- Department of Biomedical Engineering, Chung-Ang University, Seoul, Republic of Korea
Toxic heavy metal accumulation is one of anthropogenic environmental pollutions, which poses risks to human health and ecological systems. Conventional heavy metal remediation approaches rely on expensive chemical and physical processes leading to the formation and release of other toxic waste products. Instead, microbial bioremediation has gained interest as a promising and cost-effective alternative to conventional methods, but the genetic complexity of microorganisms and the lack of appropriate genetic engineering technologies have impeded the development of bioremediating microorganisms. Recently, the emerging synthetic biology opened a new avenue for microbial bioremediation research and development by addressing the challenges and providing novel tools for constructing bacteria with enhanced capabilities: rapid detection and degradation of heavy metals while enhanced tolerance to toxic heavy metals. Moreover, synthetic biology also offers new technologies to meet biosafety regulations since genetically modified microorganisms may disrupt natural ecosystems. In this review, we introduce the use of microorganisms developed based on synthetic biology technologies for the detection and detoxification of heavy metals. Additionally, this review explores the technical strategies developed to overcome the biosafety requirements associated with the use of genetically modified microorganisms.
1 Introduction
Heavy metals are elements with an atomic weight ranging from 63.5 to 200.6 g/mol and a density greater than 5 g/cm3 (Srivastava and Majumder, 2008). Certain heavy metals such as zinc (Zn), cobalt (Co), copper (Cu) and iron (Fe) are essential at a trace concentration to maintain life by involving in diverse biological functions (Appenroth, 2010; Kim et al., 2019a). Heavy metals accumulated at high concentrations can pose toxicity in living organisms by destroying membrane and DNA integrities and inhibiting the activity of proteins (Kim et al., 2015; Alrashed et al., 2021). For example, chromium (Cr), arsenic (As) and cadmium (Cd) induce DNA repair dysfunction and oxidative stresses, and thereby potentially cause cancers in human (Balali-Mood et al., 2021; Luo et al., 2021). Heavy metals inhibit enzymes by interfering with the formation of enzyme-substrate complexes, altering the functional integrity of active sites and compromising enzyme synthesis (Zackular et al., 2016; Lukowski and Dec, 2018). Interestingly, the continuous exposure to heavy metals such as As, Cd, Cu, lead (Pb), and Zn also disrupts the gut micro-ecosystem. The microbiome disruption may cause problems in immunity and resistance against bacterial infection, and eventually become vulnerable to diverse diseases (Choiniere and Wang, 2016; Zhai et al., 2019; Shao and Zhu, 2020).
For decades, human activities like farming, industrialization, and mining sharply facilitated the accumulation of heavy metals in the environment and thus threatened human wellness (Chandra et al., 2009; Nagajyoti et al., 2010; Bharagava et al., 2018) (Table 1). There have been many attempts to remediate contaminated environment, and significant attention has been paid to chemical and physical methods such as absorption, filtration, and chemical separations (Qasem et al., 2021). However, each method has its own disadvantages that limit its widespread use. For absorption, absorbents with different functional groups capturing heavy metals such as carbon, chitosan, or mineral-based absorbents are used (Qasem et al., 2021). The complex functionalization, instability, and low reusability of absorbents limit their use in industrial settings despite their high efficiency (Duan et al., 2020; Qasem et al., 2021). Membrane-based filtration is another method to separate heavy metals, which can be easily modulated by membrane pore size and permeability (Mukherjee et al., 2016). Although membrane-based filtration is easy to operate and highly efficient, frequent flux regeneration (cleaning) of membranes is necessary, which is incompatible with large-scale applications (Verma et al., 2021). Chemical methods use compounds to alter the valence state of heavy metals, such as the oxidation state, to diminish toxicity, or neutralize colloidal forces of heavy metals to precipitate and coagulate them. However, there are serious concerns about the chemical methods due to the utilization of toxic and health-harming agents to remove heavy metals; thus, chemical methods are considered non-sustainable (Bolisetty et al., 2019).
To overcome the hurdles in the remediation by chemical and physical methods, bioremediation by microorganisms is gaining attention owing to their high efficiency and sustainability. To date, microorganisms isolated from contaminated area are commonly used for bioremediation because the organisms are tolerant against toxic chemicals and often possess the ability to convert toxic compounds into non-harmful ones. However, since natural microorganisms have not evolved to detoxify heavy metals but to survive in such harsh environments, they inevitably possess low bioremediation efficiency, still more efficient than the chemical and physical methods. Thus, there is a continuous demand for enhancing microbial detoxification capacity by the genetic reconstruction of their biochemical functions. One of the hurdles in constructing high-efficient bioremediation bacteria is how to engineer their biochemical ability to improve detoxification and tolerance, as well as how to engineer their intrinsic physiology for safe management.
Heavy metal biosensors are used to achieve in situ detection and efficient bioremediation in contaminated environments (Wei et al., 2014; Yin et al., 2016; Liu et al., 2022b). Biosensors utilize biological components such as enzymes, antibodies, nucleic acids, or whole cells to selectively detect particular heavy metals and convert the biorecognition event into measurable signals (fluorescent, optical, or electrical signal) (Velusamy et al., 2022). Among those biosensors, whole-cell biosensors can offer simplicity and low cost in in situ sensing, and also provide a chassis to embed remediation modules to eliminate heavy metals (Velusamy et al., 2022).
Synthetic biology applies engineering principles similar to those used in electronics to create and build synthetic organisms (Weiss et al., 2002), allowing for overcoming biotechnological challenges in genetic engineering, biochemical synthesis, and biological computation (Na et al., 2013; Chen et al., 2020b; Ren et al., 2020; Shaker et al., 2021). Recent advances in synthetic biology have enabled microorganisms to scavenge and bio-degrade a wide range of hazardous compounds, including aromatic compounds (Xiang et al., 2021), pesticides (Bhatt et al., 2021), microplastics (Miri et al., 2022), greenhouse gases (Tran et al., 2021), etc. In addition, synthetically engineered microorganisms with enhanced tolerance against toxic chemicals have also been developed for better bioremediation (Tran et al., 2021).
Our review focuses on recent advances in the microbial detection and bioremediation of heavy metals using the technologies and approaches of synthetic biology. In addition, the technical challenges and legal regulations associated with genetically modified microorganisms (GMMs) as well as the attempts to overcome the challenges are discussed.
2 Heavy metal resistance mechanisms
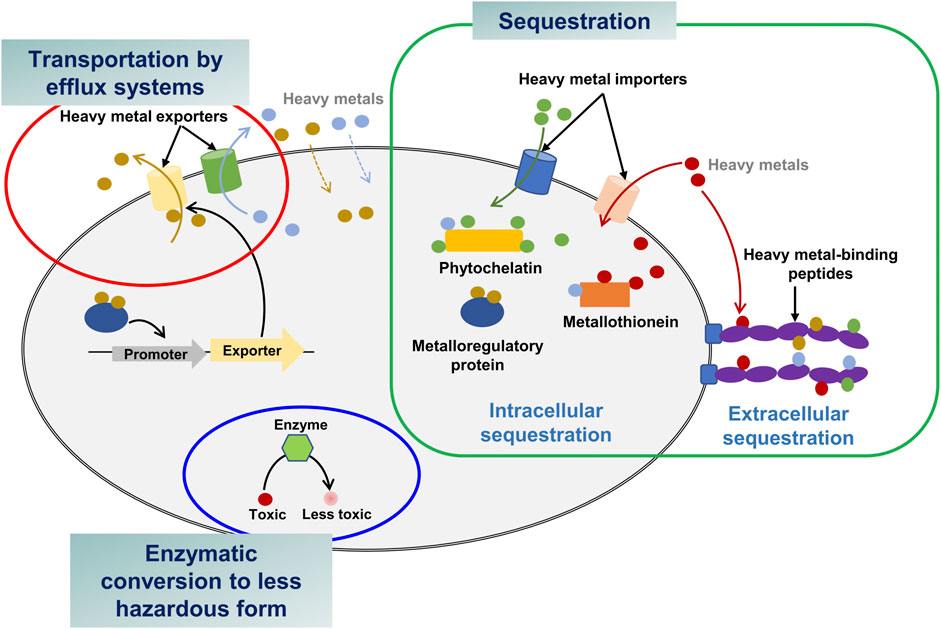
Microorganisms in contaminated areas often develop tolerance against heavy metals and hence they could be suitable bacterial hosts to engineer (Malik, 2004). There are three different resistance mechanisms: transportation via efflux pump system, intra- and extracellular sequestration, and enzymatic conversion to a less toxic form (Figure 1) (Bruins et al., 2000). Considering these mechanisms, sequestration and enzymatic conversion are widely used strategies for application in bacteria to bioremediate heavy metals. In this section, the three mechanisms are introduced with examples of bacteria that are resistant to four significant toxic heavy metals (As, Hg, Cd, and Pb) (Balali-Mood et al., 2021) (Table 2).
2.1 Transportation by efflux systems
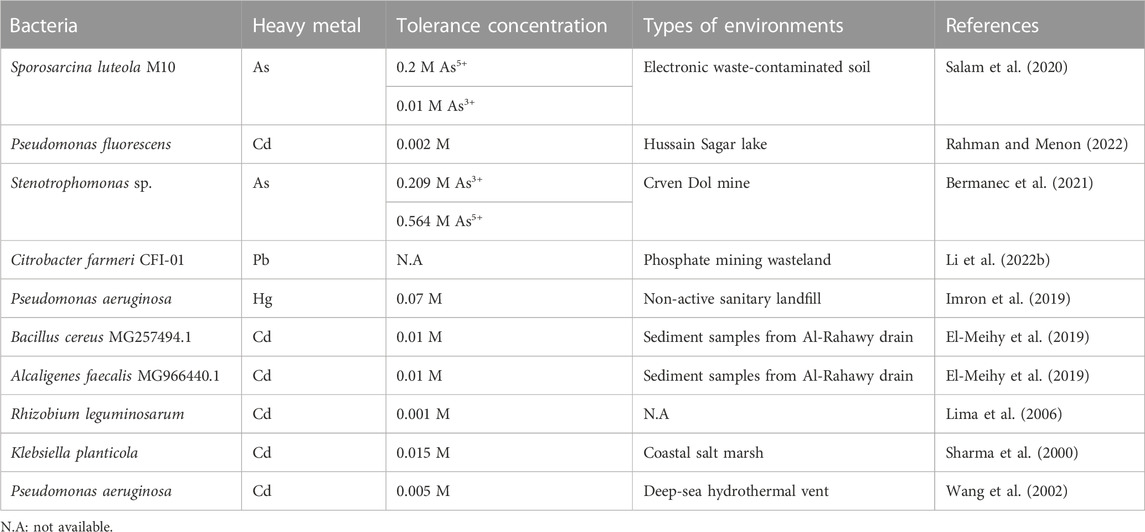
Efflux systems are found in diverse bacterial species, including Pseudomonas aeruginosa, Candida albicans, and Escherichia coli (Soto, 2013). They utilize efflux systems to export heavy metals from cytoplasm to extracellular regions in order to reduce toxicity (Bruins et al., 2000; Abdi et al., 2020). For example, As3+ is a toxic heavy metal form of As. Intracellular toxic As3+ is eradicated by the ars operon including acr3 or arsB genes encoding As3+-efflux proteins (Yang et al., 2012). Sporosarcina luteola M10 isolated from contaminated soil of electronic wastes possesses the arsB gene for As transportation, allowing the bacterium to tolerate up to 0.2 M As5+ and 0.01 M As3+ (Salam et al., 2020). Stenotrophomonas sp. and Microbacterium spp. contain diverse As-efflux systems enabling a notably high As resistance (0.564 M As5+ and 0.209 M As3+) (Bermanec et al., 2021).
The cad operon is a well-known cluster of genes such as cadABC for Cd transportation (Smith and Novick, 1972). Numerous Cd-resistant bacteria have the cadA gene, including Bacillus subtilis, Helicobacter pylori, P. putida, and Stenotrophomonas maltophilia (Jebril et al., 2022). In P. putida, Cd2+-binding CadR, a transcription factor, promoted the expression of a CadA3 efflux protein as well as CzcCBA efflux proteins, contributing to Cd resistance (Liu et al., 2021). The CzcCBA efflux system ejects intracellular Cd2+ to the outer environment in cooperation with CadA3 efflux system. P. fluorescens, recently isolated from Hussain Sagar lake, exhibited Cd-tolerance up to 0.002 M Cd2+ (Rahman and Menon, 2022). Other bacterial species, such as B. cereus MG257494.1 and Alcaligenes faecalis MG966440.1 demonstrated high tolerance up to 0.01 M Cd2+ (El-Meihy et al., 2019).
Apart from the As- and Cd-efflux systems, bacteria also possess several other efflux systems to cope with the toxicity of other heavy metals. For instance, Enterococcus hirae harbors the cop operon, which contains the copB gene encoding a Cu-efflux pump to maintain Cu homeostasis (Solioz and Stoyanov, 2003). Interestingly, the copB gene also participates in the transport of silver (Ag) to the extracellular environment (Solioz and Odermatt, 1995). E. coli possesses the zntA gene that encodes an ATP-dependent efflux system, which confers resistance to Pb and Zn (Babai and Ron, 1998). Furthermore, Cupriavidus metallidurans CH34 (previously known as Ralstonia metallidurans) has the cnrT gene downstream of the cnr operon in plasmid pMOL28, which encodes a nickel-efflux system for nickel resistance (Nies, 2003).
2.2 Intra- and extracellular sequestration
Intracellular heavy metal ions can be sequestered by metal-capturing proteins, e.g., metallothioneins, glutathione (GSH), and metallochaperones (Bazzi et al., 2020). Synechococcus sp. has a metal resistance system containing the smtA gene encoding a Cd2+- and Zn2+-binding metallothionein (Bruins et al., 2000). In Rhizobium leguminosarum, GSH-mediated Cd sequestration through GSH-Cd chelation has been identified as a novel tolerance mechanism (Lima et al., 2006). A metallochaperone, PbrD protein, in C. metallidurans CH34 is able to sequester Pb within the cells and confer protection against toxic Pb (Taghavi et al., 2009).
Extracellular sequestration is to make metal ions insoluble and accumulates them outside bacteria. For instance, when sulfate-reducing bacteria produce vast amounts of hydrogen sulfide in the extracellular environment, sulfide precipitates metal cations (Igiri et al., 2018). Hydrogen sulfide produced from Klebsiella planticola under anaerobic conditions and that from P. aeruginosa under aerobic conditions precipitates Cd2+ as CdS (Sharma et al., 2000; Wang et al., 2002). Citrobacter farmeri CFI-01, isolated from a phosphate mining wasteland, can produce soluble phosphate that precipitates Pb2+ (Li et al., 2022b).
2.3 Conversion of heavy metals into less hazardous forms
Several microorganisms have evolved mechanisms to reduce the sensitivity of cellular components to heavy metals by converting a toxic heavy metal into a less toxic form. Due to the affinity of mercury (Hg) for thiol groups, proteins with a thiol group can be inactivated by Hg. Hg resistance is a typical example of enzymatic detoxification (Misra, 1992). The mer operon of Hg-resistant bacteria such as Shigella flexneri, Staphylococcus aureus, P. stutzeri, P. aeruginosa, K. pneumoniae, Mycobacterium marinum, and Enterobacter contains the merA (mercuric ion reductase) and merB genes (organomercurial lyase). The lyase demethylates highly neurotoxic methylated mercury (MeHg) and releases toxic Hg2+, and then Hg2+ is converted to a volatile and nontoxic Hg0 by the reductase (Bruins et al., 2000; Dash and Das, 2012). The operon also contains the merT and merP genes encoding the proteins responsible for the transportation of Hg2+ from cytoplasm to outside. The cooperative Hg detoxification system effectively protects the host (Bruins et al., 2000).
