- 1Department of Bioengineering, University of Washington, Seattle, WA, United States
- 2Department of Biological and Chemical Engineering, Hongik University, Sejong, Republic of Korea
- 3Department of Microbiology and Immunology, University of North Carolina, Chapel Hill, NC, United States
- 4Department of Medicine/Division of Rheumatology, University of Washington, Seattle, WA, United States
A complex and dynamic network of interactions exists between human gastrointestinal epithelium and intestinal microbiota. Therefore, comprehending intestinal microbe-epithelial cell interactions is critical for the understanding and treatment of intestinal diseases. Primary human colonic epithelial cells derived from a healthy human donor were co-cultured with Clostridium scindens (C. scindens), a probiotic obligate anaerobe; Staphylococcus aureus (S. aureus), a facultative anaerobe and intestinal pathogen; or both bacterial species in tandem. The co-culture hanging basket platform used for these experiments possessed walls of controlled oxygen (O2) permeability to support the formation of an O2 gradient across the intestinal epithelium using cellular O2 consumption, resulting in an anaerobic luminal and aerobic basal compartment. Both the colonic epithelial cells and C. scindens remained viable over 48 h during co-culture. In contrast, co-culture with S. aureus elicited significant damage to colonic epithelial cells within 24 h. To explore the influence of the intestinal pathogen on the epithelium in the presence of the probiotic bacteria, colonic epithelial cells were inoculated sequentially with the two bacterial species. Under these conditions, C. scindens was capable of repressing the production of S. aureus enterotoxin. Surprisingly, although C. scindens converted cholic acid to secondary bile acids in the luminal medium, the growth of S. aureus was not significantly inhibited. Nevertheless, this combination of probiotic and pathogenic bacteria was found to benefit the survival of the colonic epithelial cells compared with co-culture of the epithelial cells with S. aureus alone. This platform thus provides an easy-to-use and low-cost tool to study the interaction between intestinal bacteria and colonic cells in vitro to better understand the interplay of intestinal microbiota with human colonic epithelium.
1 Introduction
The colon receives partially digested food from the small intestine and absorbs water and residual nutrients, subsequently transporting indigestible waste in the form of feces for elimination (Azzouz and Sharma, 2022). The colon is lined with a monolayer of epithelial cells that form a barrier between luminal contents and the host while maintaining an anaerobic environment within the lumen (Kim et al., 2019). Additionally, the colon has less fluid and the lowest average flow rate of the intestinal system (Harris and Scolapio, 2019). Based on these features, the colon provides an ideal growth environment for a complex microbial community that includes obligate and facultative anaerobic species and whose composition and metabolism are influenced by diet and host factors. In turn, this microbial community contributes to human metabolism through fermentation of undigested fiber (Clarke et al., 2014), production of vitamins (Guarner and Malagelada, 2003; Azzouz and Sharma, 2022), absorption of minerals (Guarner and Malagelada, 2003), and transformation of bile acids (Ridlon et al., 2014). Commensal microbiota also plays an essential role in the defense against pathogens (Cheng et al., 2020; Khan et al., 2021). Known mechanisms in this defense include direct pathogen inhibition by metabolic products of commensal microbes (Rooks and Garrett, 2016), outcompeting pathogens for nutrients and other resources that limit pathogen growth (Stubbendieck and Straight, 2016), formation of a microbial barrier in the mucosal layer (Sommer and Bäckhed, 2013), and directing the immune system towards elimination of pathogens (Baruch et al., 2021; Davar et al., 2021). Commensal microbes also interact with and regulate the colonic epithelium itself, influencing cell turnover, promoting epithelial restitution, and reorganization of tight junctions (Yu et al., 2012). In contrast, pathogenic microbes have the potential to degrade the mucus barrier and invade the mucosal epithelium; these pathogens cause or are associated with various intestine-associated diseases, including gastroenteritis, inflammatory bowel disease (IBD), pancreatitis, and cancer (Shu et al., 2023). Therefore, understanding host-microbiota interactions is essential for the preservation of human health and may lead to effective approaches to mitigate intestinal diseases.
The interactions between host and intestinal microbiota are often studied using animal models. To limit or prevent intestinal microbe colonization, animals are either raised in a germ-free environment and/or exposed to antibiotic treatment to allow control of microflora during experiments (Sivan et al., 2015). The most common animal model is the mouse due primarily to accessibility and low maintenance cost (Nguyen et al., 2015; Li and Zhang, 2022). The porcine model has a more comparable colonic structure, hormone secretion, and gut microbiota composition to the human (Heinritz et al., 2013; Roura et al., 2016), which makes the pig a popular, albeit costly, animal model. Although animal models have been commonly used in gut microbiome research for decades, ethical and cost issues, their distinct genetics, intestinal structure, microbiota composition, and immune responses impose limitations on these models for understanding human/microbiome physiology and pathophysiology (Marrero et al., 2021). In lieu of animal models, in vitro fermentation systems using reactor chambers are cost-effective platforms for intestinal microbiota studies (Li and Zhang, 2022). These batch or continuous flow systems enable precise control of physiological parameters within the system, such as pH, temperature, retention time, composition of the media, and anaerobic conditions (Payne et al., 2012). Multistage fermentation models enable the simulation of the different luminal physiological environments from the stomach to the colon in the absence of intestinal epithelium (Verhoeckx et al., 2015). However, these systems are unsuitable for assessing interactions with the host gastrointestinal tissue. To address this limitation, viable intestinal segments from animals have been integrated into fluidic systems to assemble ex vivo models, such as the Ussing chamber and the everted gut sac model, although these systems have many challenges, including their size, operational complexity, and the absence of human intestinal epithelium (Clarke, 2009; Alam et al., 2012; Rahman et al., 2021).
To replicate the interaction between human intestine and gut microbiota, in vitro models combining microfluidic platforms and co-culture of primary human intestinal cells with gut bacteria have been developed. These systems have been used to create a more complex environment for intestinal microbiota by perfusing oxygenated and deoxygenated media in separate compartments to create an O2 gradient and fluidic shear stress, as well as to prevent bacterial overgrowth and to collect secreted molecules (Tovaglieri et al., 2019; Zhang et al., 2021b). Despite their potential, the complex, sterile flow systems and enclosed structure (with inaccessible compartments) present barriers in their use. Organoids embedded in Matrigel™ formed from differentiated gastrointestinal stem cells have been used to model an internal cavity representing the gut lumen, with live bacteria inoculated through microinjection (Bartfeld et al., 2015; Williamson et al., 2018). Shortcomings of these models are the difficulty of accessing the luminal compartment, an uncontrolled luminal O2 concentration, and overgrowth of the confined microbiota. To partially address these limitations, the polarity of organoids has been reversed by disrupting the polymerization of the ECM proteins to create “luminal-out” organoids embedded in media (Co et al., 2019). Luminal-out organoids can be transferred to media containing microbes to achieve co-culture experiments, but this organoid system lacks the ability to create the low-O2 concentration needed to support anaerobic bacteria and presents further challenges in controlling and sampling the embedded tissue mimic.
Hanging basket systems have been applied to investigate host-bacteria interactions (Roodsant et al., 2020; Sasaki et al., 2020; Zhang et al., 2021a). The double-sided design of this platform, with luminal and basal compartments separated by a monolayer of mammalian gastrointestinal epithelial cells, makes it feasible to mimic the luminal-epithelial structure of intestinal tissue and eases inoculating microbiota and sampling the luminal and basal environments (Dutton et al., 2019; Šuligoj et al., 2020). Recently, a hanging basket system with an oxygenated basal reservoir and deoxygenated luminal compartment was shown to form an O2 gradient across a primary human colonic epithelium; this system supported the human epithelial cells while enabling co-culture of obligate anaerobic bacteria in the luminal compartment (Kim et al., 2019; Kim et al., 2022). This platform does not require continuous perfusion of the media and allows routine sampling and replenishment of both luminal and basal environments when used in combination with an anaerobic chamber, making it attractive and accessible to a broad group of users. However, to date, co-culture of the human epithelium with only a single strain of bacteria for a short duration (24 h) has been demonstrated with this system. In the current study, we extend the lifetime of co-culture to 48 h with the commensal bacterium Clostridium scindens and with the pathogen Staphylococcus aureus when the bacteria are co-cultured in tandem. The goal of this experimental work is to determine whether a colonic commensal obligate anaerobe can be co-cultured in the presence of living primary human colonic epithelial cells with high viability and metabolic activity, and whether this co-culture affords protection to the colonic epithelium in the presence of pathogenic bacteria.
2 Materials and methods
2.1 Fabrication of an O2 gradient cassette with a supporting collagen matrix
The O2 gradient cassette was composed of a basal fluid reservoir, a customized hanging basket (similar to a Transwell insert) placed into the basal reservoir, and an O2-impermeable machined plug for insertion into the luminal portion of the hanging basket (Supplementary Figure S1). A conventional 12-well plate was used for the basal reservoirs. The design and fabrication of the hanging basket was similar to that described previously (Gunasekara et al., 2018; Kim et al., 2019). Briefly, the hanging basket was fabricated from polycarbonate using a CNC milling machine. Next, a porous polyester (PET) membrane (0.4 µm pores, Sterlitech Cat #1300016) was attached to the bottom of the hanging basket using double-sided medical grade tape (3M, Cat #1504XL). Five minutes of plasma treatment (Harrick Plasma, Cat #PDC-32G) improved adhesion of subsequently added collagen to the basket. The basket with 200 µL neutralized collagen (rat tail, type I, Corning, Cat #354236) solution (1 mg/mL) (Kim et al., 2019; Kim et al., 2022) was incubated for 1 h at 37°C and then submerged in 3 mL crosslinking solution added to the basal reservoir for 30 min at 25°C. The crosslinking solution was 0.6 M 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC): 0.15 M N-hydroxysuccinimide (NHS): 0.1 M 2-(N-morpholino) ethanesulfonic acid (MES) buffer (pH 5.0) in water, (1:1:1 (vol). Later, the basket with partially crosslinked collagen matrix (1.1 cm2 of surface area and 2.2 mm of thickness (Gunasekara et al., 2018)) was incubated in deionized water for >18 h at 25°C to leach out residual crosslinking reagent followed by sterilization in 70% ethanol and was then air-dried in a biosafety cabinet. The basket with collagen matrix could be stored in PBS buffer at 4°C for 2–3 weeks before adding the human primary colon cells. The cells were cultured aerobically to form a confluent monolayer on the collagen/PET membrane scaffold submerged in a well of the 12-well plate (see below). For O2 gradient, an O2-impermeable plug fabricated from polycarbonate was threaded into the luminal portion of the basket. A soft Viton® fluoroelastomer O-ring (McMaster Car, Cat #1284N114) was used with the plug to form an O2-impermeable seal. A port in the plug was used for insertion of an O2 probe, and this port was sealed with ethylene propylene diene monomer (EPDM) rubber caps (McMaster-Carr, Cat #6448K117) when an O2 sensor was not in place. The O2 probe in the luminal compartment was a needle O2 probe (PreSens, NTH-PSt7/Microx4, Germany). The tip of the probe was submerged into the luminal media to approximately 2 mm above the epithelial monolayer when O2 was measured.
