- Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology, New Delhi, India
The expression of recombinant proteins by the AOX1 promoter of Komagataella phaffii is typically induced by adding methanol to the cultivation medium. Since growth on methanol imposes a high oxygen demand, the medium is often supplemented with an additional secondary carbon source which serves to reduce the consumption of methanol, and hence, oxygen. Early research recommended the use of glycerol as the secondary carbon source, but more recent studies recommend the use of sorbitol because glycerol represses PAOX1 expression. To assess the validity of this recommendation, we measured the steady state concentrations of biomass, residual methanol, and LacZ expressed from PAOX1 over a wide range of dilution rates (0.02–0.20 h−1) in continuous cultures of the Mut+ strain fed with methanol + glycerol (repressing) and methanol + sorbitol (non-repressing). We find that under these conditions, the specific PAOX1 expression rate (measured as either specific LacZ productivity or specific AOX productivity) is completely determined by the specific methanol consumption rate regardless of the type (repressing/non-repressing) of the secondary carbon source. In both cultures, the specific PAOX1 expression rate is proportional to the specific methanol consumption rate, provided that the latter is below 0.15 g/(gdw-h); beyond this threshold consumption rate, the specific PAOX1 expression rate of both cultures saturates to the same value. Analysis of the data in the literature shows that the same phenomenon also occurs in continuous cultures of Escherichia coli fed with mixtures of lactose plus repressing/non-repressing carbon sources. The specific Plac expression rate is completely determined by the specific lactose consumption rate, regardless of the type of secondary carbon source, glycerol or glucose.
Introduction
The methylotrophic yeast Komagataella phaffii, referred to earlier as Pichia pastoris (Kurtzman, 2005; Kurtzman, 2009), is a popular expression host (Schwarzhans et al., 2016; Rahimi et al., 2019a; Ergün et al., 2021). There are several reasons for this, but the most important one is that K. phaffii has an unusually strong and tightly regulated promoter which drives the expression of alcohol oxidase (AOX) in the presence of methanol (Higgins and Cregg, 1998; Ahmad et al., 2014; Gasser and Mattanovich, 2018). To be sure, K. phaffii has two alcohol oxidase genes, AOX1 and AOX2, with corresponding promoters, PAOX1 and PAOX2, but PAOX1 is used to drive recombinant protein expression since it is ∼10 times stronger than PAOX2 (Cregg et al., 1989).
In the first expression system constructed with K. phaffii, the wild-type strain was used as host, and recombinant protein was expressed under the control of PAOX1 by using methanol as inducer (Cregg et al., 1985). Although this Mut+ (methanol utilization plus) strain yielded excellent recombinant protein expression, the use of methanol as inducer led to several operational problems (Macauley-Patrick et al., 2005; Cos et al., 2006; Jahic et al., 2006; Jungo et al., 2007a; Arnau et al., 2011; Potvin et al., 2012; Yang and Zhang, 2018; García-Ortega et al., 2019; Liu et al., 2019). Indeed, methanol is inflammable which poses safety issues (Liu et al., 2022). Moreover, methanol metabolism results in high oxygen demand and heat generation, as well as excretion of toxic metabolites, such as formaldehyde, that inhibit growth (Jungo et al., 2007b; Juturu and Wu, 2018; Rahimi et al., 2019b).
The problems stemming from the use of methanol as inducer led to several strategies for reducing methanol consumption (Feng et al., 2022). One strategy was to engineer the host strain by deleting either AOX1 or both AOX1 and AOX2, thus producing the Muts (methanol utilization slow) and Mut− (methanol utilization minus) strains, respectively, whose capacity to consume methanol is substantially impaired or abolished (Chiruvolu et al., 1997). Another strategy was to introduce into the medium, in addition to the primary or inducing carbon source methanol, a secondary or non-inducing carbon source that supports growth but not induction (Ergün et al., 2021). This reduces methanol consumption due to the sparing effect of the secondary carbon source, and increases the volumetric productivity due to the enhanced cell growth derived from metabolism of the secondary carbon source (Brierley et al., 1990; Egli and Mason, 1991; Jungo et al., 2007a; Jungo et al., 2007b; Paulova et al., 2012).
The foregoing strategies have led to reduced methanol consumption, but they can also result in decreased recombinant protein expression. Recently, we found that host strain engineering decreases recombinant protein expression substantially—the specific productivities of the engineered Muts and Mut− strains are respectively 5- and 10-fold lower than that of the Mut+ strain (Singh and Narang, 2020). Since these three strains differ only with respect to their capacity for methanol consumption, the methanol consumption rate is an important determinant of the PAOX1 expression rate.
The goal of this work is to quantify the extent to which PAOX1 expression is affected by addition of a secondary carbon source to the medium. It is commonly held that this is determined by the type of the secondary carbon source. Specifically, these carbon sources have been classified as repressing or non-repressing based on the PAOX1 expression levels observed in batch cultures of the Mut− strain grown on mixtures of methanol and various secondary carbon sources (Inan and Meagher, 2001). Repressing carbon sources, such as glycerol, abolish PAOX1 expression, whereas non-repressing carbon sources, such as sorbitol, permit PAOX1 expression. The same conclusion has been reached from studies of mixed-substrate growth in fed-batch cultures (Brierley et al., 1990; Thorpe et al., 1999; Xie et al., 2005; Çelik et al., 2009; Wang et al., 2010; Gao et al., 2012; Niu et al., 2013; Carly et al., 2016; Azadi et al., 2017; Chen et al., 2017) and continuous cultures (Jungo et al., 2006; Jungo et al., 2007a; Jungo et al., 2007b; Canales et al., 2015; Berrios et al., 2017). Indeed, even though glycerol is commonly used as the secondary carbon source, the use of sorbitol has been almost unanimously recommended on the grounds that glycerol represses PAOX1 expression.