Cr exists in one of two stable oxidation states in the environment, Cr6+ and Cr3+, and Cr3+ is a less hazardous form of Cr6+ (Mitra et al., 2022). Microorganisms can enzymatically reduce Cr6+ to Cr3+ for tolerance. Under aerobic conditions, the reduction is mediated by soluble and membrane-bound reductases such as ChrR and YieF. ChrR catalyzes the reduction of Cr6+ to Cr5+, and then to Cr3+. On the other hand, the YieF enzyme can directly convert Cr6+ into Cr3+ without the intermediate formation of Cr5+. Under anaerobic conditions, anaerobic bacteria such as P. dechromaticanse and Enterobacter cloacae use membrane-bound reductases such as flavin reductases, cytochromes, and hydrogenases to carry out the reduction of Cr6+ by providing electrons from hydrogen, carbohydrates, NAD(P)H, and endogenous electrons (Lovley and Phillips, 1994; Thatoi et al., 2014).
3 Heavy metal detection
When constructing synthetic bacteria for bioremediation, the first step is to implement a heavy metal detection system for the transition from a resting state to a bioremediating state. The synthetic bacteria sensing heavy metals can also be used as a whole-cell biosensor, which can provide several benefits to overcome industrial limitations: rapid and in situ detection, eco-friendliness, and cost-effectiveness (Gupta et al., 2019; Inda and Lu, 2020).
Whole-cell biosensors harbor genetic elements/circuits that recognize a target heavy metal. Natural bacteria isolated from heavy metal-polluted areas are the source of new genetic elements for biological sensor machineries (Valls and de Lorenzo, 2002). Heavy metal-responsive transcription factors and two-component regulatory systems (TCRS) are the key components of whole-cell biosensors for heavy metal detection. Heavy metal-responsive transcription factors are the proteins that regulate the expression of target genes via heavy metals binding and TCRS consists of a sensor kinase and a response regulator. The sensor kinase senses environmental signals such as heavy metals, and the kinase in turn, activates the response regulator via phosphorylation. The phosphorylated regulator initiates the expression of target genes required for detoxification (Wang and Buck, 2012). For whole-cell biosensors, the transcription factors or response regulators are engineered to transcribe a fluorescent or bioluminescent protein in response to the presence of heavy metals in the environment.
3.1 Metal-responsive transcription factor-based biosensors
A simple genetic detection circuit can consist of sensor and transducer elements (Bereza-Malcolm et al., 2015). The sensor element is generally a transcription factor perceiving the presence of heavy metal. The signal-processing element transforms the signal from the sensor element into a detectable optical (fluorescence or pigment compounds) or electrical signal for real-time analysis (Figure 2). The signal-processing element often incorporates an amplifier, feedback loop, and/or logic gates to tune the output/input signal ratio (Figure 2) (Liu et al., 2022a).
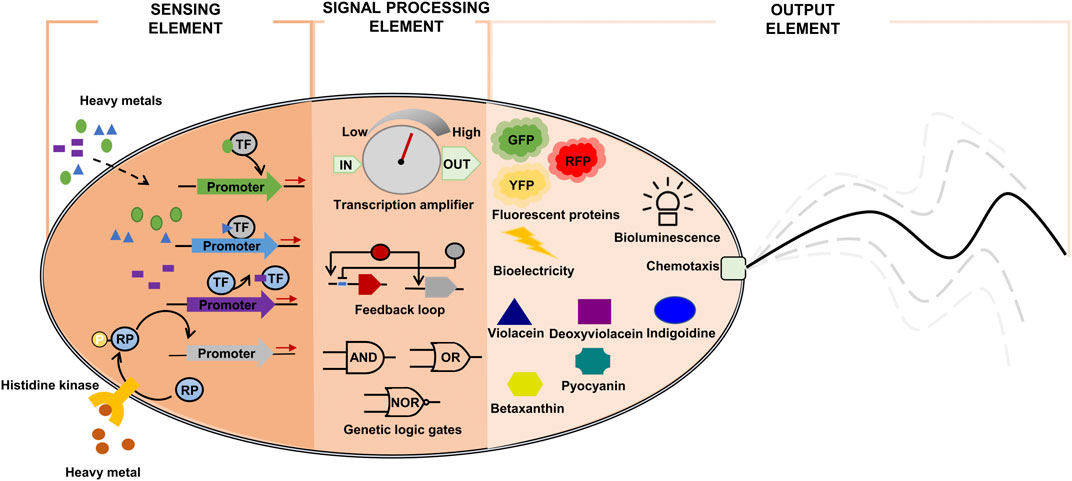
FIGURE 2. Components of synthetic bioremediation bacteria for heavy metal detection. GFP: green fluorescent protein; RFP: red fluorescent protein; RP: regulatory protein; TF: heavy metal-responsive transcription factor; YFP: yellow fluorescent protein.
The sensor element can be implemented using transcription factors that are responsive to heavy metals (Osman and Cavet, 2010). Once activated, the transcription factors initiate the production of detectable signal proteins (color, fluorescence, or electrical signals). Thus, the focus in whole-cell biosensor engineering is to implement a module of transcription factor-signal emitting protein, which can accurately transduce input signal levels (heavy metal concentrations) to proportionally output signal levels (optical or electrical signals) (Table 3).
3.1.1 As biosensor
Seven family members of metal-binding transcription factors have been identified in bacteria: ArsR-SmtB, NikR, DtxR, MerR, Fur, CopY, and CsoR-RcnR (Osman and Cavet, 2010). Each family takes part in sensing a distinct group of metals. ArsR is a repressor protein belonging to the ArsR-SmtB family. Upon binding to As, ArsR is detached from its cognitive promoter and thereby initiates the transcription of the ars operon involved in As detoxification (Busenlehner et al., 2003; Osman and Cavet, 2010).
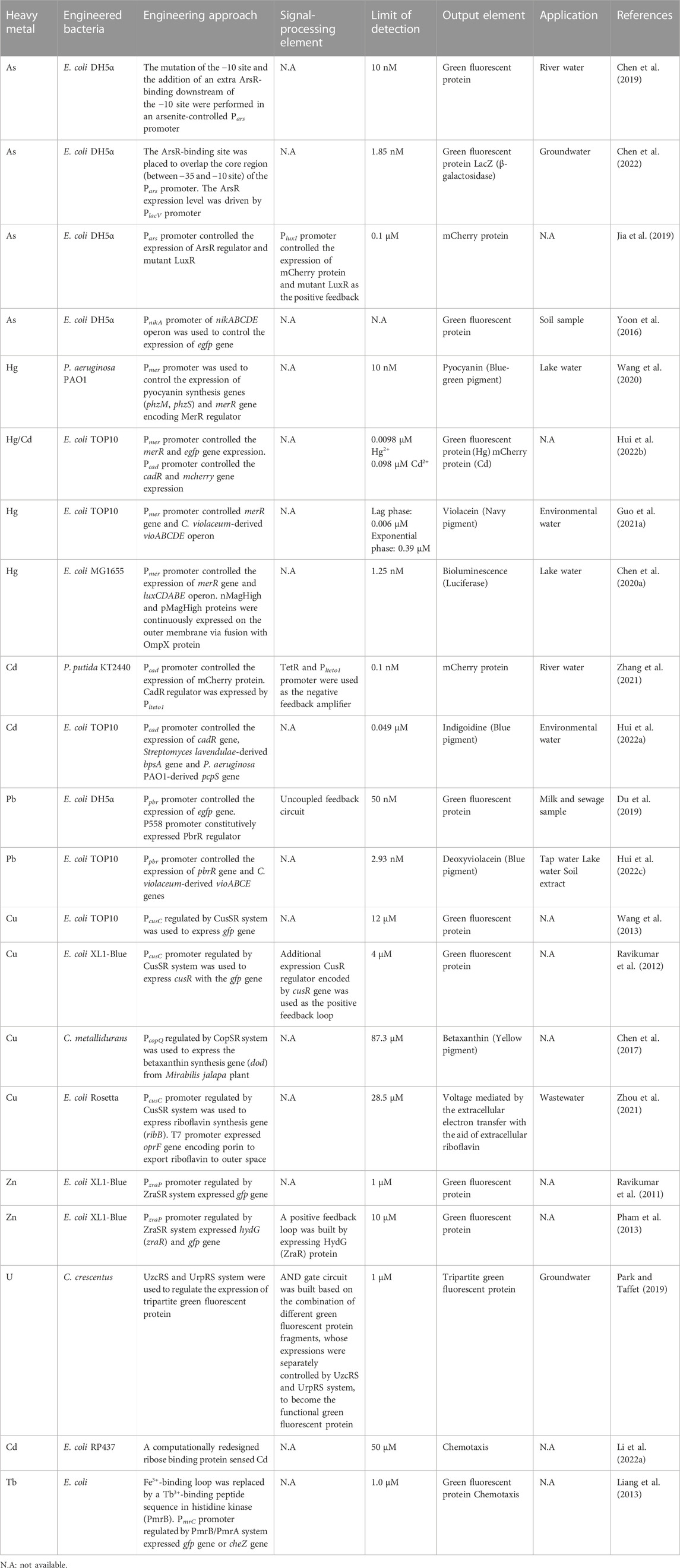
Chen et al. engineered the ArsR-responsive Pars promoter and optimized ArsR expression level to improve the limit of detection (LOD) and decrease the background signal of the biosensor based on ArsR (Figure 3A) (Chen et al., 2019). A library of mutated −10 sites of Pars promoter was generated in order to find mutant promoters showing higher sensitivity towards As as well as lower background signal than the wild-type Pars promoter. One Pars promoter variant (ParsD) containing a mutation within −10 site (GACACT) showed a medium-high fluorescent signal in response to 1 μM of As3+. In addition to the promoter engineering, the location of ArsR-binding site was further optimized to reduce background noise signal. When the second ArsR-binding site was placed downstream of the −10 site of the promoter for tight repression, the background signal was remarkably decreased from 2352 to 142, thus improving the induction ratio (fluorescence signal/background noise) from 16.8 to 179.3. Consequently, the engineered Pars promoter was able to detect As as low as 10 nM. In a following study, the ArsR-binding site was placed to overlap the core region (between −35 and −10 site) of the Pars promoter after the alignment of ArsR-binding site sequence with a library of RNA polymerase binding sites (−35 and −10 sites) and −35 site restructure in ArsR-binding site sequence (Chen et al., 2022). In addition, the ArsR expression level was also optimized. Thus, the induction ratio of the newly designed promoter was further improved up to 183.52. The implemented whole-cell biosensor could detect As concentration as low as 1.85 nM and allowed on-the-spot As identification.
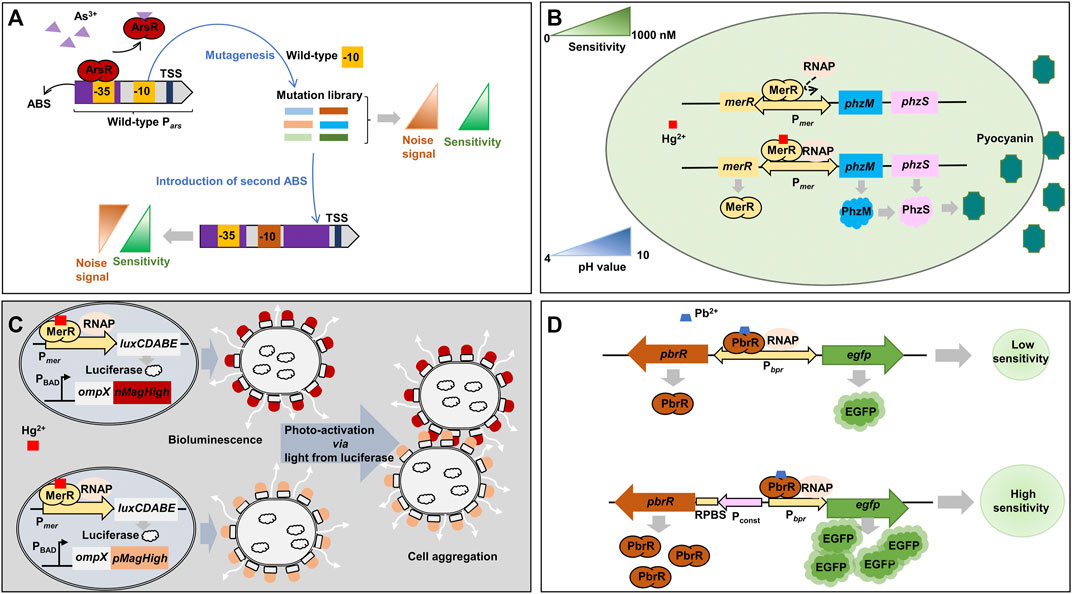
FIGURE 3. Representative synthetic biology approaches for metal-responsive transcription factor-based whole-cell biosensors. (A) Promoter engineering by screening −10 site mutations and addition of an extra ArsR-binding site (Chen et al., 2019). (B) Use of pyocyanin, a blue-green pigment, as an optical output signal by swapping the structural genes of mer operon with pyocyanin synthesis genes (Wang et al., 2020). (C) Enhancement of bioluminescent signal by biofilm formation via the cell aggregation by photoswitchable proteins, nMagHigh and pMagHigh, expressed on the outer membrane (Chen et al., 2020a). (D) Optimization of heavy metal-responsive transcription factor expression to increase biosensor sensitivity (Du et al., 2019). luxCDABE: luciferase; nMagHigh: photoswitchable protein nMagHigh; ompX: outer membrane protein X; pMagHigh: photoswitchable protein pMagHigh; ABS: ArsR-binding site; RNAP: RNA polymerase; ArsR: arsenic-sensing transcriptional regulator; EGFP: enhanced green fluorescent protein; MerR: mercury-sensing regulator; Pconst: moderate constitutive P558 promoter; Pars: arsenic-inducible promoter; PBAD: araBAD operon promoter; PbrR: lead-sensing transcriptional regulator; PhzM: phenazine-specific methyltransferase; PhzS: flavin-containing monooxygenase; Pmer: mercury-inducible promoter; Ppbr: lead-inducible promoter; RPBS: regulator protein-binding site; TSS: transcription start site.
Jia et al. incorporated a LuxR-based positive feedback into ArsR-based As biosensor to amplify output signal as well as to improve sensitivity (Jia et al., 2019). The LuxR protein lacked its N-terminal region (Δ2-162) and thus showed a constitutive transcription initiation activity even in the absence of its activator (Nistala et al., 2010). Two copies of the mutant luxR gene were incorporated into the biosensor. The first copy was under the control of Pars and the second copy was placed downstream of the mcherry gene in a bicistronic configuration regulated by PluxI. Thus, once LuxR is produced by the activation of Pars promoter in the presence of As, the LuxR triggers the positive feedback by transcribing mCherry and luxR genes via PluxI. The whole-biosensor showed a time-dependent response to As due to the positive feedback and could detect As concentration as low as 0.1 µM which was lower than the safety concentration of As in drinking water recommended by the World Health Organization.
