- 1Division of Physiology, Department of Molecular Medicine, Institute of Basic Medical Sciences, University of Oslo, Oslo, Norway
- 2Department for Immunology, Clinic for Laboratory Medicine, Oslo University Hospital, Rikshospitalet, Oslo, Norway
Tracking followed by analysis of specific point-of-interest from conventional or high-speed video recordings have been widely used for decades in various scientific disciplines such as sport, physiotherapy, and behavioral science. Another method used to characterize movement in 3D involves the use of motion capture systems, which produce files containing a collection of 3D-coordinates and corresponding timestamps. When studying animal or human movement, combining motion tracking with other recording methods–like monitoring muscle activity or sensor signals–can yield valuable insights. However, manual analysis of data from these diverse sources can be time-consuming and prone to errors. To address this issue, this article introduces a new, free, and open-source software developed in MATLAB. This software can be used as-is, or developed further to meet specific requirements. Once the coordinates are imported, multiple tools can be used for data preprocessing, such as to correct mistakes that may have occurred during tracking because of software errors or suboptimal video quality. In addition, the software can import coordinates from multiple cameras and combine them into a unified data series. With these inputs, the software can automatically calculate kinematic parameters and descriptive statistics, generate 2D and 3D animations, and analyze gait cycles, enabling swift and accurate analysis of multidimensional motion data. Moreover, the software can import electrophysiology traces and sensor signals, which can be filtered, rectified, smoothed, and correlated with the kinematic data in various ways. Thanks to its user-friendly graphical user interface, the software is easy to navigate and can be used to analyze complex movements without any need for coding skills. This versatile tool is well-suited for a wide range of experimental contexts, making it a valuable resource for researchers across diverse scientific disciplines.
Highlights
• Analyzes tracking data in 2D and 3D to evaluate motion
• Processes electrophysiology traces and sensor signals for an integrative movement analysis
• Includes multiple tools for preprocessing of data
• Features a user-friendly graphical interface, accessible without any coding knowledge
• Developed using MATLAB, but no MATLAB license is required for its use
Introduction
Tracking the movements of animals, limb joints, single cells, and other subjects in space and time is an experimental method relevant to many scientific disciplines. In bioscience, it is so widely used that it is challenging to keep track of all the species that have been studied in this way. Some examples include oikopleura (Kreneisz and Glover, 2015), tadpole (Bacqué-Cazenave et al., 2018), zebrafish (Barreiros et al., 2021), rodents (Bachmann et al., 2013), chicken (Bari et al., 2021), cats (López Ruiz et al., 2017), pigs (Boakye et al., 2020) and non-human primates (Fitzsimmons et al., 2009). In neuroscience, biomedicine, and sports science, this method is typically used to compare kinematic quantities across different conditions. It is particularly relevant for the study of locomotion, motor systems, and related defects like spinal cord injury, stroke, traumatic brain injury, Parkinson’s disease, nerve injury, and others. Video tracking is, therefore, often used in comparative studies involving different groups (e.g., wild type, KO, injured, treatment), with high frame-rate video recordings of animals and humans performing specific motor tasks (e.g., running on a treadmill). In many cases, points-of-interest (POI) are tracked on a video recording to determine their position in space and time. Alternatively, a 3D motion capture system enables direct tracking of the POI. Further analysis of these data permits an objective and quantifiable evaluation of the movement, enabling reliable inter-group comparisons.
Numerous tracking programs are available, both free and open source as well as commercial licensed software. For tracking short sequences, one option is to use the popular Java-based program ImageJ, developed by NIH, which serves as a powerful imaging platform used by scientists in many research fields (Rasband, W. S., 1997; Schneider et al., 2012). The Manual Tracking plugin in ImageJ enables the recording of coordinates and the calculation of instant speed for multiple POI (Cordelières, 2006). However, tracking POI manually can be time-consuming, particularly with high frame-rate videos. Other plugins like TrackMate (Tinevez et al., 2017), MTrack2, MultiTracker, SpotTracker, MTrackJ (Meijering et al., 2012), Speckle TrackerJ (Smith et al., 2011), and ToAST (Hegge et al., 2009) aim to provide automated, semi-automated, and manual tracking of single-particles/objects. Though these plugins can be used for motion tracking of moving animals, they may be more appropriate for time-lapse microscopy imaging. Other free programs offer automated tracking, including, Kinovea (Puig-Diví et al., 2019), OptiMouse (Ben-Shaul, 2017), Toxtrac (Rodriguez et al., 2018), Tracker (Brown and Cox, 2009; Nakamichi and Asahara, 2022), MouseActivity (Zhang et al., 2020), Traktor (Sridhar et al., 2019), idTracker (Pérez-Escudero et al., 2014), Ctrax (Branson et al., 2009), BioTracker (Mönck et al., 2018), UMATracker (Yamanaka and Takeuchi, 2018), BEMOVI (Pennekamp et al., 2015), FIMtrack (Risse et al., 2017), ezTrack (Pennington et al., 2019), Whisker-Tracker (Azarfar et al., 2018), YOLOv2 (Barreiros et al., 2021), OpenPose (Cao et al., 2021), PoseNET (Erfianto et al., 2023), AlfaPose (Mehdizadeh et al., 2021; Fang et al., 2023), DeepPoseKit (Graving et al., 2019), idtracker.ai (Romero-Ferrero et al., 2019), B-KinD (Sun et al., 2022), TRex (Walter and Couzin, 2021), DeepLabCut (Nath et al., 2019), B-SOiD (Hsu and Yttri, 2021), LEAP and SLEAP (Pereira et al., 2019; 2022), and DeepLabStream (Schweihoff et al., 2021).
In some cases, their tracking accuracy may depend on image quality, and on the use of high-contrast markers such as retroreflective beads or tape, or permanent or temporary tattoos; These can facilitate tracking and improve accuracy. The most recent programs use machine learning and deep neural networks to enable markerless tracking of single and multiple animals. The accuracy of these software packages has been validated by various studies, that have used different subjects and different programs (Mehdizadeh et al., 2021; Stenum et al., 2021; Johnson et al., 2022; Lonini et al., 2022; Washabaugh et al., 2022; John et al., 2023; Lee et al., 2023). In addition, other interesting initiatives have resulted in gathering various open-source resources, including analysis tools and downloadable datasets, under the same repositories. Examples include OpenBehaviour (https://edspace.american.edu/openbehavior/) and a GitHub repository maintained by Dr. Luca Modenese (https://github.com/modenaxe/awesome-biomechanics). Apart from open-source software, there are also multiple systems available for purchase, with some requiring specific hardware—as determined by the seller—while others do not have such requirements. Notable examples are WINanalyse (https://winanalyze.com/), ProAnalyst from Xcitex (http://www.xcitex.com/), visual 3D from C-Motion (https://c-motion.com/), Vicon (https://www.vicon.com/), Delsys (https://delsys.com/), The Motion Monitor (https://innsport.com/), and Qualisys (https://www.qualisys.com/). These systems can propose advanced motion analysis and modeling features, but they come at a cost that may not be affordable for everyone.
Preprocessing and analysis of data from 2D–3D coordinates to obtain multiple kinematic quantities, various diagrams, descriptive statistics, animations, and gait parameters can be a tedious and time-consuming process. In biomedical science, it can also be valuable to combine 2D–3D coordinates with electrophysiology and/or sensor signal recordings to analyze the function of specific muscles or determine the force applied in a defined phase of the movement. The Integration of these analyses provides an overview of the biomechanical and physiological aspects of movement. The use of spreadsheet programs for these tasks requires a great number of actions and is potentially prone to mistakes. Therefore, to make this work easier and to standardize the analysis, we created a MATLAB program called MotionAnalyser. Although MATLAB is an important research tool, a lack of coding experience may present a barrier for a significant number of users. Which is why we created a graphical user interface (GUI) aimed at providing a simple and user-friendly experience for comprehensive, accurate, fast, and reproducible analysis. The GUI is organized with task-dedicated tabs. The Coordinates tab enables preprocessing of 2D and 3D coordinates, which can then be animated in the Animation tab. Further calculations and plotting can be automated from the Kinematic tab, and descriptive statistics can be obtained from the Stat tab. In addition, other signals—such as electromyograms and force plate data—can be preprocessed and analyzed in the E-Phys and Sensor tabs. All this data can then be correlated using different mathematical models in the Correlation tab. The Stride & Gait tab enables the identification of specific limb positions relevant for the calculation of different gait cycle parameters. All data generated can be exported in multiple file formats, including a. mat file that can be reloaded to MotionAnalyser for subsequent analysis.
Materials and methods
Software requirements
MotionAnalyser runs on MATLAB R2023a or later. Source code and application installers are available at https://github.com/BoullandLab/MotionAnalyser.
Data loading
MotionAnalyser can handle any number of datasets, subject only to the maximum memory constraints of the operating system and of MATLAB. For example, a user interested in locomotion can import coordinates for the left and right legs and arms, and work with them either independently or together. Currently, the program supports loading data from spreadsheets (.xls/.xlsx), text files (.csv/.txt), and DeepLabCut (Nath et al., 2019) output files. In all other cases, data must contain appropriate headers and be organized column-wise, with timestamps being in the first column and spatial coordinates after (x, y), with the inclusion of the optional “z”coordinate for 3D. Different POI can either be imported from separate files or concatenated in a single file. All other data (EMG, sensors, etc.) follow the same schema. Data from the headers (tracked point, muscle, sensor, etc.) is used to label all tables and plots. Example data files can be downloaded together with the program (https://github.com/BoullandLab/MotionAnalyser).
