- Electrical and Computer Engineering Department, Integrated Circuits and Bioengineering Laboratory, Carnegie Mellon University, Pittsburgh, PA, United States
Biosensors translate biological events into electronic signals that quantify biological processes. They are increasingly used in in vitro diagnostics applications that leverage their ability to process small sample volumes. One recent trend has been to integrate biosensors with complementary metal-oxide-semiconductor (CMOS) chips to provide enhanced miniaturization, parallel sensing, and low power consumption at a low cost. CMOS-enabled biosensors are used in monitoring DNA hybridization, enzymatic reactions, and cell proliferation, to name a few applications. This paper explores the materials and processes used in emerging CMOS biosensors. We discuss subtractive and additive processes for creating electrodes for electrochemical sensing applications. We discuss functionalization techniques for creating bioelectronic interfaces that allow molecular events to be transduced into the electrical domain using a plurality of modalities that are readily provided by CMOS chips. Example modalities featured are optical sensing, electrochemical detection, electrical detection, magnetic sensing, and mechanical sensing.
1 Introduction
Biosensors are devices that detect chemical compounds from specific biochemical reactions or events mediated by enzymes, immunosystems, tissues, organelles, or whole cells. Biosensors translate these reactions into electrical, thermal, or optical signals (Dellweg et al., 1992). Three main components make a biosensor: one or multiple transducers, biorecognition elements (BREs), and the required signal processing systems. The transducer generates target-specific signals upon target-BRE interactions. These signals are amplified and processed and then sent to a computer for data analysis (Figure 1) (Grieshaber et al., 2008; Jang and Hassibi, 2009; Syu et al., 2018). Biosensors are widely used across multiple disciplines, from healthcare and medical diagnostics to environmental sciences.
One application of biosensors is in in vitro diagnostics (IVD) where they are used to verify the presence of biological target analytes (e.g., DNA, protein, bacteria, etc.). IVD offers the benefits of high precision alongside considerable convenience for patients, as it requires only small-scale biological samples. However, IVD tools are bulky and expensive and require trained personnel to operate. These drawbacks hinder accessibility and lengthen turnaround times. These challenges can be more problematic in developing countries and in the case of contagious diseases like the recent COVID–19 global pandemic.
In response to such cases, the concepts of decentralized healthcare and point-of-care (POC) medicine have gained great attention, resulting in the need for smaller, more cost-effective biosensors. For biosensors to be effective in POC settings, they must maintain high sensitivity and selectivity while achieving low power consumption, a small footprint, and minor sample usage (Lei et al., 2016; Thriveni and Ghosh, 2022). Complementary metal-oxide-semiconductor (CMOS) technology meets these criteria.
CMOS technology—used in building integrated circuits (ICs) and chips—allows the integration of the transducer, which may be a photodiode (PD), an ion-sensitive field-effect transistor (ISFET), a metal-oxide-semiconductor field-effect transistor (MOSFET), or a nanowire field-effect transistor (NWFET), etc., and signal processing units on a single chip. With the signal amplification and processing circuitry on the same chip as the transducer, generated signals (e.g., current and voltage) can be handled directly. In addition to making the sensor low-cost and portable, this integration reduces external noise and improves the signal-to-noise ratio (SNR) and the limit of detection (LOD) (Lei et al., 2016; Syu et al., 2018). However, to enhance sensitivity and selectivity, CMOS chips integrated into biosensors can benefit from two types of surface modification: post-CMOS micro- and nano-fabrication processes, and wet biochemical procedures.
These surface modifications are particularly important because they facilitate molecular detection at lower concentrations, enabling earlier diagnosis. Moreover, enhancing the LOD in CMOS biosensors is a step toward molecular detection using typically unused biofluids like sweat. Many biomarkers and biomolecules in human blood, such as inflammatory markers like Tumor Necrosis Factor-alpha (TNF-
Below, we provide a brief overview of how post-CMOS processes and wet biochemical procedures enhance CMOS biosensors’ performance. Circuit components and metal interconnects of ICs fabricated with CMOS technology are buried in an oxide layer (
Furthermore, to improve sensitivity and selectivity, the surface of CMOS chips may need to be functionalized with proper biological probes. For example, in affinity-based biosensing, biosensing elements such as antibodies, receptor proteins, or DNA are immobilized on the transducer’s solid surface. Target analytes interact with the capture layer and bind to the immobilized probes, and the transducer translates these biological interactions into quantifiable signals (e.g., electrical, optical, mechanical, and magnetic) (Jang and Hassibi, 2009; Lei et al., 2016; Devadhasan et al., 2015; Rogers, 2000). For example, Manickam et al. (2017) deposited (3–glycidoxypropyl) trimethoxysilane on a CMOS photodiode via chemical vapor deposition (CVD) to further immobilize synthetic DNA probes on the chip’s surface. Various techniques have been studied and employed for probe immobilization, including physical adsorption (Figure 2A), streptavidin-biotin interaction (Figure 2B), and covalent immobilization (Figure 2C). We briefly describe some of these methods below.
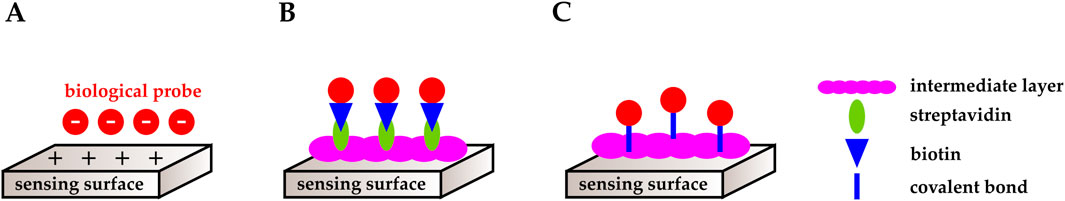
Figure 2. Schematic representation of functionalization mechanisms. (A) Physical adsorption. (B) Streptavidin-biotin complex. (C) Covalent immobilization.
Physical adsorption is a non-covalent functionalization method that relies on the ionic interactions between biological probes and the sensing surface (Figure 2A). The main advantage of this method is simplicity, as it does not require linker molecules or modifications to the capture probe and the target analyte (Nimse et al., 2014). Physical adsorption is a good method for DNA probe immobilization as DNAs are negatively charged. One example is in (Moser et al., 2018).
The use of streptavidin-biotin complexes is another non-covalent approach to functionalization. In this method, the sensing surface is functionalized with streptavidin using an intermediate layer, and the capture probe is labeled with biotin (Figure 2B). The streptavidin-biotin complex has one of the strongest non-covalent bonds, while biotin is a stable label with minimal interference with the labeled molecule’s functionality. This is a well-established technique for detecting proteins and DNA (Nimse et al., 2014; Smith et al., 2005), and an example can be found in (Crucifix et al., 2004).
Another method of functionalization employs covalent bonds. Covalent immobilization provides stronger binding and better stability compared to physical adsorption and streptavidin-biotin-based immobilization. In this method, the adherence of the biosensing element to the sensing surface is achieved through covalent bonds. Similarly to the streptavidin-biotin technique, covalent immobilization involves an intermediate layer between the sensing surface and the capture probe (Figure 2C). In biosensors with gold electrodes, for example, a common immobilization technique is modifying the electrode or the capture probes with thiol groups (RSH), which are present in cysteine amino acids, taking advantage of their strong affinity toward noble metals (Nimse et al., 2014). This technique was used in (Kuo et al., 2021; Lee et al., 2012). As another example, silanization is the preferred immobilization method in oxide sensing membranes (Pinitwong and Pijitrojana, 2015). In silanization, 3–aminopropyltriethoxysilane (APTES) and glutaraldehyde (GA) are commonly used in the crosslinking process. APTES is a self-assembled monolayer that binds to the transducer’s surface. Its amine group provides a platform for the bifunctional reagent, GA, to form covalent bonds to the capture probe (Syu et al., 2018; Nimse et al., 2014). Examples of biosensors that benefit from the APTES-GA immobilization technique are reported in (Lin C. H. et al., 2022; Zhou et al., 2021; Huang et al., 2013b).
Although advances in post-CMOS processes and wet biochemical procedures have made it possible to modify CMOS chips’ surfaces, there are challenges in this emerging field that must be addressed before moving from a research phase to full commercialization. For example, post-CMOS processes need large-scale facilities and are expensive to perform. Additionally, the biochemical procedures employed to immobilize biological probes on the CMOS biosensors include exposing the chip to harsh conditions such as low pH and elevated temperature, which may reduce chip reliability.
This paper provides a critical and comprehensive review of recently developed CMOS biosensors with surface modification and functionalization. We cover a variety of sensors and functionalization techniques. We also discuss the advantages and disadvantages of each method, the remaining challenges and shortcomings, and future trends in the field. A performance summary of 60+ surface-modified CMOS biosensors developed since the year 2003 is presented in Supplementary Tables 1–3 in the Information section of the paper.
2 CMOS electrochemical biosensing
Electrochemical sensing may be defined as the transformation of analyte-electrode interactions into a comprehensible signal (Hulanicki et al., 1991). Since the introduction of the first enzyme-electrode biochemical-based biosensor by (Clark and Lyons, 1962), electrochemical sensing has gained more attention in biosensing applications. Today, electrochemical sensing is favored because of its fast response, low cost, lack of complexity, and potential for miniaturization using nanotechnology. Additionally, electrochemical biosensing allows label-free detection, which adds to its adaptability (Li et al., 2017).
In electrochemical biosensors, the analyte resides in a gas or liquid environment that supports the analyte’s biological activity and its transport to electrodes or sensing membranes. The electrode or sensing membrane is sensitive to the target analyte and transduces the electrochemical reactions into electrical signals. This electrode is referred to as the working electrode (WE). In two-electrode electrochemical biosensors, a reference electrode (RE) provides a known stable DC voltage, and the WE potential is set/read with respect to this RE (Figures 3A, C). Another electrochemical sensing platform includes a third electrode, called the counter electrode (CE). The CE is used in Faradaic electrochemical biosensors–where a significant current is created due to the electrochemical reactions–and provides proper biasing to compensate for the voltage drop across the electrolyte’s equivalent resistance (Figures 3B, D) (Li et al., 2017).
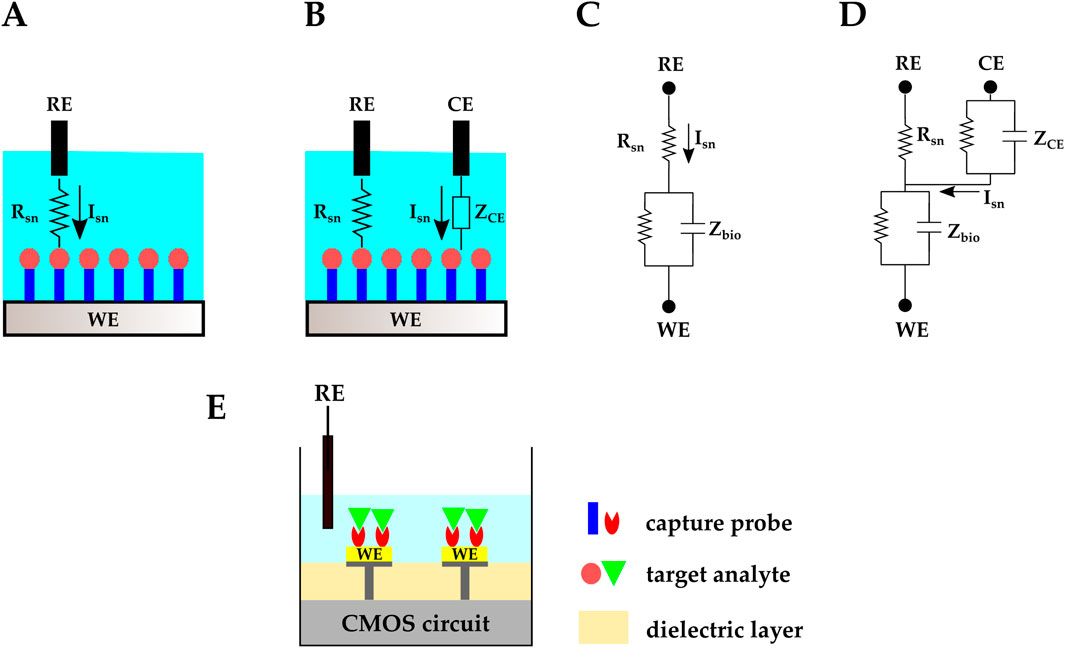
Figure 3. Schematic representation and circuit equivalent of (A, C) two-electrode and (B, D) three-electrode electrochemical biosensing. (E) CMOS electrochemical biosensor.
In conventional electrochemical biosensors, input and output electrical signals (e.g., voltage and current) are handled with a benchtop potentiostat. CMOS technology has made it possible to integrate the electrochemical biosensor’s electrodes and the readout circuit on one chip, eliminating the need for an external potentiostat (Figure 3E). Examples of CMOS potentiostats are in (Zhang et al., 2005; Honarvar Nazari and Genov, 2009; Li et al., 2021). Moreover, with CMOS technology, microelectrode array (MEA) structures can be fabricated to serve as electrodes. These MEAs provide high spatial resolution and enable high-throughput platforms with parallel sensing capabilities.
One of the main challenges in developing CMOS electrochemical biosensors is the required post-CMOS processes. Aluminum used in CMOS processes is not suitable for biosensing, and polarizable metals such as Pt and Au are not compatible with CMOS technology. Additive and subtractive procedures have been performed to create biocompatible electrodes suitable for electrochemical applications (Huang et al., 2021). It should be mentioned that fabricating REs with CMOS technology is still challenging and external Ag/AgCl REs are commonly employed in CMOS electrochemical biosensors (Zhang et al., 2019; Chang and Lu, 2020; Saengdee et al., 2021; Doi et al., 2022; Lin C. H. et al., 2022).
