- 1Department of Biotechnology and Biosciences, University of Milano-Bicocca, Milan, Italy
- 2Department of Earth and Environmental Sciences, University of Milano-Bicocca, Milan, Italy
Polyethylene (PE) is the most-produced polyolefin, and consequently, it is the most widely found plastic waste worldwide. PE biodegradation is under study by applying different (micro)organisms in order to understand the biodegradative mechanism in the majority of microbes. This study aims to identify novel bacterial species with compelling metabolic potential and strategic genetic repertoires for PE biodegradation. Pseudomonas citronellolis E5 is newly isolated from solid organic waste contaminated with plastic debris, and Rhodococcus erythropolis D4 was selected for its promising potential in biodegradable plastic determined by its genetic repertoire. P. citronellolis E5 was selected for its ability to grow on PE as the only carbon and energy source. Meaningful extracellular secreted laccase activity was also characterized for D4 during growth on PE (E5 and D4 strains have a laccase activity of (2 ± 1)×10–3 U mg−1 and (3 ± 1)×10–3 U mg−1, respectively). Despite the highest level of cell numbers recorded at 7 days of growth on PE for both strains, the patterns of the metabolic products obtained and degraded during 60 days on PE were dissimilar in the two bacteria at different sampling times. However, they mainly produced metabolites belonging to carboxylic acids and alkanes with varying numbers of carbons in the aliphatic chains. Whole-genome sequence analyses of P. citronellolis E5 compared to R. erythropolis D4 and genetic determinant prediction (by gene annotation and multiple sequence alignment with reference gene products) have been performed, providing a list of 16 and 42 gene products putatively related to different metabolic steps of PE biodegradation. Altogether, these results support insights into PE biodegradation by bacteria of the Pseudomonas and Rhodococcus genera from metabolic and genetic perspectives as a base to build up novel biotechnological platforms.
1 Introduction
In the last decades, plastic exploitation has become a relevant issue due to its exceptional production rates and the consequent difficult disposal management at the end of useful life. Among the possible fates of plastic polymers, recycling and biodegradation processes involving microorganisms are the most eco-friendly methods (Lee and Liew, 2021). Indeed, a few microorganisms can break down polymers into simpler compounds by biochemical transformations through several steps, including biodeterioration, biofragmentation of the deteriorated plastics, generating plastic fragments with lower molecular weights or oxidized molecules, assimilation, and mineralization (Danso et al., 2019; Mohanan et al., 2020). Microbial assimilation routes are activated, forming biofilms on the plastic surface and/or producing active catalytic enzymes able to oxidize the polymers. The mineralization process comprises the transport of oxidized plastic derivatives into the cells, where diverse enzymatic reactions lead to complete degradation into oxidized metabolites, including CO2 and H2O (Zhang et al., 2022; Amobonye et al., 2021). Therefore, plastic polymer biodegradation using microorganisms equipped with the necessary enzymatic arsenals is a potential strategy for addressing waste plastics.
Synthetic plastics comprehend both hydrolyzable and non-hydrolyzable polymers. Polyethylene-based plastic is considered a non-hydrolysable polyolefin and the most recalcitrant to biodegradation, persisting in the environment for a long time. Polyethylene (PE) can be distinguished in high-density (HDPE) and low-density (LDPE) according to the molecular weight, 3-dimensional structures showing different degrees of branching, and availability of functional groups on the polymer surface (Sen and Raut, 2015; Montazer et al., 2020). PE is one of the most used materials in the manufacture of single-use plastic materials and, specifically, LDPE is mainly used to make carry bags and food packaging materials.
Since the early 2000s, PE biodegradation has been under study by applying bacterial and/or fungal pure cultures isolated from different environments, microbial consortia, and invertebrate mealworms (Bonhomme et al., 2003; Ray et al., 2023). The biodegradative process is accomplished by producing modifications in the functional groups, generating oxidized oligomers or products, and altering tensile properties and molecular weight, among other properties (Soleimani et al., 2021).
One of the major bottlenecks of such a treatment strategy and in defining a chemical as biodegradable is the duration of degradation, which should be limited to 28 days as in the cases reported in the standardized biodegradation test of OECD and ISO (ready biodegradability tests, inherent biodegradability tests, and simulation tests) (Strotmann et al., 2023). For example, OECD 301 indicates that 60% of the initial concentration or calculated reference value for biochemical oxygen demand or CO2 production are considered exceedance values.
Increasing numbers of research studies are focused on isolating and identifying new potential microbial degrading strains for plastic reduction, elimination, or potential recycling. They have been isolated from various sources, including waste-contaminated soils, compost, mangrove sediments, waste of the mulch films, landfills, dumping sites, marine environments, river estuaries, remote environments (Antarctica), and sewage treatment plants (Miri et al., 2022; Thakur et al., 2023). The common goal is to evaluate the metabolic potential for polymer biodegradation, improve its oxidation and biodegradation, and comprehend in-depth the process mechanism. Polyolefins like PE showed the highest recalcitrance and required an additional effort to identify the bacteria with the highest biodegradative potential. The main bacterial genera involved in PE biodegradation are Bacillus, Pseudomonas, and Rhodococcus (Soleimani et al., 2021; Montazer et al., 2020). However, not all bacteria species can adhere to hydrophobic surfaces, often requiring polymer pretreatment to obtain hydrophobic groups on their surfaces (Soleimani et al., 2021). Bacterial members of the last two genera are noteworthy in the field of biodegradation of organic matter, xenobiotic compounds, various contaminants, and plastic polymers (Zampolli et al., 2019; Zampolli et al., 2022; Medić and Karadžić, 2022; Shilpa et al., 2023; Kim et al., 2024).
A few enzymatic functions have been associated with the PE oxidative process such as laccase-like multicopper oxidases (EC 1.10.3.2.), manganese peroxidases (EC 1.11.1.13), and lignin peroxidases (EC 1.11.1.14) (Wei and Zimmermann, 2017; Andler et al., 2022; Zampolli et al., 2023; Ray et al., 2023). The study of the PE metabolic pathway in Rhodococcus bacteria indicated a plausible scenario of genes involved in the degradation of either UV-treated or untreated PE (Santo et al., 2013; Gravouil et al., 2017; Zampolli et al., 2021; Tao et al., 2023; Mishra et al., 2024; Kim et al., 2024). Primarily, the activation of extracellular secreted genes encoding laccase-like multicopper oxidases, lipases, esterases, alcohol dehydrogenases, long-chain fatty-acid-CoA ligases, and lipid transporters in the presence of PE leads to the formation of PE oligomers, oxygenated products, or lower molecular weight isolate fragments. The subsequent involvement of membrane transporters and oxygenases, such as alkane monooxygenases/hydroxylases and cytochrome P450 hydroxylases, supports the cell entrance and the further intracellular PE oxidation, respectively (Gravouil et al., 2017; Zampolli et al., 2021; Tao et al., 2023; Mishra et al., 2024; Rong et al., 2024; Kim et al., 2024). Furthermore, genes encoding for enzymes involved in the central metabolic pathways were also upregulated, including genes of steps of β-oxidation (Tao et al., 2023; Mishra et al., 2024).
In this context, the aim of this study is the characterization of the PE biodegradative potential of two bacteria, Pseudomonas citronellolis E5 and Rhodococcus erythropolis D4, providing both metabolic characterization and identification of plausible genomic and genetic traits to aid the bioremediation of polyolefins in an eco-friendly process.
P. citronellolis E5 is a novel isolate from a solid organic waste contaminated with plastics from a waste treatment plant, and R. erythropolis D4 was previously selected for its promising potential in biodegrade plastics (Zampolli et al., 2022; Zampolli et al., 2024). The R. erythropolis D4 genome has been completely sequenced and revealed to carry genes involved in plastic removal (Zampolli et al., 2022). These two bacteria were assayed for their ability to grow on PE as the sole carbon and energy source, evaluating their laccase enzymatic activity in the extracellular environment after growth on PE and the profile of metabolic products from PE oxidation solvent-extracted and analyzed by gas chromatographic analysis coupled with mass spectrometry. Their genome analysis (whole-genome alignment, annotation search, and gene clustering against reference sequences) predicted gene products related to PE biodegradation, hinting at their genetic potential for PE mineralization.
2 Materials and methods
2.1 Bacterial strain isolation and growth conditions
The bacterial isolation process was conducted using solid waste with plastic debris and microplastics (Supplementary Figure S1) sampled from the final treatment step biowaste treatment plant in Lombardy, Italy, with the same procedure described by Zampolli et al. (2023) after shredding the waste into small particles (1 cm–100 μm). At the end of the enrichment culture process in the presence of the solid waste, the individual bacterial colonies were isolated on Luria–Bertani (LB) agar plates and screened for their capacity to grow on solid M9 mineral medium (Maniatis et al., 1982) supplemented with 1% v/v Tween80 (as dispersing agent) with added 1% w/v sterile commercial low-density (untreated) powder PE (M9-Tw-PE) (Merck Darmstadt, Germany, Mn = 1700 by GPC, Mw = 4000 by GPC, cod. 427772). The sterility of the PE powder was obtained by UV radiation for 1 h using a 15 W UV-C germicidal lamp (Gelaire Flow Laboratories, Italy). The plates were then incubated at 30°C for 4 days to check for growth of colonies. This pre-screening was repeated three times. The novel isolated bacteria were first morphologically characterized by growth on the LB agar medium and M9-PE agar plates. The Gram stain was examined under a microscope with a magnification of up to ×100 (Axiolab E re, Carl Zeiss, Germany). This preliminary screening was also applied to another plastic-degrading bacterial strain, R. erythropolis D4, selected for its plastic metabolic potential demonstrated in previous works based on genetic trait identification (Zampolli et al., 2022; Zampolli et al., 2023). R. erythropolis (identification number 8B-C2-LD D4) (Zampolli et al., 2023) and a selected bacterial strain among the seven isolates from a plastic and organic waste treatment plant, P. citronellolis (identification number A-C1-1 E5), are deposited in the Private Collection of Microbiology Laboratory - BtBs Department of University of Milano-Bicocca.
All the new isolates and R. erythropolis D4 were periodically maintained in the M9-PE at 30°C. After washing their cells in the M9 mineral medium twice, they were pre-cultured in the M9 mineral medium supplemented with 20 mM malate (M9-M) and incubated at 30°C overnight.
The bacterial cells deriving from pre-cultures in M9-M were washed and resuspended in a fresh M9, then inoculated in 20 mL flasks in the presence of 1% w/v sterile low-density powder PE (M9-PE) as the sole carbon and energy source. Each bacterial culture was inoculated from the corresponding pre-culture to obtain an initial optical density read at 600 nm (OD600) of 0.1. All cultures were incubated at 30°C under shaking (120 rpm) for up to 7 days and sampled at 0 days, 1 day, 2 days, 3 days, 4 days, and 7 days to monitor their growth by measuring OD600.
2.2 Enzymatic activity
The bacterial isolates were discriminated by their enzymatic activity by evaluating both the intracellular and extracellular laccase activity because the isolates could activate laccase-like enzymes to perform the first oxidation of PE. After the bacterial cells were grown on 100 mL M9-PE for 3 days or 20 mL M9-M for 24 h, they were harvested by centrifugation (8,000 rpm), resuspended in 50 mM potassium phosphate buffer (pH 7, PPB), vortexed for 10 min, and incubated with 50 mg mL−1 lysozyme at 30°C for 1 h. Then, the cells were lysed by sonication (10 cycles of 20 s with 20% amplitude) with a Digital Sonifier™ (BRANSON Ultrasonic Corporation, Italy), and the soluble protein fraction was obtained by centrifugation at 4,000 rpm for 10 min at 4°C.