Data preprocessing
Coordinates obtained from video tracking or motion capture are susceptible to errors, including outliers. MotionAnalyser facilitates the identification, removal, and replacement of these outliers by interpolation, nearest value, mean window, previous value, etc. (Supplementary Figure S1A). It is also possible to apply different filters, such as the root mean square, moving means, etc., to smoothen out the trajectory.
Furthermore, traces acquired from multiple cameras capturing the same object may exhibit misalignment (Supplementary Figure S1B). MotionAnalyser provides a range of tools to correct this problem. In this process, it can be an advantage to work with separated datasets. As MotionAnalyser can import different elements separately, it enables the plotting, animating and preprocessing of each element independently, and without a segment joining them (Supplementary Figure S1C). The options mentioned in the rest of this paragraph can, therefore, be applied to individual elements. For instance, to compensate for mirror images acquired by cameras in opposite positions, the x-coordinates can be inverted (Supplementary Figure S1D). Other potential misalignments of opposite cameras can also be rectified by shifting coordinates (Supplementary Figure S1E). An auto-align option can also be used with a particular reference point indicated by the user. Perspective distortions might occur in case of camera misalignment or if a subject moves closer to one of the cameras, resulting in elements of different sizes, as exaggerated in the Supplementary Figure S1F. To restore proportions, a scaling factor can be applied to the set of coordinates sought. It is also possible to focus the analysis on a specific part of the movement by selecting the first and last desired positions before trimming the dataset (Supplementary Figure S1G). Other options, such as rotation, deletion of values, renaming the different POI, and others are also available. All changes are automatically saved and used in all downstream analyses without affecting the raw files. However, when using the downsampling option, for instance, to mitigate issues with overloaded graphs (Supplementary Figure S1H), it solely impacts the display and does not alter the data used for other calculations.
Stitch
To combine two sets of coordinates recorded by cameras on the same side of a subject, MotionAnalyser enables their stitching. First, the user imports the initial set of coordinates and then imports the stitch set, which is plotted alongside the initial set (Supplementary Figures S2A, B). The user can adjust the position of the stitch coordinates and rotate them to correct for a camera misalignment. The stitching option enables the user to choose the transition point in the stitch sequence (Supplementary Figures S2C, D). Note that MotionAnalyser does not provide a function for camera calibration to correct distortions at the overlap between two cameras. It is necessary to correct this before importing the data into MotionAnalyser.
Animations
Users can animate the different elements independently or together and control various display parameters (Supplementary Video S1). For example, users can adjust the animation speed, the perspective, change the line thickness, and add or remove a marker of the desired color, so as to indicate the positions of different POI. It is also possible to normalize the movement on different planes by defining a POI as a reference. When using 2D data, an arbitrary value is assigned to the third coordinate of each POI (by default, 0). These values that can be adjusted by the user, are only used for visualization purposes, and are never used in the calculation of kinematic parameters.
Calculation of kinematic quantities
Before calculating the kinematic quantities, the user has the option to choose to set the expression of speed and acceleration as absolute values to avoid negative speeds and accelerations. The default expression of angles can be changed from degrees (default) to radians. The mathematical equations used for the calculation of the kinematic quantities are shown in the Supplementary Table S1. MotionAnalyser determines extrema values, i.e., minimum, maximum, and it calculates and plots descriptive statistics, i.e., mean, median, and mode values. These values, however, are not displayed on the Kinematic tab but can be queried and saved from the Stats tab. If there are two or more POI, MotionAnalyser can create a stick diagram of the movement. This diagram includes a segment between the POI for each time value. The stick diagram can also be normalized on the x-axis, on the y-axis, or both using a defined reference point that can be changed by the user. This enables visualization of the movement with a fixed anchoring point. This feature eliminates fluctuations, which can be useful for suppressing movements that are unrelated to the specific movement of interest, e.g., an animal swimming. Note that this normalization only affects the display, but it does not modify the calculation of kinematic variables. The same option is also available for the Animation and the Stride & Gait tabs.
Electrophysiology and sensor signal analysis
The electrophysiology and sensor signals are treated similarly, although they are displayed on different tabs to allow easy switching between them. These signals can be processed in various ways, including trimming, adjusting temporal resolution, resampling, interpolating, filtering, rectifying, and creating a signal envelope (Supplementary Table S1). Adjusting the temporal resolution should not be confused with resampling or interpolation; it simply involves changing the time array to a user-defined value. This can be particularly useful in cases where time values are missing in the raw data, but the sampling rate is known, which can occur with certain sensors. MotionAnalyser can also perform spectral analysis of the signal using Fast Fourier Transform (FFT) and can plot the power spectrum and spectrogram of the signal (Supplementary Figures S3A, B). Digital filtering is calculated using the Butterworth digital filter function with an adjustable filter order. MotionAnalyser also provides several rectification options, including mean rectification, absolute value rectification, square root rectification, and half-wave rectification (Supplementary Figures S3C, D; Supplementary Table S1). To generate a signal envelope, users can use a low-pass Butterworth filter or the Hilbert transform, both programmed with existing MATLAB functions. Alternatively, users can also use a moving average or root-mean-square of the signal with an adjustable window (Supplementary Figure S3E). In addition, users have the option to calculate the area under the curve of the signal. If required, it is always possible to revert to the original signal, allowing for experimentation with various signal processing methods.
Correlations
This tab enables the user to correlate kinematic quantities, either within elements or between different elements. This feature can be useful for plotting the cyclogram for two joint angles. In addition, the user can correlate the electrophysiology and sensor data between each other or with the kinematic quantities. In some cases, resampling or interpolation and trimming are necessary to ensure that the sampling rates and sequence duration are consistent. However, the user maintains control throughout this process, as a dialogue box provides guidance. MotionAnalyser also calculates the autocorrelation function of any of the imported signals to determine its potential rhythmicity. The cross-correlation function correlates two different signals to determine the degree of similarity and coordination between them. The phase lag and the amplitude ratio, which are calculated using MATLAB’s algorithm for FFT, are also calculated. The amplitude ratio is determined by calculating the ratio of the peak amplitude for each signal, i.e., the maximal value of the FFT for each signal. The phase lag is obtained by computing each signal’s phase at peak frequency using MATLAB’s angle function, and then taking their difference. Furthermore, MotionAnalyser offers additional methods for comparing two signals of the same duration and sampling rate: Euclidean distance, cosine similarity, Pearson’s, Spearman’s, and Kendall’s correlation coefficients. In cases where resampling or interpolating the signals is not feasible or cannot be accomplished satisfactorily, MotionAnalyser provides an alternative option for assessing the correlation of two signals using Dynamic Time Warping. This method requires the user to adjust the size of a moving window to manage the computational load in accordance with the available computing power.
Locomotion analysis
The Stride & Gait tab is specifically designed for carrying out the analysis of locomotion and represents the most interactive area of the software. Users can navigate through different limb positions from the control panel to identify the specific positions that characterize the gait cycle. A classic decomposition of the gait cycle is used by MotionAnalyser (Figure 3A), which includes a swing phase between the toe-off and heel-strike positions and a stance phase between the heel-strike and the next toe-off position (Dale, 2012). The program calculates several parameters, including the stance and swing phase durations, stride length, and duration, which are particularly useful when analyzing a defective locomotion. MotionAnalyser also plots a gait diagram. Two additional limb positions can be recorded if desired; the heel-off to calculate the heel-down duration as part of the stance phase, and the maximum toe-lift to determine the toe-lift speed and duration. To obtain reliable data, it is essential to select the heel-strike as the starting point of the gait cycle. The software can analyze an unlimited number of cycles, with each position being displayed in a distinct color that can be customized. In addition, the coordinates and corresponding time values are presented in a table for easy reference. As for the Animation and the Kinematic tabs, the movement can be normalized to a chosen point of interest on the x-axis, the y-axis, or both. The stick diagram used to identify the relevant limb position can be zoomed in and out to accommodate many gait cycles. The pan tool that appears when dragging the mouse over a background area of the diagram can be used to re-center an area of interest in the diagram.
Exporting analyzed data
After completing the analysis, users can export all analyzed data to multiple xls/xlsx/txt/csv files. It is also possible to save the data as a mat file, which enables users to further analyze the data in the MATLAB workspace, or reload the data in MotionAnalyser for a second analysis. Another advantage of the mat format is that it saves all data from position, tracking, electrophysiology, sensors, and gait into a single file. It is also possible to save all currently plotted figures. However, this function will not save previously plotted figures that were subsequently replaced. For saving sequential plots, users can save them manually one after the other, using the save option that appears on the plot when placing the mouse cursor over an empty area at the time of the plotting. Plots can be saved as image or vector graphic files. Animated models can be exported as individual AVI or animated GIF files.
Note on units of measurement
MotionAnalyser is agnostic to units of measurement. Therefore, derived quantities follow from the units of measurement of the raw data. For example, if the input file has coordinates given in mm and time in seconds, the output speed will be in mm/sec. Nevertheless, it is always possible to adjust their order of magnitude using the scaling function (e.g., mm to cm).