Most of the comparative studies cited above used constant fed-batch cultures, but these data can be difficult to interpret physiologically, because the specific growth rate decreases throughout the course of the experiment (Nieto-Taype et al., 2020). The comparative studies with continuous cultures are reviewed at length in the Discussion. Here, it suffices to note that many of these studies were performed at a fixed dilution rate
We were therefore led to study PAOX1 expression in continuous cultures of K. phaffii operated at various dilution rates with fixed concentrations of methanol + glycerol and methanol + sorbitol. To this end, we used a Mut+ strain expressing LacZ from PAOX1, but we also measured the AOX level to check the consistency of the data. We find that the specific PAOX1 expression rate (measured as either specific LacZ productivity or specific AOX productivity) is completely determined by the specific methanol consumption rate, regardless of the type (repressing/non-repressing) of the secondary carbon source.
Materials and methods
Microorganism and growth medium
A K. phaffii Mut+ strain, GS115 (his4), was procured from J. M. Cregg, Keck Graduate Institute, Claremont, CA, United States and was genetically modified to express a recombinant β-galactosidase protein. Details of the strain construction have been presented elsewhere (Singh and Narang, 2020). The resulting strain was called Mut+ (pSAOH5-T1) and was used for this study. Stock cultures were stored in 25% glycerol at −80°C.
The minimal medium composition used for shake-flask as well as chemostat cultivations was chosen such as to ensure stoichiometric limitation of the carbon and energy sources, as described in Egli and Fiechter (1981). The defined medium was supplemented with either glycerol (∼3.1 g L−1), a mixture of methanol (∼1.6 g L−1) and glycerol (∼1.5 g L−1) or a mixture of methanol (∼3.2 g L−1) and sorbitol (∼1.5 g L−1) as carbon sources. In addition, the medium contained 100 mM phosphate buffer (pH 5.5), 15.26 g NH4Cl, 1.18 g MgSO4⋅7H2O, 110 mg CaCl2⋅2H2O, 45.61 mg FeCl3, 28 mg MnSO4⋅H2O, 44 mg ZnSO4⋅7H2O, 8 mg CuSO4⋅5H2O, 8.57 mg CoCl2⋅6H2O, 6 mg Na2MoO4⋅2H2O, 8 mg H3BO3, 1.2 mg KI, 370 mg EDTA disodium salt, 2.4 mg biotin per liter. All components of the defined medium were prepared and sterilised by either filtration or autoclaving as separate stock solutions and then mixed before cultivation.
Inoculum preparation and chemostat cultivation
When required, cells were revived in a 100 ml shake flask containing 10 ml minimal medium supplemented with a suitable carbon source at 30°C and 200 rpm. These primary cultures were sub-cultured once before inoculating the reactor precultures (in the same cultivation medium as prepared for the reactor vessel), which were then used as an inoculum for the bioreactor.
Chemostat cultivations were performed using bench-scale 0.5 L mini bioreactors modified to support chemostat operation and equipped with pH, DO, temperature, level and agitation controls (Applikon Biotechnology, Netherlands) at working volumes of 0.3 L. The cultivation temperature was always maintained at 30°C and pH at 5.5 by the automatic addition of 2 M NaOH. An integrated mass flow controller ensured a constant supply of air to the reactor vessel at 80 ml min−1. Dissolved oxygen levels were monitored by a polarographic probe calibrated with respect to an air-saturated medium. Cultures were agitated to ensure fast mixing as well as aerobic conditions, such that the DO level always remained above 60%. A silicone based anti-foam agent was added to the reactor vessel as and when required to prevent foam formation and wall growth. For chemostat mode operation, the dilution rate was set by fixing the input feed flow rate, while a constant volume was maintained inside the reactor vessel by controlling the output feed flow rate via proportional control based on the on-line monitoring of the change in weight of the reactor vessel. For instance, for a dilution rate of 0.1 h−1, the input feed flow rate was fixed at 30 ml h−1 using a peristaltic pump. When the weight of the reactor vessel increased beyond the set point, the output feed pump was switched on to remove the excess volume. After inoculation, cells were grown in batch phase for some time to allow exhaustion of the initial carbon source (indicated by a rise in DO level), followed by initiating the input and output feed supplies. At any particular dilution rate, steady-state samples were withdrawn after 5-6 liquid residence times. In general, three samples were collected for each dilution rate, separated by an interval of one liquid residence time. For instance, at a dilution rate of 0.04 h−1, the first sample was taken after 150 h (6 liquid residence time), the second after 175 h (7 liquid residence time) and the third after 200 h (8 liquid residence time). Attainment of steady-state was confirmed by analysing the samples for constant dry cell weight and specific enzyme activities.
