
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 01 June 2023
Sec. Biomaterials
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1191104
This article is part of the Research Topic Organ Mimicking Technologies and Their Applications in Drug Discovery View all 7 articles
Viral and bacterial infections continue to pose significant challenges for numerous individuals globally. To develop novel therapies to combat infections, more insight into the actions of the human innate and adaptive immune system during infection is necessary. Human in vitro models, such as organs-on-chip (OOC) models, have proven to be a valuable addition to the tissue modeling toolbox. The incorporation of an immune component is needed to bring OOC models to the next level and enable them to mimic complex biological responses. The immune system affects many (patho)physiological processes in the human body, such as those taking place during an infection. This tutorial review introduces the reader to the building blocks of an OOC model of acute infection to investigate recruitment of circulating immune cells into the infected tissue. The multi-step extravasation cascade in vivo is described, followed by an in-depth guide on how to model this process on a chip. Next to chip design, creation of a chemotactic gradient and incorporation of endothelial, epithelial, and immune cells, the review focuses on the hydrogel extracellular matrix (ECM) to accurately model the interstitial space through which extravasated immune cells migrate towards the site of infection. Overall, this tutorial review is a practical guide for developing an OOC model of immune cell migration from the blood into the interstitial space during infection.
The COVID-19 pandemic has highlighted the ongoing danger posed by pathogens or microbes to our society. New disruptive research is necessary to gain a deeper understanding on how these pathogens infect specific cells in our body and the tissue damage they inflict, and to develop methods to mitigate and prevent their spread. Organs-on-chip (OOC) are microphysiological models aiming to replicate an organ’s functional unit. Over the past decade, the OOC field has experienced exponential development (Ingber, 2022). Since the first lung-on-chip was published in 2010 (Huh et al., 2010), many other OOCs and multi-organ-on-chip models have been developed. Although advances in the OOC field have been remarkable, the inclusion of cellular immune components in these systems is a recent development. The immune system plays a significant role in organ homeostasis and the inflammatory tissue response upon infection. Therefore, pathologies modeled in OOC systems without the respective cellular immune component will deviate significantly from human physiology.
Multiple reviews have summarized immunocompetent OOC models (Maharjan et al., 2020; Miller et al., 2020; Morsink et al., 2020) and the interactions between pathogens and the immune system on chip (Tang et al., 2020; Feaugas and Sauvonnet, 2021; Saygili et al., 2021). While these reviews excellently describe the state-of-the-art, the practical aspects of infection-on-chip research are not discussed in-depth.
This tutorial review aims at providing a guide for both experts and newcomers in the field on establishing an immunocompetent OOC model of the inflammatory process during infection, either by modelling inflammation with a chemoattractant or by modelling infection with a pathogen. It includes an introduction to the innate and adaptive immune system, with a focus on recruitment of innate immune cells, followed by key aspects to consider when modelling infection on a chip. Special focus is provided on integrating an endothelial and epithelial barrier on a chip using a hydrogel ECM, as well as important experimental aspects to be considered. The review also discusses incorporating immunity on-chip, mimicking infection, monitoring immune cell transmigration across the vascular wall and within the tissue and presents future perspectives on the next level of immunocompetent OOC systems.
The inflammatory cascade involves a complex interplay of various immune cells and signaling molecules interacting with non-immune cells, such as epithelial and endothelial cells. Tissue resident immune cells, the first to be activated by pathogens, release cytokines that activate the local blood vascular endothelium. The activated blood vascular endothelium increases its expression of adhesion molecules and chemokines, which attracts circulating immune cells (Maas et al., 2018). The immune cells transmigrate through the endothelial layer, cross the underlying basement membrane, and migrate through the tissue towards the site of infection (Abbas et al., 2012). More exhaustive descriptions of the mechanisms and pathways involved in immune cell transmigration can be found in other reviews (Ley et al., 2007; Abbas et al., 2012; Sturtzel et al., 2017; Maas et al., 2018).
It is essential to distinguish between the innate and adaptive immune response: the innate immune response is the first, fast response to infection, whereas the adaptive immune response is pathogen-specific and generates memory cells for resolving future infections with the same pathogen (Chaplin, 2010; Abbas et al., 2012). For an effective adaptive immune response, incorporation of the lymphatics is necessary, to allow for dendritic cell migration from the site of infection to the lymph nodes, presenting antigens to T Cells in the lymph nodes. This review will focus on the innate immune response, where innate immune cells, mainly neutrophils and monocytes, are recruited to the site of infection from the blood vasculature.
Pathogens are typically detected by their pathogen-associated molecular patterns (PAMPs) (Alper et al., 2018), which, in turn, are recognized by pattern recognition receptors (PRRs) on cells in tissues. PRR activation induces an intracellular signaling cascade that triggers pro-inflammatory actions, such as cytokine production, to attract immune cells and activate blood vessel endothelial cells (BECs). The activated endothelium attracts peripheral immune cells, which transmigrate from the blood vessel to the infection site. This transmigration cascade is a multistep process (Figure 1), which includes the capture, rolling, arrest, crawling, and transmigration of immune cells at the level of post-capillary venules (Ley et al., 2007; Abbas et al., 2012; Sturtzel et al., 2017). After crossing the microvascular endothelium, immune cells migrate through the extracellular tissue space to the site of infection.
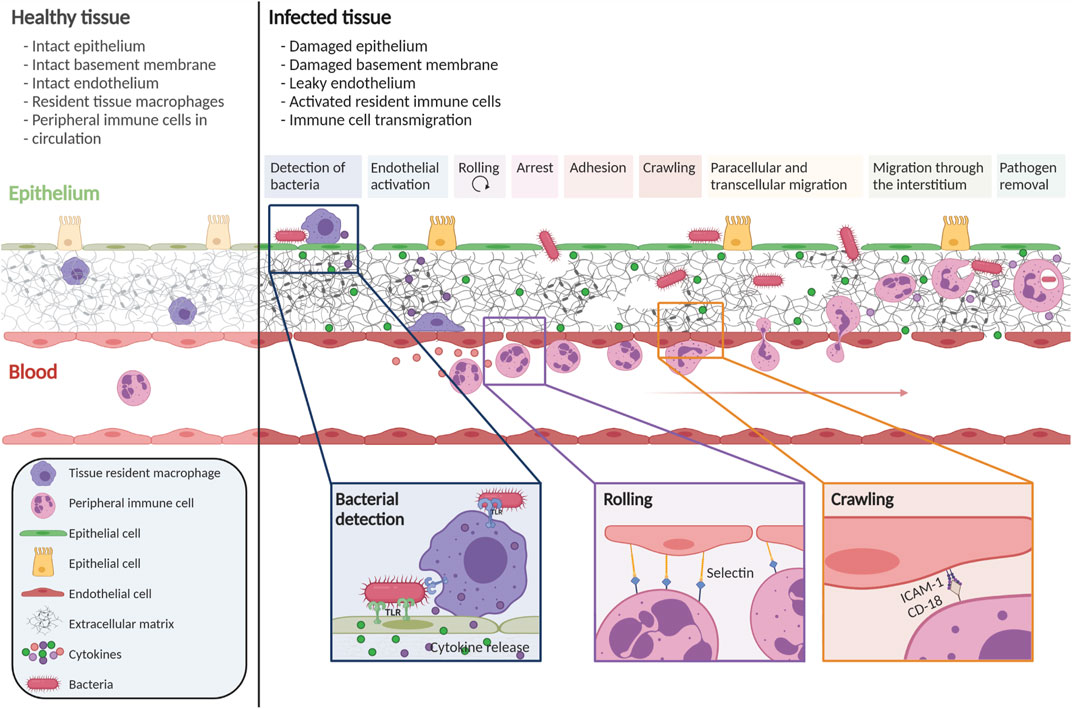
FIGURE 1. Immune cell transmigration cascade upon infection. The immune cell transmigration cascade is a multistep process. During infection, gaps in the epithelium, endothelium, and ECM are observed. Tissue-resident immune cells detect the pathogen and secrete cytokines to activate the endothelium and attract peripheral immune cells. Through multiple steps, peripheral immune cells are attracted towards the site of infection. This figure is based on numerous reviews and research publications (Ley et al., 2007; Abbas et al., 2012; Sturtzel et al., 2017; Alper et al., 2018; Maas et al., 2018). Created with BioRender.com.
To initiate the process of capture and rolling, passively flowing immune cells must come into contact with the endothelium, which occurs when BECs are activated. BEC activation arises through cytokine signaling but also through altered blood flow. In normal laminar flow conditions, the BECs are tightly aligned, and the cells are in a quiescent state. However, when the flow is disturbed and oscillatory, BECs are less aligned because they sense the flow variations, leading to BEC activation (Heo et al., 2011). In both disturbed flow and cytokine activation, activated BECs upregulate adhesion molecules like E- and P-selectins and members of the IgCAM family such as ICAM-1 amd VCAM-1, the main molecules involved in immune cell capture and rolling (Ley et al., 2007; Abbas et al., 2012; Muller, 2013; Maas et al., 2018).
When the immune cells arrest after rolling, the cells flatten to decrease their exposure to shear stress and other circulating cells (Figure 1). (Maas et al., 2018) Chemoattractant exposure, as well as binding to integrins, is essential in regulating arrest with different signals inducing firmer adhesion of the immune cell to the endothelium. Firm adhesion to the endothelium allows the cells to crawl along the endothelium to find the optimal location for transmigration across the endothelial cell wall. The majority of cells follow the paracellular transmigration route, where the cell migrates between BECs, via the cell-to-cell junctions (Muller, 2011). A smaller percentage of immune cells opt for transcellular diapedesis, in which the immune cell crosses the barrier by going through a BEC.
After reaching the extracellular tissue space, the immune cells’ ability to migrate towards the site of infection is crucial. To accomplish this, the cell tracks various chemokines through the dense extracellular matrix (ECM) network. Tissue-resident macrophages and the epithelium produce chemokines to stimulate migration. To ensure that chemokines are correctly positioned, they are bound to the ECM, often to proteoglycans in the ECM (Gill et al., 2010; O’Dwyer et al., 2018). Immune cells respond to chemokines based on factors such as chemoattractant type, concentration, and exposure time (Amulic et al., 2012). Resident cells interact with the migrating immune cells and can also produce matrix-modifying enzymes, such as matrix metalloproteinases (MMPs) and collagenases, which specifically degrade the ECM, thus potentially facilitating movement of immune cells through the tissue (Parks et al., 2004; O’Dwyer et al., 2018; Miskolci et al., 2021). The combined action of MMP production, chemokine signaling, and cell-cell interaction facilitates immune cell migration towards the site of infection in the extracellular tissue space.
In vivo studies on innate immune cell extravasation across the vascular wall have primarily focused on easily accessible structures, such as muscle, skin, and mesentery (Halin et al., 2005). Research on these tissues benefits from better imaging quality than internal tissues that are challenging to reach. Within these tissues, immune cells mostly transmigrate from the postcapillary venule (Hickey and Westhorpe, 2013; Weirather and Frantz, 2015). The specific transmigration mechanisms used by immune cells vary considerably depending on the organ of interest, the immune cell subset, and the time during inflammation caused by infection. For example, in the skin, immune cells migrate from the postcapillary venule through a large extracellular space to reach the site of infection, whereas in the lung, the air-blood barrier is very narrow, resulting in the immune cells mostly transmigrating from the capillaries into the airspace (Maas et al., 2018). Thus, the molecular mechanisms by which immune cells transmigrate depend on their microenvironment, specifically the vascular characteristics of the respective tissue. Unfortunately, live imaging of extravasation in internal organ vessels can only be carried out after a complex surgical procedure or with expensive equipment such as a two-photon microscope to perform intravital imaging (Kim et al., 2019a). Surgery not only causes stress for the animals, which may activate immune cells, but anesthesia also influences vascular dilation and immune cell activation, making it crucial to identify alternative methods to study the mechanisms of inflammation and infection in vitro (Hickey and Westhorpe, 2013).
Two main types of in vitro models for immune cell extravasation are commonly used: (1) static models and (2) basic flow chambers (Muller and Luscinskas, 2008; Salminen et al., 2020). In these models, a monolayer of BECs is cultured to confluency and immune cells are introduced either statically or under flow conditions. To generate an innate immune response, BECs and/or innate immune cells are exposed to a pro-inflammatory stimulus. In contrast, adaptive immune cells inherently extravasate, but a pro-inflammatory stimulus is necessary for activation of adaptive immune cells. The process of immune cell extravasation is visualized and analyzed using microscopy.
Static models consist of two compartments between which immune cells migrate by crossing the endothelium and then entering the second compartment. The most well-known model is the two-chamber migration assay, also known as the transwell assay (Falasca et al., 2011). In this assay, BECs are grown in a filter insert, and immune cells cross the endothelium and then squeeze through the pores of the filter insert to reach the lower compartment. Unfortunately, this system does not incorporate an ECM environment for immune cells to migrate into. A more elaborate assay, the collagen hydrogel transendothelial migration assay, was developed to address this issue. In this modified assay, an endothelial monolayer is formed on a collagen hydrogel, and immune cells migrate from the media space across the endothelial monolayer into the underlying hydrogel (Muller and Luscinskas, 2008).
In the bloodstream, immune cells are exposed to shear stress, affecting BEC function and immune cell transmigration (Brooks et al., 2002). To investigate this further, laminar flow chamber assays have been utilized to study immune cell adhesion and transmigration under flow conditions, allowing a better understanding of the extravasation cascade and the role of selectins in adhesion and rolling (Salminen et al., 2020). However, these assays involve culturing BECs on stiff and rigid substrates without an ECM hydrogel to simulate the soft interstitial space. To address this limitation, OOC models offer a more advanced platform, allowing for the co-culture of multiple cell types, including physiological fluid flow, and cell culture on soft hydrogel ECMs.
In the following sections, the development of an OOC model of infection (infection-on-chip) will be discussed. Individual components of the model are shown in Figure 2: the chip design, hydrogel ECM, microvascular endothelium, epithelium, tissue resident and circulating immune cells, and induction of inflammation with a chemotactic gradient or mimicking infection with a pathogen. These components, with practical aspects on how to establish them in the laboratory, will be elaborated on later in this review.
