
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 07 March 2024
Sec. Bioprocess Engineering
Volume 12 - 2024 | https://doi.org/10.3389/fbioe.2024.1365229
Continuous cropping obstacles seriously constrained the sustainable development of the ginseng industry. The allelopathic autotoxicity of ginsenosides is the key “trigger” of continuous cropping obstacles in ginseng. During harvest, the ginseng plants could be broken and remain in the soil. The decomposition of ginseng residue in soil is one of the important release ways of ginsenosides. Therefore, the allelopathic mechanism of ginsenosides through the decomposed release pathway needs an in-depth study. To investigate this allelopathic regulation mechanism, the integrated analysis of transcriptomics and metabolomics was applied. The prototype ginsenosides in ginseng were detected converse to rare ginsenosides during decomposition. The rare ginsenosides caused more serious damage to ginseng hairy root cells and inhibited the growth of ginseng hairy roots more significantly. By high-throughput RNA sequencing gene transcriptomics study, the significantly differential expressed genes (DEGs) were obtained under prototype and rare ginsenoside interventions. These DEGs were mainly enriched in the biosynthesis of secondary metabolites and metabolic pathways, phytohormone signal transduction, and protein processing in endoplasmic reticulum pathways. Based on the functional enrichment of DEGs, the targeted metabolomics analysis based on UPLC-MS/MS determination was applied to screen endogenous differential metabolized phytohormones (DMPs). The influence of prototype and rare ginsenosides on the accumulation of endogenous phytohormones was studied. These were mainly involved in the biosynthesis of diterpenoid, zeatin, and secondary metabolites, phytohormone signal transduction, and metabolic pathways. After integrating the transcriptomics and metabolomics analysis, ginsenosides could regulate the genes in phytohormone signaling pathways to influence the accumulation of JA, ABA, and SA. The conclusion was that the prototype ginsenosides were converted into rare ginsenosides by ginseng decomposition and released into the soil, which aggravated its allelopathic autotoxicity. The allelopathic mechanism was to intervene in the response regulation of genes related to the metabolic accumulation of endogenous phytohormones in ginseng. This result provides a reference for the in-depth study of continuous cropping obstacles of ginseng.
Ginseng (Panax ginseng C. A. Meyer) is a famous traditional Chinese medicine with high medicinal, nutritional, and healthcare value (Shen et al., 2023). After the cultivation of ginseng, the soil becomes unsuitable for ginseng continuous cropping. The growth and development are inhibited and weakened, which lead to serious root rot, fibrous root shedding, and even plant death. Therefore, the secondary planting rate of ginseng would be seriously reduced (Tagele et al., 2023). Continuous cropping obstacles in ginseng have seriously restricted the development of the ginseng industry. The recycling of land needs to be solved urgently. The reason for continuous cropping obstacles in ginseng is very complex. The changes in microbial communities and physicochemical properties of soil, the autotoxicity of allelochemicals, and the interactions of factors could cause continuous cropping obstacles (Liu et al., 2021; Chen et al., 2023a; Jia et al., 2023). The allelopathic autotoxicity refers to the inhibition of growth and development of the same plant species by allelochemicals. Ginsenosides are the main allelochemicals in ginseng and their autotoxicity plays an important role in ginseng continuous cropping. The decomposition of ginseng residue in the soil during harvesting is one of the release ways of allelochemicals, which could affect soil structure and ginseng growth directly or indirectly (Singh et al., 2021).
Phytohormones refer to trace organic compounds that are synthesized in the plant body, which have a significant regulatory effect on the growth and development of plants (Bogatek and Gniazdowska, 2014; Ohri et al., 2015). Common phytohormones include auxin, cytokinins (CKs), abscisic acid (ABA), jasmonates (JAs), salicylic acid (SA), gibberellins (GAs), ethylene class (ETH), and strigolactones (SLs). The physiological activities of phytohormones are related to their contents in plants. Studies on the composition and content changes of phytohormones in plants can help to find more interactions of phytohormones and explore possible mechanisms (Pan et al., 2010; Jiang and Asami, 2018).
By transcriptome sequencing, plant-specific key genes, function prediction, and metabolic pathways could be obtained. The adversity-related regulatory networks could be explored (Wan et al., 2022; Chen et al., 2023b). Metabolomics is the study of the changes of small molecule metabolites in a biological system before and after a perturbation during a specific physiological period (Ren et al., 2016), and the overall metabolic profile of the biological system could be characterized and the mechanisms could be clarified. Plant transcriptomics is the study of gene expression at the entire RNA transcript level in plants (Zanardo et al., 2019). Metabolomics provides more epigenetic data to support transcriptomics (Salek et al., 2015). The functional changes in genes are ultimately reflected at the metabolic level, and metabolites can amplify the expression of these genes at the functional level. Integrated transcriptomics and metabolomics analysis of biological systems can provide a more systematic and comprehensive understanding of the molecular functions and regulatory mechanisms (Wu et al., 2023; Zhang et al., 2023).
At present, the allelopathic autotoxicity mechanism of ginsenosides by decomposition as a release way needs an in-depth study. In this study, we simulated the natural decomposition process of ginseng in the soil. After decomposition, the ginsenoside composition of ginseng was analyzed comprehensively using UPLC-HRMS and the transformation was explored systematically. The allelopathic autotoxicity of ginsenosides was validated by the apparent morphology and apical cell viability of ginseng hairy roots. Transcriptomics sequencing was applied to screen the gene expression. Furthermore, a targeted metabolomics approach based on UPLC-MS/MS was applied to study the endogenous phytohormones metabolism in ginseng hairy roots. Finally, the analysis of transcriptomics and metabolomics were integrated to explore the allelopathic autotoxicity mechanism of ginseng decomposition deeply.
Methanol and acetonitrile (HPLC grade, Tedia), formic acid and acetic acid (HPLC grade, Aladdin), n-butanol (analytical grade, Tianjin Xintong Fine Chemical Co., Ltd.), phytohormone standards and internal standards (TRP, SAG, IAA-Glc, etc. Sigma-Aldrich), and ultrapure water (Milli-Q water, Merck Millipore).
Fresh ginseng root (6 years old) was collected from the plantation of Baixi Forestry, Fusong County, Baishan City, Jilin Province. It was identified by Prof. Chen Changbao of Changchun University of Chinese Medicine as Panax ginseng C. A. Meyer.
Decomposition model preparation: the main root part of ginseng was cleaned, wipe-dried, cut into small pieces, and mixed with forest soil (1:20, w/w). The model samples were placed in a ventilated area with natural light and soil humidity maintained at 10%–15%. We allowed the ginseng root to decay naturally in the forest soil for 1 year and collected the soil for further testing. Each treatment was repeated five times.
A number of fresh ginseng main root were removed and dried at 45°C, pulverized, and passed through a 60-mesh sieve for further extraction. The soil with decomposed ginseng was mixed thoroughly, dried at 45 °C, ground, and passed through a 50-mesh sieve for further extraction.
We weighed the ginseng root sample (1.0 g) and soil sample (30 g) and added 10 times the amount of 50% methanol; they were ultrasonic extracted at 50°C two times and each time for 1 h, and we combined the extracts and concentrated them under reduced pressure to 10 mL. An equal volume of water-saturated n-butanol was added and extracted three times, and the n-butanol layers were combined and concentrated under reduced pressure till they were dry. The extract was dissolved using HPLC grade methanol to 1 mL and filtered with 0.22 μm Millipore for UPLC-HRMS analysis.
Ultra-high performance liquid chromatography-quadrupole electrostatic field orbitrap high-resolution mass spectrometer (UPLC-HRMS) (Q-Exactive, Thermo-Fisher, United States) and Thermo Scientific Syncronis C18 column (100 mm × 2.1 mm, 1.7 μm) were used.
