- 1Qingdao C1 Refinery Engineering Research Center, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Sinopec Dalian (Fushun) Research Institute of Petroleum and Petrochemicals, Dalian, China
- 4Key Laboratory of Marine Chemistry Theory and Technology (Ministry of Education), College of Chemistry and Chemical Engineering, Ocean University of China, Qingdao, China
- 5Shandong Energy Institute, Qingdao, China
- 6Qingdao New Energy Shandong Laboratory, Qingdao, China
Acetogenic bacteria (acetogens) are a class of microorganisms with conserved Wood-Ljungdahl pathway that can utilize CO and CO2/H2 as carbon source for autotrophic growth and convert these substrates to acetate and ethanol. Acetogens have great potential for the sustainable production of biofuels and bulk biochemicals using C1 gases (CO and CO2) from industrial syngas and waste gases, which play an important role in achieving carbon neutrality. In recent years, with the development and improvement of gene editing methods, the metabolic engineering of acetogens is making rapid progress. With introduction of heterogeneous metabolic pathways, acetogens can improve the production capacity of native products or obtain the ability to synthesize non-native products. This paper reviews the recent application of metabolic engineering in acetogens. In addition, the challenges of metabolic engineering in acetogens are indicated, and strategies to address these challenges are also discussed.
1 Introduction
The functioning of modern society and industry heavily relies on energy, fuels, and chemicals, most of which are primarily derived from fossil fuels. The non-renewability of fossil fuels poses a potential shortage risk, and their extensive usage results in the emission of greenhouse gases, which contributes to global climate change. Renewable energy and chemicals can serve as alternatives, offering a potential solution to current environmental challenges (Jiang et al., 2021). In conventional biofuel production, food crops (first generation) or cellulosic biomass (second generation) are typically used as substrates, with microorganisms employed in the fermentation process to produce ethanol, butanol, and other chemicals (Atsumi et al., 2008a; Atsumi et al., 2008b; Lin et al., 2015; Tian et al., 2017; Birgen et al., 2019; Jeswani et al., 2020; Riaz et al., 2022). However, the production of feedstocks for first-generation biofuels relies heavily on agricultural land, which increases the risk of exacerbating food scarcities. On the other hand, second-generation biofuels using lignocellulose or waste materials as feedstocks are more sustainable and environmentally friendly. However, the feedstocks require pre-processing steps, such as degradation, making the process inefficient and costly (Xu et al., 2018). Capture and utilization of gaseous one-carbon (C1) waste feedstocks using carbon oxide (CO and/or CO2) fixing microorganisms through gas fermentation has emerged as a promising route, holding both industrial and environmental significance. Acetogenic bacteria (acetogens) are promising platform microorganisms for C1 gas fixation. They are a class of obligate anaerobic Gram-positive bacteria, that can utilize CO2/H2 or CO as energy and carbon source for autotrophic growth (Drake et al., 2006; Drake et al., 2008). The Wood-Ljungdahl pathway (WLP) is employed for CO2 fixation and acetyl-CoA production, and is coupled with energy conservation (Drake et al., 1997; Ragsdale and Pierce, 2008). Acetyl-CoA serves as a precursor for biomass synthesis or for the generation of specific products, such as acetate and ethanol. So far, over 100 acetogenic species in 23 genera have been identified that grow under a wide range of temperature and pH (Drake et al., 2006; Lee et al., 2022) Depending on the species, in addition to acetate, diverse biochemicals, including ethanol, 2,3-butanediol, lactate, or other acids and alcohols can be produced from C1 feedstocks (Kim et al., 2023). However, not all the acetogens have been investigated in details. Species from the genera of Clostridium, Acetobacterium, and Moorella are currently the most extensively studied for developing industrial applications.
In this review, the physiology and metabolism of acetogens focusing on WLP and energy conservation is firstly introduced. Recent progress in the development of genetic manipulation approaches and achievement of metabolic engineering in acetogens are summarized. Furthermore, the challenges of metabolic engineering in acetogens are outlined, and strategies aimed at overcoming these challenges are also discussed.
2 Wood-Ljungdahl pathway and energy conservation
2.1 Wood-Ljungdahl pathway
WLP is the key pathway for acetogens to fix CO2 and generate acetate, as illustrated in Figure 1 (Ljungdahl, 1986; Wood et al., 1986). WLP consists of two branches, the methyl branch and the carbonyl branch. The methyl branch is a linear metabolic branch that converts CO2 to methyl groups through six sequential steps. When acetogens autotrophically grow on CO2/H2, the first reaction of methyl branch is the reduction of CO2 to formate catalyzed by formate dehydrogenase (FDH) using two electrons. Formate then undergoes a reaction with tetrahydrofolate (THF) to form formyl-THF, catalyzed by formyl-THF synthetase (FTS) and consuming one ATP. Subsequently, formyl-THF is converted to methenyl-THF by formyl-THF cyclohydrolase (FTC), leading to the release of one H2O. The reduction of methenyl-THF to methylene-THF, and eventually to methyl-THF, occurs through the successive actions of methylene-THF dehydrogenase (MDH) and methylene-THF reductase (MTHFR), with each step involving the consumption of two electrons. In the last step of methyl branch, the methyl group of methyl-THF is transferred to a corrinoid iron-sulfur-containing protein (CoFeSP) via the methyltransferase (MT). The reducing steps of methyl branch require a total of six electrons, while the specific coenzymes carrying the reducing equivalents are diverse and still partially unrevealed (Drake and Daniel, 2004; Schuchmann and Müller, 2016). The carbonyl-branch involves a single reaction facilitated by a multi-component enzyme complex known as carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS), which catalyzes the reduction of CO2 to CO using two electrons. In the final step of WLP, the bi-functional CODH/ACS enzyme complex catalyzes the reaction that converts CO, methyl-CoFeSP and CoA into acetyl-CoA. Certain acetogens like Moorella thermoacetica and Clostridium ljungdahlii exhibit the capability of thriving using CO as sole carbon and energy source. In this situation, CO2 is generated from CO through the catalysis of CODH initially, and then enters the methyl branch (Ragsdale, 2004). Consequently, CO directly binds to the CODH/ACS. Ultimately, acetyl-CoA is phosphorylated to acetyl phosphate and further metabolized to acetate by phosphotransacetylase (PTA) and acetate kinase (ACK), releasing one molecule of ATP.
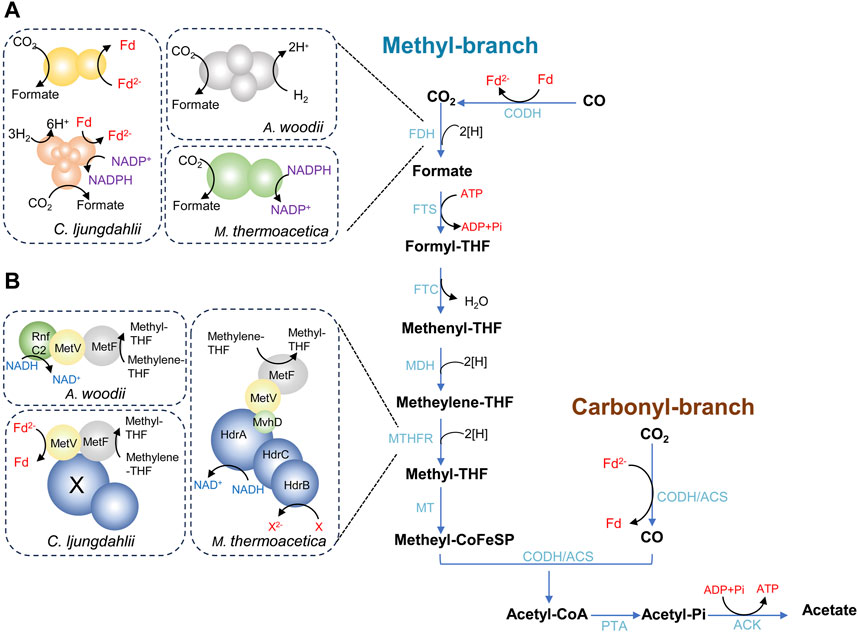
Figure 1. The Wood-Ljungdahl pathway (WLP) in acetogens. (A) shows the diversity of formate dehydrogenases; (B) shows the diversity of methylene-THF reductases. ACK, acetate kinase; ACS, acetyl-CoA synthase; CODH, CO dehydrogenase; FDH, formate dehydrogenase; FTS, formyl-THF synthetase; FTC, formyl-THF cyclohydrolase; MDH, methylene-THF dehydrogenase; MTHFR, methylene-THF reductase; MT, methyltransferase; PTA, phosphotransacetylase; THF, Tetrahydrofolate.
As described above, the fundamental chemistry of the methyl branch is the same in different acetogenic species. However, the enzymes that catalyze the reduction of CO2 to formate and the reduction of methylene-THF to methyl-THF are different, involving different coenzymes. In the step of CO2 reduction to formate, the standard redox potential (E0ʹ) of the CO2/formate couple is −432 mV (Thauer et al., 1977). Therefore, oxidation of NADH (E0ʹ = −320 mV) is not sufficient to drive this reaction. Instead, reduced ferredoxin [(ferredoxin (Fd)/reduced ferredoxin (Fd2−)) E0ʹ = −450 mV], NADPH [(NADP+/NADPH) E0ʹ = −370 mV], and H2 [(H+/H2) E0ʹ = −414 mV] all have physiological redox potentials that can potentially drive the reaction (Schuchmann and Müller, 2014). Different acetogens have evolved to use different electron donors (Figure 1A). In the model organism Acetobacterium woodii, a hydrogen-dependent CO2 reductase (HDCR) directly employs H2 as the electron donor for CO2 reduction (Schuchmann and Müller, 2013). The HDCR complex comprises a hydrogenase module (HydA2) and a formate dehydrogenase module (FdhF1 or FdhF2), linked by two electron-transferring subunits (HycB2 and HycB3). In the case of M. thermoacetica, the reduction of CO2 to formate is catalyzed by a two-subunit NADPH-dependent FDH, despite the unfavorable bioenergetics with NADPH (E0ʹ = −370 mV) (Yamamoto et al., 1983; Rosenbaum and Müller, 2023). The genome of C. ljungdahlii harbors three gene clusters encoding potential FDHs (Schuchmann and Müller, 2014). The first gene cluster (Clju_c06990–07080), encodes an FDH and an electron-bifurcating hydrogenase, and this complex putatively use either H2 or Fd2- and NADPH as electron donors (Wang et al., 2013a; Nagarajan et al., 2013; Mock et al., 2015). The second gene cluster (Clju_c20030–20040) encodes an FDH and a small ferredoxin-like protein, and this complex presumably accepts electrons from Fd2-. The third FDH is encoded by Clju_c08930, an isogene of Clju_c20040 (Schuchmann and Müller, 2014).
The MTHFR that catalyzes the reduction of methylene-THF to methyl-THF also exhibits diversity among various acetogens (Figure 1B). The MTHFR complex from A. woodii is composed of MetF, MetV, and RnfC2 in a stoichiometry of 1:1:1, with NADH serving as the electron donor (Ragsdale and Ljungdahl, 1984). In M. thermoacetica, the genes metVF encoding the MTHFR are co-transcribed together with the genes hdrCBA-mvhD. The complex MetFV-HdrABC-MvhD is hypothesized to be an electron-bifurcating enzyme, which uses NADH as reductant (Mock et al., 2014). However, the second electron acceptor is unknown yet. The MetF-MetV complex from C. ljungdahlii has been purified and characterized, indicating that the enzyme uses Fd2- as the electron donor and is not capable of electron bifurcation (Yi et al., 2021). However, a proposed energy metabolic scheme suggests that this process cannot physiologically occur, as it would result in a negative ATP gain during autotrophic acetogenesis. It is postulated that the MTHFR may assemble into a large, flexible complex involving other electron-bifurcating enzymes (Öppinger et al., 2022).
2.2 Energy conservation
Acetate kinase reaction during acetogenesis synthesizes one ATP via substrate-level phosphorylation (SLP). While, the activation of formate consumes one ATP, resulting in zero net ATP yield through SLP. This highlights a chemiosmotic energy conservation mechanism for ATP synthesis in acetogens, which relies on an Fd2--driven ion pump complex (Figure 2). There are two different ion pump complexes in acetogens, Rnf (Rhodobacter nitrogen fixation) and Ech (Energy-converting hydrogenase) (Buckel and Thauer, 2013). The Rnf complex found in A. woodii, C. ljungdahlii, and Clostridium autoethanogenum, is a ferredoxin:NAD+ oxidoreductase, and it pumps Na+ or H+ outside the cell when transferring electrons from Fd2- to NAD+ (Tremblay et al., 2012; Hess et al., 2013; Mock et al., 2015). In contrast, the Ech complex found in M. thermoacetica and Thermoanaerobacter kivui has ferredoxin:H+ oxidoreductase activity and pumps H+ (Katsyv and Müller, 2022; Rosenbaum and Müller, 2023). The transmembrane H+ or Na+ gradient created by Rnf or Ech drives the ATP synthase to generate ATP (Schoelmerich and Müller, 2019). In comparison to A. woodii, C. ljungdahlii, and C. autoethanogenum, M. thermoacetica exhibits a notable difference in the presence of cytochromes and quinones, which probably contribute to energy conservation (Gottwald et al., 1975; Rosenbaum and Müller, 2023). M. thermoacetica harbors a putative “headless” NADH dehydrogenase that may function as an H+ translocating enzyme, driving the ATP synthase by utilizing Fd2- as an electron donor and quinone as an electron acceptor (Pierce et al., 2008). The largest thermodynamic barrier in the WLP is the reduction of CO2 to CO, as the redox couple has a very low standard redox potential (E0ʹ = −520 mV) (Thauer et al., 1977). Among the identified coenzymes in the WLP, only Fd2- can provide electrons for the reaction (Schuchmann and Müller, 2014).
