- Department of Biosciences and Bioengineering, Indian Institute of Technology-Bombay, Mumbai, India
The perpetual release of natural/synthetic pollutants into the environment poses major risks to ecological balance and human health. Amongst these, contaminants of emerging concern (CECs) are characterized by their recent introduction/detection in various niches, thereby causing significant hazards and necessitating their removal. Pharmaceuticals, plasticizers, cyanotoxins and emerging pesticides are major groups of CECs that are highly toxic and found to occur in various compartments of the biosphere. The sources of these compounds can be multipartite including industrial discharge, improper disposal, excretion of unmetabolized residues, eutrophication etc., while their fate and persistence are determined by factors such as physico-chemical properties, environmental conditions, biodegradability and hydrological factors. The resultant exposure of these compounds to microbiota has imposed a selection pressure and resulted in evolution of metabolic pathways for their biotransformation and/or utilization as sole source of carbon and energy. Such microbial degradation phenotype can be exploited to clean-up CECs from the environment, offering a cost-effective and eco-friendly alternative to abiotic methods of removal, thereby mitigating their toxicity. However, efficient bioprocess development for bioremediation strategies requires extensive understanding of individual components such as pathway gene clusters, proteins/enzymes, metabolites and associated regulatory mechanisms. “Omics” and “Meta-omics” techniques aid in providing crucial insights into the complex interactions and functions of these components as well as microbial community, enabling more effective and targeted bioremediation. Aside from natural isolates, metabolic engineering approaches employ the application of genetic engineering to enhance metabolic diversity and degradation rates. The integration of omics data will further aid in developing systemic-level bioremediation and metabolic engineering strategies, thereby optimising the clean-up process. This review describes bacterial catabolic pathways, genetics, and application of omics and metabolic engineering for bioremediation of four major groups of CECs: pharmaceuticals, plasticizers, cyanotoxins, and emerging pesticides.
1 Introduction
The human population is perpetually interacting with a wide range of external chemicals, including both man-made and naturally occurring compounds. The impact of this continuous exposure can be either beneficial or detrimental to human health. While certain compounds such as pharmaceuticals, pesticides, plasticizers etc., have contributed immensely to development and sustenance (Table 1), their excessive usage has led to distribution and persistence in various ecosystems, causing disruption and toxic effects. Amongst these, “contaminants of emerging concern” (CEC) are naturally occurring or synthetic compounds which are recently detected/suspected to be present in various habitats and might significantly impact the metabolism of living organisms. The detection of such compounds can be attributed either to their recent introduction into the environment or an advancement in detection technologies. Additionally, CECs also include known contaminants with developing or poorly understood risk profiles (Sauvé and Desrosiers, 2014). Examples of CECs include compounds such as pharmaceuticals, personal care products, nanomaterials, pesticides, plasticizers, microplastics, radionuclides/rare earth elements, cyano/algal toxins and perfluorinated compounds.
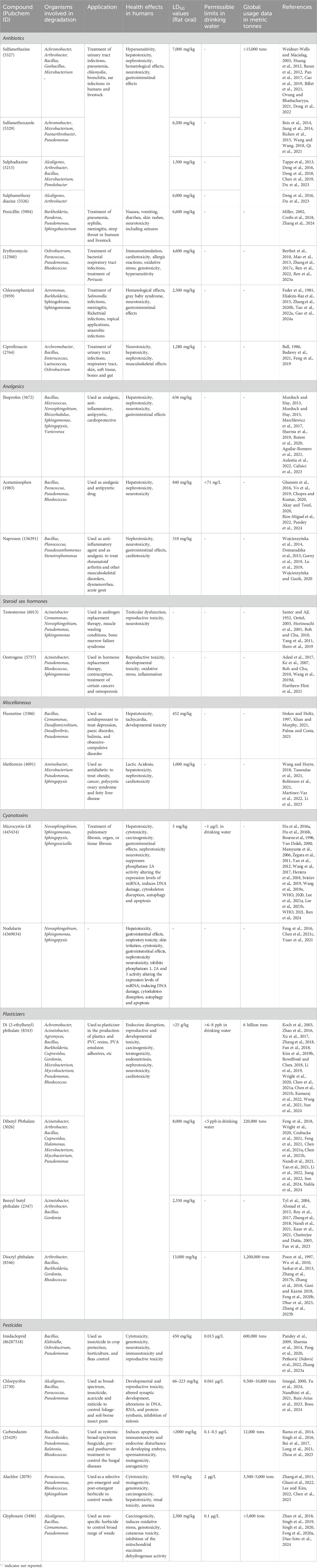
Table 1. List of contaminants of emerging concern detailing characteristics, toxic effects, applications, and microbial genus involved in degradation.
Sources of CECs in the environment can include industrial discharge, improper disposal, excretion of unmetabolized residues, improper sewage management, hospital/laboratory wastewater, agricultural run-off, or processes like eutrophication. Whereas, the prevalence of these compounds depends upon various factors such as industrial activities, agricultural practices, regulatory policies, waste management systems, and environmental conditions (Feng et al., 2023). CEC exposure to humans can occur through various routes like consumer goods, personal care products, ingestion of contaminated food and water, occupational exposure, inhalation of airborne particles and foetal exposure, amongst others, causing a variety of health effects (Feng et al., 2023). For example, long-term exposure to such contaminants has been linked to cancer, endocrine disruption, reproductive tissue damage, immune system suppression, developmental anomalies, and liver damage, amongst other health effects (Radke et al., 2020; Gonsioroski et al., 2020; Lyu et al., 2020; Syafrudin et al., 2021; Balakrishnan et al., 2022; Table 1). Additionally, CECs have been found to bioaccumulate in aquatic biota (Deere et al., 2024) and are toxic to crustaceans (Hossain et al., 2018), earthworms (Gillis et al., 2017), fish (Meador et al., 2016; Yeh et al., 2017; Picó et al., 2019) and molluscs (Canesi et al., 2022), causing ecological disruption. Therefore, removal of these compounds from various ecological compartments is a necessity.
The persistence of CECs in the environment has led to evolution of microbes to utilise them as sole source of carbon and energy (Table 1). Bioremediation involves the application of microbes to clean-up xenobiotics/pollutants from contaminated habitats and provides a desirable alternative to abiotic methods of removal due to its cost-effectiveness, efficiency, and eco-friendliness (Patel et al., 2022). Further, the application of directed genetic engineering approaches, called as “metabolic engineering,” aids in overcoming limitations associated with natural isolates (Dvorak et al., 2017). Additionally, omics techniques have emerged as essential tools for deciphering complex mechanisms underlying CEC biodegradation, which aids in enhancing the understanding of degradation pathways and designing optimal metabolic engineering strategies.
This article aims to provide a comprehensive review of microbial degradation pathways as well as the associated genes and enzymes for four major groups of contaminants of emerging concern (CECs): pharmaceuticals, plasticizers, cyanotoxins, and emerging pesticides (Table 1). Further, the application of omics techniques, including genomics, metagenomics, transcriptomics, proteomics, and metabolomics, to gain system-level insights into the metabolic pathways and regulatory mechanisms driving CEC degradation for development of efficient bioprocess has been described. Additionally, the article also highlights the importance of metabolic engineering strategies to enhance bioremediation efficiency.
2 Microbial degradation pathways and genetics
2.1 Pharmaceuticals
2.1.1 Antibiotics
Antibiotics are antibacterial agents that function by either killing (bactericidal) or inhibiting the growth (bacteriostatic) of bacteria. Antibiotics are grouped into beta-lactams, macrolides, fluoroquinolones, tetracyclines, aminoglycosides, sulfonamides, glycopeptides, oxazolidinones and carbapenems, based on their structure and mechanism of action (Etebu and Arikekpar, 2016). Large scale production, improper sewage management and disposal as well as human excretion of unmetabolized residues contribute to accumulation of these compounds in aquatic and soil ecosystems (Cycoń et al., 2019; Bilal et al., 2020).
Antibiotics have been found to occur in wastewater treatment plants (WWTPs), hospital wastewaters, as well as surface, river and groundwater across the globe. For example, antibiotic concentrations up to 14.5 μg L−1 and 64 μg L−1 (dominated by β-lactams, quinolones and sulfonamides) were detected in hospital effluents and WWTP influents, respectively, in Queensland, Australia. The concentration in surface waters and WWTP effluent was up to 2 and 3.4 μg L−1, respectively (Watkinson et al., 2009). Beta-lactam antibiotics amoxillin and penicillin G were detected at 13.3–18.47 μg L−1 and 3.12–4.75 μg L−1 in WWTP influents in Iran (Golchin et al., 2021). Various antibiotic classes such as sulfonamides (285.5–634.9 ng L−1), tetracyclines (363.4–753.3 ng L−1) and quinolones (1,355.8–1922.4 ng L−1) were detected in hospital influents in Xinjiang, China (Li et al., 2016). Sulfonamides (up to 256 ng L−1) and quinolones (up to 1,270 ng L−1) were detected at high concentrations in Wenyu river in Beijing, China (Liu et al., 2019). Erythromycin has been detected in Korean Municipal WWTP influents at a concentration of 0.4–1 μg L−1 (Sim et al., 2010) and 381 ng L−1 in River Thurso, Scotland (Nebot et al., 2015). The presence of these compounds in the environment poses a major risk due to the dissemination of antibiotic resistance genes and evolution of resistance phenotype in the microbial community (Li et al., 2015; Rolbiecki et al., 2021; Thakali et al., 2021). Additionally, antibiotic residues cause toxicity to aquatic biota and alter microbial community structure, causing ecological disruption (Ding and He, 2010; Välitalo et al., 2017).
2.1.1.1 Sulfonamides
Sulfonamide antibiotics are synthetic antimicrobial agents that are primarily used in human and veterinary medicine to combat bacterial infections. These compounds inhibit the enzyme dihydropteroate synthetase, essential for folic acid synthesis. The bacterium Bacillus cereus H38 utilises sulfamethazine as source of carbon, nitrogen and sulphur. The bacterium possesses two pathways for the catabolism of this antibiotic. In pathway I, the S-N bond is cleaved, removing SO2 and forming N-(4,6-dimethylpyrimidin-2-yl) 1,4-diphenylamine. Further, the C-N bond in this compound is cleaved to form 2-amino-4,6-dimethylpyrimidine and aniline. Pathway II proceeds via the cleavage of the N4 amine bond to form N-(3,5-dimethylpyrimidin-2-yl)-benzenesulfonamide, which is also converted to 2-amino-4,6-dimethylpyrimidine and phenyl sulphoxide upon cleavage of the S-N bond (Dong et al., 2022; Figure 1A).
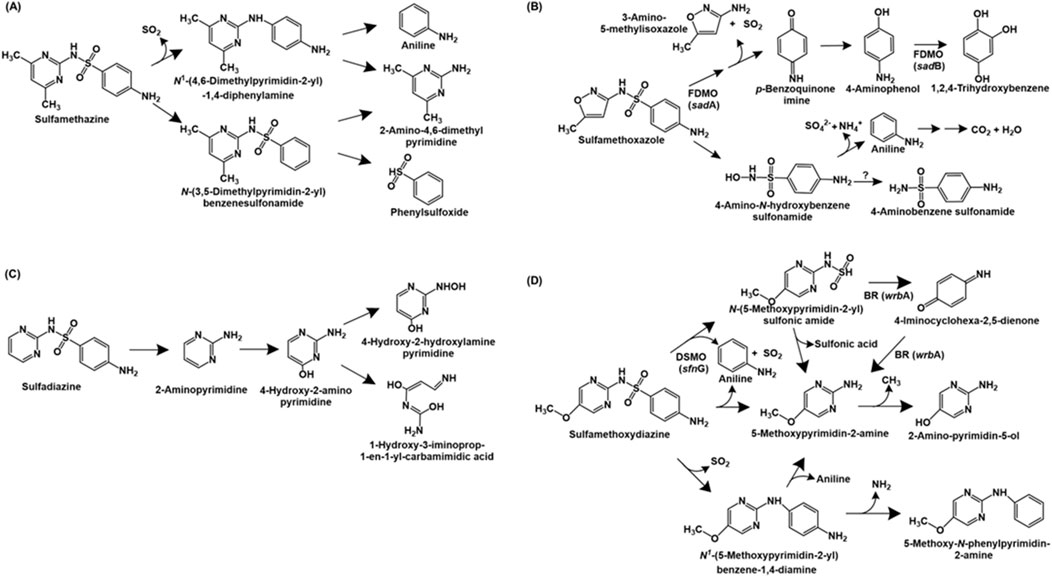
Figure 1. Bacterial degradation pathways of sulfonamide antibiotics: (A) sulfamethazine (B) sulfamethoxazole (C) sulfadiazine and (D) sulfamethoxydiazine. Gene encoding of the respective enzymes are indicated in parenthesis. Enzyme abbreviations: FDMO, flavin-dependent monoxygenase; BR, 1,4-benzoquinone reductase; DSMO, dimethylsulfone monoxygenase. Question mark indicates enzyme catalysing reaction not known.
Pseudomonas psychrophila HA-4 utilises the antibiotic sulfamethoxazole as the sole source of carbon and energy. The first step of degradation involves the hydrolysis of the compound to 4-amino-N-hydroxybenzenesulfonamide and 3-amino-5-methylisoxazole. The former undergoes deamination and desulfurization to form aniline, sulphate and ammonia. Aniline is further metabolised to carbon-di-oxide and water. Alternatively, 4-amino-N-hydroxybenzesulfonamide can be converted to 4-aminobenzenesulfonamide (Jiang et al., 2014). The metabolic pathway of sulfomethoxazole in Microbacterium sp. BR1 is initiated by ipso-hydroxylation to form an unstable intermediate which forms p-benzoquinone imine and 3-amino 5-methylisoxazole. The former is reduced to p-aminophenol, which undergoes hydroxylation to form 1,2,4-trihydroxybenzene, which might undergo ring-cleavage (Ricken et al., 2015; Ricken et al., 2017; Figure 1B).
The complete metabolic pathways for sulfadiazine and sulfametoxydiazine have been reported in Arthrobacter sp. D2 and Alcaligenes aquatillis FA, respectively. In strain D2, sulfadiazine is metabolised via the cleavage of the sulfonamide bond to form 2-aminopyrimidine, which is hydroxylated at the C-4 position to form 4-hydroxy-2-amino-pyrimidine. Subsequently, this intermediate undergoes ring-opening or hydroxylation of the amine group (to form 4-hydroxy-2-hydroxylamine-pyrimidine; Deng et al., 2016; Figure 1C). Whereas, sulfametoxydiazine metabolism in strain FA has been proposed to proceed via three different routes. In pathway I, the breakdown was initiated by the loss of aniline to form N-(5-methoxypyrimidin-2-yl) sulfonic amide. This intermediate further forms 5-methoxypyrimidin-2-amine (by loss of sulphonic acid), which forms 2-amino-pyrimidin-5-ol by loss of a methyl group. Alternatively, strain FA can directly form 5-methoxypyrimidin-2-amine by loss of aniline and SO2 (pathway II). In pathway III, the loss of SO2 from the substrate results in the generation of N1-(5-methoxypyrimidin-2-yl) benzene-1,4-diamine. This intermediate can either form 5-methoxypyrimidin-2-amine by cleavage of the C-N bond or 5-methoxy-N-phenylpyrimidin-2-amine by loss of amine group (Du et al., 2023; Figure 1D).
Three sulfonamide degradation genes sadA, sadB and sadC were identified in the genome of Microbacterium sp. strain BR1. Both sadA and sadB encoded flavin-dependent monoxygenases catalysing removal of 3-amino-5-methylisoxazole and SO2 from sulfomethoxazole and hydroxylation of 4-aminophenol to trihydroxybenzene, respectively. Whereas sadC encoded a FMN reductase involved in delivering reduced FMN to SadA and SadB. Similar homologues of sadABC have been found in the genomes of other Actinobacteria such as Paenarthrobacter sp. A01 (Cao et al., 2019), Leucobacter sulfamidivorax (Reis et al., 2019) and Arthrobacter sp. D2 and D4 (Deng et al., 2016). Aside from Actinobacteria, Alcaligenes aquatillis FA harbored three sulfametoxydiazine metabolic genes: wrbA encoding 1,4-benzoquinone reductase (involved in formation or degradation of 4-iminocyclohexa-2,5-dienone), pcaC encoding 4-carboxymuconolactone decarboxylase (involved in aromatic ring processing) and sfnG encoding dimethylsulfone monooxygenase (involved in hydroxylation of sulfametoxydiazine). Further, dfrA26 (dihydrofolate reductase) and sul2 (dihydropteroate synthetase) genes were hypothesised to be involved in resistance to sulfonamides in strain FA (Du et al., 2023).
2.1.1.2 Beta-lactams
Beta-lactam antibiotics consist of a characteristic beta-lactam ring and function by inhibiting bacterial cell wall synthesis. Although biotransformation products have been reported for various beta-lactams such as imipenem (Minerdi et al., 2016), and ampicillin (Zumstein and Helbling, 2019), the complete mineralisation pathway has been only reported for penicillin G. The penicillin G mineralisation pathway has been detailed in proteobacterial isolates belonging to the genera Burkholderia spp. (strain ABC02), Pseudomonas spp. (ABC07), Pandoraea spp. (strains ABC08 and ABC10). Initially, the enzyme beta-lactamase converts penicillin to benzylpenicilloic acid, which was acted upon by an amidase or a hydrolase type of enzyme to form phenylacetic acid. Subsequently, this intermediate is converted into acetyl-CoA and succinyl-CoA (central carbon intermediates) via the phenylacetate pathway, conserved amongst various isolates (Figure 2). The genomic analyses revealed that strain ABC07 carries two major operons for penicillin catabolism, the put and paa operon. While the put operon encodes beta-lactamase (bla), major superfamily transporter (mfs) and amidases (put1 and put2), the paa operon encodes enzymes involved in phenylacetic acid catabolism. Similar genes were also detected in strains ABC02, ABC08 and ABC10 (Crofts et al., 2018).
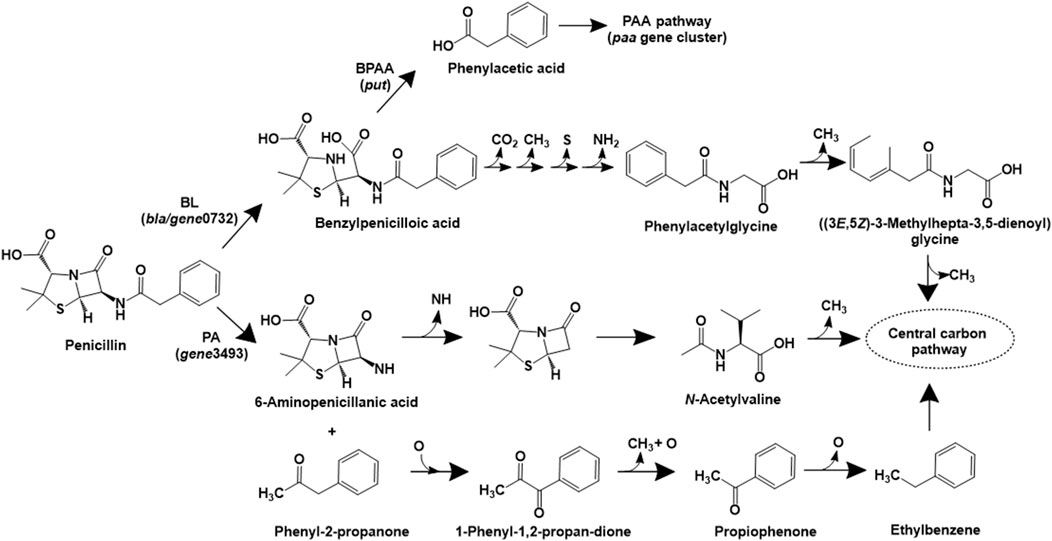
Figure 2. Bacterial degradation pathways of penicillin. Gene encoding of the respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: BL, beta-lactamase; PA, penicillin acylase; BPAA, benzylpenicilloic acid amidase.
In Sphingobacterium sp. SQW1, three different pathways for degradation of penicillin G sodium have been proposed (Zhang et al., 2024). In the intracellular pathway, penicillin is converted to benzylpenicilloic acid, which undergoes decarboxylation, demethylation, desulfurization and deamination reactions to form phenylacetylglycine. This intermediate undergoes ring opening and multiple demethylation reactions to form central carbon intermediates (Figure 2). A similar pathway involving the action of extracellular beta-lactamase (to form benzylpenicilloic acid) and multiple demethylation, desulfurization and deamination reactions has also been proposed (Zhang et al., 2024). An alternative extracellular pathway involves the action of the enzyme penicillin acylase (on penicillin) to form phenyl-2-propanone and 6-aminopenicillanic acid (6-APA) by an acylation decarboxylation reaction. The former compound undergoes oxidative dehydrogenation to form 1-phenyl-1,2-propanedione, which ultimately forms carbon-di-oxide and water. Whereas, 6-APA undergoes deamidation, hydrolysis of the beta-lactam ring, ring–opening desulfurization and demethylation to form N-acetylvaline, which undergoes demethylation and ultimately forms carbon-di-oxide and water (Figure 2). The genes encoding beta-lactamase (gene0732) and penicillin amidase (gene3493) were detected in strain SQW1 (Zhang et al., 2024).
2.1.1.3 Erythromycin
Erythromycin, a macrolide class of antibiotic, consists of a characteristic macrocyclic lactone ring and functions by inhibiting bacterial protein synthesis via binding to 50S ribosomal subunit. Two major erythromycin mineralisation pathways have been detailed in Paracoccus versutus W7. In the first pathway, the antibiotic is acted upon by the esterase EreA (Erythromycin hydrolase), leading to opening of the lactone ring. The intermediate generated (C37H70NO14) was cleaved by glucoside hydrolase, resulting in removal of the cladinose moiety. Further, dehydration followed by the action of glycoside hydrolase results in the removal of desosamine moiety. The remaining main chain compound (C21H41O9), cladinose and desosamine are metabolised via tricarboxylic acid cycle (TCA). Alternatively, erythromycin was converted to the intermediate C37H66NO12 and further acted upon by glucoside hydrolase to catalyse the removal of cladinose. The generated intermediate (C29H52NO9) is acted upon by EreA to form C29H54NO10. The action of glucoside hydrolase generates C21H41O9 and desosamine, which are funnelled into the central carbon metabolism (Ren et al., 2023a; Figure 3A). A similar erythromycin metabolism pathway has been proposed in Rhodococcus gordoniae rjjtx-2 (Ren et al., 2022).
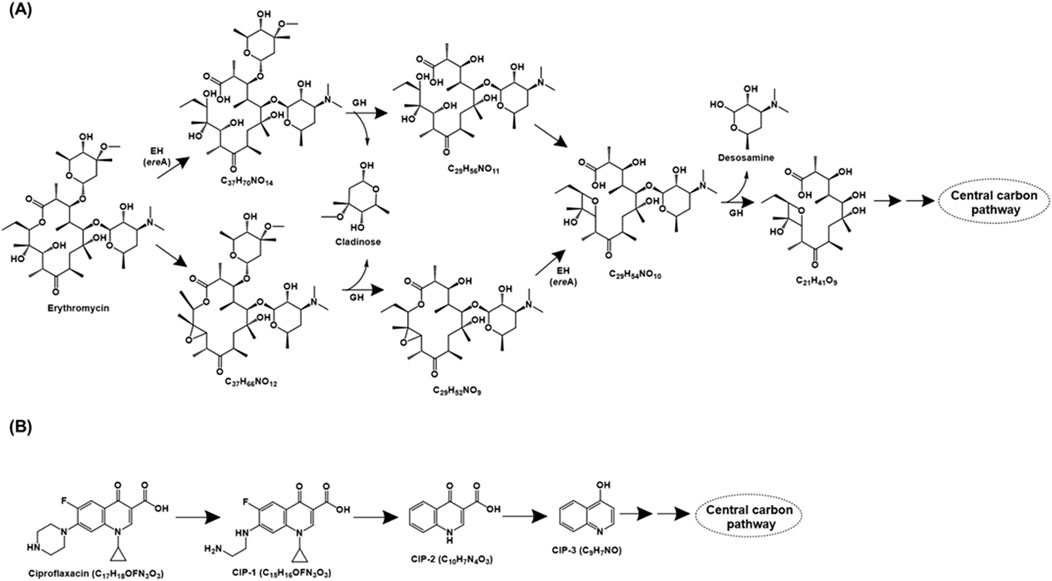
Figure 3. Bacterial degradation pathways of (A) erythromycin and (B) ciproflaxacin. Genes encoding of the respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: EH, erythromycin hydrolase; GH, glycoside hydrolase.
Literature on the genetic background of erythromycin catabolism has mainly focussed on the hydrolase ere genes, while reports about other enzymes/genes are scanty. The first erythromycin esterase gene ereA was identified in Escherichia coli (Ounissi and Courvalin, 1985) and its homologs have been detected in genus like Rhodococcus (Ren et al., 2022), Paracoccus (Ren et al., 2023a) and Providencia (Plante et al., 2003), amongst others. Similarly, type-II erythromycin hydrolase, encoded by ereB was detected in E. coli (Arthur et al., 1986) and its homologs have been frequently detected in environmental isolates such as Staphylococcus (Schmitz et al., 2000), Klebsiella and Salmonella (Fuentes et al., 2014), amongst others. The newly discovered ereC and ereD are less prevalent and have been detected in Klebsiella (Yong et al., 2009) and Riemerella (Xing et al., 2015), respectively. Interestingly, the ere homologs (except for ereD) are associated/localised on mobile genetic elements that aid in their distribution in the microbial community via horizontal gene transfer (Arthur et al., 1986; Biskri and Mazel, 2003; Yong et al., 2009).
2.1.1.4 Ciproflaxacin
Ciproflaxacin is a fluoroquinolone type of antibiotic that functions by inhibiting the bacterial enzyme DNA gyrase (topoisomerase II) and topoisomerase IV. The ciprofloxacin metabolic pathway has been proposed for consortium XG consisting of bacteria belonging to genera Achromobacter, Bacillus, Lactococcus, Ochrobactrum and Enterococcus. Ciproflaxacin (C17H18OFN3O3) degradation is initiated by the loss of the C2H2 moiety from the piperazine ring to form CIP-1 (C15H16OFN3O3). Further, the loss of piperazine moiety, cyclopropyl, and fluorine atom results in the formation of CIP-2 (C10H7N4O3), which subsequently forms CIP-3 (C9H7NO) by decarboxylation. CIP-3 was further mineralised to CO2, H2O, NH4+, NO3− and F− by the consortia XG (Figure 3B; Feng et al., 2019).
2.1.1.5 Chloramphenicol
Chloramphenicol is a broad-spectrum antibiotic that binds 50S ribosomal subunit and inhibits protein synthesis. In Aeromonas media SZW-3, chloramphenicol has been reported to be catabolised by three major pathways. The first pathway involves the cleavage of the bond between the side chain of C1 and C2, leading to the formation of p-nitrobenzoic acid, which is oxidised to form p-hydroxyaminobenzoic acid, which is further ring-cleaved (Figure 4). In pathway II, the nitro-group is sequentially reduced to an amino group, forming AMCl2. This intermediate can further be demethylated (to form CP1), dechlorinated (to form CP2) or ring-cleaved (to form Mc-AMCl2). Alternatively, chloramphenicol can undergo acetylation and sequential reduction of the nitro group to an amine, following a route similar to pathway I (Figure 4). The genomic analyses of strain SDW-3 identified genes rarD (encoding a permease that provides resistance), chloramphenicol O-acetyltransferase type B encoding gene (GE000673; involved in the acetylation of chloramphenicol), three nitro-reductases (GE003101, GE001796, GE003206; involved in biodetoxification) as well as haloacid and haloalkane dehalogenases (GE002643 and GE001139; involved in dechlorination) (Tan Z. et al., 2022).