Chromatographic conditions: mobile phase: 0.1% formic acid water (A) and acetonitrile (B); gradient elution: 0–38 min 15%–80% B, 38–40 min 80%–100% B, 40–45 min 100% B; flow rate: 300 μL/min; column temperature: 30 °C; injection volume: 5 μL.
Mass spectrometry conditions: full scan under electrospray ionization (ESI) in negative ion mode; spray voltage: 2500 V; sheath gas: 40 arb; aux gas: 15 arb; capillary temperature: 320 °C; scan range: m/z 150-2000.
A single cell line of ginseng tissue was applied to induce hairy root growth by Agrobacterium rhizogenes. The grown hairy roots were transferred several times to remove Agrobacterium rhizogenes. Then the ginseng hairy roots were inoculated into a solid medium (Meng et al., 2022). After the growth of ginseng hairy roots stabilized, 1 cm of the root tip was transferred to different groups of solid mediums for culture. Six biological replicates were set up in each group. For the G group, 1 mL of ginseng root aqueous extracts was added to the solid medium, and the total ginsenoside contents of ginsenosides in the solid medium were 0.01 mg/L, 0.1 mg/L, 1 mg/L, and 10 mg/L, respectively. For the DG group, an equal volume of aqueous extracts of decomposed ginseng products was added to the solid medium, and the total ginsenoside contents of ginsenosides in the solid medium were 0.01 mg/L, 0.1 mg/L, 1 mg/L, and 10 mg/L, respectively. For the C group, an equal volume of distilled water was added to the solid medium. The growth status of hairy roots was observed. Hairy root samples were collected after 30 days of culture and stored at −80 °C for transcriptome sequencing and metabolomics analysis.
The ginseng hairy roots were soaked in FDA-PI double stain and incubated for 30 min without light. The excess dye was removed by rinsing the root in saline. The slices were sealed with an anti-fluorescence quenching sealing solution and placed under a laser confocal microscope with excitation and detection wavelengths at 485 nm and 530 nm, respectively, to observe the cell viability of hairy roots.
We took 100 mg of ground ginseng hairy roots and added 1 mL of CTAB solution. The sample was placed in a thermostatic mixer, mixed at 60 °C for 10 min, and centrifuged at 12,000 rpm for 5 min. The supernatant was transferred to an equal volume of chloroform, mixed well, centrifuged at 12,000 rpm for 5 min, and repeated twice. The supernatant was transferred to an equal volume of LiCl solution, mixed well, placed at −20 °C for 2 h, centrifuged at 12,000 rpm for 20 min, and discarded. Ethanol of 75% was added, mixed well, and centrifuged at 12,000 rpm for 5 min, and the supernatant was discarded. The supernatant stood at room temperature for 5 min, and 30 μL of DEPC-treated water was added, mixed well with vortex, and stored at −80 °C for measurement.
The integrity of RNA was analyzed using agarose gel electrophoresis and Agilent 2100 bioanalyzer. The purity of RNA was measured by a Nano-spectrophotometer. The concentration of RNA was quantified accurately by Qubit 2.0 Fluorometer.
Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit of Illumina® (NEB, United States). The first strand of cDNA was synthesized in the M-MuLV reverse transcriptase system using the fragmented mRNA as a template. The second strand synthesis of cDNA was subsequently performed using DNA Polymerase I and RNase H. The purified double-stranded cDNA was end-repaired, A-tailed, and connected to the sequencing connector. Fragment selection was performed with AMPure XP beads software. The sorted products were PCR amplified and purified to obtain the final library. After the library was constructed, preliminary quantification was performed using a Qubit 2.0 Fluorometer, and then the insert size of the library was examined using an Agilent 2100 bioanalyzer. The effective concentration of the library was quantified accurately by qRT-PCR to ensure the quality of the library.
Transcriptome assembly was performed using Trinity. Corset was used to regroup relevant transcripts into ‘gene’ clusters (https://github.com/trinityrnaseq/trinityrnaseq). DIAMOND software was used to compare the transcript sequences after redundancy with KEGG (https://www.genome.jp/kegg/), NR (http://www.ncbi.nlm.nih.gov/), Swiss-Prot (https://www.expasy.org/resources/uniprotkb-swiss-prot), GO (https://www.geneontology.org/), COG/KOG (http://ftp.ncbi.nih.gov/pub/COG/KOG/kyva), and TrEMBL (a variety of new documentation files and the creation of TrEMBL, a computer annotated supplement to Swiss-Prot) databases. HMMER software was used to compare amino acid sequences with the Pfam (http://pfam.xfam.org/) database. The annotation information of transcripts in seven major databases was obtained. Gene expression levels were calculated by RSEM. Based on the gene length, the FPKM of each gene was calculated. Analysis of variance was performed using DESeq2, with p-values and |log2foldchange| as thresholds for significant differential expression. The enrichment analysis was performed based on the hypergeometric test. For KEGG, the hypergeometric distribution test was performed as a pathway. For GO, it was performed based on the GO term.
The hairy root was ground at a low temperature (30 Hz, 1 min). We weighed 50 mg of the ground hairy root and added 10 μL of mixed internal standard solution (100 ng/mL). Then, 1 mL of methanol/water/formic acid (15:4:1, v/v/v) was added, vortexed, and shaken for 10 min. The mixture was centrifuged at 12,000 rpm under 4 °C for 5 min. We took the supernatant, concentrated it to dryness, and redissolved it with 100 μL 80% methanol. It was filtered with 0.22 μm Millipore for UPLC-MS/MS analysis.
Ultra-high performance liquid chromatography-triple quadrupole ion trap mass spectrometer (QTRAP 6500+, AB SCIEX) and Waters ACQUITY UPLC HSS T3 C18 column (100 mm × 2.1 mm, 1.8 µm) were used to detect phytohormones.
Chromatographic conditions: mobile phase: 0.04% acetic acid in water (A) and 0.04% acetic acid in acetonitrile (B); gradient elution: 0–1 min 5% B, 1–8 min 5%–95% B, and 8–9 min 95% B; flow rate: 350 μL/min; column temperature: 40 °C; injection volume: 2 μL.
Mass spectrometry conditions: electrospray ionization (ESI) in both positive and negative ion modes. Ion spray voltage: 5500 V (positive) and −4500 V (negative); source temperature: 550°C; curtain gas: 35 psi. Phytohormones were analyzed using scheduled multiple reaction monitoring (MRM). Mass spectrometer parameters including the declustering potentials (DP) and collision energies (CE) for individual MRM transitions were done with further DP and CE optimization.
Data acquisitions were performed using Analyst 1.6.3 software (Sciex). Multiquant 3.0.3 software (Sciex) was used to quantify all metabolites. The profiles of phytohormones were compared and the contents were analyzed by orthogonal partial least square discriminant analysis (OPLS-DA) using SIMCA-P Statistics 13.0 software. A t-test was used to assess the significant difference (p < 0.05) using SPSS. The enrichment analysis of metabolic pathways was carried out using the KEGG (http://www.kegg.com/) database.
Ginsenosides in ginseng and its soil decompositions were determined by UPLC-HRMS. Figure 1 shows the total ion current chromatogram (TIC) of ginsenosides in negative ion mode. The relative retention time, precise molecular weight in MS, and the fragment ions in MS2 of the compounds detected were compared with the ginsenosides standard and data reported in the literature (Zhang et al., 2016; Chen et al., 2020). A total of 51 ginsenoside components were identified and deduced, as shown in Table 1.