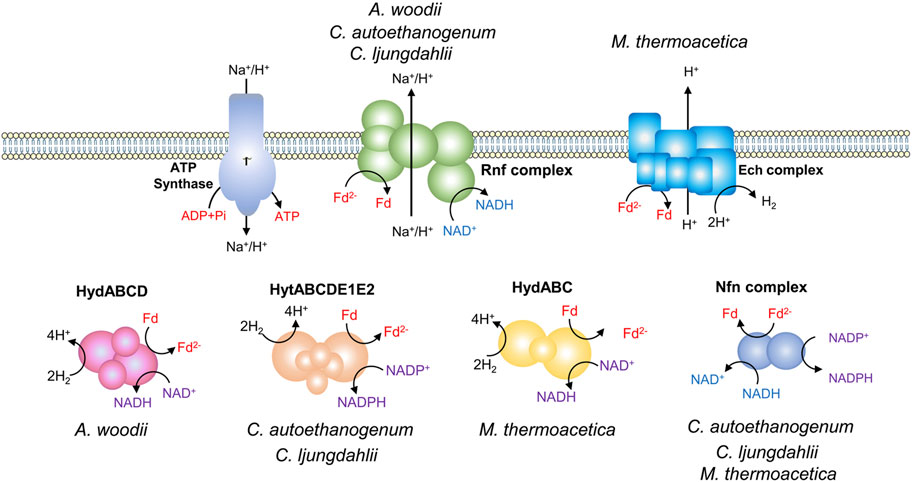
Figure 2. Energy conservation in acetogens. Different electron-bifurcating hydrogenases of acetogens: HydABCD from A. woodii, HytABCDE1E2 from C. autoethanogenum and C. ljungdahlii, and HydABC from Moorella thermoacetica. Nfn complex, the electron-bifurcating and ferredoxin-dependent transhydrogenase.
Every electron for autotrophic acetogenesis from H2 and CO2 is derived from the oxidation of hydrogen, facilitated by hydrogenases (Figure 2) (Schuchmann and Müller, 2014; Di Leonardo et al., 2022). A. woodii has a four-subunit soluble hydrogenase, HydABCD, which contains iron-sulphur centers and flavin (Schuchmann and Müller, 2012). The electron-bifurcating HydABCD catalyzes the oxidation of H2, coupling the endergonic reduction of Fd and the exergonic reduction of NAD+. Fd and NAD+ are reduced simultaneously in a 1/1 stoichiometry (Schuchmann and Müller, 2012). The electron-bifurcating hydrogenases from C. ljungdahlii and C. autoethanogenum are composed of six subunits (HytABCDE1E2) and are NADP+ and Fd-dependent. The hydrogenase forms a tight complex with the formate dehydrogenase FdhA (Wang et al., 2013a; Nagarajan et al., 2013; Mock et al., 2015). The electron-bifurcating hydrogenase of M. thermoacetica, HydABC consists of three subunits, which catalyzes the oxidation of hydrogen with NAD+ and Fd as electron acceptors (Wang et al., 2013b). Part of the Fd2− is used in the reductive reactions of acetogenesis and the remaining Fd2− can be used by the Rnf or Ech complex to generate a chemiosmotic gradient. In addition, due to the involvement of different cofactors in the energy metabolism, some acetogens such as C. ljungdahlii, C. autoethanogenum, and M. thermoacetica have an electron-bifurcating ferredoxin-dependent transhydrogenase (Nfn complex) (Huang et al., 2012; Nagarajan et al., 2013; Mock et al., 2015). It is capable of producing NADPH from Fd2− and NADH through electron bifurcation, regulating the redox pool.
In recent years, the understanding of acetogenic metabolism has gradually deepened, which supports the metabolic engineering of acetogens in principles. Meanwhile, complete genomic sequences have been published for the species including A. woodii, M. thermoacetica, C. ljungdahlii, C. autoethanogenum, and Clostridium aceticum, making genetic modification in those acetogens possible.
3 Genetic manipulation tools in acetogens
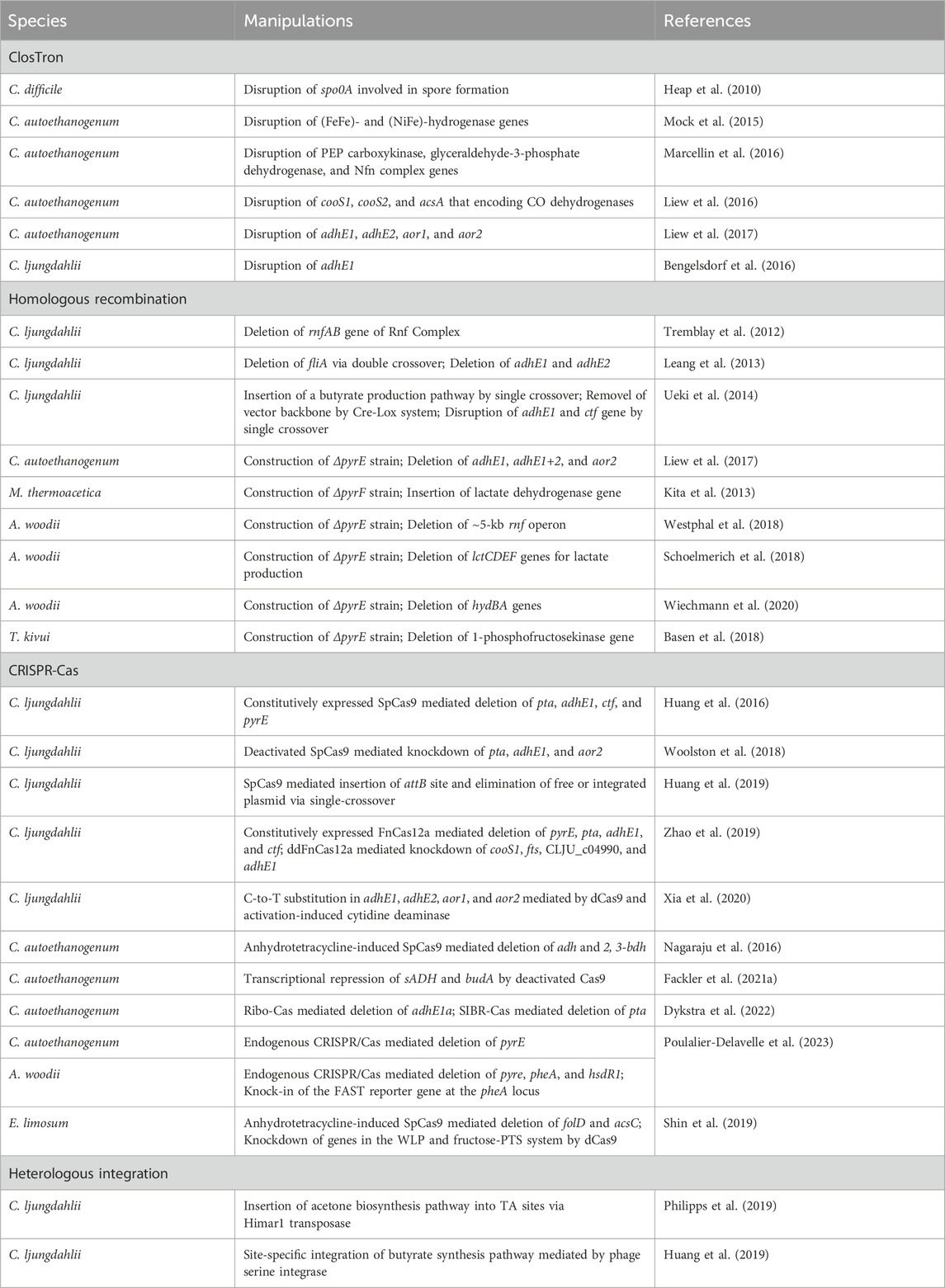
The genetic toolkit available for acetogens has significantly expanded in the past decade. In addition to plasmid-based gene expression, genome editing approaches through ClosTron, homologous recombination, CRISPR-Cas system, and heterologous integration have been developed for acetogens (Joseph et al., 2018).
3.1 DNA transfer
An efficient genetic system relies on a reliable DNA transfer method. DNA transfer in most acetogens is limited due to their Gram-positive cell wall (Bourgade et al., 2021). Currently, two usually used transformation methods for introducing foreign DNA into acetogens are electroporation and conjugation. The highly efficient electroporation of acetogens is usually described as treating early-log phase cells with cell-wall-weakening agents such as glycine or DL-threonine and osmotic stabilizers like sucrose; and then collecting and preparing the cells into competent cells; after that, electroporation is performed (Köpke et al., 2010; Kita et al., 2013; Leang et al., 2013; Straub et al., 2014; Shin et al., 2019; Zhao et al., 2019). The transformation efficiency varies depending on the species of the host cell and the type of plasmids, ranging from approximately 101–105 CFU per μg DNA (Zhao et al., 2019; Moreira et al., 2023). The DNA transfer by conjugation involves a donor bacterial strain such as Escherichia coli to transfer single-stranded DNA to the acceptor cells through cell-to-cell contact. Conjugation is applied in C. autoethanogenum (Mock et al., 2015; Nagaraju et al., 2016) and Clostridium carboxidivorans (Cheng et al., 2019). It is worth noting that thermophilic acetogen T. kivui naturally took up plasmid DNA in exponential growth phase, with a transformation frequency of up to 3.9 × 10−6 (Basen et al., 2018).
The native restriction modification (RM) systems are widespread in acetogens, which is a barrier to acetogen transformation. RM systems consist of restriction endonucleases and corresponding methyltransferases. The endonucleases recognize and cut specific DNA sequences, while the methyltransferases methylate the corresponding sequences in the host genome to protect it from cleavage (Vasu and Nagaraja, 2013). Among the four types of RM systems identified in bacteria, types I, II, and III target and cleave unmethylated DNA, while type IV recognizes DNA with foreign methylation patterns. For hosts with types I, II, or III RM systems, to circumvent the protective systems, plasmids can be methylated prior to transformation. One feasible approach is to express the native methyltransferases of the host in E. coli to create a methylation strain (Yasui et al., 2009). An example is the transformation of M. thermoacetica, where three native methyltransferases were expressed in E. coli to methylate the plasmid in order to counter the type I and type II RM systems of M. thermoacetica (Kita et al., 2013). RM systems in acetogens are often species-specific. Updated information of identified restriction endonucleases and methyltransferases is available in REBASE database (Roberts et al., 2023). In addition, conjugation is believed to allow partial evasion of the host restriction-modification barriers (Purdy et al., 2002; Cheng et al., 2019). Some acetogens like C. autoethanogenum have type IV RM systems, and plasmids constructed from dam+/dcm+ E. coli strains like DH5α will be recognized and degraded (Woods et al., 2019). To prevent this, plasmids can be constructed in E. coli strains with different dam/dcm background. One successful example is the transformation of Clostridium sp. AWRP, which has never succeeded in electroporation with dam+/dcm+ DNA, whereas achieved transformation efficiency of 3.2 × 102 CFU/μg DNA with dam−/dcm− DNA (Kwon et al., 2024). In addition to electroporation, the problem is also present in conjugative transfer. When C. autoethanogenum was the recipient, the commonly used conjugative donor strain E. coli CA434 (dam+/dcm+) exhibited a conjugation efficiency of 1.05 × 10−7 per donor cell. While the donor strain of E. coli sExpress which was obtained by introducing R-factor R702 into E. coli NEB Express strain (dam+/dcm−) had an efficiency of 7.94 × 10−5 per donor cell (Woods et al., 2019).
3.2 Plasmids for metabolic engineering in acetogens
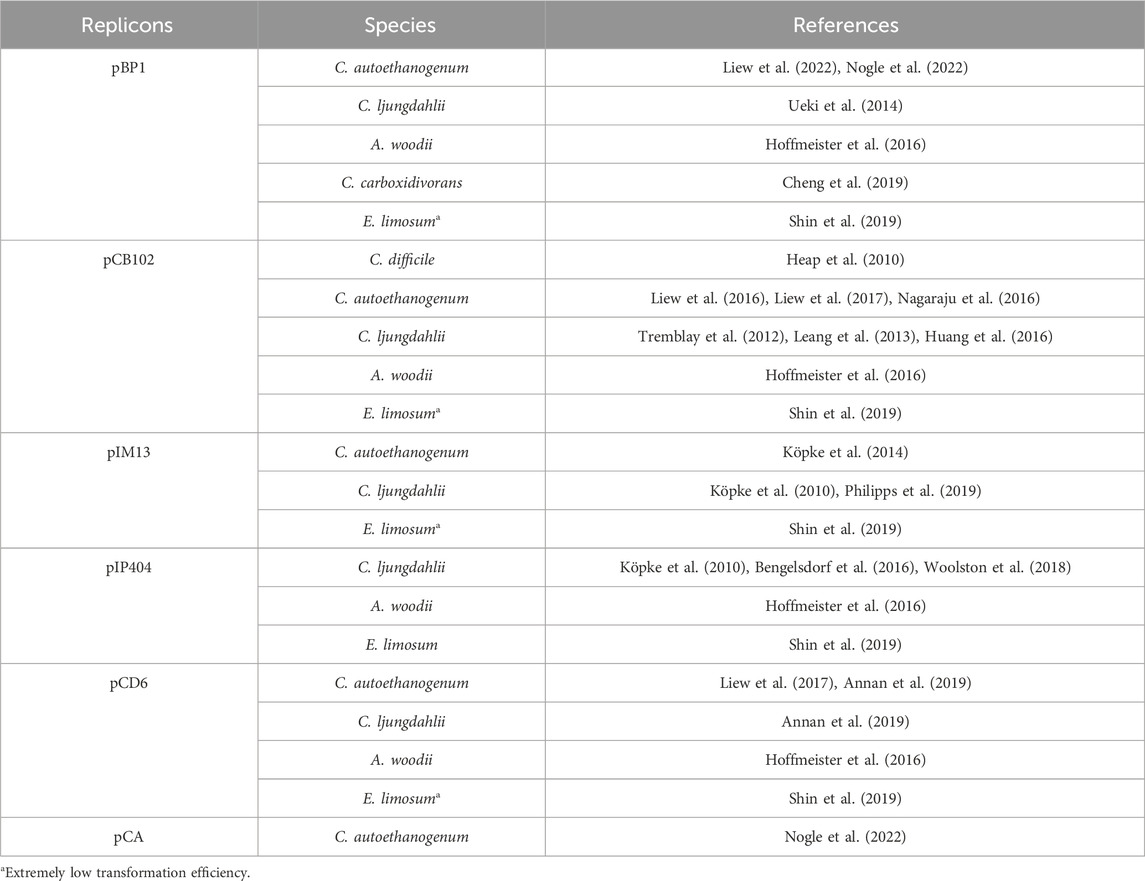
Genetic manipulation of acetogens inevitably involves the introduction of foreign DNA, which requires plasmid vectors. Acetogens are Gram-positive bacteria, whereas recombinant plasmids are typically constructed and amplified in E. coli and then “shuttled” into the target acetogens. Therefore, shuttle vectors must have both Gram-positive and Gram-negative replicons. The Gram-positive replicons are often derived from natural Gram-positive bacterial plasmids, such as pBP1 from Clostridium botulinum (van Treeck et al., 1981), pCB102 from Clostridium butyricum (Collins et al., 1985), pIM13 from Bacillus subtilis (Monod et al., 1986), pIP404 from Clostridium perfringens (Garnier and Cole, 1988), and pCD6 from Clostridium difficile (Minton et al., 2004). Recently, a low-copy native plasmid pCA from C. autoethanogenum was characterized and applied in heterologous expression, which may further facilitate physiological study and metabolic engineering of acetogens (Nogle et al., 2022). Table 1 shows a list of the Gram-positive replicons in shuttle plasmids that have been applied in acetogens. In certain cases, plasmids do not require replication and stable existence within the acetogen host. These plasmids, known as suicide vectors, have Gram-negative but not Gram-positive origins. Hence, they can be constructed and amplified in E. coli and transformed into acetogens for genomic editing (Tremblay et al., 2012). The Gram-negative replicons that are commonly used in E. coli-Acetogen shuttle vectors are ColE1, p15a, and pUC (Heap et al., 2009). In addition, antibiotic resistance genes including ermB, catP, and tetA are most extensively used selection markers for recombinant screening of acetogens. For thermophilic acetogens like M. thermoacetica, utilizing an auxotrophic mutant as a screening tool may be an optimal choice (Kita et al., 2013; Basen et al., 2018). In addition, the thermostable kanamycin resistance gene derived from Streptococcus faecalis (Trieucuot and Courvalin, 1983) has been used in thermophilic acetogens, including M. thermoacetica (Iwasaki et al., 2013) and T. kivui (Basen et al., 2018).