The transducer electrode in an electrochemical biosensor translates the target electrochemical reaction into electrical signals, which can be in the form of current or voltage. Based on the output signal type, electrochemical sensors are often classified as amperometric/voltammetric, potentiometric, and impedimetric (Lin C.-Y. et al., 2022). There is also another major category based on field-effect transistors (FETs). In the following subsections, we will discuss surface-modified CMOS electrochemical biosensors based on these classifications.
2.1 Amperometric/voltammetric biosensing
Amperometry is a Faradaic electrochemical sensing method that requires redox molecules. In this method, the WE is set at a constant voltage with respect to the RE. This controlled potential initiates oxidation or reduction reactions, resulting in an electric current (Figure 4A) that is associated with the analyte’s concentration through the Cottrell equation (Equation 1). In Equation 1, i is the current (Ampere), n is the number of electrons, F is the Faraday constant, A is the electrode’s area (
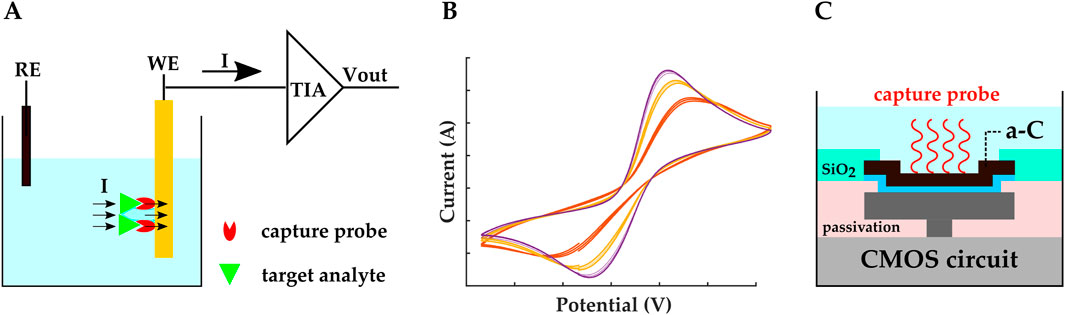
Figure 4. Schematic representation of (A) amperometric/voltammetric electrochemical biosensor. The TIA is a transimpedance amplifier. (B) An example of cyclic voltammetry I-V plot. (C) Amperometric CMOS biosensor developed by (Manickam et al., 2019a; b). Electrodes were realized by covering the exposed top metal layer with a-C. A 10 nm Ti layer was used as the adhesion layer. MB-labeled stem-loop DNA hairpin probes were covalently immobilized on the a-C electrodes.
In cyclic voltammetry (CV), the applied voltage between the WE and the RE is ramped, and plots of the resulting bidirectional current vs. the applied ramped voltage are retrieved, with oxidation and reduction peaks appearing in these I-V plots (Figure 4B). Different species lead to different peaks; hence, peaks can be studied to distinguish multiple analytes (Huang et al., 2021). Similar to amperometry, CV is a Faradaic sensing mechanism, and it requires redox molecules to be present in the solution.
Amperometry and CV are commonly used to study the exocytotic release of neurotransmitter biomolecules such as acetylcholine, dopamine (DA), glutamate, serotonin, etc. Conventional electrochemical sensors use carbon fiber microelectrodes for this application (Mosharov and Sulzer, 2005); however, carbon fiber microelectrodes allow measurements from one cell at a time, which makes the measuring process tedious and lengthy (Huang et al., 2021). In contrast, CMOS technology provides a platform for integrating thousands of MEAs on a single chip, enabling parallel sensing. For example, a three-electrode fast-scan CV (FSCV) CMOS-graphene dopamine sensor was introduced in (Nasri et al., 2017), in which carbon microfiber wires were replaced with ultrathin planar multilayer graphene sheets. To make the graphene electrodes, an epitaxial graphene film was transferred onto the CMOS chip, and after multiple lithographic and metal deposition steps, graphene microelectrodes were realized and connected to the readout circuit. In vitro measurements for a 2 μM DA release were performed to validate the sensor’s performance and sensitivities of 8–14 nA/μM were achieved for 200–400 V/s ramp rates.
Monitoring DNA hybridization is another biosensing application of amperometry and CV. For example (Manickam et al., 2019a; b), developed a CMOS three-electrode voltammetric electrochemical biosensor for molecular diagnostics (MDx) applications. This biochip included an array of
2.2 Potentiometric biosensing
In the potentiometric electrochemical sensing approach, the WE potential is measured against the RE when no significant current flows between the two electrodes (Figure 5A). The measured potential at the WE relates to the analyte concentration via the Nernst equation (Equation 2). In Equation 2,
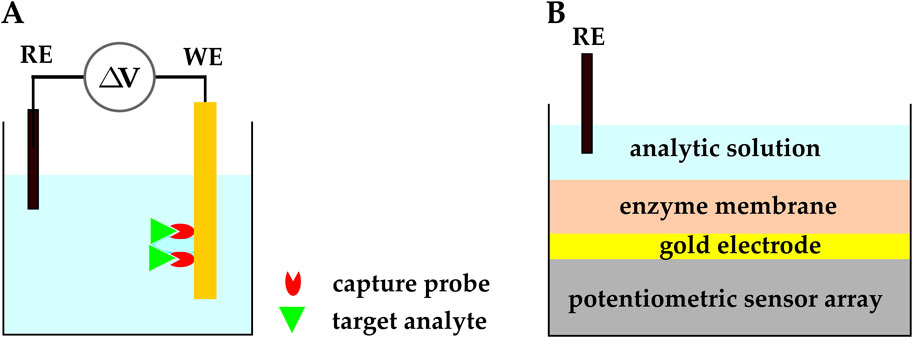
Figure 5. (A) Schematic representation of a potentiometric electrochemical biosensor.
Potentiometric electrochemical sensors may have two types of sensing electrodes: ion-selective electrodes (ISEs) and field-effect transistors (FETs). ISEs are the typical choice of electrode for potentiometric sensors, and they have been used since the 1990s. More recently, FETs have been employed as the sensing element in potentiometric sensors because of their ion sensitivity and potential for miniaturization. While CMOS FET-based biosensors have contributed a lot to the field (Ding and Qin, 2020; Walker et al., 2021; Wu et al., 2023), they are beyond the scope of this section; hence, we discuss FET-based biosensors in more detail in Section 2.4, and we focus on ISE-based biosensors in this section.
As mentioned above, potentiometric electrochemical sensors typically use ISEs and ion-selective membranes as their sensing platform. ISE-based potentiometric sensors have traditionally been used as pH sensors; however, with advances in fabrication and functionalization methods, they have found applications in biosensing and POC medicine. Recently, CMOS-based ISEs have been functionalized with biosensing elements and used to detect biomolecules such as enzymes, nucleic acids, and proteins (Ding and Qin, 2020; Walker et al., 2021; Wu et al., 2023). For example, a CMOS potentiometric biosensor with functionalized ISEs was developed for the monitoring of extracellular Adenosine
ATP plays a major role in the central nervous system as it contributes to synaptic transmission regulation and is correlated with neurodegenerative diseases and psychiatric disorders. In the presence of ATP, hydrogen peroxide (
The sensor implemented in (Doi et al., 2022) was realized by depositing gold electrodes on a CMOS charge-transfer-based potentiometric sensor array, which included
2.3 Impedimetric biosensing
Electrochemical impedance spectroscopy (EIS) is another widespread biosensing method in which the electrode-electrolyte surface is perturbed by an AC voltage or current with small enough amplitudes that the system stays in the linear region. By varying the stimulation signal frequency and studying the resultant current or voltage, the impedance of the electrochemical system can be retrieved as a function of frequency (Figure 6A) (Lei et al., 2016; Li et al., 2017; Dorledo de Faria et al., 2019; Forouhi and Ghafar-Zadeh, 2020; Lin C. H. et al., 2022). Nyquist plots are used to interpret the complex impedance of the electrochemical cell by representing the negative of the imaginary vs real parts of the impedance (Figure 6B).
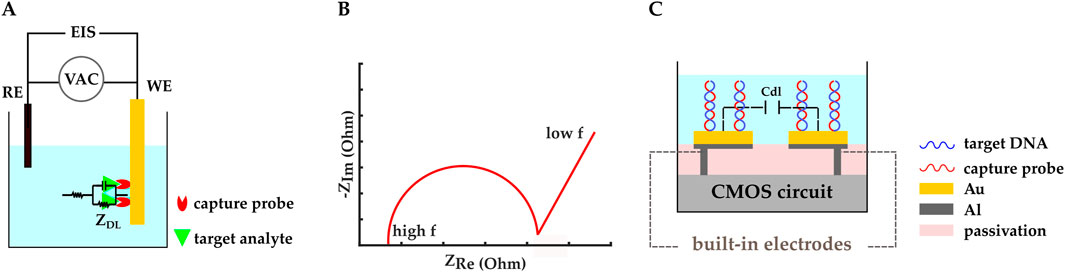
Figure 6. (A) Schematic representation of an impedimetric electrochemical biosensor. ZDL and EIS are double-layer impedance and electrochemical impedance spectroscopy, respectively. (B) An example Nyquist plot. (C) A CMOS capacitive biosensor designed by (Kuo et al., 2020). Electrodes were initially made with the top metal layer and they were covered by Au in post-processing. Thiolated microRNA–195 probes were immobilized on the gold IDE. Cdl represents the double layer capacitance.
EIS can be performed in Faradaic and non-Faradaic modes. Faradaic EIS relies on redox probes to study charge transfer resistance upon biological interactions, whereas non-Faradaic EIS does not need additional redox probes, which makes it better suited for POC applications (Lei et al., 2016; Li et al., 2017; Dorledo de Faria et al., 2019; Forouhi and Ghafar-Zadeh, 2020; Lin C. H. et al., 2022). For example, capacitive sensing is a non-Faradaic EIS method that is widely used in POC biosensors.
Capacitive sensing is commonly employed in affinity-based biosensing, where the BRE (e.g., DNA, aptamer, antibody, etc.) is immobilized on the surface of the capacitor electrodes, so the target analyte (e.g., DNA, protein, antigen, etc.) binds to the BRE. The binding event modulates the electrical double layer (EDL) and the double-layer capacitance (
A CMOS capacitive biosensor with interdigitated electrodes (IDEs) was designed, fabricated, and tested by (Kuo et al., 2020) to detect the breast cancer biomarker, microRNA–195. Gold IDEs were fabricated on the CMOS readout chip in the form of two separate combs, and complementary thiolated DNA probes were immobilized on the gold IDEs (Figure 6C). It was observed that the shift in the output frequency and the IDEs’ capacitance value increased as the microRNA-195 concentration was increased. The reported LOD, sensitivity, and dynamic range of the optimal CMOS IDE design were 0.617 fM, 645 Hz/fM, and 10 fM – 10 pM, respectively.
Another example is in (Lin C. H. et al., 2022), where a capacitive CMOS biosensor was developed to detect the Hepatitis B virus (HBV). The IDEs of the biosensor were formed by the top aluminum layer buried in the inter-metal dielectric layer and under the passivation layer. To increase the sensitivity, the passivation and inter-metal dielectric layers were thinned in a dry etching post-CMOS process. The chip’s surface was then functionalized with HBV probe DNA via an improved silanization method where 3–aminopropyldimethylethoxysilane (APDMES) was used instead of APTES. Different from APTES, each APDMES molecule reacts with one silanol group alone and prevents self-crosslinking and aggregation across the monolayer. They successfully demonstrated detection of HBV DNA hybridization with 10.8 fF/log [DNA] sensitivity in the 1–100 fM range.
In another study, Tabrizi et al. proposed a CMOS capacitive biosensor for DNA nano-mass measurement (Tabrizi et al., 2021). In this biosensor, DNA storage wells were realized by selectively etching the oxide and passivation layers to reach the top metal aluminum layer. The Al layer turned into
2.4 Field-effect transistor-based biosensing
Most bio-recognition events involve an electrostatic charge transfer, and subsequently, electric potential variations across the electrode-electrolyte system. Because FETs are sensitive to their gate potential voltage, they stand out for electrochemical biosensing applications. Moreover, FETs can be fabricated with CMOS technology (Figure 7A), offering mass production capability and low-cost manufacturing.
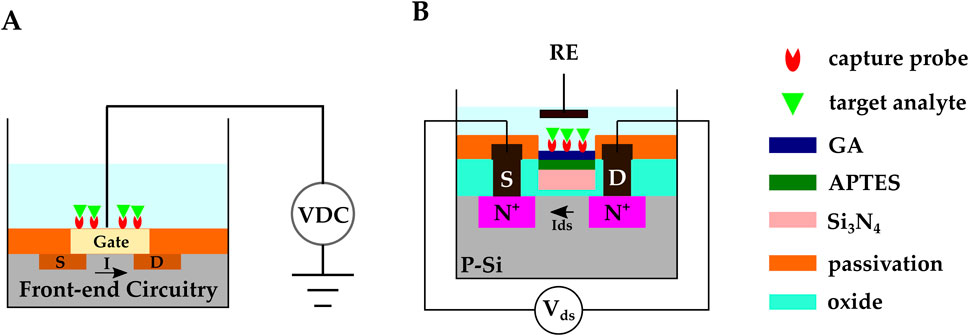
Figure 7. (A) Schematic representation of FET-based CMOS electrochemical biosensor (B) A CMOS ISFET-based biosensor developed by (Saengdee et al., 2021). Anti-HSA was covalently bonded on the
Biomolecules are detected in FET-based biosensors through two main mechanisms: 1) The FET channel surface is functionalized with the capture probes. Target species bind to the channel surface and alter the channel’s conductivity through the charge transfer process. The presence of biomolecules can be detected by studying the FET’s source-drain current (
It should be noted that standard planar gate FETs are in fact not sensitive enough to detect chemically or biologically charged species, and advances in post-CMOS processes have made it possible to fabricate FETs with special structures to achieve the required ultra-sensitivity. For example, the CMOS process was followed by a MEMS process in (Ushijima et al., 2021). These types of post-CMOS fabrication processes have allowed ISFETs (Bergveld, 1970) and nanowire FETs. These are discussed further below in Section 2.4.1, Section 2.4.2.
