
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 06 December 2023
Sec. Industrial Biotechnology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1326395
 Chengda Cui1†
Chengda Cui1† Mengke Jiang1†
Mengke Jiang1† Chengxiao Zhang1
Chengxiao Zhang1 Naxue Zhang1
Naxue Zhang1 Feng-Jie Jin1
Feng-Jie Jin1 Taihua Li1
Taihua Li1 Hyung-Gwan Lee2*
Hyung-Gwan Lee2* Long Jin1*
Long Jin1*Numerous microorganisms, including bacteria and fungus, have been identified as capable of degrading rubber. Rubber biodegradation is still understudied due to its high stability and the lack of well-defined pathways and efficient enzymes involved in microorganism metabolism. However, rubber products manufacture and usage cause substantial environmental issues, and present physical-chemical methods involve dangerous chemical solvents, massive energy, and trash with health hazards. Eco-friendly solutions are required in this context, and biotechnological rubber treatment offers considerable promise. The structural and functional enzymes involved in poly (cis-1,4-isoprene) rubber and their cleavage mechanisms have been extensively studied. Similarly, novel bacterial strains capable of degrading polymers have been investigated. In contrast, relatively few studies have been conducted to establish natural rubber (NR) degrading bacterial consortia based on metagenomics, considering process optimization, cost effective approaches and larger scale experiments seeking practical and realistic applications. In light of the obstacles encountered during the constructing NR-degrading consortia, this study proposes the utilization of multi-omics tools to discern the underlying mechanisms and metabolites of rubber degradation, as well as associated enzymes and effective synthesized microbial consortia. In addition, the utilization of omics tool-based methods is suggested as a primary research direction for the development of synthesized microbial consortia in the future.
Polyisoprene is classified as one of the eight principal categories of biopolymers that are synthesized by living microbes. NR, one of the representatives of the polyisoprene group, is widely employed among biopolymers. By coagulating and curing the latex of rubber plants, NR is transformed into an elastomer (Steinbüchel, 2001; Linos and Steinbüchel, 2005; Lynen et al., 1958). Presently, over 2,500 plant species are known to produce NR. On the contrary, the rubber tree Hevea brasiliensis, which is exclusive to subtropical regions, is the most significant commercial source of this polymer (Nair, 2010), supplying 99% of the global market. Melaleuca (Parthenium argentatum) and Russian dandelion (Taraxacum kok-saghyz) are two additional sources. Asia currently accounts for 92% of NR production, with Thailand leading the world, followed by Indonesia, Malaysia, India, Vietnam, and China.
NR is a 2-methyl-1, 3-butadiene polymer. It is a conjugated diene with double bonds in alternating locations, often known as polyisoprene. Cis-1,4-polyisoprene accounts for 90% of the dry weight of NR. Source-dependent variations exist in the molecular weight and composition of this polymer (Mooibroek and Cornish, 2000). Cis-1,4-polyisoprene rubber is a crucial biological material in contemporary society. In the context of commercial applications, this polymer typically experiences vulcanization, which modifies its molecular structure via the crosslinking of isoprene chains (Figure 1). Tires are vulcanized through the application of heat in the presence of sulfur, while latex mittens are vulcanized via peroxidation and irradiation. Rubber commonly used to construct wire coverings, mittens, and footwear is produced via this method. Moreover, in order to impart rigidity, elasticity, and longevity to unprocessed latex, a multitude of chemical constituents are incorporated into it. As a result, it finds application in industries such as manufacturing, transportation, medicine, and domestic products.
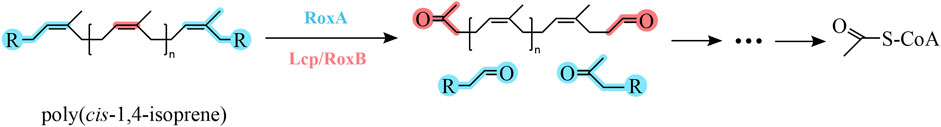
FIGURE 1. Poly (cis-1,4-isoprene) metabolic pathway of microbial degradation. Rubber oxigenases (Lcp/RoxA/RoxB) are responsible for the first cleavage step of poly (cis-1,4-isoprene) chains. The chemical structure was illustrated with ChemDraw 22.
The physical and chemical properties of NR are highly stable and difficult to undergo natural degradation, thus leading to the accumulation of large amounts of NR waste and pending disposal. Consequently, the environmental contamination of byproducts resulting from the extensive utilization of NR has emerged as a significant global concern. At this stage, incineration and landfill methods are mainly used to dispose of NR waste, and this traditional treatment method has undoubtedly caused serious pollution to the environment (Mooibroek and Cornish, 2000). It presents a significant environmental contamination problem, poses a critical risk to human health by triggering allergies and asthma, and the incineration process generates substantial quantities of atmospherically hazardous toxic gases (e.g., sulfur oxides, carbon monoxides, and cyanides) (Depaolini et al., 2017; Kasai, 2020; Chittella et al., 2021). As environmental contamination has increased, the utilization of microorganisms to degrade contaminants has emerged as a major concern. In recent years, researchers have explored the ability of some microorganisms to degrade NR in laboratory studies and practical applications and to break down rubber waste into easily degradable and environmentally benign substances.
This review focused on constructing NR degrading functional microbial consortia. We briefly reviewed its properties and biodegradation pathways, as well as present research gaps and recent achievements in NR biodegradation. Then, in comparison to single strains, we examined the benefits of employing microbial functional consortia and their potential for NR degradation. Finally, we looked at the possibility of applying multi-omics tool to develop functional microbial consortia.
The earliest report on the degradation of rubber by microbes was published in 1914, when Söhngen and Fol utilized highly pure NR as the only carbon source to demonstrate the absorption of the hydrocarbon chain by microorganisms (Söhngen and Fol, 1914). Two Actinomycetes were discovered to form colonies on the purified rubber material. Spence and Van Niel (1936) first isolated bacteria capable of degrading rubber by employing latex overlay plates. The formation of translucent halos within the opaque agar layer indicated the beginning of colony formation. The clear zone isolation technique enables the detection of bacteria that secrete extracellular enzymes during the degradation of cis-1,4-polyisoprene. This method selects NR utilizing strains based on the formation of translucent halos (hyaline zones) around colonies on opaque NR agar plates, which can be directly observed by the naked eye after the degradation action of the strain. Members of these groups usually belong to the class of Actinomycetes that can form mycelium, such as Actinobacteria, Streptomyces, and Micromonospora. In contrast, the second group of microorganisms that degrade NR do not produce any clear zones, and only the normal growth of strains can be observed on NR agar plates without forming any clear circles around the colonies. Therefore, these strains need to be in direct contact with the rubber matrix to function. These bacteria attack the rubber matrix directly, form biofilms and bind to polymers, which begin to degrade at the cell surface. Examples of this group are Nocardia, Mycobacterium, and Gordonia (Gallert, 2000).