In efforts to improve the availability of shuttle plasmids tailored for clostridial hosts and specific applications, the pMTL80000 modular plasmid system was designed (Heap et al., 2009). This system facilitates the versatile construction of Clostridium-E. coli shuttle plasmids by integrating standardized modules comprising Gram-positive and Gram-negative replicons, selection markers, and other functional modules. The plasmid series has been extensively applied in acetogens, including Clostridium species such as C. autoethanogenum (Köpke et al., 2014), C. ljungdahlii (Ueki et al., 2014), and C. carboxidivorans (Cheng et al., 2019), as well as other acetogen species like A. woodii and Eubacterium limosum (Hoffmeister et al., 2016; Shin et al., 2019).
3.3 Genome editing tools applied in acetogens
It is feasible to introduce genes into the hosts for overexpressing endogenous genes or heterologous metabolic pathways using replicable plasmids. However, maintaining plasmids for gene expressions in the host cells requires the use of antibiotics, which is unfavorable for industrial production. Moreover, there is a concern regarding the potential spread of resistance markers into the environment. In this context, several genome editing tools allowing for gene disruption and fragment insertion in acetogens have been developed (Figure 3; Table 2).
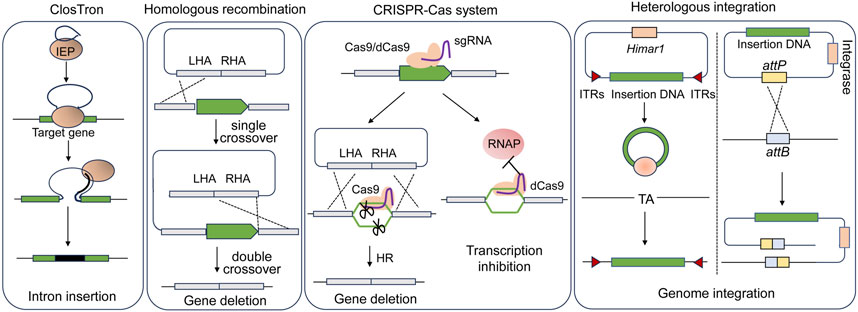
Figure 3. Scheme of genome engineering tools applied in acetogens. LHA, left homologous arm; RHA, right homologous arm; IEP, intron encoded protein; RNAP, RNA polymerase; ITRs, inverted terminal repeats.
3.3.1 ClosTron
The ClosTron, a widely applied gene disruption tool in Clostridium species, utilizes the group II Ll.LtrB intron derived from Lactococcus lactis (Heap et al., 2007; Heap et al., 2010). Facilitated by the reverse transcription activity of the intron-encoded protein LtrA, the intron integrates into the target genomic site through an RNA-mediated retrohoming mechanism (Karberg et al., 2001). To facilitate the selection of the mutants, antibiotic resistance genes are designed as a retrotransposition-activated selectable marker (RAM). Once the intron is inserted, the genes regain activity, enabling the identification of mutants through corresponding antibiotic selection (Zhong et al., 2003). ClosTron has been successfully employed in the study of acetogens, such as C. difficile (Heap et al., 2010), C. autoethanogenum (Mock et al., 2015; Liew et al., 2016; Marcellin et al., 2016; Liew et al., 2017), and C. ljungdahlii (Bengelsdorf et al., 2016). For instance, the three putative CODH genes, cooS1, cooS2 and acsA in C. autoethanogenum, were individually disrupted using ClosTron, confirming that only acsA is essential for the autotrophic growth (Liew et al., 2016). Despite its convenience in gene disruption, ClosTron does have limitations. The efficiency significantly decreases when the length of the inserted fragment exceeds 1 kb (Plante and Cousineau, 2006). Additionally, ClosTron is not suitable for studying the function of specific genes in a polycistronic gene cluster, as the insertion of the intron simultaneously silences downstream genes in the cluster (Kuehne et al., 2019; Wang et al., 2023).
3.3.2 Homologous recombination
With the improvement of transformation efficiency, gene deletion was demonstrated to be feasible by using suicide plasmids through homologous recombination (HR) in acetogens. Early attempts at HR-based gene deletion were carried out in C. ljungdahlii, resulting in successful deletion of the rnfAB (Tremblay et al., 2012) and the adhE1 and adhE2 genes that encode bi-functional aldehyde/alcohol dehydrogenases (Leang et al., 2013). Furthermore, to streamline mutant screening and allow for reusing selection markers, the counterselection marker catP and the Cre-lox system was adopted to eliminate the plasmid backbone (Ueki et al., 2014). Using this system, the butyrate synthesis pathway was integrated into the C. ljungdahlii genome. The pyrE and pyrF genes, essential for pyrimidine synthesis can serve as both positive and negative selection markers. The uracil phosphoribosyltransferase and orotate phosphoribosyltransferase encoded by pyrE and pyrF, respectively, can convert non-toxic 5-fluoroorotic acid (5-FOA) into toxic 5-fluoro-dUMP. Uracil auxotrophic strains generated by disrupting either pyrE or pyrF can be screened by the addition of uracil and 5-FOA. The auxotrophic mutant, when complemented with pyrE or pyrF through plasmids or genome integration, can be easily isolated in uracil-free medium. This approach has been successfully employed not only in mesophilic acetogens (Liew et al., 2017; Schoelmerich et al., 2018; Westphal et al., 2018; Wiechmann et al., 2020) but also in thermophilic acetogens such as M. thermoacetica and T. kivui (Kita et al., 2013; Basen et al., 2018), for which the antibiotics is not always working.
3.3.3 CRISPR-Cas
In recent years, the CRISPR-Cas gene editing system has emerged as an efficient, targeted, and scarless gene editing technique that is widely used. The application of CRISPR-Cas gene knockout systems and their derivatives has been proved successful in acetogens.
The Type II CRISPR-Cas9 system from Streptococcus pyogenes is most extensively applied (Cong et al., 2013). The Cas9 nuclease of the system is targeted by a short single guide RNA (sgRNA) and performs a double-stranded cleavage on the target DNA, which introduces a double-stranded break (DSB) on the chromosome. DSBs are repaired through homologous recombination using simultaneously introduced donor DNA as the template that contains complementary sequences to both the upstream and the downstream regions of the DSBs. In Clostridium lacking a representative recombination repair system, Cas9 is thought to function through a counterselection mechanism, where cells that undergo gene editing via occasional recombination with donor DNA survive, while unedited cells die from DSBs introduced by Cas9 (Joseph et al., 2018). The CRISPR-Cas9 gene editing tool, which utilizes a constitutive promoter to drive cas9 expression, has been successfully applied in C. ljungdahlii (Huang et al., 2016). However, conducting transformant screening and mutant screening in a single step has led to a low overall rate of positive outcomes.
In such cases, utilizing inducible promoters to control the expression of cas9 is a more favorable approach. This approach divides transformant screening and mutant screening into two steps, allowing for the screening process to be conducted in the absence or presence of inducers. This ensures that a sufficient number of colonies can be obtained in each screening process. For example, in C. autoethanogenum, a modified anhydrotetracycline-inducible promoter PIPL12 was applied to control the expression of cas9, achieving an editing efficiency of over 50% (Nagaraju et al., 2016). Similarly, the anhydrotetracycline-inducible promoter PtetO1 was employed to control cas9 expression in E. limosum, resulting in 100% editing efficiency upon induction (Shin et al., 2019). A theophylline-induced riboswitch-based system was able to efficiently control the expression of cas9 from translational level in several clostridial species (Cañadas et al., 2019). The efficiency of the Ribo-Cas system has been validated in C. autoethanogenum (Dykstra et al., 2022).
The Cas12a, also known as Cpf1, from Francisella novicida has also been successfully utilized for gene editing in C. ljungdahlii (Zhao et al., 2019). Cas12a recognizes a PAM sequence of -TTN- that is suitable for genomes with low GC content. In contrast to Cas9, the CRISPR-Cas12a system is preferred for multi-gene editing due to its requirement of only a single promoter and terminator to express multiple spacers and repeats (Zetsche et al., 2017). Patinios et al. (2021) developed a Self-splicing Intron-Based Riboswitch system to tightly control the expression of FnCas12a (SIBR-Cas) and the system has been employed in C. autoethanogenum for gene deletion (Dykstra et al., 2022).
CRISPR-Cas systems offer alternative methods for genetic modification in addition to the conventional double-strand break-homology directed repair (DSB-HR) gene editing process. The binding of deactivated Cas9 (dCas9) or DNase-deficient Cas12a (ddCas12a) to the target sites disrupts the transcription process of RNA polymerase, leading to decreased expression level of the target genes. Referred to as CRISPR interference (CRISPRi), this system has been effectively employed for gene knockdown in acetogens (Woolston et al., 2018; Shin et al., 2019; Zhao et al., 2019; Fackler et al., 2021a). For example, employing ddCas12a for CRISPRi in C. ljungdahlii led to a notable decrease of 88%–99% in the expression levels of multiple target genes (Zhao et al., 2019). Targeting the budA gene in C. autoethanogenum using dCas9 and various gRNAs resulted in a significant 75% reduction in 2,3-butanediol production (Fackler et al., 2021a). Furthermore, Xia et al. (2020) developed the Target-aid CRISPR system using deactivated Cas9 in conjunction with cytidine deaminase to facilitate precise C to T mutations at targeted sites.
Recently, the endogenous Type I-B CRISPR-Cas systems in A. woodii and C. autoethanogenum have been identified, characterized, and successfully employed for gene knockout in these organisms (Poulalier-Delavelle et al., 2023). The CRISPR-Cas system from A. woodii, with a putative PAM sequence of -CCA-, has been used to knockout the pyrE, pheA, and hsdR1 genes, as well as for the integration of the reporter gene FAST at the pheA locus. Similarly, the endogenous CRISPR-Cas system of C. autoethanogenum, with a putative PAM sequence of -TCA-, was employed to successfully knockout pyrE.
3.3.4 Heterologous integration
With regard to the introduction of large DNA fragments, targeted genomic heterologous integration has been achieved through homologous recombination based on single crossover in C. ljungdahlii (Ueki et al., 2014). However, this approach carries the potential risk of a second recombination between the homologous regions that mediate the single-crossover. The CRISPR-Cas system has shown high efficiency in gene deletion in acetogens, yet the insertion of large DNA fragments into the chromosome is challenging due to the low efficiency of homologous recombination in acetogens. Alternatively, heterologous integration can be achieved through non-targeted transposon mutagenesis, wherein transposons carrying foreign DNA are randomly inserted into the host genome. One commonly used transposon is the mariner-type Himar1 derived from Haematobia irritans, which is not species-specific and can be used in both prokaryotic and eukaryotic hosts (Lampe et al., 1996; Fu et al., 2008; Cartman and Minton, 2010). The Himar1 transposase recognizes inverted terminal repeats (ITRs) at both ends of the transposons and inserts them randomly into a -TA- target site in the genome. The Himar1 transposase system, controlled by a xylose-inducible promoter, has successfully integrated a 5-kbp acetone synthesis gene cluster into the C. ljungdahlii genome, thereby enabling the production of acetone and isopropanol (Philipps et al., 2019).
The attachment/integration (Att/Int) system, facilitated by phage serine integrases, enables precise DNA rearrangement through the phage attachment (attP) and bacterial attachment (attB) sites in the chromosome, which is a powerful tool for rapid chromosomal integration of large DNA fragments. A phage serine integrase-mediated site-specific heterologous integration system has been implemented in C. ljungdahlii (Huang et al., 2019). Initially, the attB site from bacteriophage ФCD27 was inserted into the genome using CRISPR-Cas9. Subsequently, in the modified strain, an exogenous plasmid containing the ФCD27 integrase and the attP site was integrated at the attB site. The effectiveness of this system was further validated through the integration of a 9.1-kbp exogenous butyrate synthesis pathway.
4 Achievements of metabolic engineering in acetogens
The continuous advancement of genetic tools promotes the use of metabolic engineering approaches to accelerate the development of acetogens as cell factories. Extensive efforts have been dedicated to enhancing both C1 fixation efficiency and the production of native biochemicals. The improvement of C1 fixation efficiency is primarily achieved either through the overexpression of specific enzymes in the Wood-Ljungdahl pathway (WLP) such as the selective overexpression of the four THF-dependent enzymes for the processing of formate in A woodii (Straub et al., 2014) or the introduction of alternative carbon-fixation pathways such as the glycine synthase-reductase pathway into E. limosum (Song et al., 2020). It should be noted that these results were obtained through batch fermentation, which may perform differently under continuous cultivation conditions (Pavan et al., 2022). On the other hand, the enhancement of natural product synthesis mainly revolves around the production of acetate, ethanol, and 2,3-butanediol. This was achieved by overexpressing the enzymes involved in the synthesis pathways of the desired products or by disrupting competing pathways (Banerjee et al., 2014; Straub et al., 2014; Liew et al., 2017). Furthermore, acetogens have been explored as a chassis for the production of non-natural biochemicals by introducing exogenous genes, significantly expanding their product range to include dozens of biochemicals (Figure 4; Table 3) (Fackler et al., 2021b). Notably, several of these products have demonstrated industrially relevant performance levels, with production rates in the range of g/L/h and titers at the tens of g/L level. This section will focus on the exploration of producing non-natural products through metabolic engineering of acetogens.
