
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 30 August 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.977215
This article is part of the Research Topic Microorganism and Process Engineering for Biosynthesis View all 11 articles
 Cong Jiang1
Cong Jiang1 Changwen Ye2
Changwen Ye2 Yongfeng Liu3
Yongfeng Liu3 Kuo Huang2
Kuo Huang2 Xuedeng Jiang1
Xuedeng Jiang1 Dian Zou1
Dian Zou1 Lu Li4
Lu Li4 Wenyuan Han1
Wenyuan Han1 Xuetuan Wei1*
Xuetuan Wei1*Alkaline protease has been widely applied in food, medicine, environmental protection and other industrial fields. However, the current activity and yield of alkaline protease cannot meet the demand. Therefore, it is important to identify new alkaline proteases with high activity. In this study, we cloned a potential alkaline protease gene bsp-1 from a Bacillus subtilis strain isolated in our laboratory. BSP-1 shows the highest sequence similarity to subtilisin NAT (S51909) from B. subtilis natto. Then, we expressed BSP-1 in Bacillus amyloliquefaciens BAX-9 and analyzed the protein expression level under a collection of promoters. The results show that the P43 promoter resulted in the highest transcription level, protein level and enzyme activity. Finally, we obtained a maximum activity of 524.12 U/mL using the P43 promoter after fermentation medium optimization. In conclusion, this study identified an alkaline protease gene bsp-1 from B. subtilis and provided a new method for high-efficiency alkaline protease expression in B. amyloliquefaciens.
Alkaline protease is found in all living organisms (Gupta et al., 2002). Alkaline protease has a lot of functions and it is widely applied in washing, food, textile, leather, pharmaceutical and other industries (Contesini et al., 2018; Harish and Uppuluri, 2018; Pathak and Rathod, 2018; Tavano et al., 2018; Jahangirian et al., 2019; Sharma et al., 2019; Burchacka et al., 2022). However, in recent years, the yield of alkaline protease fail to satisfy the industrial demand, causing the shortage problem of large-scale industrial enzymes (Barzkar, 2020). Therefore, developing efficient alkaline protease has great industrial prospects.
Many strains produce alkaline protease, Bacillus genus are the main producer of the enzyme in industrial production (Cai et al., 2019). Bacillus have high secretory capacity and become the main producer of the enzyme in industrial production (Westers et al., 2004). However, the yield and activity of alkaline protease in Bacillus is usually low (Zhang et al., 2020; Ouyang et al., 2021; Zhang et al., 2021; Espoui et al., 2022). To achieve high-efficiency expression of target proteins, researchers had reformed expression elements, such as signal peptides, transcription factors, and molecular chaperones (Liu et al., 2019; Suberu et al., 2019; Gong et al., 2020; Neef et al., 2021; Su et al., 2021). For instance, Gang et al. constructed a signal peptide library in Bacillus subtilis, and screened out the amylase activity up to 5086 U/mL (Fu et al., 2018). In addition, by combining the transcriptional activator Spo0A with the regulatory region of Bacillus licheniformis, Zhou et al. successfully achieved 1.46-fold increase in the yield of alkaline protease AprE (Zhou et al., 2020). In addition, since promoter is one of the most critical factors in gene expression regulation (Kumar and Bansal, 2018; Brázda et al., 2021; Cazier and Blazeck, 2021; Jensen and Galburt, 2021), its engineering also serves as an important strategy to increase microbial gene expression and metabolites production. Su et al. substituted the wild-type −10 box and −35 box of aprN promoter, resulting in significantly higher nattokinase production than those previously reported in B. subtilis (Wu et al., 2011). Liao et al. (2018) optimized the ribosome-binding site to improve β-Gal activity in B. amyloliquefaciens. Together, genetic manipulation of the above elements has been widely applied in the production of industrial enzymes.
B. subtilis has significant attributes, such as fast growth and development, strong metabolic capacity and rich products, B. subtilis has been the preferred organisms for industrial production of a variety of products (Gu et al., 2018). B. amyloliquefaciens could secrete recombinant proteins using a variety of signal peptides (Cui et al., 2018), thus it had been an efficient platform for producing various proteases (Uyar and Baysal, 2008; Matkawala et al., 2021; Meng et al., 2021). In this study, we identified an alkaline protease gene bsp-1 from B. subtilis, and then expressed it in B. amyloliquefaciens BAX-9. Subsequently, we further analyzed the expression level under different promoters. Finally, the highest expression level of alkaline protease was obtained by using the promotor P43 combined with fermentation optimization.
In this study, the TransStartFastPfu DNA polymerase was purchased from TransGen Biotech Co., Ltd. (Beijing, China). Restriction enzymes, dNTPs, T4 ligase and RNase were provided by Takara Biotechnology Co., Ltd. (Dalian, China). We purchased the Total RNA Isolation Kit and PrimeScript RT Master Mix Kit from Vazyme Biotech Co.,Ltd. (Nanjing, China). Standard alkaline protease was purchased from Sigma-Aldrich Co., LLC (St.Louis, United States). All the other chemicals were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
To express BSP-1 in B. amyloliquefaciens BAX-9, we constructed the recombinant strains following the procedure reported in our previous study (Zou et al., 2020). All strains and plasmids involved were listed in Tables 1, 2, listed all designed primers. For example, a pair of primers were designed to amplify the bsp-1 gene from B. subtilis DNA, Ppqq promoter from B. amyloliquefaciens and TamyL terminator from B. licheniformis WX-02 respectively, and then Ppqq promoter, TamyL terminator and bsp-1 gene were fused by Splicing with Overlap Extension PCR (SOE-PCR). The fused fragment and the pHY-300PLK plasmid were digested by restriction enzymes of BamHI and XbaI, and then they were ligated to obtain the expression plasmid pHY-Ppqq/BSP-1. The expression plasmid pHY-Ppqq/BSP-1 was electroporated into BAX-9 to obtain the recombinant strain. In this study, the remaining recombinant strains were constructed using the same procedure.
