- Institute of Life Technologies, School of Engineering, University of Applied Science and Arts Western Switzerland, Sion, Switzerland
This study investigates the enzymatic degradation processes of different classes of polyhydroxyalkanoates (PHAs), a group of biopolymers naturally synthesized by various microorganisms. Medium chain length PHAs (mcl-PHAs) are distinguished biopolymers due to their biodegradability and diverse material properties. Using quartz crystal microbalance measurements as a valuable tool for accurate real-time monitoring of the enzymatic degradation process, the research provides detailed kinetic data, describing the interaction between enzymes and substrates during the enzymatic degradation process. Thin films of poly-3-hydroxybutyrate (PHB) and polyhydroxyoctanoate copolymer (PHO), containing molar fractions of about 84% 3-hydroxyoctanoate and 16% 3-hydroxyhexanoate, were exposed to scl-depolymerases from Pseudomonas lemoignei LMG 2207 and recombinant mcl-depolymerase produced in Escherichia coli DH5α harboring the plasmid pMAD8, respectively. Analyses based on a heterogeneous kinetic model for the polymer degradation indicated a six-fold stronger adsorption equilibrium constant of mcl-depolymerase to PHO. Conversely, the degradation rate constant was approximately twice as high for scl-depolymerases acting on PHB. Finally, the study highlights the differences in enzyme-substrate interactions and degradation mechanisms between the investigated scl- and mcl-PHAs.
Introduction
Polyhydroxyalkanoates (PHAs) are aliphatic polyesters naturally produced by several microorganisms, including Bacillus, Pseudomonas, Azotobacter, Hydrogenomonas, and Chromatium. These microorganisms synthesize PHAs intracellularly for carbon and energy storage, typically through the fermentation of carbon-rich substrates. The accumulation process often occurs under unbalanced growth conditions such as carbon excess and limitation of essential elements like phosphorus, nitrogen, or oxygen (Raza et al., 2018). Based on the number of carbon atoms in their monomers, PHAs can be grouped into subclasses. Short-chain length PHA (scl-PHA) consists of monomers with three to five carbon atoms, while medium-chain length PHA (mcl-PHA) contains monomers with 6–14 carbon atoms.
A distinctive characteristic of PHAs lies in the vast range of physicochemical, mechanical, and thermal properties they can exhibit, owing to the broad substrate specificity of PHA polymerases and the myriad of potential monomers (Matsumoto et al., 2009; Amstutz et al., 2019). This versatility has placed PHAs as the largest group among natural polyesters (Kim et al., 2007). The advent of genetically engineered strains and innovations in carbon sources and feeding strategies further enhances our ability to produce tailored PHAs with a more recent focus on the production of mcl-PHAs and their industrial applications (Tortajada et al., 2013; Oliveira et al., 2020; Muthuraj et al., 2021; Reddy et al., 2022). Consequently, these polymers find applications in diverse fields, from bioplastics and biomaterials to medical implants and biofuels (Hazer and Steinbüchel, 2007; Brigham and Sinskey, 2012; Panchal et al., 2013; Li et al., 2016).
The proliferation of petroleum-based plastics in the environment has raised pressing concerns due to their prolonged persistence (Singh and Sharma, 2016). Both terrestrial and marine ecosystems face the mounting challenge of plastic debris accumulation, with marine regions being especially burdened by sizable aggregations known as “plastic gyres” (Derraik, 2002; Law, 2017; Drzyzga and Prieto, 2019). The widespread occurrence of microplastics, tiny fragments resulting from larger plastic breakdown or direct release, intensifies the issue, affecting a great number of species across diverse habitats (de Souza Machado et al., 2018; Hanik et al., 2019; Soo et al., 2021). Current recycling and waste management efforts address only a portion of the total plastic waste, leading to increased calls for sustainable alternatives. In this context, biobased polymers like PHAs are gaining attention. PHAs are naturally produced by microorganisms and, crucially, are biodegradable. Their ability to serve as a direct replacement for many petroleum-based plastic applications while offering a reduced environmental footprint makes them a promising candidate in the quest for sustainable materials. Although PHAs available on the market are more expensive than those derived from petroleum, they have the advantage of not necessarily being derived from nonrenewable energy and fossil raw materials. Their production is not only possible from edible substrates such as sugars or vegetable oils but also substrates obtained from agricultural and food industry waste (Khardenavis et al., 2007). From a financial and environmental point of view, a considerable potential for optimization exists for the manufacturing processes, mainly with the use of bioenergy, biocatalyst, and carbon source from waste (Chen, 2009; Chen et al., 2020). The integration of PHAs into industrial and consumer applications could not only reduce the inflow of non-degradable plastics but also provide a model for how bioengineering can offer tangible solutions to pressing global environmental issues.
However, there are still many economic and technological challenges to overcome in the field of PHAs (Wang et al., 2014). One of them is to develop unique and tailored material properties for specific applications. For example, the degradability of biopolymers is an essential property which needs to be well adapted for biomedical or packaging applications (Gumel et al., 2013; Keskin et al., 2017; Kalia et al., 2021). Biopolymers can undergo degradation through different mechanisms, which depend on the polymer structure and exposure conditions. Physical and chemical degradation are the two main types of degradation. Physical degradation results from physical changes, such as thermal embrittlement and environmental stress cracking. Chemical degradation occurs through chemical reactions, with common examples including photo-induced, thermal, thermal-oxidative, solvolytic, hydrolytic, and biological degradation. Understanding the mechanisms of biodegradation is essential for designing more sustainable and environmentally friendly materials (Meereboer et al., 2020; Silva et al., 2023). In this regard, the monitoring of the biopolymers degradation has become a key area of research.
