
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
TECHNOLOGY AND CODE article
Front. Blockchain, 15 December 2020
Sec. Blockchain for Web3 and the Metaverse
Volume 3 - 2020 | https://doi.org/10.3389/fbloc.2020.483227
This article is part of the Research TopicHealthcare and BlockchainView all 5 articles
Recent advances in technology have drastically downsized costs and implementation times for genomic services. The wide availability of low-cost genomic technologies and easy access to genomic data can significantly improve healthcare productivity and efficiency, all to the benefit of social well-being in general. For example, by creating the conditions for researchers to identify the causes of multiple diseases and contributing to the development of new drugs, we can improve the quality of life and give people, as users of genomic services, the means to positively impact their health. This article describes how blockchain technology can lay the foundations of an ecosystem that encourages users to acquire and share their genomic data in full awareness without fear of being circumvented to participate in the benefits and advances in genomic research. The starting point is Genesy, an innovative blockchain platform that transcribes genomic data, thus facilitating and, at the same time safeguarding, the users of genomic services in their relationships with parties interested in accessing and using data they own, such as research centers, pharmaceutical companies, hospitals, and geneticists. This result is obtained by exploiting blockchain technology's capabilities to notarize data and prevent their unauthorized use, and at the same time to make them objects of possible transactions between different parties. Looking ahead, the Genesy model can be generalized to promote an ecosystem, and a fair market, for all types of biomedical data.
Recent advances in sequencing technology have dramatically reduced the costs and time required to sequence DNA and RNA, and this opens up further perspectives and opportunities in genetic studies.
To give an example, Whole Genome Sequencing (WGS), a methodology that can effectively detect all variants and types of relevant DNA variants, has reduced in cost, from the 3 billion dollars of the Human Genome Project, started in 1990 and finished in 2003, to a few hundred dollars of today. This, together with discoveries on the role of junk DNA in disease development and in diseases that have so far been unexplained, heralds a new era in genetic research. Further perspectives have been opened by the coming of age of metagenomic methodologies that enable the genetic reconstruction of the characteristics of the microbial communities that populate different types of environments, such as the human gastrointestinal tract, soil, water, and air, with effects that can be either beneficial or harmful—a topic that has become all the more current and dramatic following the outbreak of the Covid-19 pandemic and the possible presence of the sars-cov-2 virus that caused it in various environments.
Therefore, genomics and as well as other areas of the biomedical field, powered up by new digital technologies, are profoundly changing practices and methods in the health sector, with the effect of significantly increasing the volumes of acquired data and of accelerating their integration in medical activities. All the same, this trend also raises new challenges in terms of privacy and data security, which, in the absence of an adequate response, risks hindering the speedy adoption and development of new tools and methodologies.
Effective and timely action in this direction would not only mitigate risks but also multiply opportunities, by laying the foundations for a marketplace of biomedical data that can equally benefit users/patients in their role of primary data owners, and, on the side of data users, the genetic research community at large, as well as public and private institutions operating in the sector, such as pharmaceutical companies, hospitals, and universities.
These requirements indicate a supporting infrastructure responding to a data space's characteristics that guarantees protection and privacy of shared, exchanged, and monetized data, in a context in which new actors can enter the game without having to go through the bureaucracy of laborious accreditation and co-optation procedures.
If there is one technology that appears to fit best with this scenario, this is the blockchain, offering a propitious convergence of technological trends. Indeed, the blockchain, after gaining worldwide interest and success through the creation of cryptocurrencies and of Bitcoin in the first place, provides a proven option to globally manage all the data that can be associated with legitimate owners and whose value can be transferred employing digital communication networks, thus paving the way for the transition from an Internet of People to an Internet of Value that encompasses the sharing and monetization of biomedical data.
Several initiatives in the area of IT infrastructure for genomics have already seized the opportunity. The purpose of this article is to start from one of these, Genesy Project (Carlini et al., 2019) (also indicated in the abbreviated form of “Genesy”) in order to identify some general criteria for the design of a blockchain-based ecosystem for the sharing of genomic data, with the potential to expand to the whole universe of biomedical data.
The remainder of this article is structured as follows. In section 2, we illustrate the Genesy ecosystem and architecture. In section 3, we quickly review some blockchain initiatives in the biomedical area, and then we focus specifically on other genomic blockchains to identify both the differences and the points in common with Genesy. In section 4, we consider the limitations of the current implementation of the Genesy model and describe future work. section 5 concludes the article.
Genesy aims to involve collaboration among users and various organizations to promote a high-level genomics ecosystem, thereby efficiently collecting and managing the large volumes of data produced in sequencing activities.
At its most basic, and at the current development stage, the Genesy architecture is given by a private blockchain composed by peer nodes, owned and managed by the company Genesy Project SRL, where user personal data, together with user phenotype information and biometric data, are stored.
These Peers host both the transactions and a NoSql document-oriented database for querying and auditing the data inserted through the blockchain. User identity is cryptographically anonymized, while phenotype information and biometric data are accessible for search purposes in the database. The genomic data are too huge to be saved on the peer node itself (a typical file output from WGS may be several gigabytes in size) and thus are saved off-chain on cloud storage and linked to the users via the blockchain through cryptographic pointers to guarantee data ownership.
For payment of services on Genesy, such as data access by data requesters like pharmaceutical companies, a connection is provided to the Stellar (Stellar, 2020) and Stripe (Stripe, 2020) networks through their APIs. Genesy starts as a private blockchain, where the main characteristics of blockchain technology that are exploited are the immutability of the ledger for data notarization and encryption-based protection against unauthorized access. Furthermore, by interfacing Genesy with a public blockchain like Stellar, which supports the XLM cryptocurrency, and a payment gateway like Stripe, users can enjoy the benefits of data monetization and streamlined access to premium services.
However, Genesy is evolving from this initial seed toward a consortium blockchain characterized by a multiplicity of participating nodes and organizations, and in fact the success of the ecosystem envisaged and promoted by Genesy partly depends on this growth process.
Indeed, we can view Genesy both as a private blockchain and as the seed of a consortium blockchain. As a matter of fact, any private blockchain can be viewed as a consortium blockchain with a single member. As trivial and obvious as this statement may seem, it is, in reality, very relevant to fully grasp the distance holding between two decidedly distinct perspectives about managing distributed resources. On the one hand, there are, in fact, intrinsically centralized ways such as those given by the standard cloud-based solutions offered by the various providers of this kind of service. By contrast, blockchains are naturally born decentralized, even in the case of private blockchains, if these are viewed and implemented as single-member consortium blockchains that may in time evolve into full-fledged multi-member consortia.
