- 1School of Clinical Medicine, Guizhou Medical University, Guiyang, Guizhou, China
- 2Organ Transplantation Department, The Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou, China
Objective: Metagenomic next-generation sequencing (mNGS) is emerging to be pivotal in infectious disease diagnosis, particularly in pulmonary infection. However, the utility of bronchoalveolar lavage fluid (BALF) versus blood mNGS remains controversial. Therefore, the current meta-analysis summarized the previous studies regarding BALF or blood mNGS in pulmonary infection, aiming to comprehensively compare the diagnostic efficiency between them.
Methods: Studies reporting paired BALF and blood mNGS data for pulmonary infection diagnosis were searched in the PubMed, Web of Science, Embase, CNKI, and Wanfang from January 2016 to March 2024.
Results: Eleven studies involving 346 assessed patients were eligible and analyzed. The pooled sensitivity, specificity, and area under the curve (AUC) of BALF mNGS were 0.94 [95% confidence interval (CI): 0.90–0.97], 0.27 (95% CI: 0.19–0.36), and 0.86 [standard error (SE): 0.06], respectively. Meanwhile, the pooled sensitivity, specificity, and AUC of blood mNGS were 0.64 (95% CI: 0.56–0.72), 0.69 (95% CI: 0.62–0.76), and 0.81 (SE: 0.05), respectively. By subgroup analyses, the AUCs of BALF mNGS and blood mNGS for viral detection were 0.70 (SE: 0.08) and 0.71 (SE: 0.08), respectively, while the AUCs of BALF mNGS and blood mNGS for nonviral (bacterial or fungal) detection were 0.83 (SE: 0.06) and 0.73 (SE: 0.08), respectively. Moreover, no threshold effect or publication bias existed, and sensitivity analysis revealed that the findings were generally robust.
Conclusion: BALF mNGS surpasses blood mNGS for total pathogen detection in pulmonary infection patients, while they share a similar efficiency for viral detection.
Systematic review registration: https://www.crd.york.ac.uk/PROSPERO/, identifier CRD42024562740.
Introduction
Lower respiratory tract infections (LRTIs) encompass series of diseases, including pneumonia, bronchitis, and chronic bronchitis, and rank as the leading cause of morbidity and mortality in adults (Feldman and Shaddock, 2019; Detelich and Kempker, 2023). Traditionally, bacteria have been considered the predominant pathogens for severe LRTIs, among which the rate of Streptococcus pneumoniae infection has been reported to approach 50% in community-acquired pneumonia (Griffin et al., 2013; Ren et al., 2021; Suaya et al., 2021). However, with an aging population, more and more patients are affected by severe viral infections, and elderly individuals bare 10 times risk to have virulent pneumonia compared to younger ones, with correspondingly higher morbidity and mortality rates from viral infections (Falsey, 2005; Wu et al., 2021). Since LRTIs become increasingly viral in nature and the specific pathogens are continuously complex, the improvement of diagnostic tools is greatly warranted, which could not only avoid multiple drug resistance but also facilitate individualized treatment (Carroll and Adams, 2016; Griffiths et al., 2017; Cilloniz et al., 2022; Eyre, 2022).
Currently, intravital culture is still the cornerstone identifying pathogenic microorganisms in infection diagnosis (Esposito, 2016; Islam, 2022). However, this method is time-consuming and tedious, and many pathogens are not culturable or difficult to culture, making it challenging for most institutions to complete the cultivation process (Jackson et al., 2020). Immunological methods are other options for pathogen detection, such as complement fixation tests, neutralization tests, enzyme-linked immunosorbent assays, immunofluorescence, and enzyme-linked immune-spot assays, which are simple to operate (Lopes-Luz et al., 2021; Yin et al., 2022). Nevertheless, due to the vast variety of pathogens, the number of antigens and antibodies developed cannot meet the clinical practice requirements in a timely manner. In addition, polymerase chain reaction (PCR) assays exhibit a high sensitivity and specificity for pathogen detection, but they can only focus on a specific kind; meanwhile, gene chip or microarray technology is only able to recognize known pathogens instead of unknown ones (Yoshida et al., 2021; Ji et al., 2023). Therefore, the exploration of novel and efficient detection technology is needed.
Along with the progress of biological technology, metagenomic next-generation sequencing (mNGS) has been introduced as a novel method in pathogen recognition, enabling nontargeted sequencing of DNA/RNA while detecting the sequence of whole pathogens (Gu et al., 2021). Benefiting from its rapid advancements, mNGS exhibits the characteristics of high-speed detection, wide coverage, and satisfied sensitivity, which can effectively make up the shortcomings of traditional culture method or PCR assay (Gu et al., 2021). Inspiringly, mNGS has been applied in the pathogenic diagnosis of pulmonary infection using bronchoalveolar lavage fluid (BALF) or blood samples, which improves pathogen identification and timely individual treatment (Chen et al., 2022; Ma et al., 2022; Tsang et al., 2022). However, the superiority between BALF and blood mNGS in pulmonary infection is controversial (Whiting et al., 2011; Boch et al., 2016; Chen et al., 2020; Chen et al., 2021). For instance, Boch T et al. discovered that the sensitivity and specificity of BALF mNGS are higher compared to blood mNGS and culture (Boch et al., 2016), while Xu Chen et al. reported that the sensitivity of BALF mNGS is higher versus blood mNGS, but no difference in specificity is discovered (Chen et al., 2020). Furthermore, Jinlian Chen et al. revealed that the BALF pathogens are highly in line with blood pathogens via mNGS detection (Chen et al., 2021).
Therefore, this meta-analysis summarized the previous studies regarding BALF or blood mNGS in pulmonary infection, aiming to comprehensively compare the diagnostic efficiency between them.
Methods
Study searching
Studies were searched among medical electronic databases from January 2016 (The approximate time that mNGS was applied in clinical practice) to March 2024 (the time that the article searching of this meta-analysis was performed) by two independent reviewers (SZ and LGF). The disputes were resolved through negotiation. The databases included PubMed, Web of Science, Embase, China National Knowledge Infrastructure (CNKI), and Wanfang. Keywords and medical subject headings terms included metagenomic sequencing, metagenomic next-generation sequencing, metagenomic second-generation sequencing, genomics, genetic diagnosis, sequencing, mNGS, pulmonary infection, PI, infection, pneumonia, bronchoalveolar lavage fluid, bronchoalveolar lavage, alveolar lavage, BALF, blood, and plasma. This meta-analysis was prepared referring to the PRISMA 2020 checklist (Page et al., 2021).
Study selection
Inclusion criteria of study selection were: i) the study had an explicit definition of pulmonary infection, which mainly consisted of symptoms, laboratory test data, imaging data, and clinicians’ final judgment; ii) the study had paired pathogen detection data of mNGS in BALF and blood; and iii) the study extracted pathogen detection data of mNGS in BALF and blood (involving true positive (TP), false positive (FP), false negative (FN), and true negative (TN)). The exclusion criteria were as follows: i) the study was a duplicate publication; ii) the study was a review, expert consensus, conference abstract, case report, letter, or dissertation; and iii) the subjects underwent multiple organ transplantation. Studies were selected by two reviewers (SZ and LGF) individually, and disagreements were settled via discussion (SZ. LGF, and YF).
Data extraction and quality assessment
Study author, publication year, setting, study type, subject numbers, and subject age were extracted and reviewed by two reviewers (SZ and LGF). In addition, the TP, FP, FN, and TN of mNGS in BALF and blood samples were recorded, which were calculated based on sample size, sensitivity, specificity, and area under the curve (AUC) if they were not provided directly. The detailed data of TP, FP, FN, and TN for viral and non-viral (bacterial or fungal) detection were also extracted if reported. The study qualities were evaluated individually by two reviewers (SZ and LGF) by Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) (Whiting et al., 2011).
Statistics
Meta-Disc version 1.4 was applied for statistical analyses. The pooled sensitivity, specificity, diagnostic odds ratio (OR), positive likelihood ratio (LR), and negative LR for the diagnosis of pathogen detection were calculated using a bivariate meta-analysis model. The summary receiver operating characteristic (sROC) curves and AUC were applied to examine the diagnostic ability. The Spearman correlation coefficient of sensitivity with 1-specificity was estimated to assess the threshold effect. The non-threshold effect heterogeneity was determined by DerSimonian and Laird random effects models using the I2 test: if P <0.05 or I2 >50%, significant heterogeneity was indicated in the pooled estimates. Moreover, subgroup analysis and sensitivity analysis (by removing each study and analyzing again) were executed to assess the potential source of heterogeneity. Deek’s test was applied for publication bias assessment. P <0.05 was indicated as significant.
