
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
TECHNOLOGY AND CODE article
Front. Blockchain, 31 July 2020
Sec. Blockchain for Web3 and the Metaverse
Volume 3 - 2020 | https://doi.org/10.3389/fbloc.2020.00032
This article is part of the Research TopicHealthcare and BlockchainView all 5 articles
 Peng Zhang1,2*
Peng Zhang1,2* Chris Downs3
Chris Downs3 Nguyen Thanh Uyen Le4
Nguyen Thanh Uyen Le4 Cory Martin3
Cory Martin3 Paul Shoemaker3
Paul Shoemaker3 Clay Wittwer3
Clay Wittwer3 Luke Mills5
Luke Mills5 Liam Kelly5
Liam Kelly5 Stuart Lackey3
Stuart Lackey3 Douglas C. Schmidt2,5
Douglas C. Schmidt2,5 Jules White2,5
Jules White2,5Significant effort is required to recruit and validate patients for research studies. Researchers are typically limited to patients that they have a physical touchpoint with (e.g., patients at VUMC). This physical access limitation reduces the research attention that patients with rare diseases with little geographic concentration and patients with disadvantaged background in rural areas receive. This paper explores the use of mobile computing and blockchain technology to provide validation of research studies and their data usage, advertisement of research studies, collection of research data, and sharing of data across studies. The paper presents key challenges of using blockchains and mobile computing to solve these issues, competing architectural approaches, and the benefits/trade-offs of each approach.
A critical component of healthcare research is finding and recruiting participants in research studies and ensuring that researchers have sufficient data to make decisions regarding patient qualification to participate in a study. For example, simple information, such as drug allergies to or a specific health condition in a patient’s medical record that may exclude them from a study, is essential to making recruitment decisions. If a single piece of important information is missing, it can lead researchers to make inappropriate or delayed decisions regarding participant selection.
As a consequence of the need for access to detailed patient information, patient recruitment typically begins in clinical settings, such as hospitals, where researchers have direct access to detailed medical record information. For example, researchers may work with a specific clinic within a medical center and educate providers about their study and the types of patients that they are looking for as participants. The clinic will have detailed medical record information and a face-to-face touchpoint with patients to facilitate recruitment of patients that meet the participation eligibility criteria of the study.
An emerging architectural model that is gaining interest is putting patients at the center of the stewardship of their medical data (Kahn et al., 2009). Patients already have the right to view and move their data between providers, so it seems a natural fit that they should have mechanisms to see and move the electronic copies of their medical data, rather than only printed copies. With a patient-centered stewardship model, patients always have direct access to their own data from all providers they have visited and can delegate access at any time. This architecture is fundamentally different from the current model (Beard et al., 2012) where patients do not have direct access to the totality of their data and must individually request portions of the data from each provider, assemble the necessary portfolio, and then deliver the combined pieces to another provider.
An early manifestation of this patient-centered stewardship model is the ability for Apple’s HealthKit (Apple, 2020) to import medical records from Epic (EPIC, 2020), which is one of the most widely used electronic medical record systems in the US, into a user’s mobile device. HealthKit directly imports data using the FHIR standard (Bender and Sartipi, 2013) into a patient’s iOS mobile device. Once on a patient’s mobile device, a patient can choose to share their health records with additional apps on the device, which may in turn deliver the data to other medical providers or researchers.
A key question that arises in this new patient-centered data stewardship model is if there are opportunities to expand how patients are recruited into research studies. In particular, given that patients now have direct control over electronic copies of their medical records and the ability to share this access with apps on their devices, can researchers recruit patients directly through those apps? With this model, researchers would produce an app that can read medical record data directly from a patient’s HealthKit records and determine if a patient potentially meets the eligibility criteria for a study. If a patient matches a research study, they could be notified of the match and given the option to directly communicate with researchers conducting the study to determine if they can participate. Moreover, they could directly transmit needed medical record information from HealthKit to the researchers to further assist their participant selection decision.
If successful, this patient-centered model could help facilitate research study recruitment in terms of recruiting cost, data management cost, and research time beyond the typical settings, such as clinics, that have access to the needed medical records to perform the preliminary stage of filtering to match patients to research studies. There are several published studies that analyze the effectiveness of recruitment for medical research, such as (Lovato et al., 1997; Gul and Ali, 2010), and they each focused on different research purposes, which created a wide variance of the cost for recruitment stage. Generally, computerized support systems would help save significant recruiting cost compared to traditional clinic-based approaches (Kaushal et al., 2003). In addition, computerized support systems have considerable potential for reducing the timeline and increase efficiency of data management process of medical research studies (Garg et al., 2005).
Another trend that is impacting patient care is the rise in production of non-traditional health-related data, such as records of self-reported meals, step counts from fitness trackers, and momentary assessments of mood or pain from patients. This data, which is typically not part of the medical record today, is increasingly demonstrating value to researchers in understanding diagnostic and disease management processes. For example, meal logs can aid researchers in understanding how effectively patients are self-managing chronic conditions, such as diabetes.
Whereas traditional medical records are directly captured through the provider in the electronic medical record system, this newer exercise tracker and other non-traditional data is typically captured through mobile devices, IoT devices in the home (e.g., wifi scales), and through online services (e.g., social networks). The data collection mechanisms span a vast array of apps, devices, and services, few of which are trusted or certified by any healthcare entities.
Now, with the new patient-centered data stewardship model, this non-traditional data is accessible side-by-side within HealthKit with traditional medical record data. This combining of both types of data in a single location offers the potential for supporting many types of innovative research, such as research on patient reported outcomes or large-scale studies of lifestyle on health.
A second interesting question related to research studies and this new patient-centered data stewardship model is if the current research data sharing and reuse model can be expanded to both incorporate this non-traditional data and put patients in control of how the data is shared with other researchers. With the current research data ownership model, patients typically do not have the ability to easily access and share the research data from them with other researchers. The lack of control of their data limits the impact that patient’s research data can have on other research studies and keeps researchers, rather than patients, in control of the data.
Since patients now have access to both their traditional health records and non-traditional health-related data on the same device, patients can potentially join research studies with little or no face-to-face interaction with researchers. In the new model, patients would feed their medical records and non-traditional data to researchers through the HealthKit conduit. Although detailed clinical studies requiring high-fidelity, close physician monitoring of health, and administration of new medications or interventions may not be possible, studies that focus on the impact of non-traditional data on health or vice-versa could be feasible without direct contact with the participant.
Moreover, if participants use HealthKit to capture and provide their medical record and non-traditional health data with researchers, it is feasible that they could simultaneously share this data with multiple research studies or redistribute previously captured data to new research studies that could benefit from it. There are certainly many studies where access to the details of how the data was collected, such as how lab tests were performed, would render this type of model ineffective. However, we posit that there are many studies, such as observational studies that research how diet affects a person’s blood sugar level or how sleep affects one’s mood, where this model is not only feasible but offers unique new research opportunities.
In this paper, we explore key research challenges to realizing this vision, although we fully acknowledge the presence of many other types of challenges, such as challenges associated with specific blockchain implementations. Through our detailed analysis of the domain-specific research challenges, we have found that Distributed Ledger Technology possesses attributes that make it a promising solution to realizing this new model for research study recruitment and sharing of research data across studies. After careful analysis of the research challenges and promising attributes of distributed ledgers, we propose an initial open architecture with a detailed set of domain-specific requirements for study participant recruitment and data sharing in the emerging patient-centered data stewardship model.
The remainder of this paper is organized as follows. Section “Motivating Healthcare Research Example” provides a motivating healthcare research example to demonstrate the need for and trends toward a patient-centric data stewardship model. Section “Challenges in Recruitment for Clinical Research” presents key challenges in clinical research recruitment today. Section “A Distributed Ledger Architecture for Research Participant Recruitment and Research Data Sharing” proposes a decentralized architecture based on Distributed Ledger Technology for facilitating data sharing in the research participant recruitment process. Section “Related Work” discusses related research on platforms for improving the recruitment process for research studies and work that leverages distributed ledger technology to felicitate healthcare data sharing. Section “Concluding Remarks” presents concluding remarks and summarizes our key lessons learned.