In the case of M9-PE cultures, the separated supernatant was filtrated to remove the residual PE using 0.45 μm filters (Millipore, Italy). The obtained cell-free supernatant fraction (CFS) was lyophilized (LIO5P Digital, 5Pascal, Italy) to maintain the enzymatic activity and resuspended in 50 mM PPB in a twenty-fifth of the initial volume. R. erythropolis D4 culture was also assessed for laccase activity.
Laccase enzymatic activity was determined as described by Zampolli et al. (2021) at 470 nm (ε: 36 mM−1 cm−1), also by adding 100 μM CuSO4. The presence of CuSO4 could enhance laccase activity, as observed by Santo et al. (2013) and Zampolli et al. (2023). One unit of enzyme activity is defined as the amount of enzyme that oxidizes 1 μmol per minute under the assay conditions. The specific activity was calculated as U mg−1 and reported as the mean of three replicates ± standard deviation (SD).
The total protein concentrations of the lyophilized CFS and cell extracts were assessed using the method of Bradford (1976) using Coomassie brilliant blue with bovine serum albumin as a standard. Protein concentrations were calculated from the standard curve by 20 μg mL−1 bovine albumin serum. All the experiments were performed in triplicate and shown as mean ± SD.
2.3 Kinetics of growth on PE
A biodegradative assay was performed to evaluate the ability to grow and degrade UV-sterilized untreated PE of the novel selected isolates, P. citronellolis strain E5 and R. erythropolis strain D4. Each bacterial culture was established in 50 mL flasks using liquid M9-PE (powder), and they were sampled at different times within a 60-day period (0 days, 3 days, 7 days, 14 days, 28 days, and 60 days) to evaluate the optical density and the total amount of bacterial cells using the colony-forming unit (CFU) count method by serially diluting 0.1 mL of each bacterial culture in a solution of M9 medium and plating the diluted suspension on LB agar medium. The plates were incubated at 30°C for 48 h. Both OD600 and CFU ml−1 data are reported as the means of three biological replicates with SD.
P. citronellolis E5 and R. erythropolis D4 were preliminarily tested for their ability to grow on metabolic intermediates of PE degradation, such as alkanes, ketones, and carboxylic acids. The growth assays were established in 20 mL flasks with 1 g/L n-dodecane, n-hexadecane, 2-hexadecanone, hexanoic acid, or dodecanoic acid to measure the optical density (each culture was established from OD600 ∼ 0.1) up to 72 h.
2.4 Biodegradation of polyethylene by GC-MS analyses
PE biodegradation was evaluated on the 20 mL M9-PE flask whole cultures, as reported in Section 2.1. M9-PE flasks without inoculum were used as abiotic controls. The biodegradation was monitored for up to 60 days (0 days, 3 days, 14 days, 28 days, and 60 days) by extracting the metabolic products from bacterial cultures with half a volume of dichloromethane (DCM) by manually shaking the separation funnel for 10 min and repeating the extraction twice. The samples were derivatized as reported in Zampolli et al. (2024) before injecting 1 µL in a 6890 N Network gas chromatograph (GC) system (J&W DB-5ms 60 m × 0.25 mm, 0.25 μm Ultra Inert GC Column, Agilent Technologies, Santa Clara, CA, United States of America) with helium at 99.99% as a carrier gas, coupled to a 5973 Network Mass Selective Detector (MSD, Agilent Technologies) at 70 eV in the scan ion monitoring mode (40–600 Da). The instrument was set as described in Zampolli et al. (2023). MSD ChemStation E.02.02.1431 (Agilent Technologies, Santa Clara, CA, United States of America) was used to analyze the resulting chromatograms. Unknown products were identified by comparing their mass spectra with the NIST11 database as long as their similarity with respect to the reference mass spectra was equal to or greater than 90%. The products obtained in each condition were classified according to different chemical classes, their peak areas were area normalized, and the proportion of different product types was calculated according to the “compound peak area/total compounds peak area × 100” (Zampolli et al., 2023). All the experiments were carried out in triplicates.
2.5 Nucleic acid extraction and manipulation
The total DNA was extracted from bacterial cells grown on M9-PE by the DNeasy UltraClean Microbial Kit (Qiagen, Italy) following the manufacturer’s instructions.
Long Range DNA Rabbit Polymerase (2.5 U μl−1, Eppendorf, Germany) was used for 16S rRNA gene amplification using the universal bacterial primers 27 F (AGAGTTTGATCCTGGCTCAG) and 1495 R (CTACGGCTACCTTGTTACGA) (Lane, 1991) (melting temperature, Tm 55°C) with the following thermocycling conditions: at 95°C for 2 min, 95°C for 30 s, specific Tm for 30 s, 72°C for 1 min and 45 s, for 30 cycles; and 72°C for 7 min. One-fifth of the PCR product was verified on 0.8% agarose gel using a 500–10,000 bp molecular ladder (Merck, Italy). Then, the PCR product was purified from the agarose gel with the kit NucleoSpin Gel and PCR Clean-up (Macherey–Nagel, Dueren, Germany), and the purified product was sequenced by the Sanger method. The nucleotide sequence of the 16S rRNA gene was obtained by comparing forward and reverse sequences, and the final gene fragment was subsequently compared to the NCBI database with BLASTn (Altschul et al., 1990) and to the SILVA database (Quast et al., 2013).
2.6 Genome sequencing and preliminary analyses
The total DNA of P. citronellolis E5 was sequenced by Illumina MiSeq v3 (2 × 300 bp; Illumina Inc., San Diego, CA). The library was obtained using Nextera XT DNA Library Preparation Kit with the standard Illumina DNA shotgun library protocol (Illumina Inc., San Diego, CA). The total reads of the P. citronellolis E5 genome resulted in 1.48 million (total base pairs around 414.4 Mbp). Trimmomatic v.0.38 (Bolger et al., 2014) was applied for the trimming process to discard the sequences with a per base sequence quality score of less than 30. The quality of the genome was evaluated using FastQC v0.11.7 (Andrews, 2010) before and after the trimming process. The resulting sequences were then assembled into contigs using Spades 3.15.5 (Bankevich et al., 2012). MeDuSa v1.6 enabled refining and assembling the contigs into scaffolds (Bosi et al., 2015), using the complete genomes of the following Pseudomonas strains as references: P. citronellolis WXP-4 (NCBI BioProject PRJNA511406), Pseudomonas putida KT2440 (PRJNA836759), and Pseudomonas hydrolytica DSWY01 (PRJNA509263).
Genome assembly quality (completeness) was determined considering the total assembly length (expected to be ∼ 7 Mb), and contiguity metrics were assessed at the contig level using QUAST 5.0.1 (Gurevich et al., 2013).
The genome was automatically annotated using the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al., 2008), Prokka (v. 1.14.6) (Seemann, 2014), and Bakta (v. 1.8.2) (Schwengers et al., 2021). The Comprehensive Antibiotic Resistance Database (CARD) and Resistance Gene Identifier (RGI; v. 6.0.3), CRISPR/Cas Finder (v. 4.2.20), and Phigaro (v. 2.3.0) were utilized to predict genes for antibiotic resistance (Alcock et al., 2020), for clustered regularly interspaced short palindromic repeats (CRISPR) (Couvin et al., 2018), and to detect and annotate prophage regions (Starikova et al., 2020). The main genomic features of P. citronellolis E5 and its comparison with R. erythropolis D4 genome were visualized by the Proksee viewer (https://proksee.ca/) (Accessed on 5 July 2024) (Grant et al., 2023).
EggNOG automatic classification was applied to evaluate the clusters of orthologous groups (COGs) for P. citronellolis E5 coding sequences (CDSs).
The genome sequencing data have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB76139.
2.7 Phylogenetic analyses
In order to evaluate the phylogenesis of P. citronellolis E5 and R. erythropolis D4, a multi-locus sequence analysis (MLSA) was carried out by the Clustal W package of MEGA-X (version 10.2) using the default parameters (Kumar et al., 2018). MLSA was used to build the concatenated sequences, then inferred for tree development using the maximum likelihood (ML) method with a gamma distribution of mutation rates with gamma optimized to 2, 100 bootstrap replicates, and the Nearest-Neighbor-Interchange algorithm. The 16S rRNA, rpoD, and ychF marker genes were selected based on annotation and similarity; the gyrB gene for the Pseudomonas genus tree and the secY gene for the Rhodococcus genus tree were also selected. They were previously identified as conserved and informative for other bacterial classifications (Adékambi et al., 2011; Orro et al., 2015).
The phylogenetic tree of 14 Pseudomonas species (including P. citronellolis E5) was built using a dataset containing the following bacterial species: P. citronellolis (8), Pseudomonas knackmussii (2), P. putida (1), Pseudomonas lalkuanensis (1) P. hydrolytica (1), and one not-categorized species. The phylogenesis of 15 Rhodococcus species (including R. erythropolis D4) was developed with the following species: R. erythropolis (6), Rhodococcus qingshengii (1), Rhodococcus ruber (2), Rhodococcus opacus (3), Rhodococcus jostii (1), and two not-specified species. The strain species were selected based on the genome sequence availability.
2.8 Genomic comparison and genetic trait prediction in P. citronellolis E5 and R. erythropolis D4
Sequence and functional comparisons were performed between P. citronellolis E5 and R. erythropolis D4 utilizing a sequence-based and functional comparison tool on the RAST server. Average nucleotide identity (ANI) was also calculated by FastANI (v. 1.3.3) and the ANI calculator (Jain et al., 2018; Yoon et al., 2017; Liang et al., 2021). Enzyme functions and metabolic pathways were predicted by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database by the KEGG Automatic Annotation Server (KAAS), providing functional annotation of genes by BLAST (Kanehisa et al., 2023).
The prediction of gene products potentially involved in PE biodegradation for P. citronellolis E5 and R. erythropolis D4 was conducted with two converging approaches. The first required the search into their genomes by annotation based on literature knowledge about PE metabolism and gene products retrieved from NCBI. The main enzymatic classes obtained by this approach were multicopper oxidases, peroxidase, lipases, esterases, alkane monooxygenases, and cytochrome P450 hydroxylases. A second method was amino acid (aa) sequence alignments carried out by Clustal Omega relying on annotation identifications and the selection of reference aa sequences (RASs) (Supplementary Table S1); 35 RASs were chosen to represent key steps of the PE biodegradative pathway, and they were clustered in a phylogenetic tree (Supplementary Figure S2). They were used in pairwise comparison with the retrieved aa sequences from the annotation search (first approach), selecting the gene products with the highest identity for comparison against the E5 and D4 genomes (second approach). The resulting gene products were also evaluated for the presence of aa translocation signal peptides using SignalP (v. 6.0) (Teufel et al., 2022) to predict their potential ability to be secreted in the extracellular environment.
The gene products obtained from these analyses were aligned by the Clustal Omega program using the default parameters of the multiple sequence alignment (MSA) tool (Sievers et al., 2011) with the same setting reported by Zampolli et al. (2024). In turn, a cluster tree was built using the selected RASs for each strain (E5 and D4). MSA served as input in MEGA software (version 10.2) by applying the maximum likelihood (ML) method (Kumar et al., 2018), with a JTT substitution matrix and a gamma distribution of mutation rates with gamma optimized to 2, inferring a phylogeny test with 50 bootstrap replicates.