Software benchmarking
To evaluate the computing speed of MotionAnalyser, we performed MATLAB benchmark tests on a Windows 10 PC that featured an Intel Core i5-6500 running at 3.2 GHz, 16 GB of RAM, and an Intel HD Graphics 530 GPU. The tests involved five input files containing 13 columns, including time, and x- and y-coordinates for six POIs, for a total of 1,300, 13,000, 130,000, 1.3 million, and 13 million values.
Software accuracy test using mathematical functions
All functions of MotionAnalyser were rigorously tested using signals generated from mathematical functions. These known mathematical properties provide a solid foundation to ensure that the outcomes of the calculations performed by MotionAnalyser are correct. For testing basic kinematic calculations, we generated two sets of data, where the x coordinate increases linearly with time, while the y coordinate increases quadratically. These respectively lead to a constant and linearly increasing speed, and a null and constant acceleration. In addition, we introduced a single outlier in one of the y-arrays. To test the accuracy of the angle calculation, we created a series of segments with known angles between them at 0, 90, and 135°. To test the gait analysis code, we created a dummy sequence of known dimensions for left and right legs. To test the treatment of electrophysiology and the sensor signal data, we created sine waves of known frequencies and amplitudes and applied successive filters while examining their effect on the FFT. We also tested the code that calculates the different correlation functions, amplitude ratio, and phase lag using two sine waves of a known phase shift of π radians and a known amplitude ratio of 2.
Software accuracy test using real data
To further test the accuracy of the calculation, we used a real dataset obtained from one of our previous studies involving a mouse swimming experiment (Züchner et al., 2018). We compared the outcome from MotionAnalyser with alternative calculation methods like ImageJ, spreadsheets, and manual measurements.
Example of use
We provide six examples of use of MotionAnalyser with real data obtained from various types of movements recorded in mice and humans. We generated all the mouse data during previous studies. For the human examples, we created the walking pattern data (Example 4) and the pendulum data (Example 5) to illustrate the potential use of MotionAnalyser. In the case of Example 6, we obtained the 3D motion capture and EMG datasets from a repository.
Example 1: Freely swimming 9-day old neonatal mice in a 210 × 165 mm pool for 20 s were filmed at 25 Hz. The movement was tracked with ImageJ and the Manual tracking plug in. This dataset comes from a previously published study (Di Bonito et al., 2015).
Example 2: Vestibular spinal test was performed on a 5-day-old neonatal mouse. This test entails placing the mouse in a device that rotates 90° from a straight position along the sagittal axis of the mouse. This simulation of falling, triggers the extension of the hindlimb on the side of the fall (Di Bonito et al., 2015). The video recording was at 200 Hz and tracking was performed as in Example 1.
Example 3: Adult mouse linear swimming, filmed from the side. The right limb joints were tracked with ImageJ as described earlier (Züchner et al., 2018). Recording and tracking parameters were similar to those described in Example 2.
Example 4: Video tracking from a human subject walking on an horizontal plane. Videos were recorded using two GoPro 7 Black cameras positioned on the left and right sides, on an axis roughly perpendicular to the line of progression. The cameras were connected to a remote control that synchronized them. The video were acquired at 60 Hz because the movements involved were slow enough. To mark the different joints on the legs, round markers of 3.5 cm diameter were cut out from a pink sheet of paper and attached with double-sided tape. Tracking was done using Kinovea.
Example 5: The leg pendulum test is sometimes used in clinics to determine the degree of spasticity of a patient (Biering-Sørensen et al., 2006; Aleksić and Popović, 2022). This is typically done with the patient sitting or lying down and the leg hanging over the edge of the couch, and then the examiner extends the leg horizontally. When released, the leg descends and rotates around the knee axis in a pendulum-like movement. This test was performed in a healthy subject combining the video camera recording at 120 Hz with a goniometer recording at 100 Hz (Moti, Denmark).
Example 6: 3D motion capture and EMG recording of a human arm during a flexion-extension. This movement involves bending the arm (flexion) and then straightening it back to the starting position (extension). The data were a subset of a larger dataset (Averta et al., 2020), case 01_10_2 in the H2 group.
Results
MotionAnalyser is built around task-oriented tabs, enabling multimodal analysis of movement based on video tracking or motion capture, electrophysiology traces, and signals from sensor systems, such as pressure sensors, goniometers, and accelerometers (Figure 1). It includes multiple features to preprocess data, analyze kinematic parameters, create animations, and calculate descriptive statistics. It also enables analysis of electrophysiology and sensor signals. Various correlation functions can be used to better understand different aspects of the movement. In addition, MotionAnalyser enables gait analysis. Once the analysis is completed, the data can be exported to various formats, including spreadsheets, vector- or image-based graphics and animations.
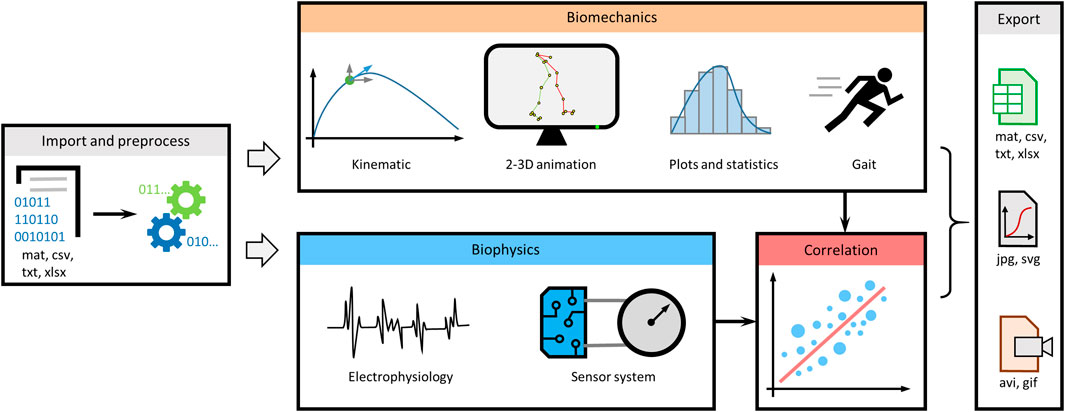
FIGURE 1. Overview diagram summarizing the different analyses that can be performed by MotionAnalyser. The program encompasses four primary analysis domains, namely, kinematics, electrophysiology, sensor signals, and stride and gait analysis. In addition, it enables correlation of results with and across each domain, allowing for comprehensive analysis.
Benchmarking MotionAnalyser on a mid-range computer showed, unsurprisingly, that some functions are slower when dealing with a high number of values, while others are less affected (Supplementary Figure S4). Loading data takes 83 s for 13 million data points (Supplementary Figure S4A). Plotting stick diagrams is the most resource-intensive operation, beginning to slow-down at 130k values before running out of memory at 13 M values (Supplementary Figure S4B). When saving data, mat files provide the highest performance; in contrast, saving csv files or spreadsheets is significantly slower (50s and 200s respectively), and the system runs out of memory with datasets of 1,3 and 13 M values. Other functions are affected differently by dataset size (Supplementary Figure S4D).
The program was further tested using synthetic data with predetermined properties. When the x-component of the array increased linearly, the program calculated a constant x-speed and x-acceleration was null, as expected (Supplementary Figures S5A, B). When the y-coordinates exhibited a parabolic increase, the program calculated a linearly increasing y-speed and a constant y-acceleration, as expected as well. Introducing a single-point perturbation in the y-array resulted in corresponding perturbations in the y-speed and y-acceleration, which are correct, as confirmed by manual calculation. Note that the two otherwise identical curves have been slightly shifted for visualization purposes. When tested on a synthetic dataset that generates known angles, MotionAnalyser correctly computed the corresponding angles (Supplementary Figure S5C). Similarly, by creating artificial arrays for the position of the toe, heel, and knee, a gait cycle was reproduced, yielding a stride length of 7 and a maximum toe-lift of 0.5 (arbitrary units), consistent with MotionAnalyser’s calculations. The gait diagram also correctly displayed stance and swing durations of 4 and 2 s respectively (Supplementary Figure S5E). To further assess the various components of the software responsible for calculating kinematic parameters, we used arrays derived from actual tracking data of a mouse hindlimb during linear swimming. The results showed that the calculated values were identical to those obtained through other means, such as ImageJ, spreadsheets, and manual calculations (Supplementary Figures S5F, G). The calculation of gait parameters from the tracking coordinates of a human subject were also identical to the manual measurements performed in Kinovea.
To evaluate the functions available for analyzing electrophysiology traces or sensor signals, we generated a signal from three sine waves at 25, 30, and 35 Hz. By applying two notch filters at 30 and 35 Hz, the original signal was correctly filtered, as evidenced by the absence of the corresponding frequencies in the power spectrum (Supplementary Figures S6A, B). The auto- and cross-correlation functions were tested with sine waves with known properties (Supplementary Figure S6C). The autocorrelation of each function showed a decaying cosine wave shape with the same period, as expected. As these two curves overlap, we slightly shifted one of them for visualization purposes (Supplementary Figure S6D). The cross-correlation correctly detected that the two functions are in antiphase (negative correlation), consistent with the calculation of a phase shift of 180° at lag 0. MotionAnalyser accurately calculated an amplitude ratio of 2, in agreement with the anticipated outcome.