As a motivating example for the exploration, we use an example of the management of a serious chronic condition that most commonly manifests in adolescent patients, namely, Type 1 Diabetes Mellitus (T1DM). T1DM is an autoimmune disease where the pancreas produces little or no insulin, which is critical to help the human body manage blood sugar levels. The treatment of this condition relies on patients to perform self-management tasks, such as self-measurement of blood glucose and self-administration of insulin, to avoid life-threatening complications (Borchers et al., 2010).
Despite physiological traits like blood glucose levels and carbohydrates intake that are commonly used as clinical indicators of how T1DM is controlled, recent studies (Mulvaney et al., 2011) have shown that psychosocial behavior in adolescent patients with T1DM can significantly affect the adherence to diabetes regimen in this population. As a result, much more diverse data, such as fatigue level, mood, location, and social context, can be collected to observe the behavior or further analyzed to provide timely intervention to poor self-management behavior (Zhang et al., 2018b). These data can easily be collected in or near real-time using Internet of Things (IoT) devices like smartphones, Bluetooth-powered glucose meters, and environmental sensors. They can complement traditional electronic health records (EHR) to provide a more comprehensive view of patient health status by including potentially influential variables from outside clinical settings (Zhang et al., 2018c).
Unlike EHR systems that have served healthcare for decades, emerging IoT-based systems that record health-related activities (such as self-observed behavior data or sensor-recorded environmental triggers) have not yet been rigorously tested and certified to integrate with high-fidelity data like provider-documented EHRs. There is a lot of distrust toward mobile app/IoT providers from physicians and certified EHR system vendors, causing delays in the data integration process. In the case of adolescents with T1DM, patients often have to maintain a journal that logs their daily diabetes management routine. The journal may locate separately from, for instance, an app that monitors daily psychosocial/behavioral traits for the same patient. It is highly likely that neither the journal nor the app data would be linked to the patient’s health records, which can create potential problems, such as inconsistencies in the medical history or misinformation, particularly when that patient changes provider.
Current healthcare systems are known to be provider-centric as forced by vendor-locked systems. These systems operate and only enable cross-system communications upon the establishment of trust relationships between vendors and providers. In the modern society where a lot of healthcare efforts are gradually becoming decentralized thanks to IoT technologies, the centralization model that is trust-dependent will become less effective and create more overhead for patients to manage care (Zhang, 2018).
Despite the importance of clinical research and continuous efforts to increase clinical research participation, many challenges exist in the recruitment process and are multi-faceted, creating barriers for researchers to complete their studies. This section discusses four such challenges, including recruiting costs, participant discovery of research studies, data reuse, and data ownership distribution.
Medical research is a long-term investment. Depending on the scope of the research, the timeline will vary. DiMasi and Grabowski estimated the average length of time from the start of clinical testing to marketing is 90.3 months in the pharmaceutical sector and 97.7 months for the biotechnology sector (DiMasi and Grabowski, 2007). Lengthy timelines directly impact the cost of capital for the medical projects and increase the financial burden for researchers and investors because it is considered as opportunity costs associated with foregone investments over the researching period.
The recruitment process alone accounts, on average, for nearly 30% of total clinical trial time (around 30 months) (Reuters, 2012). During this process, resources required to recruit and enroll participants must be sustained, including but not limited to recruitment and coordinating staff, equipment, facilities, advertisements, etc., all of which contribute to significant recruiting costs. Recruiting a large enough pool of participants to validate the statistical result of medical research has always been a difficult task for healthcare researchers. More than 81% of clinical trials are delayed because researchers cannot recruit enough participants for the studies (Nasser et al., 2011). In particular, when analyzing 374 cases at Oregon Health & Science University, 31% of clinical research studies enrolled 0–1 subject before being terminated, which creates a waste of over $1 million per year (Kitterman et al., 2011).
More recently, however, computerized support systems have proven to be advantageous in recruiting participants on a large scale at a lower cost. A study involving healthy volunteers among different recruiting methods has shown that costs per enrolled subject were lower for the EHR patient portal ($113) than letters ($559) or phone calls ($435) (Samuels et al., 2017). In addition, another study in Australia tested the effect of leveraging a technical platform (social media) in healthcare recruiting process. The results showed that the technical platform was more cost-effective, especially in the earlier stages of the studies (the cost to obtain a screened respondent: AUD$22.73 vs AUD$29.35; cost to obtain an eligible respondent: AUD$37.56 vs AUD$44.77) (Frandsen et al., 2016). These analyses show that integrating technology that can accelerate the recruitment process of medical research, which would in turn save the recruiting costs and total costs of the studies tremendously.
Recruitment of patients with a physical touchpoint leads to an institution-centric advertising model. Because clinical studies are controlled by separated institutions, participants need to put in considerable effort to find the studies that match their health status and relate to a medical condition they wish to be involved in. Popular resources include the website of National Institutes of Health (NIH) (National Institutes of Health, 2020), third-party “search engine” for proprietary market research (CenterWatch, 2020), and other tools that are not specifically designed for clinical research recruitment (e.g., Amazon Mechanical Turk; Buhrmester et al., 2016).
Most of these resources are spread across multiple information channels aiming to improve the publicity of research studies, but the distributed information may become scattered and outdated or cause confusion to potential research volunteers. Furthermore, the eligibility criteria to participate in a study can contain complex clinical terminologies that are hard to interpret by participants without advanced clinical knowledge. It is also impractical for volunteers to reach out to clinical experts for every trial they are interested in due to the large number of ongoing trials. As a result, potential volunteers may be discouraged to inquire about or participate in research studies.
Reusing and aggregating clinical data have been proven effective for facilitating the discovery of new knowledge and the processes of healthcare (Kreuzthaler et al., 2015; Chen and Butte, 2016). Recognizing these benefits, some governmental organizations including NIH (Majumder et al., 2017) and the National Science Foundation (NSF) (National Science Foundation, 2010) have started to support data sharing and openness in clinical research. In contrast, data sharing is not a popular practice as it should be in reality. There are many concerns related but not limited to the ownership of reused data, the quality of the data, and legal compliance. As the cost of recruiting patient and acquiring the data is high, researchers usually prioritize clinical workflow support, legal compliance, and their research purposes over the quality of the data for reuse. Documenting how data is acquired and transformed, storing data in a universal format, and finding accessible repositories to share the data are very time-consuming.
According to a study on biomedical data sharing (Federer et al., 2015), research subjects’ privacy is the most common reason why researchers are reluctant to share data. Other factors include publication competition, unnecessary data/manuscript audit and misuse/misinterpretation of the data. In addition, there is currently no proper mechanism to accredit researchers who contribute or share the data. In some cases, these researchers will either be included as a co-author on a publication, get recognition in the acknowledgment section of the publication, or be cited in the bibliography. Some researchers may not receive any acknowledgment for sharing their data at all.
Another data sharing concern is the loss of information and data context. Compared to the enormous number of variables present in clinical research, especially on the metadata level, data warehouses store only a fraction of the total data collected. Moreover, acquiring the core dataset alone may not be sufficient for other researchers to understand and reuse the data effectively. Although current EHR systems are designed for ease of use by researchers, many data fields still exist in unstructured format that hinder effective data sharing, and there has not been a highly reliable approach to explore this data. At the same time, inconsistency in data standards and formats in structured data also prevent researchers from sharing and learning from other data (Richesson and Krischer, 2007).
For researchers who do participate in data sharing, they are required to obtain consent from enrolled subjects for all studies. In this case, researchers may choose to request additional consent to sharing data. In practice, however, this is hard to implement as researchers are not able to foresee the purpose and results of secondary analysis that may come up much later than the time consent is obtained. In contrast to researchers’ legal compliance, patients and volunteers are much more open to data sharing. According to a study, 93% of patients were very or somewhat likely to allow their own data to be shared with university scientists, and 82% were very or somewhat likely to share with scientists in for-profit companies (Mello et al., 2018).
According to health information policies and regulations, patients possess the ownership of their health data and should be requested for consent when their data is used for secondary analysis. In current practice, patients may provide consent by physically signing a paper form or electronically signing a document online. Electronic consent forms can be used to more efficiently identify the original patient providing the consent if the forms are associated with a patient in the database. Paper-based consent forms, on the other hand, require much more effort to store (e.g., scanning and upload an electronic copy of the physical forms and manually entering data into the system) and may be lost or illegible along the process, making re-consent more difficult to establish (Taichman et al., 2016).