3 Results
3.1 Isolation of bacterial strains from plastic-contaminated organic waste
A solid organic waste rich in plastic particles was obtained from an organic waste treatment plant. It exhibited a total microbial viable count of heterotrophic bacteria of around 2 × 106 CFU mL−1. The isolation of novel plastic-degrading bacterial strains was performed by 7-day growth enrichment cultures supplemented with plastic-rich waste (Supplementary Figure S1). Thirty single bacterial colonies isolated from enrichment cultures and plated on LB agar medium were preliminarily screened for their ability to grow on PE agar plates (M9-Tw-PE). Seven isolated bacterial strains were able to grow on M9-Tw-PE plates for three subsequent repetitions. The isolates, named A, B, C, E5, F, H, and I, were characterized by a Gram stain, which revealed that A and E5 belonged to the Gram-negative group, and the remaining isolates belonged to the Gram-positive group.
Then, the isolates were tested for their ability to grow in the presence of 1% PE as the sole carbon and energy source (M9-PE). In addition, the PE degradative ability of R. erythropolis D4 was also assessed because of its intriguing potential for plastic biodegradation, evidenced in previous studies (Zampolli et al., 2022; Zampolli et al., 2023). Figure 1 shows their growth assay measuring the OD600 for up to 7 days. At the final time of growth, the maximum OD600 level was ∼ 0.6, recorded for E5 (0.57 ± 0.01) and F (0.58 ± 0.02) isolates, compared to D4 growth (0.72 ± 0.08). Instead, the minimum level of growth was registered for the B (0.35 ± 0.06) and I (0.25 ± 0.09) isolates. Therefore, the isolates A, C, E5, F, and H were selected for subsequent investigations.
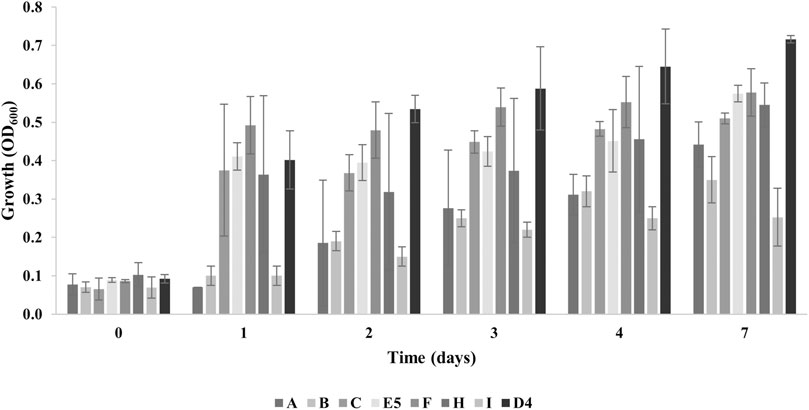
Figure 1. Growth levels of individual bacterial isolates (A, B, C, E5, F, H, and I) from plastic-contaminated organic waste compared to R. erythropolis D4 on 1% PE. The growth is presented as the mean value of the absorbance recorded at OD600 ± SD.
3.2 Screening of laccase activity during growth of plastic-degrading strains on untreated polyethylene
An enzymatic assay was used to evaluate the ability of the five selected isolated bacterial strains to secrete or produce intracellular oxidase enzymes for PE oxidation after 3 days of growth on PE, according to the hypothesis that they could activate laccase-like enzymes to perform the first oxidation of PE. The laccase activity was also assessed after the growth of the selected strains in M9-M as a reference condition. Both intracellular and CFS measurements showed a laccase activity of around one order of magnitude less than the respective conditions in M9-PE. The enzymatic assay was performed using 2,6-dimethoxyphenol (2,6-DMP) as one of the typical substrates for laccase activity both in the presence and in the absence of CuSO4 because copper is known to be a good enhancer of laccase activity (Santo et al., 2013; Zampolli et al., 2023).
Figures 2A, B show the laccase activity respectively secreted and intracellularly produced by the selected isolates A, C, E5, F, and H and R. erythropolis D4. For both cellular compartments, the highest activity was recorded in the presence of CuSO4 with a difference of at least one order of magnitude (CFS) or two to three orders of magnitude (cellular extracts) with respect to the condition of absence of CuSO4. Considering the condition with copper, the highest laccase activity was recorded in the CFS for all the tested strains. Among the bacterial isolates, the highest values were measured in CFS for E5 ((2±1) *10–3 U mg–1), F (2.3±0.1) *10–3 U mg-1), and R. erythropolis D4 ((3±1) *10–3 U mg–1) compared to intracellular values of around 10–4 U mg−1.
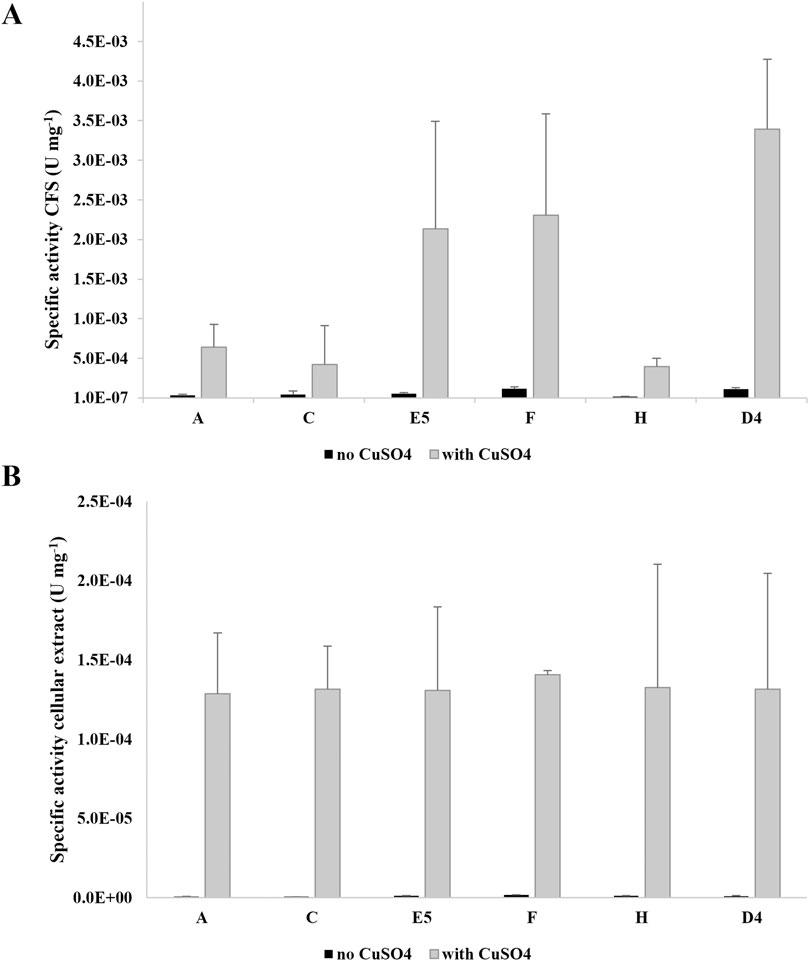
Figure 2. Specific laccase activity (U mg−1 of total proteins) of the supernatants (CFS) (A) and cellular extracts (B) of new isolates A, C, E5, F, and H and R. erythropolis D4. Laccase activity was measured in the presence of 2,6-DMP after growth on M9-PE in the absence (light gray) and in the presence of 100 µM CuSO4 (gray) added during the enzymatic assay. The laccase activity is the mean of three replicates ± SD.
These results indicated that all the isolates were able to secrete active laccase-like enzymes during growth on PE.
3.3 Molecular identification of five isolates and phylogenetic analysis of Pseudomonas citronellolis E5 and Rhodococcus erythropolis D4
The five selected isolated bacteria were identified based on 16S rRNA gene amplification and sequencing. Silva analyses revealed that the 16S rRNA gene of the A, C, E5, F, and H isolate strains shared 99% identity with Acinetobacter lwoffii, 98% identity with Priestia/Bacillus megaterium, 98% identity with P. citronellolis, 98% identity with Rhodococcus sp., and 98% identity with Bacillus sp., respectively. Thus, considering the preliminary growth assays in M9-PE, the highest values of laccase activity were reported for P. citronellolis E5 and Rhodococcus sp. F. P. citronellolis strain E5 was selected for subsequent investigations of its PE biodegradative potential because its taxonomy is distant with respect to R. erythropolis D4, chosen for its promising genetic repertoire for polyolefin degradation (Supplementary Figure S3). The phylogenesis of these two selected strains was further evaluated by an MLSA and building a phylogenetic tree using the ML method considering, respectively, 13 Pseudomonas and 14 Rhodococcus species. The selected marker genes for the two different trees were identical (16S rRNA, rpoD, and ychF) except for the gyrB gene for the Pseudomonas genus and the secY gene for Rhodococcus.
Figure 3 shows the two distinct phylogenetic trees for P. citronellolis E5 (Panel A) and R. erythropolis D4 (Panel B). The clade of the E5 strain primarily includes P. citronellolis G5.41a and P. citronellolis G5.80. The taxonomical evaluation of R. erythropolis D4 validates that its closest neighborhood includes R. jostii, R. opacus, and other R. erythropolis strains.
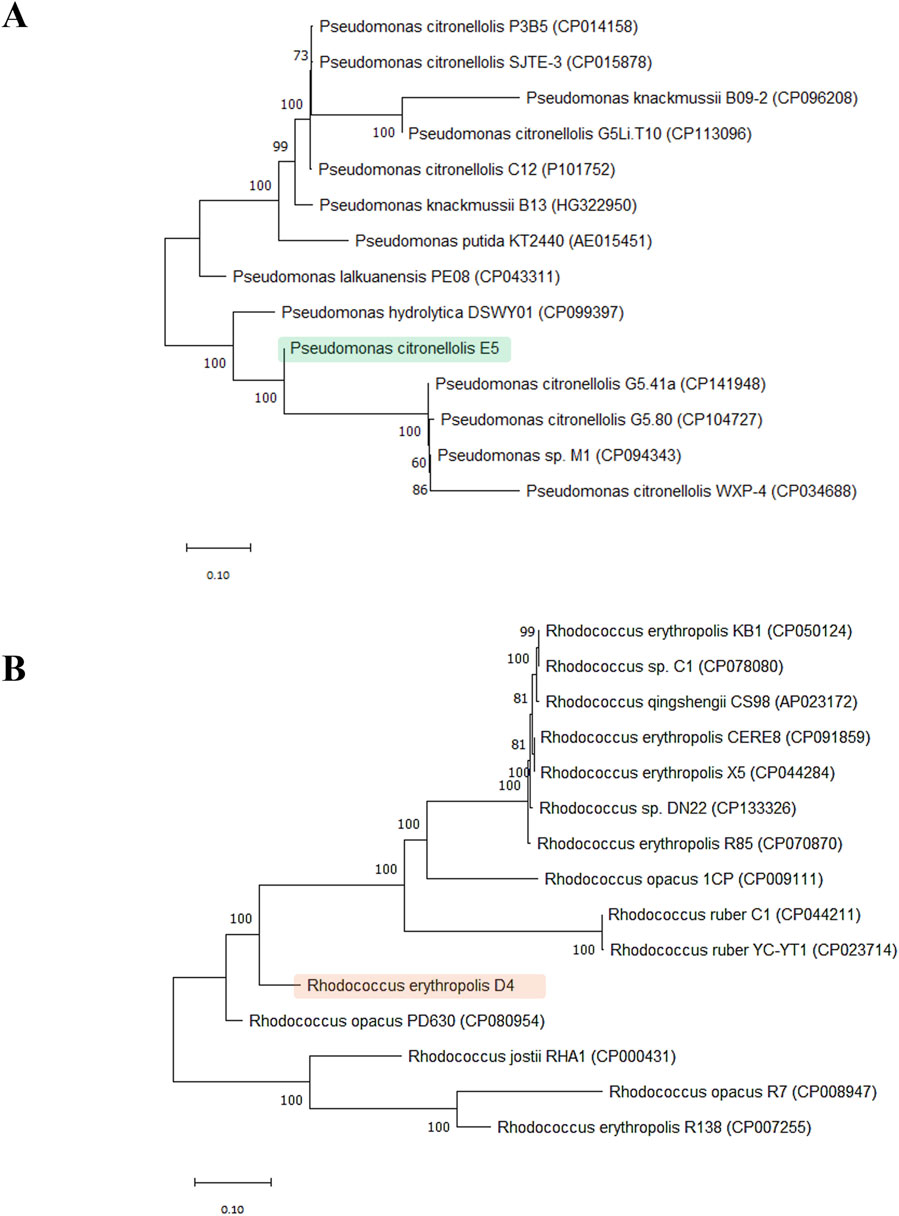
Figure 3. Phylogenetic trees. Phylogenetic analysis of P. citronellolis E5 (A) and R. erythropolis (B) based on sequence alignments with reference strains for each genus (13 Pseudomonas and 14 Rhodococcus species). The tree was constructed based on concatemer sequences of four marker genes: 16S rRNA, rpoD, ychF, and gyrB for the Pseudomonas genus and secY for the Rhodococcus genus.
