
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Biomater. Sci. , 03 October 2022
Sec. Delivery Systems and Controlled Release
Volume 1 - 2022 | https://doi.org/10.3389/fbiom.2022.1003172
This article is part of the Research Topic Women in Biomaterials Science 2022 View all 7 articles
Polymeric nanoparticles containing multiple amines and carboxylates have been frequently used in drug delivery research. Reproducible and controlled conjugation among these multifunctional biomaterials is necessary to achieve efficient drug delivery platforms. However, multiple functional groups increase the risk of unintended intramolecular/intermolecular reactions during conjugation. Herein, conjugation approaches and possible undesired reactions between multi-amine functionalized peptides, multi-carboxylate functionalized polymers, and anhydride-containing polymers [Poly(styrene-alt-maleic anhydride)-b-poly(styrene)] were investigated under different conjugation strategies (carbodiimide chemistry, anhydride ring-opening via nucleophilic addition elimination). Muti-amine peptides led to extensive crosslinking between polymers regardless of the conjugation chemistry. Results also indicate that conventional peptide quantification methods (i.e., o-phthalaldehyde assay, bicinchoninic acid assay) are unreliable. Gel permeation chromatography (GPC) provided more accurate qualitative and quantitative evidence for intermolecular crosslinking. Crosslinking densities were correlated with higher feed ratios of multifunctional peptides and carbodiimide coupling reagents. Selectively protected peptides (Lys-Alloc) exhibited no crosslinking and yielded peptide-polymer conjugates with controlled dispersity and molecular weight. Furthermore, anhydride ring-opening (ARO) nucleophilic addition elimination was successfully introduced as a facile yet robust peptide conjugation approach for cyclic anhydride-containing polymers.
Carbodiimide chemistry is commonly used for the bioconjugation of carboxylate/amine functionalized biomaterials (Biju, 2014; Shipunova et al., 2018; Elzahhar et al., 2019; Lim et al., 2019). For example, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide was used to couple a peptide (sequence: SIINFEKL) to poly(maleic anhydride-alt-I-octadecene) nanoparticles (NPs) to protect from CD8 T-cell-driven autoimmune cholangitis (Carambia et al., 2021). In another study, lipid-polymer nanoparticles were functionalized with arginine-glycine-aspartic acid (RGD) and a paclitaxel prodrug using carbodiimide chemistry (Wang et al., 2018). Additionally, the carboxyl groups on poly(lactic-co-glycolic acid/poly(styrene-alt-maleic anhydride) microparticles (PLGA/PSMA MPs) were used to conjugate an MT1-MMP-responsive peptide (sequence: GPLPLRSWGLK) for arterial chemotherapy of hepatic cancer (Davaa et al., 2017). In addition to these examples, numerous examples are found in the literature exploiting carbodiimide coupling chemistry to functionalize multifunctional biomaterials (Pan et al., 2012; Wang et al., 2014; Gandhi et al., 2016; Zhang et al., 2016; Imanparast et al., 2017; Kuo et al., 2017).
Despite their versatility, bioconjugates formed using multifunctional biomaterials (biomaterials containing multiple amine/carboxylate functional groups) must be subject to rigorous characterization, as carbodiimide coupling reagents activate numerous reaction sites. Furthermore, as carbodiimide crosslinkers are added in excess in one pot syntheses, the possibility of off-target reactions [i.e., intermolecular crosslinking (peptide as the crosslinker), intramolecular cyclizations of the peptide/polymer, and peptide conjugation to an off-target active site] is high. In most cases, though, little attention is paid to possible unintended intramolecular/intermolecular reactions because the focus is on subsequent in vitro and in vivo applications (Pan et al., 2012; Wang et al., 2014; Gandhi et al., 2016; Zhang et al., 2016; Davaa et al., 2017; Imanparast et al., 2017; Kuo et al., 2017; Wang et al., 2018; Carambia et al., 2021). For example, in a recent study evaluating the potential of redox-sensitive nanoparticles for lung cancer treatment, 3, 3′-Dithiodipropionic acid (DPA) was used as a linker to conjugate Paclitaxel (PTX) to a peptide that was later encapsulated in lipid-polymer nanoparticles. DPA contains two carboxylate functional groups, while PTX has four hydroxyl functionalities. The targeted reaction, an esterification between a carboxylate on DPA and a hydroxyl on PTX, was performed using carbodiimide chemistry. However, the conjugation was carried out, assuming that only one functional group from each reactant could participate in the reaction. As carbodiimide crosslinkers can activate all carboxylates and none of the hydroxyl groups are protected, numerous unintended products can be formed. Similar assumptions were used when conjugating PTX-DTA to amine and carboxylate-functionalized poly(ethylene glycol) (PEG). Even though this study shows promising results in vitro and in vivo, the reliability and repeatability of the results are questionable, as proper characterizations were not carried out to detect any unintended side products (Wang et al., 2018). Furthermore, conventional peptide/protein quantification assays cannot detect these side reactions but can cause significant ambiguities during in vitro and in vivo studies.
Chemical crosslinking is the most susceptible off-target reaction for the molecules herein as they have multiple functional groups (multifunctional) that can participate in carbodiimide coupling (i.e., those that bear carboxylic acid groups and amines). In nanoparticle-mediated drug delivery, nanoparticle (NP) self-assembly is highly dependent on the structural properties of the polymeric material. The packing parameter (p), defined by Israelachvili et al., combines polymer properties (v—the volume of the hydrophobic chains, a0—the optimal area of the head group, Lc—hydrophobic chain length) to predict the morphological outcome (Eq. 1) (Israelachvili et al., 1977; Blanazs et al., 2009).
Crosslinking between polymer chains can change these parameters significantly (especially v and Lc), causing uncontrolled size and morphological changes to the self-assembled nanoparticles. A nanoparticle drug delivery system with uncontrolled sizes and morphologies could lead to highly variable and poor delivery in vitro and in vivo (Alexis et al., 2008; He et al., 2010; Kaga et al., 2017). Therefore, studying these possible side reactions is crucial to ensure reproducible and controlled biomaterials development.
This work focuses on investigating the common conjugation approaches and possible undesired reactions between 1) multifunctional peptides and polymers and 2) anhydride-containing polymers to identify those that are robust and reproducible. Coupling reactions between PSMA-b-PS [Poly(styrene-alt-maleic anhydride)-b-poly(styrene)] and tartrate-resistant acid phosphatase (TRAP) binding peptide (TBP, sequence: TPLSYLKGLVTVG) (Figure 1) were selected as the model system for this study because of synthetic accessibility and application relevance (i.e., bone-targeted drug delivery) (Wang et al., 2017). A series of polymer-peptide conjugation reactions were performed to investigate the effect of multiple primary amines within TBP on bioconjugation reactions. Carbodiimide chemistry and anhydride ring opening chemistry were used for conjugations. Conjugation behaviors of a series of peptides containing 1 to 3 primary amines were investigated. The peptide series included TBP, TBP without Lysine (TBP-Lys−1), TBP with an extra lysine (TBP-Lys+1), and TBP with a protected lysine (TBP-Alloc). Furthermore, the effects on dispersity (Ð), number-average molecular weight (Mn), and weight-average molecular weight (Mw) of the polymer (PSMA-b-PS) by each of these peptides at different peptide feed ratios (10%, 15%, and 25% of carboxylate groups) were studied. Gel permeation chromatography (GPC) was used to estimate the amount of peptide conjugated, quantify the changes in polymer properties (Ð, Mn, and Mw), and predict the intramolecular and intermolecular polymer-peptide reactions.
