- 1Department of Chemistry, Oklahoma State University, Stillwater, OK, United States
- 2Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, OK, United States
- 3Department of Biochemistry and Microbiology, Oklahoma State University Center for Health Sciences, Tulsa, OK, United States
Introduction: Unique impermeability properties of the gram-negative outer cell envelope typically render these organisms intrinsically resistant to hydrophobic antibacterial compounds. Eumelanin-inspired indoylenephenyleneethynylene (EIPE) compounds possess scaffolding to which functional groups were attached to potentially provide antibacterial properties in the forms of hydrophilic (EIPE-HCl) and hydrophobic (EIPE-1) derivatives.
Methods: Standardized disk agar diffusion and microbroth dilution bioassays were employed to assess the susceptibility of disparate gram-negative and gram-positive bacterial pathogens to the two compounds. EIPE-1 mechanisms of action and intrinsic resistance were further investigated turbidimetrically in batch cultures with the aid of the gram-negative outer membrane permeabilizer compound 48/80.
Results: Hydrophobic derivative EIPE-1 exhibited a gram-positive antibacterial spectrum, while hydrophilic derivative EIPE-HCl possessed no antibacterial properties. EIPE-1 exhibited minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) values below 2.0 µg/mL against all gram-positive bacteria, including two methicillin-resistant strains. In contrast, MIC/MBC values greater than 128 µg/mL were obtained for all gram-negative bacteria examined. Susceptibility of two strains of the strict anaerobe Clostridioides difficile indicated the EIPE-1 mechanism of action does not require molecular oxygen. Turbidimetric growth curves revealed EIPE-1 induced rapid bacteriolysis of Bacillus subtilis ATCC 6633, thereby suggesting a membrane-directed modality. Lastly, the outer membrane permeabilizer compound 48/80 failed to markedly sensitize any of three phylogenetically disparate gram-negative organisms to EIPE-1.
Conclusion: These data suggest that the hydrophobic melanin-inspired derivative EIPE-1 inhibits gram-positive bacteria in a cytoplasmic membrane-directed manner independent of oxygen. Moreover, a secondary mechanism may function concomitantly with outer membrane exclusionary properties to underly the intrinsic resistance of gram-negative pathogens.
1 Introduction
The unicellular nature of bacteria coupled to their paucity of intracellular membranous ultrastructure renders their overall physiology singularly dependent on interactions at the cell envelope-environmental interface. We have reviewed this relationship from the standpoint of ionizable cell surface molecules which confer the hydrophilic polarity necessary to facilitate the association with and transport of essential solutes in aqueous environments (Wilson et al., 2001). Other research in our laboratory has emphasized physiochemical relationships between the outer gram-negative cell envelope and hydrophobic substances within the context of physiologic properties which potentiate pathogenicity (Darnell et al., 1987; Hart and Champlin, 1988; Watt et al., 2003; Ruskoski and Champlin, 2017). For example, intrinsic resistance to the hydrophobic biocide triclosan in Pseudomonas aeruginosa is due in large part to outer membrane exclusivity acting in concert with active multi-drug efflux systems (Ellison et al., 2007). Subsequent work revealed that atypical outer membrane permeability properties conferred by intrinsic cell envelope physiology is responsible for the extreme susceptibility of natural-occurring strains of Pasteurella multocida to hydrophobic substances in general and triclosan specifically (Ellison and Champlin, 2007).
Asymmetric localization of lipopolysaccharide (LPS) in the outer leaflet and the resultant absence of phospholipid bilayer expanses function with transmembrane porin selectivity for only small polar solutes to render the gram-negative outer membrane impermeable to bulky hydrophobic molecules (Nikaido, 2003; Silhavy et al., 2010). The permeability of hydrophobic molecules is further impeded by the high affinity of LPS for divalent cations which mitigate electrostatic repulsion between phosphate moieties, thereby stabilizing the outer membrane (Delcour, 2009). In contrast, gram-positive bacteria lack an outer membrane thereby exposing the cytoplasmic membrane phospholipid bilayer to hydrophobic antibiotics and other nonpolar compounds, which are then able to partition directly into the cytoplasm (Nikaido, 2003; Ellison and Champlin, 2007).
The increasing occurrence of infections by multi-drug resistant (MDR) bacteria has led the World Health Organization to establish the Global Antimicrobial Resistance and Use Surveillance System to enhance international research and surveillance capabilities (World Health Organization, 2021). MDR bacteria are extremely refractory and are emerging in disparate environments as a result of poor antibiotic stewardship resulting in a critical need for the development of efficacious antibacterial compounds (Spera et al., 2019).
Naturally occurring eumelanins are responsible for pigmentation of eyes, hair, and skin while also providing protection from harmful UV light. Eumelanin-inspired indoylenephenyleneethynylene (EIPE) derivatives possess scaffolding suitable for the attachment of disparate substituents which can be screened for antibacterial properties (Adhikari et al., 2022). This work revealed one such hydrophobic byproduct designated EIPE-1 (Figure 1) to be bactericidal for a methicillin-resistant Staphylococcus aureus (MRSA) strain. The purpose of the present study was to assess the antibacterial spectrum of the hydrophobic derivative EIPE-1 in comparison with that of a hydrophilic derivative designated EIPE-HCl to determine if their respective antibacterial properties are influenced by the disparate cell envelope ultrastructure of pathogenic bacteria which differ regarding gram reactivity.
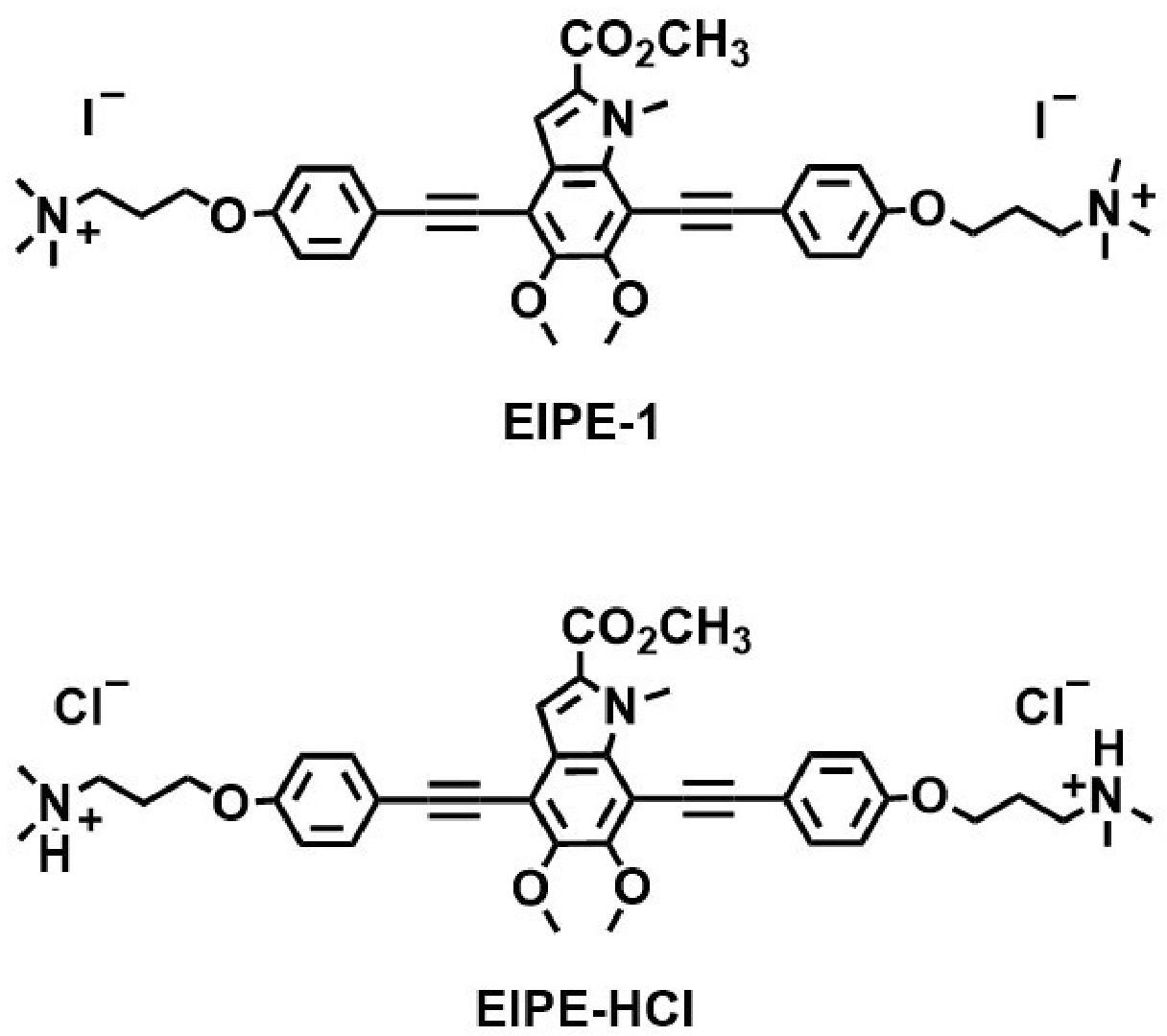
Figure 1 Molecular structures of eumelanin-inspired indoylenephenyleneethynylene (EIPE-1) and eumelanin-inspired indoylenephenyleneethynylene hydrogen chloride EIPE-HCl.