3.4 Kinetic growth of Pseudomonas citronellolis E5 and Rhodococcus erythropolis D4 on untreated polyethylene
Based on the preliminary laccase screening after growth on PE, individual growth assays of the two selected strains in the presence of 1% PE as the sole carbon and energy source were performed for up to 60 days. Figure 4 shows the kinetic growth profile of P. citronellolis E5 (panel A) and R. erythropolis D4 (panel B) by optical density at OD600 and viable cell count. Considering the bacterial cell counts, the maximum level was reached at 7 days 4.0 ± 0.4 × 108 CFU mL−1 and 5.0 ± 0.1 × 108 CFU mL−1 for E5 and D4, respectively. However, only the D4 strain evidenced growth of around one order of magnitude in 7 days. For both strains, the density of cells underwent a slight decrease after the first week of growth. The maximum OD600 was reached at 60 days and 28 days for E5 (OD600 ∼ 0.53 ± 0.2) and D4 (OD600 ∼ 0.42 ± 0.06), respectively. These variations do not correspond to the value of total viable cell count because R. erythropolis D4 is prone to aggregate during growth on hydrophobic substrates (Zampolli et al., 2014). Because PE supports the growth of these selected bacteria, other evidence of PE catabolic degradation was investigated.
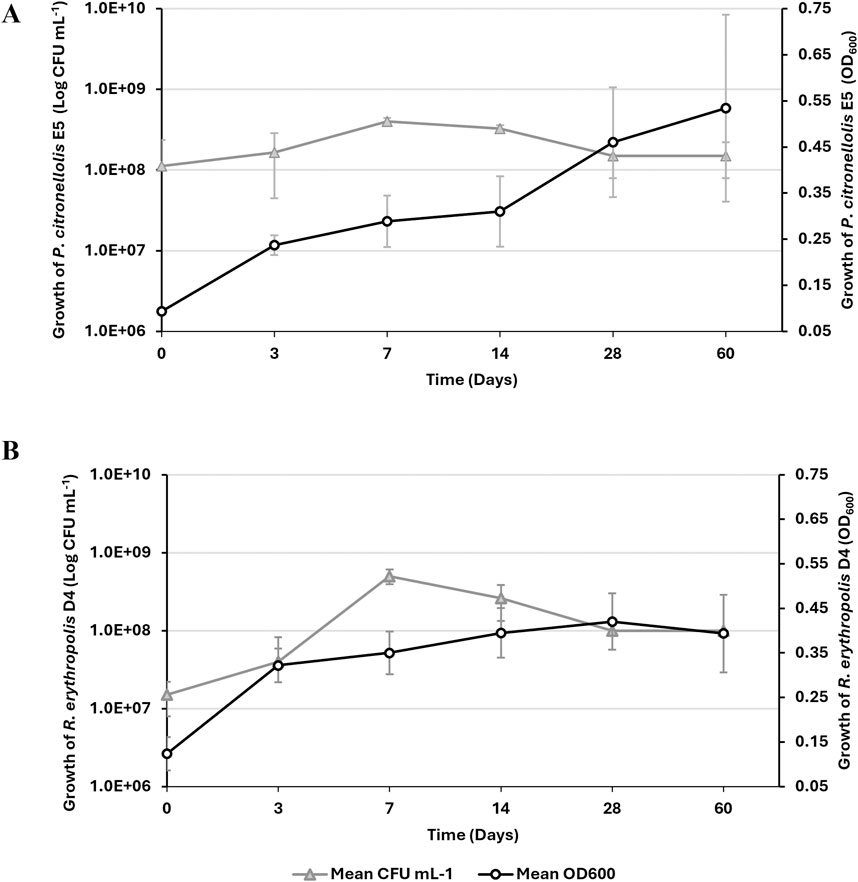
Figure 4. Growth kinetic assay of P. citronellolis E5 (A) and R. erythropolis D4 (B) on M9-PE up to 60 days. Growth is reported as the mean value of the absorbance recorded at OD600 (white circle) or counts of live bacterial cells expressed as CFU mL−1 (gray triangle) ± SD.
3.5 Biodegradation products of untreated polyethylene by GC-MS analyses
The biodegradation of PE for each M9-PE exhausted bacterial culture was recorded at 0 days, 3 days, 14 days, 28 days, and 60 days by evaluating the residual metabolic products extracted with DCM through GC-MS analyses. The resulting chromatographic profiles were compared to the ones obtained on untreated PE powder (UV-sterilized) that was not exposed to any (a)biotic degradation (no inoculum).
Unprocessed commercial PE powder (t0) was analyzed beforehand, evidencing mostly ketones with carbon chain lengths mainly ranging between C12 and C25, alkanes mainly ranging between C15 and C25, and carboxylic acids with C16 and C21 carbon chain lengths (Table 1).
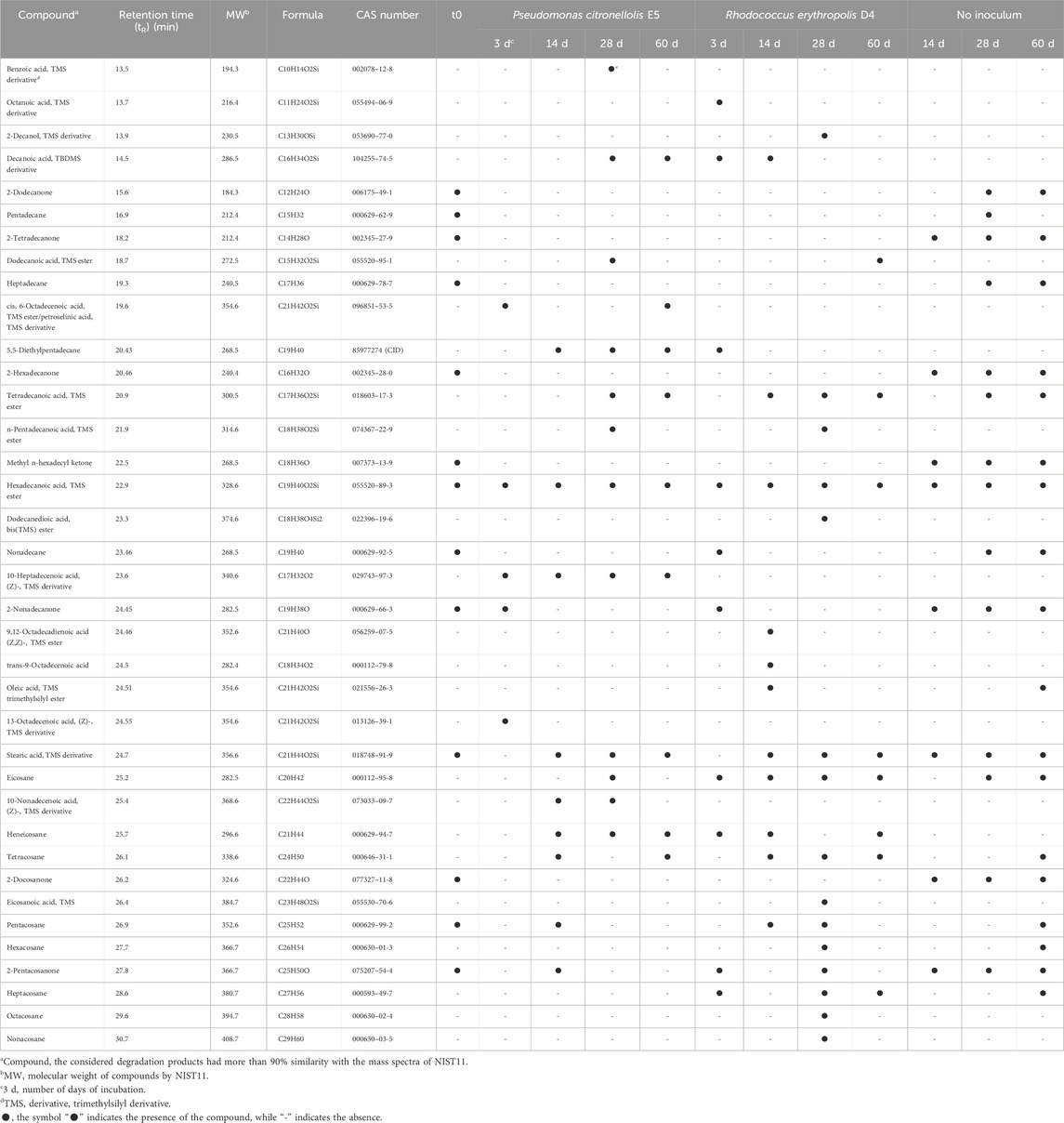
Table 1. Metabolic products detected by GC-MS analyses deriving from untreated commercial PE biodegradation by P. citronellolis E5 and R. erythropolis D4.
The compounds released by the two strains at each time point are dissimilar as well as their production pattern (Table 1; Figure 5). After a 3-day incubation in M9-PE, an increase of carboxylic acids was detected. The main difference is the presence of a higher number of carboxylic acids for P. citronellolis strain E5, showing carboxylic acids with a carbon chain length of C16 and C18 and a monounsaturated fatty acid (petroselinic acid, trimethylsilyl (TMS) derivative, TR = 19.6 min) (Table 1; Figure 5). In contrast, R. erythropolis D4 shows carboxylic acids with shorter carbon chain lengths (C8, C10, and C16). The number of ketone types decreased compared to the initial condition for both strains, while only R. erythropolis D4 produced alkane products ranging between C20 and C27 of carbon chain length. All these products indicated an initial oxidative process.
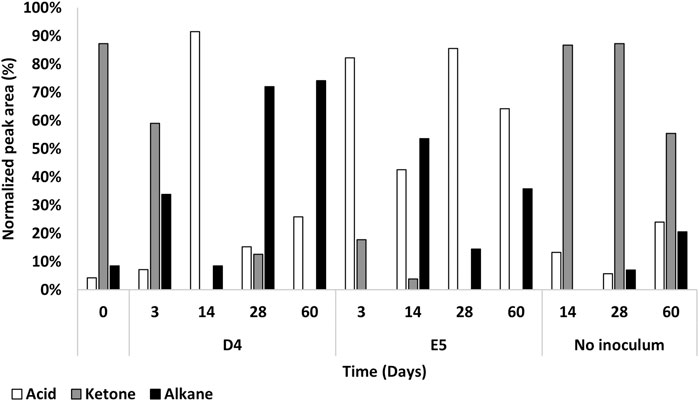
Figure 5. Percentage of normalized peak area of product types from P. citronellolis E5 and R. erythropolis D4 oxidation of PE with respect to the control (no inoculum) up to 60 days by GC-MSD analysis. The main product species after PE oxidation were carboxylic acids (white), ketones (gray), and alkanes (black).