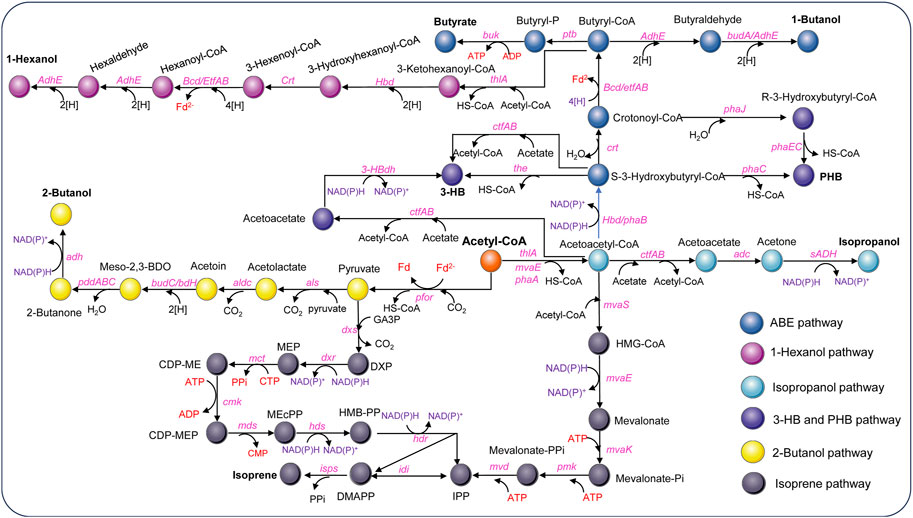
Figure 4. Metabolic pathways for non-natural product synthesis using acetyl-CoA as the starting point in acetogens [Modified from Jin et al. (2020)]. Adc, acetoacetate decarboxylase gene; adhE, aldehyde/alcohol dehydrogenase gene; adh, alcohol dehydrogenase gene; aldc, alpha-acetolactate decarboxylase gene; als, acetolactate synthase gene; bcd, butyryl-CoA-dehydrogenase gene; bdhA, butanol dehydrogenase; buk, butyrate kinase gene; budC/bdH, meso-2,3-butanediol dehydrogenase gene; cmk, CDP-ME kinase gene; crt, crotonase gene; ctfAB, CoA transferase A and B gene; dxs, DXP synthase gene; dxr, DXP reductoisomerase gene; etfAB, genes encoding electron transferring protein A&B; hbd, 3-hydroxybutyryl-CoA dehydrogenase gene; 3-HBdh, 3-HB dehydrogenase gene; hdr, HMB-PP reductase gene; hds, HMB-PP synthase gene; idi, isopentenyl pyrophosphate isomerase gene; isps, isoprene synthase gene; mct, MEP cytidylyltransferase gene; mds, MEcPP synthase gene; mvaE, acetyl-CoA acetyltransferase/HMG-CoA reductase gene; mvaK, mevalonate kinase gene; mvaS, HMG-CoA synthase gene; mvd, mevalonate diphosphate decarboxylase gene; pddABC, diol/glycerol dehydratase gene; pfor, pyruvate ferredoxin oxidoreductase; phaA, 3-ketothiolase; phaB, NAD(P)H-dependent acetoacetyl-CoA reductase gene; phaC/phaEC, encoding polyhydroxyalkanoate synthase; phaJ, (R)-enoyl-CoA hydratase gene; pmk, phosphomevalonate kinase gene; ptb, phosphotransbutylase gene; sADH, primary-secondary alcohol dehydrogenase gene; the, thioesterase gene; thlA, acetyl-CoA acetyltransferase/thiolase gene; Metabolites: HMG-CoA, hydroxymethylglutaryl-CoA; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; CDP-MEP, CDP-ME2-phosphate; MEcPP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMB-PP, 4-hydroxy-3-methylbut-2-enyl diphosphate.
4.1 Acetone and isopropanol
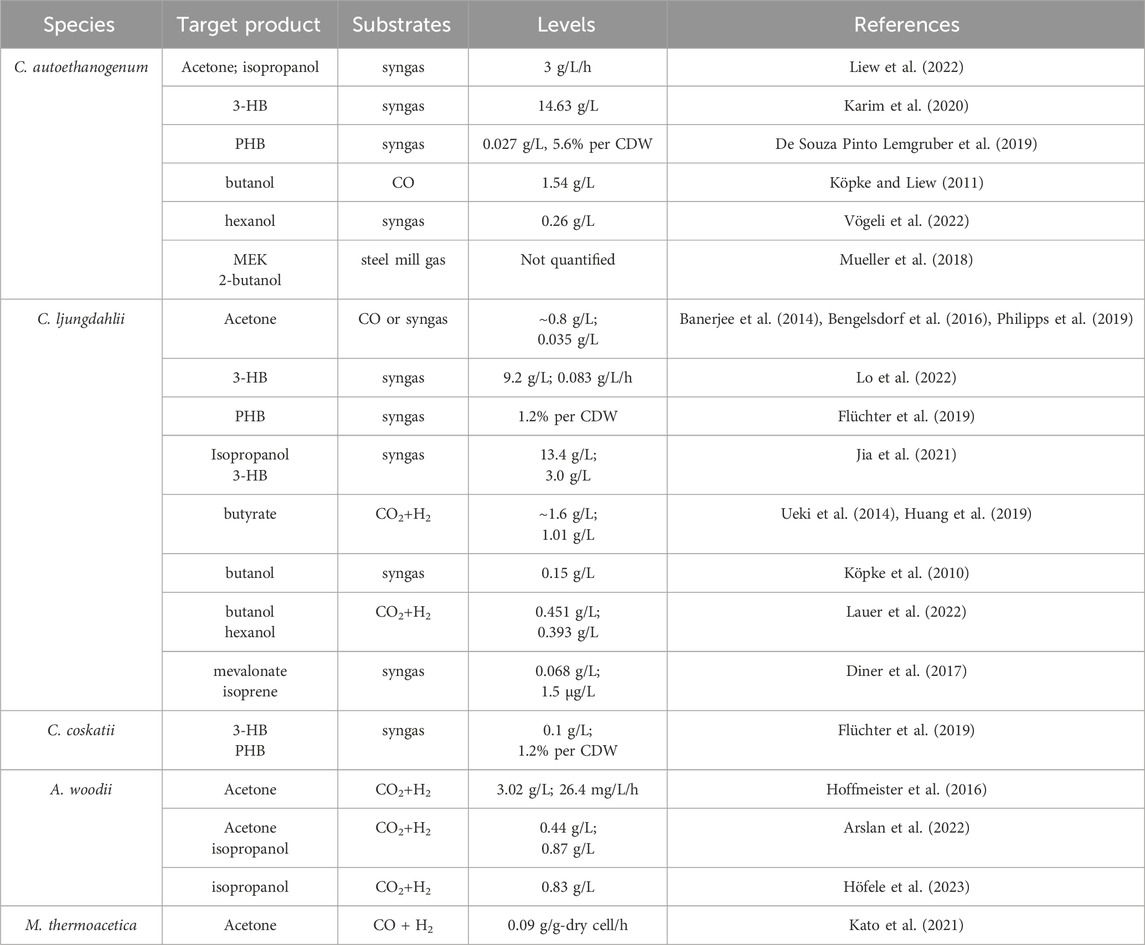
Acetone, an important commodity chemical and feedstock for poly (methyl methacrylate) production can be biosynthesized by ABE fermentation. The metabolic pathway involved has been well elucidated (Jones and Woods, 1986). Initially, two molecules of acetyl-CoA are converted to acetoacetyl-CoA by a thiolase (ThlA), and then CoA is transferred to acetate by a CoA transferase (CtfAB) to generate acetoacetate and another acetyl-CoA, and finally acetoacetate is decarboxylated by an acetoacetate decarboxylase (Adc) to generate acetone. However, ABE fermentation typically utilizes sugar as a substrate, leading to growing interest in C1 gas fermentation by acetogens. Acetone is not a natural product of any known acetogens. Proof-of-concept production of acetone has been achieved by plasmid-based heterologous expression of acetone synthesis pathways from Clostridium acetobutylicum in several acetogens, such as C. aceticum (Schiel-Bengelsdorf and Dürre, 2012), C. ljungdahlii (Banerjee et al., 2014), and A. woodii (Hoffmeister et al., 2016; Arslan et al., 2022). Studies have shown that acetone can be reduced to isopropanol by an endogenous primary-secondary alcohol dehydrogenase (sADH) in C. autoethanogenum (Köpke et al., 2014) and C. ljungdahlii (Bengelsdorf et al., 2016). The integration of acetone synthesis pathway into the genome of C. ljungdahlii resulted in the production of 0.6 mM acetone and 2.4 mM isopropanol by the engineered strain (Philipps et al., 2019). Instead, Jia et al. (2021) introduced an artificial isopropanol-producing pathway via an expression plasmid incorporating thiolase, CtfAB, Adc, and sADH from Clostridium species into C. ljungdahlii. The best-performing recombinant strain was able to produce 13.4 g/L of isopropanol in continuous gas fermentation. In addition, plasmid-based heterologous expression of the genes thlA-ctfAB-adc-sadh in A. woodii led to a maximum isopropanol production of 13.87 mM using CO2 and H2 as the carbon and energy source (Höfele et al., 2023). Recent advancements by LanzaTech have demonstrated commercial-ready levels of acetone and isopropanol production using genomically integrated strains, with continuous production rates reaching up to ∼3 g/L/h (Liew et al., 2022). Life cycle analysis confirmed a negative carbon footprint for their products. Additionally, Kato et al. successfully integrated a thermophilic acetone production pathway into the genome of M. thermoacetica, resulting in acetone yields of 0.04–0.09 g acetone/g-dry cell/h, making a notable contribution to the field of metabolic engineering of thermophilic acetogens (Kato et al., 2021).
4.2 3-HB and PHB
3-Hydroxybutyrate (3-HB) is a chiral bioproduct of interest with diverse applications in the synthesis of fine chemicals, pharmaceuticals, biofuels, and bioplastics, particularly polyhydroxybutyrate (PHB) (Tokiwa and Ugwu, 2007). 3-HB can be synthesized from acetyl-CoA through a variety of pathways. There are two main pathways proposed to generate 3-HB in acetogens (Figure 4). Both pathways involve the action of a thiolase to combine two acetyl-CoA molecules, producing acetoacetyl-CoA. However, they differ in subsequent steps. In the Hbd pathway, (S)-3-hydroxybutyryl-CoA dehydrogenase (Hbd) reduces acetoacetyl-CoA to (S)-3-hydroxybutyryl-CoA, which is then converted to 3-HB by a thioesterase catalyzed CoA removal. In contrast, the CtfAB/3-HBdh pathway initially utilizes CtfAB to convert acetoacetyl-CoA to acetoacetate by CoA transfer to acetate. Acetoacetate is then reduced by the 3-HB dehydrogenase (3-HBdh), resulting in the formation of 3-HB. For the heterologous production of 3-HB, a novel CtfAB/3-HBdh pathway consisting of the genes thlA and ctfA/B from C. acetobutylicum, as well as bdhA encoding a 3-HBdh from Clostridioides difficile 630 was introduced into C. coskatii and C. ljungdahlii through an expression plasmid (Flüchter et al., 2019). The engineered strain of C. coskatii autotrophically grown on syngas produced 0.1 g/L of 3-HB. The recombinant C. ljungdahlii strain, however, did not produce any detectable amounts of 3-HB neither under heterotrophic nor under autotrophic growth conditions. Jones et al. (2016) proposed that acetoacetate can undergo native conversion to 3-HB in C. ljungdahlii, as their acetone-generating strain, containing all the necessary genes for the CtfAB/3HBdh pathway except for 3-HBdh itself, produced significant levels of 3-HB. Subsequently, an endogenous 3-HB dehydrogenase (3-HBdh) was identified in C. ljungdahlii, which could convert acetoacetate to 3-HB in an engineered strain for isopropanol production (Jia et al., 2021). Furthermore, the introduction of the bdhA gene from C. difficile 630 via an expression plasmid into the isopropanol-producing strain facilitated the efficient co-production of isopropanol, 3-HB, and ethanol, resulting in a 3-HB titer of 3 g/L in continuous gas fermentations (Jia et al., 2021). Through in vitro prototyping and rapid biosynthetic enzyme optimization from Clostridium, the plasmid-based expression of an Hbd pathway in C. autoethanogenum demonstrated the highest performance in 3-HB production (Karim et al., 2020). In optimized fermentations, the titer of 3-HB reached 14.6 g/L, with production rates exceeding 1.5 g L−1 h−1 in a continuous system. Recently, Lo et al. (2022) identified a native (S)-3-hydroxybutyryl-CoA dehydrogenase, Hbd2 in C. ljungdahlii. In conjunction with the integration of heterologous thiolase gene, atoB from E. coli and ctfAB from C. acetobutylicum into the pyrE locus, plasmid based overexpression of both the native hbd2 and thl2 from Clostridium kluyveri significantly improved yields of 3-HB on syngas, resulting in a titer of 9.2 g/L (Lo et al., 2022).
Poly-3-hydroxybutyrate (PHB) is a biodegradable plastic naturally produced by Cupriavidus necator and microbes from Alcaligenes, Azotobacter, Bacillus, Rhizobium, and Pseudomonas (Bhagowati et al., 2015; Jin et al., 2020). PHB is synthesized through the polymerization of 3-hydroxybutyryl-CoA by PHA synthase. For the production of PHB, the PHB synthesis pathway from C. necator was expressed in C. autoethanogenum under control of the native WLP promoter. Through bioprocess optimization, a PHB yield of 5.6% per CDW with a titer of 0.027 g/L was obtained on syngas in steady-state chemostat cultures (dilution rate of 1 day−1) (De Souza Pinto Lemgruber et al., 2019). In addition, as a proof of concept, production of PHB was also achieved using recombinant C. coskatii and C. ljungdahlii strain expressing a Clostridial PHB synthesis pathway comprising the genes thlA, hbd, crt, phaJ, and phaEC (Flüchter et al., 2019) (Figure 4). The recombinant strains of both C. coskatii and C. ljungdahlii synthesized autotrophically 1.2% PHB per CDW.