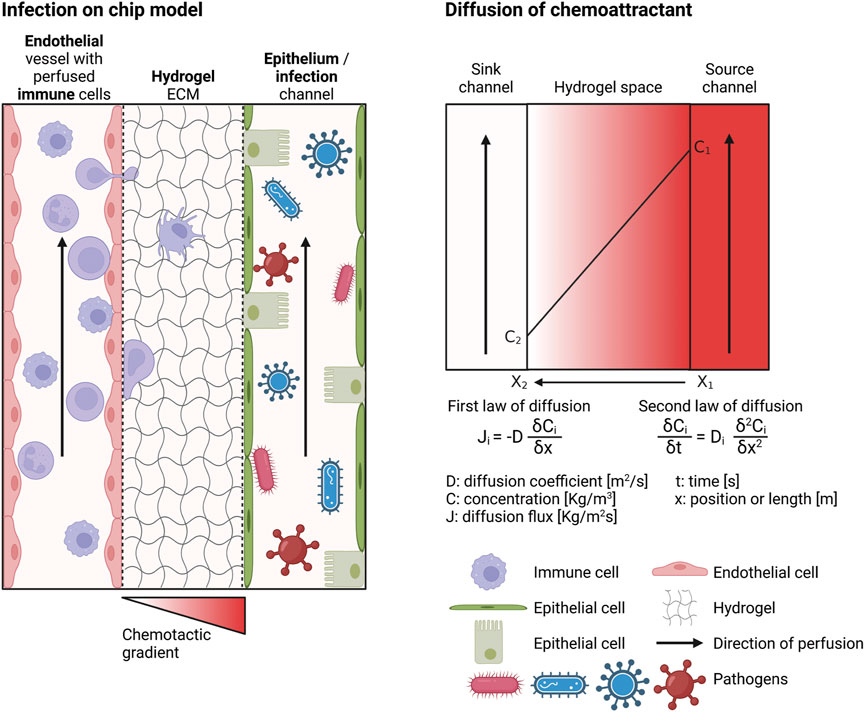
FIGURE 2. Requirements of an infection-on-chip model. The infection-on-chip schematic shows the main elements needed to create an infection-on-chip model: microvascular endothelial cells, epithelial cells, immune cells (optimally peripheral immune cells in the blood vessel and tissue resident immune cells in the ECM), a hydrogel mimicking the ECM environment, and a chemotactic gradient to induce inflammation or a pathogen to model infection. The chemoattractant diffusion is depicted (right side). Created with BioRender.com.
A microfluidic chip to model immune cell extravasation and migration during infection should be multicompartmental, to enable the creation of an endothelium-hydrogel-epithelium barrier, and perfusable, to perfuse immune cells and create a chemotactic gradient (Figure 2). Several commercialized microfluidic chips are available, such as from Mimetas, AimBiotech, and BEOnChip (Zhang and Radisic, 2017; Zommiti et al., 2022). However, a customized microfluidic chip can be developed if a specific design is desired. Figure 3 gives an overview of possible chip designs. Information concerning chip fabrication, typically using soft lithography, can be found elsewhere. (Cameron et al., 2022; Cho et al., 2022; Leung et al., 2022). Briefly, a mold is created by 3D printing or stereolithography and filled with polydimethylsiloxane (PDMS) mixed with a curing agent. Cured PDMS is post-processed and bonded to another PDMS part or a glass slide to close channel structures. The chip’s design is based on the abovementioned requirements: multi-compartmentalization with a hydrogel ECM and perfusability. Generally, designs can be categorized into two groups: chips with channels created inside the hydrogel structure and chips with a hydrogel barrier separating the microfluidic channels.
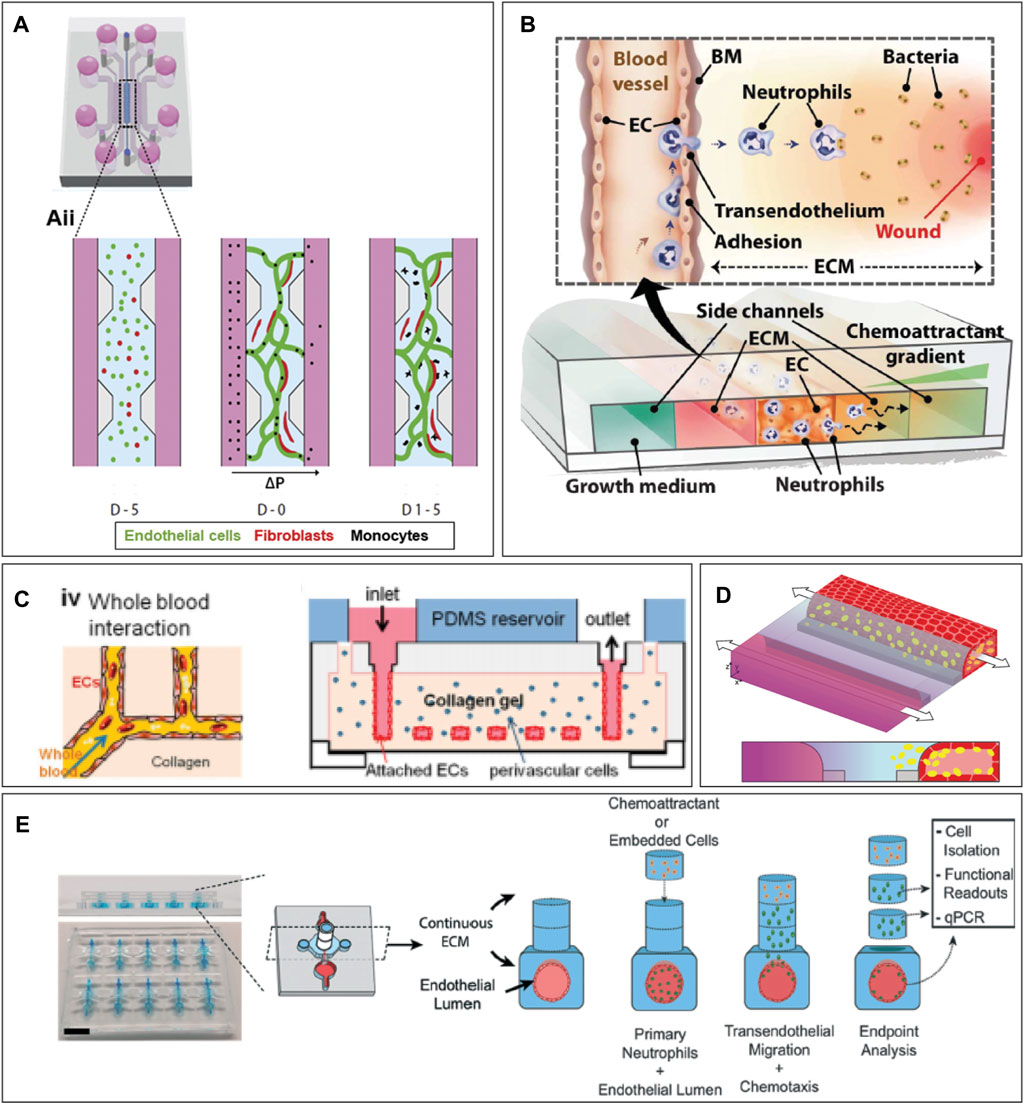
FIGURE 3. Examples of organ-on-chip designs incorporating hydrogels. (A) Pillar-based system for microvasculature formation. Image from Boussommier et al. (Boussommier-Calleja et al., 2019) (B) Neutrophil extravasation system. Image from Han et al. (Han et al., 2012) (C) Hydrogel-based chip with molded vessel structure within PDMS device. Image from Zheng et al. (Zheng et al., 2012) (D) A phaseguide separates the hydrogel from the media channel, creating a single blood vessel in the media channel. (E) Neutrophil extravasation on chip using stacks. Image from McMinn et al. (McMinn et al., 2019). Figure D was adapted from De Haan et al. (de Haan et al., 2021), under the terms of the Creative Commons Attribution License (CC BY). (https://creativecommons.org/licenses/by/4.0/).
In hydrogel-based chips, the microfluidic channels are created into the hydrogel itself, and structural material, like PDMS, is only utilized to encapsulate the structure for easier handling. To create channels inside a hydrogel, a mold can be generated out of a substrate such as PDMS, and the hydrogel is polymerized on top of the mold to create the channels (Figure 3C). (Zheng et al., 2012) These channels can be used to assess bacterial and mammalian cell chemotaxis (Cheng et al., 2007) and microvessels can be molded (Qiu et al., 2018). To create a single cylindrical blood vessel, a hydrogel is formed with a removable needle inside (Park et al., 2010; Tourovskaia et al., 2014; Zeinali et al., 2021). Such devices can be used to study immune cell extravasation from the blood vessel lumen into the hydrogel (Lee et al., 2017), to create multiple vessels inside a hydrogel with different BEC types (Hasan et al., 2015), and create hydrogel stacks on top of the blood vessel to investigate neutrophil migration distance (Figure 3E). (McMinn et al., 2019) Moreover, the hydrogel surrounding the vessel can be dried to create a stable, dense collagen construct in which endothelial cells can be grown and neutrophil extravasation can be observed (Chen et al., 2018). A sacrificial hydrogel can be used to create structures within different hydrogels. For example, agarose can be encapsulated by crosslinked gelatin hydrogels. After gelatin gelation, the agarose can be flushed out, and a cylindrical lumen remains (Bertassoni et al., 2014). Lastly, structures inside a hydrogel can be formed through laser ablation. With this method, a laser specifically ablates the regions of interest to create a unique microchannel design inside a hydrogel on chip (Brandenberg et al., 2016; Nikolaev et al., 2020).
To confine the hydrogel in one compartment, one can make use of surface tension. The hydrogel can be restricted within one channel using pillar structures (Huang et al., 2009; Bichsel et al., 2015; Adriani et al., 2017; Aizel et al., 2017; Campisi et al., 2018; Li et al., 2018; Zeinali et al., 2018; Boussommier-Calleja et al., 2019) (Figure 3A) or a channel height difference (Vulto et al., 2011; Poussin et al., 2020; de Haan et al., 2021; Riddle et al., 2022) (Figure 3D). This allows the user to create an endothelial and epithelial barrier alongside the hydrogel and observe immune cell migration through the hydrogel (Poussin et al., 2020; de Haan et al., 2021; Riddle et al., 2022). In another model, migration between the media and the gel channel is only possible via a number of smaller openings in the center of the chip (Shin et al., 2012). This model has been used to examine breast cancer metastasis as well as neutrophil transendothelial migration into the collagen hydrogel (Figure 3B). (Han et al., 2012; Bersini et al., 2014; Na et al., 2017) Lastly, a model with a suspended hydrogel can be created, for example, to model an airway-on-chip (Humayun et al., 2018).
Most designs discussed here currently do not include pump-assisted uni-directional flow. To include a pump-assisted flow of immune cells, a chip has to be adapted to be connected to a pump. This has been accomplished to investigate immune cell migration across an endothelial layer on chip (Zhang et al., 2016; Menon et al., 2017). Where one system focuses on forming a single channel with the aid of a collagen hydrogel (Menon et al., 2017), the other system engineers a biodegradable scaffold for a microvascular network from which immune cells can migrate (Zhang et al., 2016).
With the aid of surface tension, a hydrogel can be contained in one microfluidic compartment of the chip. During the design phase, it is crucial to calculate the change in contact angle necessary to maintain the hydrogel in the correct compartment. These calculations, based on the Young–Laplace equation, have been extensively described (Huang et al., 2009).
A tight connection between the pipette tip and the hydrogel inlet is recommended to simplify pipetting the hydrogel into the chip. Ensuring this tight connection involves designing the inlet size with precise dimensions matching the end of the pipette tip.
The PDMS chip material is known for its poor cellular adherence (Leung et al., 2022). Hence, surface modification of the material to allow for cell attachment is essential. Usually, the material is coated with an ECM mixture, such as collagen-fibronectin (Dabaghi et al., 2021). The hydrophobicity of PDMS results in poor attachment of ECM proteins unless further modifications are made. Thus, PDMS pre-treatment with oxygen plasma or chemicals like polydopamine or (3-aminopropyl)triethoxy silane (APTES) is recommended (Dabaghi et al., 2021). Adding a pre-treatment with ECM coating improves BEC and hydrogel adhesion to the OOC, preventing cell detachment and hydrogel contraction.
Hydrogels, as their name suggests, mainly consist of water, with a 3D polymer network forming the structure. The wide variety of available polymers with different properties enables the recapitulation of different ECM types (Liu et al., 2019). Importantly, a hydrogel’s mechanical properties can be tuned through various methods to be similar to those of human tissues. Hydrogel chemical composition and topographies can be adjusted to mimic the tissue of interest, and specific ECM components can be included if desired. An organ-specific hydrogel resembles the instructive native environment of the cells, to emulate a functional tissue environment.
The most commonly used hydrogels for on-chip culture are created from collagen, fibrinogen, Matrigel, gelatin, polyacrylamide (PA), polyethylene glycol (PEG), or hyaluronic acid (HA) (Caliari and Burdick, 2016). More recently, hydrogels comprising native tissue ECM have also been created to recapitulate the ECM of the organ of interest more accurately (Marhuenda et al., 2022). In most of the infection-on-chip models, hydrogels are the central environment between the epithelium and endothelium (Figure 2).
Immune cells, like other cells, interact with the proteins in hydrogels. On top of a 2D hydrogel surface, the cells are less constrained. On the other hand, encapsulated inside a 3D hydrogel, the cells are more constrained but have more points of adhesion to the matrix which can be used for immune cell migration, but are not necessary for immune cells to migrate (Caliari and Burdick, 2016). Figure 4 gives an overview of the hydrogel properties that influence immune cell migration.
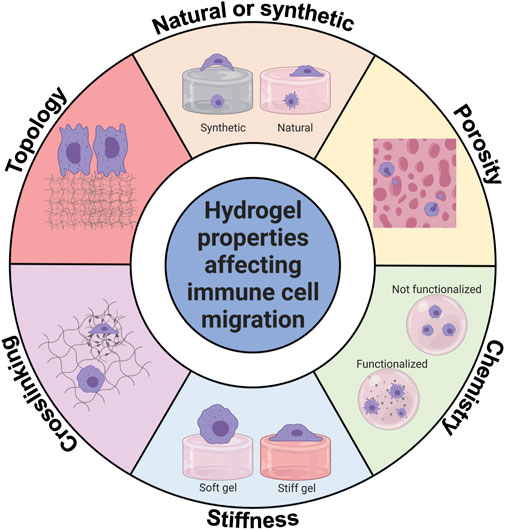
FIGURE 4. Hydrogel properties affecting innate immune cell migration. Innate immune cell migration is affected by hydrogel chemistry, type, stiffness, porosity, topology, and crosslinking. This figure shows graphically how innate immune cells can be affected. Created with BioRender.com.
Synthetic hydrogels are created from non-natural polymers, such as polyacrylamide (PA) or polyethylene glycol (PEG), while naturally derived hydrogels are produced from, for example, collagen, fibrinogen, or gelatin (Lee et al., 2017; Liu et al., 2019). While naturally derived hydrogels are considered closest to the composition of native ECM, they are susceptible to biodegradation and contraction, leading to the deformation of the intended structure (Liu et al., 2019). In contrast, synthetic hydrogels are often bioinert, meaning they do not interact with immune cells unless activated by adding specific peptides to these hydrogels.