It is therefore important to create a platform that values privacy and is able to easily trace back to the appropriate patient to re-consent, which may further encourage sharing and reuse of research data. It is also critical to ensure that data is shared and reused responsibly. Mechanisms like peer review or patient review of proposals for reusing research data can protect the subjects and the original researchers who acquired the data. With a careful design, it is possible to incorporate these desired features into the data sharing platform to allow a more flexible and direct way to obtain consent for data sharing.
How do we leverage the potential trend toward patient-centric stewardship of medical data to improve research matching, control of research data, and incorporation of non-traditional data sources accessible to mobile devices? We present an architecture that publishes or redirects research studies into a public distributed ledger that is used by researchers and research participants for finding mutual matches. The goal of this Ledger for research studies is to have a virtually centralized location for hosting and discovering research studies that is accessible from mobile apps and reduces recruitment costs. The expectation is that marketing and other costs to engage patients with the Ledger would be amortized across the thousands of studies published there and help address Challenges 3.1 and 3.2.
A second component of the approach is that individual users would download the catalog of studies and match against them directly on their mobile devices. This model would facilitate scaling up matching by not requiring researchers to already have a clinical relationship with the user and still be able to match against clinical data. Further, the patient can prospectively discover and match against studies privately, helping to address Challenge 3.2.
A final key component of the approach is that patients directly discover studies and disseminate their data to these studies. Through this model, patients control dissemination of their data, which allow them to send their data to as many studies as they wish in a self-direct manner and flat structure, enabling greater potential research data reuse. For example, a patient can provide the same set of data to ten studies that desire it without relying on the first researcher that they provide the data to share it with the other nine studies. The decision of how data, owned by the participant, is subsequently distributed is up to the participant and not the researcher that receives the data. Further, later studies that publish requests for the same data as a prior study have the potential to be matched against the same set of original participants from an earlier study and receive the original data if the participants self-provide consent.
The remainder of this section provides an overview of the key attributes of distributed ledgers and then provides an architecture for exploiting properties of distributed ledgers to design these components. The section covers both the benefits and trade-offs of the architecture.
Distributed Ledger Technology (DLT) as implemented with a Blockchain data structure was first considered by Haber and Stornetta in 1991 within their landmark paper, “How to Time-Stamp a Digital Document,” as an approach consisting of a chained data structure and a node-based distribution network (Haber and Stornetta, 1990). Faced with a future where an overwhelming majority of media would become digitized, they considered the ease with which creation and modification dates could be tampered with. As a result, a proposal was made to develop a data structure whereby a “…chain of time-stamps…” (Haber and Stornetta, 1990) consisting of the utilization of cryptographically strong hash functions would be utilized along with a consensus-based mechanism for verification within a trustless environment. This “chain” served as a starting point for the most popular data structure implementation of the Ledger called Blockchain. Along with foundational principles in peer-to-peer distribution, this also provided a framework for what was to come in 2008 when an as-yet-unidentified individual known by the name Satoshi Nakamoto distributed what would become Blockchain’s most popular implementation in the form of the paper entitled, “Bitcoin: A Peer-to-Peer Electronic Cash System” (Nakamoto, 2008). At its heart, DLT consists of two primary components: a blockchain data structure and a peer-to-peer network. In order to more fully understand these components, we will break down each in turn providing more relevant details along the way.
Within DLT, the blockchain data structure serves to represent the Ledger. As an illustrating example, Alice records a piece of data containing her name and other personal information to a text document and saves the file afterward. She would like to ensure that the information in this file is not altered by anyone with proof. Given the ease with which a digital file can be copied and modified, how might Alice certify in some provable way that her file is the original file owned by her? To expand on this scenario, another person Bob may want to perform this same task but with his name and information stored in the file. How can both versions of the document be protected against tampering and proven that they represent two distinct states entered at different points in time? This is where a blockchain data structure is useful for the purposes of creating a tamper-resistant Ledger.
Blockchain consists of n nodes that are linked together in a cryptographically protected manner. During the formation of the chain, each node consists of data provided by some client application (such as a name or other personal data) and a cryptographic hash of the data in the node that precedes it (except for the case of the root node, where no data precedes it). The hash algorithm, also called “the workhorses of modern cryptography,” (Schneier, 2004) is fundamental to this technology. Hashing algorithms have several key traits, including an input that can be of an arbitrary size, a fixed-size output space, and efficiency (Narayanan et al., 2016) with respect to computation.
Together, these properties use the information stored in the Ledger (and whatever other data might be relevant at the time of hashing – such as a timestamp) to produce a long string of letter and number combinations that represents a snapshot of that data that is computationally infeasible to reverse and also proves mathematically that the data is unaltered. If the same long string representation is embedded into the next link in the chain (along with the important source data), by hashing those bits together, a cryptographically irreversible bond can be produced from one record to another.
Within DLT, the distributed nature is commonly implemented through a peer-to-peer networking structure. More specifically, the blockchain data structure described above that serves to form the Ledger is distributed among p number of peers for the purpose of independent validation of the data in the blockchain in order to establish mathematically provable trust within an otherwise trustless environment.
Given the often-times decentralized nature of the distribution network, node identities are largely anonymous. As a result, there is a challenge in establishing trust with an anonymous party whose transactions within the Ledger look identical no matter if they are a bad or good actor. Trust is an important factor within any network whereby verifiable truth must be established that a specified bit of data has been recorded into a Ledger and has not been tampered with. As the blockchain-based Ledger has been distributed among some number of peer nodes, each individual peer holds the same exact version of that Ledger. How to establish trust within this anonymous space? What prevents bad-actors from colluding to tamper with the data in the Ledger and still certifying its original veracity? Why is it important to distribute the Ledger in the first place? The answer to these questions lies within a specific activity that typically occurs within a decentralized distribution network; namely, consensus.
Consensus mechanisms are designed to achieve agreement with respect to the veracity as it pertains to a particular activity within a system. This has been identified as “a fundamental problem of fault-tolerant distributed computing” (Fischer et al., 1985) – to achieve reliability in distributed systems, protocols are needed that enable the system as a whole to continue to function despite the failure of a limited number of components. For a Distributed Ledger, the reliability of the system is directly related to the trust within that system. The failure in the system directly relates to bad actors whose primary goal is to undermine that trust in return for personal gain. In order to achieve trust through consensus, several algorithms have been designed for this purpose including Proof of Work (Nakamoto, 2008), Proof of Stake (Buterin, 2013), and Practical Byzantine Fault Tolerance (Castro and Liskov, 1999). Each algorithm achieves consensus through different mechanisms, which have both positive and negative attributes to them (Zhang et al., 2019a), leaving the choice of which algorithm to use to the architects of the system and their stated goals.
DLT allows a user to record data in an immutable manner through the use of a blockchain data structure while also obtaining verification of that fact through the use of decentralized and distributed consensus algorithms. As a result of these two broad properties, this technology presents a compelling architecture with respect to maintaining robust transactional integrity for our solution described herein.
Figure 1 shows an architecture for a patient-centric stewardship model of research study matching and clinical data sharing. The goals of this architecture are to: (1) allow patients to perform research study matching using their health data on their local devices, (2) create immutable public descriptions of research studies and the data they consume, (3) provide patients with the ability to directly send their health data from clinical and non-traditional data sources (e.g., apps) to researchers, and (4) allow patients to control and acknowledge the sharing of their research data.