At present, researchers have found that some microorganisms with special origins have some biodegradation effect on NR, and the microbial degradation of NR has been studied by extracting strains from specific environments (Tables 1, 2). Actinomycete isolated from sludge at wastewater treatment plants could grow and multiply in liquid medium using NR as a carbon source, and it was confirmed that the isolated strains could reduce the molecular weight of NR thus achieving microbial degradation of NR; Gordonia and Actinomycete (Hiessl et al., 2012; Ali Shah et al., 2013) isolated from soil at rubber plantations were found to be effective in microbial degradation of NR; the use of insect laccase and manganese peroxidase secreted by Penicillium can also be used for the biodegradation of NR (Nayanashree and Thippeswamy, 2015b). It has been reported that Rhodococcus, the predominant species accountable for the degradation of NR, is capable of degrading organic pollutants and is therefore considered effective (Dey et al., 2020). Streptomyces have been studied by researchers to degrade non-vulcanized fresh latex and common vulcanized rubber products such as NR gloves, NR condoms and rubber car tires (Nanthini and Sudesh, 2017). Variovorax degrades ethylene glycol phthalate (Prasad, 2017) that can be used as a plasticizer for plastic and rubber products and is used as a component of NR products with improved strength. It is presumed that strains of this genus can also degrade NR. Pseudomonas has been shown to make use of NR hydrocarbons and subsequently can degrade NR (Roy et al., 2006).
The two major NR degrading enzymes rubber oxygenase (RoxA/RoxB) and latex clearing protein (Lcp) have been identified in Gram-negative and Gram-positive bacteria, respectively (Table 3). Among Gram-positive NR degrading bacteria, Lcp is responsible for the initial oxidative cleavage of cis-1,4-polyisoprene in the strains, first demonstrated in Streptomyces sp. K30. The Lcp of this strain was experimentally shown to be a type B cytochrome that cleaves cis-1,4-polyisoprene to a low molecular weight product containing isoprene with aldehyde and ketone termini. The resulting compound is further degraded to the corresponding acid by oxidoreductase OxiAB, which is encoded by the OxiAB gene and subsequently enters the β-oxidation pathway via the action of coenzyme A for eventual oxidative catabolism. Yikmis et al. found that transcription of Lcp increases in the presence of latex or synthetic cis-1,4-polyisoprene and that Lcp transported via the bisarginine transport (Tat) system is secreted (Jendrossek and Reinhardt, 2003; Yikmis et al., 2008; Imai et al., 2011; Imai et al., 2013; Sharma et al., 2018), thus confirming the role of Lcp in the degradation of NR. In Gram-negative NR degrading bacteria, including some mycobacteria, the rubber oxygenase RoxA is responsible for the degradation of cis-1,4-polyisoprene. RoxA, encoded by the roxA gene, is an exo-type and a highly representative rubber oxygenase that occupies the same important position as the Lcp. It was first isolated and confirmed in strain Xanthomonas sp. 35Y. RoxA was shown to be a key enzyme involved in the degradation of cis-1,4-polyisoprene in strain Xanthomonas sp. 35Y. RoxA first binds oxygen molecules and then cleaves the carbon-carbon double bond of cis-1,4-polyisoprene to produce isoprene with aldehyde and ketone groups at the end of the molecule (Braaz et al., 2004; Hiessl et al., 2014; Luo et al., 2014). RoxA is an extracellular dioxygenase that oxidize cis-1,4-polyisoprene to 12-oxo-4,8-dimethyltridecane-4,8-diene-1-aldehyde (ODTD) thereby degrading NR. Conversely, the rubber oxygenase RoxB is also involved in the degradation process, which degrades NR by endo-cleavage of cis-1,4-polyisoprene. It is known from the experimental results of Daisuke Kasai et al. that the LatA protein is a key enzyme for the degradation of NR in strain NBRC 109400 demonstrated by the high-throughput transposon insertion technique (Birke et al., 2013; Seidel et al., 2013; Kasai et al., 2017). The direct relatives of functional RoxB and RoxA, namely, LatA1 and LatA2, respectively, were reported to be essential for NR degradation in Rhizobacter gummiphilus NS21 (Linos et al., 2000; Ibrahim et al., 2006; Yikmis and Steinbüchel, 2012).
The rubber oxidases identified so far include the exo-type rubber oxygenase A. Nevertheless, the exo-type of reaction is accountable for its suboptimal degradation efficiency and hinders the rapid and efficient degradation of polymers. This is presumably due to the inefficient threading of the substrate chain into the enzyme (Seidel et al., 2013). The endo-type rubber oxygenase B is capable of cleaving polyisoprene to C15 oligopolyisoprene (ODTD) (Jendrossek and Birke, 2019). RoxB can endo-cleave the carbon-carbon double bond of polyisoprene to produce oligopolyisoprenes with varying degrees of polymerization (Birke et al., 2017), and this endo-type of rubber oxidase is more suitable for polyolefin degradation systems (Zhang et al., 2022). Gibson et al. (2020) screened Nocardioides WS12, which is capable of degrading isoprene, from soil near willow trees, and they found that the strain’s genome also contained genes related to the degradation of rubber and speculated that it might also possess the ability to degrade NR.
Researchers have shown through experimental studies and practical applications that the degradation ability of a single strain of bacteria for NR is limited, and the speed and efficiency are very restricted, while multiple microorganisms interacting with each other for a long period of time will give better results, so they have focused on bacterial consortia. The construction of complex consortia is no longer limited to the cultivation of a single microorganism (Nguyen et al., 2020; Castañeda Alejo et al., 2022; Joseph et al., 2022). Instead, a variety of microorganisms are co-cultured in a specific environment. Through the interactions between the strains, they directly or indirectly influence the micro-ecosystem maintaining a balance. The interrelationship is either mutually beneficial or competitive, and in general, multiple microorganisms work together to accelerate and enhance the rate of rubber degradation and stabilize their maximum metabolic functions.
Studies have shown that the degradation of pollutants by consortia is superior to that of purely cultured microorganisms (Table 4). The microorganisms in the functional consortia perform different metabolic functions and accomplish the degradation of pollutants while coping with the harsh living environment through collaboration among themselves (Araújo et al., 2020; Chen et al., 2021). There are two methods to construct functional consortia: one is the natural selection method, in which the soil and water contaminated by NR are mixed, and the suspension is extracted after static sedimentation, and NR samples are added to the suspension and incubated with iterative shaking until the weight of the NR samples no longer changes, and a naturally selected functional consortium is obtained in each generation of the culture (Nguyen et al., 2020). The other method is an artificial combination method, that is, the soil or water contaminated by NR is first screened for specificity, and the functional bacteria that can use NR as the only carbon source are screened for pure culture. A variety of bacteria with the ability to degrade NR are currently screened from the phylum Actinobacteria (Basik et al., 2021b). Later, the screened NR degrading bacteria were used for consortium construction, and the more efficient consortium for degrading NR was selected.
NR degrading bacteria screened so far have been broadly classified into two groups: the first group can produce hyaline rings on a solid medium with NR as the only carbon source; the second group can form biofilms and thus grow directly attached to the surface of NR (Hapuarachchi et al., 2016). It is important to note when constructing an artificial functional consortia: only a small fraction of microorganisms in the natural environment can grow on artificial media, so the conventional plating method alone cannot completely isolate the functional microorganisms found in an environment. These difficult to isolate microorganisms may exercise a synergistic role in the degradation of pollutants, and their absence reduces the efficiency of the functional consortia in the degradation of pollutants (Lee et al., 2013). Meanwhile, Nguyen et al. (2020) found that high microbial diversity of the consortia inhibits the degradation of NR by microorganisms. Therefore, to ensure the efficiency and to maintain the stability and harmony of functional consortia, microorganisms that are abundant in the initial environment should be incorporated alongside known functional microorganisms when constructing such consortia.