Sample collection and processing
For determination of residual substrate concentration inside the reactor, samples were withdrawn directly from the vessel. To achieve rapid biomass separation, culture samples were withdrawn using vacuum through a sampling tube attached to a 0.2-micron syringe filter and stored at −20°C until analysis. Samples for determination of biomass and enzyme activities were collected in a sampling bottle kept on ice. Biomass samples were processed immediately, while samples for measuring enzyme activities were pelleted, washed and stored at −20°C until processing.
Substrate analysis
Glycerol and sorbitol concentrations were estimated by high-performance liquid chromatography (HPLC) analysis (1100 series, Agilent Technologies, Palo Alto, United States) with detection limits of ∼1 mg/L and ∼30 mg/L. An ion-exclusion chromatography column from Phenomenex, California, United States (ROA-Organic acid H+ column, 300 × 7.8 mm, 8 µm particle size, 8% cross linkage) with a guard column (Carbo-H cartridges) was used with 5 mM H2SO4 in ultrapure water as mobile phase supplied at a constant flow rate of 0.5 ml min−1. The column chamber was maintained at 60°C and a refractive index detector was used for substrate measurement. Methanol concentrations were determined with a gas chromatograph equipped with a flame ionisation detector (GC-FID) (7890A, Agilent Technologies, Palo Alto, United States) using a HP-PLOT/Q column (30 m × 0.32 mm, 20 µm) from Agilent Technologies and nitrogen as the carrier gas. The detection limit for methanol was ∼5 mg/L.
Dry cell weight measurement
A known volume of the fermentation broth was collected and pelleted in a pre-weighed centrifuge tube. Pellets were washed twice with distilled water and then dried at 80°C to constant weight.
Cell-free extract preparation
Culture samples were collected on ice and immediately centrifuged at 4°C to collect cells. The cell pellets were washed twice with phosphate buffer (100 mM, pH 7.4) and stored at −20°C until analysis. For cell lysis, pellets were resuspended in 100 µL of chilled breaking buffer (Jungo et al., 2006). Acid-washed glass beads (0.40–0.45 mm diameter) were added to the resulting slurry followed by alternate vortexing (1 min) and resting (on ice for 1 min) steps. This cycle was repeated 4–5 times, after which the cell debris was removed by centrifugation. Cell-free extracts (supernatant) were collected in fresh tubes kept on ice and immediately used for the estimation of enzyme activities. The Bradford assay was used for the estimation of the total protein content of the cell-free extracts for which bovine serum albumin served as standard (Bradford, 1976).
β-galactosidase assay
β-galactosidase assays were performed according to the method described by Miller (1972) with modifications. Briefly, cell-free extracts were appropriately diluted and mixed with Z-buffer containing β-mercaptoethanol (Miller, 1972) and incubated at 30°C in a water-bath for 15–20 min. The reaction was started by adding ONPG and stopped by adding Na2CO3 when sufficient colour had developed. The specific β-galactosidase activity was calculated with the formula
and expressed in units mgp−1 where mgp denotes mg of total protein.
Alcohol oxidase assay
Appropriate dilutions of the cell-free extracts were used to measure alcohol oxidase activities based on the method adapted from Jungo et al. (2006). A fresh 2x stock of the assay reaction mixture containing 0.8 mM 4-aminoantipyrine, 50 mM phenolsulfonic acid, freshly prepared 4 U/ml horseradish peroxidase in potassium phosphate buffer (200 mM, pH 7.4) was prepared before setting up the assays. 100 μl of the diluted cell-free extracts were mixed with 25 µl methanol and incubated at 30°C for 10 min. After this, 100 µl of the 2x reaction mixture stock was added to the mix at time t = 0 to start the reaction and the increase in absorbance at 500 nm was monitored every 30 s for 10 min using a microplate reader (SpectraMax M2e, Molecular Devices Corporation, CA, United States). The specific alcohol oxidase activity was calculated with the formula
and reported in units mgp−1.
