
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 26 September 2023
Sec. Biomaterials
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1254356
This article is part of the Research Topic Exosomes for Drug Delivery View all 4 articles
 Haorong Chen1,2†
Haorong Chen1,2† Hanbo Yao1,2†
Hanbo Yao1,2† Jiaxin Chi1
Jiaxin Chi1 Chaowei Li1,2
Chaowei Li1,2 Yilin Liu1,2
Yilin Liu1,2 Jiayi Yang1,2
Jiayi Yang1,2 Jiaqi Yu1,2
Jiaqi Yu1,2 Jiajun Wang1,2
Jiajun Wang1,2 Yongdui Ruan1*
Yongdui Ruan1* Jiang Pi1,2,3*
Jiang Pi1,2,3* Jun-Fa Xu1,2*
Jun-Fa Xu1,2*Chemotherapy often faces some obstacles such as low targeting effects and drug resistance, which introduce the low therapeutic efficiency and strong side effects. Recent advances in nanotechnology allows the use of novel nanosystems for targeted drug delivery, although the chemically synthesized nanomaterials always show unexpected low biocompability. The emergence of exosome research has offered a better understanding of disease treatment and created novel opportunities for developing effective drug delivery systems with high biocompability. Moreover, RNA interference has emerged as a promising strategy for disease treatments by selectively knocking down or over-expressing specific genes, which allows new possibilities to directly control cell signaling events or drug resistance. Recently, more and more interests have been paid to develop optimal delivery nanosystems with high efficiency and high biocompability for drug and functional RNA co-delivery to achieve enhanced chemotherapy. In light of the challenges for developing drug and RNA co-delivery system, exosomes have been found to show very attractive prospects. This review aims to explore current technologies and challenges in the use of exosomes as drug and RNA co-delivery system with a focus on the emerging trends and issues associated with their further applications, which may contribute to the accelerated developments of exosome-based theraputics.
Exosomes are small membrane vesicles (30–150 nm) containing complex lipids, RNAs and proteins, which can be release by prokaryotes, eukaryotes, various cells, and mediate cell to cell communication in physiological and pathological conditions (Lai et al., 2022). As a subset of EV, exosomes were initially recognized as a mechanism for cells to excrete unwanted cell metabolites or other substances. However, in 1983, exosomes were firstly found in reticulocytes of sheep, and Johnstone named them exosomes in 1987 (Johnstone et al., 1987). There are three processes, including the formation of endocytic vesicles, generation of multivesicular bodies (MVBs), and release of exosomes, collectively comprise the biogenesis and development of exosomes (Han et al., 2022). Once exosomes are released from the cell, they can fuse with the plasma membrane of the recipient cell and delivers contents to the intracellular compartment, including proteins, nucleic acid, lipids and metabolites,etc., which play a crucial role in biological function (Dai et al., 2020). Exosomes thus can affect a wide range of physiological processes, such as stem cell maintenance (Batsali et al., 2020), lipid metabolism (Wang et al., 2020), aging speed (Zhang et al., 2017) immune response (Zhang and Yu, 2019), reproductive development (Esfandyari et al., 2021), cardiovascular disease (Zheng et al., 2020), infectious diseases (Schorey et al., 2015), neurodegenerative disease (Guo et al., 2021) and cancer (Paskeh et al., 2022).
RNA therapy refers to the use of nucleic acids-based molecules, which can manipulate gene expression or produce therapeutic proteins for disease treatment. The targets mainly include small interference RNA (siRNA), microRNA (miRNA), antisense oligonucleotides (ASO), and messenger RNA (mRNA) (Gupta et al., 2021). Relative to the nucleic acid therapeutics described above, although the aptamers, sarnas, and CRISPR/CAS, have been less frequently reported, they still hold great potential in the future (Yoon and Rossi, 2018; Zhu and Chen, 2018; Guo et al., 2022). Nucleic acid-based therapies have emerged as promising treatment modalities for a wide variety of diseases, these therapies allow for targeted modulation of gene expression, facilitating the treatment of both genetic and acquired conditions. Nucleic acid-based therapies have shown immense potential in providing innovative treatments for disorders such as cancer (Meng et al., 2022), infectious diseases (Xiao et al., 2019), and autoimmune diseases (Leung et al., 2010). However, for further use of nucleic acid as novel drugs for therapy, controlling pharmacokinetics (PK) parameters of small molecule drugs is still the core challenge, while there are also some remaining issue like how to improve the structural stability, reduce immunogenicity and limit the side effects of RNA molecules (Winkle et al., 2021).
Since small molecule drugs can rapidly diffuse through biological fluids, many biological barriers, and cell membranes which enable small molecules to navigate complex vascular systems and interact with virtually all tissues and cell types in the body. But situations arise such as the difficulty of the drug to breach the inevitable physiological barriers, the occurrence of adverse reactions in the blood or peripheral tissues or the loss of bioavailability or the inability to dissolve easily in the gastrointestinal tract (Zhong et al., 2021a).
Drug delivery systems (DDS) refer to the technological systems that comprehensively regulate the distribution of drugs or biomolecules in living organisms in space, time, and dose (Mainardes and Silva, 2004). Over the past two to three decades, investigators have developed viral and nonviral vectors. However, clinical success is very limited and hampered by challenges that there are no suitable delivery systems. The use of cell secretory nanovesicles, such as exosomes as vehicles for delivery application has created tremendous excitement in the field of gene therapy, likely due to their good pharmacokinetics, biocompatibility, minimal or no inherent toxicity, long plasma half-life, and intrinsic ability to cross biological barriers (Duan et al., 2021; Rezaie et al., 2022). On the one hand, single RNA therapy can effectively promote or inhibit gene expression, which has broad application prospects; On the other hand, single small-molecule drug therapy is still widely exists in clinical practice in anti-tumor drugs and other diseases. Reasonable combination chemotherapy and gene therapy and selection of exosomes as carrier lecturer are the latest idol of disease treatment development (Kobayashi et al., 2020; Zhao et al., 2020).
In this review, we describe how exosomes are produced from donor cells and absorbed by recipient cells, as well as the extraction and purification methods of exosome. Moreover, we emphatically describe the occurrence and development of exosomes as delivery systems for co-delivery of RNAs and chemotherapy drugs, as well as their potential application and current challenges for clinical therapeutic. We believe that this review would enhance our understanding of potential for exosomes as co-delivery systems of RNAs and chemotherapy drugs for novel therapeutic development, and finally promote the development of exosomes for clinical uses.
Exosomes have been regarded as descendants of the parental cell. Extracellular vesicles (EVs) are usually divided into three groups according to their size and biogenesis: exosomes (50–150 nm), microvesicles (MVs) (100–1,000 nm) and apoptotic bodies (>1,000 nm) (Jin et al., 2022). Exosomes are considered to be of endocytic origin, MVs are produced by budding and blebbing from the plasma membrane and apoptotic bodies are released by cells undergoing programmed cell death and signal cell engulfment (Colombo et al., 2014). Here, we focus on introducing the biogenesis of exosomes, how they are secreted from parent cells and how to be absorbed by recipient cells.
Understanding the biogenesis and secretion of exosomes is particularly important for the further clinical uses of exosomes. So far, multiple kinds of mechanisms involved in exosome biogenesis have been identified.
The Endosomal sorting complex required for transport (ESCRT) machinery is crucial in the biogenesis of exosomes. Defects in ESCRT machinery lead to impaired exosome secretion (Tamai et al., 2010; Tamai et al., 2012), and the development of various pathological conditions (Wei et al., 2015; Han et al., 2019). Tetraspanins CD9, CD63, and CD81 are particularly enriched in the exosomal membrane (Jeppesen et al., 2019). CD63 is abundant on late endosomes, participates in ESCRT-independent sorting of the PMEL luminal domain, and Iron loading leads to secretion of CD63 positive extracellular vesicles (Pols and Klumperman, 2009; van Niel et al., 2011; Yanatori et al., 2021). Moreover, complex lipids such as ceramide can self-associate, forming raft-like structures and contribute to the initial membrane curvature for inward budding to form ILVs (Trajkovic et al., 2008). Different Rab proteins, such as Rab5 (Gorji-Bahri et al., 2021), Rab7 (Sun et al., 2020), Rab27a (Ostrowski et al., 2010), Rab35 (Sato et al., 2008), etc. play important roles in specific steps of exosome biogenesis and secretion. Various N-ethylmaleimide-sensitive fusion attachment protein (SNAP) receptors (SNAREs) families like VAMP2/3/4/7/8 proteins are present in Ilvs (Steegmaier et al., 1999; Chen and Scheller, 2001; Pryor et al., 2004; Marshall et al., 2015; Gordon et al., 2017). Autophagy, regulated by autophagy-related genes such as Atg5 and Atg12-Atg3, determines the outcome of late endosomes and leads to late endosomal acidification (Murrow et al., 2015; Guo et al., 2017). The microenvironment surrounding the cells likewise influences exosome secretion, and can be regulated by physical signals, starvation or hyperglycemia, acidity, hypoxia, liposomes, or even circadian rhythm (Cheng et al., 2019; Debbi et al., 2022; Yeung et al., 2022). Recently, a molecular pathway linking the rigid extracellular matrix to oncogenic signaling and exosome trafficking was proposed, ultimately facilitating tumor progression (Wu et al., 2023). Exosome secretion is a complex and responsive biological process. Inhibition of a single pathway may not completely stop its secretion. Therefore, a multi-targeted approach is required to understand and manipulate exosome secretion, which involves identifying and evaluating all molecular players and pathways involved.
Exosome research has gained traction due to the observed phenotypes resulting from their entry into recipient cells, and their potential as engineered delivery vehicles in therapeutic applications. However, there is still a lack of consensus on the mechanism by which recipient cells uptake exosomes. Endocytosis is well accepted: Exosomes can be internalised by a clathrin-mediated, lipid-raft mediated, caveolin-mediated endocytosis, phagocytosis or micropinocytosis and macropinocytosis (Mulcahy et al., 2014; Gurung et al., 2021). Exosomes release their contents into recipient cells another possible entry mechanism is the direct fusion of their membranes with the cytoplasmic membrane, a process that is engaged by proteins such as SNAREs and the Rab family (Jahn and Sudhof, 1999).
At the same time, the direct contact between the exosome surface and the receptor on the receptor cell was also reported. For instance, Dendritic cell-derived exosomes can directly stimulate T cells due to carrying major histocompatibility complex (MHC) and T cell costimulatory molecules (Schorey et al., 2015). Integrins, lectins/proteoglycans and T cell immunoglobulin and mucin domain-containing protein 4, which also played an integral role in mediating exosome binding to receptor ligands of the cell (Mathieu et al., 2019). The fate of exosomes entering recipient cells is a critical aspect of pharmacokinetics. However, the ultimate destination of exosomal cargo (including potential re-secretion or degradation) remains unassessed.
Exosomes are being increasingly utilized in biomedical applications for disease prevention, diagnosis, and treatment, with rapid advancements being made towards their clinical use. In the following passages, we will briefly discuss the potential of exosomes in these areas.
The discovery, validation, and implementation of new biomarkers play a crucial role in improving clinical prognosis (He et al., 2018; Poller et al., 2018). Liquid biopsy offers several advantages, including minimal invasiveness, rapidity, and ease of dynamic monitoring (Lone et al., 2022). Taking the advantages of some properties beyond other sample sources, exosomes have emerged as a noteworthy niche in the field of liquid biopsy, attracting significant attention. While there are ongoing discussions surrounding the purity and cost of exosome extraction, they offer distinct advantages over circulating tumor cells (CTCs) and cell-free tumor DNA (ctDNA). Firstly, exosomes allow for the non-invasive collection of samples from various bodily fluids. They are relatively stable and carry a wide range of molecules, including proteins, nucleic acids, and lipids, which provide valuable insights into the tumor microenvironment. Currently, there is ongoing research exploring the potential of combining exosomal proteins, lipids, RNA, and miRNA in cancer diagnosis and prognostic assessment. In recent studies, a novel multiplex in situ detection method has been developed for exosomal miRNA and protein analysis, enabling the quantitative analysis of disease-specific miRNA and surface proteins derived from prostate cancer cells within exosomes in a single reaction (Cho et al., 2019). Exosomes can be isolated from multiple body fluids such as blood, urine, and saliva, facilitating biomarker analysis (Zhang et al., 2018). However, it is important to note that the methods for exosome isolation are still evolving, and variations in protocols may impact the quality and quantity of the obtained exosomes. Additionally, the heterogeneity of exosomes in terms of size, content, and origin poses challenges for standardization and result interpretation (Thery et al., 2018). In contrast, CTCs and ctDNA can also be isolated from peripheral blood, offering non-invasive sample collection and real-time monitoring of tumor progression and treatment response, whcih has been associated with poor prognosis in various cancer types (Yu et al., 2013; Bettegowda et al., 2014). However, CTCs exhibit molecular heterogeneity compared to primary tumor cells, which may affect their representation of the overall tumor biology. Moreover, the lack of standardized protocols for CTC isolation, detection, and characterization limits their widespread clinical application (Lin et al., 2021). Similar to CTC, the low quantities of ctDNA compared to normal cell-free DNA make its detection and quantification challenging, particularly in early-stage cancers (Yu et al., 2021). The sensitive techniques required for ctDNA detection and characterization, such as next-generation sequencing or digital PCR, restrict its accessibility in some clinical settings. Additionally, the genetic alterations detected in ctDNA may not fully capture the tumor heterogeneity, as ctDNA shedding can vary among tumor subclones (Pessoa et al., 2020).