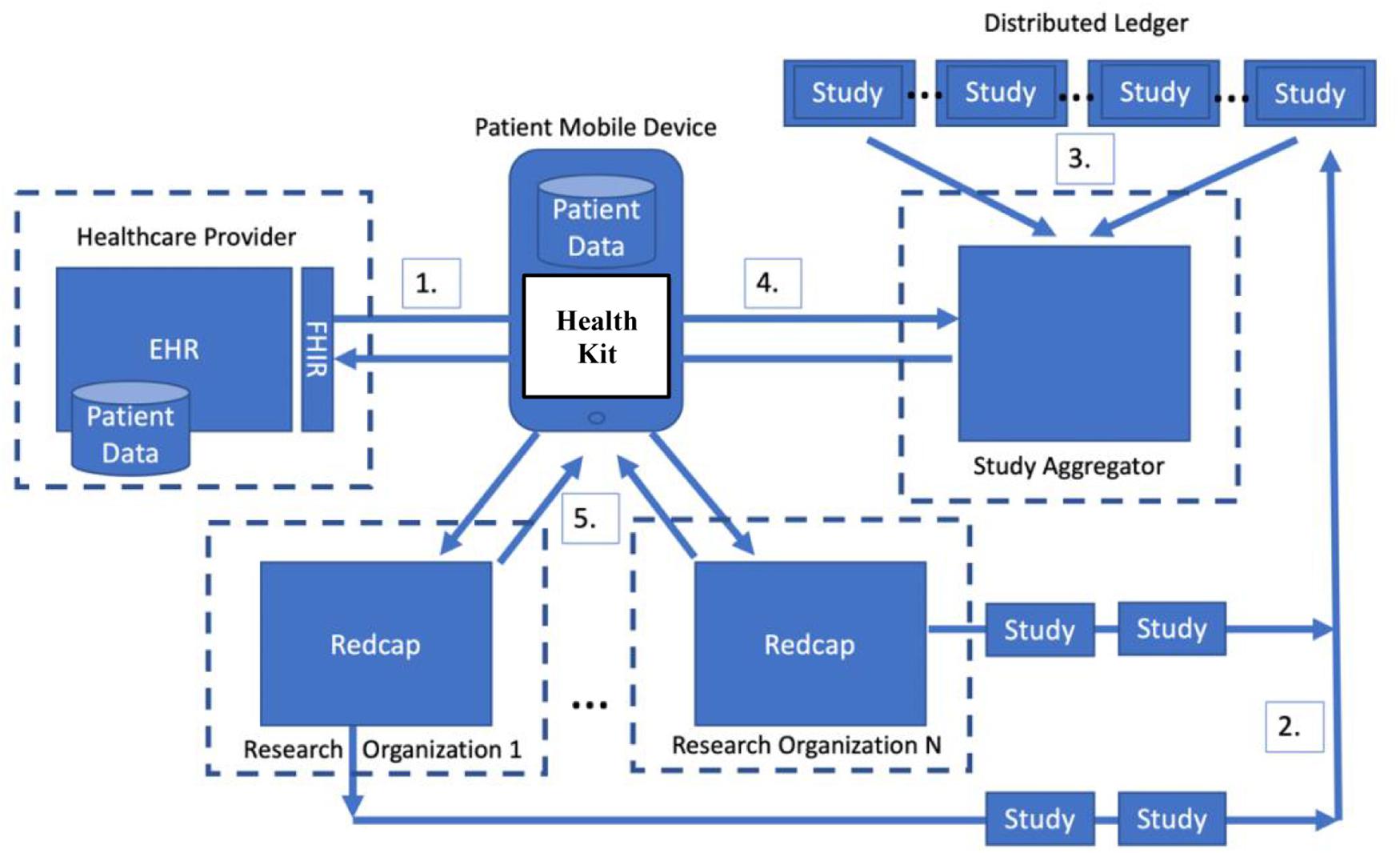
Figure 1. An architecture for enabling patient-centric stewardship of research data using a distributed ledger.
The key emerging change in the healthcare market that makes this architecture feasible is the move toward patient-centric stewardship of data on their mobile devices. As shown in Step 1 of Figure 1, patients can directly import their health data from a provider onto their mobile phone. Apple devices provide the HealthKit API and access to Epic EHRs via FHIR.
The rest of the architecture shown in Figure 1 focuses on enabling devices to discover research studies and find researchers in need of their data. The core idea is that patients have the ultimate control of where their data goes and when it may be reused in other research studies. The distributed ledger component of the architecture facilities the discovery of research studies by creating a public record of all studies and a precise description of the data consumed by each study. In order to gain access to research participant health data, researchers must publish the description of their study (Step 2) into the Ledger where it can be discovered by patient devices (Steps 3 and 4). Based on the data description, participants choose from the lists of studies that they potentially match or that would benefit from data that they have already provided to a research study in the past to share data with (Step 5).
Using a public distributed ledger for an application that facilitates both the recruitment of research participants, as well as the sharing of research data offers a number of advantages. Those advantages can be grouped into three distinct categories: data security, transaction control, and reliability. When contemplating data security for a research participant use case, it is important to note that in a public Ledger, the records of all transactions are public and immutable. That is not to say that the underlying medical data is public, but simply that the descriptions recording the access of data are public. Once a blockchain operation occurs and the transaction is recorded, that record is immutable and will propagate to all the peer nodes in the decentralized network. A study published into the blockchain cannot be retracted and will provide a permanent clear record of the data it consumed. If a research study is completed, however, it will be flagged as completed and will not be used for participant matching.
One architectural possibility would be to have research participants directly record study enrollment directly in the blockchain. An individual who agreed to participate in a research study would have a permanent record of any and every study that accessed the participant’s data. To identify a participant in a privacy-preserving manner, one approach would be storing encrypted identifying metadata to distinguish participants. Likewise, a study would be able to see what studies a participant has joined. For research studies, however, having a public record of participation is problematic because it violates privacy rules regarding research participation. To overcome this challenge, the architecture shown in Figure 1 leverages the distributed ledger only for advertising studies and recording the data that those studies consume. As shown in Step 2 of Figure 1, researchers publish a description of the study into the Ledger, but participation in studies is handled completely outside of the blockchain.
With patients stewarding their own medical data, they have the freedom to determine whether to participate in each research study. Within each study, a patient who is willing to participate would also be able to decide exactly which data to share with a particular study. This gives patients complete control over the use of their medical data. One approach would be to use the blockchain to facilitate the transfer of the data itself, but this is problematic for the same reason as recording participation in the blockchain – it would inevitably violate research participant privacy.
In the architecture shown in Figure 1, all joining of studies and sending of data is performed outside of the blockchain between the participant and researcher. Step 5 directly sends data to a research data management platform, such as REDCap (Harris et al., 2009). In other work, we have relied on direct submission of data from participants’ devices to REDCap. The key problem that this architecture overcomes is finding participants and solving the technical challenges of getting their clinical data into REDCap from their provider. Further, this architecture allows submission of data from IoT or other sources accessible to the device (e.g., Bluetooth Glucometers, Wifi Scales, etc.).
Although a participant may match a study based on an analysis done on the patient’s device, researchers may still have other criteria that are difficult or impossible to publish into the blockchain for matching. During the direct communication between the participant and the researcher, the researcher may choose not to use the participant’s data. In these cases, the data collected from the participant would need to be discarded by the researcher. A downside of the architecture is that there is no way to enforce destruction of participant data – although this is also the case in current practice. The architecture still relies on institutional controls, such as policies and Institutional Review Boards (Lincoln and Tierney, 2004), to ensure researchers act ethically.
The nature of blockchain networks provides a third important aspect: reliability. Participants and study providers must be able to trust that the chain of published research studies is valid and will not disappear. Since blockchains are a network of independent nodes, there is not a single point of failure, nor is one node able to control the entire network. Before transactions are recorded, they must be validated according to the consensus mechanism for that network. Once a transaction is validated, it is recorded and propagated to the individual nodes such that the loss of one or more nodes, or control of one or more nodes will not impact the validity of the transaction records on the entire network.
All research studies added to the blockchain include a request for participants who have a particular set of medical characteristics. Patients are notified of the availability of the study by their device and can choose to validate their medical data against the requested characteristics. If validation is successful, patients can choose to submit the validation (along with additional participation data) to the study to initiate their participation, as shown in Step 5 of Figure 1. The study provider would see a transaction indicating a successful match, along with the participation data necessary to include the patient in the study and validate the match. This chain of transactions could also include the ability for participants to monetize the use of their data, or generally for their participation, if such were a requirement. All these transactions take place directly between the participant’s device and the researcher using a standard platform, such as REDCap.
In order to expedite the matching process, studies are defined by three sets of characteristics that may be matched against: boolean conditions (ex: asthma, hypertension), enumerated characteristics (ex: hair color, relationship status), and ranged characteristics (ex: desired age range, desired weight range, how long a condition has been diagnosed). These characteristics are provided by researchers conducting the studies. These simplifications allow for primitive boolean tests to decide whether the criteria for a study match the healthcare data provided by a given patient. In order for a patient to qualify for a given study, they must have a complete (100%) satisfaction of study criteria via a simple iterative key-value boolean loop.