Calculating substrate consumption and protein productivities from the data
We are concerned with experiments in which a chemostat is fed with the primary carbon source
where
These equations were used to calculate
Results
Substrate consumption and PAOX1 expression in the presence of glycerol and sorbitol
Our goal is to study the kinetics of substrate consumption and PAOX1 expression during mixed-substrate growth on methanol + glycerol and methanol + sorbitol; however, we also characterized the substrate consumption kinetics during single-substrate growth on glycerol and sorbitol. In batch (shake-flask) cultures grown on glycerol and sorbitol, the biomass yields were quite similar (∼0.6 gdw g−1), but the maximum specific growth rates
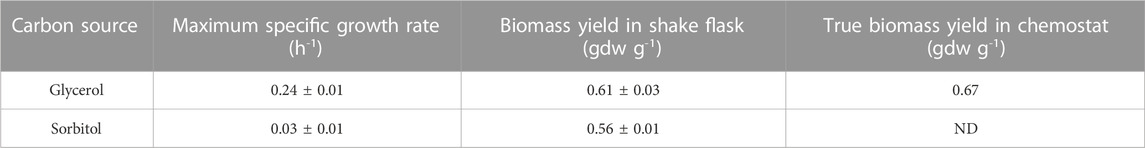
TABLE 1. Maximum specific growth rates and biomass yields during single-substrate growth of the Mut+ strain of K. phaffii on glycerol and sorbitol. The true biomass yield in the chemostat was determined by fitting the variation of the specific substrate consumption rate with
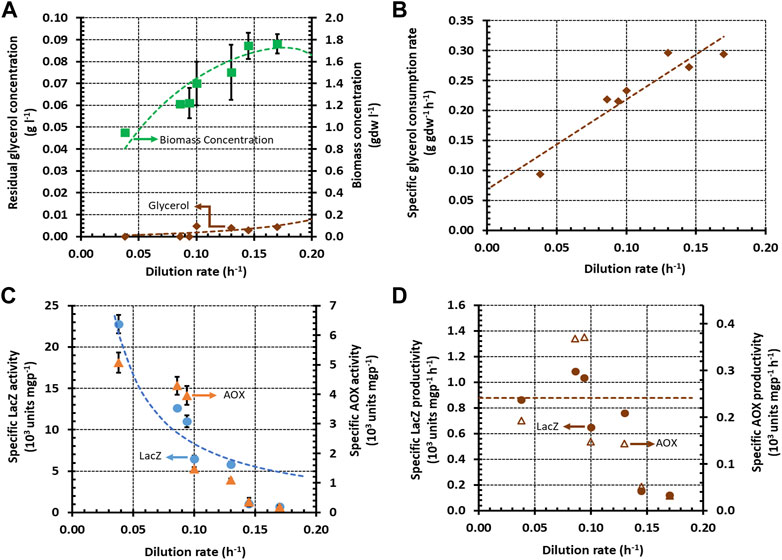
FIGURE 1. Variation of steady state concentrations and rates with the dilution rate during growth of K. phaffii strain Mut+ (pSAOH5-T1) in a chemostat fed with glycerol (∼3.1 g L−1). (A) Concentrations of biomass and residual glycerol. (B) Specific glycerol consumption rates calculated from the data in (A) using Eq. 5. (C) Specific activities of LacZ and AOX. (D) Specific Lac Z and AOX productivities calculated from the data in (C) using Eq. 6.
Substrate consumption and PAOX1 expression in the presence of mixtures
When the Mut+ strain is grown in batch cultures of methanol + glycerol and methanol + sorbitol, there is diauxic growth, but methanol is the unpreferred substrate during growth on methanol + glycerol, and the preferred substrate during growth on methanol + sorbitol (Ramón et al., 2007). Such mixtures, which display diauxic growth in batch cultures, exhibit a characteristic substrate concentration profile in continuous cultures (Egli et al., 1986; Noel and Narang, 2009) (Supplementary Figure S1A). In the dual-limited regime, which extends up to dilution rates approximately equal to the
When methanol + glycerol and methanol + sorbitol were fed to a continuous culture, the variation of the substrate concentrations with
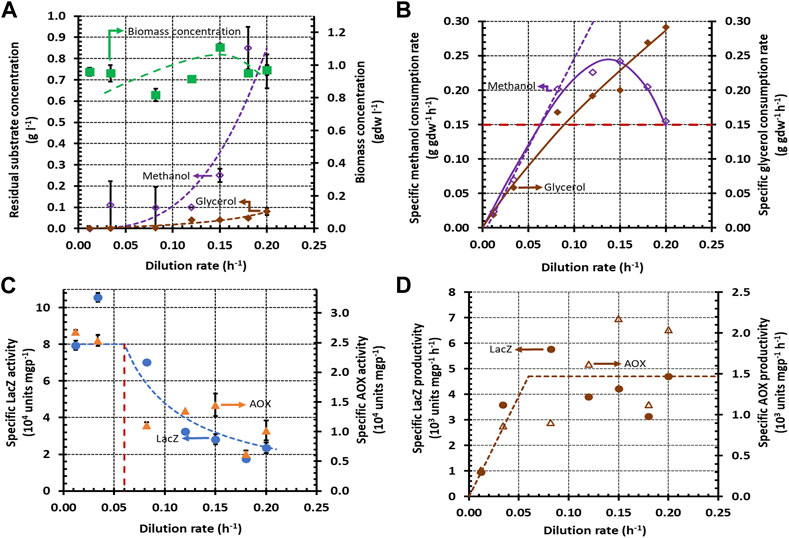
FIGURE 2. Variation of steady state concentrations with the dilution rate during growth of K. phaffii strain Mut+ (pSAOH5-T1) in a chemostat fed with a mixture of glycerol (∼1.5 g L−1) and methanol (∼1.6 g L−1). (A) Concentrations of biomass, residual glycerol, and residual methanol (B) Specific methanol and glycerol consumption rates calculated from the data in (A) using Eq. 5. The dashed line passing through the origin shows the linear increase of the specific methanol consumption rate in the dual-limited regime. The horizontal dashed line shows the threshold specific methanol consumption rate of 0.15 g gdw−1 h−1. (C) Specific activities of LacZ and AOX. (D) Specific LacZ and AOX productivities calculated from the data in (C) using Eq. 6.