2 Materials and methods
2.1 Bacterial cultures, maintenance, and cultivation conditions
Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, Serratia marcescens ATCC 8100, Salmonella typhimurium ATCC 14028, P. multocida P-1581, Bacillus subtilis ATCC 6633, and S. aureus strains ATCC 25923, ATCC 29213, and T-5706 are maintained as stock cultures in this laboratory. Escherichia coli K12 413 was provided by Dr. Gerwald Köhler (Oklahoma State University Center for Health Sciences, Tulsa, OK). All other Serratia species as well as Bacillus cereus, Staphylococcus epidermidis SK01, Enterococcus faecalis, Lactococcus lactis, and Micrococcus luteus were obtained from Dr. Sue Katz Amburn (Rogers State University, Claremore, OK). Methicillin-resistant Staphylococcus aureus strains SFL8 and SFL64 were obtained from Dr. Sallie Ruskoski (Northeastern State University, Broken Arrow, OK), while Clostridioides difficile strains R20291 and 630 were provided by Dr. George (I-Hsiu) Huang (Oklahoma State University Center for Health Sciences, Tulsa, OK).
Stock cultures were maintained at -80°C in Brain Heart Infusion (BHI; Becton, Dickinson and Co., Sparks, MD) and glycerol (15%) (Darnell et al., 1987) or at 4°C on Mueller Hinton Agar (MHA; Becton, Dickinson and Co.) plates. MHA working cultures were produced by inoculating MHA plates with stock culture cells, incubating at 37°C for about 18 h, and storing at 4°C until needed to inoculate starter cultures consisting of 20 mL volumes of Mueller Hinton Broth (MHB; Becton, Dickinson and Co.) contained in 125 mL screw-capped culture flasks or 5.0-mL volumes contained in Morton closure-capped culture tubes (18 x 150 mm). Stationary-phase inocula for test cultures were obtained by incubating starter cultures at 37°C with rotary aeration at 180 rpm for 12-15 h in an Excella E24 Incubator Shaker (New Brunswick Scientific Co., Edison, NJ). Working and starter cultures of C. difficile strains R20291 and 630 were prepared similarly using Brain Heart Infusion Agar (Becton, Dickinson and Co.) supplemented with 0.5% Difco Yeast Extract (BHIA-Yext; Becton, Dickinson and Co.) and BHI plus yeast extract (0.5%; BHI-Yext). Cultures were incubated at 37°C for 24 h in a GasPak anaerobic jar (Becton, Dickinson and Co.) or a Type A Vinyl Anaerobic Chamber (Model no. 7150000; Coy Laboratory Products, Grass Lake, MI). Oxygen was removed from all media by storing under anaerobic conditions for at least 4 h prior to use.
2.2 Synthesis of melanin-inspired compounds
Melanin-inspired compounds designated EIPE-HCl (hydrophilic) and EIPE-1 (hydrophobic) (Figure 1) were synthesized in the laboratory of Dr. Toby L. Nelson as described elsewhere (Selvaraju et al., 2015; Selvaraju et al., 2016; Adhikari et al., 2022). Stock solutions were prepared at desired concentrations in dimethyl sulfoxide (DMSO; Fisher Scientific, Fair Lawn, NJ) and stored in microcentrifuge tubes at 4°C until needed.
2.3 Disk agar diffusion bioassay
Disparate bacterial pathogens were screened for susceptibility to EIPE-HCl and EIPE-1 using a standardized disk agar diffusion bioassay as described by Clayborn et al. (2011). EIPE-HCl and EIPE-1 stock solutions (2.5 µg/µL each) were prepared in DMSO using vortex agitation. Sterile blank filter disks (6.0-mm diam; Becton Dickinson and Co.) were infused with DMSO (10 µL; control), EIPE-HCl (25 µg in 10 µL DMSO), or EIPE-1 (25 µg in 10 µL DMSO) to yield compound potencies of 25 µg per disk. Disks were dried under flowing sterile air in a biosafety cabinet before storing at 4°C until needed. Overnight starter cultures were diluted into fresh MHB to prepare test culture inocula at an OD620 of 0.025 using a Spectronic 20D optical spectrophotometer (Thermo Electron Corp., Madison, WI). Loaded disks were applied to MHA plates uniformly seeded with standardized test culture inocula and allowed to sit at 4°C for 1.0 h to allow compound diffusion in the absence of bacterial growth prior to aerobic incubation for 18 h at 37°C. C. difficile strains were uniformly spread onto BHIA-Yext plates and incubated anaerobically for 24 h at 37°C. Electronic calipers (Fisher Scientific, Waltham, MA) were employed to measure growth inhibition zone diameters from which disk diameters were subtracted.