Three aspects appear particularly relevant in this regard:
• A private blockchain lets the originating organization self-manage data access, as well as distribute access privileges to its subscribers according to any agreements, even if storage takes place on the cloud as in the case of genetic heritage in Genesy, with evident advantages at guaranteeing privacy and integrity of data compared to when access rights are transferred to other parties;
• Tamper-proof transcription of all transactions on the blockchain with the maintenance of their full history is a further, algorithmically enforced guarantee of data integrity;
• Last but not least, and most relevant indeed, private blockchains, if conceived and designed as open systems, a concept we shall delve into just below, can evolve naturally into consortium blockchains, therefore viewing them as embryonal consortia make perfect sense.
Consortium blockchains are, in this respect, full embodiments of open systems as were theorized and conceptualized as early as the 1980s (see Hewitt, 2020 for a pioneering contribution on this theme). A real-life example is given by the blockchain of the Food Trust (2020) consortium, started by IBM, with the participation to date of some of the world's major players in the agri-food business, such as Nestlé, Carrefour, Walmart, Unilever, and several other distributors and manufacturers of food products. In Food Trust, the blockchain is leveraged to track and certify products along the supply chains. There is clearly both cooperation and competition within this extensive consortium, with sub-consortia grouping entities into supply chains that compete with each other. However, there is collaboration across the boundaries of single chains, too, in that competitors share the best product tracking practices.
Genesy is in the process of producing a similar development on the less immediate but increasingly relevant theme of the genetic heritage of human and non-human organisms. To understand how this will work, we can view the Genesy blockchain as being influenced by two types of actors, namely peer nodes and service subscribers. Peer nodes are synchronized with the full blockchain, while service subscribers can be considered light-weight clients maintaining only information of their specific interest, which they update through services provided by the peer nodes they subscribe to. In the current set-up, the existing peer nodes managed by Genesy Project SRL can be subscribed both by data owners such as end-users wanting to have their DNA sequenced and then securely and privately stored and by data requesters, such as pharma companies and hospitals, wanting to pay for access to the genomes of data owners selected through phenotypic and biometric traits found on the node's database. At a certain point, another organization may provide its own independent sequencing services interested in joining the Genesy blockchain for data sharing; or an organization providing complementary biomedical data and services, such as Magnetic Resonance Imaging (MRI) scans or health monitoring services. It makes sense for such organizations to join by creating their own peer nodes added to those already present and extend the overall information made available on the blockchain. Clearly, this process can be repeated whenever chance and conditions make it feasible, thus triggering a virtuous loop of co-optation and expanding an ecosystem of biomedical data. Indeed, in an even broader outlook, Genesy can be seen as a contribution to the general objective of an Internet of Value, of which the blockchain is considered a fundamental technological enabler, where all value-adding information can be fairly shared and transferred.
As a matter of fact, while at its inception, it was expected that Genesy would have evolved into a consortium blockchain focused on human genomes, as things stand now, this is happening instead on the spur of the requests of metagenomes of microbic communities (aka microbiomes) in various environments, with concerns, according to cases, to monitor their potential toxicity or to improve their beneficial potential—a direction partly given by the covid-19 pandemic and by the growing awareness of the danger deriving from underestimating the role of pathogenic germs. Therefore, while humans still have a prominent role within the Genesy landscape, to get even fuller limelight are now the environmental metagenomes provided by organizations that want to certify the non-toxicity or the favorable features of the microbiomes populating their environments. Among such organizations, there are on the one hand restaurants, supermarkets, hospitals, nursing homes, gyms, and schools, where the potential harmfulness of the interaction between pathogenic microbial organisms and humans is a constant factor of danger, and on the other agricultural producers and livestock farms who can benefit from the presence of improved microbiomes in their crops and stock. Therefore, the Genesy blockchain is evolving into a consortium by acquiring metagenomes from the aforementioned organizations in the role of primary data owners and making them available to peer organizations whose business is to sanitize environments or improve the composition of microbic communities. Genomes of sanitized or improved microbiomes are then returned to Genesy for processing, notarization, and storage through the blockchain. Given its relevance for the agri-food business, this (meta-)genomic consortium in the making is in the course of negotiating its participation into the larger Food Trust consortium, where it will provide its specific competence to the other participants. Figure 1 illustrates the main components as well as the roles of the various stakeholders of reference in the Genesy consortium architecture.
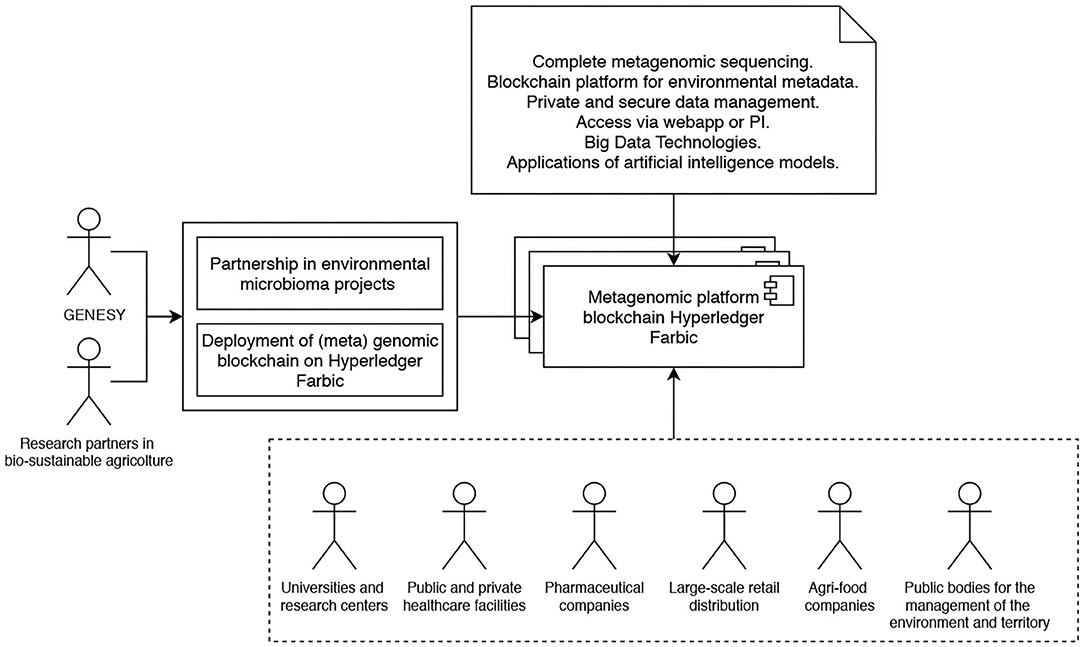
Figure 1. Overview of the main components as well as the roles of the various stakeholders of reference in the Genesy consortium architecture.