Hybrid hydrogels are created by mixing different hydrogels, for example, by mixing one natural hydrogel with another, such as collagen and fibrinogen (Sano et al., 2018). A hybrid hydrogel, consisting of a mixture of Geltrex and collagen, was used to investigate neutrophil extravasation (Riddle et al., 2022). Results showed that neutrophils extravasated to a larger extent in geltrex/collagen mix gel compared to collagen only, exemplifying the effect of hydrogel composition on neutrophil migration.
The functionalization of hydrogels with specific peptides can improve adhesion and alter cell behavior. This has been mainly observed with PEG hydrogels and the RGD peptide, a sequence of the three amino acids arginine, glycine, and aspartate. For example, implanting PEG alone induced a foreign body reaction with macrophage accumulation, which did not occur upon PEG-RGD conjugated hydrogel implantation (Lynn et al., 2010). On the other hand, naturally-derived hydrogels suffer from batch-to-batch variability, and their chemical structure can have unwanted effects on immune cells. For instance, fibrin is essential in inflammation and wound healing. In vitro, fibrin retains its chemotactic properties, attracting macrophages to the area where fibrin is present (Tanaka et al., 2019), which could influence experimental results.
Hydrogel porosity affects the mechanical structure and oxygen and nutrient diffusion. Collagen hydrogel density can be modified by altering collagen concentration in the hydrogel, which affects macrophage migration. In a dense collagen gel, macrophages migrate less than in a highly porous collagen gel (Ford et al., 2019; Pérez-Rodríguez et al., 2022). Another method of controlling density is by integrating soluble particles into a hydrogel, which, upon dissolution, leaves pores with a specific size (Lee et al., 2017). This method can be used to control pore size accurately.
Substrate stiffness is a crucial factor influencing immune cell migration. Hydrogels generally have low stiffness, which can be fine-tuned, making them optimal for assessing cell migration on different stiffness substrates. To create stiffness gradients, most studies adapt the concentration of hydrogel molecules or utilize a crosslinker (Whang and Kim, 2016).
On stiffer hydrogels, neutrophils flatten and spread more than on soft hydrogels, where they remain rounded (Oakes et al., 2009). This correlates to less total migration and more directional migration on stiffer hydrogels, whereas on soft hydrogels, neutrophils exhibit random walk behavior. When the chemoattractant fMLP is added, neutrophils on stiffer substrates migrate less and more directional than cells on a soft substrate. In another system, neutrophils are perfused through a microfluidic system with a polyacrylamide (PA) hydrogel (Jannat et al., 2010). Similar to the previous example, higher hydrogel stiffness leads to more directional migration, which could be visualized using traction force microscopy (Jannat et al., 2011).
BECs form more vessel-like structures on hydrogels with lower stiffness, whereas more MMPs are produced on higher-stiffness hydrogels (Hanjaya-Putra et al., 2010; Onken et al., 2014). Peripheral blood lymphocytes or neutrophils added to an endothelial monolayer created on soft and stiff hydrogels were found to transmigrate more on more rigid substrates (Onken et al., 2014; Lauridsen and Gonzalez, 2017).
By specific hydrogel crosslinking, a pattern of altered stiffness can be created. For instance, fibrin hydrogel can be crosslinked with blue light exposure when using a ruthenium crosslinker. A pattern can be created by only exposing specific areas to blue light (Keating et al., 2019). This technique was used to investigate macrophage migration (Hsieh et al., 2019). Again, it was observed that macrophages are flatter and less round on stiff, crosslinked gels compared to soft, not crosslinked gels. In addition, macrophages migrated more on crosslinked hydrogels than non-crosslinked ones, which correlated to increased TNF-α secretion.
Topotaxis is the directional migration of cells across a specific topology. These 3D patterns influence cellular behavior (Matellan et al., 2019). In a study with melanoma cells, it was observed that cells form long filopodia when cultured on areas with fewer pillars. In contrast, cells form short and randomly oriented protrusions in highly dense pillar areas (Park et al., 2016). A study of macrophage migration examined the effect of differently shaped collagen gels (more fibrous or more globular) and found that more macrophages transmigrated across the fibrous collagen compared to the globular collagen (Vasse et al., 2018).
In summary, hydrogel chemistry, composition, porosity, stiffness, crosslinking, and topology play a crucial role in immune cell migration and can affect the in vitro modeling of immune cell migration.
Hydrogels are produced from various precursor solutions with different properties. The production and gelation process depends on the hydrogel (Lee et al., 2017). Collagen gelation is both pH- and temperature dependent, and the hydrogel is produced from an acidic precursor solution that is neutralized (Doyle, 2016). GelMA, the modified version of gelatin, is photo-crosslinkable with UV light. Fibrinogen forms a hydrogel upon contact with thrombin within minutes, whereas collagen gelation can take between 30 min and multiple hours, depending on the gelation temperature (Liu et al., 2019). Many hydrogels (e.g., collagen, fibrin, Matrigel) require working on ice during preparation to prevent fast gelation.
The hydrogel properties of different hydrogels also have an effect on pipetting method. Due to the fast gelation time, pipetting of fibrin hydrogels has to be carried out on ice in small volumes that can immediately be transferred from the mixing tube into the chips. On the other hand, collagen gelates slower but the solution is very viscous, hence pipetting of collagen is often carried out with pipette tips with an extra wide opening. These tips are commercially available, but can also be generated in-house by cutting the end off a standard pipette tip.
More information on hydrogel production and gelation, as well as hydrogel incorporation on chip, can be found in these reviews (Caliari and Burdick, 2016; Jiang et al., 2016; Terrell et al., 2020). Depending on the chip design required for the assay, the hydrogel production requirements can play a prominent role in choosing the hydrogel for the organ-on-chip.
One of the key features of hydrogels is their permeability. It can easily be assessed using a permeability assay. A dye coupled to a fluorescent molecule with a known molecular weight (e.g., a RITC-dextran or FITC-dextran) is added, and dye diffusion into the hydrogel is monitored using timelapse microscopy. The permeability can then be calculated based on the following equation (Haase and Kamm, 2017; Ho et al., 2017; Van Duinen et al., 2017):
Papp: permeability coefficient.
CECM: concentration (measured through dye intensity) of the dye in the ECM.
VECM: volume of the ECM compartment.
AEC: surface of the monolayer where the dye interfaces with the ECM.
CVessel: concentration (measured through dye intensity) of the dye in the microchannel.
Culturing BECs on chip utilizes two main methods to model blood vessels: (1) generating a vessel-like three-dimensional structure and culturing monolayers of BECs on this defined structure (Figure 2) or (2) mixing BECs with mural cells inside a hydrogel to create self-assembled microvessels (Jeon et al., 2015; Zeinali et al., 2018; McMinn et al., 2019; Zeinali et al., 2021).
Recent advances in genetic screening, such as single-cell RNA sequencing, have shown that there is not one type but a wide variety of vascular endothelial cell (EC) subtypes specific to the vascular segment and the organ of interest. There are differences in morphology and gene expression between arterial, venous, and capillary BECs without even mentioning lymphatic ECs (Gifre-Renom et al., 2022). Moreover, BECs from different organs function differently, for example, the highly permeable liver sinusoids differ from the tight blood-brain barrier (Gifre-Renom et al., 2022). A human BEC type frequently used in research is the human umbilical vein endothelial cell (HUVEC), as these cells are widely available and easy to obtain from the umbilical cord. Although HUVECs are not microvascular cells, but venous cells, they have allowed for identification of molecular mechanisms of immune cell extravasation. Nevertheless, the choice of endothelial cell type is essential: when comparing a brain microvascular EC line to primary brain microvascular ECs, it was observed that the T Cell diapedesis was lower across primary brain microvascular ECs when compared to the brain microvascular cell line, with higher crawling distance on the primary brain microvascular ECs (Steiner et al., 2011).
Culturing BECs seems trivial, but many factors play a role in endothelial function and barrier tightness, such as confluency, cell culture media, culture substrate, shear stress, and perfusion mode.
The choice of culture material influences BEC growth. For example, in a chip with different hydrogels, HUVECs attach and grow nicely on gelatin-gelMA and alginate-gelMA but not on gelatin-alginate (Nie et al., 2018). Similarly, BECs attach more to fibrin hydrogels than to collagen I hydrogels. The most commonly used hydrogels for endothelial cell culture on chip are bovine-derived fibrin and rat tail-derived collagen I, due to their ease of use and low cost.
Cell culture media significantly affects cell morphology and function due to the presence or absence of various soluble factors (growth factors, chemokines, metabolites). For example, HUVECs cultured in M199 media lose their typical morphology only after a few passages, whereas HUVECs cultured in EGM-2 media retain their typical cobblestone morphology for up to 10 passages, most likely due to the additional growth factors present in EGM-2 media (Bala et al., 2011). Specialized media solutions have been developed for specific types of microvascular BECs, or to induce angiogenesis using a pro-angiogenic cocktail (van Duinen et al., 2019).
In the human body, BECs experience shear stress from the blood flow. Arterial BECs experience high shear stress, while venous BECs generally experience low shear stress. In vitro, shear stress can be applied in microfluidic systems, causing BECs to align along the flow direction and increase barrier tightness (Ohta et al., 2022). When applying alternating flow, this BEC alignment is not observed, indicating that this method of applying shear stress is non-physiological (Lee et al., 2019).
To experimentally validate if the endothelial barrier is intact and functional, barrier protein expression analysis and permeability assays are important tools. Junctional maturation can be assessed by immunostaining of junctional localisation of VE-cadherin, PECAM-1, JAMs, and possibly other tight junction proteins, to assess the confluency and barrier morphology. On the other hand, a permeability assay quantifies barrier tightness. The permeability of an endothelial monolayer bordering a hydrogel interface can be calculated with Eq. (1). More information on measuring endothelial permeability values in OOCs can be found elsewhere (Haase and Kamm, 2017).
Functionally, it is crucial to know the BEC type in your culture. For example, does this endothelial cell type express and relocate adhesion molecules to the cell membrane upon an infectious stimulus, and does it express known markers for the BEC type of interest? Also, while HUVEC are broadly used for their ease to culture, studies involving tissue-specific BEC are starting to emerge, to take the specificities of these cells into account, for example, in a model of pulmonary infection on chip (Bai et al., 2022). Optimizing the culture conditions mentioned in this chapter can create a favorable BEC culture environment.
In barrier organs, such as the lung, intestine, and skin, the epithelium is the first physical barrier pathogens encounter upon entry. In the infection-on-chip model, the epithelial barrier is located next to the hydrogel ECM environment and acts as the site of infection (Figure 2).
In the lung, the alveolar epithelium comprises two main cell types: alveolar type I and II cells. The thin type I cells line the alveolar sac and allow gas exchange, covering around 95% of the alveolus (Weibel, 2015). Type II cells, on the other hand, are more stem cell-like, have regenerative potential, and produce surfactant. During infection, alveolar epithelial cells secrete cytokines to activate the endothelial barrier and attract immune cells from the bloodstream (Manicone, 2009). Throughout the pathogen removal process, the epithelium is severely damaged, leading to the accumulation of liquid and cell debris in the alveolar space (Yamada et al., 2016). Multiple lung-on-chip models with an epithelial barrier have been used to study lung infection (Deinhardt-Emmer et al., 2020; Deinhardt-Emmer et al., 2021; Bai et al., 2022).
The intestinal epithelial barrier consists of a multitude of cell types. During infection, the intestinal barrier is disrupted and immune cells can migrate into the underlying tissue. This was shown in a model of T Cell migration across the intestinal epithelial barrier (de Haan et al., 2021).
The skin has a complex, multilayered cellular barrier, that is often modelled in cell culture inserts (Kosten et al., 2016). To model skin toxicity from oral exposure to metals, a multi-organ on chip with gingiva and skin tissue was developed (Koning et al., 2022). In another model, T Cell migration upon skin inflammation was investigated (Ren et al., 2021).
Epithelial cells can be obtained from primary tissue, but due to limited availability and high variability between donors, on-chip culture of these cells is challenging. In the case of alveolar epithelial cells, alveolar type II cells are highly susceptible to their environment and quickly differentiate to type I cells when placed in culture, leading to a loss of the regenerative type II population. Therefore, cell lines have been used in lung-on-chip models of infection (Huh et al., 2010; Deinhardt-Emmer et al., 2020). However, these cell lines are derived from cancerous tissue and are thus not representing the cells present in normal tissue homeostasis. Similarly, the intestinal epithelium is a complex, self-renewing environment that is often modelled with simple epithelial cell lines on chip (de Haan et al., 2021). However, recent research has shown that intestinal organoids can be grown inside a hydrogel with an in vivo-like anatomical structure, leading to a highly in vivo-like intestinal model (Nikolaev et al., 2020).
The cell culture media of epithelial cells affects their growth and differentiation. Since alveolar type II cells differentiate in standard culture media, a defined media composition has been developed to maintain alveolar type II cells (Sun et al., 2021). However, not only the epithelial cells are affected by the culture media, but BECs can sense this media too and can migrate from the endothelial compartment towards the pro-angiogenic factors in the epithelial cell compartment.
When co-culturing multiple cell types, optimizing how long cells need to form a functional barrier is necessary. For example, an immortalized alveolar epithelial cell line can take up to 3 weeks to establish a functional barrier, whereas the co-cultured BECs in the model do not need this long culture time to form a functional barrier (Sengupta et al., 2022). Moreover, the air-liquid interface (ALI) culture of pulmonary epithelial cells improves their function (Hiemstra et al., 2019), but a liquid interface is required for chemoattractant perfusion through the epithelial channel. Future studies should look into nebulization of the infectious agent to retain ALI, but this is out of the scope of the current review.
For modeling the innate immune response on chip, macrophages, monocytes or neutrophils can be cultured inside the OOC (Figure 2; Table 1). The organ of interest plays a significant role in immune cell action, as immune cells are very responsive to the other cells in their environment. Table 1 gives an overview of immune cells used on chip, their source, and isolation method. Both the innate and adaptive immune response have been modeled on chip, or a mixed population of peripheral immune cells was utilized.