2.4 Microbroth dilution bioassay
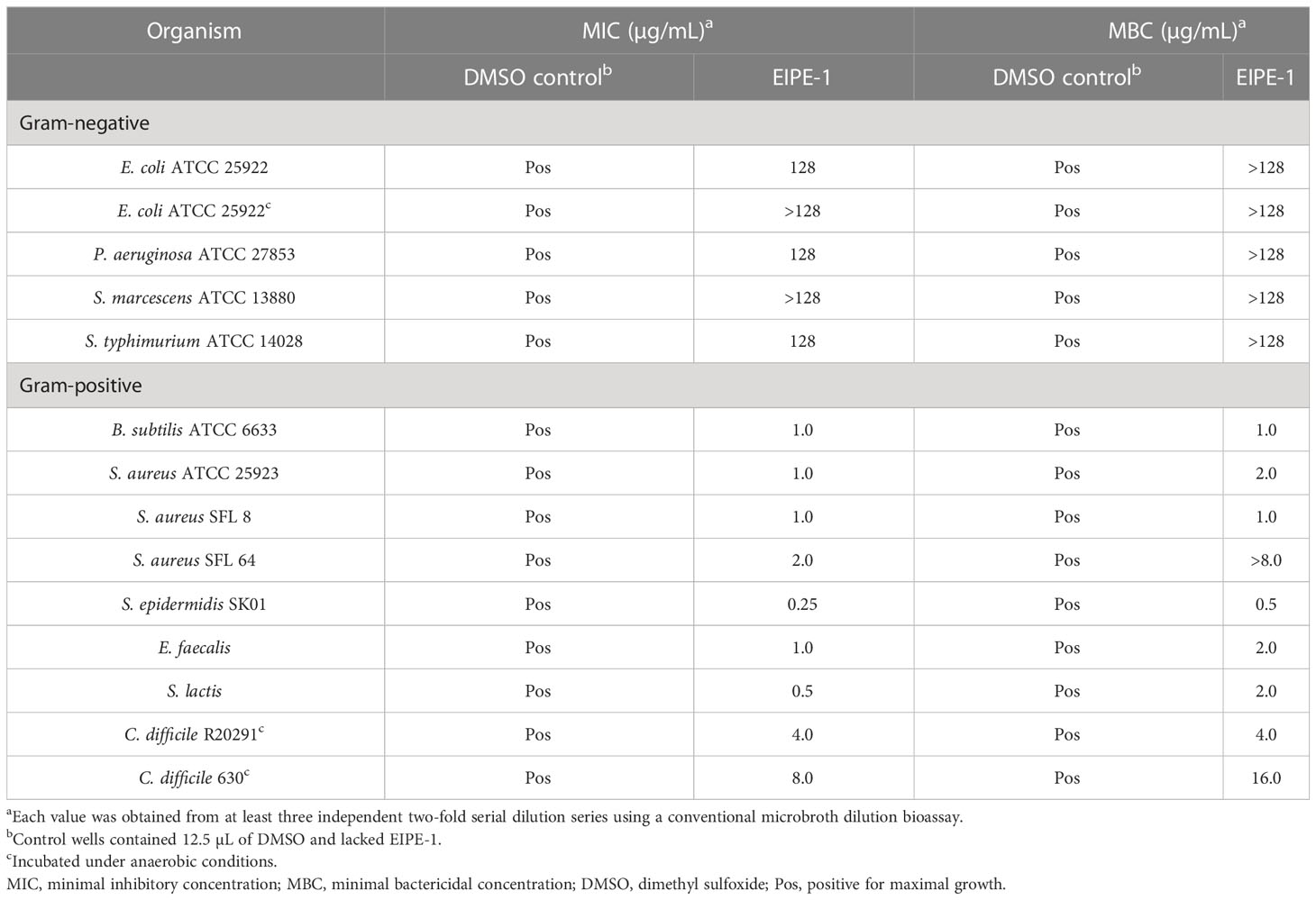
Minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) values were determined for EIPE-1 against disparate bacterial pathogens using our modification of a conventional microbroth dilution bioassay (Adhikari et al., 2022). EIPE-1 was dissolved in DMSO to a concentration of 2,560 µg/mL before diluting into sterile MHB to yield a final concentration of 128 µg/mL. Starter culture cells were used to inoculate MHB test cultures to an OD620 of 0.01 with the aid of a Spectronic 20D optical spectrophotometer. MICs were determined in microtiter plates (Fisherbrand 96 DeepWell Polypropylene Microplates; ThermoFisher Scientific, Waltham, MA) using a conventional two-fold dilution scheme beginning with an EIPE-1 stock concentration of 128 µg/mL. Assay microtiter plates were covered with an aluminum seal (Nunc Aluminum Seal; ThermoFisher Scientific, Waltham, MA) and incubated for 24 h at 37°C and 180 rpm in an Excella E24 Incubator Shaker. BHI-Yext was employed in place of MHB for C. difficile strains which were incubated anaerobically for 24 h at 37°C. MIC values were determined based on the lowest EIPE-1 concentration that completely inhibited the initiation of growth as judged by visualizing the absence of turbidity in each dilution series. MBCs were performed by transferring a small volume from each well onto a MHA plate using a 48-prong plate stamper and incubating at 37°C for 18 h. BHIA-Yext plates were employed for C. difficile strains which were incubated anaerobically at 37°C for 24 h. MBC values were determined on the basis of the lowest EIPE-1 concentration that completely inhibited macroscopic colony formation.
2.5 Batch culture growth kinetics
The effect of EIPE-1 on the batch cultural growth kinetics of selected gram-negative and gram-positive bacterial pathogens was determined using a method we have previously described (Champlin et al., 2005; Ellison and Champlin, 2007; Ellison et al., 2007) with the following modifications. EIPE-1 stock solutions were prepared in DMSO at concentrations of 100 µg/mL and 1000 µg/mL. Sterile MHB was inoculated with starter culture cells to an OD620 of 0.05 to provide test cultures. Aliquots of 5.0 mL each were transferred to culture tubes (18 x 150-mm diameter; Kimax with Morton closures). Each culture was treated with 1.0 µL of the appropriate stock solutions to reach final EIPE-1 concentrations of 0.2 µg/mL and 0.02 µg/mL, respectively. A solvent control was prepared by treating one test culture with 1.0 µL of DMSO to yield a final concentration of 0.02% (v/v), while another was left untreated to serve as a positive growth control. Excella E24 Incubator Shaker incubation was carried out at 37°C and 180 rpm while batch cultural growth was monitored turbidimetrically (OD620) at 30-min intervals using a Spectronic 20D optical spectrophotometer.
2.6 Outer membrane permeabilization
Efforts to sensitize selected gram-negative pathogens to EIPE-1 by chemically permeabilizing their outer membranes was attempted using the outer membrane permeabilizer compound 48/80 (Katsu et al., 1985) as described by Champlin et al. (2005). Batch test cultures were prepared and cultivated as described above after treatment with outer membrane permeabilizer compound 48/80 (5.0 µg/mL; Sigma-Aldridge Chemical Co., St. Louis, MO) and EIPE-1 (0.2 µg/mL) both alone and in combination to detect synergistic inhibition of batch cultural growth. The known susceptivity of P. aeruginosa to compound 48/80 sensitization to triclosan (Champlin et al., 2005; Ellison et al., 2007) was employed as a positive control. A triclosan (Irgasan DP 300; Ciba Specialty Chemical Corp., High Point, NC) stock solution was prepared in ethanol (95%) at a concentration of 2.0 µg/mL.
3 Results and discussion
3.1 Antibacterial properties screening
The screening of 14 strains of five gram-negative and 13 strains of eight gram-positive pathogenic bacteria for susceptibility to EIPE-HCl and EIPE-1 was accomplished using a standardized disk agar diffusion bioassay (Clayborn et al., 2011). Potential intrageneric disparity was examined by including multiple species of the gram-negative genus Serratia and gram-positive genus Staphylococcus. As can be seen in Table S1, all gram-negative organisms were observed to be resistant to both EPIE-HCl and EIPE-1 at a potency of 25 µg/disk. Despite also being resistant to EIPE-HCl, each of the gram-positive organisms was susceptible to EIPE-1 at the potency tested except for B. cereus, E. faecalis, and C. difficile R20291. The magnitude of values obtained with disk potencies of 25 µg was small, yet reproducible. Only slightly larger zones of inhibition were observed when selected organisms were tested with disk potencies as high as 250 µg/disk (Figure 2).