Results
Study flow and information
Based on the search strategy and databases, a total of 323 records were identified. After removing 111 duplicates, 212 records remained. Following a thorough review of the titles, abstracts, and full texts, 251 studies were excluded, leaving 11 studies in this meta-analysis (Chen et al., 2020; Chen et al., 2021; Jiang et al., 2021; Xie et al., 2021; Chien et al., 2022; Duan et al., 2022; Ma et al., 2022; Sun et al., 2022; Wei et al., 2022; Wu et al., 2022; Wang et al., 2023) (Figure 1).
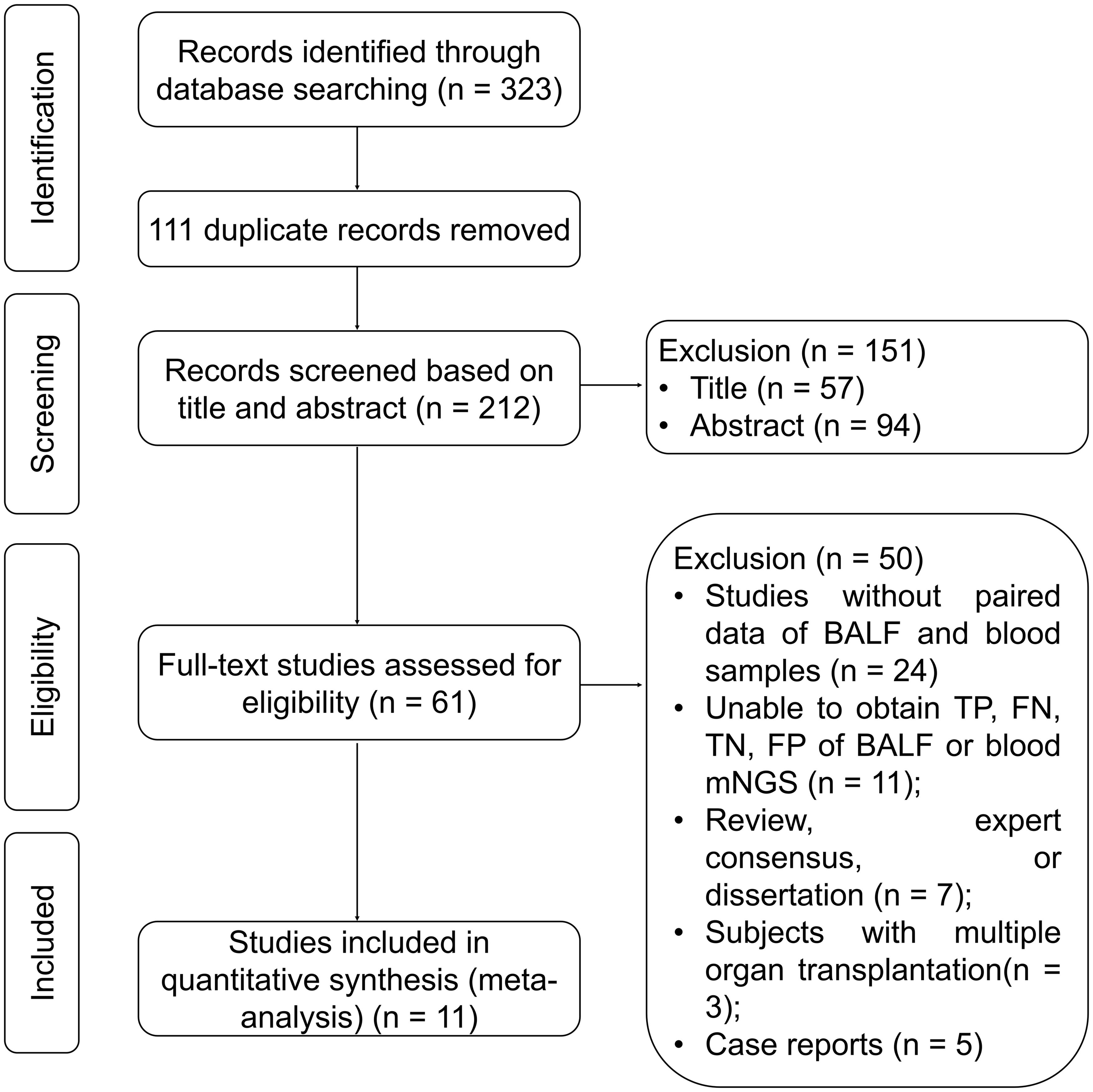
Figure 1. Study flow chart. BALF, bronchoalveolar lavage fluid; FN, false negative; FP, false positive; mNGS, metagenomic next-generation sequencing; TN, true negative; TP, true positive.
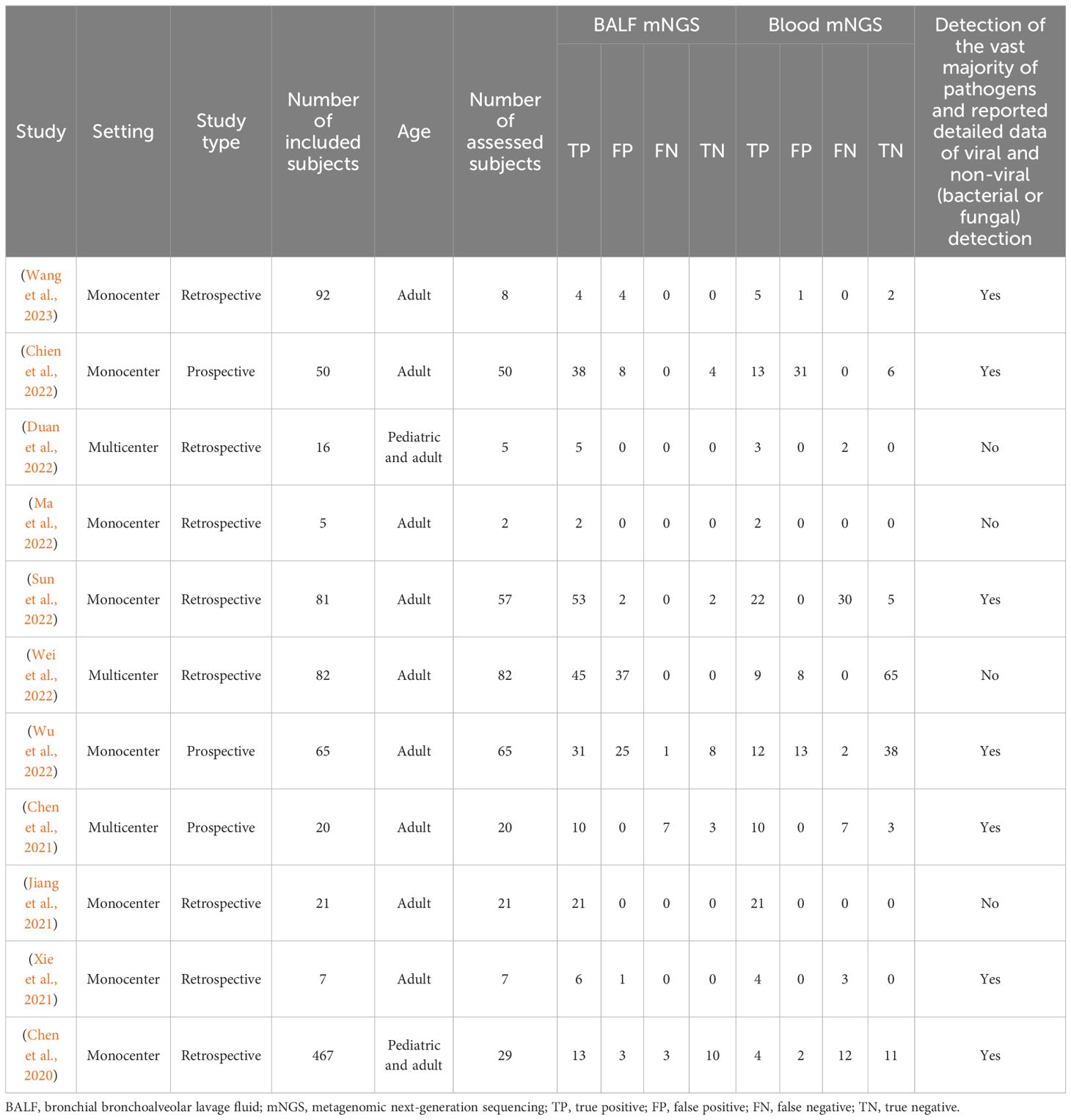
Among the 11 analyzed studies, 3 were multicenter and the other 8 were monocenter, consisting of 346 assessed patients, with the detailed information exhibited in Table 1. It was worth noting that 4 studies focused solely on a single pathogen or infrequent pathogens, lacking the result protocols for other common pathogens, while the other 7 studies included the vast majority of pathogens, but only a few of them were able to provide complete data. Therefore, this meta-analysis emphasized on the diagnostic efficiency of the overall detection rate of pathogens rather than the diagnostic efficiency of a single pathogen.