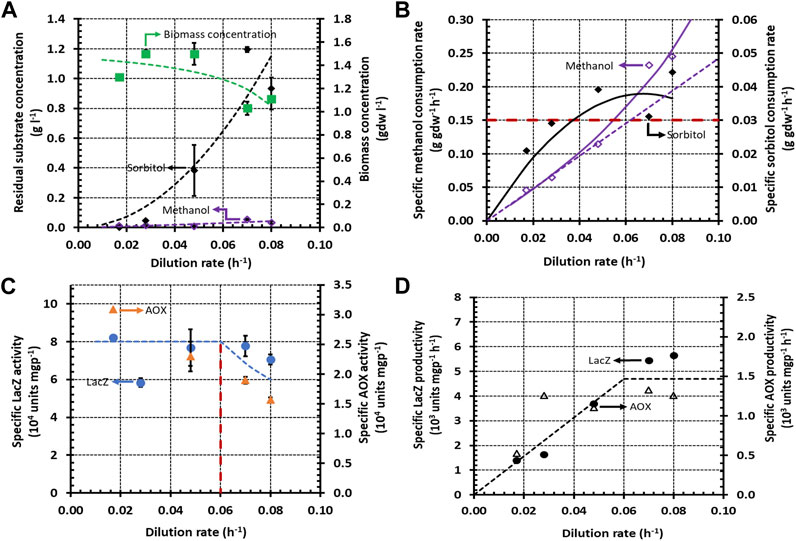
FIGURE 3. Variation of steady state concentrations with the dilution rate during growth of K. phaffii strain Mut+ (pSAOH5-T1) in a chemostat fed with a mixture of sorbitol (∼1.5 g L−1) and methanol (∼3.2 g L−1). (A) Concentrations of biomass, residual sorbitol and residual methanol. (B) Specific methanol and glycerol consumption rates calculated from the data in (A) using Eq. 5. The dashed line passing through the origin shows the linear increase of the specific methanol consumption rate in the dual-limited regime. The horizontal dashed line shows the threshold specific methanol consumption rate of 0.15 g gdw−1 h−1. (C) Specific activities of LacZ and AOX. (D) Specific LacZ and AOX productivities calculated from the data in (C) using Eq. 6.
During single-substrate growth, the specific substrate consumption rate usually increases linearly with
Although it is widely accepted that glycerol is repressing and sorbitol is non-repressing in batch cultures, we found remarkably similar specific LacZ and AOX activities and productivities in continuous cultures fed with methanol + glycerol and methanol + sorbitol. At low dilution rates (
The specific PAOX1 expression rate is a function of the specific methanol consumption rate
To test this hypothesis, we plotted the specific LacZ and AOX productivities
where
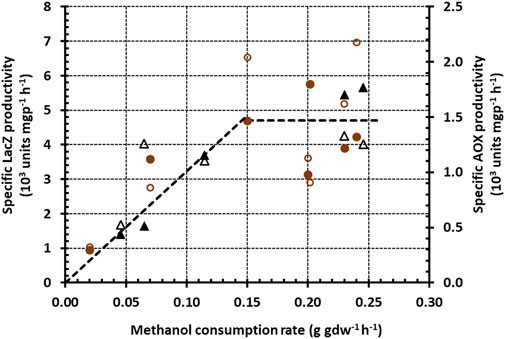
FIGURE 4. Variation of the specific LacZ (closed symbols) and AOX (open symbols) productivities with the specific methanol consumption rate during growth on methanol + glycerol (brown circles) and methanol + sorbitol (black triangles). The graph was obtained by plotting the specific methanol consumption rates in Figures 2B, 3B against the corresponding specific LacZ and AOX productivities in Figures 2D, 3D.
Discussion
Our main conclusion is that over the range of dilution rates considered in our work (0.02–0.2 h−1), the PAOX1 expression rate is completely determined by the methanol consumption rate regardless of the type of the secondary carbon source. This conclusion may appear to subvert the prevailing consensus according to which the expression rate of a promoter is strongly inhibited in the presence of repressing secondary carbon sources. However, this conclusion is based on studies with batch cultures. We show below that our conclusion is consistent with the continuous culture studies reporting the expression of not only the AOX1 promoter of K. phaffii but also the exemplary lac promoter of E. coli.
Comparison with chemostat studies of PAOX1 expression by K. phaffii
Jungo et al. (Jungo et al., 2007a; Jungo et al., 2007b) performed their mixed-substrate studies by fixing
a) The residual methanol remained negligibly small, and the biomass concentration decreased linearly.
b) The specific avidin expression rate increased hyperbolically until it reached a maximum, which was essentially the same for both mixtures.
It follows from a) that the specific methanol consumption rate, which is approximately equal to
Berrios and co-workers compared the methanol consumption and ROL production rates of the Mut+ strain at two different temperatures (22°C and 30°C) during growth on methanol, methanol + glycerol, and methanol + sorbitol (Berrios et al., 2017). These experiments were done in chemostats operated at
Comparison with chemostat studies of expression by lac promoter of E. coli
Analogous results have also been obtained in studies of lac expression in E. coli. Indeed, batch experiments with mixtures of lactose + glycerol, lactose + glucose, and lactose + glucose-6-phophate show that glycerol is non-repressing, whereas glucose and glucose-6-phosphate are repressing (Magasanik, 1970). However, when chemostat experiments were performed with these three mixtures (Smith and Atkinson, 1980), they yielded the same steady state specific β-galactosidase (LacZ) activity at all
In conclusion, the specific PAOX1 expression rate of K. phaffii appears to be completely determined by the specific methanol consumption rate regardless of the type (repressing or non-repressing) of the secondary carbon source. Analysis of the literature shows that the specific expression rate of the lac operon of E. coli is also completely determined by the specific lactose consumption rate regardless of the type of secondary carbon source. It would be interesting to explore if similar results are obtained for other microorganisms and substrate mixtures.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
AS and AN conceived and designed the research. AS conducted the experiments. AS and AN analysed the data and wrote the manuscript. All authors read and approved the manuscript.