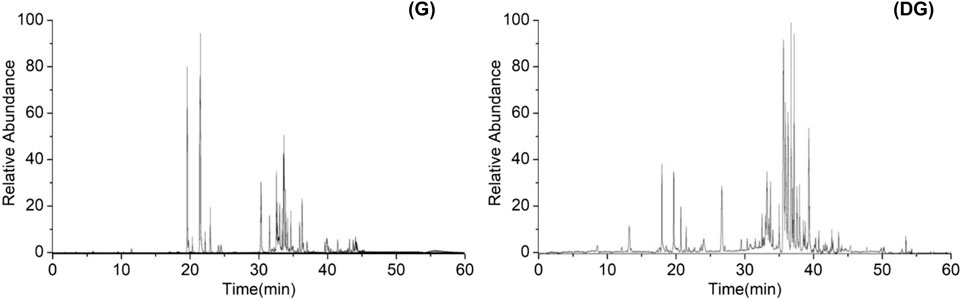
FIGURE 1. Total ion current chromatogram of ginsenosides in ginseng (G) and decomposed ginseng (DG).
The relative percentage contents of 51 ginsenosides were calculated and compared. The results showed that the contents of PPD-type ginsenosides Ra1, Rb1, Quinquefolium-R1, Ra2, Rc, Ra1-iso, Rb2, Rb3, and Pseudo-Rc1 were reduced after decomposition. Ginsenosides R4, Rs1, Rs2, Rd, Gypenoside X Ⅶ, F2, and 20(S)-Rg3 increased in content. Ginsenosides 20(S)-Rh2, 20(R)-Rh2, 20(S)-Rs3, 20(R)-Rs3, 20(R)-Rg3, and Compound K were produced. PPT-type ginsenosides NG-R1-iso, 20-Glc-Rf, NG-R1, Re, Rg1, Rf2, 20(S)-Rg2, 20(R)-Rg2, 20(S)-Rf, 20(R)-Rf, and NG-R2 were reduced after decomposition, while ginsenosides 20(S)-Rh1 and F1 increased. Ginsenosides F3 and 20(R)-Rh1 were produced. OLE-type ginsenoside Ro was decreased by decomposition. Malonyl and acetyl-substituted ginsenosides were all reduced as desubstituent transformation.
It has been reported that ginsenosides with high molecular weight and high polarity were converted to rare ginsenosides with low molecular weight and low polarity by hydrolysis of glycosidic bonds (Kim et al., 2012; Palaniyandi et al., 2016; Huang et al., 2019; Li et al., 2023). The results of this study showed that the transformation of ginsenosides by microorganisms in soil was in agreement with the reported findings. It was possible to deduce the decomposed biotransformation pathways of PPD and PPT-type ginsenosides. As shown in Supplementary Figure S1, the conversion pathways of PPD-type ginsenoside were Rb1, Rb2, Rb3, Rc→Rd→F2+Rg3→Rh2. The conversion pathways of PPT-type ginsenoside were Re→Rg2→Rh1, Re→F1, Rg1→F1+Rh1.
The growth of ginseng hairy roots was relatively slow on days 1–14, and more rapid on days 15–20, which was the optimal growth period. After 20 days, the growth was in a period of relative stability with basically no increase in weight. Compared with group C, groups G and DG gradually showed serious growth stagnation after the rapid growth period. The monthly proliferative multiple was calculated with the formula: monthly proliferative multiple=(harvest amount−inoculum amount)/inoculum amount × 100%. As shown in Supplementary Figure S2, the proliferative multiples of ginseng hairy root growth gradually decreased with the increase of ginsenosides’ concentration. The smallest proliferative multiple of ginseng hairy root growth was observed when 10 mg/L ginsenosides groups were added. The growth inhibition effect was more significant in the DG group than in the G group.
For the apparent morphological determination, the influence of ginsenoside extracts on the growth of ginseng hairy roots was compared and shown in Figure 2A. The overall growth state of hairy roots in group C was good, longer, and thicker, as well as having a yellow color and many branches. Compared to group C, the growth of ginseng hairy root was significantly inhibited in both groups G and DG. With the increase of ginsenoside concentration, the inhibitory effect increased gradually. The hairy root gradually changed from a light yellow color with many branches and dense growth to a dark yellow color with few branches and sparse growth. Therefore, the growth state showed an apoptotic tendency. With an addition of 10 mg/L ginsenosides, the growth of hairy roots was almost shriveled in both the G and DG groups. Compared to the G group, the overall growth of hairy roots in the DG group was sparser and more shriveled with fewer branches. It means that the growth of hairy roots in the DG group was inhibited more severely. The results showed that the ginsenosides extracts had significant inhibitory effects on the growth of ginseng hairy roots, and the inhibitory effect of the rare ginsenosides in decomposed ginseng was stronger.
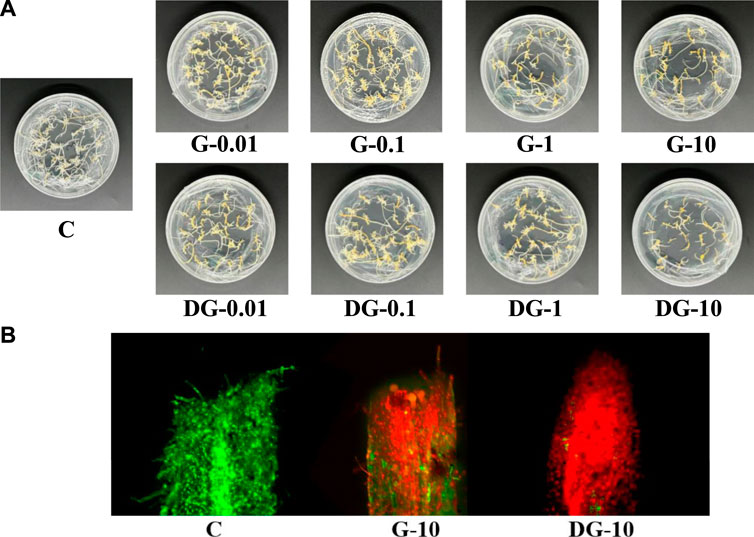
FIGURE 2. Apparent morphology of ginseng hairy root in C, G, and DG groups (A). FDA-PI fluorescence staining of ginseng hairy roots in C, G, and DG groups (B).
According to the observation of the apparent morphology of ginseng hairy roots, the G-10 and DG-10 groups presented the strongest inhibitory effects on their growth. Furthermore, the apical cell viability in these two groups was determined using confocal laser scanning microscopy. The localization of live and dead cells in the apical root was observed and is shown in Figure 2B. The apical cells were in good condition in group C. The G-10 and DG-10 groups showed different degrees of cell apoptosis. The apical cells were damaged, and the damage was more serious in the DG-10 group. It indicated that both prototype and rare ginsenosides had inhibitory effects on the apical cell of the hairy root, and the inhibitory effect of rare ginsenosides was stronger. This result was consistent with the apparent morphological changes of ginseng hairy roots.
A reference-free transcriptome was used for comparative analysis. For subsequent analysis, the transcript sequences obtained from Trinity splicing were used as reference sequences. The longest cluster sequence obtained from Corset hierarchical clustering was used as the unigenes. The length distribution of the sequence is shown in Figure 3A. The unigenes with sequence lengths greater than or equal to 2000 nt were the most abundant.