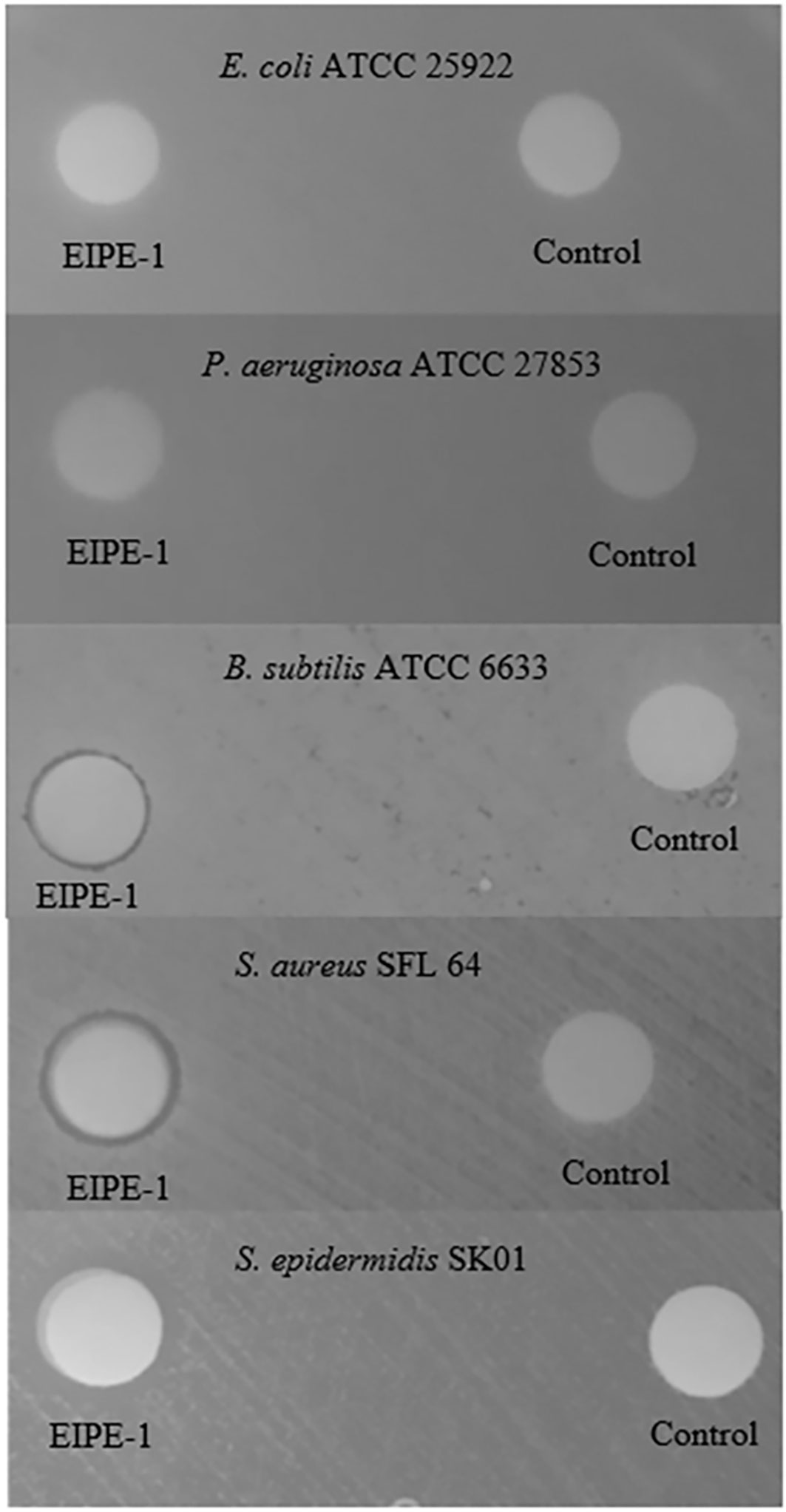
Figure 2 Disk agar diffusion bioassay for antibacterial properties of EIPE-1 for selected gram-negative and gram-positive pathogenic bacteria. Disk potencies: EIPE-1 (250 µg/disk); DMSO control (10 µL/disk).
All gram-negative and gram-positive organisms were resistant to the hydrophilic EIPE-HCl derivative at the screening potency of 25 µg/disk. These data suggest the absence of any antibacterial properties rather than an underlying role for cell envelope differences in ultrastructure between gram-negative and gram-positive bacteria with regard to intrinsic resistance. In contrast, most gram-positive bacteria were susceptible to the hydrophobic EIPE-1 while all gram-negative bacteria were resistant thereby suggesting a gram-positive spectrum of antibacterial properties. This conclusion is consistent with gram-negative bacteria being intrinsically resistant to EIPE-1 due to the impermeability of the intact outer membrane to hydrophobic substances in general (Nikaido, 2003; Champlin et al., 2005). The notion that outer cell envelope ultrastructure underlies gram-negative resistance is supported by the fact that P. multocida P-1581, which possesses an outer membrane that is atypically permeable to hydrophobic molecules (Ellison and Champlin, 2007) was slightly susceptible at a potency of 250 µg/disk (data not shown). The disk agar diffusion bioassay is a qualitative screen and cannot be used to precisely establish the efficacy of EIPE-1. This limitation was compounded by the poor diffusion properties suggested by the small magnitude of inhibition zone diameters and the absence of dose dependency on EIPE-1 disk potency (Table S1, Figure 2). These data suggested that quantitative MIC and MBC values generated by a serial broth dilution bioassay would be necessary to circumvent the apparent poor diffusibility of EIPE-1 to better establish the disparate susceptibility levels of gram-positive organisms.
3.2 Quantitative determination of EIPE-1 antibacterial properties
The quantitative determination of the EIPE-1 MIC and MBC values for selected gram-negative and gram-positive pathogenic bacteria employed a conventional microbroth dilution bioassay (Adhikari et al., 2022). Data contained in Table 1 reveal EIPE-1 MICs and MBCs of 128 µg/mL or greater for all gram-negative bacteria examined. In contrast, much lower values were obtained for gram-positive organisms with MICs ranging from 2.0 to 0.25 µg/mL with the exception of the two strictly anaerobic C. difficile strains for which the MIC and MBC values were two to six fold higher. This was possibly affected by physiological differences which required that the anaerobic organism be cultured on a richer growth medium under anaerobic conditions. C. difficile R20291 and E. faecalis exhibited much lower MIC and MBC values than gram-negative organisms in contrast to both of these gram-positive organisms exhibiting resistance in the standardized disc agar diffusion bioassay further showcases the limitation of poor diffusibility EIPE-1 possesses in agar media. Interestingly, the MBC for S. aureus SFL 64 was considerably greater than the corresponding MIC which is consistent with the enhanced refractory nature of MRSA strains. The uniformly low MIC values for gram-positive organisms in contrast with the uniformly large MIC values obtained for gram-negative bacteria further support the notion that the intrinsic EIPE-1 resistance seen in the latter group is due at least part to outer membrane exclusivity for hydrophobic solutes (Nikaido, 2003; Champlin et al., 2005). The phylogenetic disparity of the gram-negative organisms supports this conclusion because gram-negative cell envelope ultrastructure is their most notable phenotypic congruity likely to be involved in an antibacterial agent resistance mechanism.