While exosomes encapsulation therapy as a targeted drug delivery technology is still in its early stages of development, advancements in detection methods have made it possible to rapidly analyze and test specific cargo within extracellular vesicles for diagnostic purposes. This feasibility has played a crucial role in promoting the development of extracellular vesicle biomarkers. The pathological transport of exosomes throughout the course of disease development and progression holds potential for disease diagnosis, prognosis, and treatment strategie (Zhou et al., 2020; Yu et al., 2022). In addition to exosomal miRNAs, which have been extensively studied and associated with significant differences in expression profiles between disease and healthy groups, there is a vast array of other cargo present in exosomes. Although specific AUC (area under the curve) values are not provided in this table (Table 1), numerous studies have reported significant differences in exosomal miRNA expression profiles, further underscoring the potential utility of exosomes as biomarkers.
Taking into account these aforementioned advantages, exosomes have demonstrated remarkable diagnostic capabilities surpassing those of conventional biomarkers. Nevertheless, the time and cost associated with their extraction remains a topic of debate when compared to automated detection instruments. In order to fully harness the potential of exosomes in disease diagnosis, it is imperative to enhance their sensitivity and diminish the costs associated with their detection. With ongoing advanced research, it is anticipated that more cost-effective and commercially accessible exosome detection kits will become available in the foreseeable future.
The use of exosomes as vaccines holds great promise in the field of immunology. Exosomes are small extracellular vesicles that are secreted by living organisms and can play an important role in intercellular communication. As a vaccine, exosomes offer several advantages over traditional vaccines: Low toxicity and immunogenicity compared to many live vaccines and influenza vaccines; High efficiency to enter cells and release antigens for the activation and improve vaccine efficacy; Exosomes can be manipulated in terms of their composition and characteristics, allowing for flexible adjustment to transport and deliver a variety of vaccines (Santos and Almeida, 2021).
The role of communication between immune cells (macrophages, dendritic cells (DC), T cells and B cells, etc.) in the immune system is well established. Exosomes act as trucks for intercellular communication, and thus it has been increased more attention in the field of vaccine research. Exosomes isolated from LPS stimulated macrophages can stimulate the secretion of IFN-γ that appear stronger in vivo in mice in response to hepatitis B virus surface antigen (Jesus et al., 2018). DC derived exosome vaccine has great potential in metastatic melanoma and hepatocellular carcinoma (Escudier et al., 2005; Lu et al., 2017). And exosomes derived from M1 macrophages enhanced the activity of lipid calcium phosphate (LCP) nanoparticle encapsulated Trp2 vaccine and induced stronger antigen-specific cytotoxic T-cell responses through enzyme-linked immunospot (ELISPOT) assay (Cheng et al., 2017). Exosomes secreted by CD4 + T cells can significantly contribute to enhanced B cell activation and increased antibody production in the face of hepatitis B surface antigen in a dose-dependent manner (Lu et al., 2019).
In general, tumor cells can release exosomes to influence the immune system, suggesting that tumor derived exosomes may also serve as vaccines for cancer immunotherapy. Tumor derived exosomes enhance DC immunogenicity via exosome anchored peptide coated functional domains of a potent adjuvant (HMGN1) (Agrawal et al., 2017). Tumor peptide pulsed DC derived exosomes have also been identified to cause specific cytotoxic T lymphocytes as well as inhibit established mastocytoma and breast cancer growth (Whitehead et al., 2009). Tumor derived HSP70/bag-4 surface positive exosomes can stimulate NK cells to release granzyme B, which in turn triggers tumor cell apoptosis (Esteves et al., 2022). They loaded immunogenic cell death agents into α-Whey protein engineered MDA-MB-231/Luc cell-derived exosomes in vitro then activate DCS, ultimately thereby contributing to type one conventional DC maturation and CD8 + T cell activation in vitro (Huang et al., 2022).
Unlike the usual thinking of screening neoantigens from tumor cells, they derived ESAT6 from Mycobacterium tuberculosis and found that ESAT6 + exosomes could significantly contribute to the inhibition of neoantigens in B16 tumor bearing mice (Koyama et al., 2016). The number of T cells could be dose dependently stimulated to increase when the conjugation of Ag85B and ESAT6 to ubiquitin in exosomes was followed by in vivo experiments (Cheng and Schorey, 2016). Admittedly, Toxoplasma gondii exosomes could modulate macrophages to secrete inflammatory factors and elicit humoral and cellular immune responses in mice, suggesting that exosomes could serve as potential vaccine candidates against toxoplasmosis, bacteria, fungi, and viruses as well as cells after their infection can secrete exosomes (Li et al., 2018b), it is a vexing matter how to isolate and purify exosomes secreted by cells and pathogens when conducting research.
Overall, the use of exosomes as vaccines or vaccine delivery systems has significant potential to revolutionize the field of immunology and disease treatment, offering a safe and effective alternative to traditional vaccines. With ongoing research and development, we can expect to see more widespread use of exosome-based vaccines in the near future.
Exosomes have emerged as a promising drug delivery vehicle due to their unique properties. Firstly, they are naturally secreted by cells and are involved in intercellular communication, making them well-tolerated by the body with minimal toxicity (Yim et al., 2016). Secondly, they can easily penetrate cell membranes due to their small size, allowing them to efficiently deliver drugs to target cells (Hikita and Oneyama, 2022). Thirdly, they can be loaded with a wide range of therapeutic agents, such as proteins, nucleic acids, and small molecules, by modifying their surface or cargo content (Jiang et al., 2017). Additionally, they can protect their cargo from degradation and immune clearance, thus improving the stability and bioavailability of the drugs (Alhasan et al., 2014). Finally, they have the ability to target specific cells or tissues through their surface proteins, allowing for more targeted drug delivery (Salunkhe et al., 2020).
Many of the current methods for collecting exosomes are based on isolating exosomes from media obtained by classical cell culture methods. However, these methods have several limitations. For instance, they typically require large input volumes, obtained from large-scale cultures of cells, which can be time-consuming. Additionally, these methods often yield low concentrations of exosomes. In recent studies, a three-dimensional (3D) culture system using a hollow fiber bioreactor has shown potential advantages over conventional culture systems for exosome production. This 3D culture system offers higher capacity and lower cost compared to traditional methods, such as 2D-exo. It is important to note that the cargo contents of exosomes produced in 2D and 3D environments differ from each other (Haraszti et al., 2018; Wang et al., 2021). However, it should be acknowledged that 3D-exo still has some inherent limitations.
Exosomes play a crucial role as carriers for targeted drug delivery and therapy, and their purity directly impacts the function of recipient cells. Hence, in the realm of clinical diagnosis and treatment, researchers continually raise their expectations for the isolation and purification techniques of exosomes. It is important to consider the effect of rotor type, centrifugation time, and speed on exosomes since most separation methods involve centrifugation (Cvjetkovic et al., 2014). Table 2 provides a summary and comparison of several commonly used exosome isolation methods, highlighting their principles, advantages, and disadvantages. To optimize exosome production, scientists have explored the cultivation of umbilical cord-derived mesenchymal stem cells in a scalable, microcarrier-based, three-dimensional (3D) culture system. The combination of tangential flow filtration (TFF) with 3D mesenchymal stem cell culture enhances exosome production (referred to as 3D-TFF-exo) by 140-fold and improves siRNA delivery to neurons by 7-fold compared to 2D-exo (Haraszti et al., 2018). Furthermore, recent studies have demonstrated that the synergistic use of multiple methods is also beneficial for exosome separation and purification (Bellotti et al., 2021; Visan et al., 2022).
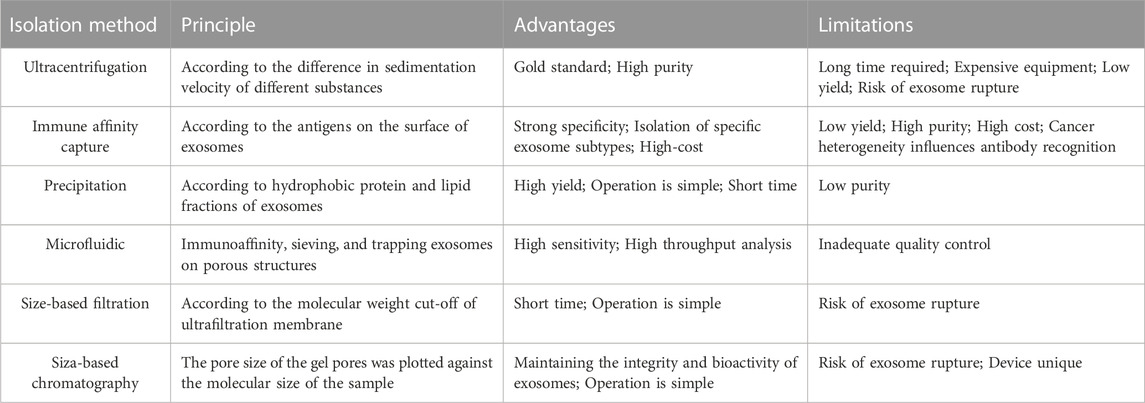
TABLE 2. The limitations and advantages of exosome isolation method (Yang et al., 2020a).
Exosomes are commonly used for delivering RNA or drugs, with two methods available for loading them, namely, preload and afterload. Preload involves delivering cargo to cells through transfection or co-incubation, followed by exosome collection. Meanwhile, afterload requires collecting exosomes from parental cells and then inducing RNA and/or drug entry by co-incubation, electroporation, or ultrasound (Ha et al., 2016). Electroporation, is found to result in extensive siRNA aggregates with much lower efficiency, highlighting the need for alternative methods in preparing siRNA loaded extracellular vesicles (Kooijmans et al., 2013). Despite the increase in exosome size after cargo loading by most methods, the protein marker remains stably expressed, demonstrating its applicability (Haney et al., 2015; Kim et al., 2016; Wang et al., 2019). Then, Improving the efficiency of exosome loading is still a priority, with previous studies showing that sonication has better loading efficacy than co-incubation and electrotransformation, making it a frontrunner in the field of loading drugs to exosomes (Haney et al., 2015; Kim et al., 2016; Gao et al., 2021; Tenchov et al., 2022). Kit-based methods have been successfully used to transfect exogenous nucleic acids into exosomes (Nie et al., 2020). Treatments such as saponin-assisted, freeze-thaw cycle, and extrusion may change the exosome morphology, biomarkers, or biological characteristics that may limit exosome development (Fuhrmann et al., 2015). Table 3 summaries the loading method of exosome.
To better target recipient cells in vitro and disease areas in vivo, researchers have developed exosome targeting technology, which can further enhancing the targeting effects of the designed exosomes and thus allow more functional application of exosomes in biomedicine.
Pre-modification involves utilizing the cellular expression machinery and natural exosome generation mechanism, allowing targeting sites to be conjugated to lamp2b. HEK293T cells were modified to produce a fusion protein consisting of Lamp2b, an exosomal protein, and a fragment of Interleukin 3 (IL3). The elevated expression of the IL3 receptor (IL3-R) in CML blasts, as compared to normal hematopoietic cells, allows it to serve as a specific receptor target in a cancer drug delivery system (Bellavia et al., 2017). In HEK293T cells, siRNA was loaded into extracellular vesicles (EVs) for delivery to SKBR3 cells. The genetically stable HEK293T cells efficiently expressed DARPin G3 on EV surface, enabling specific binding with HER2/Neu. The designed EVs successfully delivered TPD52-targeting siRNA to SKBR3 cells, leading to a remarkable 70% downregulation of gene expression (Limoni et al., 2019). The researchers utilized chondrocyte-targeting exosomes to deliver miR-140 for osteoarthritis (OA) treatment. They achieved this by fusing a chondrocyte-affinity peptide (CAP) with lysosome-associated membrane glycoprotein 2b on exosomes. CAP-exosomes efficiently encapsulated miR-140 and selectively entered chondrocytes, delivering the cargo in vitro (Liang Y. et al., 2020). In a similar manner, targeting can be accomplished by genetically modifying dendritic cells to express Lamp2b (Alvarez-Erviti et al., 2011). However, the presence of endosomal proteases during exosome formation poses a challenge as it leads to the degradation of peptides, making it difficult to achieve the desired yield of peptide-functionalized exosomes. Additionally, some studies have fused the GE11 tumor targeting polypeptide, a ligand for EGFR, to the PDGFR terminus (Ohno et al., 2013).