2.4.1 Biosensing with ion-sensitive field-effect transistors
ISFET-based electrochemical sensors are one of the common types of potentiometric biosensors. The conventional FET with a MOSFET includes a gate, a source, a drain, and a body, and the metal gate is mainly employed as the WE and BRE immobilization platform; whereas in ISFETs, an oxide functions as the sensing membrane (e.g.,
Lee et al. proposed a novel ISFET-based biosensor for DNA hybridization detection. They etched the passivation layer so that only a thin oxide layer remained covering the gate (Lee et al., 2021). The thin oxide on top of the ISFET’s gate electrode was the sensing platform, and DNA capture probes were immobilized on it via the APTES-GA method (Lin C. H. et al., 2022; Zhou et al., 2021; Huang et al., 2013b). The CMOS ISFET biosensor demonstrated HBV DNA detection in the range of 1 pM–10 nM and showed 53 mV/pH sensitivity under 100 MHz frequency.
Another ISFET-based biosensor for DNA hybridization detection was proposed by Chang et al. (Chang and Lu, 2020). The ISFET had a planar extended IDE and an external Ag/AgCl RE was used. The APTES-GA technique was employed to immobilize 5′– aminomodified HBV probe DNAs directly on the inter-metal
In another work, Saengdee et al. developed an ISFET-based biosensor for urinary albumin determination, leveraging the
Zhang et al. achieved 58 mV/pH sensitivity with a novel three-dimensional-extended-metal-gate ISFET (3D-EMG-ISFET) (Zhang et al., 2019). The ISFET had a vertically extended gate fabricated by stacked metal layers and vias. The passivation layer (
2.4.1.1 Biosensing with extended-gate ISFETs
When the analyte solution is in direct contact with the gate dielectric, any chemical reactions that occur in the fluid may affect the gate’s electrical properties and consequently modulate the ISFET’s
In a recent work, Kuo et al., (2021) successfully detected uric acid (UA) with 0.082 mg/dL LOD and 12.69 mV/(mg/dL) sensitivity with an extended-gate ISFET. The extended gate consisted of a
Sheibani et al. used a novel EG-FET to develop a wearable cortisol hormone detector from human sweat (Sheibani et al., 2021). In this work, the extended gate was an aptamer-functionalized single-layer graphene on a platinum substrate. The aptamer was linked to graphene by the 1–pyrenebutyric acid N–hydroxysuccinimide ester (PBSE) linker molecule. The extended gate and an Ag/AgCl reference electrode were exposed to the analyte solution, while the MOSFET transducer was isolated from the liquid under test (Figure 8A).
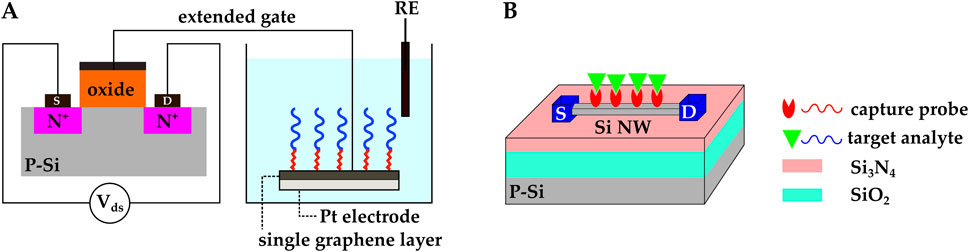
Figure 8. (A) EG-FET biosensor introduced by (Sheibani et al., 2021). The extended gate was a single-layer graphene on a platinum electrode. and it was connected to the MOSFET’s metal gate with metal vias. The extended gate and the Ag/AgCl reference electrode were exposed to the analyte solution, while the MOSFET transducer was isolated from the liquid under testing. Cortisol aptamers were immobilized on the graphene. (B) Schematic representation of a Si NW FET. The Si NW replaces the gate and the doped channel in regular MOSFETs, and the NW surface functions as the sensing membrane. Molecular probes are commonly immobilized on the NW’s surface.
2.4.2 Biosensing with Si nanowire field-effect transistors
The introduction of 1D Si nanowires (NWs) goes back to the 1990s when single-crystal Si NWs were synthesized through vapor-liquid-solid (VLS) and laser ablation methods (Lieber, 1998). A CMOS-compatible top-down method for the fabrication of Si NW was introduced later in the early 2000s (Elibol et al., 2003) and made it possible to develop CMOS Si NWFETs. Due to NW’s large surface area, NWFETs can potentially offer higher sensitivities compared to regular FETs. NWs replace the gate and the doped channel in regular MOSFETs, and the NW’s surface functions as the sensing membrane. The surface of NWs is commonly modified by covalent immobilization of molecular probes (Figure 8B). Upon absorption of the target biomolecules, the charge distribution around the NW changes. The NW’s conductivity gets modulated, and the analyte’s presence is confirmed by this modulation (Syu et al., 2018; Ambhorkar et al., 2018; Mirsian et al., 2019; Ivanov et al., 2021).
One of the major challenges in developing NWFET biosensors is to selectively functionalize the NW’s surface and to avoid functionalizing the
Yong et al. developed a polycrystalline Si NWFET (pSiNWFET) biosensor for HBV detection (Yong et al., 2021) (Figure 8B). The HBV surface antigen (HBsAg) and the HBV X protein (HBX) were successfully detected in the 5.61 fM — 0.56 pM and 3.92 fM – 0.39 pM ranges, respectively. The pSiNWFET structure consisted of a silicon substrate as the back-gate electrode, a stacked silicon oxide/silicon nitride layer as the gate dielectric, two highly doped Si areas to create source and drain, and two polysilicon NWs to form the conducting channels. HBV antibody (HBsAb) and anti-HBX were immobilized on the pSiNWFETs surface with APTES and GA. The pSiNWFETs
Krivitsky et al. designed a Si NWFET device for real-time monitoring of cancer cell’s metabolic activity and evaluating chemotherapeutic treatments (Krivitsky et al., 2019). The NWFETs surface was made sensitive to
In a recent work, a Si NW Schottky-junction FET (SiNW SJ-FETs) was introduced to detect DA in femtomolar concentrations. In a novel fabrication method, Si NWs were synthesized separately through a VLS process and later integrated into the CMOS chip by dielectrophoresis (DEP). Metal-
3 CMOS optical biosensing
In many biomedical and environmental applications, the presence of a specific molecule or nucleic acid sequence is detected through optical sensing. In optical biosensing, the illuminated element moves to an excited state and, upon relaxation, the excited electrons radiate their additional energy in the form of light. An image sensor captures the emitted light and translates it into an electrical signal, and information about the target can be retrieved from this electrical signal. Compared to charge-coupled devices, CMOS image sensors offer lower power consumption, monolithic integration of the pixel array and the auxiliary electronics, and higher temporal resolution. CMOS optical biosensors employ CMOS photodetectors as the light-to-electric signal converter, allowing the integration of the transducer and the front-end circuit on one chip to build compact and cost-effective devices (Forouhi and Ghafar-Zadeh, 2020; Sander et al., 2007). Moreover, they offer great advantages, such as enabling direct, real-time, and label-free detection of many biological and chemical substances while providing high specificity and sensitivity (Damborský et al., 2016; Lin C.-Y. et al., 2022).
In optical biosensing, labeled or label-free analytes bind to the immobilized probes upon biorecognition events and induce changes in the optical field around the sensor. Optical biosensors employ four different mechanisms for detecting target analyte-BRE binding events: direct (Figure 9A), sandwich (Figure 9B), competition (Figure 9C), and displacement (Strianese et al., 2012). In direct binding, two species are involved. They are the capture probes, immobilized on the CMOS chip surface, and the target molecules. This approach can be used if the target analyte is inherently fluorescent or is pre-labeled with a fluorescent group. The sandwich assay is used when targets have at least two binding sites to bind to the capture probe and the fluorescent tracer molecule at the same time. Capture probes are commonly immobilized on the sensor’s active region, and fluorescent tracer molecules are added to the analytic solution. After the binding event, free fluorescent tracer molecules are physically removed from the solution or optically excluded from the sensing region, so the remaining fluorescent signal is an indicator of the target biomolecule.
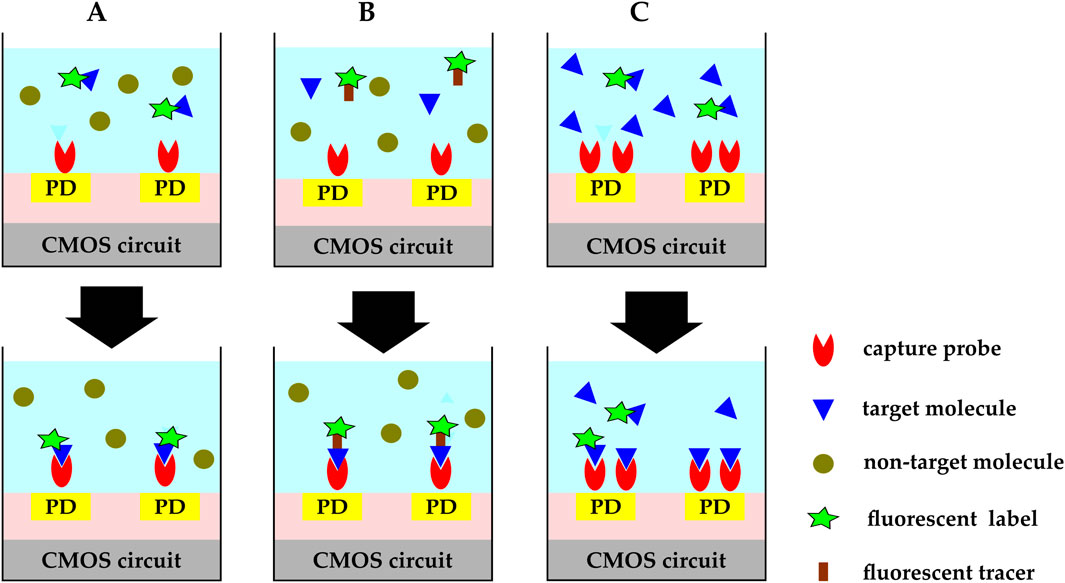
Figure 9. CMOS optical biosensing binding mechanisms:(A) direct. Capture probes are immobilized on the CMOS chip’s surface and target molecules are either inherently fluorescent or pre-labeled with a fluorescent group. (B) sandwich. Capture probes are immobilized on the sensor’s active region and fluorescent tracer molecules are added to the analytic solution. Target molecules bind to the capture probe and the fluorescent tracer molecule at the same time. Free fluorescent tracer molecules are physically removed from the solution or optically excluded from the sensing region. The remaining fluorescent signal is an indicator of the target biomolecule. (C) competitive. Capture probes are immobilized on the chip’s surface and pre-labeled target molecules are added to the sample solution while the target molecules in the analytic solution are not labeled. The added labeled molecules compete with the unlabeled molecules over the available binding sites. The fluorescent signal decreases as the concentration of the target molecules in the sample under test increases.
Direct and sandwich binding are the two most widely used binding mechanisms; however, competitive or displacement assays are more widespread for small molecules with limited binding sites. In the competitive assay technique (McCall et al., 2012), capture molecules are immobilized on the chip’s surface and labeled targets are added to the sample solution, while unlabeled target molecules already exist in the analytic solution. The added labeled molecules compete with the unlabeled molecules over the binding sites, and the fluorescent signal decreases as the concentration of the target molecules in the sample under test increases. A slightly different mechanism takes place in the displacement method explained in (Wicks and Hargrove, 2019). In this displacement technique, labeled capture molecules are immobilized on the active sites and the biosensor is exposed to pulses of samples containing the target molecules. The target molecules wash away the fluorescent labels from the upstream zone, increasing the signal level at the downstream.
Optical biosensing is commonly classified into different categories based on their source of excitation. Fluorescence (external source), chemiluminescence (chemical reaction), electrochemiluminescence (electric potential), bioluminescence (biological reaction), and thermoluminescence (heat) are some of the major categories (Anand et al., 2022). In Sections 3.1, 3.2, we focus on the two most widely used techniques, fluorescence and chemiluminescence, and we briefly touch on two other optical detection mechanisms in Section 3.3.
3.1 Fluorescence biosensing
In fluorescence biosensing the fluorescent molecules need an external excitation source; for example, a light-emitting diode (LED) or a laser. The excitation source’s radiation power is much higher than that of the emission signals, and developing optical filters that integrate with CMOS biosensors to efficiently reject excitation photons from the emission signal is a major challenge (Dandin et al., 2007; 2012). To overcome this challenge, Hong L. et al. (2015, 2017) designed a fully integrated optical system-on-chip by exploiting optical interaction with sub-wavelength metal nanostructures made of electrical interconnect layers. Instead of constructing bulky optical filters on the chip’s surface, they used the metal layers available in the CMOS technology to integrate nanoplasmonic waveguide-based filters which were capable of more than 50 dB of rejection ratio across a wide range of incident angles. The PD glass surface was functionalized with capture probes and the streptavidin-coated Qdot 800 solution was used as fluorescent tags. This surface modification allowed the detection of 48 zmol of quantum dots, reaching a fluorescence/excitation ratio of nearly −62 dB without any post-CMOS fabrication, external optical filters, lenses, or collimators.
Another key challenge in CMOS fluorescence biosensing is exposing the CMOS chips to wet processes such as DNA and protein arrays immobilization. Surface functionalization is crucial to enhance specific binding and prevent adsorption of nonspecific analytes. Stadler et al. introduced an innovative surface coating for CMOS microchips utilizing poly (ethylene glycol)methacrylate graft polymer films via silanization (Stadler et al., 2007). The polymer layer provided high loadings of functional groups for probe molecules (anti-HA and anti-FLAG) immobilization. It also completely blocked nonspecific adsorption of proteins on the chip surface.