3.3 EIPE-1 treatment in batch cultures
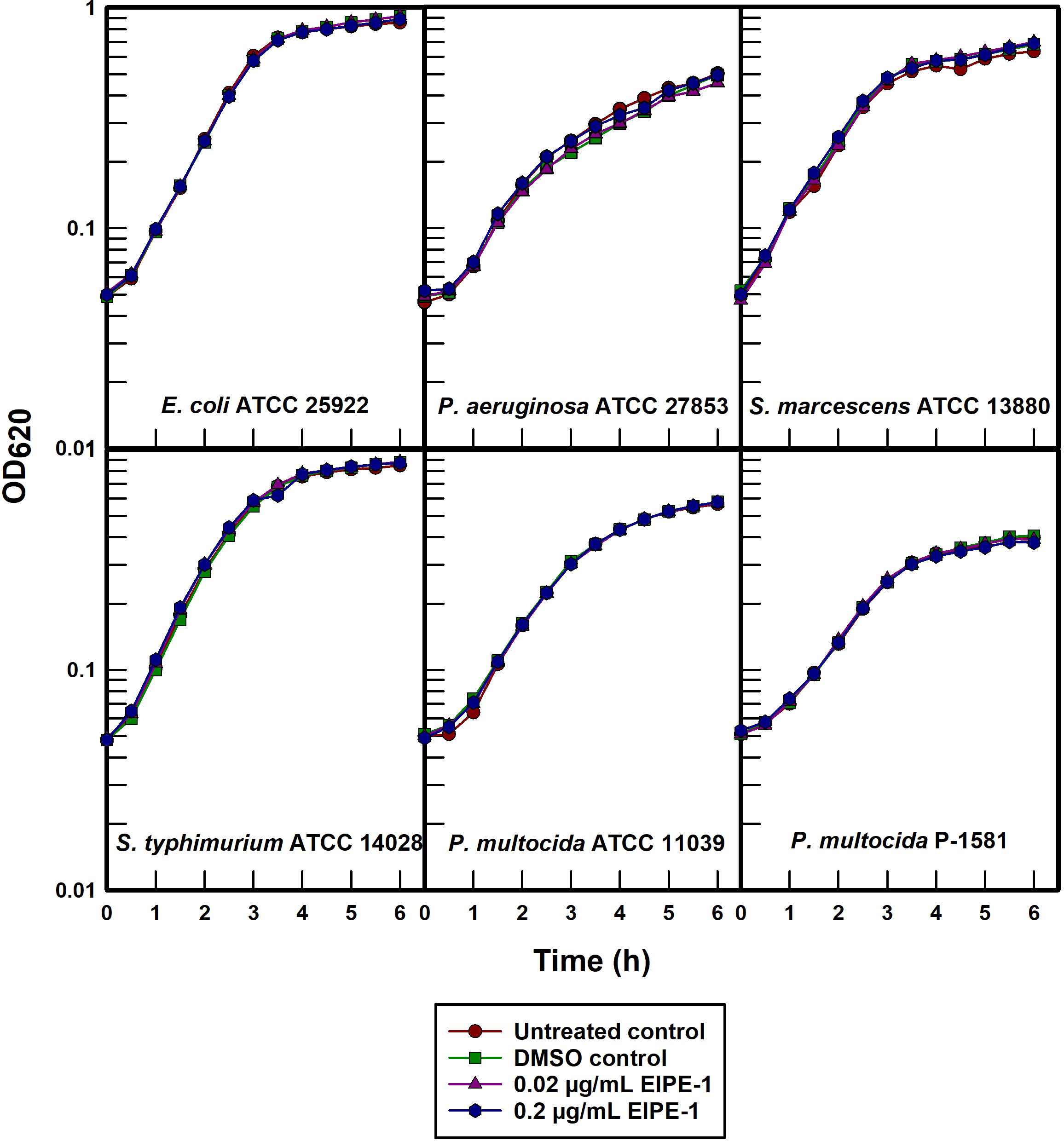
The effect of EIPE-1 on the growth kinetics of susceptible gram-positive pathogens was determined by administering EIPE-1 to batch cultures of selected organisms. This approach allowed for circumvention of the apparent poor diffusibility of EIPE-1 in agar-solidified growth media. Growth curves were constructed to initially assess the cellular and molecular mechanisms by which the compound is able to inhibit gram-positive bacterial growth. EIPE-1 concentrations of both 0.02 and 0.2 µg/mL failed to inhibit the growth of any of the five gram-negative organisms examined (Figure 3) including two cell surface hydrophobicity variants of P. multocida (Darnell et al., 1987). Because the outer membrane of P. multocida is atypically permeable to hydrophobic substances (Ellison and Champlin, 2007), this observation supports the conclusion that intrinsic resistance to EIPE-1 in gram-negative bacteria may involve an ancillary mechanism other than simple outer membrane exclusion of hydrophobic substances. Delcour (2009) posited that intrinsic antimicrobial resistance in gram-negative bacteria can be due to synergy between outer membrane impermeability and the presence of active efflux pumps, a relationship we previously demonstrated in P. aeruginosa (Ellison et al., 2007).