Post-modification is critical due to the complex process of exosome production and the variability in protein expression across different levels. Covalent coupling strategies involving chemical cross linkers and noncovalent coupling strategies utilizing hydrophobic or charge interactions are common methods for coupling targeting ligands onto exosome membranes (Liang et al., 2021). Conventional chemical reactions involve fluctuations in temperature or pressure and inappropriate changes in osmotic pressure, leading to membrane disruption or aggregation of extracellular vesicles. Click chemistry reactions are efficient as they can be performed in organic solvents and aqueous buffer solutions, with shorter reaction times. Coupling reactions do not affect the size of extracellular vesicles or their uptake into target cells, indicating that surface modifications of extracellular vesicles have appropriate reaction conditions. The exosomes, crosslinked with alkynyl groups using carbodiimide chemistry, were conjugated with the azide fluoro545 model. This coupling process does not alter the size of exosomes, nor does it affect the degree of adhesion/internalization between exosomes and receptor cells. These findings suggest that the reaction conditions employed for the modification of exosomes are gentle, preserving both the structural integrity and functional properties of the exosomes (Smyth et al., 2014). Researchers successfully achieved the loading of superparamagnetic iron oxide nanoparticles (SPIONs) and curcumin (Cur) into exosomes. They further performed click chemistry to conjugate the exosome membrane with neuropilin-1 targeting peptides (RGERPPR, RGE). This innovative approach resulted in the development of targeted exosomes specifically designed for glioma, offering combined imaging and therapeutic capabilities (Jia et al., 2018). The non-covalent modification of exosomes involves the attachment of modifiers to the surface structure of exosomes through non-covalent interactions. These modifiers can encompass lipid structures, glycosides, proteins, nucleic acids, and more. One article discusses the surface modification of exosomes using cationized pullulan to improve their liver targeting capabilities and enhance exosome-based drug delivery (Tamura et al., 2017). In another study, researchers explore a combined treatment approach employing a pH-sensitive fusogenic peptide and cationic lipids to enhance the cytosolic delivery of exosomes (Nakase and Futaki, 2015). Additionally, the use of magnetic exosomes for targeted drug delivery to specific tumor areas is investigated, potentially enhancing the effectiveness of cancer treatment (Qi et al., 2016). This study underscores the significant potential of this strategy in the field of tumor treatment.
Fundamentally, challenges such as storage, extraction, quantification, modification, and loading of exosomes hinder the development of exosomes as drug or nucleic acid delivery systems. Figure 1 shows the production process of engineered exosomes. Additionally, the in vivo transfer of exosomes and the delayed mechanism of cargo delivery through body fluids and to specific targets necessitate further consideration. The clinical applications of drug delivery vehicles, which come with their own set of challenges and limitations, require more exploration and standardization. We hope that by fostering collaboration among researchers from different disciplines, such as clinicians, cell biologists, and computer experts, we can advance exosomes hand-in-hand into the realm of clinical applications.
Small molecule drugs spearheaded the therapeutic landscape for many years, but their shortcomings also slowly unfold. The first most hindering route of drug administration is resistance mechanisms, including gene mutation, amplification, CSCs, efflux transporters, apoptosis dysregulation, and autophagy, et, which are encountered by almost all targeted anticancer drugs after some time in clinical use (Zhong et al., 2021a). Oral intake is the most commonly used route of administration because of ease of administration, high patient compliance, low cost, and flexibility in dosage form design, with compounds of low solubility resulting in poor absorption and bioavailability, insufficient solubility for intravenous administration, and development challenges resulting in increased development cost and time and a burden that is transferred to patients (frequent bolus administration) (Savjani et al., 2012). Nanoparticle—and microparticle based systems have been used to overcome the challenge of solubility, enabling small molecules to be trafficked to their site of action (Amreddy et al., 2018). Meanwhile, exosomes, have been largely developed for drug loading, have attracted increasing attention as a potentially effective drug delivery system in disease treatment due to their good biocompatibility, high stability, and good targeting.
Cancer has always been a killer of people’s continuous confrontation, and its high case fatality rate and low survival rate remain unsatisfactory. One of the reasons is that most of the current antineoplastic drugs have the disadvantages of strong cytotoxicity, poor biocompatibility and low bioavailability, leading to poor efficacy of chemotherapy.
Paclitaxel (PTX), as the first natural anticancer drug approved by FDA, has limited its clinical application due to its poor resources and low water solubility problems. Different types of nano carriers, including lipids, proteins, polymers, solid nano lotion and hybrid systems, have been reported to be used to wrap paclitaxel for cancer treatment (Khalifa et al., 2019). Macrophage derived exosomes exerted tumor growth inhibitory properties by encapsulating paclitaxel, whether in vivo or in vitro, and it was found that the encapsulated exosomes did not induce cell degeneration or necrosis in major organs (Kim et al., 2016; Wang et al., 2019). They made a bold guess that the incorporation of PTX into exosomes could not only increase their solubility but also overcome PGP mediated drug efflux, although the specific mechanism is still not well understood.
Curcumin, as a traditional Chinese herbal medicine, has anti-tumor, anti oxidant and anti-inflammatory effects, and has some efficacy in the treatment of mild and toxic COVID-19 infectious patients (Saber-Moghaddam et al., 2021). Relevant literature shows that Curcumin loaded POCA4C6 micelles have proved the rationality of further efforts to develop CPM as a treatment for breast cancer, including more detailed research on how POCA4C6 and Curcumin synergistically inhibit tumor growth and destroy BCSC (Chen et al., 2017a). Prof. Zhang’s team encapsulated curcumin into tumor or macrophage derived exosomes, which could be efficiently delivered to the brain or lungs and acted as anti-inflammatory (Sun et al., 2010; Zhuang et al., 2011). In addition, a highly efficient surface labeling technique to generate mAb exosomes loaded with romidepsin was developed, and finally its unique targeting to cancer cells and antitumor activity was confirmed (Si et al., 2020).
Doxorubicin, an antitumor broad-spectrum antibiotic used to inhibit the synthesis of RNA and DNA, has the strongest inhibitory effect on RNA. Previous literature has reported that chemically responsive nanoparticles have the potential to induce apoptosis in MCF-7 cells by enhancing DOX uptake (Zhang et al., 2022a). Yanget al used a zebrafish (Danio rerio) model, which have a fully developed and functioning BBB that has similar properties to the human BBB, to examine the efficacy of brain endothelium derived exosomes to deliver the antitumor drug doxorubicin through the blood-brain barrier (Yang et al., 2015). Doxorubicin loaded into exosomes derived from other cells similarly exhibits antitumor properties (Tian et al., 2014; Wei et al., 2019), and to promote tumor cell targeting, it can be facilitated by engineering the imDCs to express a well characterized exosomal membrane protein (lamp2b) fused to αv integrin-specific iRGD peptide (CRGDKGPDC) (Tian et al., 2014).
Loading of celastrol with exosomes improves its antitumor properties, possibly by inhibiting NF- κB The pathway (Aqil et al., 2016). Researchers have loaded aspirin into exosomes from tumor cells and gemcitabine and deferasiro co loaded into exosomes from M1 macrophages, thus showing great potential in the field of exosome loading drugs against cancer (Tran et al., 2019; Zhao et al., 2021).
The blood-brain barrier can leave brain tissue less damaged or even not by harmful substances in the circulating blood, thereby maintaining the basic stability of the brain tissue internal environment. Exosome loading of drugs has potential not only in the anti-tumor field, but also in the treatment of brain diseases due to the fact that it can cross the blood-brain barrier to reach the lesion site. In fact, most drugs cannot pass through the blood-brain barrier (Pandit et al., 2020). Exosomes are readily taken up by neuronal cells in vitro,A comparable number of exosomes were detected in the brains of PD mice after intranasal administration (Haney et al., 2015). Loading this bullet point of curcumin into exosomes, and then conjugating the exosome membrane with neuropilin-1-targeted peptide (RGERPPR,RGE)neuropilin-1-targeted peptide or the C (rgdyk) peptide had been conjugated to the exosome surface by click chemistry (Jia et al., 2018), will realize the breakthrough blood-brain barrier for the treatment of brain diseases. Loading berberine with exosomes induced overactivation of microglia, the transition from pro-inflammatory M1 type microglia to anti-inflammatory M2 type microglia, reduced neuronal apoptosis after SCI, and promoted motor function recovery (Gao et al., 2021).
The last barrier against drug treatment of disease is the gastrointestinal barrier. Either human or bovine milk exosomes can pass through the gastrointestinal barrier, making them promising oral drug delivery tools. On the one hand, the function of human milk exosomes has been largely demonstrated, but research on therapeutics to load them with drugs is still in the rising stage, and on the other hand, curcumin, paclitaxel, anthocyanins and doxorubicin can be loaded into bovine milk exosomes using appropriate solvents (Agrawal et al., 2017; Zhong et al., 2021b). This notion is not unexpected with the acidic pH could reduce degradation of exosome associated proteins (Cheng et al., 2019).
Overall, relative to free drugs, on the one hand, after reaching the disease site, drugs after loading into exosomes show stronger therapeutic effects, drug loading and longer duration (Sun et al., 2010; Kim et al., 2016) on the other hand, exosomes can cross the blood-brain barrier and gastrointestinal barrier as well as avoid being trapped by the reticuloendothelial system it is clear that exosomes must walk farther along the path of drug loading, although its starting point is later than liposomes.
RNA therapeutics offer several advantages, such as rapid production, long-term effects, and a lack of genotoxicity risk, which make them particularly useful for treating rare diseases (Whitehead et al., 2009). In contrast, exosomes have several unique characteristics, such as their small size, low immunogenicity, and customizability, which make them attractive for RNA delivery. While further research is still needed to fully explore the potential of exosomes for clinical applications, current investigations show promise for their use in the future.
miRNA is small non-coding RNA molecule that plays a critical regulatory role in gene expression (Bartel, 2009). One advantage of using miRNA-based therapies is their high specificity, allowing for targeted treatment with minimal off-target effects, however, there are also some limitations of using miRNAs as therapeutics, including delivery challenges and potential immune responses (Zhang et al., 2013). Recent research has focused on using extracellular vesicles, specifically exosomes, as a delivery system for miRNA-based therapies. There have been several studies that have demonstrated the potential of using exosomes as a delivery system for miRNA-based therapies. For instance, However, some polymeric nanoparticles that contain miR-124 are unable to migrate to the substantia nigra in order to enhance the function of dopamine neurons, so researchers have found that loading miR-124 into exosomes derived from human cord blood monocytes allows for effective transportation of miR-124 to the substantia nigra and protection of dopamine neurons (Esteves et al., 2022). Other studies have utilized exosomes collected from γδT cells, which were loaded with miR-138 and had a suppressive effect on oral squamous cell carcinoma (Li et al., 2019). Targeted modification of exosomes can also be achieved through the expression of the platelet-derived growth factor receptor’s transmembrane domain fused with a GE11 peptide in HEK293T cells (Ohno et al., 2013). Moreover, administering exosomes containing anti-mir-214 can reverse the resistance of gastric cancer to cisplatin and induce apoptosis, offering a promising approach for the treatment of cisplatin refractory gastric cancer in the future (Wang et al., 2018).
Antisense oligonucleotides (ASOs) are rapidly gaining recognition as a potent tool in gene therapy that allows for targeted regulation of gene expression at the RNA level (Kim and Rossi, 2007). However, the challenge of successfully delivering ASOs as nucleic acid therapeutics has been addressed by the discovery of exosomes. Researchers have utilized exosomes secreted by erythroid cells for the delivery of anti-mir-125b ASOs, leading to effective treatment of tumors with minimal interference from other genes (Usman et al., 2018). Moreover, exosomes loaded with anti-mir-210-asos, delivered by IFN- γ exosomes derived from stimulated human umbilical cord tissue-derived MSCs, were able to effectively alleviate symptoms of psoriasis (Zhang et al., 2022b). Exosomes capable of delivering ASOs targeting STAT6 have shown remarkable success in treating colorectal and hepatocellular carcinoma in syngeneic models, leading to inhibition of tumor growth and complete remission in 50%–80% of cases. Furthermore, these exosomes show the potential to reprogram immunosuppressive tumor-associated macrophages from a pro-tumor M2 phenotype to a pro-inflammatory M1 phenotype (Kamerkar et al., 2022).