4.3 Butyrate and butanol
Butyrate and butanol are also non-natural products that cannot be produced by wild-type C. ljungdahlii and C. autoethanogenum. In order to redirect carbon and electron flow to the production of butyrate, a fragment containing genes of thlA, hbd, crt, bcd, etfAB, ptb, and buk from C. acetobutylicum was integrated into the putative pta promoter region of C. ljungdahlii. Meanwhile, PTA-dependent acetate synthesis and a gene (Clju_c39430) encoding a homolog of CoA transferases, were disrupted. The final recombinant strain produced approximately 1.6 g/L of butyrate on CO/CO2 (Ueki et al., 2014). In addition, to validate the efficiency of the phage serine integrase-mediated genome engineering, a butyrate production pathway from C. acetobutylicum was integrated into C. ljungdahlii, enabling the production of 1.01 g/L of butyrate by fermenting syngas (Huang et al., 2019).
The production of butanol by engineered acetogens presents challenges due to requirement of additional reducing equivalents for the conversion of butyryl-CoA into butanol. In a demonstration of principle, a plasmid containing most of the C. acetobutylicum butanol synthesis pathway, encompassing thlA, hbd, crt, bcd, adhE, and bdhA genes (Figure 4) under control of the phosphotransbutyrylase operon promoter Pptb was introduced into C. ljungdahlii and a maximum amount of about 0.15 g/L butanol was detected in batch cultures of the engineered strain during mid-growth phase (Köpke et al., 2010). Another approach was carried out in C. autoethanogenum, which also expressed the C4 pathway of C. acetobutylicum including thlA, hbd, crt, bcd, etfAB genes (Köpke and Liew, 2011). The highest butanol production measured in the recombinant strain culture grown on CO was 1.54 g/L. In a recent study, an artificial biosynthesis pathway for butanol and hexanol was developed, incorporating 13 genes from C. kluyveri and C. acetobutylicum and integrated into the genome of C. ljungdahlii. In 2-L fermentations with CO2 and H2 continuous supply, the recombinant strain produced 451 mg/L of butanol, 122 mg/L of hexanol, as well as 199 mg/L of butyrate and 46 mg/L of caproate (Lauer et al., 2022). Vögeli et al. (2022) optimized and implemented the reverse β-oxidation pathway for the synthesis of C4-C6 acids and alcohols. Then engineered C. autoethanogenum strain was able to produce 1-hexanol from syngas, achieving a titer of 0.26 g/L in a 1.5 L continuous fermentation.
When utilizing 2,3-butanediol (2,3-BDO) as the substrate, the biosynthesis of 2-butanol involves two reactions: meso-2,3-BDO is converted to 2-butanone (MEK) by propanediol/glycerol dehydratase, and further reduced to 2-butanol by alcohol dehydrogenase (Adh) using NADPH. Acetogens such as C. ljungdahlii and C. autoethanogenum are capable of producing 2,3-BDO. However, the production of MEK and 2-butanol requires (R,S)-2,3-butanediol, not (R,R)-2,3-butanediol. Nevertheless, through the use of recombinant microorganisms adapted to overexpress crucial enzymes from Klebsiella species in the MEK and 2-butanol biosynthesis pathways, the production of MEK and 2-butanol was presumptively achieved (Mueller et al., 2018).
4.4 Isoprene
Isoprene is a hemiterpene (C5H8), used primarily in the production of cis-1,4-polyisoprene, the main component of natural rubber, and is also used in the creation of styrene-isoprene-styrene block copolymers, commonly employed in thermoplastic rubber and adhesives. Isopentenyl-pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) are the initial precursors for all terpenoid biosynthesis, including isoprene (Schrader and Bohlmann, 2015). There are two distinct pathways existing for the generation of IPP and DMAPP: the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, prevalent in most prokaryotes and starts with the condensation of pyruvate and glyceraldehyde-3-phosphate (GA3P), and the mevalonic acid (MVA) pathways, found in most eukaryotes, archaea, and some bacteria, which generally starts with the condensation of two acetyl-CoA molecules (Schrader and Bohlmann, 2015). In an effort to enable isoprene production from syngas, the MVA pathway was introduced into C. ljungdahlii, along with the overexpression of IPP isomerase and isoprene synthase (ipsS). However, the titer of isoprene during syngas fermentation only reached approximately 1.5 μg/L (Diner et al., 2017). Regrettably, the process of synthesizing isoprene from C1 gas is highly energy-intensive and requires additional ATP, which poses a significant challenge for acetogens with limited capacity for ATP generation. Therefore, efficient synthesis of isoprene is only possible by coupling it with other energy-generating pathways.
5 Strategies to overcome challenges in metabolic engineering of acetogens
5.1 Strategies to overcome energy limitations in acetogens
As mentioned above, efforts to genetically modify acetogens for the production of non-natural products have yielded only modest success, with low production efficiency that does not meet the demands of commercial scale production, except a few cases. This may be attributed to insufficient energy supply at autotrophic conditions, as acetogens thrive at the thermodynamic limit of life, and their ATP generation process is inherently impeded (Schuchmann and Müller, 2014). As a result, exogenous metabolic pathway modules introduced through genetic modification can only function sub-optimally under such energy constraints.
5.1.1 Improving the energetic efficiency of alcohol production through AOR route
One way to solve this problem is using an energetic advantage pathway (Charubin et al., 2018). A. woodii does not produce ethanol when growing on H2 and CO2. However, when growing on glucose, it can synthesize ethanol in addition to acetic acid, indicating the presence of metabolic pathway from acetyl-CoA to ethanol. The energy metabolism model of A. woodii gives an explanation that during acetogenesis on H2 and CO2, the net ATP gain was 0.3 (moles of ATP per mole of acetic acid). However, if the metabolic pathway is expanded to include ethanol formation from acetyl-CoA, the ATP gain (moles of ATP per mole of ethanol) in A. woodii would be negative (Mock et al., 2015). In contrast, the acetogens of Clostridia such as C. autoethanogenum, C. ljungdahlii, and C. ragsdalei, produce ethanol in addition to acetic acid when growing on H2 and CO2, due to an energy saving pathway for ethanol formation. These acetogens have two possible routes for ethanol formation (Liu et al., 2020; Zhu et al., 2020). The first route involves direct reduction of acetyl-CoA to ethanol by the bifunctional aldehyde/alcohol dehydrogenase (AdhE), with acetaldehyde as an intermediate and NAD(P)H as the reducing agent. In this route, no ATP is produced through SLP. In the second route, acetate is produced first with ATP generation through SLP, followed by its reduction to acetaldehyde by the aldehyde:ferredoxin oxidoreductases (AOR) with Fd2−. Acetaldehyde is then reduced to ethanol with NAD(P)H. Compared with the AdhE pathway, the AOR pathway generates more ATP, which is more conducive to ethanol production (Liew et al., 2017). More interestingly, AORs demonstrate less substrate specificity, as experiments reveal their ability to reduce various organic acids such as propionic acid, n-butyric acid, isobutyric acid, and n-caproic acid to their corresponding aldehydes, which suggests that the energy-saving strategy via AOR can be used in the synthesis of a variety of non-natural products in acetogens, such as butanol (Perez et al., 2013; Nissen and Basen, 2019).
5.1.2 Bioprocess combination
Another strategy is to use consortia of different organisms (Molitor et al., 2017; Charubin et al., 2018; Katsyv and Müller, 2020). The autotrophic products of acetogens are primarily the C2 compounds acetate and ethanol, which are the substrates for chain elongation via the reverse β-oxidation pathway (Angenent et al., 2016). Through the coupling of syngas fermentation with the chain elongation bioprocess using C. kluyveri (the most widely studied chain-elongating microorganism), C1 gas (CO, CO2) has been converted into n-butyrate (C4), n-caproic acid (C6), and even n-caprylic acid (C8) (Diender et al., 2016; Richter et al., 2016; Fernández-Blanco et al., 2022). For instance, a co-culture of C. aceticum and C. kluyveri at near neutral pH were carried out with continuous syngas supply and maximum concentrations of n-butyrate and n-caproate of 7.0 and 8.2 g/L, respectively, have been yielded (Fernández-Blanco et al., 2022). However, one of the major challenges for this technology is the pH mismatch between the two types of microorganisms (Ganigué et al., 2016). Therefore, the two-step bioprocess in two different reactors is an alternative route to overcome energetic limitations (Gildemyn et al., 2017) and has achieved the production of more energy-dense products such as lipids through the integrated conversion of M. thermoacetica and Yarrowia lipolytica (Hu et al., 2016), malic acid through the combined bioprocess of C. ljungdahlii and Aspergilus oryzae (Oswald et al., 2016), and polyhydroxyalkanoates through bioconversion of C. autoethanogenum and a highly enriched PHA-accumulating biomass (Lagoa-Costa et al., 2017). Recently, recombinant production of Pseudomonas aeruginosa rhamnolipids in Pseudomonas putida KT2440 was achieved on A. woodii cultures grown chemo-autotrophically with CO2 and H2 (Widberger et al., 2024).
5.1.3 Renewable reducing power supply
As the reduction of CO2 with H2 or CO as reductants is energetically limited, a potential strategy to overcome the energy limitations in acetogens is the direct supply of electrons by renewable reducing power, such as electricity (Lee et al., 2022). The process that microbes utilize electrons from an electrode to convert CO2 into multi-carbon products is known as microbial electrosynthesis (MES). Acetogens such as C aceticum, M. thermoacetica, and C. ljungdahlii, have been reported to perform MES via extracellular electron transfer (Nevin et al., 2011). Recently, Zhang et al. (2023) reported microbial electrosynthesis of acetate from CO2 using an enrichment of saline homoacetogens from Atlantic North Sea sediments under hypersaline conditions. However, it is still poorly verified if the electrons are channeled into the cell via membrane-associated redox components, nanowires, or diffusible electron shuttles like H2, flavins, or quinones. For example, extracellular electron supply induced a significant metabolic shift of C. autoethanogenum from acetate to lactate and 2,3-butanediol fermentation on fructose. Electron mediators such as methyl viologen or neutral red are necessary to facilitate electron transfer from the electrode to the cells (Kracke et al., 2016). C. carboxidivorans showed electricity-enhanced fermentation with higher alcohol production and carbon efficiency compared to open-circuit fermentation. Electron mediators such as methyl viologen and biochar facilitated electricity-driven autotrophic CO2 fixation and increased carbon efficiencies in electricity-enhanced mixotrophic fermentations (Cheng et al., 2022). C. ljungdahlii was initially reported to be able to perform direct extracellular electron transfer but later studies suggested H2 mediation is the likely mode of electron uptake (Boto et al., 2023). Hence, more research is needed to explore the underlying mechanism of extracellular electron transfer.
In addition, approaches such as using C1 liquid feedstocks (Cotton et al., 2020; Flaiz et al., 2021; Litty and Müller, 2021; Neuendorf et al., 2021) and mixotrophy (Jones et al., 2016; Maru et al., 2018; Cheng et al., 2019) are also considered to be promising strategies to overcome energy limitations in acetogens (Lee et al., 2022). However, not all the acetogens can utilize methanol or formate. Meanwhile, methanol or formate tolerance is an issue to be addressed. For sugar co-feeding with gas substrates, carbon catabolite repression (CCR) of the WL pathway may be a problem. Possible catabolite repression or native regulatory mechanisms in mixotrophy should be better studied (Hong and Zeng, 2022).
5.2 Strategies for efficient optimization of metabolic flux
The synthesis of some non-natural products is thermodynamically favorable for acetogens, such as butyrate, acetone, and isopropanol. However, the production efficiency was still low in some earlier studies, which was caused by the limited metabolic flow.
5.2.1 Genome-scale metabolic modeling
The systematic description of carbon and energy metabolism of acetogens is not only the basis for our understanding of this ancient metabolic mode, but also an important tool to guide their engineering strategy.
The generation of high-yielding strains hinges on the precise combination of enzyme species involved in the metabolic pathway and their optimal expression. In-silico assays such as genome-scale metabolic models (GEMs) are valuable for the initial analysis and estimation of pathway yield and feasibility. For example, a genome-scale model of C. autoethanogenum was used to explore alternative ATP-generating pathways. Subsequent in silico simulations identified an arginine deiminase pathway for ATP production (Valgepea et al., 2017). The OptKnock can identify gene knockout strategies that are expected to enhance the synthesis of target products (Burgard et al., 2003). Chen and Henson performed in silico metabolic engineering studies using a genome-scale reconstruction of C. ljungdahlii metabolism combined with the OptKnock computational framework to identify gene knockouts that were predicted to enhance the synthesis of these native products and non-native products, introduced through insertion of the necessary heterologous pathways (Chen and Henson, 2016). Benito-Vaquerizo et al. reported a genome-scale, constraint-based metabolic model to describe growth of a co-culture of C. autoethanogenum and C. kluyveri on syngas. Community flux balance analysis was used to gain insight into the metabolism of the two strains, their interactions, and to uncover potential strategies for producing butyrate and hexanoate (Benito-Vaquerizo et al., 2020).
5.2.2 Cell-free systems
GEMs may sometimes misjudge pathway feasibility, probably due to the lack of experimental data integration. While in vivo pathway optimization with a design-build-test approach is a time-consuming and labor-intensive process. Cell-free systems, thus, can be beneficial for pathway prototyping and rapid optimization. A high-yielding batch cell-free expression (CFE) platform was developed and optimized using C. autoethanogenum (Krüger et al., 2020). It allows for the direct application of both circular and linear DNA templates to the CFE reaction for programmable protein synthesis. The CFE platform is expected to not only enhance the protein synthesis toolkit for synthetic biology but also facilitate the rapid screening and prototyping of gene regulatory elements in non-model, industrially relevant microbes. Karim et al. built a platform for in vitro prototyping and rapid optimization of biosynthetic enzymes (iPROBE) for cell design. Cell lysates of E. coli are enriched with biosynthetic enzymes by cell-free protein synthesis and then metabolic pathways are assembled in a mix-and-match fashion to assess performance. By screening 54 different cell-free pathways for 3-HB production and optimizing a six-step butanol pathway across 205 permutations, a strong correlation (r = 0.79) between cell-free and C. autoethanogenum cellular performance was observed. The best-performing pathway was scaled up, leading to an improvement in in vivo 3-HB production in Clostridium by 20-fold to 14.63 g/L (Karim et al., 2020). The iPROBE was also applied for optimization of acetone and isopropanol metabolic flux in C. autoethanogenum (Liew et al., 2022). To increase flux to acetone and eliminate unwanted byproducts, effector genes that cause 3-HB formation were firstly identified by iPROBE and then successively knocked out. Furthermore, iPROBE was used to demonstrate the strong dependence of acetone titer on the concentration of CtfAB by varying levels of the different acetone pathway enzymes. CtfAB was titrated at concentrations ranging from 0.05 to 1.50 μM with Thl and Adc constant at 0.5 μM, increasing acetone titer from the limit of detection to 74.6 mM. These recent results indicate that cell-free systems have become an important technique for metabolic engineering efforts in acetogens.