The technological choices for implementing the Genesy architecture have been made with this evolutionary trajectory in view, resulting in the adoption of Hyperledger Fabric1, an open-source platform for the development and deployment of private and consortium blockchains. Figure 2 shows the Hyperledger infrastructure to the support of the Genesy consortium architecture (Figure 3).
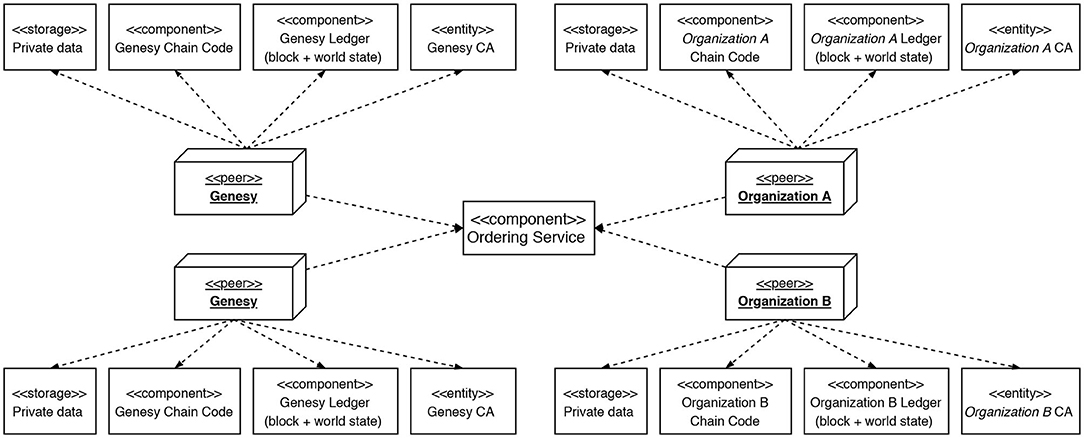
Figure 2. The Peers are nodes of the Blockchain. The Certification Authorities (CAs) are responsible for creating and managing the certificates necessary to guarantee maximum security of the whole system; each CA is entirely dedicated to an Organization, therefore no Organization can generate certificates from a CA other than its own. The Ledgers are where transactions on the Blockchain are indelibly transcribed; any Ledger can be updated only by the Peers with which it is associated and only after the transaction has been validated and approved. The Chaincode supports and implements the Smart Contracts of each Organization. Private Data are kept off-chain. Finally, the Ordering Service receives Signatures from Peers concerning new transactions. Once all Signatures have been collected, it certifies their validity and based on the policies defined in the Network authorizes the transactions and, therefore, generates a new block, whose transactions are transcribed by the Peers on the Ledgers of their competence.
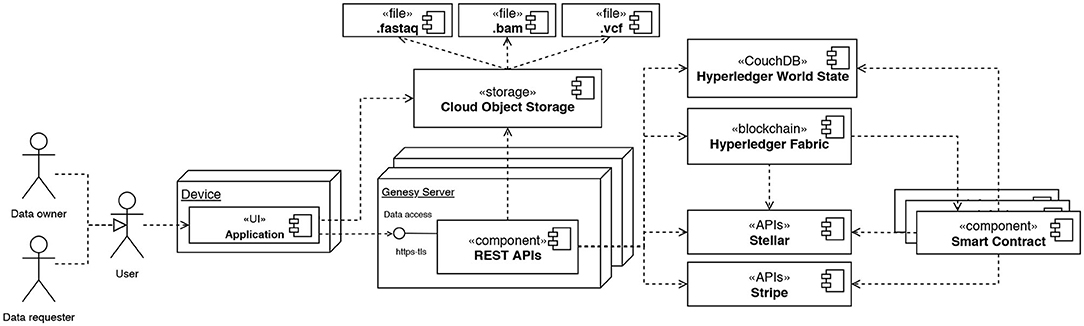
Figure 3. Deployment diagram using the UML notation modeling the high-level architecture of Genesy. Data owners and data requester can access the system via a web/smartphone application through the REST APIs provided by Genesy. The APIs put users in contact with each other, and facilitate the operations for reading, writing and transferring data.
Indeed, nothing in itself prevents the use of public blockchains for the implementation of Genesy. There are, however, some non-preferential factors, in that blockchains of this type are primarily aimed at launching the cryptocurrencies they support in the financial markets and, secondly, to install decentralized applications (dAPPS) that use cryptocurrencies to grow virtual economies (if tools, commonly known as “smart contracts,” are provided for the implementation of dAPPS).
These goals differ from Genesy's primary missions to leverage blockchain technology for the creation, certification, transcription, protection, and sharing of value-adding information in the biomedical domain, and to foster an ecosystem where organizations and people with stakes in this domain can thrive and interact. In concrete terms, this means a high computational toll for being hooked up to a public blockchain, where transactions must be propagated to all nodes involved and hence validated through computationally expensive mechanisms such as Proof-of-Work and Proof-of-Stake.
Even though public blockchains are evolving in the direction of a greater correspondence to the requirements of applications linked to specific organizational and inter-organizational contexts, at the current stage Hyperledger Fabric appears to offer all of what is needed to achieve Genesy's goals best. The features of Hyperledger Fabric that are specifically enabling and relevant from the point of view of Genesy are the following:
• Both peer nodes and light-weight clients are available and supported; peer nodes, associated with organizations, transcribe the blockchain in its entirety and manage and validate transactions, while light-weight clients choose and subscribe peer nodes to which they send transactions to have them executed on the blockchain;
• Peer nodes can be dynamically added, and the network associated with the blockchain consequently extended;
• Cryptographic protection of data via public/private keys is provided;
• Private communication between peer nodes is supported via channels, namely private “subnets” of communication between two or more specific network members, to conduct private and confidential transactions; this is particularly facilitating for the definition of agreements between the diverse organizations that participate as nodes in the blockchain; for example, a node that manages DNA profiles and allows their acquisition by data-requester such as pharmaceutical companies can privately import profiles from another organization that operates similarly and is also present as a node on the blockchain, without making the origin of the imported profiles visible to the requesting clients.