2.2 Human primary colon epithelial cell culture
Primary human colonic epithelial stem cells from the transverse colon of a cadaveric donor (male, 23 years old, RRID: CVCL_ZR41 (https://web.expasy.org/cellosaurus/CVCL_ZR41)) were isolated, expanded, and maintained on a collagen scaffold in a 6-well plate in a stem cell media (SM, Supplementary Table S1) as described previously (Wang et al., 2017; Kim et al., 2019). Cells were used between passage 7 and 12. For each experiment, the human colonic stem cells were sub-cultured from a single well of the maintenance culture into 6 hanging baskets with collagen matrix prepared as described above. The human colonic cells were expanded on the collagen matrix in the hanging basket in expansion medium (EM, Supplementary Table S1 0.5 mL placed in the luminal compartment and 3 mL placed in the basal compartment) for 6 days with media exchange every other day (Kim et al., 2019; Kim et al., 2022). After a confluent monolayer was formed on day 7, both luminal and basal media were replaced by differentiation medium without antibiotics (DM-Ab, Supplementary Table S1 0.5 mL placed in the luminal compartment and 3 mL placed in the basal compartment). On day 8, the luminal medium was replaced with 0.5 mL 10% peptone-yeast-fructose media (PYF, Supplementary Table S2) in 1× PBS.
2.3 Bacterial culture
C. scindens (ATCC 35704) was purchased from ATCC and propagated within an anaerobic chamber (Coy Laboratory Products). The bacteria were cultured in PYF medium (peptone 10 g/L, yeast extract 10 g/L, fructose 10 g/L, resazurin 1 mg/L, L-cysteine hydrochloride 0.5 g/L, CaCl2·H2O 100 mg/L, MgSO4·7H2O 50 mg/L, K2PO4 40 mg/L, KH2PO4 40 mg/L, NaHCO3 0.4 g/L, NaCl 80 mg/L, hemin (5 mg/L) and vitamin K (1 mg/L)). C. scindens was plated on PYF agar (PYF with 15 g/L agar) and cultured anaerobically at 37°C using a BD GasPak EZ pouch system (BD, Cat # 260683). Two days before each experiment a fresh subculture was generated from a single colony for co-culture experiments by culturing in the PYF medium for 48 h.
S. aureus strain Newman was kindly provided by Dr. Michelle Reniere at the University of Washington and cultured under aerobic conditions in brain-heart-infusion media (BHI, Research Products International, Cat #B11000) at 37°C. S. aureus was plated on BHI agar (BHI with 15 g/L agar), and a single colony was used to prepare a bacterial sample after 18 h of culture in BHI broth before co-culture experiments.
2.4 Co-culture experiments
After human colonic epithelial cells fully differentiated in the luminal compartment of the hanging baskets, the luminal medium was substituted with a deoxygenated medium consisting of 10% PYF in 1× PBS buffer. This co-culture medium was enriched with 200 µM cholic acid for bile acid metabolism. Simultaneously, the basal medium was refreshed with fresh DM-Ab media. For C. scindens co-culture, 5×103 colony-forming unit (CFU) in total were added to the luminal media similar to that used previously for other Clostridioides (Kim et al., 2019). The sealed co-culture or control cultures without bacteria were incubated at 37°C for 48 h. Due to its rapid growth, a minimal number of S. aureus (5 × 102 CFU in total) was added to specified samples after 24 h of co-culture with C. scindens. The samples with added S. aureus were then incubated at 37°C for an additional 24 h. The deoxygenated luminal media was 10% PYF in 1 x PBS buffer for both C. scindens and S. aureus. All inoculations were performed in an anaerobic chamber.
2.5 Measurement of epithelial cell death
To assess the viability of colonic epithelial cells cultured in the O2 gradient cassette, DM-Ab medium (3 mL) supplemented with 10 μg/mL Hoechst 33342 (Thermo Fisher, Cat #H1399) was added to the basal compartment and then the cells were incubated at 37°C for 1 h. After washing with PBS, 2 μg/mL propidium iodide (PI, Thermo Fisher, Cat # P3566) in PBS buffer (3 mL) was added to the basal compartment, and then incubated at 37°C for 20 min. Hoechst 33342 was used to label DNA in all epithelial cells, and PI was used to label the DNA of dead epithelial cells. To measure apoptosis, a dead cell apoptosis kit with Annexin V (Thermo Fisher, Cat #V13242) was used to assess the presence of phosphatidylserine on the plasma membrane of cells. The working solution of the kit contained 1 μg/mL Annexin V, 1 μg/mL PI, 1.25 mM HEPES, 7 mM NaCl, 0.05 mM EDTA, and 0.005% bovine serum albumin (BSA) in 1× binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2). 500 μL of the working solution was used for luminal staining at 25°C for 15 min, then the reaction was terminated by mixing with 2 mL binding buffer and kept on ice before imaging.
Prior to microscopic imaging, each collagen scaffold with attached cells was carefully cut from the hanging basket, inverted, and placed onto a #1 coverslip. A confocal fluorescence microscope (Olympus, Fluoview 3,000, Japan) was used to measure the fluorescence of the epithelial cells. The entire epithelial monolayer (approximately 1.1 cm2 surface area) was imaged as a 4 × 4 array with a 4× (0.16 N.A.) objective. The following fluorescence excitation/emission wavelengths were used- Hoechst 33342: Ex 405 nm, Em 430–470 nm; PI: Ex 561 nm, Em 610–670 nm; Annexin V: Ex 488 nm, Em 530–575 nm. All 16 images of each channel were stitched together to create an image of the entire well.
CellProfiler v4.0 was used to identify and calculate the image area with fluorescence intensity above an empirically set threshold to quantify the area of the epithelial monolayer positive for each of the stains. The processing protocol was modified from a previously reported method (Hinman et al., 2021). The stitched image at each emission wavelength was flat-field corrected using a Gaussian-smoothed illumination correction function (1000-pixel filter size). The corrected image was converted to binary using 2-class Otsu threshold selection. Then the area of the culture above the fluorescence threshold was calculated. The normalized PI + area was calculated as PI + area/Hoechst 33342+ area and was used as a metric for dead cells. The normalized Annexin V+ area was calculated as Annexin + area/Hoechst 33342+ area and used as a metric for apoptotic cells.
2.6 Immunofluorescence measurement of the tight junction marker ZO-1
To determine whether tight junctions among the epithelial cells were disrupted in the presence of the bacteria, methanol (4°C) was added as a fixative into the luminal compartment, and the sample was placed at −20°C for 1 h. The fixed cells were then washed in PBS and blocked with 3% BSA in PBS for 1 h at 25°C. Next, 200 µL ZO-1 primary antibody (2 μg/mL, Proteintech, Cat #21773-1-AP, RRID:AB_10733242) was added to the luminal compartment and incubated with the cells at 4°C for 18 h. The sample was then washed 3 times with IF wash buffer (0.1% BSA, 0.2% Triton-X 100, 0.05% Tween 20 in 1× PBS) and then incubated with 500 µL secondary antibody conjugated with Alexa Fluor 647 (1 μg/mL, Thermo Fisher Scientific, Cat #A27040, RRID:AB_2536101) containing 10 μg/mL Hoechst 33342 for 1 h at 25°C. The sample was then washed twice with PBS buffer. Prior to confocal microscopy, each collagen scaffold with attached cells was carefully cut from the hanging basket, inverted, and placed on a #1 coverslip. Fluorescence was then quantified using a 30× objective oil lens (1.05 N.A.). Fluorescence was measured using the following parameters- ZO-1: Ex 640 nm, Em 650–750 nm; Hoechst: Ex 405 nm, Em 430–470 nm. Five different locations (top, bottom, right, left, center) of each sample were imaged. The images were processed and analyzed within CellProfiler v4.0 following a previous protocol (Hinman et al., 2021).
2.7 Measurement of bacterial cells
The luminal contents (media + collagen scaffold) were collected and mixed in a tube with 3 mL 500 U/mL collagenase in PBS, followed by 30 min incubation at 25°C under anaerobic conditions. Next, the suspension was vortexed for 2 min, and then plated on PYF (for C. scindens) or BHI (for S. aureus) agar. The CFU on the agar plates after 24–48 h incubation anaerobically (for C. scindens) or aerobically (for S. aureus) was counted and used as a measure of the viability of the bacterial cells.
To visualize the bacterial cells while still present on the epithelial monolayer, 200 µL 1 μM SYTO 9 (Thermo Fisher, Cat #S34854) in PBS was added to the luminal compartment for 20 min. Epithelial cells were visualized by addition of Hoechst 33342 (10 μg/mL, PBS) followed by incubation for 1 h. The collagen scaffold with attached cells was carefully cut from the hanging basket, inverted, and placed on a #1 coverslip. Both bacterial and epithelial cells were imaged by confocal fluorescence microscopy using a 60× oil objective lens (1.3 N.A.) under the following parameters- Hoechst 33342: Ex 405 nm, Em 430–470 nm; SYTO9: Ex 488 nm, Em 500–520 nm.
2.8 ELISA assays of secreted cytokines
The IL-8 concentration in the basal media was measured using an interleukin 8 (IL-8) ELISA kit (Thermo Fisher Scientific, Cat #88-8086-22) following the manufacturer’s protocol. The basal media were diluted at 1:10 (control) or 1:20 (co-culture) so that the measured signals fit in the range of the standard curve provided with the kit. The MCP-1 concentration in the basal media was measured using a monocyte chemoattractant protein-1 (MCP-1) ELISA kit (Thermo Fisher Scientific, Cat #88–7399-88) following the manufacturer’s protocol.