Currently, there are studies on consortia analysis usually combined with metagenomics sequencing. In 1998, Handelsman et al. (1998) introduced the concept of metagenomics in this scenario. Metagenomics refers to the advancement of individual single studies of microorganisms to study the sum of all microbes. Metagenomics does not require the isolation and pure culture of microorganisms. It no longer focuses only on the role of a single strain, overcoming the limitations of isolation and culture methods. Metagenomics enables the exploration of non-culturable microorganisms, which were previously limited by technical methods, greatly expanding the scope of microbial research (Riesenfeld et al., 2004). In addition, the significant advantages of metagenomics are the ability to identify microbial communities with low abundance, to make more genetic resources available, to emphasize the exploitation and use of non-culturable microorganisms, and to uncover microbial activities and functions at the level of the complete community.
In recent years, the investigation of rubber biodegradation mechanisms and microbial ecology has been facilitated by the increased use of genomics, metagenomics, transcriptomics, proteomics, and multiomics, made possible by the rapid development of high-throughput sequencing (Illumina, PacBio, and Nanopore). In a study by Nawong et al. (2018), single bacterial strains and mixed culture of selected bacteria were subjected to degradation experiments on NR gloves in a mineral salt medium (MSM). Following a 30-day incubation period under identical conditions, the bacteria exclusively utilized the NR glove as a carbon source, resulting in the polymer’s degradation. The experimental results demonstrated that the weight loss rate of NR gloves from mixed bacterial culture was much higher than that of NR degraded by individual strains, with the highest weight loss rate of 9.36% for single bacterial degradation and 18.82% for the constructed consortia. Nguyen et al. (2020) isolated and cultured a batch of bacteria from waste from a Vietnamese rubber processing plant and used NR film as the only carbon source in MSM medium. After 14 days of incubation, the highest weight loss of NR films reached 34% among the six groups of bacteria consortia constructed, which greatly improved the degradation of NR.
Furthermore, by monitoring the metabolic and dynamic relationships among the functional consortia, multi-omics analysis of synthesized functional consortia could serve as a guide for subsequent manipulations of population ratios and environmental parameters (Figure 2).
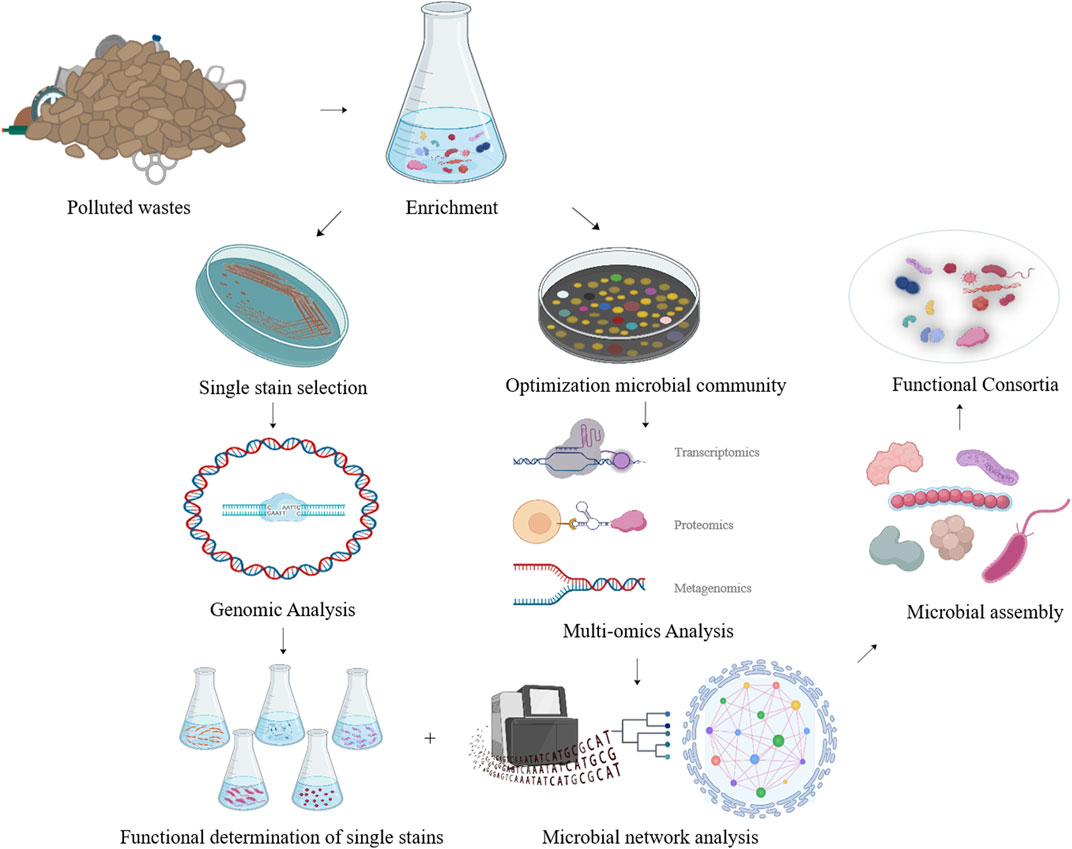
FIGURE 2. The strategy of developing efficient NR degrading microbial consortia based on multi-omics analysis of the functional microbial community achieved after enrichment.