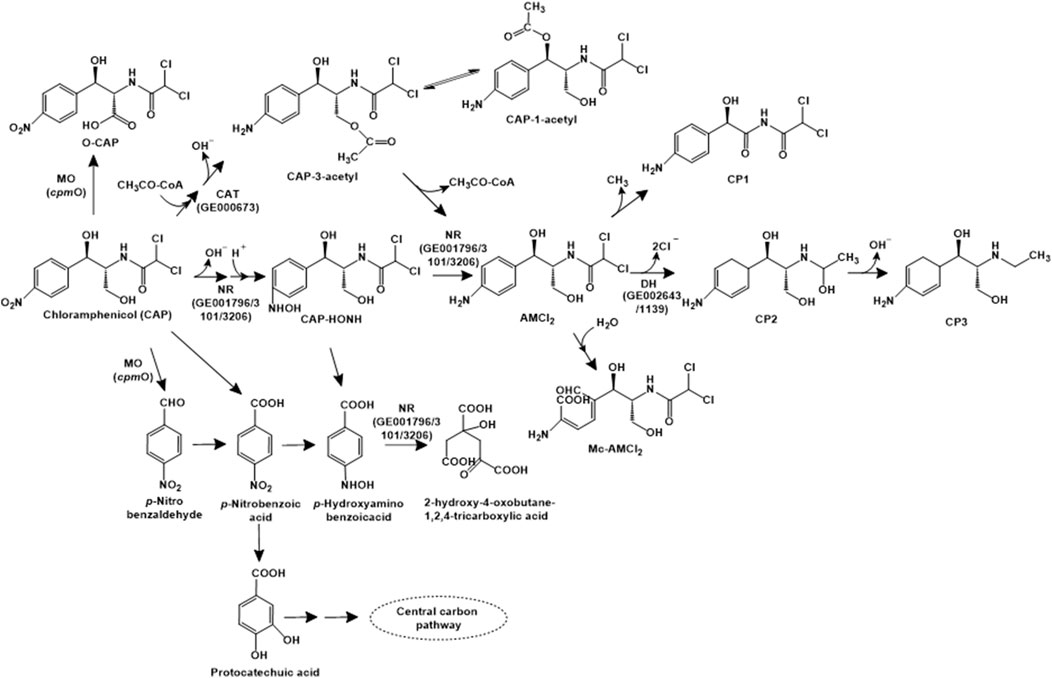
Figure 4. Bacterial degradation pathways of chloramphenicol. Enzyme abbreviations: MO, multifunctional oxidase; CAT, chloramphenicol O-acetyltransferase type B; NR, nitroreductase; DH, haloacid or haloalkane dehalogenase. Gene encoding of the respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps.
Sphingobium sp. WTD-1 has been reported to utilise this antibiotic as the sole source of carbon and energy. Three metabolic pathways for chloramphenicol have been proposed in strain WTD-1. The first pathway involves the acetylation to chloramphenicol-3-acetyl (CAP-3-acetyl), which is non-enzymatically converted to chloramphenicol-1-acetyl (CAP-1-acetyl). The second pathway involves the dehydrogenation at the C-3 hydroxyl group to form 2,2-dichloro-N-(1,1,3-trihydroxy-3-(4-nitrophenyl) propan-2-yl) acetamide (DHNOA), which is further oxidised to the dead-end metabolite 2-(2,2-dichloroacetamido)3-hydroxy-3-(4-nitrophenyl) propanoic acid (O-CAP). The third pathway involves cleavage of the C1-C2 bond to form para-nitrobenzaldehyde (PNBD), which is converted to para-nitrobenzoic acid (PNBA) and further, protocatechuic acid. This intermediate undergoes ortho- or meta-ring cleavage to form TCA cycle intermediates (Gao et al., 2024a; Figure 4). A novel multifunctional oxidase, CpmO, which carries out the oxidation of C-3 hydroxyl as well as cleavage of C1-C2 bond was identified in the genome of strain WTD-1, purified and characterised (Gao et al., 2024b).
2.1.2 Analgesics
Analgesics are pain-relieving medications that can be categorized into two groups: opioid and non-opioid. Opioid analgesics function by impacting pain perception in the brain by affecting ion channels or receptors, while non-opioid analgesics inhibit prostaglandin synthesis. Amongst these, non-opioid analgesics such as ibuprofen, acetaminophen and naproxen are widely prescribed and prevalent, and are therefore the subject of current discussion. The easy (over the counter) availability, low toxicity, extensive use, improper disposal and excretion of unmetabolized drug/associated metabolites have contributed to accumulation of these analgesics in the environment, causing adverse effects to biota, especially aquatic ecosystems (Parolini, 2020; Jan-Roblero and Cruz-Maya, 2023). For example, ibuprofen has been detected in influents (5–22 μg L−1) and effluents (0.1–2 μg L−1) of WWTPs in south-western India (Praveenkumarreddy et al., 2021). Acetaminophen (4.4–9.2 μg L−1), ibuprofen (0.8–1.2 μg L−1) and naproxen (0.5–0.9 μg L−1) have been detected at varying concentrations in municipal WWTPs in Korea (Sim et al., 2010). Acetaminophen (22.8 μg L−1) has been detected at high concentrations in the water of River Thurso, Scotland (Nebot et al., 2015). The concentration of naproxen ranged from 20–231 ng L−1 and 13–80 ng L−1 in influents and effluents, respectively of Italian WWTPs (Patrolecco et al., 2015). The occurrence of these compounds at reported concentrations causes significant toxicity to aquatic biota (Ragugnetti et al., 2011; Aguirre-Martínez et al., 2015), thereby leading to ecological disruption.
2.1.2.1 Ibuprofen
Ibuprofen [2-(p-isobutylphenyl) propionic acid] is a widely used non-steroidal anti-inflammatory drug (NSAID) that functions by inhibiting the enzyme cyclooxygenase (COX), involved in prostaglandin biosynthesis (Ghlichloo and Gerriets, 2023). The complete degradation pathway of ibuprofen has been described for Bacillus thuringiensis B1. The first step involves the aliphatic side-chain hydroxylation to form 2-hydroxyibuprofen by the action of an aliphatic monooxygenase. This intermediate is converted to 2-(4-hydroxyphenyl-) propionic acid, which is acted upon by acyl-CoA synthase/thiolase to form 1,4-hydroquinone. The action of 1,4-hydroquinone monooxygenase forms 2-hydroxy-1,4-quinol, which undergoes ortho-ring cleavage by the action of hydroxyquinol 1,2-dioxygenase to form 3-hydroxy-cis,cis-muconic acid, which is funnelled into central carbon pathway (Marchlewicz et al., 2017; Figure 5A).
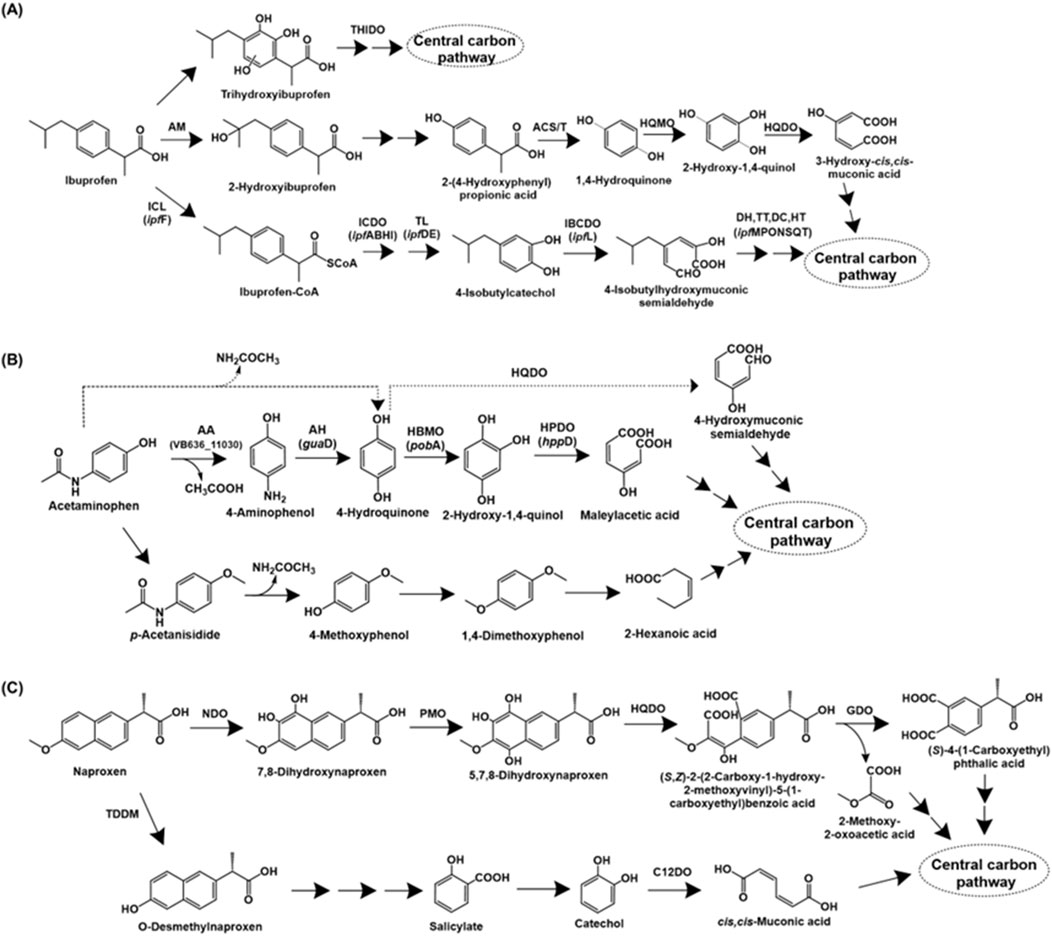
Figure 5. Bacterial degradation pathways of analgesics (A) ibuprofen (B) acetaminophen and (C) naproxen. Genes encoding respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: THIDO, trihydroxyibuprofen dioxygenase; AM, aliphatic monoxygenase; ACS/T, acyl-CoA synthase/thiolase; HQMO, 1,4-hydroquinone monoxygenase; HQDO, hydroxyquinol-1,2-dioxygenase; ICL, ibuprofen CoA ligase; ICDO, ibuprofen-CoA dioxygenase; TL, thiolase; IBCDO, isobutylcatechol dioxygenase; DH, dehydrogenase; TT, tautomerase; DC, decarboxylase; HT, hydratase; AA, M20 aminoacylase family aminohydrolase; AH, aminohydrolase or guanidine deaminase; HBMO, 4-hydroxybenzoate 3-monoxygenase; HPDO, hydroxyquinol dioxygenase or 4-hydroxyphenylpyruvate dioxygenase; TDDM, tetrahydrofolate-dependent O-demethylase; NDO, naphthalene dioxygenase; PMO, phenol monoxygenase; GDO, gentisate dioxygenase; C12DO, catechol-1,2-dioxygenase.
Alternatively, ibuprofen is also catabolised via aromatic ring hydroxylation and cleavage. Sphingomonas sp. Ibu-2 metabolises ibuprofen to ibuprofen-CoA by the action of a CoA-ligase. Further, this intermediate is converted to isobutylcatechol (upon removal of propionic acid side-chain), which is ring-cleaved via meta route (Murdoch and Hay, 2013). In Variovorax sp. strain Ibu-1, ibuprofen is metabolised via the formation of trihydroxyibuprofen, which has been proposed to undergo meta-ring cleavage to form aliphatic intermediates (Murdoch and Hay, 2015; Figure 5A).
The genomic sequence of strain Ibu-2 revealed the presence of ipfABDEF gene cluster involved in degradation, along with genes ipfHI. The genes encoded ibuprofen CoA-ligase (IpfF), ibuprofen-CoA dioxygenase (IpfABHI), thiolase (IpfD) involved in removal of acyl-CoA group and IpfE (unkown function; involved in the generation of isobutylcatechol) (Żur et al., 2018). Similar gene clusters for the conversion of ibuprofen to isobutylcatechol have been identified in Sphingopyxis granuli RW412 (Aguilar-Romero et al., 2021) and Rhizorhabdus wittichii MPO218 (Aulestia et al., 2022). Additionally, the genes for further metabolism of isobutylcatechol, that is, ipfL (4-isobutylcatechol-2,3-dioxygenase) and ipfM (hydroxymuconic semialdehyde dehydrogenase), ipfP (tautomerase), ipfO (decarboxylase), ipfN (hydratase), ipfS (hydratase), ipfQ (aldehyde dehydrogenase) and ipfT (acyl-CoA dehydrogenase) have been identified in strain MPO218. Further, the upper pathway genes were flanked by IS6100 insertion elements, indicating probable acquisition by horizontal gene transfer (Aulestia et al., 2022).
2.1.2.2 Acetaminophen
Paracetamol, also known as acetaminophen [N-(4-hydroxyphenyl)acetamide], is a commonly used analgesic and antipyretic that functions by inhibiting prostaglandin synthesis (Roberts et al., 2016). The first step of bacterial acetaminophen degradation proceeds via the action of aryl acyla midase to form 4-aminophenol, which is further converted to hydroquinone by the action of an aminohydrolase. Hydroquinone formation has also been proposed to occur directly with release of acetamide as a byproduct (Hu et al., 2013). Hydroquinone undergoes ring-cleavage by the action of a dioxygenase to form organic acids (Hu et al., 2013; Żur et al., 2018). Alternatively, hydroquinone can undergo hydroxylation to form 1,2,4-trihydroxybenzene, followed by ring-cleavage (Takenaka et al., 2003; Figure 5B). An alternate pathway for paracetamol degradation has been proposed in soil micro-organisms by Li et al., involving the methylation of paracetamol to para-acetanisidide, which is converted to 4-methoxyphenol and further, 1,4-dimethoxybenzene. This intermediate is further ring-cleaved to aliphatic intermediates (Li et al., 2014; Figure 5B).
The genetics of paracetamol degradation were detailed in the bacterium Paracoccus sp. APAP_BH8. The genes encoding a M20 aminoacylase family aminohydrolase (involved in hydrolysis of paracetamol to 4-aminophenol), guaD (guanidine deaminase for the formation of hydroquinone from 4-aminophenol), pobA (4-hydroxybenzoate-3-monooxygenase for conversion of hydroquinone to hydroquinol) and hppD (4-hydroxyphenylpyruvate dioxygenase for the ring-cleavage of hydroquinol) were identified (Pandey et al., 2024). Amidase genes involved in the conversion of paracetamol to 4-aminophenol were detected in two Pseudomonas species, with mobile genetic elements in their vicinity, indicating probable role of horizontal gene transfer. The extradiol dioxygenase genes involved in subsequent degradation were also detected in the genome (Rios-Miguel et al., 2022).
2.1.2.3 Naproxen
Naproxen [6-methoxy-alpha-methyl-2-naphthaleneacetic acid] is an NSAID, widely used as an analgesic and antipyretic. Similar to ibuprofen, naproxen functions by suppressing the cyclooxygenase (COX) enzyme activity (Ríos et al., 2022). The complete naproxen degradation pathways have been described in Stenotrophomonas maltophilia KB2 and Bacillus thuringiensis B1. In strain KB2, naproxen is dihydroxylated to 7,8-dihydroxynaproxen by the action of naphthalene dioxygenase, which has been reported to have a wide substrate range (Lee and Gibson, 1996; Selifonov et al., 1996; Phale et al., 2007). Further, the action of phenol monooxygenase generates 5,7,8-trihydroxynaproxen, which undergoes ring-fission by the action of hydroxyquinol 1,2-dioxygenase, leading to the generation of an monoaromatic intermediate. Further, the action of gentisate dioxygenase results in conversion to an aliphatic intermediate, which is funnelled into the central carbon metabolism (Wojcieszyńska et al., 2014; Figure 5C).
Naproxen degradation in Bacillus thuringiensis B1 involves the removal of methyl group to form O-desmethylnaproxen by the action of tetrahydrofolate-dependent O-demethylase. This intermediate is converted to salicylate, which subsequently forms either catechol or gentisate (dihydroxy-intermediates). The major naproxen degradation proceeds via ring-cleavage of catechol by enzyme catechol-1,2-dioxygenase; whereas, ring-cleavage by the enzymes gentisate-1,2-dioxygenase (acting on gentisate) and salicylate-1,2-dioxygenase (acting on salicylate) are minor pathways (Górny et al., 2019; Figure 5C).
2.1.3 Steroid sex hormones
In humans, steroid sex hormones are synthesised from cholesterol and can be classified as androgens, progestogens and oestrogens based on their structure and function. Androgens (such as testosterone) and oestrogens (such as oestrone: E1, 17β-oestradiol: E2, estriol: E3) regulate the development and maintenance of secondary sexual characteristics as well as the reproductive system in males and females, respectively. Whereas, progestogens (like progesterone) are essential for implantation of the embryo and maintenance of pregnancy. Aside from endogenous (naturally occurring) sex hormones, synthetic derivatives of androgens (like 19-nortestosterone), oestrogens (like 17α-ethynyloestradiol: EE2) and progestogens (like progestin) find application in agriculture, aquaculture as well as human health (Chiang et al., 2020).
Major sources of these hormones in the biosphere include human and animal excreta (Lange et al., 2002; Chang et al., 2011), use of manure and sewage derivatives as fertilizers (Kjaer et al., 2007; Hamid and Eskicioglu, 2012) and microbial transformation of phytosterols (Orrego et al., 2009). These compounds have been detected at varying concentrations in the environment. For example, the influent concentration of natural androgens was found to be 2,977 ± 739 ng L−1 (androsterone), 640 ± 263 ng L−1 (epiandrosterone) and 270 ± 132 ng L−1 (androstenedione) in WWTPs in Beijing, China (Chang et al., 2011). The concentration of Estrone (E1) was found to be 5.4–25 ng L−1 in Swiss hospital wastewater (Zhang et al., 2017a). The oestrogens Estrone (E1), 17β-estradiol (E2), and 17α-ethynylestradiol (EE2) were detected in raw sewage at concentrations up to 104, 66.9, and 5.7 ng L−1, respectively in Ontario, Canada (Atkinson et al., 2012). The persistence of these compounds in the environment impacts fish, amphibians and mammals as these hormones function as endocrine disruptors (Aris et al., 2014) and pheromone mimics (Doyle and Meeks, 2018). As compared to androgens and oestrogens, the bacterial degradation of progestogens is poorly detailed, with only biotransformation products being reported (Chiang et al., 2020).
2.1.3.1 Testosterone
The degradation of testosterone has been primarily studied in Comamonas testosteroni through the 9,10-seco pathway. The first step of this catabolic pathway involves the oxidation of the 17-hydroxyl group to a carbonyl group to form androst-4-en-3,17-dione (AD), catalysed by the enzyme 17β-hydroxysteroid dehydrogenase. Further, the action of 3-ketosteroid dehydrogenase (TesH) introduces a double bond between C-1 and C-2 to form androsta-1,4-diene-3,17-dione (ADD), which is further hydroxylated at C-9 position by the enzyme 3-ketosteroid 9α-hydroxylase to form 9α-hydroxy-androsta-1,4-diene-3,17-dione. This intermediate is unstable and forms 3-hydroxy-9,10-seconandrosta-1,3,5 (10)-triene-9,17-dione (3-HSA) upon spontaneous cleavage of the bond between C-9 and C-10 and aromatisation (Figure 6). Further, the aromatic ring is hydroxylated by the TesA1A2 monooxygenase to form the catecholic intermediate 3,4-dihydroxy-9,10-seco nandrost-1,3,5 (10)-triene-9,17-dione (3,4-DHSA), which undergoes meta ring-cleavage by the action of TesB extradiol dioxygenase to form 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1 (10),2-diene-4-oic acid (4,9-DSHA). This intermediate undergoes hydrolytic cleavage between C-5 and C-10 to produce 3aα-H-4a (3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione (HIP) and 2-hydroxyhexa-2,4-dienoic acid. The latter is further metabolised by the action of hydratase (TesE), aldolase (TesG) and a dehydrogenase (TesF). The HIP intermediate is reported for various steroid hormone degradation pathways and multiple bacteria possess a common HIP degradation pathway (Chiang et al., 2020; Figure 6). The genes involved in testosterone metabolism in C. testosteroni TA441 have been reported to be localised as a 120 kb mega cluster carrying the aromatic ring-degradation genes (tesGFEDA1A2HIJ-scdA) involved in catabolism of A and B rings and the β-oxidation gene cluster (steABCD-tesB-scdL1L2NKYM1M2FE-25–26-EC1C2GDJ-tesR) involved in HIP degradation (C and D rings). The genes encoding 3α-hydroxydehydrogenase (3α-DH) and 3-ketosteroid Δ4-5 isomerase (ksi) are localised between the two clusters. The tesR gene encoded a positive regulator of both the gene clusters (Horinouchi and Hayashi, 2023).
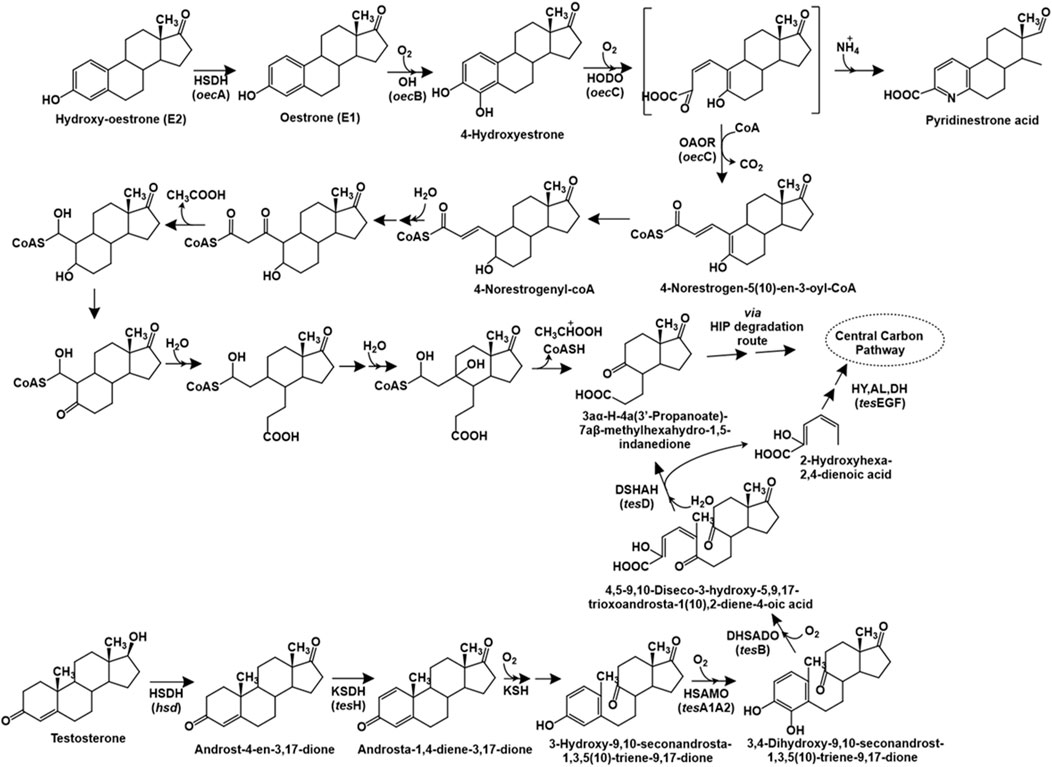
Figure 6. Bacterial degradation pathways of testosterone and oestrone. Genes encoding respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: HSDH, 17β-oestradiol dehydrogenase; OH, oestrone 4-hydroxylase; HODO, 4-hydroxyestrone 4,5-dioxygenase; OAOR, 2-oxoacid oxidoreductase; HSDH, hydroxysteroid dehydrogenase; KSDH, ketosteroid dehydrogenase; KSH, 3-ketosteroid 9α-hydroxylase; HSAMO, 3-hydroxy-9,10-secoandrosta-1,3,5 (10)-triene-9,17-dione hydroxylase; DHSADO, 3,4-dihydroxy-9,10-secoandrosta-1,3,5 (10)-triene-9,17-dione dioxygenase; DSHAH, 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1 (10),2-dien-4-oic acid hydrolase; HY, (2Z,4Z)−2-hydroxyhexa-2,4-dienoic acid hydratase; AL, aldolase; DH, acetoaldehyde dehydrogenase.
2.1.3.2 Oestrogens
The complete degradation pathway of oestrogen (E1: oestrone) has been proposed for Sphingomonas sp. strain KC8 via the 4,5-seco route (Wu et al., 2019). The first step involves the hydroxylation of oestrogen to 4-hydroxyestrone. This catecholic intermediate undergoes meta ring-cleavage by the action of 4-hydroxyestrone 4,5-dioxygenase. The product of this reaction is unstable; and undergoes abiotic recyclization (in presence of ammonium) to form pyridinestrone acid as a dead-end product. Alternatively, the enzyme 2-oxoacid oxidoreductase (belonging to indolepyruvate ferredoxin oxidoreductase family) catalyses the removal of C-4 (as CO2) and adds a coenzyme-A (CoA) moiety to the C-3 carbon to form the intermediate 4-norestrogen-5 (10)-en-3-oyl-CoA through oxidative decarboxylation, which undergoes reduction to 4-norestrogenyl-CoA. The C-2 and C-3 carbons (part of the A-ring) are removed via thiolytic β-oxidation by the action of enzymes enoyl-CoA hydaratase, β-hydroxyacyl-CoA dehydrogenase and thiolase. Further, the B-ring of oestrone undergoes hydrolytic cleavage, followed by aldolytic cleavage to remove C-1 and C-10, resulting in the formation of HIP, which is metabolised via the HIP degradation pathway (Wu et al., 2019; Chiang et al., 2020; Figure 6).
The genome of strain KC8 has been reported to harbour the gene oecA (3β,17β-hydroxysteroid dehydrogenase) and three other clusters for the metabolism of oestrogen. The cluster I carries the gene oecB encoding flavin-dependent estrone-4-hydroxylase which converts estrone to 4-hydroxyestrone. Whereas, cluster II carries the oecC gene encoding 4-hydroxyestrone-4,5-dioxygenase and other genes involved in β-oxidation. Whereas, cluster III encodes enzymes involved in C/D ring degradation (Chen et al., 2017).
2.1.4 Antidepressants
2.1.4.1 Fluoxetine
Fluoxetine [N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine] (sold under the brand name Prozac) is an antidepressant belonging to the class of selective serotonin reuptake inhibitors (SSRI). Due to its widespread application in treatment of psychiatric disorders, it has been frequently detected in aquatic ecosystems, causing toxicity to biota (Brooks et al., 2003; Shi et al., 2019; Deere et al., 2021; Ma et al., 2022). For example, fluoxetine has been detected in WWTPs and receiving waters of the Huangpu River, China at concentrations upto 42.9 ng L−1 (Wu et al., 2017). Fluoxetine and its human metabolite, norfluoxetine were detected at a concentration of 3.5–16 ng L−1 in raw wastewater and 1.2–15 ng L−1 in treated wastewater in Uppsala, Sweden (Barclay et al., 2012).
Fluoxetine catabolic pathway has been detailed for various Bacillus spp., Pseudomonas spp. and Comamonas testosteroni, which utilised it as the sole source of carbon and energy (Khan and Murphy, 2021). The fluoxetine degradation is initiated by hydrolysis of the ether bond to yield 4-(trifluoromethyl) phenol (TFMP) and 3-(methylamino)-1-phenylpropan-1-ol. The latter is utilised as the sole source of carbon and energy while TFMP was accumulated in the culture medium. However, the strains exhibited growth on TFMP as sole carbon source, which was further hydroxylated to 4-(trifluoromethyl)catechol. This intermediate was ring-cleaved via the meta pathway, as indicated by the presence of specific metabolites in the culture medium. These aliphatic intermediates undergo subsequent decarboxylation, aldolytic cleavage, hydroxylation, oxidation and a final decarboxylation to form trifluoroacetic acid, which was a dead-end product. Additionally, fluoride ion was also detected in the culture medium due to defluorination via photolytic degradation of the meta ring-cleavage product (Figure 7A; Khan and Murphy, 2021).