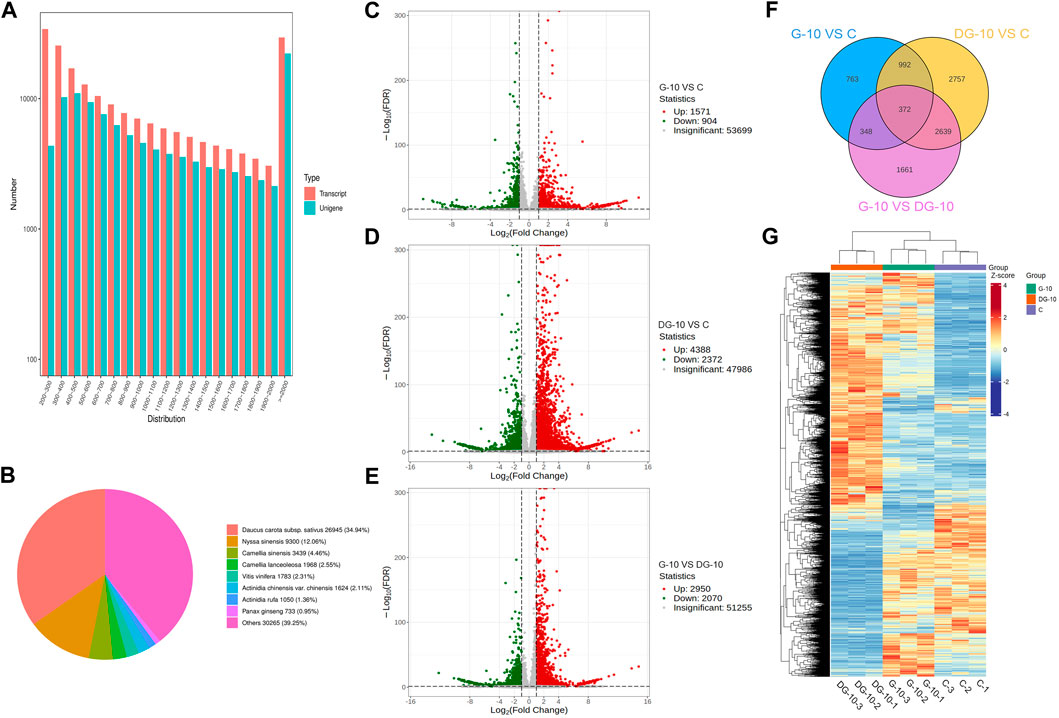
FIGURE 3. Sequence length distribution map of unigenes (A). Non-redundant homologous species distribution (B). Volcano map of DEGs screening for groups G-10 vs. C (C), DG-10 vs. C (D), and G-10 vs. DG-10 (E). Venn diagram of DEGs in ginseng hairy root between groups (F). Clustering heat map of DEGs in ginseng hairy roots between groups (G).
The unigenes sequences were aligned with KEGG, NR, Swiss-Prot, TrEMBL, COG/KOG, and GO databases using DIAMOND BLASTX software and aligned with the Pfam database using HMMER software. The annotation information of unigenes was obtained. A total of 111,001 unigenes were annotated into the database, with an annotation rate of 100%. Among them, 59,581 unigenes were annotated into the KEGG database (53.68%), 77,107 unigenes were annotated into the NR database (69.47%), 58,346 unigenes were annotated into the Swiss Prot database (52.56%), 76,893 unigenes were annotated into the TrEMBL database (69.27%), 47,116 unigenes were annotated into the KOG database (42.45%), 68,042 unigenes were annotated into the GO database (61.30%), and 45,836 unigenes were annotated into the Pfam database (41.29%). Comparative analysis showed that the most unigenes were annotated into the NR database and the least unigenes were annotated into the Pfam database.
The annotation results of the NR database are shown in Figure 3B. The most annotated species was carrot (Daucus carota) with a total of 26,945 unigenes (approximately 34.94%), and the other homologous species were Nyssa sinensis (12.06%), Camellia sinensis (4.46%), Camellia lanceoleosa (2.55%), Vitis vinifera (2.31%), Actinidia chinensis var. chinensis (2.11%), and Actinidia rufa (1.36%). This result showed that the hairy root tissue of ginseng has the greatest homology with carrot.
To explore the allelopathic mechanism of ginsenosides on hairy roots before and after ginseng decomposition, genes in the transcriptome library were analyzed for differential comparison, and DEGs were obtained. Using DESeq2, an analysis of DEGs between two groups was performed. As shown in Figures 3A, C total of 2,745 DEGs were obtained in the comparison of groups G-10 vs. C, of which 1,571 genes were upregulated and 904 genes were downregulated, significantly. As shown in Figures 3A, D total of 6,760 DEGs were obtained between groups DG-10 vs. C, of which 4,388 genes were significantly upregulated and 2,372 genes were significantly downregulated. In Figures 3A, E total of 5,020 DEGs were obtained between groups G-10 vs. DG-10, of which 2,950 genes were significantly upregulated and 2070 genes were significantly downregulated. The Venn diagram of the screened DEGs is shown in Figure 3F, which shows the overlap between groups. The hierarchical cluster analysis of DEGs in each group is shown in Figure 3G. The screened DEGs could reflect the intergroup differences between the samples. Both prototype and rare ginsenosides affected the expression of genes in ginseng hairy roots, and there was a significant difference in the expression of related genes between prototype and rare ginsenosides. Ginsenosides may be one of the main factors affecting the growth and development of ginseng hairy roots.
The enrichment of pathways of DEGs in ginsenoside-treated hairy roots was analyzed using GO and KEGG databases. The results of GO enrichment of the DEGs are shown in Figure 4A. In groups G-10 vs. C, the DEGs were mainly involved in protein refolding, protein kinase C-activating G protein-coupled receptor signaling, trehalose biosynthetic process, and mitochondrial and peroxisome fission pathways. The DEGs of the DG-10 vs. C groups were mainly involved in cellular response to hypoxia, oxygen levels, decreased oxygen levels, and phenylpropanoid biosynthetic process. The DEGs of G-10 vs. DG-10 groups were mainly involved in cellular response to oxygen levels, decreased oxygen levels, hypoxia, and heat-shock protein binding.
The results of KEGG enrichment of DEGs are shown in Figure 4B. The DEGs in groups G-10 vs. C and DG-10 vs. C were mainly involved in the biosynthesis of secondary metabolites and metabolic pathways and phytohormone signal transduction. The DEGs in groups G-10 vs. DG-10 were mainly involved in the biosynthesis of secondary metabolites and metabolic pathways and protein processing in the endoplasmic reticulum. The DEGs in the three groups’ comparison were mainly involved in the biosynthesis of secondary metabolites and metabolic pathways.
According to the results of allelopathic autotoxicity validation, the G-10 and DG-10 groups presented the most significant activities. Therefore, the 109 endogenous phytohormones in the ginseng hairy roots of these two groups were analyzed by UPLC-MS/MS. A total of 53 phytohormones were detected, and the contents are shown in Supplementary Table S1. The changes of these endogenous phytohormones between G-10 and DG-10 groups were further analyzed by targeted metabolomics strategy.
Univariate statistical analysis and multivariate statistical analysis were applied to screen DMPs in ginseng hairy roots, as shown in Table 2. The comparison of DMPs between groups was visible in the Venn diagram (Figure 5A). A total of 18 DMPs were screened out in groups G-10 vs. C. A total of 35 DMPs were screened out between groups DG-10 and C. For groups G-10 vs. DG-10, 23 DMPs were screened out. Among the three groups, a total of 9 DMPs (IAM, TRA, ICA, 2MeScZR, cZRMP, GA53, GA19, JA, and JA-Val) were screened out.