MotionAnalyser was next used on experimental data. All the plots presented in this context were generated by MotionAnalyser. The first example involves tracking of six neonatal mice (9-day-old) during a free-swimming test conducted in a confined pool. Although the trajectories of the mice were diverse, they exhibited two distinct hotspots located at opposite corners, as depicted by the heat map (Figures 2A, B). The limited observation time of 20 s and the relatively small sample size (n = 6) may have hindered the detection of other potential hotspots. The distance swam by each mouse displayed variability. Despite this, a potential trend toward short- and long-distance swimmers was observed, although a larger sample size would be necessary to validate this observation (Figure 2C). MotionAnalyser also enables the extraction of additional kinematic parameters like speed and acceleration, thereby facilitating further characterization of each mouse and the potential grouping of individuals into distinct categories.
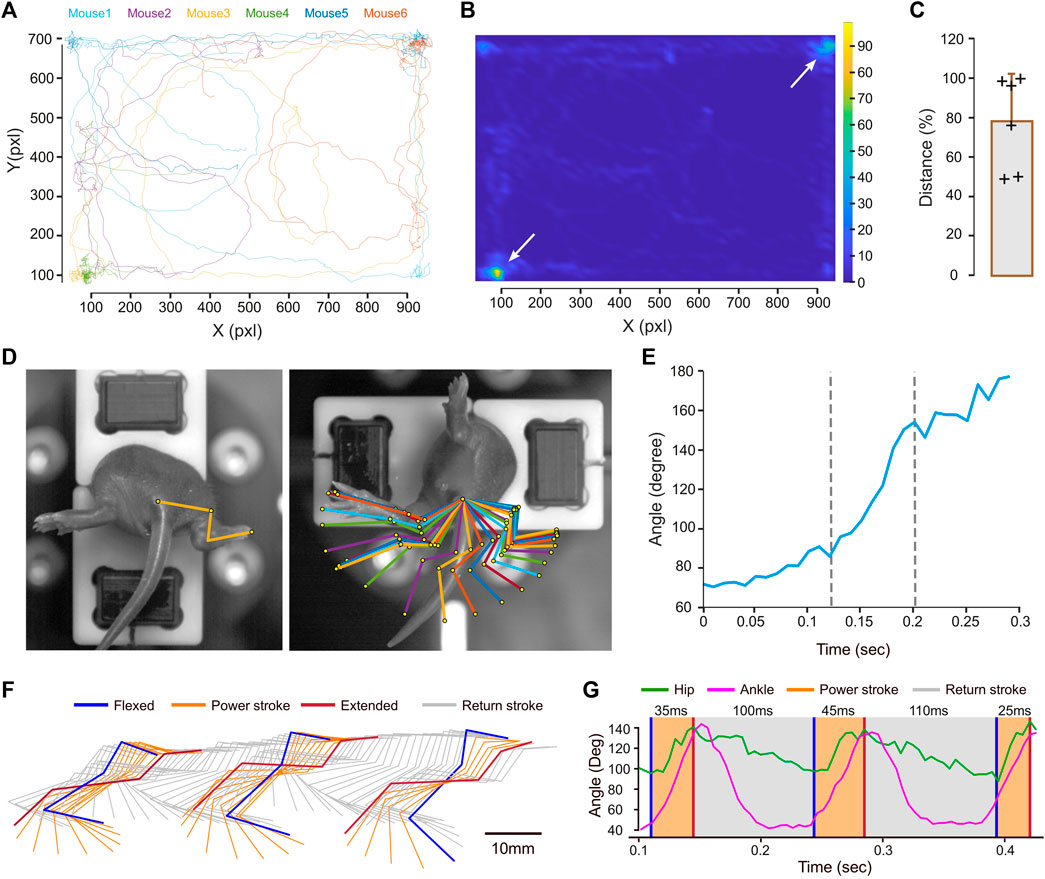
FIGURE 2. Examples illustrating the use of MotionAnalyser for analyzing animal movements. (A) Trajectory for 6 neonatal mice during free swimming. (B) Heatmap highlighting the hotspots for the trajectories depicted in (A). (C) Total displacement of each mouse. The error bar represents the standard deviation. (D) Images showing the initial and final positions of a neonatal mouse undergoing the vestibulospinal test. These are overlaid with a stick diagram depicting the sequential positions of the hindlimb during the movement. (E) Temporal angular variation of the paw-ankle-base tail angle during the vestibulospinal test. (F) Stick diagram illustrating the sequential positions of the hindlimb during a linear swim, recorded from the side of an adult mouse. The power stroke is depicted in orange, while the return stroke is represented in gray. (G) Temporal angular variation of the hip (green) and ankle (magenta) during linear swimming.
The second example illustrates the assessment of the vestibulospinal reflex in a 5-day-old mouse. The rapid rotation induced by the device holding the mouse triggers hindlimb extension (Figure 2D). A stick diagram represents the different hindlimb positions. Normalizing these positions to the coordinates of a reference point (the tail base) eliminates minor variations unrelated to limb movement, facilitating a more accurate understanding of the motion (Figure 2D). In this particular mouse, ankle extension rapidly increases in the middle of the movement. This is further supported by the plot of angle variations over time (Figure 2E). This shows that the slope of the angle opening is steeper between 125 and 200 ms, while it is less pronounced at the start and end of the movement.
The next example involves an adult mouse linear swimming test, recorded from the side to observe the movement of the hindlimb. The stick diagram, illustrates the position of the hindlimb throughout the movement (Figure 2F). Although the Stride & Gait module of the software was initially developed for analyzing gait cycles, it can be also used to evaluate other types of locomotion, such as swimming. In rodents, the swim cycle can be divided into two phases: the power, and the return, strokes (Gruner and Altman, 1980). The power stroke (depicted in orange) involves a backward extension of the hindlimb from the most flexed to the most extended hip angle position (Figures 2F, G). The return stroke (depicted in gray) involves the hindlimb moving forward at a slower pace until reaching maximal hip flexion, while the mouse glides in the water. To analyze this cycle, we used the maximal hindlimb extension following the power stroke as the entry position. When applying these criteria to three swim cycles, we found the power stroke duration to be 35, 40, and 25 ms, while the return stroke duration was significantly longer, measuring 100 and 110 ms (Figure 2G).
The fourth example shows the capability of MotionAnalyser’s to analyze human locomotion. Walking primarily involves a horizontal displacement of the legs, characterized by cycles of joint flexion and extension, as well as alternation between the left and right legs. This can be easily modeled from coordinates in the x-y-(z) plane using MotionAnalyser to generate stick diagrams and animations (Figure 3A; Supplementary Video S1). In addition, quantitative analysis of the gait cycle can also be obtained. In this example, the gait was characterized by a stance phase of almost 800 ms for a swing phase of 450 ms. The stride duration was 1.2 s, the stride length was 1.15 m, the step duration was 584 ms, and the step size was 0.66 m. The gait diagram also shows an overlap of the left and right stance phases typical for bipedal walking (Figure 3B). The profile of instant displacement, however, is not identical for all leg joints (Figure 3C). While the hip curve presents low variability, the other joints alternate peaks with almost an arrest of displacement. This corresponds to the cyclic occurrence of the swing and stance phases. The joint angle variations are consistent with the displacement. There is also a complex phase relationship between the variation of the ankle and the knee joints (Figures 3E–G). The cross-correlation coefficient of 0.8 at Lag 0 indicates a strong in-phase correlation between these angles, which aligns with the calculated phase shift of 15.5°.