Secretion of PHA depolymerases by microorganisms represents one of the possibilities for the biodegradation of extracellular PHA in the environment. While many extracellular scl-PHA depolymerases have been purified and well characterized (Handrick et al., 2001; Braaz et al., 2003; García-Hidalgo et al., 2012), only few mcl-PHA depolymerases have been described so far (Schirmer and Jendrossek, 1994; Gangoiti et al., 2012; Martínez et al., 2015). These enzymes play a crucial role in the biodegradation process, as they catalyze the breakdown of the larger polymer molecules into smaller units that can be metabolized by microorganisms. Several techniques have been developed for monitoring the enzymatic degradation of plastics, such as turbidimetric powder and film assay (Timmins et al., 1997a; Timmins et al., 1997b), Taylor dispersion analysis (Chamieh et al., 2015), and Blue-ray-based micromechanical characterization (Ceccacci et al., 2017). However, these techniques often suffer from time-consuming sample preparation and complex experimental set up. Some require the polymer to be formulated into stable suspensions or free-standing films which limits their application to specific mechanical polymer properties for monitoring the enzymatic degradation. One tool that has been proven to be useful for this purpose is the quartz crystal microbalance (QCM). Based on the inverse piezoelectric effect the QCM can be used to measure very small changes in the mass of a material in real time (Sabot and Krause, 2002; Marx, 2003). QCM sensors are commonly 5 or 10 MHz AT-cut quartz crystals with gold electrodes on both sides that vibrate in the thickness-shear mode during measurements, where the two surfaces move in an antiparallel fashion. The use of these sensors as microbalances is based on the linear relationship between the resonance frequency variation and the mass variation of the quartz crystal, which can be described by the Sauerbrey equation (Eq. 1), where Δ𝑓 and 𝑓0 represent the normalized frequency variation and the resonant frequency of the fundamental mode, respectively. The active area, the density, and the shear modulus describe the crystal used for the measurement and are represented by A, ρq, and µq, respectively. The corresponding mass variation is represented by Δm (Sauerbrey, 1959; Johannsmann, 2015).
Applying this equation to a quartz crystal with a fundamental resonance frequency (RF) of about 5 MHz, assigns a change in the RF of 1 Hz to a mass variation of about 17 ng on the sensor surface. This information is useful for designing experiments that use QCM to monitor the degradation of biodegradable plastics, such as PHA. QCMs have been used to study the enzymatic degradation of polyester films in several scientific publications. Yamashita et al., used the combination of QCM with atomic force microscopy (AFM) to study interactions between an enzyme and a biopolymer film at the molecular level Yamashita et al. (2005). In our experimental set up, a continuously circulating enzyme solution is introduced into a fluidic cell and brought in contact with a PHA coated sensors. The changes in the RF due to the degradation of PHA are monitored over time.
Various models have been developed to characterize the depolymerization kinetics of enzymes, taking into account different mechanisms, kinetics, and environmental factors. The heterogeneity of the reaction (i.e., the interaction between a soluble enzyme and an insoluble substrate in the case of PHA films) requires more complex models. The observed degradation behavior was found to be inconsistent with classical Michaelis-Menten homogenous enzymatic kinetic where both enzyme and substrate are considered as soluble. At high enzyme concentrations, the reaction rate becomes inversely proportional to the enzyme concentration. The heterogenous kinetic model proposed by Mukai et al., assumes that the depolymerase first binds to the substrate surface and then hydrolyzes polymer chains. However, if the adsorption process is faster than the rate-limiting hydrolysis, surface crowding limits the unhindered access of the enzymes catalytic domain to the substrate. This eventually leads to a decrease in the degradation rate of the poly-3-hydroxybutyrate (PHB) film when increasing the enzyme concentration (Mukai et al., 1993). This kinetic model successfully described the experimental data reported for the enzymatic degradation of PHB films by several PHB depolymerases. However, to the best of our knowledge, this approach was never applied to other classes of PHA polymers and their corresponding depolymerases.
In this study, we apply the QCM technique to model the degradation kinetics of PHA films from different PHA classes. The model we use was adapted from Mukai et al. and can be written in the form of the rate equation (Eq. 2), where υ0 represents the highest degradation rate observed for the corresponding enzyme concentration [E]0. The adsorption equilibrium constant and the degradation rate constant are represented by K et k', respectively.
We characterize mcl-PHA degradation and compare it with scl-PHA degradation by their respective depolymerases. As far as we know, this is the first detailed study of the enzymatic degradation of mcl-PHA in direct comparison of scl-PHA degradation. In this approach the QCM technique is especially useful as the mcl-PHAs are commonly highly amorphous polymers which are not readily available as free-standing films or stable polymer suspensions (Jendrossek and Handrick, 2002). Based on our experiments, we find a distinguished difference between the scl- and mcl-PHA depolymerases investigated. The results highlight the importance of understanding not only the molecular structure of the enzyme and its substrates but also their degradation mechanism. The applied analytical method allows to distinguish the parameters for adsorption equilibrium and degradation rate, providing a deeper understanding of the key features of the PHA degradation by their specific depolymerases.
Materials and methods
PHB powder was purchased from Biomer (Germany) and polyhydroxyoctanoate copolymer (PHO), containing molar fractions of about 84% 3-hydroxyoctanoate and 16% 3-hydroxyhexanoate as determined by gas chromatography, was kindly provided by Prof. Dr. Manfred Zinn (University of Applied Science and Arts Western Switzerland). All other chemicals and reagents were purchased from Sigma-Aldrich (Switzerland) and used without any further purification.
Strains, plasmid, media, and cultivation conditions
Pseudomonas lemoignei LMG 2207 was kindly provided by Prof. Dr. Dieter Jendrossek (Stuttgart University, Germany) and received on filter paper blots. The filter paper blots were stored at 4°C. The strain was used for all cultivations in the following sterilized minimal medium (adapted from Stinson and Merrick, 1974): Disodium succinate (15 mM), KH2PO4 (33 mM), Na2HPO4 × 2 H2O (33 mM), NH4Cl (18 mM), MgSO4 × 7 H2O (2 mM), FeCl3 × 6 H2O (0.037 mM) and CaCl2 × 2 H2O (0.045 mM).