At 14 days, the trend of the number of product types and normalized peak area of fatty acids and alkanes inverted for the two strains; carboxylic acids increased, and alkanes decreased in the D4 culture, while the opposite was observed for the E5 culture.
After 28 days of cultivation, the number of product types and normalized peak area of fatty acids and alkanes inverted in the two strain cultures with respect to the previous sampling time. In addition, this pattern of metabolic products was stable at the successive time point (60 days) with a few differences in the types of oxygenated products (Figure 5). This indicates an oscillatory metabolism in which every oxidized product is subsequently degraded. After 1 month, R. erythropolis D4 shows an increase in aliphatic hydrocarbons with a higher number of carbon chain lengths (up to C29) with respect to P. citronellolis E5 (up to C21), and both strains show a diversity of carboxylic acids in addition to the hexadecanoic acid and octadecanoic acid that are always present. Interestingly, the presence of benzoic acid in the P. citronellolis E5 culture extract was indicative of a potential plastic antioxidant added to the PE released from its structure after 28 days (Zampolli et al., 2023; Yao et al., 2022; Sanluis-Verdes et al., 2022).
Finally, the not-inoculated condition showed the highest percentage of ketone types in all time points, as observed in the initial condition, and the variety of oxidized products is quite stable over time. This indicates no differences in terms of chromatographic profile between the initial time and the condition without inoculum (Supplementary Figure S4).
In order to support these findings, P. citronellolis E5 and R. erythropolis D4 were preliminarily tested for the ability to grow on the most representative metabolic intermediates of PE biodegradation, such as n-dodecane, n-hexadecane, 2-hexadecanone, hexanoic acid, or dodecanoic acid, as the sole carbon and energy source. Within 72 h (the first sampling time of strain grown on M9-PE), the highest growth level of D4 strain was on 2-hexadecanone, followed by n-hexadecane and hexanoic acid. Lower growth values were reported for n-dodecane and dodecanoic acid. For P. citronellolis E5, the highest growth levels were reported in the presence of hexanoic acid and dodecanoic acid. In contrast, in the presence of the tested alkanes and ketone, E5 growth levels were lower than the other substrates (Supplementary Figure S5).
3.6 Genome sequencing of P. citronellolis E5 and comparison with R. erythropolis D4 genome
The genome of P. citronellolis E5 was completely sequenced by Illumina MiSeq v3 (2 × 300 bp), obtaining 1.48 million reads (total base pairs around 414.4 Mbp). The sequencing quality showed that the shortest reads were ∼280 bp long, and the average genome coverage was 111X. After read quality trimming, the remaining reads with 70X coverage were assembled into 244 contigs with an N50 length of 313,394 bp. Contigs were assembled into 120 scaffolds by MeDuSa v1.6 using three reference genome sequences of bacteria of the Pseudomonas genus, resulting in a 7,075,332 bp genome size and an average G-C% of around 67%.
Automated annotation analysis of P. citronellolis E5 genome sequences was obtained using the RAST server, identifying a total of 6,527 open reading frames (ORFs), in agreement with the results obtained from the automated gene prediction and annotation by Prokka and Bakta software, which predicted 6,191 ORFs and 6,275 ORFs, respectively. The general genome analysis by RAST evidenced 72 RNA genes (12 rRNAs and 60 tRNAs) and 1,648 hypothetical proteins (HP).
P. citronellolis E5 CDSs were classified by EggNOG Automatic Classification based on clusters of COGs. The assigned COGs with the highest number of CDSs were amino acid metabolism and transport (E), 10%; transcription (K), 10%; inorganic ion transport and metabolism (P), 8%; energy production and conversion (C), 7%; unknown function (S), 16% (Figure 6).
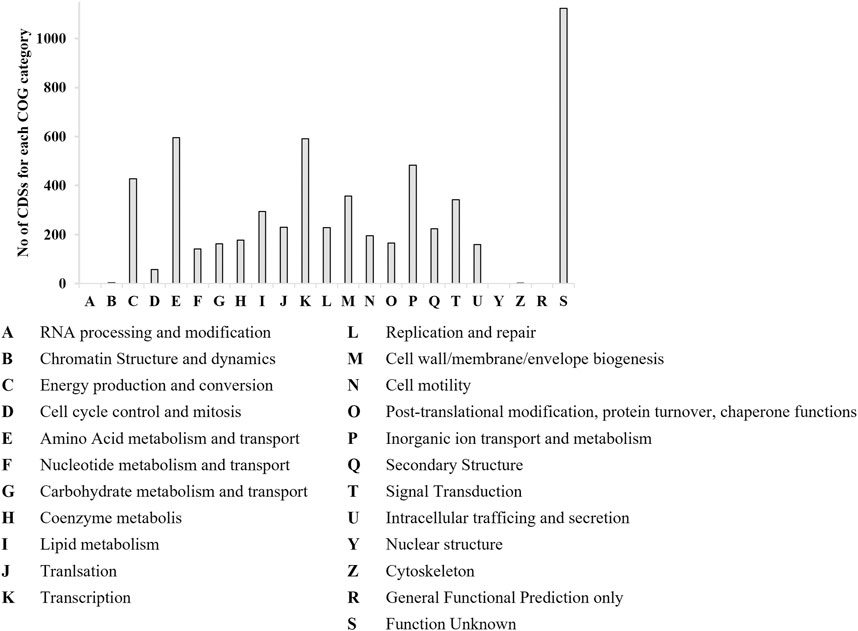
Figure 6. Number of CDSs populating each COG category by EggNOG categorization for P. citronellolis E5. The x-axis includes the abbreviations for each COG category.
The main genome features of the E5 strain are depicted in Figure 7 showing G-C content, rRNA, tRNA, and tmRNA genes encoding for phages, replication or repair, transfer, stability/defense, integration, and excision, CRISPR-Cas, and antibiotic-resistant genes (ARG). In addition, putative horizontal gene transfer (HGT) events were predicted. In the same figure, the R. erythropolis D4 genome is depicted in the first outermost ring, indicating the BLAST comparison between the two genome sequences. This representation showed that a few CDSs have a higher identity percentage than 82%; thus, comparative analyses between the P. citronellolis E5 and R. erythropolis D4 genomes were carried out. The nucleotide-level genomic similarity (ANI) computed with different methods (Jain et al., 2018; Yoon et al., 2017; Liang et al., 2021) of E5 vs. D4 genomes ranges between 65% and 68% (Figure 8). The comparative analyses obtained using the RAST server indicated that the closest neighbors of P. citronellolis E5 genome are Pseudomonas aeruginosa PAO1 (score 547), P. putida KT2440 (score 524), Pseudomonas syringae pv. phaseolicola 1448A (score 523), and Pseudomonas fluorescens PfO-1 (score 514).
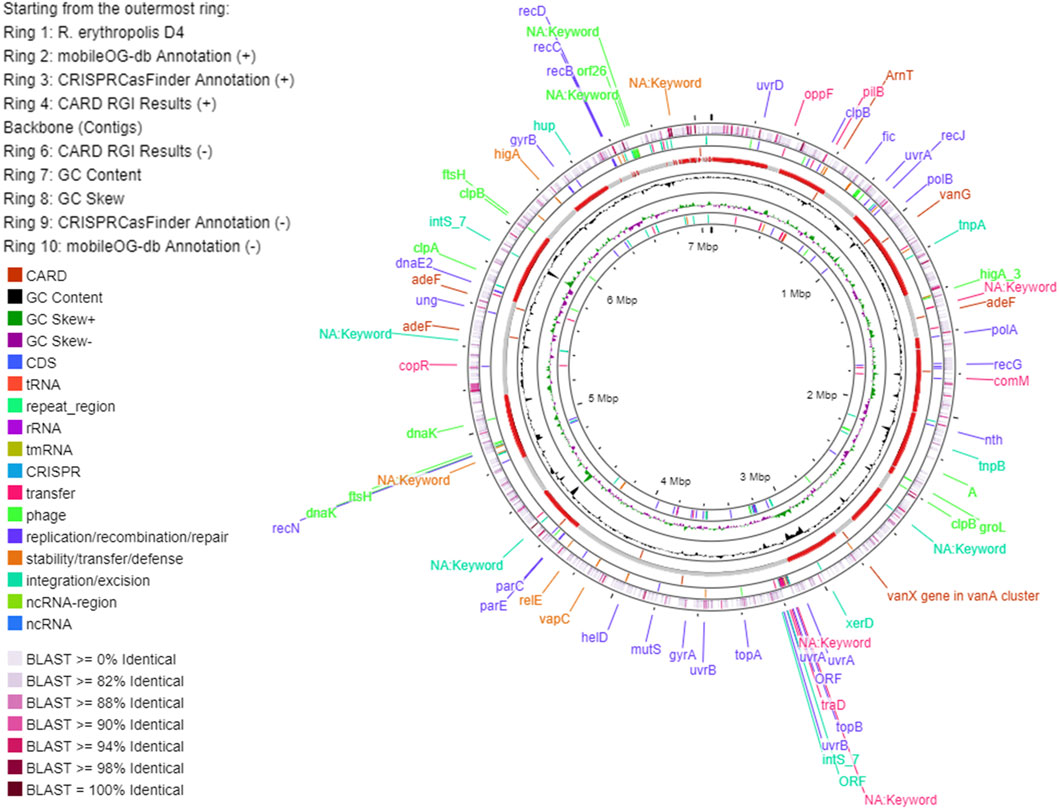
Figure 7. Circular map of P. citronellolis E5 genome predicted with Proksee viewer (https://proksee.ca/). The first outermost ring represents the BLAST comparison of the E5 genome with the genome of R. erythropolis D4 according to the sequence identity.
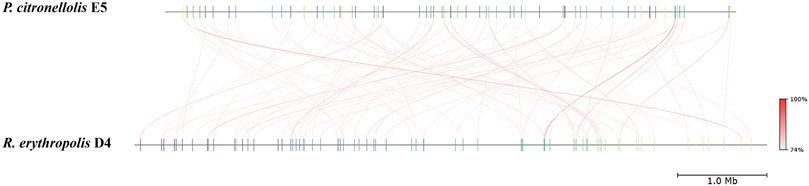
Figure 8. Pairwise comparison of P. citronellolis E5 and R. erythropolis D4 by whole-genome average nucleotide identity (ANI). Each strain genome is linearly depicted. The red lines indicate the correspondence between similar regions of the two genomes (95% threshold), thus representing the relatedness of the genome sequence. The heat map represents the ANI value for each orthologous match.
Therefore, pairwise sequence-based and functional comparisons of P. citronellolis E5 vs. R. erythropolis D4 and vs. P. aeruginosa PAO1 genomes were also carried out through RAST. As expected, the highest number of CDSs (644 genes) with more than 90% identity were shared between E5 and PAO1 genomes, which also showed 27% of shared functional gene categories. Instead, E5 vs. D4 genomes shared 17% of functional genes, and 302 genes of the E5 genome shared 50% identity compared to R. erythropolis D4.
The RAST annotated CDSs of the P. citronellolis E5 genome over 386 subsystems shared in 25 categories (Overbeek et al., 2005) are 43%, among which amino acids and derivatives (518), carbohydrates (310), cofactors, vitamins, prosthetic groups or pigments (217), protein metabolism (214), and metabolism of aromatic compounds (203) are the most populated (Figure 9). A similar amount of CDS distribution in RAST subsystem categories is observed in the D4 genome, for which 37% of CDSs are categorized, except that fatty acids, lipids, and isoprenoids is the third-most-populated category.
