- 1College of Food Science and Engineering, Harbin University, Harbin, China
- 2College of Food Science and Engineering, Tianjin University of Science and Technology, Tianjin, China
Introduction: β-glucosidase is one class of pivotal glycosylhydrolase enzyme that can cleavage glucosidic bonds and transfer glycosyl group between the oxygen nucleophiles. Lactobacillus is the most abundant bacteria in the human gut. Identification and characterization of new β-glucosidases from Lactobacillus are meaningful for food or drug industry.
Method: Herein, an acid-adapted β-glucosidase (LpBgla) was cloned and characterized from Lactobacillus paracasei. And the insight acid-adapted mechanism of LpBgla was investigated using molecular dynamics simulations.
Results and Discussion: The recombinant LpBgla exhibited maximal activity at temperature of 30°C and pH 5.5, and the enzymatic activity was inhibited by Cu2+, Mn2+, Zn2+, Fe2+, Fe3+ and EDTA. The LpBgla showed a more stable structure, wider substrate-binding pocket and channel aisle, more hydrogen bonds and stronger molecular interaction with the substrate at pH 5.5 than pH 7.5. Five residues including Asp45, Leu60, Arg120, Lys153 and Arg164 might play a critical role in the acid-adapted mechanism of LpBgla. Moreover, LpBgla showed a broad substrate specificity and potential application in the bioconversion of glycosides, especially towards the arbutin. Our study greatly benefits for the development novel β-glucosidases from Lactobacillus, and for the biosynthesis of aglycones.
Introduction
β-glucosidase (Bgla), or β-D-Glucoside glucohydrolase (3.2.1.21) is a dominant class of pivotal glycosylhydrolase enzyme that has been well-characterized in catalyzing the selective cleavage of glucosidic bonds and transferring glycosyl group between the oxygen nucleophiles (Bhatia et al., 2002). Bgla usually hydrolyzes the terminal, non-reducing β-D-glucosyl residues to release β-D-glucose from lots of glucoconjugates including glucosides, oligosaccharides and 1-O-glucosyl esters (Godse et al., 2021). Therefore, the wide range of substrates in nature contributes the ubiquitous applications of Bgla in almost all aspects of life, for example, in lots of biotechnological and industrial applications containing degradation of structural and storge polysaccharides (Bhatia et al., 2002), hydrolysis of pharmaceutical ingredients (Xiao et al., 2014), production of sugar or fuel ethanol from lignocellulosic biomass (Singhania et al., 2017), or in many crucial biological processes including cellular signaling, host-pathogen interactions, et al. (Bhatia et al., 2002).
Bgla is commonly distributed in almost all living organisms from simple bacteria to highly evolved mammals, and perform a variety of physiological roles in target host depending on their location or functions (Collins et al., 2007; Fan et al., 2011). Filamentous fungi and bacteria are known as abundant Bgla resources, in which, Bgla is mainly acted as cellulase enzyme and responsible for the catalysis of short chain oligosaccharides and cellobiose to produce glucose (Molina et al., 2016). For example, Yan et al. identified a novel Bgla from Paecilomyces thermophila, and heterologous expressed and characterized in Pichia pastoris, high enzymatic activity and yield of Bgla were obtained by high cell-density fermentation (Yan et al., 2012). A cold-adapted Bgla from a psychrotolerant bacterium Micrococcus antarcticus was cloned, over-expressed and characterized using the E. coli expression system, the enzyme exhibited maximal activity at 25°C and pH 6.5 (Fan et al., 2011). We also developed an acid-stable Bgla from Aspergillus aculeatus, characterized the enzymatic properties and probed the involved acid-denaturation mechanism using molecular dynamics simulations (Li Y. et al., 2018).
Many complex carbohydrates or degradation-resistant plant cell wall glycans from our diets can not be digested by human enzymes (Martens et al., 2011; Ndeh et al., 2017). Symbiotic bacteria inhabiting in the human gut can utilize these polysaccharides to produce low molecular weight carbohydrates or absorbable short chain fatty acids (SCFAs) (Martens et al., 2011). For example, Bacteroidetes thetaiotaomicron in human gut can utilize and depolymerize the large mannan oligosaccharides to generate mannose by the periplasmic enzymes (Cuskin et al., 2015). Moreover, the elaborate and highly specific enzyme system in gut bacteria can depolymerize the highly complex glycans, polysaccharide rhamnogalacturonan-II, by cleaving all but 1 of its 21 distinct glycosidic linkages (Ndeh et al., 2017). Liquorilactobacillus satsumensis isolated from water kefir could use the carbohydrates to produce low molecular weight glucans, and the hydrolyzed products showed prebiotic properties that could promote the abundance of Bacteroidetes and increase the contents of SCFAs, including acetate, proionate and butyrate (Tan et al., 2022). Lactobacillus are one of the most abundant bacteria in the human gut, so, Lactobacillus may have rich and robust glycosidase enzyme system. Identification and characterization of new glycosidases from Lactobacillus are meaningful for food or drug industry. Although the Bgla is one of the most widely used glycosidases, there are few Bgla have been identified or deeply studies from the Lactobacillus. Therefore, in this study, a novel Bgla gene from Lactobacillus paracasei TK1501 (LpBgla) was cloned and high-efficiently expressed in E. coli. After purified using Ni-NTA column, the enzymatic properties of LpBgla enzyme including optimum temperature, pH, stability, enzyme kinetics, and substrate specificity are systemically investigated. Furthermore, the insight acid-adapted mechanism, and the key residues involved in the catalysis of Bgla were investigated using molecular dynamics simulations. We think this work has great significance for the theoretical research and practical application.
Materials and methods
Materials
DNA Purification Kit, Quick Plasmid MiniPrep Kit were purchased from Thermofisher Scientific (Waltham, MA, United States). DNA polymerase, RNase, DNA Ligation Kit Ver.2, restriction enzymes of HindIII and XhoI were purchased from Takara (Dalian, China). Ni-NTA column and resin were purchased from Qiagen (Hilden, Germany). The substrates of β-p-Nitrophenyl-β-D-glucuronide (β-pNPG), geniposide, salicin, polydatin, heptaside and arbutin were purchased from Sigma-Aldrich (St. Louis, MO, United States). Other reagents or chemicals with high purity were derived from local sources unless noted.
Strains and plasmids
Escherichia coli (E. coli) JM109 and BL21(DE3) were cultivated using Luria-Bertani (LB) media and stored in our lab. E. coli JM109 was used as the cloning host, and BL21(DE3) was used as expression host of LpBgla gene. Lactobacillus paracasei TK1501 was screened from Chinese traditional fermented food, cultured using MRS media and stored in our lab. Plasmid used in cloning and expression of LpBgla gene was pET28a vector (Novagen, Darmstadt, Germany).
Cloning, expression and purification of LpBgla
The cloning, expression and purification of recombinant LpBgla were following and modified from previous reported methods (Jia et al., 2018; Shang et al., 2022). The LpBgla gene was amplified using the Lactobacillus paracasei TK1501 genome DNA as template, and the specific primers were LpBgla-F (5′- GGAATTCCATATGGGGGTGGTAGTATCTAACTTTC-3′) and LpBgla-R (5′- CCCAAGCTTTTATCTAAAAATCATTTTCACATACTGATG-3′). Then the fragment of LpBgla was double digested using HindIII and XhoI enzymes, and ligated into the pET28a vector with same restriction enzyme sites to construct the expression plasmid. After identified by DNA sequencing, the expression plasmid was named as pET28a-LpBgla.
The pET28a-LpBgla plasmid was transferred into the expression host E. coli BL21(DE3) to prepare recombinant strain. The colonies were identified by PCR method using LpBgla-F/LpBgla-R primers, and the positive recombinant strain was named BL21-LpBgla. Single colony of BL21-LpBgla was selected and cultured in 5 mL LB medium at 37°C overnight. Then, 2% (v/v) of the overnight-incubated bacteria solution was inoculated into 100 mL fresh LB medium, and incubated at 37°C until the OD600 was to 0.6–0.8. 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added into the solution to induce the overexpression of LpBgla at 16°C. After cultured for 16–20 h, the BL21-LpBgla cells were collected by centrifuging at 12,000 rpm 10 min, and then resuspended in lysis buffer (20 mM Tris, pH 7.4, 20 mM imidazole, 0.5 M NaCl, 1 mM DTT). A sonication process was employed to break BL21-LpBgla cells for the releasing of intracellular proteins. Subsequently, the bacteria solution was centrifuged at 12,000 rpm at 4°C for 30 min, and the supernatant was collected. We used the Ni-NTA method to purify the His-tagged LpBgla enzyme. Firstly, the supernatant of BL21-LpBgla sonication solution was gently mixed with the agarose resin and combined for 1 h at room temperature. Then the mixture solution was trapped on a column, and flowed out naturally through gravity. The resin was washed by wash buffer (20 mM Tris, pH 7.4, 40 mM imidazole, 0.5 M NaCl, 1 mM DTT), and the target LpBgla protein was eluted by elution buffer (20 mM Tris, pH 7.4, 400 mM imidazole, 0.5 M NaCl, 1 mM DTT). SDS-PAGE was used to identify the proteins during the purification process. At last, the purified LpBgla protein was dialyzed against Tris (50 mM, pH 7.4) and used for enzymatic properties characterization.