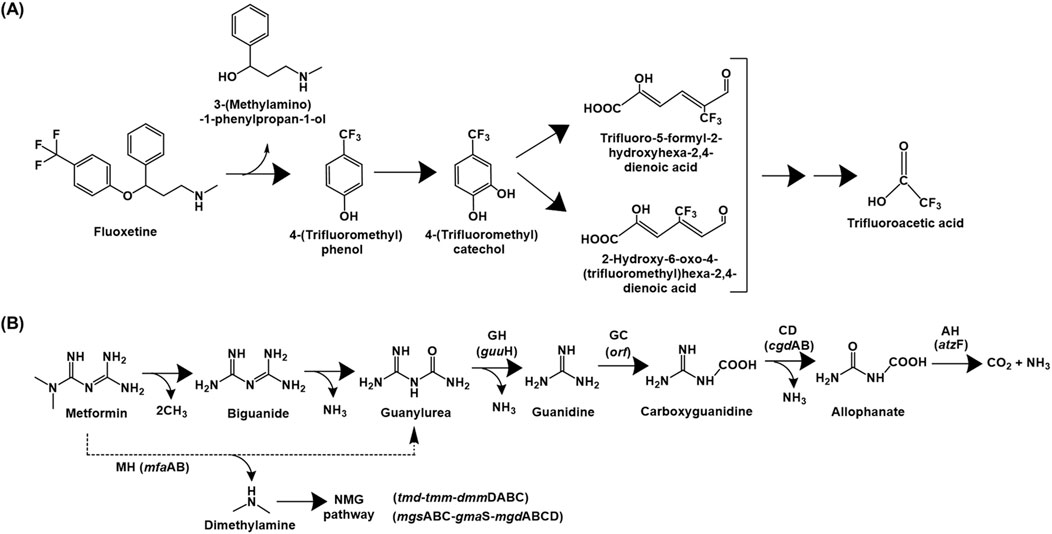
Figure 7. Bacterial degradation pathways of (A) fluoxetine and (B) metformin. Genes encoding respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: MH, metformin hydrolase; GH, guanylurea hydrolase; GC, guanidine carboxylase; CD, carboxyguanidine deaminase; AH, allophanate hydrolase; NMG pathway, N-methylglutamate pathway.
2.1.5 Antidiabetics
2.1.5.1 Metformin
Metformin (N,N-dimethylimidodicarbonimidic diamide) is a globally used first-line drug for the treatment of type-II diabetes and obesity. Its mechanism of action involves activation of the enzyme AMP-activated protein kinase, which inhibits gluconeogenesis in the liver, thereby reducing blood glucose (Pernicova and Korbonits, 2014). An approximate 70% of metformin is excreted unmetabolized through the human body, contributing significantly to its prevalence in aquatic habitats, impacting the native biota (Ambrosio-Albuquerque et al., 2021). Apart from metformin, its main breakdown product guanylurea (upon removal of dimethylamine) has been reported to accumulate as a dead-end product in surface waters, coastal waters and wastewater treatment plants globally at varying concentrations (Scheurer et al., 2009; Scheurer et al., 2012; Blair et al., 2013; Ghoshdastidar et al., 2015; Tao et al., 2018). For example, the concentration of metformin in German WWTP influents, effluents and surface waters was 111,800, 4,800 and 102 ng L−1, respectively (Trautwein et al., 2014). Whereas, in streams across the southeastern U.S., the metformin concentration was up to 16,000 ng L−1 (Bradley et al., 2016). WWTPs in Greece reported metformin concentrations up to 1,167 ng L−1 (influent) and 627 ng L−1 (effluent) (Kosma et al., 2015).
The complete mineralisation pathway of metformin has been described for the consortium of Aminobacter sp. MET and Pseudomonas mendocina MET (Martinez-Vaz et al., 2022). Metformin is converted to guanylurea via the displacement of dimethylamine (which is utilised as a carbon and nitrogen source) by Aminobacter sp. MET. Genome analyses of the strain further identified dimethylamine monooxygenase, which converts dimethylamine to methylamine via oxidation. Further, methylamine was proposed to be metabolised via the N-methyl glutamate pathway. Guanylurea was transported out of the cell by a Gdx exporter protein as it is a toxic molecule. Further, this intermediate was utilised as a sole nitrogen source by P. mendocina MET, which utilised all nitrogen atoms for growth. Guanylurea was converted to guanidine by the action of guanylurea hydrolase (GuuH), which was further converted to carboxyguanidine by the action of guanidine carboxylase (GC). This intermediate is metabolised to allophanate by the action of carboxyguanidine deaminase (CgdAB), which is converted to carbon-di-oxide and ammonia by the action of allophanate hydrolase (AtzF; Figure 7B). Alternatively, P. mendocina MET utilised metformin as a sole nitrogen source, by conversion to 1-N-bimethylguanide and further to biguanide. This metabolite is converted to guanylurea by a deamination reaction and is assimilated, as described (Martinez-Vaz et al., 2022).
Aminobacter sp. strain NyZ550 utilises metformin as a sole source of carbon, nitrogen and energy. The initial hydrolysis of metformin generates guanylurea and dimethylamine. The former accumulates as a dead-end product, while dimethylamine is utilised as a sole carbon and nitrogen source by a metabolic pathway similar to that reported in Aminobacter sp. MET. To further metabolise the guanylurea generated, Pseudomonas putida PaW340 was engineered to express guanylurea hydrolase; and both strains NyZ550 and PaW340 were co-cultured (Li et al., 2023; Figure 7B). In strain NyZ550, the genes involved in metformin metabolism were localised as three distinct clusters. Cluster I encoded the genes tmd-tmm-dmmDABC (Trimethylamine N-oxide demethylase, trimethylamine monooxygenase and dimethylamine monooxygenase), whereas cluster II encoded the genes mgsABC-gmaS-mgdABCD (N-methylglutamate synthase, γ-glutamylmethylamidesynthetase and N-methyl glutamate dehydrogenase, respectively). Both the clusters were involved in methylamine metabolism. Whereas, cluster III encoded agmatinase and hypAB (involved in loading dinickel onto agmatinase involved in metformin hydrolysis; Li et al., 2023). In Aminobacter niigataensis MD1 (isolated from activated sludge), the enzyme metformin hydrolase converts metformin to guanylurea and dimethylamine. The latter is utilised as the sole source of carbon and nitrogen via a similar route described for other isolates. Additionally, the metformin degradation gene arrangement was similar to strain NyZ550 (Chaignaud et al., 2022; Li et al., 2023).
Guanylurea metabolism in Pseudomonas mendocina GU proceeds via its hydrolytic deamination to guanidine and ammonia, which is catalysed by the enzyme guanylurea hydrolase, a novel enzyme belonging to the isochorismate hydrolase-like protein family. The bacterium utilises guanylurea but not metformin as a sole nitrogen source (Tassoulas et al., 2021). While the gene encoding guuH (encoding guanylurea hydrolase) was present separately on the chromosome, an ORF encoding guanidine carboxylase, carboxyguanidine deaminase (cgdAB) and regulatory guanidine riboswitches were clustered together. Whereas, the gene atzF (allophanate hydrolase) was localised adjacent to urea carboxylase and a transcriptional regulator (Tassoulas et al., 2021).
The genes mfaAB encoding metformin hydrolase (ureohydrolase activity) have been identified in bacteria isolated from activated sludge (Tassoulas et al., 2021). The enzyme forms an active heterocomplex that catalyses the Ni2+-dependent hydrolysis to guanylurea and methylamine (Li et al., 2024).
3 Cyanotoxins
Cyanotoxins are secondary metabolites produced by Cyanobacteria that are toxic to humans and other biota. Cyanotoxins are classified as per two main criteria: (1) mechanism of action, that is, hepatotoxins, neurotoxins, dermatotoxins, etc., and (2) chemical structure, that is, cyclic peptides (like microcystin and nodularin), alkaloids (anatoxin) or lipopolysaccharides (Ferrão-Filho and Kozlowsky-Suzuki, 2011). They are classified as emerging contaminants due to their release during extensive eutrophication/algal blooms, causing health hazards such as cancer, neuromuscular blockade, anti-acetylcholinesterase activity, anti-phosphatase activity, post synaptic cholinergic agonist activity, activation of protein kinase C, inhibition of serine/threonine protein phosphatases and inhibition of protein synthesis (Fujiki et al., 1990; Mackintosh et al., 1990; Yoshizawa et al., 1990; Codd et al., 1997; Metcalf et al., 2004; Funari and Testai, 2008; Dziga et al., 2016). Microcystins have been detected in Czech reservoir water with median and maximal concentrations of 1.5 and 18.6 μg L−1, respectively. Various cyanotoxins have been detected at varying concentrations in water reservoirs, fish tissue and aquatic plants in Nebraska, United States, highlighting their potential for bioaccumulation (Al-Sammak et al., 2014).
3.1 Microcystins and nodularin
Microcystins (MCs) are the most commonly found cyanotoxins produced by several genera of Cyanobacteria and are the most studied. MCs are cyclic heptapeptides, comprised of cyclo-(D-Ala1–X2–D-MeAsp3–Z4–Adda5–D-Glu6–Mdha7) with approximately 250 identified variants (Spoof and Catherine, 2016; Yang et al., 2020). The X and Z represent variable L-amino acids (microcystins referred as MC-XZ), MeAsp is erythro-β-methylaspartic acid, Adda is (2S, 3S, 8S, 9S) 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyl-deca-4,6-dienoic acid (β-amino acid), and Mdha is N-methyldehydroalanine (Krishnamurthy et al., 1989; Figure 8). Some examples of MC-XZ variants include MC-LR, MC-RR, MC-YR, MC-WR, MC-LY, MC-LW, MC-LF, MC-LA etc., where MC-LR is one of the most widely distributed and highly toxic variant.
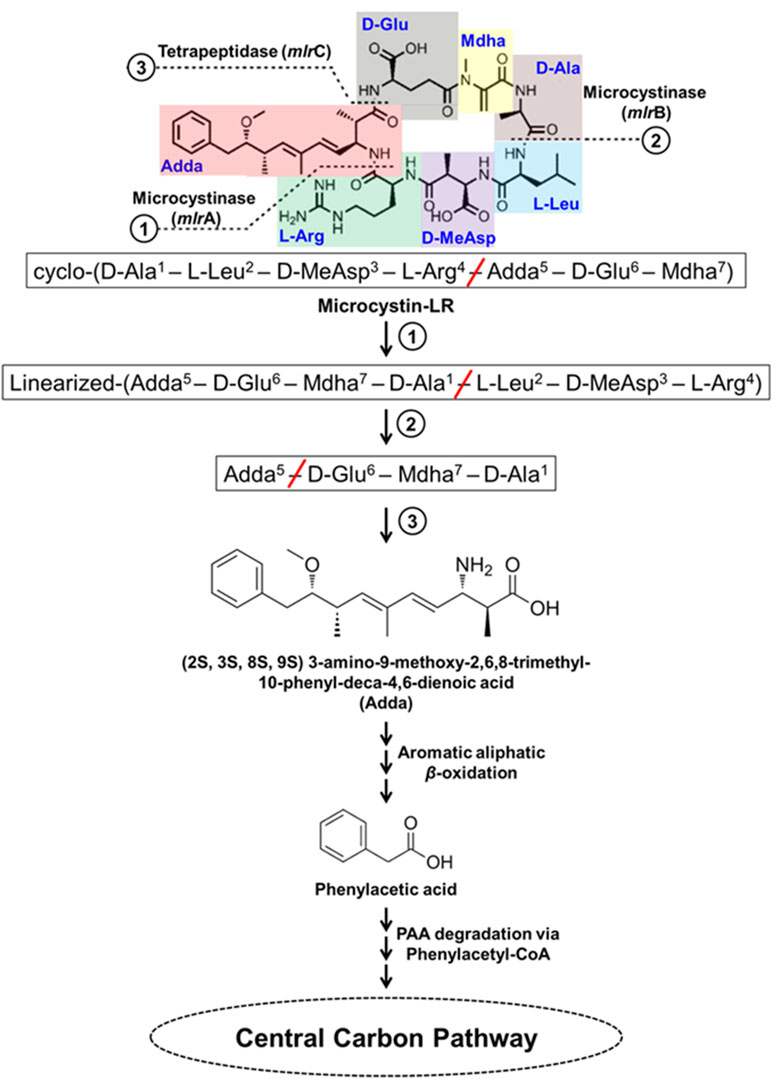
Figure 8. Bacterial degradation pathway of microcystin-LR. Gene encoding of the respective enzymes are indicated in parenthesis. The primary, secondary and tertiary cleavage sites (and corresponding metabolic steps) are indicated numerically in circles. Multiple arrows indicate multiple metabolic steps.
MC-LR degradation has been detailed for Sphingopyxis sp. YF1. The cyclic MC-LR is first linearized by cleavage of Adda-Arg peptide bond catalyzed by microcystinase which is further acted upon by linearized microcystinase cleaving the Ala-Leu peptide bond forming a tetrapeptide containing Adda. This tetrapeptide is cleaved at Adda-Glu peptide bond by tetrapeptidase forming Adda. Adda is metabolized to form aromatic aliphatic hydrocarbon (C20H26O4) by the action of aminotransferase (Figure 8). The aromatic aliphatic hydrocarbon gets converted to phenylacetic acid by the microbial β-oxidation enzymes (fatty acid-CoA ligase, acyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and thiolase) probably in four cycles of β-oxidation releasing acetyl-CoA/propanoyl-CoA in each cycle. Potential intermediates formed during β-oxidation cycles were identified as 7-methoxy-4,6-dimethyl-8-phenyloca-2,4-dienoic acid and 2-methyl-3-methoxy-4-phenylbutyric acid. Such β-oxidation of aromatic aliphatic hydrocarbons has also been reported for alkylbenzenes (Sariaslani et al., 1974; Awe et al., 2008; Nhi-Cong et al., 2010; Figure 8). Further, phenylacetic acid is activated by ligating coenzyme-A catalyzed by fatty acid-CoA ligase, a phenylacetate-CoA ligase like enzyme (PAAase), in Sphingopyxis sp. YF1 and is proposed to be degraded via phenylacetyl-CoA route. The phenylacetyl-CoA is degraded to acetyl-CoA by the action of enzymes phenylacetyl-CoA epoxidase (paaABCDE), 2-(1,2-epoxy-1,2-dihydrophenyl) acetyl-CoA isomerase (paaG), oxepin-CoA hydrolase (paaZ), 3-oxoadipyl-CoA thiolase (paaI) (Figure 8).
The genes involved in MC-LR degradation in strain YF1 include mlrBDAC cluster which converts MC-LR to Adda followed by aminotransferase and microbial β-oxidation encoding genes leading to formation of phenylacetate. Further, the paaI-paaGZ-paaABCDE gene clusters encodes enzymes for degradation of phenylacetate to acetyl-CoA. The mlr and paa clusters along with genes encoding aminotransferase and β-oxidation enzymes were located in proximity suggesting their involvement in MC-LR degradation. The mlr cluster has been observed in other MC-LR degrading microbes such as Sphingosicicella microcystinivorans B-9 and Novosphingobium sp. THN1 (Jin et al., 2018; Wang J. et al., 2019; Yang et al., 2020).
Nodularin is a cyclic pentapeptide comprising of D-MeAsp1–L-Arg2–Adda3–D-Glu4–Mdhb5, where 1st–4th amino acids are similar to 3rd–6th amino acids of MC-LR and the 5th Mdhb is N-methyldehydrobutyrine. Microcystin degrading bacteria harbouring mlrBDAC cluster have been observed to degrade nodularin, which is a pentapeptide possessing cleavage sites similar to MC-LR (Figure 8). As observed in Sphingopyxis sp. m6, during nodularin degradation mlr cluster was upregulated and products such as linearized nodularin and Adda were detected. This suggests nodularin degradation share similar enzymes/enzymatic steps (Yang et al., 2020; Yuan et al., 2021; Wei et al., 2023).
4 Plasticizers
Plasticizers are compounds used as additives to plastics to alter physical properties such as softness and flexibility. These compounds can be released into the environment during synthesis, domestic use, improper disposal or through leaching (Billings et al., 2021). Common examples of these compounds include di (2-ethylhexyl) phthalate (DEHP), dibutyl phthalate (DBP), benzyl butyl phthalate (BBP), and di-n-octyl phthalate (DnOP), which have been detected in various environmental compartments, posing significant risks to human and ecological health due to their toxicity, mutagenicity and endocrine-disrupting activity (Wang et al., 2024). In the Taihu Lake basin, China, DBP and DEHP were detected in surface waters with concentrations of 1.59 μg L−1 and 1.29 μg L−1 (mean values), respectively (Gao et al., 2019). In atmospheric samples from the North Sea, concentrations of DBP, BBP, and DEHP were found to be up to 6.6 ng L−1 (Xie et al., 2005). DEHP was found to occur at concentrations up to 18.5 μg L−1 and 0.33–97.8 μg L−1 in Taiwan river sediments (Yuan et al., 2002) and German surface waters (Fromme et al., 2002), respectively.
4.1 DEHP (Di (2-ethylhexyl) phthalate)
Di (2-ethylhexyl) phthalate (DEHP) is the most extensively used plasticizer and is a phthalate ester composed of a phthalate backbone with two 2-ethylhexyl groups attached. It is toxic, estrogenic and a potent endocrine disrupting environmental pollutant. Several bacterial genera capable of degrading DEHP have been isolated such as Gordonia, Rhodococcus, Mycrobacterium, Pseudomonas, Cupravidus, Burkholderia, Achromobacter, Agromyces, Microbacterium, Acinetobacter, Bacillus, etc (Zhao et al., 2016; Xu et al., 2017; Zhang et al., 2018; Fan et al., 2018; Li et al., 2019; Wright et al., 2020; Wang et al., 2021; Chen et al., 2021a and b; Kamaraj et al., 2022; Sun et al., 2024). The initial degradation of DEHP typically occurs through two main routes: de-esterification leading to the formation of mono-alkyl esters like mono-(2-ethyhexyl) phthalate (MEHP), or stepwise beta-oxidation of alkyl side chains resulting in DBP (Figure 9). In most of bacterial strains, DEHP is hydrolysed into MEHP by esterases, which is further converted to phthalic acid, either directly or via mono-butyl phthalate (MBP) (Ren et al., 2016; Xu et al., 2017; Nahurira et al., 2017; Fan et al., 2018; Li et al., 2019; Lamraoui et al., 2020; Zhang H. et al., 2020; Wang et al., 2021; Kamaraj et al., 2022; Hsu et al., 2023; Kou et al., 2023; Bhattacharyya et al., 2023; Dhar et al., 2023). In other strains, the alkyl side chain of DEHP is first oxidised to DBP, which is then hydrolysed to yield phthalic acid, either directly (Zhang et al., 2021; Chen et al., 2021a; Chen et al., 2022) or through intermediates like diethyl phthalate (DEP), mono-methyl phthalate (MMP) or butyl methyl phthalate (BMP), MBP (Chen et al., 2021b; Zhang et al., 2021; Figure 9).
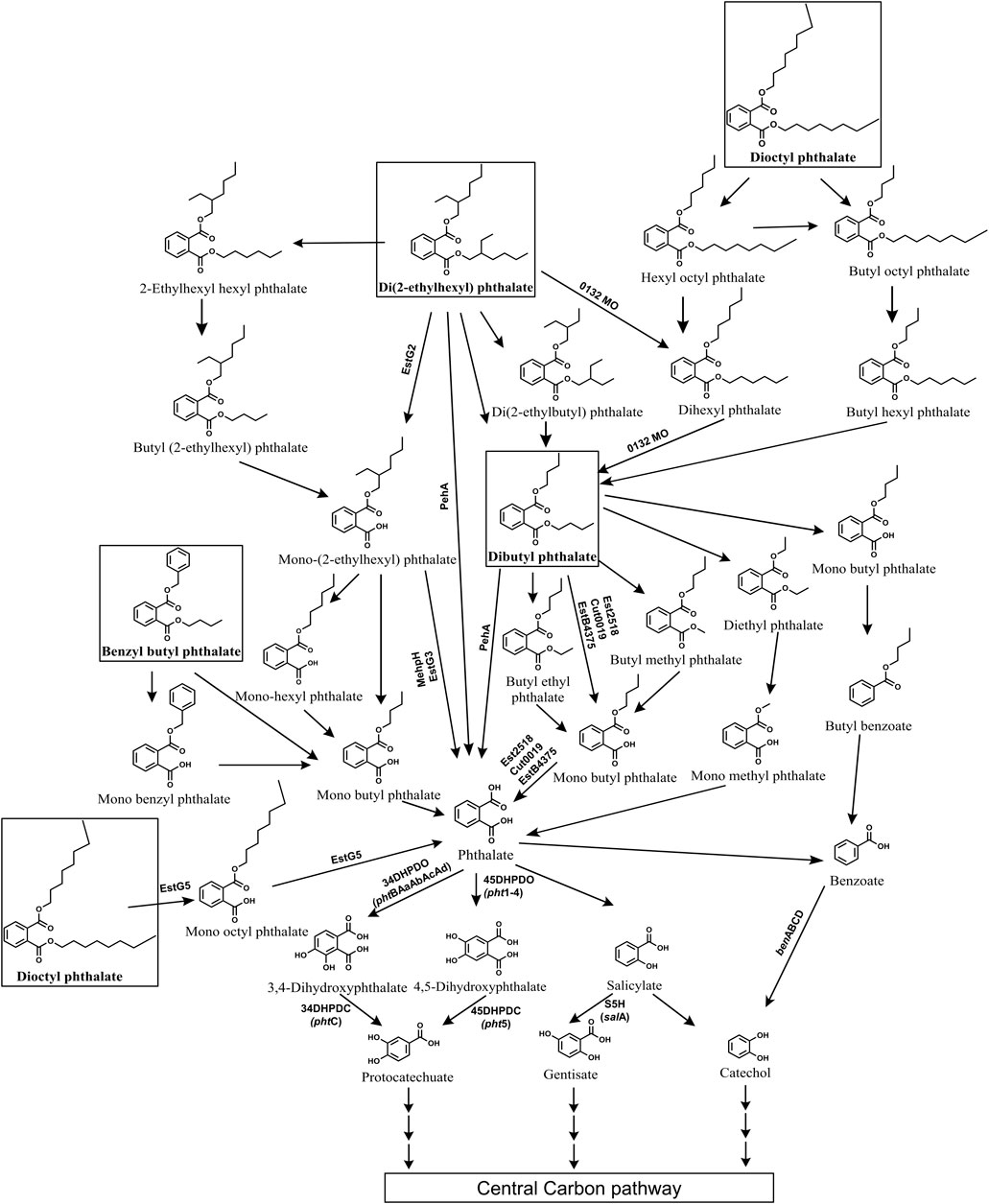
Figure 9. Bacterial degradation pathways of plasticizers: di (2-ethylhexyl) phthalate, dibutyl phthalate, benzyl butyl phthalate and di-n-octyl phthalate. Genes encoding respective enzymes are indicated in parenthesis. Multiple arrows indicate multiple metabolic steps. Enzyme abbreviations: 0132MO, 0132 Monooxygenase; EstG2, Esterase G2; EstG3, Esterase G3; EstG5, Esterase G5; Peh, phthalate ester hydrolase A; MehpH, mono ethylhexyl phthalate hydrolase; Est2518, Esterase 2,518; EstB4375, Esterase B4375; Cut0019, Esterase cut0019; 34DHPDO, 3,4-dihydroxyphthalate dioxygenase; 45DHPDO, 4,5-dihydroxyphthalate dioxygenase; 34DHPDC, 3,4-dihydroxyphthalate decarboxylase; 45DHPDC, 4,5-dihydroxyphthalate decarboxylase; S5H, salicylate-5-hydroxylase; BDO, benzoate dioxygenase.
Some strains employ both de-esterification (via MEHP) and alkyl side chain oxidation (via DBP) routes to degrade DEHP to phthalic acid (Zhao et al., 2018; Chen et al., 2021b; Chang et al., 2022). A few strains such as Rhodococcus pyridininvorans DNHP-2 exhibit alternate pathways wherein DEHP undergoes conversion to 2-ethyl hexyl benzoic acid (2EHBA), which is further converted into benzoic acid (Wang et al., 2022). In Gordonia sp. LFF, DEHP is metabolized to phthalic acid via ethylhexyl hexyl phthalate (EHHP), butyl-(2-ethylhexyl) phthalate (BEHP), MEHP, mono-hexyl phthalate (MHP), and MBP (Wang et al., 2019b). In Microbacterium sp. DEHP1 and Mycolibacterium phocacium RL-HYO1, DEHP is converted to phthalic acid through intermediates di (2-ethylbutyl) phthalate (DEBP), di-n-hexyl phthalate (DnHP), DBP, and diethyl phthalate (DEP) (Ren et al., 2021; Sun et al., 2024; Figure 9).
Further, the resulting phthalic acid is converted to protocatechuate (PCA) either via 3,4-dihydroxyphthalate (34DHP) (Fan et al., 2018; Zhao et al., 2018; Chen et al., 2021a; Chen et al., 2021b; Bhattacharyya et al., 2023) or 4,5-dihydroxyphthalate (45DHP) (Xu et al., 2017). In few strains phthalic acid is converted to salicylate and then to gentisate or catechol (Chen et al., 2007; Ren et al., 2021). While in some bacteria, phthalic acid is converted to benzoic acid and then funneled to catechol (Chen et al., 2021a; Wang et al., 2022; Sun et al., 2024). Common intermediates like PCA, gentisate and catechol are then ring cleaved by dioxygenases and subsequently funneled into the TCA cycle (Figure 9).
4.2 DBP (Dibutyl phthalate)
Dibutyl phthalate (DBP) is a plasticizer that is extensively used in the production of PVC products, such as flexible plastics, vinyl flooring, and medical devices. It exhibits severe endocrine-disrupting properties as well as liver and respiratory toxicity. Several bacterial genera such as Bacillus, Acinetobacter, Pseudomonas, Mycobacterium, Halomonas, Cupravidus, Arthrobacter, Microbacterium, among others, have been reported to degrade DBP (Feng et al., 2018; Wright et al., 2020; Feng et al., 2021; Chen et al., 2021b; Nandi et al., 2021; Li et al., 2022; Sun et al., 2024). The degradation of DBP typically begins with its hydrolysis to mono-butyl phthalate (MBP) by esterases, either directly (Kumar and Maitra, 2016; Feng et al., 2018; Xu et al., 2022; Shariati et al., 2022; Fan et al., 2023; Sun et al., 2024) or via intermediates like butyl ethyl phthalate (BEP) and butyl methyl phthalate (BMP) (Feng et al., 2021; Mondal et al., 2024). Alternatively, DBP is converted to phthalic acid through intermediates such as diethyl phthalate (DEP) or dimethyl phthalate (DMP) (Sun et al., 2019; Mondal et al., 2024; Figure 9). Alternatively, in some strains, such as Pseudomonas aeruginosa PS1 and Halomonas sp. ATBC28, DBP is converted to butyl benzoate, which is further metabolized to benzoic acid (Wright et al., 2020; Du et al., 2024).