Currently, there are several methods used to detect and analyze the ability of microorganisms to carry out the degradation of NR. The easiest and most obvious method is the weight loss measurement. To measure the weight loss of a NR sample, the entire residual rubber (including the more intact pieces and broken rubber particles) is usually separated from the rest of the culture and collected and cleaned and dried until the weight is constant (Cheng et al., 2023). The weight loss of NR is equal to the initial weight of the sample minus the residual weight. The weight of NR before degradation is recorded as w1, and the weight after washing and drying after degradation by the strain is recorded as w2: weight loss = [(w1-w2)/w1] × 100%. The advantage of the weight loss method is that it can reflect the efficiency of microbial degradation of rubber more intuitively. Theoretically, the stronger the strain’s ability to degrade NR, the higher the weight loss rate. However, the method cannot help us understand the process of microbial degradation of NR, and we cannot exclude some strains that take a long time to function. For example, some Streptomycetes take some time to grow and colonize on the surface of NR. Second, we can observe the surface structure of NR samples by scanning electron microscopy (SEM) to get an idea of the degradation ability of the microbial strains. The growth and degradation of the strain on NR and the structural changes on its surface before and after the degradation of NR by the strain can be observed using SEM. Residual NR samples are isolated from cultures and later washed, fixed and dried. Generally, at great magnification under SEM, the surface of the NR sample can be observed to show significant cracks, pits or holes compared to the initial state, which is considered to have some NR degradation (Handelsman et al., 1998). This method can visually observe the microscopic state of NR after degradation compared to the loss-in-weight method, which is clearer but does not help us to understand the mechanism of degradation. Furthermore, the color reaction can be employed to validate the degradation process of NR. The initial stage of microbial degradation of NR typically involves the oxidative cleavage of the double bond of cis-1,4-isoprene, resulting in the formation of aldehydes (Umsakul et al., 2012). The detection of aldehyde and ketone groups frequently serves as indications that NR samples have undergone some degree of microbial degradation. One of the easiest and fastest methods to detect the presence of aldehyde groups in the metabolites of a sample is the colorimetric reaction. This method mainly uses Schiff’s reagent. Schiff’s reagent does not show color when exposed to ketone groups but turns purple when exposed to aldehyde groups, which is a very sensitive reaction, and is often used to detect the formation of aldehyde groups. 2, 4-DNP produces a yellow precipitate when exposed to aldehyde and ketone groups, which can be used to detect the presence of aldehyde and ketone groups. Both reagents are often used for the preliminary detection of aldehyde groups during the degradation of NR samples. In addition, fourier transform infrared spectroscopy (FTIR), high-resolution mass spectrometry (HRMS) (i.e., LC-MS/LC-HRMS, GC-HRMS, MALDI-TOF), and nuclear magnetic resonance spectroscopy (NMR) also have a role in the degradation process of NR. The mode of degradation can be deduced from the functional group changes that occurred during the degradation of NR using FTIR spectroscopy. FTIR spectroscopy is often used to detect the creation of new functional groups and the disappearance of existing functional groups in a sample. By analyzing the changes of peaks and the intensity of peaks at specific locations in the FTIR spectra before and after the degradation of NR samples, we can know the changes of functional groups during the degradation of the samples. These data help us to verify that microorganisms have the ability to degrade NR samples and to speculate on the reactions that may occur during the degradation process. In general, the first functional groups that studies focus on are carbonyl, ester, ethylene and carbon-carbon double bonds. It has been shown that these groups usually change whenever microbial activity on the substrate surface is detected (Sudhakar et al., 2008; Nayanashree and Thippeswamy, 2015a). Additionally, studies focus on the changes of aldehyde and ketone groups. During the degradation of NR, there is a release of aldehydes and ketones. The presence of these aldehyde and ketone groups confirms the degradation of NR by the strain due to degradation. The combined observation and analysis of the above groups can collectively indicate whether the strains have a NR degradation capacity. By identifying the location and quantity of the various forms of C and H atoms within organic matter, NMR can be utilized to determine the structure of metabolites generated during the degradation. The metabolites produced during the degradation of NR can be inferred from the information from the NMR hydrogen and carbon spectra of the samples (Handelsman et al., 1998), and if the results show the presence of aldehyde and ketone groups, the degradation of NR can thus be determined.
In the majority of recent investigations, it has been observed that the biodegradation process of NR has shown a rather slow rate, despite the successful identification and isolation of a limited number of functional bacteria capable of degrading this material (Bosco et al., 2018; Joseph et al., 2022; Sarkar et al., 2022; Cheng et al., 2023). The duration of microbial culture has shown a range of 30–120 days, but assessment methods such as weight loss or SEM have only identified little deterioration. The main limitation of existing research is the absence of consistent standards and dependable techniques for accurately quantifying the efficacy of deterioration. For this reason, establishing standardized procedures for quantifying the degradation of NR is essential for future research evaluating the biodegradability of microorganisms.
In addition, many chemical additives, including vulcanization accelerators, antioxidants, retarders, and their breakdown products, are utilized in industrial rubber products, including tires. These additives are essential in commonly used rubber materials because they prolong the material’s life and durability by avoiding breakdown (Altenhoff et al., 2019; Kawakami et al., 2022). Antioxidants were shown to be the most interfering with in vitro depolymerization, and the substance’s hydrophobic nature, as well as the huge amounts of additives present, prevent considerable biodegradation rates. For this reason, it is crucial to identify specific critical additives found in common rubber in order to enhance the microbial or enzymatic degradation process, for instance by facilitating or establishing the desired pretreatment of rubber materials. Conversely, the incorporation of additive-degrading microorganisms into a functional consortia could enhance degradation capability through synergy with other bacteria in the consortia. As a result of their low enzyme production efficiency and straightforward enzyme systems, single stains are incapable of effectively combating the complex rubber degradation process. It is recommended that future research endeavors concentrate on both the discovery of novel, efficient enzymes/genes for rubber degradation and the investigation of degradation mechanisms. Furthermore, the integration of genetic engineering and enzyme engineering methodologies may facilitate the development of microorganisms that produce rubber-degrading enzymes with enhanced activities. Temperature, ultraviolet light, dissolved oxygen, humidity, pH, and a variety of biotic and abiotic factors (microbial abundance and diversity, microbial enzymatic activity, etc.) influence the microbial rubber degradation process. In order to determine the optimal conditions for rubber biodegradation, it is necessary to take into account a variety of factors, such as environmental parameters and condition of the culture. Despite our increased knowledge of enzyme activity, we know little about rubber substrate enzyme action and the microbial, molecular, and environmental variables that affect it. In order to fully understand the microbial diversity, biofilm-formation mechanisms, and biodegradation capacity of rubber waste, multi-omics studies should be conducted.
Despite the severe environmental damage caused by conventional rubber disposal methods, it is intriguing that new rubber valorization technologies that are economically and ecologically viable are being proposed and developed to meet the growing amount of rubber waste. Rubber waste is valorized through energy recovery, recycling, upcycling, or biodegradation in order to produce new, viable products or dispose of it in an environmentally friendly manner. Incineration and landfilling are currently the prevailing and efficacious approaches for managing rubber and its byproducts. For instance, a portion of the heat produced during the incineration procedure can be recovered and harnessed to generate electricity or heat. Unfortunately, traditional recycling or direct incineration has many disadvantages, namely, the tedious process of collection, sorting and cleaning, as well as limited recycling time, excessive costs, emission of harmful residues, and unsuitability for use in other products. Instead, chemical or biological methods of converting rubber waste, which are polymeric molecules, into smaller molecules and transforming these molecules into new and valuable products are emerging as an attractive and more environmentally friendly route to valorization (Stevenson et al., 2008; Andler, 2020; Bosco and Mollea, 2021). By utilizing metagenomics sequencing, one can functionally annotate related genes and assess the diversity and relative abundance of microorganisms in a given community. It was disclosed that as the microbial diversity of artificially constructed consortia decreases, the number of strains capable of degrading NR increases, thereby enhancing the community’s efficiency (Castañeda Alejo et al., 2022).
Integrating microbial consortia with multi-omics might inspire a novel rubber waste bio-upcycling technique (Figure 3). Multi-omics will illuminate rubber biodegradation metabolic pathways and microbial community interactions. The synthesis and logical design of the composition and metabolic pathways of microbial consortiums would facilitate biotransformation into a greater variety of rubber biodegradation products, thereby expanding the product line of rubber waste upcycling.