Skim milk agar medium contain 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, 1.5% Agar and 2% skim milk. To screen the protease activity, 20 µL of supernatant of the cultures were added to the wells of milk agar plates made by a hole punch. The radius of transparent circles were measured after the plates were cultured at 37°C for 12 h.
The B. amyloliquefaciens cells were inoculated into 5 ml liquid LB medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl), and incubated at 37°C for 12 h with shaking at 180 rpm. Then, a 3% (v/v) inoculum was transferred into the 50 ml alkaline protease fermentation medium (40 g/L tryptone, 20 g/L yeast extract, and 10 g/L NaCl) and cultured at 37°C for 72 h with shaking at 180 rpm.
Protein samples were prepared by 100% trichloroacetic acid (TCA) precipitation (Benabdelkamel et al., 2018). Specifically, 0.90 ml of fermentation supernatant was mixed with 0.10 ml of trichloroacetic acid, inverted 10 times to mix and incubated at 4°C for 12 h. Then, the mixture was centrifuged at 6,000 ×g for 10 min. After that, the supernatant was discarded and the pellet was washed with 0.20 ml of absolute ethanol for three times. After dried at 37°C, the pellet was dissolved with 30 µL of loading buffer containing 8 mol/L urea and 2 mol/L thiourea. At last, the samples were analyzed by SDS-PAGE electrophoresis.
1 ml of the sample was transfered to tube A (as blank control) and tube B for activity measurement, respectively. After the samples were heated at 40°C for 2 min, A was supplemented with 2 ml of trichloroacetic acid (64.70 g/L), while B was added with 1 ml of casein solution (10 g/L). Both samples were further heated at 40°C for 10 min. Then, 1 ml casein solution was added into A, and 2 ml trichloroacetic acid was added into B. The mixtures were placed at room temperature for 20 min. Subsequently, A and B were filtered and obtained supernatant, and added 5 ml of sodium carbonate solution and 1 ml of folin phenol reagent to the supernatant, mix well and heat at 40°C for 20 min, and measure the absorbance at 680 nm wavelength. The activity 1) of the dilution of sample was obtained from the standard curve. The enzymatic activity of the sample was calculated as follows:
The recombinant engineered strains were cultured for 12 h, and then the cells were collected by centrifugation and washed for transcription analysis, according to the description of Total RNA Isolation Kit and PrimeScript RT Master Mix Kit. DNA removal and RNA reverse transcription were performed simultaneously in one system. The reaction conditions which included EasyScript® RT/RI and gDNA Remover were 25°C for 10 min, 42°C for 15 min, and 85°C for 10 s. RT-PCR amplification was performed using three-steps method. Pre-denaturation: 1 cycle (95°C for 5 min); PCR reaction: 40 cycles (95°C for 30 s, 60°C for 30 s) and dissolution curve (1 min at 95°C, 1 min at 65°C). Relative quantitative analysis was performed with 16 s RNA as the internal reference gene.
Each group of experiments was designed with three independent replicates. SPSS 20.0 was used for statistical analysis, calculating the means and standard deviations, and evaluating the significance. Origin 8.5 was used to process the data and make the graphs.
We isolated a B. subtilis strain with high protease activity using skim milk plates. Then the new B. subtilis strain was named B. subtilis D7. In order to identify an alkaline protease from B. subtilis D7, we designed a primer set using the gene sequence of subtilisin NAT (S51909) from B. subtilis. With the primers, we amplified a gene fragment from B. subtilis D7. Sequencing of the gene fragment indicates that it encodes an alkaline protease (BSP-1) containing 381 amino acids (aa) (Figure 1A). Analysis of the protein sequence using SignalP 5.0 indicates a 29-aa predicted signal peptide, while the 30-77 aa and 78-275 aa are the propeptide and mature peptide, respectively (Figure 1A). We aligned the sequence of BSP-1 with four well-characterized proteases (Choi et al., 2004; Yong et al., 2004; Agrebi et al., 2009; Ku et al., 2009), indicating that it has the highest similarity to subtilisin NAT (S51909) and fibrinolytic enzyme BSF1(FJ517584) (>96% similarity) and less similar to subtilisin DJ-4 and subtilisin DFE (Figure 1B). In addition, all the proteases contain the conserved catalytic triad, including Asp-32, His-64, and Ser-221. Together, the data indicates that BSP-1 belong to the subtilisin family of serine proteases (Wei et al., 2011).