Unlike blockchains that are used to verify data integrity, the Genesy blockchain maintains, in an encrypted format, the data themselves relating to the users of the platform. This is because Genesy takes care of the entire life cycle of its users' information. To this end, the highest data security levels must be pursued, which is why Genesy does not rely on a traditional database but on the blockchain itself. Therefore, the data entered in the blockchain are not limited to hashes but also include encrypted confidential data owned by the users, who are the only ones in possession of the decryption keys to access them. The integrity hash related to entered data is always part of the ledger, being generated and recorded whenever a transaction is validated by the blockchain to verify the data's integrity every time a new block is created.
The insertion of data in the blockchain is therefore not to the disadvantage of users, but is, on the contrary, a guarantee of greater security, as nobody can change the state of the blockchain without the decryption key as well as the consent from the nodes responsible for the validation of transactions. This stems from the fact that in blockchains, unlike in standard database management systems, for an attacker to perform an alteration of sensitive data, it is not enough to modify the records in a single database, but is necessary to have control of the majority of peers and then submit a data modification transaction that must fit with the validation protocol of the blockchain.
Clearly, the only data to be entered in the blockchain will be confidential ones, such as the user's personal data, as the blockchain is not in itself conceived to be used as a database containing large data, and for other data Genesy indeed relies, as illustrated below, upon external databases (for phenotypic metadata) and cloud storage (for genetic heritage data). However, it should be added that even for data kept in the blockchain HyperLedger Fabric has implemented a significant improvement compared to the previous general state of the art by making available, for querying purposes, CouchDB, a document-oriented database that supports rich queries on the stored data.
One further aspect to be taken care of is that organizations share a common ground for operation in moving from private to consortium blockchains. This implies that the data must necessarily be of the same nature and interpretable in the same way by all organizations. This aspect is primarily dealt with by the Node.js server, a software library written in the programming language Javascript, and available in the Fabric environment, which, before forwarding the transaction to the blockchain, checks that the data are in a format that organizations are required to respect. If the data structure has been greenlighted, the server will forward the new transaction to the blockchain, where it will be executed if it is valid within the network's current state. For example, this means that organizations cannot modify data of which they are not in possession.
The Genesy blockchain is set up to seamlessly transition from private to consortium state. In fact, the only difference in terms of infrastructure requirements between private and consortium state is that, as we pointed out earlier, a private blockchain is a consortium blockchain hosting a single organization. The only action required to expand the platform from private to consortium is therefore to simply add an organization. Each organization needs a number of components necessary for its operation and, in particular, at least one Certification Authority and one Peer owned by it. There are three main steps to take to add a new organization to the network:
• Generation of the cryptographic codes necessary to create the organization;
• Creation of the physical structure of the organization, ie Certification Authority and Peer using the cryptographic encodings thus created;
• Connecting the new organization to an existing channel.
The addition of an organization to the consortium will have no impact either on the data previously entered into the blockchain or on the execution of transactions or on the already configured consensus algorithm, by virtue of Fabric's ability to support the expansion of the consortium without scalability issues.
The Hyperledger Fabric environment deals with the management of transactions and the entire blockchain, but this is not enough to ensure the functioning of the Genesy ecosystem. To support an easy interaction by the end-user, it is necessary to interface the blockchain with the outside world. To this aim, we have devised a server-side application that leverages the Node.js runtime to create a server coded in JavaScript exposing Application Programming Interfaces (APIs) to let an external client request the execution of a transaction by providing all the data necessary for its correct validation. The server also takes care of verifying the integrity and correctness of the data through the Software Development Kit (SDK) released by Hyperledger for use with programming environments such as JavaScript and Node.js, thus making possible to forward transaction data to the network according to the instructions defined within the “chain code,” which is the term used in Fabric to refer to the code used to program transactions and smart contracts. The SDK, in fact, enables connection and consequent interaction with the blockchain through a programming language.
The APIs provided by the server are closely linked to the transactions generated by the blockchain. In particular, the APIs act as a “bridge” between the external world and the blockchain. As many types of API calls are implemented as there are types of transactions envisaged by the blockchain, each call has the sole purpose of carrying out one transaction defined in the chain code. Additional operations implemented within the chain code include the communication to the applicant of the timestamp and the transaction's identification code once the latter has been carried out. The Server also takes care of verifying the integrity and correctness of the data provided, then of categorizing them as “Sensitive” or “Non-sensitive” and finally of storing the sensitive ones within the blockchain with which the Server communicates thanks to integrations developed through the Hyperledger Fabric Software Development Kit (SDK) for JavaScript.
A transaction within the network is the only operation allowed to change the status of the ledger. There can be various types of transactions in a traditional blockchain, so in Hyperledger Fabric, different kinds of transactions can be defined to fit specific application needs. The transactions are defined through the chain code instantiated on the peers, within which they get executed. The typical process of a transaction in a Hyperledger blockchain begins by sending the data necessary for validation to the Node.js server using the REST Application Programming Interfaces (API) it provides. Once the server has checked their integrity, the data will be forwarded through the SDK to the network, more precisely to the Endorsing Peers, aka the “Approval Peers,” that evaluate the transaction based on the data sent by the client. They do so by double-checking the correctness of the data and, if there are no hitches, greenlighting the transaction, having assessed its successful executability in the current state of the Ledger. If everything goes, they place a signature consent to the transaction's validity and hence send it to execution. The signatures are then forwarded through the network to the Ordering Service, which will check that all peers have validated the transaction and, according to the network's policies, will generate a new block containing the transaction. The information regarding the block is subsequently forwarded to the Committing Peers that will finally write the block containing the transaction on their ledger, thus updating the network to a new state containing a new block in the chain. Figure 4 provides a graphical representation of this transaction flow, while in Appendix instances of code snippets are given for adding, respectively, API implementation and a transaction.
While the blockchain is the heart of the platform, correct data insertion must also take into account metadata and phenotypic data maintained within a database external to the blockchain, through which the generic heritage in cloud storage (Cloud Storage Object in Figure 5) can be searched, e.g., “find all genomes of males born between 1980 and 1990 and with blue eyes.”