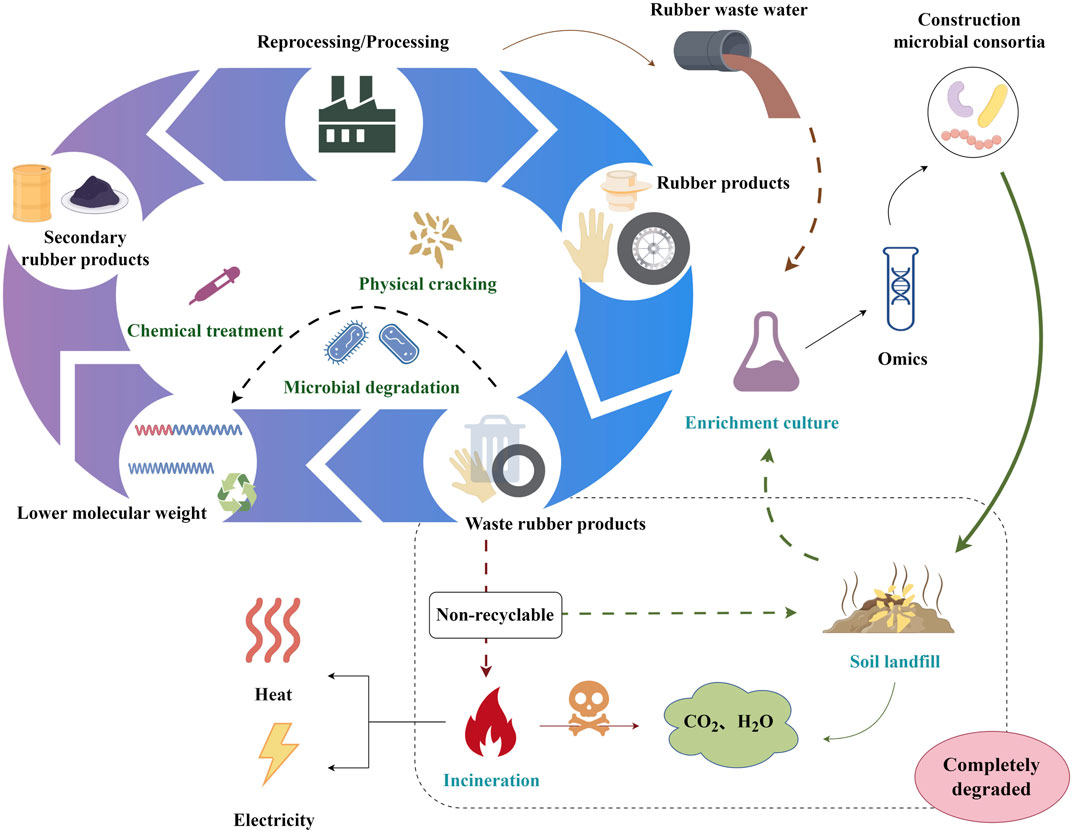
FIGURE 3. A schematic overview of the bio-upcycling of rubber waste. The figure was generated by Figdraw (www.figdraw.com).
The gradual and widespread use of NR has resulted in a growing problem of rubber waste pollution and pollutants worldwide, which is undoubtedly a huge challenge for environmental management. Traditional rubber disposal methods are not effective in solving this problem, which is becoming more serious. On the biodegradation of NR, rather, recent research has produced fresh insights. An expanding number of microorganisms implicated in the degradation process must be identified, as well as novel approaches to the sustainable dispersal of rubber wastes. NR is the most widely used rubber, and the study of its biodegradation has received a great deal of attention in the past few years. The investigation of NR degradation by microorganisms, including bacteria and fungi, has been the focus of considerable research. Despite this, progress in the study of microbial degradation of NR remains limited, particularly with regard to the identification of specific NR depolymerizing enzymes and the elucidation of NR degradation mechanisms. Recent studies have examined the limitations encountered by individual strains in their attempts to degrade NR, and progressive efforts have been made to combine several strains to develop microbial consortia. Advancements have been achieved in the developing functional consortia or synthesized co-cultures for greater efficacy. The advantages of functional microbial consortia in degrading NR and its waste surpass those of single strains. Nevertheless, the development of artificial functional consortia, which is the subject of this review, is impeded by the lack of comprehension regarding the by-products and mechanisms of NR degradation.
Currently, researchers have isolated and screened NR degrading strains from various sources and used the weight loss method to detect weight change, Schiff’s reagent to detect whether aldehyde groups are produced during the degradation of NR, SEM to observe the surface structure of NR before and after degradation, and Fourier infrared spectroscopy and nuclear magnetic resonance instruments to investigate the functional groups and motifs of NR. None of the relevant testing methods confirmed the degradation of NR by microorganisms. However, the degradation of NR by a single microorganism takes more time and has an inferior degradation rate and capacity compared to that of a complex consortia. With the advancement and development of metagenomics sequencing, we can consider combining the two. Metagenomics sequencing helps to construct the microbial composition in the consortia and discover and identify the microbial consortia with lower abundance to make more genetic resources available. It emphasizes the development and utilization of non-culturable microorganisms and discovers the functions of microorganisms from the complete community level. Therefore, it is more beneficial to explore the role of microorganisms in the degradation of NR based on metagenomics sequencing results to construct an efficient NR degradation microbial consortium.
The integration of omics/meta-omics techniques, including genomic, transcriptomic, metagenomic, and others, can facilitate the disclosure of microbial diversity. The identification of the microbial communities and critical pathways/genes involved in the degradation of NR is made possible through omics analyses, which also serve as a foundation for the development of functional consortiums. In conclusion, we deliberated on the current challenges and obstacles encountered in the field of NR biodegradation research, as well as provided some perspectives on potential areas for future investigation. At present, the development of efficient microbial consortia encounters numerous challenges stemming from the inadequate investigation of the biodegradation mechanisms underlying NR. Furthermore, future research may encounter difficulties concerning the regulation of coexistence among microorganisms and the safety and stability of synthesized microbial consortiums. The dynamic characteristics of microbial communities, biological variability, and a multitude of other uncertainties arising from microbial communities constitute obstacles for future research.
Although attention to microbial degradation of NR has continued for many years, single microorganisms that have been discovered are difficult to take on the task of treating rubber wastes. The emergence of microbial consortia has solved the survival problem of individual strains to some extent and further improved the degradation efficiency. How to construct a stable and efficient consortium is the key to solve the microbial degradation of NR. Combining the method of screening single functional strains with omics tools can not only discover more functional genes involved in the degradation of NR and clarify the metabolic pathway of microbial degradation of NR; it can also help us understand the structural composition of microorganisms in the sample. The construction of microbial consortia also faces many difficulties, such as how to rationally utilize the complex interspecies relationship between microorganisms, overcome the antagonistic effect of local microorganisms on foreign microorganisms, and achieve stable and efficient degradation goals, all of which need to be solved urgently.
CC: Conceptualization, Data curation, Writing–original draft. MJ: Conceptualization, Data curation, Writing–original draft. CZ: Data curation, Validation, Visualization, Writing–original draft. NZ: Data curation, Validation, Visualization, Writing–original draft. F-JJ: Data curation, Validation, Visualization, Writing–review and editing. TL: Data curation, Validation, Visualization, Writing–review and editing. H-GL: Resources, Supervision, Writing–review and editing. LJ: Resources, Supervision, Writing–review and editing.