The use of siRNA for targeted gene silencing has gained much attention due to its high specificity, strong efficiency, and minimal adverse effects (Dua et al., 2019). However, siRNA-based therapies also have some limitations, including potential off-target effects and poor delivery into cells. Off-target effects can lead to unintended gene knockdown, resulting in cytotoxicity or harmful side effects. Delivery into cells is another significant hurdle, as siRNAs are unable to cross the cell membrane efficiently (Kanasty et al., 2013; Subhan and Torchilin, 2019). To address these challenges, exosome-based delivery systems have emerged as a promising approach to the safe and efficient delivery of siRNA.Researchers have developed exosomes that express specific targeting molecules on their surface. For example, HEK293T cells can generate exosomes containing the targeting molecules DARPinG3 and lamp2b, fused with the fragment of IL3 produced on the surface of exosomes. When loaded with sitpd52, these exosomes are capable of downregulating target gene expression by up to 70% or inhibiting cancer cell growth in vitro and in vivo when loaded with imatinib or siBCR-ABL (Bellavia et al., 2017; Limoni et al., 2019). Unmodified exosomes may also have translational potential, studies have shown that exosomes loaded with hsiRNAs have better drug distribution relative to free hsiRNAs (Didiot et al., 2016).
To enhance the biostability of conventional siRNA, different chemical modifications have been utilized with considerable success. Alternatively, the absence of termini in circRNA offers a promising solution for developing exonuclease-resistant siRNA through a different approach. A study has shown that EVs engineered with rabies virus glycoprotein (RVG) have been effective in delivering therapeutic circRNAs specifically to the brain, resulting in functional recovery in non-human primate models of ischemic stroke (Yang et al., 2020b). LNPs, similar to those used in the COVID-19 mRNA vaccines (mRNA-1273 and BNT162b2), are also utilized for delivering circRNA vaccines and therapeutics (Wesselhoeft et al., 2019; Qu et al., 2022).
Mature mRNAs are well known for their crucial role in protein synthesis and their usefulness as transporters of genetic information to the location of the target cells. Recent research on severe acute respiratory syndrome coronavirus has found that exosomes loaded with lung mRNA encoding the spike protein of the virus demonstrated higher immune responses compared to liposomal counterparts. Notably, the exosomes remained stable even at room temperature and exhibited better in vivo distribution (Popowski et al., 2022).
LncRNAs possess a wide range of functional activities, presenting multiple avenues for therapeutic intervention, which necessitates tailoring the method of targeting to the specific mode of action of the lncRNA.A noteworthy advancement is the investigation of natural-antisense transcripts (NATs): lncRNAs transcribed in the reverse direction to coding genes, which suppress gene expression in a cis manner. Antisense oligonucleotides (ASOs) that target NATs, referred to as ‘antagoNATs’, have demonstrated remarkably promising preclinical outcomes for gene reactivation in the central nervous system (Modarresi et al., 2012). Likewise, exosomes can also be loaded with lncrnas, one article explores the regulation of exosomal lncRNA GAS5 on apoptosis of macrophages and vascular endothelial cells in atherosclerosis, which revealed that gas5 has potential therapeutic implications in the prevention of atherosclerosis (Chen et al., 2017b).
One of the primary advantages of CRISPR-Cas is its ability to target and edit DNA with high precision, enabling researchers to potentially cure genetic disorders by correcting the underlying genetic mutations responsible for the disease. At present, the CRISPR family has primarily utilized Cas9 and Cas12a for genome editing, which providing a valuable range of applications for CRISPR technology by utilizing just these two enzymes (Paul et al., 2020). Epithelial cell-derived microvesicles have been identified as safe and effective CRISPR/Cas9 carriers for treating cancer. These microvesicles demonstrate cancer cell tropism, leading to selective accumulation in tumors and exhibiting a synergistic anti-tumor effect (He et al., 2020).
Despite significant progress in the development of delivery systems for RNA therapeutics, including liposomes, polymers, and inorganic materials, delivering single RNA molecules remains a significant challenge for clinical applications. However, there is growing interest in the potential of exosomes as a superior delivery system, thanks to their low immunogenicity, excellent blood compatibility, and rapid delivery capabilities, making them an exciting area of ongoing research.
The transition from exclusively loading extracellular vesicles with RNA and drugs to jointly loading them with both demonstrates the immense potential of extracellular vesicles as carriers. Previous studies have highlighted challenges associated with separate loading of RNA and drugs, including limited transportation capacity and delivery instability. However, thanks to extensive scientific research and technological advancements, simultaneous co-loading of RNA and drugs into extracellular vesicles has emerged as a new Frontier in the field.
Currently, various cancer treatment options are available, including peer therapy, chemotherapy, hyperthermia, immunotherapy, photodynamic therapy, radiation therapy, stem cell transplantation, surgery, and targeted therapy. However, single chemotherapy drugs only target a limited range of molecules and pathways, leaving some unexpected mutations unaddressed. In this regard, non-extracellular co-delivery systems have been previously reported (Chung et al., 2021). Studies have investigated the potential of using RNAs and anticancer drugs together to enhance the efficacy of cancer treatment. Yin et al. employed multifunctional magnetic core-shell nanoparticles for simultaneous delivery of microRNA therapeutics and anticancer drugs, while Xu et al. used liposomes to co-deliver miR-101 and doxorubicin and Zhu and others developed matrix metalloproteinase 2-sensitive multifunctional polymeric micelles for tumor-specific co-delivery of siRNA and hydrophobic drugs (Zhu et al., 2014; Xu et al., 2017; Yin et al., 2018). The results of these studies showcase the potential of combining RNA and anticancer drug therapies in order to overcome chemoresistance, and provide a hopeful strategy for improving cancer treatment. Nevertheless, the utilization of this approach for combined therapeutics in a single nanocarrier is currently confined to preclinical research, mainly due to issues related to manufacturing and regulation (Eljack et al., 2022).
With the most well studied nanocarriers being mesoporous silica nanoparticles, dendrimers, polymers, liposomes, and gold nanoparticles, exosomes as carriers for co-delivery systems still many techniques stay at the laboratory research level, while exosomes have unique advantages as endogenous natural drug carriers due to their biological membrane penetration, which can improve the transport efficiency and targeting of drugs, low degradation rate, and high permeation retention effect (Zhao and Feng, 2015; Pan et al., 2019; Si et al., 2020).
Patients with colorectal cancer often develop resistance to 5-fluorouracil (5-FU), which leads to poor treatment outcomes. MiR-21 has been identified as a marker for human malignant tumors and is also associated with multiple drug resistance (Shi et al., 2010; Moridikia et al., 2018). In this study, engineered exosomes targeting HER2 were obtained by constructing HEK293 cells overexpressing HER2 antibody, and 5-FU and miR-21 inhibitor were loaded into the exosomes using electroporation and targeted to HER2-positive tumor cells. The results demonstrated that engineered exosomes effectively promoted the uptake of 5-FU-resistant HCT-116 cells and significantly downregulated miR-21 expression, leading to cell cycle arrest, reduced tumor proliferation, increased cell apoptosis, and rescued PTEN and hMSH2 expression. Furthermore, systemic administration of these exosomes significantly reduced tumor growth in a mouse model (Liang G. et al., 2020b).
In a recent study, a blood exosome-based superparamagnetic nanoparticle cluster was constructed that incorporates tumor-targeting functions into exosomes. The researchers then assembled chemotherapeutic drug doxorubicin (Dox) and cholesterol-modified single-stranded miRNA21 inhibitor (chol-miR21i) onto the exosome to create a nanoplatform that combines two different anticancer modalities. To ensure efficient release of the cargos, including RNAs, a cationic lipid-sensitive endosomolytic peptide known as L17E peptide was introduced into this exosome-based co-delivery system. By combining both drugs and RNAs, this dual therapy interferes with nuclear DNA activity and downregulates the expression of oncogenes, leading to a significant inhibition of tumor growth and fewer side effects. Overall, this study shows the potential for exosome-based therapies to deliver multiple treatment modalities for more effective cancer treatment (Zhan et al., 2020). And Figure 2 shows the effective antitumor therapy utilizing engineered exosomes with miR21i/chemo combination.
In addition, the research team also tested the use of exosomes as delivery vehicles for RNA and small molecule drugs in treating glioblastoma (GBM). They engineered the exosomes to co-load sicpla2 and metformin in an efficient manner. The resultant exos met/sicpla2 construct was shown to significantly inhibit mitochondrial energy metabolism and ATP production in GBM. The study also demonstrated that systemic administration of exos met/sicpla2 was capable of efficiently penetrating the blood-brain barrier (BBB) and accumulating specifically in GBM in patient-derived xenograft models (Zhan et al., 2022).
Exosomes there are limitations in their ability to specifically target certain recipient cell types. To address this issue, we have devised a strategy to isolate exosomes that exhibit increased binding to integrin αvβ3. This binding is facilitated by a modified version of a disintegrin and metalloproteinase 15 (A15) expressed on the exosomal membranes (A15-Exo). The A15-Exo were derived from monocyte-derived macrophages that were continuously activated by protein kinase C, resulting in cell-derived Exo exhibiting targeting properties and a 2.97-fold higher production yield. In vitro studies demonstrated that A15-Exo, co-loaded with Dox and Cho-miR159, exerted strong synergistic therapeutic effects on MDA-MB-231 cells. In vivo, vesicular delivery of miR159 and Dox effectively silenced the TCF-7 gene and exhibited improved anticancer effects without any adverse effects (Figure 3) (Gong et al., 2019).
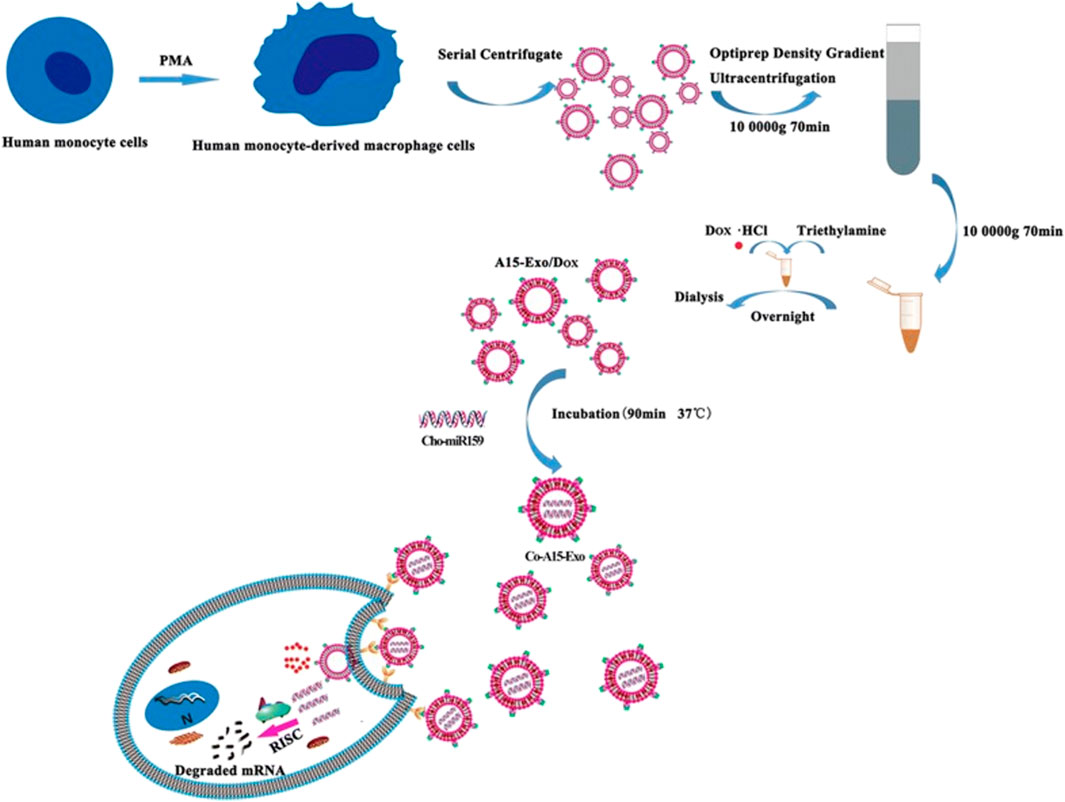
FIGURE 3. Formation and release of Dox and Cho-miR159-loaded A15-Exo (Co-A15-Exo) (Gong et al., 2019).
The combination of small molecule drugs with other therapeutic modalities that utilize unique mechanisms of action is emerging as a promising approach for overcoming drug-resistant cancers. Nucleic acids are often incorporated into such strategies due to their highly specific targeting capabilities. In order to facilitate the simultaneous delivery of drugs and nucleic acids, researchers have developed and refined smart nanocarriers that preserve the physicochemical characteristics and biological functions of both components. More importantly, successful integration of exosomes has shown promise for reversing chemoresistance.
