., Pivo, T., Humphrey, K., Leslie, R. A., and Gerba, C. P. (2019). Microbial transmission in an outpatient clinic and impact of an intervention with an ethanol-based disinfectant. Am. J. Infect. Control 47, 128–132. doi:10.1016/j.ajic.2018.06.017
Riddell, S., Goldie, S., Hill, A., Eagles, D., and Drew, T. W. (2020). The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol. J. 17, 145. doi:10.1186/s12985-020-01418-7
Ronan, V., Yeasin, R., and Claud, E. C. (2021). Childhood development and the microbiome—the intestinal microbiota in maintenance of health and development of disease during childhood development. Gastroenterology 160, 495–506. doi:10.1053/j.gastro.2020.08.065
Rosenberg, M., Azevedo, N. F., and Ivask, A. (2019a). Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 9, 6483. doi:10.1038/s41598-019-42906-3
Rosenberg, M., Ilic, K., Juganson, K., Ivask, A., Ahonen, M., Vrček, I. V., et al. (2019b). Potential ecotoxicological effects of antimicrobial surface coatings: A literature survey backed up by analysis of market reports. PeerJ 7, e6315. doi:10.7717/peerj.6315
Rosenberg, M., Visnapuu, M., Vija, H., Kisand, V., Kasemets, K., Kahru, A., et al. (2020). Selective antibiofilm properties and biocompatibility of nano-ZnO and nano-ZnO/Ag coated surfaces. Sci. Rep. 10, 13478. doi:10.1038/s41598-020-70169-w
Roslund, M. I., Parajuli, A., Hui, N., Puhakka, R., Grönroos, M., Soininen, L., et al. (2022). A Placebo-controlled double-blinded test of the biodiversity hypothesis of immune-mediated diseases: Environmental microbial diversity elicits changes in cytokines and increase in T regulatory cells in young children. Ecotoxicol. Environ. Saf. 242, 113900. doi:10.1016/j.ecoenv.2022.113900
Roslund, M. I., Puhakka, R., Nurminen, N., Oikarinen, S., Siter, N., Grönroos, M., et al. (2021). Long-term biodiversity intervention shapes health-associated commensal microbiota among urban day-care children. Environ. Int. 157, 106811. doi:10.1016/j.envint.2021.106811
Roviello, V., Scognamiglio, P. L., Caruso, U., Vicidomini, C., and Roviello, G. N. (2021). Evaluating in silico the potential health and environmental benefits of houseplant volatile organic compounds for an emerging “indoor forest bathing” approach. Int. J. Environ. Res. Public Health 19, 273. doi:10.3390/ijerph19010273
Rovira, P., McAllister, T., Lakin, S. M., Cook, S. R., Doster, E., Noyes, N. R., et al. (2019). Characterization of the microbial resistome in conventional and “raised without antibiotics” beef and dairy production systems. Front. Microbiol. 10, 1980. doi:10.3389/fmicb.2019.01980
Sahlin, K., and Medvedev, P. (2021). Error correction enables use of Oxford Nanopore technology for reference-free transcriptome analysis. Nat. Commun. 12, 2. doi:10.1038/s41467-020-20340-8
Salwiczek, M., Qu, Y., Gardiner, J., Strugnell, R. A., Lithgow, T., McLean, K. M., et al. (2014). Emerging rules for effective antimicrobial coatings. Trends Biotechnol. 32, 82–90. doi:10.1016/j.tibtech.2013.09.008
SanMiguel, A. J., Meisel, J. S., Horwinski, J., Zheng, Q., Bradley, C. W., and Grice, E. A. (2018). Antiseptic agents elicit short-term, personalized, and body site-specific shifts in resident skin bacterial communities. J. Invest. Dermatol 138, 2234–2243. doi:10.1016/j.jid.2018.04.022