2.9 Measurement of bile acid in epithelial-Clostridium scindens co-culture
After 48 h of co-culture in the presence of bile acid, both the luminal contents (media + collagen matrix) and basal media were collected separately. The luminal contents were degraded by collagenase as described above and the supernatant collected after centrifugation (10 min, 3,000×G). The luminal supernatant and basal media were assayed for bile acids by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis as described previously (Ginos et al., 2018; Navarro et al., 2020) at the University of Washington Mitochondria and Metabolism Center.
As a control, cholic or deoxycholic acid was added to S. aureus cultured in 96-well plates. Diluted S. aureus (1/106) was added to wells containing 200 µL 10% PYF (in 1× PBS) medium supplemented by 25–200 µM cholic acid or deoxycholic acid and incubated anaerobically. After 24 h at 37°C, the optical density (O.D.) at 600 nm of the medium in each well was measured.
2.10 Measurement of total enterotoxin produced by Staphylococcus aureus
The luminal contents (media + collagen scaffold) were collected and mixed with collagenase as described above. The supernatant was collected after centrifugation. The concentration of S. aureus enterotoxin in the supernatant was measured using a sandwich enzyme immunoassay kit (RIDASCREEN® SET Total, R-Biopharm AG, German) following the manufacturer’s protocol. The O.D. of each sample was measured and an O.D. of 0.15 greater than that of the negative control value was set as a threshold for a positive sample. The concentration of enterotoxin was represented as relative O.D., i.e., normalized to the positive control mean (set equal to 100%).
2.11 Statistical analyses
Origin 9 was used for statistical analyses of the data. All p values were calculated by ordinary one-way ANOVA. A significance level of 0.05 was used, and any p-value below this threshold was considered statistically significant. (*: p < 0.05, **: p < 0.01, ***: p < 0.001). In this manuscript, we define a “technical replicate” as being separate experiments on different days but using the cells of a single donor.
3 Results
3.1 Experimental overview and design
C. scindens is a bacterium often found in the human colon (White et al., 1980; MORRIS et al., 1985). C. scindens is remarkable for its 7-dehydroxylation activity that transforms primary bile acids (e.g., cholic acid) to secondary bile acids (deoxycholic acid and lithocholic acid). Secondary bile acids are reported to inhibit the growth of intestinal pathogens, such as S. aureus, supporting a role for C. scindens as a probiotic species enhancing resistance to gastrointestinal infection (Buffie et al., 2015). S. aureus is a common cause of gastrointestinal illness after consumption of S. aureus-contaminated food (CDC, 2018). The toxicity of S. aureus is derived from enterotoxins that increase the permeability of the epithelial barrier and initiate epithelial cell death (Moretó and Pérez-Bosque, 2009; Šuligoj et al., 2020). The overall goal of this study was to co-culture these two bacterial species with primary human gastrointestinal epithelial cells to demonstrate the utility of multi-species gut-on-chip models for intestinal microbiota research and the study of the interactions between the microbiome and human host.
Clostridial species such as C. scindens tolerate O2 concentrations up to 2%–3% (Morvan et al., 2021; Chia, 2023), but higher concentrations are lethal. These obligate anaerobes are able to survive in the environment of the luminal-facing differentiated colonic epithelium that exists in physiologic hypoxia of <1–3% O2. In contrast, the O2 concentration increases to 10% near the colonic epithelial cells at the crypt base, i.e., deeper in the submucosa, where the stem-cell niche produces proliferative cells to replenish the epithelial layer (Keeley and Mann, 2019; Schwerdtfeger et al., 2019). To achieve these distinct O2 conditions, a modified cell culture cassette for O2-gradient formation was previously demonstrated for co-culture of epithelial cells and anaerobes (Kim et al., 2019; Kim et al., 2022). This cassette, based on a hanging basket, possessed luminal and basal reservoirs so that both sides of the colonic epithelial cell culture were accessible. To create an O2 gradient across the tissue surface, the side walls of the luminal compartment were impervious to O2 and an O2 impermeable plug was inserted into the luminal reservoir to prevent atmospheric O2 ingress. In contrast, the basal reservoir was open to the environment. Under these conditions, a confluent monolayer of epithelial cells has been shown to consume O2 at a sufficient rate to decrease the luminal O2 to <1% in 3.75 h (Kim et al., 2019). In the current experiments, epithelial stem cells were plated on the luminal side of the hanging basket’s porous membrane in the cassette and cultured in a medium rich in growth factors (expansion medium, EM) without the luminal plug so that a high O2 tension was maintained (Figures 1A,B). After forming a confluent monolayer, the cells were fully differentiated by removal of growth factors (differentiation medium without antibiotics, DM -Ab) producing a high-resistance epithelial monolayer (TEER = 507 ± 126 Ω·cm2, n = 3, Figure 1C). The luminal DM -Ab was then replaced with a minimal nutrient medium (PYF) suitable for bacterial culture (Figure 1D). The cassette was then transferred into an anaerobic chamber (Figure 1E), the plug was installed, and the assembled cassette was placed in a standard tissue-culture incubator (Figure 1F). Under these conditions, the maximal O2 concentration in the luminal compartment was 0.6% ± 0.4% after plug installation and decreased to a steady state value of 0.4% ± 0.1% during culture due to continued O2 consumption by the epithelial cells (Supplementary Figure S2). C. scindens was subsequently inoculated onto the intestinal epithelium at 103 colony-forming unit (CFU)/ml (1,300 ± 500 cells in total). The CFU increased to 3×104 ± 0.95 × 104 CFU/mL (approximately 15,500 ± 5,000 in total, doubling time 13 h) after 48 h (p = 0.045, Supplementary Figure S3A), suggesting that C. scindens not only tolerated the luminal microenvironment but was able to replicate.
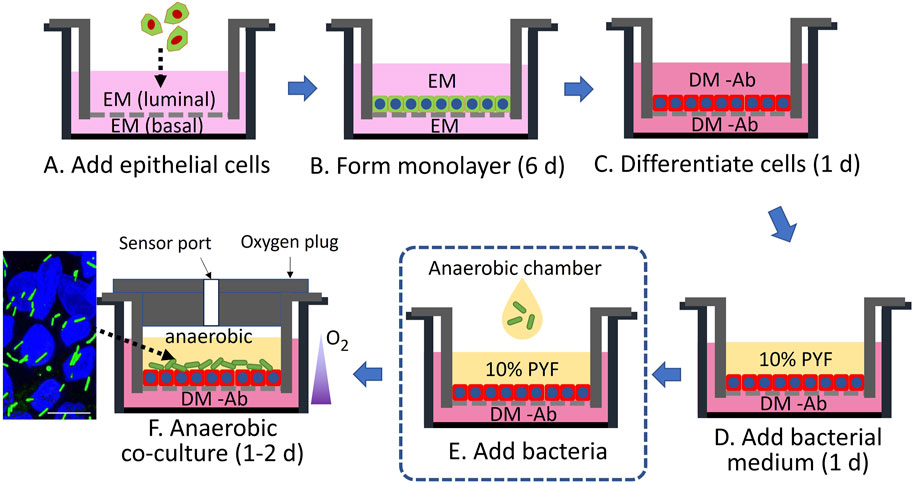
Figure 1. Schematic of co-culture of primary human colonic epithelial cells with bacteria. EM is a medium rich in growth factors to support stem cell expansion (green cells with blue nuclei), while DM -Ab is an antibiotic-free medium without stem-cell supportive growth factors to support the formation of differentiated cells (red cells with blue nuclei). PYF supports the growth of bacteria (green rods). The fluorescence micrograph (far lower left) shows the co-culture of differentiated colonic epithelial cells (Hoechst 33342-stained nuclei in blue) and C. scindens (green rods). The scale bar represents 10 µm.
3.2 Co-culture of epithelial cells with Clostridium scindens
The in vivo colonic lumen acts as a host to C. scindens with co-existence of and mutual benefit for both the mammalian and bacterial cells. C. scindens ferments fiber and metabolizes bile acids to support both the health of the microbial community as well as that of the epithelial cells (Wlodarska et al., 2017; Kang et al., 2019). In turn the epithelial cells form a tight barrier to prevent the intrusion of bacteria into the tissue and collaborate with the immune system to repel invading microbes (Chen et al., 2021). To determine whether the epithelial cells might be able to co-exist with the obligate anaerobes in the absence of an immune system, C. scindens was cultured on a confluent, differentiated epithelial cell layer. The impact on epithelial cell viability and monolayer barrier function was then assessed. When stained with propidium iodide (PI), no significant difference in epithelial cell death was observed in the presence and absence of C. scindens (9.8% ± 7.5% PI + area normalized to total Hoechst 33342+ cell area for the control vs. 10.8% ± 4.2% after 48 h of co-culture with C. scindens, p = 0.64, Figure 2A; Supplementary Figure S4A). Programmed cell death was assessed using an annexin V conjugate assay to identify phosphatidylserine in the outer membrane leaflet. No significant difference in the binding of annexin V to the epithelial cells was observed in the absence and presence of C. scindens (9.5% ± 4.0% annexin V+ area normalized to total Hoechst 33342+ cell area for the control vs. 6.1% ± 1.6% after 48 h of co-culture with C. scindens, p = 0.24, Figure 2B; Supplementary Figure S4B). Barrier integrity was measured by assaying the TEER after 48 h of culture with and without C. scindens. In both instances the TEER decreased over time but was not significantly different (control 168 ± 18 vs. 150 ± 37 Ω·cm2 with C. scindens, p = 0.45, Supplementary Figure S5A). Since TEER represents a global measurement of barrier function, the integrity of cell-cell junctions was also assessed by fluorescent staining for ZO-1, which is a constituent of tight junctions located on the cytoplasmic membrane surface (McNeil et al., 2006). When visualized by microscopy, cells with and without C. scindens co-cultured for 48 h possessed similar ZO-1 staining patterns (Figure 2C). When the area, circularity, and perimeter of the cells was measured, no significant differences were identified for the epithelial cells with and without C. scindens (area: 77 ± 19 μm2 without vs. 70 ± 15 μm2 with, p = 0.3; circularity: 0.78 ± 0.02 without vs. 0.79 ± 0.02 with, p = 0.2; perimeter: 34 ± 5 µm without vs. 32 ± 4 µm with, p = 0.3, N = 3 technical replicates, Supplementary Figure S6). These data suggest that although the number of C. scindens expanded 10-fold, the epithelial cells did not die or lose barrier integrity during the 48 h of co-culture. However, extremely high numbers of inoculated C. scindens (∼5×106 cells) did result in an increase in epithelial cell death at 48 h of co-culture (Supplementary Figure S7).