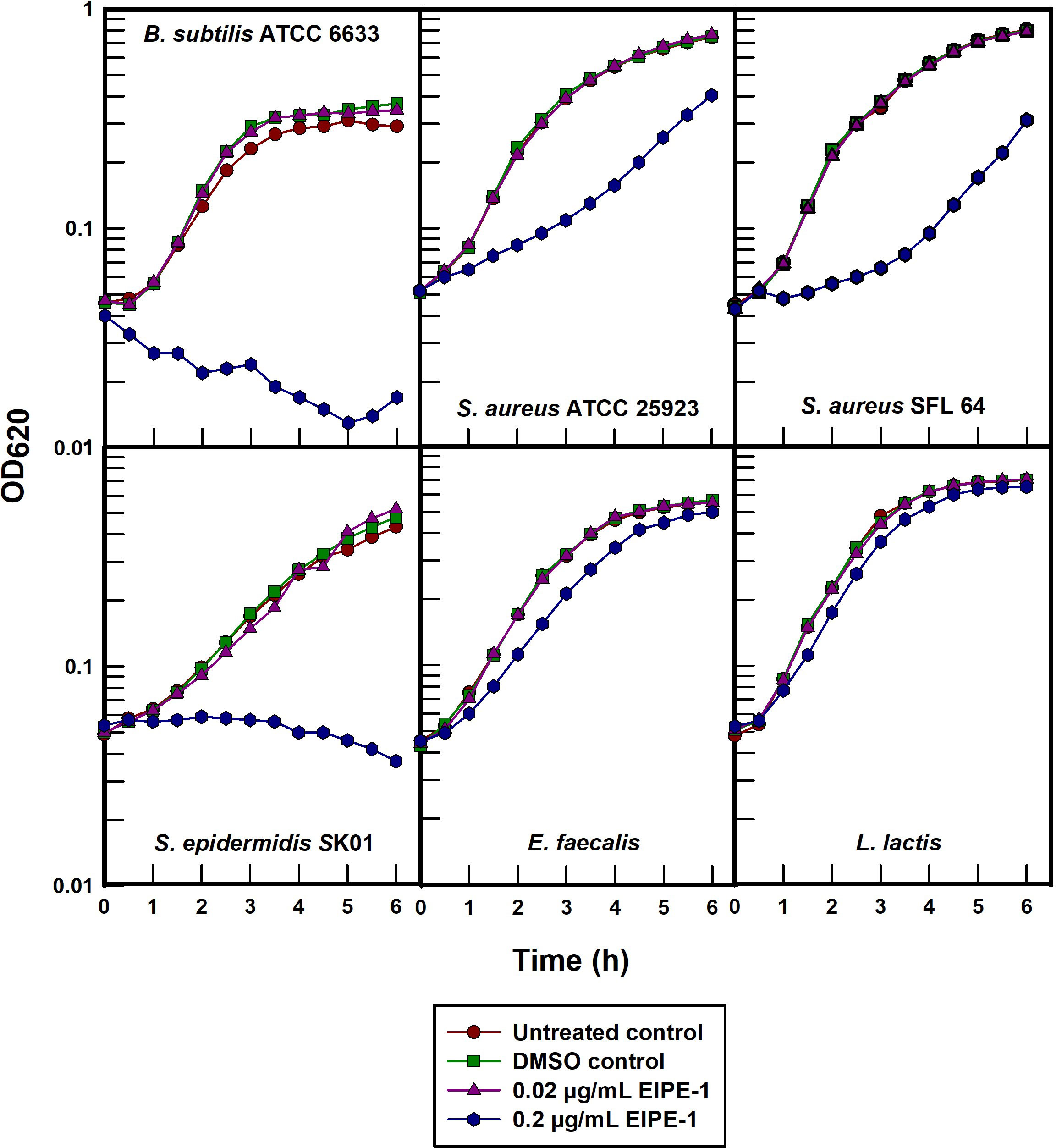
Each of the five gram-positive organisms examined were susceptible to different degrees when treated with EIPE-1 at a concentration of 0.2 µg/mL in batch cultures (Figure 4). B. subtilis ATCC 6633 was the most susceptible and can be seen to undergo bacteriolysis at the time of treatment. EIPE-1 was also bacteriolytic at this concentration for S. epidermidis SK01 although at a considerably slower rate. The growth rates of S. aureus ATCC 25923 and SFL 64 were markedly decreased while E. faecalis and S. lactis underwent only growth rate depressions in the presence of EIPE-1 at this concentration. Greater concentrations were not examined in this experiment for the purpose of minimizing potential secondary effects.
The intrinsic resistance of all the gram-negative organisms to an EPIE-1 concentration that was either growth rate inhibitory or bacteriolytic for all of the gram-positive organisms confirms its gram-positive antibacterial specificity. Moreover, the rapid bacteriolysis of B. subtilis ATCC 6633 suggests that EIPE-1 acts to inhibit the growth of susceptible bacteria by means of a cytoplasmic membrane-directed mechanism of action. The slower lysis rate observed in S. epidermidis SK01 would need to be studied further to validate that EIPE-1 is targeting its cytoplasmic membrane and not an anabolic cell process.
3.4 EIPE-1 titration of batch cultures
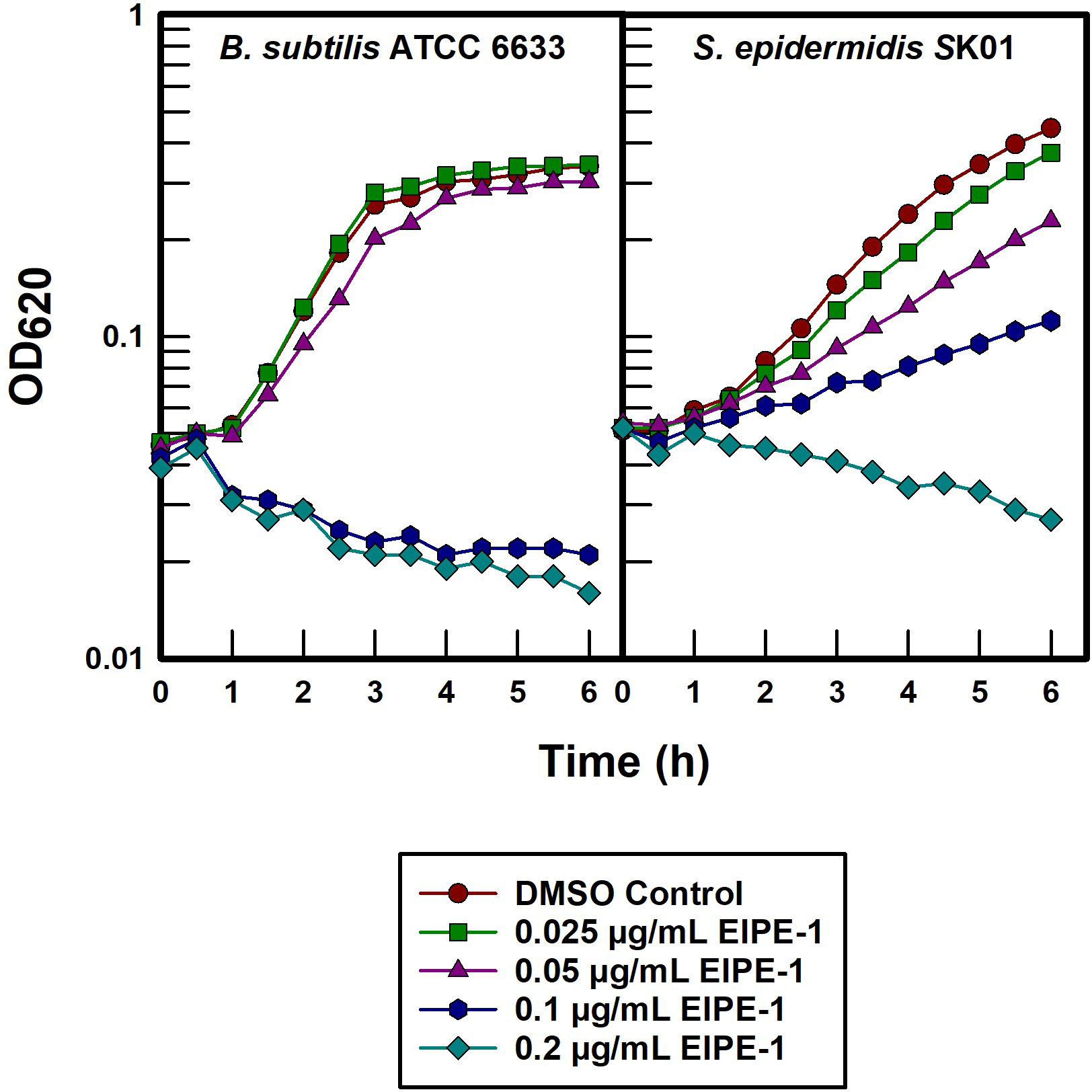
Both B. subtilis ATCC 6633 and S. epidermidis SK01 were extremely susceptible to EIPE-1 with MICs in the 0.25 to 1.0 µg/mL range (Table 1). In order to further investigate the underlying mechanism by which EIPE-1 is able to exert its antibacterial properties on gram-positive organisms, batch cultures were titrated with a serial two-fold dilution (Figure 5). B. subtilis ATCC 6633 underwent rapid bacteriolysis with 0.1 and 0.2 µg/mL concentrations, while only slight growth rate inhibition was seen at 0.05 µg/mL suggesting that a distinct concentration threshold must be achieved at which bacteriolysis is affected. In contrast, S. epidermidis SK01 was susceptible to all EIPE-1 concentrations in a dose-dependent manner with bacteriolysis only occurring with the highest concentration of 0.2 µg/mL.