β-glucosidase activity measurement
The β-glucosidase activity was assessed following previous method using β-pNPG as substrate (Li Y. et al., 2018). Briefly, 0.1 mL purified enzyme sample (0.7 mg/mL) was added into 0.5 mL working solution with 5 mM β-pNPG. After incubated at 30°C for 30 min, the reaction was terminated by addition 0.25 mL 0.2 M Na2CO3. Then the absorbance of the samples was measured using a plate reader at 410 nm. One unit (U) of LpBgla activity was designated as the amount of enzyme that produce 1 μmol p-nitrophenol per minute. The concentration of LpBgla was measured using Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, United States).
Characterization of the LpBgla
In order to investigate the optimal temperature of the LpBgla activity, we measured the enzyme activity using above standard reaction method at various temperatures of 20, 30, 40, 50, 60°C and 70°C. To access the thermostability of LpBgla, the enzyme was incubated at 20, 30, 40, 50°C and 60°C for 10, 30, 60, 90 and 120 min, the residual enzyme activity was tested using standard protocol. The initial LpBgla enzyme activity was defined as 100%.
The optimal pH of LpBgla activity was also probed by the above standard method, that is, the enzymatic reaction was performed at 30°C at different pH from 4.0 to 8.0. Meanwhile, pH stability was measured by treating the enzyme at different pH from 4.0 to 8.5 at 4°C for 60 min, and the residual enzyme activity was tested using standard protocol. The maximum LpBgla enzyme activity was defined as 100%.
Moreover, the impact of metal ions and EDTA on the activity of LpBgla was investigated. 5 mM metal ions including Mg2+, Mn2+, Fe2+, Ca2+, Zn2+, Cu2+, Fe3+ and EDTA were individually added into the reaction system. Then the LpBgla activity was measured using standard method at 30°C and pH 5.5. The relative enzyme activity of LpBgla in the absence of metal ions was defined as 100%.
Enzymatic kinetic analysis
The kinetic parameters of LpBgla were analyzed by measuring the enzyme activity at the optimum reaction conditions using different concentrations of substrate from 0.5 to 15 mM. The Km and Vmax values were determined by fitting the data to the Michaelis-Menten equation, and the kcat was calculated by kcat = Vmax/(Cenc.). All the data were processed using GraphPad prism 8.0 software.
Molecular docking and molecular dynamic (MD) simulations
The crystal structure of LpBgla was predicted and obtained from AlphaFold2 database (UniProt ID: Q03B82). 3D structure of β-pNPG was download from PubChem (http://pubchem.ncbi.nlm.nih. gov), and optimized by MOPAC (Stewart, 1990). AutoDock 4.2.6 and AutoDockTools 1.5.6 were used to conduct the molecular docking (Morris et al., 2009; Maier et al., 2015). The grid dimensions were set as 60 Å × 60 Å × 60 Å, and grid center was defined at x = 2.014, y = −13.655, z = 16.129.
To set the different pH values, the LpBgla was firstly treated using the online tool H++ (http://newbiophysics.cs.vt.edu/H++/) to calculate the pKa of each residue, then pH 5.5 or pH 7.5 environment was given to the enzyme-substrate complex, and a new pdb profile was obtained for the MD simulations. The MD simulations were performed using the GROMACS 2018.4 simulation package (Jorgensen et al., 1983), and with the force field of AMBER14SB (Mahoney and Jorgensen, 2000). Firstly, the LpBgla and β-pNPG substrate complexes were placed in a square box. Equal molecule of positive Na+ and negative Cl− were added to substitute a part of water. Rational spacing among LpBgla-β-pNPG complex and water molecules were conducted using steepest descent minimization method. Initial random velocity was made by the Maxwell distribution, and the NVT simulation time was defined as 1 ns. All the covalent bonds relating to the hydrogen atoms were constrained by LINCS algorithm (Hess, 2008). The long-range electrostatic interactions were calculated using Particle Mesh Ewald (PME) method with a grid size of 10 Å (Darden et al., 1993). The temperature was set as 333 K by V-rescale (Berendsen et al., 1984), and pressure was maintained as 1 bar under isothermalisostatic (NPT) ensemble by Parrinello-Rahman (Wang et al., 2022) method. MD simulations were carried out for 100 ns. Based on the MD trajectory, root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg) and solvent accessible surface area (SASA) values of LpBgla were calculated to analysis the structural stability with GROMACS package using gmx rms, gmx rmsf, gmx gyrate and gmx sasa programs, respectively. At last, Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) was utilized to calculate the binding free energy between the LpBgla and β-pNPG (detailed method was deposited in Supporting Material). Herein, two MD simulation calculations were carried out: one run set the environment pH as the optimal pH of LpBgla, pH 5.5; and the other run set as an alkaline environment, pH 7.5.
Substrate specificity and glycosides bioconversion of LpBgla
The catalytic specificity of LpBgla was measured with different substrates including geniposide, salicin, polydatin, esculin and arbutin. β-pNPG was used as the control substrate. Same to the β-glucosidase activity measurement, 0.1 mL purified LpBgla solution (0.7 mg/mL) was gently mixed with 0.5 mL 5 mM different substrates (dissolved in 0.1 mM PBS buffer, pH 5.5). The enzymic catalytic reaction was performed at 30°C for 30 min, and terminated by adding 0.25 mL 0.2 M Na2CO3 solution. The enzymatic activity was evaluated by detecting the production of glucose or aglycone using standard HPLC method (Yeom et al., 2012; Li Y. et al., 2018). Herein, one unit (U) enzyme activity was defined as the amount of LpBgla to produce 1 μmol glucose or aglycone per minute. Moreover, the kinetic parameters of LpBgla using each substrate were also analyzed using above methods.
β-Glucosidase can hydrolysis the β-glucosidic bond of glycosides to form aglycone and glucose. In order to analysis the bioconversion effects of LpBgla on glycosides, several different glycosides including geniposide, salicin, polydatin, esculin and arbutin were measured. The catalytic reaction was following the method of β-glucosidase activity measurement. Briefly, 0.1 mL purified LpBgla enzyme (0.7 mg/mL) was added into 0.5 mL working solution with 5 mM different glycoside samples. After incubated at 30°C for 12 h, the reaction was terminated by addition 0.25 mL 0.2 M Na2CO3. Then the reaction samples were detected by a High-performance liquid chromatography (HPLC) system using a reversed-phase chromatography NovaPak C18 column (250 × 4.6 mm, 5 μm, Waters, MA, United States) with a photodiode array detector (PDA-100, Dionex). The mobile phases were methanol with 0.1% acetic acid (phase A) and water with 0.1% acetic acid (phase B). The flow rate was 1 mL/min, and 10 μL of the samples were injected into the HPLC system. Other detailed HPLC parameters were following our previously reported method (Xie et al., 2022).
Results and discussion
Sequence identity analysis of LpBgla
In our previously reported study, the complete genome sequence of Lactobacillus paracasei TK1501 has been sequenced and deposited in the GenBank database (accession number: CP017716) (Xie et al., 2022). Based on the genome sequence of Lactobacillus paracasei TK1501, the GenBank locus of LpBgla was BKQ19_03070. Detailly, the gene of LpBgla is 2,388 bp, encodes 796 amino acids (Supplementary Table S1). Blast search in the GeneBank from NCBI showed that LpBgla exhibited the highest amino acid sequence identity of 71.34% with the Bgla from Lactobacillus casei. And the similarity of LpBgla with the Bglas from Lactobacillus perolens, Lactobacillus kimchicus, Lactobacillus paracasei and Lactobacillus rhamnosus were 56.54%, 53.45%, 33.71% and 15.91%. The structure and function of Bgla derived from Thermotoga neapolitana DSM 4359 (GenBank ID: ACM22846.1) has been analyzed in previously reported literature (Pozzo et al., 2010), which showed 28.30% amino acid sequence identity with LpBgla. The phylogenetic tree was constructed by using MEGA 6.0 biology software according to the blast sequence results, which showed that the LpBgla formed same branch, and was most closely related to β-glucosidase derived from Lactobacillus casei (Figure 1A).