Quality assessment
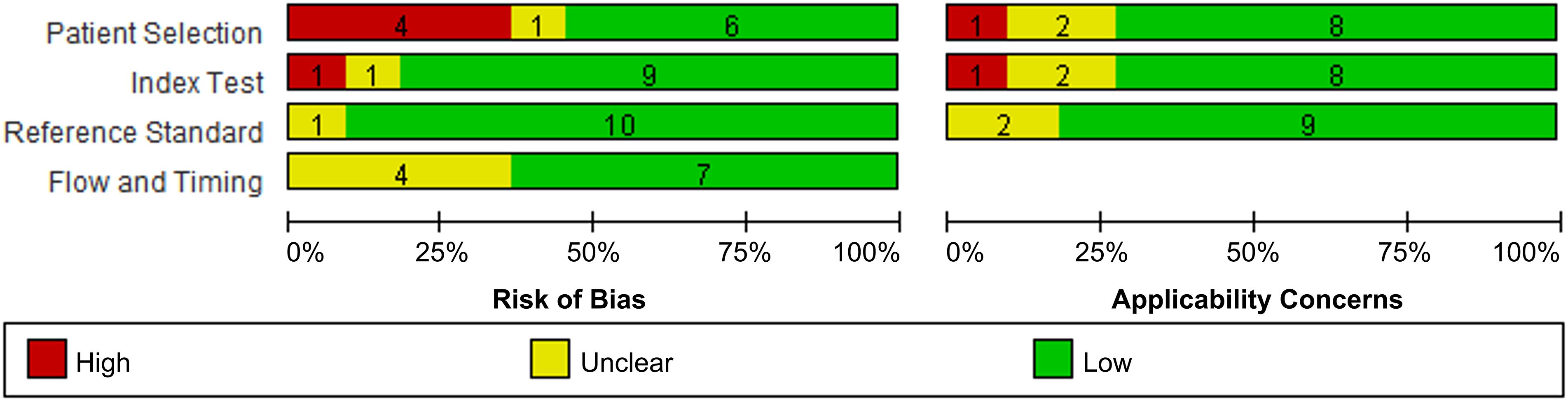
Generally, the analyzed studies showed a low risk of bias and application concerns (Figure 2). However, 4 studies and 1 study exhibited a high risk of patient selection and index test, respectively; and each 1 study disclosed corresponding application concern.
Diagnostic accuracy of BALF and blood mNGS for pulmonary infection
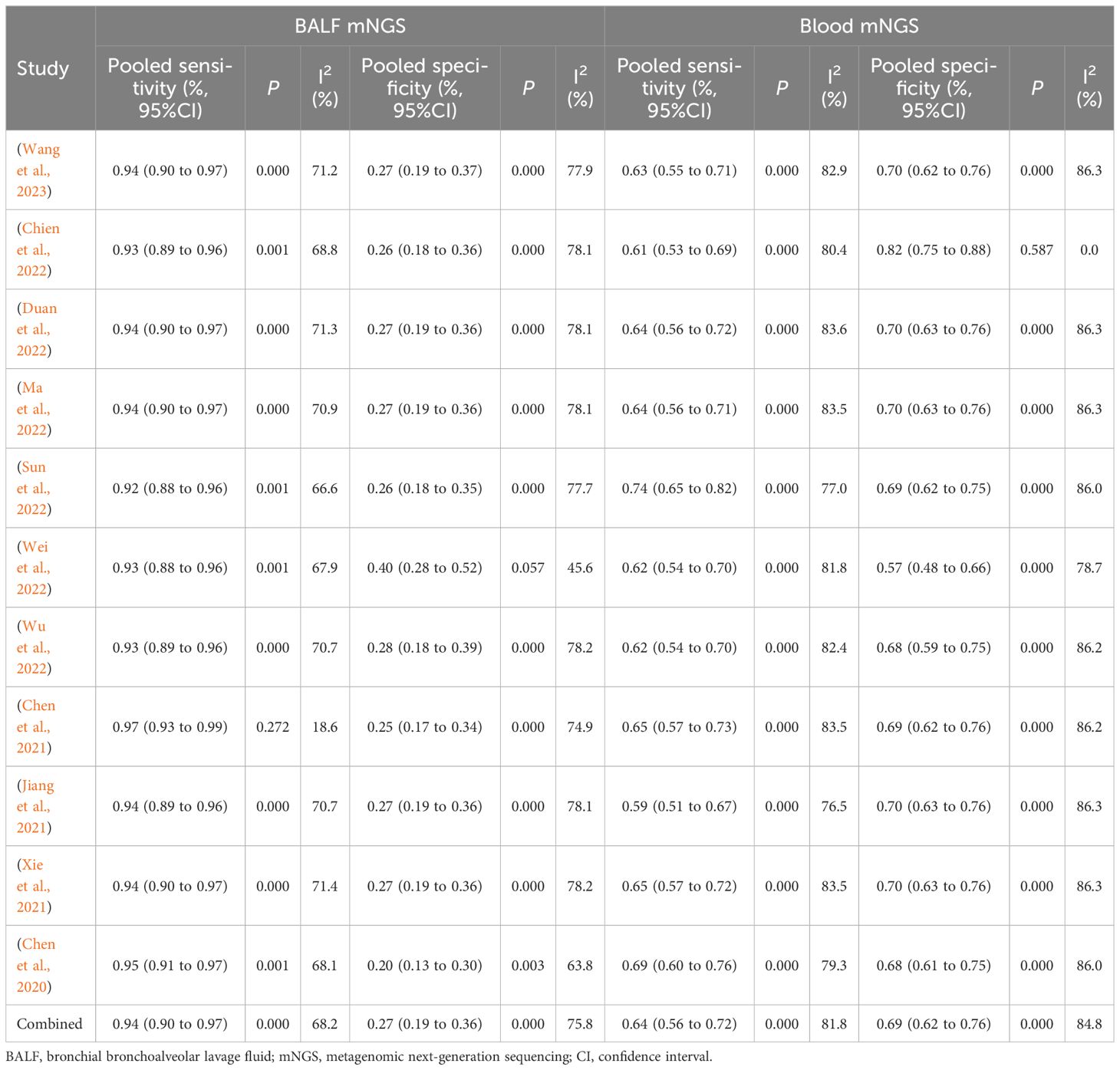
According to the eligibility criteria, all 11 studies were analyzed in this meta-analysis regarding the diagnostic accuracy of both BALF and blood mNGS for pulmonary infection. The pooled sensitivity and specificity of BALF mNGS were 0.94 (95% CI: 0.90–0.97) and 0.27 (95% CI: 0.19–0.36), respectively (Figures 3A, B). The pooled AUC of BALF mNGS was 0.86 [standard error (SE): 0.06] (Figure 3C). Moreover, the pooled diagnostic OR, positive LR, and negative LR were 11.69 (95% CI: 4.54–30.09), 1.52 (95% CI: 0.92–2.51), and 0.25 (95% CI: 0.13–0.47), respectively (Figures 3D–F).