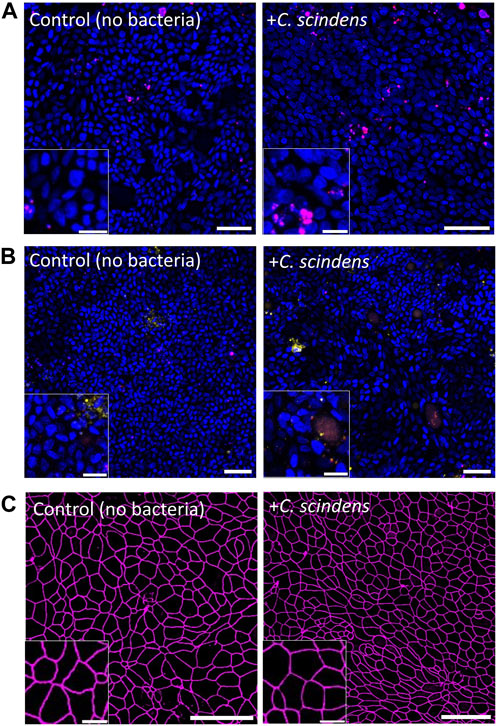
Figure 2. Co-culture of primary human colonic epithelial cells with C. scindens. (A, B) Fluorescence images of colonic epithelial cells stained with PI (red, panel (A), annexin V (yellow, panel (B), and Hoechst 33342 (blue) with and without C. scindens co-culture for 48 h. Scale bar = 50 µm with the inset scale bar = 10 µm. (C) Images of colonic epithelial cells stained for ZO-1 (magenta). Scale bar = 50 µm with the inset scale bar = 10 µm.
3.3 Measurement of epithelial cell inflammation in the presence of Clostridium scindens
IL-8 and MCP-1 are chemokines produced by epithelial cells in response to bacterial products and are often used as markers of an epithelial cell inflammatory response (Deshmane et al., 2009; La Pine and Hill, 2011). The concentrations of IL-8 and MCP-1 in the basal media were measured in the absence and presence of C. scindens co-culture after 48 h. The concentration of IL-8 was not significantly different in the two culture conditions (687 ± 149 pg/mL without vs. 789 ± 199 pg/mL with C. scindens, p = 0.38, Supplementary Figure S8A). Likewise, the concentration of MCP-1 was not significantly different (9.8 ± 2.1 pg/mL without vs. 7.4 ± 1.8 pg/mL with C. scindens, p = 0.21, Supplementary Figure S8B). The concentration of IL-8 secreted into the basal reservoir was similar to that in previous studies (300–600 pg/mL) (Wang et al., 2018b; Kim et al., 2022). Additionally, the concentration of MCP-1 was near that reported for colonic epithelial cell cultures (6.9–27 pg/mL) (Futagami et al., 2003; Fujita et al., 2010). Under these conditions, the co-culture of C. scindens did not alter the inflammatory response of the colonic cells.
3.4 Metabolism of cholic acid by Clostridium scindens
Bile acids flow from the liver into the small intestine via the bile duct. The major portion of bile acids (∼80%) are actively transported back to liver in the terminal ileum. However, ∼15% of bile acids transit through the digestive tract to reach the colon and are then metabolized by bacteria into secondary bile acids that diffuse passively across the colonic epithelium to return to the liver (Di Ciaula et al., 2017). C. scindens is known to convert primary bile acids such as cholic acid into a variety of secondary bile acids (e.g., deoxycholic acid and lithocholic acid) in the colon (White et al., 1980; Ridlon et al., 2014; Kang et al., 2019). To assess bile acid conversion by C. scindens, cholic acid (200 µM) was added in the luminal reservoir in the presence of epithelial cells with or without C. scindens and the concentrations of cholic acid and its metabolites were measured by mass spectrometry (Figure 3). In the absence of C. scindens, only cholic acid (99.37% of total luminal bile acids) was found in the luminal reservoir. In these cultures, cholic acid was also detected in the basal compartment (98.06% of total basal bile acids). In contrast, the luminal reservoir of epithelial cells co-cultured with C. scindens demonstrated a range of secondary bile acids (>99% conversion) with the predominant being apocholic acid (58.33% of total luminal bile acids), which is a byproduct of the dehydration of cholic acid (Shi et al., 2023). Other prominent metabolites identified in the luminal compartment included 3α-hydroxy-12 ketolithocholic acid (21.96% of total luminal bile acids) and deoxycholic acid (11.84% of total luminal bile acids). These two metabolites are formed via a dehydroxylation pathway of C. scindens and have been observed previously in vitro and in vivo (Marion et al., 2019; Funabashi et al., 2020). The basal compartment possessed a distinct pattern of bile acids. The predominant secondary bile acid was deoxycholic acid (50.49% of total basal bile acids) in the presence of C. scindens, but untransformed cholic acid was also present (41.61% of total basal bile acids) as were other products. The presence of the bile acids in the basal compartment was most likely due to passive diffusion as occurs in the colon in vivo (Mekhjian et al., 1979). These data suggest that molecular interactions between C. scindens-epithelial cells, such as bile acid metabolism were present in this in vitro system.
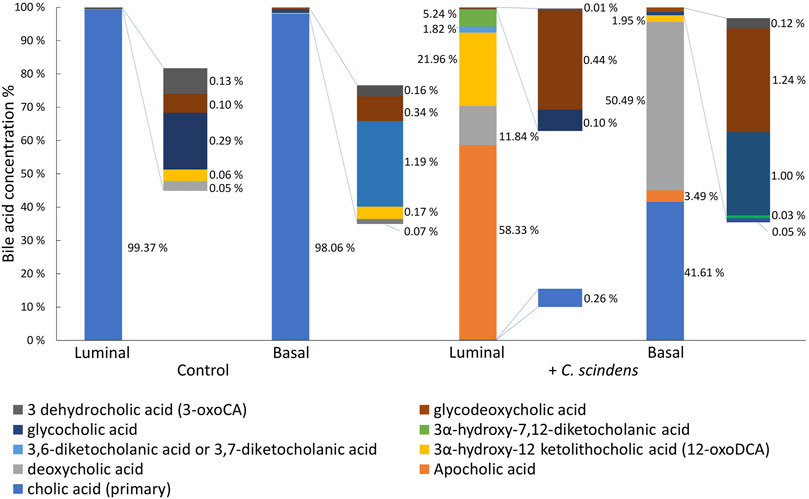
Figure 3. Formation of cholic acid metabolites. Cholic acid was added to the luminal compartment of the hanging basket with cultured epithelial cells in the absence and presence of C. scindens. The metabolites were measured in the luminal and basal compartments of the cassette by mass spectrometry.
3.5 Co-culture of colonic epithelial cells with the intestinal pathogen Staphylococcus aureus
S. aureus is a facultative anaerobe normally found on human skin and within the nasal cavities but can be an opportunistic pathogen of the colon (O'Gara, 2017). S. aureus is able to produce a range of toxins including enterotoxins (SEs), hemolysins, and leukotoxins (Kong et al., 2016) so that colonization of the colon by S. aureus can lead to severe nausea, vomiting, and diarrhea (CDC, 2018). To examine the cytotoxicity of S. aureus to colonic epithelial cells, we inoculated S. aureus onto the epithelial monolayer at 900 ± 370 CFU/mL (450 ± 190 cells in total). After 24 h of co-culture in the anaerobic cassette, S. aureus numbers increased to 1.4×108 ± 6 × 107 CFU/mL (7.0×107 ± 2.8×107 total cells), which was significantly higher than the initial inoculum (p = 0.02, Figure 4A; Supplementary Figure S3B) and represents a doubling time of ∼83 min. Growth of S. aureus on the epithelial cell culture was substantially more robust than that of C. scindens (Moretó and Pérez-Bosque, 2009; Martens et al., 2021). As the epithelial cells died as a result of co-culture with S. aureus, many nutrients may have become available to benefit the growth of S. aureus. In these co-cultures, S. aureus grew both in a dispersed manner as well as in clusters or sheets of bacteria fully covering the monolayer, suggesting the formation of biofilms.
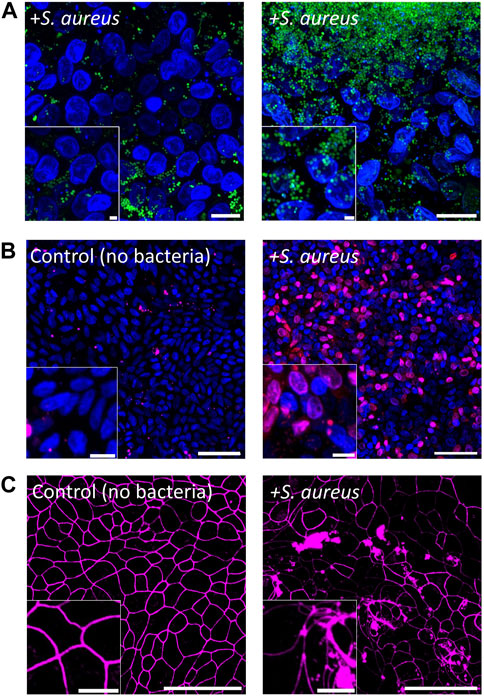
Figure 4. Co-culture of colonic epithelial cells with Staphylococcus aureus. (A) Fluorescence images of epithelial cell co-culture with S. aureus after 1 d. Colonic epithelial cell nuclei were stained with Hoechst 33342 (blue), and S. aureus was stained with SYTO9 (green). Scale bar = 10 µm and the inset scale bar is 2 µm. (B) Images of colonic epithelial cells stained with PI (red) and Hoechst 33342 (blue) cultured with and without S. aureus for 1 d. Scale bar = 50 µm and the inset scale bar = 10 µm. (C) Images of colonic epithelial cells stained with ZO-1 (magenta) when cultured with and without S. aureus. Scale bar = 50 µm and the inset scale bar = 10 µm.