Recent report has expressed doubts about the effectiveness of exosomes as a universal delivery system for RNAs, suggesting that anionic liposomes may perform better in this regard (Stremersch et al., 2016). Undoubtedly several challenges remain before meaningful clinical translation can be achieved, for example,
What are the standards for isolating, purifying, and quantifying exosomes? How should donor cells be selected, and what is the ideal cell culture environment to enable large-scale, industrial production of exosomes? While mesenchymal stem cells may be the first choice, are red blood cells, which are gene-free donor cells, the future stars?
Given that there is heterogeneity among different exosomes, how should exosomes be selected for drug or RNA loading methods? Will modifying exosomes affect their function and properties? After large-scale production of exosomes, what is the optimal long-term preservation method?
How do exosomes interact with the human internal environment, and what is their pharmacokinetic profile in body fluids?
The engineered production of exosomes involves multiple steps. Have quality control measures been put in place, and are personnel trained to ensure standardization?
Considering the aforementioned factors, it is important to address the challenges associated with co-loading RNA and drugs into exosomes. In addition to the factors mentioned earlier, achieving optimal loading capacity requires considering the appropriate ratio of drugs to RNA within the exosomes. Furthermore, it is essential to match the pH conditions that would result in the maximum release capacity for both the drugs and RNA within the exosomes. As mentioned in the literature, the researcher conducted a series of tests involving gradient concentrations of RNA and drugs, and explored different pH values in order to estimate the maximum release amount (Gong et al., 2019).
Before exosomes can be used for simultaneous RNA and drug delivery, several key issues must be addressed. A key issue is the identification and development of the optimal combination of RNA drugs for loading into extracellular vesicles, which not only requires specific targeting of disease sites, but also optimization of RNA drug ratios. Another challenge is to optimize drug release and extracellular transport mechanisms to effectively deliver drugs to target tissues. Further research is needed to modify exosomes to improve pharmacokinetics, such as half-life, biological distribution, and clearance rate.
Over the last few decades, researchers have increasingly focused on exploring the potential of exosomes as drug delivery systems. One emerging trend in cancer treatment involves utilizing combination therapy of drugs and nucleic acids delivered through exosomes. The clinicaltrials.gov resource (https://clinicaltrials.gov/ct2/home) indicates a significant number of clinical trial registrations for exosomes, including trials investigating the loading of exosomes with small molecule drugs (NCT01294072) and RNA molecules (NCT03384433). Exosomes offer several advantages as carriers of RNA and drugs. Firstly, they are naturally occurring and can be obtained from multiple cell types, providing a wide variety of targeting options while also potentially reducing immunogenicity. Secondly, exosomes are highly effective at transporting different molecules, including RNA and small molecules, in a protected manner which increases their stability and half-life. Lastly, exosomes possess tissue targeting mechanisms which allow them to deliver drugs directly to affected tissues and organs.
In summary, exosomes provide the potential to simultaneously load RNA and drugs for targeted and effective drug delivery.
HC: Writing–original draft, Writing–review and editing. HY: Writing–original draft, Writing–review and editing. JC: Writing–review and editing. CL: Writing–review and editing. YL: Writing–review and editing. JYa: Writing–review and editing. JYu: Writing–review and editing. JW: Writing–review and editing. YR: Writing–review and editing, Writing–original draft. JP: Writing–original draft, Writing–review and editing. J-FX: Writing–original draft, Writing–review and editing.
This work was supported by the National Natural Science Foundation of China (82272348, 82270013, 81870016 and 82300016), High Talent Project of Guangdong Province (2021QN02Y720), Natural Science Foundation of Guangdong Province (2023A1515030195, 2022A1515011223, and 2022A1515010525), Characteristic Innovation Project of Universities in Guangdong Province (2021KTSCX038), Key Project of Universities in Guangdong Province (2022ZDZX 2021), Innovation Team Project of Universities in Guangdong Province (2022KCXTD010), Science and Technology Project of Dongguan (20211800904782 and 20211800905542), Doctoral Initial Funding of Guangdong Medical University (4SG22209G), Discipline Construction Project of Guangdong Medical University (4SG23290G and 4SG21229GDGFY01), Open Research Fund of Songshan Lake Materials Laboratory (2021SLABFN10), Open Research Fund for Key Laboratory of Tropical Disease Control Sun Yat-sen University, Ministry of Enducation (2022kfkt01), and Youth Research Projects of Guangdong Medical University (GDMUD2022001).
We tender our apologies to those authors whose deserving research was not cited in this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agrawal, A. K., Aqil, F., Jeyabalan, J., Spencer, W. A., Beck, J., Gachuki, B. W., et al. (2017). Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 13 (5), 1627–1636. doi:10.1016/j.nano.2017.03.001
Alhasan, A. H., Patel, P. C., Choi, C. H., and Mirkin, C. A. (2014). Exosome encased spherical nucleic acid gold nanoparticle conjugates as potent microRNA regulation agents. Small 10 (1), 186–192. doi:10.1002/smll.201302143
Alipoor, S. D., Tabarsi, P., Varahram, M., Movassaghi, M., Dizaji, M. K., Folkerts, G., et al. (2019). Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Pulm. Tuberc. Dis. Markers 2019, 1907426. doi:10.1155/2019/1907426
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29 (4), 341–345. doi:10.1038/nbt.1807
Amreddy, N., Babu, A., Muralidharan, R., Panneerselvam, J., Srivastava, A., Ahmed, R., et al. (2018). Recent advances in nanoparticle-based cancer drug and gene delivery. Adv. Cancer Res. 137, 115–170. doi:10.1016/bs.acr.2017.11.003
Aqil, F., Jeyabalan, J., Agrawal, A. K., Kyakulaga, A. H., Munagala, R., Parker, L., et al. (2017). Exosomal delivery of berry anthocyanidins for the management of ovarian cancer. Food Funct. 8 (11), 4100–4107. doi:10.1039/c7fo00882a
Aqil, F., Kausar, H., Agrawal, A. K., Jeyabalan, J., Kyakulaga, A. H., Munagala, R., et al. (2016). Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp. Mol. Pathol. 101 (1), 12–21. doi:10.1016/j.yexmp.2016.05.013
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Batsali, A. K., Georgopoulou, A., Mavroudi, I., Matheakakis, A., Pontikoglou, C. G., and Papadaki, H. A. (2020). The role of bone marrow mesenchymal stem cell derived extracellular vesicles (MSC-EVs) in normal and abnormal hematopoiesis and their therapeutic potential. J. Clin. Med. 9 (3), 856. doi:10.3390/jcm9030856
Bellavia, D., Raimondo, S., Calabrese, G., Forte, S., Cristaldi, M., Patinella, A., et al. (2017). Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 7 (5), 1333–1345. doi:10.7150/thno.17092
Bellotti, C., Lang, K., Kuplennik, N., Sosnik, A., and Steinfeld, R. (2021). High-grade extracellular vesicles preparation by combined size-exclusion and affinity chromatography. Sci. Rep. 11 (1), 10550. doi:10.1038/s41598-021-90022-y
Bettegowda, C., Sausen, M., Leary, R. J., Kinde, I., Wang, Y., Agrawal, N., et al. (2014). Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6 (224), 224ra24. doi:10.1126/scitranslmed.3007094
Chen, L., Yang, W., Guo, Y., Chen, W., Zheng, P., Zeng, J., et al. (2017b). Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One 12 (9), e0185406. doi:10.1371/journal.pone.0185406
Chen, W., Li, L., Zhang, X., Liang, Y., Pu, Z., Wang, L., et al. (2017a). Curcumin: A calixarene derivative micelle potentiates anti-breast cancer stem cells effects in xenografted, triple-negative breast cancer mouse models. Drug Deliv. 24 (1), 1470–1481. doi:10.1080/10717544.2017.1381198
Chen, Y. A., and Scheller, R. H. (2001). SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2 (2), 98–106. doi:10.1038/35052017
Cheng, L., Wang, Y., and Huang, L. (2017). Exosomes from M1-polarized macrophages potentiate the cancer vaccine by creating a pro-inflammatory microenvironment in the lymph node. Mol. Ther. 25 (7), 1665–1675. doi:10.1016/j.ymthe.2017.02.007
Cheng, Y., and Schorey, J. S. (2016). Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol. Bioeng. 113 (6), 1315–1324. doi:10.1002/bit.25884
Cheng, Y., Zeng, Q., Han, Q., and Xia, W. (2019). Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 10 (4), 295–299. doi:10.1007/s13238-018-0529-4
Cho, S., Yang, H. C., and Rhee, W. J. (2019). Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis. Biosens. Bioelectron. 146, 111749. doi:10.1016/j.bios.2019.111749
Chung, S. L., Yee, M. S., Hii, L. W., Lim, W. M., Ho, M. Y., Khiew, P. S., et al. (2021). Advances in nanomaterials used in Co-delivery of siRNA and small molecule drugs for cancer treatment. Nanomater. (Basel) 11 (10), 2467. doi:10.3390/nano11102467
Colombo, M., Raposo, G., and Thery, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. doi:10.1146/annurev-cellbio-101512-122326
Cvjetkovic, A., Lotvall, J., and Lasser, C. (2014). The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 3. doi:10.3402/jev.v3.23111
Dai, J., Su, Y., Zhong, S., Cong, L., Liu, B., Yang, J., et al. (2020). Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target Ther. 5 (1), 145. doi:10.1038/s41392-020-00261-0
Debbi, L., Guo, S., Safina, D., and Levenberg, S. (2022). Boosting extracellular vesicle secretion. Biotechnol. Adv. 59, 107983. doi:10.1016/j.biotechadv.2022.107983
Didiot, M. C., Hall, L. M., Coles, A. H., Haraszti, R. A., Godinho, B. M., Chase, K., et al. (2016). Exosome-mediated delivery of hydrophobically modified siRNA for huntingtin mRNA silencing. Mol. Ther. 24 (10), 1836–1847. doi:10.1038/mt.2016.126
Dua, K., Wadhwa, R., Singhvi, G., Rapalli, V., Shukla, S. D., Shastri, M. D., et al. (2019). The potential of siRNA based drug delivery in respiratory disorders: recent advances and progress. Drug Dev. Res. 80 (6), 714–730. doi:10.1002/ddr.21571
Duan, L., Xu, L., Xu, X., Qin, Z., Zhou, X., Xiao, Y., et al. (2021). Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 13 (3), 1387–1397. doi:10.1039/d0nr07622h
Eljack, S., David, S., Faggad, A., Chourpa, I., and Allard-Vannier, E. (2022). Nanoparticles design considerations to co-deliver nucleic acids and anti-cancer drugs for chemoresistance reversal. Int. J. Pharm. X 4, 100126. doi:10.1016/j.ijpx.2022.100126
Escudier, B., Dorval, T., Chaput, N., Andre, F., Caby, M. P., Novault, S., et al. (2005). Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J. Transl. Med. 3 (1), 10. doi:10.1186/1479-5876-3-10
Esfandyari, S., Elkafas, H., Chugh, R. M., Park, H. S., Navarro, A., and Al-Hendy, A. (2021). Exosomes as biomarkers for female reproductive diseases diagnosis and therapy. Int. J. Mol. Sci. 22 (4), 2165. doi:10.3390/ijms22042165
Esteves, M., Abreu, R., Fernandes, H., Serra-Almeida, C., Martins, P. A. T., Barao, M., et al. (2022). MicroRNA-124-3p-enriched small extracellular vesicles as a therapeutic approach for Parkinson's disease. Mol. Ther. 30 (10), 3176–3192. doi:10.1016/j.ymthe.2022.06.003