To enable highly parallel detection, the authors of (Hassibi et al., 2017; Manickam et al., 2017) created a fully integrated CMOS biochip that used an inverse fluorophore assay–previously described in (Hassibi et al., 2009)–to perform parallel polymerase chain reaction (PCR). The biochip consisted of a
In a recent work (Zhu et al., 2022), created a fluorescent CMOS biosensor that can be ingested for monitoring the gastrointestinal (GI) tract. The biosensor was a single CMOS chip that integrated a 15-pixel fluorescence sensor array, a low-power wireless interface, and on-chip nanoplasmonic optical filters. All sensing and processing electronics were enclosed within a 1.2 cm
3.2 Chemiluminescence biosensing
Unlike fluorescence, chemiluminescence does not require an external excitation source, and the excitation energy is provided by the chemical reactions involved in the biological events. Not having a strong background illumination noise, chemiluminescence can have better sensitivity compared to fluorescence-based assays (Redy Keisar et al., 2024). It is also selective and enables multi-analyte parallel detection. Moreover, since light is produced inherently, no filter is needed (Selvam, 2017), which allows more compact, miniaturized, and low-cost LOC designs (Kricka, 1995). Chemiluminescence’s disadvantage compared to fluorescence measurement techniques is its higher susceptibility to changes under environmental conditions (Tokuda and Ohta, 2022).
Chemiluminescence detection methods are usually named after their luminescent-producing reagent. Luminol- and peroxyoxalate-based chemiluminescence are two of the most widely used chemiluminescence sensing techniques (Poupon-Fleuret et al., 1996; Sigvardson et al., 1984). Luminol exhibits chemiluminescence with a blue glow upon oxidation. Peroxyoxalates is an intermediate, and it transforms into 1,2–dioxetanedione. The 1,2–dioxetanedione compound decomposes into carbon dioxide (
To improve sensitivity (Sandeau et al., 2015), presented a large-area
Another study by (Joung et al., 2012) demonstrated the utility of chemiluminescence to detect human interleukin 5 (IL-5) of sub-
Another study (Hong D. et al., 2015) demonstrated further improvements in sensitivity to attomolar level detection through direct antibody immobilization on the surface of the CMOS image sensor, as well as using a simple data accumulation process with noise calibration. The chip surface was modified to introduce aldehyde groups using GA and the capture antibody was applied to the surface of the chip and incubated at
Al-Rawhani et al. developed a quadruple-mode CMOS biosensor for rapid biomarker detection. The sensor had colorimetric, chemiluminescent, surface plasmon resonance (SPR), and hydrogen ion measurement modes (Al-Rawhani et al., 2020). The CMOS chips included an array of
3.3 Beyond fluorescence and chemiluminescence biosensing
The absorption-based optical biosensor is a viable optical detection platform that relies on photon absorption instead of photon emission. Hofmann et al. examined a CMOS biochip photometer in which 100
Liu et al. developed an ingestible 6.5 mm
4 Beyond electrochemical and optical CMOS biosensing
Electrochemical and optical biosensors form the bulk of developed CMOS biosensors. However, compelling novel CMOS biosensing devices have been designed that employ other sensing mechanisms, namely, electrical, mechanical, and magnetic transduction. In this section, we review some of the recent works in CMOS biosensing in these areas.
4.1 Electrical biosensing
There is a very fine line between electrochemical and electrical transducing methods. Electrical biosensors are “devices based on measurements where no electrochemical processes take place, but the signal arises from the change of electrical properties caused by the interaction of the analytes (Hulanicki et al., 1991); ” while electrochemical biosensors transform the electrochemical effect of the analyte-electrode interaction into a measurable signal. This distinction underscores the nuanced differences between the two types of sensors in their signal generation mechanisms.
A CMOS molecular electronic chip was presented in (Hall et al., 2022) for single-molecule biosensing. Aviram et al. first used single molecules as circuit elements (Aviram and Ratner, 1974). Using this concept, Hall et al. developed a CMOS biosensor to detect DNAs, proteins, enzymes, and antibodies (Hall et al., 2022). The primary sensing element was a molecular peptide wire connecting two ruthenium (Ru) nanoelectrodes. The two nanoelectrodes had a gap of less than 30 nm and were fabricated on a 16k pixel CMOS current reader chip in post-CMOS fabrication steps. The peptide wires were loaded on each pixel and attached to the nanoelectrodes via dielectrophoresis (DEP). The Probe molecules were positioned in precisely specified sites on the wire (Figure 10A). The nominal resistance for a 25 nm long alpha-helical peptide molecular is
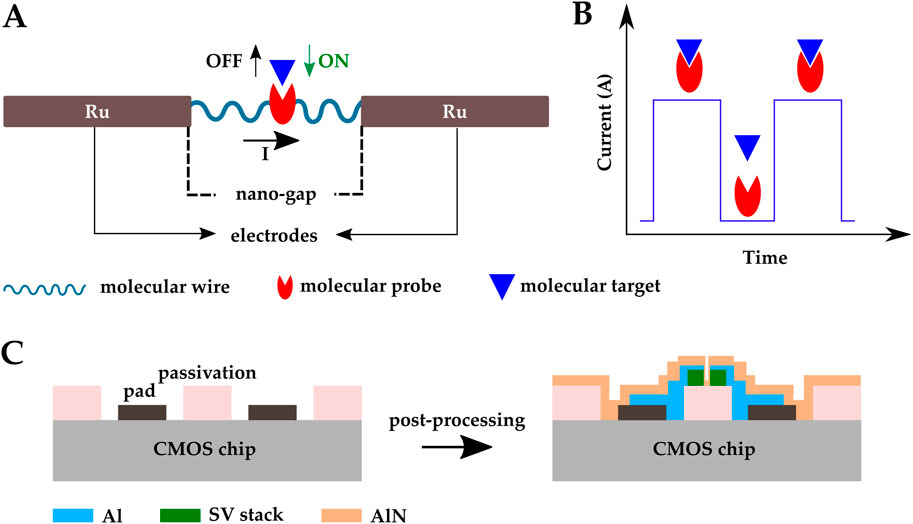
Figure 10. Schematic representation of the CMOS electrical biosensor developed by (Hall et al., 2022); (A) a peptide molecular wire connecting two Ru nanoelectrodes, (B) a series of current pulses indicating binding events happening. (C) CMOS magnetic biosensor developed by (Costa et al., 2017); SV sensors were post-fabricated on top of a CMOS IC front-end and the sensors were covered by an AlN passivation layer.
4.2 Magnetic biosensing
Biological sample metrics like blood, urine, etc are non-magnetic. Unlike electrochemical biosensors that are heavily affected by pH, ionic strength, and the temperature of their surrounding environment, magnetic sensing offers a matrix-insensitive biomarker detection platform. Immunity from matrix interference enhances the sensitivity and selectivity of the system (Zhou et al., 2021). Wang et al. and Sun et al. reported magnetic sensing with sub-ppm sensitivities (Wang et al., 2014; Sun et al., 2022). Magnetic biosensors can be based on a variation of transducers such as the Hall-effect sensor (Gambini et al., 2013; Lei et al., 2017), LC oscillator (Wang et al., 2014; Sun et al., 2022), or magneto resistive (MR) sensor (Han et al., 2006; Zhou et al., 2021; Hall et al., 2013; Costa et al., 2017).
Among magnetic sensors, giant magneto resistive spin valves (GMR SV) are CMOS compatible. These sensors are great candidates for LOC development and mass production (Adem et al., 2020). Costa et al. fabricated SV sensors on top of a CMOS IC front-end and covered the sensors with an AlN passivation layer (Figure 10C) (Costa et al., 2017). SV sensor’s fabrication included multiple steps of photo-lithography, electron beam deposition, sputtering, lift-off, and ion milling etching. The AlN passivation layer of the SV sensor array can be functionalized with biological probes for biomolecular recognition. The system achieved a magneto resistive ratio of 5.37% and successfully detected 250 nm magnetic nanoparticles.
4.3 Mechanical biosensing
Advances in microelectromechanical systems (MEMS) since the 1980s have facilitated the development of mechanical transducers. Microcantilever-based biosensors detect biological events by monitoring small displacements of the cantilever. The biosensor works by immobilizing a BRE on the cantilever surface, which captures target molecules and induces changes in the cantilever’s bent or resonance frequency. Microcantilever-based biosensors offer high sensitivity, fast response, and portability (Lavrik et al., 2004; Huang et al., 2013a; Basu et al., 2020; Li et al., 2008).
Zhao et al. demonstrated a CMOS microcantilever biosensor to detect alpha fetoprotein (AFP), a protein produced by the liver and in the yolk sac of a fetus (Zhao et al., 2021). The Si cantilever’s surface was oxidized and the AFP capture antibodies were covalently immobilized on it via APTES-GA crosslinking, while gold nanoparticles (AuNP) were functionalized with detection antibodies (Figure 11A). Detection antibodies bound to the antigen, and the antigen bound to the capture antibody immobilized on the sensor surface. This brought AuNPs in closer proximity to the sensor and induced detectable shifts in the cantilever resonance frequency. The sensor was able to detect AFP with a limit of detection (LOD) of 21 pg/mL and a linear range of 0–70 ng/mL.
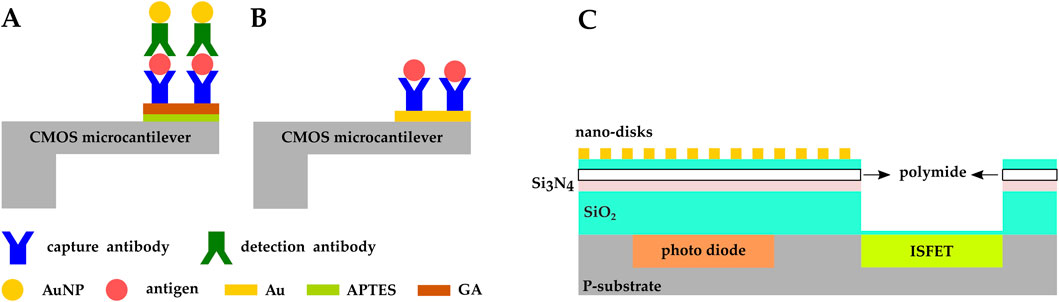
Figure 11. CMOS microcantilever-based biosensors: (A) the cantilever’s surface was oxidized and AFP capture antibodies were covalently immobilized on it via APTES-GA crosslinking. AuNPs were functionalized with detection antibodies (Zhao et al., 2021); (B) Vp capture antibodies were immobilized on the Au electrodes fabricated on the CMOS microcantilever (Wang et al., 2021). (C) CMOS multimodal biosensor developed by (Al-Rawhani et al., 2020).
In another study by the same group, a CMOS microcantilever biosensor was developed based on an alternating current electrothermal technology (ACET) configured to detect bacteria. Gold electrodes were fabricated on the CMOS microcantilever and functionalized with mercaptan self-assembled monolayer (SAM) layer, and Vibrio parahaemolyticus (Vp) capture antibodies were immobilized on the functionalized Au-patterned Si cantilever (Figure 11B). The ACE effect improved molecular aggregation around the microcantilever which led to an increase in binding probability, resulting in an LOD of
4.4 Multimodal biosensing
Monitoring cellular processes is essential in diagnostic and therapeutic research and applications. Although many cell-based biosensors (CBBs) have been developed to study cellular activities, for example, cell proliferation (Weeks et al., 2022), apoptotic cell death (Akçapınar et al., 2021; Garcia-Hernando et al., 2020), and cytotoxicity (Garcia-Hernando et al., 2020), most of the CBBs developed use only one transduction mechanism and are capable of monitoring one kind of biophysiological response (Wang et al., 2022a), while cellular activities are inherently diverse in type and scale (Hu et al., 2021). As CMOS technology allows integration of multiple sensors on one chip to monitor different types of signals (i.e., electrochemical, optical, etc.), multimodal CMOS CBBs have been introduced to capture multi-physiological cellular responses (Tokuda et al., 2006; Hu et al., 2021; Jung et al., 2021; Kumashi et al., 2021; Wang et al., 2022a).
Hu et al. introduced a multimodal CMOS biosensor array consisting of 131,072 pixels with three
Wang’s group recently reported multiple multimodal CMOS biosensors to study cellular processes. For example (Kumashi et al., 2021), performed both amperometric and impedimetric electrochemical biosensing on a 256-pixel CMOS sensor array to characterize exoelectrogens. The chip had
The same group developed another multimodal CMOS CBB (Jung et al., 2021). In this biosensor, PDs and Au electrodes were integrated in a 21,952-pixel CMOS chip for potential, optical, and four-point impedance sensing. Au electrodes were fabricated using the same method described in (Kumashi et al., 2021). Holistic on-chip cell characterization was performed on rat cardiomyocytes and single-cell resolution was achieved.
Wang et al. employed the same method as (Kumashi et al., 2021; Jung et al., 2021) to fabricate biocompatible on-chip electrodes, except they used platinum instead of gold (Wang et al., 2022b; a). Pt has lower electrode-electrolyte impedance and is a better option for impedance sensing applications. Human stem cell-derived neurogenin 2–accelerated progenitor cells (SNaPs) and mouse skeletal myoblast differentiated into muscle cells (C2C12) were encapsulated in on-chip cultured brain modeling hydrogels. Electrical tests and biological measurements successfully demonstrated multimodal CBB (Wang et al., 2022b).
