- 1Department of Biology, University of Nevada, Reno, NV, United States
- 2Center for Marine Science and Technology, Curtin University, Telfair, Moka, Mauritius
- 3Department of Psychology, University of Western Ontario, London, ON, Canada
The use of autonomous recording units (ARUs) has become an increasingly popular and powerful method of data collection for biological monitoring in recent years. However, the large-scale recordings collected using these devices are often nearly impossible for human analysts to parse through, as they require copious amounts of time and resources. Automated recognition techniques have allowed for quick and efficient analysis of these recordings, and machine learning (ML) approaches, such as deep learning, have greatly improved recognition robustness and accuracy. We evaluated the performance of two deep-learning algorithms: 1. our own custom convolutional neural network (CNN) detector (specialist approach) and 2. BirdNET, a publicly available detector capable of identifying over 6,000 bird species (generalist approach). We used audio recordings of mountain chickadees (Poecile gambeli) collected from ARUs and directional microphones in the field as our test stimulus set, with our custom detector trained to identify mountain chickadee songs. Using confidence thresholds of 0.6 for both detectors, we found that our custom CNN detector yielded higher detection compared to BirdNET. Given both ML approaches are significantly faster than a human detector and the custom CNN detector is highly accurate, we hope that our findings encourage bioacoustics practitioners to develop custom solutions for targeted species identification, especially given the availability of open-source toolboxes such as Koogu.
Introduction
Autonomous recording units (ARUs) are becoming an increasingly popular method for capturing the vocalizations of many species in the context of biological monitoring. This method is known as passive acoustic monitoring (PAM) and has resulted in the possibility for rapid culmination of large acoustic datasets due to its high accessibility and long-term applicability (Sugai et al., 2019; Ross et al., 2023). PAM further facilitates the monitoring of target species at times when humans cannot be physically present to manually record vocalizations (Ross et al., 2023). ARUs can be flexibly programmed to fit the needs of researchers, with a variety of settings that can be adjusted according to the study’s requirements (Sugai et al., 2019). Due to the large amount of data accumulated using ARUs, researchers are moving away from humans as detectors and the need for machine learning (ML) algorithms is growing. Focal recordings taken with hand-held microphones are also a common method of recording vocalizations for biological research and monitoring. These recordings, though typically shorter and more manageable, can also be lengthy and require a great deal of time to parse through. Convolution neural networks (CNN) dominate automated assessment of acoustic datasets, increasing the efficiency of biological monitoring. A variety of ML algorithms, including deep learning, have been developed and show great potential to deal with large datasets (Stowell, 2022; Xie et al., 2023).
A few studies have additionally compared the performance of a variety of machine learning detectors on a known dataset of vocalizations identified by a human observer. For instance, a study by Manzano-Rubio et al. (2022) highlighted the benefit of using readily-available and highly accessible detectors by evaluating the performances of two machine-learning detectors: BirdNET and Kaleidoscope. A study by Knight et al. (2017) compared the performance of 5 different detectors: their own CNN detector, SongScope, MonitoR, RavenPro, and Kaleidoscope. Knight et al. (2017) found that all detectors had a high rate of precision depending on an optimal threshold/confidence factor set for each detector. The detectors varied in their precision/recall abilities, with the custom-made CNN performing higher than the generalizable detectors.
To compare the performance of different machine-learning detectors, there must be a “known” test set that includes the “true” detections in the recordings. This has been seen as a limitation when using ML detectors, as “known” datasets are difficult to obtain given they require that observations (humans or ML) be 100% reliable. This can especially be difficult when using humans to determine the true dataset, as there may be significant variation among observers (Sirovic, 2016; Leroy et al., 2018). However, human detections by an expert are commonly used as the “true” or “known” dataset to which machine-learning performance is compared (Knight et al., 2017).
One solution to the limitation of acquiring a “true” dataset is to estimate the probability of detection of an observer by comparing the performance of multiple observers. One of these methods is the Huggins closed population approach (Huggins, 1989, 1991), which has been successfully used to compare human and ML detectors identifying blue whale “D calls” in Miller et al. (2023). The Huggins closed population approach, initially developed for capture/recapture biological studies, estimates the probability of detection in a “recapture” event from a known dataset (the initial “capture” event) (Huggins, 1989; 1991). The human is considered the “capture” event while the other detectors are considered “recapture” events to be compared against the human detections. This approach allows for evaluation of human performance as the machine-learning detectors may identify vocalizations that the human missed. A human “expert” must then adjudicate the detections that the machine-learning detector found, but that the human missed, to identify if these detections are true-positives or false-positives (Miller et al., 2023). Here, we used the Huggins closed population model to evaluate the performance of two ML detectors to a human detector using audio recordings containing vocalizations from a common North American passerine, the mountain chickadee (Poecile gambeli).
Mountain chickadees are nonmigratory songbirds inhabiting the montane regions of western North America (McCallum et al., 1999). Like many temperate oscine passerines, the song of mountain chickadees is typically sung by males during the breeding season and serves a dual function; to defend his territory from conspecific males and to attract females for mating (Krebs et al., 1978; Searcy, 1984; Otter et al., 1997; Christie et al., 2004). Mountain chickadees are a closed-learning species, meaning that they learn their song at their natal site during a sensitive period of development and do not produce new songs once they have crystalized their repertoire (Gammon, 2007). The song of mountain chickadees typically consists of four tonal or whistled notes with a drop in frequency between the second and third note (frequency ratio) (Wiebe and Lein, 1999; Branch and Pravosudov, 2015, 2020) (Figure 1). Despite the comparatively simplistic structure of chickadee song, previous work in this system has documented significant differences in song structure among males inhabiting high versus low elevations (Branch and Pravosudov, 2015, 2020), including variation in duration, frequency bandwidth, and time-frequency structures.
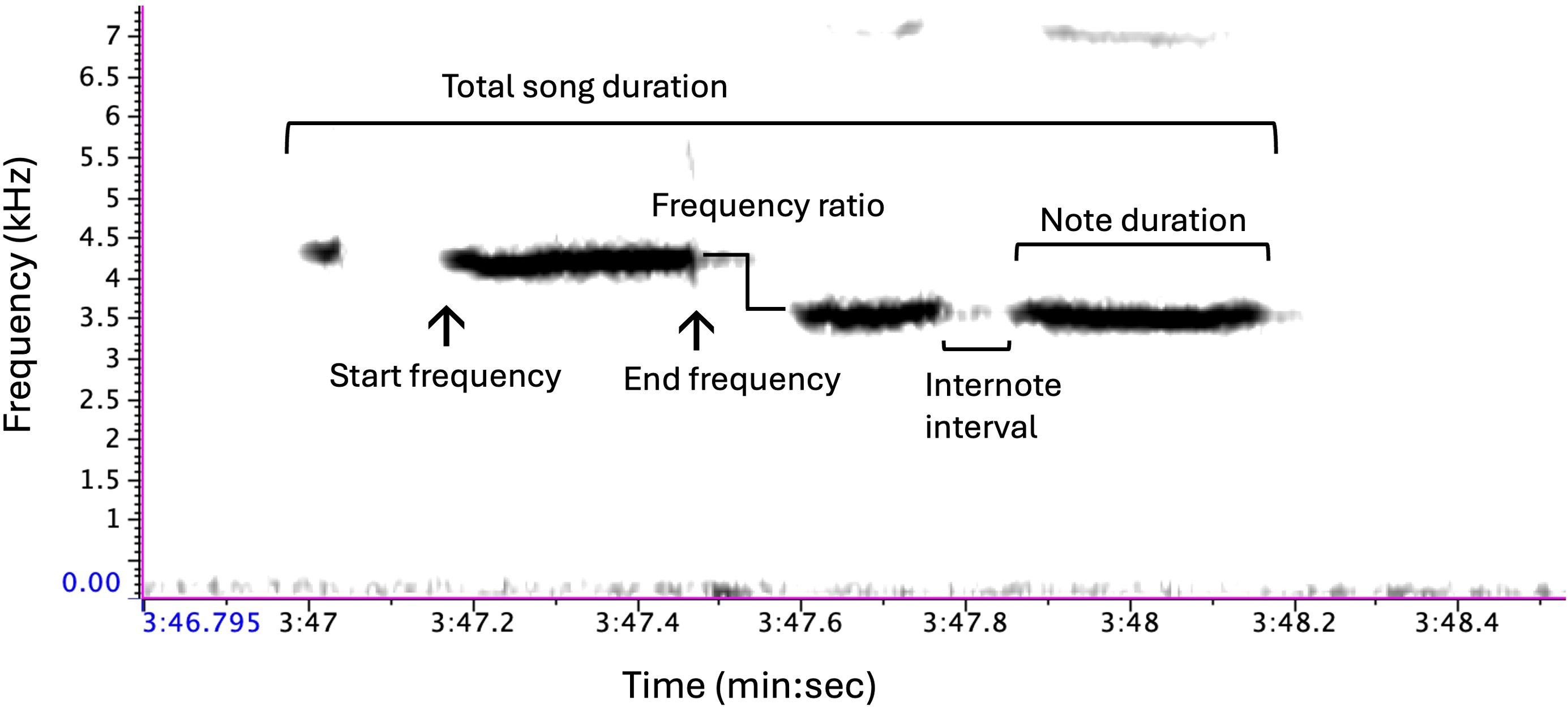
Figure 1. Schematic of typical mountain chickadee song consisting of 4 notes. The sound spectrogram (transform length of 512 points, time resolution of 11.6 ms, and frequency resolution of 86.1 Hz) represents a visualization of an acoustic signal with time (s) on the x-axis, frequency (kHz) on the y-axis, and amplitude represented by presence of black shading. Annotations have been added to the figure to highlight variation measured in each song. Created in Raven Pro 1.6.