Furthermore, the resulting phthalic acid is converted to PCA via intermediates like 34DHP (Feng et al., 2018; Liu et al., 2020; Wright et al., 2020; Nandi et al., 2021; Chen et al., 2021b) or 45DHP (Feng et al., 2021; Du et al., 2024). In certain bacterial strains, phthalic acid is converted to benzoic acid, which undergoes decarboxylation to yield catechol. For instance, Glutamibacter sp 0426, Enterobacter DNB, and Arthrobacter ZJUTW convert phthalic acid to PCA via benzoic acid (Sun et al., 2019; Liu et al., 2020; Ren et al., 2023b; Figure 9). Similarly, in Pseudomonas YJB6, phthalic acid is converted to PCA via benzoic acid, 45DHP, and catechol (Feng et al., 2021). Paenarthrobacter ureafaciens PB10 converts phthalic acid into gentisate via 4-hydroxyphthalic acid (4HP) (Shariati et al., 2022). Common intermediates like PCA, gentisate and catechol are then subjected to ring cleavage, facilitating their entry into the tricarboxylic acid (TCA) cycle (Figure 9).
4.3 BBP (Benzyl butyl phthalate)
Benzyl butyl phthalate is a plasticizer that is composed of a phthalate backbone with a benzyl group and a butyl group attached to it. It is widely used in synthesis of various industrial and consumer products like PVC pipes, rubber, adhesives, cosmetics and has been reported to demonstrate reproductive and developmental toxicity, endocrine disruption, etc. Various bacterial genera, including Bacillus, Acinetobacter, Arthrobacter, Gordonia, and others, have been reported with the ability to degrade benzyl butyl phthalate (BBP) (Chatterjee and Dutta, 2003; Zhang et al., 2018; Nandi et al., 2021; Kaur et al., 2021; Fan et al., 2023). These organisms typically employ two primary pathways for BBP degradation. In the first pathway, esterases hydrolyze the alkyl side chain of BBP, yielding mono-benzyl phthalate (MBeP), which is further metabolized to phthalic acid and benzyl alcohol (Chatterjee and Dutta, 2003; Zhang et al., 2018; Kaur et al., 2021; Fan et al., 2023; Figure 9). Alternatively, in the second pathway, the aromatic side chain of BBP undergoes hydrolysis, resulting in the formation of benzyl alcohol and mono-butyl phthalate (MBP), which is then converted to phthalic acid (Chatterjee and Dutta, 2003; Zhang et al., 2018; Nandi et al., 2021; Kaur et al., 2021; Fan et al., 2023; Figure 9). Both pathways yield benzyl alcohol, which is subsequently metabolized to catechol via benzoic acid while the resulting phthalic acid is metabolized to PCA either via benzoic acid (Zhang et al., 2018) or via 34DHP and 3,4-dihydroxybenzoic acid (Kaur et al., 2021; Figure 9).
4.4 DnOP (Di-n-octyl phthalate)
Di-n-octyl phthalate (DnOP) is a type of phthalate ester commonly used as a plasticizer in various industrial applications and is composed of two octyl groups attached to a phthalate backbone. It is known to be a potent endocrine disruptor, carcinogen and immunotoxin. Several bacterial genera capable of degrading DnOP have been identified, including Arthrobacter, Rhodococcus, Gordonia, Burkholderia, Bacillus, among others (Wu et al., 2010; Sarkar et al., 2013; Zhang et al., 2017b; Zhang et al., 2018; Gani and Kazmi, 2018; Dhar et al., 2023). In most of bacterial strains, DnOP degradation begins with the hydrolysis by diesterase to yield mono-n-octyl phthalate (MnOP) which is subsequently converted to phthalic acid (Sarkar et al., 2013; Zhang et al., 2017b; Zhang et al., 2018; Dhar et al., 2023; Figure 9). Alternatively, in a co-culture of Gordonia sp. JDC-2 and Arthrobacter sp. JDC-32, DnOP degradation occurs through sequential β-oxidation of the alkyl side chain of DnOP, leading to the formation of intermediates such as hydroxyl octyl phthalate (HOP), bis(2-oxoheptyl) phthalate (BOP), DBP and DEP. DEP is then hydrolyzed to yield MMP, which is subsequently converted to phthalic acid (Wu et al., 2010; Figure 9). Similarly, in the halotolerant consortium LF, DnOP degradation was initiated by β-oxidation of the alkyl side chain to yield intermediates such as HOP, BOP or dihexyl phthalate (DiHP), bis(2-hydroxypropyl) phthalate (BHP), DBP, and MBP (Wang et al., 2020). The resulting phthalic acid was converted to PCA either via dihydroxyphthalates (Zhang et al., 2018; Dhar et al., 2023) or benzoic acid (Zhang et al., 2018; Figure 9).
5 Pesticides
Pesticides belonging to the class of aniline derivatives, carbamates, chlorophenoxy compounds, chloroacetanilides, organochlorines, organophosphates, triazines, and neonicotinoids occur as CECs and pose major concern due to their high persistence, leachability, bioaccumulative nature and potential toxicity (Salimi et al., 2017; Khezami et al., 2024). These compounds are used in agricultural as well as non-agricultural settings for protection of crops against insects, fungi, nematodes, etc., as well as for the control of unwanted herbs and have been found to occur in various ecological compartments. For example, glyphosate was found to occur at 0.21–1.3 mg kg−1 soil in medlar planting site in Golmud, China (Jing et al., 2021). Imidacloprid and carbendazim have been detected in dust samples in China with concentrations of 25.8 ng g−1 and 35.8 ng g−1, respectively (Wang A. et al., 2019). In Italian house dust, imidacloprid and carbendazim were found to occur at concentrations between 1.6 and 39 μg g−1 and 0.08–4.9 μg g−1, respectively (Salis et al., 2017). The persistence and slow rate of natural attenuation of pesticides has led to various health and environmental issues. Majority of these compounds are mutagenic, endocrine disrupting, carcinogenic and are known to cause environmental hazards, compromised soil health and toxicity to biota, including humans (Choi et al., 2004).
5.1 Imidacloprid
Neonicotinoids are a recent class of pesticides consisting of thiacloprid, acetamiprid, imadacloprid, clothianidin, etc., used for crop protection, horticulture, and flea control. Imidacloprid is the most commonly used insecticide of the neonicotinoid group and is recognized as a contaminant of emerging concern (Selvam and Srinivasan, 2019; Petkovic Didovic et al., 2022). Low bioavailability of imidacloprid results in slow rate of natural attenuation and leads to longer half-life (∼997 days) in soil. Extensive use of imidacloprid exerts adverse effects on non-target species like fish, bees, earthworm, mice, human, etc. (Phugare et al., 2013; Pang et al., 2020).
Various bacteria including Pseudomonas, Bacillus, Klebsiella, Mycobacterium, etc., have been reported to degrade imidacloprid with various possible pathways. Among the reported routes, oxidation and nitro-reduction are two major microbial biodegradation pathways of imidacloprid (Pandey et al., 2009; Phugare et al., 2013; Pang et al., 2020; Zhang X. et al., 2023). 6-Chloronicotinic acid (6-CNA), olefinic cyclic nitroguanidine, cyclic urea, cyclic guanidine, nitroso, and nitro derivatives are major metabolites of imidacloprid nitro-reduction detected in soil and water samples. Under microaerophilic conditions, an aldehyde oxidase converts the ‘magic nitro’ group of imidacloprid to a nitrosoguanidine metabolite. The imidacloprid and/or formed product i.e., nitrosoguanidine is degraded through a more toxic nitroguanidine intermediate which is further converted into non-toxic urea metabolites (Pandey et al., 2009; Figure 10). In some microorganisms, imidacloprid is cleaved to 6-CNA by the formation of nitrosoguanidine and oxidative cleavage of guanidine residue (Phugare et al., 2013). Alternatively, imidacloprid is converted to 6-CNA via formation of 5-hydroxy and olefin metabolites by subsequent hydroxylation and dehydrogenation. 6-CNA is eventually converted to CO2 via 6-hydroxynicotinic acid (Sharma et al., 2014; Figure 10).
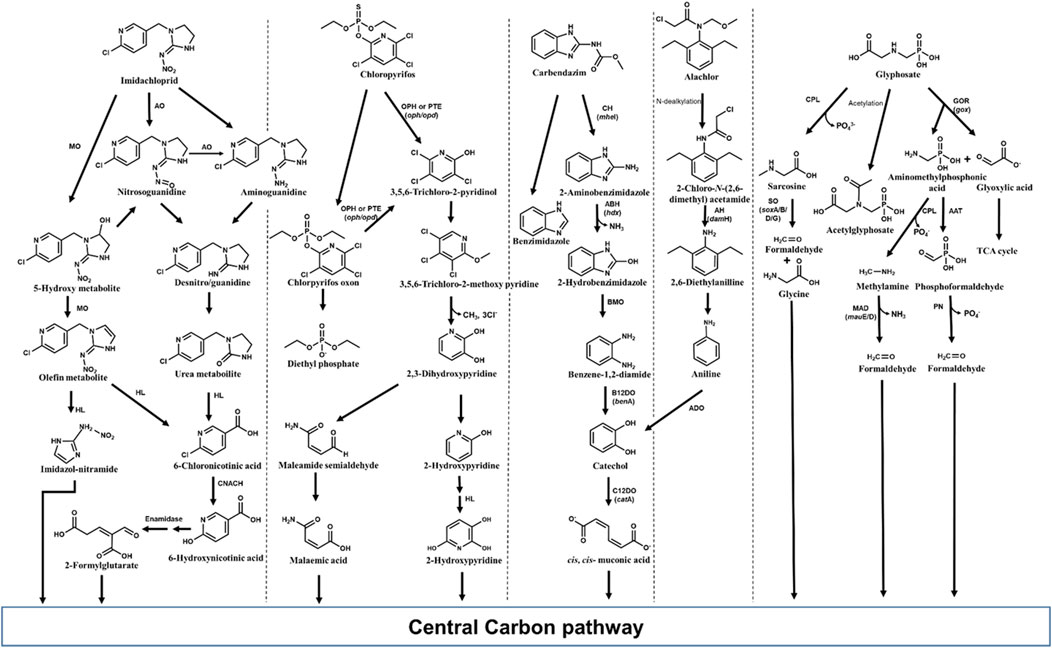
Figure 10. Metabolic pathways for degradation of various pesticides of emerging concern in bacteria. Genes encoding respective enzymes are indicated in parenthesis. Enzyme abbreviations: CPL, C-P lyase; GOR, Glyphosate oxydoreductase; ATT, AMPA aminotransferase; SO, Sarcosine oxidase; PN, Phosphonatase; MAD, methyl amine dehydrogenase; ADO, Aniline dioxygenase; C12DO, Catechol-1,2-dioxygenase; AH, amide hydrolase; CH, Carbendazim hydrolase; ABH, 2-Aminobenzimidazole hydrolase; BMO, 2-Hydrobenzimidazole monooxygenase; B12DO, Benzoate-1,2-dioxygenase; MO, monooxygenase; AO, Aldehyde oxidase; HL, Hydrolase; CNACH, 6-Chloronicotinic acid chlorohydrolase; OPH, Organophosphate hydrolase; PTE, Phosphotriesterase.
5.2 Chlorpyrifos
Chlorpyrifos, [O,O-diethyl O-(3,5,6-trichloro-2-pyridinyl)-phosphorothioate], is a broad-spectrum, chlorinated organophosphate insecticide, acaricide and miticide used to control foliage- and soil-borne insect pests on a variety of food and feed crops (Lara-Moreno et al., 2022; Bosu et al., 2024). The major health issues caused by chlorpyrifos include respiratory, immunological, reproductive, and neurological disorders in humans (Anwar et al., 2009). Although potential health risks have led to the ban of chlorpyrifos in many countries, it has been approved for limited use in densely populated countries like India, China and Bangladesh (Lara-Moreno et al., 2022). In Mexico, for example, extensive use of chlorpyrifos from 2012 to 2020 resulted in contamination of waterbodies (estuaries, drains and artesian wells) with an average concentration of 4,614 ng L−1 of chlorpyrifos (Ruiz-Arias et al., 2023). Chlorpyrifos has an average half-life of around 60–120 days in the soil, depending upon climate and soil stability (Anwar et al., 2009; Bosu et al., 2024). Various microorganisms belonging to the genera Arthrobacter, Enterobacter, Xanthomonas, Streptomyces, Stenotrophomonas, Sphingomonas, Bacillus, Synechocystis, Pseudomonas, Actinobacteria, and Klebsiella have been identified as potential chlorpyrifos degraders (Singh, 2009; Ambreen and Yasmin, 2021).
Various microorganisms are known to produce metal-dependant enzymes (hydrolases) such as organophosphorus hydrolase, phosphotriesterase (PTE), methyl parathion hydrolase and organophosphorus acid anhydrolase (OPAA) involved in chlorpyrifos bioremediation (John and Shaike, 2015; Bosu et al., 2024). Organophosphorus hydrolase effectively cleaves P–O bond in the phosphotriesters, and P–S linkage in the phosphothiolesters, yielding two major metabolites, 3,5,6-trichloro-2-pyridinol (TCP) and diethylphosphate (DETP) from chlorpyrifos. Other minor metabolites such as desethyl chlorpyrifos, chlorpyrifos oxon, desethyl chlorpyrifos oxon, and 3,5,6-trichloro-2-methoxypyridine (TMP) are also produced. Chlorpyrifos oxon, the oxidized form of chlorpyrifos, is further hydrolyzed either enzymatically or spontaneously to form diethylphosphate and TCP. The TCP can be further degraded to TMP and CO2 (Racke, 1993; John and Shaike, 2015; Figure 10).
5.3 Carbendazim
Carbendazim (methyl N-(1H-benzimidazol-2-yl) carbamate) is a systemic broad-spectrum fungicide, which is also formed as a degradation product of thiophanate-methyl and benomyl fungicides (Mazellier et al., 2003; Fang et al., 2010). Carbendazim is used worldwide as pre- and post-harvest treatment to control the Ascomycetes, Fungi imperfecti and Basidiomycetes fungal diseases on various vegetables, fruits and several other plants. Carbendazim was found to be toxic to various animals and could induce reproductive, developmental, endocrine and haematological toxicity (Rama et al., 2014; Zhou et al., 2023). Many microorganisms, predominantly bacteria such as Rhodococcus, Nocardioides, Mycobacterium, Pseudomonas, etc., have been reported to metabolize carbendazim (Singh et al., 2016; Zhou et al., 2023). Among reported microbes, few bacterial strains have been found to be efficient degraders of carbendazim. For example, Rhodococcus sp. D-1 isolated from contaminated farmland in China, could degrade 98.20% of 200 ppm carbendazim within 5 days (Bai et al., 2017). In most organisms, carbendazim degradation is initiated by its hydrolysis to 2-aminobenzimidazole (2-AB) and further, 2-hydroxybenzimidazole (2-HB) (Wang et al., 2010; Arya et al., 2015; Figure 10). Subsequently, 2-HB is converted to catechol via the formation of benzene-1,2-diamine and further channelled into TCA cycle (Singh et al., 2016; Figure 10).
5.4 Alachlor
Chloroacetanilide herbicides such as alachlor, metolachlor, and acetochlor are primary herbicides, and more than 50 million kg has been used annually in the United States (Gan et al., 2002). These herbicides are highly soluble in water and persist in soil, with residues or metabolites being detected in surface and ground water (Potter and Carpenter, 1995; Tian et al., 2021). For example, alachlor was detected in groundwaters in the United States at concentration 0.1–16.6 μg L−1, exceeding the U.S. Environmental Protection Agency (US-EPA) maximum contaminant level criteria of 2 μg L−1 (WHO, 2017).
Alachlor [2-chloro-N-(2,6-diethylphenyl)-N-(methoxymethyl]acetamide] is one of the majorly used chloroacetanilide applied as a selective pre- and post-emergent herbicide to control weeds in soybeans, peanuts, and corn crops. C-dealkylation of other chloroacetanilide herbicides like butachlor leads to formation of alachlor. Alachlor has been categorised as a human carcinogen and has been reported to mimic 17β-estradiol, thereby acting as an endocrine-disruptor (Lee and Kim, 2022). Microbes including various genera of bacteria like Paracoccus, Rhodococcus, Pseudomonas, Acinetobacter, Streptomyces, etc., and few fungi like Paecilomyces have been reported to degrade alachlor (Słaba et al., 2013; Lee and Kim, 2022; Chen et al., 2023). Alachlor is converted to 2-chloro-N-(2,6-diethylphenyl) acetamide (CDEPA) by N-dealkylation (Zhang et al., 2011; Figure 10). Various hydrolases/reductases have been reported to be involved in N-dealkylation of chloroacetanilides. For example, enzyme ChlH from Rhodococcus sp. B1 and enzymes CndB1, CndB2, and CndC1 from Sphingomonas sp. DC-6 have been reported to catalyse the N-dealkylation of alachlor as well as other chloroacetamide like acetochlor, butachlor, and pertilachlor (Chen et al., 2023; Figure 10). Subsequently, CDEPA is transformed to 2,6-diethylanilline (DEA) with the help of enzyme amidase (CmeH) or amide hydrolase (DamH). DEA is further converted to aniline which is then acted upon by aniline dioxygenase to form catechol. The formed catechol is then oxidized through an ortho-cleavage pathway to cis, cis-muconic acid and funnelled into TCA cycle (Zhang et al., 2011; Kim et al., 2013; Gao et al., 2015; Figure 10).
5.5 Glyphosate
Glyphosate is a low-molecular-weight phosphonate (non-specific organophosphate herbicide) with high aqueous solubility and mobility, which leads to rapid leaching of this compound into soil, causing contamination of water bodies. Glyphosate has been reported to cause toxicity to bacteria as well as multicellular organisms like non-target crop plants, crustaceans, molluscs and chordates including humans (cytotoxicity and genotoxicity) (Zhan et al., 2018; Singh et al., 2020). Various strains of Achromobacter, Agrobacterium, Pseudomonas, Ochrobactrum, etc., have been previously isolated from contaminated sites which can utilize glyphosate as growth substrate (Zhao et al., 2015; Zhan et al., 2018; Feng D. et al., 2020). The primary degradation products of glyphosate include aminomethylphosphonic acid (AMPA) and sarcosine, which are reported to be more toxic than the parent compound (Zhan et al., 2018; Lozano and Pizarro, 2024; Figure 10). C-P lyase removes phosphate group from glyphosate yielding sarcosine, which is cleaved by sarcosine oxidase (encoded by 7 sox genes) into glycine and formaldehyde. Both these intermediates are funnelled into microbial metabolism and biosynthetic pathways (González-Valenzuela and Dussán, 2018; Diaz-Soto et al., 2024; Figure 10). In many microorganisms, glyphosate is converted to AMPA and glyoxylate by the action of glyphosate oxidoreductase. Further, glyoxylate is metabolized to TCA cycle, whereas AMPA is either acted upon by C-P lyase to produce methylamine or by an aminotransferase to form formylphosphonate. Both the intermediates i.e., methylamine and formylphosphonate are cleaved to formaldehyde, which is used by microbes for biosynthesis (Sviridov et al., 2015; Zhao et al., 2015; Singh et al., 2019; Figure 10).
5.6 Application of OMICS and metabolic engineering to CEC degradation
The application of various omics approaches to CEC degradation can provide possible systemic-level insights into the metabolic pathways and associated regulatory mechanisms. Genomics aids in identifying key genes encoding degradative enzymes and/or the evolutionary trajectory. For example, in strain Pseudomonas sp. C5pp, the draft genome analysis revealed the presence of three gene clusters on a single contig (Supercontig-A) involved in complete Carbaryl degradation. The genome analysis further suggested acquisition of genes by horizontal gene transfer events (Trivedi et al., 2016). Genomics in conjunction with transcriptomics and proteomics aids in identifying up/downregulation of genes/proteins under target conditions. For example, in P. bharatica CSV86T, the transcription analysis showed the induction of target genes involved in benzoate (benE, benK) and glucose (gbp, oprB, glcG) utilisation (Choudhary et al., 2017). Further proteomic analysis aided in identification of Gbp and OprB as glucose binding protein and carbohydrate specific porin, respectively which are induced when the culture is grown on glucose (Basu et al., 2007; Shrivastava et al., 2011). In Methylorubrum sp. ZY-1, the integrated transcriptomic and metabolomic analyses aided in revealing degradation of pentachlorodiphenyl (PCB 118) and underlying molecular mechanisms (Wu et al., 2024). Table 2 provides a comprehensive review of the application of omics in CEC biodegradation for pharmaceuticals, cyanotoxins, plasticizers and pesticides, which are also described further.
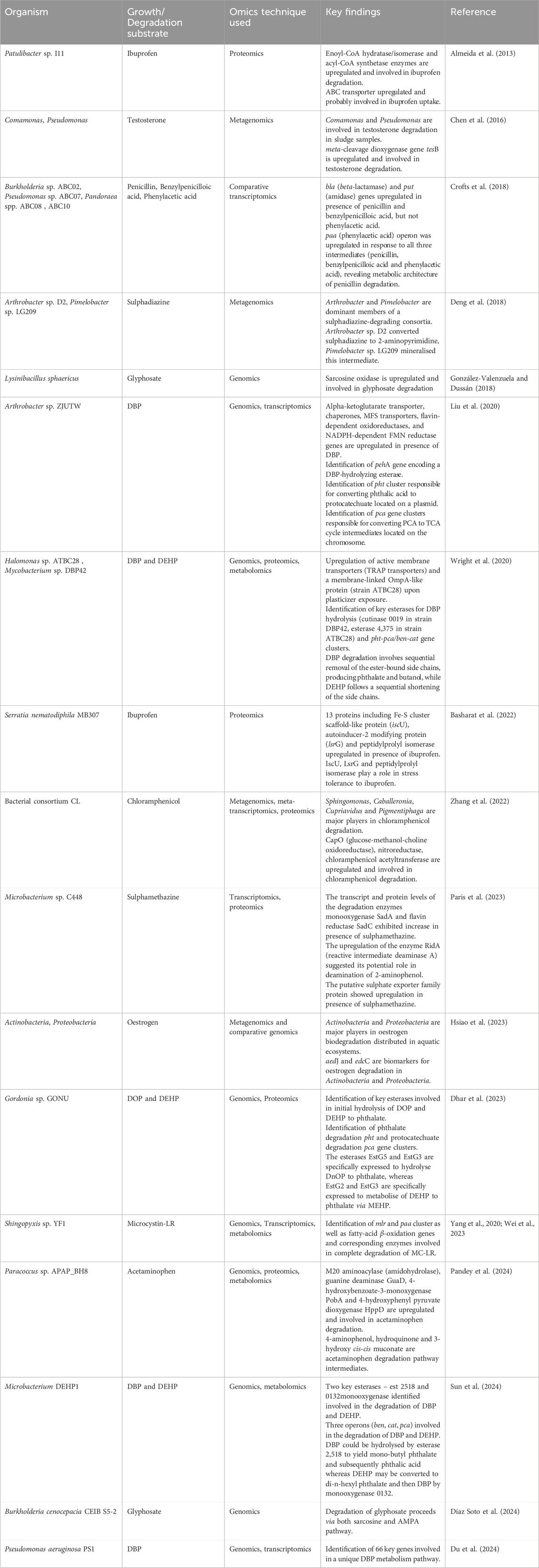
Table 2. Application of various OMICS techniques for degradation of contaminants of emerging concern.
5.7 Pharmaceuticals
Comparative transcriptomic analyses of four penicillin degrading strains revealed upregulation of genes encoding beta-lactamase (bla), penicillin amidase (put) and phenylacetic acid degradation enzymes (paa) in penicillin grown cells (as compared to alternative carbon source grown cells). In Pseudomonas sp. strain ABC07, the put operon, encoding four open reading frames: a beta-lactamase, a major facilitator family importer and two amidases (put1 and put2) was found to be upregulated in presence of penicillin and benzylpenicilloic acid, but not phenylacetic acid. Whereas, the paa operon was responsive to all three intermediates (penicillin, benzylpenicilloic acid and phenylacetic acid). Therefore, these analyses indicated the metabolic architecture of penicillin degradation in strain ABC07 (Crofts et al., 2018). A combination of metagenomic and cultivation-based techniques identified Arthrobacter and Pimelobacter as the dominant members of a sulphadiazine-degrading consortia as well as their individual roles in degradation. While Arthrobacter sp. D2 converted sulphadiazine to 2-aminopyrimidine, Pimelobacter sp. LG209 mineralised this intermediate (Deng et al., 2018). Exposure of Microbacterium sp. C448 to therapeutic and sub-therapeutic doses of sulphamethazine was assessed using transcriptomic and proteomic analyses. The transcript and protein levels of the degradation enzymes monooxygenase SadA and flavin reductase SadC exhibited increase in presence of sulphamethazine. Further, the upregulation of the enzyme RidA (reactive intermediate deaminase A) suggested its potential role in deamination of 2-aminophenol. Additionally, the putative sulphate exporter family protein showed upregulation in presence of sulphamethazine (Paris et al., 2023). An integrated multi-omics approach revealed the chloramphenicol biotransformation pathway, genes, proteins/enzymes and community structure/interactions of the activated sludge enriched consortium CL. The metagenomic analysis revealed Sphingomonas, Caballeronia, Cupriavidus and Pigmentiphaga as the major players in chloramphenicol degradation. Further, metatranscriptomic analysis revealed upregulation of specific detoxification and metabolic pathway genes such as capO, which encodes a glucose-methanol-choline oxidoreductase responsible for oxidation of C3-OH group of chloramphenicol. The proteomic analysis validated the metatranscriptomic data and the functionality of the identified enzymes such as CapO, nitroreductase and chloramphenicol acetyltransferase (Zhang et al., 2022).
The genomic and proteomic analysis of sulphamethoxazole-degrading Pseudomonas silesiensis F6a revealed six key degradation genes, deoC (2-deoxyribose 5-phosphate aldolase), narI (nitrate reductase), luxS (S-ribosyl homocysteine lyase), nuoH (NADH quinone oxidoreductase), gene 0655 (F420 dependent oxidoreductase) and gene 4,650 (amidohydrolase) involved in C-S bond cleavage, S-N bond hydrolysis and isoxazole ring-cleavage (Liu et al., 2022).
Metabolomic analyses of Sphingobacterium mizutaii S121 revealed the products of tetracycline biodegradation by the strain and the stress response mechanisms involved. Based on the analyses, two biodegradation pathways involving demethylation and one hydrolysis pathway were proposed. The levels of indole, glutamic acid and FAD, involved in regulating the activity of efflux proteins and degradation enzymes, were upregulated. Further, intracellular levels of nucleotides and amino acids were significantly increased to repair DNA/RNA and protein in response to tetracycline stress. The levels of antioxidants such as taurine and protoporphyrin IX also increased in response to the generation of reactive oxygen species due to enhanced aerobic metabolism. Under tetracycline stress, strain S121 required increased nutrients from the extracellular environment, due to which the levels of the metabolite N-palmitoyl sphingomyelin and phosphoethanolamine decreased significantly, enhancing membrane fluidity (Tan H. et al., 2022).
A metagenomic analysis of sewage samples incubated with testosterone indicated the genera Comamonas and Pseudomonas to be major players in degradation. Further, the meta-cleavage dioxygenase gene tesB was identified and exhibited a significant increase after 48 h of incubation (Chen et al., 2016). A combination of metagenomic analyses and comparative genomics revealed Actinobacteria and Proteobacteria as major players in oestrogen biodegradation distributed in aquatic ecosystems. Further, aedJ and edcC were identified as biomarkers for oestrogen degradation in Actinobacteria and Proteobacteria, respectively, with potential application for environmental detection (Hsiao et al., 2023).
Quantitative proteomics of Patulibacter sp. I11 in absence or presence of ibuprofen revealed likely proteins involved in degradation. In presence of ibuprofen, various proteins involved in aromatic degradation such as enoyl-CoA hydratase/isomerase, acyl-CoA synthetase, Rieske (2Fe-2S) domain containing were upregulated. Additionally, other proteins such as ABC transporter (probably involved in ibuprofen uptake), putative lyase, stress response protein and AMP-forming synthetase were also upregulated (Almeida et al., 2013). The stress response of Serratia nematodiphila sp. MB307 to the presence of ibuprofen was investigated using differential proteomics. Thirteen proteins were upregulated and 29 proteins were downregulated in response to ibuprofen stress. Among the upregulated proteins, Fe-S cluster scaffold-like protein IscU, autoinducer-2 modifying protein LsrG and peptidylprolyl isomerase have been implicated for their role in stress tolerance. Overall, the analyses highlighted the multifaceted stress response of strain MB307, involving a balance between protein synthesis, DNA replication, and energy production (Basharat et al., 2022).