Because each study adheres to the same language of matching criteria, relationships can be formed between the studies. A key benefit of the matching language is that it facilitates condensing the matching rules across multiple research studies into a single network of rules using the Rete algorithm (Forgy, 1989). The Rete algorithm is designed to take in a knowledge base of facts (e.g., the participants’ clinical and IoT data) and efficiently determine which rules from a set should fire (e.g., which research studies match). Each rule is defined by a set of matching conditions and an action. In the proposed architecture, the conditions are the research study matching criteria and the action is proposing to the user a possible research study is matched. The algorithm shares conditions between rules in a directed acyclic graph so that conditions are only evaluated once regardless of how many rules include the condition. For example, the condition of the participant having blood pressure above a threshold would be evaluated once, regardless of how many research studies relied on the same matching condition.
The entire body of published studies can be used to collect matching conditions and build an acyclic matching graph using Rete. A graph analyzes the necessary conditions of one study in conjunction with the sufficient conditions of another, allowing for the elimination of more complex study matching should a patient’s data deem them unqualified for a simpler study with a subset of the matching criteria. For example, if a patient fails to qualify for Study A, which requires participants to be aged 30–40, then the graph will immediately eliminate Study B which requires participants aged 33–37 with hypertension. In this way, consideration of a simple study can cascade the elimination of countless nodes/studies in the graph, drastically improving performance on patient-study matching.
The drawback of the dependency graph is the time required to generate the graph. A few considerations mitigate this cost. First, the graph need only be generated on the server, thus each mobile device does not have individual time expensed for the graph. This generation of the graph server-side is captured in Step 4 of Figure 1. Second, the proposed generation of the dependency graph is to trigger a new server-side build of the graph once daily (optimally during non-peak usage hours) to update the graph with new studies added to the blockchain and completed studies marked as no longer recruiting. As such, the dependency graph method best optimizes average-user performance– and very clearly increases scalability of the matching algorithm. The dependency graph need only be built once, and then can be shared amongst all mobile device sessions.
Although there is significant discussion on enabling patient data sovereignty using blockchains, very few of these discussions address a major fundamental problem – access to the blockchain. Interacting with a blockchain requires the setup of a node in the distributed ledger, which can be a complicated endeavor. For example, most Bitcoin (Nakamoto, 2008) users rely on a third-party wallet service (Antonopoulos, 2014) to hold their cryptocurrency, run the required distributed ledger node, and perform trades on their behalf. Despite the appearance of complete decentralization and control by the user, the user is actually dependent on the wallet service for access and is not completely in control.
A similar problem arises in using a blockchain to publish research studies. Blockchains are difficult to access from a mobile device without an intermediate service, equivalent to a wallet service for Bitcoin. Directly accessing and validating transactions on a blockchain is both time and energy consuming, which makes downloading the entire Ledger and validating it on a mobile device problematic.
The architecture shown in Figure 1 handles this access issue by introducing a Study Aggregator as shown between Steps 3 and 4. The study aggregator manages access to the distributed ledger and watches for the publication of new studies into the Ledger. When new studies are published, it validates and aggregates them into a comprehensive catalog of available studies.
A further function of the study aggregator is to use the Rete algorithm to build the acyclic research study matching graph described previously. Both interacting with the blockchain and constructing this acyclic graph are potentially expensive operations that are isolated on the server-side aggregator, where power consumption and processing power are much less problematic. Furthermore, aggregation and graph construction costs can be paid once and amortized across all mobile device accesses rather than paid on each individual device.
The downside of this approach is that it introduces a potential central point of failure and control in the system. However, there are two key reasons that this is not a significant concern. First, any number of study aggregators can be run independently by arbitrary organizations. There is no need for a single study aggregator in the system. Each research institution can run their own study aggregator and provide aggregation services to research participants’ mobile devices.
Second, the failure of an aggregator only temporarily cuts off access to the study catalog for the mobile devices currently relying on that specific aggregator. A mobile device can use multiple aggregators for redundant access or consensus. Even if one aggregator fails, a participant can discover and use other aggregators. Since the aggregator only produces a derived copy of the research matching graph, the original research study data is still immutably and reliably stored in the distributed ledger despite aggregator failures.
An additional consideration of the study aggregator is how it impacts trust, scalability, and privacy (Zhang et al., 2017). Any time that trust in the aggregator is reduced, it improves privacy at the expensive of scalability. The critical privacy and scalability tuning of the system is done in how trust relationships are established with study aggregators and how much work is offloaded to the aggregator.
The proposed architecture does not dictate how trust is established in a particular study aggregator. Our belief is that research institutions already manage the establishment of trust with research participants and are likely the best conduit to establish these trust relationships. For example, research institutions could create a trust aggregator and advertise its address on their existing websites or through face-to-face interactions with clinicians. Alternatively, non-profits organized around specific interests, such as diseases (e.g., American Cancer Society), could operate and publish aggregators.
Mobile devices rely on the acyclic matching graphs produced by the study aggregators. There is an opportunity to improve scalability and performance on the mobile device by pruning the acyclic graph at the aggregator to reduce the data transfer to the mobile device and the amount of work matching against the graph. Any pruning of the graph at the aggregator reduces the workload on the mobile device, which will be the limiting factor in the scalability of the system if the entire matching graph for every published study needs to be transferred to each mobile device.
To improve scalability, mobile devices can either: (1) send a subset of their data to an aggregator to perform intelligent pruning or (2) subscribe to aggregators that publish graphs pruned to a specific set of interests. For example, a device can send a limited set of less-sensitive and semi-anonymous data, such as age and weight, to the server and receive a pruned subset of the graph that has potentially viable studies that can be determined with further matching on the device. The benefit of this approach is that matching can be more easily scaled. The downside is that the approach inherently reduces the overall privacy of the system by requiring some set of data from the mobile device.
An alternative approach to improve scalability is to subscribe to an aggregator that publishes a pruned graph that only contains studies relevant to a specific interest. For example, an aggregator might only publish studies relevant to a specific disease. This approach also has a privacy trade-off in that subscription to the aggregator implies interest in a disease or set of diseases, which may have privacy implications (e.g., interest in cancer implies a cancer diagnosis).
In either approach, it is expected that once a match is made, the mobile device will begin direct communication with the study organization to verify the match. As part of this process, an important secondary verification will be performed, which is that the mobile device will download a description of the matching criteria directly from the research organization to ensure that the matching graph from the aggregator was accurate. If there is any discrepancy between the matching logic for the study published by the aggregator or the research study site, which would indicate tampering by one of the two entities, the mobile device will discard the match and not continue. This secondary matching is not full-proof and does indicate possible benefits to use a different aggregator than the organization operating a given study.
We performed an initial analysis of the scalability issues regarding research study matching on participants’ devices vs. on the server in terms of time and data transfer. The key scalability limitation that we found for on-device matching is shown in Figure 2. As the number of studies grows, the amount of data that has to be transmitted to the mobile device also grows. The analysis was conducted by randomly generating matching graphs representing varying numbers of studies and calculating their total size in kilobytes. We developed a compact representation of the graphs – although it is certainly possible to improve efficiency – and measured the overall amount of data that would need to be transmitted to the mobile device. As shown in the figure, the overall size of the matching graph is proportional to the number of research studies, which are expected to continually grow over time. With our test graph representation, 20,000 studies required transmitting roughly 48 megabytes to a client. Real-world studies may have more overlap in the matching conditions and there may be much more efficient representations that could lead to smaller graph sizes. This size, however, is similar in size to an average app download on a mobile device.
Figure 2 also shows the significant scalability improvement that can be achieved by sending data to the server and performing matching there. The bars labeled “Server” show the total data transfer required if the mobile device completely trusts the aggregator to perform matching on its behalf and sends data needed for matching to the server. As shown in the results, there are multiple orders of magnitude of overhead added when the mobile device does not trust the aggregator to perform matching vs. when it does.
The potential to have aggregators publish a pruned graph also illustrates a potential security issue. The mobile devices rely on the aggregator to publish an accurate graph of the studies in the blockchain. If an aggregator lies, they have the potential to perform a number of attacks from their trusted position. One potential way to overcome this issue is to use cryptographic signing of studies so that mobile devices can verify the authenticity of the study before beginning a direct interaction with a research due to a possible match. However, like any approach that relies on public key infrastructure, key distribution and trust is a significant issue. Indubitably, a set of trusted roots will be needed to provide signing chains that can be used to prove that a specific research study originated with a specific institution and researcher. The precise architecture of this distribution model is left to future work but is expected to look similar to how SSL certificates are issued for websites (Ellison et al., 1999).