3.5 Chemical outer membrane permeabilization
In view of the hydrophobic nature of EIPE-1, its gram-positive antibacterial spectrum strongly suggests that the intrinsic resistance seen in disparate gram-negative pathogens is due to exclusion by virtue of the impermeability properties of their outer cell envelopes for nonpolar molecules. To test this possibility more specifically, the outer membrane permeabilizer compound 48/80 was administered to chemically disrupt the cell surfaces of selected gram-negative pathogens (Katsu et al., 1985; Champlin et al., 2005) in an effort to synergistically sensitize the organisms to EIPE-1. Data contained in Figure 6A show only slight to no synergistic sensitization due to treatment with compound 48/80 and EIPE-1 in any of the three gram-negative test bacteria at a permeabilizer concentration that effectively sensitized P. aeruginosa to the hydrophobic biocide triclosan (Figure 6B). The absence of synergy between outer membrane permeabilization and EIPE-1 for these three gram-negative bacteria indicates that intrinsic EIPE-1 resistance in gram-negative organisms cannot be explained solely based on outer membrane impermeability to hydrophobic molecules and suggests the presence of an ancillary intrinsic resistance mechanism.
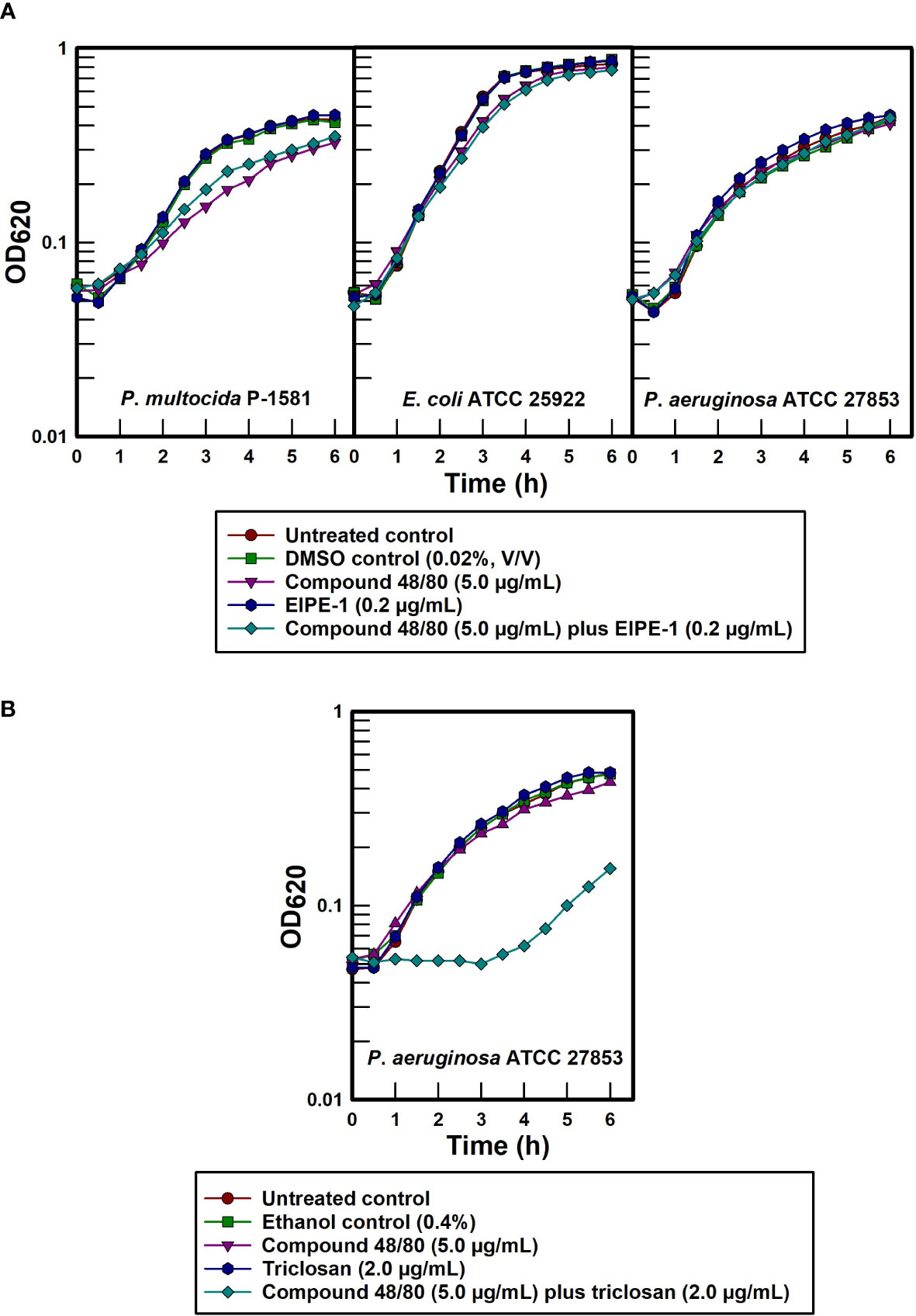
Figure 6 Effect of outer membrane permeabilization on intrinsically resistant gram-negative pathogens to EIPE-1. (A) EIPE-1 test. (B) Triclosan and P. aeruginosa control.
4 Conclusion
This investigation was designed to assess the potentially different antibacterial properties of two similar synthetic molecules which differ regarding their hydrophobicity properties, one of which had been shown to inhibit the growth of a strain of the gram-positive organism S. aureus. The specific purpose was to expand the study to other common pathogenic bacteria and determine if there was a relationship between compound polarity and the presence or absence of the gram-negative outer membrane which is generally impermeable to hydrophobic molecules. These data suggest that, in contrast with the hydrophilic melanin-inspired derivative EIPE-HCl, the hydrophobic derivative EIPE-1 possesses narrow spectrum antibacterial properties for gram-positive bacteria. Its unique core was employed as scaffolding for the attachment of a functional group that appears to affect cytoplasmic membrane disruption leading to bacteriolysis in S. epidermidis and B. subtilis which lack the protection afforded to gram-negative bacteria by virtue of their outer membrane impermeability properties. The slower bacteriolysis of S. epidermidis does not preclude the possibility that an anabolic target such as peptidoglycan biosynthesis could potentially be a mechanistic target in certain bacteria. MICs and MBCs for EIPE-1 in gram-negative organisms were on average eight-fold higher than in gram-positive organisms thereby supporting the notion that outer cell envelope ultrastructure plays a key role in determining resistance to EIPE-1. However, outer membrane exclusion of hydrophobic solutes in general does not appear to provide gram-negative cells with blanket protection from EIPE-1 in the absence of a secondary resistance mechanism as indicated by the inability of chemical permeabilization to sensitize three phylogenetically disparate gram-negative representatives. Adhikari et al. (2022) used scanning electron microscopy to observe EIPE-1 induced cell surface alterations in a strain of S. aureus. We surmise that active multi-drug efflux pumps and/or enzymes which covalently modify EIPE-1 are concomitantly involved, and a future goal of our research is to fully characterize secondary modes of intrinsic EIPE-1 resistance in gram-negative organisms. These studies are the first of their kind using these compounds, and the strong in vitro efficacy exhibited by EIPE-1 for MRSA strains, coupled with its apparent oxygen-independent membrane-directed modality, render EIPE-1 worthy of further investigation as a novel antibacterial candidate to address the increased emergence of multi-drug resistant gram-positive pathogens.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization: FRC, TLN, GAC, and EIL. Data acquisition: DRR, KN, MEs, MEb, FRC. Data curation: DRR. Formal analysis: DRR, KN, MEs, MEb, TLN, EIL, GAC, FRC. Funding acquisition: KN, FRC. Project administration: FRC. Manuscript preparation: DRR. Manuscript review and editing: TLN, EIL, GAC, FRC. All authors contributed to the article and approved the submitted version.