The genomic-proteomic-metabolomic analyses of Paracoccus sp. APAP_BH8 elucidated the genes, enzymes and metabolic pathway of acetaminophen degradation in the strain. The proteome analysis revealed the upregulation of M20 aminoacylase (amidohydrolase), guanine deaminase GuaD, 4-hydroxybenzoate-3-monoxygenase PobA and 4-hydroxyphenyl pyruvate dioxygenase HppD in presence of acetaminophen. Molecular docking studies of these enzymes with their respective substrates validated the functionality of these enzymes. Further, the metabolomic analysis revealed 4-aminophenol, hydroquinone and 3-hydroxy cis, cis-muconate as degradation pathway intermediates (Pandey et al., 2024).
Genomic and comparative transcriptome analyses of Aminobacter sp. Strain NyZ550 revealed upregulation of the genes dmm (dimethylamine monooxygenase), gmas (γ-glutamylmethylamide synthetase), mgs (N-methylglutamate synthase) and mgd (N-methylglutamate dehydrogenase) while growing on metformin, indicating the metabolism of dimethylamine. Further, the serine cycle and formate-tetrahydrofolate catabolic genes also exhibited upregulation. Importantly, the gene encoding agmatinase (putative metformin hydrolase) exhibited upregulation, highlighting its role in degradation (Li et al., 2023).
5.8 Cyanotoxins
In Sphingopyxis sp. YF1, genomic analysis revealed the presence of MC degrading cluster mlrBDAC. The MC-LR degradation products such as linearized MC-LR, tetrapeptide, Adda and its degradation intermediates, and phenylacetic acid were detected using UPLC and UPLC-ESI-MS. Further, transcriptomics and qRT-PCR analyses suggested the upregulation of mlr cluster, fatty acid β-oxidation genes and paa cluster during MC-LR degradation. Metabolomics study showed enrichment of metabolites in pantothenate and CoA biosynthesis as well as fatty-acid degradation indicating involvement of fatty acid β-oxidation in MC-LR degradation (Wei et al., 2023). In Sphingopyxis sp. m6, gene specific qPCR suggested involvement of mlr cluster in nodularin degradation and the respective enzymatic steps were identified by metabolite analysis using total ion chromatogram (Yuan et al., 2021).
5.9 Plasticizers
In Gordonia sp. GONU, genome sequencing aided in identification of key esterases involved in initial hydrolysis DOP and DEHP to phthalate as well as pht gene clusters (responsible for conversion of phthalate to protocatechuate) and pca gene clusters (responsible for conversion of protocatechuate to TCA cycle intermediates). Substrate dependent gene expression profile by qRT-PCR and protein profiling by LC-ESI-MS/MS revealed that esterases EstG5 and EstG3 are specifically expressed to hydrolyse DnOP to PA, whereas EstG2 and EstG3 are specifically expressed to metabolise DEHP to PA via MEHP (Dhar et al., 2023).
Genome sequencing of Microbacterium sp. DEHP1 identified two key esterases-est258 and monooxygenase 0132 as well as three operons (ben, cat, pca) involved in the degradation of DBP and DEHP. Genome mining and metabolite identification by GC-MS suggested that DBP could be hydrolyzed by esterase 2,518 to yield mono-butyl phthalate (MBP) and subsequently phthalic acid (PA) whereas DEHP may be converted to di-n-hexyl phthalate (DnHP) and then DBP by monooxygenase 0132. Further, metabolic profiling using UHPLC-QTOF/MS revealed that under DEHP/DBP stress, strain DEHP1 cells showed increased levels of valine (which induces production of osmoregulatory substances), glycerophospholipids (major component of cell membrane), glutathione/protoanemonin (antioxidants), and proline (key player to preserve cellular glutathione redox status by activating signaling pathway). Notably, levels of organic substances like levan and naringenin 4′-O-alpha-L-rhamnopyranoside decreased in response to DEHP stress (Sun et al., 2024).
A combined genomic and transcriptomic approach identified 66 key genes involved in two different mono-butyl phthalate-catabolism steps in Pseudomonas aeruginosa PS1. In addition to the genes encoding the metabolic pathway enzymes, most differentially expressed genes in Pseudomonas aeruginosa PS1 under DBP stress were those encoding for ABC transporters, two-component systems, biofilm formation, quorum sensing and chemotaxis (Du et al., 2024).
The genome sequencing of DBP degrading Arthrobacter sp. ZJUTW identified the presence of pehA gene encoding a DBP-hydrolyzing esterase and pht gene cluster responsible for converting phthalic acid to protocatechuate located on a plasmid, and pca gene clusters responsible for converting PCA to TCA cycle intermediates located on the chromosome. Additionally, transcriptomic analysis by RNA-seq showed the upregulation of genes encoding an alpha-ketoglutarate transporter (important for cell wall synthesis), chaperones, MFS transporters (important for DBP efflux), flavin-dependent oxidoreductases, and NADPH-dependent FMN reductase genes (Liu et al., 2020).
Proteogenomic and metabolomic analysis of Halomonas sp. ATBC28 and Mycobacterium sp. DBP42 identified key esterases for DBP hydrolysis (cutinase 0019 in strain DBP42, esterase 4,375 in strain ATBC28) and pht-pca/ben-cat gene clusters. Metabolite analysis revealed that DBP degradation involves sequential removal of the ester-bound side chains, producing phthalate and butanol, while DEHP follows a sequential shortening of the side chains. Further, in strain ATBC28, active membrane transporters (TRAP transporters 0264 and 0631) and a membrane-linked OmpA-like protein (3,348) were upregulated, potentially for detoxification and biosurfactant production, respectively (Wright et al., 2020).
5.10 Pesticides
Proteomics and metabolomics provided an enhanced understanding of alachlor biodegradation by P. marquandii. Metabolomics (by LC-MS/MS) suggested that presence of alachlor reduced the culture growth and glucose consumption rates and increased the formation of supplementary materials (UDP-glucose/galactose) and ROS scavengers (ascorbate). Proteomic analysis (2-D electrophoresis and MALDI-TOF/TOF) revealed that the presence of alachlor led to upregulation of enzymes related to energy, sugar metabolism and ROS production. Further, overexpression of cyanide hydratase implicated the key role of this enzyme in the alachlor biodegradation pathway (Szewczyk et al., 2015).
The genomic analysis of Burkholderia cenocepacia CEIB S5-2 revealed the presence of key genes involved in glyphosate degradation pathways (sarcosine and AMPA pathway), suggesting that the bacterial strain could use both routes for glyphosate degradation. Genes soxA/B/D/G encoding sarcosine oxidase enzyme involved in sarcosine pathway as well as genes gox encoding glyphosate oxidoreductase, mauE/D encoding methylamine dehydrogenase and other genes encoding aminotransferases, phosphonatase enzymes involved in AMPA pathway were present on the genome (Diaz-Soto et al., 2024). Genomic data analysis of Lysinibacillus sphaericus suggested the presence of sarcosine oxidase gene and qRT-PCR analysis showed upregulation of this gene in presence of glyphosate (González-Valenzuela and Dussán, 2018).
Degradation pathway of chlorpyrifos and glyphosate in Bacillus cereus strains AKAD 3–1 were elucidated by GC-MS based metabolomics. Analysis of the intermediate and the final metabolic products confirmed that no toxic compounds were produced during chlorpyrifos and glyphosate degradation. This indicates that the bacterium harbors the metabolic pathway for detoxification and degradation of chlorpyrifos and glyphosate into non-toxic compounds (Malla et al., 2023).
Genomic analysis of Sphingobacterium sp. InxBP1 indicated the presence of various genes encoding enzymes involved in imidacloprid degradation. For example, nitronate monooxygenase (locus id K7A41_01745), amidohydrolase family enzymes or metal-dependent hydrolases (K7A41_03835, K7A41_07535) having similarity with 6-chloronicotinic acid chlorohydrolase, and FAD dependent monooxygenase (K7A41_12,275) similar to 6-hydroxy nicotinate monooxygenase, were found to be present in the genome, indicating the potential of strain InxBP1 to degrade imidacloprid (Gautam et al., 2023).
Therefore, omics techniques provide crucial data on various factors such as genes/proteins/metabolites involved, microbial community structure/dynamics, gene expression regulation and stress response mechanisms, thereby aiding in rational design of bioremediation and metabolic engineering strategies.
Application of natural isolates for bioremediation of CECs might face limitations such as slow degradation rates, incomplete transformation into toxic by-products, reduced survivability, and presence of simple carbon sources in the environment (Nielsen, 2001; Dvořák et al., 2017; Phale et al., 2020). These limitations can be overcome by directed genetic engineering approaches, called as “metabolic engineering”. These techniques can be used to broaden metabolic diversity, enhance degradation rates, enhance physiological vigour, overcome carbon catabolite repression etc. (Dvorak et al., 2017).
Multiple reports have described metabolic engineering of CEC degradation/transformation pathways in bacteria. For example, plasmid-mediated expression of sulfonamide monooxygenase and flavin reductase rendered E. coli BL21 (DE3) resistant to sulfamethoxazole (Kim D. et al., 2019). E. coli strain W, carrying the phenylacetic acid catabolic genes (paa operon) was engineered for penicillin utilisation by expression of beta-lactamase and penicillin amidase (pga) (Crofts et al., 2018). A consortium of Aminobacter sp. NyZ550 (that converts metformin to guanylurea) and metabolically engineered P. putida PaW340 (expressing guanylurea hydrolase GuuH) was constructed for metformin mineralisation. While strain NyZ550 converted metformin to guanylurea and dimethylamine, strain PaW340 metabolised guanylurea to guanidine, which was used as nitrogen source by strain NyZ550 (Li et al., 2023). Table 3 provides a comprehensive review of the application of metabolic engineering to CEC biodegradation.
6 Conclusion and future perspectives
Contaminants of emerging concern (CECs) are a heterogeneous group of naturally occurring or synthetic compounds that pose significant risk to human and ecological health due to their unregulated release into the environment. Among these, pharmaceuticals, cyanotoxins, plasticizers and pesticides have been found to occur in diverse habitats such as WWTPs, rivers, surface waters, soil as well as the atmosphere. Measures like precise monitoring of these compounds in various habitats, tracking their transport across ecological compartments and development of stringent regulatory policies might aid in mitigating risks at the point of release. Whereas, for already contaminated habitats, microbial remediation provides an eco-friendly and cost-effective solution. Microbes have adapted to these persistent compounds by the action of broad substrate specific enzymes (biotransformation) and evolution of metabolic pathways to utilise them as growth substrate, thereby mitigating the associated risks. The application of omics reveals various pathway components such as genes, transcripts, proteins, metabolites and their complex interactions, thereby facilitating development of efficient clean-up strategies. However, the available literature on CEC biodegradation primarily focuses on biotransformation products, while reports of complete mineralisation pathways and associated enzymes are limited. Such information is crucial for metabolic engineering applications and scaling-up the bioremediation process for efficient environmental clean-up, offering potential research opportunities.
Author contributions
BS: Conceptualization, Validation, Writing–original draft, Writing–review and editing. HM: Conceptualization, Validation, Writing–original draft, Writing–review and editing. SP: Conceptualization, Validation, Writing–original draft, Writing–review and editing. TD: Conceptualization, Validation, Writing–original draft, Writing–review and editing. OI: Conceptualization, Validation, Writing–original draft, Writing–review and editing. SK: Conceptualization, Validation, Writing–original draft, Writing–review and editing. PP: Conceptualization, Supervision, Validation, Writing–review and editing, Funding acquisition, Writing–original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. PP acknowledges research grants from the Department of Science and Technology, Department of Biotechnology and Board of Research in Nuclear Science, Government of India.
Acknowledgments
HM and BS thank IIT-Bombay for the Institute Post-doctoral Fellowship. SP thanks CSIR, Government of India for Senior Research Fellowship. TD and SK thank IIT-Bombay for Senior Research Fellowship. OI thanks IIT-Bombay for Junior Research Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeel, M., Song, X., Wang, Y., Francis, D., and Yang, Y. (2017). Environmental impact of estrogens on human, animal and plant life: a critical review. Environ. Int. 99, 107–119. doi:10.1016/j.envint.2016.12.010
Aguilar-Romero, I., De la Torre-Zúñiga, J., Quesada, J. M., Haïdour, A., O’Connell, G., McAmmond, B. M., et al. (2021). Effluent decontamination by the ibuprofen-mineralizing strain, Sphingopyxis granuli RW412: metabolic processes. Environ. Pollut. 274, 116536. doi:10.1016/j.envpol.2021.116536
Aguirre-Martínez, G. V., DelValls, A. T., and Martín-Díaz, M. L. (2015). Yes, caffeine, ibuprofen, carbamazepine, novobiocin and tamoxifen have an effect on Corbicula fluminea (Müller, 1774). Ecotoxicol. Environ. Saf. 120, 142–154. doi:10.1016/j.ecoenv.2015.05.036
Ahmad, R., Verma, Y., Gautam, A. K., and Kumar, S. (2015). Assessment of estrogenic potential of di-n-butyl phthalate and butyl benzyl phthalate in vivo. Toxicol. Ind. Health. 31, 1296–1303. doi:10.1177/0748233713491803
Akay, C., and Tezel, U. (2020). Biotransformation of Acetaminophen by intact cells and crude enzymes of bacteria: a comparative study and modelling. Sci. total Environ. 703, 134990. doi:10.1016/j.scitotenv.2019.134990
Almeida, B., Kjeldal, H., Lolas, I., Knudsen, A. D., Carvalho, G., Nielsen, K. L., et al. (2013). Quantitative proteomic analysis of ibuprofen-degrading Patulibacter sp. strain I11. Biodegradation 24, 615–630. doi:10.1007/s10532-012-9610-5
Al-Sammak, M. A., Hoagland, K. D., Cassada, D., and Snow, D. D. (2014). Co-occurrence of the cyanotoxins BMAA, DABA and anatoxin-a in Nebraska reservoirs, fish, and aquatic plants. Toxins 6, 488–508. doi:10.3390/toxins6020488
Ambreen, S., and Yasmin, A. (2021). Novel degradation pathways for Chlorpyrifos and 3, 5, 6-Trichloro-2-pyridinol degradation by bacterial strain Bacillus thuringiensis MB497 isolated from agricultural fields of Mianwali, Pakistan. Pestic. Biochem. Physiol. 172, 104750. doi:10.1016/j.pestbp.2020.104750
Ambrosio-Albuquerque, E. P., Cusioli, L. F., Bergamasco, R., Gigliolli, A. A. S., Lupepsa, L., Paupitz, B. R., et al. (2021). Metformin environmental exposure: a systematic review. Environ. Toxicol. Pharmacol. 83, 103588. doi:10.1016/j.etap.2021.103588
Anwar, S., Liaquat, F., Khan, Q. M., Khalid, Z. M., and Iqbal, S. (2009). Biodegradation of chlorpyrifos and its hydrolysis product 3, 5, 6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J. Hazard. Mat. 168, 400–405. doi:10.1016/j.jhazmat.2009.02.059
Aris, A. Z., Shamsuddin, A. S., and Praveena, S. M. (2014). Occurrence of 17α-ethynylestradiol (EE2) in the environment and effect on exposed biota: a review. Environ. Int. 69, 104–119. doi:10.1016/j.envint.2014.04.011
Arthur, M., Autissier, D., and Courvalin, P. (1986). Analysis of the nucleotide sequence of the ereB gene encoding the erythromydn esterase type II. Nucleic acids Res. 14 (12), 4987–4999. doi:10.1093/nar/14.12.4987
Arya, R., Sharma, R., Malhotra, M., Kumar, V., and Sharma, A. K. (2015). Biodegradation aspects of carbendazim and sulfosulfuron: trends, scope and relevance. Curr. Med. Chem. 22, 1147–1155. doi:10.2174/0929867322666141212123449
Atkinson, S. K., Marlatt, V. L., Kimpe, L. E., Lean, D. R., Trudeau, V. L., and Blais, J. M. (2012). The occurrence of steroidal estrogens in south-eastern Ontario wastewater treatment plants. Sci. Total Environ. 430, 119–125. doi:10.1016/j.scitotenv.2012.04.069
Aulestia, M., Flores, A., Acosta-Jurado, S., Santero, E., and Camacho, E. M. (2022). Genetic characterization of the ibuprofen-degradative pathway of Rhizorhabdus wittichii MPO218. Appl. Environ. Microbiol. 88 (11), e0038822–22. doi:10.1128/aem.00388-22
Awe, S., Mikolasch, A., Hammer, E., and Schauer, F. (2008). Degradation of phenylalkanes and characterization of aromatic intermediates acting as growth inhibiting substances in hydrocarbon utilizing yeast Candida maltosa. Int. Biodeterior. Biodegrad. 62, 408–414. doi:10.1016/j.ibiod.2008.03.007
Badawy, S., Yang, Y., Liu, Y., Marawan, M. A., Ares, I., Martinez, M. A., et al. (2021). Toxicity induced by ciprofloxacin and enrofloxacin: oxidative stress and metabolism. Crit. Rev. Toxicol. 51 (9), 754–787. doi:10.1080/10408444.2021.2024496
Bai, N., Wang, S., Abuduaini, R., Zhang, M., Zhu, X., and Zhao, Y. (2017). Rhamnolipid-aided biodegradation of carbendazim by Rhodococcus sp. D-1: characteristics, products, and phytotoxicity. Sci. Total Environ. 590, 343–351. doi:10.1016/j.scitotenv.2017.03.025
Balakrishnan, A., Ponnuchamy, M., Kapoor, A., and Sivaraman, P. (2022). “Emerging contaminants in wastewater and associated treatment technologies,” in Legacy and emerging contaminants in water and wastewater: monitoring, risk assessment and remediation techniques (Cham: Springer International Publishing), 231–261. doi:10.1007/978-3-030-95443-7_11
Ball, A. P. (1986). “Overview of clinical experience with ciprofloxacin,” in Ciprofloxacin. Current topics in infectious diseases and clinical microbiology, volume 1. Editors H. C. Neu, and D. S. Reeves (Wiesbaden: Vieweg+Teubner Verlag), 85–90. doi:10.1007/978-3-663-01930-5_27
Baran, W., Adamek, E., Makowski, A., and Sobczak, A. (2012). Assessment of sulfonamides occurrence in the biosphere. Ecol. Chem. Eng. A 19 (10), 1153–1171. doi:10.2428/ecea.2012.19(10)110
Barclay, V. K., Tyrefors, N. L., Johansson, I. M., and Pettersson, C. E. (2012). Trace analysis of fluoxetine and its metabolite norfluoxetine. Part II: enantioselective quantification and studies of matrix effects in raw and treated wastewater by solid phase extraction and liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 1227, 105–114. doi:10.1016/j.chroma.2011.12.084
Basharat, Z., Moon, K. M., Foster, L. J., and Yasmin, A. (2022). Impact of azo dyes and ibuprofen on the proteome of Serratia nematodiphila sp. MB307. Curr. Protein Peptide Sci. 23 (10), 697–705. doi:10.2174/1389203723666220727142630
Basu, A., Shrivastava, R., Basu, B., Apte, S. K., and Phale, P. S. (2007). Modulation of glucose transport causes preferential utilization of aromatic compounds in Pseudomonas putida CSV86. J. Bacteriol. 189 (21), 7556–7562. doi:10.1128/jb.01235-07
Berthet, S., Charpiat, B., and Mabrut, J. Y. (2010). Erythromycin as a prokinetic agent: risk factors. J. Visc. Surg. 147 (2), e13–e18. doi:10.1016/j.jviscsurg.2010.06.001
Bhattacharyya, M., Dhar, R., Basu, S., Das, A., Reynolds, D. M., and Dutta, T. K. (2023). Molecular evaluation of the metabolism of estrogenic di (2-ethylhexyl) phthalate in Mycolicibacterium sp. Microb. Cell Fact. 22, 82. doi:10.1186/s12934-023-02096-0
Bilal, M., Mehmood, S., Rasheed, T., and Iqbal, H. M. N. (2020). Antibiotics traces in the aquatic environment: persistence and adverse environmental impact. Curr. Opin. Environ. Sci. and Health 13, 68–74. doi:10.1016/j.coesh.2019.11.005
Billet, L., Pesce, S., Rouard, N., Spor, A., Paris, L., Leremboure, M., et al. (2021). Antibiotrophy: key function for antibiotic-resistant bacteria to colonize soils—case of Sulfamethazine-degrading microbacterium sp. C448. Front. Microbiol. 12, 643087. doi:10.3389/fmicb.2021.643087
Billings, A., Jones, K. C., Pereira, M. G., and Spurgeon, D. J. (2021). Plasticisers in the terrestrial environment: sources, occurrence and fate. Environ. Chem. 18, 111–130. doi:10.1071/EN21033
Biskri, L., and Mazel, D. (2003). Erythromycin esterase gene ere (A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. agents Chemother. 47 (10), 3326–3331. doi:10.1128/aac.47.10.3326-3331.2003
Blair, B. D., Crago, J. P., Hedman, C. J., and Klaper, R. D. (2013). Pharmaceuticals and personal care products found in the Great Lakes above concentrations of environmental concern. Chemosphere 93 (9), 2116–2123. doi:10.1016/j.chemosphere.2013.07.057
Bosu, S., Rajamohan, N., Al Salti, S., Rajasimman, M., and Das, P. (2024). Biodegradation of chlorpyrifos pollution from contaminated environment-A review on operating variables and mechanism. Environ. Res. 118212, 118212. doi:10.1016/j.envres.2024.118212
Bourne, D. G., Jones, G. J., Blakeley, R. L., Jones, A., Negri, A. P., and Riddles, P. (1996). Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Appl. Environ. Microbiol. 62 (11), 4086–4094. doi:10.1128/aem.62.11.4086-4094.1996
Bradley, P. M., Journey, C. A., Button, D. T., Carlisle, D. M., Clark, J. M., and Mahler, B. J. (2016). Metformin and other pharmaceuticals widespread in wadeable streams of the southeastern United States. Environ. Technol. Lett. 3 (6), 243–249. doi:10.1021/acs.estlett.6b00170
Brooks, B. W., Foran, C. M., Richards, S. M., Weston, J., Turner, P. K., Stanley, J. K., et al. (2003). Aquatic ecotoxicology of fluoxetine. Toxicol. Lett. 142 (3), 169–183. doi:10.1016/S0378-4274(03)00066-3
Calisici, D., Yılmaz, S., and Goktas, B. (2023). Toxic, genotoxic and teratogenic effects of ibuprofen and its derivatives. Curr. Drug Targets 24, 361–370. doi:10.2174/1389450124666230104160435
Canesi, L., Balbi, T., Fabbri, R., Salis, A., and Sen, F. (2022). Green synthesis, characterization and bioactivity of biogenic zinc oxide nanoparticles. Environ. Res. 204, 111897. doi:10.1016/j.envres.2021.111897
Cao, L., Zhang, J., Zhao, R., Deng, Y., Liu, J., Fu, W., et al. (2019). Genomic characterization, kinetics, and pathways of sulfamethazine biodegradation by Paenarthrobacter sp. A01. Environ. Int. 131, 104961. doi:10.1016/j.envint.2019.104961
Chaignaud, P., Gruffaz, C., Borreca, A., Fouteau, S., Kuhn, L., Masbou, J., et al. (2022). A methylotrophic bacterium growing with the antidiabetic drug metformin as its sole carbon, nitrogen and energy source. Microorganisms 10 (11), 2302. doi:10.3390/microorganisms10112302
Chang, H., Wan, Y., Wu, S., Fan, Z., and Hu, J. (2011). Occurrence of androgens and progestogens in wastewater treatment plants and receiving river waters: comparison to estrogens. water Res. 45 (2), 732–740. doi:10.1016/j.watres.2010.08.046
Chang, T. T., Lin, Z. W., Zhang, L. Q., Liu, W. B., Zhou, Y., and Ye, B. C. (2022). Efficient biodegradation of di-(2-ethylhexyl) phthalate by a novel strain Nocardia asteroides LMB-7 isolated from electronic waste soil. Sci. Rep. 12, 15262. doi:10.1038/s41598-022-19752-x
Chatterjee, S., and Dutta, T. K. (2003). Metabolism of butyl benzyl phthalate by Gordonia sp. strain MTCC 4818. Biochem. Biophys. Res. Commun. 309, 36–43. doi:10.1016/s0006-291x(03)01513-4
Chen, F., Chen, Y., Chen, C., Feng, L., Dong, Y., Chen, J., et al. (2021a). High-efficiency degradation of phthalic acid esters (PAEs) by Pseudarthrobacter defluvii E5: performance, degradative pathway, and key genes. Sci. Total Environ. 794, 148719. doi:10.1016/j.scitotenv.2021.148719
Chen, F., Li, X., Dong, Y., Li, J., Li, Y., Li, H., et al. (2021b). Biodegradation of phthalic acid esters (PAEs) by Cupriavidus oxalaticus strain E3 isolated from sediment and characterization of monoester hydrolases. Chemosphere 266, 129061. doi:10.1016/j.chemosphere.2020.129061
Chen, G., Wang, L., Wang, M., and Hu, T. (2021c). Comprehensive insights into the occurrence and toxicological issues of nodularins. Mar. Pollut. Bull. 162, 111884. doi:10.1016/j.marpolbul.2020.111884
Chen, J., Jiang, X., Tong, T., Miao, S., Huang, J., and Xie, S. (2019). Sulfadiazine degradation in soils: dynamics, functional gene, antibiotic resistance genes and microbial community. Sci. total Environ. 691, 1072–1081. doi:10.1016/j.scitotenv.2019.07.230
Chen, J., Yang, S., Zhang, K., Chen, W., Mo, Y., and Li, L. (2022). Biochemical pathways and associated microbial process of di-2-ethyl hexyl phthalate (DEHP) enhanced degradation by the immobilization technique in sequencing batch reactor. Environ. Technol. 43, 2899–2908. doi:10.1080/09593330.2021.1909657
Chen, J. A., Li, X., Li, J., Cao, J., Qiu, Z., Zhao, Q., et al. (2007). Degradation of environmental endocrine disruptor di-2-ethylhexyl phthalate by a newly discovered bacterium, Microbacterium sp. strain CQ0110Y. Appl. Microbiol. Biotechnol. 74, 676–682. doi:10.1007/s00253-006-0700-3
Chen, S. F., Chen, W. J., Huang, Y., Wei, M., and Chang, C. (2023). Insights into the metabolic pathways and biodegradation mechanisms of chloroacetamide herbicides. Environ. Res. 229, 115918. doi:10.1016/j.envres.2023.115918
Chen, Y. L., Wang, C. H., Yang, F. C., Ismail, W., Wang, P. H., Shih, C. J., et al. (2016). Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci. Rep. 6 (1), 35386. doi:10.1038/srep35386
Chen, Y. L., Yu, C. P., Lee, T. H., Goh, K. S., Chu, K. H., Wang, P. H., et al. (2017). Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 24 (6), 712–724.e7. doi:10.1016/j.chembiol.2017.05.012
Chiang, Y. R., Wei, S. T. S., Wang, P. H., Wu, P. H., and Yu, C. P. (2020). Microbial degradation of steroid sex hormones: implications for environmental and ecological studies. Microb. Biotechnol. 13 (4), 926–949. doi:10.1111/1751-7915.13504
Choi, S. M., Yoo, S. D., and Lee, B. M. (2004). Toxicological characteristics of endocrine-disrupting chemicals: developmental toxicity, carcinogenicity, and mutagenicity. J. Toxicol.Environ. Health Part B 7, 1–23. doi:10.1080/10937400490253229
Chopra, S., and Kumar, D. (2020). Characterization, optimization and kinetics study of acetaminophen degradation by Bacillus drentensis strain S1 and waste water degradation analysis. Bioresour. Bioprocess. 7, 9–18. doi:10.1186/s40643-020-0297-x
Choudhary, A., Modak, A., Apte, S. K., and Phale, P. S. (2017). Transcriptional modulation of transport-and metabolism-associated gene clusters leading to utilization of benzoate in preference to glucose in Pseudomonas putida CSV86. Appl. Environ. Microbiol. 83 (19), 012800–e1317. doi:10.1128/AEM.01280-17
Codd, G. A., Ward, C. J., and Bell, S. G. (1997). “Cyanobacterial toxins: occurrence, modes of action, health effects and exposure routes,” in Applied Toxicology: Approaches Through Basic Science: Proceedings of the 1996 EUROTOX Congress Meeting Held in Alicante, Spain, September 22-25, 1996 (Berlin, Heidelberg: Springer Berlin Heidelberg), 399–410.