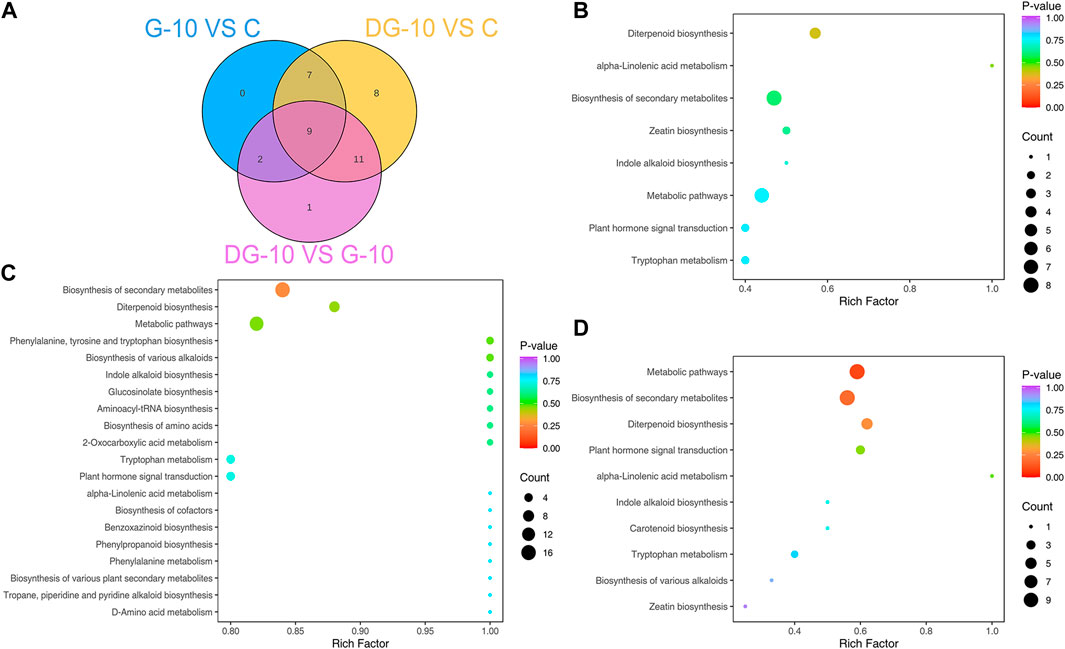
FIGURE 5. Venn diagram of DMPs in ginseng hairy root between groups (A). Enrichment plots of metabolic pathways of DMPs for groups G-10 vs. C (B), groups DG-10 vs. C (C), and groups G-10 vs. DG-10 (D).
The enrichment analysis of metabolic pathways of DMPs was performed based on the KEGG database. For groups G-10 vs. C, the metabolism of DMPs was mainly involved in eight pathways (Figure 5B), biosynthesis of secondary metabolites, diterpenoid, zeatin and metabolic pathways, phytohormone signal transduction, and tryptophan metabolism. In groups DG-10 vs. C, the DMPs were with significant enrichment in the biosynthesis of secondary metabolites, diterpenoid and metabolic pathways, tryptophan metabolism, and phytohormone signal transduction (Figure 5C). DMPs were mainly involved in 10 pathways in groups G-10 vs. DG-10, with significant enrichment also in the biosynthesis of secondary metabolites, diterpenoid and metabolic pathways, phytohormone signal transduction, and tryptophan metabolism (Figure 5D).
An integrated analysis of targeted transcriptomics of DEGs and metabolomics of DMPs was applied. The KEGG enrichment of pathways of the DEGs and DMPs is shown in Figure 6A. In groups G-10 vs. C, 7 pathways were enriched. In groups DG-10 vs. C, 23 pathways were enriched. In groups G-10 vs. DG-10, 9 pathways were enriched. For the three groups’ analysis, biosynthesis of secondary metabolites, diterpenoid and zeatin, metabolic pathways, alpha-linolenic acid, tryptophan metabolism, and phytohormone signal transduction were enriched. Furthermore, phytohormone signal transduction pathways were selected for correlation analysis.
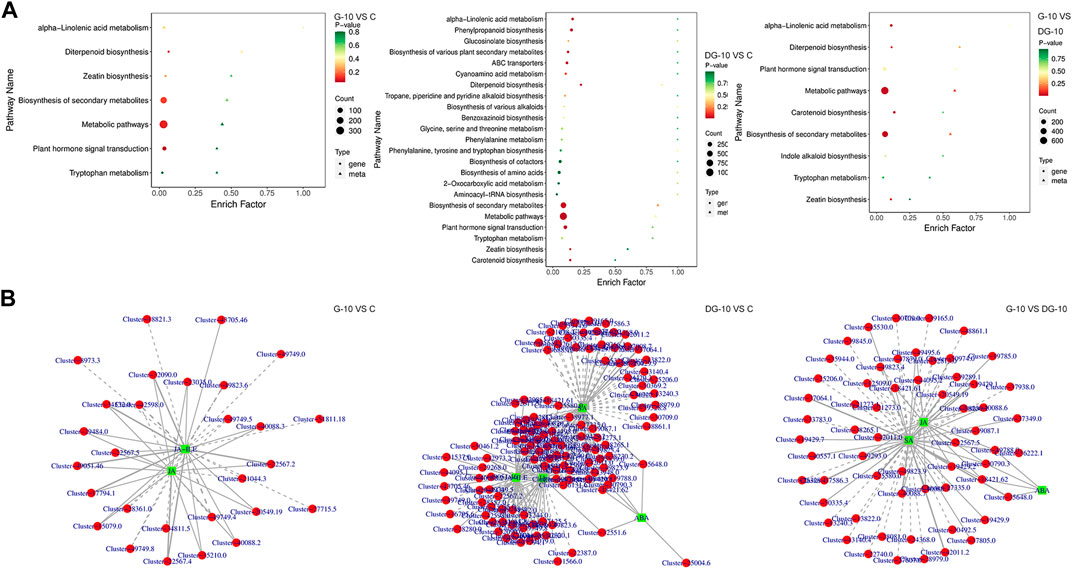
FIGURE 6. Enrichment plots of DEGs and DMPs by KEGG analysis (A). Network diagram of DEGs and DMPs correlation (B).
By correlation analysis, the DMPs and DEGs in the phytohormones signal transduction pathways with Pearson correlation coefficients greater than 0.80 and p-values less than 0.05 were selected. The network diagram of the correlation of DMPs and DEGs was constructed and shown in Figure 6B. A total of 53 DEGs and two DMPs were enriched in groups G-10 vs. C, 152 DEGs and four DMPs were enriched in groups DG-10 vs. C, and 90 DEGs and three DMPs were enriched in groups G-10 vs. group DG-10. It was observed that genes and phytohormones in ginseng hairy roots showed different accumulation patterns after the treatment of prototype and rare ginsenosides. These DEGs may be the key genes regulating the accumulation of DMPs in ginseng hairy roots. The prototype and rare ginsenosides intervened in the growth and development of ginseng hairy roots through the phytohormones signal transduction pathways.
Based on the results of integrated analysis, the relevant phytohormones and genes in the phytohormones signal transduction pathways were simultaneously mapped onto the KEGG pathway, as shown in Figure 7.
The prototype ginsenosides in ginseng were able to upregulate the gene expression of JAR1, COI1, and JAZ in the JA synthesis pathway. Whereas, MCY2 had both upregulated and downregulated gene expression. JA through JAR1 was able to promote the synthesis of JA-Ile, which acted as an activator that would promote the interaction between JAZ and COI1. COI1 inhibited the gene expression of JAZ through the ubiquitination pathway, thereby promoting JAZ degradation. As a result of JAZ degradation, it was able to control the upregulation of JAZ transcription factors, thus affecting the growth and development of plants and the tolerance to biotic and abiotic stresses. Subsequently, JAZ dissociated MCY2, which in turn interacted with DNA to participate in senescence and stress response processes. The synthesis of JA and JA-Ile could be promoted by gene expression of JAR1, COI1, JAZ, and MCY2. JA-Ile is one of the derivatives of JA. In plants, they were the key signaling compounds related to stress defense and development and were able to inhibit growth (Mouna Ghorbel). Our result showed that the intervention of prototype ginsenosides promoted the secretion of JA and JA-Ile in ginseng hairy roots, which in turn inhibited the growth and development of hairy roots.