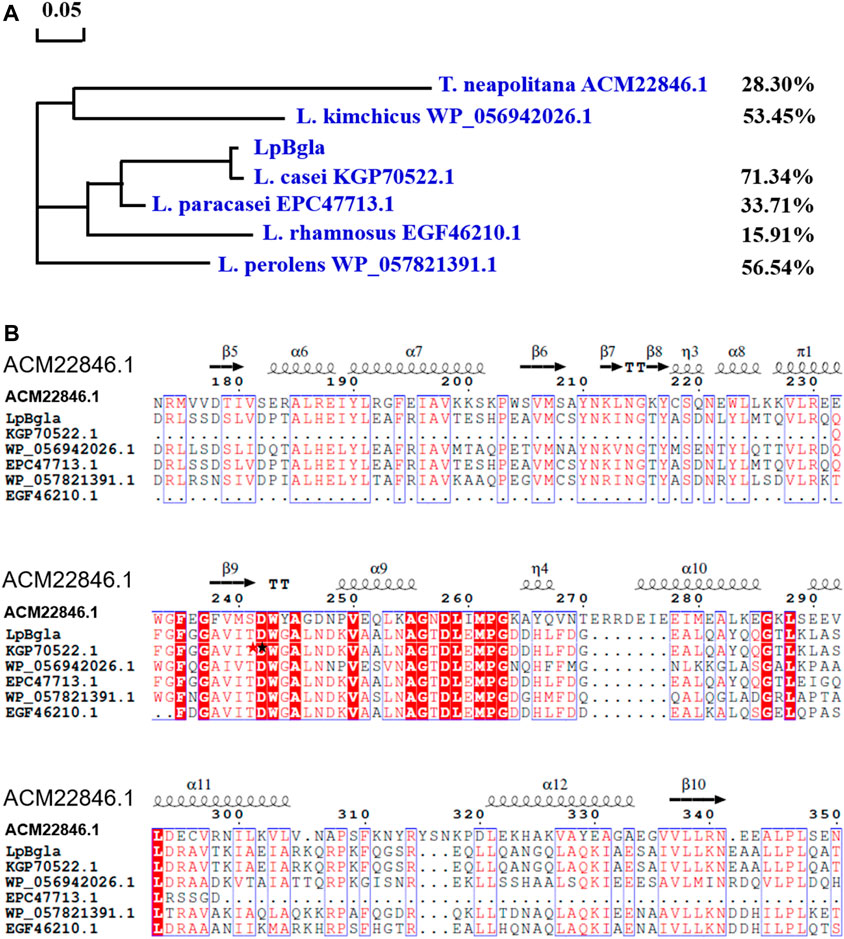
FIGURE 1. Sequence analysis of LpBgla with six Bglas from Lactobacillus casei, Lactobacillus perolens, Lactobacillus kimchicus, Lactobacillus paracasei and Lactobacillus rhamnosus and Thermotoga neapolitana. (A) Phylogenetic tree of LpBgla that constructed using MEGA 6.0. (B) Multiple alignment of LpBgla amino acid sequence with other six Bglas. The residues marked in red were strictly conserved, and boxed in blue were highly conserved. The secondary structure elements were labeled above the corresponding sequences.
Based on the blast results, the multiple sequence alignment of LpBgla was carried out using the online server Clustal X, and modified using the ESPript 3.0 (Supplementary Figure S1). Figure 1B showed the relative conserved part of the alignments, three strictly conserved fragments were identified including FxGxxxxDWxAxxxxV (residues 235–250), AGxDLxMPG (residues 255–263), and GxLxxxxL (residues 286–293). According to the crystal structure of ACM22846.1 from T. neapolitana, D242 and E458 were proved to be the essential catalytic residues, D58 and W243 were important for substrate accommodation or interaction (Pozzo et al., 2010), both of these two residues were conversed from the results of multiple sequence alignments. While in lots of other literature, two conserved residues of Glu located at the end of β-strand 4 and β-strand 7 were the general catalytic site for the β-glucosidases (Jeng et al., 2011; Kaenying et al., 2023). In line with the blast results, the identity of LpBgla with the other six Bglas ranged from 15% to 72%. Together with the results of the sequence comparison analysis, LpBgla was a novel Bgla family member.
Cloning, expression and purification of the LpBgla
We designed the specific primers of LpBgla according to the complete genome sequence of Lactobacillus paracasei TK1501. The LpBgla gene was cloned using the genome DNA as template. As shown in Supplementary Figure S2A, a specific DNA band of 2,388 bp was observed in the gel, which in line with the theoretical molecular value of LpBgla. Then the LpBgla gene was cloned into the pET28a vector to construct the expression plasmid pET28a-LpBgla. The pET28a-LpBgla was identified using HindIII or XhoI single digestion, or HindIII and XhoI double digestion, respectively. Enzymic digestion results were consisted with the theoretical prediction, which indicated that the expression pET28a-LpBgla plasmid was successfully constructed (Supplementary Figure S2B). The pET28a-LpBgla plasmid was introduced into E. coli BL21 for heterologous expression of LpBgla. The positive transformants were identified using colony PCR, the results showed that the three picked transformants were true recombinants carrying the LpBgla gene (Supplementary Figure S2C). We renamed the recombinant E. coli BL21 that transformed with pET28a-LpBgla as BL21-LpBgla.
After induced by IPTG, the LpBgla enzyme was expressed in E. coli BL21. We respectively measured the β-glucosidase activity of intracellular and extracellular parts by standard methods. The activity of intracellular section was 48.3 U/mg, and extracellular section was 1.5 U/mg, suggesting that the recombinant LpBgla protein was mainly expressed in the intracellular part. Then the LpBgla was purified using Ni-NTA affinity chromatography, as shown in Supplementary Figure S2D, a ∼85 kDa band was observed in the gel, which was almost identical to the theoretical mess of LpBgla (about 85.6 kDa). The LpBgla was also further purified and identified using size-exclusion chromatography, which proved that this enzyme existed as monomeric state (Supplementary Figure S3). These results indicated that the LpBgla from Lactobacillus paracasei TK1501 was successfully cloned and heterologous expressed in E. coli.
Enzymatic characterization of LpBgla
Temperature and pH were the key factors that affect the enzymatic activity. In order to analysis the enzymatic properties of LpBgla, we measured the β-glucosidase activities of purified LpBgla at different temperature and pH. As shown in Figure 2A, the maximum activity of LpBgla was defined as 100%, relative activities of this enzyme at 20°C–40°C were more than 60%. When the temperature was 30°C, the enzyme activity was maximized, suggesting that the optimal temperature of LpBgla was 30°C. After the LpBgla was incubated at 20°C–60°C for different times, the residual activities of all the samples were measured. In which, LpBgla still present approximate 100% of its initial activity after incubating at 20°C and 30°C for 120 min (Figure 2B), and retained over 70% activity when the temperature below 50°C (Figure 2B), indicating that LpBgla exhibited a good thermostability. LpBgla showed a general optimal temperature, which lower than some β-glucosidases from some other microorganisms, for example, Aspergillus aculeatus (70°C) (Li Y. et al., 2018), Paecilomyces thermophila (65°C) (Yan et al., 2012) and Aspergillus fumigatus Z5 (60°C) (Liu et al., 2012). The low optimal temperature maybe helpful for the application of LpBgla in the food industry.