Although the architecture has focused on scalability regarding matching, a secondary scalability concern is the metadata regarding research studies. Each research study includes data on the organization running the study, the matching criteria, the data collected by the study, and the purpose of the study. This data is not accounted for in Figure 2 and could be substantial. There are several architectural approaches to handling the scalability issues surrounding metadata that each have their own privacy-scalability trade-offs.
Our approach to handling metadata is to publish a non-blockchain address for retrieving the metadata directly from the organization hosting the study. For example, an academic institution could host web pages for each study with the metadata describing the study and include the URL for the metadata in the study description published to the blockchain. This approach eliminates the need for the aggregator to publish the catalog of metadata and reduces the data transfer to the mobile device. The aggregator only publishes the URL to retrieve the metadata and a signed hash of the study metadata that it read from the blockchain. The mobile device would compare the signed hash to the hash that it calculates for the metadata after retrieving it from the provided URL. Again, a key distribution mechanism would be needed and is not covered in the current work.
The proposed model offers an innovative framework that leverages distributed ledgers to facilitate the matching of patients to research studies by aggregating relevant healthcare data from certified EHRs that are episodic and mobile/tracking devices that are continuous. To summarize, key functional requirements of this model include the following aspects that are aligned with requirements that are specific to the healthcare/clinical research domain:
1. Preserving patient privacy: despite the use of distributed ledgers that inevitably expose some information in a distributed and shared manner, the Ledgers only serve as a conduit that enables the data exchanges between the involved parties, namely, the researchers coordinating a clinical study and the patients interested in participating in the trial. The transactions of the original health records are privacy-protected using tools, such as REDCap, that are HIPAA compliant, which is a crucial requirement of any healthcare application that involves patient data including any data exchanged via distributed ledgers (Zhang et al., 2017). Additionally, the use of cryptographic hashing on-chain assures access to data via REDCap is only granted to the intended researcher recipient.
2. Reliability of retrieved data: in order for a research study to be carried out successfully, obtaining reliable data whose owners must be properly identified is an indispensable part of the study (Davis et al., 1999). In traditional studies that involve face-to-face interactions between the participant and researcher, identifying the participant is a considerably easier task – the participant simply presents a photo identification and a set of personally identifiable information that matches the medical profile. However, in settings described in this paper, where in-person visits are not a requirement to participate in a study, verification of the identity and, in turn, the reliability of the participant data requires substantial proof. By virtue of the traceability and tamper-proof nature of distributed ledgers used by the proposed model, both identity and reliability of contributed data are easily verifiable through audit trails. If necessary, any data later on found to be unreliable or disqualified for the study can also be traced back to the original owner by the researcher in order to properly exclude the data and/or the participant from the study.
3. Data collection with minimal effort from participants: one of the key barriers of clinical trial recruitment are the frequently asked medical questions that are difficult for interested participants to understand (Cantrell and Lupinacci, 2007). Many studies already face the challenge of unable to meet a significant cohort size, and having this requirement from patients who may not be familiar with medical terminologies further turns volunteers away from participation. With the proposed model of algorithmically match patients to trials only using data available through patients’ mobile devices, patients are no longer required to understand what clinical measurements because our model enables the processing of rules pre-defined by researchers without involving patients themselves. Furthermore, patients can also be matched to multiple studies based on the data they are willing to provide without having to fill out multiple questionnaires with repeated questions. By lowering the barrier to provide data for research, participants will have an easier access to more studies and, similarly, researchers to volunteers.
4. Scalability for on-device matching: mobile devices inevitably suffer from limited space and computing power, which makes it hard for on-device matching of studies that require significant computation. Distributed ledger technology also faces scalability issues when serving healthcare applications due to the nature of the large population involved (Zhang et al., 2017). Our model proposes two options for enabling scalability on such a restrictive setting by offloading the heavyweight tasks to an aggregator service. These options do not overload either the mobile device or the underlying distributed ledgers used for data exchanges and thus provide a certain degree of scalability from the patient matching aspect.
5. Providing a more comprehensive data collection from patients upon request: patient-generated data have played an increasingly important role in clinical trials because such data is collected over time and hence captures more information about a person’s health history (Howie et al., 2014). Although medical records are important in assessing a patient’s overall conditions, patient-generated data, such as fitness, weight, or sleep data, contribute to a comprehensive picture that can provide invaluable insight to research studies. Our proposed model leverages both EHR and patient-generated data that are available on mobile devices to identify as many matches as possible. The model also allows patients to freely choose which research study their data may be shared with, and only when a patient chooses to let a study or researcher access their data on-device, it will then be delivered to the recipient.
This section presents prior research on the architectures and platforms designed to improve research study recruitment and summarizes recent work on using DLT and related technologies to enable data sharing in the healthcare space.
To date, there has been a number of efforts on providing patients, volunteers, and researchers with resources and information on clinical studies covering a large number of conditions and diseases. ClinicalTrials.gov (Zarin et al., 2011), a web-based, centralized clinical trial repository, is one of the most popular platforms where researchers register their trials publicly so that participants can easily access the study information. It is the largest clinical trial registry in the U.S. with over 300,000 trials reported. It does not contain all clinical studies, however, because some studies are not required to be registered. ResearchMatch.org (Harris et al., 2012) is another web-based, centralized platform for matching volunteers with actively recruiting trials and therefore maintains a subset of trials from ClinicalTrials.gov. ResearchMatch.org has a large number of volunteer users with their self-reported information, such as conditions and medications, that is used to provide basic trial recommendations based on a trial’s primary conditions targeted. Besana et al. (2010) proposed a domain-specific semantic ontology to represent data from patient health records and to evaluate patients’ eligibility to clinical trials. Another increasingly popular strategy to improve recruitment is the use of clinical trial alert tools that automatically apply eligibility criteria to EHRs in order to identify potential participants proactively (Heinemann et al., 2011).
Due to the increasing popularity of DLT given its unique properties, many healthcare data sharing frameworks based on distributed ledgers have been introduced in literature (Zhang et al., 2018a). For example, the MedRec system (Azaria et al., 2016) was proposed as a blockchain implementation of a healthcare data warehouse that facilitates clinical data sharing. The FHIRChain framework (Zhang et al., 2018d) was designed to enable data sharing between various healthcare data sources using the FHIR protocol and incorporated a number of key technical requirements of an interoperable healthcare service. Peterson et al. (2016) presented a healthcare blockchain with a single centralized source of trust for sharing patient data, introducing “Proof of Interoperability” based on conformance to the FHIR protocol as a means to ensure network consensus. More recently, Xia et al. (2017) described a blockchain-based system called “MeDShare” for enabling medical data sharing among cloud service providers. OpTrak, a DLT-based architecture used for exchanging and tracking opioid prescriptions is also proposed in Zhang et al. (2019b). Although these frameworks utilize distributed ledgers to provide data exchanges between different healthcare systems, they do not directly address the requirements specific to clinical studies and thus do not meet the functional requirements of our proposed model. Another study by Benchoufi and Ravaud (2017) described a smart-contract based system to collect participant consent for a clinical trial, but does not match participants to trials they may be eligible for. A framework proposed by Theodouli et al. aims to provide a patient-centric model that allows data sharing in clinical trials but does not include methodologies that algorithmically provide patients with trials they are eligible for based on their specified data to share (Theodouli et al., 2018). To the best of our knowledge, there has not been a published study that addresses all the functional requirements proposed by our model specific to the clinical trial matching domain.
Given the fundamental importance of capturing a complete picture of a patient’s healthcare history, why do researchers and medical institutions not have a universal system to share the needed research data? Currently, healthcare information is generally captured using electronic medical records by each individual provider. However, a variety of factors, ranging from data format incompatibility, differing approaches to labs, and challenges in identifying patients has led to a model where healthcare data does not flow freely between all providers.
Overcoming the challenges of healthcare data exchange is going to require allowing patients to easily control and move their data between providers and to get their non-traditional data from apps and other sources into their medical record. However, moving to a patient-centered medical data stewardship model faces immense challenges if all of the data stewardship falls solely on the medical institutions, ranging from the existing issues with data formats and labs, to additional barriers to how all patients, not just the most technically sophisticated, can durably store and authorize access to their data in a secure way (Zhang and Boulos, 2020). The underlying healthcare networks are inherently decentralized, so there is also a challenge of figuring out how to move to provide a patient-centered model without a central authority to mediate exchange and mandate decisions.