The rare ginsenosides in decomposed ginseng were able to upregulate the expression of some genes of PYR/PYL and PP2C and downregulate the expression of some genes of PYR/PYL, PP2C, and SnRK2 in the ABA synthesis pathway. The accumulation of ABA activated gene expression of PYR/PYL, which inhibited PP2C. PYR/PYL is an ABA receptor that acts at the apex of the negative regulatory pathway and controls ABA signaling by inhibiting PP2C, and PP2C inhibited the expression of SnKR2s through the dephosphorylation pathway and inactivated SnRK2. This signal synthesis pathway was involved in the process of stomatal closure and seed dormancy. The increased accumulation of ABA could impair the growth of plants (Polavarapu B Kavi Kishor).
After ginseng decomposition, the expression of some genes of PR-1 in the SA synthesis pathway was upregulated and the expression of some genes of NPR1 and TGA was downregulated. SA could indirectly promote gene expression of NPR1, which acted as a key regulator of plant species defense-activated gene expression of TGA. TGA indirectly regulated gene expression of PR-1 by interacting with NPR1 and binding to DNA. Activation of these genes promoted the secretion of SA, which resulted in the inhibition of plant growth and development (Bruno Pok Man Ngou).
Ginsenosides are a major kind of allelochemicals in ginseng. With the accumulation of ginsenosides in the soil, the microbial community and metabolic activity in the soil could change. As the microbiology of the soil altered, the growth of ginseng was affected and continuous cropping obstacles were presented (Zhang et al., 2017). Cheng ZH et al. found that ginsenosides R1, Rg3, Re, Rb2, Rb1, Rg1, Rg2, and Rd could accumulate in the rhizosphere soil through root secretion or decomposition and impede seedling emergence and growth (Cheng et al., 2015). Shen YL et al. found that the ginsenoside Rd possessed significant allelopathic activity on ginseng seed germination, seedling growth, and plant development (Shen et al., 2023). Li YL et al. found that the monomer ginsenosides Rg1, Rb1, Rh1, and their mixtures were able to disrupt the balance of fungal microbiome by stimulating potential soil-borne pathogens, leading to replanting failure of Panax notoginseng. They demonstrated that the allelopathic autotoxicity of ginsenosides was one of the reasons for continuous cropping obstacles in ginseng planting (Li et al., 2020).
In this study, prototype ginsenosides were converted into rare ginsenosides after ginseng decomposition by soil microorganisms. Both prototype and rare ginsenosides had a significant inhibitory effect on the growth of ginseng. The newly generated rare ginsenosides through decomposition had a more significant inhibitory effect on the growth of ginseng, which showed that rare ginsenosides had stronger allelopathic autotoxicity. It was reported that the relative content of prototype ginsenosides (Rb1, Rb2, and Rc) decreased, which was secreted in the rhizosphere soil by the ginseng root system, while the relative content of degraded ginsenoside products (Rd, G-VXII, and F2) increased in rhizosphere soil during ginseng growth (Li et al., 2021). Some studies showed that secretions of ginseng root could mediate negative plant-plant interactions and lead to stronger allelopathy (Cheng et al., 2015; Ding et al., 2019; Feng et al., 2022). It was demonstrated that rare ginsenosides were released through root secretion and decomposition and were then accumulated in the soil.
Phytohormones such as growth hormone, cytokinin, gibberellin, abscisic acid, ethylene, jasmonic acid, and salicylic acid played important roles in plant growth and stress responses (Zhao et al., 2023). Allelochemicals might interfere with plant growth by regulating the metabolic levels of phytohormones. Disturbances in phytohormone levels resulted in reduced metabolic activity of embryos and hindered their germination and growth (Bogatek and Gniazdowska, 2014). In this study, we found that ginsenosides could significantly affect the accumulation metabolism of endogenous phytohormones in ginseng hairy roots. The endogenous phytohormones played an important regulatory role in the allelopathic autotoxicity of ginsenosides. Some studies reported that the imbalance of accumulation of Auxin was one of the causes of allelopathic autotoxicity (Zhang et al., 2018). Accumulation of JA triggered the overactivation of defense mechanisms, which affected plant growth and development (Li et al., 2022). Peng YJ et al. found that low concentration of SA treatment stimulated the growth of some plant species, and excessive accumulation of SA inhibited plant growth and development (Peng et al., 2021). Brookbank B. P. found that ABA promoted adaptive responses to abiotic and biotic stresses, and under stress, ABA levels increased sharply and resulted in inhibition of germination and growth stagnation (Brookbank et al., 2021).
Integrated analysis of transcriptomics and metabolomics can provide more insight into biological mechanisms than these two methods applied individually (Li et al., 2016). An increasing number of studies applied combined transcriptomics and metabolomics analysis methods to correlate gene expression and metabolites to explain biological mechanisms much better (Zhang et al., 2021; Wan et al., 2022; Wu et al., 2023). In this study, we analyzed the phytohormones JA, ABA, and SA signaling pathways using integrated analysis of transcriptomics and targeted metabolomics. The allelopathy of prototype and rare ginsenosides as well as the allelopathic mechanisms of ginseng decomposition were investigated.
JA and its derivatives were able to regulate a wide range of biological processes in plant defense, growth control, and reproductive development. JA-Ile was a major form of JA-amino acid coupler that could be synthesized by recombinant JAR1 (Guranowski et al., 2007; Kitaoka et al., 2014). JAR1 was an enzyme that coupled JA to isoleucine, which was shown to play a role directly in COI1-mediated signaling (Suza and Staswick, 2008). COI1 was a key participant in the downstream process of JA biosynthesis. JA-Ile bonded to COI1 and promoted the formation of the COI1-JAZ complex, leading to the ubiquitination and subsequent degradation of JAZ (Gfeller et al., 2010). The JAZ family were transcriptional repressors of the jasmonate-responsive. The discovery of JAZ deterrents defined the core JA signaling module as COI1-JAZ-MYC2, and it allowed for a comprehensive understanding of the JA signaling pathway from phytohormone perception to transcriptional reprogramming. Upon phytohormone perception, the JAZ deterrent was degraded by the proteasome, and it released MYC2 and allowed the activation of the JA response (Chini et al., 2009). In this study, the expression of JAR1, COI1, JAZ, and MYC2 genes ultimately led to the accumulation of JA, which inhibited ginseng hairy roots’ growth and development. The JA and SA signaling pathways might activate the release of allelochemicals, as reported by Bi HH et al.
ABA is a key phytohormone for plant stress signaling. Several proteins that participated in the ABA signaling pathway had been identified, such as ABA receptors (PYR/PYL/RCAR), co-receptors PP2Cs (protein phosphatases), SnRK2 kinases (SNF1-related protein kinases), and ABI5/ABFs (transcription factors). In the ABA signal, PYR/PYL receptors interacted with and recruited PP2Cs and then released SnRK2s kinases from sequestration with PP2Cs. This allowed SnRK2s to activate downstream transcription factors of the ABA pathway (Ali et al., 2020). This signaling pathway promoted the expression of key genes in the biosynthesis of PYR/PYL, PP2C, and SnRK2s, which ultimately led to the accumulation of ABA. Furthermore, ABA affected the growth and development of ginseng hairy roots.
NPR1, an SA receptor, plays a key regulatory role in plant immune response (Ji et al., 2022). NPR1 is required for SA-mediated growth inhibition, suggesting a regulatory link between TGA factors and PR gene expression. It plays a role in the PR gene-induced SA signaling pathway. Since NPR1 does not bind to DNA, it may act through one or more transcription factors that mediate the expression of target PR genes. TGA factors play a regulatory role in PR defense gene expression through promoter-specific recruitment mediated by NPR1. TGA factors may also indirectly regulate PR gene expression by interacting with NPR1 to upregulate the expression of other DNA-binding transcription factors (Johnson et al., 2003; Yu et al., 2022). The WRKY transcription factors of the TGA family and alkaline leucine zipper proteins regulate the PR-1 promoter by binding to specific cis-elements (Pape et al., 2010). NPR1, TGA, and PR-1 gene expression of this signaling pathway induced the accumulation of SA, which affected the growth and development of ginseng hairy roots.