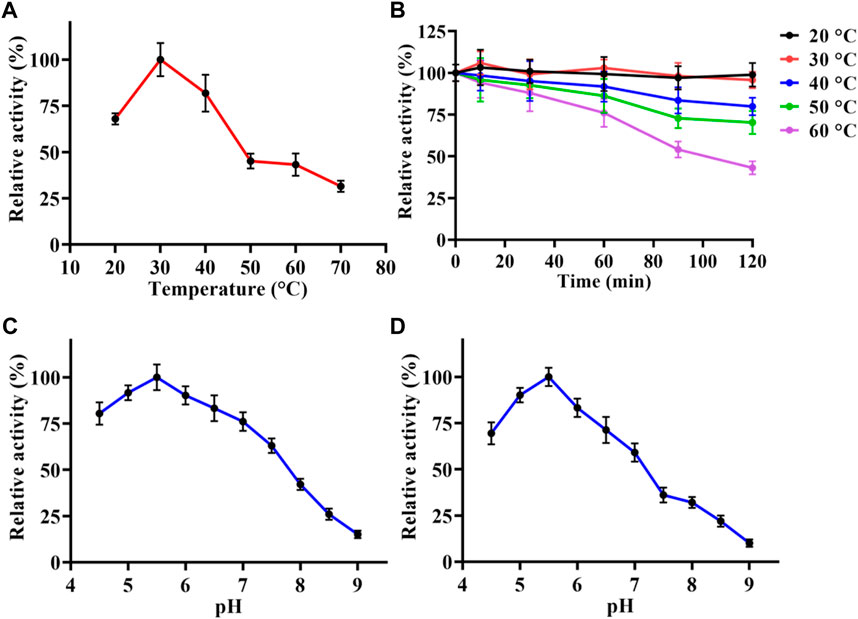
FIGURE 2. Enzymatic characterization of LpBgla. (A) Optimal temperature. (B) Thermostability analysis. (C) Optimal pH. (D) pH stability. The maximum activity of LpBgla is defined as 100% in each graph.
LpBgla exhibited higher activity in acid conditions (pH < 7.0) than alkaline (pH > 7.0), in which, the optimal pH for enzyme activity was 5.5 (Figure 2C). The optimal pH of LpBgla was similar to some other β-glucosidases from Aspergillus aculeatus (pH 5.0) (Li Y. et al., 2018), Clostridium cellulovorans (pH 6.0) (Jeng et al., 2011) and Aspergillus fumigatus (pH 6.0) (Liu et al., 2012). We also found LpBgla could maintain high activities after treated in acid conditions (pH < 7) for 60 min, while the activities sharply decreased after incubated in alkaline solutions (pH > 7) (Figure 2D). Specially, the residual activity of LpBgla was maximum when treated at pH 5.5 for 60 min (Figure 2D). All these results indicated that LpBgla was an acid stable and resistant β-glucosidase, which could be used in fermentation food, cellulose hydrolysis processes, or some other acid environment industries.
Moreover, we also investigated the effects of metal ions and EDTA on the purified LpBgla activity. As shown in Figure 3A, compared to the control, Ca2+ and Mg2+ increased the enzymatic activity to 107.3% and 112.9%, respectively. In the presence of Cu2+, Fe2+, Fe3+, Mn2+, and EDTA, the activity of LpBgla were significantly decreased (p < 0.05). Especially in the presence of Cu2+, the enzymatic activity decreased to 23.1% compared with the control (Figure 3A). Zn2+ showed negligible effects on the LpBgla activity (Figure 3A). It was indicated that several metal ions and agents, especially Cu2+, inhibited the enzymatic activity of LpBgla, suggesting that the active sites of this enzyme might have interactions with these metal ions, which made contributions to the enzymatic activity inhibition (Jeng et al., 2011; Liu et al., 2012).
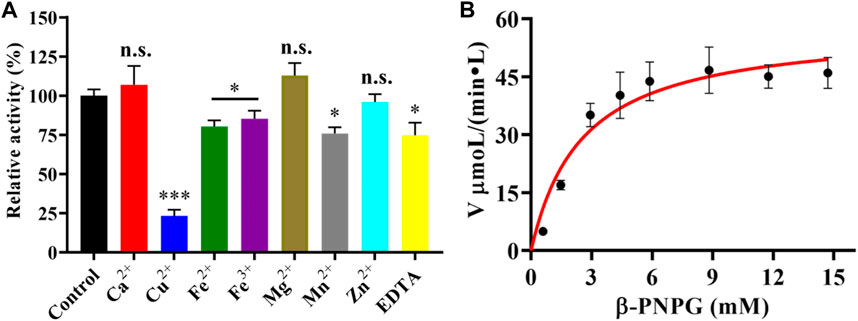
FIGURE 3. (A) Effect of various metal ions and EDTA on the enzymatic activity of LpBgla. The enzyme activity without any regents were defined as control (100%). All the values represent as Means ± SD (n = 3). n.s., not significant; *, p < 0.05; ***, p < 0.001. (B) Enzyme kinetics of LpBgla.
Kinetics analysis of LpBgla
The kinetic parameters of the purified LpBgla were determined using different concentrations of β-pNPG, and applying a nonlinear curve fit by GraphPad prism 8.0 software. The kinetics curve was shown in Figure 3B, based on the Michaelis-Menten calculations, the Km value of LpBgla was 1.44 mM, and Vmax value was 58.32 μM min−1. The kcat was calculated, and showed a high enzyme turnover of 3.98×103 s−1. Given by the kcat/Km ratio of 2.77×103 s−1 mM−1, the catalytic efficiency of LpBgla using β-pNPG as substrate was much higher than several other β-glucosidases, for example, from Aspergillus fumigatus (125.50 and 161.76 s−1 mM−1) (Liu et al., 2012), Clostridium cellulovorans (340 s−1 mM−1) (Jeng et al., 2011), Micrococcus antarcticus (1,120 s−1 mM−1) (Fan et al., 2011). Despite the monomeric nature of LpBgla, the sigmoidal character of the v0 vs. [S] remains and does not fit well to the hyperbola, it may be due to the conformational changes of enzyme’s activity center, or the substrate-induced coordination effects, or other enzyme-related factors to lead the atypical sigmoidal kinetic behavior (Porter and Miller, 2012). Similar to some previously reported monomeric enzymes with sigmoidal kinetics, for example, versatile peroxidase from Bjerkendera adusta (Siddiqui et al., 2014), cytochrome P450 enzyme 2C9 (Court et al., 2001) and glucokinase (Whittington et al., 2015) from human liver, et al., suggesting that sigmoidal kinetics are not only for oligomeric enzymes.
Structural stability analysis of LpBgla at pH 7.5 or 5.5 by MD simulations
Form the enzymatic property results, it was indicated that the LpBgla was an acid-adapted β-glucosidase, especially the optimal pH for its activity was 5.5. To investigate the involved molecular mechanism of the acid resistance of LpBgla, the structure and conformation changes of this enzyme at pH 5.5 and 7.5 were analyzed by MD simulations following our previously reported methods (Li Y. et al., 2018; Shang et al., 2022). As shown in Figure 4A, except the last 5 ns, the RMSD values of LpBgla at pH 5.5 were generally lower than that at pH 7.5 during the whole simulation time. Although the RMSF values of LpBgla at pH 5.5 and 7.5 were seemed similar, the RMSF at pH 7.5 were remarkably higher than that at pH 5.5 in residues of 60–63, 118–124, 632–640 and 736–749 (Figure 4B), indicating that the protein structure in these fragments were more flexible at pH 7.5 than at pH 5.5. Moreover, we also analyzed the average B-factor value of LpBgla residues at different pH, which would reflect the enzyme’s conformational state and structure flexibility. It was showed that the average B-factor value of LpBgla residues at pH 7.5 was higher than that at pH 5.5, suggesting that the LpBgla showed a more flexible conformation at pH 7.5 than at pH 5.5 (Figure 4C).
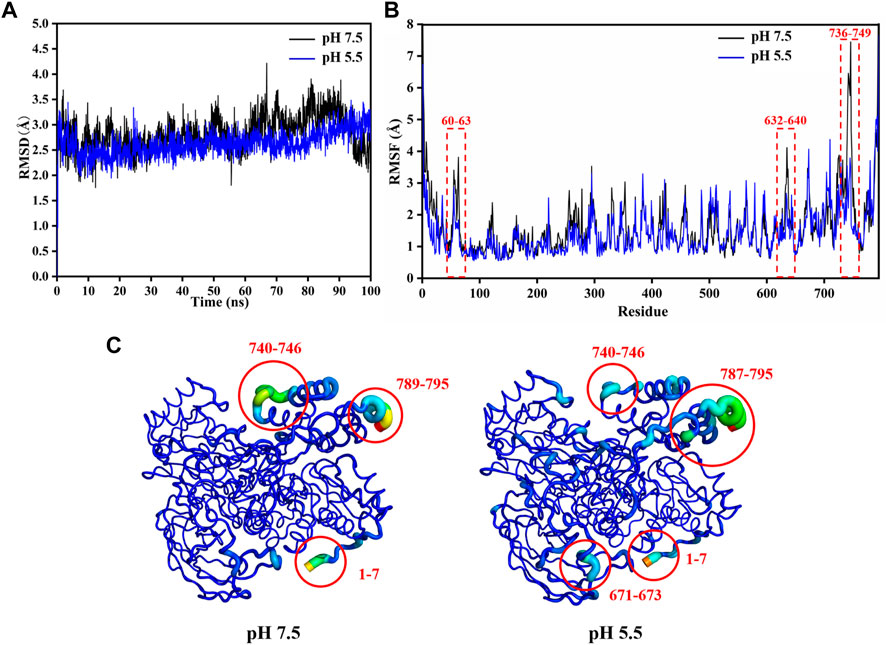
FIGURE 4. Structural stability analysis of LpBgla at different pH by MD simulations. (A) The root-mean-square deviation (RMSD) analysis. (B) The root-mean-square fluctuation (RMSF) value of each residue in the LpBgla at pH 7.5 and 5.5. (C) The B-factors of LpBgla at pH 7.5 and 5.5 binding to the substrate at the end of simulations.