This research was funded by the Korea Environment Industry and Technology Institute (KEITI) through a project to develop new, eco-friendly materials and processing technology derived from wildlife, funded by the Korea Ministry of Environment (MOE) (grant number 2021003240004) and National Research Council of Science and Technology (NST) grant by the Korea Government (MSIT) (CAP20023-200).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali Shah, A., Hasan, F., Shah, Z., Kanwal, N., and Zeb, S. (2013). Biodegradation of natural and synthetic rubbers: a review. Int. Biodeterior. Biodegr. 83, 145–157. doi:10.1016/j.ibiod.2013.05.004
Altenhoff, A. L., De Witt, J., Andler, R., and Steinbüchel, A. (2019). Impact of additives of commercial rubber compounds on the microbial and enzymatic degradation of poly (cis-1, 4-isoprene). Biodegradation 30, 13–26. doi:10.1007/s10532-018-9858-5
Andler, R. (2020). Bacterial and enzymatic degradation of poly (cis-1, 4-isoprene) rubber: novel biotechnological applications. Biotechnol. Adv. 44, 107606. doi:10.1016/j.biotechadv.2020.107606
Araújo, W. J., Oliveira, J. S., Araújo, S. C. S., Minnicelli, C. F., Silva-Portela, R. C. B., da Fonseca, M. M. B., et al. (2020). Microbial culture in minimal medium with oil favors enrichment of biosurfactant producing genes. Front. Bioeng. Biotechnol. 8, 962. doi:10.3389/fbioe.2020.00962
Atagana*, H. I., Ejechi, B. O., and Ayilumo, A. M. (1999). Fungi associated with degradation of wastes from rubber processing Industry. Environ. Monit. Assess. 55, 401–408. doi:10.1023/a:1005935032014
Basik, A. A., Nanthini, J., Yeo, T. C., and Sudesh, K. (2021a). Rubber degrading strains: microtetraspora and dactylosporangium. Polymers 13, 3524. doi:10.3390/polym13203524
Basik, A. A., Sanglier, J.-J., Yeo, C. T., and Sudesh, K. (2021b). Microbial degradation of rubber: Actinobacteria. Polymers 13, 1989. doi:10.3390/polym13121989
Berekaa, M. M., Barakaat, A., Elsayed, S. M., and Elaassar, S. A. (2005). Degradation of natural rubber by Achromobacter sp. NRB and evaluation of culture conditions. Pol. J. Microbiol./Polskie Towarzystwo Mikrobiologów = Pol. Soc. Microbiol. 54, 55–62.
Berekaa, M. M., Linos, A., Reichelt, R., Keller, U., and Steinbüchel, A. (2000). Effect of pretreatment of rubber material on its biodegradability by various rubber degrading bacteria. FEMS Microbiol. Lett. 184, 199–206. doi:10.1111/j.1574-6968.2000.tb09014.x
Birke, J., Röther, W., and Jendrossek, D. (2017). RoxB is a novel type of rubber oxygenase that combines properties of rubber oxygenase RoxA and latex clearing protein (lcp). Appl. Environ. Microbiol. 83, e00721–e00717. doi:10.1128/aem.00721-17
Birke, J., Röther, W., Schmitt, G., and Jendrossek, D. (2013). Functional identification of rubber oxygenase (RoxA) in soil and marine myxobacteria. Appl. Environ. Microbiol. 79, 6391–6399. doi:10.1128/aem.02194-13
Bode, H. B., Kerkhoff, K., and Jendrossek, D. (2001). Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2, 295–303. doi:10.1021/bm005638h
Borel, M., Kergomard, A., and Renard, M. F. (1982). Degradation of natural rubber by fungi imperfecti. Agric. Biol. Chem. 46, 877–881. doi:10.1271/bbb1961.46.877
Bosco, F., Antonioli, D., Casale, A., Gianotti, V., Mollea, C., Laus, M., et al. (2018). Biodegradation of unvulcanized natural rubber by microorganisms isolated from soil and rubber surface: a preliminary study. Bioremediat J. 22, 43–52. doi:10.1080/10889868.2018.1476454
Bosco, F., and Mollea, C. (2021). Biodegradation of natural rubber: microcosm study. Water Air Soil Pollut. 232 (6), 227. doi:10.1007/s11270-021-05171-7
Braaz, R., Fischer, P., and Jendrossek, D. (2004). Novel type of heme-dependent oxygenase catalyzes oxidative cleavage of rubber (Poly-cis-1,4-Isoprene). Appl. Environ. Microbiol. 70, 7388–7395. doi:10.1128/aem.70.12.7388-7395.2004
Bredberg, K., Erik Andersson, B., Landfors, E., and Holst, O. (2002). Microbial detoxification of waste rubber material by wood-rotting fungi. Bioresour. Technol. 83, 221–224. doi:10.1016/s0960-8524(01)00218-8
Castañeda Alejo, S. M., Tejada Meza, K., Valderrama Valencia, M. R., Arenazas Rodríguez, A. J., and Málaga Espinoza, C. J. (2022). Tire ground rubber biodegradation by a consortium isolated from an aged tire. Microorganisms 10, 1414. doi:10.3390/microorganisms10071414
Chen, H., Sun, C., Liu, R., Yuan, M., Mao, Z., Wang, Q., et al. (2021). Enrichment and domestication of a microbial consortium for degrading aniline. J. Water Process Eng. 42, 102108. doi:10.1016/j.jwpe.2021.102108
Cheng, X., Xia, M., and Yang, Y. (2023). Biodegradation of vulcanized rubber by a gut bacterium from plastic-eating mealworms. J. Hazard Mater 448, 130940. doi:10.1016/j.jhazmat.2023.130940
Chengalroyen, M. D., and Dabbs, E. R. (2013). Identification of a gene responsible for amido black decolorization isolated from Amycolatopsis orientalis. World J. Microbiol. Biotechnol. 29, 625–633. doi:10.1007/s11274-012-1218-8
Cherian, E., and Jayachandran, K. (2009). Microbial degradation of natural rubber latex by a novel species of Bacillus sp SBS25 isolated from soil. Int. J. Environ. Res. 3, 599–604.