E. coli harbors the nickel (Ni) ABC-type transporter, NikABCDE, and this operon is transcriptionally repressed by Ni via the NikR repressor (Leitch et al., 2007). Interestingly, it has been discovered that As impeded the repression activity of NikR protein and thereby As activated the transcription of the nik operon (Yoon et al., 2016). This discovery exemplifies an unpredicted interaction between heavy metals and transcription factors, which requires a deep understanding of the regulation mechanisms when constructing whole-cell biosensors.
3.1.2 Hg and Cd biosensor
The MerR transcription factor family senses several different heavy metals, for example, MerR for Hg, PbrR for Pb, CadR for Cd, CueR for Cu, ZntR for Zn, and CoaR for Co (Brown et al., 2003). P. aeruginosa PAO1 has been engineered to implement an optical biosensor for Hg detection (Figure 3B) (Wang et al., 2020). Specifically, the endogenous mer operon under the control of a Hg-responsive regulator (MerR) was engineered to transcribe a phenazine-specific methyltransferase (phzM) and a flavin-containing monooxygenase (phzS) to synthesize pyocyanin when the microorganism was exposed to Hg. Pyocyanin is a soluble blue-green pigment (optical color signal). The engineered biosensor operated stably at a wide range of pH from 4 to 10 and showed an LOD of 10 nM Hg2+.
A colorimetric biosensor for Hg detection was constructed by employing the vioABCDE operon derived from Chromobacterium violaceum, which produces violacein, a navy pigment (Guo et al., 2021a). The Pmer promoter and the Hg-responsive transcription factor-encoding merR gene obtained from the transposon Tn21 in E. coli were utilized to regulate the expression of the vioABCDE in response to Hg. Navy color could be detectable after 1 hour of induction with 8 μM Hg2+. The whole-cell biosensor in a lag phase exhibited low LOD (0.006 μM) but a narrow detection range (0–0.012 μM), whereas that in an exponential phase displayed a wider detection range (0.78–12.5 μM) but higher LOD (0.39 μM).
A dual-output signal biosensor has been developed to simultaneously detect Hg and Cd using MerR and CadR transcriptional factor (Hui et al., 2022b). The MerR was used to regulate the expression of green fluorescent protein (GFP) (green color) in response to Hg and CadR for mCherry protein (red color) in response to Cd. The sensor could quantitatively assess the concentrations of Hg and Cd within the range of 0–5 μM and 0–200 μM, respectively.
To improve the detection range towards Hg, E. coli MG1655 has been engineered based on bioluminescence (light) and light-induced artificial biofilm (Figure 3C) (Chen et al., 2020a). For the detection of Hg at very low concentration, merR gene was cloned under the control of Pmer promoter as a positive feedback loop. Once MerR is activated by Hg, the sensor continuously produces MerR as well as other downstream genes, luxCDABE, for bioluminescence. To strengthen the signal, the sensor employed nMagHigh and pMagHigh proteins, which were displayed on the cell’s surface with the aid of the circularly permutated outer membrane protein, OmpX. The two proteins were photoswitchable proteins, and once activated by light emitted from luciferase reporter protein encoded by luxCDABE, they interacted with each other and aggregated bacterial cells. The cell aggregation strengthened the bioluminescence signal. Consequently, the biosensor could detect Hg in the range of 1.25 nM and up to 1,000 nM Hg2+ (Chen et al., 2020a).
P. putida KT2440 has been engineered to improve sensitivity and specificity to Cd (Zhang et al., 2021). In the whole-cell biosensor, CadR expression was maintained at a low level by a negative feedback loop and the mcherry gene was regulated by CadR. Due to the low level of CadR, the sensor showed a very low background signal leading to greater sensitivity and better specificity.
Indigoidine, a blue pigment, has been utilized as an output signal in a Cd-sensing whole-cell biosensor (Hui et al., 2022a). The Cd-sensing system was composed of Pcad promoter and cadR gene originated from P. putida. Upon activation of CadR by Cd, Pcad promoter initiated the expression of the bpsA gene (Streptomyces lavendulae) and the pcpS gene (P. aeruginosa PAO1) for indigoidine biosynthesis. The blue color of the indigoidine could be discerned with naked eyes after 30 min of induction with 100 µM of Cd2+. Notably, this pigment-based biosensor showed no detectable background signal in contrast to the previously used fluorescence-based biosensor for Cd detection. The LOD of this indigoidine-based biosensor was 0.049 µM of Cd2+, and it was successfully applied to verify the presence of Cd in environmental water samples.
3.1.3 Pb biosensor
The Pb-sensing protein, PbrR, has a high affinity to Pb2+, but it also interacts with other divalent metal ions such as Zn2+, Cu2+, and Cd2+. Thus, PbrR has been engineered via mutagenesis for higher specificity to Pb2+ than other divalent metal ions (Jia et al., 2020b). The specificity was measured by the growth of the engineered bacterial cells by implementing the cell to produce ampicillin-resistant enzymes by the mutant PbrR under diverse metal ions. Of many mutants, a mutant (D64A/L68S) was identified to improve specificity: the affinity (growth rate) to Pb2+ of the mutant was two-fold higher than wild-type, while the affinity to other metal ions (Zn, Cu, and Cd) was comparable with wild-type.
An E. coli whole-cell biosensor for Pb detection was constructed with the PbrR-responsive promoter (Ppbr) and downstream genes of pbrR and egfp to keep PbrR at an optimal level (Figure 3D) (Du et al., 2019). In a previous study on a biosensor for Cd, the low-level expression of the transcription regulator increased sensitivity and specificity by reducing background noise (Zhang et al., 2021). However, the Pb biosensor showed an increased background signal even though the PbrR was at a low level. It could be because that the level of PbrR was too low to efficiently repress the promoter activity resulting in leaky expression of the egfp gene. When the pbrR gene was expressed by a moderate constitutive promoter and the egfp gene was still under the control of Ppbr promoter, the sensor module showed higher sensitivity within a range of 50 nM - 10 μM Pb2+. This result represents the importance of expression optimization for the reliable operation of biosensors.
Hui et al. constructed four variants of the violacein gene cluster of vioABCDE (C. violaceum) to develop multiple-pigment biosensors for detecting Pb (Hui et al., 2022c): vioABE genes for prodeoxyviolacein (green), vioABDE genes for proviolacein (blue), vioABCE genes for deoxyviolacein (purple), and vioABCDE genes for violacein (navy). For the expression of the cluster variants, the Ppbr promoter and pbrR gene obtained from C. metallidurans was utilized. The biosensors successfully produced the respective pigments upon detection of Pb. Of the biosensors, the deoxyviolacein biosensor exhibited the widest detection range (2.93–6,000 nM) and the lowest LOD (2.93 nM) among the other pigment biosensors.
3.2 Two-component regulatory system-based biosensors
Bacteria possess another machinery, called TCRS, to sense extracellular environments, such as chemical and physical changes (Bijlsma and Groisman, 2003; Casino et al., 2010). This system comprises two main components, a histidine kinase and a regulator protein. The kinase contains a periplasm-located loop formed by two transmembrane segments for receiving signals and a cytoplasm-positioned transmitter domain found within the final transmembrane segment for controlling a downstream regulator via phosphorylation or dephosphorylation. When the regulator is phosphorylated, it becomes a transcription activator and initiates the transcription of downstream genes to respond to environmental changes (Casino et al., 2010).
TCRS offers several advantages for developing whole-cell biosensors over the transcription factor-based approach. TCRS can sense extracellular as well as intracellular stimuli, but transcription factors can sense only the intracellular changes. Thus, transcription factor-based biosensors require additional modules to transmit the input signal (e.g., extracellular heavy metals) into the cytoplasm (Ulrich et al., 2005; Mimee et al., 2018). In addition, the output signals of TCRS are relatively less impacted by the levels of histidine kinase and regulator, which exempts us from optimizing the expression levels of individual biosensor component genes (Batchelor and Goulian, 2003; Shinar et al., 2007). Thus, TCRS responses are more secured from the internal noises generated in the gene expression of component genes.
3.2.1 Cu biosensor
Cu TCRS is composed of CusS for sensing Cu and CusR for regulating PcusC promoter, and this system has been widely studied and utilized to construct Cu whole-cell biosensors (Rademacher and Masepohl, 2012; Yu et al., 2022). The native CusSR TCRS of E. coli MG1655 was used to produce green fluorescence by expressing the gfp gene under Cu-responsive PcusC (Wang et al., 2013). The LOD of the system was as low as 12 µM of Cu2+. Ravikumar et al. developed a positive feedback loop by co-expressing cusR with the gfp gene via PcusC promoter to enhance Cu sensitivity (Ravikumar et al., 2012). The LOD of the biosensor was improved by 10-fold (4 μM) compared with the native system.
Chen et al. improved the specificity of a Cu biosensor based on the CopSR regulatory system, another Cu TCRS discovered in C. metallidurans. To find a promoter specific to Cu, they screened cop promoters (PcopA, PcopH, PcopT, PcopM, PcopF, PcopL, PcopQ, and PcopE) regulated by CopR in the presence of 1 mM of Cu2+, Fe3+, Co2+, Ni2+, Zn2+, or Pb2+ (Chen et al., 2017). These promoters were designed to drive the expression of the rfp gene encoding red fluorescent protein, thereby facilitating the quantitative assessment of individual promoter activity in terms of protein expression. Of the eight promoters, PcopQ showed high Cu2+-specific induction pattern and thus was employed in the Cu biosensor. For color detection, the Cu2+-specific PcopQ controlled the expression of the Mirabilis jalapa-derived dod gene (4,5-dihydroxy-phenylalanine dioxygenase), which is involved in the conversion of L-dihydroxy-phenylalanine (L-DOPA) to betaxanthin, a yellow pigment. The addition of L-DOPA in media facilitated the production of betaxanthin as a reporter.
In a study, electrical signal (voltage) was used as an output signal instead of color or fluorescence signals (Zhou et al., 2021). In E. coli, the ribB gene (3,4-dihydroxy-2-butanone-4-phosphate synthase) was transcribed for the production of riboflavin via the PcusC promoter in response to Cu (Figure 4A). In addition, a porin gene (oprF) was constitutively expressed via T7 promoter, which exports riboflavin to the outer space. The escalation of extracellular riboflavin led to the increase in voltage by the electron transfer from riboflavin to anode. The electrical biosensor showed an LOD of 28.5 µM Cu2+, which offered a quick and affordable analytical method to biosensors for the in situ monitoring of Cu in water.
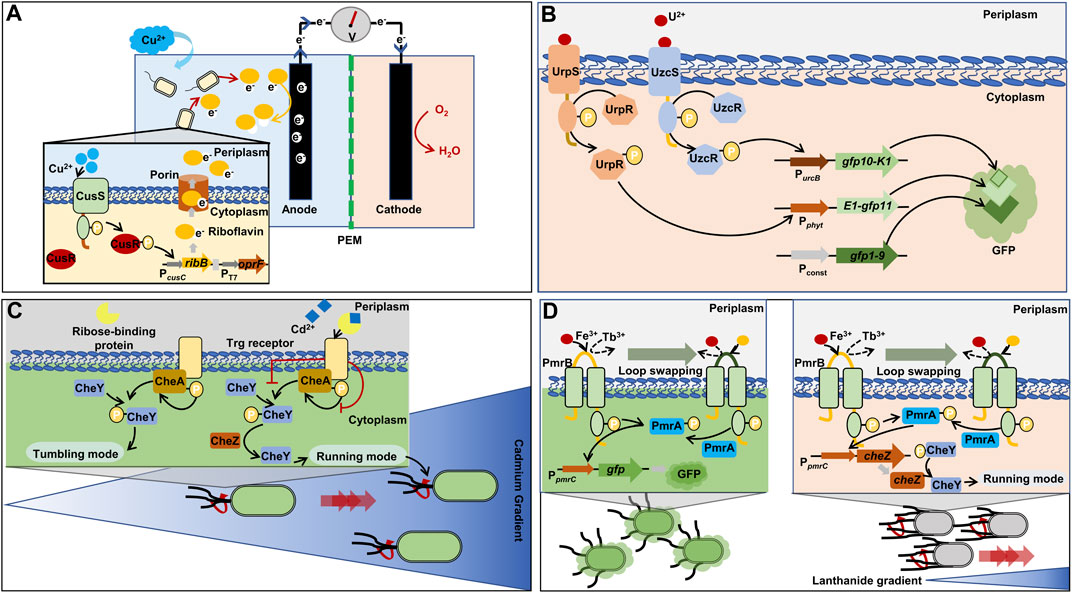
FIGURE 4. Representative synthetic biology approaches for two-component regulatory system (TCRS)-based whole-cell biosensors. (A) Rapid and inexpensive electrical detection of cooper by riboflavin as an output signal (Zhou et al., 2021). (B) Combination of two TCRS systems for the specific detection of uranium and tripartite green fluorescent protein cooperation (Park and Taffet, 2019). (C) Chemotaxis towards cadmium via retailored ribose binding protein for cadmium binding (Li et al., 2022a). (D) Formation of a lanthanide-sensing component by exchanging iron-binding motif of histidine kinase with lanthanide-binding peptide (Liang et al., 2013). cheZ: phosphatase; gfp: green fluorescent protein; gfp10: β-strand 10 of green fluorescent protein; gfp11: β-strand 11 of green fluorescent protein; gfp1-9: β-strand from 1 to 9 of green fluorescent protein; oprF: outer membrane porin F; ribB: 3,4-dihydroxy-2-butanone-4-phosphate synthase; CheA: histidine kinase; CheY: chemotaxis protein; CusR: cooper-transcriptional regulator; CusS: cooper-sensing histidine kinase; K1, E1: synthetic coiled-coils; Pconst: strong, constitutive promoter derived from rsaA gene encoding S-layer protein; PcusC: promoter derived cusC gene encoding efflux system protein and activated by phosphorylated CusR; PEM: proton exchange membrane; PmrA: iron-transcriptional regulator; PmrB: iron-sensing protein; Pphyt: promoter derived from phytase-encoded operon and activated by phosphorylated UrpR; PpmrC: promoter obtained from pmrC gene encoding phosphoethanolamine transferase and activated by phosphorylated PmrA; PT7: T7 promoter; PurcB: promoter derived from urcB gene and activated by phosphorylated UzcR; UrpR, UzcR: uranium-transcriptional regulator; UrpS, UzcS: uranium-sensing histidine kinase.
3.2.2 Zn biosensor
It was discovered that the ZraSR TCRS responded to an extracellular Zn2+ level more specifically than other metal cations (i.e., Cd2+ and Pb2+) (Leonhartsberger et al., 2001; Appia-Ayme et al., 2012). The TCRS was employed in E. coli and the gfp gene was cloned downstream of the PzraP promoter regulated by the ZraR regulator. The biosensor responded proportionally to Zn2+ and its LOD was as low as 1 µM (Ravikumar et al., 2011).
Another Zn2+-sensing system also utilized ZraSR TCRS via PzraP promoter to regulate the gfp expression in response to Zn2+. A positive feedback loop was initiated by overexpressing the zraR gene encoding ZraR regulator downstream of gfp gene to enhance the sensitivity in LB media to 10 µM (Pham et al., 2013). However, when this system was integrated into the genome of E. coli, its sensitivity was reduced to 500 µM. Despite this reduction, this genome-integrated system still was valid as a biosensor for detecting Zn2+ in heavily contaminated areas with elevated concentrations of Zn2+. The study provided evidence for the practicability of biosensors utilizing TCRS in the detection of Zn2+ across a wide range of concentrations.