Doctors also face the daunting challenge of trying to diagnose patients from a combination of symptoms and medical history. A patient’s medical record provides essential clues to a provider that help, both to diagnose patients more accurately and also help eliminate possibilities and often associated diagnostics or procedures that may expose patients to additional risk. Whenever medical information is missing, the impact can be longer, less accurate, and riskier for diagnostic processes.
This paper explores the conflicting forces that make achieving a patient-centered stewardship hard and investigates how the emerging capabilities of decentralized ledgers may help to alleviate some of these conflicts. A key goal of the work is to understand where DLT can serve a role in a patient-centered model, what problems it solves, what new problems it introduces, and what problems still remain unaddressed. Further, through the investigation, the paper analyzes distributed ledger architectural options and how they resolve conflicting forces at different levels.
The final component of the paper is a prototype architecture for using distributed ledgers to facilitate a patient-centered data stewardship model. The architecture draws insights from the detailed exploration and architectural trade-offs analysis to prescribe a set of proposed standards for using DLT in this domain. Extending upon this work, we will explore a more detailed architecture with a proof-of-concept implementation that embeds the design considerations as discussed in this paper in future work. Additionally, we recognize the critical nature of performing quantitative analyses of the architecture to facilitate the implementation of blockchain-based solutions, so we will also provide those analyses in future work.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
PZ: key author and primary researcher. CD, CM, PS, and CW: researchers and co-authors. NT: co-author. LM and LK: researchers. SL and DS: research sponsors. JW: key author, researcher, and sponsor. All authors contributed to the article and approved the submitted version.
CD, CM, PS, CW, and SL were employed by the company Solaster.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Antonopoulos, A. M. (2014). Mastering Bitcoin: Unlocking Digital Cryptocurrencies. Newton, MA: O’Reilly Media, Inc.
Apple (2020). “Apple.” Apple. Available online at: https://www.apple.com/ (accessed July 1, 2020).
Azaria, A., Ekblaw, A., Vieira, T., and Lippman, A. (2016). “Medrec: using blockchain for medical data access and permission management,” in Proceedings of the 2016 2nd International Conference on Open and Big Data (OBD) (Piscataway, NJ: IEEE), 25–30.
Beard, L., Schein, R., Morra, D., Wilson, K., and Keelan, J. (2012). The challenges in making electronic health records accessible to patients. J. Am. Med. Inform. Assoc. 19, 116–120. doi: 10.1136/amiajnl-2011-000261
Benchoufi, M., and Ravaud, P. (2017). Blockchain technology for improving clinical research quality. Trials 18, 1–5.
Bender, D., and Sartipi, K. (2013). “HL7 FHIR: an Agile and RESTful approach to healthcare information exchange,” in Proceedings of the 26th IEEE international symposium on computer-based medical systems (Piscataway, NJ: IEEE), 326–331.
Besana, P., Cuggia, M., Zekri, O., Bourde, A., and Burgun, A. (2010). “Using semantic web technologies for clinical trial recruitment,” in Proceedings of the International Semantic Web Conference (Berlin: Springer), 34–49. doi: 10.1007/978-3-642-17749-1_3
Borchers, A. T., Uibo, R., and Gershwin, M. E. (2010). The geoepidemiology of type 1 diabetes. Autoimmun. Rev. 9, A355–A365.
Buhrmester, M., Kwang, T., and Gosling, S. D. (2016). Amazon’s mechanical turk: a new source of inexpensive, yet high-quality data? Perspect. Psychol. Sci. 6, 3–5. doi: 10.1177/1745691610393980
Cantrell, M. A., and Lupinacci, P. (2007). Methodological issues in online data collection. J. Adv. Nurs. 60, 544–549. doi: 10.1111/j.1365-2648.2007.04448.x
Castro, M., and Liskov, B. (1999). “Practical byzantine fault tolerance,” in Proceedings of the Third Symposium on Operating Systems Design and Implementation OSDI, Vol. 99, Berkeley, CA, 173–186.
CenterWatch (2020). “Clinical Trial Resources.” CenterWatch. Available online at: https://www.centerwatch.com/ (accessed July 1, 2020).
Chen, B., and Butte, A. J. (2016). Leveraging big data to transform target selection and drug discovery. Clin. Pharm. Ther. 99, 285–297. doi: 10.1002/cpt.318
Davis, J. R., Nolan, V. P., Woodcock, J., and Estabrook, R. W. (1999). “Assuring data quality and validity in clinical trials for regulatory decision making,” in Workshop Report. Roundtable on Research and Development of Drugs, Biologics, and Medical Devices, Division of Health Sciences Policy, Institute of Medicine (Washington DC: National Academy Press).
DiMasi, J. A., and Grabowski, H. G. (2007). The cost of biopharmaceutical R&D: is biotech different? Manag. Decis. Econ. 28, 469–479.
Ellison, C., Frantz, B., Lampson, B., Rivest, R., Thomas, B., and Ylonen, T. (1999). SPKI Certificate Theory RFC 2693. Available online at: https://dl.acm.org/doi/book/10.17487/RFC2693 (accessed July 1, 2020).
EPIC (2020). “Epic.” Epic. Available online at: https://www.epic.com/ (accessed July 1, 2020).
Federer, L. M., Lu, Y. L., Joubert, D. J., Welsh, J., and Brandys, B. (2015). Biomedical data sharing and reuse: attitudes and practices of clinical and scientific research staff. PLoS One 10:e0129506. doi: 10.1371/journal.pone.0129506
Fischer, M. J., Lynch, N. A., and Paterson, M. S. (1985). Impossibility of distributed consensus with one faulty process. J. ACM (JACM) 32, 374–382. doi: 10.1145/3149.214121
Forgy, C. L. (1989). Rete: a fast algorithm for the many pattern/many object pattern match problem. 547–559. doi: 10.1016/b978-0-934613-53-8.50041-8
Frandsen, M., Thow, M., and Ferguson, S. G. (2016). The effectiveness of social media (Facebook) compared with more traditional advertising methods for recruiting eligible participants to health research studies: a randomized, controlled clinical trial. JMIR Res. Protoc. 5:e161. doi: 10.2196/resprot.5747
Garg, A. X., Adhikari, N. K., McDonald, H., Rosas-Arellano, M. P., Devereaux, P. J., Beyene, J., et al. (2005). Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: a systematic review. JAMA 293, 1223–1238.
Gul, R. B., and Ali, P. A. (2010). Clinical trials: the challenge of recruitment and retention of participants. J. Clin. Nursing 19, 227–233. doi: 10.1111/j.1365-2702.2009.03041.x
Haber, S., and Stornetta, W. S. (1990). “How to time-stamp a digital document,” in Proceedings of the Conference on the Theory and Application of Cryptography (Berlin: Springer), 437–455. doi: 10.1007/3-540-38424-3_32
Harris, P. A., Scott, K. W., Lebo, L., Hassan, N., Lighter, C., and Pulley, J. (2012). ResearchMatch: a national registry to recruit volunteers for clinical research. Acad. Med. J. Assoc. Am. Med. Coll. 87:66. doi: 10.1097/acm.0b013e31823ab7d2
Harris, P. A., Taylor, R., Thielke, R., Payne, J., Gonzalez, N., and Conde, J. G. (2009). Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 42, 377–381. doi: 10.1016/j.jbi.2008.08.010
Heinemann, S., Thüring, S., Wedeken, S., Schäfer, T., Scheidt-Nave, C., Ketterer, M., et al. (2011). A clinical trial alert tool to recruit large patient samples and assess selection bias in general practice research. BMC Med. Res. Methodol. 11:16. doi: 10.1186/1471-2288-11-16
Howie, L., Hirsch, B., Locklear, T., and Abernethy, A. P. (2014). Assessing the value of patient-generated data to comparative effectiveness research. Health Affairs 33, 1220–1228. doi: 10.1377/hlthaff.2014.0225
Kahn, J. S., Aulakh, V., and Bosworth, A. (2009). What it takes: characteristics of the ideal personal health record. Health Affairs 28, 369–376. doi: 10.1377/hlthaff.28.2.369
Kaushal, R., Shojania, K. G., and Bates, D. W. (2003). Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch. Int. Med. 163, 1409–1416.