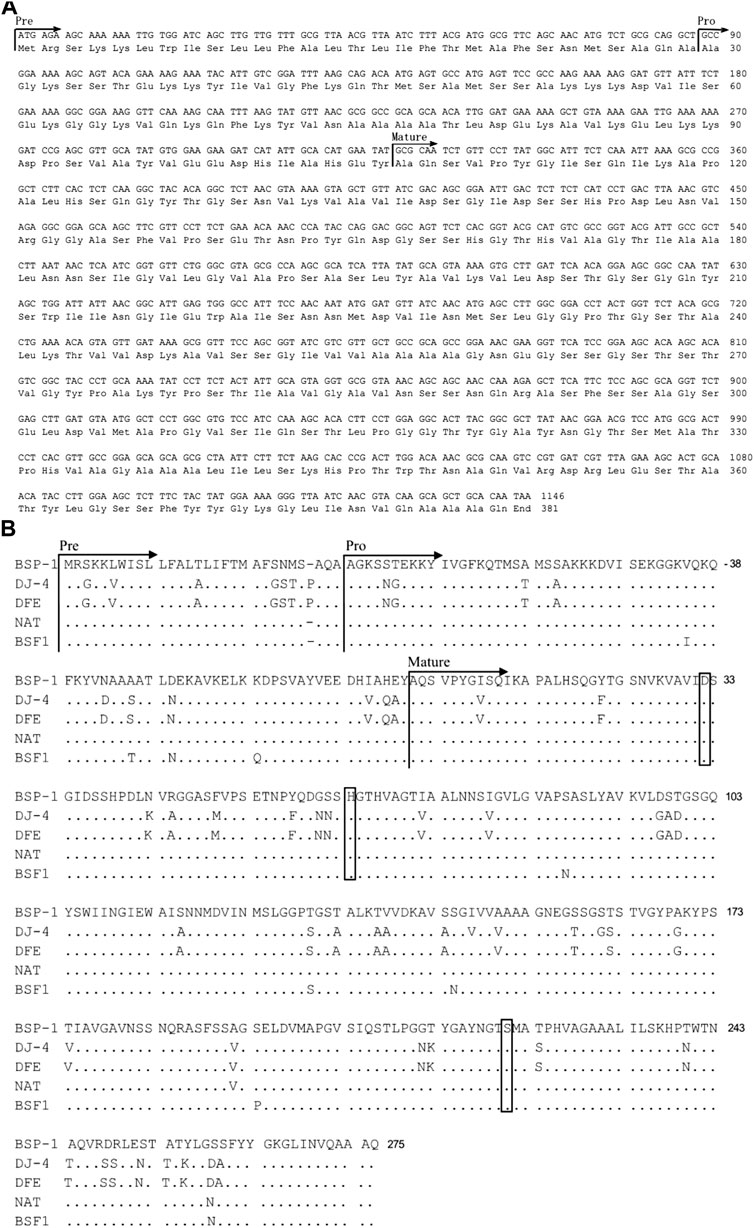
FIGURE 1. Sequence analysis of bsp-1 gene and deduced amino acids. (A) Nucleotide (upper line) and deduced amino acid (lower line) sequences of the protease BSP-1. The predicted signal peptide, propeptide and mature peptide are marked with arrows. (B) Amino acid sequence alignment of the protease BSP-1, with subtilisin (DJ-4, DFE) from B. amyloliquefaciens and subtilisin (NAT, BSF1) from B. subtilis. The signal peptide, pro-peptide, and mature peptide (mature) were indicated with arrows. “.” indicated the same residue as the first sequence. The catalytic center residues (Asp-32, His-64, and Ser-221) were boxed. The initial amino acid of the mature peptide was numbered as + 1.
B. amyloliquefaciens is considered as a safe strain that have been widely used to express industrial enzymes. B. amyloliquefaciens is rich in proteases, and can degrade heterologous proteins. Knockout of the protease genes may solve the problem. Therefore, we expressed BSP-1 in B. amyloliquefaciens BAX-9, a strain where many protease genes (epr, nprE, aprE, aprX, mpr, bpf, vpr, htrB, yktc1) have been knocked out (Chen et al., 2022). At first, the promoter of PQQ-binding-like beta-propeller repeat gene (Ppqq) from B. amyloliquefaciens was used to express BSP-1. The constructed expression plasmid, pHY-Ppqq/BSP-1, was electroporated into B. amyloliquefaciens BAX-9, generating the strain BAX-9/pHY-Ppqq/BSP-1 (Figure 2A). Subsequently, the fermentation broth of BAX-9/pHY-Ppqq/BSP-1 and the control strain (BAX-9/pHY-300) were used for SDS-PAGE analysis. As shown in Figure 2B, compared to the control strain (BAX-9/pHY-300), the target protein (30 kDa) was found in the fermentation broth of BAX-9/pHY-Ppqq/BSP-1. This result demonstrates that the bsp-1 gene was significantly expressed in BAX-9. The thickness of the band corresponds to the amount of protein present, the thicker band of the target protein also shows the feasibility and great potential of expressing the heterologous protein in B. amyloliquefaciens.
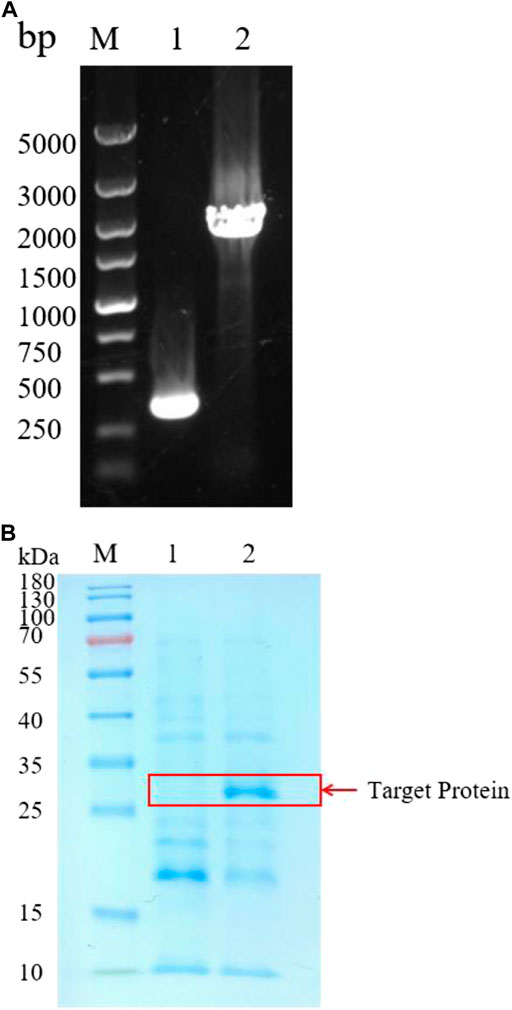
FIGURE 2. Heterologous expression of protease gene bsp-1 in B amyloliquefaciens BAX-9. (A) PCR analysis of recombinant strain colonies. M: Marker; 1-2: P BAX-9/pHY-300 and BAX-9/pHY-Ppqq/BSP-1. (B) SDS-PAGE of analysis of BAX-9/pHY-Ppqq/BSP-1. M: Marker; 1-2: BAX-9/pHY-300 and BAX-9/pHY-Ppqq/BSP-1.