Escherichia coli DH5α [supE44, ΔlacU169 (Ø80lacZΔM15), hsdR17, recA1, endA1, gyrA96, thi-1, relA1] was used as host for the expression of modified mcl-depolymerase from Pseudomonas fluorescens GK13 with the previously described plasmid pMAD8 (Ihssen et al., 2009). The preculture and agar slant was performed in LB-Amp medium: 5 g L−1 yeast extract, 10 g L−1 tryptone and 5 g L−1 NaCl, 100 mg L−1 ampicillin. The bioreactor cultivation was conducted in Terrific Broth (TB): 24 g L−1 yeast extract, 12 g L−1 tryptone, 4 mL L−1 glycerol, 0.25 mL L−1 PPG 2000, 0.017 M KH2PO4 and 0.072 M K2HPO4, 100 mg L−1 ampicillin.
Colony selection for producers of scl-depolymerases
In order to identify colonies of P. lemoignei producing extracellular scl-depolymerases, colonies were incubated on the solid media supplemented with 1.5% w/v of PHB powder at 30°C for 72 h. The selected colonies, distinguished by a circular halo around them, were used to inoculate precultures.
Preculture preparation
Pseudomonas lemoignei LMG 2207 was cultivated in shake flasks for precultures. For the preparation of the minimal medium, CaCl2 × 2 H2O, FeCl3 × 6 H2O, NH4Cl, and MgSO4 × 7 H2O were autoclaved separately and mixed at room temperature to avoid the formation of precipitates. The resulting solution was supplemented with 5 g L−1 of disodium succinate. A gas-liquid ratio of 5 was respected for all cultivations to keep sufficient microbial culture aeration. Colonies from solid media plates were selected for medium inoculation. These cultures were incubated at 30°C and 160 RPM in an orbital shaker incubator. The biomass growth was monitored by measuring the optical density at 600 nm (OD600) of 1 mL samples. The incubation was stopped once the desired OD600 was reached.
In a similar way, E. coli DH5α strain harboring the plasmid pMAD8 was streaked on LB-Amp Agar at 37°C and incubated overnight, providing single colonies to inoculate LB-Amp medium. The precultures were cultivated overnight at 37°C with 200 RPM agitation.
Batch cultivation with pH shift
For the optimization of scl-depolymerases production from P. lemoignei in a 3 L benchtop bioreactor (KLF, Bioengineering), the cultivation was carried out as reported by Terpe et al. (1999). A preculture of 200 mL with an OD600 of 1.0 was used to inoculate the main culture medium to obtain an OD600 of approximately 0.1. The fermentation was separated into biomass accumulation and depolymerase production phases, differentiated only by the pH setting of 6.3 and 7.6, respectively. The pH adjustment was realized by adding smaller volumes of NaOH 5 M or H3PO4 5 M. Furthermore, the process parameters were set to a temperature of 30°C, a stirring speed of 400 RPM, and an aeration of 2 vvm with air. The pO2 was prevented to fall below 30% by addition of O2 to the inlet gas. Samples of 5 mL were taken regularly during the whole cultivation. The OD600 of samples was measured to follow the biomass growth and to apply the pH shift during the late exponential phase. Moreover, the enzyme activity was measured on crude culture medium samples with a spectrophotometric enzyme assay.
For the expression of mcl-depolymerase by E. coli harboring the plasmid pMAD8, a stock solution of 0.17 M KH2PO4 and 0.72 M K2HPO4 was separately autoclaved and added to the main solution together with the ampicillin. Cultivation conditions in the 3 L benchtop bioreactor were set to 37°C, a pH of 7.0 (adjusted with NaOH 5 M and H3PO4 2 M), an aeration of 1 vvm, and agitation starting at 700 RPM and gradually increasing to 1,200 RPM to prevent the pO2 to drop below 30%. The bioreactor was inoculated with 100 mL of the LB-Amp preculture. The OD600 was monitored and once it reached 6, the temperature was reduced to 25°C and 0.5 mM IPTG was added for induction. After 16 h of induction, cells were harvested and isolated by centrifugation.
Spectrophotometric enzyme activity assay
To follow the production of extracellular depolymerases, the enzyme activity was monitored using the model substrate para-nitrophenyl butyrate (pNPB) according to the spectrophotometric assay described by Rios et al. (2019).
Isolation and concentration of depolymerases
For scl-depolymerases from P. lemoignei, the isolation of extracellular enzymes was performed by centrifugation of the crude culture medium at 2500 × g for 45 min at 4°C. The enzymes present in the supernatant fraction were concentrated by two consecutive ultrafiltration (UF) cycles using centrifugal concentrator tubes of 15 mL with a 50 kDa molecular weight cut-off regenerated cellulose membrane (Vivaspin Turbo 15 RC 50kDa, Sartorius). The concentrator tubes were filled with 10 mL of supernatant and centrifugated 5000 × g for 15 min at 4°C for the first concentration step and 7 min at 4°C for the second step.
For mcl-depolymerases from recombinant E. coli, the crude culture medium was centrifuged at 7000 × g for 30 min at 4°C. The pellet was resuspended in a lysis buffer consisting of 30 mM Tris-HCl (pH 8.0) and 1 mM EDTA. The cells were disrupted using a French press at 1,000 bar for two cycles, keeping the mixture on ice throughout the process. The mixture was then centrifuged at 9384 × g for 15 min at 4°C to obtain the supernatant containing the depolymerase. The solution was concentrated using a 30 kDa centrifugal concentrator tubes (Amicon, Millipore). The final step of purification was performed by preparative chromatography (Äkta start, Cytiva) using a high-flow amylose resin (n°E8022S from neb, Bioconcept Ltd.).