Fuhrmann, G., Serio, A., Mazo, M., Nair, R., and Stevens, M. M. (2015). Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control Release 205, 35–44. doi:10.1016/j.jconrel.2014.11.029
Gao, Z. S., Zhang, C. J., Xia, N., Tian, H., Li, D. Y., Lin, J. Q., et al. (2021). Berberine-loaded M2 macrophage-derived exosomes for spinal cord injury therapy. Acta Biomater. 126, 211–223. doi:10.1016/j.actbio.2021.03.018
Gong, C., Tian, J., Wang, Z., Gao, Y., Wu, X., Ding, X., et al. (2019). Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnology 17 (1), 93. doi:10.1186/s12951-019-0526-7
Gordon, D. E., Chia, J., Jayawardena, K., Antrobus, R., Bard, F., and Peden, A. A. (2017). VAMP3/Syb and YKT6 are required for the fusion of constitutive secretory carriers with the plasma membrane. PLoS Genet. 13 (4), e1006698. doi:10.1371/journal.pgen.1006698
Gorji-Bahri, G., Moghimi, H. R., and Hashemi, A. (2021). RAB5A is associated with genes involved in exosome secretion: integration of bioinformatics analysis and experimental validation. J. Cell Biochem. 122 (3-4), 425–441. doi:10.1002/jcb.29871
Guo, H., Chitiprolu, M., Roncevic, L., Javalet, C., Hemming, F. J., Trung, M. T., et al. (2017). Atg5 disassociates the V(1)V(0)-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell 43 (6), 716–730.e7. doi:10.1016/j.devcel.2017.11.018
Guo, M., Hao, Y., Feng, Y., Li, H., Mao, Y., Dong, Q., et al. (2021). Microglial exosomes in neurodegenerative disease. Front. Mol. Neurosci. 14, 630808. doi:10.3389/fnmol.2021.630808
Guo, N., Liu, J. B., Li, W., Ma, Y. S., and Fu, D. (2022). The power and the promise of CRISPR/Cas9 genome editing for clinical application with gene therapy. J. Adv. Res. 40, 135–152. doi:10.1016/j.jare.2021.11.018
Gupta, A., Andresen, J. L., Manan, R. S., and Langer, R. (2021). Nucleic acid delivery for therapeutic applications. Adv. Drug Deliv. Rev. 178, 113834. doi:10.1016/j.addr.2021.113834
Gurung, S., Perocheau, D., Touramanidou, L., and Baruteau, J. (2021). The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal 19 (1), 47. doi:10.1186/s12964-021-00730-1
Ha, D., Yang, N., and Nadithe, V. (2016). Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B 6 (4), 287–296. doi:10.1016/j.apsb.2016.02.001
Han, Q., Lv, L., Wei, J., Lei, X., Lin, H., Li, G., et al. (2019). Vps4A mediates the localization and exosome release of beta-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 457, 47–59. doi:10.1016/j.canlet.2019.04.035
Han, Q. F., Li, W. J., Hu, K. S., Gao, J., Zhai, W. L., Yang, J. H., et al. (2022). Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 21 (1), 207. doi:10.1186/s12943-022-01671-0
Haney, M. J., Klyachko, N. L., Zhao, Y., Gupta, R., Plotnikova, E. G., He, Z., et al. (2015). Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Control Release 207, 18–30. doi:10.1016/j.jconrel.2015.03.033
Hannafon, B. N., Trigoso, Y. D., Calloway, C. L., Zhao, Y. D., Lum, D. H., Welm, A. L., et al. (2016). Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 18 (1), 90. doi:10.1186/s13058-016-0753-x
Haraszti, R. A., Miller, R., Stoppato, M., Sere, Y. Y., Coles, A., Didiot, M. C., et al. (2018). Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol. Ther. 26 (12), 2838–2847. doi:10.1016/j.ymthe.2018.09.015
He, C., Jaffar Ali, D., Xu, H., Kumaravel, S., Si, K., Li, Y., et al. (2020). Epithelial cell -derived microvesicles: A safe delivery platform of CRISPR/cas9 conferring synergistic anti-tumor effect with sorafenib. Exp. Cell Res. 392 (2), 112040. doi:10.1016/j.yexcr.2020.112040
He, C., Zheng, S., Luo, Y., and Wang, B. (2018). Exosome theranostics: biology and translational medicine. Theranostics 8 (1), 237–255. doi:10.7150/thno.21945
He, J., Ren, M., Li, H., Yang, L., Wang, X., and Yang, Q. (2019). Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front. Genet. 10, 860. doi:10.3389/fgene.2019.00860
Hikita, T., and Oneyama, C. (2022). Quantification and imaging of exosomes via luciferase-fused exosome marker proteins: exoLuc system. Methods Mol. Biol. 2524, 281–290. doi:10.1007/978-1-0716-2453-1_21
Hu, X., Liao, S., Bai, H., Wu, L., Wang, M., Wu, Q., et al. (2019). Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine 40, 564–573. doi:10.1016/j.ebiom.2019.01.023
Huang, L., Rong, Y., Tang, X., Yi, K., Qi, P., Hou, J., et al. (2022). Engineered exosomes as an in situ DC-primed vaccine to boost antitumor immunity in breast cancer. Mol. Cancer 21 (1), 45. doi:10.1186/s12943-022-01515-x
Huang, Z., Zhang, Y., Zhou, J., and Zhang, Y. (2017). Urinary exosomal miR-193a can Be a potential biomarker for the diagnosis of primary focal segmental Glomerulosclerosis in children. Biomed. Res. Int. 2017, 1–6. doi:10.1155/2017/7298160
Jahn, R., and Sudhof, T. C. (1999). Membrane fusion and exocytosis. Annu. Rev. Biochem. 68, 863–911. doi:10.1146/annurev.biochem.68.1.863
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of exosome composition. Cell 177 (2), 428–445.e18. doi:10.1016/j.cell.2019.02.029
Jesus, S., Soares, E., Cruz, M. T., and Borges, O. (2018). Exosomes as adjuvants for the recombinant hepatitis B antigen:first report. Eur. J. Pharm. Biopharm. 133, 1–11. doi:10.1016/j.ejpb.2018.09.029
Ji, J., Chen, R., Zhao, L., Xu, Y., Cao, Z., Xu, H., et al. (2021). Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol. Cancer 20 (1), 58. doi:10.1186/s12943-021-01349-z
Jia, G., Han, Y., An, Y., Ding, Y., He, C., Wang, X., et al. (2018). NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 178, 302–316. doi:10.1016/j.biomaterials.2018.06.029
Jia, L., Qiu, Q., Zhang, H., Chu, L., Du, Y., Zhang, J., et al. (2019). Concordance between the assessment of Aβ42, T-tau, and P-T181-tau in peripheral blood neuronal-derived exosomes and cerebrospinal fluid. Alzheimers Dement. 15 (8), 1071–1080. doi:10.1016/j.jalz.2019.05.002
Jiang, L., Vader, P., and Schiffelers, R. M. (2017). Extracellular vesicles for nucleic acid delivery: progress and prospects for safe RNA-based gene therapy. Gene Ther. 24 (3), 157–166. doi:10.1038/gt.2017.8
Jin, X., Chen, Y., Chen, H., Fei, S., Chen, D., Cai, X., et al. (2017). Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 23 (17), 5311–5319. doi:10.1158/1078-0432.ccr-17-0577
Jin, Y., Ma, L., Zhang, W., Yang, W., Feng, Q., and Wang, H. (2022). Extracellular signals regulate the biogenesis of extracellular vesicles. Biol. Res. 55 (1), 35. doi:10.1186/s40659-022-00405-2
Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L., and Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262 (19), 9412–9420. doi:10.1016/s0021-9258(18)48095-7
Kamerkar, S., Leng, C., Burenkova, O., Jang, S. C., McCoy, C., Zhang, K., et al. (2022). Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv. 8 (7), eabj7002. doi:10.1126/sciadv.abj7002
Kanasty, R., Dorkin, J. R., Vegas, A., and Anderson, D. (2013). Delivery materials for siRNA therapeutics. Nat. Mater 12 (11), 967–977. doi:10.1038/nmat3765
Khalifa, A. M., Elsheikh, M. A., Khalifa, A. M., and Elnaggar, Y. S. R. (2019). Current strategies for different paclitaxel-loaded nano-delivery systems towards therapeutic applications for ovarian carcinoma: A review article. J. Control Release 311-312, 125–137. doi:10.1016/j.jconrel.2019.08.034
Kim, D. H., and Rossi, J. J. (2007). Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8 (3), 173–184. doi:10.1038/nrg2006
Kim, M. S., Haney, M. J., Zhao, Y., Mahajan, V., Deygen, I., Klyachko, N. L., et al. (2016). Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 12 (3), 655–664. doi:10.1016/j.nano.2015.10.012
Kobayashi, M., Sawada, K., Miyamoto, M., Shimizu, A., Yamamoto, M., Kinose, Y., et al. (2020). Exploring the potential of engineered exosomes as delivery systems for tumor-suppressor microRNA replacement therapy in ovarian cancer. Biochem. Biophys. Res. Commun. 527 (1), 153–161. doi:10.1016/j.bbrc.2020.04.076
Kooijmans, S. A. A., Stremersch, S., Braeckmans, K., de Smedt, S. C., Hendrix, A., Wood, M. J. A., et al. (2013). Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control Release 172 (1), 229–238. doi:10.1016/j.jconrel.2013.08.014
Koyama, Y., Ito, T., Hasegawa, A., Eriguchi, M., Inaba, T., Ushigusa, T., et al. (2016). Exosomes derived from tumor cells genetically modified to express Mycobacterium tuberculosis antigen: A novel vaccine for cancer therapy. Biotechnol. Lett. 38 (11), 1857–1866. doi:10.1007/s10529-016-2185-1
Lai, J. J., Chau, Z. L., Chen, S. Y., Hill, J. J., Korpany, K. V., Liang, N. W., et al. (2022). Exosome processing and characterization approaches for research and technology development. Adv. Sci. (Weinh) 9 (15), e2103222. doi:10.1002/advs.202103222
Leung, P. S., Dhirapong, A., Wu, P. Y., and Tao, M. H. (2010). Gene therapy in autoimmune diseases: challenges and opportunities. Autoimmun. Rev. 9 (3), 170–174. doi:10.1016/j.autrev.2009.10.004
Lewis, J. M., Vyas, A. D., Qiu, Y., Messer, K. S., White, R., and Heller, M. J. (2018). Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood. ACS Nano 12 (4), 3311–3320. doi:10.1021/acsnano.7b08199
Li, L., Lu, S., Liang, X., Cao, B., Wang, S., Jiang, J., et al. (2019). γδTDEs: an efficient delivery system for miR-138 with anti-tumoral and immunostimulatory roles on oral squamous cell carcinoma. Mol. Ther. Nucleic Acids 14, 101–113. doi:10.1016/j.omtn.2018.11.009
Li, S., Zhao, Y., Chen, W., Yin, L., Zhu, J., Zhang, H., et al. (2018a). Exosomal ephrinA2 derived from serum as a potential biomarker for prostate cancer. J. Cancer 9 (15), 2659–2665. doi:10.7150/jca.25201
Li, Y., Liu, Y., Xiu, F., Wang, J., Cong, H., He, S., et al. (2018b). Characterization of exosomes derived from <em>Toxoplasma gondii</em> and their functions in modulating immune responses. Int. J. Nanomedicine 13, 467–477. doi:10.2147/ijn.s151110
Liang, G., Zhu, Y., Ali, D. J., Tian, T., Xu, H., Si, K., et al. (2020b). Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnology 18 (1), 10. doi:10.1186/s12951-019-0563-2
Liang, Y., Duan, L., Lu, J., and Xia, J. (2021). Engineering exosomes for targeted drug delivery. Theranostics 11 (7), 3183–3195. doi:10.7150/thno.52570
Liang, Y., Xu, X., Li, X., Xiong, J., Li, B., Duan, L., et al. (2020a). Chondrocyte-targeted MicroRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Appl. Mater Interfaces 12 (33), 36938–36947. doi:10.1021/acsami.0c10458
Limoni, S. K., Moghadam, M. F., Moazzeni, S. M., Gomari, H., and Salimi, F. (2019). Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl. Biochem. Biotechnol. 187 (1), 352–364. doi:10.1007/s12010-018-2813-4
Lin, D., Shen, L., Luo, M., Zhang, K., Li, J., Yang, Q., et al. (2021). Circulating tumor cells: biology and clinical significance. Signal Transduct. Target Ther. 6 (1), 404. doi:10.1038/s41392-021-00817-8
Ling, H., Guo, Z., Du, S., Liao, Y., Li, Y., Ding, C., et al. (2020a). Serum exosomal miR-122-5p is a new biomarker for both acute coronary syndrome and underlying coronary artery stenosis. Biomarkers 25 (7), 539–547. doi:10.1080/1354750x.2020.1803963
Ling, H., Guo, Z., Shi, Y., Zhang, L., and Song, C. (2020b). Serum exosomal MicroRNA-21, MicroRNA-126, and PTEN are novel biomarkers for diagnosis of acute coronary syndrome. Front. Physiol. 11, 654. doi:10.3389/fphys.2020.00654
Liu, J., Yoo, J., Ho, J. Y., Jung, Y., Lee, S., Hur, S. Y., et al. (2021). Plasma-derived exosomal miR-4732-5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J. Ovarian Res. 14 (1), 59. doi:10.1186/s13048-021-00814-z