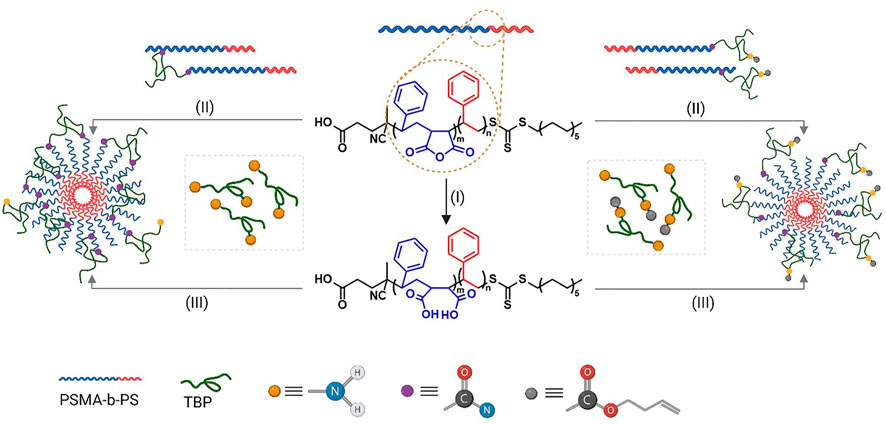
FIGURE 1. Schematic representation of PSMA-b-PS reactions with TBP under different reaction and amine-protection conditions. (I) base-catalyzed anhydride ring opening of PSMA-b-PS to yield carboxylates (II) anhydride ring opening via nucleophilic addition-elimination (TBP as the nucleophile) (III) TBP conjugation via carbodiimide chemistry.
Common solvents and reagents were purchased from commercial suppliers and used as received without additional purification unless otherwise specified. Styrene (99%, ACS grade) was purified via distillation over CaH2. Maleic anhydride (MA) was purified by recrystallization from chloroform. 2,2′-Azo-bis(isobutylnitrile) (AIBN) (initiator) was recrystallized from methanol. The chain transfer agent (CTA), 4-cyano-4-dodecylsulfanyltrithiocarbonyl sulfanyl pentanoic acid (DCT), was synthesized using a previously reported procedure (Moad et al., 2005). PSMA-b-PS was synthesized via reversible addition−fragmentation chain transfer (RAFT) polymerization by slightly modifying a previously reported method in which Styrene (Sty), MA, and DCT ([Sty]:[MA]=4:1) were dissolved in 1,4 Dioxane (128% W/W). AIBN ([AIBN]:[DCT]=1:10) was then added to the solution after restoring in ice for 30 min. After 40 min of nitrogen purging in ice, the solution was then placed in a 60°C oil bath and allowed to polymerize for 48 hours, after which the reaction was terminated by exposure to air. The reaction mixture was diluted with acetone and then added dropwise in petroleum ether. The final product was dried under vacuum (Baranello et al., 2014; Wang et al., 2017). Gel permeation chromatography (GPC) was used to determine the number average molecular weight (Mn), Weight average molecular weight (Mw), and dispersity (Ð) of the polymers.
A series of peptides consisting of 1 to 3 primary amines was synthesized to analyze efficiencies of different conjugation chemistries and possible crosslinking. The amino acid sequences were similar to TBP, but lysine was either added (TBP-Lys+1) or removed (TBP-Lys−1) to alter the number of primary amines. This addition or removal of amino acids is impractical for therapeutic use. Rather, protecting the selected functional groups using an appropriate protecting group is more appropriate. As TBP is a bone-targeting peptide, the goal was to avoid crosslinking without major manipulations in the peptide sequence. In conventional solid-phase peptide synthesis, acid-labile protecting groups prevent side reactions (Isidro-Llobet et al., 2009; Gongora-Benitez et al., 2012; Ackun-Farmmer et al., 2021). To maintain Lys primary amine protection, acid-resistant protection (Alloc protecting chemistry or TBP-Alloc) was carried out. TBP (sequence: TPLSYLKGLVTVG), TBP-Lys-1 (sequence: TPLSYLGLVTVG), TBP-Lys+1 (sequence: TPLSYLKGLVTVKG), and TBP-Alloc (sequence: TPLSYLKAllocGLVTVG) were synthesized using microwave-assisted solid-phase peptide synthesis (CEM Corp, Liberty1 synthesizer). Peptides were synthesized on Fmoc-Gly-Wang resin with O-enzotriazole-N,N,N′,N′- tetramethyl-uronium-hexafluoro-phophaste (HBTU) in DMF and 2 M N,N-diisopropylethylamine (DIEA) in NMP coupling.
After synthesis and deprotection with 5% piperazine in DMF, peptides were cleaved in a mixture of trifluoroacetic acid (TFA, 92.5%), H2O (2.5%), triisopropylsilane (TIPS, 2.5%), and 3,6-dioxa-1,8- octanedithiol (DODT, 2.5%) for 2.5 h, after which the peptide was precipitated in ice-cold diethyl ether to ensure removal of DMF and byproducts of the cleavage mixture. The resulting solid was redissolved in DMF and reprecipitated into cold ether. This washing cycle was carried out three times to remove trace impurities. The resulting peptide was purified by dialysis against water (Spectra/por MWCO 1 kDa) for 5 days with four water exchanges daily. The dialyzed product was freeze-dried and analyzed for purity using high-performance liquid chromatography (HPLC) (Supplementary Figure S2) (Shimadzu LC-20AD HPLC system, SPD-20AV UV-Vis detector, mobile phase—acetonitrile:H2O, gradient elution—acetonitrile from 5 % to 95%) and molecular weight using Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectroscopy. Peptide purity was ≥95% for peptides used in this study.
Each peptide conjugation was performed using two conjugation approaches 1) carbodiimide chemistry, a universal conjugation method for materials consisting of carboxylate and amine functionalities (Biju, 2014; Elzahhar et al., 2019), and 2) anhydride ring-opening (Nucleophilic addition-elimination), specific to anhydride containing biomaterials (e.g., the PSMA block here). Three independent reactions were carried out for each experimental condition to investigate trial-to-trial variations within the same batch. For each of these peptides (TBP, TBP-Lys−1, TBP-Lys+1, and TBP-Alloc), 10%, 15%, and 25% feed ratios of peptide were used based on the number of carboxylate groups present on the polymer chain.
The polymer was dissolved in dry DMF (25% w/v) and sonicated to ensure complete solubility. Then, excess amounts of DIC (N, N′-Diisopropylcarbodiimide, Thermo) ([DIC]:[polymer] = 1.2:1 molar ratio) and DMAP (4-Dimethylaminopyridine, Thermo) ([DMAP]:[polymer] = 1 molar ratio), were added to the polymer dissolved in DMF. The mixture was re-sonicated for 5 min to ensure that all solutes were dissolved and to activate the carboxylates on the polymer, after which the peptide (dissolved in DMF, 20% w/v) was added dropwise immediately (in 30 s). The reaction was re-sonicated at room temperature for 30 min and then stirred overnight, after which the reaction was quenched by exposing the reaction mixture to excess water and purified by dialysis (Spectra/por MWCO 6–8 kDa) for 3 days. The dialyzed mixture was then freeze-dried, and the resulting solid was analyzed by GPC to evaluate the changes in Mn, Mw, and Ð.