Protein quantification
For protein quantification, a sample volume of 1 µL was directly deposited onto the measurement pedestal of a Nanodrop spectrophotometer (DS-11 FX +, DeNovix) and measured at 280 nm. As the exact composition of the mixture of depolymerases was not known, the concentrations were calculated based on the percent extinction coefficient of bovine serum albumin provided by the device manufacturer (0.667 L g−1 cm−1).
PHA thin film preparation
PHA solutions were prepared for the coating step of thin films on quartz crystal sensors. Surface electron microscopy (SEM) was employed to support the development of a suitable coating procedure (Supplementary Figure S1). The solution was made from PHB powder or PHO film solubilized in trichloromethane (1% w/v) and heated at 50°C for 1 h to fully solubilize the polymer. A fraction of 10% v/v of toluene was added to the solutions to optimize the evaporation speed and film thickness during the spin coating process. For dynamic spin coating of quartz crystal sensors, the speed of the spin coater (Ossila Ltd.) was set at 4,000 RPM and 140 µL of polymer solution was cast on the rotating sensor center. The spinning was continued for 40 s, followed by drying the coated sensor in a vacuum oven at 37.5°C and 50 mbar for 20 min. Finally, the film was examined visually for any imperfections such as irregularities, pinholes, or scratches.
QCM measurements procedure
Experiments were carried out with a QCM (Q-1, openQCM) allowing the measurement of the RF, dissipation, and sensor temperature. The sensor type was a 5 MHz quartz crystal with a blank diameter of 13.95 mm, a frontal gold electrode diameter of 11.20 mm, an AT-fundamental cut, a quartz density of 2649.7 kg m−3, and one gold electrode on both sides. For data acquisition and real-time monitoring, the openQCM Q-1 Python Software (GUI Python version 2.0) was used.
Film stability assessment
Before any degradation experiment, the film stability was assessed by monitoring the RF variations of coated quartz sensors dispensed in circulating buffer solution. Chemical-resistant tubes (Tygon 2375-C, Saint Gobain) of 1.6 mm inner diameter, connected the measuring module to a magnetically stirred reservoir. Phosphate buffer at pH 7 was introduced into the reservoir and a constant flow was installed by a peristaltic pump, set to a flow rate of 0.5 mL min−1. Once the measuring chamber was filled, measurements were started for 1 h. For acceptable frequency variations in phosphate buffer (typically below 30 Hz), the degradation experiment was started. Otherwise, another sensor was calibrated, coated, and assessed.
Enzymatic degradation
The measuring chamber, the stirred container, and circulation tubes were kept filled with the phosphate buffer solution to avoid air bubbles. Enzyme solution was introduced into the stirred container and mixed for 1 min at 700 RPM. Afterwards, the peristaltic pump was switched on and set at a flow rate of 0.5 mL min−1. As soon as the enzyme solution circulated, measurements were started. Finally, once the enzymatic degradation was finished, measurements were kept for approximately one more hour.
Results
The degradation of thin films made of PHB and PHO was investigated using scl-depolymerases from P. lemoignei LMG 2207 and recombinant mcl-depolymerases from E. coli DH5α harboring the plasmid pMAD8, respectively. To assess the degradation process, QCM measurements were carried out. The results revealed a sigmoidal degradation rate profile, consisting of an initial acceleration phase, an inflection point representing the highest degradation rate (υ0) and a subsequent deceleration phase (Supplementary Figure S2). The raw data were analyzed to obtain the degradation rate for each experiment and are compiled in Table 1.
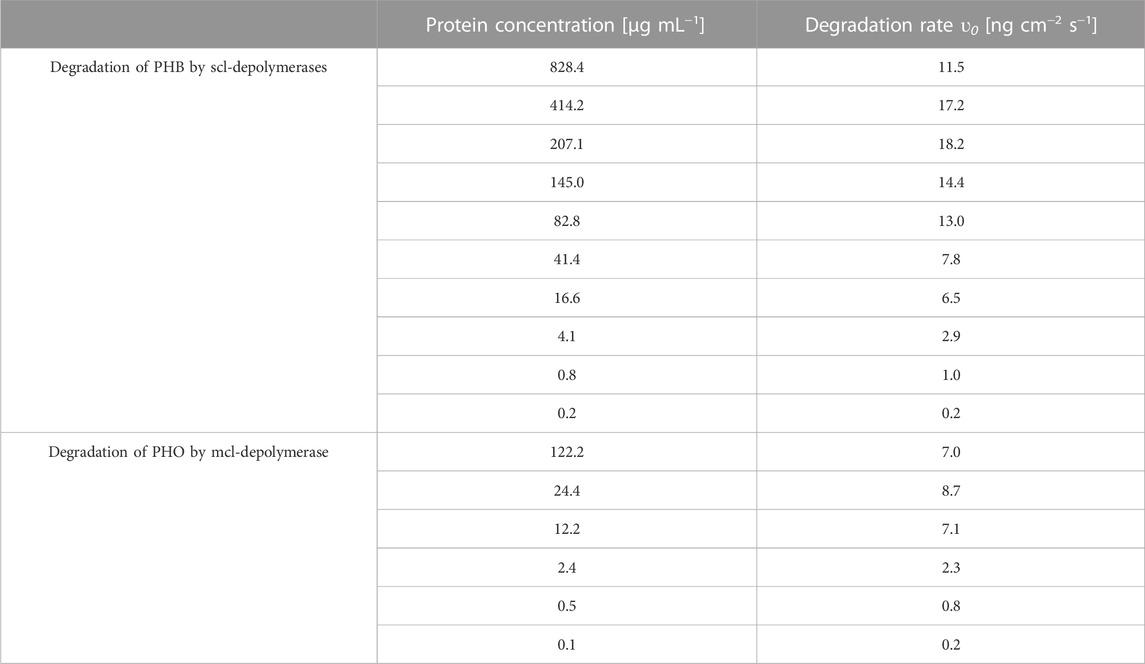
TABLE 1. Degradation rates extracted from raw data processing according to the protein concentration for all degradation experiments.