(Al-Rawhani et al., 2020) developed a CMOS biosensor for rapid biomarker detection. The sensor had four modes of detection: colorimetric, chemiluminescent, surface plasmon resonance (SPR), and hydrogen ion measurements. The CMOS chip had an array of
5 Surface modification and functionalization methods
Although CMOS technology offers many advantages for the development of biosensors, the interconnect metal available in CMOS technology, aluminum, is not suitable for biosensing applications because it is prone to corrosion and is not polarizable. Inert polarizable metals, such as gold and platinum, are not available in the standard CMOS processes. Additionally, sensor surface functionalization and probe immobilization, which play crucial roles in affinity-based detection applications, cannot be performed through CMOS technology. To address these issues, post-CMOS processes are used to fabricate biocompatible functionalized electrodes (Kuo et al., 2020). In this section, we review post-CMOS processes developed for fabricating functionalized electrodes on the CMOS chips. These processes have two main steps; the first step is fabricating the electrodes and the second step is to immobilize the capture probes on the electrodes.
5.1 Electrode fabrication
The common method to fabricate on-chip electrodes is to initially design the electrodes integrated in the CMOS chip using the Al top metal layer and to later modify the electrodes in the post-CMOS fabrication steps. This can be as simple as a one-step etching process. Zhang et al. and Tabrizi et al. removed the passivation layer on top of the Al electrodes via reactive ion etching (RIE) and chemical etching, respectively, to expose the Al electrodes (Zhang et al., 2019; Tabrizi et al., 2021). Once exposed to air, Al turns into
Polarizable metals, i.e., noble metals such as gold and platinum, are preferred for electrochemical sensing applications. In these cases, the post-CMOS process includes at least two steps; removing both the Al electrodes and the passivation layer on top of them and depositing noble metal electrodes (Au and Pt) instead. For example, Wang et al. chemically etched the Al electrodes and fabricated the Pt electrodes by sputtering (Wang et al., 2022a; b). Kumashi et al. and Jung et al. removed the Al electrodes by chemical etching and fabricated the Au electrodes using E-beam evaporation (Kumashi et al., 2021; Jung et al., 2021). Additionally, more complex processes have been developed, including multiple steps of photolithography, electron beam deposition, sputtering, lift-off, etching, DEP, PECVD, etc. Examples can be found in (Al-Rawhani et al., 2020; Hall et al., 2022; Sessi et al., 2022; Costa et al., 2017; Wang et al., 2021).
5.2 Probe immobilization
Various techniques have been studied and employed for probe immobilization, including physical adsorption, streptavidin-biotin interaction, and covalent immobilization. Physical adsorption relies on the ionic interactions between biomolecules and the transducer surface, and its main advantage is its simplicity. This method does not require linker molecules or target analyte modifications. The streptavidin-biotin complex exhibits one of the strongest noncovalent bonds. Immobilizing either streptavidin or biotin on the sensor surface and subsequently biotinylating biomolecules is a well-established technique for detecting proteins and DNA. Biotin, as a label, is known to be highly stable and has minimal impact on the function of the labeled molecule (Nimse et al., 2014; Mirsian et al., 2019; Stern et al., 2007; Hong L. et al., 2015, 2017).
Among the immobilization methods mentioned above, covalent immobilization has the highest binding strength and stability. A popular functionalization technique is to leverage the amine group (–
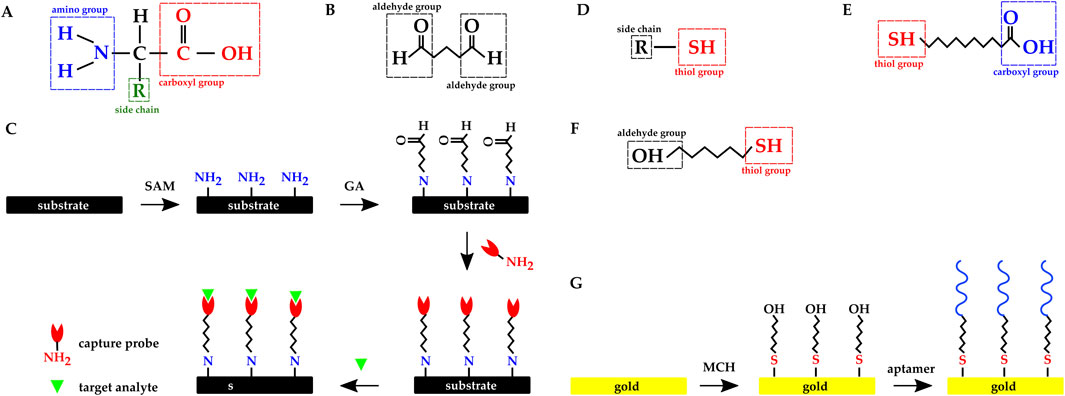
Figure 12. Chemical structure of (A) Amino acid. (B) Glutaraldehyde molecule. (C) Overview of the dielectric substrate preparation with SAM and GA molecules: amine functionalization, activation with glutaraldehyde, probe immobilization, and immunoassay. Chemical structure of (D) a molecule with thiol group. (E) 11-mercaptoundecanoic acid. (F) 6 mercapto 1 hexanol. (G) Overview of the gold electrode’s functionalization with MCH SAM and thiolated aptamer.
Glutaraldehyde (GA) is a crosslinker molecule possessing two aldehyde (–COH) functional groups (Figure 12B). To leverage the crosslinking method, an SAM with amine functional group is introduced to the sensor’s (electrodes’) surface, and GA binds to the SAM with one of GA’s binding sites. The other binding site will be available to the capture probe to bind to. Finally, the target protein will covalently bind to the capture probe (Figure 12C). SAM is chosen based on the electrode’s surface material; for example, APTES has been frequently used to functionalize dielectric surfaces (Zhao et al., 2021; Lin C. H. et al., 2022; Lee et al., 2021; Chang and Lu, 2020; Saengdee et al., 2021; Yong et al., 2021).
Gold has a strong affinity for thiol (–SH) (Figure 12D) and the SAMs used in functionalizing gold electrodes commonly have this group. Zhao et al. functionalized their gold electrodes with antibodies using 11-mercaptoundecanoic acid (11-MUA) SAM (Figure 12E) and EDC crosslinker (Zhao et al., 2021). Singh et al. used six mercapto one hexanol (MCH) (Figure 12F) SAM to functionalize their Au IDEs with thiolated aptamers (Figure 12G) (Singh et al., 2019). Kuo et al. and Lee et al. directly immobilized thiol-modified DNAs and apatamers on gold electrodes without any SAM (Kuo et al., 2020; Lee et al., 2012).
6 Discussion and future trends
CMOS technology enables the integration of transducers and readout circuits in a single chip to build biosensors; moreover, the advances in the post-processing steps that are compatible with CMOS chips have made CMOS biosensors more versatile. In this work, we reviewed recently developed CMOS biosensors with surface modifications. These biosensors use different sensing modalities for a wide variety of applications, from detecting DNA (Manickam et al., 2019a), proteins (Mirsian et al., 2019), and enzymes (Andrianova et al., 2018) to monitoring cell viability (Senevirathna et al., 2018). Here, we highlight the significance of the reviewed devices, highlighting their advantages as well as their potential limitations.
Nasri et al. (2017) and Manickam et al. (2019a,b) developed amperometric biosensors for DA and DNA detection respectively. Each of these biosensors has its own advantages and limitations. For example, through multiple steps of post-processing, Nasri et al. integrated the WE, CE, and RE in the CMOS chip, while Manickam et al. integrated the WE and CE, and used an external RE. On the other hand, Manickam et al. functionalized the biosensor’s electrodes with complementary target DNA and examined the sensors specificity, whereas Nasri et al. did not functionalize their biosensor and there is no data available on measurements in the presence of other neurotransmitters, which leaves the sensor’s specificity untested. Doi et al. (2022) used a potentiometric approach to capture spatiotemporal dynamics of ATP. They fabricated gold electrodes on their CMOS potentiometric sensor and similar to Nasri et al. (2017), this biosensor needs and external RE. Having gold electrodes in addition to the need for an external RE, increases the biosensor’s cost; however, Doi et al. functionalized their biosensor using a mixed-layered technique based on physical adsorption which could make the biosensor potentially reusable; However, its re-usability was not explored.
Kuo et al. (2020) and Lin C. H. et al. (2022) developed capacitance biosensors for DNA detection and covalently immobilized complementary probe DNAs on the electrodes. Kuo et al. fabricated gold electrodes on their biosensor while Lin et al. simply thinned the passivation layer on top of the Al electrodes. Using gold electrodes enables improved LOD and sensitivity, but it complicates the post-CMOS processing and increases the manufacturing cost. Depending on whether cost or sensitivity is prioritized, these two approaches are interchangeable.
Another category of CMOS biosensors discussed is ISFET-based biosensors. Lee et al. (2021) and Chang and Lu (2020) thinned the passivation layer on top of the ISFET gate and covalently immobilized DNA probes on the remaining thinned passivation layer. Lee et al. studied the optimized operating frequency while Chang et al. explored different electrode sizes and shapes. The latter work reported better pH sensitivity and significantly lower LOD. A more in depth look into these two biosensor’s designs would give helpful information about the electrochemistry of the biosensor and the transduction mechanism. Saengdee et al. (2020) on the other hand did not perform any post-CMOS processing and they covalently immobilized the capture antibodies on the passivation layer. As discussed above, this approach simplifies the biosensor development process but it has a negative impact on the sensitivity. Kuo et al. (2020) and Sheibani et al. (2021) fabricated gates separately, functionalized them, and wire-connected them to the CMOS FETs for UA and cortisol detection, respectively. Having an extended gate improves the ISFET’s sensitivity as the FET stays immune from the changes in the analytic environment; moreover, it makes the functionalization process easier.
We discussed NWFET-based CMOS biosensors as well (Yong et al., 2021; Krivitsky et al., 2019; Sessi et al., 2022). Because of their larger surface area, NWFETs have the potential to achieve higher sensitivities compared to ISFETs; however, NWFETs have more complex fabrication processes. Consequently, they have not been explored as much as other CMOS electrochemical biosensors.
Hassibi et al. (2017) and Manickam et al. (2017) integrated optical filters and resistive heaters with CMOS photosensors to build a portable PCR machine with continuous fluorescence detection. Zhu et al. (2022) also used a CMOS photosensor with integrated filters to built an ingestive fluorescence sensor for GI monitoring. On the other hand, Sandeau et al. (2015) and Joung et al. (2012) built CMOS chemiluminescence biosensors without having to fabricate integrated filters. They functionalized their sensors’ surfaces with appropriate probes to detect cytokines like TNF-
We also discussed CMOS biosensors that use electrical, magnetic, and mechanical transducers. The electrical CMOS biosensor developed in (Hall et al., 2022) provides real-time single molecule detection; however, the sensor development process is very complicated. Costa et al. (2017) developed a magnetic biosensor by fabricating a spin-valve on a CMOS IC. Compared to the other types of biosensors discussed, the magnetic biosensor offers better matrix-insensitivity which enhances the biosensor’s sensitivity and specificity; nevertheless, the post-processing includes multiple steps of photo-lithography, electron beam deposition, sputtering, lift-off, and ion milling which makes the sensor development process costly. Zhao et al. (2021) and Wang et al. (2021) developed cantilever-based mechanical CMOS biosensors for bacteria detection by immobilizing antibody on the oxide and gold electrodes fabricated through post-processing steps. The cantilever-based biosensors offer faster response, yet the sensitivities reported in works reviewed did not exceed that of other biosensors with different sensing modalities.
CMOS technology offers integrating multiple transducers with different sensing modalities in one chip. Having multiple transducing mechanisms simultaneously enables multi-dimensional sensing; however it should be noted that it increases the CMOS sensor design and fabrication cost, as it requires more complicated circuitry as well as larger die area. Hu et al. (2021) and Kumashi et al. (2021) used this advantage to build CMOS biosensors for monitoring cultured bacteria activity. Hu et al. integrated three PDs and two ISFETs in each pixel and used an external RE to enable imaging, pH sensing and EIS. To improve sensitivity, they fully etched away the Al ISFET gate and used the TiN layer as ISFET sensing membrane. Kumashi et al. on the other hand, fabricated gold electrodes to integrate WE, CE, and RE on the CMOS chip and enable fully-integrated amperometric and impedimetric biosensing. Wang et al. (2022a) replaced gold electrodes with Pt since it has lower electrode-electrolyte impedance. Similar to the biosensors discussed earlier, these works have different approaches to electrode design, balancing sensitivity and cost.
The works we discussed above have made major contributions to the filed. However, there are still many challenges to be addressed in order to achieve the mass production of the functionalized CMOS biosensors. For example, although post-CMOS processes such as oxide etching and metal deposition contribute to the CMOS biosensor performance improvement, they require access to micro- and nano-fabrication facilities equipped with photo lithography, etching, and metal deposition tools. In addition, process engineers must optimize the fabrication steps for each device. This can be a costly and time-consuming process as they should consider factors such as the chips’ dimensions, their minimum feature size, the thickness and material of the passivation layer to be etched, and the metal to be deposited. Some CMOS foundries offer special post-CMOS processes compatible with noble metals, by exploiting which, in-house fabrication steps can be skipped to some extent; however, it should be noted that these processes are mostly in the experimental phase and hence costly.
The other step of modifying CMOS biosensors is functionalizing the chip’s surface with the bio-recognition elements. Covalent immobilization is a widely used method for functionalizing biosensors, because it is the most stable compared to other approaches such as physical adsorption; however, the disadvantage of covalent bonding is that these bonds need to be broken for purposes such as cleaning the chips and reusing them for other applications. In these cases, the chip has to be exposed to harsh environmental conditions such as high acidity or high temperature. This can decrease the sensor’s life-time significantly.
Another disadvantage of covalent bonding is that it depends on the chemical reactions occurring between the substrate material and the SAM molecules. So, to achieve spatially selective functionalization, the sensing surfaces need to be patterned with different materials, which makes post-CMOS processing even more complicated. This issue can be addressed by using polymer-based electroactive hydrogels such as chitosan instead of SAM to enable localized functionalization. Moreover, compared to covalently bound SAMs, hydrogels are easier to remove (Buckhout-White and Rubloff, 2009; Wu et al., 2003; Tseng et al., 2014; Susanto et al., 2013; Kim et al., 2015).