The impact of S. aureus on the epithelial cells was assessed by measuring epithelial cell death and monolayer barrier integrity. Compared to control cultures, the viability of colonic epithelial cells in the presence of S. aureus was significantly decreased after 24 h of co-culture (9.8% ± 7.5% PI + area normalized to total Hoechst 33342+ cell area for the control vs. 68% ± 16% after co-culture with S. aureus, p = 0.0007, Figure 5E). Visually, a majority of the epithelial cells appeared dead in the cultures (Figure 4B). This is consistent with the measured TEER of 70 ± 12 Ω·cm2 (p < 0.001 relative to control, Supplementary Figure S5B), indicating a complete loss of barrier function in the presence of S. aureus after 24 h. To examine cell-cell tight junctions, the epithelial monolayers were stained for ZO-1, which showed a loss of cell boundaries and an appearance of large aggregates of ZO-1 in the colonic cells. This finding indicated that the intercellular tight junctions were disrupted in these cultures (Figure 4C). Further confirmation was obtained by quantification of the epithelial cells’ morphology with the area, perimeter, and circularity all significantly different compared to that of colonic cells cultured in the absence of S. aureus (area: 77 ± 19 μm2 without vs. 161 ± 43 μm2 with, p < 0.001; circularity: 0.78 ± 0.02 without vs. 0.69 ± 0.04 with, p < 0.001; perimeter: 34 ± 5 µm without vs. 54 ± 7 µm with, p < 0.001, N = 3 technical replicates, Supplementary Figure S6). The area and perimeter of the epithelial cells increased in the presence of S. aureus while the circularity decreased, in agreement with the observed distorted cell boundaries on the micrographs. Consistent with these results, the IL-8 concentration in the basal compartment of the epithelial cells was significantly increased after 24 h of S. aureus co-culture compared to that for the epithelial cells alone (531 ± 196 pg/mL vs. 1,382 ± 329 pg/mL, p = 0.02, Figure 5F). It is noteworthy that S. aureus was inoculated onto the epithelial monolayer at a 2-to-3-fold lower CFU than C. scindens. The shorter S. aureus doubling time and potentially toxin production under these conditions enabled S. aureus to expand in number rapidly, leading to a cellular inflammatory response, cell death, and destruction of the monolayer barrier integrity.
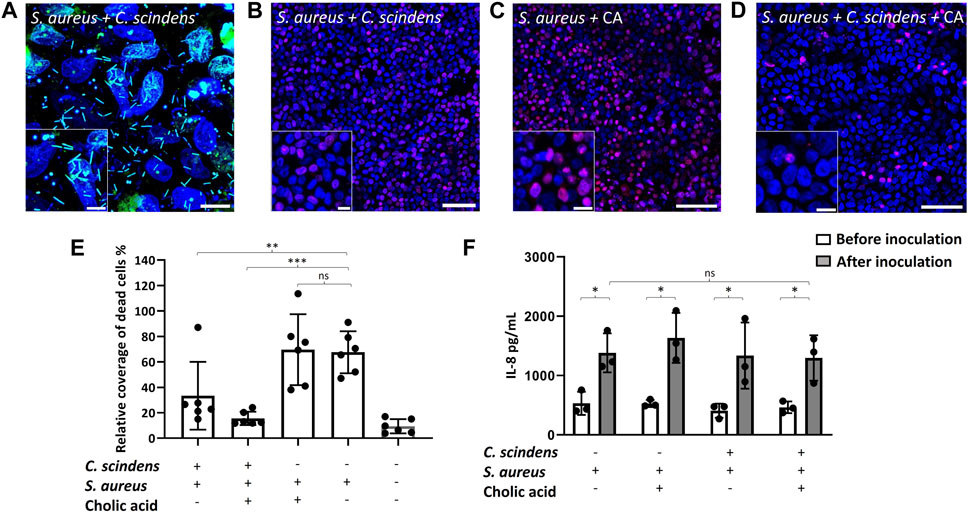
Figure 5. Co-culture of colonic epithelial cells with C. scindens and S. aureus. (A) Images of the co-culture. The nuclei of colonic epithelial cells were stained with Hoechst 33342 (blue), and S. aureus and C. scindens were stained with SYTO9 (green). Scale bar = 10 µm and the inset scale bar = 5 µm. (B–D) Images of colonic epithelial cells stained with PI (red) and Hoechst 33342 (blue) co-cultured with S. aureus + C. scindens (B), S. aureus + 200 µM cholic acid (C), and S. aureus + C. scindens + 200 µM cholic acid (D), scale bar = 50 µm and the inset scale bar = 10 µm. (E) Measurement of dead cells in the absence and presence of C. scindens, S. aureus, and/or cholic acid. The Y-axis is the PI-positive area normalized to the Hoechst 33342-positive area, N = 6 technical replicates. (F) The IL-8 concentration was measured in the basal media before (open bars) and after (closed bars) co-culturing with S. aureus, C. scindens, and/or cholic acid (200 mM). N = 3 technical replicates.
3.6 Co-culture of epithelial cells with Clostridium scindens and Staphylococcus aureus
As a commensal strain, C. scindens exerts a protective effect on the colon by converting 7α-dehydroxylating primary bile acids to form secondary bile acids which then inhibit pathogenic bacterial growth (Greathouse et al., 2015; Kang et al., 2019). To explore this defensive behavior of C. scindens, epithelial monolayers were colonized with C. scindens by co-culture for 24 h. S. aureus was then inoculated onto the C. scindens/epithelial cell co-culture (Figure 5A). After an additional 24 h of incubation, epithelial cell death, cell morphology, and secreted cytokines were measured. Pre-culture of the epithelial cells with C. scindens significantly enhanced the viability of the epithelial cells after addition of S. aureus (p = 0.02, Figures 5B,E). Remarkably, epithelial cell death in the presence of both C. scindens and S. aureus was not significantly different from epithelial cells in the absence of bacteria (p = 0.18, Figure 5E). Furthermore when ZO-1 immunofluorescence was measured, the presence of pre-cultured C. scindens also reduced the alterations in cell morphology in the co-culture with S. aureus compared to culture in the absence of C. scindens (area: 161 ± 43 μm2 with S. aureus only vs. 116 ± 30 μm2 with both, p = 0.01; circularity: 0.69 ± 0.04 with S. aureus only vs. 0.74 ± 0.03 with both, p = 0.003; perimeter: 54 ± 7 µm with S. aureus only vs. 43 ± 7 µm with both, p = 0.008, N = 3 technical replicates, Supplementary Figures S6, S10). The presence of C. scindens, however, did not significantly alter the amount of IL-8 secreted in the presence of S. aureus alone (p = 0.8, Figure 5F) suggesting that the epithelium was still inflamed by the presence of the S. aureus, but protected from death by the co-culture with C. scindens. The measured S. aureus CFU in the absence or presence of C. scindens was not significantly different (7.0×107 ± 2.8×107 without vs. 6.3×107 ± 2.4×107 with C. scindens, p = 0.83, Supplementary Figure S3B), therefore repression of S. aureus growth was not an explanation for the increased epithelial cell survival, which was affirmed when the two bacteria were co-cultured in the absence of an epithelium (Kang et al., 2019).
Secondary bile acids are reported to diminish the negative impact of S. aureus by disruption of the bacterium’s membrane (Sannasiddappa et al., 2017), and by minimizing excessive inflammation through inhibition of NF-κB and NLRP3 pathways (Zhao et al., 2023). Cholic acid (200 µM) was added to the luminal compartment during C. scindens colonization to enable the formation of secondary bile metabolites. S. aureus was then added, and epithelial cell death and IL-8 secretion were measured after 24 h. Epithelial cell death was not significantly decreased by the addition of cholic acid to the C. scindens-S. aureus mixed culture (p = 0.13, Figure 5B,D,E). IL-8 production was also not significantly altered in the presence of cholic acid relative to that without cholic acid (p = 0.92, Figure 5F). Cholic acid added to epithelial monolayers with only S. aureus (no C. scindens) did not significantly alter epithelial cell viability (p = 0.88) or IL-8 production (p = 0.46) compared to S. aureus only co-cultures (Figure 5C,E,F). The measured S. aureus CFU in the absence or presence of C. scindens + cholic acid was not significantly different (7.0×107 ± 2.8×107 without vs. 6.6×107 ± 3.8×107 with C. scindens/cholic acid, p = 0.93, Supplementary Figure S3B), so repression of S. aureus growth was again not a likely explanation for the improved epithelial cell survival. These results depart from the findings of prior studies in which bile acids repressed the growth of S. aureus (Sannasiddappa et al., 2017; Kang et al., 2019); however, we note that epithelial cells were absent from those experiments. For this reason, we cultured S. aureus in the presence of cholic acid and deoxycholic acid but without epithelial cells. We observed a significantly decreased growth rate as measured by O.D. (Supplementary Figure S9). It is likely that the epithelial cells, C. scindens, or their metabolic products other than bile acids are acting in a complex manner to impact S. aureus growth.
3.7 Measurement of Staphylococcus aureus toxin in the presence of Clostridium scindens and epithelial cells
S. aureus enterotoxins (SEs) are potent exotoxins that damage colonic epithelial cells by stimulating the secretion of cytokines and provoking inflammation (Aljasir and D’Amico, 2020; Cretenet et al., 2011). Enterotoxin B (SEB) also disrupts the integrity of epithelial barriers, initiating cell death (Martens et al., 2021). To determine whether C. scindens reduces the cytotoxicity of S. aureus by altering toxin formation, enterotoxin (SE) production by S. aureus was measured in the luminal reservoir. For simplicity, the selected assay measured total enterotoxin (SEA, SEB, SEC, and SED). Addition of C. scindens with or without cholic acid significantly diminished total SE production by S. aureus compared to cultures without C. scindens pre-culture (p = 0.007, C. scindens; p = 0.012 C. scindens + cholic acid) (Figure 6). Addition of cholic acid to the luminal reservoir of S. aureus/epithelial cell cultures in the absence of C. scindens did not significantly alter toxin production compared to the cultures without cholic acid (p = 0.5) suggesting that cholic acid itself did not modulate toxin production (Figure 6). These results indicate that the production of SE by S. aureus was suppressed when C. scindens was present, but this suppression was not reliant on the creation of secondary bile acids. Similar findings have been observed in co-cultures of S. aureus with other bacterial species in vitro (Zdenkova et al., 2016; Nogueira Viçosa et al., 2019; Rodríguez-Sánchez et al., 2022). However, it is noteworthy that this phenomenon was observed for the first time in S. aureus-C. scindens co-culture in the presence of colonic cells.