The impact of different enzyme concentrations on degradation rate was investigated. The protein concentrations tested for PHB and PHO films differed, with a wider range of 0.2–828.4 µg mL−1 for PHB and a narrower range of 0.1–122.2 µg mL−1 for PHO. Consistent with findings in the literature for scl-PHA, QCM measurements revealed that the degradation rate initially increased with increasing enzyme concentration, reaching a maximum value, and followed by a decreasing degradation rate with further increasing enzyme concentrations. The enzyme concentration corresponding to the maximum degradation rate was higher when using scl-depolymerases to degrade PHB thin films than when using mcl-depolymerases to degrade PHO thin films. According to these observations, results were analyzed using the heterogeneous degradation model, which was fitted to the observed degradation rate data through non-linear regression with the Levenberg-Marquardt algorithm. Figures 1, 2 display the observed data and the corresponding fitted models for the degradation of PHB and PHO, respectively. We like to point out, that in the case of the used experimental set-up, the observed mass loss cannot directly be correlated with the hydrolysis rate of the enzymes. Although both enzyme classes are known to primarily produce water soluble monomers, dimers, and trimers as degradation products (Abe and Doi, 1999; Handrick et al., 2004; Martínez et al., 2015), the fragmentation of the PHA polymer into non soluble degradation products would lead to their deposition on the surface and would not be observed as a frequency change in the QCM measurement. However, due to the stability testing prior to each experiment in the absence of polymer degrading enzyme, mass loss as observed by a resonance frequency increase can be attributed to the degradation of the polymer by the enzyme into soluble products. We therefore refer to the constant k' of the heterogeneous degradation model as degradation rate constant, rather than hydrolysis rate constant as originally proposed by Mukai et al. By applying the kinetic model, it was determined that the maximum degradation rate for PHB films was 17.3 ng cm−2 s−1 and occurred at a scl-depolymerases concentration of 245.6 µg mL−1. For PHO films, the maximum degradation rate was 9.5 ng cm−2 s−1 and occurred at a mcl-depolymerases concentration of 38.2 µg mL−1.
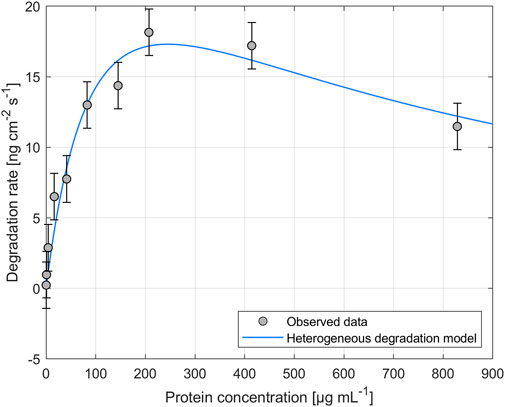
FIGURE 1. Degradation of PHB thin films exposed to depolymerases from Pseudomonas lemoignei LMG 2207 at various protein concentrations. The heterogeneous degradation model was fitted to the observed data through non-linear regression using the Levenberg-Marquardt algorithm. The degradation rate measurement for an enzyme concentration of Cprot = 82.8 µg mL−1 was carried out in triplicate and the standard deviation was used to calculate the error bars assuming a constant error.
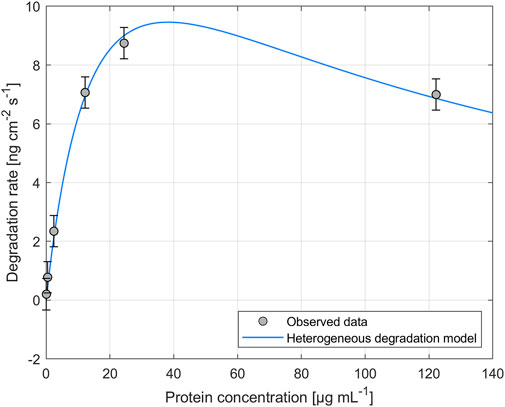
FIGURE 2. Degradation of PHO thin films exposed to recombinant mcl-depolymerases from Escherichia coli DH5α harboring the plasmid pMAD8, at various protein concentrations. The heterogeneous degradation model was fitted to the observed data through non-linear regression using the Levenberg-Marquardt algorithm. The degradation rate measurement for an enzyme concentration of Cprot = 2.4 µg mL-1 was carried out in triplicate and the standard deviation was used to calculate the error bars assuming a constant error.
The kinetic constants were obtained through non-linear regression analysis of QCM measurements and are compiled in Table 2. The adsorption equilibrium constant (K) represents the affinity of the enzyme to the substrate and reflects the initial adsorption step of the depolymerization process. The higher K value for mcl-depolymerases on PHO films compared to scl-depolymerases on PHB films indicates a stronger affinity of the mcl-depolymerases for the PHO substrate. Concerning the degradation rate constant (k'), it reflects the enzymatic degradation activity of the enzyme and represents the rate of mass reduction after adsorption. The lower k' value for mcl-depolymerases compared to scl-depolymerases suggests that mcl-depolymerases have a lower enzymatic efficiency in degrading PHO films. These results indicate that the enzyme-substrate interactions and degradation activity play key functions in the degradation of PHB and PHO films by scl- and mcl-depolymerases.
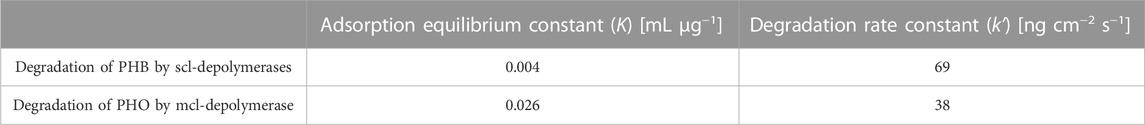
TABLE 2. Kinetic constants calculated by non-linear regression of experimentally observed data to the heterogeneous degradation model for the PHB and PHO degradation.