PSMA-b-PS and peptides were dissolved in dry DMF (20% W/V). Peptide solutions with 10%, 15%, and 25% anhydride-feeding ratios were added to the polymer solution respectively and stirred under room temperature overnight, after which the reaction was quenched by exposing the reaction mixture to excess water. The unconjugated peptide and DMF were removed using a dialysis membrane (Spectra/Por, MWCO = 6–8 kD) for 3 days. The peptide-conjugated PSMA-b-PS was then collected by freeze-drying and characterized by GPC.
GPC was used as the characterization method to evaluate the Mn, Mw, and Ð of PSMA-b-PS. DMF (0.05 M LiCl) was used as the mobile phase. GPC measurements were carried out on a Shimadzu 20A GPC system equipped with a TSKgel SuperHM-N (Tosoh Bioscience) column at 60°C at a flow rate of 0.35 ml/min using a miniDAWN TREOS light scattering detector (Wyatt Technology) and a T-rEX Refractive Index Detector (Wyatt Technology). The data was evaluated using Astra 7.3.2 software. After the peptide conjugation (by both conjugation techniques), the resulting conjugates were re-analyzed to assess the changes in molecular weight and dispersity. The formation of crossling was qualitatively deduced by comparing GPC results of the polymer and polymer-peptide conjugates (for each independent trial). For the conjugates that did not show crosslinking (TBP-Lys−1, TBP-Alloc), molar mass differences between PSMA-b-PS and the respective conjugate were used to calculate the number of conjugated peptides. As this work mainly focuses on the qualitative aspects, these calculations were done assuming that the peptide conjugation has no significant effect on the refractive index of the polymer. However, measuring dn/dc for each conjugate is suggested for high-accuracy quantitative analyses.
The OPA assay was used to quantify conjugation efficiency using primary amines present on the conjugates (KangLee and Drescher, 1978). Briefly, standards (TBP, TBP-Lys+1) and negative controls (unfunctionalized polymer, 1xPBS) were prepared in 1xPBS and concentrations calculated based on standard curves. The standards, controls, and samples (functionalized polymers) were transferred into a 96-well plate (75 µL). The OPA reagent (Thermo Scientific) was then added to each well (125 µL), samples were excited at 360 nm, and emission was measured at 455 nm (BioTek Cytation 5, Agilent). All samples, including positive controls and negative controls, were tested in 3 replicates. The number of peptides preset (primary amines) was calculated using the standard curves.
Maleic anhydride groups within PSMA-b-PS NPs provide multiple carboxylate functionalities within the shell of self-assembled NPs, which give the ability to develop biomolecule-functionalized NPs with tunable multivalency simply by controlling feed ratios (Figure 1). The peptide, TBP contains two primary-amine reactive centers, one at the amino terminus and one within the intersequence lysine side chain, where the terminal amine is more reactive due to less steric hindrance. Therefore, the polymer-peptide conjugation was carried out by controlled addition of the peptide to an activated mixture of polymer carboxylates via carbodiimide coupling. The resulting polymer-peptide conjugates were quantified by the OPA assay.
When typical peptide conjugation and quantification methods were utilized in these multifunctional peptide-polymer conjugation reactions (i.e., OPA assay), expected conjugation efficiencies were not attained (Supplementary Table S1). Furthermore, noticeable trial-to-trial inconsistencies were shown within the batches performed under the same reaction conditions (Supplementary Table S1). As the OPA assay quantifies the peptides using primary amines, the observed inefficiencies and inconsistencies suggest a phenomenon related to undesired primary amine reactions rather than the amount of peptide conjugated. To attain a better understanding of this phenomenon, the underlying chemistries were studied.
In a typical carbodiimide reaction, the coupling reagent (i.e., EDC, DCC, DIC, etc.) is added in excess to ensure high conjugation efficiency (Pan et al., 2012; Biju, 2014; Wang et al., 2014; Gandhi et al., 2016; Zhang et al., 2016; Kuo et al., 2017; Palmieri et al., 2018; Shipunova et al., 2018; Wang et al., 2018; Elzahhar et al., 2019; Lim et al., 2019; Shiu et al., 2021; Wright et al., 2021), which leads to a highly activated carboxylate environment in the reaction medium. All primary amines within the peptide can participate in nucleophilic addition-elimination regardless of relative reactivities in such environments. As a result, intramolecular and intermolecular crosslinking can occur. Intramolecular crosslinking forms two conjugates between the peptide chain and the polymer backbone, resulting in a cyclic structure. Intermolecular crosslinking adds two polymeric chains onto one peptide, resulting in high molar mass polymers and polydisperse materials. Both situations can compromise the therapeutic efficacy of the biomaterial.
Crosslinking can also interfere with the peptide quantification method, providing misleading results. When considering the OPA assay, the reagent reacts with the primary amines present on the peptide to produce a fluorometric byproduct (KangLee and Drescher, 1978; Frank et al., 1985). When the peptide sequence includes multiple primary amines, the unconjugated primary amines are used to quantify the number of peptides on the polymer chains or bioconjugates (Mu et al., 2010; Wu et al., 2012; Li et al., 2014; Ding et al., 2017; Hou et al., 2019; Papay et al., 2021; Wu et al., 2021; Liu et al., 2022). As crosslinking converts these remaining primary amines to secondary amines, the assay produces a false-negative result even though the peptide is chemically conjugated to the polymer backbone. This false-negative result can lead to the incorrect conclusions that the reaction has not succeeded (Supplementary Table S1). Moreover, assays that quantify peptides/proteins using peptide bonds (e.g., bicinchoninic acid assay) can produce false-positive results due to crosslinking. Bicinchoninic acid (BCA) assay is based on reducing Cu2+ to Cu1+ by peptide bonds, also known as the biuret reaction. Cu1+ ions are then fluorometrically characterized after complexation with BCA (Huang et al., 2010). Crosslinking results in a greater number of peptide bonds that increase Cu2+ to Cu1+ reduction. Subsequently, results suggest greater peptide conjugation. For example, the assay detects two peptides if one peptide is crosslinked via two primary amines. Therefore, inconsistent results obtained with the OPA assay may be due to crosslinking. In that case, a characterization technique that can provide information on the change in sample weight fraction, molar fraction, and dispersity after the conjugation could provide more realistic information regarding conjugation and crosslinking. Therefore, gel permeation chromatography (GPC) was used as a qualitative and quantitative analytical tool to study polymer-peptide conjugation and possible crosslinking.
Acid anhydrides and acid chlorides are at the top of the reactivity hierarchy of carboxylic acid derivatives (Bender, 1960). As the hydrophilic block of PSMA-b-PS contains cyclic anhydrides (MA), the nucleophilicity of primary amines can promote a nucleophilic addition-elimination reaction (Bender, 1960; Nagaraja et al., 2019). This nucleophilic attack results in peptide-polymer conjugates while opening the MA ring that produces carboxylates, which is necessary for the amphiphilicity of the polymer, without adding an additional step to the reaction scheme (Figure 1). Consequently, the ARO approach provides a facile yet robust conjugation approach without the need for activators and eliminates additional purification steps. Reactions were performed by mixing the polymer and the peptide dissolved in a non-nucleophilic solvent under nitrogen. The purified conjugated product (by dialysis) was analyzed qualitatively and quantitively by GPC.