As the Rg value was on behalf of the density of protein system, we compared the Rg values of LpBgla at different pH. During the whole MD simulations, the Rg values of LpBgla at pH 7.5 were thoroughly higher than that at pH 5.5 (Supplementary Figure S4A). In line with the Rg results, the SASA values of LpBgla at pH 7.5 were also higher than that at pH 5.5 during the whole 100 ns simulation time (Supplementary Figure S4B). Therefore, all the above structural and conformational analysis indicated that structure of LpBgla at pH 5.5 was more stable than that at pH 7.5.
Molecular interaction analysis between LpBgla and β-pNPG at pH 7.5 or 5.5 by MD simulations
The interaction of LpBgla with the β-pNPG was important for the catalytic process. During the whole simulation time, although the minimum distance of LpBgla to the substrate was similar either at pH 7.5 or 5.5 (Supplementary Figure S5), the contact numbers between LpBgla and the substrate were different. As shown in Figure 5A, the average contact number during the simulation between LpBgla and the substrate at pH 5.5 was 33.6 ± 1.7, which was remarkable higher than that at pH 7.5 (29.5 ± 2.6). It was suggested that at low pH, more contacts were generated between the LpBgla and substrate, and in turn might cause more interactions. We furtherly analyzed the influence of different pH on the hydrophobic and electrostatic interactions, especially of the hydrogen bonds between LpBgla and substrate. Total number of hydrogen bonds of LpBgla at pH 7.5 and 5.5 were calculated. The data showed that there were 6.4 ± 1.1 hydrogen bonds between enzyme and substrate at pH 5.5 during the 100 ns simulation time, which was more than that at pH 7.5 (5.8 ± 1.2), indicating that the interaction of LpBgla with the substrate was stronger at pH 5.5 than pH 7.5 (Figure 5B).
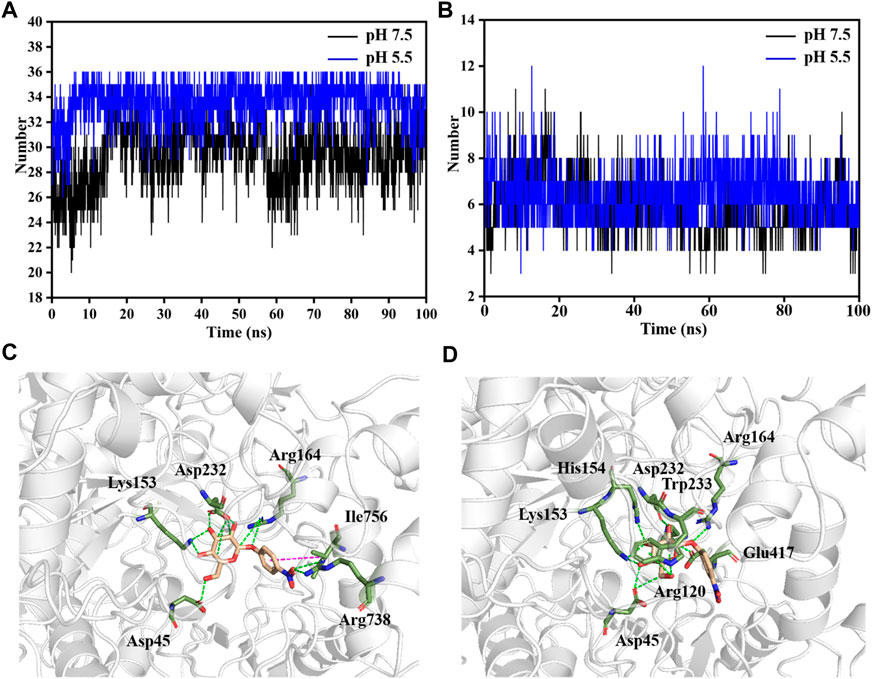
FIGURE 5. Analysis of molecular interaction between LpBgla and β-pNPG at different pH by MD simulations. (A) Average contact numbers, and (B) hydrogen bonds between LpBgla and substrate at pH 5.5 and 7.5 during the 100 ns simulations. Molecular interaction of LpBgla with substrate at (C) pH 7.5, and (D) pH 5.5 at the end of simulations. The hydrogen bonds were defined as green dashed lines, and Pi-Pi interaction was defined as magenta dashed line.
The potential key residues that involved in the molecular interaction between LpBgla and β-pNPG were also investigated at different pH conditions. In the pH 7.5 system, six residues contributed main interactions of the enzyme with substrate (Figure 5C). In which, the β-pNPG interacted with the residues of Asp45, Lys153, Arg164, Asp232 and Arg738 by forming 10 hydrogen bonds, and interacted with Ile756 by forming Pi-Pi conjugate interaction (Figure 5C, S6A). Comparatively, eight residues including Asp45, Arg120, Lys153, His154, Arg164, Asp232, Trp233 and Glu417 involved in the interaction of LpBgla with the substrate at pH 5.5, and 12 hydrogen bonds were formed during the simulations (Figure 5D, S6B). There results demonstrated that more amino acids and hydrogen bonds were formed between LpBgla and β-pNPG at pH 5.5 than pH 7.5, suggesting that the interaction of the enzyme with the substrate at pH 5.5 is stronger than that at pH 7.5.
Effects of different pH on the substrate-binding pocket of LpBgla by MD simulations
It has been proved that the structure and interactions of LpBgla at pH 7.5 and 5.5 were obviously different, so in order to verify the effects of pH on the enzyme’s catalytic efficiency, the changes of internal structure around the substrate-binding pocket of the LpBgla-β-pNPG complexes were further analyzed. As shown in Figure 6A, at pH 7.5, there was only a relative narrow channel aisle for the accessing of substrate to the catalytic center. On the contrary, after the pH was 5.5, the substrate-binding pocket was enlarged in some degree, resulting a more open space and larger channel aisle for the substrate reaching the enzyme active center (Figure 6B). These structure changes on substrate-binding pocket will not only expose the available space inside the active center, but also decrease the steric hindrance during the substrate accessing. The results fit the concept that to improve the enzymes’ activity by modifying the surrounding amino acids to enlarge their substrate-binding pocket in lots of literature (Li et al., 2022; Jia et al., 2023; Li et al., 2023). It was suggested that the substrate-binding pocket and channel aisle of LpBgla might have enlarged at pH 5.5 compared to pH 7.5.
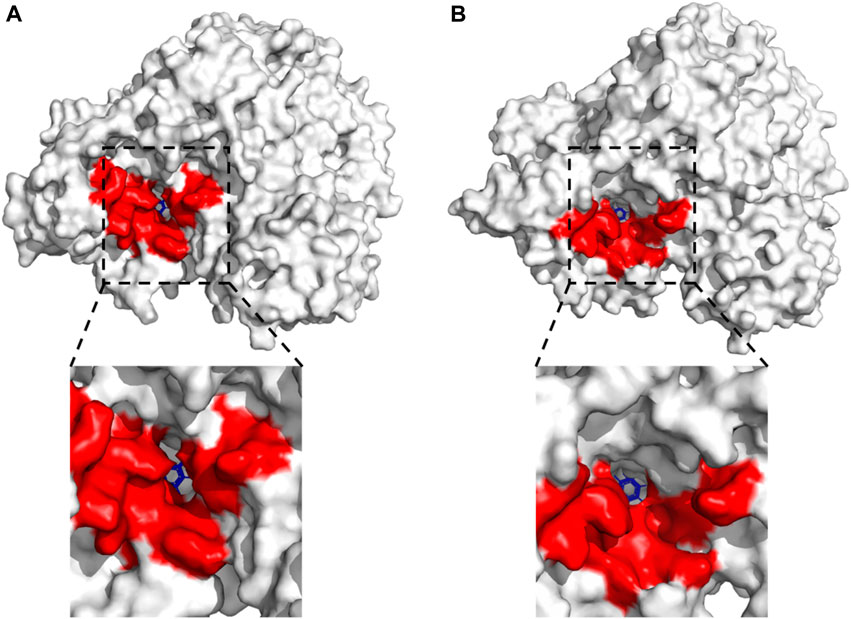
FIGURE 6. Effects of different pH on the substrate-binding pocket of LpBgla by DM simulations. Surface representation for the whole LpBgla structure at the last frame of simulations at (A) pH 7.5, and (B) pH 5.5. The below image in each panel is enlarged view of the substrate-binding pocket of LpBgla with β-pNPG.