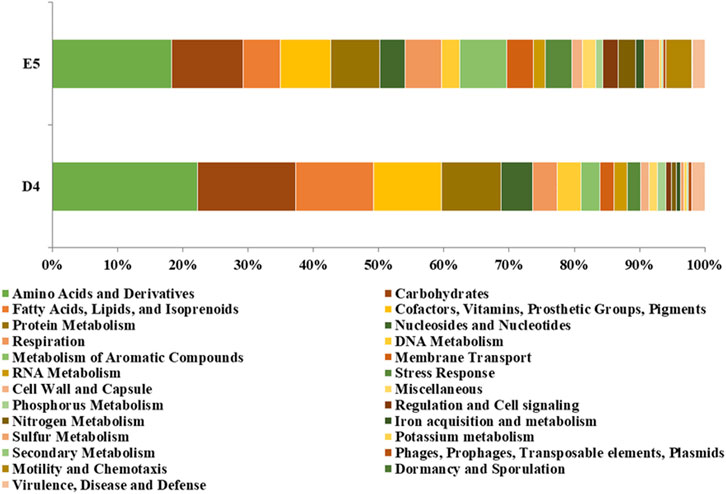
Figure 9. Percentage of CDSs in each RAST subsystem category for P. citronellolis E5 and R. erythropolis D4.
3.7 Genetic trait prediction in P. citronellolis E5 and R. erythropolis D4 genomes for PE biodegradation
The whole-genome comparative analysis of P. citronellolis E5 and R. erythropolis D4 showed distinctive traits. Two approaches to gene predictive mining were undertaken for P. citronellolis E5 and R. erythropolis D4 to predict the main functions involved in PE biodegradation. The first one involved a genome-based search focused on literature reports, knowledge about the main steps of the PE metabolism, and gene products retrieved from NCBI relying on annotation and aa sequence alignments. The annotation search extracted the following enzymatic classes: laccase-like multicopper oxidases, peroxidase, alkane monooxygenases, cytochrome P450 hydroxylases, lipases, and esterases. The second approach included the selection of 35 reference aa sequences (RASs) to frame the subsequent clusterization of gene products and to use as input against the E5 and D4 genomes (Supplementary Table S1; Supplementary Figure S2). The generated RAS tree is divided into ten clades (Supplementary Figure S2): clades I, II, III, IV, and V include (alkane) monooxygenases from different genera (Rhodococcus, Acinetobacter, Pseudomonas), clades VI and VII include respectively lipases and esterases, and clades VIII, IX, and X comprise cytochrome P450 hydroxylases, peroxidases, and multicopper oxidases-laccases, respectively. Thus, the selected 35 RASs were compared against the E5 and D4 strain genomes. The retrieved aa sequences from the annotation search (first approach) were also compared against 35 RASs. Overall, the aa sequences potentially involved in PE biodegradation according to the proposed metabolic pathway showing the highest identity percentage were selected (Supplementary Tables S2, S3). For each strain, the resulting gene products (16 for E5 and 42 for D4) deriving from these two approaches were aligned, and a cluster tree was developed showing respectively ten and twelve clades according to the RAS (aa sequence named R) clusterization (Figure 10A, B). The first metabolic step envisages laccase-like multicopper oxidases (Gravouil et al., 2017; Zampolli et al., 2021), peroxidases (Rong et al., 2024), esterases (Tao et al., 2023), or lipases (Kim et al., 2024) acting in the extracellular environment for the preliminary oxidation of PE. For this reason, only the gene products predicted to be potentially extracellular-secreted were chosen. P. citronellolis E5 evidenced two distinct multicopper oxidases (clades V and X Figure 10A), but only P1 (annotated as multicopper oxidase) showed similarity with COG2132 (multicopper oxidase with three cupredoxin domains), while P2 grouped alone (multicopper polyphenol oxidase). On the other hand, R. erythropolis D4 possesses five gene products annotated as multicopper oxidases that were distributed into clades VI and VII (Figure 10B). In addition, P6 was annotated as HP clustered with R9, suggesting that it could be an additional multicopper oxidase (clade V Figure 10B).
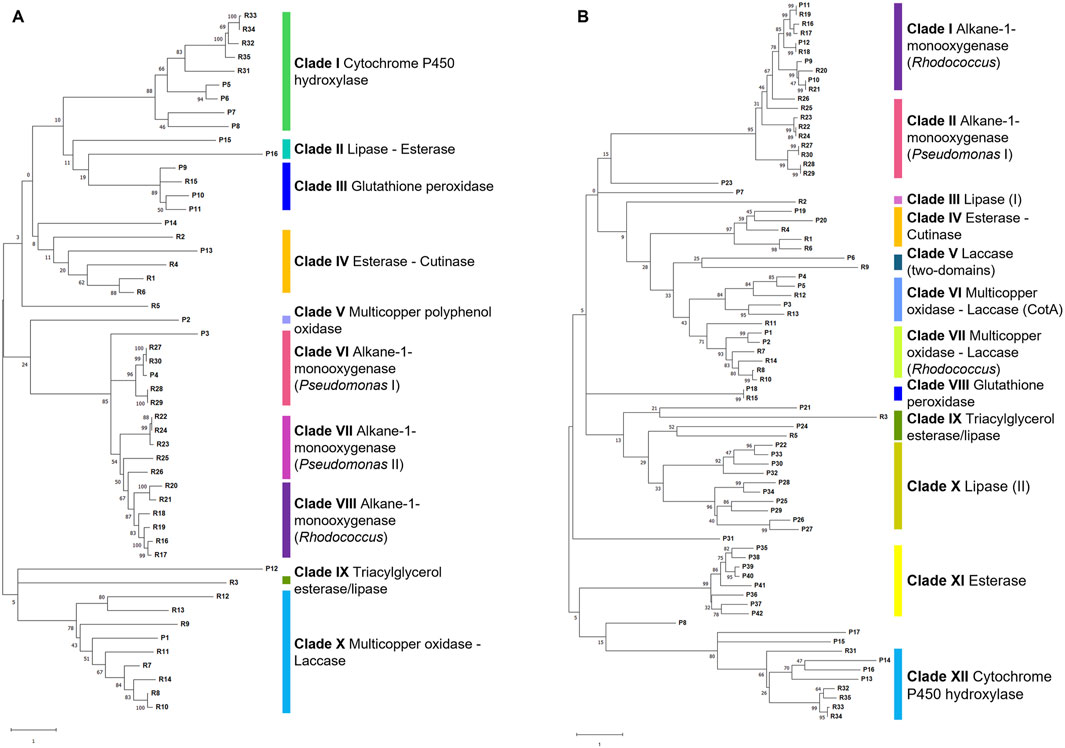
Figure 10. Cluster tree of selected gene products (named P) from P. citronellolis E5 (A) and R. erythropolis D4 (B) genomes against RAS (named R) (listed in Supplementary Table S2). Each clade is highlighted with a different color and named according to the RAS representing each clade. The clade number is in order from the top of each tree. The numbers on the branches represent the bootstrap values calculated for the ML method from the MEGA software with 50 bootstraps. Protein name abbreviations are reported in Supplementary Tables S2, S3 for P. citronellolis E5 and R. erythropolis D4, respectively.
The number of glutathione peroxidases varies for the two strains: P. citronellolis E5 and R. erythropolis D4 showed three glutathione peroxidases (COG0386, clade III Figure 10A) and one glutathione peroxidase (clade VIII Figure 10B), respectively.
Among 45 esterases and 11 lipases annotated in the E5 strain genome and 69 esterases and 29 lipases in the D4 genome, only 5 and 24 gene products were retrieved from the whole genome of, respectively, the E5 and D4 strains based on the two approaches. Only P13 of E5 and P19 and P20 of D4 share the highest identity to esterases R1, R4, and R6 (clade IV Figures 10A, B; Supplementary Table S1), and P24 of D4 is similar to triacylglycerol esterases/lipases R3 and R5 (clade IX Figure 10B; Supplementary Table S1). Regarding lipases, only clade II for the E5 tree (Figure 10A) and clade X in the D4 tree (Figure 10B) include potential lipases, even if they are clustered without RASs.
The subsequent transport of oxidized products is mainly carried out by membrane transporters (Gravouil et al., 2017; Zampolli et al., 2021). This route is feasible in the selected strains because P. citronellolis E5 and R. erythropolis D4 account for around 390 and 375 transport proteins (in the RAST server), respectively (Figure 9). Moreover, because ABC transporters have been reported to be involved in PE oxidized product transportation, the two genomes were scanned for these potential gene products, recovering 302 and 277 potential ABC transporters for the E5 and D4 strains, respectively.
The oxygenated or lower molecular weight products are then intracellularly oxidated by oxygenases such as alkane monooxygenases/hydroxylases and cytochrome P450 hydroxylases (Gravouil et al., 2017; Zampolli et al., 2021; Tao et al., 2023; Mishra et al., 2024; Rong et al., 2024). Their search showed two and four alkane monooxygenases and four and five cytochrome P450 hydroxylases, respectively, in the E5 and D4 genomes. Consistently, in E5 clusterization, only P4, an alkane monooxygenase, belongs to clade VI (alkane-1-monooxygenase - Pseudomonas I), and all four cytochrome P450 hydroxylases (P5-P8) to clade I (Figure 10A). In D4 clusterization, all four alkane monooxygenases (P9-P12) belong to clade I (alkane-1-monooxygenase - Rhodococcus), and all five cytochrome P450 hydroxylases belong to clade XII (cytochrome P450 hydroxylases) (Figure 10B).
Gene products involved in the central metabolic pathway of β-oxidation are also known to be involved in the last metabolic steps (Tao et al., 2023; Mishra et al., 2024). Both strains possess approximately 40% of the gene products required for the β-oxidation metabolism in the selected KEGG map (Supplementary Figure S6). Each functional step is associated with diverse EC numbers, including long-chain-fatty-acid-CoA ligase, oxidoreductases, enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, acetyl-CoA acyltransferase, 3-hydroxybutyryl-CoA epimerase, alcohol dehydrogenase, aldehyde dehydrogenase, and alkane 1-monooxygenase (EC: 6.2.1.3, 1.3.99.2, 4.2.1.17, 1.1.1.35, 2.3.1.16, 2.3.1.9, 5.1.2.3, 1.1.1.1, 1.2.1.3, and 1.14.15.3). A few differences between the two strains are evidenced in the β-oxidation metabolic pathway, which comprises dodecenoyl-CoA isomerase and rubredoxin reductase (EC numbers 5.3.3.8, 1.18.1.1) for E5 and fatty acyl-CoA oxidase (EC 1.3.3.6) for D4 (Supplementary Figure S6).
4 Discussion
The degradation of PE employing microorganisms is extensively investigated because this polyolefin is one of the most produced and the most recalcitrant. The identification of new bacterial isolates and investigation of their biodegradative metabolism enriches the knowledge of this current challenge.
The present work aims to characterize the PE biodegradative potential and identify plausible genomic and genetic traits of novel bacterial isolates from a solid organic waste rich in plastic debris derived from a waste treatment plant. Indeed, these kinds of samples are rich in microbial biodiversity, and the presence of diverse plastics can guarantee the identification of bacteria with metabolic capabilities (Devi et al., 2023; Mishra et al., 2024; Zampolli et al., 2024). Among other isolates, P. citronellolis E5 was isolated from organic waste rich in plastic and selected for its interesting properties toward PE oxidation. To the best of our knowledge, the E5 strain is the first isolate belonging to P. citronellolis species whose biodegradative profile toward PE was characterized by both metabolic and biodegradative assays and from the genetic and genomic point of view. The other two reports regarding P. citronellolis strains able to biodegrade polyethylene characterized their potential only at the metabolic and phenotypic levels (Pramila et al., 2012; Bhatia et al., 2014). It is interesting to notice that these other P. citronellolis strains were also isolated from municipal waste.