Discussion
The interaction strength between an enzyme and its substrate is represented by the constant K in the heterogeneous degradation model. The QCM measurements showed that the K value was six times higher for mcl-depolymerases and PHO when compared to scl-depolymerases and PHB. It indicates a stronger adsorption of mcl-depolymerase to PHO than the adsorption of scl-depolymerases to PHB. We attribute the differences in K values between these two systems to the higher hydrophobicity of the PHO polymer. Although structurally different, with a C-terminal substrate binding domain for scl-depolymerases (Behrends et al., 1996) and an N-terminal substrate binding domain for mcl-depolymerases (Gangoiti et al., 2012), both enzymes have a hydrophobic site that promotes absorption to the substrate. However, the presence of longer aliphatic side chains of the PHO polymer reduces the competitive effect of solvent absorption at the polymer surface when compared to the more polar PHB surface where water molecules will absorb to a higher extend, leading to a lower number of absorbed enzyme molecules in the equilibrium state.
The degradation rate of a biopolyester under the catalytic effect of an enzyme is expressed by the constant k' in the heterogeneous degradation model. The QCM measurements showed that the k' value was approximately twice as high for scl-depolymerases and PHB than for mcl-depolymerases and PHO. Possible explanations, apart from the different specific environments of the Serine-Histidine-Aspartate motif (i.e., catalytic triad), can be found in the different nature of the PHA-backbones. PHA hydrolysis takes place at the ester forming carbonyl groups of the polymers. For mcl-PHAs these are sterically more challenging due to their longer aliphatic side chains. This sidechain length also reduces the quality of the leaving group and the nucleophilicity of the carbonyl group.
Considering the mechanism of the degradation of PHA as proposed by Mukai et al., this leads not only to a reduced maximum degradation rate for the mcl-depolymerase when compared to the scl-depolymerases of this study but also to a lower optimal enzyme concentration since the fast adsorption accompanied by a relatively slow and rate-limiting hydrolysis process leads to faster surface crowding by the absorbed enzyme molecules on the substrate surface (Figures 1, 2).
In conclusion, we believe that these findings will support the process of developing applications for mcl-PHAs and the critical consideration of end-of-life scenarios. A low abundance of natural mcl-PHA producers in the environment, concomitant scarcity of mcl-PHA depolymerases and the observed lower degradation rate constant when compared to scl-PHA depolymerases, invite to consider new routes for monomer reutilization and upcycling. In this respect, enzymatic engineering might offer a viable solution to improve the enzymatic degradation of PHA depolymerases by addressing their substrate binding and hydrolysis profiles.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
FM: Formal Analysis, Investigation, Methodology, Software, Writing–original draft. NH: Conceptualization, Methodology, Resources, Supervision, Writing–review and editing.
Funding
The authors declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1303267/full#supplementary-material
References
Abe, H., and Doi, Y. (1999). Structural effects on enzymatic degradabilities for poly[(R)-3-hydroxybutyric acid] and its copolymers. Int. J. Biol. Macromol. 25 (1-3), 185–192. doi:10.1016/s0141-8130(99)00033-1
Amstutz, V., Hanik, N., Pott, J., Utsunomia, C., and Zinn, M. (2019). Tailored biosynthesis of polyhydroxyalkanoates in chemostat cultures. Methods Enzym. 627, 99–123. doi:10.1016/bs.mie.2019.08.018
Behrends, A., Klingbeil, B., and Jendrossek, D. (1996). Poly(3-hydroxybutyrate) depolymerases bind to their substrate by a C-terminal located substrate binding site. FEMS Microbiol. Lett. 143 (2-3), 191–194. doi:10.1111/j.1574-6968.1996.tb08479.x
Braaz, R., Handrick, R., and Jendrossek, D. (2003). Identification and characterisation of the catalytic triad of the alkaliphilic thermotolerant PHA depolymerase PhaZ7 of Paucimonas lemoignei. FEMS Microbiol. Lett. 224 (1), 107–112. doi:10.1016/s0378-1097(03)00425-7
Brigham, C. J., and Sinskey, A. J. (2012). Applications of polyhydroxyalkanoates in the medical industry. Int. J. Biotechnol. Wellness Industries 1, 52–60. doi:10.6000/1927-3037.2012.01.01.03
Ceccacci, A. C., Chen, C.-H., Hwu, E.-T., Morelli, L., Bose, S., Bosco, F. G., et al. (2017). Blu-Ray-based micromechanical characterization platform for biopolymer degradation assessment. Sensors Actuators B-chemical 241, 1303–1309. doi:10.1016/j.snb.2016.09.190
Chamieh, J., Biron, J. P., Cipelletti, L., and Cottet, H. (2015). Monitoring biopolymer degradation by taylor dispersion analysis. Biomacromolecules 16 (12), 3945–3951. doi:10.1021/acs.biomac.5b01260
Chen, G. (2009). A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38 8, 2434–2446. doi:10.1039/b812677c
Chen, G.-Q., Chen, X.-Y., Wu, F.-Q., and Chen, J.-C. (2020). Polyhydroxyalkanoates (PHA) toward cost competitiveness and functionality. Adv. Industrial Eng. Polym. Res. 3 (1), 1–7. doi:10.1016/j.aiepr.2019.11.001
Derraik, J. G. B. (2002). The pollution of the marine environment by plastic debris: a review. Mar. Pollut. Bull. 44 9, 842–852. doi:10.1016/s0025-326x(02)00220-5
de Souza Machado, A. A., Kloas, W., Zarfl, C., Hempel, S., and Rillig, M. C. (2018). Microplastics as an emerging threat to terrestrial ecosystems. Glob. Chang. Biol. 24 (4), 1405–1416. doi:10.1111/gcb.14020