Acknowledgments
The authors would like to thank Department of Biotechnology (Government of India) for funding this project (grant BT/PR13831/BBE/117/68/2015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1123703/full#supplementary-material
References
Ahmad, M., Hirz, M., Pichler, H., and Schwab, H. (2014). Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biot. 98 (12), 5301–5317. doi:10.1007/s00253-014-5732-5
Arnau, C., Casas, C., and Valero, F. (2011). The effect of glycerol mixed substrate on the heterologous production of a Rhizopus oryzae lipase in Pichia pastoris system. Biochem. Eng. J. 57, 30–37. doi:10.1016/j.bej.2011.08.004
Azadi, S., Mahboubi, A., Naghdi, N., Solaimanian, R., and Mortazavi, S. A. (2017). Evaluation of sorbitol-methanol co-feeding strategy on production of recombinant human growth hormone in Pichia pastoris. Iran. J. Pharm. Sci. 16 (4), 1555–1564.
Berrios, J., Flores, M. O., Díaz-Barrera, A., Altamirano, C., Martínez, I., and Cabrera, Z. (2017). A comparative study of glycerol and sorbitol as co-substrates in methanol-induced cultures of Pichia pastoris: Temperature effect and scale-up simulation. J. Ind. Microbiol. 44 (3), 407–411. doi:10.1007/s10295-016-1895-7
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 (1-2), 248–254. doi:10.1016/0003-2697(76)90527-3
Brierley, R. A., Bussineau, C., Kosson, R., Melton, A., and Siegel, R. S. (1990). Fermentation development of recombinant Pichia pastoris expressing the heterologous gene: Bovine lysozyme. Ann. N. Y. Acad. Sci. 589 (1), 350–362. doi:10.1111/j.1749-6632.1990.tb24257.x
Canales, C., Altamirano, C., and Berrios, J. (2015). Effect of dilution rate and methanol-glycerol mixed feeding on heterologous Rhizopus oryzae lipase production with Pichia pastoris Mut+ phenotype in continuous culture. Biotechnol. Prog. 31 (3), 707–714. doi:10.1002/btpr.2069
Carly, F., Niu, H., Delvigne, F., and Fickers, P. (2016). Influence of methanol/sorbitol co-feeding rate on pAOX1 induction in a Pichia pastoris Mut+ strain in bioreactor with limited oxygen transfer rate. J. Ind. Microbiol. Biotechnol. 43 (4), 517–523. doi:10.1007/s10295-015-1722-6
Çelik, E., Çalık, P., and Oliver, S. G. (2009). Fed-batch methanol feeding strategy for recombinant protein production by Pichia pastoris in the presence of co-substrate sorbitol. Yeast 26 (9), 473–484. doi:10.1002/yea.1679
Chen, L., Mohsin, A., Chu, J., Zhuang, Y., Liu, Y., and Guo, M. (2017). Enhanced protein production by sorbitol co-feeding with methanol in recombinant Pichia pastoris strains. Biotechnol. Bioprocess Eng. 22 (6), 767–773. doi:10.1007/s12257-017-0011-9
Chiruvolu, V., Cregg, J. M., and Meagher, M. M. (1997). Recombinant protein production in an alcohol oxidase-defective strain of Pichia pastoris in fed-batch fermentations. Enzyme Microb. Technol. 21 (4), 277–283. doi:10.1016/s0141-0229(97)00042-2
Cos, O., Ramón, R., Montesinos, J. L., and Valero, F. (2006). Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: A review. Microb. Cell Factories 5 (1), 17–20. doi:10.1186/1475-2859-5-17
Cregg, J. M., Barringer, K. J., Hessler, A. Y., and Madden, K. R. (1985). Pichia pastoris as a host system for transformations. Mol. Cell Biol. 5 (12), 3376–3385. doi:10.1128/mcb.5.12.3376-3385.1985
Cregg, J. M., Madden, K. R., Barringer, K. J., Thill, G. P., and Stillman, C. A. (1989). Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell Biol. 9 (3), 1316–1323. doi:10.1128/mcb.9.3.1316
Egli, T., Bosshard, C., and Hamer, G. (1986). Simultaneous utilization of methanol–glucose mixtures by Hansenula polymorpha in chemostat: Influence of dilution rate and mixture composition on utilization pattern. Biotechnol bioeng 28 (11), 1735–1741. doi:10.1002/bit.260281118
Egli, T., and Fiechter, A. (1981). Theoretical analysis of media used in the growth of yeasts on methanol. Microbiol 123 (2), 365–369. doi:10.1099/00221287-123-2-365
Egli, T., Lendenmann, U., and Snozzi, M. (1993). Kinetics of microbial growth with mixtures of carbon sources. Antonie Leeuwenhoek 63 (3), 289–298. doi:10.1007/bf00871224