To further improve the expression of BSP-1 in B. amyloliquefaciens, we screened different promoters driving the expression of BSP-1. The promoters include two reported high-strength promoters (P43 (Wang and Doi, 1984), PtrnQ (Song et al., 2016)) from B. subtilis and a promoter collection from B. amyloliquefaciens HZ-12. We selected the promoters of potentially highly-expressed genes in HZ-12, including ssrA (NC_014551.1: 3271400-3271038), ssrS (NC_014551.1: 2609656-2609464), rnpB (NC_014551.1: 2202786-2202405), ffs (NC_014551.1: 27641-27905), cold-shock protein gene (NC_014551.1: 1029767-1029567), PQQ protein gene (NC_014551.1: 3859752-3858532), spore coat protein gene (NC_014551.1: 1272412-1271912), hypothetical protein gene (NC_014551.1: 1272819-1272505). In addition, since B. amyloliquefaciens is highly productive of macromolecular substances such as lipopeptides, the promoters of four gene clusters (Surfactin, Iturin A, Fengycins, Bacillaene) producing lipopeptides, i.e., Psrf, Pitu, Pfen, Pbac, respectively, were also used in the study.
For efficient insertion of the promoters, we constructed a plasmid with replaceable promoter sequence by inserting the fusion fragment of bsp-1 gene and TamyL terminator into pHY300PLK, resulting in the plasmid pHY300PLK-PX/BSP-1 (Figure 3). Then, the sequences of the promoters were amplified and then inserted into pHY300PLK-PX/BSP-1. The resulting plasmids were transformed into BAX-9 to generate engineered strains for further analysis. Engineered strains containing different promoters (Psra, Psrs, Prnp, Pffs, Ppqq, Pcsp, Pscp, Phyp, Psrf, Pitu, Pfen, Pbac, P43, PtrnQ) and the control strain (BAX-9/pHY-300, BAX-9) were fermented. And the enzyme activities of these strains were preliminarily analyzed by using skim milk agar plates. As shown in Figure 4, the supernatant of the fermentation broth of the engineering strains containing P43, Pitu, Pfen, Ppqq, Psrf, Pcsp, PtrnQ and Pscp produced transparent circles of different radius (7, 7, 7, 6.50, 6, 6, 4, 1.50 mm, respectively), while other promoters (Psra, Psrs, Prnp, Pffs, Phyp, Pbac) did not produce transparent circles. The size of the transparent circle reflects the activity of BSF-1, the size of the transparent circle is positively correlated with the activity.
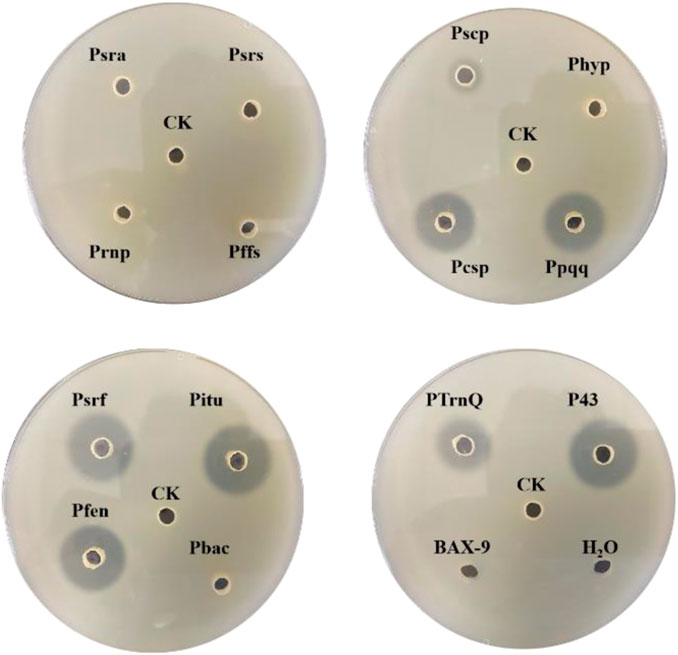
FIGURE 4. Skim milk agar plates of the fermentation supernatant of different strains. CK: BAX-9/pHY-300, Psra: BAX-9/pHY-Psra/BSP-1, Psrs: BAX-9/pHY-Psrs/BSP-1, Prnp: BAX-9/pHY-Prnp/BSP-1, Pffs: BAX-9/pHY-Pffs/BSP-1, Pcsp: BAX-9/pHY-Pcsp/BSP-1, Ppqq: BAX-9/pHY-Pscp/BSP-1, Pscp: BAX-9/pHY-Pscp/BSP-1, Phyp: BAX-9/pHY-Phyp/BSP-1, Psrf: BAX-9/pHY-Psrf/BSP-1, Pitu: BAX-9/pHY-Pitu/BSP-1, Pfen: BAX-9/pHY-Pfen/BSP-1, Pbac: BAX-9/pHY-Pbac/BSP-1, P43: BAX-9/pHY-P43/BSP-1, PtrnQ: BAX-9/pHY-PtrnQ/BSP-1.