3.2.3 Uranium (U) biosensor
Two TCRS for U sensing (UzcRS and UrpRS) have been discovered in uranium resistance bacterial Caulobacter crescentus, respectively (Park et al., 2019; Park and Taffet, 2019). For high specificity in U detection, the two TCRS systems (UzcRS and UrpRS) were employed and the two systems controlled the expression of two different fragments of whole GFP (GFP10-K1 and E1-GFP11), respectively (Figure 4B) (Cabantous et al., 2013; Park and Taffet, 2019). In the C. crescentus, the last fragment of GFP (GFP1-9) was constitutively produced. When the three different fragments were assembled, green fluorescence was emitted. Thus, the GFP fragments operated as an AND gate, and the gate was activated only when both the two TCRS systems detected U. The LOD of the U biosensor was 5 µM. In addition, the UrpRS system regulated the additional expression of the membrane protein UzcY that acts as the signal amplifier of the UzcRS system by improving the activity of UzcS to sense U. This regulation led to an increase in biosensor sensitivity, ultimately improving the sensitivity of the LOD to 1 µM.
3.2.4 Cd biosensor
Chemotaxis has been harnessed to sense heavy metal ions. Unlike other biosensors, the output of chemotaxis-employed biosensor is the movement towards a target heavy metal. Bacterial cells are able to travel to find a favorable environment: they move towards attractants such as sugars and amino acids, while they move farther away from repellents such as alcohols and heavy metal ions (Co2+, Ni2+) (Eisenbach, 2011). The chemotaxis of bacteria is also regulated by TCRS that comprises CheA (histidine kinase), CheY (response regulator), and CheZ (phosphatase) (Bi and Sourjik, 2018). When there are no attractants, CheA phosphorylates CheY, which leads to cell tumbling in place by assembling with flagellar motor. When attractants are available, CheY is dephosphorylated by CheZ and the dissociation of CheY from the flagellar motor allows for moving forward (Bi and Sourjik, 2018).
E. coli harbors the Trg receptor that mediates chemotaxis to ribose via an interaction with the periplasmic ribose-binding protein. The ribose binding protein was engineered to sense Cd ions. Consequently, the engineered E. coli host with the Cd-sensing protein showed chemotactic migration towards Cd2+ (Figure 4C) (Li et al., 2022a). The engineered chemotaxis could foster intelligent bioremediation by moving microbial cells to a contaminated site.
3.2.5 Lanthanide (Tb) biosensor
The PmrA/PmrB system, a TCRS for Fe3+-sensing in Salmonella, was engineered for the detection of lanthanide-ion (Tb3+) in E. coli (Liang et al., 2013). For sensing Tb3+, the Fe3+-binding loop in the histidine kinase of the TCRS was replaced with a Tb3+-binding peptide. The redesigned PmrA/PmrB TCRS system was capable of sensing Tb3+ at around 1.0 μM (Figure 4D). The repurposed sensing system was used to control the chemotaxis of E. coli towards Tb by regulating the expression of the CheZ protein. When CheZ was produced more due to high concentration of Tb, the engineered biosensor host could move closer/faster towards Tb than other cells producing less CheZ proteins (Bi and Sourjik, 2018).
4 Microbial remediation of heavy metals assisted by synthetic biology
In this section, microorganisms engineered for bioremediation are introduced by their remediation mechanisms. Of the three resistance mechanisms introduced in Section 2, sequestration is commonly utilized for bioremediation. Thus, constructed synthetic bacteria, to date, based on the heavy metal sequestration are introduced in this section.
4.1 Synthetic bioremediation bacteria based on extracellular sequestration
Certain bacterial species can trap heavy metals on their surface to prevent the penetration of heavy metals into the cells. The trapping is mediated via the interaction between functional groups attached on the cell membrane (amino, carboxyl, phosphate, and hydroxyl groups) and metal ions (El-Helow et al., 2000; Taniguchi et al., 2000). In addition, heavy metals can be also captured by extracellular polymeric substances secreted by bacteria, such as polysaccharides, mucopolysaccharides, and proteins (Zeng et al., 2020). However, the low inherent capture efficiency and instability of the extracellular polymeric substances prevent this approach from being broadly applied to the construction of synthetic bioremediation bacteria (Tang et al., 2021).
Recently, the advance in synthetic biology enabled us to develop metalloproteins and metal-binding peptides to exhibit improved heavy metal biosorption on the cell surface (Table 4) (Samuelson et al., 2002). The metalloproteins or metal-binding peptides were displayed on the cell membrane in the aid of membrane-anchoring proteins. In Gram-negative bacteria, outer membrane proteins (i.e., ice-nucleation protein (INP), Lpp-OmpA, OmpA, OmpC, and LamB), fimbriae/flagella, and autotransporter have been widely used as anchoring proteins. Metal-binding proteins were fused with the anchoring proteins at N- or C-terminus, or both for the membrane display (Samuelson et al., 2002; Wernerus and Stahl, 2004). In Gram-positive bacteria, staphylococcal protein A (SpA) is an effective anchoring protein to display metal-binding proteins (Wernerus et al., 2001). SpA comprises an N-terminal signal peptide (either four or five IgG-binding domains with a region rich in proline and glycine) and a C-terminal section (membrane-anchoring region) (Samuelson et al., 2002).
4.1.1 Pb bioremediation
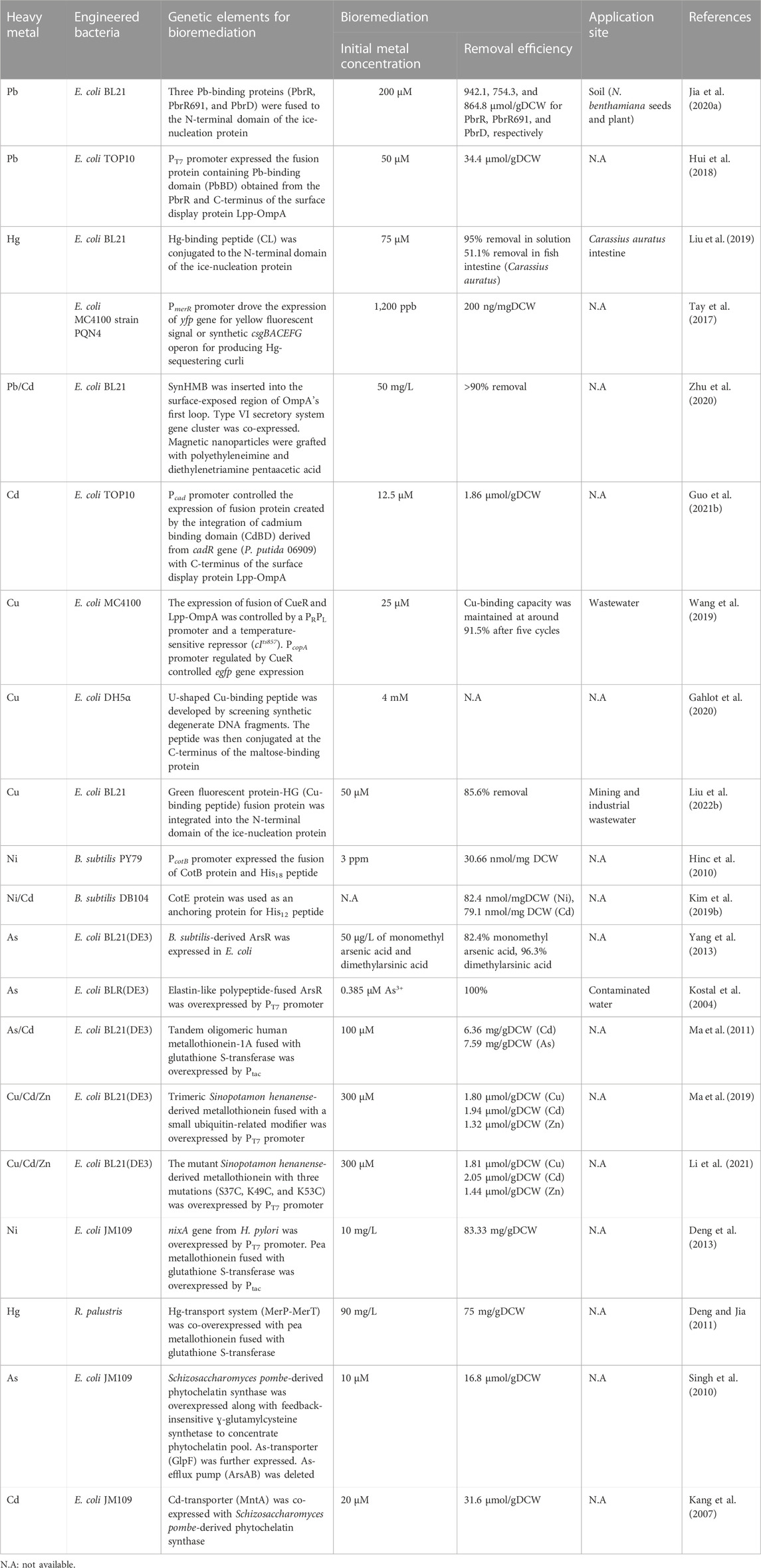
To construct Pb-capturing E. coli, three Pb-binding proteins (PbrR, PbrR691, and PbrD) derived from C. metallidurans were fused at the N-terminal domain of INP to display on the surface of E. coli BL21 (Jia et al., 2020a). When the linker between INP and a Pb-binding protein and their expression levels were optimized, the three proteins (PbrR-INP, PbrR691-INP, and PbrD-INP) showed remarkably higher Pb-adsorption capacities of 942.1, 754.3, and 864.8 mol/g dry cell weight (DCW), respectively than reported so far. When the engineered E. coli was utilized to clear Pb in soil and its evaluation was carried out with seed germination and plant growth of Nicotiana benthamiana, the germination and the growth in the decontaminated soil were improved. Specifically, when the growth was measured as biomass after 50 days of cultivation, the biomasses of the plants grown in the decontaminated soil by PbrR-INP, PbrR691-INP, and PbrD-INP were 0.74 ± 0.21 g, 1.34 ± 0.27 g, 0.73 ± 0.05 g, respectively. The biomass of the plant grown in the contaminated soil as a control was 0.10 ± 0.02 g.
The integration of the Pb-binding domain (PbBD) from the PbrR protein with the Lpp-OmpA facilitated the extracellular accumulation of Pb in E. coli (Hui et al., 2018). The PbBD presentation on cell surface enhanced Pb2+ biosorption by 1.92-fold and achieved a capacity of 34.4 μmol/gDCW. The enhanced biosorption capability of PbBD compared with the whole protein (PbrR) resulted from the superior transport and localization of the short PbBD on the bacterial surface.
4.1.2 Hg bioremediation
A synthetic Hg-binding peptide, that is rich in cysteine, has been conjugated to the N-terminal domain of INP and was expressed and displayed on the membrane of E. coli BL21 (Liu et al., 2019). With the synthetic Hg-binding peptide, the engineered E. coli could capture 95% of Hg2+ from the Hg-contaminated solution at 75 μM (Khati et al., 2012; Liu et al., 2019). As there is increasing concern about Hg pollution in the ocean and its toxicity to fish, the engineered E. coli w applied to fish to protect them from Hg. When the synthetic E. coli was fed to fish (Carassius auratus), it protected the gut microbial consortium in the fish intestine and prevented the host from Hg toxicity and cleared up to 51.1% of Hg in the intestine.
Tay et al. utilized the PmerR promoter derived from S. flexneri to drive the expression of yfp gene for yellow fluorescent signal or the synthetic csgBACEFG operon for producing Hg-sequestering curli in E. coli (Tay et al., 2017). The promoter is responsive to Hg, and curli are thin, aggregative, and extracellular fibers that adsorb Hg2+ extracellularly (Hidalgo et al., 2010). The engineered E. coli could capture Hg2+ from the environment at the removal efficiency of roughly 200 ng/mgDCW.
4.1.3 Pb and Cd bioremediation
In a study, a de novo synthetic heavy metals-binding peptide was implemented in E. coli BL21 and a chemically engineered magnetic nanoparticle was developed to efficiently capture Pb, Cd, and other heavy metals as well as to easily isolate the synthetic bioremediation bacteria (engineered E. coli) cells from decontaminated solution (Figure 5A) (Zhu et al., 2020). Firstly, a synthetic heavy metal-binding peptide, SynHMB, was inserted into the surface-exposed region within the first loop of OmpA. The type VI secretory system gene cluster of P. putida was additionally co-expressed to facilitate the display of the fusion protein (Bernal et al., 2017). Secondly, a magnetic nanoparticle was modified to contain polyethyleneimine and diethylenetriamine pentaacetic acid to interact with the engineered E. coli strain via the carboxyl groups in the nanoparticle and histidine residues of SynHMB expressed on the cell surface. The engineered E. coli strain efficiently removed over 90% of Pb and Cd from 50 mg/L of Cd2+- and 50 mg/L of Pb2+-containing solution. After decontamination, the cells were recovered using magnetic fields.
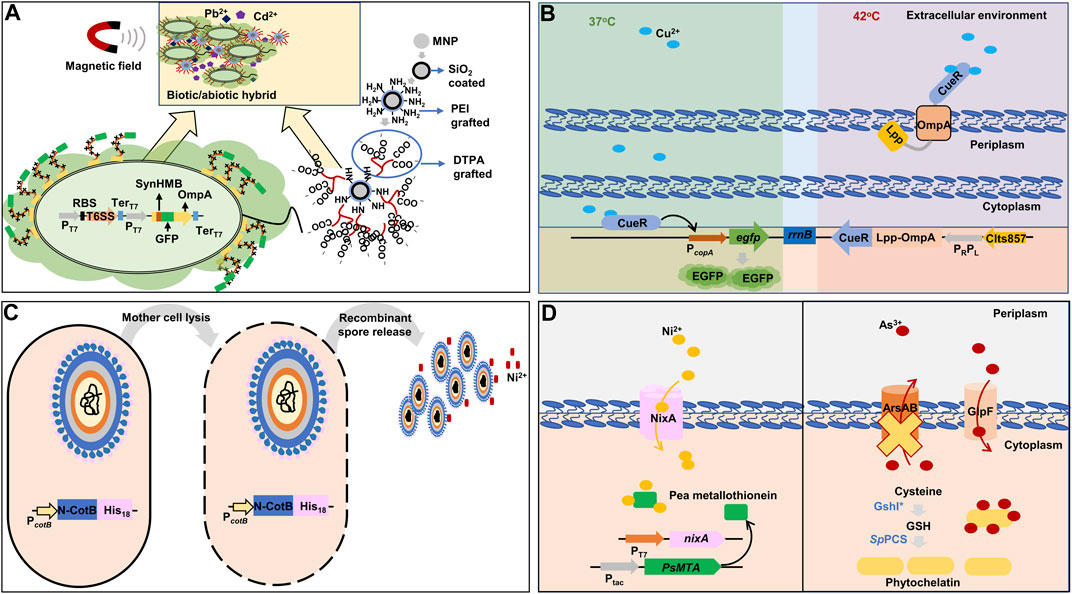
FIGURE 5. Representative synthetic biology approaches for heavy metals bioremediation in bacteria. (A) Combination of synthetic metal-binding peptide for improved extracellular sequestration of lead and cadmium, and chemically modified magnetic nanoparticles for easy recovery of bacterial cells after decontamination (Zhu et al., 2020). (B) A dual-functional synthetic bacteria for extracellular copper detection and biosorption regulated by thermal change (Wang et al., 2019). (C) Spore surface display with histidine residues to adsorbing nickel (Hinc et al., 2010). (D) Intracellular sequestration of nickel and arsenic by metallothionein and phytochelatin with the expression of respective heavy metal importers (Singh et al., 2010; Deng et al., 2013). The yellow cross denotes gene deletion. nixA: high-affinity nickel-transport protein; PsMTA: pea metallothionein; rrnB: terminator region derived from rrnB gene; ArsAB: arsenic exporter; CIts857: thermosensitive λ repressor; CueR: cooper-transcriptional regulator; DTPA: diethylenetriamine pentaacetic acid; EGFP: enhanced green fluorescent protein; Lpp: prolipoprotein; GlpF: arsenic transporter; GSH: glutathione; GshI*: feedback-insensitive ɣ-glutamylcysteine synthetase; His18: 18 histidine residues; MNP: magnetic nanoparticle; N-CotB: N-terminal domain of spore coat protein B; OmpA: outer membrane protein A; PC: constitutive promoter; PcopA: promoter derived from copA gene encoding copper exporter and activated by copper-binding CueR; PcotB: promoter derived from cotB gene encoding spore coat protein B; PEI: polyethyleneimine; PRPL: tandem promoters of λ bacteriophage; PT7: T7 promoter; Ptac: hybrids of the PlacUV5 and tryptophan (trp) operon promoter; RBS: ribosome-binding site; SpPCS: Schizosaccharomyces pombe-derived phytochelatin synthase; SynHMB: synthetic heavy metals-binding peptide; T6SS: type VI secretory system gene cluster; TerT7: T7 terminator.