Binding free energy analysis and residue contributions of LpBgla to β-pNPG at different pH by MD simulations
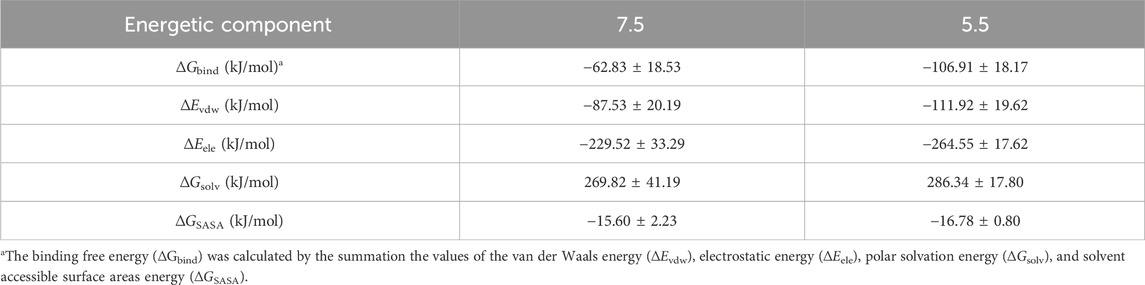
Based on the MD simulations, the binding free energy of LpBgla to the β-pNPG at pH 7.5 and pH 5.5 were calculated, and the binding free energy and contributions of its ingredients for enzyme-substrate complex were listed in Table 1. As the calculation results, electrostatic interaction and van der Waals energy in the complex were favorable for the binding (Table 1). The binding free energy between the LpBgla and β-pNPG at pH 7.5 was −62.83 ± 18.53 kJ/mol, while the binding free energy was −106.91 ± 18.17 at pH 5.5, indicating that LpBgla had a stronger binding affinity to β-pNPG at pH 5.5 than at pH 5.5 (Table 1).
Moreover, we also probed the contributions of each residue for binding free energy in LpBgla to β-pNPG at different pH. 15 residues with the highest absolute binding free energy in LpBgla were screened (Figure 7). In which, at pH 5.5, residues of Asp45, Leu60, Arg120, Lys153 and Arg164 showed significantly lower binding energy than that pH 7.5 (Figure 7), contributing the improvement on binding affinity of LpBgla to the substrate. Therefore, the binding free energy and decomposition energy calculation data were consistent to the structural and interaction results, indicating that LpBgla showed a stronger binding affinity to β-pNPG at pH 5.5 than pH 7.5, and the five residues (Asp45, Leu60, Arg120, Lys153 and Arg164) might play a critical role in the acid-adapted mechanism of LpBgla.
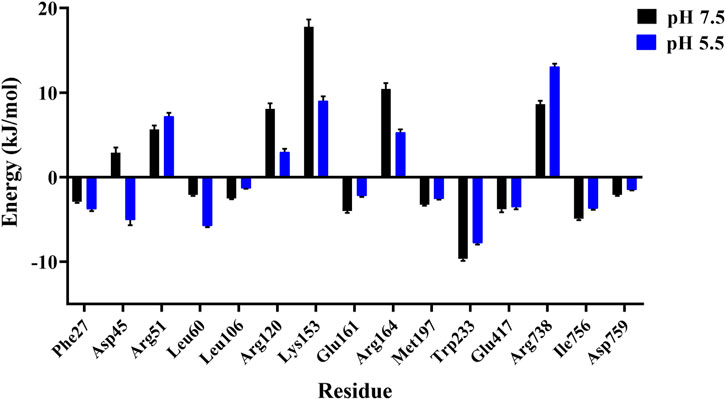
FIGURE 7. Binding free energy contribution of each residue of the LpBgla in the complex with β-pNPG at pH 7.5 and pH 5.5.
Potential application of LpBgla in glycosides bioconversion and specificity analysis
It has been reported that β-glucosidase can be used in the bioconversion of glycosides to produce aglycones (Bhatia et al., 2002; Yuksekdag et al., 2018; Xie et al., 2022). And the aglycones possess higher bioactivity than glycosides because of the reducing glycosyl groups, lower molecular weight and more easily absorbed by the gut (Harada et al., 2005; Xiao, 2017). Therefore, we explored the potential bioconversion effects of LpBgla on various glycosides, including geniposide, salicin, polydatin, esculin and arbutin. After catalysis by LpBgla, all the five glycosides conversed to lower molecular weight compounds from the HPLC results (Figure 8), indicating that LpBgla could be applied in the bioconversion of glycosides, and with a broad substrate specificity. The β-glucosidases have been grouped into three classes based on the catalytic substrate: (1) aryl-β-glucosidases, (2) true cellobiases, and (3) active on broad substrate specificity enzymes. It is was reported that most of the characterized β-glucosidases acted on a wide spectrum of substrates and belonged to the third category broad substrate specificity enzymes (Bhatia et al., 2002). For example, both of the β-glucosidases from Sulfolobus solfataricus and Pyrococcus furiosus showed a broad specific activity towards isoflavone glycosides, including daidzin, glycitin, genistin, malonyl genistin, malonyl daidzin, malonyl glycitin (Kim et al., 2012; Yeom et al., 2012). Except towards the aryl-glycosides, extracellular β-glucosidase from Paecilomyces thermophila J18 exhibited a broad substrate specificity and significantly hydrolyzed towards oligosaccharides and some β-glucans (Yang et al., 2008). Li et al., also reported a thermostable GH3 β-Glucosidase, Bgl3B from Talaromyce leycettanus with substrate specificity towards genistin and daidzin, but also with a broad substrate specificity towards oligosaccharides and polysaccharides (Li X. et al., 2018). Evern for the β-glucosidase from human, after heterogeneously expressed in Pichia pastoris, the purified enzyme showed broad substrate specificity with respect to the aglycone moiety of various aryl-glucosides, similar to the native enzyme (Berrin et al., 2002). Currently, lots of studies are underway to probe the molecular basis and mechanism of β-glucosidases’ wide substrate specificity. Identification of amino acids that occurring at the active site and substrate binding pocket are of considerable importance for revealing the enzyme’s structure-function relationships, which is also beneficial to improve the enzyme characteristics by designing and constructing mutants (Bhatia et al., 2002).
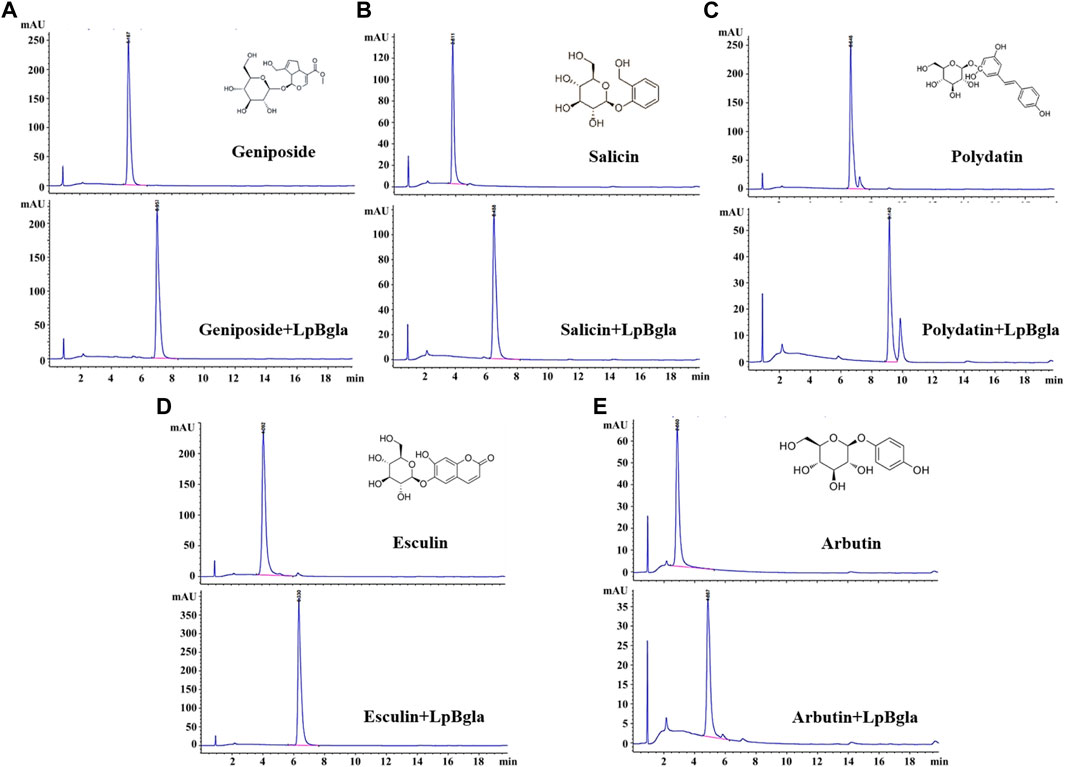
FIGURE 8. Bioconversion of various glycosides using LpBgla. (A) Geniposide. (B) Salicin. (C) Polydatin. (D) Esculin. (E) Arbutin.