Drzyzga, O., and Prieto, A. (2019). Plastic waste management, a matter for the 'community. Microb. Biotechnol. 12 (1), 66–68. doi:10.1111/1751-7915.13328
Gangoiti, J., Santos, M., Prieto, M. A., de la Mata, I., Serra, J. L., and Llama, M. J. (2012). Characterization of a novel subgroup of extracellular medium-chain-length polyhydroxyalkanoate depolymerases from actinobacteria. Appl. Environ. Microbiol. 78 (20), 7229–7237. doi:10.1128/aem.01707-12
García-Hidalgo, J., Hormigo, D., Prieto, M. A., Arroyo, M., and de la Mata, I. (2012). Extracellular production of Streptomyces exfoliatus poly(3-hydroxybutyrate) depolymerase in Rhodococcus sp. T104: determination of optimal biocatalyst conditions. Appl. Microbiol. Biotechnol. 93 (5), 1975–1988. doi:10.1007/s00253-011-3527-5
Gumel, A. M., Annuar, M. S. M., and Heidelberg, T. (2013). Current application of controlled degradation processes in polymer modification and functionalization. J. Appl. Polym. Sci. 129, 3079–3088. doi:10.1002/app.39006
Handrick, R., Reinhardt, S., Focarete, M. L., Scandola, M., Adamus, G., Kowalczuk, M., et al. (2001). A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 276 (39), 36215–36224. doi:10.1074/jbc.M101106200
Handrick, R., Reinhardt, S., Kimmig, P., and Jendrossek, D. (2004). The "intracellular" poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases. J. Bacteriol. 186 (21), 7243–7253. doi:10.1128/jb.186.21.7243-7253.2004
Hanik, N., Amstutz, V., and Zinn, M. (2019). Microplastics - from anthropogenic to natural. Chim. (Aarau) 73 (10), 841–843. doi:10.2533/chimia.2019.841
Hazer, B., and Steinbüchel, A. (2007). Increased diversification of polyhydroxyalkanoates by modification reactions for industrial and medical applications. Appl. Microbiol. Biotechnol. 74, 1–12. doi:10.1007/s00253-006-0732-8
Ihssen, J., Magnani, D., Thony-Meyer, L., and Ren, Q. (2009). Use of extracellular medium chain length polyhydroxyalkanoate depolymerase for targeted binding of proteins to artificial poly[(3-hydroxyoctanoate)-co-(3-hydroxyhexanoate)] granules. Biomacromolecules 10 (7), 1854–1864. doi:10.1021/bm9002859
Jendrossek, D., and Handrick, R. (2002). Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56, 403–432. doi:10.1146/annurev.micro.56.012302.160838
Kalia, V. C., Singh Patel, S. K., Shanmugam, R., and Lee, J.-K. (2021). Polyhydroxyalkanoates: trends and advances toward biotechnological applications. Bioresour. Technol. 326, 124737. doi:10.1016/j.biortech.2021.124737
Keskin, G., Kizil, G. V., Bechelany, M., Pochat-Bohatier, C., and Öner, M. (2017). Potential of polyhydroxyalkanoate (PHA) polymers family as substitutes of petroleum based polymers for packaging applications and solutions brought by their composites to form barrier materials. Pure Appl. Chem. 89, 1841–1848. doi:10.1515/pac-2017-0401
Khardenavis, A. A., Kumar, M. S., Mudliar, S. N., and Chakrabarti, T. (2007). Biotechnological conversion of agro-industrial wastewaters into biodegradable plastic, poly(β-hydroxybutyrate). Bioresour. Technol. 98 (18), 3579–3584. doi:10.1016/j.biortech.2006.11.024
Kim, H. W., Chung, M. G., and Rhee, Y. H. (2007). Biosynthesis, modification, and biodegradation of bacterial medium-chain-length polyhydroxyalkanoates. J. Microbiol. 45 (2), 87–97.
Law, K. L. (2017). Plastics in the marine environment. Annu. Rev. Mar. Sci. 9, 205–229. doi:10.1146/annurev-marine-010816-060409
Li, Z., Yang, J., and Loh, X. J. (2016). Polyhydroxyalkanoates: opening doors for a sustainable future. NPG Asia Mater. 8 (4), e265. doi:10.1038/am.2016.48
Martínez, V., De Santos, P. G., García-Hidalgo, J., Hormigo, D., Prieto, M. A., Arroyo, M., et al. (2015). Novel extracellular medium-chain-length polyhydroxyalkanoate depolymerase from Streptomyces exfoliatus K10 DSMZ 41693: a promising biocatalyst for the efficient degradation of natural and functionalized mcl-PHAs. Appl. Microbiol. Biotechnol. 99, 9605–9615. doi:10.1007/s00253-015-6780-1
Marx, K. A. (2003). Quartz crystal microbalance: a useful tool for studying thin polymer films and complex biomolecular systems at the solution-surface interface. Biomacromolecules 4 (5), 1099–1120. doi:10.1021/bm020116i
Matsumoto, K. i., Shozui, F., Satoh, Y., Tajima, K., Munekata, M., and Taguchi, S. (2009). Kinetic analysis of engineered polyhydroxyalkanoate synthases with broad substrate specificity. Polym. J. 41, 237–240. doi:10.1295/polymj.pj2008253
Meereboer, K. W., Misra, M., and Mohanty, A. K. (2020). Review of recent advances in the biodegradability of polyhydroxyalkanoate (PHA) bioplastics and their composites. Green Chem. 22, 5519–5558. doi:10.1039/d0gc01647k
Mukai, K., Yamada, K., and Doi, Y. (1993). Kinetics and mechanism of heterogeneous hydrolysis of poly[(R)-3-hydroxybutyrate] film by PHA depolymerases. Int. J. Biol. Macromol. 15 (6), 361–366. doi:10.1016/0141-8130(93)90054-p
Muthuraj, R., Valerio, O., and Mekonnen, T. H. (2021). Recent developments in short- and medium-chain- length Polyhydroxyalkanoates: production, properties, and applications. International Journal of Biological Macromolecules. Int. J. Biol. Macromol. 187, 422–440. doi:10.1016/j.ijbiomac.2021.07.143
Oliveira, G. H. D., Zaiat, M., Rodrigues, J. A. D., Ramsay, J. A., and Ramsay, B. A. (2020). Towards the production of mcl-PHA with enriched dominant monomer content: process development for the sugarcane biorefinery context. J. Polym. Environ. 28, 844–853. doi:10.1007/s10924-019-01637-2
Panchal, B., Bagdadi, A., and Roy, I. (2013). “Polyhydroxyalkanoates: the natural polymers produced by bacterial fermentation,” in Advances in natural polymers: composites and nanocomposites. Editors S. Thomas, P. M. Visakh, and A. P. Mathew (Berlin, Heidelberg: Springer Berlin Heidelberg), 397–421.