6 Conclusion and perspectives
In the current context of striving for carbon neutrality, acetogens hold significant potential for the sustainable production of fuels and chemicals. Their WLP enables them to efficiently convert CO2 into valuable products through fermentations, thereby mitigating carbon emissions and offering environmental benefits. The integration of synthetic biology and metabolic pathway engineering further enhances the industrial applications of acetogens. Currently, there is rapid progress in the synthetic biology and metabolic engineering of acetogens, with various metabolic components and genetic tools successfully employed across different acetogen species. Notably, CRISPR-Cas-based genetic modification techniques have been effectively utilized in several acetogen species, facilitating precise and scarless genome editing, and streamlining strain modification processes. By leveraging advanced genetic modification approaches, the incorporation of heterologous metabolic pathways has enabled the engineered acetogen strains to synthesize diverse non-natural products. Some products, such as acetone and isopropanol, have already achieved industrial-scale production, while others are undergoing laboratory research or feasibility assessments. An inherent challenge in utilizing acetogens for non-natural product synthesis lies in their limited energy gain. Strategies involving energy-efficient pathways, bioprocess integration, and microbial electrosynthesis, are being explored to address this constraint. Although notable advancements have been made in this area, continuous innovation in genetic tools is still needed for probing and manipulating acetogen metabolism, ultimately helping to overcome our dependence on the petrochemical industry.
Author contributions
J-ZZ: Writing–original draft. Y-ZL: Writing–original draft. Z-NX: Writing–original draft. H-PG: Writing–original draft. QZ: Writing–review and editing. L-CL: Writing–review and editing. F-LL: Writing–review and editing. X-QM: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (U22A20425, 32370039), Shandong Energy Institute (SEI S202104), and the Shandong Provincial Natural Science Foundation (ZR2023MC021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Angenent, L. T., Richter, H., Buckel, W., Spirito, C. M., Steinbusch, K. J. J., Plugge, C. M., et al. (2016). Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ. Sci. Technol. 50, 2796–2810. doi:10.1021/acs.est.5b04847
Annan, F. J., Al-Sinawi, B., Humphreys, C. M., Norman, R., Winzer, K., Köpke, M., et al. (2019). Engineering of vitamin prototrophy in Clostridium ljungdahlii and Clostridium autoethanogenum. Appl. Microbiol. Biotechnol. 103, 4633–4648. doi:10.1007/s00253-019-09763-6
Arslan, K., Schoch, T., Höfele, F., Herrschaft, S., Oberlies, C., Bengelsdorf, F., et al. (2022). Engineering Acetobacterium woodii for the production of isopropanol and acetone from carbon dioxide and hydrogen. Biotechnol. J. 17, e2100515. doi:10.1002/biot.202100515
Atsumi, S., Cann, A. F., Connor, M. R., Shen, C. R., Smith, K. M., Brynildsen, M. P., et al. (2008b). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311. doi:10.1016/j.ymben.2007.08.003
Atsumi, S., Hanai, T., and Liao, J. C. (2008a). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi:10.1038/nature06450
Banerjee, A., Leang, C., Ueki, T., Nevin, K. P., and Lovley, D. R. (2014). Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii. Appl. Environ. Microbiol. 80, 2410–2416. doi:10.1128/aem.03666-13
Basen, M., Geiger, I., Henke, L., and Müller, V. (2018). A genetic system for the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Appl. Environ. Microbiol. 84, e02210-17. doi:10.1128/AEM.02210-17
Bengelsdorf, F. R., Poehlein, A., Linder, S., Erz, C., Hummel, T., Hoffmeister, S., et al. (2016). Industrial acetogenic biocatalysts: a comparative metabolic and genomic analysis. Front. Microbiol. 7, 1036. doi:10.3389/fmicb.2016.01036
Benito-Vaquerizo, S., Diender, M., Parera Olm, I., Martins Dos Santos, V. A. P., Schaap, P. J., Sousa, D. Z., et al. (2020). Modeling a co-culture of Clostridium autoethanogenum and Clostridium kluyveri to increase syngas conversion to medium-chain fatty-acids. Comput. Struct. Biotechnol. J. 18, 3255–3266. doi:10.1016/j.csbj.2020.10.003
Bhagowati, P., Pradhan, S., Dash, H. R., and Das, S. (2015). Production, optimization and characterization of polyhydroxybutyrate, a biodegradable plastic by Bacillus spp. Biosci. Biotechnol. Biochem. 79, 1454–1463. doi:10.1080/09168451.2015.1034651
Birgen, C., Dürre, P., Preisig, H. A., and Wentzel, A. (2019). Butanol production from lignocellulosic biomass: revisiting fermentation performance indicators with exploratory data analysis. Biotechnol. Biofuels 12, 167. doi:10.1186/s13068-019-1508-6
Boto, S. T., Bardl, B., Harnisch, F., and Rosenbaum, M. A. (2023). Microbial electrosynthesis with Clostridium ljungdahlii benefits from hydrogen electron mediation and permits a greater variety of products. Green Chem. 25, 4375–4386. doi:10.1039/d3gc00471f
Bourgade, B., Minton, N. P., and Islam, M. A. (2021). Genetic and metabolic engineering challenges of C1-gas fermenting acetogenic chassis organisms. FEMS Microbiol. Rev. 45, fuab008. doi:10.1093/femsre/fuab008
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi:10.1016/j.bbabio.2012.07.002
Burgard, A. P., Pharkya, P., and Maranas, C. D. (2003). Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 284, 647–657. doi:10.1002/bit.10803
Cañadas, I. C., Groothuis, D., Zygouropoulou, M., Rodrigues, R., and Minton, N. P. (2019). RiboCas: a universal CRISPR-based editing tool for Clostridium. ACS Synth. Biol. 8, 1379–1390. doi:10.1021/acssynbio.9b00075
Cartman, S. T., and Minton, N. P. (2010). A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76, 1103–1109. doi:10.1128/aem.02525-09
Charubin, K., Bennett, R. K., Fast, A. G., and Papoutsakis, E. T. (2018). Engineering Clostridium organisms as microbial cell-factories: challenges & opportunities. Metab. Eng. 50, 173–191. doi:10.1016/j.ymben.2018.07.012
Chen, J., and Henson, M. A. (2016). In silico metabolic engineering of Clostridium ljungdahlii for synthesis gas fermentation. Metab. Eng. 38, 389–400. doi:10.1016/j.ymben.2016.10.002
Cheng, C., Li, W., Lin, M., and Yang, S. T. (2019). Metabolic engineering of Clostridium carboxidivorans for enhanced ethanol and butanol production from syngas and glucose. Bioresour. Technol. 284, 415–423. doi:10.1016/j.biortech.2019.03.145
Cheng, C., Shao, Y., Li, W., Liu, J., Liu, X., Zhao, Y., et al. (2022). Electricity-enhanced anaerobic, non-photosynthetic mixotrophy by Clostridium carboxidivorans with increased carbon efficiency and alcohol production. Energy Convers. Manag. 252, 115118. doi:10.1016/j.enconman.2021.115118
Collins, M. E., Oultram, J. D., and Young, M. (1985). Identification of restriction fragments from two cryptic Clostridium butyricum plasmids that promote the establishment of a replication-defective plasmid in Bacillus subtilis. J. Gen. Microbiol. 131, 2097–2105. doi:10.1099/00221287-131-8-2097
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Cotton, C. A., Claassens, N. J., Benito-Vaquerizo, S., and Bar-Even, A. (2020). Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180. doi:10.1016/j.copbio.2019.10.002
De Souza Pinto Lemgruber, R., Valgepea, K., Tappel, R., Behrendorff, J. B., Palfreyman, R. W., Plan, M., et al. (2019). Systems-level engineering and characterization of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB). Metab. Eng. 53, 14–23. doi:10.1016/j.ymben.2019.01.003
Diender, M., Stams, A. J. M., and Sousa, D. Z. (2016). Production of medium-chain fatty acids and higher alcohols by a synthetic co-culture grown on carbon monoxide or syngas. Biotechnol. Biofuels 9, 82. doi:10.1186/s13068-016-0495-0
Di Leonardo, P. F., Antonicelli, G., Agostino, V., and Re, A. (2022). Genome-scale mining of acetogens of the genus Clostridium unveils distinctive traits in [FeFe]- and [NiFe]-Hydrogenase content and maturation. Microbiol. Spectr. 10, e0101922. doi:10.1128/spectrum.01019-22
Diner, B. A., Fan, J., Scotcher Miles, C., Wells Derek, H., and Whited Gregory, M. (2017). Synthesis of heterologous mevalonic acid pathway enzymes in Clostridium ljungdahlii for the conversion of fructose and of syngas to mevalonate and isoprene. Appl. Environ. Microbiol. 84, e01723-17–e01717. doi:10.1128/AEM.01723-17
Drake, H. L., and Daniel, S. L. (2004). Physiology of the thermophilic acetogen Moorella thermoacetica. Res. Microbiol. 155, 422–436. doi:10.1016/j.resmic.2004.03.003
Drake, H. L., Daniel, S. L., Küsel, K., Matthies, C., Kuhner, C., and Braus-Stromeyer, S. (1997). Acetogenic bacteria: what are the in situ consequences of their diverse metabolic versatilities? Biofactors 6, 13–24. doi:10.1002/biof.5520060103
Drake, H. L., Gössner, A. S., and Daniel, S. L. (2008). Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125, 100–128. doi:10.1196/annals.1419.016
Drake, H. L., Küsel, K., and Matthies, C. (2006). “Acetogenic prokaryotes,” in The prokaryotes. Editors M. Dworkin, S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackebrandt (US: Springer), 354–420. doi:10.1007/0-387-30742-7_13
Dykstra, J. C., van Oort, J., Yazdi, A. T., Vossen, E., Patinios, C., van der Oost, J., et al. (2022). Metabolic engineering of Clostridium autoethanogenum for ethyl acetate production from CO. Microb. Cell Fact. 21, 243. doi:10.1186/s12934-022-01964-5
Fackler, N., Heffernan, J., Juminaga, A., Doser, D., Nagaraju, S., Gonzalez-Garcia, R. A., et al. (2021a). Transcriptional control of Clostridium autoethanogenum using CRISPRi. Synth. Biol. (Oxf) 6, ysab008. doi:10.1093/synbio/ysab008
Fackler, N., Heijstra, B. D., Rasor, B. J., Brown, H., Martin, J., Ni, Z., et al. (2021b). Stepping on the gas to a circular economy: accelerating development of carbon-negative chemical production from gas fermentation. Annu. Rev. Chem. Biomol. Eng. 12, 439–470. doi:10.1146/annurev-chembioeng-120120-021122
Fernández-Blanco, C., Veiga, M. C., and Kennes, C. (2022). Efficient production of n-caproate from syngas by a co-culture of Clostridium aceticum and Clostridium kluyveri. J. Environ. Manage 302, 113992. doi:10.1016/j.jenvman.2021.113992
Flaiz, M., Ludwig, G., Bengelsdorf, F. R., and Dürre, P. (2021). Production of the biocommodities butanol and acetone from methanol with fluorescent FAST-tagged proteins using metabolically engineered strains of Eubacterium limosum. Biotechnol. Biofuels 14, 117. doi:10.1186/s13068-021-01966-2
Flüchter, S., Follonier, S., Schiel-Bengelsdorf, B., Bengelsdorf, F. R., Zinn, M., and Dürre, P. (2019). Anaerobic production of poly(3-hydroxybutyrate) and its precursor 3-hydroxybutyrate from synthesis gas by autotrophic clostridia. Biomacromolecules 20, 3271–3282. doi:10.1021/acs.biomac.9b00342
Fu, J., Wenzel, S. C., Perlova, O., Wang, J., Gross, F., Tang, Z., et al. (2008). Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res. 36, e113. doi:10.1093/nar/gkn499
Ganigué, R., Sánchez-Paredes, P., Bañeras, L., and Colprim, J. (2016). Low fermentation pH is a trigger to alcohol production, but a killer to chain elongation. Front. Microbiol. 7, 702. doi:10.3389/fmicb.2016.00702
Garnier, T., and Cole, S. T. (1988). Identification and molecular genetic analysis of replication functions of the bacteriocinogenic plasmid pIP404 from Clostridium perfringens. Plasmid 19, 151–160. doi:10.1016/0147-619x(88)90053-4
Gildemyn, S., Molitor, B., Usack, J. G., Nguyen, M., Rabaey, K., and Angenent, L. T. (2017). Upgrading syngas fermentation effluent using Clostridium kluyveri in a continuous fermentation. Biotechnol. Biofuels 10, 83. doi:10.1186/s13068-017-0764-6