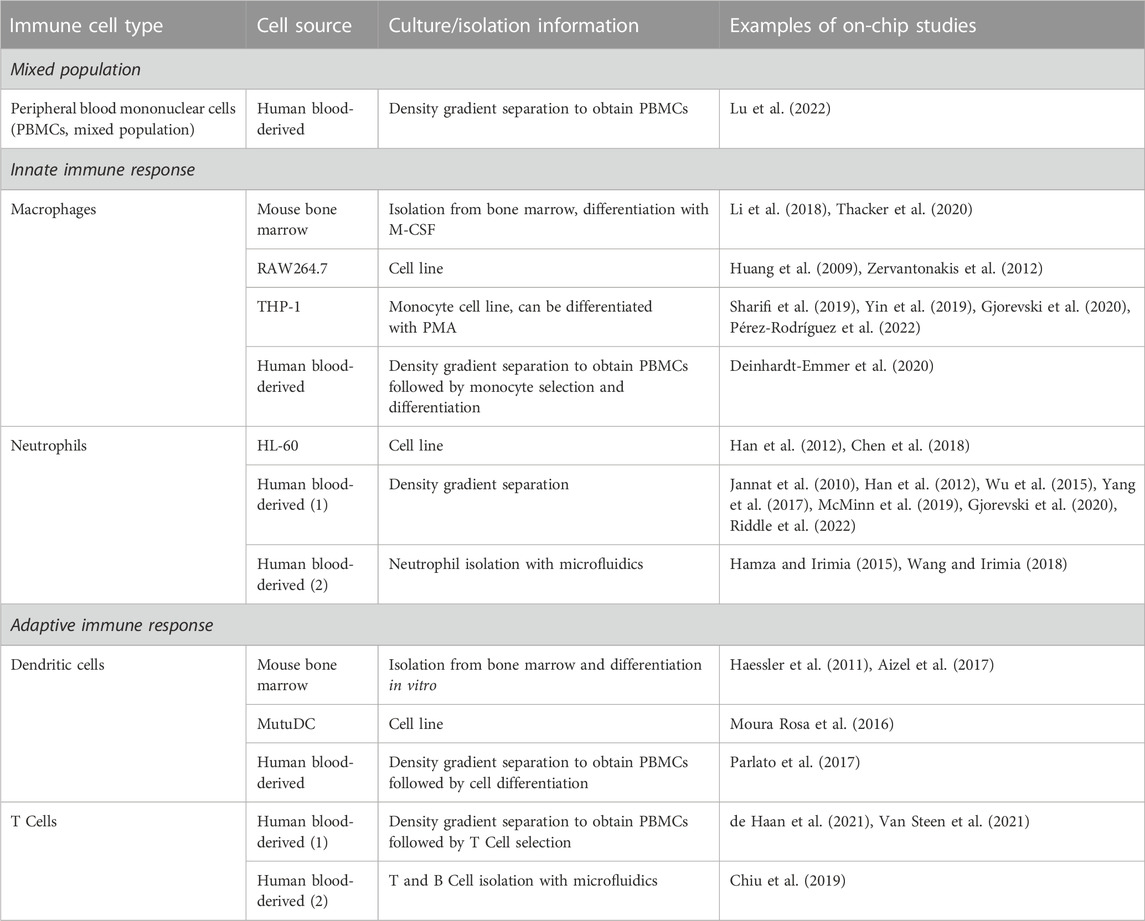
TABLE 1. Immune cell sources for on-chip culture. This table lists different types of immune cells, their source, isolation method, and examples of chips on which these cells were cultivated. This is a non-exhaustive list of examples of how immune cells were incorporated into OOC models. More methods of immune cell isolation are possible, but have not been tested on chip yet.
Even though the most physiological model would contain only freshly isolated human immune cells, the choice of cells also highly depends on the availability of specific cell types, the research question, and the organ of interest (Granton et al., 2018). Generally, combining multiple immune cell types to investigate their role during infection would be interesting, for example, by combining tissue-resident macrophages in the ECM hydrogel compartment with monocytes in the vascular compartment (Chen et al., 2023). In the infection-on-chip model, the main focus is the extravasation and migration of peripheral immune cells perfused through the blood vessel (Figure 2).
Isolation of human peripheral immune cells is traditionally carried out by gradient separation from whole blood or buffy coat, with post-processing of the cells to obtain specific populations, such as differentiated macrophages (Grievink et al., 2016; Golke et al., 2022). Recently, negative and positive selection-based methods with magnetic sorting have been developed (Chometon et al., 2020). These methods, although more costly than traditional gradient separation, achieve a higher purity of the immune cell subset of interest. Depending on the immune cell type of interest, different isolation methods can be elected (Table 1).
Like endothelial and epithelial cells, immune cells also react to their cell culture media. For example, the foreign material in fetal bovine serum (FBS) can cause monocyte activation. Hence, human autologous serum is used for immune cell culture to prevent activation (Deinhardt-Emmer et al., 2020). To visualize immune cell migration, fluorescent labelling of immune cells is standard practice. However, this chemical labeling can activate immune cells or when membrane labeling is used, limit their migration by inhibiting cell membrane motility, which is why one should label the cytoplasm or genetically modify the cells. Thus, the correct labelling solution should be used, and immune cell activation should be checked. Methods to check for immune cell activation are flow cytometry, microscopy, or assessing cytokine secretion to measure activation markers.
In the human body, immune cells in the bloodstream experience different levels of shear stress. A majority of in vitro research is carried out statically, without the perfusion of immune cells (Huang et al., 2009; Han et al., 2012; Wu et al., 2015; Aizel et al., 2017; Parlato et al., 2017; McMinn et al., 2019). For a more physiological model, immune cells should be perfused through the blood vessel channel to observe rolling, arrest, polarization, crawling and finally diapedesis across the microvascular endothelial layer into the hydrogel (Bianchi et al., 2013). Adding perfusion has a significant effect on immune cell migration in general, as shown in a study where macrophages embedded inside a hydrogel were exposed to interstitial flow or kept under static conditions (Li et al., 2018). In this model, interstitial flow increased macrophage migration inside the hydrogel. Furthermore, adding tumor cells showed that both interstitial flow and the presence of tumor cells increase macrophage motility (Lee et al., 2020). In another study, perfusing neutrophils through a microfluidic channel increased the number of neutrophils migrating into a collagen matrix towards tumor spheroids, compared to keeping the neutrophils static (Surendran et al., 2021). Additionally, T Cells, but not neutrophils, can migrate against the physiological flow in an in vitro flow chamber coated with ICAM-1 and SDF-1 or fMLP (Valignat et al., 2013). Physiological shear stress values between 2–12 dyne/cm2 were compared, and the higher the shear stress, the straighter the cells moved along or against the flow direction. Under low shear stress, more random walk behavior was observed. Although the flow rate influences migration direction, the migration speed was not affected, which was confirmed in another study (Rainger et al., 1999). Overall, these results indicate that shear stress impacts immune cell migration, showing increased migration capacity of immune cells under physiological fluid flow.
Next to the interactions with the microenvironment, cell-cell interaction is crucial for immune cell function. In a study of macrophage migration within hydrogels, co-culture with fibroblasts increased macrophage migration and led to tunnel formation (Ford et al., 2019). Hydrogel stiffness only decreased in cocultures of fibroblasts and macrophages, and remained unaltered in monocultures. Moreover, macrophages differentiated more towards an M2 phenotype in co-culture, whereas in monoculture, a mixed M1/M2 population is observed. In a study of neutrophil migration, in the presence of an IL-8 gradient, neutrophils do not migrate much, but with an endothelial layer between the neutrophils and the IL-8 gradient, neutrophil migration is significantly increased (McMinn et al., 2019). In a lymph node-on-chip system, naïve T Cells were exposed to antigen-presenting dendritic cells and binding was analyzed, showing interactions between different types of immune cells (Chiu et al., 2019). Generally, cell-cell interaction influences immune cell behavior and is an important consideration when testing immune cell migration on chip.
To attract innate immune cells, a chemical signal to direct them is necessary. This chapter focuses on solutions to generate a chemotactic gradient for immune cell migration. First, it should be mentioned that chemotaxis and haptotaxis are different processes that both take place in-vivo. Chemotaxis is defined as the directional migration of immune cells following a gradient of soluble chemokines in the environment, released from the site of infection (Petri and Sanz, 2018), while haptotaxis describes the migration of immune cells induced by a gradient of ECM-bound chemokines within the microenvironment. Here, the focus will be on chemotaxis, however, with ECM-based hydrogels, haptotaxis can also occur.
Many chemotactic agents are available, such as bacteria or viruses, factors secreted by other cells cultured in the system, or factors added to the cell culture media. For example, in an OOC, neutrophil extravasation can be induced by the presence of bacteria, pre-activated BECs, or a gradient of the bacterial peptide fMLP (Han et al., 2012; Hind et al., 2021). Here, we discuss forming a chemotactic gradient in a microfluidic device and some common chemotactic agents used for attracting immune cells.
The formation of a chemotactic gradient is crucial to investigate immune cell migration. In the absence of a gradient, when fMLP concentration is uniform, neutrophils migrate randomly in an inflammation-on-chip model using a polyacrylamide (PA) hydrogel. However, when an fMLP gradient is established, the cells move towards the higher concentration of the gradient (Jannat et al., 2010). The dominating transport mechanism in static microfluidic systems is diffusion, the random movement of particles inside a space, which eventually leads to an even spread of these particles (Figure 2). Diffusion is driven by a concentration difference of particles within a given space and is described by Fick’s first and second laws of diffusion (Figure 2). It is important to realize that the diffusion coefficient depends on the viscosity, which can vary importantly from one hydrogel to the other. It is thus essential to measure the hydrogel viscosity used and adjust the gradient’s setup accordingly. Moreover, the environment temperature and the chemoattractant’s particle size should be accounted for as well.
Many microfluidic systems use diffusion to create a chemotactic gradient (Zhao et al., 2020). A gradient can be created by simply pipetting the chemoattractant next to the hydrogel and letting it diffuse through the hydrogel or by using a pipette tip as chemottractant reservoir that is placed in one corner of the hydrogel and slowly releasing the chemoattractant, which can then diffuse through the hydrogel (Oakes et al., 2009). The drawback of using these basic gradients is that the diffusion flux (as shown in Figure 2) will decrease until the concentration is uniform throughout the entire space. To avoid this limitation, one can perfuse the chemoattractant continuously in the epithelial channel (source channel, Figure 2), while perfusing cell culture medium without chemoattractant in the endothelial channel (sink channel, Figure 2). Two main methods aimed at perfusing microfluidic channels are commonly used: pressure-driven and flow-controlled pumping systems. Tilters, also called rocking platforms, are broadly used pressure-driven systems for their simplicity as they do not require any tubings. Piston-pumps and peristaltic pumps are typical flow-controlled pumping systems.
A simple yet effective method to create flow inside a microfluidic system is by applying a pressure in a closed reservoir connected to a microfluidic channel (Luo et al., 2009). The pressure acting on the liquid contained in the reservoir induces a flow rate in the channel according to Poiseuille law. The flow rate is a linear function of the applied pressure and is limited by the fluidic resistance of the microfluidic channel (equations in Figure 5). The fluidic resistance of channels with rectangular cross-sections that are typical in microfluidics, can be calculated using the hydraulic diameter approximation. To avoid the need of tubing (de Graaf et al., 2022) to pressurize the reservoir, a hydrostatic pressure difference between the inlet and the outlet reservoirs can simply be used. One popular method that uses this approach is tilting platforms, also called rockers. They do not need much space and can thus be implemented for higher thoughput solutions (Luo et al., 2009; Yang et al., 2017; Li et al., 2018). A tilter is illustrated in Figure 5, in which the fluid level in the two reservoirs equilibrate with time. Once the levels are identical, the platform is tilted in the opposite direction, resulting in an alternating (bi-directional) flow with variable flow rate (Kim et al., 2015; Poussin et al., 2020; de Haan et al., 2021).
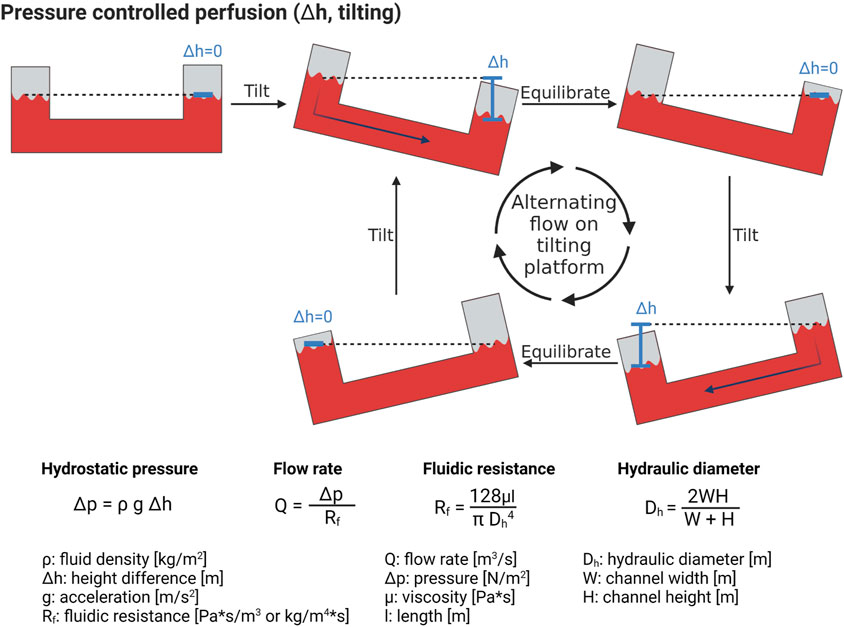
FIGURE 5. Hydrostatic pressure-induced perfusion of microfluidic systems. By tilting the microfluidic chip, a height difference in the liquid reservoirs is established, with a correlated difference in pressure, leading to fluid flow. Formulas to calculate the flow rate inside a tilting microfluidic system are presented. Created with BioRender.com.
Tilter platforms have some drawbacks, such as the limited range of flow rates than can be generated, which depends on both the platform’s tilting angle and the channel geometry. In addition, the flow is not constant and cannot be controlled. For instance, when an air bubble or cells obstruct the channel, the fluidic resistance increases, resulting in a decrease in flow rate. Finally, alternating flows are non-physiological. Recently, more evolved microfluidic systems allow for a uni-directional flow with tilter platforms (Wang and Shuler, 2018).
In contrast to pressure driven flow, controlled flows can be created by piston-pumps or peristaltic pumps, which can generate constant and uni-directional flow (Huh et al., 2010; Moura Rosa et al., 2016; Menon et al., 2017). In contrast to peristaltic pumps, piston pumps do not mechanically stress the cells, however, cells might sediment inside the piston or syringe. Peristaltic pumps do not present this limitation, however may induce hemolysis and/or cell activation due to the mechanical forces exerted on the cells (Hajipouran Benam et al., 2016; Sharifi et al., 2019).