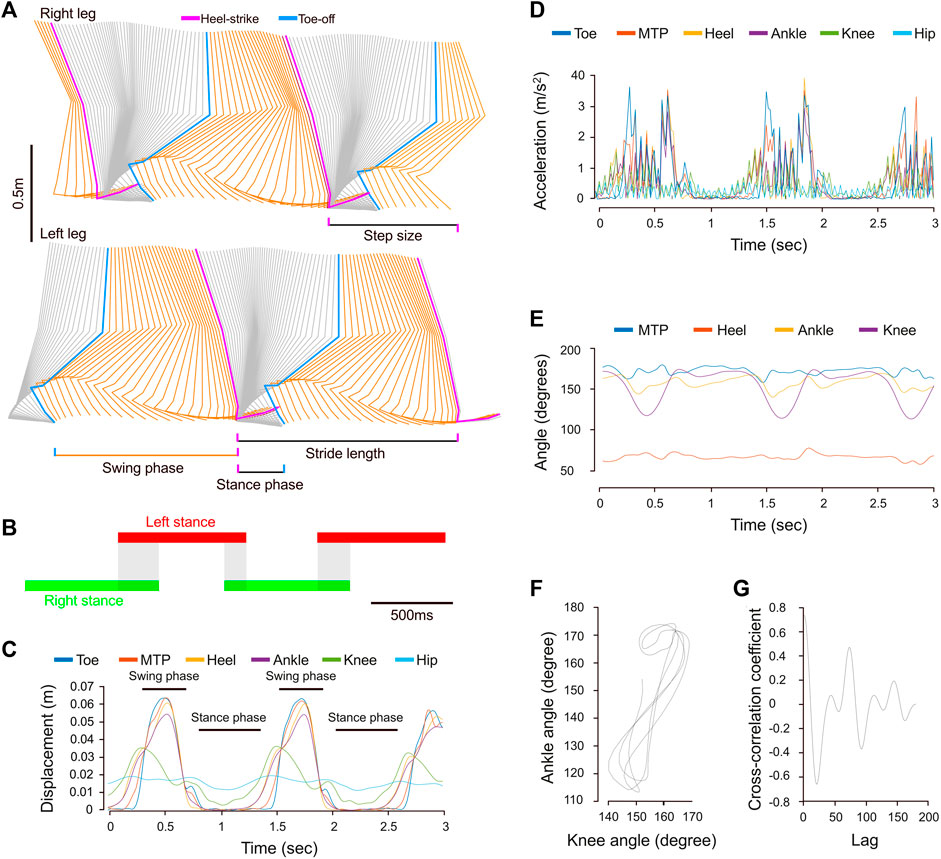
FIGURE 3. Example showcasing the utilization of MotionAnalyser for analyzing human gait. (A) Stick diagram depicting the sequential positions of the hindlimb of the right and left legs. The swing phase is depicted in orange, while the stance phase is represented in gray. (B) The gait diagram illustrating the duration of left (red) and right (green) stance phases, as well as the overlap between them (gray). (C) Temporal displacement curves for various anatomical points of the right leg. Peaks in displacement indicate the swing phase, while a halt in movement signifies the stance phase. (D) Temporal variation of the xy acceleration for the same anatomical points as in (C). (E) Temporal variation of the MTP, heel, ankle, and knee joint angles. (F) Cyclogram illustrating the relationship between the ankle and knee joint angles throughout the gait cycle. (G) Cross-correlogram displaying the relationship between ankle and knee joint angles throughout the gait cycle.
For the next example, a leg pendulum test was conducted and the leg joints were tracked, while a wireless goniometer recorded variations in the foot-knee-hip angle. The stick diagram illustrates a typical pendulum movement with a decay shape (Figure 4A). By using the Stride & Gait component of the program, the positions that correspond to the left (red) and right (green) maxima were isolated for each cycle. This allowed for the calculation of the cycle duration, which was relatively constant at 460 ms, except for the initial drop, which was faster, with 380 ms (Figure 4B). In addition, the vertical amplitude of the toe for each cycle also agreed with a typical decay process. The reliability of the goniometer—which was positioned to record variations in the knee angle—was tested against the calculated outcome obtained from video tracking. The shapes of the two curves are similar, although noticeable differences were found (Figure 4C). There were 8 degrees of disparity between baselines. Furthermore, the amplitude of the goniogram after the first minima was larger than that from the video tracking data. The cross-correlation coefficient at lag 0 was 0.99, indicating a strong similarity and signals in phase (Figure 4D). However, the correlation becomes weaker and oscillates as the lag increases, reflecting variations in the magnitude of correlation between the signals at different lags and a slight phase drift, which is already visible at 3 s (Figure 4C). These differences are likely to reflect the different modalities used to acquire the data. Tracking from video recording only accounts for a 2D movement, while the goniometer integrates the joint’s angular velocity, introducing drift.
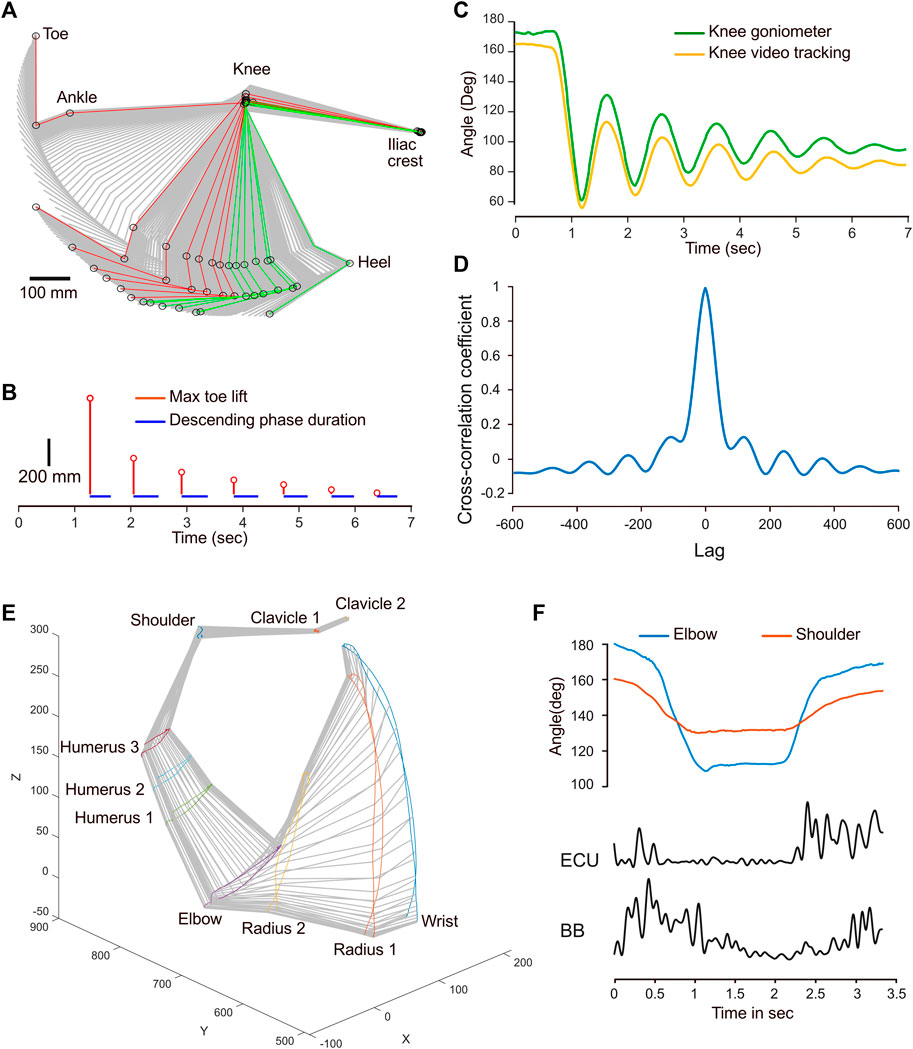
FIGURE 4. Examples of combined kinematic and EMG or goniometer signals. (A) Stick diagram illustrating the sequential positions of the hindlimb during a pendulum test. The positions corresponding to left maxima are depicted in red, while those corresponding to the right maxima are shown in green. (B) Temporal stem plot (red) showing the toe lift amplitude decay during the pendulum motion. The blue segments represent the leg’s falling phase duration. (C) Temporal angular variation of the knee joint obtained from video tracking (yellow) and a goniometer (green). (D) Cross-correlogram depicting the correlation between the knee joint angle variations obtained from video tracking and the goniometer. (E) Perspective stick diagram depicting the sequential positions of the arm and shoulder over time during a flexion-extension movement. (F) Comparison of the temporal angle variation for the elbow and shoulder joints with the EMG envelopes of the extensor carpi ulnaris (ECU) and biceps brachii (BB) muscles.
The final example demonstrates a straightforward flexion-extension movement of the arm of a healthy human subject. The coordinates from the motion capture system were imported in MotionAnalyser to create a 3D model of this movement (Figure 4E). The flexion-extension movement is primarily caused by a biphasic angle variation in the elbow and shoulder joints (Figure 4F). Processing EMG signals with MotionAnalyser for the biceps brachii and the extensor carpi ulnaris also reveals a biphasic curve, with muscle contractions occurring during the dynamic phases of flexion and extension.
Altogether, these results show that MotionAnalyser is a reliable and versatile tool that enables an integrative analysis, where motion can be confronted to electrophysiology and to sensor signals for a better understanding of the biomechanics of the movement.
Discussion
All the free and open source software packages described in the introduction implement several kinds of tracking algorithms suited for a plethora of different experimental setups and conditions. However, they mostly compute basic kinematic quantities. Derivation of higher order kinematics and its subsequent analysis is left to the researcher. Our software, instead, places itself in a somewhat opposite direction: given the position of the points of interest (which requires the tracking problem to be already solved), it computes basic and higher order kinematic variables and produces multiple plots commonly used in the analysis of motion behavior, thereby streamlining the data analysis process. Other software packages like OpenSim (Seth et al., 2018) are suitable for advanced kinematic calculations and for simulation of movements based on a skeleton model. In general, the software packages for combinatorial analysis of movement and physiological data require programming skills for installation and use. In contrast, our software aims to provide a user-friendly analysis platform that does not require coding skills. Although we found some overlap between the functions of MotionAnalyser and those of Kinovea, the latter specializes in humans and sport science, offering little generalization for animal models, and does not support 3D analysis from motion capture and gait analysis. Neither does it support the processing of electrophysiology traces and sensor signals for an integrative movement analysis. The proprietary solutions offer functionalities analogous to MotionAnalyser. Some of them are superior, for example—they integrate 3D motion capture with real-time animation and skeleton overlay, with or without muscles. They also propose different solutions to combine motion capture with external recording devices, such as force plate and EMG recording. However, they incur significant costs, do not always offer trial versions, the underlying code is often proprietary, and, in some cases are bound to a proprietary acquisition system, which further increases costs and may lack adaptability and decrease reusability of the equipment already acquired.