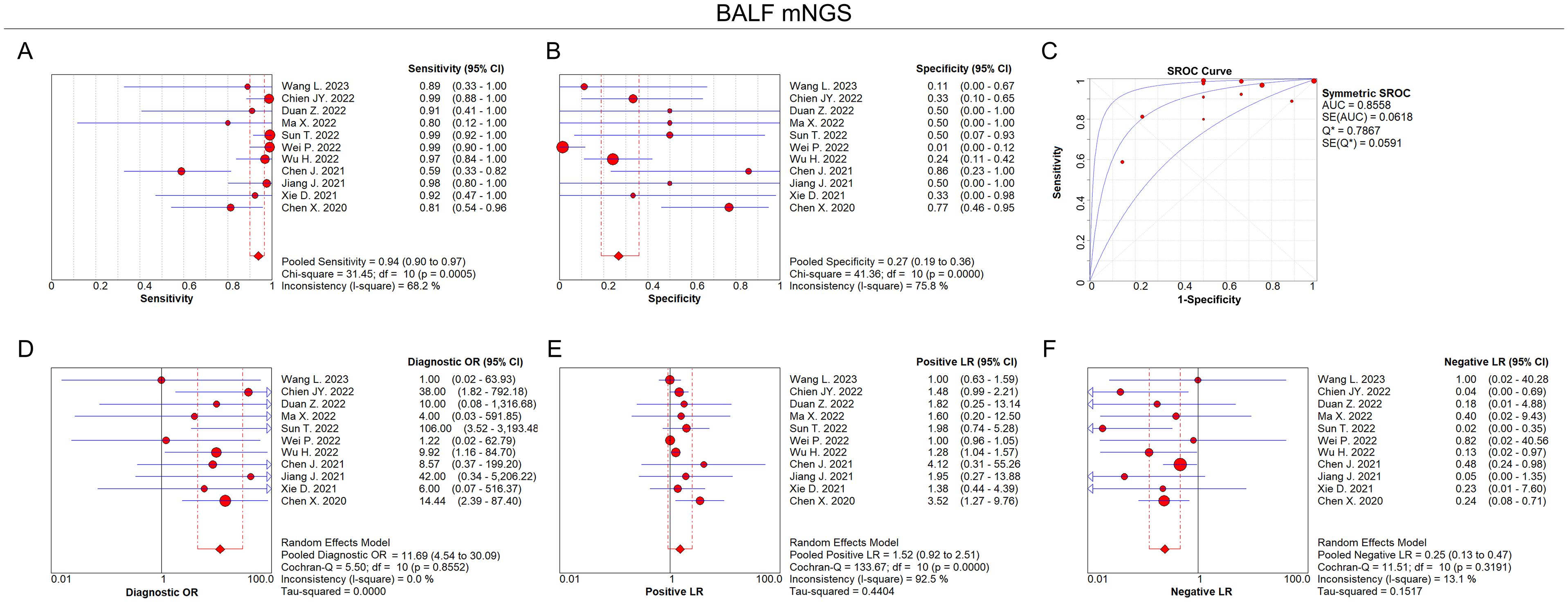
Figure 3. Diagnostic value of BALF mNGS for pulmonary infection. Pooled analysis of sensitivity (A), specificity (B), AUC of SROC curve (C), diagnostic OR (D), positive LR (E), and negative LR (F) regarding BALF mNGS for pulmonary infection diagnosis.
In terms of blood mNGS, the pooled sensitivity and specificity were 0.64 (95% CI: 0.56–0.72) and 0.69 (95% CI: 0.62–0.76), respectively (Figures 4A, B). The pooled AUC was 0.81 [standard error (SE): 0.05] (Figure 4C). In addition, the pooled diagnostic OR, positive LR, and negative LR were 8.47 (95% CI: 3.61–19.91), 2.40 (95% CI: 1.06–5.40), and 0.43 (95% CI: 0.23–0.81), respectively (Figures 4D–F).
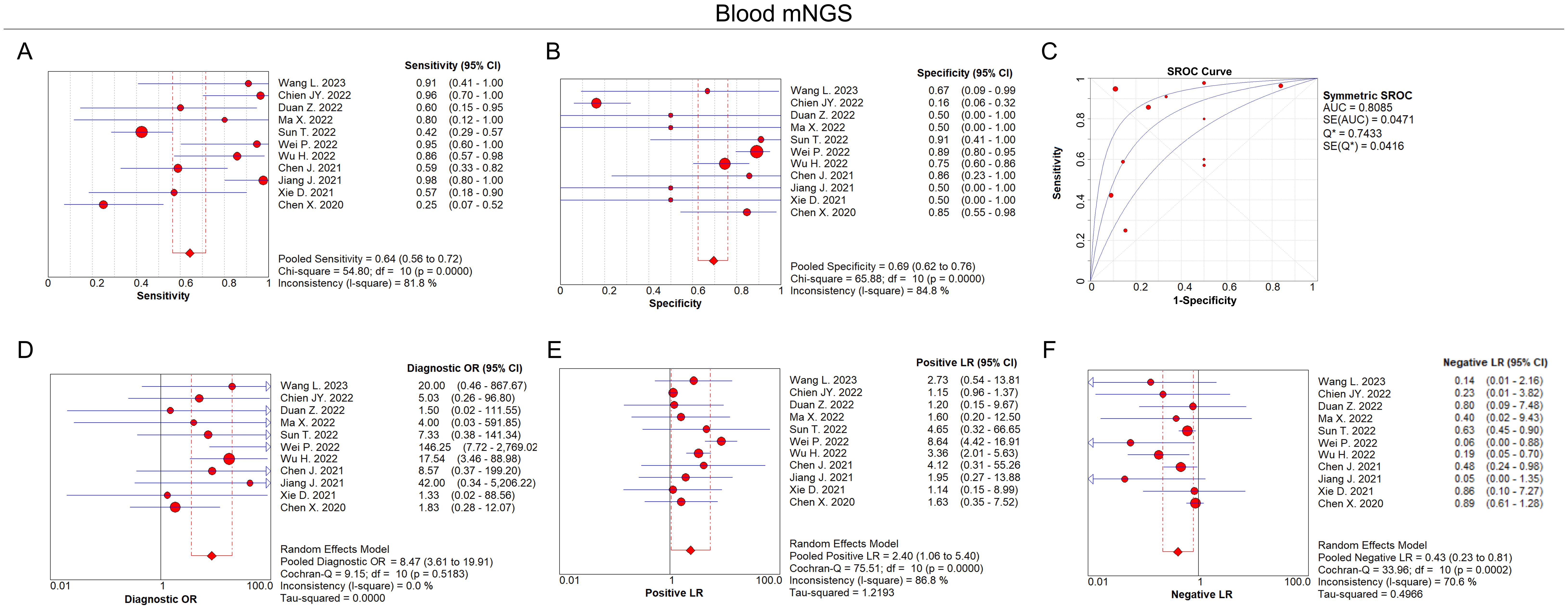
Figure 4. Diagnostic value of blood mNGS for pulmonary infection. Pooled analysis of sensitivity (A), specificity (B), AUC of SROC curve (C), diagnostic OR (D), positive LR (E), negative LR (F) in the aspect of blood mNGS on pulmonary infection diagnosis.
According to the AUC value of SROC, the diagnostic efficiency of BALF mNGS was more accurate compared to blood mNGS for pulmonary infection (0.86 versus 0.81). Meanwhile, the sensitivity of BALF mNGS was higher versus blood mNGS (0.94 versus 0.64), but the specificity of BALF mNGS was lower versus blood mNGS (0.27 versus 0.69).
BALF mNGS and blood mNGS for viral or non-viral pulmonary infection
Interestingly, 6 out of the 11 analyzed studies disclosed the detailed data of TP, FP, FN, and TN on viral and non-viral (bacterial or fungal) detection of pulmonary infection, which proceeded to detect the diagnostic accuracy of BALF and blood mNGS on viral and non-viral pulmonary infection, respectively.
For viral detection, the pooled sensitivity and specificity of BALF mNGS were 0.29 (95% CI: 0.24–0.36) and 0.81 (95% CI: 0.63–0.93), respectively (Figures 5A, B); the pooled AUC of BALF mNGS was 0.70 (SE: 0.08) (Figure 5C); and the pooled diagnostic OR, positive LR, and negative LR of BALF mNGS are shown in Supplementary Figures 1A–C. Regarding blood mNGS, the pooled sensitivity and specificity were 0.37 (95% CI: 0.30–0.45) and 0.94 (95% CI: 0.87–0.98), respectively (Figures 5D, E); the pooled AUC was 0.71 (SE: 0.08) (Figure 5F); at the same time, the pooled diagnostic OR, positive LR, and negative LR of blood mNGS are shown in Supplementary Figures 1D–F. By comparison of the AUC values, the diagnostic efficiency of BALF mNGS was fair compared with that of blood mNGS for viral pulmonary infection (0.70 versus 0.71), but both the sensitivity (0.29 versus 0.37) and specificity (0.81 versus 0.94) of BALF mNGS were lower than those of blood mNGS.