It is important to note that the mass production of CMOS biosensors that can compete with commercialized POC technologies, such as test strips, in cost, demands having a deep understanding of microelectronics and biochemistry. Avoiding post-CMOS fabrication and sample labeling, as well as building reusable biosensors can decrease the cost of the biosensors. Designing integrated sensors and sensing circuits with enhanced performance, taking advantage of sensing modalities that do not require sample labeling, and developing surface functionalization methods exploiting reversible chemical reactions are some strategies to achieve these goals.
7 Conclusion
We discussed surface-modified CMOS biosensors with different transduction mechanisms, such as electrochemical, optical, electrical, magnetic, mechanical, etc. CMOS electrochemical biosensing enables label-free detection, which reduces sample preparation steps, and allows for better integration of the sensor and sensing circuits. However, since the metal used in CMOS processes, aluminum, is not suitable for electrochemical sensing, post-CMOS processing steps are needed to fabricate working electrodes made of electrochemical-compatible material. Moreover, the development of CMOS electrochemical biosensors is facing other challenges such as Short Debye length, fabricating on-chip REs, poor specificity, and strong environmental influences that are yet to be addressed.
In CMOS optical biosensors, capture probes can be directly immobilized on the CMOS detector surface without the need for post-CMOS fabrication steps. Another advantage of optical biosensing is that it allows for both labeled and label-free detection. Between the two main optical detection methods, fluorescence-based and chemiluminescence-based, the former needs external light sources and objectives, while the latter does not (Hong L. et al., 2015; 2017; Kricka, 1995). Although developing on-chip filters is a challenging and costly task, fluorescence-based biosensors receive more attention, as fluorescence detection is the gold standard method for DNA detection. For more information on developing on-chip optical filters, the reader is encouraged to refer to (Dandin et al., 2007).
We also covered other sensing modalities for developing biosensors; i.e., electrical, magnetic, and mechanical. Magnetic biosensors’ advantage is their insensitivity to the matrix, which provides high sensitivity and specificity toward the target biomarker (Zhou et al., 2021); However, unlike electrochemical and optical biosensors, magnetic biosensors do not allow label-free detection, and it is more challenging to integrate them into a CMOS chip since they would need external permanent magnets or coils. To address the latter issue, efforts have been made to fabricate on-chip coils and LC oscillators. Furthermore, GMR SVs are compatible with CMOS technology and are promising candidates for developing fully integrated CMOS magnetic biosensors (Zhou et al., 2021). Mechanical biosensors did not emerge until the 1980s. Recently, CMOS microcantilever-based biosensors have been developed for molecular sensing applications. These biosensors offer high sensitivity, low limit of detection, fast-response, and portability (Zhao et al., 2021). Multimodal CMOS biosensors have multiple sensors with different transducing mechanisms on a single chip, making them suitable for monitoring cell activities (Tokuda et al., 2006; Al-Rawhani et al., 2020; Jung et al., 2021; Kumashi et al., 2021; Wang et al., 2022b).
Furthermore, we discussed materials and methods for modifying the surface of CMOS chips. These modifications are divided into two main categories; post-CMOS additive or subtractive fabrication processes, i.e., oxide etching and metal deposition, and biochemical processes for immobilizing the BRE, i.e., antibodies, on the chip’s surface. Finally, we discussed the future of the industry and the challenges facing the mass production of CMOS biosensors.
Author contributions
FD: Conceptualization, Investigation, Visualization, Writing–original draft, Writing–review and editing. MS: Writing–original draft, Writing–review and editing. MD: Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Carnegie Mellon.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1441430/full#supplementary-material
References
Adem, S., Jain, S., Sveiven, M., Zhou, X., O’Donoghue, A. J., and Hall, D. A. (2020). Giant magnetoresistive biosensors for real-time quantitative detection of protease activity. Sci. Rep. 10, 7941. doi:10.1038/s41598-020-62910-2
Akçapınar, R., Garipcan, B., Goodarzi, V., and Uzun, L. (2021). Designing of various biosensor devices for determination of apoptosis: a comprehensive review. Biochem. Biophysical Res. Commun. 578, 42–62. doi:10.1016/j.bbrc.2021.08.089
Al-Rawhani, M. A., Mitra, S., Barrett, M. P., Cochran, S., Cumming, D. R., Hu, C., et al. (2020). Multimodal integrated sensor platform for rapid biomarker detection. IEEE Trans. Biomed. Eng. 67, 614–623. doi:10.1109/TBME.2019.2919192
Ambhorkar, P., Wang, Z., Ko, H., Lee, S., Koo, K. I., Kim, K., et al. (2018). Nanowire-based biosensors: from growth to applications. Micromachines 9, 679. doi:10.3390/mi9120679
Anand, U., Chandel, A. K. S., Oleksak, P., Mishra, A., Krejcar, O., Raval, I. H., et al. (2022). Recent advances in the potential applications of luminescence-based, SPR-based, and carbon-based biosensors. Appl. Microbiol. Biotechnol. 106, 2827–2853. doi:10.1007/s00253-022-11901-6
Andrianova, M. S., Kuznetsov, E. V., Grudtsov, V. P., and Kuznetsov, A. E. (2018). CMOS-compatible biosensor for L-carnitine detection. Biosens. Bioelectron. 119, 48–54. doi:10.1016/j.bios.2018.07.044
Aviram, A., and Ratner, M. A. (1974). Molecular rectifiers. Chem. Phys. Lett. 29, 277–283. doi:10.1016/0009-2614(74)85031-1
Baranwal, J., Barse, B., Gatto, G., Broncova, G., and Kumar, A. (2022). Electrochemical sensors and their applications: a review. Chemosensors 10, 363. doi:10.3390/chemosensors10090363
Basu, A. K., Basu, A., and Bhattacharya, S. (2020). Micro/Nano fabricated cantilever based biosensor platform: a review and recent progress. Enzyme Microb. Technol. 139, 109558. doi:10.1016/j.enzmictec.2020.109558
Bergveld, P. (1970). Short communications: development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans. Biomed. Eng. BME 17, 70–71. doi:10.1109/TBME.1970.4502688
Buckhout-White, S. L., and Rubloff, G. W. (2009). Spatial resolution in chitosan-based programmable biomolecular scaffolds. Soft Matter 5, 3677–3681. doi:10.1039/b820356c
Cao, X., Li, L. P., Wang, Q., Wu, Q., Hu, H. H., Zhang, M., et al. (2013). Astrocyte-derived ATP modulates depressive-like behaviors. Nat. Med. 19, 773–777. doi:10.1038/nm.3162
Chang, C. F., and Lu, M. S. (2020). CMOS ion sensitive field effect transistors for highly sensitive detection of DNA hybridization. IEEE Sensors J. 20, 8930–8937. doi:10.1109/JSEN.2020.2986461
Clark, L. C., and Lyons, C. (1962). Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 102, 29–45. doi:10.1111/j.1749-6632.1962.tb13623.x
Costa, T., Cardoso, F. A., Germano, J., Freitas, P. P., and Piedade, M. S. (2017). A CMOS front-end with integrated magnetoresistive sensors for biomolecular recognition detection applications. IEEE Trans. Biomed. Circuits Syst. 11, 988–1000. doi:10.1109/TBCAS.2017.2743685
Crucifix, C., Uhring, M., and Schultz, P. (2004). Immobilization of biotinylated DNA on 2-D streptavidin crystals. J. Struct. Biol. 146, 441–451. doi:10.1016/j.jsb.2004.02.001
Damborský, P., Švitel, J., and Katrlík, J. (2016). Optical biosensors. Essays Biochem. 60, 91–100. doi:10.1042/EBC20150010
Dandin, M., Abshire, P., and Smela, E. (2007). Optical filtering technologies for integrated fluorescence sensors. Lab a Chip 7, 955–977. doi:10.1039/b704008c
Dandin, M., Abshire, P., and Smela, E. (2012). Polymer filters for ultraviolet-excited integrated fluorescence sensing. J. Micromechanics Microengineering 22, 095018. doi:10.1088/0960-1317/22/9/095018
Dellweg, H., Engels, J. W., Fox, J. L., Gierasch, L. M., Gregson, R. P., Heinritz, B., et al. (1992). Glossary for chemists of terms used in biotechnology. Pure and Appl. Chem 64, 143–168. doi:10.1351/pac199264010143
Devadhasan, J. P., Yoo, I. S., and Kim, S. (2015). Overview of CMOS image sensor use in molecular diagnostics. Curr. Appl. Phys. 15, 402–411. doi:10.1016/j.cap.2015.01.009
Ding, J., and Qin, W. (2020). Recent advances in potentiometric biosensors. TrAC - Trends Anal. Chem. 124, 115803. doi:10.1016/j.trac.2019.115803
Doi, H., Horio, T., Choi, Y. J., Takahashi, K., Noda, T., and Sawada, K. (2022). CMOS-based redox-type label-free ATP image sensor for in vitro sensitive imaging of extracellular ATP. Sensors 22, 75. doi:10.3390/s22010075
Dorledo de Faria, R. A., Dias Heneine, L. G., Matencio, T., and Messaddeq, Y. (2019). Faradaic and non-faradaic electrochemical impedance spectroscopy as transduction techniques for sensing applications. Int. J. Biosens. and Bioelectron. 5. doi:10.15406/ijbsbe.2019.05.00148
Elibol, O. H., Morisette, D., Akin, D., Denton, J. P., and Bashir, R. (2003). Integrated nanoscale silicon sensors using top-down fabrication. Appl. Phys. Lett. 83, 4613–4615. doi:10.1063/1.1630853
Enderlein, J., Ruckstuhl, T., and Seeger, S. (1999). Highly efficient optical detection of surface-generated fluorescence. Appl. Opt. 38, 724. doi:10.1364/AO.38.000724
Fellin, T., Pascual, O., and Haydon, P. G. (2006). Astrocytes coordinate synaptic networks: balanced excitation and inhibition. Physiology 21, 208–215. doi:10.1152/physiol.00161.2005
Forouhi, S., and Ghafar-Zadeh, E. (2020). Applications of cmos devices for the diagnosis and control of infectious diseases. Micromachines 11, 1003. doi:10.3390/mi11111003
Futagawa, M., Suzuki, D., Otake, R., Dasai, F., Ishida, M., and Sawada, K. (2013). Fabrication of a 128$\,\times\,$128 pixels charge transfer type hydrogen ion image sensor. IEEE Trans. Electron Devices 60, 2634–2639. doi:10.1109/TED.2013.2268208
Gambini, S., Skucha, K., Liu, P. P., Kim, J., and Krigel, R. (2013). A 10 kPixel CMOS hall sensor array with baseline suppression and parallel readout for immunoassays. IEEE J. Solid-State Circuits 48, 302–317. doi:10.1109/JSSC.2012.2224531
Garcia-Hernando, M., Calatayud-Sanchez, A., Etxebarria-Elezgarai, J., De Pancorbo, M. M., Benito-Lopez, F., and Basabe-Desmonts, L. (2020). Optical single cell resolution cytotoxicity biosensor based on single cell adhesion dot arrays. Anal. Chem. 92, 9658–9665. doi:10.1021/acs.analchem.0c00940
Gourine, A. V., Llaudet, E., Dale, N., and Spyer, K. M. (2005). ATP is a mediator of chemosensory transduction in the central nervous system. Nature 436, 108–111. doi:10.1038/nature03690
Grieshaber, D., Mackenzie, R., Vörös, J., and Reimhult, E. (2008). Electrochemical biosensors-sensor principles and architectures. Sensors 8, 1400–1458. doi:10.3390/s8031400
Hall, D. A., Ananthapadmanabhan, N., Choi, C., Zheng, L., Pan, P. P., Von Jutrzenka, C., et al. (2022). A scalable CMOS molecular electronics chip for single-molecule biosensing. IEEE Transactions on Biomedical Circuits and Systems, 1–11doi. doi:10.1109/TBCAS.2022.3211420
Hall, D. A., Gaster, R. S., Makinwa, K. A., Wang, S. X., and Murmann, B. (2013). A 256 pixel magnetoresistive biosensor microarray in 0.18 µm CMOS. IEEE J. Solid-State Circuits 48, 1290–1301. doi:10.1109/JSSC.2013.2245058
Han, S. J., Xu, L., Yu, H., Wilson, R. J., White, R. L., Pourmand, N., et al. (2006). “CMOS integrated DNA microarray based on GMR sensors,” in Technical digest - international electron devices meeting, IEDM. doi:10.1109/IEDM.2006.346887
Hassibi, A., Singh, R., Manickam, A., Sinha, R., Kuimelis, B., Bolouki, S., et al. (2017). “4.2 A fully integrated CMOS fluorescence biochip for multiplex polymerase chain-reaction (PCR) processes,” in Digest of technical papers - IEEE international solid-state circuits conference (San Francisco, CA, United States: Institute of Electrical and Electronics Engineers Inc), 60, 68–69. doi:10.1109/ISSCC.2017.7870264