While pumps are very accurate and are a good option for perfusion, there are some disadvantages to consider. First, the tubing must be sterilized and kept sterile throughout the experiment. Second, there is a high risk of bubble formation inside the tubing, which could damage the cell culture. To prevent bubbles in the OOC, bubble traps have been developed, and all media/buffers and tubing are placed inside the incubator to prevent bubble formation through changes in the humidity and temperature. Thirdly, connecting multiple OOCs to a pump can be cumbersome and time-consuming, leading to lower experimental throughput. A final consideration is the pump location: when the pump is placed inside the incubator, it is close to the chips, and the tubing can be short. However, this can cause overheating of the incubator, pose sterility issues, and the humid environment is detrimental to the pump. The pump can also be placed outside the incubator, increasing the tubing length required and, thus, the system’s complexity, especially if multiple OOCs are connected. Overall, pumps offer a reliable method to apply shear stress to an OOC, but throughput is decreased.
Chemotactic agents can be subdivided into two main categories: chemoattractant molecules, and living agents or pathogens. Importantly, when using a chemoattractant molecule, inflammation is modelled on chip, and only when adding a pathogen, infection is modelled. Of course, an infection will also cause inflammation on chip. To attract immune cells, different types of chemotactic molecules can be used. For example, these can be cytokines normally produced by other cells in the environment after infection, such as IL-2 or IL-8 (Irimia et al., 2006; Han et al., 2012; Wu et al., 2015). Pathogen-specific particles can also be used as a chemoattractant. Here, an infection with a pathogen is mimicked by inducing inflammation without needing to cultivate actual pathogens and apply the safety restrictions that accompany the use of dangerous pathogens. Some generally used molecules are PolyI:C, a viral mimic; lipopolysaccharides (LPS), a bacterial membrane saccharide; and fMLP, an immune cell-binding molecule secreted by bacteria (Jones et al., 2014; Hajipouran Benam et al., 2016; Chandrasekaran et al., 2017; Wang and Irimia, 2018). Lastly, exposure to nanoparticles can also lead to immune cell activation and migration. For all these chemotactic agents, it is crucial to choose the right concentration in which immune cells are activated but not overstimulated.
To model infection, viruses, fungi, or bacteria can be added to the infection-on-chip model. Because bacteria are still alive, interactions between the bacteria and the immune cells can be studied, which was already reported with E. Coli and tuberculosis bacteria (Huh et al., 2010; Thacker et al., 2020). Similarly, inhibiting fungal growth on chip through a neutrophil response can be investigated (Barkal et al., 2017). Various organ-on-chip systems of SARS-CoV2 infection and other viral infections have been recently developed, as described in other reviews (Chakraborty et al., 2020; Tang et al., 2020). The interactions between viruses, bacteria, and immune cells can also be studied, such as in a lung-on-chip with a double influenza and Staphylococcus aureus infection (Deinhardt-Emmer et al., 2020). Using live bacteria or viruses is more clinically relevant than a synthetic mimic but also poses more danger to the researcher handling these agents. Thus, appropriate safety measures have to be taken. One can also use cells to produce the chemoattractant within the system. For example, tumor cells secrete various agents that can attract dendritic cells, neutrophils, and macrophages (Huang et al., 2009; Parlato et al., 2017; Lee et al., 2021).
Overall, there are several options to model infection or create a chemotactic gradient, varying in complexity and type of infection.
To assess immune cell migration during infection, microscopy, specifically live cell imaging, is an essential tool. Accurate visualization of immune cell migration can be obtained with live cell imaging, where timelapse intervals and total imaging time are crucial to capture the process of interest (Figure 6A). However, there must be a balance between avoiding phototoxicity and increasing imaging frequency and length to capture all processes of interest.
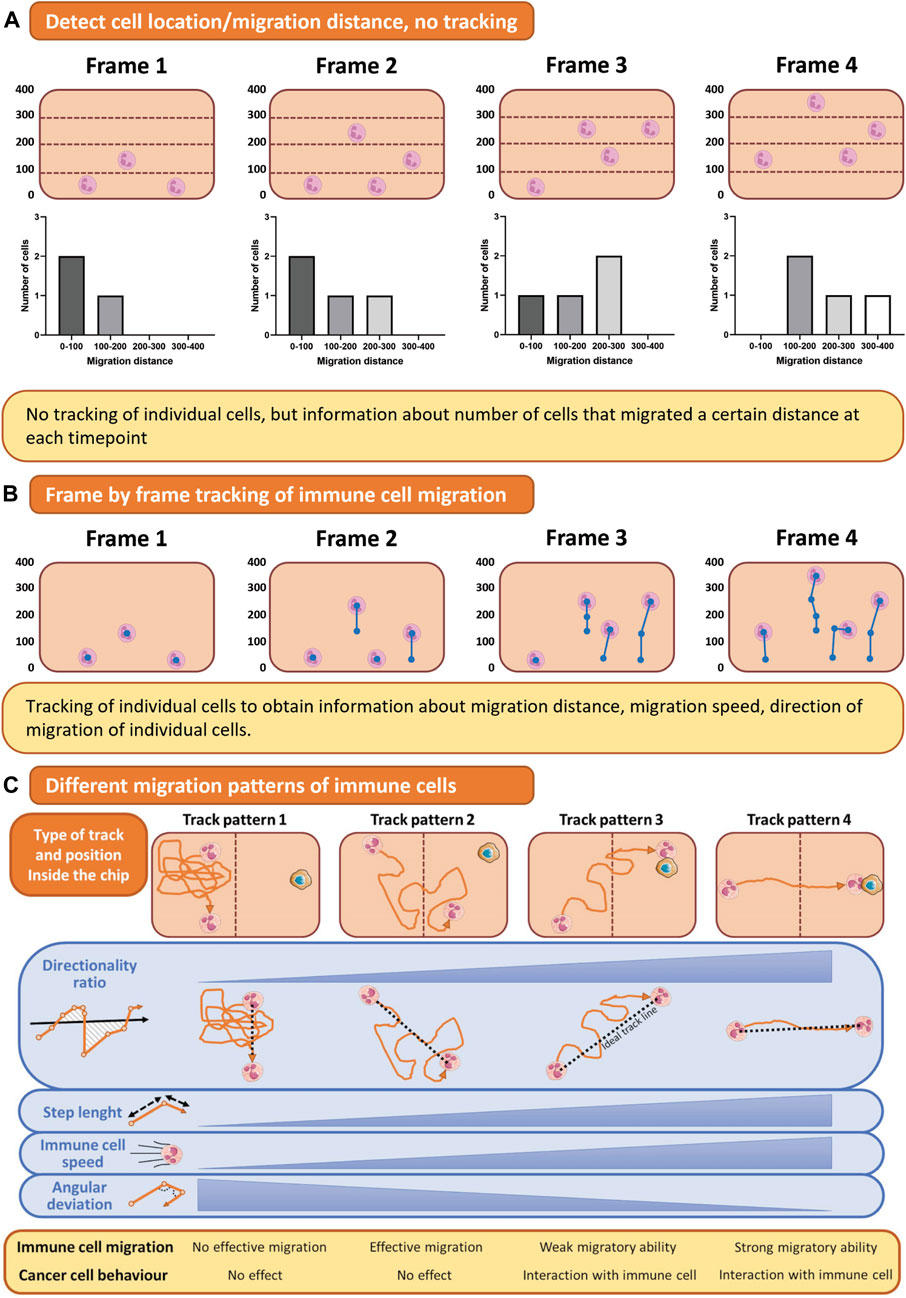
FIGURE 6. Quantification of immune cell migration. (A) For assessing immune cell location, the number of cells in a predefined area can be counted automatically. (B) Timelapse imaging of migrating immune cells results in a collection of images in which immune cells can be traced. (C) Tracking of immune cell migration shows different track patterns from which directionality, speed, and step length can be analyzed. This figure shows tracks of a single cell as an example for simple visualization purposes, but multiple cells can be tracked using these methods. Figure C was adapted from Mattei et al. (2021), under the terms of the Creative Commons Attribution License(CC BY). (https://creativecommons.org/licenses/by/4.0/).
Not only the microscopy itself but also the chip design influences the quality of the imaging and subsequent data analysis. For example, in the first lung on chip published by Huh et al. (Huh et al., 2010), neutrophils were migrating towards the site of infection, but migration dynamics could not be quantified, as it was occurring vertically across a porous membrane, and the immune cell migrated out of focus. Thus, to fully image the extravasation process, generating a model where immune cells migrate in a horizontal plane instead of vertically across a membrane is essential, as exemplified by multiple experimental designs (de Haan et al., 2021; Pérez-Rodríguez et al., 2022; Riddle et al., 2022). Live imaging of one focal plane can be carried out, and immune cell migration can be tracked.
After a timelapse video has been created, analysis of migration distance, speed, and direction is the next challenge (Figure 6). The authors refer to the following review for a more detailed overview of which parameters can be measured from a live imaging dataset of immune cell migration (Mattei et al., 2021). Traditional image analysis software such as ImageJ/FIJI or Imaris can aid in cell tracking, but these semi-automated solutions do not work for all imaged time-lapses. Thus, a tracking algorithm to track immune cell migration on chip can be developed in-house, for example, with Matlab (Pérez-Rodríguez et al., 2022). To simplify the analysis, an alternative to tracking individual immune cells is quantifying immune cells inside the hydrogel and their distance from the endothelial barrier over time (Figure 6B). With this method, no decisions on cell tracking are made, making the analysis less detailed but more robust. Next to imaging and tracking immune cells migrating horizontally in one focal plane, recent advances in spinning disk confocal microscopy have allowed for 3D timelapse imaging, to track cellular migration in all directions (Wen et al., 2021). This technique has not been applied in OOC models of infection yet, but would be of interest.
Advances in artificial intelligence (AI) have enhanced analysis methods, with the potential for more automation of (tracking) data analysis in the future (Ren et al., 2022). The first microfluidic experiments with deep learning algorithms to track cells have already been performed, such as an analysis pipeline to track bacteria in a microfluidic chip (Lugagne et al., 2020) and an algorithm to track tumor cell migration on chip (Zhang et al., 2018). In the second study, the migration direction and speed of the cells could be analyzed. So far, this technology is mainly used on relatively simple, single-cell OOC models and relies on manual adaptation during training (Ren et al., 2022). However, with more and more data becoming available, deep learning algorithms provide an immense opportunity for future analysis of immune cell migration on chip.
Current models of infection on chip have the common drawbacks of most OOC models, including only low to medium throughput, using the highly absorbing material PDMS for molding structures, and a lack of translation to clinical data. These issues must be addressed to engineer the next-generation of OOC models. To increase fabrication throughput and replace PDMS, OOCs have been fabricated with injection molding of polystyrene or similar transparent polymers, for example, to fabricate a high throughput OOC of cancer spheroid vascularization (Kim et al., 2022). Comparison of the data generated with OOCs to previously acquired data from animal studies and clinical trials is ongoing. For example, in the field of infection, many studies of immune cell extravasation have been carried out in mouse, rat, and rabbit models. With the appearance of new OOC models, it is important to investigate how these data can be compared and how both animal and OOC models translate to human diseases.
Additional complexity can be added to increase the relevance of OOC models of infection even further, for instance by mimicking both the innate and adaptive immune responses. This and other improvements may include replacing cell culture media with whole blood, adding tissue-resident immune cells, and integrating the lymphatic system.
Currently, only a subset of immune cells is perfused into a culture media or a buffer, but whole blood could be perfused to increase relevance. Not only does whole blood contain all cell types of interest, but blood also has a different viscosity and, therefore a different effect on the endothelium. Preliminary tests with whole blood perfusion in OOCs have been carried out and appear promising (Menon et al., 2017; Golomingi et al., 2022). Microfluidic devices can be modified to directly use whole blood for selective migration of neutrophils toward a site of infection. For example, whole blood was added in a microfluidic device with infected skin tissues, and neutrophil migration towards the infected skin tissues was observed (Kim et al., 2019b).
Another important part of immune surveillance is the tissue-resident immune cell population. These cells, mainly tissue-resident macrophages and T Cells, have a distinct gene expression profile and monitor the tissue microenvironment. Since these cells are present inside the tissue and their numbers are low, isolation is challenging, and source material is scarce. Further research is necessary to understand how these cells can be best isolated and cultured in vitro.
Lastly, a crucial part of the immune system, the lymphatics, is often overlooked. Generally, the main focus of infection-on-chip models is the interaction between immune cells circulating in blood vessels and the infected tissue, but for activation of the adaptive immune response, migration of dendritic cells from the site of infection to the lymph nodes to activate antigen-specific T Cells is necessary. Individual systems modeling lymph nodes or vessels on chip have been developed (Selahi et al., 2021; Shanti et al., 2021), however, to our knowledge, lymph vessels have not been incorporated into an OOC model of vasculature or other organs.
Next to infection, immune cells are also involved in other conditions, such as cancer, which adds the opportunity to utilize immunocompetent OOC models not only to model infection but also to model cancer immunotherapy. Using the human body’s natural defense system, cancer immunotherapy targets tumor tissue with the immune system, especially T Cells. Multiple studies are emerging with microfluidic models of immune cell migration towards a tumor, with known and unknown immunotherapy drugs enhancing tumor suppression by T Cells (Pavesi et al., 2017; Deng et al., 2018).
This review has discussed the generation of an infection-on-chip model to investigate infectious processes in a 3D in vitro model. By incorporating various cell types, cultured on or in hydrogel ECM environments, and perfusing the endothelial vessels, OOC models provide a more sophisticated alternative to basic 2D in vitro cultures. The modularity and versatility of OOC models enable the study of (disease) mechanisms in the presence or absence of particular aspects of the system. For example, research has demonstrated that the lack of endothelial cells prevents the migration of immune cells into a hydrogel, whereas the presence of endothelial cells significantly increases the number of immune cells that migrate (Wu et al., 2015; McMinn et al., 2019). It is impossible to study the impact of endothelium on immune cell migration in vivo as it is not feasible to completely eliminate endothelial cells from an animal. Consequently, OOC models offer an opportunity to supplement the findings from animal studies.
However, the challenge remains in translating the data obtained from these complex models to human disease. Initially, OOC models aim to replicate the disease symptoms observed in the clinic, such as drug-induced pulmonary oedema (Huh et al., 2012). Moreover, during the COVID-19 pandemic, a list of antivirals were tested in a lung-on-chip, showing that hydroxychloroquine, a drug that demonstrates efficacy against SARS-CoV2 in cell lines, did not exhibit antiviral effect on chip (Si et al., 2021). This finding translated to the clinic, where this drug proved ineffective against SARS-CoV2. Thus, to establish a correlation between research findings on infection-on-chip models and clinical data, clinical observations and readouts must be shown on chip.
Overall, the OOC field continues to develop at a fast pace with innovations occuring regularly. With new chip materials, the addition of AI, and the integration of different types of primary cells, these models will continue to evolve in the future.