Accorded to the result of the transparent circle, we selected the Pscp, Pcsp, Ppqq, Psrf, Pitu, Pfen promoters from B. amyloliquefaciens and the P43 promoter from B. subtilis for further analysis. We analyzed the proteins in the supernatant of BAX-9/pHY-Pscp/BSP-1, BAX-9/pHY-Pcsp/BSP-1, BAX-9/pHY-Ppqq/BSP-1, BAX-9/pHY-Psrf/BSP-1, BAX-9/pHY-Pitu/BSP-1, BAX-9/pHY-Pfen/BSP-1 and BAX-9/pHY-P43/BSP-1 by SDS-PAGE. As shown in Figure 5, BSP-1 was successfully expressed in all recombinant strains, and exhibited the highest level under the promoter P43, Psrf and Pfen, which was consistent with the result of the transparent circle. The data indicates that a high-strength promoter facilitates the protein expression level and thus increase the activity outcome of protein.
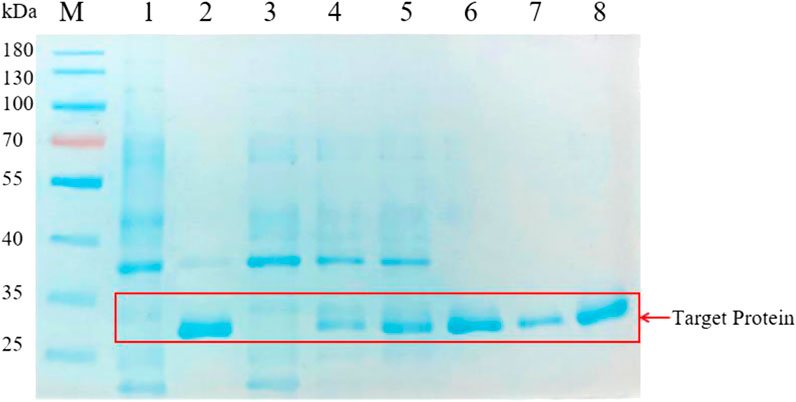
FIGURE 5. SDS-PAGE analysis of the fermentation supernatant of different strains. M: Marker; 1-8: BAX-9/pHY-300, BAX-9/pHY-P43/BSP-1, BAX-9/pHY-Pscp/BSP-1, BAX-9/pHY-Pcsp/BSP-1, BAX-9/pHY-Ppqq/BSP-1, BAX-9/pHY-Psrf/BSP-1, BAX-9/pHY-Pitu/BSP-1, BAX-9/pHY-Pfen/BSP-1.
To further analyze the effect of different promoters on the expression level of protease gene bsp-1, we measured the activity of BSP-1 and the biomass of the seven cultures at 48 h. As shown in Figure 6, the activity of BSP-1 expressed by the promoter of P43, Psrf, Pitu, and Pfen reached 115.87, 106.71, 107.25, and 107.52 U/mL respectively, improving by 4.3-fold, 4.0-fold, 4.0-fold and 4.0-fold compared to the Ppqq promoter (26.71 U/mL). The data indicated protomer sceening efficiently enhanced the activity outcome of BSP-1. In addition, the biomass of Ppqq promoter was significantly higher than that of P43, Psrf, Pitu and Pfen promoter, indicating that the enzyme activity and the biomass are not necessarily positively correlated.
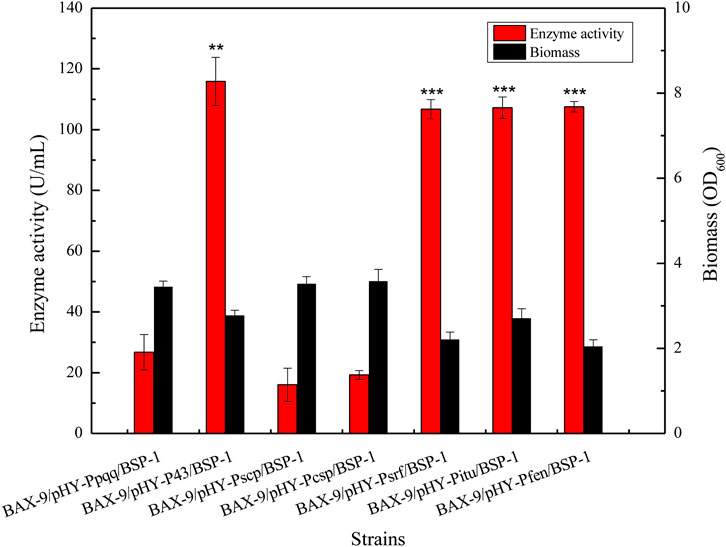
FIGURE 6. Determination of enzyme activity and biomass of recombinant strains. Asterisks show the significant difference (p < 0.05) compared with the control.
To verify the effect of promoter on gene transcription level, we established a microbial fermentation model. The total RNA of different recombinant strains was extracted and analyzed. As shown in Figure 7, the transcription levels of bsp-1 in BAX-9/pHY-P43/BSP-1, BAX-9/pHY-Pscp/BSP-1, BAX-9/pHY-Pcsp/BSP-1, BAX-9/pHY-Psrf/BSP-1, BAX-9/pHY-Pitu/BSP-1, BAX-9/pHY-Pfen/BSP-1 significantly increased compared to that in BAX-9/pHY-Ppqq/BSP-1. Among them, BAX-9/pHY-P43/BSP-1 showed the highest transcription level, 4 folds than that in BAX-9/pHY-Ppqq/BSP-1. The results of RT-PCR were consistent with the results of SDS-PAGE and enzyme activity, revealing that the efficient transcription by P43 generated high protein level and enzyme activity. The above results provide a reference to select the optimal promoter for protein expression.