Using artificial datasets, we demonstrated that MotionAnalyser is a reliable tool. In addition to data analysis, the software offers a range of features that enhance user experience. For instance, importing data from different kinds of equipment for a multimodal analysis can be challenging, especially for users with limited computer skills. MotionAnalyser has a streamlined process for import and export of data in various file formats. In addition, the software handles datasets separately, enabling users to analyze them independently while displaying them together (if desired). This feature is particularly valuable for a decomposition of limb movement, such as the gait cycle. It is also a convenient tool to simultaneously animate multiple limbs. In addition, the software proposes an extensive toolbox for data preprocessing, such as concatenation and trimming of dataset, scaling, aligning, rotating, deletion of data columns, deletion of incorrect data points, interpolation of missing points, etc. In addition, the program can recalculate the time column and resample or interpolate datasets to a desired frequency, providing a solution to a common obstacle when analyzing data from different recording devices with varying sampling rates. MotionAnalyser also proposes a set of tools for digital filtering, rectifying, smoothing, and calculating FFT.
This software can be valuable in many research fields, such as neuroscience, rehabilitation medicine, sports, robotics, and others that address questions related to movement and coordination in various types of experiments. A typical example of use is for tracking animals in a confined environment, which is a very common approach in behavioral science. Originally, we created MotionAnalyser to analyze limb movements in animals, as illustrated by the vestibulospinal and the swimming tests. Using this, it becomes easy to address problems like balance deficits or malfunction of the motor system (Bachmann et al., 2013; Di Bonito et al., 2015; Züchner et al., 2018; Toch et al., 2020). However, the concomitant use of electromyograms provides more insights into the function, as previously shown for the vestibulospinal test (Lambert et al., 2016). A combination of gait analysis with joint kinematic, electromyograms, and correlation analyses can reveal different relationships between specific parts of the movement. This approach is widely used across different disciplines like basic science (Capogrosso et al., 2016) or physical therapy (Martinez et al., 2022). Furthermore, to address a specific situation, it might be necessary to correlate movement with data from sensor systems, such as inertial measurement units in combination with a force plate (Drobnič et al., 2023). We, therefore, implemented the possibility to import and analyze data from sensor systems. The possibility to work on these different input systems within the same software allows for seamless observation of the different parameters by simply switching between the different tabs. In addition, MotionAnalyser offers a variety of mathematical correlation models that the user can choose from to compare the different kinematic parameters with electrophysiology and sensor system signals. Therefore, MotionAnalyser is a versatile tool that opens up the possibility of multimodal integrative analysis to improve the understanding of function across multiple disciplines.
In biological science and medical research, it is common to create in-house programs that are designed to be used by researchers with certain levels of computer literacy, or even by programmers themselves. In the field of movement tracking, a notable example is the development of DeepLabCut (Nath et al., 2019), a Python-based deep learning program used for tracking animals or limbs even in the absence of markers. Despite the efforts made by the DeepLabCut team to increase user-friendliness, such as providing learning materials through video tutorials and forums, it still requires time and effort for someone without substantial computer skills to establish this tracking platform. The need for more accessible software that does not require user programming skills is highlighted by recent efforts for creating an accessible open-source video analysis pipeline (Pennington et al., 2019). Because MotionAnalyser performs simpler tasks compared to machine learning-based programs, it was more straightforward to design an intuitive and user-friendly interface that necessitates only a minimal number of user-defined parameters. While it would have been simpler and quicker to code MotionAnalyser as a command line program, it was essential to develop a tool that could cater to both advanced users and to those who lack coding skills and the time to develop them. Consequently, we developed a graphical user interface (GUI), enabling users to perform analyses with minimal effort. As of the current version, the software only requires a basic level of computer literacy. There is a growing trend in the development of analysis software utilizing Python (Srinath, 2017). Python is freely available and open source, making it accessible to researchers without any associated costs. Furthermore, Python provides a wide array of libraries with diverse tools and functionalities, facilitating rapid development. However, the Python ecosystem can be quite complex to use, as it requires environment setup, library imports, installation hurdles, and other tweaks. Therefore, our choice fell on MATLAB, which—while carrying the drawback of being closed source and necessitating a license payment—has the advantage of having a straightforward installation process, while the licensing requirements do not apply to end users, thanks to its freely available runtime environment. To encourage other researchers to adopt the tool, we created a short user manual, which constitutes the base for a wiki that we are building. Additional resources include video tutorials and various example datasets. All resources are freely available for download at https://github.com/BoullandLab/MotionAnalyser. To increase the dissemination of the program, the software is released both as a .mlapp package and as a standalone installer for both Windows and Linux. Source code is available under the terms of GPLv2.
MotionAnalyser, however, has limitations that can be addressed in different ways; we will provide support for new functions and bug fixes for future releases of the program. At the moment, users deeply interested in gait analyses may find that the program is missing the option to calculate torque. Torque in human gait can be calculated using various methods, from EMG signals or by using force plates to capture the forces and moments exerted on the foot during walking or running. Given MotionAnalyser’s current ability to handle EMG and sensor signals, adding torque calculation is possibly one of the first improvements that have to be considered. Another interesting improvement could be to implement calcium imaging, which could be particularly interesting to correlate with motion. The only possible way to do that at the moment is to import values from regions of interest in the E-Phys tab. Being able to handle the image analysis directly from MotionAnalyser could be an interesting improvement. It has also been proposed to implement the analysis of audio tracks, which could be relevant in different experimental setups like the assessment of echolocation during flight maneuvers in bats (Falk et al., 2015). Yet, at the moment, the only possibility is to import the audio trace as an electrophysiogy signal. This gives access to the filtering provided in the E-Phys tab, though it may not be extensive enough for this purpose. Another improvement for a future release could be to enhance the animation tab with the possibility to fit the tracked POI to a skeletal model. A technical improvement to increase the exporting speed of video or animated gif files could also be interesting, since benchmark tests have revealed this process to be the slowest in the program. Further development of preprocessing tools could also be interesting. For instance, in the present version of the software, the dataset stitching feature primarily allows users to align and concatenate datasets based on their visual overlap. However, it does not offer camera calibration options and adjustments to compensate for distortions and smoothly blend the coordinates. Implementing these transformations in a future release of MotionAnalyser could also be considered. We encourage the use of the issue tracker of the repository to request additional features and report bugs.
Despite these limitations, MotionAnalyser is an appealing software because of its accuracy, user-friendliness, versatility, free access, and open-source code. It is designed to cater to a wide range of researchers across various disciplines, regardless of their level of computer literacy.
Data availability statement
The analysis software described in this article is free and open source. It is available for download, along with example data files, from https://github.com/BoullandLab/MotionAnalyser.
Ethics statement
The dataset showcasing animal models was from previous studies (Di Bonito et al., 2015; Züchner et al., 2018). In each case, animal experiments were approved by Ethical Committees (CIEPAL NCE/ 2014-209 in France and Forsøksdyrutvalget, FDU ID. 4591 in Norway). The datasets used to show human locomotion (Figures 3 and 4) were from a voluntary subject, who is a co-author of this article (JLB). The other datasets from human movements and EMG signals, were obtained from published databases (Averta et al., 2020).
Author contributions
Conceptualization: J-LB; Methodology: J-LB and EA; Writing–review and editing: J-LB and EA; Visualization: J-LB; Funding acquisition: J-LB. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Helse Sør-Øst RHF (2019078 and 2020033 to J-LB)
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1250102/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Examples of data preprocessing functions for adjusting a coordinate dataset. (A) Digital filtering (30 and 35 Hz) of a dummy sine signal containing components at 25, 30, and 35 Hz. (B) Fast Fourier Transform of the signal before and after sequential application of the 30 and 35 Hz Notch filters. (C) Importing coordinates for the joints of the left and right leg as a single set creates a model of the leg joints attached by a segment. Importing the left and right leg coordinates independently results in a correct representation. (D) Correction of the mirror effect by inverting x-coordinates. (E) Translation of x and/or y-coordinates to align legs on a desired point. (F) Rescaling of the dataset. (G) Trimming the dataset. (H) Reduction of segmentation density for displaying high-frequency data points.
SUPPLEMENTARY FIGURE S2 | Sequence of actions required to stitch coordinates. (A) Dataset from Camera 1 shown in grey (B) Positioning of the dataset from Camera 2 shown in orange. (B) Determination of the stitching position (green position). (D) Concatenation of the two datasets.
SUPPLEMENTARY FIGURE S3 | Examples of signal treatment options offered by MotionAnalyser. (A) Fast Fourier Transform of an EMG signal. (B) Spectrogram for the same EMG signal. (C) Raw EMG signal used in (A,B). (D) Digital filtering and rectification of the signal. (E) Signal envelope.
SUPPLEMENTARY FIGURE S4 | Benchmarking of different functions of MotionAnalyser using a mid-range computer and five different input files containing an increasing number of datapoints from 1,300 to 13 million values. (A) Time for importation of raw values. (B) Time for plotting a stick diagram. (C) Time for saving/exporting data after analysis using different file formats. (D) Time for various functions.