The constant enhancement of OOC models aims to replicate human (patho)physiology with greater precision. In this review, attention was directed towards a particular category of OOC models that imitate the innate immune cell extravasation process during infection by means of a hydrogel ECM. To model an innate immune response on chip, the following main components are needed: the barrier (microvascular endothelium, hydrogel ECM, epithelium), the (perfused) immune cell, and the chemotactic gradient mimicking the infection. With this tutorial review, practical experience for designing an infection-on-chip experiment has been summarized to encourage the further development of these models in research institutions worldwide.
LO and OG conceptualized the review topic and structure. LO wrote the original manuscript. OG and BE revised the manuscript and approved of the final version. All authors contributed to the article and approved the submitted version.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 812954. Open access funding by University of Bern.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AI, Artificial Intelligence; APTES, (3-aminopropyl)triethoxy silane; EC, Endothelial Cell; ECM, Extracellular Matrix; HA, Hyaluronic Acid; HUVEC, Human Umbilical Vein Endothelial Cell; MMP, Matrix Metalloproteinase; OOC, Organs-on-chip; PA, Polyacrylamide; PAMP, Pathogen-Associated Molecular Pattern; PDMS, Polydimethylsiloxane; PEG, Polyethylene Glycol; PRR, Pattern Recognition Receptor.
Abbas, A. K, Lichtman, A. H, and Pillai, S. Cellular and molecular immunology. 7th ed. Elsevier; Amsterdam, Netherlands, 2012.
Adriani, G, Ma, D, Pavesi, A, Kamm, R. D, and Goh, E. L. K. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab. Chip. 17, 448, 459. 2017. doi:10.1039/c6lc00638h
Aizel, K, Clark, A. G, Simon, A, Geraldo, S., Funfak, A., Vargas, P., et al. A tuneable microfluidic system for long duration chemotaxis experiments in a 3D collagen matrix. Lab. Chip. 2017;17(22):3851–3861. doi:10.1039/c7lc00649g
Alper, S, and Janssen, W. J. Lung innate immunity and inflammation methods and protocols. 1st ed. (J. M Walker, ed.). Denver, CO: Methods in molecular biology, Humana Press; Totowa, New Jersey, 2018.
Amulic, B, Cazalet, C, Hayes, G. L, Metzler, K. D, and Zychlinsky, A. Neutrophil function: From mechanisms to disease. Annu. Rev. Immunol 30, 459, 489. 2012. doi:10.1146/annurev-immunol-020711-074942
Bai, H, Si, L, Jiang, A, Belgur, C., Zhai, Y., Plebani, R., et al. Mechanical control of innate immune responses against viral infection revealed in a human lung alveolus chip. Nat. Commun 2022;13(1):1928–2017. doi:10.1038/s41467-022-29562-4
Bala, K, Ambwani, K, and Gohil, N. K. Effect of different mitogens and serum concentration on HUVEC morphology and characteristics: Implication on use of higher passage cells. Tissue Cell. 2011;43(4):216–222. doi:10.1016/j.tice.2011.03.004
Barkal, L. J, Procknow, C. L, Álvarez-Garciá, Y. R, Niu, M., Jiménez-Torres, J. A., Brockman-Schneider, R. A., et al. Microbial volatile communication in human organotypic lung models. Nat. Commun 2017;8(1):1770–1810. doi:10.1038/s41467-017-01985-4
Bersini, S, Jeon, J. S, Dubini, G, Arrigoni, C., Chung, S., Charest, J. L., et al. A microfluidic 3D invitro model for specificity of breast cancer metastasis to bone. Biomaterials. 35, 2454, 2461. 2014. doi:10.1016/j.biomaterials.2013.11.050
Bertassoni, L. E, Cecconi, M, Manoharan, V, Nikkhah, M., Hjortnaes, J., Cristino, A. L., et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab. Chip. 14, 2202, 2211. 2014. doi:10.1039/c4lc00030g
Bianchi, E, Molteni, R, Pardi, R, and Dubini, G. Microfluidics for in vitro biomimetic shear stress-dependent leukocyte adhesion assays. J. Biomech 46, 276, 283. 2013. doi:10.1016/j.jbiomech.2012.10.024
Bichsel, C. A, Hall, S. R. R, Schmid, R. A, Guenat, O. T, and Geiser, T. Primary human lung pericytes support and stabilize in vitro perfusable microvessels. Tissue Eng. - Part a. 2015;21(15-16):2166–2176. doi:10.1089/ten.tea.2014.0545
Boussommier-Calleja, A, Atiyas, Y, Haase, K, Headley, M, Lewis, C, and Kamm, R. D. The effects of monocytes on tumor cell extravasation in a 3D vascularized microfluidic model. Biomaterials. 2019;198:180–193. doi:10.1016/J.BIOMATERIALS.2018.03.005
Brandenberg, N, Lutolf, M. P, Brandenberg, N, and Lutolf, M. P. In situ patterning of microfluidic networks in 3D cell-laden hydrogels. Adv. Mater. 2016;28(34):7450–7456. doi:10.1002/ADMA.201601099
Brooks, A. R, Lelkes, P. I, and Rubanyi, G. M. Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiol. Genomics. 2002;2002(9):27–41. doi:10.1152/PHYSIOLGENOMICS.00075.2001/ASSET/IMAGES/LARGE/H70620193005
Caliari, S. R, and Burdick, J. A. A practical guide to hydrogels for cell culture. Nat. Methods. 2016;13(5):405–414. doi:10.1038/nmeth.3839
Cameron, T. C, Randhawa, A, Grist, S. M, Bennet, T., Hua, J., Alde, L. G., et al. PDMS organ-on-chip design and fabrication: Strategies for improving fluidic integration and chip robustness of rapidly prototyped microfluidic in vitro models. Micromachines. 2022;13, 1573(10). doi:10.3390/mi13101573
Campisi, M, Shin, Y, Osaki, T, Hajal, C, Chiono, V, and Kamm, R. D. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials. 2018;180:117–129. doi:10.1016/j.biomaterials.2018.07.014
Chakraborty, J, Banerjee, I, Vaishya, R, and Ghosh, S. Bioengineered in vitro tissue models to study SARS-CoV-2 pathogenesis and therapeutic validation. ACS Biomater. Sci. Eng 2020;6(12):6540–6555. doi:10.1021/ACSBIOMATERIALS.0C01226
Chandrasekaran, A, Ellett, F, Jorgensen, J, and Irimia, D. Temporal gradients limit the accumulation of neutrophils toward sources of chemoattractant. Microsystems Nanoeng 2017;3:16067–16068. doi:10.1038/micronano.2016.67
Chaplin, D. D. Overview of the immune response. J. Allergy Clin. Immunol 2010;125:S3. S23. doi:10.1016/J.JACI.2009.12.980
Chen, Z, Huang, J, Zhang, J, Xu, Z., Li, Q., Ouyang, J., et al. A storm in a teacup – a biomimetic lung microphysiological system in conjunction with a deep-learning algorithm to monitor lung pathological and inflammatory reactions. Biosens. Bioelectron 2023;219:114772. doi:10.1016/j.bios.2022.114772
Chen, Z, Tang, M, Huang, D, Jiang, W., Li, M., Ji, H., et al. Real-time observation of leukocyte-endothelium interactions in tissue-engineered blood vessel. Lab. Chip. 2018;18(14):2047. 2054. doi:10.1039/C8LC00202A
Cheng, S. Y, Heilman, S, Wasserman, M, Archer, S, Shuler, M. L, and Wu, M. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab. Chip. 2007;7(6):763–769. doi:10.1039/b618463d
Chiu, P. L, Chang, C. H, Lin, Y. L, Tsou, P. H, and Li, B. R. Rapid and safe isolation of human peripheral blood B and T lymphocytes through spiral microfluidic channels. Sci. Rep 9, 8145, 2019. doi:10.1038/s41598-019-44677-3
Cho, S, Lee, S, and Ahn, S. I. Design and engineering of organ-on-a-chip. Biomed. Eng. Lett. 2022;1:97–109. doi:10.1007/S13534-022-00258-4
Chometon, T. Q, Da Silva Siqueira, M, and Sant´anna, J. C, A protocol for rapid monocyte isolation and generation of singular human monocyte-derived dendritic cells. PLoS One. 2020;15(4):e0231132. doi:10.1371/JOURNAL.PONE.0231132
Dabaghi, M, Shahriari, S, Saraei, N, Da, K., Chandiramohan, A., Selvaganapathy, P. R., et al. Surface modification of PDMS-based microfluidic devices with collagen using polydopamine as a spacer to enhance primary human bronchial epithelial cell adhesion. Micromachines. 2021;12(2):132–211. doi:10.3390/MI12020132
de Graaf, M. N. S, Vivas, A, van der Meer, A. D, Mummery, C. L, and Orlova, V V. Pressure-driven perfusion system to control, multiplex and recirculate cell culture medium for organs-on-chips. Micromachines. 2022;13(8):1359. doi:10.3390/mi13081359
de Haan, L, Suijker, J, van Roey, R, Berges, N., Petrova, E., Queiroz, K., et al. A microfluidic 3D endothelium-on-a-chip model to study transendothelial migration of T cells in health and disease. Int. J. Mol. Sci. 2021; 22(15):8234. doi:10.3390/IJMS22158234
Deinhardt-Emmer, S, Böttcher, S, Häring, C, Giebeler, L., Henke, A., Zell, R., et al. SARS-CoV-2 causes severe epithelial inflammation and barrier dysfunction. J. Virol 2021;95, e00110-e00121. doi:10.1128/JVI.00110-21
Deinhardt-Emmer, S, Rennert, K, Schicke, E, Cseresnyés, Z., Windolph, M., Nietzsche, S., et al. Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication. 12, 025012, 2020. doi:10.1088/1758-5090/ab7073
Deng, J, Wang, E. S, Jenkins, R. W, Li, S., Dries, R., Yates, K., et al. CDK4/6 inhibition augments anti-tumor immunity by enhancing T cell activation. Cancer Discov 2018;8(2):216. 233. doi:10.1158/2159-8290.CD-17-0915
Doyle, A. D. Generation of 3D collagen gels with controlled diverse architectures. Curr. Protoc. Cell Biol 72, 1, 10. 2016. doi:10.1002/cpcb.9
Falasca, M, Raimondi, C, and Maffucci, T. Boyden chamber. Methods Mol. Biol 2011;769:87–95. doi:10.1007/978-1-61779-207-6_7
Feaugas, T, and Sauvonnet, N. Organ-on-chip to investigate host-pathogens interactions. Cell Microbiol 2021;23(7):e13336. doi:10.1111/CMI.13336
Ford, A. J, Orbach, S. M, and Rajagopalan, P. Fibroblasts stimulate macrophage migration in interconnected extracellular matrices through tunnel formation and fiber alignment. Biomaterials. 2019;209:88–102. doi:10.1016/j.biomaterials.2019.03.044
Gifre-Renom, L, Daems, M, Luttun, A, and Jones, E. A. V. Organ-specific endothelial cell differentiation and impact of microenvironmental cues on endothelial heterogeneity. Int. J. Mol. Sci 2022;23, 1477(3). doi:10.3390/IJMS23031477
Gill, S, Wight, T. N, and Frevert, C. W. Proteoglycans: Key regulators of pulmonary inflammation and the innate immune response to lung infection. Anat. Rec. Hob 2010;293(6):968. 981. doi:10.1002/AR.21094
Gjorevski, N, Avignon, B, Gérard, R, Cabon, L., Roth, A. B., Bscheider, M., et al. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab. Chip. 2020;20(18):3365–3374. doi:10.1039/D0LC00417K
Golke, T, Mucher, P, Schmidt, P, Radakovics, A., Repl, M., Hofer, P., et al. Delays during PBMC isolation have a moderate effect on yield, but severly compromise cell viability. Clin. Chem. Lab. Med 2022;60(5):701–706. doi:10.1515/CCLM-2022-0003
Golomingi, M, Kohler, J, Jenny, L, Hardy, E. T., Dobó, J., Gál, P., et al. Complement lectin pathway components MBL and MASP-1 promote haemostasis upon vessel injury in a microvascular bleeding model. Front. Immunol 2022;13. 948190, doi:10.3389/fimmu.2022.948190
Granton, E, Kim, J. H, Podstawka, J, and Yipp, B. G. The lung microvasculature is a functional immune niche. Trends Immunol 39, 890, 899. 2018. doi:10.1016/j.it.2018.09.002
Grievink, H. W, Luisman, T, Kluft, C, Moerland, M, and Malone, K. E. Comparison of three isolation techniques for human peripheral blood mononuclear cells: Cell recovery and viability, population composition, and cell functionality. Biopreserv Biobank. 2016;14(5):410–415. doi:10.1089/BIO.2015.0104/ASSET/IMAGES/LARGE/FIGURE4.JPEG
Haase, K, and Kamm, R. D. Advances in on-chip vascularization. Regen. Med 2017;12(3):285–302. doi:10.2217/rme-2016-0152
Hajipouran Benam, K, Villenave, R, and Lucchesi, C, Small airway-on-a-chip: A novel microphysiological system to study human lung inflammation in vitro. Eur. J. Immunol 2016. 13, 151–157. doi:10.1002/eji.201670200
Halin, C, Mora, J. R, Sumen, C, and Von Andrian, U. H. In vivo imaging of lymphocyte trafficking, Annu. Rev. Cell Dev. Biol 2005;21:581–603. doi:10.1146/ANNUREV.CELLBIO.21.122303.133159
Hamza, B, and Irimia, D. Whole blood human neutrophil trafficking in a microfluidic model of infection and inflammation. Lab. Chip. 2015;15(12):2625–2633. doi:10.1039/c5lc00245a
Han, S, Yan, J. J, Shin, Y, Jeon, J. J., Won, J., Eun Jeong, H., et al. A versatile assay for monitoring in vivo-like transendothelial migration of neutrophils. Lab. Chip. 2012;12(20):3861–3865. doi:10.1039/c2lc40445a
Hanjaya-Putra, D, Yee, J, Ceci, D, Truitt, R, Yee, D, and Gerecht, S. Vascular endothelial growth factor and substrate mechanics regulate in vitro tubulogenesis of endothelial progenitor cells. J. Cell Mol. Med 14, 2436, 2447. 2010. doi:10.1111/j.1582-4934.2009.00981.x