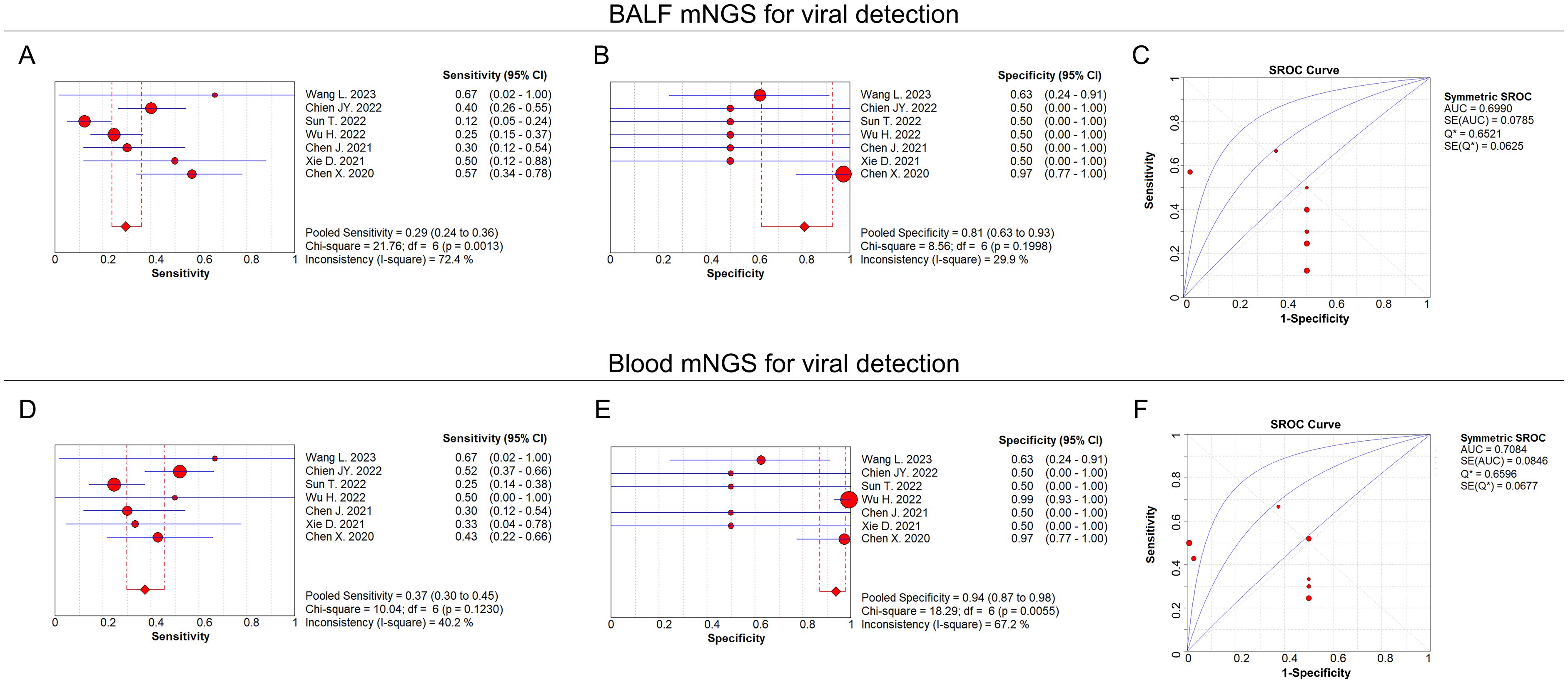
Figure 5. BALF and blood mNGS for viral detection. Pooled analysis of sensitivity (A), specificity (B), and AUC of the SROC curve (C) regarding BALF mNGS for detecting viral pulmonary infection. Pooled analysis of sensitivity (D), specificity (E), and AUC of the SROC curve (F) regarding blood mNGS for detecting viral pulmonary infection.
For non-viral (bacterial or fungal) detection, the pooled sensitivity and specificity of BALF mNGS were 0.67 (95% CI: 0.58–0.75) and 0.54 (95% CI: 0.44–0.64), respectively (Figures 6A, B); the pooled AUC of BALF mNGS was 0.83 (SE: 0.06) (Figure 6C); and the pooled diagnostic OR, positive LR, and negative LR of BALF mNGS are shown in Supplementary Figures 2A–C. As to the blood mNGS, the pooled sensitivity and specificity were 0.46 (95% CI: 0.37–0.55) and 0.64 (95% CI: 0.54–0.72), respectively (Figures 6D, E); the pooled AUC was 0.73 (SE: 0.08) (Figure 6F); moreover, information on the pooled diagnostic OR, positive LR, and negative LR of blood mNGS is shown in Supplementary Figures 2D–F.
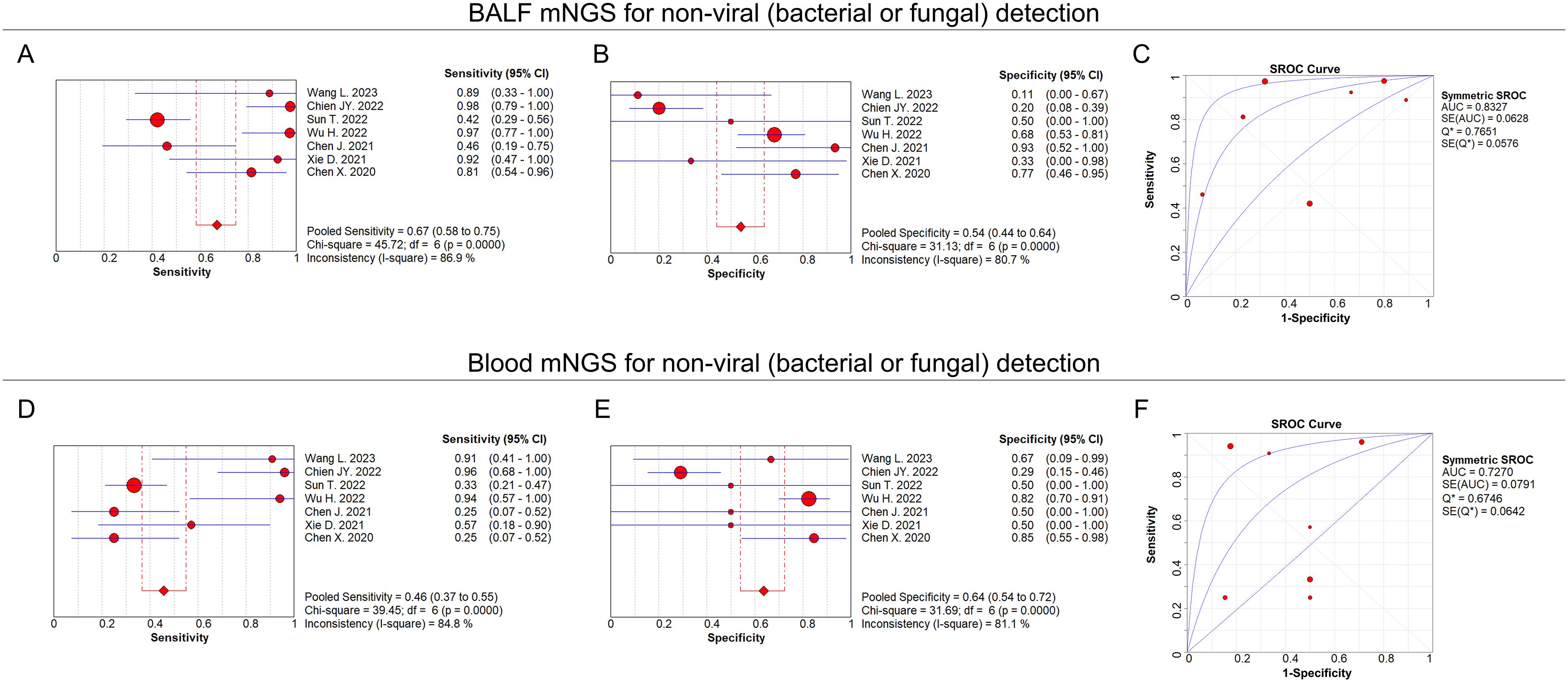
Figure 6. BALF and blood mNGS on non-viral detection. Pooled analysis of sensitivity (A), specificity (B), and AUC of the SROC curve (C) regarding BALF mNGS for detecting non-viral (bacterial or fungal) pulmonary infection. Pooled analysis of sensitivity (D), specificity (E), and AUC of the SROC curve (F) regarding blood mNGS for detecting non-viral (bacterial or fungal) pulmonary infection.