In the current study, we used audio recordings collected over several years from ARUs and handheld directional microphones targeting mountain chickadee (Poecile gambeli) song to evaluate the performance of two ML detectors. Most recent studies on the performance of deep-learning detectors have assessed performance on PAM recordings, as this is often the easiest method of collecting large amounts of data for biological monitoring. However, focal recordings using directional microphones are another popular method used in biological monitoring and research. Thus, it is valuable to compare the performance of the CNN on directional focal recordings and PAM soundscape recordings to fully assess the applicability of the CNN in the most common recording settings employed by biologists. A study by Kahl et al. (2021) found that BirdNET performed worse with soundscape recordings compared to focal recordings. Recordings from ARUs typically have lower resolutions and signal-to-noise ratios due to their non-directionality and distance from the vocalizer compared to focal recordings taken by humans. We seek to examine the findings of Kahl et al. (2021) in our own study system and add to the limited literature comparing the performance of CNNs on focal recordings compared to recordings from ARUs.
We compared the performance of two ML detectors’ performance to an expert human detector (CLB), using 1. a custom convolutional neural network (CNN) model trained using Koogu (Madhusudhana, 2023) and 2. BirdNET, an off-the-shelf generalized solution that recognizes the vocalizations of over 6000 bird species (Kahl et al., 2021). While both types of recorder stimuli will have other vocalizations present, we aimed to specifically assess the presence of mountain chickadee song and not the other vocalizations that chickadees produce (e.g. chick-a-dee or gargle calls). We predict that since the custom CNN was trained using a set of audio selections from ARUs targeting known mountain chickadee nests, that the custom CNN will outperform the generalized BirdNET detector, even when specifying mountain chickadee as the target species.
Methods
Data collection
To train and test the two song detection approaches (custom CNN and BirdNET) for male mountain chickadee songs, we used acoustic recordings from several breeding seasons (2017, 2019, 2020) at our long-term field site in northern California, Sagehen Experimental Forest, USA (Sagehen Creek Field Station, University of California Berkeley, approximately 14.5 km north of Truckee, CA). Breeding behavior and male song have been monitored and recorded annually from this population since 2013 (Branch and Pravosudov, 2015, 2020; Kozlovsky et al., 2018; Branch et al., 2019). Birds were recorded at their nests from May to July of each year using one of two approaches: 1. Swift terrestrial ARUs developed by the Cornell Lab of Ornithology’s K. Lisa Yang Center for Conservation Bioacoustics. The Swift recording device is a single unit housing a microphone and recorder. Acoustic recordings were collected at a sampling rate of 48,000 Hz and a 16-bit resolution. Or 2. A Marantz PMD661 Compact Flash Card digital recorder and Sennheiser ME – 66 unidirectional microphone with a sampling rate of 44,000 Hz and 16-bit resolution. For directional recordings, males were located auditorily and approached with microphone and recorder in hand. Each male was recorded on one day.
Chickadees are cavity-nesting birds that readily nest in human made nest boxes. At our field site, there are over 350 nest boxes across our two elevation sites, which results in approximately 100 nests per year. Swift ARUs were placed ca. 2–5 meters from active chickadee nest boxes during the nest-building and egg-laying stages of breeding. ARUs recorded from 0500 to 2000 h PST for three days at each nest box. Audio recordings from ARUs and handheld recorders were stored as. wav files on secure digital (SD) cards and uploaded daily for permanent storage.
Training the custom CNN
The dataset used to train the custom CNN model was created using audio from ARUs only. The training set consisted of 33 40-min recordings (consisting of at least 31 individual males based on nest box location) across varying times of day, from May to June 2019. From those 22 hours of ARU collected audio recordings, 246 annotated songs were pulled as positive examples of mountain chickadee song. Additional annotations representing segments of recordings that did not contain any mountain chickadee songs yielded an additional 279 annotations. CLB and an additional human observer created the training annotations by creating selections in Raven Pro 1.6 (Cornell Lab of Ornithology); mountain chickadee songs were annotated as “pos” and short sections, approximately 2 s (comparable length to mountain chickadee song and “pos” annotated selections), of the recording with no mountain chickadee song present were annotated as “neg”.
Testing and comparing detectors
Test stimuli
To test and compare the performance of the two detectors (custom CNN model and BirdNET), we used additional, nonoverlapping recordings from the same field site. For testing, we incorporated recordings from two recording devices that are traditionally used in wildlife bioacoustics, the Swift terrestrial ARUs and targeted recordings from a handheld directional microphone and digital recorder. We evaluated the performance of the detectors on recordings from ARUs compared to those from directional microphones, because they are the most common recording methods for bioacoustics data collection and can differ in audio quality. ARUs allow for constant monitoring over a long timeframe, but lack the directionality of focal recordings as they are not manned by a mobile human. Thus, to fully evaluate the capabilities of the two detectors, we chose to include recordings of both types. We used the same set of stimuli to test both detectors, which included 12 directional audio recordings (7 from 2017 and 5 from 2020) from 12 individual male mountain chickadees (identified by unique color band combinations). The directional audio recordings ranged from 0:54 seconds to 09:04 minutes. Testing stimuli also included six 40-min recordings from Swift ARUs placed by the nest boxes of six different individuals from the 2020 breeding season. For all detector performances (custom CNN model and BirdNET), we only assessed classifications of mountain chickadee songs, not calls. Each file potentially contained additional vocalizations from chickadees or other species at the field site. However, since only one type of vocalization was annotated (mountain chickadee song), the custom CNN was trained to only recognize mountain chickadee songs, and only selections for mountain chickadee songs were analyzed from the BirdNET output (BirdNET will select all vocal types of the focal species, Kahl et al., 2021).
Custom CNN detector
For data pre-processing, model construction, training and subsequent inferencing using the trained model we used Koogu (v0.7.2; Madhusudhana, 2023), an open-source framework for machine learning in bioacoustics. The underlying computing platform comprised TensorFlow (v2.13; Abadi et al., 2016) running on Python 3.10 (Python Foundation), on a HP Z-book laptop having an Intel i9-11950H and an NVIDIA RTX A4000 GPU.