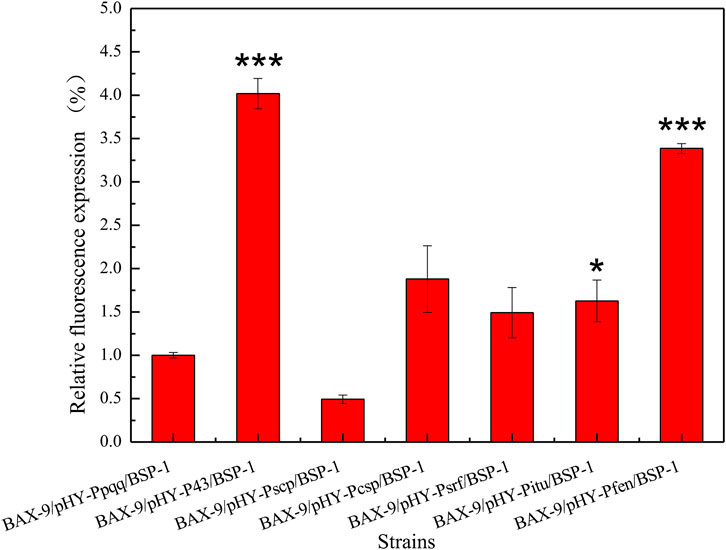
FIGURE 7. RT-PCR results of recombinant strains. Asterisks show the significant difference (p < 0.05) compared with the control.
To further improve the protease yield of the recombinant strain BAX-9/pHY-P43/BSP-1, an optimized medium (40 g/L tryptone, 20 g/L yeast extract, and 10 g/L NaCl) was selected for fermentation (Supplementary Figure S1). During the fermentation process, the activity of BSP-1 protease and the biomass of strains were determined. As shown in Figure 8, in the initial 36 h, the enzyme activity increased as the culture was growing. With the continuation of fermentation, the biomass began to decrease, and the enzyme activity continued to increase until 54 h. The enzyme activity reached a maximum of 524.12 U/mL. Finally, with the continuous decrease of biomass, the enzyme activity declined and tended to be stable. The results indicate that the accumulation of the enzyme was delayed compared to culture growth.
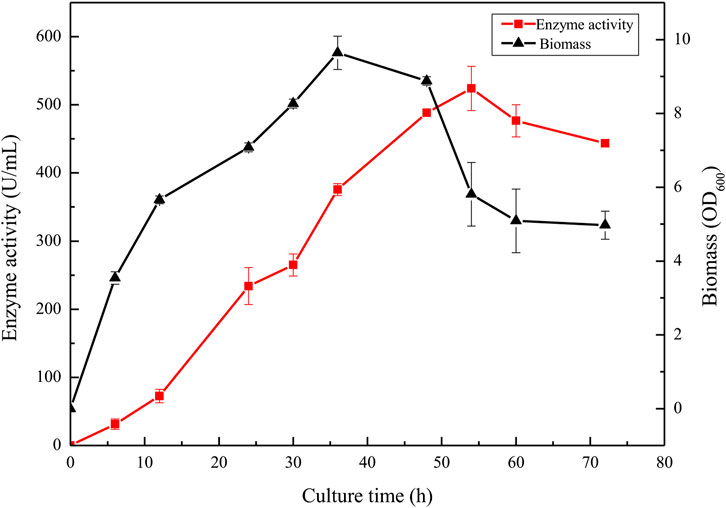
FIGURE 8. Enzyme activity and culture growth of BAX-9/pHY-P43/BSP-1 under fermentation using the optimized fermentation medium.
In conclusion, the BSP-1 protease gene from B. subtilis was identified in this study, and explored possibility of high-efficiency expression system via B. amyloliquefaciens. The enzyme activity and transcription level under the expression of different promoters Ppqq, Pscp, Pcsp, Psrf, Pitu, Pfen and P43 were analyzed, and we found P43 was the optimal promoter for heterologous expression of BSP-1 protease. Nevertheless, Psrf and Pfen show comparable expression strength to P43 and thus can be used as high-strength promoters in future. After fermentation optimization, the highest enzyme activity of the engineering strain BAX-9/pHY-P43/BSP-1 reached 524.12 U/mL. Moreover, monitoring the fermentation process also confirmed no direct correlation between enzyme activity and biomass. This study enriched the originals and hosts for protein expression, and it identified a new resource for the industrialized production of proteases.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Conceptualization, CJ and WH; Methodology, CJ; Software, CJ and CY; Validation, CJ, CY, and YL; Formal analysis, KH and XJ; Investigation, XW; Resources, XW and WH; Data curation, CJ and YL; Writing—original draft preparation, CJ; Writing—review and editing, CJ, XW, WH, DZ, and LL; Visualization, CY; Supervision, XW; Project administration, XW; Funding acquisition, XW and WH.
This study was funded by the National Natural Science Foundation of China (No. 32171423), Key Research and Development Projects of China National Tobacco Corporation (No. 110202102019), and the National Key Research and Development Program of China (No. 2019YFA0906400).