Threshold effect, publication bias, and sensitivity analyses
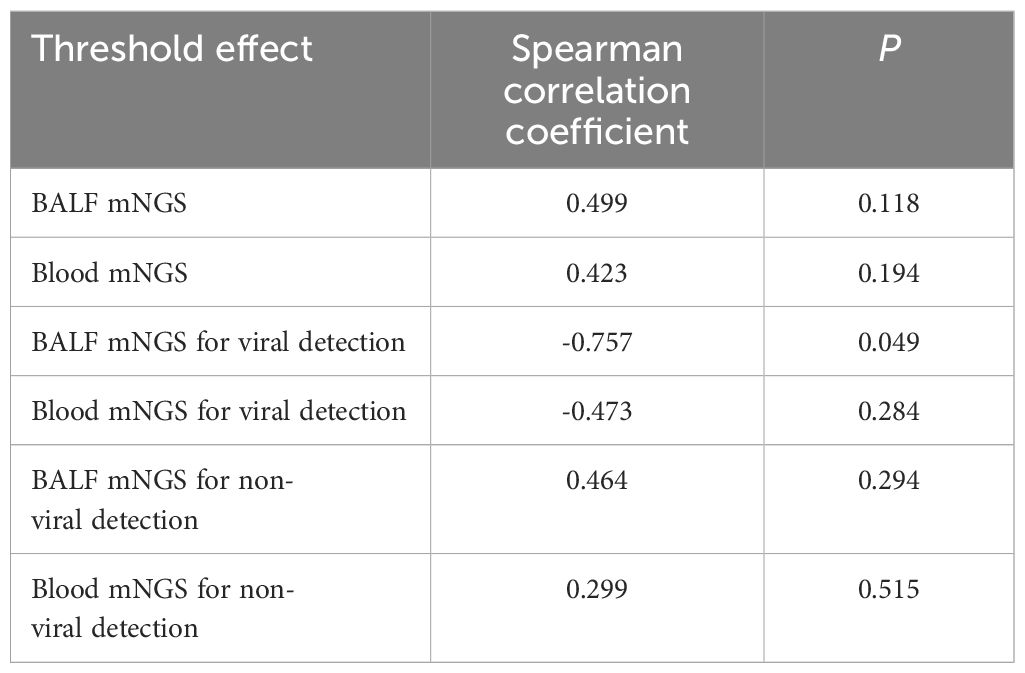
No threshold effect of diagnostic efficiency regarding BALF mNGS and blood mNGS existed in this meta-analysis (Table 2). Meanwhile, Deek’s test revealed that no publication bias existed (P=0.450 for BALF mNGS and P=0.570 for blood mNGS) (data not shown). In addition, sensitivity analysis of the meta-analysis was also performed, which revealed that the pooled diagnostic sensitivity and specificity for pulmonary infection by both BALF and blood mNGS were generally robust (Table 3). However, the study (Wei et al., 2022) affected the pooled specificity of BALF mNGS, the study (Sun et al., 2022) influenced the pooled sensitivity of blood mNGS, and the study (Chien et al., 2022) and the study (Wei et al., 2022) affected the pooled specificity of blood mNGS.
Discussion
Although mNGS has been widely utilized in the diagnosis of infectious diseases, there are still some limitations regarding its clinical application due to its relatively high cost. This is particularly true in the diagnosis of pulmonary infectious diseases, where previous meta-analyses aimed to compare the diagnostic efficacy of BALF mNGS with traditional detection methods (Chen et al., 2022). However, published studies have also reported inconsistent comparisons between blood mNGS and BALF mNGS. This article represents the first meta-analysis to compare the diagnostic accuracy of BALF mNGS and blood mNGS. By supplementing the microbiological results obtained by conventional bacterial culture methods and PCR, mNGS can provide clinicians with precise antimicrobial treatment instructions for infectious diseases (Chen et al., 2021).
BALF collection is difficult with tedious steps in clinical practice and is invasive, which is especially hard to perform in severe-illness patients and those who are unable to cooperate; otherwise, blood specimen collection is more friendly to patients (Chiu and Miller, 2019). Therefore, this study compared the diagnostic performance about these two kinds of specimens from the perspective of patients. In patients with non-systemic pulmonary infection or focal infection, BALF specimens are often used in clinical practice, while sputum specimens are relatively rarely used because they are easy to be contaminated and affected by respiratory tract and external environment; at the same time, sputum collection requires patients to collect deep sputum after coughing (Liu and Xie, 2016). However, there are few clinical studies comparing BALF and sputum specimens for mNGS, and prospective studies are needed in the future.
This meta-analysis revealed that the sensitivity of BALF mNGS was greater than that of blood mNGS (0.94 versus 0.64), but the specificity of BALF mNGS was less accurate than that of blood mNGS (0.27 versus 0.69). According to the AUC of the SROC, the diagnostic efficiency of BALF mNGS was higher versus blood mNGS (0.86 versus 0.81). The possible reasons for these results are discussed below. The sensitivity of blood mNGS may be affected by the number of days the patient was hospitalized at the time of specimen collection. Blood-mNGS has an advantage in identifying viruses. Further studies are needed to determine whether the microorganisms detected by blood mNGS, especially viruses, are from pneumonia or simply reflect unculturable microorganisms present in blood (Chien et al., 2022). False negative results for some viruses can occur with DNA sequencing but not RNA sequencing, and mNGS-based testing can provide a more comprehensive infection profile from a single test. False positives can be produced by microbes that are widespread in the environment or DNA contaminants in laboratory kits. The cutoff readings of different microorganisms in mixed infections and their clinical significance remain to be confirmed, and high detection thresholds may filter out fewer pathogens when major pathogens are detected. The sensitivity of blood mNGS may also be affected by the length of hospital stay at the time of specimen collection (Sun et al., 2022).
Mixed infection is common in immunocompromised patients, among which bacterial-fungi-viral infection is the most common type. Due to the suppression of immune function, the virus cannot be cleared in time; and once the patient is infected, common pneumonia will rapidly progress to severe pneumonia, and may even cause death (Xian et al., 2023; Rucar et al., 2024). Due to the poor effect of routine microbiological detection in the diagnosis of mixed infection, a false positivity or false negativity often appears. Therefore, mixed infection is common, but the diagnosis is difficult, and most of the diagnosis of mixed infection is based on NGS results (Dong and Zhou, 2023). Besides, the direct comparison of diagnostic performance between mNGS in BALF and blood is lacking in previous studies; thus, future comparative studies are needed.
This meta-analysis also performed a subgroup analysis regarding the pathogen types. At the same time, we also divided the pathogens of pneumonia patients into viral and non-viral pathogens for subgroup analysis. In viral pneumonia patients, both the sensitivity and specificity of blood mNGS were greater than those of BALF mNGS (0.37 versus 0.29 and 0.94 versus 0.81, respectively). In non-viral pneumonia patients, the sensitivity of BALF mNGS was more accurate than that of blood mNGS (0.67 versus 0.46), but the specificity of BALF mNGS was less accurate than that of blood mNGS (0.54 versus 0.64). According to the AUC of the SROC, the diagnostic efficiency of BALF mNGS was more accurate than that of blood mNGS (0.83 versus 0.73). These results can be attributed to the fact that viral pathogens cannot be cultured by conventional means, and imaging methods are mainly used for clinical diagnosis. On the other hand, bacterial and fungal pathogens can be diagnosed by conventional culture methods, resulting in a higher detection rate of bacterial pathogens in clinics than viral pathogens. Additionally, most papers lack independent introductions on tuberculosis and specific pathogens, such as parasites, mycoplasma, and chlamydia, which limits further subgroup analysis (Varadi et al., 2017). Similarly, most articles lack independent introductions on whether patients have undergone solid organ transplantation and have low immune function, resulting in missing data. The pathogens of lung infection in patients with solid organ transplantation and immunocompromised function differ from those in the general population. Therefore, it is meaningful to compare the results of mNGS technology in blood and BALF in solid organ transplantation and immunocompromised populations. For some viruses, if not found in BALF mNGS but detected in blood mNGS, the surface viruses are less possible to be the pathogens of lung infection; otherwise, the viruses may come from other parts of the body, enter the blood system, and spread in the blood. Some viruses commonly found in lung infections, such as EBV, can also be complications due to impaired host immunity (Tian et al., 2022).