As the decomposition of ginseng root residues in soil, its ginsenoside fractions were microbially transformed to rare ginsenosides and released into the soil. This could aggravate the allelopathic autotoxicity of ginsenosides. Transcriptomics studies showed that ginsenosides were able to significantly affect the expression of genes in the biosynthesis of secondary metabolites and metabolic pathways. Targeted metabolomics studies showed that the intervention of ginsenosides significantly affected the metabolic accumulation of endogenous phytohormones in ginseng hair roots. Integrated analysis of transcriptomics and metabolomics further revealed that ginsenosides significantly affected the DMPs and DEGs in phytohormone signal transduction. Ginsenosides presented allelopathic autotoxicity by intervening in the accumulation of JA, and decomposition enhanced the allelopathic autotoxicity by interfering with the accumulation of ABA and increasing the accumulation of SA in plants. The expression of JAR1, COI1, JAZ, MYC2, PYR/PYL, PP2C, SnRK2s, NPR1, TGA, and PR-1 genes regulated the accumulation of JA, ABA, and SA. It indicated that these DEGs were important potential candidate genes involved in continuous cropping obstacles of ginseng. The results of this study provided a useful reference for in-depth research on resolving continuous cropping obstacles in ginseng planting.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1068344.
RW: Formal Analysis, Methodology, Validation, Writing–original draft. TZ: Investigation, Methodology, Validation, Writing–review and editing. YW: Investigation, Methodology, Validation, Writing–review and editing. JD: Investigation, Writing–review and editing. YB: Validation, Writing–review and editing. XH: Investigation, Supervision, Writing–review and editing. CC: Funding acquisition, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work was financially supported by the National Natural Science Foundation of China (82073969) and the Jilin Province Science and Technology Development Plan (20220204078YY).
The authors thank CC from Changchun University of Chinese Medicine for the financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1365229/full#supplementary-material
Ali, A., Pardo, J. M., and Yun, D. J. (2020). Desensitization of ABA-signaling: the swing from activation to degradation. Front. Plant Sci. 11, 379. doi:10.3389/fpls.2020.00379
Bogatek, R., and Gniazdowska, A. (2014). ROS and phytohormons in plant-plant allelopathic interaction. Plant Signal. Behav. 2 (4), 317–318. doi:10.4161/psb.2.4.4116
Brookbank, B. P., Patel, J., Gazzarrini, S., and Nambara, E. (2021). Role of basal ABA in plant growth and development. Genes. 12 (12), 1936. doi:10.3390/genes12121936
Chen, F. H., Xie, Y. J., Jia, Q. W., Li, S. Y., Li, S. Y., Shen, N. K., et al. (2023a). Effects of the continuous cropping and soilborne diseases of Panax ginseng C. A. Meyer on rhizosphere soil physicochemical properties, enzyme activities, and microbial communities. Agronomy-Basel 13 (1), 210. doi:10.3390/agronomy13010210
Chen, L. H., Zhang, Y. B., Yang, X. W., Xu, J., Wang, Z. J., Sun, Y. Z., et al. (2023b). Application of UPLC-Triple TOF-MS/MS metabolomics strategy to reveal the dynamic changes of triterpenoid saponins during the decocting process of Asian ginseng and American ginseng. Food Chem. 424, 136425. doi:10.1016/j.foodchem.2023.136425
Chen, W., Balan, P., and Popovich, D. G. (2020). Ginsenosides analysis of New Zealand-grown forest Panax ginseng by LC-QTOF-MS/MS. J. Ginseng Res. 44 (4), 552–562. doi:10.1016/j.jgr.2019.04.007
Cheng, Z., Yang, M., Zhang, X., Xu, Y., Mei, X., Jiang, B., et al. (2015). Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. Plos One 10 (2), e0118555. doi:10.1371/journal.pone.0118555
Chini, A., Fonseca, S., Chico, J. M., Fernández-Calvo, P., and Solano, R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59 (1), 77–87. doi:10.1111/j.1365-313X.2009.03852.x
Ding, H., Ali, A., and Cheng, Z. (2019). An allelopathic role for garlic root exudates in the regulation of carbohydrate metabolism in cucumber in a hydroponic Co-culture system. Plants 9 (1), 45. doi:10.3390/plants9010045
Feng, F., Yang, C., Li, M., Zhan, S., Liu, H., Chen, A., et al. (2022). Key molecular events involved in root exudates-mediated replanted disease of Rehmannia glutinosa. Plant Physiol. biochem. 1 (172), 136–150. doi:10.1016/j.plaphy.2022.01.014
Gfeller, A., Liechti, R., and Farmer, E. E. (2010). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3 (109), cm4. doi:10.1126/scisignal.3109cm4
Guranowski, A., Miersch, O., Staswick, P. E., Suza, W., and Wasternack, C. (2007). Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 581 (5), 815–820. doi:10.1016/j.febslet.2007.01.049
Huang, X., Liu, Y., Zhang, Y., Li, S. P., Yue, H., Chen, C. B., et al. (2019). Multicomponent assessment and ginsenoside conversions of Panax quinquefolium L. roots before and after steaming by HPLC-MSn. J. Ginseng Res. 43 (1), 27–37. doi:10.1016/j.jgr.2017.08.001
Ji, C., Yang, W., Wang, Y., Su, C., Li, X., Liu, P., et al. (2022). Key residues for maintaining architecture, assembly of plant hormone SA receptor NPR1. Biochem. Biophys. Res. Commun. 613, 94–99. doi:10.1016/j.bbrc.2022.04.119
Jia, F. A., Chang, F., Guan, M., Jia, Q. A., Sun, Y., and Li, Z. (2023). Effects of rotation and Bacillus on the changes of continuous cropping soil fungal communities in American ginseng. World J. Microbiol. Biotechnol. 39 (12), 354. doi:10.1007/s11274-023-03807-w
Jiang, K., and Asami, T. (2018). Chemical regulators of plant hormones and their applications in basic research and agriculture. Biosci. Biotechnol. Biochem. 82 (8), 1265–1300. doi:10.1080/09168451.2018.1462693
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in arabidopsis. Plant Cell. 15 (8), 1846–1858. doi:10.1105/tpc.012211
Kim, S. H., Min, J. W., Quan, L.-H., Lee, S.-Y., Yang, D. U., and Yang, D. C. (2012). Enzymatic transformation of ginsenoside Rb1 by lactobacillus pentosus strain 6105 from kimchi. J. Ginseng Res. 36 (3), 291–297. doi:10.5142/jgr.2012.36.3.291
Kitaoka, N., Kawaide, H., Amano, N., Matsubara, T., Nabeta, K., Takahashi, K., et al. (2014). CYP94B3 activity against jasmonic acid amino acid conjugates and the elucidation of 12-O-β-glucopyranosyl-jasmonoyl-l-isoleucine as an additional metabolite. Phytochem 99, 6–13. doi:10.1016/j.phytochem.2013.12.019
Li, C., Xu, M., Cai, X., Han, Z., Si, J., and Chen, D. (2022). Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 23 (7), 3945. doi:10.3390/ijms23073945