A system for both biosensing and extracellular sequestration of Cd was developed (Guo et al., 2021b). The system utilized the Pcad promoter and cadR gene derived from P. putida 06909 to produce an output signal (GFP) and a sequestration agent (a fusion protein of cadmium binding domain (CdBD) and Lpp-OmpA). The CdBD was derived from the CadR of P. putida 06909 and the chimeric sequestration agent (Lpp-OmpA-CdBD) captured Cd at cell surface. The engineered cell was capable of detecting Cd at a concentration as low as 0.1 µM, and their Cd-binding capacity was 1.86 μmol/gDCW.
4.1.4 Cu bioremediation
A dual-functional E. coli that sensed and eliminated Cu ions was constructed, which switched its state from sensing to eliminating by temperature (Figure 5B) (Wang et al., 2019). Firstly, E. coli cell was designed to emit green fluorescence proportional to Cu ions in a sample. The egfp gene was placed under the PcopA promoter that was activated by the inherent Cu-binding transcription activator CueR. A fusion gene of the Cu-binding transcription activator CueR and Lpp-OmpA was constructed to capture Cu ions on the cell membrane. For the temperature-dependent transcription of the fusion gene, a PRPL promoter and a temperature-sensitive repressor (cIts857) were used: the repressor maintained its repressor activity at 37°C, but lost its activity at 42°C and, thereby, the PRPL promoter was activated. This dual-purpose E. coli cell responded specifically to Cu with a detection range of 0.01–25 μM at 37°C and exhibited Cu sequestration by CueR at 42°C. After 10 min of incubation at 42°C, the engineered strain could remove 91.5% of Cu ion in a water sample with 25 μM of Cu2+.
Lately, a putative U-shaped Cu-binding peptide was developed by screening synthetic degenerate DNA fragments, and the peptide was then conjugated at the C-terminus of the maltose-binding protein (MBP) (Gahlot et al., 2020). E. coli cells displaying the fusion of MBP and Cu-binding peptide demonstrated efficient Cu-binding and tolerance to up to 4 mM of Cu2+. This engineered cell possessed a Cu-binding capacity 32 times greater than that of the previously reported E. coli strain modified to sequester Cu2+ from Chinese wastewater (Wang et al., 2019).
A concurrent detection and biosorption bacterium has also been constructed (Liu et al., 2022b). A synthetic Cu-binding peptide, His-Gly-His-Gly-His-Gly-His (HG), was fused with GFP and the GFP-HG fusion protein was integrated into the N-terminal domain of INP for exterior display. The surface-engineered E. coli demonstrated Cu LOD down to 1 µM and adsorption up to 302.08 µmol/gDCW. In addition, since the Cu-binding peptide was fused with GFP, the binding between the peptide and Cu decreased the green fluorescence of GFP. Thus, the system could be also used as a biosensor for Cu.
4.1.5 Ni bioremediation
B. subtilis spore surface display has been used to develop protein-coated spores to adsorb Ni ions (Lin et al., 2020). CotB, a protein on the surface of Bacillus spores, was modified to contain 18 histidine residues (His18) at its C-terminus since a poly-His tag is a well-known peptide to interact with Ni ions (Figure 5C) (Hinc et al., 2010). The recombinant spores showed better Ni elimination than wild-type spores and a higher capacity of approximately 30.66 nmol/mg of recombinant spore, compared with 24.79 nmol/mg of wild-type spore. Additionally, a shorter poly-His tag containing 12 His residues increased the capacity of the recombinant spores to 82.42 nmol/mg of recombinant spores (Kim et al., 2019b).
4.2 Synthetic bioremediation bacteria based on intracellular sequestration
Another sequestration mechanism harnessed to develop synthetic bacteria for heavy metals bioremediation is intracellular sequestration. Heavy metals are transported into the cytoplasm by appropriate importers and then sequestered by metal-binding proteins/peptides (Table 4) (Diep et al., 2018). The sequestered heavy metals are often chemically converted via enzymatic processes or precipitated.
4.2.1 Intracellular sequestration by metalloregulatory proteins
Metalloregulatory proteins or heavy-metal responsive transcription factors induce the expression of proteins responsible for heavy-metal transporters, binding proteins, reducing proteins, etc. for heavy metals detoxification. Due to the excellent specificity and affinity to heavy metals, metalloregulatory proteins can be utilized to capture heavy metals inside cells.
When the ArsR proteins derived from B. subtilis were overexpressed in E. coli, the bacterial capacity to accumulate methylated As species was considerably increased (Yang et al., 2013). In another study, though ArsR proteins were overexpressed to increase accumulation capacity, the ArsR overproduction retarded cell growth significantly: the maximal OD decreased down to 25% of that of wild-type cells (Kostal et al., 2004). When elastin-like polypeptide was fused to ArsR as a solubilizing agent, cell growth was increased by two folds compared with the control while maintaining the same ability of ArsR to accumulate As3+. The synthetic bacteria overexpressing elastin-ArsR fusion protein resulted in 5- and 60-fold higher levels of As5+ and As3+ accumulation, respectively, compared with the control cells that did not express ArsR at all.
4.2.2 Intracellular sequestration by metallothioneins
Heavy metal elimination in archaea, prokaryotes, and eukaryotes is complemented by metallothioneins that are abundant in nature. Metallothioneins have a strong affinity for heavy metals because of their cysteine-rich composition: the interaction between thiol groups in the cysteine residues and heavy metal ions. The aggregation of several heavy metals (Cu, Cd, and Mg) has been enhanced by overexpressing metallothioneins in microorganisms (Yang et al., 2015).
The intracellular overabundance of metallothioneins frequently experiences instability due to solubility issue, leading to a lowered heavy metal storage capability. An efficient solution to this issue is the co-expression of metallothioneins with a soluble fusion tag such as glutathione S-transferase (GST) and MBP. When human metallothionein (hMT-1A) was overexpressed in E. coli as a fusion protein with GST, the bacterium successfully accumulated Cd and As inside cells (Ma et al., 2011). Subsequently, oligomeric tandems of hMT-1A with various repeats were constructed to further improve Cd and As accumulation capacity. The maximum Cd and As bioaccumulation was achieved by the engineered E. coli expressing the GST-fused trimeric hMT-1A protein: 6.36 mg Cd2+/gDCW and 7.59 mg As3+/gDCW.
Ma et al. also utilized a similar approach for expressing Sinopotamon henanense-derived metallothionein (ShMT) to sequester Cu, Cd and Zn in E. coli cells, but the difference was the use of small ubiquitin-related modifier (SUMO) as a solubility tag (Ma et al., 2019). Comparably, the SUMO-fused trimeric ShMT demonstrated great bioaccumulation of Cu, Cd, and Zn: 5.8-fold, 3.1-fold, and 6.7-fold higher accumulation, respectively, than those of control cells expressing only SUMO.
Li et al. engineered the metallothionein (ShMT) to improve its metal-binding ability. They conducted sequence-based alignment, structure-based molecular docking simulation, and site-directed mutagenesis to develop an efficient ShMT mutant while not modifying the folding and molecular mass observed in the best oligomeric tandem design (Ma et al., 2019; Li et al., 2021). The ShMT mutant with three mutations (S37C, K49C, and K53C), in which all the three residues were replaced with Cys, showed the maximum bioaccumulation of Cu, Cd, and Zn: 1.86 -fold, 1.71-fold, and 2.13-fold enhancement, respectively, than wild-type ShMT.
The excellent capacity for heavy metal bioaccumulation of metallothioneins is countered by their complete absence of selectivity (Ma et al., 2011; Ma et al., 2019; Li et al., 2021). Hence, for the bioaccumulation of a specific heavy metal, instead of engineering metallothioneins, appropriate transports are often co-expressed to import specific heavy metals. For selective Ni-bioaccumulation, the Ni-affinity transmembrane protein originated from H. pylori (NixA) as well as GST-fused pea metallothionein were co-expressed (Figure 5D) (Deng et al., 2013). The synthetic bacteria expressing both NixA and GST-pea metallothionein showed a maximum Ni-uptake and accumulation capacity of 83.33 mg/gDCW.
When a metallothionein (GST-fused pea metallothionein) was co-expressed with the Hg-transport system (MerP-MerT) in a photosynthetic bacterium, Rhodopseudomonas palustris, the bacterium specifically accumulated Hg, but not other metal ions in solution (Deng and Jia, 2011).
4.2.3 Intracellular sequestration by phytochelatins
Another form of cysteine-abundant peptide widely utilized to improve the capacity of bacteria to accumulate heavy metals is phytochelatins, which are found in plants or fungi (Schmoger et al., 2000). The common form of phytochelatins is (ɤ-Glu-Cys-)n-Gly (n = 1–11). Phytochelatins have a better adsorption affinity for heavy metals than metallothioneins since phytochelatins are longer and have a higher number of cysteine residues than metallothioneins, allowing them to form more stable complexes with heavy metals. Phytochelatin pools inside the cells can be boosted by increasing the level of phytochelatin precursors, cysteine, γ-glutamylcysteine, and GSH by overexpressing cysE, gshI, and gshII, respectively (Diep et al., 2018).
Singh et al. overexpressed Schizosaccharomyces pombe-derived phytochelatin synthase (SpPCS) along with feedback-insensitive ɣ-glutamylcysteine synthetase (GshI*) in E. coli, which was shown to be the tailback of GSH formation (Singh et al., 2010). The synthetic bacterium showed a 30-fold increased phytochelatin level. In addition, for the specific bioaccumulation of As, As-transporter (GlpF) was additionally expressed while the As-efflux pump (ArsAB) was deleted from the genome of E. coli (Figure 5D). The resulting strain showed 80-fold increased As accumulation compared to the wild-type strain.
In another study, for the specific bioaccumulation of Cd, the Cd-transporter (MntA) was co-expressed with SbPCS in E. coli. The engineered bacterium showed an increased Cd-specific storage capacity by 25-fold compared with the wild-type strain (Kang et al., 2007).
4.3 Difficulties in the application of synthetic biology in microbial bioremediation
The natural bacteria isolated from heavy metal-polluted sites have high potentials for bioremediation since they possess high tolerance and high removal efficiency. Though they are considered valuable platforms or chasses for synthetic bioremediation bacteria development, the current bacteria chasses used to implement genetic or regulatory elements for bioremediation are mostly laboratory model bacteria such as E. coli. The first hurdle is to isolate and cultivate natural bacteria (Bodor et al., 2020; Tran et al., 2021). Due to the complex requirements for the cultivation of isolated bacteria, it is difficult to obtain a pure bacterium and find the optimal growth conditions. As the isolated bacteria from contaminated areas are important sources of new elements for tolerance and detoxification, this difficulty hinders the advance of synthetic bioremediation bacteria construction. However, due to the advance in sequencing technologies and machine learning-based prediction models, new genes could be identified from a mixture of bacterial species without the need for bacteria isolation (Kobras et al., 2021; Konno and Iwasaki, 2023).
The second hurdle is the lack of available molecular toolkits for the non-model (isolated) bacterial organisms. This limitation hinders the construction of synthetic bacteria because natural bacteria that possess high bioremediation capability have far fewer molecular toolkits than common strains such as E. coli. Recent advances in synthetic biology toolkits that are universal, adaptable, and effective in various bacterial species could facilitate the modification of the non-model bacterial organisms, such as small regulatory RNAs (sRNAs) (Na et al., 2013), portable multiplex automated genome engineering (pORTMAGE) (Nyerges et al., 2016), clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (Citorik et al., 2014), and so on.
5 Microbial detection and remediation: Limitations and current legal regulations
5.1 Limitations of microbial detection
Although whole-cell biosensors are promising tools for in situ heavy metal detection, they also have several limitations that may hinder their effectiveness. One limitation is the possibility of false-positive results due to the lack of specificity of biosensing systems (Guo et al., 2021b). Another limitation is the relatively high LOD in complicated environmental matrices, because interferents such as organic substances, minerals, and other metals could disrupt the detection ability of biosensors (Brányiková et al., 2020).
5.1 Limitations of microbial remediation
Bioremediation has advantages over chemical and physical remediation methods in terms of efficiency, sustainability, and ecofriendliness. However, it also has several drawbacks. Firstly, it is difficult to scale up from laboratory scale to industrial scale (Diep et al., 2018). While chemical and physical methods are predictable and thus easy to scale up, bioremediation employs living organisms that are very complex systems, and thus it is difficult to predict outcomes when the scale is changed.
Secondly, bioremediation mostly takes one or two strategies to remove heavy metals, mostly sequestration (Volesky and Holan, 1995). However, chemical and physical methods can utilize various strategies to remove heavy metals, including thermal treatment, adsorption, chlorination, chemical extraction, and electrokinetics (Selvi et al., 2019).
The last difficulty is the legal limitations. The wide use of GMMs in environments triggered a debate over the legality of employing GMMs in open environments (Singh et al., 2011). U.S. Environmental Protection Agency regulations require successful pilot tests before a technology may be used (Janssen and Stucki, 2020). Besides, the release of GMMs is restricted in most European nations unless authorization is acquired, and thus the use of GMMs in environments is not widely accepted (Wesseler et al., 2022). Therefore, the sluggish deployment of GMMs is ascribed to several safety concerns, legislative restrictions, and potential threats that the public may perceive, leading to strict control. It was predicted that advanced remediation procedures might be put off for up to 10 years due to these regulations (Janssen and Stucki, 2020).
During the heavy metal bioremediation, the introduction of GMMs to the fields can raise risks since GMMs do not stay in a controlled environment. The horizontal gene transfer might quickly occur between the GMMs and indigenous microorganisms (Sobecky and Coombs, 2009), and the delivery of recombinant genes may change the genetic makeup of native microorganisms (Kumar et al., 2020). The entry of recombinant genes into other species that can propagate the gene in the natural microbial populations may also stimulate bioremediation. However, the transfer of mobile elements like transposons or vectors containing antibiotic resistance genes triggers the emergence of naturally occurring super germs resistant to antibiotics (Wang and Zhang, 2019). Additionally, the alteration in microbial communities and metabolism may produce harmful substances. Thus, the U.S. National Institutes of Health (NIH) guideline suggested that recombinant E. coli should only escape at a maximum rate of 10−8 cells (NIH, 2019).