Among bacteria with intriguing plastic-degrading capacities, bacteria of the Rhodococcus genus are well known for their degrading features against emerging contaminants, including plastic (Alvarez, 2019; Gravouil et al., 2017). R. erythropolis species present genetic traits that lead to speculation that they could efficiently contribute to PE biodegradation or reduction (Zampolli et al., 2022). Thus, R. erythropolis D4 (Zampolli et al., 2024) was selected as a comparative strain to study the biodegradation of PE in an eco-friendly process with a low toxic by-product generation.
Here, we reported a framework to study and evaluate microbial PE potential based on phenotypic observation and bioinformatic predictions viable for both Gram-negative and Gram-positive bacteria.
Among other isolates, P. citronellolis E5 and R. erythropolis D4 were assayed and proved capable of growing on untreated commercial powder PE as the sole carbon and energy source for up to 60 days, showing a high number of bacterial cells already at 7 days. Often, to study the biodegradative potential of polyolefins, the chosen polymer is subjected to pretreatment to facilitate bacterial attack. Only a few studies focused on untreated PE to evaluate the full potential of microbial cells (Devi et al., 2023; Tao et al., 2023). Biodegradative studies focusing on PE reduction or elimination showed that untreated LDPE could be degraded by different strains of Pseudomonas (Kyaw et al., 2012; Yoon et al., 2012; Montazer et al., 2020). In this case, we mimicked the condition of PE microplastic because the biodegradative growth assays were carried out with PE powder.
The secreted and intracellular laccase activity were tested to support the isolate screening for PE biodegradation. Indeed, literature reports bacteria able to activate laccase-like enzymes to perform the first oxidation of PE (Santo et al., 2013; Gravouil et al., 2017; Zampolli et al., 2021; Tao et al., 2023). In this case, all the selected bacteria were able to secrete active laccases during growth on PE, ensuring its initial attack (E5 strain showed an activity equal to ((2±1) *10−3 U mg−1, D4 strain of around (3±1) *10−3 U mg−1) (Zampolli et al., 2021; Tao et al., 2023).
In line with this data, GC-MS analysis provides direct evidence of PE depolymerization derived from the profile of their growth and the corresponding metabolic products from PE oxidation. For both strains, an oscillatory pattern of PE products was observed because every generated oxidized product (medium- and long-chain alkanes and carboxylic acids) was subsequently degraded at the next sampling time. In this regard, the preliminary growth assays on representative metabolic intermediates of PE biodegradation, including alkanes, ketones, and carboxylic acids, supported these biodegradative trends for both strains. A few differences in terms of metabolic products and detection time were evidenced by the two strains, and great differences were detected between the control (no inoculum) and the unprocessed commercial PE powder (t0). It is worth mentioning that the initial PE analyses suggested a partial oxidation of the polymer due to its production process, as already reported by Zampolli et al. (2023). A few products were similar to the one detected by Rong et al. (2024) during the degradation of a branched LDPE by Nitratireductor sp. Z-1 and Gordonia sp. Z-2 (alkanes and fatty acids). Probably, this is due to a double extraction of the supernatant of each culture using ethyl acetate and DCM; thus, they also observed short-chain alkanes, alkanols, and esters. The present work used DCM extraction. The two strains could oxidize not only the polymer chains but also all the oxidized and non-oxygenated products deriving from the PE attack. This behavior is not surprising for D4 strains because, in a previous study, the D4 strain was demonstrated to be able to utilize diverse carboxylic acids from 9 to 20 carbons in the aliphatic chain (Zampolli et al., 2024). Moreover, the R. erythropolis species is known to possess degradative abilities toward aliphatic hydrocarbons (de Carvalho and da Fonseca, 2005; Zampolli et al., 2019), and some P. citronellolis bacteria were reported to be capable of biodegrading alkanes (Koutinas et al., 2021).
In order to correlate the meaningful phenotypic data on PE with the main genomic and genetic features, the genome of P. citronellolis E5 was completely sequenced, analyzed, and compared with the genome of R. erythropolis D4 (Zampolli et al., 2024). The size and GC percentage of the E5 genome are in line with that of the other strains of P. citronellolis species (NCBI taxid 53408) and highly divergent from that of R. erythropolis D4, with an ANI below 80% (Zampolli et al., 2023).
Whole-genome alignments, annotation search, and gene clustering against reference gene products (RAS) were exploited based on literature reports and knowledge about the main steps of the PE metabolism: (i) initial attack and oxidation of PE polymer, (ii) internalization transport of oxidized products, and (iii) subsequent intracellular oxidation of oxygenated or lower molecular weight products. In relation to the main enzymatic classes involved in the first step, the classes considered included laccase-like multicopper oxidases, peroxidase, lipases, and esterases (Gravouil et al., 2017; Montazer et al., 2020; Zampolli et al., 2021; Tao et al., 2023; Mishra et al., 2024; Rong et al., 2024; Kim et al., 2024). P. citronellolis E5 exhibited a higher number of glutathione peroxidases than R. erythropolis D4, and the opposite proportion was observed for multicopper oxidases. As reported by Rong et al. (2024), glutathione peroxidases can initiate the cleavage process of C-C bonds and C-H bonds in the polymer, leading to physical PE destruction. This could be likely the case of the E5 strain that produced shorter aliphatic products than the D4 strain over time, which, in conversion, produced more medium- and long-chain alkanes potentially generated from the multicopper oxidase initial attack. In line with this, it is not possible to exclude the involvement of other oxidases for PE initial oxidation for the two strains.
A few esterases were detected in the genomes, and no lipases clustered with reference sequences for both strains. These esterases/lipases can participate in the first attack in the case of PE products with ester bonds in the oxidized backbone (Kim et al., 2024).
The subsequent oxygenated or low molecular PE fragments require the presence of transportation gene products, especially ABC transporters, for the plausible entrance into the cells (Gravouil et al., 2017; Zampolli et al., 2021). Indeed, numerous genes encoding transporters were detected in both genomes.
The main enzymatic classes potentially involved in the last step comprise intracellular enzymes, and we considered alkane monooxygenases and cytochrome P450 hydroxylases (Gravouil et al., 2017; Zampolli et al., 2021; Tao et al., 2023; Mishra et al., 2024; Rong et al., 2024; Kim et al., 2024). Bacteria belonging to the Pseudomonas and Rhodococcus genera are well known to possess these oxidative systems to degrade alkanes (Zampolli et al., 2019), and the bacteria considered showed multiple gene products potentially related to PE oxidation. Indeed, Yoon et al. (2012) and Jeon and Kim (2015) demonstrated that alkB from P. aeruginosa E4 and the alk gene cluster of P. aeruginosa E7 actively degraded LDPE. In the case of the E4 strain, this occurred even in the absence of rubredoxins and rubredoxin reductase.
These investigations provided a set for each strain of genomic traits and gene products potentially involved in PE biodegradation for each of the PE biodegradative steps, strengthening the PE mineralization potential in both selected bacteria.
5 Conclusion
In the present work, novel bacterial isolates were characterized for PE biodegradative potential. A bacterium belonging to the P. citronellolis species was selected and characterized to develop a biological eco-friendly process to eliminate PE. In addition to growth and biodegradative comparative investigations with respect to the efficient plastic-degrading R. erythropolis D4, in-depth genomic and genetic determinant exploration was performed, providing a toolbox for both Gram-negative and Gram-positive bacteria useful for multiple biotechnological applications toward PE biodegradation. Moreover, these bacterial comparative analyses pave the way for combining multiple bacteria strains to cope with the prominent issue of polyolefin degradation.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found in the article.
Author contributions
JZ: conceptualization, data curation, investigation, methodology, software, validation, visualization, writing–original draft, and writing–review and editing. EC: data curation, methodology, resources, supervision, and writing–review and editing. ML: funding acquisition, project administration, resources, and writing–review and editing. PD: conceptualization, funding acquisition, project administration, resources, supervision, writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the 2021-ATE-0460 from University of Milano-Bicocca, and it was supported by the “Micro-Val - MICROrganismi per la VALorizzazione di rifiuti della plastica” project founded by Consorzio Nazionale per la Raccolta, il Riciclo e il Recupero degli Imballaggi in Plastica (COREPLA).
Acknowledgments
We would like to acknowledge the help of Daniele Vezzini for technical support in some experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1472309/full#supplementary-material
References
Adékambi, T., Butler, R. W., Hanrahan, F., Delcher, A. L., Drancourt, M., and Shinnick, T. M. (2011). Core gene set as the basis of multilocus sequence analysis of the subclass Actinobacteridae. PLoS ONE 6, e14792. doi:10.1371/journal.pone.0014792
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517-D525–D525. doi:10.1093/nar/gkz935
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/s0022-2836(05)80360-2
Amobonye, A., Bhagwat, P., Singh, S., and Pillai, S. (2021). Plastic biodegradation: frontline microbes and their enzymes. Sci. Total. Environ. 759, 143536. doi:10.1016/j.scitotenv.2020.143536
Andler, R., Tiso, T., Andreeβen, C., Zampolli, J., D’Afonseca, V., Guajardo, C., et al. (2022). Current progress on the biodegradation of synthetic plastics: from fundamentals to biotechnological applications. Rev. Environ. Sci. Biotechnol. 21, 829–850. doi:10.1007/s11157-022-09631-2
Andrews, S. (2010). FastQC A quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/(Accessed April 10, 2023).