Crofts, T. S., Wang, B., Spivak, A., Gianoulis, T. A., Forsberg, K. J., Gibson, M. K., et al. (2018). Shared strategies for β-lactam catabolism in the soil microbiome. Nat. Chem. Biol. 14 (6), 556–564. doi:10.1038/s41589-018-0052-1
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2019). Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front. Microbiol. 10, 338. doi:10.3389/fmicb.2019.00338
Czubacka, E., Czerczak, S., and Kupczewska-Dobecka, M. (2021). The overview of current evidence on the reproductive toxicity of dibutyl phthalate. Int. J. Occup. Med. Environ. Health. 34, 15–37. doi:10.13075/ijomeh.1896.01658
Deere, J. R., Jankowski, M. D., Primus, A., Phelps, N. B., Ferrey, M., Borucinska, J., et al. (2024). Health of wild fish exposed to contaminants of emerging concern in freshwater ecosystems utilized by a Minnesota Tribal community. Integr. Environ. Assess. Manag. 20, 846–863. doi:10.1002/ieam.4822
Deere, J. R., Streets, S., Jankowski, M. D., Ferrey, M., Chenaux-Ibrahim, Y., Convertino, M., et al. (2021). A chemical prioritization process: applications to contaminants of emerging concern in freshwater ecosystems (Phase I). Sci. Total Environ. 772, 146030. doi:10.1016/j.scitotenv.2021.146030
Deng, Y., Mao, Y., Li, B., Yang, C., and Zhang, T. (2016). Aerobic degradation of sulfadiazine by Arthrobacter spp.: kinetics, pathways, and genomic characterization. Environ. Sci. and Technol. 50 (17), 9566–9575. doi:10.1021/acs.est.6b02231
Deng, Y., Wang, Y., Mao, Y., and Zhang, T. (2018). Partnership of Arthrobacter and Pimelobacter in aerobic degradation of sulfadiazine revealed by metagenomics analysis and isolation. Environ. Sci. and Technol. 52 (5), 2963–2972. doi:10.1021/acs.est.7b05913
Dhar, R., Basu, S., Bhattacharyya, M., and Dutta, T. K. (2023). Evaluation of distinct molecular architectures and coordinated regulation of the catabolic pathways of oestrogenic dioctyl phthalate isomers in Gordonia sp. Microbiol 169, 001353. doi:10.1099/mic.0.001353
Díaz-Soto, J. A., Mussali-Galante, P., Castrejón-Godínez, M. L., Saldarriaga-Noreña, H. A., Tovar-Sánchez, E., and Rodríguez, A. (2024). Glyphosate resistance and biodegradation by Burkholderia cenocepacia CEIB S5-2. Environ. Sci. Pollut. Res. 31, 37480–37495. doi:10.1007/s11356-024-33772-2
Ding, C., and He, J. (2010). Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 87, 925–941. doi:10.1007/s00253-010-2649-5
Domaradzka, D., Guzik, U., Hupert-Kocurek, K., and Wojcieszynska, D. (2015). Cometabolic degradation of naproxen by Planococcus sp. strain S5. Water Air Soil Pollut. 226, 297–298. doi:10.1007/s11270-015-2564-6
Dong, Z., Yan, X., Wang, J., Zhu, L., Wang, J., Li, C., et al. (2022). Mechanism for biodegradation of sulfamethazine by Bacillus cereus H38. Sci. Total Environ. 809, 152237. doi:10.1016/j.scitotenv.2021.152237
Doyle, W. I., and Meeks, J. P. (2018). Excreted steroids in vertebrate social communication. J. Neurosci. 38 (14), 3377–3387. doi:10.1523/JNEUROSCI.2488-17.2018
Du, H., Cheng, J. L., Li, Z. Y., Zhong, H. N., Wei, S., Gu, Y. J., et al. (2024). Molecular insights into the catabolism of dibutyl phthalate in Pseudomonas aeruginosa PS1 based on biochemical and multi-omics approaches. Sci. Total Environ. 926, 171852. doi:10.1016/j.scitotenv.2024.171852
Du, Y., Cheng, Q., Qian, M., Liu, Y., Wang, F., Ma, J., et al. (2023). Biodegradation of sulfametoxydiazine by Alcaligenes aquatillis FA: performance, degradation pathways, and mechanisms. J. Hazard. Mater. 452, 131186. doi:10.1016/j.jhazmat.2023.131186
Dvořák, P., Nikel, P., Damborský, J., and de Lorenzo, V. (2017). Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35, 845–866. doi:10.1016/j.biotechadv.2017.08.001
Dziga, D., Kokocinski, M., Maksylewicz, A., Czaja-Prokop, U., and Barylski, J. (2016). Cylindrospermopsin biodegradation abilities of aeromonas sp. isolated from rusałka lake. Toxins 8, 55. doi:10.3390/toxins8030055
Eliakim-Raz, N., Lador, A., Leibovici-Weissman, Y., Elbaz, M., Paul, M., and Leibovici, L. (2015). Efficacy and safety of chloramphenicol: joining the revival of old antibiotics? Systematic review and meta-analysis of randomized controlled trials. J. Antimicrob. Chemother. 70 (4), 979–996. doi:10.1093/jac/dku530
Etebu, E., and Arikekpar, I. (2016). Antibiotics: classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 4, 90–101.
Fan, S., Guo, J., Han, S., Du, H., Wang, Z., Fu, Y., et al. (2023). A novel and efficient phthalate hydrolase from acinetobacter sp. LUNF3: molecular cloning, characterization and catalytic mechanism. Molecules 28, 6738. doi:10.3390/molecules28186738
Fan, S., Wang, J., Li, K., Yang, T., Jia, Y., Zhao, B., et al. (2018). Complete genome sequence of Gordonia sp. YC-JH1, a bacterium efficiently degrading a wide range of phthalic acid esters. J. Biotechnol. 279, 55–60. doi:10.1016/j.jbiotec.2018.05.009
Fang, H., Wang, Y., Gao, C., Yan, H., Dong, B., and Yu, Y. (2010). Isolation and characterization of Pseudomonas sp. CBW Capab. degrading carbendazim. Biodegrad. 21, 939–946. doi:10.1007/s10532-010-9353-0
Feder, H. M. J., Osier, C., and Maderazo, E. G. (1981). Chloramphenicol: a review of its use in clinical practice. Rev. Infect. Dis. 3 (3), 479–491. doi:10.1093/clinids/3.3.479
Feng, D., Soric, A., and Boutin, O. (2020a). Treatment technologies and degradation pathways of glyphosate: a critical review. Sci. Total Environ. 742, 140559. doi:10.1016/j.scitotenv.2020.140559
Feng, N., Yang, F., Yan, H., Yin, C., Liu, X., Zhang, H., et al. (2016). Pathway for biodegrading nodularin (NOD) by Sphingopyxis sp. USTB-05. Toxins 8 (5), 116. doi:10.3390/toxins8050116
Feng, N. X., Feng, Y. X., Liang, Q. F., Chen, X., Xiang, L., Zhao, H. M., et al. (2021). Complete biodegradation of di-n-butyl phthalate (DBP) by a novel Pseudomonas sp. YJB6. Sci. Total Environ. 761, 143208. doi:10.1016/j.scitotenv.2020.143208
Feng, N. X., Yu, J., Mo, C. H., Zhao, H. M., Li, Y. W., Wu, B. X., et al. (2018). Biodegradation of di-n-butyl phthalate (DBP) by a novel endophytic Bacillus megaterium strain YJB3. Sci. Total Environ. 616, 117–127. doi:10.1016/j.scitotenv.2017.10.298
Feng, N. X., Yu, J., Xiang, L., Yu, L. Y., Zhao, H. M., Mo, C. H., et al. (2019). Co-metabolic degradation of the antibiotic ciprofloxacin by the enriched bacterial consortium XG and its bacterial community composition. Sci. Total Environ. 665, 41–51. doi:10.1016/j.scitotenv.2019.01.322
Feng, W., Deng, Y., Yang, F., Miao, Q., and Ngien, S. K. (2023). Systematic review of contaminants of emerging concern (CECs): distribution, risks, and implications for water quality and health. Water 15, 3922. doi:10.3390/w15223922
Feng, W., Wu, X., Mao, G., Zhao, T., Wang, W., Chen, Y., et al. (2020b). Neurological effects of subchronic exposure to dioctyl phthalate (DOP), lead, and arsenic, individual and mixtures, in immature mice. Environ. Sci. Poll. Res. 27, 9247–9260. doi:10.1007/s11356-019-06823-2
Ferrão-Filho, A. D. S., and Kozlowsky-Suzuki, B. (2011). Cyanotoxins: bioaccumulation and effects on aquatic animals. Mar. Drugs 9, 2729–2772. doi:10.3390/md9122729
Fromme, H., Küchler, T., Otto, T., Pilz, K., Müller, J., and Wenzel, A. (2002). Occurrence of phthalates and bisphenol A and F in the environment. Water Res. 36, 1429–1438. doi:10.1016/S0043-1354(01)00367-0
Fu, H., Ge, Y., Liu, X., Deng, S., Li, J., Tan, P., et al. (2024). Exposure to the environmental pollutant chlorpyrifos induces hepatic toxicity through activation of the JAK/STAT and MAPK pathways. Sci. Total Environ. 928, 171711. doi:10.1016/j.scitotenv.2024.171711
Fuentes, M. Á. F., Morente, E. O., Abriouel, H., Pulido, R. P., and Gálvez, A. (2014). Antimicrobial resistance determinants in antibiotic and biocide-resistant gram-negative bacteria from organic foods. Food Control 37, 9–14. doi:10.1016/j.foodcont.2013.08.041
Fujiki, H., Suganuma, M., Suguri, H., Yoshizawa, S., Takagi, K., Nakayasu, M., et al. (1990). “New tumor promoters from marine natural products,” in Marine toxins. Editors S. Hall, and G. Strichartz (Washington, DC: American Chemical Society), 232–240. doi:10.1021/bk-1990-0418.ch018
Funari, E., and Testai, E. (2008). Human health risk assessment related to cyanotoxins exposure. Crit. Rev. Toxicol. 38, 97–125. doi:10.1080/10408440701749454
Gan, J., Wang, Q., Yates, S. R., Koskinen, W. C., and Jury, W. A. (2002). Dechlorination of chloroacetanilide herbicides by thiosulfate salts. PNAS 99, 5189–5194. doi:10.1073/pnas.042105199
Gani, K. M., and Kazmi, A. A. (2018). Biochemical pathways and enhanced degradation of di-n-octyl phthalate (DOP) in sequencing batch reactor (SBR) by Arthrobacter sp. SLG-4 and Rhodococcus sp. SLG-6 isolated from activated sludge. Biodegradation 29, 117–129. doi:10.1007/s10532-018-9822-4
Gao, X., Li, J., Wang, X., Zhou, J., Fan, B., Li, W., et al. (2019). Exposure and ecological risk of phthalate esters in the Taihu Lake basin, China. Ecotoxicol. Environ. Saf. 171, 564–570. doi:10.1016/j.ecoenv.2019.01.001
Gao, Y., Chen, Y., Zhu, F., Pan, D., Huang, J., and Wu, X. (2024a). Revealing the biological significance of multiple metabolic pathways of chloramphenicol by Sphingobium sp. WTD-1. J. Hazard. Mater. 469, 134069. doi:10.1016/j.jhazmat.2024.134069
Gao, Y., Cheng, H., Song, Q., Huang, J., Liu, J., Pan, D., et al. (2024b). Characteristics and catalytic mechanism of a novel multifunctional oxidase, CpmO, for chloramphenicols degradation from Sphingobium sp. WTD-1. J. Hazard. Mater. 465, 133348. doi:10.1016/j.jhazmat.2023.133348
Gao, Y., Jin, L., Shi, H., and Chu, Z. (2015). Characterization of a novel butachlor biodegradation pathway and cloning of the debutoxylase (Dbo) gene responsible for debutoxylation of butachlor in Bacillus sp. hys-1. J. Agric. Food. Chem. 63, 8381–8390. doi:10.1021/acs.jafc.5b03326
Gautam, P., Pandey, A. K., and Dubey, S. K. (2023). Multi-omics approach reveals elevated potential of bacteria for biodegradation of imidacloprid. Environ. Res. 221, 115271. doi:10.1016/j.envres.2023.115271
Ghanem, C. I., Pérez, M. J., Manautou, J. E., and Mottino, A. D. (2016). Acetaminophen from liver to brain: new insights into drug pharmacological action and toxicity. Pharmacol. Res. 109, 119–131. doi:10.1016/j.phrs.2016.02.020
Ghani, M. U., Asghar, H. N., Nadeem, H., Shahid, M., Zeshan, M. A., Niaz, A., et al. (2022). Processes governing the environmental fates of alachlor in soil and aqueous media: a critical review. Int. J. Environ. Sci. Technol. 19 (8), 8043–8060. doi:10.1007/s13762-021-03559-w
Ghlichloo, I., and Gerriets, V. (2023). “Nonsteroidal anti-inflammatory drugs (NSAIDs),” in StatPearls (Treasure Island (FL): StatPearls Publishing).
Ghoshdastidar, A. J., Fox, S., and Tong, A. Z. (2015). The presence of the top prescribed pharmaceuticals in treated sewage effluents and receiving waters in Southwest Nova Scotia, Canada. Environ. Sci. Pollut. Res. 22, 689–700. doi:10.1007/s11356-014-3400-z
Gillis, P. L., Price, G., and Prasher, S. (2017). Lethal and sub-lethal effects of triclosan toxicity to the earthworm Eisenia fetida assessed through GC-MS metabolomics. J. Hazard. Mater. 323 (Pt A), 203–211. doi:10.1016/j.jhazmat.2016.07.022
Golchin, M., Khani, M., Sadani, M., Sadeghi, M., and Jahangiri-rada, M. (2021). Occurrence and fate of amoxicillin and penicillin G antibiotics in hospital wastewater treatment plants: a case study–Gonbad Kavous, Iran. South Afr. J. Chem. 75, 98–105. doi:10.17159/0379-4350/2021/v75a11
Gong, T., Liu, R., Che, Y., Xu, X., Zhao, F., Yu, H., et al. (2016). Engineering Pseudomonas putida KT 2440 for simultaneous degradation of carbofuran and chlorpyrifos. Microb. Biotechnol. 9, 792–800. doi:10.1111/1751-7915.12381
Gonsioroski, A., Mourikes, V. E., and Flaws, J. A. (2020). Endocrine disruptors in water and their effects on the reproductive system. Int. J. Mol. Sci. 21, 1929. doi:10.3390/ijms21061929
González-Valenzuela, L. E., and Dussán, J. (2018). Molecular assessment of glyphosate-degradation pathway via sarcosine intermediate in Lysinibacillus sphaericus. Environ. Sci. Pollut. Res. 25, 22790–22796. doi:10.1007/s11356-018-2364-9
Górny, D., Guzik, U., Hupert-Kocurek, K., and Wojcieszyńska, D. (2019). A new pathway for naproxen utilisation by Bacillus thuringiensis B1 (2015b) and its decomposition in the presence of organic and inorganic contaminants. J. Environ. Manag. 239, 1–7. doi:10.1016/j.jenvman.2019.03.034
Hamid, H., and Eskicioglu, C. (2012). Fate of estrogenic hormones in wastewater and sludge treatment: a review of properties and analytical detection techniques in sludge matrix. Water Res. 46 (18), 5813–5833. doi:10.1016/j.watres.2012.08.002
Harthern-Flint, S. L., Dolfing, J., Mrozik, W., Meynet, P., Eland, L. E., Sim, M., et al. (2021). Experimental and genomic evaluation of the oestrogen degrading bacterium Rhodococcus equi ATCC13557. Front. Microbiol. 12, 670928. doi:10.3389/fmicb.2021.670928
Herrera, N., Herrera, C., Ortíz, I., Orozco, L., Robledo, S., Agudelo, D., et al. (2018). Genotoxicity and cytotoxicity of three microcystin-LR containing cyanobacterial samples from Antioquia, Colombia. Toxicon 154, 50–59. doi:10.1016/j.toxicon.2018.09.011
Horinouchi, M., and Hayashi, T. (2023). Comprehensive summary of steroid metabolism in Comamonas testosteroni TA441: entire degradation process of basic four rings and removal of C12 hydroxyl group. Appl. Environ. Microbiol. 89 (10), e0014323–23. doi:10.1128/aem.00143-23
Horinouchi, M., Yamamoto, T., Taguchi, K., Arai, H., and Kudo, T. (2001). Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 147 (12), 3367–3375. doi:10.1099/00221287-147-12-3367
Hossain, K., Roy, K., Gadaleta, D., Ultre, M., and Benfenati, E. (2018). Chemometric modeling of aquatic toxicity of contaminants of emerging concern (CECs) in Dugesia japonica and its interspecies correlation with daphnia and fish: QSTR and QSTTR approaches. Chemosphere 280, 130652. doi:10.1016/j.chemosphere.2021.130652
Hsiao, T. H., Chen, P. H., Wang, P. H., Brandon-Mong, G. J., Li, C. W., Horinouchi, M., et al. (2023). Harnessing microbial phylum-specific molecular markers for assessment of environmental estrogen degradation. Sci. Total Environ. 896, 165152. doi:10.1016/j.scitotenv.2023.165152
Hsu, Y. S., Liu, Y. H., Lin, C. H., Tsai, C. H., and Wu, W. F. (2023). Dual bio-degradative pathways of di-2-ethylhexyl phthalate by a novel bacterium Burkholderia sp. SP4. World J. Microbiol. Biotechnol. 39, 44. doi:10.1007/s11274-022-03490-3
Hu, J., Zhang, L. L., Chen, J. M., and Liu, Y. (2013). Degradation of paracetamol by Pseudomonas aeruginosa strain HJ1012. J. Environ. Sci. Health, Part A 48 (7), 791–799. doi:10.1080/10934529.2013.744650
Hu, L., Shan, K., Lin, L., Shen, W., Huang, L., Gan, N., et al. (2016a). Multi-year assessment of toxic genotypes and microcystin concentration in northern Lake Taihu, China. Toxins 8 (1), 23. doi:10.3390/toxins8010023
Hu, Y., Chen, J., Fan, H., Xie, P., and He, J. (2016b). A review of neurotoxicity of microcystins. Environ. Sci. Pollut. Res. 23, 7211–7219. doi:10.1007/s11356-016-6073-y
Huang, H., Zhang, X. Y., Chen, T. L., Zhao, Y. L., Xu, D. S., and Bai, Y. P. (2019). Biodegradation of structurally diverse phthalate esters by a newly identified esterase with catalytic activity toward di (2-ethylhexyl) phthalate. J. Agric. Food Chem. 67, 8548–8558. doi:10.1021/acs.jafc.9b02655
Huang, M., Tian, S., Chen, D., Zhang, W., Wu, J., and Chen, L. (2012). Removal of sulfamethazine antibiotics by aerobic sludge and an isolated Achromobacter sp. S-3. J. Environ. Sci. 24 (9), 1594–1599. doi:10.1016/S1001-0742(11)60973-X
Ibero, J., Galán, B., Díaz, E., and García, J. L. (2019). Testosterone degradative pathway of Novosphingobium tardaugens. Genes 10 (11), 871. doi:10.3390/genes10110871
Jan-Roblero, J., and Cruz-Maya, J. A. (2023). Ibuprofen: toxicology and biodegradation of an emerging contaminant. Molecules 28 (5), 2097. doi:10.3390/molecules28052097
Jiang, B., Li, A., Cui, D., Cai, R., Ma, F., and Wang, Y. (2014). Biodegradation and metabolic pathway of sulfamethoxazole by Pseudomonas psychrophila HA-4, a newly isolated cold-adapted sulfamethoxazole-degrading bacterium. Appl. Microbiol. Biotechnol. 98, 4671–4681. doi:10.1007/s00253-013-5488-3
Jiang, N., Song, P., Li, X., Zhu, L., Wang, J., Yin, X., et al. (2022). Dibutyl phthalate induced oxidative stress and genotoxicity on adult zebrafish (Danio rerio) brain. J. Hazard. Mat. 424, 127749. doi:10.1016/j.jhazmat.2021.127749
Jin, H., Hiraoka, Y., Okuma, Y., Hashimoto, E. H., Kurita, M., Anas, A. R. J., et al. (2018). Microbial degradation of amino acid-containing compounds using the microcystin-degrading bacterial strain B-9. Mar. drugs 16 (2), 50. doi:10.3390/md16020050
Jing, X., Zhang, W., Xie, J., Wang, W., Lu, T., Dong, Q., et al. (2021). Monitoring and risk assessment of pesticide residue in plant-soil-groundwater systxem about medlar planting in Golmud. Environ. Sci. Pollut. Res. 28, 26413–26426. doi:10.1007/s11356-021-12403-0
John, E. M., and Shaike, J. M. (2015). Chlorpyrifos: pollution and remediation. Environ. Chem. Lett. 13, 269–291. doi:10.1007/s10311-015-0513-7
John, E. M., Varghese, E. M., and Shaike, J. M. (2020). Plasmid-mediated biodegradation of chlorpyrifos and analysis of its metabolic by-products. Curr. Microbiol. 77, 3095–3103. doi:10.1007/s00284-020-02115-y
Kamaraj, Y., Jayathandar, R. S., Dhayalan, S., Subramaniyan, S., and Punamalai, G. (2022). Biodegradation of di-(2-ethylhexyl) phthalate by novel Rhodococcus sp. PFS1 strain isolated from paddy field soil. Arch. Microbiol. 204, 21–12. doi:10.1007/s00203-021-02632-9
Kaur, R., Kumari, A., Sharma, G., Singh, D., and Kaur, R. (2021). Biodegradation of endocrine disrupting chemicals benzyl butyl phthalate and dimethyl phthalate by Bacillus marisflavi RR014. J. Appl. Microbiol. 131, 1274–1288. doi:10.1111/jam.15045
Ke, J., Zhuang, W., Gin, K. Y. H., Reinhard, M., Hoon, L. T., and Tay, J. H. (2007). Characterization of estrogen-degrading bacteria isolated from an artificial sandy aquifer with ultrafiltered secondary effluent as the medium. Appl. Microbiol. Biotechnol. 75, 1163–1171. doi:10.1007/s00253-007-0923-y
Khan, M. F., and Murphy, C. D. (2021). Bacterial degradation of the anti-depressant drug fluoxetine produces trifluoroacetic acid and fluoride ion. Appl. Microbiol. Biotechnol. 105 (24), 9359–9369. doi:10.1007/s00253-021-11675-3
Khezami, F., Gómez-Navarro, O., Barbieri, M. V., Khiari, N., Chkirbene, A., Chiron, S., et al. (2024). Occurrence of contaminants of emerging concern and pesticides and relative risk assessment in Tunisian groundwater. Sci. Total Environ. 906, 167319. doi:10.1016/j.scitotenv.2023.167319
Kim, D., Cui, R., Moon, J., Kwak, J. I., and An, Y. J. (2019a). Soil ecotoxicity study of DEHP with respect to multiple soil species. Chemosphere 216, 387–395. doi:10.1016/j.chemosphere.2018.10.163
Kim, D. W., Thawng, C. N., Lee, K., Wellington, E. M., and Cha, C. J. (2019b). A novel sulfonamide resistance mechanism by two-component flavin-dependent monooxygenase system in sulfonamide-degrading actinobacteria. Environ. Int. 127, 206–215. doi:10.1016/j.envint.2019.03.046
Kim, N. H., Kim, D. U., Kim, I., and Ka, J. O. (2013). Syntrophic biodegradation of butachlor by Mycobacterium sp. J7A and Sphingobium sp. J7B isolated from rice paddy soil. FEMS Microbiol. Lett. 344, 114–120. doi:10.1111/1574-6968.12163
Kjaer, J., Olsen, P., Bach, K., Barlebo, H. C., Ingerslev, F., Hansen, M., et al. (2007). Leaching of estrogenic hormones from manure-treated structured soils. Environ. Sci. and Technol. 41 (11), 3911–3917. doi:10.1021/es0627747
Koch, H. M., Drexler, H., and Angerer, J. (2003). An estimation of the daily intake of di (2-ethylhexyl) phthalate (DEHP) and other phthalates in the general population. Int. J. Hyg. Environ. Health. 206, 77–83. doi:10.1078/1438-4639-00205
Kosma, C. I., Lambropoulou, D. A., and Albanis, T. A. (2015). Comprehensive study of the antidiabetic drug metformin and its transformation product guanylurea in Greek wastewaters. Water Res. 70, 436–448. doi:10.1016/j.watres.2014.12.010
Kou, L., Chen, H., Zhang, X., Liu, S., Zhang, B., and Zhu, H. (2023). Biodegradation of di (2-ethylhexyl) phthalate by a new bacterial consortium. Water Sci. Technol. 88, 92–105. doi:10.2166/wst.2023.198
Krishnamurthy, T., Szafraniec, L., Hunt, D. F., Shabanowitz, J., Yates, J. R., Hauer, C. R., et al. (1989). Structural characterization of toxic cyclic peptides from blue-green algae by tandem mass spectrometry. Proc. Natl. Acad. Sci. 86, 770–774. doi:10.1073/pnas.86.3.770
Kumar, V., and Maitra, S. S. (2016). Biodegradation of endocrine disruptor dibutyl phthalate (DBP) by a newly isolated Methylobacillus sp. V29b and the DBP degradation pathway. 3 Biotech. 6, 200–212. doi:10.1007/s13205-016-0524-5
Lamraoui, I., Eltoukhy, A., Wang, J., Lamraoui, M., Ahmed, A., Jia, Y., et al. (2020). Biodegradation of di (2-ethylhexyl) phthalate by a novel Enterobacter spp. strain YC-IL1 isolated from polluted soil, Mila, Algeria. Int. J. Environ. Res. Public Health 17, 7501. doi:10.3390/ijerph17207501
Lange, I. G., Daxenberger, A., Schiffer, B., Witters, H., Ibarreta, D., and Meyer, H. H. (2002). Sex hormones originating from different livestock production systems: fate and potential disrupting activity in the environment. Anal. Chim. acta 473 (1-2), 27–37. doi:10.1016/S0003-2670(02)00748-1
Lara-Moreno, A., Morillo, E., Merchán, F., Madrid, F., and Villaverde, J. (2022). Chlorpyrifos removal in an artificially contaminated soil using novel bacterial strains and cyclodextrin. Evaluation of its effectiveness by ecotoxicity studies. Agron 12, 1971. doi:10.3390/agronomy12081971
Lee, H., and Kim, D. U. (2022). Biodegradation of Alachlor by a newly isolated bacterium: degradation pathway and product analysis. Processes 10, 2256. doi:10.3390/pr10112256
Lee, K., and Gibson, D. T. (1996). Toluene and ethylbenzene oxidation by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62 (9), 3101–3106. doi:10.1128/aem.62.9.3101-3106.1996
Lee, M., Kim, H. J., and Kim, D. (2021a). Selective and easy detection of microcystin-LR in freshwater using a bioactivated sensor based on multiwalled carbon nanotubes on filter paper. Biosens. Bioelectron. 192, 113529. doi:10.1016/j.bios.2021.113529
Lee, S., Kim, J., and Lee, J. (2021b). Colonization of toxic cyanobacteria on the surface and inside of leafy green: a hidden source of cyanotoxin production and exposure. Food Microbiol. 94, 103655. doi:10.1016/j.fm.2020.103655
Li, C., Liu, C., Li, R., Liu, Y., Xie, J., and Li, B. (2022). Biodegradation of dibutyl phthalate by the new strain Acinetobacter baumannii DP-2. Toxics 10, 532. doi:10.3390/toxics10090532
Li, C., Lu, J., Liu, J., Zhang, G., Tong, Y., and Ma, N. (2016). Exploring the correlations between antibiotics and antibiotic resistance genes in the wastewater treatment plants of hospitals in Xinjiang, China. Environ. Sci. Pollut. Res. 23, 15111–15121. doi:10.1007/s11356-016-6688-z
Li, J., Cheng, W., Xu, L., Strong, P. J., and Chen, H. (2015). Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. 22, 4587–4596. doi:10.1007/s11356-014-3665-2
Li, J., Ye, Q., and Gan, J. (2014). Degradation and transformation products of acetaminophen in soil. Water Res. 49, 44–52. doi:10.1016/j.watres.2013.11.008
Li, J., Zhang, J., Yadav, M. P., and Li, X. (2019). Biodegradability and biodegradation pathway of di-(2-ethylhexyl) phthalate by Burkholderia pyrrocinia B1213. Chemosphere 225, 443–450. doi:10.1016/j.chemosphere.2019.02.194
Li, T., Xu, Z. J., Zhang, S. T., Xu, J., Pan, P., and Zhou, N. Y. (2024). Discovery of a Ni2+-dependent heterohexameric metformin hydrolase. Nat. Commun. 15 (1), 6121. doi:10.1038/s41467-024-50409-7
Li, T., Xu, Z. J., and Zhou, N. Y. (2023). Aerobic degradation of the antidiabetic drug metformin by Aminobacter sp. strain NyZ550. Environ. Sci. and Technol. 57 (3), 1510–1519. doi:10.1021/acs.est.2c07669
Liu, T., Li, J., Qiu, L., Zhang, F., Linhardt, R. J., and Zhong, W. (2020). Combined genomic and transcriptomic analysis of the dibutyl phthalate metabolic pathway in Arthrobacter sp. ZJUTW. Biotechnol. Bioeng. 117, 3712–3726. doi:10.1002/bit.27524
Liu, X., Chen, J., Liu, Y., Wan, Z., Guo, X., Lu, S., et al. (2022). Sulfamethoxazole degradation by Pseudomonas silesiensis F6a isolated from bioelectrochemical technology-integrated constructed wetlands. Ecotoxicol. Environ. Saf. 240, 113698. doi:10.1016/j.ecoenv.2022.113698
Liu, X., Zhang, G., Liu, Y., Lu, S., Qin, P., Guo, X., et al. (2019). Occurrence and fate of antibiotics and antibiotic resistance genes in typical urban water of Beijing, China. Environ. Pollut. 246, 163–173. doi:10.1016/j.envpol.2018.12.005
Long, Z., Wang, X., Wang, Y., Dai, H., Li, C., Xue, Y., et al. (2021). Characterization of a novel carbendazim-degrading strain Rhodococcus sp. CX-1 revealed by genome and transcriptome analyses. Sci. Total Environ. 754, 142137. doi:10.1016/j.scitotenv.2020.142137
Lozano, V. L., and Pizarro, H. N. (2024). Glyphosate lessons: is biodegradation of pesticides a harmless process for biodiversity? Environ. Sci. Eur. 36, 55. doi:10.1186/s12302-024-00884-y
Lu, Z., Sun, W., Li, C., Ao, X., Yang, C., and Li, S. (2019). Bioremoval of non-steroidal anti-inflammatory drugs by Pseudoxanthomonas sp. DIN-3 isolated from biological activated carbon process. Water Res. 161, 459–472. doi:10.1016/j.watres.2019.05.065
Lyu, J., Yu, J., Zha, J., and Wang, Z. (2020). Emerging contaminants in water and their toxicological effects: a review. Sci. Total Environ. 749, 141472. doi:10.1016/j.scitotenv.2020.141472
Ma, Y., Xu, D., Li, C., Wei, S., Guo, R., Li, Y., et al. (2022). Combined toxicity and toxicity persistence of antidepressants citalopram and mirtazapine to zooplankton Daphnia magna. Environ. Sci. Pollut. Res. 29 (44), 66100–66108. doi:10.1007/s11356-022-20203-3
MacKintosh, C., Beattie, K. A., Klumpp, S., Cohen, P., and Codd, G. A. (1990). Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 264, 187–192. doi:10.1016/0014-5793(90)80245-E
Malla, M. A., Dubey, A., Kumar, A., Yadav, S., and Kumari, S. (2023). Modeling and optimization of chlorpyrifos and glyphosate biodegradation using RSM and ANN: elucidating their degradation pathways by GC-MS based metabolomics. Ecotoxicol. Environ. Saf. 252, 114628. doi:10.1016/j.ecoenv.2023.114628
Mao, F., Liu, C., He, M., Shi, W., Xue, G., and Gao, P. (2013). Isolation and identification of an erythromycin degradation bacterium and study on its biodegradation characteristics. Environ. Sci. Technol. 36 (7), 912.