SUPPLEMENTARY FIGURE S5 | Accuracy of MotionAnalyser for biomechanics analysis. (A) Dummy array of coordinates with an affine x-component and a quadratic y-component. (B) Temporal variation of x- and y-speed and acceleration calculated from the dummy arrays shown in (A). (C) Dummy array for creating segments attached at a known angle. A, known angle; MA, angle calculated by MotionAnalyser. (D) Dummy array for emulating human gait, depicting the different leg positions: heel strike (red), heel-off (green), toe-off (blue), maximum toe-lift (magenta), and intermediate positions (gray). Data are expressed as arbitrary units (Au). (E) Gait diagram generated from the dummy array (D). (F) Position of a mouse hindlimb at a specific time point during linear swimming, overlaid with the lines from three protractors centered on the hip, knee, and ankle joint. The readout from the protractor (cyan) is identical to the values calculated by MotionAnalyser (green). (G) Comparison of values calculated by MotionAnalyser using a real dataset with values calculated by ImageJ, spreadsheet calculation, or obtained by manual measurements. For better visual comparison, the velocity and acceleration values were divided by 100 and 10,000 respectively, while the gait parameters were multiplied by 10.
SUPPLEMENTARY FIGURE S6 | Accuracy of MotionAnalyser for signal treatment and correlations. (A) Digital filtering (30 and 35 Hz) of a dummy sine signal containing components at 25, 30, and 35 Hz. (B) Fast Fourier Transform of the signal before and after sequential application of the 30 and 35 Hz Notch filters. (C) Dummy sine signals of known amplitude ratio and phase shift for testing correlation functions. (D) Auto- and cross-correlation from the signals in (C). Note that the auto-correlation of f2(x) overlaps that of f3(x). For visualization purposes, f2(x) has been slightly shifted on the right.
SUPPLEMENTARY TABLE S1 | Details of the key functions used in coding MotionAnalyser, including mathematical formulas and MATLAB code.
SUPPLEMENTARY VIDEO S1 | Animated model of human walk.
References
Aleksić, A., and Popović, D. B. (2022). New scale for assessing spasticity based on the pendulum test. Comput. Methods Biomech. Biomed. Engin 25, 593–602. doi:10.1080/10255842.2021.1970144
Averta, G., Barontini, F., Catrambone, V., Haddadin, S., Handjaras, G., Held, J. P. O., et al. (2020). U-Limb: a multi-modal, multi-center database on arm motion control in healthy and post-stroke conditions. Gigascience 10, giab043. doi:10.1093/gigascience/giab043
Azarfar, A., Zhang, Y., Alishbayli, A., Miceli, S., Kepser, L., van der Wielen, D., et al. (2018). An open-source high-speed infrared videography database to study the principles of active sensing in freely navigating rodents. GigaScience 7, giy134. doi:10.1093/gigascience/giy134
Barreiros, M. O., Dantas, D. O., Silva, L. C. O., Ribeiro, S., and Barros, A. K. (2021). Zebrafish tracking using YOLOv2 and Kalman filter. Sci. Rep. 11, 3219. doi:10.1038/s41598-021-81997-9
Bachmann, L. C., Matis, A., Lindau, N. T., Felder, P., Gullo, M., and Schwab, M. E. (2013). Deep brain stimulation of the midbrain locomotor region improves paretic hindlimb function after spinal cord injury in rats. Sci. Transl. Med. 5, 208ra146. doi:10.1126/scitranslmed.3005972
Bacqué-Cazenave, J., Courtand, G., Beraneck, M., Lambert, F. M., and Combes, D. (2018). Temporal relationship of ocular and tail segmental movements underlying locomotor-induced gaze stabilization during undulatory swimming in larval Xenopus. Front. Neural Circuits 12, 95. doi:10.3389/fncir.2018.00095
Bari, M. S., Allen, S. S., Mesken, J., Cohen-Barnhouse, A. M., and Campbell, D. L. M. (2021). Relationship between range use and fearfulness in free-range hens from different rearing enrichments. Anim. (Basel) 11, 300. doi:10.3390/ani11020300
Ben-Shaul, Y. (2017). OptiMouse: a comprehensive open source program for reliable detection and analysis of mouse body and nose positions. BMC Biol. 15, 41. doi:10.1186/s12915-017-0377-3
Biering-Sørensen, F., Nielsen, J. B., and Klinge, K. (2006). Spasticity-assessment: a review. Spinal Cord. 44, 708–722. doi:10.1038/sj.sc.3101928
Boakye, M., Morehouse, J., Ethridge, J., Burke, D. A., Khattar, N. K., Kumar, C., et al. (2020). Treadmill-based gait kinematics in the yucatan mini pig. J. Neurotrauma 37, 2277–2291. doi:10.1089/neu.2020.7050
Branson, K., Robie, A. A., Bender, J., Perona, P., and Dickinson, M. H. (2009). High-throughput ethomics in large groups of Drosophila. Nat. Methods 6, 451–457. doi:10.1038/nmeth.1328
Brown, D., and Cox, A. (2009). Innovative uses of video analysis. Phys. Teach. 47, 145–150. doi:10.1119/1.3081296
Cao, Z., Hidalgo, G., Simon, T., Wei, S.-E., and Sheikh, Y. (2021). OpenPose: realtime multi-person 2D pose estimation using Part Affinity fields. IEEE Trans. Pattern Analysis Mach. Intell. 43, 172–186. doi:10.1109/tpami.2019.2929257
Capogrosso, M., Milekovic, T., Borton, D., Wagner, F., Moraud, E. M., Mignardot, J.-B., et al. (2016). A brain–spine interface alleviating gait deficits after spinal cord injury in primates. Nature 539, 284–288. doi:10.1038/nature20118
Cordelières, F. (2006). “Manual Tracking: a semi-automated tool to keep a track of movements in 2 and 3D,” in American Society for Cell Biology 46th Annual Meeting, San Diego, CA, USA.
Di Bonito, M., Boulland, J.-L., Krezel, W., Setti, E., Studer, M., and Glover, J. C. (2015). Loss of projections, functional compensation, and residual deficits in the mammalian vestibulospinal system of hoxb1-deficient mice. eNeuro 2, ENEURO.0096-15.2015. doi:10.1523/eneuro.0096-15.2015
Dale, R. B. (2012). “Clinical gait assessment,” in Physical rehabilitation of the injured athlete (Elsevier), 464–479.
Drobnič, M., Verdel, N., Holmberg, H.-C., and Supej, M. (2023). The validity of a three-dimensional motion capture system and the garmin running dynamics pod in connection with an assessment of ground contact time while running in place. Sensors (Basel) 23, 7155. doi:10.3390/s23167155
Erfianto, B., Rizal, A., and Hadiyoso, S. (2023). Empirical mode decomposition and Hilbert spectrum for abnormality detection in normal and abnormal walking transitions. Int. J. Environ. Res. Public Health 20, 3879. doi:10.3390/ijerph20053879
Falk, B., Kasnadi, J., and Moss, C. F. (2015). Tight coordination of aerial flight maneuvers and sonar call production in insectivorous bats. J. Exp. Biol. 218, 3678–3688. doi:10.1242/jeb.122283
Fang, H.-S., Li, J., Tang, H., Xu, C., Zhu, H., Xiu, Y., et al. (2023). AlphaPose: whole-body regional multi-person pose estimation and tracking in real-time. IEEE Trans. Pattern Analysis Mach. Intell. 45, 7157–7173. doi:10.1109/tpami.2022.3222784
Fitzsimmons, N. A., Lebedev, M. A., Peikon, I. D., and Nicolelis, M. A. L. (2009). Extracting kinematic parameters for monkey bipedal walking from cortical neuronal ensemble activity. Front. Integr. Neurosci. 3, 3. doi:10.3389/neuro.07.003.2009
Graving, J. M., Chae, D., Naik, H., Li, L., Koger, B., Costelloe, B. R., et al. (2019). DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife 8, e47994. doi:10.7554/elife.47994
Gruner, J. A., and Altman, J. (1980). Swimming in the rat: analysis of locomotor performance in comparison to stepping. Exp. Brain Res. 40, 374–382. doi:10.1007/bf00236146
Hegge, S., Kudryashev, M., Smith, A., and Frischknecht, F. (2009). Automated classification of Plasmodium sporozoite movement patterns reveals a shift towards productive motility during salivary gland infection. Biotechnol. J. 4, 903–913. doi:10.1002/biot.200900007
Hsu, A. I., and Yttri, E. A. (2021). B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nat. Commun. 12, 5188. doi:10.1038/s41467-021-25420-x
John, K., Stenum, J., Chiang, C.-C., French, M. A., Kim, C., Manor, J., et al. (2023). Accuracy of video-based gait analysis using pose estimation during treadmill walking versus overground walking in persons after stroke. Phys. Ther. 8, pzad121. doi:10.1093/ptj/pzad121
Johnson, C. D., Outerleys, J., and Davis, I. S. (2022). Agreement between sagittal foot and tibia angles during running derived from an open-source markerless motion capture platform and manual digitization. J. Appl. Biomech. 38, 111–116. doi:10.1123/jab.2021-0323
Kreneisz, O., and Glover, J. C. (2015). Developmental Characterization of Tail Movements in the Appendicularian Urochordate Oikopleura dioica. Brain Behav. Evol. 86, 191–209. doi:10.1159/000439517
Lambert, F. M., Bras, H., Cardoit, L., Vinay, L., Coulon, P., and Glover, J. C. (2016). Early postnatal maturation in vestibulospinal pathways involved in neck and forelimb motor control. Dev. Neurobiol. 76, 1061–1077. doi:10.1002/dneu.22375
Lee, S. Y., Park, S. J., Gim, J.-A., Kang, Y. J., Choi, S. H., Seo, S. H., et al. (2023). Correlation between Harris hip score and gait analysis through artificial intelligence pose estimation in patients after total hip arthroplasty. Asian J. Surg. S1015-9584, 00778-9. doi:10.1016/j.asjsur.2023.05.107
Lonini, L., Moon, Y., Embry, K., Cotton, R. J., McKenzie, K., Jenz, S., et al. (2022). Video-based pose estimation for gait analysis in stroke survivors during clinical assessments: a proof-of-concept study. Digit. Biomark. 6, 9–18. doi:10.1159/000520732
López Ruiz, J. R., Castillo Hernández, L., De la Torre Valdovinos, B., Franco Rodríguez, N. E., Dueñas Jiménez, J. M., Dueñas Jiménez, A., et al. (2017). Locomotion in intact and in brain cortex-ablated cats. Neuroscience 358, 37–48. doi:10.1016/j.neuroscience.2017.06.026
Martinez, C., Garbett, S., Hiromasa, K., Jackson, R., Miya, E., Miya, M., et al. (2022). Comparison of 2-D and 3-D analysis of running kinematics and actual versus predicted running kinetics. Int. J. Sports Phys. Ther. 17, 566–573. doi:10.26603/001c.34432
Mehdizadeh, S., Nabavi, H., Sabo, A., Arora, T., Iaboni, A., and Taati, B. (2021). Concurrent validity of human pose tracking in video for measuring gait parameters in older adults: a preliminary analysis with multiple trackers, viewing angles, and walking directions. J. Neuroeng Rehabil. 18, 139. doi:10.1186/s12984-021-00933-0
Meijering, E., Dzyubachyk, O., and Smal, I. (2012). “Chapter nine - methods for cell and particle tracking,” in Methods in enzymology. Editor P. M. conn (Academic Press), 183–200.