Hassibi, A., Vikalo, H., Riechmann, J. L., and Hassibi, B. (2009). Real-time DNA microarray analysis. Nucleic Acids Res. 37, e132. doi:10.1093/nar/gkp675
Hirten, R. P., Lin, K. C., Whang, J., Shahub, S., Helmus, D., Muthukumar, S., et al. (2024). Longitudinal assessment of sweat-based TNF-alpha in inflammatory bowel disease using a wearable device. Sci. Rep. 14, 2833. doi:10.1038/.s41598-024-53522-1
Hofmann, A., Meister, M., Rolapp, A., Reich, P., Scholz, F., and Schäfer, E. (2020). “Light-controlled photometer with optoelectronic CMOS biochip for quantitative PSA detection,” in Proceedings - IEEE international Symposium on Circuits and systems 2020-octob, 1–5. doi:10.1109/iscas45731.2020.9180796
Hofmann, A., Meister, M., Rolapp, A., Reich, P., Scholz, F., and Schafer, E. (2021). Light absorption measurement with a CMOS biochip for quantitative immunoassay based point-of-care applications. IEEE Trans. Biomed. Circuits Syst. 15, 369–379. doi:10.1109/TBCAS.2021.3083359
Honarvar Nazari, M., and Genov, R. (2009). “A fully differential CMOS potentiostat,” in IEEE international symposium on circuits and systems, 2177–2180. doi:10.1109/ISCAS.2009.5118228
Hong, D., Joung, H. A., Lee, D. Y., Kim, S., and Kim, M. G. (2015a). Attomolar detection of cytokines using a chemiluminescence immunoassay based on an antibody-arrayed CMOS image sensor. Sensors Actuators, B Chem. 221, 1248–1255. doi:10.1016/j.snb.2015.07.042
Hong, L., Li, H., Yang, H., and Sengupta, K. (2017). Fully integrated fluorescence biosensors on-chip employing multi-functional nanoplasmonic optical structures in CMOS. IEEE J. Solid-State Circuits 52, 2388–2406. doi:10.1109/JSSC.2017.2712612
Hong, L., McManus, S., Yang, H., and Sengupta, K. (2015b). A fully integrated CMOS fluorescence biosensor with on-chip nanophotonic filter. IEEE Symposium VLSI Circuits, Dig. Tech., C206–C207. doi:10.1109/VLSIC.2015.7231260
Hsu, C. L., Sun, A., Zhao, Y., Aronoff-Spencer, E., and Hall, D. A. (2018). “A 1620 electrochemical CMOS biosensor array with in-pixel averaging using polar modulation,” in 2018 IEEE custom integrated circuits conference, CICC 2018 (San Diego, CA, United States: Institute of Electrical and Electronics Engineers Inc), 1–4. doi:10.1109/CICC.2018.8357044
Hu, K., Arcadia, C. E., and Rosenstein, J. K. (2021). A large-scale multimodal CMOS biosensor array with 131,072 pixels and code-division multiplexed readout. IEEE Solid-State Circuits Lett. 4, 48–51. doi:10.1109/LSSC.2021.3056515
Huang, C. W., Hsueh, H. T., Huang, Y. J., Liao, H. H., Tsai, H. H., Juang, Y. Z., et al. (2013a). A fully integrated wireless CMOS microcantilever lab chip for detection of DNA from Hepatitis B virus (HBV). Sensors Actuators, B Chem. 181, 867–873. doi:10.1016/j.snb.2013.02.061
Huang, C. W., Huang, Y. J., Yen, P. W., Tsai, H. H., Liao, H. H., Juang, Y. Z., et al. (2013b). A CMOS wireless biomolecular sensing system-on-chip based on polysilicon nanowire technology. Lab a Chip 13, 4451–4459. doi:10.1039/.c3lc50798j
Huang, M., Dorta-Quiñones, C. I., Minch, B. A., and Lindau, M. (2021). On-chip cyclic voltammetry measurements using a compact 1024-electrode CMOS IC. Anal. Chem. 93, 8027–8034. doi:10.1021/acs.analchem.1c01132
Hulanicki, A., Glab, S., and Ingman, f. (1991). Chemical sensors definitions and classification. Pure and Appl. Chem 63, 1247–1250. doi:10.1351/pac199163091247
Ingle, A., Velten, A., and Gupta, M. (2019). High flux passive imaging with single-photon sensors. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern 2019-June, 6753–6762. doi:10.1109/CVPR.2019.00692
Ivanov, Y. D., Romanova, T. S., Malsagova, K. A., Pleshakova, T. O., and Archakov, A. I. (2021). Use of silicon nanowire sensors for early cancer diagnosis. Molecules 26, 3734. doi:10.3390/molecules26123734
Jang, B., Cao, P., Chevalier, A., Ellington, A., and Hassibi, A. (2009). “A CMOS fluorescent-based biosensor microarray,” in Digest of technical papers - IEEE international solid-state circuits conference. doi:10.1109/ISSCC.2009.4977495
Jang, B., and Hassibi, A. (2009). Biosensor systems in standard CMOS processes: fact or fiction? IEEE Trans. Industrial Electron. 56, 979–985. doi:10.1109/TIE.2008.2011450
Joung, H. A., Hong, D. G., and Kim, M. G. (2012). “A high sensitivity chemiluminescence-based CMOS image biosensor for the detection of human interleukin 5 (IL-5),” in Proceedings of IEEE sensors. doi:10.1109/ICSENS.2012.6411100
Jung, D., Junek, G. V., Park, J. S., Kumashi, S. R., Wang, A., Li, S., et al. (2021). A CMOS 21 952-pixel multi-modal cell-based biosensor with four-point impedance sensing for holistic cellular characterization. IEEE J. Solid-State Circuits 56, 2438–2451. doi:10.1109/JSSC.2021.3085571
Kim, E., Xiong, Y., Cheng, Y., Wu, H. C., Liu, Y., Morrow, B. H., et al. (2015). Chitosan to connect biology to electronics: fabricating the bio-device interface and communicating across this interface. Polymers 7, 1–46. doi:10.3390/polym7010001
Koizumi, S., Fujishita, K., Tsuda, M., Shigemoto-Mogami, Y., and Inoue, K. (2003). Dynamic inhibition of excitatory synaptic transmission by astrocyte-derived ATP in hippocampal cultures. Proc. Natl. Acad. Sci. U. S. A. 100, 11023–11028. doi:10.1073/pnas.1834448100
Kothari, A., Jagannath, B., Muthukumar, S., and Prasad, S. (2022). An observational study for detection and quantification of interferon-in sweat toward inflammation monitoring. Biosens. Bioelectron. X 10, 100122. doi:10.1016/.j.biosx.2022.100122
Kricka, L. J. (1995). Chemiluminescence and bioluminescence. Anal. Chem. 67, 499–502. doi:10.1021/ac00108a035
Krivitsky, V., Zverzhinetsky, M., Krivitsky, A., Hsiung, L. C., Naddaka, V., Gabriel, I., et al. (2019). Cellular metabolomics by a universal redox-reactive nanosensors array: from the cell level to tumor-on-a-chip analysis. Nano Lett. 19, 2478–2488. doi:10.1021/acs.nanolett.9b00052
Kumashi, S., Jung, D., Park, J., Tejedor-Sanz, S., Grijalva, S., Wang, A., et al. (2021). A CMOS multi-modal electrochemical and impedance cellular sensing array for massively paralleled exoelectrogen screening. IEEE Trans. Biomed. Circuits Syst. 15, 221–234. doi:10.1109/TBCAS.2021.3068710
Kuo, P. Y., Chen, Y. Y., Lai, W. H., and Chang, C. H. (2021). An extended-gate field-effect transistor applied to resistive divider integrated with the readout circuit using 180nm CMOS process for uric acid detection. IEEE Sensors J. 21, 20229–20238. doi:10.1109/JSEN.2021.3093642
Kuo, Y. H., Chen, Y. S., Huang, P. C., and Lee, G. B. (2020). A CMOS-based capacitive biosensor for detection of a breast cancer MicroRNA biomarker. IEEE Open J. Nanotechnol. 1, 157–162. doi:10.1109/OJNANO.2020.3035349
Lai, W. A., Lin, C. H., Yang, Y. S., and Lu, M. S. (2012). “Ultrasensitive detection of avian influenza virus by using CMOS impedimetric sensor arrays,” in Proceedings of the IEEE international conference on micro electro mechanical systems (MEMS), 894–897. doi:10.1109/MEMSYS.2012.6170329
Lavrik, N. V., Sepaniak, M. J., and Datskos, P. G. (2004). Cantilever transducers as a platform for chemical and biological sensors. Rev. Sci. Instrum. 75, 2229–2253. doi:10.1063/1.1763252
Lee, C., Chen, Y. W., and Lu, M. S. (2021). CMOS biosensors for the detection of DNA hybridization in high ionic-strength solutions. IEEE Sensors J. 21, 4135–4142. doi:10.1109/JSEN.2020.3031321
Lee, K. H., Choi, S., Lee, J. O., Yoon, J. B., and Cho, G. H. (2012). CMOS capacitive biosensor with enhanced sensitivity for label-free DNA detection. Dig. Tech. Pap. - IEEE Int. Solid-State Circuits Conf. 55, 120–122. doi:10.1109/ISSCC.2012.6176945
Lei, K. M., Heidari, H., Mak, P. I., Law, M. K., Maloberti, F., and Martins, R. P. (2017). A handheld high-sensitivity micro-NMR CMOS platform with B-field stabilization for multi-type biological/chemical assays. IEEE J. Solid-State Circuits 52, 284–297. doi:10.1109/JSSC.2016.2591551
Lei, K. M., Mak, P. I., Law, M. K., and Martins, R. P. (2016). CMOS biosensors for: in vitro diagnosis-transducing mechanisms and applications. Lab a Chip 16, 3664–3681. doi:10.1039/c6lc01002d
Li, H., Liu, X., Li, L., Mu, X., Genov, R., and Mason, A. J. (2017). CMOS electrochemical instrumentation for biosensor microsystems: a review. Sensors Switz. 17, 74. doi:10.3390/s17010074
Li, P., Molderez, T. R., and Verhelst, M. (2021). “A 96-channel 40nm CMOS fully-integrated potentiostat for electrochemical monitoring,” in IEEE 47th European solid state circuits conference, 167–170. doi:10.1109/ESSCIRC53450.2021.9567858
Li, Y., Vancura, C., Kirstein, K. U., Lichtenberg, J., and Hierlemann, A. (2008). Monolithic resonant-cantilever-based CMOS microsystem for biochemical sensing. IEEE Trans. Circuits Syst. I Regul. Pap. 55, 2551–2560. doi:10.1109/TCSI.2008.922027
Lieber, C. M. (1998). One-dimensional nanostructures: chemistry, physics, and applications. Solid State Commun. 107, 607–613. doi:10.1016/S0038-1098(98)00209-9
Lin, C. H., Lee, Y. C., Yang, C. M., and Lu, M. S. (2022a). Detection of DNA hybridization beyond the Debye screening length by CMOS capacitive sensors. IEEE Electron Device Lett. 43, 1319–1322. doi:10.1109/LED.2022.3182784
Lin, C.-Y., Sajal, M. S., Gilpin, Y., Dehghandehnavi, F., Batueva, A., Lin, K.-C., et al. (2022b). Cmos bioelectronics: current and future trends. Bioelectronics, 93–107. doi:10.1201/9781003263265-6
Liu, Q., Jimenez, M., Inda, M. E., Riaz, A., Zirtiloglu, T., Chandrakasan, A. P., et al. (2022). A threshold-based bioluminescence detector with a CMOS-integrated photodiode array in 65 nm for a multi-diagnostic ingestible capsule. IEEE J. Solid-State Circuits, 1–14doi. doi:10.1109/JSSC.2022.3197465
Manickam, A., Johnson, K. A., Singh, R., Wood, N., Ku, E., Cuppoletti, A., et al. (2021). “Multiplex PCR CMOS biochip for detection of upper respiratory pathogens including SARS-CoV-2,” in IEEE symposium on VLSI circuits, digest of technical papers 2021-june, 1–2. doi:10.23919/VLSICircuits52068.2021.9492353
Manickam, A., Singh, R., Mcdermott, M. W., Wood, N., Bolouki, S., Naraghi-Arani, P., et al. (2017). A fully integrated CMOS fluorescence biochip for DNA and RNA testing. IEEE J. Solid-State Circuits 52, 2857–2870. doi:10.1109/JSSC.2017.2754363
Manickam, A., You, K.-D., Wood, N., Pei, L., Liu, Y., Singh, R., et al. (2019a). “A CMOS biosensor array with 1024 3-electrode voltammetry pixels and 93dB dynamic range,” in IEEE international solid-state circuits conference, 192–194doi. doi:10.1109/ISSCC.2019.8662507
Manickam, A., You, K. D., Wood, N., Pei, L., Liu, Y., Singh, R., et al. (2019b). A CMOS electrochemical biochip with 32 32 three-electrode voltammetry pixels. IEEE J. Solid-State Circuits 54, 2980–2990. doi:10.1109/JSSC.2019.2941020
McCall, J. R., Jacocks, H. M., Baden, D. G., and Bourdelais, A. J. (2012). Development of a competitive fluorescence-based synaptosome binding assay for brevetoxins. Harmful Algae 19, 85–91. doi:10.1016/j.hal.2012.06.003
Mirsian, S., Khodadadian, A., Hedayati, M., Manzour-ol Ajdad, A., Kalantarinejad, R., and Heitzinger, C. (2019). A new method for selective functionalization of silicon nanowire sensors and Bayesian inversion for its parameters. Biosens. Bioelectron. 142, 111527. doi:10.1016/j.bios.2019.111527
Moser, N., Rodriguez-Manzano, J., Lande, T. S., and Georgiou, P. (2018). A scalable ISFET sensing and memory array with sensor auto-calibration for on-chip real-time DNA detection. IEEE Trans. Biomed. Circuits Syst. 12, 390–401. doi:10.1109/TBCAS.2017.2789161
Mosharov, E. V., and Sulzer, D. (2005). Analysis of exocytotic events recorded by amperometry. Nat. Methods 2, 651–658. doi:10.1038/nmeth782
Nasri, B., Wu, T., Alharbi, A., Gupta, M., Ranjitkumar, R., Sebastian, S., et al. (2017). 15.7 Heterogeneous integrated CMOS-graphene sensor array for dopamine detection. Dig. Tech. Pap. - IEEE Int. Solid-State Circuits Conf. 60, 268–269. doi:10.1109/ISSCC.2017.7870364