Marchlewicz, A., Guzik, U., Smułek, W., and Wojcieszyńska, D. (2017). Exploring the degradation of ibuprofen by Bacillus thuringiensis B1 (2015b): the new pathway and factors affecting degradation. Molecules 22 (10), 1676. doi:10.3390/molecules22101676
Martinez-Vaz, B. M., Dodge, A. G., Lucero, R. M., Stockbridge, R. B., Robinson, A. A., Tassoulas, L. J., et al. (2022). Wastewater bacteria remediating the pharmaceutical metformin: genomes, plasmids and products. Front. Bioeng. Biotechnol. 10, 1086261. doi:10.3389/fbioe.2022.1086261
Maruyama, T., Park, H. D., Ozawa, K., Tanaka, Y., Sumino, T., Hamana, K., et al. (2006). Sphingosinicella microcystinivorans gen. nov., sp. nov., a microcystin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 56 (1), 85–89. doi:10.1099/ijs.0.63789-0
Mazellier, P., Leroy, E., De Laat, J., and Legube, B. (2003). Degradation of carbendazim by UV/H 2 O 2 investigated by kinetic modelling. Environ. Chem. Lett. 1, 68–72. doi:10.1007/s10311-002-0010-7
Meador, J., Yeh, A., Young, G., and Gallagher, E. (2016). Contaminants of emerging concern in a large temperate estuary. Environ. Pollut. 213, 254–267. doi:10.1016/j.envpol.2016.01.088
Metcalf, J. S., Barakate, A., and Codd, G. A. (2004). Inhibition of plant protein synthesis by the cyanobacterial hepatotoxin, cylindrospermopsin. FEMS Microbiol. Lett. 235, 125–129. doi:10.1111/j.1574-6968.2004.tb09576.x
Miller, E. L. (2002). The penicillins: a review and update. J. midwifery and women's health 47 (6), 426–434. doi:10.1016/S1526-9523(02)00330-6
Minerdi, D., Zgrablic, I., Castrignanò, S., Catucci, G., Medana, C., Terlizzi, M. E., et al. (2016). Escherichia coli overexpressing a Baeyer-Villiger monooxygenase from Acinetobacter radioresistens becomes resistant to imipenem. Antimicrob. agents Chemother. 60 (1), 64–74. doi:10.1128/aac.01088-15
Mondal, T., Mondal, S., Ghosh, S. K., Pal, P., Soren, T., and Maiti, T. K. (2024). Dibutyl phthalate degradation by Paenarthrobacter ureafaciens PB10 through downstream product myristic acid and its bioremediation potential in contaminated soil. Chemosphere 352, 141359. doi:10.1016/j.chemosphere.2024.141359
Murdoch, R. W., and Hay, A. G. (2013). Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2. Microbiology 159 (Pt_3), 621–632. doi:10.1099/mic.0.062273-0
Murdoch, R. W., and Hay, A. G. (2015). The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation 26, 105–113. doi:10.1007/s10532-015-9719-4
Nahla, E., Arya, P., Maneesha, P., and Chitra, K. C. (2024). Exposure to the plasticizer dibutyl phthalate causes oxidative stress and neurotoxicity in brain tissue. Environ. Sci. Poll. Res. 31, 21399–21414. doi:10.1007/s11356-024-32604-7
Nahurira, R., Ren, L., Song, J., Jia, Y., Wang, J., Fan, S., et al. (2017). Degradation of di (2-ethylhexyl) phthalate by a novel Gordonia alkanivorans strain YC-RL2. Curr. Microbiol. 74, 309–319. doi:10.1007/s00284-016-1159-9
Nandhini, A. R., Harshiny, M., and Gummadi, S. N. (2021). Chlorpyrifos in environment and food: a critical review of detection methods and degradation pathways. Nviron. Sci. Pro. Imp. 23, 1255–1277. doi:10.1039/d1em00178g
Nandi, M., Paul, T., Kanaujiya, D. K., Baskaran, D., Pakshirajan, K., and Pugazhenthi, G. (2021). Biodegradation of benzyl butyl phthalate and dibutyl phthalate by Arthrobacter sp. via micellar solubilization in a surfactant-aided system. Water Supply 21, 2084–2098. doi:10.2166/ws.2020.347
Nebot, C., Falcon, R., Boyd, K. G., and Gibb, S. W. (2015). Introduction of human pharmaceuticals from wastewater treatment plants into the aquatic environment: a rural perspective. Environ. Sci. Pollut. Res. 22, 10559–10568. doi:10.1007/s11356-015-4234-z
Nhi-Cong, L. T., Mikolasch, A., Awe, S., Sheikhany, H., Klenk, H. P., and Schauer, F. (2010). Oxidation of aliphatic, branched chain, and aromatic hydrocarbons by Nocardia cyriacigeorgica isolated from oil-polluted sand samples collected in the Saudi Arabian Desert. J. Basic Microbiol. 50, 241–253. doi:10.1002/jobm.200900358
Nielsen, J. (2001). Metabolic engineering. Appl. Microbiol. Biotechnol. 55, 263–283. doi:10.1007/s002530000511
Oettel, M. (2003). Testosterone metabolism, dose–response relationships and receptor polymorphisms: selected pharmacological/toxicological considerations on benefits versus risks of testosterone therapy in men. Aging Male 6 (4), 230–256. doi:10.1080/13685530312331309772
Orrego, R., Guchardi, J., Hernandez, V., Krause, R., Roti, L., Armour, J., et al. (2009). Pulp and paper mill effluent treatments have differential endocrine-disrupting effects on rainbow trout. Environ. Toxicol. Chem. Int. J. 28 (1), 181–188. doi:10.1897/08-191.1
Ounissi, H., and Courvalin, P. (1985). Nucleotide sequence of the gene ereA encoding the erythromycin esterase in Escherichia coli. Gene 35 (3), 271–278. doi:10.1016/0378-1119(85)90005-8
Ovung, A., and Bhattacharyya, J. (2021). Sulfonamide drugs: structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 13 (2), 259–272. doi:10.1007/s12551-021-00795-9
Palma, T. L., and Costa, M. C. (2021). Anaerobic biodegradation of fluoxetine using a high-performance bacterial community. Anaerobe 68, 102356. doi:10.1016/j.anaerobe.2021.102356
Pan, L. J., Tang, X. D., Li, C. X., Yu, G. W., and Wang, Y. (2017). Biodegradation of sulfamethazine by an isolated thermophile–Geobacillus sp. S-07. World J. Microbiol. Biotechnol. 33, 85–88. doi:10.1007/s11274-017-2245-2
Pandey, B., Pandey, A. K., Tripathi, K., and Dubey, S. K. (2024). Biodegradation of acetaminophen: microcosm centric genomic-proteomic-metabolomics evidences. Bioresour. Technol. 401, 130732. doi:10.1016/j.biortech.2024.130732
Pandey, G., Dorrian, S. J., Russell, R. J., and Oakeshott, J. G. (2009). Biotransformation of the neonicotinoid insecticides imidacloprid and thiamethoxam by Pseudomonas sp. 1G. Biochem. Biophys. Res. Commun. 380, 710–714. doi:10.1016/j.bbrc.2009.01.156
Pang, S., Lin, Z., Zhang, W., Mishra, S., Bhatt, P., and Chen, S. (2020). Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front. Microbiol. 11, 868. doi:10.3389/fmicb.2020.00868
Paris, L., Devers-Lamrani, M., Joly, M., Viala, D., De Antonio, M., Pereira, B., et al. (2023). Effect of subtherapeutic and therapeutic sulfamethazine concentrations on transcribed genes and translated proteins involved in Microbacterium sp. C448 resistance and degradation. FEMS Microbiol. Ecol. 99 (7), fiad064. doi:10.1093/femsec/fiad064
Parolini, M. (2020). Toxicity of the Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) acetylsalicylic acid, paracetamol, diclofenac, ibuprofen and naproxen towards freshwater invertebrates: a review. Sci. Total Environ. 740, 140043. doi:10.1016/j.scitotenv.2020.140043
Patel, A. K., Singhania, R. R., Albarico, F. P. J. B., Pandey, A., Chen, C. W., and Dong, C. D. (2022). Organic wastes bioremediation and its changing prospects. Sci. Total Environ. 824, 153889. doi:10.1016/j.scitotenv.2022.153889
Patrolecco, L., Capri, S., and Ademollo, N. (2015). Occurrence of selected pharmaceuticals in the principal sewage treatment plants in Rome (Italy) and in the receiving surface waters. Environ. Sci. Pollut. Res. 22, 5864–5876. doi:10.1007/s11356-014-3765-z
Pernicova, I., and Korbonits, M. (2014). Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10 (3), 143–156. doi:10.1038/nrendo.2013.256
Petković Didović, M., Kowalkowski, T., and Broznić, D. (2022). Emerging contaminant imidacloprid in Mediterranean soils: the risk of accumulation is greater than the risk of leaching. Toxics 10, 358. doi:10.3390/toxics10070358
Phale, P. S., Basu, A., Majhi, P. D., Deveryshetty, J., Vamsee-Krishna, C., and Shrivastava, R. (2007). Metabolic diversity in bacterial degradation of aromatic compounds. Omics a J. Integr. Biol. 11 (3), 252–279. doi:10.1089/omi.2007.0004
Phale, P. S., Malhotra, H., and Shah, B. A. (2020). Degradation strategies and associated regulatory mechanisms/features for aromatic compound metabolism in bacteria. Adv. Appl. Microbiol. 112, 1–65. doi:10.1016/bs.aambs.2020.02.002
Phugare, S. S., Kalyani, D. C., Gaikwad, Y. B., and Jadhav, J. P. (2013). Microbial degradation of imidacloprid and toxicological analysis of its biodegradation metabolites in silkworm (Bombyx mori). Chem. Eng. J. 230, 27–35. doi:10.1016/j.cej.2013.06.042
Picó, Y., Belenguer, V., Corcellas, C., Díaz-Cruz, M., Eljarrat, E., Farré, M., et al. (2019). Contaminants of emerging concern in freshwater fish from four Spanish Rivers. Sci. Total Environ. 659, 1186–1198. doi:10.1016/J.SCITOTENV.2018.12.366
Plante, I., Centrón, D., and Roy, P. H. (2003). An integron cassette encoding erythromycin esterase, ere (A), from Providencia stuartii. J. Antimicrob. Chemother. 51 (4), 787–790. doi:10.1093/jac/dkg169
Poon, R., Lecavalier, P., Mueller, R., Valli, V., Procter, B. G., and Chu, I. (1997). Subchronic oral toxicity of di-n-octyl phthalate and di (2-ethylhexyl) phthalate in the rat. Food Chem. Toxicol. 35, 225–239. doi:10.1016/s0278-6915(96)00064-6
Potter, T. L., and Carpenter, T. L. (1995). Occurrence of alachlor environmental degradation products in groundwater. Environ. Sci. and Technol. 29, 1557–1563. doi:10.1021/es00006a018
Praveenkumarreddy, Y., Vimalkumar, K., Ramaswamy, B. R., Kumar, V., Singhal, R. K., Basu, H., et al. (2021). Assessment of non-steroidal anti-inflammatory drugs from selected wastewater treatment plants of Southwestern India. Emerg. Contam. 7, 43–51. doi:10.1016/j.emcon.2021.01.001
Qi, M., Liang, B., Zhang, L., Ma, X., Yan, L., Dong, W., et al. (2021). Microbial interactions drive the complete catabolism of the antibiotic sulfamethoxazole in activated sludge microbiomes. Environ. Sci. and Technol. 55 (5), 3270–3282. doi:10.1021/acs.est.0c06687
Racke, K. D. (1993). Environmental fate of chlorpyrifos. Rev. Environ. Contam. Toxicol. 131, 1–150. doi:10.1007/978-1-4612-4362-5_1
Radke, E. G., Braun, J. M., Nachman, R. M., and Cooper, G. S. (2020). Phthalate exposure and neurodevelopment: a systematic review and meta-analysis of human epidemiological evidence. Environ. Int. 137, 105408. doi:10.1016/j.envint.2019.105408
Ragugnetti, M., Adams, M. L., Guimarães, A. T., Sponchiado, G., de Vasconcelos, E. C., and de Oliveira, C. M. R. (2011). Ibuprofen genotoxicity in aquatic environment: an experimental model using Oreochromis niloticus. Water, Air, and Soil Pollut. 218, 361–364. doi:10.1007/s11270-010-0698-0
Rama, E. M., Bortolan, S., Vieira, M. L., Gerardin, D. C. C., and Moreira, E. G. (2014). Reproductive and possible hormonal effects of carbendazim. Regul. Toxicol. Pharm. 69, 476–486. doi:10.1016/j.yrtph.2014.05.016
Reis, A. C., Kolvenbach, B. A., Chami, M., Gales, L., Egas, C., Corvini, P. F. X., et al. (2019). Comparative genomics reveals a novel genetic organization of the sad cluster in the sulfonamide-degrader ‘Candidatus Leucobacter sulfamidivorax’strain GP. BMC genomics 20, 885–923. doi:10.1186/s12864-019-6206-z
Reis, P. J., Reis, A. C., Ricken, B., Kolvenbach, B. A., Manaia, C. M., Corvini, P. F., et al. (2014). Biodegradation of sulfamethoxazole and other sulfonamides by Achromobacter denitrificans PR1. J. Hazard. Mater. 280, 741–749. doi:10.1016/j.jhazmat.2014.08.039
Ren, J., Ni, S., Shen, Y., Niu, D., Sun, R., Wang, C., et al. (2022). Characterization of the erythromycin degradation pathway and related enzyme in Rhodococcus gordoniae rjjtx-2. J. Clean. Prod. 379, 134758. doi:10.1016/j.jclepro.2022.134758
Ren, J., Qi, X., Zhang, J., Niu, D., Shen, Y., Yu, C., et al. (2023a). Biodegradation efficiency and mechanism of erythromycin degradation by Paracoccus versutus W7. J. Environ. Manag. 332, 117372. doi:10.1016/j.jenvman.2023.117372
Ren, J., Xu, C., Shen, Y., Li, C., Dong, L., Huhe, T., et al. (2023b). Environmental factors induced macrolide resistance genes in composts consisting of erythromycin fermentation residue, cattle manure, and maize straw. Environ. Sci. Pollut. Res. 30 (24), 65119–65128. doi:10.1007/s11356-023-27087-x
Ren, L., Jia, Y., Ruth, N., Qiao, C., Wang, J., Zhao, B., et al. (2016). Biodegradation of phthalic acid esters by a newly isolated Mycobacterium sp. YC-RL4 and the bioprocess with environmental samples. Environ. Sci. Poll. Res. 23, 16609–16619. doi:10.1007/s11356-016-6829-4
Ren, L., Wang, G., Huang, Y., Guo, J., Li, C., Jia, Y., et al. (2021). Phthalic acid esters degradation by a novel marine bacterial strain Mycolicibacterium phocaicum RL-HY01: characterization, metabolic pathway and bioaugmentation. Sci. Total Environ. 791, 148303. doi:10.1016/j.scitotenv.2021.148303
Ren, Y., Wang, J., Guo, W. W., Chen, J. W., Xu, L. Z., Wu, Z. W., et al. (2024). PKM2/Hif-1α signal suppression involved in therapeutics of pulmonary fibrosis with microcystin-RR but not with pirfenidone. Toxicon 247, 107822. doi:10.1016/j.toxicon.2024.107822
Ricken, B., Fellmann, O., Kohler, H. P. E., Schäffer, A., Corvini, P. F. X., and Kolvenbach, B. A. (2015). Degradation of sulfonamide antibiotics by Microbacterium sp. strain BR1–elucidating the downstream pathway. New Biotechnol. 32 (6), 710–715. doi:10.1016/j.nbt.2015.03.005
Ricken, B., Kolvenbach, B. A., Bergesch, C., Benndorf, D., Kroll, K., Strnad, H., et al. (2017). FMNH2-dependent monooxygenases initiate catabolism of sulfonamides in Microbacterium sp. strain BR1 subsisting on sulfonamide antibiotics. Sci. Rep. 7 (1), 15783. doi:10.1038/s41598-017-16132-8
Ríos, A. L. M., Gutierrez-Suarez, K., Carmona, Z., Ramos, C. G., and Oliveira, L. F. S. (2022). Pharmaceuticals as emerging pollutants: case naproxen an overview. Chemosphere 291, 132822. doi:10.1016/j.chemosphere.2021.132822
Rios-Miguel, A. B., Smith, G. J., Cremers, G., van Alen, T., Jetten, M. S., den Camp, H. J. O., et al. (2022). Microbial paracetamol degradation involves a high diversity of novel amidase enzyme candidates. Water Res. X 16, 100152. doi:10.1016/j.wroa.2022.100152
Roberts, E., Nunes, V. D., Buckner, S., Latchem, S., Constanti, M., Miller, P., et al. (2016). Paracetamol: not as safe as we thought? A systematic literature review of observational studies. Ann. rheumatic Dis. 75 (3), 552–559. doi:10.1136/annrheumdis-2014-206914
Robinson, A., Tassoulas, L., Martinez Vaz, B., Aukema, K., and Wackett, L. (2021). Metformin (glucophage) biodegradation: insights from microbiome and biochemical analyses. FASEB J. 35. doi:10.1096/fasebj.2021.35.S1.04286
Roh, H., and Chu, K. H. (2010). A 17β-Estradiol-utilizing bacterium, Sphingomonas strain KC8: Part I - characterization and abundance in wastewater treatment plants. Environ. Sci. and Technol. 44 (13), 4943–4950. doi:10.1021/es1001902
Rolbiecki, D., Harnisz, M., Korzeniewska, E., Buta, M., Hubeny, J., and Zieliński, W. (2021). Detection of carbapenemase-producing, hypervirulent Klebsiella spp. in wastewater and their potential transmission to river water and WWTP employees. Int. J. Hyg. Environ. Health 237, 113831. doi:10.1016/j.ijheh.2021.113831
Rowdhwal, S. S. S., and Chen, J. (2018). Toxic effects of di-2-ethylhexyl phthalate: an overview. Biomed. Res. Int. 2018, 1–10. doi:10.1155/2018/1750368
Roy, N. M., Zambrzycka, E., and Santangelo, J. (2017). Butyl benzyl phthalate (BBP) induces caudal defects during embryonic development. Environ. Toxicol. Pharmacol. 56, 129–135. doi:10.1016/j.etap.2017.09.009
Ruiz-Arias, M. A., Medina-Díaz, I. M., Bernal-Hernández, Y. Y., Barrón-Vivanco, B. S., González-Arias, C. A., Romero-Bañuelos, C. A., et al. (2023). The situation of chlorpyrifos in Mexico: a case study in environmental samples and aquatic organisms. Environ. Geochem. Health. 45, 6323–6351. doi:10.1007/s10653-023-01618-4
Rutere, C., Knoop, K., Posselt, M., Ho, A., and Horn, M. A. (2020). Ibuprofen degradation and associated bacterial communities in hyporheic zone sediments. Microorganisms 8 (8), 1245. doi:10.3390/microorganisms8081245
Salimi, M., Esrafili, A., Gholami, M., Jonidi Jafari, A., Rezaei Kalantary, R., Farzadkia, M., et al. (2017). Contaminants of emerging concern: a review of new approach in AOP technologies. Environ. Monit. Assess. 189, 414–422. doi:10.1007/s10661-017-6097-x
Salis, S., Testa, C., Roncada, P., Armorini, S., Rubattu, N., Ferrari, A., et al. (2017). Occurrence of imidacloprid, carbendazim, and other biocides in Italian house dust: potential relevance for intakes in children and pets. J. Environ. Sci. Health. Part B 52, 699–709. doi:10.1080/03601234.2017.1331675
Santer, M., and Ajl, S. J. (1952). Steroid metabolism by a species of Pseudomonas: II. Direct evidence for the breakdown of testosterone. J. Biol. Chem. 199 (1), 85–89. doi:10.1016/S0021-9258(18)44813-2
Sariaslani, F. S., Harper, D. B., and Higgins, I. J. (1974). Microbial degradation of hydrocarbons. Catabolism of 1-phenylalkanes by Nocardia salmonicolor. Biochem. J. 140, 31–45. doi:10.1042/bj1400031
Sarkar, J., Chowdhury, P. P., and Dutta, T. K. (2013). Complete degradation of di-n-octyl phthalate by Gordonia sp. strain Dop5. Chemosphere 90, 2571–2577. doi:10.1016/j.chemosphere.2012.10.101
Sauvé, S., and Desrosiers, M. (2014). A review of what is an emerging contaminant. Chem. Central J. 8, 15. doi:10.1186/1752-153X-8-15
Scheurer, M., Michel, A., Brauch, H. J., Ruck, W., and Sacher, F. (2012). Occurrence and fate of the antidiabetic drug metformin and its metabolite guanylurea in the environment and during drinking water treatment. Water Res. 46 (15), 4790–4802. doi:10.1016/j.watres.2012.06.019
Scheurer, M., Sacher, F., and Brauch, H. J. (2009). Occurrence of the antidiabetic drug metformin in sewage and surface waters in Germany. J. Environ. Monit. 11 (9), 1608–1613. doi:10.1039/B909311G
Schmitz, F. J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Köhrer, K., et al. (2000). Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45 (6), 891–894. doi:10.1093/jac/45.6.891
Selifonov, S. A., Grifoll, M., Eaton, R. W., and Chapman, P. J. (1996). Oxidation of naphthenoaromatic and methyl-substituted aromatic compounds by naphthalene 1, 2-dioxygenase. Appl. Environ. Microbiol. 62 (2), 507–514. doi:10.1128/aem.62.2.507-514.1996
Selvam, V., and Srinivasan, S. (2019). Neonicotinoid poisoning and management. Indian J. Crit. Care Med. 23 (Suppl. 4), S260–S262. doi:10.5005/jp-journals-10071-23308
Shariati, S., Ebenau-Jehle, C., Pourbabaee, A. A., Alikhani, H. A., Rodriguez-Franco, M., Agne, M., et al. (2022). Degradation of dibutyl phthalate by paenarthrobacter sp. shss isolated from saravan landfill, hyrcanian forests, Iran. Biodegradation 33, 59–70. doi:10.1007/s10532-021-09966-7
Sharma, K., Kaushik, G., Thotakura, N., Raza, K., Sharma, N., and Nimesh, S. (2019). Fate of ibuprofen under optimized batch biodegradation experiments using Micrococcus yunnanensis isolated from pharmaceutical sludge. Int. J. Environ. Sci. Technol. 16, 8315–8328. doi:10.1007/s13762-019-02400-9
Sharma, S., Singh, B., and Gupta, V. K. (2014). Assessment of imidacloprid degradation by soil-isolated Bacillus alkalinitrilicus. Environ. Monit. Assess. 186, 7183–7193. doi:10.1007/s10661-014-3919-y
Shi, W., Han, Y., Guan, X., Rong, J., Su, W., Zha, S., et al. (2019). Fluoxetine suppresses the immune responses of blood clams by reducing haemocyte viability, disturbing signal transduction and imposing physiological stress. Sci. Total Environ. 683, 681–689. doi:10.1016/j.scitotenv.2019.05.308
Shrivastava, R., Basu, B., Godbole, A., Mathew, M. K., Apte, S. K., and Phale, P. S. (2011). Repression of the glucose-inducible outer-membrane protein OprB during utilization of aromatic compounds and organic acids in Pseudomonas putida CSV86. Microbiology 157 (5), 1531–1540. doi:10.1099/mic.0.047191-0
Sim, W. J., Lee, J. W., and Oh, J. E. (2010). Occurrence and fate of pharmaceuticals in wastewater treatment plants and rivers in Korea. Environ. Pollut. 158 (5), 1938–1947. doi:10.1016/j.envpol.2009.10.036
Singh, B. K. (2009). Organophosphorus-degrading bacteria: ecology and industrial applications. Nat. Rev. Microbiol. 7, 156–164. doi:10.1038/nrmicro2050
Singh, S., Kumar, V., Gill, J. P. K., Datta, S., Singh, S., Dhaka, V., et al. (2020). Herbicide glyphosate: toxicity and microbial degradation. Int. J. Environ. Res. Public Health 17, 7519. doi:10.3390/ijerph17207519
Singh, S., Kumar, V., and Singh, J. (2019). Kinetic study of the biodegradation of glyphosate by indigenous soil bacterial isolates in presence of humic acid, Fe (III) and Cu (II) ions. J. Environ. Chem. Eng. 7, 103098. doi:10.1016/j.jece.2019.103098
Singh, S., Singh, N., Kumar, V., Datta, S., Wani, A. B., Singh, D., et al. (2016). Toxicity, monitoring and biodegradation of the fungicide carbendazim. Environ. Chem. Lett. 14, 317–329. doi:10.1007/s10311-016-0566-2
Słaba, M., Szewczyk, R., Piątek, M. A., and Długoński, J. (2013). Alachlor oxidation by the filamentous fungus Paecilomyces marquandii. J. Hazard. Mat. 261, 443–450. doi:10.1016/j.jhazmat.2013.06.064
Smegal, D. C. (2000). Human health risk assessment chlorpyrifos. US Environmental protection agency, office of prevention, pesticides and toxic substances, office of pesticide programs, health effects division. Washington, DC: US Government Printing Office, 1–131.