Audio recordings were resampled to a sampling frequency of 16 kHz and band-pass filtered to suppress energies outside of the range 2968–5188 Hz. Then, the recordings were split into 1.8 s long segments with an overlap of 1.3 s between consecutive segments, and the waveform amplitudes were normalized to occur in the range [−1.0, 1.0]. Spectrograms of each segment were computed using a 32 ms Hann window with 50% overlap between frames, resulting in time and frequency resolutions of 16 ms and 31.25 Hz, respectively. The spectrograms were clipped along the frequency axis to only retain portions between 2968 Hz and 5188 Hz, resulting in model inputs having dimensions of 72 × 111 (height × width). The clip length and bandwidth (for filtering and clipping) values were chosen based on a statistical assessment of the training set annotations’ durations (1.27 s ± 0.17) and frequency bounds (lower: 3295.43 Hz ± 138.27; upper: 4601.21 Hz ± 172.45), respectively. The chosen values ensured that the songs were well-contained (plus a little cushioning on all sides) within what formed independent inputs to the model. Corresponding to some of the longer-duration annotations, the chosen segment length and overlap settings resulted in more than one spectrogram per annotation. Overall, the input preparation step generated 287 spectrograms containing the target songs (or parts thereof) and 427 spectrograms without the target songs.
We chose a quasi-DenseNet (Madhusudhana et al., 2021) architecture considering its computational efficiency and ability to train well with few samples. The model consisted of a 12-filter 3×3 pre-convolution layer followed by four quasi-dense blocks having 2, 2, 4 and 2 layers per block with a growth rate of 8. The final quasi-dense block was connected to a 32-node fully connected layer which was followed by a 2-node fully connected layer with sigmoid activation. To improve model generalization, dropout (Srivastava et al., 2014), with a rate of 0.05, were used. The model was trained over 80 epochs using the Adam optimizer (Kinga and Adam, 2015) with a mini-batch size of 128. Training considered 90% of the training samples while the remaining 10% were used for evaluating the model through the training process. Training losses were weighted appropriately to address class imbalance in the training inputs. The learning rate was set to an initial value of 0.01 and was successively reduced by a factor of 10 at epochs 30, 50 and 70.
BirdNET
BirdNET (https://github.com/kahst/BirdNET-Analyzer) is an existing audio analyzer that uses machine learning to process and classify avian vocalizations of over 6,000 different species worldwide (Kahl et al., 2021). In addition, the BirdNET detector and species list can be accessed using a personal computer, so that large .wav audio files can be processed. BirdNET detections are reported as .txt selection tables, openable in Raven Pro 1.6. The BirdNET-Analyzer has been trained to detect and identify the full range of mountain chickadee vocalizations.
Testing with custom CNN detector
Folders of .wav files with annotations, stored as selection tables, for each detector (custom CNN, BirdNET) were created in RavenPro 1.6. Audio files in the test dataset were subjected to the same pre-processing and preparation scheme as the training set. Positive class test inputs were determined using the same rules as the positive class training inputs. Having no explicitly annotated noise sections, those inputs that did not have any temporal overlap with any mountain chickadee song annotations were considered as negative class test samples. Contrary to the training phase, model inputs during testing were generated with a segment overlap of 1.6 s. To avoid reporting multiple detections for an underlying song, contiguous segments with segment-level scores above a detection threshold (0.6) were combined using the inferencing protocol defined in Madhusudhana et al. (2024). Combined detections were written out in Raven Pro.txt selection table format for subsequent analyses.
Testing with BirdNET
BirdNET-Analyzer version 2.4 was run on a 2020 MacBook Pro with a M1 chip and 8 GB memory and accessed via https://github.com/kahst/BirdNET-Analyzer#setup-macos. All .wav test files were included in a single folder on the desktop and commands were run in the terminal. The following commands were used to run detections, –min_conf was set to 0.6 and –slist was used to specify species_list.txt as “Poecile gambeli_Mountain Chickadee.” All other options were left as defaults, see https://github.com/kahst/BirdNET-Analyzer?tab=readme-ov-file#technical-details.
Performance assessment
Preliminary assessment
To determine the confidence thresholds used for the custom CNN detections, we assessed inputs for which the detector produced accurate detections of mountain chickadee song. We used this data to assess the precision and recall performance at different thresholds (0.6 and 0.9) produced by the custom CNN detector (see Supplementary Material). Precision is the proportion of detections that are true positives (mountain chickadee song, in this study) and recall is the proportion of detections detected (or recalled) from the total number of vocalizations (mountain chickadee songs) present (Knight et al., 2017). Precision and recall are of the most widely used metrics for assessing detector performance (e.g., Raghavan et al., 1989; Knight et al., 2017; Miller et al., 2023), Based on these preliminary assessments, a detector threshold of 0.6 was chosen as it yielded an optimal precision versus recall trade-off. This was matched for BirdNET, as we found 0.6 to also be a suitable threshold in this detector and wanted to match performance as closely as possible.
Double-observer assessment
The Huggins closed population approach estimates the probability of detection in a “recapture” event compared to a known initial “capture” event (Huggins, 1989). Therefore, we analyzed the probability that the detectors (recaptures) detected all the songs in the dataset determined by the human observer (initial capture event) using this approach. Capture history was created by making a column for each detection made by each detector (custom CNN, BirdNET, human). An additional column was created identifying the reconciliation for each additional detection made by the machine-learning detectors but not the human (false positive or true positive), adjudicated by the “expert judge” (SMH). Test inputs which the custom CNN detector identified as above the set threshold were considered to contain mountain chickadee songs. The expert (SMH) judged the annotated detections paired with the spectrograms in Raven Pro 1.6 as either “true positives” or “false positives”. True positives were identified as all detections where SMH and automated detector agreed, and false positives were identified as all detections where SMH did not agree with the automated detector. “False negatives” were identified as songs that any of the detectors missed. These reconciliations were annotated in selection tables in Raven Pro 1.6, and the data was transferred to an excel file containing all detections (see Supplementary Material).
We inspected the effect of signal-to-noise ratio (SNR) as a potential covariate in detection rates. This was done by inspecting the spectrograms for each detection. Vocalizations were classified into three SNR categories, depending on the ratio of relative strength of the song signal and background noise, calculated in decibels (dB). Songs with a SNR of >10 dB, thus containing a medium-strength signal with little to no background noise or a strong signal with low- to medium- strength background noise were classified as “high” SNR songs. Songs with a SNR of 3-10 dB and thus containing medium-strength signals with medium background noise or strong signals with heavy background noise were classified as having “medium” SNRs. Songs with an SNR below 3 dB and thus containing a weak signal or a medium signal but heavy background noise, were classified as having “low” SNRs. We also classified signals with an interfering vocalization by another individual or species as having a low SNR due to its high interference with the performance of the detectors. For examples of each SNR level, see Supplementary Material.
The capture history (each true positive detection) for each detector was incorporated to create Huggins closed population model estimates of probability of detection, where “human” was considered the first capture occasion, and custom CNN and BirdNET were considered the second and third capture occasions, or re-captures. Models were created using software package MARK (White and Burnham, 1999) via RMark (Laake, 2013) in R version 4.3.3 (R Core Team, 2023).
Five Huggins models for estimating the probability of detections were considered for custom CNN and BirdNET. The models with the lowest AICc values (Akaike’s Information Criterion; Burnham and Anderson, 2002) were selected as the most supported models. The first model assumed that the detectability varied between the detectors. The second model included SNR as a covariate, and detectability was modeled separately for each combination of detectors and SNR levels. The third model assumed the detectability of the human analyst and each automated detector to be the same. The fourth model included recording type (Swift ARU or directional recording) as a covariate, and detectability was modeled separately for each combination of detectors and recording types. Finally, the fifth model included both recording type and SNR as covariates, and detectability was modeled separately for each combination of detectors, SNR levels, and recording types. R code for producing the Huggins double-observer models can be accessed in Supplementary Materials.