Schiavone, A., Pugliese, N., Samarelli, R., Cumbo, C., Minervini, C. F., Albano, F., et al. (2022). Factors affecting the quality of bacterial genomes assemblies by canu after nanopore sequencing. Appl. Sci. Switz. 12, 3110. doi:10.3390/app12063110
Scotti, E., Boué, S., Sasso, G., Zanetti, F., Belcastro, V., Poussin, C., et al. (2017). Exploring the microbiome in health and disease. Toxicol. Res. Appl. 1, 239784731774188. doi:10.1177/2397847317741884
Selway, C. A., Mills, J. G., Weinstein, P., Skelly, C., Yadav, S., Lowe, A., et al. (2020). Transfer of environmental microbes to the skin and respiratory tract of humans after urban green space exposure. Environ. Int. 145, 106084. doi:10.1016/j.envint.2020.106084
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533. doi:10.1371/journal.pbio.1002533
Shapira, T., Monreal, I. A., Dion, S. P., Buchholz, D. W., Imbiakha, B., Olmstead, A. D., et al. (2022). A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605, 340–348. doi:10.1038/s41586-022-04661-w
Shi, L. D., Xu, Q. J., Liu, J. Y., Han, Z. X., Zhu, Y. G., and Zhao, H. P. (2021). Will a non-antibiotic metalloid enhance the spread of antibiotic resistance genes: The selenate story. Environ. Sci. Technol. 55, 1004–1014. doi:10.1021/acs.est.0c05698
Soininen, L., Roslund, M. I., Nurminen, N., Puhakka, R., Laitinen, O. H., Hyöty, H., et al. (2022). Indoor green wall affects health-associated commensal skin microbiota and enhances immune regulation: A randomized trial among urban office workers. Sci. Rep. 12, 6518. doi:10.1038/s41598-022-10432-4
Stenseth, N. C., Dharmarajan, G., Li, R., Shi, Z. L., Yang, R., and Gao, G. F. (2021). Lessons learnt from the COVID-19 pandemic. Front. Public Health 9, 694705. doi:10.3389/fpubh.2021.694705
Stevens, E. J., Bates, K. A., and King, K. C. (2021). Host microbiota can facilitate pathogen infection. PLoS Pathog. 17, e1009514. doi:10.1371/journal.ppat.1009514
Stoler, N., and Nekrutenko, A. (2021). Sequencing error profiles of Illumina sequencing instruments. Nar. Genom Bioinform 3, lqab019. doi:10.1093/nargab/lqab019
Tan, K. T., Slevin, M. K., Meyerson, M., and Li, H. (2022). Identifying and correcting repeat-calling errors in nanopore sequencing of telomeres. Genome Biol. 23, 180. doi:10.1186/s13059-022-02751-6
Tanaka, M., and Nakayama, J. (2017). Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 66, 515–522. doi:10.1016/j.alit.2017.07.010
Tischer, C., Kirjavainen, P., Matterne, U., Tempes, J., Willeke, K., Keil, T., et al. (2022). Interplay between natural environment, human microbiota and immune system: A scoping review of interventions and future perspectives towards allergy prevention. Sci. Total Environ. 821, 153422. doi:10.1016/j.scitotenv.2022.153422
Tran, P., Kopel, J., Ristic, B., Marsh, H., Fralick, J., and Reid, T. (2022). Antimicrobial seleno-organic coatings and compounds acting primarily on the plasma membrane: A review. Adv. Redox Res. 4, 100031. doi:10.1016/j.arres.2022.100031
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi:10.1016/j.tig.2018.05.008
Vassallo, A., Kett, S., Purchase, D., and Marvasi, M. (2021). Antibiotic-resistant genes and bacteria as evolving contaminants of emerging concerns (e-CEC): Is it time to include evolution in risk assessment? Antibiot. (Basel) 10, 1066. doi:10.3390/antibiotics10091066