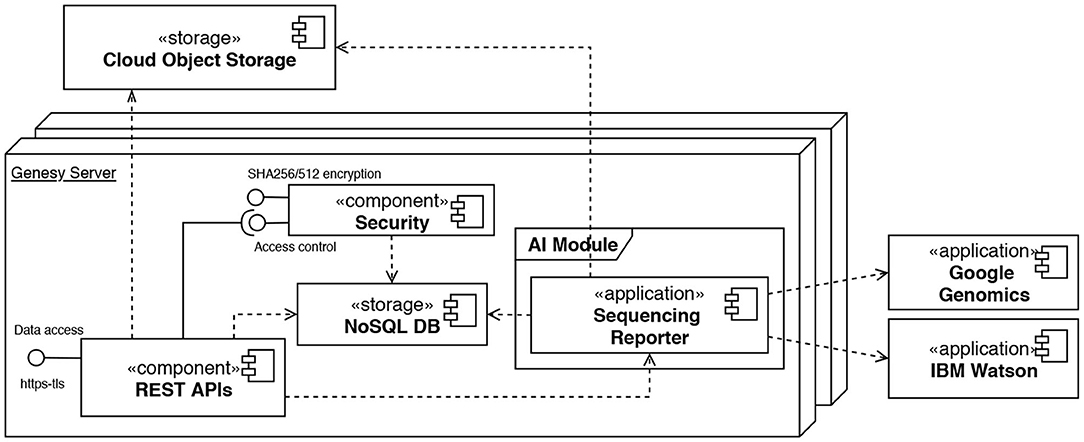
Figure 5. Deployment diagram using the UML notation modeling the architecture of a Genesy peer. Internally, the APIs rely on a security component to manage access control (“Security”) and a local storage (“NoSQL DB”) to store the users' access metadata. A further component, namely the “Sequencing Reporter” is responsible to analyze the data produced by sequencing and creating reports.
Therefore, to push data into the platform, it is not enough to define transactions executed within the blockchain. It is also necessary to differentiate between the sensitive data that will be inserted in the blockchain and the metadata that will be inserted in the external database. The connecting server handles this by supporting the interaction between the user and the platform.
User insertion of new data triggers, in fact, a sequence of actions whereby the connecting server identifies sensitive data, encrypts them to make them readable only by the inserting user, and forwards them to the blockchain server. The blockchain server validates and carries out the transaction, then enters the user data in the register by creating a user ID that is returned as a receipt of a successful transaction to the connecting server. In turn, the latter acknowledges the transaction as carried out and forwards the metadata and user ID to the server that manages the external database to write them in. The data owner will then be able to reconstruct, whenever the need arises, the relationship between metadata and sensitive data in virtue of the corresponding user ID, and will be the only one who can do so, in that their encryption is based on a user-defined password.
Access to a user's data by another user is always carried out through the blockchain. This is a necessary step to check that the requesting user is permitted to access such data. To this aim, the connecting server forwards the request to the blockchain server, verifying whether the data to which the requesting user has the right to access include those for which the request is made. Only in this case is their display allowed.
The economic model of Genesy must be seen as an evolving system, rather than as a static picture. If Genesy evolves into a consortium blockchain as a consequence of the addition of new peer nodes corresponding to as many organizations capable of producing value-adding information of a biomedical nature, then further economic models will be put into practice since each of such organizations will have its own way to generate revenues and returns for itself and its stakeholders. On the other hand, the approach described here identifies several technological options, general enough to be effectively reused and adapted to the possible variations of the initial business model.
So, how does Genesy make money as of today? In two ways, by selling services to primary users, turning them into data owners, and selling data to data requesters. Primary users, such as individuals who wish to have their DNA sequenced, can use the Stripe payment gateway to pay by credit card (in euros, dollar, or any other supported fiat currency) Genesy for the subscribed services. Data requesters, such as pharmaceutical companies, hospitals, and universities, can similarly use Stripe (and bank transfers) to pay for access to genomic data residing on the Genesy, prior consent formally granted by data owners, who receive compensation in consequence. In fact, Stellar allows for the creation of utility tokens for payments on platforms dedicated to the sale of specific products and services, and this possibility has been exploited for Genesy by creating a token for the use of the genomic services offered therein. The user can purchase Genesy services either by paying in fiat currency through Stripe or the Genesy tokens. Genesy tokens are, in turn, distributed to the data owners by Genesy in return for their willingness to give access to their data to requesters. In this way, rewarded data owners can monetize their data into a free or discounted subscription of additional services, which generate further data. The integration between Genesy and Stellar is aptly supported by Stellar, which provides SDKs (Software Development Kits) and REST APIs to interact with Fabric networks. Thus, a particular payment can be made by invoking the stellar APIs in a Fabric smart contract and then complete the asset transfer as agreed upon in the Fabric network. This is illustrated in Figure 6.
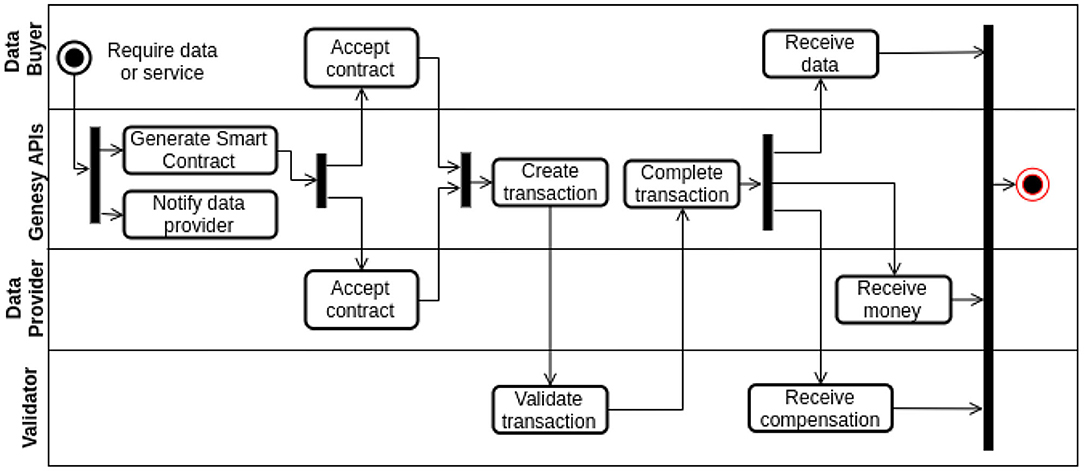
Figure 6. Activity Diagram in UML notation depicting a general transaction in Genesy. A data provider is a company or the data owner who had previously sequenced and shared her DNA and who has interesting traits that the data buyer (i.e., the data requester) wants to analyze further. A data buyer is a customer wanting to have her DNA sequenced and then securely and privately stored. The same schema also applies when both data buyer and data provider are companies wanting to exchange data or services, for example when a pharma company (data buyer) requests data or a service to a DNA sequencing provider (data provider), to provide, say, different solutions to their customers.