Results
We investigated the relative detection performances of the two automated detectors: custom CNN (threshold 0.6) and BirdNET (threshold 0.6). We also assessed the performance of a human analyst to consider any detections that the machine-learning detectors made but that the human missed. The human analyst identified 509 total songs out of the 520 known songs across all the test audio recordings, resulting in a detection rate of 97.9%. The custom CNN reported 489 detections and BirdNET reported 313 detections. According to the results of the Huggins closed population model, where detectability varied between the detectors, the custom CNN identified 93.6% of the total number of songs (487/520), with two false-positive detections. BirdNET detected 60% of the songs (313/520). Thus, the custom CNN detector detected more songs compared to the BirdNET detector.
The best Huggins model for the probability of detection (i.e. had the lowest AICc value) was the model that included SNR and Recording type as covariates (see Table 1), indicating that SNR and recording type together affected the relative performance of the detectors. See Table 2 for a summary of each comparison.
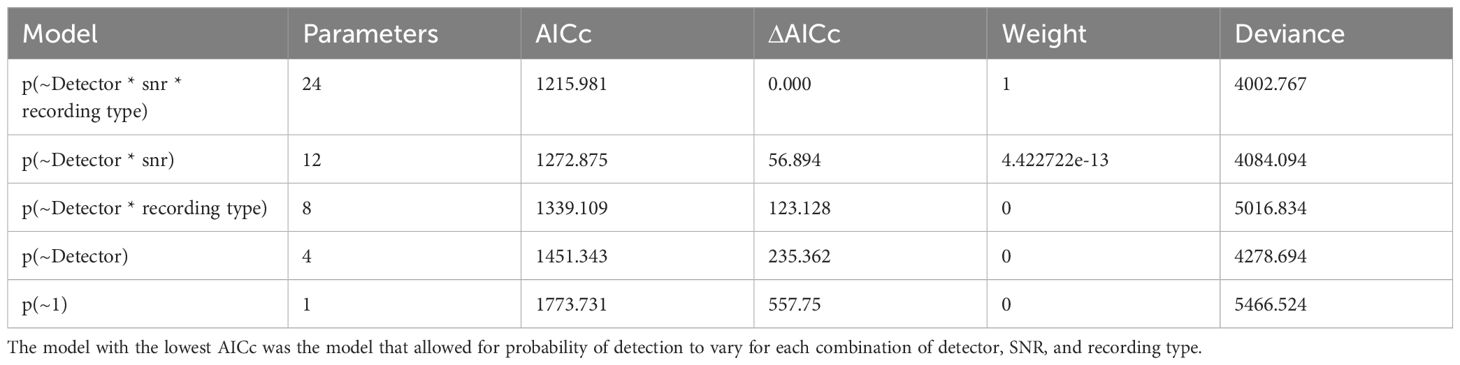
Table 1. AICc and deviance for the five different Huggins double-observer models of probability of detection.
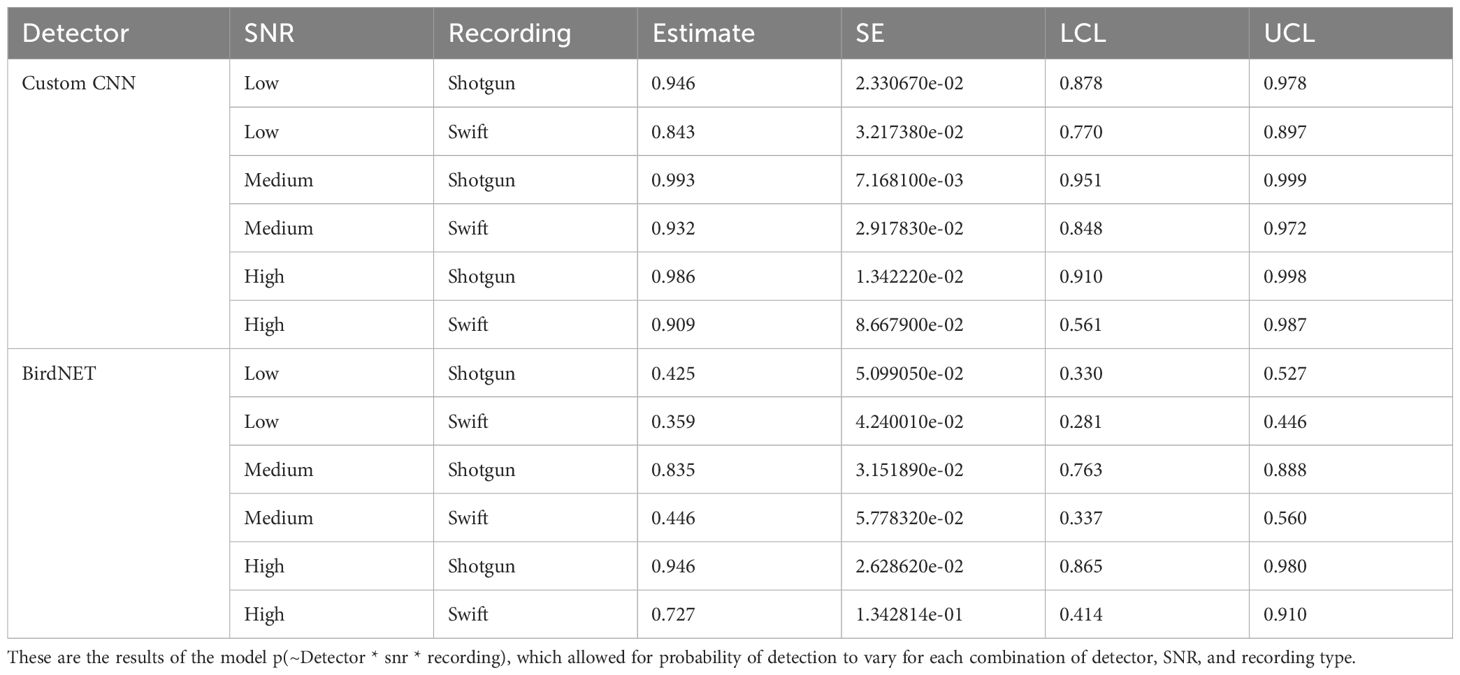
Table 2. Estimates of probability of detection estimates with standard error (SE), and lower (LCL) and upper (UCL) 95% confidence limits for the Huggins closed population mark-recature model for the custom CNN and BirdNET, signal to noise ratio, and recording type.
According to the Huggins model where each detector and SNR levels were considered separately, the custom CNN detector detected the least number of songs in the low SNR category, with an 88.7% rate of detectability compared to 97.2% and 97.6% for medium and high SNR, respectively (see Table 3). BirdNET also performed worst in the low SNR category, with 38.7% detectability of low SNR songs, 70% detectability of medium SNR songs, and 91.7% detectability of high SNR songs. See Figure 2.