When analyzing the peptide conjugates with multiple primary amines (i.e., TBP and TBP-Lys+1), a significant mass distribution shift from Gaussian was observed regardless of the feed ratio (Figures 2A,B). Moreover, a secondary peak towards the high molecular weight region was observed for the 25% feed ratio of TBP and all the feed ratios of TBP-Lys+1. These data suggest an inversely proportional relationship of peptide conjugate dispersity with the number of primary amines on the peptide. However, the expected trend for the desired peptide conjugation was a gradual shift toward higher molecular weight proportional to the feed ratio. The Ð values obtained from chromatograms show 2 to 4-fold increases in Ð for TBP and TBP-Lys+1 conjugates, supporting the qualitative observations above (Supplementary Tables S2, S3). The expected molecular weight for 10% TBP conjugation was 53 kDa, yet the observed Mn and Mw are 60 kDa and 186 kDa, respectively (Supplementary Table S2). Even though Mn is comparable with the expected molecular weight, Mw shows significant deviations (>3-fold increase), indicating a substantial impact on the weight fraction of polymer chains. These data suggest crosslinking between polymer chains, resulting in high molecular weight crosslinked polymer networks. With higher feed ratios, greater deviations (for both Mn and Mw) from the expected molecular weight were observed (Supplementary Table S2), suggesting the peptide was consumed as a crosslinker. A similar trend was observed for the polymer-peptide conjugates obtained from TBP-Lys+1. Moreover, the addition of Lys to the sequence exhibits more significant changes in molecular weight compared to TBP (Supplementary Table S3, Figures 3A,B), which was expected as crosslinking density increases when the crosslinker consists of more reactive centers.
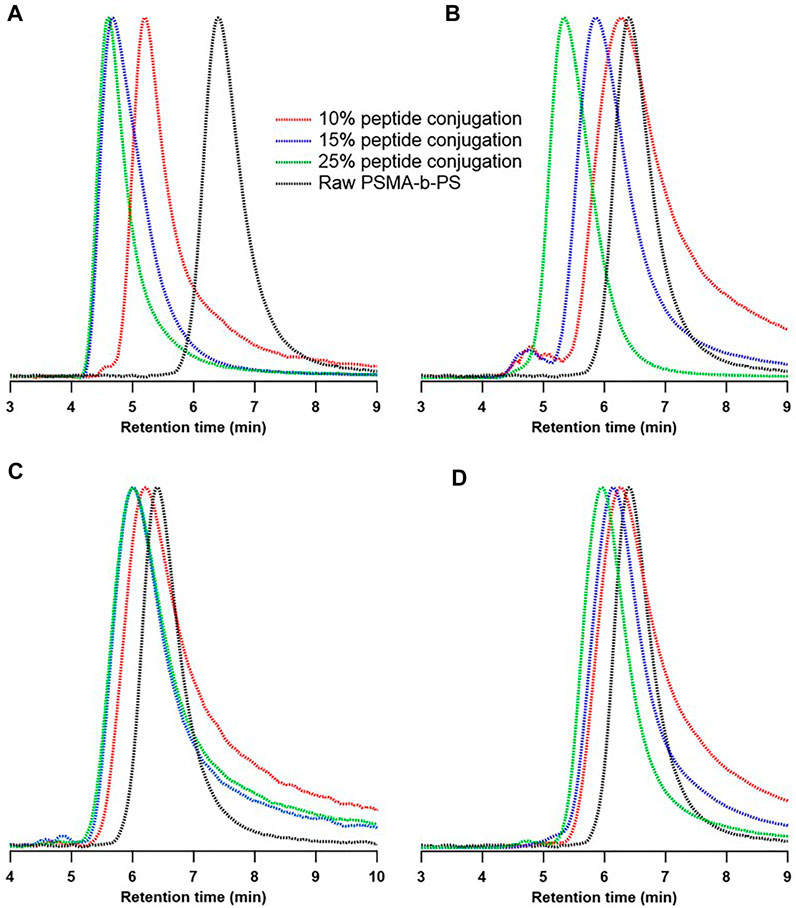
FIGURE 2. GPC chromatograms for the TBP conjugated PSMA-b-PS obtained from ARO. (A) TBP (B) TBP-Lys+1 (C) TBP-Lys−1 (D) TBP-Alloc.
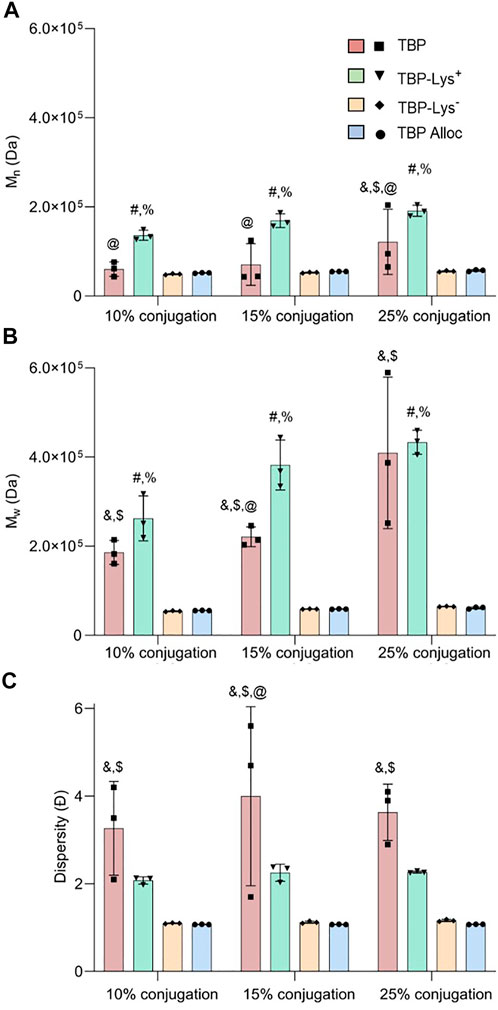
FIGURE 3. Variation of (A) weight average molecular weight (Mw), (B) number average molecular weight (Mn), and (C) dispersity (Ð) of peptide-functionalized polymers concerning the peptide type for ARO conjugation. & (TBP–TBP-Lys−1), $ (TBP–TBP-Alloc), # (TBP-Lys+1–TBP-Lys−1), % (TBP-Lys+1–TBP-Alloc), @ (TBP–TBP-Lys+1) p < 0.05 indicates significance evaluated by two-way ANOVA with Tukey’s multiple comparisons test, N = 3.
TBP-Lys−1 and TBP-Alloc exhibited more controlled conjugation reactions regardless of feed ratio. The Gaussian nature of PSMA-b-PS molecular weight distribution was preserved after the conjugation, and no secondary peaks were visible on GPC chromatograms (Figures 2C,D). Ð values for these conjugates did not show significant deviations from the polymer compared to TBP and TBP-Lys+1 (Supplementary Tables S4, S5). This indicates a uniform distribution of peptides among polymer chains without crosslinking. The average number of conjugated peptides was estimated using the molecular weight differences obtained by GPC data. TBP-Lys−1 and TBP-Alloc showed close correlations with the expected numbers for 10% and 15% feed ratios (Supplementary Tables S4, S5). However, the percent conjugation did not exceed 20% for TBP-Lys−1 and 15% percent for TBP-Alloc, even at the 25% feed ratio. This implies a limitation for ARO reaction with increasing steric hindrance at the reaction center.