5.2 Current technical attempts to resolve the biosafety issues
There is an urgent demand for additional technologies to ensure the safe use of GMMs. Introducing genetic safeguards into engineered bacteria is examples of overcoming these restrictions (Moe-Behrens et al., 2013). For instance, toxin and antitoxin have been utilized to prevent horizontal gene transfer. The gene encoding toxin is harbored in a plasmid while the gene encoding for antitoxin is harbored in the genome (Paul et al., 2005). When the plasmid is transferred to another bacterial species that do not harbor the antitoxin gene, the bacteria will be killed by the toxin. However, mutations within the toxin gene may allow the successful transfer of the plasmid. To avoid the mutational escape, two or three separate systems can be introduced in the plasmid and the genome of synthetic bacteria. An example of this approach is that two doc genes encoding toxic Doc proteins and one ecoRI gene encoding a restriction endonuclease were cloned in three different plasmids. This approach prevents the mutational escape and the escape rate was below 10−9 when tested with E. coli and P. putida (Xue et al., 2022a; Xue et al., 2022b).
Controlled replication of plasmids is another method for safeguarding recombinant genes (Li et al., 2023). RepL, a replication protein that primarily regulates plasmid copy number, was introduced to a plasmid as a plasmid amplifier. The presence of an inducer for RepL expression increased the plasmid number, while the plasmid number was limited without an inducer. Furthermore, the conditional replication origin ColE2-P9 was introduced into the plasmid to control plasmid replication. The ColE2-P9 requires the initiator protein Rep that was introduced into the host’s chromosome (Yagura et al., 2006). Thus, when the plasmid was transferred to other bacteria, its replication was not initiated and its copy number was limited.
There is another strategy to control the growth of synthetic bacteria (Ronchel and Ramos, 2001). The asd gene engineered to be activated by XylS, which is essential for cell wall synthesis, was introduced into the genome of P. putida while the inherent native asd gene was disrupted. Another gene, a toxin gef gene that was engineered to be negatively regulated by XylS, was also introduced into the genome. When 3-methyl benzoate is not supplied to the host, which is an activator of XylS, the synthetic bacteria do not generate cell wall. Furthermore, the synthetic bacteria start produce a toxin protein from the gef gene and consequently dies because the cell does not have the antitoxin gene.
6 Conclusion and future perspectives
Heavy metal pollution is now becoming a global problem due to the numerous threats to human health and environment. Physical and chemical remediation have been widely applied to the elimination of hazardous heavy metals at an industrial scale. However, due to the concerns about sustainability, environmental friendliness, high cost, and the slow process, microbial remediation has gained attention as an alternative and innovative approach remediating heavy metals.
Non-model bacterial species predominate in nature and certain bacterial species may exhibit special metabolic features useful for bioremediation. Recent advances in synthetic biology could provide new genetic toolkits to upgrade the bioremediation capability of the microorganisms not only for laboratory bacterial species but also for non-model (isolated) microorganisms. However, operational reclamation of heavy metal-contaminated areas cannot be accomplished using a particular methodology. Physical, chemical, and biological methods must be combined to be entirely successful and comprehensive remediation, and this should be inspired the convergence of various fields.
Microorganisms can be an integrated platform or chassis of a heavy metal detection module and a remediation module, and the two modules can be linked to work in an orchestrated manner. Thus, they are highly effective in bioremediation compared with conventional methods in which detection and remediation are separated.
Future advance in engineering technologies with synthetic biology principles would pave the way for enhanced remediation platforms to safeguard human health and environments.
Author contributions
DN, TDT, and WL defined the conception and designed the manuscript. TDT and WL wrote and revised the manuscript. TDT contributed to figures and tables editing. WL contributed to tables and information collection. DN supervised and revised the manuscript. All authors contributed to this review article and approved the submitted version.
Funding
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIT) (NRF-2022M3A9B6082687 and No. NRF-2023R1A2C1008156). This research was supported by the Chung-Ang University Young Scientist Scholarship in 2021.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Razik, M. A., Azmy, A. F., Khairalla, A. S., and Abdelghani, S. (2020). Metal bioremediation potential of the halophilic bacterium, Halomonas sp. strain WQL9 isolated from Lake Qarun, Egypt. Egypt. J. Aquat. Res. 46, 19–25. doi:10.1016/j.ejar.2019.11.009
Abdi, S. N., Ghotaslou, R., Ganbarov, K., Mobed, A., Tanomand, A., Yousefi, M., et al. (2020). Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug Resist. 13, 423–434. doi:10.2147/Idr.S228089
Alrashed, M., Tabassum, H., Almuhareb, N., Almutlaq, N., Alamro, W., Alanazi, S. T., et al. (2021). Assessment of DNA damage in relation to heavy metal induced oxidative stress in females with recurrent pregnancy loss (RPL). Saudi J. Biol. Sci. 28, 5403–5407. doi:10.1016/j.sjbs.2021.05.068
Appenroth, K. J. (2010). What are "heavy metals" in Plant Sciences? Acta Physiol. Plant. 32, 615–619. doi:10.1007/s11738-009-0455-4
Appia-Ayme, C., Hall, A., Patrick, E., Rajadurai, S., Clarke, T. A., and Rowley, G. (2012). ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem. J. 442, 85–93. doi:10.1042/BJ20111639
Babai, R., and Ron, E. Z. (1998). An Escherichia coli gene responsive to heavy metals. FEMS Microbiol. Lett. 167, 107–111. doi:10.1111/j.1574-6968.1998.tb13215.x
Balali-Mood, M., Naseri, K., Tahergorabi, Z., Khazdair, M. R., and Sadeghi, M. (2021). Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 12, 643972. doi:10.3389/fphar.2021.643972
Batchelor, E., and Goulian, M. (2003). Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U. S. A. 100, 691–696. doi:10.1073/pnas.0234782100
Bazzi, W., Abou Fayad, A. G., Nasser, A., Haraoui, L. P., Dewachi, O., Abou-Sitta, G., et al. (2020). Heavy metal toxicity in armed conflicts potentiates AMR in A. Baumannii by selecting for antibiotic and heavy metal co-resistance mechanisms. Front. Microbiol. 11, 68. doi:10.3389/fmicb.2020.00068
Bereza-Malcolm, L. T., Mann, G., and Franks, A. E. (2015). Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synth. Biol. 4, 535–546. doi:10.1021/sb500286r
Bermanec, V., Paradžik, T., Kazazić, S. P., Venter, C., Hrenović, J., Vujaklija, D., et al. (2021). Novel arsenic hyper-resistant bacteria from an extreme environment, Crven Dol mine, Allchar, North Macedonia. J. Hazard. Mater. 402, 123437. doi:10.1016/j.jhazmat.2020.123437
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi:10.1038/ismej.2016.169
Bharagava, R. N., Saxena, G., Mulla, S. I., and Patel, D. K. (2018). Characterization and identification of recalcitrant organic pollutants (ROPS) in tannery wastewater and its phytotoxicity evaluation for environmental safety. Arch. Environ. Con. Tox. 75, 259–272. doi:10.1007/s00244-017-0490-x
Bhatt, P., Bhatt, K., Sharma, A., Zhang, W. P., Mishra, S., and Chen, S. H. (2021). Biotechnological basis of microbial consortia for the removal of pesticides from the environment. Crit. Rev. Biotechnol. 41, 317–338. doi:10.1080/07388551.2020.1853032
Bi, S., and Sourjik, V. (2018). Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 45, 22–29. doi:10.1016/j.mib.2018.02.002
Bijlsma, J. J. E., and Groisman, E. A. (2003). Making informed decisions: Regulatory interactions between two-component systems. Trends Microbiol. 11, 359–366. doi:10.1016/S0966-842X(03)00176-8
Bodor, A., Bounedjoum, N., Vincze, G. E., Kis, A. E., Laczi, K., Bende, G., et al. (2020). Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Bio. 19, 1–22. doi:10.1007/s11157-020-09522-4
Bolisetty, S., Peydayesh, M., and Mezzenga, R. (2019). Sustainable technologies for water purification from heavy metals: Review and analysis. Chem. Soc. Rev. 48, 463–487. doi:10.1039/c8cs00493e
Brányiková, I., Lucáková, S., Kuncová, G., Trögl, J., Synek, V., Rohovec, J., et al. (2020). Estimation of Hg(II) in soil samples by bioluminescent bacterial bioreporter E. coli ARL1, and the effect of humic acids and metal ions on the biosensor performance. Sensors 20, 3138. doi:10.3390/s20113138
Brown, N. L., Stoyanov, J. V., Kidd, S. P., and Hobman, J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. doi:10.1016/S0168-6445(03)00051-2
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotox. Environ. Safe. 45, 198–207. doi:10.1006/eesa.1999.1860
Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143. doi:10.1016/S0168-6445(03)00054-8
Cabantous, S., Nguyen, H. B., Pedelacq, J. D., Koraichi, F., Chaudhary, A., Ganguly, K., et al. (2013). A new protein-protein interaction sensor based on tripartite split-GFP association. Sci. Rep. 3, 2854. doi:10.1038/srep02854
Casino, P., Rubio, V., and Marina, A. (2010). The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 20, 763–771. doi:10.1016/j.sbi.2010.09.010
Chandra, R., Bharagava, R. N., Yadav, S., and Mohan, D. (2009). Accumulation and distribution of toxic metals in wheat (Triticum aestivum L.) and Indian mustard (Brassica campestris L.) irrigated with distillery and tannery effluents. J. Hazard. Mater. 162, 1514–1521. doi:10.1016/j.jhazmat.2008.06.040
Chen, F., Warnock, R. L., Van Der Meer, J. R., and Wegner, S. V. (2020a). Bioluminescence-triggered photoswitchable bacterial adhesions enable higher sensitivity and dual-readout bacterial biosensors for mercury. ACS Sens. 5, 2205–2210. doi:10.1021/acssensors.0c00855
Chen, P.-H., Lin, C., Guo, K.-H., and Yeh, Y.-C. (2017). Development of a pigment-based whole-cell biosensor for the analysis of environmental copper. RSC Adv. 7, 29302–29305. doi:10.1039/c7ra03778c
Chen, S. Y., Wei, W. P., Yin, B. C., Tong, Y. B., Lu, J. J., and Ye, B. C. (2019). Development of a highly sensitive whole-cell biosensor for arsenite detection through engineered promoter modifications. ACS Synth. Biol. 8, 2295–2302. doi:10.1021/acssynbio.9b00093
Chen, S. Y., Zhang, Y., Li, R., Wang, B., and Ye, B. C. (2022). De novo design of the ArsR regulated Pars promoter enables a highly sensitive whole-cell biosensor for arsenic contamination. Anal. Chem. 94, 7210–7218. doi:10.1021/acs.analchem.2c00055
Chen, Y., Banerjee, D., Mukhopadhyay, A., and Petzold, C. J. (2020b). Systems and synthetic biology tools for advanced bioproduction hosts. Curr. Opin. Biotech. 64, 101–109. doi:10.1016/j.copbio.2019.12.007
Choiniere, J., and Wang, L. (2016). Exposure to inorganic arsenic can lead to gut microbe perturbations and hepatocellular carcinoma. Acta. Pharm. Sin. B 6, 426–429. doi:10.1016/j.apsb.2016.07.011
Chung, J. Y., Yu, S. D., and Hong, Y. S. (2014). Environmental source of arsenic exposure. J. Prev. Med. Public Health 47, 253–257. doi:10.3961/jpmph.14.036
Citorik, R. J., Mimee, M., and Lu, T. K. (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145. doi:10.1038/nbt.3011
Dash, H. R., and Das, S. (2012). Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegrad. 75, 207–213. doi:10.1016/j.ibiod.2012.07.023
Deng, X., He, J., and He, N. (2013). Comparative study on Ni2+-affinity transport of nickel/cobalt permeases (NiCoTs) and the potential of recombinant Escherichia coli for Ni2+ bioaccumulation. Bioresour. Technol. 130, 69–74. doi:10.1016/j.biortech.2012.11.133
Deng, X., and Jia, P. (2011). Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour. Technol. 102, 3083–3088. doi:10.1016/j.biortech.2010.10.051
Diep, P., Mahadevan, R., and Yakunin, A. F. (2018). Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng. Biotechnol. 6, 157. doi:10.3389/fbioe.2018.00157
Du, R. X., Guo, M. Z., He, X. Y., Huang, K. L., Luo, Y. B., and Xu, W. T. (2019). Feedback regulation mode of gene circuits directly affects the detection range and sensitivity of lead and mercury microbial biosensors. Anal. Chim. Acta. 1084, 85–92. doi:10.1016/j.aca.2019.08.006
Duan, C. Y., Ma, T. Y., Wang, J. Y., and Zhou, Y. B. (2020). Removal of heavy metals from aqueous solution using carbon-based adsorbents: A review. J. Water Process Eng. 37, 101339. doi:10.1016/j.jwpe.2020.101339