Raza, Z. A., Abid, S., and Banat, I. M. (2018). Polyhydroxyalkanoates: characteristics, production, recent developments and applications. Int. Biodeterior. Biodegrad. 126, 45–56. doi:10.1016/j.ibiod.2017.10.001
Reddy, V. U. N., Ramanaiah, S. V., Reddy, M. V., and Chang, Y.-C. (2022). Review of the developments of bacterial medium-chain-length polyhydroxyalkanoates (mcl-PHAs). Bioengineering 9 (5), 225. doi:10.3390/bioengineering9050225
Rios, N. S., Mendez-Sanchez, C., Arana-Peña, S., Rueda, N., Ortiz, C., Gonçalves, L. R. B., et al. (2019). Immobilization of lipase from Pseudomonas fluorescens on glyoxyl-octyl-agarose beads: improved stability and reusability. Biochim. Biophys. Acta BBA - Proteins Proteomics 1867, 741–747. doi:10.1016/j.bbapap.2019.06.005
Sabot, A., and Krause, S. (2002). Simultaneous quartz crystal microbalance impedance and electrochemical impedance measurements. Investigation into the degradation of thin polymer films. Anal. Chem. 74 14, 3304–3311. doi:10.1021/ac0200724
Sauerbrey, G. (1959). Verwendung von schwingquarzen zur wagung dunner schichten und zur mikrowagung. Z. Fur Phys. 155 (2), 206–222. doi:10.1007/Bf01337937
Schirmer, A., and Jendrossek, D. (1994). Molecular characterization of the extracellular poly(3-hydroxyoctanoic acid) [P(3HO)] depolymerase gene of Pseudomonas fluorescens GK13 and of its gene product. J. Bacteriol. 176 (22), 7065–7073. doi:10.1128/jb.176.22.7065-7073.1994
Silva, R. R. A., Marques, C. S., Arruda, T. R., Teixeira, S. C., and de Oliveira, T. V. (2023). Biodegradation of polymers: stages, measurement, standards and prospects. Macromol 3 (2), 371–399. doi:10.3390/macromol3020023
Singh, P., and Sharma, V. P. (2016). Integrated plastic waste management: environmental and improved health approaches. Procedia Environ. Sci. 35, 692–700. doi:10.1016/j.proenv.2016.07.068
Soo, C. L., Sabana, S., Chen, C. A., and Hii, Y. S. (2021). Understanding microplastics in aquatic ecosystems – a mini review. Borneo J. Mar. Sci. Aquac. (BJoMSA) 5 (2), 63–69. doi:10.51200/bjomsa.v5i2.3386
Stinson, M. W., and Merrick, J. M. (1974). Extracellular enzyme secretion by Pseudomonas lemoignei. J. Bacteriol. 119 (1), 152–161. doi:10.1128/jb.119.1.152-161.1974
Terpe, K., Kerkhoff, K., Pluta, E., and Jendrossek, D. (1999). Relationship between succinate transport and production of extracellular poly(3-hydroxybutyrate) depolymerase in Pseudomonas lemoignei. Appl. Environ. Microbiol. 65 (4), 1703–1709. doi:10.1128/AEM.65.4.1703-1709.1999
Timmins, M. R., Gilmore, D. F., Lotti, N., Scandola, M., Fuller, R. C., and Lenz, R. W. (1997a). A spectrophotometric method for detection of enzymatic degradation of thin polymer films. J. Environ. Polym. Degrad. 5 (1), 1–15. doi:10.1007/BF02763564
Timmins, M. R., Lenz, R. W., and Clinton Fuller, R. (1997b). Heterogeneous kinetics of the enzymatic degradation of poly(β-hydroxyalkanoates). Polymer 38 (3), 551–562. doi:10.1016/S0032-3861(96)00530-7
Tortajada, M., da Silva, L. F., and Prieto, M. A. (2013). Second-generation functionalized medium-chain-length polyhydroxyalkanoates: the gateway to high-value bioplastic applications. Int. Microbiol. 16, 1–15. doi:10.2436/20.1501.01.175
Wang, Y., Yin, J., and Chen, G. Q. (2014). Polyhydroxyalkanoates, challenges and opportunities. Curr. Opin. Biotechnol. 30, 59–65. doi:10.1016/j.copbio.2014.06.001
Keywords: polyhydroxyalkanoates, depolymerase enzymes, quartz crystal microbalance, degradation kinetics, biodegradable polymers, enzymatic degradation
Citation: Millan F and Hanik N (2023) Degradation kinetics of medium chain length Polyhydroxyalkanoate degrading enzyme: a quartz crystal microbalance study. Front. Bioeng. Biotechnol. 11:1303267. doi: 10.3389/fbioe.2023.1303267
Received: 27 September 2023; Accepted: 01 December 2023;
Published: 14 December 2023.
Edited by:
George Guo-Qiang Chen, Tsinghua University, ChinaReviewed by:
Sebastian L. Riedel, Berlin Technical University of Applied Sciences, GermanySanja Jeremic, University of Belgrade, Serbia
Copyright © 2023 Millan and Hanik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nils Hanik, bmlscy5oYW5pa0BoZXZzLmNo
 Fabien Millan
Fabien Millan Nils Hanik
Nils Hanik