The platform manages the tokens through two accounts, i.e., two separate distribution nodes, the first and most important being the Central Bank that generates new tokens at need. Secondly, there is a Treasury with the role of managing all tokens so far created and, therefore, in circulation. The Central Bank generates new tokens in only two cases, i.e., when new ones are purchased through Stripe; or when genomic data are shared, and 20% of the turnover is returned to the purchased data owners through the generation of new tokens. This grants a return to all the parties involved: the data owner from the gain derived from providing the data to Genesy from providing the service; and the data purchaser from getting access to useful data. Users can store their tokens within the Treasury, enabling transfer to other users easily, quickly, and at no cost through an exchange transaction via the blockchain. It is also possible to exchange Genesy tokens for euros by selling them to Genesy SRL, which purchases them via the Stripe platform. Once this is done, Genesy SRL transfers the tokens back from the Treasury to the Central Bank, making them available again. This process of buying back the tokens is useful because it avoids having to create countless tokens with a finite life cycle, while in this way, they are effectively put back into circulation. This compensation model future evolution is that the Genesy coins upgrade from utility tokens into full-fledged security tokens. As such, they will be allowed to move outside of the Genesy ecosystem and be exchanged against cryptocurrencies such as Stellar's native XLM and fiat currencies.
As far as privacy and control of data are concerned, users may revoke access to their sensitive data at any time, without explanation. Note that giving access to one's data does not, by all means, amount to making them visible in their entirety, a situation that would thwart the protection granted by the blockchain, and hence the whole business model on which Genesy hinges, because, at that point, there would be nothing to prevent anyone who gains access from extracting the whole data from their owners. Rather, inbound data requests go through a workflow to identify the relevant minimal subsets of the data in storage, typically 0.2–0.5% of a WGS. Such requests are triggered by queries from business users (pharma companies, universities, private research centers, etc.) that input them into a Web app that funnels them via APIs in JSON format to a Web server. The queries may contain parameters such as reference gene panel, sex, geographic area, previous pathologies. In general, biometric and phenotypic parameters are used, as well as specific genetic research keys. A process is then activated to read through the metadata and the huge genomic files (BAM, VCF, etc.). The results are saved, in CSV or JSON format, in a customer's dedicated area and accessible through Identity and Access Management (IAM) services available on the blockchain platform.
On the other hand, the longer time the DNA is shared, the higher the profit. Time of data sharing is measured by internal meters, and the proceeds will be shared by all the accounts of users sharing their DNA. This means that even if one's DNA is not analyzed, he/she could still earn. The immediate compensation for the general sharing of DNA and the future variable compensation linked to the analyses performed on the platform, pharmaceutical, and research companies may have a special interest in certain types of DNA owned by specific individuals. They will never see the name of the data owners, who will be notified by Genesy if they may want to contact these organizations for further analysis and revenue. Needless to say, the last word is up to the data owners.
Genetic data are private data of their originating owners and therefore subject to regulations aimed at guaranteeing their protection and privacy, first and foremost the General Data Protection Regulation (GDPR) promulgated by the European Union in 2018, which has impacted the processing of personal data at all levels and in all sectors. Genesy has been designed in compliance with the GDPR, by taking advantage of the cryptographic protection offered by the blockchain and adding further protection measures, such as making access to data via the Web dependent on the verification and certification of secure communication protocols, as well as on the identification of the server providing the service and client station from which data are accessed. Furthermore, sensitive data entered within the Genesy blockchain are protected through the Advanced Encryption Standard, a block encryption algorithm of proven effectiveness and worldwide adoption, particularly in the version with 256-bit keys (AES-256). In addition to enabling key-based encryption and decryption, AES-256 is considered quantum resistant (see for instance Rao et al., 2017 for an analysis supporting this view) and for this reason fits perfectly with the blockchain, by meeting the security needs of a context where data are permanent and hence could be stolen at any time by an attacker with quantum computing power. Indeed, quantum computing power would still not be enough for an attacker to decrypt data protected by a quantum-resistant cryptographic algorithm in good time. Therefore, since sensitive data in Genesy are encrypted and decrypted via AES-256, they are suitably guarded even in the face of the coming of age of quantum computing.
Since Genesy's economic model provides for the development of a secondary market of data offered by data originators to other parties, such data transfers need obviously to be normed to the safeguard of all stakeholders and in compliance with the indications given on the subject by the GDPR. This is done by having the data owner sign an informed consent document on the data that she is willing to disclose, which is one of the ways sanctioned by the GDPR to disclose personal data.
A definitely more controversial issue, which embraces the relationship between blockchain and GDPR and consequently also concerns Genesy, is attributable to the right to the erasure of data. In fact, the GDPR is based on the assumption that data can be modified or deleted where necessary to comply with the legal requirements enshrined in articles 16 and 17. By contrast, a salient feature of the blockchain is precisely the non-erasability of the information to guarantee its integrity and increase trust in the network. This tension between blockchain and GDPR is subject to evolution, especially concerning the interpretative clarifications on the applicability of the GDPR in the various technological contexts in which data are generated and stored. As for the blockchain, a debate is underway whether the data typically stored on a distributed ledger, such as public keys and transactional data, qualify as personal data for the GDPR (European Parliamentary Research Service, 2019). In particular, the question is whether personal data that have been encrypted or hashed still qualify as personal data. While waiting for these questions to receive definitive answers, Genesy is, in any case, able to guarantee the deletion of the data attributable to the genetic heritage of its subscribers, as these are kept off-chain in the cloud. On the other hand, only confidential personal data are kept on-chain and thus encrypted for their protection, therefore falling within the ongoing debate referred to above.
Blockchain technology (Zheng et al., 2017) has recently gained massive attention from the media and companies worldwide, mostly because of its capability to record tamper-proof transactions across peer-to-peer networks of computers. Since its widespread adoption in financial services related to cryptocurrency transactions, enthusiasm for the use of blockchain technology has embraced other sectors, including healthcare, as the fundamental elements underlying blockchain technology, namely its decentralized and encrypted way of distributing, sharing, and storing information, appear interesting for health data (Gordon et al., 2017), inclusive genomic data (Shabani, 2019; Thiebes et al., 2020). From these premises, several projects and proposals for applications of blockchains to healthcare have recently sprouted.