Nucleophilic addition-elimination reactions propagate through a tetrahedral intermediate, generating a higher activation barrier when bulky groups are present (Klein, 2017). TBP-Lys−1 is less sterically hindered than TBP-Alloc as it does not contain Lys and Alloc protecting group. At higher feed ratios (15% and 25%), TBP-Lys−1 conjugations resulted in 14 and 19 peptides per polymer chain (expected 15 and 25, respectively), while TBP-Alloc exhibited maximum conjugation at 14 peptides per polymer chain. Also, when more peptides are added to the polymer chain, the hydrophilic block becomes more sterically hindered and eventually saturates. According to the results we obtained from this work, PSMA-b-PS exhibited saturation after adding 19 (TBP-Lys−1) and 14 peptides (TBP-Alloc) per chain. Nevertheless, additional kinetic experiments are required to conclude the limitations on this conjugation approach.
In addition to the statistically significant molecular weight (Mn and Mw) and dispersity increases shown in TBP and TBP-Lys+1, larger standard deviations among replicates were observed (Figures 3A,C). Generally, an increased standard error was observed with increasing feed ratios (Figures 3A,B). This trend provides evidence of an uncontrolled reaction occurring in the medium regardless of controlled reaction conditions. As TBP and TBP-Lys+1 act as crosslinkers, the outcome of the reaction becomes more unpredictable when greater amounts of peptide are in the feed. Ð values suggest TBP-Lys+1 has better control than TBP, even with greater primary amines (Figure 3C). As Ð is the ratio between Mw and Mn, closer Mn and Mw values provide comparatively smaller Ð values. Because of the increased number of reaction centers, TBP-Lys+1 conjugates have larger values for both Mn and Mw than TBP (Figures 3A,B). Therefore, the lower Ð values obtained for TBP-Lys+1 do not indicate a controlled conjugation reaction. Therefore, close attention should be given to possible side reactions when standardizing synthetic/bioconjugation protocols.
TBP-Lys−1 and TBP-Alloc showed excellent trial-to-trial reproducibility with all feed ratios explored (Figures 3A–C). As the extra reaction centers are eliminated or protected, the only possible reaction pathway is conjugation without crosslinking. The results suggest incorporating a selective protection strategy for multiple reaction centers, even though it requires additional deprotection and purification steps. These findings highlight the importance of paying close attention to possible undesired reactions and performing comprehensive material characterization before proceeding to in vitro or in vivo studies. In addition, the ARO peptide conjugation pathway provides a facile yet robust conjugation method that can be adopted for anhydride-containing biomaterials. No coupling reagents (EDC, DIC, DCC, etc.) or buffering were required for the reaction, eliminating the related purification steps. Therefore, this reaction pathway provides a facile, hassle-free peptide conjugation approach for material containing cyclic anhydrides. However, it is essential to highlight that the reactivity can be impacted by the peptide, polymer composition, and secondary structure. Moreover, the peptide density and distribution on the nanoparticle are crucial factors for target-specific binding and will be a focus in future studies that incorporate analytical techniques, including nanoparticle tracking analysis and in vitro and in vivo bioassays (Ashby et al., 2015; Vestad et al., 2017; Newman et al., 2018).
Even though peptide conjugation via ARO nucleophile addition-elimination provides a facile conjugation approach (with selective protection), it is not compatible with biomaterials that lack cyclic anhydrides. As carbodiimide coupling is the most common approach in such cases, (Israelachvili et al., 1977; Moad et al., 2005; Blanazs et al., 2009; Pan et al., 2012; Baranello et al., 2014; Biju, 2014; Wang et al., 2014; Gandhi et al., 2016; Zhang et al., 2016; Davaa et al., 2017; Imanparast et al., 2017; Kuo et al., 2017; Shipunova et al., 2018; Wang et al., 2018; Elzahhar et al., 2019; Lim et al., 2019; Ackun-Farmmer et al., 2021; Carambia et al., 2021) it is beneficial to investigate the effect of these undesired reactions in carbodiimide-activated environments. The conventional approach of carbodiimide coupling for biomaterials uses an aqueous medium due to solubility and compatibility. (Nam et al., 2008; Fischer, 2010; Mansuroğlu and Mustafaeva, 2012; Biju, 2014; Werengowska-Ciećwierz et al., 2015) However, in aqueous media, water can act as a competitor for the desired nucleophile leading to hydrolysis of carbodiimide intermediate. This interference can lead to ambiguities. Therefore, the competing nucleophile was eliminated using a non-nucleophilic organic solvent, N,N-Dimethylformamide (DMF). Furthermore, shifting the solvent characteristics from polar protic to polar aprotic increases the nucleophile's potential energy, thus enhancing the rate of the reaction. (Klein, 2017) MA groups of PSMA-b-PS were ring-opened to obtain carboxylates before the reactions to eliminate the ARO conjugation pathway (Figure 1).
Similar to the ARO conjugation approach, TBP-Lys−1 and TBP-Alloc carbodiimide conjugates did not indicate crosslinking (Supplementary Figure S1). Molecular weights of the conjugates were similar to the expected values, and no significant increases in Ð values were found (Supplementary Tables S6, S7). For the ARO nucleophilic addition-elimination, the number of conjugated peptides did not reach the expected number at higher feed ratios, especially at 25%. This trend was more evident for TBP-Alloc (Supplementary Table S5). However, this limit was not shown for any feed ratio with the carbodiimide approach (Supplementary Table S7). As the carbodiimide reaction proceeds through an activated intermediate, the electrophilicity of the carboxylate center is enhanced. It facilitates a less energetic nucleophilic attack even with a more sterically hindered nucleophile (TBP-Alloc). The polar aprotic solvent (DMF) does not energetically stabilize the nucleophile. Therefore, the nucleophile decreases the potential energy by participating in the addition-elimination reaction.
No significant trial-to-trial variations were observed (Figures 4A–C) for Mn, Mw, and Ð for TBP-Lys−1 and TBP-Alloc carbodiimide conjugates. These data indicate controlled and consistent reactions for peptides with one nucleophilic center. The Ð values show a slight increase compared to ARO peptide conjugates, which is expected due to the increased reaction rates in the carbodiimide-activated medium. Activation of the electrophilic center with carbodiimide crosslinkers is not nucleophile specific. Therefore, the rate of undesired nucleophilic attacks also increases, producing more unanticipated products. This is evident from the results for TBP-polymer conjugates. Compared with the ARO approach, carbodiimide conjugates show much more significant (>5-fold increase) increases in weight fractions (Supplementary Table S8). Furthermore, highly significant trial-to-trial variations were observed for Mn, Mw, and Ð at higher feed ratios (Figures 4A–C). These results suggest that more attention should be given to the chemical nature (i.e., multiple reaction centers, intramolecular cyclization, competing nucleophiles, etc.) of the starting materials and possible off-target reactions when using activated reaction media.