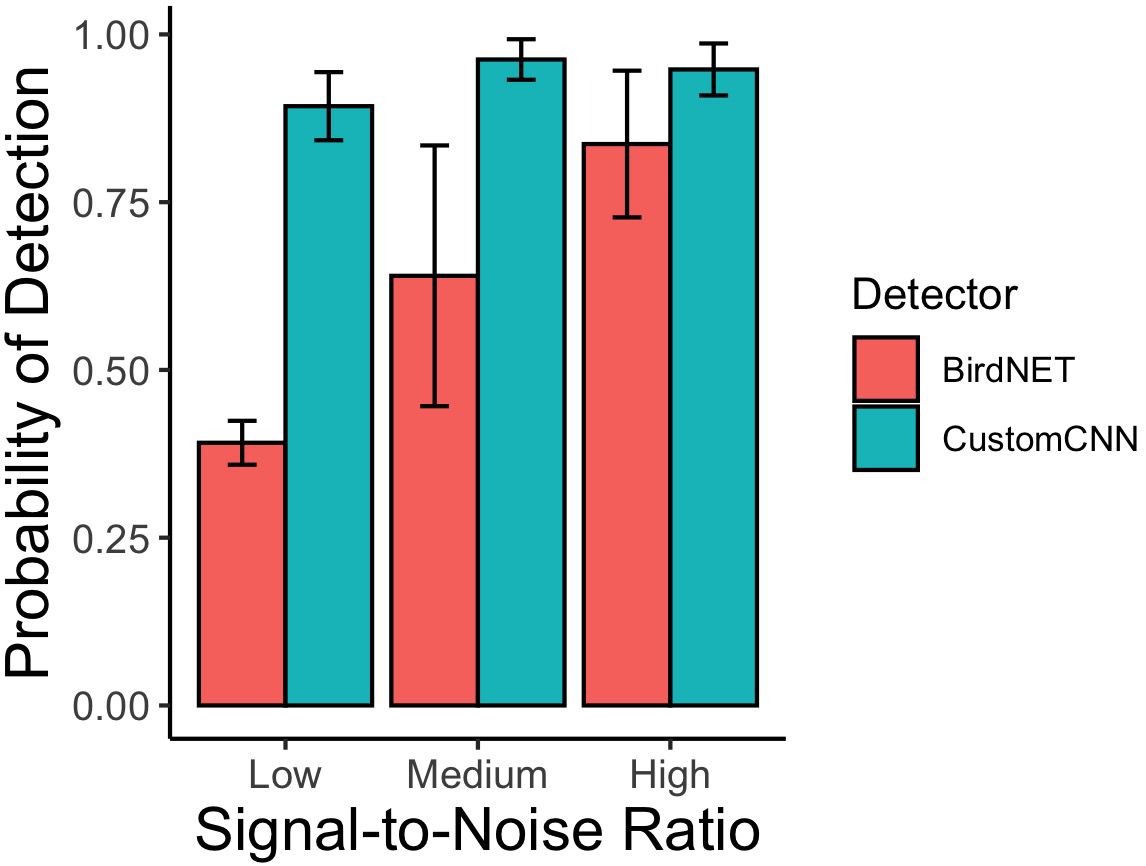
Figure 2. Detection probability of mountain chickadee songs as a function of signal-to-noise ratio (SNR) from the adjudicated detections of the custom CNN detector and BirdNET. Bars and whiskers represent the mean and standard error calculated across recording type (ARU and directional microphone) performances (package ggplot2, stat = “summary” in R Core Team).
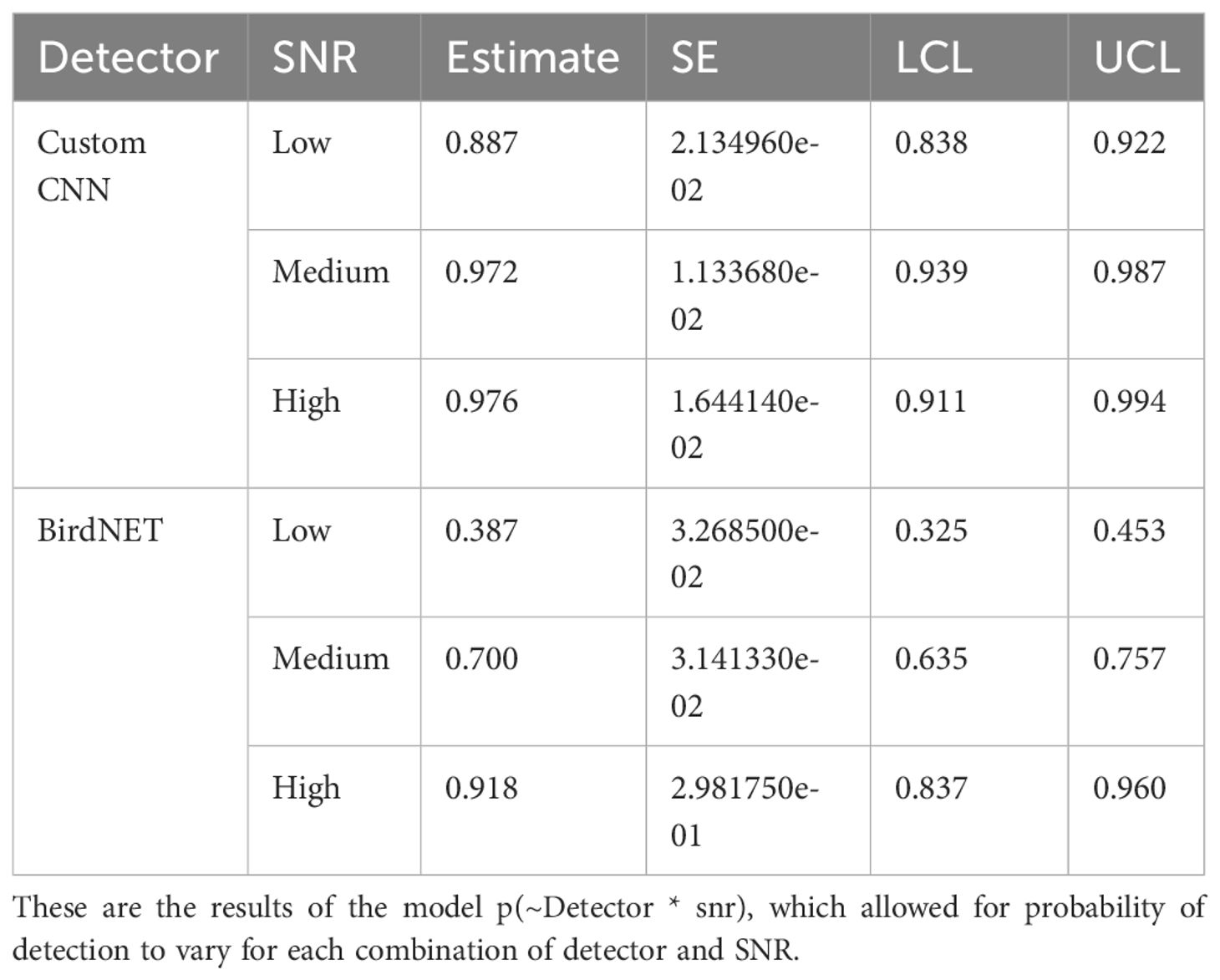
Table 3. Estimates of probability of detection estimates with standard error (SE), and lower (LCL) and upper (UCL) 95% confidence limits for the Huggins closed population mark-recapture model for the custom CNN, BirdNET, and signal to noise ratio.
Lastly, we found that the custom CNN performed worse with the Swift ARU recordings (87.8% detectability) compared to the directional microphone recordings (97.7% detectability) (see Table 4). Similarly, BirdNET performed worse with the Swift ARU recordings (40.8% detectability) compared to the directional microphone recordings (73.6% detectability). See Figure 3.