Chittella, H., Yoon, L. W., Ramarad, S., and Lai, Z. W. (2021). Rubber waste management: a review on methods, mechanism, and prospects. Polym. Degrad. Stab. 194, 109761. doi:10.1016/j.polymdegradstab.2021.109761
Depaolini, A. R., Bianchi, G., Fornai, D., Cardelli, A., Badalassi, M., Cardelli, C., et al. (2017). Physical and chemical characterization of representative samples of recycled rubber from end-of-life tires. Chemosphere 184, 1320–1326. doi:10.1016/j.chemosphere.2017.06.093
Dey, S., Kumar, H., Sinha, S. K., Goud, V. V., and Das, S. (2020). Bacterial biofilm-based nitrate and phosphate removal from rubber latex wastewater for sustainable water usage. Water Environ. J. 34, 170–182. doi:10.1111/wej.12515
Gallert, C. (2000). Degradation of latex and of natural rubber by Streptomyces strain La 7. Syst. Appl. Microbiol. 23, 433–441. doi:10.1016/s0723-2020(00)80075-2
Gibson, L., Larke-Mejía, N. L., and Murrell, J. C. (2020). Complete genome of isoprene degrading Nocardioides sp. WS12. Microorganisms 8, 889. doi:10.3390/microorganisms8060889
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., and Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249. doi:10.1016/s1074-5521(98)90108-9
Hapuarachchi, S. N. S., Kariyapper, S. R., Gunawardana, M., Egodage, S. M., and Ariyadasa, T. U. (2016). Biodegradation of natural rubber latex by a novel bacterial species isolated from soil. 2016 Moratuwa Eng. Res. Conf. (MERCon), 293–296. doi:10.1109/mercon.2016.7480156
Heisey, R. M., and Papadatos, S. (1995). Isolation of microorganisms able to metabolize purified natural rubber. Appl. Environ. Microbiol. 61, 3092–3097. doi:10.1128/aem.61.8.3092-3097.1995
Hesham, A.E.-L., Mohamed, N. H., Ismail, M. A., and Shoreit, A. A. M. (2015). Degradation of natural rubber latex by new Streptomyces labedae strain ASU-03 isolated from Egyptian soil. Microbiology 84, 351–358. doi:10.1134/s0026261715030078
Hiessl, S., Böse, D., Oetermann, S., Eggers, J., Pietruszka, J., and Steinbüchel, A. (2014). Latex clearing protein—an oxygenase cleaving poly(cis-1,4-isoprene) rubber at the cis double bonds. Appl. Environ. Microbiol. 80, 5231–5240. doi:10.1128/aem.01502-14
Hiessl, S., Schuldes, J., Thürmer, A., Halbsguth, T., Bröker, D., Angelov, A., et al. (2012). Involvement of two latex-clearing proteins during rubber degradation and insights into the subsequent degradation pathway revealed by the genome sequence of Gordonia polyisoprenivorans strain VH2. Appl. Environ. Microbiol. 78, 2874–2887. doi:10.1128/aem.07969-11
Ibrahim, E. M. A., Arenskötter, M., Luftmann, H., and Steinbüchel, A. (2006). Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic Actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl. Environ. Microbiol. 72, 3375–3382. doi:10.1128/aem.72.5.3375-3382.2006
Imai, S., Ichikawa, K., Muramatsu, Y., Kasai, D., Masai, E., and Fukuda, M. (2011). Isolation and characterization of Streptomyces, Actinoplanes, and Methylibium strains that are involved in degradation of natural rubber and synthetic poly(cis-1,4-isoprene). Enzyme Microb. Technol. 49, 526–531. doi:10.1016/j.enzmictec.2011.05.014
Imai, S., Yoshida, R., Endo, Y., Fukunaga, Y., Yamazoe, A., Kasai, D., et al. (2013). Rhizobacter gummiphilus sp. nov., a rubber-degrading bacterium isolated from the soil of a botanical garden in Japan. J. Gen. Appl. Microbiol. 59, 199–205. doi:10.2323/jgam.59.199
Jendrossek, D., and Birke, J. (2019). Rubber oxygenases. Appl. Microbiol. Biotechnol. 103, 125–142. doi:10.1007/s00253-018-9453-z
Jendrossek, D., and Reinhardt, S. (2003). Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol. Lett. 224, 61–65. doi:10.1016/s0378-1097(03)00424-5
Joseph, A., Gupta, P., De, G., Lal, M., Meena, M. K., Singh, L. P., et al. (2022). Biodegradation of natural rubber by fungi and bacteria. Nat. Environ. Pollut. Technol. 21, 1039–1048. doi:10.46488/nept.2022.v21i03.010
Kasai, D. (2020). Poly (cis-1, 4-isoprene)-cleavage enzymes from natural rubber-utilizing bacteria. Biosci. Biotechnol. Biochem. 84, 1089–1097. doi:10.1080/09168451.2020.1733927
Kasai, D., Imai, S., Asano, S., Tabata, M., Iijima, S., Kamimura, N., et al. (2017). Identification of natural rubber degradation gene in Rhizobacter gummiphilus NS21. Biosci. Biotechnol. Biochem. 81, 614–620. doi:10.1080/09168451.2016.1263147
Kawakami, T., Sakai, S., Obama, T., Kubota, R., Inoue, K., and Ikarashi, Y. (2022). Characterization of synthetic turf rubber granule infill in Japan: rubber additives and related compounds. Sci. Total Environ. 840, 156716. doi:10.1016/j.scitotenv.2022.156716
Krishnaswamy, V., and Ahongsangbam, N. (2017). A study on mineralisation of poly cis 1,4 isoprene (NR) and synthetic rubber gloves (SRG) by the bacterial consortium. Ann. Appl. Microbiol. Biotechnol. J. 1, 1–10. doi:10.36876/aamb.1005
Lee, D.-J., Show, K.-Y., and Wang, A. (2013). Unconventional approaches to isolation and enrichment of functional microbial consortium – a review. Bioresour. Technol. 136, 697–706. doi:10.1016/j.biortech.2013.02.075
Linos, A., Reichelt, R., Keller, U., and Steinbüchel, A. (2000). A Gram-negative bacterium, identified as Pseudomonas aeruginosa AL98, is a potent degrader of natural rubber and synthetic cis-1,4-polyisoprene. FEMS Microbiol. Lett. 182, 155–161. doi:10.1111/j.1574-6968.2000.tb08890.x
Linos, A., and Steinbüchel, A. (2005). Biodegradation of natural and synthetic rubbers. Biopolym. Online Biol. Chem. Biotechnol. Appl. 2. doi:10.1002/3527600035.bpol2010
Luo, Q., Hiessl, S., Poehlein, A., Daniel, R., and Steinbüchel, A. (2014). Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl. Environ. Microbiol. 80, 3895–3907. doi:10.1128/aem.00473-14
Lynen, F., Eggerer, H., Henning, U., and Kessel, I. (1958). Farnesyl-pyrophosphat und 3-Methyl-Δ3-butenyl-1-pyrophosphat, die biologischen Vorstufen des Squalens. Zur Biosynthese der Terpene, III. Angew. Chem. 70, 738–742. doi:10.1002/ange.19580702403
Mohamed, N., Ismail, M., Abdel-Mageed, W., and Shoreit, A. (2017). Biodegradation of natural rubber latex of Calotropis procera by two endophytic fungal species. J. Bioremediat. Biodegr. 8, 380. doi:10.4172/2155-6199.1000380
Mooibroek, H., and Cornish, K. (2000). Alternative sources of natural rubber. Appl. Microbiol. Biotechnol. 53, 355–365. doi:10.1007/s002530051627
Muralidharan, M., and Krishnaswamy, V. (2016). Artificial rubber mineralization by co-cultured bacterial strains. Int. J. Biol. Res. 4, 105. doi:10.14419/ijbr.v4i2.6414