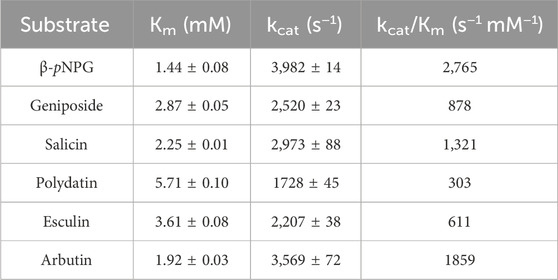
To further analysis the substrate specificity of LpBgla on different glycosides, the enzyme kinetics of LpBgla on geniposide, salicin, polydatin, esculin and arbutin were analyzed. In a general enzymatic reaction case, the index of kcat often referred to as the catalytic rate of an enzyme, a higher kcat value implies a higher catalytic rate. Km is used to reflect the affinity between the enzyme and substrate, in which, a lower Km value indicates a higher affinity of enzyme to the substrate. The kcat/Km can be used to reflect one enzyme’s catalytic efficiency, or used as a specificity constant to compare the catalytic efficiency of one enzyme to different substrates (Eisenthal et al., 2007). The corresponding kinetic parameters were listed in Table 2, kcat values of LpBgla on geniposide, salicin, polydatin, esculin and arbutin were 2,520, 2,973, 1728, 2,207 and 3,569 s−1, respectively, indicating that the LpBgla showed the highest enzymatic rate on arbutin among these five substrates, while lower than β-pNPG. The Km values of LpBgla on geniposide, salicin, polydatin, esculin and arbutin were 2.87, 2.25, 5.71, 3.61 and 1.92 mM, respectively, suggesting the affinity between LpBgla and arbutin was the strongest among these five substrates. The kcat/Km values of LpBgla on the above five substrates were 878, 1,321, 303, 611 and 1859 s−1 mM−1, respectively (Table 2). In line with the substrate affinity results, the catalytic efficiencies of LpBgla towards arbutin was the highest among these five substrates, which was about 67.2% compared to the β-pNPG (Table 2).
Conclusion
In this study, the LpBgla was cloned and characterized from Lactobacillus paracasei TK1501. The recombinant LpBgla was an acid-adapted enzyme that exhibited maximal activity at temperature of 30°C and pH 5.5, and the enzymatic activity was inhibited by Cu2+, Mn2+, Zn2+, Fe2+, Fe3+ and EDTA. The Km value of LpBgla was 1.44 mM, and the catalytic efficiency given by the kcat/Km ratio was 2.76×103 s−1 mM−1. MD simulations indicated that LpBgla showed more stable structure, and wider substrate-binding pocket and channel aisle at pH 5.5 than pH 7.5. The molecular interaction between LpBgla and β-pNPG at pH 5.5 was stronger than pH 7.5 due to the additional hydrogen bonds and lower binding free energy. Five residues including Asp45, Leu60, Arg120, Lys153 and Arg164 might play a critical role in the acid-adapted mechanism of LpBgla. Moreover, LpBgla showed a broad substrate specificity and potential application in the bioconversion of glycosides, especially towards the arbutin.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YX: Conceptualization, Funding acquisition, Investigation, Methodology, Writing–original draft. XY: Formal Analysis, Investigation, Methodology, Writing–original draft. CL: Formal Analysis, Investigation, Methodology, Writing–original draft. SW: Funding acquisition, Project administration, Supervision, Writing–original draft. LJ: Project administration, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of Heilongjiang Province in China (No. LH2022C055), Higher Education Teaching Reform Research Project in Heilongjiang Province (No. SJGY20210525), and National College Students Innovation and Entrepreneurship Training Program (No. 202310234002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1334695/full#supplementary-material
References
Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. V., Dinola, A., and Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. doi:10.1063/1.448118
Berrin, J. G., Mclauchlan, W. R., Needs, P., Williamson, G., Puigserver, A., Kroon, P. A., et al. (2002). Functional expression of human liver cytosolic β-glucosidase in Pichia pastoris: insights into its role in the metabolism of dietary glucosides. Eur. J. Biochem. 269, 249–258. doi:10.1046/j.0014-2956.2001.02641.x
Bhatia, Y., Mishra, S., and Bisaria, V. S. (2002). Microbial β-glucosidases: cloning, properties, and applications. Crit. Rev. Biotechnol. 22, 375–407. doi:10.1080/07388550290789568
Collins, C. M., Murray, P. G., Denman, S., Morrissey, J. P., Byrnes, L., Teeri, T. T., et al. (2007). Molecular cloning and expression analysis of two distinct β-glucosidase genes, bg1 and aven1, with very different biological roles from the thermophilic, saprophytic fungus Talaromyces emersonii. Mycol. Res. 111, 840–849. doi:10.1016/j.mycres.2007.05.007
Court, M. H., Duan, S. X., Hesse, L. M., Venkatakrishnan, K., and Greenblatt, D. J. (2001). Cytochrome P-450 2B6 is responsible for interindividual variability of propofol hydroxylation by human liver microsomes. Anesthesiology 94, 110–119. doi:10.1097/00000542-200101000-00021
Cuskin, F., Lowe, E. C., Temple, M. J., Zhu, Y., Cameron, E., Pudlo, N. A., et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169. doi:10.1038/nature13995
Darden, T., York, D., and Pedersen, L. (1993). Particle Mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. doi:10.1063/1.464397
Eisenthal, R., Danson, M. J., and Hough, D. W. (2007). Catalytic efficiency and kcat/KM: a useful comparator? Trends Biotechnol. 25, 247–249. doi:10.1016/j.tibtech.2007.03.010
Fan, H. X., Miao, L. L., Liu, Y., Liu, H. C., and Liu, Z. P. (2011). Gene cloning and characterization of a cold-adapted β-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microb. Technol. 49, 94–99. doi:10.1016/j.enzmictec.2011.03.001
Godse, R., Bawane, H., Tripathi, J., and Kulkarni, R. (2021). Unconventional β-glucosidases: a promising biocatalyst for industrial Biotechnology. Appl. Biochem. Biotechnol. 193, 2993–3016. doi:10.1007/s12010-021-03568-y
Harada, T., Masuda, S., Arii, M., Adachi, Y., Nakajima, M., Yadomae, T., et al. (2005). Soy isoflavone aglycone modulates A hematopoietic response in combination with soluble.BETA.-Glucan: scg. Biol. Pharm. Bull. 28, 2342–2345. doi:10.1248/bpb.28.2342
Hess, B. (2008). P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122. doi:10.1021/ct700200b
Jeng, W. Y., Wang, N. C., Lin, M. H., Lin, C. T., Liaw, Y. C., Chang, W. J., et al. (2011). Structural and functional analysis of three β-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis. J. Struct. Biol. 173, 46–56. doi:10.1016/j.jsb.2010.07.008
Jia, L., Wang, C., Zhang, S., Yuan, Z., Lu, F., and Liu, Y. (2023). Structure-guided protein engineering to reconstruct the biocatalyst properties for efficient L-3,4-dihydroxyphenylalanine production. Chem. Eng. J. 11, 143894–211481. doi:10.1016/j.cej.2023.143894
Jia, L., Wang, W., Shang, J., Zhao, W., Wei, W., Wang, Y., et al. (2018). Highly efficient soluble expression, purification and characterization of recombinant Aβ42 fromEscherichia coli. RSC Adv. 8, 18434–18441. doi:10.1039/c8ra00042e
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., and Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. doi:10.1063/1.445869