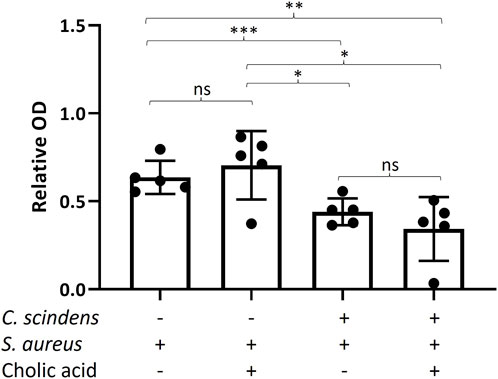
Figure 6. Measurement of total enterotoxin produced by S. aureus in the absence and presence of C. scindens ± cholic acid. The Y-axis is the relative optical density (O.D., 420 nm) normalized to an enterotoxin standard, N = 5 technical replicates. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
4 Discussion
Culture innovations were made to maintain the viability of epithelial cells in culture with bacteria for 48 h when using the O2 gradient cassette. In contrast, prior cultures of the epithelial cells in the O2 gradient cassette were heavily damaged at 24 h (4, 7). Additionally, prior reports used only a single, probiotic, nonpathogenic bacteria due to the very short lifespan of the epithelial cells under the conditions used. In this study, the obligate anaerobic commensal C. scindens was co-cultured with primary human colonic epithelial cells under an O2 gradient. The O2 gradient enabled survival of the obligate anaerobic bacteria in the anaerobic microenvironment while providing adequate O2 concentration for the support of the human cells. Compatibility between the two species (human and microbe) over a 48-h period was shown by the high cell viability, excellent barrier function, appropriate cell morphology, and continued cytokine production. The extended epithelial cell lifetime enabled pre-colonization of the epithelium with a probiotic (C. scindens) followed by introduction of a pathogen strain (S. aureus) to mimic the sequence of events occurring during intestinal infection in vivo. The O2 gradient cassette is a simple, inexpensive, easy-to-use device that does not require a flow system so that this culture system with extended lifetime is accessible to nonexperts.
When the intestinal pathogen S. aureus was inoculated in the luminal compartment at lower density than C. scindens under comparable conditions, greater than 60% of the epithelial cells died within 24 h. When S. aureus was inoculated after establishing an epithelial cells-C. scindens co-culture, death of the epithelial cells was greatly reduced and was not significantly different compared with the amount of cell death in the absence of bacteria. These results suggest that the toxicity of S. aureus was reduced in the presence of C. scindens. Although the growth of S. aureus was not noticeably repressed in the presence of C. scindens, the concentration of enterotoxin decreased by nearly 50%. C. scindens has been reported in multiple in vitro studies and an animal study to have an antimicrobial effect on intestinal pathogenic strains (Buffie et al., 2015; Greathouse et al., 2015; Kang et al., 2019; Zhao et al., 2023). The co-culture results here demonstrate a similar outcome in the presence of primary human colon cells within our in vitro platform. These data suggest that the platform could be used to expand the study of antimicrobial effects of intestinal probiotics in an environment more closely mimicking the in vivo colonic environment.
Bile acids are known to play important roles in modulating the intestinal microbiome. C. scindens converts primary bile acids (cholic and chenodeoxycholic acid) to secondary bile acids (e.g., deoxycholic acid and lithocholic acid) in the intestine (Greathouse et al., 2015; Kang et al., 2019). Furthermore, secondary bile acids affect lipid membranes similarly to detergent (Kumar et al., 2020) and are believed to serve an antimicrobial role within the intestinal microbiome (Ridlon et al., 2016). In the current study, although C. scindens was shown to convert cholic acid to secondary bile acids in the luminal medium, the growth of S. aureus was not significantly affected by these bile acid products. This finding was in contrast to prior antimicrobial studies using bile acid supplemented medium, which demonstrated reduced S. aureus growth (Supplementary Figure S9). Of note is that these studies did not utilize C. scindens for bile acid conversion and epithelial cells were not present. One explanation for the different outcomes could be the transport of secondary bile acids from the luminal to the basal compartment across the epithelial monolayer. In the intestine, >95% of bile acids are re-absorbed and then delivered to the liver through the portal vein (Ticho et al., 2019). In the current studies, the majority of deoxycholic acid, which has a stronger antimicrobial effect on S. aureus compared to other secondary bile acids (Kang et al., 2019), was detected in the basal media. Thus, the decreased concentration of bile acid in the luminal reservoir may have reduced its antimicrobial impact on S. aureus. In the presence of the cholic acid in the cultures the dominant species in the basal medium was deoxycholic acid while the apocholic acid was the majority metabolite in the luminal compartment. Apocholic acid is an uncommon bile acid found in human feces and known to be a carcinogenic byproduct resulting from the mild dehydration of cholic acid facilitated by bacterial dehydrogenases (Shi et al., 2023). While there are no reports of apocholic acid production by C. scindens, our findings suggest that the deoxygenation pathway of intestinal bacteria may be influenced by the presence of host cells, a phenomenon not observed in single strain cultures.
Enterotoxin is known to trigger an immune response that leads to damage of intestinal mucosal cells and diarrhea (Fayyaz et al., 2022). At least 20 types of S. aureus enterotoxins with a molecular size of ∼25 kDa (220–240 amino acids) have been identified (Pinchuk et al., 2010). The production of enterotoxins is dependent on a variety of parameters, including temperature, pH, and the presence of other microorganisms (Cretenet et al., 2011; Perales-Adán et al., 2018). Recent studies have shown that probiotic isolates can alter biochemical conditions to repress S. aureus enterotoxin production in bacterial co-culture in vitro without significantly inhibiting the growth of S. aureus (Aljasir and D’Amico, 2020). Similar results were observed here in the presence of colonic epithelial cells. Further studies on the role of probiotics in altering the luminal environment in the presence of human epithelium and pathogenic bacteria will be valuable in gaining a better understanding of the interplay of how these cells impact enterotoxin production and activity.
In the colon, stem cells at the base of the intestinal crypts serve to replenish the epithelium as the differentiated cells undergo apoptosis within a few days of migrating to the luminal surface (Wang et al., 2018a). This process maintains a self-regenerating epithelial monolayer believed to be influenced by intestinal microbiota (Donaldson et al., 2016). The primary colonic epithelial monolayer used in this study was composed of fully differentiated cells. In the absence of co-cultured bacteria, this monolayer maintains a high TEER barrier for only 2–3 days once fully differentiated due to the natural lifetime of differentiated colonic epithelial cells and a lack of proliferative cells that would otherwise maintain the monolayer (Wang et al., 2017; Wang et al., 2018a). After this time the TEER drops as the cells begin to die without replacement. The change in TEER over time is consistent with previous studies using soft scaffolds (Gunasekara et al., 2018; Kim et al., 2019; Speer et al., 2019). Future studies are planned with long-lived (>30 days) 2D and 3D crypt models that maintain a stem-cell niche to continually support the epithelium (Wang et al., 2018a; Wang et al., 2022). These models are expected to sustain epithelial integrity for significantly longer duration and enable the study of more complex microbiota and epithelial interactions.
In this study, fully differentiated, primary human colonic epithelial cells were inoculated with either intestinal probiotic (C. scindens), pathogenic (S. aureus), or both bacterial species and co-cultured under an O2 gradient. The O2 gradient produced the necessary anaerobic environment for the obligate and facultative anaerobic bacteria while maintaining the viability and function of the mammalian cells. The epithelial monolayer retained viability and barrier integrity in the presence of C. scindens for 48 h but was degraded within 24 h in the presence of a smaller inoculum of S. aureus. Pre-culturing the epithelium with C. scindens reduced the concentration of enterotoxin in subsequent co-culture with S. aureus and improved the survival of the epithelial cells but did not inhibit the growth of S. aureus. The O2-gradient cassette provided an effective and economical platform to study the interactions of intestinal microbiota with primary human cells in an environment more closely mimicking the colonic environment and provides an easy-to-use system for studies of the gut microbiome.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by University of Washington and University of North Carolina review boards. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
HW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. RK: Conceptualization, Writing–review and editing. YW: Conceptualization, Writing–review and editing. KF: Conceptualization, Writing–review and editing. CS: Conceptualization, Writing–review and editing. NA: Conceptualization, Writing–review and editing, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing–original draft. RT: Conceptualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health under R01DK120606.
Acknowledgments
We would like to thank Lauren Shull at the University of Washington for providing materials, and University of Washington Mitochondria and Metabolism Center for technical assistance.
Conflict of interest
NA, YW, CS. have a financial interest in Altis Biosystems LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1382389/full#supplementary-material
References
Alam, M. A., Al-Jenoobi, F. I., and Al-Mohizea, A. M. (2012). Everted gut sac model as a tool in pharmaceutical research: limitations and applications. J. Pharm. Pharmacol. 64, 326–336. doi:10.1111/j.2042-7158.2011.01391.x
Aljasir, S. F., and D’amico, D. J. (2020). The effect of protective cultures on Staphylococcus aureus growth and enterotoxin production. Food Microbiol. 91, 103541. doi:10.1016/j.fm.2020.103541
Azzouz, L. L., and Sharma, S. (2022). “Physiology, large intestine,” in StatPearls (Treasure Island (FL): StatPearls Publishing LLC).
Bartfeld, S., Bayram, T., Van De Wetering, M., Huch, M., Begthel, H., Kujala, P., et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6. doi:10.1053/j.gastro.2014.09.042
Baruch, E. N., Youngster, I., Ben-Betzalel, G., Ortenberg, R., Lahat, A., Katz, L., et al. (2021). Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609. doi:10.1126/science.abb5920
Buffie, C. G., Bucci, V., Stein, R. R., Mckenney, P. T., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi:10.1038/nature13828
CDC (2018). Staphylococcal (staph) food poisoning. Available at: https://www.cdc.gov/foodsafety/diseases/staphylococcal.html.