Nair, K. P. P. (2010). The agronomy and economy of important tree crops of the developing world. Agron. Econ. Important Tree Crops Dev. World 30, 313–351. doi:10.1016/C2010-0-64818-8
Nanthini, J., and Sudesh, K. (2017). Biodegradation of natural rubber and natural rubber products by Streptomyces sp. strain CFMR 7. J. Polym. Environ. 25, 606–616. doi:10.1007/s10924-016-0840-1
Nawong, C., Umsakul, K., and Sermwittayawong, N. (2018). Rubber gloves biodegradation by a consortium, mixed culture and pure culture isolated from soil samples. Braz. J. Microbiol. 49, 481–488. doi:10.1016/j.bjm.2017.07.006
Nayanashree, G., and Thippeswamy, B. (2015a). Biodegradation of natural rubber by laccase and manganese peroxidase enzyme of Bacillus subtilis. Environ. Process. 2, 761–772. doi:10.1007/s40710-015-0118-y
Nayanashree, G., and Thippeswamy, B. (2015b). Natural rubber degradation by laccase and manganese peroxidase enzymes of Penicillium chrysogenum. Int. J. Environ. Sci. Technol. 12, 2665–2672. doi:10.1007/s13762-014-0636-6
Nguyen, L. H., Nguyen, H. D., Tran, P. T., Nghiem, T. T., Nguyen, T. T., Dao, V. L., et al. (2020). Biodegradation of natural rubber and deproteinized natural rubber by enrichment bacterial consortia. Biodegradation 31, 303–317. doi:10.1007/s10532-020-09911-0
Prasad, B. (2017). Biodegradation of ortho-dimethyl phthalate by a binary culture of Variovorax sp. BS1 and Achromobacter denitrificans. Int. J. Environ. Sci. Technol. 14, 2575–2582. doi:10.1007/s13762-016-0942-2
Riesenfeld, C. S., Schloss, P. D., and Handelsman, J. (2004). Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552. doi:10.1146/annurev.genet.38.072902.091216
Roy, R. V., Das, M., Banerjee, R., and Bhowmick, A. K. (2006). Comparative studies on crosslinked and uncrosslinked natural rubber biodegradation by Pseudomonas sp. Bioresour. Technol. 97, 2485–2488. doi:10.1016/j.biortech.2005.09.024
Sarkar, B., Gupta, A. M., Shah, M. P., and Mandal, S. (2022). Poly-cis-isoprene degradation by Nocardia sp. BSTN01 isolated from industrial waste. Appl. Biochem. Biotechnol. 194, 3333–3350. doi:10.1007/s12010-022-03854-3
Sato, S., Honda, Y., Kuwahara, M., Kishimoto, H., Yagi, N., Muraoka, K., et al. (2004). Microbial scission of sulfide linkages in vulcanized natural rubber by a white rot basidiomycete, ceriporiopsis subvermispora. Biomacromolecules 5, 511–515. doi:10.1021/bm034368a
Schade, A. L. (1937). Observations on a monascus isolated from rubber. Mycologia 29, 295–302. doi:10.2307/3754286
Seidel, J., Schmitt, G., Hoffmann, M., Jendrossek, D., and Einsle, O. (2013). Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proc. Natl. Acad. Sci. U.S.A. 110, 13833–13838. doi:10.1073/pnas.1305560110
Sharma, V., Siedenburg, G., Birke, J., Mobeen, F., Jendrossek, D., and Prakash, T. (2018). Metabolic and taxonomic insights into the Gram-negative natural rubber degrading bacterium Steroidobacter cummioxidans sp. nov., strain 35Y. PLoS one 13, e0197448. doi:10.1371/journal.pone.0197448
Singh, L. (2017). Biodegradation of synthetic dyes: a mycoremediation approach for degradation/decolourization of textile dyes and effluents. J. Appl. Biotechnol. Bioeng. 3, 430–435. doi:10.15406/jabb.2017.03.00081
Söhngen, N. L., and Fol, J. G. (1914). Decomposition of rubber by micro-organisms. J. Soc. Chem. Industry 33, 430–436.
Spence, D., and Van Niel, C. B. (1936). Bacterial decomposition of the rubber in Hevea latex. Rubber Chem. Technol. 9, 633–643. doi:10.5254/1.3539496
Steinbüchel, A. (2001). Perspectives for biotechnological production and utilization of biopolymers: metabolic engineering of polyhydroxyalkanoate biosynthesis pathways as a successful example. Macromol. Biosci. 1, 1–24. doi:10.1002/1616-5195(200101)1:1<1::aid-mabi1>3.0.co;2-b
Stevenson, K., Stallwood, B., and Hart, A. G. (2008). Tire rubber recycling and bioremediation: a review. Bioremediat. J. 12, 1–11. doi:10.1080/10889860701866263
Sudhakar, M., Doble, M., Murthy, P. S., and Venkatesan, R. (2008). Marine microbe-mediated biodegradation of low- and high-density polyethylenes. Int. Biodeterior. Biodegr. 61, 203–213. doi:10.1016/j.ibiod.2007.07.011
Tsuchii, A., and Takeda, K. (1990). Rubber-degrading enzyme from a bacterial culture. Appl. Environ. Microbiol. 56, 269–274. doi:10.1128/aem.56.1.269-274.1990
Umsakul, K., Watcharakul, S., Hodgson, B., Chumeka, W., and Tanrattanakul, V. (2012). Biodegradation of a blended starch/natural rubber foam biopolymer and rubber gloves by Streptomyces coelicolor CH13. Electron. J. Biotechnol. 15, 8. doi:10.2225/vol15-issue1-fulltext-10
Vivod, R., Andler, R., Oetermann, S., Altenhoff, A.-L., Seipel, N., Holtkamp, M., et al. (2019). Characterization of the latex clearing protein of the poly(cis-1,4-isoprene) and poly(trans-1,4-isoprene) degrading bacterium Nocardia nova SH22a. J. Gen. Appl. Microbiol. 65, 293–300. doi:10.2323/jgam.2019.01.003
Watcharakul, S., Röther, W., Birke, J., Umsakul, K., Hodgson, B., and Jendrossek, D. (2016). Biochemical and spectroscopic characterization of purified Latex Clearing Protein (Lcp) from newly isolated rubber degrading Rhodococcus rhodochrous strain RPK1 reveals novel properties of Lcp. BMC Microbiol. 16, 92. doi:10.1186/s12866-016-0703-x
Yikmis, M., Arenskötter, M., Rose, K., Lange, N., Wernsmann, H., Wiefel, L., et al. (2008). Secretion and transcriptional regulation of the latex-clearing protein, lcp, by the rubber-degrading bacterium Streptomyces sp. strain K30. Appl. Environ. Microbiol. 74, 5373–5382. doi:10.1128/aem.01001-08
Yikmis, M., and Steinbüchel, A. (2012). Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl. Environ. Microbiol. 78, 4543–4551. doi:10.1128/aem.00001-12
Keywords: natural rubber, polyisoprene, rubber-degrading bacteria, microbial consortia, metagenomics, omics
Citation: Cui C, Jiang M, Zhang C, Zhang N, Jin F-J, Li T, Lee H-G and Jin L (2023) Assembly strategies for rubber-degrading microbial consortia based on omics tools. Front. Bioeng. Biotechnol. 11:1326395. doi: 10.3389/fbioe.2023.1326395
Received: 23 October 2023; Accepted: 27 November 2023;
Published: 06 December 2023.
Edited by:
Ka Yu Cheng, CSIRO Land and Water, AustraliaReviewed by:
Bekir Engin Eser, Aarhus University, DenmarkCopyright © 2023 Cui, Jiang, Zhang, Zhang, Jin, Li, Lee and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Jin, aXNhY2NraW1AYWx1bW5pLmthaXN0LmFjLmty; Hyung-Gwan Lee, dHJ1c3RpbkBrcmliYi5yZS5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.