YL was employed by GeneMind Biosciences Company Limited. CY and KH were employed by China National Tobacco Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.977215/full#supplementary-material
Agrebi, R., Haddar, A., Hmidet, N., Jellouli, K., Manni, L., and Nasri, M. (2009). BSF1 fibrinolytic enzyme from a marine bacterium Bacillus subtilis A26: Purification, biochemical and molecular characterization. Process Biochem. 44 (11), 1252–1259. doi:10.1016/j.procbio.2009.06.024
Barzkar, N. (2020). Marine microbial alkaline protease: An efficient and essential tool for various industrial applications. Int. J. Biol. Macromol. 161, 1216–1229. doi:10.1016/j.ijbiomac.2020.06.072
Benabdelkamel, H., Masood, A., Alanazi, I. O., and Alfadda, A. A. (2018). Comparison of protein precipitation methods from adipose tissue using difference gel electrophoresis. Electrophoresis 39 (14), 1745–1753. doi:10.1002/elps.201800124
Brázda, V., Bartas, M., and Bowater, R. P. (2021). Evolution of diverse strategies for promoter regulation. Trends Genet. 37 (8), 730–744. doi:10.1016/j.tig.2021.04.003
Burchacka, E., Pięta, P., and Łupicka-Słowik, A. (2022). Recent advances in fungal serine protease inhibitors. Biomed. Pharmacother. 146, 112523. doi:10.1016/j.biopha.2021.112523
Cai, D., Rao, Y., Zhan, Y., Wang, Q., and Chen, S. (2019). Engineering Bacillus for efficient production of heterologous protein: Current progress, challenge and prospect. J. Appl. Microbiol. 126 (6), 1632–1642. doi:10.1111/jam.14192
Cazier, A. P., and Blazeck, J. (2021). Advances in promoter engineering: Novel applications and predefined transcriptional control. Biotechnol. J. 16 (10), 2100239. doi:10.1002/biot.202100239
Chen, W., Li, L., Ye, C., Zhao, Z., Huang, K., Zou, D., et al. (2022). Efficient production of extracellular alkaline protease in Bacillus amyloliquefaciens by host strain construction. Lwt. Food Sci. Technol. 163, 113620. doi:10.1016/j.lwt.2022.113620
Choi, N.-S., Chang, K.-T., Jae Maeng, P., and Kim, S.-H. (2004). Cloning, expression, and fibrin (ogen) olytic properties of a subtilisin DJ-4 gene from Bacillus sp. DJ-4. FEMS Microbiol. Lett. 236 (2), 325–331. doi:10.1111/j.1574-6968.2004.tb09665.x
Contesini, F. J., Melo, R. R. d., and Sato, H. H. (2018). An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 38 (3), 321–334. doi:10.1080/07388551.2017.1354354
Cui, W., Han, L., Suo, F., Liu, Z., Zhou, L., and Zhou, Z. (2018). Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 34 (10), 145. doi:10.1007/s11274-018-2531-7
Espoui, A. H., Larimi, S. G., and Darzi, G. N. (2022). Optimization of protease production process using bran waste using Bacillus licheniformis. Korean J. Chem. Eng. 39 (3), 674–683. doi:10.1007/s11814-021-0965-3
Fu, G., Liu, J., Li, J., Zhu, B., and Zhang, D. (2018). Systematic screening of optimal signal peptides for secretory production of heterologous proteins in Bacillus subtilis. J. Agric. Food Chem. 66 (50), 13141–13151. doi:10.1021/acs.jafc.8b04183
Gong, J.-S., Ye, J.-P., Tao, L.-Y., Su, C., Qin, J., Zhang, Y.-Y., et al. (2020). Efficient keratinase expression via promoter engineering strategies for degradation of feather wastes. Enzyme Microb. Technol. 137, 109550. doi:10.1016/j.enzmictec.2020.109550
Gu, Y., Xu, X., Wu, Y., Niu, T., Liu, Y., Li, J., et al. (2018). Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab. Eng. 50, 109–121. doi:10.1016/j.ymben.2018.05.006
Gupta, R., Beg, Q., and Lorenz, P. (2002). Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59 (1), 15–32. doi:10.1007/s00253-002-0975-y
Harish, B., and Uppuluri, K. B. (2018). Microbial serine protease inhibitors and their therapeutic applications. Int. J. Biol. Macromol. 107, 1373–1387. doi:10.1016/j.ijbiomac.2017.09.115
Jahangirian, H., Azizi, S., Rafiee-Moghaddam, R., Baratvand, B., and Webster, T. J. (2019). Status of plant protein-based green scaffolds for regenerative medicine applications. Biomolecules 9 (10), 619. doi:10.3390/biom9100619
Jensen, D., and Galburt, E. A. (2021). The context-dependent influence of promoter sequence motifs on transcription initiation kinetics and regulation. J. Bacteriol. 203 (8), e00512–00520. doi:10.1128/JB.00512-20
Ku, T. W., Tsai, R. L., and Pan, T. M. (2009). A simple and cost-saving approach to optimize the production of subtilisin NAT by submerged cultivation of Bacillus subtilis natto. J. Agric. Food Chem. 57 (1), 292–296. doi:10.1021/jf8024198
Kumar, A., Bansal, M., Ghosh, P. K., and Yadav, K. L. (2018). MWCNT/TiO2 hybrid nano filler toward high-performance epoxy composite. Ultrason. Sonochem. 76051, 37–46. doi:10.1016/j.ultsonch.2017.09.005
Liao, Y., Wang, B., Ye, Y., and Pan, L. (2018). Determination and optimization of a strong promoter element from Bacillus amyloliquefaciens by using a promoter probe vector. Biotechnol. Lett. 40 (1), 119–126. doi:10.1007/s10529-017-2449-4
Liu, Y., Shi, C., Li, D., Chen, X., Li, J., Zhang, Y., et al. (2019). Engineering a highly efficient expression system to produce BcaPRO protease in Bacillus subtilis by an optimized promoter and signal peptide. Int. J. Biol. Macromol. 138, 903–911. doi:10.1016/j.ijbiomac.2019.07.175
Matkawala, F., Nighojkar, S., Kumar, A., and Nighojkar, A. (2021). Microbial alkaline serine proteases: Production, properties and applications. World J. Microbiol. Biotechnol. 37 (4), 63. doi:10.1007/s11274-021-03036-z
Meng, Y., Yao, Z., Le, H. G., Lee, S. J., Jeon, H. S., Yoo, J. Y., et al. (2021). Characterization of a salt-resistant fibrinolytic protease of Bacillus licheniformis HJ4 isolated from Hwangseokae jeotgal, a traditional Korean fermented seafood. Folia Microbiol. (Praha). 66 (5), 787–795. doi:10.1007/s12223-021-00878-w
Neef, J., van Dijl, J. M., and Buist, G. (2021). Recombinant protein secretion by Bacillus subtilis and lactococcus lactis: Pathways, applications, and innovation potential. Essays Biochem. 65 (2), 187–195. doi:10.1042/EBC20200171
Ouyang, X., Liu, Y., Qu, R., Tian, M., Yang, T., Zhu, R., et al. (2021). Optimizing protein-glutaminase expression in Bacillus subtilis. Curr. Microbiol. 78 (5), 1752–1762. doi:10.1007/s00284-021-02404-0
Pathak, A. P., and Rathod, M. G. (2018). A review on alkaline protease producers and their biotechnological perspectives. Ind. J. Geo-Mar. Sci. 47 (6), 1113–1119.