Nimse, S. B., Song, K., Sonawane, M. D., Sayyed, D. R., and Kim, T. (2014). Immobilization techniques for microarray: challenges and applications. Sensors Switz. 14, 22208–22229. doi:10.3390/s141222208
Pinitwong, W., and Pijitrojana, W. (2015). “Development of ion sensitive field effect transistor for urinary microalbumin detection by silanization,” in ECTI-CON 2015 - 2015 12th international conference on electrical engineering/electronics, computer, telecommunications and information technology (Hua Hin, Thailand: Institute of Electrical and Electronics Engineers Inc.). doi:10.1109/ECTICon.2015.7206967
Pires, N. M. M., Dong, T., Hanke, U., and Hoivik, N. (2014). Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors Switz. 14, 15458–15479. doi:10.3390/s140815458
Poupon-Fleuret, C., Steghens, J.-P., and Bernengo, J.-C. (1996). Luminol chemiluminescence-based porphyrin assays without hydrogen peroxide: a spectral study of mechanism and enhancement. Analyst 121, 1539. doi:10.1039/an9962101539
Redy Keisar, O., Pevzner, A., Fridkin, G., Shelef, O., Shabat, D., and Ashkenazi, N. (2024). Highly sensitive chemiluminescence sensors for the detection and differentiation of chemical warfare agents. Anal. Methods 16, 1736–1740. doi:10.1039/d3ay02054a
Rogers, K. R. (2000). Principles of affinity-based biosensors. Mol. Biotechnol. 14, 109–130. doi:10.1385/MB:14:2.109
Saengdee, P., Promptmas, C., Thanapitak, S., Srisuwan, A., Pankiew, A., Thornyanadacha, N., et al. (2020). Optimization of 3-aminopropyltriethoxysilane functionalization on silicon nitride surface for biomolecule immobilization. Talanta 207, 120305. doi:10.1016/j.talanta.2019.120305
Saengdee, P., Thanapitak, S., Ongwattanakul, S., Srisuwan, A., Pankiew, A., Thornyanadacha, N., et al. (2021). A silicon nitride ion sensitive field effect transistor-based immunosensor for determination of urinary albumin. Electrochem. Sci. Adv. 2. doi:10.1002/elsa.202100078
Sandeau, L., Vuillaume, C., Contié, S., Grinenval, E., Belloni, F., Rigneault, H., et al. (2015). Large area CMOS bio-pixel array for compact high sensitive multiplex biosensing. Lab a Chip 15, 877–881. doi:10.1039/c4lc01025f
Sander, D., Dandin, M., Ji, H., Nelson, N., and Abshire, P. (2007). “Low-noise CMOS fluorescence sensor,” in Proceedings - IEEE international symposium on circuits and systems (New Orleans, LA, United States: Institute of Electrical and Electronics Engineers Inc.), 2007–2010. doi:10.1109/iscas.2007.378431
Selvam, S. (2017). “3 - imaging biomaterial-associated inflammation,” in Monitoring and evaluation of biomaterials and their performance in vivo. Editor R. J. Narayan (Woodhead Publishing), 47–68. doi:10.1016/B978-0-08-100603-0.00003-1
Senevirathna, B. P., Lu, S., Dandin, M. P., Basile, J., Smela, E., and Abshire, P. A. (2018). Real-time measurements of cell proliferation using a lab-on-CMOS capacitance sensor array. IEEE Trans. Biomed. Circuits Syst. 12, 510–520. doi:10.1109/TBCAS.2018.2821060
Sessi, V., Ibarlucea, B., Seichepine, F., Klinghammer, S., Ibrahim, I., Heinzig, A., et al. (2022). Multisite dopamine sensing with femtomolar resolution using a CMOS enabled aptasensor chip. Front. Neurosci. 16, 875656. doi:10.3389/fnins.2022.875656
Sheibani, S., Capua, L., Kamaei, S., Akbari, S. S. A., Zhang, J., Guerin, H., et al. (2021). Extended gate field-effect-transistor for sensing cortisol stress hormone. Commun. Mater. 2, 10. doi:10.1038/s43246-020-00114-x
Sigvardson, K. W., Kennish, J. M., and Birks, J. W. (1984). Peroxyoxalate chemiluminescence detection of polycyclic aromatic amines in liquid chromatography. Anal. Chem. 56, 1096–1102. doi:10.1021/ac00271a011
Singh, N. K., Thungon, P. D., Estrela, P., and Goswami, P. (2019). Development of an aptamer-based field effect transistor biosensor for quantitative detection of Plasmodium falciparum glutamate dehydrogenase in serum samples. Biosens. Bioelectron. 123, 30–35. doi:10.1016/j.bios.2018.09.085
Smith, C. L., Milea, J. S., and Nguyen, G. H. (2005). Immobilization of nucleic acids using biotin-strept(avidin) systems. Berlin, Heidelberg: Springer Berlin Heidelberg, 63–90. doi:10.1007/128_017
Stadler, V., Beyer, M., König, K., Nesterov, A., Torralba, G., Lindenstruth, V., et al. (2007). Multifunctional CMOS microchip coatings for protein and peptide arrays. J. Proteome Res. 6, 3197–3202. doi:10.1021/pr0701310
Stern, E., Klemic, J. F., Routenberg, D. A., Wyrembak, P. N., Turner-Evans, D. B., Hamilton, A. D., et al. (2007). Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature 445, 519–522. doi:10.1038/.nature05498
Stradiotto, N. R., Yamanaka, H., and Zanoni, M. V. B. (2003). Electrochemical sensors: a powerful tool in analytical chemistry. J. Braz. Chem. Soc. 14, 159–173. doi:10.1590/s0103-50532003000200003
Strianese, M., Staiano, M., Ruggiero, G., Labella, T., Pellecchia, C., and D’Auria, S. (2012). “Fluorescence-based biosensors,” in Methods in molecular biology, 193–216. doi:10.1007/978-1-61779-806-1{_}9
Sun, J.-H., Ling, B., Kaiser, M. A.-A., and Sideris, C. (2022). A drift-compensated magnetic spectrometer for point-of-care wash-free immunoassays using a concurrent dual-frequency oscillator. Institute of Electrical and Electronics Engineers IEEE, 173–176. doi:10.1109/esscirc55480.2022.9911522
Susanto, H., Samsudin, A. M., Rokhati, N., and Widiasa, I. N. (2013). Immobilization of glucose oxidase on chitosan-based porous composite membranes and their potential use in biosensors. Enzyme Microb. Technol. 52, 386–392. doi:10.1016/j.enzmictec.2013.02.005
Syu, Y.-C., Hsu, W.-E., and Lin, C.-T. (2018). Review—field-effect transistor biosensing: devices and clinical applications. ECS J. Solid State Sci. Technol. 7, Q3196–Q3207. doi:10.1149/2.0291807jss
Tabrizi, H. O., Forouhi, S., Ghafar-Zadeh, M., Magierowski, S., and Ghafar-Zadeh, E. (2021). “CMOS capacitive DNA nano-mass measurement for DNA storage application,” in Canadian conference on electrical and computer engineering (ON, Canada: Institute of Electrical and Electronics Engineers Inc). doi:10.1109/CCECE53047.2021.9569194
Thriveni, G., and Ghosh, K. (2022). Advancement and challenges of biosensing using field effect transistors. Biosensors 12, 647. doi:10.3390/bios12080647
Tokuda, T., and Ohta, J. (2022). DNA optical readout methods. New York, NY: Springer, 589–600. doi:10.1007/978-1-4614-3447-4_10
Tokuda, T., Yamamoto, A., Kagawa, K., Nunoshita, M., and Ohta, J. (2006). A CMOS image sensor with optical and potential dual imaging function for on-chip bioscientific applications. Sensors Actuators, A Phys. 125, 273–280. doi:10.1016/j.sna.2005.08.023
Tseng, T. T., Chang, C. F., and Chan, W. C. (2014). Fabrication of implantable, enzyme-immobilized glutamate sensors for the monitoring of glutamate concentration changes in vitro and in vivo. Molecules 19, 7341–7355. doi:10.3390/.molecules19067341
Ushijima, E., Fujimoto, S., and Nakazato, K. (2021). Local reaction fields on a CMOS sensor array utilizing a magneto-Archimedes levitation–based magnetic particle-arrangement method. Sensors Actuators, A Phys. 323, 112655. doi:10.1016/j.sna.2021.112655
van der Spiegel, J., Lauks, I., Chan, P., and Babic, D. (1983). The extended gate chemically sensitive field effect transistor as multi-species microprobe. Sensors Actuators 4, 291–298. doi:10.1016/0250-6874(83)85035-5
Walker, N. L., Roshkolaeva, A. B., Chapoval, A. I., and Dick, J. E. (2021). Recent advances in potentiometric biosensing. Curr. Opin. Electrochem. 28, 100735. doi:10.1016/j.coelec.2021.100735
Wang, A. Y., Sheng, Y., Li, W., Jung, D., Junek, G., Park, J., et al. (2022a). “A CMOS cellular interface array for digital physiology featuring high-density multi-modal pixels and reconfigurable sampling rate,” in Digest of technical papers - IEEE international solid-state circuits conference 2022-february, 202–204. doi:10.1109/ISSCC42614.2022.9731629
Wang, A. Y., Sheng, Y., Li, W., Jung, D., Junek, G. V., Liu, H., et al. (2022b). A multimodal and multifunctional CMOS cellular interfacing array for digital physiology and pathology featuring an ultra dense pixel array and reconfigurable sampling rate. IEEE Transactions on Biomedical Circuits and Systems, 1–18doi. doi:10.1109/TBCAS.2022.3224064
Wang, H., Mahdavi, A., Park, J., Chi, T., Butts, J., Hookway, T. A., et al. (2014). “Cell culture and cell based sensor on CMOS,” in IEEE 2014 biomedical circuits and systems conference, BioCAS 2014 - proceedings (Lausanne, Switzerland: Institute of Electrical and Electronics Engineers Inc), 468–471. doi:10.1109/BioCAS.2014.6981764
Wang, L., Fu, D., Liu, X., Zhao, J., Zhao, J., Yuan, Q., et al. (2021). Highly sensitive biosensor based on a microcantilever and alternating current electrothermal technology. J. Micromechanics Microengineering 31, 015009. doi:10.1088/1361-6439/abcae6
Weeks, R., Zhou, X., Yuan, T. L., and Zhang, J. (2022). Fluorescent biosensor for measuring ras activity in living cells. J. Am. Chem. Soc. 144, 17432–17440. doi:10.1021/jacs.2c05203
Wicks, S. L., and Hargrove, A. E. (2019). Fluorescent indicator displacement assays to identify and characterize small molecule interactions with RNA. Methods 167, 3–14. doi:10.1016/j.ymeth.2019.04.018
Wu, J., Liu, H., Chen, W., Ma, B., and Ju, H. (2023). Device integration of electrochemical biosensors. Nat. Rev. Bioeng. 1, 346–360. doi:10.1038/s44222-023-00032-w
Wu, L. Q., Yi, H., Li, S., Rubloff, G. W., Bentley, W. E., Ghodssi, R., et al. (2003). Spatially selective deposition of a reactive polysaccharide layer onto a patterned template. Langmuir 19, 519–524. doi:10.1021/la026518t
Yong, S. K., Shen, S. K., Chiang, C. W., Weng, Y. Y., Lu, M. P., and Yang, Y. S. (2021). Silicon nanowire field-effect transistor as label-free detection of hepatitis b virus proteins with opposite net charges. Biosensors 11, 442. doi:10.3390/.bios11110442
Zafar, H., Channa, A., Jeoti, V., and Stojanović, G. M. (2022). Comprehensive review on wearable sweat-glucose sensors for continuous glucose monitoring. Sensors 22, 638. doi:10.3390/s22020638
Zhang, J., Huang, Y., Trombly, N., Yang, C., and Mason, A. (2005). Electrochemical array microsystem with integrated potentiostat. IEEE Sensors, 385–388. doi:10.1109/ICSENS.2005.1597716
Zhang, J., Rupakula, M., Bellando, F., Garcia Cordero, E., Longo, J., Wildhaber, F., et al. (2019). Sweat biomarker sensor incorporating picowatt, three-dimensional extended metal gate ion sensitive field effect transistors. ACS Sensors 4, 2039–2047. doi:10.1021/acssensors.9b00597
Zhao, J., Wang, L., Fu, D., Zhao, D., Wang, Y., Yuan, Q., et al. (2021). Gold nanoparticles amplified microcantilever biosensor for detecting protein biomarkers with high sensitivity. Sensors Actuators, A Phys. 321, 112563. doi:10.1016/j.sna.2021.112563
Zhou, X., Mai, E., Sveiven, M., Pochet, C., Jiang, H., Huang, C. C., et al. (2021). A 9.7-nT, 704-ms magnetic biosensor front-end for detecting magneto-relaxation. IEEE J. Solid-State Circuits 56, 2171–2181. doi:10.1109/JSSC.2020.3043669
Keywords: complementary metal-oxide-semiconductor (CMOS), biosensor, immobilization, post-CMOS process, transduction, lab-on-a-chip (LOC)
Citation: Dehghandehnavi F, Sajal MS and Dandin M (2024) Surface-modified CMOS biosensors. Front. Bioeng. Biotechnol. 12:1441430. doi: 10.3389/fbioe.2024.1441430
Received: 31 May 2024; Accepted: 09 October 2024;
Published: 06 November 2024.
Edited by:
Zhugen Yang, Cranfield University, United KingdomReviewed by:
Roopa Gowda, ASM America, Inc., United StatesHao Zhuang, University of California, Berkeley, United States
Copyright © 2024 Dehghandehnavi, Sajal and Dandin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc Dandin, bWRhbmRpbkBhbmRyZXcuY211LmVkdQ==
 Fahimeh Dehghandehnavi
Fahimeh Dehghandehnavi Md. Sakibur Sajal
Md. Sakibur Sajal Marc Dandin
Marc Dandin