Hasan, A, Paul, A, Memic, A, and Khademhosseini, A. A multilayered microfluidic blood vessel-like structure. Biomed. Microdevices. 17, 88, 2015. doi:10.1007/s10544-015-9993-2
Heo, K. S, Fujiwara, K, and Abe, J. I. Disturbed-flow-mediated vascular reactive oxygen species induce endothelial dysfunction. Circ. J 75, 2722, 2730. 2011. doi:10.1253/circj.CJ-11-1124
Hickey, M. J, and Westhorpe, C. L. V. Imaging inflammatory leukocyte recruitment in kidney, lung and liver-challenges to the multi-step paradigm. Immunol. Cell Biol 91, 281, 289. 2013. doi:10.1038/icb.2012.83
Hiemstra, P. S, Tetley, T. D, and Janes, S. M. Airway and alveolar epithelial cells in culture. Eur. Respir. J 2019;54, 1900742(5). doi:10.1183/13993003.00742-2019
Hind, L. E, Giese, M. A, Schoen, T. J, Beebe, D. J, Keller, N, and Huttenlocher, A. Immune cell paracrine signaling drives the neutrophil response to A. fumigatus in an infection-on-a-chip model. Cell Mol. Bioeng 2021;14(2):133–145. doi:10.1007/s12195-020-00655-8
Ho, Y. T, Adriani, G, Beyer, S, Nhan, P. T, Kamm, R. D, and Kah, J. C. Y. A facile method to probe the vascular permeability of nanoparticles in nanomedicine applications. Sci. Rep 2017;7(1):707–713. doi:10.1038/s41598-017-00750-3
Hsieh, J. Y, Keating, M. T, Smith, T. D, Meli, V. S, Botvinick, E. L, and Liu, W. F. Matrix crosslinking enhances macrophage adhesion, migration, and inflammatory activation. Apl. Bioeng 2019;3(1):016103. doi:10.1063/1.5067301
Huang, C. P, Lu, J, Seon, H, Lee, A. P., Flanagan, L. A., Kim, H. Y., et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab. Chip. 2009;9(12):1740–1748. doi:10.1039/b818401a
Huh, D, Leslie, D. C, Matthews, B. D, Fraser, J. P., Jurek, S., Hamilton, G. A., et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med 2012;4, 159ra147(159). doi:10.1126/scitranslmed.3004249
Huh, D, Matthews, B. D, Mammoto, A, Montoya-Zavala, M, Yuan Hsin, H, and Ingber, D. E. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi:10.1126/science.1188302
Humayun, M, Chow, C. W, and Young, E. W. K. Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab. Chip. 2018;18(9):1298–1309. doi:10.1039/c7lc01357d
Ingber, D. E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet 2022;23(8):467. 491. doi:10.1038/S41576-022-00466-9
Irimia, D, Liu, S. Y, Tharp, W. G, Samadani, A, Toner, M, and Poznansky, M. C. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab. Chip. 2006;6(2):191–198. doi:10.1039/b511877h
Jannat, R, Dembo, M, Hammer, D, and Hammer, D. A. Neutrophil adhesion and chemotaxis depend on substrate mechanics. J. Phys. Condens. Matter, 2010;22, 194117(19). doi:10.1088/0953-8984/22/19/194117
Jannat, R. A, Dembo, M, and Hammer, D. A. Traction forces of neutrophils migrating on compliant substrates. Biophys. J 2011;101(3):575–584. doi:10.1016/j.bpj.2011.05.040
Jeon, J. S, Bersini, S, Gilardi, M, Dubini, G., Charest, J. L., Moretti, M., et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. U. S. A 112, 214, 219. 2015. doi:10.1073/pnas.1417115112
Jiang, W, Li, M, Chen, Z, and Leong, K. W. Cell-laden microfluidic microgels for tissue regeneration. Lab. Chip. 16, 4482, 4506. 2016. doi:10.1039/c6lc01193d
Jones, C. N, Hoang, A. N, Dimisko, L, Hamza, B, Martel, J, and Irimia, D. Microfluidic platform for measuring neutrophil chemotaxis from unprocessed whole blood. J. Vis. Exp 2014;(88):51215–51216. doi:10.3791/51215
Keating, M, Lim, M, Hu, Q, and Botvinick, E. Selective stiffening of fibrin hydrogels with micron resolution via photocrosslinking. Acta Biomater 87, 88, 96. 2019. doi:10.1016/j.actbio.2019.01.034
Kim, J. J, Ellett, F, Thomas, C. N, Jalali, F., Anderson, R. R., Irimia, D., et al. A microscale, full-thickness, human skin on a chip assay simulating neutrophil responses to skin infection and antibiotic treatments. Lab. Chip. 2019;19(18):3094. 3103. doi:10.1039/C9LC00399A
Kim, J. Y, Fluri, D. A, Kelm, J. M, Hierlemann, A, and Frey, O. 96-Well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J. Lab. Autom 20, 274, 282. 2015. doi:10.1177/2211068214564056
Kim, Y, Ko, J, Shin, N, Park, S., Lee, S., Kim, S., et al. All-in-one microfluidic design to integrate vascularized tumor spheroid into high-throughput platform. Biotechnol. Bioeng 119, 3678, 3693. 2022. doi:10.1002/bit.28221
Kim, Y. M, Jeong, S, Choe, Y. H, and Hyun, Y. M. Two-photon intravital imaging of leukocyte migration during inflammation in the respiratory system. Acute Crit. Care. 2019;34(2):101. 107. doi:10.4266/ACC.2019.00542
Koning, J. J, Rodrigues Neves, C. T, Schimek, K, Thon, M., Spiekstra, S. W., Waaijman, T., et al. A multi-organ-on-chip approach to investigate how oral exposure to metals can cause systemic toxicity leading to langerhans cell activation in skin. Front. Toxicol 2022;3:824825. doi:10.3389/FTOX.2021.824825
Kosten, I. J, Spiekstra, S. W, De Gruijl, T. D, and Gibbs, S. MUTZ-3 Langerhans cell maturation and CXCL12 independent migration in reconstructed human gingiva. ALTEX - Altern Anim Exp. 2016;33(4):423–434. doi:10.14573/ALTEX.1510301
Lauridsen, H. M, and Gonzalez, A. L. Biomimetic, ultrathin and elastic hydrogels regulate human neutrophil extravasation across endothelial-pericyte bilayers. PLoS One. 2017;12(2):01713866-e171419. doi:10.1371/journal.pone.0171386
Lee, D. W, Choi, N, and Sung, J. H. A microfluidic chip with gravity-induced unidirectional flow for perfusion cell culture. Biotechnol. Prog 2019;35(1):e2701. doi:10.1002/BTPR.2701
Lee, J, Kim, S. E, Moon, D, and Doh, J. A multilayered blood vessel/tumor tissue chip to investigate T cell infiltration into solid tumor tissues. Lab. Chip. 2021;21(11):2142–2152. doi:10.1039/D1LC00182E
Lee, S. H, Shim, K. Y, Kim, B, and Sung, J. H. Hydrogel-based three-dimensional cell culture for organ-on-a-chip applications. Biotechnol. Prog 2017;33(3):580–589. doi:10.1002/btpr.2457
Lee, S. W. L, Seager, R. J, Litvak, F, Spill, F., Sieow, J. L., Leong, P. H., et al. Integrated in silico and 3D in vitro model of macrophage migration in response to physical and chemical factors in the tumor microenvironment. Integr. Biol 2020;12(4):90. 108. doi:10.1093/INTBIO/ZYAA007
Leung, C. M, de Haan, P, Ronaldson-Bouchard, K, Kim, G. A., Ko, J., Rho, H. S., et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim. 2022;2(1):33–29. doi:10.1038/s43586-022-00118-6
Ley, K, Laudanna, C, Cybulsky, M. I, and Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol 2007;7(9):678–689. doi:10.1038/nri2156
Li, R, Serrano, J. C, Xing, H, Lee, T. A., Azizgolshani, H., Zaman, M., et al. Interstitial flow promotes macrophage polarization toward an M2 phenotype. Mol. Biol. Cell. 29, 1927, 1940. 2018. doi:10.1091/mbc.E18-03-0164
Liu, H, Wang, Y, Cui, K, Guo, Y, Zhang, X, and Qin, J. Advances in hydrogels in organoids and organs-on-a-chip. Adv. Mater. 2019;31(50):1902042–1902128. doi:10.1002/adma.201902042
Lu, R. X. Z, Lai, B. F. L, Rafatian, N, Gustafson, D., Campbell, S. B., Banerjee, A., et al. Vasculature-on-a-chip platform with innate immunity enables identification of angiopoietin-1 derived peptide as a therapeutic for SARS-CoV-2 induced inflammation. Lab. Chip. 2022;22(6):1171–1186. doi:10.1039/D1LC00817J
Lugagne, J. B, Lin, H, and Dunlop, M. J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol 2020;16, e1007673(4). doi:10.1371/JOURNAL.PCBI.1007673
Luo, Y, Qin, J, and Lin, B. Methods for pumping fluids on biomedical lab-on-a-chip. Front. Biosci 14, 3913, 3924. 2009. doi:10.2741/3500
Lynn, A. D, Kyriakides, T. R, and Bryant, S. J. Characterization of the in vitro macrophage response and in vivo host response to poly(ethylene glycol)-based hydrogels. J. Biomed. Mater Res. - Part A. 2010;93(3):941–953. doi:10.1002/jbm.a.32595
Maas, S. L, Soehnlein, O, and Viola, J. R. Organ-specific mechanisms of transendothelial neutrophil migration in the lung, liver, kidney, and aorta. Front. Immunol 9, 2739, 2018. doi:10.3389/fimmu.2018.02739
Maharjan, S, Cecen, B, and Zhang, Y. S. 3D immunocompetent organ-on-a-chip models. Small Methods. 2020;4, 2000235(9). doi:10.1002/smtd.202000235
Manicone, A. M. Role of the pulmonary epithelium and inflammatory signals in acute lung injury. Expert Rev. Clin. Immunol 2009;5(1):63. 75. doi:10.1586/1744666x.5.1.63
Marhuenda, E, Villarino, A, Narciso, M, Elowsson, L., Almendros, I., Westergren-Thorsson, G., et al. Development of a physiomimetic model of acute respiratory distress syndrome by using ECM hydrogels and organ-on-a-chip devices. Front. Pharmacol 2022;13:945134. doi:10.3389/fphar.2022.945134
Matellan, C, del, R. , and Hernández, A. E. Engineering the cellular mechanical microenvironment – from bulk mechanics to the nanoscale. J. Cell Sci 132, jcs229013, 2019. doi:10.1242/jcs.229013
Mattei, F, Andreone, S, Mencattini, A, De Ninno, A., Businaro, L., Martinelli, E., et al. Oncoimmunology meets organs-on-chip. Front. Mol. Biosci 2021;8:627454. doi:10.3389/fmolb.2021.627454
McMinn, P. H, Hind, L. E, Huttenlocher, A, and Beebe, D. J. Neutrophil trafficking on-a-chip: An: In vitro, organotypic model for investigating neutrophil priming, extravasation, and migration with spatiotemporal control. Lab. Chip. 2019;19(21):3697–3705. doi:10.1039/c9lc00562e
Menon, N. V, Tay, H. M, Wee, S. N, Li, K. H. H, and Hou, H. W. Micro-engineered perfusable 3D vasculatures for cardiovascular diseases. Lab. Chip. 2017;17(17):2960–2968. doi:10.1039/c7lc00607a
Miller, C. P, Shin, W, Ahn, E. H, Kim, H. J, and Kim, D. H. Engineering microphysiological immune system responses on chips. Trends Biotechnol 2020;38(8):857–872. doi:10.1016/J.TIBTECH.2020.01.003
Miskolci, V, Klemm, L. C, and Huttenlocher, A. Cell migration guided by cell–cell contacts in innate immunity. Trends Cell Biol 2021;31(2):86–94. doi:10.1016/J.TCB.2020.11.002
Morsink, M. A. J, Willemen, N. G. A, Leijten, J, Bansal, R, and Shin, S. R. Immune organs and immune cells on a chip: An overview of biomedical applications. Micromachines. 2020;11, 849(9). doi:10.3390/MI11090849
Moura Rosa, P, Gopalakrishnan, N, Ibrahim, H, Haug, M, and Halaas, Ø The intercell dynamics of T cells and dendritic cells in a lymph node-on-a-chip flow device. Lab. Chip. 16, 3728, 3740. 2016. doi:10.1039/c6lc00702c
Muller, W. A. Getting leukocytes to the site of inflammation. Vet. Pathol 50, 7, 22. 2013. doi:10.1177/0300985812469883
Muller, W. A, and Luscinskas, F. W. Assays of transendothelial migration in vitro. Methods Enzymol 2008;443:155–176. doi:10.1016/S0076-6879(08)02009-0
Muller, W. A. Mechanisms of leukocyte transendothelial migration. Annu. Rev. Pathol. Mech. Dis 6, 323, 344. 2011. doi:10.1146/annurev-pathol-011110-130224
Na, K, Lee, M, Shin, H. W, and Chung, S. In vitro nasal mucosa gland-like structure formation on a chip. Lab. Chip. 17, 1578, 1584. 2017. doi:10.1039/c6lc01564f
Nie, J, Gao, Q, Wang, Y, Zeng, J., Zhao, H., Sun, Y., et al. Vessel-on-a-chip with hydrogel-based microfluidics. Small. 14, 1802368, 2018. doi:10.1002/smll.201802368
Nikolaev, M, Mitrofanova, O, Broguiere, N, Geraldo, S., Dutta, D., Tabata, Y., et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nat. 2020;585(7826):574–578. doi:10.1038/s41586-020-2724-8
Oakes, P. W, Patel, D. C, Morin, N. A, Zitterbart, D. P., Fabry, B., Reichner, J. S., et al. Neutrophil morphology and migration are affected by substrate elasticity. Blood. 2009;114(7):1387–1395. doi:10.1182/blood-2008-11-191445
O’Dwyer, D. N, Gurczynski, S. J, and Moore, B. B. Pulmonary immunity and extracellular matrix interactions. Matrix Biol 73, 122, 134. 2018. doi:10.1016/j.matbio.2018.04.003
Ohta, M, Sakamoto, N, Funamoto, K, Wang, Z, Kojima, Y, and Anzai, H. A review of functional analysis of endothelial cells in flow chambers. J. Funct. Biomater 2022;13(3):92. doi:10.3390/JFB13030092
Onken, M. D, Mooren, O. L, Mukherjee, S, Shahan, S. T, Li, J, and Cooper, J. A. Endothelial monolayers and transendothelial migration depend on mechanical properties of the substrate. Cytoskeleton. 71, 695, 706. 2014. doi:10.1002/cm.21203
Park, J. H, Chung, B. G, Lee, W. G, Kim, J., Brigham, M. D., Shim, J., et al. Microporous cell-laden hydrogels for engineered tissue constructs. Biotechnol. Bioeng 106, 138, 148. 2010. doi:10.1002/bit.22667
Park, J. S, Kim, D. H, Kim, H. N, Wang, C. J., Kwak, M. K., Hur, E., et al. Directed migration of cancer cells guided by the graded texture of the underlying matrix. Nat. Mater. 15, 792, 801. 2016. doi:10.1038/nmat4586