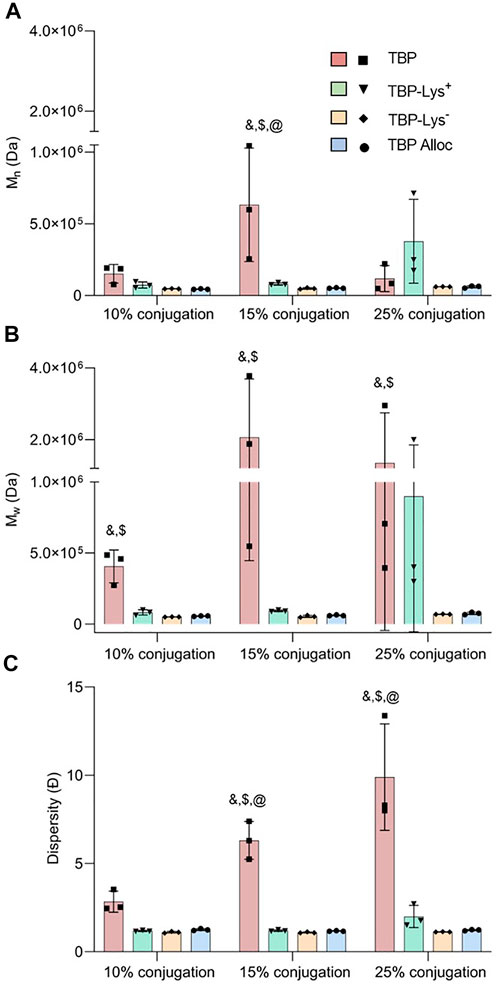
FIGURE 4. Variation of (A) weight average molecular weight (Mw), (B) number average molecular weight (Mn), and (C) dispersity (Ð) of peptide-functionalized polymers concerning the peptide type for carbodiimide conjugation in DMF. & (TBP–TBP-Lys−1), $ (TBP–TBP-Alloc), (TBP–TBP-Lys+1) p < 0.05 indicates significance evaluated by two-way ANOVA with Tukey’s multiple comparisons test, N = 3.
Interestingly, TBP-Lys+1 exhibited a more controlled reaction than TBP (Figure 4C) for 10% and 15% feed ratios. Similar control was also obtained based on Ð values of ARO conjugates (Figure 3C, Supplementary Table S9), yet larger conjugates were observed (Figures 3A,B). This could result from two competing side reactions: intramolecular cyclization and crosslinking. As TBP-Lys+1 possesses extra reaction centers and a bulkier molecular structure, intramolecular nucleophilic attacks are energetically favored versus intermolecular crosslinking when carboxylates on the polymer chain are activated. The greater the number of activated carboxylates present, the greater the possibility of intermolecular crosslinking, as more molecules can surpass the activation barrier, which is supported by Mn and Mw values at 25% feed ratio (Figures 4A,B). Further experiments were carried out using increased chemical equivalents of carbodiimide crosslinker to provide more activated carboxylates while keeping the peptide feed ratios at 10% and 15%. At increased carbodiimide crosslinker feed ratios (4.0 eq.), there are notable increases in Mn, Mw, and Ð (Supplementary Table S10). Highly significant increases in molecular weights were observed when the reaction medium contained excess activated carboxylates and more substantial amounts of peptide (Figures 5A,B). In addition, the resulting conjugates exhibited a great degree of polydispersity, suggesting greater intermolecular crosslinking (Figure 5C).
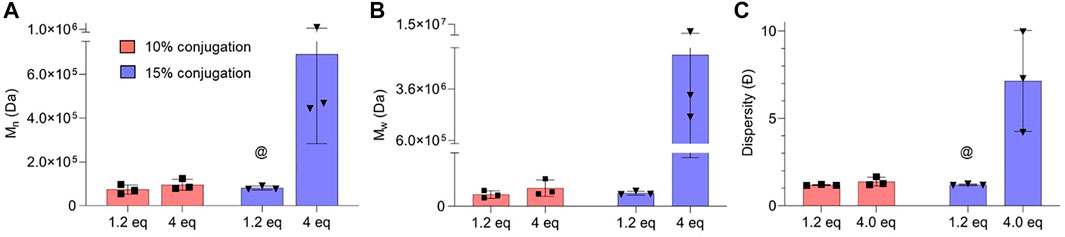
FIGURE 5. Variation of (A) weight average molecular weight (Mn), (B) number average molecular weight (Mw), and (C) dispersity (Ð) of peptide-functionalized polymers concerning the chemical equivalents (eq) of carbodiimide crosslinker. @ (1.2–4.0 eq) p < 0.05 indicates significance evaluated by two-way ANOVA with Tukey’s multiple comparisons test, N = 3.
Results of this study suggest that both ARO electrophilic substitution-elimination and carbodiimide approaches facilitated efficient conjugation in selectively protected multifunctional peptide-polymer systems. ARO provides a facile yet robust conjugation approach for cyclic anhydride-containing biomaterials by eliminating additional purification steps and carbodiimide crosslinkers. This reaction demonstrated more sensitivity towards the steric hindrance of the reaction center, exhibiting decreased efficiencies at higher feed ratios (i.e., >15% for the selected peptides in this study). However, bioconjugation reactions involving multifunctional peptides and polymeric materials demand extra attention in synthesis and characterization due to the increased tendency of undesired reactions. Intermolecular crosslinking was one of the most probable undesired reactions in such cases. When crosslinking occurs, indirect peptide quantification methods (i.e., OPA assay, BCA assay, etc.) fail to provide correct information regarding the peptide conjugation. Furthermore, undesired crosslinking makes the outcome of in vitro/in vivo applications unpredictable. Therefore, incorporating a secondary characterization method is necessary for similar bioconjugation reactions. GPC showed good potential as a secondary characterization method to acquire more convincing information regarding peptide conjugation and crosslinking. However, this method’s accuracy can vary according to the properties of selected biomaterials. Regardless of the reactivity of multiple reaction centers, crosslinking occurred at increased feed ratios of activators and peptides. Expected peptide conjugates were achieved (independent of the conjugation method) by protecting unwanted reaction centers. Therefore, selective protection is recommended for similar conjugation approaches to avoid undesired reactions. Furthermore, it is advisable to prevent indirect characterization methods that detect functional groups resulting from the reaction because those detectable functional groups can result from an undesired reaction.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
IC and DB designed the experiments. IC, YL, EA-S, and BX contributed to the synthesis of polymer-peptide conjugates and experimental data collection. IC analyzed the data. IC and DB wrote the manuscript. All authors edited and revised the manuscript.
This work was supported by grants from the National Science Foundation (NSF) (CBET1450987 and DMR2103553); National Institutes of Health (NIH) (R01 AR064200, R01 AR056696, P30 AR06955, R21 AR081063 (to DB) and S10 OD030302); Orthopaedic Research and Education Foundation Grant 20-072 (to DB), Orthopaedic Trauma Association Grant 6272 (to DB), and the University of Rochester Medical Center Department of Orthopaedics Goldstein Award (to DB).