Gottwald, M., Andreesen, J. R., LeGall, J., and Ljungdahl, L. G. (1975). Presence of cytochrome and menaquinone in Clostridium formicoaceticum and Clostridium thermoaceticum. J. Bacteriol. 122, 325–328. doi:10.1128/jb.122.1.325-328.1975
Heap, J. T., Kuehne, S. A., Ehsaan, M., Cartman, S. T., Cooksley, C. M., Scott, J. C., et al. (2010). The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. doi:10.1016/j.mimet.2009.10.018
Heap, J. T., Pennington, O. J., Cartman, S. T., Carter, G. P., and Minton, N. P. (2007). The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70, 452–464. doi:10.1016/j.mimet.2007.05.021
Heap, J. T., Pennington, O. J., Cartman, S. T., and Minton, N. P. (2009). A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85. doi:10.1016/j.mimet.2009.05.004
Hess, V., Schuchmann, K., and Müller, V. (2013). The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biol. Chem. 288, 31496–31502. doi:10.1074/jbc.m113.510255
Höfele, F., Schoch, T., Oberlies, C., and Dürre, P. (2023). Heterologous production of isopropanol using metabolically engineered Acetobacterium woodii strains. Bioeng. (Basel) 10, 1381. doi:10.3390/bioengineering10121381
Hoffmeister, S., Gerdom, M., Bengelsdorf, F. R., Linder, S., Flüchter, S., Öztürk, H., et al. (2016). Acetone production with metabolically engineered strains of Acetobacterium woodii. Metab. Eng. 36, 37–47. doi:10.1016/j.ymben.2016.03.001
Hong, Y., and Zeng, A. P. (2022). Biosynthesis based on one-carbon mixotrophy. Adv. Biochem. Eng. Biotechnol. 180, 351–371. doi:10.1007/10_2021_198
Hu, P., Chakraborty, S., Kumar, A., Woolston, B., Liu, H., Emerson, D., et al. (2016). Integrated bioprocess for conversion of gaseous substrates to liquids. Proc. Natl. Acad. Sci. U. S. A. 113, 3773–3778. doi:10.1073/pnas.1516867113
Huang, H., Chai, C., Li, N., Rowe, P., Minton, N. P., Yang, S., et al. (2016). CRISPR/Cas9-Based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth. Biol. 5, 1355–1361. doi:10.1021/acssynbio.6b00044
Huang, H., Chai, C., Yang, S., Jiang, W., and Gu, Y. (2019). Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab. Eng. 52, 293–302. doi:10.1016/j.ymben.2019.01.005
Huang, H., Wang, S., Moll, J., and Thauer, R. K. (2012). Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J. Bacteriol. 194, 3689–3699. doi:10.1128/jb.00385-12
Iwasaki, Y., Kita, A., Sakai, S., Takaoka, K., Yano, S., Tajima, T., et al. (2013). Engineering of a functional thermostable kanamycin resistance marker for use in Moorella thermoacetica ATCC39073. FEMS Microbiol. Lett. 343, 8–12. doi:10.1111/1574-6968.12113
Jeswani, H. K., Chilvers, A., and Azapagic, A. (2020). Environmental sustainability of biofuels: a review. Proc. Math. Phys. Eng. Sci. 476, 20200351. doi:10.1098/rspa.2020.0351
Jia, D., He, M., Tian, Y., Shen, S., Zhu, X., Wang, Y., et al. (2021). Metabolic engineering of gas-fermenting Clostridium ljungdahlii for efficient Co-production of isopropanol, 3-hydroxybutyrate, and ethanol. ACS Synth. Biol. 10, 2628–2638. doi:10.1021/acssynbio.1c00235
Jiang, W., Hernández Villamor, D., Peng, H., Chen, J., Liu, L., Haritos, V., et al. (2021). Metabolic engineering strategies to enable microbial utilization of C1 feedstocks. Nat. Chem. Biol. 17, 845–855. doi:10.1038/s41589-021-00836-0
Jin, S., Bae, J., Song, Y., Pearcy, N., Shin, J., Kang, S., et al. (2020). Synthetic biology on acetogenic bacteria for highly efficient conversion of C1 gases to biochemicals. Int. J. Mol. Sci. 21, 7639. doi:10.3390/ijms21207639
Jones, D. T., and Woods, D. R. (1986). Acetone-butanol fermentation revisited. Microbiol. Rev. 50, 484–524. doi:10.1128/mr.50.4.484-524.1986
Jones, S. W., Fast, A. G., Carlson, E. D., Wiedel, C. A., Au, J., Antoniewicz, M. R., et al. (2016). CO2 fixation by anaerobic non-photosynthetic mixotrophy for improved carbon conversion. Nat. Commun. 7, 12800. doi:10.1038/ncomms12800
Joseph, R. C., Kim, N. M., and Sandoval, N. R. (2018). Recent developments of the synthetic biology toolkit for Clostridium. Front. Microbiol. 9, 154. doi:10.3389/fmicb.2018.00154
Karberg, M., Guo, H., Zhong, J., Coon, R., Perutka, J., and Lambowitz, A. M. (2001). Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19, 1162–1167. doi:10.1038/nbt1201-1162
Karim, A. S., Dudley, Q. M., Juminaga, A., Yuan, Y., Crowe, S. A., Heggestad, J. T., et al. (2020). In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat. Chem. Biol. 16, 912–919. doi:10.1038/s41589-020-0559-0
Kato, J., Takemura, K., Kato, S., Fujii, T., Wada, K., Iwasaki, Y., et al. (2021). Metabolic engineering of Moorella thermoacetica for thermophilic bioconversion of gaseous substrates to a volatile chemical. Amb. Express 11, 59. doi:10.1186/s13568-021-01220-w
Katsyv, A., and Müller, V. (2020). Overcoming energetic barriers in acetogenic C1 conversion. Front. Bioeng. Biotechnol. 8, 621166. doi:10.3389/fbioe.2020.621166
Katsyv, A., and Müller, V. (2022). A purified energy-converting hydrogenase from Thermoanaerobacter kivui demonstrates coupled H+-translocation and reduction in vitro. J. Biol. Chem. 298, 102216. doi:10.1016/j.jbc.2022.102216
Kim, J. Y., Lee, M., Oh, S., Kang, B., Yasin, M., and Chang, I. S. (2023). Acetogen and acetogenesis for biological syngas valorization. Bioresour. Technol. 384, 129368. doi:10.1016/j.biortech.2023.129368
Kita, A., Iwasaki, Y., Sakai, S., Okuto, S., Takaoka, K., Suzuki, T., et al. (2013). Development of genetic transformation and heterologous expression system in carboxydotrophic thermophilic acetogen Moorella thermoacetica. J. Biosci. Bioeng. 115, 347–352. doi:10.1016/j.jbiosc.2012.10.013
Köpke, M., Gerth, M. L., Maddock, D. J., Mueller, A. P., Liew, F., Simpson, S. D., et al. (2014). Reconstruction of an acetogenic 2,3-butanediol pathway involving a novel NADPH-dependent primary-secondary alcohol dehydrogenase. Appl. Environ. Microbiol. 80, 3394–3403. doi:10.1128/aem.00301-14
Köpke, M., Held, C., Hujer, S., Liesegang, H., Wiezer, A., Wollherr, A., et al. (2010). Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U. S. A. 107, 13087–13092. doi:10.1073/pnas.1004716107
Köpke, M., and Liew, F. (2011). Recombinant microorganism and methods of production thereof. U.S. Patent Application Publication. 20110236941A1.
Kracke, F., Virdis, B., Bernhardt, P. V., Rabaey, K., and Krömer, J. O. (2016). Redox dependent metabolic shift in Clostridium autoethanogenum by extracellular electron supply. Biotechnol. Biofuels 9, 249. doi:10.1186/s13068-016-0663-2
Krüger, A., Mueller, A. P., Rybnicky, G. A., Engle, N. L., Yang, Z. K., Tschaplinski, T. J., et al. (2020). Development of a clostridia-based cell-free system for prototyping genetic parts and metabolic pathways. Metab. Eng. 62, 95–105. doi:10.1016/j.ymben.2020.06.004
Kuehne, S. A., Rood, J. I., and Lyras, D. (2019). Clostridial genetics: genetic manipulation of the pathogenic clostridia. Microbiol. Spectr. 7. doi:10.1128/microbiolspec.GPP3-0040-2018
Kwon, H. J., Lee, J., Kwon, S. J., and Lee, H. S. (2024). Development of a genetic engineering toolbox for syngas-utilizing acetogen Clostridium sp. AWRP. Microb. Cell Fact. 23, 6. doi:10.1186/s12934-023-02272-2
Lagoa-Costa, B., Abubackar, H. N., Fernández-Romasanta, M., Kennes, C., and Veiga, M. C. (2017). Integrated bioconversion of syngas into bioethanol and biopolymers. Bioresour. Technol. 239, 244–249. doi:10.1016/j.biortech.2017.05.019
Lampe, D. J., Churchill, M. E., and Robertson, H. M. (1996). A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15, 5470–5479. doi:10.1002/j.1460-2075.1996.tb00930.x
Lauer, I., Philipps, G., and Jennewein, S. (2022). Metabolic engineering of Clostridium ljungdahlii for the production of hexanol and butanol from CO2 and H2. Microb. Cell Fact. 21, 85. doi:10.1186/s12934-022-01802-8
Leang, C., Ueki, T., Nevin, K. P., and Lovley, D. R. (2013). A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl. Environ. Microbiol. 79, 1102–1109. doi:10.1128/aem.02891-12
Lee, H., Bae, J., Jin, S., Kang, S., and Cho, B. K. (2022). Engineering acetogenic bacteria for efficient one-carbon utilization. Front. Microbiol. 13, 865168. doi:10.3389/fmicb.2022.865168
Liew, F., Henstra, A. M., Kӧpke, M., Winzer, K., Simpson, S. D., and Minton, N. P. (2017). Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab. Eng. 40, 104–114. doi:10.1016/j.ymben.2017.01.007
Liew, F., Henstra, A. M., Winzer, K., Köpke, M., Simpson, S. D., and Minton, N. P. (2016). Insights into CO2 fixation pathway of Clostridium autoethanogenum by targeted mutagenesis. mBio 7, e00427-16. doi:10.1128/mBio.00427-16
Liew, F. E., Nogle, R., Abdalla, T., Rasor, B. J., Canter, C., Jensen, R. O., et al. (2022). Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat. Biotechnol. 40, 335–344. doi:10.1038/s41587-021-01195-w
Lin, P. P., Mi, L., Morioka, A. H., Yoshino, K. M., Konishi, S., Xu, S. C., et al. (2015). Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 31, 44–52. doi:10.1016/j.ymben.2015.07.001
Litty, D., and Müller, V. (2021). Butyrate production in the acetogen Eubacterium limosum is dependent on the carbon and energy source. Microb. Biotechnol. 14, 2686–2692. doi:10.1111/1751-7915.13779
Liu, Z. Y., Jia, D. C., Zhang, K. D., Zhu, H. F., Zhang, Q., Jiang, W. H., et al. (2020). Ethanol metabolism dynamics in Clostridium ljungdahlii grown on carbon monoxide. Appl. Environ. Microbiol. 86, e00730-20. doi:10.1128/AEM.00730-20
Ljungdahl, L. G. (1986). The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40, 415–450. doi:10.1146/annurev.micro.40.1.415
Lo, J., Humphreys, J. R., Magnusson, L., Wachter, B., Urban, C., Hebdon, S. D., et al. (2022). Acetogenic production of 3-Hydroxybutyrate using a native 3-Hydroxybutyryl-CoA Dehydrogenase. Front. Microbiol. 13, 948369. doi:10.3389/fmicb.2022.948369
Marcellin, E., Behrendorff, J. B., Nagaraju, S., DeTissera, S., Segovia, S., Palfreyman, R. W., et al. (2016). Low carbon fuels and commodity chemicals from waste gases–Systematic approach to understand energy metabolism in a model acetogen. Green Chem. 18, 3020–3028. doi:10.1039/c5gc02708j
Maru, B. T., Munasinghe, P. C., Gilary, H., Jones, S. W., and Tracy, B. P. (2018). Fixation of CO2 and CO on a diverse range of carbohydrates using anaerobic, non-photosynthetic mixotrophy. FEMS Microbiol. Lett. 365. doi:10.1093/femsle/fny039
Minton, N., Carter, G., Herbert, M., O’keeffe, T., Purdy, D., Elmore, M., et al. (2004). The development of Clostridium difficile genetic systems. Anaerobe 10, 75–84. doi:10.1016/j.anaerobe.2003.11.003
Mock, J., Wang, S., Huang, H., Kahnt, J., and Thauer, R. K. (2014). Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J. Bacteriol. 196, 3303–3314. doi:10.1128/jb.01839-14
Mock, J., Zheng, Y., Mueller, A. P., Ly, S., Tran, L., Segovia, S., et al. (2015). Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J. Bacteriol. 197, 2965–2980. doi:10.1128/jb.00399-15
Molitor, B., Marcellin, E., and Angenent, L. T. (2017). Overcoming the energetic limitations of syngas fermentation. Curr. Opin. Chem. Biol. 41, 84–92. doi:10.1016/j.cbpa.2017.10.003
Monod, M., Denoya, C., and Dubnau, D. (1986). Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 167, 138–147. doi:10.1128/jb.167.1.138-147.1986
Moreira, J. P. C., Heap, J. T., Alves, J. I., and Domingues, L. (2023). Developing a genetic engineering method for Acetobacterium wieringae to expand one-carbon valorization pathways. Biotechnol. Biofuels Bioprod. 16, 24. doi:10.1186/s13068-023-02259-6
Mueller, A. P., Koepke, M., and Nagaraju, S. (2018). Recombinant microorganisms and uses therefor. U.S. Patent No US9890384 B2. Washington, DC: U.S. Patent and Trademark Office.