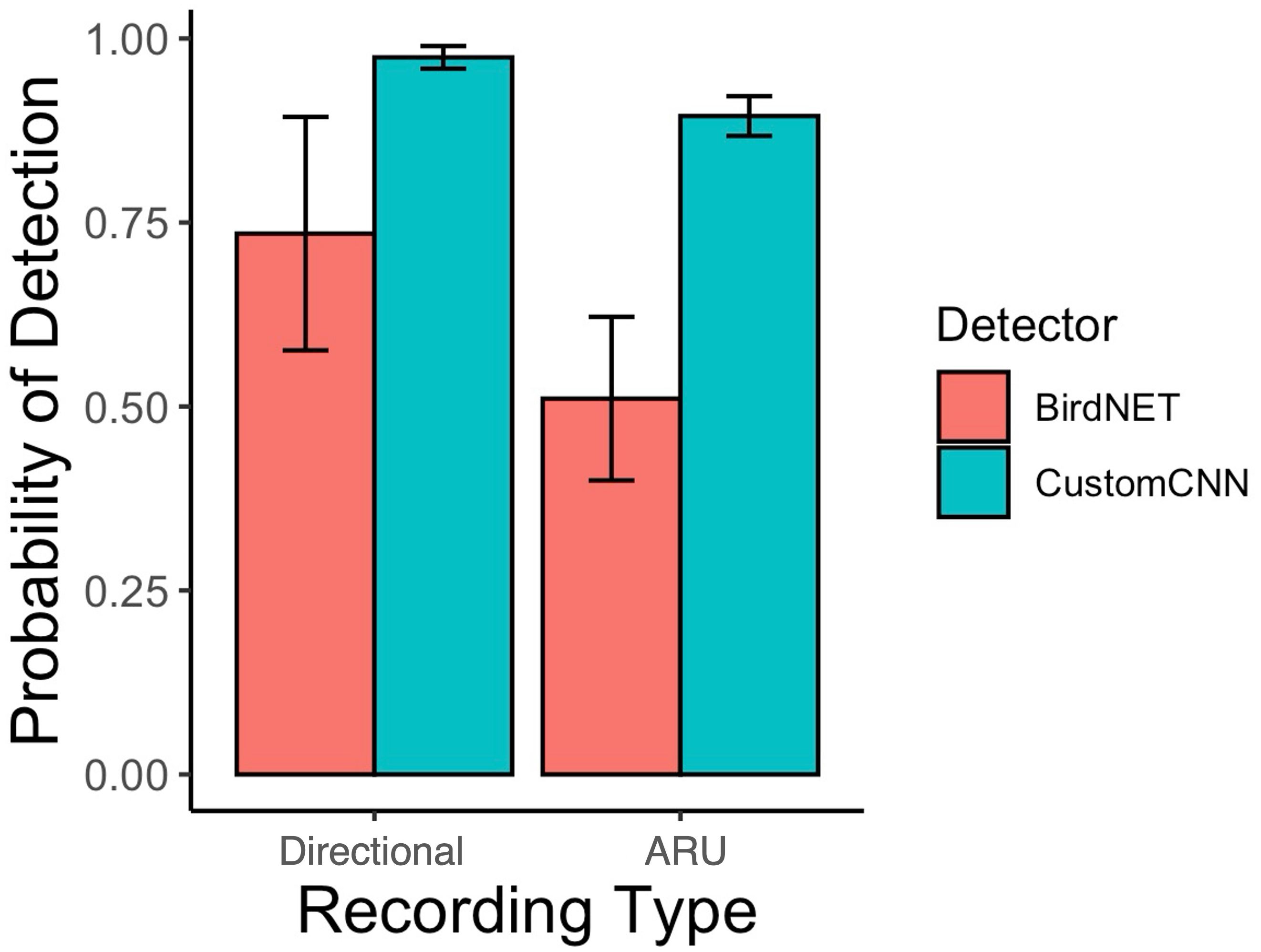
Figure 3. Detection probability of mountain chickadee songs as a function of recording type from the adjudicated detections of the custom CNN detector, and BirdNET. Bars and whiskers represent the mean and standard error calculated across all signal-to-noise ratio performances (package ggplot 2, stat = “summary” in R Core Team).
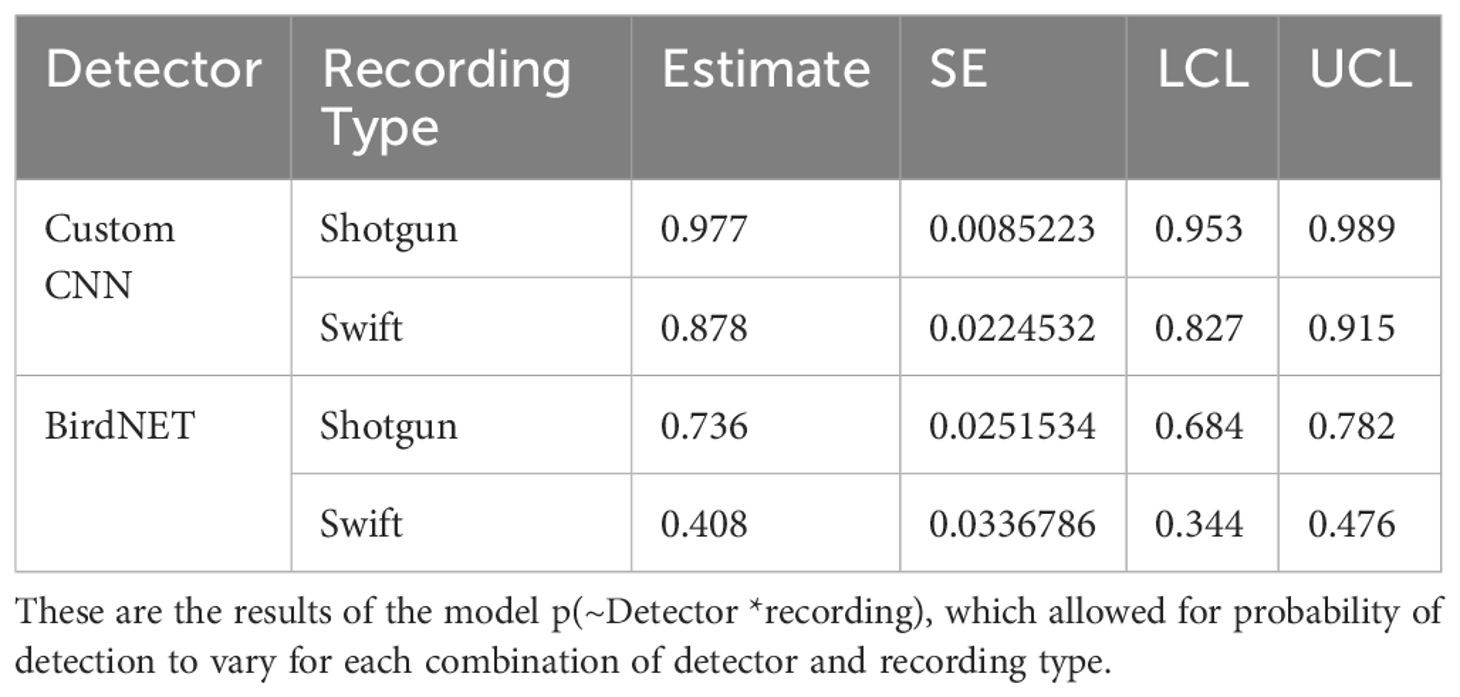
Table 4. Estimates of probability of detection estimates with standard error (SE), and lower (LCL) and upper (UCL) 95% confidence limits for the Huggins closed population capture-recapture model for the custom CNN and BirdNET, and recording type.
Discussion
Previous studies have found that custom deep-learning algorithms are able to detect focal vocalizations at a very high rate of precision (e.g., Knight et al., 2017; Miller et al., 2023). Our own custom CNN detector performed at a very high-level with 93.6% recall, indicating its proficiency in identifying the mountain chickadee songs from our study population. When using the model that included recording type as a covariate, we found that the custom CNN performed comparatively worse with the Swift ARU recordings compared to the directional recordings, however, still relatively high; 87.8% detectability. This was also a much stronger performance than that of the BirdNET detector which dropped to 40.8% detectability in the same category. However, our initial goal was to create a detector that can parse through lengthy recordings, as they can take hours to process depending on length and number of target vocalizations present. The difference in performance of both BirdNET and our custom CNN for the ARU compared to directional recordings needs to be further explored, as ARU recordings demand the use of machine-learning detectors. Additional studies should focus on comparing the performance of ML detectors using focal versus PAM recordings to establish optimal settings that result in maximized performance for PAM recordings.
Overall, we found that a low signal to noise ratio had a strong negative impact on both ML detectors’ abilities to identify the target vocalizations. This was expected, as the ML detectors rely on SNR to identify vocalizations of interest. Songs with interfering noise and weak signals leading to a low SNR score should indeed be more difficult to detect. Our findings corroborate the findings of Pérez-Granados (2023a), who found that BirdNET’s detection performance generally decreased with distance. Our findings thus further highlight the limitations of using detectors with long-range recordings and researchers should be wary when relying on detectors to identify vocalizations from low SNR/long distance recordings.