Lone, S. N., Nisar, S., Masoodi, T., Singh, M., Rizwan, A., Hashem, S., et al. (2022). Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 21 (1), 79. doi:10.1186/s12943-022-01543-7
Lu, J., Wu, J., Xie, F., Tian, J., Tang, X., Guo, H., et al. (2019). CD4(+) T cell-released extracellular vesicles potentiate the efficacy of the HBsAg vaccine by enhancing B cell responses. Adv. Sci. (Weinh) 6 (23), 1802219. doi:10.1002/advs.201802219
Lu, Y., Duan, Y., Xu, Q., Zhang, L., Chen, W., Qu, Z., et al. (2020). Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J. Cell Mol. Med. 24 (2), 1311–1318. doi:10.1111/jcmm.14783
Lu, Z., Zuo, B., Jing, R., Gao, X., Rao, Q., Liu, Z., et al. (2017). Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 67 (4), 739–748. doi:10.1016/j.jhep.2017.05.019
Lv, C. Y., Ding, W. J., Wang, Y. L., Zhao, Z. Y., Li, J. H., Chen, Y., et al. (2018). A PEG-based method for the isolation of urinary exosomes and its application in renal fibrosis diagnostics using cargo miR-29c and miR-21 analysis. Int. Urol. Nephrol. 50 (5), 973–982. doi:10.1007/s11255-017-1779-4
Mainardes, R. M., and Silva, L. P. (2004). Drug delivery systems: past, present, and future. Curr. Drug Targets 5 (5), 449–455. doi:10.2174/1389450043345407
Marshall, M. R., Pattu, V., Halimani, M., Maier-Peuschel, M., Muller, M. L., Becherer, U., et al. (2015). VAMP8-dependent fusion of recycling endosomes with the plasma membrane facilitates T lymphocyte cytotoxicity. J. Cell Biol. 210 (1), 135–151. doi:10.1083/jcb.201411093
Mathieu, M., Martin-Jaular, L., Lavieu, G., and Thery, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21 (1), 9–17. doi:10.1038/s41556-018-0250-9
Meng, F., Wang, J., and Yeo, Y. (2022). Nucleic acid and oligonucleotide delivery for activating innate immunity in cancer immunotherapy. J. Control Release 345, 586–600. doi:10.1016/j.jconrel.2022.03.045
Min, L., Zhu, S., Chen, L., Liu, X., Wei, R., Zhao, L., et al. (2019). Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: A comparison with plasma total miRNAs. J. Extracell. Vesicles 8 (1), 1643670. doi:10.1080/20013078.2019.1643670
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30 (5), 453–459. doi:10.1038/nbt.2158
Moridikia, A., Mirzaei, H., Sahebkar, A., and Salimian, J. (2018). MicroRNAs: potential candidates for diagnosis and treatment of colorectal cancer. J. Cell Physiol. 233 (2), 901–913. doi:10.1002/jcp.25801
Mulcahy, L. A., Pink, R. C., and Carter, D. R. (2014). Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3. doi:10.3402/jev.v3.24641
Murrow, L., Malhotra, R., and Debnath, J. (2015). ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17 (3), 300–310. doi:10.1038/ncb3112
Nakano, T., Chen, I. H., Wang, C. C., Chen, P. J., Tseng, H. P., Huang, K. T., et al. (2019). Circulating exosomal miR-92b: its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am. J. Transpl. 19 (12), 3250–3262. doi:10.1111/ajt.15490
Nakase, I., and Futaki, S. (2015). Combined treatment with a pH-sensitive fusogenic peptide and cationic lipids achieves enhanced cytosolic delivery of exosomes. Sci. Rep. 5, 10112. doi:10.1038/srep10112
Nie, H., Xie, X., Zhang, D., Zhou, Y., Li, B., Li, F., et al. (2020). Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 12 (2), 877–887. doi:10.1039/c9nr09011h
Ohno, S., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21 (1), 185–191. doi:10.1038/mt.2012.180
Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Raposo, G., Savina, A., et al. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12 (1), 19–30. doi:10.1038/ncb2000
Pan, B., Qin, J., Liu, X., He, B., Wang, X., Pan, Y., et al. (2019). Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front. Genet. 10, 1096. doi:10.3389/fgene.2019.01096
Pandit, R., Chen, L., and Gotz, J. (2020). The blood-brain barrier: physiology and strategies for drug delivery. Adv. Drug Deliv. Rev. 165-166, 1–14. doi:10.1016/j.addr.2019.11.009
Paskeh, M. D. A., Entezari, M., Mirzaei, S., Zabolian, A., Saleki, H., Naghdi, M. J., et al. (2022). Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 15 (1), 83. doi:10.1186/s13045-022-01305-4
Paul, B., Montoya, G., and Crispr-Cas12a, (2020). CRISPR-Cas12a: functional overview and applications. Biomed. J. 43 (1), 8–17. doi:10.1016/j.bj.2019.10.005
Pessoa, L. S., Heringer, M., and Ferrer, V. P. (2020). ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 155, 103109. doi:10.1016/j.critrevonc.2020.103109
Poller, W., Dimmeler, S., Heymans, S., Zeller, T., Haas, J., Karakas, M., et al. (2018). Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur. Heart J. 39 (29), 2704–2716. doi:10.1093/eurheartj/ehx165
Pols, M. S., and Klumperman, J. (2009). Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315 (9), 1584–1592. doi:10.1016/j.yexcr.2008.09.020
Popowski, K. D., Moatti, A., Scull, G., Silkstone, D., Lutz, H., Lopez de Juan Abad, B., et al. (2022). Inhalable dry powder mRNA vaccines based on extracellular vesicles. Matter 5 (9), 2960–2974. doi:10.1016/j.matt.2022.06.012
Pryor, P. R., Mullock, B. M., Bright, N. A., Lindsay, M. R., Gray, S. R., Richardson, S. C., et al. (2004). Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5 (6), 590–595. doi:10.1038/sj.embor.7400150
Qi, H., Liu, C., Long, L., Ren, Y., Zhang, S., Chang, X., et al. (2016). Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano 10 (3), 3323–3333. doi:10.1021/acsnano.5b06939
Qu, L., Yi, Z., Shen, Y., Lin, L., Chen, F., Xu, Y., et al. (2022). Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185 (10), 1728–1744.e16. doi:10.1016/j.cell.2022.03.044
Rezaie, J., Feghhi, M., and Etemadi, T. (2022). A review on exosomes application in clinical trials: perspective, questions, and challenges. Cell Commun. Signal 20 (1), 145. doi:10.1186/s12964-022-00959-4
Rodriguez, M., Bajo-Santos, C., Hessvik, N. P., Lorenz, S., Fromm, B., Berge, V., et al. (2017). Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 16 (1), 156. doi:10.1186/s12943-017-0726-4
Saber-Moghaddam, N., Salari, S., Hejazi, S., Amini, M., Taherzadeh, Z., Eslami, S., et al. (2021). Oral nano-curcumin formulation efficacy in management of mild to moderate hospitalized coronavirus disease-19 patients: an open label nonrandomized clinical trial. Phytother. Res. 35, 2616–2623. doi:10.1002/ptr.7004
Salunkhe, S., Dheeraj, , Basak, M., Chitkara, D., and Mittal, A. (2020). Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: strategies and significance. J. Control Release 326, 599–614. doi:10.1016/j.jconrel.2020.07.042
Santos, P., and Almeida, F. (2021). Exosome-based vaccines: history, current state, and clinical trials. Front. Immunol. 12, 711565. doi:10.3389/fimmu.2021.711565
Sato, M., Sato, K., Liou, W., Pant, S., Harada, A., and Grant, B. D. (2008). Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27 (8), 1183–1196. doi:10.1038/emboj.2008.54
Savjani, K. T., Gajjar, A. K., and Savjani, J. K. (2012). Drug solubility: importance and enhancement techniques. ISRN Pharm. 2012, 1–10. doi:10.5402/2012/195727
Schorey, J. S., Cheng, Y., Singh, P. P., and Smith, V. L. (2015). Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 16 (1), 24–43. doi:10.15252/embr.201439363
Sehrawat, T. S., Arab, J. P., Liu, M., Amrollahi, P., Wan, M., Fan, J., et al. (2021). Circulating extracellular vesicles carrying sphingolipid cargo for the diagnosis and dynamic risk profiling of alcoholic hepatitis. Hepatology 73 (2), 571–585. doi:10.1002/hep.31256
Shi, G. H., Ye, D. W., Yao, X. D., Zhang, S. L., Dai, B., Zhang, H. L., et al. (2010). Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol. Sin. 31 (7), 867–873. doi:10.1038/aps.2010.48
Si, Y., Kim, S., Zhang, E., Tang, Y., Jaskula-Sztul, R., Markert, J. M., et al. (2020). Targeted exosomes for drug delivery: biomanufacturing, surface tagging, and validation. Biotechnol. J. 15 (1), e1900163. doi:10.1002/biot.201900163
Smyth, T., Petrova, K., Payton, N. M., Persaud, I., Redzic, J. S., Graner, M. W., et al. (2014). Surface functionalization of exosomes using click chemistry. Bioconjug Chem. 25 (10), 1777–1784. doi:10.1021/bc500291r
Steegmaier, M., Klumperman, J., Foletti, D. L., Yoo, J. S., and Scheller, R. H. (1999). Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol. Biol. Cell 10 (6), 1957–1972. doi:10.1091/mbc.10.6.1957
Stremersch, S., Vandenbroucke, R. E., Van Wonterghem, E., Hendrix, A., De Smedt, S. C., and Raemdonck, K. (2016). Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control Release 232, 51–61. doi:10.1016/j.jconrel.2016.04.005
Subhan, M. A., and Torchilin, V. P. (2019). Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl. Res. 214, 62–91. doi:10.1016/j.trsl.2019.07.006
Sun, C., Wang, P., Dong, W., Liu, H., Sun, J., and Zhao, L. (2020). LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging (Albany NY) 12 (11), 10427–10440. doi:10.18632/aging.103268
Sun, D., Zhuang, X., Xiang, X., Liu, Y., Zhang, S., Liu, C., et al. (2010). A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 18 (9), 1606–1614. doi:10.1038/mt.2010.105
Tamai, K., Shiina, M., Tanaka, N., Nakano, T., Yamamoto, A., Kondo, Y., et al. (2012). Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 422 (2), 377–385. doi:10.1016/j.virol.2011.11.009
Tamai, K., Tanaka, N., Nakano, T., Kakazu, E., Kondo, Y., Inoue, J., et al. (2010). Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 399 (3), 384–390. doi:10.1016/j.bbrc.2010.07.083
Tamura, R., Uemoto, S., and Tabata, Y. (2017). Augmented liver targeting of exosomes by surface modification with cationized pullulan. Acta Biomater. 57, 274–284. doi:10.1016/j.actbio.2017.05.013
Tenchov, R., Sasso, J. M., Wang, X., Liaw, W. S., Chen, C. A., and Zhou, Q. A. (2022). Exosomes─Nature’s lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS Nano 16 (11), 17802–17846. doi:10.1021/acsnano.2c08774
Thery, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7 (1), 1535750. doi:10.1080/20013078.2018.1535750
Tian, Y., Li, S., Song, J., Ji, T., Zhu, M., Anderson, G. J., et al. (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35 (7), 2383–2390. doi:10.1016/j.biomaterials.2013.11.083
Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., et al. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319 (5867), 1244–1247. doi:10.1126/science.1153124
Tran, P. H. L., Wang, T., Yin, W., Tran, T. T. D., Barua, H. T., Zhang, Y., et al. (2019). Development of a nanoamorphous exosomal delivery system as an effective biological platform for improved encapsulation of hydrophobic drugs. Int. J. Pharm. 566, 697–707. doi:10.1016/j.ijpharm.2019.06.028
Usman, W. M., Pham, T. C., Kwok, Y. Y., Vu, L. T., Ma, V., Peng, B., et al. (2018). Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 9 (1), 2359. doi:10.1038/s41467-018-04791-8
van Niel, G., Charrin, S., Simoes, S., Romao, M., Rochin, L., Saftig, P., et al. (2011). The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21 (4), 708–721. doi:10.1016/j.devcel.2011.08.019
Visan, K. S., Lobb, R. J., Ham, S., Lima, L. G., Palma, C., Edna, C. P. Z., et al. (2022). Comparative analysis of tangential flow filtration and ultracentrifugation, both combined with subsequent size exclusion chromatography, for the isolation of small extracellular vesicles. J. Extracell. Vesicles 11 (9), e12266. doi:10.1002/jev2.12266
Wang, N., Li, X., Zhong, Z., Qiu, Y., Liu, S., Wu, H., et al. (2021). 3D hESC exosomes enriched with miR-6766-3p ameliorates liver fibrosis by attenuating activated stellate cells through targeting the TGFβRII-SMADS pathway. J. Nanobiotechnology 19 (1), 437. doi:10.1186/s12951-021-01138-2