Nagarajan, H., Sahin, M., Nogales, J., Latif, H., Lovley, D. R., Ebrahim, A., et al. (2013). Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 12, 118. doi:10.1186/1475-2859-12-118
Nagaraju, S., Davies, N. K., Walker, D. J., Köpke, M., and Simpson, S. D. (2016). Genome editing of Clostridium autoethanogenum using CRISPR/Cas9. Biotechnol. Biofuels 9, 219. doi:10.1186/s13068-016-0638-3
Neuendorf, C. S., Vignolle, G. A., Derntl, C., Tomin, T., Novak, K., Mach, R. L., et al. (2021). A quantitative metabolic analysis reveals Acetobacterium woodii as a flexible and robust host for formate-based bioproduction. Metab. Eng. 68, 68–85. doi:10.1016/j.ymben.2021.09.004
Nevin, K. P., Hensley, S. A., Franks, A. E., Summers, Z. M., Ou, J., Woodard, T. L., et al. (2011). Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl. Environ. Microbiol. 77, 2882–2886. doi:10.1128/aem.02642-10
Nissen, L. S., and Basen, M. (2019). The emerging role of aldehyde:ferredoxin oxidoreductases in microbially-catalyzed alcohol production. J. Biotechnol. 306, 105–117. doi:10.1016/j.jbiotec.2019.09.005
Nogle, R., Nagaraju, S., Utturkar, S. M., Giannone, R. J., Reynoso, V., Leang, C., et al. (2022). Clostridium autoethanogenum isopropanol production via native plasmid pCA replicon. Front. Bioeng. Biotechnol. 10, 932363. doi:10.3389/fbioe.2022.932363
Öppinger, C., Kremp, F., and Müller, V. (2022). Is reduced ferredoxin the physiological electron donor for MetVF-type methylenetetrahydrofolate reductases in acetogenesis? A hypothesis. Int. Microbiol. 25, 75–88. doi:10.1007/s10123-021-00190-0
Oswald, F., Dörsam, S., Veith, N., Zwick, M., Neumann, A., Ochsenreither, K., et al. (2016). Sequential mixed cultures: from syngas to malic acid. Front. Microbiol. 7, 891. doi:10.3389/fmicb.2016.00891
Patinios, C., Creutzburg, S. C. A., Arifah, A. Q., Adiego-Pérez, B., Gyimah, E. A., Ingham, C. J., et al. (2021). Streamlined CRISPR genome engineering in wild-type bacteria using SIBR-Cas. Nucleic Acids Res. 49, 11392–11404. doi:10.1093/nar/gkab893
Pavan, M., Reinmets, K., Garg, S., Mueller, A. P., Marcellin, E., Köpke, M., et al. (2022). Advances in systems metabolic engineering of autotrophic carbon oxide-fixing biocatalysts towards a circular economy. Metab. Eng. 71, 117–141. doi:10.1016/j.ymben.2022.01.015
Perez, J. M., Richter, H., Loftus, S. E., and Angenent, L. T. (2013). Biocatalytic reduction of short-chain carboxylic acids into their corresponding alcohols with syngas fermentation. Biotechnol. Bioeng. 110, 1066–1077. doi:10.1002/bit.24786
Philipps, G., de Vries, S., and Jennewein, S. (2019). Development of a metabolic pathway transfer and genomic integration system for the syngas-fermenting bacterium Clostridium ljungdahlii. Biotechnol. Biofuels 12, 112. doi:10.1186/s13068-019-1448-1
Pierce, E., Xie, G., Barabote, R. D., Saunders, E., Han, C. S., Detter, J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573. doi:10.1111/j.1462-2920.2008.01679.x
Plante, I., and Cousineau, B. (2006). Restriction for gene insertion within the Lactococcus lactis Ll.LtrB group II intron. RNA 12, 1980–1992. doi:10.1261/rna.193306
Poulalier-Delavelle, M., Baker, J. P., Millard, J., Winzer, K., and Minton, N. P. (2023). Endogenous CRISPR/Cas systems for genome engineering in the acetogens Acetobacterium woodii and Clostridium autoethanogenum. Front. Bioeng. Biotechnol. 11, 1213236. doi:10.3389/fbioe.2023.1213236
Purdy, D., O’Keeffe, T. A., Elmore, M., Herbert, M., McLeod, A., Bokori-Brown, M., et al. (2002). Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46, 439–452. doi:10.1046/j.1365-2958.2002.03134.x
Ragsdale, S. W. (2004). Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39, 165–195. doi:10.1080/10409230490496577
Ragsdale, S. W., and Ljungdahl, L. G. (1984). Purification and properties of NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Acetobacterium woodii. J. Biol. Chem. 259, 3499–3503. doi:10.1016/s0021-9258(17)43122-x
Ragsdale, S. W., and Pierce, E. (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898. doi:10.1016/j.bbapap.2008.08.012
Riaz, S., Mazhar, S., Abidi, S. H., Syed, Q., Abbas, N., Saleem, Y., et al. (2022). Biobutanol production from sustainable biomass process of anaerobic ABE fermentation for industrial applications. Arch. Microbiol. 204, 672. doi:10.1007/s00203-022-03284-z
Richter, H., Molitor, B., Diender, M., Sousa, D. Z., and Angenent, L. T. (2016). A narrow pH range supports butanol, hexanol, and octanol production from syngas in a continuous Co-culture of Clostridium ljungdahlii and Clostridium kluyveri with in-line product extraction. Front. Microbiol. 7, 1773. doi:10.3389/fmicb.2016.01773
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2023). REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 51, D629–D630. doi:10.1093/nar/gkac975
Rosenbaum, F. P., and Müller, V. (2023). Moorella thermoacetica: a promising cytochrome-and quinone-containing acetogenic bacterium as platform for a CO2-based bioeconomy. Green Carbon 1, 2–13. doi:10.1016/j.greenca.2023.06.002
Schiel-Bengelsdorf, B., and Dürre, P. (2012). Pathway engineering and synthetic biology using acetogens. FEBS Lett. 586, 2191–2198. doi:10.1016/j.febslet.2012.04.043
Schoelmerich, M. C., Katsyv, A., Sung, W., Mijic, V., Wiechmann, A., Kottenhahn, P., et al. (2018). Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4587–4595. doi:10.1111/1462-2920.14412
Schoelmerich, M. C., and Müller, V. (2019). Energy conservation by a hydrogenase-dependent chemiosmotic mechanism in an ancient metabolic pathway. Proc. Natl. Acad. Sci. U. S. A. 116, 6329–6334. doi:10.1073/pnas.1818580116
Schrader, J., and Bohlmann, J. (2015). Biotechnology of isoprenoids. Switzerland: Springer International Publishing. doi:10.1007/978-3-319-20107-8
Schuchmann, K., and Müller, V. (2012). A bacterial electron-bifurcating hydrogenase. J. Biol. Chem. 287, 31165–31171. doi:10.1074/jbc.m112.395038
Schuchmann, K., and Müller, V. (2013). Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342, 1382–1385. doi:10.1126/science.1244758
Schuchmann, K., and Müller, V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821. doi:10.1038/nrmicro3365
Schuchmann, K., and Müller, V. (2016). Energetics and application of heterotrophy in acetogenic bacteria. Appl. Environ. Microbiol. 82, 4056–4069. doi:10.1128/aem.00882-16
Shin, J., Kang, S., Song, Y., Jin, S., Lee, J. S., Lee, J. K., et al. (2019). Genome engineering of Eubacterium limosum using expanded genetic tools and the CRISPR-cas9 system. ACS Synth. Biol. 8, 2059–2068. doi:10.1021/acssynbio.9b00150
Song, Y., Lee, J. S., Shin, J., Lee, G. M., Jin, S., Kang, S., et al. (2020). Functional cooperation of the glycine synthase-reductase and Wood-Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proc. Natl. Acad. Sci. U. S. A. 117, 7516–7523. doi:10.1073/pnas.1912289117
Straub, M., Demler, M., Weuster-Botz, D., and Dürre, P. (2014). Selective enhancement of autotrophic acetate production with genetically modified Acetobacterium woodii. J. Biotechnol. 178, 67–72. doi:10.1016/j.jbiotec.2014.03.005
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bact. Rev. 41, 100–180. doi:10.1128/mmbr.41.1.100-180.1977
Tian, L., Perot, S. J., Hon, S., Zhou, J., Liang, X., Bouvier, J. T., et al. (2017). Enhanced ethanol formation by Clostridium thermocellum via pyruvate decarboxylase. Microb. Cell Fact. 16, 171. doi:10.1186/s12934-017-0783-9
Tokiwa, Y., and Ugwu, C. U. (2007). Biotechnological production of (R)-3-hydroxybutyric acid monomer. J. Biotechnol. 132, 264–272. doi:10.1016/j.jbiotec.2007.03.015
Tremblay, P. L., Zhang, T., Dar, S. A., Leang, C., and Lovley, D. R. (2012). The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. mBio 4, e00406–e00412. doi:10.1128/mBio.00406-12
Trieucuot, P., and Courvalin, P. (1983). Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5''-aminoglycoside phosphotransferase Type-III. Gene 23, 331–341. doi:10.1016/0378-1119(83)90022-7
Ueki, T., Nevin, K. P., Woodard, T. L., and Lovley, D. R. (2014). Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii. mBio 5, e01636–e01614. doi:10.1128/mBio.01636-14
Valgepea, K., Loi, K. Q., Behrendorff, J. B., Lemgruber, R. S. P., Plan, M., Hodson, M. P., et al. (2017). Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum. Metab. Eng. 41, 202–211. doi:10.1016/j.ymben.2017.04.007
van Treeck, U., Schmidt, F., and Wiedemann, B. (1981). Molecular nature of a streptomycin and sulfonamide resistance plasmid (pBP1) prevalent in clinical Escherichia coli strains and integration of an ampicillin resistance transposon (TnA). Antimicrob. Agents Chemother. 19, 371–380. doi:10.1128/aac.19.3.371
Vasu, K., and Nagaraja, V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72. doi:10.1128/mmbr.00044-12
Vögeli, B., Schulz, L., Garg, S., Tarasava, K., Clomburg, J. M., Lee, S. H., et al. (2022). Cell-free prototyping enables implementation of optimized reverse β-oxidation pathways in heterotrophic and autotrophic bacteria. Nat. Commun. 13, 3058. doi:10.1038/s41467-022-30571-6
Wang, D., You, M., Qiu, Z., Li, P., Qiao, M., and Xu, C. (2023). Development of an efficient ClosTron system for gene disruption in Ruminiclostridium papyrosolvens. Appl. Microbiol. Biotechnol. 107, 1801–1812. doi:10.1007/s00253-023-12427-1
Wang, S., Huang, H., Kahnt, J., Mueller, A. P., Köpke, M., and Thauer, R. K. (2013a). NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386. doi:10.1128/jb.00678-13
Wang, S., Huang, H., Kahnt, J., and Thauer, R. K. (2013b). A reversible electron-bifurcating ferredoxin- and NAD-dependent [FeFe]-hydrogenase (HydABC) in Moorella thermoacetica. J. Bacteriol. 195, 1267–1275. doi:10.1128/jb.02158-12
Westphal, L., Wiechmann, A., Baker, J., Minton, N. P., and Müller, V. (2018). The Rnf complex is an energy-coupled transhydrogenase essential to reversibly link cellular NADH and ferredoxin pools in the acetogen Acetobacterium woodii. J. Bacteriol. 200, e00357-18. doi:10.1128/JB.00357-18
Widberger, J., Wittgens, A., Klaunig, S., Krämer, M., Kissmann, A. K., Höfele, F., et al. (2024). Recombinant production of Pseudomonas aeruginosa rhamnolipids in P. putida KT2440 on Acetobacterium woodii cultures grown chemo-autotrophically with carbon dioxide and hydrogen. Microorganisms 12, 529. doi:10.3390/microorganisms12030529
Wiechmann, A., Ciurus, S., Oswald, F., Seiler, V. N., and Müller, V. (2020). It does not always take two to tango: “Syntrophy” via hydrogen cycling in one bacterial cell. ISME J. 14, 1561–1570. doi:10.1038/s41396-020-0627-1
Wood, H. G., Ragsdale, S. W., and Pezacka, E. (1986). The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol. Rev. 2, 345–362. doi:10.1111/j.1574-6968.1986.tb01865.x
Woods, C., Humphreys, C. M., Rodrigues, R. M., Ingle, P., Rowe, P., Henstra, A. M., et al. (2019). A novel conjugal donor strain for improved DNA transfer into Clostridium spp. Anaerobe 59, 184–191. doi:10.1016/j.anaerobe.2019.06.020
Woolston, B. M., Emerson, D. F., Currie, D. H., and Stephanopoulos, G. (2018). Rediverting carbon flux in Clostridium ljungdahlii using CRISPR interference (CRISPRi). Metab. Eng. 48, 243–253. doi:10.1016/j.ymben.2018.06.006
Xia, P. F., Casini, I., Schulz, S., Klask, C. M., Angenent, L. T., and Molitor, B. (2020). Reprogramming acetogenic bacteria with CRISPR-targeted base editing via deamination. ACS Synth. Biol. 9, 2162–2171. doi:10.1021/acssynbio.0c00226
Xu, R., Zhang, K., Liu, P., Han, H., Zhao, S., Kakade, A., et al. (2018). Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 269, 557–566. doi:10.1016/j.biortech.2018.08.118
Yamamoto, I., Saiki, T., Liu, S. M., and Ljungdahl, L. G. (1983). Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J. Biol. Chem. 258, 1826–1832. doi:10.1016/s0021-9258(18)33062-x
Yasui, K., Kano, Y., Tanaka, K., Watanabe, K., Shimizu-Kadota, M., Yoshikawa, H., et al. (2009). Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 37, e3. doi:10.1093/nar/gkn884
Yi, J., Huang, H., Liang, J., Wang, R., Liu, Z., Li, F., et al. (2021). A heterodimeric reduced-ferredoxin-dependent methylenetetrahydrofolate reductase from syngas-fermenting Clostridium ljungdahlii. Microbiol. Spectr. 9, e0095821. doi:10.1128/Spectrum.00958-21
Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., et al. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34. doi:10.1038/nbt.3737
Zhang, X., Arbour, T., Zhang, D., Wei, S., and Rabaey, K. (2023). Microbial electrosynthesis of acetate from CO2 under hypersaline conditions. Environ. Sci. Ecotechnol 13, 100211. doi:10.1016/j.ese.2022.100211
Zhao, R., Liu, Y., Zhang, H., Chai, C., Wang, J., Jiang, W., et al. (2019). CRISPR-Cas12a-Mediated gene deletion and regulation in Clostridium ljungdahlii and its application in carbon flux redirection in synthesis gas fermentation. ACS Synth. Biol. 8, 2270–2279. doi:10.1021/acssynbio.9b00033
Zhong, J., Karberg, M., and Lambowitz, A. M. (2003). Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 31, 1656–1664. doi:10.1093/nar/gkg248
Keywords: acetogenic bacteria, synthetic biology, metabolic engineering, C1 gases, gas fermentation, cell factory
Citation: Zhang J-Z, Li Y-Z, Xi Z-N, Gao H-P, Zhang Q, Liu L-C, Li F-L and Ma X-Q (2024) Engineered acetogenic bacteria as microbial cell factory for diversified biochemicals. Front. Bioeng. Biotechnol. 12:1395540. doi: 10.3389/fbioe.2024.1395540
Received: 04 March 2024; Accepted: 28 June 2024;
Published: 11 July 2024.
Edited by:
Ethan I. Lan, National Yang Ming Chiao Tung University, TaiwanReviewed by:
Kaspar Valgepea, University of Tartu, EstoniaPaul Lin, National Yang Ming Chiao Tung University, Taiwan
Copyright © 2024 Zhang, Li, Xi, Gao, Zhang, Liu, Li and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Zhang, emhhbmdxdWFuLmZzaHlAc2lub3BlYy5jb20=; Xiao-Qing Ma, bWF4cUBxaWJlYnQuYWMuY24=
 Jun-Zhe Zhang
Jun-Zhe Zhang Yu-Zhen Li1,2
Yu-Zhen Li1,2 Fu-Li Li
Fu-Li Li Xiao-Qing Ma
Xiao-Qing Ma