Confidence scores can be employed as filters to impact the number of detections by detectors (Pérez-Granados, 2023b). Scores closer to 1 increase the chances that the detected vocalization will belong to the focal species, while scores closer to 0 increases the chance that the detector will detect all vocalizations, but with reduced accuracy (Pérez-Granados, 2023b). Studies by Katz et al. (2016) and Knight et al. (2017) found that performance varies widely with score threshold and should be used as a tool to select an optimal threshold depending on detector performance evaluation. For our study, we employed confidence scores of 0.6 for both BirdNET and our custom CNN, because a confidence score of 0.6 was determined to be the optimum precision/recall for our detectors, with a high accuracy of detection (nearly 100% accuracy). This means that the detectors only identified songs with a confidence threshold of 0.6 and failed to identify songs below that confidence threshold. Our set confidence score impacted the failure of both detectors to detect songs with low SNR as these songs did not reach the confidence criterion set. This is an important trade-off to consider as our threshold resulted in highly accurate song detections (precision), but reduced our chances of identifying songs with low signal power (recall). As discussed in Bota et al. (2023), the confidence score employed depends on the goals of the researchers. If the goal is to detect all instances of song present, then a lower confidence score might be the best choice (Bota et al., 2023). However, if the goal is to detect songs accurately and eliminate noise and false positives, then a higher confidence score may be the best choice (Bota et al., 2023). We chose an intermediate confidence score of 0.6 based on our analysis of the optimal precision/recall of the custom CNN detector (see Supplementary Material). We would like to highlight the importance of considering the impact of setting confidence scores when fine-tuning detectors to fit the needs of the relevant study.
We found that our custom deep-learning CNN outperforms the generalist detector, BirdNET, for our dataset. This finding supports the findings of Knight et al. (2017) who found that their custom CNN outperformed more generalizable detectors such as RavenPro and Kaleidoscope. This could be due to a number of reasons. One major explanation for why the CNN performed better than BirdNET is that the custom CNN was trained with recordings from the same study system as our test set. Mountain chickadee song varies across space (Branch and Pravosudov, 2015, 2020) and our custom CNN was trained to specifically detect the songs of our population. Thus, the custom CNN was tailored to the specific population, and song patterns therein, of this study. It is therefore likely that our custom CNN may perform worse when applied to other populations, since song varies geographically (Branch and Pravosudov, 2015). We suggest that Koogu CNN models should be trained specifically with the vocalizations of the target population to maximize performance. This was not done with BirdNET, as we aimed to test an “off the shelf” generalized machine learning detector compared to a specifically-tailored detector. It is possible that BirdNET would outperform our custom CNN in detecting the songs of other populations. Finally, our custom CNN was not only trained using our study populations’ songs, but was also trained to exclude the background noise specific to our study area. A study by Ventura et al. (2024) found that focusing on background modelling resulted in improved performance of their custom CNN. Thus, future studies should consider background noise as a potential covariate for training their custom CNNs.
When developing custom solutions such as the one we have developed in this study, a concern that bioacoustics practitioners must often contend with is the level of expertise and effort required. While the ready availability of open-source bioacoustics ML toolboxes such as Koogu, OpenSoundscape (Lapp et al., 2023), and Ketos (Kirsebom et al., 2021) help towards addressing some of the concerns, the low-code nature of Koogu presents a gentle learning curve for non-expert programmers. Given its abstraction of data (audio and annotations) processing, model-building, and training processes as parametric functional interfaces, Koogu facilitates rapid development and testing, allowing users to quickly experiment with different parameters. Consequently, toolboxes like Koogu are already widely used for developing bioacoustics solutions (e.g., Miller et al., 2023; Suresh et al., 2023; Madhusudhana et al., 2024; Owens et al., 2024).
In conclusion, we supported our predictions and found evidence that our custom deep-learning CNN model performs at a high rate of detectability, outperforming an existing, generalist model, BirdNET. This indicates that the Koogu deep-learning algorithm can be trained to specialize in specific vocalizations of a specific population, yielding higher detectability, in contrast to BirdNET, which has been trained to detect the vocalizations of over 6000 species (Kahl et al., 2021). We recognize that BirdNET can be tailored to better fit certain species and populations if needed, but that requires expertise and time that reduces the value of using a readily available, generalist detector. We suggest that once a researcher requires this specificity, it will be more effective to train a CNN model with the population-specific vocalizations for the study system. Although BirdNET is an enormously powerful tool for identifying a wide range of avian vocalizations, the use of custom CNN deep-learning algorithms allows for specific identification of a species or population, which is often of great interest to researchers studying individual variation in animal vocalizations. Post-processing, including cutoffs for amplitude ratios, can further the utility of custom CNNs, such that individual animals can be detected with high accuracy and efficiency. The deep-learning Python program, Koogu, can be applied to other species to allow for the detection of vocalizations relevant to specific research questions and focal study systems, such as the work on blue whale D calls (Miller et al., 2023). Advancements of this kind allow researchers to ask questions we previously could not answer, enhancing the power of bioacoustics monitoring and moving the field forward.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
To the best of our knowledge, no birds were harmed by the collection of this data. All applicable national and institutional guidelines for the use of animals were followed. All procedures were approved by the UNR IACUC ethics committee in accordance with the UNR IACUC protocol (00046), under California Department of Fish and Wildlife Permit SC-5210 (DocID: D-0019571790-9).
Author contributions
SH: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. SM: Conceptualization, Data curation, Methodology, Software, Validation, Writing – review & editing. CB: Conceptualization, Methodology, Supervision, Writing – review & editing, Funding acquisition, Visualization.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Thank you to Ginny Halterman for providing annotations used to train the custom mountain chickadee song detector. Thank you to Brian S. Miller and Nat Kelly for their advice and R code. Thank you to two reviewers whose comments and suggestions greatly improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbirs.2024.1425463/full#supplementary-material
References
Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., et al. (2016). Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint 1603.04467. doi: 10.48550/arXiv.1603.04467
Bota G., Manzano-Rubio R., Catalán L., Gómez-Catasús J., Pérez-Granados C. (2023). Hearing to the unseen: AudioMoth and BirdNET as a cheap and easy method for monitoring cryptic bird species. Sensors 23, 7176. doi: 10.3390/s23167176
Branch C. L., Pitera A. M., Kozlovsky D. Y., Bridge E. S., Pravosudov V. V. (2019). Smart is the new sexy: female mountain chickadees increase reproductive investment when mated to males with better spatial cognition. Ecol. Lett. 22, 897–903. doi: 10.1111/ele.13249
Branch C. L., Pravosudov V. V. (2015). Mountain chickadees from different elevations sing different songs: acoustic adaptation, temporal drift or signal of local adaptation? R Soc. Open Sci. 2, 150019. doi: 10.1098/rsos.150019
Branch C. L., Pravosudov V. V. (2020). Variation in song structure along an elevation gradient in a resident songbird. Behav. Ecol. Sociobiol. 74, 9. doi: 10.1007/s00265-019-2786-5
Burnham K. P., Anderson D. R. (2002). Model selection and multimodel inference: a practical information-theoretic approach. 2nd edition (New York: Springer-Verlag).
Christie P. J., Mennill D. J., Ratcliffe L. M. (2004). Pitch shifts and song structure indicate male quality in the dawn chorus of black-capped chickadees. Behav. Ecol. Sociobiol. 55, 341–348. doi: 10.1007/s00265-003-0711-3
Gammon D. E. (2007). “How postdispersal social environment may influence acoustic variation in birdsong,” in Ecology and behavior of chickadees and titmice an integrated approach. Ed. Otter K. (Oxford University Press, Oxford UK), 183–197.
Huggins R. M. (1989). On the statistical analysis of capture experiments. Biometrika 76, 133–140. doi: 10.1093/biomet/76.1.133
Huggins R. M. (1991). Some practical aspects of a conditional likelihood approach to capture experiments. Biometrics 47, 725. doi: 10.2307/2532158
Kahl S., Wood C. M., Eibl M., Klinck H. (2021). BirdNET: A deep learning solution for avian diversity monitoring. Ecol. Informat. 61, 101236. doi: 10.1016/j.ecoinf.2021.101236
Katz J., Hafner S. D., Donovan. T. (2016). Assessment of error rates in acoustic monitoring with the R package monitoR. Bioacoustics 25, 177–196. doi: 10.1080/09524622.2015.1133320
Kinga D. P., Ba J. L. (2015). Adam: a method for stochastic optimization. In International conference on learning representations (ICLR). 5, p. 6.