Pursuing this direction, Genecoin (2019), a service providing blockchain-based archiving of genomic data, samples and backs up DNA by sending the user a kit to collect a spit sample to be forwarded to a DNA sequencing provider that in turn digitizes the extracted genome, and then encrypts it and stores it in the Bitcoin blockchain, which is used as a permanent cloud backup for that data. However, this approach has some potential disadvantages compared to traditional data storage methods since health data, like genomic data, tend to be very large and difficult to replicate on each node, thus significantly influencing the speed and scalability of a fully distributed system such as a blockchain, all well-known issues that concern the scalability of initial prototypes of blockchain-based applications (Croman et al., 2016; Angraal et al., 2017).
Alternately, instead of storing actual patient data, several proposals advocate the blockchain as a method to manage access control, where health data is stored off-chain and may be secured, corrected, and erased as appropriate, especially for security and privacy concerns. In contrast, only the metadata containing pointers to those data are stored on-chain for checking the authenticity and accuracy of the off-chain medical records (Esposito et al., 2018; Vazirani et al., 2019). This approach is substantially the same, also adopted by Genesy, but enriched, as illustrated above, in a significant way with a mechanism of monetization and leverage of genomic data to enable the bootstrap and development of a market for their exchange. However, several other proposals along this line have been put forward and are discussed below.
The Zenome project (Kulemin et al., 2017) leverages the Ethereum blockchain to support a decentralized database of genomic information. The platform enables the management of genomic data by preserving their privacy and making it possible to profit from the sale of access to different parts of the genome; it also provides an extensible interface for making genetic services work natively. Zenome provides for four entities that have different roles and are engaged in a variety of interactions within the system functionalities, namely: (i) nodes supplying storage and computing capacity made available to users of the platform; (ii) end-users who upload individual genetic data to the platform and who eventually use the genetic services provided by (iii) genetics service providers on the platform exploiting genetic data as part of their business; and (iv) analysts, data scientists, scientific organizations, etc., that are interested in analyzing the genetic information present on the platform.
Encrypgen (2019) is another genomic ecosystem that incorporates blockchain technology to create a monetized platform with which users can securely store and manage their DNA profile, maintaining the privacy and the ability to make a profit from selling access to different parts of the genome. Thus, on the one hand, the platform lets people share their data with other parties in return for a payment in tokens. On the other hand, it stores all genomes privately, in the cloud, and on personal servers, while a “bespoke” blockchain maintains lightweight metadata about the files kept off-chain and serves as an audit trail of transactions.
Finally, Shivom (2020) is a blockchain-based genomic ecosystem that acts as a bridge between users' DNA data and the value-adding actors who can leverage such data for research purposes. It enables individuals to interact with healthcare services and enterprises to boost medical and pharmaceutical research globally, all the while protecting patient confidentiality. To this aim, it offers a library of open data pipelines to let researchers perform analysis on the platform yet keeps data owners fully anonymous.
While the previous platforms are based on public or bespoke blockchains, a highly flexible framework for permissioned blockchains such as Hyperledger Fabric may be all the better suited to handle a genomic marketplace by meeting adaptively the various requirements that may arise in such a complex and highly dynamic environment (Gordon et al., 2017). For example, it may be necessary to identify researchers who require access to genomic data so that the owners of the data are fully aware of who they are and of the organizations to which they belong.
To this end, network access must be limited to data buyers whose identity has been verified. Therefore, a blockchain that supports permissioned access can play an important role in handling consent. Furthermore, a large, decentralized data marketplace requires smart contract functionality and high transaction throughput, which can easily be achieved by private blockchains in contrast to public blockchains. The ability to write transactions to the blockchain is limited to a group of permissioned validator nodes. Although this makes private blockchains more centralized and less dependable, storing data securely on the cloud may effectively counteract the threats.
Among all the projects of genomic ecosystems, Nebula (Grishin et al., 2018) is the one closer to Genesy. Nebula is a distributed platform for the generation, sharing, and analysis of genomic data with the overall goal to accelerate the global availability of genomic data and facilitate their access by combining a decentralized system design, privacy-preserving technologies, and an equitable compensation model. The benefits that Nebula aims to realize include full control over proprietary data, transparency, and privacy, as well as a radical optimization of the costs for data transfer and sequencing. Nebula is based on Exonum (2020), an extensible open-source framework for the application and customization of private and permissioned blockchains. Like Genesy, its main players are data owners and buyers. The former are public or private entities or institutions that store genomic data and control their access and transfer. The latter are researchers and the organizations they belong to, who wish to acquire access rights to genomic data from their owners. However, Genesy and Nebula differ substantially in their business model and, generally speaking, underlying socio-economic vision. Nebula supports a mechanism to move sequencing costs from data owners, such as end-users and biobanks, to data buyers, such as pharma and biotech companies. Therefore, data purchasers are enabled to query the files in the Nebula database to identify interesting profiles and offer matching individuals to pay sequencing costs to obtain access to and analyze their genomic data, based on information on their medical conditions and other traits, particularly phenotypic data, obtained primarily through survey questions. On the one hand, this approach lowers the initial entry costs to users interested in genomic services. On the other hand, it gives control over the data to corporate subjects rather than data originators. By contrast, Genesy endows users with the full mastery of their data and therefore favors the development of a market and an ecosystem based on individual data owners, with the promise of significant returns for them derived from the economic fairness in the use of genomic information, and with a general outlook that leads to growing education and awareness of the state and evolution of their health.
Genesy's main current limitation is its private blockchain status, which we discussed in the previous sections. However, there we also indicated that Genesy is based on an open governance model and therefore, the co-optation of new organizations is contemplated, foreseen, and actively sought; as a matter of fact, such an evolution is in the course of happening, as illustrated in the paper, in the context of the sharing of metagenomic data of microbial communities. In this context, Genesy will also make use of developments that favor cooperation, such as the feature of the new major release of Hyperledger Fabric (2020), which provides for stakeholders to define, cooperatively and in a decentralized manner, the smart contracts through which they intend to establish business relationships. Therefore, in the trajectory toward a consortium network in support of a dynamic ecosystem, it will be a question of maintaining the advantages of efficiency and specialization of a private blockchain while encouraging the maximum degree of collaboration and fairness.
Soon, there is also the alignment of the descriptive metadata of the genetic heritage with the international standards of the genomic sector, such as in particular, those developed within the initiatives of the Genomic Standard Consortium (Genomic Standard Consortium, 2020). This alignment will include users' phenotypic data through surveys and interviews and genetic markers detected in genomic sequencing to grant maximum meta-information for data selection to the interesting part.