El-Helow, E. R., Sabry, S. A., and Amer, R. M. (2000). Cadmium biosorption by a cadmium resistant strain of Bacillus thuringiensis: Regulation and optimization of cell surface affinity for metal cations. Biometals 13, 273–280. doi:10.1023/A:1009291931258
El-Meihy, R. M., Abou-Aly, H. E., Tewfike, T. A., El-Alkshar, E. A., and Youssef, A. M. (2019). Characterization and identification of cadmium-tolerant bacteria isolated from contaminated regions in Egypt. Biocatal. Agric. Biotechnol. 21, 101299. doi:10.1016/j.bcab.2019.101299
Gahlot, D. K., Taheri, N., Mahato, D. R., and Francis, M. S. (2020). Bioengineering of non-pathogenic Escherichia coli to enrich for accumulation of environmental copper. Sci. Rep. 10, 20327. doi:10.1038/s41598-020-76178-z
Guo, Y., Hui, C. Y., Liu, L. S., Chen, M. P., and Huang, H. Y. (2021a). Development of a bioavailable Hg(II) sensing system based on MerR-regulated visual pigment biosynthesis. Sci. Rep. 11, 13516. doi:10.1038/s41598-021-92878-6
Guo, Y., Hui, C. Y., Zhang, N. X., Liu, L. S., Li, H., and Zheng, H. J. (2021b). Development of cadmium multiple-signal biosensing and bioadsorption systems based on artificial cad operons. Front. Bioeng. Biotech. 9, 585617. doi:10.3389/fbioe.2021.585617
Gupta, N., Renugopalakrishnan, V., Liepmann, D., Paulmurugan, R., and Malhotra, B. D. (2019). Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 141, 111435. doi:10.1016/j.bios.2019.111435
Ha, E., Basu, N., Bose-O'reilly, S., Dorea, J. G., Mcsorley, E., Sakamoto, M., et al. (2017). Current progress on understanding the impact of mercury on human health. Environ. Res. 152, 419–433. doi:10.1016/j.envres.2016.06.042
Hidalgo, G., Chen, X. C., Hay, A. G., and Lion, L. W. (2010). Curli produced by Escherichia coli PHL628 provide protection from Hg(II). Appl. Environ. Microb. 76, 6939–6941. doi:10.1128/Aem.01254-10
Hinc, K., Ghandili, S., Karbalaee, G., Shali, A., Noghabi, K. A., Ricca, E., et al. (2010). Efficient binding of nickel ions to recombinant Bacillus subtilis spores. Res. Microbiol. 161, 757–764. doi:10.1016/j.resmic.2010.07.008
Hui, C. Y., Guo, Y., Gao, C. X., Li, H., Lin, Y. R., Yun, J. P., et al. (2022a). A tailored indigoidine-based whole-cell biosensor for detecting toxic cadmium in environmental water samples. Environ. Technol. Inno. 27, 102511. doi:10.1016/j.eti.2022.102511
Hui, C. Y., Guo, Y., Li, H., Chen, Y. T., and Yi, J. (2022b). Differential detection of bioavailable mercury and cadmium based on a robust dual-sensing bacterial biosensor. Front. Microbiol. 13, 846524. doi:10.3389/fmicb.2022.846524
Hui, C. Y., Guo, Y., Yang, X. Q., Zhang, W., and Huang, X. Q. (2018). Surface display of metal binding domain derived from PbrR on Escherichia coli specifically increases lead(II) adsorption. Biotechnol. Lett. 40, 837–845. doi:10.1007/s10529-018-2533-4
Hui, C. Y., Guo, Y., Zhu, D. L., Li, L. M., Yi, J., and Zhang, N. X. (2022c). Metabolic engineering of the violacein biosynthetic pathway toward a low-cost, minimal-equipment lead biosensor. Biosens. Bioelectron. 214, 114531. doi:10.1016/j.bios.2022.114531
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018, 1–16. doi:10.1155/2018/2568038
Imron, M. F., Kurniawan, S. B., and Soegianto, A. (2019). Characterization of mercury-reducing potential bacteria isolated from Keputih non-active sanitary landfill leachate, Surabaya, Indonesia under different saline conditions. J. Environ. Manage. 241, 113–122. doi:10.1016/j.jenvman.2019.04.017
Inda, M. E., and Lu, T. K. (2020). Microbes as biosensors. Annu. Rev. Microbiol. 74, 337–359. doi:10.1146/annurev-micro-022620-081059
Janssen, D. B., and Stucki, G. (2020). Perspectives of genetically engineered microbes for groundwater bioremediation. Environ. Sci. Process Impacts 22, 487–499. doi:10.1039/c9em00601j
Jebril, N., Boden, R., and Braungardt, C. (2022). Cadmium resistant bacteria mediated cadmium removal: A systematic review on resistance, mechanism and bioremediation approaches. IOP Conf. Ser. Earth Environ. Sci. 1002, 012006. doi:10.1088/1755-1315/1002/1/012006
Jia, X., Bu, R., Zhao, T., and Wu, K. (2019). Sensitive and specific whole-cell biosensor for arsenic detection. Appl. Environ. Microbiol. 85, e00694–e00619. doi:10.1128/AEM.00694-19
Jia, X., Li, Y., Xu, T., and Wu, K. (2020a). Display of lead-binding proteins on Escherichia coli surface for lead bioremediation. Biotechnol. Bioeng. 117, 3820–3834. doi:10.1002/bit.27525
Jia, X. Q., Ma, Y. B., Bu, R. R., Zhao, T. T., and Wu, K. (2020b). Directed evolution of a transcription factor PbrR to improve lead selectivity and reduce zinc interference through dual selection. Amb. Express 10, 67. doi:10.1186/s13568-020-01004-8
Kang, S. H., Singh, S., Kim, J. Y., Lee, W., Mulchandani, A., and Chen, W. (2007). Bacteria metabolically engineered for enhanced phytochelatin production and cadmium accumulation. Appl. Environ. Microbiol. 73, 6317–6320. doi:10.1128/AEM.01237-07
Khati, W., Ouali, K., Mouneyrac, C., and Banaoui, A. (2012). Metallothioneins in aquatic invertebrates: Their role in metal detoxification and their use in biomonitoring. Aquat. Toxicol. 18, 784–794. doi:10.1016/j.egypro.2012.05.094
Kim, H. S., Kim, Y. J., and Seo, Y. R. (2015). An overview of carcinogenic heavy metal: Molecular toxicity mechanism and prevention. J. Cancer Prev. 20, 232–240. doi:10.15430/JCP.2015.20.4.232
Kim, J. J., Kim, Y. S., and Kumar, V. (2019a). Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem. Med. Bio. 54, 226–231. doi:10.1016/j.jtemb.2019.05.003
Kim, W., Kim, D., Back, S., Lee, Y. S., Abari, A. H., and Kim, J. (2019b). Removal of Ni2+ and Cd2+ by surface display of polyhistidine on Bacillus subtilis spore using CotE anchor protein. Biotechnol. Bioprocess Eng. 24, 375–381. doi:10.1007/s12257-018-0467-2
Kobras, C. M., Fenton, A. K., and Sheppard, S. K. (2021). Next-generation microbiology: From comparative genomics to gene function. Genome Biol. 22, 123. doi:10.1186/s13059-021-02344-9
Konno, N., and Iwasaki, W. (2023). Machine learning enables prediction of metabolic system evolution in bacteria. Sci. Adv. 9, eadc9130. doi:10.1126/sciadv.adc9130
Kostal, J., Yang, R., Wu, C. H., Mulchandani, A., and Chen, W. (2004). Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Appl. Environ. Microbiol. 70, 4582–4587. doi:10.1128/AEM.70.8.4582-4587.2004
Kumar, A., Kumar, A., Singh, R., Singh, R., Pandey, S., Rai, A., et al. (2020). “Genetically engineered bacteria for the degradation of dye and other organic compounds,” in Abatement of environmental pollutants: Trends and strategies (UK: Elsevier), 331–350.
Leitch, S., Bradley, M. J., Rowe, J. L., Chivers, P. T., and Maroney, M. J. (2007). Nickel-specific response in the transcriptional regulator, Escherichia coli NikR. J. Am. Chem. Soc. 129, 5085–5095. doi:10.1021/ja068505y
Leonhartsberger, S., Huber, A., Lottspeich, F., and Böck, A. (2001). The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J. Mol. Biol. 307, 93–105. doi:10.1006/jmbi.2000.4451
Li, H., Zhang, C., Chen, X., You, H., and Lai, L. (2022a). Tailoring Escherichia coli chemotactic sensing towards cadmium by computational redesign of ribose-binding protein. mSystems 7, e0108421. doi:10.1128/msystems.01084-21
Li, J., Cui, M., Zhao, J., Wang, J., and Fang, X. (2023). A self-amplifying plasmid based ultrasensitive biosensor for the detection of As(Ⅲ) in water. Biosens. Bioelectron. 221, 114937. doi:10.1016/j.bios.2022.114937
Li, X., Ren, Z., Crabbe, M. J. C., Wang, L., and Ma, W. (2021). Genetic modifications of metallothionein enhance the tolerance and bioaccumulation of heavy metals in Escherichia coli. Ecotoxicol. Environ. Saf. 222, 112512. doi:10.1016/j.ecoenv.2021.112512
Li, Y., Guo, S., Zheng, Y., Yu, J., Chi, R., and Xiao, C. (2022b). Bioimmobilization of lead in phosphate mining wasteland by isolated strain Citrobacter farmeri CFI-01. Environ. Pollut. 307, 119485. doi:10.1016/j.envpol.2022.119485
Liang, H., Deng, X., Bosscher, M., Ji, Q., Jensen, M. P., and He, C. (2013). Engineering bacterial two-component system PmrA/PmrB to sense lanthanide ions. J. Am. Chem. Soc. 135, 2037–2039. doi:10.1021/ja312032c
Lima, A. I. G., Corticeiro, S. C., and Figueira, E. M. D. P. (2006). Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzyme Microb. Tech. 39, 763–769. doi:10.1016/j.enzmictec.2005.12.009
Lin, P., Yuan, H., Du, J., Liu, K., Liu, H., and Wang, T. (2020). Progress in research and application development of surface display technology using Bacillus subtilis spores. Appl. Microbiol. Biotechnol. 104, 2319–2331. doi:10.1007/s00253-020-10348-x
Liu, C. J., Yu, H., Zhang, B. C., Liu, S. L., Liu, C. G., Li, F., et al. (2022a). Engineering whole-cell microbial biosensors: Design principles and applications in monitoring and treatment of heavy metals and organic pollutants. Biotechnol. Adv. 60, 108019. doi:10.1016/j.biotechadv.2022.108019
Liu, H. Z., Zhang, Y., Wang, Y. S., Xie, X. B., and Shi, Q. S. (2021). The connection between Czc and Cad systems involved in cadmium resistance in Pseudomonas putida. Int. J. Mol. Sci. 22, 9697. doi:10.3390/ijms22189697
Liu, M. R., Kakade, A., Liu, P., Wang, P., Tang, Y., and Li, X. K. (2019). Hg2+-binding peptide decreases mercury ion accumulation in fish through a cell surface display system. Sci. Total. Environ. 659, 540–547. doi:10.1016/j.scitotenv.2018.12.406
Liu, M. R., Li, Z. N., Chen, Z. J., Qi, X. E., Yang, L., and Chen, G. (2022b). Simultaneous biodetection and bioremediation of Cu2+ from industrial wastewater by bacterial cell surface display system. Int. Biodeter. Biodegr. 173, 105467. doi:10.1016/j.ibiod.2022.105467
Lovley, D. R., and Phillips, E. J. (1994). Reduction of chromate by Desulfovibrio vulgaris and its c(3) cytochrome. Appl. Environ. Microbiol. 60, 726–728. doi:10.1128/aem.60.2.726-728.1994
Lukowski, A., and Dec, D. (2018). Influence of Zn, Cd, and Cu fractions on enzymatic activity of arable soils. Environ. Monit. Assess. 190, 278. doi:10.1007/s10661-018-6651-1
Luo, L., Wang, B., Jiang, J. W., Fitzgerald, M., Huang, Q., Yu, Z., et al. (2021). Heavy metal contaminations in herbal medicines: Determination, comprehensive risk assessments, and solutions. Front. Pharmacol. 11, 595335. doi:10.3389/fphar.2020.595335
Ma, W., Li, X., Wang, Q., Ren, Z., Crabbe, M. J. C., and Wang, L. (2019). Tandem oligomeric expression of metallothionein enhance heavy metal tolerance and bioaccumulation in Escherichia coli. Ecotoxicol. Environ. Saf. 181, 301–307. doi:10.1016/j.ecoenv.2019.06.022
Ma, Y., Lin, J., Zhang, C., Ren, Y., and Lin, J. (2011). Cd(II) and As(III) bioaccumulation by recombinant Escherichia coli expressing oligomeric human metallothioneins. J. Hazard. Mater. 185, 1605–1608. doi:10.1016/j.jhazmat.2010.10.051
Malik, A. (2004). Metal bioremediation through growing cells. Environ. Int. 30, 261–278. doi:10.1016/j.envint.2003.08.001
Matta, G., and Gjyli, L. (2016). Mercury, lead and arsenic: Impact on environment and human health. J. Chem. Pharm. Sci. 9, 718–725.
Mimee, M., Nadeau, P., Hayward, A., Carim, S., Flanagan, S., Jerger, L., et al. (2018). An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918. doi:10.1126/science.aas9315
Miri, S., Saini, R., Davoodi, S. M., Pulicharla, R., Brar, S. K., and Magdouli, S. (2022). Biodegradation of microplastics: Better late than never. Chemosphere 286, 131670. doi:10.1016/j.chemosphere.2021.131670
Misra, T. K. (1992). Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid 27, 4–16. doi:10.1016/0147-619x(92)90002-R
Mitra, S., Chakraborty, A. J., Tareq, A., Bin Emran, T., Nainu, F., Khusro, A., et al. (2022). Impact of heavy metals on the environment and human health: Novel therapeutic insights to counter the toxicity. J. King Saud. Univ. Sci. 34, 101865. doi:10.1016/j.jksus.2022.101865
Moe-Behrens, G. H., Davis, R., and Haynes, K. A. (2013). Preparing synthetic biology for the world. Front. Microbiol. 4, 5. doi:10.3389/fmicb.2013.00005
Mukherjee, R., Bhunia, P., and De, S. (2016). Impact of graphene oxide on removal of heavy metals using mixed matrix membrane. Chem. Eng. J. 292, 284–297. doi:10.1016/j.cej.2016.02.015
Na, D., Yoo, S. M., Chung, H., Park, H., Park, J. H., and Lee, S. Y. (2013). Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174. doi:10.1038/nbt.2461
Nagajyoti, P. C., Lee, K. D., and Sreekanth, T. V. M. (2010). Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 8, 199–216. doi:10.1007/s10311-010-0297-8
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi:10.1016/S0168-6445(03)00048-2
NIH (2019). NIH guidelines for research involving recombinant or synthetic nucleic acid molecules. [Online]. US: National Institute of Health, Office of Science Policy. Available: https://osp.od.nih.gov/wp-content/uploads/2019_NIH_Guidelines.htm (Accessed January 14th, 2023).