Mönck, H. J., Jörg, A., von Falkenhausen, T., Tanke, J., Wild, B., Dormagen, D., et al. (2018). BioTracker: an open-source computer vision framework for visual animal tracking. arXiv:1803.07985.
Nakamichi, R., and Asahara, H. (2022). Motion-capture analysis of mice using a video recorded on an iPhone camera. Bio Protoc. 12, e4539. doi:10.21769/bioprotoc.4539
Nath, T., Mathis, A., Chen, A. C., Patel, A., Bethge, M., and Mathis, M. W. (2019). Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176. doi:10.1038/s41596-019-0176-0
Pennekamp, F., Schtickzelle, N., and Petchey, O. L. (2015). BEMOVI, software for extracting behavior and morphology from videos, illustrated with analyses of microbes. Ecol. Evol. 5, 2584–2595. doi:10.1002/ece3.1529
Pennington, Z. T., Dong, Z., Feng, Y., Vetere, L. M., Page-Harley, L., Shuman, T., et al. (2019). ezTrack: an open-source video analysis pipeline for the investigation of animal behavior. Sci. Rep. 9, 19979. doi:10.1038/s41598-019-56408-9
Pereira, T. D., Aldarondo, D. E., Willmore, L., Kislin, M., Wang, S. S.-H., Murthy, M., et al. (2019). Fast animal pose estimation using deep neural networks. Nat. Methods 16, 117–125. doi:10.1038/s41592-018-0234-5
Pereira, T. D., Tabris, N., Matsliah, A., Turner, D. M., Li, J., Ravindranath, S., et al. (2022). SLEAP: a deep learning system for multi-animal pose tracking. Nat. Methods 19, 486–495. doi:10.1038/s41592-022-01426-1
Pérez-Escudero, A., Vicente-Page, J., Hinz, R. C., Arganda, S., and de Polavieja, G. G. (2014). idTracker: tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 11, 743–748. doi:10.1038/nmeth.2994
Puig-Diví, A., Escalona-Marfil, C., Padullés-Riu, J. M., Busquets, A., Padullés-Chando, X., and Marcos-Ruiz, D. (2019). Validity and reliability of the Kinovea program in obtaining angles and distances using coordinates in 4 perspectives. PLOS ONE 14, e0216448. doi:10.1371/journal.pone.0216448
Rasband, W. S. (1997). ImageJ U. S. National institutes of health; bethesda, MD, USA. Available at: http://rsb.info.nih.gov/ij/.
Risse, B., Berh, D., Otto, N., Klämbt, C., and Jiang, X. (2017). FIMTrack: an open source tracking and locomotion analysis software for small animals. PLOS Comput. Biol. 13, e1005530. doi:10.1371/journal.pcbi.1005530
Rodriguez, A., Zhang, H., Klaminder, J., Brodin, T., Andersson, P. L., and Andersson, M. (2018). ToxTrac: a fast and robust software for tracking organisms. Methods Ecol. Evol. 9, 460–464. doi:10.1111/2041-210x.12874
Romero-Ferrero, F., Bergomi, M. G., Hinz, R. C., Heras, F. J. H., and de Polavieja, G. G. (2019). idtracker.ai: tracking all individuals in small or large collectives of unmarked animals. Nat. Methods 16, 179–182. doi:10.1038/s41592-018-0295-5
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Schweihoff, J. F., Loshakov, M., Pavlova, I., Kück, L., Ewell, L. A., and Schwarz, M. K. (2021). DeepLabStream enables closed-loop behavioral experiments using deep learning-based markerless, real-time posture detection. Commun. Biol. 4, 130. doi:10.1038/s42003-021-01654-9
Seth, A., Hicks, J. L., Uchida, T. K., Habib, A., Dembia, C. L., Dunne, J. J., et al. (2018). OpenSim: simulating musculoskeletal dynamics and neuromuscular control to study human and animal movement. PLOS Comput. Biol. 14, e1006223. doi:10.1371/journal.pcbi.1006223
Smith, T. M., Berk, A. S., and Upadhyay, H. (2011). Severe hypothermia in a patient with spinal cord injury without radiological abnormality. J. Emerg. Trauma Shock 4, 421–424. doi:10.4103/0974-2700.83878
Sridhar, V. H., Roche, D. G., and Gingins, S. (2019). Tracktor: image-based automated tracking of animal movement and behaviour. Methods Ecol. Evol. 10, 815–820. doi:10.1111/2041-210x.13166
Stenum, J., Rossi, C., and Roemmich, R. T. (2021). Two-dimensional video-based analysis of human gait using pose estimation. PLoS Comput. Biol. 17, e1008935. doi:10.1371/journal.pcbi.1008935
Sun, J. J., Ryou, S., Goldshmid, R. H., Weissbourd, B., Dabiri, J. O., Anderson, D. J., et al. (2022). “Self-supervised keypoint discovery in behavioral videos,” in Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2022, New Orleans, LA, USA, 18-24 June 2022, 2161–2170.
Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., et al. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90. doi:10.1016/j.ymeth.2016.09.016
Toch, M., Harris, A., Schakman, O., Kondratskaya, E., Boulland, J.-L., Dauguet, N., et al. (2020). Onecut-dependent Nkx6.2 transcription factor expression is required for proper formation and activity of spinal locomotor circuits. Sci. Rep. 10, 996. doi:10.1038/s41598-020-57945-4
Walter, T., and Couzin, I. D. (2021). TRex, a fast multi-animal tracking system with markerless identification, and 2D estimation of posture and visual fields. eLife 10, e64000. doi:10.7554/elife.64000
Washabaugh, E. P., Shanmugam, T. A., Ranganathan, R., and Krishnan, C. (2022). Comparing the accuracy of open-source pose estimation methods for measuring gait kinematics. Gait Posture 97, 188–195. doi:10.1016/j.gaitpost.2022.08.008
Yamanaka, O., and Takeuchi, R. (2018). UMATracker: an intuitive image-based tracking platform. J. Exp. Biol. 221, jeb182469. doi:10.1242/jeb.182469
Zhang, C., Li, H., and Han, R. (2020). An open-source video tracking system for mouse locomotor activity analysis. BMC Res. Notes 13, 48. doi:10.1186/s13104-020-4916-6
Keywords: software, motion, integrated, electrophysiology, sensors, analysis, open source
Citation: Annavini E and Boulland J-L (2023) Integrated software for multi-dimensional analysis of motion using tracking, electrophysiology, and sensor signals. Front. Bioeng. Biotechnol. 11:1250102. doi: 10.3389/fbioe.2023.1250102
Received: 30 June 2023; Accepted: 13 October 2023;
Published: 22 November 2023.
Edited by:
Rafael Gonzalez-Cano, University of Granada, SpainReviewed by:
Francis A. M. Manno, East Carolina University, United StatesAlberto Ambesi Impiombato, Psychogenics, United States
Copyright © 2023 Annavini and Boulland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Luc Boulland, ai5sLmJvdWxsYW5kQG1lZGlzaW4udWlvLm5v
 Eis Annavini
Eis Annavini Jean-Luc Boulland
Jean-Luc Boulland