The new frontier of medicine is personalized medicine, where doctors will recommend the most effective treatment plans and medicines based on our bio-medical data. In fact, an ever-growing set of advanced technologies will contribute to making those data a priceless mine of information. For example, through the use of low-pass sequencing technologies, we can now analyze whole DNA at an affordable cost, and soon we will be able to discover more and more about our past, present, and future. This, in turn, will boost the development of personalized medicine that can manage patients' health much more effectively than traditional medical practice. The more will be analyzed, the faster the development will be.
Our model envisions a state of the art blockchain technologies along with cloud computing to lay the bases of a new ecosystem where data originators (namely, the people of this planet) have mastership and control over their own genomic data. Thus, they can get rewarded for sharing them via monetary and health benefits. This does not hold just for genomic data but can be extended to all kinds of personal bio-medical information, giving it its fair value, to the advantage of genomics research and healthcare research in general.
FC and RC contributed to the conception and the prototyping of the project and provided feedback for the manuscript drafts. RP, SD, and FZ contributed to the design and writing of the study. All authors contributed to the manuscript revision, read, and approved the submitted version. A shorter version of the article has been accepted at the Second International Workshop on Blockchain Applications and Theory (BAT 2020).
FC and RC are founding partners of Genesy Project Ltd., while RP is the company Scientific Advisor© (but not employed).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. ^Available online at https://www.hyperledger.org/projects/fabric.
Angraal, S., Krumholz, H. M., and Schulz, W. L. (2017). Blockchain technology: applications in health care. Circul. Cardiovasc. Qual. Outcomes 10:e003800. doi: 10.1161/CIRCOUTCOMES.117.003800
Carlini, R., Carlini, F., Dalla Palma, S., and Pareschi, R. (2019). “Genesy: a blockchain-based platform for dna sequencing,” in Proceedings of the 2nd Distributed Ledger Technology Workshop (DLT 2019) (Pisa: CEUR), 68–72.
Croman, K., Decker, C., Eyal, I., Gencer, A. E., Juels, A., Kosba, A., et al. (2016). “On scaling decentralized blockchains,” in International Conference on Financial Cryptography and Data Security (Barbados: Springer), 106–125.
Encrypgen (2019). Encrypgen. Available online at: https://encrypgen.com/ (accessed October 22, 2019).
Esposito, C., De Santis, A., Tortora, G., Chang, H., and Choo, K.-K. R. (2018). Blockchain: a panacea for healthcare cloud-based data security and privacy? IEEE Cloud Comput. 5, 31–37. doi: 10.1109/MCC.2018.011791712
European Parliamentary Research Service (2019). Blockchain and the General Data Protection Regulation. Available online at: https://www.europarl.europa.eu/RegData/etudes/STUD/2019/634445/EPRS_STU(2019)634445_EN.pdf.
Exonum (2020). Exonum. Available online at: www.exonum.com
Food Trust (2020). Food Trust. Available online at: https://www.ibm.com/blockchain/solutions/food-trust
Genecoin (2019). Genecoin. Available online at: http://genecoin.me/ (accessed October 22, 2019).
Genomic Standard Consortium (2020). Available online at: https://gensc.org/
Gordon, W., Wright, A., and Landman, A. (2017). Blockchain in health care: decoding the hype. N. Engl. J. Med. Catal.
Grishin, D., Obbad, K., Estep, P., Quinn, K., Zaranek, S. W., Zaranek, A. W., et al. (2018). Accelerating genomic data generation and facilitating genomic data access using decentralization, privacy-preserving technologies and equitable compensation. Blockchain Healthcare Tdy. 1, 1–23. doi: 10.30953/bhty.v1.34
Hewitt, C. (2020). Offices are open systems. ACM Trans. Inform. Syst. 4, 271–287. doi: 10.1145/214427.214432
Hyperledege Fabric (2020). Hyperledger Fabric (version 2.0). Available online at: https://hyperledger-fabric.readthedocs.io/en/release-2.0/whatsnew.html
Kulemin, N., Popov, S., and Gorbachev, A. (2017). The Zenome Project: Whitepaper Blockchain-Based Genomic Ecosystem. Zenome.
Rao, S., Mahto, D., Yadav, D. K., and Khan, D. A. (2017). The AES-256 cryptosystem resists quantum attacks. Int. J. Adv. Res. Comput. Sci. 8, 404–408.
Shabani, M. (2019). Blockchain-based platforms for genomic data sharing: a de-centralized approach in response to the governance problems? J. Am. Med. Inform. Assoc. 26, 76–80. doi: 10.1093/jamia/ocy149
Shivom (2020). Shivom. Available online at: https://www.shivom.io/
Stellar (2020). Stellar - An Open Network for Money. Available online at: https://www.stellar.org/
Stripe (2020). Stripe - A Complete Payments Platform. Available online at: https://stripe.com
Thiebes, S., Kannengießer, N., Schmidt-Kraepelin, M., and Sunyaev, A. (2020). “Beyond data markets: opportunities and challenges for distributed ledger technology in genomics,” in Hawaii International Conference on System Sciences (Maui, HI).
Vazirani, A. A., O'Donoghue, O., Brindley, D., and Meinert, E. (2019). Implementing blockchains for efficient health care: systematic review. J. Med. Internet Res. 21:e12439. doi: 10.2196/12439
Zheng, Z., Xie, S., Dai, H., Chen, X., and Wang, H. (2017). “An overview of blockchain technology: architecture, consensus, and future trends,” 2017 IEEE International Congress on Big Data (BigData Congress) (Honlulu, HI), 557–564.
Keywords: blockchain, genomics, health care, DNA, sequencing
Citation: Carlini F, Carlini R, Dalla Palma S, Pareschi R and Zappone F (2020) The Genesy Model for a Blockchain-Based Fair Ecosystem of Genomic Data. Front. Blockchain 3:483227. doi: 10.3389/fbloc.2020.483227
Received: 15 July 2019; Accepted: 24 November 2020;
Published: 15 December 2020.
Edited by:
Victoria L. Lemieux, University of British Columbia, CanadaReviewed by:
Andrea Vitaletti, Sapienza University of Rome, ItalyCopyright © 2020 Carlini, Carlini, Dalla Palma, Pareschi and Zappone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Remo Pareschi, cmVtby5wYXJlc2NoaUB1bmltb2wuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.