Parks, W. C, Wilson, C. L, and López-Boado, Y. S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol 2004;4(8):617–629. doi:10.1038/NRI1418
Parlato, S, De Ninno, A, Molfetta, R, Toschi, E., Salerno, D., Mencattini, A., et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep 2017;7(1):1093. doi:10.1038/S41598-017-01013-X
Pavesi, A, Tan, A. T, Koh, S, Chia, A., Colombo, M., Antonecchia, E., et al. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017;2, e89762(12). doi:10.1172/JCI.INSIGHT.89762
Pérez-Rodríguez, S, Borau, C, García-Aznar, J. M, and Gonzalo-Asensio, J. A microfluidic-based analysis of 3D macrophage migration after stimulation by Mycobacterium, Salmonella and Escherichia. BMC Microbiol 2022;22(1):211–214. doi:10.1186/s12866-022-02623-w
Petri, B, and Sanz, M. J. Neutrophil chemotaxis. Cell Tissue Res 2018;371(3):425–436. doi:10.1007/S00441-017-2776-8
Poussin, C, Kramer, B, Lanz, H. L, Van den Heuvel, A., Laurent, A., Olivier, T., et al. 3D human microvessel-on-a-chip model for studying monocyte-to-endothelium adhesion under flow - application in systems toxicology. ALTEX. 2020;37(1):47–63. doi:10.14573/altex.1811301
Qiu, Y, Ahn, B, Sakurai, Y, Hansen, C. E., Tran, R., Mimche, P. N., et al. Microvasculature-on-a-chip for the long-term study of endothelial barrier dysfunction and microvascular obstruction in disease. Nat. Biomed. Eng 2, 453, 463. 2018. doi:10.1038/s41551-018-0224-z
Rainger, G. E, Buckley, C. D, Simmons, D. L, and Nash, G. B. Neutrophils sense flow-generated stress and direct their migration through α(v)β3-integrin. Am. J. Physiol. - Hear Circ. Physiol. 1999;276(3 45-3). doi:10.1152/AJPHEART.1999.276.3.H858
Ren, J, Wang, N, Guo, P, Fan, Y, Lin, F, and Wu, J. Recent advances in microfluidics-based cell migration research. Lab. Chip. 2022;22(18):3361–3376. doi:10.1039/D2LC00397J
Ren, X, Getschman, A. E, Hwang, S, Volkman, B. F., Klonisch, T., Levin, D., et al. Investigations on T cell transmigration in a human skin-on-chip (SoC) model. Lab. Chip. 2021;21(8):1527–1539. doi:10.1039/D0LC01194K
Riddle, R. B, Jennbacken, K, Hansson, K. M, and Harper, M. T. Endothelial inflammation and neutrophil transmigration are modulated by extracellular matrix composition in an inflammation-on-a-chip model. Sci. Rep 2022;12(1):6855–6914. doi:10.1038/s41598-022-10849-x
Salminen, A. T, Allahyari, Z, Gholizadeh, S, McCloskey, M. C., Ajalik, R., Cottle, R. N., et al. In vitro studies of transendothelial migration for biological and drug discovery. Front. Med. Technol 2020;2:600616. doi:10.3389/FMEDT.2020.600616
Sano, E, Mori, C, Nashimoto, Y, Yokokawa, R, Kotera, H, and Torisawa, Y. S. Engineering of vascularized 3D cell constructs to model cellular interactions through a vascular network. Biomicrofluidics. 12, 042204, 2018. doi:10.1063/1.5027183
Saygili, E, Yildiz-Ozturk, E, Green, M. J, Ghaemmaghami, A. M, and Yesil-Celiktas, O. Human lung-on-chips: Advanced systems for respiratory virus models and assessment of immune response. Biomicrofluidics. 2021;15, 021501(2). doi:10.1063/5.0038924
Selahi, A, Fernando, T, Chakraborty, S, Muthuchamy, M, Zawieja, D. C, and Jain, A. Lymphangion-chip: A microphysiological system which supports co-culture and bidirectional signaling of lymphatic endothelial and muscle cells. Lab. Chip. 2021;22(1):121–135. doi:10.1039/D1LC00720C
Sengupta, A, Roldan, N, Kiener, M, Froment, L., Raggi, G., Imler, T., et al. A new immortalized human alveolar epithelial cell model to study lung injury and toxicity on a breathing lung-on-chip system. Front. Toxicol 4, . 2022):840606–840618. doi:10.3389/ftox.2022.840606
Shanti, A, Hallfors, N, Petroianu, G. A, Planelles, L, and Stefanini, C. Lymph nodes-on-chip: Promising immune platforms for pharmacological and toxicological applications. Front. Pharmacol 2021;12:711307. doi:10.3389/fphar.2021.711307
Sharifi, F, Htwe, S. S, Righi, M, Liu, H., Pietralunga, A., Yesil-Celiktas, O., et al. A foreign body response-on-a-chip platform. Adv. Healthc. Mater. 8, 1801425, . 2019. doi:10.1002/adhm.201801425
Shin, Y, Han, S, Jeon, J. S, Yamamoto, K., Zervantonakis, I. K., Sudo, R., et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc 7, 1247, 1259. 2012. doi:10.1038/nprot.2012.051
Si, L, Bai, H, Rodas, M, Cao, W., Oh, C. Y., Jiang, A., et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat. Biomed. Eng 2021;5(8):815–829. doi:10.1038/s41551-021-00718-9
Steiner, O, Coisne, C, Engelhardt, B, and Lyck, R. Comparison of immortalized bEnd5 and primary mouse brain microvascular endothelial cells as in vitro blood-κBrain barrier models for the study of T cell extravasation. J. Cereb. Blood Flow. Metab 2011;31(1):315–327. doi:10.1038/jcbfm.2010.96
Sturtzel, C. 2017, Advances in experimental medicine and biology 1003 the immunology of cardiovascular homeostasis and pathology. In: S Sattler, and T Kennedy, eds. Springer, Berlin, Germany
Sun, D, Evans, L, Perrone, F, Sokleva, V., Lim, K., Rezakhani, S., et al. A functional genetic toolbox for human tissue-derived organoids. Elife. 2021;10. e67886, doi:10.7554/ELIFE.67886
Surendran, V, Rutledge, D, Colmon, R, and Chandrasekaran, A. A novel tumor-immune microenvironment (TIME)-on-Chip mimics three dimensional neutrophil-tumor dynamics and neutrophil extracellular traps (NETs)-mediated collective tumor invasion. Biofabrication. 2021;13, 035029(3). doi:10.1088/1758-5090/ABE1CF
Tanaka, R, Saito, Y, Fujiwara, Y, Jo, J, and Tabata, Y. Preparation of fibrin hydrogels to promote the recruitment of anti-inflammatory macrophages. Acta Biomater 89, 152, 165. 2019. doi:10.1016/j.actbio.2019.03.011
Tang, H, Abouleila, Y, Si, L, Ortega-Prieto, A. M., Mummery, C. L., Ingber, D. E., et al. Human organs-on-chips for virology. Trends Microbiol 2020;28(11):934–946. doi:10.1016/J.TIM.2020.06.005
Terrell, J. A, Jones, C. G, Kabandana, G. K. M, and Chen, C. From cells-on-a-chip to organs-on-a-chip: Scaffolding materials for 3D cell culture in microfluidics. J. Mater Chem. B. 2020;8(31):6667–6685. doi:10.1039/D0TB00718H
Thacker, V V., Dhar, N, Sharma, K, Barrile, R, Karalis, K, and McKinney, J. D. A lung-on-chip model of early Mycobacterium tuberculosis infection reveals an essential role for alveolar epithelial cells in controlling bacterial growth. Elife. 2020;9:e59961-e59973. doi:10.7554/eLife.59961
Tourovskaia, A, Fauver, M, Kramer, G, Simonson, S, and Neumann, T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med 2014;239(9):1264–1271. doi:10.1177/1535370214539228
Haessler, U, Pisano, M, Wu, M, and Swartz, M. A. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc. Natl. Acad. Sci. U. S. A 108, 5614, 5619. 2011. doi:10.1073/pnas.1014920108
Valignat, M. P, Theodoly, O, Gucciardi, A, Hogg, N, and Lellouch, A. C. T lymphocytes orient against the direction of fluid flow during LFA-1-mediated migration. Biophys. J 2013;104(2):322. 331. doi:10.1016/J.BPJ.2012.12.007
Van Duinen, V, Van Den Heuvel, A, Trietsch, S. J, Lanz, H. L., van Gils, J. M., van Zonneveld, A. J., et al. 96 perfusable blood vessels to study vascular permeability in vitro. Sci. Rep 2017;7(1):18071–18111. doi:10.1038/s41598-017-14716-y
van Duinen, V, Zhu, D, Ramakers, C, van Zonneveld, A. J, Vulto, P, and Hankemeier, T. Perfused 3D angiogenic sprouting in a high-throughput in vitro platform. Angiogenesis. 2019;22(1):157–165. doi:10.1007/S10456-018-9647-0
Van Steen, A. C. I, Kempers, L, Schoppmeyer, R, Blokker, M., Beebe, D. J., Nolte, M. A., et al. Transendothelial migration induces differential migration dynamics of leukocytes in tissue matrix. J. Cell Sci 2021;134, jcs258690(21). doi:10.1242/jcs.258690
Vasse, G. F, Kühn, P. T, Zhou, Q, Bhusari, S. A., Reker-Smit, C., Melgert, B. N., et al. Collagen morphology influences macrophage shape and marker expression in vitro. J. Immunol. Regen. Med 1, 13, 20. 2018. doi:10.1016/j.regen.2018.01.002
Vulto, P, Podszun, S, Meyer, P, Hermann, C, Manz, A, and Urban, G. A. Phaseguides: A paradigm shift in microfluidic priming and emptying. Lab. Chip. 11, 1596, 2011. doi:10.1039/c0lc00643b
Wang, X, and Irimia, D. Neutrophil chemotaxis in one droplet of blood using microfluidic assays. Methods Mol. Biol., 2018;1749:351–360. doi:10.1007/978-1-4939-7701-7_25
Wang, Y. I, and Shuler, M. L. UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems. Lab. Chip. 2018;18(17):2563–2574. doi:10.1039/C8LC00394G
Weibel, E. R. On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med 2015;191(5):504–513. doi:10.1164/rccm.201409-1663OE
Weirather, J, and Frantz, S. Role of the innate immune system in ischemic heart failure. Inflamm. Hear Fail. 2015, 5: 19–38. doi:10.1016/B978-0-12-800039-7.00002-5
Wen, C, Miura, T, Voleti, V, Yamaguchi, K., Tsutsumi, M., Yamamoto, K., et al. 3deecelltracker, a deep learning-based pipeline for segmenting and tracking cells in 3d time lapse images. Elife. 2021;10. e59187, doi:10.7554/ELIFE.59187
Whang, M, and Kim, J. Synthetic hydrogels with stiffness gradients for durotaxis study and tissue engineering scaffolds. Tissue Eng. Regen. Med 2016;13(2):126–139. doi:10.1007/s13770-016-0026-x
Wu, X, Newbold, M. A, and Haynes, C. L. Recapitulation of in vivo-like neutrophil transendothelial migration using a microfluidic platform†. Analyst. 2015;140(15):5055–5064. doi:10.1039/C5AN00967G
Yamada, M, Fujino, N, and Ichinose, M. Inflammatory responses in the initiation of lung repair and regeneration: Their role in stimulating lung resident stem cells. Inflamm. Regen 2016;36(1):15–18. doi:10.1186/s41232-016-0020-7
Yang, K, Wu, J, Xu, G, Xie, D., Peretz-Soroka, H., Santos, S., et al. A dual-docking microfluidic cell migration assay (D2-Chip) for testing neutrophil chemotaxis and the memory effect. Integr. Biol. (United Kingdom). 2017;9(4):303–312. doi:10.1039/c7ib00037e
Yin, F, Zhu, Y, Zhang, M, Yu, H, Chen, W, and Qin, J. A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol. Vitr 54, 105, 113. 2019. doi:10.1016/j.tiv.2018.08.014
Zeinali, S, Bichsel, C. A, Hobi, N, Funke, M., Marti, T. M., Schmid, R. A., et al. Human microvasculature-on-a chip: Anti-neovasculogenic effect of nintedanib in vitro. Angiogenesis. 2018;21(4):861–871. doi:10.1007/s10456-018-9631-8
Zeinali, S, Thompson, E. K, Gerhardt, H, Geiser, T, and Guenat, O. T. Remodeling of an in vitro microvessel exposed to cyclic mechanical stretch. Apl. Bioeng 5, 026102, 2021. doi:10.1063/5.0010159
Zervantonakis, I. K, Hughes-Alford, S. K, Charest, J. L, Condeelis, J. S, Gertler, F. B, and Kamm, R. D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. U. S. A 109, 13515, 13520. 2012. doi:10.1073/pnas.1210182109
Zhang, B, Montgomery, M, Chamberlain, M. D, Ogawa, S., Korolj, A., Pahnke, A., et al. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 15, 669, 678. 2016. doi:10.1038/nmat4570
Zhang, B, and Radisic, M. Organ-on-a-chip devices advance to market. Lab. Chip. 2017;17(14):2395–2420. doi:10.1039/C6LC01554A
Zhang, Z, Chen, L, Humphries, B, Brien, R., Wicha, M. S., Luker, K. E., et al. Morphology-based prediction of cancer cell migration using an artificial neural network and a random decision forest. Integr. Biol. (Camb). 2018;10(12):758. 767. doi:10.1039/C8IB00106E
Zhao, W, Zhao, H, Li, M, and Huang, C. Microfluidic devices for neutrophil chemotaxis studies. J. Transl. Med 2020;18(1):168–219. doi:10.1186/s12967-020-02335-7
Zheng, Y, Chen, J, and Craven, M, In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl. Acad. Sci 109, 9342–9347. 2012. doi:10.1073/pnas.1201240109
Keywords: infection, organ-on-chip, infection-on-chip, endothelium, immune cells, hydrogel ECM, chemoattractant, inflammation
Citation: Van Os L, Engelhardt B and Guenat OT (2023) Integration of immune cells in organs-on-chips: a tutorial. Front. Bioeng. Biotechnol. 11:1191104. doi: 10.3389/fbioe.2023.1191104
Received: 21 March 2023; Accepted: 10 May 2023;
Published: 01 June 2023.
Edited by:
Xiuli Zhang, Soochow University, ChinaReviewed by:
Ting-Yuan Tu, National Cheng Kung University, TaiwanCopyright © 2023 Van Os, Engelhardt and Guenat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivier T. Guenat, b2xpdmllci5ndWVuYXRAdW5pYmUuY2g=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.