Egli, T., and Mason, C. A. (1991). Mixed substrates and mixed cultures. Biotechnol 18, 173–201. doi:10.1016/b978-0-7506-9188-8.50015-2
Ergün, B. G., Berrios, J., Binay, B., and Fickers, P. (2021). Recombinant protein production in Pichia pastoris: From transcriptionally redesigned strains to bioprocess optimization and metabolic modelling. FEMS Yeast Res. 21 (7), foab057. doi:10.1093/femsyr/foab057
Feng, A., Zhou, J., Mao, H., Zhou, H., and Zhang, J. (2022). Heterologous protein expression enhancement of Komagataella phaffii by ammonium formate induction based on transcriptomic analysis. Biochem. Eng. J. 185, 108503. doi:10.1016/j.bej.2022.108503
Gao, M. J., Li, Z., Yu, R. S., Wu, J. R., Zheng, Z. Y., Shi, Z. P., et al. (2012). Methanol/sorbitol co-feeding induction enhanced porcine interferon-α production by P. pastoris associated with energy metabolism shift. Bioprocess Biosyst. Eng. 35 (7), 1125–1136. doi:10.1007/s00449-012-0697-1
García-Ortega, X., Cámara, E., Ferrer, P., Albiol, J., Montesinos-Seguí, J. L., and Valero, F. (2019). Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol-free GAP promoter. Where do we stand? N. Biotechnol. 53, 24–34. doi:10.1016/j.nbt.2019.06.002
Gasser, B., and Mattanovich, D. (2018). A yeast for all seasons–Is Pichia pastoris a suitable chassis organism for future bioproduction? FEMS Microbiol. Lett. 365 (17), fny181. doi:10.1093/femsle/fny181
Higgins, D. R., and Cregg, J. M. (1998). “Introduction to Pichia pastoris,” in Pichia protocols (Totowa, New Jersey, United States: Humana Press), 1–15.
Inan, M., and Meagher, M. M. (2001). Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J. Biosci. Bioeng. 92 (6), 585–589. doi:10.1016/s1389-1723(01)80321-2
Jahic, M., Veide, A., Charoenrat, T., Teeri, T., and Enfors, S. O. (2006). Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol. Prog. 22 (6), 1465–1473. doi:10.1021/bp060171t
Jungo, C., Marison, I., and von Stockar, U. (2007a). Mixed feeds of glycerol and methanol can improve the performance of Pichia pastoris cultures: A quantitative study based on concentration gradients in transient continuous cultures. J. Biotechnol. 128 (4), 824–837. doi:10.1016/j.jbiotec.2006.12.024
Jungo, C., Rérat, C., Marison, I. W., and von Stockar, U. (2006). Quantitative characterization of the regulation of the synthesis of alcohol oxidase and of the expression of recombinant avidin in a Pichia pastoris Mut+ strain. Enzyme Microb. Technol. 39 (4), 936–944. doi:10.1016/j.enzmictec.2006.01.027
Jungo, C., Schenk, J., Pasquier, M., Marison, I. W., and von Stockar, U. (2007b). A quantitative analysis of the benefits of mixed feeds of sorbitol and methanol for the production of recombinant avidin with Pichia pastoris. J. Biotechnol. 131 (1), 57–66. doi:10.1016/j.jbiotec.2007.05.019
Juturu, V., and Wu, J. C. (2018). Heterologous protein expression in Pichia pastoris: Latest research progress and applications. ChembioChem 19 (1), 7–21. doi:10.1002/cbic.201700460
Kurtzman, C. P. (2009). Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J. Ind. Microbiol. Biotechnol. 36 (11), 1435–1438. doi:10.1007/s10295-009-0638-4
Kurtzman, C. P. (2005). Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int. J. Syst. Evol. Microbiol. 55 (2), 973–976. doi:10.1099/ijs.0.63491-0
Liu, B., Zhao, Y., Zhou, H., and Zhang, J. (2022). Enhancing xylanase expression of Komagataella phaffii induced by formate through Mit1 co-expression. Bioprocess Biosyst. Eng. 45 (9), 1515–1525. doi:10.1007/s00449-022-02760-6
Liu, W. C., Inwood, S., Gong, T., Sharma, A., Yu, L. Y., and Zhu, P. (2019). Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit. Rev. Biotechnol. 39 (2), 258–271. doi:10.1080/07388551.2018.1554620
Macauley-Patrick, S., Fazenda, M. L., McNeil, B., and Harvey, L. M. (2005). Heterologous protein production using the Pichia pastoris expression system. Yeast 22 (4), 249–270. doi:10.1002/yea.1208
Magasanik, B. (1970). “Glucose effects: Inducer exclusion and repression,” in The lactose operon. Editors J. Beckwith, and D. Zipser (New York: Cold Spring Harbor Press), 189–220.