Funding
A portion of this work was supported by intramural funds provided by the Oklahoma State University Center for Health Sciences to FRC and the National Institute of General Medical Sciences of the National Institutes of Health under award P20GM103447 to KN and FRC. These sponsors were not involved with study design; data collection, analysis, or interpretation; manuscript preparation; or publication decisions.
Acknowledgments
We are grateful to Drs. Sue Katz Amburn (Rogers State University), Sallie Ruskoski (Northeastern State University), Gerwald Köhler (Oklahoma State University Center for Health Sciences), and I-Hsiu (George) Huang (Oklahoma State University Center for Health Sciences) for their generous gifts of stock cultures of organisms not routinely maintained in our laboratory.
Conflict of interest
The authors EIL and FRC declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2023.1253097/full#supplementary-material
References
Adhikari S., Essandoh M. A., Starr W. C., Sah P., La Force C. N., Eleshy R. G., et al. (2022). Eumelanin-inspired antimicrobial with biocidal activity against methicillin-resistant Staphylococcus aureus. ACS Appl. Bio. Mater. 5 (2), 545–551. doi: 10.1021/acsabm.1c01036
Champlin F. R., Ellison M. L., Bullard J. W., Conrad R. S. (2005). Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents. 26 (2), 159–164. doi: 10.1016/j.ijantimicag.2005.04.020
Clayborn A. B., Toofan S. N., Champlin F. R. (2011). Influence of methylation on the antibacterial properties of triclosan in Pasteurella multocida and Pseudomonas aeruginosa variant strains. J. Hosp. Infect. 77 (2), 129–133. doi: 10.1016/j.jhin.2010.09.021
Darnell K. R., Hart M. E., Champlin F. R. (1987). Variability of cell surface hydrophobicity among Pasteurella multocida somatic serotype and Actinobacillus lignieresii strains. J. Clin. Microbiol. 25 (1), 67–71. doi: 10.1128/jcm.25.1.67-71.1987
Delcour A. H. (2009). Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794 (5), 808–816. doi: 10.1016/j.bbapap.2008.11.005
Ellison M. L., Champlin F. R. (2007). Outer membrane permeability for nonpolar antimicrobial agents underlies extreme susceptibility of Pasteurella multocida to the hydrophobic biocide triclosan. Vet. Microbiol. 124 (3-4), 310–318. doi: 10.1016/j.vetmic.2007.04.038
Ellison M. L., Roberts A. L., Champlin F. R. (2007). Susceptibility of compound 48/80-sensitized Pseudomonas aeruginosa to the hydrophobic biocide triclosan. FEMS Microbiol. Lett. 269 (2), 295–300. doi: 10.1111/j.1574-6968.2007.00640.x
Hart M. E., Champlin F. R. (1988). Susceptibility to hydrophobic molecules and phospholipid composition in Pasteurella multocida and Actinobacillus lignieresii. Antimicrob. Agents Chemother. 32 (9), 1354–1359. doi: 10.1128/AAC.32.9.1354
Katsu T., Shibata M., Fujita Y. (1985). Increased permeability of the outer membrane of Escherichia coli induced by the dimer in compound 48/80. Chem. Pharm. Bull. (Tokyo). 33 (2), 893–895. doi: 10.1248/cpb.33.893
Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 (4), 593–656. doi: 10.1128/MMBR.67.4.593-656.2003
Ruskoski S. A., Champlin F. R. (2017). Cell surface physiology and outer cell envelope impermeability for hydrophobic substances in Burkholderia multivorans. J. Med. Microbiol. 66 (7), 965–971. doi: 10.1099/jmm.0.000532
Selvaraju S., Adhikari S., Hopson R. A., Dai S., Rheingold A. L., Borunda M. F., et al. (2016). Effects of structural variations on the optical and electronic properties of eumelanin-inspired small molecules. J. Mater. Chem. C. 4 (18), 3995–3999. doi: 10.1039/C5TC03982G
Selvaraju S., Sachinthani K. A. N., Hopson R. A., McFarland F. M., Guo S., Rheingold A. L., et al. (2015). Eumelanin-inspired core derived from vanillin: A new building block for organic semiconductors. Chem. Commun. 51 (14), 2957–2959. doi: 10.1039/C4CC09011J
Silhavy T. J., Kahne D., Walker S. (2010). The bacterial cell envelope. Cold. Spring Harb. Perspect. Biol. 2 (5), 1–18. doi: 10.1101/cshperspect.a000414
Spera A. M., Esposito S., Pagliano P. (2019). Emerging antibiotic resistance: carbapenemase- Producing enterobacteria. New bad bugs, still no new drugs. Infez. Med. 27 (4), 357–364.
Watt J. M., Wade M. M., Holman S. C., Wilson W. W., Keil D. E., Pruett S. B., et al. (2003). Influence of serotype A capsulation on cell surface physiologic factors in Pasteurella multocida. Colloids Surf B: Biointerfaces. 28 (2–3), 227–238. doi: 10.1016/S0927-7765(02)00139-X
Wilson W. W., Wade M. M., Holman S. C., Champlin F. R. (2001). Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods 43 (3), 153–164. doi: 10.1016/S0167-7012(00)00224-4
World Health Organization (2021). Global antimicrobial resistance and use surveillence system (GLASS). Report. Vol. 4. Available at: https://www.who.int/publications/i/item/9789240027336.
Keywords: bacterial cell envelope, hydrophobic antibacterial, eumelanin-inspired compound, gram-positive cell envelope, gram-negative cell envelope, biocide resistance
Citation: Reed DR, Nehmzow K, Essandoh MA, Ebqa'ai MA, Nelson TL, Lutter EI, Cook GA and Champlin FR (2023) Relationship between cell envelope ultrastructure and the antibacterial properties of a novel hydrophobic eumelanin-inspired derivative. Front. Bacteriol. 2:1253097. doi: 10.3389/fbrio.2023.1253097
Received: 04 July 2023; Accepted: 28 July 2023;
Published: 17 August 2023.
Edited by:
Myron Christodoulides, University of Southampton, United KingdomReviewed by:
Roshni R. Kharadi, Corteva Agriscience, United StatesSujeet Kumar, The Ohio State University, United States
Copyright © 2023 Reed, Nehmzow, Essandoh, Ebqa'ai, Nelson, Lutter, Cook and Champlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franklin R. Champlin, ZnJhbmtsaW4uY2hhbXBsaW5Ab2tzdGF0ZS5lZHU=
 Daniel R. Reed
Daniel R. Reed Katherine Nehmzow3
Katherine Nehmzow3 Mohammad A. Ebqa'ai
Mohammad A. Ebqa'ai Toby L. Nelson
Toby L. Nelson Erika I. Lutter
Erika I. Lutter Franklin R. Champlin
Franklin R. Champlin