Although mNGS has gained widespread use in the diagnosis of infectious diseases, its clinical application is still subject to some doubts due to its relatively expensive nature as a new technology. Specifically, in the diagnosis of pulmonary infectious diseases, previous meta-analyses aimed to compare the diagnostic efficacy of BALF mNGS with that of traditional detection methods. However, published studies have also found inconsistent comparisons between blood and BALF. A previous meta-analysis was published on the application of second-generation sequencing technology in the diagnosis of pulmonary infection, mainly comparing the application of second-generation sequencing technology and conventional culture technology in the diagnosis of pulmonary infection. For critically ill patients with no clear pathogen diagnosis from routine testing, early selection of mNGS is recommended or even earlier when diagnosis is not possible. Although this meta-analysis compares mNGS with conventional detection methods, there is no doubt that the diagnostic performance of mNGS is superior to that of conventional methods due to its advanced nature. The current meta-analysis further compared the diagnostic performance of mNGS among different samples (blood and BALF) and explores its application in clinical practice, enabling clinicians to choose the most appropriate detection methods. On the one hand, selecting the most appropriate methods can reduce the burden on patients and minimize the occurrence of excessive medical treatment (Chen et al., 2022).
There are certain limitations in this meta-analysis. For instance, although this meta-analysis discusses the comparative analysis of the clinical application of blood and alveolar lavage fluid mNGS, it concludes that the diagnostic efficacy of alveolar lavage fluid is superior to that of blood. Pulmonary infection is a disease that changes and develops rapidly, and it exhibits different characteristics at different stages. For instance, when pulmonary infection progresses to sepsis and pathogens enter the blood, the detection rate of mNGS will naturally increase. Additionally, for blood-borne pulmonary infection, mNGS may be detected earlier in the blood than in the alveolar lavage fluid referring to our experience. Different pathogens have different causes of bacterial entry into the blood or treatment. Therefore, determining when to conduct mNGS detection is a problem that must be addressed. However, the continuous and dynamic use of mNGS to detect blood and alveolar lavage will impose a significant financial burden and some adverse effects on patients (Sun et al., 2022). Furthermore, we used clinical comprehensive diagnosis as the gold standard, including pathogen culture, imaging detection, nucleic acid detection, etc. As a result, there were significant differences in diagnosis among the included papers, resulting in substantial heterogeneity. At the same time, only a portion of the articles explained whether antibiotics and antiviral drugs were used before detection, but the original data and specific situation could not be provided. Microbial contamination also complicates the interpretation of mNGS results, and the rich genetic background of the human host limits the amount of pathogen gene sequences (Chen et al., 2020; Wei et al., 2022).
mNGS offers significant advantages and potential in detecting certain pathogens, particularly in cases of low pathogen DNA load and atypical symptoms. For instance, Dan Xie et al. proposed that mNGS could be utilized for diagnosing Pneumocystis jirovecii pneumonia (PJP) in kidney transplant recipients, specifically for psittacosis, pneumocystis pneumonia, and other specific pathogenic microorganisms (Xie et al., 2021). This study provides guidance for PJP prophylaxis and immunosuppressant use in renal transplant recipients during and after acute rejection treatment. Juan Jiang et al. suggested that simultaneous detection of mNGS in BALF and blood samples obtained from the same patient among ordinary PJP patients was highly consistent with the detection of Pneumocystis jirovecii (Jiang et al., 2021). Therefore, when bronchoscopy is ineffective, mNGS in blood samples may be a viable option for PJP patients. Xiaoxu Ma et al. demonstrated that peripheral blood mNGS could be used in diagnosing invasive pulmonary aspergillosis (Ma et al., 2022). Zhimei Duan et al. believed that mNGS had greater potential in diagnosing psitsitosis fever, and BALF samples, in particular, were a promising diagnostic sample in all kinds of samples (Duan et al., 2022). However, in patients who cannot tolerate bronchoscopy, blood samples can still be used as an alternative method for early diagnosis of psitsitosis with mNGS, even though the pathogen DNA load in the samples is lower than that in BALF. For these particular pathogens, are the microorganisms detected by mNGS clinically significant pathogens or colonizing microorganisms? Blood samples may help distinguish pneumocystis colonization from infection. The pathogen is present in the respiratory tract of healthy individuals, so interpreting the mNGS results from BALF specimens can sometimes be challenging. Therefore, identifying PJP in blood samples by mNGS may be more convincing, as it should not be present in the blood of healthy individuals (Hammarstrom et al., 2019).
Conclusively, this meta-analysis summarizes that BALF mNGS is superior to blood mNGS for pathogenic detection in pulmonary infection patients, especially for non-viral (bacterial or fungal) pathogens. Interestingly, for viral detection, BALF mNGS exhibits similar diagnostic efficiency to blood mNGS in pulmonary infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZS: Data curation, Investigation, Visualization, Writing – original draft. GL: Data curation, Supervision, Validation, Visualization, Writing – review & editing. YN: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Thanks to the Natural Science Foundation of Guizhou Province (Natural Science)-2k[2023] general 380 support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2024.1440288/full#supplementary-material
Supplementary Figure 1 | Other information on BALF and blood mNGS for viral detection.
Supplementary Figure 2 | Other information on BALF and blood mNGS for non-viral detection.
References
Boch T., Spiess B., Cornely O. A., Vehreschild J. J., Rath P. M., Steinmann J., et al. (2016). Diagnosis of invasive fungal infections in haematological patients by combined use of galactomannan, 1,3-beta-D-glucan, Aspergillus PCR, multifungal DNA-microarray, and Aspergillus azole resistance PCRs in blood and bronchoalveolar lavage samples: results of a prospective multicentre study. Clin. Microbiol. Infect. 22, 862–868. doi: 10.1016/j.cmi.2016.06.021
Carroll K. C., Adams L. L. (2016). Lower respiratory tract infections. Microbiol. Spectr. 4, 0029-2016. doi: 10.1128/microbiolspec.DMIH2-0029-2016
Chen J., Zhao Y., Shang Y., Lin Z., Xu G., Bai B., et al. (2021). The clinical significance of simultaneous detection of pathogens from bronchoalveolar lavage fluid and blood samples by metagenomic next-generation sequencing in patients with severe pneumonia. J. Med. Microbiol. 70, 867–873. doi: 10.1099/jmm.0.001259
Chen S., Kang Y., Li D., Li Z. (2022). Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in patients with pulmonary infections: Systematic review and meta-analysis. Int. J. Infect. Dis. 122, 867–873. doi: 10.1016/j.ijid.2022.07.054
Chen X., Ding S., Lei C., Qin J., Guo T., Yang D., et al. (2020). Blood and bronchoalveolar lavage fluid metagenomic next-generation sequencing in pneumonia. Can. J. Infect. Dis. Med. Microbiol. 2020, 6839103. doi: 10.1155/2020/6839103
Chien J. Y., Yu C. J., Hsueh P. R. (2022). utility of metagenomic next-generation sequencing for etiological diagnosis of patients with sepsis in intensive care units. Microbiol. Spectr. 10, e0074622. doi: 10.1128/spectrum.00746-22
Chiu C. Y., Miller S. A. (2019). clinical metagenomics. Nat. Rev. Genet. 20, 341–355. doi: 10.1038/s41576-019-0113-7
Cilloniz C., Pericas J. M., Rojas J. R., Torres A. (2022). Severe infections due to respiratory viruses. Semin. Respir. Crit. Care Med. 43, 60–74. doi: 10.1055/s-0041-1740982
Detelich J. F., Kempker J. A. (2023). Respiratory infections. Clin. Chest Med. 44, 509–517. doi: 10.1016/j.ccm.2023.03.007