Nieto-Taype, M. A., Garcia-Ortega, X., Albiol, J., Montesinos-Seguí, J. L., and Valero, F. (2020). Continuous cultivation as a tool toward the rational bioprocess development with Pichia Pastoris cell factory. Front. Bioeng. Biotechnol. 8, 632. doi:10.3389/fbioe.2020.00632
Niu, H., Jost, L., Pirlot, N., Sassi, H., Daukandt, M., Rodriguez, C., et al. (2013). A quantitative study of methanol/sorbitol co-feeding process of a Pichia pastoris Mut+/pAOX1-lacZ strain. Microb. Cell factories 12 (1), 33–38. doi:10.1186/1475-2859-12-33
Noel, J. T., and Narang, A. (2009). Gene regulation in continuous cultures: A unified theory for bacteria and yeasts. B Math. Biol. 71 (2), 453–514. doi:10.1007/s11538-008-9369-3
Paulová, L., Hyka, P., Branská, B., Melzoch, K., and Kovar, K. (2012). Use of a mixture of glucose and methanol as substrates for the production of recombinant trypsinogen in continuous cultures with Pichia pastoris Mut+. J. Biotechnol. 157 (1), 180–188. doi:10.1016/j.jbiotec.2011.10.010
Pfeffer, M., Maurer, M., Köllensperger, G., Hann, S., Graf, A. B., and Mattanovich, D. (2011). Modeling and measuring intracellular fluxes of secreted recombinant protein in Pichia pastoris with a novel 34 S labeling procedure. Microb. Cell Factories 10 (1), 47.
Pirt, S. J. (1965). The maintenance energy of bacteria in growing cultures. Proc. R. Soc. B 163 (991), 224–231. doi:10.1098/rspb.1965.0069
Potvin, G., Ahmad, A., and Zhang, Z. (2012). Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: A review. Biochem. Eng. J. 64, 91–105. doi:10.1016/j.bej.2010.07.017
Rahimi, A., Hosseini, S. N., Javidanbardan, A., and Khatami, M. (2019a). Continuous fermentation of recombinant Pichia pastoris Mut+ producing HBsAg: Optimizing dilution rate and determining strain-specific parameters. Food Bioprod. Process. 118, 248–257. doi:10.1016/j.fbp.2019.09.011
Rahimi, A., Hosseini, S. N., Karimi, A., Aghdasinia, H., and Mianroodi, R. A. (2019b). Enhancing the efficiency of recombinant Hepatitis B surface antigen production in Pichia pastoris by employing continuous fermentation. Biochem. Eng. J. 141, 112–119. doi:10.1016/j.bej.2018.10.019
Ramón, R., Ferrer, P., and Valero, F. (2007). Sorbitol co-feeding reduces metabolic burden caused by the overexpression of a Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 130 (1), 39–46. doi:10.1016/j.jbiotec.2007.02.025
Schwarzhans, J. P., Wibberg, D., Winkler, A., Luttermann, T., Kalinowski, J., and Friehs, K. (2016). Non-canonical integration events in Pichia pastoris encountered during standard transformation analysed with genome sequencing. Sci. Rep. 6, 38952. doi:10.1038/srep38952
Singh, A., and Narang, A. (2020). The Mut+ strain of Komagataella phaffii (Pichia pastoris) expresses PAOX1 5- and 10-times faster than Muts and Mut− strains: Evidence that formaldehyde or/and formate are true inducers of PAOX1. Appl. Microbiol. Biotechnol. 104 (18), 7801–7814. doi:10.1007/s00253-020-10793-8
Smith, S. S., and Atkinson, D. E. (1980). The expression of β-galactosidase by Escherichia coli during continuous culture. Arch. Biochem. Biophys. 202 (2), 573–581. doi:10.1016/0003-9861(80)90464-6
Thorpe, E. D., d'Anjou, M. C., and Daugulis, A. J. (1999). Sorbitol as a non-repressing carbon source for fed-batch fermentation of recombinant Pichia pastoris. Biotechnol. Lett. 21 (8), 669–672. doi:10.1023/A:1005585407601
Wang, Z., Wang, Y., Zhang, D., Li, J., Hua, Z., Du, G., et al. (2010). Enhancement of cell viability and alkaline polygalacturonate lyase production by sorbitol co-feeding with methanol in Pichia pastoris fermentation. Bioresour. Technol. 101 (4), 1318–1323. doi:10.1016/j.biortech.2009.09.025
Xie, J., Zhou, Q., Du, P., Gan, R., and Ye, Q. (2005). Use of different carbon sources in cultivation of recombinant Pichia pastoris for angiostatin production. Enzyme Microb. Technol. 36 (2-3), 210–216. doi:10.1016/j.enzmictec.2004.06.010
Keywords: Komagataella phaffii (Pichia pastoris), recombinant protein, glycerol, sorbitol, methanol, methanol consumption rate
Citation: Singh A and Narang A (2023) PAOX1 expression in mixed-substrate continuous cultures of Komagataella phaffii (Pichia pastoris) is completely determined by methanol consumption regardless of the secondary carbon source. Front. Bioeng. Biotechnol. 11:1123703. doi: 10.3389/fbioe.2023.1123703
Received: 14 December 2022; Accepted: 22 March 2023;
Published: 05 April 2023.
Edited by:
Pau Ferrer, Autonomous University of Barcelona, SpainReviewed by:
Jianguo Zhang, University of Shanghai for Science and Technology, ChinaSarma VRK Mutturi, Central Food Technological Research Institute (CSIR), India
Patrick Fickers, University of Liège, Belgium
Copyright © 2023 Singh and Narang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atul Narang, YW5hcmFuZ0BkYmViLmlpdGQuYWMuaW4=
†Present address: Anamika Singh, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
 Anamika Singh
Anamika Singh Atul Narang
Atul Narang