The authors also wish to thank Clyde Overby for intellectual discussions regarding the work as well as technical assistance, Dr. Brittany Abraham for helpful edits, and Prof. James McGrath for the use of the Zetasizer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbiom.2022.1003172/full#supplementary-material
Ackun-Farmmer, M. A., Soto, C. A., Lesch, M. L., Byun, D., Yang, L., Calvi, L. M., et al. (2021). Reduction of leukemic burden via bone-targeted nanoparticle delivery of an inhibitor of C-chemokine (C-C motif) ligand 3 (CCL3) signaling. FASEB J. 35 (4), e21402. doi:10.1096/fj.202000938rr
Alexis, F., Pridgen, E., Molnar, L. K., and Farokhzad, O. C. (2008). Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 5 (4), 505–515. doi:10.1021/mp800051m
Ashby, J., Duan, Y., Ligans, E., Tamsi, M., and Zhong, W. (2015). High-throughput profiling of nanoparticle-protein interactions by fluorescamine labeling. Anal. Chem. 87 (4), 2213–2219. doi:10.1021/ac5036814
Baranello, M. P., Bauer, L., and Benoit, D. S. (2014). Poly(styrene-alt-maleic anhydride)-based diblock copolymer micelles exhibit versatile hydrophobic drug loading, drug-dependent release, and internalization by multidrug resistant ovarian cancer cells. Biomacromolecules 15 (7), 2629–2641. doi:10.1021/bm500468d
Bender, M. L. (1960). Mechanisms of catalysis of nucleophilic reactions of carboxylic acid derivatives. Chem. Rev. 60 (1), 53–113. doi:10.1021/cr60203a005
Biju, V. (2014). Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 43 (3), 744–764. doi:10.1039/c3cs60273g
Blanazs, A., Armes, S. P., and Ryan, A. J. (2009). Self-assembled block copolymer aggregates: From micelles to vesicles and their biological applications. Macromol. Rapid Commun. 30 (4-5), 267–277. doi:10.1002/marc.200800713
Carambia, A., Gottwick, C., Schwinge, D., Stein, S., Digigow, R., Seleci, M., et al. (2021). Nanoparticle-mediated targeting of autoantigen peptide to cross-presenting liver sinusoidal endothelial cells protects from CD8 T-cell-driven autoimmune cholangitis. Immunology 162 (4), 452–463. doi:10.1111/imm.13298
Davaa, E., Lee, J., Jenjob, R., and Yang, S. G. (2017). MT1-MMP responsive doxorubicin conjugated poly(lactic-co-glycolic acid)/poly(styrene-alt-maleic anhydride) core/shell microparticles for intrahepatic arterial chemotherapy of hepatic cancer. ACS Appl. Mat. Interfaces 9 (1), 71–79. doi:10.1021/acsami.6b08994
Ding, H., Gangalum, P. R., Galstyan, A., Fox, I., Patil, R., Hubbard, P., et al. (2017). HER2-positive breast cancer targeting and treatment by a peptide-conjugated mini nanodrug. Nanomedicine Nanotechnol. Biol. Med. 13 (2), 631–639. doi:10.1016/j.nano.2016.07.013
Elzahhar, P., Belal, A. S. F., Elamrawy, F., Helal, N. A., and Nounou, M. I. (2019). Bioconjugation in drug delivery: Practical perspectives and future perceptions. Methods Mol. Biol. 2000, 125–182. doi:10.1007/978-1-4939-9516-5_11
Fischer, M. J. (2010). “Amine coupling through EDC/NHS: A practical approach,” in Surface plasmon resonance (Chram: Springer), 55–73.
Frank, C., Church, D. H. P., GeorgeCatignaniSwaisgood, L. H E., and Swaisgood, H. E. (1985). An o-phthalaldehyde spectrophotometric assay for proteinases. Anal. Biochem. 146 (2), 343–348. doi:10.1016/0003-2697(85)90549-4
Gandhi, S., Arami, H., and Krishnan, K. M. (2016). Detection of cancer-specific proteases using magnetic relaxation of peptide-conjugated nanoparticles in biological environment. Nano Lett. 16 (6), 3668–3674. doi:10.1021/acs.nanolett.6b00867
Gongora-Benitez, M., Mendive-Tapia, L., Ramos-Tomillero, I., Breman, A. C., Tulla-Puche, J., and Albericio, F. (2012). Acid-labile cys-protecting groups for the fmoc/tBu strategy: Filling the gap. Org. Lett. 14 (21), 5472–5475. doi:10.1021/ol302550p
He, C., Hu, Y., Yin, L., Tang, C., and Yin, C. (2010). Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 31 (13), 3657–3666. doi:10.1016/j.biomaterials.2010.01.065
Hou, G., Chen, X., Li, J., Ye, Z., Zong, S., and Ye, M. (2019). Physicochemical properties, immunostimulatory activity of the Lachnum polysaccharide and polysaccharide-dipeptide conjugates. Carbohydr. Polym. 206, 446–454. doi:10.1016/j.carbpol.2018.09.067
Huang, T., Long, M., and Huo, B. (2010). Competitive binding to cuprous ions of protein and BCA in the bicinchoninic acid protein assay. Open Biomed. Eng. J. 4, 271–278. doi:10.2174/1874120701004010271
Imanparast, F., Faramarzi, M. A., Vatannejad, A., Paknejad, M., Deiham, B., Kobarfard, F., et al. (2017). mZD7349 peptide-conjugated PLGA nanoparticles directed against VCAM-1 for targeted delivery of simvastatin to restore dysfunctional HUVECs. Microvasc. Res. 112, 14–19. doi:10.1016/j.mvr.2017.02.002
Isidro-Llobet, A., Alvarez, M., and Albericio, F. (2009). Amino acid-protecting groups. Chem. Rev. 109 (6), 2455–2504. doi:10.1021/cr800323s
Israelachvili, J. N., Mitchell, D. J., and Ninham, B. W. (1977). Theory of self-assembly of lipid bilayers and vesicles. Biochimica Biophysica Acta - Biomembr. 470 (2), 185–201. doi:10.1016/0005-2736(77)90099-2
Kaga, S., Truong, N. P., Esser, L., Senyschyn, D., Sanyal, A., Sanyal, R., et al. (2017). Influence of size and shape on the biodistribution of nanoparticles prepared by polymerization-induced self-assembly. Biomacromolecules 18 (12), 3963–3970. doi:10.1021/acs.biomac.7b00995
KangLee, S. D. G. D., and Drescher, D. G. (1978). Fluorometric amino-acid analysis with o-phthaldialdehyde (OPA). Int. J. Biochem. 9 (7), 457–467. doi:10.1016/0020-711x(78)90075-7
Klein, D. R. (2017). “Alkyl halides: Nucleophilic substitution and elimination reactions,” in Organic chemistry. 3 ed. (WileyPLUS).