Vassallo, A., Kett, S., Purchase, D., and Marvasi, M. (2022). The bacterial urban resistome: Recent advances. Antibiot. (Basel) 11, 512. doi:10.3390/antibiotics11040512
Vernocchi, P., del Chierico, F., and Putignani, L. (2016). Gut microbiota profiling: Metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 7, 1144. doi:10.3389/fmicb.2016.01144
Vijayan, P., Chithra, P. G., Abraham, P., George, J. S., Maria, H. J., Sreedevi, T., et al. (2022). Nanocoatings: Universal antiviral surface solution against COVID-19. Prog. Org. Coat. 163, 106670. doi:10.1016/j.porgcoat.2021.106670
Vincent, M., Hartemann, P., and Engels-Deutsch, M. (2016). Antimicrobial applications of copper. Int. J. Hyg. Environ. Health 219, 585–591. doi:10.1016/j.ijheh.2016.06.003
Visnapuu, M., Rosenberg, M., Truska, E., Nõmmiste, E., Šutka, A., Kahru, A., et al. (2018). UVA-induced antimicrobial activity of ZnO/Ag nanocomposite covered surfaces. Colloids Surf. B Biointerfaces 169, 222–232. doi:10.1016/j.colsurfb.2018.05.009
Wang, X., Lan, B., Fei, H., Wang, S., and Zhu, G. (2021). Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils. J. Hazard Mater 411, 124848. doi:10.1016/j.jhazmat.2020.124848
Warnes, S. L., Little, Z. R., and Keevil, C. W. (2015). Human coronavirus 229E remains infectious on common touch surface materials. mBio 6, e01697–e01615. doi:10.1128/mBio.01697-15
Willis, A. D. (2019). Rarefaction, alpha diversity, and statistics. Front. Microbiol. 10, 2407. doi:10.3389/fmicb.2019.02407
Wu, Z., Chan, B., Low, J., Chu, J. J. H., Hey, H. W. D., and Tay, A. (2022). Microbial resistance to nanotechnologies: An important but understudied consideration using antimicrobial nanotechnologies in orthopaedic implants. Bioact. Mater 16, 249–270. doi:10.1016/j.bioactmat.2022.02.014
Xiao, T., and Zhou, W. (2020). The third generation sequencing: The advanced approach to genetic diseases. Transl. Pediatr. 9, 163–173. doi:10.21037/tp.2020.03.06
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi:10.1038/nature11053
Yin, X., Yang, Y., Deng, Y., Huang, Y., Li, L., Chan, L. Y. L., et al. (2022). An assessment of resistome and mobilome in wastewater treatment plants through temporal and spatial metagenomic analysis. Water Res. 209, 117885. doi:10.1016/j.watres.2021.117885
Zhou, E., Qiao, D., Yang, Y., Xu, D., Lu, Y., Wang, J., et al. (2020). A novel Cu-bearing high-entropy alloy with significant antibacterial behavior against corrosive marine biofilms. J. Mater Sci. Technol. 46, 201–210. doi:10.1016/j.jmst.2020.01.039
Keywords: antimicrobial coating, diversity, microbiota, dysbiosis, homeostasis, resistome, microbiome, metagenome
Citation: Mäki A, Salonen N, Kivisaari M, Ahonen M and Latva M (2023) Microbiota shaping and bioburden monitoring of indoor antimicrobial surfaces. Front. Built Environ. 9:1063804. doi: 10.3389/fbuil.2023.1063804
Received: 07 October 2022; Accepted: 30 January 2023;
Published: 07 February 2023.
Edited by:
Hasim Altan, Prince Mohammad bin Fahd University, Saudi ArabiaReviewed by:
Armin Asghari Alamdari, Koç University, TürkiyeCopyright © 2023 Mäki, Salonen, Kivisaari, Ahonen and Latva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anita Mäki, YW5pdGEucy5tYWtpQGdtYWlsLmNvbQ==; Martti Latva, bWFydHRpLmxhdHZhQHNhbWsuZmk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.