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi:10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi:10.1089/cmb.2012.0021
Bhatia, M., Girdhar, A., Tiwari, A., and Nayarisseri, A. (2014). Implications of a novel Pseudomonas species on low density polyethylene biodegradation: an in vitro to in silico approach. SpringerPlus 3, 497. doi:10.1186/2193-1801-3-497
Bolger, A. M., Lohse, M., and Bjoern Usade, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3, 2114–2120. doi:10.1093/bioinformatics/btu170
Bonhomme, S., Cuer, A., Delort, A.-M., Lemaire, J., Sancelme, M., and Scott, G. (2003). Environmental biodegradation of polyethylene. Polym. Degrad. Stab. 81, 441–452. doi:10.1016/S0141-3910(03)00129-0
Bosi, E., Donati, B., Galardini, M., Brunetti, S., Sagot, M.-F., Lió, P., et al. (2015). MeDuSa: a multi-draft based scaffolder. Bioinformatics 31, 2443–2451. doi:10.1093/bioinformatics/btv171
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi:10.1016/0003-2697(76)90527-3
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46 (W1), W246–W251. doi:10.1093/nar/gky425
Danso, D., Chow, J., and Streit, W. R. (2019). Plastics: environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 85, 010955–19. doi:10.1128/AEM.01095-19
de Carvalho, C. C. C. R., and da Fonseca, M. M. R. (2005). The remarkable Rhodococcus erythropolis. Appl. Microbiol. Biotechnol. 67, 715–726. doi:10.1007/s00253-005-1932-3
Devi, D., Gupta, K. K., Chandra, H., Sharma, K. K., Sagar, K., Mori, E., et al. (2023). Biodegradation of low-density polyethylene (LDPE) through application of indigenous strain Alcaligenes faecalis ISJ128. Environ. Geochem. Health. 45, 9391–9409. doi:10.1007/s10653-023-01590-z
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi:10.1093/nar/gkad326
Gravouil, K., Ferru-Clement, R., Colas, S., Heyle, R., Kadri, L., Bourdeau, L., et al. (2017). Transcriptomics and lipidomics of the environmental strain Rhodococcus ruber point out consumption pathways and potential metabolic bottlenecks for polyethylene degradation. Environ. Sci. Technol. 51, 5172–5181. doi:10.1021/acs.est.7b00846
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi:10.1093/bioinformatics/btt086
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. doi:10.1038/s41467-018-07641-9
Jeon, H. J., and Kim, M. N. (2015). Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 103, 141–146. doi:10.1016/j.ibiod.2015.04.024
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., and Ishiguro-Watanabe, M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. doi:10.1093/nar/gkac963
Kim, Y.-J., Kim, Y. H., Shin, Y. R., Choi, S. Y., Park, J. A., Kim, H. O., et al. (2024). Efficient biodegradation of low-density polyethylene by Pseudomonas plecoglossicida SYp2123 was observed through FTIR and FESEM analysis. Biotechnol. Bioprocess Eng. 29, 743–750. doi:10.1007/s12257-024-00108-3
Koutinas, M., Kyriakou, M., Andreou, K., Hadjicharalambous, M., Kaliviotis, E., Pasias, D., et al. (2021). Enhanced biodegradation and valorization of drilling wastewater via simultaneous production of biosurfactants and polyhydroxyalkanoates by Pseudomonas citronellolis SJTE-3. Bioresour. Technol. 340, 125679. doi:10.1016/j.biortech.2021.125679
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. E35, 1547–1549. doi:10.1093/molbev/msy096
Kyaw, B. M., Champakalakshmi, R., Sakharkar, M. K., Lim, C. S., and Sakharkar, K. R. (2012). Biodegradation of low-density polythene (LDPE) by Pseudomonas species. Indian J. Microbiol. 52, 411–419. doi:10.1007/s12088-012-0250-6
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic acid techniques in bacterial systematics. Editors E. Stackebrandt, and M. Goodfellow (New York, NY: John Wiley and Sons), 115–175.
Lee, A., and Liew, M. S. (2021). Tertiary recycling of plastics waste: an analysis of feedstock, chemical and biological degradation methods. J. Mater. Cycles Waste Manag. 23, 32–43. doi:10.1007/s10163-020-01106-2
Liang, Q., Liu, C., Xu, R., Song, M., Zhou, Z., Li, H., et al. (2021). fIDBAC: a platform for fast bacterial genome identification and typing. Front. Microbiol. 12, 723577. doi:10.3389/fmicb.2021.723577
Maniatis, T., Fritsch, E. F., and Sambrook, J. (1982). Molecular cloning: a laboratory manual. New York, USA: Cold Spring Harbor Laboratory.
Medić, A. B., and Karadžić, I. M. (2022). Pseudomonas in environmental bioremediation of hydrocarbons and phenolic compounds key catabolic degradation enzymes and new analytical platforms for comprehensive investigation. World J. Microbiol. Biotechnol. 38, 165. doi:10.1007/s11274-022-03349-7
Miri, S., Saini, R., Davoodi, S. M., Pulicharla, R., Brar, S. K., and Magdouli, S. (2022). Biodegradation of microplastics: better late than never. Chemosphere 286, 131670. doi:10.1016/j.chemosphere.2021.131670
Mishra, R., Chavda, P., Kumar, R., Pandit, R., Joshi, M., Kumar, M., et al. (2024). Exploring genetic landscape of low-density polyethylene degradation for sustainable troubleshooting of plastic pollution at landfills. Sci. Total Environ. 912, 168882–169697. doi:10.1016/j.scitotenv.2023.168882
Mohanan, N., Montazer, Z., Sharma, P. K., and Levin, D. B. (2020). Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 11, 580709. doi:10.3389/fmicb.2020.580709
Montazer, Z., Najafi, M. B. H., and Levin, D. B. (2020). Challenges with verifying microbial degradation of polyethylene. Polymers 12, 123. doi:10.3390/polym12010123
Orro, A., Cappelletti, M., D’Ursi, P., Milanesi, L., Di Canito, A., Zampolli, J., et al. (2015). Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance. PloS ONE 10, e0139467. doi:10.1371/journal.pone.0139467
Overbeek, R., Begley, T., Butler, R. M., Choudhuri, J. V., Chuang, H. Y., Cohoon, M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. doi:10.1093/nar/gki866
Pramila, R., Padmavathy, K., Ramesh, K. V., and Mahalakshm, K. (2012). Brevibacillus parabrevis, Acinetobacter baumannii and Pseudomonas citronellolis - potential candidates for biodegradation of low density polyethylene (LDPE). J. Bacteriol. Res. 4, 9–14. doi:10.5897/JBR12.003
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (Database issue), D590–D596. doi:10.1093/nar/gks1219
Ray, A. S., Rajasekaran, M., Uddin, M., and Kandasamy, R. (2023). Laccase driven biocatalytic oxidation to reduce polymeric surface hydrophobicity: an effective pre-treatment strategy to enhance biofilm mediated degradation of polyethylene and polycarbonate plastics. Sci. Total Environ. 904, 166721. doi:10.1016/j.scitotenv.2023.166721
Rong, Z., Ding, Z.-H., Wu, Y.-H., and Xu, X.-W. (2024). Degradation of low-density polyethylene by the bacterium Rhodococcus sp. C-2 isolated from seawater. Sci. Total Environ. 907, 167993. doi:10.1016/j.scitotenv.2023.167993
Sanluis-Verdes, A., Colomer-Vidal, P., Rodriguez-Ventura, F., Bello-Villarino, M., Spinola-Amilibia, M., Ruiz-Lopez, E., et al. (2022). Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. Nat. Commun. 13, 5568. doi:10.1038/s41467-022-33127-w
Santo, M., Weitsman, R., and Sivan, A. (2013). The role of the copper-binding enzyme - laccase - in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegr. 84, 204–210. doi:10.1016/j.ibiod.2012.03.001
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7, 000685. doi:10.1099/mgen.0.000685
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi:10.1093/bioinformatics/btu153
Sen, S. K., and Raut, S. (2015). Microbial degradation of low density polyethylene (LDPE): a review. J. Environ. Chem. Eng. 3, 462–473. doi:10.1016/j.jece.2015.01.003
Shilpa, , Basak, N., and Meena, S. S. (2023). Biodegradation of low density polythene (LDPE) by a novel strain of Pseudomonas aeruginosa WD4 isolated from plastic dumpsite. Biodegradation 35, 641–655. doi:10.1007/s10532-023-10061-2
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Soleimani, Z., Gharavi, S., Soudi, M., and Moosavi-Nejad, Z. (2021). A survey of intact low-density polyethylene film biodegradation by terrestrial Actinobacterial species. Int. Microbiol. 24, 65–73. doi:10.1007/s10123-020-00142-0
Starikova, E. V., Tikhonova, P. O., Prianichnikov, N. A., Rands, C. M., Zdobnov, E. M., Ilina, E. N., et al. (2020). Phigaro: high-throughput prophage sequence annotation. Bioinformatics 36, 3882–3884. doi:10.1093/bioinformatics/btaa250
Strotmann, U., Thouand, G., Pagga, U., Gartiser, S., and Heipieper, H. J. (2023). Toward the future of OECD/ISO biodegradability testing new approaches and developments. Appl. Microbiol. Biotechnol. 107, 2073–2095. doi:10.1007/s00253-023-12406-6
Tao, X., Ouyang, H., Zhou, A., Wang, D., Matlock, H., Morgan, J. S., et al. (2023). Polyethylene degradation by a Rhodococcous strain isolated from naturally weathered plastic waste enrichment. Environ. Sci. Technol. 57, 13901–13911. doi:10.1021/acs.est.3c03778
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi:10.1038/s41587-021-01156-3
Thakur, B., Singh, J., Singh, J., Angmo, D., and Vig, A. P. (2023). Biodegradation of different types of microplastics: molecular mechanism and degradation efficiency. Sci. Total Environ. 877, 162912. doi:10.1016/j.scitotenv.2023.162912
Wei, R., and Zimmermann, W. (2017). Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb. Biotechnol. 10, 1308–1322. doi:10.1111/1751-7915.12710
Yao, C., Xia, W., Dou, M., Du, Y., and Wu, J. (2022). Oxidative degradation of UV-irradiated polyethylene by laccase-mediator system. J. Hazard. Mater. 440, 129709. doi:10.1016/j.jhazmat.2022.129709
Yoon, M. G., Jeon, J. H., and Kim, M. N. (2012). Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J. Bioremed. Biodegr. 3, 145. doi:10.4172/2155-6199.1000145
Yoon, S. H., Ha, S. M., Lim, J. M., Kwon, S. J., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 110, 1281–1286. doi:10.1007/s10482-017-0844-4
Zampolli, J., Collina, E., Lasagni, M., and Di Gennaro, P. (2014). Biodegradation of variable-chain-length n-alkanes in Rhodococcus opacus R7 and the involvement of an alkane hydroxylase system in the metabolism. Amb. Express 4, 73. doi:10.1186/s13568-014-0073-4
Zampolli, J., Mangiagalli, M., Vezzini, D., Lasagni, M., Ami, D., Natalello, A., et al. (2023). Oxidative degradation of polyethylene by two novel laccase-like multicopper oxidases from Rhodococcus opacus R7. Environ. Technol. Innov. 32, 103273. doi:10.1016/j.eti.2023.103273
Zampolli, J., Orro, A., Manconi, A., Ami, D., Natalello, A., and Di Gennaro, P. (2021). Transcriptomic analysis of Rhodococcus opacus R7 grown on polyethylene by RNA seq. Sci. Rep. 11, 21311. doi:10.1038/s41598-021-00525-x
Zampolli, J., Orro, A., Vezzini, D., and Di Gennaro, P. (2022). Genome-based exploration of Rhodococcus species for plastic-degrading genetic determinants using bioinformatic analysis. Microorganisms 10, 1846. doi:10.3390/microorganisms10091846
Zampolli, J., Vezzini, D., Brocca, S., and Di Gennaro, P. (2024). Insights into the biodegradation of polycaprolactone through genomic analysis of two plastic-degrading Rhodococcus bacteria. Front. Microbiol. 14, 1284956. doi:10.3389/fmicb.2023.1284956
Zampolli, J., Zeaiter, Z., Di Canito, A., and Di Gennaro, P. (2019). Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus. Appl. Microbiol. Biotechnol. 103, 1069–1080. doi:10.1007/s00253-018-9539-7
Keywords: polyethylene biodegradation, Rhodococcus erythropolis, Pseudomonas citronellolis, genome analysis, laccase activity, gene clusters
Citation: Zampolli J, Collina E, Lasagni M and Di Gennaro P (2024) Insights into polyethylene biodegradative fingerprint of Pseudomonas citronellolis E5 and Rhodococcus erythropolis D4 by phenotypic and genome-based comparative analyses. Front. Bioeng. Biotechnol. 12:1472309. doi: 10.3389/fbioe.2024.1472309
Received: 29 July 2024; Accepted: 08 November 2024;
Published: 12 December 2024.
Edited by:
Tajalli Keshavarz, University of Westminster, United KingdomCopyright © 2024 Zampolli, Collina, Lasagni and Di Gennaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrizia Di Gennaro, cGF0cml6aWEuZGlnZW5uYXJvQHVuaW1pYi5pdA==
 Jessica Zampolli
Jessica Zampolli Elena Collina
Elena Collina Marina Lasagni
Marina Lasagni Patrizia Di Gennaro
Patrizia Di Gennaro