Chen, Y., Cui, W., Li, X., and Yang, H. (2021). Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front. Immunol. 12, 761981. doi:10.3389/fimmu.2021.761981
Cheng, W. Y., Wu, C. Y., and Yu, J. (2020). The role of gut microbiota in cancer treatment: friend or foe? Gut 69, 1867–1876. doi:10.1136/gutjnl-2020-321153
Chia, A. R. (2023). “The biological characterization and development of molecular engineering tools for Clostridium scindens,”. Master of Science in Engineering (Baltimore, MD, United States: Johns Hopkins University).
Clarke, G., Stilling, R. M., Kennedy, P. J., Stanton, C., Cryan, J. F., and Dinan, T. G. (2014). Minireview: gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28, 1221–1238. doi:10.1210/me.2014-1108
Clarke, L. L. (2009). A guide to Ussing chamber studies of mouse intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1151–G1166. doi:10.1152/ajpgi.90649.2008
Co, J. Y., Margalef-Català, M., Li, X., Mah, A. T., Kuo, C. J., Monack, D. M., et al. (2019). Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520.e4. doi:10.1016/j.celrep.2019.01.108
Cretenet, M., Even, S., and Le Loir, Y. (2011). Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. Dairy Sci. Technol. 91, 127–150. doi:10.1007/s13594-011-0014-9
Davar, D., Dzutsev, A. K., Mcculloch, J. A., Rodrigues, R. R., Chauvin, J. M., Morrison, R. M., et al. (2021). Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602. doi:10.1126/science.abf3363
Deshmane, S. L., Kremlev, S., Amini, S., and Sawaya, B. E. (2009). Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interferon Cytokine Res. 29, 313–326. doi:10.1089/jir.2008.0027
Di Ciaula, A., Garruti, G., Lunardi Baccetto, R., Molina-Molina, E., Bonfrate, L., Wang, D. Q., et al. (2017). Bile acid physiology. Ann. Hepatol. 16 (Suppl. 1), S4–S14. doi:10.5604/01.3001.0010.5493
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi:10.1038/nrmicro3552
Dutton, J. S., Hinman, S. S., Kim, R., Wang, Y., and Allbritton, N. L. (2019). Primary cell-derived intestinal models: recapitulating physiology. Trends Biotechnol. 37, 744–760. doi:10.1016/j.tibtech.2018.12.001
Fayyaz, K., Nawaz, A., Olaimat, A. N., Akram, K., Farooq, U., Fatima, M., et al. (2022). Microbial toxins in fermented foods: health implications and analytical techniques for detection. J. Food Drug Anal. 30, 523–537. doi:10.38212/2224-6614.3431
Fujita, K., Ewing, C. M., Getzenberg, R. H., Parsons, J. K., Isaacs, W. B., and Pavlovich, C. P. (2010). Monocyte chemotactic protein-1 (MCP-1/CCL2) is associated with prostatic growth dysregulation and benign prostatic hyperplasia. Prostate 70, 473–481. doi:10.1002/pros.21081
Funabashi, M., Grove, T. L., Wang, M., Varma, Y., Mcfadden, M. E., Brown, L. C., et al. (2020). A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570. doi:10.1038/s41586-020-2396-4
Futagami, S., Hiratsuka, T., Tatsuguchi, A., Suzuki, K., Kusunoki, M., Shinji, Y., et al. (2003). Monocyte chemoattractant protein 1 (MCP-1) released from Helicobacter pylori stimulated gastric epithelial cells induces cyclooxygenase 2 expression and activation in T cells. Gut 52, 1257–1264. doi:10.1136/gut.52.9.1257
Ginos, B. N. R., Navarro, S. L., Schwarz, Y., Gu, H., Wang, D., Randolph, T. W., et al. (2018). Circulating bile acids in healthy adults respond differently to a dietary pattern characterized by whole grains, legumes and fruits and vegetables compared to a diet high in refined grains and added sugars: a randomized, controlled, crossover feeding study. Metabolism 83, 197–204. doi:10.1016/j.metabol.2018.02.006
Greathouse, K. L., Harris, C. C., and Bultman, S. J. (2015). Dysfunctional families: Clostridium scindens and secondary bile acids inhibit the growth of Clostridium difficile. Cell Metab. 21, 9–10. doi:10.1016/j.cmet.2014.12.016
Guarner, F., and Malagelada, J. R. (2003). Gut flora in health and disease. Lancet 361, 512–519. doi:10.1016/s0140-6736(03)12489-0
Gunasekara, D. B., Speer, J., Wang, Y., Nguyen, D. L., Reed, M. I., Smiddy, N. M., et al. (2018). A monolayer of primary colonic epithelium generated on a scaffold with a gradient of stiffness for drug transport studies. Anal. Chem. 90, 13331–13340. doi:10.1021/acs.analchem.8b02845
Harris, C., and Scolapio, J. S. (2019). Initial evaluation and care of the patient with new-onset intestinal failure. Gastroenterol. Clin. North Am. 48, 465–470. doi:10.1016/j.gtc.2019.08.001
Heinritz, S. N., Mosenthin, R., and Weiss, E. (2013). Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr. Res. Rev. 26, 191–209. doi:10.1017/s0954422413000152
Hinman, S. S., Huling, J., Wang, Y., Wang, H., Bretherton, R. C., Deforest, C. A., et al. (2021). Magnetically-propelled fecal surrogates for modeling the impact of solid-induced shear forces on primary colonic epithelial cells. Biomaterials 276, 121059. doi:10.1016/j.biomaterials.2021.121059
Kang, J. D., Myers, C. J., Harris, S. C., Kakiyama, G., Lee, I. K., Yun, B. S., et al. (2019). Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol. 26, 27–34.e4. doi:10.1016/j.chembiol.2018.10.003
Keeley, T. P., and Mann, G. E. (2019). Defining physiological normoxia for improved translation of cell physiology to animal models and humans. Physiol. Rev. 99, 161–234. doi:10.1152/physrev.00041.2017
Khan, I., Bai, Y., Zha, L., Ullah, N., Ullah, H., Shah, S. R. H., et al. (2021). Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front. Cell Infect. Microbiol. 11, 716299. doi:10.3389/fcimb.2021.716299
Kim, R., Attayek, P. J., Wang, Y., Furtado, K. L., Tamayo, R., Sims, C. E., et al. (2019). An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 12, 015006. doi:10.1088/1758-5090/ab446e
Kim, R., Wang, Y., Sims, C. E., and Allbritton, N. L. (2022). A platform for Co-culture of primary human colonic epithelium with anaerobic probiotic bacteria. Front. Bioeng. Biotechnol. 10, 890396. doi:10.3389/fbioe.2022.890396
Kong, C., Neoh, H. M., and Nathan, S. (2016). Targeting Staphylococcus aureus toxins: a potential form of anti-virulence therapy. Toxins (Basel) 8, 72. doi:10.3390/toxins8030072
Kumar, Y., Yadav, R., and Bhatia, A. (2020). Can natural detergent properties of bile acids be used beneficially in tackling coronavirus disease-19? Future Virol. 15, 779–782. doi:10.2217/fvl-2020-0210
La Pine, T. R., and Hill, H. R. (2011). “148 - host defense mechanisms against bacteria,” in Fetal and neonatal physiology. Editors R. A. POLIN, W. W. FOX, and S. H. ABMAN Fourth Edition (Philadelphia: W.B. Saunders).