Kirsebom O. S., Frazao F., Padovese B., Sakib S., Matwin S. (2021). Ketos—A deep learning package for creating acoustic detectors and classifiers. J. Acoustical Soc. America 150, A164–A164. doi: 10.1121/10.0007998
Knight E. C., Hannah K. C., Foley G. J., Scott C. D., Brigham R. M., Bayne E. (2017). Recommendations for acoustic recognizer performance assessment with application to five common automated signal recognition programs. Avian Conserv. Ecol. 12, 14. doi: 10.5751/ACE-01114-120214
Kozlovsky D. Y., Branch C. L., Pitera A. M., Pravosudov V. V. (2018). Fluctuations in annual climatic extremes are associated with reproductive variation in resident mountain chickadees. R Soc. Open Sci. 5, 171604. doi: 10.1098/rsos.171604
Krebs J. R., Ashcroft R., Webber. M. I. (1978). Song repertoires and territory defense. Nature 271, 539–542. doi: 10.1038/271539a0
Laake J. L. (2013). RMark: An R Interface for Analysis of Capture-Recapture Data with MARK. AFSC Processed Rep. 2013-01. Seattle WA 98115: Alaska Fish. Sci. Cent., NOAA, Natl. Mar. Fish. Serv., 25.
Lapp S., Rhinehart T., Freeland-Haynes L., Khilnani J., Syunkova A., Kitzes J. (2023). OpenSoundscape: An open-source bioacoustics analysis package for Python. Methods Ecol. Evol. 14, 2321–2328. doi: 10.1111/2041-210X.14196
Leroy E. C., Thomisch K., Royer J., Boebel O., Van Opzeeland I. (2018). On the reliability of acoustic annotations and automatic detections of Antarctic blue whale calls under different acoustic conditions. J. Acoustical Soc. America. 144, 740–754. doi: 10.1121/1.5049803
Madhusudhana S., Klinck H., Symes L. B. (2024). Extensive data engineering to the rescue: building a multi-species katydid detector from unbalanced, atypical training datasets. Phil. Trans. R. Soc B 379, 20230444. doi: 10.1098/rstb.2023.0444
Madhusudhana S., Shiu Y., Klinck H., Fleishman E., Liu X., Nosal E. M., et al. (2021). Improve automatic detection of animal call sequences with temporal context. J. R. Soc. Interface 18, 20210297. doi: 10.1098/rsif.2021.0297
Manzano-Rubio R., Bota G., Brotons L., Soto-Largo E., Pérez-Granados C. (2022). Low-cost open-source recorders and ready-to-use machine learning approaches provide effective monitoring of threatened species. Ecol. Inf. 72, 101910. doi: 10.1016/j.ecoinf.2022.101910
McCallum D. A., Grundel R., Dahlsten D. L. (1999). “Mountain chickadee (Poecile gambeli),” In Birds of the world. Cornell lab of ornithology. Eds. Poole A. F., Gill F. B., number 453. (Philadelphia, Pennsylvania, USA: Academy of Natural Sciences; Washington, D.C., USA: American Ornithologists' Union).
Miller B. S., Madhusudhana S., Aulich M. G., Kelly N. (2023). Deep learning algorithm outperforms experience human observer at detection of blue whale D-calls: a double observer analysis. Remote Sens. Ecol. Conserv. 9, 104–116. doi: 10.1002/rse2.297
Otter K., Chruszcz B., Ratcliffe L. (1997). Honest advertisement and song output during the dawnchorus of black-capped chickadees. Behav. Ecol. 8, 167–173. doi: 10.1093/beheco/8.2.167
Owens A. F., Hockings K. J., Imron M. A., Madhusudhana S., Niun M. A., Setia T. M., et al. (2024). Automated detection of Bornean white-bearded gibbon (Hylobates albibarbis) vocalisations using an open-source framework for deep learning. bioRxiv, 2024–2004. doi: 10.1101/2024.04.15.589517
Pérez-Granados C. (2023a). BirdNET: applications, performance, pitfalls and future opportunities. Ibis 165, 1068–1075. doi: 10.1111/ibi.13193
Pérez-Granados C. (2023b). A first assessment of BirdNET performance at varying distances: A playback experiment. Ardeola 70, 221–233. doi: 10.13157/arla.70.2.2023.sc1
R Core Team. (2023). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. R version 4.3.3. Available online at: https://www.R-project.org/.
Raghavan V., Bollmann P., Jung. G. S. (1989). A critical investigation of recall and precision as measures of retrieval system performance. ACM Trans. Inf. Syst. 7, 205–229. doi: 10.1145/65943.65945
Ross S. R. J., O’Connell D. P., Deichmann J. L., Desjonquères C., Gasc A., Phillips J. N., et al. (2023). Passive acoustic monitoring provides a fresh perspective on fundamental ecological questions. Funct. Ecol. 37, 959–975. doi: 10.1111/1365-2435.14275
Searcy W. A. (1984). Song repertoire size and female preferences in song sparrows. Behav. Ecol. Sociobiol. 14, 281–286. doi: 10.1007/BF00299499
Sirovic A. (2016). Variability in the performance of the spectrogram correlation detector for north-East Pacific blue whale calls. Bioacoustics 25, 145–160. doi: 10.1080/09524622.2015.1124248
Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. (2014). Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958.
Stowell D. (2022). Computational bioacoustics with deep learning: a review and roadmap. PeerJ 10, e13152. doi: 10.7717/peerj.13152
Sugai L. S. M., Silva T. S. F., Ribeiro J. W. Jr., Llusia D. (2019). Terrestrial passive acoustic monitoring: review and perspectives. BioScience 69, 15–25. doi: 10.1093/biosci/biy147
Suresh N., Thomas B., Wang S., Madhusudhana S., Ramesh V., Klinck H. (2023). Deep learning for large scale conservation bioacoustics—A demonstration on the Malabar whistling thrush and the dhole. J. Acoustical Soc. America 154, A22–A23. doi: 10.1121/10.0022665
Ventura T. M., Ganchev T. D., Pérez-Granados C., de Oliveira A. G., de S. G., Pedroso G., et al. (2024). The importance of acoustic background modelling in CNN-based detection of the neotropical White-lored Spinetail (Aves, Passeriformes, Furnaridae). Bioacoustics 33, 103–121. doi: 10.1080/09524622.2024.2309362
White G. C., Burnham K. P. (1999). Program MARK: survival estimation from populations of marked animals. Bird Stud 46, S120–S139. doi: 10.1080/00063659909477239
Wiebe M. O., Lein M. R. (1999). Use of song types by mountain chickadees (Poecile gambeli). Wilson Bull. 111, 368–375.
Keywords: automated detector, mountain chickadee, machine-learning, passive acoustics, double-observer
Citation: Haley SM, Madhusudhana S and Branch CL (2024) Comparing detection accuracy of mountain chickadee (Poecile gambeli) song by two deep-learning algorithms. Front. Bird Sci. 3:1425463. doi: 10.3389/fbirs.2024.1425463
Received: 29 April 2024; Accepted: 05 August 2024;
Published: 28 August 2024.
Edited by:
Cristian Pérez-Granados, University of Alicante, SpainReviewed by:
Jiangjian Xie, Beijing Forestry University, ChinaGerard Bota Cabau, Forest Technology Center of Catalonia (CTFC), Spain
Copyright © 2024 Haley, Madhusudhana and Branch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofia M. Haley, c29maWFoQHVuci5lZHU=
 Sofia M. Haley
Sofia M. Haley Shyam Madhusudhana
Shyam Madhusudhana Carrie L. Branch
Carrie L. Branch