Dong Z. L., Zhou K. H. (2023). Application progress of metagenomic next-generation sequencing in pulmonary infection after organ transplantation. J. Clin. Pulmonary Med. 28, 1757–1761. doi: 10.3969/j.issn.1009–6663.2023.11.029
Duan Z., Gao Y., Liu B., Sun B., Li S., Wang C., et al. (2022). The application value of metagenomic and whole-genome capture next-generation sequencing in the diagnosis and epidemiological analysis of psittacosis. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.872899
Esposito S. (2016). Infectious diseases: pathophysiology, diagnostics and prevention. Int. J. Mol. Sci. 17 (9), 1464. doi: 10.3390/ijms17091464
Eyre D. W. (2022). Infection prevention and control insights from a decade of pathogen whole-genome sequencing. J. Hosp Infect. 122, 180–186. doi: 10.1016/j.jhin.2022.01.024
Falsey A. R. (2005). Respiratory syncytial virus infection in elderly and high-risk adults. Exp. Lung Res. 31 Suppl 1, 77. doi: 10.1056/NEJMoa043951
Feldman C., Shaddock E. (2019). Epidemiology of lower respiratory tract infections in adults. Expert Rev. Respir. Med. 13, 63–77. doi: 10.1080/17476348.2019.1555040
Griffin M. R., Zhu Y., Moore M. R., Whitney C. G., Grijalva C. G. (2013). U.S. hospitalizations for pneumonia after a decade of pneumococcal vaccination. N Engl. J. Med. 369, 155–163. doi: 10.1056/NEJMoa1209165
Griffiths C., Drews S. J., Marchant D. J. (2017). Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin. Microbiol. Rev. 30, 277–319. doi: 10.1128/CMR.00010-16
Gu W., Deng X., Lee M., Sucu Y. D., Arevalo S., Stryke D., et al. (2021). Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124. doi: 10.1038/s41591-020-1105-z
Hammarstrom H., Grankvist A., Broman I., Kondori N., Wenneras C., Gisslen M., et al. (2019). Serum-based diagnosis of Pneumocystis pneumonia by detection of Pneumocystis jirovecii DNA and 1,3-beta-D-glucan in HIV-infected patients: a retrospective case control study. BMC Infect. Dis. 19, 658. doi: 10.1186/s12879–019-4289–4
Islam T. (2022). Infectious diseases surveillance update. Lancet Infect. Dis. 22, 952. doi: 10.1016/S1473-3099(22)00381-4
Jackson C. D., Burroughs-Ray D. C., Summers N. A. (2020). Clinical guideline highlights for the hospitalist: 2019 American thoracic society/Infectious diseases society of America update on community-Acquired pneumonia. J. Hosp Med. 15, 743–745. doi: 10.12788/jhm.3444
Ji C., Zhou L., Chen Y., Fang X., Liu Y., Du M., et al. (2023). Microfluidic-LAMP chip for the point-of-care detection of gene-deleted and wild-type African swine fever viruses and other four swine pathogens. Front. Vet. Sci. 10. doi: 10.3389/fvets.2023.1116352
Jiang J., Bai L., Yang W., Peng W., An J., Wu Y., et al. (2021). Metagenomic next-generation sequencing for the diagnosis of pneumocystis jirovecii pneumonia in non-HIV-infected patients: A retrospective study. Infect. Dis. Ther. 10, 1733–1745. doi: 10.1007/s40121-021-00482-y
Liu Q., Xie Z. X. (2016). Collection, storage and results interpretation of sputum specimen. Chin. J. Gen. Practitioners 15, 734–736. doi: 10.3760/cma.j.issn.1671–7368.2016.09.022
Lopes-Luz L., Mendonca M., Bernardes Fogaca M., Kipnis A., Bhunia A. K., Buhrer-Sekula S. (2021). Listeria monocytogenes: review of pathogenesis and virulence determinants-targeted immunological assays. Crit. Rev. Microbiol. 47, 647–666. doi: 10.1080/1040841X.2021.1911930
Ma X., Zhang S., Xing H., Li H., Chen J., Li H., et al. (2022). Invasive pulmonary aspergillosis diagnosis via peripheral blood metagenomic next-generation sequencing. Front. Med. (Lausanne) 9. doi: 10.3389/fmed.2022.751617
Page M. J., McKenzie J. E., Bossuyt P. M., Boutron I., Hoffmann T. C., Mulrow C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372, n71. doi: 10.1136/bmj.n71
Ren J., Kang J. B., Ma Y. P., Zhang J. H., Dong C. X., Kang J. M., et al. (2021). Pathogen distribution and antimicrobial resistance among lower respiratory tract infections in patients with hematological Malignancies. Zhonghua Nei Ke Za Zhi 60, 875–879. doi: 10.3760/cma.j.cn112138–20201228–01056
Rucar A., Totet A., Le Govic Y., Demey B., Damiani C. (2024). Pulmonary co-infections by Pneumocystis jirovecii and Herpesviridae: a seven-year retrospective study. Ann. Clin. Microbiol. Antimicrob. 23, 8. doi: 10.1186/s12941-023-00663-2
Suaya J. A., Fletcher M. A., Georgalis L., Arguedas A. G., McLaughlin J. M., Ferreira G., et al. (2021). Identification of Streptococcus pneumoniae in hospital-acquired pneumonia in adults. J. Hosp Infect. 108, 146–157. doi: 10.1016/j.jhin.2020.09.036
Sun T., Liu Y., Cai Y., Zhai T., Zhou Y., Yang B., et al. (2022). A paired comparison of plasma and bronchoalveolar lavage fluid for metagenomic next-generation sequencing in critically ill patients with suspected severe pneumonia. Infect. Drug Resist. 15, 4369–4379. doi: 10.2147/IDR.S374906
Tian X., Duan W., Zhang X., Wu X., Zhang C., Wang Z., et al. (2022). Metagenomic next-generation sequencing reveals the profile of viral infections in kidney transplant recipients during the COVID-19 pandemic. Front. Public Health 10. doi: 10.3389/fpubh.2022.888064
Tsang H. F., Yu A. C. S., Jin N., Yim A. K. Y., Leung W. M. S., Lam K. W., et al. (2022). The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: case reports and literature review. Expert Rev. Mol. Diagn. 22, 575–582. doi: 10.1080/14737159.2022.2071607
Varadi L., Luo J. L., Hibbs D. E., Perry J. D., Anderson R. J., Orenga S., et al. (2017). Methods for the detection and identification of pathogenic bacteria: past, present, and future. Chem. Soc. Rev. 46, 4818–4832. doi: 10.1039/C6CS00693K
Wang L., Li S., Qin J., Tang T., Hong J., Tung T. H., et al. (2023). Clinical diagnosis application of metagenomic next-generation sequencing of plasma in suspected sepsis. Infect. Drug Resist. 16, 891–901. doi: 10.2147/IDR.S395700
Wei P., Wu L., Li Y., Shi J., Luo Y., Wu W., et al. (2022). Metagenomic next-generation sequencing for the detection of pathogenic microorganisms in patients with pulmonary infection. Am. J. Transl. Res. 14, 6382–6388.
Whiting P. F., Rutjes A. W., Westwood M. E., Mallett S., Deeks J. J., Reitsma J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536. doi: 10.7326/0003-4819-155-8-201110180-00009
Wu H., Cheng H., Fang Y., Wang X., Xie W. (2022). Application value of metagenomic next generation sequencing technology in etiological diagnosis of severe pneumonia. J. Xinjiang Med. Univ. 45, 1301–1305.
Wu Y., Goplen N. P., Sun J. (2021). Aging and respiratory viral infection: from acute morbidity to chronic sequelae. Cell Biosci. 11, 112. doi: 10.1186/s13578-021-00624-2
Xian Y., Duan Z. Q., Li H., Bi X. G., Zhang K. X. (2023). Characteristics of infection pathogens and risk of death after kidney trans-plantation. Chin. J. Infection Control 22, 539–546.
Xie D., Xu W., You J., Yuan X., Li M., Bi X., et al. (2021). Clinical descriptive analysis of severe Pneumocystis jirovecii pneumonia in renal transplantation recipients. Bioengineered 12, 1264–1272. doi: 10.1080/21655979.2021.1911203
Yin J., Wang A., Zhou J., Chen Y., Liang C., Zhu X., et al. (2022). Establishment of an immunological method for detection of bluetongue virus by fluorescence-linked immunosorbent assay. Microbiol. Spectr. 10, e0142922. doi: 10.1128/spectrum.01429-22
Yoshida K., Hatachi T., Okamoto Y., Aoki Y., Kyogoku M., Moon Miyashita K., et al. (2021). Application of multiplex polymerase chain reaction for pathogen identification and antibiotic use in children with respiratory infections in a PICU. Pediatr. Crit. Care Med. 22, e644–e6e8. doi: 10.1097/PCC.0000000000002794
Keywords: pulmonary infection, bronchoalveolar lavage fluid, blood, metagenomic next-generation sequencing, diagnostic efficiency
Citation: Sun Z, Liang G and Niu Y (2024) Comparison between metagenomic next-generation sequencing in bronchoalveolar lavage fluid and blood for the diagnostic performance of respiratory tract infection: a meta-analysis. Front. Bacteriol. 3:1440288. doi: 10.3389/fbrio.2024.1440288
Received: 29 May 2024; Accepted: 29 July 2024;
Published: 15 August 2024.
Edited by:
Zhuo Ma, Albany College of Pharmacy and Health Sciences, United StatesReviewed by:
Hien Nguyen, UC Davis Health, United StatesYulia Rosa Saharman, University of Indonesia, Indonesia
Copyright © 2024 Sun, Liang and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulin Niu, bml1eXVsaW5ubkAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Zhou Sun
Zhou Sun Guofu Liang1†
Guofu Liang1† Yulin Niu
Yulin Niu