Spoof, L., and Catherine, A. (2016). “Appendix 3: tables of microcystins and nodularins,” in Handbook of cyanobacterial monitoring and cyanotoxin analysis (Wiley), 526–537. doi:10.1002/9781119068761.app3
Stokes, P. E., and Holtz, A. (1997). Fluoxetine tenth anniversary update: the progress continues. Clin. Ther. 19 (5), 1135–1250. doi:10.1016/S0149-2918(97)80066-5
Sun, R., Wang, L., Jiao, Y., Zhang, Y., Zhang, X., Wu, P., et al. (2019). Metabolic process of di-n-butyl phthalate (DBP) by Enterobacter sp. DNB-S2, isolated from Mollisol region in China. Environ. Pollut. 255, 113344. doi:10.1016/j.envpol.2019.113344
Sun, Y., Zhang, Y., Ma, Y., Xin, R., Li, X., and Niu, Z. (2024). Exploring the potential of a new marine bacterium associated with plastisphere to metabolize dibutyl phthalate and bis (2-ethylhexyl) phthalate by enrichment cultures combined with multi-omics analysis. Environ. Pollut. 342, 123146. doi:10.1016/j.envpol.2023.123146
Svirčev, Z., Lalić, D., Bojadžija Savić, G., Tokodi, N., Drobac Backović, D., Chen, L., et al. (2019). Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Archives Toxicol. 93, 2429–2481. doi:10.1007/s00204-019-02524-4
Sviridov, A. V., Shushkova, T. V., Ermakova, I. T., Ivanova, E. V., Epiktetov, D. O., and Leontievsky, A. A. (2015). Microbial degradation of glyphosate herbicides. Appl. Biochem. Microbiol. 51, 188–195. doi:10.7868/s0555109915020221
Syafrudin, M., Kristanti, R. A., Yuniarto, A., Hadibarata, T., Rhee, J., Al-Onazi, W. A., et al. (2021). Pesticides in drinking water—a review. Int. J. Environ. Res. public health 18, 468. doi:10.3390/ijerph18020468
Szewczyk, R., Soboń, A., Słaba, M., and Długoński, J. (2015). Mechanism study of alachlor biodegradation by Paecilomyces marquandii with proteomic and metabolomic methods. J. Hazard. Mater. 291, 52–64. doi:10.1016/j.jhazmat.2015.02.063
Takenaka, S., Okugawa, S., Kadowaki, M., Murakami, S., and Aoki, K. (2003). The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl. Environ. Microbiol. 69 (9), 5410–5413. doi:10.1128/AEM.69.9.5410-5413.2003
Tan, H., Kong, D., Li, Q., Zhou, Y., Jiang, X., Wang, Z., et al. (2022b). Metabolomics reveals the mechanism of tetracycline biodegradation by a Sphingobacterium mizutaii S121. Environ. Pollut. 305, 119299. doi:10.1016/j.envpol.2022.119299
Tan, Z., Yang, X., Chen, L., Liu, Y., Xu, H. J., Li, Y., et al. (2022a). Biodegradation mechanism of chloramphenicol by Aeromonas media SZW3 and genome analysis. Bioresour. Technol. 344, 126280. doi:10.1016/j.biortech.2021.126280
Tao, Y., Chen, B., Zhang, B. H., Zhu, Z. J., and Cai, Q. (2018). Occurrence, impact, analysis and treatment of metformin and guanylurea in coastal aquatic environments of Canada, USA and Europe. Adv. Mar. Biol. 81, 23–58. doi:10.1016/bs.amb.2018.09.005
Tappe, W., Herbst, M., Hofmann, D., Koeppchen, S., Kummer, S., Thiele, B., et al. (2013). Degradation of sulfadiazine by Microbacterium lacus strain SDZm4, isolated from lysimeters previously manured with slurry from sulfadiazine-medicated pigs. Appl. Environ. Microbiol. 79 (8), 2572–2577. doi:10.1128/AEM.03636-12
Tassoulas, L. J., Robinson, A., Martinez-Vaz, B., Aukema, K. G., and Wackett, L. P. (2021). Filling in the gaps in metformin biodegradation: a new enzyme and a metabolic pathway for guanylurea. Appl. Environ. Microbiol. 87 (11), e03003–e03020. doi:10.1128/AEM.03003-20
Thakali, O., Malla, B., Tandukar, S., Sthapit, N., Raya, S., Furukawa, T., et al. (2021). Release of antibiotic-resistance genes from hospitals and a wastewater treatment plant in the Kathmandu valley, Nepal. Water 13, 2733. doi:10.3390/w13192733
Tian, Z., Wark, D. A., Bogue, K., and James, C. A. (2021). Suspect and non-target screening of contaminants of emerging concern in streams in agricultural watersheds. Sci. Total Environ. 795, 148826. doi:10.1016/j.scitotenv.2021.148826
Trautwein, C., Berset, J. D., Wolschke, H., and Kümmerer, K. (2014). Occurrence of the antidiabetic drug Metformin and its ultimate transformation product Guanylurea in several compartments of the aquatic cycle. Environ. Int. 70, 203–212. doi:10.1016/j.envint.2014.05.008
Trivedi, V. D., Jangir, P. K., Sharma, R., and Phale, P. S. (2016). Insights into functional and evolutionary analysis of carbaryl metabolic pathway from Pseudomonas sp. strain C5pp. Sci. Rep. 6 (1), 38430. doi:10.1038/srep38430
Tyl, R. W., Myers, C. B., Marr, M. C., Fail, P. A., Seely, J. C., Brine, D. R., et al. (2004). Reproductive toxicity evaluation of dietary butyl benzyl phthalate (BBP) in rats. Reprod. Toxicol. 18, 241–264. doi:10.1016/j.reprotox.2003.10.006
Välitalo, P., Kruglova, A., Mikola, A., and Vahala, R. (2017). Toxicological impacts of antibiotics on aquatic micro-organisms: a mini-review. Int. J. Hyg. Environ. Health 220, 558–569. doi:10.1016/j.ijheh.2017.02.003
Van Dolah, F. M. (2000). Marine algal toxins: origins, health effects, and their increased occurrence. Environ. health Perspect. 108 (Suppl. 1), 133–141. doi:10.1289/ehp.00108s1133
Vo, H. N. P., Le, G. K., Nguyen, T. M. H., Bui, X. T., Nguyen, K. H., Rene, E. R., et al. (2019). Acetaminophen micropollutant: historical and current occurrences, toxicity, removal strategies and transformation pathways in different environments. Chemosphere 236, 124391. doi:10.1016/j.chemosphere.2019.124391
Wang, A., Mahai, G., Wan, Y., Jiang, Y., Meng, Q., Xia, W., et al. (2019c). Neonicotinoids and carbendazim in indoor dust from three cities in China: spatial and temporal variations. Sci. Total Environ. 695, 133790. doi:10.1016/j.scitotenv.2019.133790
Wang, G. S., and Hoyte, C. (2018). Review of biguanide (metformin) toxicity. J. intensive care Med. 34 (11-12), 863–876. doi:10.1177/0885066618793385
Wang, J., Wang, C., Li, Q., Shen, M., Bai, P., Li, J., et al. (2019a). Microcystin-LR degradation and gene regulation of microcystin-degrading Novosphingobium sp. THN1 at different carbon concentrations. Front. Microbiol. 10, 1750. doi:10.3389/fmicb.2019.01750
Wang, J., and Wang, S. (2018). Microbial degradation of sulfamethoxazole in the environment. Appl. Microbiol. Biotechnol. 102, 3573–3582. doi:10.1007/s00253-018-8845-4
Wang, L., Gan, D., Gong, L., Zhang, Y., Wang, J., Guan, R., et al. (2022). Analysis of the performance of the efficient di-(2-ethylhexyl) phthalate-degrading bacterium Rhodococcus pyridinovorans DNHP-S2 and associated catabolic pathways. Chemosphere 306, 135610. doi:10.1016/j.chemosphere.2022.135610
Wang, P., Gao, J., Zhao, Y., Zhang, M., and Zhou, S. (2021). Biodegradability of di-(2-ethylhexyl) phthalate by a newly isolated bacterium Achromobacter sp. RX. Sci. Total Environ. 755, 142476. doi:10.1016/j.scitotenv.2020.142476
Wang, W., Zhang, Z., Ni, H., Yang, X., Li, Q., and Li, L. (2012). Decolorization of industrial synthetic dyes using engineered Pseudomonas putida cells with surface-immobilized bacterial laccase. Microb. Cell Fact. 11, 75–14. doi:10.1186/1475-2859-11-75
Wang, Y., Sun, Q., Li, Y., Wang, H., Wu, K., and Yu, C. P. (2019d). Biotransformation of estrone, 17β-estradiol and 17α-ethynylestradiol by four species of microalgae. Ecotoxicol. Environ. Saf. 180, 723–732. doi:10.1016/j.ecoenv.2019.05.061
Wang, Y., Xu, L., Wang, J., and Zhao, Q. (2017). Application of microcystin LR in the preparation of drugs for preventing and treating pulmonary fibrosis. Available at: https://pubchem.ncbi.nlm.nih.gov/patent/CN-106620647-A.
Wang, Y., Zhan, W., Ren, Q., Cheng, S., Wang, J., Ma, X., et al. (2019b). Biodegradation of di-(2-ethylhexyl) phthalate by a newly isolated Gordonia sp. and its application in the remediation of contaminated soils. Sci. Total Environ. 689, 645–651. doi:10.1016/j.scitotenv.2019.06.459
Wang, Y., Zhang, Y., Luo, Q., Zeng, X., and Lu, Y. (2020). Di-n-octyl phthalate degradation by a halotolerant bacterial consortium LF and its application in soil. Environ. Technol. 41, 345–351. doi:10.1080/09593330.2020.1713903
Wang, Z., Li, G., Wu, Q., Liu, C., Shen, J., and Yan, W. (2019e). Microcystin-LR exposure induced nephrotoxicity by triggering apoptosis in female zebrafish. Chemosphere 214, 598–605. doi:10.1016/j.chemosphere.2018.09.103
Wang, Z., Liang, G., Jiang, S., Wang, F., Li, H., Li, B., et al. (2024). Understanding the environmental impact and risks of organic additives in plastics: a call for sustained research and sustainable solutions. Emerg. Contam. 10, 100388. doi:10.1016/j.emcon.2024.100388
Wang, Z., Wang, Y., Gong, F., Zhang, J., Hong, Q., and Li, S. (2010). Biodegradation of carbendazim by a novel actinobacterium Rhodococcus jialingiae djl-6-2. Chemosphere 81, 639–644. doi:10.1016/j.chemosphere.2010.08.040
Watkinson, A. J., Murby, E. J., Kolpin, D. W., and Costanzo, S. D. (2009). The occurrence of antibiotics in an urban watershed: from wastewater to drinking water. Sci. total Environ. 407 (8), 2711–2723. doi:10.1016/j.scitotenv.2008.11.059
Wei, J., Pengji, Z., Zhang, J., Peng, T., Luo, J., and Yang, F. (2023). Biodegradation of MC-LR and its key bioactive moiety Adda by Sphingopyxis sp. YF1: comprehensive elucidation of the mechanisms and pathways. Water Res. 229, 119397. doi:10.1016/j.watres.2022.119397
Weidner-Wells, M. A., and Macielag, M. J. (2003). “Antibacterial agents, sulfonamides,” in Kirk-othmer encyclopedia of chemical Technology (Wiley). doi:10.1002/0471238961.1921120606152505.a01.pub2
WHO (2017). Guidelines for drinkingwater quality fourth edition incorporating the first addendum. Who Chron. 38, 104–108.
WHO (2020). Cyanobacterial toxins: microcystins. Background document for development of WHO Guidelines for drinking-water quality and Guidelines for safe recreational water environments. Geneva: World Health Organization. (WHO/HEP/ECH/WSH/2020.6). License: CC BY-NCSA 3.0 IGO.
WHO (2021). Guidelines on recreational water quality. Volume 1: coastal and fresh waters. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
Wojcieszyńska, D., Domaradzka, D., Hupert-Kocurek, K., and Guzik, U. (2014). Bacterial degradation of naproxen–undisclosed pollutant in the environment. J. Environ. Manag. 145, 157–161. doi:10.1016/j.jenvman.2014.06.023
Wojcieszyńska, D., and Guzik, U. (2020). Naproxen in the environment: its occurrence, toxicity to nontarget organisms and biodegradation. Appl. Microbiol. Biotechnol. 104 (5), 1849–1857. doi:10.1007/s00253-019-10343-x
Wright, R. J., Bosch, R., Gibson, M. I., and Christie-Oleza, J. A. (2020). Plasticizer degradation by marine bacterial isolates: a proteogenomic and metabolomic characterization. Environ. Sci. Technol. 54, 2244–2256. doi:10.1021/acs.est.9b05228
Wu, K., Lee, T. H., Chen, Y. L., Wang, Y. S., Wang, P. H., Yu, C. P., et al. (2019). Metabolites involved in aerobic degradation of the A and B rings of estrogen. Appl. Environ. Microbiol. 85 (3), 022233–e2318. doi:10.1128/AEM.02223-18
Wu, M., Xiang, J., Chen, F., Fu, C., and Xu, G. (2017). Occurrence and risk assessment of antidepressants in Huangpu River of Shanghai, China. Environ. Sci. Pollut. Res. 24, 20291–20299. doi:10.1007/s11356-017-9293-x
Wu, X., Liang, R., Dai, Q., Jin, D., Wang, Y., and Chao, W. (2010). Complete degradation of di-n-octyl phthalate by biochemical cooperation between Gordonia sp. strain JDC-2 and Arthrobacter sp. strain JDC-32 isolated from activated sludge. J. Hazard. Mat. 176, 262–268. doi:10.1016/j.jhazmat.2009.11.022
Wu, Y., Zhu, M., Ouyang, X., Qi, X., Guo, Z., Yuan, Y., et al. (2024). Integrated transcriptomics and metabolomics analyses reveal the aerobic biodegradation and molecular mechanisms of 2, 3′, 4, 4′, 5-pentachlorodiphenyl (PCB 118) in Methylorubrum sp. ZY-1. Chemosphere 356, 141921. doi:10.1016/j.chemosphere.2024.141921
Xie, Z., Ebinghaus, R., Temme, C., Caba, A., and Ruck, W. (2005). Atmospheric concentrations and air–sea exchanges of phthalates in the North Sea (German Bight). Atmos. Environ. 39, 3209–3219. doi:10.1016/j.atmosenv.2005.02.021
Xing, L., Yu, H., Qi, J., Jiang, P., Sun, B., Cui, J., et al. (2015). ErmF and ereD are responsible for erythromycin resistance in Riemerella anatipestifer. PLoS One 10 (6), e0131078. doi:10.1371/journal.pone.0131078
Xu, J., Lu, Q., de Toledo, R. A., and Shim, H. (2017). Degradation of di-2-ethylhexyl phthalate (DEHP) by an indigenous isolate Acinetobacter sp. SN13. Int. Biodeter. Biodeg. 117, 205–214. doi:10.1016/j.ibiod.2017.01.004
Xu, W. J., Wan, Q., Wang, W. F., Wang, Y., Feng, F. Y., Cheng, J. J., et al. (2022). Biodegradation of dibutyl phthalate by a novel endophytic Bacillus subtilis strain HB-T2 under in-vitro and in-vivo conditions. Environ. Technol. 43, 1917–1926. doi:10.1080/09593330.2020.1858181
Yan, H., Wang, H., Wang, J., Yin, C., Ma, S., Liu, X., et al. (2012). Cloning and expression of the first gene for biodegrading microcystin LR by Sphingopyxis sp. USTB-05. J. Environ. Sci. 24 (10), 1816–1822. doi:10.1016/S1001-0742(11)61016-4
Yan, Y., Zhu, F., Zhu, C., Chen, Z., Liu, S., Wang, C., et al. (2021). Dibutyl phthalate release from polyvinyl chloride microplastics: influence of plastic properties and environmental factors. Water Res. 204, 117597. doi:10.1016/j.watres.2021.117597
Yang, F., Huang, F., Feng, H., Wei, J., Massey, I. Y., Liang, G., et al. (2020). A complete route for biodegradation of potentially carcinogenic cyanotoxin microcystin-LR in a novel indigenous bacterium. Water Res. 174, 115638. doi:10.1016/j.watres.2020.115638
Yang, Y. Y., Pereyra, L. P., Young, R. B., Reardon, K. F., and Borch, T. (2011). Testosterone-mineralizing culture enriched from swine manure: characterization of degradation pathways and microbial community composition. Environ. Sci. and Technol. 45 (16), 6879–6886. doi:10.1021/es2013648
Yeh, A., Marcinek, D., Meador, J., and Gallagher, E. (2017). Effect of contaminants of emerging concern on liver mitochondrial function in Chinook salmon. Aquat. Toxicol. 190, 21–31. doi:10.1016/j.aquatox.2017.06.011
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. agents Chemother. 53 (12), 5046–5054. doi:10.1128/aac.00774-09
Yoshizawa, S., Matsushima, R., Watanabe, M. F., Harada, K. I., Ichihara, A., Carmichael, W. W., et al. (1990). Inhibition of protein phosphatases by microcystis and nodularin associated with hepatotoxicity. J. Cancer Res. Clin. Oncol. 116, 609–614. doi:10.1007/BF01637082
Yuan, M., Ding, Q., Sun, R., Zhang, J., Yin, L., and Pu, Y. (2021). Biodegradation of nodularin by a microcystin-degrading bacterium: performance, degradation pathway, and potential application. Toxins 13 (11), 813. doi:10.3390/toxins13110813
Yuan, S. Y., Liu, C., Liao, C. S., and Chang, B. V. (2002). Occurrence and microbial degradation of phthalate esters in Taiwan river sediments. Chemosphere 49, 1295–1299. doi:10.1016/S0045-6535(02)00495-2
Žegura, B., Štraser, A., and Filipič, M. (2011). Genotoxicity and potential carcinogenicity of cyanobacterial toxins–a review. Mutat. Res. 727 (1-2), 16–41. doi:10.1016/j.mrrev.2011.01.002
Zhan, H., Feng, Y., Fan, X., and Chen, S. (2018). Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 102, 5033–5043. doi:10.1007/s00253-018-9035-0
Zhang, H., Lin, Z., Liu, B., Wang, G., Weng, L., Zhou, J., et al. (2020a). Bioremediation of di-(2-ethylhexyl) phthalate contaminated red soil by Gordonia terrae RL-JC02: characterization, metabolic pathway and kinetics. Sci. Total Environ. 733, 139138. doi:10.1016/j.scitotenv.2020.139138
Zhang, J., Gan, W., Zhao, R., Yu, K., Lei, H., Li, R., et al. (2020b). Chloramphenicol biodegradation by enriched bacterial consortia and isolated strain Sphingomonas sp. CL5. 1: the reconstruction of a novel biodegradation pathway. Water Res. 187, 116397. doi:10.1016/j.watres.2020.116397
Zhang, J., Li, X., Klümper, U., Lei, H., Berendonk, T. U., Guo, F., et al. (2022). Deciphering chloramphenicol biotransformation mechanisms and microbial interactions via integrated multi-omics and cultivation-dependent approaches. Microbiome 10 (1), 180. doi:10.1186/s40168-022-01361-5
Zhang, J., Zhang, C., Zhu, Y., Li, J., and Li, X. (2018). Biodegradation of seven phthalate esters by Bacillus mojavensis B1811. Int. Biodeterior. Biodegrad. 132, 200–207. doi:10.1016/j.ibiod.2018.04.006
Zhang, J., Zheng, J. W., Liang, B., Wang, C. H., Cai, S., Ni, Y. Y., et al. (2011). Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J. Agric. Food Chem. 59, 4614–4621. doi:10.1021/jf104695g
Zhang, K., Liu, Y., Luo, H., Chen, Q., Zhu, Z., Chen, W., et al. (2017b). Bacterial community dynamics and enhanced degradation of di-n-octyl phthalate (DOP) by corncob-sodium alginate immobilized bacteria. Geoderma 305, 264–274. doi:10.1016/j.geoderma.2017.06.009
Zhang, K., Wu, X., Luo, H., Wang, W., Yang, S., Chen, J., et al. (2021). Biochemical pathways and enhanced degradation of dioctyl phthalate (DEHP) by sodium alginate immobilization in MBR system. Water Sci. Technol. 83, 664–677. doi:10.2166/wst.2020.605
Zhang, K., Zhao, Y., and Fent, K. (2017a). Occurrence and ecotoxicological effects of free, conjugated, and halogenated steroids including 17α-hydroxypregnanolone and pregnanediol in Swiss wastewater and surface water. Environ. Sci. and Technol. 51 (11), 6498–6506. doi:10.1021/acs.est.7b01231
Zhang, Q., Qiu, C., Jiang, W., Feng, P., Xue, X., Bukhari, I., et al. (2023b). The impact of dioctyl phthalate exposure on multiple organ systems and gut microbiota in mice. Heliyon 9, e22677. doi:10.1016/j.heliyon.2023.e22677
Zhang, S., Liu, Y., Mohisn, A., Zhang, G., Wang, Z., and Wu, S. (2024). Biodegradation of penicillin G sodium by Sphingobacterium sp. SQW1: performance, degradation mechanism, and key enzymes. J. Hazard. Mater. 468, 133485. doi:10.1016/j.jhazmat.2024.133485
Zhang, W., Qiu, L., Gong, A., and Yuan, X. (2017c). Isolation and characterization of a high-efficiency erythromycin A-degrading Ochrobactrum sp. strain. Mar. Pollut. Bull. 114 (2), 896–902. doi:10.1016/j.marpolbul.2016.10.076
Zhang, X., Huang, Y., Chen, W. J., Wu, S., Lei, Q., Zhou, Z., et al. (2023a). Environmental occurrence, toxicity concerns, and biodegradation of neonicotinoid insecticides. Environ. Res. 218, 114953. doi:10.1016/j.envres.2022.114953
Zhao, H., Tao, K., Zhu, J., Liu, S., Gao, H., and Zhou, X. (2015). Bioremediation potential of glyphosate-degrading Pseudomonas spp. strains isolated from contaminated soil. J. Gen. Appl. Microbiol. 61, 165–170. doi:10.2323/jgam.61.165
Zhao, H. M., Du, H., Lin, J., Chen, X. B., Li, Y. W., Li, H., et al. (2016). Complete degradation of the endocrine disruptor di-(2-ethylhexyl) phthalate by a novel Agromyces sp. MT-O strain and its application to bioremediation of contaminated soil. Sci. Total Environ. 562, 170–178. doi:10.1016/j.scitotenv.2016.03.171
Zhao, H. M., Hu, R. W., Chen, X. X., Chen, X. B., Lü, H., Li, Y. W., et al. (2018). Biodegradation pathway of di-(2-ethylhexyl) phthalate by a novel Rhodococcus pyridinivorans XB and its bioaugmentation for remediation of DEHP contaminated soil. Sci. Total Environ. 640, 1121–1131. doi:10.1016/j.scitotenv.2018.05.334
Zhou, T., Guo, T., Wang, Y., Wang, A., and Zhang, M. (2023). Carbendazim: ecological risks, toxicities, degradation pathways and potential risks to human health. Chemosphere 314, 137723. doi:10.1016/j.chemosphere.2022.137723
Zumstein, M. T., and Helbling, D. E. (2019). Biotransformation of antibiotics: exploring the activity of extracellular and intracellular enzymes derived from wastewater microbial communities. Water Res. 155, 115–123. doi:10.1016/j.watres.2019.02.024
Żur, J., Piński, A., Marchlewicz, A., Hupert-Kocurek, K., Wojcieszyńska, D., and Guzik, U. (2018). Organic micropollutants paracetamol and ibuprofen—toxicity, biodegradation, and genetic background of their utilization by bacteria. Environ. Sci. Pollut. Res. 25, 21498–21524. doi:10.1007/s11356-018-2517-x
Keywords: biodegradation, pharmaceuticals, plasticizers, cyanotoxins, pesticides, omics, metabolic pathways, metabolic engineering
Citation: Shah BA, Malhotra H, Papade SE, Dhamale T, Ingale OP, Kasarlawar ST and Phale PS (2024) Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation. Front. Bioeng. Biotechnol. 12:1470522. doi: 10.3389/fbioe.2024.1470522
Received: 25 July 2024; Accepted: 05 September 2024;
Published: 19 September 2024.
Edited by:
Segaran P. Pillai, United States Department of Health and Human Services, United StatesReviewed by:
Shaw Gargis, Centers for Disease Control and Prevention (CDC), United StatesFederica Spina, University of Turin, Italy
Copyright © 2024 Shah, Malhotra, Papade, Dhamale, Ingale, Kasarlawar and Phale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prashant S. Phale, cHBoYWxlQGlpdGIuYWMuaW4=
†These authors have contributed equally to this work and share first authorship
 Bhavik A. Shah
Bhavik A. Shah Harshit Malhotra
Harshit Malhotra Sandesh E. Papade
Sandesh E. Papade Tushar Dhamale
Tushar Dhamale Omkar P. Ingale
Omkar P. Ingale Sravanti T. Kasarlawar
Sravanti T. Kasarlawar Prashant S. Phale
Prashant S. Phale