Kaenying, W., Tagami, T., Suwan, E., Pitsanuwong, C., Chomngam, S., Okuyama, M., et al. (2023). Structural and mutational analysis of glycoside hydrolase family 1 Br2 β-glucosidase derived from bovine rumen metagenome. Heliyon 9, e21923. doi:10.1016/j.heliyon.2023.e21923
Kim, B. N., Yeom, S. J., Kim, Y. S., and Oh, D. K. (2012). Characterization of a β-glucosidase from Sulfolobus solfataricus for isoflavone glycosides. Biotechnol. Lett. 34, 125–129. doi:10.1007/s10529-011-0739-9
Li, C., Gao, X., Qi, H., Zhang, W., Li, L., Wei, C., et al. (2023). Substantial improvement of an epimerase for the synthesis of D-allulose by biosensor-based high-throughput microdroplet screening. Angew. Chem. Int. Ed. Engl.: 62, e202216721. doi:10.1002/anie.202216721
Li, T., Chu, X., Yuan, Z., Yao, Z., Li, J., Lu, F., et al. (2022). Biochemical and structural properties of a high-temperature-active laccase from Bacillus pumilus and its application in the decolorization of food dyes. Foods 11, 1387. doi:10.3390/foods11101387
Li, X., Xia, W., Bai, Y., Ma, R., Yang, H., Luo, H., et al. (2018a). A novel thermostable GH3 β-glucosidase from Talaromyce leycettanus with broad substrate specificity and significant soybean isoflavone glycosides-hydrolyzing capability. Biomed. Res. Int. 2018, 4794690. doi:10.1155/2018/4794690
Li, Y., Hu, X., Sang, J., Zhang, Y., Zhang, H., Lu, F., et al. (2018b). An acid-stable β-glucosidase from Aspergillus aculeatus: gene expression, biochemical characterization and molecular dynamics simulation. Int. J. Biol. Macromol. 119, 462–469. doi:10.1016/j.ijbiomac.2018.07.165
Liu, D., Zhang, R., Yang, X., Zhang, Z., Song, S., Miao, Y., et al. (2012). Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microb. Cell Fact. 11, 25. doi:10.1186/1475-2859-11-25
Mahoney, M. W., and Jorgensen, W. L. (2000). A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 112, 8910–8922. doi:10.1063/1.481505
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., and Simmerling, C. (2015). ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. doi:10.1021/acs.jctc.5b00255
Martens, E. C., Lowe, E. C., Chiang, H., Pudlo, N. A., Wu, M., Mcnulty, N. P., et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221. doi:10.1371/journal.pbio.1001221
Molina, G., Contesini, F. J., De Melo, R. R., Sato, H. H., and Pastore, G. M. (2016). in New and future developments in microbial Biotechnology and bioengineering. Editor V. K. Gupta (Amsterdam: Elsevier), 155–169.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi:10.1002/jcc.21256
Ndeh, D., Rogowski, A., Cartmell, A., Luis, A. S., Basle, A., Gray, J., et al. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70. doi:10.1038/nature21725
Porter, C. M., and Miller, B. G. (2012). Cooperativity in monomeric enzymes with single ligand-binding sites. Bioorg. Chem. 43, 44–50. doi:10.1016/j.bioorg.2011.11.001
Pozzo, T., Pasten, J. L., Karlsson, E. N., and Logan, D. T. (2010). Structural and functional analyses of β-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J. Mol. Biol. 397, 724–739. doi:10.1016/j.jmb.2010.01.072
Shang, J., Yue, S., Zeng, F., Chen, Y., and Jia, L. (2022). Heterologous expression, characterization and evolution prediction of a diaphorase from Geobacillus sp. Y4.1MC1. Y4.1MC1. Biotechnol. Lett. 44, 101–112. doi:10.1007/s10529-021-03215-w
Siddiqui, K. S., Ertan, H., Charlton, T., Poljak, A., Daud Khaled, A. K., Yang, X., et al. (2014). Versatile peroxidase degradation of humic substances: use of isothermal titration calorimetry to assess kinetics, and applications to industrial wastes. J. Biotechnol. 178, 1–11. doi:10.1016/j.jbiotec.2014.03.002
Singhania, R. R., Patel, A. K., Pandey, A., and Ganansounou, E. (2017). Genetic modification: a tool for enhancing beta-glucosidase production for biofuel application. Bioresour. Technol. 245, 1352–1361. doi:10.1016/j.biortech.2017.05.126
Stewart, J. J. (1990). MOPAC: a semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 4, 1–103. doi:10.1007/BF00128336
Tan, L. L., Ngiam, J. J., Sim, E. S. Z., Conway, P. L., and Loo, S. C. J. (2022). Liquorilactobacillus satsumensis from water kefir yields α-glucan polysaccharides with prebiotic and synbiotic qualities. Carbohydr. Polym. 290, 119515. doi:10.1016/j.carbpol.2022.119515
Wang, F., Ge, X., Yuan, Z., Zhang, X., Chu, X., Lu, F., et al. (2022). Insights into the mechanism for the high-alkaline activity of a novel GH43 β-xylosidase from Bacillus clausii with a promising application to produce xylose. Bioorg. Chem. 126, 105887. doi:10.1016/j.bioorg.2022.105887
Whittington, A. C., Larion, M., Bowler, J. M., Ramsey, K. M., Bruschweiler, R., and Miller, B. G. (2015). Dual allosteric activation mechanisms in monomeric human glucokinase. Proc. Natl. Acad. Sci. U. S. A. 112, 11553. doi:10.1073/pnas.1506664112
Xiao, J. (2017). Dietary flavonoid aglycones and their glycosides: which show better biological significance? Crit. Rev. Food Sci. Nutr. 57, 1874–1905. doi:10.1080/10408398.2015.1032400
Xiao, J., Muzashvili, T. S., and Georgiev, M. I. (2014). Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol. Adv. 32, 1145–1156. doi:10.1016/j.biotechadv.2014.04.006
Xie, Y., Wang, Y., Han, Y., Zhang, J., Wang, S., Lu, S., et al. (2022). Complete genome sequence of a novel Lactobacillus paracasei TK1501 and its application in the biosynthesis of isoflavone aglycones. Foods 11, 2807. doi:10.3390/foods11182807
Yan, Q., Hua, C., Yang, S., Li, Y., and Jiang, Z. (2012). High level expression of extracellular secretion of a β-glucosidase gene (PtBglu3) from Paecilomyces thermophila in Pichia pastoris. Protein Expr. Purif. 84, 64–72. doi:10.1016/j.pep.2012.04.016
Yang, S., Jiang, Z., Yan, Q., and Zhu, H. (2008). Characterization of a thermostable extracellular β-glucosidase with activities of exoglucanase and transglycosylation from Paecilomyces thermophila. J. Agric. Food Chem. 56, 602–608. doi:10.1021/jf072279+
Yeom, S. J., Kim, B. N., Kim, Y. S., and Oh, D. K. (2012). Hydrolysis of isoflavone glycosides by a thermostable β-glucosidase from Pyrococcus furiosus. J. Agric. Food Chem. 60, 1535–1541. doi:10.1021/jf204432g
Keywords: β-glucosidase, acid-adapted, enzymatic property, molecular dynamics simulations, bioconversion
Citation: Xie Y, Yan X, Li C, Wang S and Jia L (2024) Characterization and insight mechanism of an acid-adapted β-Glucosidase from Lactobacillus paracasei and its application in bioconversion of glycosides. Front. Bioeng. Biotechnol. 12:1334695. doi: 10.3389/fbioe.2024.1334695
Received: 07 November 2023; Accepted: 15 January 2024;
Published: 24 January 2024.
Edited by:
Jeong Chan Joo, Catholic University of Korea, Republic of KoreaReviewed by:
Ismael Bustos-Jaimes, National Autonomous University of Mexico, MexicoRufeng Wang, Shanghai University of Traditional Chinese Medicine, China
Copyright © 2024 Xie, Yan, Li, Wang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longgang Jia, bG9uZ2dhbmdqaWFAdHVzdC5lZHUuY24=; Shumei Wang, d2FuZ3NodW1laTI0QDEyNi5jb20=
 Yufeng Xie1,2
Yufeng Xie1,2 Shumei Wang
Shumei Wang Longgang Jia
Longgang Jia