Sharma, M., Gat, Y., Arya, S., Kumar, V., and Kumar, A. (2019). A review on microbial alkaline protease: An essential tool for various industrial approaches. Ind. Biotechnol. New. Rochelle. N. Y). 15 (2), 69–78. doi:10.1089/ind.2018.0032
Song, Y., Nikoloff, J. M., Fu, G., Chen, J., Li, Q., Xie, N., et al. (2016). Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. Plos One 11 (7), e0158447. doi:10.1371/journal.pone.0158447
Su, L., Li, Y., and Wu, J. (2021). Efficient secretory expression of Bacillus stearothermophilus α/β-cyclodextrin glycosyltransferase in Bacillus subtilis. J. Biotechnol. 331, 74–82. doi:10.1016/j.jbiotec.2021.03.011
Suberu, Y., Akande, I., Samuel, T., Lawal, A., and Olaniran, A. (2019). Cloning, expression, purification and characterisation of serine alkaline protease from Bacillus subtilis RD7. Biocatal. Agric. Biotechnol. 20, 101264. doi:10.1016/j.bcab.2019.101264
Tavano, O. L., Berenguer-urcia, A., Secundo, F., and Fernandez-afuente, R. (2018). Biotechnological applications of proteases in food technology. Compr. Rev. Food Sci. Food Saf. 17 (2), 412–436. doi:10.1111/1541-4337.12326
Uyar, F., and Baysal, Z. (2008). Production and characterization of neutral and alkaline protease from different Bacillus subtilis strains. Chem. Asian. J. 20 (6), 4523–4530.
Wang, P.-Z., and Doi, R. H. (1984). Overlapping promoters transcribed by bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J. Biol. Chem. 259 (13), 8619–8625. doi:10.1016/S0021-9258(17)39775-2
Wei, X., Luo, M., Xu, L., Zhang, Y., Lin, X., Kong, P., et al. (2011). Production of fibrinolytic enzyme from Bacillus amyloliquefaciens by fermentation of chickpeas, with the evaluation of the anticoagulant and antioxidant properties of chickpeas. J. Agric. Food Chem. 59 (8), 3957–3963. doi:10.1021/jf1049535
Westers, L., Westers, H., and Quax, W. J. (2004). Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Biochimica Biophysica Acta - Mol. Cell Res. 1694 (1-3), 299–310. doi:10.1016/j.bbamcr.2004.02.011
Wu, S.-M., Feng, C., Zhong, J., and Huan, L.-D. (2011). Enhanced production of recombinant nattokinase in Bacillus subtilis by promoter optimization. World J. Microbiol. Biotechnol. 27 (1), 99–106. doi:10.1007/s11274-010-0432-5
Yong, P., Yang, X. J., Lu, X., and Zhang, Y. Z. (2004). Cloning and expression of a fibrinolytic enzyme (subtilisin DFE) gene from Bacillus amyloliquefaciens DC-4 in Bacillus subtilis. Res. Microbiol. 155 (3), 167–173. doi:10.1016/j.resmic.2003.10.004
Zhang, K., Su, L., and Wu, J. (2020). Recent advances in recombinant protein production by Bacillus subtilis. Annu. Rev. Food Sci. Technol. 11, 295–318. doi:10.1146/annurev-food-032519-051750
Zhang, Q., Wu, Y., Gong, M., Zhang, H., Liu, Y., Lv, X., et al. (2021). Production of proteins and commodity chemicals using engineered Bacillus subtilis platform strain. Essays Biochem. 65 (2), 173–185. doi:10.1042/ebc20210011
Zhou, C., Zhou, H., Fang, H., Ji, Y., Wang, H., Liu, F., et al. (2020). Spo0A can efficiently enhance the expression of the alkaline protease gene aprE in Bacillus licheniformis by specifically binding to its regulatory region. Int. J. Biol. Macromol. 159, 444–454. doi:10.1016/j.ijbiomac.2020.05.035
Keywords: alkaline protease, bacillus amyloliquefaciens, promoter screening, recombinant expression, fermentation optimization
Citation: Jiang C, Ye C, Liu Y, Huang K, Jiang X, Zou D, Li L, Han W and Wei X (2022) Genetic engineering for enhanced production of a novel alkaline protease BSP-1 in Bacillus amyloliquefaciens. Front. Bioeng. Biotechnol. 10:977215. doi: 10.3389/fbioe.2022.977215
Received: 24 June 2022; Accepted: 08 August 2022;
Published: 30 August 2022.
Edited by:
Qi Xianghui, Jiangsu University, ChinaReviewed by:
Meijuan Xu, Jiangnan University, ChinaCopyright © 2022 Jiang, Ye, Liu, Huang, Jiang, Zou, Li, Han and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuetuan Wei, d2VpeHVldHVhbkBtYWlsLmh6YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.