Kuo, R., Saito, E., Miller, S. D., and Shea, L. D. (2017). Peptide-conjugated nanoparticles reduce positive Co-stimulatory expression and T cell activity to induce tolerance. Mol. Ther. 25 (7), 1676–1685. doi:10.1016/j.ymthe.2017.03.032
Li, C., Liu, F., Gong, Y., Wang, Y., Xu, H., Yuan, F., et al. (2014). Investigation into the Maillard reaction between ɛ-polylysine and dextran in subcritical water and evaluation of the functional properties of the conjugates. LWT - Food Sci. Technol. 57 (2), 612–617. doi:10.1016/j.lwt.2014.01.039
Lim, M., Lee, W., Bang, G., Lee, W. J., Park, Y., Kwon, Y., et al. (2019). Synthesis of far-red- and near-infrared-emitting Cu-doped InP/ZnS (core/shell) quantum dots with controlled doping steps and their surface functionalization for bioconjugation. Nanoscale 11 (21), 10463–10471. doi:10.1039/c9nr02192b
Liu, X., Xue, F., Li, C., and Adhikari, B. (2022). Physicochemical properties of films produced using nanoemulsions stabilized by carboxymethyl chitosan-peptide conjugates and application in blueberry preservation. Int. J. Biol. Macromol. 202, 26–36. doi:10.1016/j.ijbiomac.2021.12.186
Mansuroğlu, B., and Mustafaeva, Z. (2012). Characterization of water-soluble conjugates of polyacrylic acid and antigenic peptide of FMDV by size exclusion chromatography with quadruple detection. Mater. Sci. Eng. C 32 (2), 112–118. doi:10.1016/j.msec.2011.10.004
Moad, G., Chong, Y. K., Postma, A., Rizzardo, E., and Thang, S. H. (2005). Advances in RAFT polymerization: The synthesis of polymers with defined end-groups. Polymer 46 (19), 8458–8468. doi:10.1016/j.polymer.2004.12.061
Mu, L., Zhao, M., Yang, B., Zhao, H., Cui, C., and Zhao, Q. (2010). Effect of ultrasonic treatment on the graft reaction between soy protein isolate and gum acacia and on the physicochemical properties of conjugates. J. Agric. Food Chem. 58 (7), 4494–4499. doi:10.1021/jf904109d
Nagaraja, A., Jalageri, M. D., Puttaiahgowda, Y. M., Raghava Reddy, K., and Raghu, A. V. (2019). A review on various maleic anhydride antimicrobial polymers. J. Microbiol. Methods 163, 105650. doi:10.1016/j.mimet.2019.105650
Nam, K., Kimura, T., and Kishida, A. (2008). Controlling coupling reaction of EDC and NHS for preparation of collagen gels using ethanol/water co‐solvents. Macromol. Biosci. 8 (1), 32–37. doi:10.1002/mabi.200700206
Newman, M. R., Russell, S. G., Schmitt, C. S., Marozas, I. A., Sheu, T.-J., Puzas, J. E., et al. (2018). Multivalent presentation of peptide targeting groups alters polymer biodistribution to target tissues. Biomacromolecules 19 (1), 71–84. doi:10.1021/acs.biomac.7b01193
Palmieri, G., Tate, R., Gogliettino, M., Balestrieri, M., Rea, I., Terracciano, M., et al. (2018). Small synthetic peptides bioconjugated to hybrid gold nanoparticles destroy potentially deadly bacteria at submicromolar concentrations. Bioconjug. Chem. 29 (11), 3877–3885. doi:10.1021/acs.bioconjchem.8b00706
Pan, L., He, Q., Liu, J., Chen, Y., Ma, M., Zhang, L., et al. (2012). Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 134 (13), 5722–5725. doi:10.1021/ja211035w
Papay, Z. E., Magramane, S., Kiraly, M., Szalkai, P., Ludanyi, K., Horvath, P., et al. (2021). Optimization and development of albumin-biopolymer bioconjugates with solubility-improving properties. Biomedicines 9 (7), 737. doi:10.3390/biomedicines9070737
Shipunova, V. O., Zelepukin, I. V., Stremovskiy, O. A., Nikitin, M. P., Care, A., Sunna, A., et al. (2018). Versatile platform for nanoparticle surface bioengineering based on SiO2-binding peptide and proteinaceous Barnase*Barstar interface. ACS Appl. Mat. Interfaces 10 (20), 17437–17447. doi:10.1021/acsami.8b01627
Shiu, S. T., Lee, W. F., Chen, S. M., Hao, L. T., Hung, Y. T., Lai, P. C., et al. (2021). Effect of different bone grafting materials and mesenchymal stem cells on bone regeneration: A micro-computed tomography and histomorphometric study in a rabbit calvarial defect model. Int. J. Mol. Sci. 22 (15), 8101. doi:10.3390/ijms22158101
Vestad, B., Llorente, A., Neurauter, A., Phuyal, S., Kierulf, B., Kierulf, P., et al. (2017). Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 6 (1), 1344087. doi:10.1080/20013078.2017.1344087
Wang, G., Wang, Z., Li, C., Duan, G., Wang, K., Li, Q., et al. (2018). RGD peptide-modified, paclitaxel prodrug-based, dual-drugs loaded, and redox-sensitive lipid-polymer nanoparticles for the enhanced lung cancer therapy. Biomed. Pharmacother. 106, 275–284. doi:10.1016/j.biopha.2018.06.137
Wang, H., Dong, C., Zhao, P., Wang, S., Liu, Z., and Chang, J. (2014). Lipid coated upconverting nanoparticles as NIR remote controlled transducer for simultaneous photodynamic therapy and cell imaging. Int. J. Pharm. X. 466 (1-2), 307–313. doi:10.1016/j.ijpharm.2014.03.029
Wang, Y., Newman, M. R., Ackun-Farmmer, M., Baranello, M. P., Sheu, T. J., Puzas, J. E., et al. (2017). Fracture-targeted delivery of beta-catenin agonists via peptide-functionalized nanoparticles augments fracture healing. ACS Nano 11 (9), 9445–9458. doi:10.1021/acsnano.7b05103
Werengowska-Ciećwierz, K., Wiśniewski, M., Terzyk, A. P., and Furmaniak, S. (2015). The chemistry of bioconjugation in nanoparticles-based drug delivery system. Adv. Condens. Matter Phys. 2015, 1–27. doi:10.1155/2015/198175
Wright, T. A., Rahman, M. S., Bennett, C., Johnson, M. R., Fischesser, H., Ram, N., et al. (2021). Hydrolytically stable maleimide-end-functionalized polymers for site-specific protein conjugation. Bioconjug. Chem. 32 (11), 2447–2456. doi:10.1021/acs.bioconjchem.1c00487
Wu, J., Chen, H., Zhou, L., Liu, W., Zhong, J., and Liu, C. (2021). An insight into heat-induced gelation of whey protein isolate–lactose mixed and conjugate solutions: Rheological behavior, microstructure, and molecular forces. Eur. Food Res. Technol. 247 (7), 1711–1724. doi:10.1007/s00217-021-03741-x
Wu, K., Yang, J., Liu, J., and Kopecek, J. (2012). Coiled-coil based drug-free macromolecular therapeutics: In vivo efficacy. J. Control. Release 157 (1), 126–131. doi:10.1016/j.jconrel.2011.08.002
Keywords: carbodiimide chemistry, peptide conjugation, anhydride-containing polymers, nanoparticles, multifunctional biomaterials, bioconjugation
Citation: Chandrasiri I, Liu Y, Adjei-Sowah E, Xiao B and Benoit DSW (2022) Reproducible and controlled peptide functionalization of polymeric nanoparticles. Front. Front. Biomater. Sci. 1:1003172. doi: 10.3389/fbiom.2022.1003172
Received: 25 July 2022; Accepted: 06 September 2022;
Published: 03 October 2022.
Edited by:
Cristina Scielzo, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Arul Prakash Francis, Saveetha Dental College and Hospitals, IndiaCopyright © 2022 Chandrasiri, Liu, Adjei-Sowah, Xiao and Benoit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danielle S. W. Benoit, ZGFuaWVsbGUuYmVub2l0QHJvY2hlc3Rlci5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.