Li, C., and Zhang, X. (2022). Current in vitro and animal models for understanding foods: human gut-microbiota interactions. J. Agric. Food Chem. 70, 12733–12745. doi:10.1021/acs.jafc.2c04238
Marion, S., Studer, N., Desharnais, L., Menin, L., Escrig, S., Meibom, A., et al. (2019). In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 10, 481–503. doi:10.1080/19490976.2018.1549420
Marrero, D., Pujol-Vila, F., Vera, D., Gabriel, G., Illa, X., Elizalde-Torrent, A., et al. (2021). Gut-on-a-chip: mimicking and monitoring the human intestine. Biosens. Bioelectron. 181, 113156. doi:10.1016/j.bios.2021.113156
Martens, K., Seys, S. F., Alpizar, Y. A., Schrijvers, R., Bullens, D. M. A., Breynaert, C., et al. (2021). Staphylococcus aureus enterotoxin B disrupts nasal epithelial barrier integrity. Clin. Exp. Allergy 51, 87–98. doi:10.1111/cea.13760
Mcneil, E., Capaldo, C. T., and Macara, I. G. (2006). Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 17, 1922–1932. doi:10.1091/mbc.e05-07-0650
Mekhjian, H. S., Phillips, S. F., and Hofmann, A. F. (1979). Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig. Dis. Sci. 24, 545–550. doi:10.1007/bf01489324
Moretó, M., and Pérez-Bosque, A. (2009). Dietary plasma proteins, the intestinal immune system, and the barrier functions of the intestinal mucosa. J. Anim. Sci. 87, E92–E100. doi:10.2527/jas.2008-1381
Morris, G. N., Winter, J., Cato, E. P., Ritchie, A. E., and Bokkenheuser, V. D. (1985). Clostridium scindens sp. nov., a human intestinal bacterium with desmolytic activity on corticoids. Int. J. Syst. Evol. Microbiol. 35, 478–481. doi:10.1099/00207713-35-4-478
Morvan, C., Folgosa, F., Kint, N., Teixeira, M., and Martin-Verstraete, I. (2021). Responses of Clostridia to oxygen: from detoxification to adaptive strategies. Environ. Microbiol. 23, 4112–4125. doi:10.1111/1462-2920.15665
Navarro, S. L., Levy, L., Curtis, K. R., Elkon, I., Kahsai, O. J., Ammar, H. S., et al. (2020). Effect of a flaxseed lignan intervention on circulating bile acids in a placebo-controlled randomized, crossover trial. Nutrients 12, 1837. doi:10.3390/nu12061837
Nguyen, T. L., Vieira-Silva, S., Liston, A., and Raes, J. (2015). How informative is the mouse for human gut microbiota research? Dis. Model Mech. 8, 1–16. doi:10.1242/dmm.017400
Nogueira Viçosa, G., Vieira Botelho, C., Botta, C., Bertolino, M., Fernandes De Carvalho, A., Nero, L. A., et al. (2019). Impact of co-cultivation with Enterococcus faecalis over growth, enterotoxin production and gene expression of Staphylococcus aureus in broth and fresh cheeses. Int. J. Food Microbiol. 308, 108291. doi:10.1016/j.ijfoodmicro.2019.108291
O’Gara, J. P. (2017). Into the storm: chasing the opportunistic pathogen Staphylococcus aureus from skin colonisation to life-threatening infections. Environ. Microbiol. 19, 3823–3833. doi:10.1111/1462-2920.13833
Payne, A. N., Zihler, A., Chassard, C., and Lacroix, C. (2012). Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 30, 17–25. doi:10.1016/j.tibtech.2011.06.011
Perales-Adán, J., Rubiño, S., Martínez-Bueno, M., Valdivia, E., Montalbán-López, M., Cebrián, R., et al. (2018). LAB bacteriocins controlling the food isolated (Drug-Resistant) staphylococci. Front. Microbiol. 9, 1143. doi:10.3389/fmicb.2018.01143
Pinchuk, I. V., Beswick, E. J., and Reyes, V. E. (2010). Staphylococcal enterotoxins. Toxins 2, 2177–2197. doi:10.3390/toxins2082177
Rahman, S., Ghiboub, M., Donkers, J. M., Van De Steeg, E., Van Tol, E. A. F., Hakvoort, T. B. M., et al. (2021). The progress of intestinal epithelial models from cell lines to gut-on-chip. Int. J. Mol. Sci. 22, 13472. doi:10.3390/ijms222413472
Ridlon, J. M., Harris, S. C., Bhowmik, S., Kang, D. J., and Hylemon, P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39. doi:10.1080/19490976.2015.1127483
Ridlon, J. M., Kang, D. J., Hylemon, P. B., and Bajaj, J. S. (2014). Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338. doi:10.1097/mog.0000000000000057
Rodríguez-Sánchez, S., Ramos, I. M., Rodríguez-Pérez, M., Poveda, J. M., Seseña, S., and Palop, M. L. (2022). Lactic acid bacteria as biocontrol agents to reduce Staphylococcus aureus growth, enterotoxin production and virulence gene expression. LWT 170, 114025. doi:10.1016/j.lwt.2022.114025
Roodsant, T., Navis, M., Aknouch, I., Renes, I. B., Van Elburg, R. M., Pajkrt, D., et al. (2020). A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front. Cell Infect. Microbiol. 10, 272. doi:10.3389/fcimb.2020.00272
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi:10.1038/nri.2016.42
Roura, E., Koopmans, S. J., Lallès, J. P., Le Huerou-Luron, I., De Jager, N., Schuurman, T., et al. (2016). Critical review evaluating the pig as a model for human nutritional physiology. Nutr. Res. Rev. 29, 60–90. doi:10.1017/s0954422416000020
Sannasiddappa, T. H., Lund, P. A., and Clarke, S. R. (2017). In vitro antibacterial activity of unconjugated and conjugated bile salts on Staphylococcus aureus. Front. Microbiol. 8, 1581. doi:10.3389/fmicb.2017.01581
Sasaki, N., Miyamoto, K., Maslowski, K. M., Ohno, H., Kanai, T., and Sato, T. (2020). Development of a scalable coculture system for gut anaerobes and human colon epithelium. Gastroenterology 159, 388–390.e5. doi:10.1053/j.gastro.2020.03.021
Schwerdtfeger, L. A., Nealon, N. J., Ryan, E. P., and Tobet, S. A. (2019). Human colon function ex vivo: dependence on oxygen and sensitivity to antibiotic. PLoS One 14, e0217170. doi:10.1371/journal.pone.0217170
Shi, J., Lei, Y., Wu, J., Li, Z., Zhang, X., Jia, L., et al. (2023). Antimicrobial peptides act on the rumen microbiome and metabolome affecting the performance of castrated bulls. J. Anim. Sci. Biotechnol. 14, 31. doi:10.1186/s40104-023-00832-5
Shu, L. Z., Ding, Y. D., Xue, Q. M., Cai, W., and Deng, H. (2023). Direct and indirect effects of pathogenic bacteria on the integrity of intestinal barrier. Ther. Adv. Gastroenterol. 16, 175628482311764. doi:10.1177/17562848231176427
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi:10.1126/science.aac4255
Sommer, F., and Bäckhed, F. (2013). The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi:10.1038/nrmicro2974
Speer, J. E., Gunasekara, D. B., Wang, Y., Fallon, J. K., Attayek, P. J., Smith, P. C., et al. (2019). Molecular transport through primary human small intestinal monolayers by culture on a collagen scaffold with a gradient of chemical cross-linking. J. Biol. Eng. 13, 36. doi:10.1186/s13036-019-0165-4
Stubbendieck, R. M., and Straight, P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155. doi:10.1128/jb.00275-16
Šuligoj, T., Vigsnæs, L. K., Abbeele, P. V. D., Apostolou, A., Karalis, K., Savva, G. M., et al. (2020). Effects of human milk oligosaccharides on the adult gut microbiota and barrier function. Nutrients 12, 2808. doi:10.3390/nu12092808
Ticho, A. L., Malhotra, P., Dudeja, P. K., Gill, R. K., and Alrefai, W. A. (2019). Intestinal absorption of bile acids in health and disease. Compr. Physiol. 10, 21–56. doi:10.1002/cphy.c190007
Tovaglieri, A., Sontheimer-Phelps, A., Geirnaert, A., Prantil-Baun, R., Camacho, D. M., Chou, D. B., et al. (2019). Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 7, 43. doi:10.1186/s40168-019-0650-5
Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., et al. (2015). The Impact of Food Bioactives on Health: in vitro and ex vivo models. Springer International Publishing.
Wang, Y., Disalvo, M., Gunasekara, D. B., Dutton, J., Proctor, A., Lebhar, M. S., et al. (2017). Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol. Gastroenterol. Hepatol. 4, 165–182.e7. doi:10.1016/j.jcmgh.2017.02.011
Wang, Y., Kim, R., Gunasekara, D. B., Reed, M. I., Disalvo, M., Nguyen, D. L., et al. (2018a). Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell Mol. Gastroenterol. Hepatol. 5, 113–130. doi:10.1016/j.jcmgh.2017.10.007
Wang, Y., Kim, R., Hwang, S. J., Dutton, J., Sims, C. E., and Allbritton, N. L. (2018b). Analysis of interleukin 8 secretion by a stem-cell-derived human-intestinal-epithelial-monolayer platform. Anal. Chem. 90, 11523–11530. doi:10.1021/acs.analchem.8b02835
Wang, Y., Sims, C. E., and Allbritton, N. L. (2022). Human 2D crypt model for assaying intestinal stem cell proliferation and differentiation. Anal. Chem. 94, 9345–9354. doi:10.1021/acs.analchem.2c00905
White, B. A., Lipsky, W. R. L., Fricke, R. J., and Hylemon, P. B. (1980). Bile acid induction specificity of 7 alpha-dehydroxylase activity in an intestinal Eubacterium species. Steroids 35, 103–109. doi:10.1016/0039-128x(80)90115-4
Williamson, I. A., Arnold, J. W., Samsa, L. A., Gaynor, L., Disalvo, M., Cocchiaro, J. L., et al. (2018). A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol. Gastroenterol. Hepatol. 6, 301–319. doi:10.1016/j.jcmgh.2018.05.004
Wlodarska, M., Luo, C., Kolde, R., D’Hennezel, E., Annand, J. W., Heim, C. E., et al. (2017). Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 22, 25–37.e6. doi:10.1016/j.chom.2017.06.007
Yu, L. C., Wang, J. T., Wei, S. C., and Ni, Y. H. (2012). Host-microbial interactions and regulation of intestinal epithelial barrier function: from physiology to pathology. World J. Gastrointest. Pathophysiol. 3, 27–43. doi:10.4291/wjgp.v3.i1.27
Zdenkova, K., Alibayov, B., Karamonova, L., Purkrtova, S., Karpiskova, R., and Demnerova, K. (2016). Transcriptomic and metabolic responses of Staphylococcus aureus in mixed culture with Lactobacillus plantarum, Streptococcus thermophilus and Enterococcus durans in milk. J. Ind. Microbiol. Biotechnol. 43, 1237–1247. doi:10.1007/s10295-016-1794-y
Zhang, J., Hernandez-Gordillo, V., Trapecar, M., Wright, C., Taketani, M., Schneider, K., et al. (2021a). Coculture of primary human colon monolayer with human gut bacteria. Nat. Protoc. 16, 3874–3900. doi:10.1038/s41596-021-00562-w
Zhang, J., Huang, Y. J., Yoon, J. Y., Kemmitt, J., Wright, C., Schneider, K., et al. (2021b). Primary human colonic mucosal barrier crosstalk with super oxygen-sensitive faecalibacterium prausnitzii in continuous culture. Med 2, 74–98.e9. doi:10.1016/j.medj.2020.07.001
Keywords: intestine, gut microbiome, multi-species culture, oxygen gradient, bile acid, enterotoxin, interactions
Citation: Wang H, Kim R, Wang Y, Furtado KL, Sims CE, Tamayo R and Allbritton NL (2024) In vitro co-culture of Clostridium scindens with primary human colonic epithelium protects the epithelium against Staphylococcus aureus. Front. Bioeng. Biotechnol. 12:1382389. doi: 10.3389/fbioe.2024.1382389
Received: 05 February 2024; Accepted: 28 March 2024;
Published: 12 April 2024.
Edited by:
Nic Davis Leipzig, University of Akron, United StatesReviewed by:
Mindy Engevik, Medical University of South Carolina, United StatesShaohua Wang, Ohio University, United States
Copyright © 2024 Wang, Kim, Wang, Furtado, Sims, Tamayo and Allbritton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy L. Allbritton, bmxhbGxickB1dy5lZHU=
 Hao Wang1
Hao Wang1 Raehyun Kim
Raehyun Kim Kathleen L. Furtado
Kathleen L. Furtado Rita Tamayo
Rita Tamayo Nancy L. Allbritton
Nancy L. Allbritton