Wang, P., Wang, H., Huang, Q., Peng, C., Yao, L., Chen, H., et al. (2019). Exosomes from M1-polarized macrophages enhance paclitaxel antitumor activity by activating macrophages-mediated inflammation. Theranostics 9 (6), 1714–1727. doi:10.7150/thno.30716
Wang, W., Zhu, N., Yan, T., Shi, Y. N., Chen, J., Zhang, C. J., et al. (2020). The crosstalk: exosomes and lipid metabolism. Cell Commun. Signal 18 (1), 119. doi:10.1186/s12964-020-00581-2
Wang, X., Zhang, H., Bai, M., Ning, T., Ge, S., Deng, T., et al. (2018). Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol. Ther. 26 (3), 774–783. doi:10.1016/j.ymthe.2018.01.001
Wei, H., Chen, J., Wang, S., Fu, F., Zhu, X., Wu, C., et al. (2019). <p>A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro</p>. Int. J. Nanomedicine 14, 8603–8610. doi:10.2147/ijn.s218988
Wei, J. X., Lv, L. H., Wan, Y. L., Cao, Y., Li, G. L., Lin, H. M., et al. (2015). Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology 61 (4), 1284–1294. doi:10.1002/hep.27660
Wei, P., Wu, F., Kang, B., Sun, X., Heskia, F., Pachot, A., et al. (2020b). Plasma extracellular vesicles detected by Single Molecule array technology as a liquid biopsy for colorectal cancer. J. Extracell. Vesicles 9 (1), 1809765. doi:10.1080/20013078.2020.1809765
Wei, S., Peng, L., Yang, J., Sang, H., Jin, D., Li, X., et al. (2020a). Exosomal transfer of miR-15b-3p enhances tumorigenesis and malignant transformation through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric cancer. J. Exp. Clin. Cancer Res. 39 (1), 32. doi:10.1186/s13046-019-1511-6
Wesselhoeft, R. A., Kowalski, P. S., Parker-Hale, F. C., Huang, Y., Bisaria, N., and Anderson, D. G. (2019). RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74 (3), 508–520.e4. doi:10.1016/j.molcel.2019.02.015
Whitehead, K. A., Langer, R., and Anderson, D. G. (2009). Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8 (2), 129–138. doi:10.1038/nrd2742
Winkle, M., El-Daly, S. M., Fabbri, M., and Calin, G. A. (2021). Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 20 (8), 629–651. doi:10.1038/s41573-021-00219-z
Wu, B., Liu, D. A., Guan, L., Myint, P. K., Chin, L., Dang, H., et al. (2023). Stiff matrix induces exosome secretion to promote tumour growth. Nat. Cell Biol. 25 (3), 415–424. doi:10.1038/s41556-023-01092-1
Wu, T., Chen, Y., Du, Y., Tao, J., Li, W., Zhou, Z., et al. (2018). Circulating exosomal miR-92b-5p is a promising diagnostic biomarker of heart failure with reduced ejection fraction patients hospitalized for acute heart failure. J. Thorac. Dis. 10 (11), 6211–6220. doi:10.21037/jtd.2018.10.52
Xiao, Q., Guo, D., and Chen, S. (2019). Application of CRISPR/Cas9-Based gene editing in HIV-1/AIDS therapy. Front. Cell Infect. Microbiol. 9, 69. doi:10.3389/fcimb.2019.00069
Xu, F., Liao, J. Z., Xiang, G. Y., Zhao, P. X., Ye, F., Zhao, Q., et al. (2017). MiR-101 and doxorubicin codelivered by liposomes suppressing malignant properties of hepatocellular carcinoma. Cancer Med. 6 (3), 651–661. doi:10.1002/cam4.1016
Yanatori, I., Richardson, D. R., Dhekne, H. S., Toyokuni, S., and Kishi, F. (2021). CD63 is regulated by iron via the IRE-IRP system and is important for ferritin secretion by extracellular vesicles. Blood 138 (16), 1490–1503. doi:10.1182/blood.2021010995
Yang, D., Zhang, W., Zhang, H., Zhang, F., Chen, L., Ma, L., et al. (2020a). Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics 10 (8), 3684–3707. doi:10.7150/thno.41580
Yang, L., Han, B., Zhang, Z., Wang, S., Bai, Y., Zhang, Y., et al. (2020b). Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation 142 (6), 556–574. doi:10.1161/circulationaha.120.045765
Yang, T., Martin, P., Fogarty, B., Brown, A., Schurman, K., Phipps, R., et al. (2015). Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 32 (6), 2003–2014. doi:10.1007/s11095-014-1593-y
Yang, T. T., Liu, C. G., Gao, S. C., Zhang, Y., and Wang, P. C. (2018). The serum exosome derived MicroRNA-135a, -193b, and -384 were potential alzheimer's disease biomarkers. Biomed. Environ. Sci. 31 (2), 87–96. doi:10.3967/bes2018.011
Yeung, C. C., Dondelinger, F., Schoof, E. M., Georg, B., Lu, Y., Zheng, Z., et al. (2022). Circadian regulation of protein cargo in extracellular vesicles. Sci. Adv. 8 (14), eabc9061. doi:10.1126/sciadv.abc9061
Yim, N., Ryu, S. W., Choi, K., Lee, K. R., Lee, S., Choi, H., et al. (2016). Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 7, 12277. doi:10.1038/ncomms12277
Yin, P. T., Pongkulapa, T., Cho, H. Y., Han, J., Pasquale, N. J., Rabie, H., et al. (2018). Overcoming chemoresistance in cancer via combined MicroRNA therapeutics with anticancer drugs using multifunctional magnetic core-shell nanoparticles. ACS Appl. Mater Interfaces 10 (32), 26954–26963. doi:10.1021/acsami.8b09086
Yoon, S., and Rossi, J. J. (2018). Therapeutic potential of small activating RNAs (saRNAs) in human cancers. Curr. Pharm. Biotechnol. 19 (8), 604–610. doi:10.2174/1389201019666180528084059
Yu, D., Li, Y., Wang, M., Gu, J., Xu, W., Cai, H., et al. (2022). Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 21 (1), 56. doi:10.1186/s12943-022-01509-9
Yu, M., Bardia, A., Wittner, B. S., Stott, S. L., Smas, M. E., Ting, D. T., et al. (2013). Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339 (6119), 580–584. doi:10.1126/science.1228522
Yu, W., Hurley, J., Roberts, D., Chakrabortty, S. K., Enderle, D., Noerholm, M., et al. (2021). Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann. Oncol. 32 (4), 466–477. doi:10.1016/j.annonc.2021.01.074
Zhan, Q., Yi, K., Cui, X., Li, X., Yang, S., Wang, Q., et al. (2022). Blood exosomes-based targeted delivery of cPLA2 siRNA and metformin to modulate glioblastoma energy metabolism for tailoring personalized therapy. Neuro Oncol. 24 (11), 1871–1883. doi:10.1093/neuonc/noac071
Zhan, Q., Yi, K., Qi, H., Li, S., Li, X., Wang, Q., et al. (2020). Engineering blood exosomes for tumor-targeting efficient gene/chemo combination therapy. Theranostics 10 (17), 7889–7905. doi:10.7150/thno.45028
Zhang, J., Liu, S. C., Luo, X. H., Tao, G. X., Guan, M., Yuan, H., et al. (2016). Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J. Clin. Lab. Anal. 30 (6), 1116–1121. doi:10.1002/jcla.21990
Zhang, L., and Yu, D. (2019). Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 1871 (2), 455–468. doi:10.1016/j.bbcan.2019.04.004
Zhang, P., Zhou, X., He, M., Shang, Y., Tetlow, A. L., Godwin, A. K., et al. (2019a). Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 3 (6), 438–451. doi:10.1038/s41551-019-0356-9
Zhang, W., Jiang, X., Bao, J., Wang, Y., Liu, H., and Tang, L. (2018). Exosomes in pathogen infections: A bridge to deliver molecules and link functions. Front. Immunol. 9, 90. doi:10.3389/fimmu.2018.00090
Zhang, W., Lin, J., Shi, P., Su, D., Cheng, X., Yi, W., et al. (2022b). Small extracellular vesicles derived from MSCs have immunomodulatory effects to enhance delivery of ASO-210 for psoriasis treatment. Front. Cell Dev. Biol. 10, 842813. doi:10.3389/fcell.2022.842813
Zhang, Y., Kim, M. S., Jia, B., Yan, J., Zuniga-Hertz, J. P., Han, C., et al. (2017). Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 548 (7665), 52–57. doi:10.1038/nature23282
Zhang, Y., Li, K., Han, X., Chen, Q., Shao, L., and Bai, D. (2022a). A photochemical-responsive nanoparticle boosts doxorubicin uptake to suppress breast cancer cell proliferation by apoptosis. Sci. Rep. 12 (1), 10354. doi:10.1038/s41598-022-14518-x
Zhang, Y., Wang, Z., and Gemeinhart, R. A. (2013). Progress in microRNA delivery. J. Control Release 172 (3), 962–974. doi:10.1016/j.jconrel.2013.09.015
Zhang, Y., Zhang, Y., Yin, Y., and Li, S. (2019b). Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol. Res. Pract. 215 (8), 152466. doi:10.1016/j.prp.2019.152466
Zhang, Z., Liu, X., Yang, X., Jiang, Y., Li, A., Cong, J., et al. (2023). Identification of faecal extracellular vesicles as novel biomarkers for the non-invasive diagnosis and prognosis of colorectal cancer. J. Extracell. Vesicles 12 (1), e12300. doi:10.1002/jev2.12300
Zhao, J., and Feng, S. S. (2015). Nanocarriers for delivery of siRNA and co-delivery of siRNA and other therapeutic agents. Nanomedicine (Lond) 10 (14), 2199–2228. doi:10.2217/nnm.15.61
Zhao, L., Gu, C., Gan, Y., Shao, L., Chen, H., and Zhu, H. (2020). Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control Release 318, 1–15. doi:10.1016/j.jconrel.2019.12.005
Zhao, R., Zhang, Y., Zhang, X., Yang, Y., Zheng, X., Li, X., et al. (2018b). Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol. Cancer 17 (1), 68. doi:10.1186/s12943-018-0817-x
Zhao, Y., Zheng, Y., Zhu, Y., Zhang, Y., Zhu, H., and Liu, T. (2021). M1 macrophage-derived exosomes loaded with gemcitabine and deferasirox against chemoresistant pancreatic cancer. Pharmaceutics 13 (9), 1493. doi:10.3390/pharmaceutics13091493
Zhao, Z., Yang, Y., Zeng, Y., and He, M. (2016). A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab. Chip 16 (3), 489–496. doi:10.1039/c5lc01117e
Zhao, Z. H., Chen, Z. T., Zhou, R. L., Zhang, X., Ye, Q. Y., and Wang, Y. Z. (2018a). Increased DJ-1 and alpha-synuclein in plasma neural-derived exosomes as potential markers for Parkinson's disease. Front. Aging Neurosci. 10, 438. doi:10.3389/fnagi.2018.00438
Zheng, D., Huo, M., Li, B., Wang, W., Piao, H., Wang, Y., et al. (2020). The role of exosomes and exosomal MicroRNA in cardiovascular disease. Front. Cell Dev. Biol. 8, 616161. doi:10.3389/fcell.2020.616161
Zhong, J., Xia, B., Shan, S., Zheng, A., Zhang, S., Chen, J., et al. (2021b). High-quality milk exosomes as oral drug delivery system. Biomaterials 277, 121126. doi:10.1016/j.biomaterials.2021.121126
Zhong, L., Li, Y., Xiong, L., Wang, W., Wu, M., Yuan, T., et al. (2021a). Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct. Target Ther. 6 (1), 201. doi:10.1038/s41392-021-00572-w
Zhou, B., Xu, K., Zheng, X., Chen, T., Wang, J., Song, Y., et al. (2020). Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target Ther. 5 (1), 144. doi:10.1038/s41392-020-00258-9
Zhu, G., and Chen, X. (2018). Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 134, 65–78. doi:10.1016/j.addr.2018.08.005
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., et al. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18 (1), 74. doi:10.1186/s12943-019-1000-8
Zhu, L., Perche, F., Wang, T., and Torchilin, V. P. (2014). Matrix metalloproteinase 2-sensitive multifunctional polymeric micelles for tumor-specific co-delivery of siRNA and hydrophobic drugs. Biomaterials 35 (13), 4213–4222. doi:10.1016/j.biomaterials.2014.01.060
Keywords: engineered exosomese, drug, RNA, co-delivery system, combination therapies
Citation: Chen H, Yao H, Chi J, Li C, Liu Y, Yang J, Yu J, Wang J, Ruan Y, Pi J and Xu J-F (2023) Engineered exosomes as drug and RNA co-delivery system: new hope for enhanced therapeutics?. Front. Bioeng. Biotechnol. 11:1254356. doi: 10.3389/fbioe.2023.1254356
Received: 07 July 2023; Accepted: 05 September 2023;
Published: 26 September 2023.
Edited by:
Zhicheng Wang, Fudan University, ChinaReviewed by:
Shi-Cong Tao, Shanghai Jiao Tong University, ChinaCopyright © 2023 Chen, Yao, Chi, Li, Liu, Yang, Yu, Wang, Ruan, Pi and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongdui Ruan, cnVhbnlvbmdkdWl0YW5neGlhQDE2My5jb20=; Jiang Pi, amlhbmdwaUBnZG11LmVkdS5jbg==; Jun-Fa Xu, eHVqdW5mYUBnZG11LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.