Kitterman, D. R., Cheng, S. K., Dilts, D. M., and Orwoll, E. S. (2011). The prevalence and economic impact of low-enrolling clinical studies at an academic medical center. Acad. Med. J. Assoc. Am. Med. Coll. 86:1360. doi: 10.1097/acm.0b013e3182306440
Kreuzthaler, M., Schulz, S., and Berghold, A. (2015). Secondary use of electronic health records for building cohort studies through top-down information extraction. J. Biomed. Inform. 53, 188–195. doi: 10.1016/j.jbi.2014.10.010
Lincoln, Y. S., and Tierney, W. G. (2004). Qualitative research and institutional review boards. Qual. Inq. 10, 219–234. doi: 10.1177/1077800403262361
Lovato, L. C., Hill, K., Hertert, S., Hunninghake, D. B., and Probstfield, J. L. (1997). Recruitment for controlled clinical trials: literature summary and annotated bibliography. Controll. Clin. Trials 18, 328–352. doi: 10.1016/s0197-2456(96)00236-x
Majumder, M. A., Guerrini, C. J., Bollinger, J. M., Cook-Deegan, R., and McGuire, A. L. (2017). Sharing data under the 21st century cures act. Genet. Med. 19, 1289–1294. doi: 10.1038/gim.2017.59
Mello, M. M., Lieou, V., and Goodman, S. N. (2018). Clinical trial participants’ views of the risks and benefits of data sharing. New Engl. J. Med. 378, 2202–2211. doi: 10.1056/nejmsa1713258
Mulvaney, S. A., Hood, K. K., Schlundt, D. G., Osborn, C. Y., Johnson, K. B., Rothman, R. L., et al. (2011). Development and initial validation of the barriers to diabetes adherence measure for adolescents. Diabet. Res. Clin. Pract. 94, 77–83. doi: 10.1016/j.diabres.2011.06.010
Nakamoto, S. (2008). Bitcoin: A Peer-to-peer Electronic Cash System. Available online at: https://bitcoin.org/en/bitcoin-paper (accessed July 1, 2020).
Narayanan, A., Bonneau, J., Felten, E., Miller, A., and Goldfeder, S. (2016). Bitcoin and Cryptocurrency Technologies: a Comprehensive Introduction. Princeton, NJ: Princeton University Press.
Nasser, M. N., Grady, D., and Balke, C. W. (2011). Commentary: improving participant recruitment in clinical and translational research. Acad. Med. J. Assoc. Am. Med. Coll. 86:1334. doi: 10.1097/acm.0b013e3182302831
National Institutes of Health (2020). “NIH Clinical Center: Patient Recruitment at the NIH Clinical Center.” National Institutes of Health. Available online at: https://www.cc.nih.gov/recruit (accessed July 1, 2020).
National Science Foundation (2010). NSF Data Sharing Policy 2010. Available online at: http://www.nsf.gov/bfa/dias/policy/dmp.jsp (accessed July 1, 2020).
Peterson, K., Deeduvanu, R., Kanjamala, P., and Boles, K. (2016). A blockchain-based approach to health information exchange networks. 1, 1–10.
Richesson, R. L., and Krischer, J. (2007). Data standards in clinical research: gaps, overlaps, challenges and future directions. J. Am. Med. Inform. Assoc. 14, 687–696. doi: 10.1197/jamia.m2470
Samuels, M. H., Schuff, R., Beninato, P., Gorsuch, A., Dursch, J., Egan, S., et al. (2017). Effectiveness and cost of recruiting healthy volunteers for clinical research studies using an electronic patient portal: a randomized study. J. Clin. Transl. Sci. 1, 366–372. doi: 10.1017/cts.2018.5
Schneier, B. (2004). Cryptanalysis of MD5 and SHA: Time for a New Standard. Computerworld 19. Available online at: https://www.schneier.com/essays/archives/2004/08/cryptanalysis_of_md5.html (accessed August 19, 2004).
Taichman, D. B., Backus, J., Baethge, C., Bauchner, H., De Leeuw, P. W., Drazen, J. M., et al. (2016). Sharing clinical trial data: a proposal from the international committee of medical journal editors. Ann. Int. Med. 164, 505–506.
Theodouli, A., Arakliotis, S., Moschou, K., Votis, K., and Tzovaras, D. (2018). “On the design of a Blockchain-based system to facilitate Healthcare Data Sharing,” in Proceedings of the 2018 17th IEEE International Conference On Trust, Security And Privacy In Computing And Communications/12th IEEE International Conference On Big Data Science And Engineering (TrustCom/BigDataSE) (Piscataway, NJ: IEEE), 1374–1379.
Xia, Q. I., Sifah, E. B., Asamoah, K. O., Gao, J., Du, X., and Guizani, M. (2017). MeDShare: trust-less medical data sharing among cloud service providers via blockchain. IEEE Access 5, 14757–14767. doi: 10.1109/access.2017.2730843
Zarin, D. A., Tse, T., Williams, R. J., Califf, R. M., and Ide, N. C. (2011). The ClinicalTrials. gov results database—update and key issues. New Engl. J. Med. 364, 852–860. doi: 10.1056/nejmsa1012065
Zhang, P. (2018). Architectures and Patterns for Moving Towards the Use of High-frequency, Low-fidelity Data in Healthcare. Ph.D. dissesrtation, Vanderbilt University, Nashville, TN.
Zhang, P., and Boulos, M. N. K. (2020). “Blockchain solutions for healthcare,” in Precision Medicine for Investigators, Practitioners and Providers, eds J. Faintuch, and S. Faintuch (Cambridge, MA: Academic Press), 519–524. doi: 10.1016/b978-0-12-819178-1.00050-2
Zhang, P., Schmidt, D., White, J., and Mulvaney, S. (2018b). “Towards precision behavioral medicine with IoT: iterative design and optimization of a self-management tool for type 1 diabetes,” in Proceedings of the 2018 IEEE International (Piscataway, NJ: IEEE), 64–74.
Zhang, P., Schmidt, D. C., White, J., and Dubey, A. (2019a). “Consensus mechanisms and information security technologies,” in Advances in Computers, Vol. 115, eds S. Kim, G. Chandra Deka, and P. Zhang (Amsterdam: Elsevier), 181–209. doi: 10.1016/bs.adcom.2019.05.001
Zhang, P., Schmidt, D. C., White, J., and Lenz, G. (2018a). “Blockchain technology use cases in healthcare,” in Advances in computers, Vol. 111, eds P. Raj, and G. C. Deka (Amsterdam: Elsevier), 1–41. doi: 10.1016/bs.adcom.2018.03.006
Zhang, P., Stodghill, B., Pitt, C., Briody, C., Schmidt, D. C., White, J., et al. (2019b). Optrak: tracking opioid prescriptions via distributed ledger technology. Int. J. Inform. Syst. Soc. Change 10, 45–61. doi: 10.4018/ijissc.2019040104
Zhang, P., Walker, M. A., White, J., Schmidt, D. C., and Lenz, G. (2017). “Metrics for assessing blockchain-based healthcare decentralized apps,” in Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom) (Piscataway, NJ: IEEE), 1–4.
Zhang, P., White, J., and Schmidt, D. (2018c). “Architectures and patterns for leveraging high-frequency, low-fidelity data in the healthcare domain,” in Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI) (Piscataway, NJ: IEEE), 463–464.
Keywords: patient-centered, data, data sharing, interoperability, blockchain, distributed ledger technology, research studies
Citation: Zhang P, Downs C, Le NTU, Martin C, Shoemaker P, Wittwer C, Mills L, Kelly L, Lackey S, Schmidt DC and White J (2020) Toward Patient-Centered Stewardship of Research Data and Research Participant Recruitment With Blockchain Technology. Front. Blockchain 3:32. doi: 10.3389/fbloc.2020.00032
Received: 12 July 2019; Accepted: 26 June 2020;
Published: 31 July 2020.
Edited by:
Victoria L. Lemieux, The University of British Columbia, CanadaReviewed by:
Andrea Vitaletti, Sapienza University of Rome, ItalyCopyright © 2020 Zhang, Downs, Le, Martin, Shoemaker, Wittwer, Mills, Kelly, Lackey, Schmidt and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhang, cGVuZy56aGFuZ0B2YW5kZXJiaWx0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.