Li, L., Wang, Y., Zhao, M., Wang, K., Sun, C., Zhu, L., et al. (2021). Integrative transcriptome analysis identifies new oxidosqualene cyclase genes involved in ginsenoside biosynthesis in Jilin ginseng. Genomics 113 (4), 2304–2316. doi:10.1016/j.ygeno.2021.05.023
Li, M., Ma, M., Wu, Z., Liang, X., Zheng, Q., Li, D., et al. (2023). Advances in the biosynthesis and metabolic engineering of rare ginsenosides. Appl. Microbiol. Biotechnol. 107 (11), 3391–3404. doi:10.1007/s00253-023-12549-6
Li, S., Todor, A., and Luo, R. (2016). Blood transcriptomics and metabolomics for personalized medicine. Comput. Struct. Biotechnol. J. 14, 1–7. doi:10.1016/j.csbj.2015.10.005
Li, Y., Dai, S., Wang, B., Jiang, Y., Ma, Y., Pan, L., et al. (2020). Autotoxic ginsenoside disrupts soil fungal microbiomes by stimulating potentially pathogenic microbes. Appl. Environ. Microbiol. 86 (9), 001300–e220. doi:10.1128/aem.00130-20
Liu, S., Wang, Z. Y., Niu, J. F., Dang, K. K., Zhang, S. K., Wang, S. Q., et al. (2021). Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 463 (1-2), 427–446. doi:10.1007/s11104-021-04911-2
Meng, X. R., Zhang, T., Chen, C. B., Li, Q., and Liu, J. W. (2022). Regulatory network of ginsenoside biosynthesis under Ro stress in the hairy roots of Panax ginseng revealed by RNA sequencing. Front. Bioeng. Biotechnol. 10, 1006386. doi:10.3389/fbioe.2022.1006386
Ohri, P., Bhardwaj, R., Bali, S., Kaur, R., Jasrotia, S., Khajuria, A., et al. (2015). The common molecular players in plant hormone crosstalk and signaling. Curr. Protein Pept. Sci. 16 (5), 369–388. doi:10.2174/1389203716666150330141922
Palaniyandi, S. A., Son, B. M., Damodharan, K., Suh, J. W., and Yang, S. H. (2016). Fermentative transformation of ginsenoside Rb1 from Panax ginseng C. A. Meyer to Rg3 and Rh2 by Lactobacillus paracasei subsp. tolerans MJM60396. Biotechnol. Bioprocess Eng. 21 (5), 587–594. doi:10.1007/s12257-016-0281-7
Pan, X., Welti, R., and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5 (6), 986–992. doi:10.1038/nprot.2010.37
Pape, S., Thurow, C., and Gatz, C. (2010). The arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1. Plant Physiol. 154 (4), 1805–1818. doi:10.1104/pp.110.165563
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi:10.1146/annurev-arplant-081320-092855
Ren, S. C., Shao, Y. P., Zhao, X. J., Hong, C. S., Wang, F. B., Lu, X., et al. (2016). Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol. Cell. Proteomics 15 (1), 154–163. doi:10.1074/mcp.M115.052381
Salek, R. M., Neumann, S., Schober, D., Hummel, J., Billiau, K., Kopka, J., et al. (2015). COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access. Metabolomics 11 (6), 1587–1597. doi:10.1007/s11306-015-0810-y
Shen, Y. L., Fan, M. L., Li, G., Li, Q., Meng, X. R., Wang, E. P., et al. (2023). Allelopathic effect of ginsenoside Rd on ginseng seeds and seedlings. Appl. Ecol. Environ. Res. 21, 059–072. doi:10.15666/aeer/2101_059072
Singh, A. A., Rajeswari, G., Nirmal, L. A., and Jacob, S. (2021). Synthesis and extraction routes of allelochemicals from plants and microbes: a review. Rev. Anal. Chem. 40 (1), 293–311. doi:10.1515/revac-2021-0139
Suza, W. P., and Staswick, P. E. (2008). The role of JAR1 in Jasmonoyl-l-isoleucine production during Arabidopsis wound response. Planta 227 (6), 1221–1232. doi:10.1007/s00425-008-0694-4
Tagele, S. B., Kim, R.-H., Jeong, M., Lim, K., Jung, D.-R., Lee, D., et al. (2023). Soil amendment with cow dung modifies the soil nutrition and microbiota to reduce the ginseng replanting problem. Front. Plant Sci. 14, 1072216. doi:10.3389/fpls.2023.1072216
Wan, H., Qian, J., Zhang, H., Lu, H., Li, O., Li, R., et al. (2022). Combined transcriptomics and metabolomics analysis reveals the molecular mechanism of salt tolerance of huayouza 62, an elite cultivar in rapeseed (Brassica napus L.). Int. J. Mol. Sci. 23 (3), 1279. doi:10.3390/ijms23031279
Wu, W., Luo, X., Wang, Y., Xie, X., Lan, Y., Li, L., et al. (2023). Combined metabolomics and transcriptomics analysis reveals the mechanism underlying blue light-mediated promotion of flavones and flavonols accumulation in Ligusticum chuanxiong Hort. microgreens. J. Photochem. Photobiol. B 242, 112692. doi:10.1016/j.jphotobiol.2023.112692
Yu, X., Cui, X., Wu, C., Shi, S., and Yan, S. (2022). Salicylic acid inhibits gibberellin signaling through receptor interactions. Mol. Plant 15 (11), 1759–1771. doi:10.1016/j.molp.2022.10.001
Zanardo, L. G., de Souza, G. B., and Alves, M. S. (2019). Transcriptomics of plant-virus interactions: a review. Theor. Exp. Plant Physiol. 31 (1), 103–125. doi:10.1007/s40626-019-00143-z
Zhang, A. H., Lei, F. J., Fu, J. F., Zhou, R. J., and Zhang, L. X. (2017). Influence of exogenous ginsenosides on new forest soil microbial communities. Zhongguo Zhongyao Zazhi 42 (24), 4756–4761. doi:10.19540/j.cnki.cjcmm.2017.0213
Zhang, M., Zou, X., Du, Y., Pan, Z., He, F., Sun, Y., et al. (2023). Integrated transcriptomics and metabolomics reveal the mechanism of alliin in improving hyperlipidemia. Foods 12 (18), 3407. doi:10.3390/foods12183407
Zhang, W., Lu, L. Y., Hu, L. Y., Cao, W., Sun, K., Sun, Q. B., et al. (2018). Evidence for the involvement of auxin, ethylene and ROS signaling during primary root inhibition of arabidopsis by the allelochemical benzoic acid. Plant Cell. Physiol 59 (9), 1889–1904. doi:10.1093/pcp/pcy107
Zhang, X., Ma, R., Liu, X., Jiang, X., and Wang, L. (2016). Simultaneous determination of ginsenoside Rg1, Re and notoginsenoside R1 in human plasma by LC-MS/MS and its application in a pharmacokinetic study in Chinese volunteers. Biomed. Chromatogr. 30 (12), 1915–1921. doi:10.1002/bmc.3766
Zhang, Z., Chen, Q., Tan, Y., Shuang, S., Dai, R., Jiang, X., et al. (2021). Combined transcriptome and metabolome analysis of alfalfa response to thrips infection. Genes. 12 (12), 1967. doi:10.3390/genes12121967
Keywords: ginsenoside, decomposition, transcriptomics, metabolomics, integrated analysis, allelopathic autotoxicity
Citation: Wang R, Zhou T, Wang Y, Dong J, Bai Y, Huang X and Chen C (2024) Exploring the allelopathic autotoxicity mechanism of ginsenosides accumulation under ginseng decomposition based on integrated analysis of transcriptomics and metabolomics. Front. Bioeng. Biotechnol. 12:1365229. doi: 10.3389/fbioe.2024.1365229
Received: 04 January 2024; Accepted: 12 February 2024;
Published: 07 March 2024.
Edited by:
Bikram Basak, Hanyang University, Republic of KoreaReviewed by:
Sailendra Singh, Migal—Galilee Research Institute, IsraelCopyright © 2024 Wang, Zhou, Wang, Dong, Bai, Huang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Huang, aHVhbmd4aW5yb3NlQDE2My5jb20=; Changbao Chen, Y2NiQGNjdWNtLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.