Nistala, G. J., Wu, K., Rao, C. V., and Bhalerao, K. D. (2010). A modular positive feedback-based gene amplifier. J. Biol. Eng. 4, 4. doi:10.1186/1754-1611-4-4
Nyerges, A., Csorgo, B., Nagy, I., Balint, B., Bihari, P., Lazar, V., et al. (2016). A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. P. Natl. Acad. Sci. U. S. A. 113, 2502–2507. doi:10.1073/pnas.1520040113
Osman, D., and Cavet, J. S. (2010). Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27, 668–680. doi:10.1039/b906682a
Pan, J. L., Plant, J. A., Voulvoulis, N., Oates, C. J., and Ihlenfeld, C. (2010). Cadmium levels in Europe: Implications for human health. Environ. Geochem. Health 32, 1–12. doi:10.1007/s10653-009-9273-2
Park, D. M., Overton, K. W., and Jiao, Y. Q. (2019). The UzcRS two-component system in Caulobacter crescentus integrates regulatory input from diverse auxiliary regulators. Mol. Microbiol. 111, 678–699. doi:10.1111/mmi.14180
Park, D. M., and Taffet, M. J. (2019). Combinatorial sensor design in Caulobacter crescentus for selective environmental uranium detection. ACS Synth. Biol. 8, 807–817. doi:10.1021/acssynbio.8b00484
Paul, D., Pandey, G., and Jain, R. K. (2005). Suicidal genetically engineered microorganisms for bioremediation: Need and perspectives. Bioessays 27, 563–573. doi:10.1002/bies.20220
Pham, V. D., Ravikumar, S., Lee, S. H., Hong, S. H., and Yoo, I.-K. (2013). Modification of response behavior of zinc sensing HydHG two-component system using a self-activation loop and genomic integration. Bioprocess. Biosyst. Eng. 36, 1185–1190. doi:10.1007/s00449-012-0845-7
Prakash, S., and Verma, A. K. (2021). Arsenic: It's toxicity and impact on human health. Int. J. Biol. Innov. 3, 38–47. doi:10.46505/IJBI.2021.3102
Qasem, N. a. A., Mohammed, R. H., and Lawal, D. U. (2021). Author correction: Removal of heavy metal ions from wastewater: A comprehensive and critical review. npj Clean Water 4, 52. doi:10.1038/s41545-021-00144-z
Rademacher, C., and Masepohl, B. (2012). Copper-responsive gene regulation in bacteria. Microbiology 158, 2451–2464. doi:10.1099/mic.0.058487-0
Rahman, K., Menon, U., V, S., and J, R. (2022). Removal of cadmium by heavy metal–resistant bacteria isolated from Hussain Sagar Lake—Hyderabad. Biomass Convers. Biorefinery 12, 1703–1713. doi:10.1007/s13399-021-01354-8
Ravikumar, S., Pham, V. D., Lee, S. H., Yoo, I.-K., and Hong, S. H. (2012). Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop. J. Ind. Microbiol. Biotechnol. 39, 861–868. doi:10.1007/s10295-012-1096-y
Ravikumar, S., Yoo, I.-K., Lee, S. Y., and Hong, S. H. (2011). A study on the dynamics of the zraP gene expression profile and its application to the construction of zinc adsorption bacteria. Bioprocess. Biosyst. Eng. 34, 1119–1126. doi:10.1007/s00449-011-0562-7
Ren, J., Lee, J., and Na, D. (2020). Recent advances in genetic engineering tools based on synthetic biology. J. Microbiol. 58, 1–10. doi:10.1007/s12275-020-9334-x
Ronchel, M. C., and Ramos, J. L. (2001). Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl. Environ. Microbiol. 67, 2649–2656. doi:10.1128/AEM.67.6.2649-2656.2001
Salam, M., Varma, A., Chaudhary, D., and Aggarwal, H. (2020). Novel arsenic resistant bacterium Sporosarcina luteola M10 having potential bioremediation properties. J. Microbiol. Exp. 8, 213–218. doi:10.15406/jmen.2020.08.00311
Samuelson, P., Gunneriusson, E., Nygren, P. A., and Stahl, S. (2002). Display of proteins on bacteria. J. Biotechnol. 96, 129–154. doi:10.1016/s0168-1656(02)00043-3
Schmoger, M. E., Oven, M., and Grill, E. (2000). Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 122, 793–801. doi:10.1104/pp.122.3.793
Selvi, A., Rajasekar, A., Theerthagiri, J., Ananthaselvam, A., Sathishkumar, K., Madhavan, J., et al. (2019). Integrated remediation processes toward heavy metal removal/recovery from various environments-A review. Front. Environ. Sci. 7, 66. doi:10.3389/fenvs.2019.00066
Shaker, B., Ahmad, S., Lee, J., Jung, C. J., and Na, D. (2021). In silico methods and tools for drug discovery. Comput. Biol. Med. 137, 104851. doi:10.1016/j.compbiomed.2021.104851
Shao, M. M., and Zhu, Y. (2020). Long-term metal exposure changes gut microbiota of residents surrounding a mining and smelting area. Sci. Rep. 10, 4453. doi:10.1038/s41598-020-61143-7
Sharma, P. K., Balkwill, D. L., Frenkel, A., and Vairavamurthy, M. A. (2000). A new Klebsiella planticola strain (Cd-1) grows anaerobically at high cadmium concentrations and precipitates cadmium sulfide. Appl. Environ. Microbiol. 66, 3083–3087. doi:10.1128/AEM.66.7.3083-3087.2000
Shinar, G., Milo, R., Martínez, M. R., and Alon, U. (2007). Input–output robustness in simple bacterial signaling systems. Proc. Natl. Acad. Sci. U. S. A. 104, 19931–19935. doi:10.1073/pnas.0706792104
Singh, J. S., Abhilash, P. C., Singh, H. B., Singh, R. P., and Singh, D. P. (2011). Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene 480, 1–9. doi:10.1016/j.gene.2011.03.001
Singh, S., Kang, S. H., Lee, W., Mulchandani, A., and Chen, W. (2010). Systematic engineering of phytochelatin synthesis and arsenic transport for enhanced arsenic accumulation in E. coli. Biotechnol. Bioeng. 105, 780–785. doi:10.1002/bit.22585
Smith, K., and Novick, R. P. (1972). Genetic studies on plasmid-linked cadmium resistance in Staphylococcus aureus. J. Bacteriol. 112, 761–772. doi:10.1128/jb.112.2.761-772.1972
Sobecky, P. A., and Coombs, J. M. (2009). Horizontal gene transfer in metal and radionuclide contaminated soils. Methods Mol. Biol. 532, 455–472. doi:10.1007/978-1-60327-853-9_26
Solioz, M., and Odermatt, A. (1995). Copper and silver transport by CopB-ATPase in membrane-vesicles of Enterococcus hirae. J. Biol. Chem. 270, 9217–9221. doi:10.1074/jbc.270.16.9217
Solioz, M., and Stoyanov, J. V. (2003). Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27, 183–195. doi:10.1016/S0168-6445(03)00053-6
Soto, S. M. (2013). Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4, 223–229. doi:10.4161/viru.23724
Srivastava, N. K., and Majumder, C. B. (2008). Novel biofiltration methods for the treatment of heavy metals from industrial wastewater. J. Hazard. Mater. 151, 1–8. doi:10.1016/j.jhazmat.2007.09.101
Suhani, I., Sahab, S., Srivastava, V., and Singh, R. P. (2021). Impact of cadmium pollution on food safety and human health. Curr. Opin. Toxicol. 27, 1–7. doi:10.1016/j.cotox.2021.04.004
Sundseth, K., Pacyna, J. M., Pacyna, E. G., Pirrone, N., and Thorne, R. J. (2017). Global sources and pathways of mercury in the context of human health. Int. J. Environ. Res. Public Health 14, 105. doi:10.3390/ijerph14010105
Taghavi, S., Lesaulnier, C., Monchy, S., Wattiez, R., Mergeay, M., and Van Der Lelie, D. (2009). Lead(II) resistance in Cupriavidus metallidurans CH34: Interplay between plasmid and chromosomally-located functions. Ant. Van Leeuwenhoek 96, 171–182. doi:10.1007/s10482-008-9289-0
Tang, C. J., Chen, X., Feng, F., Liu, Z. G., Song, Y. X., Wang, Y. Y., et al. (2021). Roles of bacterial cell and extracellular polymeric substance on adsorption of Cu(II) in activated sludges: A comparative study. J. Water Process Eng. 41, 102094. doi:10.1016/j.jwpe.2021.102094
Taniguchi, J., Hemmi, H., Tanahashi, K., Amano, N., Nakayama, T., and Nishino, T. (2000). Zinc biosorption by a zinc-resistant bacterium, Brevibacterium sp. strain HZM-1. Appl. Microbiol. Biotechnol. 54, 581–588. doi:10.1007/s002530000415
Tay, P. K. R., Nguyen, P. Q., and Joshi, N. S. (2017). A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synth. Biol. 6, 1841–1850. doi:10.1021/acssynbio.7b00137
Thatoi, H., Das, S., Mishra, J., Rath, B. P., and Das, N. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J. Environ. Manage 146, 383–399. doi:10.1016/j.jenvman.2014.07.014
Tran, K. M., Lee, H. M., Thai, T. D., Shen, J., Eyun, S. I., and Na, D. (2021). Synthetically engineered microbial scavengers for enhanced bioremediation. J. Hazard. Mater. 419, 126516. doi:10.1016/j.jhazmat.2021.126516
Ulrich, L. E., Koonin, E. V., and Zhulin, I. B. (2005). One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13, 52–56. doi:10.1016/j.tim.2004.12.006
Valls, M., and De Lorenzo, V. (2002). Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 26, 327–338. doi:10.1016/s0168-6445(02)00114-6
Velusamy, K., Periyasamy, S., Kumar, P. S., Rangasamy, G., Pauline, J. M. N., Ramaraju, P., et al. (2022). Biosensor for heavy metals detection in wastewater: A review. Food Chem. Toxicol. 168, 113307. doi:10.1016/j.fct.2022.113307
Verma, B., Balomajumder, C., Sabapathy, M., and Gumfekar, S. P. (2021). Pressure-driven membrane process: A review of advanced technique for heavy metals remediation. Processes 9, 752. doi:10.3390/pr9050752
Volesky, B., and Holan, Z. R. (1995). Biosorption of heavy-metals. Biotechnol. Progr. 11, 235–250. doi:10.1021/bp00033a001
Wang, B., Barahona, M., and Buck, M. (2013). A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron. 40, 368–376. doi:10.1016/j.bios.2012.08.011
Wang, B. J., and Buck, M. (2012). Customizing cell signaling using engineered genetic logic circuits. Trends Microbiol. 20, 376–384. doi:10.1016/j.tim.2012.05.001
Wang, C. L., Ozuna, S. C., Clark, D. S., and Keasling, J. D. (2002). A deep-sea hydrothermal vent isolate, Pseudomonas aeruginosa CW961, requires thiosulfate for Cd2+ tolerance and precipitation. Biotechnol. Lett. 24, 637–641. doi:10.1023/A:1015043324584
Wang, D., Zheng, Y. N., Fan, X. S., Xu, L. N., Pang, T., Liu, T., et al. (2020). Visual detection of Hg2+ by manipulation of pyocyanin biosynthesis through the Hg2+-dependent transcriptional activator MerR in microbial cells. J. Biosci. Bioeng. 129, 223–228. doi:10.1016/j.jbiosc.2019.08.005
Wang, F., and Zhang, W. (2019). Synthetic biology: Recent progress, biosafety and biosecurity concerns, and possible solutions. Int. J. Biosaf. Biosecurity 1, 22–30. doi:10.1016/j.jobb.2018.12.003
Wang, W., Jiang, F., Wu, F., Li, J., Ge, R., Li, J., et al. (2019). Biodetection and bioremediation of copper ions in environmental water samples using a temperature-controlled, dual-functional Escherichia coli cell. Appl. Microbiol. Biotechnol. 103, 6797–6807. doi:10.1007/s00253-019-09984-9
Wei, W., Liu, X. Z., Sun, P. Q., Wang, X., Zhu, H., Hong, M., et al. (2014). Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ. Sci. Technol. 48, 3363–3371. doi:10.1021/es4046567
Weiss, R., Homsy, G. E., and Knight, T. F. (2002). “Toward in vivo digital circuits,” in Evolution as computation. Editors L. F. Landweber, and E. Winfree (Berlin, Heidelberg: Springer), 275–295.
Wernerus, H., Lehtio, J., Teeri, T., Nygren, P. A., and Stahl, S. (2001). Generation of metal-binding Staphylococci through surface display of combinatorially engineered cellulose-binding domains. Appl. Environ. Microbiol. 67, 4678–4684. doi:10.1128/Aem.67.10.4678-4684.2001
Wernerus, H., and Stahl, S. (2004). Biotechnological applications for surface-engineered bacteria. Biotechnol. Appl. Biochem. 40, 209–228. doi:10.1042/BA20040014
Wesseler, J., Kleter, G., Meulenbroek, M., and Purnhagen, K. P. (2022). EU regulation of genetically modified microorganisms in light of new policy developments: Possible implications for EU bioeconomy investments. Appl. Econ. Perspect. Policy, 1–21. doi:10.1002/aepp.13259
Xiang, L., Li, G. Q., Wen, L., Su, C., Liu, Y., Tang, H. Z., et al. (2021). Biodegradation of aromatic pollutants meets synthetic biology. Synth. Syst. Biotechnol. 6, 153–162. doi:10.1016/j.synbio.2021.06.001
Xue, Y. B., Du, P., Shendi, A. a. I., and Yu, B. (2022a). Mercury bioremediation in aquatic environment by genetically modified bacteria with self-controlled biosecurity circuit. J. Clean. Prod. 337, 130524. doi:10.1016/j.jclepro.2022.130524
Xue, Y., Qiu, T., Sun, Z., Liu, F., and Yu, B. (2022b). Mercury bioremediation by engineered Pseudomonas putida KT2440 with adaptationally optimized biosecurity circuit. Environ. Microbiol. 24, 3022–3036. doi:10.1111/1462-2920.16038
Yagura, M., Nishio, S. Y., Kurozumi, H., Wang, C. F., and Itoh, T. (2006). Anatomy of the replication origin of plasmid ColE2-P9. J. Bacteriol. 188, 999–1010. doi:10.1128/JB.188.3.999-1010.2006
Yang, H. C., Fu, H. L., Lin, Y. F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi:10.1016/B978-0-12-394390-3.00012-4
Yang, T., Chen, M.-L., and Wang, J.-H. (2015). Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological or environmental significance. Trac. Trends Anal. Chem. 66, 90–102. doi:10.1016/j.trac.2014.11.016
Yang, T., Liu, J. W., Gu, C., Chen, M. L., and Wang, J. H. (2013). Expression of arsenic regulatory protein in Escherichia coli for selective accumulation of methylated arsenic species. ACS Appl. Mater. Interfaces 5, 2767–2772. doi:10.1021/am400578y
Yin, K., Lv, M., Wang, Q. N., Wu, Y. X., Liao, C. Y., Zhang, W. W., et al. (2016). Simultaneous bioremediation and biodetection of mercury ion through surface display of carboxylesterase E2 from Pseudomonas aeruginosa PA1. Water Res. 103, 383–390. doi:10.1016/j.watres.2016.07.053
Yoon, Y., Kang, Y., Chae, Y., Kim, S., Lee, Y., Jeong, S. W., et al. (2016). Arsenic bioavailability in soils before and after soil washing: The use of Escherichia coli whole-cell bioreporters. Environ. Sci. Pollut. Res. 23, 2353–2361. doi:10.1007/s11356-015-5457-8
Yu, L., Cao, Q., Chen, W., Yang, N., Yang, C.-G., Ji, Q., et al. (2022). A novel copper-sensing two-component system for inducing Dsb gene expression in bacteria. Sci. Bull. 67, 198–212. doi:10.1016/j.scib.2021.03.003
Zackular, J. P., Moore, J. L., Jordan, A. T., Juttukonda, L. J., Noto, M. J., Nicholson, M. R., et al. (2016). Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat. Med. 22, 1330–1334. doi:10.1038/nm.4174
Zeng, W., Zhang, S., Xia, M., Wu, X., Qiu, G., and Shen, L. (2020). Insights into the production of extracellular polymeric substances of Cupriavidus pauculus 1490 under the stimulation of heavy metal ions. RSC Adv. 10, 20385–20394. doi:10.1039/C9RA10560C
Zhai, Q. X., Cen, S., Jiang, J. C., Zhao, J. X., Zhang, H., and Chen, W. (2019). Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: A pilot study of Chinese children. Environ. Res. 171, 501–509. doi:10.1016/j.envres.2019.01.060
Zhang, G. B., Hu, S. T., and Jia, X. Q. (2021). Highly sensitive whole-cell biosensor for cadmium detection based on a negative feedback circuit. Front. Bioeng. Biotechnol. 9, 799781. doi:10.3389/fbioe.2021.799781
Zhou, T., Li, R., Zhang, S., Zhao, S., Sharma, M., Kulshrestha, S., et al. (2021). A copper-specific microbial fuel cell biosensor based on riboflavin biosynthesis of engineered Escherichia coli. Biotechnol. Bioeng. 118, 210–222. doi:10.1002/bit.27563
Keywords: heavy metals, synthetic biology, bioremediation, whole-cell biosensor, genetically engineered microorganisms
Citation: Thai TD, Lim W and Na D (2023) Synthetic bacteria for the detection and bioremediation of heavy metals. Front. Bioeng. Biotechnol. 11:1178680. doi: 10.3389/fbioe.2023.1178680
Received: 03 March 2023; Accepted: 04 April 2023;
Published: 13 April 2023.
Edited by:
Xuehua Wan, Nankai University, ChinaReviewed by:
Gonzalo Durante-Rodriguez, Margarita Salas Center for Biological Research, Spanish National Research Council (CSIC), SpainChang-ye Hui, Shenzhen Prevention and Treatment Center for Occupational Diseases, China
Copyright © 2023 Thai, Lim and Na. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dokyun Na, Ymxpc3N6ZW5AY2F1LmFjLmty
†These authors have contributed equally to this work
 Thi Duc Thai
Thi Duc Thai Wonseop Lim
Wonseop Lim Dokyun Na
Dokyun Na